Abstract
Protein kinase D (PKD) signaling plays a critical role in the regulation of DNA synthesis, proliferation, cell survival, adhesion, invasion/migration, motility, and angiogenesis. To date, relatively little is known about the potential role of PKD in the development and/or progression of human colorectal cancer (CRC). We evaluated the expression of different PKD isoforms in CRC and investigated the antitumor activity of PKD inhibitors against human CRC. PKD2 was the dominant isoform expressed in human colon cancer cells. PKD3 expression was also observed but PKD1 expression, at both the RNA and protein levels, was not detected. Suppression of PKD using the small molecule inhibitors, CRT0066101 and kb-NB142-70, resulted in low micromolar in vitro antiproliferative activity against multiple human CRC cell lines. Drug treatment was associated with dose-dependent suppression of PKD2 activation. Incubation with CRT0066101 resulted in G2/M phase arrest and induction of apoptosis in human CRC cells. Further studies showed that CRT0066101 treatment gave rise to a dose-dependent increase in expression of cleaved PARP and activated caspase-3, in addition to inhibition of AKT and ERK signaling, and suppression of NF-κB activity. Transfection of PKD2-targeted siRNAs resulted in similar effects on downstream pathways as observed with small molecule inhibitors. Daily administration of CRT0066101 resulted in significant inhibition of tumor growth in HCT116 xenograft nude mice. Taken together, our studies show that PKD plays a significant role in mediating growth signaling in CRC and may represent a novel chemotherapeutic target for the treatment of CRC.
Keywords: protein kinase D, human colorectal cancer, apoptosis, NF-κB, chemotherapy target
Introduction
Colorectal cancer (CRC) is a major public health problem in the U.S. and globally. In the U.S., it is the second leading cause of cancer mortality (1). For 50 years, the main chemotherapeutic treatment was the fluoropyrimidine 5-fluorouracil (5-FU). From 1996 to 2004, 3 new anticancer agents were approved, which include the oral fluoropyrimidine capecitabine, the topoisomerase I inhibitor irinotecan, and the platinum analog oxaliplatin. Since 2004, the FDA has approved 5 molecular targeting agents, including the anti-epidermal growth factor receptor antibodies cetuximab and panitumumab, the anti-vascular endothelial growth factor inhibitors bevacizumab and ziv-aflibercept, and a small molecule inhibitor that targets multiple tyrosine kinases, regorafenib. Significant advances have been made in chemotherapy treatment options for patients with metastatic disease, such that improvements in 2-year survival are now being reported, with median overall survival rates of 21–24 months (2–4). Despite these advances, none of the currently available treatment options have impacted on 5-year overall survival, and cellular drug resistance remains a significant obstacle to successful chemotherapy (5). Thus, identification of novel signaling pathways and targets that mediate the growth and proliferation of CRC is critically important for discovering and developing novel therapeutic agents with enhanced antitumor activity that overcome drug resistance and improve overall quality of life.
Protein kinase D (PKD) is a subfamily of serine/threonine kinases of the calcium/calmodulin-dependent kinase superfamily, comprised of 3 different isoforms, PKD1, PKD2, and PKD3 (6). This signaling pathway functions downstream of protein kinase C (PKC), G protein coupled receptors, and tyrosine kinase receptors. PKD can be activated in a PKC-dependent as well as a PKC-independent manner. In turn, activated PKD phosphorylates a wide range of downstream targets at specific sites, subsequently regulating their activity and/or subcellular localization (7). It was been well-documented that PKD plays a critical role in the regulation of several important cellular processes, such as DNA synthesis, proliferation, cell survival, adhesion, invasion/migration, motility, and angiogenesis (8). Moreover, PKD signaling has been implicated in several human tumors, including breast, pancreatic, and prostate cancer, and glioblastoma. For example, PKD2 and PKD3 appear to be highly expressed in breast cancer (9). PKD1 expression level is elevated in human ductal adenocarcinoma of the pancreas compared to normal pancreatic tissues (10). PKD2 has been shown to be an important mediator of induction of various angiogenic factors in human pancreatic cancer cells and in the angiogenic response of the host vasculature (11). Expression of PKD1 and PKD3 has been shown to be elevated in human prostate carcinoma tissue compared to normal prostate epithelial tissue, and advanced stage tumors were found to have increased PKD3 nuclear accumulation (12). PKD2 was also highly expressed in both low-grade and high-grade human gliomas (13).
Given the importance of PKD in tumor biology, investigators have focused on developing novel small molecule inhibitors targeting PKD. Several such agents with anticancer activity have been identified, including CID755673, kb-NB142-70 and CRT0066101 (14–16). CID755673 is a non-ATP competitive pan-PKD inhibitor that was discovered in a high throughput screening campaign. The IC50 of CID755673 for PKD1, PKD2, and PKD3 was 182, 280, and 227 nmol/L, respectively, using an established radiometric kinase assay. This compound blocked PMA-induced activation of PKD1 in human prostate cancer LNCaP cells. Moreover, CID755673 displayed inhibitory effects on cell proliferation, migration, and invasion in prostate cancer cells (16). A series of structural analogs of CID755673 was subsequently developed, including kb-NB142-70, kb-NB165-09, kb-NB165-31, kb-NB165-92, and kb-NB184-02. These agents exhibited at least 2-fold higher potency and improved kinase selectivity when compared to the parent compound (15). The most potent analog, kb-NB142-70, inhibited PKD1, PKD2, and PKD3 enzymatic activity with an IC50 of 28, 59, and 53 nmol/L, respectively. kb-NB142-70 also significantly inhibited cell proliferation, migration, and invasion. Unfortunately, kb-NB142-70 did not exhibit in vivo antitumor activity due to rapid metabolism (17). CRT0066101 is a small molecule PKD-specific inhibitor developed by investigators in the U.K., and it exhibited in vitro antitumor activity in human pancreatic cancer cells. CRT0066101 significantly suppressed neurotensin-induced PKD1/2 activation, blocked NF-κB mediated cellular proliferation and survival, and induced apoptosis. Moreover, CRT0066101 inhibited Panc-1 cell growth in in vivo xenograft mouse models (14). In addition to CID755673, kb-NB142-70 and CRT0066101, several other pan-PKD inhibitors have been reported in the literature (18, 19).
In the present study, we investigated PKD isoform expression in CRC, evaluated the therapeutic efficacy of targeting PKD in human CRC, and determined its potential molecular mechanisms of action. We present both in vitro and in vivo evidence showing that CRT0066101 has cytotoxic as well as antitumor activity against human CRC model systems. These findings provide evidence that PKD may represent a potential target for CRC chemotherapy.
Materials and Methods
Chemicals and reagents
CRT0066101 was kindly provided by Dr. Sushovan Guha and Cancer Research Technology Inc. For in vitro use, the drug was resuspended in dimethyl sulfoxide (DMSO, Sigma, USA) while it was resuspended in 5% sterile dextrose solution for in vivo studies. CID755673 and kb-NB142-70 were synthesized as previously described (15). The DMSO concentration never exceeded 0.1% in any experiment. This dose had no effect on cell growth nor did it affect protein expression. WST-1 was purchased from Roche Diagnostics (Indianapolis, IN). Phorbol 12-myristate 13-acetate (PMA) and other chemicals were obtained from Sigma. The following siRNAs were synthesized by Dharmacon Research (ThermoScientific; Lafayette, CO): siPKD2 – 5′-UGAGACACCUUCACUUCAA-’3 (#D-004197-05); siPKD3 – 5′-GGGAGAGUGUUACCAUUGA-’3 (#D-005029-06); siCon - 5′-GGAUACUGCCAAUCUCUAGG-’3.
Tissue culture and human CRC cell lines
Normal human colon epithelial CCD 841 CoN and FHC cell lines and the human cancer RKO cell line were obtained from ATCC. HCT116 p53 (+/+) and p53 (−/−) cell lines were kindly provided by Dr. Bert Vogelstein (20). H630 and H630R1 cells have been maintained in our laboratory after being originally obtained from Dr. Adi Gazdar (21). All cell lines, with the exception of FHC, were maintained in RPMI-1640 (Invitrogen; Carlsbad, CA) with 10% (v/v) fetal bovine serum at 37°C in a humidified incubator with 5% CO2. FHC cells were maintained according to ATCC guidelines. HCT116 and RKO cells were authenticated by STR profiling at the University of Pittsburgh Cell Culture and Cytogenetics Facility (August 2013). Cells were tested monthly for mycoplasma by the MycoAlert Mycoplasma detection assay (Cambrex BioScience).
Cell viability assay
Human CRC cells were plated in 96-well plates at a density of 800–1500 cells/well. On the following day, cells were incubated with various concentrations of PKD inhibitors for 72 hours. Cell viability was determined by the WST-1 assay. The IC50 value was defined as the drug concentration that inhibits 50% cell growth compared with the untreated controls and calculated by Graphpad Prism 6.0 software.
Clonogenic assay
HCT116 and RKO cells were seeded in 6-well plates at density of 400 cells/well. On the following day, cells were exposed to various concentrations of CRT0066101 for 24 hours, after which time, the growth medium was then replaced. After 10–14 days, cell colonies were fixed with trypan blue solution (75% methanol/25% acetic acid/0.25% trypan blue) for 15 minutes, washed, and air-dried before counting colonies >50 cells.
siRNA transfection
Cells were plated at a density of 2 × 105 cells/well. On the following day, siRNAs (10 nM) were complexed with Lipofectamine2000 (LF; Invitrogen) in serum-free RPMI-1640 medium and added to the plated cells. After 48 hours, cells were processed for Western blot analysis or for flow cytometry.
Western blot analysis
Cell lysate protein concentrations were determined using the DC Protein Assay (Bio-Rad; Hercules, CA). Equal amounts of protein (30 μg) from each cell lysate were resolved on SDS-PAGE using the method of Laemmli and transferred onto 0.45 μm nitrocellulose membranes (Bio-Rad). Membranes were blocked and incubated overnight with primary antibodies at 4°C. The following antibodies were used in the experiments: anti-p-PKD2 ((Ser876) #07-385; Upstate), anti-PKD2 (#07-488, Upstate), anti-PKD1 (gift from Dr. Peter Storz), anti-PKD3 (#5655; Cell Signaling), anti-p-ERK (#sc-7383; Santa Cruz Biotechnology), anti-ERK (#sc-94; Santa Cruz Biotechnology), anti-p-AKT (#9542; Cell Signaling), anti-AKT (#9272; Cell Signaling), anti-PARP (#9542; Cell Signaling), anti-cleaved caspase-3 (#9661; Cell Signaling), anti-GAPDH (#47724; Santa Cruz Biotechnology), anti-β-actin (#4970; Cell Signaling) and anti-α-tubulin (EMD Biosciences). Detection of β-actin, GAPDH, or α-tubulin was routinely used as protein loading controls. After multiple TBST washes (1×TBS, 0.1% Tween-20), membranes were incubated with corresponding horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) for 1 hour at room temperature. Proteins were detected by the enhanced chemiluminescence method (SuperSignal West Pico substrate; Pierce; Rockford, IL). Quantitation of signal intensities was performed by densitometry on a Xerox scanner using ImageJ software.
RNA extracts and real-time qRT-PCR analysis
Total RNA was extracted by the guanidine isothiocyanate/phenol/chloroform method (Trizol; Invitrogen). The integrity and purity of the RNA was determined by UV spectrophotometry at OD260/OD280. The first-strand cDNA was synthesized using 1.0 μg total RNA and the iScript™ Reverse Transcription Supermix for real-time quantitative polymerase chain reaction (qRT-PCR) (Bio-Rad; Hercules, CA). PCR was performed in triplicate using the SsoFast™ Probes Supermix (Bio-Rad) in a final reaction volume of 10 μL with gene-specific primer/probe sets, and a standard thermal cycling procedure (40 cycles) on a Bio-Rad CFX96™ Real-Time PCR System. PKD1, PKD2, PKD3 and 18S RNA levels were assessed using TaqMan Gene Expression real-time PCR assays (Applied Biosystems; assay ID: Hs00177037_m1, ID: Hs00212828_m1, ID: Hs00178657_m1 and Hs03928990_g1, respectively). A cDNA array of 24 human colon tumors (Colon Cancer cDNA Array III; Origene, Rockville, MD) was also analyzed for PKD isoform expression (normalized by β-actin RNA levels; assay ID: Hs01060665_g1). Results were expressed as the threshold cycle (Ct). The relative quantification of the target transcripts was determined by the comparative Ct method (ΔΔCt) according to the manufacturer’s protocol. The 2−ΔΔCt method was used to analyze the relative changes in gene expression. Control experiments were conducted without reverse transcription to confirm that the total RNA was not contaminated with genomic DNA.
Flow cytometry
HCT116 and RKO cells were seeded in 6-well plates at a density of 4×105 cells/well. After exposure to CRT0066101 for 24 hours, cells were harvested, washed twice with PBS, resuspended with 1×binding buffer, stained with FITC Annexin V Apoptosis Detection Kit (BD Biosciences), and analyzed on the BD Accuri C6 Flow Cytometer (BD Accuri Cytometers Inc.) at the UPCI Cytometry Facility. For cell cycle analysis, cells were washed with PBS and fixed with 70% ethanol overnight. Cells were washed with PBS, and then treated with 1 mg/mL RNase A for 30 minutes at 37°C. Cells were incubated with propidium iodide (1 mg/mL) for 45 minutes before detection by flow cytometry.
NF-κB activity assay
HCT116 and RKO cells were seeded in 24-well plates at a density of 1×105 cells/well. On the following day, pGL3-Luc-NF-κB or pGL3-Luc DNA (0.5 μg) was co-transfected with 0.1 μg of Renilla luciferase plasmid DNA into cells using Lipofectamine 2000 according to the manufacturer’s protocol (Invitrogen). The Renilla luciferase plasmid DNA was used as an internal control for transfection efficiency. After 6 hours, transfection medium was changed, and on the following day, cells were incubated for 2 hours with various concentrations of PKD inhibitors followed by addition of 50 ng/mL TNF-α for an additional 5 hours. Firefly luciferase values were normalized with renilla activity and the reporter assays were performed in triplicate. To generate RKO cells that stably express NF-κB-driven luciferase, RKO cells were transfected with pGL3-Luc-NF-κB. After 48 hours, 400 ug/mL hygromycin B was added to the cells. After two weeks of antibiotic selection, the heterogeneous RKO cells were incubated for 2 hours with various concentrations of PKD inhibitors followed by the presence or absence of 100 nM PMA for an additional 5 hours. Firefly luciferase activity was then determined as described above and was normalized to total soluble protein.
Xenograft mouse model experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. HCT116 cells (<70% confluent) were harvested, and 5×106 cells in 0.1 mL of medium were implanted subcutaneously on the back of athymic nude female mice. When the tumor size reached approximately 100 mm3, mice were randomized into the following groups (5 mice per group): (A) control (vehicle; 5% dextrose); (B) 40 mg/kg, (C) 80 mg/kg and (D) 120 mg/kg CRT0066101 (dissolved in 5% dextrose) administered orally once daily. Therapy was administered for 3 weeks, and animals were sacrificed on day 21 after treatment with CRT0066101. Tumor volume was measured as V = 1/2ab2, in which “a” and “b” represents length and width of tumor (22). Tumor volumes were monitored 3 times per week. At the time the animals were euthanized, half of the tumor tissue was fixed with formalin and paraffin-embedded for immunohistochemistry. The other half was snap-frozen in liquid nitrogen and stored at −80°C. Tissue slides were processed by the Department of Pathology Development Laboratory and the Tissue and Research Pathology Services at the University of Pittsburgh for Ki-67 (#9027; Cell Signaling), in situ TUNEL staining (APOPTAG Peroxidase kit; Chemicon), p-ERK (#4370; Cell Signaling), and M30 (#12140322001; Roche).
Statistical analysis
Data were expressed as the mean ± SD. Statistical analysis of the data was performed using one-way ANOVA (SPSS software). P<0.05 was considered statistically significant.
Results
PKD expression in CRC
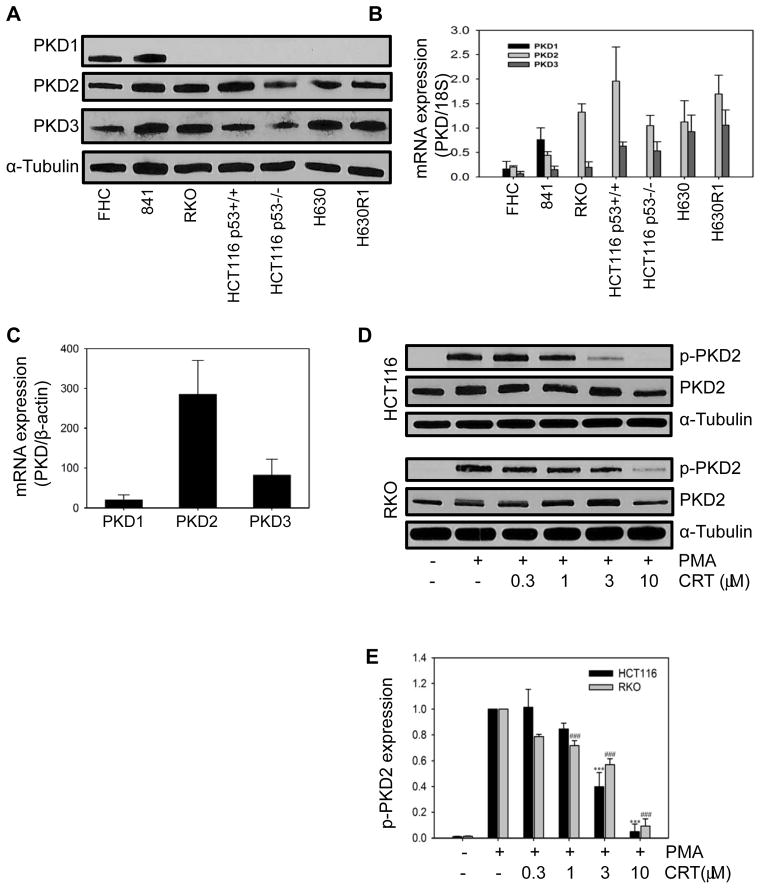
We analyzed the expression of the 3 different PKD isoforms in two normal epithelial colon cell lines (FHC and 841) and three human CRC cancer cell lines. As shown in Fig. 1A and B, PKD1 was only expressed in normal colon cells and not in CRC cells. In contrast, PKD2 and PKD3 were expressed, at the protein and mRNA level, in all cell lines. To provide further support for the differential level of expression of the respective PKD RNA isoforms in human CRC, we used the CellMiner web-based program to interrogate RNA transcript patterns in the NCI-60 cell panel (23). The 7 human CRC cell lines in the panel are COLO205, HCC2998, HCT116, HCT15, HT29, KM12, and SW620. The level of PKD1 RNA expression was significantly reduced below the median transcript expression (Z score =0) in all cell lines, while PKD3 RNA expression was somewhat more variable (Fig. S1). In sharp contrast, expression of PKD2 RNA was elevated in 5 of 7 cell lines. A search of the Broad-Novartis Cancer Cell Line Encyclopedia database (24), which contains genetic information on 1,000 cell lines, revealed that human CRC cell lines express the lowest PKD1 mRNA levels among all the cell lines (Fig. S2). PKD2 expression in the 61 CRC cell lines was slightly higher than the mean mRNA levels (8.36 versus 8.26; RMA, log2) while PKD3 expression was somewhat lower than the mean (8.07 versus 8.42; RMA, log2). To provide further support for the potential clinical relevance of PKD expression, we performed qPCR analysis on cDNAs obtained from 24 human colon tumors (Origene cDNA array). After estimation of all PKD isoforms, we observed a nearly identical RNA expression pattern with PKD2 being the dominantly expressed isoform (Fig. 1C). Taken together, this data suggests that PKD2 is the most abundant isoform in CRC.
Fig. 1. Expression of PKD in human CRC cell lines.
Normal colon epithelial cells and CRC cells were harvested and processed for Western blot analysis (A) and qRT-PCR analysis (B). (C) qRT-PCR analysis of cDNAs from 24 human colon cancers. (D) Cells were incubated with CRT0066101 for 24 hours, followed by stimulation with 100 nmol/L PMA for 30 minutes. Cells were immediately processed for Western blot analysis. (E) Expression values represent the mean ± SD from 3 individual experiments. Densitometry measurements of p-PKD2 after treatment with PMA alone were normalized to 1.
We next investigated the effect of pan-PKD inhibitors on PKD activation in the human CRC cell lines. Given the high level of PKD2 expression in HCT116 and RKO and the availability of a specific p-PKD2 antibody, we monitored phosphorylation of the PKD2 isoform by Western blot analysis. Cells were incubated with PKD inhibitors followed by stimulation with PMA, a known activator of the PKC/PKD pathway. As shown in Fig. 1D and E, treatment with CRT0066101 significantly inhibited PKD2 phosphorylation in a dose-dependent manner. The highest concentration of CRT0066101 (10 μmol/L) almost completely blocked PKD2 activation in HCT116 and RKO cells. The concentration that inhibited 50% of PKD2 activation was 2 and 3 μmol/L, respectively. Treatment of RKO cells with a different PKD inhibitor, kb-NB142-70, also decreased p-PKD2 levels, but to a much lesser extent than CRT0066101 (Fig. S3A; IC50, 38 μmol/L).
Effect of PKD inhibition on growth of human CRC cells
To determine the potential impact of PKD suppression on cell growth, a panel of CRC cell lines was treated with different PKD inhibitors. Cells were exposed to CRT0066101, CID755673, and kb-NB142-70 for 72 hours, and cell proliferation was determined by the WST-1 assay. CRT0066101 exhibited low μmol/L IC50 values against all CRC cell lines in the panel (Table 1). kb-NB142-70 had similar inhibitory effects on cell growth albeit with slighter higher IC50 values. In contrast, CID755673 was significantly less potent, as had been shown previously observed with different human cancer model systems (16). Growth inhibition was similar in both p53+/+ and p53−/− HCT116 cells suggesting that the cytotoxic effects of these molecules are mediated through p53-independent pathways. These compounds were also able to maintain their inhibitory activity in cells that are resistant to both chemotherapy and radiation therapy. Of note, the 5-FU-resistant H630R1 cells were as sensitive to the PKD inhibitors as parental H630 cells. In addition to the WST-1 assay, we utilized the clonogenic assay to determine the effect of CRT0066101 on HCT116 and RKO clonogenic growth. CRT0066101 effectively decreased colony number in a dose-dependent manner (Fig. S4).
Table 1.
Effect of PKD Inhibitors on Cell Proliferation
| Cell Lines | PKD Inhibitor IC50 (μmol/L)
|
||
|---|---|---|---|
| CRT0066101 | kb-NB142-70 | CID755673 | |
| RKO | 0.90 ± 0.17 | 2.82 ± 0.67 | 24.68 ± 5.50 |
| HCT116 (p53+/+) | 0.77 ± 0.25 | 2.69 ± 0.07 | 15.51 ± 2.55 |
| HCT116 (p53−/−) | 1.01 ± 0.06 | 3.08 ± 0.38 | 17.52 ± 1.80 |
| H630 | 1.28 ± 0.18 | 4.33 ± 2.18 | 46.7 ± 27.65 |
| H630R1 | 1.56 ± 0.34 | 8.35 ± 4.31 | 35.28 ± 5.42 |
IC50 values represent the concentration of drug that suppressed cell growth by 50%.
Values represent the mean ± S.D. from 3–5 determinations.
Effect of PKD inhibition on cell cycle distribution and apoptosis
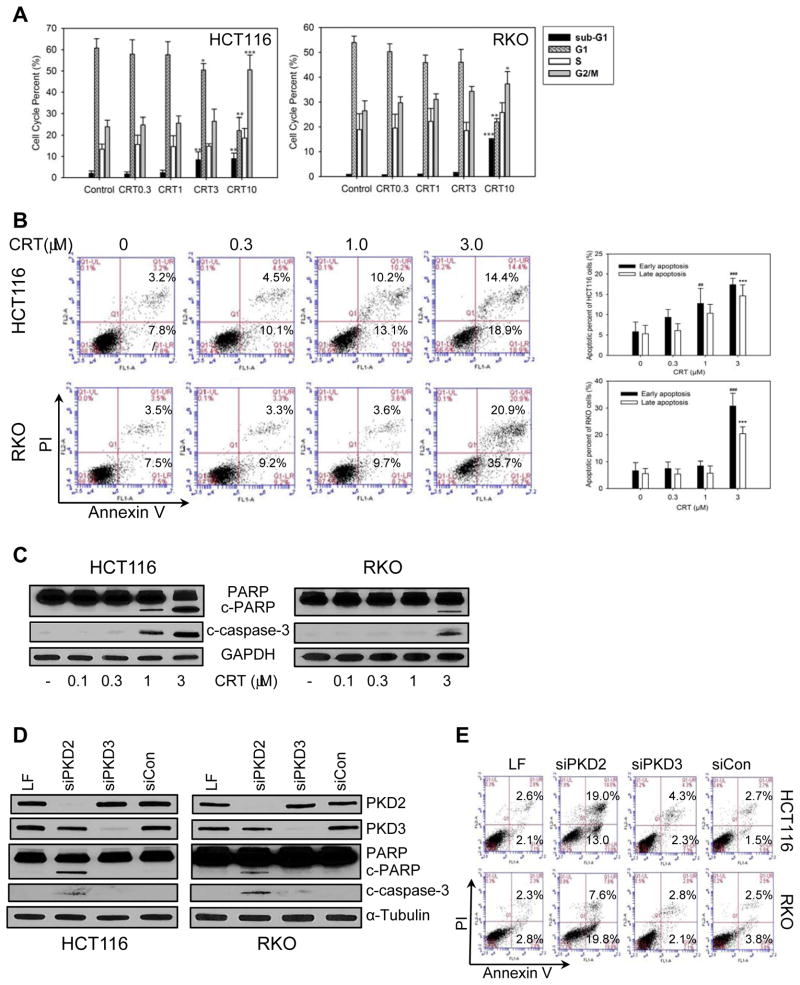
Given the growth inhibitory activity of CRT0066101 on human CRC cells, we evaluated the potential mechanisms of action of PKD inhibition in these cells. To determine the effect of CRT0066101 on distribution of the cell cycle, HCT116 and RKO cells were treated with CRT0066101 for 24 hours, stained with PI, and analyzed by flow cytometry. As shown in Fig. 2A, CRT0066101 blocked cell cycle progression at the G2/M phase in a dose-dependent manner. Similar results were obtained with kb-NB142-70 (Fig. S3 B and C). This block at the G2/M checkpoint coincided with a decrease in the fraction of cells in the G1 population. We also observed a significant increase in the sub-G1 phase in both cell lines suggesting that CRT0066101 may induce apoptosis of human CRC cells. To further investigate the potential impact of this agent on inducing apoptosis, cells were treated with CRT0066101 for 24 hours, stained with FITC-Annexin V and PI and analyzed by flow cytometry. As shown in Fig. 2B, CRT0066101 significantly induced apoptosis in both cell lines in a dose-dependent manner. A different molecule kb-NB142-70 induced slightly lower levels of apoptosis (Fig. S3 D and E). We then investigated the effect of drug treatment on the expression of other markers of apoptosis. As determined by Western blot analysis, CRT0066101 treatment (1–3 μmol/L) resulted in cleavage of both PARP and caspase-3, respectively (Fig. 2C). Since the PKD inhibitors suppress all PKD isoforms, we transfected isoform-specific siRNAs into CRC cells to identify which isoform might be responsible for the growth inhibitory activity of the small molecule inhibitors. Each siRNA specifically and potently suppressed only the intended targeted isoform (Fig. 2D). Knockdown of PKD2, but not PKD3, resulted in cleavage of PARP and caspase-3. Further, transfection of PKD2 siRNA induced apoptosis as determined by flow cytometry whereas PKD3 siRNA transfection had no effect on Annexin V/PI staining (Fig. 2E).
Fig. 2. Effect of PKD inhibition on cell cycle distribution and apoptosis.
HCT116 and RKO cells were incubated with various concentrations of CRT0066101 (0.3–10 μmol/L) for 24 hours. (A) Cell cycle distribution was detected by flow cytometry. Cycle percentages represent the mean ± SD from 3 individual experiments using HCT116 and RKO cells. (B) Apoptotic cells were detected by flow cytometry. Apoptotic cell percentages represent the mean ± SD from 3 individual experiments. (C) Expression of cleaved-PARP and cleaved caspase-3 was determined by Western Blot analysis after 24 hours exposure to CRT0066101. ##: p<0.01, ###:<0.001 early apoptosis vs. control, ***: p<0.001 late apoptosis vs. control. (D) Cells were transfected with 10 nM siRNAs for 48 hours, and expression of cleaved PARP and caspase-3 was analyzed by Western blot. (E) Cells were transfected with 10 nM siRNAs for 48 hours and processed for apoptotic cell detection by flow cytometry.
Effect of CRT0066101 on key cellular survival pathways
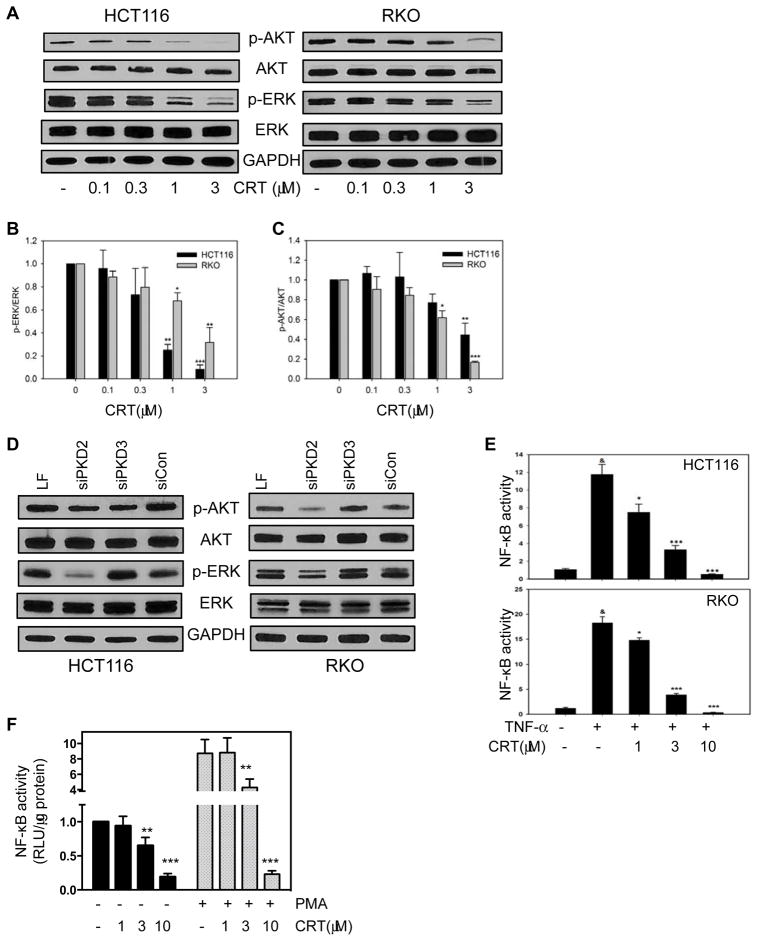
The PI3K-AKT, MAPK, and NF-κB signaling pathways are well-established survival pathways in human CRC. Constitutive activation of each of these pathways, as a result of point mutations, has been shown to increase cancer cell proliferation and drug resistance (25). It has also been demonstrated that each of these pathways are downstream of PKD signaling (26). We next determined whether any of these pathways might represent active downstream components of PKD signaling in CRC cancer cells. We treated human HCT116 and RKO cells with CRT0066101 for 24 hours, and then determined expression of p-AKT, p-ERK, total AKT and total ERK by Western blot analysis. CRT0066101 treatment resulted in a significant reduction in expression of p-AKT and p-ERK, with no effect on total AKT and ERK protein levels (Fig. 3A–C). Transfection of PKD2 siRNA resulted in a similar decrease in p-AKT expression in RKO cells and a decrease in p-ERK expression in both cell lines (Fig. 3D). In contrast, treatment with the PKD3 siRNA did not alter these pathways. We next detected the effect of PKD inhibition on NF-κB activity. HCT116 and RKO cells were transiently transfected with a luciferase plasmid under the control of the NF-κB response element. After incubation of PKD inhibitors for 2 hours, cells were stimulated by TNF-α (50 ng/mL) for an additional 5 hours. NF-κB activity was determined by the dual luciferase assay. As shown in Fig. 3E, CRT0066101 effectively suppressed, in a dose-dependent manner, the activation of NF-κB. Significant inhibition was observed after treatment with 1 μmol/L CRT0066101. In contrast, higher doses of kb-NB142-70 were required to obtain similar suppression of NF-κB activity (Fig. S3F). To determine whether PKD suppression can alter basal expression of NF-κB, RKO cells were stably transfected with the NF-κB-driven luciferase plasmid. Addition of CRT0066101 to these cells in the absence of an inducer resulted in a dose-dependent inhibition of basal NF-κB activity (Fig. 3F). While TNF-α activates the NF-κB pathway, it does not directly induce the PKC/PKD pathway (data not shown). Thus, cells pre-incubated with CRT0066101 were stimulated with PMA, a known PKD stimulator. As shown in Fig. 3F, PMA-induced NF-κB activation was suppressed with the PKD inhibitor. Taken together, our findings suggest that PKD inhibition and in particular PKD2, results in suppression of key signaling pathways in human CRC, including AKT, ERK, and NF-κB.
Fig. 3. Effect of CRT0066101 on downstream cell survival pathways.
(A) Cells were exposed to CRT0066101 for 24 hours, and protein expression was determined by Western blot analysis. (B) Semi-quantitative analysis of Western blots for p-ERK/total ERK. (C) Semi-quantitative analysis of Western blots for p-AKT/total AKT. Values represent the mean ± S.D. from at least 3 individual blots. (E) HCT116 and RKO cells were transiently transfected with a luciferase plasmid under the control of a NF-κB response element. On the following day, cells were treated with CRT0066101 for 2 hours, and then stimulated with TNF-α for an additional 5 hours. NF-κB activity was determined by the dual-luciferase assay. (F) RKO cells stably expressing NF-κB-luciferase were treated with CRT0066101 for 2 hours, and then stimulated with PMA (100 nM) for an additional 5 hours. Luciferase values represent the mean ± S.D. from 3–5 separate experiments. *: p<0.05, **: p<0.01, ***: p<0.001 vs. untreated cells (B, C), TNF-α stimulated cells (E), unstimulated cells (-PMA) (F), or PMA-stimulated cells (F). &: p<0.001 vs untreated cells (E).
Antitumor activity of PKD small molecule inhibitors
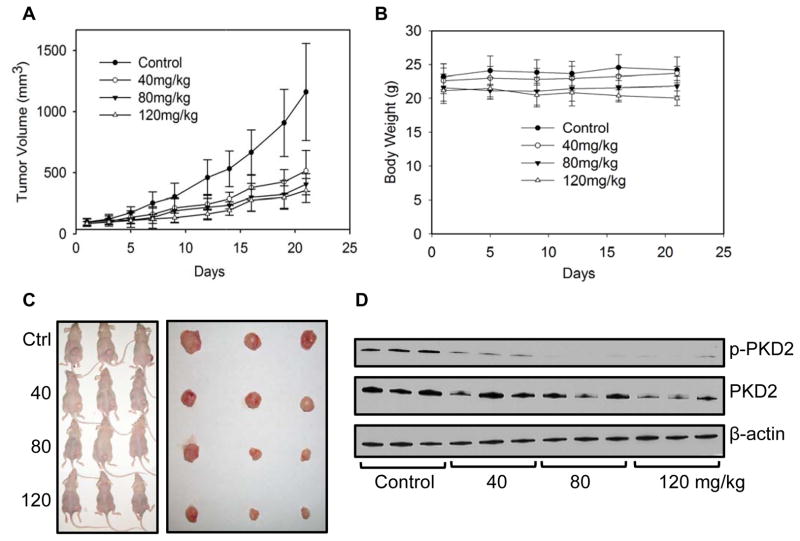
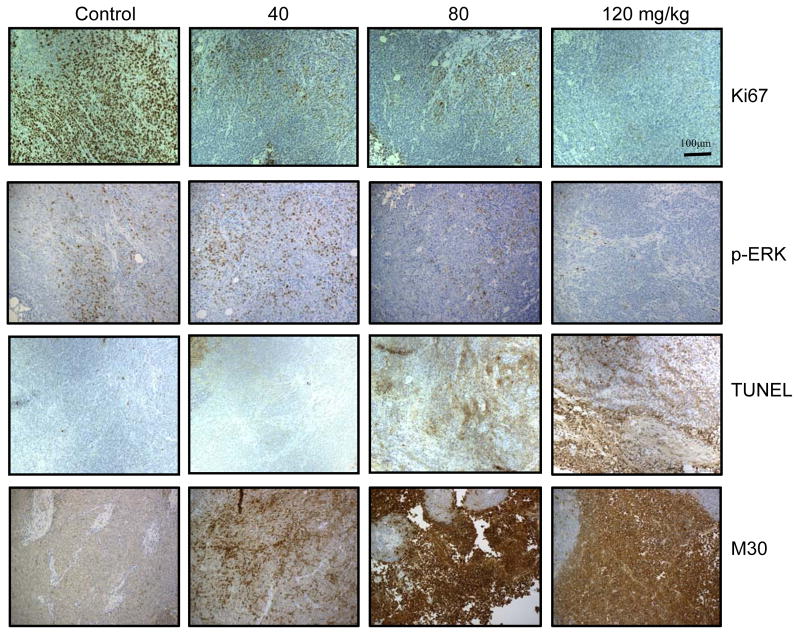
Our in vitro results suggest that PKD plays an active role in CRC growth and proliferation. As CRT0066101 is an orally bioavailable PKD inhibitor, we further evaluated the in vivo antitumor activity of CRT0066101 using HCT116 tumor-bearing athymic nude mice. Mice were administered different oral doses of CRT0066101 (40 mg/kg, 80 mg/kg, and 120 mg/kg) once daily for 3 weeks. At the end of the 21-day treatment period, tumor volume was decreased 55.6%, 65.2%, and 69.5%, respectively, as compared to control, vehicle-treated mice (Fig. 4A, C). While the reduction in mean tumor volume appeared to be dose-dependent, this effect was not statistically significant. As compared to the vehicle-treated group, significant antitumor activity for each dose was achieved on day 16, day 12, and day 9, respectively. No gross toxicities were evident as determined by body weight, even at the highest dose (Fig. 4B). We also determined the potential effect of drug treatment on PKD2 activation within the tumor xenografts. As seen in Fig. 4D, treatment with CRT066101 resulted in a dose-dependent suppression of p-PKD2 in the xenograft tumors. Tumor xenografts were examined for additional markers of growth and apoptosis. For these studies, Ki67 expression and the level of TUNEL and M30 staining was investigated. Ki67 is a well-established marker for cell proliferation. Treatment with the lowest CRT0066101 dose used in this experiment, 40 mg/kg, resulted in a significant reduction in Ki67 expression (Fig. 5). In addition, expression levels of p-ERK were investigated. We observed a dose-dependent inhibition of p-ERK expression which correlated with our in vitro results. The TUNEL (terminal transferase-mediated dUTP nick end-labeling) assay detects DNA fragmentation as a result of apoptotic signaling cascades. CRT0066101 treatment significantly increased TUNEL staining in a dose-dependent manner (Fig. 5). To verify that the increased TUNEL staining signified apoptosis and not necrosis, staining for M30, a caspase-cleaved cytokeratin 18, was performed. As shown in Fig. 5, CRT0066101 administration significantly increased M30 staining suggesting that the PKD inhibitor induced apoptosis in our in vivo CRC model.
Fig. 4. Effect of CRT0066101 on in vivo tumor growth.
Athymic nude mice bearing HCT116 tumor xenografts were administered daily vehicle control (dextrose) or CRT0066101 at 40, 80, and 120 mg/kg orally for 3 weeks (5 mice per group). Tumor volume (A) and body weight (B) were measured 3 times per week. (C) Mouse and excised tumor images after treatment with CRT0066101. (D) Protein expression in xenografts was determined after 3 weeks of CRT0066101 treatment. Tumors were harvested 2 hours after the last dose.
Fig. 5. In vivo biological activity of CRT0066101.
Tumors were fixed in formalin and paraffin-embedded. Slides were stained for Ki67, p-ERK, TUNEL, and M30 expression (x200) as described in the Methods section.
Discussion
It is now well-established that PKD signaling mediates several key cellular processes, including DNA synthesis, proliferation, survival, adhesion, invasion/migration, motility, and angiogenesis (8). This pathway has also been implicated in a broad range of human tumors. Moreover, there is evidence suggesting that different PKD isoforms may be associated with specific cancers (27). In the literature, relatively little is known about the precise role of PKD in CRC. For this reason, we initially determined the respective mRNA and protein expression levels of each PKD isoform in normal human colon and CRC cell lines. PKD1 expression was detectable only in the normal colon cell lines. However, significant expression of PKD2 and PKD3 was observed in all cell lines. These findings were validated in a search of the CellMiner and CCLE databases and subsequently confirmed with expression analysis in human colon tumor samples. The absence of PKD1 in CRC was somewhat puzzling as it had been previously reported that PKD1 is abundantly expressed in mouse intestinal cells and that expression of this particular isoform is necessary for DNA synthesis and cellular migration (28). It is conceivable that the loss of PKD1 expression may be the result of epigenetic silencing as normal intestinal cells transform into cancer cells. In support of this possibility, investigators have also identified loss of PKD1 expression in gastric and breast cancer resulting from epigenetic regulatory mechanisms (9, 29). With this in mind, we attempted to re-express PKD1 with the use of DNA methyltransferase inhibitors (decitabine and RG108) but were unsuccessful in doing so (data not shown). The CCLE database states that HCT116 cells have a point mutation in PKD1 (S625N) and RKO cells have two point mutations (G779D; C853R). However, at this time, it is unclear as to the potential impact of these discrete point mutations on protein expression and/or stability.
The development and evaluation of small molecule inhibitors directed against novel targets, such as PKD, would appear to be critically important for identifying new therapeutic options that can ultimately improve patient response and outcome. To date, several PKD inhibitors have been developed with activity against a wide variety of tumor types (8). Our studies reveal that two of these inhibitors, kb-NB142-70 and CRT0066101, exhibited significant activity against a wide range of human CRC cell lines. Interestingly, these PKD inhibitors equally suppressed growth of multidrug resistant cells as well as p53-deficient cells suggesting that PKD inhibitors may be able to overcome chemoresistance and radioresistance mechanisms.
With respect to the potential mode of action, our studies show that PKD inhibition resulted in cell cycle arrest at the G2/M phase, a finding that was previously demonstrated in human prostate cancer cells (15). Similarly, Kienzle et al. demonstrated that PKD1 and PKD2 are necessary in the G2 phase of the cell cycle in HeLa cells (30). Interestingly, siRNA knockdown of PKD3 resulted in accumulation of prostate cancer cells in G0/G1 phase (12). Studies by Azoitei et al. showed that glioblastoma cells accumulate in G0/G1 phase after PKD2 suppression (13). Thus, in addition to cell-specific expression, the different PKD isoforms may play different roles in mediating cell division and proliferation.
Using our human CRC model systems, we observed that PKD inhibition, through the use of either small molecule inhibitors or siRNAs, resulted in significant induction of apoptosis. This effect was detected by both Annexin V/PI staining and Western blot analysis of cleaved PARP and caspases-3. In addition to induction of apoptotic pathways, PKD inhibition suppressed Akt and ERK signaling. Previously, PKD has been shown to mediate the MEK/ERK/RSK pathway and promote cell proliferation through a stimulatory effect on GPCR (31). The pro-proliferative effects of PKD in cancer cells are associated with activation of both ERK1/2 and AKT. In human prostate cancer, PKD3 modulated both the extent and duration of ERK1/2 activation (12). In the present study, we showed that the small molecule inhibitor CRT0066101 strongly suppressed activation of ERK in human CRC. This agent also significantly blocked AKT activation. Transfection of PKD2 siRNA resulted in similar effects on these two downstream pathways. Other key regulators of cell survival, proliferation, and motility are the NF-κB transcription factors. Previous studies have shown that PKD is a mediator of NF-κB induction in a variety of cells exposed to GPCR agonists or oxidative stress (32). PKD2 gene silencing dramatically blocked LPA-stimulated NF-κB promoter activity in non-transformed human colonic epithelial NCM460 cells (33). However, they observed no decrease in ERK signaling. PKD2 has also been implicated in mediating NF-kB activation by Bcr-Abl in myeloid leukemia cells (34). In keratinocytes, in which phorbol esters are major tumor promoters, PKDs stimulate proliferation and prevent differentiation. These findings suggest that PKDs and, in particular, PKD2 may play an important role in phorbol ester-sensitive tumors, such as skin tumors and colon cancer. PMA induces NF-κB activation, and PKDs has been shown to promote cell survival through activating NF-κB signaling pathway in response to oxidative stress (8). Thus, it is conceivable that the pro-survival effects of PKD2 in CRC cells, in response to PMA treatment, may be partly attributable to NF-κB activation. Herein, we confirmed that suppression of PKD with small molecule inhibitors leads to significant inhibition of NF-κB activity in CRC. Taken together, these three key survival pathways, AKT, ERK, and NF-κB, each of which are located downstream of PKD signaling, appear to be critical for the cytotoxic activity of PKD inhibitors in human CRC.
To date, several PKD inhibitors have been studied in in vivo animal models. As one example, kb-NB142-70, demonstrated significantly improved in vitro activity compared to the structurally related CID755673, but exhibited no antitumor activity in vivo, which is most likely due to rapid metabolism (17). Alternatively, a naphthyridine-based PKD inhibitor showed PKD inhibition and suppression of PKD-dependent downstream pathways in an in vivo rat model (35). The PKD inhibitors kb-NB142-70 and CRT0066101 displayed similar activities in our in vitro models, and CRT0066101 also demonstrated significant antitumor activity in two pancreatic animal models (14). Our data confirm the antitumor activity of CRT0066101 and further demonstrate that colon cancer xenografts are responsive to in vivo inhibition of the PKD pathway. With the CRC models that were used in this study, we observed greater tumor suppression at lower doses than previously reported suggesting that colon cancer may be more susceptible to blockage of this growth-mediating pathway. However, given the critical roles of PKDs in both endothelial and epithelial cells, a major concern is that the use of pan-PKD inhibitors may lead to off-target effects. Several PKD inhibitors have demonstrated such off-target effects against other kinases (19, 35). It is conceivable that the orally bioavailable inhibitor CRT0066101 may bind to and inhibit other kinases. Notably, the 2-(4-aminopyrimidin-2-yl)phenol moiety in CRT0066101 is also present in small molecule kinase inhibitors targeting the proto-oncogene serine/threonine-protein kinase (Pim-1), checkpoint kinase 2, inhibitor of NF-κB kinase subunit beta, tropomyosin-related kinases and others (36–39). However, specific PKD isoform targeting with either siRNAs or antisense molecules can suppress in vivo tumor growth (11, 40). Thus, the development of small molecule inhibitors and/or nucleic acid-based molecules, such as siRNAs, targeting specific PKD isoforms is warranted.
In conclusion, we report that small molecule inhibitors directed against PKD signaling exhibited potent in vitro cytotoxic and in vivo antitumor activity. The biological effects of these agents appear to be mediated by inhibition of PKD2 activation, G2/M phase arrest, induction of apoptosis, and inhibition of AKT, ERK, and NF-κB signaling pathways. These studies are important as they provide support for the potential role of PKD as a novel target for cancer chemotherapy. Moreover, they provide a rational basis for the design and development of novel agents that may act either alone or in combination with presently available anticancer agents to enhance clinical activity and/or overcome cellular drug resistance.
Supplementary Material
Acknowledgments
Financial Information: Research funds provided by a grant from NIH/NCI, P30-CA147904 (Recipient: N.E. Davidson)
The authors thank Drs. Shaoyu Wu and Wei Yang for excellent technical assistance and Ms. Kara George Rosenker for the preparation of kb-NB142-70. This project used the UPCI Tissue and Research Pathology Services and the Cytometry Facility that are supported in part by award P30CA047904.
Footnotes
The authors disclose no potential conflicts of interest.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Chu E. An update on the current and emerging targeted agents in metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11:1–13. doi: 10.1016/j.clcc.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Kohne CH, Lenz HJ. Chemotherapy with targeted agents for the treatment of metastatic colorectal cancer. Oncologist. 2009;14:478–88. doi: 10.1634/theoncologist.2008-0202. [DOI] [PubMed] [Google Scholar]
- 4.Lucas AS, O’Neil BH, Goldberg RM. A decade of advances in cytotoxic chemotherapy for metastatic colorectal cancer. Clin Colorectal Cancer. 2011;10:238–44. doi: 10.1016/j.clcc.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–8. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- 7.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology. 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaValle CR, George KM, Sharlow ER, Lazo JS, Wipf P, Wang QJ. Protein kinase D as a potential new target for cancer therapy. BBA Rev Cancer. 2010;1806:183–92. doi: 10.1016/j.bbcan.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eiseler T, Doppler H, Yan IK, Goodison S, Storz P. Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 2009;11:R13. doi: 10.1186/bcr2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trauzold A, Schmiedel S, Sipos B, Wermann H, Westphal S, Roder C, et al. PKCmu prevents CD95-mediated apoptosis and enhances proliferation in pancreatic tumour cells. Oncogene. 2003;22:8939–47. doi: 10.1038/sj.onc.1207001. [DOI] [PubMed] [Google Scholar]
- 11.Azoitei N, Pusapati GV, Kleger A, Moller P, Kufer R, Genze F, et al. Protein kinase D2 is a crucial regulator of tumour cell-endothelial cell communication in gastrointestinal tumours. Gut. 2010;59:1316–30. doi: 10.1136/gut.2009.206813. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Deng F, Singh SV, Wang QMJ. Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKC epsilon/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 2008;68:3844–53. doi: 10.1158/0008-5472.CAN-07-5156. [DOI] [PubMed] [Google Scholar]
- 13.Azoitei N, Kleger A, Schoo N, Thal DR, Brunner C, Pusapati GV, et al. Protein kinase D2 is a novel regulator of glioblastoma growth and tumor formation. Neuro Oncol. 2011;13:710–24. doi: 10.1093/neuonc/nor084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harikumar KB, Kunnumakkara AB, Ochi N, Tong Z, Deorukhkar A, Sung B, et al. A novel small-molecule inhibitor of protein kinase D blocks pancreatic cancer growth in vitro and in vivo. Mol Cancer Ther. 2010;9:1136–46. doi: 10.1158/1535-7163.MCT-09-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavalle CR, Bravo-Altamirano K, Giridhar KV, Chen J, Sharlow E, Lazo JS, et al. Novel protein kinase D inhibitors cause potent arrest in prostate cancer cell growth and motility. BMC Chem Biol. 2010;10:5. doi: 10.1186/1472-6769-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharlow ER, Giridhar KV, LaValle CR, Chen J, Leimgruber S, Barrett R, et al. Potent and selective disruption of protein kinase D functionality by a benzoxoloazepinolone. J Biol Chem. 2008;283:33516–26. doi: 10.1074/jbc.M805358200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Clausen DM, Beumer JH, Parise RA, Egorin MJ, Bravo-Altamirano K, et al. In vitro cytotoxicity, pharmacokinetics, tissue distribution, and metabolism of small-molecule protein kinase D inhibitors, kb-NB142-70 and kb-NB165-09, in mice bearing human cancer xenografts. Cancer Chemother Pharmacol. 2013;71:331–44. doi: 10.1007/s00280-012-2010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamber GG, Meredith E, Zhu QM, Yan WL, Rao C, Capparelli M, et al. 3,5-Diarylazoles as novel and selective inhibitors of protein kinase D. Bioorg Med Chem Lett. 2011;21:1447–51. doi: 10.1016/j.bmcl.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 19.Sharlow ER, Wilson GM, Close D, Leimgruber S, Tandon M, Reed RB, et al. Discovery of diverse small molecule chemotypes with cell-based PKD1 inhibitory activity. PLoS One. 2011;6:e25134. doi: 10.1371/journal.pone.0025134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 21.Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, et al. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710–8. [PubMed] [Google Scholar]
- 22.Wei N, Liu GT, Chen XG, Liu Q, Wang FP, Sun H. H1, a derivative of tetrandrine, exerts anti-MDR activity by initiating intrinsic apoptosis pathway and inhibiting the activation of Erk1/2 and Akt1/2. Biochem Pharmacol. 2011;82:1593–603. doi: 10.1016/j.bcp.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 23.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guha S, Tanasanvimon S, Sinnett-Smith J, Rozengurt E. Role of protein kinase D signaling in pancreatic cancer. Biochem Pharmacol. 2010;80:1946–54. doi: 10.1016/j.bcp.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borges S, Storz P. Protein kinase D isoforms: new targets for therapy in invasive breast cancers? Expert Rev Anticancer Ther. 2013;13:895–8. doi: 10.1586/14737140.2013.816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinnett-Smith J, Rozengurt N, Kui R, Huang C, Rozengurt E. Protein kinase D1 mediates stimulation of DNA synthesis and proliferation in intestinal epithelial IEC-18 cells and in mouse intestinal crypts. J Biol Chem. 2011;286:511–20. doi: 10.1074/jbc.M110.167528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim M, Jang HR, Kim JH, Noh SM, Song KS, Cho JS, et al. Epigenetic inactivation of protein kinase D1 in gastric cancer and its role in gastric cancer cell migration and invasion. Carcinogenesis. 2008;29:629–37. doi: 10.1093/carcin/bgm291. [DOI] [PubMed] [Google Scholar]
- 30.Kienzle C, Eisler SA, Villeneuve J, Brummer T, Olayioye MA, Hausser A. PKD controls mitotic Golgi complex fragmentation through a Raf-MEK1 pathway. Mol Biol Cell. 2013;24:222–33. doi: 10.1091/mbc.E12-03-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinnett-Smith J, Zhukova E, Hsieh N, Jiang X, Rozengurt E. Protein kinase D potentiates DNA synthesis induced by Gq-coupled receptors by increasing the duration of ERK signaling in swiss 3T3 cells. J Biol Chem. 2004;279:16883–93. doi: 10.1074/jbc.M313225200. [DOI] [PubMed] [Google Scholar]
- 32.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–30. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu TT, Leung WY, Moyer MP, Strieter RM, Rozengurt E. Protein kinase D2 mediates lysophosphatidic acid-induced interleukin 8 production in nontransformed human colonic epithelial cells through NF-kappaB. Am J Physiol Cell Physiol. 2007;292:C767–77. doi: 10.1152/ajpcell.00308.2006. [DOI] [PubMed] [Google Scholar]
- 34.Mihailovic T, Marx M, Auer A, Van Lint J, Schmid M, Weber C, et al. Protein kinase D2 mediates activation of nuclear factor kappaB by Bcr-Abl in Bcr-Abl+ human myeloid leukemia cells. Cancer Res. 2004;64:8939–44. doi: 10.1158/0008-5472.CAN-04-0981. [DOI] [PubMed] [Google Scholar]
- 35.Meredith EL, Ardayfio O, Beattie K, Dobler MR, Enyedy I, Gaul C, et al. Identification of orally available naphthyridine protein kinase D inhibitors. J Biol Chem. 2010;53:5400–21. doi: 10.1021/jm100075z. [DOI] [PubMed] [Google Scholar]
- 36.Caldwell JJ, Welsh EJ, Matijssen C, Anderson VE, Antoni L, Boxall K, et al. Structure-based design of potent and selective 2-(quinazolin-2-yl)phenol inhibitors of checkpoint kinase 2. J Med Chem. 2011;54:580–90. doi: 10.1021/jm101150b. [DOI] [PubMed] [Google Scholar]
- 37.Ekambaram R, Enkvist E, Vaasa A, Kasari M, Raidaru G, Knapp S, et al. Selective bisubstrate inhibitors with sub-nanomolar affinity for protein kinase Pim-1. Chem Med Chem. 2013;8:909–13. doi: 10.1002/cmdc.201300042. [DOI] [PubMed] [Google Scholar]
- 38.Nagarajan S, Doddareddy M, Choo H, Cho YS, Oh KS, Lee BH, et al. IKKbeta inhibitors identification part I: homology model assisted structure based virtual screening. Bioorg Med Chem. 2009;17:2759–66. doi: 10.1016/j.bmc.2009.02.041. [DOI] [PubMed] [Google Scholar]
- 39.Park H, Chi O, Kim J, Hong S. Identification of novel inhibitors of tropomyosin-related kinase A through the structure-based virtual screening with homology-modeled protein structure. J Chem Inf Model. 2011;51:2986–93. doi: 10.1021/ci200378s. [DOI] [PubMed] [Google Scholar]
- 40.LaValle CR, Zhang LY, Xu SP, Eiseman JL, Wang QJ. Inducible silencing of protein kinase D3 inhibits secretion of tumor-promoting factors in prostate cancer. Mol Cancer Ther. 2012;11:1389–99. doi: 10.1158/1535-7163.MCT-11-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.