Abstract
Pol III promoters such as U6 are commonly used to express small RNAs, including small interfering RNA, short hairpin RNA, and guide RNA, for the clustered regularly interspaced short palindromic repeats genome-editing system. However, whether the small RNAs were precisely expressed as desired has not been studied. Here, using deep sequencing to analyze small RNAs, we show that, for mouse U6 promoter, sequences immediately upstream of the putative initiation site, which is often modified to accommodate the restriction enzyme sites that enable easy cloning of small RNAs, are critical for precise transcription initiation. When the promoter is kept unmodified, transcription starts precisely from the first available A or G within the range of positions −1 to +2. In addition, we show that transcription from another commonly used pol III promoter, H1, starts at multiple sites, which results in variability at the 5′ end of the transcripts. Thus, inaccuracy of 5′ end of small RNA transcripts might be a common problem when using these promoters to express small RNAs based on currently believed concepts. Our study provides general guidelines for minimizing the variability of initiation, thereby enabling more accurate expression of small RNAs.
Introduction
Because of the ease of silencing target genes, small interfering RNA (siRNA)/short hairpin RNA (shRNA) is currently one of the most commonly used tools for biomedical research, and particular siRNAs/shRNAs are also being explored as therapeutic candidates in a number of diseases. To provide long-term suppression of target genes, siRNA/shRNAs are commonly expressed with pol III promoters partly because of their defined sites for transcription initiation and termination.1,2,3,4,5,6,7,8 While the two siRNA strands can be transcribed separately to generate the siRNA duplex,3,4,6 shRNAs are transcribed as a single transcript with a hairpin structure,1,2,5,7,8 which is processed by Dicer into siRNA duplexes.5,9
Clustered regularly interspaced palindromic repeats (CRISPR)-Cas9 system is a natural mechanism in bacteria and archaea to defend against foreign DNA.10,11 The type II prokaryotic CRISPR-Cas9 endonuclease system has been recently applied as a new strategy for targeted genome engineering in which a short chimeric guide RNA (gRNA) guides Cas9 nuclease to edit genomic target sites.12,13,14,15,16,17,18,19,20 Compared with other genome-editing technologies, such as zinc finger nuclease and transcription activator-like effector nuclease, this new technology can be used to edit any genome target site with ease. The Cas9 enzyme being invariant, the only requirement for editing any chosen genomic site is the expression of specific small guide RNAs.12,13,14,15,16,17,18,19,20
Expressing small RNAs with desired sequence is a critical step for both siRNA/shRNA and CRISPR-Cas9 technologies. For siRNA, expressing two small RNAs with the exact designed sequences is critical to ensure loading of the desired strand into the RNA-induced silencing complex and to maintain specificity for the target gene. For shRNA, the 5′ end of shRNA transcripts is also critical, because the transcripts form a stem loop structure that is processed by Dicer, which measures 21 to 22 nt from the 5′ end of the transcript and performs a cleavage to generate the mature siRNA duplex.21,22 Even one nucleotide difference at the 5′ end of the transcript will generate a different siRNA duplex. For gRNAs of the CRISPR system, the first ~20-nt sequence of the guide RNA transcript defines the CRISPR target.10,13,14,15,16,18,19 Thus, precision at the 5′ end of these small RNA transcripts is critical for these technologies to work properly.
RNA pol III promoters, such as U6 and H1, are commonly used to express these small RNAs. It is believed that mouse U6 promoter transcription starts at the +1 position (23 nt after the TATA box), with G as the preferred initiation nucleotide.23,24,25 However, the exact U6 transcription initiation site has not been rigorously studied. Here, we show that the initiation of small RNAs driven by the mouse U6 promoter is often not at the presumed initiation site and is affected by the surrounding sequence. In addition, we found that the transcription initiation site of another commonly used promoter, H1, is generally variable. Thus, variability in the initiation site, which results in imprecision at the 5′ end of small RNA transcripts, might be a common problem. Our study provides general guidelines for using the pol III promoters to express small RNAs accurately.
Results
U6 transcription initiation is not always at the +1 position
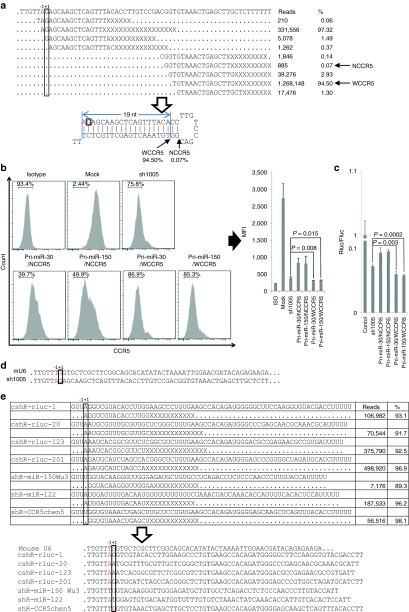
sh1005 is an shRNA that efficiently targets CCR5 expression26,27,28,29,30,31,32 and has been approved for a clinical trial aiming to treat HIV. However, U6-driven shRNAs showed high toxicity when used in T cells.27,33 Because miRNA-based shRNA mitigates the toxicity of conventional shRNA and also might have better functionality,34,35 we converted its presumed mature sequence (GGUGUAAACUGAGCUUGCUCUU)26,30 into miRNA-based shRNAs using pri-miR-30 or pri-miR-150 as backbone (see Supplementary Figure S1a). Surprisingly, both miRNA-based shRNAs showed significantly decreased functionality compared with the original sh1005 (see Supplementary Figure S1b). To understand how the original sh1005 and sh1005 in the miRNA backbone are processed by the endogenous miRNA machinery and why miRNA-based shRNAs are less efficient, we transfected the constructs into 293FT cells and sequenced the small RNAs. The miRNA-based shRNAs were processed precisely as predicted to generate the presumed mature sh1005 sequence (NCCR5; see Supplementary Figure S1c). To our surprise, the predominant mature product generated from sh1005 was UGUAAACUGAGCUUGCUCUUU (termed WCCR5), not its presumed mature sequence GGUGUAAACUGAGCUUGCUCUU (termed NCCR5; Figure 1a and Supplementary Table S1 and S2). To test whether this is the cause of the compromised functionality, we constructed two new miRNA-based shRNAs to express the WCCR5 sequence (see Supplementary Figure S1d). The processing of these miRNA-based shRNAs was confirmed by small RNA sequencing, and the results showed that they were processed exactly as predicted (see Supplementary Figure S1e). The functionality of these two new miRNA-based shRNAs (WCCR5) was higher than that of the previous miRNA-based shRNAs (NCCR5) and slightly higher than that of sh1005 (Figure 1b). The functionality of shRNAs was further confirmed by luciferase reporter assay (Figure 1c). These data show that the presumptive mature siRNA sequence according to the current conception can be significantly in error, and knowing the exact small RNA products generated from shRNAs is critical for designing optimal miRNA-based shRNAs.
Figure 1.
Transcription from the U6 promoter does not necessarily start from the +1 (G) position. (a) sh1005 was transfected into 293FT cells, and small RNAs were analyzed by deep sequencing. Representative reads of sequenced small RNAs are shown (all small RNAs sequenced for sh1005 can be found in Supplementary Tables S2 and S3). “X” indicates the positions at which these reads might end. The putative initiation site of transcription is boxed, and the real initiation site is marked in red. The Dicer cleavage site of the presumed mature sequences (NCCR5) and the real mature sequences (WCCR5) are indicated by arrows. (b) CCR5 knockdown efficiency of shRNAs in TZM-bl cells. TZM-bl cells were transfected with shRNAs and analyzed for CCR5 expression by flow cytometry 72 hours later. The bar graph on the right represents averages (±SD) of triplicates. MFI, mean fluorescence intensity. (c) The functionality of sh1005 and its miRNA-based shRNA counterpart was tested with the dual-luciferase assay, which was performed 24 hours after cotransfection of shRNAs with a pCHECK2 vector containing a 2x repeat of a sequence in the 3′ UTR that is fully complementary to the target sequence. The ratio of Renilla luciferase (Rluc, reporter) to firefly luciferase (Fluc, internal control) normalized to the negative control (mock vector) is shown. Error bar, 1 SD (d) Comparison of the initiation site of the natural mouse U6 small RNA with that of sh1005. (e) The transcription initiation site for seven randomly selected shRNAs was determined by analyzing the small RNAs after transfection. The initiation site is determined according to the 5′ end of the predominant sequenced small RNAs generated from 5′ arm of shRNA. All sequenced small RNAs can be found in Supplementary Tables S1 and S2. UTR, untranslated region.
It has been shown that, unlike the 3′ end of mature products of shRNA that is subject to extensive modification after its biogenesis, the 5′ end of mature products is generally protected from modification36,37,38,39,40 and therefore, the 5′ end of mature products generated from the 5′ arm of shRNA should faithfully indicate the original transcription initiation site. It is interesting that, judging from the predominant mature products generated from the 5′ arm of sh1005, the initiation site of transcription for sh1005 is at the –1 (A) position, not the generally accepted +1 (G) position for the mouse U6 promoter (Figure 1a,d). The single-nucleotide shift of the sh1005 transcription initiation site extended the stem by 1 bp (Figure 1a, lower panel). Since Dicer measures 21–22 nt from the 5′ end of the transcript and performs a cleavage to generate the mature siRNA duplex,21,22 this extension may have caused a shift in the Dicer cleavage site and thus may explain why the mature siRNA sequence is not the presumed one. However, it appears that variability of transcription initiation might be a common problem in using the U6 promoter to express small RNAs. A recent report in which the U6 promoter was used to express miR-30 also showed that the initiation site is not precise and that the actual dominant mature product generated is not miR-30 (69% of the reads were 3 nt shorter at the 5′ end).41 The precision of the initiation site is critical for expressing shRNA or miRNA mimics to generate the desired mature sequences. Thus, we wanted to determine the exact initiation site of the U6 promoter. We first randomly selected seven shRNAs available in the laboratory, transfected them into 293FT, and sequenced the small RNAs. The results showed that the initiation site was not always at the +1 (G) position and that it followed a pattern: if the –1 position is an A, the initiation site is at –1, whereas if the –1 position is a T, the initiation site is at +1 (G, Figure 1e). This pattern is reminiscent of the native mouse U6 sequence, which has the sequence GTTT upstream of the +1 (G) position (Figure 1d). Thus, it appears that if the –1 position happens to be a T, as in the native U6 promoter, the initiation site will be at +1 (G). If the –1 position is changed to A, the initiation site will be changed to the –1 position, suggesting that the nucleotides upstream of the initiation sites can affect transcription initiation.
Transcription initiation is affected by the sequence around the initiation site
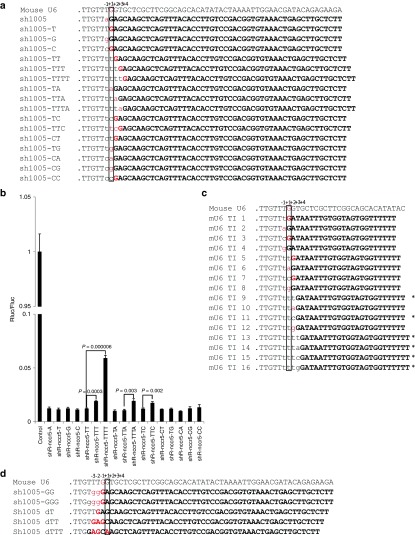
To understand how the surrounding sequence affects U6 promoter initiation site selection, we systematically investigated the effect of nucleotide alterations around the +1 position on the transcription initiation of sh1005 by inserting different nucleotides upstream, as shown in Figure 2a. The constructs were transfected into 293FT cells separately, and small RNA products were sequenced to determine the initiation site. Transcription initiation site selection appeared to follow a simple rule: transcription starts from the first A or G beginning at the –1 position (Figure 2a). It appears that transcription can also start from the first C, but with much less efficiency (Table 1). According to read abundances, transcription efficiency appears to decrease if A/G occurs at, or downstream of, the +3 position (Table 1). To confirm the effect of a change in initiation site on transcription efficiency, we measured the functionality of shRNAs, which decreased significantly if the initiation site was at the +3 position, as is the case for shR-nccr5-TTT, -TTTA, or -TTC, and the functionality was reduced further if the initiation site was at the +4 position (shR-nccr5-TTTT; Figure 2b). The results appear to be consistent with small RNA cloning data, suggesting that transcription efficiency appears to decrease if initiation begins at the +3 position or further downstream.
Figure 2.
Analysis of the transcription initiation site driven by the U6 promoter. (a) The transcription initiation site for sh1005 serial mutations. The predominant sequenced small RNAs can be found in Table 1. Lowercase letters indicate the nucleotides replacing the A at the –1 position of sh1005. The presumed initiation site of transcription is boxed, and the real initiation site is marked red. The original sh1005 is marked bold. (b) The functionality of sh1005 serial mutations. The dual-luciferase assay was performed as in Figure 1c. (c) The transcription initiation site for small RNAs (~21 nt in length) in which the nucleotides around the initiation site are different. Lowercase letters indicate nucleotides inserted between the –2 position of the U6 promoter and the universal sequence GATAATTTGTGGTAGTGGTT. Asterisks indicates constructs for which an insufficient number of reads were sequenced to determine the exact initiation site. The predominant small RNAs can be found in Supplementary Figure S2b. (d) The sequence immediately upstream of the –1 site affects transcription initiation. The initiation sites of the indicated sh1005 mutations with altered sequences immediately upstream of the –1 site are marked red. The predominant sequenced small RNAs can be found in Table 2.
Table 1. Representative small RNA reads cloned from sh1005 serial mutations in Figure 2a.
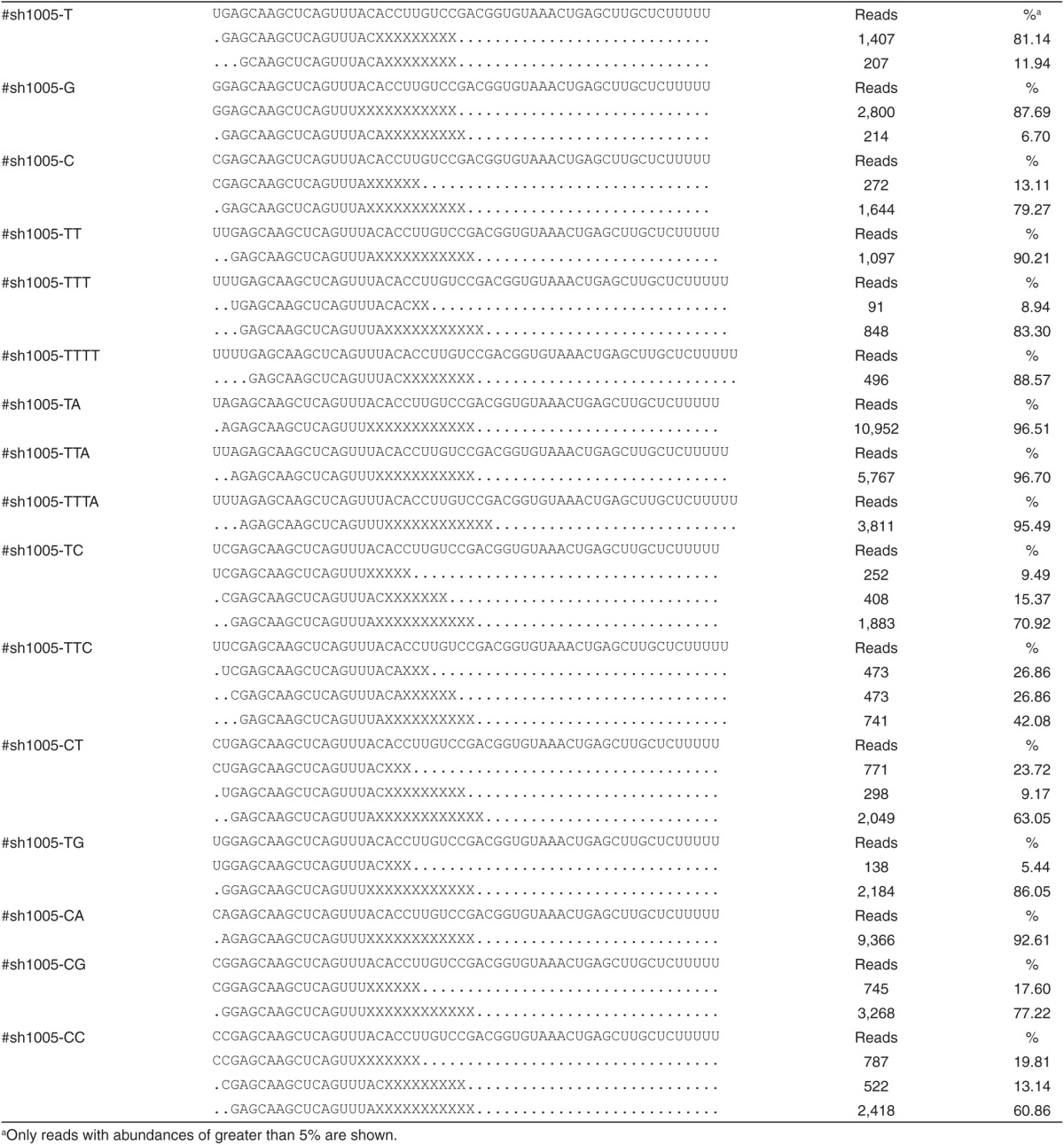
In previous experiments, we relied on mature shRNA products to determine transcription initiation sites. Loading of the mature shRNA sequence into the RNA-induced silencing complex might introduce bias because of the strong sequence preference of the process. Thus, we designed a series of constructs expressing a small RNA (GAUAAUUUGUGGUAGUGGUU) with different sequences around position +1. The small RNA is ~21 nt in length, so we could sequence it with our regular small RNA sequencing method. The constructs were transfected into 293FT cells separately, and the small RNAs were sequenced. The results were consistent with the experiments done with sh1005. Transcription did not start until there was an A or G, and the initiating A or G had to be within the range of positions –1 to +2 for efficient transcription (Figure 2c and Supplementary Figure S2). We were not able to sequence many reads for any of the constructs in which A or G was at the +3 or +4 positions, suggesting that the number of transcripts decreased if the A or G was at, or downstream of, position +3 (Figure 2c and Supplementary Figure S2).
Thus, sequence changes around the putative initiation site, ranging from positions –1 to +4, can affect transcription initiation. To understand how the sequence upstream of the −1 site affects transcription initiation, the T at positions –2 and –3 was changed to G by point mutation (shR1005-GG and shR-1005-GGG; Figure 2d). Compared with shR1005-G (Figure 2a), the transcription initiation of shR1005-GG and shR1005-GGG started at multiple sites (Figure 2d). We further deleted the Ts at positions –1, –2, and –3 (Figure 2d). Compared with sh1005-T (Figure 2a) and sh1005 dT, in which the transcription initiation is precise, the transcription of sh1005 dTT and sh1005 dTTT started at multiple sites (Figure 2d and Table 2). These data suggest that the sequence immediately upstream of the –1 site can affect initiation, and two continuous Ts immediately upstream of the –1 site are required for accurate initiation of mouse U6-driven small RNA expression. Thus, the sequence immediately upstream of the –1 site affects transcription initiation by the mouse U6 promoter.
Table 2. Representative small RNA reads cloned from sh1005 serial mutations in Figure 2d.

In summary, we identified a new region in the mouse U6 promoter—the sequence around the putative initiation site ranging from position –3 to +4—that affects the precision and efficiency of transcription initiation. Our results suggest the following guidelines for using the mouse U6 promoter to generate the desired small RNA sequence: the initiation nucleotide can be either A or G, and it should be within the range of positions –1 to +2. It is also critical to maintain the sequence immediately upstream of the initiation site, especially a continuous sequence of Ts, and avoid inserting A, G, or even C upstream of the desired initiation site.
Applying the new guidelines to improve the design of shRNA and gRNA
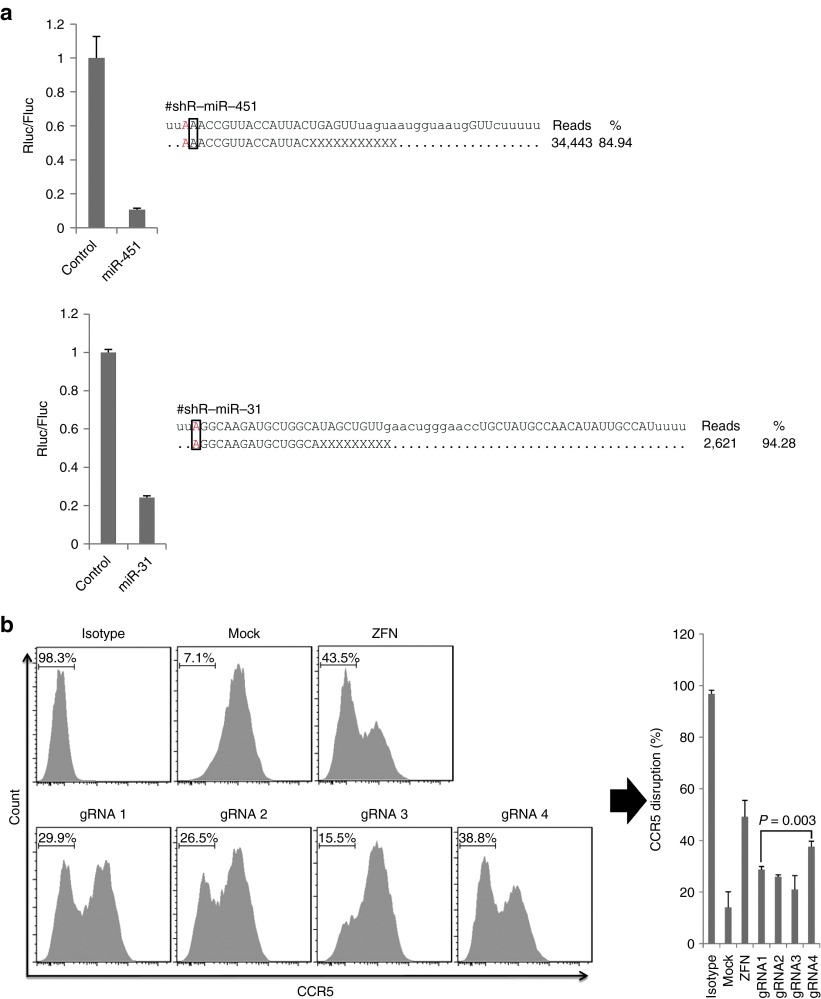
The starting nucleotide for natural miRNAs or potent siRNAs is often U or A, which is not easy to convert into shRNAs driven by the U6 promoter, at least according to the previously held concept. With our findings on the mouse U6 promoter initiation site, it should be easy to design shRNA or miRNA mimics starting with nucleotide A. To test this, we designed two shRNAs mimicking miR-451 and miR-31, both starting with nucleotide A, and both were highly potent in repressing targets (Figure 3a). To confirm that the miRNA mimics were processed as predicted, both constructs were transfected into 293FT cells, and the small RNAs were sequenced. The mature products were exactly the same as for the miRNAs (Figure 3a). Thus, improved knowledge of the U6 promoter enables the design of shRNA or miRNA mimics starting with nucleotide A, and the desired sequence can be generated as designed.
Figure 3.
Applying the new finding to the design of miRNA mimics and gRNAs starting with A. (a) The functionality of miR-451 and miR-31 mimics (left) was tested as in Figure 1c. The predominant mature products generated from these two mimics are shown. All small RNAs sequenced can be found in Supplementary Tables S1 and S2. The mature miRNA sequences designated in miRBase are shown as uppercase letters. (b) The knockout efficiency of gRNAs starting with A or G, with zinc finger nuclease (ZFN) serving as the positive control. TZM-bl cells were cotransfected with gRNA constructs and Cas9-expressing plasmid and analyzed for CCR5 expression by flow cytometry 72 hours later. The bar graph on the right represents the averages (±SD) of triplicates. gRNA, guide RNA.
Because it can knock out any gene with ease, the recently developed genome-editing technology, based on the CRISPR-Cas system, promises to revolutionize the biomedical research field as much or more than RNAi. However, the target-site selection is limited by the requirements that NGG should be the sequence downstream of the target site and that the initiation site must be G if the U6 promoter is used to transcribe the gRNA. Now, with our new finding about U6 initiation site selection, we can choose target sites starting with either A or G, which doubles the number of possible target sites. Four gRNAs targeting the CCR5 gene were chosen, two of them initiated with A and two initiated with G, to test gene-knockout efficiency. The gRNA constructs targeting CCR5 were cotransfected with a Cas9-expressing plasmid into TZM-bl cells. Knockout efficiency was determined by flow cytometry 72 hours later, and all gRNAs were found to be functional (Figure 3b). It is noteworthy that the gRNA4 construct with the highest functionality is actually initiated with A (Figure 3b and Supplementary Table S3). Thus, improved knowledge of the U6 promoter doubles the number of choices of target sites for CRISPR-Cas system–mediated genome-editing technology.
Transcription of other pol III promoters
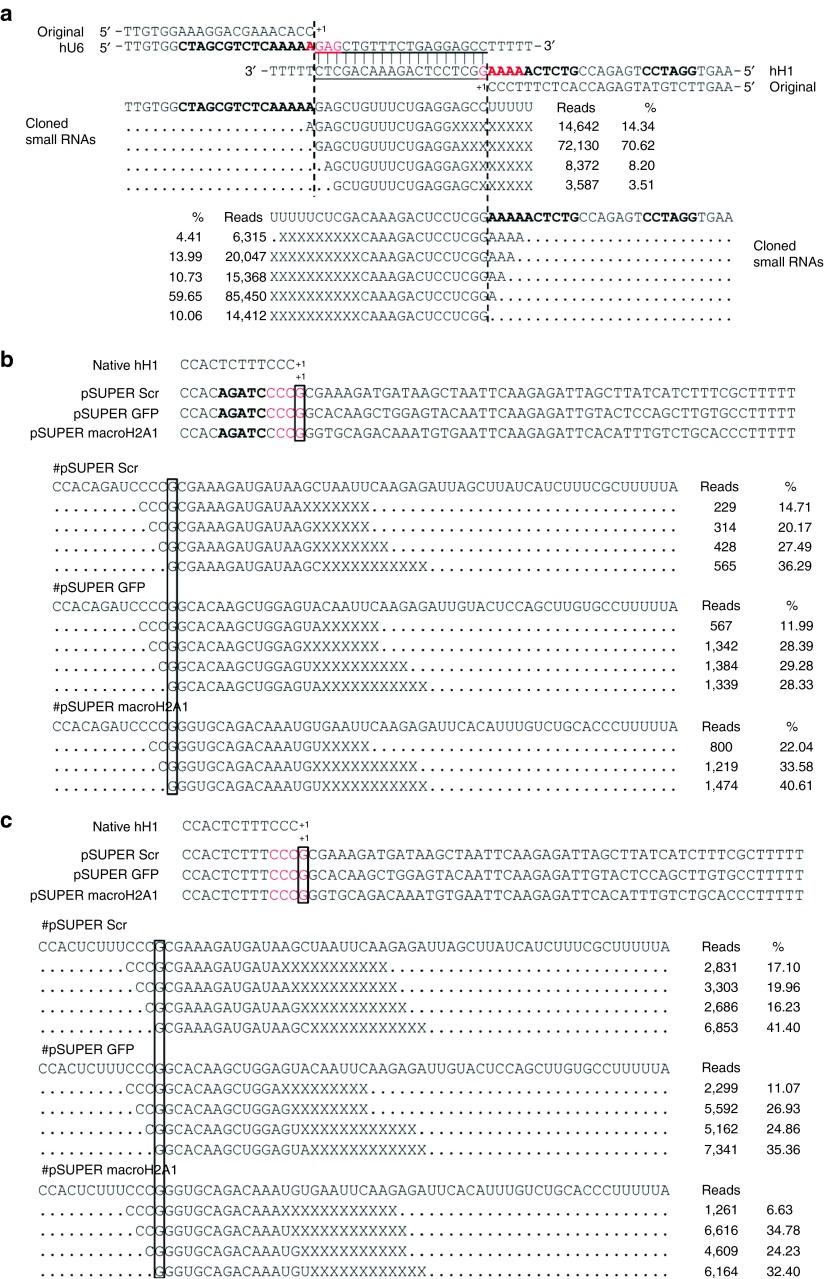
Until this point, all experiments were based on the mouse U6 promoter. Next, we checked the transcription initiation site of two different pol III promoters. V45 pHIPPY PGL3 luciferase is a plasmid using the human U6 and H1 promoters to drive transcription of two small RNAs of ~22 nt in length from opposite directions to generate an siRNA duplex targeting the luciferase gene.42 We sequenced the resulting small RNAs to determine the initiation sites for these promoters. As shown in Figure 4a, the putative initiation sites for both promoters were not correct, as the transcripts actually started from multiple sites. The reason for this variability might be that the sequences upstream of the initiation sites in this construct were altered to accommodate the restriction enzyme site and termination signals (TTTTT) for both promoters (Figure 4a). This possibility would be consistent with our previous conclusion that the sequence immediately upstream of the initiation site contributes to the accuracy of initiation. To confirm this hypothesis, we sequenced small RNAs generated from several shRNAs expressed by the human U6 promoter (pLKO.1 vector) in which the sequence upstream of the initiation site was kept unaltered. As shown in Supplementary Figure S3a, the initiation site for these shRNAs was precisely where predicted, suggesting that maintaining intact the sequence upstream of the initiation site is critical for the precision of initiation of the human U6 promoter, just like what we show for mouse U6 promoter (Figure 2). We also sequenced small RNAs generated from several shRNAs driven by the H1 promoter (pSUPER vector) in which the sequences upstream of the initiation sites were altered. As shown in Figure 4b, transcription started from multiple sites in all cases. However, after we corrected the mutated upstream sequence by point mutation, the transcription still started from multiple sites (Figure 4c). Thus, it appears that transcription driven by the H1 promoter is generally variable, which results in inaccuracy at the 5′ end of the transcripts. It is noteworthy that the transcription initiation constantly started at multiple sites ranging from −3 to +1 position (Figure 4a–c).
Figure 4.
The transcription driven by the H1 promoter is generally variable. (a) Small RNAs generated from V45 pHIPPY PGL3-transfected cells. The putative siRNA sequences are underlined. The dotted line indicates the presumed initiation site, and bold letters represent the mutated sequence. (b) The initiation site of shRNAs driven by the human H1 promoter with an altered sequence upstream of the initiation site. (c) The initiation site of the shRNAs from Figure 4b with the original H1 promoter. shRNA, short hairpin RNA; siRNA, small interfering RNA.
Discussion
It has been commonly accepted that transcription from the U6 promoter starts at the +1 position (23 nt downstream of the TATA box), with G as the preferred initiation nucleotide. In this study, we systematically investigated transcription initiation of the mouse U6 promoter and identified a new region, the sequence around the putative initiation site ranging from position –3 to +4, which affects transcription initiation accuracy and the efficiency of the U6 promoter. Transcription can start with A or G, ranging from the –1 to the +2 positions. This new knowledge provides general guidelines for using the U6 promoter to express small RNAs such as siRNAs/shRNAs and gRNAs for CRISPR.
A previous study showed that the transcription initiation of a yeast tRNA promoter prefers purine (both A and G).43 Our study also showed that the transcription initiation of U6 promoter prefers purine, suggesting that the preference for purine as the initiation nucleotide might be a conserved phenomenon across species. However, for some other pol III promoters, such as H1, the transcription can start with pyrimidine (Figure 4), suggesting that preference for purine as the initiation nucleotide might not be a universal rule for pol III promoters.
Our study also showed that the sequences around the putative initiation site in the U6 promoter are important for precision of transcription initiation; however, the sequences are commonly altered to accommodate a restriction enzyme recognition sequence in order to simplify the cloning procedure, because their effect on transcription initiation was not known previously. In fact, when we checked the sequence upstream of the initiation site in commercially available vectors that use the U6 promoter to express small RNAs, we found that, in all cases, the sequence upstream of the initiation site had been changed (see Supplementary Figure S3b). Such changes are likely to result in inaccuracy of the 5′ end of transcripts. In addition, we showed that transcription initiation using another commonly used promoter, H1, which has been used in several commercially available vectors (see Supplementary Figure S4), is generally variable (Figure 4). Thus, inaccuracy of the 5′ end of transcripts driven by the pol III promoter might be a common problem. In addition to yielding small RNA products that are not the desired sequences, thus affecting on-target functionality, such inaccuracy is also likely to increase off-target effects caused by undesired small RNA products. Thus, our study provides guidelines for using the pol III promoter to transcribe accurately, which might increase on-target functionality and minimize off-target effects. Although the sequence upstream of the putative initiation site in commercially available vectors was modified, the sequence in some freely available vectors, such as pLL3.7, pLB, and pLKO.1, which can be easily obtained from Addgene, was not changed. These vectors can be used to express small RNAs precisely.
It appears that the initiation constantly starts from multiple sites ranging from −3 to +1 position for H1 promoter (Figure 4). The information might be used to increase the chance to design potent shRNAs. Currently, it is still impossible to predict the potency of shRNA. By purposely using H1 promoter to generate shRNA transcripts with different 5′ ends from a single construct, which will be processed by Dicer into distinct siRNA duplexes because Dicer can make a cleavage by measuring the distance from the 5′ ends,22 the chances of getting a potent shRNA will be significantly increased since one nucleotide change might change the potency dramatically, although the off-targets might also be increased due to multiple mature siRNAs might be generated.
It has been commonly accepted that the termination signal for pol III promoter-driven transcription is a simple four to six-residue poly-T,44 which is currently used as the terminal signal for almost all small RNA expression cassettes driven by the pol III promoter. However, we found that poly-T might not be an efficient termination signal. miR-155, which has five continuous Ts in the middle of its pri-miRNA, can be efficiently expressed using the U6 promoter, as abundant mature miR-155 can be cloned at a level similar to miR-223, which does not have a poly-T signal in the middle of its pri-miRNA sequence (see Supplementary Figure S5). This finding suggests that the poly-T signal in miR-155 does not terminate transcription efficiently at this site and is consistent with a recent report in which Nielson et al.45 found that the poly-T signal does not cause termination by itself but rather causes a pause by Pol III, and termination requires an additional signal from the RNA secondary structure. Thus, the currently accepted concept of pol III promoter-driven transcription initiation and termination is incomplete, which often results in expression of small RNAs from these promoters that do not have the designed sequence.
It is surprising that no systematic studies have been done to provide guidelines on how to use pol III promoters appropriately, although these promoters have been commonly used to express siRNA and shRNA since 2002. In most cases, the pol III product sequence is assumed based on current concepts of Pol III transcription initiation. Very few studies have validated the exact sequence experimentally by sequencing. As our study shows, the exact mature product of sh1005, which has been approved for a clinical trial, is not the presumed sequence, and transcription does not initiate from the presumed initiation site (Figure 1a). A recent report in which U6 promoter was used to express miR-30 also showed that the initiation site is not precise.41 Thus, a significant number of previous studies those used pol III promoter to express small RNAs will need to be revisited, because the putative small RNA sequences expressed might actually be incorrect.
Materials and methods
Plasmids. All the small RNA constructs were designed as oligos and inserted into pLL3.7 at restriction sites HpaI and XhoI. The inserted sequences are listed in Supplementary Table S3. V45 pHIPPY PGL3 luciferase, shRNA constructs driven by the human U6 promoter (pLKO.1), and the human H1 promoter (pSUPER vector) were obtained from Addgene.
Dual luciferase assay. 293FT cells (Invitrogen, Carlsbad, CA) were cultured according to the manufacturer's instructions. The day before transfection, 293FT cells were trypsinized and diluted to 105 cells/ml and seeded in 96-well plates in a volume of 100 µl/well. shRNA construct (0.1 µg) and psiCHECK2 harboring the target sequence (0.1 µg) were cotransfected into 293FT cells with lipofectamine 2000 per the manufacturer's instructions. The Dual-Glo luciferase assay (Promega, Madison, MI) was performed 24 hours later.
Small RNA sequencing. Small RNA libraries were constructed and sequenced in a similar manner as described previously46,47 but with major modifications to improve adaptor ligation efficiency and thereby reduce ligation bias. The same method has been used in our recent study.36 A manuscript describing the detailed methods is in preparation. Briefly, 28 hours after the constructs were transfected into 293FT cells, the small RNAs were purified with the miRNeasy kit (Qiagen, Valencia, CA) per the manufacturer's instructions. Small RNA (50 ng) was ligated with 3′ and 5′ linkers (with barcode) using an improved ligation method that was optimized comprehensively to minimize the ligation bias between different small RNAs. The ligated small RNAs were reverse transcribed and amplified with the KAPA library amplification kit (KAPA Biosystems, Woburn, MA) for 10 cycles, and the library sequenced using the Illumina HiSeq2000, San Diego, CA. All reads that were sequenced only once were discarded to lower the noise level. The sequenced small RNAs are included in Supplementary Table S1.
Flow cytometry analysis of CCR5 expression. Seventy-two hours after transfection of the constructs into TZM-bl cells (with lipofectamine 2000 per the manufacturer's instructions), the cells were detached and stained with CCR5 antibody conjugated with APC (BD Pharmingen, San Jose, CA) and subjected to flow cytometry analysis. The pLL3.7 vector used to express small RNAs has a green fluorescent protein marker, which can serve as a marker for cells that are transfected; thus, green fluorescent protein–positive cells were gated by their expression of CCR5 and analyzed.
Statistical analysis. Student's t-test (two-tailed, assuming equal variances on all experimental data sets) was used to compare two groups of independent samples.
SUPPLEMENTARY MATERIAL Figrue S1. The processing of miRNA-based shRNAs. Figure S2. The small RNAs generated from the constructs in Figure 2c. Figure S3. Human U6 promoter. Figure S4. Commercially available vectors using the H1 promoter. Figure S5. The mature miRNAs generated from the indicated pri-miRNAs. Table S1. Small RNAs generated from various constructs. Table S2. Simplified data from Supplementary Table S1. Table S3. The oligo sequences that were inserted into pLL3.7 vectors.
Acknowledgments
The authors thank Jessica Montoya for technical support and Dianne Mitchell and the staff of the genomic core facility at Texas Tech University Health Science Center for their assistance with DNA sequencing.
The authors declare that they have no conflict of interest.
Supplementary material
The processing of miRNA-based shRNAs.
The small RNAs generated from the constructs in Figure 2c.
Human U6 promoter.
Commercially available vectors using the H1 promoter.
The mature miRNAs generated from the indicated pri-miRNAs.
Small RNAs generated from various constructs.
Simplified data from Supplementary Table S1.
The oligo sequences that were inserted into pLL3.7 vectors.
References
- Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, et al. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Winer I, Engelke DR. Effective expression of small interfering RNA in human cells. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, et al. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, et al. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–472. doi: 10.1038/cr.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, et al. Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Maser RL, Calvet JP, Pederson T. U6 small nuclear RNA is transcribed by RNA polymerase III. Proc Natl Acad Sci USA. 1986;83:8575–8579. doi: 10.1073/pnas.83.22.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goomer RS, Kunkel GR. The transcriptional start site for a human U6 small nuclear RNA gene is dictated by a compound promoter element consisting of the PSE and the TATA box. Nucleic Acids Res. 1992;20:4903–4912. doi: 10.1093/nar/20.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Cathomen T. RNA guides genome engineering. Nat Biotechnol. 2013;31:208–209. doi: 10.1038/nbt.2527. [DOI] [PubMed] [Google Scholar]
- An DS, Donahue RE, Kamata M, Poon B, Metzger M, Mao SH, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci USA. 2007;104:13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kamata M, Kittipongdaja P, Chen KN, Kim S, Pang S, et al. Characterization of a potent non-cytotoxic shRNA directed to the HIV-1 co-receptor CCR5. Genet Vaccines Ther. 2009;7:8. doi: 10.1186/1479-0556-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata M, Liu S, Liang M, Nagaoka Y, Chen IS. Generation of human induced pluripotent stem cells bearing an anti-HIV transgene by a lentiviral vector carrying an internal murine leukemia virus promoter. Hum Gene Ther. 2010;21:1555–1567. doi: 10.1089/hum.2010.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Kamata M, Chen KN, Pariente N, An DS, Chen IS. Inhibition of HIV-1 infection by a unique short hairpin RNA to chemokine receptor 5 delivered into macrophages through hematopoietic progenitor cell transduction. J Gene Med. 2010;12:255–265. doi: 10.1002/jgm.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood. 2010;115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds GP, Johnstone HA, Millington ML, Boyd MP, Burke BP, Breton LR. The use of cell-delivered gene therapy for the treatment of HIV/AIDS. Immunol Res. 2010;48:84–98. doi: 10.1007/s12026-010-8169-7. [DOI] [PubMed] [Google Scholar]
- Ringpis GE, Shimizu S, Arokium H, Camba-Colón J, Carroll MV, Cortado R, et al. Engineering HIV-1-resistant T-cells from short-hairpin RNA-expressing hematopoietic stem/progenitor cells in humanized BLT mice. PLoS One. 2012;7:e53492. doi: 10.1371/journal.pone.0053492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D, et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14:494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wu Y, Choi JG, Wu H. Lower and upper stem-single-stranded RNA junctions together determine the Drosha cleavage site. Proc Natl Acad Sci USA. 2013;110:20687–20692. doi: 10.1073/pnas.1311639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Mani V, Hammond SM. Deep sequencing of microRNA precursors reveals extensive 3' end modification. RNA. 2011;17:1795–1803. doi: 10.1261/rna.2713611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, et al. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ye C, Ramirez D, Manjunath N. Alternative processing of primary microRNA transcripts by Drosha generates 5' end variation of mature microRNA. PLoS One. 2009;4:e7566. doi: 10.1371/journal.pone.0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T, et al. Selective stabilization of mammalian microRNAs by 3' adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, et al. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaykas A, Moon RT. A plasmid-based system for expressing small interfering RNA libraries in mammalian cells. BMC Cell Biol. 2004;5:16. doi: 10.1186/1471-2121-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zecherle GN, Whelen S, Hall BD. Purines are required at the 5' ends of newly initiated RNAs for optimal RNA polymerase III gene expression. Mol Cell Biol. 1996;16:5801–5810. doi: 10.1128/mcb.16.10.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Sakonju S, Brown DD. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980;19:27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–1580. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The processing of miRNA-based shRNAs.
The small RNAs generated from the constructs in Figure 2c.
Human U6 promoter.
Commercially available vectors using the H1 promoter.
The mature miRNAs generated from the indicated pri-miRNAs.
Small RNAs generated from various constructs.
Simplified data from Supplementary Table S1.
The oligo sequences that were inserted into pLL3.7 vectors.