ABSTRACT
It is not currently possible to predict the probability of whether a woman with a chlamydial genital infection will develop pelvic inflammatory disease (PID). To determine if specific biomarkers may be associated with distinct chlamydial pathotypes, we utilized two Chlamydia muridarum variants (C. muridarum Var001 [CmVar001] and CmVar004) that differ in their abilities to elicit upper genital tract pathology in a mouse model. CmVar004 has a lower growth rate in vitro and induces pathology in only 20% of C57BL/6 mouse oviducts versus 83.3% of oviducts in CmVar001-infected mice. To determine if chemokine and cytokine production within 24 h of infection is associated with the outcome of pathology, levels of 15 chemokines and cytokines were measured. CmVar004 infection induced significantly lower levels of CXCL1, CXCL2, tumor necrosis factor alpha (TNF-α), and CCL2 in comparison to CmVar001 infection with similar rRNA (rs16) levels for Chlamydiae. A combination of microRNA (miRNA) sequencing and quantitative real-time PCR (qRT-PCR) analysis of 134 inflammation-related miRNAs was performed 24 h postinfection to determine if the chemokine/cytokine responses would also be reflected in miRNA expression profiles. Interestingly, 12 miRNAs (miR-135a-5p, miR298-5p, miR142-3p, miR223-3p, miR299a-3p, miR147-3p, miR105, miR325-3p, miR132-3p, miR142-5p, miR155-5p, and miR-410-3p) were overexpressed during CmVar004 infection compared to CmVar001 infection, inversely correlating with the respective chemokine/cytokine responses. To our knowledge, this is the first report demonstrating that early biomarkers elicited in the host can differentiate between two pathological variants of chlamydiae and be predictive of upper tract disease.
IMPORTANCE
It is apparent that an infecting chlamydial population consists of multiple genetic variants with differing capabilities of eliciting a pathological response; thus, it may be possible to identify biomarkers specific for a given virulence pathotype. miRNAs are known to regulate genes that in turn regulate signaling pathways involved in disease pathogenesis. Importantly, miRNAs are stable and can reflect a tissue response and therefore have the potential to be biomarkers of disease severity. Currently, with respect to chlamydial infections, there is no way to predict whether an infected patient is more or less likely to develop PID. However, data presented in this study indicate that the expression of a specific miRNA profile associated with a virulent variant early in the infection course may be predictive of an increased risk of pelvic inflammatory disease, allowing more aggressive treatment before significant pathology develops.
INTRODUCTION
Chlamydia trachomatis infections in women differ greatly in the nature and intensity of disease. A major conundrum with respect to chlamydial infections is why some women develop clinical disease while the majority remain asymptomatic. Clearly, there can be multiple reasons for this, including host genetic differences (1), the use of oral contraceptives (2, 3), the stage of the menstrual cycle in which infection occurs (4), and the presence of other risk factors such as bacterial vaginosis (5), but it is more likely that a combination of factors is responsible. Another possible explanation is that the infecting population consists of multiple genetic variants, with the pathological phenotype being dependent upon the numerical representation of variants in the infecting population. Miyairi and colleagues recognized differences among serovars of C. trachomatis, reporting that ocular serovars had longer developmental cycles and lower growth rates than genital serovars, and related this to the pathotype (6). Along the same line, Kari et al. tested two different isolates of C. trachomatis serovar A and observed that one isolate which had smaller plaques, slower growth, and increased sensitivity to gamma interferon (IFN-γ) had a lower peak number of organisms with shorter infection and lesser pathology upon conjunctival infection of nonhuman primates (7). Thus, it is clear that even within a given serovar, there can be significant variations in virulence.
The concept that variants within populations exist naturally was presented by Ramsey and colleagues in their examination of the Nigg and Weiss strains of C. muridarum which differ in virulence (8). We reported a similar observation using plaque-purified C. caviae isolated from a conjunctival swab from an infected guinea pig (9). In an elegant study, Sturdevant and colleagues infected mice with C. trachomatis serovar D and found that mice resolved infections at vastly different times, from 10 to 77 days (10). They found that when mice were infected with chlamydiae isolated from mice at 10 days and 49 days postinfection, the infection courses were consistently shorter for the day 10 isolate and longer for the day 49 isolate. Moreover, the day 49 isolate produced more pathology in the upper genital tract than the day 10 isolate. The authors concluded that the “serovar D parental stock was a mixture of organisms varying in virulence for the mouse” (10). The study results clearly indicate that infecting populations consist of mixtures of variants with different pathological capabilities and that the outcome of the infection is dependent upon the virulence of those variants within the population.
Currently, there is no way to determine whether a woman infected with Chlamydia is at risk for upper genital tract disease. It would be advantageous if one could identify specific biomarkers that would help to predict the pathological outcome of the infection early in the disease process to enable proactive treatment for the prevention of pelvic inflammatory disease (PID). MicroRNAs (miRNAs) are abundant and evolutionarily conserved noncoding small (~22-nucleotide) oligonucleotide molecules. They are immune modulators that serve as an important link between innate and adaptive immune responses (11). Dysregulation of miRNA expression has been linked to cardiovascular diseases, cancer, infectious metabolic diseases, and other diseases (12–14). Most importantly, miRNAs have been shown to regulate chemokine cytokine responses during different bacterial infections (15, 16). In addition, several studies have suggested that miRNAs could be a new class of biomarkers for diagnostic and therapeutic purposes under different pathological conditions (17, 18). Thus, we hypothesized that different pathological variants of C. muridarum would elicit different miRNA profiles and that such profiles would be predictive of the development or lack of development of upper genital tract disease. In this study, we characterized two chlamydial variants (C. muridarum Var001 [CmVar001] and CmVar004) that differ in their abilities to induce upper genital tract pathology in mice by examining microRNA (miRNA) expression profiles induced by these variants and their relationship to chemokine/cytokine responses within 24 h of infection.
RESULTS
In vitro growth characteristics of chlamydial variants.
We characterized plaque-purified variants of C. muridarum derived from genital tract swabs of infected mice. These variants differed in their plaque size and in vitro growth characteristics (Table 1). Growth curve analysis of variants showed different phenotypes, with CmVar001 being the fastest growing variant followed by CmVar002, while CmVar001.1, CmVar003, and CmVar004 had lower growth rates. The rate of elementary body (EB) generation per developmental cycle was calculated by dividing the number of infectious progeny from supernatants at 36 h by the number of EBs used to infect the cultures (multiplicity of infection = 1). Similarly, the EB generation rate was higher for CmVar001 and CmVar002 than for CmVar001.1, CmVar003, and CmVar004. Interestingly, there was a 6-fold difference in EB production rates between CmVar001 and CmVar004, and the doubling time results showed that the 2-fold EB increase time for CmVar004 was 136 min versus 78 min for CmVar001, indicating that, in vitro, CmVar004 grows slower than CmVar001. While, with the exception of the plaque size, the data were not evaluated statistically, as only two growth curve experiments were performed, nevertheless, the data do suggest phenotypic differences among the variants.
TABLE 1 .
In vitro growth phenotype of C. muridarum M variantsa
| C. muridarum variant | Plaque size (mm) | EB generation rate (fold increase) | Rate of 2-fold EB increase (min) |
|---|---|---|---|
| CmVar001 | 1.05 ± 0.29 | 25.4 ± 3.8 | 78 |
| CmVar001.1 | 0.96 ± 0.08 | 14.4 ± 3.1 | 121 |
| CmVar002 | 0.97 ± 0.25 | 22.3 ± 1.4 | 87 |
| CmVar003 | 0.38 ± 0.25* | 8.8 ± 0.7 | 128 |
| CmVar004 | 0.55 ± 0.16* | 3.8 ± 2.2 | 136 |
*, P < 0.00001 according to a two-tailed t test compared to CmVar001.
In vivo disease phenotype of infection with chlamydial variants.
Since the above data demonstrated that the variants have different growth phenotypes in vitro, it was important to determine whether they would also differ in vivo in a genital infection of mice. C57BL/6 and BALB/c mice were infected genitally with 3 × 105 inclusion-forming units (IFU), and the course of the infection was monitored by measuring the number of IFU in genital tract swabs. No statistical significant differences in the bacterial loads were noted during the primary course of infection in C57BL/6 mice infected with C. muridarum variants CmVar001 and CmVar004 (2-factor [day, group] analysis of variance with repeated measures), but a longer infection was observed in mice infected with CmVar001 (see Fig. S1A in the supplemental material). In contrast, CmVar001 elicited a significantly higher bacterial burden and longer infection in BALB/c mice than in mice infected with CmVar004 (P < 0.0001) (see Fig. S1B). Furthermore, there were no statistical significant differences between the results determined for mice infected with CmVar001.1, CmVar002, or CmVar003 and those determined for mice infected with CmVar004 or CmVar001.
An important measure of disease outcome is the development of hydrosalpinx, which is associated with infertility. Hence; we assessed the presence of hydrosalpinx 35 days after infection in each of two groups of five C57BL/6 mice infected with one of the variants (Table 2). Because of major differences in hydrosalpinx development in mice infected with CmVar001 versus those infected with CmVar004, the experiment with those groups was repeated twice. Importantly, there was a significant difference with respect to the development of hydrosalpinx between mice infected with the CmVar001 and CmVar004 variants (chi-square test with Yates correction, P < 0.0001), with only 4 of 15 mice infected with CmVar004 developing unilateral or bilateral hydrosalpinx compared to 14 of 15 mice infected with CmVar001. Similarly, 6 of 30 (20%) oviducts in CmVar004-infected mice were positive for hydrosalpinx in contrast to 25 of 30 (83.3%) oviducts in mice infected with CmVar001. A similar difference was observed in BALB/c mice. Mice infected with the CmVar001.1, CmVar003, and CmVar002 variants showed an intermediate phenotype with regard to the production of hydrosalpinx, although all of the results were statistically significantly different from those determined for the mice infected with CmVar001 (chi-square test with Yates correction, P < 0.01). In addition, CmVar001 also elicited significantly more hydrosalpinx formation than did the original C. muridarum parent stock from which the variants were derived (chi-square test with Yates correction, P < 0.007) (Table 2).
TABLE 2 .
In vivo phenotype of chlamydial variantsa
| C. muridarum variant | No. of mice positive for hydrosalpinx/total no. of mice (%) |
No. of oviducts positive for hydrosalpinx/total no. of oviducts (%) |
||
|---|---|---|---|---|
| C57BL/6 | BALB/c | C57BL/6 | BALB/c | |
| CmVar001 | 14/15 (93.3) | 5/5 (100) | 25/30 (83.3) | 10/10 (100) |
| CmVar001.1 | 3/5 (60) | 3/10 (30)† | ||
| CmVar002 | 4/5 (80) | 4/10 (40)** | ||
| CmVar003 | 4/5 (80) | 4/10 (40)** | ||
| CmVar004 | 4/15 (26.7)† | 6/30 (20)# | 5/10 (50)* | |
| Parent stock | 5/8 (62.5) | 7/16 (44)† | ||
P values compared to CmVar001 using the chi-square test with the Yates correction (one tailed) in comparison to CmVar001 are indicated as follows: *, P < 0.05; **, P < 0.02; †, P < 0.008; #, P < 0.0001.
The significant difference in the development of upper genital tract disease raised the issue of whether CmVar001 and CmVar004 were reaching the upper tract at the same rate. Therefore, C57BL/6 mice were infected intravaginally, and on day 10, oviducts and the upper portion of the uterine horns were processed for the isolation and enumeration of chlamydiae. Similar numbers of chlamydiae were found in the oviducts and upper uterine horns of CmVar001- and CmVar004-infected mice, suggesting that the bacterial load is not the cause of the reduced pathology observed in CmVar004-infected mice (Fig. 1). We observed that 9 animals gave positive CmVar004 results in upper tract isolations and that all 10 animals gave positive CmVar001 results in the upper genital tract. We chose to use only CmVar001 and CmVar004 for further experiments because they represented the opposite extremes in growth rates and pathology induction of the variants.
FIG 1 .
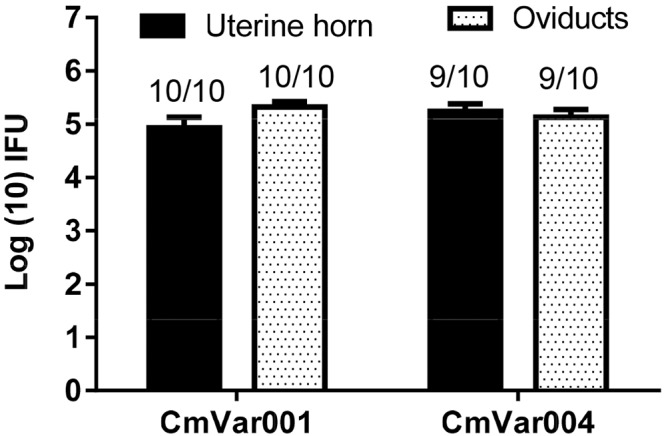
Upper genital tract isolations of CmVar001 and CmVar004 variants indicate similar numbers of IFU in oviducts and upper uterine horns. C57BL/6 mice were infected at 3 × 105 IFU, and cervical swabs were collected once every 3 days to determine the IFU on a McCoy cell monolayer. On day 10, mice were euthanized to collect oviducts and samples of an upper portion of uterine horn and of tissue were homogenized, sonicated, and plated on HeLa cell monolayers for determination of IFU. Log10 IFU/ml values were calculated from genital swabs and tissue isolations, and the averages of the results determined for 10 animals were plotted as means ± standard deviations. The numbers above the bars indicate the ratios of the number of mice positive for unilateral or bilateral hydrosalpinx to the total number of mice. Cervical swab data indicated that all mice were infected in the lower genital tract (data not shown). No statistical significant differences between CmVar001 and CmVar004 upper tract isolations were observed by a two-way analysis of variance (ANOVA).
Differential expression of chemokines and cytokines during chlamydial infection.
Production of specific chemokines and cytokines is inherent in the development of a pathological response to chlamydial infection. Because there were differences in the courses of infection and the development of pathology during CmVar001 and CmVar004 infection, we expected to observe a difference in the chemokine and cytokine profiles elicited by each variant. To evaluate the chemokine and cytokine profile in the cervix, we analyzed cervical tissue at 24 h postinfection for the expression of 15 key chemokines and cytokines (tumor necrosis factor alpha [TNF-α], interleukin-1β [IL-1β], IL-6, IL-10, CXCL10, CXCL1, CXCL2, CCL1, CCL2, CCL3, CCL4, CCL5, IFN-γ, IFN-β, and transforming growth factor β [TGF-β]) which we previously showed to be upregulated 24 h after chlamydial infection (19). C57BL/6 mice were infected intracervically rather than intravaginally since intracervical inoculation places a large number of chlamydiae directly at the target tissue and results in a synchronous infection for the first developmental cycle (~42 to ~48 h). We performed an initial screen and determined that animals infected with CmVar001 and CmVar004 had similar levels of chlamydial rRNA (rs16) (data not shown) 24 h postinfection. We observed significantly lower levels of CXCL1, CXCL2, TNF-α, CCL1, and CCL2 in cervical tissue from mice infected with CmVar004 than in cervical tissue from mice infected with CmVar001 (Fig. 2A) (P < 0.05 according to a one-tailed t test). A trend toward decreased expression of IL-1β, CXCL10, and IL-6 was also observed with CmVar004 infection, though it was not statistically significant (data not shown). In addition, the other chemokines and cytokines measured were similarly expressed in CmVar001 and CmVar004 (data not shown). These data indicated that CmVar004 induced lower levels of important molecules for the initiation of the inflammatory response (TNF-α, CXCL1, CXCL2) than CmVar001, correlating with the relative pathogenicities of each variant.
FIG 2 .
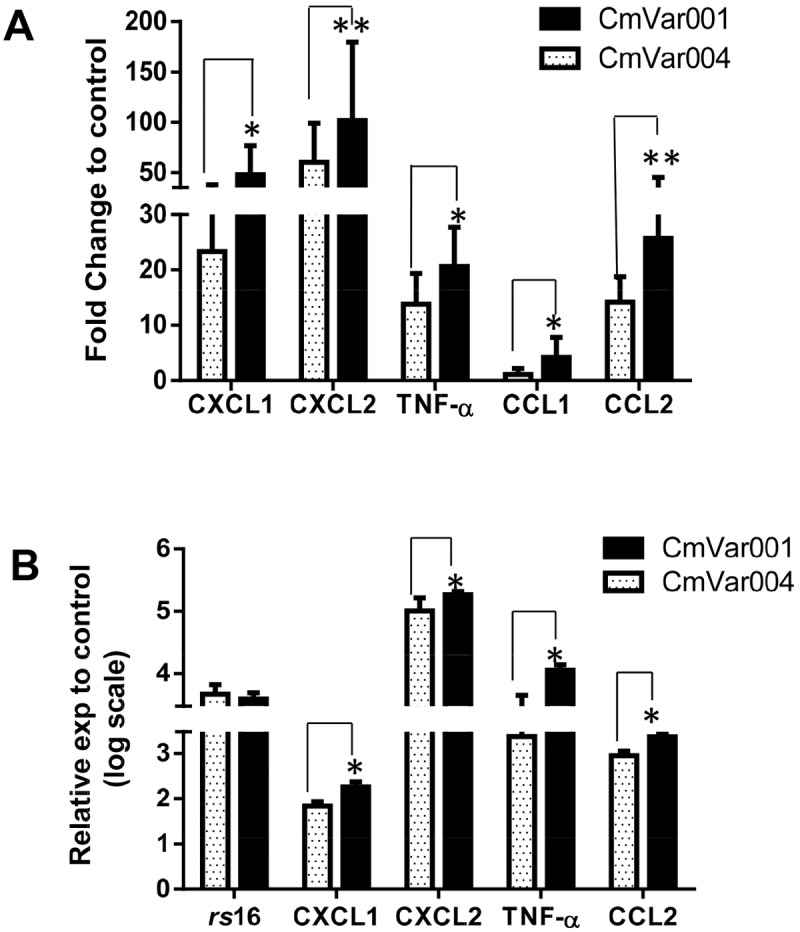
Chemokine and cytokine expression analyses from mice and BM1.11 cells. C57BL/6 mice were infected with 1 × 107 IFU of CmVar001 or CmVar004 variants by intracervical inoculation. (A) Expression levels of chemokine and cytokine transcripts are plotted as fold change relative to control levels, and only CmVar001 and CmVar004 gene results that were statistically significantly different are shown in the graph. Data are from an average of 9 animals per group where CmVar004 infection had lower expression of CXCL1, CXCL2, TNF-α, and CCL1 than CmVar001 infection (*, P < 0.05, one-tailed t test). We observed that CXCL2 results were significant at 10% (**, P < 0.09) and that CCL2 results were significant at 6% (**, P < 0.06). (B) Mouse oviduct epithelial cells (BM1.11) were infected with CmVar001 and CmVar004 and variants. The graph is a representation of three individual experiments, and data represent an average of three samples per each group (*, P < 0.01, one-tailed t test). exp, expression.
Since it is likely that these molecules are produced by the infection of epithelial cells, we wanted to determine if we would see a similar phenomenon in vitro in the infection of mouse genital tract epithelial cells. Mouse oviduct epithelial cells (Bm1.11) were infected with CmVar001 and CmVar004 variants at a multiplicity of infection of 1. At 24 h postinfection, RNA was isolated and expression levels of various chemokines and cytokines were determined. The data strongly supported the in vivo results, in that CmVar004 induced significantly lower levels of CXCL1, CXCL2, TNF-α, and CCL2 with similar levels of rs16 than did CmVar001 (Fig. 2B) (P < 0.05). Therefore, both the in vitro and in vivo chemokine and cytokine expression data correlated with the pathological outcomes of the attenuated CmVar004 and the virulent CmVar001 variants. These data indicate that the early chemokine and cytokine responses elicited in mice within the first developmental cycle of chlamydial infection by defined virulent and attenuated variants are predictive of the outcome of the disease, resulting in more and less upper tract pathology, respectively.
Differential expression of miRNA(s) during chlamydial infection.
In mammalian cells, miRNAs play a pivotal role in regulation of different processes such as development, metabolism, and inflammation as well as in modulating chemokine and cytokine expression (15, 21–26). However, the role inflammation-related miRNAs play during chlamydial pathogenesis and their correlation to chemokine and cytokine expression has not been studied. The population of miRNAs elicited by CmVar001, CmVar004, and sham inoculation of control mice (4 C57BL/6 mice for each group) infected by the intracervical route was first explored by relatively low-coverage Illumina MiSeq sequencing. This small-RNA sequencing and bioinformatic analysis identified 2,935 uniquely mapped mouse miRNAs (data not shown). Using stringent cutoff values, differential expression analysis comparing the CmVar001- and CmVar004-infected mice to the sham-inoculated mice identified five (CmVar004 versus controls) and four (CmVar001 versus controls) miRNAs that were significantly differentially expressed (Table 3). Two miRNAs, miR223-3p and miR18a-5p, were common to CmVar001 and CmVar004. Further, differential expression analysis of CmVar001 versus CmVar004 showed that miR223-3p was overexpressed in CmVar004-infected mice (Table 3). The relatively low-coverage small-RNA sequencing analysis results presented above provided evidence that CmVar001 and CmVar004 elicit different host miRNA responses.
TABLE 3 .
DEseq of miRNAs discovered by sequencinga
| Comparison and miRNA | MRC | Log fold change | P value | FDR | |
|---|---|---|---|---|---|
| Control versus CmVar001 | CmVar001 | Controls | |||
| mmu-miR-223-3p | 78.00 | 29.73 | −1.39 | 0.0000 | 0.0003 |
| mmu-miR-203-3p* | 430.87 | 121.22 | −1.83 | 0.0000 | 0.0003 |
| mmu-miR-18a-5p* | 22.29 | 5.64 | −1.98 | 0.0000 | 0.0021 |
| mmu-miR-215-5p | 12.51 | 0.00 | 0.0000 | 0.0000 | |
| Control vs CmVar004 | CmVar004 | Controls | |||
| mmu-miR-98-5p | 137.96 | 57.49 | −1.26 | 0.0000 | 0.0001 |
| mmu-miR-21a-3p | 146.06 | 56.68 | −1.37 | 0.0000 | 0.0000 |
| mmu-miR-155-5p | 34.77 | 8.91 | −1.96 | 0.0000 | 0.0001 |
| mmu-miR-18a-5p* | 23.50 | 5.41 | −2.12 | 0.0000 | 0.0001 |
| mmu-miR-223-3p* | 175.75 | 28.51 | −2.62 | 0.0000 | 0.0000 |
| CmVar001 versus CmVar004 | CmVar001 | CmVar004 | |||
| mmu-miR-223-3p | 61.00 | 100.00 | 1.00 | 0.0002 | 0.04 |
| mmu-miR-203-3p | 300.00 | 100.00 | −2.00 | 0.0001 | 0.02 |
MRC, mean read count; FDR, false discovery rate. Cutoff of FDR < 0.05; minimum 10 reads; upper fold change = 1; *, lower fold change = −1. DEseq, differential expression levels.
In order to confirm sequencing results and also focus on the miRNAs involved in immunopathogenesis, we measured the expression profile of 134 miRNAs by real-time PCR, at 24 h after intracervical infection, using 5 C57BL/6 mice per group. We observed differential expression of miRNAs during CmVar001 and CmVar004 infection in comparison to sham-inoculated control mice. There were distinct differences in the types and levels of the miRNAs expressed in mice infected by CmVar001 and CmVar004 compared to controls. Data analyses of miRNA for 1.5-fold or higher differences between CmVar001 and CmVar004 indicated that 12 miRNAs were expressed at higher levels with CmVar004 infection than with CmVar001. Among the 12, we observed that 10 miRNAs (miR-135a-5p, miR298-5p, miR142-5p, miR223-3p, miR147-3p, miR105, miR132-3p, miR142-3p, miR155-5p, and miR-410-3p) were upregulated in mice infected with the avirulent CmVar004 variant in comparison to mice infected with the virulent CmVar001 variant (Fig. 3 and Table 4). Two miRNAs, miR-299a-3p and miR-325-3p, were downregulated in both infections; however, the downregulation was significantly greater in animals infected with CmVar001. As shown in Table 4, miR-155-5p, miR-142-3p, miR-132-3p, miR105, miR223-3p, and mir-147-3p regulate chemokine and cytokine responses as well as modulating Toll-like receptor 2 (TLR2) expression under different pathological conditions (15, 21–25). These data indicate that miRNAs expressed at higher levels during CmVar004 infection are associated with the downregulation of chemokine and cytokine expression in mice, resulting in lesser upper genital tract pathology during a CmVar004 infection. Conversely, the low expression level of these inflammatory miRNAs in mice infected with the virulent CmVar001 variant is associated with the increased incidence of hydrosalpinx in those mice.
FIG 3 .
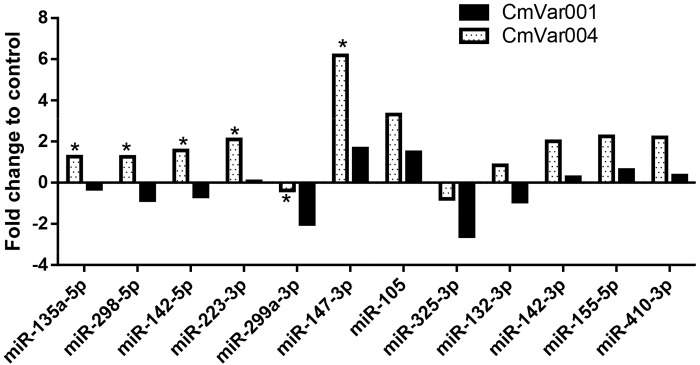
miRNA expression analyses from mice. C57BL/6 mice were infected with 1 × 107 IFU of CmVar001 or CmVar004 variants by intracervical inoculation, and the miRNA expression profile was measured as described in Materials and Methods. Results represent an average of 5 to 7 animals per group; statistical significance was determined by the Wilcoxon rank sum one-tailed test (Bonferroni correction), with P < 0.05 being considered significant. Data analyses of miRNA for determinations of differences between CmVar004 and CmVar001 results at a level of 1.5-fold or higher indicate that 12 miRNAs were expressed at higher levels with CmVar004 infection than with CmVar001 infection and that the results for 6 miRNAs were statistically significant as well.
TABLE 4 .
In vivo expression profile of miRNAa
| microRNA | Fold increase (CmVar004 > CmVar001) | One-tailed test P value (CmVar004 > CmVar001) | Related pathway(s) and direction of regulation (source) |
|---|---|---|---|
| miR-135a-5p | 1.55 | 0.003 | ND |
| miR-299a-3p | 1.63 | 0.05 | ND |
| miR-155-5p | 1.64 | N.S. | CXCL1↓ (15) |
| miR-142-3p | 1.75 | N.S. | IRAK1 (23) |
| miR-132-3p | 1.76 | N.S. | IRAK4↓, TNF-α (50) |
| miR-325-3p | 1.81 | N.S. | ND |
| miR-105 | 1.83 | N.S. | TLR2↓, TNF-α, IL-6 (21) |
| miR-410-3p | 1.86 | N.S. | ND |
| miR-223-3p | 2.04 | 0.05 | IL-1B↓, IL-6↓, TNF-α (22) |
| miR-298-5p | 2.10 | 0.02 | ND |
| miR-142-5p | 2.23 | 0.02 | ND |
| miR-147-3p | 4.54 | 0.05 | TLR2↓, TNF-α, IL-6 (24) |
N.S., not significant; ND, not determined.
Comparative genome sequencing of chlamydial variants.
High-quality draft genomes were obtained for each C. muridarum plaque variant by deep sequencing (see Table S1 in the supplemental material). A draft assembly of the conserved C. muridarum plasmid was also recovered for each plaque variant, excluding the possibility that the attenuated growth and virulence phenotypes were caused by plasmid-free C. muridarum (27). Using the published C. muridarum Nigg genome (Genbank accession no. AE002160) as a reference, comparative genome analysis identified 52 high-quality single nucleotide polymorphisms (SNPs) and insertions and deletions (indels), supported by very deep sequence coverage (>2,600×), within the draft plaque variant genomes, including synonymous and nonsynonymous changes, likely resulting in several frameshift mutations (Table 5; see also Table S2). The pattern for 18 of these SNPs or indels within or between predicted genes across the plaque variants correlates with the observed phenotypes of differential growth rates and gross pathology (Table 5; see also Table S2). Nine of these SNPs and indels occur within the ompA gene (TC0052) that encodes the major outer membrane protein (MOMP). These mutations cause an amino acid deletion within the MOMP third variable domain (VD3) in the CmVar001 variant (Table 5 and data not shown). In the CmVar004 variant, these mutations cause a double amino acid deletion and a glycine-to-cysteine substitution within MOMP VD4 (Table 5 and data not shown). The glycine-to-cysteine substitution may impact MOMP structure and intramolecular complexing through disulfide cross-linking (28, 29). Phenotype-correlating mutations across plaque variants are observed in several other genes, including TC0412 (homolog of CT135), TC0155 [3′(2′),5′-bisphosphate nucleotidase], and TC0682 (tetraacyldisaccharide 4′-kinase) (Table 5; see also Table S2). Phenotype-correlating mutations are also found within several genes with no known function (annotated as conserved hypothetical genes); this suggests that these genes may have functions that are related to the observed variant phenotypes. Notably, disruption of CT135 was recently shown to reduce infectivity in a C. trachomatis mouse model of infection (30). Thus, comparative genomic analysis of plaque-purified variants identified mutations that may explain the virulence characteristics of variants in a particular stock.
TABLE 5 .
SNPs and indels identified in variants with potential phenotype correlation relative to the Nigg genome (Genbank accession no. AE002160)a
| SNP location | Gene locant: protein encoded | Gene start (NT) | Gene stop (NT) | CmVar001 |
CmVar001.1 |
CmVar002 |
CmVar003 |
CmVar004 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NT change | AA change | NT change | AA change | NT change | AA change | NT change | AA change | NT change | AA change | ||||
| 58847 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | C→A | G→C | C→A | G→C | ||||||
| 58899 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | A→ | DEL | A→ | DEL | ||||||
| 58900 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | A→ | DEL | A→ | DEL | ||||||
| 58901 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | G→ | DEL | G→ | DEL | ||||||
| 58902 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | C→ | DEL | C→ | DEL | ||||||
| 58903 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | T→ | DEL | T→ | DEL | ||||||
| 59061 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | T→ | DEL | T→ | DEL | T→ | DEL | ||||
| 59062 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | G→ | DEL | G→ | DEL | G→ | DEL | ||||
| 59063 | TC_0052: major outer membrane protein, porin | 58653 | 59816 | T→ | DEL | T→ | DEL | T→ | DEL | ||||
| 187432 | TC_0155: 3′(2′),5′-bisphosphate nucleotidase, putative | 187163 | 188212 | G→A | H→Y | G→A | H→Y | G→A | H→Y | ||||
| 187984 | TC_0155: 3′(2′),5′-bisphosphate nucleotidase, putative | 187163 | 188212 | C→T | G→S | ||||||||
| 381736 | Intergenic: NA | NA | NA | T→A | NA→NA | ||||||||
| 473191 | TC_0412: conserved hypothetical protein | 472757 | 473854 | →A | FS | →A | FS | ||||||
| 473705 | TC_0412: conserved hypothetical protein | 472757 | 473854 | G→ | FS | G→ | FS | G→ | FS | ||||
| 798169 | TC_0668: conserved hypothetical protein | 797014 | 798240 | T→G | F→V | ||||||||
| 814976 | TC_0682: tetraacyldisaccharide 4′-kinase | 814899 | 816008 | C→A | V→L | C→A | V→L | C→A | V→L | ||||
| 1004276 | TC_0867: conserved hypothetical protein | 1002965 | 1004440 | T→C | S→P | T→C | S→P | T→C | S→P | T→C | S→P | ||
| 1062351 | Intergenic | NA | NA | C→A | NA→NA | C→A | NA→NA | C→A | NA→NA | ||||
Mutations common to all variants are excluded (see Table S1 in the supplemental material for the entire list). Chlamydial variants are arranged from highest growth rate to lowest (left to right). Gross pathology results also followed this pattern. FS, frameshift; DEL, deletion; INS, insertion; NA, not applicable (intergenic); NT change, nucleotide change; AA change, amino acid change.
DISCUSSION
Chlamydial genital infections, as in many infectious diseases, vary greatly in the clinical outcome of the disease, ranging from subclinical infection with little or no sequelae to full-blown cervicitis followed by pelvic inflammatory disease resulting in tubal obstruction, ectopic pregnancy, or infertility. Using the C. muridarum mouse model of genital infection, we undertook studies to expand upon the concept that a given chlamydial population consists of variants with different levels of pathogenic potential. In support of this concept, 5 plaque-purified isolates from mice infected with the less virulent Nigg strain were characterized for their pathology-inducing capabilities. Interestingly, even though the choosing of the plaques for evaluation was purely random, a range of pathology-associated phenotypes was identified. While there was little difference in the infection courses, two variants, CmVar001 and CmVar004, differed dramatically in the ability to elicit hydrosalpinx. This ability was reflected in their plaque size, growth rate, and chemokine profile, with the more virulent variant having an increased growth rate and increased production of proinflammatory chemokines.
Deep genome sequencing of these variants identified patterns of mutations that correlated with these observed growth and virulence phenotypes, but no definitive conclusions can be drawn linking a specific mutation to the virulence phenotype. Thus, even though the initial infection population of chlamydiae was relatively attenuated, variants were present in that population with a wide range of pathogenic potential. It is interesting that a virulent variant was contained in the parent population but that virulence was not reflected in the overall pathological phenotype of the parent population. Whether the pathogenicity of a given population represents the sum total or the average of the pathogenicities of its constituent variants cannot be concluded at this point. What is clear is that the virulence phenotype of a population does not necessarily reflect the virulence phenotype of all of the individual variants within that population. Using C. caviae in the guinea pig model of chlamydial genital infection, we previously observed that when a wild-type variant, capable of eliciting a strong inflammatory response, and a less virulent and slower-growing variant eliciting significantly less inflammation are inoculated together into the animal, the outcome of the infection resembles that of the virulent variant (31). Moreover, even though equal numbers of the variants are inoculated, the wild-type variant becomes dominant by the end of the infection (31). This suggested that the composition of a chlamydial population is dynamic and constantly in flux, governed by host genetics and microenvironment of the infection site, among other factors.
If the composition of individual variants within a chlamydial population defines the potential pathological outcome of the disease, the identification of specific variants associated with more-severe disease might aid in determining the risk of upper tract disease. However, while it is technically feasible to genotype a chlamydial population from a swab (32–34), it is not yet logistically and economically feasible to use this methodology in a clinic setting. Moreover, we are still a long way from being able to definitively state which gene or group of genes determines chlamydial virulence capability. It is currently more practical to determine if there is a particular host factor(s) that demonstrates the potential for more-severe disease, particularly a more intense acute inflammatory response. The number of polymorphonuclear leukocytes (PMNs) present in a cervical scraping is an inexact measure and has been of limited diagnostic use for PID. Nevertheless, it is interesting that the virulent CmVar001 variant results in the expression of significantly greater levels of key inflammatory cytokines and chemokines, including TNF-α, CXCL1, CXCL2, and CCL1, than the attenuated CmVar004 variant both in vitro and in vivo. Importantly, these differences were noted at 24 h after infection, when chlamydiae were still well within their first developmental cycle and when PMNs were just beginning to reach the infected cells in the epithelium (35). The reason for this inflammatory difference between the virulent and attenuated variants is not known. It could be related to the higher growth rate and more productive infection associated with the virulent CmVar001 variant; however, if that were the case, it would be difficult to establish parameters to predict the possibility of different degrees of upper genital tract pathology, especially because of dilution effects of the population of multiple variants and the very strong likelihood that individuals are infected with differing numbers of organisms. Moreover, unlike our experimental models, women are likely exposed multiple times in an ongoing relationship.
An alternative set of biomarkers is that of host miRNA differentially expressed in response to infection. A recent report on chlamydial conjunctival infection from human samples showed differential expression of miRNAs (36). However, it did not show any evidence of specific miRNAs in predicting pathology, as the study compared samples from individuals with scarring and those without inflammation in contrast to an early conjunctivitis stage of trachoma. Our preliminary sequencing of miRNAs in mouse cervical tissues elicited by CmVar004 and CmVar001 infections demonstrated differential expression levels, and differences were observed in chemokine and cytokine responses. With this evidence, we then focused on the expression of 134 inflammation-related miRNAs in order to determine if the differences seen in chemokine and cytokine responses between the CmVar004 and CmVar001 variants would also be reflected in specific quantitative real-time PCR (qRT-PCR) miRNA expression profiles within 24 h of infection. Interestingly, 12 different miRNAs (miR-135a-5p, miR-298-5p, miR-142-5p, miR-223-3p, miR-299a-3p, miR-147-3p, miR-105, miR-325-3p, miR-132-3p, miR-142-3p, miR155-5p, and miR-410-3p) were overexpressed following infection with the attenuated variant, CmVar004, in contrast to infection with the virulent variant, CmVar001. The results for six of these (miR-135a-5p, miR299a-3p, miR223-3p, miR298-5p, miR142-5p, and miR147-3p) are also statistically significant. The results with respect to differential expression of miR223-3p were also supported by the sequencing data. To our knowledge, this is the first report demonstrating that differences in the early miRNA profile may be predictive of the outcome of the infection with regard to upper genital tract pathology. Because of the large number of miRNAs with multiple regulatory functions in the immune response, it may be possible to define a particular profile from a genital tract specimen early in the course of infection that will be prognostic for the severity of disease, allowing physicians to treat those infections more aggressively.
The significant upregulation of 12 different miRNAs by the attenuated variant in comparison to the virulent variant supports our observation of a lower inflammatory chemokine and cytokine response elicited by the avirulent variant. These results suggest that the interaction of the attenuated variant with the host cell initiates a pathway(s) that results in the production of miRNAs that downregulate inflammatory responses. Conversely, the virulent variant may also be initiating pathways that suppress the expression of those miRNAs, resulting in increased inflammatory chemokine and or cytokine expression. Among those known miRNAs that are upregulated in infection with the avirulent variant, miR-155, miR-142-3p, miR-147-3p, miR-105, and miR-132 have been shown to downregulate chemokines CXCL1, TNF-α, and IL-6 and also to modulate TLR2 expression under different pathological conditions (15, 16, 24). We also observed a lower CXCL1 and TNF-α response in infection with the attenuated variant in addition to the CXCL2, CCL1, and CCL2 results noted. It is interesting that the activation of the TLR2 pathway, in contrast to the TLR4 pathway, is associated with the pathological response in chlamydial genital infections (37). It is possible that activation of certain miRNAs by the avirulent variant blocks or downregulates the TLR2 pathway, resulting in a lower pathological response. Nevertheless, the exact mechanism(s) by which chlamydiae initiate these processes, including the role of the observed pattern of mutations within the variant genomes, remains to be determined.
Several reports have described the role for miRNAs in regulating the chemokine and cytokine responses in bacterial induced inflammation. For example, molecules such as lipopolysaccharide (LPS), TNF-α, and IFN-γ have been shown to induce miRNA expression (26, 38, 39). During Helicobacter pylori infection, overexpression of miR-155 reduces the expression of IL-8 and growth-related oncogene–α (GRO-α) (15) in human gastric epithelial mucosa cells. A total of 28 miRNAs are upregulated and 2 are downregulated in patients with Mycobacterium tuberculosis infection in comparison to healthy individuals. Among those, miR-144* is possibly inhibitory for TNF-α and IFN-γ production and T-cell proliferation, suggesting the involvement of miRNA in antituberculosis immunity (16). miR-142-3p downregulated interleukin-1 receptor-associated kinase 1 (IRAK-1) expression, resulting in downregulation of NF-κB, TNF-α, and IL-6 during Mycobacterium bovis bacillus Calmette-Guérin (BCG) infection in mouse macrophages (23). In a brain inflammation model, miR-132 was found to have an anti-inflammatory effect by targeting mRNA of acetyl cholinesterase enzyme, which results in increased levels of acetyl choline, an important inhibitor of peripheral inflammation (25). In miR-223−/− mouse macrophages, significantly elevated levels of IL-1β, IL-6, and TNF-α were observed in comparison to those seen with wild-type macrophages during LPS stimulation, indicating that miR-223 plays a role in anti-inflammatory processes (22). miR-147 is induced by stimulation of multiple TLRs, and its overexpression in mouse macrophages decreased TNF-α and IL-6 expression induced by TLR2, TLR3, and TLR4 ligands [PAM3CSK4, poly(I ⋅ C), and LPS], indicating that miR-147 functions in a negative-feedback mechanism to regulate inflammation (24). In addition to this, miR-105 has also been shown to modulate TLR2 expression in human keratinocytes (21). All these reports indicate that miRNAs play a regulatory role in chemokine and cytokine expression.
While we have presented data to demonstrate that the miRNA profile is able to distinguish between virulent and attenuated variants in our model, the more important point is that regardless of the specific variants, it is possible that the miRNA profile may be able to serve as a predicative biomarker for disease severity. From the viewpoint of human medicine, the nature of the actual variant causing the disease is irrelevant; it is more important to have a means to determine if there is an increased risk of severe outcome so that appropriate therapy can be initiated before irreversible pathology occurs. Moreover, the predictive value of biomarkers such as miRNAs early in the infection course may be in their use as a measure to determine the effectiveness of a chlamydial vaccine. Clearly, it is not feasible to use PID or infertility as an outcome for the measure of vaccine effectiveness; rather, the miRNA profile may have value as a correlate for PID or infertility or the lack thereof.
In summary, we have presented evidence that an infecting chlamydial population consists of multiple genetic variants with distinct virulence phenotypes. Moreover, infection with virulent versus attenuated phenotypes can be detected early in the course of the infection by the level of expression of specific miRNAs that can regulate various aspects of the immune response and correlate with the chemokine and cytokine response elicited by the variants. Importantly, the miRNA expression profile assessed at 24 h after infection is predictive of the eventual development of hydrosalpinx, the major pathological outcome for chlamydial infection in females. While 24 h is not a practical time point to use in analyses of human infections, the results do suggest that observable differences in the miRNA response can be detected early in the infection course. Considering the impact of miRNAs on the regulation of the immune response and the association of specific miRNAs with infection by defined chlamydial pathotypes, they may hold promise as early-stage biomarkers to be used for prognostics in chlamydial genital infections.
MATERIALS AND METHODS
Cell lines.
Oviduct epithelial cells (Bm1.11) were provided by Raymond Johnson (Indiana University, Indianapolis, IN) and were cultured in Ham’s F-12 media supplemented with 10% fetal bovine serum (FBS), 12.5 ng/ml human recombinant keratinocyte growth factor (Sigma-Aldrich, St. Louis, MO), 2 mM GlutaMAX (Invitrogen), and 50 μg/ml gentamicin. HeLa cells and McCoy cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS, 1 mM glutamine, 100 μM nonessential amino acids (Invitrogen), and 50 μg/ml gentamicin.
Chlamydial infection and experimental animals.
Mice were infected with C. muridarum strain Nigg, which has been passaged in our laboratory since 1977, when it was obtained from the American Type Culture Collection as a yolk sac preparation. The Nigg stock, as reported by Ramsey and coworkers, consists of multiple variants, some of which have the same set of SNPs as seen in the genome sequencing of the variants obtained in vivo in this study (K. H. Ramsey, personal communication). To obtain chlamydial variants, nude mice were infected with 3 × 105 inclusion-forming units (IFU) of C. muridarum and swabs were collected on day 6 and day 9. These swabs were plated for plaques, and random plaques were chosen to be subjected to subplaque production and expansion. CmVar001.1, CmVar003, and CmVar004 were collected from the day 6 swab, and CmVar002 and CmVar001 variants were collected from the day 9 swab from the same mouse. In vitro growth curve analysis was performed as described previously (8) to determine the differences in growth phenotype. Eight-week-old C57BL/6 and BALB/c mice were purchased from Jackson Laboratories (Bar Harbor, ME) and Harlan-Sprague Dawley (Indianapolis, IN), respectively. Mice were given a 2.5-mg subcutaneous injection of medroxyprogesterone (Depo-Provera; Upjohn, Kalamazoo, MI) 7 days prior to infection to place the mice in a state of anestrus. Mice were infected intravaginally with 3 × 105 IFU (40) or intracervically with 1 × 107 IFU as described previously (19). Cervices were collected from mice infected intracervically for total RNA isolation 24 h after infection. To monitor the course of infection, swabs were collected from mice infected intravaginally every 3 days for determination of IFU. On day 35, all mice were euthanized for gross pathology observation. All protocols were approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences.
Chlamydial culture.
Chlamydiae were quantified from the genital tract using a Dacro swab and then cultured in McCoy cells according to standard procedures. In order to quantify the number of chlamydiae from tissues, oviducts and a small portion of the upper uterine horn were dissected and placed in 1 ml of 2-sucrose-phosphate buffer transport medium with 0.1 mg/ml gentamicin, 0.2 mg/ml vancomycin, and 2.5 µg/ml of amphotericin B (Fungizone). The tissues were homogenized and then sonicated for 1 min. After centrifugation to pellet the large-cell debris, the supernatants were diluted with Eagle’s minimal essential medium supplemented with 10% fetal bovine serum (FBS) and 5% glucose (pH 7.4) and inoculated onto confluent HeLa cell monolayers for the determination of IFU. Chlamydial inclusions were stained with C. muridarum-specific immune serum and with a fluorescent secondary anti-mouse IgG antibody (Alexa Fluor 488).
Genome sequencing.
Genomic DNAs extracted from plaque-purified C. muridarum variants were subjected to library preparation and next-generation whole-genome sequencing (paired end, 100 bp) using a single channel of an Illumina HiSeq 2500 flow cell (1/8 channel per variant). Sequence reads were assembled using CLC Genomics Workbench software (version 6.0.2) by mapping reads to the C. muridarum Nigg (Genbank accession no. AE002160) reference genome (similarity fraction = 0.85) (41) (see Table S1). For each sequenced isolate, consensus contigs were extracted and ordered into a pseudomolecule, using the reference genome for contig order. Gene identification and annotation were performed as previously described (42). SNPs and indels between C. muridarum variants and the reference C. muridarum genome were identified as previously described using a NUCmer-based bioinformatics pipeline (43–45).
Total RNA isolation and expression analysis of miRNA and mRNA transcripts.
Total RNA was isolated from cervices and cell samples by using an miRNeasy kit (Qiagen) after 24 h of infection with CmVar004 or CmVar001. Animals in a control group were sham inoculated. The concentration of RNA was determined by Nanodrop, and absorbance at 260 and 280 and absorbance at 260 and 230 were taken into consideration to check for RNA quality. RNA was reverse transcribed with SuperScript III enzyme (Invitrogen) according to the manufacturer’s instructions using random hexamer and dT for priming. Quantitative PCR was performed on samples using IQ-SYBR mix (Bio-Rad) and a CFX96 PCR detection system (Bio-Rad). Levels of chlamydial rRNA (rs16) were measured to determine chlamydial growth, and samples that showed rs16 levels similar to those determined for CmVar004 and CmVar001 were chosen for chemokine and cytokine (9 individual animals per [control, CmVar004, or CmVar001] group) analyses and miRNA analyses (4 to 5/group). All primers were designed using Beacon Design software (Bio-Rad).
To analyze miRNAs by sequencing, total RNAs of 4 samples from each group were subjected to small-RNA sequencing. To analyze miRNA by sequencing, total RNA was size selected and sequenced. Total RNA quality was assessed using an RNA Nano chip on an Agilent BioAnalyzer 2100 instrument (Agilent Technologies) prior to library preparation. Sequencing libraries were constructed using a DGE Small RNA Library Preparation kit (Illumina). Library preparations were validated by the use of a BioAnalyzer 2100 system using High Sensitivity DNA chips (Agilent Technologies) prior to sequencing. Small-RNA sequencing was performed using an Illumina MiSeq instrument. miRNAs were mapped to the mouse reference genome (GRCm38.70) using Bow tie v 0.12.9 (maximum number of mismatches = 2) (46). The number of reads mapped to each miRNA was counted by HTSeq against the annotated mouse miRNA gff3 file downloaded from miRBase (release 20) (47). Data normalization and differential gene expression analyses were performed using DESeq (48) and EdgeR (49), independently comparing C. muridarum CmVar004-infected and CmVar001-infected replicates to uninfected controls. Up- and downregulated miRNAs were filtered for significantly differentially expressed miRNAs using the following parameters: a cutoff false-discovery rate (FDR) = <0.05; minimum read count = 10; an upper fold change value of 1; and a lower fold change value of −1. To confirm the miRNA sequencing results and also to focus on immune-related miRNAs, we measured miRNA expression levels for 5 samples per group to run miRNA arrays from Qiagen that contained 134 different miRNAs related to inflammation (combination of inflammatory response and autoimmunity and immunopathogenesis miScript miRNA PCR arrays). Data analyses were performed using Web-based software provided by SABiosciences. miRNA expression analyses were performed during CmVar004 and CmVar001 infection with respect to control animals, and small nucleolar RNAs (SNORDs) were also used as internal controls (SNORD 61, 68, 72, 95, and 96A), which had the least variability of all the samples with respect to CmVar004 and CmVar001 infection. Data are presented as a clustergram and indicate fold change differences in regulation (up or down) of miRNAs. Data obtained by comparisons to the control were used to determine the fold change differences between CmVar004 and CmVar001 with respect to expression of individual miRNAs by using the one-tailed t test.
Statistical analysis.
Data are presented as the mean ± 1 standard deviation. The course of the infection among variants was analyzed with a 2-factor (days, group) analysis of variance with repeated measures. Statistical analyses for chemokine and cytokine data were carried out only with the measurements that showed a higher fold increase with the CmVar001 variant than with the CmVar004 variant, and data were analyzed by one-tailed t test. MicroRNAs that showed fold increases in mice that were infected with CmVar004 in comparison to those infected with CmVar001 were analyzed for statistical significance by a nonparametric one-tailed (with Bonferroni correction) Wilcoxon rank sum test.
Nucleotide sequence accession numbers.
The whole genome shotgun project has been deposited at DDBJ/EMBL/GenBank under accession numbers JNON00000000, JNOO00000000, JNOP00000000, JNOQ00000000, and JNOR00000000.
SUPPLEMENTAL MATERIAL
In vivo course of infection of chlamydial variants (CmVar004 and CmVar001) in two different strains of mice (C57BL/6 and BALB/c mice). Download
Alignment of C. muridarum MOMP (ompA) from Nigg, CmVar001, and CmVar004. Download
Sequencing metrics for plaque variants and the original source culture. Reads are mapped onto the published C. muridarum Nigg genome (Genbank accession no. ).
Comparative genome sequences of C. muridarum variants.
ACKNOWLEDGMENTS
This research was supported by a Marion B. Lyon New Scientist Development Award (L.Y.), the Arkansas Children’s Hospital Research Institute and Arkansas Biosciences Institute (R.G.R. and L.Y.), and U19 AI084044 from the NIAID, NIH (J.R., A.T.M., and R.G.R.).
Footnotes
Citation Yeruva L, Myers GSA, Spencer N, Creasy HH, Adams NE, Maurelli AT, McChesney GR, Cleves MA, Ravel J, Bowlin A, Rank RG. 2014. Early microRNA expression profile as a prognostic biomarker for the development of pelvic inflammatory disease in a mouse model of chlamydial genital infection. mBio 5(3):e01241-14. doi:10.1128/mBio.01241-14.
REFERENCES
- 1. Miyairi I, Ziebarth J, Laxton JD, Wang X, van Rooijen RN, Williams RW, Lu L, Byrne GI, Cui Y. 2012. Host genetics and Chlamydia disease: prediction and validation of disease severity mechanisms. PLoS One 7:e33781. 10.1371/journal.pone.0033781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barron AL, Pasley JN, Rank RG, White HJ, Mrak RE. 1988. Chlamydial salpingitis in female guinea pigs receiving oral contraceptives. Sex. Transm. Dis. 15:169–173. 10.1097/00007435-198807000-00013 [DOI] [PubMed] [Google Scholar]
- 3. Kinghorn GR, Waugh MA. 1981. Oral contraceptive use and prevalence of infection with Chlamydia trachomatis in women. Br. J. Vener. Dis. 57:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rank RG, Sanders MM, Kidd AT. 1993. Influence of the estrous cycle on the development of upper genital tract pathology as a result of chlamydial infection in the guinea pig model of pelvic inflammatory disease. Am. J. Pathol. 142:1291–1296 [PMC free article] [PubMed] [Google Scholar]
- 5. Marrazzo JM, Wiesenfeld HC, Murray PJ, Busse B, Meyn L, Krohn M, Hillier SL. 2006. Risk factors for cervicitis among women with bacterial vaginosis. J. Infect. Dis. 193:617–624. 10.1086/500149 [DOI] [PubMed] [Google Scholar]
- 6. Miyairi I, Mahdi OS, Ouellette SP, Belland RJ, Byrne GI. 2006. Different growth rates of Chlamydia trachomatis biovars reflect pathotype. J. Infect. Dis. 194:350–357. 10.1086/505432 [DOI] [PubMed] [Google Scholar]
- 7. Kari L, Whitmire WM, Carlson JH, Crane DD, Reveneau N, Nelson DE, Mabey DC, Bailey RL, Holland MJ, McClarty G, Caldwell HD. 2008. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J. Infect. Dis. 197:449–456. 10.1086/525285 [DOI] [PubMed] [Google Scholar]
- 8. Ramsey KH, Sigar IM, Schripsema JH, Denman CJ, Bowlin AK, Myers GS, Rank RG. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect. Immun. 77:3284–3293. 10.1128/IAI.00147-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Binet R, Bowlin AK, Maurelli AT, Rank RG. 2010. Impact of azithromycin resistance mutations on the virulence and fitness of Chlamydia caviae in guinea pigs. Antimicrob. Agents Chemother. 54:1094–1101. 10.1128/AAC.01321-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sturdevant GL, Kari L, Gardner DJ, Olivares-Zavaleta N, Randall LB, Whitmire WM, Carlson JH, Goheen MM, Selleck EM, Martens C, Caldwell HD. 2010. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect. Immun. 78:3660–3668. 10.1128/IAI.00386-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Connell RM, Rao DS, Baltimore D. 2012. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 30:295–312. 10.1146/annurev-immunol-020711-075013 [DOI] [PubMed] [Google Scholar]
- 12. Dai R, Ahmed SA. 2011. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl. Res. 157:163–179. 10.1016/j.trsl.2011.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ha TY. 2011. MicroRNAs in human diseases: from autoimmune diseases to skin, psychiatric and neurodegenerative diseases. Immune Netw. 11:227–244. 10.4110/in.2011.11.5.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. 2010. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 10:111–122. 10.1038/nri2708 [DOI] [PubMed] [Google Scholar]
- 15. Xiao B, Liu Z, Li BS, Tang B, Li W, Guo G, Shi Y, Wang F, Wu Y, Tong WD, Guo H, Mao XH, Zou QM. 2009. Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response. J. Infect. Dis. 200:916–925. 10.1086/605443 [DOI] [PubMed] [Google Scholar]
- 16. Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X. 2011. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol. Immunol. 48:1084–1090. 10.1016/j.molimm.2011.02.001 [DOI] [PubMed] [Google Scholar]
- 17. Eulalio A, Schulte L, Vogel J. 2012. The mammalian microRNA response to bacterial infections. RNA Biol. 9:742–750. 10.4161/rna.20018 [DOI] [PubMed] [Google Scholar]
- 18. Ryu MS, Langkamp-Henken B, Chang SM, Shankar MN, Cousins RJ. 2011. Genomic analysis, cytokine expression, and microRNA profiling reveal biomarkers of human dietary zinc depletion and homeostasis. Proc. Natl. Acad. Sci. U. S. A. 108:20970–20975. 10.1073/pnas.1117207108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rank RG, Lacy HM, Goodwin A, Sikes J, Whittimore J, Wyrick PB, Nagarajan UM. 2010. Host chemokine and cytokine response in the endocervix within the first developmental cycle of Chlamydia muridarum. Infect. Immun. 78:536–544. 10.1128/IAI.00772-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derbigny WA, Kerr MS, Johnson RM. 2005. Pattern recognition molecules activated by Chlamydia muridarum infection of cloned murine oviduct epithelial cell lines. J. Immunol. 175:6065–6075 [DOI] [PubMed] [Google Scholar]
- 21. Benakanakere MR, Li Q, Eskan MA, Singh AV, Zhao J, Galicia JC, Stathopoulou P, Knudsen TB, Kinane DF. 2009. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J. Biol. Chem. 284:23107–23115. 10.1074/jbc.M109.013862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H, Li H, Wang G, Evans AR, Safe S, Wu C, Zhou B. 2012. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 125:2892–2903. 10.1161/CIRCULATIONAHA.111.087817 [DOI] [PubMed] [Google Scholar]
- 23. Xu G, Zhang Z, Wei J, Zhang Y, Zhang Y, Guo L, Liu X. 2013. microR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis (Edinb) 93:606–611. 10.1016/j.tube.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 24. Liu G, Friggeri A, Yang Y, Park YJ, Tsuruta Y, Abraham E. 2009. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 106:15819–15824. 10.1073/pnas.0901216106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O’Neill LA. 2009. Boosting the brain’s ability to block inflammation via microRNA-132. Immunity 31:854–855. 10.1016/j.immuni.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 26. O’Neill LA, Sheedy FJ, McCoy CE. 2011. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11:163–175. 10.1038/nri2957 [DOI] [PubMed] [Google Scholar]
- 27. O’Connell CM, Ingalls RR, Andrews CW, Jr, Scurlock AM, Darville T. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179:4027–4034. 10.4049/jimmunol.179.6.4027 [DOI] [PubMed] [Google Scholar]
- 28. Sun G, Pal S, Sarcon AK, Kim S, Sugawara E, Nikaido H, Cocco MJ, Peterson EM, de la Maza LM. 2007. Structural and functional analyses of the major outer membrane protein of Chlamydia trachomatis. J. Bacteriol. 189:6222–6235. 10.1128/JB.00552-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bavoil P, Ohlin A, Schachter J. 1984. Role of disulfide bonding in outer membrane structure and permeability in Chlamydia trachomatis. Infect. Immun. 44:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sturdevant GL, Carlson JH, Whitmire WM, Zhou B, Song L, Caldwell HD. 20 December 2013. Infectivity of urogenital Chlamydia trachomatis plasmid-deficient, CT135-null, and double-deficient strains in female mice. Pathog. Dis. 10.1111/2049-632X.12121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rank RG, Bowlin AK, Tormanen KI, Wang Y, Maurelli AT. 2012. Effect of the inflammatory response upon the in vivo competition between two chlamydial variants in the guinea pig model of inclusion conjunctivitis. Infect. Immun. 80:612–619. 10.1128/IAI.06054-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Seth-Smith HM, Harris SR, Skilton RJ, Radebe FM, Golparian D, Shipitsyna E, Duy PT, Scott P, Cutcliffe LT, O’Neill C, Parmar S, Pitt R, Baker S, Ison CA, Marsh P, Jalal H, Lewis DA, Unemo M, Clarke IN, Parkhill J, Thomson NR. 2013. Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture. Genome Res. 23:855–866. 10.1101/gr.150037.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seth-Smith HM, Harris SR, Scott P, Parmar S, Marsh P, Unemo M, Clarke IN, Parkhill J, Thomson NR. 2013. Generating whole bacterial genome sequences of low-abundance species from complex samples with IMS-MDA. Nat. Protoc. 8:2404–2412. 10.1038/nprot.2013.147 [DOI] [PubMed] [Google Scholar]
- 34. Putman TE, Suchland RJ, Ivanovitch JD, Rockey DD. 2013. Culture-independent sequence analysis of Chlamydia trachomatis in urogenital specimens identifies regions of recombination and in-patient sequence mutations. Microbiology 159:2109–2117. 10.1099/mic.0.070029-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rank RG, Whittimore J, Bowlin AK, Wyrick PB. 2011. In vivo ultrastructural analysis of the intimate relationship between polymorphonuclear leukocytes and the chlamydial developmental cycle. Infect. Immun. 79:3291–3301. 10.1128/IAI.00200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Derrick T, Roberts Ch, Rajasekhar M, Burr SE, Joof H, Makalo P, Bailey RL, Mabey DC, Burton MJ, Holland MJ. 2013. Conjunctival MicroRNA expression in inflammatory trachomatous scarring. PLoS Negl. Trop. Dis. 7:e2117. 10.1371/journal.pntd.0002117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Darville T, O’Neill JM, Andrews CW, Jr, Nagarajan UM, Stahl L, Ojcius DM. 2003. Toll-like receptor-2, but not Toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J. Immunol. 171:6187–6197. 10.4049/jimmunol.171.11.6187 [DOI] [PubMed] [Google Scholar]
- 38. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 104:1604–1609. 10.1073/pnas.0610731104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922. 10.1038/nature06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barron AL, White HJ, Rank RG, Soloff BL, Moses EB. 1981. A new animal model for the study of Chlamydia trachomatis genital infections: infection of mice with the agent of mouse pneumonitis. J. Infect. Dis. 143:63–66. 10.1093/infdis/143.1.63 [DOI] [PubMed] [Google Scholar]
- 41. Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397–1406. 10.1093/nar/28.6.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galens K, Orvis J, Daugherty S, Creasy HH, Angiuoli S, White O, Wortman J, Mahurkar A, Giglio MG. 2011. The IGS standard operating procedure for automated prokaryotic annotation. Stand. Genomics Sci. 4:244–251. 10.4056/sigs.1223234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Delcher AL, Phillippy A, Carlton J, Salzberg SL. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30:2478–2483. 10.1093/nar/30.11.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eppinger M, Radnedge L, Andersen G, Vietri N, Severson G, Mou S, Ravel J, Worsham PL. 2012. Novel plasmids and resistance phenotypes in Yersinia pestis: unique plasmid inventory of strain Java 9 mediates high levels of arsenic resistance. PLoS One 7:e32911. 10.1371/journal.pone.0032911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc. Natl. Acad. Sci. U. S. A. 109:E2010–E2017. 10.1073/pnas.1207359109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42:D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nahid MA, Yao B, Dominguez-Gutierrez PR, Kesavalu L, Satoh M, Chan EKL. 1 February 2013. Regulation of TLR2-mediated tolerance and cross-tolerance through IRAK4 modulation by miR-132 and miR-212. J. Immunol. 190(3):1250–1263. 10.4049/jimmunol.1103060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo course of infection of chlamydial variants (CmVar004 and CmVar001) in two different strains of mice (C57BL/6 and BALB/c mice). Download
Alignment of C. muridarum MOMP (ompA) from Nigg, CmVar001, and CmVar004. Download
Sequencing metrics for plaque variants and the original source culture. Reads are mapped onto the published C. muridarum Nigg genome (Genbank accession no. ).
Comparative genome sequences of C. muridarum variants.


