Abstract
Myxobacteria are social microbes that exhibit complex multicellular behaviors. By use of fluorescent reporters, we show that Myxococcus xanthus isolates produce long narrow filaments that are enclosed by the outer membrane (OM) and contain proteins. We show that these OM tube (OMT) structures are produced at surprisingly high levels when cells are placed in liquid medium or buffer without agitation. OMTs can be long and easily exceed multiple cell lengths. When viewed by transmission electron microscopy, their morphology varies between tubes and chain-like structures. Intermediate-like structures are also found, suggesting that OMTs may transition between these two morphotypes. In support of this, video epifluorescence microscopy found that OMTs in solution dynamically twist and jiggle. On hard surfaces, myxobacteria glide, and upon cell-cell contact, they can efficiently exchange their OM proteins and lipids by a TraAB-dependent mechanism. Although the structure of OMTs hints at a possible role as conduits for exchange, evidence is presented to the contrary. For example, abundant OMT production occurs in traA or traB mutants and when cells are grown in liquid medium, yet transfer cannot occur under these conditions. Thus, genetic and environmental conditions that promote OMT production are incongruent with OM exchange.
INTRODUCTION
Myxobacteria are social microbes that depend on cell-cell interactions during different stages of their life cycle, including gliding motility and fruiting body development (1). A behavior in Myxococcus xanthus organisms that we recently described is their ability to exchange or transfer outer membrane (OM) proteins and lipids between cells (2–4). In contrast, cytoplasmic and inner membrane proteins are not transferred. Transfer requires cell-cell contact on a hard surface, and physical cell movement by gliding motility significantly increases the efficiency of exchange (3, 5). Transfer does not occur in liquid or when cells are densely packed but poorly aligned. Recently, the traAB operon was discovered as a genetic determinant for transfer (2). Unlike most protein transport systems, which unidirectionally transfer cargo from one cell to another (6), the TraA and TraB proteins are required in both donor and recipient cells, indicating that transfer is bidirectional. Because fluorescently labeled lipids are also exchanged by a TraAB-dependent mechanism, the model for transfer invokes the transient fusion of the OM, resulting in the exchange of content between cells (2). TraA functions as a cell surface adhesin and as a molecular recognition determinant that identifies other cells that express the same or similar traA alleles to coordinate social behaviors (2, 7, 8). We have also suggested that TraA functions as a fusogen to catalyze OM fusion between cells (2, 8). The function of TraB is less clear, although it contains an OmpA domain that is predicted to bind to the cell wall.
To monitor OM exchange in live cells, we developed a fluorescent reporter that fuses an OM lipoprotein signal and sorting sequence to mCherry (SSOM-mCherry) (3). This red reporter allows visualization of transfer to live recipient cells labeled with the green fluorescent protein (GFP) localized in the cytoplasm, which itself does not transfer. Similarly, lipid transfer can be visualized with fluorescent lipid dyes (2). During microscopic examination of cells harboring SSOM-mCherry, we discovered long tubular filaments emanating from M. xanthus cells. In the myxobacterial literature, there have been numerous reports of M. xanthus cells producing extracellular appendages or material (9–18). However, the chemical composition, function, and relationship of these structures have only been partly elucidated, and many unanswered questions remain. Here, we report the identification of OM tubes (OMTs) and the investigation of their composition, structure, and origin and their possible relationship to OM exchange.
MATERIALS AND METHODS
Strains, plasmids, and media.
Bacterial strains used in this study are listed in Table 1. M. xanthus was routinely grown in CTT medium (1% Casitone; 1 mM KH2PO4; 8 mM MgSO4; 10 mM Tris-HCl, pH 7.6) in the dark at 33°C. Nutrient levels of Casitone were sometimes reduced to 1/2 or 1/4 of usual levels. As needed for selection, the medium was supplemented with kanamycin (Km; 50 μg/ml), oxytetracycline (Tc; 15 μg/ml), or streptomycin (Sm; 600 μg/ml). TPM buffer (10 mM Tris-HCl, pH 7.6; 1 mM KH2PO4; 8 mM MgSO4) was used to wash cells or for starvation treatment. For strain construction, routine methods were used for electroporation and for generalized transduction of Mx4 and Mx8 (3). Escherichia coli was cultured at 37°C in LB medium that was supplemented with Km (50 μg/ml), ampicillin (100 μg/ml), or Sm (100 μg/ml) when necessary.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant feature(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1 | Cloning vector, Kmr | Life Technologies |
| pDP1 | PpilA-SSIM-mCherry in pCR2.1, Kmr | 3 |
| pDP302 | PpilA-SST1-mCherry in pCR2.1, Kmr | This study |
| pXW2 | PpilA-SSOM-mCherry in pCR2.1, Kmr | 3 |
| pXW6 | PpilA-SSOM-mCherry in pKSAT, Smr | 3 |
| Strains | ||
| DH5α | E. coli cloning strain | Lab collection |
| DK1622 | A+ S+, wild-type M. xanthus | 19 |
| DK8601 | A− S−, ΔpilA aglB1, Tcr | 20 |
| DK8606 | A− S−, ΔpilA aglB1 PpilA-gfp, Kmr Tcr | 20 |
| DK396 | A− S−, aglT194→NS pilR119→NS traA227P→L (multiple other mutations) | 2 |
| DK8615 | A+ S−, ΔpilQ | 19 |
| DZ1 | A− S− tan variant (multiple mutations) | 21 |
| DW700 | A+ S−, ΔpilA::Tc PpilA-gfp, Kmr Tcr | 3 |
| DW701 | A− S+, PpilA-gfp aglB1 dsp-1693, Kmr | 3 |
| DW702 | A− S−, DW701 ΔpilA::Tc, Kmr Tcr | 3 |
| DW709 | A+ S−, ΔpilA PpilA-gfp, Kmr | This study |
| DW1041 | A+ S+, DK1622(pXW2) | 3 |
| DW1042 | A− S+, aglB1, DK1217(pXW2), Kmr | 3 |
| DW1047 | A− S−, DK8601(pXW2), Kmr | 3 |
| DW1048 | A− S−, DK8601(pDP1), Kmr | 3 |
| DW1053 | A+S−, DK10410 ΔpilA(pXW2) | This study |
| DW1056 | A− S−, DK8606(pXW6), Kmr Smr Tcr | 3 |
| DW1165 | A− S−, DK1693, aglB1 dsp-1693 ΔpilA::Tc PpilA-gfp (pXW6) Kmr Smr Tcr | This study |
| DW1411 | A− S−, PpilA− SSOM–mCherry, DK8601(pXW6), Smr Tcr | 2 |
| DW1412 | A− S−, traA::Km, DW1411, Kmr Smr Tcr | 2 |
| DW1413 | A− S−, traB::Km, DW1411, Kmr Smr Tcr | 2 |
| DW1414 | A+ S−, PpilA-gfp ΔpilA::Tc | 2 |
| DW1477 | A− S−, DK8601(pDP302) | This study |
| HW-1 | Wild-type M. fulvus | 7 |
| LS1191 | A+ S+, esg258 MXAN_4265::Km, Kmr | L. Shimkets |
| Mxx23 | Wild-type M. xanthus | 7 |
To construct a fluorescent periplasmic reporter, the mCherry gene was fused to the type I signal sequence (SS) from cglF (22). To do this, the PpilA–type II–SSOM cassette from pXW2 (3) was replaced with a PpilA–type I–SScglF cassette. Specifically, pXW2 was digested with HindIII and XbaI, and a PCR-generated PpilA–type I–SScglF fragment was cloned into those sites. The primers used for amplification were PpilA-HindIII-FW (5′-GGAAGCTTGGTACCGAGTCTACTAGTGGATCCGTCATGTTGGACGAGGTC-3′) and XbaI-cglF-SS-PpilA-SD-Rev (5′-CACATCTAGAGGCGCTCGCGACCGTGGGGAGAACGAACGAAGCGCACAGGGCAAGTGAGGCGAGGGCTTTCATCACTGCCTCCCGGGGGTCCTCAGAGAAGGTTG-3′). Restriction sites are underlined, and the 5′ region in the latter primer contains 57 bases from cglF. The resulting plasmid was called pDP302.
Fluorescent staining of cells.
Cell membranes were routinely stained with the fluorescent lipophilic dye DilC (component H or A in the tracer sampler kit from Life Technologies/Molecular Probes) by mixing cells and dyes and incubating for 0.5 to 2 h as described previously (2). Other dyes within the sampler kit also worked to various degrees on M. xanthus cells. The stained cells were observed either directly or after washing in TPM buffer.
A polymyxin B (PMB) BODIPY FL fluorescent conjugate (Life Technologies/Molecular Probes) was used to stain lipopolysaccharide (LPS). M. xanthus cells were suspended in magnesium-free Tris phosphate buffer (i.e., 10 mM Tris-HCl, pH 7.6; 1 mM KH2PO4), and the PMB conjugate was added to a final concentration of 1 μM. After incubation for 15 min, the cell suspensions were microscopically observed using a fluorescein isothiocyanate (FITC) filter cube. The magnesium-free version of TPM was found to improve staining (data not shown).
Treatment with inhibitors.
To test for inhibition of OM tube production, M. xanthus cells were treated with the bacteriostatic antibiotic chloramphenicol (Cm) or spectinomycin (Spec) at various concentrations. The respiration chain inhibitor sodium azide and the ATP uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma-Aldrich) were also tested at various concentrations. For these experiments, fresh colonies, 1 or 2 days old, were directly harvested from CTT plates, placed in a microcentrifuge tube with TPM buffer, and vortexed for a minute. Cells were pelleted by centrifugation for 1 min, the supernatant was discarded, and the cells were resuspended with various solutions.
Microscopy.
Transmission electron microscopy (TEM) was conducted on a Hitachi 7000 instrument. Before examination, M. xanthus cultures were grown in an unshaken flask for 4 to 18 h at 33°C. A small aliquot of cells was then pipetted onto a carbon-coated grid. The drop was allowed to remain for 30 s before excess fluid was removed, and the grid was gently washed with distilled water. Cells were then negatively stained with 2% uranyl acetate (wt/vol) for 1 to 2 s, and the grid was then blotted dry.
Light and epifluorescence microscopy was essentially done as described previously (2, 3). Generally, cell aliquots were placed on glass slides under a micro-cover glass and viewed in the middle of the field, away from the cover edge, with a 100× oil immersion phase-contrast objective lens. A Texas Red filter cube was used to visualize red fluorescent reporters, and a FITC filter cube was used to visualize green fluorescent reporters. To test for the presence of SSOM-mCherry in slime trails, an A-motile strain (DW1053) was first incubated overnight without shaking and then spotted on thin pads (0.3 ml of 0.9% agarose in 1/4× CTT) that had been prepared on glass slides. The resulting slides were incubated in a humid chamber at 33°C, and the cells were microscopically examined at 4, 8, 24, and 48 h for fluorescent trails.
RESULTS
OM tube (OMT) discovery.
While investigating OM exchange in M. xanthus with the SSOM-mCherry OM lipoprotein reporter (3), we observed by epifluorescence microscopy narrow tubular-like structures projecting out from cells (Fig. 1). Interestingly, when viewed by phase-contrast, differential interference contrast, or dark field microscopy these filaments were rarely detected (Fig. 1 and data not shown). These OMT structures were observed to be both cell bound and unbound. Cell-attached OMTs were frequently found at the poles, although a peritrichous distribution was also common (Fig. 1C). In initial observations, OMTs were found to be abundant in some cultures, whereas other cultures seemed to produce few OMTs. Upon examination of conditions, we found that OMTs became more abundant and longer when cultures were grown without shaking (submerge culture) for a couple of hours to days (Fig. 1A and B). Although OMTs were abundant after 2 h incubations, their relative abundance and lengths increased with the age of the culture. For instance, a 2-day-old culture produced OMTs whose numbers were in excess of the number of cells in the culture (Fig. 1B). OMTs were also detected to various degrees in bacteria from shaking cultures (300 rpm), in which they were found to be consistently shorter and less abundant, presumably because shearing forces had broken them off from the cells (Fig. 1C). Last, we note that OMT detection assays used here are qualitative and OMT numbers vary among fields of view even under a single wet-mount slide.
FIG 1.
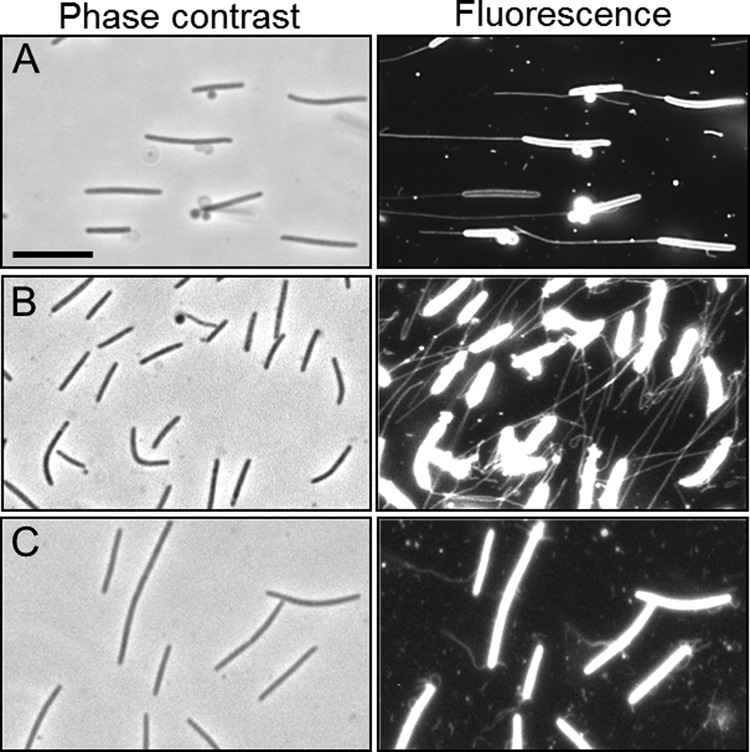
OMT production in M. xanthus. (A) A phase-contrast micrograph of a culture that had been incubated without shaking for 1 day reveals no OMTs. A fluorescence micrograph of the same field reveals long OM tubes (SSOM-mCherry reporter in DW1047; aglB1 ΔpilA). (B) Prolonged static incubation (2 days) results in abundant OMTs (SSOM-mCherry reporter in DW1165; ΔpilA aglB1 dsp-1693). The dsp mutation also contributes to higher levels of OMT. (C) A culture that had been grown in liquid medium with shaking and was harvested at mid-log phase shows short and broken OMTs (DW1047). Bar, 8 μm.
The visualization of OMTs by the OM lipoprotein reporter implied that they were composed of OM components, including lipids. To directly test for lipids in OMTs, a strain that did not contain the SSOM-mCherry reporter was stained with a fluorescent lipid dye similar to one we previously used to stain M. xanthus OMs (2). The OMTs were indeed labeled by this lipid dye (DilC) (Fig. 2A), indicating the presence of membranes. The length of OMTs varied over a considerable range. For instance, short (<0.2-μm) to long (>40-μm) (Fig. 2A) OMTs were observed by fluorescence microscopy. In a number of cases, particularly after overnight static incubations, bright fluorescent spheres, which appeared to be vesicles, were sometimes found associated with OMTs (Fig. 1A and 2B). In some cases, multiple vesicles were associated with a single OMT, appearing as beads on a string. Such vesicles could be located midshaft or at the tips. At present, it is unclear if these vesicles are produced by the OMTs or are extracellular vesicles that bind OMTs, or whether the vesicles move along these filaments. Since the vesicles were more prevalent in older cultures, we suspect that their production was stress induced. Their physiological relevance, if any, is currently unknown.
FIG 2.
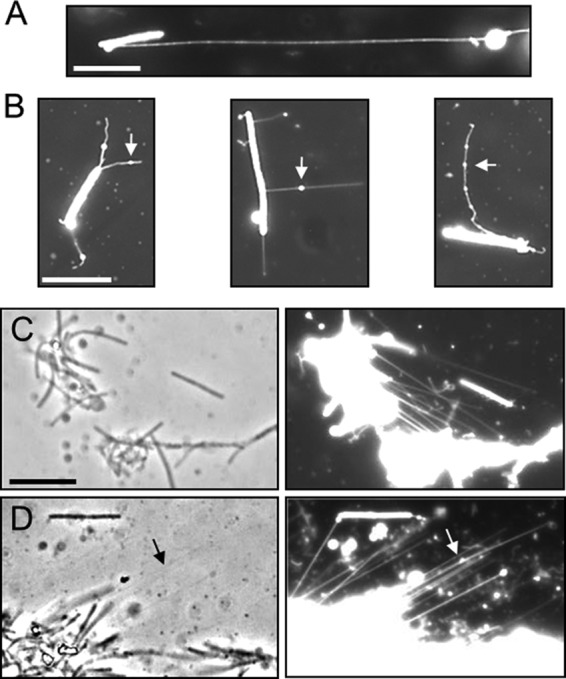
OMTs detected by lipophilic dye (DilC) fluorescent staining. (A) An OMT that exceeds the length of the cell >6-fold (DK8601; aglB1 ΔpilA). (B) Vesicle-like structures (arrows) associated with OMTs (DW1047). (C) Two clumps of cells appear to be connected by OMTs (compare left and right panels) (DK8615; ΔpilQ). (D) OMT bundles are faintly visible by phase contrast (arrow) and appear to anchor a cell clump to the glass slide (right panel; DK8615). Bars, 8 μm.
In a few cases, OMTs appeared to link cells together or even to hold two clumps of cells together (Fig. 1B and 2C). In other examples, OMTs anchored cells to the glass slide (Fig. 2D), even in the presence of thermal currents (see Movie S1 in the supplemental material). The OMTs were also faintly visible in some phase-contrast micrographs (e.g., Fig. 2D, arrow), apparently because they formed bundles or were tightly aligned with neighboring OMTs.
OMT ultrastructure.
To gain insight into their ultrastructure, we used uranyl acetate negative staining and TEM to visualize OMTs. Interestingly, TEM analysis showed that the morphology of OMTs varied. Some OMTs appeared to be composed of rings or links strung together in chains (Fig. 3A and B). In other cases they appeared as tubes without links (Fig. 3C). Both morphotypes were frequently seen by TEM. When found as chains, the OMTs varied in length from 1 to >75 links. What appeared to be individual links in these images had a structure that was similar to OM vesicles produced by M. xanthus (23, 24). Consequently we cannot distinguish between OM vesicles and single OMT links and whether they are different. The width of the OMTs was ∼50 nm, making them considerably thicker than type IV pili (∼6 nm), and they are clearly distinct from pili, as the strain used for TEM was a ΔpilA mutant (25). Interestingly, in some cases the chain links appeared to be in the process of unwinding (or winding) (Fig. 3D). These observations suggested to us that OMTs may twist and/or untwist around themselves and could exist in equilibrium between these states. Consistent with this idea, in wet mounts OMTs are dynamic structures and can be seen oscillating and twisting (see Movies S2 and S3 in the supplemental material). It is also possible that links within chains may rupture or fuse, resulting in tube structures. When visualized in wet mounts by epifluorescence microscopy, OMTs could be seen detaching from cells, apparently as a result of heat and current forces generated by the microscope (e.g., see Movie S1 in the supplemental material). Finally, OMT fragments, as seen by epifluorescence microscopy and TEM, are frequently seen unassociated from cells (e.g., Fig. 3A), suggesting that their binding affinity for cells is relatively low.
FIG 3.
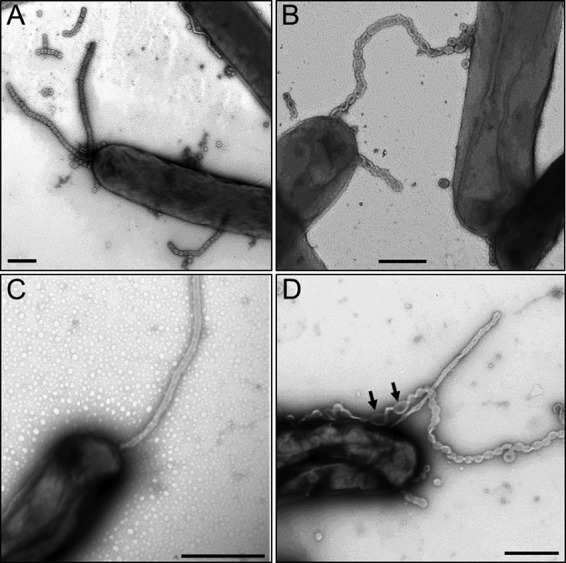
Transmission electron micrographs of OMTs in M. xanthus DW1165. (A) Chain-like OMTs protruding from the cell pole and sides and pieces that were broken off in the extracellular milieu. Vesicles or single links are present that are attached to cells or are unattached. (B) An OMT touches two cells. (C) Straight OMT emanating from cell pole. (D) Straight and chained OMTs. Arrows mark a region that is apparently untwisting. Bars, 0.5 μm.
OMTs are derived from the OM.
OMTs contain lipid and lipoprotein constituents (Fig. 1 and 2) and appear to originate from the OM (Fig. 3B). To investigate whether OMTs are also composed of inner membranes, we tested whether the inner membrane marker SSIM-mCherry (lipoprotein) labeled OMTs (3). In contrast to what was found with SSOM-mCherry (Fig. 1), the SSIM-mCherry reporter did not label OMTs (Fig. 4A) (DW1048), suggesting that OMTs are not composed of the inner membrane. As a control, when the same DW1048 culture was subsequently colabeled with DilC, OMTs were easily observed (Fig. 4A). To test whether OMTs could be labeled with a periplasmic marker, we constructed an mCherry fusion to a type I signal sequence derived from the CglF protein (22). As expected, this reporter (SST1-mCherry) labeled the cell envelope (Fig. 4A, inset); however, OMTs were not labeled. Again as a control, when this culture was colabeled with DilC, OMTs were readily detected (Fig. 4A).
FIG 4.
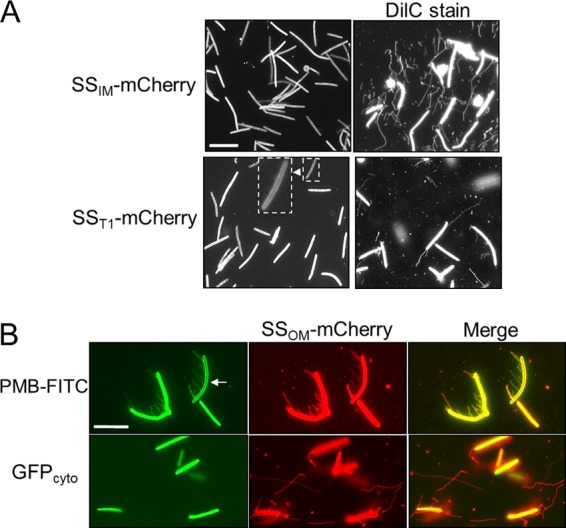
The ability of different cell markers to label OMTs in M. xanthus. (A) The left panels show that the SSIM-mCherry (DW1048; aglB1 ΔpilA) and SST1-mCherry (DW1477; aglB1 ΔpilA) reporters do not label OMTs. (Inset) SST1-mCherry localizes to the cell envelope. Right panels show same cultures stained with DilC. (B) Polymyxin B (PMB) conjugated to FITC labels LPS on the cell envelope (see the underexposed cell [arrow]) and OMTs revealed by colocalization with the SSOM-mCherry reporter in the merged image (DW1047). Cytoplasmic GFP does not label OMTs (DW1165). Bars, 8 μm.
Our above results suggest that OMTs contain lipopolysaccharide (LPS). To test this, cells were incubated with the LPS-specific probe polymyxin B (PMB) conjugated to a FITC fluorescent tag. PMB-FITC indeed labeled cell filaments that colocalize with the SSOM-mCherry reporter (Fig. 4B, merged image). In contrast, a GFP reporter that is cytoplasmically localized did not label OMTs (Fig. 4B), indicating that OMTs do not contain cytoplasmic material.
OMTs are produced by environmental isolates.
To test whether OMT production was an anomaly of our laboratory strain or a general property of Myxococcus spp., we examined a panel of 16 environmental isolates that we had used in a prior study (7). As shown in representative images in Fig. 5, OMTs were indeed detected in all of the isolates, including in a Myxococcus fulvus species. We conclude that OMTs are a general property of M. xanthus isolates and, probably, of other myxobacterial species.
FIG 5.
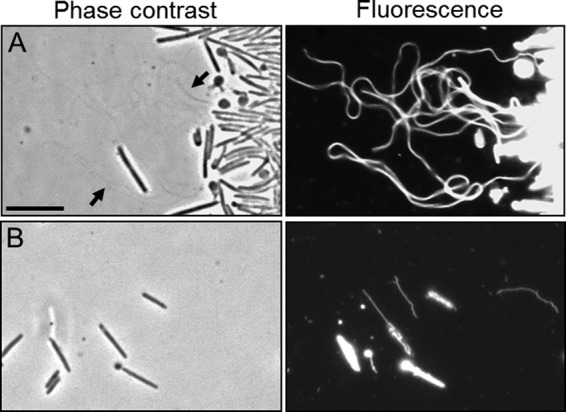
Environmental isolates produce OMTs. Strains were stained with the lipophilic dye DilC. (A) M. xanthus Mxx23 (wild type). Bundled OMTs are faintly visible by phase-contrast microscopy (arrow). (B) M. fulvus HW-1 (wild type). Note that dye staining of environmental isolates was heterogeneous; some cells stained and some did not. Bar, 8 μm.
Recently, OM tube-like structures have been reported in divergent bacterial species, including Bacillus subtilis, Francisella novicida, Salmonella enterica serovar Typhimurium, and Delftia sp. strain Cs1-4 (26–29). To extend and compare our findings, we surveyed distantly related species for OMTs by again using the lipophilic fluorescent dye assay used here to stain M. xanthus OM material. We examined E. coli and Pseudomonas aeruginosa strains under a variety of growth conditions in liquid medium or from agar plates. Using this method, OM vesicles were readily seen; however, no OMTs were detected in these species (data not shown). We also conducted limited tests with Klebsiella pneumoniae and Rhodobacter sphaeroides, using conditions that were similar to those used for M. xanthus, and again we did not detect OMTs in these species (data not shown). These results indicate that OMT production is limited, at least under our assay conditions, to a subset of bacteria.
OMT growth conditions and rate.
Next, we cultivated M. xanthus under a variety of conditions to look for environmental factors that may induce or repress OMT formation. The following conditions did not significantly alter OMT levels: temperature (23°C, 33°C, or 37°C), growth phase (log phase versus stationary phase versus development), or nutrient conditions (TPM versus 1/2× CTT or CTT) (Fig. 6A and data not shown). Interestingly, the one condition that reduced OMT levels was growth on agar plates. That is, colonies that were 3 to 5 days old produced fewer OMTs (see below). In addition, when single cells or small rafts of cells swarmed out from a colony, no OMTs were detected (data not shown), suggesting that OMTs are not constituents of slime trails, which are characteristically seen as phase-bright tracks left behind gliding M. xanthus bacteria on an agar surface (12). However, we note that if a liquid culture, which produces OMTs, was pipetted onto an agar pad at a low cell density, individual cells could be separated from groups of cells, and OMTs were indeed seen inside the droplet zone for hours to a day after transfer, but again we did not observe OMTs from cells that had emerged from the edge of inoculum spots (data not shown). As described below, the older cells that are at high cell density within an established colony produce some OMTs.
FIG 6.
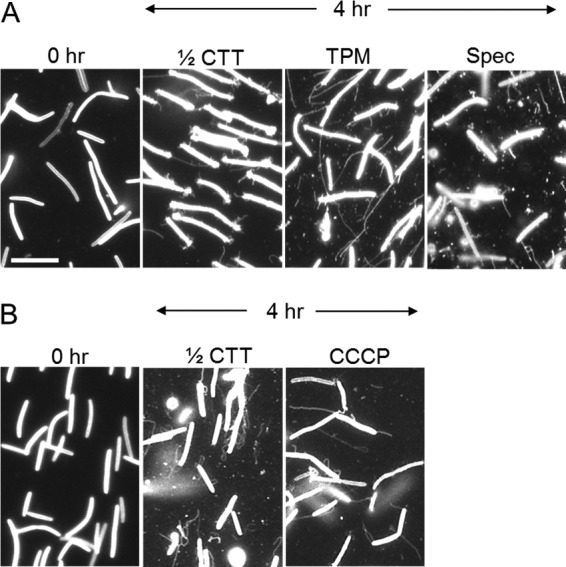
OMTs are produced in the presence of metabolic inhibitors and in starvation buffer (TPM). (A) Cells harvested directly from a CTT plate, vortexed and centrifuged contain few OMTs (0 h; DW1047). Static incubation in the indicated media induces OMT production. Spectinomycin (Spec; 150 μg/ml) was added to 1/2× CTT. (B) OMTs in the presence of CCCP (25 μg/ml; strain DW1165). Bar, 8 μm.
As cells that were harvested from a colony produced fewer OMTs, we followed their growth in liquid medium. Thus, by observing and measuring the length of OMTs at 0, 1, 2, 4, and 8 h after transfer to liquid medium, we estimated that OMTs grow at a rate in the range of 1 to 3 μm/h (e.g., Fig. 6). We also note that when we observed OMTs, we did so in the middle of the cover glass field, where cells were most dispersed, and that for unknown reasons, cells at the edge of the cover glass tended to have more OMTs.
OMTs are produced in the presence of antibiotics and inhibitors.
To test whether OMT production requires active cell metabolism, we treated cells with a panel of bacteriostatic antibiotics and poisons that block translation and energy production. For these experiments, cells were directly harvested from fresh or log-phase growing cells, e.g., 3- to 5-day-old colonies, where few OMTs are produced, and were vortexed and centrifuged to shear and remove any OMTs present (Fig. 6, 0 h). Cells were then placed in medium or buffer in the absence or presence of the following inhibitors: chloramphenicol (40 and 80 μg/ml), spectinomycin (100 and 150 μg/ml), sodium azide (0.02, 0.06, 0.2, and 0.5%), and CCCP (15, 25, and 50 μM) (Fig. 6 and data not shown). Under all of these conditions, OMTs were still made at levels roughly similar to those of the untreated controls. Therefore, active translation, proton motive force, and high cellular ATP levels are not critical for OMT production within the experimental time frame (0 to 8 h).
The SST1-mCherry reporter does not transfer.
Interestingly, the cellular reporters that labeled OMTs (SSOM-mCherry and lipophilic dyes) can be transferred by a TraAB-dependent mechanism, and the reporters that did not label OMTs (GFPCyto and SSIM-mCherry) are not transferred (2, 3). To further investigate these relationships and to determine whether a targeted periplasmic protein can be exchanged, we tested for SST1-mCherry transfer. However, under a variety of conditions, no transfer of SST1-mCherry was detected, even when the ratio of donor to recipient cells was increased to 10:1 or 20:1 and incubation times were extended to 10 or 17 h (Fig. 7A and data not shown). As a positive control, SSOM-mCherry exhibited robust transfer, as GFP (green)-labeled recipients obtained the red reporter and became yellow in the merged micrograph (Fig. 7B). This result extends the mentioned correlation, as SST1-mCherry does not label OMTs (Fig. 4A) and does not transfer. We note that although the SST1-mCherry reporter did not transfer, the CglE and CglF proteins, which contain type I signal sequences, did transfer (2, 22). One plausible explanation for this difference is that the CglE/F proteins could be associated with the OM, while SST1-mCherry is soluble in the periplasm or associated with the inner membrane.
FIG 7.
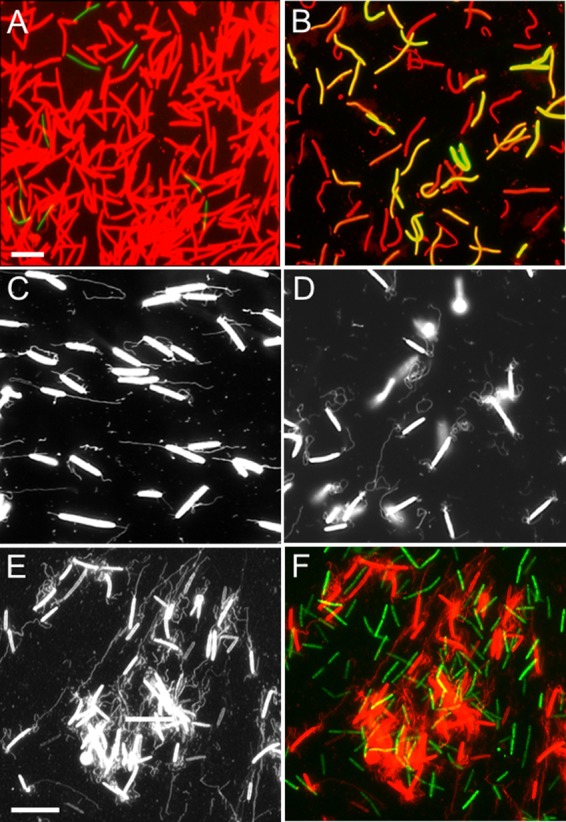
OM exchange and OMT production are distinct processes. (A) A 20:1 ratio of SST1-mCherry donor (DW1477) and GFPCyto-labeled recipient (DW1414; ΔpilA) cells was incubated on TPM agar at high cell density (3 × 109 CFU/ml) for 10 h. Cells were harvested, washed, and microscopically examined as described previously (3). (B) As for panel A, except that donor strain (DW1047) cells contained SSOM-mCherry and were mixed with the recipient (DW1414) cells at a 1:1 ratio. Both panels A and B show green and red merged fluorescent images. Bar in panel A represents 8 μm for panels A and B. (C) traA (DW1412) mutant cells make OMTs. (D) traB (DW1413) mutant cells make OMTs. (E and F) Donor (DW1411; PpilA-SSOM-mCherry) and recipient (DW709; PpilA-gfp) cells were mixed at 1:1 ratios at high cell density in liquid and incubated for 4 h. (E) High-contrast black-and-white image for OMT visualization by the SSOM-mCherry reporter. (F) Identical field, except that it is a colored image merged with an image of GFP-labeled recipient cells, revealing no OM exchange. Bar in panel E represents 8 μm for panels C to F.
OM exchange and OMTs are separate.
Next we tested whether OMTs could serve as a conduit for OM exchange (1–3) by first examining mutants that are not capable of transferring OM proteins and lipids. Importantly, in traA and traB mutant backgrounds OMTs were abundantly produced (Fig. 7C and D), demonstrating that the only known host proteins required for OM exchange are not required for OMT production. In a prior study, we found that TraAB overexpression resulted in M. xanthus cells adhering together side by side, end to end, and in chains, which occurred independently of OMTs (2), showing that TraAB-dependent cell-cell adhesion does not depend on OMT production and thus are separate processes. In addition, we have found that OM exchange does not occur in liquid (3, 20), while in contrast, OMTs are abundantly produced in liquid. Figure 7E and F illustrates this point that OMT production is distinct from OM exchange. That is, in liquid, where abundant OMTs are made, OM exchange does not occur, as the GFP-labeled recipient cells do not obtain the SSOM-mCherry reporter.
The size, quantity, and structural detail of OMTs (Fig. 1 to 3) suggest that there might be specific cellular factors or proteins responsible for their production. To address this and to possibly gain insight into their function, we tested additional candidate mutants for OMT defects. First, a number of single and double mutants defective in motility (pilA, pilR, pilQ, mglA, aglT, and aglB) were tested. In all mutant strains tested, including nonmotile mutants, abundant OMTs were produced (Fig. 1 and data not shown). Similar to the above results found in liquid, this finding indicates that motility is not required for OMT production and again differs from OM exchange, which requires motility (3, 20). We further surveyed other candidate mutants for defects in OMTs, including fibril (dsp or dif), LPS O antigen or lipid A (lpxC), and iso-branched-chain fatty acid (iso-15:0; esg [MXAN_4265])-defective mutants, and again found that all of these mutants produced OMTs around wild-type levels (Fig. 1 and data not shown). However, we note that the dsp mutant appeared to overproduce OMTs (Fig. 1B and data not shown). Last, the DZ1 and DK396 strains, which have been extensively propagated in the laboratory and mutagenized and contain numerous mutations that make them asocial (e.g., lack of motility, phase tan-locked mutation, developmental defects, and others) (2, 21), still make OMTs (data not shown). Thus, although high levels of OMT production would appear to come at a high metabolic cost, mutations that block OMT production do not easily occur during extended laboratory propagation.
DISCUSSION
Here we showed that Myxococcus spp. produce long tubular filaments that emanate from cell bodies and poles. We called these structures OMTs because they contain OM proteins and are enclosed by the OM. Although M. xanthus has served as a model organism for the past 50 years, OMTs have been largely overlooked. We believe the primary reason for this is that under standard conditions, OMTs are not seen by light microscopy, and routine TEM detection is difficult because these structures are fragile and their levels are typically low in shaker flask cultures or agar colonies. However, the SSOM-mCherry reporter easily facilitates their detection and shows that OMTs are abundant cell surface appendages.
Over the years, investigators have described different myxobacterial appendages (9, 11, 17, 25, 30). Two of these structures, type IV pili and fibrils, are clearly not related to OMTs, which are produced in pil and dsp mutants (Fig. 3). Aside from pili and fibrils, M. xanthus produces thin extracellular filaments, which for the lack of a better term have been called “slime” excretions (10, 18, 31, 32). Fluegel first visualized slime threads 50 years ago by light microscopy and India ink staining (32). The term “trail” is sometimes also associated with slime, because myxobacteria produce phase-bright tracks when they glide (12); however, their possible relationship to the aforementioned slime excretion has not been demonstrated. Since the chemical composition and ultrastructure of slime and slime trails are poorly understood, their possible relationship to OMTs is unknown.
Ramaswamy and colleagues described an M. xanthus mutant (SR200) that produces polar chain-like structures that are visible by TEM (16). Their structure had the same size and ultrastructure as the OMTs described here (Fig. 3A). However, in contrast to our findings, that team reported that their polar chains were present only in the SR200 mutant, not in wild-type cells or other mutants. Based on our findings, we suspect that those cells that were scored as negative were not grown in a static state for a long enough time for OMTs to be abundantly produced and easily detected by EM. The SR200 mutant was isolated based on its defect in calcofluor white binding and is defective in fibril/exopolysaccharide production. Consistent with their finding, we found that a dsp mutant overproduced OMTs (Fig. 1B), suggesting that fibrils may partly mask or block OMT production. In even earlier studies in the Shimkets and McCurdy labs, it was found that wild-type M. xanthus isolates produce polar structures that look identical to those reported here (13, 15, 33). Based on their TEM observations, McCurdy and colleagues called the structures polar LPS extrusions, although no direct evidence was provided that they consist of LPS.
The discovery of OMTs sparked our interest because they might provide mechanistic insight for how OM proteins and lipids are exchanged between cells (2, 4). In principle, OMTs could serve as conduits to fuse adjoining cells for OM content exchange. In eukaryotes, and to a lesser degree in bacteria, there is precedence for membrane tubes playing such a role (27, 34, 35). Furthermore, because OMTs are narrow (∼50 nm in diameter), their tips are highly curved, a topology that destabilizes lipids and promotes membrane fusion (36). In addition, we also found a correlation between cellular markers that labeled OMTs and those that were exchanged, whereas markers that did not label OMTs were not transferred.
Although the OMT model provides a structure to explain OM exchange, we found inconsistencies with this idea. First, conditions that promotes robust OMT production, i.e., static liquid growth (Fig. 1 to 3), actually block OM exchange. Therefore, the production of high levels of OMTs is clearly not sufficient to promote OM exchange (Fig. 7E and F). Second, TraA and TraB, which are absolutely required for OM exchange (2, 7), are not required for OMT production (Fig. 7C and D). Third, in a large-scale forward genetic screen for factors required for OM exchange, we did not identify any gene required for OMT production (A. Dey and D. Wall, unpublished data), indicating that these processes are distinct. Fourth, TraAB overexpression causes cells to directly adhere together independent of OMTs (2). Fifth, we have prepared crude OMT/OM vesicle preparations from a strain labeled with SSOM-mCherry and added them back to live M. xanthus cells under conditions that promote OM exchange (3, 20); however, SSOM-mCherry uptake by live cells or Cgl/Tgl stimulation did not occur (data not shown).
Recently, the Auer group generated vivid three-dimensional images that similarly show M. xanthus OMTs (OM vesicle chains) (14). Their studies are corroborative with our studies, although we note one difference made in the conclusions. Remis et al. concluded that OMTs are induced by biofilm formation, i.e., submerged culture growth (14). In contrast, we found that biofilm formation was not required. Instead, cells simply need to be in a static unstructured liquid environment for their production (Fig. 1 to 3). In addition, some of the strains that we used are pilus and/or fibril defective and, therefore, are defective in biofilm formation, yet they produced considerable amounts of OMTs (Fig. 1B).
After the original submission of the manuscript, the Mignot group reported the use of the SSOM-mCherry reporter to similarly visualize OM tubes in M. xanthus (37). By use of the described fluorescent lipid reporters and time-lapse video microscopy, they found that in one case, following direct cell-cell contact and lipid transfer, the cells physically separated from each other by gliding motility and appeared to produce an OMT connecting the cells. In addition, those authors suggested that the fluorescent lipid was weakly transferred from one cell to the other via the OMT. In contrast, they reported that they were unable to detect transfer of fluorescent proteins by OMTs, though they could visualize direct cell-to-cell transfer. For the reasons noted above, we suspect that if the interpretation of weak lipid transfer by an OMT was correct, it might be the by-product of myxobacteria spontaneously producing OMTs. Since OMTs are abundant, on occasion it is possible that cells become transiently connected by OMTs, which could allow transfer of limited material. Clearly, a key experiment to resolve whether OMTs are required for OM exchange would be to identify the cellular machinery involved in their production, assuming that specific components exist, and then to directly test whether OMT deficient mutants can or cannot undergo OM exchange.
For a number of reasons, during the course of our studies we began to wonder whether there are dedicated cellular components involved in OMT production. First, OMT production in M. xanthus resembles OM vesicle production in other Gram-negative bacteria, such as E. coli and P. aeruginosa, and to date, OM vesicle production appears not to involve the function of dedicated proteins, though small molecules can induce their formation, as can defects in OM-cell wall attachment (38, 39). Second, when cells were treated with various metabolic inhibitors (Fig. 6), we observed no block in OMT formation, suggesting that their production may occur independently of specific protein function. Thus, alternatively, OMTs might be the by-product of OM dynamics that sloughs off membrane material to perhaps relieve membrane stress (39). Third, in our pilot screen of candidate mutants, including those involved in OM biogenesis and extensively propagated lab strains, we found no strain that was notably defective in OMT production. Because OMT production results in a substantial loss of cellular material and OMT functions are presumably nonessential, we expected that if such mutants were possible, they would easily arise under laboratory propagation. Hopefully, by a combination of genetic and microscopy approaches, future studies will elucidate how OMTs are produced and what their role is in the biology of myxobacteria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Zhaojie Zhang for assistance with electron microscopy and Larry Shimkets for a strain.
This work was supported by NSF grant MCB-848141 and NIH grant GM101449 to D.W.
Footnotes
Published ahead of print 3 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00850-13.
REFERENCES
- 1.Pathak DT, Wei X, Wall D. 2012. Myxobacterial tools for social interactions. Res. Microbiol. 163:579–591. 10.1016/j.resmic.2012.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pathak DT, Wei X, Bucuvalas A, Haft DH, Gerloff DL, Wall D. 2012. Cell contact-dependent outer membrane exchange in myxobacteria: genetic determinants and mechanism. PLoS Genet. 8:e1002626. 10.1371/journal.pgen.1002626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, Pathak DT, Wall D. 2011. Heterologous protein transfer within structured myxobacteria biofilms. Mol. Microbiol. 81:315–326. 10.1111/j.1365-2958.2011.07710.x [DOI] [PubMed] [Google Scholar]
- 4.Nudleman E, Wall D, Kaiser D. 2005. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science 309:125–127. 10.1126/science.1112440 [DOI] [PubMed] [Google Scholar]
- 5.Wall D, Wu SS, Kaiser D. 1998. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J. Bacteriol. 180:759–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng TT, Tyler BM, Setubal JC. 2009. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9(Suppl 1):S2. 10.1186/1471-2180-9-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pathak DT, Wei X, Dey A, Wall D. 2013. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 9:e1003891. 10.1371/journal.pgen.1003891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wall D. 2014. Molecular recognition in myxobacterial outer membrane exchange: functional, social and evolutionary implications. Mol. Microbiol. 91:209–220. 10.1111/mmi.12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konovalova A, Petters T, Sogaard-Andersen L. 2010. Extracellular biology of Myxococcus xanthus. FEMS Microbiol. Rev. 34:89–106. 10.1111/j.1574-6976.2009.00194.x [DOI] [PubMed] [Google Scholar]
- 10.Yu R, Kaiser D. 2007. Gliding motility and polarized slime secretion. Mol. Microbiol. 63:454–467. 10.1111/j.1365-2958.2006.05536.x [DOI] [PubMed] [Google Scholar]
- 11.Dworkin M. 1999. Fibrils as extracellular appendages of bacteria: their role in contact-mediated cell-cell interactions in Myxococcus xanthus. Bioessays 21:590–595. [DOI] [PubMed] [Google Scholar]
- 12.Burchard RP. 1982. Trail following by gliding bacteria. J. Bacteriol. 152:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnold JW, Shimkets LJ. 1988. Cell surface properties correlated with cohesion in Myxococcus xanthus. J. Bacteriol. 170:5771–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. 2014. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ. Microbiol. 16:598–610. 10.1111/1462-2920.12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacRae TH, McCurdy D. 1976. Evidence for motility-related fimbriae in the gliding microorganism Myxococcus xanthus. Can. J. Microbiol. 22:1589–1593. 10.1139/m76-234 [DOI] [PubMed] [Google Scholar]
- 16.Ramaswamy S, Dworkin M, Downard J. 1997. Identification and characterization of Myxococcus xanthus mutants deficient in calcofluor white binding. J. Bacteriol. 179:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu W, Yang Z, Lux R, Zhao M, Wang J, He X, Shi W. 2012. Direct visualization of the interaction between pilin and exopolysaccharides of Myxococcus xanthus with eGFP-fused PilA protein. FEMS Microbiol. Lett. 326:23–30. 10.1111/j.1574-6968.2011.02430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducret A, Valignat MP, Mouhamar F, Mignot T, Theodoly O. 2012. Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc. Natl. Acad. Sci. U. S. A. 109:10036–10041. 10.1073/pnas.1120979109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wall D, Kolenbrander PE, Kaiser D. 1999. The Myxococcus xanthus pilQ (sglA) gene encodes a secretin homolog required for type IV pilus biogenesis, social motility, and development. J. Bacteriol. 181:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall D, Kaiser D. 1998. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc. Natl. Acad. Sci. U. S. A. 95:3054–3058. 10.1073/pnas.95.6.3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zusman DR, Krotoski DM, Cumsky M. 1978. Chromosome replication in Myxococcus xanthus. J. Bacteriol. 133:122–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathak DT, Wall D. 2012. Identification of the cglC, cglD, cglE, and cglF genes and their role in cell contact-dependent gliding motility in Myxococcus xanthus. J. Bacteriol. 194:1940–1949. 10.1128/JB.00055-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahnt J, Aguiluz K, Koch J, Treuner-Lange A, Konovalova A, Huntley S, Hoppert M, Sogaard-Andersen L, Hedderich R. 2010. Profiling the outer membrane proteome during growth and development of the social bacterium Myxococcus xanthus by selective biotinylation and analyses of outer membrane vesicles. J. Proteome Res. 9:5197–5208. 10.1021/pr1004983 [DOI] [PubMed] [Google Scholar]
- 24.Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. 2012. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology 158:2742–2752. 10.1099/mic.0.060343-0 [DOI] [PubMed] [Google Scholar]
- 25.Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10. 10.1046/j.1365-2958.1999.01339.x [DOI] [PubMed] [Google Scholar]
- 26.McCaig WD, Koller A, Thanassi DG. 2013. Production of outer membrane vesicles and outer membrane tubes by Francisella novicida. J. Bacteriol. 195:1120–1132. 10.1128/JB.02007-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600. 10.1016/j.cell.2011.01.015 [DOI] [PubMed] [Google Scholar]
- 28.Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. 2011. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS One 6:e20725. 10.1371/journal.pone.0020725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galkina SI, Romanova JM, Bragina EE, Tiganova IG, Stadnichuk VI, Alekseeva NV, Polyakov VY, Klein T. 2011. Membrane tubules attach Salmonella Typhimurium to eukaryotic cells and bacteria. FEMS Immunol. Med. Microbiol. 61:114–124. 10.1111/j.1574-695X.2010.00754.x [DOI] [PubMed] [Google Scholar]
- 30.Kearns DB, Campbell BD, Shimkets LJ. 2000. Myxococcus xanthus fibril appendages are essential for excitation by a phospholipid attractant. Proc. Natl. Acad. Sci. U. S. A. 97:11505–11510. 10.1073/pnas.210448597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolgemuth C, Hoiczyk E, Kaiser D, Oster G. 2002. How myxobacteria glide. Curr. Biol. 12:369–377. 10.1016/S0960-9822(02)00716-9 [DOI] [PubMed] [Google Scholar]
- 32.Fluegel W. 1963. Simple method for demonstrating myxobacterial slime. J. Bacteriol. 85:1173–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobson WJ, McCurdy HD, MacRae TH. 1979. The function of fimbriae in Myxococcus xanthus. II. The role of fimbriae in cell-cell interactions. Can. J. Microbiol. 25:1359–1372 [DOI] [PubMed] [Google Scholar]
- 34.Davis DM, Sowinski S. 2008. Membrane nanotubes: dynamic long-distance connections between animal cells. Nat. Rev. Mol. Cell Biol. 9:431–436. 10.1038/nrm2399 [DOI] [PubMed] [Google Scholar]
- 35.Rechavi O, Goldstein I, Kloog Y. 2009. Intercellular exchange of proteins: the immune cell habit of sharing. FEBS Lett. 583:1792–1799. 10.1016/j.febslet.2009.03.014 [DOI] [PubMed] [Google Scholar]
- 36.Wickner W, Schekman R. 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15:658–664. 10.1038/nsmb.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducret A, Fleuchot B, Bergam P, Mignot T. 2013. Direct live imaging of cell-cell protein transfer by transient outer membrane fusion in Myxococcus xanthus. eLife 2:e00868. 10.7554/eLife.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schertzer JW, Whiteley M. 2012. A bilayer-couple model of bacterial outer membrane vesicle biogenesis. mBio 3:e00297–11. 10.1128/mBio.00297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


