Abstract
Liquid chromatography-electrospray ionization-infrared multiphoton dissociation (IRMPD) mass spectrometry was developed to investigate the distributions of intrastrand crosslinks formed between cisplatin and two oligodeoxynucleotides (ODNs), d(A1T2G3G4G5T6A7C8C9C10A11T12) (G3-D) and its analog d(A1T2G3G4G5T6T7C8C9C10A11T12) (G3-H), that have been reported to adopt different secondary structures in solution. Based on the formation of site-specific fragment ions upon IRMPD, two isobaric crosslink products were differentiated for each ODN. The preferential formation of G3G4 and G4G5 crosslinks was determined as a function of reaction conditions, including incubation temperature and presence of metal ions. G3-D consistently exhibited a greater preference for formation of the G4G5 crosslink compared to the G3-H ODN. The ratio of G3G4:G4G5 crosslinks increased for both G3-D and G3-H at higher incubation temperatures or when metal salts were added. Comparison of the IRMPD fragmentation patterns of the unmodified ODNs and the intramolecular platinated crosslinks indicated that backbone cleavage was significantly suppressed near the crosslink.
Keywords: tandem mass spectrometry, cisplatin, oligodeoxynucleotides, photodissociation, hairpin
Introduction
Hairpins, characterized by DNA engaged in intrastrand base pairing, are one of the simplest secondary structural elements of nucleic acids and have proven to play important roles in a number of biological processes, including DNA replication, the regulation of gene transcription and DNA recombination [1-9]. The thermodynamics and kinetics of the formation of DNA hairpins and their interactions with other ligands have been extensively studied using spectroscopic methods such as absorption [10], fluorescence [11-14], circular dichroism [15, 16], NMR [17-19], X-ray crystallography [20] and gel electrophoresis [21]. Unraveling the recognition and interactions of DNA with drugs or other ligands remains a key interest, and there are unresolved questions about the degree of conformational changes of DNA that occur during ligand binding. A number of studies have sought to elucidate the kinetics and thermodynamics of ligand binding to hairpin DNA (as well as other conformations of DNA) [8, 9]. Hairpin structures offer a compelling target due to the significant differences in electrostatic environment and accessibility between the stem and more open loop regions.
There has been particular interest in the reactions of platinum complexes with various DNA structures, including hairpins. Within the cellular environment, the most widely used anticancer drug, cis-diamminedichloroplatinum (II) (cisplatin), is hydrolyzed and then reacts with cellular DNA. The predominant products are 1,2-intrastrand crosslinks involving adjacent bases, primarily as cis-[Pt(d(GpG))(NH3)2] (60-65% abundance) or cis-[Pt(d(ApG))(NH3)2] (20-25% abundance) [22-24]. Coordination of platinum by DNA is known to induce considerable bending and unwinding of the DNA and may even cause conversion of hairpin and duplex structures into other structures. Chow et al. reported that interaction of cisplatin occurred preferentially with guanines in the loop region of hairpins, an outcome mediated by loop flexibility and “deformability” [25]. The most extensive effort to probe the role of hairpins in the crosslinking of DNA by cisplatin has been undertaken by the Marzilli group [26-29]. They employed NMR spectroscopy to elucidate the structural features of the complexes formed between cisplatin or metal ions with the self-complementary DNA sequence G3-D (d(A1T2G3G4G5T6A7C8C9C10A11T12)), which exists as a duplex or hairpin in solution, and its analog G3-H (d(A1T2G3G4G5T6T7C8C9C10A11T12)), which adopts a hairpin structure in solution [27-29]. Marzilli et al. showed that the intrastrand crosslinking of cisplatin to G3-D depended on the leaving ligand as well as other factors; for example, the G4G5 intrastrand crosslinked product greatly predominated over G3G4 products (~25:1) upon reaction with cis-Pt(NH3)2Cl2 in which G3, G4, and G5 are used to designate the positions of the crosslinked guanines. In contrast, both intrastrand crosslinked G3G4 and G4G5 products were equally favored (1:1 G3G4:G4G5) upon reaction of G3-D with cis-Pt(NH3)2(H2O)22+. Above and beyond the specific G crosslinking sites, the reaction of the G3-D and G3-H oligomers with cisplatin resulted in the interconversion of secondary structures (e.g. from duplexes to hairpins or hairpins to weaker hairpins and coils). Based on results from denaturing gel electrophoresis experiments and NMR spectroscopy, Marzilli et al. proposed that the G3-D sequence favored formation of an intrastrand G4-G5 crosslinked hairpin over an intrastrand G3-G4 coil upon reaction with cisplatin [27]. With A7 of G3-D replaced by T7 in the G3-H sequence, the resulting central TT mismatch destabilized the duplex form and ultimately resulted in a ratio of intrastrand crosslinked G4G5 to G3G4 weak hairpin products of about 3:1 and without a significant dependence on the nature of the leaving ligand of the cisplatin reactant (cis-Pt(NH3)2Cl2 versus cis-Pt(NH3)2(H2O)22+) [28]. It was also reported that the addition of Zn2+ stabilized the duplex form and slightly shifted the distribution of G3G4 and G4G5 products for the G3-H sequence, going from 28:72 G3G4:G4G5 in the absence of Zn2+ to 33:67 in the presence of Zn2+ [28]. A similar shift was not noted for G3-D.
Tandem mass spectrometry (MS/MS) has evolved as a compelling strategy for sequencing nucleic acids and determining their structural modifications [30-40]. Several groups have reported the successful characterization of cisplatin adducts [35-40] which is one focus of the present study. For example, Egger et al. elucidated the kinetics and the preferred binding sites of cisplatin (at GG and GTG sites) in double-stranded DNA oligonucleotides [41]. Nyakas et al. demonstrated that the enhanced cleavage of the 3’-C-O bond next to a GG site in oligodeoxyribonucleotides was catalyzed by the adjacent cisplatin adduction site, as revealed by the formation of a highly characteristic w ion [42]. This same group also elucidated the fragmentation pathways of platinated quadruplex DNA [43] and RNA [44]. More recently, our group reported a comparison of MS/MS methods, including CID, IRMPD (10.6 μm), ETD, NETD and UVPD (193 nm) as well as hybrid MS/MS processes [45], termed ETcaD, ET-IRMPD and ET-UVPD, for characterization of DNA/cisplatin adducts [46]. Based on the latter systematic study, it was concluded that IRMPD offered the best characteristics for pinpointing the cisplatin adduction sites in the fragment-rich spectra obtained for the DNA/cisplatin adducts [46].
There have been few studies employing MS/MS for the characterization of DNA hairpins and none for adducts of hairpins. Based on comparison of the MS/MS patterns of three isomeric 15-mer ODNs, Mo et al. confirmed that the fragmentation patterns of the ODNs varied depending on whether they adopted hairpin or random structures in solution, thus implying that conformational differences were retained after transfer of the complexes by ESI to the gas phase [47]. The same group correlated the gas-phase hydrogen/deuterium exchange kinetics of hairpin ODNs with their predicted stabilities in solution [48]. Fabris et al. reported that the degree of backbone fragmentation of nucleic acids could be inhibited by specific base-pairing interactions (via masking or shielding certain regions from cleavage), thus showing that the higher-order structures of nucleic acids influenced the resulting MS/MS behavior [49].
The present study was motivated by our interest in capitalizing on the specificity of MS/MS strategies to differentiate isomeric DNA structures, including those arising from the reactions of ligands with different DNA conformations in solution. As a platform for this objective, we focus on demonstrating the ability to characterize cisplatin adducts produced upon reaction of two ODNs, G3-D (d(A1T2G3G4G5T6A7C8C9C10A11T12)) and its analog G3-H (d(A1T2G3G4G5T6T7C8C9C10A11T12)) that were the benchmarks of Marzilli's series of studies described above [26-29]. We explore the use of LC-MS/MS to evaluate the products of the reactions of each ODN with cisplatin and in the presence of different metal ions (Mg2+ and Zn2+) in solution. Although the resulting products are characterized in the gas phase, they are formed in solution using typical conditions for cisplatin reactions, and the resulting crosslinked products are secured by covalent bonds, meaning that the adduction sites are not labile during ESI transport to the gas phase. We utilize IRMPD to distinguish between the two types of DNA/cisplatin adducts for each sequence and demonstrate the ability to monitor the preferential formation of G3G4 and G4G5 products as a function of reaction conditions.
Experimental
Materials and Reagents
Single strand oligodeoxynucleotides, G3-D (d(ATG GGT ACC CAT) and G3-H (d(ATG GGT TCC CAT)) were purchased from Integrated DNA Technologies (Coralville, IA, USA) on the 1 μmol scale and used without further purification. Stock solutions were prepared in water at approximately 1 mM and the exact concentration was determined using a Nanodrop ND-1000 spectrophotometer at 260 nm (Wilmington, DE, USA) with the extinction coefficients provided by the manufacturer. Each oligodeoxynucleotide was diluted to approximately 10 μmol/L prior to LC-MS/MS analysis. HPLC grade acetonitrile, ammonium acetate (NH4OAc) and water (Fisher Scientific, Pittsburg, PA, USA) were used for preparing LC mobile phase. cis-Diamminedichloroplatinum (II) (cDDP) was purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). Zinc acetate dihydrate (Zn(OAc)2·2H2O) and Magnesium Acetate Tetrahydrate (Mg(OAc)2·4H2O) were purchased from Fisher Scientific (Pittsburgh, PA, USA).
Crosslinking reaction of cisplatin
After the cDDP solution (500 μmol/L) was incubated with water for 24 h at 37 °C to form the reactive species [Pt(NH3)2(H2O)2]2+, it was mixed with the corresponding oligonucleotide in a molar ratio of 3:1 (120 μmol/L:40 μmol/L) and then incubated with ammonium acetate (90 mmol/L) for another 2 hours at 37 °C (or 5 days at 4 °C as described for some of the experiments). Prior to mass spectrometric analysis, the reaction mixture was diluted with water to yield a final concentration of approximately 10 μmol/L.
Mass Spectrometry and Liquid Chromatography
Mass spectrometric analysis was performed on a Bruker Daltonics HCTUltra quadrupole ion trap mass spectrometer (Bremen, Germany). The ion charge control (ICC) target was set to 75,000 with a maximum accumulation time of 100 ms. The ion source parameters were set as follows: dry temperature 100 °C, nebulizer pressure 12 psi, and dry gas flow 6 L/min for operation in the negative ESI mode. IRMPD was performed using a Synrad (Mukilteo, WA) 50 W CO2 laser at a wavelength of 10.6 μm. Typical IRMPD conditions included an isolation width of 4.0 m/z, and a q value of 0.1 with the laser operating at a power level of 9-10% (relative to 50 W). The irradiation time was adjusted to optimize the amount of fragmentation observed, typically between 50 and 150 ms. Liquid chromatography was carried out using a Dionex UltiMate 3000 system (Sunnyvale, CA) similar to that previously described [46].
Results and Discussion
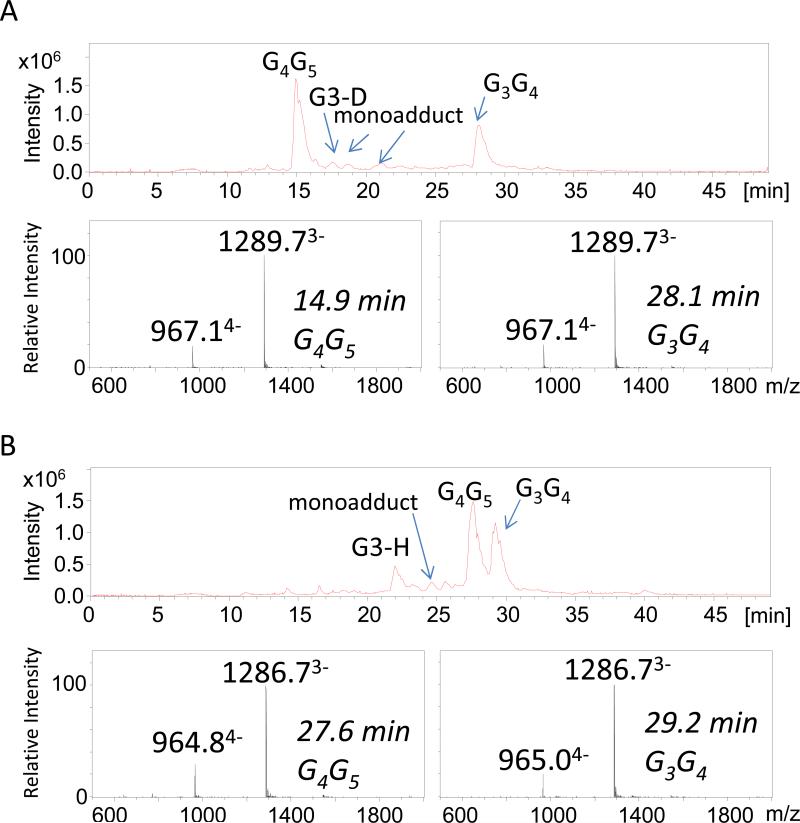
Figure 1 shows the total ion chromatograms for the ODN/cisplatin incubates. The mass spectra of the platinated 12-mer single-stranded oligonucleotides G3-D and G3-H provided proof of successful adduct formation. In the G3G4G5 sequence, the N7 position of the central guanine residue G4 is the most nucleophilic site and the preferential site of attack of the monofunctional complex, [Pt(NH3)2ClH2O]+. In the negative mode ESI mass spectra, triply and quadruply charged products ions consistent with ODN/cisplatin intrastrand crosslinks were observed predominantly (Figure 1A). Two isobaric products of the G3-D/cisplatin intrastrand adducts, G4G5 and G3G4, eluted at 14.9 and 28.1 minutes, respectively (Figure 1A). The ions of m/z 965 and 1287 were assigned as the [M - 4H]4− and [M - 3H]3− ions of the corresponding G3-H/cisplatin intrastrand crosslinks. Two isobaric products of the G3-H/cisplatin intrastrand adducts, G4G5 and G3G4, eluted at 27.6 and 29.2 minutes, respectively (Figure 1B). (Differentiation of the isobaric products for each ODN is described in the next section). The stacking interaction between A7 and G4 in the structure of G3-D, which is presumed to stabilize the hairpin form of G4G5, may contribute to the significant difference in retention times between the G4G5 and G3G4 adducts for G3-D (in contrast to the very similar retention times for the two isobaric crosslinks for G3-H). Interstrand crosslinks were not observed to any appreciable extent. Monoadducts, which are known to be precursors in the crosslinking process involving the cisplatin ligands, were detected as low abundance products. Cisplatin reacts with the N7 position of guanine residues to form the initial monoadducts, but the second step of the crosslinking reaction is efficient and rapidly converts the monoadducts to stable crosslinks upon reaction of a second guanine residue. Thus, only a small amount of the monoadducts appeared in the chromatographic profiles (see Figure 1). As elucidated by the MS/MS patterns described below, the main products were 1,2-intrastrand crosslinks involving adjacent bases (>90% of products) [22-24]. 1,3-intrastrand crosslinks or interstrand crosslinks involving non-adjacent guanines are typically far less favored, and in fact were not detected in the present study. Furthermore, for the ODNs studied the guanine residues on opposite strands (assuming the ODNs may form some duplex-like structures in solution) are so far apart that they are not positioned to allow effective cisplatin interstrand crosslinking. Low abundances of the unreacted single strands were also observed for each ODN in Figure 1. For these reactions, sodium and potassium adducts which normally congest the spectra of ODNs and reduce detection sensitivity were inconsequential because the high concentration of ammonium ions in the mobile phase displaced the sodium and potassium ions.
Figure 1.
Total ion chromatogram (TIC) for each eluting species and the associated ESI mass spectra for (A) the G3-D/cisplatin incubate and the (B) the G3-H/cisplatin incubate. Each cisplatin/ODN incubated was reacted for two hours at 37 °C .
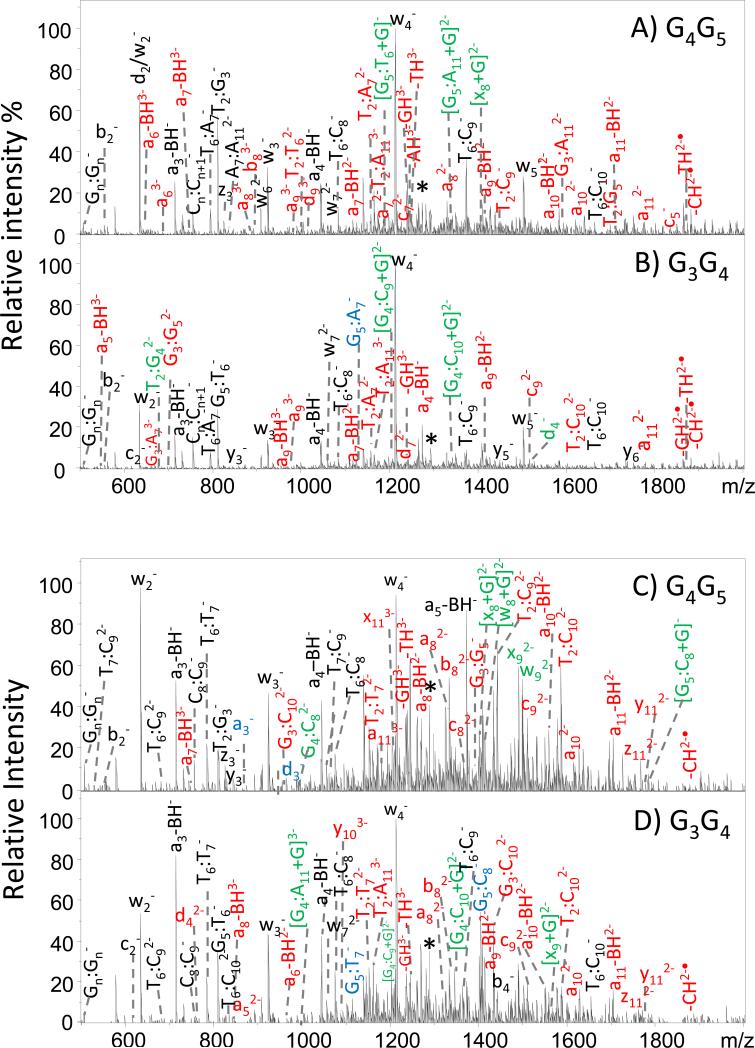
Despite having different retention times upon chromatographic separation, the two expected intrastrand crosslinked products for G3-D or G3-H have identical masses, and their differentiation relies on the generation of unique MS/MS patterns for each adduct. We have previously explored the fragmentation behavior of DNA/cisplatin adducts using an array of ion activation methods [46]. IRMPD resulted in rich fragmentation patterns with respect to production of the most diverse array of fragments, thus allowing the cisplatin adduction sites to be pinpointed [46]. Based on that prior benchmark study, IRMPD was employed in the present investigation for the analysis of cisplatin intrastrand crosslinks corresponding to the major peaks in the chromatograms in Figure 1. Representative IRMPD spectra are displayed in Figure 2 for the 3- charge state of the G3-D/cisplatin and G3-H/cisplatin intrastrand crosslinks, and expanded regions of the mass spectra are shown in Supplemental Figure 2. All fragment ions were assigned according to the nomenclature of McLuckey [30, 31] as illustrated in Supplemental Figure 1. The IRMPD spectra were dominated by abundant a – B and w ions arising from the cleavage of the 3’- C-O bond after initial base loss and internal fragment ions (the latter denoted by Bx:By in which the Bx and By indicate the positions of the nucleobases that are incorporated in the internal products). Supplemental Tables 1 and 2 summarize the m/z values and abundances of the fragment ions in tabular format for each crosslinked ODN, and Supplemental Table 3 shows the IRMPD mass spectral data for the corresponding unmodified ODNs.
Figure 2.
IRMPD spectra for (A) the G4G5 cisplatin crosslink of G3-D; (B) the G3G4 cisplatin crosslink of G3-D ; (C) the G4G5 cisplatin crosslink of G3-H, and (D) the G3G4 cisplatin crosslink of G3-H. Fragment ions in red and in green contain the Pt modification. Fragment ions in black and in blue are the Pt-free fragments. Unique fragments for each adducts that allowed them to be identified are labeled in green and blue. Precursor ions are noted with an asterisk.
The discrimination of G3G4 and G4G5 crosslinks is a challenge due to the minor differences in the primary structures of the crosslinks. As a result, they share a large number of identical fragment ions and chromatographic separation is essential to facilitate differentiation. Although many of the fragment ions produced by the G3G4 and G4G5 isomers have the same m/z values, there are a few key fragment ions that allow them to be confidently distinguished by IRMPD. In Figure 2, the fragments marked in green (bearing the Pt modification) and in blue (without the platinum modification) are important for localizing the Pt modification and site of crosslink. Expansions of some of these key diagnostic ions are shown in Supplemental Figure 2. In Figures 2A and 2B (for ODN G3-D), the G3G4 crosslink produces several Pt-containing ions, including T2:G4 (m/z 684.4), G4:C9 (m/z 1204.7), G4:C10 (m/z 1349.1) and d4 (m/z 1522.1), whereas the G4G5 crosslink produces G5:G6 (m/z 1189.3), G5:G11 (m/z 1341.7), and x8 (m/z 1403.3) as diagnostic Pt-containing ions. These fragment ions are specific to each of the two isomers. For the G3-H ODN (Figures 2C and 2D), upon IRMPD the following unique Pt-containing ions allow differentiation of the isomeric crosslinks: x8 (m/z 1399.7), w8 (m/z 1408.2), x9 (m/z 1489.7), w9 (m/z 1498.2), and internal ion G5:G8 (m/z 1783.1) for the G4G5 crosslink and G4:G11 (m/z 999.3), G4:G10 (m/z 1344.2), and x9 (m/z 1564.7) for the G3G4 crosslink. Among the platinum-bearing fragment ions that allow the modification to be identified, the key ones arise from cleavage of bonds next to the adduction sites, including [G5:T6+Pt(NH3)2+G]− (m/z 1189.3) for the G4G5 crosslink and [G4:C10+Pt(NH3)2+G]− (m/z 1349.1) for the G3G4 crosslink of G3-D. These ions confirm the existence of a covalent bond between the Pt and guanine bases. A few ions in Figure 2 cannot be readily assigned without further information due to having m/z values that are consistent with several possible structures. However, once the platinum modification site is located based on the other cisplatin-containing fragment ions, most of these types of fragments can be assigned. For example for the G4G5 adduct of G3-H, the ion of m/z 810.4 could initially be assigned as T2:G3− or G5:T6−. After the Pt modification was located at the G4G5 site, this ion was confirmed as T2:G3−. Based on the MS/MS spectra, the platination site for each crosslink were identified.
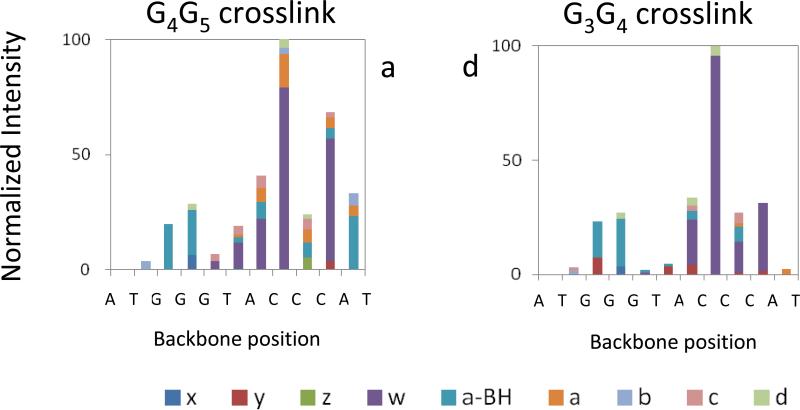
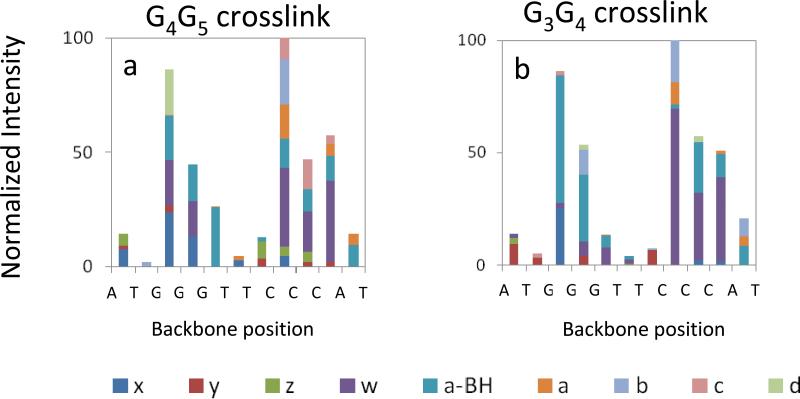
In order to examine trends in the preferred dissociation pathways of the different DNA crosslink structures, the distributions of all the series of fragments arising from cleavage at each backbone bond site were graphically tabulated for the IRMPD spectra, and the corresponding stacked bar graphs are summarized in Figure 3 and Figure 4. The IRMPD spectra corresponding to the unmodified ODNs are summarized in bar graph form in Supplemental Figure 3. In terms of cleavage of the number of backbone cleavage sites (eleven possible sites for these ODNs), the fragment ion series produced upon IRMPD cover the entire (for platinated G3-H: 11 out of 11 backbone cleavages) or almost entire (for platinated G3-D, unmodified G3-D and G3-H: 10 out of 11 backbone cleavages) ODN sequence, thus providing nearly full sequence coverage. Both the G3G4 and G4G5 structures of each ODN show a non-uniform cleavage pattern upon IRMPD, with a notable preference for cleavage at backbone sites 8 (between C8 and C9) and 10 (between C10 and A11). The non-uniform fragmentation pattern indicates that the backbone bonds are not equally prone to dissociation. The bulky crosslinked Pt modification seems to be responsible for a masking effect at backbone bonds 3, 4 and 5 for G3-D and 5 for G3-H (Figures 3 and 4) relative to the fragmentation of the unmodified ODNs (Supplemental Figure 3). For both unmodified and crosslinked ODNs, the backbone cleavages at positions 2 and 6 are disfavored owing to the low reactivity of the thymine base (i.e. dissociation of deprotonated ODNs typically is initiated by base loss, followed by backbone cleavage to form a – B and w ions. Thymine loss is disfavored, thus suppressing formation of the corresponding a – B and w ions originating from thymine positions).[50, 51] For the intrastrand crosslinks of G3-H, cleavage at the 3 and 4 backbone positions is almost as prominent as cleavages at the 8 and 10 positions, even for the G3G4 adduct. In the sequence of G3-H, the A7 of G3-D is replaced with T7. A7 was located inside the presumed hairpin loop and stacked above G4 for G3-D. The stacking interaction between A7 and G4 stabilizes the hairpin form of G3-D [26], whereas in the structure of G3-H, there is no possibility of such stacking interaction. If there is even some retention of stacking interactions in the gas phase, this might explain why cleavages at the 3 and 4 positions are less prevalent for G3-D compared to G3-H. Some evidence for retention of hairpin structure in the gas phase has been reported previously, either based on the formation of specific fragment ions observed uniquely for hairpins [47], based on hydrogen/deuterium exchange experiments [48], or based on ion mobility measurements [52].
Figure 3.
Relative abundances of IRMPD fragment ions at each backbone cleavage site for [G3-D + Pt(NH3)2 - 5H]3− : (a) all fragments of G4G5 crosslink; (b) all fragments of G3G4 crosslink.
Figure 4.
Relative abundances of IRMPD fragment ions at each backbone cleavage site for [G3-H + Pt(NH3)2 - 5H]3− : (a) all fragments of G4G5 crosslink; (b) all fragments of G3G4 crosslink.
Both unmodified ODNs display a significant preference for cleavage at the 5 backbone position (between G5 and T6) upon IRMPD (Supplemental Figure 3), a cleavage site that was not favored for the intrastrand crosslinks. This result highlights the suppression of cleavage near the platination site for the G3G4 and G4G5 crosslinks. The unmodified ODNs primarily produce conventional a – B and w ions upon IRMPD, in contrast to the crosslinks which yield a greater array of fragment types, including less common x, y, z, a and d ions, a result that again demonstrates the significant impact of platination on the fragmentation pathways of the ODNs. Interestingly, the crosslinks favored generation of w2 and w4 ions, ions that arise from backbone cleavages remote from the platination sites (at backbone position 10 for w2 and position 8 for w4). These w ions are not predominant for the unmodified ODNs. The IRMPD spectra for the crosslinks also revealed the presence of more numerous or more abundant internal ions, such as T6:C8 of m/z 1083 for G4G5 of G3-D (Figure 2A, cleavage at backbone positions 5 and 8), T6:C8 of m/z 1074 for G3G4 of G3-H (Figure 2D, cleavage at backbone positions 5 and 8) and T6:C7 of m/z 785 for G4G5 of G3-H (Figure 2C, cleavage at backbone positions 5 and 7, respectively), than observed for the unmodified ODNs.
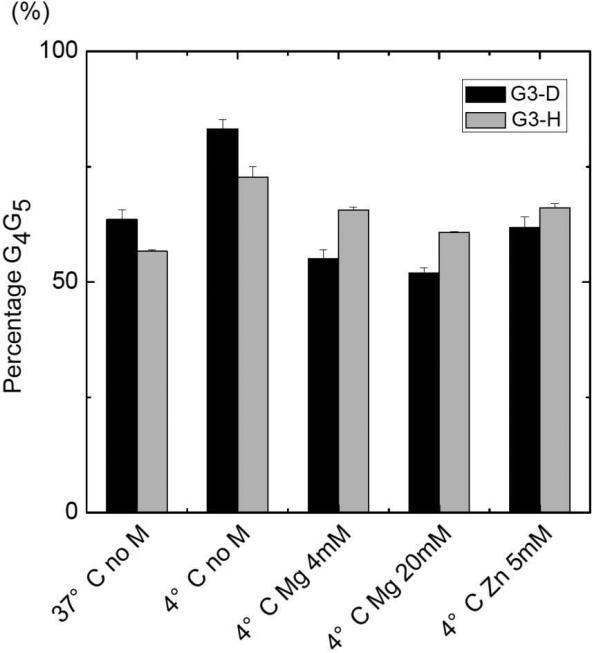
Based on the integration of the chromatographic peaks and the assignment of G3G4 or G4G5 structures from the IRMPD MS/MS spectra, the distribution of the crosslinks was determined (see examples of chromatographic traces in Figure 1). The results are illustrated in bar graph form in Figure 5 for the two ODNs as a function of the temperature of the solution and the presence of metal ions (Zn2+ or Mg2+). In general, reaction of each ODN with cisplatin at 4 °C yielded more monoadducts and unreacted ODNs and correspondingly lower abundances of intrastrand crosslinks compared to the reactions undertaken at 37 °C. Moreover, the portion of the G4G5 crosslinks relative to G3G4 crosslinks was lower at 37 °C than at 4 °C for both G3-D and G3-H, presumably because the higher temperature destabilized the hairpin forms in solution due to disruption of the hydrogen-bonds of the base pairs (i.e. partial melting at the higher temperature). For example, for G3-D, the G3G4:G4G5 product ratio increased from 17:83 at 4 °C to 38:62 at 37 °C, indicating greater production of the G3G4 species (or suppression of the G4G5 product). For G3-H, the G3G4:G4G5 product ratio increased from 29:71 at 4 °C to 45:55 at 37 °C, likewise indicating enhanced generation of the G3G4 product at the higher temperature.
Figure 5.
Effect of temperature and metal ions on the DNA/cisplatin products(G4G5 adducts). M = metal. All the solutions containing metal ions were run at 4 °C. All DNA/cisplatin reactions were run for 2 hours (at 37 °C) or for 5 days (at 4 °C). The standard deviation error bars are based on triplicate results.
Metal salts are known to play a role in the adoption of higher order structures of ODNs because the adduction of metal ions can shield negatively charged phosphate backbone groups and thereby modulate repulsive forces arising from more compact secondary structures (such as hairpins) [53, 54]. In the present study, the addition of Zn2+ or Mg2+ reduced the preference for formation of G4G5 crosslinks relative to G3G4 crosslinks, resulting in an average G3G4:G4G5 product ratio of 46:54 for G3-D (compared to 17:83 in the absence of metals) and 37:63 for G3-H (compared to 29:71 in the absence of metals). In each case, the presence of the metal enhanced the portion of G3G4 products, similar to the temperature effect noted above. The observed decrease in the formation of platinated G4G5 crosslinks confirms that incipient hairpin structures (ones that preferentially lead to G4G5 crosslinks) are diminished in the presence of M2+ or for reactions undertaken at elevated temperature.
In Marzilli's extensive studies of the cisplatin reactions of G3-D, it was reported that the presence of Zn2+ changed the G3G4:G4G5 ratio slightly to from 28:72 to 33:67, while the addition of Mg2+ had no effect. Addition of Mg2+ or Zn2+ did not change the crosslinked product ratio of the G3-D reactions [28, 29]. Based on our LC-ESI-MS results, in the absence of Zn2+ or Mg2+ the G3G4:G4G5 ratio was 27:73 for G3-H or 17:83 for G3-D. We found in every case that addition of a metal salt to the cisplatin/ODN solutions caused a modest reduction in the preference for formation of the G4G5 crosslink and concomitant increase in the abundance of the G3G4 crosslink, as described and rationalized above.
Conclusions
This work highlights the application of a LC-MS/MS-based method for characterization of cisplatin/oligonucleotide crosslinks in the absence or presence of different metal ions (Mg2+ and Zn2+) in solution and evaluation of the distribution of isobaric crosslinks arising from changes in solution conditions. IRMPD proved successful for differentiation of the two types of DNA/cisplatin adducts for each ODN. Both ODNs exhibited a preference for formation of the G4G5 crosslink over the G3G4 crosslink, and this preference was diminished at higher incubation temperatures or in the presence of auxiliary metals, presumably due to partial disruption of the secondary structure of the DNA in solution.
Supplementary Material
Acknowledgment
Funding from NIH (RO1 GM65956) and the Welch Foundation (F1155) is gratefully acknowledged.
References
- 1.Crews S, Ojala D, Posakony J, Nishiguchi J, Attardi G. Nucleotide-sequence of a region of human mitochondrial-DNA containing the precisely identified origin of replication. Nature. 1979;277(5693):192–198. doi: 10.1038/277192a0. [DOI] [PubMed] [Google Scholar]
- 2.Roth DB, Menetski JP, Nakajima PB, Bosma MJ, Gellert M. V(D)J recombination: Broken DNA molecules with covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70(6):983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 3.Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human-disease form hairpin structures in-vitro. Cell. 1995;81(4):533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 4.Mariappan SVS, Catasti P, Chen X, Ratliff R, Moyzis RK, Bradbury EM, Gupta G. Solution structures of the individual single strands of the fragile X DNA triplets (GCC)(n)center dot(GGC)(n). Nucleic Acids Research. 1996;24(4):784–792. doi: 10.1093/nar/24.4.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varani G. Exceptionally stable nucleic-acid hairpins. Annual Review of Biophysics and Biomolecular Structure. 1995;24:379–404. doi: 10.1146/annurev.bb.24.060195.002115. [DOI] [PubMed] [Google Scholar]
- 6.Wadkins RM. Targeting DNA secondary structures. Current Medicinal Chemistry. 2000;7(1):1–15. doi: 10.2174/0929867003375461. [DOI] [PubMed] [Google Scholar]
- 7.Metzler R, Ambjornsson T, Hanke A, Zhang YL, Levene S. Single DNA conformations and biological function. Journal of Computational and Theoretical Nanoscience. 2007;4(1):1–49. [Google Scholar]
- 8.Nguyen B, Wilson WD. The Effects of Hairpin Loops on Ligand-DNA Interactions. Journal of Physical Chemistry B. 2009;113(43):14329–14335. doi: 10.1021/jp904830m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lah J, Drobnak I, Dolinar M, Vesnaver G. What drives the binding of minor groove-directed ligands to DNA hairpins. Nucleic Acids Research. 2008;36(3):897–904. doi: 10.1093/nar/gkm1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez B, Baumruk V, Leulliot N, Gouyette C, Huynh-Dinh T, Ghomi M. Thermodynamic and structural features of ultrastable DNA and RNA hairpins. Journal of Molecular Structure. 2003;65:167–74. [Google Scholar]
- 11.Jung JM, Van Orden A. Folding and unfolding kinetics of DNA hairpins in flowing solution by multiparameter fluorescence correlation spectroscopy. Journal of Physical Chemistry B. 2005;109(8):3648–3657. doi: 10.1021/jp0453515. [DOI] [PubMed] [Google Scholar]
- 12.Grunwell JR, Glass JL, Lacoste TD, Deniz AA, Chemla DS, Schultz PG. Monitoring the Conformational Fluctuations of DNA Hairpins Using Single-Pair Fluorescence Resonance Energy Transfer. Journal of the American Chemical Society. 2001;123(18):4295–4303. doi: 10.1021/ja0027620. [DOI] [PubMed] [Google Scholar]
- 13.Orden AV, Jung J. Fluorescence correlation spectroscopy for probing the kinetics and mechanisms of DNA hairpin formation. Biopolymers. 2008;89(1):1–16. doi: 10.1002/bip.20826. [DOI] [PubMed] [Google Scholar]
- 14.Kim J, Doose S, Neuweiler H, Sauer M. The initial step of DNA hairpin folding: a kinetic analysis using fluorescence correlation spectroscopy. Nucleic Acids Research. 2006;34(9):2516–2527. doi: 10.1093/nar/gkl221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaumot J, Eritja R, Navea S, Gargallo R. Classification of nucleic acids structures by means of the chemometric analysis of circular dichroism spectra. Analytica Chimica Acta. 2009;642(1–2):117–126. doi: 10.1016/j.aca.2008.12.052. [DOI] [PubMed] [Google Scholar]
- 16.Johnson WC. CD of nucleic acids. In: Berova N, Nakanishi K, Woody RW, editors. Circular Dichroism. Wiley-VCH; 2000. [Google Scholar]
- 17.Avizonis DZ, Kearns DR. Structural characterization of d(CAACCCGTTG) and d(CAACGGGTTG) mini-hairpin loops by heteronuclear NMR -The effects of purines versus pyrimidines in DNA hairpins. Nucleic Acids Research. 1995;23(7):1260–1268. doi: 10.1093/nar/23.7.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lam SL, Chi LM. Use of chemical shifts for structural studies of nucleic acids. Progress in Nuclear Magnetic Resonance Spectroscopy. 2010;56(3):289–310. doi: 10.1016/j.pnmrs.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh M, Kumar NV, Varshney U, Chary KVR. Structural characterisation of a uracil containing hairpin DNA by NMR and molecular dynamics. Nucleic Acids Research. 1999;27(19):3938–3944. doi: 10.1093/nar/27.19.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerman SB, Pheiffer BH. Direct demonstration that the ethanol-induced transition of DNA is between the A-form and B-form - X-ray-diffraction study. Journal of Molecular Biology. 1979;135(4):1023–1027. doi: 10.1016/0022-2836(79)90526-6. [DOI] [PubMed] [Google Scholar]
- 21.Shubsda M, Goodisman J, Dabrowiak JC. Characterization of hairpin-duplex interconversion of DNA using polyacrylamide gel electrophoresis. Biophysical Chemistry. 1999;76(2):95–115. doi: 10.1016/s0301-4622(98)00217-8. [DOI] [PubMed] [Google Scholar]
- 22.Fichtinger-Schepman AMJ, Van der Veer JL, Den Hartog JHJ, Lohman PHM, Reedijk J. Adducts of the antitumor drug cis-diamminedichloroplatinum(II) with DNA: formation, identification, and quantitation. Biochemistry. 1985;24(3):707–713. doi: 10.1021/bi00324a025. [DOI] [PubMed] [Google Scholar]
- 23.Fichtinger-Schepman AMJ, van Oosterom AT, Lohman PHM, Berends F. cis-Diamminedichloroplatinum(II)-induced DNA Adducts in Peripheral Leukocytes from Seven Cancer Patients: Quantitative Immunochemical Detection of the Adduct Induction and Removal after a Single Dose of cis-Diamminedichloroplatinum(II). Cancer Research. 1987;47(11):3000–3004. [PubMed] [Google Scholar]
- 24.Mangrum JB, Farrell NP. Excursions in polynuclear platinum DNA binding. Chemical Communications. 2010;46(36):6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meroueh M, Kjellstrom J, Martensson KSM, Elmroth SKC, Chow CS. Reactions of platinum(II) complexes with a DNA hairpin, d(CGCGTTGTTCGCG): structural characterization and kinetic studies. Inorganica Chimica Acta. 2000;297(1-2):145–155. [Google Scholar]
- 26.Iwamoto M, Mukundan S, Marzilli LG. DNA adduct formation by platinum anticancer drugs - insight into an unuaual GPG intrastrand cross-link in a hairpin-like DNA oligonucleotide using NMR and distance geometry methods. Journal of the American Chemical Society. 1994;116(14):6238–6244. [Google Scholar]
- 27.Yohannes PG, Zon G, Doetsch PW, Marzilli LG. DNA hairpin formation in adducts with platinum anticancer drugs - gel-eletrophoresis provides new information and a caveat. Journal of the American Chemical Society. 1993;115(12):5105–5110. [Google Scholar]
- 28.Villanueva JM, Jia X, Yohannes PG, Doetsch PW, Marzilli LG. Cisplatin (cis-Pt(NH3)(2)Cl-2) and cis- Pt(NH3)(2)(H2O)(2) (2+) intrastrand cross-linking reactions at the telomere GGGT DNA sequence embedded in a duplex, a hairpin, and a bulged duplex: Use of Mg2+ and Zn2+ to convert a hairpin to a bulged duplex. Inorganic Chemistry. 1999;38(26):6069–6080. doi: 10.1021/ic990603f. [DOI] [PubMed] [Google Scholar]
- 29.Jia X, Zon G, Marzilli LG. Multinuclear NMR investigation of zinc(2+) binding to a dodecamer oligodeoxyribonucleotide: insights from carbon-13 NMR spectroscopy. Inorganic Chemistry. 1991;30(2):228–239. [Google Scholar]
- 30.McLuckey S, Van Berkel G, Glish G. Tandem Mass Spectrometry of Small, Multiply Charged Oligonucleotides. Journal of The American Society for Mass Spectrometry. 1992;3(1):60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 31.McLuckey SA, Habibi-Goudarzi S. Decompositions of multiply charged oligonucleotide anions. Journal of the American Chemical Society. 1993;115(25):12085–12095. [Google Scholar]
- 32.Wu J, McLuckey SA. Gas-phase fragmentation of oligonucleotide ions. International Journal of Mass Spectrometry. 2004;237(2-3):197–241. [Google Scholar]
- 33.Cerny RL, Tomer KB, Gross ML, Grotjahn L. Fast atom bombardment combined with tandem mass spectrometry for determining structures of small oligonucleotides. Analytical Biochemistry. 1987;165(1):175–182. doi: 10.1016/0003-2697(87)90217-x. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Wan KX, Ramanathan R, Taylor JS, Gross ML. Structure and fragmentation mechanisms of isomeric T-rich oligodeoxynucleotides: a comparison of four tandem mass spectrometric methods. Journal of the American Society for Mass Spectrometry. 1998;9(7):683–691. doi: 10.1016/S1044-0305(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhang QR, Yu ET, Kellersberger KA, Crosland E, Fabris D. Toward building a database of bifunctional probes for the MS3D investigation of nucleic acids structures. Journal of The American Society for Mass Spectrometry. 2006;17(11):1570–1581. doi: 10.1016/j.jasms.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Le Pla RC, Ritchie KJ, Henderson CJ, Wolf CR, Harrington CF, Farmer PB. Development of a Liquid Chromatography–Electrospray Ionization Tandem Mass Spectrometry Method for Detecting Oxaliplatin–DNA Intrastrand Cross-Links in Biological Samples. Chemical Research in Toxicology. 2007;20(8):1177–1182. doi: 10.1021/tx700088j. [DOI] [PubMed] [Google Scholar]
- 37.Beck JL, Colgrave ML, Ralph SF, Sheil MM. Electrospray ionization mass spectrometry of oligonucleotide complexes with drugs, metals, and proteins. Mass Spectrometry Reviews. 2001;20(2):61–87. doi: 10.1002/mas.1003. [DOI] [PubMed] [Google Scholar]
- 38.Anichina J, Zhao Y, Hrudey SE, Schreiber A, Li X-F. Electrospray Ionization Tandem Mass Spectrometry Analysis of the Reactivity of Structurally Related Bromo-methyl-benzoquinones toward Oligonucleotides. Analytical Chemistry. 2011;83(21):8145–8151. doi: 10.1021/ac201646z. [DOI] [PubMed] [Google Scholar]
- 39.Barry JP, Vouros P, Vanschepdael A, Law SJ. Mass AND Sequence Verification of modified oligonucleotides using electrospray tandem mass spectrometry. Journal of Mass Spectrometry. 1995;30(7):993–1006. [Google Scholar]
- 40.Iannitti-Tito P, Weimann A, Wickham G, Sheil MM. Structural analysis of drug-DNA adducts by tandem mass spectrometry. Analyst. 2000;125(4):627–633. doi: 10.1039/a908920i. [DOI] [PubMed] [Google Scholar]
- 41.Egger AE, Hartinger CG, Ben Hamidane H, Tsybin YO, Keppler BK, Dyson PJ. High Resolution Mass Spectrometry for Studying the Interactions of Cisplatin with Oligonucleotides. Inorganic Chemistry. 2008;47(22):10626–10633. doi: 10.1021/ic801371r. [DOI] [PubMed] [Google Scholar]
- 42.Nyakas A, Eymann M, Schurch S. The Influence of Cisplatin on the Gas-Phase Dissociation of Oligonucleotides Studied by Electrospray Ionization Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2009;20(5):792–804. doi: 10.1016/j.jasms.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 43.Stucki SR, Nyakas A, Schurch S. Tandem mass spectrometry of platinated quadruplex DNA. Journal of Mass Spectrometry. 2011;46(12):1288–1296. doi: 10.1002/jms.2019. [DOI] [PubMed] [Google Scholar]
- 44.Nyakas A, Stucki SR, Schurch S. Tandem Mass Spectrometry of Modified and Platinated Oligoribonucleotides. Journal of The American Society for Mass Spectrometry. 2011;22(5):875–887. doi: 10.1007/s13361-011-0106-z. [DOI] [PubMed] [Google Scholar]
- 45.Smith SI, Brodbelt JS. Hybrid Activation Methods for Elucidating Nucleic Acid Modifications. Analytical Chemistry. 2011;83(1):303–310. doi: 10.1021/ac102411a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Z, Shaw JB, Brodbelt JS. Comparison of MS/MS Methods for Characterization of DNA/Cisplatin Adducts. Journal of The American Society for Mass Spectrometry. 2012;24(2):265–272. doi: 10.1007/s13361-012-0532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mo JJ, Hakansson K. Characterization of nucleic acid higher order structure by high-resolution tandem mass spectrometry. Analytical and Bioanalytical Chemistry. 2006;386(3):675–681. doi: 10.1007/s00216-006-0614-z. [DOI] [PubMed] [Google Scholar]
- 48.Mo JJE, Todd GC, Hakansson K. Characterization of Nucleic Acid Higher Order Structure by Gas-Phase H/D Exchange in a Quadrupole-FT-ICR Mass Spectrometer. Biopolymers. 2009;91(4):256–264. doi: 10.1002/bip.21134. [DOI] [PubMed] [Google Scholar]
- 49.Fabris D, Kellersberger KA, Wilhide JA. Higher-order structure of nucleic acids in the gas phase: Top-down analysis of base-pairing interactions. International Journal of Mass Spectrometry. 2012;312(0):155–162. doi: 10.1016/j.ijms.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan KX, Gross J, Hillenkamp F, Gross ML. Fragmentation mechanisms of oligodeoxynucleotides studied by H/D exchange and electrospray ionization tandem mass spectrometry. Journal of the American Society for Mass Spectrometry. 2001;12(2):193–205. doi: 10.1016/S1044-0305(00)00208-7. [DOI] [PubMed] [Google Scholar]
- 51.Premstaller A, Huber CG. Factors determining the performance of triple quadrupole, quadrupole ion trap and sector field mass spectrometer in electrospray ionization mass spectrometry. 2. Suitability for de novo sequencing. Rapid Communications in Mass Spectrometry. 2001;15(13):1053–1060. doi: 10.1002/rcm.338. [DOI] [PubMed] [Google Scholar]
- 52.Shammel Baker E, Dupuis N, Bowers MT. DNA Hairpin, Pseudoknow, and Cruciform Stability in a Solvent-Free Environment. J. Phys. Chem. B. 2009;113:1722–1727. doi: 10.1021/jp807529m. [DOI] [PubMed] [Google Scholar]
- 53.Anastassopoulou J. Metal–DNA interactions. Journal of Molecular Structure. 2003;651–653:19–26. [Google Scholar]
- 54.Langlais M, Tajmirriahi HA, Savoie R. Raman spectroscopic study of the effects of Ca2+, Mg2+, Zn2+, and Cd2+ ions on calf thymus DNA: binding sites and conformational changes. Biopolymers. 1990;30(7-8):743–752. doi: 10.1002/bip.360300709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.