ABSTRACT
The amikacin resistance gene aphA6 was first detected in the nosocomial pathogen Acinetobacter baumannii and subsequently in other genera. Analysis of 133 whole-genome sequences covering the taxonomic diversity of Acinetobacter spp. detected aphA6 in the chromosome of 2 isolates of A. guillouiae, which is an environmental species, 1 of 8 A. parvus isolates, and 5 of 34 A. baumannii isolates. The gene was also present in 29 out of 36 A. guillouiae isolates screened by PCR, indicating that it is ancestral to this species. The Pnative promoter for aphA6 in A. guillouiae and A. parvus was replaced in A. baumannii by PaphA6, which was generated by use of the insertion sequence ISAba125, which brought a −35 sequence. Study of promoter strength in Escherichia coli and A. baumannii indicated that PaphA6 was four times more potent than Pnative. There was a good correlation between aminoglycoside MICs and aphA6 transcription in A. guillouiae isolates that remained susceptible to amikacin. The marked topology differences of the phylogenetic trees of aphA6 and of the hosts strongly support its recent direct transfer within Acinetobacter spp. and also to evolutionarily remote bacterial genera. Concomitant expression of aphA6 must have occurred because, contrary to the donors, it can confer resistance to the new hosts. Mobilization and expression of aphA6 via composite transposons and the upstream IS-generating hybrid PaphA6, followed by conjugation, seems the most plausible mechanism. This is in agreement with the observation that, in the recipients, aphA6 is carried by conjugative plasmids and flanked by IS that are common in Acinetobacter spp. Our data indicate that resistance genes can also be found in susceptible environmental bacteria.
IMPORTANCE
We speculated that the aphA6 gene for an enzyme that confers resistance to amikacin, the most active aminoglycoside for the treatment of nosocomial infections due to Acinetobacter spp., originated in this genus before disseminating to phylogenetically distant genera pathogenic for humans. Using a combination of whole-genome sequencing of a collection of Acinetobacter spp. covering the breadth of the known taxonomic diversity of the genus, gene cloning, detailed promoter analysis, study of heterologous gene expression, and comparative analysis of the phylogenetic trees of aphA6 and of the bacterial hosts, we found that aphA6 originated in Acinetobacter guillouiae, an amikacin-susceptible environmental species. The gene conferred, upon mobilization, high-level resistance to the new hosts. This work stresses that nonpathogenic bacteria can act as reservoirs of resistance determinants, and it provides an example of the use of a genomic library to study the origin and dissemination of an antibiotic resistance gene to human pathogens.
INTRODUCTION
Acinetobacter spp. are increasingly responsible for severe nosocomial infections, in particular, in intensive care units (1). Acinetobacter ventilator-associated pneumonia, urinary tract infections, and bacteremia are difficult to cure, at least in part because strains are often resistant to multiple antimicrobial agents. Although Acinetobacter baumannii is the most prevalent pathogen in the genus, other species have been shown to be responsible for human infections, such as Acinetobacter pittii and Acinetobacter nosocomialis and, to a lesser extent, Acinetobacter ursingii, Acinetobacter haemolyticus, Acinetobacter lwoffii, and Acinetobacter parvus (2). Improvement of identification will likely indicate that additional Acinetobacter spp. can also be isolated in clinical settings.
In A. baumannii, chromosomally encoded β-lactamases, basal expression of efflux pumps, and low membrane permeability are responsible for broad intrinsic resistance. This species also has the remarkable ability to become resistant to numerous antibiotics by horizontal acquisition of mobile genetic elements, plasmids (3), transposons (4), resistance islands (5), or a combination of all (6).
Aminoglycosides, together with β-lactams and quinolones, are the major drug classes used in therapy of Acinetobacter infections. Despite side effects, ototoxicity and nephrotoxicity, and increasing resistance, the aminoglycosides retain therapeutic value, in particular because of their bactericidal activity and the synergy they display with β-lactams.
There are four known mechanisms of resistance to aminoglycosides in bacterial human pathogens: (i) decreased intracellular accumulation of the antibiotic (7); (ii) modification of the target (8); (iii) trapping of the drug (9); (iv) enzymatic modification of the drug (10), primarily through N-acetylation, O-nucleotidylation, or O-phosphorylation, which is the most common mechanism. Since amikacin, because of an (S)-4-amino-2-hydroxybutyrate side chain moiety substituted at the N-1 position of the 2-deoxystreptamine ring, is inactivated in Gram-negative bacteria only by 6′-acetyltransferases type I and 3′-phosphotransferases types VI and VII (10), it remains one of the most active aminoglycosides for the treatment of infections due to Acinetobacter spp. (11).
Aminoglycoside 3′-phosphotransferase type VI [Aph(3′)-VI] was first reported in A. baumannii, and the structural gene for the enzyme, aphA6, was carried by self-transferable plasmids (12). Subsequent dissemination of this resistance determinant in various members of the family Enterobacteriaceae and in Pseudomonas aeruginosa is due to dissemination of the 5.2-kb Tn1528 composite transposon delimited by two copies of IS15-Δ (13). More recently, it has been reported that, in A. baumannii, aphA6 is also part of the transposon TnaphA6, in which it is immediately flanked by two copies of ISAba125 (14). The low G+C content of aphA6, 33 mol%, and the link with an insertion sequence widely spread among Acinetobacter spp. suggest an origin in this genus for the gene.
We have analyzed the genomes of 133 strains of Acinetobacter spp., covering the breadth of the known taxonomic diversity of the genus, which allowed to us to build a robust phylogeny of the entire genus (M. Touchon, J. Cury, E. J. Yoon, L. Krizova, G. Cerqueira, C. Murphy, M. Feldgarden, J. Wortman, D. Clermont, T. Lambert, C. Grillot-Courvalin, A. Nemec, P. Courvalin, E. P. Rocha, submitted for publication). This new evolutionary tree was used to assess pending taxonomic issues and sample the genus for key mechanisms generating genetic variability, and it allowed the detection of genes for numerous class D β-lactamases and Acinetobacter-derived cephalosporinases (15).
We have systematically screened these genomes for the presence of genes encoding aminoglycoside-modifying enzymes, and we report the origin of aphA6 in the chromosome of Acinetobacter guillouiae, an environmental species distantly related to A. baumannii and rarely responsible for human infections, and this gene’s intra- and intergenus dissemination.
RESULTS
The aphA6 gene is intrinsic to A. guillouiae.
Analysis of the whole-genome sequences of 133 strains of Acinetobacter allowed us to detect the aphA6 gene in 5 out of 34 A. baumannii strains, 1 of 8 A. parvus strains, and both A. guillouiae strains (Fig. 1). The level of sequence similarity between the deduced Aph(3′)-VI proteins of the various species was always higher than the average sequence similarity across all homologs shared by the two species (Table 1). As an example, the Aph(3′)-VI proteins of A. guillouiae NIPH 991 and A. parvus CIP 102159 had 99.3% sequence similarity, whereas their homologs had on average only 89.2% similarity. In both A. guillouiae strains, aphA6 was located in large contigs that also carried genes for ribosomal proteins S1, S6, S18, S20, S21, and L9, indicating a chromosomal location for aphA6. Thirty-six additional A. guillouiae isolates obtained from a wide spectrum of ecosystems, including human clinical specimens as well as soil and water samples from various natural locations, were screened for the presence of aphA6 by PCR, and 29 were found to harbor the gene (Table 2). The high prevalence of aphA6 among these ecologically diverse populations of A. guillouiae (82%) can be taken as evidence that the gene is intrinsic to this species.
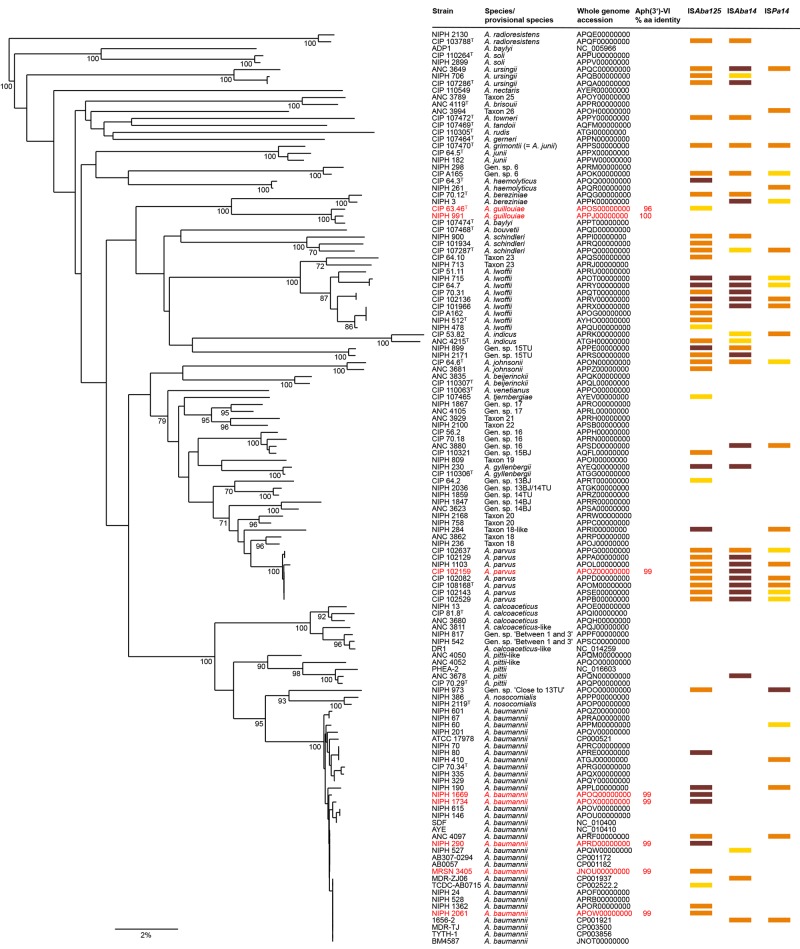
FIG 1 .
Neighbor-joining tree based on a partial (861-bp) sequence of the rpoB gene. Bootstrap values (≥70%) obtained after 1,000 replicates are given at the nodes. Bar, 2% sequence divergence. The percent amino acid identity is relative to A. guillouiae NIPH 991. The sequences were compared using the neighbor-joining method and a simple matching cost matrix. The accession numbers of the genome sequences from which the data were retrieved are indicated in the third column. Red, genome with aphA6. The distribution of the insertion sequences indicated at the top is represented with a color code: white, absence of IS; yellow, single copy; orange, a few (2 to 9) copies; brown, numerous (≥10) copies. The high similarity of the rpoB sequences of A. guillouiae and A. baylyi CIP 107474T is likely a result of the transfer of the complete rpoB gene from A. guillouiae to this A. baylyi strain (31).
TABLE 1 .
Similarities between homologs and Aph(3′)-VI proteins
| Species and strain | % similarity with homolog or Aph(3′)-VI proteina
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
A. guillouiae |
A. parvus |
A. baumannii |
||||||
| CIP 63.46T | NIPH 991 | CIP 102159 | NIPH 1669 | NIPH 1734 | NIPH 2061 | NIPH 290 | MRSN 3405 | |
| A. guillouiae | ||||||||
| CIP 63.46T | 98.8 | 89.5 | 89.1 | 89.1 | 89.1 | 89.2 | 89.2 | |
| NIPH 991 | 97.4 | 89.2 | 89.1 | 89.1 | 89.1 | 89.1 | 89.0 | |
| A. parvus | ||||||||
| CIP 102159 | 98.1 | 99.3 | 90.6 | 90.6 | 90.6 | 90.6 | 90.6 | |
| A. baumannii | ||||||||
| NIPH 1669 | 97.6 | 99.2 | 99.5 | 99.3 | 99.3 | 99.4 | 99.4 | |
| NIPH 1734 | 97.6 | 99.2 | 99.5 | 100.0 | 99.3 | 99.3 | 99.3 | |
| NIPH 2061 | 97.6 | 99.2 | 99.5 | 100.0 | 100.0 | 99.3 | 99.3 | |
| NIPH 290 | 97.6 | 99.2 | 99.5 | 100.0 | 100.0 | 100.0 | 99.8 | |
| MRSN 3405 | 98.1 | 99.3 | 100.0 | 99.7 | 99.7 | 99.7 | 99.7 | |
The average similarities between homologous proteins (values in lightface) or with Aph(3′)-VI (values in boldface); similarity was defined as unique pairwise reciprocal best hits with at least 80% similarity in amino acid sequence and less than 20% difference in protein length. Analysis of orthology was made for every pair of genomes.
TABLE 2 .
A. guillouiae strains studied
| Strain | Specimen, country,a yr of isolation | aphA6b | MIC (μg/ml)c |
Reference | ||
|---|---|---|---|---|---|---|
| Kanamycin | Amikacin | Gentamicin | ||||
| ANC 4134d | Meadow mud, CZ, 2011 | + | >32 | 8 | 1 | This study |
| NIPH 2408d | Raw milk, CZ | + | >32 | 0.5 | 0.5 | 31 |
| NIPH 820 | Unknown | + | 32 | 4 | 0.5 | 31 |
| NIPH 2127 | Wound (human), SE, 1980 | + | 32 | 4 | 0.5 | 31 |
| NIPH 682d | Blood (human), CZ, 1997 | + | 32 | 2 | 1 | 31 |
| ANC 3935d | Forest soil, CZ, 2011 | + | 32 | 2 | 0.5 | This study |
| ANC 4146d | Forest soil, CZ, 2011 | + | 32 | 2 | 0.5 | This study |
| ANC 4181 | Pond mud, CZ, 2011 | + | 16 | 2 | 0.5 | This study |
| ANC 3814d | Freshwater sediment, CZ, 2009 | + | 16 | 1 | 0.5 | This study |
| CIP 63.46Td,e | Sewage, before 1951 | + | 16 | 1 | 0.5 | 31 |
| ANC 4111d | Freshwater sediment, CZ, 2011 | + | 8 | 0.5 | 0.25 | This study |
| ANC 4170 | Creek mud, CZ, 2011 | + | 8 | 2 | 0.5 | This study |
| NIPH 2272 | Contact lens, SE, 1980 | + | 8 | 1 | 1 | 31 |
| ANC 4125 | Meadow mud, CZ, 2011 | + | 8 | 1 | 1 | This study |
| ANC 3626d | Forest soil, CZ, 2007 | + | 8 | 1 | 0.5 | 31 |
| ANC 4145 | Forest soil, CZ, 2011 | + | 8 | 1 | 0.5 | This study |
| ANC 3826 | Freshwater sediment, CZ, 2009 | + | 8 | 1 | 0.5 | This study |
| ANC 3827 | Freshwater sediment, CZ, 2009 | + | 8 | 1 | 0.5 | This study |
| NIPH 2169 | Sputum (human), DK, 1988–1989 | + | 8 | 1 | 0.25 | 31 |
| ANC 3812 | Freshwater sediment, CZ, 2009 | + | 4 | 0.5 | 0.5 | This study |
| ANC 3679d | Forest soil, CZ, 2008 | + | 4 | 1 | 0.5 | This study |
| ANC 4136 | Freshwater sediment, CZ, 2011 | + | 2 | 1 | 1 | This study |
| ANC 4143 | Freshwater sediment, CZ, 2011 | + | 2 | 1 | 1 | This study |
| NIPH 2689 | Activated sludge plant, AU, 1995 | + | 2 | 0.5 | 0.5 | 31 |
| ANC 4133d | Creek mud, CZ, 2011 | + | 2 | 0.5 | 0.5 | This study |
| NIPH 2529 | Blood (human), NL, 1999 | + | 1 | 0.5 | 0.5 | 31 |
| ANC 4258d | Creek mud, CZ, 2012 | + | 1 | 0.5 | 0.5 | This study |
| NIPH 991d,e | Ear swab (human), CZ, 1998 | + | 1 | 0.5 | 0.25 | 31 |
| NIPH 769 | Barley field soil, UK, 1993–1994 | + | 0.5 | 0.5 | 0.5 | 31 |
| ANC 4184 | Creek mud, CZ, 2011 | + | 0.5 | 0.5 | 0.25 | This study |
| ANC 4140d,f | Meadow mud, CZ, 2011 | + | 1 | 0.5 | 0.5 | This study |
| NIPH 2273d | Urine (human), SE, 1980 | − | 0.5 | 0.5 | 0.5 | 31 |
| NIPH 2525 | Freshwater with sediment, DK, 1997 | − | 0.5 | 0.5 | 0.25 | 31 |
| NIPH 2536 | Eye (cat), NL, 2001 | − | 1 | 0.5 | 0.5 | 31 |
| NIPH 2681 | Feces (human), NL, 2000 | − | 1 | 0.5 | 0.5 | 31 |
| ANC 3834d | Freshwater sediment, CZ, 2009 | − | 0.5 | 0.5 | 0.5 | This study |
| ANC 3911d | Forest soil, CZ, 2010 | − | 0.5 | 0.5 | 0.5 | This study |
| NIPH 2680 | Feces (human), NL, 2000 | − | 0.5 | 0.25 | 0.25 | 31 |
Country abbreviations: AU, Australia; CZ, Czech Republic; DK, Denmark; NL, The Netherlands; SE, Sweden; UK, United Kingdom.
Presence (+) or absence (−) of the gene, detected by PCR.
Strain analyzed by qRT-PCR; the promoter and aphA6 gene were sequenced.
Strains for which the whole genome was sequenced. Information for such strains is shown in boldface.
Strain with truncated aphA6 gene.
Genomic environment of aphA6.
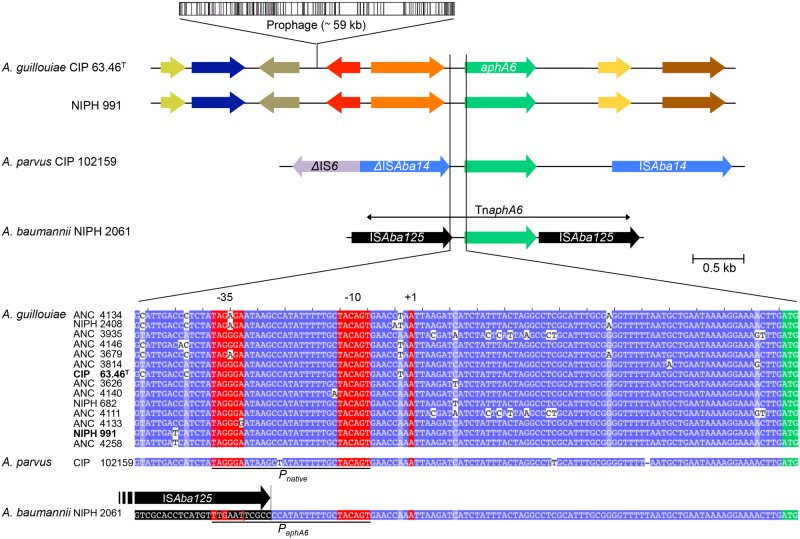
The A. guillouiae and A. parvus strains had a nearly identical 153-bp sequence upstream from aphA6 that included the Pnative promoter (Fig. 2). The promoter was located from bp 104 to bp 75 upstream from the start codon of aphA6, and the −35 and −10 sequences were separated by 17 bp. The aphA6 gene in the five A. baumannii isolates was part of a 3.1-kb composite transposon, TnaphA6, which consists of two 1.1-kb directly oriented copies of ISAba125 bracketing the aphA6 gene, as described for multiply antibiotic-resistant A. baumannii clinical strains (14). The 93 bp between the end of the right inverted repeat (IRR) of the upstream ISAba125 copy and the beginning of aphA6 in the five A. baumannii isolates were identical to those in the A. guillouiae and A. parvus strains (Fig. 2; see also Table S1 in the supplemental material). Insertion of the left ISAba125 copy in A. baumannii disrupted the Pnative promoter between the −35 and −10 sequences and brought a new −35 sequence, generating the strong PaphA6 hybrid promoter. This promoter has recently been described as PNDM-1 upstream from blaNDM-1 (16); this region has been shown convincingly to be the result of a fusion, which likely occurred in A. baumannii, between the promoter of aphA6, designated PaphA6, the first 19 bp of aphA6, and the N-terminal portion of the blaNDM-1 gene (17).
FIG 2 .
Genomic environment and promoters of aphA6. (Top) Arrows indicate open reading frames and sense of transcription. Open box, prophage; colored arrows, ORFs; green arrow, aphA6; purple arrow, IS6; blue arrow, ISAba14; black arrow, ISAba125; double-headed arrow, TnaphA6. (Bottom) Sequence alignment of aphA6 promoters from 14 A. guillouiae, A. parvus CIP 102159, and A. baumannii NIPH 2061 isolates. Identity between the sequences is indicated in blue. The Pnative and PaphA6 hybrid promoters are underlined; the −35 and −10 sequences and the transcriptional start site (+1) as determined by 5′ RACE-PCR are in red boxes; the start codon of aphA6 is in a green box. Boldface indicates the whole genome sequence of A. guillouiae. The promoter of A. baumannii NIPH 2061 was determined previously (16).
The two A. guillouiae strains have a nearly identical 100-kb region of DNA (similar gene content and order) flanking the aphA6 gene, except for the presence of a large prophage integrated 2.5 kb upstream from aphA6 in A. guillouiae CIP 63.46T (Fig. 2). The genomic context of the aphA6 gene in the A. guillouiae strains was determined by PCR mapping over ca. 4.5 kb (see Fig. S1 in the supplemental material). The 12 strains studied had aphA6 flanking regions indistinguishable from those in strains NIPH 991 and CIP 63.46T. No large insertions or deletions in the noncoding regions were detected. Strains ANC 4133 and ANC 4140 had undergone DNA rearrangements, deletions, or large insertions downstream from the first open reading frame (ORF) on the right side of aphA6. The conserved environment of the gene in A. guillouiae is consistent with its ancestral integration in the chromosome of this species. In the seven aphA6-negative A. guillouiae strains, only the upstream ORF was found, indicating loss of the gene following various deletion events (see Fig. S1).
In A. parvus CIP 102159, aphA6 was part of a small contig (ca. 1.8 kb) flanked by 53-bp and 43-bp truncated copies of ISAba14 located, respectively, 154 bp upstream and 844 bp downstream from the gene (Fig. 2; see also Table S1 in the supplemental material). In order to determine the right and left regions flanking this small fragment, the sequences upstream and downstream from aphA6 were determined by using thermal asymmetric interlaced PCR (TAIL-PCR). The presence of the adjacent ISAba14 was confirmed, with the left copy, beginning at position 343, truncated, whereas the right copy was complete (1,282 bp) (see Fig. S2 in the supplemental material). Comparative analysis revealed 21 base changes between the two IS. Conventional PCR with primers 102159-Fi and 102159-Ri (see Table S2 and Fig. S2 in the supplemental material) complementary to sequences outside the ΔISAba14-aphA6-ISAba14 region confirmed the TAIL-PCR results by amplification of the expected 4,198-bp fragment but also of a 1,140-bp fragment (see Fig. S2). Sequence determination of the latter revealed ΔISAba14, the result of the fusion of the 5′ portion of the left copy with the 3′ portion of the right copy, generated by recombination at position 510 based on the substitutions between the two IS. In addition, a circular form containing aphA6 and ISAba14 with the expected mutations of the two copies, which represented the excision product following the homologous recombination event, was also found by using inverse PCR carried out with primers 102159-R1 and 102159-F1 (see Table S2). PCR amplification with primers 102159-Fi and 102159-Ri (see Table S2 and Fig. S2) on genomic DNA from six aphA6-negative A. parvus strains confirmed the presence of a ΔISAba14 target for integration of the gene. In summary, analysis of the genomic environment of aphA6 in A. parvus allowed detection of the gene integrated in the chromosome and when excised, and also to identify the target IS element for integration.
Resistance conferred by Aph(3′)-VI.
In contrast to other Aph(3′) proteins from Gram-negative bacteria, the type VI enzyme confers resistance, not only to kanamycin but also to amikacin (12). Throughout the whole-genome analysis, it was found that A. baumannii strains NIPH 290, NIPH 1669, NIPH 1734, and MRSN 3405 encode additional aminoglycoside-modifying enzymes, such as Ant(2″)-Ia and Aph(3′)-I, whose substrates include kanamycin. In addition, A. baumannii NIPH 290 and MRSN 3405 harbored five and two aphA6 copies, respectively (see Table S1 in the supplemental material). A. baumannii NIPH 2061, which had a single copy of aphA6 and no other aminoglycoside resistance gene, was thus selected for further studies. As expected, this strain had high MICs of kanamycin, 1,024 µg/ml, and amikacin, 128 µg/ml. In contrast, A. guillouiae CIP 63.46T and NIPH 991 had low kanamycin MICs, of 16 and 1 µg/ml, respectively, and remained susceptible to amikacin, with MICs of 1 and 0.5 µg/ml, respectively (18, 19). The aminoglycoside MICs against A. parvus CIP 102159 could not be determined due to extremely slow growth of the strain. The susceptibilities to kanamycin and amikacin of the 29 additional aphA6-containing A. guillouiae strains were determined (Table 2), and the strains did not produce other aminoglycoside-modifying enzymes that might affect amikacin activity (data not shown). The MICs of kanamycin against the strains ranged from 0.5 to >32 µg/ml, but all the strains remained susceptible (MICs ≤8 µg/ml) to amikacin (18, 19).
Two possibilities could account for the unexpected susceptibility of the aphA6-carrying A. guillouiae strains: (i) mutations affecting the enzyme activity and (ii) low levels of gene expression.
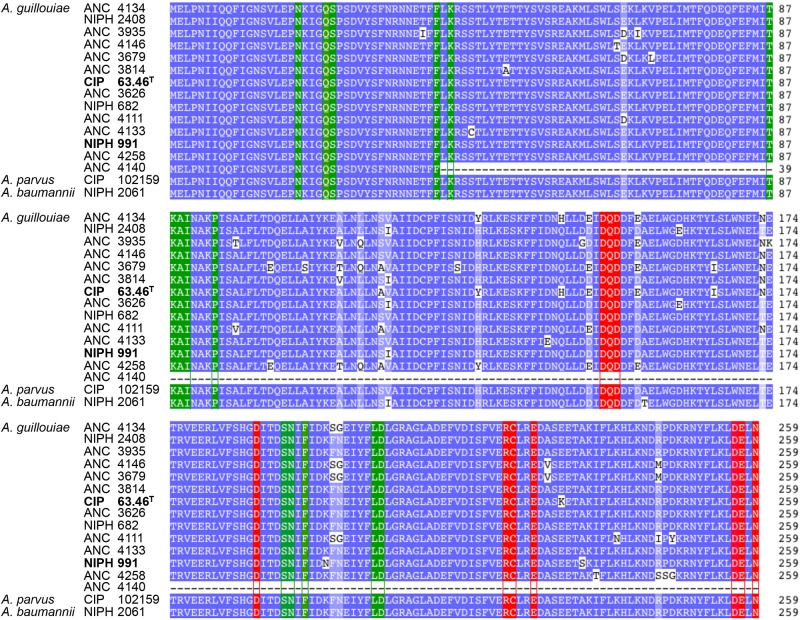
The sequences of the aphA6 genes in the five A. baumannii strains were nearly identical (Table 1). The deduced sequences of Aph(3′)-VI from the two A. guillouiae strains and of A. baumannii NIPH 2061 were compared, together with the deduced sequence of A. parvus and of 12 additional A. guillouiae strains (Fig. 3). Despite various substitutions, we did not find any mutations in the residues that were part of the active site (20).
FIG 3 .
Comparison of Aph(3′)-VI sequences. Multiple alignment of the deduced sequences of Aph(3′)-VI from A. guillouiae and A. parvus CIP 102159 and strain NIPH 2061, representatives of the five identical A. baumannii genes. Amino acids are shaded according to percent identity; darker amino acids are more highly conserved. Red box, antibotic binding site; green box, ATP binding site (20).
Resistance correlates with aphA6 expression.
The level of aphA6 expression in the same 14 A. guillouiae isolates was determined by quantitative reverse transcriptase PCR (qRT-PCR), normalized to that of rpoB, and compared with the kanamycin and amikacin MICs (Fig. 4). The A. baumannii strains had aphA6 expression levels 102- to 106-fold higher than those of the A. guillouiae strains. There was good correlation between the aminoglycoside MICs and the aphA6 expression levels in the A. guillouiae strains, and the kanamycin MICs were always higher than those of amikacin. A. guillouiae ANC 4134, NIPH 2408, and ANC 3679 had the same mutation, G→A, in the Pnative −35 sequence (Fig. 2), very high MICs of kanamycin (Table 2), and their genes were the most highly expressed (Fig. 4). Interestingly, A. guillouiae ANC 4134, which had respective kanamycin and amikacin MICs of >32 and 8 µg/ml, expressed aphA6 at a level intermediate between the levels for the A. guillouiae and A. baumannii strains. There was good agreement between kanamycin and amikacin MICs.
FIG 4 .
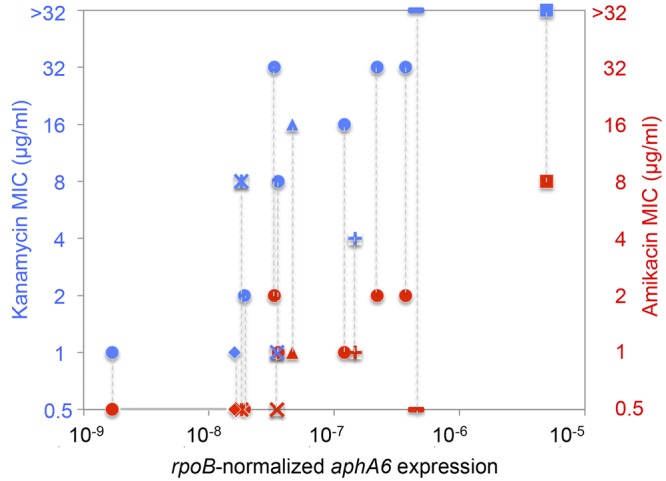
Aminoglycoside MICs and aphA6 expression. The MICs of kanamycin (blue) and amikacin (red) against 14 A. guillouiae strains were determined by agar dilution, and rpoB-normalized aphA6 expression was measured by qRT-PCR in duplicate in two independent experiments. ■, ANC 4134; ♦, NIPH 991; ▲, CIP 63.46T; —, NIPH 2408; X, ANC 4140; +, ANC 3679; *, ANC 4133; ●, remaining A. guillouiae strains.
To compare the strength of the Pnative and PaphA6 promoters, the genes from A. guillouiae CIP 63.46T and NIPH 991 and that of A. baumannii NIPH 2061 (Table 3) were cloned under the control of the Pnative promoter or that of PaphA6 in shuttle vector pAT747 (pUC18ΩoripWH1266) into Escherichia coli TOP10 and transferred to aminoglycoside-susceptible A. baumannii BM4587.
TABLE 3 .
Susceptibility to aminoglycosides
| Strain/plasmida | MIC (μg/ml)b |
|
|---|---|---|
| Kanamycin | Amikacin | |
| E. coli TOP10 | 2 | 1 |
| E. coli TOP10/pAT747 (pUC18Ωori pWH1266) | 2 | 1 |
| A. baumannii BM4587 | 2 | 2 |
| A. baumannii BM4587/pAT747 (pUC18Ωori pWH1266) | 2 | 2 |
| A. guillouiae CIP 63.46T | 16 | 1 |
| E. coli TOP10/pAT747ΩPnativeaphA6 A. guillouiae CIP 63.46T | 512 | 32 |
| E. coli TOP10/pAT747ΩPaphA6aphA6 A. guillouiae CIP 63.46T | 2,048 | 128 |
| A. baumannii BM4587/pAT747ΩPnativeaphA6 A. guillouiae CIP 63.46T | 1,024 | 128 |
| A. baumannii BM4587/pAT747ΩPaphA6aphA6 A. guillouiae CIP 63.46T | 4,096 | 1,024 |
| A. guillouiae NIPH 991 | 1 | 0.5 |
| E. coli TOP10/pAT747ΩPnativeaphA6 A. guillouiae NIPH 991 | 32 | 4 |
| E. coli TOP10/pAT747ΩPaphA6aphA6 A. guillouiae NIPH 991 | 256 | 16 |
| A. baumannii BM4587/pAT747ΩPnativeaphA6 A. guillouiae NIPH 991 | 512 | 64 |
| A. baumannii BM4587/pAT747ΩPaphA6aphA6 A. guillouiae NIPH 991 | 2,048 | 256 |
| A. parvus CIP 102159 | NGc | NG |
| E. coli TOP10/pAT747ΩPnativeaphA6 A. parvus CIP 102159 | 2,048 | 256 |
| A. baumannii BM4587/pAT747ΩPnativeaphA6 A. parvus CIP 102159 | 4,096 | 1,024 |
| A. baumannii NIPH 2061 | 1,024 | 128 |
| E. coli TOP10/pAT747ΩPaphA6aphA6 A. baumannii NIPH 2061 | 2,048 | 128 |
| A. baumannii BM4587/pAT747ΩPaphA6aphA6 A. baumannii NIPH 2061 | 4,096 | 1,024 |
There were no changes in aminoglycoside resistance of E. coli TOP10 or A. baumannii BM4587 after acquisition of pAT747 (Table 3). The genes from A. guillouiae were expressed and functional under the control of their respective Pnative promoter in both E. coli and A. baumannii, where they conferred resistance not only to kanamycin but also to amikacin. However, they conferred much higher levels of resistance (4 to 8 times higher) when expressed from the hybrid PaphA6. The MICs of kanamycin and amikacin were 4 to 16 times higher against E. coli and A. baumannii harboring the constructs with aphA6 from CIP 63.46T compared to those containing the gene from NIPH 991, reflecting the difference in levels of resistance of the respective A. guillouiae strains (Table 3). Since this difference was also observed in plasmids for which the genes were expressed from PaphA6, it is likely that it was not due to the fact that the CIP 63.46T gene was expressed at two-fold-higher levels (Fig. 4).
As expected, the MICs of kanamycin were always higher than those of amikacin. Amikacin is a derivative of kanamycin A that has an aminohydroxybutyric acid substitution at position 1 of the deoxystreptamine ring, which partially blocks modification at the 3′- and 2″-hydroxyl and 3-amino groups.
Unexpectedly, the MICs of both drugs were always higher against the A. baumannii than the E. coli hosts, despite the fact that the plasmid copy number, determined by qPCR, was much more elevated in E. coli (ca. 250 copies per genome equivalent) than in A. baumannii (ca. 20 copies). This confirmed the notion of the importance of the bacterial host background for expression of resistance, and it constitutes additional indirect evidence for the origin of aphA6 in an Acinetobacter species.
The aphA6 gene from A. parvus was also readily expressed in both new hosts, and more so in A. baumannii (Table 3). Heterologous expression of aphA6 from A. guillouiae NIPH 991 and A. parvus CIP 102159 was as efficient as that from A. baumannii NIPH 2061, as judged by the levels of resistance achieved.
Dissemination of aphA6.
Phylogenetic trees of aphA6 containing Acinetobacter spp. were constructed by using the partial sequence (861 bp) of the rpoB (Fig. 1) or aphA6 gene (see Fig. S3 in the supplemental material). The rpoB tree was congruent with the results of the previous analysis (Fig. 1), presenting three distinct clades of the respective species (see Fig. S3A). However, that of aphA6 generated a single group (see Fig. S3B), which was consistent with the high degree of sequence similarity observed between the enzymes of the three species (Table 1). The marked differences in topologies of the trees strongly support horizontal transfer of the aphA6 gene within Acinetobacter spp.
Together with Aac(6'), the Aph(3′) proteins are the most widely distributed aminoglycoside-modifying enzymes in human bacterial pathogens (10). We have comparatively analyzed the various types (isozyme forms) and subtypes (resistance profiles conferred) of Aph(3′) enzymes found in Gram-positive and -negative bacteria (see Fig. S4 in the supplemental material). This allowed us to clearly distinguish the known types and subtypes and also to identify two new types, VIII in A. rudis and IX in A. gerneri, which are closely related to but distinct from type VI (our unpublished data). The Aph(3′)-VI proteins, one of the most widely spread types, were monophyletic with more than 92.7% sequence identity and were intertwined in various genera, confirming recent horizontal transfer of the corresponding genes, not only among Acinetobacter spp. but also to phylogenetically remote Gram-negative bacteria, such as Enterobacteriaceae and P. aeruginosa (see Fig. S4 in the supplemental material). Analysis of the sequences surrounding aphA6 (see Fig. S5 in the supplemental material) indicated that the gene was generally carried by self-transferable plasmids and often flanked by an IS or remnants of an IS, most often ISAba125 and ISAba14, and less frequently ISPa14; the three genetic elements are commonly found in the genomes of Acinetobacter spp., in particular in that of A. parvus, where aphA6 reversibly integrates in the chromosome and could represent an intermediate in transfer (Fig. 1).
DISCUSSION
The structural gene aphA6 is of clinical importance in Gram-negative bacteria since it encodes an enzyme, Aph(3′)-VI, which modifies amikacin, the most potent aminoglycoside in therapy of infections due to Acinetobacter spp. The gene, which was first detected in A. baumannii (12), has disseminated to numerous members of the family Enterobacteriaceae and to P. aeruginosa (see Fig. S4 in the supplemental material) by a combination of plasmid transfer (3) and transposition (4). We suggested an origin in this genus for aphA6, based on the base composition of the gene and on its physical link with an insertion sequence common in Acinetobacter spp.
The gene was part of the chromosome of the two A. guillouiae isolates in this study (see Table S1 in the supplemental material) and is present in the vast majority of the additional strains of this species screened by PCR (Table 2), confirming that it is ancestral to A. guillouiae. This raises the question of the physiological role of Aph(3′)-VI insofar as the corresponding gene is poorly expressed (Table 3 and Fig. 4) and can be lost (Table 2). Since it was not found in the other Acinetobacter spp., with the exception of a single strain of A. parvus and of the A. baumannii strains where, as already mentioned, it is acquired and part of transposable elements (Fig. 2; see also Fig. S5 in the supplemental material) (13, 14), this confirms that A. guillouiae is a likely source for aphA6.
The reservoir species is one of the most prevailing Acinetobacter species in natural environments, as revealed in the current prospective study focused on cultivable Acinetobacter strains in soil and water ecosystems in the Czech Republic which are not directly associated with human activity (A. Nemec and L. Krizova, unpublished data).
The mechanism for intra- and intergeneric transfer of the amikacin resistance gene from A. guillouiae to other Acinetobacter spp., and to members of the family Enterobacteriaceae and P. aeruginosa, remains unknown. However, and importantly, A. guillouiae is occasionally found also in human clinical specimens, which makes direct horizontal acquisition from this species to human pathogens conceivable. During this process, concomitant expression of the gene must have occurred, since it confers high-level resistance in the new hosts (Table 3) (12). A possible two-step scenario would be mobilization and expression of aphA6 by the formation of a composite transposon, the upstream copy of the IS element generating the potent hybrid promoter PaphA6 (Fig. 2), followed by lateral transfer by plasmid conjugation. ISAba125 suggests itself as a prominent candidate for this dual role since, in addition to providing a −35 promoter sequence (Fig. 2) (16), it is widely spread in Acinetobacter spp., including A. guillouiae (Fig. 1), and commonly associated with aphA6 in the recipients (see Fig. S5 in the supplemental material) (14). Decryption of the silent aac(6′)-Ij gene intrinsic to Acinetobacter sp. 13 by IS18 led to resistance to amikacin (21), and that of the ampC gene leading to cephalosporin resistance following insertion of ISAba125 has been reported in A. baumannii (22).
It has been proposed that the only transfer of a blaOXA gene for a class D β-lactamase within the Acinetobacter genus was from genomic species 6 to A. parvus (15). Remarkably, recipient A. parvus, despite the fact it has one of the smallest genomes among Acinetobacter spp., contains the 14 key type IV pilus and competence-associated components (M. Touchon et al., submitted for publication), and transformation could thus be a mechanism for acquisition of foreign genetic information. The aphA6 gene in A. parvus is part of the remnant of a composite transposon with two terminal direct copies of ISAba14 (Fig. 2) that are able to mediate excision and integration of the gene. This structure is compatible with integration of aphA6 by homologous recombination between the two IS following transformation. Alternatively, aphA6 could have been spread by transduction, since we found, upstream from the gene in A. guillouiae CIP 63.46T, the remnants of a large prophage; we recently showed that prophages are very common in Acinetobacter species (M. Touchon et al., submitted for publication). Whatever the actual mechanism of transfer, it must have been a recent direct event, since the promoters and regions upstream from aphA6 are perfectly conserved in A. guillouiae, A. parvus, and A. baumannii (Fig. 2).
The recently described metallo-β-lactamase NDM-1 confers resistance to all β-lactams except aztreonam. It has been convincingly shown that blaNDM-1 is a chimeric gene resulting from a very recent in-frame fusion between the N-terminal portion of aphA6 extending to PaphA6 and a β-lactamase gene that occurred in an Acinetobacter sp. (17). It has also been proposed that worldwide dissemination of blaNDM-1 is, at least in part, due to control of its expression by the strong hybrid promoter PaphA6, which has broad-host-range activity (17). The blaNDM-1 and aphA6 genes are often physically linked on plasmids in Gram-negative bacteria, and their dissemination could therefore be the result of coselection.
It has been proposed that certain antibiotic resistance genes in human bacterial pathogens originated from antibiotic-producing microorganisms, which have to protect themselves against suicide (23). However, there are a few examples of species reservoirs of resistance genes in which, like aphA6 in A. guillouiae, the gene is not (or poorly) expressed, e.g., Acinetobacter radioresistens for blaOXA-23 (24), Kluyvera spp. for blaCTX for CTX-M-type extended-spectrum β-lactamases (25), and Shewanella algae for quinolone resistance determinant qnrA (26). Based on the data presented here, it thus appears that such genes should be searched for, not only in resistant (27) but also in environmental susceptible nonpathogenic bacteria.
MATERIALS AND METHODS
Bacterial strains.
The whole-genome sequences of 133 Acinetobacter strains, including 13 genomes that are reported in GenBank RefSeq (28), 118 strains from the collection of A. Nemec (designated NIPH or ANC) and of the Institut Pasteur (designated CIP), A. baumannii MRSN 3405 (29), and BM4587 (30) were studied (Fig. 1). The strains were selected to reflect the currently known breadth of the diversity of the genus Acinetobacter at the species level (15). In addition, 36 strains of A. guillouiae, chosen to represent different sources, including human and animal specimens as well as water and soil samples obtained from natural ecosystems, were screened for the presence of the aphA6 gene (Table 2). Of these 36 strains, 15 were from the original study that described A. guillouiae (31), while 21 were selected from an as-yet-unpublished taxonomic study focused on the distribution of cultivable Acinetobacter strains in the natural ecosystems of the Czech Republic (Nemec and Krizova, unpublished). The 21 environmental strains were identified as A. guillouiae based on comprehensive phenotypic testing, rpoB gene comparative analysis, and whole-cell matrix-assisted laser desorption ionization–time of flight mass spectrometry profiling (31, 32; Nemec and Krizova, unpublished). E. coli One Shot TOP10 (Invitrogen) and A. baumannii BM4587 were used as recipients for cloning the aphA6 gene. Bacteria were grown according to their physiological requirements, at 30°C to 37°C, in brain heart infusion broth and agar (Difco Laboratories).
Antimicrobial susceptibility testing.
MICs were determined by microdilution in Mueller-Hinton broth (Bio-Rad) or dilution in Mueller-Hinton II agar (BBL BD) (18, 19).
Sequence analysis.
The aphA genes inferred from whole-genome sequences obtained in this work or retrieved from GenBank were compared with known sequences by using the BLAST tool of the national Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov) (33). Multiple-sequence alignments and the amino acid identity calculation of the deduced protein sequences were carried out by using the MUSCLE program (multiple sequence comparison by log-expectation; http://www.ebi.ac.uk/Tools/msa/muscle/). Comparative nucleotide sequence analysis of the RNA polymerase β-subunit (rpoB) gene was performed according to methods described by Nemec et al. (32). Similarity calculations and cluster analyses were carried out using Bionumerics 6.6 software with default parameters for the region corresponding to nucleotide positions 2915 to 3775 of the rpoB coding region of A. baumannii CIP 70.34T. The promoter and coding regions of aphA6 in 12 of the 36 additional A. guillouiae strains were amplified with the aphA6-up200 and aphA6-down80 primers (see Table S2 in the supplemental material), the products were sequenced with primers aphA6-up200, aphA6-down80, aphA6-R, and aphA6-F2, and the sequences were analyzed with the GCG sequence analysis software package (version 10.1; Genetics Computer Group, Madison, WI).
DNA manipulation and recombinant DNA techniques.
Genomic DNA was extracted as described previously (15). DNA amplification was performed in a GeneAmp PCR 9700 system (PerkinElmer Cetus) with Phusion high-fidelity DNA polymerase (Thermo Scientific). The PCR products were purified with a QIAquick PCR purification kit (Qiagen). Digestion with restriction endonucleases (New England Biolabs), ligation with T4 DNA ligase (New England Biolabs), and transformation with recombinant plasmid DNA were performed by standard methods. Plasmid DNA was purified with a Nucleospin plasmid miniprep kit (Macherey-Nagel). Nucleotide sequencing was carried out with a CEQ 8000 DNA analysis system automatic sequencer (Beckman Instruments).
Identification of the aphA6 gene.
The list of putative homologs between pairs of genomes was determined with blastp based on the sequence of Aph(3′)-VI (34). The presence of aphA6 in the additional A. guillouiae strains was determined by PCR using primers aph3-VIA1 and -VIA2 (see Table S2 in the supplemental material) (35). The reactions were performed in a final volume of 20 µl containing 10 µl of Taq PCR master mix (Qiagen), 0.2 µM of each primer, and 1.5 µl of a DNA suspension obtained by alkaline lysis. The amplification reactions were performed with the following parameters: 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, and 60 s at 72°C.
RNA isolation and qRT-PCR.
Total RNA was extracted from exponentially growing bacterial cells (optical density at 600 nm, ≈0.8) by using TRIzol reagent (Invitrogen) (30). RNA samples were treated using the Turbo DNA-free kit (Applied Biosystems) to remove any genomic DNA carryover. Expression of the aphA6 and rpoB genes was quantified by qRT-PCR using a LightCycler RNA amplification kit with SYBR green I (Roche Diagnostics) with the following cycle profile: 1 cycle at 95°C for 30 s, followed by 45 cycles at 95°C for 5 s, 56°C for 10 s, and 72°C for 20 s. The primers used are listed in Table S2 in the supplemental material. The transcriptome of aphA6 was normalized to that of rpoB, and each experiment was performed in duplicate at least twice independently.
Construction of A. baumannii-E. coli shuttle vector pAT747.
A 1,349-bp fragment containing the replication origin of plasmid pWH1266 (36) was amplified with primers ori-pWH-AflIII-F and ori-pWH-AflIII-R (see Table S2 in the supplemental material), digested by AflIII, and ligated to AflIII-linearized pUC18 to generate pAT747, which was transformed into E. coli TOP10 cells with selection on medium containing ampicillin at 100 µg/ml.
Cloning of aphA6 genes.
The 1,060-bp fragment that included 175 bp upstream from the −35 motif of the aphA6 promoter was amplified from A. baumannii NIPH 2061 with primers ISaba125-BamHI-F and aphA6-XbaI-R (see Table S2 in the supplemental material). Similarly, 1,091-bp fragments encompassing 195 bp upstream from the −35 sequence of the promoter were amplified from A. guillouiae CIP 63.46T and NIPH 991 by using aphA6-guillouiae-BamHI-F and aphA6-XbaI-R primers (see Table S2), and the BamHI-XbaI-digested products were ligated to BamHI-XbaI-linearized pAT747. The 1,081-bp fragment was amplified from A. parvus CIP 102159 by using aphA6-parvus-F and aphA6-XbaI-R primers (see Table S2) and digested by XbaI, and the fragment including 168 bp upstream from the −35 sequence of the promoter was ligated to XbaI-linearized pAT747. Plasmids pAT747ΩPaphA6aphA6 were constructed by successive PCR. First, the 295-bp PaphA6 fragment was amplified from A. baumannii NIPH 2061 with primers ISaba125-BamHI-F and aphA6-comp-R, and the 795-bp aphA6 genes from A. guillouiae NIPH 991 and CIP 63.46T were amplified with primers aphA6-comp-F and aphA6-XbaI-R. In the second step, the two products were linked in 1,076-bp fragments by overlapping PCR using primer pair ISaba125-BamHI and aphA6-XbaI-R to obtain PaphA6aphA6, which was digested with BamHI and XbaI and cloned into pAT747. The plasmids were transformed into E. coli TOP10 with selection on medium containing ampicillin at 100 µg/ml and kanamycin at 25 µg/ml and electrotransformed into A. baumannii BM4587 cells with selection on medium containing ticarcillin at 30 µg/ml and kanamycin at 25 µg/ml. The orientations and sequences of all the inserts were verified by using forward and reverse universal primers.
Determination of the transcriptional start site.
5′-rapid amplification of cDNA ends PCR (5′ RACE-PCR) was carried out with the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer’s instruction, using nested aphA6 gene-specific primers aphA6-R and aphA6-R2 (see Table S2 in the supplemental material). The resulting amplified fragments were purified, cloned into pCR-Blunt (Invitrogen), and sequenced.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Genomic context of aphA6 in A. guillouiae strains. (A) Map of the 4,459-bp region containing the aphA6 gene in A. guillouiae NIPH 991 and CIP 63.46T. Open arrows represent coding sequences and direction of transcription. Arrowheads indicate the 5′ end position and orientation of the primers (see Table S2). Horizontal lines depict the PCR products corresponding to overlapping amplified fragments. Numbering starts from the end of the ORF 1,591 bp upstream from the aphA6 gene. The sizes of the PCR products are indicated. (B) PCR map of A. guillouiae strains. +, present; −, absent; ---, not detected. Download
Genomic environment and excision of aphA6 in A. parvus CIP 102159. Arrows indicate open reading frames and sense of transcription. Colored arrows, ORFs; green arrow, aphA6; blue arrow, ISAba14; purple arrow, ΔIS6; thick black line, identity between A. guillouiae and A. parvus strains; vertical bars in ISAba14, base substitutions. PCR primer abbreviations: Fi, 102159-Fi; Ri, 102159-Ri; F1, 102159-F1; R1, 102159-R1. Download
Molecular phylogeny of rpoB and aphA6 sequences. Phylogenetic trees of aphA6-containing Acinetobacter were constructed using a partial sequence (861 bp) of rpoB (A) or that of the aphA6 gene (B). Multiple sequence alignments with MUSCLE v. 3.7 (39) were performed, and the phylogenetic trees were reconstructed using the maximum likelihood method implemented in the PhyML program (v. 3.0 aLRT) with the GTR matrix (40). GenBank accession numbers for each protein are indicated in brackets. The trees were rooted using A. gerneri and A. rudis as outgroups. We performed 100 bootstrap experiments on the sequences to assess the robustness of the topology. Boldface indicates that the whole genome of the strain was sequenced. Download
Molecular phylogeny of Aph(3′) proteins. Roman numbers indicate the type of enzyme. The list of putative homologs of Aph(3′)-VI was determined using blastp against Aph(3′)-VI of A. guillouiae NIPH 991 (GenBank accession number ENV16316) and all nonredundant GenBank CDS translations with e-values lower than 10−120 and manual curation to avoid redundancy. The GenBank accession number for each protein is indicated in brackets. The phylogeny of the Aph(3′) proteins was explored by construction of multiple sequence alignments with MUSCLE v. 3.7 (39). The unrooted phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v. 3.0 aLRT) with the WAG matrix and a gamma correction for variable evolutionary rates (40). We performed 100 bootstrap experiments on the sequences to assess the robustness of the topology. Bold, strains for which the whole genome was sequenced; *, previously designated Aph(3′)-VIII (41). Download
Genomic environment of aphA6. (Left) Black, Acinetobacter spp.; gray, Enterobacteriaceae, P. aeruginosa, and Alcaligenes faecalis; bold, strains for which the whole genome was sequenced; ND, not determined. (Right) Arrows indicate open reading frames and sense of transcription. Green arrow, aphA6; other colored arrows, IS elements; open arrows, ORFs; Δ, partially deleted. Download
Genomic contexts of aphA6 genes in draft genomes
Primers used in this study
ACKNOWLEDGMENTS
We thank Martina Maixnerova for help with susceptibility testing.
E.-J.Y. was supported by an unrestricted grant from Reckitt-Benckiser, A.N. and L.K. were supported by grant 13-26693 S from the Czech Science Foundation, and M.T. was supported by European Research Council grant EVOMOBILOME 281605. This project was funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN272200900018C.
Footnotes
Citation Yoon E, Goussard S, Touchon M, Krizova L, Cerqueira G, Murphy C, Lambert T, Grillot-Courvalin C, Nemec A, Courvalin P. 2014. Origin in Acinetobacter guillouiae and dissemination of the aminoglycoside-modifying enzyme Aph(3′)-VI. mBio 5(5):e01972-14. doi:10.1128/mBio.01972-14.
ADDENDUM IN PROOF
The article listed in the text as “submitted for publication” has since been accepted: M. Touchon, J. Cury, E. J. Yoon, L. Krizova, G. Cerqueira, C. Murphy, M. Feldgarden, J. Wortman, D. Clermont, T. Lambert, C. Grillot-Courvalin, A. Nemec, P. Courvalin, E. P. Rocha, Genome Biol. Evol., in press.
REFERENCES
- 1. Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 5:939–951. 10.1038/nrmicro1789 [DOI] [PubMed] [Google Scholar]
- 2. Turton JF, Shah J, Ozongwu C, Pike R. 2010. Incidence of Acinetobacter species other than A. baumannii among clinical isolates of Acinetobacter: evidence for emerging species. J. Clin. Microbiol. 48:1445–1449. 10.1128/JCM.02467-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldstein FW, Labigne-Roussel A, Gerbaud G, Carlier C, Collatz E, Courvalin P. 1983. Transferable plasmid-mediated antibiotic resistance in Acinetobacter. Plasmid 10:138–147. 10.1016/0147-619X(83)90066-5 [DOI] [PubMed] [Google Scholar]
- 4. Devaud M, Kayser FH, Bächi B. 1982. Transposon-mediated multiple antibiotic resistance in Acinetobacter strains. Antimicrob. Agents Chemother. 22:323–329. 10.1128/AAC.22.2.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fournier PE, Vallenet D, Barbe V, Audic S, Ogata H, Poirel L, Richet H, Robert C, Mangenot S, Abergel C, Nordmann P, Weissenbach J, Raoult D, Claverie JM. 2006. Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7. 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams MD, Chan ER, Molyneaux ND, Bonomo RA. 2010. Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54:3569–3577. 10.1128/AAC.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45:3375–3380. 10.1128/AAC.45.12.3375-3380.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galimand M, Courvalin P, Lambert T. 2003. Plasmid-mediated high-level resistance to aminoglycosides in Enterobacteriaceae due to 16S rRNA methylation. Antimicrob. Agents Chemother. 47:2565–2571. 10.1128/AAC.47.8.2565-2571.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magnet S, Smith TA, Zheng R, Nordmann P, Blanchard JS. 2003. Aminoglycoside resistance resulting from tight drug binding to an altered aminoglycoside acetyltransferase. Antimicrob. Agents Chemother. 47:1577–1583. 10.1128/AAC.47.5.1577-1583.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lambert T. 2010. Aminoglycosides and gram-negative bacteria, p 225–242. In Courvalin P, Leclercq R, Rice LB. (ed), Antibiogram. ASM Press, Washington, DC [Google Scholar]
- 11. Nemec A, Dolzani L, Brisse S, van den Broek P, Dijkshoorn L. 2004. Diversity of aminoglycoside-resistance genes and their association with class 1 integrons among 2 strains of pan-European Acinetobacter baumannii clones. J. Med. Microbiol. 53:1233–1240. 10.1099/jmm.0.45716-0 [DOI] [PubMed] [Google Scholar]
- 12. Lambert T, Gerbaud G, Courvalin P. 1988. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3′-aminoglycoside phosphotransferase. Antimicrob. Agents Chemother. 32:15–19. 10.1128/AAC.32.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lambert T, Gerbaud G, Courvalin P. 1994. Characterization of transposon Tn1528, which confers amikacin resistance by synthesis of aminoglycoside 3′-O-phosphotransferase type VI. Antimicrob. Agents Chemother. 38:702–706. 10.1128/AAC.38.4.702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nigro SJ, Post V, Hall RM. 2011. Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66:1504–1509. 10.1093/jac/dkr163 [DOI] [PubMed] [Google Scholar]
- 15. Périchon B, Goussard S, Walewski V, Krizova L, Cerqueira G, Murphy C, Feldgarden M, Wortman J, Clermont D, Nemec A, Courvalin P. 2014. Identification of 50 class D beta-lactamases and 65 Acinetobacter-derived cephalosporinases in Acinetobacter spp. Antimicrob. Agents Chemother. 58:936–949. 10.1128/AAC.01261-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dortet L, Nordmann P, Poirel L. 2012. Association of the emerging carbapenemase NDM-1 with a bleomycin resistance protein in Enterobacteriaceae and Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:1693–1697. 10.1128/AAC.05583-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Toleman MA, Spencer J, Jones L, Walsh TR. 2012. blaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56:2773–2776. 10.1128/AAC.06297-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Clinical and Laboratory Standards Institute . 2012. Performance standards for antimicrobial susceptibility testing, vol M100-MS22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 19. European, Committee on Antimicrobial Susceptibility Testing . 2014. Breakpoint tables for interpretation of MICs and zone diameters, version 4.0. http://eucast.org/clinical_breakpoints/. Accessed 18 January 2014
- 20. Fong DH, Berghuis AM. 2002. Substrate promiscuity of an aminoglycoside antibiotic resistance enzyme via target mimicry. EMBO J. 21:2323–2331. 10.1093/emboj/21.10.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rudant E, Courvalin P, Lambert T. 1998. Characterization of IS18, an element capable of activating the silent aac(6')-Ij gene of Acinetobacter sp. 13 strain BM2716 by transposition. Antimicrob. Agents Chemother. 42:2759–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamidian M, Hancock DP, Hall RM. 2013. Horizontal transfer of an ISAba125-activated ampC gene between Acinetobacter baumannii strains leading to cephalosporin resistance. J. Antimicrob. Chemother. 68:244–245. 10.1093/jac/dks345 [DOI] [PubMed] [Google Scholar]
- 23. Courvalin P, Weisblum B, Davies J. 1977. Aminoglycoside-modifying enzyme of an antibiotic-producing bacterium acts as a determinant of antibiotic resistance in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 74:999–1003. 10.1073/pnas.74.3.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poirel L, Figueiredo S, Cattoir V, Carattoli A, Nordmann P. 2008. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52:1252–1256. 10.1128/AAC.01304-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Humeniuk C, Arlet G, Gautier V, Grimont P, Labia R, Philippon A. 2002. Beta-lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045–3049. 10.1128/AAC.46.9.3045-3049.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poirel L, Rodriguez-Martinez JM, Mammeri H, Liard A, Nordmann P. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523–3525. 10.1128/AAC.49.8.3523-3525.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Spanogiannopoulos P, Waglechner N, Koteva K, Wright GD. 2014. A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria. Proc. Natl. Acad. Sci. U. S. A. 111:7102–7107. 10.1073/pnas.1402358111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pruitt KD, Tatusova T, Maglott DR. 2007. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35:D61–D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, St Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, Waterman PE. 2013. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J. Infect. Dis. 208:1142–1151. 10.1093/infdis/jit293 [DOI] [PubMed] [Google Scholar]
- 30. Coyne S, Guigon G, Courvalin P, Périchon B. 2010. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob. Agents Chemother. 54:333–340. 10.1128/AAC.01037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nemec A, Musílek M, Sedo O, De Baere T, Maixnerová M, van der Reijden TJ, Zdráhal Z, Vaneechoutte M, Dijkshoorn L. 2010. Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int. J. Syst. Evol. Microbiol. 60:896–903. 10.1099/ijs.0.013656-0 [DOI] [PubMed] [Google Scholar]
- 32. Nemec A, Krizova L, Maixnerova M, van der Reijden TJ, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162:393–404. 10.1016/j.resmic.2011.02.006 [DOI] [PubMed] [Google Scholar]
- 33. Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martin P, Jullien E, Courvalin P. 1988. Nucleotide sequence of Acinetobacter baumannii aphA-6 gene: evolutionary and functional implications of sequence homologies with nucleotide-binding proteins, kinases and other aminoglycoside-modifying enzymes. Mol. Microbiol. 2:615–625. 10.1111/j.1365-2958.1988.tb00070.x [DOI] [PubMed] [Google Scholar]
- 35. Vila J, Ruiz J, Navia M, Becerril B, Garcia I, Perea S, Lopez-Hernandez I, Alamo I, Ballester F, Planes AM, Martinez-Beltran J, de Anta TJ. 1999. Spread of amikacin resistance in Acinetobacter baumannii strains isolated in Spain due to an epidemic strain. J. Clin. Microbiol. 37:758–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hunger M, Schmucker R, Kishan V, Hillen W. 1990. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene 87:45–51. 10.1016/0378-1119(90)90494-C [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Genomic context of aphA6 in A. guillouiae strains. (A) Map of the 4,459-bp region containing the aphA6 gene in A. guillouiae NIPH 991 and CIP 63.46T. Open arrows represent coding sequences and direction of transcription. Arrowheads indicate the 5′ end position and orientation of the primers (see Table S2). Horizontal lines depict the PCR products corresponding to overlapping amplified fragments. Numbering starts from the end of the ORF 1,591 bp upstream from the aphA6 gene. The sizes of the PCR products are indicated. (B) PCR map of A. guillouiae strains. +, present; −, absent; ---, not detected. Download
Genomic environment and excision of aphA6 in A. parvus CIP 102159. Arrows indicate open reading frames and sense of transcription. Colored arrows, ORFs; green arrow, aphA6; blue arrow, ISAba14; purple arrow, ΔIS6; thick black line, identity between A. guillouiae and A. parvus strains; vertical bars in ISAba14, base substitutions. PCR primer abbreviations: Fi, 102159-Fi; Ri, 102159-Ri; F1, 102159-F1; R1, 102159-R1. Download
Molecular phylogeny of rpoB and aphA6 sequences. Phylogenetic trees of aphA6-containing Acinetobacter were constructed using a partial sequence (861 bp) of rpoB (A) or that of the aphA6 gene (B). Multiple sequence alignments with MUSCLE v. 3.7 (39) were performed, and the phylogenetic trees were reconstructed using the maximum likelihood method implemented in the PhyML program (v. 3.0 aLRT) with the GTR matrix (40). GenBank accession numbers for each protein are indicated in brackets. The trees were rooted using A. gerneri and A. rudis as outgroups. We performed 100 bootstrap experiments on the sequences to assess the robustness of the topology. Boldface indicates that the whole genome of the strain was sequenced. Download
Molecular phylogeny of Aph(3′) proteins. Roman numbers indicate the type of enzyme. The list of putative homologs of Aph(3′)-VI was determined using blastp against Aph(3′)-VI of A. guillouiae NIPH 991 (GenBank accession number ENV16316) and all nonredundant GenBank CDS translations with e-values lower than 10−120 and manual curation to avoid redundancy. The GenBank accession number for each protein is indicated in brackets. The phylogeny of the Aph(3′) proteins was explored by construction of multiple sequence alignments with MUSCLE v. 3.7 (39). The unrooted phylogenetic tree was reconstructed using the maximum likelihood method implemented in the PhyML program (v. 3.0 aLRT) with the WAG matrix and a gamma correction for variable evolutionary rates (40). We performed 100 bootstrap experiments on the sequences to assess the robustness of the topology. Bold, strains for which the whole genome was sequenced; *, previously designated Aph(3′)-VIII (41). Download
Genomic environment of aphA6. (Left) Black, Acinetobacter spp.; gray, Enterobacteriaceae, P. aeruginosa, and Alcaligenes faecalis; bold, strains for which the whole genome was sequenced; ND, not determined. (Right) Arrows indicate open reading frames and sense of transcription. Green arrow, aphA6; other colored arrows, IS elements; open arrows, ORFs; Δ, partially deleted. Download
Genomic contexts of aphA6 genes in draft genomes
Primers used in this study