In addition to promoting integrin activation at the immunological synapse, SKAP55 links T cell antigen receptor-associated signaling molecules to the cytoskeleton and promotes integrin-independent adhesion via the T cell receptor.
Abstract
The T cell receptor (TCR) triggers the assembly of “SLP-76 microclusters,” which mediate signals required for T cell activation. In addition to regulating integrin activation, we show that Src kinase–associated phosphoprotein of 55 kD (SKAP55) is required for microcluster persistence and movement, junctional stabilization, and integrin-independent adhesion via the TCR. These functions require the dimerization of SKAP55 and its interaction with the adaptor adhesion and degranulation-promoting adaptor protein (ADAP). A “tandem dimer” containing two ADAP-binding SKAP55 Src homology 3 (SH3) domains stabilized SLP-76 microclusters and promoted T cell adhesion via the TCR, but could not support adhesion to integrin ligands. Finally, the SKAP55 dimerization motif (DM) enabled the coimmunoprecipitation of the Rap1-dependent integrin regulator Rap1-GTP–interacting adaptor molecule (RIAM), the recruitment of talin into TCR-induced adhesive junctions, and “inside-out” signaling to β1 integrins. Our data indicate that SKAP55 dimers stabilize SLP-76 microclusters, couple SLP-76 to the force-generating systems responsible for microcluster movement, and enable adhesion via the TCR by mechanisms independent of RIAM, talin, and β1 integrins.
Introduction
T cells interact with antigen-presenting cells via junctions known as immunological synapses. Although transient interactions trigger T cell receptor (TCR)-proximal responses, sustained conjugate formation is required for optimal cytokine production, proliferation, differentiation, and memory function (Mempel et al., 2004; Henrickson et al., 2008; Zheng et al., 2008). After antigen recognition, the TCR recruits the tyrosine kinase ζ-chain associated phosphoprotein of 70 kD (ZAP-70), giving rise to multivalent signaling complexes referred to as microclusters (Acuto et al., 2008; Smith-Garvin et al., 2009; Balagopalan et al., 2010; Bunnell, 2010). The Src homology 2 (SH2) domain–containing leukocyte protein of 76 kD (SLP-76), which is essential for T cell development and activation, nucleates structures that rapidly form alongside activated TCR microclusters (Bunnell et al., 2002; Nguyen et al., 2008). These “SLP-76 microclusters” are stabilized by multivalent interactions among their constituent adaptors and effectors, including linker of activated T cells (LAT), Grb2, Sos1, Grb2-related adaptor downstream of Shc (Gads), Nck, Vav1, and PLCγ1 (Barda-Saad et al., 2005; Braiman et al., 2006; Bunnell et al., 2006; Houtman et al., 2006; Balagopalan et al., 2007; Sylvain et al., 2011). SLP-76 microclusters are hubs of tyrosine phosphorylation whose persistence is correlated with TCR-induced increases in calcium entry, Erk activation, and CD69 expression (Singer et al., 2004; Campi et al., 2005; Bunnell et al., 2006; Lasserre et al., 2011). In addition, SLP-76 microclusters are transported to the center of the immune synapse, where they appear to be down-modulated via retrograde actin flows, myosin contractility, and microtubule-directed movement (Bunnell et al., 2002; Nguyen et al., 2008; Babich et al., 2012). Although SLP-76 microclusters incorporate multiple actin-binding proteins and regulators, the precise mechanisms by which they are coupled to actin are not well understood (Barda-Saad et al., 2005; Barda-Saad et al., 2010; Lasserre et al., 2010; Sylvain et al., 2011). Nevertheless, these systems initiate and sustain antigen recognition in the context of the immune synapse, and may apply the forces that permit the TCR to overcome significant steric barriers and to act as a mechanosensor (Valitutti et al., 1995; Seminario and Bunnell, 2008; Wang and Reinherz, 2012).
The SLP-76 SH2 domain binds to the adhesion and degranulation-promoting adaptor protein (ADAP), an important regulator of integrin activation and cytoskeletal reorganization (Geng et al., 1999; Raab et al., 1999; Krause et al., 2000; Peterson et al., 2001; Griffiths and Penninger, 2002; Medeiros et al., 2007; Burbach et al., 2011). The interaction of the SLP-76 SH2 domain with ADAP stabilizes SLP-76 microclusters and recruits ADAP into these structures (Pauker et al., 2011; Coussens et al., 2013). The adaptor protein Src kinase–associated phosphoprotein of 55 kD (SKAP55) is essential for the pro-adhesive functions of ADAP, and binds directly to a conserved proline-rich region in ADAP via its Src homology 3 (SH3) domain (Schraven et al., 1997; Marie-Cardine et al., 1998a; Wang et al., 2003, 2007; Kliche et al., 2006). Although SKAP55 binds regulator of adhesion and polarity in leukocytes (RapL), a regulator of the αL integrin leukocyte function–associated antigen 1 (LFA-1; αLβ2), the manner in which SKAP55 engages Rap1-GTP–interacting adaptor molecule (RIAM), a regulator of integrin β chains, remains elusive (Ménasché et al., 2007). Consequently, the mechanisms by which the TCR regulates adhesion via immunologically relevant integrins such as very late antigen 4 (VLA-4; α4β1) and very late antigen 5 (VLA-5; α5β1) remain unresolved (Seminario et al., 1998; Kliche et al., 2006). Even though it is widely assumed that SKAP55 primarily acts through its effects on integrins, we and others have shown that the TCR can function as a fast-acting adhesion receptor (Nguyen et al., 2008). Therefore, it possible that SKAP55 also influences conjugate formation by promoting the efficient coupling of TCR and SLP-76 microclusters to the underlying actin cytoskeleton via associated adaptors, such as ADAP and RIAM (Lafuente et al., 2004; Pauker et al., 2011).
In the context of immune synapses, multiple adhesive and co-stimulatory molecules contribute to T cell activation, making it difficult to dissect the pathways operating downstream of a particular receptor. This presents a significant complication, as integrins recruited into physiological immune synapses and lipid bilayer–induced synapses (e.g. LFA-1, VLA-4) signal through ADAP and SKAP55 (Grakoui et al., 1999; Hunter et al., 2000; Mittelbrunn et al., 2004; Baker et al., 2009). Here, we use plate-bound antibodies specific for the TCR to establish that SKAP55 is an integral component of TCR-induced SLP-76 microclusters, and is required for their persistence and centripetal movement. We also show that SKAP55 regulates T cell spreading downstream of the TCR, facilitates the formation of stable contacts, and promotes adhesion via the TCR in the absence of integrin ligands. These processes require a functional SKAP55 dimer and can be supported by an artificial “tandem dimer” (TD) consisting of two SKAP55 SH3 domains coupled via a flexible linker. These data demonstrate that SKAP55 regulates cluster dynamics and TCR-based spreading and adhesion by multimerizing ADAP. However, the TD cannot support the transmission of “inside-out” signals from the TCR to β1 integrins and cannot bind to RIAM and talin, which play crucial roles in T cell adhesion via integrins. Further, we show that the deletion of an N-terminal dimerization motif (DM) within SKAP55 prevents the formation of SKAP55–RIAM–talin complexes and eliminates the recruitment of talin into the TCR-induced adhesive junctions within which SLP-76 microclusters are nucleated. Thus, we have identified distinct structural elements that enable SKAP55 to independently regulate adhesion via the TCR and the transmission of local inside-out signals from the TCR to integrins.
Results
The recruitment of SKAP55 into signalling microclusters is dependent on SLP-76 and ADAP
The constitutive association of SKAP55 with ADAP enables these adaptors to participate in a variety of biological processes (Wu et al., 2002; Wang et al., 2003, 2007; Burbach et al., 2008, 2011). Because all SKAP55 molecules associate with ADAP, which engages SLP-76 directly when tyrosine phosphorylated, we hypothesized that SKAP55 would function within SLP-76 microclusters (Marie-Cardine et al., 1998a; Pauker et al., 2011). Thus, we plated SLP-76–deficient Jurkat cells (J14 cells) that had been stably reconstituted with SLP-76.YFP (J14.SY cells) on stimulatory antibody (OKT3)-coated and glass surfaces that had been BSA blocked to minimize the deposition of serum components on the glass. T cells settling onto these substrates are activated via the TCR (Fig. S1 A), develop signaling microclusters, and undergo cytoskeletal rearrangements that convert the responding cells into flat adherent cells within 3–5 min. Under these conditions, endogenous SKAP55 colocalized with SLP-76 in TCR-induced microclusters (Fig. 1 A). Similarly, an exogenous SKAP55 chimera tagged with the red fluorescent protein TagRFP-Turbo (TRT) entered and co-migrated with SLP-76 microclusters (Video 1). To display the movement of the SLP-76– and SKAP55-containing microclusters, movies were compressed into maximum-over-time (MOT) images that depict persistent, mobile microclusters as radial spokes extending from the periphery to the center of the contact (Fig. 1 B). SKAP55 was cytoplasmic in SLP-76–deficient J14 cells expressing monomeric YFP (mYFP), but clustered when coexpressed with exogenous SLP-76.YFP (Fig. 1 C). Although ADAP increases the half-life of SKAP55, it is possible to express exogenous SKAP55 in the absence of ADAP (Fig. S1 B; Huang et al., 2005). Using ADAP-deficient JDAP cells, we demonstrated that the recruitment of SKAP55 into persistent and mobile microclusters also requires ADAP (Fig. 1 D). To display microcluster persistence and directionality, we created kymographs depicting lateral movement over time. In these images the long, slanted traces represent persistent and mobile microclusters, whereas shorter, vertical traces represent labile, immobile microclusters (Fig. 1 E, top). SLP-76 microclusters did not undergo sustained movements in the absence of ADAP, even when exogenous SKAP55.mCFP was present. Reconstitution with exogenous ADAP restored normal microcluster dynamics, and enabled the comigration of SKAP55 with SLP-76 (Fig. 1 E). Thus, SKAP55 recruitment to and movement with signaling microclusters requires both SLP-76 and ADAP.
Figure 1.
SKAP55 is recruited into microclusters by SLP-76 and ADAP. (A) Immunofluorescent staining for endogenous SKAP55 in J14.SY cells fixed 5 min after plating on anti-CD3 coated, BSA-blocked stimulatory coverslips. (B) J14.SY cells expressing either SKAP55.TRT (top) or TRT (bottom) were stimulated on coverslips and imaged for 5 min. MOT projections depict the trajectories of microclusters over time (n = 20). (C) MOT images of J14 cells coexpressing SKAP55.mRFP1 and either mYFP (top) or SLP-76.mYFP (bottom) (n = 3). (D) MOT images of ADAP-deficient JDAP cells coexpressing SKAP55.3×Flag.mCFP, SLP-76.YFP, and either 3×Flag.TRT (top) or TRT.3×Flag.TRT.ADAP-120 (bottom) (n = 4). (E) Kymographs depicting the lateral movements of SLP-76, SKAP55, and ADAP microclusters along a narrow region of interest spanning the center of the cell. Bars: (A–D) 10 µm; (E) 5 µm × 60 s.
SKAP55 is required for the stabilization of SLP-76 microclusters and TCR-initiated contacts
To determine if SKAP55 influences the dynamics of SLP-76 microclusters, we performed transient knockdown experiments using a SKAP55-specific shRNA expression vector incorporating a monomeric red fluorescent protein 1 (mRFP1) reporter. J14.SY cells transfected with the knockdown vector or a control mRFP1 expression vector were isolated using a YFP+/RFP+ gate and imaged on stimulatory coverslips. The knockdown cells had fewer SLP-76 microclusters, and these structures were both labile and immobile (Fig. S1, C and D; and Video 2). Differences in microcluster persistence, displacement, and peak speed were quantified by manual microcluster tracing, which confirmed the significance of the alterations in microcluster dynamics (Table 1; ±SEM in three or more experiments). These changes were also depicted by compiling average traces for each cell, and deriving a single composite kymograph for each condition, weighing each cell equally (Fig. S1 E). In addition, the time-averaged fraction of SLP-76.YFP intensity falling within microclusters was reduced upon SKAP55 knockdown (Table 1); this parameter reflects microcluster number, relative brightness, and survival over time (Table 1). SKAP55-deficient cells also formed less stable contacts with the stimulatory substrate over time. To quantitate this phenotype, we used an automated segmentation algorithm acting on the cytoplasmic mRFP1 signal to track contact boundaries. Regions of boundary growth (red) and retraction (blue) were defined by comparison with images captured 30 s earlier; regions that remained constant were labeled in gray (Fig. S1 F; and Videos 3 and 4). Consistent with our observations, the fluctuation of contact, defined as the mean over time of the dynamic area (red + blue) per current area (red + gray), increased after SKAP55 knockdown (Fig. S1, F and G). These changes were associated with increases in the extent and duration of periodic cycles of contact growth and retraction (Fig. S1 H; color indicates rate of growth or retraction along the cell perimeter). Because these analyses were computationally intensive, we also categorized cell boundaries as stable or unstable by blinded manual scoring. This approach also revealed a significant destabilization of contact boundaries in cells lacking SKAP55 (Fig. S1, I and J).
Table 1.
Impact of SKAP55 mutants on SLP-76 microcluster dynamics
| SKAP55 chimera | Persistence | P | Inward movement | P | Max speed | P | % SLP-76 in clusters | P | Clusters tracked per diameter | P | Totalcells | n (experiments) |
| s | µm | nm/s | ||||||||||
| mRFP1 | 210.5 ± 7.20 | 3.0 ± 0.2 | 147.7 ± 6.1 | 62.0 ± 2.8 | 15.1 ± 0.48 | 46 | 19 | |||||
| KD | 115.8 ± 38.2 | ** | 0.6 ± 0.1 | ** | 57.8 ± 8.6 | ** | 27.3 ± 6.9 | ** | 12.5 ± 1 | *, †† | 10 | 3 |
| WT | 215.7 ± 7.52 | 3.4 ± 0.3 | 150 ± 8.6 | 72.1 ± 2.3 | 17.2 ± 0.87 | * | 29 | 15 | ||||
| WR | 129.3 ± 18.9 | **, †† | 1.3 ± 0.3 | **, †† | 127.8 ± 31.9 | 37.6 ± 6.6 | **, †† | 13.6 ± 1.8 | 8 | 3 | ||
| SH3 | 150.5 ± 16.4 | **, †† | 1.4 ± 0.5 | **, †† | 104.6 ± 23.4 | *, † | 41.9 ± 15.4 | *, †† | 9.6 ± 1.5 | **, †† | 10 | 5 |
| ΔPH | 222.9 ± 2.1 | 2.8 ± 0.2 | 154.2 ± 9.6 | 77.3 ± 2.9 | 15.3 ± 0.33 | 10 | 3 | |||||
| 3YF | 169 ± 16.4 | *, †† | 2.3 ± 0.3 | 153.5 ± 17.4 | 66.5 ± 5.1 | 16.4 ± 0.98 | 10 | 4 | ||||
| ΔDM | 175.9 ± 23.3 | 1.5 ± 0.2 | **, †† | 137.7 ± 15.1 | 44.9 ± 5.2 | **, †† | 13.3 ± 1.5 | † | 10 | 3 | ||
| DM | 133.1 ± 5.9 | **, †† | 1.1 ± 0.1 | **, †† | 101.6 ± 6.2 | **, † | 32 ± 4.0 | **, †† | 14 ± 2 | 10 | 3 | |
| TD | 227.2 ± 11.6 | 2.5 ± 0.2 | 129.1 ± 23.8 | 73.1 ± 1.1 | 14 ± 0.95 | † | 10 | 3 |
SLP-76 microcluster dynamics in J14.SY cells expressing each of the indicated constructs were measured on a per cell basis by manual tracing of microcluster trajectories from SLP-76 kymographs. Values for each experiment were obtained by averaging these per cell values, weighing each cell equally. KD, knockdown. Values displayed in the table are mean ± SEM for the indicated number of experiments. From parental J14.SY cells expressing mRFP1: *, P < 0.05; **, P < 0.01. From J14.SY cells expressing SKAP55.WT.mRFP1: †, P < 0.05; ††, P < 0.01.
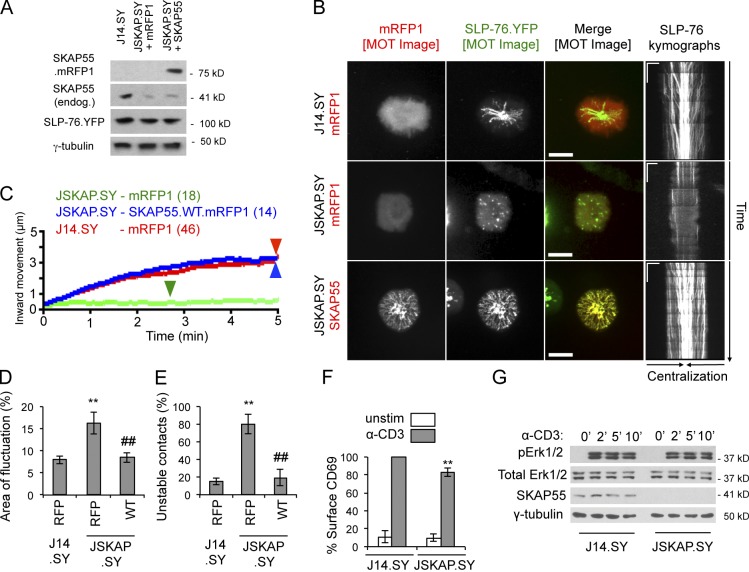
To verify that the phenotypes observed upon transient knockdown were caused by the specific loss of SKAP55, rather than off-target effects of the hairpin, we derived SKAP55-deficient J14.SY cells (JSKAP.SY) using a lentivirally encoded SKAP55-specific shRNA. These cells were transfected with vectors encoding either mRFP1 or a hairpin-resistant SKAP55.mRFP1 chimera (Fig. 2 A). In contrast to J14.SY cells, JSKAP.SY cells produced immobile and short-lived SLP-76 microclusters (Fig. 2, B and C; Table 2; and Video 5). The reexpression of SKAP55 restored optimal microcluster dynamics, confirming that SKAP55 is an integral component of SLP-76 microclusters (Fig. 2, B and C; Table 2; and Video 6). Finally, reconstitution reduced boundary fluctuation in TCR-induced contacts, and reduced the fraction of contacts scored as “unstable,” confirming that SKAP55 also regulates the junctions formed in response to solo TCR ligation (Fig. 2, D and E).
Figure 2.
SKAP55 is required for microcluster persistence and movement, and for contact stability. (A) Confirmation of the efficacy of SKAP55 knockdown and add-back in stable SKAP55 knockdown cells (JSKAP.SY) with or without transient reconstitution (n = 3). (B) J14.SY and JSKAP.SY cells expressing mRFP1 or SKAP55.mRFP1 were stimulated on coverslips and imaged for at least 5 min. Representative MOT images and kymographs are shown. Bars: (MOT images) 10 µm; (kymographs) 5 µm × 60 s. See Table 2 for microcluster properties and experiment numbers. (C) Composite microcluster traces for the conditions examined in B. Numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (D) Fraction of the contact area engaged in fluctuation (see Fig. S1, F and G; n = 3). (E) Fraction of cells scored as displaying unstable contacts (see Fig. S1, I and J; n = 5). (F) Surface expression of CD69 in J14.SY and JSKAP.SY cells, after normalization to a TCR-stimulated J14.SY control (n = 4). (G) Kinetics of Erk1/2 phosphorylation in J14.SY and JSKAP.SY stimulated for the indicated time points (n = 3). Error bars indicate mean ± SEM. From parental J14.SY cells (with or without mRFP1): **, P < 0.01. From JSKAP.SY (with or without mRFP1): ##, P < 0.01.
Table 2.
Impact of SKAP55 mutants on SLP-76 microcluster dynamics in JSKAP.SY
| SKAP55 chimera | Persistence | P | Inward movement | P | Max speed | P | % SLP-76 in clusters | P | Clusters tracked per diameter | P | Total cells | n (experiments) |
| s | µm | nm/s | ||||||||||
| mRFP1 | 103.9 ± 35.2 | †† | 0.89 ± 0.13 | †† | 84.04 ± 15.6 | †† | 25.8 ± 6.8 | †† | 11.7 ± 1.2 | † | 18 | 7 |
| WT | 235.3 ± 13.5 | ** | 3.27 ± 0.47 | ** | 131.1 ± 10.5 | ** | 67.3 ± 2.9 | ** | 15.4 ± 1 | * | 14 | 6 |
| Hom | 221.2 ± 14.8 | ** | 3.29 ± 0.13 | ** | 188.4 ± 25.9 | **, † | 73.7 ± 0.4 | ** | 15.9 ± 0.57 | * | 10 | 3 |
| ΔDM | 111.3 ± 23.8 | †† | 0.88 ± 0.09 | †† | 74.6 ± 18.4 | †† | 19.0 ± 7.2 | †† | 4.8 ± 0.92 | **, †† | 9 | 3 |
| TD | 203.4 ± 20.9 | 2.95 ± 0.14 | ** | 129.5 ± 12.6 | 65.1 ± 2.8 | ** | 13.5 ± 1.3 | 10 | 3 |
SLP-76 microcluster dynamics in JSKAP.SY cells were measured as in Table 1. From JSKAP.SY cells expressing mRFP1: *, P < 0.05; **, P < 0.01. From JSKAP.SY with SKAP55.WT.mRFP1 add-back: †, P < 0.05; ††, P < 0.01.
Defects in SLP-76 microcluster persistence and movement have been correlated with significant reductions in TCR-proximal signals, including calcium entry and CD69 up-regulation (Bunnell et al., 2006; Sylvain et al., 2011). Although the loss of SKAP55 clearly impacted microcluster properties and TCR-induced cytoskeletal rearrangements, the suppression of endogenous SKAP55 did not dramatically impact TCR-induced increases in CD69 expression or Erk activation (Fig. 2, F and G).
The SKAP55 SH3 domain is necessary but insufficient for recruitment into and stabilization of SLP-76 microclusters
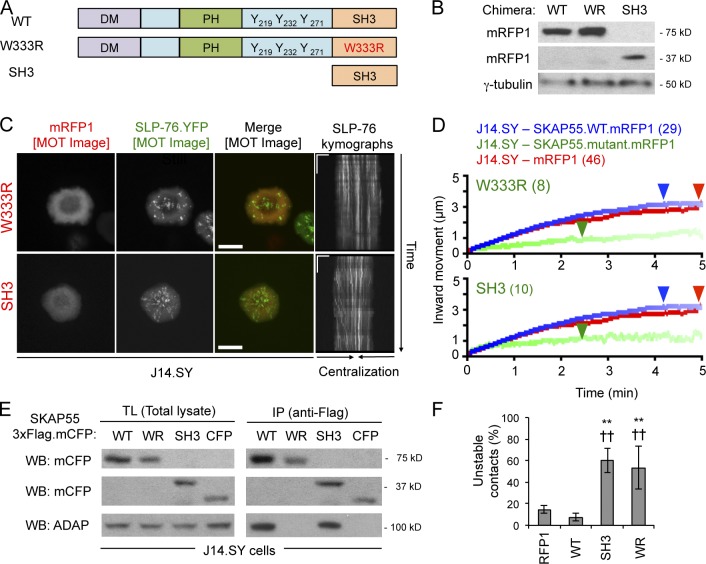
Consistent with the important role of ADAP in the recruitment of SKAP55 into SLP-76 microclusters (Fig. 1, D and E), an ADAP nonbinding SKAP55 SH3 domain mutant chimera (W333R; “WR”) failed to cocluster with SLP-76 when overexpressed in J14.SY cells (Fig. 3, A–C). Relative to cells expressing either a WT chimera or an mRFP1 control, cells expressing the W333R mutant exhibited short-lived and immobile microclusters (Fig. 3, B–D; Video 7; and Table 1). The SLP-76 microclusters produced in cells expressing the W333R mutant also incorporated less SLP-76, but did not show significant decreases in peak microcluster speed (Table 1). An isolated SKAP55 SH3 domain (“SH3”) behaved as a dominant-negative for microcluster dynamics even though it interacted with ADAP (Fig. 3, A–E; and Table 1). Finally, the W333R mutant and the isolated SH3 domain reduced the stability of TCR-induced contacts (Fig. 3 F). Thus, regions in the N terminus of SKAP55 are critical for its function.
Figure 3.
The SKAP55 SH3 domain is necessary but not sufficient for stabilization of SLP-76 microclusters. (A) Domain structures of the wild-type (WT), SH3 mutant (W333R, “WR”), and SH3-only SKAP55 chimeras (SH3). (B) mRFP1 expression levels in J14.SY cells transiently transfected with the indicated SKAP55.mRFP1 chimeras (n = 3). (C) Cells from B were stimulated, imaged, and presented as in Fig. 2 B. See Table 1 for microcluster properties and experiment numbers. Bars: (MOT images) 10 µm; (kymographs) 5 µm × 60 s. (D) Composite microcluster traces for the conditions examined in C. Numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (E) SKAP55.3×Flag.mCFP chimeras were expressed in J14.SY cells, immunoprecipitated with anti-Flag, and Western blotted for mCFP or ADAP (n = 4). (F) Fraction of cells scored as displaying unstable contacts (n = 4). Error bars indicate mean ± SEM. From parental J14.SY cells expressing mRFP1: **, P < 0.01. From J14.SY cells expressing SKAP55.WT.mRFP1: ††, P < 0.01.
The SKAP55 Pleckstrin homology (PH) domain and linker tyrosines play minimal roles in SLP-76 microcluster dynamics and contact stability downstream of the TCR
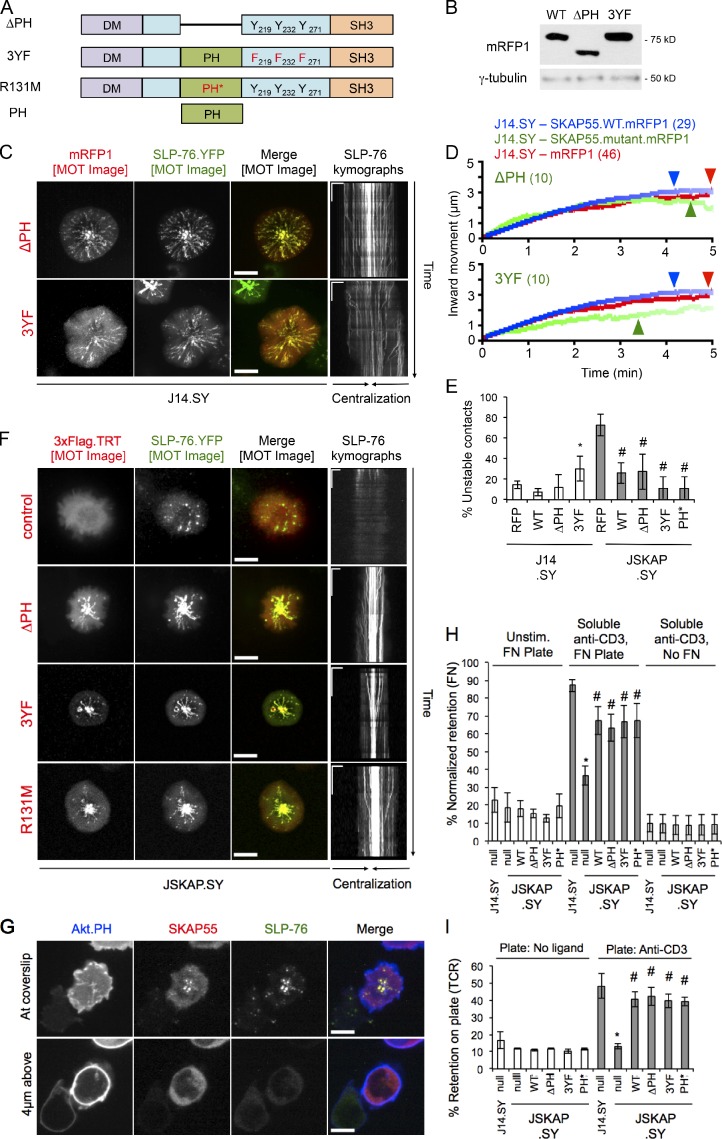
Within the central portion of SKAP55, two regions have been implicated in intermolecular interactions: a PH domain and a tyrosine-phosphorylated linker region (Wu et al., 2002; Hornbeck et al., 2004; Swanson et al., 2008; Fig. 4 A). Nevertheless, SKAP55 chimeras lacking the PH domain (ΔPH) or the linker tyrosines (Y219F/Y232F/Y271F, “3YF”) entered SLP-76 microclusters (Fig. 4, A–C). Only the 3YF chimera exhibited even a mild (but significant) dominant-negative effect on microcluster dynamics or TCR-mediated contact stability (Fig. 4, D and E; and Table 1). Reconstitution of JSKAP.SY cells with matched levels of shRNA-resistant wild-type, 3YF, ΔPH, or inositide nonbinding (R131M, PH*) SKAP55 chimeras all improved SLP-76 microcluster dynamics relative to cells expressing only 3×Flag.TRT, and all three mutant chimeras stabilized contacts in JSKAP.SY cells (Fig. 4, E and F; and Fig. S2 A; Swanson et al., 2008). SKAP55 did not recruit to the plasma membrane at surfaces outside of SLP-76 microclusters, either at or above the stimulatory coverslip, even though the phosphatidylinositol-3,4,5-trisphosphate–binding PH domain of Akt clearly labeled these membranes (Fig. 4 G). Furthermore, an isolated SKAP55 PH domain chimera was not recruited to the plasma membrane and remained distributed throughout the cytoplasm and nucleus after TCR ligation (Fig. S2 B). Thus, in our model system, the PH domain and the linker tyrosines of SKAP55 are neither necessary nor sufficient for either the recruitment of SKAP55 to the plasma membrane or the entry into and stabilization of SLP-76 microclusters by SKAP55.
Figure 4.
The SKAP55 PH domain and linker tyrosines are dispensable for recruitment into and stabilization of SLP-76 microclusters, and for T cell adhesion. (A) Domain structures of the ΔPH, 3YF, R131M, and PH-alone SKAP55 chimeras. (B) J14.SY cells were transiently transfected with the indicated SKAP55.mRFP1 chimeras; expression levels were determined by Western blotting (n = 3). (C) Cells from B were stimulated, imaged, and presented as in Fig. 2 B. See Table 1 for microcluster properties and experiment numbers. (D) Composite microcluster traces for the conditions examined in C; numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (E) Fraction of cells scored as displaying unstable contacts (n = 4). (F) JSKAP.SY cells transiently transfected with the indicated SKAP55.mRFP1 chimeras were stimulated, imaged, and presented as in Fig. 2 B (n = 3). (G) JSKAP.SY cells transfected with Akt.PH.mCFP and SKAP55.mRFP1 chimeras were imaged as in Fig. 2 B. Still images acquired at and 4 µm above the coverslip are shown (n = 3). (H) Fractional retention of unstimulated or TCR-stimulated cells on fibronectin-coated coverslips was calculated by dividing the post-wash signal by the prewash signal and normalizing to the corresponding PMA control (n = 3). (I) Fractional retention of T cells on uncoated or anti-CD3 coated coverslips was calculated by dividing the post-wash signal by the prewash signal (n = 3). Bars: (C and F, MOT images; and G) 10 µm; (C and F, kymographs) 5 µm × 60 s. Error bars indicate mean ± SEM. From parental J14.SY cells (with or without mRFP1): *, P < 0.05. From JSKAP.SY (with or without mRFP1): #, P < 0.05.
SKAP55 mediates the inside-out pathways that enable integrin-dependent adhesion after TCR ligation (Wang et al., 2003; Kliche et al., 2006). However the role of the SKAP55 PH domain in the transmission of these signals remains controversial (Kliche et al., 2006; Burbach et al., 2011). In the current study, the WT, ΔPH, 3YF, and R131M SKAP55 chimeras reconstituted TCR-induced adhesion to fibronectin (Fig. 4 H), confirming that neither the PH domain nor the linker tyrosines of SKAP55 are required for the activation of the predominant fibronectin-binding integrins expressed on Jurkat T cells (α4β1/VLA-4 and α5β1/VLA-5; Seminario et al., 1998). Because the loss of SKAP55 destabilized the contacts formed in response to solo TCR ligation (Fig. 2), we also examined whether adhesion via the TCR was impaired in the absence of SKAP55 (Nguyen et al., 2008; Chapman et al., 2012). In fact, the loss of SKAP55 reduced TCR-dependent adhesion to the basal levels observed in the absence of immobilized stimulatory antibodies. Reconstitution with matched levels of shRNA-resistant wild-type, ΔPH, 3YF and R131M chimeras significantly attenuated this defect (Figs. 4 I and S2 C). Thus, the cluster stabilizing and pro-adhesive functions of SKAP55 require the interaction with ADAP and an unidentified structural element N-terminal to the PH domain.
SKAP55 homologue (SKAP-Hom) is functionally redundant with SKAP55 downstream of the TCR
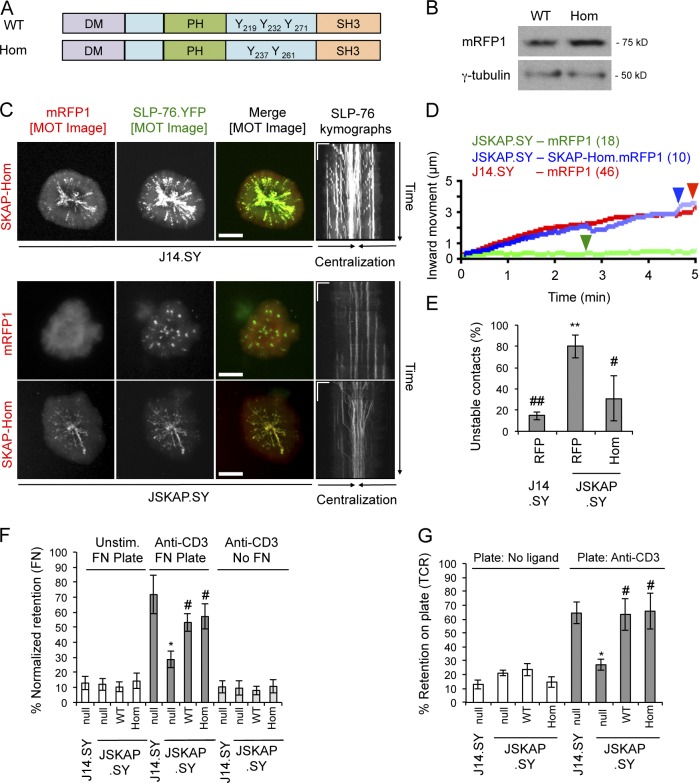
SKAP55 and SKAP-Hom possess similar domain structures (Fig. 5 A). Although SKAP-Hom is not expressed in Jurkat T cells, it has been observed in primary murine T cells (Marie-Cardine et al., 1998b; Jo et al., 2005). On this basis it has been suggested that SKAP-Hom cannot compensate for the loss of SKAP55 (Jo et al., 2005; Heng et al., 2008). However, when expressed in J14.SY cells, an mRFP1-tagged SKAP-Hom chimera entered and co-migrated with TCR-induced SLP-76 microclusters (Fig. 5, B and C). In addition, when expressed in JSKAP.SY cells, the SKAP-Hom chimera reconstituted normal microcluster dynamics and reduced the fraction of cells exhibiting unstable contacts (Fig. 5, D and E; Table 2; and Fig. S2 D). Furthermore, the SKAP-Hom chimera restored TCR-elicited adhesion to fibronectin and adhesion via the TCR itself. (Fig. 5, F and G; and Fig. S2 E). Whereas our studies used SKAP55 and SKAP-Hom chimeras expressed at similar levels, the study that argued for the nonequivalence of these proteins did not evaluate their relative abundances. Our data suggest that SKAP-Hom only fails to compensate for the loss of SKAP55 in primary T cells because it is not expressed at sufficient levels (Marie-Cardine et al., 1998b; Heng et al., 2008).
Figure 5.
SKAP-Hom heterodimerizes with and is functionally redundant with SKAP55. (A) Domain structures of the SKAP55 and SKAP-Hom chimeras. (B) Cells were lysed and Western blotted to determine relative expression of the SKAP55 (WT) and SKAP-Hom (Hom) chimeras (n = 3). (C) J14.SY and JSKAP.SY cells transiently expressing mRFP1 or SKAP-Hom.mRFP1 were stimulated, imaged, and presented as in Fig. 2 B. See Tables 1 and 2 for microcluster properties and experiment numbers. Bars: (MOT and conventional images) 10 µm; (kymographs) 5 µm × 60 s. (D) Composite microcluster traces for the conditions examined in C; numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (E) Fraction of cells scored as displaying unstable contacts (n = 3). (F and G) Fractional retention on fibronectin-coated or anti-CD3-coated coverslips, as described in Fig. 4 (n = 3). Error bars indicate mean ± SEM. From parental J14.SY cells (with or without mRFP1): *, P < 0.05; **, P < 0.01. From JSKAP.SY (with or without mRFP1): #, P < 0.05; ##, P < 0.01.
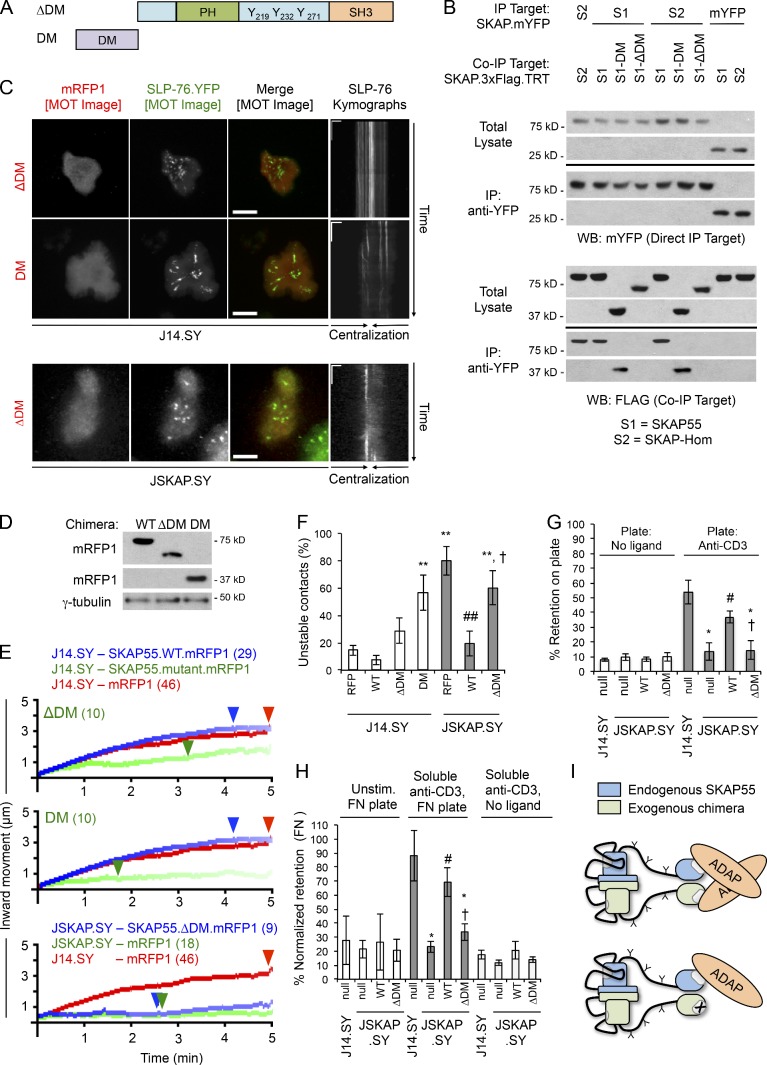
The SKAP55 DM is required for recruitment to and stabilization of SLP-76 microclusters, and for adhesion to both TCR and integrin ligands
Because SKAP-Hom can substitute for SKAP55, the N-terminal DM observed in both proteins could contribute to microcluster stabilization and T cell adhesion. Secondary structure prediction and molecular modeling indicated that the DM motif of SKAP55 forms a homodimer, as in SKAP-Hom (Fig. S3, A–D; Swanson et al., 2008; Zhang, 2008; Roy et al., 2010). To test this, we created deletion mutants lacking the entire DM (ΔDM) or retaining only the DM motif (Fig. 6 A). Co-immunoprecipitations confirmed that the N-terminal DM motif of SKAP55 is both necessary and sufficient for dimer formation (Fig. 6 B). Finally, both SKAP55 and the isolated SKAP55 DM motif captured SKAP-Hom, which is consistent with the conservation of key hydrophobic residues within the dimerization interface (Fig. S3, A and E). Therefore, SKAP55 and SKAP-Hom are likely to form functional heterodimers if coexpressed in vivo.
Figure 6.
The SKAP55 DM is required for microcluster entry, microcluster stabilization, and the pro-adhesive functions of SKAP55. (A) Domain structures of the ΔDM and DM alone SKAP55 chimeras. (B) Differentially tagged SKAP55 (S1) and SKAP-Hom (S2) chimeras were coexpressed in E6.1 Jurkat cells. Immunoprecipitations were performed with anti-YFP antibody. Total lysates and immunoprecipitates were blotted with anti-YFP or anti-Flag (n = 4). (C) J14.SY and JSKAP.SY cells transiently expressing the ΔDM and DM alone chimeras were stimulated, imaged, and presented as in Fig. 2 B. See Tables 1 and 2 for microcluster properties and experiment numbers. Bars: (MOT and conventional images) 10 µm; (kymographs) 5 µm × 60 s. (D) Cells were lysed and Western blotted to determine relative expression of the WT, ΔDM, and DM-only chimeras (n = 3). (E) Composite microcluster traces for the conditions in C; numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (F) Fraction of cells scored as displaying unstable contacts (n = 5 or more). (G and H) Fractional retention on anti-CD3–coated or fibronectin-coated coverslips, as in Fig. 4 (n = 3). (I) Proposed mechanism by which the SKAP55 dimer stabilizes SLP-76 microclusters downstream of TCR ligation. Error bars indicate mean ± SEM. From parental J14.SY cells (with or without mRFP1): *, P < 0.05; **, P < 0.01. From JSKAP.SY (with or without mRFP1): #, P < 0.05; ##, P < 0.01. From JSKAP.SY with SKAP55.WT.mRFP add-back: †, P < 0.05.
Upon overexpression, the SKAP55 ΔDM mutant remained cytoplasmic and weakly destabilized TCR-induced SLP-76 microclusters (Fig. 6, C–E; and Table 1). In contrast, the isolated SKAP55 DM motif potently antagonized microcluster dynamics (Fig. 6, C–E; Table 1; and Video 8). Furthermore, the isolated DM motif destabilized TCR-dependent contacts to a greater extent than the ΔDM chimera (Fig. 6 F). However, in the absence of endogenous SKAP55, an shRNA-resistant ΔDM chimera failed to restore microcluster dynamics (Fig. 6, C and E; Video 9; Table 2; and Fig. S2 F). Similarly, reconstitution with the ΔDM chimera failed to restore either TCR-induced adhesion to fibronectin or adhesion via the TCR (Fig. 6, G and H; and Fig. S2 G). Thus, the SKAP55 DM motif is required for the adhesive function of the TCR and for TCR-mediated increases in integrin adhesion. Thus, we hypothesized that the most potent dominant-negative SKAP55 chimeras, including the SH3 domain mutant (W333R) and the isolated DM motif destabilize SLP-76 microclusters by heterodimerizing with endogenous SKAP55 and preventing the multimerization of ADAP (Fig. 6 I).
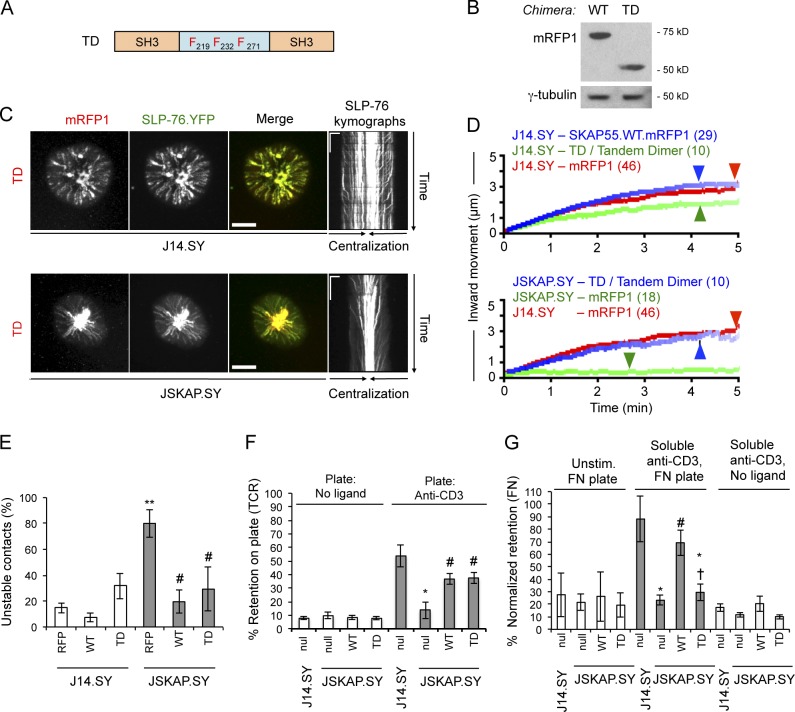
A TD composed of linked SKAP55 SH3 domains stabilizes SLP-76 microclusters and promotes adhesion to TCR ligands, but not to integrin ligands
Because the SKAP55 DM motif could stabilize SLP-76 microclusters by dimerizing ADAP and increasing overall microcluster avidity or by recruiting additional effector proteins, we generated an artificial TD consisting of two SKAP55 SH3 domains linked by a 3YF linker region (Fig. 7 A). Unlike the isolated SH3 domain (Fig. 3), this chimera entered SLP-76 microclusters and had no significant impact on SLP-76 microcluster dynamics upon overexpression in J14.SY cells (Fig. 7, C and D; and Table 1). Although the TD weakly destabilized TCR-initiated contacts upon overexpression in J14.SY cells, this effect was not significant (Fig. 7 E). In reconstitution studies, the hairpin-resistant TD restored optimal SLP-76 microcluster dynamics (Fig. 7, C and D; Video 10; Table 2; and Fig. S2 H). Remarkably, reconstitution with the TD increased the stability of TCR-induced contacts and promoted adhesion via the TCR nearly as well as wild-type SKAP55, but could not support TCR-mediated increases in adhesion to the α4β1 and α5β1 integrin ligand fibronectin (Fig. 7, E–G; and Fig. S2 G; Shimizu et al., 1990; Arroyo et al., 1992; Springer, 1994; Seminario et al., 1998). Thus, the dimerization of the SKAP55 SH3 domain is sufficient to support microcluster stability and adhesion via the TCR, whereas the DM motif plays a distinct and critical role in the regulation of adhesion via β1 integrins (Fig. 7 G).
Figure 7.
An artificial SKAP55 TD is sufficient for recruitment to and stabilization of SLP-76 microclusters, and for adhesion to TCR but not integrin ligands. (A) Domain structure of the SKAP55 TD chimera. (B) J14.SY cells were transiently transfected with WT and TD chimeras and blotted to confirm comparable expression levels (n = 3). (C) J14.SY and JSKAP.SY cells expressing the TD chimera were stimulated, imaged, and presented as in Fig. 2 B. See Tables 1 and 2 for microcluster properties and experiment numbers. Bars: (conventional images) 10 µm; (kymographs) 5 µm × 60 s. (D) Composite microcluster traces for the conditions examined in C; numbers in parentheses indicate the total number of cells examined. Line intensity corresponds to the fraction of microclusters surviving; arrowheads identify points of half-maximal microcluster dissociation. (E) Fraction of cells scored as displaying unstable contacts (n = 6). (F and G) Fractional retention on anti-CD3–coated or fibronectin-coated coverslips, as described in Fig. 4 (n = 3). Error bars indicate mean ± SEM. From parental J14.SY cells (with or without mRFP1): *, P < 0.05; **, P < 0.01. From JSKAP.SY (with or without mRFP1): #, P < 0.05. From JSKAP.SY with SKAP55.WT.mRFP1 add-back: †, P < 0.05.
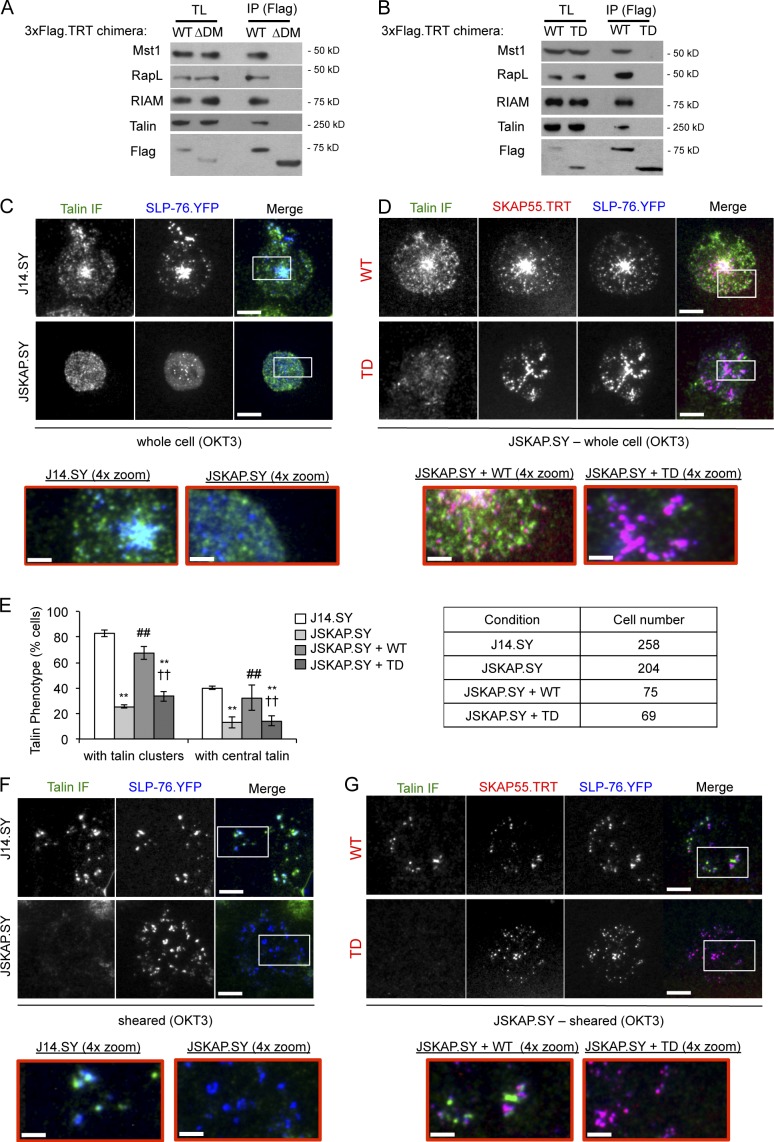
The DM motif is required for the interaction of SKAP55 with the integrin-activating molecule RIAM and for the recruitment of talin into TCR-induced adhesive junctions
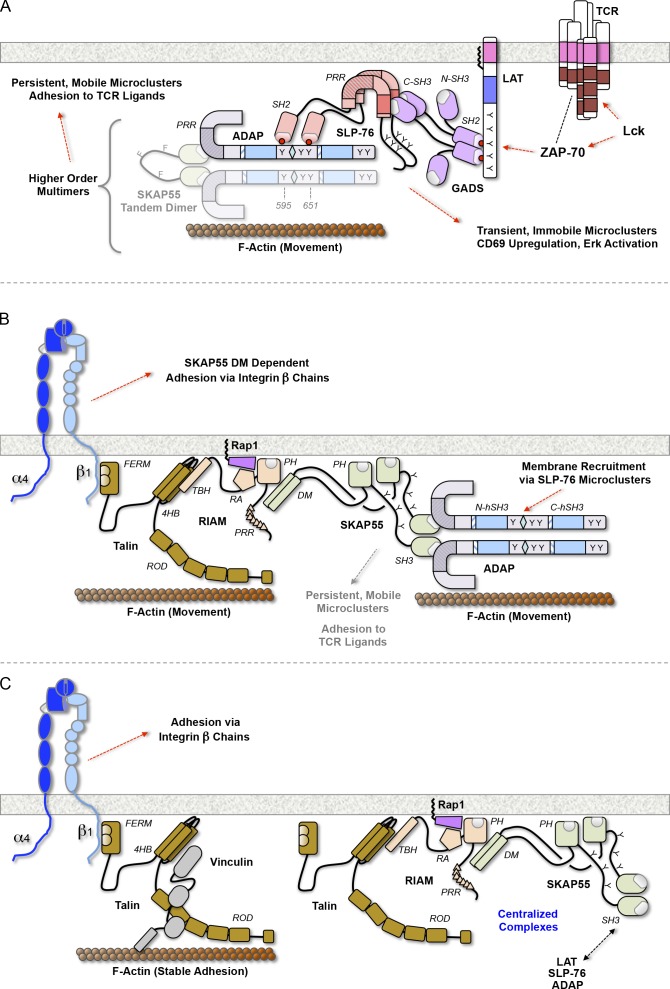
The SKAP55 DM motif regulates LFA-1 affinity via a complex incorporating RapL, macrophage stimulating 1 (Mst1), and Rap1 (Raab et al., 2010). As expected, RapL coimmunoprecipitated with full-length SKAP55 and was not captured with the ΔDM chimera (Fig. 8 A). However, this complex acts via the αL subunit, and cannot explain the ability of SKAP55 to facilitate adhesion to fibronectin. We found that SKAP55 also requires its DM motif to capture RIAM, a Rap1-dependent adaptor molecule that interacts with talin to facilitate integrin activation via a motif present in all integrin β chains (Fig. 8 A; Han et al., 2006; Ménasché et al., 2007; Watanabe et al., 2008; Lee et al., 2009). None of these integrin-activating proteins coimmunoprecipitated with the SKAP55 TD (Fig. 8 B).
Figure 8.
The SKAP55 DM is required for binding to RapL and RIAM, and governs the recruitment of talin into TCR-induced contacts. (A and B) 3×Flag.TRT-tagged SKAP55 wild-type (WT), DM deleted (ΔDM), and TD chimeras were expressed in J14.SY cells and immunoprecipitated via the Flag epitope. Blots of total lysates and immunoprecipitates are shown (n = 3). (C and D) Images of SLP-76.YFP (blue) and endogenous talin immunofluorescence (green) in J14.SY or JSKAP.SY cells stimulated on anti-CD3–coated plates and fixed after 10 min. Boxed regions were magnified 4× (shown below) to show microclusters at higher resolution (n = 3 or more). (D) JSKAP.SY cells were transfected with wild-type (WT) or TD SKAP55.3×Flag.TRT chimeras (red) before stimulation. (E) The frequency of cells exhibiting talin clustering and the central accumulation of talin were scored manually, using the images acquired above. (F and G) Cells were prepared as in C and D, but after fixation and before immunofluorescent staining cell bodies were sheared away, leaving cellular material only within the areas of tight contact between the cell and stimulatory substrate. Bars: (main panels) 10 µm; (enlarged panels) 2.5 µm. Error bars indicate mean ± SEM. From parental J14.SY cells: **, P < 0.01. From JSKAP.SY cells: ##, P < 0.01. From JSKAP.SY reconstituted with SKAP55.WT.3×Flag.TRT: ††, P < 0.01.
In J14.SY cells stimulated through the TCR, endogenous talin entered distinct structures found alongside peripheral SLP-76 microclusters, but also accumulated in the center of the contact, where it overlapped with a centralized pool of SLP-76 microclusters (Fig. 8 C and Fig. S4 A). In JSKAP.SY cells, talin clusters were more uniformly distributed, and the accumulation of talin in the center of the contact was reduced significantly (Fig. 8, C and E; and Fig. S4 B). These defects in talin localization were eliminated upon reconstitution with wild-type SKAP55, but not with the TD (Fig. 8, D and E; and Fig. S4, C and D). Because SKAP55 regulates T cell adhesion via the TCR (Figs. 4–7), we postulated that SKAP55 regulates the integrity of the adhesive junctions formed by the TCR itself. These junctions are subdomains within the plasma membrane that form intimate contacts with the underlying substrate and are substantially resistant to extraction via mechanical disruption (Harder and Kuhn, 2000; Bunnell et al., 2001, 2002). To examine whether SLP-76 and talin are retained within these junctions, we sheared away the bodies of cells fixed after stimulation on immobilized TCR ligands. The central accumulations of SLP-76 and talin observed in J14.SY cells were eliminated by this procedure, which indicates that this pool is not in intimate contact with the substrate. Nevertheless, SLP-76 and talin were retained in adherent complexes in the contact periphery and partially overlapped with one another. However, SLP-76, but not talin, was retained within the adhesive junctions formed by JSKAP.SY cells. Upon reconstitution with wild-type SKAP55, but not with the TD, the retention of talin at sites adjacent to SLP-76 microclusters was restored (Fig. 8 G). These data suggest that the SKAP55 DM motif governs the recruitment of talin into TCR-dependent adhesive junctions and the recruitment of talin into a central pool that is not tethered to the stimulatory substrate.
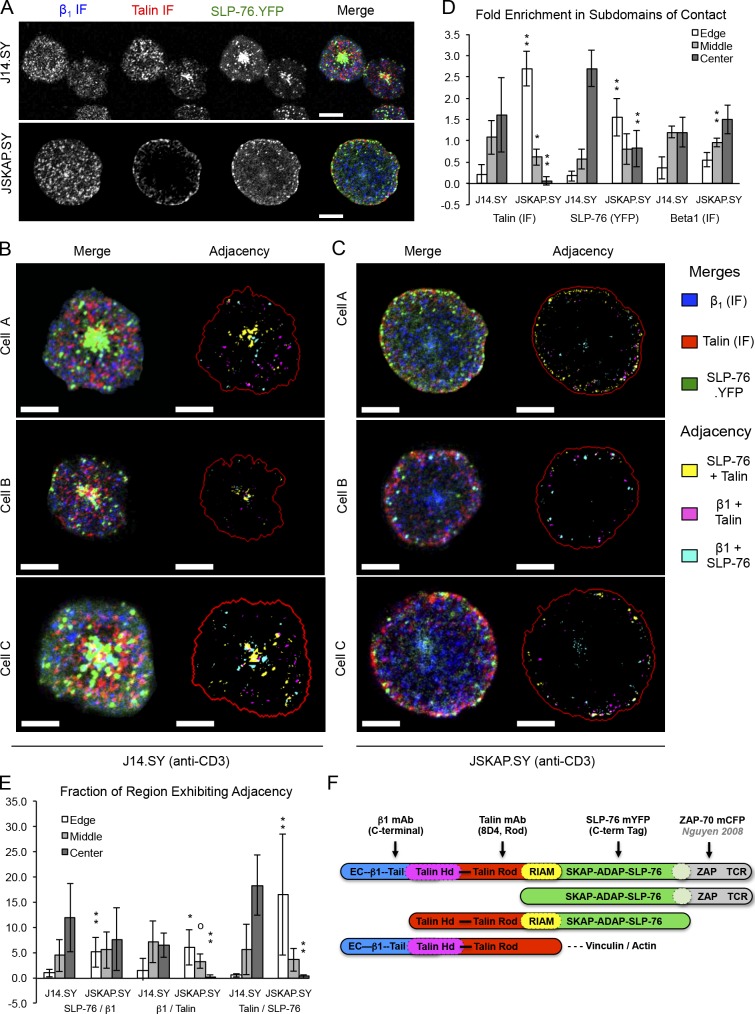
SKAP55 governs the distribution of SLP-76 and talin microclusters within TCR-induced contacts, but does not alter the distribution of β1 integrins
Talin interacts with the cytoplasmic tails of integrin β-chains and promotes the conversion of integrins into high-affinity states; therefore, SKAP55 and talin might be expected to control the localization of β1 integrins. Nevertheless, immunofluorescent stains revealed that β1 integrin clusters were distributed throughout the contact, whether SKAP55 was present or not (Fig. 9, A–C). In contrast, the loss of SKAP55 reduced the central clustering of talin and SLP-76, and increased the appearance of these proteins at the edge of the contact (Fig. 9, A, C, and D). Remarkably, the distributions of β1 integrin, talin, and SLP-76 were substantially exclusive of one another, with overlap primarily occurring at the margins of adjacent structures (Fig. 9, B–D). Because similar mutually exclusive arrangements have been observed for microclusters whose components clearly interact with one another (e.g. ZAP-70 and SLP-76 microclusters), these structures may represent distinct functional modules that interact with one another at their boundaries (Nguyen et al., 2008; Sherman et al., 2011).
Figure 9.
SKAP55 is dispensable for β1 integrin clustering downstream of TCR ligation. (A) J14.SY and JSKAP.SY cells were stimulated on anti-CD3–coated plates and fixed after 10 min. Immunofluorescent images of endogenous β1 integrin (blue) and endogenous talin (red), and direct images of SLP-76.YFP (green) are shown in greyscale and as a pseudocolored merged image (n = 3). (B and C) Higher-resolution merged images of representative J14.SY (B, left) or JSKAP.SY (C, left) cells prepared as in A. SLP-76, talin, and β1 integrin microclusters were identified algorithmically and used to derive regions of adjacency (right panels). The pseudocoloring scheme is indicated on the right. (n = 3). Bars, 10 µm. (D) Enrichment of talin, SLP-76, and β1 clustered areas in distinct domains of the contact (edge, middle, and center), relative to the entire contact. (E) Fraction of each domain of the contact occupied by the indicated regions of adjacency. For D and E, data are shown ±SD (error bars) for six cells acquired in three independent experiments. From parental J14.SY cells (with or without mRFP1): °, P < 0.10; *, P < 0.05; **, P < 0.01. (F) Diagram showing the binding sites of the fluorescent probes used in A–E.
To identify regions of potential interaction among adjacent structures, we used an automated thresholding algorithm to identify SLP-76, talin, and β1 microclusters. Regions of pairwise “adjacency” (see Materials and methods) were pseudocolored yellow (SLP-76 near talin), magenta (talin near β1), or cyan (β1 near SLP-76), merged, and labeled to indicate the boundary of the contact (Fig. 9, B and C). Microcluster enrichments in the edge of the contact, the center of the contact, and the intervening “middle” region were calculated by normalizing cluster density in the region of interest by the corresponding cluster density in the entire contact (Fig. 9 D). Similarly, we calculated the fraction of each domain engaged in adjacency (Fig. 9 E). In J14.SY cells, regions of adjacency between SLP-76 and talin (yellow) were predominantly found in the center of the contact, with the remainder found in the middle region, rather than the edge of the contact. In contrast, adjacency between β1 integrin and talin (magenta) was more evenly distributed between the middle region and the center of the contact. In the absence of SKAP55, these distributions were significantly altered, such that the regions of adjacency between SLP-76 and talin and between β1 integrin and talin shifted toward the outer margin of the contact.
Discussion
The mechanisms by which SKAP-family adaptors are integrated into receptor proximal signaling networks are not yet well understood. Here, we show that SKAP55 is recruited into SLP-76 microclusters via ADAP, and is retained within the TCR-induced junctions that anchor T cells to stimulatory substrates. In addition, SKAP55 regulates the persistence and movement of SLP-76 microclusters, stabilizes the boundaries of TCR-induced contacts, and facilitates T cell adhesion via the TCR and via β1 integrins. Although microcluster persistence is commonly associated with efficient T cell activation, SKAP55 was not required for TCR-dependent increases in CD69 expression or Erk activation, establishing that these outputs do not require the formation of persistent microclusters. These data require a reevaluation of prior observations that linked microcluster persistence to the efficacy of TCR-induced Erk activation and CD69 up-regulation, and suggest that these defects were caused by the impaired recruitment or function of key effector molecules, rather than a nonspecific reduction in the cohesion of SLP-76 microclusters (Bunnell et al., 2006; Sylvain et al., 2011).
We also show that the SKAP55 PH domain and the predominant tyrosine phosphorylation sites in SKAP55 are not required for normal SLP-76 microcluster dynamics, for adhesion via the TCR, or for adhesion via β1 integrins. Instead, these functions all require an N-terminal dimerization (DM) motif similar to that observed in SKAP-Hom (Swanson et al., 2008). The DM motifs of SKAP55 and SKAP-Hom form both homotypic and heterotypic dimers; further, contrary to prior studies, our data indicate that SKAP-Hom is functionally redundant with SKAP55 in T cells (Jo et al., 2005). Finally, we show that the DM motif influences T cell adhesion via two distinct mechanisms, involving either the oligomerization of microcluster components and the generation of adhesive junctions by the TCR, or the coupling of the DM motif to effector proteins responsible for the activation of β1 integrins.
Based on the data presented in Figs. 3–7, we conclude that overexpressed SKAP55 mutants reduce microcluster persistence and movement to the extent that they prevent the dimerization of endogenous SKAP55 SH3 domain ligands. Consistent with this hypothesis, SKAP55 mutants do not restore normal microcluster dynamics unless capable of dimerizing endogenous ADAP (Fig. S5 A). According to this model, the dominant-negative impacts of the monovalent SKAP55 SH3 domain and the SKAP55 ΔDM mutant result from the sequestration of SH3 domain ligands from endogenous SKAP55, whereas the potent dominant-negative impacts of the full-length SKAP55 SH3 mutant and the isolated DM motif reflect their ability to perturb endogenous SKAP55 dimers (Figs. 6 I and S5 B).
The SKAP55 TD supports microcluster persistence, microcluster movement, the stabilization of TCR-induced contacts, and adhesion via the TCR. However, it does not support integrin activation downstream of the TCR, and does not interact with regulators of integrin activation, such as RIAM, talin, RapL, and Mst1. Thus, the integrin-activating ability of SKAP55 represents a distinct functional module that can be ablated without impacting the dynamics of SLP-76 microclusters. In contrast, we were unable to identify mutations that decouple the persistence and movement of SLP-76 microclusters from contact stabilization and adhesion via the TCR.
SKAP55 participates in the regulation of integrin affinity through distinct Rap1-containing complexes: one incorporates RapL and acts on αL integrins, the other incorporates RIAM and talin and acts though integrin β chains (Ménasché et al., 2007; Raab et al., 2010; Kliche et al., 2012). Our data demonstrate that the DM motif of SKAP55 is required for the association of RIAM and talin with SKAP55, for TCR-induced adhesion via β1 integrins, for the retention of talin in adhesive junctions adjacent to SLP-76 microclusters, and for the colocalization of talin and SLP-76 in nonadherent centralized microclusters (Fig. 10 B). In the absence of SKAP55, integrin-mediated adhesion is impaired, talin is eliminated from TCR-induced junctions, and the central clusters of talin and SLP-76 are lost. Finally, talin and SLP-76 redistribute to the outermost edge of the contact in the absence of SKAP55.
Figure 10.
Distinct roles of SKAP55 domains in microcluster stabilization, adhesion via the TCR, and integrin regulation. (A) SKAP55-dependent multimers enable microcluster stabilization and movement, and are required for adhesion via the TCR and via integrins. Microcluster persistence requires interactions capable of joining LAT, Gads, SLP-76, and ADAP into a minimal signaling complex. Constituents of stable microclusters (e.g., ADAP), may interact directly or indirectly with the actin cytoskeleton to promote T cell adhesion and/or microcluster movement. (B) The SKAP55 DM motif couples SLP-76 microclusters to RIAM and talin, enabling the conformational regulation of integrins via their β chains. (C) Models depicting the maturation or termination of the integrin-activating complex in B. Domains are abbreviated as follows: 4HB, four helix bundle; CC, coiled-coil region; FERM, FAK, ezrin, radixin, and moesin domain; PRR, proline-rich region; RA, Ras-association domain; hSH3, helically extended Src-homology 3 domain; TBH, talin-binding helix.
SLP-76, talin, and β1 integrins participate in interconnected complexes. Nevertheless, these structures were distinguishable and adjacent from one another. This pattern is consistent with the sizes of the complexes involved, assuming their constituents adopt extended conformations, as diagrammed in Fig. 9 F (Shaw, 2006; Kanchanawong et al., 2010; Margadant et al., 2011). However, triple adjacency was rarely observed, which suggests that complexes bridging SLP-76 to β1 integrins are not maintained for long periods of time. Instead, we suggest that SLP-76 and talin are initially recruited to the outermost edges of TCR-induced contacts through SKAP55-independent interactions with Abi/WAVE complexes (Zipfel et al., 2006; Nolz et al., 2007). Subsequently, we expect talin to interact with SLP-76 in a SKAP55- and Rap1-dependent manner, giving rise to activated clusters of β1 integrins in the regions lagging the edge of the contact (Fig. 10 B). Finally, we expect vinculin to displace SKAP55 and SLP-76 from fully activated integrins, whereas nonengaged SLP-76 and talin complexes are extracted from the cell surface and transported to the center of the contact (Fig. 10 C; Lee et al., 2013).
In contrast to the DM motif, the PH domain of SKAP55 proved dispensable for the persistence and movement of SLP-76 microclusters, for the stabilization of TCR-induced contacts, and for adhesion via both the TCR and β1 integrins (Fig. S5 C). In addition, SKAP55 was not recruited to the plasma membranes of TCR-stimulated Jurkat T cells, despite constitutively high levels of phosphatidylinositol-3,4,5-trisphosphate in these cells, as confirmed through the use of the PH domain of Akt as a positive control (Seminario et al., 2004). Our data are consistent with prior studies in Jurkat T cells, which also failed to identify a role for the PH domain of SKAP55 in TCR-induced adhesion to integrin ligands (Kliche et al., 2006). Although several studies have reported the recruitment of SKAP55 into an insoluble membrane fraction in Jurkat cells, SKAP55 may, in fact, be rendered insoluble through interactions with large signaling complexes, or through interactions with insoluble actin polymers (Ménasché et al., 2007; Burbach et al., 2008, 2011; Raab et al., 2011). Nevertheless, our observations appear to be at odds with the roles of the SKAP55 PH domain in synaptic localization and integrin activation in primary murine T cells (Burbach et al., 2011). Because the isolated SKAP55 PH domain appears to be incapable of binding the plasma membrane, we propose that this domain normally engages lipids only once SKAP55 has been recruited into microclusters via ADAP, whereupon lipid recognition by the PH domain promotes the conversion of SKAP55 into a “open” conformation that liberates the DM motif to engage integrin-activating ligands (Fig. S5 D). Whatever the source of these differences, our data indicate that the PH domain is not an integral component of the systems that activate integrins in response to TCR ligation, but may instead regulate these functions in primary T cells.
Our data suggest SKAP55 regulates microcluster dynamics, contact stability, and TCR adhesion by multimerizing ADAP, SLP-76, Gads, and LAT into higher-order complexes that couple TCR-induced microclusters to the pro-adhesive and force-generating systems of the actin cytoskeleton (Fig. 10, A and B). In their incorporation of tyrosine kinase-dependent scaffolds that impact the organization of the cytoskeleton, SLP-76 microclusters resemble focal adhesions and podosomes (Burgstaller and Gimona, 2004; Johansson et al., 2004; Evans and Matsudaira, 2006; Gomez et al., 2006; Block et al., 2008; Gimona et al., 2008; Ha et al., 2008; Machesky et al., 2008; Oikawa et al., 2008; Ory et al., 2008; Albiges-Rizo et al., 2009; Carrizosa et al., 2009; Oikawa and Takenawa, 2009; Sage et al., 2012). Based on these similarities and on the coupling of SLP-76 microclusters to the actin cytoskeleton, the orientation of actin filaments above SLP-76 microclusters, the association of SLP-76 microclusters with adhesive structures, and the adjacency of SLP-76 microclusters to integrins, we propose that SLP-76 microclusters function as lymphocyte-specific podosomes (Bunnell et al., 2001, 2002; Nguyen et al., 2008; Bunnell, 2010; van den Dries et al., 2013). In this manner, SLP-76 microclusters may couple integrin-mediated adhesions to local actin polymerization, generating both the “downwardly directed” forces required to overcome glycocalyx repulsion and the lateral shear forces required for efficient TCR mechanotransduction (Seminario and Bunnell, 2008; Martínez-Martín et al., 2011; Wang and Reinherz, 2012).
Materials and methods
Cell lines and transfections
Jurkat T cells were maintained in RPMI-1640 (BioWhittaker) supplemented with 10% FBS, 20 mM glutamine, and 10 µg/ml ciprofloxacin. SLP-76–deficient Jurkat cells (J14 cells) were a gift of A. Weiss (University of California, San Francisco, San Francisco, CA). ADAP-deficient Jurkat cells (JDAP cells) were a gift of R. Wange (National Institutes of Health, Bethesda, MD). J14 cells stably reconstituted with SLP-76.YFP (J14.SY cells) have been described previously (Bunnell et al., 2006). J14.SY cells lacking SKAP55 (JSKAP.SY cells) were generated by lentiviral transduction and selection in puromycin. J14.SY and JSKAP.SY lines stably expressing SKAP55 chimeras tagged with a red fluorescent protein were created by lentiviral transduction and sorting for double-positive cells with matched brightness via flow cytometry. Transient transfections were performed as described previously (Bunnell et al., 2006). Human 293T renal epithelial cells were cultured in DMEM (Cellgro) supplemented with 10% FBS, 20 mM l-glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin.
Lentivirus production and lentiviral transduction
24 h before transfection, 293T cells were replated so that on the day of transfection they were ∼65–75% confluent. For each 10-cm plate, 18 µl of FuGENE 6 transfection reagent (Roche) was incubated with 142 µl of serum-free DMEM for 5 min. Concurrently, 6 µg of the lentiviral expression and/or knockdown vectors was combined with 1.5 µg of the packaging plasmid psPAX2 and 0.5 µg of the pseudotyping plasmid pMD2.G in 40 µl of serum-free DMEM. These mixes were combined and incubated for 30 min at room temperature, and were added to 293T cells in a dropwise fashion. After 12–15 h, the transfection cocktail was replaced with fresh media, and the lentiviral supernatants were collected 66–72 h after transfection. Jurkat cells were transduced by combining equal parts rapidly dividing Jurkat cells, fresh lentiviral supernatants, and complete RPMI. For fluorescent chimeras, the efficiency of transduction was assessed in fixed cells after 48 h, and was typically 50–90%.
SKAP55 cloning and mutagenesis
Vectors encoding mRFP1 and TRT were obtained from R. Tsien (University of California, Davis, La Jolla, CA) and subcloned into “n1-type” vectors (Takara Bio Inc.) by PCR. Where specified, sequences encoding triple FLAG epitope tags (3×Flag) were subcloned 5′ to the encoded fluorescent protein. The mYFP-n1 vector containing the monomerizing A206K mutation was modified by the addition of a triple Myc epitope tag (3×Myc) 5′ to the fluorescent protein via overlap extension PCR. Unless otherwise noted, SKAP55 and associated mutants were created by overlap extension PCR and subcloned into the mRFP1-n1 or 3×Flag.TRT-n1 vectors at the XhoI and AgeI sites. Homo sapiens SKAP55 was isolated from E6.1 Jurkat T cell cDNA by PCR with primers SCB267 and SCB269. SKAP55 deletion and truncation mutants were generated as follows: ΔDM, by removing residues Q2-G60 with primers SCB405 and SCB269; and ΔPH, by removing residues V108-L212 with primers NS729, NS730, SCB267, and SCB269. The monovalent SH3 domain was amplified with primers MO805 and SCB269. The DM motif was amplified with SCB267 and MO802. Homo sapiens SKAP-Hom was amplified from a commercial cDNA (Thermo Fisher Scientific) using MO808 and MO811. Point mutations were generated by QuikChange PCR mutagenesis using the indicated primers: W333R with NS727/NS728; 3YF in a stepwise fashion using NS732/NS733 (Y219F), NS734/NS735 (Y232F), and NS736/NS737 (Y271F). The SKAP55 TD was created in two steps: first, a monovalent SKAP55 SH3 domain was cloned into mRFP1-n1 using MO805/SCB269; second, a fragment containing the linker-3YF-SH3 was obtained using MO812/SCB269 and a SKAP55.3YF template; the latter product was cut with BspEI and AgeI and subcloned into SKAP55.SH3.mRFP1 at an AgeI site between the SH3 domain and mRFP1. The top-performing hairpin, which targets the linker region between the PH and SH3 domains, was identified from a panel of pLKO.1-based shRNA expression vectors targeting SKAP55 (Thermo Fisher Scientific; target sequence, 5′-CCAGATGAAGAGCATGATCTA-3′). SKAP55 chimeras were rendered immune to the SKAP55 shRNA by PCR overlap extension mutagenesis with primers MO834, MO835, SCB267, and SCB269, which introduced silent point mutations in the SKAP55 linker region between the PH and SH3 domains. For transient knockdown experiments, the cassette driving the expression of this hairpin was PCR amplified using RB608/RB610, cut with SpeI and NheI, and subcloned into a SpeI site that had been introduced upstream the cytomegalovirus promoter of an mRFP1-n1 expression vector. For generation of stable cell lines, we first generated a pLEX-MCS based lentiviral expression vector encoding 3×Flag.TRT-n1, but lacking the IRES-puromycin resistance cassette. The indicated SKAP55 chimeras were cloned into this vector using NheI and AgeI. Please refer to Table S2 for sequences of primers used in PCR mutagenesis and cloning.
Antibodies and Western blotting
Cell lysates were prepared by lysing Jurkat cells in lysis buffer containing 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 10 mM NaF, 1% Triton X-100, 1 mM Na3VO4, and the Complete Protease Inhibitor Cocktail (Roche). Western blotting was conducted with antibodies directed against ADAP (BD), β1 integrin (EP1041Y; Abcam), CD69-PE/Cy7 (No. 310912; BioLegend), Phospho Erk1/2 (No. 4370; Cell Signaling Technology), total Erk 1/2 (No. 4695; Cell Signaling Technology), Flag (M2; Sigma-Aldrich), RFP (ACT-CM-MRRFP10; Allele), GFP (JL-8; Takara Bio Inc.), γ-tubulin (Sigma-Aldrich), Mst1 (Epitomics), RapL (Sigma-Aldrich), RIAM (Epitomics), SKAP55 (gift from B. Schraven [University of Magdeburg, Magdeburg, Germany] and S. Kliche [Otto von Guerike University, Magdeburg, Germany]; targets SKAP55 DM), SKAP55 (targets SKAP55 PH-SH3 linker; BD), and talin (8D4; Sigma-Aldrich). Anti-SKAP55 (BD) and anti-talin (Sigma-Aldrich) were used in immunofluorescence studies. Immunoprecipitations were performed by prebinding anti-YFP (ab290; Abcam) or anti-Flag (M2; Sigma-Aldrich) to beads coated with protein A or protein G (Thermo Fisher Scientific) for 1 h at room temperature. Antibody-bound beads were washed in lysis buffer and immunoprecipitations were performed for 14–18 h at 4°C.
Sample preparation for microscopy
Jurkat T cells were stimulated on glass-bottomed 96-well plates that were pretreated with 0.01% poly-l-lysine, coated with 10 µg/ml OKT3, and blocked with 1% BSA to limit the deposition of serum components during imaging. Each cell was imaged, beginning within 2 min of injecting the responding cells into the well, and only one field was imaged per well. Live cells were imaged at 37°C in RPMI 1640 (supplemented with 10% FBS and 1% l-glutamine) buffered with 25 mM Hepes, pH 7.4. Each cell used in analysis was imaged for no fewer than 5 min. For immunofluorescence assays, cells were stimulated in 37°C in RPMI 1640 (supplemented with 10% FBS and 1% l-glutamine) buffered with 25 mM Hepes, pH 7.4, and subsequently fixed and stained as described previously (Bunnell et al., 2003). In brief, cells were fixed in 1× DPBS (cellgro; Corning) with 1% PFA (Sigma-Aldrich) and washed three times in PFN buffer (1× DPBS with 8% calf serum, 0.1% saponin, and 0.02% sodium azide). Cells were then blocked in PFN/blocking buffer (PFN supplemented with 2% goat serum), washed, and stained with relevant primary antibodies for 1 h at room temperature in PFN/blocking buffer. After washing, cells were incubated with Alexa Fluor 568– or 647–conjugated secondary antibodies (Invitrogen), as noted in Table S1. Each wash step involved three exchanges of PFN buffer.
Image acquisition
Imaging runs were performed on a spinning-disc confocal microscope as described previously (Bunnell et al., 2006; Nguyen et al., 2008; Sylvain et al., 2011). In brief, images were acquired using a modified live-cell imaging system (UltraVIEW; PerkinElmer) consisting of a 5-line laser launch (442/488/514/568/647 nm; Prairie Technologies), a spinning-disc confocal head (CSU-10; Yokogawa Corporation of America), an Axiovert 200M stand (Carl Zeiss), a 40× Plan-Neofluar oil immersion objective lens (NA 1.3; Carl Zeiss), and either a charge-coupled device (CCD) camera (Orca-II-ER; Hamamatsu Photonics), or an intensified CCD (ICCD) camera (XR MEGA-10; Stanford Photonics). The latter camera was coupled via a 2.5× expanding lens to provide resolution comparable to the Orca-II-ER. Temperature was maintained at 37°C using an air blower (Nevtek) and a lens heater. Image acquisition was controlled using proprietary PerkinElmer software or the open source Micro-Manager software package (Edelstein et al., 2010). For Fig. 4 F and Fig. 9 (A–C), images were acquired using a confocal microscope (A1R; Nikon) with a 60× oil immersion objective lens (NA 1.4). Samples were maintained at 37°C using a stage heater. NIS-Elements AR software (Nikon) was used to capture images in real time. Please refer to microscopy spreadsheet (Table S1) for further details.
Post-acquisition image processing and presentation
Image analysis was performed using iVision (Biovision Technologies) or ImageJ (National Institutes of Health). In all cases, representative images are shown. Where merged images are shown, the pseudocoloring scheme is always indicated by the colors of the corresponding text labels. SLP-76 microcluster dynamics were quantitated by generating two kymographs per cell. The trajectories of SLP-76 microclusters arising within the first 2 min of the imaging run were manually traced to calculate microcluster displacement (inward movement), persistence, and maximum speed. These parameters were used to derive means for each cell, for each experiment, and for each condition. In addition, average traces were generated for each cell. These traces were compiled to yield composite kymographs displaying microcluster position and survival as a function of time. The microcluster half-life indicated on these composite kymographs is the point at which one half of the SLP-76 microcluster traces have dissipated. The incorporation of SLP-76 into microclusters was quantitated separately: the cell boundary was identified and the mean background outside the cell was subtracted, then the mean background inside the cell was determined, clusters were defined as structures with brightness 4× greater than the cellular background, and clustered SLP-76 was calculated as the total background-subtracted intensity within the clustered area divided by the total background-subtracted intensity within the cell. The means and statistical tests shown in all figures and tables (except for Fig. 9, D and E) were calculated by compiling the means of independent experiments, each of which incorporated multiple cells. The total number of cells examined across all experiments are shown for Tables 1 and 2, all composite kymographs, and for Fig. 8 E. All data are shown as means ± SEM unless otherwise indicated. For analyses of microcluster position within the contact and for microcluster adjacency analyses, microcluster masks were developed using a segmentation algorithm sensitive to local intensity maxima. Regions of adjacency were identified by dilating the SLP-76 and talin masks by ∼300 nm and defining the intersections of dilated and undilated masks as regions of adjacency. Cell boundary masks were developed using an algorithm that acts on the sum of all channels. Distinct subdomains of the contact were defined by iteratively eroding the cell boundary mask until fixed fractions of the contact area remain (“edge,” outermost 25% of the contact area; “center,” innermost 25%; “middle,” the intervening 50%). The fractions of each domain occupied by microclusters or regions of adjacency were determined, and relative enrichments were calculated by normalizing to the fraction of the entire contact occupied by the corresponding mask.
iVision scripts
To analyze microcluster persistence, movement, stoichiometry, and max speeds, kymographs in the x and y dimensions were generated and scaled using the “Make Kymo2” or “Make Kymo 3 −SPI+OPT” scripts, depending on the settings used during image acquisition (see Table S1). Clusters that formed within the first 2 min during image acquisition were manually traced and compiled using the “Manual Kymo tracer” script, and the stoichiometry of clustering was determined using the “Find clustered Brightness 2” script. Max speeds were calculated using the “Max Speeds” script. Individual traces from kymographs in the x and y directions were compiled to yield a composite trace for each cell using the “Average trace compiler” script. Composite traces for all cells per condition were also compiled using the “Average trace compiler” script. The resulting composite microcluster traces were assembled into figures using the “RGB line Output NS” script and then running the “Scale bars µm” script. Scale bars were added to all other images using the “Add scale bars” script. To assess the relative adjacencies of SLP-76, talin, and β1 integrin, the images were first analyzed using the “Three color autosegment” script followed by the “Autosegment subroutine” script. To assess boundary fluctuation over time, the “Boundary fluctuation v1 MJO” script was used. Numerical outputs were then copied into Excel (Microsoft).
Statistical analyses
All data are presented as means ± SEM unless otherwise noted. The number of experiments is indicated in all cases unless otherwise noted. Please note that for all quantified imaging experiments the numbers of experiments and cells are presented in Tables 1 and 2. Statistical significances were calculated in Excel using a two-tailed Student’s t test for unpaired samples with similar variances.
Plate bound adhesion assays
Jurkat cells were labeled with 4 µM 2′7′-bis(2-carboxyethyl), 5(-6) carboxyfluorescein (BCECF-AM; Molecular Probes) in 1× PBS (no calcium or magnesium) for 15 min at 37°C. Labeled cells were plated at a density of 2.5 × 105 cells per well in 96-well glass-bottom plates. After the cells settled for the indicated times, BCECF fluorescence was read on a SpectraMax plate reader (488 ex/530 em, 515 nm cutoff). Nonadherent cells were removed by gentle pipetting in prewarmed 1× HBSS, and BCECF retention was determined. For adhesion to fibronectin, plates were treated with 0.01% poly-l-lysine, left uncoated or coated with 10 µg/ml fibronectin (Gibco), and blocked with 1% BSA for 1 h at 37°C. Before plating, labeled cells in 1× HBSS were left unstimulated, stimulated for 30 min with 2 µg/ml OKT3 (anti-CD3ε, BioExpress), or stimulated for 30 min with 50 ng/ml PMA. After 30 min on plates, fractional retention was determined and normalized to the PMA control. For adhesion via the TCR, unstimulated cells were plated in wells treated with 0.01% poly-l-lysine, left uncoated or coated with 10 µg/ml OKT3, and blocked with 1% BSA. Fractional retention was determined after a 7-min incubation in complete media (Nguyen et al., 2008; Chapman et al., 2012). Standard deviations, standard errors, and statistical significances were calculated with Excel, using a two-tailed Student’s t test for unpaired samples with similar variances.
Bioinformatics and structural modeling
The secondary structures of the SKAP55 and SKAP-Hom DM motifs were predicted using the prediction server at www.predictprotein.org (Rost and Sander, 1993, 1994; Rost, 1996). The sequences of the DM motifs of SKAP55 and SKAP-Hom across vertebrate species were aligned using the constraint-based multiple protein alignment tool COBALT (Papadopoulos and Agarwala, 2007). The structure of the SKAP55 DM-PH module (spanning M1-S213) was predicted using the I-Tasser threading server, with the existing SKAP-Hom DM-PH structure as a constraint (2OTX, chain A; Swanson et al., 2008; Zhang, 2008; Roy et al., 2010, 2012). Of five models obtained, four preserved the overall topology of the SKAP-Hom DM-PH module, and possessed C-scores ranging from −1.10 to −2.36. The fifth model is unlikely to be valid, as it perturbed highly conserved elements within the PH domain; reflecting this, the C-score for this model was very low (−4.54). All molecular views were generated using Swiss-PdbViewer (http://spdbv.vital-it.ch/; Guex and Peitsch, 1997).
Online supplemental material
Video 1 depicts the recruitment of SKAP55.3×Flag.TRT (red) into SLP-76 microclusters upon TCR ligation. SLP-76 microcluster dynamics and boundary fluctuation in SKAP55 knockdown J14.SY (Fig. S1) in addition to methodology with regards to how unstable contacts were manually scored. Video 2 depicts the J14.SY cell expressing SKAP55.shRNA.mRFP1 depicted in Fig. S1. Videos 3 and 4 depict boundary fluctuation analysis of J14.SY cells expressing either the SKAP55 shRNA (Video 2) or control mRFP1 plasmid (Video 3). Areas of boundary fluctuation (growth and retraction) are shown in red and blue, respectively. Video 5 depicts a J14.SY stably deficient in endogenous SKAP55 (termed JSKAP.SY), expressing control mRFP1, while Video 6 depicts a JSKAP.SY cells transiently reconstituted with exogenous SKAP55.mRFP1. Furthermore, we show expression levels of all SKAP55 chimeras used in imaging and functional assays (Fig. S2) demonstrating comparable expression among all exogenous constructs. Video 7 depicts a J14.SY cell overexpressing the SKAP55.W333R.mRFP1 chimera. Our modeling data indicates that SKAP55, like SKAP-Hom, dimerizes via an N-terminal α-helical bundle (Fig. S3). Video 8 depicts a J14.SY cell overexpressing the SKAP55.DM.mRFP1 chimera while Video 9 shows a JSKAP.SY cell reconstituted with SKAP55.ΔDM.mRFP1. In comparison, Video 10 depicts a JSKAP.SY cell expressing the SKAP55.TD.mRFP1 chimera. Line scans depict endogenous talin and SLP-76.YFP position relative to one another in a diametrical slice of J14.SY or JSKAP.SY cells reconstituted with WT or TD chimeras (Fig. S4). Our model of how SKAP55 stabilizes SLP-76 microclusters upon TCR ligation is shown in Fig. S5. Table S1 indicates all conditions used in acquisition of images. Table S2 indicates the sequences of primers used in PCR mutagenesis and cloning. iVision scripts used in image analysis and quantitation, and instructions for their use are available for download as a ZIP file. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201305088/DC1.
Supplementary Material
Acknowledgments
The authors thank Dr. Burkhart Schraven and Dr. Stefanie Kliche for providing us with antibodies specific for the SKAP55 DM motif. In addition, we thank Ken Nguyen, Hector Flores, and Jared Hawkins for preparing SKAP55 mutants and for performing initial imaging studies involving SKAP55. We thank Michael Fray, Bridget Larkin, Joanne Russo, James Sampson, and Dr. Maria-Cristina Seminario for critical reading and helpful discussion of the manuscript.
All research was supported by a Brain and Immuno-Imaging Award from the Dana Foundation, a Scientist Development Grant from the American Heart Association (0635546T), and a grant from the NIAID (NIH R01 AI076575-01). In addition, we recognize the W.M. Keck Foundation and the Eshe Fund for their generous support of core facilities at Tufts. Finally, we thank Dr. Alenka Lovy for her assistance with confocal microscopy, and Dr. Robert Jackson for access to the Imaging Center affiliated with the Tufts Center for Neuroscience Research (P30 NS047243).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used in this paper:
- ADAP
- adhesion and degranulation-promoting adaptor protein
- CCD
- charge-coupled device
- DM
- dimerization motif
- Gads
- Grb2-related adaptor downstream of Shc
- ICCD
- intensified CCD
- LAT
- linker of activated T cells
- LFA-1
- leukocyte function–associated antigen 1
- MOT
- maximum-over-time
- mRFP1
- monomeric red fluorescent protein
- Mst1
- macrophage stimulating 1
- mYFP
- monomeric YFP
- PH
- Pleckstrin homology
- RapL
- regulator of adhesion and polarity in leukocytes
- RIAM
- Rap1-GTP–interacting adaptor molecule
- SH2
- Src homology 2
- SH3
- Src homology 3
- SKAP55
- Src kinase–associated phosphoprotein of 55 kD
- SKAP-Hom
- SKAP55 homologue
- SLP-76
- SH2 domain–containing leukocyte protein of 76 kD
- TCR
- T cell receptor
- TD
- tandem dimer
- TRT
- TagRFP-Turbo
- VLA-4
- very late antigen 4
- VLA-5
- very late antigen 5
References
- Acuto O., Di Bartolo V., Michel F. 2008. Tailoring T-cell receptor signals by proximal negative feedback mechanisms. Nat. Rev. Immunol. 8:699–712 10.1038/nri2397 [DOI] [PubMed] [Google Scholar]
- Albiges-Rizo C., Destaing O., Fourcade B., Planus E., Block M.R. 2009. Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122:3037–3049 10.1242/jcs.052704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo A.G., Sánchez-Mateos P., Campanero M.R., Martín-Padura I., Dejana E., Sánchez-Madrid F. 1992. Regulation of the VLA integrin-ligand interactions through the β1 subunit. J. Cell Biol. 117:659–670 10.1083/jcb.117.3.659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babich A., Li S., O’Connor R.S., Milone M.C., Freedman B.D., Burkhardt J.K. 2012. F-actin polymerization and retrograde flow drive sustained PLCγ1 signaling during T cell activation. J. Cell Biol. 197:775–787 10.1083/jcb.201201018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R.G., Hsu C.J., Lee D., Jordan M.S., Maltzman J.S., Hammer D.A., Baumgart T., Koretzky G.A. 2009. The adapter protein SLP-76 mediates “outside-in” integrin signaling and function in T cells. Mol. Cell. Biol. 29:5578–5589 10.1128/MCB.00283-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L., Barr V.A., Sommers C.L., Barda-Saad M., Goyal A., Isakowitz M.S., Samelson L.E. 2007. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Mol. Cell. Biol. 27:8622–8636 10.1128/MCB.00467-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopalan L., Coussens N.P., Sherman E., Samelson L.E., Sommers C.L. 2010. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb. Perspect. Biol. 2:a005512 10.1101/cshperspect.a005512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda-Saad M., Braiman A., Titerence R., Bunnell S.C., Barr V.A., Samelson L.E. 2005. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nat. Immunol. 6:80–89 10.1038/ni1143 [DOI] [PubMed] [Google Scholar]
- Barda-Saad M., Shirasu N., Pauker M.H., Hassan N., Perl O., Balbo A., Yamaguchi H., Houtman J.C., Appella E., Schuck P., Samelson L.E. 2010. Cooperative interactions at the SLP-76 complex are critical for actin polymerization. EMBO J. 29:2315–2328 10.1038/emboj.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block M.R., Badowski C., Millon-Fremillon A., Bouvard D., Bouin A.P., Faurobert E., Gerber-Scokaert D., Planus E., Albiges-Rizo C. 2008. Podosome-type adhesions and focal adhesions, so alike yet so different. Eur. J. Cell Biol. 87:491–506 10.1016/j.ejcb.2008.02.012 [DOI] [PubMed] [Google Scholar]
- Braiman A., Barda-Saad M., Sommers C.L., Samelson L.E. 2006. Recruitment and activation of PLCgamma1 in T cells: a new insight into old domains. EMBO J. 25:774–784 10.1038/sj.emboj.7600978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell S.C. 2010. Multiple microclusters: diverse compartments within the immune synapse. Curr. Top. Microbiol. Immunol. 340:123–154 10.1007/978-3-642-03858-7_7 [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Kapoor V., Trible R.P., Zhang W., Samelson L.E. 2001. Dynamic actin polymerization drives T cell receptor-induced spreading: a role for the signal transduction adaptor LAT. Immunity. 14:315–329 10.1016/S1074-7613(01)00112-1 [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Hong D.I., Kardon J.R., Yamazaki T., McGlade C.J., Barr V.A., Samelson L.E. 2002. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158:1263–1275 10.1083/jcb.200203043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell S.C., Barr V.A., Fuller C.L., Samelson L.E. 2003. High-resolution multicolor imaging of dynamic signaling complexes in T cells stimulated by planar substrates. Sci. STKE. 2003:PL8 10.1126/stke.2003.177.pl8 [DOI] [PubMed] [Google Scholar]
- Bunnell S.C., Singer A.L., Hong D.I., Jacque B.H., Jordan M.S., Seminario M.C., Barr V.A., Koretzky G.A., Samelson L.E. 2006. Persistence of cooperatively stabilized signaling clusters drives T-cell activation. Mol. Cell. Biol. 26:7155–7166 10.1128/MCB.00507-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach B.J., Srivastava R., Medeiros R.B., O’Gorman W.E., Peterson E.J., Shimizu Y. 2008. Distinct regulation of integrin-dependent T cell conjugate formation and NF-kappa B activation by the adapter protein ADAP. J. Immunol. 181:4840–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbach B.J., Srivastava R., Ingram M.A., Mitchell J.S., Shimizu Y. 2011. The pleckstrin homology domain in the SKAP55 adapter protein defines the ability of the adapter protein ADAP to regulate integrin function and NF-kappaB activation. J. Immunol. 186:6227–6237 10.4049/jimmunol.1002950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller G., Gimona M. 2004. Actin cytoskeleton remodelling via local inhibition of contractility at discrete microdomains. J. Cell Sci. 117:223–231 10.1242/jcs.00839 [DOI] [PubMed] [Google Scholar]
- Campi G., Varma R., Dustin M.L. 2005. Actin and agonist MHC-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202:1031–1036 10.1084/jem.20051182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrizosa E., Gomez T.S., Labno C.M., Klos Dehring D.A., Liu X., Freedman B.D., Billadeau D.D., Burkhardt J.K. 2009. Hematopoietic lineage cell-specific protein 1 is recruited to the immunological synapse by IL-2-inducible T cell kinase and regulates phospholipase Cgamma1 Microcluster dynamics during T cell spreading. J. Immunol. 183:7352–7361 10.4049/jimmunol.0900973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman N.M., Yoder A.N., Houtman J.C. 2012. Non-catalytic functions of Pyk2 and Fyn regulate late stage adhesion in human T cells. PLoS ONE. 7:e53011 10.1371/journal.pone.0053011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coussens N.P., Hayashi R., Brown P.H., Balagopalan L., Balbo A., Akpan I., Houtman J.C., Barr V.A., Schuck P., Appella E., Samelson L.E. 2013. Multipoint binding of the SLP-76 SH2 domain to ADAP is critical for oligomerization of SLP-76 signaling complexes in stimulated T cells. Mol. Cell. Biol. 33:4140–4151 10.1128/MCB.00410-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans J.G., Matsudaira P. 2006. Structure and dynamics of macrophage podosomes. Eur. J. Cell Biol. 85:145–149 10.1016/j.ejcb.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Geng L., Raab M., Rudd C.E. 1999. Cutting edge: SLP-76 cooperativity with FYB/FYN-T in the up-regulation of TCR-driven IL-2 transcription requires SLP-76 binding to FYB at Tyr595 and Tyr651. J. Immunol. 163:5753–5757 [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., Linder S. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20:235–241 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Gomez T.S., McCarney S.D., Carrizosa E., Labno C.M., Comiskey E.O., Nolz J.C., Zhu P., Freedman B.D., Clark M.R., Rawlings D.J., et al. 2006. HS1 functions as an essential actin-regulatory adaptor protein at the immune synapse. Immunity. 24:741–752 10.1016/j.immuni.2006.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. 1999. The immunological synapse: a molecular machine controlling T cell activation. Science. 285:221–227 10.1126/science.285.5425.221 [DOI] [PubMed] [Google Scholar]
- Griffiths E.K., Penninger J.M. 2002. Communication between the TCR and integrins: role of the molecular adapter ADAP/Fyb/Slap. Curr. Opin. Immunol. 14:317–322 10.1016/S0952-7915(02)00334-5 [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Ha V.L., Bharti S., Inoue H., Vass W.C., Campa F., Nie Z., de Gramont A., Ward Y., Randazzo P.A. 2008. ASAP3 is a focal adhesion-associated Arf GAP that functions in cell migration and invasion. J. Biol. Chem. 283:14915–14926 10.1074/jbc.M709717200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., Ginsberg M.H. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796–1806 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Harder T., Kuhn M. 2000. Selective accumulation of raft-associated membrane protein LAT in T cell receptor signaling assemblies. J. Cell Biol. 151:199–208 10.1083/jcb.151.2.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng T.S., Painter M.W., and Immunological Genome Project Consortium 2008. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9:1091–1094 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- Henrickson S.E., Mempel T.R., Mazo I.B., Liu B., Artyomov M.N., Zheng H., Peixoto A., Flynn M.P., Senman B., Junt T., et al. 2008. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol. 9:282–291 10.1038/ni1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P.V., Chabra I., Kornhauser J.M., Skrzypek E., Zhang B. 2004. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 4:1551–1561 10.1002/pmic.200300772 [DOI] [PubMed] [Google Scholar]
- Houtman J.C., Yamaguchi H., Barda-Saad M., Braiman A., Bowden B., Appella E., Schuck P., Samelson L.E. 2006. Oligomerization of signaling complexes by the multipoint binding of GRB2 to both LAT and SOS1. Nat. Struct. Mol. Biol. 13:798–805 10.1038/nsmb1133 [DOI] [PubMed] [Google Scholar]
- Huang Y., Norton D.D., Precht P., Martindale J.L., Burkhardt J.K., Wange R.L. 2005. Deficiency of ADAP/Fyb/SLAP-130 destabilizes SKAP55 in Jurkat T cells. J. Biol. Chem. 280:23576–23583 10.1074/jbc.M413201200 [DOI] [PubMed] [Google Scholar]
- Hunter A.J., Ottoson N., Boerth N., Koretzky G.A., Shimizu Y. 2000. Cutting edge: a novel function for the SLAP-130/FYB adapter protein in beta 1 integrin signaling and T lymphocyte migration. J. Immunol. 164:1143–1147 [DOI] [PubMed] [Google Scholar]
- Jo E.K., Wang H., Rudd C.E. 2005. An essential role for SKAP-55 in LFA-1 clustering on T cells that cannot be substituted by SKAP-55R. J. Exp. Med. 201:1733–1739 10.1084/jem.20042577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.W., Lye M.H., Barthel S.R., Duffy A.K., Annis D.S., Mosher D.F. 2004. Eosinophils adhere to vascular cell adhesion molecule-1 via podosomes. Am. J. Respir. Cell Mol. Biol. 31:413–422 10.1165/rcmb.2004-0099OC [DOI] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., Waterman C.M. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliche S., Breitling D., Togni M., Pusch R., Heuer K., Wang X., Freund C., Kasirer-Friede A., Menasche G., Koretzky G.A., Schraven B. 2006. The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell. Biol. 26:7130–7144 10.1128/MCB.00331-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliche S., Worbs T., Wang X., Degen J., Patzak I., Meineke B., Togni M., Moser M., Reinhold A., Kiefer F., et al. 2012. CCR7-mediated LFA-1 functions in T cells are regulated by 2 independent ADAP/SKAP55 modules. Blood. 119:777–785 10.1182/blood-2011-06-362269 [DOI] [PubMed] [Google Scholar]
- Krause M., Sechi A.S., Konradt M., Monner D., Gertler F.B., Wehland J. 2000. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J. Cell Biol. 149:181–194 10.1083/jcb.149.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., Carman C.V., Freeman G.J., Berezovskaya A., Constantine E., Springer T.A., Gertler F.B., Boussiotis V.A. 2004. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 7:585–595 10.1016/j.devcel.2004.07.021 [DOI] [PubMed] [Google Scholar]
- Lasserre R., Charrin S., Cuche C., Danckaert A., Thoulouze M.I., de Chaumont F., Duong T., Perrault N., Varin-Blank N., Olivo-Marin J.C., et al. 2010. Ezrin tunes T-cell activation by controlling Dlg1 and microtubule positioning at the immunological synapse. EMBO J. 29:2301–2314 10.1038/emboj.2010.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasserre R., Cuche C., Blecher-Gonen R., Libman E., Biquand E., Danckaert A., Yablonski D., Alcover A., Di Bartolo V. 2011. Release of serine/threonine-phosphorylated adaptors from signaling microclusters down-regulates T cell activation. J. Cell Biol. 195:839–853 10.1083/jcb.201103105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Lim C.J., Puzon-McLaughlin W., Shattil S.J., Ginsberg M.H. 2009. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284:5119–5127 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.S., Anekal P., Lim C.J., Liu C.C., Ginsberg M.H. 2013. Two modes of integrin activation form a binary molecular switch in adhesion maturation. Mol. Biol. Cell. 24:1354–1362 10.1091/mbc.E12-09-0695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky L., Jurdic P., Hinz B. 2008. Grab, stick, pull and digest: the functional diversity of actin-associated matrix-adhesion structures. Workshop on invadopodia, podosomes and focal adhesions in tissue invasion. EMBO Rep. 9:139–143 10.1038/sj.embor.7401162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margadant F., Chew L.L., Hu X., Yu H., Bate N., Zhang X., Sheetz M. 2011. Mechanotransduction in vivo by repeated talin stretch-relaxation events depends upon vinculin. PLoS Biol. 9:e1001223 10.1371/journal.pbio.1001223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Cardine A., Hendricks-Taylor L.R., Boerth N.J., Zhao H., Schraven B., Koretzky G.A. 1998a. Molecular interaction between the Fyn-associated protein SKAP55 and the SLP-76-associated phosphoprotein SLAP-130. J. Biol. Chem. 273:25789–25795 10.1074/jbc.273.40.25789 [DOI] [PubMed] [Google Scholar]
- Marie-Cardine A., Verhagen A.M., Eckerskorn C., Schraven B. 1998b. SKAP-HOM, a novel adaptor protein homologous to the FYN-associated protein SKAP55. FEBS Lett. 435:55–60 10.1016/S0014-5793(98)01040-0 [DOI] [PubMed] [Google Scholar]
- Martínez-Martín N., Fernández-Arenas E., Cemerski S., Delgado P., Turner M., Heuser J., Irvine D.J., Huang B., Bustelo X.R., Shaw A., Alarcón B. 2011. T cell receptor internalization from the immunological synapse is mediated by TC21 and RhoG GTPase-dependent phagocytosis. Immunity. 35:208–222 10.1016/j.immuni.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros R.B., Burbach B.J., Mueller K.L., Srivastava R., Moon J.J., Highfill S., Peterson E.J., Shimizu Y. 2007. Regulation of NF-kappaB activation in T cells via association of the adapter proteins ADAP and CARMA1. Science. 316:754–758 10.1126/science.1137895 [DOI] [PubMed] [Google Scholar]
- Mempel T.R., Henrickson S.E., Von Andrian U.H. 2004. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 427:154–159 10.1038/nature02238 [DOI] [PubMed] [Google Scholar]
- Ménasché G., Kliche S., Chen E.J., Stradal T.E., Schraven B., Koretzky G. 2007. RIAM links the ADAP/SKAP-55 signaling module to Rap1, facilitating T-cell-receptor-mediated integrin activation. Mol. Cell. Biol. 27:4070–4081 10.1128/MCB.02011-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M., Molina A., Escribese M.M., Yáñez-Mó M., Escudero E., Ursa A., Tejedor R., Mampaso F., Sánchez-Madrid F. 2004. VLA-4 integrin concentrates at the peripheral supramolecular activation complex of the immune synapse and drives T helper 1 responses. Proc. Natl. Acad. Sci. USA. 101:11058–11063 10.1073/pnas.0307927101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K., Sylvain N.R., Bunnell S.C. 2008. T cell costimulation via the integrin VLA-4 inhibits the actin-dependent centralization of signaling microclusters containing the adaptor SLP-76. Immunity. 28:810–821 10.1016/j.immuni.2008.04.019 [DOI] [PubMed] [Google Scholar]
- Nolz J.C., Medeiros R.B., Mitchell J.S., Zhu P., Freedman B.D., Shimizu Y., Billadeau D.D. 2007. WAVE2 regulates high-affinity integrin binding by recruiting vinculin and talin to the immunological synapse. Mol. Cell. Biol. 27:5986–6000 10.1128/MCB.00136-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Takenawa T. 2009. PtdIns(3,4)P2 instigates focal adhesions to generate podosomes. Cell Adhes. Migr. 3:195–197 10.4161/cam.3.2.7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Itoh T., Takenawa T. 2008. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol. 182:157–169 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory S., Brazier H., Pawlak G., Blangy A. 2008. Rho GTPases in osteoclasts: orchestrators of podosome arrangement. Eur. J. Cell Biol. 87:469–477 10.1016/j.ejcb.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Papadopoulos J.S., Agarwala R. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 23:1073–1079 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Pauker M.H., Reicher B., Fried S., Perl O., Barda-Saad M. 2011. Functional cooperation between the proteins Nck and ADAP is fundamental for actin reorganization. Mol. Cell. Biol. 31:2653–2666 10.1128/MCB.01358-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson E.J., Woods M.L., Dmowski S.A., Derimanov G., Jordan M.S., Wu J.N., Myung P.S., Liu Q.H., Pribila J.T., Freedman B.D., et al. 2001. Coupling of the TCR to integrin activation by Slap-130/Fyb. Science. 293:2263–2265 10.1126/science.1063486 [DOI] [PubMed] [Google Scholar]
- Raab M., Kang H., da Silva A., Zhu X., Rudd C.E. 1999. FYN-T-FYB-SLP-76 interactions define a T-cell receptor zeta/CD3-mediated tyrosine phosphorylation pathway that up-regulates interleukin 2 transcription in T-cells. J. Biol. Chem. 274:21170–21179 10.1074/jbc.274.30.21170 [DOI] [PubMed] [Google Scholar]
- Raab M., Wang H., Lu Y., Smith X., Wu Z., Strebhardt K., Ladbury J.E., Rudd C.E. 2010. T cell receptor “inside-out” pathway via signaling module SKAP1-RapL regulates T cell motility and interactions in lymph nodes. Immunity. 32:541–556 10.1016/j.immuni.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Smith X., Matthess Y., Strebhardt K., Rudd C.E. 2011. SKAP1 protein PH domain determines RapL membrane localization and Rap1 protein complex formation for T cell receptor (TCR) activation of LFA-1. J. Biol. Chem. 286:29663–29670 10.1074/jbc.M111.222661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B. 1996. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266:525–539 10.1016/S0076-6879(96)66033-9 [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584–599 10.1006/jmbi.1993.1413 [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 19:55–72 10.1002/prot.340190108 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Yang J., Zhang Y. 2012. COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40(Web Server issue):W471–W477 10.1093/nar/gkr1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage P.T., Varghese L.M., Martinelli R., Sciuto T.E., Kamei M., Dvorak A.M., Springer T.A., Sharpe A.H., Carman C.V. 2012. Antigen recognition is facilitated by invadosome-like protrusions formed by memory/effector T cells. J. Immunol. 188:3686–3699 10.4049/jimmunol.1102594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraven B., Marie-Cardine A., Koretzky G. 1997. Molecular analysis of the fyn-complex: cloning of SKAP55 and SLAP-130, two novel adaptor proteins which associate with fyn and may participate in the regulation of T cell receptor-mediated signaling. Immunol. Lett. 57:165–169 10.1016/S0165-2478(97)00053-9 [DOI] [PubMed] [Google Scholar]
- Seminario M.C., Bunnell S.C. 2008. Signal initiation in T-cell receptor microclusters. Immunol. Rev. 221:90–106 10.1111/j.1600-065X.2008.00593.x [DOI] [PubMed] [Google Scholar]
- Seminario M.C., Sterbinsky S.A., Bochner B.S. 1998. Beta 1 integrin-dependent binding of Jurkat cells to fibronectin is regulated by a serine-threonine phosphatase. J. Leukoc. Biol. 64:753–758 [DOI] [PubMed] [Google Scholar]
- Seminario M.C., Precht P., Bunnell S.C., Warren S.E., Morris C.M., Taub D., Wange R.L. 2004. PTEN permits acute increases in D3-phosphoinositide levels following TCR stimulation but inhibits distal signaling events by reducing the basal activity of Akt. Eur. J. Immunol. 34:3165–3175 10.1002/eji.200425206 [DOI] [PubMed] [Google Scholar]
- Shaw A.S. 2006. Lipid rafts: now you see them, now you don’t. Nat. Immunol. 7:1139–1142 10.1038/ni1405 [DOI] [PubMed] [Google Scholar]
- Sherman E., Barr V., Manley S., Patterson G., Balagopalan L., Akpan I., Regan C.K., Merrill R.K., Sommers C.L., Lippincott-Schwartz J., Samelson L.E. 2011. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity. 35:705–720 10.1016/j.immuni.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Van Seventer G.A., Horgan K.J., Shaw S. 1990. Regulated expression and binding of three VLA (beta 1) integrin receptors on T cells. Nature. 345:250–253 10.1038/345250a0 [DOI] [PubMed] [Google Scholar]
- Singer A.L., Bunnell S.C., Obstfeld A.E., Jordan M.S., Wu J.N., Myung P.S., Samelson L.E., Koretzky G.A. 2004. Roles of the proline-rich domain in SLP-76 subcellular localization and T cell function. J. Biol. Chem. 279:15481–15490 10.1074/jbc.M313339200 [DOI] [PubMed] [Google Scholar]
- Smith-Garvin J.E., Koretzky G.A., Jordan M.S. 2009. T cell activation. Annu. Rev. Immunol. 27:591–619 10.1146/annurev.immunol.021908.132706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer T.A. 1994. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 76:301–314 10.1016/0092-8674(94)90337-9 [DOI] [PubMed] [Google Scholar]
- Swanson K.D., Tang Y., Ceccarelli D.F., Poy F., Sliwa J.P., Neel B.G., Eck M.J. 2008. The Skap-hom dimerization and PH domains comprise a 3′-phosphoinositide-gated molecular switch. Mol. Cell. 32:564–575 10.1016/j.molcel.2008.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvain N.R., Nguyen K., Bunnell S.C. 2011. Vav1-mediated scaffolding interactions stabilize SLP-76 microclusters and contribute to antigen-dependent T cell responses. Sci. Signal. 4:ra14 10.1126/scisignal.2001178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valitutti S., Dessing M., Aktories K., Gallati H., Lanzavecchia A. 1995. Sustained signaling leading to T cell activation results from prolonged T cell receptor occupancy. Role of T cell actin cytoskeleton. J. Exp. Med. 181:577–584 10.1084/jem.181.2.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dries K., Schwartz S.L., Byars J., Meddens M.B., Bolomini-Vittori M., Lidke D.S., Figdor C.G., Lidke K.A., Cambi A. 2013. Dual-color superresolution microscopy reveals nanoscale organization of mechanosensory podosomes. Mol. Biol. Cell. 24:2112–2123 10.1091/mbc.E12-12-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.H., Reinherz E.L. 2012. The structural basis of αβ T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol. Rev. 250:102–119 10.1111/j.1600-065X.2012.01161.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Moon E.Y., Azouz A., Wu X., Smith A., Schneider H., Hogg N., Rudd C.E. 2003. SKAP-55 regulates integrin adhesion and formation of T cell-APC conjugates. Nat. Immunol. 4:366–374 10.1038/ni913 [DOI] [PubMed] [Google Scholar]
- Wang H., Liu H., Lu Y., Lovatt M., Wei B., Rudd C.E. 2007. Functional defects of SKAP-55-deficient T cells identify a regulatory role for the adaptor in LFA-1 adhesion. Mol. Cell. Biol. 27:6863–6875 10.1128/MCB.00556-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N., Bodin L., Pandey M., Krause M., Coughlin S., Boussiotis V.A., Ginsberg M.H., Shattil S.J. 2008. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin αIIbβ3. J. Cell Biol. 181:1211–1222 10.1083/jcb.200803094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Fu J., Shen S.H. 2002. SKAP55 coupled with CD45 positively regulates T-cell receptor-mediated gene transcription. Mol. Cell. Biol. 22:2673–2686 10.1128/MCB.22.8.2673-2686.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 9:40 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Jin B., Henrickson S.E., Perelson A.S., von Andrian U.H., Chakraborty A.K. 2008. How antigen quantity and quality determine T-cell decisions in lymphoid tissue. Mol. Cell. Biol. 28:4040–4051 10.1128/MCB.00136-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel P.A., Bunnell S.C., Witherow D.S., Gu J.J., Chislock E.M., Ring C., Pendergast A.M. 2006. Role for the Abi/wave protein complex in T cell receptor-mediated proliferation and cytoskeletal remodeling. Curr. Biol. 16:35–46 10.1016/j.cub.2005.12.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.