Abstract
The next generation of QTL (quantitative trait loci) mapping populations have been designed with multiple founders, where one to a number of generations of intercrossing are introduced prior to the inbreeding phase to increase accumulated recombinations and thus mapping resolution. Examples of such populations are Collaborative Cross (CC) in mice and Multiparent Advanced Generation Inter-Cross (MAGIC) lines in Arabidopsis. The genomes of the produced inbred lines are fine-grained random mosaics of the founder genomes. In this article, we present a novel framework for modeling ancestral origin processes along two homologous autosomal chromosomes from mapping populations, which is a major component in the reconstruction of the ancestral origins of each line for QTL mapping. We construct a general continuous time Markov model for ancestral origin processes, where the rate matrix is deduced from the expected densities of various types of junctions (recombination breakpoints). The model can be applied to monoecious populations with or without self-fertilizations and to dioecious populations with two separate sexes. The analytic expressions for map expansions and expected junction densities are obtained for mapping populations that have stage-wise constant mating schemes, such as CC and MAGIC. Our studies on the breeding design of MAGIC populations show that the intercross mating schemes do not matter much for large population size and that the overall expected junction density, and thus map resolution, are approximately proportional to the inverse of the number of founders.
Keywords: Arabidopsis multiparent recombinant inbred lines (AMPRIL), Collaborative Cross (CC), quantitative trait loci (QTL) mapping resolution, identity by descent (IBD), Multiparent Advanced Generation Inter-Cross (MAGIC), multiparental populations, MPP
DISSECTING the genetic architecture of complex traits is a central task in many fields such as animal breeding and crop science. There are mainly two approaches to identify the underlying quantitative trait locus/loci (QTL): QTL mapping in experimental breeding populations and association or linkage disequilibrium mapping in natural and constructed diversity panels (Cavanagh et al. 2008). Although association mapping among apparently unrelated individuals can greatly improve the genetic resolution of causative variants by exploring historical recombinations, it has a major disadvantage of possibly spurious associations due to unknown population structure if not accounted for appropriately (Voight and Pritchard 2005). Although well-controlled QTL mapping populations do not have such a problem, the traditional population designs such as recombinant inbred lines (RILs) and nearly isogenic lines (NILs) have low mapping resolution because of few captured recombinations and low segregation probability of QTL among only few founder lines. We use abbreviations like RIL and NIL in a rather loose way to refer to single and multiple lines as well a population as a whole.
To overcome the limitations of traditional biparental QTL mapping and association mapping, the next generations of mapping populations have been designed with multiple founders, where from one to a number of generations of intercrossing are introduced prior to the inbreeding phase to increase accumulated recombinations and thus mapping resolution. These population resources include the mouse Collaborative Cross (CC) (Churchill et al. 2004), the Arabidopsis MAGIC lines (Kover et al. 2009), and the Arabidopsis multiparent RIL (AMPRIL) (Huang et al. 2011).
The genome of an individual sampled from mapping populations is a random mosaic of founder origins, and it is necessary to model the ancestral origins and reconstruct them for downstream QTL mapping. For example, the reconstruction outputs such as posterior probabilities of ancestral origins at putative QTL are used as genetic predictors in the mixed model of QTL mapping (Huang et al. 2011). The primary goal of this article is to construct a general model of ancestral origins along two homologous chromosomes that can be applied for both traditional and next generation mapping populations.
Modeling of ancestral origins was pioneered by Haldane and Waddington (1931) in terms of two-locus diplotype (two ordered haplotypes) probabilities. Their recurrence relations were derived for 2-way RILs by selfing or sibling (brother–sister) mating and for NILs by repeated backcrossing. The closed-form solutions were given mainly for the final completely homozygous lines. Johannes and Colome-Tatche (2011) derived the two-locus diplotype probabilities for 2-way RILs by selfing. Broman (2012a) extended the approach of Haldane and Waddington (1931) to 2n-way RILs by sibling mating and obtain only numerical recipes for calculating two-locus diplotype probabilities. The haplotype probabilities for 2n-way RILs have been calculated by Broman (2005) and Teuscher and Broman (2007) and those for advanced inter-cross populations by Darvasi and Soller (1995), Winkler et al. (2003), and Broman (2012b).
For next generation mapping populations, the complexity of modeling ancestral origins along two autosomal chromosomes increases very fast with the number of founders. For example, for four-way RILs by sibling mating, there are 700 two-locus diplotype states even after accounting for various symmetries (Broman 2012a). To reduce this complexity, we assume that each founder contributes on average equally to the sample’s genomes. Thus founders’ identities do not matter, and we need only to model the change of ancestral origins along chromosomes.
This work builds on the theory of junctions in inbreeding (Fisher 1949, 1954). A junction is defined as a boundary point on chromosomes where two distinct ancestral origins meet, and the boundary points that occur at the same location along multiple chromosomes are counted as a single junction. Two chromosomes at a locus are identical by descent (IBD) if they have the same ancestral origins. A junction as defined on two homologous chromosomes is called internal when the two pairs of chromosome segments before and after the junction are either both IBD or both non-IBD; otherwise we call a junction external. Fisher (1949, 1954) and Bennett (1953, 1954) developed the theory of junctions for repeated sibling, selfing, and parent–offspring mating. Stam (1980) extended the theory to a finite random mating population without selfing by using recurrence relations of three-gene IBD probabilities, instead of Fisher’s elaborate generation matrix for various mating types. The author obtained the expected density (per morgan) of external junctions on two chromosomes at any generation.
We generalize the Stam approach to model the ancestral origins in mapping populations. The expected densities for all junction types are systematically investigated, rather than only external junctions. The breeding population size and mating schemes may vary from one generation to the next. The number of offspring may be non-Poisson distributed or even fixed, and there may be self-fertilizations.
The closed-form expressions for the map expansion R, that is, the expected junction density on one chromosome, and the overall expected density ρ on two homologous chromosomes are derived in our approach. The latter can be used as a measure of QTL map resolution, since both simulation and analytic studies (Darvasi and Soller 1995; Weller and Soller 2004) have shown that mapping resolution is inversely proportional to ρ in traditional mapping populations. The other factors affecting map resolution include sample size and QTL effects. Thus, we may compare different QTL mapping populations or breeding designs for a particular population type, in terms of ρ.
In this article, we build a general theoretical framework for modeling ancestral original processes along two homologous autosomal chromosomes and apply it to study breeding designs of mapping populations; however, we do not use it to infer ancestral origins from observed molecular marker data. In the Methods section, we describe a continuous time Markov model where the rate matrix can be deduced from various expected junction densities. To evaluate the theoretical predictions for expected junction densities, various types of breeding populations with multiple stages of constant mating schemes are simulated. In addition, the breeding designs of MAGIC populations are studied, by varying the number of founders and the intercross mating schemes, from the aspect of overall expected junction density ρ and thus mapping resolution. Finally, we discuss the model assumptions such as Markov approximations and random mating schemes.
Methods
Continuous time Markov chain
Consider a diploid population founded at generation 0, and the generations are nonoverlapping. The founder population consists of L fully inbred or L/2 outbred founders. The mating scheme ℳt describes how the population of size Nt at generation t produces the next generation. We first focus on two homologous autosomes from monoecious populations without selfing or equivalently dioecious populations with equal numbers of females and males. The monoecious populations with random selfing can be easily derived from nonselfing populations. See Supporting Information, Table S1 for a list of symbols used in this article.
For an individual randomly sampled from a breeding population, we model the ancestral origin processes along its two homologous chromosomes by a continuous time Markov chain, the time parameter being genetic distance along chromosomes. We assign a unique founder genome label (FGL) to the whole genome of each inbred founder or to each of the two haploid genomes of each outbred founder. The ancestral origins on two chromosomes at a single locus are represented by an ordered pair, each taking one of L ≥ 2 possible FGLs. The ancestral origins at the leftmost locus follow the stationary distribution of the reversible Markov chain, and thus the ancestral origin process does not depend on the direction of chromosomes.
Due to the Markov assumption, it is sufficient to consider two-locus diplotypes and their marginal single-locus genotypes. We assume that founders are exchangeable so that the distribution of ancestral origins does not depend on the founders’ identities. As a result, single-locus genotypes and two-locus diplotypes of ancestral origins are reduced into IBD configurations, and each of the L FGLs has equal probability to be the ancestor of genes that are IBD. Let (ab), (abc), and (abcd) be two-, three-, and four-gene IBD configurations, respectively. Set a = 1, and b, c, … are set in order. If the focus gene is IBD to a previous gene, the same integer label is assigned. Otherwise, one plus the maximum of the previous gene labels is assigned. We use genes and gene labels interchangeably if there are no ambiguities.
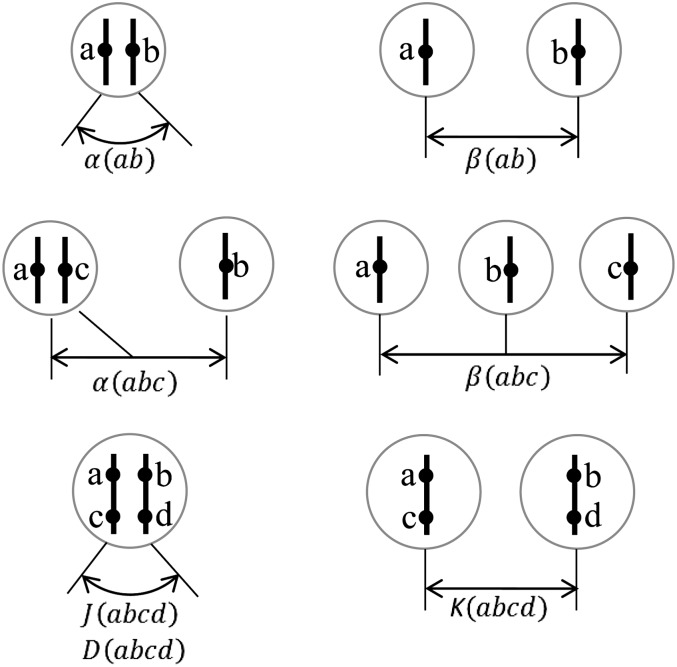
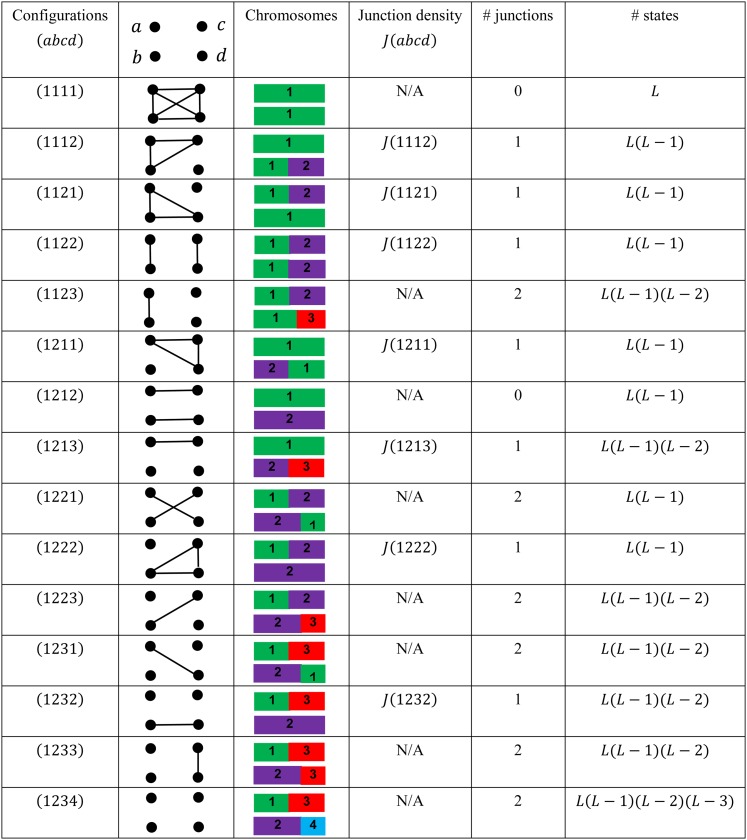
Let D(abcd) be the two-locus probability of the IBD configuration (abcd) given that a(b) is the maternal (paternal) derived gene at one locus, c(d) is the maternal (paternal) derived gene at the other locus, and haplotypes ac and bd are in a single individual (Figure 1). For the number of FGLs L ≥ 4, there are 15 two-locus IBD configurations (Figure 2), the same as single-locus configurations in a pair of individuals (Nadot and Vayssiex 1973). In the limit that the genetic distance δ between two loci (in morgans) goes to zero, there is at most one junction between two loci. Thus D(abcd) = 0 as δ → 0 for the six IBD configurations (1123), (1221), (1223), (1231), (1233), and (1234) with more than one junction between two loci (Figure 2).
Figure 1.
Quantities defined for two-, three-, and four-gene IBD configurations. Solid circles denote genes on chromosomes.
Figure 2.
The seven types of junctions and their relations to the 15 two-locus IBD configurations. Different colors represent different ancestral chromosomes. The haplotype ac is maternally derived, and bd is paternally derived. The junction types and their densities are defined only for IBD configurations with one junction between two loci. The last column shows the number of two-locus ancestral origin states given the junction type and the number L ≥ 3 of genome origins and in total L4 possible states of ancestral origins at two loci. The number of junctions for the configuration (1122) is 1 because the two breakpoints are duplicated copies and thus at the same point along chromosomes.
Since a pair of ancestral origins along two chromosomes can change or remain the same, the probabilities for the remaining nine IBD configurations are related as
| (1a) |
| (1b) |
where α(11) and α(12) are the marginal probabilities of IBD and non-IBD at one locus, respectively. Thus, we need only to consider the seven IBD configurations with exactly one junction between two loci, excluding the two no-junction configurations (1111) and (1212), at the limit δ → 0. The relation between two-locus IBD configurations and the seven types of junctions is shown in Figure 2.
Let J(abcd) be the expected number of junctions of type (abcd) per morgan, and it does not depend on the genetic distance δ. According to the theory of the continuous time Markov chain (Norris 1997), the two-locus IBD configuration probability D(abcd) = J(abcd)δ as δ → 0, and the rate matrix of ancestral origin processes is fully determined by J(abcd) for the seven junction types. Since the junction densities do not depend on the direction of chromosomes, we have J(1112) = J(1211) and J(1121) = J(1222). In addition, the two haplotypes ac and bd are exchangeable for autosomes, and we have J(1211) = J(1222) and J(1213) = J(1232).
The map expansion R as a marginal expected junction density on one chromosome is given by
| (2) |
on the maternally derived chromosome, which is the same as the expected density, J(1112) + J(1122) + J(1211) + J(1213) on the paternally derived chromosome (Figure 2), due to the symmetry between the two autosomal chromosomes. The overall expected junction density ρ on two chromosomes in a single individual is given by the sum over the seven expected junction densities. It holds that
| (3) |
since the junctions of type (1122) are double counted. Both R and ρ converge to J(1122) as t → ∞ for the case of complete inbreeding when only junctions of type (1122) exist.
In summary, under the assumption of founder symmetry the model of ancestral origin processes can be described by the initial distribution at the leftmost site of chromosomes via the one-locus non-IBD probability α(12) and the transition rate matrix of continuous time Markov chain that can be deduced from the three independent expected junction densities J(1232), J(1222), and J(1122) (Norris 1997). We may replace one of the three densities by R or ρ, according to Equations 2 and 3. The recurrence relations of these expected densities are given after the next section on the non-IBD probabilities.
Recurrence relations for single-locus non-IBD probabilities
The calculation of the probabilities for four-gene IBD configurations at two loci necessitates the introduction of the probabilities for two- and three-gene IBD configurations at a single locus. In nonselfing populations, it matters whether two homologous genes are in a single individual. Let α(ab) be the single-locus probability of the IBD configuration (ab) given that the homologous genes a and b are in a single individual and β(ab) be the probability given that two genes are in distinct individuals (Figure 1). There are two two-gene IBD configurations, (12) and (11), and they are related by α(11) = 1 − α(12) and β(11) = 1 − β(12).
Without selfing, two homologous genes in one individual at generation t must come from two individuals of the previous generation. The recurrence relations for the two-gene non-IBD probabilities are given by
| (4a) |
| (4b) |
where st is the coalescence probability that two genes come from a single individual of the previous generation t − 1 given that they are in distinct individuals at generation t, and the fraction 1/2 refers to the probability that the two genes are the copies of a single gene given that they are in a single individual.
We denote by α(abc) the probability of the IBD configuration (abc) given that two particular homologous genes are in one individual and the third gene is in another individual and by β(abc) the probability given that each gene is in one of three distinct individuals (Figure 1). We set two genes a and c in a single individual and gene b in another (Figure 1). This is the case in deriving the expected densities in the next section, where the configuration probability αt−1(abc) of the previous generation t − 1 contributes in forms of new junctions to Jt(abcd) [except Jt(1122)] in the current generation.
Let qt be the coalescence probability that one particular gene comes from one individual and the other two genes come from another individual of the previous generation t − 1 given that the three genes are in distinct individuals at generation t. The recurrence relations for the three-gene non-IBD probabilities are given by
| (5a) |
| (5b) |
where the factor 2 in Equation 5a denotes that two possible pairs of genes come from a single individual of the previous generation, and they are the pair a and b and the pair c and b, excluding the pair a and c because of nonselfing; the factor 3 in Equation 5b denotes that three possible pairs of genes come from a single individual of the previous generation. The term in parentheses (1 − st − 2qt) is the probability that three genes come from three distinct individuals of the previous generation t − 1 given that they are in distinct individuals at generation t, the probability 1 − st that one pair of genes comes from two distinct individuals minus the probability 2qt that the third gene and either gene of the pair come from a single individual of the previous generation.
In addition to the two-gene IBD configurations, there is only one independent three-gene IBD configuration (abc) (Cockerham 1971). The probability α(122) is necessary in deriving the expected density J(1222), and it can obtained from the relation
| (6) |
where αt(1_2) = αt(12) is the marginal non-IBD probability between genes a and c. Since αt(112) = αt(122) due to the symmetry between genes a and c in a single individual, we obtain αt(122) = (αt(12) − αt(123))/2.
Recurrence relations for expected junction densities
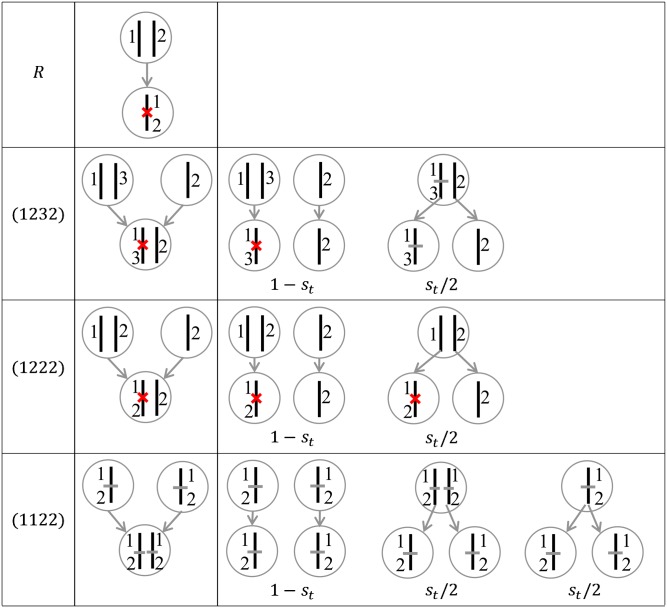
The accumulation of recombination events over many generations leads to genetic map expansion R, which is defined as the expected junction density (per morgan) on a randomly chosen autosomal chromosome. The derivation of the recurrence relation for R directly follows Fisher’s theory of junction (Fisher 1954): a new junction is formed whenever a recombination event occurs between two homologous chromosomes that are non-IBD at the location of a crossover (Figure 3). Thus we have (MaCleod et al. 2005, appendix by P. Stam)
| (7) |
which shows clearly how the inbreeding slows down the map expansion.
Figure 3.
Schematics of the recurrence relations for the map expansion R and the expected junction densities of the types (1232), (1222), and (1122). Red x’s denote the newly formed junctions in generation t, and gray horizontal bars denote the existing junctions. The center column show the junctions in a single individual of the current generation, and the right column shows the junctions in two distinct individuals of the current generation.
To proceed for two homologous chromosomes, we define K(abcd) as the expected junction density given that the haplotypes ac and bd are in two distinct individuals, in addition to the expected junction density J(abcd) given that the two haplotypes are in a single individual (Figure 1). The contributions to junctions in the current generation come from either existing junctions at the previous generation or a new junction via a crossover event. The recurrence relations of J(abcd) and K(abcd) are analogous to Equations 4a and 4b for α(12) and β(12), in terms of the contribution or surviving of existing junctions at the previous generation. We thus focus on the formation of a new junction.
The schematic illustrations of the recurrence relations for junction types (1232), (1222), and (1122) are shown in Figure 3. The formation of junction type (1232) involves three chromosomes: two parental chromosomes of haplotype ac involving in the crossover and the parental chromosome of haplotype bd. A new junction of type (1232) is formed whenever the three homologous chromosomes at the location of the crossover have the IBD configuration (123). We thus have
| (8a) |
| (8b) |
where the first term on the right-hand side of Equation 8b has only the contribution of the existing junctions for the scenario that the two haplotypes ac and bd come from a single individual of the previous generation.
The recurrence relations for the junctions of type (1222) can be similarly obtained. If the two haplotypes ac and bd come from two distinct individuals of the previous generation, a new junction of type (1222) is formed whenever the three homologous chromosomes at the location of the crossover have the IBD configuration (122). And if they come from a single individual of the previous generation, a new junction of type (1222) involving two homologous parent chromosomes is formed at rate α(12). We obtain
| (9a) |
| (9b) |
which are the same as those obtained by Stam (1980) if we replace αt−1(122) by αt−1(123) according to Equation 6.
For continuous modeling of chromosomes, it is impossible that independent recombination events occur at exactly the same location, and thus a new junction of type (1122) can be formed only by duplicating a chromosome segment. We have
| (10a) |
| (10b) |
where the first term on the right-hand side of Equation 10b refers to the scenario that the two haplotypes ac and bd are the copies of a single chromosome segment of the previous generation. Thus overall density ρ in Equation 3 does not depend on three-gene non-IBD probabilities according to Equation 7 and Equations 10a and 10b.
We focus on calculating the three less complicated densities R, J(1232), and J(1122), and they depend on the non-IBD probabilities α(12) and α(123). The basic model parameters are the two-gene coalescence probability st and the three-gene coalescence probability qt, and they are determined by the population size Nt−1 and the mating scheme ℳt−1 of the previous generation.
Monoecious populations with random selfing
In monoecious populations with random selfing, all the homologous genes are equivalent, and as a result it is not necessary to distinguish whether two homologous genes are in a single individual or in two distinct individuals. It is a Wright–Fisher ideal population but with variable size. We redefine st and qt as unconditional coalescence probabilities regardless of two or three homologous genes being in distinct individuals or not at generation t. Thus st is also the probability that an individual at generation t is produced by selfing. According to Equations 4b and 5b, we have
| (11a) |
| (11b) |
where αt(12) and βt(123) are redefined as unconditional probabilities regardless of the distribution of two or three homologous genes among individuals at generation t.
The recurrence relation for R in Equation 7 remains the same. We similarly redefine J(1232) and J(1122) as unconditional expected junction densities, and their recurrence relations are revised as
| (12a) |
| (12b) |
according to Equations 8b and 10b.
Simulation of breeding populations
To evaluate the theoretical predictions of the expected junction densities, we simulate various mapping populations. Assuming that there is no natural selection, we simulate the genome ancestral origins of a breeding individual by first simulating a breeding pedigree according to the breeding design Ω = {L, Nt, ℳt} and then dropping a FGL on the pedigree. A unique FGL is assigned to the whole genome of each complete inbred founder or to the haploid gamete of each outbred founder. Each descendant gamete is specified as a list of FGL segments determined by chromosomal crossovers. In a diploid breeding population, each gamete consists of a pair of homologous chromosomes of 1 M in length. The number of crossovers between a pair of homologous chromosomes follows a Poisson distribution.
For a mapping population with the particular breeding design, the realized junction densities and IBD probabilities are saved for all individuals in any generation in each simulation replicate, and they are averaged over in total 104 replicates. The breeding pedigrees are fixed or vary across replicates, according to the nature of the mating scheme ℳt. Various mating schemes are used in simulating breeding pedigrees. We use pairing to refer to the intercross mating scheme in 2n-way (n ≥ 1) RILs, where individuals in parent populations are assigned to exclusive pairs, and each pair produces one offspring. The gender is alternative female and male for a dioecious population. Selfing and sibling are used to produce inbred lines at inbreeding stage.
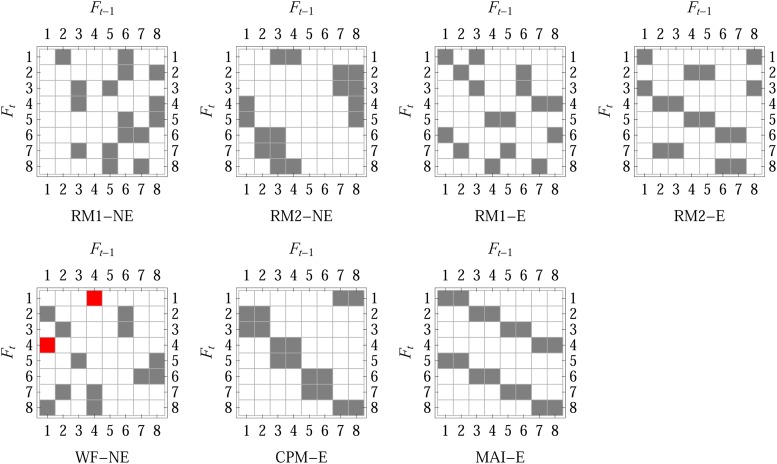
To study the breeding design of MAGIC populations, we introduce seven intercross mating schemes. We denote by RM1 the random mating where each sampling of two randomly chosen distinct individuals produces one offspring, by RM2 the random mating where each sampling of two randomly chosen distinct individuals produces two offspring, and by WF the Wright–Fisher type random mating where each sampling of two randomly chosen individuals—distinct or not—produces one offspring. In addition, we denote by CPM the circular pair mating where each individual mates with its right neighbor to produce one offspring (Kimura and Crow 1963) and by MAI the maximum avoidance of inbreeding mating scheme (Wright 1921). We combine these mating schemes with -NE if each parent contributes a Poisson-distributed number of gametes to the next generation, and -E if each parent contributes exactly two gametes. Thus, we have five random mating schemes, RM1-NE, RM2-NE, RM1-E, RM2-E, and WF-NE, and two regular mating schemes CPM-E and MAI-E, and they are illustrated in Figure 4.
Figure 4.
Intercross mating schemes represented in the format of M-matrix (Boucher and Nagylaki 1988) for a constant population of size eight. Gray entry mij denotes that individual j of the previous generation t − 1 contributes one gamete to its offspring i, and red entries show two-gamete contributions (selfing).
Results
Multistage breeding populations
In the definition of the recurrence relations the mating scheme ℳt and the population size Nt are allowed to vary from one generation to the next. In that case no closed-form analytic expressions for the expected junction densities can be derived. However, experimental breeding (animal or plant) populations for QTL mapping can usually be divided into multiple stages, each stage having constant mating schemes. Thus, we may obtain closed-form solutions for multistage breeding populations by linking results via the initial conditions of each subsequent stage.
We adopt the conceptual framework of the multistage breeding designs introduced by Valdar et al. (2003) in the simulation studies of QTL mapping populations. The design has three stages of mixing, intercross, and inbreeding, and we use subscripts F, I, and II for them, respectively. From the model perspective, the stages are not fundamentally different, and some of them may be merged or dropped. For convenience, and in correspondence to many actual breeding populations, we choose our mixing stage to consist of one generation, where NF founders are mated under scheme ℳF, leading to population F1. These founders are chosen in an attempt to maximize genetic diversity.
The F1 population is intercrossed by mating scheme ℳI for U generations. The intercross stage F1 ∼ FU+1 introduces accumulative recombination events and creates outbred individuals whose genomes are fine-grained mosaics of founder genomes. However, an outbreeder’s genomes are heterogeneous and unique. To produce immortal lines by the paradigm of genotype once and phenotype many times, an inbreeding stage is often followed. The last intercross population FU+1 is inbred by mating scheme ℳII for V generations. To generate nearly fully inbred lines at the last generation g = U + V + 1, the number V of inbreeding generations is usually ≥6 for selfing plants and ≥20 for sibling mating animals.
In the multistage framework, the breeding design can be represented by L, U, V, NF, NI, NII, ℳF, ℳI, and ℳII. It holds that NF = L if founders are completely inbred that are assumed in the following for RIL and MAGIC populations. The population size NI at the intercross stage may be fully determined by NF if, for example, the mixing mating scheme ℳF is diallel crosses (Table 1). The population size NII at the inbreeding stage is 1 for ℳII = selfing and 2 for ℳII = sibling.
Table 1. Coalescence probabilities for mating schemes.
| Mating scheme | Coalescence probability |
Population size |
||
|---|---|---|---|---|
| ℳt−1 | st | qt | Nt−1 | Nt |
| Pairing | 0 | 0 | Even | Nt−1/2 |
| Selfing | 1 | 0 | 1 | Nt−1 |
| Sibling | 1/2 | 0 | 2 | Nt−1 |
| Half diallel | ≥3 | |||
| Full diallel | ≥3 | Nt−1(Nt−1 − 1) | ||
| RM1-NE | st(1 − st) | ≥2 | ≥2 | |
| RM2-NE | ≥2 | ≥4 even | ||
| RM1-E | st | ≥2 | Nt−1 | |
| RM2-E | st | ≥2 even | Nt−1 | |
| WF-NE | st(1 − st) | ≥1 | ≥1 | |
The coalescence probabilities qt are set to zero implicitly for random-selfing populations of size 1 and nonselfing populations of size 2. The coalescence probabilities for the diallel crosses are given only for monoecious populations. In the full diallel, all possible combinations of two different parents are crossed, whereas the half diallel excludes reciprocal crosses. In the coalescence probabilities st for RM2-NE and RM2-E, 1/(Nt − 1) refers to the probability that the parents of the two genes come from a single mating-pair sampling (that results in two offspring), and 1/Nt−1 refers to the probability that the two genes come from the same parent given that their parents come from different samplings.
Constant random mating populations
The closed-form solutions of the recurrence relations in our model can be obtained for each stage of breeding populations, where the mating scheme ℳ and thus the coalescence probabilities s and q are constant. The dependences of the coalescence probabilities on various mating schemes are given in Table 1. We derive explicit expressions for both random-selfing populations and nonselfing populations.
First, consider autosomes in nonselfing populations with population size N ≥ 2. We obtain explicit expressions by solving the linear recurrence relations of order 1 in the Methods section. The eigenvalues of the transition matrix for the two- and three-gene non-IBD probabilities are denoted by
| (13a) |
| (13b) |
where λ1 > λ2, and λ3 ≥ λ4. Set q and βt(123) to zero for N = 2. The explicit expressions are given in the following forms,
| (14a) |
| (14b) |
| (14c) |
| (14d) |
| (14e) |
where the constant coefficients C1, … , C12 are not independent, and they are determined by the eight initial conditions α0(12), β0(12), α0(123), β0(123), J0(1122), K0(1122), J0(1232), and K0(1232). Their expressions are given in File S1. The map expansion Rt in Equation 14c is obtained by accumulative summing of the non-IBD probability αt(12) in Equation 14a. The constant term in Equation 14d is the same as the constant term in Equation 14c since Rt = Jt(1122) in the case of complete inbreeding (that is, t → ∞). The map expansion Rt in Equation 14c has been derived by Stam in the appendix of MaCleod et al. (2005).
Next consider monoecious populations with random selfing. The (largest) eigenvalues λ1 and λ3 for the two- and three-gene non-IBD probabilities are given by
| (15a) |
| (15b) |
respectively. The coalescence probability q and the initial condition α0(123) are set to zero for N = 1. From the recurrence relations of random-selfing populations in the Methods section, we obtain
| (16a) |
| (16b) |
| (16c) |
| (16d) |
| (16e) |
where α0(12), α0(123), R0, J0(1122), and J0(1232) are the initial conditions, and the initial population is chosen so that α0(12) and α0(123) do not depend on the distribution of two or three genes among individuals. The first terms in Equations 16d and 16e refer to the surviving junctions of types (1122) and (1232), respectively.
The results for random-selfing and nonselfing populations can be unified under the following assumptions. If the population size N is large and thus s is small, the eigenvalues λ2 and λ4 are small, and the involved terms can be ignored. Furthermore, if the dependences of the initial conditions on the distributions of genes or haplotypes among individuals are small, the results for nonselfing populations in Equations 14a–14e are simplified to those for random selfing monoecious populations in Equations 16a–16e, where the largest eigenvalues λ1 and λ3 are given in Equations 13a and 13b.
Further approximations can be obtained if the population size N is very large and thus s is very small. In this case, the three-gene coalescence probability q = s + O(s2) as s → 0 (Table 1), where the probability that three genes come from a single individual of the previous generation (multiple collisions) is ignored. Thus, the eigenvalues λ1 = 1 − s/2 + O(s2) according to Equations 13a and 15a, and λ3 = 1 − 3s/2 + O(s2) according to Equations 13b and 15b. This is consistent with the coalescent theory in a large random mating population where the effective population size Ne is given by 1/s and the coalescence rate of three genes is given by 3/(2Ne).
2n-way RIL
RIL populations are a central type of breeding population from which many other types of populations can be derived with regard to the ancestral origin process along chromosomes. For example, one funnel in a CC population is an 8-way RIL population (Churchill et al. 2004), the maize nested associated mapping (NAM) population is a collection of independent 2-way RIL populations obtained by crossing a reference line to a set of diversity lines (Buckler et al. 2009), and the AMPRIL population is a collection of six independent 4-way RILs (Huang et al. 2011). In the framework of multistage breeding populations, the breeding design of RILs is L = NF = 2n, ℳF = ℳI = pairing, and ℳII = selfing or sibling. The founders are assumed to be completely inbred. The number of intercross (pairing) generations U = n − 1 for selfing, U = 0 for 2-way sibling mating, and U = n − 2 for 2n-way (n ≥ 2) sibling mating. We focus on the inbreeding stage and refer to the subscript 0 in the initial conditions in Equations 14a–14e and Equations 16a–16e as generation U + 1.
Since inbreeding is completely avoided at the intercross stage of RILs, the initial conditions at generation U + 1 for the inbreeding stage can be obtained straightforwardly. It holds that αU+1(12) = 1, JU+1(1122) = 0, and JU+1(1232) = RU+1 = U since each generation produces on average one junction per morgan in the case of no inbreeding. Putting these initial conditions into Equations 16a–16e and noting that the coalescence probability s = 1 for selfing at the inbreeding stage (Table 1), we obtain the results at the last generation g = U + V + 1 as shown in Table 2A for RILs by selfing.
Table 2. Results for 2n-way RILs on autosomes at the last generation g = U + V + 1, where U = n − 1 for selfing, U = 0 for n = 1 sibling, and U = n − 2 for n ≥ 2 sibling.
| Quantity | Theoretical prediction |
|---|---|
| A. 2n-way selfing | |
| αg(12) | |
| Jg(1232) | |
| Rg | |
| ρg | |
| B. 2-way sibling | |
| αg(12) | |
| Jg(1232) | 0 |
| Rg | |
| ρg | |
| C. 2n-way (n ≥ 2) sibling | |
| αg(12) | |
| Jg(1232) | |
| Rg | |
| ρg | |
The eigenvalues and
For autosomes in RIL populations obtained by sibling mating, we need extra initial conditions. It holds that KU+1(1122) = 0 and KU+1(1232) = U. In addition, βU+1(12) = 1/2, αU+1(123) = 0, and set βU+1(123) = 0 for 2-way sibling mating; βU+1(12) = 1 and αU+1(123) = βU+1(123) = 1 for 2n-way (n ≥ 2) sibling mating. Putting all these initial conditions into Equations 14a–14e and noting that the coalescence probability s = 1/2 for sibling at the inbreeding stage (Table 1), we obtain the results at the last generation g = U + V + 1 as shown in Table 2, B and C.
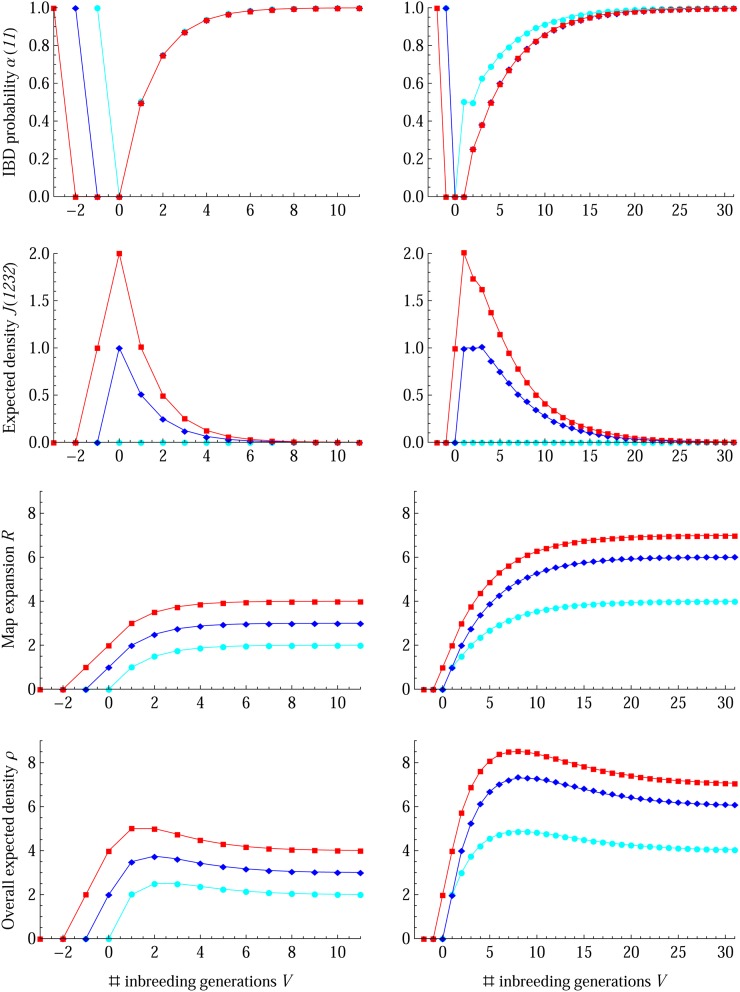
Figure 5 shows that the closed-form solutions in Table 2 fit very well with the forward simulation results for two-, four-, and eight-way RILs by selfing or sibling. The differences between analytic and simulation results are small; see the left column of Figure 6 for the case of eight-way RILs by sibling mating; they may be due to the sampling error in a limited number of simulation replicates (104), but also due to the Markov approximation of ancestral origin processes. In contrast to the monotonic increasing of the map expansion R, a mode exists for the expected density J(1232) and the overall expected density ρ, since the survival rate of old junctions remains the same and the gain rate of new junctions decreases when the population is getting more inbred, as shown in recurrence Equation 8b.
Figure 5.
Results of 2n-way RILs by selfing (left column) and sibling for autosomes (right column). The cyan circles, blue diamonds, and red rectangles denote the simulation results for n = 1, 2, and 3, respectively, and lines denote the theoretical results.
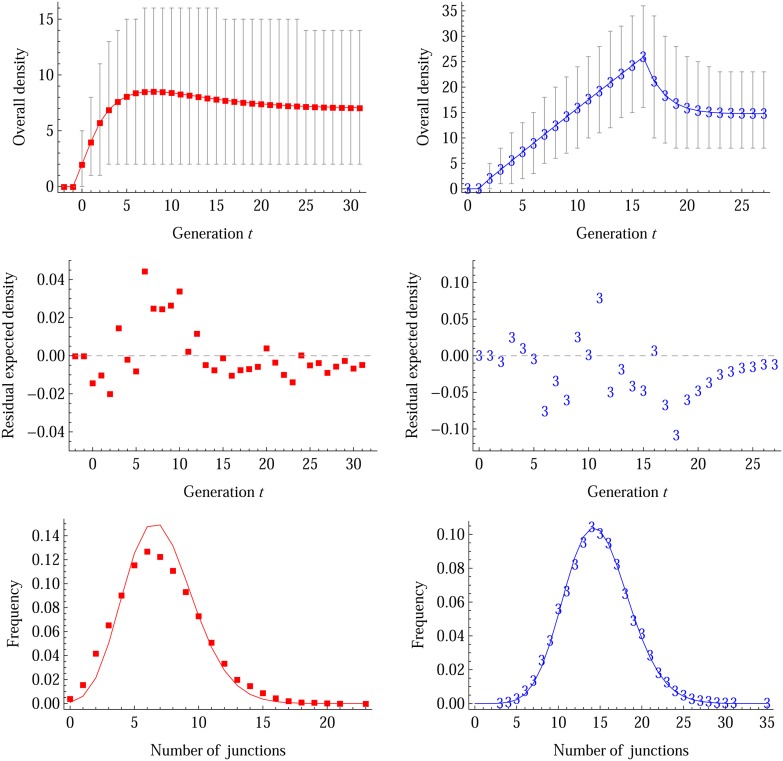
Figure 6.
Simulation evaluations on the distribution of overall junction density. The left column shows the results for eight-way RILs by sibling mating, and the right column shows those for the MAGIC population with NI = 100 and ℳI = (3) RM1-E. In the top row, the solid lines are the theoretical expected densities, the plot markers (red dots and blue “3”) are the means of the simulation results, and the error bars refer to the intervals between the 2.5% and 97.5% quantiles derived from 104 simulation replicates. Note that the error bar shows the variance of the overall junction density, but not the variance of the mean overall junction density (ρ). In the middle row, the residual expected densities refer to the simulated expectations subtracted by the theoretical values. In the bottom row, the solid lines refer to the theoretical Poisson distributions for the number of junctions within 1 M in length at the last generation, and the plot markers show the simulated values.
The map expansions R for 2n-way (n ≥ 1) RILs by selfing or sibling on autosomes, shown in Table 2, equal those given by Broman (2012a), although time origins are different. The AMPRIL population is a special RIL by selfing with n = 2 and V = 3, producing heterogeneous inbred lines (Huang et al. 2011). The overall expected density of this population at the last generation has been calculated to be ρ = 3.625 by using the inheritance vectors for the breeding pedigree, which is consistent with our results in Table 2A.
To guide the design of RIL populations in optimizing QTL map resolution, we calculate the three largest overall expected junction densities ρ and the corresponding inbreeding generations V according to the analytic expressions in Table 2. As shown in Table S2, for the 4-way RIL by selfing in the AMPRIL population, a larger overall expected density ρ = 3.75 would have been obtained if we reduce one generation of selfing (V = 2). In short, the F3 population has largest ρ for 2-way RILs by selfing and the F4 population for 4- or 8-way RILs by selfing; whereas for 2n-way (1 ≤ n ≤ 5) RILs by sibling mating the F9 population has largest ρ.
NIL
We can also derive the results for a biparental NIL population, although it does not fit in our framework. The two homologous chromosomes in a NIL population are no longer symmetric, and we need five independent expected junction densities. Suppose that the hybrid in each generation backcrosses with the fully inbred father parent, and thus there are no junctions on the paternally derived chromosome so that J(1122) = J(1213) = J(1211) = 0. The non-IBD probabilities in a biparental NIL population are the same as those for a two-way RIL by selfing, and the map expansion on the maternally derived chromosome of the hybrid is thus given by Rg = 2[1 − (1/2)V] at the last generation g = U + V + 1 with U = 0 (see Table 2) since Equation 7 still holds. It holds that Jg(1232) = 0 and Jg(1222) = Jg(1121) = Rg/2.
MAGIC populations
The MAGIC population is an advanced breeding population. It generalizes the advanced intercross lines (AILs) (Darvasi and Soller 1995) and the heterogeneous stock (HS) population (Mott et al. 2000) by attaching an inbreeding stage to produce (nearly) completely inbred lines. MAGIC can also be regarded as a RIL population with a modified intercross mating scheme, ℳI, from pairing to random mating. In the following, we apply our model to study the breeding design of MAGIC populations by varying the number NF of inbred founders or the intercross mating scheme ℳI, in terms of the map expansion R and the overall expected junction density ρ that are used as measures of map resolution in QTL mapping populations.
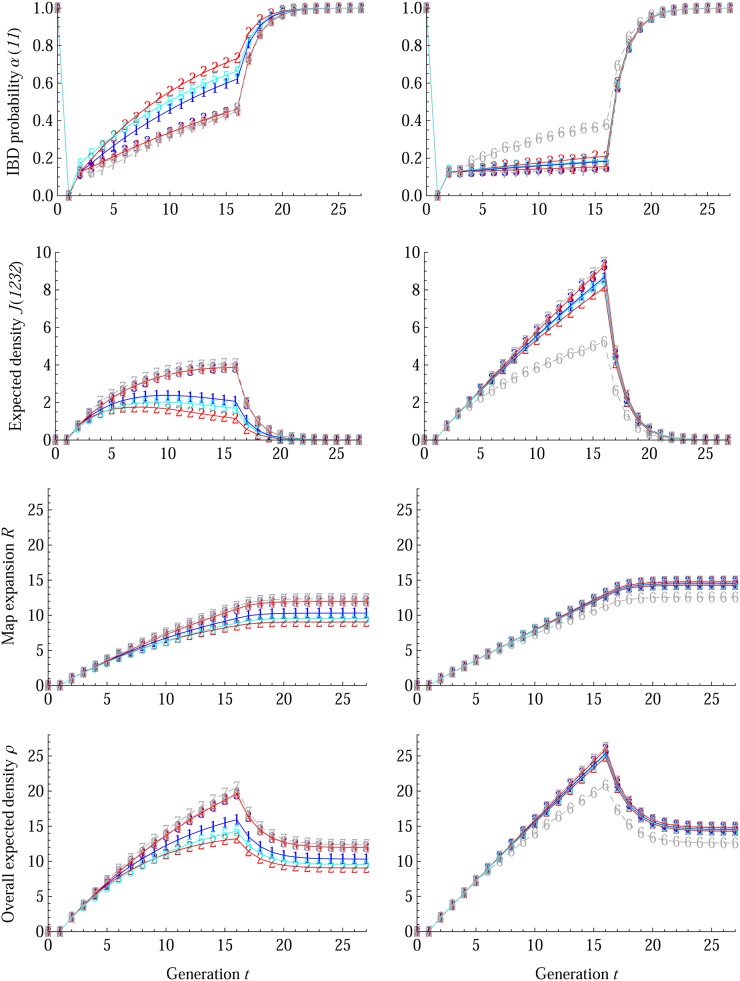
We first study the effects of the intercross mating scheme ℳI by both the forward simulations and the theoretical recurrence relations described in the Methods section. In total, seven intercross mating schemes ℳI are included: (1) RM1-NE, (2) RM2-NE, (3) RM1-E, (4) RM2-E, (5) WF-NE, (6) CPM-E, and (7) MAI-E. There are no theoretical results for the two regular mating schemes (CPM-E and MAI-E), since our model does not apply to them. We set ℳII = selfing for the inbreeding stage, since it is feasible for MAGIC populations used in Arabidopsis and many important crops. For the mixing stage, we set NF = 8, ℳF = RM1-NE, and NI = 8 or 100 for each of two sets of simulations. We use RM1-NE as the mixing random mating scheme instead of commonly used diallel crosses, so that the intercross population size NI can be chosen arbitrarily (Table 1).
Figure 7 shows the effects of the intercross mating scheme ℳI on the IBD probability α(11), the expected density J(1232), the map expansion R, and the overall expected junction density ρ. Their sharp changes indicate the transition from the intercross stage to the inbreeding stage in the breeding design. Theoretical results fit the simulations very well, and the differences are negligible; see the right column of Figure 6 for the MAGIC population with NI = 100 and ℳI = (3) RM1-E. The effects of intercross scheme ℳI are small in the beginning, and we thus consider the effects only after 10 generations. As shown in the left column of Figure 7 for NI = 8, the ranking of intercross scheme ℳI on R and ρ is RM2-NE < WF-NE < RM1-NE < RM1-E = RM2-E < CPM-E < MAI-E. The right column of Figure 7 show similar ranking for NI = 100: CPM-E ≪ RM2-NE < WF-NE < RM1-NE < RM1-E = RM2-E < MAI-E, except that CPM-E is far smaller and all other mating schemes are similar. The ranking of intercross scheme ℳI on the IBD probability α(11) is inversed, which is reasonable since the map expansion is an accumulation of the non-IBD probability α(12) (see Equation 7).
Figure 7.
Breeding designs on the intercross mating scheme ℳI in MAGIC populations with NF = 8, ℳF = RM1-NE, and ℳII = selfing. The intercross population size NI = 8 in the left column and NI =100 in the right column. The simulation results are denoted by integer markers for ℳI = (1) RM1-NE, (2) RM2-NE, (3) RM1-E, (4) RM2-E, (5) WF-NE, (6) CPM-E, and (7) MAI-E, respectively. The solid lines represent the theoretical results for the random intercross schemes, and dashed lines connect simulation results for the regular schemes (6) CPM-E and (7) MAI-E to guide one’s eyes. The blue color is for (1) RM1-NE and (3) RM1-E, red is for (2) RM2-NE and (4) RM2-E, cyan is for (5) WF-NE, and gray is for (6) CPM-E and (7) MAI-E.
The results of Figure 7 indicate that random mating schemes with equal contributions are better than those with nonequal contributions, and MAI-E is slightly better. They are consistent with those of the simulation studies of Rockman and Kruglyak (2008) on recombinant inbred AIL populations, where there are two inbred founders. However, the differentiations of intercross scheme ℳI (except CPM-E) diminish with increasing intercross population size. Thus, we derive below closed-form solutions for the map expansion R and the overall density ρ in MAGIC populations, assuming that the intercross population size NI is large and that ℳII = selfing at the inbreeding stage. We denote by sF (or sI) the constant coalescence probabilities at the mixing (or intercross) stage.
The closed-form solutions of R and ρ can be obtained by applying Equation 3 and Equations 16c and 16d to the intercross stage with the initial conditions at generation 2 and then to the selfing inbreeding stage with the initial conditionals at generation U + 1. Here we assume that the intercross population size NI is large so that α2(12) ≈ β2(12) and Equations 16c and 16d are good approximations for the nonselfing intercross mating scheme ℳI. In the founder population, α0(12) = 0, β0(12) = 1, and J0(1122) = K0(1122) = R0 = 0. The initial conditions at generation 2 can be obtained: α2(12) ≈ sI/2 + (1 − sI)(1 − sF), R2 = 1, and J2(1122) = 0 according to the recurrence relations for a nonselfing mixing mating scheme ℳF. We obtain for the last generation g = U + V + 1,
| (17) |
| (18) |
where U ≥ 1, and the eigenvalue λ1 of the intercross stage is given by Equation 13a for nonselfing intercross mating ℳI and otherwise by Equation 15a. Both R and ρ converge to those of 2n-way (n ≥ 2) RILs by selfing when α2(12) = 1 and λ1 → 1 (Table 1).
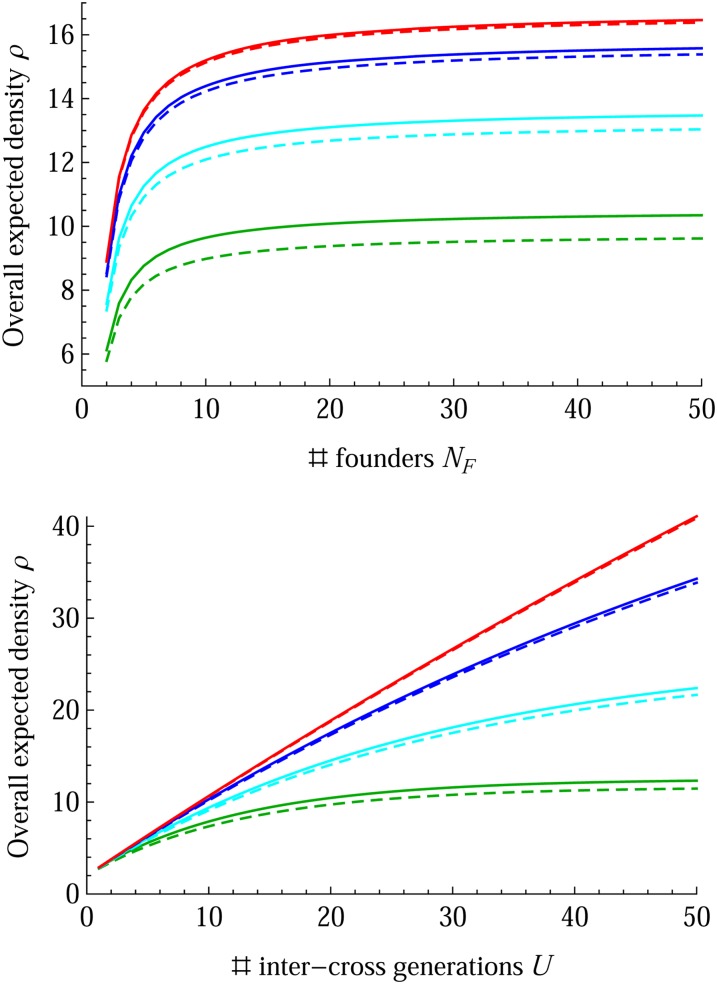
The expected densities of R and ρ in Equations 17 and 18 show that they are linearly related to α2(12) and thus approximately linearly related to 1/NF, the inverse of the number of founders according to the coalescence probability sF in Table 1, and that they are also asymptotically linearly related to the number U of intercross generations when the intercross population size NI increases and thus λ1 → 1. Figure 8 shows that ρ becomes flat when NF > ∼10 and that the approximation of ρ in Equation 18 slightly underestimates those obtained from the recurrence relations, but it gets very accurate as the intercross population size NI increases up to ∼50.
Figure 8.
Breeding designs for the number of founders NF and the number of intercross generations U in MAGIC populations. The overall expected density ρg is at the last generation g = U + V + 1. The basic design is ℳF = RM1-NE, ℳI = RM1-E, ℳII = selfing, and V = 6. Top, U = 15; bottom, NF = 8. The line colors green, cyan, blue, and red denote the results for NI = 4, 8, 20, and 50, respectively. The solid lines represent the results from the recurrence relations, and the dashed lines show those from the approximation of Equation 18.
Discussion
We have presented a general framework for modeling ancestral origin processes in a wide range of diploid breeding populations such as RILs, AILs, HSs, and MAGICs. For a new type of breeding population, we may apply the recurrence relations or deduce closed-form solutions if the design is stage-wise. The model framework may also be applied to natural populations if a recently founded population exists. In this scenario, the model parameters such as the coalescence probabilities or the effective population size are usually unknown, and they have to be estimated jointly with the ancestral origins from the genetic marker data.
The closed-form solutions of the expected junction densities are obtained for 2n-way RILs by selfing or sibling for any natural number n ≥ 1. From these expected densities, the rate matrix of the continuous time Markov chain of ancestral origin processes can be deduced and then the diplotype probabilities. Broman (2012a) extended the approach of Haldane and Waddington (1931) to 4- and 8-way RIL populations from sibling mating, but that approach does not lead to explicit expressions for two-locus diplotype probabilities.
One key assumption of our approach is the symmetry of founders in breeding populations. It helps to reduce model complexity greatly, so that our model can be applied to an arbitrary number of founders, whereas the complexity of the approach by Haldane and Waddington (1931) and Broman (2012a) increases very fast with the number of founders. Meanwhile, the approximation does not affect the results on junction densities where only changes of founder origins matter. The assumption is valid for MAGIC populations with random mating schemes and for 2n-way (n ≥ 1) RILs by selfing, but not valid for 2n-way (n ≥ 2) RILs by sibling mating due to the initial exclusive pairwise mating. However, this violation is not critical since individual samples are collected at the last generation and their ancestral genomes have been well mixed by many generations of random chromosomal segregations during inbreeding.
The second key assumption is the Markov property of ancestral origin processes along chromosomes. Therefore our model reduces to the IBD model of Stam (1980), where IBD and non-IBD tracts are exponentially distributed. However, even under the model of the sequential Markov coalescence (McVean and Cardin 2005; Marjoram and Wall 2006), the IBD tract length is not exponentially distributed, because the transition rate depends on the total branch length of the local tree, that is, the coalescent time in the case of two chromosomes. The nonexponential distribution for the length of IBD tracts has been modeled by Martin and Hospital (2011) in two-way RILs by sibling mating and by Chapman and Thompson (2003) in random mating populations, and their results show that the deviations from nonexponential distributions are acceptable, particularly for large populations.
The deviation from the Markov property is reflected in the variances of junction densities. The occurrence of junctions along two homologous chromosomes is an inhomogeneous process according to the continuous time Markov model. However, for the case of a nearly complete inbred individual at the last generation, the distribution of the overall junction density follows approximately a Poisson distribution with mean ρ. As shown in Figure 6 for eight-way RILs by sibling mating, the simulated variance (∼10) of the overall junction density is larger than the theoretical variance (∼7); whereas for the MAGIC population with NI = 100 and ℳI = (3) RM1-E, the theoretical Poisson distribution fits very well with the simulated distribution. The goodness of fit depends very little on the intercross random mating scheme, but it becomes slightly worse for small intercross population size NI = 8.
Finally, the breeding design assumes a single random mating population with variable size and no population structure. In contrast to the above two assumptions, this assumption does affect the expected junction densities. The breeding mating schemes of selfing and sibling and the mixing mating schemes of diallel crosses are regarded as special kinds of random mating. Essentially, a pair of individuals in the same generation is assumed to be statistically equivalent in terms of ancestral origins. For regular mating schemes such as CPM or subdivided populations, a set of recurrence relations is needed to account for different distances between two individuals, as shown by Kimura and Crow (1963) in calculating two-gene IBD probabilities.
The approximation of random mating implies the lack of natural selection since the founder population. Our model does not apply to those parts of genomes in breeding populations that are under artificial or natural selection. Still, even here we can use our model as a null model to investigate the strength of selection. Furthermore, we can try to use our model for describing parts of the breeding process like the inbreeding stage.
Liu et al. (2010) described a hidden Markov model (HMM) for ancestral inference in complex pedigrees with inbreeding from genetic marker data, where the inbreeding model is integrated into the Lander–Green algorithm. Their prior inbreeding model is specially built for sibling mating, and it is a discrete time Markov process. They model the two-locus diplotype probabilities D(abcd) between neighbor markers under an assumption of small intermarker distance (<0.001 M). Similarly, Liu et al. (2010) reduced the diplotype probabilities into three basic probabilities, where the diplotype probability D(1232) was calculated through the recurrence relations of the probabilities of three and four distinct genes among two siblings.
An HMM is under development for reconstructing ancestral origins from marker data, by using the present model of ancestral origin processes as the prior distribution. Then, we will be able to evaluate our general approach with both the specially designed model of Liu et al. (2010) and a relatively simple HMM approach such as that in R/happy (Mott et al. 2000) that does not account for the joint pattern of recombination breakpoints in two homologous chromosomes.
Supplementary Material
Acknowledgments
The authors have no competing financial interests. This research was supported by the Stichting Technische Wetenschappen (STW) - Technology Foundation, which is part of the Nederlandse Organisatie voor Wetenschappelijk Onderzoek - Netherlands Organisation for Scientific Research, and is partly funded by the Ministry of Economic Affairs, under grant no. STW-Rijk Zwaan project 12425.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.163006/-/DC1.
Communicating editor: B. S. Yandell
Literature Cited
- Bennett J. H., 1953. Junctions in inbreeding. Genetica 26: 392–406 [DOI] [PubMed] [Google Scholar]
- Bennett J. H., 1954. The distribution of heterogeneity upon inbreeding. J. R. Stat. Soc. Ser. B Stat. Methodol. 16: 88–99 [Google Scholar]
- Boucher W., Nagylaki T., 1988. Regular systems of inbreeding. J. Math. Biol. 26: 121–142 [DOI] [PubMed] [Google Scholar]
- Broman K., 2005. The genomes of recombinant inbred lines. Genetics 169: 1133–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2012a Genotype probabilities at intermediate generations in the construction of recombinant inbred lines. Genetics 190: 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K. W., 2012b Haplotype probabilities in advanced intercross populations. G3 2: 199–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler E. S., Holland J. B., Bradbury P. J., Acharya C. B., Brown P. J., et al. , 2009. The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Cavanagh C., Morell M., Mackay I., Powell W., 2008. From mutations to magic: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11: 215–221 [DOI] [PubMed] [Google Scholar]
- Chapman N., Thompson E., 2003. A model for the length of tracts of identity by descent in finite random mating populations. Theor. Popul. Biol. 64: 141–150 [DOI] [PubMed] [Google Scholar]
- Churchill G., Airey D., Allayee H., Angel J., Attie A., et al. , 2004. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36: 1133–1137 [DOI] [PubMed] [Google Scholar]
- Cockerham C., 1971. Higher order probability functions of identity of alleles by descent. Genetics 69: 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi A., Soller M., 1995. Advanced intercross lines, an experimental population for fine genetic-mapping. Genetics 141: 1199–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R., 1949. The Theory of Inbreeding. Oliver & Boyd, London [Google Scholar]
- Fisher R., 1954. A fuller theory of junctions in inbreeding. Heredity 8: 187–197 [Google Scholar]
- Haldane J., Waddington C., 1931. Inbreeding and linkage. Genetics 16: 357–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Paulo M.-J., Boer M., Effgen S., Keizer P., et al. , 2011. Analysis of natural allelic variation in Arabidopsis using a multiparent recombinant inbred line population. Proc. Natl. Acad. Sci. USA 108: 4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes F., Colome-Tatche M., 2011. Quantitative epigenetics through epigenomic perturbation of isogenic lines. Genetics 188: 215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Crow J. F., 1963. On maximum avoidance of inbreeding. Genet. Res. 4: 399–415 [Google Scholar]
- Kover P. X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I. M., et al. , 2009. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E. Y., Zhang Q., McMillan L., Pardo-Manuel de Villena F., Wang W., 2010. Efficient genome ancestry inference in complex pedigrees with inbreeding. Bioinformatics 26: i199–i207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MaCleod, A. K., C. S. Haley, and J. A. Woolliams, 2005. Marker densities and the mapping of ancestral junctions. Genet. Res. 85: 69–79 [DOI] [PubMed] [Google Scholar]
- Marjoram P., Wall J. D., 2006. Fast “coalescent” simulation. BMC Genet. 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin O. C., Hospital F., 2011. Distribution of parental genome blocks in recombinant inbred lines. Genetics 189: 645–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G., Cardin N., 2005. Approximating the coalescent with recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1387–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott R., Talbot C., Turri M., Collins A., Flint J., 2000. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 97: 12649–12654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadot R., Vayssiex G., 1973. Algorithme du calcul des coefficients d’identite. Biometrics 29: 347–359 [Google Scholar]
- Norris, J. R., 1997 Markov Chains Cambridge University Press, Cambridge, UK/London/New York. [Google Scholar]
- Rockman M. V., Kruglyak L., 2008. Breeding designs for recombinant inbred advanced intercross lines. Genetics 179: 1069–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P., 1980. The distribution of the fraction of the genome identical by descent in finite random mating populations. Genet. Res. 35: 131–155 [Google Scholar]
- Teuscher F., Broman K. W., 2007. Haplotype probabilities for multiple-strain recombinant inbred lines. Genetics 175: 1267–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Flint J., Mott R., 2003. QTL fine-mapping with recombinant-bred heterogeneous stocks and in vitro heterogeneous stocks. Mamm. Genome 14: 830–838 [DOI] [PubMed] [Google Scholar]
- Voight B. F., Pritchard J. K., 2005. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 1: e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller J., Soller M., 2004. An analytical formula to estimate confidence interval of qtl location with a saturated genetic map as a function of experimental design. Theor. Appl. Genet. 109: 1224–1229 [DOI] [PubMed] [Google Scholar]
- Winkler C., Jensen N., Cooper M., Podlich D., Smith O., 2003. On the determination of recombination rates in intermated recombinant inbred populations. Genetics 164: 741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S., 1921. Systems of mating. II. The effects of inbreeding on the genetic composition of a population. Genetics 6: 124–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.