Abstract
Thiol peroxidases are critical enzymes in the redox control of cellular processes that function by reducing low levels of hydroperoxides and regulating redox signaling. These proteins were also shown to regulate genome stability, but how their dysfunction affects the actual mutations in the genome is not known. Saccharomyces cerevisiae has eight thiol peroxidases of glutathione peroxidase and peroxiredoxin families, and the mutant lacking all these genes (∆8) is viable. In this study, we employed two independent ∆8 isolates to analyze the genome-wide mutation spectrum that results from deficiency in these enzymes. Deletion of these genes was accompanied by a dramatic increase in point mutations, many of which clustered in close proximity and scattered throughout the genome, suggesting strong mutational bias. We further subjected multiple lines of wild-type and ∆8 cells to long-term mutation accumulation, followed by genome sequencing and phenotypic characterization. ∆8 lines showed a significant increase in nonrecurrent point mutations and indels. The original ∆8 cells exhibited reduced growth rate and decreased life span, which were further reduced in all ∆8 mutation accumulation lines. Although the mutation spectrum of the two independent isolates was different, similar patterns of gene expression were observed, suggesting the direct contribution of thiol peroxidases to the observed phenotypes. Expression of a single thiol peroxidase could partially restore the growth phenotype of ∆8 cells. This study shows how deficiency in nonessential, yet critical and conserved oxidoreductase function, leads to increased mutational load and decreased fitness.
Keywords: thiol peroxidase, mutation, genome stability, Saccharomyces cerevisiae
BEING hereditary material, DNA is propagated from generation to generation while maintaining genome stability in the face of random mutations. Genome instability results mainly from DNA damage and error-prone DNA synthesis, and it is crucial for any organism to properly maintain its genome and neutralize damage to it (Nishant et al. 2010). There are various types of agents that can cause DNA damage by modifying the structural properties of DNA, and oxidative stress is one of the best-studied factors that may affect DNA stability. Aerobic organisms inevitably produce reactive oxygen species (ROS) as a consequence of incomplete reduction of molecular oxygen during respiration, as well as by other metabolic processes. ROS may cause oxidative damage by reacting with and affecting functions of many cellular metabolites and macromolecules, including proteins, lipids, and DNA (van Loon et al. 2010). Continuous exposure of intracellular milieu to ROS is implicated in the etiology of various diseases, such as diabetes, neurodegeneration, and atherosclerosis. ROS are also involved in cancer development by increasing the mutation rate via oxidative DNA damage and genomic instability (Marnett 2000; Reddy et al. 2009). More than a hundred different types of oxidative DNA damage have been described; among them, oxidative conversion of guanine to 7,8-dihydro-8-oxoguanine (8-oxo-G) is the most prominent modification. 8-oxo-G leads to misincorporation of adenine in place of guanine, resulting in the CG→AT transversion. Spontaneous deamination of methylated cytosine is a hydrolytic modification, leading to thymine substitution, and CG→TA transitions are driven by oxidation of methylated cytosine (van Loon et al. 2010). These modifications serve as markers of oxidative DNA damage and are the most common point mutations in a variety of human genetic diseases, including cancer (Cooke et al. 2003; van Loon et al. 2010).
Organisms have evolved a wide variety of defense mechanisms to prevent or limit the consequences of oxidative damage. They include two independent mechanisms of DNA repair, base excision repair and nucleotide excision repair (Degtyareva et al. 2008), as well as antioxidant defense systems, such as superoxide dismutase, catalase, and peroxiredoxin (Prx), which scavenge ROS (Huang et al. 2003). These and other systems collaborate in protecting cells from genomic instability induced by oxidative stress (Kryston et al. 2011). Recent studies suggest that antioxidant defense is the first line of defense against oxidative DNA damage. For example, thiol-dependent oxidoreductases prevent large-scale chromosomal rearrangements, and their deficiency leads to an increase in the mutation rate (Iraqui et al. 2009; Tang et al. 2009). These enzymes include ubiquitous thiol peroxidases containing a catalytic cysteine (Cys) residue that becomes oxidized in the process of scavenging hydrogen peroxide, alkyl hydroperoxides, and other peroxide forms (Wong et al. 2002). Most organisms have multiple Prxs, e.g., five Prxs occur in the yeast Saccharomyces cerevisiae, 10 in the plant Arabidopsis thaliana, and six in mammals (Park et al. 2000; Horling et al. 2003; Rhee et al. 2005). Three glutathione peroxidases (Gpxs), members of another family of thiol peroxidases, were also identified in yeast, and eight such enzymes occur in mammals (Inoue et al. 1999; Brigelius-Flohe 2006). Like Prxs, Gpxs catalyze the reduction of hydrogen peroxide and other hydroperoxides, but most often use glutathione as an electron donor for these reactions (Brigelius-Flohe 2006). Both Prxs and Gpxs occur in various cellular compartments, show tissue-specific expression in mammalian cells, and differ in abundance (Brigelius-Flohe 1999; Muller et al. 2007; Gerashchenko et al. 2012).
Despite proposed roles of these antioxidant enzymes in genome stability, there is no direct information on the genome-wide mutation process and DNA stability when these functions are compromised. A recent study screened a yeast knockout library and identified individual peroxidases as mutation suppressors (Huang et al. 2003). Following this observation, additional studies examined mutation rates for cells lacking individual and multiple Prxs, showing that the Tsa1 null yeast cells had the strongest mutator phenotype. These studies assessed mutation rates for small marker loci such as canavanine (CAN1) and uracil (URA3), which are commonly used for fluctuation assays but have limitations for interpreting the richness, diversity, and specificity of mutation phenotypes when applied to entire genomes (Wong et al. 2004; Iraqui et al. 2009; Tang et al. 2009). Currently, information on the genome-wide mutational spectra of thiol peroxidase null cells is lacking.
To begin addressing the functions of thiol peroxidases in genome stability, we previously developed a model using the budding yeast S. cerevisiae, in which we deleted all eight thiol peroxidases (∆8 cells) and found that deficiency in these enzymes disrupts hydrogen peroxide-dependent transcriptional regulation (Fomenko et al. 2011). In the present study, we developed an independent ∆8 strain and assessed how deficiency in these enzymes in two different isolates affects cellular fitness and genome stability. We also subjected ∆8 and wild-type (WT) cells to a long-term mutation accumulation (MA) experiment. Genome sequencing of ∆8 isolates as well as various MA lines revealed that thiol peroxidases are important factors for genome maintenance. We describe several hypermutability regions in a pattern of clustered mutations in ∆8 cells, which are extended up to 150 kb. These data suggest that the lack of thiol-based redox control leads to a distinct mutator phenotype, directly implicating thiol peroxidases in genome stability.
Materials and Methods
S. cerevisiae strains
Construction of a mutant strain lacking all eight thiol peroxidases was reported previously (Fomenko et al. 2011). Briefly, it was obtained by mating Prx null (∆5Prx) (Wong et al. 2004) and Gpx null (∆3Gpx) (Avery and Avery 2001) strains. The resulting diploids were analyzed by tetrad dissection; several hundred spores were checked on Yeast Nitrogen Base selection plates for histidine, leucine, uracil, and methionine auxotrophy and for G418 resistance. The growing colonies were selected, and we performed a second PCR screening for each locus with the use of HIS3, URA3, and KanMX markers. Initially, we isolated colonies lacking six or seven peroxidases, named ∆6 and ∆7, followed by deletion of the remaining one or two peroxidases to yield ∆8 cells. Details regarding the strains used can be found in Supporting Information, Table S1. Both WT cells and multiple Gpx and Prx mutants were in the BY4741 background. Yeast strains were grown on yeast extract–peptone–dextrose (YPD) medium. For respiratory growth, cells were grown on YPD containing 2% glycerol, ethanol, or lactic acid.
Phenotypic characterization of the peroxidase null strain
After growing cells for 2 days on YPD plates, individual colonies were picked for both WT and ∆8 strains, each inoculated in 10 ml of YPD liquid media and cultured overnight at 30° with moderate shaking. Overnight cultures were diluted to an OD600 of 0.02. Cell growth was monitored by OD600 measurements. YPD plates and liquid culture were supplemented with 20 µg/ml phleomycin for TSA1-expressed strains. Data from individual growth curves were used to calculate cell division time. For replicative aging experiments, 20 individual virgin daughter cells were collected for both WT and ∆8 cells. New buds from these original daughter cells were separated and discarded, as they were formed, using a dissecting microscope. This process continued until cells ceased dividing.
Establishment of MA lines and whole-genome sequencing
MA lines were initiated from isogenic lines of WT and ∆8 strains, which were passed through 50 single-cell bottlenecks for a total of ∼1500 cell divisions for WT lines and ∼800 divisions for ∆8 lines over a course of 1 year. We kept the original, initial isogenic WT and ∆8 cells frozen at −80°, which were subsequently sequenced together with the MA lines. At the end of the bottleneck process, cells from the 1-year-old ∆8 and WT lines were centrifuged and kept frozen at −80°. DNA from each line was extracted using MasterPure yeast DNA purification kit (Epicenter) according to the manufacturer’s protocol. For sequencing, genomic DNA samples were prepared according to the standard Illumina protocols for library preparation. Initial cells and all MA lines were paired-end sequenced with 75-bp reads on an Illumina Hi-Sequation 2000 platform.
Assembly and detection of base substitutions and small indels
Yeast reference genome was downloaded from the National Center for Biotechnology Information web site (http://ftp.ncbi.nih.gov). Sequencing reads were aligned to the reference genome using a variety of tools. The best results were obtained using Burrows–Wheeler Aligner, which utilizes a backtracking method to search for inexact matches and provides a mapping quality score for each read (Li and Durbin 2009). Further analysis was conducted using the Genome Analysis Toolkit (GATK) framework (McKenna et al. 2010) according to the program’s manual. Comparisons with the reference genome, as well as pairwise comparisons of initial vs. MA lines, were performed to identify point mutation substitutions and small indels. The following additional criteria were implemented: (i) the nucleotide in question should have been covered by at least three reads and (ii) the GATK score for this position should have been >10 (which corresponds to P = 0.001 probability of misidentification). The data were further analyzed using a SHORE toolkit (Schneeberger et al. 2009), which was designed to allow a higher tolerance for diverged regions. The reported base substitutions and structural variants were then compared with the GATK output, and read alignments were manually verified with Tablet software (Milne et al. 2010). For each base substitution or indel candidate, location on the chromosome was analyzed. An annotation file downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org/) was used to determine coding and intergenic noncoding regions.
Calculation of mutation rates and analysis of mutation clusters
Mutation rate was calculated by dividing the number of detected mutations by the total number of bases in the reference genome sequence and by the number of generations that each strain was propagated. For the initial ∆8 strain, the number of generations is unknown, so we calculated the mutation frequencies only. Standard errors were calculated as the square root of mutation rate divided by the number of nucleotides in the genome and the number of generations (Denver et al. 2004). For MA lines 2A and 2B, mutations that were common to both lines were considered to have occurred in each of the lines. Mutation numbers for different mutation types were calculated by further normalizing mutations from AT base pairs by the AT fraction of the genome and mutations from GC base pairs by the GC fraction of the genome. Clusters were analyzed according to a previously described method (Michaelson et al. 2012). Details of the analysis can be found in the figure legend (Figure 3).
Figure 3.
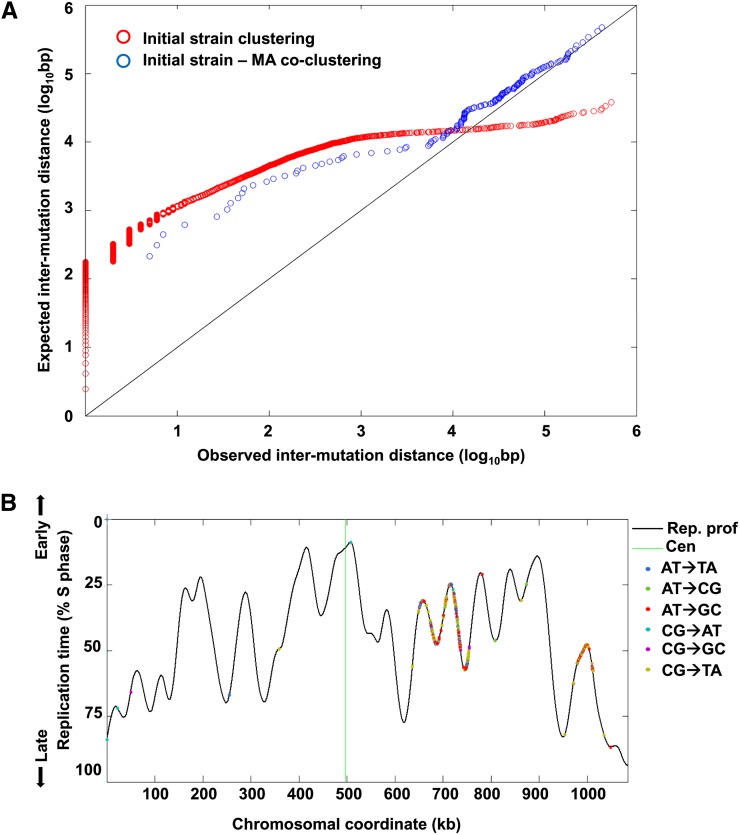
Mutation clustering in ∆8 cells. (A) Observed vs. expected distances between adjacent mutations. Observed distances are the distances between consecutive mutations on the same chromosome in the initial stain. To calculate the expected distances between mutations, we generated 100 sets of randomized genomic positions, each with the same number of loci as the number of mutations in the initial strain. We further calculated the distances between adjacent loci in each randomized set (as we did for the observed mutations) and averaged the distances across the 100 randomized iterations. Both observed and expected distances were sorted by increasing distance and plotted against each other (red circles). The diagonal represents equal expected and observed distances. As can be seen, most mutations were much closer to each other than expected by chance while mutational clusters were themselves very distant from other clusters, leading to the long tail of higher-than-expected intramutation distances. This indicates a very strong clustering of mutations along the genome. The distances between mutations in the MA lines and the nearest mutations in the initial strain were also calculated and compared to a set of 100 randomized genomic positions, each with the same number of loci as the number of mutations in the MA lines, as above. The observed vs. expected distances between MA line and initial strain mutations are shown in blue circles. Mutations in the MA lines tended to fall closer to initial strain mutation clusters than expected by chance. (B) An example of mutation clusters on chromosome VII of the original ∆8 strain. Shown is the DNA replication timing profile of the chromosome (black line; high values represent early replication, low values late replication; peaks correspond to replication origins and valleys to replication termini). The different mutation types are shown as dots along the replication profile. Two very tight mutation clusters can be observed (at ∼640–755 kb and at ∼980–1010 kb) with almost no mutations outside these two clusters. Similar plots for all chromosomes, with the locations of mutations in both the initial strain and the MA lines, are shown in Figure S2B.
Spot assays for genotoxin sensitivity
Resistance of WT and ∆8 strains to different genotoxic agents was assessed by spot assays. Ten microliters of WT, ∆3Gpx, and ∆5Prx cultures at the OD600 of 0.3 were serially diluted to fresh YPD plates containing indicated amounts of campothecin, phleomycin, benomyl, or hydroxyurea. Cells were grown for 3 days at 30°, and pictures were taken. For the ∆8 strain, the initial OD600 was 0.6 since the number of cells of this strain was approximately 2-fold lower in the absence of treatment.
Messenger RNA isolation and fragmentation for transcriptome analyses
Frozen yeast pellets were cryo-grinded in a BioSpec mini bead beater. Lysis was performed with solutions from a RNAqueous mini kit (LifeTech). Isolation of total RNA proceeded according to the kit manual. Messenger RNA (mRNA) was purified from total RNA using an mRNA Direct Kit (LifeTech) according to the manufacturer’s guidelines. Fragmentation of mRNA was achieved by a 30-min incubation in 1 mM EDTA, 500 mM NaHCO3, pH 9.2, at 95°.
Precipitation of RNA and DNA
The following components were added to the initial sample: 1/10 volume of ammonium acetate (Ambion), 5 µl of glycogen (5 mg/ml, Ambion), and 2.5 volumes of absolute ethanol. The mixture was incubated for 1 hr at −20°, and nucleic acids were precipitated by a 15-min centrifugation at 20,000 × g. This method was used to precipitate DNA and RNA after all enzymatic reactions.
Sequencing library preparation
mRNA fragments were treated with T4 polynucleotide kinase (Thermo Scientific) for 1 hr in 10 µl total reaction volume. RNA was loaded on a 15% TBE–urea polyacrylamide gel (Invitrogen). The band corresponding to 50–70 nt was cut out of the gel and crushed with a disposable pestle (Kimble Chase), and RNA was eluted during a 3-hr incubation at 37° in 0.3 ml of elution buffer (20 mM Tris–HCl, pH 7.0, 2 mM EDTA, 1/10 volume of 3 M ammonium acetate, 40 U/ml Superase-In). Gel particles were eliminated by a Corning Costar spin-X 0.22-µm column. RNA was precipitated. One hundred nanograms of 3′ adapter per sample were ligated by T4 RNA ligase 2 truncated KQ or K227Q (New England Biolabs). Reaction products were precipitated, and reverse transcription was set up as follows. RNA pellet was dissolved in 11.5 µl of water, and 4 pmol of reverse transcription primer was added along with 1 µl of dNTP mix (10 mM each). The mixture was incubated for 5 min at 65° and then chilled on ice. Two microliters of DTT and 4 µl of First Strand Buffer (refer to manufacturer’s protocols for composition) were added along with 0.5 µl of SuperScript II and 0.5 µl Superase-In to the total volume of 20 µl. Reaction then continued for 30 min at 42°, 1 min at 65°, and 5 min at 80°. RNA was degraded in the presence of 80 mM NaOH at 95° for 30 min and then neutralized by the same amount of HCl. Reaction products were precipitated and run on 10% TBE–urea polyacrylamide gel (Invitrogen), and the band corresponding to the transcriptase extended product was cut off. DNA was eluted as described above, precipitated, and later used in a CircLigase II reaction (20 µl total volume, according to the manufacturer’s instructions; Epicentre). Products of ligation were used in the final PCR without additional precipitations or purifications. PCR was set up as follows: 10 pmol reverse and forward primers (Table S3), 10 µl High Fidelity buffer, 0.5 µl Phusion polymerase (New England Biolabs), 1–4 µl of ligation product, and water up to 50 µl. PCR mix was subjected to 6–10 cycles of amplification (94° for 15 sec, 55° for 10 sec, 65° for 10 sec). Cycling was finalized for 2 min at 65°. PCR products were precipitated and run on 8% TBE polyacrylamide gel, and the band corresponding to the amplified products was cut off. DNA was eluted as described above, precipitated, and sent for sequencing on an Illumina HiSequation 2000 platform. The data analysis was described previously (Gerashchenko et al. 2012).
Expression of TSA1 in WT and mutant lines
We used phleomycin (bleMX6) containing plasmid pCEV-G1-Ph (Vickers et al. 2013) to express the TSA1 in WT and ∆8 strains. The entire coding sequence of TSA1 (from start to stop codon) was amplified with specific primers containing 5′-NotI and 3′-PacI restriction sites and cloned into a plasmid for expression under the control of yeast TEF1 promoter. For yeast transformation, overnight cultures were diluted and grown until mid-log phase (OD600 = 0.6). One milliliter of each culture was used for transformation with an empty vector as control and the TSA1-containing construct for expression using a lithium acetate method. After transformation, cells were incubated for 3 hr at 30° and spread onto plates containing 20 µg/ml phleomycin. Colonies were chosen after 5 days and restreaked onto new plates containing either 40 µg/ml phleomycin or 100 µg/ml zeocin since the bleMX6 cassette provided resistance to both antibiotics (Vickers et al. 2013). Colonies, which were grown on both plates, were used for subsequent experiments.
Results
Deletion of eight thiol peroxidases leads to growth and life-span defects
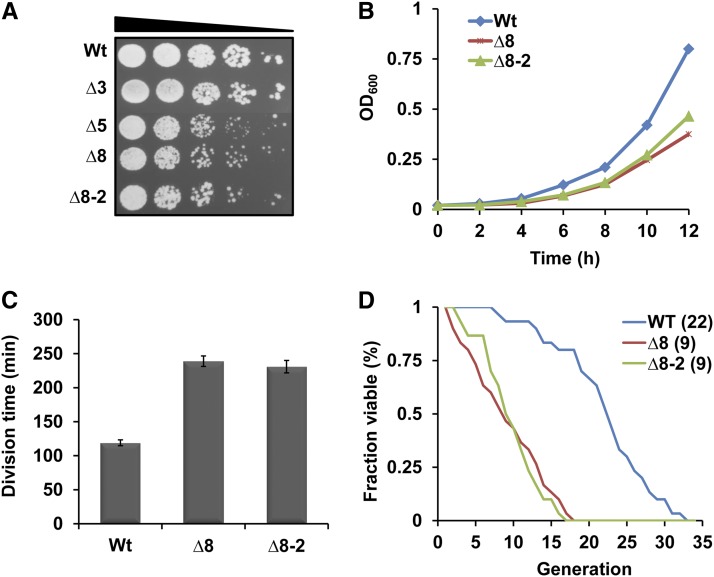
Combined knockout of eight thiol peroxidases (Table S1), prepared from cells lacking three Gpxs (∆3) and five Prxs (∆5), was not lethal in S. cerevisiae cells (Fomenko et al. 2011), but led to several phenotypic changes (Figure 1). Thiol peroxidase deficiency yielded an approximately twofold decrease in growth rate (Figure 1C). We also measured the replicative life span of ∆8 and WT strains. The mean life span of ∆8 cells was nine divisions, approximately 2-fold lower compared to the 22 divisions observed in the WT strain (Figure 1D).
Figure 1.
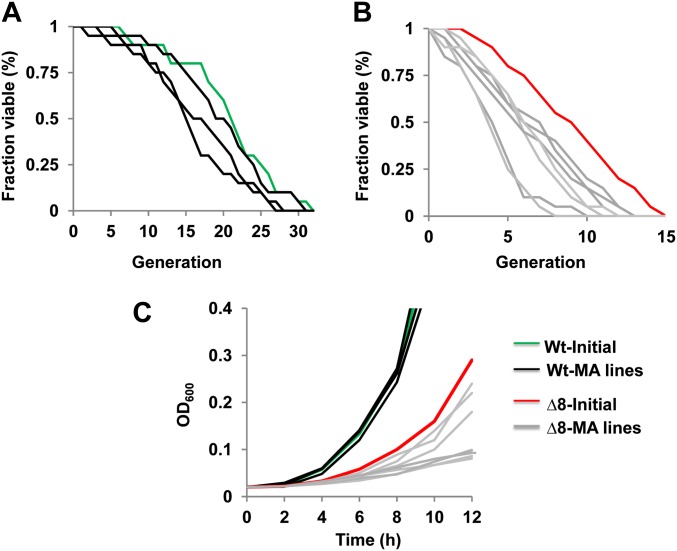
Deletion of eight thiol peroxidases causes growth defects and reduces life span in yeast. (A) Decreased colony growth of ∆8 cells. Overnight cultures of WT, ∆3, ∆5, and two independent ∆8 isolates were diluted to an OD600 of 0.3. Then, 5 µl of 10-fold dilution of each cell suspension was spotted onto YPD plates and photographed 3 days later. (B) Reduced cell growth of ∆8 cells. Growth of WT and ∆8 cells in liquid YPD medium was monitored by measuring the OD600. (C) Increased division time of ∆8 cells. Division time (in minutes) was calculated for each strain and plotted. (D) Replicative life span of WT and ∆8 cells. Numbers in brackets next to each strain indicate mean life span. The y-axis represents the fraction of survivors (fraction of mother cells capable of budding) at each generation (x-axis).
The genome of the thiol peroxidase null strain is highly unstable
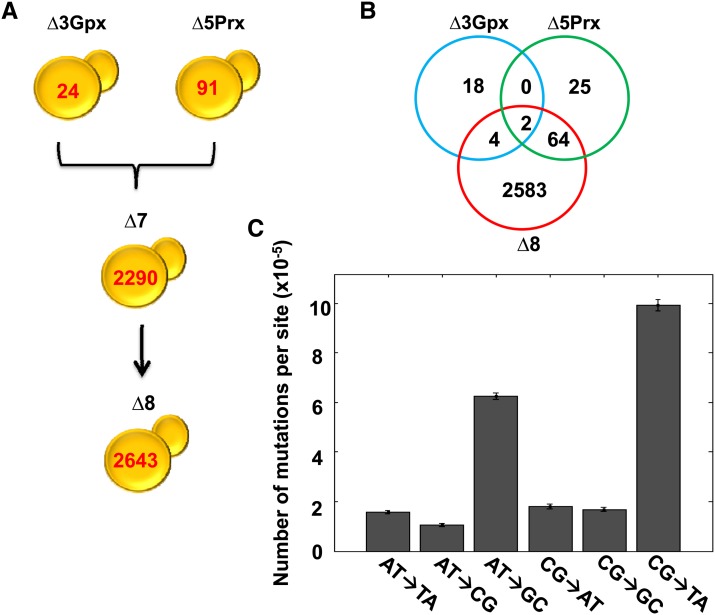
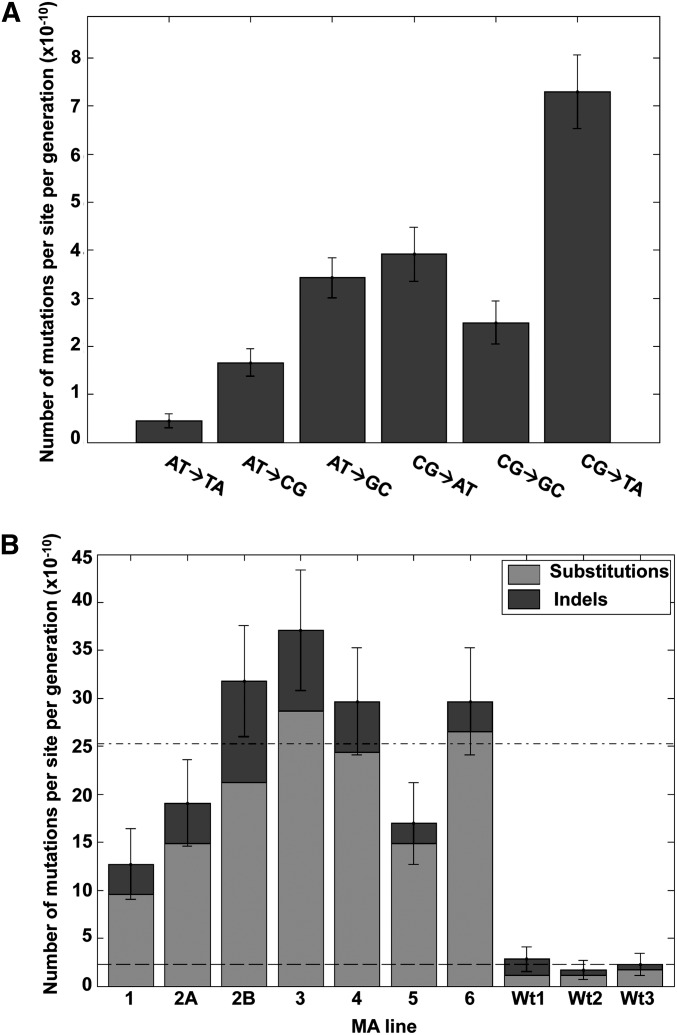
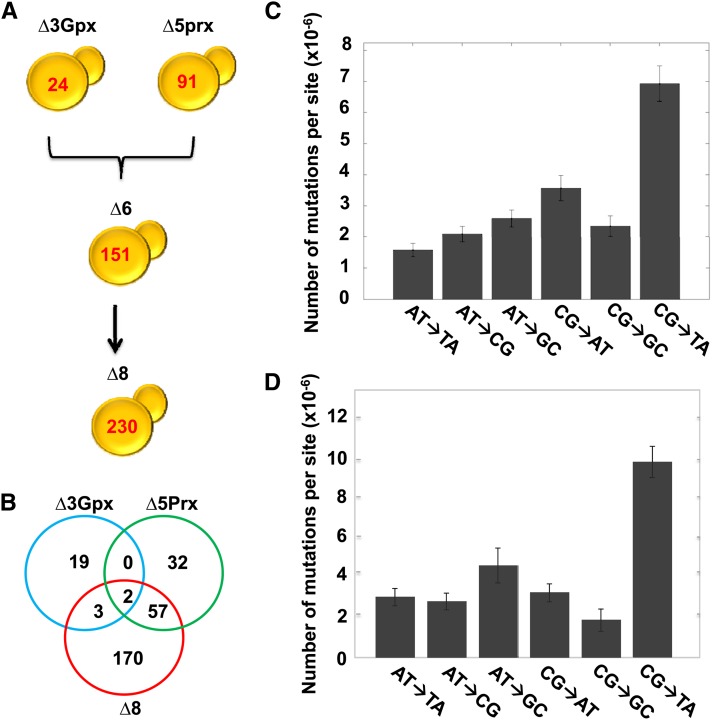
To assess the role of thiol peroxidases in genome stability, we sequenced and analyzed the genomes of ∆3, ∆5, and ∆8 (∆8 initial) strains and compared them with the genome of isogenic WT (WT initial) cells. This procedure revealed 2633 point mutations in ∆8 relative to WT cells (Figure 2A). Most of them occurred during and after construction of this strain, since only 24 mutations were observed in ∆3 cells and 91 mutations in ∆5 cells compared to the reference WT genome (Figure 2B). In the initial ∆8 cells, CG→TA and AT→GC transitions were much more common than other mutation types (Figure 2C). We examined the locations of point mutations and found that 1161 mutations were in noncoding regions. Among 1472 mutations within coding sequences, 1041 were silent (Table 1 and File S1). The initial ∆8 strain also had 11 mutations found in noncoding regions (9 base substitutions and 2 insertions) of the mitochondrial genome. To test for a possible change in mitochondrial function, we analyzed the respiratory function by growing cells on ethanol, glycerol, and lactic acid and found that the ∆8 strain can utilize these substrates (Figure S1). Mutations found in the coding region of the genome were distributed among 261 genes, with 149 genes having more than one mutation. Most genes with multiple mutations coded for cell-wall proteins rich in multiple tandem repeats. A putative chloride transporter (YHL008C) had the highest number of mutations (196 mutations), followed by the flocculation gene FLO1 (YAR050) with 108 mutations and the PRM7 (YDL039C) pheromone-regulated protein with 39 mutations (File S1). Among 90 small deletions and 60 small insertions (up to three bases) that were found in ∆8 strain, 33 were in coding regions, with the remaining 117 in noncoding regions (Table 1 and File S1). The highest number of mutations (point mutations and short indels) was observed on chromosome 7, which had 672 mutations, and the lowest on chromosome 10, which featured only 6 mutations (Figure S2A). This difference was much greater than that expected based on the difference in chromosome length (see below).
Figure 2.
Quantification of mutations in multiple thiol peroxidase mutants. (A) Mutational landscape of thiol peroxidase mutants. Genome sequencing revealed 24 mutations in ∆3, 91 in ∆5, 2290 in ∆7 (all peroxidases lacking except for Gpx2), and 2643 in ∆8 cells. (B) Mutations identified in the mutant strains represented by Venn diagram. Mutation distribution is shown for three strains: ∆3 Gpx (blue), ∆5 Prx (green), and ∆8 strain (red). The number of mutations was calculated using the GATK procedure, and the common mutations were identified by comparing the sets of mutations among the strains. (C) Number of mutations of each type in the original ∆8 strain. Number of mutations was calculated by dividing the number of detected mutations by the total number of bases in the reference genome sequence.
Table 1. Mutations observed in the original ∆8 strain compared to isogenic WT cells.
| No. | Fraction | |
|---|---|---|
| Type of substitution | ||
| Transition | ||
| A:T > G:C | 956 | 0.36 |
| G:C > A:T | 942 | 0.36 |
| Transversion | ||
| A:T > T:A | 239 | 0.09 |
| A:T > C:G | 164 | 0.06 |
| G:C > T:A | 171 | 0.07 |
| G:C > C:G | 161 | 0.06 |
| Total | 2633 | |
| Substitutions | ||
| Position | ||
| Noncoding | 1161 | 0.44 |
| Coding | 1472 | 0.56 |
| Coding sequences | ||
| Nonsense | 4 | 0.0015 |
| Missense | 427 | 0.29 |
| Silent | 1041 | 0.71 |
| Indels | ||
| Noncoding | 117 | 0.78 |
| Coding | 33 | 0.22 |
Distribution of 2633 point mutations and 150 indels identified in the initial ∆8 strain. The entire list of mutations, together with positions and nucleotide changes, is shown in File S1.
We observed a striking mutation clustering in the ∆8 strain: many mutations were in close proximity to each other, while individual mutation clusters were separated by long sequence stretches with almost no mutations (Figure 3A). The largest cluster occurred on chromosome 7 and extended ∼150 kb. This clustering pattern was indicative of a strong mutational bias operating in this strain. Mutation clusters did not show any bias with respect to mutation spectrum; rather, they included all types of mutations, and clusters were not associated with any particular time of replication (Figure 3B and Figure S2B), nor did they correspond to the origin of replication or locations of Tys/LTRs or transfer RNAs (data not shown). However, we found that mutations tended to have repeats in closer proximity than expected (Figure S3), indicating that tandem repeat-associated mutations are a contributing factor to the genomic distribution of mutations in the initial mutant. Finally, we performed pulse-field gel electrophoresis (PFGE) to identify the large-scale genome rearrangements. PFGE revealed no apparent changes in chromosome size of the ∆8 strain compared to WT cells (Figure S4).
Decreased fitness of MA thiol peroxidase null cells
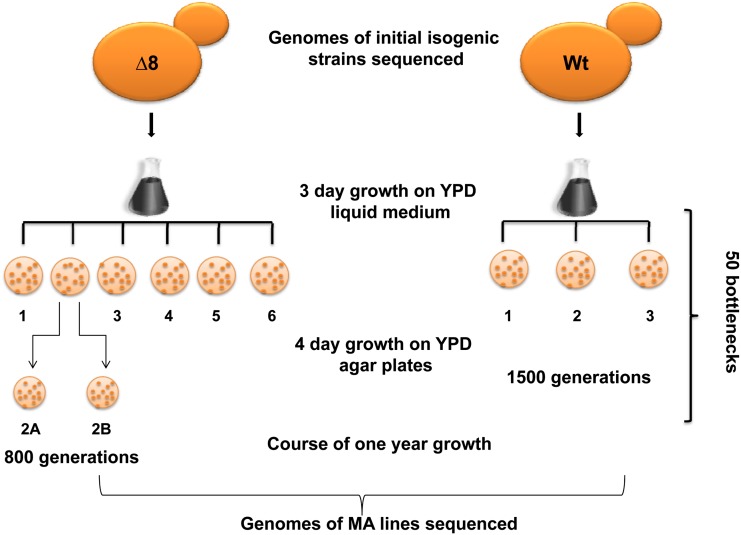
The large number of mutations observed in ∆8 cells offered an opportunity to use this strain to examine the process of MA and the relationship between mutation accumulation and cell fitness. Thus, we subjected the ∆8 strain to a year-long MA experiment, in which several separate MA lines, derived from the same culture of ∆8 cells (∆8 initial cells), were cultured in parallel with several lines of the isogenic WT strain (also derived from the same WT culture and designated WT initial cells), each passing through 50 (once per week) single-cell bottleneck selections (Figure 4). Each bottleneck step consisted of a 3-day growth of single colonies in liquid culture followed by 4 days colonial growth on YPD agar plates. We calculated the total number of divisions as 1500 for WT and 800 for ∆8 cells. The experiment was initiated with six lines of ∆8 and three lines of WT cells. After 100 generations, one of ∆8 lines was divided into two parallel lines and named MA line 2A and 2B, thereby resulting in a total of 7 ∆8 lines.
Figure 4.
Experimental design of the mutation accumulation experiment. The ∆8 strain was obtained by mating ∆3 and ∆5 cells, followed by deletion of the remaining peroxidase gene (Gpx2). The genomes of initial WT and ∆8 cells were sequenced, and cells (seven MA lines for ∆8 and three MA lines for WT cells) were subjected to a bottleneck process: colonies were grown for 4 days from an individual cell on solid media followed by growth for 3 days in liquid culture. This process was repeated 50 times over the course of 1 year, followed by genome sequencing of final cell populations. One of ∆8 lines was split into two lines following 100 generations, after which they were treated as independent MA lines.
To assess how the long-term MA experiment affected cell fitness, we compared life span and growth rate of initial and MA lines after the long-term growth of WT and ∆8 cells. Interestingly, both replicative life span and growth rate were decreased in all independent ∆8 MA lines (at least in two lines, P < 0.05), while this effect was not observed in WT MA lines (Figure 5). However, there was no correlation between the observed number of mutations in each line and the degree to which life span and growth rate were decreased. For example, the mutant MA line 3 accumulated 36 mutations (both point mutations and indels) and its mean replicative life span was six divisions, whereas the mutant MA line 1 had 12 mutations with the mean replicative life span of four divisions (Figure 5 and Table S2).
Figure 5.
Decreased fitness of MA lines. Twenty individual virgin daughter cells were collected for corresponding WT and ∆8 MA lines and analyzed by using a microscope with a micromanipulator to determine replicative life span. Statistical analysis of the life-span data were performed using a Wilcoxon rank-sum test. P-value data for this experiment are in Table S2. (A) Replicative life span of WT initial (green line) and WT MA lines (black lines). (B) Replicative life span of ∆8 initial (red line) and ∆8 MA lines (gray lines). (C) Growth rate of initial and MA ∆8 lines.
Thiol peroxidase null MA lines exhibit high point mutation and indel rates
We analyzed mutation rates and spectra of yeast cells by sequencing the genomes of initial strains and ∆8 and WT lines subjected to the MA procedure. A total of 132 point mutations were observed in ∆8 MA lines, but only 7 point mutations in WT MA lines. Among the 132 mutations, 49 were in noncoding regions and 83 in coding sequences, including 52 missense, 3 nonsense, and 28 silent mutations (Table 2 and Figure S5A). We identified 5 common mutations between MA lines 2A and 2B (which were separated from the common line 2 after 100 generations) and confirmed that all 5 mutations appeared prior to the separation of lines by sequencing the PCR-amplified fragments from mutated regions. The remaining 9 mutations for MA line 2A and 15 mutations for MA line 2B occurred independently and showed no overlap. The data suggest that the occurrence of 5 mutations did not drive common mutations during the rest of the experiment. No overlaps in mutations were observed among other MA lines. Mutations showed a tendency to occur in proximity to the mutation clusters in the ∆8 initial strain and co-clustering was still significantly stronger than expected by chance (Figure 3A). The overall transition rate was twofold higher than the transversion rate (Ts/Tv). The specific mutation distribution for MA lines was very similar to that of the ∆8 initial strain, but the CG→AT transversion rate was higher during the long-term growth (Figure 6A). The observed mutational pattern suggested that oxidative DNA damage had a major role. Oxidative deamination of cytosine, which leads to thymine substitution, and the high oxidation potential of guanine to 8-oxoguanine that is mismatched to adenine were in agreement with our observations (Ossowski et al. 2010; Thiviyanathan et al. 2008).
Table 2. Mutations observed in WT and ∆8 MA lines following the long-term MA experiment.
| WT | ∆8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Substitutions | 1 | 2 | 3 | 1 | 2A | 2B | 3 | 4 | 5 | 6 |
| Position | ||||||||||
| Noncoding | 1 | 0 | 0 | 4 | 7 | 10 | 8 | 6 | 4 | 10 |
| Nonsense | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 |
| Missense | 1 | 0 | 2 | 2 | 2 | 6 | 14 | 8 | 10 | 10 |
| Silent | 0 | 2 | 1 | 2 | 4 | 4 | 6 | 8 | 0 | 4 |
| Indels | ||||||||||
| Position | ||||||||||
| Noncoding | 1 | 1 | 0 | 3 | 3 | 9 | 7 | 4 | 2 | 1 |
| Coding | 2 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 2 |
| All mutations | 5 | 3 | 4 | 12 | 18 | 30 | 36 | 27 | 16 | 28 |
| Mutation rate (×10−10) | 2.8 | 1.7 | 2.2 | 12.7 | 19 | 31.8 | 37.1 | 29.6 | 16.9 | 29.6 |
| Mean (×10−10) | 2.2 | 25.3 | ||||||||
Figure 6.
Observed mutation rates for MA lines. (A) Observed total mutation rates per site/per generation after the long-term growth of MA lines. Shown is the average for all MA lines. (B) The mutation rate and standard error per MA line. Horizontal dashed lines are the average mutation rate (base substitutions and indels) for the ∆8 and the WT lines.
Among the seven point mutations identified in WT MA lines, four were in coding regions and three in untranslated regions (Table 2 and Figure S6A). The average base substitution rate was 15-fold higher in ∆8 MA lines (19.9 × 10−10 per site per generation for ∆8 MA lines and 1.3 × 10−10 for WT lines). We also calculated the MA line-specific base substitution mutation rates for each ∆8 line, which varied from 9.59 × 10−10 to 28.6 × 10−10 (Figure 6B).
The genomes of MA lines were further analyzed to identify small indels of 1–3 bp, identifying 17 insertions and 18 deletions in ∆8 MA lines. The majority of indels were in noncoding areas, with six occurring in coding regions. Of 29 intergenic indels, 10 were found in telomeric regions (Table 2 and Figure S5, B and C). We also identified 3 single-base-pair deletions in the mitochondrial genome of three MA lines. As mentioned above, there were substitutions and small indels identified in the mitochondrial genome of the initial ∆8 strain, and several more accumulated during the long-term growth of the MA lines. However, mitochondria were still functional in all ∆8 MA lines at the end of the 1-year growth, indicating that the observed mutations did not interfere with the respiratory function in any of the MA lines (Figure S1). Further analysis of the genomes of WT MA lines revealed a total of 5 small insertions, including 3 in coding regions, whereas we did not detect small deletions (Table 2 and Figure S6B). Only one WT line harbored a single double-nucleotide insertion in the mitochondrial genome, which also occurred in the noncoding region. We did not observe mutational hotspots during the long-term growth of WT or ∆8 MA lines. The average rates of short indels were estimated at 5.3 × 10−10 for ∆8 and 0.94 × 10−10 for WT lines, and the individual indel mutation rates varied from 2.1 × 10−10 to 10.6 × 10−10 per site per generation in ∆8 MA lines (Figure 6B and Table 2).
Characterization of an independent ∆8 isolate
Genome instability was a striking phenotype of the ∆8 mutant, but a possibility remained that this mutator phenotype was the result of a single mutational catastrophe during preparation of the ∆8 mutant rather than the result of thiol peroxidase deficiency. To address this possibility, we prepared an independent ∆8 isolate from ∆3 and ∆5 cells, this time with a minimal number of generations, and further sequenced the genome of this strain. The new mutant also had a very strong mutator phenotype. We detected 230 point mutations and 15 small indels in this mutant, which corresponded to ∼2 mutations per generation during strain construction; this can be compared with the observed 2.2 mutations per 1500 generations in WT cells (Figure 7 and File S1). We characterized several phenotypes of this new mutant and found its overall behavior was similar to that of the original ∆8 strain, including reduced growth rate and shortened replicative life span (Figure 1). The mutation spectrum of the new isolate was also similar to that of the previous isolate, and CG→TA transitions were much more common than other mutation types (Figure 7D). There were 20 commonly mutated genes, with the majority of these genes inherited from the ∆5 cells during the mating process. These mutated genes featured nucleotide substitution mutations except for the metallochaperone Mtm1, which had a frameshift in the first isolate and a substitution mutation in the second isolate. It is difficult to evaluate the functional consequences of mutated genes, if any, for the observed phenotypes (File S1).
Figure 7.
Characterization of mutations in an independent ∆8 isolate. (A–C) Quantification of mutations and characterization of the second ∆8 mutant are as shown in Figure 2 for the first ∆8 mutant. (D) Mutations found in ∆7 were subtracted from the first ∆8 mutant strain, and the spectrum of remaining mutations was analyzed.
An additional consistent observation with the new isolate was that mutations tended to be in close proximity to each other (Figure S7). Overall, these data showed that deficiency in eight thiol peroxidases leads to a strong mutator phenotype in yeast cells, characterized by point mutations and indels and pointing to a critical role of these proteins in genome stability.
Deletion of eight thiol peroxidases modulates sensitivity of cells to DNA damage
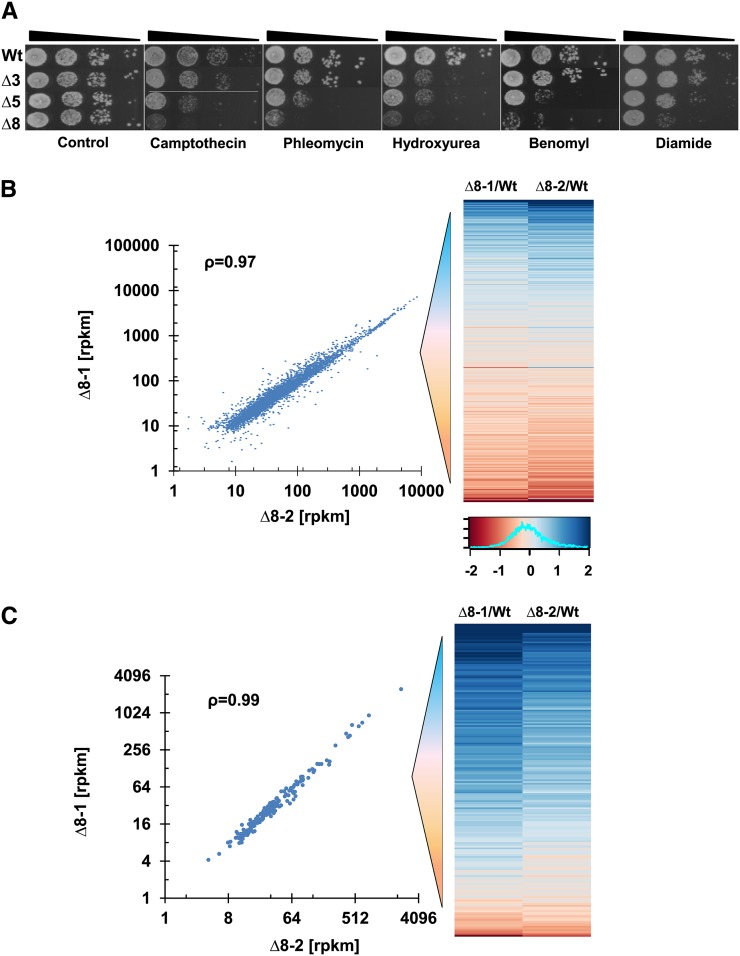
To test whether the impaired proliferation and the observed mutator phenotype of ∆8 cells were related to DNA replication and repair, we examined the sensitivity of cells to genotoxic stresses. Cells were treated with camptothecin and hydroxyurea, which are commonly used as replication inhibitors, and with benomyl, which affects the function of microtubules, causing improper chromosome segregation during cell division and delayed DNA replication (van Loon et al. 2010; Sheltzer et al. 2011). In addition, we treated cells with phelomycin, which induces double-strand breaks that are repaired by homologous recombination (Reiter et al. 1972). In all cases, the ∆5Prxs and ∆8 strains, but not the ∆3Gpxs strain, displayed significant sensitivity to these drugs, suggesting that the observed slow proliferation phenotype could be related to the defect in both replication and recombinational repair processes in the absence of peroxiredoxins (Figure 8A). Consistent with this observation, the new ∆8 isolate showed the same sensitivity to all these drugs (Figure S8). It is known that in most cancer types replication stress causes an aggressive mutator phenotype (Loeb 2001). These observations support the idea that thiol peroxidases have critical roles in the DNA damage response and in ROS control. Accordingly, deficiency in these enzymes leads to a strong mutator phenotype by delaying replication.
Figure 8.
Spot assay and comparison of gene expression data. (A) Sensitivity of ∆8 and WT cells to DNA damage agents. In the control panel, ∆8 and ∆5 cells were used with a twofold higher cell density compared to other strains to observe similar growth. Ten-fold dilution of WT and ∆8 cells grown on YPD plates containing 15 µg/ml camptothecin (replication inhibitor), 150 mM hydroxyurea (replication inhibitor), 0.3 µg/ml phleomycine (double-strand break inducer), or 15 µg/ml benomyl (microtubule poison). (B) Comparison of expression of 4493 genes in ∆8 isolates with the expression graded as log2 of the fold increase/decrease in the heat map on the right, ranked from high (blue) to low (red). Mean rpkm (reads per kilobase transcript per million reads) values of two repetitions for each ∆8 isolate were calculated, the values <10 in both isolates were eliminated, and Spearman correlation (ρ) was calculated. Expression distribution of genes for both ∆8 isolates is shown by a histogram at the bottom right. (C) Analysis of genes known to regulate stability of the nuclear genome.
Consistent transcriptome landscape of ∆8 cells
Even though there was a 10-fold difference in mutations and a large difference in the number cell divisions between the two ∆8 strains, both independent isolates were similar with regard to growth characteristics, drug sensitivity, and life span, suggesting that the observed phenotypic changes were largely due to thiol peroxidase deficiency. We further examined this possibility by analyzing gene expression via RNA-seq. The numerous mutations that independently emerged in two ∆8 mutant strains did not result in significant differences in their gene expression, as the expression of 4493 genes was highly correlated (ρ = 0.97) (Figure 8A and File S2). At the same time, the expression of 90% genes varied between −0.73 and 0.99 (Log2) in both ∆8 isolates, and 10% of genes increased or decreased expression at least twofold, compared to WT cells (Figure 8A). We were particularly interested in the transcriptional regulation of DNA repair genes as it could provide further molecular insights into the basis for the mutator phenotype of ∆8 cells. A total of 183 such genes are known to maintain stability of the yeast nuclear genome (File S2). We found that the expression of these genes was well correlated (ρ = 0.99), and most of them were upregulated in both ∆8 isolates compared to WT cells (Figure 8B). Among these genes, ubiquitin-dependent DNA repair genes, which are involved in the nonhomologous end-joining pathway that repairs double-strand breaks, were top hits. Recently, it was shown that this pathway promotes DNA repair, supporting cell survival following DNA repair (Kato et al. 2014). Perhaps, the error-prone double-strand break repair in the repetitive regions initiates mutations in ∆8 cells, leading to the observed clustered mutation pattern.
Overexpression of the abundant thiol peroxidase Tsa1 partially restores the growth phenotype
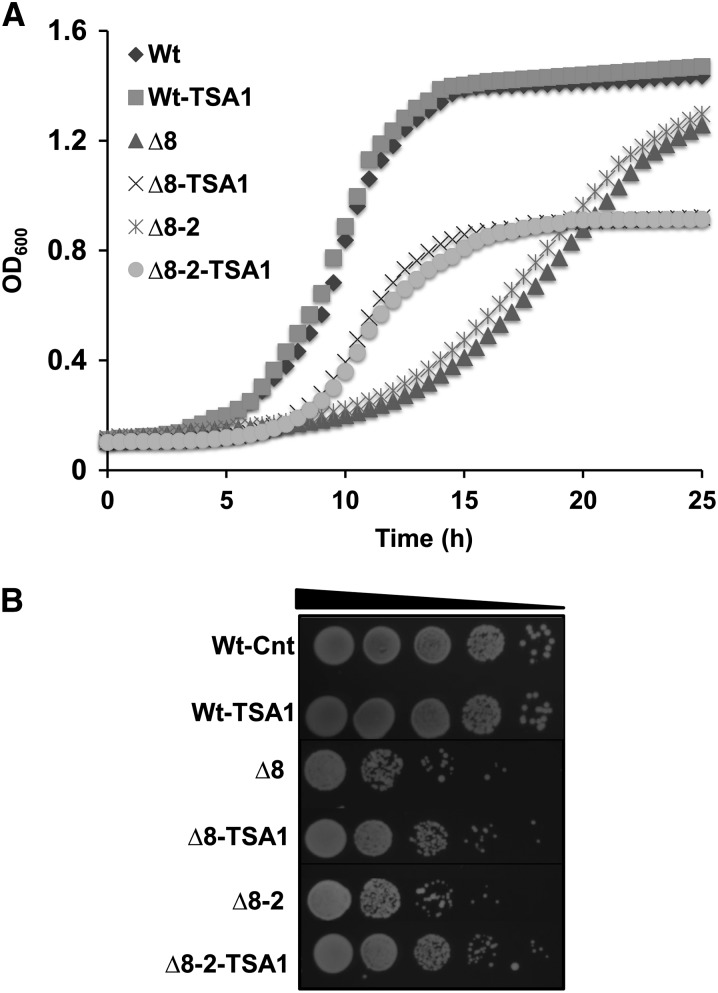
To further characterize the consequences of deleting eight thiol peroxidase genes, we tested the complementation of growth of mutant cells by overexpressing Tsa1 under the constitutive yeast TEF1 promoter. Tsa1 was previously described as a highly abundant peroxidase that suppresses genome instability (Iraqui et al. 2009). In WT cells, its expression did not affect cell growth. The slow growth of thiol peroxidase null strains was partially restored in the logarithmic phase by Tsa1 expression, but both populations attained a lower level of cell density than either WT or control ∆8 cells (Figure 9A). Thus, expression of Tsa1 only partially rescued the growth of ∆8 cells. We also confirmed the restoration of growth by spotting equal amounts of cells onto YPD plates and examining them 3 days later (Figure 9B).
Figure 9.
Expression of TSA1 in ∆8 cells. (A) Reduced cell growth of ∆8 cells was partially rescued by Tsa1 expression. Growth of WT and ∆8 cells expressing (or not) TSA1 in liquid YPD medium containing 20 µg/ml phleomycin was monitored by measuring the OD600 every 30 min. Mean values of two independent measurements for each point are shown. (B) Colony growth of Tsa1-expressing ∆8 cells. Overnight cultures of WT and two independent ∆8 isolates as well as the corresponding strains expressing Tsa1 were diluted to an OD600 of 0.3. Then, 5 µl of 10-fold dilution of each cell suspension were spotted onto YPD plates containing 20 µg/ml and photographed 3 days later.
Discussion
It has been ∼2.5 billion years since the great oxygenation event (GOE). The cost of living in an oxygen-rich environment is currently the subject of intense research. Evidence indicates that, in addition to evolving ways to utilize O2, organisms developed defense systems against dangers posed by ROS and oxidative stress in general. Thiol peroxidases are the enzymes that clear low levels of hydroperoxides from cells, but they may differ from other antioxidant proteins since they are also found in anaerobes. In fact, they occur in nearly every living organism, indicating that they evolved prior to the GOE (Edgar et al. 2012). Thus, additional, presumably more ancient, noncanonical roles of peroxidases might not be known.
In the current study, we performed whole-genome sequencing to characterize the impact of thiol peroxidase deficiency on genome stability, following the deletion of all Prxs (five genes) and Gpxs (three genes). These ∆8 cells were viable, but manifested several phenotypes indicative of decreased fitness, such as slow growth, shorter life span, and decreased colony size in two independent ∆8 isolates. We cannot completely exclude the possibility that some of these phenotypes were affected by nonperoxidase mutations, but the striking similarity between the two independent preparations of ∆8 mutants, which did not share common mutations, argues against this possibility. It should be noted, however, that, because it is difficult to generate a clean (isogenic to wild type) ∆8 strain, nonperoxidase mutations may play some role. Regardless, our key finding was that both isolates had a very strong mutator phenotype, leading to the accumulation of numerous mutations in their genomes that were different in the two ∆8 mutants and suggesting that it is the absence of thiol peroxidases that compromises genome integrity.
To further address the role of thiol peroxidases in genome stability, we designed a controlled experiment, wherein WT (three independent lines) and ∆8 cells (seven independent lines) were subjected to a parallel, long-term MA experiment, characterized by 50 weekly bottlenecks. After this long-term growth, we compared the replicative life span and growth characteristics for all experimental MA lines. Interestingly, there was a reduction in fitness in all ∆8 MA lines for both life span and growth rate. This experiment showed that all seven parallel ∆8 MA lines became increasingly vulnerable during growth in the absence of thiol peroxidases. Since thiol peroxidases are highly conserved proteins and occur in nearly all living organisms, these results suggest that the presence of these enzymes is required for protection and recovery of populations from natural collapse, subject to environmental and genetic challenges.
We sequenced the genomes of initial and MA ∆8 and WT strains and compared their mutation spectra. All ∆8 lines had significantly more mutations than any of the WT lines: the average mutation rate (base substitutions and indels) of the mutant MA lines was 11-fold higher than that of the WT MA lines. We observed 0.0027 mutations per generation (12 mutations/three lines/1500 generations) for WT and 0.03 mutations per generation (167 mutations/seven lines/800 generations) for the ∆8 mutant. The mutation rate for WT was consistent with a previous measurement (Lynch et al. 2008). The variation among mutation rates for each MA line could be an indicator of the influence of new mutations. There were almost 200 genes having missense and/or frameshift mutations in the initial ∆8 strain, which possibly interfered with the function of these proteins, including some DNA repair enzymes, thereby affecting the mutation rate. This issue highlights the difficulty in determining a single mutation rate as it changes over time.
We did not identify recurrent mutations among MA lines. One interesting observation in the ∆8 strains was the finding of long clusters of mutations. Proximity of these mutations suggested that clustered events could reduce the deleterious effects of random mutations or that mutations in the regions that harbor nonessential genes are preserved and enriched, while mutations in more critical regions lead to lethality and so could not be recovered in our bottleneck selection design. A similar mutation clustering was recently reported in another yeast experiment (Roberts et al. 2012). This study observed nonrandom biased clustered mutations near chromosome breakpoints, which had a common motif targeted by specific deaminases. Our results also indicate that there could be a different intrinsic mechanism for the observed mutation distribution in the thiol peroxidase null strain in parallel to random selection, resulting in such an extensive mutation pattern.
To test for general association of mutations in the initial strain and tandem repeats, we analyzed the distribution of distances between mutations and repeats, comparing them to the distances expected by chance (Figure 3 and Figure S3). It is known that these repetitive DNA sequences are highly unstable and mutate at high rates, including a high frequency of double-strand DNA breaks (Bzymek and Lovett 2001; Gemayel et al. 2010). It appears that the observed long stretches of mutation clusters in ∆8 cells could be initiated at tandem repeats initiated by double-strand DNA breaks. Subsequently, error-prone repair could introduce additional mutations in these regions. Apparently, this observation is also applicable to the second ∆8 isolate since clustering was also observed in this mutant. A second line of evidence from gene expression data also supports this hypothesis as most of the double-strand DNA break repair genes were upregulated in both ∆8 strains.
Our genotoxic assays and transcriptome analyses also suggest additional mechanisms by which thiol peroxidases may maintain genome stability. The ∆8 strains are very sensitive to replication inhibitors and microtubule poisons, which delay DNA replication and cellular proliferation, respectively. It appears that DNA damage occurs in the absence of thiol peroxidases as a consequence of oxidative stress, even if this stress is relatively mild. We hypothesize that thiol peroxidases interact with transcription factors to activate or repress gene expression. In the absence of thiol peroxidases, transcription regulation of these cells is compromised. This may be an additional factor that contributes to the high mutator phenotype in ∆8 cells. A global regulatory role of peroxidases in transcriptional signaling is consistent with this idea (Fomenko et al. 2011). Finally, we showed that overexpression of a major thiol peroxidase Tsa1 was able to partially rescue the growth phenotype of ∆8 cells.
We described the mutational landscape of cells lacking thiol peroxidases and showed that these enzymes have important roles in maintaining genome integrity due to counteracting the damaging effects of low levels of peroxides and regulating DNA repair genes. Our genome analyses suggested that, in the original ∆8 strain, deletion of all eight thiol peroxidase genes coincided with a rapid accumulation of many mutations (while very few mutations were detected in ∆3 and ∆5 mutants), but subsequently changes accumulated in the genome that slowed down the mutation rate (based on the mutation rate of ∆8 cells revealed by our long-term MA experiment). In the independent ∆8 isolate, the mutation rate was also very high, but not as high (even considering the few cell divisions). Perhaps, a mutational catastrophe occurred during meiosis that led to ∆7 cells, prior to preparation of ∆8 cells. Indeed, the number of mutations from this step to create ∆8 was similar to that observed in the second ∆8 isolate and MA lines (Figure 7D).
It was shown previously that yeast cells can withstand the rate of ∼10−3 mutations per gene per cell division (Herr et al. 2011). Above this threshold, cells eventually die, and only the cells with suppressor mutations that decrease the rate of error-prone replication escape from extinction. Such suppressors may tolerate substantial increase in mutation rate before losing viability (Herr et al. 2011). We speculate that the observed reduced mutation rate in ∆8 MA lines reflects the fact that these cells acquired important modifier loci to suppress the high mutation rate of the ∆8 cells. There is also evidence for a further reduction in mutation rate during strain construction: lines 2A/B had 5 mutations during the first 100 generations of the MA procedure, whereas line 2A had only 9 additional and line 2B 15 additional mutations in the remaining 700 generations (for line 2A, this is a significant reduction; Fisher’s exact, P = 0.025). This implies that selection might have acted during the MA procedure and that the mutation rate declined, perhaps also partially affecting the spectrum of mutations. Cells with deleterious mutations were eliminated following each bottleneck, and cells with the acquired lower mutator phenotype were selected. In the end, we observed lower-than-expected (based on the comparison of WT and ∆8 genomes) numbers of mutations in MA lines following the long-term growth experiment, although these numbers were still much higher than mutations in the WT MA cells.
We presented the first genome-wide mutation spectrum of cells as a consequence of thiol peroxidase deficiency, which uncovered an important function of these enzymes in genome stability. There are several reports showing that mice lacking Prdx1, an ortholog of yeast Tsa1p, are highly prone to the development of various malignancies (Neumann et al. 2003; Egler et al. 2005). Our study supports the idea that thiol peroxidases are key protectors against DNA damage and genome instability and can serve as tumor suppressors under conditions of oxidative stress. These genes are conserved in all three domains of life and are present in nearly all living organisms, but often they are not essential when knocked out in various organisms. Understanding the mechanism by which they contribute to genome stability offers insights into their functions, conservation in the three domains of life, and essential roles in DNA maintenance and protection against cancer.
Supplementary Material
Acknowledgments
We thank Shamil Sunyaev and Paz Polak for discussion and Siming Ma for help with RNAseq data analysis. This work was supported by National Institutes of Health grant GM065204 to V.N.G.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.169243/-/DC1.
Communicating editor: J. A. Nickoloff
Literature Cited
- Avery A. M., Avery S. V., 2001. Saccharomyces cerevisiae expresses three phospholipid hydroperoxide glutathione peroxidases. J. Biol. Chem. 276: 33730–33735. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R., 1999. Tissue-specific functions of individual glutathione peroxidases. Free Radic. Biol. Med. 27: 951–965. [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohe R., 2006. Glutathione peroxidases and redox-regulated transcription factors. Biol. Chem. 387: 1329–1335. [DOI] [PubMed] [Google Scholar]
- Bzymek M., Lovett S. T., 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98: 8319–8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke M. S., Evans M. D., Dizdaroglu M., Lunec J., 2003. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 17: 1195–1214. [DOI] [PubMed] [Google Scholar]
- Degtyareva N. P., Chen L., Mieczkowski P., Petes T. D., Doetsch P. W., 2008. Chronic oxidative DNA damage due to DNA repair defects causes chromosomal instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 28: 5432–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Green E. W., Zhao Y., van Ooijen G., Olmedo M., et al. , 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egler R. A., Fernandes E., Rothermund K., Sereika S., de Souza-Pinto N., et al. , 2005. Regulation of reactive oxygen species, DNA damage, and c-myc function by peroxiredoxin 1. Oncogene 24: 8038–8050. [DOI] [PubMed] [Google Scholar]
- Fomenko D. E., Koc A., Agisheva N., Jacobsen M., Kaya A., et al. , 2011. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. USA 108: 2729–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R., Vinces M. D., Legendre M., Verstrepen K. J., 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu. Rev. Genet. 44: 445–477. [DOI] [PubMed] [Google Scholar]
- Gerashchenko M. V., Lobanov A. V., Gladyshev V. N., 2012. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 109: 17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr A. J., Ogawa M., Lawrence N. A., Williams L. N., Eggington J. M., et al. , 2011. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 7: e1002282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horling F., Lamkemeyer P., Konig J., Finkemeier I., Kandlbinder A., et al. , 2003. Divergent light-, ascorbate-, and oxidative stress-dependent regulation of expression of the peroxiredoxin gene family in Arabidopsis. Plant Physiol. 131: 317–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D., 2003. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA 100: 11529–11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Matsuda T., Sugiyama K., Izawa S., Kimura A., 1999. Genetic analysis of glutathione peroxidase in oxidative stress response of Saccharomyces cerevisiae. J. Biol. Chem. 274: 27002–27009. [DOI] [PubMed] [Google Scholar]
- Iraqui I., Kienda G., Soeur J., Faye G., Baldacci G., et al. , 2009. Peroxiredoxin Tsa1 is the key peroxidase suppressing genome instability and protecting against cell death in Saccharomyces cerevisiae. PLoS Genet. 5: e1000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Nakajima K., Ui A., Muto-Terao Y., Ogiwara H., et al. , 2014. Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell 53: 617–630. [DOI] [PubMed] [Google Scholar]
- Kryston T. B., Georgiev A. B., Pissis P., Georgakilas A. G., 2011. Role of oxidative stress and DNA damage in human carcinogenesis. Mutat. Res. 711: 193–201. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A., 2001. A mutator phenotype in cancer. Cancer Res. 61: 3230–3239. [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett L. J., 2000. Oxyradicals and DNA damage. Carcinogenesis 21: 361–370. [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J. J., Shi Y., Gujral M., Zheng H., Malhotra D., et al. , 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne I., Bayer M., Cardle L., Shaw P., Stephen G., et al. , 2010. Tablet: next generation sequence assembly visualization. Bioinformatics 26: 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H., 2007. Trends in oxidative aging theories. Free Radic. Biol. Med. 43: 477–503. [DOI] [PubMed] [Google Scholar]
- Neumann C. A., Krause D. S., Carman C. V., Das S., Dubey D. P., et al. , 2003. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature 424: 561–565. [DOI] [PubMed] [Google Scholar]
- Nishant K. T., Wei W., Mancera E., Argueso J. L., Schlattl A., et al. , 2010. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 6: e1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. G., Cha M. K., Jeong W., Kim I. H., 2000. Distinct physiological functions of thiol peroxidase isoenzymes in Saccharomyces cerevisiae. J. Biol. Chem. 275: 5723–5732. [DOI] [PubMed] [Google Scholar]
- Reddy V. P., Zhu X., Perry G., Smith M. A., 2009. Oxidative stress in diabetes and Alzheimer’s disease. J. Alzheimers Dis. 16: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter H., Milewskiy M., Kelley P., 1972. Mode of action of phleomycin on Bacillus subtilis. J. Bacteriol. 111: 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S. G., Chae H. Z., Kim K., 2005. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38: 1543–1552. [DOI] [PubMed] [Google Scholar]
- Roberts S. A., Sterling J., Thompson C., Harris S., Mav D., et al. , 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol. Cell 46: 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger, K., J. Hagmann, S. Ossowski, N. Warthmann, S. Gesing et al, 2009 Simultaneous alignment of short reads against multiple genomes. Genome Biol. 10: R98. [DOI] [PMC free article] [PubMed]
- Sheltzer J. M., Blank H. M., Pfau S. J., Tange Y., George B. M., et al. , 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. M., Siu K. L., Wong C. M., Jin D. Y., 2009. Loss of yeast peroxiredoxin Tsa1p induces genome instability through activation of the DNA damage checkpoint and elevation of dNTP levels. PLoS Genet. 5: e1000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiviyanathan V., Somasunderam A., Volk D. E., Hazra T. K., Mitra S., et al. , 2008. Base-pairing properties of the oxidized cytosine derivative, 5-hydroxy uracil. Biochem. Biophys. Res. Commun. 366: 752–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon B., Markkanen E., Hubscher U., 2010. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair (Amst.) 9: 604–616. [DOI] [PubMed] [Google Scholar]
- Vickers C. E., Bydder S. F., Nielsen L. K., 2013. Dual gene expression cassette vectors with antibiotic selection markers for engineering in Saccharomyces cerevisiae. Microb. Cell Fact. 12: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C. M., Zhou Y., Ng R. W., Kung Hf H. F., Jin D. Y., 2002. Cooperation of yeast peroxiredoxins Tsa1p and Tsa2p in the cellular defense against oxidative and nitrosative stress. J. Biol. Chem. 277: 5385–5394. [DOI] [PubMed] [Google Scholar]
- Wong C. M., Siu K. L., Jin D. Y., 2004. Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J. Biol. Chem. 279: 23207–23213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.