Abstract
Genome duplication is thought to be central to the evolution of morphological complexity, and some polyploids enjoy a variety of capabilities that transgress those of their diploid progenitors. Comparison of genomic sequences from several tetraploid (AtDt) Gossypium species and genotypes with putative diploid A- and D-genome progenitor species revealed that unidirectional DNA exchanges between homeologous chromosomes were the predominant mechanism responsible for allelic differences between the Gossypium tetraploids and their diploid progenitors. Homeologous gene conversion events (HeGCEs) gradually subsided, declining to rates similar to random mutation during radiation of the polyploid into multiple clades and species. Despite occurring in a common nucleus, preservation of HeGCE is asymmetric in the two tetraploid subgenomes. At-to-Dt conversion is far more abundant than the reciprocal, is enriched in heterochromatin, is highly correlated with GC content and transposon distribution, and may silence abundant A-genome-derived retrotransposons. Dt-to-At conversion is abundant in euchromatin and genes, frequently reversing losses of gene function. The long-standing observation that the nonspinnable-fibered D-genome contributes to the superior yield and quality of tetraploid cotton fibers may be explained by accelerated Dt to At conversion during cotton domestication and improvement, increasing dosage of alleles from the spinnable-fibered A-genome. HeGCE may provide an alternative to (rare) reciprocal DNA exchanges between chromosomes in heterochromatin, where genes have approximately five times greater abundance of Dt-to-At conversion than does adjacent intergenic DNA. Spanning exon-to-gene-sized regions, HeGCE is a natural noninvasive means of gene transfer with the precision of transformation, potentially important in genetic improvement of many crop plants.
Keywords: cotton, homeologous gene conversion, polyploidy, heterochromatin
GENOME duplication is a potentially rich source of genes with new (Stephens 1951; Ohno 1970) or modified functions (Lynch and Conery 2000), and is thought to be central to the evolution of morphological complexity (Freeling and Thomas 2006). Genome doubling may confer advantages to a polyploid (Comai 2005), via mechanisms such as increased gene dosage, “intergenomic heterosis” conferred by multiple alleles in a polyploid nucleus, or the evolution of novel gene functions (neofunctionalization) (Stephens 1951; Ohno 1970). Over time, duplicated genes may evolve subdivisions of ancestral functions (subfunctionalization) (Lynch and Force 2000) that render them interdependent. Subfunctionalization may sometimes lead to neofunctionalization (He and Zhang 2005).
Polyploids have been suggested to enjoy a variety of capabilities that transgress those of their diploid progenitors. For example, the notion that polyploids may adapt better than diploids to environmental extremes has been suggested, based on both their geographic distribution (Muntzing 1936; Love and Love 1949; Stebbins 1950; Grant 1971) and on an inferred abundance of paleopolyploidizations near the Cretaceous–Tertiary extinction (Fawcett et al. 2009). A variety of morphological, physiological, and gene expression changes have been associated with polyploidy. Experimental data are available to evaluate causality of only a few such cases in specific adaptations of polyploids, with more data needed but offering some support (as recently reviewed in Madlung 2013).
Angiosperms (flowering plants) are an outstanding model for studying consequences of genome duplication salient to higher eukaryotes. All angiosperms are paleopolyploid (Bowers et al. 2003; Jiao et al. 2011), and their abundance of multiple independent genome duplications (Paterson et al. 2010) provides “natural replicates” for a variety of investigations. Study of the genes from three rounds of ancient whole genome duplications in Arabidopsis reveals a short phase of function relaxation followed by diversifying selection (Guo et al. 2013). The ability to study multiple independent genome duplications in a lineage also permits inference of the temporal orders and rates at which different duplication-associated events/mechanisms occur. Their larger genome sizes and smaller effective population sizes than microbes that have experienced genome duplication such as yeast (Gu et al. 2003; Christoffels et al. 2004; Scannell et al. 2006) and Paramecium (Aury et al. 2006), makes angiosperms more appropriate for studying consequences of genome duplication in higher eukaryotes (Lynch et al. 2001; Lynch 2006).
The ability to “synthesize” newly polyploid plants by artificial crosses and chromosomal manipulation using colchicine, has revealed striking immediate reactions of genomes to duplication. These reactions include loss and restructuring of low-copy DNA sequences (Song et al. 1995; Feldman et al. 1997; Ozkan et al. 2001; Shaked et al. 2001; Kashkush et al. 2002; Ozkan et al. 2002), activation of genes and retrotransposons (O’Neill et al. 2002; Kashkush et al. 2003), gene silencing (Chen and Pikaard 1997a, 1997b; Comai et al. 2000; Lee and Chen 2001), and subfunctionalization of gene expression patterns (Adams et al. 2003, 2004). Gene silencing in the allopolyploid hybrid between Arabidopsis thaliana and Cardaminopsis arenosa is arguably related to defense response against transposons (Comai et al. 2000). Changes of 24-nt siRNA and DNA methylation levels in Arabidopsis hybrids are greatest at loci for which two parents differ substantially (Groszmann et al. 2011; Greaves et al. 2011). Chromosome rearrangement and reactivation of transposable elements are well known when plants are under “genomic stress,” which includes formation of polyploids (McClintock 1983). Instability of hybrid genomes has been attributed to bursts of transposition in both animals and plants; cytosine demethylation and deacetylation of lysine residues on histones may be responsible (Fontdevila 2005).
However, to learn whether immediate reactions of genomes to duplication provide raw material for the beginnings of adaptation or are merely symptoms of imminent extinction, it is necessary to investigate naturally formed polyploids that have survived the test of time. The extinction hypothesis seems generally more likely, given that unreduced gametes are produced more or less continuously by organisms but only a tiny fraction result in successful lineages. For example, dramatic early-generation mutations in synthetic Brassica napus (Pires et al. 2004) are not paralleled in naturally occurring forms (Rana et al. 2004).
A particularly intriguing example of possible advantages associated with polyploidy comes from the cotton genus, Gossypium, in which two diploids and two tetraploids have each been independently domesticated for production of the same product, seedborne epidermal fibers. A-genome diploids native to Africa, and Mexican D-genome diploids diverged ∼5–10 MYA (Senchina et al. 2003) . They were reunited ∼1–2 MYA by trans-oceanic dispersal to the New World of a maternal A-genome propagule resembling Gossypium herbaceum (Wendel 1989), hybridization with a native D-genome species resembling G. raimondii, and chromosome doubling. The nascent AtDt allopolyploid spread throughout the American tropics and subtropics, diverging into at least three subclades and five species, with two of those species (G. hirsutum and G. barbadense) being independently domesticated.
In India, where scientific improvement programs are active for both ploidies, tetraploid (“AtDt” genome) cottons consistently have substantially higher yield and superior fiber qualities than A-genome diploids (Anonymous 1997). Remarkably, the majority of genetic variation among tetraploid cottons has been ascribed to chromosomes from the D-genome diploid progenitor that does not produce spinnable fiber, suggesting that postpolyploidy selection for superior fiber yield and quality of tetraploid cottons has preferentially operated upon the Dt genome (Jiang et al. 1998; Rong et al. 2007).
In the present study, sequencing and careful comparison of several tetraploid Gossypium species and genotypes and representatives of their putative progenitor genomes reveals that homeologous gene conversion events (HeGCEs) account for the vast majority of allelic differences between polyploid cottons and their diploid progenitors. High survivorship of alleles that were converted shortly after polyploid formation is suggested to reflect both a rapid rate of conversion at that time and also adaptive significance of many resulting alleles. A second cadre of converted alleles are closely associated with domestication, suggesting a mechanism by which chromosomes from the D-genome diploid progenitor that does not produce spinnable fiber may have come to account for the majority of genetic variation in fiber characteristics among tetraploid cottons. In partial summary, these data suggest that HeGCEs are an early and important mechanism by which genomes adapt to the duplicated state and may also contribute to plant domestication and crop improvement.
Methods
Read mapping and single nucleotide variation detection
Sequences of cotton D-genome v2 (G. raimondii), A-genome (G. herbaceum), and a tetraploid genome (G. hirsutum, Acala ‘Maxxa’) are from Paterson et al. (2012). G. herbaceum is sequenced with read depth of 32× and Acala ‘Maxxa’ with read depth of 82×. Sequences of three additional tetraploid genomes (G. hirsutum GA120R1B3 30×, G. hirsutum race yucatanese 15×; and G. mustelinum 46×) are from Illumina sequencing of paired-end libraries. Reads are aligned to the reference genome using Burrows-Wheeler Aligner (BWA) (Li and Durbin 2009). Single nucleotide variants (SNVs) between D- and A-genomes are called with Samtools/bcftools (Li et al. 2009; Li 2011) using reads with mapping quality >30 and base quality >30. Raw SNVs between progenitor genomes are further filtered by keeping those with read depth between 4 and 60. Raw SNVs for the tetraploid species are further filtered by keeping those with read depth between 7 and twice the average effective read coverage. After aligning reads from the tetraploid genome to the reference genome, alleles are assigned to each subgenome by referring to the parental alleles.
Detection of converted alleles
SNVs from the two progenitor genomes are identified by aligning reads from the A-genome to the reference D-genome. For each of the SNV sites, the orthologous alleles in the tetraploid genomes are sorted into subsets corresponding to the respective parental subgenomes (Supporting Information, Figure S6). A locus that differs between the diploid progenitors but for which a tetraploid shows only the allele from one progenitor (i.e., is monomorphic) is regarded as converted, if meeting the following criteria. Assuming that alleles at a locus follow a binomial distribution when sampling tetraploid DNA, read coverage of 7× is necessary to keep the false positive rate under 0.0078. To remove false positives caused by undersampling, we filtered out monomorphic alleles with read coverage under 7. False-positive converted alleles can also be derived from deletion of a progenitor allele in a subgenome. In this event, read coverage of deleted sites would average half that of the sites that show both parental alleles. Average mapped read coverage for the four tetraploid cotton species studied are shown in Figure S7. Reduction of read coverage by half is not observed in either of the conversion categories, compared to sites that show both parental alleles.
Quality control
To rule out the possibility that inferred conversions are due to the At subgenome being more divergent than the Dt from the reference genome, the effect of relaxing the value of editing distance (−n flag in BWA) was investigated. If the sequence divergence was a major factor, the magnitude of conversion bias would be reduced after relaxing the editing distance. The default value for −n flag is 0.04. The estimated mean nucleotide divergence between A- and D-genomes is ∼0.64%, ranging from 1.13e-5 to 0.012. Theoretically, the default value is sufficiently large to allow the mapping of At-genome reads even for most diverged regions. Figure S8A shows the proportion of allele changes resulting from relaxing the value of −n flag to 0.8. The magnitude of conversion bias is not decreased, but increased. At-to-Dt converted alleles became the most common one, presumably due to an increase of mapped reads from the Dt subgenome after allowing more mismatches. That is, the relative frequency of At alleles declined under the SNP calling threshold because more Dt-subgenome reads aligned to the reference. To test this, we reduced the frequency threshold of calling a heterozygous SNP (Figure S8B). We observed an increase of unchanged alleles and decrease of both converted types. Expectedly, the log ratio of the two conversion types also increased (Figure S9). We also tried reads with mapping quality from 1 to 30 with increments of 5; all of the tests show similar results. We used reads with mapping quality >20 for this study.
We further investigated possible artifacts of read alignment by assessing the sequence divergence between the A- and D-genomes near HeGCE sites. We divided the genome into nonoverlapping 10-kb bins. For each bin, we calculated the number of At-to-Dt converted alleles and the nucleotide divergence between the A- and D-genomes. The nucleotide divergence in each bin is normalized by the proportion of sites covered by reads to remove the variation of reads coverage. The Pearson correlation coefficient of nucleotide divergence and At-to-Dt conversions is 0.089, which indicates only weak positive correlations (Figure S10A). To compare with other types of allele changes, we also looked into the correlation between Dt-to-At converted sites and unchanged sites. The other two types also show similar levels of correlations (r = 0.0613 for Dt-to-At converted alleles; r = 0.0979 for unchanged alleles) (Figure S10, B and C).
Evaluation of gene function impact
Gene function impact of allele changes is measured relative to the cotton reference gene models as described (Paterson et al. 2012), by the following four categories: (a) Altered translation initiation site − allele changes in the first codon of a coding sequence that leads to an amino acid other than methionine; (b) altered splicing sites − allele changes disrupting the “GT–AG” conserved sites flanking introns; (c) introduced stop codons − allele changes introducing premature stop codons into the normal protein coding sequence; and (d) altered stop codons − allele changes altering stop codon to encode an amino acid.
Estimation of the size of biased-conversion tracks
Due to the large variance of measurement of the length of conversion tracks in base pairs (bp), we use number of continuous At-to-Dt conversion alleles as a measure. We searched for continuous tracks of conversion alleles both in the genome and in Highly-biased conversion region (HC). If we assume random mutation follows a Poisson distribution, the length of continuous mutations should follow an exponential distribution. Both genome-wide and HC conversion tracks show longer continuous tracks than expected (Figure S2). Conversion tracks in the HC show an excess of long ones (four to six alleles) and a deficiency of shorter ones (fewer than three alleles) than the genome-wide set. From the distribution, the average length of conversion tracks across the genome is 3.75 continuous alleles. Genome-wide, the average distance between three continuous conversion alleles is 320.3 bp and 412.6 bp for four. Estimation of the average length of conversion tracks across the genome is (320.3/3 + 412.6/4) × 3.75/2 = 394 bp. In HC, the average length of conversion tracks is 4.32 continuous alleles, with the average length of four continuous At-to-Dt conversion alleles being 600.8 bp and 647.3 bp for five. Estimation of the average length of conversion tracks in HC is (600.8/4 + 647.3/5) × 4.32/2 = 604 bp.
Results
Unidirectional DNA exchanges between homeologous loci were the predominant mutational mechanism in the nascent Gossypium polyploid
DNA recombination, typically by reciprocal exchanges between homologous chromosomes (“crossing over”), is a central element of eukaryotic transmission genetics and is also implicated in repair of highly deleterious DNA double-strand breaks (DSBs). Reciprocal exchanges are often accompanied by tracts of unidirectional, local DNA exchanges known as “gene conversion.” Most models proposed to account for homologous DSB repair (synthesis-dependent strand annealing, classic double-strand break repair, and break-induced replication, although not single strand annealing) (Helleday et al. 2007), also predict the occurrence of tracts of unidirectional gene conversion, even when reciprocal crossing over occurs.
Building on rich evidence of concerted evolution in tandemly repeated sequences such as ribosomal RNA genes (Eickbush and Eickbush 2007) and multigene families such as primate olfactory receptor genes (Sharon et al. 1999), we recently showed gene conversion to have occurred in the past ∼400,000 years between duplicated rice genes that diverged from a common ancestor 70 million years ago (MYA) (Wang et al. 2009). While this is an extreme case, nonrandom similarity between duplicated genes widely distributed across other genomes (Chapman et al. 2006; Wang et al. 2007) suggests the phenomenon to be widespread.
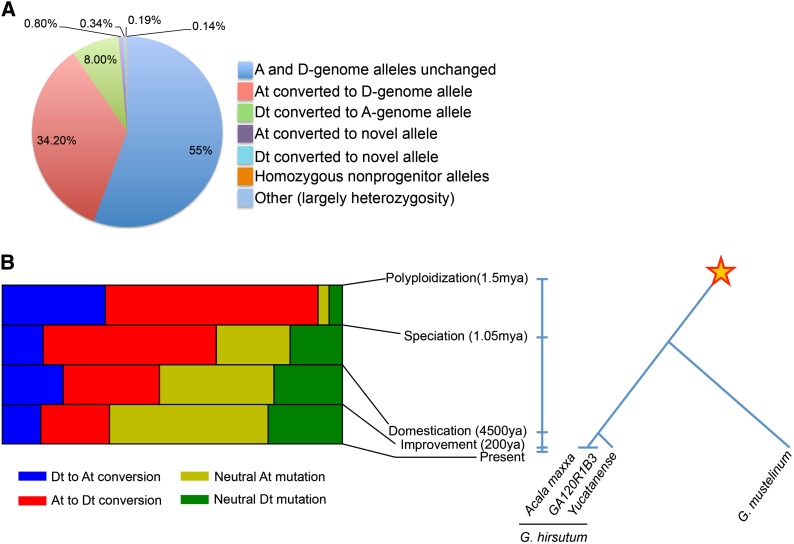
During the 1–2 MY since its formation, unidirectional DNA exchanges between homeologous chromosomes have greatly outnumbered random mutations in AtDt tetraploid cotton. Mapping of 38× Illumina coverage from the A-genome species G. herbaceum to the reference D-genome v2 (G. raimondii) (Paterson et al. 2012) revealed 2,145,177 SNVs between the two, with 60% remaining unchanged in an AtDt tetraploid, G. hirsutum cultivar Acala ‘Maxxa.’ Among the 40% of changed sites, 25% now have only D-genome alleles and 10.6% have only A-genome alleles, with only ∼4.4% having new mutations. SNVs inferred (by methods previously described) (Paterson et al. 2012) to confer striking changes of gene function are even more biased, with 45% of sites changed, 34.2% to D-genome alleles and 8% to A-genome alleles (Figure 1A).
Figure 1.
Allelic changes in polyploid cotton. (A) Allelic changes causing striking inferred protein changes between At and Dt subgenomes of tetraploid (G. hirsutum) Acala ‘Maxxa.’ (B) Evolutionary history of intergenomic conversion in polyploid cotton. The proportion of At and Dt neutral (intergenic) mutations and conversions in both HC and LC are shown in each window in time. The star marks AD allopolyploidization occurring ∼1–2 MYA. See Figure S1 for raw data.
To infer the levels and patterns of occurrence of gene conversions following polyploid cotton formation (Figure 1B), we used a parsimony-based method. For comparison with Acala ‘Maxxa,’ we resequenced another G. hirsutum cultivar (GA120R1B3) from a different US production region (i.e., separated by <200 years), a wild G. hirsutum (race yucatanese) separated by ∼4500 years, and G. mustelinum, the tetraploid cotton species most divergent from G. hirsutum separated by ∼1 million yr. If a converted allele is shared by two lineages, we assume that the event occurred in the common ancestor rather than independently in each lineage. Indeed, most conversions were present in all four genomes (85.03–87.54%; Figure 1B), outnumbering omnipresent random mutations by approximately eightfold (χ2 = 23,599, 87,473 for Dt, At, P<0.001. Parsimony implies that these HeGCEs occurred prior to speciation in the nascent Gossypium polyploid.
When the polyploid Gossypium lineage diverged into multiple species, HeGCE appears to have been abating (Figure 1B). This is inferred based on the observation that HeGCEs account for only ∼62% (4921) of polymorphisms among the divergent species G. hirsutum and G. mustelinum, ∼47% between wild and cultivated forms of G. hirsutum, and 33% between G. hirsutum cultivars from different production regions (Figure 1B and Figure S1A). Consideration of the genomic distribution of HeGCEs in these various taxa suggests that it abates sooner in heterochromatin than in euchromatin (Figure S1B).
Asymmetric evolution of the polyploid cotton subgenomes
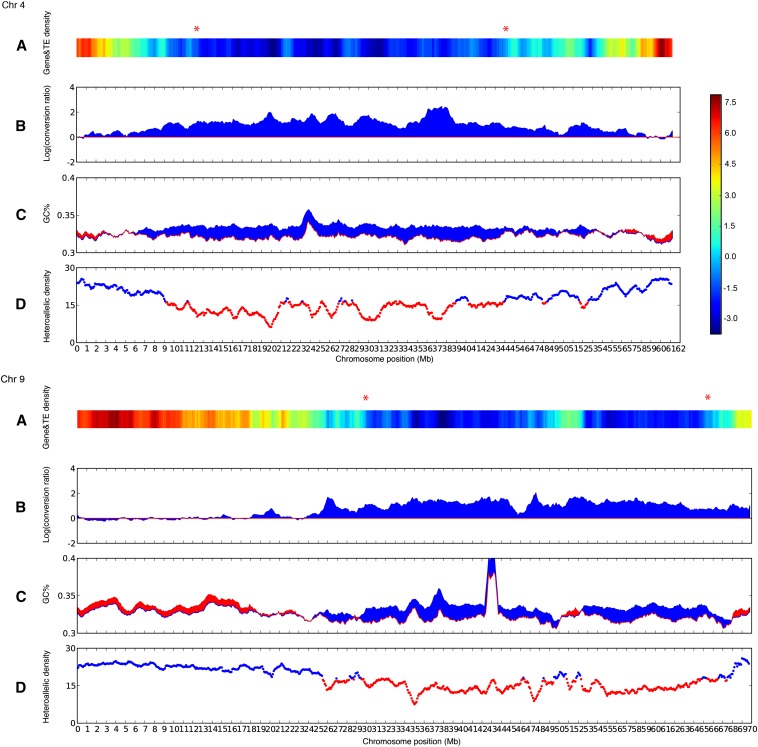
Despite inhabiting a common nucleus, the evolution of the cotton At and Dt subgenomes differs in several striking ways. First, At-to-Dt conversion is enriched in heterochromatin (Figure 2). Euchromatin, localized in the terminal regions of cotton chromosomes, shows largely similar rates of the two conversion types (log-likelihood ratio ∼0). Occasional 1-Mb bins with Dt-to-At biased conversion (log-likelihood ratio <0) unanimously reside in euchromatin.
Figure 2.
Distributions of biased conversion, heterozygosity, and GC content across chromosomes 4 and 9. Chromosomes are divided into 1-Mb bins using sliding windows with 100-kb step size. (A) Heatmap of gene and transposon density across chromosomes. Log ratio of Z-scores of gene vs. transposon density in each bin is shown, with blue marking transposon-rich and red marking gene-rich regions. Red asterisks mark inferred heterochromatin boundaries based on densities of genes and transposons. (B) Genomic distribution of log-likelihood ratio of At-to-Dt converted alleles to Dt-to-At converted alleles in tetraploid cotton, calculated for each bin. Red line denotes log-likelihood ratio of zero. (C) Comparison of GC% between cotton A- and D-genomes. Blue bins indicate GC% of D > A genome; other bins are red. (D) Genomic distribution of heteroallelic density (i.e., with at least one allele differing from the other three) in tetraploid cotton (Acala ‘Maxxa’). Heteroallelic density is calculated for each bin, corrected by read coverage, with below-average density indicated by red lines and average or higher density by blue lines. See Figure S3 for other chromosomes.
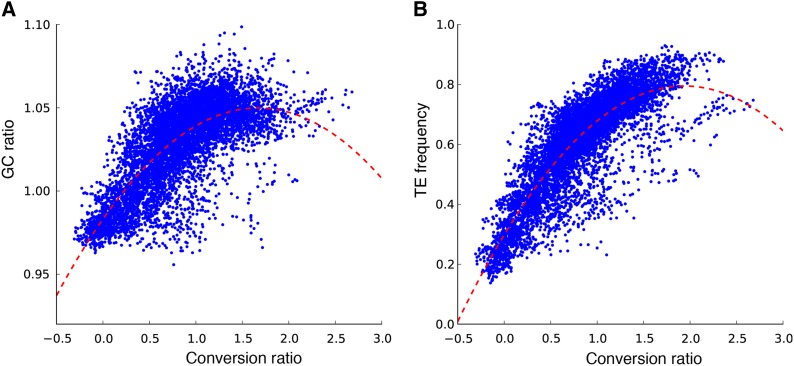
As in many other eukaryotes (Duret and Galtier 2009) cotton gene conversion is GC biased, and this bias is closely related to the divergent evolution of the At and Dt subgenomes. The D-genome has higher GC content than the A-genome in the heterochromatin where At-to-Dt conversion is enriched, and the A-genome has higher GC content than the D-genome in the euchromatin where Dt-to-At conversion is enriched (Figure 2). The high correlation of tetraploid cotton gene conversion with the GC ratio between the two progenitor genomes (R2 = 0.506) (Figure 3A), may explain the 30% more A-to-G and T-to-C mutations in the D-genome than the A-genome since their divergence (Rong et al. 2012).
Figure 3.
Genome-wide correlations between conversion, GC% ratio of D- to A-genome, and transposon frequency (per base pair), for 1-Mbp bins using 100-kbp sliding windows. Conversion ratio is measured by log-likelihood ratio following Figure 2B. GC% ratio and transposon density are calculated for each bin. (A) Correlation between conversion and GC% ratios (R2 = 0.506). Data are best fitted with the quadratic function y = −0.02392x2 + 0.07992x + 0.9831. (B) Correlation between conversion ratios and transposon frequency (R2 = 0.684). Data are best fitted with the quadratic function y = −0.1322x2 + 0.5126x + 0.2981.
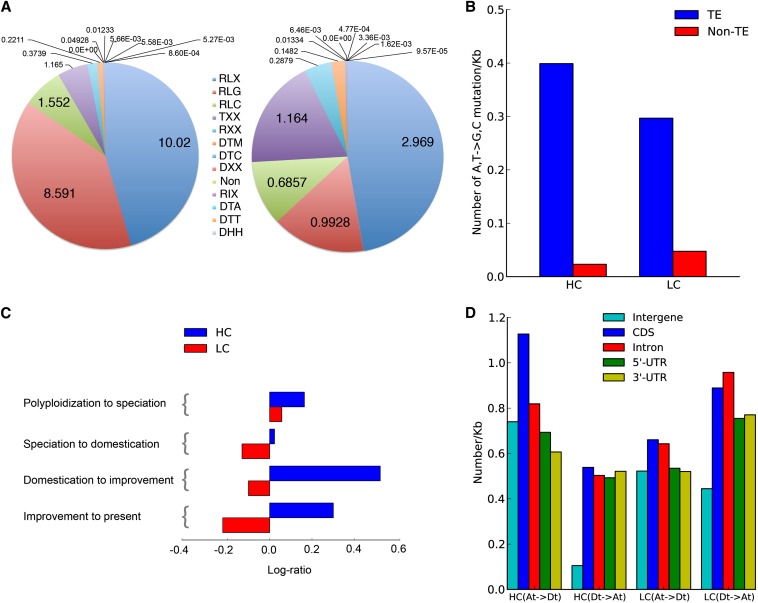
To investigate genomic features related to HeGCEs, we compared regions with highly (HC)- or little (LC)-biased conversion. Across the genome, the log-ratio of At-to-Dt vs. Dt-to-At conversions per 1-Mb bin averages 0.80 (SD = 0.46). Log ratios above 1.72 (mean + 2 SD) indicate largely At-to-Dt conversion (HC), and below −0.12 (mean − 2 SD) indicate Dt to At (LC). HC overwhelmingly locates in heterochromatin and LC in euchromatin. Heterochromatic At-to-Dt conversion is highly correlated with transposon distribution (R2 = 0.684) (Figure 3B) and enriched for potential DNA methylation sites suitable for transposon silencing (Slotkin and Martienssen 2007). Long terminal repeat (LTR), particularly gypsy-type, retrotransposons are overwhelmingly enriched in HC [>70% of all types of transposable elements, and 10.02% of total length (in base pairs) of HC vs. 2.97% of LC] (Figure 4A). A,T-to-G,C conversion, providing potential DNA methylation sites, is more frequent in transposable element (TE) than non-TE regions across the genome (Figure 4B). Further, in HC, A,T-to-G,C significantly outnumber G,C-to-A,T conversions for At to Dt (50.04 vs. 34.69%), but not Dt to At (39.95 vs. 41.26%). In LC, A,T-to-G,C conversions are similar to G,C to A,T (43.97 vs. 38.13%) for At to Dt but differ for Dt to At (47.35 vs. 32.52%) (Table 1).
Figure 4.
Intergenomic conversion of genes and transposons. (A) Proportion (% by total base pair length) of each transposon type in HC and LC. Transposon types are: RLX, LTR retrotransposon; RLG, Gypsy; TXX, transposon (either transposon or retrotransposon); RLC, Copia; RXX, retrotransposons; DTM, mutator; DTC, CACTA; DXX, DNA transposons; RIX, LINE; Non, others; DTA, hAT; DTT, Tc1-Mariner; DHH, Helitron. See Figure S4 for counts. (B) Proportion of A,T-to-G,C conversion in transposons in HC vs. LC. (C) Phylogenetic distribution of the log ratio of nonsynonymous to synonymous conversions in HC and LC. Conversions are more gene-enriched in HC than LC. See Figure S5 for raw data. (D) Density of converted alleles in each functional region.
Table 1. Number and proportion (%) of conversion types in HC and LC.
| HC (At → Dt) | HC (Dt → At) | LC (At → Dt) | LC (Dt → At) | |
|---|---|---|---|---|
| A → G | 6932 (19.63) | 1047 (16.62) | 2147 (15.90) | 2908 (18.88) |
| T → C | 6828 (19.33) | 993 (15.76) | 2249 (16.65) | 3020 (19.61) |
| G → A | 5067 (14.35) | 989 (15.70) | 2034 (15.06) | 1843 (11.97) |
| C → T | 5029 (14.24) | 935 (14.84) | 2091 (15.48) | 1821 (11.82) |
| T → G | 2011 (5.69) | 244 (3.87) | 772 (5.72) | 675 (4.38) |
| A → C | 1902 (5.39) | 233 (3.70) | 770 (5.70) | 690 (4.48) |
| G → C | 1771 (5.01) | 407 (6.46) | 822 (6.09) | 927 (6.02) |
| C → G | 1755 (4.97) | 373 (5.92) | 780 (5.78) | 935 (6.07) |
| C → A | 1091 (3.09) | 352 (5.59) | 483 (3.58) | 656 (4.26) |
| G → T | 1064 (3.01) | 323 (5.13) | 541 (4.01) | 688 (4.47) |
| A → T | 936 (2.65) | 200 (3.18) | 415 (3.07) | 603 (3.92) |
| T → A | 929 (2.63) | 203 (3.22) | 402 (2.98) | 636 (4.13) |
In heterochromatic regions where reciprocal DNA exchanges are rare, some conversions might simply be neutral or slightly deleterious relics not yet purged due to inefficient selection, as is true of retroelement insertions (Paterson et al. 2009) and other rearrangements (Bowers et al. 2005). Longer persistence of harmful mutations in HC than LC may explain an increasing ratio (in HC) of nonsynonymous to synonymous mutations across the phylogenetic tree of the four Gossypium species studied (Figure 4C). Indeed, the overall ratio of nonsynonymous to synonymous mutations is larger in heterochromatin than euchromatin (Fisher’s exact test, P = 1.334e-6), and heterochromatin conversion tracks are longer than the genome-wide average (Figure S2). All detected conversion tracks in the genome and HC region are listed in Table S1.
Gene-altering conversions are of widespread importance
Strong evidence suggests gene-altering conversions to be of widespread functional importance, perhaps serving as an alternative to reciprocal DNA exchanges to form new allele combinations in heterochromatic regions. Heterochromatic Dt-to-At conversions are approximately five times more frequent in genes than adjacent intergenic DNA (Figure 4D; Fisher’s exact test, P = 0.0001) and At-to-Dt conversions are also gene enriched (P = 0.0157). Conversions more frequently restored gene functions in cotton heterochromatin than euchromatin (Fisher’s exact test, P = 0.0008). Among 59 HC and 206 LC alleles that experienced striking mutations in the A-genome since its divergence from the F-genome [using the D-genome as outgroup (16), including premature mutations (32 HC and 80 LC), splice site alterations (22 and 89), translation initiation alterations (3 and 19), and stop codon losses (2 and 18)], At-to-Dt conversion found in Acala ‘Maxxa’ restored function to 45.8% (27) of HC vs. only 22.3% (46) of LC SNVs. Recent evidence of widespread gene conversion in the centromere cores of maize (Shi et al. 2010) and Arabidopsis (Yang et al. 2012) further supports the importance of this mechanism.
Discussion
Whole genome comparison of four naturally occurring tetraploid cottons and representatives of their progenitor genomes reveals extensive DNA exchanges accumulated during the past 1–2 MY since polyploid formation. Remarkably, the two constituent “subgenomes” of tetraploid cotton have experienced very different evolution while residing within a common nucleus, with more than twice as many conversions of At-to-Dt alleles than the reciprocal. The bias is unlikely to be caused by incomplete lineage sorting. Genetic diversity within both A- and D-genomes are estimated to be ∼10 times less than the diversity between the two genomes (Van Deynze et al. 2009). Comparison of At and Dt subgenomes of tetraploid cotton to their corresponding diploid progenitors shows small and comparable sequence divergence (Cronn et al. 2002). G. raimondii is quite narrow genetically, so there would not be much lineage sorting on the D-side (Cronn et al. 2002). The A-genome is a little more heterogeneous when considering the two A-genome species (G. arboreum and G. herbaceum) (Cronn et al. 2002). Given these considerations and keeping in mind the 5–7 MY of divergence between the A- and D-genome diploids (Senchina et al. 2003), it seems improbable that there would be segregation for many common alleles.
A key advantage of the cotton system is that polyploids have survived in the wild for 1 MY or more, effectively ruling out the hypothesis that HeGCEs are a symptom of a dysfunctional genome destined for extinction. Extensive gene conversion in the centromere cores of maize and Arabidopsis has been described using mapping populations, in which only the first several generations are observed (Shi et al. 2010; Yang et al. 2012). On the contrary, some studies show that noncrossover gene conversion is relatively rare compared to crossover-associated gene conversion in A. thaliana hybrids (Lu et al. 2012; Wijnker et al. 2013). Cytogenetic studies of recently and naturally formed polyploid species, Tragopogon miscellus and resynthesized B. napus reveal a prolonged phase of genomic instability coupled with chromosome rearrangement and translocation (Xiong et al. 2011; Chester et al. 2012). However, in each of these systems and other synthetic or recent polyploids, it is hard to assert whether the sorts of rapid responses to polyploidy that are observed (chromosome rearrangement and translocation) are the beginnings of adaptation or symptoms of pending extinction.
Frequent conversion of mostly heterochromatic At alleles by GC-rich Dt DNA may have helped to silence abundant A-genome-derived retrotransposons, perhaps stabilizing the early polyploid. It seems intuitive that D-genome alleles, from the progenitor native to the New World habitat of the polyploid, may confer many adaptations that are lacking from the Old World A-genome. However, this explanation of enrichment for At-to-Dt conversions is not consistent with the strong heterochromatic bias observed for these conversions. The approximately two times larger physical size of the A- than the D-genomes is largely due to retrotransposons, mostly in heterochromatin (Paterson et al. 2012) and some having spread to the Dt genome following polyploidy (Zhao et al. 1998). Bursts of retrotransposition following hybridization (McClintock 1983; Fontdevila 2005) can cause many DSBs (Gasior et al. 2006) that are fatal to cells if not repaired (Van Gent et al. 2001). More quickly than random mutations, GC-biased gene conversion may have provided the At genome with (Dt derived) targets for DNA methylation-based transposon silencing. Transcripts from At-derived retrotransposons may enter the RNAi pathway for RNA-dependent DNA methylation (Slotkin et al. 2009; Groszmann et al. 2011). An existing example similar to this process is the Drosophila P cytotype female that produces a repressor protein and piRNA to inhibit P-element transposition in gametes (Brennecke et al. 2008). Correlation between DNA methylation and gene conversion implies that the two are mechanistically related (Colot et al. 1996) by an as-yet-unknown process. In primates, biased gene conversion is shown as a major force for stabilizing constitutively methylated CpG islands (Cohen et al. 2011).
Meiotic recombination tends to be concentrated in small regions on chromosomes known as “recombination hotspots.” Recombination is initiated by introduction of double-strand breaks to the hotspot alleles. When a DSB occurs at a hotspot that is heterozygous with an inactive (“cold”) hotspot allele, the hotspot alleles are replaced with cold alleles by gene conversion during DNA repair. The process will cause a rapid loss of the recombination hotspot in the genome. The existence of recombination hotspots is therefore considered a “hotspot-conversion paradox” (Boulton et al. 1997). The paradox predicts small numbers of hotspots in the region with a high conversion rate. Consistent with this prediction, the high rate of conversion in the heterochromatin region identified in this study might be partially explained by the paradox.
The ability to “copy” existing alleles via HeCGEs may expedite the ability of polyploids to evolve new or more exaggerated phenotypes, by achieving allele dosages that exceed those of their progenitors. For example, Dt-to-At conversion steadily declines following polyploid formation, but accelerates during cotton domestication and improvement (Figure 1B and Figure S1). This may provide a mechanism to explain the long-standing irony that so many QTL for yield and quality of tetraploid cotton map to chromosomes derived from an ancestor (D) that lacks spinnable fibers (Jiang et al. 1998). This genetic process may complement paramutations that copy epigenetic information. In Arabidopsis hybrids, for example, methylation levels of one parental allele change to match the other (Greaves et al. 2011). Our findings show that nonreciprocal exchange of both genetic and epigenetic information may be important to the integrity of hybrid genomes.
HeCGEs may have practical value in crop improvement. The length of cotton conversion tracts averages 455 bp, ranging from 279 to 3650 bp (see Methods), somewhat longer than in mammals (Chen et al. 2007) and often spanning entire exons or occasional genes. Cotton and many other neopolyploid crops are genetically depauperate and occasional crosses with exotic or synthetic polyploids for crop improvement may be prone to bursts of transposition, DSBs, and conversion. A fascinating area for further study is whether occasional “successes” in extracting valuable alleles from exotic germplasm (e.g., Campbell et al. 2011) might have occurred by gene conversion rather than crossing over. Introgression by gene conversion might solve the widespread challenge of extracting desirable alleles from exotic germplasm while leaving behind nearby undesirable ones, i.e., with the precision of transformation but by a natural noninvasive means.
While their early evolution involved extensive intergenomic exchange, modern tetraploid cottons show strict disomy (Kimber 1961). The gradual decline of intergenomic conversion during the course of polyploid cotton evolution (Figure 1B) may have been due in part to fewer DSBs as the nascent polyploid was stabilized, perhaps by silencing of abundant A-genome-derived retrotransposons. The evolutionary success of a newly formed polyploid may require a delicate balance between genomic-stress induced novel variation (McClintock 1983) and stabilization via such quantitative factors as we suggest or qualitative factors such as the ph1 locus enforcing pairing specificity of wheat (Griffiths et al. 2006).
Data Access
The G. raimondii genome sequence is in the National Center for Biotechnology Information (NCBI) with BioProject accession PRJNA171262. Other sequences are available at the NCBI short read archive for G. herbaceum (accession F1-1, SRA061660), G. hirsutum (accession GA120R1B3, SRA068148), G. hirsutum (race yucatanese, SRA068479), and G. mustelinum (SRA068485).
Supplementary Material
Acknowledgments
The authors appreciate financial support from the US National Science Foundation (IIP-0917856, IIP-1127755 to A.H.P.); United States Department of Agriculture [Agricultural Research Service (ARS)-58-6402-7-241, 58-6402-1-644, and 58-6402-1-645 to D.G.P. and ARS 6402-21310-003-00 to B.E.S.); Bayer Crop Science; the Consortium for Plant Biotechnology Research; and Cotton, Inc. (A.H.P.).
Footnotes
Communicating editor: J. Schimenti
Literature Cited
- Adams K. L., Cronn R., Percifield R., Wendel J. F., 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100(8): 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Percifield R., Wendel J. F., 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168(4): 2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anonymous, 1997 Zonal Coordinators Annual Report of All India Coordinated Cotton Improvement Project. Central Institute for Cotton Research, Coimbatore, India.
- Aury J. M., Jaillon O., Duret L., Noel B., Jubin C., et al. , 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444(7116): 171–178. [DOI] [PubMed] [Google Scholar]
- Boulton A., Myers R. S., Redfield R. J., 1997. The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl. Acad. Sci. USA 94: 8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. E., Chapman B. A., Rong J., Paterson A. H., 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422(6930): 433–438. [DOI] [PubMed] [Google Scholar]
- Bowers J. E., Arias M. A., Asher R., Avise J. A., Ball R. T., et al. , 2005. Comparative physical mapping links conservation of microsynteny to chromosome structure and recombination in grasses. Proc. Natl. Acad. Sci. USA 102(37): 13206–13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Malone C. D., Aravin A. A., Sachidanandam R., Stark A., et al. , 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322(5906): 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B., Chee P. W., Lubbers E., Bowman D. T., Meredith W. R., et al. , 2011. Genetic improvement of the Pee Dee cotton germplasm collection following seventy years of plant breeding. Crop Sci. 51: 955–968. [Google Scholar]
- Chapman B. A., Bowers J. E., Feltus F. A., Paterson A. H., 2006. Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA 103(8): 2730–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. M., Cooper D. N., Chuzhanova N., Ferec C., Patrinos G. P., 2007. Gene conversion: mechanisms, evolution and human disease. Nat. Rev. Genet. 8(10): 762–775. [DOI] [PubMed] [Google Scholar]
- Chen Z. J., Pikaard C. S., 1997a Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11(16): 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Pikaard C. S., 1997b Transcriptional analysis of nucleolar dominance in polyploid plants: biased expression/silencing of progenitor rRNA genes is developmentally regulated in Brassica. Proc. Natl. Acad. Sci. USA 94(7): 3442–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M., Gallagher J. P., Symonds V. V., Veruska Cruz da Silva A., Mavrodiev E. V., et al. , 2012. Extensive chromosomal variation generated in a recently formed polyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A., Koh E. G. L., Chia J. M., Brenner S., Aparicio S., et al. , 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol. Biol. Evol. 21(6): 1146–1151. [DOI] [PubMed] [Google Scholar]
- Cohen N. M., Kenigsberg E., Tanay A., 2011. Primate CpG islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell 145(5): 773–786. [DOI] [PubMed] [Google Scholar]
- Colot V., Maloisel L., Rossignol J. L., 1996. Interchromosomal transfer of epigenetic states in Ascobolus: transfer of DNA methylation is mechanistically related to homologous recombination. Cell 86(6): 855–864. [DOI] [PubMed] [Google Scholar]
- Comai L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6(11): 836–846. [DOI] [PubMed] [Google Scholar]
- Comai L., Tyagi A. P., Winter K., Holmes-Davis R., Reynolds S. H., et al. , 2000. Phenotypic instability and rapid gene silencing in newly formed arabidopsis allotetraploids. Plant Cell 12(9): 1551–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R. C., Small R. L., Haselkorn T., Wendel J. F., 2002. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am. J. Bot. 89(4): 707–725. [DOI] [PubMed] [Google Scholar]
- Duret L., Galtier N., 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10: 285–311. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Eickbush D. G., 2007. Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175(2): 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J. A., Maere S., Van de Peer Y., 2009. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 106: 5737–5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A. A., et al. , 1997. Rapid elimination of low-copy DNA sequences in polyploid wheat: a possible mechanism for differentiation of homoeologous chromosomes. Genetics 147(3): 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontdevila A., 2005. Hybrid genome evolution by transposition. Cytogenet. Genome Res. 110(1–4): 49–55. [DOI] [PubMed] [Google Scholar]
- Freeling M., Thomas B. C., 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16(7): 805–814. [DOI] [PubMed] [Google Scholar]
- Gasior S. J., Wakeman T. P., Xu B., Deininger P. L., 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357(5): 1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V., 1971. Plant Speciation, Ed. 1 Columbia University Press, New York. [Google Scholar]
- Greaves I. K., Groszmann M., Ying H., Taylor J. M., Peacock W. J., et al. , 2011. Trans chromosomal methylation in Arabidopsis hybrids. Proc. Natl. Acad. Sci. USA 109(9): 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S., Sharp R., Foote T. N., Bertin I., Wanous M., et al. , 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Groszmann M., Greaves I. K., Albertyn Z. I., Scofield G. N., W. J. Peacock et al, 2011. Changes in 24-nt siRNA levels in Arabidopsis hybrids suggest an epigenetic contribution to hybrid vigor. Proc. Natl. Acad. Sci. USA 108(6): 2617–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z. L., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., et al. , 2003. Role of duplicate genes in genetic robustness against null mutations. Nature 421(6918): 63–66. [DOI] [PubMed] [Google Scholar]
- Guo H., Lee T. H., Wang X., Paterson A. H., 2013. Function relaxation followed by diversifying selection after whole-genome duplication in flowering plants. Plant Physiol. 162: 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. L., Zhang J. Z., 2005. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169(2): 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T., Lo J., van Gent D. C., Engelward B. P., 2007. DNA double-strand break repair: from mechanistic understanding to cancer treatment. DNA Repair (Amst.) 6(7): 923–935. [DOI] [PubMed] [Google Scholar]
- Jiang C., Wright R. J., K. M. EI-Zik, and A. H. Paterson, 1998. Polyploid formation created unique avenues for response to selection in Gossypium (cotton). Proc. Natl. Acad. Sci. USA 95(8): 4419–4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345): 97–100. [DOI] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A., 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160(4): 1651–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K., Feldman M., Levy A. A., 2003. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 33(1): 102–106. [DOI] [PubMed] [Google Scholar]
- Kimber G., 1961. Basis of the Diploid-like meiotic behaviour of polyploid cotton. Nature 191: 98–99. [Google Scholar]
- Lee H. S., Chen Z. J., 2001. Protein-coding genes are epigenetically regulated in Arabidopsis polyploids. Proc. Natl. Acad. Sci. USA 98(12): 6753–6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21): 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14): 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16): 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love A., Love D., 1949. The geobotanical significance of polyploidy. Portugaliae Acta (Suppl): 273–352. [Google Scholar]
- Lu P., Han X., Qi J., Yang J., Wijeratne Asela J., et al. , 2012. Analysis of Arabidopsis genome-wide variations before and after meiosis and meiotic recombination by resequencing Landsberg erecta and all four products of a single meiosis. Genome Res. 22: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2006. The origins of eukaryotic gene structure. Mol. Biol. Evol. 23(2): 450–468. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494): 1151–1155. [DOI] [PubMed] [Google Scholar]
- Lynch M., Force A., 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154(1): 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., O’Hely M., Walsh B., Force A., 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159(4): 1789–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock, B., 1983 The significance of responses of the genome to challenge. Essential Readings in Evolutionary Biology. Johns Hopkins University Press, Baltimore.
- Muntzing A., 1936. The evolutionary significance of autopolyploidy. Hereditas 21: 363–378. [Google Scholar]
- O’Neill R. J., O’Neill M. J., Graves J. A., 2002. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393(6680): 68–72. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1970 Evolution by Gene Duplication. Springer-Verlag, Berlin. [Google Scholar]
- Ozkan H., Levy A. A., Feldman M., 2001. Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13(8): 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H., Levy A. A., Feldman M., 2002. Rapid differentiation of homeologous chromosomes in newly-formed allopolyploid wheat. Isr. J. Plant Sci. 50: S65–S76. [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Freeling M., Tang H., Wang X., 2010. Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 61: 349–372. [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., et al. , 2012. Repeated polyploidization of Gossypium and the evolution of spinnable cotton fibers. Nature 492: 423–427. [DOI] [PubMed] [Google Scholar]
- Pires J. C., Zhao J. W., Schranz M. E., Leon E. J., Quijada P. A., et al. , 2004. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. Lond. 82(4): 675–688. [Google Scholar]
- Rana D., Boogaart T., O’Neill C. M., Hynes L., Bent E., et al. , 2004. Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40(5): 725–733. [DOI] [PubMed] [Google Scholar]
- Rong J., Abbey C., Bowers J. E., Brubaker C. L., Chang C., et al. , 2004. A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166(1): 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J., Wang X., Schulze S. R., Compton R. O., Williams-Coplin T. D., et al. , 2012. Types, levels and patterns of low-copy DNA sequence divergence, and phylogenetic implications, for Gossypium genome types. Heredity (Edinb) 108(5): 500–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Byrne K. P., Gordon J. L., Wong S., Wolfe K. H., 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440(7082): 341–345. [DOI] [PubMed] [Google Scholar]
- Senchina D. S., Alvarez I., Cronn R. C., Liu B., Rong J. K., et al. , 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20(4): 633–643. [DOI] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A. A., 2001. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13(8): 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D., Glusman G., Pilpel Y., Khen M., Gruetzner F., et al. , 1999. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics 61(1): 24–36. [DOI] [PubMed] [Google Scholar]
- Shi J., Wolf S. E., Burke J. M., Presting G. G., Ross-lbarra J., et al. , 2010. Widespread gene conversion in centromere cores. PLoS Biol. 8(3): e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R. K., Martienssen R., 2007. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8: 272–285. [DOI] [PubMed] [Google Scholar]
- Slotkin R. K., Vaughn M., Borges F., Tanurdzic M., Becker J. D., et al. , 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K. M., Lu P., Tang K. L., Osborn T. C., 1995. Rapid genome change in Synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl. Acad. Sci. USA 92(17): 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G. L., 1950. Variation and Evolution in Plants, Columbia University Press, New York. [Google Scholar]
- Stephens S., 1951. Possible significance of duplications in evolution. Adv. Genet. 4: 247–265. [DOI] [PubMed] [Google Scholar]
- Van Deynze A., Stoffel K., Lee M., Wilkins T. A., Kozik A., et al. , 2009. Sampling nucleotide diversity in cotton. BMC Plant Biol. 9: 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gent D. C., Hoeijmakers J. H. J., Kanaar R., 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2(3): 196–206. [DOI] [PubMed] [Google Scholar]
- Wang X., Tang H., Bowers J. E., Feltus F. A., Paterson A. H., 2007. Extensive concerted evolution of rice paralogs and the road to regaining independence. Genetics 177(3): 1753–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Tang H., Bowers J. E., Paterson A. H., 2009. Comparative inference of illegitimate recombination between rice and sorghum duplicated genes produced by polyploidization. Genome Res. 19(6): 1026–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., 1989. New World tetraploid cottons contain old-world cytoplasm. Proc. Natl. Acad. Sci. USA 86(11): 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnker E., James G. V., Ding J., Becker F., Klasen J. R., et al. , 2013. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2: e01426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z., Gaeta R. T., Pires J. C., 2011. Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Yuan Y., Wang L., Li J., Wang W., et al. , 2012. Great majority of recombination events in Arabidopsis are gene conversion events. Proc. Natl. Acad. Sci. USA 109(51): 20992–20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. P., Si Y., Hanson R. E., Crane C. F., Price H. J., et al. , 1998. Dispersed repetitive DNA has spread to new genomes since polyploid formation in cotton. Genome Res. 8(5): 479–492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.