Abstract
Pleuropulmonary blastoma is a rare childhood malignancy of lung mesenchymal cells that can remain dormant as epithelial cysts or progress to high-grade sarcoma. Predisposing germline loss-of-function DICER1 variants have been described. We sought to uncover additional contributors through whole exome sequencing of 15 tumor/normal pairs, followed by targeted resequencing, miRNA analysis and immunohistochemical analysis of additional tumors. In addition to frequent biallelic loss of TP53 and mutations of NRAS or BRAF in some cases, each case had compound disruption of DICER1: a germline (12 cases) or somatic (3 cases) loss-of-function variant plus a somatic missense mutation in the RNase IIIb domain. 5p-Derived microRNA (miRNA) transcripts retained abnormal precursor miRNA loop sequences normally removed by DICER1. This work both defines a genetic interaction landscape with DICER1 mutation and provides evidence for alteration in miRNA transcripts as a consequence of DICER1 disruption in cancer.
Introduction
Pleuropulmonary blastoma (PPB) is a rare, aggressive sarcoma arising from mesenchymal cells of the lung during early childhood. The classic initial stage of PPB (mean age at presentation 10 months) features dilated airspaces lined by lung epithelium (Type I PPB). The mesenchymal cells within the walls of the cysts have the potential to transform into high-grade, sarcoma-forming cystic and solid (Type II PPB) or purely solid (Type III PPB) masses by 3–4 years of age;1 however, not all cysts naturally progress to life-threatening sarcoma. Germline loss-of-function variants in DICER1 have been described in familial PPB,2 and these patients variably show increased risk for ovarian Sertoli–Leydig tumors, renal cystic nephromas, nodular hyperplasia and carcinoma of the thyroid gland and an assortment of other rare extrapulmonary benign or malignant neoplastic conditions, thus implicating DICER1 as a tumor suppressor.2, 3, 4, 5, 6, 7, 8, 9 Surprisingly, DICER1 expression is reportedly lost in tumor-associated epithelium in some cases, but retained in the tumor mesenchyme.2 We sought to uncover additional and cooperating genetic events driving PPB progression in tumor mesenchyme and to investigate molecular consequences of DICER1 mutation.
Results
DICER1 incurs biallelic disruption in PPB
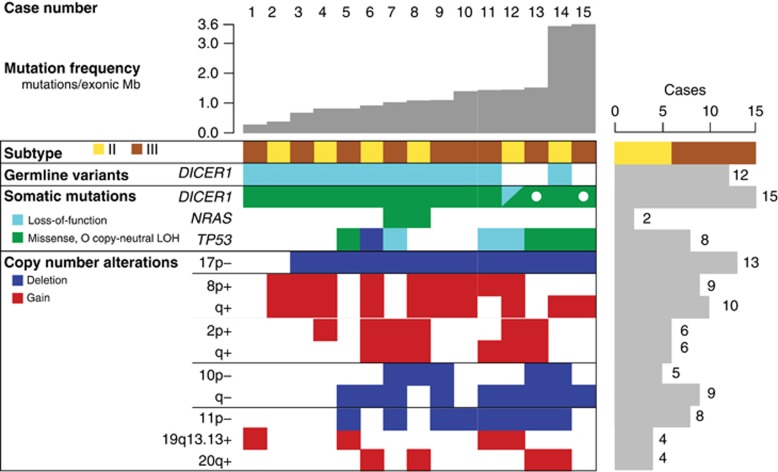
Analysis of exome sequence data from mesenchymal tissue from 15 PPBs (6 Type II, 9 Type III; Supplementary Table 1) with paired normal DNA (88 × mean coverage of 18 863 genes) uncovered 1.1 exonic mutations per megabase (0.85 non-silent). Despite the young age of PPB patients, these mutation rates are more consistent with adult cancers than pediatric malignancies.10 The two cases with the highest mutation rates, 3.6 mutations per Mb, had loss-of-function mutations in DNA repair genes: BTBD12/SLX4 p.L1621fs in a Type III PPB and PARP1 c.1159+1A>G in a Type II PPB that recurred following chemotherapy. In total, 623 somatic mutations were found in 568 genes (Supplementary Table 2), of which only three were mutated at significant frequency (q⩽0.1, MutSig v1.511): DICER1, TP53 and NRAS (Figure 1, Table 1 and Supplementary Table 3).
Figure 1.
Matrix of frequent copy-number alterations and significantly mutated genes derived from exome sequence data in each case. Cases are in columns and genetic alterations are in rows with events color-coded as indicated. The loss-of-function category includes nonsense, splice-site, insertion and deletion mutations. Copy-neutral LOH refers to chromosome- or arm-level loss-of-heterozygosity without a change in copy number (for example, loss of the chromosome containing the wild-type allele and duplication of the chromosome containing the mutant allele), as shown in Figure 3.
Table 1. Genes with significant somatic mutation frequencies.
| Gene | Description | Mutations | Patients (n=15) | Unique sites | Missense | Nonsense, splice site or frameshift | q-value |
|---|---|---|---|---|---|---|---|
| DICER1 | Dicer 1, ribonuclease type III | 16 | 15 | 7 | 15 | 1 | 7.8 × 10−11 |
| TP53 | Tumor protein p53 | 7 | 7 | 7 | 4 | 3 | 1.2 × 10−10 |
| NRAS | Neuroblastoma RAS viral (v-ras) oncogene homolog | 2 | 2 | 2 | 2 | 0 | 2.6 × 10−4 (COSMIC only) |
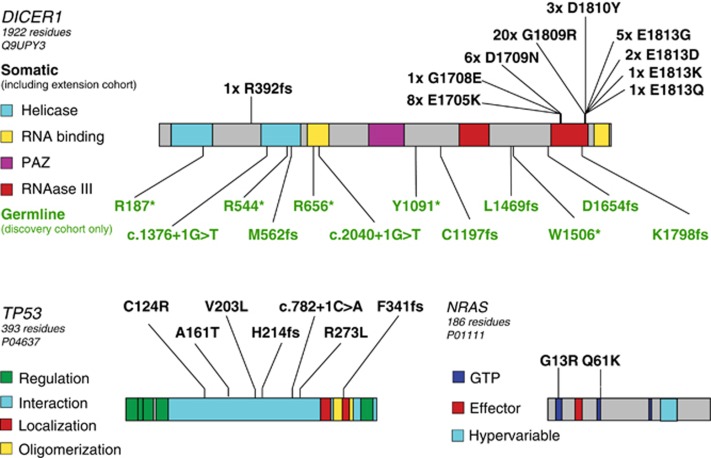
Somatic DICER1 missense mutations were found in all 15 cases by exome sequence analysis, and in an additional 32 of 34 PPBs by targeted sequencing of an extension cohort (Supplementary Table 4). Nearly all of these somatic mutations clustered in the RNase IIIb domain (Figure 2), in some cases affecting amino acids identical to those reported in ovarian Sertoli–Leydig tumors,12 a tumor seen in association with familial PPB. The single somatic mutation outside this region was in a case without a germline variant. This tumor had two somatic events, a 10 bp frameshift insertion and an RNase IIIb missense mutation. The most frequent mutation, p.Gly1809Arg, was seen in seven of nine Type III PPBs by exome sequencing and in 13 extension cases. Notably, this mutation has not been reported in any other malignancy to date (Catalogue of Somatic Mutations in Cancer v.6813), suggesting it may be characteristic of progressive PPB.
Figure 2.
Location of somatic mutations and germline variants in significantly mutated genes. Protein domains are as annotated from the UniProt record indicated under each gene name. Somatic mutations are indicated by black text above the protein model, whereas germline variants are indicated by green text below the protein model. Mutations detected in the extension cohort using targeted resequencing are included in the counts of somatic DICER1 mutations.
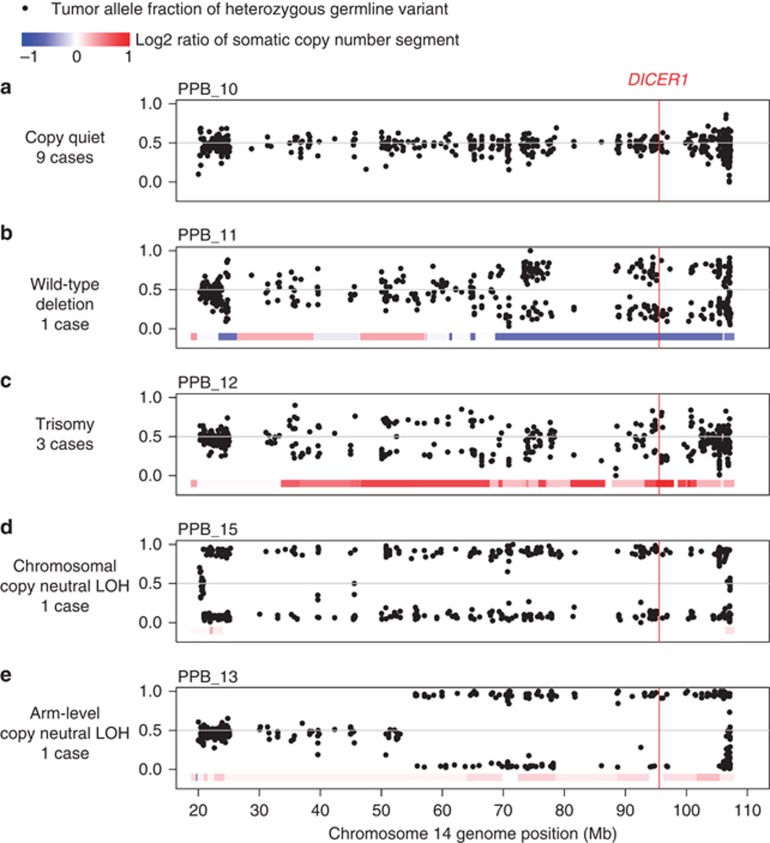
In addition to somatic missense mutations in DICER1, each of the 15 whole exome sequencing cases also had loss of the second DICER1 allele through one of several genetic mechanisms (Figures 3a and e). Germline loss-of-function DICER1 sequence variants were found in 12 of 15 cases: nine without further copy-number alterations (Figure 3a), one with somatic deletion of the predisposing germline allele (Figure 3b) and two with somatic, arm-level gain of 14q that duplicated either the somatic missense or germline loss-of-function allele (Figure 3c). One case without a predisposing germline variant had two somatic mutations: a 10 bp frameshift insertion and an RNase IIIb missense mutation that was also duplicated by copy-number gain (similar to Figure 3c). The remaining two cases without a predisposing germline allele had copy neutral loss-of-heterozygosity at the DICER1 locus owing to somatic uniparental disomy of all (Figure 3d; PPB_15) or part (Figure 3e; PPB_13, 14q21->qter) of chromosome 14. Sanger sequencing of one case confirmed that the germline and somatic mutations occurred in trans (Supplementary Figure 1), consistent with allele fractions observed in the tumor sequence data (0.2 germline loss-of-function variant, 0.68 somatic missense mutant). These data suggest that DICER1 requires biallelic disruption, functioning as a unique variant of a two-hit tumor suppressor rather than through haploinsufficiency. However, retention of some DICER1 activity appears to be necessary to ensure survival of PPB cells, as no cases harbored two complete loss-of-function variants and mutant alleles were often duplicated. Therefore, the focused hotspot region of missense mutation in DICER1 appears to be functionally distinct from simple loss-of-function variants found through the gene body.
Figure 3.
Somatic chromosome 14 copy-number segments and allele fractions of variants heterozygous in matched normal. Data points are the tumor allele fractions of heterozygous, non-reference alleles in the matched normal sample. Somatic deletion of the non-reference allele results in tumor allele fractions near 0, whereas gain of the non-reference base or deletion of the reference base results in values approaching 1. Colored bars represent copy-number states inferred from fractional coverage values. Each panel depicts different combinations of copy-number alteration and loss-of-heterozygosity detected by whole exome sequencing. (a) Copy quiet: Compound germline loss-of-function (LOF) and somatic RNase IIIb missense mutation without further copy-number alteration (nine cases). (b) Wild-type deletion: Deletion of wild-type allele resulting in hemizygosity for the somatic RNase IIIb missense mutation (PPB_11). (c) Trisomy: Copy-number gain of chromosome 14 resulting in duplication of RNase IIIb mutant allele and retention of germline LOF allele (two cases). An additional case has duplication of the germline LOF allele and retention of the RNase IIIb mutant allele (PPB_5). (d) Chromosomal copy neutral loss-of-heterozygosity: Copy-neutral loss of wild-type allele and duplication of entire chromosome 14 containing the somatic RNase IIIb missense mutation (PPB_15). (e) Arm-level copy neutral loss-of-heterozygosity: Copy-neutral loss of wild-type allele and duplication of 14q containing the somatic RNase IIIb missense mutation (PPB_13).
DICER1 RNase IIIb domain mutations lead to defective cleavage of 5p miRNAs from the pre-miRNA loop sequence
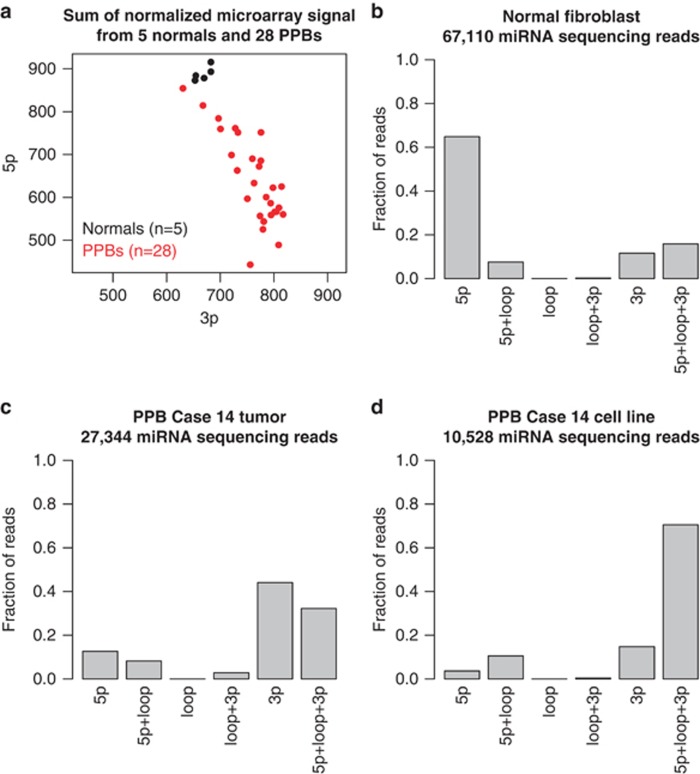
Double-stranded precursor miRNAs (pre-miRNAs) are normally processed by DICER1 into three maturation products: two potentially functional units derived from either the 5-prime (5p) or 3-prime (3p) arm of the precursor (mature 5p and 3p miRNAs) and the hairpin loop. In vivo studies on mouse mesenchymal stem cells transfected with human DICER1 (hsDICER) mutant constructs14 had shown that the RNase IIIa domain of DICER1 serves a role in the removal of pre-miRNA loop sequences from the 3p mature miRNA. The RNase IIIb domain cleaves the 5p mature miRNA from the hairpin loop. Our microarray analysis of miRNA expression levels found that primary PPB tumors had significant reduction in expression of 5p-derived miRNAs (Figure 4a and Supplementary Figure 2), consistent with reported DICER1 mutant expression profiles in Sertoli–Leydig tumors and in mouse hsDICER1 constructs.12,14,15 We subsequently confirmed this observation by quantitative polymerase chain reaction (PCR) of seven miRNAs in 8–13 samples (Supplementary Figure 3 and Supplementary Table 5).
Figure 4.
(a) Relative expression level of 5p- and 3p-derived miRNAs in 5 normal lung and 28 PPB tissues as measured by total microarray intensities. Normal tissues (black) had higher expression of 5p miRNA compared with 3p miRNA, whereas all PPBs (red) appeared to have lower overall expression of 5p miRNAs compared with normal tissues. (b–d) The fraction of reads with start and end points confined to functional units of 1595 miRNA primary transcripts. 5p and 3p Regions as annotated by miRbase build 19. Hairpin loop regions were defined as the genome segment between 5p and 3p regions. 5p+Loop+3p denotes reads that include 5p, loop and 3p sequence (that is, precursor miRNAs). The fraction of reads corresponding to these regions in each miRNA individually is provided as Supplementary Figure 6 and Supplementary Table 9. (b) In normal tissues, reads were primarily derived from mature 5p and 3p miRNAs sequenced, with higher expression from 5p miRNA compared with 3p miRNA, consistent with the microarray data. (c) In addition to skewed ratio of 5p and 3p miRNA expression compared with normal tissues, primary PPB tissue had increased fraction of reads containing 5p and loop sequences (5p+loop), and full-length, pre-miRNAs (5p+loop+3p) (Supplementary Figure 4). (d) Owing to the presumed depletion of normal cells, the presence of extended 5p miRNAs and pre-miRNAs is even clearer in a PPB cell line derived from the tissue displayed in (c).
Because of the concern that oligonucleotide probes used for expression arrays may still hybridize to 5p miRNAs with loop sequence attached, we sought additional sequence confirmation of the specific defect in DICER1 function. Direct sequencing of miRNA from primary tumor and cell line from PPB case 14 uncovered retention of pre-miRNA loop sequences joined to 5p miRNAs compared with a normal lung fibroblast cell line (Figures 4b and d and Supplementary Figure 4), providing direct evidence that these mutations result in an inability to cleave the 5p end of pre-miRNA hairpins and lead to retention of pre-miRNA loop sequences in DICER mutant cancers.
Additional drivers of PPB development
Loss of TP53 occurred in 13 of 15 whole exome sequencing cases (Figure 1 and Tables 1 and 2). Five cases had deletion of one 17p allele, seven had deletion of one allele and mutation of the remaining allele and one had homozygous deletion of TP53 owing to an arm-level loss of 17p and a focal 130 kb deletion. While deletion of TP53 appears to occur early, most point mutations are subclonal to DICER1 mutations, suggesting that only a sub-population of PPB cells are malignant clones completely lacking TP53. Analysis of TP53 expression by immunohistochemistry in 27 tumors confirmed the presence of subclonal cell populations with strong, diffuse staining in nine cases (Supplementary Figure 5). TP53 expression by immunohistochemistry was consistent with molecular status in 23 of 27 cases (Supplementary Table 6).
Table 2. Chromosome regions of significant somatic copy-number alteration.
| Chromosome arm | Gain or loss | Genes | Amplification cases | Amplification q-value | Deletion cases | Deletion q-value | Candidate targets |
|---|---|---|---|---|---|---|---|
| 2p | Gain | 924 | 6 | 3.0 × 10−5 | 0 | 0.89 | ALK, MYCN, REL |
| 2q | Gain | 1556 | 6 | 3.1 × 10−5 | 0 | 0.89 | |
| 8p | Gain | 580 | 9 | 4.0 × 10−9 | 1 | 0.82 | |
| 8q | Gain | 859 | 10 | 4.8 × 10−12 | 0 | 0.89 | MYC |
| 10q | Loss | 1268 | 0 | 0.90 | 9 | 3.5 × 10−6 | PTEN, SUFU |
| 11p | Loss | 862 | 0 | 0.90 | 8 | 0.0024 | WT1 |
| 17p | Loss | 683 | 0 | 0.90 | 13 | 0 | TP53, MAP2K4 |
| 20q | Gain | 753 | 4 | 0.0023 | 0 | 0.97 | |
| 19q13.13 | Focal gain | 196 | 4 | 4.5 × 10−4 | 0 | — | Zinc-finger cluster |
Two activating NRAS missense mutations that we identified in PPB have been widely reported in other tumors (p.Gly13Arg and p.Gln61Lys).13 An additional case harbored an in-frame 9 bp insertion in the BRAF kinase domain (p.472_473insTVY), similar to activating insertions reported in pilocytic astrocytoma16 and papillary thyroid cancer.17 These mutations implicate increased RAS signaling as a feature of PPB development, consistent with mutation and expression profiling of other pediatric solid tumors.18
Analysis of copy-number variation using GISTIC219 uncovered nine loci of significant copy-number variation (q⩽0.1; Table 2 and Supplementary Tables 7 and 8). Eight of these correspond to whole chromosome (2+, 8+) or arm-level (10q−, 11q−, 17p−, 20q+) alterations. Oncogenes driven by amplification in other cancers include MYC on chromosome 8 and ALK, MYCN and REL on chromosome 2.20 Tumor suppressors on frequently lost chromosome arms include PTEN and SUFU on 10q; WT1 on 11p; and TP53 and MAP2K4 on 17p.20 In addition to TP53, ATM, BCOR and SUFU had hemizygous loss-of-function mutations, implicating them as additional targets of inactivation in PPB. Four cases had a focal amplification of 7.35 Mb on 19q13.13, containing 196 genes of which >20% encode zinc-finger proteins. The only gene in this region listed in the Cancer Gene Census20 is an unlikely amplification target; tumor suppressor CEBPA, a transcription factor that inhibits cyclin-dependent kinases 2 and 4.21 This region is also significantly amplified in esophageal squamous cell carcinomas,22 although the exact target of this event remains unknown. This focal event may be indicative of a more complex structural event not captured by our whole exome sequencing assay, particularly as PPB karyotypes harbor frequent translocations including two cases with 19q13 translocations recorded in the International PPB Registry (http: //www.ppbregistry.org).
Discussion
We describe here an extensive genomic analysis of the rare embryonal tumor pleuropulmonary, identifying predisposing loss-of-function DICER1 mutations coupled with partial loss-of-function missense somatic DICER1 point mutations leading to defective cleavage of 5p-derived miRNAs with retention of loop sequences. TP53 is deleted and/or mutated in 13/15 cases and appears to be an early event following DICER1 mutation in tumor cells. Copy-number changes in critical genomic regions including PTEN, ATM and WT1 are common. The identification of RAS pathway mutations suggests that activation of Ras signaling may be one additional element that underlies progression from cysts to tumors. Combined, these analyses confirm a multistep genetic pathogenesis in the lung mesenchymal cells of solid PPB, similar to that seen in many adult tumors.
miRNA-seq analysis showed the effects of a somatic RNase IIIb mutation on miRNA precursor molecules with correct cleavage of mature miRNAs originating from the 3p arm and near total absence of correct cleavage of mature miRNAs originating from the 5p arm. Interestingly, one of the miRNAs whose 5p arm was correctly cut was miR-451, a known target of Ago2 and not DICER1.23
Previous reports have shown gene expression profiles of DICER1 RNase IIIb mutant cells and tumors indicate derepression of genes normally regulated by let-7 family and others producing an oncogenic expression profile.14,15 We hypothesize that the retention of loop sequence results in premature degradation of the 5p-derived transcripts, as the ratio of 5p to 3p miRNAs was lower in DICER1 mutant tumors compared with normal cells. Whether the retention of the loop leads to degradation or improper loading of the miRNA onto the RNA-induced silencing complex (RISC), or both, is unclear and was not investigated in this study.
Altered miRNA profiles and resulting gene derepression may not be the only tumorigenic mechanism in PPB. DICER1 knockdown has been shown to decrease double-strand break repair and this has been postulated as a potential mechanism by which DICER1 mutations may contribute to cancer.24,25 The effect of RNase IIIb mutations on other small RNA categories, including those with a role in DNA repair, has not been studied. PPB does appear to have a higher mutation rate (more on par with glioma and breast carcinoma) than other pediatric cancers.10,13 However, given that TP53 mutations were also present in the large majority of PPBs, it would be premature to attribute this high mutation rate to an effect of mutant DICER1 on DNA repair alone.
Finally, biallelic mutations leading to complete loss of DICER1 function were not seen in any of the 49 cases in this report, suggesting that one or more of the 3p mature miRNAs generated from the retained RNase IIIa activity in DICER1 appear to be necessary to ensure survival of PPB cells. If tumor cells are more reliant on the cellular quantity of expressed 3p miRNAs for survival than normal cells, then targeted antisense molecules to mature 3p miRNAs may be an effective therapeutic approach.
Materials and methods
Cohort
Families were ascertained through the International PPB Registry (http: //www.ppbregistry.org). All research subjects provided written consent to the molecular and family history studies as approved by the Human Research Protection Offices at Washington University and Children's National Medical Centers in St Louis, MO, USA and Washington, DC, USA, respectively. Blood and saliva specimens were collected as a source of genomic DNA through family visits made by the research team or mailed to Washington University by the research subject or his/her health-care professional. DNA was extracted from peripheral blood lymphocytes or saliva using standard protocols. Snap frozen or formalin-fixed, paraffin-embedded tumor tissue was collected from treating institutions whenever available. All diagnoses of PPB were confirmed by pathologic review by DAH and LPD.
Exome sequencing and analysis
Exome sequencing was performed by the Broad Institute Genomics Platform and analyzed using previously a described pipeline.26 Briefly, reads were aligned using bwa,27 followed by indel realignment and quality score recalibration using the Genome Analysis Toolkit.28 Somatic mutations were detected using muTect29 and indelocator, then annotated using Oncotator (http: //www.broadinstitute.org/oncotator). MutSig v.1.5 was used to detect significantly mutated genes.10 Copy-number calls were derived from relative fractional sequence coverage of each exon compared with a panel of normals. GISTIC2 was used to detect significant copy-number alterations.19 These and other tools used for exome sequence analysis are described at http: //www.broadinstitute.org/cancer/cga.
miRNA expression profiling
Whole snap frozen tumor tissue from 17 Type II (including two replicates) and 11 Type III PPBs and adjacent normal tissue from five lungs (three patients) was used for the miRNA microarray assay. Four 10 μm sections were cut with cryotome. One additional 5 μm slide was cut and stained with hemtoxylin and eosin to assess tumor content and the percentage of necrosis. Total RNA including small RNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Purified total RNA was then quantified using NanoDrop ND2000 (NanoDrop Technologies, Wilmington, DE, USA). RNA quality was evaluated with Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). Only RNA with RNA integrity number >6.0 was used for miRNA microarray assay.
The miRNA expression profiling was performed on GeneChip miRNA 2.0 Arrays containing 15 644 probe sets (Affymetrix, Santa Clara CA,USA). Briefly, total RNA (300 ng) was labeled with FlashTag Biotin HSR RNA Labeling Kit (Genisphere Inc., Hatfield, PA, USA) according to the manufacturer's protocol. Samples were applied to the array, hybridized overnight and washed using the GeneChip Hybridization, Wash and Stain Kit (Affymetrix). Array probe intensities were normalized using the Robust Multichip Average function within the R (http: //www.r-project.org) package 'affy'.30 To annotate 5p- and 3p-derived sequences, Affymetrix probe identifiers were mapped from miRbase build 10.1 to build 19 names. R v. 2.15.1, including heatmap.2 and RColorBrewer libraries, was used to calculate median and total probe intensities, and to cluster and visualize normalized expression values (Supplementary Figure 2).
Quantitative real-time PCR
Total RNA (300 ng) was reverse transcribed using the Qiagen miScript II RT Kit (Qiagen) according to the manufacturer's protocol. The mature miRNA was then analyzed by real-time PCR using miScript PCR System (Qiagen) on the StepOnePlus Real-Time PCR System (Applied Biosystems, Grand Island, NY, USA). miRNA expression was analyzed using 2−ΔΔCt method. Relative mature miRNA expression was determined in reference to the internal small nuclear RNA control SNORA73A. In statistical analysis, the results were presented as mean±s.e.m., and assessed differences between the two groups using Student's t-test.
Phasing of DICER1 germline variant and somatic mutation
We used an allele-specific primer design to determine the cis–trans relationships of germline and somatic mutations. The patient studied was heterozyous for a germline mutation c. 4407_4410delTTCT (normal germline allele sequence: 5′-GCTTTTGTAAAGAAAATCTCTCTTTCTCCTTTTTCAAC-3'). The deleted 4 bases in the primer set are underlined. This patient's tumor harbored an additional somatic mutation, c. 5125G>A. To amplify the germline wild-type allele, we used a forward allele-specific primer (5′-GCTTTTGTAAAGAAAATCTCTCTTTCT-3′) and a reverse primer (5′-CCACTATGCCGTCAGAACTC-3′). PCR was performed in a final volume of 25 μl containing 50 ng DNA, 5 pmol of each primer, 2 μl dNTP, 2.5 μl of 10 × PCR buffer and 2.5 U ExTaq (Takara Bio, Otsu, Shiga, Japan). The cycling profile included an initial step at 94 °C for 5 min first, followed by 10 cycles including a denaturation step at 94 °C for 55 s, annealing step at 62 °C for 30 s and an elongation step at 72 °C for 3 min; then by 30 cycles, which consisted of a denaturation step at 94 °C for 55 s, annealing step at 57.5 °C for 45 s and an elongation step at 72 °C for 3 min; and a final extension step at 72 °C for 10 min. The sequencing reactions were performed using BigDye Sequencing Kit (v. 3.1) according to the manufacturer's protocol (Applied Biosystems, Grand Island, NY, USA). Data were collected using an Applied Biosystems 3130xl Genetic Analyzer (Applied Biosystems, Grand Island, NY, USA).
Validation set ion torrent sequencing and analysis
Formalin-fixed, paraffin-embedded PPB tumors from 34 cases were used for the validation set. Pure tumor areas were identified by hematoxylin and eosin slide and 5 mm cores were taken from the representative area for each tumor. Following paraffin removal, DNA was extracted using the Maxwell 16 formalin-fixed, paraffin-embedded Tissue LEV DNA Purification Kit (Promega, Madison, WI, USA) on the Maxwell 16 Instrument (Promega). DNA quality and quantity was assessed using a Nanodrop (ThermoFisher, Wilmington, DE, USA) and Qubit (Qubit 2.0; Life Technologies, Grand Island, NY, USA), respectively. Samples with bioanalyzer results yielding a narrow peak indicating a loss of amplicons after library preparation were interpreted as poor quality. Sequencing was performed on an Ion Torrent Personal Genome Machine (Life Technologies, Carlsbad, CA, USA) using standard protocols. A custom multiplex PCR panel was designed for the coding regions of DICER1 and TP53 with an average amplicon length <200 bp (Custom Ampliseq; Life Techologies). Ten nanograms of starting DNA was used for the initial PCR amplification. The resulting amplicons were then barcoded and ligated to adaptors (Ion Ampliseq Library Kit 2.0; Life Technologies). Templates were prepared using the ION PGM Template OT2 200 Prep Kit and the One Touch 2 System (Life Technologies). Sequencing was performed on a 318 chip (ION PGM Sequencing 200 Kit v.2; Life Technologies) with an average of six samples per chip plus one each positive and negative control samples to achieve an average depth of coverage of 500 filtered reads. Signal processing, mapping and quality control was performed with Torrent Suite v.3.4.1 (Life Technologies). Variant calls were made using Torrent Variant Caller Plugin 3.4 choosing somatic mutation workflow and default settings, call P-value <0.05. BAM files containing raw reads were reviewed using Integrative Genomics Viewer v.2.3.31,32 Variants were annotated with Alamut mutation HT software (Interactive Biosoftware, Rouen, France) and named using HUGO nomenclature for DICER1 transcript NM_177438.2 and TP53 transcript NM_000546.5. For both DICER1 and TP53, nonsense, frameshift and canonical splice-site mutations were considered loss of function. SIFT (Sorting Intolerant From Tolerant) was used to assess the potential significance of predicted novel missense amino-acid substitutions.33 TP53 mutations were additionally compared with the IARC TP53 Mutation Database Release 16 (R16, November 2012; TP53 somatic mutation data set, http://p53.iarc.fr). We required a minimum ABQV ⩾20. We used allele depth/filtered depth to calculate allele frequency. We required a minimum number of 100 reads total for each locus and minimum 10 reads of each unique variant (except known SNPs). TP53 mutations classified as loss-of-function and missense mutations with SIFT scores predictive of deleterious effects are listed in Supplementary Table 6. Allele frequencies from the common SNP c.215C>G; p.Pro72Arg were included, when present, as an assessment for loss of heterozygosity.
miRNA-Seq
miRNA-enriched fractions separated from larger RNAs (>200 nt) were prepared using miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Enriched miRNA was quantitated on the Agilent 2100 Bioanalyzer using the Small RNA Kit (Agilent). The miRNA library preparation was performed using the Ion Total RNA-Seq Kit v.2 following standard protocols (Life Technologies). Approximately 100 ng of small RNA was used for the initial hybridization and ligation procedure. Reverse transcription was then performed and products were purified using the Magnetic Bead Cleanup Module (Life Technologies). The cDNA was then further amplified and purified to yield the final library product. Templates were prepared using the ION PGM Template OT2 200 Prep Kit and the One Touch 2 System (Life Technologies). Sequencing of the miRNA was performed on a 316 chip (ION PGM Sequencing 200 Kit v.2; Life Technologies). Signal processing, mapping and quality control was performed with Torrent Suite v.3.4.2 (Life Technologies). Reads were aligned to miRNA hairpin sequence reference from miRBase (miRBase.org, Manchester, UK) and to the hg19/GRCh37 human genome reference sequence.
The Genome Analysis Toolkit DepthOfCoverage tool was used to determine the read coverage at each base across 1595 miRNA primary transcripts annotated by miRbase build 19. Coverage values were then mapped to their percent location along the primary transcript and normalized to a total fractional coverage for each miRNA (that is, the sum of fractional coverage across each primary transcript equals one). These fractional coverage distributions were then summed across all miRNAs and plotted as Supplementary Figure 4. Genome coordinates from miRbase denoting the location of miRNA 5p- and 3p-derived miRNAs within a primary transcript were converted to percent locations for each miRNA and the distribution plotted as a key to index the coverage distribution plots (see row 4 on Supplementary Figure 4). miRNAs with both 5p (black) and 3p (red) elements were depicted separately from miRNAs without these annotations (blue).
To count the number of reads corresponding to each annotated miRNA component, we used the R package ‘rbamtools' version 2.0 to extract the mapping position of each read in the bam file that fell within an miRNA. To remove adapter sequence and to account for uncertainty in the annotation of the exact 5p and 3p coordinates, we trimmed 12 bp from the start and 6 bp from the end of each read. We then counted the number of reads with trimmed start and end points located in all possible combinations of 5p, loop, 3p and unannotated regions.
Immunohistochemistry for p53
Immunohistochemistry for p53 was performed on 4 μm sections of each of the validation set tumors using the same block from which cores were taken for DNA extraction. Using an automated stainer (BondMax; Leica Biosystems, Buffalo Grove, IL, USA), anti-p53 antibody (clone Y5, prediluted, rabbit monoclonal; Biocare Medical, Concord, CA, USA) was applied for 10 min incubation following antigen retrieval in ER1 solution (citrate based) for 30 min (Leica, Buffalo Grove, IL, USA). Stains were reviewed by three pathologists (DAH, CTR and LPD) and the percentage of tumor cell nuclei with strong staining was recorded and compared with TP53 mutation status (Supplementary Figure 5 and Supplementary Table 6). Strongly positive nuclei were considered to be likely associated with missense mutations leading to delayed degradation.
Acknowledgments
Sequence data used for this analysis are available from dbGaP under accession phs000543. The work was conducted as part of the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Health Institute in Mexico. We would like to acknowledge the technical expertise and data generation efforts of The Broad Institute Genomics Platform and Cancer Genome Analysis group. TJP was supported by a Canadian Institutes of Health Research Fellowship. DAH is supported by National Cancer Institute (R01CA143167), and National Heart, Lung and Blood Institute (R01HL109265 and R01HL111190). We thank the members of the Ogawa Laboratory for shared insights during reporting of genomic findings of PPB.34
MM is a paid consultant for and equity holder in Foundation Medicine, a genomics-based oncology diagnostics company.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Hill DA, Jarzembowski JA, Priest JR, Williams G, Schoettler P, Dehner LP. Type I pleuropulmonary blastoma: pathology and biology study of 51 cases from the international pleuropulmonary blastoma registry. Am J Surg Pathol. 2008;32:282–295. doi: 10.1097/PAS.0b013e3181484165. [DOI] [PubMed] [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965–965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Bahubeshi A, Hamel N, Pasini B, Asioli S, Baynam G, et al. Extending the phenotypes associated with DICER1 mutations. Hum Mutat. 2011;32:1381–1384. doi: 10.1002/humu.21600. [DOI] [PubMed] [Google Scholar]
- Frio TBA. Dicer1 mutations in familial multinodular goiter with and without ovarian Sertoli–Leydig cell tumors. JAMA. 2011;305:68–77. doi: 10.1001/jama.2010.1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade I, Bacchelli C, Davies H, Murray A, Abbaszadeh F, Hanks S, et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48:273–278. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- Doros LA, Rossi CT, Yang J, Field A, Williams GM, Messinger Y, et al. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1–renal sarcoma Mod Pathole-pub ahead of print 31 January 2014; doi: 10.1038/modpathol.2013.242 [DOI] [PMC free article] [PubMed]
- Doros L, Yang J, Dehner L, Rossi CT, Skiver K, Jarzembowski JA, et al. DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer. 2012;59:558–560. doi: 10.1002/pbc.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehner LP, Jarzembowski JA, Hill DA. Embryonal rhabdomyosarcoma of the uterine cervix: a report of 14 cases and a discussion of its unusual clinicopathological associations. Mod Pathol. 2012;25:602–614. doi: 10.1038/modpathol.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz KAP, Pacheco MC, Yang J, Williams GM, Messinger Y, Hill DA, et al. Ovarian sex cord-stromal tumors, pleuropulmonary blastoma and DICER1 mutations: a report from the International Pleuropulmonary Blastoma Registry. Gynecol Oncol. 2011;122:246–250. doi: 10.1016/j.ygyno.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes Nature 2013[Internet]. 16 June 2013 [cited 18 June 2013] advance online publication. Available at: : http://www.nature.com/nature/journal/vaop/ncurrent/full/nature12213.html . [DOI] [PMC free article] [PubMed]
- Getz G, Höfling H, Mesirov JP, Golub TR, Meyerson M, Tibshirani R, et al. Comment on ‘The Consensus Coding Sequences of Human Breast and Colorectal Cancers'. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- Heravi-Moussavi A, Anglesio MS, Cheng S-WG, Senz J, Yang W, Prentice L, et al. Recurrent somatic DICER1mutations in nonepithelial ovarian cancers. N Engl J Medi 2012366234–242. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtan AM, Lu V, Bhutkar A, Sharp PA. In vivo structure–function analysis of human Dicer reveals directional processing of precursor miRNAs. RNA. 2012;18:1116–1122. doi: 10.1261/rna.032680.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anglesio MS, Wang Y, Yang W, Senz J, Wan A, Heravi-Moussavi A, et al. Cancer-associated somatic DICER1 hotspot mutations cause defective miRNA processing and reverse-strand expression bias to predominantly mature 3p strands through loss of 5p strand cleavage. J Pathol. 2013;229:400–409. doi: 10.1002/path.4135. [DOI] [PubMed] [Google Scholar]
- Eisenhardt AE, Olbrich H, Röring M, Janzarik W, Anh TNV, Cin H, et al. Functional characterization of a BRAF insertion mutant associated with pilocytic astrocytoma. Int J Cancer. 2011;129:2297–2303. doi: 10.1002/ijc.25893. [DOI] [PubMed] [Google Scholar]
- Hou P, Liu D, Xing M. Functional characterization of the T1799-1801del and A1799-1816ins BRAF mutations in papillary thyroid cancer. Cell Cycle. 2007;6:377–379. doi: 10.4161/cc.6.3.3818. [DOI] [PubMed] [Google Scholar]
- Shukla N, Ameur N, Yilmaz I, Nafa K, Lau C-Y, Marchetti A, et al. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Iakova P, Wilde M, Welm A, Goode T, Roesler WJ, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Yoshida K, Shiraishi Y, Shimamura T, Sato Y, Nishimura R, et al. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014;74:2742–2749. doi: 10.1158/0008-5472.CAN-13-2470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.