Abstract
SUMMARY
The OXA β-lactamases were among the earliest β-lactamases detected; however, these molecular class D β-lactamases were originally relatively rare and always plasmid mediated. They had a substrate profile limited to the penicillins, but some became able to confer resistance to cephalosporins. From the 1980s onwards, isolates of Acinetobacter baumannii that were resistant to the carbapenems emerged, manifested by plasmid-encoded β-lactamases (OXA-23, OXA-40, and OXA-58) categorized as OXA enzymes because of their sequence similarity to earlier OXA β-lactamases. It was soon found that every A. baumannii strain possessed a chromosomally encoded OXA β-lactamase (OXA-51-like), some of which could confer resistance to carbapenems when the genetic environment around the gene promoted its expression. Similarly, Acinetobacter species closely related to A. baumannii also possessed their own chromosomally encoded OXA β-lactamases; some could be transferred to A. baumannii, and they formed the basis of transferable carbapenem resistance in this species. In some cases, the carbapenem-resistant OXA β-lactamases (OXA-48) have migrated into the Enterobacteriaceae and are becoming a significant cause of carbapenem resistance. The emergence of OXA enzymes that can confer resistance to carbapenems, particularly in A. baumannii, has transformed these β-lactamases from a minor hindrance into a major problem set to demote the clinical efficacy of the carbapenems.
INTRODUCTION
The simplest classification for β-lactamases has been based on molecular structure (1); in this scheme, there are four major classes. Classes A to C have been well documented as both chromosomally encoded and plasmid-mediated enzymes (2). The class D β-lactamases have been much more elusive and, for the most part, were identifiable only as plasmid-encoded β-lactamases in Gram-negative bacteria. These early enzymes were essentially penicillinases, which, unlike the class A β-lactamases, could hydrolyze and confer resistance to oxacillin as well as penicillin, hence the name oxacillinases and the prefix OXA. When the substrate profile of the early OXA enzymes was considered by the scheme reported by Bush et al., these enzymes were designated 2d (3).
EARLY OXA β-LACTAMASES
Hedges et al. (4) demonstrated that plasmid-encoded β-lactamases in Gram-negative bacilli could be divided into two groups, the ubiquitous TEM enzymes and another, small group that was distinguishable by its ability to hydrolyze oxacillin. Unlike the TEM enzymes, this group had a heterogeneous substrate profile and was encoded by a much narrower range of plasmids. These β-lactamases had properties similar to those found on a plasmid (R1818) identified by Datta and Kontomichalou (5), which was later renamed R46 (6). These enzymes had a lower specific activity against penicillin than the TEM β-lactamases. On the other hand, they had much higher activity against oxacillin and methicillin. Their emergence presumably coincided with the widespread introduction of flucloxacillin and methicillin for the treatment of staphylococcal infections. They were also relatively less effective against first-generation cephalosporins and are poorly inhibited by major class A β-lactamase inhibitors such as clavulanic acid.
Closely related β-lactamases were originally distinguished by their isoelectric points. Sykes and Matthew (7) identified three distinct oxacillinases, named OXA-1, OXA-2, and OXA-3. The first two possessed a characteristic shared by most of the members of this class, in that they were inhibited by chloride ions (8). The first gene to be sequenced was from plasmid R46 and was actually blaOXA-2 (9, 10). The first blaOXA-1 gene to be sequenced was part of a transposon carried on plasmid RGN238 (11). Compared with the OXA-2 β-lactamase, there was only about 48% homology. There was, however, a degree of conservation around the active-site serine at position 71 and a leucine at position 179.
These were the first motifs that were identified in the OXA enzymes, and they have become an important characteristic in the examination of these enzymes. The location of the blaOXA-1 gene was in a Tn21-derived transposon, inserted between the aad gene encoding aminoglycoside resistance and its promoter. Interestingly, the rather dissimilar blaOXA-2 gene was found in exactly the same position, which suggests some strong evolution pressure not just for the presence of these genes in the bacterial cell but also for their carriage within this specific transposon. Subsequent OXA enzymes have been given a unique number based on the chronological order in which they were discovered, and later, these numbers were verified by their unique amino acid sequence by the Lahey Clinic (http://www.lahey.org/Studies/), where the complete list of OXA β-lactamases can be found. Closely related numbers do not reflect similarities but merely the order in which they were reported.
A new group of plasmid-borne β-lactamases was found in Pseudomonas aeruginosa (12). They were called PSE (Pseudomonas-specific enzymes) because they were considered to be specific to Pseudomonas aeruginosa (13). There were four main enzymes, PSE-1 to PSE-4. The genes were transferable to Escherichia coli both in the laboratory (14) and in clinical isolates (13). The designation of these enzymes as Pseudomonas specific was clearly incorrect; however, it was only when the genes were sequenced that it was found that they were not a homogenous group. All enzymes were active against the anti-Pseudomonas penicillin carbenicillin, but Philippon et al. (15) noticed that one of them (PSE-2) was also able to hydrolyze oxacillin. When the genes were sequenced, PSE-2 was closely related to the OXA β-lactamases, but the other PSE enzymes were not, despite a similar substrate profile. PSE-2 had only about 36% homology to OXA-2 but was much closer in structure to OXA-5 (16). There was great diversity among the five enzymes studied by this group, which suggested that these enzymes came from distant sources and had evolved from an ancestral gene a long time ago. Because of the close structural similarity of PSE-2 with the OXA β-lactamases, it was later proposed that this enzyme should be reclassified as OXA-10 (17).
OXA EXTENDED-SPECTRUM β-LACTAMASES
The role of the class D OXA enzymes was considered to be secondary to the much more ubiquitous class A enzymes, particularly in conferring resistance to the penicillins. Some of them had been identified originally in Pseudomonas aeruginosa. In the 1980s, this organism was responding well to the third-generation cephalosporin ceftazidime. However, a strain of Pseudomonas aeruginosa was identified, which had been isolated from a patient in Turkey in October 1991 (17). The bacterium was multiresistant, particularly to ceftazidime. This resistance was transferable, and when sequenced, the enzyme had 2 amino acid changes from the OXA-10 enzyme, altering an arginine at amino acid 143 to serine and a glycine at position 157 to aspartate. Although the OXA-10 β-lactamase did provide weak hydrolysis of cefotaxime, ceftriaxone, and aztreonam (18), the mutation considerably increased the binding of ceftazidime to the active site. The new enzyme, designated OXA-11, was the first example of an OXA enzyme that had become, through mutation, an extended-spectrum β-lactamase (ESBL).
Besides OXA-11, there have been several more ESBLs derived from OXA-10, namely, OXA-13, OXA-14 (19), OXA-16 (20), OXA-17 (21), OXA-19 (22), and OXA-28 (23). All of these ESBLs have been found in Pseudomonas aeruginosa. They possess up to nine variations from the parental enzyme. The effect of these changes from the parental enzyme on the function of the protein and, thus, the resistance profile has not been studied. It is interesting to note that the glycine-to-aspartate mutation at position 157 found in OXA-11 is also found in four of these OXA-10 ESBL derivatives. OXA-14 has this mutation only, indicating that this is the crucial mutation for high-level ceftazidime resistance (19). The reason why this mutation affects ceftazidime resistance is not clear: it certainly affects the kinetics, but the mutation does not appear to directly influence the active site (24); rather, it affects the dimerization of the enzyme, and this may have a profound effect on the hydrolysis of ceftazidime by increasing the bonding of cephalosporin.
The reason why other OXA-10-derived ESBLs have emerged is less easy to identify. There are two OXA-10-like ESBLs; both have the glycine-to-aspartate substitution at position 157, but OXA-11 has the additional asparagine-to-serine mutation at position 143, and OXA-16 has the alanine-to-threonine mutation at position 127. Neither of these mutations is found in any of the other OXA-10-like ESBLs, suggesting that they are not the precursors to these enzymes and also, perhaps, that they are not so important in cephalosporin resistance. OXA-19 has nine mutations compared to OXA-10, and many of the later OXA-10-like ESBLs (OXA-28, -35, -145, and -147) have most of these mutations, suggesting that this has been the most successful group. One OXA-10-like ESBL, OXA-17, has a single asparagine-to-serine mutation at position 73. This is near the active site and confers resistance to cefotaxime and ceftriaxone but virtually no resistance to ceftazidime (21).
The OXA-2-like ESBLs are much more diverse. There are six of them, and each of them has a single mutation from the parental β-lactamase. In only two of them is the mutation at the same position: in OXA-15, there is an aspartate-to-glycine substitution at position 149 (25), and in OXA-36, there is an aspartate-to-tyrosine mutation at position 149. The dissimilarity between the substituted amino acids glycine and tyrosine suggests that the loss of aspartate is the crucial aspect of the hydrolysis of cephalosporins, rather than the acquisition of either of these substituted amino acids.
Although often crucial to the resistance profile of Pseudomonas aeruginosa, the OXA-derived ESBLs have hardly been identified in other species, so they do not appear to be spreading. One enzyme, OXA-21, was found in a strain of Acinetobacter baumannii, and this was the first report of an OXA β-lactamase sequence in this species (26). The clinical impact of these enzymes is largely unknown, as so few epidemiological studies have been performed (18), but the role of OXA ESBLs does not appear to be very significant compared with the role of pandemic CTX-M ESBLs (18).
OXA-23-LIKE β-LACTAMASES
The first group of carbapenem-resistant OXA-type β-lactamases to be identified in A. baumannii was the OXA-23 group. The OXA-23 β-lactamase was first identified in an A. baumannii isolate collected in Edinburgh, United Kingdom, in 1985, interestingly the same year that imipenem was first approved for use (27). The isolate was found to have MICs of imipenem of 16 mg liter−1, and the resistance phenotype was transferable, indicating a plasmid location (28). The sequence was published in 2000 (29), and subsequently, as shown in Table 1 and Fig. 1, 18 further alleles of the blaOXA-23 gene have since been identified. The genes for this group of enzymes are frequently plasmid-borne and have been found in many Acinetobacter species as well as species belonging to the Enterobacteriaceae (Table 1). The discovery of several blaOXA-23-like genes (blaOXA-23, blaOXA-102, blaOXA-103, blaOXA-105, blaOXA-133, and blaOXA-134) on the chromosome of Acinetobacter radioresistens isolates indicates that this species is the likely natural source of this enzyme group (30–32). Comparison of the GC content of a gene with that of the whole genome of an organism provides evidence for whether that gene originates from that organism, as the two GC contents are likely to be similar if this is the case. The blaOXA-23-like genes from Acinetobacter radioresistens have a GC content of 37.5%, which is lower than the average for the A. baumannii genome (33), suggesting that they have been recently acquired by A. baumannii.
TABLE 1.
Carbapenem-hydrolyzing OXA-type carbapenemasesa
| Enzyme group | Enzyme(s) | No. of enzymes in group | Location(s) | Host species |
|---|---|---|---|---|
| OXA-23-like | OXA-23, OXA-27, OXA-49, OXA-73, OXA-102, OXA-103, OXA-105, OXA-133, OXA-134, OXA-146, OXA-165–OXA-171, OXA-225, OXA-239 | 19 | C and P | A. baumannii, A. junii, A. radioresistens, A. pittii, Proteus mirabilis, Acinetobacter phenon 5, Acinetobacter phenon 6/ct 13TU, A. nosocomialis, Acinetobacter genomic species 10/11, A. lwoffii, Klebsiella pneumoniae, A. baylyi |
| OXA-40-like | OXA-40, OXA-25, OXA-26, OXA-72, OXA-139, OXA-160, OXA-207 | 7 | C and P | A. baumannii, A. haemolyticus, A. pittii, A. baylyi, Pseudomonas aeruginosa, A. calcoaceticus, K. pneumoniae |
| OXA-51-like | OXA-51, OXA-64–OXA-71, OXA-75–OXA-80, OXA-82–OXA-84, OXA-86–OXA-95, OXA-98–OXA-100, OXA-104, OXA-106–OXA-113, OXA-115–OXA-117, OXA-120–OXA-128, OXA-130–OXA-132, OXA-138, OXA-144, OXA-148–OXA-150, OXA-172–OXA-180, OXA-194–OXA-197, OXA-200–OXA-203, OXA-206, OXA-208, OXA-216, OXA-217, OXA-219, OXA-223, OXA-241, OXA-242, OXA-248–OXA-250, OXA-254 | 95 | C and P | A. baumannii, A. nosocomialis, Enterobacter cloacae, Escherichia coli, K. pneumoniae |
| OXA-58-like | OXA-58, OXA-96, OXA-97, OXA-164 | 4 | C and P | A. baumannii, A. pittii, A. nosocomialis, Acinetobacter phenon 6/ct 13TU, A. junii, Acinetobacter genomic species 9, A. bereziniae, A. calcoaceticus, A. radioresistens, E. cloacae, Comamonas testosteroni, E. coli, K. pneumoniae, Delftia acidovorans |
| OXA-134a-like | OXA-134a, OXA-186–OXA-191 | 7 | C | A. lwoffii |
| OXA-143-like | OXA-143, OXA-182, OXA-231, OXA-253, OXA-255 | 5 | P | A. baumannii, A. pittii |
| OXA-213 | OXA-213 | 17 | C | A. calcoaceticus |
| OXA-214-like | OXA-214, OXA-215 | 5 | C | A. haemolyticus |
| OXA-211-like | OXA-211, OXA-212, OXA-309 | 6 | C | A. johnsonii |
| OXA-229-like | OXA-228–OXA-230, OXA-257 | 8 | C | A. bereziniae |
| OXA-235-like | OXA-235–OXA-237, OXA-278 | 7 | C | A. schindleri |
| OXA-48-like | OXA-48, OXA-48b, OXA-162, OXA-163, OXA-181, OXA-199, OXA-204, OXA-232, OXA-244, OXA-245, OXA-247 | 11 | C and P | E. cloacae, K. pneumoniae, E. coli, Shewanella xiamenensis, Citrobacter freundii, Serratia marcescens, Providencia rettgeri, Klebsiella oxytoca, Enterobacter sakazakii, A. baumannii |
FIG 1.
Maximum likelihood amino acid tree of the OXA-type β-lactamases found in Acinetobacter species. For enzymes for which a designation was not available, the species and strain designation are used instead. Due to its large size, the OXA-51 enzyme group has been collapsed so that the other enzyme groups can be more easily seen. Relationships between the OXA-51-like enzymes are shown in Fig. 5. The tree was implemented in SeaView version 4.2.5 (172) with PhyML using a Le and Gascuel (LG) model (173). Support was estimated by using approximate likelihood ratio tests, the results of which label the branches. The tree was visualized by using FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/) and is midpoint rooted.
Kinetic studies have been performed on three OXA-23-like enzymes: OXA-23, OXA-27, and OXA-146 (Table 2) (34–36). The kinetic properties determined vary considerably between studies, even for the same enzyme, making it difficult to make valid comparisons. The enzymes are able to hydrolyze oxyiminocephalosporins, aminopenicillins, piperacillin, oxacillin, and aztreonam in addition to the carbapenems (34, 37). Among the carbapenems, OXA-23 has a much higher turnover rate for imipenem than for meropenem, ertapenem, or doripenem (36). Despite the relatively low turnover rates for carbapenems displayed by these enzymes, it appears that they counter this through the presence of a “hydrophobic bridge” across the top of the active site, formed by phenylalanine 110 and methionine 221 (35), as the presence of a corresponding bridge in OXA-40 has been shown to be responsible for tight binding of the carbapenems (38, 39). Recently, it was demonstrated that OXA-146 has an expanded hydrolytic spectrum that includes ceftazidime, which is not hydrolyzed by the other class D OXA-type carbapenemases, while retaining its activity against the carbapenems (35). This is the first observation of an OXA-type carbapenemase with ESBL properties, which may prove to be problematic in the future. This difference in spectrum is due to the duplication of an alanine residue at position 220, within the β5-β6 loop of the enzyme. This duplication pushes the methionine 221 out of a position where it can form the hydrophobic bridge, thus removing steric clashes between methionine 221 and ceftazidime and aztreonam.
TABLE 2.
Enzyme kinetic measurements illustrating the ability of the OXA-type β-lactamases to hydrolyze carbapenems
| Enzyme | Group | Imipenem |
Meropenem |
Reference | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Vmax (%) | Km (μM) | kcat/Km ratio (s−1 mM−1) | kcat (s−1) | Vmax (%) | Km (μM) | kcat/Km ratio (s−1 mM−1) | |||
| OXA-23 | 23 | 0.35 | 4.8 | 73 | 0.068 | ≤1 | ≥68,000 | 36 | ||
| OXA-23 | 23 | 0.49 | 0.204 | 0.0024 | 35 | |||||
| OXA-27 | 23 | 0.1a | 20 | 0.04a | 15 | 34 | ||||
| OXA-146 | 23 | 0.043 | 0.046 | 0.00093 | 35 | |||||
| OXA-40 | 40 | 4a | 20 | 2a | 775 | 41 | ||||
| OXA-40 | 40 | 0.1 | 6.5 | 15 | 42 | |||||
| OXA-40 | 40 | 1 | 0.58 | 0.00172 | 0.37 | 0.0099 | 0.037 | 43 | ||
| OXA-40 | 40 | 2.1 | 0.752 | 0.0028 | 35 | |||||
| OXA-25 | 40 | 3a | 11 | 0.4a | 12 | 34 | ||||
| OXA-26 | 40 | 2.4a | 3 | 0.4a | 3 | 34 | ||||
| OXA-51 | 51 | 3.1b | 11 | 45 | ||||||
| OXA-69 | 51 | 0.1 | 3,600 | 0.03 | 0.06 | 4,500 | 0.01 | 46 | ||
| OXA-58 | 58 | 0.1 | 7.5 | 13.5 | <0.01 | 0.075 | <0.15 | 51 | ||
| OXA-58 | 58 | 0.22 | 1.3 | 169 | 49 | |||||
| OXA-134 | 134 | 0.1 | 10 | 10 | 0.05 | 250 | 0.2 | 56 | ||
| OXA-143 | 143 | 0.05 | 5 | 0.01 | 0.3 | 100 | 0.003 | 58 | ||
| OXA-229 | 229 | 0.09 | 30 | 3 | <0.01 | 0.03 | <0.1 | 72 | ||
| OXA-235 | 235 | 0.07 | 0.19 | 0.00034 | 0.117 | 0.016 | 0.0073 | 43 | ||
| OXA-48 | 48 | 2 | 14 | 145 | 0.1 | 200 | 0.5 | 62 | ||
| OXA-48 | 48 | 4.8 | 13 | 370 | 0.07 | 11 | 6 | 64 | ||
| OXA-163 | 48 | 0.03 | 530 | 0.05 | 0.07 | 2,200 | 0.03 | 64 | ||
| OXA-181 | 48 | 4 | 40 | 100 | 0.1 | 70 | 1 | 69 | ||
| OXA-232 | 48 | 1 | 35 | 30 | 0.1 | 125 | 1 | 69 | ||
Percent relative to penicillin G.
Percent relative to ampicillin.
The production of OXA-23 by an A. baumannii strain is enough to confer resistance to the carbapenems. As shown in Table 3, when a low-copy-number plasmid vector carrying a cloned blaOXA-23 gene is put into sensitive A. baumannii type strain CIP 70.10, MICs of the carbapenems are increased to 16 μg ml−1, the breakpoint for considering strains to be resistant (40). However, when the blaOXA-23 gene is carried in a strain that also expresses the AdeABC efflux pump, the MICs are increased to >32 μg ml−1. This indicates that, unlike some of the other OXA-type carbapenemases, strains do not require other resistance mechanisms to work in synergy with OXA-23 to be carbapenem resistant, although high levels of resistance are achieved only when there are other mechanisms present.
TABLE 3.
Carbapenem MICs for wild-type and transformed recipient strains encoding OXA-type β-lactamasesa
| Strain | Enzyme group | MIC (μg ml−1) |
Reference | |
|---|---|---|---|---|
| Imi | Mer | |||
| A. baumannii CIP 70.10 | 0.25 | 0.25 | 40 | |
| A. baumannii BM4547 | 0.5 | 0.5 | 40 | |
| A. baumannii ATCC 17978 | 0.125 | 0.5 | 36 | |
| A. baumannii CIP 70.10 + OXA-23 | 23 | 16 | 16 | 40 |
| A. baumannii BM4547 + OXA-23 | 23 | >32 | >32 | 40 |
| A. baumannii ATCC 17978 + OXA-23 | 23 | 16 | 64 | 36 |
| A. baumannii CLA-1 ΔOXA-40 | 2 | 4 | 40 | |
| A. baumannii 17978 | 0.25 | 0.5 | 43 | |
| A. baylyi | 0.125 | 0.25 | 43 | |
| A. baumannii CLA-1 OXA-40 | 40 | >32 | >32 | 40 |
| A. baumannii 17978 + OXA-40 | 40 | >32 | >32 | 43 |
| A. baumannii CIP 70.10 + OXA-40 | 40 | 4 | 4 | 40 |
| A. baumannii BM4547 + OXA-40 | 40 | 8 | 8 | 40 |
| A. baylyi + OXA-40 | 40 | 32 | 32 | 43 |
| A. nosocomialis ATCC 17903 | 0.12 | 0.25 | 161 | |
| A. baumannii ATCC 15151 | 0.5 | 159 | ||
| A. nosocomialis ATCC 17903 + OXA-138 | 51 | 32 | 32 | 161 |
| A. baumannii ATCC 15151 + OXA-82 | 51 | 32 | 159 | |
| A. baumannii CIP 70.10 + OXA-58 | 58 | 2 | 2 | 40 |
| A. baumannii BM4547 + OXA-58 | 58 | 32 | 32 | 40 |
| A. baumannii CIP 70.10 + OXA-97 | 58 | 2 | 2 | 54 |
| E. coli TOP10 | 0.06 | 0.06 | 56 | |
| E. coli TOP10 + OXA-134 | 134 | 0.5 | 0.5 | 56 |
| A. baumannii ATCC 19606 | 0.19 | 0.19 | 58 | |
| A. baumannii ATCC 19606 + OXA-143 | 143 | 32 | 32 | 58 |
| E. coli TOP10 | 0.12 | 71 | ||
| E. coli TOP10 + OXA-211 | 211 | 0.25 | 71 | |
| E. coli TOP10 + OXA-213 | 213 | 0.25 | 71 | |
| E. coli TOP10 + OXA-214 | 214 | 0.12 | 71 | |
| E. coli TOP10 | 0.06 | 0.03 | 72 | |
| E. coli TOP10 + OXA-229 | 229 | 0.12 | 0.06 | 72 |
| A. baylyi + OXA-235 | 235 | 1 | 2 | 43 |
| A. baumannii 17978 + OXA-235 | 235 | 3 | 4 | 43 |
| K. pneumoniae CIP53153 | 0.06 | 0.06 | 62 | |
| E. coli DH10B | 0.06 | 0.06 | 62 | |
| E. coli TOP10 | 0.12 | 0.01 | 64 | |
| E. coli HB4 | 0.25 | 0.25 | 69 | |
| E. coli J53 | 0.06 | 0.03 | 200 | |
| E. coli HB101 | 0.12 | 0.03 | 65 | |
| K. pneumoniae CIP53153 + OXA-48 | 48 | 2 | 0.25 | 62 |
| E. coli DH10B + OXA-48 | 48 | 2 | 0.25 | 62 |
| E. coli TOP10 + OXA-48 | 48 | 0.5 | 0.1 | 64 |
| E. coli HB101 + OXA-48 | 48 | 0.5 | 0.006–0.12 | 65 |
| E. coli TOP10 + OXA-163 | 48 | 0.25 | 0.02 | 64 |
| E. coli J53 + OXA-163 | 48 | 0.5 | 0.03 | 200 |
| E. coli TOP10 + OXA-181 | 48 | 0.5 | 0.06b | 99 |
| E. coli HB4 + OXA-181 | 48 | >32 | 32 | 69 |
| E. coli TOP10 + OXA-204 | 48 | 0.5 | 0.06b | 100 |
| E. coli TOP10 + OXA-232 | 48 | 0.25 | 0.06 | 69 |
| E. coli HB4 + OXA-232 | 48 | 24 | 8 | 69 |
| E. coli HB101 + OXA-245 | 48 | 2 | 0.12 | 65 |
| E. coli J53 + OXA-247 | 48 | 0.5 | 0.03 | 200 |
E. coli strain HB4 lacks the porins OmpF and OmpC. Imi, imipenem; Mer, meropenem.
The same value was found for OXA-48 in these studies.
OXA-40/24-LIKE β-LACTAMASES
The second group of OXA-type β-lactamases from A. baumannii to be identified was the OXA-40 group. The founding member of this group, OXA-24, which was subsequently renamed OXA-40, was identified in isolates in 1997, which were part of an outbreak in Spain (41). A further 6 enzyme variants have since been discovered (Table 1 and Fig. 1). While initially found only in isolates of A. baumannii, more recent reports have identified blaOXA-40-like genes on plasmids in other Acinetobacter species as well as in P. aeruginosa and Klebsiella pneumoniae (Table 1). The GC content of the blaOXA-40-like genes is 34.1%, which is considerably lower than the average for the A. baumannii genome, indicating that these genes are very unlikely to have originated from within this species (Table 4) (33).
TABLE 4.
Diversity of blaOXA genes found in Acinetobacter species and the housekeeping genes from two MLST schemesc
| Gene | No. of alleles | π (JC) | πN (JC) | πS (JC) | πN/πS ratio | % GC |
|---|---|---|---|---|---|---|
| blaOXA genes | ||||||
| blaOXA-51-like | 95 | 0.01789 | 0.01062 | 0.04395 | 0.241638 | 39.2 |
| blaOXA-23-like | 16a | 0.00663 | 0.00462 | 0.01401 | 0.329764 | 37.5 |
| blaOXA-40-like | 7 | 0.00242 | 0.00311 | 0 | 34.1 | |
| blaOXA-58-like | 4 | 0.00178 | 0.00229 | 0 | 37.4 | |
| blaOXA-143-like | 5 | 0.05782 | 0.02823 | 0.17205 | 0.164080 | 34.4 |
| blaOXA-235-like | 7 | 0.05389 | 0.02825 | 0.14726 | 0.191838 | 45.8 |
| blaOXA-213-like | 11b | 0.09899 | 0.04043 | 0.34439 | 0.117396 | 36.7 |
| blaOXA-214-like | 5 | 0.00802 | 0.00282 | 0.02633 | 0.107102 | 41.1 |
| blaOXA-211-like | 6 | 0.04659 | 0.02752 | 0.11580 | 0.237651 | 45.7 |
| blaOXA-229-like | 8 | 0.01263 | 0.00788 | 0.03003 | 0.262404 | 38.3 |
| Housekeeping genes from Oxford MLST scheme | ||||||
| gltA | 54 | 0.03454 | 0.00409 | 0.16852 | 0.024270 | 41 |
| gyrB | 98 | 0.04628 | 0.00468 | 0.23992 | 0.019507 | 41.6 |
| gdhB | 100 | 0.05113 | 0.01387 | 0.22013 | 0.063008 | 36.5 |
| recA | 64 | 0.06156 | 0.00368 | 0.38409 | 0.009581 | 43.2 |
| cpn60 | 50 | 0.03184 | 0.00582 | 0.13351 | 0.043592 | 37.5 |
| gpi | 183 | 0.08073 | 0.01152 | 0.35216 | 0.032712 | 41.3 |
| rpoD | 72 | 0.03508 | 0.00900 | 0.15189 | 0.059253 | 39.4 |
| Mean | 0.04874 | 0.00752 | 0.23575 | 0.035989 | 40.07 | |
| Housekeeping genes from Pasteur MLST scheme | ||||||
| cpn60 | 56 | 0.05097 | 0.00484 | 0.25524 | 0.018963 | 37.6 |
| fusA | 58 | 0.04641 | 0.01566 | 0.17275 | 0.090651 | 44.7 |
| gltA | 55 | 0.06654 | 0.01098 | 0.33502 | 0.032774 | 41.1 |
| pyrG | 29 | 0.09052 | 0.00183 | 0.56740 | 0.003225 | 40.8 |
| recA | 58 | 0.10513 | 0.00898 | 0.38394 | 0.023389 | 43.3 |
| rplB | 36 | 0.05028 | 0.00862 | 0.19302 | 0.044659 | 44.2 |
| rpoB | 56 | 0.05566 | 0.00986 | 0.23000 | 0.042870 | 43.7 |
| Mean | 0.06650 | 0.00868 | 0.30534 | 0.036647 | 42.2 |
Nucleotide sequences for blaOXA-102, blaOXA-103, and blaOXA-105 were not available.
Nucleotide sequences for 6 genes were not available.
Only the nucleotide sequence for blaOXA-134a was available for this group, so it was excluded from the analyses. (Based on data accessed from two Acinetobacter databases [http://pubmlst.org/abaumannii/ and http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html] and the NCBI database [http://www.ncbi.nlm.nih.gov/] on 22 August 2013.) JC, Jukes-Cantor.
The kinetic properties of three members of the OXA-40 group have been determined: OXA-40, OXA-25, and OXA-26 (Table 2) (34, 35, 41–43). In general, the enzymes can hydrolyze the penicillins but appear to show weak activity against cephalosporins and the carbapenems. However, as shown in Table 2, the estimates of the kinetic parameters vary considerably. Of the three enzymes, OXA-40 demonstrated the highest activity against the carbapenems. The publication of the crystal structure of OXA-40 in 2007 began to demonstrate the basis for the substrate specificity of the enzyme (38), and this has been followed more recently by crystal structures of OXA-40 bound to β-lactamase inhibitors and to the carbapenem doripenem (39, 44). The structure solved by Santillana et al. (38) was the first to identify a hydrophobic bridge formed by tyrosine 112 and methionine 223 (OXA-40 numbering), which restricts access to the active site of the enzyme. This feature has since been identified in other OXA-type β-lactamases such as OXA-23 (35). By performing site-directed mutagenesis of tyrosine 112 and methionine 223, the authors of that study provided evidence that these residues are responsible for orienting the carbapenems in the active site, with the mutated enzymes demonstrating a reduced ability to hydrolyze imipenem (38, 44).
Isolates that carry an OXA-40-like β-lactamase are typically resistant to the carbapenems. In a study where the blaOXA-40 gene was insertionally inactivated, the bacterium became more sensitive to the carbapenems, penicillins, and cephalosporins, demonstrating the role that this enzyme plays in increasing resistance to the β-lactams in general (Table 3) (40). Similarly, insertion of the cloned blaOXA-40 gene on low-copy-number plasmid pAT801 by transformation into carbapenem-sensitive clinical strains also increases the MIC to >32 mg liter−1 (Table 3) (40). However, as with blaOXA-23 described above, when blaOXA-40 is inserted into a sensitive laboratory strain, the MIC is increased only to a moderate 4 mg liter−1, which increases to 8 mg liter−1 when inserted into the same strain that also expresses the AdeABC efflux pump. These data demonstrate that for OXA-40, as for OXA-23, a range of resistance mechanisms work in concert to achieve high levels of carbapenem resistance.
OXA-51-LIKE β-LACTAMASES
The largest group of OXA-type β-lactamases identified to date are the OXA-51-like β-lactamases. The founding member of the group, OXA-51, was first identified in A. baumannii isolates from Argentina isolated in 1996 (45). These enzymes are intrinsic to A. baumannii and are naturally found on the chromosome of this species, which has led to the identification of 95 enzyme variants to date (Table 1). The huge number of variants, alone, is an indication that these chromosomally encoded enzymes have been under considerable selective pressure from antibiotic use, and these enzymes are not benign and do play a role in resistance. Only two of the OXA-51-like enzymes have had their kinetic properties studied: OXA-51 and OXA-69 (Table 2) (45, 46). While the two enzymes differed in their affinities for both imipenem and meropenem, both enzymes demonstrated only weak hydrolytic activity toward the carbapenems. Despite this weak activity, experiments have shown that when enzymes from this group are expressed in vivo, they can dramatically increase the carbapenem MIC for an isolate, thus conferring resistance. As shown in Table 3, when OXA-51-like enzymes were expressed in sensitive A. baumannii and Acinetobacter nosocomialis strains, the carbapenem MICs were increased to 32 μg ml−1, a level of resistance comparable to that conferred by the acquired OXA-type carbapenemases in similar experiments. While further work is required to determine whether all members of the OXA-51-like group are able to confer carbapenem resistance, the enzymes of this group remain a major concern, as they present the possibility that all A. baumannii isolates may be capable of becoming resistant to the carbapenems.
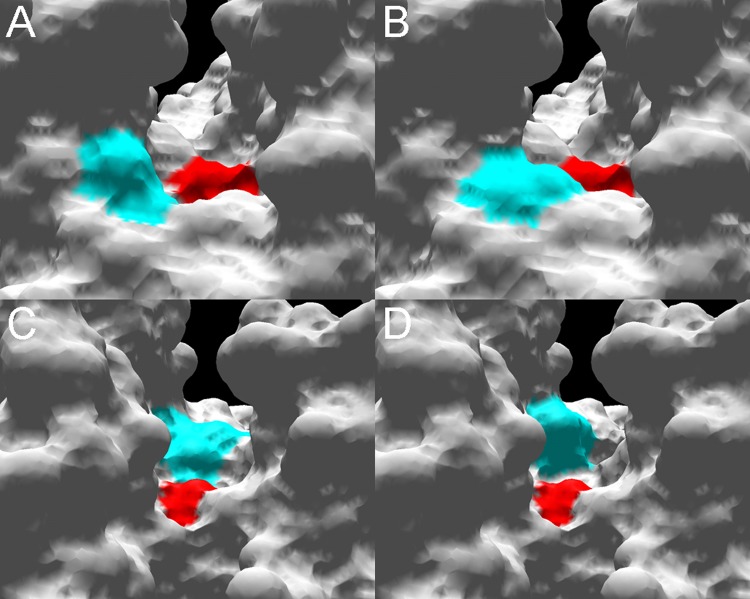
The crystal structure for an OXA-51-like enzyme has not been solved, but structural modeling and sequence analysis can be used to identify structural elements involved in activity toward the carbapenems. A recent study that conducted in silico docking of β-lactam substrates into a molecular model of OXA-51 showed that imipenem had the weakest interaction with the enzyme, with ceftazidime showing the strongest interaction (47). However, these data are inconsistent with kinetic data where hydrolysis of ceftazidime was not detected (45, 46). A further study has demonstrated that the hydrolysis of oxacillin by OXA-51 is thermodynamically more favorable than the hydrolysis of imipenem and that a larger conformational change in the tertiary structure of the enzyme is detected when bound to imipenem than when bound to oxacillin (48), which is consistent with previous kinetic data (45, 46). Analyses of molecular models of the OXA-51-like enzymes indicate that there are likely to be differences between the hydrolytic profiles of these enzymes. As shown in Fig. 2, amino acid changes at positions predicted to be located within the active-site cleft of the enzyme are predicted to alter the shape of the cleft, which may impact β-lactam binding and hydrolysis. Unlike the OXA-40, OXA-23, and OXA-58 β-lactamases, which either have been shown or are predicted to have a hydrophobic bridge across the active-site cleft of the enzyme (35, 38, 39, 49), modeling of the OXA-51-like enzymes suggests that this feature is absent (Fig. 3).
FIG 2.
Molecular surface models of the active-site cleft of the OXA-51-like enzymes. Structural models were created by using the online protein homology/analogy recognition engine (phyre) (http://www.sbg.bio.ic.ac.uk/∼phyre/), using the crystal structure for OXA-40/24 reported by Santillana et al. (38) as a backbone. Using local numbering, structures were viewed in Swiss-PdbViewer DeepView version 4 (http://spdbv.vital-it.ch/). (A) View down the active-site cleft of OXA-66 with the active-site serine 80 (red) and an isoleucine at position 129 (blue); (B) view down the active-site cleft of OXA-83 with the active-site serine 80 (red) and a leucine at position 129 (blue); (C) view down the active-site cleft of OXA-66 (from the opposite orientation to panels A and B) with the active-site serine 80 (red) and a leucine at position 167 (blue); (D) view down the active-site cleft of OXA-82 with the active-site serine 80 (red) and a valine at position 167 (blue).
FIG 3.
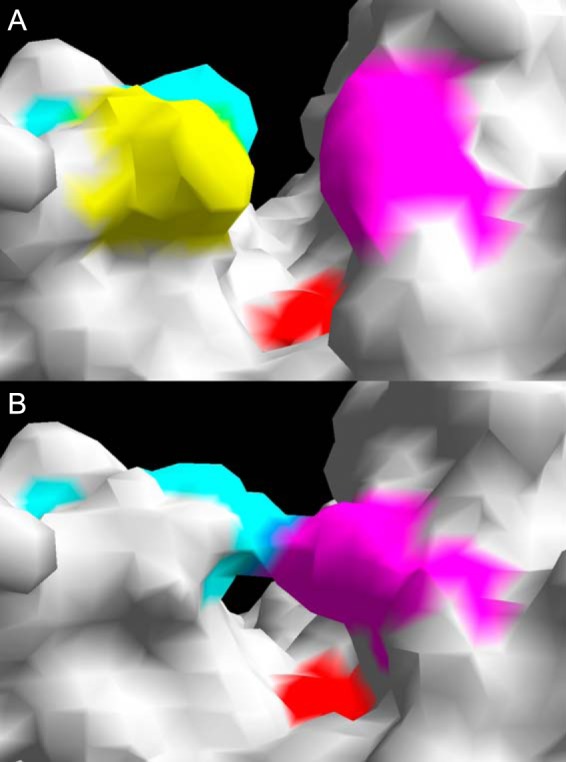
Molecular surface models of the active-site cleft of OXA-66 (A) and OXA-40 (B), showing the absence of a hydrophobic bridge in OXA-66. Using local numbering, the active-site serine 80 is shown in red. (A) Leucine 110 is in yellow, phenylalanine 111 is in blue, and tryptophan 222 is in pink; (B) tyrosine 112 is in blue, and methionine 223 is in pink.
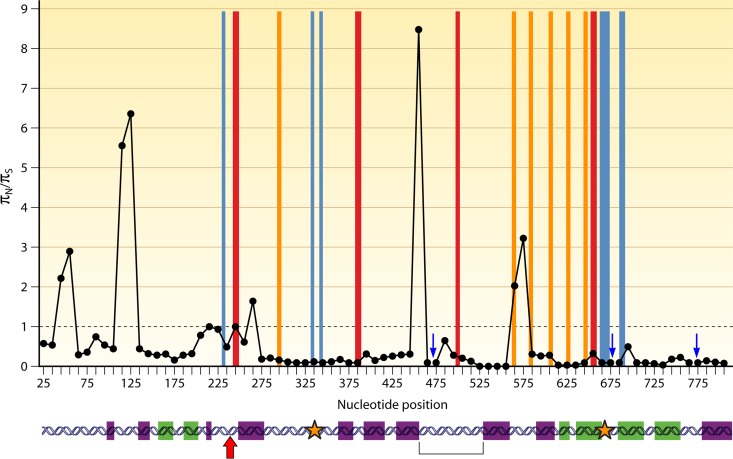
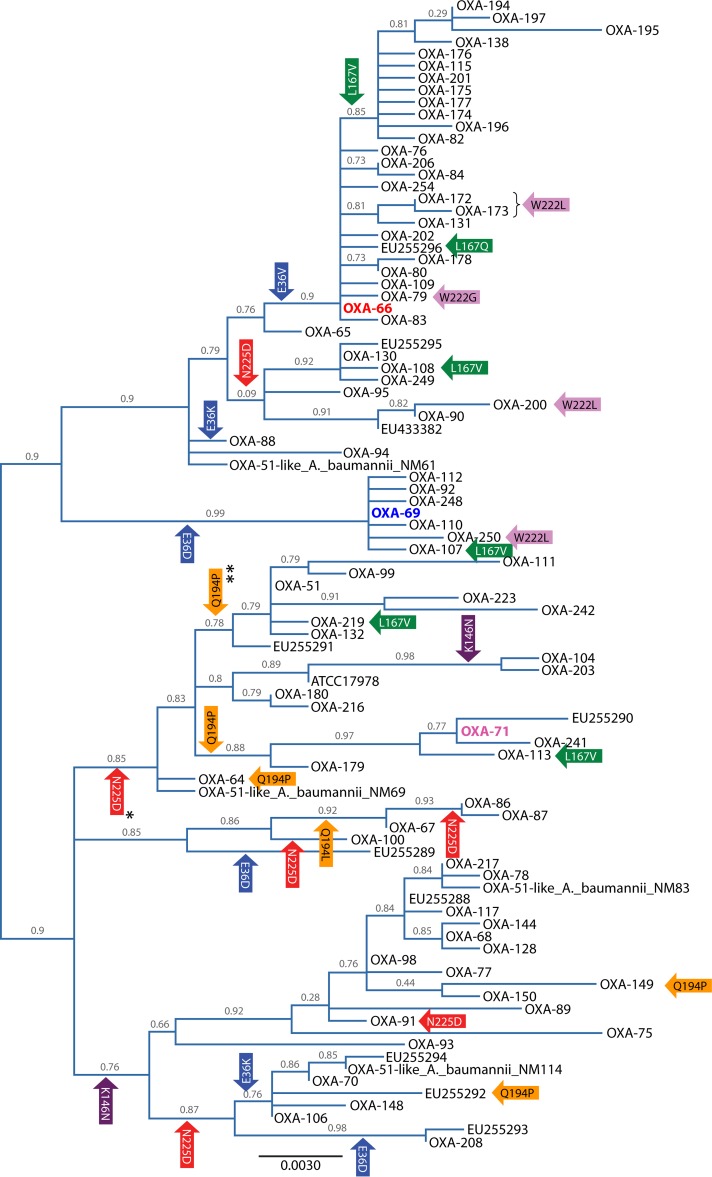
Sequence analysis of the blaOXA-51-like genes provides evidence that regions of these genes are preferentially altered under the selective pressure of antibiotic usage (Fig. 4). Evidence for positive selection, as indicated by a πN/πS ratio (the ratio of nonsynonymous to synonymous nucleotide polymorphisms) of >1, can be detected between amino acid positions 64 and 97, which surround the active-site serine residue at position 81; between amino acid positions 144 and 160, at the start of the Ω-loop; and between amino acid positions 180 and 200, which are located in regions corresponding to those suggested to be important for dimer formation in OXA-13 (50). In addition, there are regions, indicated by blue arrows in Fig. 4, where a πN/πS ratio could not be determined, as the polymorphisms coded for only nonsynonymous changes, which also indicates that they may be under positive selection. These include amino acid positions 147 to 167, spanning most of the Ω-loop, and amino acid positions 214 to 237, which include residues at positions 217 to 219 corresponding to active-site residues in OXA-40 (38) and a number of residues that correspond to those involved in carbapenem specificity in OXA-40, including position 222, which forms one half of the hydrophobic bridge (38). In addition to the πN/πS ratios, evidence for selection can be demonstrated by looking at the amino acid phylogeny of the enzymes, as shown in Fig. 5. There are six polymorphic amino acid positions that are located in more than one clade of the phylogeny, suggesting that these sites may be under selection. Five of these positions are in the regions described above as likely to be important for enzyme function, such as positions 146 and 167 in the Ω-loop, position 194 in the region important for dimer formation, and positions 222 and 225 in the region predicted to be involved in carbapenem specificity. The remaining polymorphic site at position 36 is not known to be involved in enzyme function according to previous data for other OXA-type enzymes (38, 50) but is located in a region demonstrating high πN/πS ratios in Fig. 4 and, as such, warrants further investigation as to the potential role that residues at this position may play. Together, these data indicate that different OXA-51-like enzymes are likely to vary in their spectrum of activity, and further characterization of the different enzyme variants is required to determine this.
FIG 4.
Regions of the OXA-51-like enzymes with evidence of selection. πN/πS, ratio of nonsynonymous to synonymous polymorphisms, with a ratio of >1 providing evidence for positive selection. Ratios were calculated with DNAsp version 5 (174) in sliding windows 50 nucleotides long, with a step size of 10 nucleotides. The horizontal gray line indicates a πN/πS ratio of 1; red boxes indicate predicted active-site elements (38); blue boxes indicate predicted sites involved in specificity for the carbapenems (38); orange boxes indicate predicted sites involved in dimer formation (50); blue arrows indicate regions where a πN/πS ratio could not be determined, as these regions contained only nonsynonymous polymorphisms; purple boxes indicate α-helices; green boxes indicate β-sheets; the red arrow indicates the active-site serine; orange stars indicate the location of residues forming the hydrophobic bridge in OXA-40 (38); and the bracket indicates the location of the Ω-loop.
FIG 5.
Maximum likelihood nucleotide tree of the blaOXA-51-like genes intrinsic to A. baumannii. For enzymes for which a designation was not available, the strain designation or gene accession number (http://www.ncbi.nlm.nih.gov/) is used instead. The tree was implemented in SeaView version 4.2.5 (172) with PhyML using a general time-reversible (GTR) model (173). Support was estimated by using approximate likelihood ratio tests, the results of which label the branches. The tree was visualized by using FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/) and is midpoint rooted. The gene coding for the enzyme representative of global clone 1 isolates, OXA-66, is highlighted in red; the gene for OXA-69, representative of global clone 2 isolates, is highlighted in blue; and the gene for OXA-71, representative of global clone 3 isolates, is highlighted in pink. Colored arrows represent genes or branches containing polymorphisms that are observed in more than one position within the tree. The arrows are color coded by the amino acid position in the enzyme that the location of the polymorphism codes for and are labeled with the amino acid change from the consensus that results from the polymorphism. *, except for OXA-223 and OXA-242; **, except for OXA-242.
OXA-58-LIKE β-LACTAMASES
The first member of the OXA-58 group of enzymes was identified in France in 2003. It was found in a multidrug-resistant A. baumannii clinical isolate that also demonstrated carbapenem resistance (51). Analysis of the enzyme kinetics of OXA-58 revealed properties similar to those found in other OXA-type enzymes in A. baumannii (Table 2), with weak activity against the carbapenems and penicillin and an ability to hydrolyze cefpirome and cephalothin but not ceftazidime, cefotaxime, or cefepime (51). As seen with OXA-40, enzyme kinetics for OXA-58 measured by different groups gave different results, with OXA-58 demonstrating a high rate of catalysis but a low substrate affinity for imipenem in a study by Verma et al. (49) compared with the data generated by Poirel et al. (51) (Table 2). Mutation of the lysine residue to an alanine at position 86 in OXA-58 resulted in an almost complete loss of function of the enzyme, and it has been shown that carbamylation of this lysine is required for enzyme activity (49). Similarly to OXA-40 and OXA-23, it is thought that the more closed and hydrophobic nature of the active-site cleft on OXA-58 is responsible for its ability to hydrolyze the carbapenems (49). As shown in Table 3, the insertion by transformation of a cloned blaOXA-58 gene on a low-copy-number vector into a sensitive A. baumannii strain increased resistance but not to levels considered resistant (40). However, transformation into a reference A. baumannii strain with increased expression levels of the AdeABC efflux system produced a resistant isolate, demonstrating that clinically significant levels of resistance with this enzyme require multiple resistance mechanisms to work in concert. Due to the many resistance mechanisms possessed by A. baumannii, isolates of this species carrying blaOXA-58-like genes are frequently identified with high levels of carbapenem resistance (52).
Despite it being 10 years since the first discovery of OXA-58, only a further three variants of the enzyme have been identified. An A. baumannii isolate from Singapore isolated in 1996 was found to encode OXA-96 (53), OXA-97 was encoded by A. baumannii isolates collected between 2001 and 2005 in Tunisia (54), and OXA-164 was identified in isolates recovered from a patient in Germany (55). The reasons for this are unclear, although it may be due in part to a lack of sequence data. The GC content of the blaOXA-58-like genes falls below that of the A. baumannii core genome (Table 4) (33), indicating their likely import from another bacterial species into A. baumannii, as seen with the blaOXA-23-like genes (30). Should this import have been a recent event, this may also explain the lack of diversity observed in this enzyme group.
OXA-134a β-LACTAMASE
Following the identification of A. radioresistens as the natural host of the OXA-23-like enzymes, and as A. baumannii is the natural host for the OXA-51-like enzymes, further studies have been conducted to identify naturally occurring OXA-type enzymes belonging to other Acinetobacter species. One such study identified OXA-134a from an Acinetobacter lwoffii isolate (56). Screening of further A. lwoffii isolates has revealed a further six enzyme variants, OXA-186 to OXA-191, and has shown that the genes for this group of enzymes are universal in this species (56, 57). The relationships between the amino acid sequences of the OXA-134a-like enzymes are shown in Fig. 1.
In A. lwoffii, the blaOXA-134a-like genes do not appear to be very highly expressed, as the isolates are not resistant to any β-lactam antibiotics. However, when the genes coding for OXA-134a and OXA-187 were cloned into E. coli, they conferred reduced susceptibility to the carbapenems (Table 3) and most cephalosporins and conferred resistance to the penicillins and penicillin–β-lactamase inhibitor combinations (56). The enzyme kinetic measurements for OXA-134a demonstrated it to be a relatively weak hydrolyzer of the carbapenems (Table 2). To date, enzymes from this group have been found only in A. lwoffii and are not presenting as a clinical problem. However, they represent a reservoir of enzyme variants that may have the potential to evolve to confer higher levels of β-lactam resistance and be transferred to other species, as has happened with the OXA-23-like enzymes.
OXA-143 β-LACTAMASE
In 2004, three A. baumannii isolates that were resistant to almost all β-lactam antibiotics, including the carbapenems, but were negative by PCR for known acquired carbapenem-hydrolyzing β-lactamases were identified in patients from Brazil. Further investigation identified a new enzyme, named OXA-143 (58). As with other acquired OXA-type β-lactamases in A. baumannii, the enzyme kinetics for OXA-143 showed low levels of carbapenem hydrolysis (Table 2), but the enzyme conferred high levels of resistance to the carbapenems when the gene coding for it was transformed into a previously sensitive A. baumannii reference strain (Table 3) (58). Analysis of the nucleotide sequences of the blaOXA-143-like genes indicates that they are under a greater degree of positive selection than the multilocus sequence typing (MLST) housekeeping genes of A. baumannii, as would be expected if these genes are under antibiotic selection (Table 4). Together, these data indicate that, as with the other acquired OXA-type carbapenemases, this group of enzymes is capable of conferring high levels of carbapenem resistance when expressed in isolates alongside other resistance mechanisms and are likely contributing to carbapenem resistance in clinical isolates.
To date, this group of enzymes has been identified only in A. baumannii and Acinetobacter pittii isolates from Brazil and South Korea and only on plasmids (58–61). The GC content of the enzymes is much lower than that for the A. baumannii genome as a whole (Table 4), which, along with their exclusive plasmidic location, strongly suggests that these enzymes originated from another bacterial species. As shown in Fig. 1, the amino acid sequences of these enzymes are similar to those of other Acinetobacter-derived OXA-type β-lactamases, indicating that another Acinetobacter species is the original host organism. Note that in Fig. 1, the OXA-143-like enzymes are very closely related to the OXA-40-like enzymes, and it may even be the case that the OXA-143-like enzymes are the progenitors of the OXA-40-like enzymes. As larger numbers of sequences of both of these enzyme groups become available, the relationship between these two enzyme groups will be clarified.
OXA-48 β-LACTAMASE
The other carbapenem-hydrolyzing OXA-type enzymes discussed in this article are largely restricted to Acinetobacter species, in particular A. baumannii. However, in 2001, a Klebsiella pneumoniae isolate was obtained from a patient in Istanbul, Turkey, which was found to be multidrug resistant, including resistance to the carbapenems. In this isolate, a new OXA-type β-lactamase was identified and named OXA-48 (62). This enzyme and its variants are now widespread in K. pneumoniae and other Enterobacteriaceae and have now been reported in A. baumannii as well (63), and they represent one of the most concerning developments in carbapenem resistance in the last decade.
Analysis of the enzyme kinetics of OXA-48 showed that the enzyme has a low level of hydrolytic activity against the carbapenems, with much greater activity against imipenem than against meropenem (Table 2) (62). In addition, when the enzyme was cloned into sensitive E. coli reference strains, it resulted in only modest increases in MICs of the carbapenems (Table 3) (62, 64, 65). Since 2001, 10 more named OXA-48 variants have been identified (Table 1), although it should be noted that a number of related unnamed enzymes have been identified in Shewanella species, thought to be the original host genus for this group of enzymes (66–68). Four of these enzymes, OXA-162, OXA-163, OXA-181, and OXA-232, have had their kinetic properties measured, and while OXA-163 appears to be a very poor hydrolyzer of the carbapenems, OXA-181 and OXA-232 appear broadly similar to OXA-48 in their activity, with OXA-232 perhaps demonstrating better hydrolysis of the penicillins, although cross-study comparisons may not be very reliable (Table 2) (64, 69). Interestingly, the lack of activity against the carbapenems in OXA-163 is associated with an ability to hydrolyze ceftazidime and aztreonam, which is not detectable in OXA-48, and an increased ability to hydrolyze cefotaxime and cefepime over OXA-48, making this enzyme more similar to an ESBL than to a carbapenem-hydrolyzing oxacillinase (64). As with OXA-48, the carbapenem MICs obtained when seven of the variants were cloned into sensitive E. coli strains showed only moderate increases (Table 3). However, when two of these variants, OXA-181 and OXA-232, were cloned into sensitive E. coli strains that also lacked the porins OmpF and OmpC, carbapenem MICs were significantly increased, demonstrating that the presence of OXA-48-like enzymes alongside other resistance mechanisms can confer high levels of carbapenem resistance (69).
The crystal structure of OXA-48 has been solved and demonstrates that the enzyme has a different mechanism for carbapenem hydrolysis compared to that of the other carbapenem-hydrolyzing oxacillinases (70). Unlike OXA-40 and OXA-23, OXA-48 does not possess a hydrophobic bridge across the active site, demonstrating that other features must be responsible for the ability to hydrolyze carbapenems. The tertiary structure of OXA-48 is more similar to that of OXA-10 and OXA-13 than it is to that of OXA-40 or OXA-23, and comparisons of the sequences and structures of OXA-48 and OXA-13 indicate that small changes to the amino acid residues both within the active-site region and in the β5-β6 loop of OXA-48 are likely to be responsible for the enzyme's ability to hydrolyze the carbapenems, with the β5-β6 loop providing a hydrophilic environment harboring water molecules that are required for hydrolysis (70). In addition, the active-site cleft of OXA-48 contains a larger hydrophobic region than that in OXA-13, which allows the hydroxyethyl group of meropenem greater mobility. This ensures that the antibiotic can be brought into proximity with a water molecule for deacylation of the antibiotic (70). These features demonstrate that OXA-48 has followed a different evolutionary trajectory from the OXA-type carbapenemases common in Acinetobacter species to develop the ability to hydrolyze the carbapenems.
OTHER CARBAPENEM-HYDROLYZING OXA-TYPE β-LACTAMASES
Recent studies aiming to identify the naturally occurring OXA-type enzymes of other Acinetobacter species have successfully identified a number of new enzyme groups. These new groups include the OXA-211-like enzymes from Acinetobacter johnsonii, the OXA-213-like enzymes from Acinetobacter calcoaceticus, the OXA-214-like enzymes from Acinetobacter haemolyticus, the OXA-229-like enzymes from Acinetobacter bereziniae, and the OXA-235-like enzymes from Acinetobacter schindleri (Table 1 and Fig. 1).
The parent enzymes from each of these five groups have been cloned into sensitive type strains to examine their contribution toward β-lactam resistance, including carbapenem resistance. In all five cases, the enzymes increased the MICs of the penicillins for the strains to high levels, had little or no effect on cephalosporin MICs, and slightly increased carbapenem MICs (Table 3) (43, 71, 72). Two of the enzymes, OXA-229 and OXA-235, have had their kinetic properties measured and in both cases hydrolyze carbapenems at low levels, below those of the acquired carbapenemases OXA-40 and OXA-58 (43, 72). In addition, the specific activities of OXA-211, OXA-213, and OXA-214 for the carbapenems have been measured, and all three enzymes demonstrated better activity against imipenem than OXA-51 although not as much as OXA-23 (71). Analyses of the nucleotide sequences of the genes for the enzymes from these five groups demonstrate that they are under similar levels of positive selection as the other acquired carbapenem-hydrolyzing oxacillinases (Table 4). This suggests that these enzymes are likely to have been evolving under antibiotic selective pressure. Together, these data indicate that the enzymes from these five groups are able to hydrolyze the carbapenems and represent a reservoir of enzyme variants that could, under carbapenem selective pressure, evolve to confer clinically significant levels of carbapenem resistance and be transferred to other members of the Acinetobacter genus.
GENETIC LOCATION, MOBILIZATION, AND CONTROL
ISAba1
Insertion sequence (IS) elements play a major role in the mobilization and expression of OXA-type β-lactamases. This is particularly evident with the OXAs found in Acinetobacter species, where insertion sequences are frequently identified in association with an OXA β-lactamase gene (Fig. 6). The most prevalent of these insertion sequences is ISAba1. The first description of ISAba1 was from A. baumannii isolates in 2001 (73), although sequences corresponding to this element had been identified previously (74). The IS element was located upstream of the blaADC genes, and it has been shown that in this location, the IS element drives blaADC gene expression, which confers resistance to cephalosporin antibiotics (75). The transposase gene in ISAba1 is formed from two overlapping open reading frames and requires a frameshift during translation to form a complete transcript coding for the functional transposase (76). The transposition frequency of ISAba1 in E. coli has been reported to be (2.1 ± 0.7) × 10−7 transconjugants per recipient, or (1.8 ± 5.0) × 10−4 transconjugants per recipient when a frameshift mutation was introduced, suggesting that conditions under which frameshifts during transcription are likely to occur are likely to greatly increase the transposition frequency (76). This is supported by a study that demonstrated ISAba1 mobilization in response to the presence of the frameshift mutagen acriflavine (77). ISAba1 has since been identified in association with genes for a number of OXA-type beta-lactamases, such as the blaOXA-23-like genes, blaOXA-51-like genes, blaOXA-58-like genes, and blaOXA-235-like genes (Fig. 6) (43, 78, 79). When ISAba1 is located upstream of blaOXA genes, it tends to increase their expression to a level that confers resistance to the carbapenems. The location of ISAba1 25 bp upstream of blaOXA-23 provides a promoter sequence that drives transcription of the gene (80, 81). Similarly, when ISAba1 is located 7 bp upstream of the blaOXA-51-like genes, it provides a promoter that can increase blaOXA-51-like gene expression levels by 50-fold (82). This increase in expression levels raised the MICs of the carbapenems for the isolate and appears to have been selected by the use of carbapenems to treat the patient from whom the isolates were recovered (82). The MICs of the carbapenems for A. baumannii isolates overexpressing blaOXA-51-like genes due to an ISAba1 insertion have been shown to be the same as those for isolates expressing acquired OXA-type carbapenemases (83), indicating that all A. baumannii isolates could become carbapenem resistant through insertion of this promiscuous IS element.
FIG 6.
Mobile genetic elements associated with blaOXA genes. Commonly occurring features are color coded. (A and B) See reference 81; (C) see reference 94; (D) see reference 149; (E) see reference 175; (F) see reference 101; (G) see reference 159; (H) see reference 93; (I) see reference 176; (J) ISAba1 upstream of the blaOXA-51-like gene (78) and on one occasion found downstream as well (dashed box), see reference 204; (K) see reference 177; (L) see reference 178; (M) see reference 95; (N) see reference 179; (O) see reference 95; (P) see reference 79; (Q) see reference 90; (R) see reference 149; (S) see reference 79; (T) see reference 92; (U) see reference 79; (V) see reference 89; (W) see reference 180; (X and Y) see reference 79; (Z) see reference 91; (AA) see reference 96; (AB) see reference 64; (AC) see reference 99; (AD) see reference 97; (AE) see reference 98; (AF) see reference 64; (AG) see reference 100; (AH) see reference 69; (AI) see reference 43; (AJ) see reference 181; (AK) see reference 25; (AL) see reference 22.
In addition to its role in expression of blaOXA genes, ISAba1 also appears to be responsible for their mobilization, and a number of different transposons containing ISAba1 either singly or in multiple copies have been identified (Fig. 6). An example of these transposons is Tn2006, where two oppositely oriented copies of ISAba1 bracket a blaOXA-23 gene. A 9-bp target site duplication is found before the right inverted repeat of the upstream IS element and after the right inverted repeat of the downstream IS element, which indicates that the whole structure is mobilized as one unit (81). The identification of ISAba1 within a number of different transposons carrying blaOXA genes, in addition to other antibiotic resistance determinants, as well as its widespread nature in a number of Acinetobacter species (84) indicate the important role that this IS element plays in genomic plasticity within the acinetobacters.
ISAba3
The insertion sequence ISAba3 is very commonly found in association with blaOXA-58-like genes, often with one copy upstream and one copy downstream of the β-lactamase gene, forming a composite transposon (85–88). The element, which contains a single open reading frame coding for its transposase, was first identified in an A. baumannii isolate in 2003 (51). When located upstream and in the opposite orientation to a blaOXA gene, the element provides a promoter that drives blaOXA gene expression, in a manner similar to that of ISAba1. The lack of target site duplications either upstream or downstream of the ISAba3 elements suggests that this insertion sequence may be involved in disseminating blaOXA genes through a process of homologous recombination rather than through transposition (79). In many cases, the ISAba3 element found upstream of blaOXA-58-like genes is interrupted by another insertion sequence, such as ISAba2 (79), IS1008 (89), IS18 (79), and ISAba825 (90) (Fig. 6). In these instances, these chimeric elements often contain hybrid promoters that drive the expression of the β-lactamase gene. For example, the ISAba2/ISAba3 element contains the −35 region of the promoter within the right inverted repeat of ISAba2, while the −10 region is located within ISAba3. These chimeric promoters vary in the levels of expression of the blaOXA-58-like gene that they drive, suggesting that the use of the carbapenems is selecting for these more complicated genetic structures (89, 90).
Other Insertion Sequences in Acinetobacter
While ISAba1 and ISAba3 elements are commonly found in association with blaOXA genes in Acinetobacter species, there are many other IS elements that have been identified less frequently (Fig. 6). The insertion sequence ISAba125 has been identified in association with blaOXA-58 and appears to have been responsible for the duplication of this gene within an A. pittii isolate (91). This kind of gene duplication mediated by insertion sequences appears to alter the carbapenem MIC for an isolate. A study of three isolates obtained in Rome that contained one, two, or three copies of blaOXA-58 found that these isolates had imipenem MICs of 16, 32, and 128 mg liter−1, respectively (92). The duplication of the blaOXA-58 genes appears to have been mediated by IS26 elements that bracketed the ISAba2/ISAba3-blaOXA-58-ISAba3 unit. The insertion sequence ISAba4 has been found 25 bp upstream of blaOXA-23, where it provides a promoter, along with a fragment corresponding to the right inverted repeat of ISAba4 downstream of the same blaOXA-23 gene. It has been proposed that this structure mobilizes the blaOXA-23 gene through one-ended transposition, and this structure has been named Tn2007 (81). The insertion sequences ISAba9 and ISAba10 have both been found to be inserted into the middle of ISAba1 elements upstream of blaOXA genes. The chimeric ISAba9/ISAba1 element was located upstream of blaOXA-51, where the hybrid promoter increased the expression level of the blaOXA-51 gene by 8-fold over the level obtained with ISAba1 alone (93). Similarly, the chimeric ISAba10/ISAba1 element was found upstream of blaOXA-23, where the hybrid promoter increased the expression level of the blaOXA-23 gene by between 2- and 5-fold and increased carbapenem MICs by 2- to 8-fold compared to the levels with ISAba1 alone (94). In addition to providing promoters to increase the expression of blaOXA genes, some insertion sequences have been found to insert into the middle of blaOXA genes, disrupting their function. The insertion sequences ISAba15 and ISAba19 have been identified to be inserted into blaOXA-66 and blaOXA-78, respectively (95). To date, this has been detected very rarely, suggesting that in most instances, it is more beneficial for the bacteria to carry a gene coding for a functional OXA-51-like enzyme.
Insertion Sequences Associated with blaOXA-48-like Genes
When the blaOXA-48 gene was first identified, it was found in association with an upstream IS1999 element (62). It was later demonstrated that there is a second IS1999 element downstream of the blaOXA-48 gene, and this composite transposon was named Tn1999 (96). This transposon was shown to be able to mobilize the blaOXA-48 gene in E. coli although at a low frequency of <1 × 10−7 transconjugants per recipient. Further variants of Tn1999 have been identified, which contain an IS1R element inserted into the upstream IS1999 element (named Tn1999.2) (97) or an IS1R element inserted into the upstream IS1999 element and a second IS1R element inserted downstream of blaOXA-48 (named Tn1999.3) (Fig. 6) (98). As is seen with the chimeric elements in Acinetobacter species, the chimeric IS1R/IS1999 element contains a hybrid promoter, and when cloned into a sensitive E. coli strain, it resulted in a 2-fold-higher rate of hydrolysis of imipenem than that with just IS1999 alone (97). In this strain, this change was not significant, but it could improve survival in a strain that also restricts the influx of carbapenems.
Other blaOXA-48-like genes have been found on plasmids in association with other insertion sequences (Fig. 6). The blaOXA-163 gene is located downstream of an ISEcl4 element, a variant of IS4321 (64). However, unlike the Tn1999 family of transposons, the ISEcl4 element does not provide a promoter for blaOXA-163 gene expression; rather, transcription is driven by the original chromosomal promoter. The blaOXA-181, blaOXA-204, and blaOXA-232 genes are associated with ISEcp1 elements but each in slightly different arrangements (69, 99, 100). The blaOXA-181 gene is located downstream of a complete ISEcp1 element that forms a transposon named Tn2013, which mobilizes the gene through a one-ended transposition mechanism (99). The blaOXA-204 gene is located on a similar transposon, called Tn2016, that differs from Tn2013 by an ISkpn15 element inserted into the upstream ISEcp1 sequence (100). In the case of Tn2016, the ISkpn15/ISEcp1 chimeric element does not contain a hybrid promoter, and the expression of the gene is likely to be derived from the promoter located within ISEcp1. In the case of blaOXA-232, much of the upstream ISEcp1 element has been lost, along with the upstream target site duplication, although the 3′ fragment of ISEcp1 that remains contains the intact promoter that drives the expression of the blaOXA-232 gene (69). It has been speculated that this deletion of much of the ISEcp1 element has stabilized the blaOXA-232 gene on the plasmid, as it can no longer be mobilized by an ISEcp1 transposase (69). The three completely separate groups of insertion sequences with which blaOXA-48-like genes are associated may indicate that blaOXA-48-like genes have been mobilized from their original location in Shewanella species on several occasions.
XerC/XerD
Unlike the other OXA-type β-lactamases found in Acinetobacter species, the blaOXA-40-like genes are rarely found in association with insertion sequences, with only one report of an association with an upstream IS4 family element (101). Analysis of the nucleotide sequence surrounding a plasmid-located blaOXA-40 gene identified the presence of two conserved inverted repeats on either side of the gene, separated from one another by a 6-bp variable region (102). These sequences are homologous to the target sites for XerC and XerD recombinases, which are usually involved in converting chromosome and plasmid dimers into monomers (103). It is likely that the blaOXA-40-like genes are mobilized through a process of site-directed recombination mediated by XerC and XerD, and blaOXA-40-like genes flanked by XerC/XerD target sites have been identified in a number of different plasmid backgrounds (101, 104, 105). In addition, an 11-kb plasmid carrying two copies of blaOXA-72, belonging to the blaOXA-40-like group, has been found in a number of isolates from Lithuania (106), with a likely role for the XerC/XerD recombinases in duplicating the gene.
ISCRs
ISCRs (CR stands for common region) are an unusual type of insertion sequence, often found in class 1 integrons, that lack inverted repeats and transpose through rolling-circle replication (107). Some of the ESBL OXA-type enzymes have been found in association with ISCR elements (108–112). While ISCR elements contain promoters that can be used for downstream gene expression, this does not appear to be a common role that they play in relation to the ESBL blaOXA genes. The blaOXA-18 gene appears to have been mobilized by two ISCR19 elements to insert into an aac(6′)-Ib, where the promoter in the 5′ conserved sequence can be used for blaOXA-18 expression (108). Similarly, blaOXA-45 is bracketed by two ISCR5 elements but utilizes a promoter located in a fragment of an IS1247 element located just upstream (109). ISCR elements in association with ESBL OXA-type β-lactamases have not been reported very frequently, and this may be due to a combination of these enzymes not being so frequently identified and researches overlooking the presence of ISCR elements when they do identify an OXA-type ESBL.
Plasmids
Most of the groups of OXA-type β-lactamases have been identified on plasmids, with the exception of the more recently identified groups of OXAs intrinsic to some Acinetobacter species (Table 1). In A. baumannii, these plasmids are many and varied in size and genetic content but can be broadly grouped based upon their replicase gene sequences (113). Table 5 summarizes the data from all studies to date that have analyzed blaOXA gene content and identified plasmids based upon this typing scheme. While there are a number of different plasmid groups on which genes for the acquired OXA-type β-lactamases of the OXA-23-like, OXA-40-like, and OXA-58-like groups can be located, their genes are almost always found in association with a group 6 plasmid, either by being carried directly on it or by being located in an isolate that also carries a group 6 plasmid (114). It has been suggested that the plasmids belonging to group 6 are self-transmissible and that they may represent a system for general plasmid mobilization and conjugation in A. baumannii (113, 114).
TABLE 5.
OXA-type β-lactamases and plasmid groupsa carried by A. baumannii isolates.
| β–Lactamase | Plasmid rep gene group(s) | Reference(s) |
|---|---|---|
| OXA-23 | 1, 6 | 113, 114, 165, 202 |
| OXA-40 | 2, 6, 12 | 105, 106, 113, 114 |
| OXA-72 | 2 | 203 |
| OXA-58 | 1, 2, 3, 4, 6, 8, 10, 12, 14 | 113, 114, 203 |
| OXA-235 | 2, 6 | 43 |
As defined by Bertini et al. (113).
In contrast to the widespread nature of the Acinetobacter blaOXA genes on many different plasmids, the blaOXA-48 gene appears to be located on a single plasmid type (115). The plasmid, which is of the IncL/M type, has a broad host range among the Enterobacteriaceae and was shown to transfer between species at quite a high frequency of 3.3 × 10−5 transconjugants per recipient. This plasmid carrying blaOXA-48 has been identified in several species and has been circulating for at least a decade, indicating that the spread of this plasmid is largely responsible for the spread of the blaOXA-48 gene (115, 116).
EPIDEMIOLOGY AND CLONALITY
Geographical Dissemination
Resistance to the carbapenems has increased globally in the last decade, and this is caused in part by the spread of OXA-type β-lactamases. Data from the United States show that the percentage of A. baumannii isolates resistant to imipenem has increased from an average of ∼10% between 1999 and 2005 to 48% in 2008, and meropenem resistance has increased from ∼19% to 57.4% over the same time period (117). Similarly, a study analyzing 274 A. baumannii isolates obtained between May 2008 and June 2009 from 16 countries, 14 of which were European countries, found that 47.1% and 45.2% of isolates were resistant to imipenem and meropenem, respectively (118). While the general trend is for an increase in the percentage of resistant isolates, the different blaOXA genes that are partly responsible for this are not uniformly disseminated globally.
The first blaOXA-23 gene was identified in Europe (37). However, the second and third genes within this group were identified in Singapore (34) and China (NCBI accession number AY288523) (unpublished data), giving an early indication that this group of β-lactamases was widespread. Subsequent studies have identified isolates carrying blaOXA-23-like genes from all over the world (40, 119–123). Of all of the acquired OXA-type β-lactamases, those of the blaOXA-23-like group currently appear to be the most widespread and are particularly prevalent in Africa (123), South America (120, 124, 125), and East and Southeast Asia (53, 126). While not as prevalent as the blaOXA-23-like genes, the blaOXA-58-like genes are also globally distributed (53, 126–129). However, they are detected at very high frequencies in southern European Mediterranean countries such as Italy (130), Greece (131), and Turkey (132). Until recently, the blaOXA-40-like genes appeared to be restricted largely to the Iberian Peninsula, where they have been the predominant acquired OXA-type β-lactamase (133–135). However, reports have identified blaOXA-40-like genes in more distant locations, such as the United States (136) and Asia (31), and in particular, there are increasingly reports of these enzymes being identified on plasmids in Taiwan (137, 138). It may be that a more recent transfer onto plasmids by this enzyme group has limited its spread to date. In contrast, the blaOXA-143-like enzymes, which are closely related to the blaOXA-40-like enzymes, have been identified only in Brazil (58, 61, 125). However, this apparent discrepancy may be explained by the close cultural connections between South America, Portugal, and Spain. The first blaOXA-48 gene was detected in an isolate from Turkey (62), and blaOXA-48-like genes appear to have spread within Turkey (139), India (140), and some North African countries (141, 142) but have also been identified in European countries and Russia (see reference 143 for a comprehensive list). Until very recently, the blaOXA-48-like genes seemed not to have spread to America or Southeast Asia. However, the first reports of blaOXA-48 genes in the United States (144) and Japan (145) were published in 2013, and it seems likely that this group of enzymes will successfully spread globally in the near future.
Association of blaOXA Genes with Global Lineages
With the rapid improvements to DNA sequencing technology and the associated decreasing cost, it is becoming more feasible to study epidemiology through whole-genome sequencing and comparative genomics. While this approach has not yet become standard practice due to the cost still remaining relatively high compared to the costs of other typing methods, a few studies have taken a comparative genomics approach, which has shed light upon the different antibiotic resistance phenotypes of the bacteria by identifying blaOXA genes and the supporting genetic elements that they carry (84, 146–149). The largest of these studies, analyzing 136 Acinetobacter genomes, identified the presence of blaOXA genes and ISAba1 in a number of different Acinetobacter species and spread across a number of different evolutionary lineages with A. baumannii (84). As more genome sequences become available, more in-depth analyses associating specific resistance genes with different genetic backgrounds will be possible.
The more common methods currently used for analyzing the underlying genetic differences among isolates are multilocus sequence typing (MLST) and multilocus sequences analysis (MLSA). Typically, these methods involve sequencing of fragments of seven housekeeping genes from each isolate and then either assigning the identified alleles a number, resulting in each isolate receiving a seven-number code known as its sequence type (ST), as is the case with MLST, or performing direct analyses on the nucleotide sequences obtained from the seven genes, as is the case with MLSA. These approaches are useful as they identify the underlying genetic relatedness between isolates and can be used to compare isolates obtained from any location and at any time point. Two MLST schemes exist for A. baumannii, the Oxford scheme (150) and the Pasteur scheme (151). It should be noted that the gpi gene included in the Oxford scheme is not always present (152; B. A. Evans, unpublished data). This may be due to its genetic location within the capsule locus in A. baumannii (153), a region of the genome that is likely to be under selection, which may result in deletions and recombination. This is supported by evidence that the gpi locus as well as the gyrB locus (another gene used in the Oxford scheme), which is located next to the capsule locus, have undergone horizontal gene transfer (154). A recent analysis of the data available in the databases for both A. baumannii MLST schemes found that nearly 60% of 496 isolates belong to one of 26 clones (isolates belonging to the same clonal complex and found in more than one country), 18 of which were found on more than one continent (155). The largest of these clones was represented by clonal complex 92 (CC92) (Oxford scheme)/CC2 (Pasteur scheme). This corresponded to the previously identified international clone II (156). In terms of acquired OXA-type β-lactamases, isolates from this group were found to carry blaOXA-23-like, blaOXA-40-like, and blaOXA-58-like genes. In contrast, the second largest clone, represented by CC109 (Oxford scheme)/CC1 (Pasteur scheme), corresponding to international clone I (156), was found to carry only blaOXA-23-like and blaOXA-58-like genes, while isolates from CC187 (Oxford scheme)/CC3 (Pasteur scheme), corresponding to international clone III (157), were found to encode only blaOXA-58-like genes. Other identified clones also carried specific combinations of acquired blaOXA genes, but CC92/CC2 was the only clone that appeared to harbor all three major enzyme groups. This may partly explain the success of this widespread clonal group. The A. baumannii clones identified also largely correspond with blaOXA-51-like gene clustering (158), as might be expected, as these genes are naturally found on the A. baumannii chromosome. However, for a few isolates, there was a mismatch between the clonal complex to which they belonged and the blaOXA-51-like gene that they carried. This mismatch, combined with the fact that blaOXA-51-like genes have been identified on plasmids and in other bacterial species (88, 159–162), demonstrates that these genes can be horizontally transferred and, as such, are not a reliable method for identifying the underlying bacterial genotype. Another typing method that delineated A. baumannii isolates into sequence groups (SGs) also utilizes the blaOXA-51-like genes, casting doubt on its reliability for determining the underlying isolate genotype (163).
Unlike the Acinetobacter OXA-type β-lactamases, genes coding for the OXA-48 enzyme have spread much more rapidly to far more bacterial species and genera (Table 1). Therefore, for the blaOXA-48-like enzymes, it seems as if it is the spread of plasmids rather than the spread of clonal lineages that is responsible for the spread of the gene (115, 116). However, the spread of a clone carrying the blaOXA-48 gene has been identified (164), and it remains to be seen whether more such clones will emerge in the near future, particularly if more structures that reduce the mobility of the gene, such as that surrounding blaOXA-232, are identified (69). Ultimately, whether the prevalence of certain blaOXA genes is due to their hitchhiking with a particularly successful clonal lineage or whether the success of certain clonal lineages is due to the blaOXA genes that they possess is not clear.
High-Resolution Typing
One of the most common methods used for fine-scale typing of bacterial isolates is pulsed-field gel electrophoresis (PFGE). Unlike MLST, PFGE can identify small differences between isolates within a clonal outbreak, and as such, it is very useful for local epidemiological purposes. While PFGE data are often consistent with MLST data (154), it is not unusual for isolates to have the same MLST sequence type but to produce different PFGE patterns (152, 165, 166). Another commonly used method is repetitive sequence-based PCR (rep-PCR) using the DiversiLab system (52), and the discriminative power of this system is similar to that of PFGE. The use of these techniques and comparing them with blaOXA gene content illustrated the degree of mobility that the blaOXA genes have, where isolates with the same PFGE or rep-PCR profile vary in their blaOXA gene content (137, 138, 167) or isolates with different profiles can have the same blaOXA gene content (152, 167, 168). The use of such techniques with high discriminatory power is useful in outbreak situations, where, when used in conjunction with blaOXA gene screening, they can be used to detect small changes in local epidemiology, such as the acquisition of a resistance gene or conversion between one allele of a resistance gene and another (55, 169).
FUTURE CONCERNS
Until the discovery of the OXA-23 β-lactamase, the OXA enzymes were a relatively minor group of plasmid-encoded β-lactamases, which were active predominantly against the penicillins. The introduction of the carbapenems and the rise of Acinetobacter baumannii has, for the OXA enzymes, opened “Pandora's box.” There has been an explosion of new OXA enzymes, many closely related to each other. For the first time, the chromosomal origins of the OXA enzymes in both A. baumannii and other Acinetobacter species became evident. The migration of these genes onto transposons has allowed them to become the predominant mechanism of resistance to carbapenems in A. baumannii and a major contributor of resistance in the Enterobacteriaceae. This, of course, was unpredictable and could not be expected. Now that we are aware of the diversity and potential of these enzymes, we must try and determine some strategies to control them, particularly in the absence of sufficient novel anti-Gram-negative antibiotics. The πN/πS ratio for each group of OXA enzymes indicates positive selection and signifies that these enzymes are likely currently still evolving. This presumably is a result of our use of carbapenems. We have virtually no understanding of how individual carbapenems have influenced the evolution of individual OXA enzyme groups, and this is a massive gap in our knowledge, as it would enable us to alter therapy in an attempt to curb the emergence of new variants. We do, however, have evidence to suggest that some drugs that are not supposed to select these enzymes in Acinetobacter baumannii, such as ertapenem, can do so in laboratory experiments and possibly may do so in the clinic (170, 171). Carbapenems that do not kill A. baumannii, whether or not they contain an active OXA carbapenemase, could provide very powerful selective environments for the spread and evolution of these genes. Not all carbapenems are the same, and until we know the impact of individual carbapenems on the population, it will be impossible to stem the evolution of these genes.
So what of the future? The ability of the OXA β-lactamases to confer carbapenem resistance has already had a huge impact on the ability to treat Gram-negative infections. The current situation suggests that the problem caused by OXA carbapenemases is only set to increase. It is unlikely that the introduction of new carbapenems alone, especially in the absence of data on their impact on current enzymes, will be able to curb this tide. It is possible that an OXA β-lactamase inhibitor administered with new or even current carbapenems could overcome strains with these resistance genes. However, the huge diversity of blaOXA genes that have already evolved would suggest that some blaOXA genes may already be resistant to new inhibitors or may rapidly evolve to be so.
In A. baumannii, the diversity of genes is so great that it is unlikely that the OXA carbapenemases will ever be controlled, as the situation with these genes is now very similar to that of the ESBLs in the Enterobacteriaceae. However, infections caused by Acinetobacter baumannii are generally limited to severely ill, immunocompromised, nosocomial patients. The emergence of OXA carbapenemases in the Enterobacteriaceae, particularly Klebsiella spp., is of major significance because these bacteria are true pathogens that are able to infect immunocompetent individuals. So far, a relatively small number of OXA carbapenemases have emerged in the Enterobacteriaceae, and their numbers should, and perhaps could, be reduced or eradicated. This would prolong the active life of the carbapenems, but very careful management of carbapenems would be needed to achieve this. Failure runs the risk that the diversity of the current OXA carbapenemases will increase so that no new drugs directed toward OXA β-lactamases would be effective. As carbapenems are often considered the last resort for treatment that does not involve the much more toxic antibiotic colistin, preservation of carbapenem efficacy is essential for continued clinical success against severe Gram-negative infections.
Biographies

Benjamin A. Evans is a Senior Lecturer in Microbiology at Anglia Ruskin University, Cambridge, United Kingdom. He received his B.Sc. Honors in Microbiology and Infection from the University of Edinburgh in 2005, followed by a Ph.D. in Medical Microbiology from the same Institute in 2009. Following postdoctoral positions at the University of Manchester from 2009 to 2011 and at the University of Liverpool from 2011 to 2012, he joined Anglia Ruskin University as a Lecturer in 2012, followed by appointment as Senior Lecturer in 2013. His research spans three main areas: antibiotic resistance mechanisms and epidemiology, the evolution and impact of natural transformation in bacteria, and the use of next-generation sequencing approaches to study bacterial population diversity. In the seven years since starting his Ph.D. in 2006, he has published 15 journal articles and presented 9 conference papers.

Sebastian G. B. Amyes obtained his B.Sc., Ph.D., and D.Sc. from University College London. He became Assistant Professor at the Medical School of the University of Edinburgh in 1977, Associate Professor in 1988, and full Professor of Microbial Chemotherapy in 1992. He won the annual Young Scientist prizes of the Royal Pharmaceutical Society in 1985 and of the Pathological Society in 1988. He was awarded the honorary degree of Doctor honoris causa from the Semmelweis Medical University in Budapest in 2004. His recent research has concentrated on carbapenem resistance in Acinetobacter baumannii and the spread of extended-spectrum beta-lactamases in the Enterobacteriaceae. He was the first to identify OXA carbapenemases in A. baumannii. He has obtained 60 research grants, published more than 510 papers and major conference communications, and supervised 51 Ph.D. students. He has been an editor of five journals and a member of government committees relating to antibiotic resistance.
REFERENCES
- 1.Ambler RP. 1980. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289:321–331. 10.1098/rstb.1980.0049 [DOI] [PubMed] [Google Scholar]
- 2.Gutkind GO, Di Conza J, Power P, Radice M. 2013. β-Lactamase-mediated resistance: a biochemical, epidemiological and genetic overview. Curr. Pharm. Des. 19:164–208. 10.2174/138161213804070320 [DOI] [PubMed] [Google Scholar]
- 3.Bush K, Jacoby GA, Medeiros AA. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211–1233. 10.1128/AAC.39.6.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedges RW, Datta N, Kontomichalou P, Smith JT. 1974. Molecular specificities of R factor-determined β-lactamases: correlation with plasmid compatibility. J. Bacteriol. 117:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta N, Kontomichalou P. 1965. Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208:239–241. 10.1038/208239a0 [DOI] [PubMed] [Google Scholar]
- 6.Meynell E, Datta N. 1966. The relation of resistance transfer factors to the F-factor (sex-factor) of Escherichia coli K12. Genet. Res. 7:134–140. 10.1017/S0016672300009538 [DOI] [PubMed] [Google Scholar]
- 7.Sykes RB, Matthew M. 1976. The β-lactamases of gram-negative bacteria and their role in resistance to β-lactam antibiotics. J. Antimicrob. Chemother. 2:115–157. 10.1093/jac/2.2.115 [DOI] [PubMed] [Google Scholar]
- 8.Dale JW, Smith JT. 1974. R-factor-mediated β-lactamases that hydrolyze oxacillin: evidence for two distinct groups. J. Bacteriol. 119:351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutcliffe JG. 1978. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc. Natl. Acad. Sci. U. S. A. 75:3737–3741. 10.1073/pnas.75.8.3737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dale JW, Godwin D, Mossakowska D, Stephenson P, Wall S. 1985. Sequence of the OXA2 β-lactamase: comparison with other penicillin-reactive enzymes. FEBS Lett. 191:39–44. 10.1016/0014-5793(85)80989-3 [DOI] [PubMed] [Google Scholar]
- 11.Ouellette M, Bissonnette L, Roy PH. 1987. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 beta-lactamase gene. Proc. Natl. Acad. Sci. U. S. A. 84:7378–7382. 10.1073/pnas.84.21.7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacoby GA. 1974. Properties of R plasmids determining gentamicin resistance by acetylation in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 6:239–252. 10.1128/AAC.6.3.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medeiros AA, Hedges RW, Jacoby GA. 1982. Spread of a “Pseudomonas-specific” beta-lactamase to plasmids of enterobacteria. J. Bacteriol. 149:700–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedges RW, Matthew M. 1979. Acquisition by Escherichia coli of plasmid-borne β-lactamases normally confined to Pseudomonas spp. Plasmid 2:269–278. 10.1016/0147-619X(79)90045-3 [DOI] [PubMed] [Google Scholar]
- 15.Philippon AM, Paul GC, Jacoby GA. 1983. Properties of PSE-2 β-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 24:362–369. 10.1128/AAC.24.3.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couture F, Lachapelle J, Levesque RC. 1992. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol. Microbiol. 6:1693–1705. 10.1111/j.1365-2958.1992.tb00894.x [DOI] [PubMed] [Google Scholar]
- 17.Hall LM, Livermore DM, Gur D, Akova M, Akalin HE. 1993. OXA-11, an extended-spectrum variant of OXA-10 (PSE-2) β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 37:1637–1644. 10.1128/AAC.37.8.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657–686. 10.1128/CMR.18.4.657-686.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danel F, Hall LM, Gur D, Livermore DM. 1995. OXA-14, another extended-spectrum variant of OXA-10 (PSE-2) beta-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1881–1884. 10.1128/AAC.39.8.1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Danel F, Hall LM, Gur D, Livermore DM. 1998. OXA-16, a further extended-spectrum variant of OXA-10 beta-lactamase, from two Pseudomonas aeruginosa isolates. Antimicrob. Agents Chemother. 42:3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danel F, Hall LM, Duke B, Gur D, Livermore DM. 1999. OXA-17, a further extended-spectrum variant of OXA-10 beta-lactamase, isolated from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:1362–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mugnier P, Casin I, Bouthors AT, Collatz E. 1998. Novel OXA-10-derived extended-spectrum beta-lactamases selected in vivo or in vitro. Antimicrob. Agents Chemother. 42:3113–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poirel L, Girlich D, Naas T, Nordmann P. 2001. OXA-28, an extended-spectrum variant of OXA-10 beta-lactamase from Pseudomonas aeruginosa and its plasmid- and integron-located gene. Antimicrob. Agents Chemother. 45:447–453. 10.1128/AAC.45.2.447-453.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danel F, Paetzel M, Strynadka NC, Page MG. 2001. Effect of divalent metal cations on the dimerization of OXA-10 and -14 class D β-lactamases from Pseudomonas aeruginosa. Biochemistry 40:9412–9420. 10.1021/bi0025969 [DOI] [PubMed] [Google Scholar]
- 25.Danel F, Hall LM, Gur D, Livermore DM. 1997. OXA-15, an extended-spectrum variant of OXA-2 beta-lactamase, isolated from a Pseudomonas aeruginosa strain. Antimicrob. Agents Chemother. 41:785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vila J, Navia M, Ruiz J, Casals C. 1997. Cloning and nucleotide sequence analysis of a gene encoding an OXA-derived beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 41:2757–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon JA. 1985. Imipenem/cilastatin: the first carbapenem antibiotic. Drug Intell. Clin. Pharm. 19:895–899 [PubMed] [Google Scholar]
- 28.Scaife W, Young HK, Paton RH, Amyes SG. 1995. Transferable imipenem-resistance in Acinetobacter species from a clinical source. J. Antimicrob. Chemother. 36:585–586. 10.1093/jac/36.3.585 [DOI] [PubMed] [Google Scholar]
- 29.Donald HM, Scaife W, Amyes SGB, Young HK. 2000. Sequence analysis of ARI-1, a novel OXA β-lactamase, responsible for imipenem resistance in Acinetobacter baumannii 6B92. Antimicrob. Agents Chemother. 44:196–199. 10.1128/AAC.44.1.196-199.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252–1256. 10.1128/AAC.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendes RE, Bell JM, Turnidge JD, Castanheira M, Jones RN. 2009. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63:55–59. 10.1093/jac/dkn434 [DOI] [PubMed] [Google Scholar]
- 32.Boo TW, Crowley B. 2009. Detection of blaOXA-58 and blaOXA-23-like genes in carbapenem-susceptible Acinetobacter clinical isolates: should we be concerned? J. Med. Microbiol. 58:839–841. 10.1099/jmm.0.008904-0 [DOI] [PubMed] [Google Scholar]
- 33.Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JM, Raoult D, Medigue C, Weissenbach J, Cruveiller S. 2008. Comparative analysis of acinetobacters: three genomes for three lifestyles. PLoS One 3:e1805. 10.1371/journal.pone.0001805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afzal-Shah M, Woodford N, Livermore DM. 2001. Characterization of OXA-25, OXA-26, and OXA-27, molecular class D β-lactamases associated with carbapenem resistance in clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 45:583–588. 10.1128/AAC.45.2.583-588.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaitany KC, Klinger NV, June CM, Ramey ME, Bonomo RA, Powers RA, Leonard DA. 2013. Structures of the class D carbapenemases OXA-23 and OXA-146: mechanistic basis of activity against carbapenems, extended-spectrum cephalosporins and aztreonam. Antimicrob. Agents Chemother. 57:4848–4855. 10.1128/AAC.00762-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CA, Antunes NT, Stewart NK, Toth M, Kumarasiri M, Chang M, Mobashery S, Vakulenko SB. 2013. Structural basis for carbapenemase activity of the OXA-23 β-lactamase from Acinetobacter baumannii. Chem. Biol. 20:1107–1115. 10.1016/j.chembiol.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paton R, Miles RS, Hood J, Amyes SGB. 1993. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2:81–88. 10.1016/0924-8579(93)90045-7 [DOI] [PubMed] [Google Scholar]
- 38.Santillana E, Beceiro A, Bou G, Romero A. 2007. Crystal structure of the carbapenemase OXA-24 reveals insights into the mechanism of carbapenem hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 104:5354–5359. 10.1073/pnas.0607557104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider KD, Ortega CJ, Renck NA, Bonomo RA, Powers RA, Leonard DA. 2011. Structures of the class D carbapenemase OXA-24 from Acinetobacter baumannii in complex with doripenem. J. Mol. Biol. 406:583–594. 10.1016/j.jmb.2010.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heritier C, Poirel L, Lambert T, Nordmann P. 2005. Contribution of acquired carbapenem-hydrolyzing oxacillinases to carbapenem resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:3198–3202. 10.1128/AAC.49.8.3198-3202.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bou G, Oliver A, Martinez-Beltran J. 2000. OXA-24, a novel class D β-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44:1556–1561. 10.1128/AAC.44.6.1556-1561.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heritier C, Poirel L, Aubert D, Nordmann P. 2003. Genetic and functional analysis of the chromosome-encoded carbapenem-hydrolyzing oxacillinase OXA-40 of Acinetobacter baumannii. Antimicrob. Agents Chemother. 47:268–273. 10.1128/AAC.47.1.268-273.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higgins PG, Perez-Llarena FJ, Zander E, Fernandez A, Bou G, Seifert H. 2013. OXA-235, a novel class D beta-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 57:2121–2126. 10.1128/AAC.02413-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bou G, Santillana E, Sheri A, Beceiro A, Sampson JM, Kalp M, Bethel CR, Distler AM, Drawz SM, Pagadala SRR, van den Akker F, Bonomo RA, Romero A, Buynak JD. 2010. Design, synthesis, and crystal structures of 6-alkylidene-2′-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase. J. Am. Chem. Soc. 132:13320–13331. 10.1021/ja104092z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brown S, Young HK, Amyes SGB. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15–23. 10.1111/j.1469-0691.2004.01016.x [DOI] [PubMed] [Google Scholar]
- 46.Heritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174–4179. 10.1128/AAC.49.10.4174-4179.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tiwari V, Nagpal I, Subbarao N, Moganty RR. 2012. In-silico modeling of a novel OXA-51 from β-lactam-resistant Acinetobacter baumannii and its interaction with various antibiotics. J. Mol. Model. 18:3351–3361. 10.1007/s00894-011-1346-3 [DOI] [PubMed] [Google Scholar]
- 48.Tiwari V, Moganty RR. 24 July 2013, posting date Conformational stability of OXA-51 β-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J. Biomol. Struct. Dyn. [Epub ahead of print.] 10.1080/07391102.2013.819789 [DOI] [PubMed] [Google Scholar]
- 49.Verma V, Testero SA, Amini K, Wei W, Liu J, Balachandran N, Monoharan T, Stynes S, Kotra LP, Golemi-Kotra D. 2011. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter baumannii. J. Biol. Chem. 286:37292–37303. 10.1074/jbc.M111.280115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pernot L, Frenois F, Rybkine T, L'Hermite G, Petrella S, Delettre J, Jarlier V, Collatz E, Sougakoff W. 2001. Crystal structures of the class D β-lactamase OXA-13 in the native form and in complex with meropenem. J. Mol. Biol. 310:859–874. 10.1006/jmbi.2001.4805 [DOI] [PubMed] [Google Scholar]
- 51.Poirel L, Marque S, Heritier C, Segonds C, Chabanon G, Nordmann P. 2005. OXA-58, a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:202–208. 10.1128/AAC.49.1.202-208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higgins P, Dammhayn C, Hackel M, Seifert H. 2010. Global spread of carbapenem-resistant Acinetobacter baumannii. J. Antimicrob. Chemother. 65:233–238. 10.1093/jac/dkp428 [DOI] [PubMed] [Google Scholar]
- 53.Koh TH, Sng LH, Wang GC, Hsu LY, Zhao Y. 2007. IMP-4 and OXA β-lactamases in Acinetobacter baumannii from Singapore. J. Antimicrob. Chemother. 59:627–632. 10.1093/jac/dkl544 [DOI] [PubMed] [Google Scholar]
- 54.Poirel L, Mansour W, Bouallegue O, Nordmann P. 2008. Carbapenem-resistant Acinetobacter baumannii isolates from Tunisia producing the OXA-58-like carbapenem-hydrolyzing oxacillinase OXA-97. Antimicrob. Agents Chemother. 52:1613–1617. 10.1128/AAC.00978-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Higgins PG, Schneiders T, Hamprecht A, Seifert H. 2010. In vivo selection of a missense mutation in adeR and conversion of the novel blaOXA-164 gene into blaOXA-58 in carbapenem-resistant Acinetobacter baumannii isolates from a hospitalized patient. Antimicrob. Agents Chemother. 54:5021–5027. 10.1128/AAC.00598-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Figueiredo S, Poirel L, Seifert H, Mugnier P, Benhamou D, Nordmann P. 2010. OXA-134, a naturally occurring carbapenem-hydrolyzing class D beta-lactamase from Acinetobacter lwoffii. Antimicrob. Agents Chemother. 54:5372–5375. 10.1128/AAC.00629-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turton JF, Hyde R, Martin K, Shah J. 2012. Genes encoding OXA-134-like enzymes are found in Acinetobacter lwoffii and A. schindleri and can be used for identification. J. Clin. Microbiol. 50:1019–1022. 10.1128/JCM.06173-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins PG, Poirel L, Lehmann M, Nordmann P, Seifert H. 2009. OXA-143, a novel carbapenem-hydrolyzing class D beta-lactamase in Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:5035–5038. 10.1128/AAC.00856-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim CK, Lee Y, Lee H, Woo GJ, Song W, Kim MN, Lee WG, Jeong SH, Lee K, Chong Y. 2010. Prevalence and diversity of carbapenemases among imipenem-nonsusceptible Acinetobacter isolates in Korea: emergence of a novel OXA-182. Diagn. Microbiol. Infect. Dis. 68:432–438. 10.1016/j.diagmicrobio.2010.07.014 [DOI] [PubMed] [Google Scholar]
- 60.Gionco B, Pelayo JS, Venancio EJ, Cayo R, Gales AC, Carrara-Marroni FE. 2012. Detection of OXA-231, a new variant of blaOXA-143, in Acinetobacter baumannii from Brazil: a case report. J. Antimicrob. Chemother. 67:2531–2532. 10.1093/jac/dks223 [DOI] [PubMed] [Google Scholar]
- 61.Mostachio AK, Levin AS, Rizek C, Rossi F, Zerbini J, Costa SF. 2012. High prevalence of OXA-143 and alteration of outer membrane proteins in carbapenem-resistant Acinetobacter spp. isolates in Brazil. Int. J. Antimicrob. Agents 39:396–401. 10.1016/j.ijantimicag.2012.01.021 [DOI] [PubMed] [Google Scholar]
- 62.Poirel L, Heritier C, Tolun V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goncalves D, Cecilio P, Ferreira H. 2013. First detection of OXA-48-like producing Acinetobacter baumannii in the faecal flora of nursing home residents in Northern Portugal, abstr eP748 Abstr. 23rd Eur. Congr. Clin. Microbiol. Infect. Dis., Berlin, Germany [Google Scholar]
- 64.Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. 2011. OXA-163, an OXA-48-related class D beta-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 55:2546–2551. 10.1128/AAC.00022-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oteo J, Hernandez JM, Espasa M, Fleites A, Saez D, Bautista V, Perez-Vazquez M, Fernandez-Garcia MD, Delgado-Iribarren A, Sanchez-Romero I, Garcia-Picazo L, Miguel MD, Solis S, Aznar E, Trujillo G, Mediavilla C, Fontanals D, Rojo S, Vindel A, Campos J. 2013. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 68:317–321. 10.1093/jac/dks383 [DOI] [PubMed] [Google Scholar]
- 66.Poirel L, Heritier C, Nordmann P. 2004. Chromosome-encoded Ambler class D beta-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob. Agents Chemother. 48:348–351. 10.1128/AAC.48.1.348-351.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Potron A, Poirel L, Nordmann P. 2011. Origin of OXA-181, an emerging carbapenem-hydrolyzing oxacillinase, as a chromosomal gene in Shewanella xiamenensis. Antimicrob. Agents Chemother. 55:4405–4407. 10.1128/AAC.00681-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zong Z. 2012. Discovery of blaOXA-199, a chromosome-based blaOXA-48-like variant, in Shewanella xiamenensis. PLoS One 7:e48280. 10.1371/journal.pone.0048280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, Nordmann P. 2013. Genetic and biochemical characterisation of OXA-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int. J. Antimicrob. Agents 41:325–329. 10.1016/j.ijantimicag.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 70.Docquier JD, Calderone V, De Luca F, Benvenuti M, Giuliani F, Bellucci L, Tafi A, Nordmann P, Botta M, Rossolini GM, Mangani S. 2009. Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases. Chem. Biol. 16:540–547. 10.1016/j.chembiol.2009.04.010 [DOI] [PubMed] [Google Scholar]
- 71.Figueiredo S, Bonnin RA, Poirel L, Duranteau J, Nordmann P. 2012. Identification of the naturally occurring genes encoding carbapenem-hydrolysing oxacillinases from Acinetobacter haemolyticus, Acinetobacter johnsonii, and Acinetobacter calcoaceticus. Clin. Microbiol. Infect. 18:907–913. 10.1111/j.1469-0691.2011.03708.x [DOI] [PubMed] [Google Scholar]
- 72.Bonnin RA, Ocampo-Sosa AA, Poirel L, Guet-Revillet H, Nordmann P. 2012. Biochemical and genetic characterization of carbapenem-hydrolyzing beta-lactamase OXA-229 from Acinetobacter bereziniae. Antimicrob. Agents Chemother. 56:3923–3927. 10.1128/AAC.00257-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corvec S, Caroff N, Espaze E, Giraudeau C, Drugeon H, Reynaud A. 2003. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 52:629–635. 10.1093/jac/dkg407 [DOI] [PubMed] [Google Scholar]
- 74.Segal H, Thomas R, Elisha BG. 2003. Characterization of class 1 integron resistance gene cassettes and the identification of a novel IS-like element in Acinetobacter baumannii. Plasmid 49:169–178. 10.1016/S0147-619X(03)00011-8 [DOI] [PubMed] [Google Scholar]
- 75.Heritier C, Poirel L, Nordmann P. 2006. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 12:123–130. 10.1111/j.1469-0691.2005.01320.x [DOI] [PubMed] [Google Scholar]
- 76.Mugnier PD, Poirel L, Nordmann P. 2009. Functional analysis of insertion sequence ISAba1, responsible for genomic plasticity of Acinetobacter baumannii. J. Bacteriol. 191:2414–2418. 10.1128/JB.01258-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopes BS, Hamouda A, Findlay J, Amyes SGB. 2011. Effect of frameshift mutagen acriflavine on control of resistance genes in Acinetobacter baumannii. J. Med. Microbiol. 60:211–215. 10.1099/jmm.0.025544-0 [DOI] [PubMed] [Google Scholar]
- 78.Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, Pitt TL. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72–77. 10.1111/j.1574-6968.2006.00195.x [DOI] [PubMed] [Google Scholar]
- 79.Poirel L, Nordmann P. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442–1448. 10.1128/AAC.50.4.1442-1448.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Segal H, Jacobson RK, Garny S, Bamford CM, Elisha BG. 2007. Extended −10 promoter in ISAba-1 upstream of blaOXA-23 from Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3040–3041. 10.1128/AAC.00594-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corvec S, Poirel L, Naas T, Drugeon H, Nordmann P. 2007. Genetics and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-23 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:1530–1533. 10.1128/AAC.01132-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Figueiredo S, Poirel L, Croize J, Recule C, Nordmann P. 2009. In vivo selection of reduced susceptibility to carbapenems in Acinetobacter baumannii related to ISAba1-mediated overexpression of the natural blaOXA-66 oxacillinase gene. Antimicrob. Agents Chemother. 53:2657–2659. 10.1128/AAC.01663-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Evans BA, Hamouda A, Amyes SG. 2013. The rise of carbapenem-resistant Acinetobacter baumannii. Curr. Pharm. Des. 19:223–238. 10.2174/138161213804070285 [DOI] [PubMed] [Google Scholar]
- 84.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tsakris A, Ikonomidis A, Poulou A, Spanakis N, Vrizas D, Diomidous M, Pournaras S, Markou F. 2008. Clusters of imipenem-resistant Acinetobacter baumannii clones producing different carbapenemases in an intensive care unit. Clin. Microbiol. Infect. 14:588–594. 10.1111/j.1469-0691.2008.01996.x [DOI] [PubMed] [Google Scholar]
- 86.Poirel L, Lebessi E, Heritier C, Patsoura A, Foustoukou M, Nordmann P. 2006. Nosocomial spread of OXA-58-positive carbapenem-resistant Acinetobacter baumannii isolates in a paediatric hospital in Greece. Clin. Microbiol. Infect. 12:1138–1141. 10.1111/j.1469-0691.2006.01537.x [DOI] [PubMed] [Google Scholar]
- 87.Zarrilli R, Casillo R, Di Popolo A, Tripodi MF, Bagattini M, Cuccurullo S, Crivaro V, Ragone E, Mattei A, Galdieri N, Triassi M, Utili R. 2007. Molecular epidemiology of a clonal outbreak of multidrug-resistant Acinetobacter baumannii in a university hospital in Italy. Clin. Microbiol. Infect. 13:481–489. 10.1111/j.1469-0691.2006.01675.x [DOI] [PubMed] [Google Scholar]
- 88.Leski TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, Stenger DA, Taitt CR, Vora GJ. 2013. Identification of blaOXA-51-like, blaOXA-58, blaDIM-1, and blaVIM carbapenemase genes in hospital Enterobacteriaceae isolates from Sierra Leone. J. Clin. Microbiol. 51:2435–2438. 10.1128/JCM.00832-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen TL, Wu RCC, Shaio MF, Fung CP, Cho WL. 2008. Acquisition of a plasmid-borne bla(OXA-58) gene with an upstream IS1008 insertion conferring a high level of carbapenem resistance to Acinetobacter baumannii. Antimicrob. Agents Chemother. 52:2573–2580. 10.1128/AAC.00393-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ravasi P, Limansky AS, Rodriguez RE, Viale AM, Mussi MA. 2011. ISAba825, a functional insertion sequence modulating genomic plasticity and blaOXA-58 expression in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:917–920. 10.1128/AAC.00491-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evans BA, Hamouda A, Towner KJ, Amyes SGB. 2010. Novel genetic context of multiple blaOXA-58 genes in Acinetobacter genospecies 3. J. Antimicrob. Chemother. 65:1586–1588. 10.1093/jac/dkq180 [DOI] [PubMed] [Google Scholar]
- 92.Bertini A, Poirel L, Bernabeu S, Fortini D, Villa L, Nordmann P, Carattoli A. 2007. Multicopy blaOXA-58 gene as a source of high-level resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2324–2328. 10.1128/AAC.01502-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Figueiredo S, Poirel L, Papa A, Koulourida V, Nordmann P. 2009. Overexpression of the naturally occurring blaOXA-51 gene in Acinetobacter baumannii mediated by novel insertion sequence ISAba9. Antimicrob. Agents Chemother. 53:4045–4047. 10.1128/AAC.00292-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee Y, Kim C-K, Lee H, Jeong SH, Yong D, Lee K. 2011. A novel insertion sequence, ISAba10, inserted into ISAba1 adjacent to the blaOXA-23 gene and disrupting the outer membrane protein gene carO in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:361–363. 10.1128/AAC.01672-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zander E, Higgins PG, Fernandez-Gonzalez A, Seifert H. 2013. Detection of intrinsic blaOXA-51-like by multiplex PCR on its own is not reliable for the identification of Acinetobacter baumannii. Int. J. Med. Microbiol. 303:88–89. 10.1016/j.ijmm.2012.12.007 [DOI] [PubMed] [Google Scholar]
- 96.Aubert D, Naas T, Heritier C, Poirel L, Nordmann P. 2006. Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J. Bacteriol. 188:6506–6514. 10.1128/JB.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carrer A, Poirel L, Eraksoy H, Cagatay AA, Badur S, Nordmann P. 2008. Spread of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Istanbul, Turkey. Antimicrob. Agents Chemother. 52:2950–2954. 10.1128/AAC.01672-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Giani T, Conte V, Di Pilato V, Aschbacher R, Weber C, Larcher C, Rossolini GM. 2012. Escherichia coli from Italy producing OXA-48 carbapenemase encoded by a novel Tn1999 transposon derivative. Antimicrob. Agents Chemother. 56:2211–2213. 10.1128/AAC.00035-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. 2011. Characterization of OXA-181, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 55:4896–4899. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Potron A, Nordmann P, Poirel L. 2013. Characterization of OXA-204, a carbapenem-hydrolyzing class D beta-lactamase from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 57:633–636. 10.1128/AAC.01034-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Acosta J, Merino M, Viedma E, Poza M, Sanz F, Otero JR, Chaves F, Bou G. 2011. Multidrug-resistant Acinetobacter baumannii harboring OXA-24 carbapenemase, Spain. Emerg. Infect. Dis. 17:1064–1067. 10.3201/eid/1706.091866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D'Andrea MM, Giani T, D'Arezzo S, Capone A, Petrosillo N, Visca P, Luzzaro F, Rossolini GM. 2009. Characterization of pABVA01, a plasmid encoding the OXA-24 carbapenemase from Italian isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 53:3528–3533. 10.1128/AAC.00178-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Summers DK, Sherratt DJ. 1988. Resolution of ColE1 dimers requires a DNA sequence implicated in the three-dimensional organization of the cer site. EMBO J. 7:851–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merino M, Acosta J, Poza M, Sanz F, Beceiro A, Chaves F, Bou G. 2010. OXA-24 carbapenemase gene flanked by XerC/XerD-like recombination sites in different plasmids from different Acinetobacter species isolated during a nosocomial outbreak. Antimicrob. Agents Chemother. 54:2724–2727. 10.1128/AAC.01674-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Grosso F, Quinteira S, Poirel L, Novais A, Peixe L. 2012. Role of common blaOXA-24/OXA-40-carrying platforms and plasmids in the spread of OXA-24/OXA-40 among Acinetobacter species clinical isolates. Antimicrob. Agents Chemother. 56:3969–3972. 10.1128/AAC.06255-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Povilonis J, Seputiene V, Krasauskas R, Juskaite R, Miskinyte M, Suziedelis K, Suziedeliene E. 2013. Spread of carbapenem-resistant Acinetobacter baumannii carrying a plasmid with two genes encoding OXA-72 carbapenemase in Lithuanian hospitals. J. Antimicrob. Chemother. 68:1000–1006. 10.1093/jac/dks499 [DOI] [PubMed] [Google Scholar]
- 107.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Naas T, Namdari F, Bogaerts P, Huang TD, Glupczynski Y, Nordmann P. 2008. Genetic structure associated with blaOXA-18, encoding a clavulanic acid-inhibited extended-spectrum oxacillinase. Antimicrob. Agents Chemother. 52:3898–3904. 10.1128/AAC.00403-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li H, Walsh TR, Toleman MA. 2009. Molecular analysis of the sequences surrounding blaOXA-45 reveals acquisition of this gene by Pseudomonas aeruginosa via a novel ISCR element, ISCR5. Antimicrob. Agents Chemother. 53:1248–1251. 10.1128/AAC.00480-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Villa L, Carattoli A. 2005. Integrons and transposons on the Salmonella enterica serovar Typhimurium virulence plasmid. Antimicrob. Agents Chemother. 49:1194–1197. 10.1128/AAC.49.3.1194-1197.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tosini F, Visca P, Luzzi I, Dionisi AM, Pezzella C, Petrucca A, Carattoli A. 1998. Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob. Agents Chemother. 42:3053–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Navia MM, Ruiz J, Vila J. 2002. Characterization of an integron carrying a new class D beta-lactamase (OXA-37) in Acinetobacter baumannii. Microb. Drug Resist. 8:261–265. 10.1089/10766290260469516 [DOI] [PubMed] [Google Scholar]
- 113.Bertini A, Poirel L, Mugnier PD, Villa L, Nordmann P, Carattoli A. 2010. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:4168–4177. 10.1128/AAC.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Towner KJ, Evans B, Villa L, Levi K, Hamouda A, Amyes SG, Carattoli A. 2011. Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing class D beta-lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55:2154–2159. 10.1128/AAC.01661-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562. 10.1128/AAC.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 18(31):pii=20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549 [DOI] [PubMed] [Google Scholar]
- 117.Rhomberg PR, Jones RN. 2009. Summary trends for the Meropenem Yearly Susceptibility Test Information Collection Program: a 10-year experience in the United States (1999-2008). Diagn. Microbiol. Infect. Dis. 65:414–426. 10.1016/j.diagmicrobio.2009.08.020 [DOI] [PubMed] [Google Scholar]
- 118.Nordmann P, Picazo JJ, Mutters R, Korten V, Quintana A, Laeuffer JM, Seak JC, Flamm RK, Morrissey I. 2011. Comparative activity of carbapenem testing: the COMPACT study. J. Antimicrob. Chemother. 66:1070–1078. 10.1093/jac/dkr056 [DOI] [PubMed] [Google Scholar]
- 119.Coelho JM, Turton JF, Kaufmann ME, Glover J, Woodford N, Warner M, Palepou MF, Pike R, Pitt TL, Patel BC, Livermore DM. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and southeast England. J. Clin. Microbiol. 44:3623–3627. 10.1128/JCM.00699-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Villegas MV, Kattan JN, Correa A, Lolans K, Guzman AM, Woodford N, Livermore D, Quinn JP. 2007. Dissemination of Acinetobacter baumannii clones with OXA-23 carbapenemase in Colombian hospitals. Antimicrob. Agents Chemother. 51:2001–2004. 10.1128/AAC.00226-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou H, Yang Q, Yu YS, Wei ZQ, Li LJ. 2007. Clonal spread of imipenem-resistant Acinetobacter baumannii among different cities of China. J. Clin. Microbiol. 45:4054–4057. 10.1128/JCM.00343-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Evans BA, Hamouda A, Abbasi SA, Khan FA, Amyes SGB. 2011. High prevalence of un-related multidrug-resistant Acinetobacter baumannii isolates in Pakistani military hospitals. Int. J. Antimicrob. Agents 37:580–581. 10.1016/j.ijantimicag.2011.01.023 [DOI] [PubMed] [Google Scholar]
- 123.Olaitan AO, Berrazeg M, Fagade OE, Adelowo OO, Alli JA, Rolain JM. 2013. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase, Nigeria. Int. J. Infect. Dis. 17:e469–e470. 10.1016/j.ijid.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 124.Stietz MS, Ramirez MS, Vilacoba E, Merkier AK, Limansky AS, Centron D, Catalano M. 2013. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I-III. Infect. Genet. Evol. 14:294–301. 10.1016/j.meegid.2012.12.020 [DOI] [PubMed] [Google Scholar]
- 125.Climaco EC, Oliveira ML, Pitondo-Silva A, Oliveira MG, Medeiros M, Lincopan N, da Costa Darini AL. 2013. Clonal complexes 104, 109 and 113 playing a major role in the dissemination of OXA-carbapenemase-producing Acinetobacter baumannii in Southeast Brazil. Infect. Genet. Evol. 19:127–133. 10.1016/j.meegid.2013.06.024 [DOI] [PubMed] [Google Scholar]
- 126.Wang H, Guo P, Sun H, Wang H, Yang Q, Chen M, Xu Y, Zhu Y. 2007. Molecular epidemiology of clinical isolates of carbapenem-resistant Acinetobacter spp. from Chinese hospitals. Antimicrob. Agents Chemother. 51:4022–4028. 10.1128/AAC.01259-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Marque S, Poirel L, Heritier C, Brisse S, Blasco MD, Filip R, Coman G, Naas T, Nordmann P. 2005. Regional occurrence of plasmid-mediated carbapenem-hydrolyzing oxacillinase OXA-58 in Acinetobacter spp. in Europe. J. Clin. Microbiol. 43:4885–4888. 10.1128/JCM.43.9.4885-4888.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Coelho J, Woodford N, Afzal-Shah M, Livermore D. 2006. Occurrence of OXA-58-like carbapenemases in Acinetobacter spp. collected over 10 years in three continents. Antimicrob. Agents Chemother. 50:756–758. 10.1128/AAC.50.2.756-758.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peleg AY, Franklin C, Walters LJ, Bell JM, Spelman DW. 2006. OXA-58 and IMP-4 carbapenem-hydrolyzing β-lactamases in an Acinetobacter junii blood culture isolate from Australia. Antimicrob. Agents Chemother. 50:399–400. 10.1128/AAC.50.1.399-400.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Migliavacca R, Espinal P, Principe L, Drago M, Fugazza G, Roca I, Nucleo E, Bracco S, Vila J, Pagani L, Luzzaro F. 2013. Characterization of resistance mechanisms and genetic relatedness of carbapenem-resistant Acinetobacter baumannii isolated from blood, Italy. Diagn. Microbiol. Infect. Dis. 75:180–186. 10.1016/j.diagmicrobio.2012.11.002 [DOI] [PubMed] [Google Scholar]
- 131.Gogou V, Pournaras S, Giannouli M, Voulgari E, Piperaki ET, Zarrilli R, Tsakris A. 2011. Evolution of multidrug-resistant Acinetobacter baumannii clonal lineages: a 10 year study in Greece (2000-09). J. Antimicrob. Chemother. 66:2767–2772. 10.1093/jac/dkr390 [DOI] [PubMed] [Google Scholar]
- 132.Kulah C, Mooij MJ, Comert F, Aktas E, Celebi G, Ozlu N, Rijnsburger MC, Savelkoul PH. 2010. Characterisation of carbapenem-resistant Acinetobacter baumannii outbreak strains producing OXA-58 in Turkey. Int. J. Antimicrob. Agents 36:114–118. 10.1016/j.ijantimicag.2010.03.017 [DOI] [PubMed] [Google Scholar]
- 133.Ruiz M, Marti S, Fernandez-Cuenca F, Pascual A, Vila J. 2007. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin. Microbiol. Infect. 13:1192–1198. 10.1111/j.1469-0691.2007.01825.x [DOI] [PubMed] [Google Scholar]
- 134.Quinteira S, Grosso F, Ramos H, Peixe L. 2007. Molecular epidemiology of imipenem-resistant Acinetobacter haemolyticus and Acinetobacter baumannii isolates carrying plasmid-mediated OXA-40 from a Portuguese hospital. Antimicrob. Agents Chemother. 51:3465–3466. 10.1128/AAC.00267-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Villalon P, Valdezate S, Medina-Pascual MJ, Carrasco G, Vindel A, Saez-Nieto JA. 2013. Epidemiology of the Acinetobacter-derived cephalosporinase, carbapenem-hydrolysing oxacillinase and metallo-beta-lactamase genes, and of common insertion sequences, in epidemic clones of Acinetobacter baumannii from Spain. J. Antimicrob. Chemother. 68:550–553. 10.1093/jac/dks448 [DOI] [PubMed] [Google Scholar]
- 136.Lolans K, Rice TW, Munoz-Price LS, Quinn JP. 2006. Multicity outbreak of carbapenem-resistant Acinetobacter baumannii isolates producing the carbapenemase OXA-40. Antimicrob. Agents Chemother. 50:2941–2945. 10.1128/AAC.00116-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lu PL, Doumith M, Livermore DM, Chen TP, Woodford N. 2009. Diversity of carbapenem resistance mechanisms in Acinetobacter baumannii from a Taiwan hospital: spread of plasmid-borne OXA-72 carbapenemase. J. Antimicrob. Chemother. 63:641–647. 10.1093/jac/dkn553 [DOI] [PubMed] [Google Scholar]
- 138.Kuo SC, Yang SP, Lee YT, Chuang HC, Chen CP, Chang CL, Chen TL, Lu PL, Hsueh PR, Fung CP. 2013. Dissemination of imipenem-resistant Acinetobacter baumannii with new plasmid-borne blaOXA-72 in Taiwan. BMC Infect. Dis. 13:319. 10.1186/1471-2334-13-319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Alp E, Percin D, Colakoglu S, Durmaz S, Kurkcu CA, Ekincioglu P, Gunes T. 2013. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae in a tertiary university hospital in Turkey. J. Hosp. Infect. 84:178–180. 10.1016/j.jhin.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 140.Lascols C, Hackel M, Marshall SH, Hujer AM, Bouchillon S, Badal R, Hoban D, Bonomo RA. 2011. Increasing prevalence and dissemination of NDM-1 metallo-beta-lactamase in India: data from the SMART study (2009). J. Antimicrob. Chemother. 66:1992–1997. 10.1093/jac/dkr240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Carrer A, Poirel L, Yilmaz M, Akan OA, Feriha C, Cuzon G, Matar G, Honderlick P, Nordmann P. 2010. Spread of OXA-48-encoding plasmid in Turkey and beyond. Antimicrob. Agents Chemother. 54:1369–1373. 10.1128/AAC.01312-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lahlaoui H, Poirel L, Barguellil F, Moussa MB, Nordmann P. 2012. Carbapenem-hydrolyzing class D beta-lactamase OXA-48 in Klebsiella pneumoniae isolates from Tunisia. Eur. J. Clin. Microbiol. Infect. Dis. 31:937–939. 10.1007/s10096-011-1389-5 [DOI] [PubMed] [Google Scholar]
- 143.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J. Antimicrob. Chemother. 67:1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- 144.Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. 2013. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob. Agents Chemother. 57:130–136. 10.1128/AAC.01686-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nagano N, Endoh Y, Nagano Y, Toyama M, Matsui M, Shibayama K, Arakawa Y. 2013. First report of OXA-48 carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in Japan from a patient returned from Southeast Asia. Jpn. J. Infect. Dis. 66:79–81. 10.7883/yoken.66.79 [DOI] [PubMed] [Google Scholar]
- 146.Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Adams MD, Chan ER, Molyneaux ND, Bonomo RA. 2010. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:3569–3577. 10.1128/AAC.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J. Bacteriol. 190:8053–8064. 10.1128/JB.00834-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhu L, Yan Z, Zhang Z, Zhou Q, Zhou J, Wakeland EK, Fang X, Xuan Z, Shen D, Li QZ. 2013. Complete genome analysis of three Acinetobacter baumannii clinical isolates in China for insight into the diversification of drug resistance elements. PLoS One 8:e66584. 10.1371/journal.pone.0066584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 43:4382–4390. 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Karah N, Haldorsen B, Hermansen NO, Tveten Y, Ragnhildstveit E, Skutlaberg DH, Tofteland S, Sundsfjord A, Samuelsen O. 2011. Emergence of OXA-carbapenemase- and 16S rRNA methylase-producing international clones of Acinetobacter baumannii in Norway. J. Med. Microbiol. 60:515–521. 10.1099/jmm.0.028340-0 [DOI] [PubMed] [Google Scholar]
- 153.Kenyon JJ, Hall RM. 2013. Variation in the complex carbohydrate biosynthesis loci of Acinetobacter baumannii genomes. PLoS One 8:e62160. 10.1371/journal.pone.0062160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hamouda A, Evans BA, Towner KJ, Amyes SG. 2010. Characterization of epidemiologically unrelated Acinetobacter baumannii isolates from four continents by use of multilocus sequence typing, pulsed-field gel electrophoresis, and sequence-based typing of blaOXA-51-like genes. J. Clin. Microbiol. 48:2476–2483. 10.1128/JCM.02431-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Karah N, Sundsfjord A, Towner K, Samuelsen O. 2012. Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15:237–247. 10.1016/j.drup.2012.06.001 [DOI] [PubMed] [Google Scholar]
- 156.Dijkshoorn L, Aucken H, Gerner-Smidt P, Janssen P, Kaufmann ME, Garaizar J, Ursing J, Pitt TL. 1996. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J. Clin. Microbiol. 34:1519–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.van Dessel H, Dijkshoorn L, van der Reijden T, Bakker N, Paauw A, van den Broek P, Verhoef J, Brisse S. 2004. Identification of a new geographically widespread multiresistant Acinetobacter baumannii clone from European hospitals. Res. Microbiol. 155:105–112. 10.1016/j.resmic.2003.10.003 [DOI] [PubMed] [Google Scholar]
- 158.Evans BA, Hamouda A, Towner KJ, Amyes SG. 2008. OXA-51-like beta-lactamases and their association with particular epidemic lineages of Acinetobacter baumannii. Clin. Microbiol. Infect. 14:268–275. 10.1111/j.1469-0691.2007.01919.x [DOI] [PubMed] [Google Scholar]
- 159.Chen TL, Lee YT, Kuo SC, Hsueh PR, Chang FY, Siu LK, Ko WC, Fung CP. 2010. Emergence and distribution of plasmids bearing the blaOXA-51-like gene with an upstream ISAba1 in carbapenem-resistant Acinetobacter baumannii isolates in Taiwan. Antimicrob. Agents Chemother. 54:4575–4581. 10.1128/AAC.00764-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hu WS, Yao SM, Fung CP, Hsieh YP, Liu CP, Lin JF. 2007. An OXA-66/OXA-51-like carbapenemase and possibly an efflux pump are associated with resistance to imipenem in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3844–3852. 10.1128/AAC.01512-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee YT, Turton JF, Chen TL, Wu RCC, Chang WC, Fung CP, Chen CP, Cho WL, Huang LY, Siu LK. 2009. First identification of bla(OXA-51-like) in non-baumannii Acinetobacter spp. J. Chemother. 21:514–520. 10.1179/joc.2009.21.5.514 [DOI] [PubMed] [Google Scholar]
- 162.Lee YT, Kuo SC, Chiang MC, Yang SP, Chen CP, Chen TL, Fung CP. 2012. Emergence of carbapenem-resistant non-baumannii species of Acinetobacter harboring a blaOXA-51-like gene that is intrinsic to A. baumannii. Antimicrob. Agents Chemother. 56:1124–1127. 10.1128/AAC.00622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Turton JF, Gabriel SN, Valderrey C, Kaufmann ME, Pitt TL. 2007. Use of sequence-based typing and multiplex PCR to identify clonal lineages of outbreak strains of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:807–815. 10.1111/j.1469-0691.2007.01759.x [DOI] [PubMed] [Google Scholar]
- 164.Potron A, Kalpoe J, Poirel L, Nordmann P. 2011. European dissemination of a single OXA-48-producing Klebsiella pneumoniae clone. Clin. Microbiol. Infect. 17:E24–E26. 10.1111/j.1469-0691.2011.03669.x [DOI] [PubMed] [Google Scholar]
- 165.Izdebski R, Fiett J, Hryniewicz W, Gniadkowski M. 2012. Molecular analysis of Acinetobacter baumannii isolates from invasive infections in 2009 in Poland. J. Clin. Microbiol. 50:3813–3815. 10.1128/JCM.02271-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sonnevend A, Ghazawi A, Al Munthari N, Pitout M, Hamadeh MB, Hashmey R, Girgis SK, Sheikh FA, Al Haj M, Nagelkerke N, Pal T. 2013. Characteristics of epidemic and sporadic strains of Acinetobacter baumannii isolated in Abu Dhabi hospitals. J. Med. Microbiol. 62:582–590. 10.1099/jmm.0.055681-0 [DOI] [PubMed] [Google Scholar]
- 167.Schleicher X, Higgins PG, Wisplinghoff H, Korber-Irrgang B, Kresken M, Seifert H. 2013. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005-2009). Clin. Microbiol. Infect. 19:737–742. 10.1111/1469-0691.12026 [DOI] [PubMed] [Google Scholar]
- 168.Lee HY, Chang RC, Su LH, Liu SY, Wu SR, Chuang CH, Chen CL, Chiu CH. 2012. Wide spread of Tn2006 in an AbaR4-type resistance island among carbapenem-resistant Acinetobacter baumannii clinical isolates in Taiwan. Int. J. Antimicrob. Agents 40:163–167. 10.1016/j.ijantimicag.2012.04.018 [DOI] [PubMed] [Google Scholar]
- 169.Zander E, Chmielarczyk A, Heczko P, Seifert H, Higgins PG. 2013. Conversion of OXA-66 into OXA-82 in clinical Acinetobacter baumannii isolates and association with altered carbapenem susceptibility. J. Antimicrob. Chemother. 68:308–311. 10.1093/jac/dks382 [DOI] [PubMed] [Google Scholar]
- 170.Brink AJ, Feldman C, Grolman DC, Muckart D, Pretorius J, Richards GA, Senekal M, Sieling W. 2004. Appropriate use of the carbapenems. S. Afr. Med. J. 94:857–861 http://blues.sabinet.co.za/WebZ/Authorize?sessionid=0:autho=pubmed:password=pubmed2004&/AdvancedQuery?&format=F&next=images/ejour/m_samj/m_samj_v94_n10_a21.pdf [PubMed] [Google Scholar]
- 171.Amyes SG, Walsh FM, Bradley JS. 2007. Best in class: a good principle for antibiotic usage to limit resistance development? J. Antimicrob. Chemother. 59:825–826. 10.1093/jac/dkm059 [DOI] [PubMed] [Google Scholar]
- 172.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224. 10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 173.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 174.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 175.Zhou H, Zhang T, Yu D, Pi B, Yang Q, Zhou J, Hu S, Yu Y. 2011. Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55:4506–4512. 10.1128/AAC.01134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Al-Hassan L, El Mehallawy H, Amyes SG. 2013. Diversity in Acinetobacter baumannii isolates from paediatric cancer patients in Egypt. Clin. Microbiol. Infect. 19:1082–1088. 10.1111/1469-0691.12143 [DOI] [PubMed] [Google Scholar]
- 177.Lopes BS, Al-Hassan L, Amyes SG. 2012. ISAba825 controls the expression of the chromosomal blaOXA-51-like and the plasmid borne blaOXA-58 gene in clinical isolates of Acinetobacter baumannii isolated from the USA. Clin. Microbiol. Infect. 18:E446–E451. 10.1111/j.1469-0691.2012.03979.x [DOI] [PubMed] [Google Scholar]
- 178.Post V, Hamidian M, Hall RM. 2012. Antibiotic-resistant Acinetobacter baumannii variants belonging to global clone 1. J. Antimicrob. Chemother. 67:1039–1040. 10.1093/jac/dkr586 [DOI] [PubMed] [Google Scholar]
- 179.Lopes BS, Evans BA, Amyes SG. 2012. Disruption of the blaOXA-51-like gene by ISAba16 and activation of the blaOXA-58 gene leading to carbapenem resistance in Acinetobacter baumannii Ab244. J. Antimicrob. Chemother. 67:59–63. 10.1093/jac/dkr415 [DOI] [PubMed] [Google Scholar]
- 180.Zarrilli R, Vitale D, Di Popolo A, Bagattini M, Daoud Z, Khan AU, Afif C, Triassi M. 2008. A plasmid-borne blaOXA-58 gene confers imipenem resistance to Acinetobacter baumannii isolates from a Lebanese hospital. Antimicrob. Agents Chemother. 52:4115–4120. 10.1128/AAC.00366-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zander E, Seifert H, Higgins PG. 2014. Insertion sequence IS18 mediates overexpression of blaOXA-257 in a carbapenem-resistant Acinetobacter bereziniae isolate. J. Antimicrob. Chemother. 69:270–271. 10.1093/jac/dkt313 [DOI] [PubMed] [Google Scholar]
- 182.Naas T, Namdari F, Reglier-Poupet H, Poyart C, Nordmann P. 2007. Panresistant extended-spectrum β-lactamase SHV-5-producing Acinetobacter baumannii from New York City. J. Antimicrob. Chemother. 60:1174–1176. 10.1093/jac/dkm366 [DOI] [PubMed] [Google Scholar]
- 183.De Champs C, Poirel L, Bonnet R, Sirot D, Chanal C, Sirot J, Nordmann P. 2002. Prospective survey of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolated in a French hospital in 2000. Antimicrob. Agents Chemother. 46:3031–3034. 10.1128/AAC.46.9.3031-3034.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Da Silva GJ, Quinteira S, Bertolo E, Sousa JC, Gallego L, Duarte A, Peixe L. 2004. Long-term dissemination of an OXA-40 carbapenemase-producing Acinetobacter baumannii clone in the Iberian Peninsula. J. Antimicrob. Chemother. 54:255–258. 10.1093/jac/dkh269 [DOI] [PubMed] [Google Scholar]
- 185.Merkier AK, Centron D. 2006. blaOXA-51-type β-lactamase genes are ubiquitous and vary within a strain in Acinetobacter baumannii. Int. J. Antimicrob. Agents 28:110–113. 10.1016/j.ijantimicag.2006.03.023 [DOI] [PubMed] [Google Scholar]
- 186.Brown S, Amyes SGB. 2005. The sequences of seven class D β-lactamases isolated from carbapenem-resistant Acinetobacter baumannii from four continents. Clin. Microbiol. Infect. 11:326–329. 10.1111/j.1469-0691.2005.01096.x [DOI] [PubMed] [Google Scholar]
- 187.Vahaboglu H, Budak F, Kasap M, Gacar G, Torol S, Karadenizli A, Kolayli F, Eroglu C. 2006. High prevalence of OXA-51-type class D β-lactamases among ceftazidime-resistant clinical isolates of Acinetobacter spp.: co-existence with OXA-58 in multiple centres. J. Antimicrob. Chemother. 58:537–542. 10.1093/jac/dkl273 [DOI] [PubMed] [Google Scholar]
- 188.Tsakris A, Ikonomidis A, Spanakis N, Pournaras S, Bethimouti K. 2007. Identification of a novel blaOXA-51 variant, blaOXA-92, from a clinical isolate of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:348–349. 10.1111/j.1469-0691.2006.01598.x [DOI] [PubMed] [Google Scholar]
- 189.Evans BA, Brown S, Hamouda A, Findlay J, Amyes SG. 2007. Eleven novel OXA-51-like enzymes from clinical isolates of Acinetobacter baumannii. Clin. Microbiol. Infect. 13:1137–1138. 10.1111/j.1469-0691.2007.01828.x [DOI] [PubMed] [Google Scholar]
- 190.Bogaerts P, Naas T, Wybo I, Bauraing C, Soetens O, Pierard D, Nordmann P, Glupczynski Y. 2006. Outbreak of infection by carbapenem-resistant Acinetobacter baumannii producing the carbapenemase OXA-58 in Belgium. J. Clin. Microbiol. 44:4189–4192. 10.1128/JCM.00796-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Lee K, Kim MN, Choi TY, Cho SE, Lee S, Whang DH, Yong D, Chong Y, Woodford N, Livermore DM. 2009. Wide dissemination of OXA-type carbapenemases in clinical Acinetobacter spp. isolates from South Korea. Int. J. Antimicrob. Agents 33:520–524. 10.1016/j.ijantimicag.2008.10.009 [DOI] [PubMed] [Google Scholar]
- 192.Sevillano E, Gallego L, Garcia-Lobo JM. 2009. First detection of the OXA-40 carbapenemase in P. aeruginosa isolates, located on a plasmid also found in A. baumannii. Pathol. Biol. (Paris) 57:493–495. 10.1016/j.patbio.2008.05.002 [DOI] [PubMed] [Google Scholar]
- 193.Chu YW, Cheung TK, Chu MY, Lo JY, Dijkshoorn L. 2009. OXA-23-type imipenem resistance in Acinetobacter baumannii in Hong Kong. Int. J. Antimicrob. Agents 34:285–286. 10.1016/j.ijantimicag.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 194.McGann P, Milillo M, Clifford RJ, Snesrud E, Stevenson L, Backlund MG, Viscount HB, Quintero R, Kwak YI, Zapor MJ, Waterman PE, Lesho EP. 2013. Detection of New Delhi metallo-beta-lactamase (encoded by blaNDM-1) in Acinetobacter schindleri during routine surveillance. J. Clin. Microbiol. 51:1942–1944. 10.1128/JCM.00281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Turton JF, Baddal B, Perry C. 2011. Use of the accessory genome for characterization and typing of Acinetobacter baumannii. J. Clin. Microbiol. 49:1260–1266. 10.1128/JCM.02335-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lopes BS, Gould IM, Opazo AF, Amyes SG. 2012. The resistance profile of Acinetobacter baumannii strains isolated from the Aberdeen Royal Infirmary. Int. J. Antimicrob. Agents 39:361–362. 10.1016/j.ijantimicag.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 197.Zander E, Nemec A, Seifert H, Higgins PG. 2012. Association between beta-lactamase-encoding blaOXA-51 variants and DiversiLab rep-PCR-based typing of Acinetobacter baumannii isolates. J. Clin. Microbiol. 50:1900–1904. 10.1128/JCM.06462-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Matsui M, Suzuki S, Suzuki M, Arakawa Y, Shibayama K. 2013. Rapid discrimination of Acinetobacter baumannii international clone II lineage by pyrosequencing SNP analyses of blaOXA-51-like genes. J. Microbiol. Methods 94:121–124. 10.1016/j.mimet.2013.05.014 [DOI] [PubMed] [Google Scholar]
- 199.Kasap M, Torol S, Kolayli F, Dundar D, Vahaboglu H. 2013. OXA-162, a novel variant of OXA-48 displays extended hydrolytic activity towards imipenem, meropenem and doripenem. J. Enzyme Inhib. Med. Chem. 28:990–996. 10.3109/14756366.2012.702343 [DOI] [PubMed] [Google Scholar]
- 200.Gomez S, Pasteran F, Faccone D, Bettiol M, Veliz O, De Belder D, Rapoport M, Gatti B, Petroni A, Corso A. 2013. Intrapatient emergence of OXA-247: a novel carbapenemase found in a patient previously infected with OXA-163-producing Klebsiella pneumoniae. Clin. Microbiol. Infect. 19:E233–E235. 10.1111/1469-0691.12142 [DOI] [PubMed] [Google Scholar]
- 201.Zhou Z, Du X, Wang L, Yang Q, Fu Y, Yu Y. 2011. Clinical carbapenem-resistant Acinetobacter baylyi strain coharboring blaSIM-1 and blaOXA-23 from China. Antimicrob. Agents Chemother. 55:5347–5349. 10.1128/AAC.00425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bonnin RA, Rotimi VO, Al Hubail M, Gasiorowski E, Al Sweih N, Nordmann P, Poirel L. 2013. Wide dissemination of GES-type carbapenemases in Acinetobacter baumannii isolates in Kuwait. Antimicrob. Agents Chemother. 57:183–188. 10.1128/AAC.01384-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Di Nocera PP, Rocco F, Giannouli M, Triassi M, Zarrilli R. 2011. Genome organization of epidemic Acinetobacter baumannii strains. BMC Microbiol. 11:224. 10.1186/1471-2180-11-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Espinal P, Marti S, Soto S, Dominguez M, Campos J, Vila J. 2009. Characterization of the resistance mechanism in carbapenem resistant Acinetobacter baumannii clinical isolates. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., presentation C1-1885 [Google Scholar]