Abstract
Colorectal cancer (CRC) is the second leading cause of cancer deaths in the U.S and Western world. Despite increased screening and advances in treatment, the mortality rate (ca. 50,000/year) and high national health-care burden for CRC are likely to remain high unless an effective non-invasive screening test for CRC is instituted for a large segment of the population. Blood-based protein biomarkers hold great promise for early disease diagnosis and personalized medicine; yet robust and reproducible multiplexing platforms and methodologies have lagged behind their genomic counterparts.
Here, we report the development of a novel, multiplexed, hybrid immunoassay for CRC that is formatted on barcoded VeraCode™ micro-beads, which have until now only been used for genomic assays. The method combines a sandwich immunoassay format for detection of serum protein biomarkers with an antigen assay for autoantibody detection. The serum protein biomarkers CEA and GDF15 as well as autoantibodies to the p53 tumor associated antigen (TAA) were used to exemplify the method. This multiplex biomarker panel was configured to run on Illumina’s holographically barcoded VeraCode™ micro-bead platform, which is capable of measuring hundreds of analytes simultaneously in a single well from small volumes of blood (<50 μL) using a 96-well industry standard microtiter plate. This novel use of the VeraCode™ micro-bead platform translates into a potentially low volume, high throughput, multiplexed assay for CRC, for the purposes of biomarker validation, as well as patient screening, diagnostics and prognostics. In an evaluation of a 186 patient sera training set (CRC and normal), we obtained a diagnostic sensitivity of 54% and a specificity of 98%. We anticipate that by expanding and refining the biomarkers in this initial panel, and performing more extensive clinical validations, such an assay could ultimately provide a basis for CRC population screening to complement the more invasive, expensive and low throughput (but highly sensitive and specific) colonoscopy.
Keywords: biomarker, multiplex immunoassay, autoantibodies, tumor-associated antigens, colorectal cancer
1. Introduction
1.1 Need for an Early, Non-Invasive Diagnostic Assay for Colorectal Cancer
Colorectal cancer (CRC) constitutes the second most diagnosed cancer, with an estimated 150,000 new cases and 50,000 CRC-related deaths per year in the US (Howlader et al., 2012). Nearly half of those newly diagnosed with CRC die within five years, largely due to late-stage detection of the disease. An individual’s lifetime risk of developing CRC is 6%, with over 90% of the cases occurring after the age of 50 (Davies et al., 2005). Consequently, the American Cancer Society recommends screening every five years for the over 75 million Americans over the age of 50.
Currently, the gold standard for CRC screening is the colonoscopy. Although a very effective method for diagnosing CRC and detecting precancerous polyps, insufficient capacity of this low throughput test for population-wide screening, along with cost, discomfort and inconveniences associated with the procedure, resulted in the screening of only 21-34% of recommended individuals as of 2004 (Subramanian et al., 2004; Vijan et al., 2004). Alternatives to the colonoscopy, such as the fecal occult blood test (FOBT), sigmoidoscopy, and barium enema are also available, but they also each have severe deficiencies and are not considered to be as effective as the colonoscopy (Rex et al., 2009). In particular, the widely used FOBT has a high rate of false positives (~80%) (Ahlquist, 1997; Doolittle et al., 2001; Davies et al., 2005) as well as a low sensitivity for cancer and pre-malignant lesions.
Another approach to reducing the high mortality rate of CRC is to perform an inexpensive and non-invasive screen as part of a standard general physical examination for the appropriate population groups (e.g. persons over 50), which could detect a large fraction of patients who would normally be missed due to non-compliance. Improved fecal tests are being developed, for instance, based on molecular profiling of DNA such as the Exact Science Pre-Gen Plus™ (Berger et al., 2006); however, such tests have not been widely accepted by the medical community, potentially due to the emphasis on home-collection of fecal samples (Woolf, 2004). Yet diagnosing CRC at an early stage is indispensable as the 5-year survival rate is around 90% when caught at the localized stage (SEER Summary Staging) and drops to 70% with regional metastasis and 12% with distant metastasis (Howlader et al., 2012). Therefore, an early, noninvasive screen for CRC which can complement the colonoscopy is urgently needed.
1.2 Serological Assays for Cancer Detection
In contrast to fecal based CRC screening, blood testing based on detection of multiple biomarkers provides a minimally-invasive, more patient friendly method of pre-screening for CRC. One such approach is based on detection of aberrant methylation of CpG-islands (CGI-methylation) in freely circulating DNA in blood. Epigenomics is developing Epi ProColon, a blood-based test based on detection of methylation markers in Septin9 (Toth et al., 2012). Serum proteins and autoantibodies against tumors-associated antigens (TAAs) in blood also comprise a potential source of valuable CRC biomarkers. A 2011 review found 63 studies on the serological diagnosis of CRC with more than 50 TAAs and other serum protein biomarkers in development (Creeden et al., 2011). Autoantibodies to TAAs have been detected in patient’s blood even in the early stages of cancer (Chapman et al., 2008). Furthermore, autoantibody biomarkers have several advantages over other serum biomarkers, including long-term stability and “the inherent amplification of signals provided by the host’s own immune system to low levels of tumor-associated antigens in early disease” (Anderson and LaBaer, 2005; Storr et al., 2006). However, any single autoantibody biomarker rarely exceeds 15% diagnostic sensitivity (Zhang et al., 2003; Casiano et al., 2006; Belousov et al., 2008), thereby highlighting the need to discover and clinically validate large panels or signatures of TAAs, in multiplex, as well as to combine autoantibodies with other serum biomarker types such as circulating proteins, to achieve both sensitive and specific cancer diagnosis.
1.3 Success of Multiplexed Bio-Assays in Genomics
In the genomics realm, highly parallelized and multiplexed bio-assay technologies such as high density DNA microarrays/micro-bead arrays (Fodor et al., 1991; Chee et al., 1996; Gunderson et al., 2004), massively parallel DNA sequencing (Margulies et al., 2005; Bentley et al., 2008), microfluidic chips (Dettloff et al., 2008) and bead suspension “arrays” (Fulton et al., 1997; Lin et al., 2009) have revolutionized the ability of physicians to provide personalized medical care. These technologies offer the ability to simultaneously screen large numbers of analytes using only small sample volumes, providing for highly effective discovery, validation and clinical assay of biomarkers for disease diagnosis and prognosis as well as for the prediction of therapeutic efficacy. Major successes include genome-wide gene expression profiling which has led to a new understanding of cellular control pathways and powerful multiplexed diagnostic/prognostic tools such as for predicting breast cancer recurrence (e.g. the Amsterdam 70-gene signature (van ’t Veer et al., 2002) currently used in Agendia’s FDA-approved MammaPrint® microarray assay).
1.4 Application of VeraCode™ Technology for Multiplex Serological Immunoassay of Colorectal Cancer
The utilization of multiplexing and multi-marker signatures for protein-based serological assays holds great promise in the realm of cancer diagnostics and prognostics, yet lags behind its genomic counterpart. Multiplexed bead-based immunoassays have until now been essentially limited to the Luminex (Austin, TX) xMAP® technology (Fulton et al., 1997), which has been used for example to detect antibodies directed against both viral proteins (Opalka et al., 2003) and parasitic antigens (Fouda et al., 2006), as well as pneumococcal (Schlottmann et al., 2006) and meningococcal polysaccharides (de Voer et al., 2008). Here, we report the development of a novel protein-based serological immunoassay platform using Illumina’s VeraCode™ micro-bead technology. The VeraCode™ system differs from such existing multiplexed bead platforms in that it uses digital, 24-bit holographic barcoding for nearly unlimited potential coding capacity, instead of analog coding with embedded fluorophores, whose broad spectral emissions and spectral overlap limit the coding capacity (currently at 500 for FLEXMAP 3D® coding system by Luminex). Furthermore, the VeraCode™ system uses a hydrophilic bio-friendly glass bead surface for low non-specific binding and, instead of a hydrophobic polymeric (e.g. polystyrene) bead surface which can mediate background in serological assays (Waterboer et al., 2006). Finally, since the VeraCode™ barcoding is not based fluorescence, 2-color fluorescence analyte readout is more readily implemented on the VeraCode™ system for maximum flexibility.
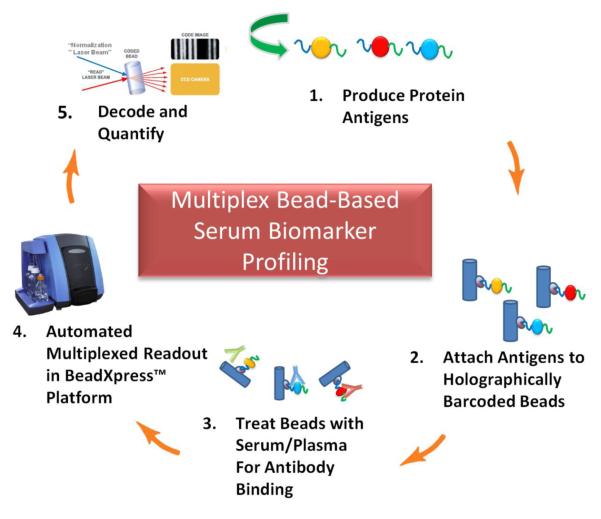
By adapting the VeraCode™ digital holographic bead technology and BeadXpress™ reader, originally developed by Illumina (San Diego, CA) for genomic applications (up to 384-plex) (Lin et al., 2009), we have developed a novel, high sensitivity, high throughput and reproducible multiplex immunoassay approach requiring very low blood sample volumes. The overall approach is exemplified diagrammatically in Figure 1 for detection of autoantibodies to TAAs. We attach recombinant proteins (antigens) to VeraCode™ beads using standard chemistries and then perform serum autoantibody screening from patient blood. Alternatively, in cases where an antigen is difficult to produce recombinantly, or otherwise difficult to obtain, it can be produced on-demand with cell-free protein expression and rapidly in situ purified onto VeraCode™ beads for subsequent assay. Likewise, sandwich immunoassays are performed by using a capture antibody instead of an antigen on the beads and using an anti-analyte detection antibody (not depicted in Figure 1). In either case, VeraCode™ beads can be fluorescently read to detect the bound serum autoantibody or protein biomarker, and decoded using the BeadXpress™ reader to determine the particular antigen or capture antibody present on the bead. We show proof-of-concept in CRC for using a hybrid multiplexed VeraCode™ assay which combines a sandwich immunoassay format for detection of serum protein (non-antibody) biomarkers with an autoantibody assay of TAAs.
Figure 1. Multiplexed Serum Biomarker Profiling on VeraCode™ Beads.
Candidate biomarker proteins are expressed in vitro and then attached to the VeraCode beads. Each protein TAA is attached to a uniquely coded bead. Various beads are combined and treated with serum/plasma, then detected by an anti-[human IgG] fluorescently labeled antibody, and finally “read” and decoded via the BeadXpress™ reader.
2. Materials and Methods
2.1 Supplies and Reagents
EDC (1-(3-dimethylaminopropyl)-3-ethyl-carbodiimide HCl), Sulfo-NHS (N-Hydroxysulfosuccinimide), MES (2-(N-Morpholino)ethanesulfonic Acid), EZ-Link Amine-PEO3-Biotin, EZ-Link-Sulfo-NHS-LC-Biotin, hydroxylamine and streptavidin were purchased from Thermo-Fisher-Pierce (Rockford, IL). The PURExpress™ In Vitro Protein Synthesis Kit was from New England Biolabs (Ipswich, MA). The TNT® T7 Quick for PCR DNA Rabbit Reticulocyte Cell-Free Expression Lysate was from Promega (Madison, WI). The DyLight 649 AffiniPure Mouse Anti-Human IgG, DyLight 649 AffiniPure Goat Anti-Human IgG and HRP Conjugated Mouse Anti-Human IgG antibodies were from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA). The Streptavidin R-Phycoerythrin Conjugate and the recombinant human MAP4K4 protein were from Invitrogen (Carlsbad, CA). Clones from the Human ORFeome Collection were purchased from Open Biosystems/Thermo-Fisher (Huntsville, AL). The pETBlue-2 vector was from (EMD Biosciences, Inc., San Diego, CA). PD SpinTrap G-25 Columns were from GE Healthcare Life Sciences (Pittsburgh, PA). Carboxyl-terminated VeraCode™ beads were from Illumina (San Diego, CA). 400 μL capacity Ultrafree-MC Micro-Centrifuge Filter Units, Pore Size 0.45 μm Durapore PVDF Membrane and the Mouse Anti-Carcinoembryonic Antigen (CEA) Capture Antibody, Clone 1105, were from Millipore (Billerica, MA). The Mouse Anti-Carcinoembryonic Antigen (CEA) Detection Antibody, Clone 26/3/13 and recombinant human cyclin B1 (CCNB1) protein were from Abcam (Cambridge, MA). The CEA standard protein and ELISA were from GenWay Biotech (San Diego, CA). The Mouse Anti-GDF15 Capture Antibody, Clone 147627, the Biotinylated Goat Anti-GDF15 Affinity Purified Polyclonal Detection Antibody, and the Recombinant Human GDF15 Standard Protein were from R&D Systems (Minneapolis, MN). Nunc-Immuno 96-Well Polystyrene Microtiter Plates, PolySorp, were from Thermo-Fisher Scientific (Waltham, MA). SureBlue TMB 1-Component Microwell Peroxidase Substrate was from KPL (Gaithersburg, Maryland). Recombinant human TP53 (p53) protein was from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant human IGF2BP2/IMP-2/p62 protein was from Sino Biological (Beijing, China).
2.2 Recombinant and Cell-Free Proteins (Candidate TAAs)
Human recombinant proteins were purchased commercially (see 2.1 Supplies and Reagents). For cell-free protein expression, clones from the Human ORFeome Collection were used as the source for the ORFs. Standard Gateway® recombination cloning was performed (Walhout et al., 2000) (Invitrogen, Carlsbad, CA) to transfer the ORFs into a custom T7 driven cell-free protein expression vector containing a C-terminal streptavidin binding affinity tag (SBP-Tag; (Keefe et al., 2001)) and an N-terminal VSV-G epitope tag. Expression reactions were performed using one of two transcription/translation coupled systems, the Rabbit Reticulocyte Lysate (TNT® T7 Quick for PCR DNA), or the PURExpress® E. coli based reconstituted system, all according to the manufacturer’s instructions. The expression plasmid used was a derivative of the pETBlue-2 vector containing the aforementioned tags and sequences for Gateway® cloning.
2.3 Attachment of Recombinant Proteins and Capture Antibodies to VeraCode™ Beads
Commercial recombinant proteins, which are supplied in a variety of formats, concentrations and buffers, were passed over a PD SpinTrap G-25 Column to remove potentially incompatible buffer components (e.g. Tris buffer or residual glutathione used in purifying GST fusion proteins) and to unify the buffer conditions. In the case where proteins were supplied lyophilized, they were first dissolved to 1 μg/μL in water and then supplemented to 1X PBS, pH 7.5, from a 5X stock before column purification. The PD SpinTrap G-25 columns were performed according to the manufacturer’s instructions (equilibration in 300 μL 1X PBS; 70-130 μL loading of the manufacturer supplied or reconstituted protein). Following the desalting (buffer exchange), 1/4th volume of 5X PBS was added to the eluate to ensure an adequate buffering capacity of the protein for the subsequent bead attachment steps. Note that for optimal results with some proteins (e.g. p53 and MAP4K4), the column buffer exchange step was omitted and the manufacturer supplied proteins were simply supplemented to 1X PBS (either from a 5X stock or as detailed above for lyophilized proteins). Note that while a comprehensive analysis of all possible buffer conditions was not done, some proteins (e.g. antibodies) coupled more efficiently to the VeraCode™ beads using a MES buffering system (0.1 M MES, pH 4.7, 0.9 % NaCl) instead of PBS. In this case, MES buffer replaced the PBS in the aforementioned steps. Recombinant protein concentration used for subsequent bead attachment was typically 0.1 μg/μL in the corresponding buffer (if this concentration was not possible based on how the protein was supplied by the manufacturer, concentration was kept as high as reasonably possible). Capture antibodies to be coupled to VeraCode™ beads were not desalted, but were simply supplemented to 1X concentrated MES Buffer and used at 0.5 μg/μL for subsequent bead attachment.
Recombinant proteins and antibodies were attached to carboxyl-terminated VeraCode™ beads by a 2-step method. VeraCode™ beads are 240×28 micron, holographically encoded, glass micro-cylinders with a carboxylated surface chemistry. First, 10,000 to 40,000 VeraCode™ beads were washed 3× 800 μL with MES Buffer (0.1 M MES, pH 4.7, 0.9 % NaCl) by sequential mixing, pelleting the beads by brief and gentle spinning (or allowing beads to settle by gravity) and removing the supernatant (wash buffer) by manual pipetting, being careful not to lose the bead pellet. All washes were performed in this manner unless otherwise indicated. After discarding the final wash, 200 μL of Sulfo-NHS Buffer (1 mg/mL in MES Buffer; prepared immediately prior to use) was added to each washed bead pellet. Beads were mixed immediately and briefly. 200 μL of EDC Buffer (1 mg/mL in MES Buffer; prepared immediately prior to use) was immediately added to each sample (containing both beads and Sulfo-NHS Buffer) and immediately mixed to combine. Following incubation for 1 hr with mixing (all extended mixing steps for VeraCode™ beads were done at 1,200 rpm on a VorTemp 56 shaker, Labnet International Inc., Edison, NJ), the beads were then washed 3× 800 μL briefly with MES Buffer and then 1× 800 μL quickly with 1X PBS (for proteins or antibodies prepared in MES Buffer, this PBS wash was omitted). The protein coupling reaction immediately followed, in which 10-40 μg of the previously prepared recombinant protein or 100 μg of antibody was added to the beads, mixed, and incubated for 1 hr with mixing (a comprehensive titration analysis was not performed due to the wide range of protein classes and wide range of concentrations at which they were supplied by the manufacturers, however, the amounts added are believed to be sufficient to saturate the bead surface, as using a calculation of 2.5 mg/m2 binding capacity of a solid non-porous surface as reported for avidin and 15 mg/m2 for antibodies (Plant et al., 1991), we estimate 40,000 beads can bind a maximum of roughly 2-10 μg). Beads were then spun down, and the protein solution was removed. The beads were washed 2× 800 μL briefly with BSA Block (1% BSA [w/v] in TBS-T; TBS = 50 mM Tris, pH 7.5, 200 mM NaCl; TBS-T contains 0.05% [v/v] Tween-20) before discarding the wash and incubation with an additional 400 μL of BSA Block for 30 min. Beads were then washed briefly 1x with 800 μL of PBS-1M NaCl, 1× 30 min with 400 μL of PBS-1M NaCl (with shaking) and then 2 times briefly with 800 μL TBS-T. Beads were stored in TBS-T at 4°C.
2.4 Expression and Attachment of Cell-free Produced Proteins to VeraCode™ Beads
For optimal performance, we used an indirect method of coating VeraCode™ beads with biotin followed by streptavidin. Streptavidin beads were then used to in situ capture/purify cell-free produced proteins carrying the SBP-Tag (Keefe et al., 2001), directly from the crude expression reaction. First, a vial of 20,000 carboxyl-terminated VeraCode™ beads was washed 5× 400 μL with MES Buffer (0.1 M MES, pH 4.7, 0.9 % NaCl). After discarding the final wash, 200 μL of MES Buffer was added to each washed bead pellet. To this, 25 μL of 48 mM EZ-Link Amine-PEO3-Biotin stock was added. Beads were mixed immediately and briefly. Next, 25 μL of EDC Buffer (100 mg/mL in water; prepared immediately prior to use) was immediately added to each sample (containing both beads and Biotin-Amine Linker), mixed, and incubated for 1 hr with mixing. Beads were then spun down, and the reaction solution was removed. The beads were washed 4× 400 μL (5 min each) with Quench Buffer (10 mM hydroxylamine in PBS-T; prepared immediately prior to use; PBS-T is standard PBS buffer with 0.05% [v/v] Tween-20) before discarding the wash and incubation with an additional 400 μL of Quench Buffer for 30 min. Beads were then further washed briefly 2x with 400 μL of PBS containing 1M NaCl (first wash brief and then leaving in the second wash for 1 hr with mixing). Finally, beads were washed 4× 400 μL briefly with TBS-T. Beads were stored, protected from light, in TBS-T at 4°C.
Before coating with Streptavidin, Biotin-VeraCode™ beads were pre-treated 2× 5 min using 400 μL of BSA Block with mixing. After removing the Block, 250 μL Streptavidin solution (1 mg/mL in BSA Block) was added and incubated for 30 min with mixing. After removing this solution, beads were washed 3× 400 μL with TBS-T, followed by 5 min washes of 3× 400 μL with TBS containing 1M NaCl. Finally, beads were washed briefly 3× 400 μL with TBS and stored at +4°C in this buffer.
TAAs were expressed as proteins containing a C-terminal SBP-Tag (Keefe et al., 2001) using a cell-free system according to the manufacturer’s instructions (Rabbit Reticulocyte or PURExpress™; see 2.2 of Materials and Methods). 25 μL of cell-free protein expression reaction was mixed with an equal volume of BSA Block and clarified by 1 min in a standard micro-centrifuge (15,000 rpm) followed by passing through a 0.45 micron pore size spin filtration device (400 μL capacity Ultrafree-MC Micro-Centrifuge Filter Units, Pore Size 0.45 μm Durapore PVDF Membrane). The aforementioned streptavidin VeraCode™ beads were pelleted, briefly washed 3× 400 μL in TBS-T followed by 2× 5 min each with BSA Block. Next, the diluted cell-free protein expression reaction was added and mixed 30 min for protein capture (note that this amount of cell-free protein expression reaction is used for a minimum of 500 beads and a maximum of 5,000 beads). Protein capture was followed by 4× 400 μL brief washes with TBS-T before the beads were re-suspended to their original concentration in TBS-T. Beads were stored in TBS-T at 4°C protected from light.
2.5 Biotin Labeled Detection Antibodies for VeraCode Sandwich Immunoassays
While the biotin labeled anti-GDF15 antibody used in the VeraCode™ assays was from a commercial source (see 2.1 Supplies and Reagents), the anti-CEA antibody used in the VeraCode™ assays was biotin labeled in-house as follows: The commercial antibody as supplied (see 2.1 Supplies and Reagents) was prepared to 1 mg/mL (100 μg used) and supplemented to 100 mM sodium bicarbonate from a 1M stock. A high concentration 10 mM stock of EZ-Link-Sulfo-NHS-LC-Biotin was prepared fresh and the appropriate volume immediately added to the antibody to yield a 15-fold molar excess. The reaction was carried out for 30 min with gentle mixing. The reaction was then quenched by adding 1/9th volume of 200 mM glycine in 200 mM sodium bicarbonate and 200 mM NaCl and subsequently mixing for 15 min. To avoid losses in the subsequent desalting column, a BSA carrier was then added from a 10% (w/v) stock to yield a final 0.05% (w/v). To remove unreacted biotin, the reaction mix was then desalted on PD SpinTrap G-25 columns. The PD SpinTrap G-25 columns were performed according to the manufacturer’s instructions (equilibration in 300 μL of TBS). Following the desalting (buffer exchange), 1/9th volume of 10X TBS was added to the eluate to ensure an adequate buffering capacity.
2.6 Serological Assay on VeraCode™ Beads
Colorectal cancer and normal sera/plasma samples were from Asterand Inc. (Detroit, MI), ProMedDx, LLC (Norton, MA), the Ontario Institute of Cancer Research (OICR) and Analytical Biological Services Inc. (Wilmington, DE). Colorectal cancer patient samples were an approximate 50:50 distribution of a) stage T2 or T3 (AJCC staging) non-metastatic and b) stage T3 or T4 metastatic.
To perform a multiplexed bead experiment, beads with the different proteins and/or capture antibodies, each identifiable by a unique holographic barcode, were pooled into a round bottom 96-well polypropylene microtiter plate. Kitting was done according to Illumina’s (San Diego, CA) standard protocol except that TBS-T was used at all kitting steps and 30 min is allowed for beads to settle into wells (typically 30-50 beads of each species per well). Human serum/plasma samples (diluted at 1/50 in BSA Block for TAA validation studies or diluted 1/10 for the hybrid 3-Plex p53 TAA and GDF15/CEA sandwich immunoassay) were added at 100 μL/well and shaken for 30 min. Samples were removed and beads were washed 6 × 250 μL briefly with BSA Block. For TAA validation studies, beads were then probed with 100 μL of an Anti-Human IgG Fluorescent (DyLight 649) Secondary Antibody diluted to 10 μg/mL in BSA Block. Probing was for 30 min with mixing. The probe solution was removed and discarded, and the beads washed 6× 250 μL briefly with TBS-T. The final wash solution was discarded, leaving the bead pellets and a small residual liquid volume in the wells of the readout plate (~70 μL). Beads were scanned using the BeadXpress™ reader (Illumina, San Diego, CA). For the aforementioned hybrid 3-plex assay, biotin labeled anti-GDF15 (0.05 μg/mL) and anti-CEA (1 μg/mL) antibodies were first added (together) in BSA Block immediately after the serum/plasma (and subsequent wash) step. Probing was for 30 min with mixing. The probe solution was removed and discarded, and the beads washed 6× 250 μL briefly with TBS-T. Probing with the Anti-Human IgG Fluorescent (DyLight 649) Secondary Antibody and subsequent washing was then performed as described above. Finally, the beads were probed with Streptavidin R-Phycoerythrin for 30 min with mixing at 10 μg/mL in BSA Block, washed 6× 250 μL briefly with TBS-T and scanned in the BeadXpress™ reader as described above. Straight sandwich immunoassays for GDF15 and CEA (but no TAA detection) were performed in the same manner except the Anti-Human IgG Fluorescent (DyLight 649) Secondary Antibody probing was omitted.
2.7 ELISA Analysis of TAAs and Circulating Non-Antibody Protein Biomarkers
The p53 autoantibody (TAA) ELISA was performed similar to published reports (Zhang et al., 2003). Briefly, the recombinant protein was diluted to 0.5 ng/μL in PBS and 100 μL used to passively coat each well of a 96-well polystyrene microtiter plate (Nunc-Immuno 96-Well Plates, PolySorp). Plates were then washed with TBS-T and pre-treated with BSA Block. Sera/plasma samples were diluted to 1/100 in BSA Block and 100 μL added to the wells for 30 min incubation. Detection was with an HRP conjugated mouse anti-human IgG antibody followed by development with SureBlue TMB 1-Component Microwell Peroxidase Substrate. The CEA sandwich ELISA was performed according to the manufacturer instructions (see 2.1 Supplies and Reagents).
3. Results
3.1 Attaching Proteins to VeraCode™ Beads
Recombinant proteins were directly and covalently attached to VeraCode™ beads using standard carbodiimide (EDC) chemistries to link amine groups on the proteins to the carboxyl groups on the beads. In the case of cell-free expressed proteins, they were affinity captured directly from the crude expression reactions by their C-terminal SBP-Tag (Keefe et al., 2001) onto streptavidin coated VeraCode™ beads. For preparation of streptavidin coated VeraCode™ beads, optimal results (data not shown) were obtained by first attaching a biotin-amine linker to the carboxyl beads using the aforementioned carbodiimide chemistry, followed by attachment of (tetrameric) streptavidin to the biotinylated beads. With either recombinant or cell-free proteins, successful attachment of the proteins to the beads is readily verified (quality controlled) by detection of epitope or fusion tags present in the proteins. An example of this quality control measure is shown in Supplementary Figure 1 with the p53 and MAP4K4 proteins. Detection of recombinant proteins was via a GST fusion tag in this case and cell-free proteins via their N-terminal VSV-G epitope tag. With the recombinant proteins, signal to background ratios were 250:1 and 125:5 for p53 and MAP4K4 respectively, and for the cell-free proteins 34:1 and 87:1 (note that all DNA clones used to produce cell-free proteins were sequence verified).
3.2 Validate Ability to Detect TAAs on VeraCode™ Beads by Comparison to ELISA
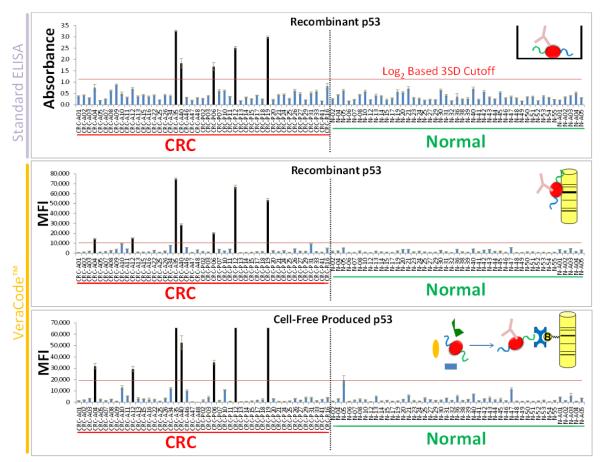
First, human p53 (TP53) (Koziol et al., 2003; Saleh et al., 2004; Nozoe et al., 2007; Reuschenbach et al., 2009) was validated as a positive control TAA using a conventional ELISA to detect autoantibodies in the serum/plasma of 47 healthy (normal) and 47 colorectal cancer patient samples (94 total patient samples) (Figure 2, top panel). To calculate cutoffs, the ELISA values were log2-transformed (to achieve better Gaussian distribution of the data) and the standard deviation across the normal patient cohort was calculated. A diagnostic scoring cutoff set at 3 standard deviations above the mean for the normal patient cohort yielded 11% sensitivity for colorectal cancer detection at 100% specificity with these samples. This method of setting cutoffs is commonly used for autoantibody immunoassays (e.g. (Liu et al., 2009)).
Figure 2. Validate VeraCode™ Immunoassay Using the p53 TAA and Comparing to ELISA with Colorectal Cancer Patients.
Results from ELISA (top panel) were compared to results obtained from VeraCode™ beads coated with either purified recombinant p53 (middle panel) or cell-free produced p53 which was in situ purified on the beads (bottom panel). Protein TAAs were bound to the VeraCode™ carboxyl beads as depicted in the inset diagrams and the beads used to assay patient serum/plasma for the presence of autoantibodies. MFI= Mean Fluorescence Intensity of the BeadXpress™ instrument readout. Individual patient samples are denoted on the x-axis whereby the prefix CRC = Colorectal Cancer and N = Normal (Healthy Individuals). The overall CRC and normal patient cohorts are also labeled below the x-axis. Error bars represent standard deviation of replicate wells in the ELISA or replicate beads in the VeraCode™ assay. The red horizontal lines on the graphs indicate the diagnostic scoring cutoffs (based on log2 data). The black vertical bars are positive samples.
Next, to technically validate the VeraCode™ bead assay using the p53 TAA, we evaluated the data obtained from screening the same patient cohort against beads to which either purified recombinant p53 or cell-free produced p53 was attached (Figure 2, middle and bottom panels, respectively). The cutoff and scoring was done as with the ELISA. The error bars represent the intra-assay bead-to-bead variance in fluorescence intensity within each sample-protein pair (i.e. variance of replicate beads). Results from ELISA were compared to results obtained from VeraCode™ beads. All 5 colorectal cancer samples which scored positive in the ELISA also score positive on both VeraCode™ bead assays (with both recombinant and cell-free p53 protein). In addition, two additional hits in the CRC cohort were detected by the VeraCode™ assay (same two patients detected with both recombinant and cell-free proteins) but 100% specificity versus the normal patients was maintained.
3.3 Reproducibility of Multiplex VeraCode™ Immunoassays
In order to establish intra-assay precision, we performed the multiplex bead assay on triplicate samples of four CRC and four normal patient sera/plasma in a 96-well plate. Two TAAs were used in this multiplexed experiment: The p53 control (discussed earlier) and Cyclin B1 (Koziol et al., 2003; Chen et al., 2007; Reuschenbach et al., 2009). Each of the three replicate wells of each sample contained approximately 50 beads per TAA. Two previously known p53-positive sera (based on ELISA and VeraCode™ data in Figure 2) were chosen for this experiment, whereas their sero-reactivity against CyclinB1 was not known a priori (i.e. positives not necessarily expected based on low diagnostic sensitivity of individual TAAs). Results are shown in Supplementary Figure 2. An average intra-assay CV of 10% across all samples and proteins was achieved (see error bars in Supplementary Figure 2 for more detail). The diagnostic scoring cutoff for p53 was calculated based on the normal samples as discussed earlier, however, for maximum stringency, the calculations were done before averaging the MFI values of the replicate samples (MFI = Mean Fluorescence Intensity; i.e. mean of all beads within one sample per TAA). With this, the scoring cutoff accounts for variance across the sample replicates. Of note, using this cutoff, previously known p53-positive samples were correctly detected in this VeraCode™ bead experiment, with no false positives (neither in CRC nor normal samples).
Likewise, to define inter-assay reproducibility, we ran and compared three independent assays (on three different days) on the same lot of p53 VeraCode™ beads using a 94-member training set of plasma/serum samples (47 normal and 47 CRC) (Supplementary Figure 3). Again, the scoring cutoff was determined by the non-averaged, replicate MFI values, resulting in a more stringent analysis (i.e. MFI values of the three different assay runs for the normal patient samples were not averaged before determining cutoff). Results are shown in Supplementary Figure 3. Despite this stringent cutoff, all five previously known p53-positive samples (based on ELISA in Figure 2) remained positive on the VeraCode™ beads in this rigorous inter-assay setting. Furthermore, the two additional p53-positives picked up only by the VeraCode™ assay (and not ELISA) shown in Figure 2 (single assay), also remained positive in this rigorous inter-assay setting. The average inter-assay CV was 20% across all sample-protein pairs (see error bars in Supplementary Figure 3 for more detail).
Finally, as an additional metric of inter-assay reproducibility, linear regression analysis of two separate assay runs of the 94 samples for two different TAAs (assayed in multiplex) showed R2 values ≥0.96 in both cases (Supplementary Figure 4; p53, as well as insulin-like growth factor 2 mRNA binding protein 2 [IGF2BP2] (Reuschenbach et al., 2009)).
3.4 Improved Sensitivity for CRC Using Hybrid Multiplexed VeraCode™ Assay of Autoantibodies and Serum Proteins
Next, to show compatibility of the VeraCode™ based assay with multiple biomarker classes and to increase the overall diagnostic sensitivity for CRC beyond TAAs alone, we combined three distinct biomarkers, i) autoantibodies against the aforementioned p53 TAA, as well as detection of serum levels of ii) carcinoembryonic antigen (CEA) and iii) the cytokine GDF15. While CEA is very well known as a CRC biomarker for disease monitoring and prognostics, its diagnostic use alone or as part of a biomarker panel is currently under investigation (Creeden et al., 2011; Su et al., 2012). GDF15 (MIC-1) is perhaps not as well as established for CRC, however, several recent studies suggest it may be useful as a prognostic and diagnostic marker (Xue et al., 2010; Wallin et al., 2011; Brown et al., 2012).
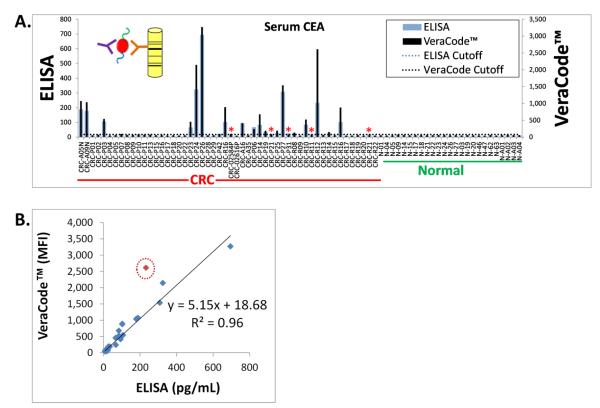
First, we sought to demonstrate that sandwich immunoassay detection of non-antibody serum protein biomarkers could also be achieved on the VeraCode™ system. As with the p53 TAA, this was done by comparison to conventional ELISA. In this case CEA was used as a model system (with 52 CRC and 25 normal serum/plasma samples). For the VeraCode™ assay, an anti-CEA capture antibody was attached to the bead surface. Following incubation with the serum/plasma samples to capture the CEA, detection was with a biotin labeled anti-CEA antibody followed by a fluorescently labeled streptavidin. ELISA (sandwich immunoassay format) was simply performed using a commercially available kit (see Materials and Methods). Results are shown in Figure 3. Clear agreement is apparent between the two assays as seen by the overlaid bar graphs in Figure 3A. By setting a cutoff for each assay based on the normal patient cohort (as previously described) in order to score positive CEA “hits”, 90% concordance was observed between the ELISA and VeraCode™ assays in the CRC patient cohort, with 100% specificity in both cases in the normal patient cohort.
Figure 3. Concordance of ELISA and VeraCode™ for Detection of Circulating Protein Markers Such as CEA in CRC.
(A.) Results from ELISA (blue bars) were compared to results obtained from VeraCode™ beads (black bars). Both assays were formatted as a sandwich immunoassay whereby serum/plasma CEA protein was bound to the ELISA plate or bead surface by a monoclonal anti-CEA capture antibody, followed by detection using a labeled anti-CEA antibody targeting a different epitope than the capture antibody, as depicted in the inset diagram for the VeraCode™ beads. MFI= Mean Fluorescence Intensity of the BeadXpress™ instrument readout. Individual patient samples are denoted on the x-axis whereby the prefix CRC = Colorectal Cancer and N = Normal (Healthy Individuals). The overall CRC and normal patient cohorts are also labeled below the x-axis. The black dotted and blue horizontal lines on the graph indicate the diagnostic scoring cutoffs for each assay (based on log2 data). The red asterisks represent the only discordant hits, which were borderline positive or negative CRC samples that fell extremely close to the cutoffs. (B.) Linear regression analysis comparing VeraCode™ bead results, plotted on the y-axis in MFI, with ELISA results, plotted on the x-axis in pg/mL. The R2 value is 0.96 excluding one outlier (0.90 if the outlier, noted by the red circle, is included).
To assess quantitative concordance of the signal magnitudes of each assay, a linear regression was performed as shown in Figure 3B. An R2 value of 0.9 was obtained in this case with one clear outlier which yielded an abnormally high signal in the VeraCode™ assay. Eliminating this outlier yields an R2 value of 0.96.
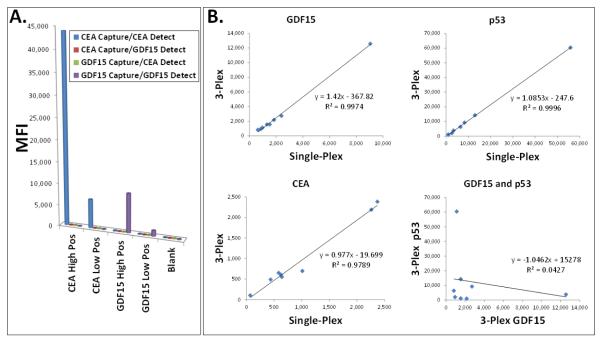
Next, in order to assay these two distinct biomarker types (autoantibodies to TAAs and non-antibody serum proteins) in multiplex, we formatted a novel hybrid assay on the VeraCode™ platform. p53 TAA beads for autoantibody detection were configured as before. For detection of the non-antibody serum proteins, the beads were configured for a sandwich immunoassay by attaching capture antibodies for CEA and GDF15 on different barcoded bead species. Following incubation of the pooled beads with the serum/plasma samples (to capture either anti-p53 autoantibody, CEA or GDF15), detection of bound autoantibody was with a fluorescently labeled monoclonal mouse anti-[human IgG] secondary antibody, which was chosen for its lack of cross-reactivity with mouse IgG (i.e. the CEA and GDF15 capture and detection antibodies). Detection of the bound CEA and GDF15 proteins was with corresponding biotin labeled detection antibodies followed by a fluorescently labeled streptavidin. Importantly, owing to the unique 2-color fluorescent readout capabilities of the BeadXpress™ reader, autoantibody detection and CEA/GDF15 detection could be achieved with different colors (DyLight™ 649 at 670 nm emissions and R-Phycoerythrin at 578 nm emissions, respectively). This adds an extra measure of assurance that if any cross-reaction between the autoantibody and sandwich immunoassay systems were to occur, it would not generate a signal (e.g. if the anti-[human IgG] were to cross-react with the CEA or GDF15 beads, this could be distinguished from true CEA or GDF15 signal on the basis of the fluorescence color). Nonetheless, a critical first step was to confirm that these three biomarkers could indeed be multiplexed without cross-reaction or interference among the various capture and detection agents. As a first step, since recombinant protein standards are available for CEA and GDF15, the standards were spiked into BSA Block buffer (see Materials and Methods) to create high and low positive samples in the VeraCode™ assay. A series of single-plex measurements were performed to test all possible permutations of capture antibody bead species, analyte (CEA or GDF15) and detection antibody. Results are shown in Figure 4A. As seen, a positive, dose dependent signal was only observed in cases where the correct capture and detection antibodies were matched with the correct analyte, and no signal when mismatched (the blank, corresponding to buffer without analyte, also yielded no signal).
Figure 4. Development of a Hybrid Multiplex Assay for Autoantibodies (TAAs) and Circulating Proteins.
A hybrid VeraCode™ Bead™ assay for multiplexed detection of serum autoantibodies to the p53 TAA (antigen-antibody assay format) as well as detection of the circulating serum proteins CEA and GDF15 (sandwich immunoassay format) was developed. (A.) To verify the sandwich immunoassay portion of the assay could be multiplexed, recombinant CEA or GDF15 standard proteins were spiked into buffer to create high and low positive samples (see x-axis; blank is just buffer). All possible combinations of capture antibody beads, detection antibody and analyte (CEA or GDF15 samples) were tested in a series of single-plex assays to verify lack of cross-reaction. (B.) To verify the p53 autoantibody assay could be fully multiplexed with the CEA and GDF15 sandwich immunoassays, a 3-plex VeraCode Bead™ assay was performed using a range of positive and negative sera/plasma for each analyte, and the results for each biomarker from the 3-plex assay (y-axis) were plotted by linear regression against the single-plex results (x-axis). To show the specificity of this metric, a linear regression between 3-plex measurements of GDF15 and p53 autoantibody, in which no correlation is expected, was also performed (lower right).
Since for the p53 autoantibody detection there is no standard protein which can be used (since the analyte is the serum autoantibody), the full 3-plex assay (CEA, GDF15 and p53 autoantibody) was compared to single-plex measurements for a variety of known positive and negative samples spanning a range signals, in order to confirm multiplexing compatibility. A linear regression analysis between the single-plex and 3-plex assays is shown in Figure 4B, yielding an R2 value ≥0.98 for all 3 biomarkers. To show the specificity of this metric, a linear regression between 3-plex measurements of GDF15 and p53 autoantibody, in which no correlation is expected, yields an R2 value of 0.04.
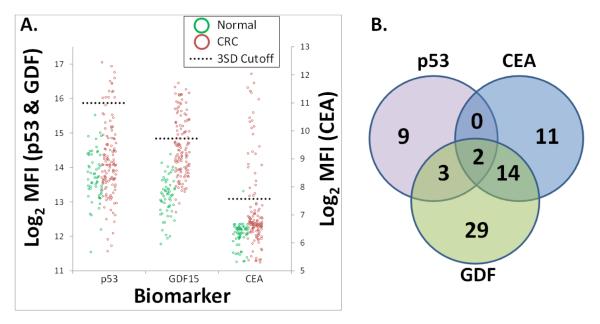
Finally, in a culmination of these efforts, the full 3-plex assay was performed on 186 CRC and normal patient serum/plasma samples (59 normal and 127 CRC) (Figure 5A). Using the aforementioned cutoff and scoring method, individually, CEA, GDF15 and the p53 TAA were 21%, 38% and 11% sensitive and 98%, 100% and 100% specific, respectively. Composite sensitivity and specificity of all 3 biomarkers in the multiplexed assay was 54% and 98%, respectively and biomarker overlap (or lack thereof) is shown in the Venn Diagram in Figure 5B. Notably, while partial redundancy is observed, each biomarker detects several CRC patients that the other biomarkers do not (9, 11 and 29 unique patients for p53, CEA and GDF15, respectively).
Figure 5. Diagnostic Performance of a Three Marker CRC Assay Combining Autoantibody (TAA) and Circulating Protein Detection.
(A.) 3-plex recombinant TAA and soluble protein assay on VeraCode™ beads was performed 186 patient samples (59 normal and 127 CRC). Protein TAAs were bound directly to the VeraCode™ carboxyl beads and used to assay patient serum/plasma for the presence of autoantibodies, while capture antibodies were bound to VeraCode™ carboxyl beads to assay for circulating protein markers. Individual markers are denoted on the x-axis. Individual patient samples are denoted by green circles (healthy individuals) and red circles (colorectal cancer patients). MFI= Mean Fluorescence Intensity of the BeadXpress™ instrument readout (log2 data shown). The black dotted lines indicate the diagnostic scoring cutoffs, set at 3 standard deviations above the mean of the normal patients. CEA, GDF15 and the p53 TAA were 21%, 38% and 11% sensitive and 98%, 100% and 100% specific, respectively. Composite sensitivity and specificity of all 3 biomarkers in the multiplexed assay was 54% and 98%, respectively. (B.) Venn diagram demonstrating the number of “hits”, or CRC patients which tested positive, specific to each biomarker. The overlap of circles indicates patients that were detected by more than one biomarker.
4. Discussion
Here we demonstrate the novel adaptation of Illumina’s multiplexed, genomic, VeraCode™ micro-bead technology for high-throughput immunoassay and validation of two classes of serological biomarkers: autoantibodies to TAAs (see Figure 1) and circulating non-antibody proteins, using colorectal cancer (CRC) as a model system. We have created a multiplexed “hybrid” assay for the simultaneous detection of these two classes of serological biomarkers. To our knowledge, this is the first report of use of the VeraCode™ micro-beads as a protein/immunoassay platform. The potential advantages of this assay include its requirement for only a small volume of blood, the ability to multiplex and perform this in a high-throughput manner, and the ability to add in new biomarkers to eventually achieve a higher level of sensitivity while maintaining a high specificity for CRC diagnosis. Our goal is to continue to add to and refine our 3-marker CRC panel, thereby creating an effective CRC diagnostic screening test, which would be predicted to have excellent compliance due to its non-invasive nature. This approach could be used as a targeted population-wide screening test (for people over 50), or could eventually replace the colonoscopy altogether, assuming that the appropriate level of sensitivity and specificity is achieved by the expansion of our CRC biomarker panel. Another use for this novel protein-based platform could be for the high-throughput clinical validation studies which are urgently needed for the constant stream of newly reported putative serological biomarkers. For example, emerging proteomic techniques such as high density protein microarrays (Hudson et al., 2007; Babel et al., 2009; Anderson et al., 2011) have greatly accelerated the pace at which candidate TAAs are currently being discovered. However, a major bottleneck is the rigorous clinical validation of these candidates in order to establish their true clinical utility and significance. A high throughput validation method is desperately needed for testing the plethora of discovered or partially validated serological biomarkers, such as TAAs, which are being reported for various cancers with potential use in diagnostics (Reuschenbach et al., 2009; Creeden et al., 2011). When moving to clinical studies on very large and diverse patient populations, it would be desirable to screen as many candidate TAAs as practical, since diagnostic performance of biomarkers under these rigorous conditions cannot always be predicted (in fact, a great many biomarkers fail at this stage). Furthermore, it is increasingly clear that due to the heterogeneity of human cancers, panels or signatures of biomarkers, including different classes of biomarkers, will be required for optimal diagnostic performance in the ultimate clinical assay. The VeraCode™ bead-based, multiplexed, solid-phase immunoassay method reported here is ideally suited both for clinical validation and diagnostic detection of serological biomarker panels or signatures, including autoantibodies against TAAs as well as non-antibody protein biomarkers.
Technical validation of the tumor biomarker assay itself is a critical step in the development of clinical test (Marchio et al., 2011). We first validated the VeraCode™ technology for serological immunoassays by comparison to the gold standard and clinically accepted ELISA method. For detection of autoantibodies against TAAs, VeraCode™ results obtained using both a commercial recombinant or a cell-free produced p53 protein compared well to the ELISA data (96% “hit” concordance in CRC) confirming the validity of the method. Indeed, the only discordance occurred where the VeraCode™ immunoassays were able to reproducibly detect two additional low-positive, statistically valid CRC hits (4% increase in diagnostic sensitivity). This increased sensitivity is likely the result of decreased background in the normal patient samples relative to the p53-positive samples, particularly with the recombinant protein (see Figure 2 middle panel). A basis for this low background may be the relatively “bio-friendly”, hydrophilic glass bead surface as opposed to the hydrophobic polystyrene ELISA plates.
As additional technical validation, it should be noted that the overall diagnostic sensitivity of the p53 VeraCode™ assay for CRC (15% in above experiments) is in excellent agreement with literature reports (average of 8% and maximum of 24% sensitive in systematic survey (Reuschenbach et al., 2009)). Finally, intra- and inter-assay CVs of the VeraCode™ TAA assays were strong at 10% (48 total data points) and 20% (282 total data points), respectively. While the inter-assay CVs were acceptable, future improvements in reproducibility may be achieved with the development of rigorous assay-to-assay normalization controls and with better mixing approaches for the large and relatively dense 240 micron glass beads (cylinders), which tend to settle quickly and may result in poor and inconsistent mixing and binding kinetics.
Likewise, the VeraCode™ system was also technically validated against ELISA for detection of non-antibody circulating protein biomarkers using a sandwich immunoassay format. In this case, the CRC biomarker CEA was used as a model system. Here, 94% hit concordance was seen between the two assay types in 52 CRC samples (and quantitative correlation of R2 = 0.9 when a linear regression is performed between the assays). Not surprisingly, the only discordant hits were borderline positive or negative CRC samples that fell extremely close to the cutoffs (see red asterisks in Figure 3A), as the consistently low background in the normal patients resulted in a very low scoring cutoff (both assays show 100% specificity against normal samples).
Next, by combining the most robust TAA observed in our studies, p53, with sandwich immunoassay based quantification of the well-known CRC biomarker CEA, and the cytokine GDF15 in a hybrid multiplexed assay, we achieved a composite diagnostic sensitivity and specificity of 54% and 98%, respectively (186 samples CRC and normal). Thus, we demonstrate the ability to measure, in multiplex, two distinctly different biomarker types using different assay formats, simultaneously, on the VeraCode™ beads. As with the TAAs alone, the additive benefit of combing multiple biomarkers stems from the lack of complete redundancy, with each biomarker detecting several patients (9 to 29) which the others did not, and with no single biomarker exceeding 38% sensitivity (GDF15).
It is important to emphasize that while the particular biomarkers used here were chosen to exemplify the immunoassay method, the clinical studies performed here were only preliminary, retrospective validation studies on a particular cohort of CRC and normal patient samples, and that the results of these studies would need further validation using larger patient cohorts, as well as non-target disease controls (e.g. inflammatory bowel disease and cancers other than CRC) and ultimately, blinded studies and prospective clinical studies. In the future, , it is expected that the CRC biomarker panel would not only expand, but would be refined through elimination of biomarkers as further studies are performed using the VeraCode™ immunoassay methods presented here. For example, GDF15 is a stress-induced cytokine and in addition to CRC has been shown to be a biomarker for a variety of conditions such as heart disease (reviewed in (Wollert and Kempf, 2012)) and worsening albuminuria in patients with type 2 diabetes (Hellemons et al., 2012).
In conclusion, we have demonstrated the VeraCode™ bead platform provides the basis for a robust, sensitive, accurate and high throughput test for multiplexed biomarker detection, as well as for the eventual clinical diagnostic assay which could be employed for biomarker signatures or panels. We anticipate that addition of more biomarkers to the assay could ultimately provide the necessary diagnostic performance for non-invasive population-wide CRC screening which could complement the expensive, slower and more invasive colonoscopy.
Supplementary Material
Highlights.
Created a multiplexed immunodiagnostic protein platform on Illumina’s VeraCode beads.
We detect circulating proteins and autoantibodies to TAAs on VeraCode beads.
Our hybrid multiplexed assay detects CEA, GDF15 and autoantibodies to p53 in CRC.
Our assay achieved 54% sensitivity and 98% specificity for CRC.
Eventually, this assay could be used to screen for CRC to complement colonoscopy.
Acknowledgements
Grant Support This work was funded in part by a Phase I and II SBIR grants (R43/R44 CA137948) from the National Institutes of Health to AmberGen Incorporated.
Financial Support: NIH Grant R43/R44 CA137948.
This project was funded by AmberGen, in part using SBIR grant funds from the National Institutes of Health (see Financial Support, above).
Abbreviations
- TAAs
tumor associated antigens
- CRC
colorectal cancer
- CEA
carcinoembryonic antigen
- GDF15
growth differentiation factor 15
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ Contributions HPO, KJR and MJL contributed to project conception and design. HPO, AA, ZL, ZW, KIB and MJL contributed to development of methodology and analysis and interpretation of data. AA, ZL, ZW and KIB were responsible for data acquisition. HPO, KJR and MJL were responsible for writing and review of the manuscript.
Disclosure of Potential Conflicts of Interest: HPO, AA, ZL, ZW, KJR and MJL are current employees of AmberGen Incorporated. KIB is a former employee of AmberGen. AmberGen is a developer of commercial diagnostic assays.
References
- Ahlquist DA. Fecal occult blood testing for colorectal cancer. Can we afford to do this? Gastroenterol Clin North Am. 1997;26:41–55. doi: 10.1016/s0889-8553(05)70282-x. [DOI] [PubMed] [Google Scholar]
- Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–33. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KS, Sibani S, Wallstrom G, Qiu J, Mendoza EA, Raphael J, Hainsworth E, Montor WR, Wong J, Park JG, Lokko N, Logvinenko T, Ramachandran N, Godwin AK, Marks J, Engstrom P, Labaer J. Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J Proteome Res. 2011;10:85–96. doi: 10.1021/pr100686b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babel I, Barderas R, Diaz-Uriarte R, Martinez-Torrecuadrada JL, Sanchez-Carbayo M, Casal JI. Identification of tumor-associated autoantigens for the diagnosis of colorectal cancer in serum using high density protein microarrays. Mol Cell Proteomics. 2009;8:2382–95. doi: 10.1074/mcp.M800596-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belousov PV, Kuprash DV, Sazykin AY, Khlgatian SV, Penkov DN, Shebzukhov YV, Nedospasov SA. Cancer-associated antigens and antigen arrays in serological diagnostics of malignant tumors. Biochemistry (Mosc) 2008;73:562–72. doi: 10.1134/s000629790805009x. [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara ECM, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–9. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger BM, Schroy PC, 3rd, Rosenberg JL, Lai-Goldman M, Eisenberg M, Brown T, Rochelle RB, Billings PR. Colorectal cancer screening using stool DNA analysis in clinical practice: early clinical experience with respect to patient acceptance and colonoscopic follow-up of abnormal tests. Clin Colorectal Cancer. 2006;5:338–43. doi: 10.3816/CCC.2006.n.003. [DOI] [PubMed] [Google Scholar]
- Brown DA, Hance KW, Rogers CJ, Sansbury LB, Albert PS, Murphy G, Laiyemo AO, Wang Z, Cross AJ, Schatzkin A, Danta M, Srasuebkul P, Amin J, Law M, Breit SN, Lanza E. Serum macrophage inhibitory cytokine-1 (MIC-1/GDF15): a potential screening tool for the prevention of colon cancer? Cancer Epidemiol. Biomarkers Prev. 2012;21:337–46. doi: 10.1158/1055-9965.EPI-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiano CA, Mediavilla-Varela M, Tan EM. Tumor-associated antigen arrays for the serological diagnosis of cancer. Mol Cell Proteomics. 2006;5:1745–59. doi: 10.1074/mcp.R600010-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman CJ, Murray A, McElveen JE, Sahin U, Luxemburger U, Tureci O, Wiewrodt R, Barnes AC, Robertson JF. Autoantibodies in lung cancer: possibilities for early detection and subsequent cure. Thorax. 2008;63:228–33. doi: 10.1136/thx.2007.083592. [DOI] [PubMed] [Google Scholar]
- Chee M, Yang R, Hubbell E, Berno A, Huang XC, Stern D, Winkler J, Lockhart DJ, Morris MS, Fodor SP. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–4. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin P, Qiu S, Peng XX, Looi K, Farquhar MG, Zhang JY. Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int. J. Oncol. 2007;30:1137–44. [PubMed] [Google Scholar]
- Creeden J, Junker F, Vogel-Ziebolz S, Rex D. Serum tests for colorectal cancer screening. Mol Diagn Ther. 2011;15:129–41. doi: 10.1007/BF03256403. [DOI] [PubMed] [Google Scholar]
- Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer. 2005;5:199–209. doi: 10.1038/nrc1569. [DOI] [PubMed] [Google Scholar]
- de Voer RM, van der Klis FR, Engels CW, Rijkers GT, Sanders EA, Berbers GA. Development of a fluorescent-bead-based multiplex immunoassay to determine immunoglobulin G subclass responses to Neisseria meningitidis serogroup A and C polysaccharides. Clin Vaccine Immunol. 2008;15:1188–93. doi: 10.1128/CVI.00478-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettloff R, Yang E, Rulison A, Chow A, Farinas J. Nucleic acid amplification of individual molecules in a microfluidic device. Anal. Chem. 2008;80:4208–13. doi: 10.1021/ac800339w. [DOI] [PubMed] [Google Scholar]
- Doolittle BR, Emanuel J, Tuttle C, Costa J. Detection of the mutated K-Ras biomarker in colorectal carcinoma. Exp Mol Pathol. 2001;70:289–301. doi: 10.1006/exmp.2001.2364. [DOI] [PubMed] [Google Scholar]
- Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–73. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- Fouda GG, Leke RF, Long C, Druilhe P, Zhou A, Taylor DW, Johnson AH. Multiplex assay for simultaneous measurement of antibodies to multiple Plasmodium falciparum antigens. Clin Vaccine Immunol. 2006;13:1307–13. doi: 10.1128/CVI.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR., Jr. Advanced multiplexed analysis with the FlowMetrix system. Clin. Chem. 1997;43:1749–56. [PubMed] [Google Scholar]
- Gunderson KL, Kruglyak S, Graige MS, Garcia F, Kermani BG, Zhao C, Che D, Dickinson T, Wickham E, Bierle J, Doucet D, Milewski M, Yang R, Siegmund C, Haas J, Zhou L, Oliphant A, Fan JB, Barnard S, Chee MS. Decoding randomly ordered DNA arrays. Genome Res. 2004;14:870–7. doi: 10.1101/gr.2255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemons ME, Mazagova M, Gansevoort RT, Henning RH, de Zeeuw D, Bakker SJ, Lambers-Heerspink HJ, Deelman LE. Growth-differentiation factor 15 predicts worsening of albuminuria in patients with type 2 diabetes. Diabetes Care. 2012;35:2340–6. doi: 10.2337/dc12-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975-2009 (Vintage 2009 Populations) National Cancer Institute; Bethesda, MD: 2012. [Google Scholar]
- Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17494–9. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 2001;23:440–6. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, Chan EK, Tan EM. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin. Cancer Res. 2003;9:5120–6. [PubMed] [Google Scholar]
- Lin CH, Yeakley JM, McDaniel TK, Shen R. Medium- to high-throughput SNP genotyping using VeraCode microbeads. Methods Mol. Biol. 2009;496:129–42. doi: 10.1007/978-1-59745-553-4_10. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang P, Li Z, Xu W, Dai L, Wang K, Zhang J. Evaluation of tumour-associated antigen (TAA) miniarray in immunodiagnosis of colon cancer. Scand. J. Immunol. 2009;69:57–63. doi: 10.1111/j.1365-3083.2008.02195.x. [DOI] [PubMed] [Google Scholar]
- Marchio C, Dowsett M, Reis-Filho JS. Revisiting the technical validation of tumour biomarker assays: how to open a Pandora’s box. BMC Med. 2011;9:41. doi: 10.1186/1741-7015-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozoe T, Yasuda M, Honda M, Inutsuka S, Korenaga D. Clinicopathologic significance in serum presence of anti-p53 antibody in patients with colorectal carcinoma. Hepatogastroenterology. 2007;54:1422–5. [PubMed] [Google Scholar]
- Opalka D, Lachman CE, MacMullen SA, Jansen KU, Smith JF, Chirmule N, Esser MT. Simultaneous quantitation of antibodies to neutralizing epitopes on virus-like particles for human papillomavirus types 6, 11, 16, and 18 by a multiplexed luminex assay. Clin. Diagn. Lab. Immunol. 2003;10:108–15. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant AL, Locascio-Brown L, Haller W, Durst RA. Immobilization of binding proteins on nonporous supports. Comparison of protein loading, activity, and stability. Appl. Biochem. Biotechnol. 1991;30:83–98. doi: 10.1007/BF02922025. [DOI] [PubMed] [Google Scholar]
- Reuschenbach M, von Knebel Doeberitz M, Wentzensen N. A systematic review of humoral immune responses against tumor antigens. Cancer Immunol. Immunother. 2009;58:1535–44. doi: 10.1007/s00262-009-0733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM. American college of gastroenterology guidelines for colorectal cancer screening 2008. Am. J. Gastroenterol. 2009;104:739–50. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- Saleh J, Kreissler-Haag D, Montenarh M. p53 autoantibodies from patients with colorectal cancer recognize common epitopes in the N- or C-terminus of p53. Int. J. Oncol. 2004;25:1149–55. [PubMed] [Google Scholar]
- Schlottmann SA, Jain N, Chirmule N, Esser MT. A novel chemistry for conjugating pneumococcal polysaccharides to Luminex microspheres. J. Immunol. Methods. 2006;309:75–85. doi: 10.1016/j.jim.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Storr SJ, Chakrabarti J, Barnes A, Murray A, Chapman CJ, Robertson JF. Use of autoantibodies in breast cancer screening and diagnosis. Expert Rev Anticancer Ther. 2006;6:1215–23. doi: 10.1586/14737140.6.8.1215. [DOI] [PubMed] [Google Scholar]
- Su BB, Shi H, Wan J. Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol. 2012;18:2121–6. doi: 10.3748/wjg.v18.i17.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Klosterman M, Amonkar MM, Hunt TL. Adherence with colorectal cancer screening guidelines: a review. Prev. Med. 2004;38:536–50. doi: 10.1016/j.ypmed.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Toth K, Sipos F, Kalmar A, Patai AV, Wichmann B, Stoehr R, Golcher H, Schellerer V, Tulassay Z, Molnar B. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PloS one. 2012;7:e46000. doi: 10.1371/journal.pone.0046000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vijan S, Inadomi J, Hayward RA, Hofer TP, Fendrick AM. Projections of demand and capacity for colonoscopy related to increasing rates of colorectal cancer screening in the United States. Aliment. Pharmacol. Ther. 2004;20:507–15. doi: 10.1111/j.1365-2036.2004.01960.x. [DOI] [PubMed] [Google Scholar]
- Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, van den Heuvel S, Vidal M. GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–92. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wallin U, Glimelius B, Jirstrom K, Darmanis S, Nong RY, Ponten F, Johansson C, Pahlman L, Birgisson H. Growth differentiation factor 15: a prognostic marker for recurrence in colorectal cancer. Br. J. Cancer. 2011;104:1619–27. doi: 10.1038/bjc.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterboer T, Sehr P, Pawlita M. Suppression of non-specific binding in serological Luminex assays. J. Immunol. Methods. 2006;309:200–4. doi: 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Wollert KC, Kempf T. Growth differentiation factor 15 in heart failure: an update. Curr Heart Fail Rep. 2012;9:337–45. doi: 10.1007/s11897-012-0113-9. [DOI] [PubMed] [Google Scholar]
- Woolf SH. A smarter strategy? Reflections on fecal DNA screening for colorectal cancer. N Engl J Med. 2004;351:2755–8. doi: 10.1056/NEJMe048259. [DOI] [PubMed] [Google Scholar]
- Xue H, Lu B, Zhang J, Wu M, Huang Q, Wu Q, Sheng H, Wu D, Hu J, Lai M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J Proteome Res. 2010;9:545–55. doi: 10.1021/pr9008817. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Casiano CA, Peng XX, Koziol JA, Chan EK, Tan EM. Enhancement of antibody detection in cancer using panel of recombinant tumor-associated antigens. Cancer Epidemiol. Biomarkers Prev. 2003;12:136–43. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.