Abstract
Evading apoptosis is a hallmark of B-cell chronic lymphocytic leukemia (CLL) cells and an obstacle to current chemotherapeutic approaches. Inhibiting histone deacetylase (HDAC) has emerged as a promising strategy to induce cell death in malignant cells. We have previously reported that the HDAC inhibitor MGCD0103 induces CLL cell death by activating the intrinsic pathway of apoptosis. Here, we show that MGCD0103 decreases the autophagic flux in primary CLL cells. Activation of the PI3K/AKT/mTOR pathway, together with the activation of caspases, and to a minor extent CAPN1, resulting in cleavage of autophagy components, were involved in MGCD0103-mediated inhibition of autophagy. In addition, MGCD0103 directly modulated the expression of critical autophagy genes at the transcriptional level that may contribute to autophagy impairment. Besides, we demonstrate that autophagy is a pro-survival mechanism in CLL whose disruption potentiates cell death induced by anticancer molecules including HDAC and cyclin-dependent kinase inhibitors. In particular, our data highlight the therapeutic potential of MGCD0103 as not only an inducer of apoptosis but also an autophagy suppressor in both combination regimens with molecules like flavopiridol, known to induce protective autophagy in CLL cells, or as an alternative to circumvent undesired immunomodulatory effects seen in the clinic with conventional autophagy inhibitors.
Keywords: autophagy, B-cell chronic lymphocytic leukemia, histone deacetylase inhibitor, MGCD0103, caspases, calpains
Introduction
B-cell chronic lymphocytic leukemia (CLL) results from a monoclonal expansion of CD5+/CD19+/CD23+ B lymphocytes that accumulate in lymphatic organs.1 Circulating CLL cells are mainly arrested in G0/G1 and fail to undergo apoptosis. This pool of malignant cells is fed by a proportion of CLL cells, ranging from 0.1 to 1% per day, that proliferate in the bone marrow and lymph nodes.2 Despite considerable improvements in CLL chemoimmunotherapy, conventional drugs fail to cure CLL.
Histone deacetylase (HDAC) inhibitors have the ability to reverse epigenetic alterations associated with cancer initiation/development and chemoresistance, and to induce tumor cell death.3, 4 Inhibitors of zinc-dependent HDACs (classes I, II, IV) have emerged as promising cytotoxic agents and are currently being evaluated in several clinical trials for the treatment of cancer.3
Besides their pro-apoptotic effect, HDAC inhibitors can also induce autophagy,5, 6, 7 a ‘self-eating' process whereby proteins and organelles are sequestered in autophagosomes, and delivered to lysosomes for digestion.8 Whereas autophagy operates as a housekeeping process to promote cell survival, autophagy defects have been found in human cancers, suggesting its tumor-suppressive role.9 Nevertheless, the role of autophagy in cancer remains controversial,6, 10, 11 and little is known about autophagy in CLL.12, 13
We have previously shown that MGCD0103, a selective inhibitor of HDACs 1, 2, 3 and 11, triggers in primary CLL cells a death amplification loop involving caspases and calpains.14
Here, we show that MGCD0103 inhibits autophagy in primary CLL cells while operating at three levels: activation of the phosphatidylinositide 3-kinase/AKT/mammalian target of rapamycin (PI3K/AKT/mTOR) pathway, caspase- and calpain-1-mediated cleavage of autophagy components and downregulation of autophagy gene transcription. Finally, we demonstrate that basal autophagy functions in CLL as a pro-survival mechanism whose disruption potentiates cell death.
Materials and methods
Patients, cell separation and culture conditions
Subjects were CLL patients (n=34) and healthy donors (n=3) seen at the ‘Centre Hospitalier de Luxembourg'. The clinical features of patients are summarized in Supplementary Table 1. The experiments were approved by the ‘Comité National d'Ethique de Recherche' and all subjects signed an informed consent according to the Declaration of Helsinki.
Peripheral blood mononuclear cells (PBMCs) were isolated using lymphocyte separation medium (MP Biomedicals, Eindhoven, The Netherlands). PBMCs and Mec-1 cells were cultured in Iscove's modified Dulbecco's medium (Lonza, Verviers, Belgium), MCF-7 cells, JVM-2 and JVM-3 cells in RPMI-1640 medium (Lonza) and HeLa cells in Dulbecco's modified Eagle's medium, all containing 10% fetal bovine serum. The cells were incubated at 37 °C in an atmosphere of 5% CO2.
Reagents and antibodies
MGCD0103 was provided by MethylGene Inc. (Quebec, Canada). Valproic acid (VPA; Depakine IV, Sanofi Aventis) and bortezomib (Velcade IV, Millennium Pharmaceuticals, Inc.) were purchased from the hospital's pharmacy. Q-VD-OPh and Z-VEID-fmk were from R&D Systems (Abingdon, UK). PD151746 and calpeptin were purchased from Merck Chemicals Ltd (Overijse, Belgium). Chloroquine, 3-methyladenine (3-MA), bafilomycin A1, flavopiridol, TRAIL, Actinomycin D and anti-β-actin (ACTB) antibody were from Sigma-Aldrich (Diegem, Belgium). Anti-sequestosome-1 (SQSTM1/p62) antibody was from BD Biosciences (Erembodegem, Belgium). All other antibodies were from Cell Signaling Technology (Leiden, The Netherlands).
Cytotoxicity assay
PBMCs were seeded in 96-well cell culture plates and assays were performed in triplicate. Cell viability was measured after 6 h (when flavopiridol was used) or 48 h of incubation using the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Eke, Belgium). Combination experiments done with an autophagy inhibitor and an HDAC inhibitor were carried out at nonconstant ratios, keeping constant the concentration of the former while varying the doses of the latter.
MGCD0103 and VPA were used at concentrations that encompass the clinically relevant concentrations.14, 15, 16 Chloroquine concentrations were fixed to 5 and 10 μmol/l, thus matching plasma concentrations.17 The 3-MA is not administered in clinic and was used at concentrations that resulted in 70 and 55% of cell viability after a 48- h treatment (0.3 and 0.6 mmol/l, respectively).
When flavopiridol was used, human serum was added to the culture medium instead of bovine serum (there is a significant flavopiridol protein binding in human serum, contrarily to bovine serum), and a short treatment time was chosen (6 h) in order to better reproduce the clinical conditions.18 A combination of flavopiridol and MGCD0103 was done with a constant ratio design.
Drug combination analysis was performed following the median-effect method19 using CompuSyn software (Combosyn, Inc., Paramus, NJ, USA). Combination index (CI)<1, CI=1 and CI>1 indicate synergism, additive effect, and antagonism, respectively. For the 6-h exposure experiments, CI values could not be determined as the LC50 (lethal concentration 50%) of the drugs was not reached. Therefore, the percent of cell viability was used to determine whether drug combination enhanced or decreased cell death, as recommended by Chou.19
Immunoblotting analysis
Cells were lysed in RIPA buffer (Merck Chemicals Ltd) supplemented with protease (Roche, Vilvoorde, Belgium) and phosphatase inhibitor (Sigma-Aldrich) cocktails. SDS–polyacrylamide gel electrophoresis and immunoblotting were done using standard procedures. ImageQuant TL 7.0 software (GE Healthcare, Zeist, The Netherlands) was used for protein quantification.
Analysis of autophagy by flow cytometry
Autophagy was monitored with the Cyto-ID Autophagy Detection Kit (Enzo Life Sciences, Antwerpen, Belgium) using a 488-nm excitable fluorescent probe that selectively stains autophagic compartments. Data were acquired on a BD FACSCanto flow cytometer (BD Biosciences) and were analyzed using the BD FACSDiva software (BD Biosciences).
Analysis of cell viability by flow cytometry
PBMC viability was analyzed according to changes in forward and side scatters, or using Annexin V-APC (BD Biosciences) and propidium iodide (Sigma-Aldrich) staining, as previously described.14
Autophagy PCR array assay
The Human Autophagy RT2 Profiler PCR Array (SABiosciences, Venlo, The Netherlands) was used for autophagy gene expression profiling in primary CLL cells. Detection was performed using a ViiA 7 Real-Time PCR System (Applied Biosystems, Halle, Belgium). The data were analyzed by the ΔΔCt method, using The PCR Array Data Analysis Web-based software (Qiagen, Venlo, The Netherlands).
siRNA transfections
The 8 × 106 primary CLL cells were transfected with the Amaxa nucleofection system (Lonza) using either Cell Line Nucleofector Kit V and program U016 or Human B Cell Nucleofector Kit and program U015. Small interference RNAs (siRNAs) were from Qiagen. For single and double transfections, 350 and 250 pmol of each siRNA was used, respectively.
HeLa cells were transfected with 100 pmol of siRNA using Lipofectamine 2000 (Life Technologies, Gent, Belgium).
Real-time reverse transcription-PCR
Total RNA was extracted using miRNeasy kit (Qiagen), and real-time PCR was performed using Reverse Transcriptase Core kit (Eurogentec, Seraing, Belgium) and Power SYBR Green PCR Master Mix (Applied Biosystems). The primers were purchased from Eurogentec. Detection was performed using a ViiA 7 Real-Time PCR System (Applied Biosystems).
Statistical analysis
Statistical analysis was carried out using a two-sided t-test, with a P-value of <0.05 considered statistically significant.
Results
MGCD0103 decreases the levels of autophagy-related proteins in primary CLL cells
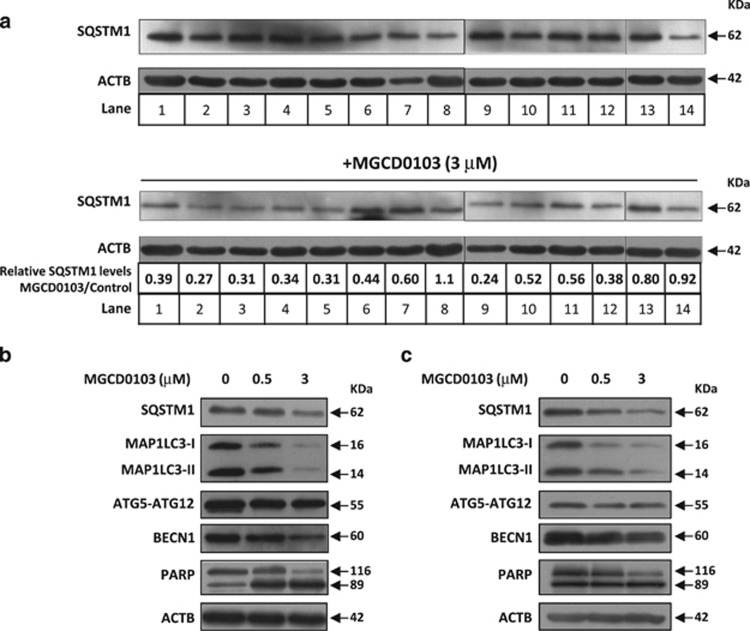
To investigate whether MGCD0103 modulates autophagy in CLL cells, we first analyzed its effect on the levels of SQSTM1 protein, a known autophagy substrate.20 CLL cells from 14 patients were treated during 24 h with MGCD0103 (3 μmol/l) according to previously described considerations.14
MGCD0103 considerably reduced SQSTM1 levels in 11 out of 14 CLL samples (Figure 1a). The differences observed were likely because of interindividual variability in patients, as CLL samples were randomly chosen from patients with diverse Rai stages and prognostic factors (Supplementary Table 1).
Figure 1.
MGCD0103 decreases the levels of autophagy-related proteins in primary CLL cells. Immunoblotting analysis of protein extracts from PBMCs incubated in the absence or presence of MGCD0103 (0.5 or 3 μmol/l, as indicated) for 24 h. Autophagy-related proteins are indicated, as well as ACTB that was used as a loading control. (a) Shown are representative blots from the samples of 14 patients. The expression levels of SQSTM1 in MGCD0103-treated cells relative to the control and normalized to ACTB levels are indicated. (b, c) Shown are two representative blots from eight independent experiments.
We also assessed the expression levels of the phosphatidylethanolamine-conjugated form of microtubule-associated protein 1 light chain (MAP1LC3-II), required for vesicle elongation and autophagosome formation, the autophagy-related regulator ATG5 (as ATG5-ATG12 conjugate) and beclin-1 (BECN1), required for early steps of autophagy. Similar to SQSTM1, MGCD0103 decreased the levels of above-mentioned proteins (Figures 1b and c). This effect was already apparent at the clinically relevant dose of 0.5 μmol/l and mirrored apoptosis levels (see poly (ADP-ribose) polymerase cleavage).
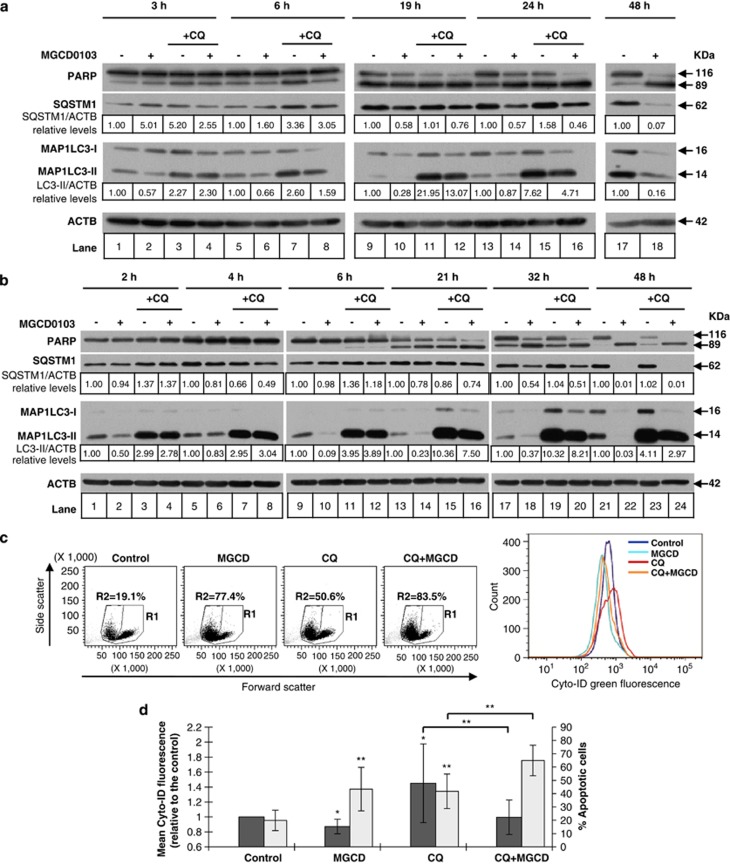
MGCD0103 decreases the autophagic flux in primary CLL cells
Reduced levels of SQSTM1 and MAP1LC3-II may indicate either an induction or a decrease in the autophagic flux.
To assess the autophagic flux, time course analysis of MAP1LC3-II and SQSTM1 expression was performed (Figures 2a and b). When cells were exposed to MGCD0103 (3 μmol/l), the decrease in MAP1LC3-II levels could be observed at early time points (after 2–3 h). This decrease did not result from an increased MAP1LC3 turnover as no accumulation of MAP1LC3-II was detected in the presence of chloroquine that prevents autophagosome–lysosome fusion. At the time points analyzed, MAP1LC3-II levels were lower in MGCD0103-treated cells than in untreated cells, even in the presence of chloroquine. Interestingly, SQSTM1 levels significantly decreased only after 19–48 h, several hours after the drop in MAP1LC3-II, and they were not restored by chloroquine, suggesting that they were reduced by an autophagy-independent mechanism. Similar results were obtained with the autophagosome–lysosome fusion inhibitor, Bafilomycin A1 (Supplementary Figure 1a). These data suggest that MGCD0103 inhibits the autophagic flux in primary CLL cells, an effect that was also observed at the clinically relevant dose of 0.5 μmol/l (Supplementary Figure 1b).
Figure 2.
MGCD0103 decreases the autophagic flux in primary CLL cells. (a, b) The effect of MGCD0103 (3 μmol/l) on autophagy was assessed by immunoblotting in the presence of chloroquine in time course experiments. ACTB was used as a loading control. Two representative blots (a and b) from four independent experiments are shown. The expression levels of proteins in treated cells relative to the corresponding untreated control and normalized to ACTB levels are indicated. (c, d) Autophagy was assessed by flow cytometry using the Cyto-ID green autophagy dye. For the left panel in (c), single-cell lymphocyte population was gated in R1 (live and apoptotic cells), whereas apoptotic cells were gated in R2 on the basis of cell shrinkage (decreased in forward scatter (FSC)) and increase in granularity (increase in side scatter (SSC)). Shown on the right panel of (c) are the distributions of the dye green fluorescence (channel FL1) within each R1 population. Dot plots and Cyto-ID green fluorescence intensity from a representative experiment are shown in (c). (d) Results are represented by histograms as mean Cyto-ID green fluorescence of treated cells relative to control cells (dark bars). The percentage of apoptotic cells represents the percentage of cells in R2 (bright bars). The mean values±s.d. (n=8) are shown. Values are compared with the corresponding control value. *P<0.05 and **P<0.01.
Further confirmation of this finding was obtained by flow cytometry using Cyto-ID fluorescent marker of autophagic compartments (Figures 2c and d, and Supplementary Figures 2a and b). As expected, chloroquine generated an increase in fluorescence intensity because of autophagosome accumulation. Either alone or in the presence of chloroquine, MGCD0103 decreased the fluorescence intensity of the dye, suggesting that it reduced autophagosome formation in primary CLL cells. Thus, MGCD0103 decreased the amount of autophagic vacuoles while increasing the percentage of apoptotic cells.
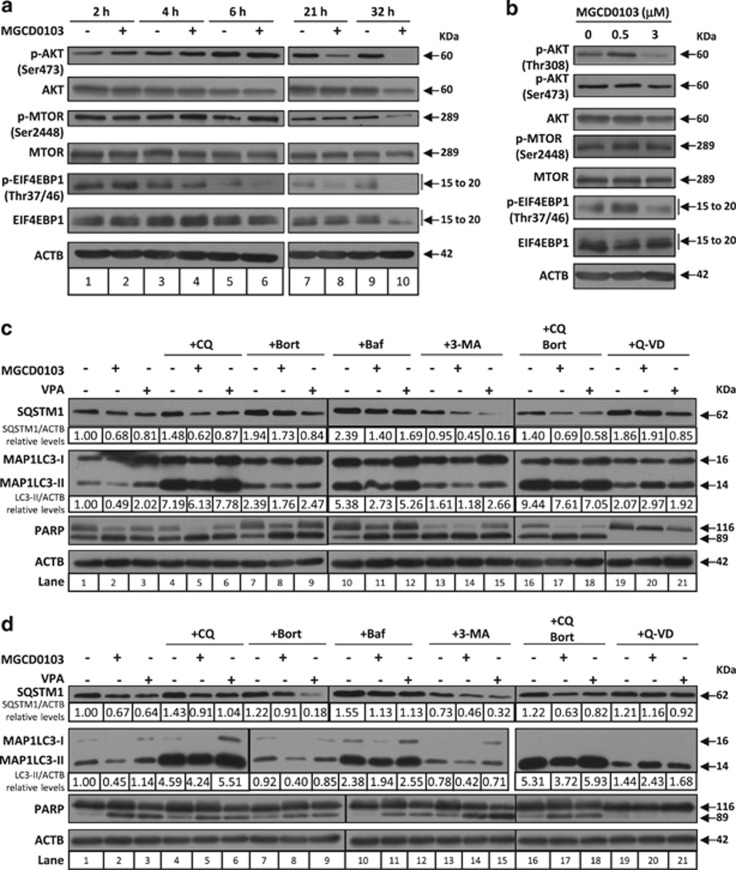
MGCD0103 inhibits autophagy through activation of the PI3K/AKT/mTOR pathway and caspases
To analyze the molecular mechanisms behind MGCD0103-mediated inhibition of autophagy in primary CLL cells, we first investigated the involvement of the PI3K/AKT/mTOR pathway, a major signaling cascade that regulates autophagy.21 To this end, we examined the phosphorylation of AKT and mTOR, as well as that of the mTORC1 substrate, the eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1). Accumulation of p-AKT (Ser473), p-mTOR and p-EIF4EBP1 (Thr37/46) was observed at early time points (between 2 and 6 h depending on the protein) following treatment with 3 μmol/l MGCD0103 (Figure 3a, see also Supplementary Table 2 for blot quantification), thus consistent with MGCD0103 mediating the early activation of the PI3K/AKT/mTOR pathway. Loss of these phosphorylated forms, already observed after 6 h for p-AKT and 4 h for p-EIF4EBP1, is observed in all cases at later time points (21 and 32 h).
Figure 3.
MGCD0103 inhibits autophagy in primary CLL cells through activation of the PI3K/AKT/mTOR pathway and caspases. (a, b) The effect of MGCD0103 on the PI3K/AKT/mTOR pathway was assessed in primary CLL cells by immunoblotting through the analysis of the phosphorylation status of key regulators. (a) Time course experiments were performed with 3 μmol/l of MGCD0103 and (b) the effect of the 24-h treatment was analyzed using two concentrations of MGCD0103 (0.5 and 3 μmol/l, as indicated). Relative protein quantification is available in Supplementary Table 2. Blots from three independent experiments are shown. (c, d) MGCD0103 decreases the autophagic flux in primary CLL cells in a caspase-dependent manner. Cells were incubated for 24 h alone or in the presence of MGCD0103 (3 μmol/l) or VPA (4 mmol/l), with or without chloroquine (CQ; 20 μmol/l), bafilomycin A1 (Baf; 20 nmol/l), 3-MA (1 mmol/l), bortezomib (Bort; 2 nmol/l) or Q-VD-OPh (Q-VD; 10 μmol/l). Caspase and proteasome inhibitors were added to PBMCs 1 h before and autophagy inhibitors 1 h after MGCD0103 or VPA. Two representative blots (c and d) from five independent experiments are shown. The levels of indicated proteins were analyzed by immunoblotting, using ACTB as a loading control. The expression levels of proteins in treated cells relative to the untreated control and normalized to ACTB levels are indicated.
To gain more insight into the later-stage impact of MGCD0103 on the PI3K/AKT/mTOR pathway, we next investigated its activation after a 24-h treatment using concentrations of 0.5 and 3 μmol/l. Whereas increased phosphorylation of AKT (Thr308), mTOR, and EIF4EBP1 was still observed in cells treated with 0.5 μmol/l, global dephosphorylation occurred with 3 μmol/l of MGCD0103 (Figure 3b and Supplementary Table 2).
Altogether, these results demonstrate that MGCD0103 can induce, in a time and dose-dependent manner, the activation of the PI3K/AKT/mTOR cascade. The early activation of this pathway may explain the rapid MGCD0103-mediated inhibition of autophagy in primary CLL cells (Figures 2a and b). However, prolonged incubation with MGCD0103 (3 μmol/l) amplified autophagy inhibition (Figures 2a and b), whereas the PI3K/AKT/mTOR pathway was by then almost completely inhibited—an event that normally activates autophagy—suggesting that another mechanism impairs autophagy at late time points. We, therefore, analyzed the effects of autophagy blockade, as well as proteasome and caspase inhibition on the levels of SQSTM1 and MAP1LC3-II in primary CLL cells after a 24-h incubation with MGCD0103 (3 μmol/l; Figures 3c and d). As previously shown (Figure 2), MGCD0103 reduced the levels of both proteins even in the presence of chloroquine (compare lanes 1 and 2, and lanes 4 and 5). Besides, the loss of SQSTM1 and MAP1LC3-II bands was not prevented by the proteasome inhibitor, bortezomib. Failure of chloroquine to inhibit MGCD0103-induced decrease in SQSTM1 and MAP1LC3-II levels was also observed with two other autophagy inhibitors, bafilomycin A1 (compare lanes 10 and 11) and 3-MA (autophagy initiation inhibitor; lanes 13 and 14), ruling out a molecule-dependent effect of chloroquine. Similarly, simultaneous inhibition of autophagy and the proteasome did not prevent the loss of SQSTM1 and MAP1LC3-II in response to MGCD0103. In contrast, the pan-caspase inhibitor Q-VD-OPh completely prevented the decrease in protein levels elicited by MGCD0103 (compare lanes 19 and 20). Altogether, these results suggest that MGCD0103-mediated autophagy inhibition results from the activation of the PI3K/AKT/mTOR pathway followed by caspase-mediated degradation of autophagy-related proteins. This effect is not a common property of HDAC inhibitors as VPA, an HDAC I and IIa inhibitor, did not decrease the autophagic flux in primary CLL cells, as assessed by the levels of MAP1LC3-II in the presence of chloroquine and bafilomycin A1 (Figures 3c and d).
Contrarily to other autophagy-related proteins, MAP1LC3 has been proposed to be neither a caspase nor a calpain-1 substrate in vitro.22 Our data show that MAP1LC3-I and MAP1LC3-II levels decrease in MGCD0103-treated primary CLL cells (Figures 1b and c). One mechanism that may contribute to the drop in MAP1LC3-I is the observed reduction in ATG4B amounts, the enzyme that generates MAP1LC3-I (Supplementary Figures 3a and b). The decrease in ATG4B levels was incompletely inhibited by Q-VD-OPh, suggesting that it is partly caspase dependent. The reduced levels of MAP1LC3-II were likely because of the drop in ATG5 levels observed in the presence of MGCD0103 (Figures 1b and 5b).
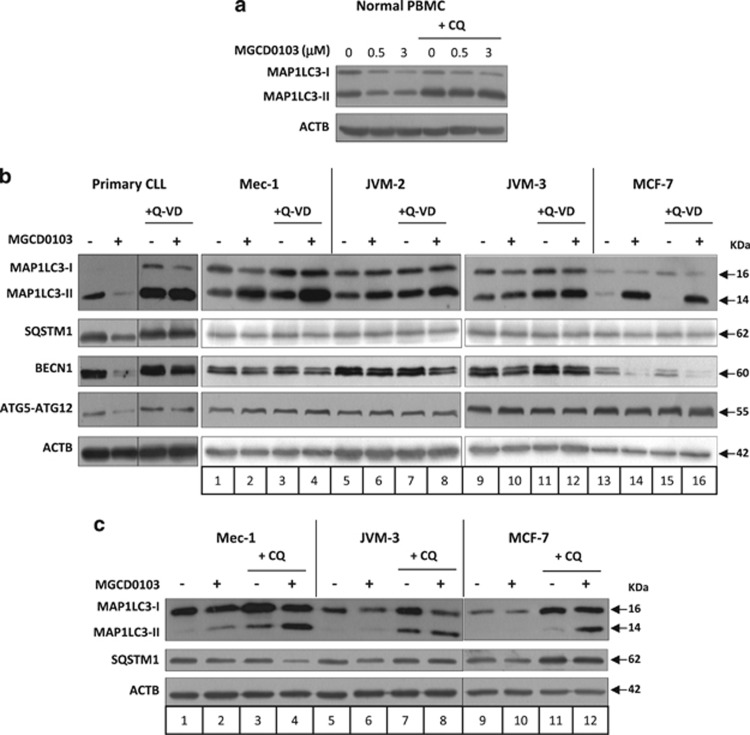
Figure 5.
MGCD0103-induced inhibition of autophagy is specific to primary CLL cells. (a) PBMCs from healthy donors (n=3) were incubated with or without MGCD0103 (0.5 or 3 μmol/l) and/or chloroquine (CQ; 20 μmol/l) for 24 h. (b, c) The effect of MGCD0103 on autophagy was assessed in cell lines. Cells were incubated with or without MGCD0103 (3 μmol/l) and/or Q-VD-OPh (10 μmol/l for CLL patient PBMCs and 20 μmol/l for cell lines) for 24 h (PBMC, JVM-2 and JVM-3) or 48 h (Mec-1 and MCF-7), and/or chloroquine (CQ; 40 μmol/l) for 24 h. The levels of indicated proteins were analyzed by immunoblotting using ACTB as a loading control. Representative blots from at least three independent experiments are shown in each panel.
MGCD0103 modulates the expression of key autophagy regulators at the transcriptional level
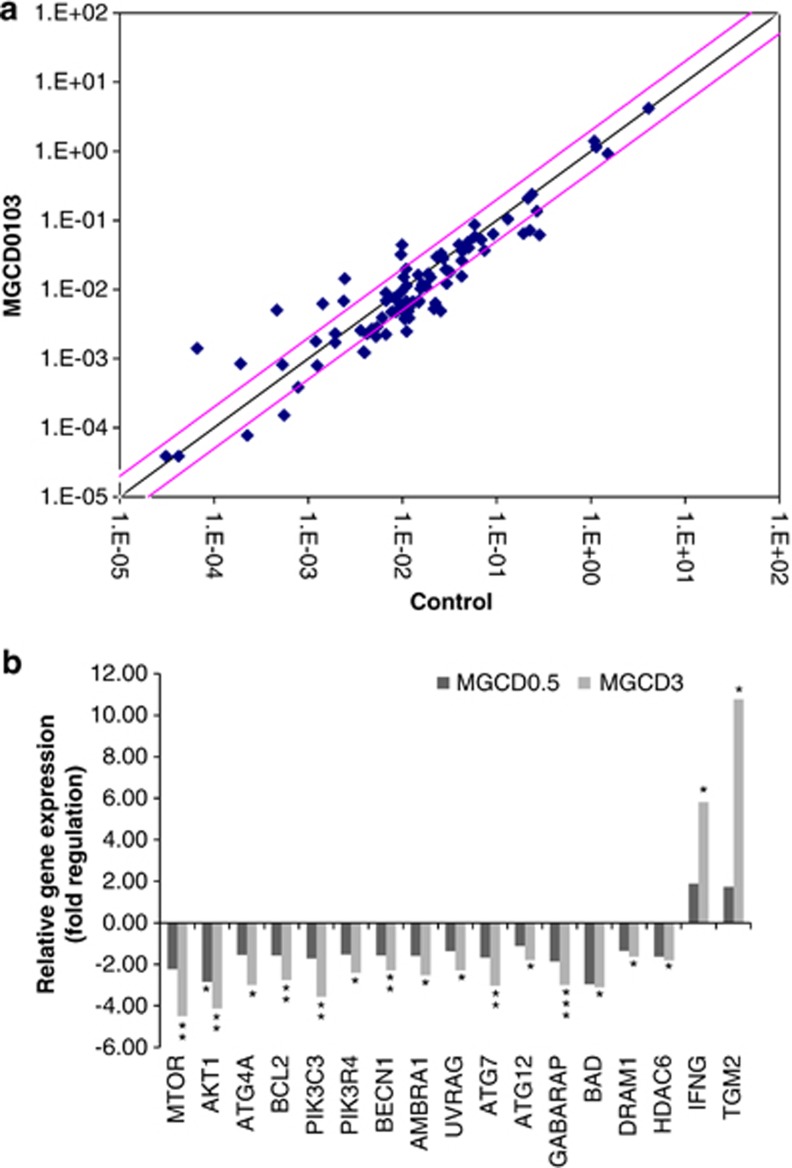
To investigate whether modulation of autophagy-related genes could account for the effects of MGCD0103 on autophagy in primary CLL cells, a screening for gene expression was performed using an autophagy PCR array. A 24-h treatment with MGCD0103 (3 μmol/l) reduced the expression of multiple genes involved in autophagy (Figure 4a). In particular, PIK3C3, PIK3R4, BECN1, AMBRA1 and UVRAG, key components of the BECN1–VPS34 complex,21 were significantly downregulated, as were ATG7, ATG12, GABARAP, BAD, DRAM1 and HDAC6, the positive regulators of autophagy (Figure 4b). Expression levels of mTOR and AKT1 also decreased. MGCD0103 increased the expression of IFNG (interferon-γ), an activator of ‘defense' autophagy,23 and TGM2 (transglutaminase type 2), previously associated with either autophagy inhibition24 or completion.25 Similar trends were observed with MGCD0103 at 0.5 μmol/l (Figure 4b). The decreased mRNA levels did not result from a global RNA degradation, as demonstrated by experiments using the RNA synthesis inhibitor Actinomycin D (Supplementary Figure 4). Altogether, these data demonstrate that MGCD0103 may also operate at the transcriptional level to inhibit autophagy in primary CLL cells.
Figure 4.
MGCD0103 modulates autophagy gene expression at the transcriptional level in primary CLL cells. Screening for autophagy gene expression in PBMCs incubated with or without MGCD0103 for 24 h was performed using an autophagy PCR array. Results represent the mean values of four independent experiments. (a) The scatter plot graphs the expression level (2-ΔCt) of each gene in the control sample versus the MGCD0103 (3 μmol/l) sample. The black line indicates fold changes (2-ΔΔCt) of 1. The pink lines indicate a fold change of 2 in gene expression. (b) Histograms represent the fold up- or downregulation of relative gene expression in primary CLL cells incubated with MGCD0103 (0.5 and 3 μmol/l) for 24 h. When fold change values are >1, the fold regulation is equal to the fold change. When fold change values are <1, the fold regulation is the negative inverse of the fold change. *P<0.05, **P<0.01 and ***P<0.001.
MGCD0103-induced inhibition of autophagy is specific to primary CLL cells
We next tested the selectivity of MGCD0103 toward primary CLL cells by comparing its ability to inhibit autophagy in PBMCs from healthy donors, and in the CLL-derived cell lines Mec-1 and JVM-3, the mantle cell lymphoma cell line JVM-2 and the breast cancer cell line MCF-7.
In normal PBMCs, a reduction in the levels of MAP1LC3-II upon MGCD0103 treatment was observed that was alleviated by the presence of chloroquine (Figure 5a), suggesting that MGCD0103 did not modulate the autophagic flux.
In the cell lines, and in contrast to primary CLL cells, MGCD0103 increased the levels of MAP1LC3-II and failed to decrease ATG5-ATG12 levels (Figure 5b). A slight decrease in SQSTM1 levels was observed in Mec-1 and MCF-7 cells, and MGCD0103 treatment decreased BECN1 expression in all cell lines. However, the impact of MGCD0103 in these cells persisted in the presence of Q-VD-OPh, indicating that it was caspase independent (Figure 5b). Moreover, when treated with MGCD0103+chloroquine, the cell lines showed increased levels of MAP1LC3-II and restoration of SQSTM1 amounts (in JVM-3 and MCF-7; Figure 5c). These results demonstrate that, contrarily to primary CLL cells, MGCD0103 induces autophagy in cell lines.
Activation of CAPN1 is involved in the decreased autophagic flux in MGCD0103-treated cells from some CLL samples
We previously demonstrated that MGCD0103 induces the caspase-mediated activation of calpains in primary CLL cells.14 Calpains can cleave ATG5, thus provoking a switch from autophagy to apoptotic cell death.26, 27 To understand the role of calpains in MGCD0103-induced inhibition of autophagy, we used two inhibitors of the major ubiquitous calpains, calpain-1 (CAPN1) and calpain-2 (CAPN2): PD151746 (PD), which specifically inhibits CAPN1, and calpeptin that inhibits both CAPN1 and CAPN2. The effects of these inhibitors on MGCD0103-mediated decrease in autophagy proteins levels varied from one CLL sample to another (Supplementary Figure 5). However, in at least some cases, CAPN1 was involved in the cleavage of autophagy-related proteins in MGCD0103-treated CLL cells (Supplementary Figure 5b). Variations in the levels of CAPN1 and CAPN2 mRNA, as assessed by real-time reverse transcription-PCR (data not shown), could not explain the differences observed between patient samples.
We then investigated in more detail the protease-mediated cleavage of SQSTM1, as its degradation has been widely used as a hallmark of autophagy activation. In MGCD0103-treated CLL cells, the pattern of SQSTM1 cleavage products included bands of ca. 30 and 37 KDa (Supplementary Figures 6a and b). Presence of the 30-KDa band was insensitive to PD151746 (Supplementary Figure 6a, lane 3 and Supplementary Figure 6b, lanes 3 and 4), whereas it was considerably blocked by caspase-6 (CASP6) inhibitor Z-VEID-fmk (Supplementary Figure 6b, lanes 5 and 6), consistent with the previously reported role of CASP6 in SQSTM1 cleavage.22 Besides, Q-VD-OPh significantly reduced the levels of the 30- and the 37-KDa fragments (Supplementary Figures 6a and b), indicating that SQSTM1 cleavage in CLL cells involves not only CAPN1 and CASP6, but also other caspases. Evidence that the observed bands are specific SQSTM1 cleavage products is given in Supplementary Figures 6c and d.
A model for MGCD0103-mediated inhibition of autophagy in primary CLL cells is illustrated in Supplementary Figure 7.
Inhibition of autophagy decreases CLL cell viability
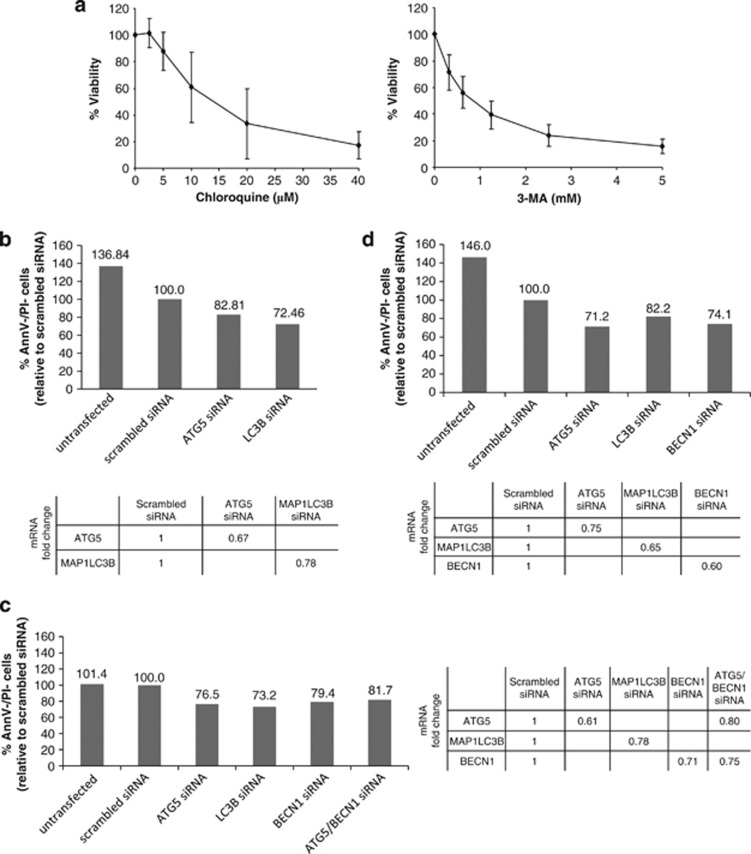
Our data suggest that autophagy inhibition might decrease CLL cell survival. To test this hypothesis, we first treated CLL cells with late- or early-stage inhibitors of autophagy (chloroquine and 3-MA, respectively). Both drugs decreased CLL cell viability in a dose-dependent manner (Figure 6a), suggesting that basal autophagy is a survival mechanism in primary CLL cells. To confirm this finding, siRNA-mediated knockdown of key autophagy genes was performed. In agreement with previous reports,28, 29 primary CLL cells were highly refractory to transfections, probably owing to their quiescent nature. Nevertheless, in three out of seven CLL samples analyzed, introduction of either ATG5, MAP1LC3B, BECN1 or ATG5/BECN1 siRNAs resulted in decreased target gene expression (ranging from 22 to 40% when compared with cells treated with scrambled siRNAs), together with decreased cell viability (Figures 6b–d). These results confirm the prosurvival effect of basal autophagy in primary CLL cells.
Figure 6.
Inhibition of autophagy decreases primary CLL cell viability. (a) PBMCs from CLL patients (n=5) were incubated with varying concentrations of chloroquine or 3-MA for 48 h. Viability was determined by a CCK-8 colorimetric assay. Each sample was run in triplicate and was normalized to cells incubated without drug. Data represent the mean values±s.d. (b–d) PBMCs from CLL patients were transfected with autophagy gene-specific or nontargeting scrambled siRNA as indicated. Viability of CLL cells was assessed by flow cytometry following Annexin V-APC/PI staining. Results obtained with three different patient samples at (b) 24 h, (c) 48 h and (d) 72 h after transfection are shown. Viable cells (Annexin V-APC neg/PI neg cells) are represented and expressed relative to scrambled siRNA control, set at 100% viability. Expression levels of targeted genes were analyzed by real-time reverse transcription (RT)-PCR and were normalized to the 28S ribosomal RNA level of the same sample. RT-PCR values are expressed as relative fold change to the scrambled siRNA condition.
The combination of autophagy and HDAC inhibitors potentiates CLL cell death
In order to evaluate the therapeutic potential of regimens combining autophagy and HDAC inhibitors in the treatment of CLL, we tested the cytotoxicity of either MGCD0103 or VPA in combination with chloroquine or 3-MA. Hence, we followed the median-effect method19 using two concentrations of each of the autophagy inhibitors and various concentrations of the HDAC inhibitors, chosen based on the considerations detailed under Materials and Methods.
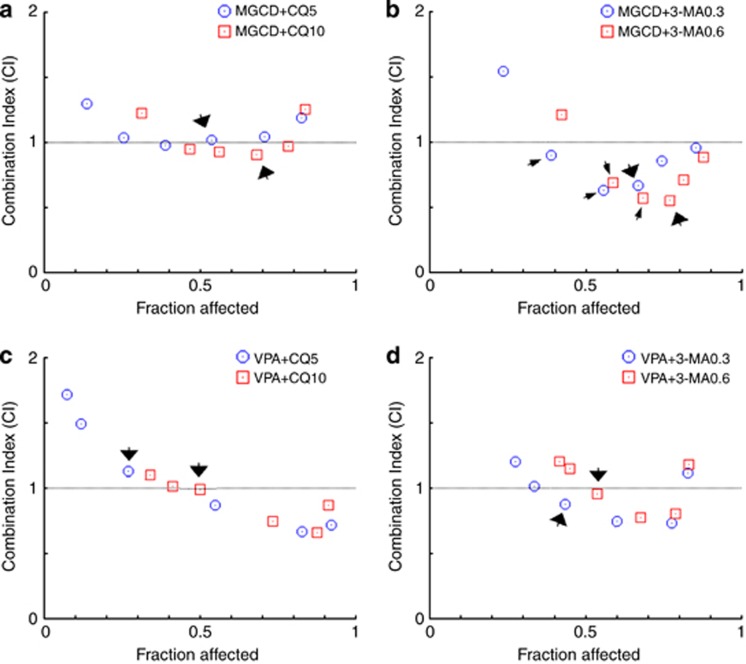
When MGCD0103 was used at the clinically relevant concentration of 0.4 μmol/l, its association with chloroquine displayed nearly additive effect (0.9<CI<1.1) on the inhibition of cell viability (arrows in Figure 7a), whereas its combination with 3-MA led to a clear synergism (0.3<CI<0.7) (big arrows in Figure 7b). Importantly, this synergism was also observed at lower concentrations of MGCD0103 (0.1 and 0.2 μmol/l) (small arrows). Finally, combinations of VPA (at the clinically relevant concentration of 1 mmol/l) with 0.3 and 0.6 mmol/l of 3-MA displayed a slightly synergistic to nearly additive effect (0.85<CI<0.9) respectively (arrows in Figure 7d), whereas its association with chloroquine was at best nearly additive (arrows in Figure 7c). Supplementary Table 3 recapitulates the CI values of the drug combinations discussed above and their biological description.
Figure 7.
The combination of HDAC inhibitors and autophagy inhibitors synergistically/additively kills CLL cells at clinically relevant concentrations. Combination analyses were performed following the median-effect method.19 PBMCs from CLL patients (n=5) were exposed to an HDAC inhibitor (MGCD0103 (MGCD) or VPA) and an autophagy inhibitor (chloroquine (CQ) or 3-MA) simultaneously for 48 h. Various concentrations of HDAC inhibitors (shown as blue dots or red squares, and increasing from left to right along the x axis) were combined with fixed doses of CQ (5 and 10 μmol/l) or 3-MA (0.3 and 0.6 mmol/l). (a, b) The concentrations of MGCD0103 used for these experiments were (in μmol/l) 0.05, 0.1, 0.2, 0.4, 0.8 and 1.6, and (c, d) those of VPA were (in mmol/l) 0.25, 0.5, 1, 2, 4 and 8. CIs for different levels of cell death (fraction affected) were calculated using the CompuSyn software. CI<1, CI=1 and CI>1 indicate synergism, additive effect and antagonism, respectively.
Collectively, these results demonstrate that inhibition of autophagy may synergize or act additively with HDAC inhibitors at clinically achievable concentrations to kill CLL cells.
The combination of MGCD0103 with flavopiridol considerably enhances CLL cell death
Our data suggest that MGCD0103 could enhance the efficiency of therapeutic molecules, such as the cyclin-dependent kinase (CDK) inhibitor flavopiridol, that induce a protective autophagic response in primary CLL cells.30
To test this hypothesis, we treated CLL cells with combinations of MGCD0103 and flavopiridol during 6 h—a time period close to the clinically effective schedule of administration of flavopiridol in CLL patients.18 Although neither drug had a significant effect on cell survival when used at clinically relevant concentrations (0.4 μmol/l (MGCD0103) and 1.5 μmol/l (flavopiridol)), under these conditions, their combination clearly decreased cell viability (Table 1, P(flavopiridol versus flavopiridol+MGCD0103)=0.03; P(MGCD0103 versus flavopiridol+MGCD0103)=0.01).
Table 1. Viability (%) of chronic lymphocytic leukemia (CLL) cells after a 6-h treatment with MGCD0103 and flavopiridol alone or in combination.
|
MGCD0103 (μM) |
||||||
|---|---|---|---|---|---|---|
| 0 | 0.05 | 0.1 | 0.2 | 0.4 | 0.8 | |
| Flavopiridol (μM) | ||||||
| 0 | 100 | 104.12±6.51 | 96.91±8.16 | 101.15±10.50 | 97.33±9.30 | 86.74±14.42 |
| 0.1875 | 92.90±14.70 | 78.16±19.59 | ||||
| 0.375 | 96.22±14.51 | 80.59±20.37 | ||||
| 0.75 | 89.32±16.90 | 79.88±23.77 | ||||
| 1.5 | 92.89±11.64 | 70.68±14.71 | ||||
| 3 | 83.42±9.21 | 68.70±16.50 | ||||
Peripheral blood mononuclear cells (PBMCs) from CLL patients (n=5) were incubated with various concentrations of MGCD0103 and flavopiridol, either alone or simultaneously, for 6 h. Combination experiments were done using a constant ratio design. Viability was determined by a CCK-8 colorimetric assay. Each sample was run in triplicate and was normalized to cells incubated without drug. Data represent the mean values±s.d.
These data demonstrate that the association of MGCD0103 with the autophagy inducer flavopiridol potentiates CLL cell death.
Discussion
Autophagy may promote either cell survival or cell death; however, its role in CLL remains unclear. In this study, we found that MGCD0103 decreases the autophagic flux in primary CLL cells. In addition, we provided evidence that basal autophagy functions in CLL as a pro-survival mechanism whose disruption potentiates cell death induced by HDAC and CDK inhibitors.
We previously reported that MGCD0103 triggers a cell death amplification loop involving caspases and calpains in primary CLL cells.14 The present report shows that MGCD0103 inhibits autophagy through the activation of the PI3K/AKT/mTOR pathway followed by activation of caspases and CAPN1, leading to the degradation of autophagy-related proteins in primary CLL cells. Besides, our data suggest that MGCD0103-induced autophagy blockade may also result from transcriptional downregulation of key regulators.
The existence of an interplay between apoptosis and autophagy has been well documented.22, 27, 31, 32, 33 Indeed, the caspase-mediated cleavage of BECN1 inhibited BECN1-induced autophagy and generated pro-apoptotic BECN1 fragments that triggered cytochrome c release from mitochondria.22, 33 Similarly, release of cytochrome c was induced following calpain-mediated generation of an ATG5 fragment.27 In line with these observations, MGCD0103-induced cleavage of BECN1 and ATG5 observed in the present study may be part of the death amplification loop activated in primary CLL cells.
The ATG5 gene product is a crucial proteolytic target for mechanisms aiming to disrupt/modulate autophagy. Using a cell-free system, Yousefi et al.27 have shown that ATG5 can be cleaved by calpains, but not caspases 3, 6, 7, 8 and 9. However, our results obtained ex vivo suggest that caspases are the major proteases responsible for MGCD0103-induced ATG5 cleavage in primary cells, either directly or, as observed in some CLL patients, through activation of CAPN1. These findings suggest that a caspase different from the aforementioned caspases could cleave ATG5 in a calpain-independent manner.
The role of autophagy in CLL has remained controversial. Thus, resistance to dasatinib has been correlated to autophagy induction,34 and cell death was induced in experimental systems where autophagy was inhibited either by chloroquine or expression of miR-130a.12 On the other hand, treatment of CLL cells with dexamethasone induced autophagic cell death.13 More recently, it was shown that many stimuli can induce autophagy in CLL cells; however, only endoplasmic reticulum stress-inducing agents induced protective autophagy.30 Consistent with previous reports,12 we found that pharmacological inhibitors of autophagy decreased CLL cell viability in a dose-dependent manner. In addition, the loss of viability observed in cells where partial knockdown of ATG5, BECN1 and MAP1LC3B was achieved by siRNA confirmed the survival-promoting effect of basal autophagy in primary CLL cells. Moreover, we demonstrated that inhibition of autophagy potentiates HDAC inhibitor-induced cell death in CLL cells at clinically relevant concentrations. Although chloroquine and 3-MA may have off-target effects, our data suggest that targeting autophagy may enhance the therapeutic effects of HDAC inhibitors in CLL, consistent with previous findings in other cancer models.10, 35
Autophagy plays a central role in the immune response to cancer,11, 36, 37 and the immunomodulatory effects of autophagy inhibitors (chloroquine and hydroxychloroquine) has been exploited in the treatment of autoimmune disorders.38, 39 Unfortunately, the immunomodulatory properties of these inhibitors have an adverse effect on antitumor immunity.11, 40 Thus, clinically promising chemotherapeutical strategies based on combinations of autophagy inhibitors with conventional anticancer drugs currently face the limitation that autophagy inhibition is not restricted to cancer cells but also affects immune cells.
Chloroquine has previously been shown to neutralize ex vivo the protective autophagy elicited in CLL cells by flavopiridol,30 a CDK inhibitor that induces high response rates in heavily pretreated CLL patients, including high-risk patients.41, 42 Unlike chloroquine, we found that the autophagy-inhibiting properties of MGCD0103 specifically target CLL cells but not the other leukemic and nonleukemic cells, including normal PBMCs. Importantly, MGCD0103 counteracted autophagy, a mechanism of resistance to cell death elicited by the CDK inhibitor, and potentiated the cytotoxic effect of flavopiridol. This finding opens new horizons to circumvent the immunomodulatory effects of chloroquine and hydroxychloroquine.
Recently, Tenovin-6, an inhibitor of the NAD(+)-dependent class III HDACs, has been shown to inhibit the late stages of autophagy in CLL cells.43 Suppression of autophagy by inhibitors of zinc-dependent HDACs had hitherto been reported only in cardiomyocytes44 and, more recently, in myeloid leukemia where the pro-survival role of autophagy was also documented.45 Instead, HDAC inhibitors have been described to induce autophagy by a variety of mechanisms including transcriptional induction of MAP1LC35 and BECN1,46 inactivation of mTOR signaling,5, 7 induction of endoplasmic reticulum stress response7 and production of reactive oxygen species.47 Although MGCD0103 induced autophagy in cancer cell lines, it decreased the autophagic flux in primary CLL cells. As observed with other anticancer drugs such as metformin,48, 49 autophagy modulation by HDAC inhibitors appears to be cell type dependent. Moreover, our data underline the well-documented notion that cell lines do not recapitulate all the features of the original primary tumors,50, 51, 52 a caveat that must be borne in mind when studying the pathology of CLL.
Autophagy regulation by HDAC inhibitors can occur at the epigenetic level through modulation of autophagy gene expression, as we and others showed.5, 53 However, targets of HDACs include nonhistone proteins.54 In this context, additional mechanisms whereby HDACs may contribute to autophagy include HDAC6-mediated deacetylation of cortactin, enhancing autophagosome-lysosome fusion,55 or the regulation, by HDAC6, of the transport of aggregates and autophagic machinery components to the aggresome.56 MGCD0103 is not an HDAC6 inhibitor; however, we cannot rule out that MGCD0103-mediated downregulation of HDAC6 expression (Figure 4b) may contribute to the regulation of autophagy in primary CLL cells.
Because SQSTM1 protein is incorporated into autophagosomes and degraded by autophagy,20, 57, 58 its degradation has been widely used as a hallmark of autophagy activation.20, 57, 59 Here, we showed that SQSTM1 can be degraded by caspases and CAPN1 independently of autophagy, in agreement with the results of Norman et al.22 and Pham et al.60 These findings are in line with the myriad of interacting partners and cellular functions of SQSTM1 beyond its role in autophagy61 and should prompt researchers to caution in drawing conclusions when interpreting SQSTM1 levels.
The contribution of autophagy to microenvironmental signals and CLL progression in the proliferation niches remains to be explored. Similarly, further investigation will be necessary to evaluate whether the role of autophagy in CLL is dependent on the stage of the disease, the genetic aberration profile or the expression of some prognostic markers. Addressing these questions will help understanding how CLL patients may best benefit from a therapeutic strategy involving alteration of autophagy.
Acknowledgments
This work was supported by Télévie (grant no. 7.4527.09). We thank MethylGene Inc. (Quebec, Canada) for supplying MGCD0103, Professor Stephen Senn, Dr Fred Fack, Dr Emilia Mahoney, Tony Kaoma and Marwan Sleiman.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- Burger JA. Targeting the microenvironment in chronic lymphocytic leukemia is changing the therapeutic landscape. Curr Opin Oncol. 2012;24:643–649. doi: 10.1097/CCO.0b013e3283589950. [DOI] [PubMed] [Google Scholar]
- Rikiishi H. Autophagic and apoptotic effects of HDAC inhibitors on cancer cells. J Biomed Biotechnol. 2011;2011:830260. doi: 10.1155/2011/830260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True O, Matthias P. Interplay between histone deacetylases and autophagy—from cancer therapy to neurodegeneration. Immunol Cell Biol. 2012;90:78–84. doi: 10.1038/icb.2011.103. [DOI] [PubMed] [Google Scholar]
- Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA. 2012;109:6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy. 2010;6:1057–1065. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- Gordy C, He YW. The crosstalk between autophagy and apoptosis: where does this lead. Protein Cell. 2012;3:17–27. doi: 10.1007/s13238-011-1127-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew JS, Medina EC, Esquivel JA, 2nd, Mahalingam D, Swords R, Kelly K, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14:2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend KN, Hughson LR, Schlie K, Poon VI, Westerback A, Lum JJ. Autophagy inhibition in cancer therapy: metabolic considerations for antitumor immunity. Immunol Rev. 2012;249:176–194. doi: 10.1111/j.1600-065X.2012.01141.x. [DOI] [PubMed] [Google Scholar]
- Kovaleva V, Mora R, Park YJ, Plass C, Chiramel AI, Bartenschlager R, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- Laane E, Tamm KP, Buentke E, Ito K, Kharaziha P, Oscarsson J, et al. Cell death induced by dexamethasone in lymphoid leukemia is mediated through initiation of autophagy. Cell Death Differ. 2009;16:1018–1029. doi: 10.1038/cdd.2009.46. [DOI] [PubMed] [Google Scholar]
- El-Khoury V, Moussay E, Janji B, Palissot V, Aouali N, Brons NH, et al. The histone deacetylase inhibitor MGCD0103 induces apoptosis in B-cell chronic lymphocytic leukemia cells through a mitochondria-mediated caspase activation cascade. Mol Cancer Ther. 2010;9:1349–1360. doi: 10.1158/1535-7163.MCT-09-1000. [DOI] [PubMed] [Google Scholar]
- Atmaca A, Al-Batran SE, Maurer A, Neumann A, Heinzel T, Hentsch B, et al. Valproic acid (VPA) in patients with refractory advanced cancer: a dose escalating phase I clinical trial. Br J Cancer. 2007;97:177–182. doi: 10.1038/sj.bjc.6603851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung V, Richard N, Brady H, Maier A, Kelter G, Heise C. Histone deacetylase inhibitor MGCD0103 synergizes with gemcitabine in human pancreatic cells. Cancer Sci. 2011;102:1201–1207. doi: 10.1111/j.1349-7006.2011.01921.x. [DOI] [PubMed] [Google Scholar]
- Di Trani L, Savarino A, Campitelli L, Norelli S, Puzelli S, D'Ostilio D, et al. Different pH requirements are associated with divergent inhibitory effects of chloroquine on human and avian influenza A viruses. Virol J. 2007;4:39. doi: 10.1186/1743-422X-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Lin TS, Dalton JT, Wu D, Phelps MA, Fischer B, et al. Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood. 2007;109:399–404. doi: 10.1182/blood-2006-05-020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6:1042–1056. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Akar U, Ozpolat B, Mehta K, Fok J, Kondo Y, Lopez-Berestein G. Tissue transglutaminase inhibits autophagy in pancreatic cancer cells. Mol Cancer Res. 2007;5:241–249. doi: 10.1158/1541-7786.MCR-06-0229. [DOI] [PubMed] [Google Scholar]
- D'Eletto M, Farrace MG, Rossin F, Strappazzon F, Giacomo GD, Cecconi F, et al. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ. 2012;19:1228–1238. doi: 10.1038/cdd.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia HG, Zhang L, Chen G, Zhang T, Liu J, Jin M, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010;6:61–66. doi: 10.4161/auto.6.1.10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Van Bockstaele F, Pede V, Naessens E, Van Coppernolle S, Van Tendeloo V, Verhasselt B, et al. Efficient gene transfer in CLL by mRNA electroporation. Leukemia. 2008;22:323–329. doi: 10.1038/sj.leu.2405007. [DOI] [PubMed] [Google Scholar]
- Serafini M, Naldini L, Introna M. Molecular evidence of inefficient transduction of proliferating human B lymphocytes by VSV-pseudotyped HIV-1-derived lentivectors. Virology. 2004;325:413–424. doi: 10.1016/j.virol.2004.04.038. [DOI] [PubMed] [Google Scholar]
- Mahoney E, Lucas DM, Gupta SV, Wagner AJ, Herman SE, Smith LL, et al. ER stress and autophagy: new discoveries in the mechanism of action and drug resistance of the cyclin-dependent kinase inhibitor flavopiridol. Blood. 2012;120:1262–1273. doi: 10.1182/blood-2011-12-400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–20729. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein L, Soulieres D, Johnston JB, Aloyz R. p53 and autophagy contribute to dasatinib resistance in primary CLL lymphocytes. Leuk Res. 2011;35:99–102. doi: 10.1016/j.leukres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- Lopez G, Torres K, Lev D. Autophagy blockade enhances HDAC inhibitors' pro-apoptotic effects: potential implications for the treatment of a therapeutic-resistant malignancy. Autophagy. 2011;7:440–441. doi: 10.4161/auto.7.4.14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19:144–152. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Capan E, Zhao Y, Zhao J, Stolz D, Watkins SC, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- Abarientos C, Sperber K, Shapiro DL, Aronow WS, Chao CP, Ash JY. Hydroxychloroquine in systemic lupus erythematosus and rheumatoid arthritis and its safety in pregnancy. Expert Opin Drug Saf. 2011;10:705–714. doi: 10.1517/14740338.2011.566555. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Silverman E, Bargman JM. The role of antimalarial agents in the treatment of SLE and lupus nephritis. Nat Rev Nephrol. 2011;7:718–729. doi: 10.1038/nrneph.2011.150. [DOI] [PubMed] [Google Scholar]
- Choi KS. Autophagy and cancer. Exp Mol Med. 2012;44:109–120. doi: 10.3858/emm.2012.44.2.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian BA, Grever MR, Byrd JC, Lin TS. Flavopiridol in the treatment of chronic lymphocytic leukemia. Curr Opin Oncol. 2007;19:573–578. doi: 10.1097/CCO.0b013e3282efb9da. [DOI] [PubMed] [Google Scholar]
- Lin TS, Ruppert AS, Johnson AJ, Fischer B, Heerema NA, Andritsos LA, et al. Phase II study of flavopiridol in relapsed chronic lymphocytic leukemia demonstrating high response rates in genetically high-risk disease. J Clin Oncol. 2009;27:6012–6018. doi: 10.1200/JCO.2009.22.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum SF, Groves MJ, James J, Murray K, Appleyard V, Prescott AR, et al. Dysregulation of autophagy in chronic lymphocytic leukemia with the small-molecule Sirtuin inhibitor Tenovin-6. Sci Rep. 2013;3:1275. doi: 10.1038/srep01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci USA. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankov MV, El Khatib M, Kumar Thakur B, Heitmann K, Panayotova-Dimitrova D, Schoening J, et al. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy Leukemia 2013. e-pub ahead of print 12 September 2013; doi: 10.1038/leu.2013.264 [DOI] [PMC free article] [PubMed]
- Cao Q, Yu C, Xue R, Hsueh W, Pan P, Chen Z, et al. Autophagy induced by suberoylanilide hydroxamic acid in Hela S3 cells involves inhibition of protein kinase B and up-regulation of Beclin 1. Int J Biochem Cell Biol. 2008;40:272–283. doi: 10.1016/j.biocel.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Li J, Liu R, Lei Y, Wang K, Lau QC, Xie N, et al. Proteomic analysis revealed association of aberrant ROS signaling with suberoylanilide hydroxamic acid-induced autophagy in Jurkat T-leukemia cells. Autophagy. 2010;6:711–724. doi: 10.4161/auto.6.6.12397. [DOI] [PubMed] [Google Scholar]
- Ben Sahra I, Laurent K, Giuliano S, Larbret F, Ponzio G, Gounon P, et al. Targeting cancer cell metabolism: the combination of metformin and 2-deoxyglucose induces p53-dependent apoptosis in prostate cancer cells. Cancer Res. 2010;70:2465–2475. doi: 10.1158/0008-5472.CAN-09-2782. [DOI] [PubMed] [Google Scholar]
- Dowling RJ, Zakikhani M, Fantus IG, Pollak M, Sonenberg N. Metformin inhibits mammalian target of rapamycin-dependent translation initiation in breast cancer cells. Cancer Res. 2007;67:10804–10812. doi: 10.1158/0008-5472.CAN-07-2310. [DOI] [PubMed] [Google Scholar]
- van Staveren WC, Solis DY, Hebrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: experimental models for cancer cells in situ? For cancer stem cells. Biochim Biophys Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- van Helden SF, van Leeuwen FN, Figdor CG. Human and murine model cell lines for dendritic cell biology evaluated. Immunol Lett. 2008;117:191–197. doi: 10.1016/j.imlet.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Huszthy PC, Daphu I, Niclou SP, Stieber D, Nigro JM, Sakariassen PO, et al. In vivo models of primary brain tumors: pitfalls and perspectives. Neuro Oncol. 2012;14:979–993. doi: 10.1093/neuonc/nos135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn MY, Ahn SG, Yoon JH. Apicidin, a histone deaceylase inhibitor, induces both apoptosis and autophagy in human oral squamous carcinoma cells. Oral Oncol. 2011;47:1032–1038. doi: 10.1016/j.oraloncology.2011.07.027. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Yao TP. Quality control autophagy: a joint effort of ubiquitin, protein deacetylase and actin cytoskeleton. Autophagy. 2010;6:555–557. doi: 10.4161/auto.6.4.11812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao TP. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Kominami E, Tanaka K, Komatsu M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy. 2008;4:1063–1066. doi: 10.4161/auto.6826. [DOI] [PubMed] [Google Scholar]
- Buchser WJ, Laskow TC, Pavlik PJ, Lin HM, Lotze MT. Cell-mediated autophagy promotes cancer cell survival. Cancer Res. 2012;72:2970–2979. doi: 10.1158/0008-5472.CAN-11-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VC, Pitti R, Anania VG, Bakalarski CE, Bustos D, Jhunjhunwala S, et al. Complementary proteomic tools for the dissection of apoptotic proteolysis events. J Proteome Res. 2012;11:2947–2954. doi: 10.1021/pr300035k. [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT. p62: a versatile multitasker takes on cancer. Trends Biochem Sci. 2012;37:230–236. doi: 10.1016/j.tibs.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.