PrPSc forms micrometer long amyloidic strings that can congregate in clusters and webs at the surface of living cells.
Abstract
Mammalian prions refold host glycosylphosphatidylinositol-anchored PrPC into β-sheet–rich PrPSc. PrPSc is rapidly truncated into a C-terminal PrP27-30 core that is stable for days in endolysosomes. The nature of cell-associated prions, their attachment to membranes and rafts, and their subcellular locations are poorly understood; live prion visualization has not previously been achieved. A key obstacle has been the inaccessibility of PrP27-30 epitopes. We overcame this hurdle by focusing on nascent full-length PrPSc rather than on its truncated PrP27-30 product. We show that N-terminal PrPSc epitopes are exposed in their physiological context and visualize, for the first time, PrPSc in living cells. PrPSc resides for hours in unexpected cell-surface, slow moving strings and webs, sheltered from endocytosis. Prion strings observed by light and scanning electron microscopy were thin, micrometer-long structures. They were firmly cell associated, resisted phosphatidylinositol-specific phospholipase C, aligned with raft markers, fluoresced with thioflavin, and were rapidly abolished by anti-prion glycans. Prion strings and webs are the first demonstration of membrane-anchored PrPSc amyloids.
Introduction
Prions (Prusiner, 1982) spread by refolding host glycoprotein PrPC into corrupt β-sheet–rich PrPSc (Bolton et al., 1982; Prusiner et al., 1990). PrPC is a small, multifunctional (Aguzzi et al., 2008; Biasini et al., 2012) cell surface protein with a C-terminal glycosylphosphatidylinositol (GPI) moiety (Stahl et al., 1987), two N-glycosylation sites, and a disulfide bond (Turk et al., 1988). Although covalently identical (Oesch et al., 1985; Basler et al., 1986; Stahl et al., 1993), the PrP isoforms have distinct properties (Meyer et al., 1986): in its mature form, PrPSc has a C-terminal protease-resistant core (PrP27-30; Fig. 1 A), precipitates in detergents, forming amyloids such as prion rods (Prusiner et al., 1983), resists cleavage of its GPI by phosphatidylinositol-specific phospholipase C (PIPLC; Stahl et al., 1990), and fails to react natively with antibodies (Abs) to PrP27-30 (core Abs; Kitamoto et al., 1987; Serban et al., 1990), with some exceptions (Korth et al., 1997).
Figure 1.
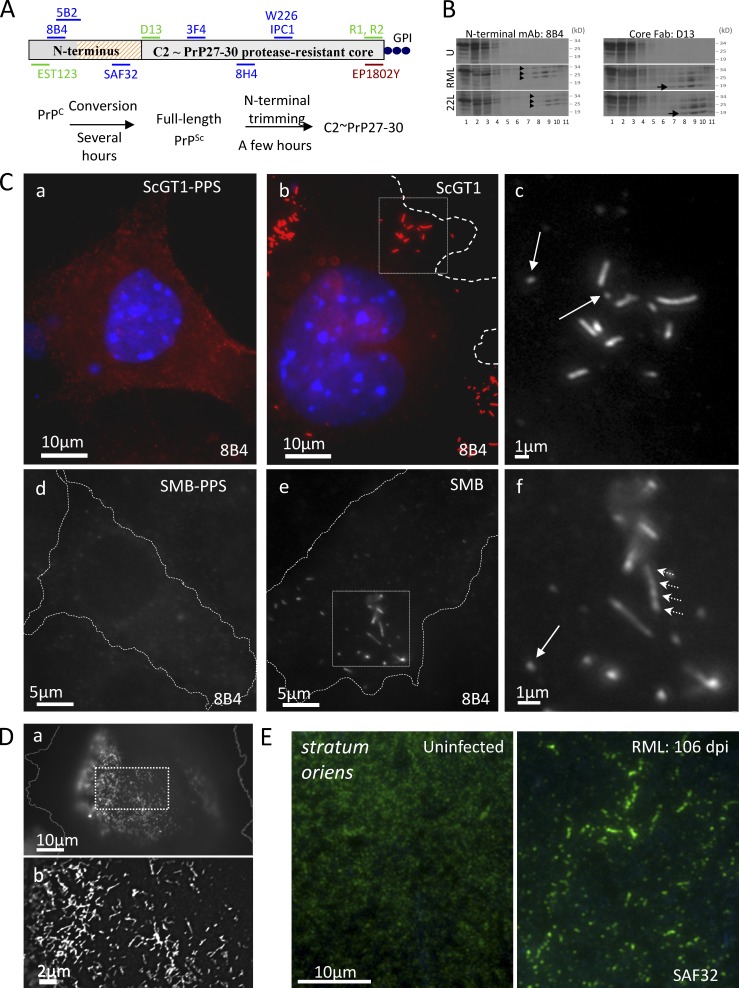
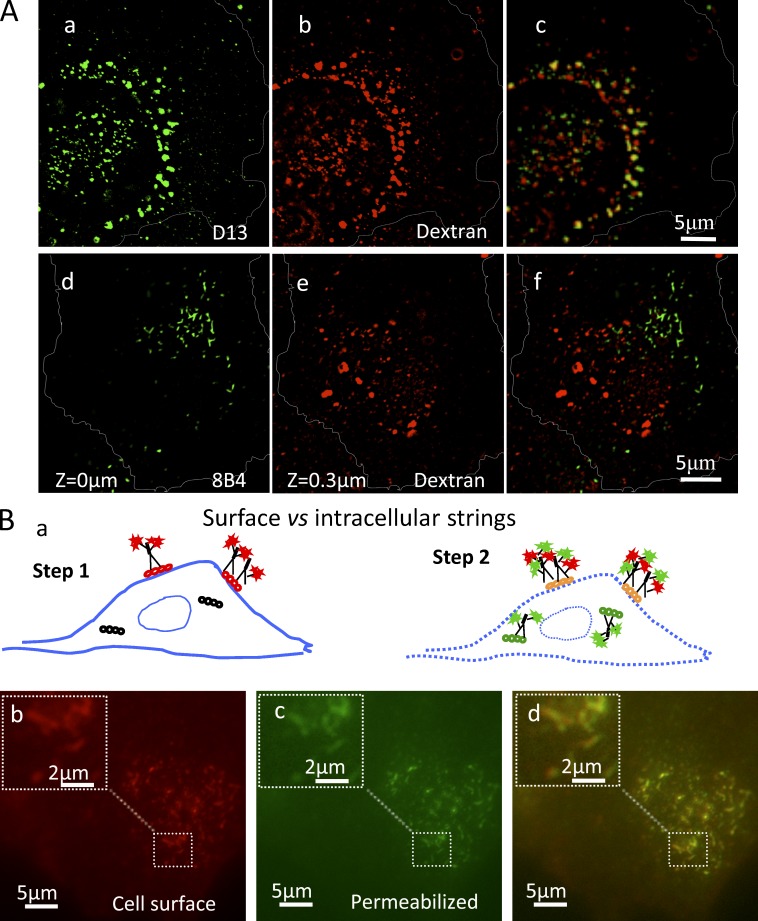
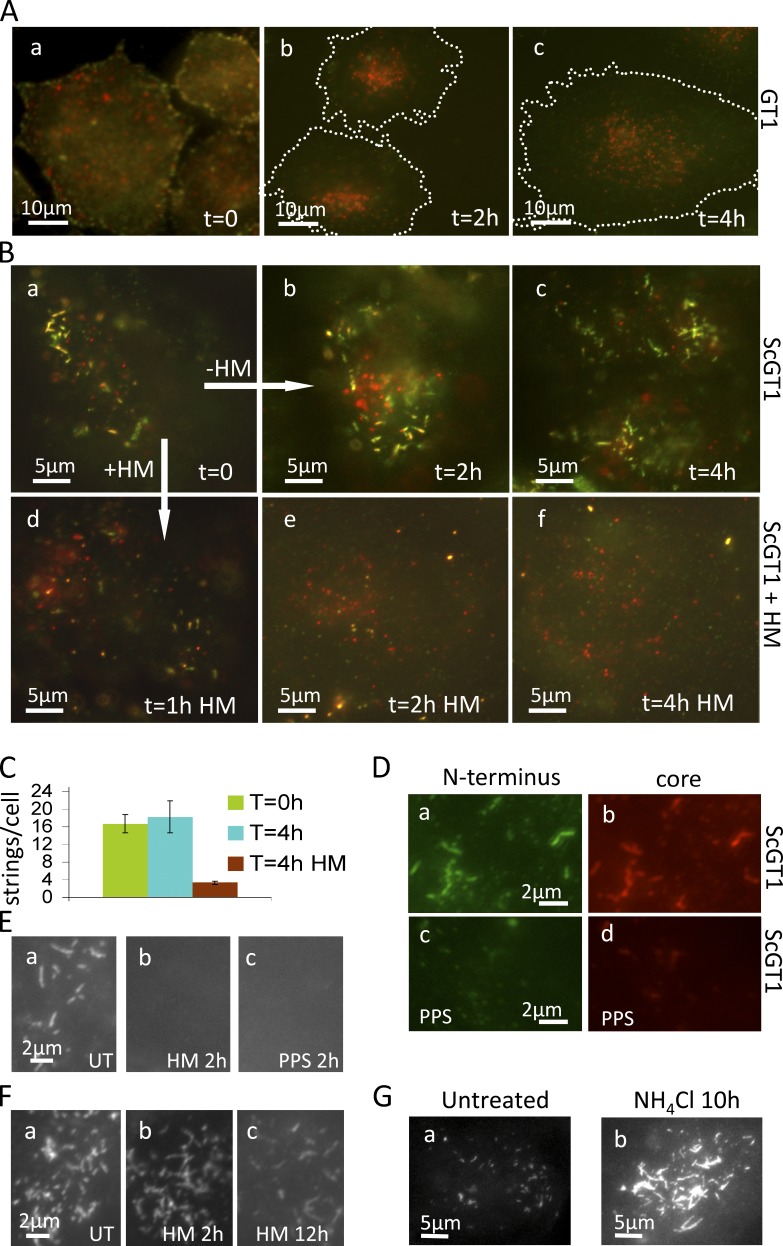
N-proximal PrP Abs decorate clusters of micrometer-long strings and webs in infected cells and hippocampus. (A) Abs and Fabs used in this study; summary of PrPSc metabolism. Hatched, octarepeats (aa 51–90); blue, mouse mAbs; green, humanized Fabs. EP1802Y is a rabbit mAb. (B) Lysates of uninfected GT1 (U) and of infected ScGT1/RML and ScGT1/22L cells were fractionated by sedimentation through sucrose-Sarkosyl gradients (top, fraction 1). Fractions were analyzed in WB developed with N-terminal mAb 8B4 or core Fab D13. Arrowheads show the glycoforms of highly aggregated FL PrPSc (fractions 8–11). Arrow (right) shows the 19-kD unglycosylated version of PrP27-30, which is not recognized by 8B4 (left; see also Fig. S2 C). PrP27-30 and FL PrPSc aggregates are absent from GT1 lysate. See also Fig. S1 H. (C) Subconfluent infected or PPS-cured ScGT1 (a–c) and SMB (d–f) cells were fixed, permeabilized, stained with 8B4 (red; a and b), counterstained with DAPI (blue; a and b), and examined by fluorescence microscopy with a 100× objective. Cells were outlined using the corresponding DIC pictures (Fig. S2 E). Boxes in b and e are enlarged in c and f, respectively. In infected cells, 8B4 decorates clusters of strings up to 5 µm long; arrows show short strings. The seemingly intense PrPC signal observed in ScGT1-PPS compared with that in ScGT1 is discussed in the Immunofluorescence section of Materials and methods. (D) ScGT1 growth-arrested by a 6-d MMC/BrdU treatment were stained with mAb SAF32 using the FA-denaturing protocol. Webs of strings cover large parts of the cell periphery. (b) Enlarged, 3D deconvolved version of box in a. (E) SAF32 labeling of cryosections of uninfected and RML infected hippocampus stratum oriens of FVB mice at 106 dpi.
Most PrPC is GPI anchored to plasma membrane (PM) rafts (Harmey et al., 1995; Taraboulos et al., 1995) and caveolae (Peters et al., 2003), but topological variants have been described previously (Hegde et al., 1998). In infected cells, a minority of PrPC molecules is slowly converted into PrPSc (Borchelt et al., 1990; Taraboulos et al., 1990a). Prion conversion may occur in rafts (Taraboulos et al., 1995; Kaneko et al., 1997; Naslavsky et al., 1997; Bate et al., 2004), on the cell surface (Goold et al., 2011; Godsave et al., 2013), or in endosomes (Caughey and Raymond, 1991; Borchelt et al., 1992; Godsave et al., 2008; Marijanovic et al., 2009). It produces a full-length (FL) PrPSc intermediate, which is then N-proximally truncated within a few hours (Caughey et al., 1991; Taraboulos et al., 1992; Taguchi et al., 2009) by acidic proteases to yield a “C2” (Chen et al., 1995) fragment similar to PrP27-30 (Dron et al., 2010). Infected brains (Hope et al., 1986) and cells contain variable proportions of PrPC, FL PrPSc, and PrP27-30 (Dron et al., 2010).
Cell-associated prions have been assumed to comprise a size continuum of poorly defined FL PrPSc/PrP27-30 oligomers. However, the existence of membrane-anchored PrPSc aggregates has never been demonstrated, and the configuration of PrPSc in the infected cell remains unknown.
Whereas PrP27-30 persists in the endocytic system for days (Taraboulos et al., 1990b; McKinley et al., 1991; Arnold et al., 1995; Jeffrey et al., 1996; Veith et al., 2009), the subcellular localization of FL PrPSc is unknown. This is a crucial issue because, although often quantitatively minor, FL PrPSc is the immediate product of the poorly understood conversion event. More generally, the field suffers from the current inability to visualize prions in live cells, a task made exceedingly difficult by the limited native accessibility of PrP27-30 epitopes.
Here we focused on FL PrPSc and especially on its unstructured N terminus (aa 23–89). The extent to which N-terminal epitopes are natively exposed has remained vague, but ELISA with octarepeat Abs (Yam et al., 2010) and measurement of PrPSc avidity to immobilized copper (Dron et al., 2010) suggest that N-proximal determinants are accessible in FL PrPSc, at least in detergents.
We studied FL PrPSc in several mouse cells, including Min6, a pancreatic β cell line (Miyazaki et al., 1990) whose susceptibility to prion infection is reported here for the first time. We now show that N-proximal Abs (N-Abs) do react natively with cell surface FL PrPSc in its physiological context. This permits us, for the first time, to visualize prions in live cells.
Using immunofluorescence microscopy, we find that FL PrPSc forms micrometer-long cell-surface “strings,” which can assemble in clusters and vast webs comprising FL PrPSc and PrP27-30 but no detectable PrPC. Strings are firmly cell associated; resist PIPLC; align with the raft sphingolipid, GM1, and with caveolin-1; and fluoresce with thioflavin T (ThT), suggesting amyloids. Correlative light microscopy (LM)/immunogold scanning electron microscopy (SEM) shows that FL PrPSc follows thin tracks on the cell surface. FL PrPSc stays in strings for hours before endocytosis, sheltered from premature hydrolysis. Strings rapidly fragment and internalize when cells are exposed to anti-prion sulfated glycans, revealing new mechanistic facets of these important agents. We also observe strings in the hippocampus of scrapie-infected mice.
Results
Peculiar strings and webs decorated by N-proximal PrP Abs
We used nondenaturing immunofluorescence microscopy with N-Abs (Fig. 1 A) to visualize FL PrPSc found in infected cells (Fig. 1 B and Fig. S1 H). Unexpectedly, N-terminal mAb 8B4 decorated string-like objects in ScGT1 (Fig. 1 C, b and c) and SMB (Fig. 1 C, e and f) cells, both infected with the Rocky Mountain Laboratory (RML) strain of mouse-adapted scrapie, but not in three types of negative control—cells cured from prions with pentosan polysulfate (PPS; Fig. 1 C, a and d; and Fig. S2 A); uninfected GT1-1 cells (Fig. S2 B), and CHO cells refractory to prion infection (Fig. S1 G)—or in cells labeled in the presence of competing 8B4-specific peptide (Fig. S2 D). Strings were also decorated by N-terminal mAbs SAF32 (Fig. 2 D), 5B2 (unpublished data), and Fab EST123 (Fig. 4 B). There were also strings in 22L-infected ScGT1 (ScGT1/22L; Fig. S1 E), N2a/RML (Fig. S1 F), and Min6/RML (Fig. 2 G); ME7-infected CAD/ME7 (Fig. S3 H); and primary mouse dorsal root ganglion (DRG) cells co-cultured with ScGT1 (Fig. S1 I). Abundant strings also appeared in the hippocampus stratum oriens of mice after RML infection. Figs. 1 E and S1 J show cryosections at 106 d after inoculation (dpi).
Figure 2.
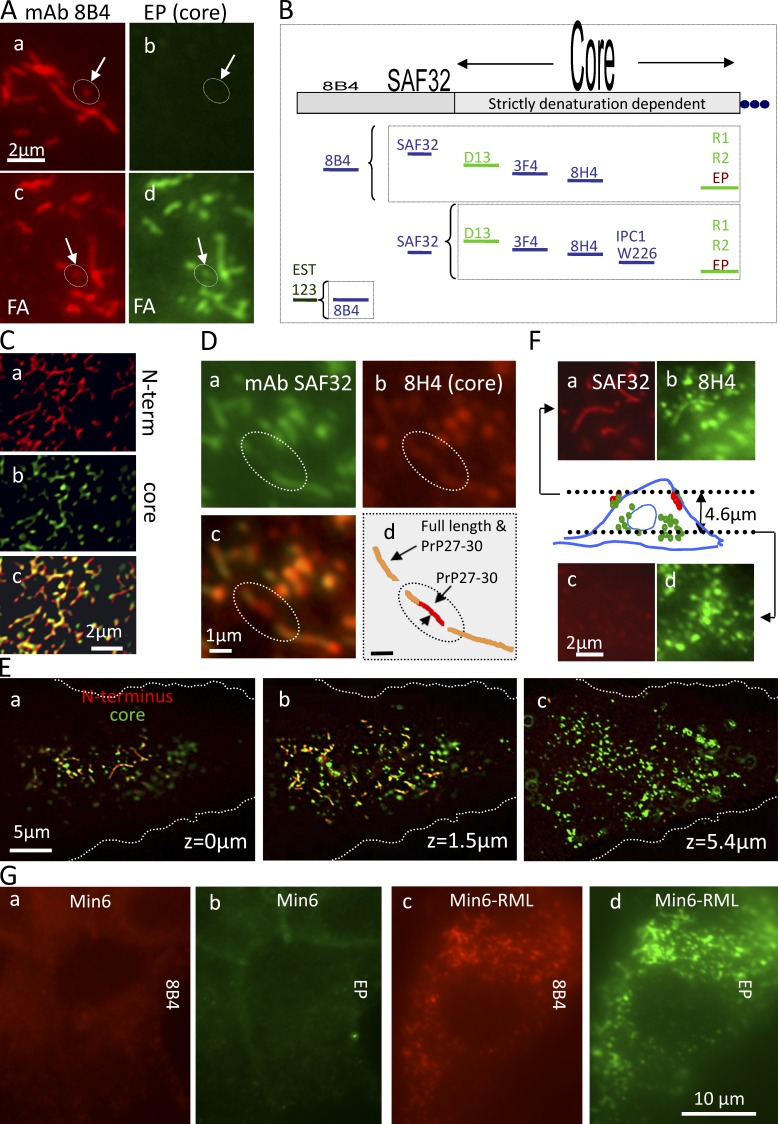
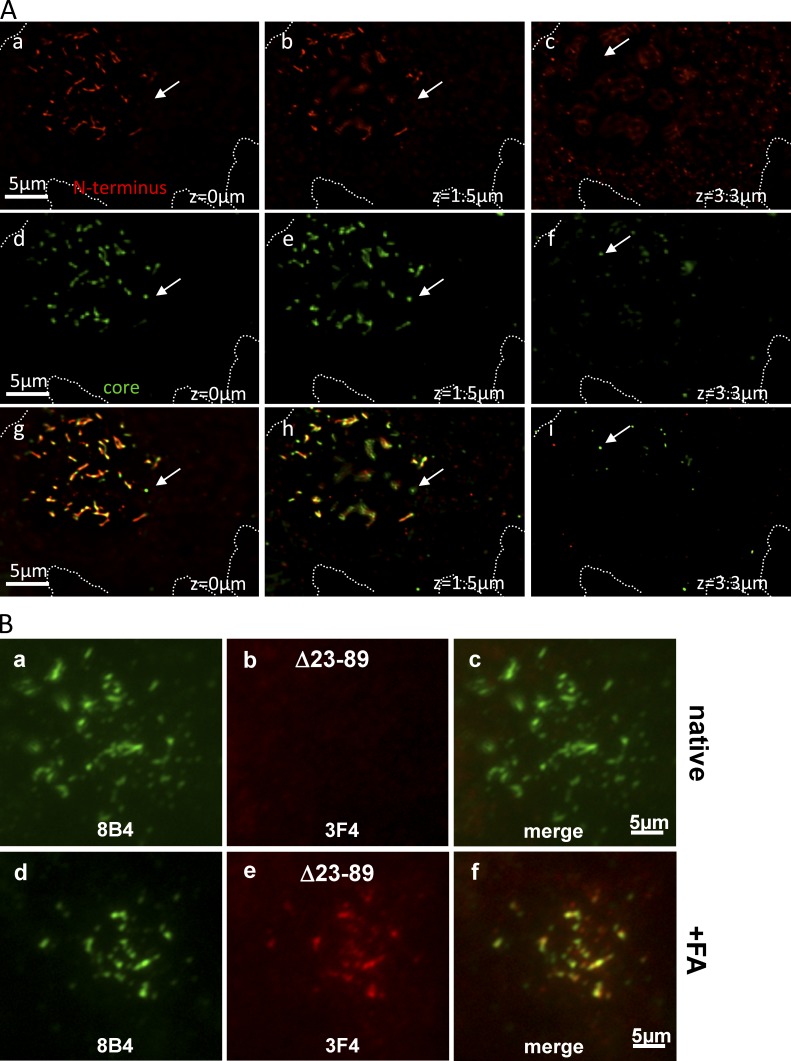
Core Abs stain strings only after denaturation: strings contain PrPSc but no detectable PrPC. (A) Fixed ScGT1 costained with N-terminal mAb 8B4 (red) and with core mAb EP1802Y (green). In c and d, cells were denatured with FA before staining. After FA, EP1802Y largely codecorates strings (circles; arrows, short strings) outlined by 8B4 (compare c with d); there was no detectable EP1802Y staining of strings in nondenatured cells (a and b). (B) Pairs of N- and core Abs used to costain strings in this study. Core Abs stained strings only after denaturation, whereas N-Abs decorated them natively. Nevertheless, denaturation still increased the staining intensity of 8B4 and SAF32 (see Fig. S3 F). The dependence of epitopes on denaturation is roughly indicated by their print size on top of the PrP map. (C) In growth-arrested ScGT1, webs costain with SAF32 and core 8H4 after FA denaturation. Variable relative red and green intensities indicate varied proportions of FL PrPSc/PrP27-30 along strings. Images were 3D deconvolved. (D) ScGT1 were costained with SAF32 (a) and with 8H4 (b) after FA denaturation. Two strings codecorated by both Abs (d, orange) are bridged by a PrP27-30 strand (circled; d, red). (E and F) PrPSc strings do not coincide with cytoplasmic PrP27-30 deposits. ScGT1 were costained with SAF32 and core 8H4 using FA denaturation. z-stacks were acquired (z = 0 is dorsal). (E) 3D-deconvolved images are shown for three planes a total of 5.4 µm apart (colors superimposed). Notice that strings stain in orange and are concentrated in the dorsal plane (a and b). In contrast, abundant cytoplasmic PrP27-30 stains exclusively in green (b and c). (F) A detail of the cell in E. The scheme depicts strings staining with both Abs (red and green) near the top of the cell (z = 0 µm) and intracellular PrP27-30 (green; z = 4.6 µm), which is negative for N-terminal staining. (G) PrPSc strings are codecorated with 8B4 and EP1802Y in Min6/RML but are absent from uninfected cells (FA denaturation protocol).
Figure 4.
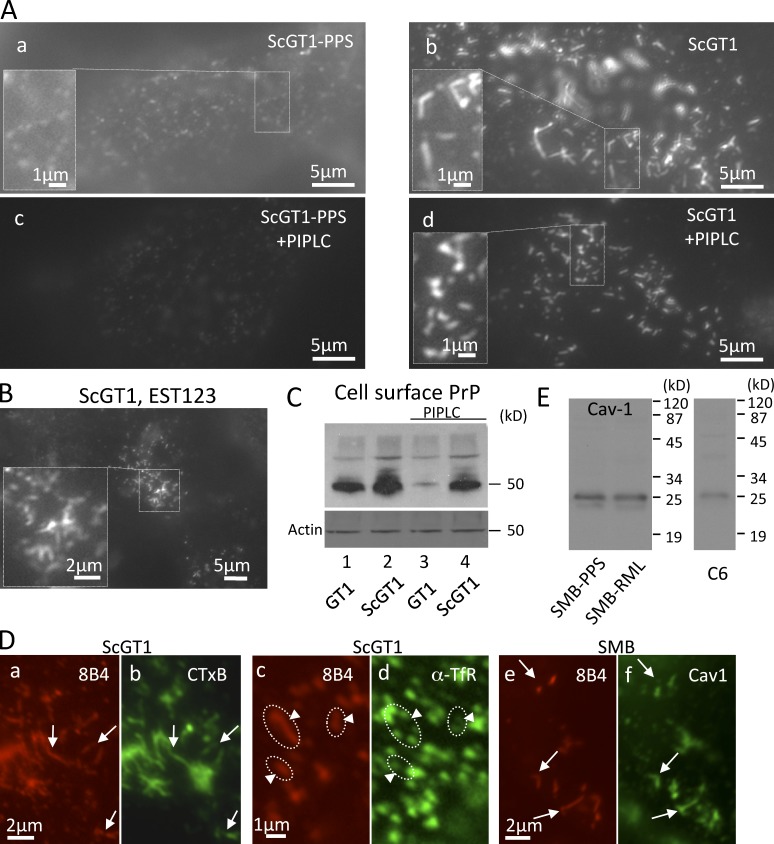
PrPSc strings on the surface of living cells resist extraction by PIPLC and colocalize with GM1 and caveolin. (A) Live ScGT1 and ScGT1-PPS cooled to 8°C were stained with 8B4 followed by RRX secondary Fabs. (c and d) Cells were treated with PIPLC (1 U/ml; for 1 h at 8°C) before staining. (B) Live ScGT1 were labeled at 8°C with Fab EST123 followed by secondary Fab (monovalent, to prevent Ab-induced PrP patching). (C) GT1 and ScGT1 cells chilled to 8°C were treated or not with PIPLC and incubated with 8B4, and cell-bound mAb was then quantified in a WB developed with secondary Abs coupled to HRP. In infected cells, PIPLC reduced the amount of cell surface–bound 8B4 by ∼50% (lanes 2 and 4). (D, a–d) On the surface of live ScGT1 cells, 8B4-decorated strings (a and c, red) largely overlapped with CTxB (b, arrows) but not with TfR (d, circles and arrowheads). (e and f) Colocalization of 8B4 strings (red) with caveolin 1 (green) in fixed and permeabilized SMB cells treated with GdnSCN. (E) SMB contain caveolin-1. Positive control: C6 glioma.
The labeled structures ranged from dots (Fig. 1 C, c and f, arrows) to strings typically ≤5 µm long (but see Fig. S1 A, b). Their thickness was not optically resolved (resolution ≤300 nm, but see Fig. 6 E). Strings were sometimes sinuous (Fig. S1 A), branching (Fig. 1 D), or interrupted (Fig. S1 A and Fig. 2 D) and often organized in a few clusters/cell (Fig. 1 C, b and e). Strings were occasionally observed on intercellular connections (Figs. S1 D and S3 G). Immunostaining typically fluctuated along strings (Fig. 1 C, f, arrowheads; Fig. S1 A, b; and see Fig. 6 B).
Figure 6.
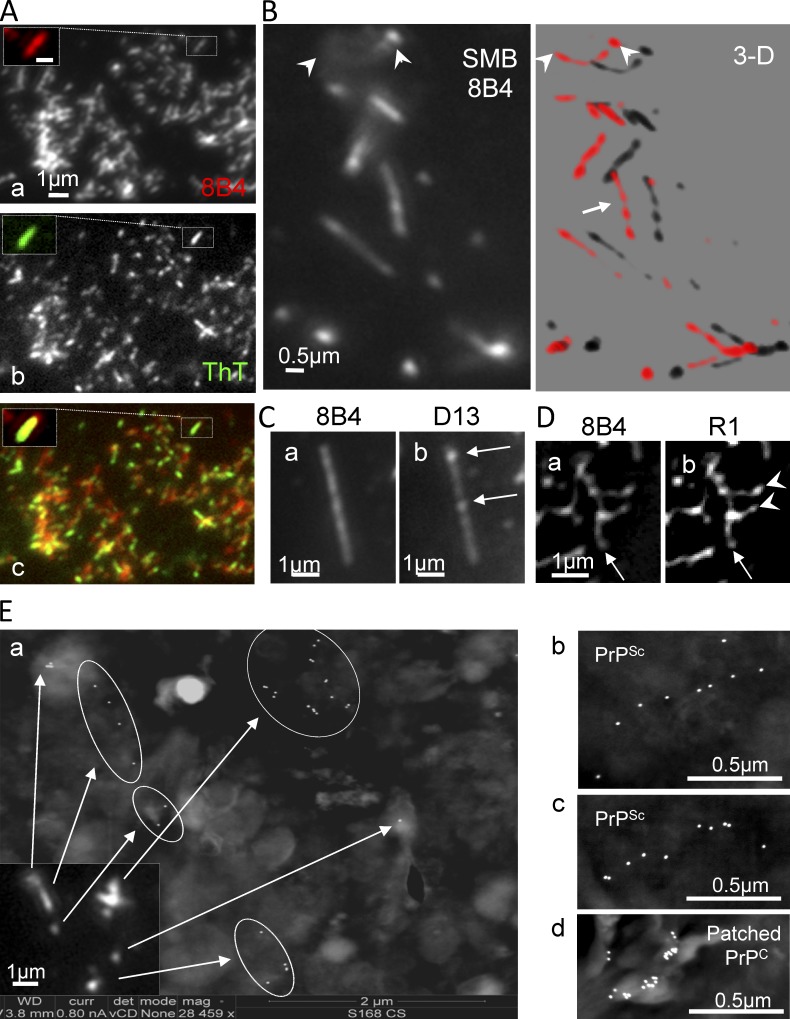
Strings and webs stain with ThT and have a beaded structure: LM/SEM. (A) Live growth-arrested ScGT1 were stained at 8°C with 8B4 followed by RRX secondary Fab. PM sheets were then transferred to glass slides, fixed, stained with ThT, and examined for the fluorescence shift indicative of amyloids. ThT staining (b, inset, green) colocalized with 8B4-labeled strings (a); merge shown in c. Bar: (a, inset) 0.5 µm. (B–D) Quasi periodic immunostaining of strings/webs. Fixed SMB (B) and fixed/FA-treated ScGT1 (C and D) cells were stained with the indicated Abs. Both N- and core Abs had a punctuated distribution hinting at a “beaded chain.” In some cases, individual foci were especially prominent with one Ab but not with the other (C, b, arrows), suggesting local differences in the ratio of FL/trimmed PrPSc. (B) Several z-planes 300 nm apart (such as the one in the left panel) were acquired. 3D reconstructions show quasi-periodic immunostaining (right, arrow) of strings crossing the focal plane (arrowheads). The distance between consecutive peaks is 300–600 nm. (D) In many instances, strings organized in webs of various complexities. Some strings appeared to serve as organizing “stems” (arrow), which sprouted ramifications (D, b, arrowheads). (E) Correlative LM/SEM. Live ScGT1 (a–c) growing on ITO-coated coverglass slides were treated with PIPLC and then stained with 8B4 followed by fluorescent and 18-nm immunogold labels before fixation (Materials and methods). (d) ScGT1-PPS (not treated with PIPLC) were labeled as above, except that a clustering Ab was used to produce PrPC patches on the cell surface. Cells were fixed, visualized by immunofluorescence (a, inset), postfixed, and further processed for SEM-BSe analysis without metal coating. (a) Strings seen by immunofluorescence (inset) correlated well with thin lines of immunogold in SEM-BSe (arrows and circles). Additional examples of strings are shown in b and c. In contrast, patched PrPC appeared as unorganized clumps of immunogold (d).
In ScGT1, the abundance of strings increased in the days after seeding, and strings were present in up to 80–100% of cells after 2–3 wk. Strings appeared more rapidly and were more abundant in cells seeded without trypsin (Fig. S1 B and see Materials and methods section Optimization of strings). Strikingly, strings disappeared within 1–2 d of the cells reaching confluence; they were then replaced by small dots (Fig. S1 C). Here, we used subconfluent cultures unless stated otherwise. When subconfluent ScGT1 were growth arrested with mitomycin C (MMC)/BrdU, string prevalence gradually reached 100% and cells developed cell-wide webs (Fig. 1 D).
Strings/webs contain PrPSc and PrP27-30 but no PrPC
In ScGT1 (Fig. 2, A–D; and Fig. S3, A–D) and Min6/RML (Fig. 2 G and Fig. S2 F), core Abs codecorated the strings (Fig. 2 A, c and d) and webs (Fig. 2, C and G) that were outlined by N-Abs, but only after denaturation with formic acid (FA; Kitamoto et al., 1987; Fig. 2 A); strings thus contain PrPSc but no detectable PrPC (Serban et al., 1990; Taraboulos et al., 1990b). Guanidine thiocyanate (GdnSCN) also exposed core epitopes in strings, albeit less efficiently (Fig. S3, B and D; and see Materials and methods). No strings were elicited by denaturation in uninfected cells (Fig. S3 E). Even though N-Abs detected native PrPSc, denaturation still increased their staining intensity, but without revealing new strings (Fig. S3 F). Core-proximal SAF32 epitopes were more dependent on denaturation (∼6× increase) than N-proximal 8B4 (∼2×; Fig. S3 F and illustrated in Fig. 2 B). Accordingly, in brain cryosections, an antigen retrieval step enhanced SAF32 immunostaining (Fig. 1 E and Fig. S1 J).
Strings also contained variable amounts of trimmed PrP27-30, as shown by the variable relative staining intensities of N- versus core Abs (see Fig. 6 C). Occasionally, two seemingly unconnected 8B4-decorated strings were bridged by a PrP27-30 strand (Fig. 2 D, circle). Yet, we have never detected strings composed solely of PrP27-30. Importantly, both strings and PrP 27-30 stores seem to contain similar types of PrPSc, as similar [GdnSCN] concentrations were required to expose the core D13 epitope in strings and in endosomes (Fig. S3 I).
Contrary to the perinuclear (Fig. 2, E and F), endosomal (Fig. 3 A) PrP 27–30, strings resided at the cell periphery and did not localize with internalized dextran (Fig. 3 A, d–f) nor with other intracellular markers and tracers (Table S1), suggesting a surface localization.
Figure 3.
Strings are cell surface structures. (A) Unlike most PrP27-30, FL PrPSc strings do not reside in dextran-positive endosomes. ScGT1 were incubated with a fixable Alexa 555–dextran (2 h; 250 µg/ml; red) to saturate the endosomal system, fixed, permeabilized, denatured with 3 M GdnSCN, and stained with either core D13 (a–c) or N-terminal mAb 8B4 (d–f). Within the cell (a–c), most D13 colocalized with dextran (c), consistent with endocytic PrP27-30 localization. The dorsal z = 0 plane (d) contains strings but no dextran. Internalized dextran was first seen in the contiguous z = 300 nm plane within the cell interior (e). The two images are superposed in f. Images sharpened by 2D nearest neighbor deconvolution. (B) In chilled ScGT1, strings are found exclusively on the cell surface. (Step 1) Live ScGT1 were stained at 8°C with 8B4 followed by RRX secondary Fabs (red). (Step 2) Cells were fixed and permeabilized, blocked with unlabeled anti-mouse Fabs, and restained with 8B4 followed by a secondary IgG (green). No additional strings structures were observed in the green (c) versus the red (b) channels, indicating no intracellular strings.
Live detection of cell surface PIPLC-resistant strings
Having shown that N-terminal mAbs react with native FL PrPSc, we turned to determine if strings reside on the cell surface. Incubating live ScGT1 (on ice) with 8B4 decorated abundant strings (Fig. 3 B, b). Restaining with 8B4 after fixation/permeabilization did not reveal intracellular strings (Fig. 3 B, d). In chilled ScGT1, PrPSc strings are thus cell surface structures. Staining live cells with N-Fab EST123 (Fig. 4 B) confirmed that strings were not induced by Ab-mediated “patching” (Mayor et al., 1994).
Clusters of strings were specific to infected ScGT1 cells (Fig. 4 A, b). In addition, both infected and cured cells displayed disperse PrP punctae on a diffuse background, which occasionally loosely aligned in structures much less compact than PrPSc strings (Fig. 4 A, a, inset). In contrast to PrPC (Fig. 4 A, c), strings resisted release with PIPLC (Fig. 4 A, d), substantiating their PrPSc nature (Stahl et al., 1990). They also resisted harsh salt, pH, or polyanion extraction (Table 1), indicating a tight cell association, conceivably through GPI anchoring.
Table 1.
Extraction conditions withstood by strings in live cells at 4°C
| No. | Extraction | Rationale of use |
| 1 | 100 mM NaOAc, pH 5.5, 5 min | Disrupt pH-dependent interactions |
| 2 | 100 mM Na2CO3, pH 11, 3 min | Disrupt pH-dependent interactions |
| 3 | 10 µg/ml PPS, 30 min, or 500 µg/ml PPS, 1 h, or 1 µg/ml HM, 90 min (in PBS) | Compete with interactions with sulfated glycosaminoglycans |
| 4 | 10 µg/ml PPS in 100 mM Na2CO3, 3 min | Combines conditions 2 and 3 |
| 5 | 2 M NaCl, 2 min | Disrupts electrostatic interactions including interactions with GAGs |
| 6 | 0.1% Triton X-100 in PBS, 5 min | Does not extract GPI-APs (Mayor and Maxfield, 1995) |
(Nos. 1–5) Live ScGT1 cells on Permanox slides were chilled to ice temperature, treated as detailed, and then fixed and stained with 8B4, except for HM-treated cells, which were immunostained on ice before fixation. (No. 6) For the Triton X-100 treatment, cells chilled to 4°C were first immunostained, then incubated with Triton X-100, and then fixed.
By quantifying cell-surface 8B4 binding, we found that, in subconfluent ScGT1, PIPLC-resistant PrPSc comprises ∼1/2 of all surface FL PrP, the rest being PrPC and possibly PIPLC-sensitive PrPSc (Fig. 4 C). The actual level of FL PrPSc might be even higher, as only ∼1/2 of 8B4 epitopes are natively accessible (Fig. S3 F).
We reasoned that if PrPSc is tethered to PM rafts, then rafts might align or coalesce to follow strings, as these dynamic 3–5-nm nanodomains (Simons and Gerl, 2010) tend to rearrange when their components (e.g., GPI-anchored proteins [GPI-APs]) are clustered (Harder et al., 1998; Sharma et al., 2004).
Indeed, the GM1 ligand cholera toxin subunit B (CTxB) codecorated strings on the surface of live ScGT1 (Fig. 4 D, a and b), whereas strings segregated away from nonraft transferrin receptor (Fig. 4 D, c and d) and clathrin (Table S1). Moreover, in SMB cells, strings colocalized with caveolin-1 (Fig. 4 D, e and f). These results strongly suggest that PrPSc within strings is anchored to raft-like PM domains, plausibly through its GPI moiety.
Strings are slow moving objects, suggesting PrPSc oligomers
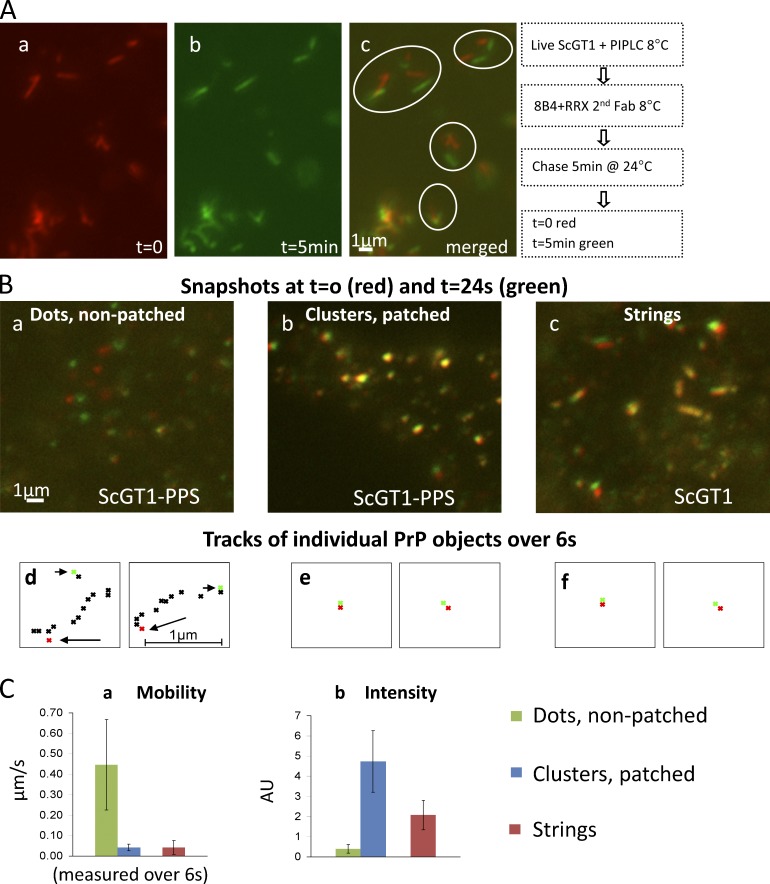
We set out to track strings in live cells. PIPLC-treated ScGT1 were labeled with 8B4 and followed by time-lapse microscopy at 24°C (Video 1). Within 5 min, individual strings had diverged from their original location (Fig. 5 A, red) by up to 3 µm (green), while retaining their identity. At 24°C, strings are thus stable, slow moving (0–3 µm/5 min) structures.
Figure 5.
Live staining: FL PrPSc strings are slowly mobile, resembling Ab-clustered PrPC patches. (A) ScGT1 were treated with PIPLC, labeled with 8B4/secondary Fab (all at 8°C), and followed by time-lapse microscopy at 24°C. Snapshots at t = 0 (red) and t = 5 min (green) are superimposed in c. (B) ScGT1-PPS (a and b) and ScGT1 (c) cells were labeled with 8B4 followed by secondary Fab at 8°C. (b) PrPC was patched with a tertiary Ab. Cells were then imaged every 400 ms for 24 s at 24°C. Snapshots at t = 0 s (red) and t = 24 s (green) are superimposed. Three types of PrP objects were followed: small dots in cells not patched with the tertiary Ab (a, uninfected), large Ab-induced clusters (b, uninfected), and strings (c, infected). (d–f) Tracks of individual PrP objects over 6 s. Starting point in red (arrow) and end point in green (arrowhead). Many small 8B4 dots (a) were too mobile to allow tracking for the 6-s period. (C) Mean mobility (a) and fluorescence intensity (b) of the three object types measured over 6 s. Both large Ab-induced clusters (blue columns) and strings (red) were mostly immobile, whereas smaller dots (green) in nonpatched samples were very mobile (∼0.4 µm/s).
GPI-APs are typically very mobile on the cell surface (approximately several micrometers per second [Sheets et al., 1997]), unless clustered (Goswami et al., 2008). We compared the mobility of strings to that of free and of Ab-clustered surface PrPC. Live GT1 labeled with 8B4/fluorescent Fabs displayed diffuse staining with occasional small PrPC dots (Fig. 5 B, a). To produce PrPC patches (Fig. 5, B and C, patched), we used tertiary polyclonal Abs (see Materials and methods). Cells were followed by time-lapse microscopy (Videos 2–4). Fig. 5 B shows an overlay of snapshots at t = 0 (red) and t = 24 s (green; tracks shown in Fig. 5 B, d–f). Whereas spontaneous PrPC dots were very mobile (0.4 µm/s; Fig. 5 C, a), both PrPSc strings and PrPC patches were almost immobilized. This supports the idea that strings contain aggregates of GPI-anchored PrP. Furthermore, FRAP studies of immunolabeled strings showed no evidence of longitudinal mobility of PrPSc in strings (Fig. S4 A). Importantly, photobleaching did not appear to sever the strings, as the nonbleached fluorescent portions remained spatially associated throughout the experiment (Fig. S4 A, arrows).
Structural details of strings: Membrane-bound amyloids
Given the amyloidogenic propensity of PrPSc in detergents, we asked whether strings might contain amyloids. ThT (Naiki et al., 1989) allows visualization of single amyloid fibrils (Ban et al., 2003). To improve the signal/noise ratio, we stained isolated PM sheets rather than whole cells. Strings labeled with 8B4 on live ScGT1 were transferred to glass coverslips using a tear-off procedure (Sanan and Anderson, 1991; Fig. 6 A, a). Subsequent ThT staining (Fig. 6 A, b) almost perfectly coincided with 8B4 in strings (Fig. 6 A, c). In many cases, the ThT staining was punctuated (Fig. S4 D). ThT was specific to PrPSc because it did not stain Ab-clustered PrPC patches in uninfected cells (Fig. S4 B, compare c and d) nor FA-denatured strings (Naiki et al., 1989; Fig. S4 C, d). This suggests that PrPSc forms amyloids when attached to membranes in its natural context (and not only in detergent lysates).
We next revisited the intriguing fluctuation of PrP immunostaining along strings (Fig. 1 C, f, arrowheads; and Fig. S1 A, b). In unsaturated micrographs, strings were clearly punctuated with both N- and core Abs even in FA-denatured samples (Fig. 6, B–D). This beaded pattern was also seen in webs (Fig. 6 D) where lateral strings (arrowheads) seem to sprout out of a master strand (arrow). 3D deconvolution revealed a beaded configuration in strings penetrating the focal plane (Fig. 6 B, arrowheads). The relative intensity of individual foci with N- and core Abs was uneven along strings (Fig. 6 C, arrows, compare a and b), suggesting variable PrP27-30 and FL PrPSc ratios.
For a more detailed view of how FL PrPSc is distributed along strings on the cell surface, we devised a method of positional correlation of immunofluorescent LM/SEM. Cells were grown on indium-tin oxide (ITO)–coated glass slides. This transparent layer is conductive enough to permit SEM imaging of immunogold-labeled cells without further metal coating (Pluk et al., 2009). Positional fluorescence/SEM correlation was achieved using a custom designed gold nanoruler built underneath the ITO coating and visible by both LM and SEM (Fig. S4 E, arrows). Live chilled PIPLC-treated ScGT1 were immunolabeled with 8B4 followed by a mixed secondary detection including both fluorescent and gold labels (Materials and methods). Cells were first visualized by fluorescence and differential interference contrast (DIC; Fig. S4 E), and then postfixed and imaged by SEM using a back-scattered electron (BSe) detector. Immunofluorescent strings correlated well with stretches of immunogold detected by SEM (Fig. 6 E, a; and Fig. S4 F). By BSe SEM, immunogold-labeled PrP in strings (Fig. 6 E, a–c) were aligned in micrometer-long threads within an estimated resolution of 40 nm (Materials and methods, Correlative LM/SEM), significantly better than the 300-nm resolution provided by LM. In contrast, Ab-patched PrPC on the surface of cured cells appeared as unorganized clusters of immunogold (Fig. 6 E, d).
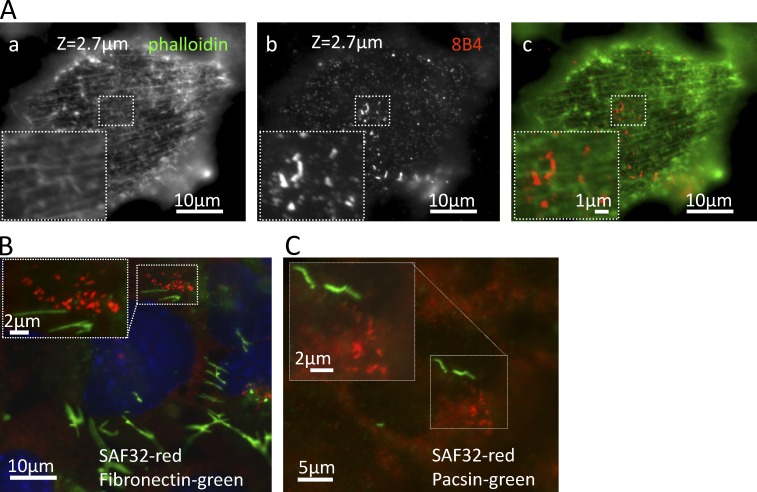
Given the strict alignment of strings observed in SEM we asked whether a cellular structure may guide strings. By immunofluorescence, we found no evidence for actin or microtubules guiding strings structure, as neither fiber aligned with strings (Fig. 7 A and Fig. S5 A) and short treatments with cytochalasin D or nocodazole (Fig. S5, B and C) or prolonged incubations on ice (Table 1) failed to disrupt strings. Longer exposure to higher nocodazole concentrations (1 µM for 1 h), however, largely dispersed strings in ScGT1 (unpublished data). Whether this is a direct effect of microtubule disruption remains to be seen. We also found no coalignment of strings with fibronectin threads (Fig. 7 B) nor with micrometer-long strands of the membrane adaptor pacsin 2 (Hansen et al., 2011; Fig. 7 C).
Figure 7.
Strings do not align with actin fibers or other fibrillar structures. (A) Fixed, permeabilized ScGT1 cells were stained with 8B4 (b and c [red]) and costained with phalloidin to stain actin fibers (a and c [green]). We found no colocalization or alignment between the strings and actin fibers. We also failed to discern higher scale organizations in fibers that would correspond to string clusters. (B and C) Cells were fixed, denatured with GdnSCN, stained with SAF32, and costained with either anti-fibronectin (B) or anti-pacsin 2 (C) Abs. No colocalization was detected.
Transmission electron microscopy (TEM) analysis of sections of prion-infected cells failed to identify recognizable cellular or extracellular structures in association with PM clusters of FL PrP (Fig. S5 D). Interestingly, PrP clusters were occasionally associated with membrane protrusions reminiscent of macropinocytic processes (Fig. S5 D, e and g).
Fl PrPSc stays in strings for >4 h, avoiding internalization
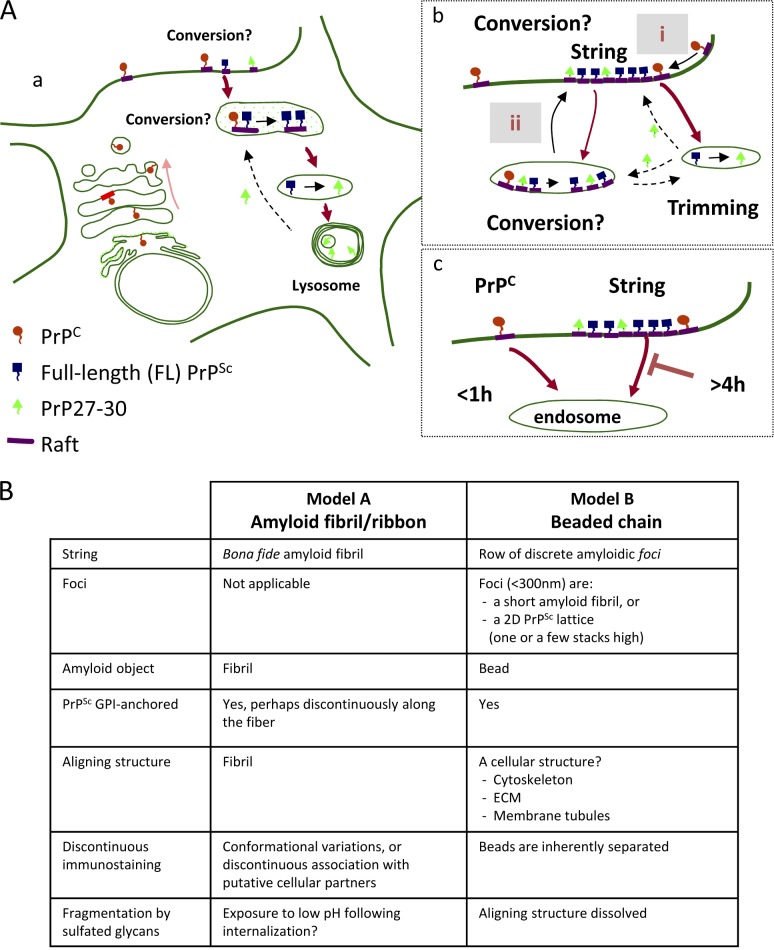
Because strings contain FL PrPSc, they must be located at or downstream of PrPSc synthesis, but upstream of endocytosis and trimming (see Fig. 10 A), providing a transit site for nascent FL PrPSc.
Figure 10.
Models. (A, a) Map of PrPSc metabolism and trafficking in infected cells. FL PrPSc is formed on rafts either on the cell surface or in an endocytic compartment. FL PrPSc is then N-proximally truncated into PrP27-30 by acidic proteases and accumulates in endolysosomes. (b) Strings acquire PrPSc either by converting cell surface PrPC (alternative i) or by importing it from intracellular conversion compartments (ii). PrP27-30 is probably recycled from hydrolytic endosomes (dashed arrows). However, PrP27-30 could also be generated by in-string truncation on the cell surface. (c) Whereas PrPC is rapidly endocytosed in subconfluent GT1, PrPSc is delayed in cell surface strings for >4 h. (B) Models of string structure. (Model A) Strings are amyloid fibrils/ribbons anchored to the cell surface. (Model B) Strings are composed of PrPSc foci aligned by yet unknown cellular structures (e.g., ECM or membrane elements; see Discussion).
To determine if FL PrPSc lingers in this transit site before internalization, we pulse-labeled live subconfluent GT1 (Fig. 8 A) and ScGT1 (Fig. 8 B) for 20 min at 37°C with a mixture of 8B4 and RRX secondary Fabs (red) followed by a 2- or 4-h unlabeled chase. To test if 8B4-labeled PrP still resided on the surface at the end of the chase, cells were chilled and incubated with cy2 secondary Fabs (Fig. 8, A and B, green). In GT1, PrPC was largely internalized after 2 h, as expected (Sunyach et al., 2003; notice the intracellular red dots and the lack of green signal in Fig. 8 A, b and c). In contrast, ScGT1 strings remained almost unchanged in number even after a 4-h chase and were still on the cell surface as they labeled in green (Fig. 8, B [b and c] and quantified in C). Thus, FL PrPSc stays in cell surface strings for >4 h. In other experiments strings gradually disappear from the cell surface between 8 and 18 h after immunolabeling (unpublished data). Importantly, strings appeared to be shorter lived in confluent cells as well as in more crowded regions of the dish, consistent with their low abundance under these conditions (Fig. S1 C).
Figure 8.
Strings are long lived (>4 h) but are rapidly fragmented and eliminated by sulfated glycans. Strings are more stable in growth-arrested cells. (A and B) Live GT1 (A) and ScGT1 (B) were stained (37°C for 20 min) with a mixture of 8B4 and RRX secondary Fabs (red), chased in unlabeled medium for 0–4 h, and then cooled on ice and labeled again with cy2 secondary Fabs (green) to reveal remnant 8B4-labeled PrP on the cell surface. PrPC rapidly internalized as shown by the intracellular red dots and the lack of green in A (b and c). In contrast, strings were still on the cell surface of ScGT1 after 4 h (B, a–c). In B (d–f), 1 µg/ml HM was included in the chase. Remarkably, strings were fragmented after 1 h (B, d) and disappeared thereafter (e and f). Notice the different scale bars in A and B: A shows entire cells, roughly outlined by the dashed line; whereas B focuses on string clusters. (C) Quantification cy2-labeled (cell surface) strings from the experiment shown in B. ScGT1 at t = 0 h and t = 4 h (with or without HM). For each time point, strings were counted out of a z-stack of four randomly chosen fields with three to five cells/field. Error bars show standard error. (D) ScGT1 untreated or treated with PPS (5 µg/ml; 1 h) were fixed, FA denatured, and stained with N-terminal mAb 8B4 (a and c) and core mAb EP1802Y (b and d). PPS treatment eliminated strings as visualized by both N-terminal mAb 8B4 and core mAb EP1802Y (c and d). Subconfluent (E) or growth-arrested (F) ScGT1 were treated with 1 µg/ml HM or 5 µg/ml PPS for the indicated times, fixed, denatured, and immunostained with 8B4 (E) or SAF32 (F). Most growth-arrested cells still had strings after 2 h of HM treatment, and 10–20% of cells retained some strings even after 12-h incubation with HM (F, c). (G) Abundant and intensely stained (mAb 8B4) strings in ScGT1 cells treated for 10 h with 20 mM NH4Cl.
Compared with PrPC, FL PrPSc is thus sheltered from premature internalization and exposure to hydrolytic endosomes (Caughey and Raymond, 1991; Taraboulos et al., 1992).
Many sulfated glycans (e.g., PPS [Caughey and Raymond, 1993]) and dextran-based heparan-mimetic HM5004 (HM; Schonberger et al., 2003) are potent prion inhibitors, but their mode of action is poorly understood (Ben-Zaken et al., 2003; Hijazi et al., 2005; Horonchik et al., 2005). When 1 µg/ml HM was included in the chase medium of the 8B4 pulse-labeled ScGT1, labeled strings rapidly fragmented (Fig. 8 B, a and d) and subsequently disappeared (Fig. 8, B [e and f] and quantified in C). HM and PPS did not merely reduce N-proximal epitopes, but rather completely obliterated the strings, as no strings were observed with either N-terminal mAbs 8B4 (Fig. 8 D, a and c) or core mAbs (Fig. 8 D, b and d) after denaturation. This presumably required an active metabolism because strings remained intact even at much higher PPS concentrations at 4°C (Table 1).
Of note, strings were much more resistant to sulfated glycans in growth-arrested cells, with some strings and webs remaining detectable with 8B4 even after 12-h incubation with 1 µg/ml HM (Fig. 8 F, compare with nonarrested cells in E). The increased stability of strings in growth-arrested cells may explain the vast abundance of strings in these cells (Fig. 1 D).
In some cases, strings/webs are the major cellular PrPSc store
Our analysis at the single cell level allowed us to refine previous biochemical studies, which suggested that FL PrPSc is quantitatively internalized and trimmed (Taraboulos et al., 1992). Here we found that the ratio of PrPSc in cell surface strings versus intracellular PrP27-30 varied among individual cells. In some cells (Fig. 9 A), there was little cytoplasmic PrP27-30 and most PrPSc was clearly concentrated in strings (compare with Fig. 2 E). More generally, in ScGT1, the proportion of strings gradually increased upon growth arrest: 3 wk after a MMC/BrdU treatment (Materials and methods), there was almost no PrPSc outside of strings (unpublished data). This parallels biochemical findings that the ratio of FL PrPSc/truncated PrP27-30 varies among cell types, with post-mitotic neurons having a high proportion of FL PrPSc (Dron et al., 2010).
Figure 9.
In some cells, strings are the major PrPSc store. (A) In some cells, strings are the major PrPSc store. ScGT1 were costained with N-terminal mAb 8B4 and core EP1802Y using the FA-denaturing protocol. z-stacks were acquired, images were processed by 3D deconvolution, and three focal planes are shown (see z-values). Images obtained with N- (a–c, red) and core (d–f, green) Abs are superimposed in g–i. The cell has little PrP27-30 in cytoplasmic stores (arrows); most cellular PrPSc (including PrP27-30) is in strings and stains with both Abs. (B) An abridged PrP lacking the N terminus can access strings. ScGT1 stably expressing the abridged PrP Δ23-89 version were fixed and costained with mAbs 8B4 (green) and 3F4 (red) with or without prior denaturation with FA (as indicated). 3F4 recognizes Δ23-89 PrP (constructed on the MHM2 platform) but not endogenous mouse PrP, whereas N-terminal mAb 8B4 does not recognize the abridged Δ23-89 PrP. In some cells, 3F4 codecorated the 8B4 strings (d–f), but only after denaturation (compare b and e), indicating that the abridged PrP are present in strings in their PrPSc form. N-proximal sequences are thus not necessary (in cis) to guide PrP to strings.
It is thought that PrPSc is trimmed in endosomes. A 10-h treatment with the lysosomotropic amine NH4Cl robustly increased the abundance of strings and their immunofluorescence intensity. Typically, 10–20% of cells became overloaded with intensely stained strings (Fig. 8 G). 5% of cells became covered with large webs (unpublished data) similar to those found in growth-arrested cells, with the rest of the population exhibiting a smaller but still significant increase. Whether this was directly caused by inhibition of acid proteases or by other effects of long NH4Cl treatment (e.g., possible trafficking jams) remains to be seen.
If PrP27-30 is generated in endosomes, its presence in cell surface strings suggests recycling events (Fig. 10 A, b). We found that a 3F4-tagged abridged Δ23-89 PrP version lacking the N terminus (Rogers et al., 1993) can localize to strings in a prion, denaturation-dependent configuration (Fig. 9 B) when expressed in ScGT1 cells. Thus, the lack of the N terminus does not prevent PrP27-30 from joining strings containing FL PrPSc (Fig. 10 A, b), at least in cells that also produce endogenous, wild-type FL PrP.
Discussion
N-proximal epitopes natively exposed: A new vista into prion biology
Using Abs to the N-proximal epitopes of PrP we show that FL PrPSc organizes in micrometer-long cell surface strings and vast webs, profoundly influencing its cell biology. Strings are the first evidence of GPI-anchored PrPSc amyloids. Their presence in infected brains reinforces their relevance to prion biology. The finding that N-proximal PrPSc epitopes are exposed in their physiological environment enabled us to visualize and track prions in live cells (Figs. 5 and 8). It should now be possible to sort living cells according to their prion status. The rapid response of strings to prion-inhibiting agents (Fig. 8 B) should aid genetic and pharmacological screens. Proteomic discovery of native PrPSc interactors should be facilitated.
As strings do not react natively with core mAbs/Fabs, they contain little if any PrPC. They do, however, contain PrPSc because they stain intensely with core Abs after denaturation, resist release from the cell surface by PIPLC, fluoresce with ThT, and resist extraction with FA (Goold et al., 2011). Furthermore, judging by the [GdnSCN] required to expose the D13 epitope, strings contain PrPSc species broadly analogous to those found in endosomes. Thus, strings do not appear to contain large amounts of protease-sensitive PrPSc, as these species tend to be revealed by core Abs at lower [GndSCN] concentrations (Tzaban et al., 2002).
Because strings align with GM1 and cav-1, they are probably GPI tethered to the PM, a conclusion supported by their tight attachment to the cell surface (Table 1) and their mobility profile, which is similar to that of Ab-clustered PrPC (Fig. 5, B and C). Incidentally, it is well known that clustering cell-surface GPI APs, including PrPC (Mouillet-Richard et al., 2000), can lead to abnormal signaling (e.g., through clustering of raft-resident Src [Suzuki et al., 2007]). Whether or not membrane-anchored PrPSc in strings elicits abnormal signaling remains to be seen. Of note, prion-infected cells and brains have elevated levels of phosphorylated Src (Nixon, 2005).
Structure of strings
Immunofluorescence indicates that strings are micrometer long, are linear but occasionally branching objects, are of undetermined width (limited by the optical resolution), and have an inhomogeneous PrP distribution. Correlative immunofluorescence/SEM improves the resolution to <40 nm (Materials and methods, Correlative LM/SEM). Both SEM and TEM confirm that FL PrPSc forms clusters on the surface of infected cells.
ThT staining hints that strings contain amyloids, but little is known of amyloid configurations of membrane-anchored GPI-APs. A GPI-anchored Sup35 yeast prion was shown to propagate on the cell surface (Speare et al., 2010), but its amyloidic characteristics have not been reported. Models of strings (Fig. 10 B) need to explain three salient findings: predominant linearity of individual strings with occasional branching and networking, discontinuous staining, and the presumed membrane anchoring of PrPSc.
By analogy to prion rods, one possibility is that strings are amyloid fibrils or ribbons (Fig. 10 B, Model A). Fibrils can occasionally branch (Harper et al., 1997), explaining PrPSc webs. Interestingly, even fibrils assembled from homogeneous precursors can stain discontinuously, e.g., fluctuating immunostaining along fibrils of anchorless PrP (Novitskaya et al., 2006; Makarava et al., 2009) and discontinuous ThT staining along fibrils of β2-microglobulin (Ban et al., 2003) or amylin (Patil et al., 2011). It remains to be seen if the punctuated staining of strings reflects structural heterogeneities or a discontinuous association with unknown cellular components. As amyloid filaments are β-helically twisted, anchoring of each individual PrPSc molecule would impose large topologic constraints on the membrane (perhaps even disrupting the bilayer continuity). Alternatively, unanchored GPI moieties might associate with hydrophobic structures within strings (e.g., lipids [Klein et al., 1998; Wille et al., 2009]) or a portion of PrPSc molecules in strings may lack the GPI moiety altogether (Borchelt et al., 1993).
Another possibility is that strings represent discrete PrPSc foci aligned by unknown cellular structures (Fig. 10 B, Model B). Foci could be short amyloid fibrils (e.g., “rods” are <200 nm long), protofibrils (Harper et al., 1997), or 2D PrPSc lattices, as described for PrP27-30 (Wille et al., 2002). Plausibly, lattices should be only a few stacks high, allowing many PrPSc molecules to be both GPI anchored to the PM and accessible to mAb 8B4. The TEM results suggest that FL PrP is immediately adjacent to the surface of infected cells (Fig. S5 D). What cellular architecture could possibly align PrPSc foci? ECM threadlike components are one possibility. Heparan sulfate–binding regions of PrP N terminus (Warner et al., 2002) could conceivably hold foci onto ECM guides, but this interaction should withstand a strong PPS, HM, or NaCl challenge in chilled cells (Table 1).
Tubulation driven by protein-membrane scaffolding could provide another aligning mechanism. Multimeric raft ligands can induce micrometer-long membrane tubules in cells and in liposomes, e.g., SV40 and Shiga toxin B (Römer et al., 2007; Ewers et al., 2010). Being a multimeric raft component, PrPSc could perhaps also induce membrane tubulations. Interestingly, SAF32-labeled PrPSc was found in PM invaginations in neurites in the hippocampus (Godsave et al., 2013).
A groundbreaking atomic force microscopy study visualized assemblies of 0.5–4-µm-long fibrillar protrusions covering the ScN2a surface (Wegmann et al., 2008). Whether or not these structures contain PrP was not established, but their length and kinetics of appearance (days after plating) are reminiscent of strings. Further work will establish whether these atomic force microscopy protrusions relate to strings.
Where do strings acquire their PrPSc content?
One possibility is that strings produce their own PrPSc. Indeed, PrP amyloid fibrils can accrete and convert PrPC. It is not yet known whether 2D PrPSc lattices can also expand laterally by templating PrPC. The prolonged residence of strings on the cell surface (Fig. 8 B), where PrPC is abundant, could provide opportunities for the conversion. However, import of FL PrPSc from distal compartments, perhaps together with PrP27-30, is also possible.
The generation of C2/PrP27-30 is thought to involve acidic proteases; trimmed PrP27-30 could be recycled from endosomes (Fig. 10 A, b, dotted arrows). Indeed, the N terminus was not required for the trafficking of an abridged Δ23–88 PrP to strings (Fig. 9 B), at least in cells coexpressing wild-type FL PrP. String PrP27-30 may also originate from cell surface digestion. Examples of extracellular acidic proteolysis include macrophages resolving cholesterol plaques (Haka et al., 2009) and osteoclasts attacking mineralized bone.
Delayed PrPSc internalization
Compared with PrPC, the internalization of strings was strikingly delayed (Fig. 8, A–C; and Fig. 10 A, c, red arrows), perhaps contributing to the large amount of cell surface FL PrPSc observed in the brain (Jeffrey et al., 1996; Bouzamondo-Bernstein et al., 2004; Dron et al., 2010). It will be important to establish if the aggregation of PrPSc plays a role in its prolonged cell surface residence.
How strings are eventually removed from the cell surface remains to be established. Our failure to observe intracellular strings suggests that they fragment before endocytosis. However, they could also disassemble soon after internalization, as artificial PrP fibrils, for instance, can rapidly disintegrate at pH 4.5 (Qi et al., 2012). Whether or not strings internalize via LRP1 (Jen et al., 2010), via raft-dependent mechanisms, or by other ways remains to be determined. Clathrin-coated pits are limited to a 200-nm cargo (Conner and Schmid, 2003) and thus could accommodate single PrPSc foci but not whole strings. Whole string internalization would require alternative processes such as macropinocytosis (Wadia et al., 2008). It is also possible that strings switch between various endocytic mechanisms depending on the cell physiology, for instance, in response to changes in the local cell density, as shown for viral endocytosis (Snijder et al., 2009).
It will be important to determine if the delayed PrPSc internalization affects the biology of prions. In the mouse brain, the half-life of RML PrPSc (a protease-resistant strain) is only 1.5 d (Mallucci et al., 2003; Safar et al., 2005). Presumably, protease-sensitive strains such as transmissible mink encephalopathy drowsy (Bessen and Marsh, 1992) are even more labile. For such strains, the organization of PrPSc in strings and the prevention of premature lysosomal challenge could be crucial.
Our findings raise several questions. What is the ultrastructure of strings and webs? Are they aligned by cellular elements, and are these elements organized ad hoc by prion infection? How are strings internalized? Why are strings so sensitive to sulfated glycans? How significant is the raft engagement by strings to prion pathogenesis? More generally, it is tempting to ask whether cell surface strings/webs illustrate general phenomena related to the aggregation or clustering of membrane proteins. Such situations could include functional amyloids (Berson et al., 2003; Huff et al., 2003), clustering of PrPC by its Aβ ligand (Freir et al., 2011; Resenberger et al., 2011), controlled homotypic interaction of membrane proteins in the ER (Lingwood et al., 2009), and perhaps clustering of cellular receptors by multimeric viral and bacterial ligands.
Materials and methods
Reagents
Cell culture reagents were obtained from Biological Industries except for OptiMEM, Neurobasal medium, and the B27 supplement (Invitrogen). Tissue culture dishes and chamber slides were purchased from Thermo Fisher Scientific. HM was provided by D. Barritault (Université Paris XII, Créteil, France). Cross-linkable Alexa 555–dextran, Cascade blue dextran, G418 sulfate, Alexa Fluor 488 and Alexa Fluor 555 CTxB, LysoTracker DND-99, BODIPY, and PIPLC were obtained from Invitrogen. N-Glycosidase F (PNGase F) was obtained from New England BioLabs, Inc. Fluorescent aerolysin (FLAER) was purchased from Protox Biotech. Phalloidin-tetramethylrhodamine, DAPI, Hoechst 33258, MMC, BrdU, and other reagents were purchased from Sigma-Aldrich.
PrP Abs
We used the following PrP Abs and dilutions for immunofluorescence: mAb 8H4 (5 µg/ml) and the N-terminal mAb 8B4 (2–5 µg/ml; Li et al., 2000) obtained from Alicon. SAF32 (2 µg/ml; Demart et al., 1999; Perrier et al., 2004; 189720) was obtained from Cayman Chemical. Core mAb 3F4 (5 µg/ml; Kascsak et al., 1987) was purchased from EMD Millipore (MAB1562). It binds to residues Met108 and Met111 (Rogers et al., 1991) in chimeric mouse/hamster MHM2-PrP but does not recognize the wild-type mouse PrP endogenous to GT1 cells. Mouse mAb IPC1 (Canello et al., 2008) was a gift from R. Gabizon (Hadassah University Hospital, Jerusalem, Israel). IPC1 was obtained by immunizing PrP0/0 mice with recombinant mouse PrP. It is available from Sigma-Aldrich (p5999). Mouse mAb W226 (Petsch et al., 2011) was a kind gift from L. Stitz (Friedrich-Loeffler-Institut, Tuebingen, Germany). It was produced from the immunization of PrP0/0 mice with phosphotungstic acid-purified mouse prions. Humanized PrP Fabs D13, R1, and R2 (10 µg/ml) (Williamson et al., 1998) and EST123 (10–20 µg/ml; Peretz et al., 2001; Leclerc et al., 2003) were purchased from InPro Biotechnologies. Of note, the EST123 epitope appeared to be fixation sensitive and reacted only on living cells. Octarepeat mAb 5B2 (2 µg/ml; Li et al., 2000) was purchased from Santa Cruz Biotechnology, Inc. Rabbit mAb EP1802Y (2 µg/ml) was purchased from Epitomics.
Epitopes recognized by PrP Abs
The following epitopes were recognized by PrP Abs: EST123: residues 29–37; 8B4: 37–44; SAF32: octarepeats; D13: 96–106; 3F4: M109 and M112 in MHM2; IPC1: 144–152; W226: 145–153; R1 and R2: 225–231; and EP1802Y: 214–230.
Other primary Abs
Rab9 (SC28573), Rab7 (SC10767), EEA1 (SC6414), calnexin (SC6465), BiP (SC13968), Rab5A (SC309), Rab4A (SC312), Rab11 (SC9020), GM130 (SC30100), clathrin HC (SC6579), and M6PR/IGFIIR (SC25462) were obtained from Santa Cruz Biotechnology, Inc. Lamp1/CD107a (clone 1D4B; 121602) and transferrin receptor/CD71 (113809) rat mAbs were obtained from BioLegend. β-Tubulin mouse mAb (clone TBN06, IgG3, MS-1226-P0) was obtained from Lab Vision. Caveolin-1 antiserum (610060) was obtained from BD. Fibronectin (F3648) and pacsin 2 (SAB1300127) antisera were obtained from Sigma-Aldrich.
Working batches of primary Abs were diluted in 1% BSA in PBS, supplemented with 0.1% thimerosal to prevent microorganism contamination.
Secondary Abs
Secondary Abs including subtype-specific anti-mouse IgG, Fab fragments, and gold coupled anti-HRP were obtained from Jackson ImmunoResearch Laboratories, Inc., except the Alexa-coupled Abs (Invitrogen) and HRP-coupled anti-mouse Fab (Abcam). When necessary, we used secondary Abs that had been cross-absorbed against IgG from other species, to prevent cross-labeling. Great care was taken to control this issue.
Cells and prion strains
Cell lines were maintained in DMEM-OptiMEM (1:1) with 5% heat-inactivated FBS, penicillin, streptomycin, and glutamine (D/O medium) and were typically split at 1:4 using 0.05% wt/vol trypsin (in 0.004% wt/vol EDTA in PBS) once a week. For immunofluorescence, cells were seeded on 8-well Permanox or coverglass bottom chamber slides.
GT1-1 (herein called GT1) are immortalized mouse hypothalamic neurons (Mellon et al., 1990) susceptible to prion infection (Schätzl et al., 1997). ScGT1 and ScGT1/22L are GT1 cells chronically infected with the RML or 22L (Nishida et al., 2000) strains of mouse-adapted scrapie, respectively. SMB cells persistently infected with the Chandler scrapie isolate (Clarke and Haig, 1970) and their PPS-cured counterpart, and the 22L strain of mouse-adapted scrapie were purchased from the TSE Resource Center (Veterinary Laboratories Agency Weybridge). Infected cells were cured by exposure to 5 µg/ml PPS for 1 mo; we detected no protease-resistant or insoluble PrP in these cells for up to 3 yr after curing (Fig. S2 A). ScN2a-cav are chronically RML-infected mouse N2a cells stably expressing human caveolin-1 under G418 selection (Naslavsky et al., 1999). GT1-Δ23-89 stably express an abridged mouse PrP (lacking the N terminus) on the MHM2 platform under G418 selection (Scott et al., 1992). CAD (cath.a differentiated) cells (Qi et al., 1997) are a variant of the central nervous system catecholaminergic line cath.a. Their infectibility with RML and ME7 has been reported previously (Mahal et al., 2007). Min6 is a mouse pancreatic β line (Miyazaki et al., 1990) whose susceptibility to prion infection is reported here for the first time.
DRGs (Fig. S1 I) were dissected from embryonic mice as described previously (Backström et al., 2000). In brief, the dissected ganglia from 15–16-d embryonic C57BL/6 mice were dissociated by several passages through a constricted Pasteur pipette and the cells were seeded into 35-mm Petri dishes (1.5 × 105 cells/dish; Corning) coated with collagen (Vitrogen 100; Collagen Biomaterials). The cells were then grown in a culture medium based on serum-free Neurobasal medium and B27 supplement (1:50), l-glutamine (1:100), gentamicin sulfate (15 µg/ml; all obtained from Gibco), and nerve growth factor (1 ng/ml; Sigma-Aldrich) for 6 d. ScGT1 cells were then added and cocultivated for 5 d in the same medium. The cells were then fixed, permeabilized, denatured with GdnSCN, and stained with mAb 8B4.
Optimization of strings
In ScGT1 cells, the abundance of strings increased in the days after seeding, as long as the culture stayed subconfluent; however, strings disappeared at confluence (Fig. S1 C, b). To prepare ScGT1 for string immunofluorescence, it is crucial to start with a subconfluent ScGT1 stock and to thoroughly disperse cells on seeding. Strings disappear upon trypsinization and slowly reappear in the following days. In cells split with trypsin, abundant strings are routinely obtained when ScGT1 cells are seeded at 5–10% confluence and grown for 2–3 wk. In cells split without trypsin (0.05% wt/vol EDTA in PBS), strings are already present the day after seeding and the proportion of string-positive cells often surpasses 90% after 5 d. Spitting cells without trypsin was especially important for obtaining strings when using glass-bottomed slides. We found that NH4Cl administered for 10 h before the experiment often increased the abundance and intensity of strings (Fig. 8 G).
Cytostatic regimen
GT1 and ScGT1 were treated with MMC (0.04 µg/ml, 6 h) followed by MMC (0.02 µg/ml)/BrdU (5 µM) for 5d and refreshed with medium without MMC/BrdU. 6 d after starting the treatment, string prevalence was 90–100%; 3 wk later cells were covered by vast PrPSc webs and had almost no intracellular PrP27-30. A variety of substrates, such as polylysine, matrigel, gelatin, and Cell-Tak, did not increase (and in some cases decreased) strings. Of note, strings were abundant in ScGT1 cells seeded on ITO-coated slides (see Correlative LM/SEM section).
In SMB cells, strings were more abundant in D/O with 5% FBS than in M199 with 10% FBS and 5% newborn calf serum (the medium recommended by the supplier). Similar to ScGT1, SMB cells also lost strings after reaching confluence. In ScN2a-cav cells, strings were especially visible in cultures growth arrested by a 7-d treatment with the minor groove binding agent distamycin A (10 µM; Fig. S1 F).
PIPLC treatment of live cells at low temperature
Cells grown on multiwell slides or 60-mm plates were cooled to 8°C, washed twice with cold PBS, and incubated with PIPLC (1–2 U/ml in PBS) for 1 h at 8°C.
Immunofluorescence
Fixed cells.
Cells grown on multiwell slides were fixed (45 min at RT) with 12% formalin in PBS or with 4% formaldehyde in PBS, with indistinguishable results, quenched (1% NH4Cl wt/vol in PBS), and permeabilized with Triton X-100 (either 0.01% or 0.1% vol/vol, with indistinguishable results, in PBS, at RT for 5 min). Strings were also decorated with N-Abs when cells were fixed stringently with formalin (12%, 90 min) and post-fixed with MeOH at −20°C to prevent the potential Ab-induced patching of GPI-APs (Mayor et al., 1994; Harder et al., 1998; Tanaka et al., 2010), or when they were fixed only with MeOH.
In some experiments, formalin-fixed cells were then denatured either with 98% FA (7 min at RT) or 3 M GdnSCN (5 min at RT) to unmask PrPSc epitopes and then thoroughly rinsed (see In situ denaturation).
Cells were then blocked with BSA (1% wt/vol in PBS for 1 h at RT). All primary and secondary Abs were diluted in blocking buffer (1% wt/vol BSA in PBS) and cleared by ultracentrifugation (100,000 g for 15 min at 4°C) before use. Cells were incubated with primary Abs (either 2 h at RT or 18 h at 4°C, with indistinguishable results) followed by secondary Abs for 45 min at RT.
To stain cells with phalloidin, they were fixed with formaldehyde at 37°C (to preserve both the integrity of the actin cytoskeleton and the phalloidin binding). Cells stained for microtubules were fixed with methanol (10 min at −20°C; Fig. S5 A).
To stain nuclei, DAPI (10 µg/ml) or Hoechst 33342 (1 µg/ml) were mixed either with the secondary Abs or included in the mounting solution. Cells were mounted in N2-saturated 80% vol/vol glycerol and 100 mM Tris-Cl, pH 9, supplemented with 5% wt/vol N-propyl gallate (Giloh and Sedat, 1982).
In ScGT1 and in cured ScGT1-PPS monolayers, we observed that strings are often most abundant in flat, nondividing cells. In contrast, PrPC is usually more abundant in round and dividing cells. For this reason, ScGT1 cells chosen for their abundant strings often appear to have less immunolabeled PrPC than their cured ScGT1-PPS counterparts (e.g., Fig. 1 C, a and b). Compounding this bias, we also found that the region of the PM that carries strings is sometimes depressed (in terms of focal plane) relative to the surrounding PM regions. Thus, micrographs of strings often to do not capture surface PrPC to the correct extent.
In situ denaturation
Since the discovery that PrPSc is poorly immunoreactive (Serban et al., 1990), a variety of techniques have been developed to retrieve its epitopes. FA (Kitamoto et al., 1987) and hydrolytic autoclaving (Kitamoto et al., 1992) are among the most efficient means of exposing PrPSc in brain tissue. In contrast, immunofluorescent studies of prions in cultured cells have most often used guanidine salts, such as 3 M GdnSCN or 6 M GdnHCl (Taraboulos et al., 1990b).
Here we found that these two chaotropes do not efficiently expose core PrP27-30 epitopes within strings. Using 3 M GdnSCN, core Abs stained only intermittently along the strings delineated by N-Abs (Fig. S3 B). In contrast, FA resulted in a much more continuous staining of strings by core Abs (e.g., compare the patterns of Fab D13 immunostaining in Fig. S3 D).
Cell surface labeling/clustering with PrP Abs
For labeling at low temperature, cells were rinsed with ice-cold PBS and incubated with the primary Abs diluted in 1% BSA in PBS (1 h, either on ice or at 8°C with indistinguishable results), followed by secondary Abs diluted in 1% BSA in PBS (45 min, either on ice or at 8°C). When needed, CTxB (2–6 µg/ml) was added to the latter step; in this case both the toxin and the secondary Ab were resuspended in 0.2% BSA. Secondary Fab fragments were used to minimize cell surface patching. To induce cell surface clusters, we used either polyclonal secondary Abs (Fig. S4 B) or secondary Fabs in conjunction with a tertiary polyclonal Ab (Fig. 5, B and C; and Fig. 6 E, d). Cells were then fixed and examined by fluorescent microscopy and SEM.
Live tracking of PrPC and strings using time-lapse microscopy
For the experiments described in Fig. 5 (A and B) and Videos 1–4, chilled cells were cell surfaced labeled with 8B4 followed by Fab-RRX as described in the previous paragraph. To produce PrPC patches (Fig. 5, B and C, patched), we used tertiary polyclonal Abs at a concentration adjusted so that the integrated 8B4 intensity in patches (Fig. 5 B, b) was about twice that in PrPSc strings (Fig. 5 B, c, per 300 nm/unit length; and Fig. 5 C, b, histogram; n = 28 for each object type). The 1/2 immunostaining ratio was chosen to account for the reduced native 8B4 reactivity in strings (Fig. S3 F). Labeled cells were then placed in full D/O medium (without phenol red, with 10 mM Hepes, pH 7.3), gradually warmed to 24°C and then examined by time-lapse microscopy.
Pulse-chase immunofluorescence labeling experiments
In Fig. 8 (A and B), the cells were immunolabeled for 20 min at 37°C in full D/O medium including a mixture of 8B4 and RRX secondary Fabs. To prepare this Ab mixture, Abs (25 µg/ml 8B4 and 300 µg/ml RRX-coupled secondary Fabs) were incubated together for 30 min on ice in 1% BSA in saline. They were then cleared by a high speed centrifugation (100,000 g for 15 min at 4°C), diluted 1:10 in full D/O medium and applied to the cells. After chase periods in unlabeled medium, the cells were cooled to ice temperature and labeled with Cy2 secondary Fab (30 µg/ml) to restain 8B4 still residing on the cell surface.
Competing mAb 8B4 with a specific peptide
The PrP specificity of mAb 8B4 staining of strings was verified using the synthetic peptide N-CRYPGQGSPGRYPGQGSPG-COOH (Sigma-Aldrich), which contains a duplicate of the established epitope RYPGQGSPG (Li et al., 2000). 8B4 and SAF32 (2 µg/ml) were mixed with increasing concentrations of the peptide (0–8 µg/ml) in PBS + 1% BSA and incubated for 1 h at RT before cell immunolabeling (Fig. S2 D).
Staining live cells with tracers and Abs
Cells growing on multiwell slides (either Permanox or coverglass bottomed slides) were incubated with the following tracers (added to the cell medium at 37°C) before microscopic examination (Table S1): fixable Alexa 555 dextran or Cascade blue dextran (250 µg/ml, 2–5 h), LysoTracker (100 nM, 45 min), BODIPY (to stain neutral lipids in lipid droplets; 5 µg/ml), and Alexa 488–labeled aerolysin (25 nM; see Table S1).
Cell lysis and enzymatic treatments
Cells were lysed and prepared for Western immunoblotting essentially as described previously (Naslavsky et al., 1997; Tzaban et al., 2002). Cells were scraped off the dish in ice-cold PBS, spun down in a microcentrifuge (10 min, 4°C, gav = 260 g), and then resuspended and lysed with ice-cold lysis buffer (0.5% Triton X-100, 0.25% sodium deoxycholate, 150 mM NaCl, 10 mM Tris-HCl, pH 7.5, and 10 mM EDTA). Lysates were immediately cleared in a microcentrifuge (5 min, 4°C, gav = 5,000), and biochemical analyses were performed on the supernatant. Protein concentrations were measured using the Bradford assay (Bio-Rad Laboratories) and samples were typically normalized for their protein content before loading on SDS-PAGE gels. Protease-resistant PrPSc was defined as the PrP fraction resistant to proteolysis catalyzed by proteinase K (20 µg/ml, 37°C, 30 min). Proteolysis was stopped by adding PMSF to a final concentration of 2 mM followed by a 20-min incubation on ice. Deglycosylation of samples with PNGase F was performed as per the manufacturer’s instructions.
Gel electrophoresis and Western blots (WBs)
Samples were supplemented with Laemmli loading buffer ×4 (containing β-mercaptoethanol) and incubated at 95°C for 10 min before loading on 12.5% polyacrylamide (acrylamide/bisacrylamide, 29:1) gels and electrophoresis. Samples were electro-transferred to PVDF membranes in a Tris/glycine transfer buffer (Towbin et al., 1979) containing 0.001% wt/vol Sarkosyl instead of SDS. Membranes were blocked with 1% fat UHT milk (30 min at RT), rinsed with TBST (10 mM Tris HCl, pH 7.4, 150 mM NaCl, and 0.3% Tween 20) and sequentially incubated with the primary Abs (2 h at RT or 12 h at 4°C) and the secondary (30 min at RT) Abs, both in TBST. Membranes were then rinsed extensively in TBST, developed with ECL reagents (100 mM Tris, pH 8.5, 0.05% H2O2, 1.25 mM luminol, and 0.2 mM p-coumaric acid) for 1 min, and imaged. For WB with either 8B4 or SAF32, two high salt rinsing steps (each 10 min at 4°C; 450 mM NaCl, 0.3% Tween 20, and 30 mM Tris-Cl, pH 8) were added after incubation with the primary Ab. After two further rinses in regular salt conditions, PVDF membranes were reblocked with 1% fat UHT milk. In some experiments membranes were stripped from both primary and secondary Abs with 300 mM NaOH (5 min at RT), and then rinsed with DDW, equilibrated with TBST, blocked, and reprobed with other Abs.
Velocity sedimentation in sucrose gradients
The entire procedure was performed on ice. 7 × 106 cells were scraped off the plate in ice-cold Hepes buffer (20 mM Hepes, pH 7.4, 150 mM NaCl, 0.1 mM CaCl2, and 1 mM MgCl2), and then lysed for 30 min (on ice) in 350 µl Hepes buffer containing 2% wt/vol N-octyl glucopyranoside. Lysates were then cleared in a microcentrifuge (5 min, 4°C, gav = 5,000). Supernatants were brought to 1% Sarkosyl, incubated for 30 min on ice, and then loaded on top of a 10–60% sucrose step gradient in TNS (10 mM Tris, pH 7.5, 150 mM NaCl, and 1% Sarkosyl). Gradients were built in polyallomer (11 × 34 mm) tubes from 300 µl of each of the following sucrose concentrations: 10, 15, 20, 25, 30, and 60% in TNS. The gradients were spun for 1 h at 4°C at 55,000 rpm (gav = 225,000 g) in a TLS-55 rotor of an Optima TL ultracentrifuge (Beckman Coulter). Eleven fractions (180 µl each) were collected from the top of the tube.
Fab blocking procedure
In the experiment described in Fig. 3 B, we used 8B4 twice to detect two different pools of PrP. Living cells on ice were surface stained with 8B4 followed by a secondary Fab-RRX (step 1). Step 2 was as follows: cells were fixed with formaldehyde, permeabilized, and postfixed with methanol (−20°C), reblocked with BSA, and then incubated with an excess of unlabeled anti-mouse Fabs. 8B4 was then used again to detect intracellular PrP (followed this time by secondary Abs coupled to Alexa 488). This Fab-blocking procedure (adapted from Sunyach et al. [2003]) was performed to minimize the reaction of the secondary Ab used after fixation (in step 2) with cell-surface 8B4 mAb remaining from step 1.
PM tear-off and ThT staining
PM sheets of ScGT1 or of PPS-cured cells were prepared as described previously (Sanan and Anderson, 1991). Cells grown on glass coverslips were immunolabeled on ice with mAb 8B4 followed by RRX secondary Fab and then pressed for 20–30 s onto polylysine-coated glass coverslips. The polylysine-coated coverslips were detached, rinsed in ice-cold PBS, and immediately fixed with 12% formalin in PBS (15 min on ice followed by 30 min at RT), and rinsed with PBS. They were then incubated for 10 min at RT in 0.05% wt/vol ThT freshly made in 50% EtOH. Coverslips were then dipped twice in 80% EtOH and rinsed 3× in a large volume of ddH2O followed by a 1.5-h incubation in PBS at 4°C before mounting. In Fig. S4, fixed coverslips were incubated in 98% FA (at RT for 2 min) and then thoroughly rinsed before ThT staining.
Microscopy and image analysis
Cells or PM tear-off preparations were examined with a microscope (Axiovert 200M; Carl Zeiss) equipped with a motorized z-drive, a motorized XY stage, and a CoolSnap HQ cooled CCD camera (Princeton Instruments). Microscopy was performed at RT. We used an NA = 1.3, 100× Plan Neofluar oil objective (except in Figs. S3 I and S1 B, where NA = 1.4, 63× Plan Apochromat, and NA = 1.3, 40× Plan Neofluar, objectives were used, respectively), in conjunction with High Efficiency filter sets (Carl Zeiss). Images were acquired and processed using the MetaMorph software (Molecular Devices). In many cases, z-stacks at 0.2–0.3-µm intervals were acquired. In some cases (indicated in the relevant figures), nearest neighbor 2D deconvolution (MetaMorph) was used to enhance the signal/noise ratio. In other cases, z-stack images were 3D deconvolved by a blind deconvolution protocol using the Huygens software from Scientific Volume Imaging. To detect the fluorescence shift of ThT, we used the 47 High Efficiency filter set (excitation 436/25 nm, emission 480/40 nm; Carl Zeiss). Long exposures (up to 15 s) were needed to detect ThT fluorescence of strings.
In some figures, the cell perimeter was outlined using DIC images (Fig. 1 C, a, b, d, and e; Fig. S2 E; and Fig. 2 E), or filipin fluorescence (Fig. 3 A; filipin staining not depicted).
FRAP.
FRAP experiments were performed in an LSM710 confocal microscope (Carl Zeiss) equipped with a 37°C incubator using an NA = 1.4, 63× Plan Neofluar objective. Live SMB cells labeled with 8B4/RRX secondary Fab were photobleached and then observed for up to 30 min by time-lapse microscopy. Images were acquired using the Zen software.
Correlative LM/SEM
Correlation LM/SEM was conducted on cells grown on ITO-coated glass. This transparent layer permits both LM and SEM imaging and is conductive enough to permit SEM imaging of immunogold-labeled cells without further metal coating (Pluk et al., 2009). The slides were custom built by the Center for Nanoscience and Nanotechnology (The Hebrew University of Jerusalem). Positional LM/SEM correlation was achieved by a gold nanoruler deposited on the slides, which was visible in both microscopy modes. The microscope glasses (0.17–0.19-mm thick; IBIDI) were coated by evaporation with a 10-nm titanium layer followed by a 50-nm gold layer. Mask-assisted optical UV lithography and wet etching were then used to create a numbered titanium-gold topographic nanoruler, which was visualized by both LM and SEM (Fig. S4). Slides were then cleaned by ultrasonication and overlaid with a transparent 40-nm ITO conductive layer (by sputtering). The slides were then mounted with self-adhesive 8 wells (sticky wells; IBIDI) and sterilized with 70% ethanol.
Cells were grown on these 8-chambered ITO-coated slides. Live ScGT1 and ScGT1-PPS cells cooled to 8°C were incubated sequentially with mAb 8B4, HRP-labeled secondary anti-mouse Fabs from goat, and goat anti-HRP Ab coupled to 18-nm gold. This immunogold staining was followed by donkey anti-goat Fab (in ScGT1 cells) or whole IgG (for PrPC patching in ScGT1-PPS cells) coupled to RRX. The HRP-labeled secondary Fab served as a spacer to enhance immunogold labeling of the cell surface PrP. In some cases the cells were treated with PIPLC (2 U/ml in PBS; 8°C for 1 h) before immunolabeling. Cells were then fixed (4% formaldehyde in 0.1 M sodium cacodylate for 20 min on ice, and then 1 h at RT), rinsed with 0.1 M sodium cacodylate, and imaged with a 1.3 NA oil, 100× objective for fluorescence and DIC. Slides were also imaged by phase microscopy at lower magnifications for orientation purposes. The cells were then postfixed with 2% formaldehyde/2.5% glutaraldehyde (0.1 M sodium cacodylate, for 1 h at RT), rinsed thoroughly, and postfixed with 1% OsO4 and 1.5% potassium ferricyanide (0.1 M sodium cacodylate, for 1 h at RT), thoroughly rinsed, dehydrated in a graded ethanol series, and then gradually substituted with Freon 114. Keeping the slides in Freon overnight at 4°C facilitated the removal of the sticky wells. Freon remnants were evaporated in a chemical hood.
Samples were imaged in a Magellan 400L Extra High Resolution SEM equipped with a Schottky field emission gun (Center for Nanosciences and Nanotechnology). Colloidal gold (18 nm) was detectable using BSe at magnifications as low as 5,000. For this purpose, we used a variable contrast solid-state BSe detector (vCD) at slow (≥3 µs/well) high resolution scans. Samples were imaged in immersion mode with a landing energy of 5 kV and a beam current of 800 pA at working distances of 3.5–4 mm (Figs. 6 E and S4 F). The size of the gold particles determined at higher magnifications corresponded to monodisperse 18-nm gold as expected. In our hands, the nanoruler was absolutely required for LM/SEM correlative imaging of immunolabeled objects of micrometer and submicrometer length (such as PrPSc strings).
The spatial resolution of the SEM experiments is <40 nm limited by the length of the Ab bridge (12 nm for IgGs: 8B4 and gold-labeled tertiary Ab, 7 nm for the bridging Fab, and 9 nm gold radius [Hermann et al., 1996]). However, examining straight strings suggests that the actual resolution is even better (Fig. 6 E).
Tissue and cell processing for cryosectioning
Animals and preparation of tissue.
Prion-infected and control brain tissues were provided by H. Wille and S.B. Prusiner (University of California, San Francisco, San Francisco, CA). Wild-type FVB mice were injected intracerebrally with 30 µl of 1% brain homogenate from mice infected with RML prions or from normal mice as controls. RML-infected mice were killed at 104–106 dpi and control mice were killed at 116 dpi. Onset of overt prion disease occurs at ∼120 dpi. Mice were killed according to the Public Health Service (National Institutes of Health) Guide for the Care and Use of Laboratory Animals and transcardially perfused with fixative containing 2% PFA and 0.2% glutaraldehyde in PHEM buffer (25 mM Hepes, 10 mM EGTA, 60 mM Pipes, and 2 mM MgCl2, pH 7.2), as described previously (Mironov et al., 2003). Brains were postfixed in the same fixative for 16–24 h, transferred to 1% PFA in PHEM buffer for 7 d, and then stored in 0.5% PFA in PHEM.
Preparation of cells.
Min6 and Min6/RML cells growing on 90-mm dishes were fixed by addition to the culture medium of an equal volume of 4% PFA in 2× PHEM buffer for 24 h at RT. The fixative was replaced with PBS containing 0.15 M glycine (PBSG) and cells were washed twice with PBSG. The PBSG was then replaced by 5 ml of prewarmed 1% gelatin in PBS and left for 30 min at 37°C. Cells were scraped with a rubber policeman, pelleted in an Eppendorf tube, resuspended into 1 ml of prewarmed 12% wt/vol gelatin in 0.1 M of phosphate buffer, pH 7.2 (at 37°C), and then pelleted again. The gelatin was allowed to set at 4°C.
Cryosectioning.
Blocks of tissue, 1 mm3, were cut from the hippocampus CA1 region or from gelatin-embedded cell pellets. These were rinsed first with PBS, then with PBS containing 0.02 M glycine, transferred into 2.3 M sucrose as cryoprotectant, and rotated end over end for 16 h at 4°C. Blocks were then placed onto metal pins for cryosectioning and frozen in liquid nitrogen (Peters et al., 2006). Cryosectioning was performed at −110°C using an Ultratome UCT with cryo-attachment (Leica). Sections for cryoimmunogold electron microscopy, 70- or 200-nm thick, were picked up from the diamond knife with 1% methyl cellulose 25CP and 1.15 M sucrose solution, and transferred to formvar/carbon-coated copper grids (hexagonal grid bars, spaced 200 µm across). Sections for immunofluorescence were 200- or 500-nm thick and were picked up using 2.3 M sucrose and placed on glass slides.
For SAF32 immunofluorescence of brain tissue (Fig. 1 E), we performed an antigen retrieval step before blocks were infiltrated with sucrose. The blocks were rinsed successively with PBS, PBSG, and 0.01 M sodium citrate, pH 6.0 (antigen retrieval solution), and then heated at 95°C in de-gassed antigen retrieval solution for 30 min. The blocks were then allowed to cool slowly (20 min) and rinsed with PBS before transfer into sucrose solution.
Immunofluorescence labeling of cryosections.
Cryosections were rinsed twice with PBS and five times for 3 min with PBSG. Non-specific Ab binding was then blocked with PBSG containing 0.5% cold fish gelatin and 0.1% BSA (gel + BSA) for 15 min. Sections were incubated with 1 µg/ml SAF32 in PBS containing gel + BSA for 1 h at RT. Slides were rinsed five times for 3 min with PBSG before incubating for 30 min with Alexa 488–conjugated goat anti–mouse IgG (Invitrogen) in PBS containing gel + BSA. Sections were washed five times for 3 min with PBSG and then mounted in Vectashield (Vector Laboratories) containing 2 µg/ml DAPI. Images were made using an AxioObserver Z1 inverted microscope (Carl Zeiss), equipped with an ORCA AF Black and White CCD camera (Hamamatsu Photonics) and axiovision software (Carl Zeiss), which adds false colors to the images.
Immunolabeling for TEM.
Grids were placed with sections facing down on plates of 2% gelatin in a 37°C incubator for 30 min. Grids were then floated with the sections facing down onto drops of washing and Ab solutions: PBSG, 5× 2 min; gel + BSA in PBSG, 5 min; SAF32 Ab at 1 µg/ml in PBS containing gel + BSA, 1 h; PBSG, 5× 3 min; PBSG containing 1% BSA, 3 min; bridging Ab rabbit anti–mouse IgG (Dako) in PBS containing 1% BSA, 30 min; PBSG, 5× 2 min; PBSG containing 0.1% BSA, 3 min; Protein A conjugated to 10-nm gold particles (Utrecht Medical Center) in PBS containing 1% BSA, 20 min; PBS, 3× 5 s, followed by 6×, 3 min; 1% glutaraldehyde in PBS, 5 min; milliQ water, 10× 2 min; 0.6% uranyl acetate in 1.8% methyl cellulose, pH 4.8, 5 min. Grids were then picked up in wire loops, excess uranyl solution was drained off, and grids were allowed to dry. Electron microscopy was performed using a CM10 microscope (FEI) at 80 kV.
Recommendations for optimal string detection
In our hands, the following considerations need to be met to obtain satisfying immunofluorescent images of strings. (a) The best is to adhere to the culture conditions outlined in the Optimization of strings section, as a basis for further improvement. (b) To detect strings using core Abs, it is essential to use FA rather than GdnSCN, as the latter chaotrope yields a dotted staining pattern that is difficult to distinguish from the strong cytoplasmic PrP27-30 perinuclear stores. (c) To detect strings on live cells, 8B4 and EST123 are the Abs of choice because they react well with nondenatured strings. In contrast, with SAF32, the signal/noise ratio obtained in native staining on the surface of live cells is suboptimal. (d) To detect strings in fixed and denatured cells, SAF32 and 8B4 can be used interchangeably. They can also be used together in costaining experiments because they are of different IgG subclasses. (e) Finally, strings are best detected using high quantum yield fluorophores, in conjunction with high efficiency filters (such as the High Efficiency series) and high numerical aperture objectives.
Online supplemental material
Fig. S1 shows the presence of PrP strings and webs in a variety of cell systems and in the mouse hippocampus. Fig. S2 demonstrates the specificity of detection of prion strings with mAb 8B4 to infected cells. It also includes WB analysis of PrP GT1/ScGT1/ScGT1-PPS and in Min6 cells. Fig. S3 illustrates and compares the potency of FA and GdnSCN to reveal epitopes along strings. It includes a micrograph of strings in CAD/ME7 cells. Fig. S4 deals with structural features of strings including FRAP analysis of longitudinal diffusion along strings in live ScGT1 and the specificity of ThT staining of PM sheets to prion strings, and illustrates the LM/SEM correlative approach. Fig. S5 shows that strings do not coalign with microtubules and are impervious to short-term action of nocodazole or cytochalasin Videos 1–4 display time-lapse microscopy of PrPSc strings and of PrPC on the surface of live GT1 and ScGT1 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201308028/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201308028.dv.
Supplementary Material
Acknowledgments
We thank S. Ben-Yehuda, S. Even-Ram, R. Gabizon, I. Gahali-Sass, E. Metzer, U. Sass, O. Schonberger, and H. Wille for useful discussions; H. Falk, L. Friedkin, N. Moshkowitz, and T. Mosevitzky for excellent technical assistance; S. Johansson for advice on fibronectin; L. Stitz for the W226 mAb; and D. Barritault for HM. P.J. Peters and S.F. Godsave are grateful to H. Wille and S. Prusiner for providing prion-infected and control brain tissue. We are grateful to E. Blayvas, G. Chechelinsky, S. Eliav, D. Greental, N. Mazurski, and I. Popov (Center for Nanoscience and Nanotechnology, The Hebrew University of Jerusalem [HUJI]) for excellent assistance with the SEM and the nanofabrication of ITO/gold nanoruler slides and to H. Glickstein and Y. Feinstein-Rotkopf (Core facility, Faculty of Medicine, HUJI) for their help with the TEM and confocal microscopy.
This work was supported in part by a USAID American Schools and Hospitals Abroad grant for the procurement of an LSM710 confocal microscope at the Faculty of Medicine (HUJI). This study was supported by grants from the Israel Science Foundation (to A. Taraboulos), the US-Israel Binational Science Foundation (to G.A. Carlson and A. Taraboulos), European Commision FP7 project No. 222887 PRIORITY (to C. Korth, P.J. Peters, K. Kristensson, and A. Taraboulos), and the Horwitz Foundation (to A. Taraboulos). S. Karniely was supported by a Valazzi-Pikovski Fellowship for postdoctoral researchers. C. Korth was supported by the Joint Programme for Neurodegenerative Disease Research DEMTEST (BMBF 01ED1201B).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Ab
- antibody
- BSe
- back-scattered electron
- CTxB
- cholera toxin subunit B
- dpi
- days after inoculation
- DIC
- differential interference contrast
- DRG
- dorsal root ganglia
- FA
- formic acid
- FL
- full-length
- GdnSCN
- guanidine thiocyanate
- GPI
- glycosylphosphatidylinositol
- GPI-AP
- GPI-anchored protein
- HM
- heparan-mimetic HM5004
- ITO
- indium-tin oxide
- LM
- light microscopy
- MMC
- mitomycin C
- N-Abs
- N-proximal Abs
- PIPLC
- phosphatidylinositol-specific phospholipase C
- PM
- plasma membrane
- PPS
- pentosan polysulfate
- RML
- Rocky Mountain Laboratory
- SEM
- scanning electron microscopy
- TEM
- transmission electron microscopy
- ThT
- thioflavin T
- WB
- Western blot
References
- Aguzzi A., Baumann F., Bremer J. 2008. The prion’s elusive reason for being. Annu. Rev. Neurosci. 31:439–477 10.1146/annurev.neuro.31.060407.125620 [DOI] [PubMed] [Google Scholar]
- Arnold J.E., Tipler C., Laszlo L., Hope J., Landon M., Mayer R.J. 1995. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J. Pathol. 176:403–411 10.1002/path.1711760412 [DOI] [PubMed] [Google Scholar]
- Backström E., Chambers B.J., Kristensson K., Ljunggren H.G. 2000. Direct NK cell-mediated lysis of syngenic dorsal root ganglia neurons in vitro. J. Immunol. 165:4895–4900 [DOI] [PubMed] [Google Scholar]
- Ban T., Hamada D., Hasegawa K., Naiki H., Goto Y. 2003. Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J. Biol. Chem. 278:16462–16465 10.1074/jbc.C300049200 [DOI] [PubMed] [Google Scholar]
- Basler K., Oesch B., Scott M., Westaway D., Wälchli M., Groth D.F., McKinley M.P., Prusiner S.B., Weissmann C. 1986. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 46:417–428 10.1016/0092-8674(86)90662-8 [DOI] [PubMed] [Google Scholar]
- Bate C., Salmona M., Diomede L., Williams A. 2004. Squalestatin cures prion-infected neurons and protects against prion neurotoxicity. J. Biol. Chem. 279:14983–14990 10.1074/jbc.M313061200 [DOI] [PubMed] [Google Scholar]
- Ben-Zaken O., Tzaban S., Tal Y., Horonchik L., Esko J.D., Vlodavsky I., Taraboulos A. 2003. Cellular heparan sulfate participates in the metabolism of prions. J. Biol. Chem. 278:40041–40049 10.1074/jbc.M301152200 [DOI] [PubMed] [Google Scholar]
- Berson J.F., Theos A.C., Harper D.C., Tenza D., Raposo G., Marks M.S. 2003. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J. Cell Biol. 161:521–533 10.1083/jcb.200302072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen R.A., Marsh R.F. 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 66:2096–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini E., Turnbaugh J.A., Unterberger U., Harris D.A. 2012. Prion protein at the crossroads of physiology and disease. Trends Neurosci. 35:92–103 10.1016/j.tins.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton D.C., McKinley M.P., Prusiner S.B. 1982. Identification of a protein that purifies with the scrapie prion. Science. 218:1309–1311 10.1126/science.6815801 [DOI] [PubMed] [Google Scholar]
- Borchelt D.R., Scott M., Taraboulos A., Stahl N., Prusiner S.B. 1990. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 110:743–752 10.1083/jcb.110.3.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt D.R., Taraboulos A., Prusiner S.B. 1992. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 267:16188–16199 [PubMed] [Google Scholar]
- Borchelt D.R., Rogers M., Stahl N., Telling G., Prusiner S.B. 1993. Release of the cellular prion protein from cultured cells after loss of its glycoinositol phospholipid anchor. Glycobiology. 3:319–329 10.1093/glycob/3.4.319 [DOI] [PubMed] [Google Scholar]
- Bouzamondo-Bernstein E., Hopkins S.D., Spilman P., Uyehara-Lock J., Deering C., Safar J., Prusiner S.B., Ralston H.J., III, DeArmond S.J. 2004. The neurodegeneration sequence in prion diseases: evidence from functional, morphological and ultrastructural studies of the GABAergic system. J. Neuropathol. Exp. Neurol. 63:882–899 [DOI] [PubMed] [Google Scholar]
- Canello T., Engelstein R., Moshel O., Xanthopoulos K., Juanes M.E., Langeveld J., Sklaviadis T., Gasset M., Gabizon R. 2008. Methionine sulfoxides on PrPSc: a prion-specific covalent signature. Biochemistry. 47:8866–8873 10.1021/bi800801f [DOI] [PubMed] [Google Scholar]
- Caughey B., Raymond G.J. 1991. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 266:18217–18223 [PubMed] [Google Scholar]
- Caughey B., Raymond G.J. 1993. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 67:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caughey B., Raymond G.J., Ernst D., Race R.E. 1991. N-terminal truncation of the scrapie-associated form of PrP by lysosomal protease(s): implications regarding the site of conversion of PrP to the protease-resistant state. J. Virol. 65:6597–6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S.G., Teplow D.B., Parchi P., Teller J.K., Gambetti P., Autilio-Gambetti L. 1995. Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270:19173–19180 10.1074/jbc.270.32.19173 [DOI] [PubMed] [Google Scholar]
- Clarke M.C., Haig D.A. 1970. Evidence for the multiplication of scrapie agent in cell culture. Nature. 225:100–101 10.1038/225100a0 [DOI] [PubMed] [Google Scholar]
- Conner S.D., Schmid S.L. 2003. Regulated portals of entry into the cell. Nature. 422:37–44 10.1038/nature01451 [DOI] [PubMed] [Google Scholar]
- Demart S., Fournier J.G., Creminon C., Frobert Y., Lamoury F., Marce D., Lasmézas C., Dormont D., Grassi J., Deslys J.P. 1999. New insight into abnormal prion protein using monoclonal antibodies. Biochem. Biophys. Res. Commun. 265:652–657 10.1006/bbrc.1999.1730 [DOI] [PubMed] [Google Scholar]
- Dron M., Moudjou M., Chapuis J., Salamat M.K., Bernard J., Cronier S., Langevin C., Laude H. 2010. Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J. Biol. Chem. 285:10252–10264 10.1074/jbc.M109.083857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers H., Romer W., Smith A.E., Bacia K., Dmitrieff S., Chai W., Mancini R., Kartenbeck J., Chambon V., Berland L., et al. 2010. GM1 structure determines SV40-induced membrane invagination and infection. Nat. Cell Biol. 12:11–18 10.1038/ncb1999 [DOI] [PubMed] [Google Scholar]
- Fivaz M., Vilbois F., Thurnheer S., Pasquali C., Abrami L., Bickel P.E., Parton R.G., van der Goot F.G. 2002. Differential sorting and fate of endocytosed GPI-anchored proteins. EMBO J. 21:3989–4000 10.1093/emboj/cdf398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freir D.B., Nicoll A.J., Klyubin I., Panico S., Mc Donald J.M., Risse E., Asante E.A., Farrow M.A., Sessions R.B., Saibil H.R., et al. 2011. Interaction between prion protein and toxic amyloid β assemblies can be therapeutically targeted at multiple sites. Nat. Commun. 2:336 10.1038/ncomms1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloh H., Sedat J.W. 1982. Fluorescence microscopy: reduced photobleaching of rhodamine and fluorescein protein conjugates by n-propyl gallate. Science. 217:1252–1255 10.1126/science.7112126 [DOI] [PubMed] [Google Scholar]
- Godsave S.F., Wille H., Kujala P., Latawiec D., DeArmond S.J., Serban A., Prusiner S.B., Peters P.J. 2008. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J. Neurosci. 28:12489–12499 10.1523/JNEUROSCI.4474-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsave S.F., Wille H., Pierson J., Prusiner S.B., Peters P.J. 2013. Plasma membrane invaginations containing clusters of full-length PrPSc are an early form of prion-associated neuropathology in vivo. Neurobiol. Aging. 34:1621–1631 10.1016/j.neurobiolaging.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goold R., Rabbanian S., Sutton L., Andre R., Arora P., Moonga J., Clarke A.R., Schiavo G., Jat P., Collinge J., Tabrizi S.J. 2011. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat. Commun. 2:281 10.1038/ncomms1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 135:1085–1097 10.1016/j.cell.2008.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haka A.S., Grosheva I., Chiang E., Buxbaum A.R., Baird B.A., Pierini L.M., Maxfield F.R. 2009. Macrophages create an acidic extracellular hydrolytic compartment to digest aggregated lipoproteins. Mol. Biol. Cell. 20:4932–4940 10.1091/mbc.E09-07-0559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Howard G., Nichols B.J. 2011. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J. Cell Sci. 124:2777–2785 10.1242/jcs.084319 [DOI] [PubMed] [Google Scholar]
- Harder T., Scheiffele P., Verkade P., Simons K. 1998. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 141:929–942 10.1083/jcb.141.4.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmey J.H., Doyle D., Brown V., Rogers M.S. 1995. The cellular isoform of the prion protein, PrPc, is associated with caveolae in mouse neuroblastoma (N2a) cells. Biochem. Biophys. Res. Commun. 210:753–759 10.1006/bbrc.1995.1723 [DOI] [PubMed] [Google Scholar]
- Harper J.D., Lieber C.M., Lansbury P.T., Jr 1997. Atomic force microscopic imaging of seeded fibril formation and fibril branching by the Alzheimer’s disease amyloid-beta protein. Chem. Biol. 4:951–959 10.1016/S1074-5521(97)90303-3 [DOI] [PubMed] [Google Scholar]
- Hegde R.S., Mastrianni J.A., Scott M.R., DeFea K.A., Tremblay P., Torchia M., DeArmond S.J., Prusiner S.B., Lingappa V.R. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science. 279:827–834 10.1126/science.279.5352.827 [DOI] [PubMed] [Google Scholar]
- Hermann R., Walther P., Müller M. 1996. Immunogold labeling in scanning electron microscopy. Histochem. Cell Biol. 106:31–39 10.1007/BF02473200 [DOI] [PubMed] [Google Scholar]
- Hijazi N., Kariv-Inbal Z., Gasset M., Gabizon R. 2005. PrPSc incorporation to cells requires endogenous glycosaminoglycan expression. J. Biol. Chem. 280:17057–17061 10.1074/jbc.M411314200 [DOI] [PubMed] [Google Scholar]
- Hope J., Morton L.J., Farquhar C.F., Multhaup G., Beyreuther K., Kimberlin R.H. 1986. The major polypeptide of scrapie-associated fibrils (SAF) has the same size, charge distribution and N-terminal protein sequence as predicted for the normal brain protein (PrP). EMBO J. 5:2591–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horonchik L., Tzaban S., Ben-Zaken O., Yedidia Y., Rouvinski A., Papy-Garcia D., Barritault D., Vlodavsky I., Taraboulos A. 2005. Heparan sulfate is a cellular receptor for purified infectious prions. J. Biol. Chem. 280:17062–17067 10.1074/jbc.M500122200 [DOI] [PubMed] [Google Scholar]
- Huff M.E., Balch W.E., Kelly J.W. 2003. Pathological and functional amyloid formation orchestrated by the secretory pathway. Curr. Opin. Struct. Biol. 13:674–682 10.1016/j.sbi.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Jeffrey M., Goodsir C.M., Fowler N., Hope J., Bruce M.E., McBride P.A. 1996. Ultrastructural immuno-localization of synthetic prion protein peptide antibodies in 87V murine scrapie. Neurodegeneration. 5:101–109 10.1006/neur.1996.0014 [DOI] [PubMed] [Google Scholar]
- Jen A., Parkyn C.J., Mootoosamy R.C., Ford M.J., Warley A., Liu Q., Bu G., Baskakov I.V., Moestrup S., McGuinness L., et al. 2010. Neuronal low-density lipoprotein receptor-related protein 1 binds and endocytoses prion fibrils via receptor cluster 4. J. Cell Sci. 123:246–255 10.1242/jcs.058099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Vey M., Scott M., Pilkuhn S., Cohen F.E., Prusiner S.B. 1997. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc. Natl. Acad. Sci. USA. 94:2333–2338 10.1073/pnas.94.6.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kascsak R.J., Rubenstein R., Merz P.A., Tonna-DeMasi M., Fersko R., Carp R.I., Wisniewski H.M., Diringer H. 1987. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61:3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T., Ogomori K., Tateishi J., Prusiner S.B. 1987. Formic acid pretreatment enhances immunostaining of cerebral and systemic amyloids. Lab. Invest. 57:230–236 [PubMed] [Google Scholar]
- Kitamoto T., Shin R.W., Doh-ura K., Tomokane N., Miyazono M., Muramoto T., Tateishi J. 1992. Abnormal isoform of prion proteins accumulates in the synaptic structures of the central nervous system in patients with Creutzfeldt-Jakob disease. Am. J. Pathol. 140:1285–1294 [PMC free article] [PubMed] [Google Scholar]
- Klein T.R., Kirsch D., Kaufmann R., Riesner D. 1998. Prion rods contain small amounts of two host sphingolipids as revealed by thin-layer chromatography and mass spectrometry. Biol. Chem. 379:655–666 10.1515/bchm.1998.379.6.655 [DOI] [PubMed] [Google Scholar]
- Korth C., Stierli B., Streit P., Moser M., Schaller O., Fischer R., Schulz-Schaeffer W., Kretzschmar H., Raeber A., Braun U., et al. 1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 390:74–77 10.1038/36337 [DOI] [PubMed] [Google Scholar]
- Leclerc E., Peretz D., Ball H., Solforosi L., Legname G., Safar J., Serban A., Prusiner S.B., Burton D.R., Williamson R.A. 2003. Conformation of PrP(C) on the cell surface as probed by antibodies. J. Mol. Biol. 326:475–483 10.1016/S0022-2836(02)01365-7 [DOI] [PubMed] [Google Scholar]
- Li R., Liu T., Wong B.S., Pan T., Morillas M., Swietnicki W., O’Rourke K., Gambetti P., Surewicz W.K., Sy M.S. 2000. Identification of an epitope in the C terminus of normal prion protein whose expression is modulated by binding events in the N terminus. J. Mol. Biol. 301:567–573 10.1006/jmbi.2000.3986 [DOI] [PubMed] [Google Scholar]
- Lingwood D., Schuck S., Ferguson C., Gerl M.J., Simons K. 2009. Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J. Biol. Chem. 284:12041–12048 10.1074/jbc.M900220200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahal S.P., Baker C.A., Demczyk C.A., Smith E.W., Julius C., Weissmann C. 2007. Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. USA. 104:20908–20913 10.1073/pnas.0710054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarava N., Ostapchenko V.G., Savtchenko R., Baskakov I.V. 2009. Conformational switching within individual amyloid fibrils. J. Biol. Chem. 284:14386–14395 10.1074/jbc.M900533200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallucci G., Dickinson A., Linehan J., Klöhn P.C., Brandner S., Collinge J. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 302:871–874 10.1126/science.1090187 [DOI] [PubMed] [Google Scholar]
- Marijanovic Z., Caputo A., Campana V., Zurzolo C. 2009. Identification of an intracellular site of prion conversion. PLoS Pathog. 5:e1000426 10.1371/journal.ppat.1000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Maxfield F.R. 1995. Insolubility and redistribution of GPI-anchored proteins at the cell surface after detergent treatment. Mol. Biol. Cell. 6:929–944 10.1091/mbc.6.7.929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor S., Rothberg K.G., Maxfield F.R. 1994. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 264:1948–1951 10.1126/science.7516582 [DOI] [PubMed] [Google Scholar]
- McKinley M.P., Taraboulos A., Kenaga L., Serban D., Stieber A., DeArmond S.J., Prusiner S.B., Gonatas N. 1991. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Invest. 65:622–630 [PubMed] [Google Scholar]
- Mellon P.L., Windle J.J., Goldsmith P.C., Padula C.A., Roberts J.L., Weiner R.I. 1990. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 5:1–10 10.1016/0896-6273(90)90028-E [DOI] [PubMed] [Google Scholar]
- Meyer R.K., McKinley M.P., Bowman K.A., Braunfeld M.B., Barry R.A., Prusiner S.B. 1986. Separation and properties of cellular and scrapie prion proteins. Proc. Natl. Acad. Sci. USA. 83:2310–2314 10.1073/pnas.83.8.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov A., Jr, Latawiec D., Wille H., Bouzamondo-Bernstein E., Legname G., Williamson R.A., Burton D., DeArmond S.J., Prusiner S.B., Peters P.J. 2003. Cytosolic prion protein in neurons. J. Neurosci. 23:7183–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J., Araki K., Yamato E., Ikegami H., Asano T., Shibasaki Y., Oka Y., Yamamura K. 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology. 127:126–132 10.1210/endo-127-1-126 [DOI] [PubMed] [Google Scholar]
- Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J.L., Lehmann S., Launay J.M., Kellermann O. 2000. Signal transduction through prion protein. Science. 289:1925–1928 10.1126/science.289.5486.1925 [DOI] [PubMed] [Google Scholar]
- Naiki H., Higuchi K., Hosokawa M., Takeda T. 1989. Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177:244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Stein R., Yanai A., Friedlander G., Taraboulos A. 1997. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem. 272:6324–6331 10.1074/jbc.272.10.6324 [DOI] [PubMed] [Google Scholar]
- Naslavsky N., Shmeeda H., Friedlander G., Yanai A., Futerman A.H., Barenholz Y., Taraboulos A. 1999. Sphingolipid depletion increases formation of the scrapie prion protein in neuroblastoma cells infected with prions. J. Biol. Chem. 274:20763–20771 10.1074/jbc.274.30.20763 [DOI] [PubMed] [Google Scholar]
- Nishida N., Harris D.A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., Lehmann S. 2000. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 74:320–325 10.1128/JVI.74.1.320-325.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.R. 2005. Prion-associated increases in Src-family kinases. J. Biol. Chem. 280:2455–2462 10.1074/jbc.M410883200 [DOI] [PubMed] [Google Scholar]
- Novitskaya V., Makarava N., Bellon A., Bocharova O.V., Bronstein I.B., Williamson R.A., Baskakov I.V. 2006. Probing the conformation of the prion protein within a single amyloid fibril using a novel immunoconformational assay. J. Biol. Chem. 281:15536–15545 10.1074/jbc.M601349200 [DOI] [PubMed] [Google Scholar]
- Oesch B., Westaway D., Wälchli M., McKinley M.P., Kent S.B., Aebersold R., Barry R.A., Tempst P., Teplow D.B., Hood L.E., et al. 1985. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 40:735–746 10.1016/0092-8674(85)90333-2 [DOI] [PubMed] [Google Scholar]
- Patil S.M., Mehta A., Jha S., Alexandrescu A.T. 2011. Heterogeneous amylin fibril growth mechanisms imaged by total internal reflection fluorescence microscopy. Biochemistry. 50:2808–2819 10.1021/bi101908m [DOI] [PubMed] [Google Scholar]
- Peretz D., Williamson R.A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I.R., Legname G., Wormald M.R., Rudd P.M., et al. 2001. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 412:739–743 10.1038/35089090 [DOI] [PubMed] [Google Scholar]
- Perrier V., Solassol J., Crozet C., Frobert Y., Mourton-Gilles C., Grassi J., Lehmann S. 2004. Anti-PrP antibodies block PrPSc replication in prion-infected cell cultures by accelerating PrPC degradation. J. Neurochem. 89:454–463 10.1111/j.1471-4159.2004.02356.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P.J., Mironov A., Jr, Peretz D., van Donselaar E., Leclerc E., Erpel S., DeArmond S.J., Burton D.R., Williamson R.A., Vey M., Prusiner S.B. 2003. Trafficking of prion proteins through a caveolae-mediated endosomal pathway. J. Cell Biol. 162:703–717 10.1083/jcb.200304140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters P.J., Bos E., Griekspoor A. 2006. Cryo-immunogold electron microscopy. Curr. Protoc. Cell Biol. Chapter 4:Unit 4.7. [DOI] [PubMed] [Google Scholar]
- Petsch B., Müller-Schiffmann A., Lehle A., Zirdum E., Prikulis I., Kuhn F., Raeber A.J., Ironside J.W., Korth C., Stitz L. 2011. Biological effects and use of PrPSc- and PrP-specific antibodies generated by immunization with purified full-length native mouse prions. J. Virol. 85:4538–4546 10.1128/JVI.02467-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli F., Lehmann S., Maridonneau-Parini I. 2005. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur. J. Neurosci. 21:2063–2072 10.1111/j.1460-9568.2005.04049.x [DOI] [PubMed] [Google Scholar]
- Pluk H., Stokes D.J., Lich B., Wieringa B., Fransen J. 2009. Advantages of indium-tin oxide-coated glass slides in correlative scanning electron microscopy applications of uncoated cultured cells. J. Microsc. 233:353–363 10.1111/j.1365-2818.2009.03140.x [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. 1982. Novel proteinaceous infectious particles cause scrapie. Science. 216:136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., McKinley M.P., Bowman K.A., Bolton D.C., Bendheim P.E., Groth D.F., Glenner G.G. 1983. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell. 35:349–358 10.1016/0092-8674(83)90168-X [DOI] [PubMed] [Google Scholar]
- Prusiner S.B., Scott M., Foster D., Pan K.M., Groth D., Mirenda C., Torchia M., Yang S.L., Serban D., Carlson G.A., et al. 1990. Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell. 63:673–686 10.1016/0092-8674(90)90134-Z [DOI] [PubMed] [Google Scholar]
- Qi Y., Wang J.K., McMillian M., Chikaraishi D.M. 1997. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17:1217–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Moore R.A., McGuirl M.A. 2012. Dissociation of recombinant prion protein fibrils into short protofilaments: implications for the endocytic pathway and involvement of the N-terminal domain. Biochemistry. 51:4600–4608 10.1021/bi300201e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resenberger U.K., Harmeier A., Woerner A.C., Goodman J.L., Müller V., Krishnan R., Vabulas R.M., Kretzschmar H.A., Lindquist S., Hartl F.U., et al. 2011. The cellular prion protein mediates neurotoxic signalling of β-sheet-rich conformers independent of prion replication. EMBO J. 30:2057–2070 10.1038/emboj.2011.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M., Serban D., Gyuris T., Scott M., Torchia T., Prusiner S.B. 1991. Epitope mapping of the Syrian hamster prion protein utilizing chimeric and mutant genes in a vaccinia virus expression system. J. Immunol. 147:3568–3574 [PubMed] [Google Scholar]
- Rogers M., Yehiely F., Scott M., Prusiner S.B. 1993. Conversion of truncated and elongated prion proteins into the scrapie isoform in cultured cells. Proc. Natl. Acad. Sci. USA. 90:3182–3186 10.1073/pnas.90.8.3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer W., Berland L., Chambon V., Gaus K., Windschiegl B., Tenza D., Aly M.R., Fraisier V., Florent J.C., Perrais D., et al. 2007. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 450:670–675 10.1038/nature05996 [DOI] [PubMed] [Google Scholar]
- Sabharanjak S., Sharma P., Parton R.G., Mayor S. 2002. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell. 2:411–423 10.1016/S1534-5807(02)00145-4 [DOI] [PubMed] [Google Scholar]
- Safar J.G., DeArmond S.J., Kociuba K., Deering C., Didorenko S., Bouzamondo-Bernstein E., Prusiner S.B., Tremblay P. 2005. Prion clearance in bigenic mice. J. Gen. Virol. 86:2913–2923 10.1099/vir.0.80947-0 [DOI] [PubMed] [Google Scholar]
- Sanan D.A., Anderson R.G. 1991. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J. Histochem. Cytochem. 39:1017–1024 10.1177/39.8.1906908 [DOI] [PubMed] [Google Scholar]
- Schätzl H.M., Laszlo L., Holtzman D.M., Tatzelt J., DeArmond S.J., Weiner R.I., Mobley W.C., Prusiner S.B. 1997. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 71:8821–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonberger O., Horonchik L., Gabizon R., Papy-Garcia D., Barritault D., Taraboulos A. 2003. Novel heparan mimetics potently inhibit the scrapie prion protein and its endocytosis. Biochem. Biophys. Res. Commun. 312:473–479 10.1016/j.bbrc.2003.10.150 [DOI] [PubMed] [Google Scholar]
- Scott M.R., Köhler R., Foster D., Prusiner S.B. 1992. Chimeric prion protein expression in cultured cells and transgenic mice. Protein Sci. 1:986–997 10.1002/pro.5560010804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serban D., Taraboulos A., DeArmond S.J., Prusiner S.B. 1990. Rapid detection of Creutzfeldt-Jakob disease and scrapie prion proteins. Neurology. 40:110–117 10.1212/WNL.40.1.110 [DOI] [PubMed] [Google Scholar]
- Sharma P., Varma R., Sarasij R.C., Ira K., Gousset G., Krishnamoorthy M., Rao, Mayor S. 2004. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 116:577–589 10.1016/S0092-8674(04)00167-9 [DOI] [PubMed] [Google Scholar]
- Sheets E.D., Lee G.M., Simson R., Jacobson K. 1997. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 36:12449–12458 10.1021/bi9710939 [DOI] [PubMed] [Google Scholar]
- Simons K., Gerl M.J. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11:688–699 10.1038/nrm2977 [DOI] [PubMed] [Google Scholar]
- Snijder B., Sacher R., Rämö P., Damm E.M., Liberali P., Pelkmans L. 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 461:520–523 10.1038/nature08282 [DOI] [PubMed] [Google Scholar]
- Speare J.O., Offerdahl D.K., Hasenkrug A., Carmody A.B., Baron G.S. 2010. GPI anchoring facilitates propagation and spread of misfolded Sup35 aggregates in mammalian cells. EMBO J. 29:782–794 10.1038/emboj.2009.392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl N., Borchelt D.R., Hsiao K., Prusiner S.B. 1987. Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell. 51:229–240 10.1016/0092-8674(87)90150-4 [DOI] [PubMed] [Google Scholar]
- Stahl N., Borchelt D.R., Prusiner S.B. 1990. Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry. 29:5405–5412 10.1021/bi00474a028 [DOI] [PubMed] [Google Scholar]
- Stahl N., Baldwin M.A., Teplow D.B., Hood L., Gibson B.W., Burlingame A.L., Prusiner S.B. 1993. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 32:1991–2002 10.1021/bi00059a016 [DOI] [PubMed] [Google Scholar]
- Stuermer C.A., Langhorst M.F., Wiechers M.F., Legler D.F., Von Hanwehr S.H., Guse A.H., Plattner H. 2004. PrPc capping in T cells promotes its association with the lipid raft proteins reggie-1 and reggie-2 and leads to signal transduction. FASEB J. 18:1731–1733 [DOI] [PubMed] [Google Scholar]
- Sunyach C., Jen A., Deng J., Fitzgerald K.T., Frobert Y., Grassi J., McCaffrey M.W., Morris R. 2003. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 22:3591–3601 10.1093/emboj/cdg344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K.G., Fujiwara T.K., Sanematsu F., Iino R., Edidin M., Kusumi A. 2007. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: single-molecule tracking study 1. J. Cell Biol. 177:717–730 10.1083/jcb.200609174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi Y., Shi Z.D., Ruddy B., Dorward D.W., Greene L., Baron G.S. 2009. Specific biarsenical labeling of cell surface proteins allows fluorescent- and biotin-tagging of amyloid precursor protein and prion proteins. Mol. Biol. Cell. 20:233–244 10.1091/mbc.E08-06-0635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K.A., Suzuki K.G., Shirai Y.M., Shibutani S.T., Miyahara M.S., Tsuboi H., Yahara M., Yoshimura A., Mayor S., Fujiwara T.K., Kusumi A. 2010. Membrane molecules mobile even after chemical fixation. Nat. Methods. 7:865–866 10.1038/nmeth.f.314 [DOI] [PubMed] [Google Scholar]
- Taraboulos A., Rogers M., Borchelt D.R., McKinley M.P., Scott M., Serban D., Prusiner S.B. 1990a. Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA. 87:8262–8266 10.1073/pnas.87.21.8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Serban D., Prusiner S.B. 1990b. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol. 110:2117–2132 10.1083/jcb.110.6.2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Raeber A.J., Borchelt D.R., Serban D., Prusiner S.B. 1992. Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell. 3:851–863 10.1091/mbc.3.8.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S.B. 1995. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J. Cell Biol. 129:121–132 10.1083/jcb.129.1.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76:4350–4354 10.1073/pnas.76.9.4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk E., Teplow D.B., Hood L.E., Prusiner S.B. 1988. Purification and properties of the cellular and scrapie hamster prion proteins. Eur. J. Biochem. 176:21–30 10.1111/j.1432-1033.1988.tb14246.x [DOI] [PubMed] [Google Scholar]
- Tzaban S., Friedlander G., Schonberger O., Horonchik L., Yedidia Y., Shaked G., Gabizon R., Taraboulos A. 2002. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry. 41:12868–12875 10.1021/bi025958g [DOI] [PubMed] [Google Scholar]
- Veith N.M., Plattner H., Stuermer C.A., Schulz-Schaeffer W.J., Bürkle A. 2009. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy. Eur. J. Cell Biol. 88:45–63 10.1016/j.ejcb.2008.08.001 [DOI] [PubMed] [Google Scholar]
- Wadia J.S., Schaller M., Williamson R.A., Dowdy S.F. 2008. Pathologic prion protein infects cells by lipid-raft dependent macropinocytosis. PLoS ONE. 3:e3314 10.1371/journal.pone.0003314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner R.G., Hundt C., Weiss S., Turnbull J.E. 2002. Identification of the heparan sulfate binding sites in the cellular prion protein. J. Biol. Chem. 277:18421–18430 10.1074/jbc.M110406200 [DOI] [PubMed] [Google Scholar]
- Wegmann S., Miesbauer M., Winklhofer K.F., Tatzelt J., Muller D.J. 2008. Observing fibrillar assemblies on scrapie-infected cells. Pflugers Arch. 456:83–93 10.1007/s00424-007-0433-x [DOI] [PubMed] [Google Scholar]
- Wille H., Michelitsch M.D., Guenebaut V., Supattapone S., Serban A., Cohen F.E., Agard D.A., Prusiner S.B. 2002. Structural studies of the scrapie prion protein by electron crystallography. Proc. Natl. Acad. Sci. USA. 99:3563–3568 10.1073/pnas.052703499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wille H., Bian W., McDonald M., Kendall A., Colby D.W., Bloch L., Ollesch J., Borovinskiy A.L., Cohen F.E., Prusiner S.B., Stubbs G. 2009. Natural and synthetic prion structure from X-ray fiber diffraction. Proc. Natl. Acad. Sci. USA. 106:16990–16995 10.1073/pnas.0909006106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R.A., Peretz D., Pinilla C., Ball H., Bastidas R.B., Rozenshteyn R., Houghten R.A., Prusiner S.B., Burton D.R. 1998. Mapping the prion protein using recombinant antibodies. J. Virol. 72:9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam A.Y., Gao C.M., Wang X., Wu P., Peretz D. 2010. The octarepeat region of the prion protein is conformationally altered in PrP(Sc). PLoS ONE. 5:e9316 10.1371/journal.pone.0009316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.