Abstract
Over the past decade, bioprinting has emerged as a promising patterning strategy to organize cells and extracellular components both in two and three dimensions (2D and 3D) to engineer functional tissue mimicking constructs. So far, tissue printing has neither been used for 3D patterning of mesenchymal stem cells (MSCs) in multiphase growth factor embedded 3D hydrogels nor been investigated phenotypically in terms of simultaneous differentiation into different cell types within the same micropatterned 3D tissue constructs. Accordingly, we demonstrated a biochemical gradient by bioprinting nanoliter droplets encapsulating human MSCs, bone morphogenetic protein 2 (BMP-2), and transforming growth factor β1 (TGF- β1), engineering an anisotropic biomimetic fibrocartilage microenvironment. Assessment of the model tissue construct displayed multiphasic anisotropy of the incorporated biochemical factors after patterning. Quantitative real time polymerase chain reaction (qRT-PCR) results suggested genomic expression patterns leading to simultaneous differentiation of MSC populations into osteogenic and chondrogenic phenotype within the multiphasic construct, evidenced by upregulation of osteogenesis and condrogenesis related genes during in vitro culture. Comprehensive phenotypic network and pathway analysis results, which were based on genomic expression data, indicated activation of differentiation related mechanisms, via signaling pathways, including TGF, BMP, and vascular endothelial growth factor.
Keywords: 3D bioprinting, micropatterning, biomimetic tissue platforms, functional tissue models, tissue interfaces, genomic expression analysis
Introduction
Recent advances in tissue engineering have enabled engineered 3D tissue structures for various applications, including regenerative medicine and in vitro biomimetic functional tissue platforms.1−16 Engineered 3D tissue models that mimic native tissues have emerged and started playing an important role in drug discovery and development.17−19 The need for complex microengineered 3D methods for tissue engineering applications is well accepted due to the limitations of 2D systems in effectively representing the complex tissue environment.20−22 Current 3D tissue scaffolding methods present shortcomings due to lack of control over spatial and temporal control over cell seeding and extracellular matrix (ECM) composition.4,13,22−24 To engineer 3D biomimetic multiphase complex tissue structures such as tissue interfaces, it is critical to have control over the microenvironment components, including cellular, extracellular, and biological factor gradients in microscale. As a result of recent advances in stem cell biology, it is possible to create microenvironments which can direct controlled differentiation of cells and, hence, facilitate reorganization of the bioengineered structures toward a specific tissue phenotype.25
Designs involving single-phasic,26,27 dual-phasic,28 and continuous-gradation29 scaffolds have been developed. However, the small scale of the tissue interfaces (Figure 1A,B), which range from 50 μm to 2 mm in length (depending on tissue, species and age),30,31 presents significant challenges in engineering the microscale anisotropy observed in extracellular, biochemical, and cellular composition.32 Therefore, in the case of interface tissue engineering, there is an unmet need for advanced biomanufacturing methods to mimic the intricate microscale 3D anisotropic environment with precision. Bioprinting can overcome the limitations of existing tissue scaffolding methods in interface tissue engineering by providing control over encapsulation and patterning of the cells and the accompanying ECM components in microscale.23,33
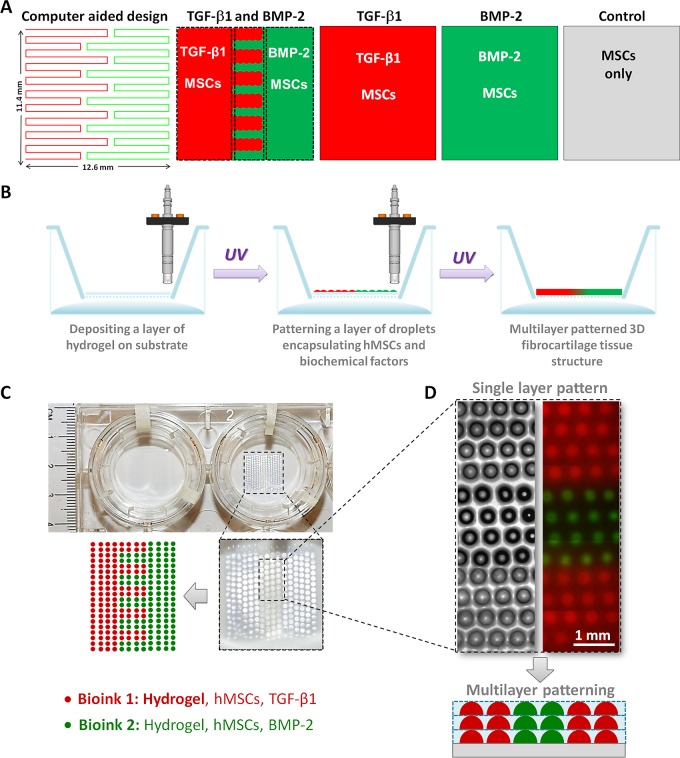
Figure 1.
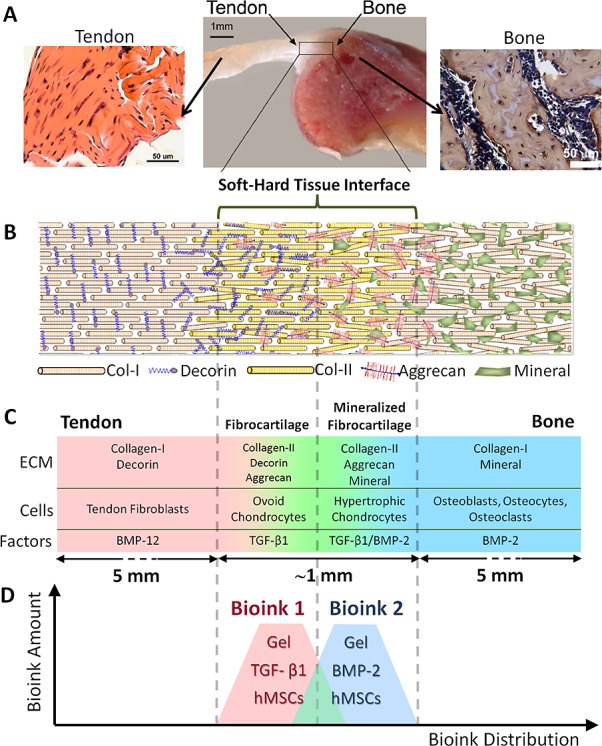
Complex anisotropic organization of the fibrocartilage phase at the bone–tendon interface. (A) Both tendon and bone tissues are rich in collagen type I with significant difference in cellular composition. Tendon (soft tissue) attaches to bone (hard tissue) through an anisotropic insertion site: the fibrocartilage phase. Scale bars in the figures represent 50 μm, 1 mm, and 50 μm, respectively. (B) Fibrocartilage phase presents an intricate gradation in terms of structure, ECM components, cells, and biological factors in a diminutive space (∼1–2 mm in length). (C) Fibrocartilage phase is composed of four continuous and intertwined phases: (i) tendon (proper), (ii) fibrocartilage, (iii) mineralized fibrocartilage, and (iv) bone. (D) We mimicked the native fibrocartilage phase via 3D micropatterning nanoliter gel encapsulated hMSCs, ECM components, and compositions of biochemical factors, BMP-2 and TGF-β1 in a multiphase pattern. Tendon and bone junction photograph was adapted from Wopenka et al.,50 copyright 2008, and reproduced with permission of Society for Applied Spectroscopy.
Bioprinting involves the use of computer-aided transfer processes for patterning and assembling living and nonliving materials with a prescribed 2D or 3D organization to produce bioengineered structures serving in regenerative medicine, drug discovery, and basic cell biology studies.34 Bioprinting, biomanufacturing, and multilayer fabrication methods have been used to control cell patterning and seeding.4,11,33,35−45 In this study, we expand the use of microscale bioprinting to facilitate engineering of the complex anisotropic fibrocartilage tissue phase via nanoliter encapsulation and printing of MSCs along with biochemical factors and ECM components (Figure 1C,D). MSCs were used in this study as they are the common progenitors of musculoskeletal tissues, including bone and cartilage.46,47
Phenotypic characterization of cells within in vitro culture models by genomic expression analysis provides essential generic indicators and high-content biomarkers for drug testing and for studying the impact of uncharacterized perturbations on cells.17,48,49 In this work, we investigated the state of the patterned cells within the 3D multiphasic tissue constructs in terms of a comprehensive genomic expression analysis, which is directly relevant to development of new in vitro functional models and their use in drug discovery. Through genomic expression analysis, we demonstrate the potential of bioprinting in engineering functional biomimetic multiphase 3D tissue models, such as the fibrocartilage phase at the soft and hard tissue interface.
Experimental Section
Micropatterning and in Vitro Culture of Fibrocartilage Phase
A computer-aided design of the bioprinting pathway was first generated (Figure 2A) for each experimental group. Microdroplets of Bioinks were generated in a sterile laminar flow hood under controlled humidity by cell-encapsulating droplet generation system developed in our laboratory.9,33,51,52 Using the valve-based droplet ejector setup, the Bioink droplets composed of cell encapsulating hydrogel were printed on gel-coated substrates (Transwell permeable culture inserts, Corning Inc., Figure 2B). The nominal droplet size was around 300 μm in diameter following deposition on the substrate (Figure 2C,D). The interdroplet distance was determined by the size of the droplets residing on the substrate, which was around 700 μm measured from center to center (Figure 2D). Multiple layers of these droplets were printed and photo-cross-linked layer-by-layer using ultraviolet light (UV) at a power setting of 6.9 mW/cm2 for 30 s based on earlier work.4 Bioprinted and photo-cross-linked multiple layers were merged forming a seamless and continuous 3D tissue structure (Figure 2D). Methacrylated gelatin precursor solution (5%) with photoinitiator for photo-cross-linking (0.5%, Irgacure 2959) was used as the major constituent of the Bioink, as we described previously.4 Diffusion and integration of the phases were assessed using the fluorescent Rhodamine B (red; 0.04 mM; 479 Da) and Dextran-Alexa Fluor 488 (green; 0.01 mM; 10 kDa). To bioprint the fibrocartilage phase, hydrogel solution was supplemented with human MSCs (hMSCs, Lonza) at a concentration of 106 cells per milliliter of hydrogel and growth factors (BMP-2 at 20 ng per mL of hydrogel; TGF-β1 at 10 ng per mL of hydrogel; human recombinant growth factors from R&D Systems). To maintain cellular viability during bioprinting, Bioink was supplemented with 10% culture medium and the pH was neutralized to 7.0 using 0.1 M sodium hydroxide solution. hMSCs were mixed into the Bioink and patterned in microdroplets with BMP-2 and TGF- β1 growth factors in single phase or multiphase pattern representing the fibrocartilage phase. We evaluated four different Bioink compositions to investigate the effect of bioprinting based patterning on engineered fibrocartilage phase: (1) multiphase TGF-β1 and BMP-2 patterning with hMSCs, (2) single phase TGF-β1 patterning with hMSCs, (3) single phase BMP-2 patterning with hMSCs, and (4) control (no growth factors, hMSCs only). The culture medium was composed of α-MEM (Sigma), 10% MSC-qualified-FBS (Invitrogen), 60 U/mL Pen-Strep (Invitrogen), and 2.5 μg/mL Fungizone (Sigma), based on our earlier work.6,13,14,16 The culture medium was changed every two days, and all samples were maintained at 37 °C, 5% CO2, 95–99% relative humidity (to prevent dehydration) throughout the experiment (up to 36 days). To evaluate cell viability in the bioprinted constructs, cells were stained with fluorescent dyes of calcein-AM and propidium iodide (Live-Dead assay, Invitrogen) after bioprinting was completed.
Figure 2.
Micropatterning and bioprinting the anisotropic 3D fibrocartilage phase. (A) A computer-aided drawing and a path map of the bioprinter was developed. Experimental groups and corresponding Bioink compositions were determined. (B) Nanoliter droplets encapsulating hMSCs, ECM components, and growth factors were deposited on a cell culture insert covered with a thin layer of hydrogel. The lower part of the insert was filled with DPBS to keep the deposited nanoliter droplets hydrated. Each layer was photo-cross-linked using UV light to stabilize the structure. (C) The patterned nanoliter droplets initially form a single layer structure on the insert. (D) Single layer design and layer-by-layer deposition can be repeated to achieve a multilayered 3D tissue construct in a single phase or multiphase format. When multiple layers of these droplets are printed and cross-linked, they merge forming a seamless and continuous 3D tissue structure.
Quantitative RT-PCR Analysis
Extraction and isolation of mRNA was performed separately and individually for (i) control, (ii) single phase BMP-2, (iii) single phase TGF-β, and (iv) multiphase BMP-2 and TGF-β groups using the TRIzol reagent and following the manufacturer’s RNA isolation protocol (Invitrogen). qRT-PCR array analysis was used to assess the differentiation of hMSCs to bone, cartilage, tendon, adipose, and muscle phenotypes after 14, 21, and 36 days of culture. Genomic expression analysis was performed using the Human Mesenchymal Stem Cell RT2 Profiler PCR Array (PAHS-082Z, SABiosciences, Qiagen, Valencia, CA) for the expression of 84 key genes according to manufacturer’s instructions utilizing Roche LightCycler 480 instrument. The data generated were analyzed using the SABioscience software. Normalization was performed using arithmetic mean utilizing housekeeping genes (ACTB, B2M, GAPDH, HPRT1, RPLP0). The genes were initially categorized in terms of stemness markers, MSC-specific markers, and other genes associated with MSCs. Stemness markers were grouped as FGF2, INS, LIF, POU5F1, SOX2, TERT, WNT3A, ZFP42. MSC-specific markers were categorized as ALCAM, ANPEP, BMP2, CASP3, CD44, ENG, ERBB2, FUT4, FZD9, ITGA6, ITGAV, KDR, MCAM, NGFR, NT5E, PDGFRB, PROM1, THY1, VCAM1. Other genes associated with MSCs were ANXA5, BDNF, BGLAP, BMP7, COL1A1, CSF2, CSF3, CTNNB1, EGF, FUT1, GTF3A, HGF, ICAM1, IFNG, IGF1, IL10, IL1B, IL6, ITGB1, KITLG, MITF, MMP2, NES, NUDT6, PIGS, PTPRC, SLC17A5, TGFB3, TNF, VEGFA, VIM, VWF. MSC differentiation markers were categorized in four main groups: (i) genes involved in osteogenesis and chondrogenesis, namely, BMP2, BMP4, BMP6, COL1A1, ERBB2, FGF10, GDF6, HDF, IGF1, IL10, IL6, KDR, LIF, RUNX2, SOX9, TBX5, TGFB1, TGFB3, VEGFA, WNT3A; (ii) genes involved in adipogenesis, namely, PPARG, RHOA, RUNX2; (iii) genes involved in myogenesis, namely, JAG1, NOTCH1; and (iv) genes involved in tenogenesis, namely, BMP2, GDF15, SMAD4, TGFB1.
Pathway and Network Analysis
Comprehensive network and pathway analyses were performed using the qRT-PCR data and GeneGo Metacore Software and Database. For GeneGo Metacore pathway and network analysis, fold regulation data obtained from qRT-PCR results were used. To eliminate the noise level data points, intensity levels and fold change data were compared for each sample at each time point. Fold change values were determined relative to control group, which included only hMSCs and did not include any growth factors. According to this comparison, the genes were categorized in the following groups: (i) single phase BMP-2, (ii) single phase TGF-β, and (iii) multiphase BMP-2 and TGF-β. Before analysis, the general threshold value was set as 1.3 and the p-value was set as 0.01 for all time points and samples. Next, GO processes were determined for day 14 (Table S1 in the Supporting Information), day 21 (Table S2 in the Supporting Information), and day 36 (Table S3 in the Supporting Information) for all three categories. Similarly, pathway maps were determined for day 14 (Figure S1 and Table S4 in the Supporting Information), day 21 (Figure S2 and Table S5 in the Supporting Information), and day 36 (Figure S3 and Table S6 in the Supporting Information). Process networks were obtained for day 14 (Table S7 in the Supporting Information), day 21 (Table S8 in the Supporting Information), and day 36 (Table S9 in the Supporting Information). Finally, map folders (Figure S4 in the Supporting Information) were determined for all time points using GeneGo software.
Results and Discussion
Anisotropy and Multiphase Patterning of Engineered 3D Fibrocartilage Tissue Model
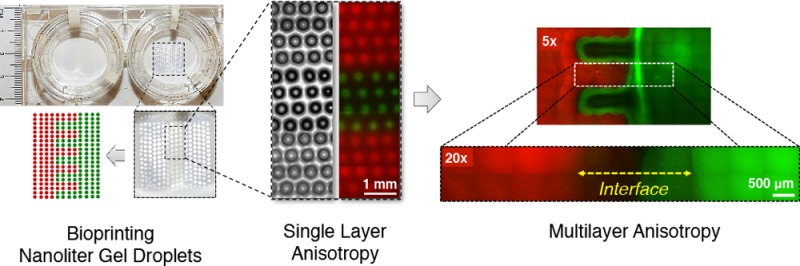
We studied the distinctness and integration of the bioprinted phases by using large molecular weight fluorescent dyes, Rhodamine B (red) and Dextran-Alexa Fluor 488 (green), where red color represents the TGF-β1 phase and the green color represents the BMP-2 phase (Figure 2A–C). The printed multiphase hydrogel structure representing an anisotropic tissue unit displayed boundaries between the individual droplets immediately after printing (Figure 2D). The dyes were considered to mimic the embedded growth factors in different phases in constructs, and they were employed to visualize the anisotropy after patterning (Figure 3A). Release and delivery of growth factors from hydrogel carriers have been extensively studied for applications in tissue engineering and regenerative medicine.555−557 In this study, our aim was to retain the growth factors in the patterned hydrogel constructs together with the cells, which would assist in differentiation of embedded stem cells toward osteogenic and chondrogenic phenotypes in the patterned structures. Imaging after patterning indicated a limited integration and a gradient between the two adjacent phases, and a distinction was still present between the bulk of two phases (Figure 3A). In a smaller scale, the boundaries were observed to fade and smooth transitions emerged between the two phases after the multilayer printing process was completed (Figure 3A inset). The transition region in the tissue construct (indicated by the dashed line in the Figure 3A inset) was observed to be around ∼1–2 mm in length, which mimics the native fibrocartilage interface region. A similar integration pattern between the phases was considered to be present in the case of growth factor and cell patterning.
Figure 3.
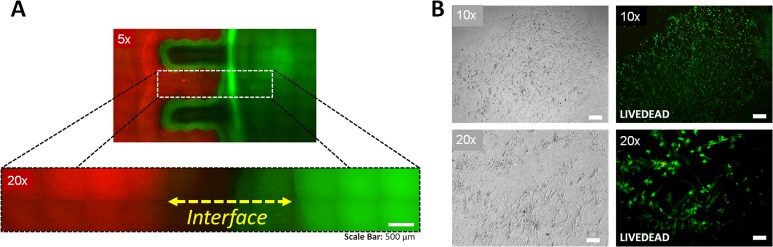
Micropatterned anisotropic fibrocartilage phase. (A) The predesigned architecture was realized via depositing and patterning nanoliter droplets encapsulating hMSCs and compositions of TGF-β1 and BMP-2. Red color zone represents the TGF-β1 patterned section, and green color represents the BMP-2 patterned section. Red to green color transition region in the engineered construct (indicated by dashed line) was observed to be around ∼1–2 mm in length, which mimics the native fibrocartilage interface region. Scale bar represents 500 μm of length. (B) Viability of hMSCs was assessed via a live/dead assay one day after deposition and micropatterning of nanoliter droplets at various magnifications using phase contrast and fluorescent microscopy. Cell viability was greater than 90%, indicated by green colored cells. Cells displayed typical healthy morphology in the bioprinted constructs. Scale bars represent 500 μm of length for the upper images, and 100 μm of length for the lower images.
Most biomaterials and scaffolding approaches result in mismatch of compositional properties at the tissue interface due to the lack of the physiological anisotropy.59,60 Soft–hard tissue interfaces between tendon, ligament, cartilage, and bone are complex, and they are composed of four main zones: (i) soft tissue proper, (ii) fibrocartilage, (iii) mineralized fibrocartilage, and (iv) bone, in a microscale intricate organization.50 Mimicking the functional integration site of soft tendon tissue to rigid bone tissue can be attained by regenerating the fibrocartilage phase, which requires population by multiple cell types and associated ECM heterogeneity similar to a native tissue interface.2 An anisotropic and stratified structure is essential to mimic the mechanical, compositional and cellular features of the tissue interface. The transition occurs in a microscopic space (50 μm to ∼1–2 mm, depending on species and age)30,31 with dramatic change in cellular, ECM, and biological factor composition (Figure 1), which could be mimicked using bioprinting method.
Morphological Organization and Characterization of Embedded Cells in Multiphase Patterned Tissue Structure
In an earlier study, we presented an extensive genomic analysis of stem cell markers in bioprinted stem cells, which infer the proliferation potential of the printed cells.51 In this study, to test the viability of the cells after patterning, we performed calcein-AM and propidium iodide based viability assay on the cells. Cell viability was observed to be greater than 90% after micropatterning (Figure 3B) in the engineered fibrocartilage phase. This result indicated that bioprinting did not significantly affect cell survival, which is consistent with our earlier studies.9,33,51,52 Cells displayed typical healthy morphology generally observed in hydrogels (Figure 3B).
Genomic Expression Analysis on Single Phase and Multiphase Patterned Tissue Models
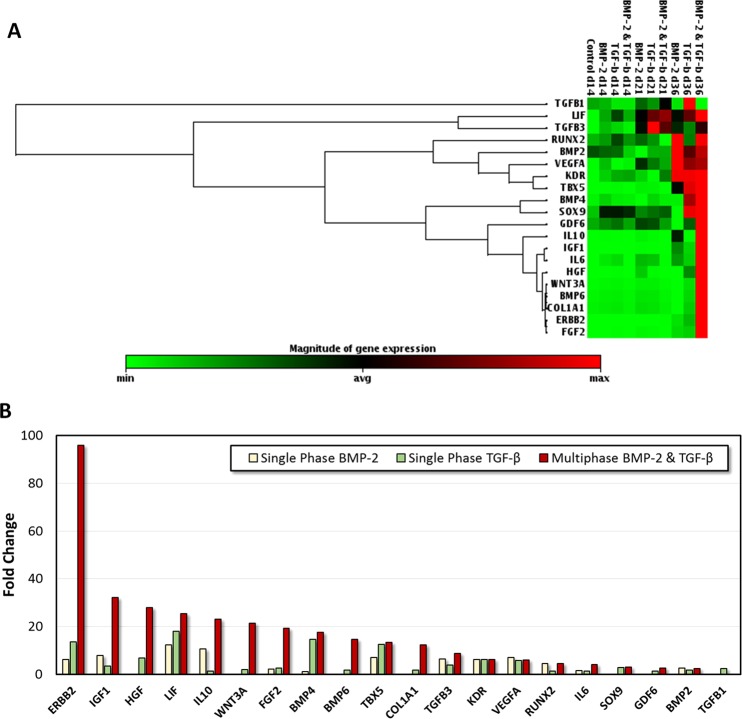
Quantitative RT-PCR genomic expression analysis results demonstrated that most of the osteogenesis and chondrogenesis related genes analyzed were simultaneously upregulated in multiphase BMP-2 and TGF-β1 patterned constructs, especially after long-term culture (Figure 4A). Fold change values were observed to be higher for most osteogenesis and chondrogenesis related genes in the multiphase BMP-2 and TGF-β construct compared to single phase constructs after 36 days in culture (Figure 4B). A number of tendon, muscle, and adipose tissue related genes were also expressed at lower upregulation values (Figure 5).
Figure 4.
Osteogenesis and chondrogenesis related genes in single phase and multiphase samples for days 14, 21, and 36. (A) Red color intensity in osteogenesis and chondrogenesis related genes in BMP-2 and TGF-β1 patterned groups, especially after 36 days of culture, suggests simultaneous expression of these two phenotypes in the engineered tissue constructs. (B) Fold changes were higher for most osteogenesis and chondrogenesis related genes in the multiphase BMP-2 and TGF-β tissue construct compared to single phase constructs after 36 days in culture.
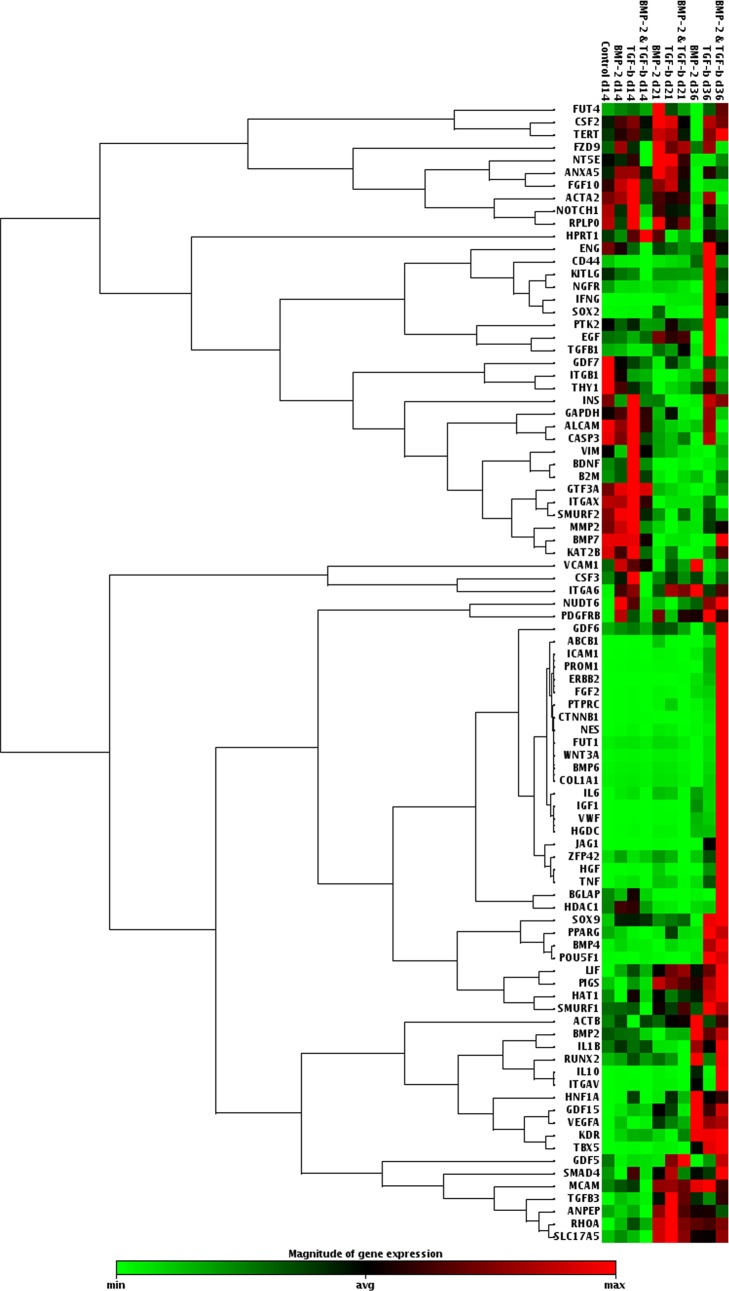
Figure 5.
Genomic expression analysis results presented as a clustergram after three different culture durations (days 14, 21, and 36) for both single phase and multiphase patterning.
Various growth factors, including BMP-2 and TGF-β superfamily factors, have previously been immobilized in combination with ECM components by utilizing the inherent binding ability of these cytokines and ECM components.55,56 These factors were used in combination with bioprinting to form 2D structures to study the response and differentiation of cells, which were seeded postprinting.57,58 In this study we incorporated BMP-2 and TGF-β1 in photo-cross-linkable gelatin based hydrogel matrix in combination with hMSCs as a Bioink to form multiphase 3D tissue models. With this 3D tissue structure, we studied phenotypic differentiation and genomic expression of embedded hMSCs toward bone and cartilage, mimicking the fibrocartilage phase in skeletal system.
Phenotypic Pathway and Network Analysis Based on Genomic Expression Data
General pathway analysis was performed on genomic expression data to obtain a comprehensive list of all the differentiation related pathways involved in the engineered tissue model, which is an approach directly related to drug discovery and development. Activation of differentiation related mechanisms, via signaling pathways, including TGF, Wnt, BMP, and vascular endothelial growth factor (VEGF), were analyzed and presented for qRT-PCR results obtained at day 14 (Figure S1 in the Supporting Information), day 21 (Figure S2 in the Supporting Information), and day 36 (Figure S3 in the Supporting Information), with references to the relevant literature. The specific genes identified in each analysis and the statistical p-values calculated by the GeneGo software are presented in Tables S1–S9 in the Supporting Information. These results demonstrated that, at all the time points, differentiation related pathways were activated in the engineered fibrocartilage tissues via bone and cartilage related signaling pathways, including TGF, Wnt, BMP, and VEGF (Figures S1–S4 in the Supporting Information). In the light of the various pathways observed in this study, future studies are needed that focus on specific relevant pathways involved in differentiation of hMSCs in engineered interface tissues. The approach and the results presented in this work are directly relevant to development of new in vitro functional models based on stem cells and their use in drug discovery.
Summary and Conclusions
We present the application of emerging bioprinting technology in engineering anisotropic multiphase 3D tissue models with potential impact in in vitro drug testing, discovery, and development. We designed a biochemical gradient with microscale gels encapsulating hMSCs and growth factors in an organization that aims to mimic the native fibrocartilage phase. Quantitative RT-PCR analysis showed that the hMSCs displayed an upregulation of osteogenesis and chondrogenesis related genes simultaneously in the 3D fibrocartilage model. Phenotypic pathway and network analysis results were presented based on the genomic expression data obtained from the model. Bioprinted microscale anisotropic tissue structures can potentially be utilized as functional in vitro 3D tissue models and platforms for high-throughput pharmaceutical testing and validation studies. Functional tissue models coupled with comprehensive genomic expression analysis on high-content biomarkers via bioinformatics data mining tools open new venues in drug testing and discovery. These methods and platforms would ultimately allow the use of a patient’s own cells for generating personalized in vitro functional tissue models as testbeds for assessing drug candidates and therapeutics.
Acknowledgments
This work was conducted with support from Harvard Catalyst, The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award 8UL1TR000170-05 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health. Utkan Demirci (U.D.) acknowledges that this material is based in part upon work supported by the National Science Foundation under NSF CAREER Award Number 1150733 and NIH R21HL112114. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Supporting Information Available
Pathway maps, map folder results, tables of GO processes, tables of pathway maps, and tables of process networks. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): Utkan Demirci (U.D.) is a founder of, and has an equity interest in, DXNow, a company that is developing microfluidic and imaging technologies for point-of-care diagnostic solutions. U.D.’s interests were reviewed and are managed by the Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Langer R.; Vacanti J. P. Tissue Engineering. Science 1993, 2605110920–926. [DOI] [PubMed] [Google Scholar]
- Mikos A. G.; Herring S. W.; Ochareon P.; Elisseeff J.; Lu H. H.; Kandel R.; Schoen F. J.; Toner M.; Mooney D.; Atala A.; Van Dyke M. E.; Kaplan D.; Vunjak-Novakovic G. Engineering complex tissues. Tissue Eng. 2006, 12123307–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasoglu S.; Gurkan U. A.; Wang S.; Demirci U. Manipulating biological agents and cells in micro-scale volumes for applications in medicine. Chem. Soc. Rev. 2013, 42135788–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Fan Y.; Xu F.; Erkmen B.; Urkac E. S.; Parlakgul G.; Bernstein J.; Xing W.; Boyden E. S.; Demirci U. Simple precision creation of digitally specified, spatially heterogeneous, engineered tissue architectures. Adv. Mater. 2013, 2581192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Tasoglu S.; Kavaz D.; Demirel M. C.; Demirci U. Emerging technologies for assembly of microscale hydrogels. Adv. Healthcare Mater. 2012, 12149–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Golden R.; Kishore V.; Riley C. P.; Adamec J.; Akkus O. Immune and inflammatory pathways are involved in inherent bone marrow ossification. Clin. Orthop. Relat. Res. 2012, 47092528–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Inci F.; Mullick O.; Gurkan U. A.; Sung Y.; Kavaz D.; Li B.; Denkbas E. B.; Demirci U. Release of magnetic nanoparticles from cell-encapsulating biodegradable nanobiomaterials. ACS Nano 2012, 686640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Finley T. D.; Turkaydin M.; Sung Y.; Gurkan U. A.; Yavuz A. S.; Guldiken R. O.; Demirci U. The assembly of cell-encapsulating microscale hydrogels using acoustic waves. Biomaterials 2011, 32317847–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.; Ceyhan E.; Gurkan U. A.; Demirci U. Statistical modeling of single target cell encapsulation. PLoS One 2011, 67e21580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Wu J.; Wang S.; Durmus N. G.; Gurkan U. A.; Demirci U. Microengineering methods for cell-based microarrays and high-throughput drug-screening applications. Biofabrication 2011, 33034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Sridharan B.; Durmus N. G.; Wang S.; Yavuz A. S.; Gurkan U. A.; Demirci U. Living bacterial sacrificial porogens to engineer decellularized porous scaffolds. PLoS One 2011, 64e19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Beyazoglu T.; Hefner E.; Gurkan U. A.; Demirci U. Automated and adaptable quantification of cellular alignment from microscopic images for tissue engineering applications. Tissue Eng., Part C 2011, 176641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Kishore V.; Condon K. W.; Bellido T. M.; Akkus O. A scaffold-free multicellular three-dimensional in vitro model of osteogenesis. Calcif. Tissue Int. 2011, 885388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Krueger A.; Akkus O. Ossifying bone marrow explant culture as a three-dimensional mechanoresponsive in vitro model of osteogenesis. Tissue Eng., Part A 2011, 173–4417–28. [DOI] [PubMed] [Google Scholar]
- Gurkan U. A.; Cheng X.; Kishore V.; Uquillas J. A.; Akkus O. Comparison of morphology, orientation, and migration of tendon derived fibroblasts and bone marrow stromal cells on electrochemically aligned collagen constructs. J. Biomed. Mater. Res., Part A 2010, 9441070–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan U. A.; Gargac J.; Akkus O. The sequential production profiles of growth factors and their relations to bone volume in ossifying bone marrow explants. Tissue Eng., Part A 2010, 1672295–306. [DOI] [PubMed] [Google Scholar]
- Peck Y.; Wang D.-A. Three-dimensionally engineered biomimetic tissue models for in vitro drug evaluation: delivery, efficacy and toxicity. Expert Opin. Drug Delivery 2013, 103369–83. [DOI] [PubMed] [Google Scholar]
- Chang R.; Emami K.; Wu H.; Sun W. Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model. Biofabrication 2010, 24045004. [DOI] [PubMed] [Google Scholar]
- Prestwich G. D. Evaluating drug efficacy and toxicology in three dimensions: using synthetic extracellular matrices in drug discovery. Acc. Chem. Res. 2008, 411139–48. [DOI] [PubMed] [Google Scholar]
- Birgersdotter A.; Sandberg R.; Ernberg I. Gene expression perturbation in vitro - A growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005, 155405–412. [DOI] [PubMed] [Google Scholar]
- Tibbitt M. W.; Anseth K. S. Hydrogels as Extracellular Matrix Mimics for 3D Cell Culture. Biotechnol. Bioeng. 2009, 1034655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geckil H.; Xu F.; Zhang X. H.; Moon S.; Demirci U. Engineering hydrogels as extracellular matrix mimics. Nanomedicine 2010, 53469–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert P. Materials science. Printing cells. Science 2007, 3185848208–9. [DOI] [PubMed] [Google Scholar]
- Khademhosseini A.; Langer R.; Borenstein J.; Vacanti J. P. Microscale technologies for tissue engineering and biology. Proc. Natl. Acad. Sci. U.S.A. 2006, 10382480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab K.; Norotte C.; Marga F.; Murphy K.; Vunjak-Novakovic G.; Forgacs G. Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2010, 22022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodeo S. A.; Suzuki K.; Deng X. H.; Wozney J.; Warren R. F. Use of recombinant human bone morphogenetic protein-2 to enhance tendon healing in a bone tunnel. Am. J. Sports Med. 1999, 274476–88. [DOI] [PubMed] [Google Scholar]
- Mutsuzaki H.; Sakane M.; Nakajima H.; Ito A.; Hattori S.; Miyanaga Y.; Ochiai N.; Tanaka J. Calcium-phosphate-hybridized tendon directly promotes regeneration of tendon-bone insertion. J. Biomed. Mater. Res., Part A 2004, 702319–27. [DOI] [PubMed] [Google Scholar]
- Cooper J. A.; Lu H. H.; Ko F. K.; Freeman J. W.; Laurencin C. T. Fiber-based tissue-engineered scaffold for ligament replacement: design considerations and in vitro evaluation. Biomaterials 2005, 26131523–32. [DOI] [PubMed] [Google Scholar]
- Sant S.; Hancock M. J.; Donnelly J. P.; Iyer D.; Khademhosseini A. Biomimetic gradient hydrogels for tissue engineering. Can. J. Chem. Eng. 2010, 886899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R. R.; Misol S. Tendon and Ligament Insertion. A Light and Electron Microscopic Study. J. Bone Jt. Surg., Am. Vol. 1970, A 5211–20. [PubMed] [Google Scholar]
- Wang I. N. E.; Mitroo S.; Chen F. H.; Lu H. H.; Doty S. B. Age-dependent changes in matrix composition and organization at the ligament-to-bone insertion. J. Orthop. Res. 2006, 2481745–55. [DOI] [PubMed] [Google Scholar]
- Lu H. H.; Subramony S. D.; Boushell M. K.; Zhang X. Z. Tissue Engineering Strategies for the Regeneration of Orthopedic Interfaces. Ann. Biomed. Eng. 2010, 3862142–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.; Hasan S. K.; Song Y. S.; Xu F.; Keles H. O.; Manzur F.; Mikkilineni S.; Hong J. W.; Nagatomi J.; Haeggstrom E.; Khademhosseini A.; Demirci U. Layer by Layer Three-Dimensional Tissue Epitaxy by Cell-Laden Hydrogel Droplets. Tissue Eng., Part C 2010, 161157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabien G.; Mironov V.; Nakamura M. Bioprinting is coming of age: report from the International Conference on Bioprinting and Biofabrication in Bordeaux (3B′09). Biofabrication 2010, 21010201. [DOI] [PubMed] [Google Scholar]
- Nakamura M.; Kobayashi A.; Takagi F.; Watanabe A.; Hiruma Y.; Ohuchi K.; Iwasaki Y.; Horie M.; Morita I.; Takatani S. Biocompatible inkjet printing technique for designed seeding of individual living cells. Tissue Eng. 2005, 1111–121658–66. [DOI] [PubMed] [Google Scholar]
- Boland T.; Xu T.; Damon B.; Cui X. Application of inkjet printing to tissue engineering. Biotechnol. J. 2006, 19910–7. [DOI] [PubMed] [Google Scholar]
- Barron J. A.; Wu P.; Ladouceur H. D.; Ringeisen B. R. Biological laser printing: a novel technique for creating heterogeneous 3-dimensional cell patterns. Biomed. Microdevices 2004, 62139–47. [DOI] [PubMed] [Google Scholar]
- Ceyhan E.; Xu F.; Gurkan U. A.; Emre A. E.; Turali E. S.; El Assal R.; Acikgenc A.; Wu C. A. M.; Demirci U. Prediction and control of number of cells in microdroplets by stochastic modeling. Lab Chip 2012, 12224884–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmus N. G.; Tasoglu S.; Demirci U. Bioprinting Functional droplet networks. Nat. Mater. 2013, 126478–9. [DOI] [PubMed] [Google Scholar]
- Gurkan U. A.; Sung Y.; El Assal R.; Xu F.; Trachtenberg A.; Kuo W.; Demirci U. Bioprinting anisotropic stem cell microenvironment. J. Tissue Eng. Regener. Med. 2012, 6S1366–6. [Google Scholar]
- Tasoglu S.; Demirci U. Bioprinting for stem cell research. Trends Biotechnol. 2013, 31110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Celli J.; Rizvi I.; Moon S.; Hasan T.; Demirci U. A three-dimensional in vitro ovarian cancer coculture model using a high-throughput cell patterning platform. Biotechnol. J. 2011, 62204–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Sridharan B.; Wang S. Q.; Gurkan U. A.; Syverud B.; Demirci U. Embryonic stem cell bioprinting for uniform and controlled size embryoid body formation. Biomicrofluidics 2011, 10.1063/1.3580752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Wu J. H.; Wang S. Q.; Durmus N. G.; Gurkan U. A.; Demirci U. Microengineering methods for cell-based microarrays and high-throughput drug-screening applications. Biofabrication 2011, 10.1088/1758-5082/3/3/034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P.; Riminucci M.; Gronthos S.; Robey P. G. Bone marrow stromal stem cells: Nature, biology, and potential applications. Stem Cells 2001, 193180–92. [DOI] [PubMed] [Google Scholar]
- Prockop D. J. Marrow stromal cells as steam cells for nonhematopoietic tissues. Science 1997, 276530971–4. [DOI] [PubMed] [Google Scholar]
- Gunther E. C.; Stone D. J.; Rothberg J. M.; Gerwien R. W. A quantitative genomic expression analysis platform for multiplexed in vitro prediction of drug action. Pharmacogenomics J. 2005, 52126–34. [DOI] [PubMed] [Google Scholar]
- Hughes T. R.; Marton M. J.; Jones A. R.; Roberts C. J.; Stoughton R.; Armour C. D.; Bennett H. A.; Coffey E.; Dai H.; He Y. D.; Kidd M. J.; King A. M.; Meyer M. R.; Slade D.; Lum P. Y.; Stepaniants S. B.; Shoemaker D. D.; Gachotte D.; Chakraburtty K.; Simon J.; Bard M.; Friend S. H. Functional discovery via a compendium of expression profiles. Cell 2000, 1021109–26. [DOI] [PubMed] [Google Scholar]
- Wopenka B.; Kent A.; Pasteris J. D.; Yoon Y.; Thomopoulos S. The tendon-to-bone transition of the rotator cuff: a preliminary Raman spectroscopic study documenting the gradual mineralization across the insertion in rat tissue samples. Appl. Spectrosc. 2008, 62121285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.; Kim Y.-G.; Dong L.; Lombardi M.; Haeggstrom E.; Jensen R.V.; Hsiao L.-L.; Demirci U. Drop-on-Demand Single Cell Isolation and Total RNA Analysis. PLoS One 2011, 63e17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F.; Moon S.J.; Emre A.E.; Turali E.S.; Song Y.S.; Hacking S.A.; Nagatomi J.; Demirci U. A droplet-based building block approach for bladder smooth muscle cell (SMC) proliferation. Biofabrication 2010, 10.1088/1758-5082/2/1/014105. [DOI] [PubMed] [Google Scholar]
- Rodeo S. A.; Arnoczky S. P.; Torzilli P. A.; Hidaka C.; Warren R. F. Tendon-healing in a bone tunnel. A biomechanical and histological study in the dog. J. Bone Jt. Surg. Am. 1993, 75121795–803. [DOI] [PubMed] [Google Scholar]
- Silva M. J.; Thomopoulos S.; Kusano N.; Zaegel M. A.; Harwood F. L.; Matsuzaki H.; Havlioglu N.; Dovan T. T.; Amiel D.; Gelberman R. H. Early healing of flexor tendon insertion site injuries: Tunnel repair is mechanically and histologically inferior to surface repair in a canine model. J. Orthop. Res. 2006, 245990–1000. [DOI] [PubMed] [Google Scholar]
- Rider C. C. Heparin/heparan sulphate binding in the TGF-beta cytokine superfamily. Biochem. Soc. Trans. 2006, 34, 458–60. [DOI] [PubMed] [Google Scholar]
- Ruppert R.; Hoffmann E.; Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur. J. Biochem. 1996, 2371295–302. [DOI] [PubMed] [Google Scholar]
- Miller E. D.; Phillippi J. A.; Fisher G. W.; Campbell P. G.; Walker L. M.; Weiss L. E. Inkjet Printing of Growth Factor Concentration Gradients and Combinatorial Arrays Immobilized on Biologically-Relevant Substrates. Comb. Chem. High Throughput Screening 2009, 126604–18. [DOI] [PubMed] [Google Scholar]
- Phillippi J. A.; Miller E.; Weiss L.; Huard J.; Waggoner A.; Campbell P. Microenvironments engineered by inkjet bioprinting spatially direct adult stem cells toward muscle- and bone-like subpopulations. Stem Cells 2008, 261127–34. [DOI] [PubMed] [Google Scholar]
- Lee S.-H.; Shin H. Matrices and scaffolds for delivery of bioactive molecules in bone and cartilage tissue engineering. Adv. Drug Delivery Rev. 2007, 594-5339–359. [DOI] [PubMed] [Google Scholar]
- Tessmar J. K.; Goepferich A. M. Matrices and scaffolds for protein delivery in tissue engineering. Adv. Drug Delivery Rev. 2007, 594-5274–291. [DOI] [PubMed] [Google Scholar]
- Drury J. L.; Mooney D. J. Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 2003, 24244337–4351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.