Abstract
The FHIT gene at FRA3B is one of the earliest and most frequently altered genes in the majority of human cancers. It was recently discovered that the FHIT gene is not the most fragile locus in epithelial cells, the cell of origin for most Fhit-negative cancers, eroding support for past claims that deletions at this locus are simply passenger events that are carried along in expanding cancer clones, due to extreme vulnerability to DNA damage rather than to loss of FHIT function. Indeed, recent reports have reconfirmed FHIT as a tumor suppressor gene with roles in apoptosis and prevention of the epithelial–mesenchymal transition. Other recent works have identified a novel role for the FHIT gene product, Fhit, as a genome “caretaker.” Loss of this caretaker function leads to nucleotide imbalance, spontaneous replication stress, and DNA breaks. Because Fhit loss-induced DNA damage is “checkpoint blind,” cells accumulate further DNA damage during subsequent cell cycles, accruing global genome instability that could facilitate oncogenic mutation acquisition and expedite clonal expansion. Loss of Fhit activity therefore induces a mutator phenotype. Evidence for FHIT as a mutator gene is discussed in light of these recent investigations of Fhit loss and subsequent genome instability.
Keywords: Common fragile sites, FRA3B/FHIT, Replication stress, Genome instability, Mutator phenotype
Introduction
Common fragile sites (CFSs) are large genomic loci that display chromosomal instability in response to genotoxic or replicative stress [1]. This chromosomal instability, most often displayed as metaphase breaks, occurs at CFSs before instability at other genomic regions is observed. Because many fragile sites map to loci non-randomly altered in cancers, it was thought that these CFSs might harbor genes that contribute to cancer development through genomic alteration [2]. Indeed, the tumor suppressor gene FHIT spans the most fragile CFS in human lymphoblasts, FRA3B; recent mapping of CFSs in epithelial and fibroblast-derived cells has shown that activation of specific fragile loci varies in cells of different tissues [3] and the FRA3B/FHIT locus is not necessarily the most fragile locus in non-hematopoietic tissues [3–5].
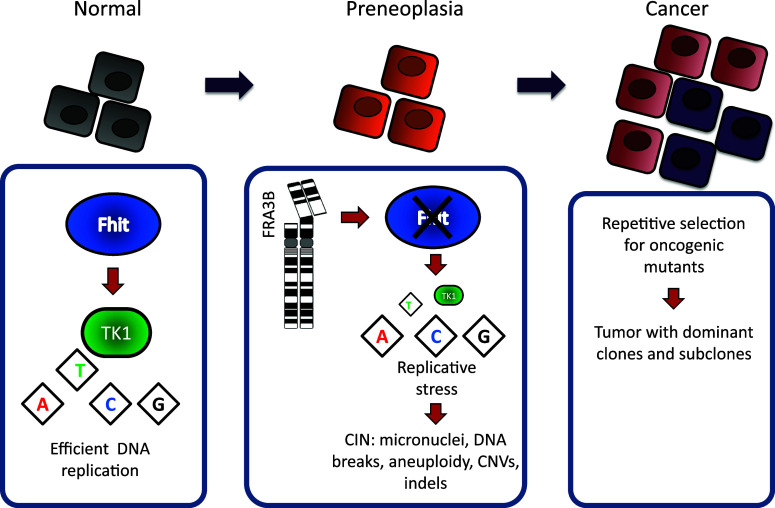
Alterations at FRA3B, which frequently result in loss of Fhit protein expression, are associated with benign, preneoplastic, and malignant lesions of many human organs. Since the discovery of this first gene at a fragile locus, nearly 1,000 reports concerning the FHIT gene or gene product have been published and many include evidence of the role of Fhit protein in tumor suppression and correlation of loss of Fhit expression with various clinical features of various types of cancer, such as poor prognosis, invasiveness, and poor outcome [6]. Very recently, several reports described a role for Fhit in preventing epithelial–mesenchymal transition (EMT), an important morphologic change that occurs in cancers as they become invasive [35]. However, the most recent function to be assigned to Fhit, and perhaps its most significant cancer-associated function, is the discovery that Fhit is a genome “caretaker”, whose loss induces global genome instability [7]. When Fhit activity is lost, cells exhibit spontaneous replication stress, increased levels of single-stranded DNA (ssDNA), double-strand DNA (dsDNA) breaks, and chromosomal instability. Replication fork stabilization by Fhit protein is effected through positive regulation of thymidine kinase 1 (TK1), an enzyme necessary for the salvage pathway of thymidine triphosphate (TTP) pool biosynthesis. Adequate TTP synthesis is necessary to maintain DNA replication fork stability.
Fhit loss also increases the fragility of other CFS [4] and it has been shown that loss of Fhit caretaker function leads to a mutator phenotype, with allele copy number alterations, expression changes, and exome mutations in Fhit-deficient cells and tissues [7, 8], increasing the likelihood of progression to cancer. There were some earlier hints of involvement of Fhit in a caretaker function due to close study of sebaceous tumors, glandular tumors that occur in sweat and oil glands of humans, often near the eye. A group of very observant German ophthalmologists first observed loss of Fhit expression in periocular sebaceous gland carcinoma [9, 10] in patients with Muir–Torre syndrome (a type of tumor thought to be due exclusively to inactivation of mismatch repair genes such as the MSH2) and suggested that loss of Fhit then initiates further mutations. Thus, the Fhit protein, encoded by a gene at a highly unstable genomic locus, paradoxically regulates global genome instability. In this review we briefly summarize old and new research concerning the Fhit suppressor functions and then describe the more recent discovery of the Fhit caretaker function.
Alterations at the FRA3B/FHIT locus
The FHIT gene, located at 3p14.2, is a large ten-exon gene [11]. Though the gene is ~2 Mb in length, FHIT messenger RNA is only 1.1 kb with exons 5–9 encoding a small protein of 147 amino acids. The core of FRA3B overlaps FHIT exons 4–6 and the majority of FRA3B alterations occur within this region [12]. Aberrant transcripts of FHIT are detected at a high frequency in cancer cells, many of them due to deletions within the FRA3B core region, and reduction or absence of Fhit protein expression occurs in nearly 50 % of all cancers. Silencing of Fhit expression can also occur by CpG methylation within the promoter, and this mode of Fhit expression silencing is a frequent occurrence in cancer cells. Fhit loss occurs most commonly in epithelial-derived cancers of the lung, esophagus, breast, throat, stomach, skin, pancreas, cervix, and kidney; though there are examples of Fhit loss in many other types of cancer.
There are also numerous reports of genetic alterations at the FHIT locus and loss of Fhit protein expression in preneoplasias, suggesting a tumor suppressive role for Fhit in the early stages of cancer development [13–15]. FHIT gene alterations and loss of Fhit protein expression occurs in up to 90 % of precancerous lesions of squamous cell carcinoma of the lung [15], and is even observed in the histologically normal bronchial epithelium of chronic smokers; thus, Fhit loss may be an initiating event in the development of lung tumors of carcinogen-exposed individuals [16, 17]. The FHIT gene is highly sensitive to environment carcinogens, especially carcinogens present in cigarette smoke, so it is not surprising that Fhit is lost in early pre-cancer cells of lung squamous cell carcinoma. In sporadic breast cancer, Fhit loss also occurs in the precancerous stages, primarily at the pre-invasive dysplastic stage with estimates of Fhit loss occurring in as many as 70 % of ductal in situ carcinomas and loss of heterozygosity (LOH) at the FHIT locus in as many as 25 % of intraductal hyperplasias [13, 18]. Fhit loss is also very common in the precancerous lesions of the esophagus [8, 19], cervix [20, 21], oral cavity [22], and other organ tissues [23–25], suggesting that in many different types of cancer, Fhit plays an important role in suppressing the formation of the early preneoplastic and premalignant lesions.
FHIT as a tumor suppressor gene
Due to the exquisite fragility seen at FRA3B and other CFSs, it has been argued that alterations at CFSs are passenger events in cancer development. However, there have been many reports that support findings that the FHIT gene is a tumor suppressor. First, Fhit protein is lost or reduced in most cancers. Second, Fhit knockout mice are much more susceptible to cancer induction than wild-type mice. Fhit-/- mice not only develop more carcinogen-induced tumors but also develop more spontaneous tumors than wild-type control mice [26, 27]. Over-expression of Fhit in Fhit-negative cancer cell lines suppresses tumorigenicity of xenografts in mice, and FHIT viral gene therapy prevents and reverses carcinogen-induced tumors in a gastric cancer mouse model [28]. It has been shown that a major mechanism of tumor suppression by Fhit is the promotion of apoptosis. One of the few Fhit-interacting proteins discovered is ferredoxin reductase. Fhit interacts with and stabilizes ferredoxin reductase in the mitochondria during oxidative stress [29–31]. This interaction results in increased reactive oxygen species production, causing caspase-3 activation and apoptosis. It has also been reported that Fhit can increase calcium release from the mitochondria, offering an additional means of promotion of apoptosis [32].
Fhit also suppresses EMT as another tumor suppressor mechanism [33, 34]. A recent study demonstrated that Fhit silencing in bronchial cells induced over-expression of two primary EMT-associated targets, MMP-9 and vimentin [35]. In this study, Fhit silencing increased cell invasion dependent on Slug, a member of the Snail family of transcription factors known to be key mediators of EMT. The authors determined that EGFR was the receptor orchestrating the Src/ERK/Slug pathway modulated by Fhit. This recent work strengthens previous reports that Fhit expression minimizes the invasiveness and metastatic potential of cancer cells. Gaudio and colleagues [36] have recently discovered a novel Fhit-interacting protein, Annexin 4. Their work demonstrates that over-expression of Fhit prevents Annexin 4 translocation to the plasma membrane from the cytosol following paclitaxel treatment. This result suggests that Fhit and paclitaxel act synergistically to increase apoptosis in tumor cells, both in vitro and in vivo. Fhit negativity is an accepted marker of poor prognosis in cancer patients, and this work suggests a mechanism through which Fhit loss could contribute to a poor prognosis.
FHIT as a genome caretaker
CFS fragility is cell type specific
The mechanisms of fragility at FRA3B and other CFSs have remained elusive for many years. CFS instability has historically been attributed to the fact that they harbor sequences with a propensity for forming secondary structures that could hinder replication fork movement. This would lead to fork collapse and dsDNA breaks. However, Letessier and colleagues [3] recently demonstrated that the fragility of FRA3B does not result from replication stress-induced fork stalling but instead from a scarcity of replication initiation events. They showed that in lymphoblastoid cells, but not in fibroblasts, initiation events are absent from the FRA3B core and are instead only present on the fringes of the fragile region. This forces replication forks coming from the flanking regions to cover approximately 700 kb in order to complete replication. These genomically distant origins were shown to fire in mid-S phase, leaving the fragile core incompletely replicated upon fork slowing. Notably, FRA3B instability is not found in all cells types but only in cells showing the pattern of sparse initiation origins. The FRA3B locus of NHEK epithelial cells shows Repli-seq data that would suggest it is less fragile in these cells than in lymphoblasts [4]. Thus, it has been proposed that CFSs are the latest initiation-poor regions to complete replication in a given cell type. Repli-seq data for three of the top epithelial fragile regions examined in the Huebner lab [4] support this model of fragility dependent on placement of DNA replication origins in cells derived from specific tissues [3]. FRA16D, another fragile site that encompasses the tumor suppressor gene WWOX, shows similar distributions of replication origins in both epithelial and lymphoblast cells [4]. In contrast, FRA2I/2q33 shows sparse origins in epithelial cells but adequate origins in lymphoblast cells. Therefore, depending on the CFS in question, epithelial cells show either differences or similarities in active CFSs when compared with lymphoblasts. FRA16D is among the most active in both cell types and thus is among the most sensitive to replication stress in cells of epithelial and lymphoblast origin. Expression of the Wwox protein is very frequently reduced or absent in human solid tumors, particularly breast cancers [37] (see R. Aqeilan chapter in this issue).
Fhit loss increases CFS fragility
In addition to paucity of initiation events, it has recently been demonstrated that loss of Fhit increases the fragility of other CFS [4]. In MCF10A cells, knock down of FHIT expression resulted in a twofold increase in chromosomal breaks and gaps. Induction of several novel fragile loci was also observed, including 7q22, 9p22, 1q44, and 2q23 [4]. This increase in fragility occurred in the absence of exogenous replication stress, strengthening the argument for Fhit loss as the causative stress. CFSs are frequently altered in cancer so it is likely that Fhit loss increases the occurrence of CFS alterations in cancer cells. Because of its exquisite fragility, it has been argued that loss of FHIT expression is a passenger event during clonal expansion rather than a cancer driver [38]. However, breast and other epithelial cancers show reduced or absent expression of FHIT and yet the FRA3B/FHIT locus is apparently not the most fragile CFS in breast epithelial cells [4]. This finding, combined with the observation that Fhit loss increases fragile breaks at other CFSs, is consistent with the idea that loss of FHIT expression is selected for during cancer initiation or progression.
Fhit-negative cells show increased chromosomal instability
Genome instability is a hallmark of sporadic cancers, which expedites tumorigenesis and provides DNA damage hotspots for mutations [39]. Genome instability includes chromosomal instability, microsatellite instability, and point mutations. Chromosomal instability represents the most commonly observed cancer-driving instability and can be subdivided into structural or numerical chromosomal irregularities [40]. Examples of structural chromosomal instability are translocations, inversions, deletions, and duplications. Numerical instability includes aneuploidy, triploidy, and tetraploidy. In addition to increased DNA breaks at CFSs, Fhit-deficient cells demonstrate other forms of genome instability [4, 7, 8]. Fhit-silenced MCF10A cells show >2-fold increased micronucleus formation vs. shCtrl cells. This illustrates that the loss of Fhit protein activity in epithelial cells is sufficient to cause an increase in chromosomal instability, even in the absence of exogenous genotoxic stress [4]. Furthermore, mouse kidney [MK] cells from Fhit−/− mice show twofold increased spontaneous sister chromatid exchange (SCE) frequency vs. +/+ MK cells [4]. Fhit−/− MK cells and mouse embryo fibroblasts (MEFs) also show increased insertions/deletions and chromosomal aneuploidy, including chromosome gains and losses [8]. Copy number variations (CNVs), spanning >10 kb, were not seen in late passage +/+ MEF cell lines but numerous CNVs occurred in post-senescent −/− cells, including acquired gains within chromosome band 10D2 encompassing the murine Mdm2 gene, an oncogene frequently involved in cellular transformation. This amplification was accompanied by ~4-fold increase in Mdm2 mRNA expression. CNVs were also detected in −/− weanling tail tissue, illustrating existence of genome instability in normal tissue not related to development of preneoplastic clones. After silencing FHIT in transformed MCF10A breast epithelial cells, chromosomal losses or gains at 3p14.2, 7q34, 9q21.3, 15q22.2, 16q23.3, and 3p21.2, which carry tumor suppressor genes or oncogenes, were observed [4]. For example, the RORA gene at FRA15A (15q22.2) was deleted after FHIT silencing. The RORA gene is expressed in normal breast, prostate, and ovarian epithelium, is frequently induced upon cellular stress, and is inactivated in cancers that arise from these organs [41] (see D. Smith chapter in this issue).
Chromosomal instability in Fhit-negative cells correlated with pro-survival mRNA and protein expression changes: an in vitro study
Fhit−/− mouse kidney accumulated genetic alterations due to loss of Fhit caretaker function and, additionally, showed striking changes in expression of the p53–p21 cell cycle control pathway [8]. Between passage 7 and passage 12, p53 protein expression increased dramatically in these −/− cells, while p21 protein disappeared abruptly. This was in accord with mRNA data, indicating that at late passages in −/− cells, Cdkn1a (p21) mRNA can be down tenfold. This suggests that the overexpressed p53 was mutated, as p21 is an important downstream p53 target. Indeed, the absence of p21 expression was due to a single base substitution in Trp53, a G to C mutation in the DNA-binding domain of p53, which likely decreased p53-mediated Cdkn1a gene transactivation and thus p21 protein expression. These pro-survival expression changes resulted in increased clonogenicity of Fhit−/− kidney and MEF cells at late passage, both with and without carcinogen treatment. Microscopic examination of morphology and lack of β-galactosidase staining of −/− clones suggested that clonogenic late passage Fhit−/− cells acquired resistance to carcinogen-induced apoptosis and cell cycle arrest. Interestingly, Fhit−/− kidney and MEF cells recruited different resistance mechanisms in response to carcinogen treatment. The surviving −/− kidney cells showed down-regulated caspase 3 and reduced cleaved PARP in the complete absence of p21 expression due to mutant p53. MEF −/− cells instead demonstrated a functional p53–p21 pathway but instead up-regulated Survivin protein while maintaining steady anti-apoptotic Mcl1. Thus, the kidney cells silenced apoptotic signals while MEF cells activated anti-apoptotic signals.
Fhit loss-induced replication stress causes chromosome instability in Fhit-negative cells
Several models have been proposed as common mechanisms for the origin of genome instability, including oxidative stress, telomere erosion, impaired DNA repair, and chromosome segregation errors; however, these forms likely do not contribute to the initiation of instability but rather to ongoing instability as they are seen in more advanced lesions [42]. The prevailing hypothesis for the origin of genome instability in preneoplastic cells is that defects in DNA replication result in DNA breaks that, when incorrectly repaired, produce chromosomal changes [43]. Thus, it is important to define the molecular source of replication stress that initiates genome instability. Oncogene activation can cause replication stress, chromosomal instability, and promote tumorigenesis, and has been proposed as a mechanistic basis of genome instability [40]. However, oncogene-induced replication stress is probably not the initiating event. For example, oncogene activation is achieved through various mechanisms that involve chromosome alterations, including translocations that change expression of the oncogene, duplications that increase the oncogene copy number, point mutations within the oncogene that increase its activity, deletion of a negative regulator, or epigenetic changes that affect gene expression. Also, because many oncogenes that induce senescence require a second genetic “hit” to uncouple mitogenic signaling from the senescence barrier [44], it seems probable that some degree of genetic instability and heterogeneity must precede oncogene activation. The FHIT gene has been called the “weakest link” in the genome [28], making it a first target for inactivation in cells undergoing transformation and its deletion a strong candidate initiator of genomic instability [28, 45, 46]. Alterations at FRA3B/FHIT can be caused by normal metabolic processes or by exposure to thus-far-undefined environmental stresses. Increased CNVs, amplification of known oncogenes, and deletion of possible tumor suppressors in multiple Fhit-deficient models supports the proposal that FHIT loss leads to genomic instability and contributes to cancer development.
Fhit loss was shown to induce genome instability, indicating that Fhit has a genome caretaker function. It has now been shown that the genome caretaker function of Fhit prevents spontaneous replication stress [7]. Mechanistically, the replication stress in Fhit-deficient cells is caused by a decrease in thymidine triphosphate (TTP) pools: silencing Fhit expression leads to a moderate decrease in TTP, a reduction sustained in stably Fhit-silenced cells. Nucleotide imbalance is a well-known source of replication stress, which causes polymerase stalling thereby hindering replication fork progression. Following TTP decrease, Fhit-negative cells exhibit multiple markers of replication stress and DNA damage, including phosphorylated ATR, phosphorylated H2AX, 53BP1 foci, stalled replication forks, and dsDNA breaks. Interestingly, the DNA damage caused by Fhit loss-induced replication stress is “checkpoint blind” in that, despite phosphorylation of ATR, downstream Chk1 does not become phosphorylated and cells do not exhibit cell cycle arrest. Thymidine supplementation rescues the replication defects and suppresses dsDNA breaks in Fhit-silenced cells. Fhit up-regulates TK1 enzyme expression for the timely production of TTP during S phase via the thymidine salvage pathway. TK1 deficiency is therefore the proposed mechanism for replication stress in Fhit-silenced cells. Interestingly, the Tk1 knockout mouse also exhibits evidence of replication stress and chromosome instability. Examination of B lymphocytes and erythrocytes from Tk1−/− mice revealed a dramatic decrease in TTP levels, activation of the replication stress checkpoint kinase Chk1, and phosphorylation of the DNA damage marker H2AX [47]. In another study it was shown that Tk1−/− reticulocytes and normochromatic erythrocytes had a five- and eightfold increase in spontaneous micronuclei formation, respectively [48]. These mice are also partially immune-deficient, as are the Fhit−/− mice [27]. These findings are in accord with the Fhit/TK1 model where loss of Fhit causes reduced TK1, insufficient TTP synthesis, and subsequent replication stress and genome instability.
Replication stress causes genome instability and cellular transformation
Studies in yeast models demonstrated that decreased dNTP levels lead to increased mutagenesis through an increase in genomic instability [49]. It has also been shown that decreased dNTP levels are the cause of the DNA damage seen during oncogene-induced DNA replication stress and that restoration of cellular dNTP levels by addition of exogenous nucleotides is sufficient to suppress the DNA damage response [50]. However, in many cases, including oncogene-induced replication stress, cells exit the cell cycle and become senescent in response to this dNTP imbalance. A number of recent reports have attempted to define the mechanism of oncogene-induced senescence bypass and subsequent cellular transformation. It has been shown that when the cell culture medium is supplemented with exogenous nucleosides, melanocytes expressing oncogenic BRAF or NRAS can bypass senescence [51]. These investigators also demonstrated that either ectopic expression of RRM2, an enzyme involved in nucleotide biosynthesis, or exogenous nucleoside supplementation could overcome senescence. Because these cells maintain oncogene expression, the authors concluded that the cells that had bypassed senescence could then become transformed. Whereas oncogenes cause a dramatic decrease in dNTP levels, Fhit loss causes only moderate TTP reduction. This creates optimum DNA damage to induce global genome instability, because it is sufficient to negatively affect DNA synthesis but does not block cell cycle progression and cause senescence. This global genome instability thus drives Fhit-deficient early preneoplastic cells to neoplastic progression through cycles of mutation and selective clonal expansion.
Fhit loss and the mutator phenotype hypothesis
FHIT inactivation causes a mutator genotype/phenotype
Normal proliferating cells possess robust mechanisms to ensure genomic integrity with each cell division. These mechanisms include DNA replication factories that copy DNA with remarkable fidelity, DNA repair pathways to correct any replication mistakes or repair DNA damage caused by environmental and endogenous agents, and mitotic checkpoints that promote faithful segregation of sister chromatids to daughter cells. As a result, proliferating cells are able to limit the number of spontaneous mutations to <1 mutation per 10 billion nucleotides per cell division depending on the cell type [52]. In contrast, cancer cell genomes often carry thousands of mutations, with different cells within a tumor harboring synonymous and unique mutations. In order to account for the large numbers of mutations in cancer cells, Lawrence Loeb first postulated that cancer cells express a “mutator phenotype”, that is they have an elevated mutation rate compared to nonmalignant cells [53]. The development of a mutator phenotype is hypothesized to be an early step in tumorigenesis, due to the necessity of multiple cooperating oncogenic mutations to initiate and sustain the neoplastic process [54]. An elevated mutation rate would generate mostly random, nearly neutral mutations; however, upon acquisition of an oncogenic, tumor-initiating mutation, selective clonal expansion would ensue capturing previously occurring passenger mutations within the given cell lineage. Throughout tumorigenesis, cancer-driving mutations occur at random and multiple rounds of clonal selection generate a tumor made up of a predominant clone and several subclones. Thus, the mutator phenotype is integral to the clonal evolution of cancer, facilitating the acquisition of cancer hallmarks [55].
So how do normal or precancerous cells acquire a mutator phenotype? The model proposes that random genome-wide DNA damaging events, either from environmental or endogenous sources, produce random mutations. By chance, a mutation within a genome maintenance gene, such as a DNA repair gene, would destabilize the genomic integrity of the affected mutant cell [54]. There are probably hundreds of potential “DNA caretaker” genes that might act as a mutator gene if randomly activated or inactivated, so long as a mutation elevates the mutation rate without significantly reducing cellular fitness. Cancers that require multiple mutation “hits” are predicted to be more dependent on a mutator phenotype as the probability of acquiring the necessary cooperating mutations is significantly increased when the mutation rate is elevated [53]. In support of the mutator phenotype, many familial cancer syndromes are caused by inherited mutations in DNA caretaker genes. It is undisputed that tumor cells of most cancers have accumulated thousands of mutations and even tens of thousands, but it has been argued that the increased proliferation rate and longevity of tumor cells is sufficient to account for the number of mutations present in cancer cells, without postulating a mutator phenotype [56]. Indeed, the time between carcinogen exposure and clinical appearance of a tumor can take more than 20 years. Nevertheless, estimates of mutation rates in cells following the cancer-initiating event are 200-fold higher than the rate in normal cells, suggesting that in the early stages of tumorigenesis, cells express a mutator phenotype [52, 57].
A main criticism of the mutator phenotype is the low frequency of sightings of inactivated or activated mutator genes in sporadic cancers. Many cancer genomes have now been sequenced and each year the number of sequenced cancer genomes steadily increases. These large genomic studies have revealed a puzzling trend: many cancer genomes have thousands even hundreds of thousands of mutations, yet there are relatively few mutations involving known DNA caretaker genes [40]. This has led some to argue that genomic instability in cancers is not caused by a mutator phenotype but rather by oncogene-induced DNA replication stress. However, with the finding that Fhit has a DNA caretaker function through support of DNA replication [7], the FHIT gene is predicted to be a candidate mutator gene for a large fraction of sporadic cancers, as FHIT deletions do occur in the early stages of cancer development and are among the most frequent mutations in cancers, as confirmed by the many large cancer genome sequencing projects.
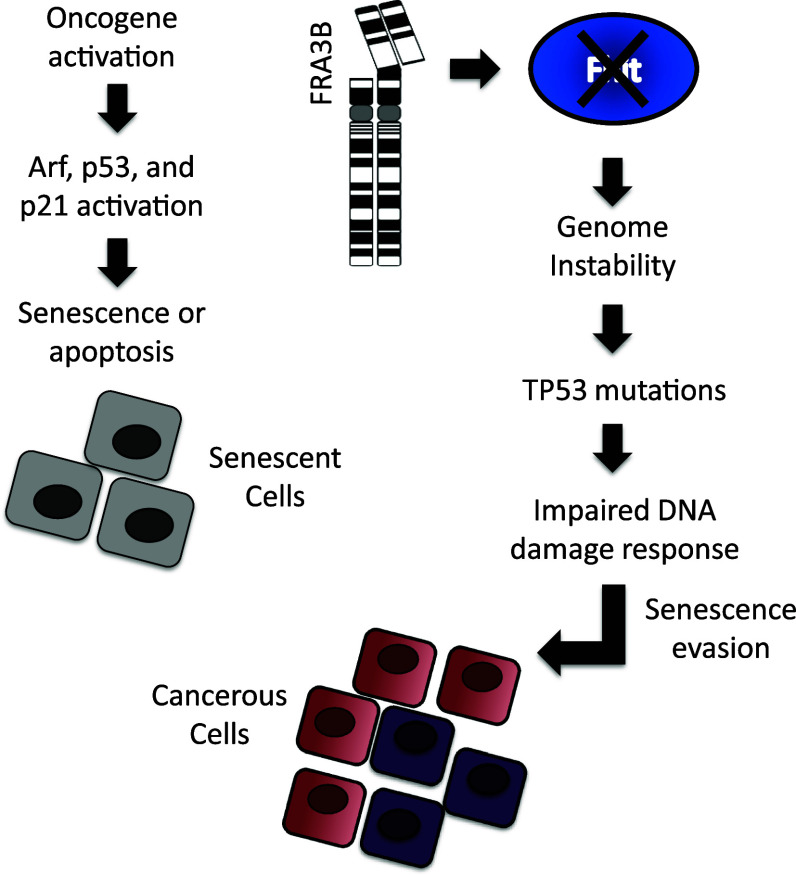
Fhit loss specifically causes chromosome instability, yet cancers also exhibit thousands of base-substitution mutations and small insertions and deletions. Thus, for FHIT to be a good candidate mutator gene, Fhit inactivation must also account for the presence of these point mutations and small insertions and deletions. New evidence now suggests that replication stress may be a source of both point mutations and small insertions and deletions. Studies in yeast, mammalian cells, and cancer cells have begun to reveal that chromosome alterations caused by replication stress can produce a mutator phenotype. Poli et al. [58] investigated DNA replication in different mutant yeast strains under chronic replication stress. Interestingly, they found that yeast mutants that have spontaneous DNA breaks and a chromosome instability phenotype adjusted to chronic replication stress by increasing dNTP pools [58]. Davidson et al. [59] made a similar observation, and further showed that the elevated dNTP pools increased the mutation rate and conferred a mutator phenotype. It is possible that a similar adaptation occurs in Fhit-deficient cells where there is an initial shortage of TTP and replication stress, but then a later expansion of the pools leading to a mutator phenotype. Separate studies in yeast have shown that break-induced replication at collapsed replication forks is error prone, producing clusters of base-substitution mutations and small insertions and deletions [60]. Repair of DSBs by non-homologous end joining can also produce small insertions and deletions [61], and repair by homologous recombination repair of DSBs, though considered to be error free, has now been shown to introduce clustered point mutations and small insertions and deletions [62]. There is also evidence for replication stress-induced mutations in cancer cells from several sequenced cancer genomes. These studies have revealed the presence of non-random, clustered mutations in cancer genomes [63]. The pattern of local hypermutation and mutation clusters suggests that they arose simultaneously, and because they frequently map to chromosome breakpoints it is likely they were generated during repair of a DSB or a collapsed replication fork [64]. Finally, a recent study showed that premalignant colon adenomas exhibited a mutator phenotype, and the authors concluded that replication stress was a major contributor to the elevated mutation frequency [52]. Finally, exome DNAs of Fhit−/− mouse liver tissue and kidney cells show more small insertions and deletions than exome DNAs from wild-type mice, with and without carcinogen treatment [8]. The collective evidence suggests that replication stress can produce a mutator phenotype, in accord with the proposal that FHIT is a strong candidate caretaker gene, loss of which causes a mutator phenotype (Fig. 1).
Fig. 1.
FHIT loss as a mechanism for evasion of oncogene-induced senescence
Fhit loss is a mechanism for bypass of oncogene-induced senescence
In normal and preneoplastic cells, oncogene-induced DNA damage results in cellular senescence and apoptosis, mediated by DNA damage checkpoint pathways. DNA damage checkpoints form a tumorigenesis barrier preventing the development of cancerous lesions from precancerous cells [45]. p53 and ATM proteins are central to the senescence barrier to tumorigenesis, and without mutational inactivation or experimental manipulation of these or other key components within the checkpoint pathway, oncogenes fail to transform cells [44]. Oncogene activation also induces senescence in a DNA damage-independent way through the Arf-p53 pathway [65]. It is unlikely that genomic instability would develop in senescent cells, yet it is still commonly suggested that oncogene-induced genomic alterations facilitate mutational inactivation of checkpoint proteins [42]. This is also unlikely, as evidence indicates that senescence is irreversible. Despite the tumorigenesis barrier that senescence poses in response to oncogene activation, tumorigenic cells do acquire resistance to senescence. In fact, it is the senescence barrier that is believed to be a major selective force for the highly frequent mutations in TP53 and ATM found in cancer cells [46]. Indeed, the oncogenes that cause DNA damage and senescence also transform cells in vitro and promote tumorigenesis in mouse models. So how do we reconcile these apparent contradictions? An important point to make here is that in the in vitro models, oncogenes are activated in a very high number of cells and yet transformation occurs in fewer than 1 in 105 cells. Furthermore, transgenic oncogenes that are ectopically expressed in mice induce clonal rather than systemic tumors [66]. This is in contrast to human cancers, where oncogenes are not ectopically activated but become activated only in tumor cells. These observations confirm that driver mutations only work in the right genetic and environmental context or “soil” to support clonal expansion, underlying the importance of having a degree of genome instability and heterogeneity prior to oncogene activation [66]. This is also in accord with the hypothesis that senescence is an irreversible process, and that pre-existing, non-senescent cells are selected for clonal expansion [67].
The extent of genetic heterogeneity in cells prior to oncogene activation is unknown, but recent findings conclude that heterogeneity is widespread in normal cells, increases with age and is a strong predictor of disease [68–70]. Thus, it is possible that mutations that impair the DNA damage checkpoints may be present in some cells prior to oncogene-induced senescence. Such mutations only have a selective advantage in cells accumulating DNA damage, and thus would not be detectable in the early preneoplastic stages of tumorigenesis until after senescence evasion and clonal expansion. Evidence for this comes from follow-up studies of the DNA caretaker function of Fhit. In primary cells established from Fhit +/+ and Fhit−/− mice, we have discovered that the genome instability caused by Fhit loss can lead to inactivation of the p53 pathway allowing evasion of senescence and resistance to genotoxic agents [8], though it is likely that other pathways to evasion of senescence will be observed when multiple in vitro cell lines are examined in detail. In MEFs, Fhit−/− cells incurred a chromosome alteration resulting in amplification of the Mdm2 gene. This amplification was selected for as cells underwent senescence and resulted in a rapid immortalization of Fhit−/− MEFs. In a separate model system, mouse kidney epithelial cells established from Fhit +/+ and Fhit−/− mice were subcultured for several weeks or treated with a genotoxic drug to induce apoptosis. The Fhit−/− cells were more resistant to genotoxin treatment and resistant clones contained a Trp53 mutation within the DNA-binding domain leading to complete p21 silencing. The importance of these findings is that Fhit loss-induced genome instability produces spontaneous mutations, which randomly affect the p53 pathway, and under selective pressure, p53 checkpoint-resistant clones emerge. These findings do not diminish the importance of oncogenes, as oncogenes drive tumorigenesis. Instead, our work resolves the problem of oncogene-induced senescence. Oncogenes require inactivation of the p53 pathway to transform cells. They contribute to the selective force for TP53 mutations, but evidence is lacking that they can induce TP53 mutations. Fhit loss in mouse cells can provide the “soil” that allows p53 inactivation as a result of genome instability. Thus, Fhit loss and subsequent oncogene activation likely work in concert, with loss of Fhit inducing mutations, and oncogenes inducing the senescence barrier that selects for p53 (or other tumor suppressor pathway) inactivation (Fig. 2).
Fig. 2.
FHIT loss-induced genome instability model for tumorigenic clonal expansion. Deletions in FHIT alleles occur due to FRA3B fragility causing loss of Fhit protein expression. Fhit loss triggers replication stress, followed by stress-induced chromosomal instability. Chromosomal instability facilitates the acquisition of oncogenic mutations followed by selective clonal expansion
Conclusions
Genomic instability is observed in human pre-cancerous lesions, and concurrent loss of Fhit expression has been detected in precancerous lesions. This suggests that Fhit loss is one of the earliest changes to occur in the preneoplastic process. We have concluded that Fhit is a genome “caretaker”, and that loss of this caretaker function initiates the onset of genome instability and cancer development. An important feature of the Fhit loss-induced genomic instability model is that cells acquire replication stress-induced chromosomal alterations without DNA damage response activation, in contrast to observations in oncogene-activated cells, possibly because the replication defects caused by Fhit loss fall below the threshold level needed to fully activate the S and G2 checkpoints. This hypothesis is supported by the fact that aphidicolin induces fragile site expression by slowing or stalling replication forks, yet fragile sites are routinely detected in metaphase chromosomes, which indicates a failure of the S and G2 checkpoints to block mitotic entry despite the presence of damaged loci. Studies have also suggested that eukaryotic cells lack a checkpoint surveillance mechanism to ensure completion of DNA replication before mitotic entry [71]. Thus, it is possible that DNA replication is incomplete in Fhit-deficient cells because of slower fork progression, and as cells pass through mitosis, under-replicated chromosomes either break or fail to properly segregate. In theory, without DNA damage checkpoint activation, Fhit-deficient cells could continue to proliferate for years, and over time accumulate extensive genome alterations. Over these many cell generations, Fhit-deficient cells could generate significant mutational diversity and cell heterogeneity, as is the case with Fhit−/− mice. Indeed, Fhit−/− mice develop normally and live long lives, making Fhit inactivation an ideal target to initiate genome instability without compromising fitness at the cellular and organism levels. Thus, Fhit loss would provide the “soil” for the emergence of preneoplastic clones under selective pressure. This is consistent with the recent finding that clonal somatic chromosome anomalies increase with age in the normal population [68, 69] and is consistent with the fact that cancer risk increases with age. It is also consistent with the enhanced susceptibility of Fhit−/− mice to spontaneous hyperplastic lesions and tumors and highly enhanced susceptibility to carcinogen-induced tumors [27].
The finding that Fhit loss-induced genomic alterations were a consequence of TTP pool depletion provides an intriguing twist to the ongoing narrative of chromosome fragile sites: the fragile FHIT gene product may protect fragile sites. For example, there is a class of chromosome fragile sites, the folate-sensitive fragile sites, which are unstable under conditions that cause thymidylate depletion, including culturing in medium deficient in thymidine or folate. Thymidine supplementation rescues the fragility of these sites [72]. Furthermore, folate deficiency causes chromosome instability in human and mouse blood cells [73, 74]. These findings independently establish that an insufficient supply of TTP can cause chromosome instability at specific loci, and that the salvage pathway, via TK1 activity, is a required source of TTP to support DNA synthesis. Interestingly, folate is an important nutrient that serves as a cofactor for TTP synthesis. Studies have shown that folate deficiency correlates with several types of cancer, linking TTP availability and tumorigenesis. This is consistent with our findings that loss of Fhit decreases TTP availability and promotes tumorigenesis [75]. Because Fhit-deficient cells have increased fragility at CFSs and Fhit silencing induced several previously unidentified fragile loci, this may be evidence for a novel class of chromosome fragile sites, the so-called Fhit-sensitive fragile sites that are induced, ironically, by a loss of the fragile FHIT gene product.
References
- 1.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 2.Yunis JJ, Soreng AL. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]
- 3.Letessier A, Millot GA, Koundrioukoff S, Lachagès AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 4.Hosseini SA, Horton S, Saldivar JC, Miuma S, Stampfer MR, Heerema NA, Huebner K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Gene Chromosome Cancer. 2013;52(11):1017–1029. doi: 10.1002/gcc.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Tallec B, Millot GA, Blin ME, Brison O, Dutrillaux B, Debatisse M. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013;4(3):420–428. doi: 10.1016/j.celrep.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Saldivar JC, Shibata H, Huebner K. Pathology and biology associated with the fragile FHIT gene and gene product. J Cell Biochem. 2010;109:858–865. doi: 10.1002/jcb.22481. [DOI] [PubMed] [Google Scholar]
- 7.Saldivar JC, Miuma S, Bene J, Hosseini SA, Shibata H, Sun J, Wheeler LJ, Mathews CK, Huebner K. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet. 2012;8:e1003077. doi: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miuma S, Saldivar JC, Karras JR, Waters CE, Paisie CA, Wang Y, Jin V, Sun J, Druck T, Zhang J, Huebner K. Fhit deficiency-induced global genome instability promotes mutation and clonal expansion. PLoS ONE. 2013;8(11):e80730. doi: 10.1371/journal.pone.0080730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holbach LM, von Moller A, Decker C, Jünemann AG, Rummelt-Hofmann C, Ballhausen WG. Loss of fragile histidine triad (FHIT) expression and microsatellite instability in periocular sebaceous gland carcinoma in patients with Muir-Torre syndrome. Am J Ophthalmol. 2002;134(1):147–148. doi: 10.1016/S0002-9394(02)01434-4. [DOI] [PubMed] [Google Scholar]
- 10.Goldberg M, Rummelt C, Foja S, Holbach LM, Ballhausen WG. Different genetic pathways in the development of periocular sebaceous gland carcinomas in presumptive Muir-Torre syndrome patients. Hum Mutat. 2006;27(2):155–162. doi: 10.1002/humu.20281. [DOI] [PubMed] [Google Scholar]
- 11.Huebner K, Garrison PN, Barnes LD, Croce CM. The role of the FHIT/FRA3B locus in cancer. Annu Rev Genet. 1998;32:7–31. doi: 10.1146/annurev.genet.32.1.7. [DOI] [PubMed] [Google Scholar]
- 12.Durkin SG, Ragland RL, Arlt MF, Mulle JG, Warren ST, Glover TW. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc Natl Acad Sci USA. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guler G, Uner A, Guler N, Han SY, Iliopoulos D, McCue P, Huebner K. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55:471–478. doi: 10.1111/j.1440-1827.2005.01855.x. [DOI] [PubMed] [Google Scholar]
- 14.Michael D, Beer DG, Wilke CW, Miller DE, Glover TW. Frequent deletions of FHIT and FRA3B in Barrett’s metaplasia and esophageal adenocarcinomas. Oncogene. 1997;15:1653–1659. doi: 10.1038/sj.onc.1201330. [DOI] [PubMed] [Google Scholar]
- 15.Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti M, et al. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58:5032–5037. [PubMed] [Google Scholar]
- 16.Mao L, Lee JS, Kurie JM, Fan YH, Lippman SM, Lee JJ, Ro JY, Broxson A, Yu R, Morice RC, et al. Clonal genetic alterations in the lungs of current and former smokers. J Natl Cancer Inst. 1997;89:857–862. doi: 10.1093/jnci/89.12.857. [DOI] [PubMed] [Google Scholar]
- 17.Ahmadian M, Wistuba II, Fong KM, Behrens C, Kodagoda DR, Saboorian MH, Shay J, Tomlinson GE, Blum J, Minna JD, et al. Analysis of the FHIT gene and FRA3B region in sporadic breast cancer, preneoplastic lesions, and familial breast cancer probands. Cancer Res. 1997;57:3664–3668. [PubMed] [Google Scholar]
- 18.Kitamura A, Yashima K, Okamoto E, Andachi H, Hosoda A, Kishimoto Y, Shiota G, Ito H, Kaibara N, Kawasaki H. Reduced Fhit expression occurs in the early stage of esophageal tumorigenesis: no correlation with p53 expression and apoptosis. Oncology. 2001;61:205–211. doi: 10.1159/000055376. [DOI] [PubMed] [Google Scholar]
- 19.Mori M, Mimori K, Shiraishi T, Alder H, Inoue H, Tanaka Y, Sugimachi K, Huebner K, Croce CM. Altered expression of Fhit in carcinoma and precarcinomatous lesions of the esophagus. Cancer Res. 2000;60:1177–1182. [PubMed] [Google Scholar]
- 20.Birrer MJ, Hendricks D, Farley J, Sundborg MJ, Bonome T, Walts MJ, Geradts J. Abnormal Fhit expression in malignant and premalignant lesions of the cervix. Cancer Res. 1999;59:5270–5274. [PubMed] [Google Scholar]
- 21.Butler D, Collins C, Mabruk M, Leader MB, Kay EW. Loss of Fhit expression as a potential marker of malignant progression in preinvasive squamous cervical cancer. Gynecol Oncol. 2002;86:144–149. doi: 10.1006/gyno.2002.6712. [DOI] [PubMed] [Google Scholar]
- 22.Yuge T, Nibu K, Kondo K, Shibahara J, Tayama N, Sugasawa M. Loss of FHIT expression in squamous cell carcinoma and premalignant lesions of the larynx. Ann Otol Rhinol Laryngol. 2005;114:127–131. doi: 10.1177/000348940511400208. [DOI] [PubMed] [Google Scholar]
- 23.Luan X, Ramesh KH, Cannizzaro LA. FHIT gene transcript alterations occur frequently in myeloproliferative and myelodysplastic diseases. Cytogenet Cell Genet. 1998;81:183–188. doi: 10.1159/000015025. [DOI] [PubMed] [Google Scholar]
- 24.Ozkara SK, Corakçi A. FHIT expression in neoplastic, hyperplastic, and normal endometrium. Int J Gynecol Cancer. 2005;15:1081–1088. doi: 10.1111/j.1525-1438.2005.00265.x. [DOI] [PubMed] [Google Scholar]
- 25.Velickovic M, Delahunt B, Grebe SK. Loss of heterozygosity at 3p14.2 in clear cell renal cell carcinoma is an early event and is highly localized to the FHIT gene locus. Cancer Res. 1999;59:1323–1326. [PubMed] [Google Scholar]
- 26.Fong LY, Fidanza V, Zanesi N, Lock LF, Siracusa LD, Mancini R, Siprashvili Z, Ottey M, Martin SE, Druck T, et al. Muir-Torre-like syndrome in Fhit-deficient mice. Proc Natl Acad Sci USA. 2000;97:4742–4747. doi: 10.1073/pnas.080063497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rüdiger T, McCue PA, Croce CM, Huebner K. The tumor spectrum in FHIT-deficient mice. Proc Natl Acad Sci USA. 2001;98:10250–10255. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huebner K, Croce CM. FRA3B and other common fragile sites: the weakest links. Nat Rev Cancer. 2001;1:214–221. doi: 10.1038/35106058. [DOI] [PubMed] [Google Scholar]
- 29.Okumura H, Ishii H, Pichiorri F, Croce CM, Mori M, Huebner K. Fragile gene product, Fhit, in oxidative and replicative stress responses. Cancer Sci. 2009;100:1145–1150. doi: 10.1111/j.1349-7006.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichiorri F, Okumura H, Nakamura T, Garrison PN, Gasparini P, Suh SS, Druck T, McCorkell KA, Barnes LD, Croce CM, et al. Correlation of fragile histidine triad (Fhit) protein structural features with effector interactions and biological functions. J Biol Chem. 2009;284:1040–1049. doi: 10.1074/jbc.M806638200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapasso F, Pichiorri F, Gaspari M, Palumbo T, Aqeilan RI, Gaudio E, Okumura H, Iuliano R, Di Leva G, Fabbri M, et al. Fhit interaction with ferredoxin reductase triggers generation of reactive oxygen species and apoptosis of cancer cells. J Biol Chem. 2008;283:13736–13744. doi: 10.1074/jbc.M709062200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Rimessi A, Marchi S, Fotino C, Romagnoli A, Huebner K, Croce CM, Pinton P, Rizzuto R. Intramitochondrial calcium regulation by the FHIT gene product sensitizes to apoptosis. Proc Natl Acad Sci USA. 2009;106:12753–12758. doi: 10.1073/pnas.0906484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jayachandran G, Sazaki J, Nishizaki M, Xu K, Girard L, Minna JD, Roth JA, Ji L. Fragile histidine triad-mediated tumor suppression of lung cancer by targeting multiple components of the Ras/Rho GTPase molecular switch. Cancer Res. 2007;67:10379–10388. doi: 10.1158/0008-5472.CAN-07-0677. [DOI] [PubMed] [Google Scholar]
- 34.Joannes A, Bonnomet A, Bindels S, Polette M, Gilles C, Burlet H, Cutrona J, Zahm JM, Birembaut P, Nawrocki-Raby B. Fhit regulates invasion of lung tumor cells. Oncogene. 2010;29:1203–1213. doi: 10.1038/onc.2009.418. [DOI] [PubMed] [Google Scholar]
- 35.Joannes A, Grelet S, Duca L, Gilles C, Kileztky C, Dalstein V, Birembaut P, Polette M, Nawrocki-Raby B. Fhit regulates EMT targets through an EGFR/Src/ERK/Slug signaling axis in human bronchial cells. Mol Cancer Res. 2014 doi: 10.1158/1541-7786.MCR-13-0386-T. [DOI] [PubMed] [Google Scholar]
- 36.Gaudio E, Paduano F, Spizzo R, Ngankeu A, Zanesi N, Gaspari M, Ortuso F, Lovat F, Rock J, Hill GA, Kaou M, Cuda G, Aqeilan RI, Alcaro S, Croce CM, Trapasso F. Fhit delocalizes annexin a4 from plasma membrane to cytosol and sensitizes lung cancer cells to paclitaxel. PLoS ONE. 2013;8(11):e78610. doi: 10.1371/journal.pone.0078610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: clinical significance and future implications. Exp Biol Med. 2014;239(3):253–263. doi: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- 38.Le Beau MM, Drabkin H, Glover TW, Gemmill R, Rassool FV, McKeithan TW, Smith DI. An FHIT tumor suppressor gene? Genes Chromosome Cancer. 1998;21:281–289. doi: 10.1002/(SICI)1098-2264(199804)21:4<281::AID-GCC1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 39.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability–an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–228. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- 41.Zhu Y, McAvoy S, Kuhn R, Smith DI. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25:2901–2908. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- 42.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 43.Aguilera A, Gómez-González B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 44.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 45.Bartkova J, Horejsí Z, Koed K, Krämer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 46.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 47.Austin WR, Armijo AL, Campbell DO, Singh AS, Hsieh T, Nathanson D, Herschman HR, Phelps ME, Witte ON, Czernin J, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209:2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dobrovolsky VN, McGarrity LJ, VonTungeln LS, Mittelstaedt RA, Morris SM, Beland FA, Heflich RH. Micronucleated erythrocyte frequency in control and azidothymidine-treated Tk +/+, Tk ± and Tk−/− mice. Mutat Res. 2005;570:227–235. doi: 10.1016/j.mrfmmm.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Holmberg C, Fleck O, Hansen HA, Liu C, Slaaby R, Carr AM, Nielsen O. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev. 2005;19:853–862. doi: 10.1101/gad.329905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aird KM, Zhang R (2014) Nucleotide metabolism, oncogene-induced senescence and cancer, Cancer Letters (In press) [DOI] [PMC free article] [PubMed]
- 51.Aird KM, Zhang G, Li H, Tu Z, Bitler BG, Garipov A, Wu H, Wei Z, Wagner SN, Herlyn M, Zhang R. Suppression of nucleotide metabolism underlies the establishment and maintenance of oncogene-induced senescence. Cell Rep. 2013;3(4):1252–1265. doi: 10.1016/j.celrep.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolaev SI, Sotiriou SK, Pateras IS, Santoni F, Sougioultzis S, Edgren H, Almusa H, Robyr D, Guipponi M, Saarela J, et al. A single-nucleotide substitution mutator phenotype revealed by exome sequencing of human colon adenomas. Cancer Res. 2012;72:6279–6289. doi: 10.1158/0008-5472.CAN-12-3869. [DOI] [PubMed] [Google Scholar]
- 53.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11:450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loeb LA. Mutator phenotype in cancer: origin and consequences. Semin Cancer Biol. 2010;20:279–280. doi: 10.1016/j.semcancer.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bodmer W, Bielas JH, Beckman RA. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–3560. doi: 10.1158/0008-5472.CAN-07-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Poli J, Tsaponina O, Crabbé L, Keszthelyi A, Pantesco V, Chabes A, Lengronne A, Pasero P. dNTP pools determine fork progression and origin usage under replication stress. EMBO J. 2012;31:883–894. doi: 10.1038/emboj.2011.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson MB, Katou Y, Keszthelyi A, Sing TL, Xia T, Ou J, Vaisica JA, Thevakumaran N, Marjavaara L, Myers CL, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31:895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iraqui I, Chekkal Y, Jmari N, Pietrobon V, Fréon K, Costes A, Lambert SA. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 2012;8:e1002976. doi: 10.1371/journal.pgen.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, Lee SE. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012;8:e1003026. doi: 10.1371/journal.pgen.1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deem A, Keszthelyi A, Blackgrove T, Vayl A, Coffey B, Mathur R, Chabes A, Malkova A. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 66.Nicholson JM, Duesberg P. On the karyotypic origin and evolution of cancer cells. Cancer Genet Cytogenet. 2009;194:96–110. doi: 10.1016/j.cancergencyto.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 67.Sarkisian CJ, Keister BA, Stairs DB, Boxer RB, Moody SE, Chodosh LA. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9:493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 68.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young MA, Larson DE, Sun CW, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, et al. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, Pasero P, Lisby M, Haber JE, Aragón L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science. 2007;315:1411–1415. doi: 10.1126/science.1134025. [DOI] [PubMed] [Google Scholar]
- 72.Sutherland GR. Heritable fragile sites on human chromosomes I. Factors affecting expression in lymphocyte culture. Am J Hum Genet. 1979;31:125–135. [PMC free article] [PubMed] [Google Scholar]
- 73.Blount BC, Ames BN. DNA damage in folate deficiency. Baillieres Clin Haematol. 1995;8:461–478. doi: 10.1016/S0950-3536(05)80216-1. [DOI] [PubMed] [Google Scholar]
- 74.MacGregor JT, Schlegel R, Wehr CM, Alperin P, Ames BN. Cytogenetic damage induced by folate deficiency in mice is enhanced by caffeine. Proc Natl Acad Sci USA. 1990;87:9962–9965. doi: 10.1073/pnas.87.24.9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rampersaud GC, Bailey LB, Kauwell GP. Relationship of folate to colorectal and cervical cancer: review and recommendations for practitioners. J Am Diet Assoc. 2002;102:1273–1282. doi: 10.1016/S0002-8223(02)90281-6. [DOI] [PubMed] [Google Scholar]