Abstract
We report that the paired homeodomain transcription factor Pax6 is imported into the nucleus by the Karyopherin β family member Karyopherin 13 (Kap13). Pax6 was identified as a potential cargo for Kap13 by a yeast two-hybrid screen. Direct binding of Pax6 to Kap13 was subsequently confirmed by in vitro assays with recombinant proteins, and binding in vivo was shown by coimmunoprecipitation. Ran-dependent import of Pax6 by Kap13 was shown to occur by using a digitonin-permeabilized cells assay. Kap13 binds to Pax6 via a nuclear localization sequence (NLS), which is located within a segment of 80 amino acid residues that includes the homeodomain. Kap13 showed reduced binding to Pax6 when either region located at each end of the homeodomain (208 to 214 and 261 to 267) was deleted. The paired-type homeodomain transcription factor family includes more than 20 members. All members contain a region similar to the NLS found in Pax6 and are therefore likely to be imported by Kap13. We confirmed this hypothesis for Pax3 and Crx, which bind to and are imported by Kap13.
Trafficking of proteins and nucleic acids in or out of the cell nucleus is restricted by the nuclear envelope. The passage across the nuclear envelope occurs through the nuclear pores, which allow unrestricted traffic of molecules below a certain size limit, which is 40 to 60 kDa for proteins. Proteins and nucleic acids above the size limit (and some below the limit) are transported across the nuclear envelope via a complex system of soluble carriers (5, 7, 50, 55, 59, 68, 77, 85, 86, 94). Each carrier is responsible for the import or export of one or more subsets of proteins or nucleic acids. The carriers bind their cargoes either in the cytoplasm or in the nucleus, dock them to components of the nuclear pore complexes, and assist their passage across the nuclear envelope (8, 80). The carriers are generally known as karyopherins (Kaps), importins, exportins, or transportins.
Many proteins are imported into the nucleus via the classical import pathway mediated by a heterodimer formed by Kap β1 and one of the six members of the Kap α family. All proteins entering this pathway contain segments known as classical monopartite or classical bipartite nuclear localization sequences (NLS), which consist of one cluster of positively charged amino acid residues or two clusters separated by approximately 10 to 12 residues, respectively (23, 47, 78). The NLS is recognized by Kap α, which in turn binds Kap β. Kap β mediates the transient docking of the import complex to the nuclear pore components. The assembly and disassembly of the import complexes is controlled by the GTPase Ran. RanGTP binds in the nucleus to Kap β and mediates the release of the cargo, allowing the carriers to be recycled to the cytoplasm and the cargoes to exert their functions unimpeded at their proper subcellular localization (21, 62, 64). Ran cycles between the GTP- and GDP-bound forms, and its interactions are regulated by other proteins, including RanBP1, RCC1, RanGAP, and p10 (NTF2) (69).
Proteins that do not contain a classical NLS are imported into the nuclei by one of the approximately 20 members of the Kap β family. Kap βs have a molecular size of 95 to 125 kDa and share a region of homology at the N-terminal end (82). Proteins entering via a nonclassical pathway bind directly to a Kap β family member; no Kap α is required. Some members of the Kap β family (exportins) mediate export of proteins from the nuclei via a nuclear export signal. Unlike the import complexes, exportin-cargo complexes are stabilized by the high RanGTP concentration present in the nuclei, while complexes between importins and their cargoes disassemble in the presence of nuclear RanGTP.
Proteins and RNAs that represent either import or export cargoes have been identified for many Kaps (5, 7, 50, 55, 59, 68, 77, 85, 86, 94). For most Kaps few cargoes have been identified, though many more are likely to exist.
Kap13, also referred to as importin 13 (63), is a member of the Kap β family. Kap13 is ubiquitously expressed, as indicated by Northern blot analysis of human and rat tissues (96). Ubiquitous expression of Kap13 is also indicated by the large diversity of tissues and cell types from which expressed sequence tags have been obtained, as listed in the UniGene database. Here we report that Kap13 mediates nuclear import of paired-type homeodomain transcription factors and that the import is achieved via an NLS that includes the paired-type homeodomain.
Paired-type homeodomain transcription factors belong to the large category of homeodomain transcription factors that control development and differentiation (33). The homeodomain factors contain a conserved 60-amino-acid DNA binding motif referred to as the homeodomain. The homeodomain has a conserved helix-turn-helix structure composed of three helices (24, 75, 76, 92). Homeodomain transcription factors are organized into classes based on sequence similarity (20). The paired class of homeodomain proteins is characterized by the presence of a serine at position 9 of helix III, in addition to a 128-amino-acid DNA binding domain referred to as the paired domain, located upstream of the homeodomain (12). A distinct category of homeodomains consists of paired-like homeodomains. They share 55 to 75% similarity with the paired homeodomains, do not have a paired domain, and do not contain a serine at position 9 of helix III, but rather a lysine or glutamine. Hereafter we refer collectively to paired and paired-like homeodomains as paired-type homeodomain proteins. The paired-type homeodomain proteins form a monophyletic group, characterized by the presence of at least five out of six highly conserved residues of the homeodomain: P26, D27, E32, R44, Q46, and A54 (32). There are approximately 170 human homeodomain-containing proteins, of which 26 belong to the paired-type category. We report that three paired-type homeodomain transcription factors are imported into the nucleus by Kap13, and we speculate that most or all members of this family are imported via the same pathway.
MATERIALS AND METHODS
cDNA constructs.
The Kap13 (GenBank accession no. AF267987) cDNA was obtained by PCR from human brain cDNA (Clontech, Palo Alto, Calif.). For bacterial expression, Kap13 was subcloned into pQE30 (Qiagen, Valencia, Calif.) by using the BamHI and HindIII sites. Nucleotides 922 to 2889 of Kap13 were cloned into the yeast two-hybrid vector pAS2-1 (Clontech) by using the EcoRI site. Human full-length Pax6 (isoform A) was obtained as an expressed sequence tag clone (GenBank no. AI337595) from the I.M.A.G.E Consortium and was used as a template for all further constructs of Pax6, unless otherwise specified. Human Crx, Prh, and Pax6 were subcloned into the bacterial expression vector pET41a (Novagen, Madison, Wis.) as glutathione S-transferase (GST) (N-terminal) and His-tag (C-terminal) fusions via the SpeI and HindIII sites. The mouse Pax3 cDNA (nucleotide segment 382 to 1197) was cloned as Crx and Pax6 but utilized the SpeI and NotI sites. (The mouse and human Pax3 primary sequences are 100% identical in the NLS region and 98% identical overall.) Pax6 deletion constructs were subcloned from a yeast two-hybrid clone into the prey vector pVP16 (45) by utilizing the NotI and EcoRI sites. Mouse and human Pax6 have identical amino acid sequences. Enhanced green fluorescent protein was subcloned via PCR from pEGFP-C1 (Clontech) into pET41a by using the SpeI and KpnI sites. Bacterially expressed Pax6 deletion constructs having N-terminal GST-green fluorescent protein (GFP) and C-terminal His-tag fusions were created by subcloning Pax6 fragments from the yeast two-hybrid clone into pET41a-GFP by using the HindIII and XhoI sites. The mouse RanBP1 cDNA was subcloned into pGEX6P-1 (Amersham Biosciences, Piscataway, N.J.) by using the BamHI and XhoI sites. Human Ran was subcloned into pQE30 via the BamHI and HindIII sites. Kap13 was cloned as an N-terminal myc-tagged construct into the pCS2-MT (79) vector by using the EcoRI and XbaI sites. Pax6 wild-type and Pax6 mutant PCR products (BglII, XhoI) were cloned into the BamHI and SalI sites of pEGFP-Cl. All PCR amplifications were done with the Advantage HF2 polymerase (Clontech) except for Prh, which was done with GC 2 polymerase (Clontech). All constructs were verified by sequencing.
Yeast two-hybrid screen.
The yeast two-hybrid screen was essentially performed as described previously (Yeast Protocols Handbook [Clontech] and reference 6). The plasmid pAS2-1-Kap13 (922 to 2889) was transformed into the yeast strain Y190. Transformants were selected and checked for autotransactivation by growth on medium lacking histidine, followed by a β-galactosidase assay. The growth on medium lacking histidine was suppressed by addition of 3-AT (Sigma, St. Louis, Mo.). A large-scale library transformation was done with a random primed, nondirectionally cloned cDNA library from the murine cell line EML (erythroid-myeloid-lymphoid) which contained an estimated number of 1.33 million independent clones in the correct reading frame before amplification (91). The library was amplified twice. Transformants (5 × 106) were plated and screened on 150-mm-diameter plates with medium lacking leucine-tryptophan-histidine. Colonies were picked and checked for β-galactosidase production by using a filter assay with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. Plasmid purification was done from the positive clones, and a second round of interaction screening was performed to confirm the interactions. The inserts from the positive clones were sequenced. The interaction screens using Pax6 deletion constructs were done by using essentially the same methodology as that described above.
Protein expression and purification.
For bacterial protein expression cultures were grown at 37°C to an optical density at 600 nm of ∼1.0 and then were shifted to 18°C. After the temperature equilibrated, the cultures were induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside and were grown overnight with shaking at 170 rpm. Recombinant proteins were purified on glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, N.J.) or Ni-nitrilotriacetic acid agarose (Qiagen). Kap13 and pQE30-Ran were expressed in the strain M15[pREP4] (Qiagen). The pET41a constructs were expressed in strain BL21(DE3) Codon Plus RIL (Stratagene, La Jolla, Calif.). All other proteins were expressed in the BL21 strain (Novagen).
Nuclear import assays.
Import reactions were performed at room temperature essentially as described previously (46, 65). HeLa cells were grown on 12-mm-diameter glass coverslips. Cells were permeabilized with 35 μg of digitonin (Roche)/ml for 5 min on ice. Recombinant Kaps (1 μM) and cargoes (1 μM) in transport buffer (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 250 mM sucrose, 2 mM dithiothreitol) were overlaid on the cells. A Ran mix (3 μM RanGDP, 0.2 μM NTF2 [67], 0.2 μM RanBP1, and 0.2 μM RNA1p [26]) was added along with an energy-regenerating system (5 mM creatine phosphate, 0.25 mM each ATP and GTP, and 1 U of creatine kinase). Cells were fixed by using 3% paraformaldehyde for 20 min and were mounted by using Vectashield (Vector Laboratories, Burlingame, Calif.). Cargoes that did not contain GFP were labeled with fluorescein isothiocyanate (FITC; Sigma) according to a protocol recommended by Pierce (Rockford, Ill.). Import reactions were visualized by fluorescence microscopy with an Olympus BX60 microscope and a ×40 0.75-numerical-aperture objective. All pictures within an experiment were taken under identical exposure conditions with an Optronics NTSC digital camera and Flashpoint acquisition software (Integrated Technologies, Indianapolis, Ind.). Quantitation of nuclear fluorescence intensity was done on digital images by measuring a total of 20 to 40 nuclei from 4 to 7 images by using the software ImageJ (http://rsb.info.nih.gov/ij/).
GST pull-downs.
GST fusion proteins (1 to 2 μg) were bound to glutathione-Sepharose 4B in a solution containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 200 mM KCl, 10% glycerol, 1% Triton X-100 for 1 h at 4°C or for 20 min at room temperature. Bound proteins were washed three times in a solution of 50 mM sodium phosphate, 600 mM NaCl, 10% glycerol followed by two washes in binding buffer (20 mM HEPES-KOH [pH 7.3], 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 1 mM dithiothreitol, 10% glycerol, 0.1% Tween 20). Bacterial lysates containing induced Kap13 were prepared in binding buffer without Tween 20. The lysate (200 μl) was incubated with beads at 4°C for 1 h. Supernatants were removed and beads were washed four times with 0.5 ml of washing buffer (binding buffer supplemented with 150 mM NaCl). Proteins were eluted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, boiled for 10 min, and analyzed by SDS-10% PAGE. For the pull-down experiments done with purified Kap13 (1 to 2 μg; see Fig. 3, 5, and 8), binding of GST proteins occurred in the presence of 50 μg of bovine serum albumin, the binding buffers contained 10 μg of bovine serum albumin, and the washing buffer was supplemented with 200 mM NaCl (for Fig. 3 and 5; 150 mM NaCl for Fig. 8). Where applicable, 15 μg of RanGTP was added to pull-down assays. Ran was loaded with GTP as described previously (58). The amount of bound Kap13 shown in Fig. 5 was quantitated by using the ImageJ software.
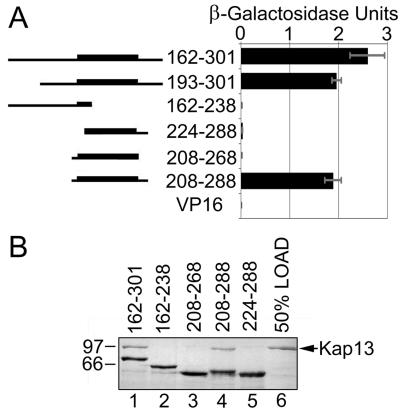
FIG. 3.
The Pax6 residues 208 to 288 are necessary and sufficient for binding to Kap13. (A) Yeast two-hybrid interaction screens using Kap13 and deletion constructs of Pax6. The interaction was measured by using a β-galactosidase assay. The homeodomain (210 to 269) is represented by a thicker line in the left diagram. The minimal segment that interacts with Kap13 spans amino acids 208 to 288. VP16, which is fused to all constructs, does not produce any transactivation by itself. (B) GST-GFP-Pax6 segments were expressed in E. coli, immobilized on glutathione-Sepharose, and incubated with Kap13 expressed and purified from E. coli. The proteins retained on the beads were analyzed by SDS-PAGE followed by Coomassie staining. The minimal Pax6 segment that binds Kap13 is 208 to 288 (lanes 3). Lane 6 shows 50% of the amount of Kap13 incubated with the Pax6 segments.
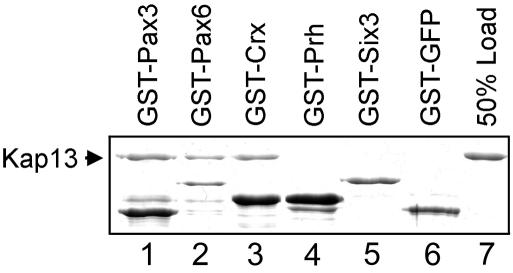
FIG. 8.
Kap13 binds Pax3, Pax6, and Crx (lanes 1 to 3) but not Six3, Prh, or GST-GFP (lanes 4 to 6) in a GST pull-down assay. The homeodomain factors were expressed as GST fusions in E. coli, purified, and immobilized on glutathione-Sepharose. Purified recombinant Kap13 was incubated with the immobilized homeodomain factors. The proteins retained on the beads after washing were analyzed by SDS-PAGE followed by Coomassie staining. Lane 7 shows 50% of the Kap13 that was used in lanes 1 to 6.
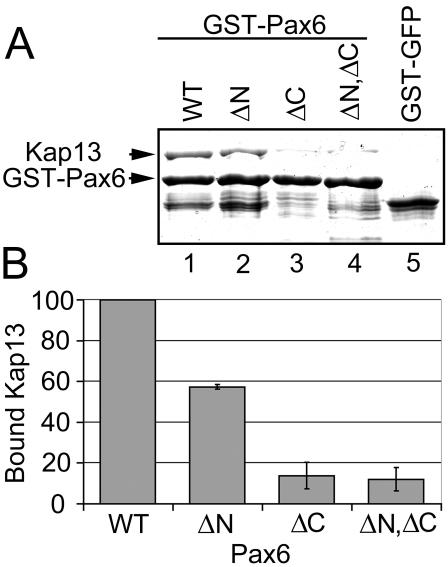
FIG. 5.
Binding of Pax6 to Kap13 is reduced when either basic cluster of Pax6, located at either end of the homeodomain, is deleted. Pax6 constructs lacking nucleotides that encode residues 208 to 214 (denoted ΔN) or 261 to 267 (ΔC) were expressed in E. coli. These constructs along with wild-type Pax6 (WT) were expressed as GST fusions, purified, immobilized on glutathione-Sepharose beads, and incubated with recombinant Kap13. (A) Pax6 has reduced binding to Kap13 when these regions are deleted (compare lanes 2, 3, and 4 to lane 1). Lane 5, Kap13 does not bind to the control GST-GFP construct. (B) The deletion of amino acids 208 to 214 decreases binding to approximately 60% compared to that of the wild type. Deletion of amino acids 261 to 267 leads to an even stronger reduction, as does the deletion of both regions. Error bars indicate standard errors of the means for three independent experiments.
Mutagenesis.
Pax6 internal deletions were created by using the Quick Change XL Site-Directed Mutagenesis kit (Stratagene) as directed by the manufacturer. Primers (forward, 5′ CAAATGCGACTTCAGCTGAAGACATCCTTTACCCAAGAGCAAATTG 3′; reverse, 5′ CAATTTGCTCTTGGGTAAAGGATGTCTTCAGCTGAAGTCGCATTTG 3′ and forward, 5′ GCAAGAATACAGGTATGGTTTTCTAATGAAGAAAAACTGAGG 3′; reverse, 5′ CCTCAGTTTTTCTTCATTAGAAAACCATACCTGTATTCTTGC 3′) were used to generate the Pax6 deletions of residues 208 to 214 and 261 to 267, respectively.
Coimmunoprecipitation.
293T cells were grown in 10-cm-diameter dishes and were transfected with 24 μg of plasmid with Lipofectamine 2000 (Invitrogen) as specified by the manufacturer. Cells were harvested by using a cell scraper and were centrifuged at 1,000 × g for 5 min, and supernatants were discarded. Cell pellets were resuspended in 200 μl of immunoprecipitation buffer (150 mM NaCl, 20 mM Tris-HCl [pH 7.4], 1% NP-40) supplemented with 10 μl of protease inhibitor cocktail (P8849; Sigma) and 65 U of DNase. RanGAP (20 μg) was added to the lysates and was incubated for 15 min at 37°C to hydrolyze endogenous RanGTP. Lysates were centrifuged at 10,000 × g at 4°C for 1 h. Supernatants were removed and the pellets were discarded. Protein G Sepharose beads (40 μl; Amersham) was added to the lysates and was incubated on a rotating wheel at 4°C for 1 h to preclear the lysates. Lysates were centrifuged at 14,000 × g in a microcentrifuge at 4°C, supernatants were removed, and preclearing was repeated. Hybridoma supernatant (50 μl) containing anti-Pax6 immunoglobulin G was added to each lysate and was incubated at 4°C on a rotating wheel for 2 h. Protein G Sepharose beads (30 μl; Amersham) was added to the lysates and was incubated on a rotating wheel at 4°C for 1 h. Lysates were pulse centrifuged and the supernatants were removed. Beads were washed four times with wash buffer (250 mM NaCl, 20 mM Tris-HCl [pH 7.4], 0.2% NP-40). Proteins were eluted from the beads with SDS-PAGE sample buffer, boiled for 10 min, and subjected to SDS-PAGE on a 7.5% gel followed by transfer to nitrocellulose. The myc-tagged Kap13 was detected by using a 1:200 dilution of the c-myc antibody (9E10; Santa Cruz, Santa Cruz, Calif.). GFP-Pax6 was detected by using a 1:20 dilution of Pax6 hybridoma supernatant. The Pax6 hybridoma monoclonal antibody developed by Atsushi Kawakmai was obtained from the Developmental Studies Hybridoma Bank created under the auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, The University of Iowa, Iowa City.
Structural analysis.
The homeodomain region of Pax6 was threaded through the crystal structure of the Drosophila Prd homeodomain bound to DNA (1Fjl-A.PDB, 1Fjl-B.PDB, and 1Fjl-C.PDB) (95) using a Swiss-Model (first approach) (72). Among the known homeodomain three-dimensional structures, Drosophila Paired homeodomain has the most similar primary structure to Pax6. Structure visualization, distance determination, and manipulation were done by using a Deep View SwissPdb Viewer (38), and final images were rendered by using a persistence of vision ray tracer.
Sequence analysis.
The segment 208 to 288 of Pax6 was used in a query to search the Homo sapiens protein database at by using BlastP (2). The search was done by using the default values, except the search results were not filtered for low-complexity regions, and the expect value was set to 10,000. The Blosum80 algorithm was used. The results were used to compare the region 208 to 288 of Pax6 to other paired-type homeodomain proteins.
The consensus between Pax6, Pax3, and Crx consists of residues that are conserved between the three factors based on physical and chemical properties. These residues are considered potentially important for binding to Kap13. Residues at each position that are not conserved between the three factors were excluded from the consensus.
The consensus written in PROSITE syntax RKXXRXRTXFTXXQ[LI]EXLEXXFX[RK]TXYPD [VI][YF]XREX[LV]AX[RK]XXLXEXR[IV]QVWFXNRRA[KR]XR was used to search the protein database at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_pattinprot.html (19).
RESULTS
Pax6 was identified as a potential cargo for Kap13 by using the yeast two-hybrid system.
A yeast two-hybrid screen using Kap13 as bait was performed in order to identify potential import cargoes. The interactions detected by the yeast two-hybrid system occur in the nuclei of yeast cells, where Ran is present mostly in the GTP-bound form. RanGTP dissociates the imported cargoes from their carriers, and therefore interactions of intact Kaps with their import cargoes are unlikely to be detectable in this system (21, 62, 64). However, if the Ran-interacting domain is removed from the Kaps, the dissociating effect of RanGTP is abolished (10, 73).
Kap13 contains 963 amino acids and has a computed molecular size of 108 kDa. The first 100 to 300 amino acid residues of Kaps are responsible for binding RanGTP, while the rest of the amino acids are generally believed to be involved in cargo binding (10, 68, 73). Our yeast two-hybrid assay was performed by using the segment of amino acid residues 307 to 963, which does not contain the putative Ran binding domain of Kap13.
Seventeen positive clones were obtained, all of which encoded the paired homeodomain transcription factor Pax6. The 17 clones represent two independent founding clones from the library. Fifteen of the clones contained the same region of Pax6, amino acid residues 162 to 301, while the remaining two clones contained segment 193 to 301. All clones include the paired homeodomain (residues 210 to 269).
Immobilized recombinant Pax6 binds Kap13 in a RanGTP-dependent manner.
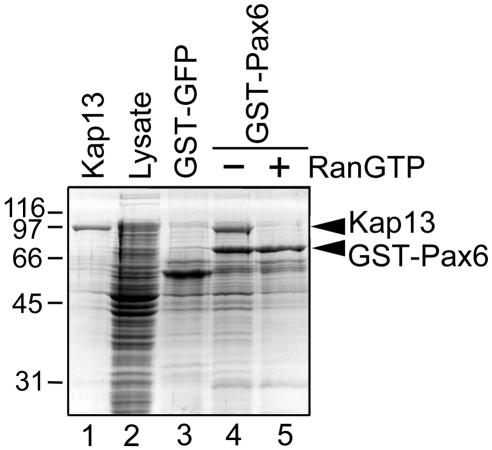
In order to verify that binding of Pax6 to Kap13 is direct and not mediated by yeast proteins, a pull-down assay was done with purified immobilized Pax6. Pax6 was expressed in Escherichia coli as a GST fusion and was immobilized on glutathione-Sepharose beads. The beads were incubated with a bacterial lysate that expressed Kap13 and were subsequently washed. The retained proteins were analyzed by SDS-PAGE. Kap13 is indeed retained by the immobilized Pax6 in the absence but not in the presence of RanGTP, as expected for an import cargo (Fig. 1).
FIG. 1.
Immobilized Pax6 retains Kap13 from a bacterial lysate, and the binding is abolished by RanGTP. GST-Pax6 purified from E. coli was immobilized on glutathione-Sepharose beads and incubated with lysate from E. coli that expressed Kap13. The bound proteins were eluted with SDS-PAGE sample buffer and were subjected to SDS-PAGE followed by Coomassie staining. The lysate is shown in lane 2; Kap13 cannot be distinguished from the bacterial proteins. The incubation was done in the absence (lane 4) or presence (lane 5) of RanGTP. Lane 3 demonstrates that the control construct GST-GFP does not bind Kap13; lane 1 illustrates Kap13 purified from a bacterial lysate similar to that shown in lane 2.
Kap13 mediates Ran-dependent import of Pax6 into the nuclei of digitonin-permeabilized cells.
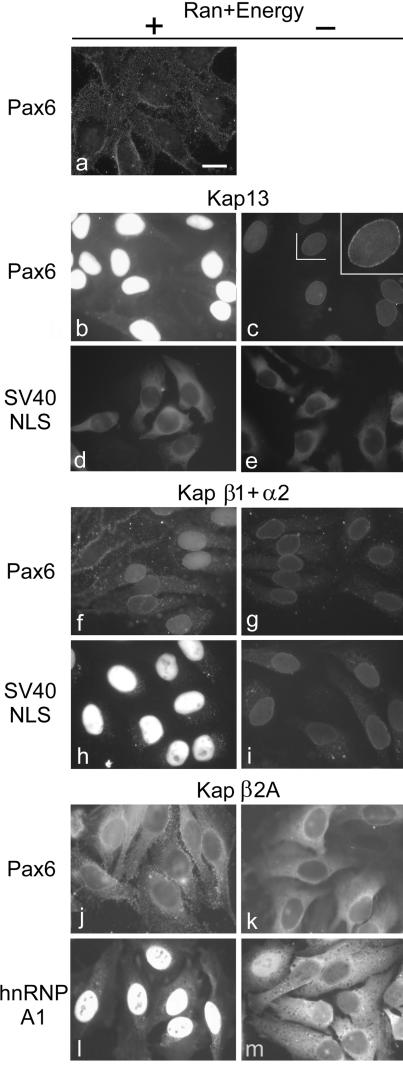
HeLa cells were grown on glass coverslips and were treated with digitonin in conditions that permeabilize the plasma membranes but leave intact the nuclear membrane (1). This assay allows reconstitution of nuclear import by using recombinant purified factors without interference from endogenous Kaps. The treatment allows soluble cytoplasmic proteins to leak out of the cell and be washed away. In addition, permeabilization allows recombinant purified proteins intracellular access when overlaid on the cells. As shown in Fig. 2b, Pax6 is imported into the nuclei of the permeabilized cells when Kap13 is present together with Ran, RanBP1, RanGAP, NTF2, and an energy regenerating system. No import of Pax6 is detectable if Kap13 is absent (Fig. 2a). In the absence of Ran and the energy regenerating system, Kap13 does not mediate import of Pax6 but only mediates docking at the nuclear envelope (Fig. 2c, inset). Both the Ran system and the energy regenerating system are necessary for import of Pax6 (data not shown).
FIG. 2.
Kap13 mediates the import of Pax6 into the nuclei of digitonin-permeabilized cells. HeLa cells were permeabilized with digitonin, incubated for 10 min with the import mixtures, washed, and fixed. Only the cargo imported into the nuclei is retained after washing.FITC-labeled GST-Pax6 is imported into the nuclei in the presence of Kap13, Ran, RanBP1, RanGAP, NTF2, and an energy regenerating system (b), but it is not imported in the absence of Kap13 (a) or in the absence of Ran and the energy regenerating system (c). A rim staining that indicates binding to the nuclear pore complexes is visible in the last case (c, inset). (d and e) Kap13 does not import a GST-GFP construct containing the classical NLS of SV40 T antigen. (f and g) Kap β1 and Kap α2 induce weak import of Pax6 in some cells, rim staining in others, and no import in other cells in the presence of Ran and energy and rim staining in some cells in the absence of Ran and energy. (h and i) As a positive control, Kap β1 and Kap α2 induce strong Ran- and energy-dependent import of a GST-GFP construct containing the classical NLS. (j and k) Pax6 is not imported into the nuclei by Kap β2A. (l and m) Positive control. Kap β2A mediates strong Ran- and energy-dependent import of GST-GFP-hnRNP A1. Scale bar, 20 μm.
The Kap β1 and α2 heterodimer mediates weak import of Pax6 in some cells and docking at the nuclear envelope, but no import was detectable in others. No docking or import is visible in the remaining cells (Fig. 2f). A GST-GFP construct including the NLS of the simian virus 40 (SV40) T antigen was used as positive control for Kap β1- and α2-mediated import.
Kap13 does not mediate import of the SV40 T antigen NLS (Fig. 2d). To date the only Kaps shown to be able to bind directly and mediate import of the classical NLS are Kaps α1 to -6 (49).
Kap β2A (transportin) does not produce any detectable import of Pax6 but mediates, as expected, a robust import of its known cargo hnRNP A1 (Fig. 2j to m) (11, 29, 74).
Amino acid residues 208 to 288 of Pax6 are sufficient for interaction with Kap13.
As mentioned above, the yeast two-hybrid screen identified two independent clones representing overlapping regions of Pax6. These regions contain the entire homeodomain and a portion of the transactivation domain. To further narrow down the region of Pax6 that interacts with Kap13, deletion constructs were created and interaction assays were performed by using the yeast two-hybrid system (Fig. 3A). Based on this assay the minimal region of Pax6, which interacts with Kap13, encompasses amino acid residues 208 to 288. This region contains the entire homeodomain of Pax6 (amino acid residues 210 to 269).
These findings, which cannot exclude the influence of other yeast proteins, were verified by in vitro binding of purified recombinant proteins (Fig. 3B). Segments of Pax6 fused to GST-GFP were expressed in E. coli and were immobilized on glutathione-Sepharose beads. The beads were incubated with recombinant purified Kap13, and the bound proteins were analyzed by SDS-PAGE (Fig. 3B). With similar results to the yeast two-hybrid interaction assays, the GST pull-down assays detected that the minimal region of Pax6 necessary for binding to Kap13 spans residues 208 to 288.
Kap13 recognizes an NLS which includes the homeodomain of Pax6.
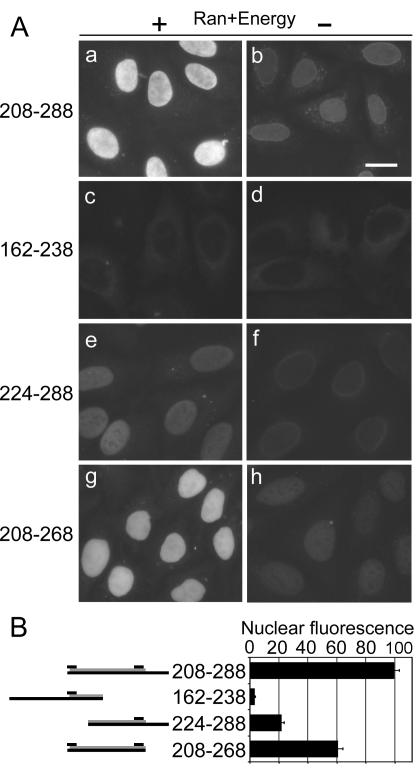
The ability of segment 208 to 288 of Pax6 to function as an NLS was tested by using a digitonin-permeabilized cell assay. The segment was expressed in E. coli as a fusion construct with GST-GFP, purified, and used for import assays in conjunction with recombinant Kap13. Kap13 imports the segment into the nuclei in a Ran- and energy-dependent manner as expected (Fig. 4A, frames a and b). The segment 224 to 288 confers import activity that is approximately 20% of that conferred by the segment 208 to 288 (Fig. 4A, frame e versus a). Segment 162 to 238 has virtually no NLS activity (Fig. 4A, frame c). Residues 269 to 288, which extend beyond the homeodomain, also contribute to the NLS function as shown by a 40% decrease of NLS activity in the absence of the segment (Fig. 4A, frame g versus a). While the segments 224 to 288 and 208 to 268 interact with Kap13 as indicated by the import assays, although they do so more weakly than segment 208 to 288, their affinity for Kap13 is insufficient to detect interactions in the binding assays (Fig. 4 versus Fig. 3).
FIG. 4.
The segment 208 to 288, which contains the whole homeodomain of Pax6, is sufficient for its nuclear import by Kap13. Import assays in digitonin-permeabilized cells were performed by using GST-GFP fusions of Pax6 segments. Import assays were performed as described in the legend to Fig. 2. (A) Fluorescence microscopy images. (B, left side) Diagram of the Pax6 segment. The gray bars represent the homeodomain, and the upper short black bars indicate the positions of the two basic clusters. (B, right side) Average fluorescence intensity of the nuclei, in arbitrary units, for experiments represented in frames c, e, g, and i of panel A. Error bars indicate standard errors of the means. The import of segment 224 to 288, which contains the C-terminal but not the N-terminal basic cluster, is reduced to 20% compared to that of the segment 208 to 288, which includes both basic clusters (frame e versus frame a, P = 1.2 × 10−22, Student t test, two-tailed). The import of the segment 162 to 238, which contains the N-terminal but not the C-terminal cluster, is reduced to negligible levels (c versus a, P = 2.7 × 10−19). The segment 269 to 288, which extends beyond the C-terminal basic cluster, also contributes to the NLS activity—in its absence the import is decreased by 40% compared to that of the segment 208 to 268 (g versus a, P = 2.6 × 10−8). The experiment was repeated with similar results.
Binding of Pax6 to Kap13 is reduced when either basic cluster (208 to 214 or 261 to 267) of Pax6, located at each end of the homeodomain, is deleted.
Pax6 constructs lacking nucleotides that encode amino acids 208 to 214 or 261 to 267 were expressed in E. coli. These constructs, along with wild-type Pax6, were expressed as GST fusions, purified, immobilized on glutathione-Sepharose beads, and incubated with recombinant Kap13. Pax6 has reduced binding to Kap13 when these regions are deleted from Pax6. Figure 5 shows that the deletion of amino acids 208 to 214 decreased binding to approximately 60% compared to that of the wild type. Deletion of amino acids 261 to 267 led to an even stronger reduction, as did the deletion of both regions. These basic clusters are part of the homeodomain, which spans amino acid residues 210 to 269. These data, together with those presented in Fig. 3 and 4, support the conclusion that at least part of the homeodomain is directly involved in Kap13 binding.
Kap13 interacts in vivo with Pax6 but not with the Pax6 mutant lacking regions 208 to 214 and 261 to 267.
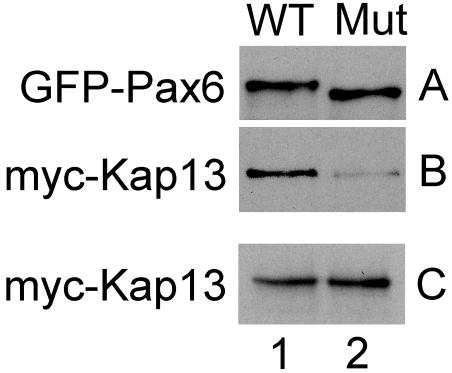
Constructs for expression of Myc-tagged Kap13 and GFP-tagged Pax6 were cotransfected into 293T cells. Twenty-four hours posttransfection, transfected cells were harvested and lysates were used for coimmunoprecipitation (see Material and Methods). Wild-type and mutant Pax6 were immunoprecipitated equally well by the anti-Pax6 monoclonal antibody immobilized on protein G Sepharose beads (Fig. 6A, columns 1 and 2). Kap13 coimmunoprecipitated with wild-type Pax6 but not with the mutant Pax6 (Fig. 6B, columns 1 and 2). Panel C shows that Kap13 was equally expressed in both lysates (columns 1 and 2). No signal for Myc-Kap13 or GFP-Pax6 was detected in cells that were not transfected with the respective plasmids (data not shown).
FIG. 6.
Myc-Kap13 interacts in vivo with Pax6 but not with the Pax6 mutant (Mut) lacking regions 208 to 214 and 261 to 267. Lysates from cells cotransfected with constructs for expression of Myc-Kap13 and GFP-Pax6 were used for coimmunoprecipitation. (A) Wild-type (WT) and mutant Pax6 were immunoprecipitated equally well by the anti-Pax6 monoclonal antibody immobilized on protein G Sepharose beads. (B) Kap13 coimmunoprecipitated with wild-type Pax6 but not with the mutant Pax6. (C) Kap13 was expressed equally in both lysates.
Two other homeodomain proteins, Pax3 and Crx, which contain segments similar to the Pax6 NLS, bind to and are imported into the nuclei by Kap13.
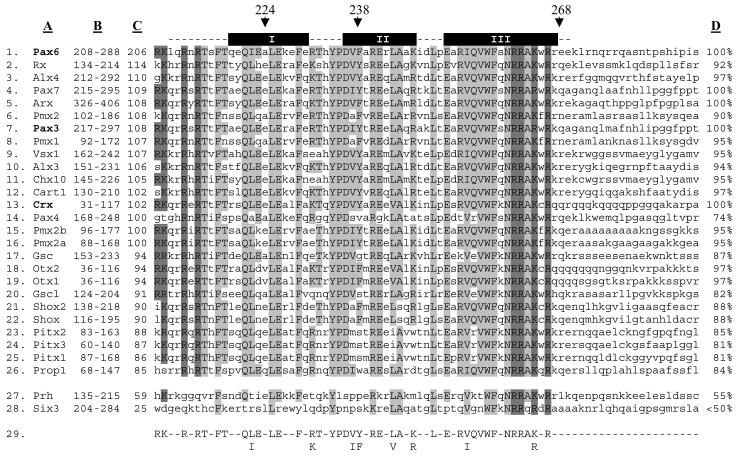
All paired-type transcription factors show similarity to the NLS region of Pax6 (Fig. 7). The high degree of conservation suggested that Kap13 might function as an import carrier for other members of the family. In order to verify this hypothesis we tested Pax3 and Crx for binding to Kap13. We also tested two non-paired-type homeodomain proteins, Prh and Six3, which have lower similarity to Pax6 in the NLS region (Fig. 7, positions 27 and 28).
FIG. 7.
The paired-type homeodomain family contains a segment similar to the Pax6 domain recognized by Kap13. The paired-type homeodomain transcription factors are listed in positions 1 to 26. Two non-paired-type transcription factors, which do not interact with Kap13, are listed in positions 27 to 28. Position 29 shows the consensus formed between the three paired-type homeodomain proteins shown to be imported by Kap13 (Pax6, Pax3, and Crx, which are in bold). Column A lists the homeodomain transcription factors. Column B shows the position of sequence represented in the figure. Column C shows the level of similarity each homeodomain transcription factor has with the segment 208 to 288 of Pax6, as given by the score computed by the Blosum80 algorithm. The homeodomain is depicted as dashed lines above the Pax6 sequence; the alpha helices are denoted by black boxes labeled I, II, and III. Arrows denote positions 224, 238, and 268 of Pax6, which are mentioned in the legends to Fig. 3 and 4. Column D shows the percentage of similarity each protein has to the consensus (position 29). Amino acids that are identical with the consensus are in capital letters and are shaded. The darker shade designates the basic amino acids at each end of the homeodomain. Amino acids that are not represented in the consensus are in lowercase.
The homeodomain proteins Pax6, Pax3, Crx, Prh, and Six3 were expressed in E. coli as GST fusions, immobilized on glutathione-Sepharose beads, and incubated with recombinant Kap13. Pax6, Pax3, and Crx bind to Kap13, while Prh and Six3 do not (Fig. 8).
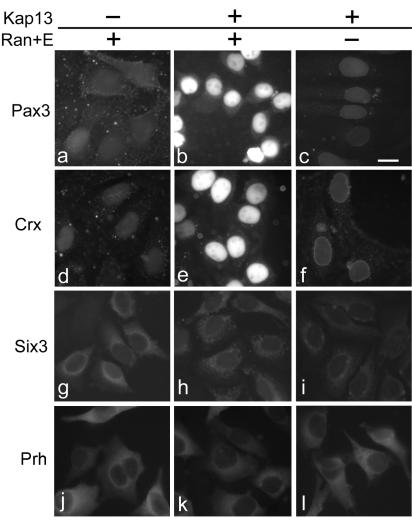
The potential cargoes were tested in import assays by using digitonin-permeabilized cells. Kap13 mediates import of Pax3 and Crx in a Ran- and energy-dependent manner, while Prh and Six3 are not imported by Kap13 (Fig. 9).
FIG. 9.
Kap13 mediates nuclear import of Pax3 and Crx but not Six3 or Prh in digitonin-permeabilized cells. FITC-labeled GST-Pax3 and GST-Crx are imported into the nuclei by Kap13 in the presence (b and e) but not in the absence (c and f) of Ran, RanBP1, RanGAP, NTF2, and an energy-regenerating system. Pax3 and Crx are not imported into the nuclei in the absence of Kap13 (a and d). Kap13 does not mediate import of FITC-labeled GST-Six3 (h) or GST-Prh (k). Bar, 20 μm.
DISCUSSION
This study shows that Kap13 mediates nuclear import of three members of the paired-type homeodomain transcription factor family: Pax6, Pax3, and Crx.
Kap13 binds to the segment 208 to 288 of Pax6 and mediates its entry into the nucleus. This region overlaps with the homeodomain (210 to 269), which is not surprising when one takes into account that for 90% of the proteins for which both the NLS and the DNA-binding region are known the motifs overlap (18). Figure 4 shows that the region 162 to 238 of Pax6 that contains the N-terminal basic cluster cannot promote import in digitonin-permeabilized cells. Thus, Kap13 cannot utilize this region as an NLS alone. The region 224 to 288, which includes the C-terminal cluster, cannot be utilized as an efficient NLS either, because it is only imported 20% as efficiently as the segment 208 to 288 is, which includes both basic clusters. In addition, the direct binding assays (Fig. 5) show that Kap13 binds 40% less efficiently to Pax6 when the region 208 to 214 is deleted, and the reduction is even more pronounced when the region 261 to 267 is deleted. This directly shows that each of these regions has a significant contribution to binding to Kap13. The fact that removal of amino acids 208 to 214 from Pax6 reduces binding to Kap13 by 40% but exclusion of amino acids 208 to 223 reduces import by 80% compared to the 208 to 288 construct suggests that amino acids within segment 215 to 223 also contribute to binding to Kap13. In addition, or alternatively, the two basic residues in segment 201 to 207 may replace and partially compensate for the deleted basic residues in segment 208 to 214. Taken together, the data show that Kap13 recognizes an NLS that is located within a segment of 80 amino acid residues that includes the homeodomain of Pax6 and that both segments of the homeodomain, 208 to 214 and 261 to 267, are important for binding to Kap13, but neither is sufficient to confer the binding and import capacity of the complete NLS.
It remains possible that other Kaps may also mediate import of Pax6 in addition to Kap13 in vivo. These potential Kaps may utilize the same NLS that Kap13 does or, alternatively, may utilize different sequences.
Glaser et al. noted that the regions located at the N-terminal (residues 207 to 212) and C-terminal (residues 261 to 276) ends of the Pax6 homeodomain resemble the monopartite and bipartite classical NLS, respectively (37). Our data show that the Kap α2 and β1 heterodimer, which imports the monopartite and bipartite classical NLS efficiently, cannot mediate efficient import of Pax6. In addition, our import assay data show that the import capacity of the 80-residue NLS is fourfold higher than the sum of the import activities of the segments 162 to 238 and 224 to 288, which separately contain the two basic elements (Fig. 4). This observation indicates that the two basic elements are not independent and redundant NLS but that they act in a cooperative manner, probably in conjunction with other residues, to form the NLS of Pax6.
The important contributions of the homeodomain and, in particular, the N- and C-terminal basic clusters for NLS activity are supported by previous studies on Pax6 and other paired-type homeodomain proteins. It was shown that the segment LKRKLQR of quail Pax6 (equivalent to positions 206 to 212 in human Pax6) is necessary for nuclear import in vivo. For another member of the paired-type homeodomain family, Vsx-1, it was shown that the deletion of the segment QKRKKRR, also equivalent to 206 to 212 of human Pax6, impairs its nuclear localization (54). Mutations of N- or C-terminal basic amino acids of Cart-1 reduce import, and if both basic regions are mutated the import is eliminated completely (30). The region containing amino acid residues 35 to 107 of Crx, which contains the entire homeodomain, was shown to be required for nuclear import (15). It should be mentioned that none of the above-mentioned studies identified the Kap responsible for the nuclear import of the paired-type homeodomain factors.
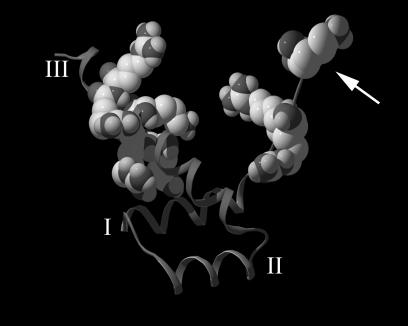
Figure 10 depicts the three-dimensional structure of the Pax6 homeodomain, inferred by threading the Pax6 sequence through the known three-dimensional structure of Prd bound to DNA (95). The structure is composed of a helix-turn-helix comprised of three alpha helices. The model in Fig. 10 shows that the two basic clusters at each end of the homeodomain are much closer in space than anticipated from the primary sequence: the minimal distance between the two basic clusters, which occurs between the side chains of R214 and K264, is approximately 8 Å (for R212 and R262 it is approximately 16 Å). This distance is considerably smaller than the 65 Å that separates two amino acids that are 46 residues apart in an alpha helix. If Pax6 binds to Kap13 in a fashion similar to that of Pax6 binding to DNA, it might suggest that the 46 intervening residues between the N- and C-terminal basic clusters have a structural role of exposing the two basic clusters in the proper position and orientation for contact with Kap13. It is, however, quite possible that Kap13 makes contact with residues within the region 208 to 288 besides the N- and C-terminal basic clusters, and these residues could have significant contributions to the NLS activity. Additionally, the idea that the intervening 46 residues could be simply replaced by any sequence without affecting the NLS function cannot be excluded. However, this would be unexpected, because the homeodomain is likely to have a complex tertiary structure even when not bound to DNA.
FIG. 10.
Three-dimensional structure of the Pax6 homeodomain generated by threading the Pax6 sequence through the known crystal structure of the Drosophila Prd homeodomain bound to DNA (DNA not shown). The basic residues located at the extremities of the homeodomain are represented as space-filling structures. The helical segments represent the three alpha helices of the homeodomain and are denoted by I, II, and III. The conformation shows that the basic clusters at each end of the homeodomain are much closer in space than would be expected from the primary sequence. The arrow points to the N-terminal end of the homeodomain.
Taking into account that Kap13 binds to a region containing the homeodomain of Pax6 and that this region is highly conserved within the paired-type homeodomain family, we tested other family members for binding to Kap13 and for Kap13-mediated import. Indeed, other paired-type family members do bind and are imported into the nuclei of digitonin-permeabilized cells. Figure 7 lists 26 human paired-type homeodomain transcription factors in decreasing order of similarity to the Pax6 region 208 to 288. The basic clusters (208 to 214 or 261 to 267), shown to be directly important for Kap13 binding, are highly similar among these factors, as is the rest of the homeodomain. In order to reveal which residues may be essential for the NLS activity we developed a consensus based on the three factors that we showed to be imported by Kap13. The three factors used to build the consensus are not those most similar to Pax6, as shown by the list of scores in column C of Fig. 7. Residues from the consensus are potentially important for binding to Kap13. These residues are highly conserved among the paired-type family. As column D of Fig. 7 shows, most of the paired-type homeodomain family members have an 85% or greater similarity to the consensus, which suggests that most paired-type homeodomain proteins may bind to Kap13 and be imported into the nuclei by Kap13.
The fact that we did not find any other paired-type homeodomain proteins in our yeast two-hybrid screen can be explained by a combination of factors. It is possible that other paired-type homeodomain proteins were not present in the cell type used to generate the library for the yeast two-hybrid screening—paired-type homeodomain proteins have a narrow developmental and cell type expression. Alternatively, the fusion proteins in yeast are toxic or do not fold correctly (17).
It does not appear that Kap13 mediates the import of homeodomain proteins in general, as indicated by the fact that the homeodomain factors Prh and Six3, which do not belong to the paired-type family, do not interact with and are not imported by Kap13. Prh is only 55% similar to the above-mentioned consensus, and Six3 is less than 50% similar. Prh contains, in conserved form, the region (261 to 267) of Pax6, which abolished binding to Kap13 when deleted. This further shows that although this region is important for binding to Kap13, Pax6 and other paired-type factors contain additional elements that confer binding to Kap13 and these elements are absent from Prh. It remains to be investigated if Kap13 can mediate the import of other classes of homeodomain-containing proteins.
Interestingly, segment 269 to 288 of Pax6, which extends beyond the homeodomain, is not conserved among paired-type homeodomain proteins but yet enhances binding to Kap13 and significantly increases the nuclear import efficiency in digitonin-permeabilized cells. It remains a possibility that the corresponding region in other family members confers enhanced binding to Kap13 due to similar three-dimensional features despite the lack of any obvious similarity at the primary structure level. Another possibility is that the import enhancement conferred by this region is limited to Pax6. If this is the case, it remains unclear what the physiological relevance of the enhanced import would be because Pax3 and Crx, which do not show any similarity within this region, are clearly imported into the nuclei by Kap13 in digitonin-permeabilized cells.
A previous study reported that Kap α2 binds in vitro to Pax2, Pax3, Pax5, Pax6, and Pax8 (51). The binding was tested by using Pax factors expressed in a reticulocyte lysate system and not purified proteins. It was not investigated if Kap β1 can bind to the complex, and if Kap β1 and α2 can mediate the import of the Pax factors into nuclei. As shown in Fig. 2, we found that Kap α2 and β1 mediate weak import in a fraction of the cell population. Pax2, Pax5, and Pax8 do not belong to the paired-type homeodomain family—they do not encode a functional homeodomain but instead encode only a remnant N-terminal half of the homeodomain (53, 93) and thus are unlikely to be cargoes for Kap13.
A previous study reported that Kap13 acts as an import factor for SUMO-1/sentrin-conjugating enzyme UBC9 and for MGN (a protein shown to play multiple roles in Drosophila embryogenesis) in complex with RBM8 (Y14), an MGN-binding protein which is loaded onto mRNA as a result of the splicing reaction. It was also found that Kap13 has export activity towards the translation initiation factor eIF1A (63). UBC9 does not contain any basic cluster that is similar to the homeodomain NLS. RBM8, which is the member of the RBM8-MGN complex that makes direct contact with Kap13, contains two large domains of high charge density (8 out of 12 and 13 out of 23 basic residues), but they are situated more than 100 residues apart and do not show spacing patterns similar to the basic clusters of the homeodomain NLS. In conclusion, the two previously identified cargoes imported by Kap13 do not contain sequences similar to the NLS identified by this study. It is possible, however, that the three-dimensional structures of these proteins contain elements that are similar to the NLS of the homeodomain factors. Alternatively, these proteins may bind to different segments of Kap13. An intriguing question remains as to whether there is any functional connection among the paired-type homeodomain factors UBC9 and MGN, which would explain their nuclear import via the same pathway.
Pax6 is expressed in the developing central nervous system and various ocular tissues and is considered the master control factor for the morphogenesis and evolution of the eye (34, 56, 70, 93). Loss-of-function mutants of Pax6 lack eyes, and ectopic expression of Pax6 can direct ectopic eye development in Xenopus and Drosophila organisms (3, 16, 39, 88). Pax3 is among the first transcription factors expressed in the embryo, being a principal regulator of neurogenic and myogenic progenitor cell specification, migration, and embryonic segmentation (9, 61). Crx plays a crucial role in photoreceptor differentiation and development, and it is also expressed in cells within the pineal gland (14, 27, 31, 57).
Interestingly, many of the paired-type homeodomain proteins are associated with genetic diseases. Haploinsufficiency of Pax6 causes the human genetic disorder Aniridia, which results in the clinical phenotype of hypoplasia of the iris (71, 89). Mutations of Pax6 also cause Peter's Anomaly, which causes defects of the anterior chamber of the eye and results in corneal opacity (41). Mutations of Pax6 cause the Small Eye phenotype in mice (36, 43, 44). Pax3 haploinsufficiency causes Waardenburg Syndrome, characterized by a wide bridge of the nose, pigmentary disturbances, and in some cases cochlear deafness (4, 66, 87). Mutations in Crx result in Leber Congenital Amaurosis and Cone-Rod Dystrophy 2, both causing visual impairment (27, 28, 83).
As mentioned, some syndromes caused by paired-type homeodomain factors are due to haploinsufficiency effects (22), which means that the correct nuclear concentration of these factors is crucial for normal development. This suggests that any aberration that disrupts nuclear import of these factors could also create disease phenotypes. For instance, any aberration that disrupts Pax6 import could cause Aniridia (35, 81). The Pax6 mutation R208W causes Aniridia (40, 42) and disrupts Pax6 nuclear transport in Cos-7 cells (37). Another study, however, found no change in Pax6 (R208W) localization in quail cells (13). In addition, there have been reports that disease-causing mutations in Crx and Pitx2 affect nuclear import and therefore may contribute to the disease phenotype (25, 52). Our work suggests that mutations within the paired-type homeodomain could impair nuclear import, thereby contributing to the etiology of disease.
Acknowledgments
We are grateful to Elias Coutavas for technical assistance and critical reading of the manuscript and to Maria Georgescu for help with the yeast two-hybrid assay. We thank Peter Scambler for the Pax3 cDNA (60), Aira Kimura for the Crx cDNA (48), Guillermo Oliver for the Six3 cDNA (97), Ivan Topisirovic and Biljana Culjkovic for the Prh cDNA (90), Yuh Min Chook and Nabeel Yaseen for Ran system cofactors, Shickwann Tsai for the EML library used for the yeast two-hybrid, and Tarik Soliman for constructs. We thank Miran Yoon and Marc Kirschner for critical reading of the manuscript. We are grateful to the reviewers for their helpful comments and suggestions.
This work was supported in part by NIH grant R01 GM057569 to A.R.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altmann, C. R., R. L. Chow, R. A. Lang, and A. Hemmati-Brivanlou. 1997. Lens induction by Pax-6 in Xenopus laevis. Dev. Biol. 185:119-123. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin, C. T., C. F. Hoth, J. A. Amos, E. O. da-Silva, and A. Milunsky. 1992. An exonic mutation in the HuP2 paired domain gene causes Waardenburg's syndrome. Nature 355:637-638. [DOI] [PubMed] [Google Scholar]
- 5.Barry, D. M., and S. R. Wente. 2000. Nuclear transport: never-ending cycles of signals and receptors. Essays Biochem. 36:89-103. [DOI] [PubMed] [Google Scholar]
- 6.Bartel, P., and S. Fields. 1997. The yeast two-hybrid system. Oxford University Press, New York, N.Y.
- 7.Bayliss, R., A. H. Corbett, and M. Stewart. 2000. The molecular mechanism of transport of macromolecules through nuclear pore complexes. Traffic 1:448-456. [DOI] [PubMed] [Google Scholar]
- 8.Bednenko, J., G. Cingolani, and L. Gerace. 2003. Nucleocytoplasmic transport: navigating the channel. Traffic 4:127-135. [DOI] [PubMed] [Google Scholar]
- 9.Blake, J., and M. R. Ziman. 2003. Aberrant PAX3 and PAX7 expression. A link to the metastatic potential of embryonal rhabdomyosarcoma and cutaneous malignant melanoma? Histol. Histopathol. 18:529-539. [DOI] [PubMed] [Google Scholar]
- 10.Bogerd, H. P., R. E. Benson, R. Truant, A. Herold, M. Phingbodhipakkiya, and B. R. Cullen. 1999. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J. Biol. Chem. 274:9771-9777. [DOI] [PubMed] [Google Scholar]
- 11.Bonifaci, N., J. Moroianu, A. Radu, and G. Blobel. 1997. Karyopherin beta-2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94:5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bopp, D., M. Burri, S. Baumgartner, G. Frigerio, and M. Noll. 1986. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 47:1033-1040. [DOI] [PubMed] [Google Scholar]
- 13.Carrière, C., S. Plaza, J. Caboche, C. Dozier, M. Bailly, P. Martin, and S. Saule. 1995. Nuclear localization signals, DNA binding, and transactivation properties of quail Pax-6 (Pax-QNR) isoforms. Cell Growth Differ. 6:1531-1540. [PubMed] [Google Scholar]
- 14.Chen, S., Q. L. Wang, Z. Nie, H. Sun, G. Lennon, N. G. Copeland, D. J. Gilbert, N. A. Jenkins, and D. J. Zack. 1997. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19:1017-1030. [DOI] [PubMed] [Google Scholar]
- 15.Chen, S., Q. L. Wang, S. Xu, I. Liu, L. Y. Li, Y. Wang, and D. J. Zack. 2002. Functional analysis of cone-rod homeobox (CRX) mutations associated with retinal dystrophy. Hum. Mol. Genet. 11:873-884. [DOI] [PubMed] [Google Scholar]
- 16.Chow, R. L., C. R. Altmann, R. A. Lang, and A. Hemmati-Brivanlou. 1999. Pax6 induces ectopic eyes in a vertebrate. Development 126:4213-4222. [DOI] [PubMed] [Google Scholar]
- 17.Coates, P. J., and P. A. Hall. 2003. The yeast two-hybrid system for identifying protein-protein interactions. J. Pathol. 199:4-7. [DOI] [PubMed] [Google Scholar]
- 18.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Combet, C., C. Blanchet, C. Geourjon, and G. Deléage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 291:147-150. [DOI] [PubMed] [Google Scholar]
- 20.Coulier, F., C. Popovici, R. Villet, and D. Birnbaum. 2000. MetaHox gene clusters. J. Exp. Zool. 288:345-351. [DOI] [PubMed] [Google Scholar]
- 21.Dasso, M., and R. T. Pu. 1998. Nuclear transport: run by Ran? Am. J. Hum Genet. 63:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Elia, A. V., G. Tell, I. Paron, L. Pellizzari, R. Lonigro, and G. Damante. 2001. Missense mutations of human homeoboxes: a review. Hum. Mutat. 18:361-374. [DOI] [PubMed] [Google Scholar]
- 23.Dingwall, C., J. Robbins, S. M. Dilworth, B. Roberts, and W. D. Richardson. 1988. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J. Cell Biol. 107:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esposito, G., F. Fogolari, G. Damante, S. Formisano, G. Tell, A. Leonardi, R. Di Lauro, and P. Viglino. 1996. Analysis of the solution structure of the homeodomain of rat thyroid transcription factor 1 by 1H-NMR spectroscopy and restrained molecular mechanics. Eur. J. Biochem. 241:101-113. [DOI] [PubMed] [Google Scholar]
- 25.Fei, Y., and T. E. Hughes. 2000. Nuclear trafficking of photoreceptor protein crx: the targeting sequence and pathologic implications. Investig. Ophthalmol. Vis. Sci. 41:2849-2856. [PubMed] [Google Scholar]
- 26.Floer, M., and G. Blobel. 1996. The nuclear transport factor karyopherin beta binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J. Biol. Chem. 271:5313-5316. [DOI] [PubMed] [Google Scholar]
- 27.Freund, C. L., C. Y. Gregory-Evans, T. Furukawa, M. Papaioannou, J. Looser, L. Ploder, J. Bellingham, D. Ng, J. A. Herbrick, A. Duncan, S. W. Scherer, L. C. Tsui, A. Loutradis-Anagnostou, S. G. Jacobson, C. L. Cepko, S. S. Bhattacharya, and R. R. McInnes. 1997. Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91:543-553. [DOI] [PubMed] [Google Scholar]
- 28.Freund, C. L., Q. L. Wang, S. Chen, B. L. Muskat, C. D. Wiles, V. C. Sheffield, S. G. Jacobson, R. R. McInnes, D. J. Zack, and E. M. Stone. 1998. De novo mutations in the CRX homeobox gene associated with Leber congenital amaurosis. Nat. Genet. 18:311-312. [DOI] [PubMed] [Google Scholar]
- 29.Fridell, R. A., R. Truant, L. Thorne, R. E. Benson, and B. R. Cullen. 1997. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J. Cell Sci. 110:1325-1331. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa, K., T. Iioka, M. Morishita, A. Yamaguchi, H. Shindo, H. Namba, S. Yamashita, and T. Tsukazaki. 2002. Functional domains of paired-like homeoprotein Cart1 and the relationship between dimerization and transcription activity. Genes Cells 7:1135-1147. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa, T., E. M. Morrow, and C. L. Cepko. 1997. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91:531-541. [DOI] [PubMed] [Google Scholar]
- 32.Galliot, B., C. de Vargas, and D. Miller. Evolution of homeobox genes: Q50 Paired-like genes founded the Paired class. 1999. Dev. Genes Evol. 209:186-197. [DOI] [PubMed] [Google Scholar]
- 33.Gehring, W. J. 1987. Homeo boxes in the study of development. Science 236:1245-1252. [DOI] [PubMed] [Google Scholar]
- 34.Gehring, W. J. 1996. The master control gene for morphogenesis and evolution of the eye. Genes Cells 1:11-15. [DOI] [PubMed] [Google Scholar]
- 35.Glaser, T., L. Jepeal, J. G. Edwards, S. R. Young, J. Favor, and R. L. Maas. 1994. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat. Genet. 7:463-471. [DOI] [PubMed] [Google Scholar]
- 36.Glaser, T., J. Lane, and D. Housman. 1990. A mouse model of the Aniridia-Wilms tumor deletion syndrome. Science 250:823-827. [DOI] [PubMed] [Google Scholar]
- 37.Glaser, T., D. Walton, J. Cai, J. Epstein, L. Jepeal, and R. L. Maas. 1995. Pax6 mutations in Aniridia, p. 51-82. In J. R. Wiggs (ed.), Molecular genetics of ocular disease, Wiley-Liss, Inc., New York, N.Y.
- 38.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 39.Halder, G., P. Callaerts, and W. J. Gehring. 1995. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267:1788-1792. [DOI] [PubMed] [Google Scholar]
- 40.Hanson, I. M., A. Seawright, K. Hardman, S. Hodgson, D. Zaletayev, G. Fekete, and V. van Heyningen. 1993. PAX6 mutations in aniridia. Hum. Mol. Genet. 2:915-920. [DOI] [PubMed] [Google Scholar]
- 41.Hanson, I. M., J. M. Fletcher, T. Jordan, A. Brown, D. Taylor, R. J. Adams, H. H. Punnett, and V. van Heyningen. 1994. Mutations at the PAX6 locus are found in heterogeneous anterior segment malformations including Peters' anomaly. Nat. Genet. 6:168-173. [DOI] [PubMed] [Google Scholar]
- 42.Hanson, I., A. Churchill, J. Love, R. Axton, T. Moore, M. Clarke, F. Meire, and V. van Heyningen. 1999. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum. Mol. Genet. 8:165-172. [DOI] [PubMed] [Google Scholar]
- 43.Hill, R. E., J. Favor, B. L. Hogan, C. C. Ton, G. F. Saunders, I. M. Hanson, J. Prosser, T. Jordan, N. D. Hastie, and V. van Heyningen. 1991. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature 354:522-525. [DOI] [PubMed] [Google Scholar]
- 44.Hogan, B. L., G. Horsburgh, J. Cohen, C. M. Hetherington, G. Fisher, and M. F. Lyon. 1986. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J. Embryol. Exp. Morphol. 97:95-110. [PubMed] [Google Scholar]
- 45.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jäkel, S., and D. Görlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 48.Kimura, A., D. Singh, E. F. Wawrousek, M. Kikuchi, M. Nakamura, and T. Shinohara. 2000. Both PCE-1/RX and OTX/CRX interactions are necessary for photoreceptor-specific gene expression. J. Biol. Chem. 275:1152-1160. [DOI] [PubMed] [Google Scholar]
- 49.Kohler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Görlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komeili, A., and E. K. O'Shea. 2001. New perspectives on nuclear transport. Annu. Rev. Genet. 35:341-364. [DOI] [PubMed] [Google Scholar]
- 51.Kovac, C. R., A. Emelyanov, M. Singh, N. Ashouian, and B. K. Birshtein. 2000. BSAP (Pax5)-importin alpha 1 (Rch1) interaction identifies a nuclear localization sequence. J. Biol. Chem. 275:16752-16757. [DOI] [PubMed] [Google Scholar]
- 52.Kozlowski, K., and M. A. Walter. 2000. Variation in residual PITX2 activity underlies the phenotypic spectrum of anterior segment developmental disorders. Hum. Mol. Genet. 9:2131-2139. [DOI] [PubMed] [Google Scholar]
- 53.Krauss, S., T. Johansen, V. Korzh, and A. Fjose. 1991. Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature 353:267-270. [DOI] [PubMed] [Google Scholar]
- 54.Kurtzman, A. L., and N. Schechter. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98:5602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lei, E. P., and P. A. Silver. 2002. Protein and RNA export from the nucleus. Dev. Cell. 2:261-272. [DOI] [PubMed] [Google Scholar]
- 56.Lewis, E. B. 1992. The 1991 Albert Lasker Medical Awards. Clusters of master control genes regulate the development of higher organisms. JAMA 267:1524-1531. [PubMed] [Google Scholar]
- 57.Li, X., S. Chen, Q. Wang, D. J. Zack, S. H. Snyder, and J. Borjigin. 1998. A pineal regulatory element (PIRE) mediates transactivation by the pineal/retina-specific transcription factor CRX. Proc. Natl. Acad. Sci. USA 95:1876-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lounsbury, K. M., A. L. Beddow, and I. G. Macara. 1994. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J. Biol. Chem. 269:11285-11290. [PubMed] [Google Scholar]
- 59.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Magnaghi, P., C. Roberts, S. Lorain, M. Lipinski, and P. J. Scambler. 1998. HIRA, a mammalian homologue of Saccharomyces cerevisiae transcriptional co-repressors, interacts with Pax3. Nat. Genet. 20:74-77. [DOI] [PubMed] [Google Scholar]
- 61.Mansouri, A. 1998. The role of Pax3 and Pax7 in development and cancer. Crit. Rev. Oncog. 9:141-149. [DOI] [PubMed] [Google Scholar]
- 62.Melchior, F., B. Paschal, J. Evans, and L. Gerace. 1993. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 123:1649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mingot, J. M., S. Kostka, R. Kraft, E. Hartmann, and D. Görlich. 2001. Importin 13: a novel mediator of nuclear import and export. EMBO J. 20:3685-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore, M. S., and G. Blobel. 1993. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365:661-663. [DOI] [PubMed] [Google Scholar]
- 65.Moore, M. S., and G. Blobel. 1992. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell 69:939-950. [DOI] [PubMed] [Google Scholar]
- 66.Morell, R., T. B. Friedman, S. Moeljopawiro, Hartono, Soewito, and J. H. Asher, Jr. 1992. A frameshift mutation in the HuP2 paired domain of the probable human homolog of murine Pax-3 is responsible for Waardenburg syndrome type 1 in an Indonesian family. Hum. Mol. Genet. 1:243-247. [DOI] [PubMed] [Google Scholar]
- 67.Moroianu, J., M. Hijikata, G. Blobel, and A. Radu. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 92:6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 69.Nishimoto, T. 2000. Upstream and downstream of ran GTPase. Biol. Chem. 381:397-405. [DOI] [PubMed] [Google Scholar]
- 70.Nishina, S., S. Kohsaka, Y. Yamaguchi, H. Handa, A. Kawakami, H. Fujisawa, and N. Azuma. 1999. PAX6 expression in the developing human eye. Br J. Ophthalmol. 83:723-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O'Donnell, F. E., Jr., and H. R. Pappas. 1982. Autosomal dominant foveal hypoplasia and presenile cataracts. A new syndrome. Arch. Ophthalmol. 100:279-281. [DOI] [PubMed] [Google Scholar]
- 72.Peitsch, M. C. 1996. ProMod and Swiss-Model: internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 73.Plafker, S. M., and I. G. Macara. 2000. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pollard, V., W. M. Michael, S. Nakielny, M. C. Siomi, and G. Dreyfuss. 1996. A novel receptor-mediated nuclear protein import pathway. Cell 86:985-994. [DOI] [PubMed] [Google Scholar]
- 75.Qian, Y. Q., M. Billeter, G. Otting, M. Muller, W. J. Gehring, and K. Wuthrich. 1989. The structure of the Antennapedia homeodomain determined by NMR spectroscopy in solution: comparison with prokaryotic repressors. Cell 59:573-580. [DOI] [PubMed] [Google Scholar]
- 76.Qian, Y. Q., K. Furukubo-Tokunaga, D. Resendez-Perez, M. Muller, W. J. Gehring, and K. Wuthrich. 1994. Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain. J. Mol. Biol. 238:333-345. [DOI] [PubMed] [Google Scholar]
- 77.Quimby, B. B., and A. H. Corbett. 2001. Nuclear transport mechanisms. Cell Mol. Life Sci. 58:1766-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 79.Roth, M. B., A. M. Zahler, and J. A. Stolk. 1991. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol. 115:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 81.Schedl, A., A. Ross, M. Lee, D. Engelkamp, P. Rashbass, V. van Heyningen, and N. D. Hastie. 1996. Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell 86:71-82. [DOI] [PubMed] [Google Scholar]
- 82.Strom, A. C., and K. Weis. 2001. Importin-beta-like nuclear transport receptors. Genome Biol. 2:3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Swain, P. K., S. Chen, Q. L. Wang, L. M. Affatigato, C. L. Coats, K. D. Brady, G. A. Fishman, S. G. Jacobson, A. Swaroop, E. Stone, P. A. Sieving, and D. J. Zack. 1997. Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron 19:1329-1336. [DOI] [PubMed] [Google Scholar]
- 84.Swaroop, A., Q. L. Wang, W. Wu, J. Cook, C. Coats, S. Xu, S. Chen, D. J. Zack, and P. A. Sieving. 1999. Leber congenital amaurosis caused by a homozygous mutation (R90W) in the homeodomain of the retinal transcription factor CRX: direct evidence for the involvement of CRX in the development of photoreceptor function. Hum. Mol. Genet. 8:299-305. [DOI] [PubMed] [Google Scholar]
- 85.Sweitzer, T. D., D. C. Love, and J. A. Hanover. 2000. Regulation of nuclear import and export. Curr. Top. Cell. Regul. 36:77-94. [DOI] [PubMed] [Google Scholar]
- 86.Talcott, B., and M. S. Moore. 1999. Getting across the nuclear pore complex. Trends Cell Biol. 9:312-318. [DOI] [PubMed] [Google Scholar]
- 87.Tassabehji, M., A. P. Read, V. E. Newton, R. Harris, R. Balling, P. Gruss, and T. Strachan. 1992. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature 355:635-636. [DOI] [PubMed] [Google Scholar]
- 88.Tomarev, S. I., P. Callaerts, L. Kos, R. Zinovieva, G. Halder, W. Gehring, and J. Piatigorsky. 1997. Squid Pax-6 and eye development. Proc. Natl. Acad. Sci. USA 94:2421-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ton, C. C., H. Hirvonen, H. Miwa, M. M. Weil, P. Monaghan, T. Jordan, V. van Heyningen, N. D. Hastie, H. Meijers-Heijboer, M. Drechsler, et al. 1991. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell 67:1059-1074. [DOI] [PubMed] [Google Scholar]
- 90.Topisirovic, I., B. Culjkovic, N. Cohen, J. M. Perez, L. Skrabanek, and K. L. Borden. 2003. The proline-rich homeodomain protein, PRH, is a tissue-specific inhibitor of eIF4E-dependent cyclin D1 mRNA transport and growth. EMBO J. 322:689-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsai, S., S. Bartelmez, E. Sitnicka, and S. Collins. 1994. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 8:2831-2841. [DOI] [PubMed] [Google Scholar]
- 92.Tsao, D. H., J. M. Gruschus, L. H. Wang, M. Nirenberg, and J. A. Ferretti. 1995. The three-dimensional solution structure of the NK-2 homeodomain from Drosophila. J. Mol. Biol. 251:297-307. [DOI] [PubMed] [Google Scholar]
- 93.Walther, C., and P. Gruss. 1991. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development 113:1435-1449. [DOI] [PubMed] [Google Scholar]
- 94.Weis, K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112:441-451. [DOI] [PubMed] [Google Scholar]
- 95.Wilson, D. S., B. Guenther, C. Desplan, and J. Kuriyan. 1995. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82:709-719. [DOI] [PubMed] [Google Scholar]
- 96.Zhang, C., N. B. Sweezey, S. Gagnon, B. Muskat, D. Koehler, M. Post, and F. Kaplan. 2000. A novel karyopherin-beta homolog is developmentally and hormonally regulated in fatal lung. Am. J. Respir. Cell Mol. Biol. 22:451-459. [DOI] [PubMed] [Google Scholar]
- 97.Zhu, C. C., M. A. Dyer, M. Uchikawa, H. Kondoh, O. V. Lagutin, and G. Oliver. 2002. Six3-mediated auto repression and eye development requires its interaction with members of the Groucho-related family of co-repressors. Development 129:2835-2849. [DOI] [PubMed] [Google Scholar]