Abstract
Listeria monocytogenes is a gram-positive facultative intracellular bacterium, which replicates in the cytoplasm of myeloid cells. Interferon β (IFNβ) has been reported to play an important role in the mechanisms underlying Listeria disease. Although studies in murine cells have proposed the bacteria-derived cyclic-di-AMP to be the key bacterial immunostimulatory molecule, the mechanism for IFNβ expression during L. monocytogenes infection in human myeloid cells remains unknown. Here we report that in human macrophages, Listeria DNA rather than cyclic-di-AMP is stimulating the IFN response via a pathway dependent on the DNA sensors IFI16 and cGAS as well as the signalling adaptor molecule STING. Thus, Listeria DNA is a major trigger of IFNβ expression in human myeloid cells and is sensed to activate a pathway dependent on IFI16, cGAS and STING.
Keywords: innate immunity, interferon beta, Listeria monocytogenes
Introduction
The innate immune system is capable of detecting foreign DNA and triggers early protective responses. In addition, microbial and host DNA may also stimulate activities with pathological consequences for the host (Paludan & Bowie, 2013). More than ten DNA sensors have been proposed and suggested to act in highly cell type-specific manners (Paludan & Bowie, 2013). The proposed DNA sensors include interferon (IFN) inducible 16 (IFI16) and DDX41, but notably cyclic GMP-AMP (cGAMP) synthetase (cGAS) is now well characterized (Unterholzner et al, 2010; Zhang et al, 2011; Sun et al, 2013). Common to all these proteins is that they stimulate expression of IFNβ via a pathway proceeding through the protein stimulator of IFN genes (STING), the kinase TBK1 and the transcription factor IFN regulatory factor 3 (Paludan & Bowie, 2013). IFI16 is a member of the PYHIN family of proteins, which also includes AIM2 and murine p204 (Paludan & Bowie, 2013). IFI16 is mainly localized in the nucleus, but a small cellular pool of IFI16 localizes to the cytoplasm through a mechanism controlled by acetylation of the nuclear localization signal in the protein (Li et al, 2012). IFI16 is able to directly interact with DNA, and to induce IFNα/β expression and inflammasome activation (Unterholzner et al, 2010; Kerur et al, 2011). DDX41 is expressed in dendritic cells, capable of interacting with DNA, and reported to be involved in early responses to both viral and bacterial infections (Zhang et al, 2011). Recently, DDX41 has also been proposed to be a sensor for cyclic dinucleotides (CDNs) (Parvatiyar et al, 2012). A major breakthrough in the field of DNA sensing was the identification of cGAS, which upon DNA binding produces the non-canonical CDN 2′3′-cGAMP, a cellular secondary messenger which directly binds STING and stimulates IFN expression (Ablasser et al, 2013; Diner et al, 2013; Gao et al, 2013; Sun et al, 2013; Wu et al, 2013). The interplay between cGAS and other proposed DNA sensors is still not known.
Listeria monocytogenes is an intracellular gram-positive bacteria and the causative agent of listeriosis. Listeriosis usually occurs in the setting of pregnancy, immunosuppression or extremes of age (Barbuddhe & Chakraborty, 2009). L. monocytogenes infects a wide range of cell types, including macrophages (Hamon et al, 2006). Following phagocytosis by macrophages, L. monocytogenes secretes the cytolysin listeriolysin O (LLO) which disrupts the vacuolar membrane (Cossart et al, 1989). This allows L. monocytogenes to escape into the cytoplasm where replication occurs in the more favourable growth conditions. This step is considered of major importance since L. monocytogenes strains without functional LLO are avirulent.
Following infection of innate immune cells with L. monocytogenes, a variety of cytokines, including type I IFNs, are produced. IFNα/β induction is important for antiviral defence, but IFNAR−/− mice exhibit increased resistance to L. monocytogenes infection, perhaps due to decreased IFN-induced apoptosis in IFNAR−/− macrophages (Stockinger et al, 2002; Auerbuch et al, 2004; Carrero et al, 2004; O'Connell et al, 2004). Studies mainly performed in murine cells have demonstrated that induction of IFNα/β and cytokines during L. monocytogenes infection is the result of recognition of both bacterial cells wall components by Toll-like receptors (TLR) and additional bacteria-derived molecules by intracellular TLR-independent-sensing mechanisms (Leber et al, 2008; Stockinger et al, 2009; Yang et al, 2010; Abdullah et al, 2012). Cytosolic delivery of DNA derived from L. monocytogenes is a potent activator of type I IFN induction (Stetson & Medzhitov, 2006). In addition, CDNs released from L. monocytogenes are also potent stimulators of IFN induction (Woodward et al, 2010; Burdette et al, 2011). CDNs are recognized by STING and DDX41 and have been proposed to be major contributors to the induction of IFN during Listeria infection in the murine system (Burdette et al, 2011; Sauer et al, 2011; Parvatiyar et al, 2012; Schwartz et al, 2012). However, recent reports indicate that human cells are less responsive to bacterial CDNs than murine cells, due to the differential ability of human and murine STING to bind the 3′5′-linked bacterial CDNs (Ablasser et al, 2013; Conlon et al, 2013; Diner et al, 2013; Gao et al, 2013; Zhang et al, 2013), hence leaving open the question as to how human myeloid cells sense L. monocytogenes infection and induce IFNα/β expression.
To investigate the role of IFI16, DDX41 and cGAS in the sensing of L. monocytogenes DNA, we utilized human primary monocyte-derived macrophages and THP1-derived cell lines generated to stably express shRNAs targeting the proposed sensors. Our results demonstrate that, unlike what has been reported for murine macrophages (Schwartz et al, 2012), induction of IFNβ expression by human macrophages does not correlate with overexpression of the multidrug efflux pump MdrT, which transports the CDN cyclic-di-AMP into the cytoplasm (Crimmins et al, 2008). In contrast, in these cells, Listeria genomic DNA was found to be the major pathogen-associated molecular pattern (PAMP) in Listeria extracts and to induce IFNβ expression in a manner dependent on IFI16, DDX41, cGAS and STING. During L. monocytogenes infection, the bacteria-induced IFNβ response was dependent on IFI16, cGAS and STING, but not DDX41. Confocal microscopy revealed that IFI16 and cGAS associated with synthetic and bacterial DNA in the cytoplasm and also co-localized with STING foci after stimulation with DNA or infection with L. monocytogenes but not after treatment with CDNs. Thus, Listeria DNA is a major trigger of IFNβ expression in human myeloid cells and is sensed by IFI16 and cGAS to activate the STING pathway.
Results
Induction of IFNβ expression by L. monocytogenes in human myeloid cells
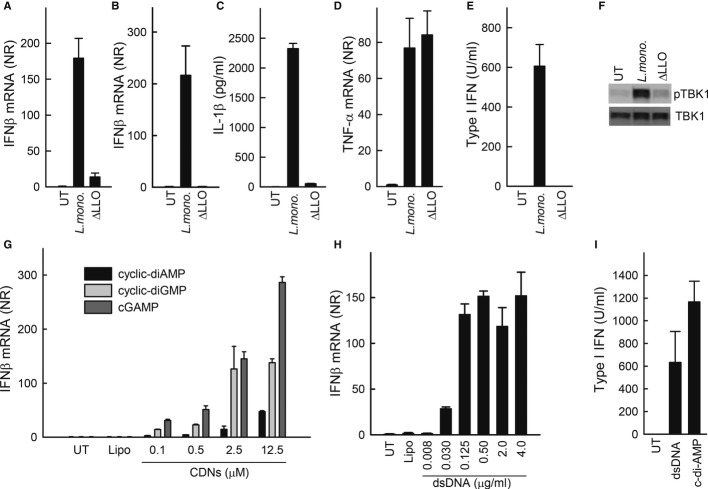
The induction of type I IFN by L. monocytogenes infection has been well established (Stetson & Medzhitov, 2006; Stockinger et al, 2002; Woodward et al, 2010). During infection, macrophages are an important producer of IFNα/β (Stockinger et al, 2009; Solodova et al, 2011), resulting in apoptosis and ultimately facilitation of bacterial infection (Stockinger et al, 2002; Auerbuch et al, 2004; Rayamajhi et al, 2010). Several studies have demonstrated that both L. monocytogenes DNA and CDNs stimulate IFNβ induction (Burdette et al, 2011; Chan et al, 2006; Parvatiyar et al, 2012; Sauer et al, 2011; Schwartz et al, 2012; Stetson et al, 2006), and it has recently been reported that cGAMP is a second messenger in DNA signalling (Wu et al, 2013). To examine whether THP1 cells responded to L. monocytogenes, the cells were infected with increasing amounts of the bacteria, and accumulation of IFNβ mRNA was measured. We observed a dose-dependent induction of IFNβ expression between multiplicity of infection (MOI) 1 and 60 (Supplementary Fig S1A), and the expression was not affected by extensive washing of the bacteria prior to infection of the macrophages (Supplementary Fig S1B). IFNβ mRNA started to accumulate between 2 and 4 h after infection and remained increased for several hours (Supplementary Fig S1C). The bacteria were observed in both vacuoles and the cytoplasm as detected by electron microscopy (Supplementary Fig S1D), and did productively infect the macrophage-like cells (Supplementary Fig S1E). It has previously been reported that Listeria-induced type I IFN expression in murine cells is dependent on bacterial escape into the cytoplasm (Leber et al, 2008). As shown in Fig1A and B, stimulation of IFNβ expression by L. monocytogenes in both human monocyte-derived macrophages (hMDM)s and PMA-differentiated THP1 cells was abrogated if the bacterium was unable to express LLO and hence reach the cytoplasm. Accumulation of IL-1β protein in the culture supernatant was also dependent on bacterial escape into the cytoplasm, whereas TNF-α expression occurred independent of LLO (Fig1C and D), both findings confirming previous reports (Leber et al, 2008; Sauer et al, 2010). The IFN response was also observed at the level of bioactivity as measured by stimulation of IFNAR-driven gene expression by the culture supernatant (Fig1E), and induction of the IFN-stimulated genes CXCL10 and MxA1 (Supplementary Fig S2A and B). At the level of signalling, infection with wild-type (WT) L. monocytogenes but not the LLO-deletion mutant stimulated phosphorylation of TBK1 (Fig1F), which is essential for induction of type I IFN expression via cytosolic sensing pathways.
Figure 1. IFNβ induction by Listeria monocytogenes, DNA and cyclic dinucleotides (CDNs) in human macrophages.
A–E Human primary macrophages (A) and PMA-differentiated THP1 macrophages (B–E) were infected with 25 multiplicity of infection (MOI) of WT L. monocytogenes or listeriolysin O (LLO)-deficient (ΔLLO) L. monocytogenes (strain LO28). (A, B, D) The cells were lysed 6 h post-treatment, and total RNA was isolated or (C, E) supernatants were isolated 6 and 18 h post-infection. IFNβ and TNF-α mRNA, and levels was determined by RT-qPCR, IL-1β protein levels were determined by ELISA (6 h) and type I IFN bioactivity was determined by the HEK-BLUE IFN assay (18 h).
F Lysates from PMA-differentiated THP1 cells treated as indicated for 6 h were subjected to Western blotting with antibodies specific for phosphor and total TBK1.
G, H PMA-differentiated THP1 macrophages were transfected with increasing concentration of (G) CDNs or (H) dsDNA. Total RNA was isolated 6 h post-treatment and IFNβ mRNA levels were determined by RT-qPCR.
I Supernatants from PMA-differentiated THP1 cells stimulated with dsDNA (2 μg) or cyclic-di-AMP (12.5 μM) for 6 h were analysed for IFN bioactivity.
Data information: Data represent mean ± SD of duplicates, representative of at least three independent experiments.
Since both DNA and CDNs have been proposed as Listeria PAMPs stimulating IFNβ expression (Burdette et al, 2011; Chan et al, 2006; Parvatiyar et al, 2012; Sauer et al, 2011; Schwartz et al, 2012; Stetson et al, 2006), we wanted to evaluate how PMA-differentiated THP1 cells responded to these stimuli. We found the THP1 cells to respond to CDNs in a dose-dependent manner, with cGAMP stimulation giving rise to the strongest IFNβ expression, followed by cyclic-di-GMP, and cyclic-di-AMP, which is the CDN produced by L. monocytogenes, being the least potent stimulus (Fig1G). Likewise, the THP1 cells were found to respond potently to dsDNA with a saturated IFNβ response observed after stimulation with 0.125 μg/ml (Fig1H). Both CDNs and DNA induced IFN bioactivity (Fig1I).
It has recently been reported that the L. monocytogenes strain LO28 used for the experiments presented in Fig1 naturally overexpresses the multidrug efflux pump MdrT, which correlates with high induction of IFNβ in murine macrophages (Schwartz et al, 2012). By using murine bone marrow-derived macrophages (BMMs), we were able to confirm that the LO28 strain with high MdrT expression induced significantly more IFNβ than the 10403s strain (Fig2A). Moreover, in the murine system, LO28 mutants with either a deletion of the chromosomal copy of MdrT or a reintroduction of the TetR family transcriptional repressor BrtA lacking in LO28 lost the ability to potently induce IFNβ expression (Fig2A). Interestingly, in hMDMs, 10403s was a much more potent inducer of IFNβ expression than LO28, and modulation of MdrT expression in this strain did not alter the ability to activate innate immune responses (Fig2B and Supplementary Fig S3). In THP1 and U937 cells, bacteria-induced IFNβ expression was also observed to be independent of MdrT expression (Fig2C and D).
Figure 2. IFNβ induction by Listeria monocytogenes in human macrophages does not correlate with expression of the multidrug efflux pump MdrT.
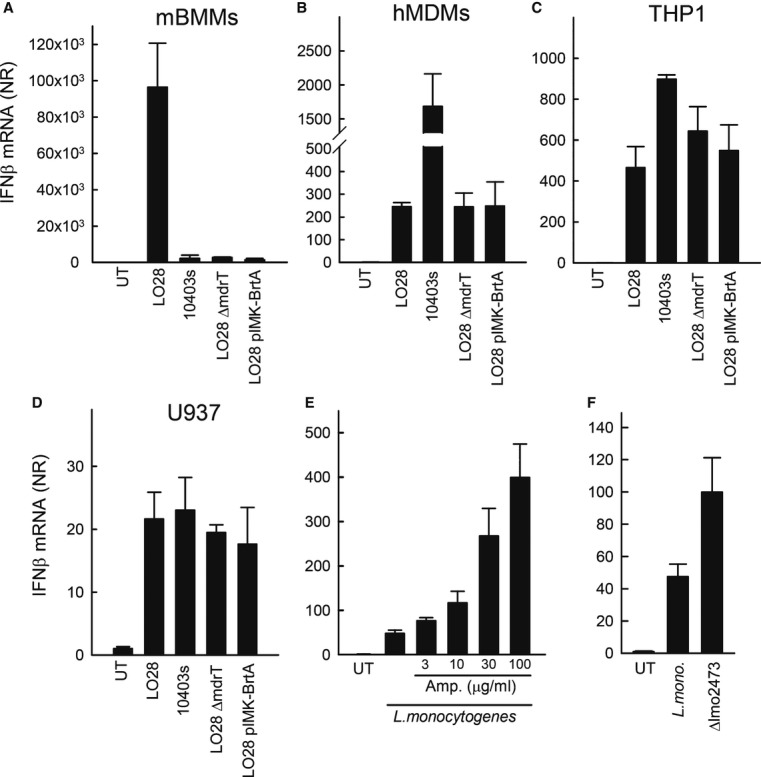
A–D Mouse bone marrow-derived macrophages (BMMs) (A), Human primary monocyte-derived macrophages (B), PMA-differentiated THP1 cells (C) or PMA-differentiated U937 cells (D) were infected with the L. monocytogenes strains LO28 and 10403s or LO28 mutants with altered expression of the multidrug efflux pump MdrT (MOI 25).
E PMA-differentiated THP1 cells were infected with L. monocytogenes strain LO28 for 2 h and treated with ampicillin in the indicated concentrations.
F PMA-differentiated THP1 cells were infected with L. monocytogenes strain 10403s or the derived mutant Δlmo2473.
Data information: Total RNA was isolated 6 h p.i. and RT-qPCR analysis for IFNβ was performed. Data represent mean ± SD of duplicates, representative of at least three independent experiments.
Previous reports have demonstrated that stimulation of bacteriolysis in the cytosol of macrophages triggers inflammasome signalling dependent on the DNA sensor AIM2 (Sauer et al, 2010). To test the impact of bacteriolysis in the macrophage cytosol on IFNβ induction, we evaluated the effect of the β-lactam ampicillin. As shown in Fig2E, treatment with ampicillin 2 h post-infection led to a dose-dependent elevation in bacteria-induced expression of IFNβ mRNA. Finally, infection with a L. monocytogenes mutant lacking the lmo2473 gene, and which lyses with increased frequency in the macrophage cytosol (Sauer et al, 2010), induced a stronger IFNβ response than did the WT bacteria (Fig2F).
Collectively, these data demonstrate that induction of IFNβ by L. monocytogenes in human myeloid cells, which respond to both DNA and CDNs, is dependent on bacterial escape into the cytoplasm and correlates with bacteriolysis in the cytoplasm, but does not correlate with expression of the efflux pumps MdrT, capable of pumping CDNs into the cytoplasm.
Induction of IFNβ by L. monocytogenes correlates with IFI16 expression
THP1 cells exhibit a monocyte-like phenotype and can be differentiated into a macrophage-like phenotype by treatment with PMA. During the course of these studies, we observed that differentiated THP1 cells respond to DNA with significantly higher IFNβ expression as compared to undifferentiated THP1 cells (Supplementary Fig S4). Interestingly, when challenging the THP1 cells with L. monocytogenes, we found that unlike the PMA-differentiated cells, the undifferentiated cells were unable to produce IFNβ in response to the bacterial challenge (Fig3A). These data indicate that a factor is present in differentiated THP1 cells, which leads to enhanced cellular IFN expression during L. monocytogenes infection.
Figure 3. Induction of IFNβ by Listeria monocytogenes infection correlates with expression of IFI16.
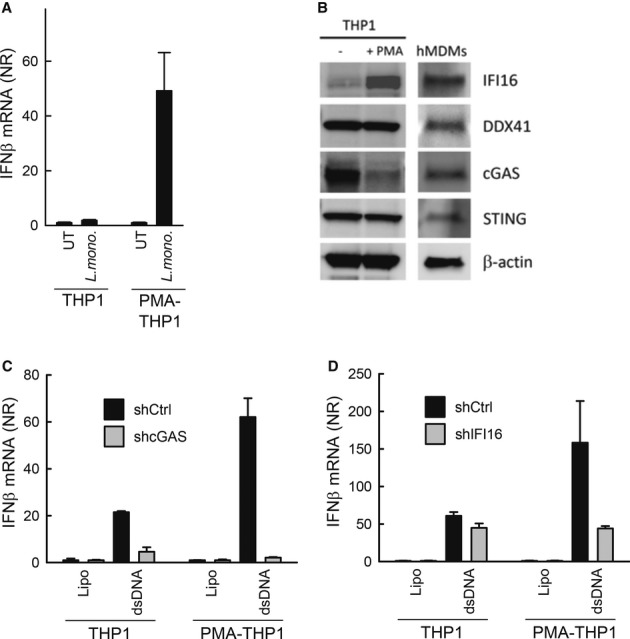
A THP1 cells either undifferentiated or PMA differentiated were infected with L. monocytogenes (strain LO28, multiplicity of infection, MOI 25). Total RNA was isolated 6 h p.i. and RT-qPCR analysis for IFNβ was performed.
B Whole lysates from THP1 cells and hMDMs were subjected to Western blotting using antibodies against IFI16, cGAS, DDX41, STING and β-actin.
C, D THP1-shCtrl, THP1-shIFI16 and THP1-shcGAS were seeded and either differentiated with PMA or left untreated prior to stimulation with dsDNA (2 μg/ml) for 6 h. Total RNA was isolated and IFNβ mRNA induction was determined by RT-qPCR.
Data information: Data in (A), (C) and (D) represent mean ± SD of duplicates, representative of 2–3 independent experiments.
In order to examine the expression of proteins of the DNA/CDN pathway in the cell line and primary cells used in this study, we prepared whole-cell extracts from undifferentiated THP1 cells, PMA-differentiated THP1 cells and hMDMs. The extracts were subjected to Western blotting, and expression of IFI16, DDX41, cGAS and STING was monitored. We observed that DDX41, cGAS and STING were expressed in both cell types (Fig3B). While expression of DDX41 and STING was not affected by the PMA-differentiation procedure in the THP1 cells, the levels of cGAS decreased dramatically during the differentiation procedure (Fig3B). By contrast, IFI16 was not expressed at significant levels in the undifferentiated THP1 cells, but was expressed in hMDMs and also at high levels in THP1 cells after PMA differentiation (Fig3B). When examining for the role of IFI16 and cGAS in DNA-driven IFNβ expression in the parental and PMA-differentiated THP1 cells, we found that cGAS was essential for DNA-driven IFN expression irrespective of the differentiation stage of the cells, despite the pronounced downregulation of cGAS expression in PMA-differentiated cells (Fig3C). By contrast, a significant role for IFI16 was seen only in differentiated THP1 cells (Fig3D). Thus, the expression of IFI16 correlates with the ability of cells to induce IFNβ expression in response to L. monocytogenes infection.
IFNβ induction by bacterial DNA and L. monocytogenes infection is dependent on IFI16 and cGAS
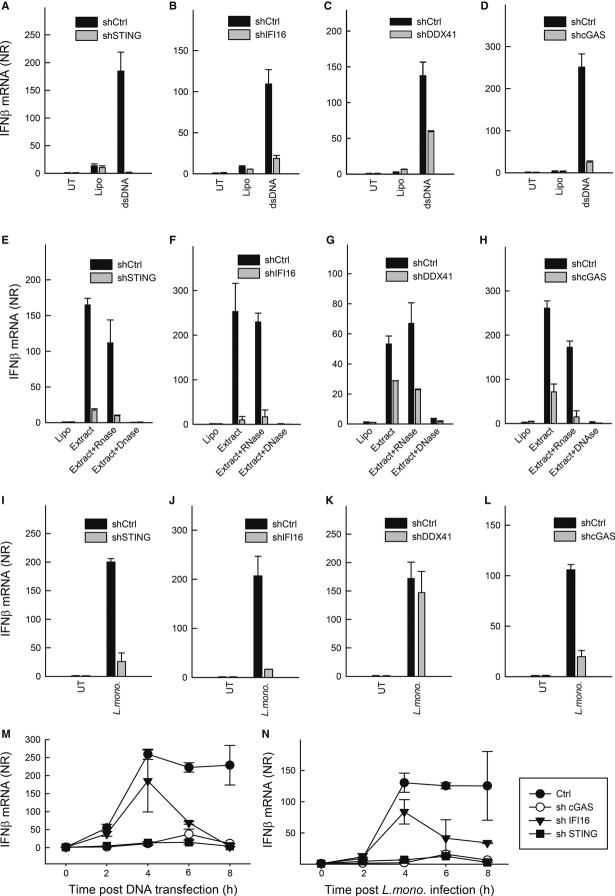
Listeria monocytogenes DNA can be sensed by the PYHIN family member AIM2, leading to activation of the inflammasome (Rathinam et al, 2010; Sauer et al, 2010). To investigate the roles for IFI16, DDX41 and cGAS in sensing of L. monocytogenes infection, we used previously established THP1 cell lines with stable shRNA-mediated knock-down of these DNA sensors and their downstream adaptor STING (Horan et al, 2013; Jakobsen et al, 2013). Consistent with a role for IFI16, DDX41, cGAS and STING in DNA-sensing pathways, IFNβ induction following DNA transfection was impaired in all knock-down cells compared to shCTRL-THP1 (Fig4A–D) but not in response to Sendai virus infection (Jakobsen et al, 2013).
Figure 4. IFNβ induction by Listeria monocytogenes extracts is dependent on IFI16, cGAS and STING.
A–L PMA-differentiated THP1 macrophages transduced with lentivirus-encoding control shRNA or a shRNA sequence targeted to cGAS, IFI16, DDX41 or STING were transfected with dsDNA (2 μg/ml) (A–D), L. monocytogenes (strain LO28, multiplicity of infection, MOI 25) total extracts or extracts pre-treated with either RNase or DNase (E-H), or infected with L. monocytogenes (I-L). The cells were lysed 6 h post-treatment, and IFNβ mRNA induction was determined by qPCR.
M, N The Ctrl and gene-targeted shRNA cell lines were treated with DNA and L. monocytogenes as above. Total RNA was harvested at the indicated time points, and IFNβ mRNA induction was determined by qPCR.
Data information: Data represent mean ± SD of duplicates, representative of 2–3 independent experiments.
Given the above findings, and the reports on STING and DDX41 being involved in both CDN- and DNA-sensing pathways (Burdette et al, 2011; Holm et al, 2012; Parvatiyar et al, 2012), we were interested in further clarifying Listeria DNA as an IFN-inducing PAMP in human myeloid cells. For this purpose, THP1-shCTRL, THP1-shIFI16, THP1-shDDX41, THP1-shcGAS and THP1-shSTING cells were transfected with L. monocytogenes extracts which either had not been further treated, or had been treated with RNase or DNase (Supplementary Fig S5A). Transfection of PMA-differentiated THP1-shCTRL cells with L. monocytogenes extracts not treated with nucleases stimulated a robust induction of IFNβ (Supplementary Fig S5B). This response was not affected by RNase treatment but was abrogated if the extracts had been treated with DNAse prior to transfection (Supplementary Fig S5B). Moreover, using an assay for activation of IFN expression by small heat-resistant molecules, previously used as an assay for CDN activity (Wu et al, 2013), we found that L. monocytogenes extracts did not contain small heat-resistant molecules in concentrations high enough to induce significant IFNβ expression in PMA-differentiated THP1 cells (Supplementary Fig S5C). Importantly, the IFNβ response induced by L. monocytogenes extracts was largely abrogated in PMA-differentiated THP1-shIFI16, THP1-shcGAS and THP1-shSTING cells, and partly reduced in THP1-shDDX41 cells (Fig4E–H). Importantly, when challenging the THP1-derived PMA-differentiated cell lines with L. monocytogenes, we found that cells with knock-down of IFI16, cGAS or STING, but not cells with knock-down of DDX41 exhibited reduced induction of IFNβ expression in response to the infection (Fig4I–L). Despite the lack of a role for DDX41 in L. monocytogenes-induced IFNβ expression, the cells with knock-down of DDX41 did, similarly to cells with knock-down of STING, but unlike the cells with knock-down of IFI16 or cGAS, exhibit reduced IFNβ expression after stimulation with cyclic-di-AMP (Supplementary Fig S6). Finally, we examined the requirement for cGAS, IFI16 and STING over time in stimulation of IFNβ expression by DNA or L. monocytogenes. For both stimuli, the expression of IFNβ was dependent on cGAS and STING regardless the time points examined (Fig4M and N). Interestingly, IFI16 was required for late but not early induction of IFNβ expression by both DNA and L. monocytogenes.
Thus, stimulation of IFNβ expression by L. monocytogenes in human myeloid cells is dependent on IFI16, cGAS and STING, and although we did confirm a role for DDX41 in signalling in response to synthetic DNA and CDNs, expression of this protein was not essential for evoking IFN responses to L. monocytogenes infection.
In order to look into the signalling events involved in STING-driven IFN responses, we evaluated the activation of TBK1 in the THP1-derived cell lines with knock-down of IFI16, cGAS, DDX41 or STING. As shown in Fig5, L. monocytogenes-induced phosphorylation of TBK1, which occurs immediately downstream of STING (Tanaka & Chen, 2012), was compromised in cells with reduced expression of IFI16, cGAS or STING but not DDX41.
Figure 5. L. monocytogenes infection in human myeloid cells stimulates phosphorylation of TBK1 in a manner dependent on IFI16, cGAS and STING.
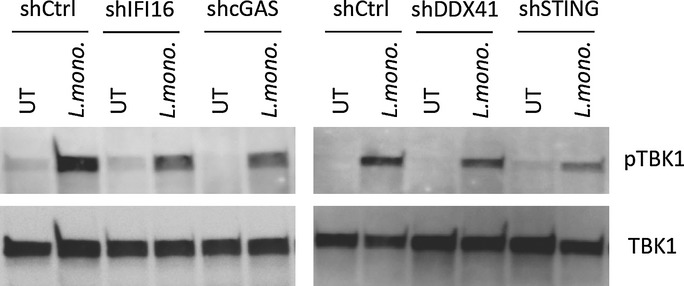
PMA-differentiated THP1 macrophages transduced with lentivirus-encoding control shRNA or a shRNA sequence targeted to cGAS, IFI16, DDX41 or STING were infected with L. monocytogenes (strain LO28, MOI 25). Whole-cell lysates were generated 3 h post-infection, and levels of phospho- and total TBK1 were determined by Western blotting.
Cumulatively, these results suggest that IFI16, cGAS and STING are essential for induction of IFNβ after L. monocytogenes infection in human macrophages.
IFI16 and cGAS co-localize with DNA in the cytoplasm and are selectively recruited to DNA-activated STING signalsomes
To investigate whether IFI16 engages with DNA and STING, we transfected cells with DNA or c-di-AMP. The macrophages were fixed 4 h post-treatment and stained for IFI16 and STING. In mock-treated cells, IFI16 exhibited a predominantly nuclear distribution, with some diffuse distribution to the cytosol, whilst STING staining was detected widely in the cytoplasm (Fig6A, upper panels). As reported previously, cytosolic IFI16 relocalized to punctuate structures in DNA-treated cells co-localizing with DNA and STING (Fig6A). In cells treated with c-di-AMP, we also observed abundant STING foci formation, but IFI16 was not detected in these structures and retained the same staining pattern as in the mock-treated cells (Fig6B). Following infection with L. monocytogenes, DNA could be visualized in the cytoplasm 4 h p.i. utilizing DAPI staining (Fig6C). Importantly, we observed co-localization between IFI16 and cytosolic DNA, consistent with the role of IFI16 as a DNA sensor. Furthermore, redistribution of STING to the site of bacterial DNA and IFI16 accumulation was observed in the infected cells (Fig6C). Quantification of STING foci formation above background level revealed that dsDNA transfection led to redistribution of STING in about 50% of cells, c-di-AMP transfection in about 75% of cells and infection with L. monocytogenes-induced formation of STING foci in about 30% of cells. When staining for cGAS in cells stimulated with DNA or infected with L. monocytogenes, we observed partial co-localization between cGAS and STING (Supplementary Fig S7A), similar to what was observed when staining with anti-IFI16 (Fig6A). This was not observed after stimulation with c-di-AMP. The redistribution of cGAS was spatially related to the location of both synthetic and bacterial DNA with partial overlap (Supplementary Fig S7B).
Figure 6. IFI16 and STING co-localize with DNA in the cytoplasm after Listeria monocytogenes infection.
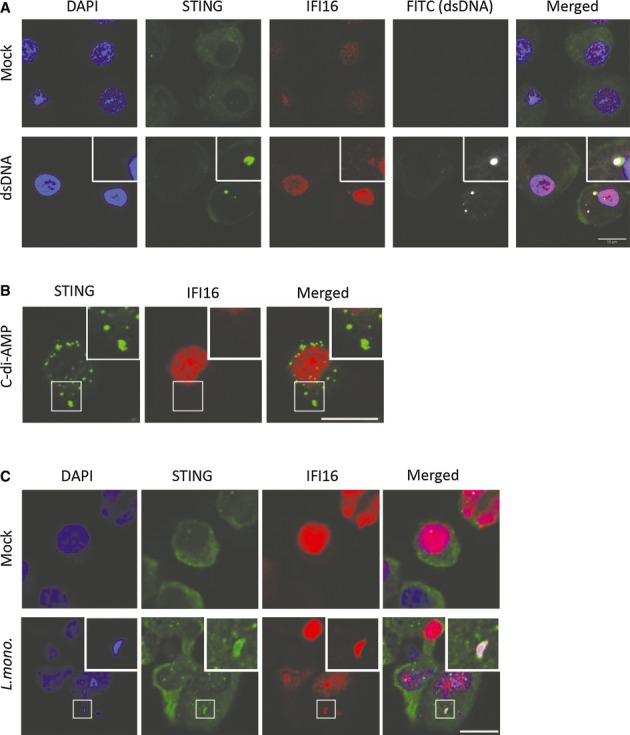
A–C PMA-differentiated THP1 macrophages were (A) transfected with 2 μg/ml of FITC-labelled dsDNA and (B) 3.6 μM of cyclic-di-AMP, or (C) infected with L. monocytogenes (strain LO28, MOI25). Four hours post-treatment, the cells were fixed and stained with anti-IFI16 and anti-STING specific antibodies. DNA was visualized with DAPI (blue).
All together, these results suggest that the bacterial DNA is the major IFN-inducing PAMP in human macrophages during L. monocytogenes infection and this stimulates a pathway dependent on IFI16, cGAS and STING. Furthermore, the data demonstrate a role for IFI16 in relation to the cGAS-cGAMP-STING axis, potentially by acting as DNA sensor in the amplification phase of the signalling events eventually leading to the pathological IFNβ response in Listeria-infected individuals.
Discussion
Listeria monocytogenes is an important human pathogen which infects macrophages by escaping from the endosomal environment into the cytoplasm for replication (Cossart et al, 1989). The pathology of listeriosis involves excess immune activation, and it has been reported that the type I IFN system is a major player in stimulation of this response (Stockinger et al, 2002; Auerbuch et al, 2004; Carrero et al, 2004; O'Connell et al, 2004). Knowledge on how L. monocytogenes triggers IFN responses in human macrophages is thus of central importance. It is known that the immune sensing of L. monocytogenes stimulating IFNβ expression occurs in the cytoplasm (Leber et al, 2008), but the Listeria IFN-inducing PAMP in human cells and the cellular-sensing pathways involved are not known. While it is well established that the bacterium harbours two potent PAMPs potentially exposed to the cytoplasm, namely genomic DNA and c-di-AMP, largely all published data have been generated in murine systems. Previous work has demonstrated that murine cells respond very potently to bacterial CDNs (McWhirter et al, 2009; Jin et al, 2011; Sauer et al, 2011) and that the ability of L. monocytogenes to induce type I IFN correlates with the activity of the multidrug efflux pump MdrT, which transports CDNs into the cytoplasm (Woodward et al, 2010; Schwartz et al, 2012). However, recent data on the activation of the DNA-dependent STING-TBK1-IRF3 pathway have revealed large differences on activation of human and murine STING by CDNs (Cavlar et al, 2013; Conlon et al, 2013; Diner et al, 2013). In this work, we demonstrate that induction of IFN expression by L. monocytogenes in human myeloid cells does not correlate with MdrT activity, does correlate with bacteriolysis in the macrophage cytosol, and that bacterial DNA is the most potent IFN-inducing PAMP in bacterial extracts. Moreover, bacterial DNA localized to the cytoplasm during infection and co-localized with IFI16, cGAS and STING. Finally, expression of IFI16, cGAS and STING was essential for bacteria-induced type I IFN expression. Hence, bacterial genomic DNA is the predominant IFN-inducing PAMP in human myeloid cells during L. monocytogenes and triggers the host response through a pathway dependent on IFI16, cGAS and STING (Supplementary Fig S8).
A key finding of the present work is the identification of DNA as the main IFN-inducing PAMP in human myeloid cells during L. monocytogenes infection, as opposed to the key role for bacterial CDNs in murine cells. One likely explanation for this is that human STING is less responsive to bacterial CDNs with 3′5′ linkage and binds the endogenous 2′3′-cGAMP with more than 500 times higher affinity than cyclic-di-GMP (Zhang et al, 2013). However, based on the existing literature, it cannot be excluded that other factors may also contribute to explanation of the present findings. This idea is at least partly supported by the fact that THP1 STING differs from reference human STING at four positions including an arginine at position 232 as opposed to a histidine in reference STING, and consequently displays a responsiveness to 3′5′-linked CDNs which is intermediary between murine STING and reference human STING (Diner et al, 2013). In addition, mice do not have a clear IFI16 orthologue, and given the data presented in this work, it is possible that IFI16 is essential to achieve full responsiveness to DNA. Alternatively, human cells may be more efficient than murine cells in the process of sensing and degrading bacterial components in the cytoplasm, hence allowing exposure of DNA to immune sensors. Randow and associates have recently described cytosolic systems that detect bacteria to stimulate their degradation (Thurston et al, 2009). Similarly, this laboratory has reported that sensing of herpesvirus DNA in the cytoplasm requires a cytoplasmic system to sense and degrade the viral capsid (Horan et al, 2013). Finally, it is also possible that the bacteria actively expose its DNA to the host cytoplasm for its own benefit and do so more efficiently in human cells. This phenomenon has been described for Mycobacteria tuberculosis, which uses the ESX-1 secretion system to achieve this (Manzanillo et al, 2012).
About a dozen of DNA sensors have been proposed (Paludan & Bowie, 2013), and IFI16, DDX41 and cGAS have been reported to stimulate a signalling pathway dependent on STING (Unterholzner et al, 2010; Zhang et al, 2011; Sun et al, 2013). It has been discussed as to whether the proposed DNA sensors act in cell type-dependent manners or whether they act in concert to induce IFN responses (Paludan & Bowie, 2013). In this study, we found that undifferentiated THP1 cells express high amounts of cGAS and DDX41 but only very little IFI16, whereas differentiated THP1 cells express high amounts of IFI16 and DDX41 but less cGAS. hMDMs expressed all three proposed DNA sensors at high levels. Interestingly, we found that L. monocytogenes infection induced IFNβ expression only in the differentiated THP1 cells and the hMDMs, hence correlating with expression of IFI16. Moreover, we found that IFI16 co-localized with cytosolic DNA and STING foci in L. monocytogenes-infected cells. When testing for the role of DNA sensors in driving IFNβ repression in PMA-differentiated THP1 cells infected with L. monocytogenes, we found IFI16 and cGAS to be essential, whereas knock-down of DDX41 did not affect this response, although a partial role for DDX41 was found after stimulation with synthetic DNA. Therefore, in a cell type expressing IFI16, DDX41 and cGAS, reduction of the levels of particularly IFI16 or cGAS significantly reduced IFN expression induced by either synthetic DNA or genomic bacterial DNA. These data argue against the DNA sensors acting in cell type-dependent pathways, but rather that they act in concert to stimulate the STING-TBK1-IRF3 pathway. In this respect, it was interesting to observe IFI16 to be essential for sustained but not initial IFNβ expression, and that both cGAS and IFI16 co-localize with STING after stimulation with DNA but not with c-di-AMP. These data could suggest a role for IFI16 in amplification of cGAS-dependent DNA-driven immune responses. At present, a molecular explanation for IFI16 how works in DNA-driven IFN responses remains to be provided, although a structural explanation of how DNA binding exposes the signalling pyrin domain has been reported (Jin et al, 2012). With the present identification of the requirement for both IFI16 and cGAS in DNA-driven IFN responses in PMA-differentiated THP1 cells, there is an urgent need to understand the interplay between these two proteins, including whether IFI16 acts upstream of, parallel to, or downstream in the cGAS pathway, and also to resolve the potential temporal aspect of the requirement for IFI16 in DNA-stimulated innate immune responses. DDX41, which was first described to be a DNA sensor, has subsequently been demonstrated to bind CDNs and stimulate the STING pathway (Zhang et al, 2011; Parvatiyar et al, 2012). Together with the knowledge on cGAS producing the STING-activating endogenous CDN 2′3′ cGAMP upon DNA binding (Sun et al, 2013; Wu et al, 2013), it is tempting to speculate that DDX41 acts in the DNA-stimulated pathway by amplifying STING activation at the level of cGAMP.
In conclusion, we demonstrate that bacterial genomic DNA is the main PAMP inducing IFNβ expression during L. monocytogenes infection in human myeloid cells and that this proceeds through a pathway dependent on both of the two DNA sensors IFI16 and cGAS as well as the downstream adaptor molecule STING (Supplementary Fig S8). The present work raises questions concerning the mechanistic interplay between IFI16 and cGAS in the innate DNA-sensing pathway, and also points towards a molecular understanding of Listeria bacteriolysis as being important for further understanding of the immune-pathological mechanisms of listeriosis.
Materials and Methods
Ethics statement
C57BL/6 mice were bred and maintained under specific pathogen-free conditions in the animal facilities at the University of Aarhus in accordance with the national guidelines for animal care and use. The animals were sacrificed by cervical dislocation and used to generate bone marrow-derived cells. Human macrophages were generated from monocytes purified from buffy coats collected at Aarhus University Hospital Blood bank from anonymous donors. Ethics approval was granted from the ethical committee of County of Central Jutland (permission no. M-20110199).
Cells
THP1 and U937 cells (ATCC) were cultured as non-adherent monocyte-like cells in normal growth media (RPMI containing 10% FCS, 600 μg/ml glutamine, 200 IU/ml penicillin and 100 μg/ml streptomycin (Gibco)). THP1 and U937 cells were differentiated into macrophage-like cells by addition of 150 and 50 nM Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich), respectively. Differentiation of BMMs was achieved by culture of murine bone marrow cells for 7 d in the presence of granulocyte-macrophage colony-stimulating factor (Sigma-Aldrich), and harvest of adherent cells as previously described (Rasmussen et al, 2011). Differentiation of human monocytes into hMDMs was achieved by culture for 6–8 days in the presence of macrophage colony-stimulating factor (Sigma-Aldrich) as previously described (Horan et al, 2013). The HEK-BLUE IFNα/β reporter cell line (InvivoGen) was cultivated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% foetal bovine serum (FBS) and selection markers.
shRNA-mediated silencing
The lentiviral shRNA expression plasmid pLKO.1 was utilized for the studies described here (OpenBiosystems, Huntsville, AL). Cell lines with stable knock-down of STING, IFI16, cGAS and DDX41 were generated as described previously (Horan et al, 2013; Jakobsen et al, 2013). The targeting shRNA sequence was IFN-inducible protein 16 (IFI16) clone ID TCRN0000019079; stimulator of IFN genes (Sting) clone ID TCRN000163296; Sting clone ID TCRN0000161345; dead box helicase 41 (Ddx41) clone ID TRCN0000104013; and cyclic GMP-AMP synthase (cGAS) clone ID TRCN0000146282. The control shRNA vector was empty-vector pLKO.1 with an 18-nt shuttle sequence instead of the hairpin sequences. The transduced cells were under selection with puromycin (1 μg/ml), and resistant cells were used for experiments, hence generating polyclonal cell populations. Efficiency of knock-down was verified by Western blot analysis with specific antibodies utilizing β-actin as a loading control, and was performed for all thawed cell lines prior to use in experiments. The cells were used for experiments not more than 1 month after thawing.
Bacteria
The bacteria used were WT L. monocytogenes strain LO28 and the LLO-deletion mutant (Kocks et al, 1992), 10403s serotype 1/2a, 10403s Δlmo2473, LO28 serotype 1/2c, LO28 ΔmdrT and LO28+ pIMK− (Sauer et al, 2010; Schwartz et al, 2012). For propagation of bacteria, single colonies were inoculated into 5 mL of BHI (brain–heart infusion) media and incubated overnight at 37°C without shaking. Optical density of the culture was measured at 600 nm. An OD600 of 1.0 were considered equivalent to approximately 2 × 109 CFU/ml. THP1-macrophage-like cells were infected with a MOI 1–60, in normal growth media, without penicillin and streptomycin. One hour after infection, the medium was replaced by medium containing 50 μg/ml gentamycin (Sandoz, Cat.no. 010256) to kill extracellular bacteria. After another hour, the medium was once more replaced to now contain 10 μg/ml gentamycin. For qPCR analysis, cells were lysed 6 h p.i. and RNA extracted utilizing a Roche RNA extraction kit, according to manufacturer's instructions, or cells were fixed 4 h p.i. for confocal as described in the following section.
Preparation of and stimulation with L. monocytogenes extracts
Listeria monocytogenes 5-ml overnight cultures were pelleted, resuspended in 400 μl PBS and sonicated for 2 min with cooling using Diagenode Bioruptor BCD-200. Cleared extracts were prepared and left untreated or treated with RNase (100 μg/ml) or DNase (100 unit/ml) for 45 min as described (Stetson et al, 2006). For transfection of THP1-macrophage-like cells, 5 μl of L. monocytogenes extracts were complexed with 2 μl of Lipofectamine 2000 (Invitrogen) and transfected into THP1 cells according to manufacturer's instructions. Six h p.i. cells were lysed and RNA extracted as described above.
L. monocytogenes growth assay
THP1 cells (3.5 × 106) were plated in monolayer on 12-mm round coverslips in 60-mm petri dishes containing 5 ml of medium supplemented with 150 nM PMA. After overnight incubation, the medium was replaced with PMA-free medium, and the cells were left to incubate overnight. The cells were infected with 106 bacteria per petri dish for 1 h, and the cell monolayer was washed three times with 37°C PBS followed by addition of preheated media supplemented with 10 μg/ml gentamycin. After the indicated time points, the cells were lysed by placing coverslips in duplicate in 5 ml of sterile distilled water. Tubes were vortexed for 30 s and dilutions were plated onto BHI agar plates. After overnight incubation at 37°C, the number of colonies was counted and number of colonies per coverslip was calculated.
Electron microscopy
The cells were washed twice in PBS and fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.2 overnight at 4°C. Subsequently, the cells were washed in 0.1 M cacodylate buffer, pH 7.2 and embedded in 2% agarose. This was followed by post-fixation in 1% osmium in 0.1 M cacodylate buffer, pH 7.2 for 60 min, washed as above and subsequently in maleate buffer, pH 5.2. The agarose embedded cells were én bloc stained with 0.5% uranylacetate in maleate buffer for 60 min. The cells were washed again, dehydrated in graded alcohols, transferred to propyleneoxide 3 × 10 min, followed by overnight incubation in 50% propylenoxide and 50% Epon (TAAB, Berkshire, England) and finally infiltrated with Epon. Sections with a thickness of 50–70 nm were cut for electron microscopy with a Reichert Jung Ultracut E microtome. The sections were stained with saturated uranyl acetate for 10 min and Pb-citrate for 2 min. Ultrastructural analysis was performed with a Phillips 100 CM electron microscope.
Transfection with CDNs
For transfection of THP1 cells (either undifferentiated or PMA-differentiated) with cyclic-di-AMP, cyclic-di-GMP or 3′5′ cyclic-GMP-AMP, 2 μl of Lipofectamine 2000 were complexed with cyclic-di-AMP following the instructions of the manufacturer for DNA transfection. Transfections were performed according to manufacturer's instructions to reach final concentrations of 3.6–50 μM as specified in for the individual data set. Six hours post-transfection, cells were lysed and RNA extracted as above or cells were fixed for confocal as described in the following section.
Assay for activation of IFN pathway by CDNs
Listeria monocytogenes extracts were centrifuged at 65,000 g for 15 min, the supernatants were treated with DNase, heated to 95°C for 5 min and centrifuged at 12,000 g for 5 min. Supernatants were collected. 1.5 × 105 PMA-differentiated THP1 cells were stimulated with 20 μl of c-di-AMP in the indicated concentrations, heat-resistant supernatants from L. monocytogenes extracts (in the presence or absence of spiking with synthetic c-di-AMP) in 300 μl of a digitonin-based permeabilization buffer (50 mM HEPES, pH 7, 100 mM KCl, 3 mM MgCl2, 0.1 mM DTT, 85 mM sucrose, 0.2% BSA, 1 mM ATP) containing 10 μg/ml digitonin. After incubation at 37°C for 30 min, the permeabilization buffer was replaced by RPMI with 10% FCS and l-Glutamine and the cells were incubated for another 4 h at 37°C. Expression of IFNβ and β-actin was measured by RT-qPCR.
Western blotting
Whole-cell extracts were denatured in XT Sample buffer and XT Reducing Agent and subjected to SDS–PAGE (Bio-Rad). IFI16 was detected with mouse anti-IFI16 (sc-8023, Santa Cruz), DDX41 rabbit anti-DDX41 (SAB-2100554, Sigma-Aldrich), cGAS with rabbit anti-MB21D1 (Sigma-Aldrich), STING with mouse anti-STING (MAB7169, R&D Systems), phosphor-TBK1/NAK (rabbit mAb #3504S, Cell Signaling), anti-TBK1/NAK (rabbit mAb #5483S, Cell Signaling) and β-actin with anti-β-actin-HRP (Ab9900, Abcam). Secondary antibodies used were peroxidase-conjugated F(ab′)2 donkey anti-mouse IgG (H+L) and peroxidase-conjugated Affinipure F(ab′)2 donkey anti-rabbit IgG (H+L) (both from Jackson ImmunoResearch).
IFN bioassay
The presence of bioactive human type I IFN in supernatants from cells was measured using the HEK-Blue IFNα/β reporter cell line (InvivoGen). The cell line expresses a fully active type I IFN pathway, generated through stable transfection of hSTAT2 and IRF9 genes into HEK293 cells. Furthermore, it harbours a reporter gene, expressing the secreted embryonic alkaline phosphatase (SEAP), under the control of the IFN-inducible ISG54 promoter. Detection of released type I IFN was performed according to the manufacturer's recommendations. Briefly, 50,000 HEK-BLUE cells were added to 80 μl of supernatant in a 96-well plate. After 24 h of incubation at 37°C, 20 μl of supernatants from HEK-BLUE cells were added to 180 μl Quanti-Blue reagent (InvivoGen). The colorimetric reaction was measured at 620 nm and presented as means of biological triplicates (U/ml) ± SD.
ELISA for IL-1β
Levels of IL-1β protein were measured by ELISA (DY201-05, R&D Systems). Instructions of the manufacturer were followed.
RT-PCR and primers
Gene expression was determined by real-time PCR, using TaqMan detection systems (Applied Biosystems). Expression levels were normalized to β-actin expression and data presented as the fold induction over untreated controls for each phenotype. Data represent the mean ± SD from biological replicates. IFNβ, Applied Biosystems TaqMan Assay Hs01077958_s1; β-actin, Applied Biosystems TaqMan Assay Hs99999903_m1; CXCL10, Applied Biosystems TaqMan Assay Hs00171042_m1; MxA1, Applied Biosystems Taqman Assay Hs00895608_m1, TNF-α, Applied Biosystems Taqman Assay Hs01113624_g1.
Confocal microscopy
For visualization of IFI16, following infection with bacteria at indicated times, cells were fixed and permeabilized with methanol at −20°C and labelled with antibodies against IFI16 (N-terminal, Santa Cruz sc-8023) or STING (Imgenex IMG-6485A). For visualization of IFI16 and STING following transfection with cyclic-di-AMP, and for all staining for cGAS, THP1 cells were fixed with 4% formaldehyde and permeabilized with 0.2% Triton X-100. The antibodies used were rabbit anti-cGAS (HPA031700 SIGMA, Anti-MB21D1) and sheep anti-STING (R&D Systems, AF6516). Images were acquired on Zeiss LSM 710 confocal microscope, using a 63× 1.4 oil lens. Image processing was performed using Zen 2010 (Zeiss) and ImageJ.
Statistical analysis
The data are shown as mean ± SD. The statistical significance was determined by two-tailed Student's t-test or Wilcoxon rank sum test.
Acknowledgments
The technical assistance of Kirsten Stadel Petersen and Herdis Berg Johansen is greatly appreciated.
Author contributions
KH and SRP conceived and designed the experiments. KH, TP, AL, SEJ, SHR, SBJ, RN, KAH and MRJ performed the experiments. KH and SRP analysed the data. JHL, TD and MRJ contributed reagents/materials/analysis tools. SRP wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbuddhe SB, Chakraborty T. Listeria as an enteroinvasive gastrointestinal pathogen. Curr Top Microbiol Immunol. 2009;337:173–195. doi: 10.1007/978-3-642-01846-6_6. [DOI] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. EMBO J. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CW, Crafton E, Fan HN, Flook J, Yoshimura K, Skarica M, Brockstedt D, Dubensky TW, Stins MF, Lanier LL, Pardoll DM, Housseau F. Interferon-producing killer dendritic cells provide a link between innate and adaptive immunity. Nat Med. 2006;12:207–213. doi: 10.1038/nm1352. [DOI] [PubMed] [Google Scholar]
- Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–5225. doi: 10.4049/jimmunol.1300097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Vicente MF, Mengaud J, Baquero F, Perez-Diaz JC, Berche P. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect Immun. 1989;57:3629–3636. doi: 10.1128/iai.57.11.3629-3636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crimmins GT, Herskovits AA, Rehder K, Sivick KE, Lauer P, Dubensky TW, Jr, Portnoy DA. Listeria monocytogenes multidrug resistance transporters activate a cytosolic surveillance pathway of innate immunity. Proc Natl Acad Sci U S A. 2008;105:10191–10196. doi: 10.1073/pnas.0804170105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, Tuschl T, Patel DJ. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, Rixon FJ, Herold BC, Fitzgerald KA, Paludan SR. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan KA, Hansen K, Jakobsen MR, Holm CK, Waggoner L, West JA, Unterholzner L, Iversen MB, Soby S, Thomson M, Jensen SB, Rasmussen SB, Ellermann-Eriksen S, Kurt-Jones EA, Landolfo S, Melchjorsen J, Bowie AG, Damania B, Fitzgerald KA, Paludan SR. Proteasomal degradation of herpes simplex virus capsids in macrophage releases DNA to the cytosol for recognition by DNA sensors. J Immunol. 2013;190:2311–2319. doi: 10.4049/jimmunol.1202749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, Jin T, Laustsen A, Hansen K, Ostergaard LJ, Fitzgerald KA, Xiao T, Mikkelsen JG, Mogensen TH, Paludan SR. IFI16 senses DNA forms of the retroviral replication cycle and controls HIV-1 replication. PNAS. 2013;110:E4571–E4580. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Hill KK, Filak H, Mogan J, Knowles H, Zhang B, Perraud AL, Cambier JC, Lenz LL. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595–2601. doi: 10.4049/jimmunol.1100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin TC, Perry A, Jiang JS, Smith P, Curry JA, Unterholzner L, Jiang ZZ, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. Structures of the HIN domain: DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 2012;36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 Acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–531. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Leber JH, Crimmins GT, Raghavan S, Meyer-Morse NP, Cox JS, Portnoy DA. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. PNAS. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, Fitzgerald KA, Hayakawa Y, Vance RE. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, Miller JF, Cheng G. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu Z, Modlin RL, Liu YJ, Cheng G. The helicase DDX41 recognizes the bacterial secondary messenger cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of autophagy by alpha-herpesviruses in myeloid cells is mediated by cytoplasmic viral DNA through a mechanism dependent on stimulator of IFN genes. J Immunol. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFN-alphabeta enables Listeria monocytogenes to suppress macrophage activation by IFN-gamma. J Exp Med. 2010;207:327–337. doi: 10.1084/jem.20091746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Witte CE, Zemansky J, Hanson B, Lauer P, Portnoy DA. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe. 2010;7:412–419. doi: 10.1016/j.chom.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect Immun. 2011;79:688–694. doi: 10.1128/IAI.00999-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz KT, Carleton JD, Quillin SJ, Rollins SD, Portnoy DA, Leber JH. Hyperinduction of host beta interferon by a Listeria monocytogenes strain naturally overexpressing the multidrug efflux pump MdrT. Infect Immun. 2012;80:1537–1545. doi: 10.1128/IAI.06286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solodova E, Jablonska J, Weiss S, Lienenklaus S. Production of IFN-beta during Listeria monocytogenes infection is restricted to monocyte/macrophage lineage. PLoS ONE. 2011;6:e18543. doi: 10.1371/journal.pone.0018543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stockinger S, Materna T, Stoiber D, Bayr L, Steinborn R, Kolbe T, Unger H, Chakraborty T, Levy DE, Muller M, Decker T. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- Stockinger S, Kastner R, Kernbauer E, Pilz A, Westermayer S, Reutterer B, Soulat D, Stengl G, Vogl C, Frenz T, Waibler Z, Taniguchi T, Rulicke T, Kalinke U, Muller M, Decker T. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois C, Jin T, Xiao T, Fitzgerald P, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Zhang ZQ, Yuan B, Bao MS, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–235. doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.