Abstract
Purpose
Metastasis heterogeneity presents a significant obstacle to the development of targeted cancer therapeutics. In this study, we sought to establish from a large series of human melanoma metastases whether there exists a determined pattern in tumor cellular heterogeneity that may guide the development of future targeted immunotherapies.
Experimental Design
From a cohort of 1514 patients with metastatic melanoma, biopsies were procured over a 17 year period from 3086 metastatic tumors involving various anatomic sites. To allow specific tumor cell profiling, we utilized established immunohistochemical methods to perform semi-quantitative assessment for a panel of prototypic melanocyte differentiation antigens (MDAs) including gp100, MART-1, and tyrosinase (TYR). To gain insight into the endogenous host immune response against these tumors, we further characterized tumor cell expression of MHC I and MHC II and, also, the concomitant CD4+ and CD8+ T cell infiltrate.
Results
Tumor cell profiling for MDA expression demonstrated an anatomic site-specific pattern of antigen expression that was highest in brain, intermediate in soft tissues/lymph nodes, and lowest in visceral metastases. Hierarchical clustering analysis supported that melanoma metastases have a phylogenetically determined, rather than a stochastic, pattern of antigen expression that varies by anatomic site. Further, TYR expression was more frequently lost in metastatic sites outside of the brain and was uniquely correlated with both endogenous CD8+ and CD4+ T cell infiltrate.
Conclusion
Site-specific antigen heterogeneity represents a novel attribute for human melanoma metastases that should be considered in future therapy development and when assessing the responsiveness to antigen specific immunotherapies.
Keywords: Melanoma, metastases, antigen, immunoediting
Introduction
Cancer metastases can demonstrate cellular heterogeneity between synchronous tumors (interlesional heterogeneity) and within individual tumors (intralesional heterogeneity) (1). This phenomenon is thought to be driven by the stochastic genetic instability of tumor cell clones combined with the non-stochastic selective pressures of the host and tumor microenvironment (1-3). Early evidence for metastatic tumor heterogeneity in animal models (4, 5) has been substantiated by more recent high resolution deep sequencing analyses of human tumors (6). Metastasis heterogeneity, thus, presents a significant obstacle to the current developmental paradigm for highly targeted molecular and immune based therapeutics for patients with metastatic cancer. Studies of cutaneous melanoma provide a distinctive opportunity to gain further insight into the nature of human metastasis heterogeneity. Melanoma metastases have been shown to have a high mutation frequency (7, 8), diverse phenotype (4, 9), a clinically diffuse dissemination pattern (10), and a unique ability to elicit spontaneous antigen specific host immune responses (11, 12). As highly targeted immune therapies are currently in development for the treatment of metastatic melanoma, an improved understanding of metastasis heterogeneity is critical to assessing potential tumor susceptibility in future clinical studies.
To allow facile, reproducible, and specific profiling of the tumor cells within individual metastases, we utilized established immunohistochemical methods to perform semi-quantitative assessment for a panel of prototypic melanocyte differentiation antigens (MDAs) including gp100 (13, 14), MART-1 (13, 14), and tyrosinase (TYR) (15). These tumor lineage antigens serve as favorable profiling markers given their high expression in normal melanocytes and primary melanoma tumors, but heterogeneous expression in metastatic lesions (16-19). T cell recognition and clearance of MDA expressing cells (immunoediting) has been implicated as a putative mechanism for tumor antigen heterogeneity among melanoma metastases (20-23). To gain insight into the relationship between antigen expression in metastases and endogenous host immune response, we further characterized tumor cell expression of MHC I and II and, also, the concomitant CD4+ and CD8+ T cell infiltrate within the tumors.
Methods
Study Population
Between May 1, 1995 and August 7, 2012, 3234 patients with metastatic cutaneous melanoma were evaluated in the Surgery Branch of the National Cancer Institute. 1514 patients underwent protocol associated metastatic tumor biopsies to obtain a total of 3483 specimens. 3086 of these biopsies were diagnostic for metastatic melanoma and further characterized by an immunohistochemistry panel of tumor markers including: gp100, MART-1, tyrosinase, MHC class I, and MHC class II. 2886 of the evaluable biopsies were fine-needle aspirates (FNA), 303 were frozen sections, and 25 were not specified and thus excluded from the analysis. Biopsies were further considered non-evaluable and removed from the analysis for the following reasons: non-diagnostic for melanoma, absence of quantitative antigen expression data, or medical history that was not consistent with cutaneous melanoma.
All information relating to the biopsies was collected in a prospective fashion beginning in 1995. The site of the biopsy was established from the pathology report and clinical notes. The distinction between subcutaneous, lymph node (LN), and other soft-tissue metastases is often difficult to make clinically. To maintain consistency and accuracy in denoting site of metastases, biopsies from specified lymph node basins (groin, axilla, iliac, intra-abdominal, and cervical) or clinically obvious lymph nodes were classified as lymph node metastases. All other non-nodal soft tissue lesions were categorized as soft tissue/subcutaneous (ST/SQ) metastases. A site specific analysis was performed on all sites for which greater than 20 evaluable biopsies were available. Limited sample size for other sites (bone, adrenal, omentum, parotid, kidney and pancreas) precluded their inclusion in the current analysis.
Tissue Procurement
Tumor samples and biopsies were obtained on clinical research protocols approved by the Institutional Review Board and U.S. Food and Drug Administration. All patients gave informed consent for biopsy and analysis of their tumors in accordance with the Declaration of Helsinki. Tissue samples were prepared using two methods. First, multiple radial passage FNAs were performed using a 22- or 23-gauge needle to aspirate in situ lesions or freshly resected tumors. The aspirate was diluted in RPMI medium 1640 (GIBCO BRL, Grand Island, NY) and cytospins were prepared. Second, samples of surgically resected tumor specimens were prepared by freezing in OCT compound (Miles, Elkhart, IN). Frozen tissue was then sectioned and evaluated for the presence of tumor using hematoxylin and eosin staining. Evaluation of antigen expression by FNA has been described previously to correlate highly with antigen expression evaluated by frozen section (14, 24).
Immunohistochemistry
Throughout the duration of the study, all staining was performed by a single cytopathology technician (P.A.F.). Melanoma antigen immunoreactivity of tumor cells in the biopsy specimens was assessed on acetone-fixed cytospins or frozen tissue sections by using antibodies against gp100 (clone HMB45), MART-1 (clone M2-7C10), and tyrosinase (clone T311). A negative sample control (purified myeloma protein, mouse IgG1 kappa; Organon Teknika Corporation, Durham, NC) and positive controls (melanoma cell lines) were performed throughout the study period. A modified avidin-biotin procedure (Vector Laboratories, Burlingame, CA) was performed, with either 3,3-diaminobenzidine or, in the case of highly pigmented samples, 3-amino-9-ethylcarbazole as the chromogen. MHC expression of tumor cells in the biopsy specimens was assessed using antibodies against MHC class I (Clone W6/32) and MHC class II (Clone 4A12). Lesions were categorized as negative (no positive cells), 1-25% (one positive cell or more), 25-50%, 50-75%, and >75% according to the percentage of cells expressing a given antigen. Lymphocytic infiltrate in the biopsy specimens was assessed by using antibodies against CD4 and CD8 (Coulter Becton-Dickinson). Infiltrate was graded on a 0-3+ scale. All specimens were prospectively assessed by a consistent group of board certified cytopathologists in the Department of Cytopathology/NIH using established standardized criteria and the results were serially recorded in a prospectively established database.
Statistical Analysis
For descriptive analytic purposes, the antigen expression trend for a staining distribution was determined using the slope resulting from a simple linear regression of the percent of tumors with a given expression level vs. the categorized expression levels, identified as 0=absent, 1=1-25%, 2=25-50%, 3=50-75%, and 4≥75%. The associations between the slopes so obtained and the ordered CD4+ or CD8+ T cell infiltration levels were determined by a Spearman correlation. The correlation would be interpreted primarily according to the magnitude of the correlation coefficient: ∣r∣ >0.70 is strong correlation; 0.50 < ∣r∣ < 0.70 is moderately strong correlation; 0.3 < ∣r∣ < 0.5 is weak to moderately strong correlation, and if ∣r∣ <0.3, then this would be considered weak correlation. The p-values associated with correlation coefficients are tests of whether r=0 and are of less importance. As these correlations were done based on the slopes for a T cell infiltrate category, and there were only 4 categories for each T cell infiltrate (0 to 3+), they should be interpreted as approximate indicators of the strength of the relationship. Associations between expression levels or between expression levels and infiltrate scores were determined using a Jonckheere-Terpstra test for trend (25). A small p-value would indicate that the two measures were related to one another. In addition, a marginal homogeneity test was used to assess the discordance between paired categorical expression levels between two compared levels. The fractions without expression were compared between antigens in a pairwise fashion using McNemar’s test for paired categorical data. A Kruskal-Wallis test was used to determine if the expression levels or lymphocytic infiltrates varied according to site. For those expression levels or infiltrates in which there was found to be significant variation by site, individual pairwise tests of sites were also performed using a Cochran-Armitage test for trend (26). Hierarchical cluster analysis utilizing between group linkage was performed using DendroUPGMA (27). The program calculates the root-mean-square deviation (RMSD) between pairs of sets of variables, transforms these coefficients into distances and makes a clustering using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) algorithm (28). Unrooted dendrograms were generated to illustrate the clustering of anatomic sites with similar antigen expression. The length of branching is inversely proportional to the antigen similarities between sites. Analyses were performed using SAS version 9.3 (SAS Institute, Cary NC), StatXact 9 (Cytel, Inc., Cambridge MA), or STATA 12.0/IC statistical software (StataCorp, College Station TX). In view of the large number of exploratory analyses performed, p<0.005 would be considered statistically significant while 0.005 < p <0.05 would be considered a strong trend.
Results
Human melanoma metastases demonstrate interlesional heterogeneity in MDA and MHC expression
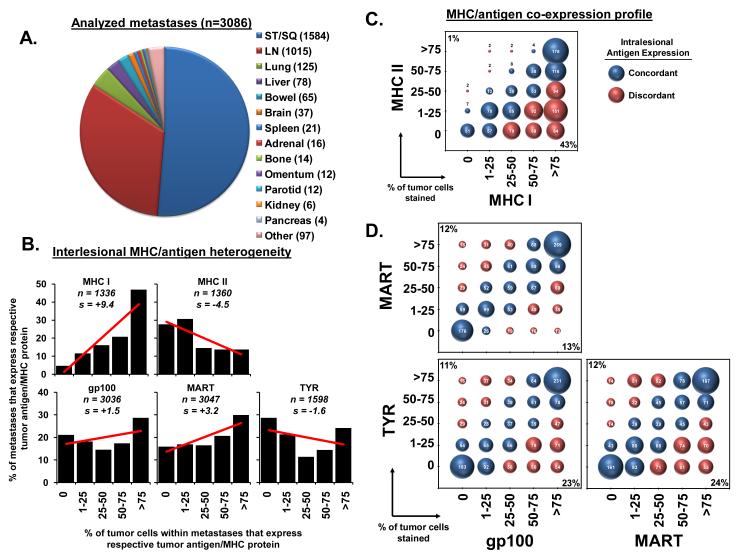
From a cohort of 1514 patients with metastatic melanoma, biopsies were procured over a 17 year period from 3086 metastatic tumors involving various anatomic sites (Figure 1A). The most commonly biopsied sites included soft tissue and subcutaneous (ST/SQ, n=1584) followed by lymph nodes (LN, n=1015). However, other frequently sampled metastatic sites included lung (n=125), liver (n=78), bowel (n=65), brain (n=37), and spleen (n=21).
Figure 1.
Melanoma metastases demonstrate interlesional heterogeneity. (A) The distribution of lesions by anatomic site. The number of biopsies for a given site is indicated (n). Abbreviations: ST/SQ (soft tissue/subcutaneous), LN (lymph node). (B) The staining distribution of MHC and MDA expression across all biopsies is shown; “n” indicates the number of biopsies stained for a given antigen; “s” indicates the slope of the linear regression line. (C) The co-expression profile of the 1319 lesions stained for both MHCI and II. The diameter of the bubble is proportionate to the sample size displayed within each individual bubble. For descriptive purposes, lesions were defined as having concordant (blue) or discordant (red) expression of MHC I and II. The percent of lesions with MHC II expression discordantly higher than MHC I is shown in left upper corner, the percent of lesions with MHC II expression discordantly lower is shown in the right lower corner. (D) The 1561 lesions stained for all three MDAs are displayed as in (C).
Initial comparative analysis across all metastatic lesions demonstrated interlesional heterogeneity in major histocompatibility complex (MHC) and MDA expression ranging from complete absence of antigen to uniformly high expression (>75% of tumor cells) within individual metastases (Figure 1B). The pattern of staining for MHC I (n=1336) and MHC II (n=1360) differed significantly (p<0.0001 by the marginal homogeneity test). Simple linear regression analysis of the percentage of tumors within a given expression level versus the expression level category demonstrated strong inverse trends towards a higher percentage of metastases staining strongly for MHC I (slope=+9.4) and a lower percentage for MHC II (slope=−4.5). The frequency of metastases with complete absence of staining for MHC I and MHC II was also notably different (5% and 28%, respectively, p<0.0001 by McNemar’s test). In contrast to the highly skewed expression for the MHC molecules, the staining for gp100 (n=3036), MART (n=3047), and TYR (n=1598) was found to be more evenly distributed among the metastases (Figure 1B). Pairwise comparison of the interlesional expression pattern for the three MDAs revealed that there was a substantial association between expression levels (p<0.0001 by Jonckheere-Terpstra test), but variability among the MDAs was also noted. TYR expression demonstrated a trend toward lower percentages of stained cells (slope=−1.6), while gp100 (slope=+1.5) and MART (slope=+3.2) both trended toward greater frequencies of stained cells. Differences between antigen expression were further illustrated by significant variability in the percentage of metastases with complete absence of staining for MART, gp100, and TYR (16%, 21%, 28% respectively, p<0.0001 for each pairwise comparison of the three MDAs using McNemar’s test).
To better define the relationship between antigen markers within individual metastases, we next determined the concordance and discordance frequency between MHC I and II expression and, also, between the three MDAs. Concordant expression between two markers was defined as having <25% of the tumor cells within a single lesion differ in their expression of the two markers. Conversely, discordant expression was defined as having >25% of the cells differ in expression. Co-expression plots revealed that MHC I and MHC II expression (n=1319) were concordant in 56% of lesions (Figure 1C). Metastases with low frequencies of MHC I positive cells consistently possessed low frequencies of MHC II positive cells and lesions with a high frequency of MHC II positive cells concomitantly expressed a high degree of MHC I staining. However, consistent with their inversely skewed expression pattern, MHC II expression in metastases was rarely seen in the absence of MHC I (1% of tumors), with the majority of discordant lesions expressing higher MHC class I when compared to MHC II (43% of tumors). Next, we compared the synchronous expression of the MDAs within individual metastases (n=1561) (Figure 1D). Co-expression plots revealed a strong linear association between each of the antigen pairs (all 3 comparisons: p <0.0001 by Jonckheere-Terpstra test). These strong concordant relationships were not surprising given that gp100, MART and TYR expression share a common transcriptional pathway involving the microphthalmia-associated transcription factor (MITF) (29, 30). However, we did observe metastases with discordant antigen expression: 25% in MART vs. gp100, 34% in gp100 vs. TYR, and 36% in MART vs. TYR. The lesions with discordant MART and gp100 expression were evenly distributed between having high MART and low gp100 (12%) and low MART and high gp100 expression (13%). In contrast, the co-expression plots for TYR revealed that the discordant lesions demonstrated absence of TYR twice as often as gp100 (23% vs. 11%) and MART (24% vs. 12%). Cumulatively, these findings demonstrated that melanoma metastases have significant interlesional heterogeneity in MHC and MDA expression with discordant MDA expression in approximately one third of lesions, and more frequent absence of TYR expression when compared to MART and gp100.
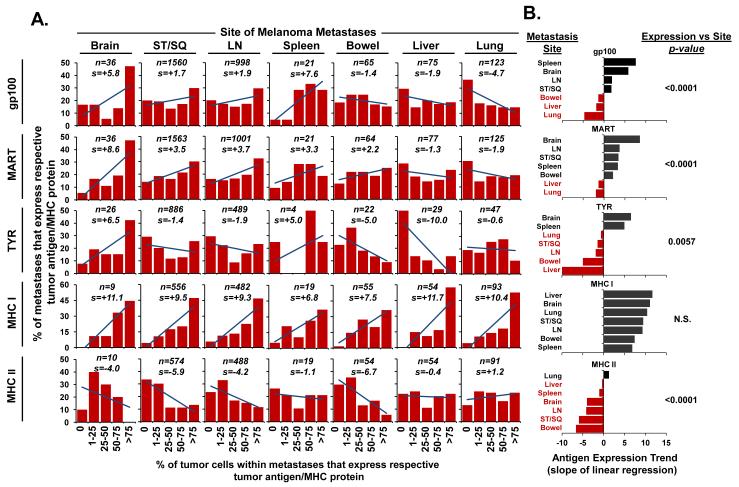
MDA and MHC II expression in metastases demonstrate a site-specific pattern
To determine if the observed tumor heterogeneity varied by location of the metastases in the host, we next compared the MDA and MHC expression pattern in lesions from the seven most frequently biopsied anatomic sites (ST/SQ, LN, lung, liver, bowel, brain, and spleen) (Figure 2A). Analysis for each of the MDAs revealed site specific antigen variability (gp100: p<0.0001, MART: p<0.0001, TYR: p=0.0057). Trend analysis of the antigen distribution patterns was used to rank the expression associated with individual anatomic sites (Figure 2B). Brain metastases consistently demonstrated high expression of each of the MDAs based upon positive skewed antigen trends (gp100: slope=+5.8; MART: slope=+8.6; TYR: slope=+6.5). In contrast, liver and lung metastases demonstrated lower expression of each of the MDAs with consistently negative antigen trends (gp100: slope=−1.9 and −4.7; MART: slope=−1.3 and −1.9; TYR: slope=−10.0 and −0.6, for liver and lung respectively). Individual pairwise comparison of liver, lung, and bowel metastases further established that each of these sites had lower MDA expression compared to brain metastases (p<0.05) (Supplementary Table 1). Antigen expression was more variable for ST/SQ and LN metastases which demonstrated a trend toward higher percentages of stained cells for gp100 (slope=+1.7 and +1.9, respectively) and MART (slope=+3.5 and +3.7, respectively), but lower expression of TYR (slope=−1.4 and −1.9, respectively). Individual pairwise comparison of ST/SQ and LN metastases demonstrated that each of these sites also had higher gp100 and MART expression compared to liver and lung metastases (p<0.05) (Supplementary Table 1). Tumor cell expression of MHC class I was high across all sites (slope range: +6.8 to +11.7) with no significant site specific variability (p=0.41). MHC II expressing tumor cells were less commonly found in the metastases, however, its staining profile did vary by anatomic site (p<0.0001). Lung metastases demonstrated the highest expression of MHC II (slope=+1.2), whereas ST/SQ (slope=−5.9) and bowel (slope=−6.7) lesions showed the lowest expression. From these findings, we concluded that MDA and MHC II expression in metastases varied significantly by anatomic site.
Figure 2.
MDA and MHC II expression demonstrate a site-specific pattern. (A) The distribution of antigen expression for each anatomic site; “n” indicates the number of biopsies stained for a given antigen; “s” indicates the slope of the linear regression line. (B) Ranking of antigen expression trend by metastatic site. Red coding of sites indicate negative antigen trends. The results are presented from high positive to low or negative slope for display purposes and are not tested for this trend as it was not a pre-specified hypothesis. The p-values indicate the probability that no variability exists across all sites as determined by the Kruskal-Wallis test.
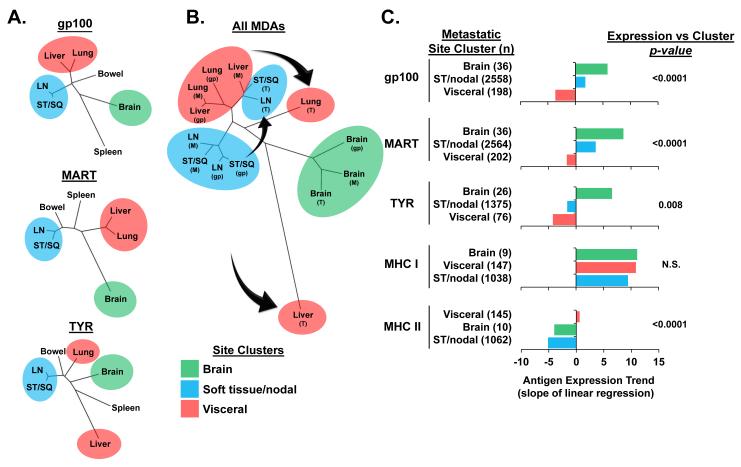
MDA expression in melanoma metastases cluster in a site-specific phylogenetic pattern
We next investigated whether the site specific antigen heterogeneity in the metastases had a phylogenetically determined, rather than a stochastic, pattern. Hierarchical clustering of the individual metastatic sites was performed based upon their antigen expression profiles to determine if there were antigenic similarities between anatomic sites (Figure 3A, Supplementary Table 2). Dendrogram branch analyses for each of the MDAs revealed that ST/SQ and LN metastases had a similar antigen profile and formed a discrete “soft tissue/nodal” cluster which was distinct from a “visceral” organ cluster that included lung and liver metastases. Brain metastases consistently segregated independently and were divergent from the other two main clusters. Bowel and spleen metastases did not show a consistent clustering pattern. Analysis of the sites based upon the cumulative profile of all three MDAs revealed that the brain expression of MART, gp100, and TYR shared a similar pattern and clustered as a single group (Figure 3B, Supplementary Table 3). Further, ST/SQ and LN metastasis, again, formed a discrete soft tissue/nodal cluster based upon shared expression of gp100 and MART which was distinct from a visceral cluster that included lung and liver metastases expressing the same antigens. Interestingly, the TYR expression in ST/SQ and LN metastases did not cluster with the other two MDAs, but had an expression pattern that was more akin to gp100 and MART expression in visceral metastases. In addition, the TYR expression in lung and liver tumors segregated independently and away from the main visceral cluster. Based upon these findings, we concluded that antigen expression in melanoma metastases clustered in a site specific phylogenetic pattern, but TYR expression in soft tissue/nodal and visceral sites clustered independently from gp100 and MART. To determine the magnitude and significance of these antigen profile differences, we performed statistical comparisons of the expression patterns for these unique site clusters (Figure 3C, Supplementary Figure 1). Analysis for each of the MDAs confirmed cluster specific antigen variability (gp100: p<0.0001, MART: p<0.0001, TYR: p=0.008). Trend analysis of the antigen distribution patterns was used to rank the expression associated with the individual metastatic site clusters. Consistent with their independent clustering, brain metastases demonstrated positive antigen trends for all three MDAs which were higher in magnitude compared to both the soft tissue/nodal and visceral metastases. In turn, soft tissue/nodal tumors had higher MDA expression when compared to visceral tumors, which had negative antigen trends for all MDAs. Consistent with their independent clustering pattern by dendrogram analysis, the TYR expression in soft tissue/nodal metastases had a negative antigen trend that was not statistically different from visceral metastases by pairwise comparison (p=0.16 by Cochran-Armitage test) (Supplementary Table 4). In sum, these findings supported a non-stochastic stratified pattern of MDA expression that was highest in brain metastases, intermediate in soft tissue/nodal metastases, and lowest in visceral metastases. Notably, the expression profile for gp100 and MART were similar, but TYR antigen expression in sites outside of the brain was distinctly lower than the other two antigens.
Figure 3.
MDA expression in melanoma metastases cluster in a site specific phylogenetic pattern. (A) Dendrogram branch analyses based upon expression of individual MDAs in metastases of differing anatomic sites. (B) Dendrogram branch analyses of cumulative MDA expression in metastases. Arrows indicate the divergence of TYR expression from the clustering of gp100 and MART. Abbreviations: gp (gp100), M (MART), T (TYR). (C) Ranking of antigen expression trend by metastatic cluster site. Clustered sites are defined as: brain, ST/nodal (ST/SQ and LN), and visceral (lung and liver). The ordered results are presented from high positive to low or negative slope for display purposes and are not tested for this trend as it was not a pre-specified hypothesis. The p-values indicate the probability that no variability exists across the clustered sites as determined by the Kruskal-Wallis test.
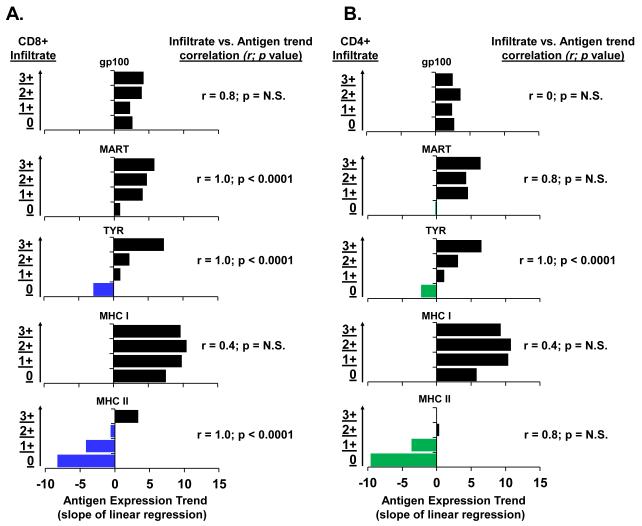
TYR is uniquely correlated with both CD4+ and CD8+ T cell infiltrate
To explore if the observed loss and variation in TYR expression may be a result of host immune targeting, we next sought to define the relationship between antigen expression with CD8+ and CD4+ T cell infiltration. Lymphocytic infiltrate was categorically scored from 0 (no infiltrate) to 3+ (high infiltrate). The antigen expression trend for each level of CD8+ and CD4+ lymphocytic infiltrate was determined (Supplementary Figure 2) and used to rank the expression associated with each degree of T cell infiltration. Based on overall expression levels, there were strong ordered associations between CD8+ T cell infiltration and MART, TYR, and MHC II expression in the metastases (each r=1.00; p<0.0001; Figure 4A). Interestingly, TYR was the only marker that demonstrated a strong association with CD4+ T cell infiltration (r=1.00; p<0.0001; Figure 4B). We concluded that the exclusive correlation of TYR expression with both CD8+ and CD4+ T cell infiltrate could be consistent with immunoediting as the mechanism for the selective loss of TYR among the metastases.
Figure 4.
TYR is uniquely correlated with both CD4+ and CD8+ T cell infiltrate. (A) Correlation between antigen expression trend and degree of CD8 infiltrate (y-axis). (B) Correlation between antigen expression trend and degree of CD4 infiltrate (y-axis). Blue (CD8) and green (CD4) coding of bars indicates negative antigen trends. Results presented are the Spearman correlation coefficient and a p-value for a test of whether r=0.
Discussion
From the prospective analyses of over 3000 human melanoma metastases, we demonstrate an anatomic site-specific pattern of melanocyte differentiation antigen expression with the highest levels seen in brain, intermediate levels in soft tissues/lymph nodes, and lowest levels in visceral metastases. Although heterogeneity of metastases has been well described (16-19), to our knowledge, a determined pattern of tumor antigen expression based upon anatomic site of metastasis has not been previously demonstrated in humans. Conceptually, metastatic heterogeneity is thought to result from stochastic genetic and epigenetic events under selective pressure exerted by the host and tumor microenvironment. This theory is based upon our understanding of the metastatic process as described originally by Paget’s “seed and soil” hypothesis (31), and refined by work from Fidler, and others, in animal models (1, 32). Site-specific interactions between the “seed” and “soil” could partially explain the tumor heterogeneity at different anatomic sites found in our study. In fact, the observation that brain metastases have the highest MDA expression is particularly consistent with early studies in nude mice which demonstrated that melanoma brain metastases were uniformly pigmented compared to the variable pigmentation of tumors observed at other metastatic sites (4). In contrast to these immune deficient animal models, here we sought to explore the interplay of the immune response with metastatic heterogeneity. Immunoediting, whereby antigen expressing cells are selectively eliminated or antigens are reversibly lost as an escape mechanism has been demonstrated in several animal models (20-23, 33, 34). However, the in vivo association of specific tumor antigens and MHC expression with natural endogenous immune responses has not been clearly demonstrated human. In a large cohort of melanoma metastases, we observed a significant association between tumor expression of MHC II (not MHC I) and CD8 infiltrate. We hypothesize that this counterintuitive finding suggests an important role for CD4+ T helper cell interaction with MHC II in the tumor microenvironment. This CD4+ “help” may facilitate CD8+ immune responses against endogenous tumor antigens. Although direct evidence for this phenomenon is beyond the observational aspect of this study, CD4 help has been demonstrated in numerous viral and tumor models.(35, 36) We believe that this observation in human tumors provides a biologically relevant association between these variables that may be further explored in future therapies involving CD4+ adoptive immunotherapy or vaccine efforts.
We also report that TYR expression is absent twice as often in melanoma metastases compared with gp100 and MART. Further, the discordant and selective loss of TYR was uniquely correlated with the levels of both endogenous CD8+ and CD4+ infiltrating T cells, suggesting that TYR expression in metastases may be naturally and selectively edited by antigen specific T cells. MART expression, although correlated with CD8, was not correlated with CD4 possibly suggesting the importance of a polyclonal T-cell population in immunoediting. It is interesting that in our antigen expression cluster analysis, brain metastases demonstrated no variation in TYR expression when compared with the profile of the other antigens. This finding would suggest that the process driving differential TYR loss is mitigated within the brain, potentially consistent with the immune-privileged status of the central nervous system. Interestingly, experimental evidence in animal models of melanoma has established immunoediting as the mechanism behind the preferential and site specific loss of TYR compared with other MDAs. Our findings are remarkably consistent with those reported in a double-transgenic MT-ret/AAD mouse model, which recapitulates the natural history of human melanoma through the spontaneous development of cutaneous and visceral tumors (37). The authors reported that TYR and tyrosinase-related protein 2 (TRP2) expression were markedly reduced in both liver and lung tumors when compared to cutaneous tumors. Further, they noted a concomitant natural induction of CD8+ T cells specific for both TYR and TRP2, suggesting that the visceral tumors, rather than the cutaneous tumors, were preferentially subjected to immunoediting by antigen specific T cells. In another murine study, a vaccine targeting normal melanocytes induced polyclonal T cell responses that resulted in the generation of B16 melanoma escape variants which grew aggressively in vivo, became amelanotic, and preferentially lost expression of TYR and TRP2, but maintained expression of other melanoma-associated antigens, such as gp100 (38); a finding that directly parallels our human observations.
Collectively, our current profiling of human metastasis antigen expression and immune infiltrate provides compelling evidence for a non-stochastic distribution of antigen expression based upon anatomic site. Although our study focused on the well-described MDAs, these results may warrant future investigation into the site-specific expression of a wide variety of antigens including mutated antigens and cancer-testis antigens, as well as the intratumoral heterogeneity of these antigens in individual metastases. The broad implications of our study would further support that clinicians utilizing immunotherapeutics – and potentially other targeted therapies – should consider that target assessment within metastasis is highly contingent on the anatomic site of biopsy. Ideally, biopsies should be obtained from the sites of disease anticipated to be most influential on the eventual outcome of the patient. Further, tracking antigen loss in tumors should be performed on the same lesion to avoid the confounding variable of interlesional heterogeneity. In sum, the cumulative findings in this report support that future clinical effort utilizing targeted immune therapies must account for site specific heterogeneity when assessing their use and impact on metastases.
Supplementary Material
Statement of Translational Relevance.
Metastasis heterogeneity presents a significant obstacle to the current developmental paradigm for highly targeted molecular and immune based cancer therapeutics. We sought to establish from a large series of human melanoma metastases whether a determined pattern in cellular heterogeneity exists that may guide future clinical efforts. By profiling for a panel of prototypic melanocyte lineage antigens, we found a non-stochastic site-specific pattern of expression in metastases that was highest in brain, intermediate in soft tissues/lymph nodes, and lowest in visceral sites. Tyrosinase demonstrated a unique expression profile with more frequent loss and an exclusive correlation with both endogenous CD8+ and CD4+ T cell infiltrate. We believe site-specific antigen heterogeneity represents a novel attribute for human melanoma metastases that should be considered when assessing the responsiveness to antigen specific therapies.
Acknowledgement
This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
Disclosures: There are no commercial or financial disclosures.
References
- 1.Fidler IJ. Biological heterogeneity of cancer: implication to therapy. Hum Vaccin Immunother. 2012;8:1141–2. doi: 10.4161/hv.19643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langley RR, Fidler IJ. Tumor cell-organ microenvironment interactions in the pathogenesis of cancer metastasis. Endocr Rev. 2007;28:297–321. doi: 10.1210/er.2006-0027. [DOI] [PubMed] [Google Scholar]
- 3.Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649–69. doi: 10.1158/0008-5472.CAN-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fidler IJ, Gruys E, Cifone MA, Barnes Z, Bucana C. Demonstration of multiple phenotypic diversity in a murine melanoma of recent origin. J Natl Cancer Inst. 1981;67:947–56. [PubMed] [Google Scholar]
- 5.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–5. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 6.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prickett TD, Wei X, Cardenas-Navia I, Teer JK, Lin JC, Walia V, et al. Exon capture analysis of G protein-coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet. 2011;43:1119–26. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozlowski JM, Hart IR, Fidler IJ, Hanna N. A human melanoma line heterogeneous with respect to metastatic capacity in athymic nude mice. J Natl Cancer Inst. 1984;72:913–7. [PubMed] [Google Scholar]
- 10.Leong SP, Gershenwald JE, Soong SJ, Schadendorf D, Tarhini AA, Agarwala S, et al. Cutaneous melanoma: a model to study cancer metastasis. J Surg Oncol. 2011;103:538–49. doi: 10.1002/jso.21816. [DOI] [PubMed] [Google Scholar]
- 11.Boon T, van der Bruggen P. Human tumor antigens recognized by T lymphocytes. J Exp Med. 1996;183:725–9. doi: 10.1084/jem.183.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robbins PF, Kawakami Y. Human tumor antigens recognized by T cells. Curr Opin Immunol. 1996;8:628–36. doi: 10.1016/s0952-7915(96)80078-1. [DOI] [PubMed] [Google Scholar]
- 13.Fetsch PA, Marincola FM, Filie A, Hijazi YM, Kleiner DE, Abati A. Melanoma-associated antigen recognized by T cells (MART-1): the advent of a preferred immunocytochemical antibody for the diagnosis of metastatic malignant melanoma with fine-needle aspiration. Cancer. 1999;87:37–42. [PubMed] [Google Scholar]
- 14.Fetsch PA, Steinberg SM, Riker AI, Marincola FM, Abati A. Melanoma antigen expression in serial fine-needle aspiration samples in patients with metastatic malignant melanoma participating in immunotherapy clinical trials: a preliminary look. Cancer. 2001;93:409–14. doi: 10.1002/cncr.10138. [DOI] [PubMed] [Google Scholar]
- 15.Fetsch PA, Riker AI, Marincola FM, Abati A. Tyrosinase immunoreactivity in fine-needle aspiration samples of metastatic malignant melanoma. Cancer. 2000;90:252–7. doi: 10.1002/1097-0142(20000825)90:4<252::aid-cncr9>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 16.Blessing K, Sanders DS, Grant JJ. Comparison of immunohistochemical staining of the novel antibody melan-A with S100 protein and HMB-45 in malignant melanoma and melanoma variants. Histopathology. 1998;32:139–46. doi: 10.1046/j.1365-2559.1998.00312.x. [DOI] [PubMed] [Google Scholar]
- 17.Cormier JN, Hijazi YM, Abati A, Fetsch P, Bettinotti M, Steinberg SM, et al. Heterogeneous expression of melanoma-associated antigens and HLA-A2 in metastatic melanoma in vivo. Int J Cancer. 1998;75:517–24. doi: 10.1002/(sici)1097-0215(19980209)75:4<517::aid-ijc5>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 18.de Vries TJ, Fourkour A, Wobbes T, Verkroost G, Ruiter DJ, van Muijen GN. Heterogeneous expression of immunotherapy candidate proteins gp100, MART-1, and tyrosinase in human melanoma cell lines and in human melanocytic lesions. Cancer Res. 1997;57:3223–9. [PubMed] [Google Scholar]
- 19.de Vries TJ, Smeets M, de Graaf R, Hou-Jensen K, Brocker EB, Renard N, et al. Expression of gp100, MART-1, tyrosinase, and S100 in paraffin-embedded primary melanomas and locoregional, lymph node, and visceral metastases: implications for diagnosis and immunotherapy. A study conducted by the EORTC Melanoma Cooperative Group. J Pathol. 2001;193:13–20. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH729>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 22.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–46. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235–71. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 24.Marincola FM, Hijazi YM, Fetsch P, Salgaller ML, Rivoltini L, Cormier J, et al. Analysis of expression of the melanoma-associated antigens MART-1 and gp100 in metastatic melanoma cell lines and in in situ lesions. J Immunother Emphasis Tumor Immunol. 1996;19:192–205. doi: 10.1097/00002371-199605000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Hollander M, W. D. Nonparametric Statistical Methods. Second Edition John Wiley and Sons, Inc.; New York: 1990. pp. 189–269. [Google Scholar]
- 26.Agresti A. Categorical Data Analysis. John Wiley and Sons, Inc.; New York: 1990. pp. 79–129. [Google Scholar]
- 27.Garcia-Vallve S, Puigbo P. DendroUPGMA: A dendrogram construction utility. 2009 http://genomes.urv.cat/UPGMA/index.php?entrada=Example1.
- 28.Garcia-Vallve S, Palau J, Romeu A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol Biol Evol. 1999;16:1125–34. doi: 10.1093/oxfordjournals.molbev.a026203. [DOI] [PubMed] [Google Scholar]
- 29.Du J, Miller AJ, Widlund HR, Horstmann MA, Ramaswamy S, Fisher DE. MLANA/MART1 and SILV/PMEL17/GP100 are transcriptionally regulated by MITF in melanocytes and melanoma. Am J Pathol. 2003;163:333–43. doi: 10.1016/S0002-9440(10)63657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou L, Panthier JJ, Arnheiter H. Signaling and transcriptional regulation in the neural crest-derived melanocyte lineage: interactions between KIT and MITF. Development. 2000;127:5379–89. doi: 10.1242/dev.127.24.5379. [DOI] [PubMed] [Google Scholar]
- 31.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1889;8:98–101. [PubMed] [Google Scholar]
- 32.Fidler IJ. Tumor heterogeneity and the biology of cancer invasion and metastasis. Cancer Res. 1978;38:2651–60. [PubMed] [Google Scholar]
- 33.Landsberg J, Kohlmeyer J, Renn M, Bald T, Rogava M, Cron M, et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 2012;490:412–6. doi: 10.1038/nature11538. [DOI] [PubMed] [Google Scholar]
- 34.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 35.Antony PA, Piccirillo CA, Akpinarli A, Finkelstein SE, Speiss PJ, Surman DR, et al. CD8+ T cell immunity against a tumor/self-antigen is augmented by CD4+ T helper cells and hindered by naturally occurring T regulatory cells. J Immunol. 2005;174:2591–2601. doi: 10.4049/jimmunol.174.5.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–6. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 37.Lengagne R, Graff-Dubois S, Garcette M, Renia L, Kato M, Guillet JG, et al. Distinct role for CD8 T cells toward cutaneous tumors and visceral metastases. J Immunol. 2008;180:130–7. doi: 10.4049/jimmunol.180.1.130. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Perez L, Kottke T, Diaz RM, Ahmed A, Thompson J, Chong H, et al. Potent selection of antigen loss variants of B16 melanoma following inflammatory killing of melanocytes in vivo. Cancer Res. 2005;65:2009–17. doi: 10.1158/0008-5472.CAN-04-3216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.