Abstract
Background
The human gastric pathogen Helicobacter pylori is a paradigm for chronic bacterial infections. Its persistence in the stomach mucosa is facilitated by several mechanisms of immune evasion and immune modulation, but also by an unusual genetic variability which might account for the capability to adapt to changing environmental conditions during long-term colonization. This variability is reflected by the fact that almost each infected individual is colonized by a genetically unique strain. Strain-specific genes are dispersed throughout the genome, but clusters of genes organized as genomic islands may also collectively be present or absent.
Results
We have comparatively analysed such clusters, which are commonly termed plasticity zones, in a high number of H. pylori strains of varying geographical origin. We show that these regions contain fixed gene sets, rather than being true regions of genome plasticity, but two different types and several subtypes with partly diverging gene content can be distinguished. Their genetic diversity is incongruent with variations in the rest of the genome, suggesting that they are subject to horizontal gene transfer within H. pylori populations. We identified 40 distinct integration sites in 45 genome sequences, with a conserved heptanucleotide motif that seems to be the minimal requirement for integration.
Conclusions
The significant number of possible integration sites, together with the requirement for a short conserved integration motif and the high level of gene conservation, indicates that these elements are best described as integrating conjugative elements (ICEs) with an intermediate integration site specificity.
Keywords: Plasticity zone, Helicobacter pylori, Integrating conjugative element, Type IV secretion system, Horizontal gene transfer
Background
Infections with the human gastric pathogen H. pylori are paradigmatic examples of chronic, or persistent, bacterial infections in the face of a constant immune response [1]. H. pylori infections are usually contracted during early childhood and persist for the lifetime of the host, but most infected individuals develop only mild gastric inflammation without overt symptoms. Nevertheless, a substantial fraction of infected persons develops more severe consequences, making H. pylori the principal cause of (symptomatic) chronic active gastritis and peptic ulcer disease, and a major risk factor for development of gastric adenocarcinoma and mucosa-associated lymphoid tissue (MALT) lymphoma [2,3]. For survival and persistent growth in the presence of a constant immune response and in an environment which is changing considerably over decades of infection, permanent adaptation of the bacteria is thought to be required [4]. Such adaptive processes may include regulatory mechanisms acting on gene expression, but also reversible or irreversible genome changes. For instance, it has been shown that strains isolated from patients with atrophic gastritis [5] or marginal zone B-cell MALT lymphoma [6] have reduced genomes in comparison to gastritis or ulcer strains, and a strain isolated from a gastric cancer patient had lost further genes in comparison to a strain isolated previously from the same patient during atrophic gastritis [7]. That genome plasticity plays a role in bacterial persistence is further supported by the observation that natural transformation competence, which is upregulated upon DNA stress [8], promotes persistent colonization in mice [9].
Allelic diversity caused by high mutation rates and frequent recombination events is a striking property of H. pylori strains. Genetic fingerprints of individual strains obtained by multilocus sequence typing of housekeeping genes have indicated that clonal transmission is likely to occur, but is followed by a rapid adaptation to the new host, so that H. pylori isolates from different subjects are almost always unique [4]. On the other hand, while recombination events generating allelic diversity are frequent, genome changes involving gain or loss of genes seem to be rare [10]. Nevertheless, on the level of gene content, evidence has been presented that H. pylori is a species with an open pan-genome, in which each individual isolate contains a distinct set of non-core, or strain-specific, genes [6,11-13]. Comparative analysis of the first sequenced H. pylori genomes suggested that these strain-specific genes are often located in genomic regions that had previously been termed plasticity zones or plasticity regions, a designation originally used to describe a particular genetic locus with high variation between the first two H. pylori genome sequences [14]. However, with the availability of more sequencing data and more complete H. pylori genome sequences, it became clear that parts of the plasticity regions are usually organized as genomic islands that may be integrated in one of several different genetic loci. Furthermore, they generally contain complete sets of genes required to produce type IV secretion machineries, as well as genes encoding different DNA-processing proteins [11,15,16], suggesting that they are actually mobile genetic elements capable of horizontal gene transfer between bacterial cells, and that they might be best described as conjugative transposons or integrating conjugative elements (ICEs).
The actual plasticity of these islands partly derives from the fact that gene rearrangements, insertions or deletions may have occurred within them, but it is not clear whether they also carry variable passenger genes. Interestingly, intrahost variation among genes of the plasticity zones, including deletions in a type IV secretion system gene, has been found for sequential isolates obtained from a duodenal ulcer patient over a course of 10 years [17]. Although several candidate genes of these plasticity regions have been suggested as disease markers, e.g. dupA for duodenal ulcer [18,19], or jhp950 for marginal zone B-cell MALT lymphoma [20], the functions of the plasticity zones are currently not well-understood.
To address the question of plasticity zone prevalence, and of their genetic diversity, we have performed a comparative analysis of these genome islands from a larger number of H. pylori genome sequences, including newly determined genome sequences of nine additional strains from different backgrounds. We show that these elements have a high prevalence throughout all populations, and that gene evolution within the elements is not congruent with the rest of the genomes. The wide variety of integration loci together with a conserved sequence motif at each integration site suggests an integration mechanism that depends on a short recognition motif in the DNA sequence only.
Results
Prevalence of plasticity regions in the H. pylori population
We have reported previously that H. pylori strain P12 contains three genome regions with similarity to the prototypical plasticity zones, but only one of them (PZ2) corresponds to the originally described locus, whereas the other two regions (PZ1 and PZ3) have a genetic organization typical for genome islands and contain genes for type IV secretion systems that might make them capable of self-transfer [11]. In comparison, the original two genome sequences (strains 26695 and J99) contain only truncated and highly rearranged portions of these genome islands (Additional file 1: Figure S1). As reported previously, the most conserved type IV secretion system genes fall into one of two distinct groups, which have been termed either tfs3 and tfs3a/b[16], or tfs3 and tfs4[11]. In accordance with Ref. [11], where conserved tfs3 genes have been shown not to be more closely related to tfs4 genes than to the respective comB genes encoding the type IV secretion system used for natural transformation, we consider tfs3 and tfs4 here as independent systems. Moreover, since there is evidence for horizontal gene transfer of the corresponding islands [11,16], but not for transposition within a strain, we propose to use the term integrating conjugative elements (ICE) and refer to individual islands as ICEHptfs3 or ICEHptfs4, respectively. A comparison of different designations of the islands and associated type IV secretion systems is given in Table 1. To determine the occurrence of ICEHptfs3 and ICEHptfs4 elements in the H. pylori population and the degree of variation among them, we performed a comparative sequence analysis of these elements from 36 completely sequenced H. pylori genomes available in public databases (Table 2).
Table 1.
Comparison of plasticity zone mobile genetic element and associated type IV secretion system (T4SS) designations
| Element designation used in this study | T4SS designation used in this study | Element designation used in [[16]] | T4SS designation used in [[16] | Element designation used in [[11] |
|---|---|---|---|---|
| ICEHptfs3 |
TFS3 |
TnPZ type 2 |
TFS3 |
PZ3 |
| ICEHptfs4a |
TFS4a |
TnPZ type 1b |
TFS3b |
PZ1 |
| ICEHptfs4b |
TFS4b |
TnPZ type 1 |
TFS3a |
n.a. |
| ICEHptfs4c | TFS4c | n.a. | n.a. | n.a. |
n.a., not applicable.
Table 2.
Properties of ICE elements in strains with complete genome sequences
| Strain | ICE type | Integration site (P12) | Pos. LJ | Pos. RJ | Size (kb) 14 | Complete T4SS? |
|---|---|---|---|---|---|---|
|
52 |
none |
|
|
|
|
|
|
B38 |
none2 |
|
|
|
|
|
|
F16 |
none |
|
|
|
|
|
|
HPAG1 |
none |
|
|
|
|
|
|
Sat464 |
none |
|
|
|
|
|
|
v225d |
none |
|
|
|
|
|
|
26695 |
ICEHptfs33 |
hpp12_981 |
1049829 |
473989 |
(16.0) |
N |
|
26695 |
ICEHptfs4a/4b3 |
hpp12_13285 |
1071598 |
464996 |
(18.3) |
N |
|
35A |
ICEHptfs4a |
hpp12_92-91 |
359215 |
309788 |
(10.0)15 |
N |
|
51 |
ICEHptfs3/4a3 |
hpp12_999 |
none |
1034232 |
(32.2) |
N |
|
83 |
ICEHptfs3 |
hpp12_65 |
7908512 |
106931 |
(27.8) |
N |
|
83 |
ICEHptfs4b |
hpp12_1495 |
1522267 |
150317212 |
(19.1) |
N |
|
908, 2017, 2018
1
|
ICEHptfs4a/b4 |
hpp12_995-9796 |
99180112 |
none |
(14.6) |
N |
|
B8 |
ICEHptfs3 |
hpp12_439-438 |
487322 |
526844 |
39.516 |
Y |
|
B8 |
ICEHptfs4a |
hpp12_1380-5S-rRNA5,7 |
52870812 |
452245 |
(37.0) |
N |
|
Cuz20 |
ICEHptfs4b |
hpp12_210-211 |
266516 |
227821 |
38.5 |
Y |
|
ELS37 |
ICEHptfs3 |
hpp12_511-5128 |
884907 |
838572 |
46.3 |
Y |
|
ELS37 |
ICEHptfs4b |
hpp12_511-5128 |
838326 |
none |
(2.0) |
N |
|
F30 |
ICEHptfs4a |
hpp12_92-91 |
1239533 |
1287710 |
(10.0)15 |
N |
|
F32 |
ICEHptfs3 |
hpp12_312-313 |
328469 |
1058181 |
(4.1 + 25.5)17 |
N |
|
F57 |
ICEHptfs4a |
hpp12_92-91 |
152065 |
103732 |
(10.0)15 |
N |
|
F57 |
ICEHptfs4b |
hpp12_259 |
323634 |
284294 |
39.3 |
Y |
|
G27 |
ICEHptfs4b |
hpp12_1009-1010 |
1085072 |
1045702 |
39.4 |
Y |
|
Gambia 94/24 |
ICEHptfs3 |
hpp12_15085 |
1473904 |
1521243 |
47.3 |
Y |
|
Gambia 94/24 |
ICEHptfs4a/b4 |
hpp12_994-5S-rRNA |
106932212 |
none |
(35.2) |
N |
|
HUP-B14 |
ICEHptfs3 |
hpp12_1365 |
135565613 |
135565613 |
(10.8) |
N |
|
India 7 |
ICEHptfs3 |
hpp12_599 |
752074 |
798006 |
45.9 |
Y |
|
India 7 |
ICEHptfs4a |
hpp12_1391-152810 |
none |
none |
(7.3) |
N |
|
J99 |
ICEHptfs33 |
hpp12_444-445 |
104487813 |
104487813 |
(16.7) |
N |
|
J99 |
ICEHptfs4a/b4 |
hpp12_994-5S-rRNA |
none |
none |
(25.3) |
N |
|
Lithuania 75 |
ICEHptfs3 |
hpp12_1508 |
1516637 |
none |
(34.8) |
N |
|
Lithuania 75 |
ICEHptfs4c |
n.a. (plasmid integration) |
352813 |
352813 |
(10.1) |
N |
|
P12 |
ICEHptfs3 |
hp1354 |
1424780 |
1394778 |
(30.0) |
N |
|
P12 |
ICEHptfs4a |
hp0464 |
452023 |
492769 |
40.7 |
Y |
|
PeCan4 |
ICEHptfs3 |
hpp12_1528-15235 |
1530039 |
1536824 |
(6.8) |
N |
|
PeCan4 |
ICEHptfs4a |
hpp12_1528-15235, 11 |
1578142 |
1537082 |
41.1 |
Y |
|
PeCan18 |
ICEHptfs3 |
hpp12_440-439 |
1015120 |
1064481 |
49.416 |
Y |
|
PeCan18 |
ICEHptfs4a |
hpp12_994-5S-rRNA |
1067535 |
none |
(3.1) |
N |
|
Puno120 |
ICEHptfs3 |
hpp12_994-5S-rRNA |
1004976 |
none |
(6.8 + 26.6)18 |
N |
|
Puno135 |
ICEHptfs3/4b3 |
hpp12_994-5S-rRNA |
1014870 |
1059997 |
(45.1) |
Y |
|
Shi112 |
ICEHptfs3 |
hpp12_226-225 |
281418 |
232869 |
48.616 |
Y |
|
Shi112 |
ICEHptfs4b |
hpp12_1380-5S-rRNA |
1412827 |
1451480 |
38.7 |
Y |
|
Shi169 |
ICEHptfs4b |
hpp12_211-210 |
240310 |
201136 |
39.2 |
Y |
|
Shi417 |
ICEHptfs3 |
hpp12_1510 |
1546576 |
1591512 |
(44.9) |
Y |
|
Shi417 |
ICEHptfs4b |
hpp12_1126-1125 |
1186887 |
1147709 |
39.2 |
Y |
|
Shi470 |
ICEHptfs4b |
hpp12_495 |
874710 |
913872 |
39.2 |
Y |
|
SJM180 |
ICEHptfs3 |
hpp12_454-453 |
1413941 |
none |
(23.2) |
N |
|
SJM180 |
ICEHptfs4a |
hpp12_1364-13659 |
1371932 |
141618012 |
(24.1) |
N |
|
SNT49 |
ICEHptfs4b |
hpp12_65 |
61216 |
100646 |
39.4 |
Y |
|
South Africa 7 |
ICEHptfs4c |
hpp12_1366 |
1568674 |
1527381 |
41.316 |
Y |
|
South Africa 7 |
ICEHptfs4b |
hpp12_943-944 |
934499 |
973788 |
39.3 |
Y |
|
XZ274 |
ICEHptfs4a |
hpp12_92-91 |
162178 |
111739 |
(10.0)15 |
N |
| XZ274 | ICEHptfs4b | hpp12_776 | 653446 | 612019 | (41.4)16 | Y |
1Strains 908, 2017 and 2018 are sequential isolates from a single patient [17] and do not show major differences in their ICEHptfs4 sequences. However, note that GenBank entries EF195724.1, EF195725.1 and EF195726.1 describe ICEHptfs3 clusters in these strains [17] that are not present in the genome sequences.
2HELPY0971 is possibly a vestige of hpp12_1321/pz7.
3resulting from insertions of 2-3 genomic islands and subsequent rearrangements.
4containing ICEHPtfs4a-type genes close to the left junction, and ICEHptfs4b-type genes close to the right junction.
5associated with genome rearrangement in comparison to strain P12.
6associated with deletion of hpp12_980 to hpp12_995 (5’) including one copy of 5S-23S-rRNA.
7associated with a recombination between the two 5S-23S-rRNA loci (including hpp12_1381-1384).
8partial duplication of both genes; ICEHptfs3 inserted into truncated ICEHptfs4b.
9within a restriction-modification system inserted into this region.
10integrated together with a 0.9 kb fragment of ICEHptfs3 and a putative toxin-antitoxin system.
11integration of ICEHptfs4a into remnant of ICEHptfs4b, which is in turn integrated into truncated ICEHptfs3.
12irregular integration, using internal AAGAATG motif.
13left and right junctions coincide due to irregular integration.
14numbers in parentheses indicate incomplete ICE elements.
15disrupted by a chromosomal inversion from hpp12_92 to hpp12_128.
16size of ICE increased by IS element insertion.
17interrupted by a chromosomal rearrangement between hpp12_312 and hpp12_1044 (including babC deletion).
18original integration probably in hpp12_994-5S-rRNA locus; from there, relocation of 26.6 kb fragment via internal AAGAATG motifs into hpp12_1510; 1.4 kb duplication (containing xerT) in both loci.
We found that only 6 out of these 36 strains do not contain ICEHptfs3 or ICEHptfs4 islands or fragments thereof (Table 2). Among the remaining 30 strains, 19 harbour ICEHptfs3 islands, 6 of which seem to have complete gene sets, and 27 harbour ICEHptfs4 islands, 12 of which are complete. There are 3 strains with two different ICEHptfs4 elements, and 16 strains which have at least parts of both ICEHptfs3 and ICEHptfs4. Three strains (strains 51, SJM180 and Puno135) contain hybrid arrangements of ICEHptfs3 and ICEHptfs4 islands, but these seem to result from DNA rearrangements after integration of two independent genome islands (see below). Thus, each complete or truncated island can be assigned to either the ICEHptfs3 or the ICEHptfs4 type. Within the ICEHptfs3 group, two distinct variants can be discriminated, which differ by the presence (e.g., strain PeCan18) or absence (e.g., strain B8) of the pz21-pz23 genes (Figure 1A). In contrast, three variants of ICEHptfs4, defined by orthologous, but variant sets of genes at both ends of the genome islands, or in their central regions, can be distinguished and are termed here ICEHptfs4a, ICEHptfs4b and ICEHptfs4c, respectively (Figure 1B; Table 1). The third subtype, ICEHptfs4c, was only found in strain SouthAfrica7, which belongs to the hpAfrica2 population (see below), and as a plasmid-borne fragment in strain Lithuania75. Both types of genome island seem to vary considerably in size between strains (Table 2), but this is often due to small deletions within the islands or to insertion of IS elements; therefore, complete ICEHptfs3 islands have “standard” sizes of about 37.5, or 46 kb, depending on the presence of pz21-23 orthologs, while complete ICEHptfs4a, ICEHptfs4b and ICEHptfs4c usually comprise about 41, 39.5, and 39.5 kb, respectively (Figure 1A, B).
Figure 1.
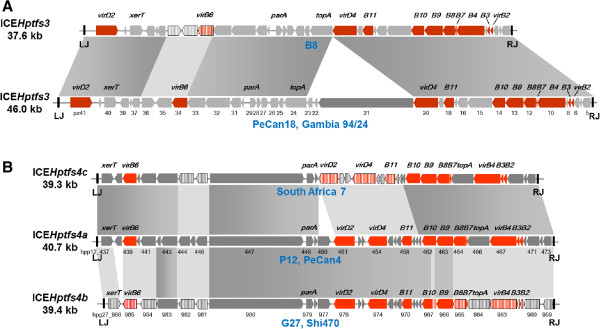
Gene arrangement of prototypical ICEHptfs3 (A) and ICEHptfs4 (B) islands. Genes encoding type IV secretion system components are drawn as red arrows, and other genes as grey arrows. Regions with high nucleotide sequence similarity are connected by dark grey bars, and regions with low to intermediate levels of similarity by light grey bars. Hatched arrows indicate orthologous, but clearly distinct gene variants. Typical sizes of the corresponding elements are indicated on the left. ICEHptfs3 elements differ by the presence or absence of pz21-pz23 genes (according to the nomenclature of [15]) and by several distinct variants of the pz34, pz35, and/or pz36 genes. However, variations within these two regions do not correlate with each other and were thus not considered for ICEHptfs3 subclassification. In contrast, ICEHptfs4 islands are further subclassified into ICEHptfs4a, ICEHptfs4b and ICEHptfs4c groups according to the presence of orthologous gene variants. Note that the polymorphic genes hpp12_446/hpg27_981 and hpp12_444-445/hpg27_982 could not clearly be assigned to ICEHptfs4a or ICEHptfs4b and were thus not considered for classification of ICEHptfs4 subtypes. LJ, left junction; RJ, right junction.
Geographic distribution of ICEHptfs3 and ICEHptfs4 islands
It is well-established that H. pylori strains cluster into distinct populations according to their geographic origin when multilocus sequence typing using partial sequences of seven housekeeping genes is employed [21-23]. In contrast to this allelic variability, which suggests a common evolution of H. pylori and humans, consistent gene content profiles of individual populations could not be found, with the exception of one hypothetical gene (jhp914) present only in strains from the hpAfrica1 population [24]. Interestingly, comparison of gene content microarray data [24] with ICEHptfs4 composition suggests that most hpAfrica1 strains contain ICEHptfs4a genes close to the left junctions and in the mid region (jhp947-jhp951; hp1000-hp1006; Additional file 1: Figure S1), but ICEHptfs4b genes close to the right junctions (jhp917-jhp924; Additional file 1: Figure S1), while hpEurope strains variably contain these genes. Since there are only three hpAfrica1 strains among the 36 complete genome sequences analysed (strains 908, 2017 and 2018 were isolated from the same patient and are very similar), we decided to determine draft genome sequences of three further strains originating from Western Africa, as well as of six strains isolated in Europe, five of which had been tested positive for the presence of an ICEHptfs4a-type or an ICEHptfs4b-type virB4 gene (data not shown). Sequence analysis revealed that all strains except one (196A) contain at least 37 kb of ICEHptfs3 and/or ICEHptfs4 sequences (Table 3).
Table 3.
Properties of ICE elements identified in draft genome sequences
| Strain | Population 1 | ICE type | Integration site (P12) | Motif | Pos. LJ 4 | Size (kb) 5 | Complete T4SS (Y/N) |
|---|---|---|---|---|---|---|---|
|
196A |
hpEurope |
none |
|
|
|
n.a. |
|
|
166 |
hpEurope |
ICEHptfs4c |
hpp12_1518-1519 |
AAAGAATG |
1613471 |
39.6 |
Y |
|
175 |
hpEurope |
ICEHptfs3 |
hpp12_13663 |
TAAGAATG |
1440427 |
(10.8) |
N |
|
175 |
hpEurope |
ICEHptfs4b |
hpp12_120 |
GAAGAATG |
126992 |
(39.0)6 |
N |
|
175 |
hpEurope |
ICEHptfs4c |
hpp12_1510 |
TAAGAATG |
1602176 |
39.3 |
Y |
|
328 |
hpEurope |
ICEHptfs4a |
hpg27_335 |
AAAGAATA |
366213 |
(2.3) |
N |
|
328 |
hpEurope |
ICEHptfs4b |
hpp12_1365 |
AAAGAATG |
1436629 |
40.2 |
Y |
|
ATCC43526 |
hpEurope |
ICEHptfs3/4a2 |
hpp12_1508 |
TAAGAATG |
1598758 |
(47.5) |
N |
|
ATCC43526 |
hpEurope |
ICEHptfs4a |
hpp12_189-188 |
TAAGAATG |
191853 |
(22.6) |
N |
|
P1 |
hpEurope |
ICEHptfs3 |
hpp12_746-745 |
AAACAATA |
800162 |
(13.3) |
N |
|
P1 |
hpEurope |
ICEHptfs4b |
hpp12_1366 |
TAAGAATG |
1439080 |
39.4 |
Y |
|
1_17C |
hpAfrica1 |
ICEHptfs4a/b |
hpp12_994-5S-rRNA |
|
1054197 |
(37.6) |
N |
|
6_17A |
hpAfrica1 |
ICEHptfs4a/b |
hpp12_994-5S-rRNA |
|
1054197 |
(37.7) |
N |
|
6_28C |
hpAfrica1 |
ICEHptfs4a |
hpp12_994-5S-rRNA |
|
1054197 |
(1.6) |
N |
| 6_28C | hpAfrica1 | ICEHptfs4b | hpp12_438 | AAAGAATG | 453993 | (35.5) | N |
1inferred from the Neighbor-joining tree shown in Figure 2.
2resulting from insertion of two genome islands and rearrangements associated with IS element insertion and two copies of pz21/hpp12_447-like genes.
3associated with a genome rearrangement between hpp12_1366 and hpp12_1298.
4genomic position of AAGAATG motif in strain P12.
5numbers in parentheses indicate incomplete ICE elements.
6contains 28 kb of prophage-related sequences.
To examine possible variations in plasticity zone distribution among phylogeographic groups, we first constructed a phylogenetic tree based on MLST gene sequences, using all 36 fully sequenced strains, the nine strains sequenced in this study, and 345 reference strains from the MLST database (Figure 2). No correlation between phylogeographic groups and the presence or absence of either ICEHptfs3 or ICEHptfs4 could be found. However, all hpAfrica1 strains contain truncated versions of ICEHptfs4b or of an ICEHptfs4a/b variant similar to the hpAfrica1 strains mentioned above (Tables 2 and 3). We then calculated Neighbor-joining phylogenetic trees using conserved ICEHptfs3 or ICEHptfs4 gene sequences (concatenated virB9, virB11 and virD4 sequences) and compared them with an MLST-derived tree (Figure 3A, B). Interestingly, ICEHptfs4ab genes clustered in a similar way as housekeeping gene sequences did, except for a much closer relationship of these genes than of housekeeping genes between hpAfrica2 strain SouthAfrica7 and other populations (Figure 3B; Additional file 2: Figure S2). In contrast, ICEHptfs3 sequences formed at least three strongly divergent clades that were not congruent with the MLST population structure. These clades seem to correspond to (1) the hspAmerind population; (2) a mixture of hspEAsia and hpAsia2 populations; and (3) a mixture of hpEurope and hpAfrica1 populations (Figure 3B; Additional file 2: Figure S2). However, the number of ICEHptfs3-positive strains analysed may be too low to definitely draw conclusions from this observation.
Figure 2.
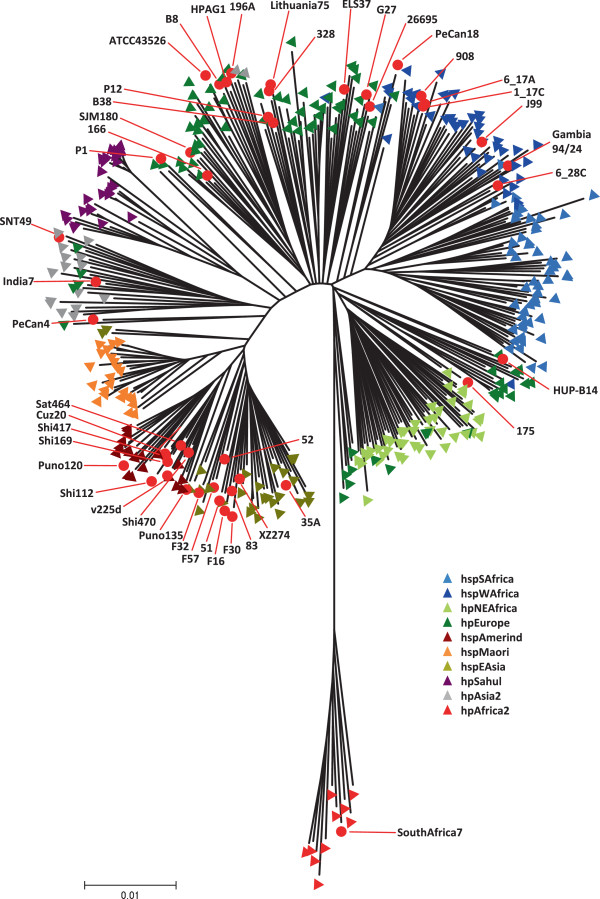
Phylogeography of the analysed strains. The Neighbor-joining tree was calculated with concatenated MLST sequences from 345 reference strains from the H. pylori MLST database ( http://pubmlst.org/helicobacter/) and from all strains analysed in this study. MLST database phylogeography assignments are indicated by coloured triangles, and locations of sequenced strains are indicated by red dots.
Figure 3.
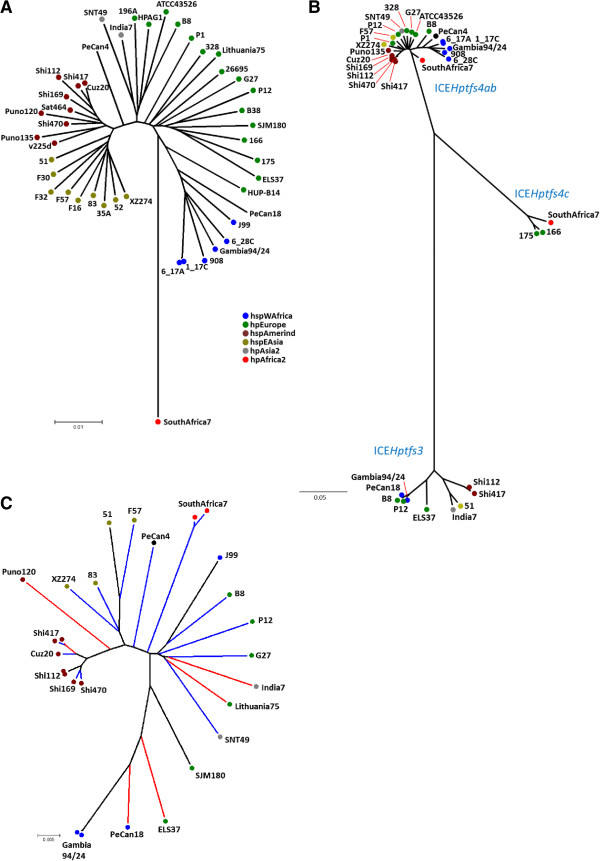
Neighbor-joining analysis of type IV secretion system gene sequences. (A) Phylogenetic tree calculated with MLST sequences for fully sequenced strains only, with phylogeography assignments based on the Neighbor-joining tree shown in Figure 2. Note that unequivocal classification of strains PeCan4 and PeCan18 was not possible. (B) Phylogenetic tree calculated from concatenated virB9, virB11 and virD4 ortholog sequences of all ICEHptfs3 and ICEHptfs4 islands. (C) Neighbor-joining tree calculated from DNA sequences of methylase/helicase (hpp12_447/pz21) orthologs. Orthologs associated with ICEHptfs3 elements are marked by blue branch lines, and orthologs associated with ICEHptfs4 elements by red branch lines. Black lines indicate hybrid elements or the presence of two different elements in the same strain. Colouring of individual strains by phylogeographic origin is shown according to the tree in Figure 2.
Identification of conserved and ICE type-specific genes
Since both ICEHptfs3 and ICEHptfs4 islands contain genes for complete type IV secretion systems and may coexist in a single strain, an open question is whether individual genes or groups of genes from one type of island have the capacity to complement deficiencies in the other. Sequence comparisons showed that each of the type IV secretion apparatus components is clearly distinguishable between the different types (and partly between subtypes) of islands, with amino acid sequence similarities ranging from 40% to 80% (Table 4). This is also true for putative DNA processing or segregation proteins such as XerT, ParA, TopA or VirD2 (but not for the putative methylase/helicase PZ21 (OrfQ)/HPP12_447; see below), suggesting that the individual secretion systems might be sufficiently divergent to be incompatible.
Table 4.
Amino acid similarities and identities between ICE Hptfs4a -encoded proteins and proteins from ICE Hptfs3 and ICE Hptfs4b/c islands
| Gene P12 | Size (aa) | Identity/similarity ICE Hptfs4b 1 | Identity/similarity ICE Hptfs4c 1 | Orthologous gene on ICE Hptfs3 | Identity/similarity ICE Hptfs3 1 | Putative function |
|---|---|---|---|---|---|---|
|
hpp12_437 |
357 |
56/73 |
98/98 |
hpb8_521/pz40 |
63/76 |
XerT |
|
hpp12_438 |
227 |
missing |
95/97 |
hpb8_524/pz37 |
77/83 |
|
|
hpp12_439 |
432 |
32/49 |
93/95 |
hpb8_527/pz34 |
23/42
2
|
VirB6 |
|
hpp12_440 |
92 |
missing |
93/96 |
missing |
- |
|
|
hpp12_441 |
466 |
40/60 |
96/97 |
hpb8_526/pz35 |
23/46
2
|
|
|
hpp12_442/443 |
737 |
94/95 |
94/95 |
hpb8_543/pz15 |
32/50 |
|
|
hpp12_444/445 |
464 |
28/46 |
97/993 |
hpb8_529/pz32 |
26/53
4
|
|
|
hpp12_446 |
340 |
28/43 |
95/973 |
hpb8_530/pz31 |
30/47 |
|
|
hpp12_447 |
2808 |
94/96 |
92/95 |
pz21 |
89/93 |
|
|
hpp12_448 |
218 |
98/99 |
97/99 |
hpb8_532/pz29 |
67/81 |
ParA |
|
hpp12_449 |
94 |
98/100 |
94/94 |
hpb8_533/pz28 |
37/69 |
|
|
hpp12_450 |
392 |
92/96 |
missing |
missing |
- |
|
|
hpp12_451 |
637 |
93/95 |
35/51 |
hpb8_519,517/pz41 |
35/56 |
VirD2 |
|
hpp12_452 |
104 |
97/98 |
n.d.5 |
missing |
- |
|
|
hpp12_453 |
93 |
98/100 |
n.d.5 |
missing |
- |
|
|
hpp12_454 |
575 |
98/99 |
62/77 |
hpb8_538/pz20 |
50/66 |
VirD4 |
|
hpp12_455 |
170 |
98/98 |
46/60
6
|
hpb8_538/pz206 |
32/50 |
|
|
hpp12_456 |
96 |
97/98 |
n.d.5 |
missing |
- |
|
|
hpp12_457 |
151 |
97/97 |
35/51 |
missing |
- |
|
|
hpp12_458 |
313 |
99/99 |
58/74 |
hpb8_540/pz18 |
42/64 |
VirB11 |
|
hpp12_459 |
99 |
98/100 |
n.d.5 |
missing |
- |
|
|
hpp12_460 |
87 |
93/96 |
n.d.5 |
missing |
- |
|
|
hpp12_461 |
97 |
97/97 |
91/93 |
missing |
- |
|
|
hpp12_462 |
421 |
80/84 |
92/95 |
hpb8_544/pz14 |
53/69 |
VirB10 |
|
hpp12_463 |
510 |
97/98 |
94/97 |
hpb8_545/pz13 |
47/66 |
VirB9 |
|
hpp12_464 |
389 |
55/73 |
98/99 |
hpb8_546/pz12 |
38/62 |
VirB8 |
|
hpp12_465 |
38 |
55/75 |
55/75 |
hpb8_547/pz11 |
44/58 |
VirB7 |
|
hpp12_466 |
677 |
45/62 |
94/97 |
hpb8_537/pz24 |
45/61 |
TopA |
|
hpp12_467 |
807 |
44/63 |
96/97 |
hpb8_548/pz10 |
38/58 |
VirB4 |
|
hpp12_468 |
88 |
54/75 |
95/97 |
hpb8_550/pz8 |
39/58 |
VirB3 |
|
hpp12_469 |
100 |
42/63 |
93/97 |
hpb8_551/pz7 |
30/45 |
VirB2 |
|
hpp12_470/471 |
508 |
34/54 |
94/96 |
hpb8_528/pz33 |
35/51 |
|
|
hpp12_472 |
97 |
missing |
90/93 |
missing |
- |
|
| hpp12_473 | 259 | 34/57 | 92/93 | hpb8_554/pz5 | 37/63 |
1numbers printed in normal face correspond to >90% identity (identical genes), and numbers in bold face to 40-85% similarity.
2genes hpb8_526 and pz35, as well as hpb8_527 and pz34 share only 61/73% and 54/70% identity/similarity, respectively, to each other, but are equally similar to hpp12_441 and hpp12_439, respectively.
3some ICEHptfs4c islands contain the ICEHptfs4b versions with lower similarities in these sites.
4similarities confined to short regions only.
5no significant similarity detectable, but gene with similar size and orientation present.
6ICEHptfs4c and ICEHptfs3 islands contain fusions of hpp12_454 and hpp12_455.
To define further common ICE gene products and to identify ICE-type-specific genes, we performed similarity searches with all other amino acid sequences as well. The results show that nine further, hypothetical ICEHptfs4a genes have similar counterparts in ICEHptfs3-type islands (Table 4). Interestingly, orthologs of the conserved hypothetical genes hpb8_524 or hpp12_438 are present in ICEHptfs3, ICEHptfs4a and ICEHptfs4c islands, but absent from ICEHptfs4b islands. Because of their sequence similarities, we speculate that these hypothetical genes have additional conserved functions for genome island maintenance and/or transfer. In contrast, genes that are specific for either type of genome island might be cargo proteins of the respective mobile genetic elements, fulfilling more specific roles. Such specific genes for ICEHptfs4 islands are hpp12_440 (present only on ICEHptfs4a and ICEHptfs4c islands), hpp12_450/hpg27_977 (which is specifically absent in ICEHptfs4c islands), hpp12_452, hpp12_453, hpp12_456, hpp12_459-461, and hpp12_472 (Table 4). Specific genes of ICEHptfs3 islands include hpb8_522, hpb8_523, hpb8_525, hpb8_531, hpb8_534, hpb8_535, hpb8_539, hpb8_541, hpb8_542, hpb8_549, hpb8_552, pz22 and pz23. Interestingly, ICEHptfs3 islands in some strains have insertions of specific genes encoding Fic domain-containing or JHP940-like proteins (Additional file 3: Figure S3).
The putative DNA methylase/helicase gene pz21(orfQ)/hpp12_447 may be found associated with either ICEHptfs3 or ICEHptfs4 islands. In striking contrast to the above-mentioned divergence between orthologous ICEHptfs3 and ICEHptfs4 genes, the methylase/helicase orthologs present on ICEHptfs3 (e.g., pz21) and on ICEHptfs4a/b/c islands (e.g., hpp12_447) are highly conserved (90-98% similarity), indicating an evolutionary pressure for this gene which is distinct from other genes on the genome islands. A Neighbor-joining tree of pz21/hpp12_447 orthologs shows a certain clustering according to geographic origin, but this clustering is clearly independent of gene association with either ICEHptfs3 or ICEHptfs4 (Figure 3C). Indeed, in cases where both ICEHptfs3 and ICEHptfs4 methylase/helicase orthologs are present in a single strain (Shi112, Shi417, Gambia94/24), these orthologs are always more similar to each other than to ICEHptfs3 or ICEHptfs4 orthologs of geographically related strains, and even more similar than two ICEHptfs4 methylase/helicase orthologs present in a single strain (SouthAfrica7) are to each other (Figure 3C). Because of these high sequence similarities, homologous recombination between ICEHptfs3 and ICEHptfs4 methylase/helicase orthologs is possible. By analysing the gene arrangements of the hybrid ICEHptfs3-ICEHptfs4 elements mentioned above, we could identify situations where such recombination events seem to have occurred indeed after integration of one ICE element into another (Additional file 4: Figure S4).
Analysis of ICE integration sites
Originally, the plasticity zone was found located at a distinct position within H. pylori genomes (i.e., between the ftsZ gene (hp0979) and one copy of the 5S-23S rRNA genes) [14]. However, analysis of strain P12, Shi470 and G27 genome sequences showed that ICEHptfs3 and ICEHptfs4 elements are able to integrate as well into different genomic locations, in a manner similar to conjugative transposons or genome islands [11,16]. To examine further variations in integration sites, we compared the sequences of ICE integration sites and duplicated junction motifs in all genome sequences with recognizable left and/or right ICEHptfs3 and ICEHptfs4 junctions. In addition to 12 different sites described previously [16], we identified further 28 chromosomal sites and one plasmid site where complete or partial ICEHptfs3 or ICEHptfs4 elements can be integrated (Tables 2 and 3; Figure 4). Although these integration sites cluster in certain genome regions, such as the originally identified ICE integration locus (plasticity zone 2 in P12), the left border region of ICEHptfs4a, or a locus containing several restriction-modification system genes (hpp12_1364-1366), there is no obvious general preference for ICE integration. We also did not observe different patterns of ICEHptfs3 versus ICEHptfs4 integration sites; in fact, some integration sites are used by either ICEHptfs3 or ICEHptfs4 (Figure 4).
Figure 4.
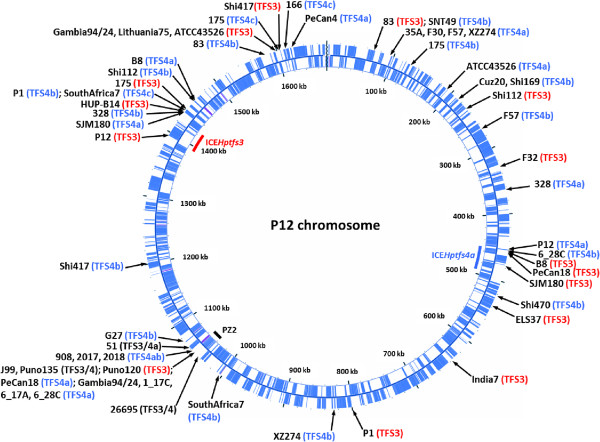
Integration sites of all ICEHptfs3 and ICEHptfs4 islands mapped onto the genome of strain P12. Positions of these elements as well as of plasticity zone 2 (PZ2) in the genome of P12 are shown within the circle. Each arrow indicates an individual ICEHptfs3 and/or ICEHptfs4 integration site. Note that the integration sites shown for strains where one island is integrated into another are not indicative of their genomic location in comparison to the main genome (for example, ICEHptfs3 of strain PeCan18 is inserted into a ICEHptfs4a fragment and therefore shown at 456 kb, but the ICEHptfs4a fragment is in fact integrated in the PZ2 region at 1059 kb in this strain).
All islands with detectable junctions contained the conserved sequence motif AAGAATG [11,16], and this motif is always present in the corresponding empty sites of PZ-free strains (albeit sometimes mutated), suggesting that it represents a minimal requirement for integration of ICEHptfs3 and ICEHptfs4 elements. To determine whether additional sequences are required to form an integration site, we compared the sequences of the flanking regions of ICEHptfs3 and ICEHptfs4 separately (Figure 5; Additional file 5: Figure S5). There is a certain preference for A or T close to the left junctions of both ICEHptfs3 and ICEHptfs4 islands (-1 to -3 or -1 to -6), but the alignment revealed no significant consensus sequences otherwise. However, there seems to be a stronger preference of A at the -1 position (resulting in AAAGAATG motifs) in ICEHptfs4 than in ICEHptfs3 islands. Furthermore, the low prevalence of the last G at the right junctions of ICEHptfs3 islands may even suggest that only six bases (AAGAAT) are used by ICEHptfs3 islands.
Figure 5.
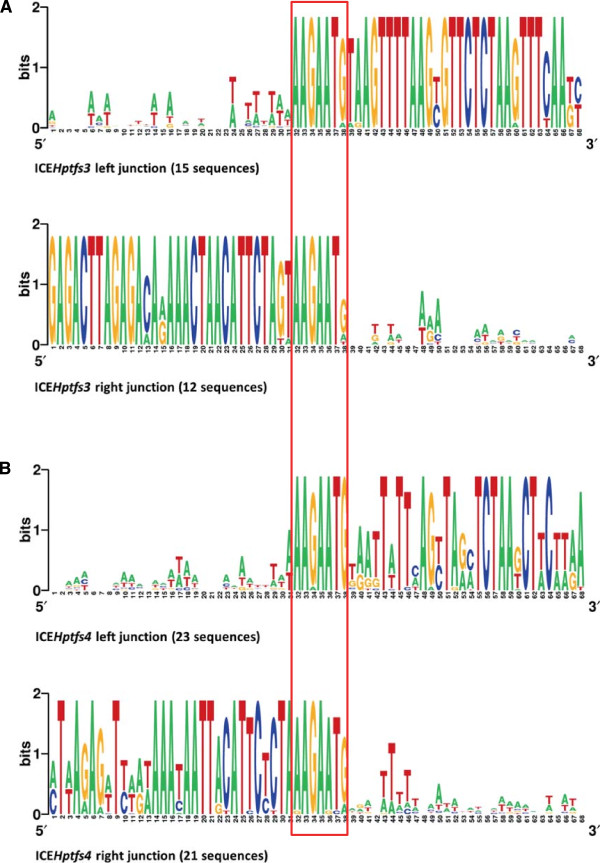
Comparative analysis of integration sites. Sequence logos for nucleotide sequences around ICEHptfs3(A) or ICEHptfs4(B) integration sites were generated using Weblogo [43]. The level of sequence conservation is indicated by the height of the letters (with a maximum of 2 bits at each position).
Identification of a unique ICEHptfs4 variant in the hpAfrica1 population
Since deletions of single genes or different sets of genes are frequent for both ICEHptfs3 and ICEHptfs4 islands (Table 2), we checked whether these occur randomly or at conserved sites. Deletions found within ICEHptfs3 variants range from small deletions (pz26 and pz27) to loss of major parts of the island (Additional file 3: Figure S3A), and mostly seem to occur at random positions and without conserved sequence motifs (data not shown). However, we also identified several cases where ICEHptfs3 truncation sites are flanked by AAGAATG motifs, suggesting that recombination events similar to ICE integration resulted in some deletions (Additional file 3: Figure S3A). For ICEHptfs4 islands, we found certain deletions that are more frequent. For example, four hspEAsia strains (35A, F30, F57, XZ274) have identical truncations of their ICEHptfs4a islands (Additional file 3: Figure S3B). These elements also have identical integration sites (Figure 4) and are accompanied by a common genome rearrangement [25], suggesting that the observed truncations reflect the situation in a common ancestor of all four strains. In fact, these truncated versions are the only ICEHptfs4a remnants that we found in hspEAsia or hspAmerind strains; all other complete or truncated variants in these populations are of the ICEHptfs4b type. A second common truncation was found in all hspWAfrica strains (908/2017/2018, Gambia94/24, 1_17C, 6_17A, 6_28C) and involved a loss of several genes close to the right junctions of their ICEHptfs4b or ICEHptfs4a/b islands, including the 5’ regions of the respective virB4 genes (Additional file 3: Figure S3B). The same deletion occurs in hspWAfrica strain J99, where the corresponding virB4 gene (jhp917/918) is also known as dupA[18]. All these ICEHptfs4b islands have their right junctions deleted and are furthermore inserted at the same genome position (Tables 2 and 3), flanked on the truncation site by jhp916, jhp915 and jhp914 orthologs (Figure 6A). A closer inspection of the right border revealed that truncations have occurred at a CATTCTT (or AAGAATG on the reverse strand) motif which is conserved in the virB4 genes of ICEHptfs4b (but not ICEHptfs4a) islands. Interestingly, those ICEHptfs4b variants which contain ICEHptfs4a genes close to their left borders, all have another small truncation of about 300 bp at their left junctions, which also has occurred at a conserved CATTCTT motif upstream of the xerT gene (Additional file 3: Figure S3B), indicating that these islands have integrated in an irregular fashion, producing irregular left junctions (ILJ) and irregular right junctions (IRJ; Figure 6A). Since the nearby jhp914 gene has previously been reported to be specifically present in the hpAfrica1 population [24], we asked whether this truncated right border might be a general signature of hpAfrica1 strains. To test this hypothesis, we performed a BLAST search of draft genome sequences with a 260 bp query sequence spanning the right border of J99 (including the IRJ). Of 78 retrieved draft genome sequences having the same IRJ, 64 also contained the jhp914 gene (data not shown). Furthermore, we checked a panel of H. pylori strains isolated in Nigeria for the presence of the irregular ICEHptfs4b right border (Figure 6B). PCR analysis with primers specific to virB4 and jhp914, respectively (Figure 6A), confirmed that 14 out of 19 strains from this population were positive for a similar gene arrangement in this locus and thus for an IRJ (Figure 6B, and data not shown).
Figure 6.
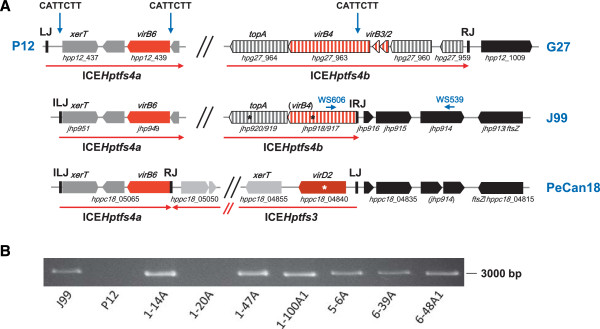
A truncated version of ICEHptfs4 in hspWAfrica strains. (A) Most hspWAfrica strains (exemplified here by J99) have an ICEHptfs4 variant composed of ICEHptfs4a genes (compared here with P12) close to the left junction and ICEHptfs4b genes (compared here with G27) close to the right junction of the island. In these strains, the left part of the island is shortened by 350 bp at a CATTCTT motif upstream of xerT, and the right part by approximately 3850 bp at a CATTCTT motif within ICEHptfs4b virB4, generating irregular left and right junctions (ILJ and IRJ). In strain PeCan18, the ICEHptfs4a fragment has probably been integrated in a similar manner, using irregular integration at the same chromosomal position, but the majority of ICEHptfs4a seems to have been deleted subsequently by (regular) integration of an ICEHptfs3 at the same internal virB4 motif and another internal CATTCTT motif upstream of ICEHptfs4a virB6. Gene colouring is as in Figure 1, and asterisks denote frameshift or nonsense mutations (B) PCR analysis of the ICEHptfs4b right junction in H. pylori strains from Nigeria. PCR was performed from chromosomal DNA of the indicated strains with primers WS606 and WS539 (see Figure 6A).
Discussion
The unusual genetic heterogeneity of H. pylori has been well-documented in terms of allelic diversity, establishing it as a species with a very high population recombination rate, and allowing for different populations from different geographic regions to be identified [4]. MLST analysis of these populations has revealed important insights into the coevolution of H. pylori and humans, and into migration events of human populations, but relatively little is known about bacterial population-specific properties on a genomic level. Striking differences in the presence or absence of putative host interaction genes have been reported for East Asian H. pylori strains in comparison to European strains [12], and many divergent genes were found to evolve under positive selection between East Asian and non-Asian strains [12,26]. Previous comparative analysis of a small number of H. pylori genome sequences indicated that many strain-specific genes are located either at potential genome rearrangement sites or within the plasticity zones [11]. However, for those plasticity zone regions that are organized in ICEHptfs3 or ICEHptfs4 islands as described here, identification of further novel genes seems unlikely. Instead, the gene content of a given type of ICEHptfs3 or ICEHptfs4 island is, apart from the variable presence of JHP940- or Fic domain protein-encoding genes, highly conserved, strongly suggesting that these elements are autonomous elements with fixed contents rather than true regions of genome plasticity. Nevertheless, partial truncations, insertions of restriction-modification systems, IS elements or even distinct genome islands, and associated rearrangements [25] are frequent within both types of ICE and result in a considerable amount of variation. Rearrangements between ICEHptfs3 and ICEHptfs4 elements may be facilitated by recombination events within pz21/hpp12_447 (methylase/helicase) orthologs present on both types of islands. Apart from that, ICEHptfs3 and ICEHptfs4 islands are clearly distinct and do not seem to exchange individual genes. The fact that pz21/hpp12_447 orthologs are the only genes with high similarity between ICEHptfs3 and ICEHptfs4 elements, indicates that these orthologs are either frequently exchanged between both types of island, or that they are subject to strong selective pressures.
Interestingly, certain regions of both ICEHptfs3 and ICEHptfs4 islands are much more variable than others. For instance, we were able to identify 3, 5, and 4 distinct clades, respectively, for the pz34, pz35 and pz36 orthologs on ICEHptfs3 elements (data not shown), whereas all other ICEHptfs3 genes are more conserved. However, similar to the variability of hpp12_444/445 and hpp12_446 orthologs among ICEHptfs4 islands, where two clades each can be distinguished (data not shown), no clear correlation of these different clades with individual geographic groups could be found. Likewise, the three different subtypes of ICEHptfs4 islands which are characterized by orthologous, but distinct sets of genes, do not seem to be restricted to certain geographic groups. We also performed a preliminary analysis of two further hpAfrica2 strain genome sequences [27] and one hpSahul strain genome sequence [13] that were published after completion of our comparative analysis. Both hpAfrica2 strains contain one full-length ICEHptfs4b element, and the hpSahul strain harbours a full-length ICEHptfs4b and a partial ICEHptfs3 element (data not shown), which further supports the notion that these elements are present in all phylogeographic groups. The modular structure of ICEHptfs4 islands indicates that parts of these elements can easily be exchanged, and that all variants may coexist in a given H. pylori population. Indeed, ICEHptfs4a, b and c islands all have some common genes which may be used for exchange of modules. However, it is striking that all members of ICEHptfs4b subtypes consistently lack hpp12_438 orthologs and that hybrid elements between different ICEHptfs4 subtypes do not occur. An exception is the combination of ICEHptfs4a (left) with ICEHptfs4b (right), which seems to occur in hpAfrica1 strains only, and always in a truncated version. These restrictions on modular exchange suggest that there is a selective pressure on maintenance of cognate left and right ICEHptfs4 ends, for example by an inability of hybrid elements to be excised and/or transferred. The presence of ICEHptfs3-like islands in other Helicobacter species, such as H. cetorum[16,28] and H. suis[29], indicates that these elements were acquired a long time ago (i.e., before the cag pathogenicity island, which is absent in hpAfrica2 strains and was acquired more than 60000 years ago [30]). Whereas microdiversity within cag pathogenicity island genes correlates with microdiversity in housekeeping genes, this is not the case for ICEHptfs3 or ICEHptfs4 genes, which shows again that these islands are subject to more frequent horizontal gene transfer.
Horizontal gene transfer of typical ICEs involves several steps [31]: first, the element is usually excised from the chromosome by a recombinase to generate a circular intermediate; second, this circular form is transferred from the donor to a recipient cell by conjugation; and third, the ICE integrates into the recipient cell chromosome via site-specific or unspecific recombination. In the case of ICEHptfs4, the first step is dependent on the XerT recombinase [11], and the second on the VirD2 relaxase [32], both of which are encoded on the ICE. It is likely, but has not been shown yet, that the ICE-encoded type IV secretion system is responsible for the conjugative transfer process. It is also currently unclear whether the XerT recombinase catalyzes integration of the ICE into the recipient cell chromosome as well. An interesting finding of this study was the presumptive minimal requirement for integration of both ICEHptfs3 and ICEHptfs4 islands, the sequence motif AAGAATG (or possibly AAGAAT for ICEHptfs3), as suspected previously [11,16]. Thus, the total number of possible insertion sites might be limited only by the number of these motifs in intergenic regions or in non-essential genes. In total, we identified more than 40 different integration sites, but the total number of possible integration sites might be significantly higher, given that AAGAATG sequences are found approximately 550 times within individual H. pylori genomes (data not shown). Many well-characterized ICEs integrate into a unique position in the host cell genome (the primary attachment site), often in the 3’ regions of tRNA loci [31]. In the absence of primary attachment sites, these elements are sometimes capable of integrating into secondary sites with much less specificity, but this may result in ICE immobility or even toxicity for the host cell [33]. In contrast, other ICE-like elements, which are often termed conjugative transposons, have very low integration site specificities, with as many as 100,000 possible integration sites in a given host strain [34,35]. In this regard, ICEHptfs3 and ICEHptfs4 seem to integrate with an intermediate specificity, but still with the potential to insert into coding regions and thereby to disrupt essential genes. Possible integration sites are also located on the ICE elements themselves, and we found several cases where one ICE is integrated into another. We could also identify situations where these internal sites were used for irregular ICE integration, associated with truncation of the left and/or right ICE ends, and possibly an incapability of these elements to excise.
Finally, despite the presence of genes encoding host interaction factors such as JHP940 [36], or correlated with disease outcome, such as dupA[18], the (potentially different) functions of ICEHptfs3 and ICEHptfs4 islands are currently unclear. In our analysis, a total of 18 strains were positive for dupA (the ICEHptfs4b virB4 gene), and 12 additional strains were found positive for ICEHptfs4a or ICEHptfs4c virB4 genes, which are likely to have the same functions. Because of this, and since not all of these strains have complete ICEs or even complete type IV secretion systems, testing for the presence of the dupA gene alone, and correlations of dupA with pathology is probably not useful. It has been shown that a more complete analysis of type IV secretion system genes is more significant as a virulence marker [19]. Therefore, future correlation studies should determine the presence of the complete set of genes.
Conclusions
Taken together, our comparative analysis reinforces the notion that major parts of the H. pylori plasticity zones described earlier should in fact be considered as mobile genetic elements with conserved gene content, rather than regions of genome plasticity. Although horizontal gene transfer of complete ICEHptfs3 or ICEHptfs4 elements remains to be demonstrated experimentally, the number of different integration sites indicates a considerable mobility, possibly also within individual H. pylori genomes. In this regard, these elements differ from the cag pathogenicity island, for which only one integration site is known (although rearrangements may occur). The high prevalence and wide distribution of these ICEs throughout all H. pylori populations suggest that they might provide an as yet unknown fitness benefit to their hosts.
Methods
Draft genome sequencing of H. pylori strains
To select H. pylori strains for draft genome sequencing, chromosomal DNA was prepared from a panel of laboratory strains or of clinical isolates, using a QIAamp DNA mini kit, and analysed by PCR with primer pair DupA-WXF (5′-GATATACCATGGATGAGTTCYRTAYTAACAGAC-3′) and JHP0919R2 (5′-GCCCACCAGTTGCAAAAACAAATGAAC-3′) [37], or with primer pair WS393 (5′-TATGGTATCAGGGCATACC) and WS394 (5′-GTTCTTTGAGATACTCAGG-3′) for the presence of ICEHptfs4b or ICEHptfs4a virB4, respectively. Based on this analysis, we selected 3 virB4-positive strains isolated in Western Africa, 5 virB4-positive strains isolated in Europe, and one virB4-negative strain isolated in Europe for genome sequencing.
Whole genomic DNA was isolated from bacteria that were subjected to minimal passage, using Qiagen Genomic‒tip 100/G columns and the Genomic DNA Buffer Set (Qiagen). Genomic DNA was processed to generate 3 kb mate pair libraries, which were sequenced with 50 bp paired-end reads on an Illumina HiSeq 2000 platform (GATC, Konstanz, Germany). This resulted in 24-60 million reads per genome, which were cured from PCR replicates and mapped to a reference sequence consisting of concatenated ICEHptfs3 (strain B8), ICEHptfs4a (strain P12), and ICEHptfs4b (strain G27) sequences, using BWA [38] with default parameters. Unmapped reads were assembled de novo using Velvet [39], and ICE elements were identified by BLAST searches ( http://blast.ncbi.nlm.nih.gov/Blast.cgi). Gaps within ICE elements were closed by Sanger sequencing.
Software tools for analysis of H. pylori genome sequences
For comparative analysis, we evaluated all complete H. pylori genome sequences available in GenBank at the time of initiation of the study. We used multilocus sequence typing analysis to assign all strains to the populations and subpopulations described previously [21]. To do so, partial nucleotide sequences of the housekeeping genes atpA, efp, mutY, ppa, trpC, ureI and yphC were concatenated for each strain and aligned with the corresponding sequences of 345 reference strains from the MLST database ( http://pubmlst.org/helicobacter), using the Muscle algorithm within MEGA5.2 [40]. All phylogenetic trees were constructed and tested by neighbor joining with MEGA5.2, using the Kimura 2-parameter model of nucleotide substitution, and 1,000 bootstrap replications. ICE elements were identified in complete or draft genome sequences using BLAST search and visualization with the Artemis Comparison Tool [41]. A chromosomal map of strain P12 was generated using CGView [42], and WebLogo [43] was used to display sequence alignments of ICE border regions.
Genetic analysis of hpAfrica1 strains
Genomic DNA of H. pylori strains was prepared using a QIAamp DNA mini kit. For MLST analysis, the housekeeping genes atpA, efp, mutY, ppa, trpC, ureI and yphC were partially amplified by PCR, using the primer sets described in the MLST database ( http://pubmlst.org/helicobacter), and the PCR products were sequenced. Sequences were trimmed to the required sizes, concatenated and analyzed for clustering, as described above. For examination of the right junctions of ICEHptfs4 islands, PCR fragments were amplified with a PANScript DNA polymerase (PAN Biotech, Aidenbach, Germany) under standard conditions in the presence of 3 mM MgCl2 and at an annealing temperature of 52°C, using primers WS606 (5′-AGCAATAAAACGCTTAAAAGTCTC-3′) and WS539 (5′-ATGTCCAGTAAGGAATTTGTC-3′), and subsequently analyzed by gel electrophoresis.
GenBank accession numbers
The accession numbers for the ICEHptfs3 and ICEHPtfs4 sequences determined in thuis study are as follows: 166_ICEHptfs4c [GenBank:KF861855]; 175_ICEHptfs3 [GenBank:KF861857]; 175_ICEHptfs4b [GenBank:KF861858]; 175_ICEHptfs4c [GenBank:KF861859]; 328_ICEHptfs4a [GenBank:KF861860]; 328_ICEHptfs4b [GenBank:KF861861]; ATCC43526_ICEHptfs3/4a [GenBank:KF861862]; ATCC43526_ICEHptfs4a [GenBank:KF861863]; P1_ICEHptfs3 [GenBank:KF861854]; P1_ICEHptfs4b [GenBank:KF861856]; 1_17C_ICEHptfs4b [GenBank:KF861864]; 6_17A_ICEHptfs4b [GenBank:KF861865]; 6_28C_ICEHptfs4b [GenBank:KF861866]. Sequences of other ICE elements can be found in GenBank under the strain designations and at the genome positions shown in Table 1.
Availability of supporting data
The phylogenetic trees shown in Figures 2 and 3 have been deposited in TreeBASE and can be accessed under http://purl.org/phylo/treebase/phylows/study/TB2:S15635.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WF conceived of and participated in the design of the study, analysed sequence data and wrote the manuscript. UB carried out the molecular genetic studies. BK and CS participated in sequence analysis. SIS participated in strain isolation and selection. RH participated in the design of the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Gene arrangement of the plasticity zones in H. pylori strains 26695 and J99. Both strains contain highly rearranged truncated versions, presumably resulting from consecutive integration of 2-3 islands (ICEHptfs3, ICEHptfs4a, ICEHptfs4b) and subsequent rearrangements, some of which are associated with insertion elements (IS605), as indicated.
Neighbor-joining trees of conserved type IV secretion genes. (A) Phylogenetic tree calculated from concatenated virB9, virB11 and virD4 ortholog sequences of ICEHptfs3 elements. (B) Phylogenetic tree calculated from concatenated virB9, virB11 and virD4 ortholog sequences of ICEHptfs4 elements.
Alignments of truncated ICEHptfs3 and ICEHptfs4 elements. (A) ICEHptfs3 elements are shown in comparison to ICEHptfs3 from strain PeCan18 (gene designation according to [15]). Additional specific genes inserted in certain elements only are shown in blue. fic, gene encoding Fic family protein similar to conserved hypothetical proteins found in Neisseria spp.; fic*, gene encoding Fic family protein similar to H. pylori chromosome-encoded proteins (e.g., JHP651). (B) Truncated ICEHptfs4a, ICEHptfs4b and ICEHptfs4c elements are shown in comparison to the complete elements found in strains P12 and G27, respectively. Asterisks within genes indicate frameshift or nonsense mutations; blue arrows indicate truncations or rearrangements at internal AAGAATG motifs.
Evidence for homologous recombination between methylase/helicase orthologs in hybrid ICEHptfs3/ICEHptfs4 elements. (A) The gene arrangement of a hybrid ICE element in strain SJM180 is compared with the ICEHptfs4a element from strain P12 and the ICEHptfs3 element from strain PeCan18. Note that the putative DNA methylase/helicase ortholog might originate from either ICEHptfs3 (pz21) or ICEHptfs4a (hpp12_447). Gene colouring is analogous to Figure 1 and Additional file 3: Figure S3, and frameshift or nonsense mutations are indicated by asterisks. (B) Hypothetical steps for generation of the SJM180 gene arrangement shown in (A). First, insertion of an ICEHptfs3 element (red) into an already integrated ICEHptfs4a element (blue) generates a composite element. Subsequently, homologous recombination between the pz21 and hpp12_447 orthologs and an independent truncation close to the ICEHptfs4a right junction result in the deletions observed. Similar recombination events might have generated the hybrid ICE element arrangements in strains 51, Puno 135 and J99 (data not shown).
Border sequences of ICEHptfs3 (A) and ICEHptfs4 (B) elements. Genome positions within the respective sequences are indicated; sequences of the islands are printed in italics, and the duplicated integration motifs in bold and red.
Contributor Information
Wolfgang Fischer, Email: fischer@mvp.uni-muenchen.de.
Ute Breithaupt, Email: breithaupt@uni-muenchen.de.
Beate Kern, Email: kern_b@mvp.uni-muenchen.de.
Stella I Smith, Email: stellaismith@yahoo.com.
Carolin Spicher, Email: spicher@mvp.uni-muenchen.de.
Rainer Haas, Email: haas@mvp.uni-muenchen.de.
Acknowledgements
This work was supported by an ERA-NET PathoGenoMics3 grant (HELDIVPAT) and by DFG grant HA 2697/12-1 to RH. We thank Evelyn Weiss for expert technical assistance, and Muinah A. Fowora and Lino E. Torres for assistance during H. pylori strain screening.
References
- Monack DM, Mueller A, Falkow S. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, Mardis ER, Engstrand LG, Gordon JI. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc Natl Acad Sci USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiberge JM, Boursaux-Eude C, Lehours P, Dillies MA, Creno S, Coppée JY, Rouy Z, Lajus A, Ma L, Burucoa C, Ruskoné-Foumestraux A, Courillon-Mallet A, De Reuse H, Boneca IG, Lamarque D, Mégraud F, Delchier JC, Médigue C, Bouchier C, Labigne A, Raymond J. From array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma. BMC Genomics. 2010;11:368. doi: 10.1186/1471-2164-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakis M, Chen SL, Karam SM, Engstrand L, Gordon JI. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci USA. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer MS, Fero J, Salama NR. DNA damage triggers genetic exchange in Helicobacter pylori . PLoS Pathog. 2010;6:e1001026. doi: 10.1371/journal.ppat.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer MS, Cohen IE, Sessler TH, Fero J, Salama NR. Natural Competence Promotes Helicobacter pylori Chronic Infection. Infect Immun. 2013;81:209–215. doi: 10.1128/IAI.01042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Stack A, Josenhans C, Niehus E, Dietrich G, Correa P, Fox JG, Falush D, Suerbaum S. Genomic changes during chronic Helicobacter pylori infection. J Bacteriol. 2006;188:249–254. doi: 10.1128/JB.188.1.249-254.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W, Windhager L, Rohrer S, Zeiller M, Karnholz A, Hoffmann R, Zimmer R, Haas R. Strain-specific genes of Helicobacter pylori : genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38:6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, Takahashi N, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC Microbiol. 2011;11:104. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Wise MJ, Tay CY, Windsor HM, Marshall BJ, Peacock C, Perkins T. Comparative Analysis of the Full Genome of Helicobacter pylori Isolate Sahul64 Identifies Genes of High Divergence. J Bacteriol. 2014;196:1073–1083. doi: 10.1128/JB.01021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori . Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Velapatino B, Mukhopadhyay AK, Cahuayme L, Bussalleu A, Combe J, Gilman RH, Berg DE. Cluster of type IV secretion genes in Helicobacter pylori 's plasticity zone. J Bacteriol. 2003;185:3764–3772. doi: 10.1128/JB.185.13.3764-3772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Lee W, Subramaniam D, Anant S, Herrera P, Cabrera L, Balqui J, Barabas O, Kalia A, Gilman RH, Berg DE. Helicobacter pylori 's plasticity zones are novel transposable elements. PLoS ONE. 2009;4:e6859. doi: 10.1371/journal.pone.0006859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvi A, Devi SM, Ahmed I, Hussain MA, Rizwan M, Lamouliatte H, Mégraud F, Ahmed N. Microevolution of Helicobacter pylori type IV secretion systems in an ulcer disease patient over a ten-year period. J Clin Microbiol. 2007;45:4039–4043. doi: 10.1128/JCM.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori . Gastroenterology. 2005;128:833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SW, Sugimoto M, Shiota S, Graham DY, Yamaoka Y. The intact dupA cluster is a more reliable Helicobacter pylori virulence marker than dupA alone. Infect Immun. 2012;80:381–387. doi: 10.1128/IAI.05472-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehours P, Dupouy S, Bergey B, Ruskoné-Foumestraux A, Delchier JC, Rad R, Richy F, Tankovic J, Zerbib F, Mégraud F, Ménard A. Identification of a genetic marker of Helicobacter pylori strains involved in gastric extranodal marginal zone B cell lymphoma of the MALT-type. Gut. 2004;53:931–937. doi: 10.1136/gut.2003.028811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Mégraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadström T, Suerbaum S, Achtman M. An African origin for the intimate association between humans and Helicobacter pylori . Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moodley Y, Linz B, Yamaoka Y, Windsor HM, Breurec S, Wu JY, Maady A, Bernhöft S, Thiberge JM, Phuanukoonnon S, Jobb G, Siba P, Graham DY, Marshall BJ, Achtman M. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. Gain and loss of multiple genes during the evolution of Helicobacter pylori . PLoS Genet. 2005;1:e43. doi: 10.1371/journal.pgen.0010043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Kawai M, Yahara K, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. Birth and death of genes linked to chromosomal inversion. Proc Natl Acad Sci USA. 2011;108:1501–1506. doi: 10.1073/pnas.1012579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SS, Valk PL, McClain MS, Shaffer CL, Metcalf JA, Bordenstein SR, Cover TL. Comparative genomic analysis of East Asian and non-Asian Helicobacter pylori strains identifies rapidly evolving genes. PLoS ONE. 2013;8:e55120. doi: 10.1371/journal.pone.0055120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SS, Bertoli MT, Kersulyte D, Valk PL, Tamma S, Segal I, McClain MS, Cover TL, Berg DE. Genome Sequences of Three hpAfrica2 Strains of Helicobacter pylori . Genome Announc. 2013;1:e00729–13. doi: 10.1128/genomeA.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Rossi M, Berg DE. Sequence Divergence and Conservation in Genomes of Helicobacter cetorum Strains from a Dolphin and a Whale. PLoS One. 2013;8:e83177. doi: 10.1371/journal.pone.0083177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermoote M, Vandekerckhove TT, Flahou B, Pasmans F, Smet A, De Groote D, Van Criekinge W, Ducatelle R, Haesebrouck F. Genome sequence of Helicobacter suis supports its role in gastric pathology. Vet Res. 2011;42:51. doi: 10.1186/1297-9716-42-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbermann P, Josenhans C, Moodley Y, Uhr M, Stamer C, Vauterin M, Suerbaum S, Achtman M, Linz B. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6:e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- Grove JI, Alandiyjany MN, Delahay RM. Site-specific Relaxase Activity of a VirD2-like Protein Encoded within the tfs4 Genomic Island of Helicobacter pylori . J Biol Chem. 2013;288:26385–26396. doi: 10.1074/jbc.M113.496430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menard KL, Grossman AD. Selective pressures to maintain attachment site specificity of integrative and conjugative elements. PLoS Genet. 2013;9:e1003623. doi: 10.1371/journal.pgen.1003623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AP, Mullany P. A modular master on the move: the Tn 916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–258. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Mullany P, Williams R, Langridge GC, Turner DJ, Whalan R, Clayton C, Lawley T, Hussain H, McCurrie K, Morden N, Allan E, Roberts AP. Behavior and target site selection of conjugative transposon Tn916 in two different strains of toxigenic Clostridium difficile . Appl Environ Microbiol. 2012;78:2147–2153. doi: 10.1128/AEM.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DJ, Park KS, Kim JH, Yang SH, Yoon JY, Han BG, Kim HS, Lee SJ, Jang JY, Kim KH, Kim MJ, Song JS, Kim HJ, Park CM, Lee SK, Lee BI, Suh SW. Helicobacter pylori proinflammatory protein up-regulates NF-κB as a cell-translocating Ser/Thr kinase. Proc Natl Acad Sci USA. 2010;107:21418–21423. doi: 10.1073/pnas.1010153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein NR, Argent RH, Marx CK, Patel SR, Robinson K, Atherton JC. Helicobacter pylori dupA is polymorphic, and its active form induces proinflammatory cytokine secretion by mononuclear cells. J Inf Dis. 2010;202:261–269. doi: 10.1086/653587. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene arrangement of the plasticity zones in H. pylori strains 26695 and J99. Both strains contain highly rearranged truncated versions, presumably resulting from consecutive integration of 2-3 islands (ICEHptfs3, ICEHptfs4a, ICEHptfs4b) and subsequent rearrangements, some of which are associated with insertion elements (IS605), as indicated.
Neighbor-joining trees of conserved type IV secretion genes. (A) Phylogenetic tree calculated from concatenated virB9, virB11 and virD4 ortholog sequences of ICEHptfs3 elements. (B) Phylogenetic tree calculated from concatenated virB9, virB11 and virD4 ortholog sequences of ICEHptfs4 elements.
Alignments of truncated ICEHptfs3 and ICEHptfs4 elements. (A) ICEHptfs3 elements are shown in comparison to ICEHptfs3 from strain PeCan18 (gene designation according to [15]). Additional specific genes inserted in certain elements only are shown in blue. fic, gene encoding Fic family protein similar to conserved hypothetical proteins found in Neisseria spp.; fic*, gene encoding Fic family protein similar to H. pylori chromosome-encoded proteins (e.g., JHP651). (B) Truncated ICEHptfs4a, ICEHptfs4b and ICEHptfs4c elements are shown in comparison to the complete elements found in strains P12 and G27, respectively. Asterisks within genes indicate frameshift or nonsense mutations; blue arrows indicate truncations or rearrangements at internal AAGAATG motifs.
Evidence for homologous recombination between methylase/helicase orthologs in hybrid ICEHptfs3/ICEHptfs4 elements. (A) The gene arrangement of a hybrid ICE element in strain SJM180 is compared with the ICEHptfs4a element from strain P12 and the ICEHptfs3 element from strain PeCan18. Note that the putative DNA methylase/helicase ortholog might originate from either ICEHptfs3 (pz21) or ICEHptfs4a (hpp12_447). Gene colouring is analogous to Figure 1 and Additional file 3: Figure S3, and frameshift or nonsense mutations are indicated by asterisks. (B) Hypothetical steps for generation of the SJM180 gene arrangement shown in (A). First, insertion of an ICEHptfs3 element (red) into an already integrated ICEHptfs4a element (blue) generates a composite element. Subsequently, homologous recombination between the pz21 and hpp12_447 orthologs and an independent truncation close to the ICEHptfs4a right junction result in the deletions observed. Similar recombination events might have generated the hybrid ICE element arrangements in strains 51, Puno 135 and J99 (data not shown).
Border sequences of ICEHptfs3 (A) and ICEHptfs4 (B) elements. Genome positions within the respective sequences are indicated; sequences of the islands are printed in italics, and the duplicated integration motifs in bold and red.


