Abstract
The stringent response is a conserved global regulatory mechanism that is related to the synthesis of (p)ppGpp nucleotides. Gram-positive bacteria, such as Staphylococcus aureus, possess three (p)ppGpp synthases: the bifunctional RSH (RelA/SpoT homolog) protein, which consists of a (p)ppGpp synthase and a (p)ppGpp hydrolase domain, and two truncated (p)ppGpp synthases, designated RelP and RelQ. Here, we characterized these two small (p)ppGpp synthases. Biochemical analyses of purified proteins and in vivo studies revealed a stronger synthetic activity for RelP than for RelQ. However, both enzymes prefer GDP over GTP as the pyrophosphate recipient to synthesize ppGpp. Each of the enzymes was shown to be responsible for the essentiality of the (p)ppGpp hydrolase domain of the RSH protein. The staphylococcal RSH-hydrolase is an efficient enzyme that prevents the toxic accumulation of (p)ppGpp. Expression of (p)ppGpp synthases in a hydrolase-negative background leads not only to growth arrest but also to cell death. Transcriptional analyses showed that relP and relQ are strongly induced upon vancomycin and ampicillin treatments. Accordingly, mutants lacking relP and relQ showed a significantly reduced survival rate upon treatments with cell wall-active antibiotics. Thus, RelP and RelQ are active (p)ppGpp synthases in S. aureus that are induced under cell envelope stress to mediate tolerance against these conditions.
INTRODUCTION
Bacteria can quickly adapt to various environmental conditions, by the use of a wide range of sensory and regulatory systems. The stringent response is a highly conserved regulatory mechanism that is provoked by nutrient limitations. Subsequently, the bacteria synthesize (p)ppGpp, which is known as a nutritional alarmone, by using ATP as a pyrophosphate donor to phosphorylate GTP or GDP. In Escherichia coli and in related Gram-negative bacteria, the production and degradation of (p)ppGpp are governed by two homologous enzymes: the (p)ppGpp synthase RelA and the bifunctional enzyme SpoT, which possesses (p)ppGpp synthase and hydrolase activities. Upon amino acid deprivation, RelA synthase activity provokes the accumulation of (p)ppGpp in the bacterial cell and causes the downregulation of genes involved in macromolecular biosynthesis (i.e., translational machinery, nucleotide biosynthesis, and transport) and upregulation of genes coding for enzymes of amino acid biosynthesis and transport (1). The bifunctional SpoT protein is described as an enzyme that responds to other nutritional stresses, such as limitations in fatty acids (2, 3), iron (4), and carbon (5). In the past, it was postulated that in contrast to Gram-negative enterics, most bacteria possess only one enzyme for the synthesis of (p)ppGpp, designated RSH, for RelA/SpoT homolog.
In many Gram-positive bacteria, in particular firmicutes, RSH is responsible for (p)ppGpp accumulation upon amino acid deprivation as well as for (p)ppGpp degradation under nutrient-rich conditions (6–9). Recently it was shown, that Bacillus subtilis and Streptococcus mutans possess two additional enzymes with (p)ppGpp synthase activity, designated small alarmone synthases SAS1/SAS2 (10) and RelP/RelQ (11), respectively. These enzymes lack the hydrolase domain and the C-terminal regulatory domain of RSH proteins. For B. subtilis and S. mutans, these enzymes were shown to be active (p)ppGpp synthases in vitro; however, their activities and functions in vivo are still unclear, and strains lacking these enzymes have hardly any phenotypes (12–14). Because RelP and RelQ enzymes lack sensory domains, one can assume that they are mostly regulated at the transcriptional level and that their function is restricted to certain, so far ill-defined, stress conditions. For example, transcription of ywaC, which codes for one of the two Bacillus synthases, was induced upon alkaline shock conditions (10).
RelP/Q homologs have also been identified in the genome of the major human pathogen Staphylococcus aureus (10, 11). The S. aureus RelP (RelPSau) protein is 60% identical to the S. mutans RelP (RelPSmu) protein, and the RelQSau protein is 78% identical to the RelQSmu protein. These proteins show several conserved homologies of important domains for synthetic activity of the well-characterized RSH protein of Streptococcus equisimilis (15). In particular, homologies in a region surrounding the tyrosine residue 308, which is responsible for GTP/GDP binding to the enzyme, and the glutamate residue 323, which coordinates the catalytic Mg2+ and serves as the GTP/GDP 3′-OH proton acceptor, implicates them as potentially active (p)ppGpp synthases.
Here we aimed to decipher the function of these currently uncharacterized enzymes. We show that each enzyme contributes to the essentiality of the hydrolase activity of RSH, which is very efficient in S. aureus and prevents the toxic accumulation of (p)ppGpp. We observed transcriptional induction of relP and relQ after diverse cell wall stress stimuli, such as vancomycin and ampicillin. Mutants lacking relP and relQ possess a significantly reduced survival rate upon treatments with cell wall-active antibiotics.
MATERIALS AND METHODS
Strains and plasmids.
S. aureus strain HG001 (see Table S1 in the supplemental material), an 8325 derivate with a repaired rsbU gene, was used as the strain background to generate the mutants indicated under “Mutagenesis strategies.” For complementation and overexpression studies, full-length relP, relQ, and rsh, harboring a mutation in the hydrolase domain referred to as rshHyd, were cloned into an anhydrotetracyclin (ATc)-inducible vector pCG 248 (see Table S1). For relP and relQ cloning, the oligonucleotides are described in Table S2 in the supplemental material. The hydrolase mutation in rshHyd was constructed to substitute for histidine and aspartate of the conserved HD domain with alanine and serine. This mutation was introduced by overlapping PCR employing oligonucleotides described in Table S2.
Growth conditions and media.
Strains were grown in CYPG (10 g/liter Casamino Acids, 10 g/liter yeast extract, 5 g/liter NaCl, 0.5% glucose, 0.06 M phosphoglycerate) (16), in Mueller-Hinton (MH) broth for antibiotic resistance experiments, or in a chemically defined medium (CDM) (17). For strains carrying erythromycin, tetracycline, and chloramphenicol resistance genes, antibiotics were used only in precultures at a concentration of 5 μg/ml for erythromycin and tetracycline and 10 μg/ml for chloramphenicol. Bacteria from an overnight culture were diluted to an initial optical density at 600 nm (OD600) of 0.05 in fresh medium and grown with shaking (220 rpm) at 37°C to the desired growth phase. For the induction of a stringent response, strains were treated with mupirocin, an isoleucyl tRNA synthase inhibitor (0.5 μg/ml), for 1 h.
For glucose and iron starvation assays, bacteria were grown in CYPG to the midexponential growth phase (OD600 of 0.5). Afterwards, the bacteria were filtered over a 0.22-μm-pore filter, applying vacuum, washed twice with sterile phosphate-buffered saline (PBS), and resuspended in an equal volume of CYPG medium with or without 55.6 mM methyl-α-d-glucopyranoside (α-MGP) (Sigma-Aldrich) for glucose starvation and 600 μM 2,2′-dipyridyl (Sigma-Aldrich) for iron starvation. Bacteria were further incubated for 1, 2, and 3 h with shaking (222 rpm) at 37°C and were harvested at the indicated time points for RNA isolation.
Mutagenesis strategies.
The markerless relP single mutant, relQ single mutant, relP relQ double mutant, and the relP relQ rsh triple mutant [referred to as (p)ppGpp0] were obtained using the ATc-inducible suicide mutagenesis vector pKOR1 (see Table S1 in the supplemental material). First, single mutants of relP, with deletion of nucleotides (nt) 450 to 536 (Δ450–536), and relQ (Δ343–429) were generated by deletions of regions containing known conserved domains responsible for the synthetic activities. Afterwards, the relQ mutation was introduced into the relP single mutant, resulting in the markerless relP relQ double mutant. Finally the rsh mutation, with a deletion of the whole enzymatic N terminus, was introduced into the double mutant. The construction of an rsh mutation was only possible in a relP- or relQ-negative background (see Results). The deletions in relP (SAOUHSC_02811), relQ (SAOUHSC_00942), and rsh (SAOUHSC_1742) were introduced by overlapping PCRs employing the oligonucleotides described in Table S2 in the supplemental material. The amplicons were cloned into pKOR1 using the Gateway cloning system (Invitrogen). The resulting plasmids pCG229 (Δ450–536 in relP), pCG230 (Δ343–429 in relQ), and pCG263 (Δ249–951 in rsh) were verified by sequencing and transformed into restriction-deficient strain RN4220 (18), from which they were transduced into the indicated S. aureus strains. Mutagenesis was performed as described previously (19). Mutation of relP, relQ, and rsh in strain HG001 was verified by sequence analysis of PCR amplicons.
The conditional rsh mutant was generated using the pMUTIN4 vector (20), as described previously (8). The mutation was transduced into S. aureus strain HG001-229 (relP single mutant) or HG001-230 (relQ single mutant) using Φ11 lysates of strain RN4220-55 (8). Transductants were verified by PCR and pulsed-field gel electrophoresis (PFGE).
The HG001 vraR mutant was generated by the transduction of vraR::Tn bursa aurealis using Φ11 lysates of strain USA300 JE2 NE554 acquired from the Nebraska transposon mutant library.
RNA isolation and Northern blot analysis.
RNA isolation and Northern blot analysis were performed as described previously (21). Briefly, bacteria were lysed in 1 ml of TRIzol reagent (Invitrogen Life Technologies, Karlsruhe, Germany) with 0.5 ml zirconia-silica beads (0.1-mm diameter) in a high-speed homogenizer (Savant Instruments, Farmingdale, NY). RNA was isolated as described in the instructions provided by the manufacturer of TRIzol (Life Technologies). For the detection of specific transcripts, digoxigenin (DIG)-labeled probes were generated using a DIG-labeling PCR kit following the manufacturer's instructions (Roche Biochemicals). The sequences of the oligonucleotides used for probe generation were described previously (8) or are indicated in Table S2 in the supplemental material.
Detection of (p)ppGpp accumulation in S. aureus cells.
The cells were grown in a modified low-phosphate CDM (0.4 mM phosphate, buffered at pH 7.2 with 40 mM MOPS [morpholinepropanesulfonic acid]) to an OD600 of 0.5, at which point [32P]H3PO4 (50 μCi/ml) was added to the cultures. After 3 h of growth, the cells were shifted into CDM containing 0.5 μg/ml mupirocin for an additional 30 min. For the extraction of nucleotides, 200 μl ice-cold 2 M formic acid was added per ml of culture followed by incubation for 30 min on ice. The bacteria were lysed with 0.5 ml zirconia-silica beads (0.1-mm diameter) in a high-speed homogenizer, and the extracts were centrifuged at 12,000 × g for 5 min at 4°C. An equal volume of phenol-chloroform-isoamyl alcohol (50:49:1 [vol/vol/vol]) saturated with deionized water was added to the resulting supernatant. The mixture was agitated and centrifuged at 12,000 × g for 5 min at 4°C. Ten microliters of the resulting supernatant was spotted onto a polyethyleneimine-cellulose thin-layer sheet (Macherey-Nagel). For the one-dimensional (1D) thin-layer chromatography (TLC) analysis, 1.2 M KH2PO4 (pH 3.5) was used as the chromatographic solvent.
Purification of RelP and RelQ.
For synthesis of N-terminal His tag fusion proteins of RelP and RelQ, XhoI- and BamHI-digested PCR fragments (for the oligonucleotides used, see Table S2 in the supplemental material) were amplified from S. aureus strain HG001 and cloned into the XhoI-BamHI sites of the protein expression vector pET15b (Novagen), yielding pCG121-RelP and pCG122-RelQ, respectively. The verified plasmids were transformed in E. coli strain BL21 (Promega) for further protein expression experiments. His tagged RelP and RelQ fusion proteins were purified on a Ni-nitrilotriacetic acid (NTA) affinity column (GE Healthcare Bio-Sciences Corp.) using imidazole for elution. For removal of the imidazole and for concentration, the proteins were ultrafiltrated with ultracentrifugal filter devices (Millipore, Amicon). Thereafter, the proteins were diluted in storage buffer containing reaction buffer-glycerol (1:1 [vol/vol]) and stored at −20°C.
In vitro assay of (p)ppGpp synthase activity.
The assay for (p)ppGpp synthase activity was performed as described previously (10). Briefly, a reaction mixture with a final volume of 25 μl contained 10 μCi/ml [γ-32P]ATP ((3,000 Ci mmol−1; PerkinElmer), 2 mM ATP, 2 mM GTP or GDP, 0.5 μg purified protein, and reaction buffer (50 mM Tris-acetate [pH 7.8], 3.3 mM magnesium acetate, 60 mM potassium acetate, 30 mM ammonium acetate, 1 mM dithiothreitol). The reaction was performed for 1 h at 30°C and was stopped by the addition of 1 μl of 88% formic acid. After addition of 12.5 μl of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol/vol]) to the mixture, it was mixed briefly and then centrifuged at 10,000 × g for 5 min at 4°C. The aqueous phase was transferred to another tube for storage at −20°C. For TLC, 5 μl was spotted onto a polyethyleneimine-cellulose sheet (Macherey-Nagel), and 1.5 M KH2PO4 (pH 3.5) was used as the chromatographic solvent.
RESULTS
RelP and RelQ are responsible for the essentiality of the RSH hydrolase domain of S. aureus.
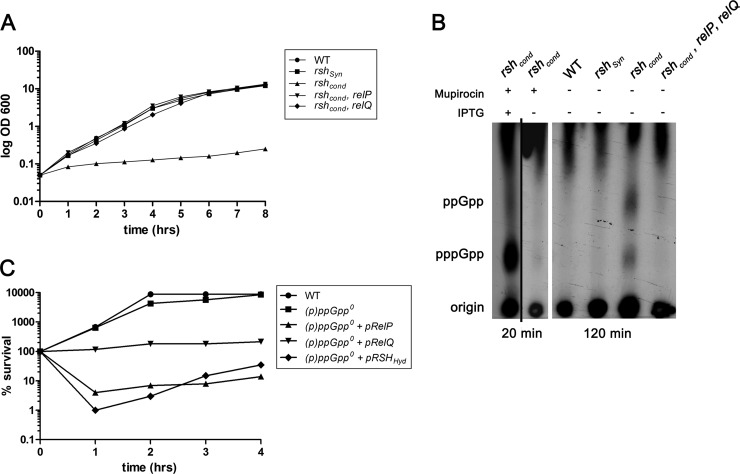
Previous studies showed that the RSH protein is essential for growth in S. aureus (8, 22). The essentiality of RSH could be constrained to an essential hydrolase domain of the protein, since a mutant defective only in the synthase domain was viable (8). Thus, we hypothesized that rsh mutants accumulate (p)ppGpp due to the activity of the two additional synthases RelP and RelQ. To verify this hypothesis, we constructed relP and relQ mutants, which were not impaired in growth. A conditional rsh mutation (rshcond) was introduced into the relP and relQ single mutants by using pMUTIN4, which is a plasmid that integrates into the chromosomal rsh gene, thus creating a Pspac-rsh fusion in which rsh transcription can be controlled by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside). The growth curves showed that the conditional rshcond mutant was not able to grow without the inducer IPTG, whereas the relP rshcond and relQ rshcond double mutants were able to grow comparably to the wild-type and rshSyn mutant strains (Fig. 1A). Thus, deletion of either relP or relQ restores the viability of an rsh mutant. We conclude that in a relP- or relQ-negative background, the hydrolytic activity of RSH is no longer essential, since the activity of one (p)ppGpp synthase is not sufficient to accumulate toxic amounts of (p)ppGpp molecules in the bacterial cell. This could be confirmed by thin-layer chromatography showing residual (p)ppGpp accumulation in the conditional rshcond mutant during growth without IPTG (Fig. 1B). The detected accumulation could be eliminated by the additional deletion of relP and relQ. Accordingly, in contrast to wild-type bacteria, an rsh knockout strain could be readily obtained in a relP- and relQ-negative background (see “Mutagenesis strategies” in Materials and Methods). This triple mutant strain is devoid of any pppGpp synthase and hydrolase activity and is designated (p)ppGpp0.
FIG 1.
The essentiality of the RSH-hydrolase to prevent toxic accumulation of (p)ppGpp. (A) Growth of wild-type strain HG001, the rshsyn mutant, the conditional rshcond mutant, the rshcond relP double mutant, and the rshcond relQ double mutant in CYPG medium without the inducer IPTG. Growth curves represent the means of four independent experiments. (B) Detection of (p)ppGpp accumulation in the conditional rshcond mutant due to RelP and/or RelQ by TLC. Strains were grown with IPTG to the early exponential phase and shifted to different conditions: Extended incubation for 2 h without mupirocin and without IPTG resulted in accumulation of ppGpp and pppGpp in the conditional rshcond strain but not in the triple mutant (rshcond relP relQ) strain or strains with active hydrolase (wild type or rshSyn mutant). As a control, the stringent response was induced for 20 min with mupirocin (0.5 μg/ml) in the conditional rshcond mutant grown with and without IPTG. (C) Overexpression of (p)ppGpp synthases from an ATc-inducible promoter in a (p)ppGpp0 strain showed decreased abilities to form colonies on TSA plates. Wild-type, (p)ppGpp0, (p)ppGpp0 plus pRelP, (p)ppGpp0 plus pRelQ, and (p)ppGpp0 plus pRSHHyd strains were grown to the exponential growth phase (OD600 of 0.5, time 0), followed by a further incubation with 0.2 μg/ml ATc. At indicated time points, percent survival was calculated by counting the number of cells able to form colonies, normalized to CFU before ATc treatment (time 0). CFU counts represent the means of three independent experiments.
To deepen the understanding of toxic accumulation of (p)ppGpp in the staphylococcal cell, we overexpressed each of the three (p)ppGpp synthases RelP, RelQ, and RSHHyd (with a nonfunctional hydrolase domain) in the (p)ppGpp0 strain (see Fig. S1 in the supplemental material). Therefore, we used a multicopy plasmid with an anhydrotetracycline (ATc)-inducible promoter. A strong induction of rshHyd and relP resulted in a fast and severe drop in CFU within 1 h (Fig. 1C), whereas relQ only showed growth inhibition. These experiments clearly indicate that the hydrolase domain of the RSH protein is essential to prevent an otherwise toxic accumulation of (p)ppGpp molecules in the staphylococcal cell, caused by the synthetic activities of the three (p)ppGpp synthases.
The RSH-hydrolase of S. aureus is highly efficient in preventing the toxic accumulation of (p)ppGpp in the cell.
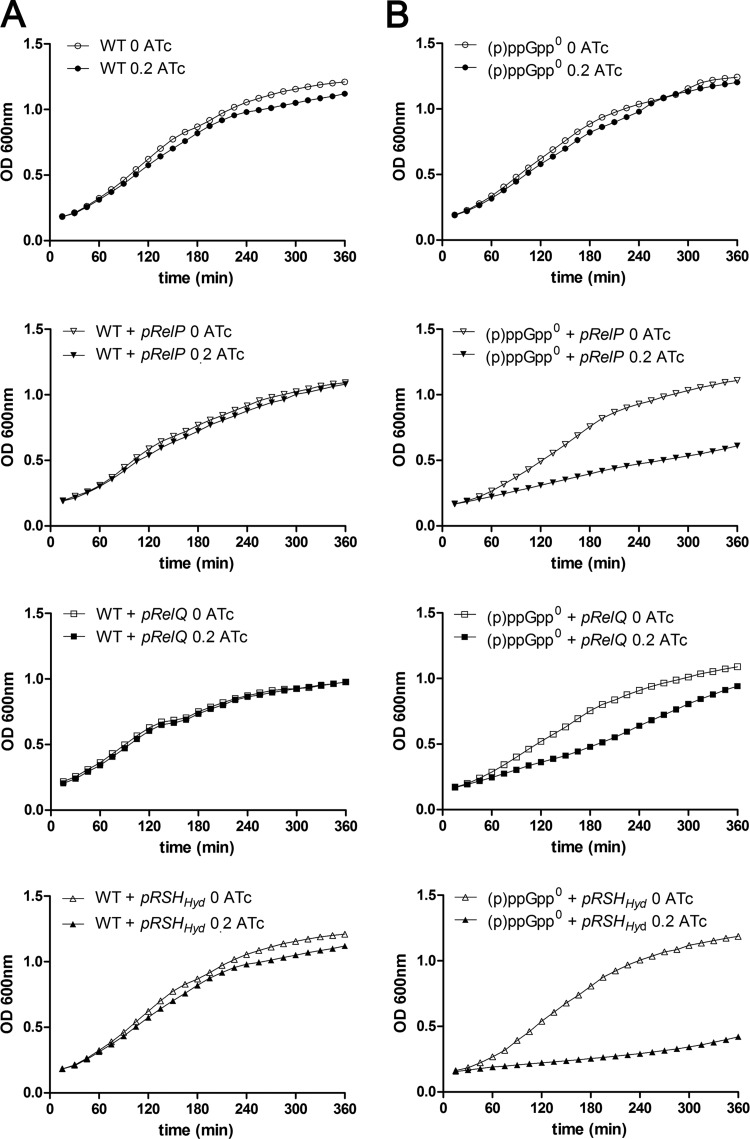
To investigate the efficiency of staphylococcal (p)ppGpp RSH-hydrolase activity, we overexpressed the three synthases in the hydrolase-negative (p)ppGpp0 strain and in the hydrolase-positive wild-type strain. The growth analyses indicated that, in the hydrolase-negative background, overexpression of either relP, relQ, or rshHyd resulted in growth inhibition (Fig. 2), and again, relQ showed the weakest effect on growth. However, when the synthases were expressed in the wild-type strain, no inhibition was observed. Therefore, a single copy of rsh in the bacterial chromosome is sufficient to prevent toxic accumulation of (p)ppGpp, even when RelP, RelQ, or RSH is overexpressed on an inducible multicopy plasmid, which indicates high efficiency for the S. aureus RSH-hydrolase.
FIG 2.
The staphylococcal RSH-hydrolase is very efficient to degrade (p)ppGpp molecules. Shown are growth of the WT, WT plus pRelP, WT plus pRelQ, and WT plus pRSHHyd strains (A) and growth of the (p)ppGpp0, (p)ppGpp0 plus pRelP, (p)ppGpp0 plus pRelQ, and (p)ppGpp0 plus pRSHHyd strains (B) to the exponential growth phase, followed by further incubation with (filled symbols) or without (open symbols) 0.2 μg/ml ATc. Growth curves represent the means of four independent experiments.
Direct evidence for (p)ppGpp synthase activity of RelP and RelQ.
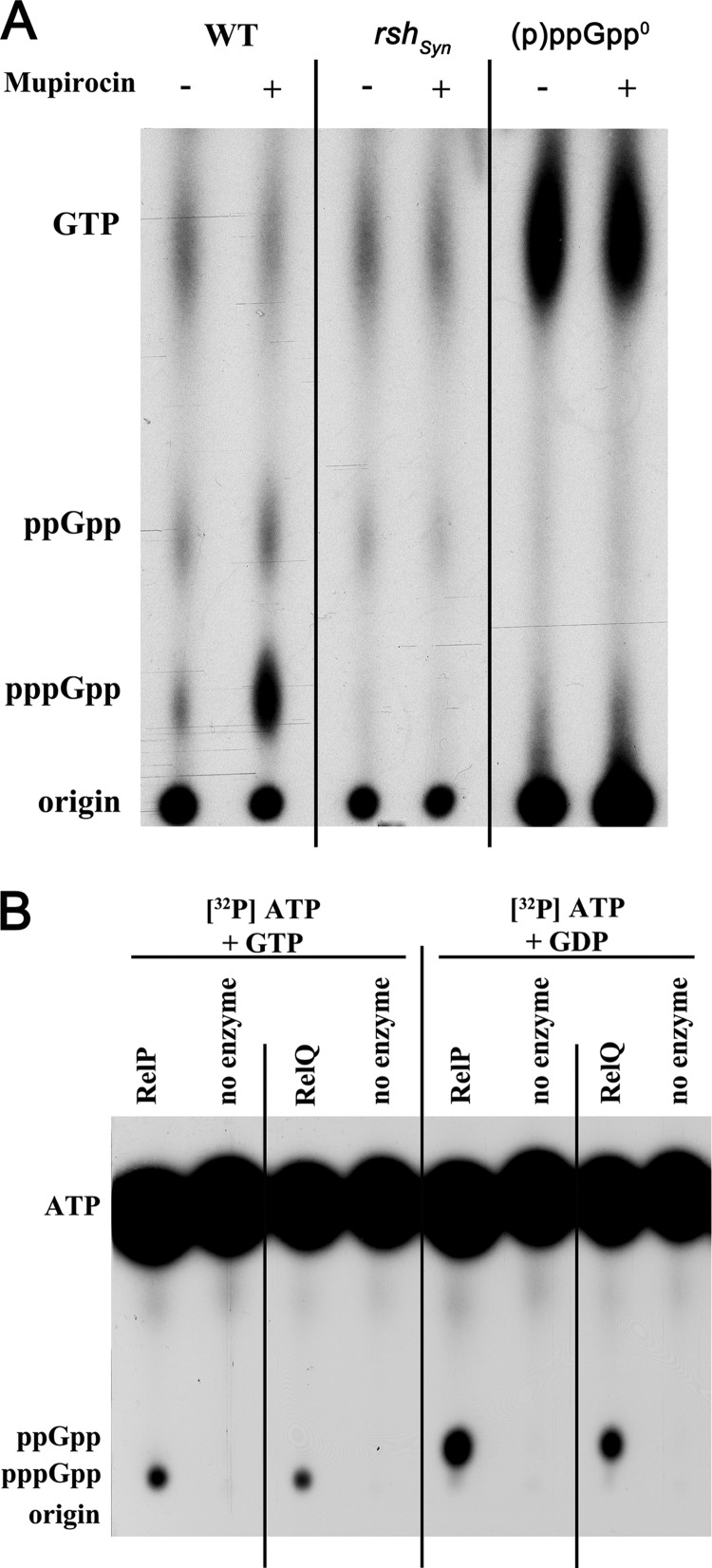
In a previous study we could demonstrate by using mupirocin to mimic isoleucine starvation that wild-type S. aureus was proficient in synthesizing and accumulating pppGpp and ppGpp (8). A mutant defective in the (p)ppGpp synthase domain of RSH (rshSyn) showed no synthesis or accumulation of these molecules under this condition. The results indicated that only the RSH protein of S. aureus responds to amino acid limitation, with no detectable contribution of RelP or RelQ. However, a slight residual signal for ppGpp was still detectable in the rshSyn mutant, which was independent of amino acid limitation (8) and was not detectable in the (p)ppGpp0 triple mutant (Fig. 3A). This finding confirms that the ppGpp that was detected in the rshSyn mutant was synthesized by RelP and/or RelQ. Notably, compared to the other analyzed strains, a strong accumulation of GTP could be detected in the ppGpp0 mutant.
FIG 3.
In vivo and in vitro evidence for (p)ppGpp synthetic activities of RelP and RelQ. (A) Detection of (p)ppGpp synthesis after amino acid deprivation in vivo. 32P-labeled nucleotides of formic acid extracts of S. aureus were detected by thin-layer chromatography. Strain HG001 (WT), the rshsyn mutant, and the rshsyn relP relQ [(p)ppGpp0] triple mutant were grown to the exponential phase, followed by the addition of mupirocin (0.5 μg/ml) for 20 min. (B) Characterization of the (p)ppGpp synthase activities of RelP and RelQ in vitro. Purified RelP or RelQ proteins (0.5 μg) were assayed for (p)ppGpp synthase activity in the presence of [γ-32P]ATP, 2 mM ATP, and either 2 mM GTP or 2 mM GDP. Reaction mixtures were analyzed by 1D thin-layer chromatography. The positions of the origin and signals corresponding to pppGpp, ppGpp, and ATP are indicated.
For further analysis, RelP and RelQ were purified, and their enzymatic activities were assayed using [γ-32P]ATP in the presence of GTP or GDP, respectively. Separation of the (p)ppGpp molecules by thin-layer chromatography (TLC) revealed that both enzymes synthesized (p)ppGpp and that RelP had a slightly higher activity (Fig. 3B). The in vitro synthase activity assay also indicated that RelP and RelQ proteins synthesize ppGpp from GDP more efficiently than pppGpp from GTP. Thus, S. aureus possesses two additional functional (p)ppGpp synthases, whereby RelP seems to be more active than RelQ in vitro and in vivo.
Overexpression of either relP, relQ, or rsh leads to a similar characteristic stringent response.
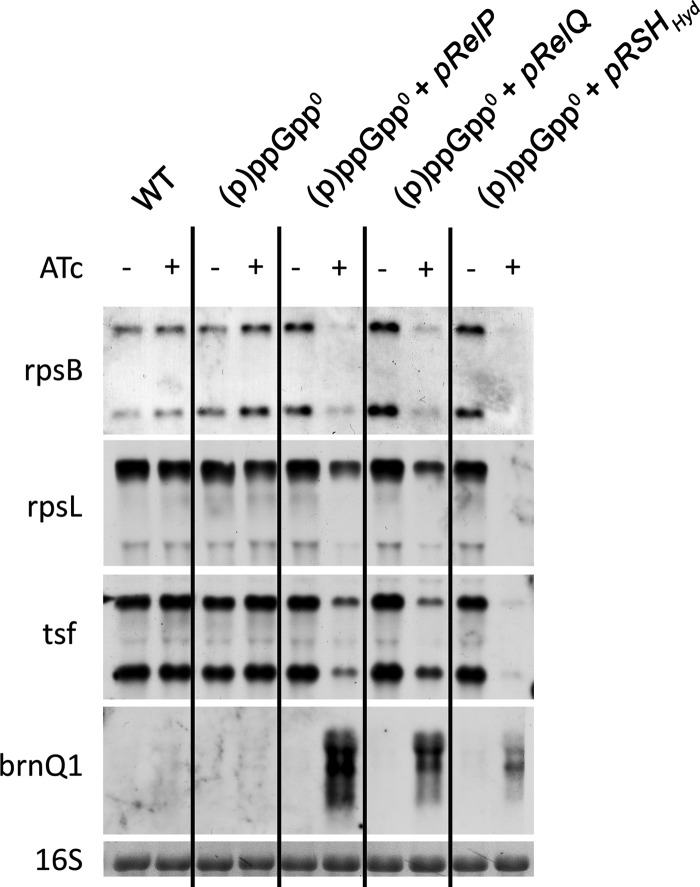
We overexpressed each of the synthases (RelP, RelQ, and RSH without hydrolase) on an ATc-inducible plasmid in the (p)ppGpp0 mutant (see Fig. S1 in the supplemental material) and analyzed the expression of selected hallmark genes after induction. The expression of each of the synthases led to transcriptional repression of genes involved in the synthesis of cellular macromolecules: i.e., genes coding for ribosomal proteins (rpsB and rpsL) and a translational elongation factor (tsf) (Fig. 4). These genes represent some of the hallmark genes that are typically influenced by the RSH-mediated stringent response and are repressed upon amino acid deprivation (8, 23). brnQ1, which codes for a branched-chain amino acid transporter, showed transcriptional induction once a (p)ppGpp synthase was present. This gene is tightly and negatively controlled by another regulator, the CodY repressor. CodY senses the branched-chain amino acid pool and the intracellular GTP concentration and is activated by the binding of these cofactors (24–26). Previously, we showed that the stringent response imposed by amino acid limitation deactivates the CodY repressor in S. aureus (8, 23). Here, this interaction can be observed just by the expression of any of the (p)ppGpp synthases without changes in the availability of branched-chain amino acids.
FIG 4.
Induction of a stringent response by overexpression of relP and relQ. Shown is (p)ppGpp synthase-dependent repression or induction of target genes. Wild-type (WT) strain HG001, the (p)ppGpp0 mutant, and the (p)ppGpp0 mutant complemented with pRelP, pRelQ, or pRSHHyd synthases were grown in CYPG to the exponential growth phase followed by further incubation with (+) or without (−) ATc (0.2 μg/ml) for 1 h. RNA was hybridized with digoxigenin-labeled PCR fragments. The 16S rRNA detected in the ethidium bromide-stained gels is indicated as loading control in the bottom lane.
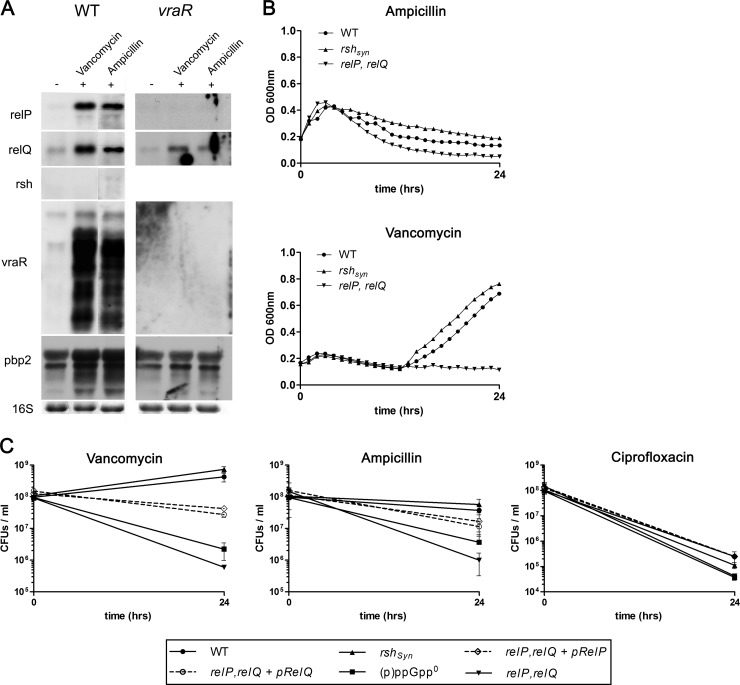
relP and relQ are strongly induced upon cell wall stress stimuli.
Since RelP and RelQ appeared to be short (p)ppGpp synthases with no regulatory C terminus compared to the RSH protein, we assumed they were regulated at the transcriptional level. Therefore, we examined several conditions as potential signals for relP or relQ activation. In E. coli, glucose and iron deprivation led to a SpoT-mediated (p)ppGpp accumulation (4, 5). We performed Northern blot analyses of logarithmic-phase cultures that had been treated with methyl-α-d-glucopyranoside to induce glucose starvation and with 2,2′-dipyridyl for iron starvation. However, these experiments revealed no inducible effects on relP or relQ transcription (data not shown). Next, we screened published microarray studies of S. aureus for putative signals. RelP was found to be induced upon cell wall stress, mediated by cell wall-active antibiotics and as a core member of the VraS/R system (27, 28). This regulatory system is strongly influenced by cell wall-active antibiotics and causes increased resistance to β-lactam antibiotics and vancomycin. We confirmed that relP and vraR transcription is strongly enhanced upon vancomycin and ampicillin treatments in the wild type, whereas these inductions could not been detected in a vraR-negative background. (Fig. 5A). Furthermore, relQ was also strongly induced by cell wall-active antibiotics. This induction was still seen in the vraR mutant, although to a lesser extent. This indicates that RelQ is also involved in the VraS/R-mediated cell wall stress stimulon. However, the rsh gene showed no induction upon both treatments, indicating a minor role of the RSH protein in the response to cell wall stress.
FIG 5.
RelP and RelQ synthases mediate tolerance to cell wall-active antibiotics. (A) Upregulation of genes upon vancomycin and ampicillin treatments. Wild-type strain HG001 and its isogenic vraR mutant were grown in CYPG medium to the exponential growth phase (OD600 of 0.5), followed by the addition of vancomycin (3.12 μg/ml, corresponding to 5× the MIC) and ampicillin (9.4 μg/ml, corresponding to 6× the MIC) for 30 min. RNA was hybridized with digoxigenin-labeled PCR fragments. The 16S rRNA detected in the ethidium bromide-stained gels as a loading control is indicated at the bottom. (B) Growth behavior of strains upon treatments with vancomycin and ampicillin. Strains were grown to the exponential phase in Mueller-Hinton (MH) broth and subsequently treated with 3.12 μg/ml vancomycin or 9.4 μg/ml ampicillin for 24 h. (C) Time-kill curves of strains in the presence of vancomycin, ampicillin, and ciprofloxacin. Strains grown to the exponential growth phase (OD600 of 0.5, time 0) were treated with 3.12 μg/ml (5× the MIC) vancomycin, 9.4 μg/ml (6× the MIC) ampicillin, and 2.5 μg/ml (10× the MIC) ciprofloxacin for 24 h. Viable counts were determined by plating known dilutions of the samples on tryptic soy agar (TSA) plates before antibiotic treatments (time 0) and after 24 h. Time-kill curves represent the means of four independent experiments. The levels of significance after 24 h were determined by the two-tailed Student's t test. After vancomycin treatment, the significance level was P < 0.005 for the comparisons of the WT versus the relP relQ mutant and the WT versus the (p)ppGpp0 mutant. After ampicillin treatment, the significance level was P < 0.05 for the comparisons of the WT versus the relP relQ mutant and the WT versus the (p)ppGpp0 mutant. There were no significant differences upon ciprofloxacin treatment.
RelP and RelQ are involved in the adaptation to cell envelope stress.
(p)ppGpp molecules, as effectors of the stringent response, are molecules that contribute to the survival of bacteria under nonfavorable growth conditions. Previous studies revealed a contribution of (p)ppGpp to increased β-lactam resistance in E. coli and S. aureus (29, 30). To further show this contribution, we analyzed the growth and survival of strains lacking rshSyn, relP, and/or relQ under cell envelope stress conditions employed by vancomycin and ampicillin. Addition of 5× MIC of vancomycin (3.12 μg/ml) resulted in growth inhibition with a slight decrease in the OD. After 12 h, the wild type and rshSyn mutant were able to recover. In contrast the relP relQ double mutant showed no regrowth (Fig. 5B). Treatment with ampicillin (6× MIC [9.4 μg/ml]) showed a stronger decrease in the OD of the mutant lacking relP and relQ compared to the wild type or the rshSyn mutant strain.
Furthermore, time-kill kinetic studies revealed that strains lacking relP and relQ were more efficiently killed by vancomycin and ampicillin than strains harboring the two additional (p)ppGpp synthases (Fig. 5C). Interestingly, complementation of the relP relQ mutants indicated that one of the two synthases was sufficient to partially recover survival. Accordingly, relP and relQ single mutants showed slight but not significant differences in survival compared to the wild-type strain (data not shown).
To verify that the relP- and relQ-dependent tolerance against cell wall-active antibiotics was specific and not mediated by general growth inhibition, we also performed a control experiment using ciprofloxacin, which is a bactericidal antibiotic that interferes with replication by inhibiting the bacterial gyrase. relP and relQ mediated no increased tolerance against this gyrase inhibitor. Therefore, we conclude that both additional (p)ppGpp synthases RelP and RelQ possess an essential role for survival under cell envelope stress conditions.
DISCUSSION
Two small (p)ppGpp synthases, RelP and RelQ, mediate tolerance to cell wall stress conditions.
The stringent response and its effector molecules were discovered decades ago (31). Besides significant progress in understanding how these small molecules mediate such a wide range of effects, many basic questions are still open. Specifically, the roles of the recently discovered small, truncated versions of (p)ppGpp synthases in B. subtilis and Streptococcus mutans are largely unclear (10, 11). Here we characterized these additional versions of putative (p)ppGpp synthases in S. aureus and elucidated their functional roles in the adaptation to cell envelope stress conditions. Mutants lacking both small synthases were less able to survive high doses of cell wall-active antibiotics than strains harboring these synthases. This result was in accordance with those obtained for Enterococcus faecalis, where the basal levels of RelQ-mediated (p)ppGpp accumulation could be shown to contribute to an increased tolerance against vancomycin (12).
How does (p)ppGpp accumulation in the cell lead to pronounced antibiotic tolerance? A leading hypothesis implicates the inactivity of antibiotic targets in growth-arrested cells as the mechanism for mediating this tolerance (32, 33). In E. coli, for instance, antibiotic tolerance was linked to slow growth and reduced autolysis phenotypes, which were dependent on a functional RelA (29, 34). In a recent study that investigated the heterogeneous expression of methicillin resistance in S. aureus, it was shown that a highly resistant derivative of such a culture harbored a stop mutation in the rsh gene. This subclone had a growth defect that was presumably due to (p)ppGpp overexpression. When screened for repressor mutations that recovered normal growth and antibiotic-sensitive phenotypes, a single point mutation in relQ was revealed (30). These results demonstrate that (p)ppGpp molecules play an important role in the high-level resistance toward methicillin in MRSA strains that is most likely linked to the slow-growth phenotype imposed by (p)ppGpp.
However, our results cannot be explained by a growth arrest phenotype mediated by (p)ppGpp accumulation, since the analyzed relP and relQ mutants showed no growth differences compared to the more tolerant rshSyn or wild-type strains. We assume that (p)ppGpp possesses a more active role in S. aureus in mediating tolerance against cell wall stress, such as targeting components of cell wall biosynthesis or the cell wall turnover process. Survival upon ciprofloxacin treatment, which targets cell replication, was not significantly affected by RelP and RelQ, and this result is in accordance with results that have become more apparent in recent years showing that antibiotic tolerance depends rather on adaptive responses than on growth arrest and target inactivity per se. For Escherichia coli and Pseudomonas aeruginosa, the passive influence of (p)ppGpp molecules on their increased tolerance toward antibiotics by a slowing of growth could be excluded (35). This work nicely shows that the stringent response directly curtails the production of pro-oxidant molecules in bacterial cells and on the other hand induces antioxidant defenses. A recent study implicated superoxide dismutases of S. aureus and E. faecalis in tolerance to vancomycin (36). In the future, it would be interesting to examine whether these enzymes are under the regulatory control of a RelP/RelQ-mediated stringent response.
The transcriptional induction of relP and relQ upon treatment with cell wall-active antibiotics.
Upon treatment with vancomycin and ampicillin, relP and relQ transcription was strongly induced. relP, which was annotated as SACOL2518 at that time, was described as one of the few members of the “core” cell wall stress stimulon (CWSS) of S. aureus (27), which is controlled by the activity of the two-component system VraS/R. This two-component system, which was initially discovered and described by Kuroda et al. (28, 37), responds to the damage in the cell wall structure or inhibition of cell wall biosynthesis and is responsible for increased resistance toward cell wall-active antibiotics. In our study, we confirmed that relP and vraS/R transcription is strongly induced upon cell wall stress and also showed the induction of relQ, encoding the second small (p)ppGpp synthase. The transcriptional induction of relP is strongly VraS/R dependent, and this regulator is also necessary for full relQ induction. The observation that relQ is still responsive in a vraR mutant may explain why relQ is not a core member of the cell wall stress stimulon. Further microarray analyses indicated a slight induction of relP transcription in response to low-pH conditions (38) and a characteristic stringent response transcription pattern under alkaline shock conditions (39). We confirmed that expression of relP is pH dependent (data not shown). Since vraS/R transcription was also enhanced, we assumed that increased relP expression was due to cell wall stress, induced by low or high pH, rather than occurred through direct activation.
The LiaR/S two-component system displays the corresponding homolog to the staphyloccocal VraS/R system in other Gram-positive bacteria; however, no indication as to whether it also controls relQ or relP in these organisms is evident. Nevertheless, the relP homolog (ywaC) is also strongly activated by cell envelope stress in B. subtilis (40). Here the transcriptional induction of ywaC is under the regulation of σM, which is a well-described alternative sigma factor in B. subtilis that contributes to cell envelope stress responses. In S. aureus, the extracytoplasmic function (ECF) sigma factor σS is described as the homologous sigma factor that also responds to cell wall-active antibiotics or detergents (41, 42). Data for the σS regulon are lacking, and σS binding motifs are not defined. However, promoter sequences of relP and relQ rather predict the regulation of both genes by the essential housekeeping factor σA than by alternative sigma factors.
The essentiality of the RSH-hydrolase in preventing the toxic accumulation of (p)ppGpp in S. aureus.
Here we report direct evidence suggesting that the staphylococcal RSH-hydrolase is essential due to the toxic accumulation of (p)ppGpp in the presence of constitutive, low-level RelP and RelQ synthase activities. The essentiality of the RSH-hydrolase is unique for S. aureus and could be based on two assumptions. Either the staphylococcal RelP and RelQ enzymes possess stronger synthetic activities than their homologs in other bacteria, or S. aureus is more sensitive regarding the accumulation of (p)ppGpp molecules in the cell. Nevertheless, the need for detoxification was emphasized by the indicated strong RSH-hydrolase activity that was shown in the overexpression assays (Fig. 1B and Fig. 2).
Surprisingly, overexpression of (p)ppGpp synthases can lead to death of the bacteria, which was indicated by a severe drop of the CFU within 1 h. Only the two stronger synthases RelP and RSH showed such a phenotype, whereas RelQ, with its lower synthetic activity, only inhibited growth. This finding led to the assumption that the intracellular concentration of (p)ppGpp determines the lytic or bacteriostatic outcome of the cell. However, why the predicted (p)ppGpp accumulation in S. aureus leads to cell lysis is still unknown. A first hint stems from a study in E. coli that showed that controlled induction of the relA gene inhibited phospholipid and peptidoglycan synthesis, which in turn could lead to enhanced cell lysis (43).
Do RelP and RelQ elicit a characteristic stringent response?
Analysis of purified proteins revealed that both enzymes are able to catalyze (p)ppGpp synthesis with a higher activity of RelP, which is in accordance with results described for S. mutans (11). Furthermore, we demonstrated that both enzymes preferred the synthesis of ppGpp over that of pppGpp in vitro as well as in vivo. Whether there is a difference in the stringent responses elicited by ppGpp versus pppGpp in firmicutes remains unclear. Recently, it could be shown for E. coli (46) that ppGpp is more potent than pppGpp in vitro as well as in vivo. Our preliminary experiments indicate that transcriptional induction of RSH versus RelP/Q in a (p)ppGpp0 strain may impose a similar response with regard to some selected hallmark genes (Fig. 4). However, one can assume that under more physiological conditions, the impact of the enzymes may differ substantially. This is supported by the observation that the cell wall stress stimulon (27, 28) has little overlap with the stringent response imposed by amino acid limitation or mupirocin (23, 44, 45). Thus, whether there is a quantitative or qualitative difference between the outcomes of RSH versus RelP/Q activity in S. aureus needs further evaluation.
In summary, three (p)ppGpp synthases are active in S. aureus. Whereas RSH seems to be necessary to address amino acid starvation imposed, e.g., after phagocytosis (23), RelP and RelQ are involved in mediating cell envelope stress tolerance.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by grants to C. Wolz from the Deutsche Forschungsgemeinschaft TR34 and to T. Geiger from the Fortüne Program (2068-0-0) of the Medical Faculty of the University of Tübingen.
Footnotes
Published ahead of print 13 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01201-13.
REFERENCES
- 1.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68:1128–1148. 10.1111/j.1365-2958.2008.06229.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battesti A, Bouveret E. 2009. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191:616–624. 10.1128/JB.01195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62:1048–1063. 10.1111/j.1365-2958.2006.05442.x [DOI] [PubMed] [Google Scholar]
- 4.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56:958–970. 10.1111/j.1365-2958.2005.04601.x [DOI] [PubMed] [Google Scholar]
- 5.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]
- 6.Eymann C, Homuth G, Scharf C, Hecker M. 2002. Bacillus subtilis functional genomics: global characterization of the stringent response by proteome and transcriptome analysis. J. Bacteriol. 184:2500–2520. 10.1128/JB.184.9.2500-2520.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kazmierczak KM, Wayne KJ, Rechtsteiner A, Winkler ME. 2009. Roles of rel in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39. Mol. Microbiol. 72:590–611. 10.1111/j.1365-2958.2009.06669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geiger T, Goerke C, Fritz M, Schafer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78:1873–1883. 10.1128/IAI.01439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemos JA, Brown TA, Jr, Burne RA. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431–1440. 10.1128/IAI.72.3.1431-1440.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67:291–304. 10.1111/j.1365-2958.2007.06018.x [DOI] [PubMed] [Google Scholar]
- 11.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. 2007. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65:1568–1581. 10.1111/j.1365-2958.2007.05897.x [DOI] [PubMed] [Google Scholar]
- 12.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191:2248–2256. 10.1128/JB.01726-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemos JA, Nascimento MM, Lin VK, Abranches J, Burne RA. 2008. Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190:5291–5299. 10.1128/JB.00288-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Natori Y, Tagami K, Murakami K, Yoshida S, Tanigawa O, Moh Y, Masuda K, Wada T, Suzuki S, Nanamiya H, Tozawa Y, Kawamura F. 2009. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p)ppGpp synthetase genes, relA, yjbM, and ywaC. J. Bacteriol. 191:4555–4561. 10.1128/JB.00263-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117:57–68. 10.1016/S0092-8674(04)00260-0 [DOI] [PubMed] [Google Scholar]
- 16.Novick RP. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587–636. 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- 17.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191:2953–2963. 10.1128/JB.01492-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 19.Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58–63. 10.1016/j.plasmid.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 20.Vagner V, Dervyn E, Ehrlich SD. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097–3104. 10.1099/00221287-144-11-3097 [DOI] [PubMed] [Google Scholar]
- 21.Goerke C, Campana S, Bayer MG, Doring G, Botzenhart K, Wolz C. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304–1311. 10.1128/IAI.68.3.1304-1311.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gentry D, Li T, Rosenberg M, McDevitt D. 2000. The rel gene is essential for in vitro growth of Staphylococcus aureus. J. Bacteriol. 182:4995–4997. 10.1128/JB.182.17.4995-4997.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geiger T, Francois P, Liebeke M, Fraunholz M, Goerke C, Krismer B, Schrenzel J, Lalk M, Wolz C. 2012. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8:e1003016. 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blagova EV, Levdikov VM, Tachikawa K, Sonenshein AL, Wilkinson AJ. 2003. Crystallization of the GTP-dependent transcriptional regulator CodY from Bacillus subtilis. Acta Crystallogr. D Biol. Crystallogr. 59:155–157. 10.1107/S0907444902018358 [DOI] [PubMed] [Google Scholar]
- 25.Handke LD, Shivers RP, Sonenshein AL. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798–806. 10.1128/JB.01115-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belitsky BR, Sonenshein AL. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224–1236. 10.1128/JB.01780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobral RG, Jones AE, Des Etages SG, Dougherty TJ, Peitzsch RM, Gaasterland T, Ludovice AM, de Lencastre H, Tomasz A. 2007. Extensive and genome-wide changes in the transcription profile of Staphylococcus aureus induced by modulating the transcription of the cell wall synthesis gene murF. J. Bacteriol. 189:2376–2391. 10.1128/JB.01439-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- 29.Tuomanen E, Tomasz A. 1986. Induction of autolysis in nongrowing Escherichia coli. J. Bacteriol. 167:1077–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb. Drug Resist. 19:153–159. 10.1089/mdr.2013.0053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cashel M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J. Biol. Chem. 244:3133–3141 [PubMed] [Google Scholar]
- 32.Levin BR, Rozen DE. 2006. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4:556–562. 10.1038/nrmicro1445 [DOI] [PubMed] [Google Scholar]
- 33.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5:48–56. 10.1038/nrmicro1557 [DOI] [PubMed] [Google Scholar]
- 34.Goodell W, Tomasz A. 1980. Alteration of Escherichia coli murein during amino acid starvation. J. Bacteriol. 144:1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. 10.1126/science.1211037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ladjouzi R, Bizzini A, Lebreton F, Sauvageot N, Rince A, Benachour A, Hartke A. 2013. Analysis of the tolerance of pathogenic enterococci and Staphylococcus aureus to cell wall active antibiotics. J. Antimicrob. Chemother. 68:2083–2091. 10.1093/jac/dkt157 [DOI] [PubMed] [Google Scholar]
- 37.Kuroda M, Kuwahara-Arai K, Hiramatsu K. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485–490. 10.1006/bbrc.2000.2277 [DOI] [PubMed] [Google Scholar]
- 38.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407–8423. 10.1128/JB.186.24.8407-8423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson KL, Roux CM, Olson MW, Luong TT, Lee CY, Olson R, Dunman PM. 2010. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol. Med. Microbiol. 60:208–250. 10.1111/j.1574-695X.2010.00736.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eiamphungporn W, Helmann JD. 2008. The Bacillus subtilis sigma(M) regulon and its contribution to cell envelope stress responses. Mol. Microbiol. 67:830–848. 10.1111/j.1365-2958.2007.06090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. 2008. Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One 3:e3844. 10.1371/journal.pone.0003844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller HK, Carroll RK, Burda WN, Krute CN, Davenport JE, Shaw LN. 2012. The extracytoplasmic function sigma factor σS protects against both intracellular and extracytoplasmic stresses in Staphylococcus aureus. J. Bacteriol. 194:4342–4354. 10.1128/JB.00484-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodionov DG, Ishiguro EE. 1995. Direct correlation between overproduction of guanosine 3′,5′-bispyrophosphate (ppGpp) and penicillin tolerance in Escherichia coli. J. Bacteriol. 177:4224–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, Olson PD, Projan SJ, Dunman PM. 2006. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J. Bacteriol. 188:6739–6756. 10.1128/JB.00609-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reiss S, Pane-Farre J, Fuchs S, Francois P, Liebeke M, Schrenzel J, Lindequist U, Lalk M, Wolz C, Hecker M, Engelmann S. 2012. Global analysis of the Staphylococcus aureus response to mupirocin. Antimicrob. Agents Chemother. 56:787–804. 10.1128/AAC.05363-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res. 41:6175–6189. 10.1093/nar/gkt302 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.