Abstract
Recent evidences indicated Nrf2 is more potent than Nrf1 in the activation of antioxidant genes. However, the roles of Nrf proteins in the regulation of copper-responsive transcription have not been well addressed. We took the toxicogenomic approach and the present network and Gene Ontology analyses results showed that Nrf1 and Nrf2 are distinctively involved in copper-responsive transcriptional regulation in HepG2 transcriptome. Cells deficient in either Nrf1 or Nrf2 were more susceptible to copper exposure than wild type cells. Nrf1 and Nrf2 null cells were transfected with the luciferase reporters containing either ARE(s) or a combination of ARE(s) and MREs, and then treated with copper. In Nrf2-null (Nrf2−/−) cells, copper did not activate transcription of reporter genes, whereas Nrf1 deficiency did not affect copper-inducible activation. Ectopic expression of Nrf2 restored copper-inducible transcription in Nrf2−/− cells. However, the changes in the intrinsic mRNA levels of MT-1 in Nrf null cells following copper treatment showed that Nrf1 and Nrf2 equally contributed to MT-1 activation after 4 h, while Nrf1involved more than Nrf2 following 24 h exposure. These results suggest that while Nrf2 is crucial for MRE/ARE-mediated transcription in response to copper, Nrf1 may activate MT-1 expression by a mechanism different from that Nrf2 employs.
Keywords: Nrf1, Nrf2, ARE, transcription, copper, metallothionein-1
Introduction
The transition metal copper is an essential trace element which serves as a cofactor for enzymes involved in a variety of biological processes (Valko et al. 2005). Exposure to elevated concentrations of copper, however, can induce intracellular oxidative stress responses because of its redox potential (Song et al. 2009). Copper also has a high affinity for sulfhydryl residues that can lead to oxidative damage of cellular components (Letelier et al. 2005; Scandalios 1997; Valko et al. 2005). Copper-induced stress activates cellular responses that ultimately affect the transcription of detoxification genes, control the cellular redox status, and protect against oxidative damage (Song et al. 2009). The transcription of some of these genes is regulated through antioxidant response elements (AREs); cis-acting sequences in the promoter regions of target genes (Favreau and Pickett 1995; Xie et al. 1995). One group of transcription factors that bind to AREs, Nrf1 and Nrf2, are members of the Cap’n’Collar basic leucine zipper family. These transcription factors are ubiquitously expressed and regulate the transcription of antioxidant and Phase II metabolizing enzymes in response to oxidative stress and xenobiotic exposures (Jaiswal 2004; McMahon et al. 2001; Venugopal and Jaiswal 1996). Ectopic expression of either Nrf1 or Nrf2 increases the expression of ARE-driven reporter genes (Nguyen et al. 2000; Venugopal and Jaiswal 1996).
Nrf2 is a prominent factor in activation of ARE-mediated gene expression, compared to Nrf1 (Ma 2013; Niture et al. 2010; Venugopal and Jaiswal 1996). The role of Nrf2 in regulating ARE-mediated transcription has been elucidated in studies using Nrf2 null (Nrf2−/−) mice (Chanas et al. 2002; McMahon et al. 2001; Nguyen et al. 2000). The role of Nrf1 in the activation of ARE-mediated transcription, however, has not been completely resolved. Nrf1-deficient (Nrf1−/−) cells showed reduced levels of glutathione and γ-glutamylcysteine synthetase (γ-GCS) expression (Chan et al. 1998; Kwong et al. 1999). Conditional knockout mice with a hepatocyte-specific Nrf1 deletion have profound levels of oxidative stress, as well as decreased expression of several ARE-dependent genes (Xu et al. 2005). These studies suggest that Nrf1 has an overlapping function with Nrf2 in the regulation of antioxidant and detoxifying genes, however, Nrf1 is likely to have functions independent of Nrf2 (Ohtsuji et al. 2008).
The roles of Nrf1 and Nrf2 in controlling metal-inducible transcription have not been well addressed. A group recognized distinct roles of Nrf1 and Nrf2 in the activation of ARE-dependent genes, and reported that the expressions of metallothionein -1 (MT-1) and MT-2 genes were dependent on Nrf1 but not Nrf2 (Ohtsuji et al. 2008).
A toxicogenomic approach was used to get comprehensive understanding on a mechanistic link between Nrf proteins and copper toxicity. The present functional network and Gene Ontology (GO) analyses using copper-responsive genes present in the HepG2 transcriptome (Song et al. 2009) showed the differential functions of Nrf1 and Nrf2 in copper-responsive transcriptional regulation. The present cytotoxicity assay results showed that Nrf1−/− and Nrf2−/− fibroblasts were more susceptible to copper toxicity than wild type cells. Copper exposure significantly increased ARE and/or MRE-driven reporter gene expression in Nrf1−/− cells, but not in Nrf2−/− cells, while copper-responsive MT-1 expression was more dependent on Nrf1 than Nrf2 following 24-h exposure. These results suggest that while Nrf2 is the most prominent factor for MRE/ARE-mediated transcription in response to copper, copper-responsive MT-1 transcription following 24-h exposure is more dependent on Nrf1, and Nrf1 mediates it by a mechanism different from that Nrf2 uses.
Material and Methods
Cell Culture
Nrf1 and Nrf2 null cell lines, Nrf1−/− and Nrf2−/−, were derived from liver fibroblasts of mice containing targeted deletions of the respective genes (Kwong et al. 1999). The wild type cell line (Nrf1+/+/Nrf2+/+) was derived from liver fibroblasts obtained from the parental mouse strain. Cells were maintained in complete DMEM/F-12 medium supplemented with 15% fetal calf serum, 2 mM L-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, 100 μM non-essential amino acids, and 150 μM 2-mercaptoethanol (Invitrogen/Life Technologies, Carlsbad, CA) in a humidified incubator at 37°C with 5% CO2. Cytotoxicity assays were performed as described by Shokri et al. (Shokri et al. 2000).
Plasmid constructs
The level of copper-inducible transcription was determined by measuring the activity of luciferase produced in mouse liver fibroblasts transfected with various reporter plasmids: p42Luc contains the minimal mouse MT-1 promoter (−42 to +62); p150Luc includes the region of the mouse MT-1 gene from −150 to +62, which contains four functional metal response elements (MREs) and one ARE (Dalton et al. 1994); and pARE4Luc consists of four tandem copies of the USF/ARE from the mouse MT-1 gene inserted into p42Luc. The p42Luc plasmid contains a TATA box but lacks metal- or stress-responsive regulatory elements (i.e., negative control). A reporter gene containing one copy of the ARE from the human γ-GCS light chain promoter was also used (pGCSLuc) (Chan and Kwong 2000). The Nrf2 expression plasmid, pEF/Nrf2, was previously described. (Chan and Kwong 2000).
Transient transfection and reporter gene assays
For transient transfection studies, cells were plated at a density of 8.0 × 104 cells/well in 24-well culture plates, and grown for 18–24 h before transfection. Cells were then washed with phosphate buffered saline and then transfected with 650 ng/well of the reporter plasmid and 160 ng/well of the control plasmid pSV-βgal (Promega, Madison, WI) using Lipofectin, according to the manufacturer’s instructions (Invitrogen/Life Technologies). In experiments using the Nrf2-expression plasmid, cells were transfected with 650 ng/well of reporter plasmid, 160 ng/well of pSV-βgal, and 300 ng/well of pEF/Nrf2. Following this incubation, the transfection mixture was removed, replaced with complete medium, and the cells were allowed to recover overnight. Copper (300 μM, final concentration) was then added and the incubation continued for an additional 24 h. Cell lysates were prepared and then luciferase and β-galactosidase activities were determined using Luciferase and β-Galactosidase Enzyme Assay Systems, respectively, according to manufacturer’s instructions (Promega). All assays were performed in triplicate and luciferase activities were normalized to β-galactosidase activity.
Real time quantitative RT-PCR
The levels of MT-1 and metal response transcription factor 1 (MTF-1) mRNA were determined using real time RT-PCR. Total RNA was isolated using the SV Total RNA Isolation System according to manufacturer instructions (Promega). Reverse transcriptase reactions and PCR were performed using QuantiTect SYBR Green RT-PCR kits (Qiagen, Inc., Valencia, CA). Real time RT-PCR was performed using an ABI Prism 7000 thermal cycler (Invitrogen/Life Technologies). Sequences of the primers used to amplify mouse β-actin, MT-1, and MTF-1 are presented in Supplemental Table 1. Samples were subjected to an initial incubation at 50°C for 30 min, followed by denaturation at 95°C for 15 min, and then 40 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. At the end of the 40 cycles, samples were subjected to melting analyses to confirm amplification specificity of the products.
Fold-change in MT-1 and MTF-1 mRNA levels was determined by the ΔΔCt relative quantification method. The amount of MT-1 and MTF-1 mRNA in each sample was normalized to that of β-actin mRNA. RNA isolated from cells treated with 300 μM copper for 4 or 24 h was compared to RNA isolated from untreated cells.
Functional network and Gene Ontology analyses
The Ingenuity Pathway Analysis (IPA) platform was used to identify significant functional networks from the IPA library of pathways (http://www.ingenuity.com). Previously, transcriptome data was obtained for HepG2 cells treated with 100, 200, 400, or 600 μM copper sulfate for 4, 8, 12, or 24 h (Song et al. 2009). The initial expression data matrix for IPA has been previously described (GEO accession number GSE9539) (Song et al. 2009). Upon uploading the expression data matrix, focus genes (network eligible molecules) were identified based on cutoff values (≥ 1.5-fold change) and overlaid onto a global molecular network developed from information in the Ingenuity Knowledge Base. Networks of focus genes were then algorithmically generated based on their connectivity, and subsequently analyzed to identify the biological functions that were most significant to the genes in the network. IPA calculated a significance score for each network using a p-value calculation and the score was displayed as the negative log of that p-value. Therefore, networks with scores of 2 or higher had at least a 99% confidence of not being generated by random chance alone, and were considered as significant.
GO analysis of the genes in networks of Nrf1 and Nrf2 was performed using the Gene Ontology Enrichment Analysis Software Toolkit (GOEAST, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences)(Zheng and Wang 2008). Significantly enriched GO categories were identified through Batch-Genes Analysis under GOEAST Advance Tools.
Promoter analysis
The web-based program PAINT (Promoter Analysis and Interaction Network Toolset, Ver. 4.0-pre) (Vadigepalli et al. 2003) was used to identify the copper-responsive genes containing putative Nrf1 and/or Nrf2 binding sites in their promoters among all Nrf1 and Nrf2 target genes. In PAINT, Upstreamer fetches 5,000 base pairs of DNA sequence upstream of the transcription start site for each gene, and then TF Retriever uses the TRANSFAC Public databases to find transcription factor binding sites on upstream sequences. The MATCH filter option was set to ‘Minimize false positives’, and core similarity threshold was 1.00.
Statistical analysis
Statistical analysis was performed using StatView software (SAS Institute, Cary NC). The results are presented as the mean ± standard mean error. The significance of mean differences was detected by analysis of variance (ANOVA) followed by Fisher’s Protected Least Squares Difference post hoc test for individual comparisons. The criterion for statistical significance was set at p < 0.05.
Results
Identification of functional networks of Nrf1 or Nrf2-regulated genes using the HepG2 copper transcriptome
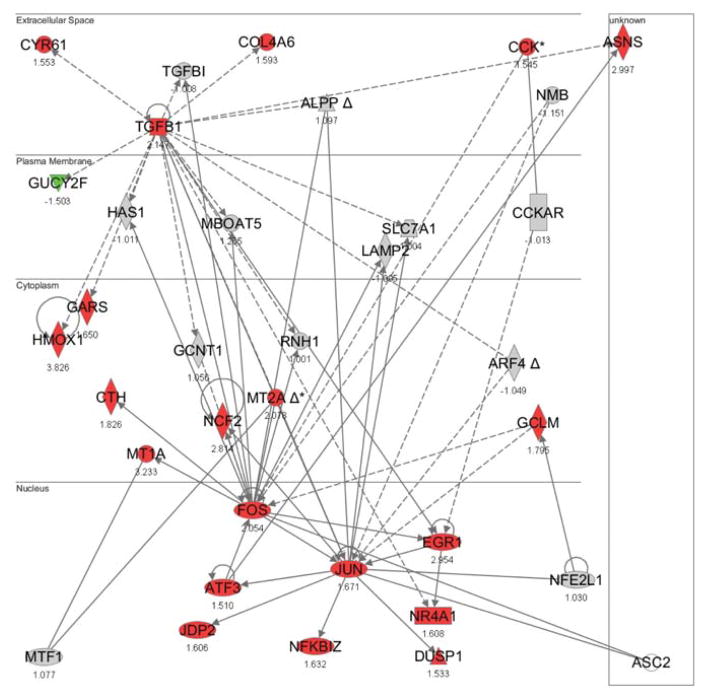
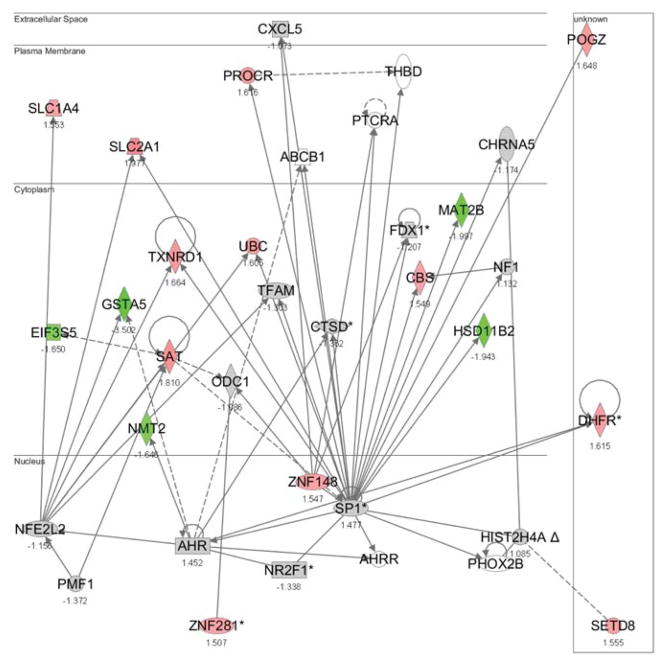
The downstream activities of Nrf1 and Nrf2 with copper-responsive genes in HepG2 transcriptome (Song et al. 2009) were investigated to confirm differential functions between Nrf1 and Nrf2. Ingenuity Pathways Analysis was used to identify significant functional networks of copper-responsive genes under each copper-exposure condition. Significant functional networks were filtered to find networks having Nrf1 or Nrf2 as one of their focus genes. Using this approach, five significant functional networks for Nrf1 and four for Nrf2 were identified. For Nrf1, the functional networks were mapped with genes that were differentially expressed after 8 h exposures to 100, 200 or 600 μM copper (one network per each condition), 12 h exposure to 600 μM copper (one network), and 24 h exposure to 600 μM copper (one network) (Fig. 1, Suppl. Figs. 1 – 4). Functional networks for Nrf2 showed the interactions between the differentially expressed genes following exposure to higher concentrations of copper; 8 or 24 h exposure to 400 μM copper, and 8 or 24 h exposure to 600 μM copper (Fig. 2, Suppl. Figs. 5 – 7). The top functions relevant to the focus genes in each functional network are summarized in Table 1. The genes in Nrf1 networks were associated with cardiovascular system development and function, cell-to-cell signaling and interaction, cellular compromise, connective tissue disorders, gene expression, DNA replication, recombination and repair, lipid metabolism, and dermatological diseases and conditions (Fig. 1). Those in the Nrf2 networks were related with cardiovascular disease, metabolic disease, cell death, respiratory disease, hematological disease, gene expression, DNA replication, recombination and repair, and cancer (Fig. 2).
Figure 1. Significant functional network with Nrf1 as one of its focus genes.
Functional networks were identified through IPA and a network with the highest score is shown. Data used in these analyses were from Song et al. (Song et al. 2009). Genes in red (upregulated) or in green (downregulated) were differentially regulated by 200 μM of copper (8 h exposure). (NFE2L1 = Nrf1)
Figure 2. Significant functional network with Nrf2 as one of its focus genes.
Functional networks were identified through IPA and a representative network is shown. Data used in these analyses were from Song et al. (Song et al. 2009). Genes in red (upregulated) or in green (downregulated) were differentially regulated by 400 μM of copper (24 h exposure). (NFE2L2 = Nrf2)
Table 1.
Top functions of each significant functional network for Nrf1 or Nrf2
| Experimental Condition | Score | Number of Focus Molecules | Top Functions | |
|---|---|---|---|---|
| Nrf1 | 100 μM 8 h | 17 | 11 | Cardiovascular System Development and Function, Cell-To-Cell Signaling and Interaction, Cellular Compromise |
| 200 μM 8 h | 38 | 21 | Cell Death, Connective Tissue Disorders, Cellular Compromise | |
| 600 μM 8 h | 4 | 14 | Gene Expression, DNA Replication, Recombination, and Repair, Molecular Transport | |
| 600 μM 12 h | 4 | 18 | Lipid Metabolism, Small Molecule Biochemistry, Connective Tissue Development and Function | |
| 600 μM 24 h | 26 | 35 | Gene Expression, Cellular Compromise, Dermatological Diseases and Conditions | |
| Nrf2 | 400 μM 8 h | 9 | 15 | Cardiovascular Disease, Metabolic Disease, Cell Death |
| 400 μM 24 h | 8 | 17 | Molecular Transport, Small Molecule Biochemistry, Cell Death | |
| 600 μM 8 h | 10 | 22 | Respiratory Disease, Cell Death, Hematological Disease | |
| 600 μM 24 h | 7 | 21 | DNA Replication, Recombination, and Repair, Gene Expression, Cancer |
Gene Ontology and promoter analyses for Nrf1 and Nrf2 target genes
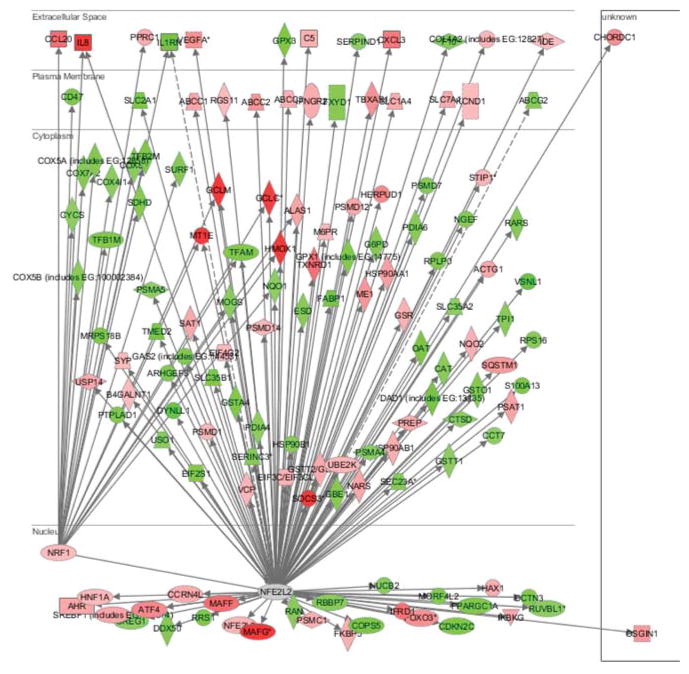
Differentially expressed genes following 4 h exposure to 400 μM copper (Song et al. 2009) were uploaded into IPA and Nrf1 or Nrf2 centered networks generated to further investigate the differential functions between Nrf1 and Nrf2, (Fig. 3). The Nrf1 network contained seven upregulated and 18 downregulated genes, and the Nrf2 network had 63 upregulated and 61 downregulated genes. Five upregulated genes, glutamate-cysteine ligase, catalytic subunit (GCLC), glutamate-cysteine ligase, modifier subunit (GCLM), heme oxygenase-1 (HMOX1), interleukin 8 (IL8) and metallothionein 1E (MT1E) were targets genes for both Nrf1 and Nrf2. For the downregulated genes, the common target genes for both Nrf1 and Nrf2 were interleukin 1 receptor antagonist (IL1RN), mannosyl-oligosaccharide glucosidase (MOGS), NAD(P)H dehydrogenase, quinone 1 (NQO1), proteasome subunit, alpha type, 5 (PSMA5) and transcription factor A, mitochondrial (TFAM) (Table 2).
Figure 3. Nrf1- or Nrf2-centered network with copper-responsive genes.
Nrf1- or Nrf2-centered networks were generated by uploading the genes differentially expressed following 4 h exposure to 400 μM copper using IPA. Data used in this analysis were from Song et al. (Song et al. 2009). Genes in red were upregulated and those in green were downregulated by copper. (NFE2L2 = Nrf2)
Table 2.
Copper-responsive genes in Nrf1 or Nrf2 networks
| Up-regulated Genes | Down-regulated Genes | ||
|---|---|---|---|
|
| |||
| Nrf1 network | Nrf2 network | Nrf1 network | Nrf2 network |
| CCL20 | ABCC1 N,A | ALAS1 | ABCG2 A |
| GCLC N | ABCC2 A | CD47 A | ACTG1 |
| GCLM | ABCC3 A | COX4I1 A | ARHGEF3 A |
| HMOX1 N,A | AHR | COX5A | CAT |
| IL8 A | ALAS1 | COX5B | CCT7 A |
| MT1E | ATF4 A | COX7A2 | CDKN2C |
| PPRC1 A | B4GALNT1 | COX8A | COPS5 |
| C5 | CYCS | CREG1 A | |
| CCRN4L | IL1RN | CTSD | |
| CHORDC1 | MOGS | DAD1 | |
| COL4A2 A | NFE2L2 A | DCTN3 A | |
| CXCL3 A | NQO1 N,A | DDX50 A | |
| EIF3C/EIF3CL A | PSMA5 A | DYNLL1 | |
| EIF4G2 | SDHD A | EIF2S1 A | |
| FKBP5 A | SURF1 | ESD N,A | |
| FOXO3 A | TFAM A | F10 A | |
| GCLC N | TFB1M | FABP1 | |
| GCLM | VEGFA A | FXYD1 | |
| GSR A | G6PD A | ||
| GSTT2/GSTT2B | GAS2 | ||
| HAX1 | GBE1 A | ||
| HERPUD1 | GPX1 A | ||
| HMOX1 N,A | GPX3 | ||
| HNF1A | GSTA4 | ||
| HSP90AA1 A | GSTO1 | ||
| HSP90AB1 | GSTT1 | ||
| IDE A | HSP90B1 | ||
| IFNGR2 | IL1RN | ||
| IFRD1 A | MOGS | ||
| IKBKG | MORF4L2 | ||
| IL8 A | MRPS18B A | ||
| KCND1 | NGEF N | ||
| M6PR | NQO1 N,A | ||
| MAFF A | NUCB2 A | ||
| MAFG | OAT | ||
| ME1 N | PDIA4 A | ||
| MT1E | PDIA6 | ||
| NARS A | PPARGC1A A | ||
| NFE2L3 | PSMA4 A | ||
| NQO2 | PSMA5 A | ||
| NRF1 | PSMD7 | ||
| OSGIN1 A | PTPLAD1 | ||
| PREP | RAN A | ||
| PSAT1 | RARS N,A | ||
| PSMC1 A | RBBP7 N | ||
| PSMD1 A | RPLP0 | ||
| PSMD12 | RPS16 A | ||
| PSMD14 A | RRS1 | ||
| RGS11 | RUVBL1 | ||
| SAT1 A | S100A13 | ||
| SLC1A4 A | SEC23A A | ||
| SLC7A11 | SERINC3 | ||
| SOCS3 A | SERPIND1 A | ||
| SQSTM1 | SLC2A1 N | ||
| SREBF1 A | SLC35A2 A | ||
| STIP1 A | SLC35B1 A | ||
| SYP A | TFAM A | ||
| TBXAS1 | TMED2 A | ||
| TXNRD1 A | TPI1 A | ||
| UBE2K | USO1 | ||
| USP14 | VSNL1 A | ||
| VCP A | |||
| VEGFA A | |||
|
| |||
| 7 (5) | 63 (5) | 18 (5) | 61 (5) |
Genes written in bold letters are common target genes of Nrf1 and Nrf2.
Numbers at the bottom row are the numbers of Nrf1 or Nrf2 target genes which were up- or downregulated by copper (400 μM, 4 h), and numbers in parenthesis are the numbers of common targets genes of Nrf1 and Nrf2.
Gene with AP-1 binding site(s) on the promoter region
Gene with NF-E2 binding site(s) on the promoter region
Promoter analysis using PAINT was performed to identify genes with Nrf1 or Nrf2 binding sites. The lists of target genes of Nrf1 or Nrf2 were uploaded into PAINT, and those with the putative AP-1 or NF-E2 binding site were identified. CNC-bZIP factors are known to bind to the NF-E2/AP-1 element as heterodimers together with small Maf proteins (Kwong et al. 1999). Therefore, genes with AP-1 and/or NF-E2 binding sequences in their promoter regions can interact directly with Nrf1 or Nrf2 in their responses to copper. For the Nrf1 network, four out of seven upregulated genes and eight out of 18 downregulated genes had AP-1 and/or NF-E2 binding sequences. For the Nrf2 network, 32 out of 63 upregulated genes and 33 out of 61 downregulated genes had AP-1 and/or NF-E2 binding sequences (Table 2).
To interpret the differences in the target genes into a biological and functional context, Gene Ontology analyses were performed for the target genes of Nrf1 or Nrf2 using GOEAST (Zheng and Wang 2008). The enriched GO categories identified for either Nrf1 or Nrf2 only are summarized (Tables 3 and 4). The enriched GO categories for the upregulated genes in Nrf1 network were mostly related with positive regulation of angiogenesis, regulation of blood pressure and response to reactive oxygen species, whereas those for the upregulated genes in Nrf2 network were associated with protein folding, positive regulation of cell division, protein oligomerization, activation of NF-κB-inducing kinase activity, cell redox homeostasis, lipid storage (Table 3). For the downregulated genes, the enriched GO categories for Nrf1 target genes included regulation of embryonic development, positive regulation of blood coagulation, positive regulation of cell division, opsonization and endoplasmic reticulum unfolded protein response, whereas those for Nrf2 target genes had regulation of Rho protein signal transduction, glucose metabolic process, translational elongation, ribosome biogenesis and ER to Golgi vesicle-mediated transport (Table 4). The differences between the enriched GO categories identified for Nrf1 or Nrf2 further support the differential functions of Nrf1 and Nrf2.
Table 3.
Differential enriched GO categories for Nrf1 and Nrf2 target genes up-regulated by copper
| Ontology | Enriched GO Categories | |
|---|---|---|
| Nrf1 Target Genes | Nrf2 Target Genes | |
| Biological process | Negative regulation of sequence-specific DNA binding transcription factor activity | Protein folding |
| Regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ATP catabolic process | |
| Negative regulation of DNA binding | Positive regulation of cell division | |
| Positive regulation of angiogenesis | M/G1 transition of mitotic cell cycle | |
| Regulation of blood pressure | Positive regulation of transcription, DNA-dependent | |
| Regulation of blood vessel size | Protein oligomerization | |
| Response to endogenous stimulus | Aggresome assembly | |
| Response to reactive oxygen species | Regulation of interferon-gamma-mediated signaling pathway | |
| Cellular response to xenobiotic stimulus | Activation of NF-kappaB-inducing kinase activity | |
| Cell redox homeostasis | ||
| Cellular chloride ion homeostasis | ||
| B cell homeostasis | ||
| Lipid storage | ||
| Molecular function | Growth factor activity | |
| Sequence-specific DNA binding | ||
| Insulin binding | ||
| Proteasome binding | ||
| Unfolded protein binding | ||
| ATPase activity | ||
| Thioredoxin-disulfide reductase activity | ||
| Glutathione disulfide oxidoreductase activity | ||
Table 4.
Differential enriched GO categories for Nrf1 and Nrf2 target genes down-regulated by copper
| Ontology | Enriched GO Categories | |
|---|---|---|
| Nrf1 Target Genes | Nrf2 Target Genes | |
| Biological process | Regulation of embryonic development | Regulation of Rho protein signal transduction |
| Positive regulation of blood coagulation | Glucose metabolic process | |
| Positive regulation of cell division | Glutathione metabolic process | |
| Positive regulation of endocytosis | Translational elongation | |
| Opsonization | Arginyl-tRNA aminoacylation | |
| Respiratory electron transport chain | Protein folding | |
| Endoplasmic reticulum unfolded protein response | Cell redox homeostasis | |
| Ribosome biogenesis | ||
| ER to Golgi vesicle-mediated transport | ||
| Intracellular protein transport | ||
| Molecular function | Growth factor activity | Rho guanyl-nucleotide exchange factor activity |
| Heme binding | Structural constituent of ribosome | |
| Cytochrome-c oxidase activity | Unfolded protein binding | |
| Tumor necrosis factor receptor binding | ||
| Calcium ion binding | ||
| NADP binding | ||
| Threonine-type endopeptidase activity | ||
| Mannosyl-oligosaccharide glucosidase activity | ||
| Glucose-6-phosphate dehydrogenase activity | ||
| Glutathione peroxidase activity | ||
| Protein disulfide oxidoreductase activity | ||
| Glutathione transferase activity | ||
| Sugar:hydrogen symporter activity | ||
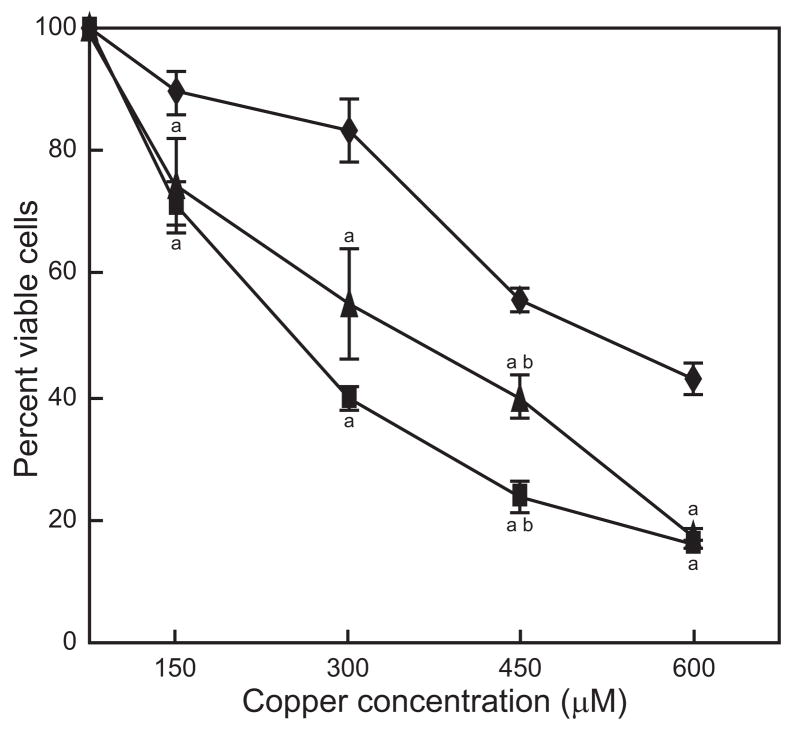
Effect of copper on Nrf1−/− and/Nrf2−/− cell viability
Cytotoxicity experiments were performed to determine the levels of sensitivity to copper for Nrf1−/− and/Nrf2−/− cells. Nrf1−/− and Nrf2−/− cells were significantly more sensitive than wild type cells when exposed to copper for 24 h at all concentrations tested (Fig. 4). LC50s for wild type, Nrf1−/− and/Nrf2−/− cells were approximately 520 μM, 350 μM and 250 μM, respectively. The number of viable cells was lower for Nrf2−/− cells compared to Nrf1−/− cells. However, the difference was significant only at 450 μM copper.
Figure 4. Effect of copper on cell viability.
Wild type (◆), Nrf1−/− (▲) and Nrf2−/− (■) cells were exposed to copper for 24 h. Data are expressed as mean ± standard mean error (n = 3) relative to the number of viable wild type cells grown in the absence of added copper. “a”, indicates significantly different from wild type cells; “b”, indicates significant difference between Nrf1−/− and Nrf2−/−.
Effect of copper on metal and oxidative stress-responsive transcription in Nrf1−/− and Nrf2−/− cells
The ability of copper to induce MT and oxidative stress-responsive transcription in the absence of Nrf1 or Nrf2 was investigated using luciferase reporter genes whose expression were regulated by portions of the MT-1 promoter, or concatenated copies of AREs (Dalton et al. 1994). Previous experiments showed that exposing COS-7 cells transfected with p150Luc and pARE4Luc reporters to 400 μM copper for 24 h resulted in increased expression of luciferase (unpublished observations). Wild type, Nrf1−/− and Nrf2−/− cells were exposed to 300 μM copper for 24 h. The 300 μM concentration was selected because the difference in cell viability between Nrf1−/− and/Nrf2−/− cells was the largest at this concentration of copper while the lethality is still less than 20 % for wild type cells (Fig. 4).
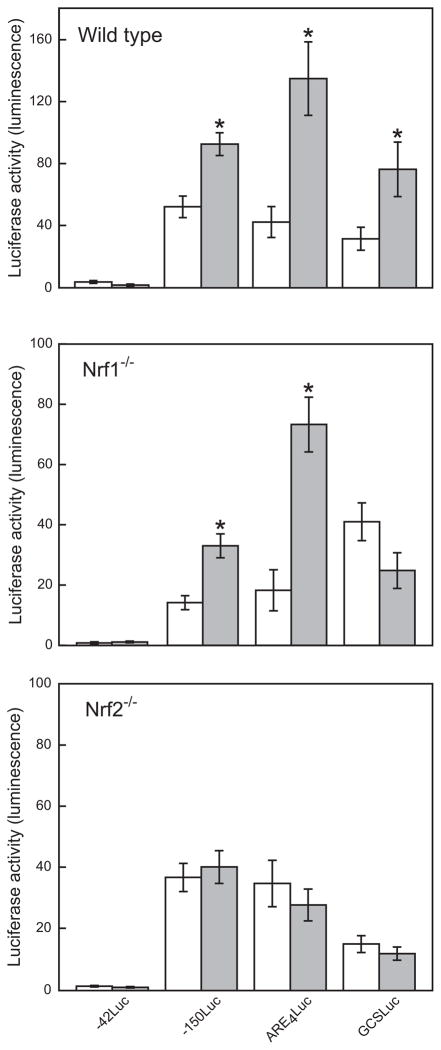
Copper did not significantly induce transcription of p42Luc reporter gene in any of the cell lines examined (Fig. 5). Significant increases in transcriptional activity were observed in wild type cells transfected with the p150Luc, pARE4Luc or pGCSLuc reporters (Fig 5). In Nrf1−/− cells, the basal levels of expression for p150Luc and pARE4Luc were reduced approximately 4-fold and 2-fold, respectively, compared to those observed in wild type cells (Fig 5). However, copper exposure caused significant increases in expressions of p150Luc and pARE4Luc reporters, 2.3-fold and 4.1-fold, respectively, but did not significantly affect the expression of pGSCLuc. The magnitude of the fold-increase in reporter genes expression following copper exposure in Nrf1−/− cells was comparable to that observed in wild type cells. These results suggest that Nrf2 can partially compensate for the absence of Nrf1 in Nrf1−/− cells to induce the expression of p150Luc and pARE4Luc.
Figure 5. Effect of copper on luciferase reporter gene induction.
Wild type, Nrf1−/− or Nrf2−/− cells were transfected with reporter genes p42Luc, p150Luc, pARE4Luc, or pGCSLuc, and then exposed to 0 (white bars) or 300 μM copper for 24 h (gray bars). The luciferase activity was then measured and normalized to the amount of β-galactosidase activity. Data are expressed as mean ± standard mean error. “*” indicates significantly different from cells not exposed to copper (open white bars), by ANOVA, p < 0.05, n = 3 observations.
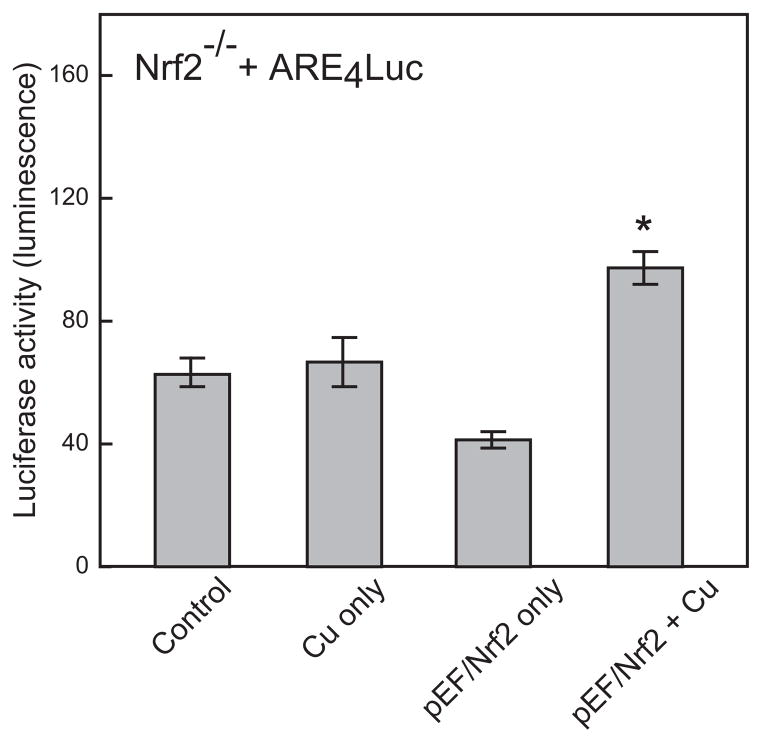
In Nrf2−/− cells, the basal level of reporter gene expression was not significantly affected, with the exception of the pGCSLuc vector. Contrary to the observations with Nrf1−/− cells, copper exposure did not cause any change in the levels of expression of p150Luc and pARE4Luc in Nrf2−/− cells (Fig. 5). When Nrf2−/− cells were co-transfected with pARE4Luc and the Nrf2 expression vector, pEF/Nrf2, copper-inducible expression of ARE-based reporter gene was partially restored (Fig. 6). These observations demonstrated that Nrf2 is a dominant factor in activation of MRE and/or ARE-mediated gene expression following copper exposure.
Figure 6. Effect of expressing Nrf2 in Nrf2−/− cells on copper-inducible transcription.
Nrf2−/− cells were transfected with the Nrf2 expression plasmid, pEF/Nrf2 and pARE4Luc, and then exposed to 300 μM copper for 24 h. The luciferase activity was then measured and normalized to the amount of β-galactosidase enzyme activity. Data are expressed as mean ± standard mean error. “*” indicates significantly different from cells exposed to copper only, by ANOVA, p < 0.05, n = 3 observations.
Effect of copper on MT-1 and MTF-1 RNA levels in Nrf1−/− and Nrf2−/− cells
To determine the involvement of Nrf1 and Nrf2 in the regulation of copper-inducible transcription of MT-1, changes in the steady-state mRNA levels of MT-1 were measured using real-time RT-PCR in wild type and Nrf-null cells following copper exposure (300 μM copper for 4 and 24 h) (Table 5). In wild type cells, copper exposure significantly increased MT-1 mRNA levels, 6.02-fold after 4 h and 3.12-fold after 24 h, compared to those in untreated cells. In Nrf1−/− cells, copper produced a significant 3.27-fold increase in MT-1 steady state mRNA level relative to untreated cells after 4 h treatment. Consistent with the results of reporter gene assays, copper caused significant increases in MT-1 mRNA levels in wild type and Nrf1−/− cells following 4 h exposure, whereas MT-1 mRNA upregulation was only 1.20-fold and not significant after 24 h. That is, the level of steady state MT-1 mRNA returned to control levels after 24 h in Nrf1−/− cells. In Nrf2−/− cells, contrary to the reporter gene assay results, copper exposure significantly upregulated MT-1 mRNA levels 3.14-fold after 4 h and 1.87-fold after 24 h, compared to those in untreated cells.
Table 5.
Steady state levels of mouse MT-1 and MTF-1 mRNA
| Cell type | Metallothionein-1 | MTF-1 | ||
|---|---|---|---|---|
| No metal | Copper | |||
| 4 h | 24 h | |||
| Wild-type | 1.00 ± 0.87 | 6.02 ± 0.65a | 3.12 ± 0.58a | 1.00 ± 0.26 |
| Nrf1−/− | 1.00 ± 0.78 | 3.27 ± 0.81ab | 1.20 ± 0.44c | 1.18 ± 0.15 |
| Nrf2−/− | 1.00 ± 0.54 | 3.14 ± 0.42ab | 1.87 ± 0.90a | 0.92 ± 0.63 |
Significantly different from untreated cells
Significantly different from wild type cells (4 hr copper treatment)
Significantly different from wild type cells (24 hr copper treatment)
Data are expressed as mean ± standard mean error (n = 3)
One of the reasons for the inconsistency between reporter gene assay and real-time RT-PCR results might be that endogenous MT-1 promoter contains much longer DNA sequences upstream from the region that p150Luc has, and Nrf1 can bind with some putative binding site(s) in that upstream region. And it would be noteworthy that the reporter assay results represent the accumulated protein during the treatment, while the real-time qPCR results indicate the temporal mRNA levels at the specific time points.
The increase in MT-1 steady state mRNA levels in wild type cells was approximately twice those observed in Nrf1−/− and Nrf2−/− cells after a 4 hour exposure to copper. The steady state level of MT-1 mRNA was lower following 24 h exposure in the three cell lines. Basal expression of MT-1 decreased in Nrf1−/− and Nrf2−/− cells, but was not significantly different from levels in wild type cells (data not shown).
Because MT-1 expression is mediated primarily through interaction of the transcription factor MTF-1 and MREs (Radtke et al. 1993), the levels of MTF-1 mRNA were also determined. There was no significant difference in MTF-1 mRNA levels between Nrf1−/−, Nrf2−/− and wild type cells. These results indicate that changes in the levels of copper-inducible transcription in Nrf-null cells were not due to any changes in MTF-1 levels.
Taken all together, these results showed that Nrf1 contributed more to copper-responsive expression of MT-1 than Nrf2 did following longer exposure like 24 h, while both Nrf proteins involved equally after 4 h exposure. The results also demonstrated that Nrf1 might mediate copper-responsive MT-1 transcription by a mechanism different from that Nrf2 used, suggesting that Nrf1 and Nrf2 are distinctively involved in copper-responsive MT-1 expression.
Discussion
The CNC-bZIP transcription factors, Nrf1 and Nrf2, play important roles in the protection against oxidative stress by regulating the transcription of a variety of stress-responsive genes. To characterize the differential functions of Nrf1 and Nrf2, their downstream activities with copper-responsive genes in the HepG2 transcriptome were investigated (Song et al. 2009). Significant functional networks for Nrf1 were mapped with the genes differentially expressed at both low and high concentrations of copper (100, 200 and 600 μM); while significant functional networks for Nrf2 contained genes differentially expressed at high concentrations of copper (400 and 600 μM). The prevalent biological functions of Nrf1 networks were associated with development and various cellular processes under homeostatic and stressed conditions. For the Nrf2 networks, most biological functions were associated with diseases and cell death (Table 1). The functional network analysis showed clear differences between Nrf1 and Nrf2 in their activities with copper-responsive genes. These results are consistent with previous analysis of HepG2 copper transcriptome, where lower concentrations of copper (100 and 200 μM) affected genes associated with physiological adaptive responses and higher concentrations (400 and 600 μM) induced toxicological responses (Song et al. 2009). The functional network analysis suggests that although both Nrf1 and Nrf2 regulate oxidative stress responsive genes, Nrf2 is crucial for maintaining cellular homeostasis under severe stress conditions, whereas Nrf1 is indispensable for countering steady-state stress under normal homeostatic conditions.
Using GO analysis of the target genes in each Nrf network, we investigated the divergent roles of Nrf1 and Nrf2 in exerting the copper-induced responses in hepatocyte cell line (HepG2). Among 25 Nrf1 target genes and 124 Nrf2 target genes, the two networks shared only 10 common target genes (Table 2). To place the differences in the target genes into a biological and functional context, GO analyses were performed. Clear differences between the enriched GO categories identified for Nrf1 or Nrf2 target genes were observed (Tables 3 and 4). This observation is consistent with a previous report showing Nrf1 contributed to the maintenance of CNS homeostasis by regulating target genes distinct from Nrf2 (Kobayashi et al. 2011).
Early studies in knockout mice demonstrated that Nrf1 was critical in fetal liver erythropoiesis (Chan et al. 1998). Nrf1 seems to be a more primitive and less specialized regulator of oxidative stress, whereas Nrf2 seems to have diversified away from Nrf1 by specializing in the inducible regulation of a much more diverse set of cytoprotective genes, but has lost many of the former associations with hematopoiesis (Maher and Yamamoto 2010). Recently, Chorley et al. identified a novel role for Nrf2 in retinoid signaling (Chorley et al. 2012). These studies demonstrated the divergent roles of Nrf1 and Nrf2 and further support the present functional network and GO analyses results.
Kwong et al. demonstrated that Nrf1−/− cells are sensitive to oxidative stressors (Kwong et al. 1999). Likewise, Nrf2-deficient mice are highly susceptible to chemical-induced toxicity, carcinogenesis and oxidative burden (Kensler et al. 2007). Consistent with previous studies, Nrf1−/− and Nrf2−/− cells were hypersensitive to copper exposure, compared to wild type cells (Fig. 4). The sensitivity of both cell lines may be due to the lack of Nrf1- or Nrf2-mediated transcription of genes involved in attenuating oxidative stress through the detoxification and elimination of reactive oxygen species and electrophiles (Chan and Kan 1999; Nguyen et al. 2009).
To investigate the roles of Nrf1 and Nrf2 in regulating copper-inducible transcription, Nrf1 and Nrf2 null cells were transfected with luciferase reporter genes whose expressions were regulated by portions of the MT-1 promoter, concatenated copies of AREs or a single ARE. Copper significantly induced transcription of all reporter genes in wild type cells (Fig. 5). This observation is consistent with a previous study that shows the upregulation of MT transcription following exposure to copper is mediated by AREs, as well as MREs (Mattie and Freedman 2004). In the absence of Nrf2, however, copper exposure did not induce reporter gene expression, suggesting that Nrf2 is a major regulator of copper-inducible transcription, and mediates it via AREs and/or MREs. Restoration of ARE-mediated transcription following ectopic expression of Nrf2 in Nrf2−/− cells further supports the prominent role of this transcription factor in regulating copper-inducible transcription (Fig. 6).
On the contrary, copper induced transcription of p150Luc and pARE4Luc in Nrf1−/− cells (Fig. 5), which suggests that Nrf2 compensates the loss of Nrf1 function, or that Nrf1 has a minor role in antioxidant defense. Ohtsuji et al. (2008) observed that Nrf1 deficiency resulted in upregulation of some Nrf2-target genes. Accordingly, they suggested that the lack of Nrf1 activates Nrf2, as a backup defense against endogenous stressors (Ohtsuji et al. 2008).
Contribution of Nrfs to the regulation of MT-1 expression has not been well addressed. The present reporter gene assay results showed that copper induced p150Luc transcription in Nrf1−/− cells, but not in Nrf2−/− cells. Since p150Luc contains one ARE, as well as four MREs, these results suggest that Nrf2 may be involved in regulating copper-inducible MT-1 transcription via a metal responsive pathway as well. The involvement of Nrf2 in regulating copper-inducible MT transcription was unexpected since MT transcription is dependent on interaction between MTF-1 and MRE’s (Radtke et al. 1993). MRE’s have been shown to mediate oxidative stress-responsive activation of MT-1 transcription, however, the mechanism of this response has not been clearly resolved (Dalton et al. 1994).
In wild type and Nrf2−/− cells, copper exposure significantly increased MT-1 mRNA levels after both 4 and 24 h. In Nrf1−/− cells copper exposure significantly increased MT-1 mRNA levels only after 4 h, and the copper-induced increase in MT-1 expression was diminished to no-metal control level after 24 h exposure. The increased levels of MT-1 mRNA in Nrf null cells were about halves of those observed in wild type cells. Ohtsuji et al. (2008) showed that MT-1 expression decreased in Nrf1 deficient mouse liver compared with that in control mice. They also observed that Nrf1 and Nrf2 bound the MT-1 ARE with comparable affinity, but Nrf1 preferentially activated transcription through the ARE. It should be noted that the experiments of Ohtsuji et al. (2008) did not involve copper treatment.
The present qPCR results were inconsistent with those of Ohtsuji et al. (2008) in that Nrf2 deficiency affected the activation of MT-1 expression following copper treatment, decreasing the upregulation of MT-1expression into half of that observed in wild type cells after 4 h exposure. This implied that MT-1 expression was intimately dependent on Nrf2 as well following short copper treatment like 4 h. The effect of Nrf1 deficiency was clear following 24 h exposure to copper. The level of steady state MT-1 mRNA returned to untreated control levels after 24 h in Nrf1−/− cells, suggesting that Nrf1 contributed more to copper-responsive expression of MT-1 than Nrf2 did following longer exposure like 24 h. These results may imply that Nrf2 is the prominent transcription factor interacting with −150 to +62 region, while Nrf1 may interact with some putative binding site(s) in the region upstream of −150 in MT-1 promoter.
Multiple microarray analyses between wild-type and Nrf2-deficient mice identified numerous Nrf2-regulated genes which are involved in electrophiles conjugation, antioxidative response, glutathione (GSH) homeostasis, production of reducing equivalents and proteasome pathways (Osburn and Kensler 2008; Thimmulappa et al. 2002).
In the present study, Nrf1 or Nrf2 centered networks generated through IPA using HepG2 transcriptome (Song et al., 2009) showed that the target genes of Nrf1 and Nrf2 included NAD(P)H:quinone reductase (NQO1), glutathione S-transferase A4 (GSTA4), glutathione peroxidase 1 (GPX1), glutamate-cysteine ligase, catalytic subunit (GCLC), heme oxygenase 1 (HMOX1) and thioredoxin reductase 1 (TXNRD1), which were well known to be regulated mainly by Nrf2-dependent mechanism (Jennings et al. 2013; Thimmulappa et al. 2002).
A couple of previous studies demonstrated that expressions of some of these genes were modulated by metals via Nrf2 in HepG2 cells (Abdelhamid et al. 2010; Kim 2012). In addition, Gong and Cederbaum demonstrated that Nrf2 plays a key role in the adaptive response against increased oxidative stress caused by CYP2E1 in HepG2 cells (Gong and Cederbaum 2006). Furthermore, investigations using Nrf2 knockout mice clearly demonstrated the importance of this transcription factor in the protection against drug-induced stress across multiple organs (Copple et al. 2008; Jennings et al. 2013). Therefore, Nrf2-mediated cytoprotective adaptive responses are prevalent and powerful mechanisms for the various types of cells, particularly those associated with detoxification and exposed to the external environment, to cope with oxidative or electrophilic stress (Osburn and Kensler 2008).
Conclusions
The results of our study clearly demonstrated that Nrf1 and Nrf2 differentially regulate copper-inducible transcription. Nrf2 was a dominant factor in activation of ARE and/or MRE-mediated transcription in response to copper. Nrf1 and Nrf2 equally contributed to MT-1 activation after 4 h, while Nrf1involved more than Nrf2 following 24 h exposure, and may mediate it by a mechanism different from that Nrf2 employs. Functional network and GO analyses corroborate the differential functions of Nrf1 and Nrf2 in the regulation of copper-responsive transcription.
Supplementary Material
Nrf null cells were more susceptible to copper exposure than wild type cells.
Copper did not activate ARE-mediated transcription in Nrf2-null (Nrf2−/−) cells.
Nrf2 is crucial for MRE/ARE-mediated transcription in response to copper.
Nrf1 and Nrf2 equally contributed to copper-responsive MT-1 expression after 4 h, while Nrf1involved more than Nrf2 following 24 h exposure.
Nrf1 and Nrf2 have distinctive roles in copper-responsive transcription.
Acknowledgments
The authors would like to thank Dr. Glen Andrews (University of Kansas Medical Center) for providing the mouse MT-1-based Luc reporter plasmids and Dr. Jefferson Y. Chan (University of California-Irvine) for the Nrf1−/− and Nrf2−/− cell lines. This work was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health (Z01ES102046) and by Natural Science Research Institute of Gangneung-Wonju National University (to C.-H. Lee).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelhamid G, Anwar-Mohamed A, Elmazar MM, El-Kadi AO. Modulation of NAD(P)H:quinone oxidoreductase by vanadium in human hepatoma HepG2 cells. Toxicology in vitro: an international journal published in association with BIBRA. 2010;24:1554–1561. doi: 10.1016/j.tiv.2010.06.017. [DOI] [PubMed] [Google Scholar]
- Chan JY, Kwong M. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim Biophys Acta. 2000;1517:19–26. doi: 10.1016/s0167-4781(00)00238-4. [DOI] [PubMed] [Google Scholar]
- Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf1, results in anemia and embryonic lethality in mice. EMBO Journal. 1998;17:1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Kan YW. Nrf2 is essential for protection against acute pulmonary injury in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12731–12736. doi: 10.1073/pnas.96.22.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanas SA, Jiang Q, McMahon M, McWalter GK, McLellan LI, Elcombe CR, Henderson CJ, Wolf CR, Moffat GJ, Itoh K, Yamamoto M, Hayes JD. Loss of Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. The Biochemical journal. 2002;365:405–416. doi: 10.1042/BJ20020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley BN, Campbell MR, Wang X, Karaca M, Sambandan D, Bangura F, Xue P, Pi J, Kleeberger SR, Bell DA. Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res. 2012;40:7416–7429. doi: 10.1093/nar/gks409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copple IM, Goldring CE, Kitteringham NR, Park BK. The Nrf2-Keap1 defence pathway: role in protection against drug-induced toxicity. Toxicology. 2008;246:24–33. doi: 10.1016/j.tox.2007.10.029. [DOI] [PubMed] [Google Scholar]
- Dalton T, Palmiter RD, Andrews GK. Transcriptional induction of the mouse metallothionein-I gene in hydrogen peroxide-treated Hepa cells involves a composite major late transcription factor/antioxidant response element and metal response promoter elements. Nucleic acids research. 1994;22:5016–5023. doi: 10.1093/nar/22.23.5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau LV, Pickett CB. The rat quinone reductase antioxidant response element. Identification of the nucleotide sequence required for basal and inducible activity and detection of antioxidant response element-binding proteins in hepatoma and non-hepatoma cell lines. The Journal of biological chemistry. 1995;270:24468–24474. doi: 10.1074/jbc.270.41.24468. [DOI] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology. 2006;43:144–153. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- Jennings P, Limonciel A, Felice L, Leonard MO. An overview of transcriptional regulation in response to toxicological insult. Archives of toxicology. 2013;87:49–72. doi: 10.1007/s00204-012-0919-y. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Kim GNK, Eui-Su, Kwon Young-In, Jang Hae-Dong. Potential mechanism of kaempferol against Cu2+-induced oxidative stress through chelating activity and regulation of nuclear factor-erythroid-2-related factor 2 signaling. Food Science and Biotechnology. 2012;21:1469–1475. [Google Scholar]
- Kobayashi A, Tsukide T, Miyasaka T, Morita T, Mizoroki T, Saito Y, Ihara Y, Takashima A, Noguchi N, Fukamizu A, Hirotsu Y, Ohtsuji M, Katsuoka F, Yamamoto M. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells. 2011;16:692–703. doi: 10.1111/j.1365-2443.2011.01522.x. [DOI] [PubMed] [Google Scholar]
- Kwong M, Kan YW, Chan JY. The CNC basic leucine zipper factor, Nrf1, is essential for cell survival in response to oxidative stress-inducing agents. Role for Nrf1 in ? -GCSl and gss expression in mouse fibroblasts. J Biol Chem. 1999;274:37491–37498. doi: 10.1074/jbc.274.52.37491. [DOI] [PubMed] [Google Scholar]
- Letelier ME, Lepe AM, Faundez M, Salazar J, Marin R, Aracena P, Speisky H. Possible mechanisms underlying copper-induced damage in biological membranes leading to cellular toxicity. Chem Biol Interact. 2005;151:71–82. doi: 10.1016/j.cbi.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Ma Q. Role of nrf2 in oxidative stress and toxicity. Annual review of pharmacology and toxicology. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher J, Yamamoto M. The rise of antioxidant signaling--the evolution and hormetic actions of Nrf2. Toxicol Appl Pharmacol. 2010;244:4–15. doi: 10.1016/j.taap.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Mattie MD, Freedman JH. Copper-inducible transcription: regulation by metal- and oxidative stress responsive pathways. Am J Physiol Cell Physiol. 2004;286:C293–301. doi: 10.1152/ajpcell.00293.2003. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. The Journal of biological chemistry. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niture SK, Kaspar JW, Shen J, Jaiswal AK. Nrf2 signaling and cell survival. Toxicol Appl Pharmacol. 2010;244:37–42. doi: 10.1016/j.taap.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M, Katsuoka F, Kobayashi A, Aburatani H, Hayes JD, Yamamoto M. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J Biol Chem. 2008;283:33554–33562. doi: 10.1074/jbc.M804597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osburn WO, Kensler TW. Nrf2 signaling: an adaptive response pathway for protection against environmental toxic insults. Mutation research. 2008;659:31–39. doi: 10.1016/j.mrrev.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein I promoter. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scandalios JG. Oxidative stress and the molecular biology of antioxidant defenses. Cold Spring Harbor Laboratory Press; Plainview, N.Y: 1997. [Google Scholar]
- Shokri F, Heidari M, Gharagozloo S, Ghazi-Khansari M. In vitro inhibitory effects of antioxidants on cytotoxicity of T-2 toxin. Toxicology. 2000;146:171–176. doi: 10.1016/s0300-483x(00)00172-4. [DOI] [PubMed] [Google Scholar]
- Song MO, Li J, Freedman JH. Physiological and toxicological transcriptome changes in HepG2 cells exposed to copper. Physiological genomics. 2009;38:386–401. doi: 10.1152/physiolgenomics.00083.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer research. 2002;62:5196–5203. [PubMed] [Google Scholar]
- Vadigepalli R, Chakravarthula P, Zak DE, Schwaber JS, Gonye GE. PAINT: a promoter analysis and interaction network generation tool for gene regulatory network identification. Omics: a journal of integrative biology. 2003;7:235–252. doi: 10.1089/153623103322452378. [DOI] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12:1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- Venugopal R, Jaiswal AK. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Belinsky M, Xu Y, Jaiswal AK. ARE- and TRE-mediated regulation of gene expression. Response to xenobiotics and antioxidants. J Biol Chem. 1995;270:6894–6900. doi: 10.1074/jbc.270.12.6894. [DOI] [PubMed] [Google Scholar]
- Xu Z, Chen L, Leung L, Yen TS, Lee C, Chan JY. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci U S A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic acids research. 2008;36:W358–363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.