Abstract
Formation of multi-component signaling complex necrosomes is essential for tumor necrosis factor α (TNF)-induced programmed necrosis (also called necroptosis). However, the mechanisms of necroptosis are still largely unknown. We isolated a TNF-resistant L929 mutant cell line generated by retrovirus insertion and identified that disruption of the guanine nucleotide-binding protein γ 10 (Gγ10) gene is responsible for this phenotype. We further show that Gγ10 is involved in TNF-induced necroptosis and Gβ2 is the partner of Gγ10. Src is the downstream effector of Gβ2γ10 in TNF-induced necroptosis because TNF-induced Src activation was impaired upon Gγ10 knockdown. Gγ10 does not affect TNF-induced activation of NF-κB and MAPKs and the formation of necrosomes, but is required for trafficking of necrosomes to their potential functioning site, an unidentified subcellular organelle that can be fractionated into heterotypic membrane fractions. The TNF-induced Gβγ-Src signaling pathway is independent of RIP1/RIP3 kinase activity and necrosome formation, but is required for the necrosome to function.
Keywords: Gβγ complex, Src kinase, necroptosis, necrosome, heterotypic membrane, RIP1, RIP3
Introduction
In contrast to apoptosis, necrosis was often considered merely as a kind of chaotic and non-regulated cell death before mechanistic studies began to reveal that necrosis can also be regulated under certain circumstances1,2. Necrotic cell death has been implicated in a variety of physiopathological processes, such as host responses to genotoxic stresses and other adverse environmental factors such as bacterial and viral infections, as well as in pathogenesis of various diseases such as atherosclerosis, Crohn's syndrome, acute pancreatitis and neurodegenerative diseases3,4,5,6. Hence, it is conceivable that a thorough investigation to decipher the mechanisms of necrosis will provide us more therapeutic strategies for the treatment of various diseases. A broad range of extracellular stimuli, such as H2O2, DNA alkylating agent MNNG and cytokine tumor necrosis factor α (TNF), can induce necrosis7,8. Among these, TNF-induced receptor-interacting protein 1 (RIP1)- and RIP3-dependent necrotic cell death (named necroptosis) has been intensively studied.
TNF is a pleiotropic cytokine that regulates diverse signaling pathways including apoptosis (caspase-dependent cell death) and necroptosis (caspase-independent cell death)9. TNF can induce necroptosis in certain cell types such as murine fibrosarcoma L929 cells, cells of hematopoietic origin and epithelial cells of the gastrointestinal tract10,11. Inhibition of caspases or overexpression of RIP3 can switch TNF-induced apoptosis to necroptosis in some cell types12. It is known that TNF induces apoptosis or necroptosis by engaging TNF receptor 1 (TNFR1). When TNF binds to TNFR1, TNFR1 becomes trimerized, leading to the recruitment of the downstream effectors to form signaling complex I. TNF-induced gene expression is primarily mediated by NF-κB and MAP kinase pathways downstream of complex I, which contains TNFR1, TNFR1-associated death domain (TRADD), TNFR-associated factor 2 (TRAF2), RIP1 and inhibitor of κB kinase (IKK) complex13. Upon TNFR1 endocytosis, TRADD interacts with Fas-associated death domain (FADD), which subsequently recruits RIP1 and caspase-8 to form complex II, leading to the induction of caspase-dependent apoptosis13. When RIP3 is highly expressed in a given cell, it can incorporate into complex II to form the necrosome (also known as complex IIb), which triggers necrotic cell death12,14,15. Recently, mixed lineage kinase domain-like (MLKL) has been identified as a novel component of the necrosome14,16,17,18. Knockdown of MLKL abolished TNF-induced necroptosis without affecting the interaction between RIP1 and RIP314,16. Moreover, phosphorylation of MLKL by RIP3 has been suggested to be critical for necrotic signaling14.
Heterotrimeric guanine nucleotide-binding protein (G protein) or Gαβγ complex plays a central role in the G protein coupled-receptor (GPCR) signaling pathway. In early studies, Gα was considered the major player of this complex while Gβγ was regarded merely as a docking platform for Gα. The first evidence indicating that Gβγ functions not only as a scaffold but also as a signal transducer/activator came from the study of the activation of muscarinic-gated potassium channels in chicken embryonic atrial cells19. More recently, Gβγ has also been revealed to participate in the activation of a variety of signaling pathways including the cAMP/PKA, PI3K, calcium, Src (Rous sarcoma oncogene) kinase and GIRK pathways20,21,22,23,24. Gβγ dimer within the heterotrimeric G protein complex is composed of Gβ and Gγ subunits. As the isoforms of Gβs or Gγs share a high degree of sequence homology, it was proposed that they might function redundantly. However, a growing body of evidence suggests that each distinct Gβ or Gγ isoform may intrinsically possess unique biological functions25,26. In addition, different Gβ and Gγ combinations also seem to perform distinctive functions27.
In order to identify novel molecules that regulate TNF-induced necroptosis, we performed a systematic screening for phenotypes of defective necroptosis in L929 cells carrying gene mutations introduced by random retrovirus insertion. As a result, we successfully identified several novel protein components that are involved in TNF-induced necroptosis28,29,30,31. Gγ10 is one of the genes identified to be required for TNF-induced necroptosis. Knockdown of Gγ10, but not the other γ subunits, rendered the L929 cells resistant to TNF-induced cell death. Further study revealed that Gα subunits are not involved in necroptosis and a specific β subunit Gβ2 is required for TNF-induced necroptosis. The participation of the Gβ2γ10 complex in TNF-induced necroptosis is supported by the data that Gβγ inhibitor suppressed TNF-induced necroptosis. Although it is required for TNF-induced necroptosis, Gγ10 is not involved in the assembly of necrosomes. Src activation is downstream of the Gβ2γ10 complex in TNF-induced necroptosis and Src appears to play a role in necrosome trafficking, which is apparently required for necrosomes to execute necroptosis.
Results
Gγ10 is specifically required for TNF-induced necroptosis.
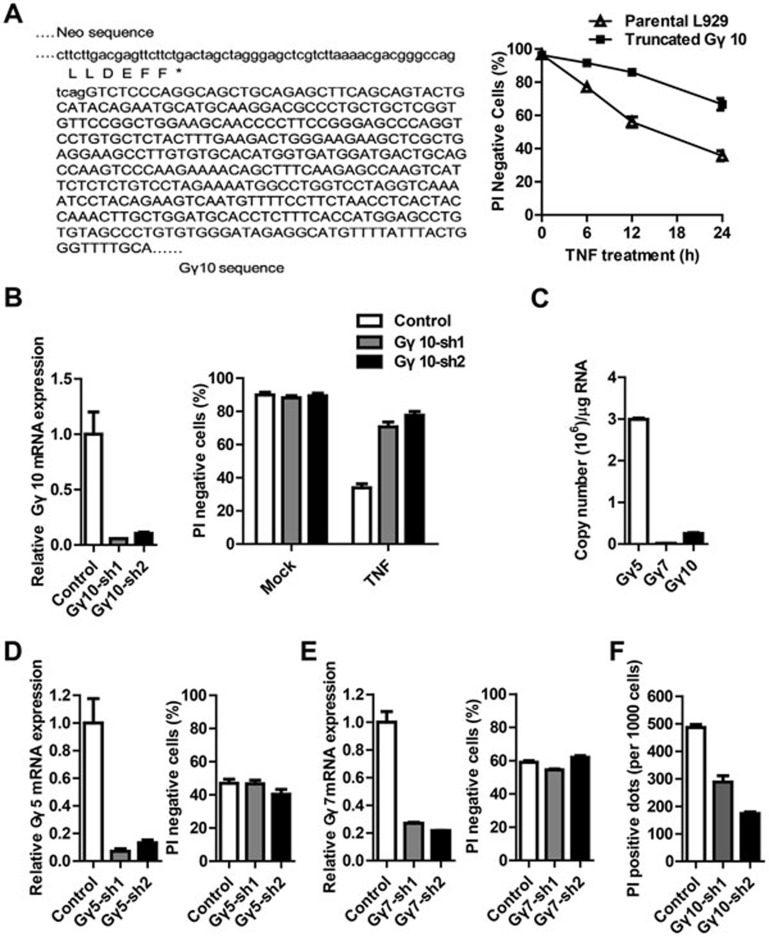
In order to find new genes involved in TNF-induced necroptosis, we performed a functional screening among mutants of murine fibrosarcoma L929 cells generated by random retrovirus insertion as described previously28,29,30,31. One of the resistant lines was found to carry a disrupted Gγ10 gene resulting from the insertion of the retroviral genome into the intron between the first and second exons of Gγ10 (Figure 1A). This mutant L929 cell line, named truncated Gγ10, is resistant to TNF-induced death when compared to the parental L929 cells (Figure 1A). To confirm the role of Gγ10 in TNF-induced necroptosis, we used shRNAs to knock down Gγ10 gene in L929 cells. As shown in Figure 1B and Supplementary information, Figure S1A, two Gγ10-specific shRNAs can effectively reduce Gγ10 mRNA level, and the reduction of Gγ10 expression resulted in a resistance to TNF-induced cell death.
Figure 1.
Gγ10 is required for TNF-induced necroptosis. (A) A TNF-resistant L929 mutant cell line generated by retrovirus insertion was isolated and the insertion site was identified by 3′ RACE. The insertion is in the Gγ10 gene and the cell line was named truncated Gγ10. The sequence of the fused Gγ10 (uppercase) and neo transgene-containing DNA fragment is shown. The amino acid sequence of the neo gene is shown under its cDNA sequence. The parental and truncated Gγ10 cells were treated with TNF (10 ng/ml) for different time periods. Cell survival rate was determined using propidium iodide (PI) exclusion (right panel). (B) L929 cells were infected with lentivirus encoding Gγ10 shRNA or control shRNA. After 48 h, knockdown efficiency of Gγ10 was examined by qRT-PCR. Gγ10-knockdown cells were treated with TNF (10 ng/ml) for 12 h and cell viability was measured as in A. (C) The mRNA levels of Gγ5, Gγ7 and Gγ10 in L929 cells were measured by absolute quantification RT-PCR. (D, E) L929 cells were infected with lentivirus encoding Gγ5 or Gγ7 shRNA. Knockdown efficiency and TNF-induced cell death were determined as in B. (F) Peritoneal macrophages were infected with lentivirus encoding Gγ10 shRNA or control shRNA for 5 days and then challenged with LPS (100 ng/ml) plus zVAD (20 μM) for 30 h. PI-positive dots (dead cells) were counted under a microscope. The average numbers of dead cells per 1 000 cells obtained from at least three different views of about 500 cells per view are shown. Data in A, B and D-F depict mean ± SEM of one representative experiment of three or more. See also Supplementary information, Figure S1.
The mouse Gγ family contains 12 members, which are very similar in amino acid sequences. Sequence alignment of Gγ family members showed that Gγ5 and Gγ7 are more similar to Gγ10 than the other Gγ family members (Supplementary information, Figure S1B). The mRNA level of Gγ10 is much lower than that of Gγ5, and much higher than that of Gγ7 in L929 cells (Figure 1C). We knocked down Gγ5 and Gγ7 and found that their knockdown had no effect on TNF-induced cell death (Figure 1D and 1E). These data suggest that Gγ10, but not the other Gγ family members, is involved in TNF-induced necroptosis in L929 cells.
As lipopolysaccharide (LPS), the cell wall component of Gram-negative bacteria, plus pan caspase inhibitor N-Benzyloxycarbonyl-Val-Ala-Asp(O-Me) fluoromethyl ketone (zVAD) can induce RIP3-dependent necroptosis of peritoneal macrophages12,32, we tested whether Gγ10 is required for the necroptosis of macrophages. As shown in Figure 1F and Supplementary information, Figure S1C, knockdown of Gγ10 reduced LPS plus zVAD-induced macrophage death. Knockdown Gγ10 also partially inhibited TNF plus cycloheximide (CHX) plus zVAD-indued MEF necroptosis (Supplementary information, Figure S1D). Thus, Gγ10 participates in RIP3-dependent necroptosis in different cell types.
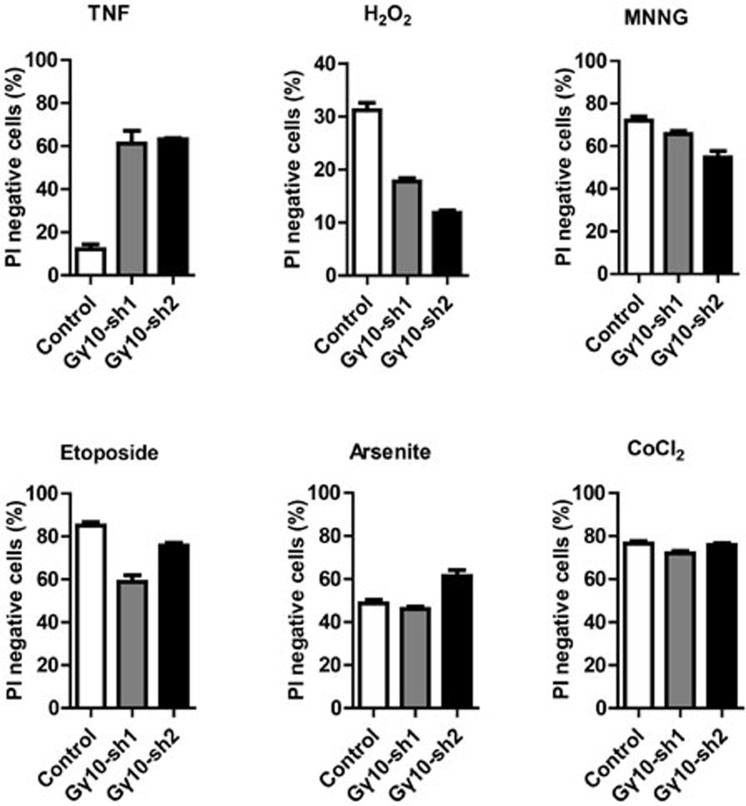
We next tested whether Gγ10 is also involved in cell death induced by other stimuli in L929 cells. H2O2, DNA alkylating agent MNNG, topoisomerase inhibitor Etoposide, arsenite and CoCl2 were tested. As shown in Figure 2, cell death induced by these stimuli either was enhanced or remained unchanged in Gγ10-knockdown cells. Thus, Gγ10 is selectively involved in TNF-induced necroptosis.
Figure 2.
Gγ10 is selectively involved in TNF-induced necroptosis. Control and Gγ10-knockdown L929 cells were treated with either TNF (10 ng/ml) for 10 h, H2O2 (3 mM) for 4 h, MNNG (0.5 mM) for 30 h, Etoposide (50 μM) for 30 h, Arsenite (25 μM) for 16 h or CoCl2 (50 mM) for 30 h. Cell viability was measured using PI exclusion. Data depict mean ± SEM of one representative experiment of three or more.
The Gβγ complex containing Gγ10 and Gβ2 participates in TNF-induced necroptosis
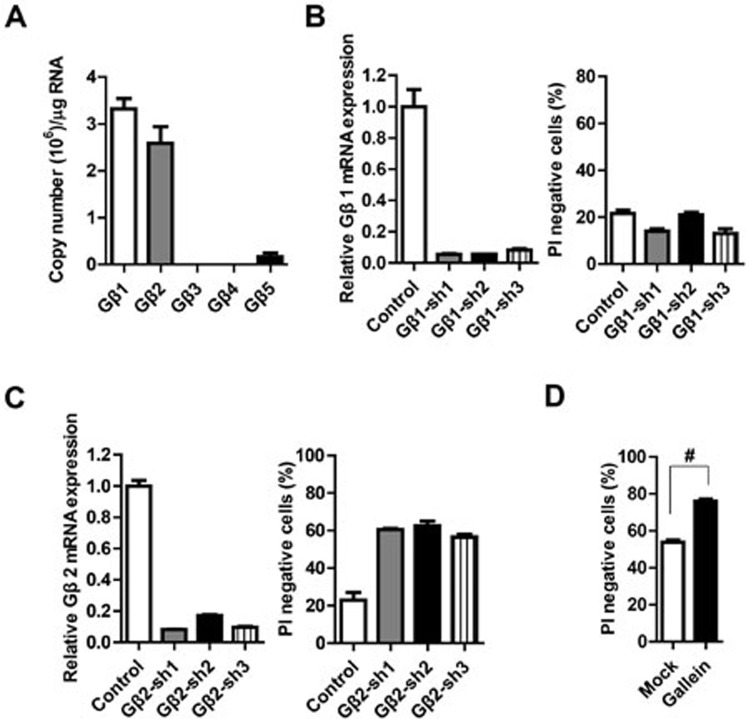
As nearly all Gγ family members require the interaction with Gβ family members to function properly33,34, Gβ family members likely also participate in TNF-induced necroptosis. The expression levels of several Gβ family members were measured in L929 cells. Similar to Gγ family members, the expression abundance of Gβ protein family members varies in L929 cells. Gβ1 and Gβ2 are highly expressed while Gβ3 and Gβ4 are undetectable and Gβ5 is expressed at a low level in L929 cells (Figure 3A). As Gβ1 and Gβ2 are highly expressed in L929 cells, we used shRNAs to knock down their expression and examined whether any of these two Gβ proteins is required for TNF-induced necroptosis in L929 cells. As shown in Figure 3B, Gβ1 knockdown did not affect TNF-induced necroptosis; in contrast Gβ2 knockdown blocked TNF-induced necroptosis (Figure 3C). In order to further confirm the result, we knocked out Gβ2 in L929 using the CRISPR/Cas9-mediated gene targeting method35,36. As shown in Supplementary information, Figure S2A, Gβ2 expression in the four knockout cell lines was almost completely eliminated and TNF-induced necroptosis was inhibited. This result reinforces that Gβ2 is involved in TNF-induced necroptosis. It is worth to note that the amino acid sequences of Gβ1 and Gβ2 are highly similar (Supplementary information, Figure S2B), but Gβ1 and Gβ2 appear to behave distinctively in necroptosis. As the expression of Gβ3 and Gβ4 is undetectable in L929 cells and Gβ5 is less similar to Gβ2 than Gβ1, we did not further test the involvement of these β subunits in necroptosis.
Figure 3.
Gβ2 is required for TNF-induced necroptosis in L929 cells. (A) The mRNA expression levels of Gβ family members in L929 cells were measured by absolute quantification RT-PCR. (B, C) L929 cells were infected with lentivirus encoding Gβ1 or Gβ2 shRNA. Knockdown efficiency and TNF-induced cell death were determined as described in Figure 1B. (D) L929 cells were pretreated with or without Gβγ complex inhibitor gallein (75 μM) for 2 h and then challenged with TNF (10 ng/ml) for 10 h. Cell viability was measured using PI exclusion. #P < 0.01, t test. Data in B-D depict mean ± SEM of one representative experiment of three or more. See also Supplementary information, Figure S2.
When overexpressed in 293T cells, Gγ10 was shown to interact with Gβ2 (Supplementary information, Figure S2C), which is consistent with a previous study37. To test whether the Gγ and Gβ subunits act as a Gβγ dimer to regulate necroptosis, we treated the cells with a Gβγ complex inhibitor, gallein, to inhibit the function of Gβγ complex38. Compared to the control, cells pretreated with gallein were more resistant to TNF-induced necroptosis (Figure 3D). However, gallein may not be an effective inhibitor as the inhibition of necroptosis with gallein was less efficient than the knockdown of Gγ10 or Gβ2. Indeed, it was reported that gallein only blocked the interaction of Gβγ with some but not all effectors39. Nonetheless, the inhibitory effect of gallein suggests that the Gγ10- and Gβ2-containing Gβγ complex is responsible for TNF-induced necroptosis.
Gα subunits are not required for TNF-induced necroptosis
As the Gβγ complex interacts with Gα subunits and regulates their GTPase activity, it needs to be evaluated whether Gα subunits are involved in TNF-induced necroptosis. Gα subunits were divided into four major classes (Gαs, Gαi, Gα12/13 and Gαq) based on their sequence homologies40. Different Gα subunit classes have different functions.
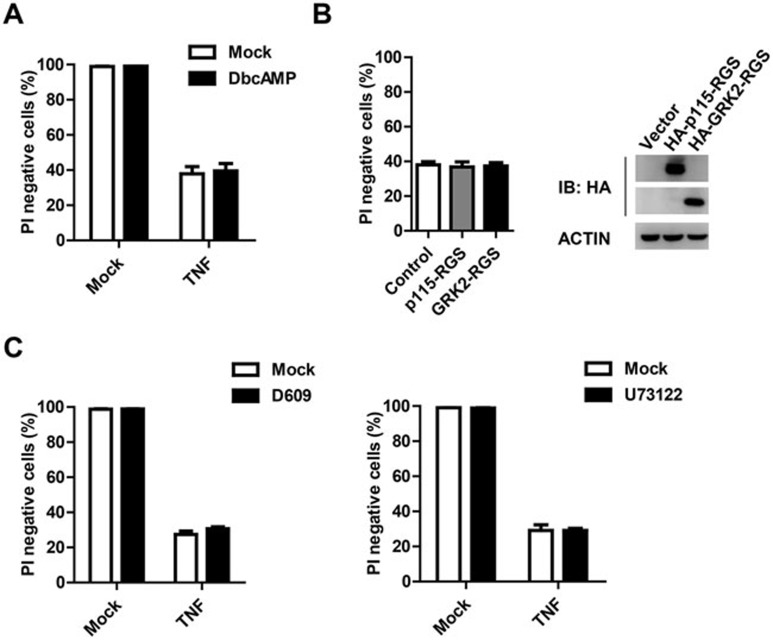
Activation of Gαs or Gαi can activate or inhibit adenylate cyclase activity, respectively, which can influence cellular cAMP production. Studies from several research groups have revealed that TNF treatment did not affect cAMP production in L929 cells and other cell lines41,42,43, indicating that TNF does not activate Gαs or Gαi. Because cAMP is one of the most important downstream effectors of Gαs and Gαi, whether cAMP can influence TNF toxicity has also been studied. Some studies showed that cAMP could protect cells from TNF-induced cell death while others argued against this conclusion42,44. To determine whether cAMP plays any role in our system, L929 cells were pretreated with the cAMP mimic drug dbcAMP and then challenged with TNF. As shown in Figure 4A, dbcAMP did not affect TNF-induced cell death in L929 cells.
Figure 4.
Gα subunits are not required for TNF-induced necroptosis in L929 cells. (A) Cells were pretreated with or without cAMP-mimic drug, dbcAMP (100 μM) overnight and then challenged with TNF (10 ng/ml) for 12 h. Cell viability was measured using PI exclusion. (B) Cells were infected with lentivirus encoding HA-p115-RGS or HA-GRK2-RGS. After 48 h, part of the cells were harvested for immunoblotting with anti-HA and anti-actin antibodies, and other parts of the cells were challenged with TNF (10 ng/ml) for 12 h before cell viability was measured using PI exclusion. (C) Cells were pretreated with PLC inhibitor D609 (20 μg/ml) for 30 min or U73122 (2 μM) for 45 min and then challenged with TNF (10 ng/ml) for 8 h. Cell viability was measured using PI exclusion. Data in A-C depict mean ± SEM of one representative experiment of two or three.
Overexpression of RhoGEF-p115 RGS domain can selectively inhibit the activity of the Gα12/13 subunits45,46. We found that overexpression of RhoGEF-p115 RGS domain had no effect on TNF-induced necroptosis (Figure 4B), while it suppressed lysophosphatidic acid (LPA)-induced Gα12/13-dependent stress fiber formation (Supplementary information, Figure S3A)47,48,49.
GRK2-RGS domain overexpression can inhibit Gαq activity45,46, but had no effect on TNF-induced necroptosis (Figure 4B). The inhibition of Gαq activity by GRK2-RGS domain overexpression was demonstrated by the results that such overexpression inhibited ATP-induced calcium mobilization (Supplementary information, Figure S3B), which was known to be Gαq-dependent43. A major downstream effector of Gαq is phospholipase C (PLC)50. Pretreatment of L929 cells with PLC inhibitor D609 or U73122 did not affect TNF-induced necroptosis (Figure 4C).
Taken together, these results suggest that Gαs, Gαi, Gα12/13 and Gαq are unlikely to be involved in TNF-induced necroptosis.
Src is activated by TNF stimulation in a Gγ10-dependent manner
Previous studies have shown that cAMP, PI3K, GIRK channel, calcium and Src pathways can act downstream of the Gβγ complex20,21,22,23,24,51. Among these pathways, we found that the cAMP pathway is not involved in TNF-induced necroptosis in L929 cells (Figure 4A). By using the PI3K inhibitor LY294002, we found that inhibition of PI3K did not affect TNF-induced necroptosis (Supplementary information, Figure S4A). As for GIRK channel activity19, no study has been done to address the relationship between cellular potassium concentration and TNF-induced cell death. We found that the potassium concentration did not significantly change during TNF stimulation, and that Gγ10 knockdown did not affect potassium concentration before and after TNF stimulation (Supplementary information, Figure S4B). Intracellular calcium concentration has been reported to affect TNF-induced cell death29,52. As the Gβγ complex can modulate N-type calcium channels53,54, we examined whether intracellular calcium concentration was affected by Gγ10 knockdown and found that Gγ10 knockdown did not affect calcium concentration (Supplementary information, Figure S4C).
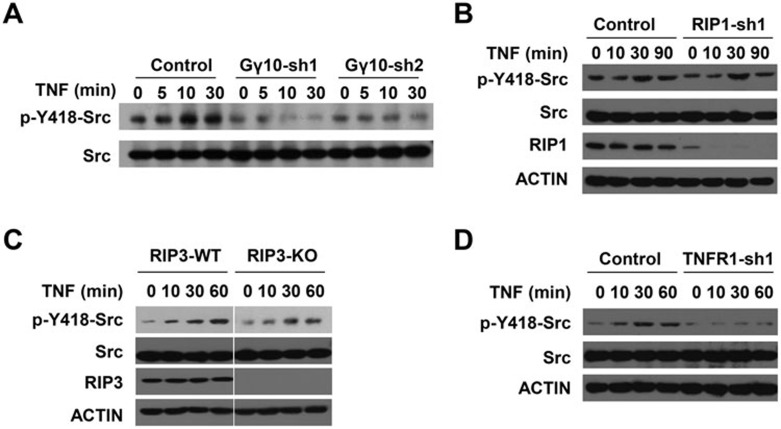
Src kinase activity can be regulated by the Gβγ complex22,55,56,57,58, and activation of Src by TNF stimulation has been reported59,60. We examined whether TNF-induced activation of Src was compromised when Gγ10 was knocked down in L929 cells. Consistent with previous studies60, the phosphorylation level of Tyr418 in mouse Src, a marker of Src kinase activation, was increased in L929 cells after TNF treatment (Figure 5A). Interestingly, this activation was diminished in Gγ10-knockdown cells (Figure 5A).
Figure 5.
Src is activated in L929 cells by TNF stimulation in a Gγ10-dependent but necrosome-independent manner. (A) Control and Gγ10-knockdown L929 cells were treated with TNF (10 ng/ml) for the indicated time periods. Cells were then harvested and the expression levels of the total Src kinase and Tyr418-phosphorylated Src in whole-cell extract were examined by immunoblotting with anti-Src and anti-phospho-Tyr418-Src antibodies. (B-D) The same as in A except that control and RIP1-knockdown, RIP3-knockout, or TNFR1-knockdown L929 cells were used. See also Supplementary information, Figure S3.
We next determined whether the activation of Gγ10-Src signaling is dependent on RIP1 and RIP3, and found that TNF-induced Src activation was not affected by RIP1 knockdown or RIP3 knockout (Figure 5B and 5C). To ensure that TNF-induced Src activation is dependent on TNFR1, we analyzed Src phosphorylation in TNFR1-knockdown cells. Knockdown of TNFR1 reduced TNF-induced Src phosphorylation (Figure 5D) and cell death (Supplementary information, Figure S4D). Taken together, these results suggest that TNF-induced Src activation is not a downstream event of the necrosome formation.
Src kinase is a downstream effector of Gβ2γ10 complex in TNF-induced necroptosis
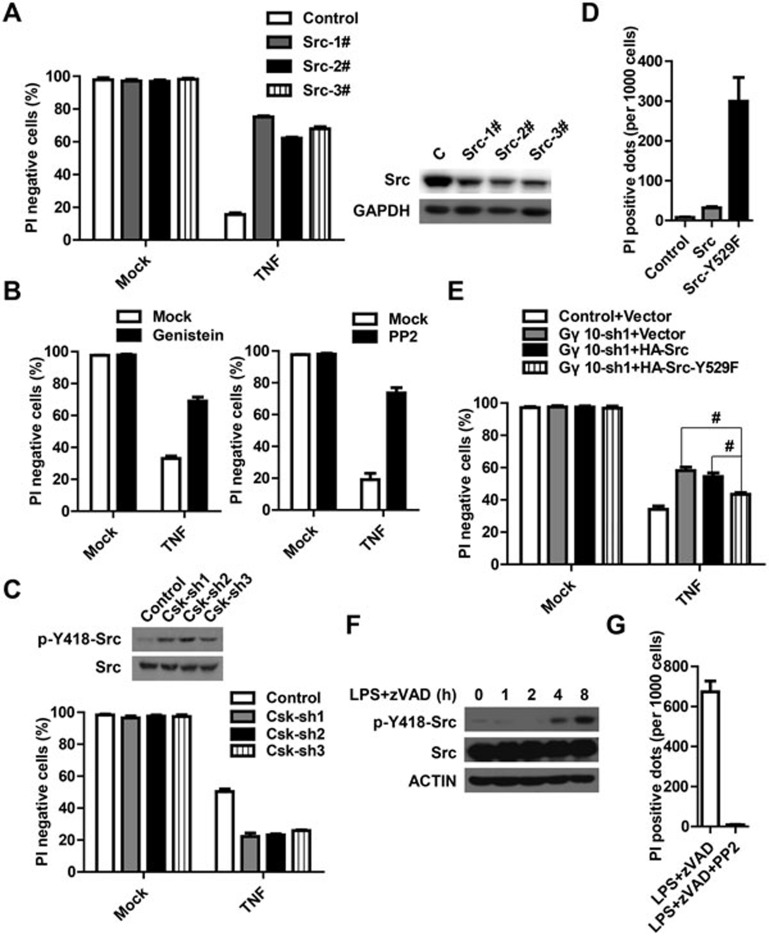
As Gγ10 is required for the upregulation of Src kinase activity induced by TNF, Src likely serves as the signal effector downstream of Gβ2γ10 in TNF-induced necroptosis. To test this hypothesis, we examined whether Src inactivation could inhibit necrotic cell death as the Gγ10 knockdown did. L929 cells were infected with control or Src shRNA-encoding lentivirus and the cell lysates were analyzed for Src protein levels. By an unknown reason, an efficient knockdown of Src resulted in cell death and we only obtained viable cells with ∼50% reduction of Src protein levels. Decrease in Src protein levels reduced TNF-induced cell death (Supplementary information, Figure S5A), reminiscent of what was observed in Gγ10-knockdown cells. However, the resistance to TNF-induced cell death was less obvious in Src-knockdown cells than in Gγ10-knockdown cells, which is most likely due to the less effective knockdown of Src compared with that of Gγ10. In order to confirm this result, CRISPR/Cas9-mediated gene targeting was used to knock out Src in L929 cells. We were unable to knock out Src gene in both alleles in L929 cells, but several knockout cell lines showed significant reduction of Src expression (Figure 6A). Consistent with Src-knockdown experiments, Src gene knockout also decreased TNF-induced necroptosis (Figure 6A). To further confirm the involvement of Src in TNF-induced necroptosis, we pretreated cells with the general tyrosine kinase inhibitor Genistein or selective Src kinase family inhibitor PP2, and found that both inhibitors attenuated TNF-induced cell death (Figure 6B, Supplementary information, Figure S5B and S5C).
Figure 6.
Src kinase activity is required for TNF-induced necroptosis. (A) The Src protein level was analyzed in control and Src-knockout cell lines. The control and three Src-knockout cell lines were named as Control, Src-1#, Src-2# and Src-3#, respectively. The cells were challenged with TNF (10 ng/ml) and cell viability was measured. (B) Cells were pretreated with or without general tyrosine kinase inhibitor Genistein (100 μM) for 2 h or Src kinase family-specific inhibitor PP2 (2 μM) for 6 h. Cells were then challenged with TNF for 10 h. Cell viability was measured. (C) Parts of Control and Csk-knockdown cells were harvested and the cell extracts were subjected to western blot to examine the levels of Src and pTyr418-Src, and other parts of Control and Csk-knockdown cells were treated with TNF for 8 h. Cell viability was measured. (D) L929 cells were infected with control lentivirus or the indicated lentivirus expressing WT Src or constitutively active mutant Src-Y529F for 48 h. PI-positive dots (dead cells) were counted under a microscope. The average numbers of dead cells per 1 000 cells obtained from at least three different views of about 500 cells per view are shown. (E) Control or Gγ10-knockdown L929 cells were infected with lentivirus-expressing Vector, HA-Src (WT) or HA-Src-Y592F. A low dose of lentivirus was selected to adjust the expression of Src-Y529F to a modest level so that Src-Y529F-expressing cells did not undergo spontaneous necroptosis in the absence of TNF stimulation. A total of 48 h after lentivirus infection, cells were challenged with TNF for 10 h. Cell viability was measured. #P < 0.01, t test. (F) Peritoneal macrophages were treated with LPS (100 ng/ml) plus zVAD (20 μM) for different periods of time. Tyr418 phosphorylation level of Src was measured. (G) Peritoneal macrophages were pretreated with or without PP2 (2 μM) for 6 h and then challenged with LPS (100 ng/ml) plus zVAD (20 μM) for 20 h. PI-positive dots (dead cells) were counted under a microscope. The average numbers of dead cells per 1 000 cells obtained from at least three different views of about 500 cells per view are shown. Data in A-E, and G depict mean ± SEM of one representative experiment of three or more. See also Supplementary information, Figure S4.
It has been reported that the basal activity of Src is usually low as its kinase activity is suppressed in quiescent cells61. Phosphorylation of Tyr529 in Src by tyrosine kinase Csk contributes to the suppression of the kinase activity of Src62. Knockout or knockdown of Csk leads to the upregulation of Src kinase activity63,64. Indeed, Csk knockdown in L929 cells upregulated the phosphorylation level of Tyr418 in Src, indicating an elevated Src kinase activity (Figure 6C and Supplementary information, Figure S5D). In supporting the positive role of Src in TNF-induced necroptosis, Csk-knockdown cells were more sensitive to TNF-induced necroptosis than control L929 cells (Figure 6C). In order to further decipher the role of activated Src in necroptosis, a constitutively active mutant Src-Y529F65 was introduced into L929 cells. Consistently, spontaneous necroptosis significantly increased in cells overexpressing Src-Y529F when compared to control or wild-type (WT) Src-overexpressing cells (Figure 6D and Supplementary information, Figure S5E). Spontaneous necroptosis driven by Src-Y529F overexpression was induced in a RIP3-dependent manner (Supplementary information, Figure S5F). More importantly, as shown in Figure 6E and Supplementary information, Figure S5G, Src-Y529F but not WT Src can partially reverse Gγ10 knockdown-induced TNF resistance. In this experiment, to avoid the bias originating from the spontaneous necroptosis induced by Src-Y529F overexpression, we adjusted the expression level of Src-Y529F by reducing the amount of lentivirus so that Src-Y529F-expressing cells did not undergo spontaneous necroptosis in the absence of TNF stimulation (Figure 6E).
As LPS plus zVAD can induce RIP3-dependent peritoneal macrophage necroptosis12,32, we next attempted to study whether Src is involved in this signaling pathway. LPS plus zVAD activated Src kinase in a time-dependent manner and treatment with the Src kinase inhibitor PP2 abolished LPS plus zVAD-induced macrophage necroptosis (Figure 6F and 6G), suggesting that Src kinase plays an important role in necroptosis induced by different stimuli in different cell types. However, LPS alone did not induce cell death in macropahges though it was sufficient to induce Src activation, suggesting that a combined effect of Src activation and caspase inhibition is required for macrophage necroptosis (Supplementary information, Figure S5H). Collectively, our data demonstrated that Src kinase is a downstream effector of Gβ2γ10-mediated necroptosis.
Gγ10 does not affect the signaling downstream of complex I and the formation of necrosomes but is required for the translocation of RIP1/RIP3/MLKL-containing necrosomes into a heterotypic membrane fraction.
It is well established that activation of the NF-κB and MAPK pathways is the downstream event of complex I formation13,66,67. We found that Gγ10 knockdown did not affect TNF-induced phosphorylation of NF-κB (p65), ERK, JNK and p38 (Supplementary information, Figure S6A), indicating that Gγ10 has no effect on complex I signaling.
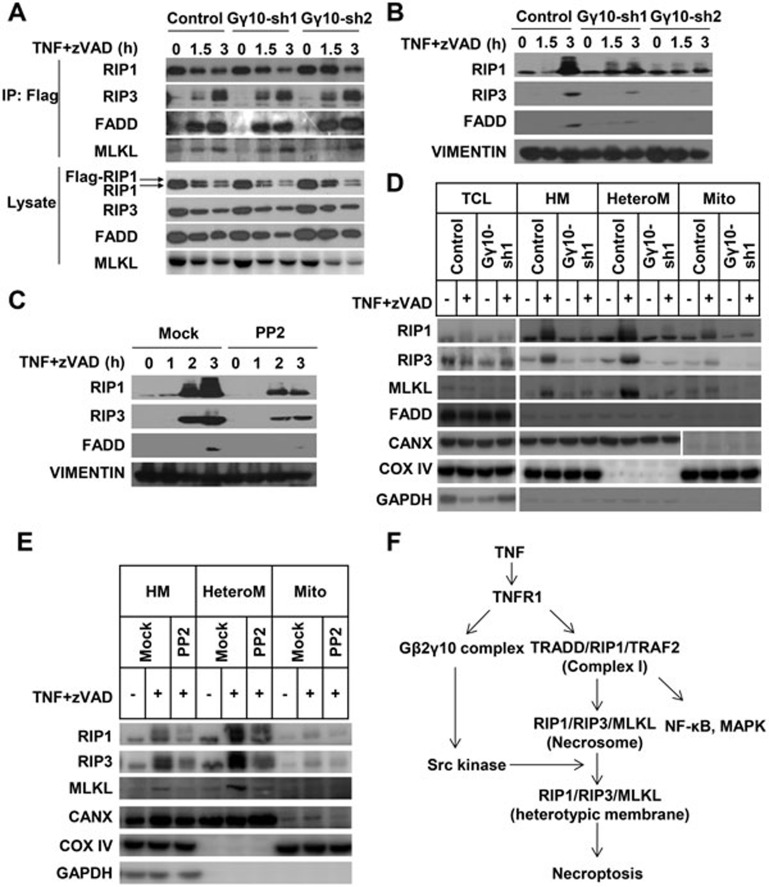
As the formation of necrosomes is the hallmark of TNF-induced necroptosis, we investigated whether Gγ10 plays any role in the assembly of necrosomes. In order to effectively immunoprecipitate RIP1, we knocked in a flag tag into one allele of the RIP1 gene by homology-directed recombination to express N-terminal flag-tagged RIP1 in L929 cells. We have determined that flag-RIP1 can be incorporated into necrosomes as WT RIP1 and that the knock-in L929 line shows a similar sensitivity to TNF as the parental L929 line in terms of necroptosis (data not shown). As some components of necrosomes can be cleaved by caspase(s), TNF plus zVAD treatment was used68. Knockdown of Gγ10 or pretreatment with Src kinase inhibitor reduced TNF plus zVAD-induced necroptosis in flag-RIP1 L929 cells (Supplementary information, Figure S6B and S6C). Flag-RIP1 was immunoprecipitated with anti-flag antibody beads, and the co-immunoprecipitated components of necrosomes were analyzed. As shown in Figure 7A, RIP3, FADD and MLKL were co-immunoprecipitated with flag-RIP1 regardless of whether Gγ10 was knocked down or not. Consistent with this observation, treatment with the Src kinase inhibitor PP2 did not affect TNF-induced necrosome formation (Supplementary information, Figure S6D).
Figure 7.
Gγ10 is dispensible for necrosome formation but is required for accumulation of RIP1 and RIP3 in the heterotypic membrane fraction during necroptosis. (A) Flag-RIP1-knock-in L929 cells were infected with lentivirus-expressing Gγ10 shRNA or control shRNA. Cells were challenged with TNF (10 ng/ml) plus zVAD (20 μM) for the indicated time periods. Whole-cell lysates were subjected to immunoprecipitation using anti-Flag M2 beads. The protein abundance of RIP1, RIP3, FADD and MLKL in the immunoprecipitates and total-cell lysates were analyzed by immunoblotting with corresponding antibodies. (B) L929 cells were challenged with TNF (10 ng/ml) plus zVAD (20 μM) for the indicated time periods. The Triton X-100-insoluble fractions of control or Gγ10-knockdown cells were isolated as described in Materials and Methods. The collected insoluble fractions were dissolved with 1% SDS and then were analyzed for the indicated protein by immunoblotting. (C) Cells were pretreated with PP2 (2 μM) for 2 h and then processed as in B. (D) Gγ10-knockdown and control cells were challenged with TNF (10 ng/ml) plus zVAD (20 μM) for 3 h. The indicated fractions were isolated as described in Materials and Methods and then were dissolved with 1% SDS. The quantities of the indicated proteins were analyzed by immunoblotting with antibodies against RIP1, RIP3, MLKL, FADD, CANX, COX IV and GAPDH. CANX is an ER and MAM marker protein. COX IV is a mitochondrial marker protein. HM, heavy membrane; HeteroM, heterotypic membrane; Mito, mitochondria. (E) Cells were pretreated with PP2 (2 μM) for 4 h and then processed as in D. (F) A proposed model of the pathways in TNF-induced necroptosis. The TNF-activated Gβ2γ10-Src pathway acts downstream of TNFR1 but parallels the complex I-necrosome pathway. Src regulates the trafficking of necrosomes to a special subcellular organelle that can be fractionated into the heterotypic membrane fraction. Translocation of necrosomes to this organelle is required for necrosomes to execute necroptosis. See also Supplementary information, Figure S5.
It has been reported that necrosomes assemble upon TNF stimulation and then increasingly accumulate in Triton X-100 (or NP-40)-insoluble cellular fraction69,70. As necrosome formation was unaffected by Gγ10 knockdown, we next examined whether Gγ10 knockdown would affect the accumulation of necrosomes in the detergent-insoluble fraction. As shown in Figure 7B, while RIP1 and RIP3 proteins accumulated in Triton X-100-insoluble fraction in control cells, this enrichment was reduced in Gγ10-knockdown cells. Consistently, PP2 treatment also reduced the accumulation of RIP1 and RIP3 in Triton X-100-insoluble fraction (Figure 7C).
It was shown in a recent study that TNF induces an enrichment of RIP3 in the Triton X-100-insoluble portion of heavy membrane (HM) fraction69. As crude mitochondria exist in the HM fraction, we sought to further investigate the role of mitochondria in necrosome signal transduction. Indeed, we found that TNF plus zVAD treatment led to an enrichment of RIP1, RIP3 and MLKL in the HM fraction (Figure 7D). We further fractionated the HM fraction into pure mitochondrial and heterotypic membrane fractions using a published method71,72. The heterotypic membrane fraction is also called microsome (μS)71. The transmission electron microscopy pictures of mitochondrial and heterotypic membrane fractions were shown in Supplementary information, Figure S6E. The heterotypic membrane fraction may contain endoplasmic reticulum (ER), Golgi and mitochondrial associated membranes (MAM). TNF plus zVAD induced an enrichment of RIP1, RIP3 and MLKL in the heterotypic membrane fraction but not in the pure mitochondrial fraction (Figure 7D). Interestingly, knockdown of Gγ10 significantly blocked TNF-induced accumulation of RIP1, RIP3 and MLKL in the HM and the heterotypic membrane fractions (Figure 7D). FADD was not enriched in HM and the heterotypic membrane fraction, suggesting that FADD was dissociated from the necrosomes during the translocation (Figure 7D). Furthermore, when cells were treated with the Src kinase inhibitor PP2, the accumulation of RIP1, RIP3 and MLKL in the HM and heterotypic membrane fractions was also inhibited (Figure 7E), suggesting that the Gβ2γ10-Src signaling regulates the translocation of RIP1/RIP3/MLKL-containing necrosomes to the heterotypic membrane fraction.
Discussion
By employing random retrovirus insertion, we uncovered a novel role of the Gβγ complex in TNF-induced necrotic cell death. We provide genetic evidence that Gγ10 and Gβ2 are selectively involved in TNF-induced necroptosis. Furthermore, we show that Src kinase is a Gβ2γ10-downstream effector in TNF-induced necroptosis. The Gβ2γ10-Src signaling pathway likely acts in parallel to the signaling pathway of TNFR1-mediated complex I and necrosome formation, but functions to regulate the translocation of RIP1/RIP3/MLKL-containing necrosomes to an unidentified subcellular organelle that can be fractionated into a heterotypic membrane fraction. A proposed model of how Gβ2γ10-Src signaling functions in necroptosis is shown in Figure 7F.
The Gβγ complex has been recognized as a critical regulator in diverse signaling pathways53,55,73. It has been shown to participate in unfolded protein response (UPR) and O3-induced cell death in Arabidopsis74,75. A specific Gβγ complex Gβ2γ2, but not the other forms of the Gβγ complex, can mediate apoptosis induced by familial Alzheimer's disease-associated mutant of amyloid precursor protein27. Pertussis toxin (PTX), an exotoxin produced by bacterium Bordetella pertussis, can inhibit TNF-induced necroptosis41,76. PTX inhibits Gα activity through ADP ribosylation, which prevents Gα and, as a result, heterotrimeric G proteins from interacting with GPCRs. When heterotrimeric G proteins cannot be activated by GPCRs, Gβγ complex cannot be released from the heterotrimer, and therefore PTX can also affect signaling pathways mediated by Gβγ55,77,78,79. As Gα subunits are not involved in TNF-induced necroptosis (Figure 4A-4C), it is possible that PTX would reduce the basal-free Gβγ level and thus inhibits TNF-induced cell death via diminishing the signaling mediated by Gβγ.
It is known that activation of Gαβγ heterotrimeric complexes leads to the release of Gβγ dimers. To date there is almost no publication suggesting that TNF activates Gαβγ heterotrimers. We also failed to detect G protein subunits in TNFR1 complexes before and after TNF stimulation in L929 cells (data not shown). Therefore, it is likely that either TNF induces a weak activation of Gαβγ, which cannot be detected, or TNF just cannot activate Gαβγ at all. If TNF does not activate Gαβγ, a question remains as to how TNF activates Gβ2γ10 to upregulate Src activity. The regulation of Gβ2γ10 by TNF is difficult to be explained by classical models of G protein activation. As our data undoubtedly demonstrate the involvement of Gβ2γ10 in TNF-induced necroptosis, TNF likely regulates Gβγ dimer activity through a currently unrevealed mechanism, which may even do not require activation of the Gαβγ complex. The mechanism of TNF-induced Gβ2γ10 activation awaits further investigation.
Src kinase was first discovered as a cellular ortholog of V-Src in Rous sarcoma virus (RSV)80,81. Src kinase has been shown to contain several domains, including N-terminal myristoylated domain, SH3 domain, SH2 domain and kinase domain82. Its kinase activity is regulated by tyrosine phosphorylation induced by itself and other kinases such as Csk. In resting cells, Tyr529 in Src is phosphorylated by Csk and thus Src remains in an inactive form63. Src kinase has been reported to be involved in a wide spectrum of signaling pathways including the receptor tyrosine kinase, GPCR and steroid receptor signaling pathways82. The role of Src in cell death can be either inhibiting or promoting. Src inhibits Fas-induced apoptosis through Erk1-/2-dependent degradation of the death accelerator Bik83. Src also phosphorylates human caspase-8 at Tyr380 to inhibit caspase-8 activity, but this is not the case in mice as mouse caspase-8 lacks Tyr38084. The promoting effect of Src in cell death was observed in zVAD-induced L929 cell death. It was shown that knockdown of Src or inhibition of Src kinase activity decreased zVAD-induced cell death85. JNK and ERK were shown to act downstream of Src in zVAD-induced cell death85. However, inhibiting JNK or ERK had little effect on TNF-induced necroptosis in L929 cells (Supplementary information, Figure S7A). Furthermore, inhibition of Src kinase activity did not influence JNK and ERK activation induced by TNF (Supplementary information, Figure S7B). Therefore, the downstream events of Src activation appear to be stimulus-dependent. The Src-mediated signaling pathways are quite different in zVAD- and TNF-induced cell death.
Although many reports have indicated that TNF activates Src kinase59,60,86, the mechanisms of Src activation have not been well studied. PKC was reported to regulate TNF-induced Src activation87. However, our results showed that a pretreatment with PKC inhibitor did not affect TNF-induced Src activation in L929 cells (Supplementary information, Figure S8A). Furthermore, PKC inhibitor did not render cells resistant to TNF-induced necroptosis88 (Supplementary information, Figure S8B). Nevertheless, our data demonstrate that the Gβγ complex acts downstream of the TNFR1 in mediating Src kinase activation (Figure 5).
The heterotypic membrane fraction was shown to play an important role in apoptosis71. Crudely prepared mitochondria were highly sensitive to BID-induced cytochrome C release, while highly purified mitochondria were less sensitive. Chipuk et al.71 showed that heterotypic membranes contaminated in the crude mitochondria preparation could restore mitochondrial sensitivity to BID stimulation. Here, we show that RIP1/RIP3 necrosomes translocate to the heterotypic membrane fraction and that the Gγ10-Src pathway is required for this translocation. The close correlation between the accumulation of RIP1/RIP3 in heterotypic membranes accumulation and necroptosis suggests the requirement of this translocation in cell death; however, the precise subcellular location into which RIP1/RIP3 necrosomes need to be translocated is still unknown as the heterotypic membrane fraction contains membranes of different types (Supplementary information, Figure S5E). For example, the heterotypic membranes include MAM, which is believed to serve as the contact site of the ER and mitochondrion, and has been suggested as the signaling juncture that facilitates calcium and lipid transfers between organelles89,90. Other membrane structures in the heterotypic membrane fraction might also serve as platforms where necrosomes execute cell death.
Among the components of the TNF-induced necroptosis pathway, RIP3 and its binding partner RIP1 have been proven to be the central regulators11,12,15. In addition, the important roles of MLKL and PGAM5 in necroptosis have also been uncovered recently14,16,69. However, it still remains largely unknown how a nascently assembled necrosome transmits the death signal. Our study reveals a parallel pathway required for the death signal transduction of necrosomes (Figure 7F). The Gβ2γ10-Src pathway is activated by TNF and its receptor engagement. Activation of the Gβ2γ10-Src pathway is required for necrosome translocation. How the Gβ2γ10-Src pathway controls necrosome trafficking and how necrosomes function in the heterotypic membrane fraction await future studies.
Materials and Methods
Plasmid construction
The genes of interest were amplified by PCR using specific primers and cloned into the BamHI and SmaI sites of pLV-EF1α-MCS-IRES-Puro vector (Biosettia, San Diego, CA, USA) or the BamHI and XhoI sites of pBOBE vector using the ExoIII-assisted ligase-free cloning method as described elsewhere91. For Lentivirus-based shRNA expression vectors, the DNA oligos encoding shRNA sequences were designed and cloned into the expression vector pLV-H1-EF1α-puro using the single oligonucleotide RNAi technology developed by Biosettia as described elsewhere12. All lentiviral-shRNA vectors were constructed following the manufacturer's protocol. The shRNA target sequences are listed in Supplementary information, Table S1.
Immunoprecipitation
Cells rinsed once with cold PBS were lysed in cold lysis buffer (20 mM Tris-HCl pH 7.5, 120 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Sodium pyrophosphate, 1 mM β-Glycerophosphate, 1 mM Na3VO4, 1 mM PMSF, 1× Protease inhibitor mix (Sigma)) as described elsewhere92. The soluble fractions of cell lysates were isolated by centrifugation at 20 000× g for 30 min at 4 °C in a centrifuger. Immunoprecipitation was performed using anti-flag M2 beads, or anti-HA beads as described previously93.
Antibodies
Mouse anti-RIP1 antibodies were obtained from BD Biosciences (San Jose, CA, USA). RIP3 polyclonal antibody was raised in rabbits using Escherichia coli-expressed GST-RIP3 (287-387 amino acids)12. FADD polyclonal antibody was raised in rabbits using E. coli-expressed full-length FADD. p38 antibody was generated as described94. p65, p-p65, p-p38, JNK, p-JNK, ERK, p-ERK, Src (36D10), pY416-Src antibodies were obtained from Cell Signaling Inc. (Beverly, MA, USA). Mouse anti-HA (F-7), mouse anti-GAPDH (6C5) and mouse anti-β-actin (C4) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). Mouse anti-flag antibody (M2) was obtained from Sigma (Saint Louis, MO, USA). Rabbit anti-vimentin antibody (10366-1-AP) was obtained from Proteintech Company (Chicago, IL, USA).
RNA extraction and real-time PCR analysis
RNA extractions and real-time PCR analyses were performed as described previously12. Total RNA was isolated from the indicated cells using TRIZOL reagent (Invitrogen) according to the manufacturer's instructions. In all, 1.5 μg total RNA was used to prepare cDNA using oligo(dT)12 as a primer. RNA expression levels were normalized to an internal control, GAPDH. Real-time PCR was performed in CFX96 RealTime System (Bio-Rad). For Gγ and Gβ mRNA absolute quantification, RT-PCR products were purified and quantified, and series were diluted as templates for a standard curve. The real-time primers are listed in Supplementary information, Table S2.
CRISPR/Cas9-mediated gene knockout
The method used for CRISPR/Cas9-mediated gene knockout was described in35,36. In brief, Cas9-target sites for the indicated genes were designed in http://www.genome-engineering.org/crispr/?page_id=41. Then, synthetic nucleotides containing target sites were sub-cloned into gRNA_Cloning Vector (Addgene). Then gRNA vector was co-transfected with hCas9 (Addgene) into L929 cells. After G418 selection, cells were isolated as single clones. The protein quantity of each single clone was analyzed. The Cas9-target sites are: Gβ2-target site-1: TCATCTGAATTCGCCCCAC; Gβ2-target site-2: CTGTGCCTACGCCCCCTCA; Src-target site: GCCGCGGGCGGCACGAAGG.
Cell culture
HEK293T cell line and mouse fibrosarcoma L929 cell line were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 4 mM L-glutamine, 100 IU penicillin and 100 mg/ml streptomycin at 37 °C in a humidified incubator containing 5% CO2.
Virus packaging
Recombinant lentiviruses were packaged in 293T cells in the presence of helper plasmids (pMDLg, pRSV-REV and pVSV-G) using a calcium phosphate precipitation method. The transfected cells were cultured at 37 °C for 48 h and the virus was then collected for infection.
Reagents
PP2, Genistein, dbcAMP, LY294002, PI were obtained from Sigma-Aldrich Chemicals. zVAD was obtained from Calbiochem (San Diego, CA, USA). Gallein was obtained from Tocris Bioscience (Bristol, UK). Fluo-3 AM was obtained from Invitrogen (Molecular Probes, Eugene, OR, USA). Mouse TNF was obtained from eBioscience (San Diego, CA, USA). Asante Potassium Green 1 (APG1) AM was obtained from TEFLABS (Austin, TX, USA).
Determination of calcium concentration
Determination of calcium concentration was performed following the manuals. In brief, cells were treated with or without TNF for the indicated times. Before being harvested for FACS, cells were loaded with 4 μg of Fluo-3 acetoxymethyl (AM) per ml in the presence of 4 mM probenecid (Sigma, Saint Louis, MO, USA) at 37 °C for 30 min. FACS was performed at 488 nm excitation, and the fluorescence of fluo-3 was collected at 530 nm. Then, the mean fluorescent values of viable cells were considered as the relative calcium concentrations. For experiment of ATP-induced calcium mobilization, cells were pre-incubated with Fluo-3 as above and then harvested for FACS. After assay by FACS for 25 s, ATP was added to stimulate cells and then continued the measurement by FACS. FACS data were analyzed by FlowJo software and processed with photoshop.
Immunofluorescence imaging
Cells were fixed with freshly prepared 4% paraformaldehyde (PFA) in PBS. The fixed cells were then permeabilized in 0.2% Triton X-100/PBS, blocked with 3% BSA in PBS, stained with anti-HA (mouse, 1:100, Santa Cruz) and labeled with goat-anti-mouse AlexaFluor 594 (1:1 000, Invitrogen) and Oregon Green Phalloidin (Invitrogen). All images were captured and processed using identical settings in the Zeiss LSM 780 laser scanning confocal microscope with a 100×/1.49 NA oil objective. Duplicate cultures were examined, and similar results were obtained in at least three independent experiments.
Determination of potassium concentration
Determination of potassium concentration was performed following the fluorescent ion indicator handbook of TEFLABS. In brief, cells were treated with 2 μM Asante Potassium Green 1 (APG1) AM and 100 ppm Pluronic F-127 for 60 min at 37 °C. Flow cytometry analysis was performed at 488 nm excitation, and the fluorescence of fluo-3 was collected at 530 nm. Then, the mean fluorescent values of viable cells were considered as the relative potassium concentrations.
Measurement of cell survival rate
Cell survival rates were determined by flow cytometry as described previously12. Plasma membrane integrity was tested by the ability of cells to exclude PI. Cells were trypsinized, collected by centrifugation and resuspended in PBS containing 5 μg/ml PI. The level of PI incorporation was quantified by flow cytometer (BD, FACSCalibur). Cell size was evaluated by forward-angle light scattering. PI-negative cells with a normal size were considered as living cells. For spontaneous necroptosis and peritoneal macrophage experiment, PI was directly added in the medium and incubated for 10 min. Cell numbers and PI-positive dots were counted afterward by microscopy.
Isolation of Triton X-100-insoluble fragments
Isolation of Triton X-100-insoluble fragments was performed as described elsewhere69. Cells were isolated after washing twice with PBS and harvested by scraping. After centrifugation at 500× g for 3 min, cell pellet was resuspended in 1 ml lysis buffer. After 30-min incubation on ice, the cells were centrifuged at 20 000× g for 15 min. Cell pellet was washed with lysis buffer for four times and centrifuged again at 20 000× g for 3 min. Pellet was resuspended in SDS sample buffer (70 mM Tris-Cl pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, 0.01% Bromphenol Blue in lysis buffer) and then sonicated before analysis.
Fractionation of L929 cells
L929 cells were fractionated and HM, mitochondria and heterotypic membrane were isolated as described previously71 with little modification. In brief, about 3 × 108 cells were washed with cold PBS and harvested into 50-ml tubes. Cells were homogenized by glass homogenizer (Kontes, Vineland, NJ, USA) in homogenization buffer (225 mM mannitol, 75 mM sucrose, 0.1 mM EGTA and 30 mM Tris-HCl pH 7.4). The lysate was centrifuged at 800× g to remove nuclei. The supernatant was centrifuged at 100 000× g for 10 min at 4 °C and the subsequent pellet was collected as HM. For further purification, HM was resuspended and loaded on 20% percoll and centrifuged at 10 000× g for 30 min at 4 °C. Mitochondria were recovered from the bottom of the gradient; the heterotypic membranes were collected from the upper gradient. Each fraction was quantified and the indicated proteins were analyzed by western blot. For investigating the composition of the fractions, the freshly prepared mitochondria fraction and heterotypic membrane fraction were subjected to transmission electron microscopy (JEM 2100 HC, JEOL USA) by the conventional method.
Acknowledgments
This work was supported by the National Basic Research Program of China (973 Program; 2013CB944903 and 2014CB541804), the National Natural Science Foundation of China (31330047, 91029304, 81061160512 and 31221065), the Hi-Tech Research and Development Program of China (863 program; 2012AA02A201), the 111 Project (B12001) and the Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University (SKLCSB2012KF003).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Related to Figure 1.
Gβ2 is required for TNF induced necroptosis in L929 cells, related to Figure 3.
Overexpression p115-RGS domain and GRK2-RGS domain can inhibit Gα dependent signal pathways, related to Figure 4.
Src is activated in L929 cells by TNF stimulation in a Gγ10 dependent but necrosome independent manner, related to Figure 5.
Src kinase is involved in necroptosis, related to Figure 6.
Gγ10 plays no role in necrosome formation but is required for accumulation of RIP1 and RIP3 in the heterotypic membrane fraction during the process of necroptosis, related to Figure 7.
TNF induced ERK and JNK activation does not depend on Src and is not required for TNF induced necroptosis in L929 cells, related to Discussion.
PKC inhibitor has no effect on TNF induced Src kinase activation and necroptosis in L929 cells, related to Discussion.
References
- Kawahara A, Ohsawa Y, Matsumura H, Uchiyama Y, Nagata S. Caspase-independent cell killing by Fas-associated protein with death domain. J Cell Biol. 1998;143:1353–1360. doi: 10.1083/jcb.143.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Vandenabeele P, Beyaert R, Declercq W, Fiers W. Tumour necrosis factor-induced necrosis versus anti-Fas-induced apoptosis in L929 cells. Cytokine. 1997;9:801–808. doi: 10.1006/cyto.1997.0252. [DOI] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Vandenabeele P. Necrosis, a well-orchestrated form of cell demise: signalling cascades, important mediators and concomitant immune response. Biochim Biophys Acta. 2006;1757:1371–1387. doi: 10.1016/j.bbabio.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Han J, Zhong CQ, Zhang DW. Programmed necrosis: backup to and competitor with apoptosis in the immune system. Nat Immunol. 2011;12:1143–1149. doi: 10.1038/ni.2159. [DOI] [PubMed] [Google Scholar]
- Lin J, Li H, Yang M, et al. A role of RIP3-mediated macrophage necrosis in atherosclerosis development. Cell Rep. 2013;3:200–210. doi: 10.1016/j.celrep.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yu SW, Wang H, Poitras MF, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992;267:5317–5323. [PubMed] [Google Scholar]
- Fiers W, Beyaert R, Declercq W, Vandenabeele P. More than one way to die: apoptosis, necrosis and reactive oxygen damage. Oncogene. 1999;18:7719–7730. doi: 10.1038/sj.onc.1203249. [DOI] [PubMed] [Google Scholar]
- Vercammen D, Beyaert R, Denecker G, et al. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Wang L, Miao L, et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Zhang DW, Shao J, Lin J, et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science. 2009;325:332–336. doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang H, Wang Z, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Cho YS, Challa S, Moquin D, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Jitkaew S, Cai Z, et al. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci USA. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang Z, Ren J, et al. Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res. 2013;23:994–1006. doi: 10.1038/cr.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Czabotar PE, Hildebrand JM, et al. The Pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- Evans RM, Zamponi GW. Presynaptic Ca2+ channels--integration centers for neuronal signaling pathways. Trends Neurosci. 2006;29:617–624. doi: 10.1016/j.tins.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Leopoldt D, Hanck T, Exner T, Maier U, Wetzker R, Nurnberg B. Gbetagamma stimulates phosphoinositide 3-kinase-gamma by direct interaction with two domains of the catalytic p110 subunit. J Biol Chem. 1998;273:7024–7029. doi: 10.1074/jbc.273.12.7024. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Hawes BE, van Biesen T, Luttrell DK, Lansing TJ, Lefkowitz RJ. Role of c-Src tyrosine kinase in G protein-coupled receptor- and Gbetagamma subunit-mediated activation of mitogen-activated protein kinases. J Biol Chem. 1996;271:19443–19450. doi: 10.1074/jbc.271.32.19443. [DOI] [PubMed] [Google Scholar]
- Tang WJ, Gilman AG. Type-specific regulation of adenylyl cyclase by G protein beta gamma subunits. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- Whiteway M, Hougan L, Dignard D, et al. Function of the STE4 and STE18 genes in mating pheromone signal transduction in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 1988;53 Pt 2:585–590. doi: 10.1101/sqb.1988.053.01.067. [DOI] [PubMed] [Google Scholar]
- Kolesnikov AV, Rikimaru L, Hennig AK, et al. G-protein betagamma-complex is crucial for efficient signal amplification in vision. J Neurosci. 2011;31:8067–8077. doi: 10.1523/JNEUROSCI.0174-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwindinger WF, Betz KS, Giger KE, Sabol A, Bronson SK, Robishaw JD. Loss of G protein gamma 7 alters behavior and reduces striatal alpha(olf) level and cAMP production. J Biol Chem. 2003;278:6575–6579. doi: 10.1074/jbc.M211132200. [DOI] [PubMed] [Google Scholar]
- Giambarella U, Yamatsuji T, Okamoto T, et al. G protein betagamma complex-mediated apoptosis by familial Alzheimer's disease mutant of APP. EMBO J. 1997;16:4897–4907. doi: 10.1093/emboj/16.16.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li Q, Xie C, et al. Beta-actin is required for mitochondria clustering and ROS generation in TNF-induced, caspase-independent cell death. J Cell Sci. 2004;117:4673–4680. doi: 10.1242/jcs.01339. [DOI] [PubMed] [Google Scholar]
- Ono K, Wang X, Han J. Resistance to tumor necrosis factor-induced cell death mediated by PMCA4 deficiency. Mol Cell Biol. 2001;21:8276–8288. doi: 10.1128/MCB.21.24.8276-8288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang N, Zhou H, et al. Distinct roles of basal steady-state and induced H-ferritin in tumor necrosis factor-induced death in L929 cells. Mol Cell Biol. 2005;25:6673–6681. doi: 10.1128/MCB.25.15.6673-6681.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Ono K, Kim SO, Kravchenko V, Lin SC, Han J. Metaxin is required for tumor necrosis factor-induced cell death. EMBO Rep. 2001;2:628–633. doi: 10.1093/embo-reports/kve135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liang Y, Shao F, Wang X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proc Natl Acad Sci USA. 2011;108:20054–20059. doi: 10.1073/pnas.1116302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Kalyanaraman V, Gautam N. Differential ability to form the G protein betagamma complex among members of the beta and gamma subunit families. J Biol Chem. 1996;271:7141–7146. doi: 10.1074/jbc.271.12.7141. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Neer EJ. G protein beta gamma subunits. Annu Rev Pharmacol Toxicol. 1997;37:167–203. doi: 10.1146/annurev.pharmtox.37.1.167. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K, Kunsch C, Bonner LM, Robishaw JD. Isolation of cDNA clones encoding eight different human G protein gamma subunits, including three novel forms designated the gamma 4, gamma 10, and gamma 11 subunits. J Biol Chem. 1995;270:21765–21771. doi: 10.1074/jbc.270.37.21765. [DOI] [PubMed] [Google Scholar]
- Lehmann DM, Seneviratne AM, Smrcka AV. Small molecule disruption of G protein beta gamma subunit signaling inhibits neutrophil chemotaxis and inflammation. Mol Pharmacol. 2008;73:410–418. doi: 10.1124/mol.107.041780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Smrcka AV. Understanding molecular recognition by G protein betagamma subunits on the path to pharmacological targeting. Mol Pharmacol. 2011;80:551–557. doi: 10.1124/mol.111.073072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62:544–552. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- Branellec D, De Cremoux P, Barreau P, Calvo F, Chouaib S. Tumor necrosis factor-mediated cell lysis in vitro: relationship to cAMP accumulation and guanine nucleotide-binding proteins. Eur J Immunol. 1992;22:963–967. doi: 10.1002/eji.1830220413. [DOI] [PubMed] [Google Scholar]
- Wang J, Tang R, Lv M, Zhang J, Shen B. Selective unresponsiveness to the inhibition of p38 MAPK activation by cAMP helps L929 fibroblastoma cells escape TNF-alpha-induced cell death. Mol Cancer. 2010;9:6. doi: 10.1186/1476-4598-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock VP, Lofthouse EJ, Jupp OJ, Gauld SB, Anderson HM, MacEwan DJ. Selective down-regulation of the G(q)alpha/G11alpha G-protein family in tumour necrosis factor-alpha induced cell death. Mol Cell Biochem. 2000;206:67–74. doi: 10.1023/a:1007066409645. [DOI] [PubMed] [Google Scholar]
- Uboldi AD, Savage N. The adenylate cyclase activator forskolin partially protects L929 cells against tumour necrosis factor-alpha-mediated cytotoxicity via a cAMP-independent mechanism. Cytokine. 2002;19:250–258. [PubMed] [Google Scholar]
- Arai K, Maruyama Y, Nishida M, et al. Differential requirement of G alpha12, G alpha13, G alphaq, and G beta gamma for endothelin-1-induced c-Jun NH2-terminal kinase and extracellular signal-regulated kinase activation. Mol Pharmacol. 2003;63:478–488. doi: 10.1124/mol.63.3.478. [DOI] [PubMed] [Google Scholar]
- Carman CV, Parent JL, Day PW, et al. Selective regulation of Galpha(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem. 1999;274:34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- Le Page SL, Bi Y, Williams JA. CCK-A receptor activates RhoA through G alpha 12/13 in NIH3T3 cells. Am J Physiol Cell Physiol. 2003;285:C1197–1206. doi: 10.1152/ajpcell.00083.2003. [DOI] [PubMed] [Google Scholar]
- Kranenburg O, Poland M, van Horck FP, Drechsel D, Hall A, Moolenaar WH. Activation of RhoA by lysophosphatidic acid and Galpha12/13 subunits in neuronal cells: induction of neurite retraction. Mol Biol Cell. 1999;10:1851–1857. doi: 10.1091/mbc.10.6.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- Rochdi MD, Watier V, La Madeleine C, Nakata H, Kozasa T, Parent JL. Regulation of GTP-binding protein alpha q (Galpha q) signaling by the ezrin-radixin-moesin-binding phosphoprotein-50 (EBP50) J Biol Chem. 2002;277:40751–40759. doi: 10.1074/jbc.M207910200. [DOI] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Rebois RV, Hebert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SK, Fung KP, Choy YM, Lee CY. Slow increase in intranuclear and cytosolic free calcium concentrations in L929 cells is important in tumour necrosis factor-alpha-mediated cell death. Oncology. 1997;54:55–62. doi: 10.1159/000227662. [DOI] [PubMed] [Google Scholar]
- Ikeda SR. Voltage-dependent modulation of N-type calcium channels by G-protein beta gamma subunits. Nature. 1996;380:255–258. doi: 10.1038/380255a0. [DOI] [PubMed] [Google Scholar]
- Meir A, Bell DC, Stephens GJ, Page KM, Dolphin AC. Calcium channel beta subunit promotes voltage-dependent modulation of alpha 1 B by G beta gamma. Biophys J. 2000;79:731–746. doi: 10.1016/S0006-3495(00)76331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan AN, Tiruppathi C, Smrcka AV, Malik AB, Minshall RD. Gbetagamma activation of Src induces caveolae-mediated endocytosis in endothelial cells. J Biol Chem. 2004;279:48055–48062. doi: 10.1074/jbc.M405837200. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Della Rocca GJ, van Biesen T, Luttrell DK, Lefkowitz RJ. Gbetagamma subunits mediate Src-dependent phosphorylation of the epidermal growth factor receptor. A scaffold for G protein-coupled receptor-mediated Ras activation. J Biol Chem. 1997;272:4637–4644. doi: 10.1074/jbc.272.7.4637. [DOI] [PubMed] [Google Scholar]
- Nino G, Hu A, Grunstein JS, et al. G Protein betagamma-subunit signaling mediates airway hyperresponsiveness and inflammation in allergic asthma. PLoS One. 2012;7:e32078. doi: 10.1371/journal.pone.0032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt JM, Stork PJ. Galpha and Gbeta gamma require distinct Src-dependent pathways to activate Rap1 and Ras. J Biol Chem. 2002;277:43024–43032. doi: 10.1074/jbc.M204006200. [DOI] [PubMed] [Google Scholar]
- Lee CW, Lin CC, Lin WN, et al. TNF-alpha induces MMP-9 expression via activation of Src/EGFR, PDGFR/PI3K/Akt cascade and promotion of NF-kappaB/p300 binding in human tracheal smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2007;292:L799–812. doi: 10.1152/ajplung.00311.2006. [DOI] [PubMed] [Google Scholar]
- Huang S, Dudez T, Scerri I, et al. Defective activation of c-Src in cystic fibrosis airway epithelial cells results in loss of tumor necrosis factor-alpha-induced gap junction regulation. J Biol Chem. 2003;278:8326–8332. doi: 10.1074/jbc.M208264200. [DOI] [PubMed] [Google Scholar]
- Zheng XM, Resnick RJ, Shalloway D. A phosphotyrosine displacement mechanism for activation of Src by PTPalpha. EMBO J. 2000;19:964–978. doi: 10.1093/emboj/19.5.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole PA, Shen K, Qiao Y, Wang D. Protein tyrosine kinases Src and Csk: a tail's tale. Curr Opin Chem Biol. 2003;7:580–585. doi: 10.1016/j.cbpa.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Imamoto A, Soriano P. Disruption of the csk gene, encoding a negative regulator of Src family tyrosine kinases, leads to neural tube defects and embryonic lethality in mice. Cell. 1993;73:1117–1124. doi: 10.1016/0092-8674(93)90641-3. [DOI] [PubMed] [Google Scholar]
- Nada S, Yagi T, Takeda H, et al. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell. 1993;73:1125–1135. doi: 10.1016/0092-8674(93)90642-4. [DOI] [PubMed] [Google Scholar]
- Kmiecik TE, Shalloway D. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell. 1987;49:65–73. doi: 10.1016/0092-8674(87)90756-2. [DOI] [PubMed] [Google Scholar]
- Goetze S, Xi XP, Kawano Y, et al. TNF-alpha-induced migration of vascular smooth muscle cells is MAPK dependent. Hypertension. 1999;33:183–189. doi: 10.1161/01.hyp.33.1.183. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ke Q, Yang Y, et al. Cardiomyocytes overexpressing TNF-alpha attract migration of embryonic stem cells via activation of p38 and c-Jun amino-terminal kinase. FASEB J. 2003;17:2231–2239. doi: 10.1096/fj.03-0030com. [DOI] [PubMed] [Google Scholar]
- Feng S, Yang Y, Mei Y, et al. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, et al. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, McStay GP, Bharti A, et al. Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell. 2012;148:988–1000. doi: 10.1016/j.cell.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE. Phospholipid synthesis in a membrane fraction associated with mitochondria. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- Inglese J, Koch WJ, Touhara K, Lefkowitz RJ. G beta gamma interactions with PH domains and Ras-MAPK signaling pathways. Trends Biochem Sci. 1995;20:151–156. doi: 10.1016/s0968-0004(00)88992-6. [DOI] [PubMed] [Google Scholar]
- Wang S, Narendra S, Fedoroff N. Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. Proc Natl Acad Sci USA. 2007;104:3817–3822. doi: 10.1073/pnas.0611735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. Different signaling and cell death roles of heterotrimeric G protein alpha and beta subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell. 2005;17:957–970. doi: 10.1105/tpc.104.029603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepburn A, Boeynaems JM, Fiers W, Dumont JE. Modulation of tumor necrosis factor-alpha cytotoxicity in L929 cells by bacterial toxins, hydrocortisone and inhibitors of arachidonic acid metabolism. Biochem Biophys Res Commun. 1987;149:815–822. doi: 10.1016/0006-291x(87)90440-2. [DOI] [PubMed] [Google Scholar]
- Burns DL. Subunit structure and enzymic activity of pertussis toxin. Microbiol Sci. 1988;5:285–287. [PubMed] [Google Scholar]
- Minshall RD, Tiruppathi C, Vogel SM, et al. Endothelial cell-surface gp60 activates vesicle formation and trafficking via G(i)-coupled Src kinase signaling pathway. J Cell Biol. 2000;150:1057–1070. doi: 10.1083/jcb.150.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov YV, Jones NC, Olwin BB. Regulation of myogenesis by fibroblast growth factors requires beta-gamma subunits of pertussis toxin-sensitive G proteins. Mol Cell Biol. 1998;18:5780–5787. doi: 10.1128/mcb.18.10.5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalloway D, Coussens PM, Yaciuk P. Overexpression of the c-src protein does not induce transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1984;81:7071–7075. doi: 10.1073/pnas.81.22.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeya T, Hanafusa H. Structure and sequence of the cellular gene homologous to the RSV src gene and the mechanism for generating the transforming virus. Cell. 1983;32:881–890. doi: 10.1016/0092-8674(83)90073-9. [DOI] [PubMed] [Google Scholar]
- Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–7909. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- Lopez J, Hesling C, Prudent J, et al. Src tyrosine kinase inhibits apoptosis through the Erk1/2-dependent degradation of the death accelerator Bik. Cell Death Differ. 2012;19:1459–1469. doi: 10.1038/cdd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursi S, Rufini A, Stagni V, et al. Src kinase phosphorylates Caspase-8 on Tyr380: a novel mechanism of apoptosis suppression. EMBO J. 2006;25:1895–1905. doi: 10.1038/sj.emboj.7601085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Chiu LY, Maa MC, Wang JS, Chien CL, Lin WW. zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy. 2011;7:217–228. doi: 10.4161/auto.7.2.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Chen JJ, Chen CC. c-Src-dependent tyrosine phosphorylation of IKKbeta is involved in tumor necrosis factor-alpha-induced intercellular adhesion molecule-1 expression. J Biol Chem. 2003;278:9944–9952. doi: 10.1074/jbc.m208521200. [DOI] [PubMed] [Google Scholar]
- Huang WC, Chen JJ, Inoue H, Chen CC. Tyrosine phosphorylation of I-kappa B kinase alpha/beta by protein kinase C-dependent c-Src activation is involved in TNF-alpha-induced cyclooxygenase-2 expression. J Immunol. 2003;170:4767–4775. doi: 10.4049/jimmunol.170.9.4767. [DOI] [PubMed] [Google Scholar]
- O'Connell MA, Kelleher D, Liskamp RM, Hall N, O'Neill LA, Long A. TNF-mediated cytotoxicity of L929 cells: role of staurosporine in enhancement of cytotoxicity and translocation of protein kinase C isozymes. Cytokine. 1997;9:83–92. doi: 10.1006/cyto.1996.0140. [DOI] [PubMed] [Google Scholar]
- Rusinol AE, Cui Z, Chen MH, Vance JE. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J Biol Chem. 1994;269:27494–27502. [PubMed] [Google Scholar]
- Hayashi T, Su TP. Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell. 2007;131:596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- Li C, Evans RM. Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res. 1997;25:4165–4166. doi: 10.1093/nar/25.20.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhang N, Zhang D, et al. Determinants that control the distinct subcellular localization of p38alpha-PRAK and p38beta-PRAK complexes. J Biol Chem. 2008;283:11014–11023. doi: 10.1074/jbc.M709682200. [DOI] [PubMed] [Google Scholar]
- Zhou H, Zheng M, Chen J, et al. Determinants that control the specific interactions between TAB1 and p38alpha. Mol Cell Biol. 2006;26:3824–3834. doi: 10.1128/MCB.26.10.3824-3834.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XN, Wang XK, Wu SQ, et al. Phosphorylation of Raptor by p38beta participates in arsenite-induced mammalian target of rapamycin complex 1 (mTORC1) activation. J Biol Chem. 2011;286:31501–31511. doi: 10.1074/jbc.M111.233122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1.
Gβ2 is required for TNF induced necroptosis in L929 cells, related to Figure 3.
Overexpression p115-RGS domain and GRK2-RGS domain can inhibit Gα dependent signal pathways, related to Figure 4.
Src is activated in L929 cells by TNF stimulation in a Gγ10 dependent but necrosome independent manner, related to Figure 5.
Src kinase is involved in necroptosis, related to Figure 6.
Gγ10 plays no role in necrosome formation but is required for accumulation of RIP1 and RIP3 in the heterotypic membrane fraction during the process of necroptosis, related to Figure 7.
TNF induced ERK and JNK activation does not depend on Src and is not required for TNF induced necroptosis in L929 cells, related to Discussion.
PKC inhibitor has no effect on TNF induced Src kinase activation and necroptosis in L929 cells, related to Discussion.