Abstract
DNA exonucleases, enzymes that hydrolyze phosphodiester bonds in DNA from a free end, play important cellular roles in DNA repair, genetic recombination and mutation avoidance in all organisms. This article reviews the structure, biochemistry and biological functions of the 17 exonucleases currently identified in the bacterium Escherichia coli. These include the exonucleases associated with DNA polymerases I (polA), II (polB) and III (dnaQ/mutD), Exonucleases I (xonA/sbcB), III (xthA), IV, VII (xseAB), IX (xni/xgdG) and X (exoX), the RecBCD, RecJ, and RecE exonucleases, SbcCD endo/exonuclease, the DNA exonuclease activities of RNase T (rnt) and Endonuclease IV (nfo) and TatD. These enzymes are diverse in terms of substrate specificity and biochemical properties and have specialized biological roles. Most of these enzymes fall into structural families with characteristic sequence motifs, and members of many of these families can be found in all domains of life.
Keywords: DNase, DNA repair, mutation, homologous recombination, genome rearrangements, DNA polymerase, abasic sites, mismatch repair, proofreading, double-strand break repair, dsDNA, ssDNA, metal binding, mutator, lagging strand, flap endonuclease, base excision repair, replication gap repair, tandem repeats
INTRODUCTION TO DNA EXONUCLEASES
Nucleases are enzymes that hydrolyze phosphodiester bonds of nucleic acids. Early in the classification of nuclease activity, a distinction was made whether such enzymes cleaved internally in the chain (“endonucleases”) or from the end in a step-wise manner (“exonucleases”), borrowing a system analogous to that for proteases. In practice, a nuclease can be tested for its requirement for an end by comparing linear vs. circular DNA substrates, with exonucleases cleaving the former but not the latter. This classification has been complicated by the fact that certain nucleases require an end to initiate degradation, but cleave DNA internally to yield oligonucleotide products. By the practical definition, these are exonucleases; formally, they are endonucleases—the field has used the term “endo/exonucleases” to refer to this class of nucleases. Some endonucleases that will cleave circular DNA also possess intrinsic exonuclease activity, hydrolyzing linear substrates in a stepwise fashion (for example Exonuclease III/Endonuclease II and Endonuclease IV) and are therefore both true exonucleases and true endonucleases.
This article will focus on DNA exonucleases from E. coli, including the endo/exonucleases, and their roles in DNA metabolism. For information about endonucleases and their activities, the reader is directed to the articles on DNA repair. Prior to the discussion of the biochemistry, structure and function individual E. coli exonucleases, I will introduce some concepts and terminology regarding the discovery and classification of exonucleases from E. coli.
Discovery of Exonucleases
In the current literature, there are 17 exonucleases identified in E. coli (Table 1). These exonucleases were discovered by several means. Beginning in the early 1960s, efforts were made to purify exonuclease activities biochemically and to categorize their properties. As the nucleases were discovered, they were identified by numbers (Exonuclease I, II etc.). The genes corresponding to these were, in some cases, found by reverse genetics and named corresponding to the activity name (xon for exonuclease one, xth for exonuclease three, etc.). Some of these nuclease activities are associated with subunits of DNA polymerases: for example, Exonuclease II is the 3′ to 5′ proofreading activity of DNA polymerase I (the product of the polA gene); Exonuclease VI is associated with the 5′ to 3′ exonuclease of the same enzyme. Other activities were initially identified for their genetic function as in specific biological pathways and later discovered to be exonucleases. These are often named for the phenotypic properties they affect, for example the RecBCD and RecJ nuclease, involved in recombination, and SbcB and SbcCD nuclease, playing antirecombinational roles, named for “suppressor of RecBCD. In the post-genomic era, a number of nuclease activities have been identified from predictions based on sequence similarity to known exonucleases (Exonucleases IX and X).
Table 1.
DNA exonucleases of E. coli
| Enzyme | Gene(s) | Preferred substrate | Polarity | Other features | Biological role in DNA metabolism |
|---|---|---|---|---|---|
| ExoI | xonA (=sbcB) | ssDNA | 3′ -5′ | processive; stimulated by SSB | recombination, damage repair, mutation avoidance, MMR |
| ExoIII | xthA (=xth) | dsDNA | 3′ - 5′ | acts at nicks and ends, abasic endonuclease; 3′ phosphatase; can remove damaged 3′ residues | base excision repair, repair of oxidative and ionizing radiation damage |
| ExoIV | orn?2 | Olignucleo- tides | 3′ -5′ | may be identical to RNA oligoribonuclease2 | ? nucleotide recycling? |
| ExoVII | xseA, xseB | ssDNA | 3′ - 5′, 5′ - 3′’ | heterodimer, processive, oligonucleotide products, metal cofactor-independent | damage repair, mutation avoidance, MMR |
| ExoVIII | recE | dsDNA | 5′ - 3′ | processive, encoded by cryptic Rac prophage | recombination, damage repair |
| ExoIX | ygdG (=xni) | ? | ? | in 5′ nuclease/FEN-1 family3 | ? |
| ExoX | exoX | ssDNA, dsDNA | 3′ - 5′ | distributive | mutation avoidance, MMR |
| DNA polI 5′ Exo (ExoVI) | polA | ssDNA | 5′ - 3′ | flap endonuclease | replication (Okazaki fragment maturation) |
| DNA Pol I 3′ Exo (ExoII) | polA | ssDNA | 3′ - 5′ | proofreading | |
| DNA Pol II 3′ Exo | polB | ssDNA | 3′ - 5′ | proofreading | |
| DNA Pol III 3′ Exo | dnaQ (=mutD) | ssDNA | 3′ - 5′ | subunit | proofreading |
| RecBCD | recB, recC, recD | dsDNA, ssDNA | 3′ - 5′, 5′ - 3′ | ExoV, highly processive, ATP-dependent, oligonucleotide products, helicase activity; Chi recognition | recombination, DSB repair, restriction of bacteriophage |
| RecJ | recJ | ssDNA | 5′ - 3′ | processive; stimulated by SSB | recombination, damage repair, mutation avoidance, MMR |
| RNase T (Exo T) | rnt | ssDNA | 3′ - 5′ | activity on DNA > RNA, homodimer, distributive | repair?3 |
| SbcCD | sbcC, sbcD | dsDNA | 3′ - 5′ > 5′ - 3′ | ATP-dependent, processive | repair |
| Endonucleas e IV exonuclease activity | nfo | dsDNA | 3′ -5′ | Zn2+ enzyme; can digest damaged 3′ residues; also possesses endonuclease 5′ to AP sites and other damaged residues | BER, NIR, repair of oxidative and ionizing radiation damage |
| TatD (Exonuclease XI) | tatD | dsDNA > ssDNA | 3′ - 5′ | Mg2+ dependent DNase, unclear whether endonuclease or exonuclease activity | ?, E. coli YcfH and YjjV are paralogs; yeast Tat-D is orthologous, with 3′ to 5′ Exo/Endo activity. |
Abbreviations: Pol, polymerase; Exo, exonuclease, dsDNA, double-strand DNA; ssDNA single-strand DNA; MMR, mismatch repair; DSB, double-strand break; BER, base excision repair; NIR, nucleotide incision repair
Eukaryotic oligoribonuclease has activity on oligodeoxynucleotides making it likely that E. coli oligoribonuclease is identical to exonuclease IV.
RNase T can act as a high-copy suppressor of some DNA repair defects of RecJ− ExoI− ExoVII− strains, has potent DNase activity and so could play a redundant function in DNA metabolism.
Biochemical properties of exonucleases
Exonucleases can be further distinguished by substrate specificity, reaction products and other enzymatic properties. Some enzymes show strong specificity; others are more ambivalent. Most of the exonucleases that degrade DNA will not degrade RNA, although there are examples of enzymes with dual specificity (e. g. RNase T found later to have robust DNase activity). Many double-strand DNA (dsDNA) specific exonucleases will only degrade one of the two strands of the duplex and therefore show a distinct polarity of degradation (3′ to 5′ or 5′ to 3′). Most single-strand DNA (ssDNA) specific-exonucleases also exhibit polarity of digestion. Some dsDNA exonucleases require 5′ phosphates; others will degrade molecules with 5′ OH ends.
Exonucleases are classified by the products of the reaction (mononucleotides vs. oligonucleotides) and whether released products contain 5′ or 3′ phosphate residues. Some exonucleases will bind substrate and execute a series of hydrolysis events before dissociation; this is termed “processivity”, which can be quantified by substrate competition experiments that assay how many nucleotides are released per a single binding event. Processivity in the many thousands of nucleotides is not uncommon. On the other hand, other exonucleases are “distributive” and release only a single nucleotide for each binding.
Structural families and exonuclease mechanism
In their function as hydrolases, exonucleases bind metal cation cofactors that assist catalysis. Most E. coli enzymes employ Mg2+ in this role. These metal ions are coordinated by conserved aspartate, glutamate or histidine residues that define certain structural families of evolutionarily related enzymes. Most of these families can be found in all three domains of life, archaea, eubacteria and eukaryotes. Except for the metal-binding, architectural residues, the overall sequence conservation among nucleases in these families is weak. Although it is possible to assign an unknown function to a structural family by iterative sequence searches, note that this is not predictive of substrate specificity or other enzymatic properties, nor even of nuclease activity. Indeed, as is described below, nuclease superfamilies include non-nuclease members such as phosphatases.
The largest structural family, with 8 members in E. coli, are the 3′ to 5′ exonucleases in the DEDD family, named for conserved aspartate and glutamate residues in the active site; this family has also been termed the DnaQ superfamily (134) (Figure 1A). This family is highly diverged and found in all domains of life, with little sequence conservation except the metal binding sites. In this family within E. coli and Salmonella are the proofreading exonuclease activity of DNA polymerase III, encoded by the DnaQ, epsilon subunit, and the exonuclease domain of DNA polymerases I and II. Exonuclease I, X and RNase T are likewise in this family, as is RNase D and oligoribonuclease (ORN).
Figure 1.
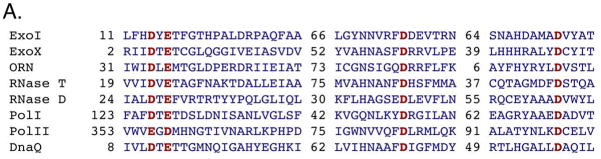
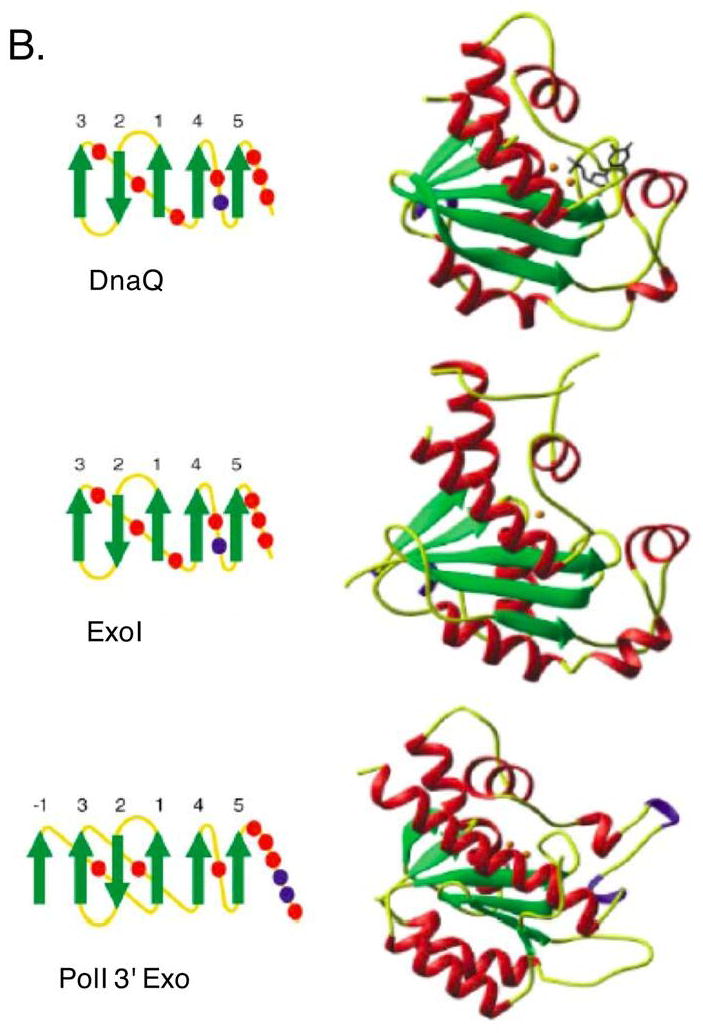
A. E. coli’s DnaQ superfamily members. Shown are aligned amino acid sequences of exonuclease I, exonuclease X, oligoribonuclease, RNase D, RNase T, and the 3′ exonucleases of DNA polymerases I, II and III. Conserved acid residues are shown in bold and comprise metal coordination residues for those proteins with determined three-dimensional structure. Numbers refer to amino acid residues not shown. B. Structure of three members of the DnaQ/DEDD superfamily: DnaQ, ExoI and Polymerase I 3′ exonuclease domain. Figure republished with permission from Hamdan et al. 2002.
The 3′ exonuclease domain of DNA polymerase I was one of the first DNA exonuclease structures determined by X-ray crystallography. The exonuclease site was surprisingly distant from the polymerase site and appeared to interact with single-strand DNA. Two metal cations were revealed in the active site (14, 15), establishing a new paradigm for phosphodiester bond hydrolysis. One metal was deduced to promote formation of the attacking hydroxide ion that will hydrolyze the phosphodiester bond; the other stabilizes the pentacoordinated transition state and/or oxyanion leaving-group. The amino acid residues interacting with the two metal ions are those seen conserved in this family, which serve to establish the architecture of the attacking water molecule, single-strand DNA substrate and metal ions.
Although the primary sequences of this family are highly diverged, their overall topologies and structures are quite similar (Figure 1B). There are X-ray crystallographic structures for DnaQ (103), ExoI (22), RNase T (292), RNase D (291) ORN (39), Polymerases I (14, 15) and II (Brunzell, PDB ID1q8i, unpublished). The common structural feature can be incorporated in a variety of quaternary arrangements: this group includes monomeric proteins (ExoI, RNaseD, the Pol I, Pol II exonucleases), homodimers (RNase T, ORN, ExoX) as well as members of larger complexes (DnaQ).
A second superfamily of nucleases includes two of E. coli’s exonucleases (RecB of the RecBCD nuclease and Exonuclease VIII) in addition to bacteriophage lambda exonuclease, Mrr-like restriction endonucleases and the archaeal Holliday junction resolvase from Pyrococcus furiosus. Note that this group includes true endonucleases, true exonucleases and endo/exonucleases as well as structure-specific enzymes. This family is defined by three motifs, with conserved glutamate and aspartate residues, defining the “endonuclease fold” (10). This fold is also found in defunct, inactive nucleases such as RecC, where it may play a structural role (210).
A number of exonucleases share structure with phosphatases and comprise a larger family of phosphoesterases. This includes RecJ, a member of the DHH family (for a conserved motif in the protein). The DHH family, which includes the polyphosphatase PPX1, is found in all three domains of life (9, 204, 239). This group particularly expanded in the archaeal species, which harbor multiple members, including Methanococcus jannashii with six DHH family members and Archaeoglobus fulgidus with seven. Likewise, SbcCD, a coiled coil protein with endonuclease and exonuclease activity is related to a family of repair enzymes such as the eukaryotic Rad50/Mre11 and T4 bacteriophage gp46/47. SbcD, the nuclease subunit, shares conserved residues (DXH-(25)-GDXXD-(-25)-GNHD) with a family of serine/threonine phosphatases, including lambda phosphatase (226, 284). In both nucleases and phosphatases, the conserved motifs of these phosphoesterases define metal binding sites involved in catalysis or substrate binding.
Biological role of exonucleases
Exonucleases play key cellular roles in mutation avoidance and genome stability, DNA repair and recombination, which will be discussed as each exonuclease is introduced below. Although no two exonucleases in E. coli are exactly alike in their properties, the biological roles of exonucleases are often obscured by functional redundancy. This is especially a problem with the single-strand DNA-specific exonucleases, with some phenotypes not apparent until four members are knocked out.
E. COLI EXONUCLEASES: PROPERTIES, STRUCTURE AND FUNCTION
The proofreading exonucleases associated with DNA polymerases I, II and III
Three of E. coli DNA polymerases have associated 3′ to 5′ exonuclease activities. In DNA polymerases I and II, the exonuclease activity resides on a domain within the polymerase subunit; in DNA Polymerase III, the exonuclease is encoded by a separate subunit, epsilon, the product of the dnaQ (mutD) gene. These associated 3′ to 5′ activities increase the fidelity of DNA synthesis (reviewed in (137)).
DNA polymerase III (Pol III) performs the bulk of DNA replication of E. coli. The holoenzyme it is composed of 10 distinct subunits, including the polymerase core, processivity clamp and clamp loader complex (see module 4.4.2). The DNA polymerase III core (176, 179, 218) consists of three subunits: 130 kD alpha, encoded by dnaE, in which the polymerase activity resides, 27 kD epsilon, encoded by dnaQ/mutD gene with the 3′ to 5′ exonuclease activity and the small 9 kD theta protein, encoded by the holE gene. The holE gene is non-essential (229) and the role of the theta in the complex is still incompletely understood, although theta does appear to stabilize the epsilon subunit in vitro and in vivo (237, 240). The DnaQ protein consists of two domains connected by a flexible linker: the N-terminal domain possesses the exonuclease activity and the C-terminal domain interacts with alpha and theta (197). The structure of the DnaQ exonuclease domain shows similarity with other members of the DEDD superfamily (103). The alpha and epsilon subunits interact not only physically but also mutually stimulate each other’s activities in vitro (175, 237).
Mutations isolated in the dnaQ gene, such as the classic dnaQ49 and mutD5 “mutator” alleles, cause a greatly elevated spontaneous mutation rate due to increased replication errors and saturation of mismatch repair capabilities (217). The dnaQ49 allele, which causes temperature-sensitivity on rich salt-free media (110) appears to affect the interaction with alpha and theta (117), whereas these interactions are preserved in mutD5. Mutations in two of the metal-binding carboxylates, dnaQ926, in the first conserved motif of the exonuclease active site, produces a strain that is essentially inviable due to the catastrophic accumulation of mutations (so-called “error catastrophe”). This strain survives only in combination with compensating antimutator alleles in the alpha, dnaE-encoded, polymerase subunit or with overproducers of the MutL mismatch repair protein (85).
The DNA Polymerase I (Pol I) 3′ to 5′ exonuclease activity, was purified as Exonuclease II from E. coli and later found to reside within the same 103 kD protein as the polymerase activity (148). Polymerase I is the most abundant DNA polymerase in E. coli (at approximately 400/cell) and is the polymerase believed to mature Okazaki fragments produced by Polymerase III-mediated lagging strand synthesis, including the removal of the RNA primer. Two domains in Polymerase I can be separated by proteolysis (126): a small N-terminal fragment containing the 5′ to 3′ exonuclease activity (see below) and the large C-terminal fragment (also known as the Klenow fragment) which possesses both polymerase and 3′ to 5′ exonuclease activities. The 3′ to 5′ exonuclease activity of DNA polymerase I bound to single-strand DNA was one of the first exonucleases to be characterized structurally ((14, 15), see discussion above). The polymerase and 3′ to 5′ active sites are quite distant at 35 Angstroms. The exonuclease binds to 3′ single-strand DNA, and 3 nucleotides are displaced from the polymerase active site. Two metal ions occupy the active site, coordinated by conserved residues that comprise the motifs that define the DEDD/DnaQ superfamily (16, 69).
Although by analogy to other proofreading activities associated with DNA polymerases, the 3′ exonuclease in Polymerase I should contribute to replication fidelity, this has been difficult to establish genetically. Inactivation of the Pol I 3′ exonuclease causes a modest, 2–4 fold increase in specific lacZ reversion rates. This is apparent in an orientation-specific manner, confirming the expectation that Pol I errors arise primarily on the lagging strand (177). In this study, deficiency in the 3′ exonuclease of Pol I did not strongly enhance the mutator phenotype conferred by defects in the Polymerase III holoenzyme, suggesting that Pol I does not readily replace Pol III in the processing of misincorporation errors.
DNA Polymerase II (Pol II) of E. coli also possesses an associated 3′ proofreading exonucleases. The polB gene is induced seven-fold by DNA damage via the SOS response (20), although is found at levels comparable to Polymerase III (about 30–50 molecules per cell) in uninduced cells (203). Of the three SOS DNA polymerases (Polymerases II, IV and V), the 90 kD Polymerase II is the only one with an intrinsic proofreading exonuclease. Pol II has a low rate of misincorporation in vitro and the loss of the proofreading activities elevates mutations 13–240-fold (29). In vitro, Polymerase II can polymerize DNA opposite abasic sites (21) and is required for efficient replication restart in UV-irradiated E. coli (206). Pol II is also involved in error-prone bypass of acetylaminofluorene lesions in vivo (191). In vivo, in the absence of mismatch repair and in the presence of an antimutator allele of DNA polymerase III (dnaE915), inactivation of Polymerase II’s exonuclease activity increases mutagenesis, as assayed by mutation to rifampicin resistance in rpoB; this is especially apparent at one specific G to A mutational hotspot (205). Mutations in F′ lacZ that occur during lactose selection (so-called “adaptive mutations”) are also elevated by inactivation of Pol II proofreading (87). Loss of Pol II proofreading has a synergistic effect on mutation rates in combination with mutator Pol III alleles in dnaQ and dnaE, indicating that Polymerase II proofreading can process mispairs created by Polymerase III during chromosomal replication (12). The spectrum of mutations in rpoB exhibited by polB mutants also supports the idea that there is polymerase switching between Pol III and Pol II during DNA replication, in which Pol II plays a largely antimutagenic role (60).
The 5′ exo/endonuclease activity of DNA polymerase I
A second hydrolase activity, associated with DNA polymerase I, specific for 5′ ends, was discovered by two groups (72, 127) and was known initially as Exonuclease VI. Proteolysis showed this activity resides in the small N-terminal domain of the protein, separated from the large fragment (also known as the Klenow fragment (126)) containing the 3′ exonuclease and polymerase activity of enzyme (221, 222). The 5′ nuclease activity is distributive (247), and limited relative to 3′ exonuclease (221, 222). The 5′ exonuclease, coupled to the DNA polymerase activity, affords Pol I its “nick translation” activity, wherein 5′ to 3′ polymerization at a nick is accompanied by 5′ to 3′ digestion of the DNA ahead of the polymerase (149). Mutations in the 5′ to 3′ exonuclease produce lethality under certain growth conditions but not those in the DNA polymerase or 3′ to 5′ exonuclease activities (reviewed in (149)).
The 5′ exonuclease activity of Pol I from Thermus aquaticus and E. coli is not a simple exonuclease but rather a structure-specific endonuclease activity, which cleaves the junction between a 5′ single-strand and duplex (172, 173). In the intact enzyme, polymerase activity I at a nick could displace DNA ahead of the enzyme, resulting in a 5′ flap that is then cleaved by the 5′ endo/exonuclease. The nuclease activity, although endonucleolytic in nature, requires a free 5′ end to thread through the enzyme (173). This “flappase” activity is rather inefficient on long 5′ ssDNA tails, assuring that is specific to the types of substrates encountered during its role in Okazaki fragment maturation during DNA replication (see below). Both Taq and Eco Pol I cleave between the two paired bases closest to the 5′ end (173, 270) and Taq Pol I cleavage is most efficient when a primer strand is juxtaposed to the flap as would be presented during Okazaki fragment processing. DNA polymerase I binds to the beta processivity clamp (163), but the consequences of this interaction on 5′ cleavage activity is not yet reported.
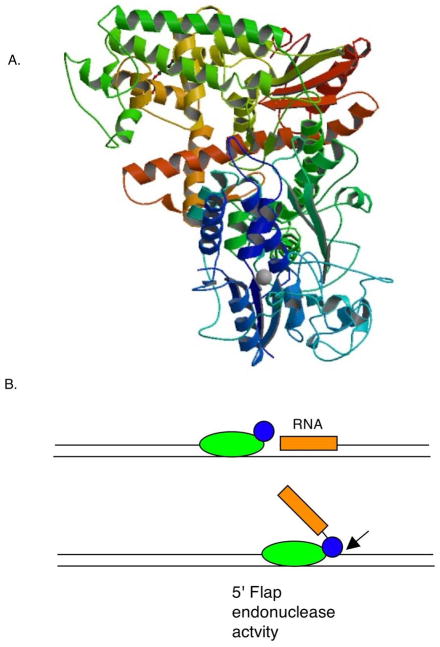
The 5′ exonuclease has motifs characteristic of a large group of nucleases found in prokaryotes and eukaryotes, the Rad2/XPG family and is structurally and functionally equivalent to the DNase IV/Fen-1 (“Flap endonuclease 1”) enzyme of mammals (156, 211), RAD27/RTH1 of yeast (231) and bacteriophage T4 RNase H (187) and T5 5′ exonuclease (32). The structure of Taq DNA polymerase I (Figure 2A) indicated three metal binding sites with two of these in close proximity (125). The residues that define these two sites are highly conserved (99) and when mutated are defective in cleavage activity (269). Evident in the structures of this class of enzymes is the presence of a helical arch in the protein that creates a hole that can accommodate single-strand but not double-strand DNA (32).
Figure 2.
A. Structure of Taq DNA polymerase I, showing 5′ exonuclease domain in blue and cyan. Image from RCSB PDB (www.pdb.org), PDB ID: 1TAQ (Kim, Y., S. H. Eom, J. Wang, D. S. Lee, S. W. Suh, and T. A. Steitz. 1995. Crystal structure of Thermus aquaticus DNA polymerase. Nature 376:612–616). B. Okazaki maturation by Polymerase I. Polymerization at a gap causes displacement of lagging strand RNA primer (orange), which is cleaved by 5′ flap endonuclease activity.
Mutants in the 5′ exonuclease activity of Pol I, polAex1− (133) were originally isolated as hyper-recombinational and subsequently found to be mildly sensitive to UV and methyl methanesulfonate (MMS) and temperature-sensitive for growth on rich medium. In this mutant, polymerase activity is normal and the 5′ to 3′ exonuclease activity is specifically reduced. Okazaki fragment maturation is delayed in these mutants (252), suggesting a role for PolI in removal of RNA primers and formation of ligatable DNA chains (Figure 2B). Supporting the idea that the PolA 5′ exonuclease is the functional equivalent of the eukaryotic FEN1/RAD27 function, the N-terminal domain of polA can partially complement rad27 mutants of Saccharomyces cerevisiae (238) and the yeast gene can partially complement polA107Ex-mutants (196).
The polA gene, under certain growth conditions, can be completely deleted but such mutants are lethal in combination with those in the xni/xgdG gene, a paralogous gene to the 5′ exonuclease domain of polymerase I (91), see below.
Mutants in the 5′ exonuclease of DNA polymerase I are also sensitive to peroxide (19). The 5′ exonuclease activity has also been implicated in the repair of daughter strand gaps that are formed after UV irradiation, a process known as “post-replication repair”. Gap-filling is defective in polA546, defective in 5′ to 3′ exonuclease activity (225).
Deficiency in the 5′ exonuclease of DNA polymerase I causes an increase in certain types of mutation (185, 190), as was established first for its yeast counterpart, RAD27 (RTH1). This includes expansions of simple sequence repeats (116), expansions of CTG/CAG trinucleotide repeats (89) and duplication mutations at short sequence repeats (250). Displacement of 5′ DNA ends by DNA polymerase during Okazaki fragment maturation, coupled with the failure to be cleaved by the 5′ flappase activity and ligation to the growing chain can lead to the observed duplication mutations.
Exonuclease I
Exonuclease I (ExoI) was the first exonuclease to be purified and characterized from E. coli (146, 147); it was identified as a Mg2+-dependent 3′ to 5′ exonuclease, digesting DNA to mononucleotides. It has potent activity, degrading up to 10,000 nucleotides/min and is strongly specific for ssDNA. ExoI dissociates upon encountering dsDNA (25). It is active as a 55 kD monomer (200). ExoI is a member of the DnaQ superfamily and its structure (22) is highly similar to the 3′ to 5′ exonucleases active site of the Klenow fragment of DNA polymerase I. ExoI is not widespread in bacterial genomes; it appears to be restricted to the gamma-proteobacteria.
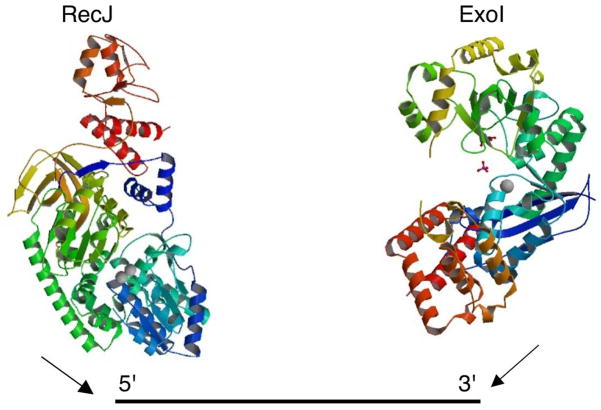
Exonuclease I is a processive exonuclease (247). Its structure indicates an extended binding site encompassing 12 residues (22, 23) and appears have the potential to encircle its substrate, a simple way to achieve processivity (Figure 3). The active site is contained in a deep positively charged groove in the structure that is wide enough only to contain ssDNA, explaining its strong specificity for single-strands.
Figure 3.
Structures of single-strand DNA specific exonucleases RecJ exonuclease and Exonuclease I (Images from RCSB PDB (www.pdb.org) ID: 2ZXP Wakamatsu, T., Y. Kitamura, Y. Kotera, N. Nakagawa, S. Kuramitsu, and R. Masui. 2010. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J Biol Chem 285:9762–9769 and PDB ID: 1FXX Breyer, W. A., and B. W. Matthews. 2000. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol 7:1125–1128). RecJ digests ssDNA 5′ to 3′ and ExoI 3′ to 5′. Both produce mononucleotide products.
The single-strand DNA binding protein, SSB, stimulates digestion of DNA by ExoI (169, 214), partly by removal of substrate secondary structure that would impede the enzyme, but also by recruitment through specific physical interactions (169, 215). ExoI binds to the C-terminal domain of SSB, the site of its interactions with a number of DNA metabolic enzymes (169). A peptide derived from the C terminal domain of SSB binds to ExoI through interactions in the pocket between its exonuclease and SH3-like domain (169).
ExoI is a part of a network of replication and repair proteins interacting with SSB (27), which includes RecJ exonuclease, Topoisomerase I, RecO recombination protein, RecG and RecQ helicases. Preparations of SSB are notoriously contaminated with trace amounts of ExoI. This led to the discovery that ExoI can stimulate RecA-mediated DNA strand exchange between linear duplex and circular single-strand molecules, when the 5′ ends are heterologous (13, 132). ExoI processively digests the displaced 3′ strand from the linear duplex and drives the reaction forward by degradation of the competitor strand for pairing. (RecJ exonuclease, with opposite polarity to ExoI, has similar properties, see below.) The recruitment of ExoI, and other repair factors, to gaps in DNA through interactions with SSB may constitute a tool-kit for gap-repair.
Mutations in structural gene for exonuclease I, sbcB, were originally isolated by their ability to suppress UV-sensitivity and recombination deficiency of recBC mutant strains (named for suppressor of recBC)(139, 141). The suppressor is attributed to the accumulation of long 3′ ssDNA tails (that would normally be degraded by ExoI) that permit the alternative recombination pathway, the RecFOR pathway, to operate. In these strains, co-suppressor mutations in the endo/exonuclease SbcCD have accumulated and contribute to suppressor phenotype (see below). Null mutations in the gene for Exo I (alternatively named “xonA”) were isolated by loss of nuclease activity directly and are not as effective for suppression of recBCD (139, 248, 271). SbcB-type alleles have been found to retain ability to bind 3′ ends and protect from degradation (248); presumably they act as better suppressors that XonA-type alleles by blocking the access of other exonucleases to 3′ ssDNA ends. Supporting this idea, an sbcB allele mimics phenotypes of double mutants in Exonuclease I and VII (258).
In addition to this antirecombinational activity, ExoI can promote recombination by the RecBCD pathway, by “end-blunting” (248, 249), trimming 3′ ends to provide blunt ends required for RecBCD digestion and RecA loading. It has also been implicated in postsynaptic DNA processing of recombination intermediates (90, 256).
The absence of ExoI also promotes a number of mutations and genetic rearrangements including frameshifts in repetitive sequence runs (261), mutations templated by synthesis in a quasi-palindrome (81, 258), deletions at short direct repeats (2, 28, 84), and RecA-independent recombination reactions between short regions of homology (82). These can be explained by the ability of ExoI to scavenge displaced 3′ ssDNA ends that are intermediates of misalignment reactions that lead to mutations or genetic rearrangements. ExoI is one of the four exonucleases that promote mismatch repair (26, 50, 256). In many of the functions of exonuclease I listed above, the enzyme is often redundant with other exonucleases, including RecJ, Exonuclease VII, Exonuclease X, and SbcCD (see below).
Exonuclease III
Exonuclease III (ExoIII) was identified as an exonuclease activity associated with a phosphatase (208, 209). Exonuclease III is a Mg2+-dependent 3′ to 5′ exonuclease on dsDNA, releasing 5′ phosphomononucleotides as degradation products. The enzyme will not degrade ssDNA. ExoIII will degrade only one strand of the duplex in the 3′ to 5′ direction such that extensive ssDNA-tailed molecules or fully ssDNA is produced. Exonuclease III does not require a dsDNA blunt end and will degrade from a nick on a circular molecule in the 3′ to 5′ direction, producing single-strand gaps. ExoIII is a distributive enzyme, dissociating frequently during the course of digestion (247).
ExoIII will remove a 3′ phosphate in a Mg2+-dependent manner, releasing inorganic orthophosphate. It will remove other 3′ residues damaged by radiation or oxidation, such as 3′ phosphoglycolate, 3′ phosphoaldehyde and urea N-glycosides (68, 106, 138, 183, 193). Exonuclease III also possesses endonuclease activity on apurinic/apyrimidinic DNA (265), an activity originally purified as “endonuclease II”. The incision at abasic sites is 5′ to the abasic residue. ExoIII is the most abundant abasic endonuclease in E. coli, accounting for 80% of the activity (266). ExoIII also possesses RNase H activity, degrading the RNA residues of a RNA:DNA hybrid molecule (266). Each of these activities is believed to be manifest from a single active site. ExoIII is therefore a key enzyme for the repair of depurination events caused by spontaneous cleavage of the N-glycosidic bond (158) and for repair of the abasic sites that are the intermediates in the base excision repair (BER) pathways of damaged nucleotide bases.
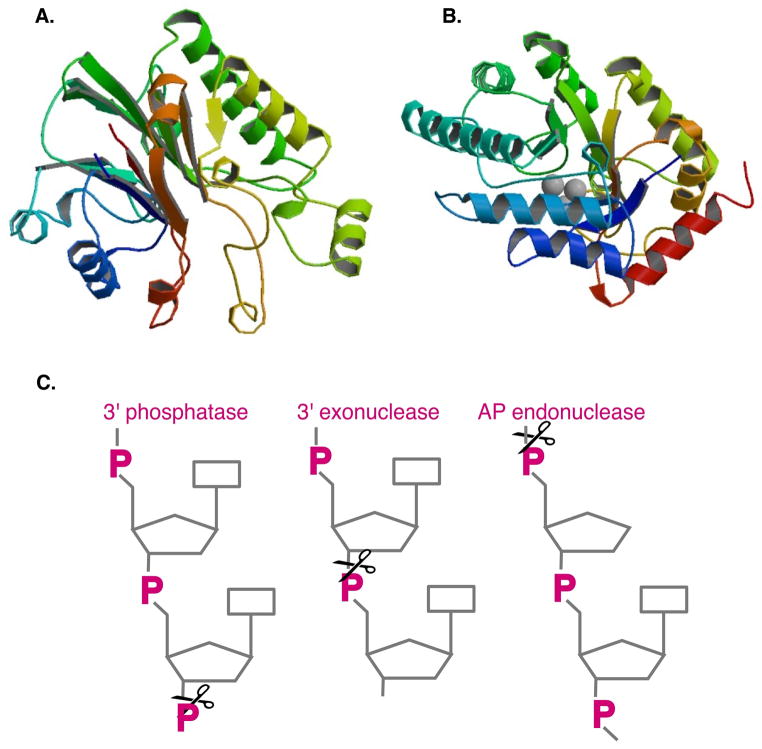
Exonuclease III orthologs are found in many organisms including mammals (67). The crystal structure of E. coli Exonuclease III (Figure 4A) shows a single metal ion active site with catalytic aspartate and histidine residues, suggested to be the single site for all the activities of the enzyme (184). Conserved residues among ExoIII and other AP endonucleases cluster in this region and the overall structure of the protein is similar to DNase I and E. coli RNase H.
Figure 4.
A. Structure of Exonuclease III (Image from RCSB PDB (www.pdb.org) PDB ID: 1AKO Mol, C. D., C. F. Kuo, M. M. Thayer, R. P. Cunningham, and J. A. Tainer. 1995. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 374:381–386). B. Structure of Endonuclease IV, showing 3 Zn2+ ions (PDB ID: 1QTW Hosfield, D. J., Y. Guan, B. J. Haas, R. P. Cunningham, and J. A. Tainer. 1999. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98:397–408). C. Activities of Exonuclease III and Endonuclease IV: 3′ phosphatase, 3′ to 5′ exonuclease, AP (abasic) endonuclease.
Mutants in exonuclease III, xthA (or xth), were isolated by direct microassay screens of E. coli lysates (181) and shown to co-defective in endonuclease II activity (272). Mutants in xthA were defective in repair of strand breaks produced by gamma-irradiation (219) and in base excision repair, a process whose intermediate is abasic sites. Mutants in dUTPase, (dut) that incorporate large amounts of uracil into DNA are inviable in the presence of additional mutations in exonuclease III; this inviability can be relieved by a mutation in uracil N-glycosylase (ung), that convert uracil residues to abasic sites (245). Mutants in xthA are hypersensitive to hydrogen peroxide (66) and near -UV light (213). Exonuclease III is controlled by the stationary phase and general stress response sigma factor, RpoS (212), and is required for the resistance to peroxide that is induced as cells begin to enter to stationary phase. Exonuclease III is also required for resistance to chain-terminating residue, 3′ azidothymidine, and is most likely the enzyme that removes this blocked 3′ residue in vivo (Cooper and Lovett, unpublished).
The exonuclease activity of Endonuclease IV
Endonuclease IV (EndoIV) is a Zn2+-dependent apurinic endonuclease, cleaving DNA at abasic sites (153, 159). Exonuclease III, also a potent AP endonuclease, accounts for about 80% of the cleavage activity, whereas EndoIV constitutes the bulk of the remaining activity (59, 266). Unlike ExoIII, EndoIV can also incise 5′ to oxidized residues such as 5-hydroxyuracil, 5,6-dihydrothymine and 2,6-diamino-4-hydroxy-5-N methylformamidopyrimidine (“Fapy”) residues (114). In a process known as “nucleotide incision repair” (NIR), this incision would allow DNA polymerase I to displace and cleave a flap with the damaged residue. Although it had been reported to have negligible exonuclease activity, EndoIV does indeed possess an intrinsic 3′ to 5′ exonuclease activity, in addition to its endonuclease activity, detected for both E. coli and Thermotoga maritima enzymes (124). Substrates with recessed 3′ ends were found to be preferred substrates for its exonuclease, which is highly sensitive to ionic strength, metal ions, EDTA and reducing agents (124) expaining why early attempts to assay exonuclease may have failed. This activity can process nicks to gaps but is reduced when the 5′ residue is an abasic deoxyribose, as would be produced by EndoIV AP endonuclease activity (95). Endo IV can release 3′ phosphates and phosphoglycoaldehydes with a Km of 50–100 nM (152).
The crystal structure of Endonuclease IV (Figure 4B) indicates the presence of three Zn2+ atoms, which generate the attacking nucleophile by deprotonation of water, stabilize the transition state and the leaving group (111, 115). Mutational analysis shows that incision and exonuclease depend on common residues (95). This exonuclease activity, as that of ExoIII, mostly likely plays a role in the removal of 3′ blocking lesions in bacterial cells, including 3′ phosphates and phosphoglycolates (152). Although EndoIV plays a secondary role to ExoIII in the processing of hydrogen peroxide-induced lesions, Endonuclease IV appears to be the primary processor of 3′ lesions induced by bleomycin treatment (151), tert-butyl hydroperoxide, and nitric oxides (59, 151, 195).
Mutants in nfo, the gene for EndoIV, are sensitive to alkyating agents such as MMS, mitomycin C, and to the oxidants tert-butyl hydroperoxide and bleomycin. The sensitivity to tertbutyl hydroperoxide and bleomycin is more severe than that demonstrated by mutants in Exonuclease III (xthA) suggesting some specialization of EndoIV for the processing of free radical lesions. In combination with mutations in xthA, an nfo mutation further enhances the killing by hydrogen peroxides, MMS and gamma rays (59). Endonuclease IV is induced by superoxide generators such as paraquat, plumbagin, menadione and phenazine methosulfate; induction was more extensive in sodAB mutants lacking superoxide dismutase (33). The nfo gene is regulated as part of the SoxRS-dependent response to superoxide stress (3, 97, 251).
Exonuclease IV
Exonuclease IV activity has been described in one paper (118) and the gene encoding this activity has never been identified. Exonuclease IV was characterized by its stronger activity on short DNA oligonucleotides than on native or denatured DNA of longer length. Its products were 5′ phosphorylated mononucleotides.
One likely possibility is that Exonuclease IV is identical to oligoribonuclease (ORN), the product of the orn gene, an Mg2+-dependent 3′ to 5′ RNA exonuclease with preference for short oligonucleotides of 5 nucleotides or less in length (63, 275, 281, 286). ORN is a member of the DEDD DnaQ superfamily of 3′ exonucleases, which includes Exonucleases I, X, RNase T, RNase D and the proofreading exonucleases of DNA polymerases I, II and III. The orn gene is essential for viability of E. coli (92) and is presumed to be required for mRNA decay and nucleotide recycling. ORN is functional as a dimer (281), similar to RNase T (155) and Exonuclease X (Viswanathan and Lovett, unpublished). Activity on DNA has not been reported, but ORN, like RNase T, may degrade both RNA and DNA oligomers. The likelihood that ORN is DNA Exonuclease IV is encouraged by the fact that E. coli ORN will hydrolyze a 5′ nitrophenol ester of thymidine (deoxyribose) monophosphate (274) and the human homolog of ORN, Sfn, degrades both RNA and DNA oligonucleotides (192).
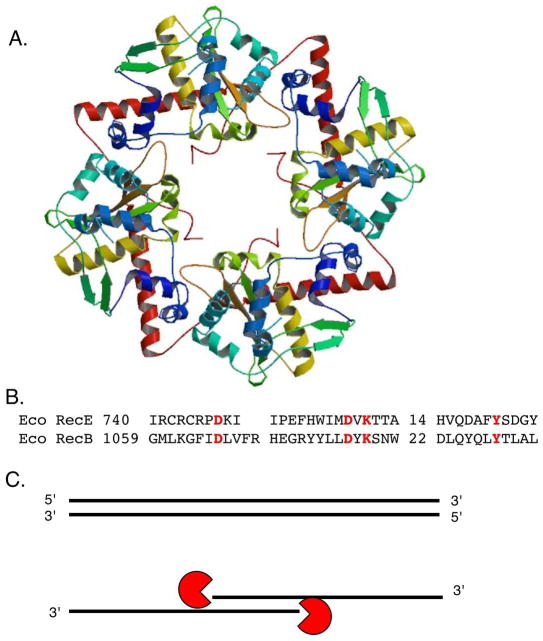
RecBCD (Exonuclease V)
The RecBCD nuclease, originally termed Exonuclease V, is a multifunctional enzyme combining ATP-dependent DNA helicase activity with Mg2+-dependent endonuclease activity (see review (76)). It is a heterotrimer composed of two large subunits, RecB (134 kD) and RecC (129 kD) and one small subunit, RecD (67 kD). RecBCD requires a blunt end to initiate unwinding and degradation (123, 201, 244) and in the presence of Mg2+ and ATP is an extraordinarily potent exonuclease, degrading hundreds to thousand of bases per second. Products of digestion are oligonucleotides and RecBCD digests both strands of the duplex. In limiting Mg2+ conditions, the nuclease activity is suppressed and the enzyme acts primarily as a helicase, nicking the DNA occasionally. The nuclease active site is present in the C-terminal domain of RecB that catalyzes cleavage of both strands (264, 276, 277) and both the RecB and RecD subunits act as motors to translocate the complex on ssDNA (77, 78, 234, 242). In the absence of the RecD subunit, the nuclease activity is diminished; the RecBC complex retains weak helicase activity, although it is less processive (136, 198).
Considerable interest in RecBCD concerns its ability to react to specific octomeric DNA sequences, known as Chi sites, which alter the properties of the enzyme in vitro and in vivo. When the enzyme encounters Chi (5′ GCTGGTGG 3′), its helicase activity becomes attenuated, the strandedness of its nuclease activity is altered and a RecA-loading function is revealed. DNA is unwound by two helicase motors, in the RecB and RecD subunits, operating on the 5′ and 3′ ended strands, respectively (77, 242). Initially, when RecBCD loads on a linear duplex molecule, the RecD motor translocates faster than the RecB motor, which extrudes a loop from the complex as the enzyme moves along DNA. Forward progress of the complex is arrested when the RecC subunit recognizes the Chi site; the RecD motor slows or disengages and a slow-moving complex is powered predominately by the RecB subunit (232, 233). Interaction with Chi switches the dispensation of the DNA strands relative to the nuclease active site in RecB (6, 79, 80). In the absence of Chi, the nuclease activity is directed predominantly to the 3′ ended strand, whereas after Chi recognition, the 5′ ending strand is targeted. After Chi recognition, the product of the reaction is a 3′ single-strand tailed duplex molecule. Encountering of Chi also causes RecBCD to load RecA onto the emergent 3′ single strand (7). The RecB subunit C-terminal domain, which by itself makes stable complexes with RecA, is required for RecA loading (5, 43, 235) and this activity is essential for recombination in vivo (11).
The nuclease domain of RecB is a member of structurally related endonucleases, including restriction enzymes (EcoRV and PvuII), the Vsr endonuclease involved in very short patch excision repair, lambda bacteriophage exonuclease, Pyrococcus Holliday junction resolvase and eukaryotic DNA2 protein (10). The RecB and RecD subunits possess motifs characteristic of Superfamily 1 (SF1) helicase proteins (228).
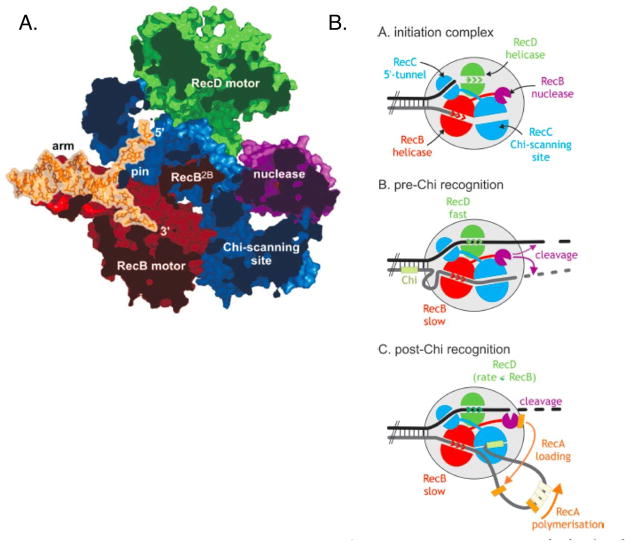
The structure of E. coli heterotrimeric RecBCD complex with partially unwound duplex oligonucleotide (Figure 5A) provided key insights in the complex behavior of the enzyme (227). The RecC protein contributes a number of architectural features including two channels for the 5′ and 3′ unwound DNA strands and a pin structure, which appears to split the duplex at the entrance to the two channels. The RecB and RecD subunits are disposed to provide motors to feed the 3′ and 5′ strands, respectively, to a single nuclease activity site in the rear of the complex, with a single Ca2+ ion bound. Residues that contribute to Chi recognition in RecC cluster in the 3′ channel preceding the nuclease domain. The RecC subunit itself surprising resembles a defunct SF1 family helicase, and is proposed to use these features as a Chi-scanning activity. Presumably, an encounter with a Chi-containing 3′ strand causes it to be bound in the Chi-scanner region, preventing its access to the nuclease site; this allows the 5′ strand to contact the nuclease site more freely (Figure 5B). RecC also possesses a defunct nuclease domain, similar to the active nuclease domain in RecB; this is proposed to act as a hoop into which the 5′ single-strand is fed (210).
Figure 5.
A. Structure of RecBCD complex with DNA. B. Activities of the enzyme, before and after Chi recognition. Figure reproduced with permission from Dillingham and Kowalczykowski 2008.
In vivo, one of the roles of the RecBCD nuclease is to defend against viral infection and replication. Bacteriophage with linear genomes or which possess linear ends during their replication (such as during rolling-circle replication) commonly overcome RecBCD digestion by encoding a specific inhibitor of RecBCD (the most well-studied of which is the Gam protein of bacteriophage lambda (188). RecBCD also inhibits rolling circle replication of bacteriophage or plasmids that lack Chi sequences. The presence of Chi sequences promotes the accumulation of high molecular weight DNA, linear concatemers whose ends are protected by RecA (61, 279).
Chi sequences are over-represented in the E. coli genome in one orientation relative to the progression of the replication fork (8, 18) and occur about once every 4 kb. The probability that RecBCD will recognize a single Chi element is about 30% in vitro (80, 243); in vivo, multiple Chi sites can additively protect linear DNA from degradation in a RecA-dependent fashion (143). Chi sequences not only aid in the protection of DNA but also stimulate homologous recombination in their vicinity (144). RecBCD-dependent recombination is proposed to repair spontaneous double strand breaks that occur in chromosomes during replication (129, 142). Cultures of mutants in recBC have lower viabilities; only about 30% of the population will form visible colonies (30).
RecBCD is required for double-strand break repair and recombination that arises from ds-ended substrates, such as those presented during conjugation and generalized transduction. (See (199)). Mutants in recB or recC have 100–1000 fold reduced inheritance of genetic markers after congugation or transduction (112). Mutants in the third subunit, recD, do not show reduction in recombination (4, 17)--in fact, they are hyper-recombinational--adding to confusion about the role of this subunit in the function of the complex. In recD mutants in which the nuclease activity of RecBCD is lost (4, 17), recombination becomes dependent on alternative 5′ to 3′ exonucleases such as RecJ exonuclease or exonuclease VII (71, 161, 167). Loss of RecJ, ExoI and ExoVII causes lethality in recD strains (70). In recD mutants, recombination still requires function of recB and recC (167) presumably to unwind DNA and to load RecA on the emerging ssDNA. In the absence of RecD subunit, RecBC loads RecA constitutively, even in the absence of Chi sequences (42).
RecJ exonuclease
The properties of RecJ exonuclease are similar to Exonuclease I, with reverse polarity: it degrades ssDNA in a 5′ to 3′ polarity in a reaction that requires Mg2+ (166). It is a processive monomeric exonuclease of 60 kD, degrading DNA to mononucleotides at a maximal rate of several thousand nucleotides/min (104, 166) and a Kd of 10 nM. RecJ does not require a terminal 5′ phosphate and will digest equally well DNA terminating in 5′ OH. RecJ has no activity on blunt dsDNA and requires at least 6 unpaired bases to bind and to initiate degradation (104). Once bound to a ssDNA -tailed molecule, RecJ can digest into a dsDNA region to a limited extent but most often terminates digestion at the ds/ssDNA boundary (104). As with ExoI, SSB stimulates the ability of RecJ to bind and to digest DNA (104). Although interactions between E. coli RecJ and SSB proteins have not been yet demonstrated, the heterologous T4 gp32 ssDNA binding protein did not stimulate nuclease activity, suggesting a specific interaction is responsible for stimulation. Direct interaction between Haemophilus influenzae RecJ and SSB has been established, between the catalytic core domain of RecJ and the C-terminal domain of SSB (224).
RecJ is the best studied member of the DHH family of phosphoesterases (9, 239) and RecJ orthologs are found in virtually all bacterial genomes with the notable exception of Mycoplasmas and Mycobacteria. Initially five (9)) and later seven motifs (204, 239) were found to define this class and mutational studies have verified the essentiality of residues in the seven motifs for nuclease activity in vitro and biological function in vivo (239). Motif 5 of seven (204), (not identified in the earlier five-motif comparison (9)), with conserved asparagine, appears to be present specifically in nuclease members of this family. The structure of a C-terminally truncated Thermus themophilus RecJ shows a C-shaped molecule, with a narrow cleft into which ssDNA but not dsDNA can access the metal ion active site at the wall of this cleft (273). Only one Mn2+ ion was found in the structure, but the location of conserved aspartate residues in its vicinity suggest the possibility of a second metal binding site, with the metal recruited or stabilized by DNA binding. Indeed, the recent structure of the fully intact Tth RecJ (263), with four domains shows two metal-ions (Figure 3). Domain III, partially lacking in the earlier structure, has similarity to the OB-fold, explaining the higher affinity of the intact enzyme for ssDNA (263). The O-structure of the enzyme, with the hole bounded by Domain I (containing the active site metal ions), Domain II (with nuclease-specific motif 5 residues at its periphery) and OB-fold Domain III would allow it completely encircle DNA, accounting for the processivity of RecJ.
RecJ was identified as a gene required for the so-called “RecFOR” and “RecE” pathways of recombination (94, 109, 165), pathways that operate independently of the RecBCD nuclease. Both RecBCD-independent pathways employ many of the same functions (RecFOR, RecJ, RecQ and RuvABC). The RecFOR pathway is believed to be specialized for recombination at gaps in DNA (230). Although normally inefficient in catalyzing recombination from double-strand ends in E. coli, increasing the levels of 3′ DNA ends by inactivation of 3′ exonucleases, ExoI and SbcCD, or increasing expression of 3′ end-generating RecE/ExoVIII makes it more active on dsDNA substrates. (Note that in most other bacterial species, which naturally lack ExoI, the RecFOR pathway may operate more efficiently on dsDNA substrates for recombination.) In the genetic backgrounds recBC sbcBC (eliminating ExoI and SbcCD) or recBC sbcA (activating expression of RecET), deficiency in recJ leads to a 1000–10,000-fold reduction in conjugational recombination; recJ deficiency also eliminates the residual recombination seen in recBC mutants with a concomitant loss of cell viability (165). In wild-type cells, recJ is required for recombination of plasmid alleles via a pathway that also includes RecFOR (130). The interpretation of these genetic properties suggest that RecJ provides presynaptic DNA processing activity equivalent to RecBCD, for production of recombinogenic ssDNA. In wild-type strains, RecBCD is specialized for degradation from a linear end; RecJ is specialized for degradation from a single-strand gap. In combination with a helicase like RecQ, RecJ can lengthen ssDNA gapped regions or produce 3′ ssDNA tailed recombinogenic molecules from double-strand ends, in a manner comparable to the RecBCD nuclease. In a reconstituted in vitro system containing RecA and RecOR, RecJ provides the nuclease activity to permit recombination between homologous dsDNA linear and dsDNA supercoiled circular molecules; RecF and SSB are nonessential, although stimulatory to the reaction (105). Inclusion of RecQ stimulates RecJ’s exonucleolytic processing but destablizes heteroduplex production formation. Unlike RecBCD, RecJ does not possess RecA loading activity, which is contributed by the RecFOR proteins for both the RecFOR and RecE pathways of recombination.
RecJ is required, along with ExoI, for efficient recombination via the RecBCD pathway, with the double mutant reduced about 10-fold for Hfr conjugal inheritance in E. coli (261), a 30-fold reduction in Salmonella for transduction with limiting homologies (180) and approximately 4-fold reduction for lambda recombination (207) in E. coli. Like ExoI, it may also promote recombination by the RecBCD pathway by presynaptic end-blunting (248, 249) and by post-synaptic stabilization of joint heteroduplex molecules (90, 261). In vitro, RecJ can accelerate RecA-promoted branch migration, by removal of the competing strand for pairing, and can help RecA bypass regions of nonhomology in strand exchange (53).
After UV irradiation, the combined action of RecJ exonuclease and RecQ helicase activities can result in degradation of nascent DNA. This is more pronounced in mutants of RecA (40) and its loading factors RecFOR, which fail to protect gapped regions of the chromosome (56). Both RecJ and RecQ are required for rapid recovery of DNA synthesis following UV-irradiation and prevent potentially mutagenic bypass by error-prone polymerases (54). This degradation may also help to prevent replication fork reversal (55).
As a ssDNA-specific exonuclease, RecJ shares overlapping functions with other such enzymes including Exonucleases I, VII and X. Any one of these four enzymes that can catalyze the excision reaction during mismatch repair of replication errors (26, 50, 256). RecJ plays a redundant role with other single-strand DNA exonucleases for UV survival, particularly with fellow 5′ to 3′ ssdNA exonuclease ExoVII (248, 261). Either RecJ or ExoI are required to process lesions produced by gamma-irradiation to activate the regulatory SOS response to DNA damage (220). RecJ is also implicated, along with the other ssDNA exonucleases, in avoidance of a number of genetic rearrangements and recombination between short homologies. Over 90% of the deletions between short direct DNA repeats are aborted by the combined action of RecJ, ExoI and ExoX (84). Gene conversion between 80 bp homologies, which occurs independent of RecA, is stimulated almost 1000-fold by the loss of both RecJ and ExoI. The mechanism of these recombinational events is not known but appears to be targeted to the replication fork, since recombination is more efficient with exogenous ssDNA that is complementary to the lagging-strand replication template relative to that of the leading strand (82). The quadruple RecJ, ExoI, ExoVII and ExoX mutant is cold-sensitive for growth and the lethality can be alleviated by blocks in the early steps in the mismatch repair pathway, suggesting that mismatch repair in the absence of ssDNA degradation is somehow toxic (26). The quadruple mutant also shows a massive increase in RecA-independent crossover recombination at very short homologies at permissive temperature, leading to the suggestion that the cold-sensitive lethality is caused by catastrophic genetic rearrangements that result from the abnormal accumulation of ssDNA in these cells (82).
The recJ gene is found in an operon downstream of the disulfide bond isomerase gene, dsbC (182). Mutations in dsbC (originally denoted xprA) have polar effects on recJ expression (164). Expression of the operon appear is activated by cell envelope stress, in a sigmaE-dependent manner (98). RecJ levels are very low in the cell, estimated at 5 molecules per cell and expression is kept low by poor translation (100, 166).
SbcCD exo/endonuclease
The two subunit SbcCD nuclease is a combined ATPase/endo/exonuclease protein in a family that includes bacteriophage T4 gp46/47 nuclease and the eukaryotic Mre11 Rad50 Xrs2/Nbs1 complex (57). The SbcC subunit is an SMC-like coiled coil protein with an intrinsic ATPase; the nuclease activity resides in the SbcD subunit (48, 49). In vitro, SbcCD is an ATP-independent single-strand DNA endonuclease and an ATP-dependent 3′ to 5′ exonuclease (45, 47, 48). SbcCD is a structure-specific nuclease, showing preference for hairpin secondary structures in DNA, which it cleaves in an ATP-dependent fashion close to the unpaired tip (48). SbcCD can also cleave 4-strand cruciforms and can open hairpin-capped ends.
In vivo, SbcCD will cleave at secondary structures formed by inverted repeats, or palindromic DNA sequences, producing double-strand breaks (83). These breaks can be repaired by recombination between sister chromosomes (83) or by single-strand annealing at homologies flanking the break (28). SbcCD is also required to repair breaks made by restriction endonuclease activity (58) and its exonuclease activity may process the ends of broken chromosomes. In addition, its SMC character may bind and coordinate the two ends of broken DNA molecules to assist in their repair. SbcCD can remove a streptavidin/biotin moiety at a 5′ end and may therefore remove, by its endonuclease activity, covalently attached or tightly bound proteins that interfere with end processing (46). SbcCD’s 3′ exonuclease activity may digest 3′ tailed substrates in vivo, explaining its discovery as a function inhibitory to the RecFOR-mediated recombination of double-strand ended substrates (160). Mutations in SbcCD produce synergistic UV-sensitivity in combination with other ssDNA-specific exonucleases, particularly with the 3′ to 5′ exonucleases, ExoI and ExoVII (248). Loss of SbcCD and Exonuclease I blocks the “reckless” DNA degradation and anucleate cell formation provoked by RecBCD in UV-irradiated recA recombination mutants (278). This confirms that Exonuclease I and SbcCD can remove single-stranded tails (“end-blunting”) on DNA in vivo to permit RecBCD loading and subsequent DNA degradation.
SbcCD expression increases as cells starve and enter the stationary phase of growth and is at least partially under RpoS regulation, although this may be indirect (62). The SMC-like SbcC proteins localize to the replication fork as visualized as a GFP fusion, whereas the SbcD nuclease subunits appear evenly distributed through the cytoplasm (62).
Exonuclease VII
Exonuclease VII (ExoVII) possesses two subunits, a large catalytic subunit (XseA) at 53 kD and a smaller subunit (XseB) at 10 kD, with the stoichometry 1: 4 (254). The genes in E. coli are encoded by unlinked loci, xseA and xseB (253); in other bacteria, the genes are found in a single operon. Catalytic activity is found in the large subunit, whereas the small subunits are believed to regulate the activity of the enzyme. Exonuclease VII activity is strongly single-strand DNA-specific and possesses dual polarity, degrading both 3′ and 5′ ends, in a processive reaction (37, 38). It can digest DNA containing thymine-dimers. It can degrade a 3′ end as short as 4 nucleotides, and can digest into the duplex region, probably the result of its processivity and “breathing” of the duplex to form single-stranded termini. The products of the reaction are oligonucleotides, primarily 4–12 nucleotides in length (37), indicating an endonucleolytic mechanism, although the enzyme is classified as an exonuclease for its requirement for a free ssDNA end. ExoVII is the only DNA exonuclease activity from E. coli that does not require exogenous Mg2+ ion; it is active even in 8 mM EDTA (38). Because of the seeming absence of metal ion requirement and stimulation by phosphate, it is possible that ExoVII is a phosphorylase rather than a hydrolase enzyme, with phosphate rather than water attacking the phosphodiester bond. However, the Thermotoga enzyme does require Mg2+ and conserved aspartate residues, reminiscent of a metal binding site (145) and found invariant among xseA orthologs, including E. coli. Presumably, the E. coli enzyme is purified with a very tightly bound metal ion and is a true hydrolase. In contrast to ExoI and RecJ, ExoVII is inhibited by SSB (Cooper and Lovett, unpublished results).
Exonuclease VII is a very well conserved enzyme and xseA orthologs are found in almost every bacterial genome, as well as a few archaeal species. (Because of their small size and lack of catalytic activity, xseB orthologs are more difficult to identify.) Despite its prevalence, Exonuclease VII is the one of the most poorly understood exonucleases of E. coli. A number of residues in both subunits are conserved and are similar to the charged residue motifs found conserved in other hydrolytic metal-dependent exonucleases. The N-terminal domain of ExoVII resembles the OB fold, characteristic of single-strand nucleic acid binding proteins (145). There is no crystal structure for XseA to provide clues about the mechanism; a XseB structure can be found (Protein Data Bank, PDB ID:1VP7 “Crystal structure of Exodeoxyribonuclease VII small subunit from Bordetella pertussis at 2.40 A resolution”, Joint Center for Structural Genomics) resembling a “paper clip” of alpha helices. The sedimentation coefficient and Stokes radius of the enzyme suggests an elongated structure (38). It is not clear how the XseB subunits interact with the larger XseA catalytic subunit, nor is it known how they regulate the activity of the enzyme.
Mutants in exonuclease VII are hyper-recombinational and weakly sensitive to nalidixic acid and UV irradiation (36). For UV irradiation, exonuclease VII deficiency is strongly synergistic with recJ (261). It has been suggested that ExoVII may participate in the mechanism of “long-patch excision repair” of UV damage, suggested to be recombinational in nature (51). Specifically, a role for RecJ and/or ExoVII in widening gaps caused by UV blocks to DNA synthesis on the lagging strand been suggested, which would promote nucleation of RecA and recombinational gap filling repair (261). In contrast with the redundant role of ExoVII and RecJ for repair, there is no normal requirement for ExoVII in homologous recombination measured after conjugation (36, 261). This may be because recombinational substrates are SSB-bound; ExoVII is inhibited by SSB (Cooper and Lovett, unpublished results), whereas RecJ is not (104). Mutants in exonuclease VII are hyper-recombinational (36), perhaps due to the accumulation of lesions normally repaired by ExoVII. Mutations in xseA modestly enhance the temperature-sensitivity promoted by 5′ exonuclease-deficient alleles of Polymerase I, polA480 and polA546 (35), suggesting a potential role in processing displaced ends during Okazaki fragment maturation.
ExoVII is one of the four ssDNA exonucleases that can mediate mismatch repair in E. coli (26, 256). ExoVII seems to abort frameshift and template-switch mutations in a manner partially redundant with 3′ exonuclease, ExoI (81, 258, 261). By itself, deficiency in Exonuclease VII stimulates frameshift mutations 6–7 fold; in combination with ExoI frameshift mutations are stimulated 12–30 fold. Stimulation is seen for both +1 and −1 frameshift mutations. Some of this effect may be due to constitutive SOS induction and induction of mutagenic polymerase, DinB, in these strains, since accumulation of −1 frameshifts is alleviated by mutations that block SOS induction such as lexAind− and recA and by dinB (107). However, DinB is highly selective for effects on −1 frameshifts (262) and the ExoI and ExoVII effects are equally strong on assays detecting +1 frameshifts (261), suggesting an additional effect on mutagenesis.
Similar effects are seen for a mutational hotspot in thyA in which mutations are templated by synthesis within an inverted repeat structure (81, 258). Loss of ExoVII stimulates these events about 7-fold, which is enhanced to 40-fold by the additional loss of ExoI. These effects have been explained by the role for the exonucleases in scavenging displaced 3′ ends that result from stalled replication. In the absence of degradation, these strands misalign, causing the template-switch that leads to mutation in the presence of an inverted DNA repeat nearby.
Expression of xseA in E. coli appears to be repressed by cAMP response protein, CRP, during nutritional downshifts (113) and may be regulated by the macrophage response transcriptional regulator, SlyA, in Salmonella (236). Interestingly, in Neisseria meningitidis, xseB is up-regulated when the bacterium encounters human host cells, leading to an increase in resistance to UV light and alkylating agents (186).
Exonuclease VIII/RecE
Exonuclease VIII is encoded by the recE gene of the cryptic lambdoid Rac prophage of E. coli K-12. It possesses processive Mg2+-dependent 5′ to 3′ exonuclease activity on dsDNA (119, 120, 140) and is the functional equivalent of the lambda exonuclease (“Exo”, Red alpha) required for bacteriophage recombination. RecE is a much larger protein than lambda Exo (866 compared to 226 amino acids), although the exonuclease activity appears to reside in its approximately 300 amino acid C-terminal domain (34, 41, 170, 189). RecE belongs to an evolutionary-related extended group of nucleases, including restriction endonucleases, RecB and lambda exonuclease (10, 34). Although there is little overall sequence similarity, RecB, lambda Exo and RecE share similar residues in their active sites (Figure 6B), with RecE more closely related to RecB than to lambda Exo. Unlike lambda Exo, which requires a 5′ phosphate for its exonuclease activity, RecE exonuclease is not affected by the absence of a 5′ phosphate (120). Like lambda exonuclease, RecE is highly processive, degrading the 5′ strand dsDNA to mononucleotides (120), with a catalytic rate of 19 nucleotide/second and a Kd of 70 nM (282). RecE has no activity on ssDNA nor on dsDNA circles (120, 140).
Figure 6.
A. Structure of RecE (Image from RCSB PDB (www.pdb.org) PDB ID: 3H4R Zhang, J., X. Xing, A. B. Herr, and C. E. Bell. 2009. Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure 17:690–702). B. Alignment of active site regions of RecE and RecB nucleases. C. 5′ to 3′ exonuclease activity of RecE on dsDNA.
The crystal structure of RecE C-terminal domain shows it to be a tetrameric toroid (Figure 6A), with a central cavity large enough to surround dsDNA substrate at its front and a ssDNA product at its rear (280). This structure nicely explains RecE’s processivity on linear dsDNA substrates and lack of endonuclease activity. Similar to lambda Exo, which associates with the recombination annealing protein, Red beta, RecE protein functions in conjunction with the RecT annealing protein (44, 101, 102, 194). Both functions are required for recombination (44), even if the DNA was pre-resected (189). Specific protein interactions exist between these pairs and the heterologous pairs (Red alpha/RecT; RecE/Red beta) do not support recombination (189). This interaction allows coupling of RecE degradation of the 5′ strand to the loading of RecT on the resulting ssDNA tail to promote annealing of complementary strands (101) or strand invasion of a homologous DNA duplex (102). RecET-orthologs can be seen in many bacteriophage genomes; the Gifsy-1 prophage of Salmonella enterica serovar typhimurium LT2 strains contains RecET orthologs (150).
RecE was discovered by its ability to restore recombination function to Red mutants of lambda or to E. coli cells deficient in the RecBCD nuclease (141, 246). The recE and recT genes can restore function to lambda by recombination between the cryptic prophage and a Red− mutant lambda genome, producing a phage known as “lambda reverse” (96, 285). The Rac prophage has been lost in certain E. coli K-12 lineages, (e. g. AB1157) (121, 122). Although the recET genes are not normally expressed, zygotic induction of the genes transferred by conjugation into E. coli lacking the Rac prophage can suppress the recombination deficiency of recBCD mutants, explaining the prophage’s name (Rac for “Recombination Activation”) (168). Mutations in the Rac prophage known as sbcA (“suppressor of recBC”), produce a stable suppression of recombination deficiency of recBC mutants by driving constitutive expression of the RecET genes (41, 88, 94, 121, 131, 140, 246, 268). RecET will promote RecA-independent recombination of plasmids and lambda bacteriophage (86, 93, 241); for chromosomal recombination measured after conjugation, the RecET pathway requires additional recombination functions including RecA, RecFOR, RecJ and RecQ (94, 165, 171, 174). Interest in the RecET and lambda Red pathways has been revived because of their ability to catalyze recombination between short homologies and their practical use in bacterial genetic engineering (“recombineering” or “ET cloning”) (reviewed in (52, 255, 283)). The RecET pathway is a recombination system capable of repairing of broken dsDNA molecules by recombinational, two-ended repair (128, 241) with a homologous partner.
Exonuclease IX
Exonuclease IX (ExoIX) is encoded by the ygdG gene and possess sequence similarity to the 5′ exonuclease domain of DNA polymerase I (216). Despite its name, exonuclease activity has yet to be demonstrated for the E. coli protein. The reported 3′ to 5′ exonuclease activity of the protein (223) appears to be a consequence of Exonuclease III contamination of the preparation (108). Although the Staphylococcus aureus orthologous protein, Sau FEN, has flap endonuclease and 5′ exonuclease activity, the E. coli ExoIX enzyme does not (1). Because the E. coli protein lacks the conserved residues of the proposed metal binding site II (1), it may not possess cleavage activity or may required additional factors. E. coliExonuclease IX protein binds single-strand DNA binding protein, SSB and to the histone-like protein H-NS (1), suggesting a potential role in DNA metabolism.
Mutations in xni (ygdG), the structural gene for exonuclease IX, are synthetically lethal with those in polymerase I (91), implicating the protein in Okazaki fragment maturation. Mutations in xni do not have any detected synthetic phenotypes on recombination, UV or oxidative damage repair in combination with mutations in other single-strand exonucleases, ExoI, ExoVII, ExoX or RecJ (162).
Exonuclease X
Exonuclease X was discovered as an open reading frame with predicted similarity to the DnaQ family of 3′ to 5′ exonucleases. Purification and characterization of the 25 kD protein demonstrated it possessed Mg2+-dependent 3′ to 5′ exonuclease activity on both single-strand and dsDNA substrates, with a preference for the former (260). Despite the fact that the enzyme is distributive, it has a fairly rapid catalytic rate of 1400 nt/min/monomer on ssDNA. The Kd of the protein for ssDNA is 2 nM, confirming a very strong affinity for ssDNA. RNA does not act as a competitive inhibitor of the exonuclease activity, suggesting the protein is specific for DNA substrates.
Exonuclease X is a member of the DnaQ or DEDD superfamily and is most homologous to the proofreading activity of PolC from gram-positive bacteria and to the mammalian TREX1 protein implicated in autoimmune disorders, with which it shares many mechanistic features (157). ExoX purifies as a homodimer, similar to RNase T, another member of the family (Viswanathan and Lovett, unpublished). There is no crystal structure of the protein, although structures for TREX1 may provide some insights (24, 65).
A mutant in Exonuclease X had little phenotype on its own but effects can be seen in combination with other ssDNA exonucleases. Loss of ExoX with ExoI in recD mutants of E. coli causes a 100-fold decrease in recombinants detected after conjugation, suggesting a role in recombination secondary to ExoI, revealed when the nuclease activity of RecBCD is impaired (70). ExoX, in combination with RecJ and ExoI, plays a role in promoting genetic stability by aborting deletion between tandem repeated sequences. In the absence of these three exonucleases, deletion rate between adjacent 100 bp repeats is elevated about 20-fold (84). The quadruple mutant in RecJ, ExoI, ExoVII and ExoX is UV-sensitive, cold-sensitive for growth and exhibits a modest mutator phenotype, epistatic to MutS, suggesting a deficiency in mismatch repair (26). Confirming this, cell extracts from RecJ ExoI VII X mutant are devoid of mismatch repair capacity (256). Consistent with the known polarity of these enzymes, either RecJ or Exo VII can support mismatch repair from a nick 5′ from the mismatch whereas ExoI, ExoX and, to a lesser extent, ExoVII can support repair from 3′ direction (50, 256). The ssExo mutants are sensitive to 2-aminopurine, which increases the load of mismatches in the cell; this sensitivity can be suppressed by mutations blocking earlier steps in mismatch repair, including MutL, MutS or UvrD (26). This finding and the relatively weak mutator phenotype have been attributed to the notion that attempts at mismatch correction in the absence of exonucleases is lethal.
The mechanism of cold-lethality of the strain is not fully understood, but it can be relieved fully by mutations in the DNA helicase UvrD and partially by upstream mutations in the mismatch repair pathway, MutS, MutH and MutL (26). The displacement of ssDNA by UvrD, in part in response to mismatch correction, appears to be lethal in the absence of ssDNA exonucleases. It does not appear merely to be due to the induction of the SOS response since mutations in recA, lexA and sulA did not alleviate the lethality (Sutera and Lovett, unpublished results). The quadruple RecJ ExoI ExoVII ExoX mutant also exhibits extraordinarily high rates of recA-independent recombination at short sequence homologies, 20–70 fold elevated relative to wild-type strains at 25 bp of homology (82). The cold-sensitive phenomenon could be related to increased recombination rates, promoted by UvrD unwinding of DNA, contributing to a catastrophic destabilization of the genome.
RNase T (Exonuclease T)
RNase T was initially identified as a RNA exonuclease removing the terminal AMP residue of tRNA molecules (73–75); it also removes the two terminal residues during the maturation of 5S rRNA (154). The gene encoding RNase T was identified as a high-copy suppressor of the UV sensitivity in mutants of ssDNA exonucleases, RecJ ExoI and ExoVII (259), implicating a potential role as a DNA exonuclease. RNase T appeared to compensate specifically for ExoI in such strains, since it did not suppress recJ xseA double mutants. RNase T overexpression also appeared to support UV survival of strains deficient in nucleotide excision repair, uvrA or uvrC (259).
RNase T has robust exonuclease activity on long ssDNA substrates, degrading in a 3′ to 5′ polarity dependent on Mg2+ or Mn2+ (257). Only very weak degradation was observed on dsDNA. Its Km for ssDNA was 5 nM in contrast to a reported Km for tRNA of 14 μM. Although its mechanism was nonprocessive, RNase T could extensively degrade ssDNA, as much as 30,000 bases on a single molecule (257), in comparison to its limited degradation of tRNA or rRNA (290). (For RNA, cytosine residues in particular appear to inhibit degradation (290)). Higher affinity for ssDNA over RNA was confirmed in a second study using oligonucleotide substrates ((289). RNase T will produce blunt ends from 3′ ssDNA overhangs, such as those produced by restriction endonucleases (289). It has been reported that like its RNase activity (155), RNase T’s DNase activity requires dimerization (289).
RNase T is a member of the DEDD DnaQ superfamily of 3′ exonucleases (135). Mutation in these conserved motifs are required for RNase activity (287). A crystal structure confirms the resemblance to proofreading 3′ exonucleases and to other members of this family such as ORN and RNase D (292). The catalytic and substrate binding residues are contributed by separate molecules at the dimer interface, explaining why dimer formation is essential for function (287, 288, 292).
TatD (Exonuclease XI?)
TatD, the fourth gene in the twin-arginine transport system operon (Tat) of E. coli, has been reported to have Mg2+-dependent DNase activity (267) and has been termed named “Exonuclease XI” in one study (31). Orthologs are found in all three domains of life, including organisms that lack the twin-arginine protein secretory pathway. E. coli possesses two paralogs to TatD, YjjV, and YcfH. TatD, but not the two paralogs, has been classified as a horizontally transferred gene of E. coli (64). The yeast Tat-D protein, expressed in E. coli, has been characterized as an endonuclease and exonuclease with a 3′ to 5′ polarity (202). It will cleave both single and double-strand DNA, with a preference for the latter, and has an acidic pH optimum (pH 5). Neither TatD nor its paralogs has any effect on the secretion of proteins by Tat pathway (267). However, another study found a role for TatD in quality control of FeS proteins that are translocated by the Tat pathway, promoting rapid turnover of misfolded substrates (178), a property difficult to reconcile with proposed nuclease activity of the protein. TatD mutants of E. coli have been reported to exhibit a two-fold increase in the number of constitutive RecA-GFP foci visualized in growing, a property shared by mutants of other 3′ exonucleases such as Exonucleases III, VII, and X (31).
SUMMARY AND CONCLUSIONS
E. coli possesses 15 proteins with verified exonuclease activity and two others, Exonuclease IX (YgdG) and TatD, that are suspected exonucleases because they are orthologous to known exonucleases. All but one activity (Exonuclease IV) have defined genetic basis and there is reason to believe that Exonuclease IV is the product of the orn gene. Many of these functions are vital to mutation avoidance, genome stability, DNA repair or genetic recombination. Some exonucleases are essential for viability (DnaQ, Pol I 5′ exo and ORN), whereas loss of others is lethal in combinations. The biochemical properties of these enzymes including polarity, processivity, substrate specificity and protein interactions are suited to their biological role. Some exonucleases appear to have redundant functions in vivo and phenotypes are not evident until multiple exonucleases are eliminated. There is structural information for most of the E. coli exonucleases or their orthologs in thermophilic eubacteria.
Most exonucleases fall into structural superfamilies with members in all domains of life, suggesting that these activities evolved quite early in the history of life and play important biological roles. The DEDD/DnaQ family has been especially prolific and E. coli possesses 8 members of this family, including DNA proofreading exonucleases associated with DNA polymerases and stand-alone DNase, RNase or dual DNase/RNase activities. RecJ, Exonuclease III, Endonuclease IV and SbcCD appear to be ancient activities. Is E. coli especially blessed with exonucleases? It is difficult to say whether E. coli possesses more exonucleases than other bacterial species since systematic biochemical characterization of exonuclease activities has only taken place with E. coli. There are likely to be other exonucleases in bacteria that remain to be discovered and certainly new exonuclease activities in E. coli will be forthcoming.
Acknowledgments
Unpublished work cited was supported by NIH grants GM51753 and GM43889 to STL. I thank Steve Kowalczykowski and Katsumi Morimatsu for comments on the manuscript.
Bibliography
- 1.Allen LM, Hodskinson MRG, Sayers JR. Active site substitutions delineate distinct classes of eubacterial flap endonuclease. Biochem J. 2009;418:285–292. doi: 10.1042/BJ20081637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allgood ND, Silhavy TJ. Escherichia coli xonA (sbcB) mutants enhance illegitimate recombination. Genetics. 1991;127:671–680. doi: 10.1093/genetics/127.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amábile-Cuevas CF, Demple B. Molecular characterization of the soxRS genes of Escherichia coli: two genes control a superoxide stress regulon. Nucleic Acids Res. 1991;19:4479–4484. doi: 10.1093/nar/19.16.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson DG, Churchill JJ, Kowalczykowski SC. A single mutation, RecB(D1080A,) eliminates RecA protein loading but not Chi recognition by RecBCD enzyme. J Biol Chem. 1999;274:27139–27144. doi: 10.1074/jbc.274.38.27139. [DOI] [PubMed] [Google Scholar]
- 6.Anderson DG, Kowalczykowski SC. The recombination hot spot chi is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes & Development. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 7.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 8.Arakawa K, Uno R, Nakayama Y, Tomita M. Validating the significance of genomic properties of Chi sites from the distribution of all octamers in Escherichia coli. Gene. 2007;392:239–246. doi: 10.1016/j.gene.2006.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- 10.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold DA, Kowalczykowski SC. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J Biol Chem. 2000;275:12261–12265. doi: 10.1074/jbc.275.16.12261. [DOI] [PubMed] [Google Scholar]
- 12.Banach-Orlowska M, I, Fijalkowska J, Schaaper RM, Jonczyk P. DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli. Mol Microbiol. 2005;58:61–70. doi: 10.1111/j.1365-2958.2005.04805.x. [DOI] [PubMed] [Google Scholar]
- 13.Bedale WA, Inman RB, Cox MM. A reverse DNA strand exchange mediated by recA protein and exonuclease I. The generation of apparent DNA strand breaks by recA protein is explained. J Biol Chem. 1993;268:15004–15016. [PubMed] [Google Scholar]
- 14.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 15.Beese LS, Steitz TA. Structural basis for the 3′-5′ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernad A, Blanco L, Lázaro JM, Martín G, Salas M. A conserved 3′----5′ exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989;59:219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- 17.Biek DP, Cohen SN. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 19.Boling M, Adler H, Masker W. Restoration of viability to an Escherichia coli mutant deficient in the 5′----3′ exonuclease of DNA polymerase I. J Bacteriol. 1984;160:706–710. doi: 10.1128/jb.160.2.706-710.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonner CA, Hays S, McEntee K, Goodman MF. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- 22.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol. 2000;7:1125–1128. doi: 10.1038/81978. [DOI] [PubMed] [Google Scholar]
- 23.Brody RS, Doherty KG, Zimmerman PD. Processivity and kinetics of the reaction of exonuclease I from Escherichia coli with polydeoxyribonucleotides. J Biol Chem. 1986;261:7136–7143. [PubMed] [Google Scholar]
- 24.Brucet M, Querol-Audí J, Serra M, Ramirez-Espain X, Bertlik K, Ruiz L, Lloberas J, Macias MJ, Fita I, Celada A. Structure of the dimeric exonuclease TREX1 in complex with DNA displays a proline-rich binding site for WW Domains. J Biol Chem. 2007;282:14547–14557. doi: 10.1074/jbc.M700236200. [DOI] [PubMed] [Google Scholar]
- 25.Brutlag D, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. 36. A proofreading function for the 3′ leads to 5′ exonuclease activity in deoxyribonucleic acid polymerases. J Biol Chem. 1972;247:241–248. [PubMed] [Google Scholar]
- 26.Burdett V, Baitinger C, Viswanathan M, Lovett ST, Modrich P. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc Natl Acad Sci USA. 2001;98:6765–6770. doi: 10.1073/pnas.121183298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butland G, Peregrín-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 28.Bzymek M, Saveson CJ, Feschenko VV, Lovett ST. Slipped misalignment mechanisms of deletion formation: in vivo susceptibility to nucleases. J Bacteriol. 1999;181:477–482. doi: 10.1128/jb.181.2.477-482.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai H, Yu H, McEntee K, Kunkel TA, Goodman MF. Purification and properties of wild-type and exonuclease-deficient DNA polymerase II from Escherichia coli. J Biol Chem. 1995;270:15327–15335. doi: 10.1074/jbc.270.25.15327. [DOI] [PubMed] [Google Scholar]
- 30.Capaldo FN, Ramsey G, Barbour SD. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J Bacteriol. 1974;118:242–249. doi: 10.1128/jb.118.1.242-249.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Centore RC, Lestini R, Sandler SJ. XthA (Exonuclease III) regulates loading of RecA onto DNA substrates in log phase Escherichia coli cells. Mol Microbiol. 2008;67:88–101. doi: 10.1111/j.1365-2958.2007.06026.x. [DOI] [PubMed] [Google Scholar]
- 32.Ceska TA, Sayers JR, Stier G, Suck D. A helical arch allowing single-stranded DNA to thread through T5 5′-exonuclease. Nature. 1996;382:90–93. doi: 10.1038/382090a0. [DOI] [PubMed] [Google Scholar]
- 33.Chan E, Weiss B. Endonuclease IV of Escherichia coli is induced by paraquat. Proc Natl Acad Sci USA. 1987;84:3189–3193. doi: 10.1073/pnas.84.10.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang HW, Julin DA. Structure and function of the Escherichia coli RecE protein, a member of the RecB nuclease domain family. J Biol Chem. 2001;276:46004–46010. doi: 10.1074/jbc.M108627200. [DOI] [PubMed] [Google Scholar]
- 35.Chase JW, Masker WE. Deoxyribonucleic acid repair in Escherichia coli mutants deficient in the 5′----3′ exonuclease activity of deoxyribonucleic acid polymerase I and exonuclease VII. J Bacteriol. 1977;130:667–675. doi: 10.1128/jb.130.2.667-675.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chase JW, Richardson CC. Escherichia coli mutants deficient in exonuclease VII. J Bacteriol. 1977;129:934–947. doi: 10.1128/jb.129.2.934-947.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chase JW, Richardson CC. Exonuclease VII of Escherichia coli. Mechanism of action. J Biol Chem. 1974;249:4553–4561. [PubMed] [Google Scholar]
- 38.Chase JW, Richardson CC. Exonuclease VII of Escherichia coli. Purification and properties. J Biol Chem. 1974;249:4545–4552. [PubMed] [Google Scholar]
- 39.Chin KH, Yang CY, Chou CC, Wang AHJ, Chou SH. The crystal structure of XC847 from Xanthomonas campestris: a 3′-5′ oligoribonuclease of DnaQ fold family with a novel opposingly shifted helix. Proteins. 2006;65:1036–1040. doi: 10.1002/prot.21148. [DOI] [PubMed] [Google Scholar]
- 40.Chow KH, Courcelle J. RecBCD and RecJ/RecQ initiate DNA degradation on distinct substrates in UV-irradiated Escherichia coli. Radiat Res. 2007;168:499–506. doi: 10.1667/RR1033.1. [DOI] [PubMed] [Google Scholar]
- 41.Chu CC, Templin A, Clark AJ. Suppression of a frameshift mutation in the recE gene of Escherichia coli K-12 occurs by gene fusion. J Bacteriol. 1989;171:2101–2109. doi: 10.1128/jb.171.4.2101-2109.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churchill JJ, Anderson DG, Kowalczykowski SC. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes & Development. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Churchill JJ, Kowalczykowski SC. Identification of the RecA protein-loading domain of RecBCD enzyme. J Mol Biol. 2000;297:537–542. doi: 10.1006/jmbi.2000.3590. [DOI] [PubMed] [Google Scholar]
- 44.Clark AJ, Sharma V, Brenowitz S, Chu CC, Sandler S, Satin L, Templin A, Berger I, Cohen A. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J Bacteriol. 1993;175:7673–7682. doi: 10.1128/jb.175.23.7673-7682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connelly JC, de Leau ES, Leach DR. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Connelly JC, de Leau ES, Leach DR. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair (Amst) 2003;2:795–807. doi: 10.1016/s1568-7864(03)00063-6. [DOI] [PubMed] [Google Scholar]
- 47.Connelly JC, de Leau ES, Okely EA, Leach DR. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 48.Connelly JC, Kirkham LA, Leach DR. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Connelly JC, Leach DR. The sbcC and sbcD genes of Escherichia coli encode a nuclease involved in palindrome inviability and genetic recombination. Genes Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 50.Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 51.Cooper PK, Hanawalt PC. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 53.Corrette-Bennett SE, Lovett ST. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. J Biol Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 54.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. DNA damage-induced replication fork regression and processing in Escherichia coli. Science. 2003;299:1064–1067. doi: 10.1126/science.1081328. [DOI] [PubMed] [Google Scholar]
- 56.Courcelle J, Hanawalt PC. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol Gen Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- 57.Cromie GA, Connelly JC, Leach DR. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell. 2001;8:1163–1174. doi: 10.1016/s1097-2765(01)00419-1. [DOI] [PubMed] [Google Scholar]
- 58.Cromie GA, Leach DR. Recombinational repair of chromosomal DNA double-strand breaks generated by a restriction endonuclease. Mol Microbiol. 2001;41:873–883. doi: 10.1046/j.1365-2958.2001.02553.x. [DOI] [PubMed] [Google Scholar]
- 59.Cunningham RP, Saporito SM, Spitzer SG, Weiss B. Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol. 1986;168:1120–1127. doi: 10.1128/jb.168.3.1120-1127.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Curti E, McDonald JP, Mead S, Woodgate R. DNA polymerase switching: effects on spontaneous mutagenesis in Escherichia coli. Mol Microbiol. 2009;71:315–331. doi: 10.1111/j.1365-2958.2008.06526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dabert P, Ehrlich SD, Gruss A. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc Natl Acad Sci USA. 1992;89:12073–12077. doi: 10.1073/pnas.89.24.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darmon E, Lopez-Vernaza MA, Helness AC, Borking A, Wilson E, Thacker Z, Wardrope L, Leach DR. SbcCD regulation and localization in Escherichia coli. J Bacteriol. 2007;189:6686–6694. doi: 10.1128/JB.00489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datta AK, Niyogi K. A novel oligoribonuclease of Escherichia coli. II. Mechanism of action. J Biol Chem. 1975;250:7313–7319. [PubMed] [Google Scholar]
- 64.Davids W, Zhang Z. The impact of horizontal gene transfer in shaping operons and protein interaction networks--direct evidence of preferential attachment. BMC Evol Biol. 2008;8:23. doi: 10.1186/1471-2148-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Silva U, Choudhury S, Bailey SL, Harvey S, Perrino FW, Hollis T. The crystal structure of TREX1 explains the 3′ nucleotide specificity and reveals a polyproline II helix for protein partnering. J Biol Chem. 2007;282:10537–10543. doi: 10.1074/jbc.M700039200. [DOI] [PubMed] [Google Scholar]
- 66.Demple B, Halbrook J, Linn S. Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J Bacteriol. 1983;153:1079–1082. doi: 10.1128/jb.153.2.1079-1082.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Demple B, Herman T, Chen DS. Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci USA. 1991;88:11450–11454. doi: 10.1073/pnas.88.24.11450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Demple B, Johnson A, Fung D. Exonuclease III and endonuclease IV remove 3′ blocks from DNA synthesis primers in H2O2-damaged Escherichia coli. Proc Natl Acad Sci U S A. 1986;83:7731–7735. doi: 10.1073/pnas.83.20.7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derbyshire V, Freemont PS, Sanderson MR, Beese L, Friedman JM, Joyce CM, Steitz TA. Genetic and crystallographic studies of the 3′,5′-exonucleolytic site of DNA polymerase I. Science. 1988;240:199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- 70.Dermić D. Functions of multiple exonucleases are essential for cell viability, DNA repair and homologous recombination in recD mutants of Escherichia coli. Genetics. 2006;172:2057–2069. doi: 10.1534/genetics.105.052076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dermić D, Zahradka D, Petranović M. Exonuclease requirements for recombination of lambda-phage in recD mutants of Escherichia coli. Genetics. 2006;173:2399–2402. doi: 10.1534/genetics.106.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deutscher MP, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXIX. Hydrolysis of deoxyribonucleic acid from the 5′ terminus by an exonuclease function of deoxyribonucleic acid polymerase. J Biol Chem. 1969;244:3029–3037. [PubMed] [Google Scholar]
- 73.Deutscher MP, Marlor CW. Purification and characterization of Escherichia coli RNase T. J Biol Chem. 1985;260:7067–7071. [PubMed] [Google Scholar]
- 74.Deutscher MP, Marlor CW, Zaniewski R. Ribonuclease T: new exoribonuclease possibly involved in end-turnover of tRNA. Proc Natl Acad Sci USA. 1984;81:4290–4293. doi: 10.1073/pnas.81.14.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Deutscher MP, Marlor CW, Zaniewski R. RNase T is responsible for the end-turnover of tRNA in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:6427–6430. doi: 10.1073/pnas.82.19.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 78.Dillingham MS, Webb MR, Kowalczykowski SC. Bipolar DNA translocation contributes to highly processive DNA unwinding by RecBCD enzyme. J Biol Chem. 2005;280:37069–37077. doi: 10.1074/jbc.M505520200. [DOI] [PubMed] [Google Scholar]
- 79.Dixon DA, Kowalczykowski SC. Role of the Escherichia coli recombination hotspot, chi, in RecABCD-dependent homologous pairing. J Biol Chem. 1995;270:16360–16370. doi: 10.1074/jbc.270.27.16360. [DOI] [PubMed] [Google Scholar]
- 80.Dixon DA, Kowalczykowski SC. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 81.Dutra BE, Lovett ST. Cis and trans-acting effects on a mutational hotspot involving a replication template switch. J Mol Biol. 2006;356:300–311. doi: 10.1016/j.jmb.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 82.Dutra BE, V, Sutera A, Lovett ST. RecA-independent recombination is efficient but limited by exonucleases. Proc Natl Acad Sci USA. 2007;104:216–221. doi: 10.1073/pnas.0608293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eykelenboom JK, Blackwood JK, Okely E, Leach DRF. SbcCD Causes a Double-Strand Break at a DNA Palindrome in the Escherichia coli Chromosome. Mol Cell. 2008;29:644–651. doi: 10.1016/j.molcel.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 84.Feschenko VV, Rajman LA, Lovett ST. Stabilization of perfect and imperfect tandem repeats by single-strand DNA exonucleases. Proc Natl Acad Sci USA. 2003;100:1134–1139. doi: 10.1073/pnas.0233122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fishel RA, James AA, Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981;294:184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- 87.Foster PL, Gudmundsson G, Trimarchi JM, Cai H, Goodman MF. Proofreading-defective DNA polymerase II increases adaptive mutation in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:7951–7955. doi: 10.1073/pnas.92.17.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fouts KE, Wasie-Gilbert T, Willis DK, Clark AJ, Barbour SD. Genetic analysis of transposon-induced mutations of the Rac prophage in Escherichia coli K-12 which affect expression and function of recE. J Bacteriol. 1983;156:718–726. doi: 10.1128/jb.156.2.718-726.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 90.Friedman-Ohana R, Cohen A. Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc Natl Acad Sci USA. 1998;95:6909–6914. doi: 10.1073/pnas.95.12.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fukushima S, Itaya M, Kato H, Ogasawara N, Yoshikawa H. Reassessment of the in vivo functions of DNA polymerase I and RNase H in bacterial cell growth. J Bacteriol. 2007;189:8575–8583. doi: 10.1128/JB.00653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gillen JR, Clark AJ. The RecE pathway of bacterial recombination. In: Grell RF, editor. Mechanisms in Recombination. Plenum Press; New York, NY: 1974. pp. 123–136. [Google Scholar]
- 94.Gillen JR, Willis DK, Clark AJ. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Golan G, Ishchenko AA, Khassenov B, Shoham G, Saparbaev MK. Coupling of the nucleotide incision and 3′-->5′ exonuclease activities in Escherichia coli endonuclease IV: Structural and genetic evidences. Mutat Res. 2009 doi: 10.1016/j.mrfmmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 96.Gottesman MM, Gottesman ME, Gottesman S, Gellert M. Characterization of bacteriophage lambda reverse as an Escherichia coli phage carrying a unique set of host-derived recombination functions. J Mol Biol. 1974;88:471–487. doi: 10.1016/0022-2836(74)90496-3. [DOI] [PubMed] [Google Scholar]
- 97.Greenberg JT, Monach P, Chou JH, Josephy PD, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guisbert E, V, Rhodius A, Ahuja N, Witkin E, Gross CA. Hfq modulates the sigmaE-mediated envelope stress response and the sigma32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol. 2007;189:1963–1973. doi: 10.1128/JB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gutman PD, Minton KW. Conserved sites in the 5′-3′ exonuclease domain of Escherichia coli DNA polymerase. Nucleic Acids Res. 1993;21:4406–4407. doi: 10.1093/nar/21.18.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Haggerty TJ, Lovett ST. IF3-mediated suppression of a GUA initiation codon mutation in the recJ gene of Escherichia coli. J Bacteriol. 1997;179:6705–6713. doi: 10.1128/jb.179.21.6705-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc Natl Acad Sci USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hamdan S, Carr PD, Brown SE, Ollis DL, Dixon NE. Structural basis for proofreading during replication of the Escherichia coli chromosome. Structure. 2002;10:535–546. doi: 10.1016/s0969-2126(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 104.Han ES, Cooper DL, Persky NS, Sutera VA, Whitaker RD, Montello ML, Lovett ST. RecJ exonuclease: substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. doi: 10.1093/nar/gkj503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Handa N, Morimatsu K, Lovett ST, Kowalczykowski SC. Reconstitution of initial steps of dsDNA break repair by the RecF pathway of E. coli. Genes & Development. 2009;23:1234–1245. doi: 10.1101/gad.1780709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Henner WD, Grunberg SM, Haseltine WA. Enzyme action at 3′ termini of ionizing radiation-induced DNA strand breaks. J Biol Chem. 1983;258:15198–15205. [PubMed] [Google Scholar]
- 107.Hersh MN, Morales LD, Ross KJ, Rosenberg SM. Single-strand-specific exonucleases prevent frameshift mutagenesis by suppressing SOS induction and the action of DinB/DNA polymerase IV in growing cells. J Bacteriol. 2006;188:2336–2342. doi: 10.1128/JB.188.7.2336-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hodskinson MRG, Allen LM, Thomson DP, Sayers JR. Molecular interactions of Escherichia coli ExoIX and identification of its associated 3′-5′ exonuclease activity. Nucleic Acids Res. 2007;35:4094–4102. doi: 10.1093/nar/gkm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Horii Z, Clark AJ. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 110.Horiuchi T, Maki H, Sekiguchi M. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol Gen Genet. 1978;163:277–283. doi: 10.1007/BF00271956. [DOI] [PubMed] [Google Scholar]
- 111.Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 112.Howard-Flanders P, Theriot L. Mutants of Escherichia coli K-12 defective in DNA repair and in genetic recombination. Genetics. 1966;53:1137–1150. doi: 10.1093/genetics/53.6.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hutchings MI, Drabble WT. Regulation of the divergent guaBA and xseA promoters of Escherichia coli by the cyclic AMP receptor protein. FEMS Microbiol Lett. 2000;187:115–122. doi: 10.1111/j.1574-6968.2000.tb09146.x. [DOI] [PubMed] [Google Scholar]
- 114.Ischenko AA, Saparbaev MK. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature. 2002;415:183–187. doi: 10.1038/415183a. [DOI] [PubMed] [Google Scholar]
- 115.Ivanov I, Tainer JA, McCammon JA. Unraveling the three-metal-ion catalytic mechanism of the DNA repair enzyme endonuclease IV. Proc Natl Acad Sci USA. 2007;104:1465–1470. doi: 10.1073/pnas.0603468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast RTH1 5′ to 3′ exonuclease for the stability of simple repetitive DNA. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 117.Jonczyk P, Nowicka A, Fijałkowska IJ, Schaaper RM, Cieśla Z. In vivo protein interactions within the Escherichia coli DNA polymerase III core. J Bacteriol. 1998;180:1563–1566. doi: 10.1128/jb.180.6.1563-1566.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jorgensen SE, Koerner JF. Separation and characterization of deoxyribonucleases of Escherichia coli B. I. Chromatographic separation and properties of two deoxyribo-oligonucleotidases. J Biol Chem. 1966;241:3090–3096. [PubMed] [Google Scholar]
- 119.Joseph JW, Kolodner R. Exonuclease VIII of Escherichia coli. I. Purification and physical properties. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- 120.Joseph JW, Kolodner R. Exonuclease VIII of Escherichia coli. II. Mechanism of action. J Biol Chem. 1983;258:10418–10424. [PubMed] [Google Scholar]
- 121.Kaiser K, Murray NE. On the nature of sbcA mutations in E. coli K 12. Mol Gen Genet. 1980;179:555–563. doi: 10.1007/BF00271745. [DOI] [PubMed] [Google Scholar]
- 122.Kaiser K, Murray NE. Physical characterisation of the “Rac prophage” in E. coli K12. Mol Gen Genet. 1979;175:159–174. doi: 10.1007/BF00425532. [DOI] [PubMed] [Google Scholar]
- 123.Karu AE, MacKay V, Goldmark PJ, Linn S. The recBC deoxyribonuclease of Escherichia coli K-12. Substrate specificity and reaction intermediates. J Biol Chem. 1973;248:4874–4884. [PubMed] [Google Scholar]
- 124.Kerins SM, Collins R, McCarthy TV. Characterization of an endonuclease IV 3′-5′ exonuclease activity. J Biol Chem. 2003;278:3048–3054. doi: 10.1074/jbc.M210750200. [DOI] [PubMed] [Google Scholar]
- 125.Kim Y, Eom SH, Wang J, Lee DS, Suh SW, Steitz TA. Crystal structure of Thermus aquaticus DNA polymerase. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 126.Klenow H, Henningsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci USA. 1970;65:168–175. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klett RP, Cerami A, Reich E. Exonuclease VI, a new nuclease activity associated with E. coli DNA polymerase. Proc Natl Acad Sci USA. 1968;60:943–950. doi: 10.1073/pnas.60.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kobayashi I, Takahashi N. Double-stranded gap repair of DNA by gene conversion in Escherichia coli. Genetics. 1988;119:751–757. doi: 10.1093/genetics/119.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kolodner R, Fishel RA, Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kolodner R, Hall SD, Luisi-DeLuca C. Homologous pairing proteins encoded by the Escherichia coli recE and recT genes. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 132.Konforti BB, Davis RW. ATP hydrolysis and the displaced strand are two factors that determine the polarity of RecA-promoted DNA strand exchange. J Mol Biol. 1992;227:38–53. doi: 10.1016/0022-2836(92)90680-i. [DOI] [PubMed] [Google Scholar]
- 133.Konrad EB, I, Lehman R. A conditional lethal mutant of Escherichia coli K12 defective in the 5′ leads to 3′ exonuclease associated with DNA polymerase I. Proc Natl Acad Sci USA. 1974;71:2048–2051. doi: 10.1073/pnas.71.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koonin EV. A conserved ancient domain joins the growing superfamily of 3′-5′ exonucleases. Curr Biol. 1997;7:R604–606. doi: 10.1016/s0960-9822(06)00311-3. [DOI] [PubMed] [Google Scholar]
- 135.Koonin EV, Deutscher MP. RNase T shares conserved sequence motifs with DNA proofreading exonucleases. Nucleic Acids Res. 1993;21:2521–2522. doi: 10.1093/nar/21.10.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Korangy F, Julin DA. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry. 1993;32:4873–4880. doi: 10.1021/bi00069a024. [DOI] [PubMed] [Google Scholar]
- 137.Kornberg A, Baker TA. DNA replication. 2. W. H. Freeman; New York, NY: 1992. [Google Scholar]
- 138.Kow YW, Wallace SS. Exonuclease III recognizes urea residues in oxidized DNA. Proc Natl Acad Sci USA. 1985;82:8354–8358. doi: 10.1073/pnas.82.24.8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kushner SR, Nagaishi H, Clark AJ. Indirect suppression of recB and recC mutations by exonuclease I deficiency. Proc Natl Acad Sci USA. 1972;69:1366–1370. doi: 10.1073/pnas.69.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kushner SR, Nagaishi H, Clark AJ. Isolation of exonuclease VIII: the enzyme associated with sbcA indirect suppressor. Proc Natl Acad Sci USA. 1974;71:3593–3597. doi: 10.1073/pnas.71.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kushner SR, Nagaishi H, Templin A, Clark AJ. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci USA. 1971;68:824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol Mol Biol Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kuzminov A, Schabtach E, Stahl FW. Chi sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Larrea AA, I, Pedroso M, Malhotra A, Myers RS. Identification of two conserved aspartic acid residues required for DNA digestion by a novel thermophilic Exonuclease VII in Thermotoga maritima. Nucleic Acids Res. 2008;36:5992–6003. doi: 10.1093/nar/gkn588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lehman IR. The deoxyribonucleases of Escherichia coli. I. Purification and properties of a phosphodiesterase. J Biol Chem. 1960;235:1479–1487. [PubMed] [Google Scholar]
- 147.Lehman IR, Nussbaum AL. The deoxyribonucleases of Escherichia coli. V. On the specificity of exonuclease I (phosphodiesterase) J Biol Chem. 1964;239:2628–2636. [PubMed] [Google Scholar]
- 148.Lehman IR, Richardson CC. The deoxyribonucleases of Escherichia coli. IV. An exonuclease activity present in purified preparations of deoxyribonucleic acid polymerase. J Biol Chem. 1964;239:233–241. [PubMed] [Google Scholar]
- 149.Lehman IR, Uyemura DG. DNA polymerase I: essential replication enzyme. Science. 1976;193:963–969. doi: 10.1126/science.781842. [DOI] [PubMed] [Google Scholar]
- 150.Lemire S, Figueroa-Bossi N, Bossi L. A singular case of prophage complementation in mutational activation of recET orthologs in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190:6857–6866. doi: 10.1128/JB.00769-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Levin JD, Demple B. In vitro detection of endonuclease IV-specific DNA damage formed by bleomycin in vivo. Nucleic Acids Res. 1996;24:885–889. doi: 10.1093/nar/24.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Levin JD, Johnson AW, Demple B. Homogeneous Escherichia coli endonuclease IV. Characterization of an enzyme that recognizes oxidative damage in DNA. J Biol Chem. 1988;263:8066–8071. [PubMed] [Google Scholar]
- 153.Levin JD, Shapiro R, Demple B. Metalloenzymes in DNA repair. Escherichia coli endonuclease IV and Saccharomyces cerevisiae Apn1. J Biol Chem. 1991;266:22893–22898. [PubMed] [Google Scholar]
- 154.Li Z, Deutscher MP. The tRNA processing enzyme RNase T is essential for maturation of 5S RNA. Proc Natl Acad Sci USA. 1995;92:6883–6886. doi: 10.1073/pnas.92.15.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li Z, Zhan L, Deutscher MP. Escherichia coli RNase T functions in vivo as a dimer dependent on cysteine 168. J Biol Chem. 1996;271:1133–1137. doi: 10.1074/jbc.271.2.1133. [DOI] [PubMed] [Google Scholar]
- 156.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 157.Lindahl T, Barnes DE, Yang YG, Robins P. Biochemical properties of mammalian TREX1 and its association with DNA replication and inherited inflammatory disease. Biochem Soc Trans. 2009;37:535–538. doi: 10.1042/BST0370535. [DOI] [PubMed] [Google Scholar]
- 158.Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- 159.Ljungquist S. A new endonuclease from Escherichia coli acting at apurinic sites in DNA. J Biol Chem. 1977;252:2808–2814. [PubMed] [Google Scholar]
- 160.Lloyd RG, Buckman C. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J Bacteriol. 1985;164:836–844. doi: 10.1128/jb.164.2.836-844.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lloyd RG, Porton MC, Buckman C. Effect of recF, recJ, recN, recO and ruv mutations on ultraviolet survival and genetic recombination in a recD strain of Escherichia coli K12. Mol Gen Genet. 1988;212:317–324. doi: 10.1007/BF00334702. [DOI] [PubMed] [Google Scholar]
- 162.Lombardo M-J, Aponyi I, Ray MP, Sandigursky M, Franklin WA, Rosenberg SM. xni-deficient Escherichia coli are proficient for recombination and multiple pathways of repair. DNA Repair (Amst) 2003;2:1175–1183. doi: 10.1016/s1568-7864(03)00135-6. [DOI] [PubMed] [Google Scholar]
- 163.Lopez de Saro F, O’Donnell M. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc Natl Acad Sci U S A. 2001;98:8376–8380. doi: 10.1073/pnas.121009498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lovett ST, Clark AJ. Cloning of the Escherichia coli recJ chromosomal region and identification of its encoded proteins. J Bacteriol. 1985;162:280–285. doi: 10.1128/jb.162.1.280-285.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lovett ST, Clark AJ. Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol. 1984;157:190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lovett ST, Kolodner RD. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:2627–2631. doi: 10.1073/pnas.86.8.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lovett ST, Luisi-DeLuca C, Kolodner RD. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics. 1988;120:37–45. doi: 10.1093/genetics/120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Low B. Restoration by the rac locus of recombinant forming ability in recB - and recC - merozygotes of Escherichia coli K-12. Mol Gen Genet. 1973;122:119–130. doi: 10.1007/BF00435185. [DOI] [PubMed] [Google Scholar]
- 169.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc Natl Acad Sci USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Luisi-DeLuca C, Clark AJ, Kolodner RD. Analysis of the recE locus of Escherichia coli K-12 by use of polyclonal antibodies to exonuclease VIII. J Bacteriol. 1988;170:5797–5805. doi: 10.1128/jb.170.12.5797-5805.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Luisi-DeLuca C, Lovett ST, Kolodner RD. Genetic and physical analysis of plasmid recombination in recB recC sbcB and recB recC sbcA Escherichia coli K-12 mutants. Genetics. 1989;122:269–278. doi: 10.1093/genetics/122.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lundquist RC, Olivera BM. Transient generation of displaced single-stranded DNA during nick translation. Cell. 1982;31:53–60. doi: 10.1016/0092-8674(82)90404-4. [DOI] [PubMed] [Google Scholar]
- 173.Lyamichev V, Brow MA, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- 174.Mahdi AA, Lloyd RG. Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol Gen Genet. 1989;216:503–510. doi: 10.1007/BF00334397. [DOI] [PubMed] [Google Scholar]
- 175.Maki H, Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc Natl Acad Sci USA. 1987;84:4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Maki H, Kornberg A. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the alpha subunit, devoid of nuclease activities. J Biol Chem. 1985;260:12987–12992. [PubMed] [Google Scholar]
- 177.Makiela-Dzbenska K, Jaszczur M, Banach-Orlowska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol Microbiol. 2009;74:1114–1127. doi: 10.1111/j.1365-2958.2009.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Matos CF, Di Cola A, Robinson C. TatD is a central component of a Tat translocon-initiated quality control system for exported FeS proteins in Escherichia coli. EMBO Rep. 2009;10:474–479. doi: 10.1038/embor.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.McHenry CS, Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. J Biol Chem. 1979;254:1748–1753. [PubMed] [Google Scholar]
- 180.Miesel L, Roth JR. Evidence that SbcB and RecF pathway functions contribute to RecBCD-dependent transductional recombination. J Bacteriol. 1996;178:3146–3155. doi: 10.1128/jb.178.11.3146-3155.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Milcarek C, Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. I. Isolation and characterization of mutants for exonuclease 3. J Mol Biol. 1972;68:303–318. doi: 10.1016/0022-2836(72)90215-x. [DOI] [PubMed] [Google Scholar]
- 182.Missiakas D, Georgopoulos C, Raina S. The Escherichia coli dsbC (xprA) gene encodes a periplasmic protein involved in disulfide bond formation. EMBO J. 1994;13:2013–2020. doi: 10.1002/j.1460-2075.1994.tb06471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mitzel-Landbeck L, Schutz G, Hagen U. In vitro repair of radiation-induced strand breaks in DNA. Biochim Biophys Acta. 1976;432:145–153. doi: 10.1016/0005-2787(76)90156-8. [DOI] [PubMed] [Google Scholar]
- 184.Mol CD, Kuo CF, Thayer MM, Cunningham RP, Tainer JA. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 185.Morel P, Reverdy C, Michel B, Ehrlich SD, Cassuto E. The role of SOS and flap processing in microsatellite instability in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:10003–10008. doi: 10.1073/pnas.95.17.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Morelle S, Carbonnelle E, Matic I, Nassif X. Contact with host cells induces a DNA repair system in pathogenic Neisseriae. Mol Microbiol. 2005;55:853–861. doi: 10.1111/j.1365-2958.2004.04426.x. [DOI] [PubMed] [Google Scholar]
- 187.Mueser TC, Nossal NG, Hyde CC. Structure of bacteriophage T4 RNase H, a 5′ to 3′ RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins. Cell. 1996;85:1101–1112. doi: 10.1016/s0092-8674(00)81310-0. [DOI] [PubMed] [Google Scholar]
- 188.Murphy KC. Lambda Gam protein inhibits the helicase and chi-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes & Development. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 190.Nagata Y, Mashimo K, Kawata M, Yamamoto K. The roles of Klenow processing and flap processing activities of DNA polymerase I in chromosome instability in Escherichia coli K12 strains. Genetics. 2002;160:13–23. doi: 10.1093/genetics/160.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nguyen LH, Erzberger JP, Root J, Wilson DM. The human homolog of Escherichia coli Orn degrades small single-stranded RNA and DNA oligomers. J Biol Chem. 2000;275:25900–25906. doi: 10.1074/jbc.M002672200. [DOI] [PubMed] [Google Scholar]
- 193.Niwa O, Moses RE. Synthesis by DNA polymerase I on bleomycin-treated deoxyribonucleic acid: a requirement for exonuclease III. Biochemistry. 1981;20:238–244. doi: 10.1021/bi00505a002. [DOI] [PubMed] [Google Scholar]
- 194.Noirot P, Kolodner RD. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273:12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 195.Nunoshiba T, DeRojas-Walker T, Tannenbaum SR, Demple B. Roles of nitric oxide in inducible resistance of Escherichia coli to activated murine macrophages. Infect Immun. 1995;63:794–798. doi: 10.1128/iai.63.3.794-798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ohnishi G, Daigaku Y, Nagata Y, Ihara M, Yamamoto K. Saccharomyces cerevisiae RAD27 complements its Escherichia coli homolog in damage repair but not mutation avoidance. Genes Genet Syst. 2004;79:183–187. doi: 10.1266/ggs.79.183. [DOI] [PubMed] [Google Scholar]
- 197.Ozawa K, Jergic S, Park AY, Dixon NE, Otting G. The proofreading exonuclease subunit epsilon of Escherichia coli DNA polymerase III is tethered to the polymerase subunit alpha via a flexible linker. Nucleic Acids Res. 2008;36:5074–5082. doi: 10.1093/nar/gkn489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Palas KM, Kushner SR. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J Biol Chem. 1990;265:3447–3454. [PubMed] [Google Scholar]
- 199.Persky NS, Lovett ST. Mechanisms of recombination: lessons from E. coli. Crit Rev Biochem Mol Biol. 2008;43:347–370. doi: 10.1080/10409230802485358. [DOI] [PubMed] [Google Scholar]
- 200.Prasher DC, Conarro L, Kushner SR. Amplification and purification of exonuclease I from Escherichia coli K12. J Biol Chem. 1983;258:6340–6343. [PubMed] [Google Scholar]
- 201.Prell A, Wackernagel W. Degradation of linear and circular DNA with gaps by the recBC enzyme of Escherichia coli. Effects of gap length and the presence of cell-free extracts. Eur J Biochem. 1980;105:109–116. doi: 10.1111/j.1432-1033.1980.tb04480.x. [DOI] [PubMed] [Google Scholar]
- 202.Qiu J, Yoon JH, Shen B. Search for apoptotic nucleases in yeast: role of Tat-D nuclease in apoptotic DNA degradation. J Biol Chem. 2005;280:15370–15379. doi: 10.1074/jbc.M413547200. [DOI] [PubMed] [Google Scholar]
- 203.Qiu Z, Goodman MF. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem. 1997;272:8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- 204.Rajman LA, Lovett ST. A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J Bacteriol. 2000;182:607–612. doi: 10.1128/jb.182.3.607-612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rangarajan S, Gudmundsson G, Qiu Z, Foster PL, Goodman MF. Escherichia coli DNA polymerase II catalyzes chromosomal and episomal DNA synthesis in vivo. Proc Natl Acad Sci USA. 1997;94:946–951. doi: 10.1073/pnas.94.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Rangarajan S, Woodgate R, Goodman MF. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;96:9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Razavy H, Szigety SK, Rosenberg SM. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in chi-stimulated recombination in vivo. Genetics. 1996;142:333–339. doi: 10.1093/genetics/142.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Richardson CC, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. Purification of the enzyme and characterization of the phosphatase activity. J Biol Chem. 1964;239:242–250. [PubMed] [Google Scholar]
- 209.Richardson CC, I, Lehman R, Kornberg A. A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. II. Characterization of the exonuclease activity. J Biol Chem. 1964;239:251–258. [PubMed] [Google Scholar]
- 210.Rigden DJ. An inactivated nuclease-like domain in RecC with novel function: implications for evolution. BMC Struct Biol. 2005;5:9. doi: 10.1186/1472-6807-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Robins P, Pappin DJ, Wood RD, Lindahl T. Structural and functional homology between mammalian DNase IV and the 5′-nuclease domain of Escherichia coli DNA polymerase I. J Biol Chem. 1994;269:28535–28538. [PubMed] [Google Scholar]
- 212.Sak B, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci U S A. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sammartano LJ, Tuveson RW. Escherichia coli xthA mutants are sensitive to inactivation by broad-spectrum near-UV (300- to 400-nm) radiation. J Bacteriol. 1983;156:904–906. doi: 10.1128/jb.156.2.904-906.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sandigursky M, Franklin WA. Escherichia coli single-stranded DNA binding protein stimulates the DNA deoxyribophosphodiesterase activity of exonuclease I. Nucleic Acids Res. 1994;22:247–250. doi: 10.1093/nar/22.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein-protein interactions between the Escherichia coli single-stranded DNA-binding protein and exonuclease I. Radiat Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 216.Sayers JR. Computer aided identification of a potential 5′-3′ exonuclease gene encoded by Escherichia coli. J Theor Biol. 1994;170:415–421. doi: 10.1006/jtbi.1994.1202. [DOI] [PubMed] [Google Scholar]
- 217.Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Seeberg E, Steinum AL. Repair of x-ray-induced deoxyribonucleic acid single-strand breaks in xth mutants of Escherichia coli. J Bacteriol. 1980;141:1424–1427. doi: 10.1128/jb.141.3.1424-1427.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Serment-Guerrero J, Breña-Valle M, Espinosa-Aguirre JJ. In vivo role of Escherichia coli single-strand exonucleases in SOS induction by gamma radiation. Mutagenesis. 2008;23:317–323. doi: 10.1093/mutage/gen017. [DOI] [PubMed] [Google Scholar]
- 221.Setlow P, Brutlag D, Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. I. A proteolytic fragment containing the polymerase and 3′ leads to 5′ exonuclease functions. J Biol Chem. 1972;247:224–231. [PubMed] [Google Scholar]
- 222.Setlow P, Kornberg A. Deoxyribonucleic acid polymerase: two distinct enzymes in one polypeptide. II. A proteolytic fragment containing the 5′ leads to 3′ exonuclease function. Restoration of intact enzyme functions from the two proteolytic fragments. J Biol Chem. 1972;247:232–240. [PubMed] [Google Scholar]
- 223.Shafritz KM, Sandigursky M, Franklin WA. Exonuclease IX of Escherichia coli. Nucleic Acids Res. 1998;26:2593–2597. doi: 10.1093/nar/26.11.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sharma R, Rao DN. Orchestration of Haemophilus influenzae RecJ exonuclease by interaction with single-stranded DNA-binding protein. J Mol Biol. 2009;385:1375–1396. doi: 10.1016/j.jmb.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 225.Sharma RC, Smith KC. Role of DNA polymerase I in postreplication repair: a reexamination with Escherichia coli delta polA. J Bacteriol. 1987;169:4559–4564. doi: 10.1128/jb.169.10.4559-4564.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 227.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 228.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 229.Slater SC, Lifsics MR, O’Donnell M, Maurer R. holE, the gene coding for the theta subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (epsilon-subunit) mutant. J Bacteriol. 1994;176:815–821. doi: 10.1128/jb.176.3.815-821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Smith KC, Wang TV, Sharma RC. recA-dependent DNA repair in UV-irradiated Escherichia coli. J Photochem Photobiol B. 1987;1:1–11. doi: 10.1016/1011-1344(87)80002-7. [DOI] [PubMed] [Google Scholar]
- 231.Sommers CH, Miller EJ, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 232.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD enzyme switches lead motor subunits in response to chi recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–654. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 234.Spies M, Dillingham MS, Kowalczykowski SC. Translocation by the RecB motor is an absolute requirement for {chi}-recognition and RecA protein loading by RecBCD enzyme. J Biol Chem. 2005;280:37078–37087. doi: 10.1074/jbc.M505521200. [DOI] [PubMed] [Google Scholar]
- 235.Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 236.Stapleton MR, V, Norte A, Read RC, Green J. Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J Biol Chem. 2002;277:17630–17637. doi: 10.1074/jbc.M110178200. [DOI] [PubMed] [Google Scholar]
- 237.Studwell-Vaughan PS, O’Donnell M. DNA polymerase III accessory proteins. V. Theta encoded by holE. J Biol Chem. 1993;268:11785–11791. [PubMed] [Google Scholar]
- 238.Sun X, Wu P, Zheng L, Thrower D, Partikian A, Qiu J, Shen B. Suppression of Saccharomyces cerevisiae rad27 null mutant phenotypes by the 5′ nuclease domain of Escherichia coli DNA polymerase I. Curr Genet. 2002;41:379–388. doi: 10.1007/s00294-002-0323-x. [DOI] [PubMed] [Google Scholar]
- 239.Sutera VA, Han ES, Rajman LA, Lovett ST. Mutational analysis of the RecJ exonuclease of Escherichia coli: identification of phosphoesterase motifs. J Bacteriol. 1999;181:6098–6102. doi: 10.1128/jb.181.19.6098-6102.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Taft-Benz S, Schaaper R. The theta subunit of Escherichia coli DNA polymerase III: a role in stabilizing the epsilon proofreading subunit. J Bacteriol. 2004;186:2774–2780. doi: 10.1128/JB.186.9.2774-2780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Takahashi NK, Kusano K, Yokochi T, Kitamura Y, Yoshikura H, Kobayashi I. Genetic analysis of double-strand break repair in Escherichia coli. J Bacteriol. 1993;175:5176–5185. doi: 10.1128/jb.175.16.5176-5185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Taylor AF, Smith GR. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature. 2003;423:889–893. doi: 10.1038/nature01674. [DOI] [PubMed] [Google Scholar]
- 243.Taylor AF, Smith GR. RecBCD enzyme is altered upon cutting DNA at a chi recombination hotspot. Proc Natl Acad Sci USA. 1992;89:5226–5230. doi: 10.1073/pnas.89.12.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 245.Taylor AF, Weiss B. Role of exonuclease III in the base excision repair of uracil-containing DNA. J Bacteriol. 1982;151:351–357. doi: 10.1128/jb.151.1.351-357.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Templin A, Kushner SR, Clark AJ. Genetic analysis of mutations indirectly suppressing recB and recC mutations. Genetics. 1972;72:105–115. [PMC free article] [PubMed] [Google Scholar]
- 247.Thomas KR, Olivera BM. Processivity of DNA exonucleases. J Biol Chem. 1978;253:424–429. [PubMed] [Google Scholar]
- 248.Thoms B, Borchers I, Wackernagel W. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. J Bacteriol. 2008;190:179–192. doi: 10.1128/JB.01052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Thoms B, Wackernagel W. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J Bacteriol. 1998;180:5639–5645. doi: 10.1128/jb.180.21.5639-5645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 251.Tsaneva IR, Weiss B. soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Uyemura D, Eichler DC, Lehman IR. Biochemical characterization of mutant forms of DNA polymerase I from Escherichia coli. II. The polAex1 mutation. J Biol Chem. 1976;251:4085–4089. [PubMed] [Google Scholar]
- 253.Vales LD, Rabin BA, Chase JW. Isolation and preliminary characterization of Escherichia coli mutants deficient in exonuclease VII. J Bacteriol. 1983;155:1116–1122. doi: 10.1128/jb.155.3.1116-1122.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Vales LD, Rabin BA, Chase JW. Subunit structure of Escherichia coli exonuclease VII. J Biol Chem. 1982;257:8799–8805. [PubMed] [Google Scholar]
- 255.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 256.Viswanathan M, Burdett V, Baitinger C, Modrich P, Lovett ST. Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J Biol Chem. 2001;276:31053–31058. doi: 10.1074/jbc.M105481200. [DOI] [PubMed] [Google Scholar]
- 257.Viswanathan M, Dower KW, Lovett ST. Identification of a potent DNase activity associated with RNase T of Escherichia coli. J Biol Chem. 1998;273:35126–35131. doi: 10.1074/jbc.273.52.35126. [DOI] [PubMed] [Google Scholar]
- 258.Viswanathan M, Lacirignola JJ, Hurley RL, Lovett ST. A novel mutational hotspot in a natural quasipalindrome in Escherichia coli. J Mol Biol. 2000;302:553–564. doi: 10.1006/jmbi.2000.4088. [DOI] [PubMed] [Google Scholar]
- 259.Viswanathan M, Lanjuin A, Lovett ST. Identification of RNase T as a high-copy suppressor of the UV sensitivity associated with single-strand DNA exonuclease deficiency in Escherichia coli. Genetics. 1999;151:929–934. doi: 10.1093/genetics/151.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Viswanathan M, Lovett ST. Exonuclease X of Escherichia coli. A novel 3′-5′ DNase and DnaQ superfamily member involved in DNA repair. J Biol Chem. 1999;274:30094–30100. doi: 10.1074/jbc.274.42.30094. [DOI] [PubMed] [Google Scholar]
- 261.Viswanathan M, Lovett ST. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wakamatsu T, Kitamura Y, Kotera Y, Nakagawa N, Kuramitsu S, Masui R. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J Biol Chem. 2010;285:9762–9769. doi: 10.1074/jbc.M109.096487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang J, Chen R, Julin DA. A single nuclease active site of the Escherichia coli RecBCD enzyme catalyzes single-stranded DNA degradation in both directions. J Biol Chem. 2000;275:507–513. doi: 10.1074/jbc.275.1.507. [DOI] [PubMed] [Google Scholar]
- 265.Weiss B. Endonuclease II of Escherichia coli is exonuclease III. J Biol Chem. 1976;251:1896–1901. [PubMed] [Google Scholar]
- 266.Weiss B. Exodeoxyribonucleases of Escherichia coli. In: Boyer PB, editor. The Enzymes. Academic Press; New York, NY: 1981. pp. 203–231.pp. 14 [Google Scholar]
- 267.Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T. TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J Biol Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 268.Willis DK, Satin LH, Clark AJ. Mutation-dependent suppression of recB21 recC22 by a region cloned from the Rac prophage of Escherichia coli K-12. J Bacteriol. 1985;162:1166–1172. doi: 10.1128/jb.162.3.1166-1172.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Xu Y, Derbyshire V, Ng K, Sun XC, Grindley ND, Joyce CM. Biochemical and mutational studies of the 5′-3′ exonuclease of DNA polymerase I of Escherichia coli. J Mol Biol. 1997;268:284–302. doi: 10.1006/jmbi.1997.0967. [DOI] [PubMed] [Google Scholar]
- 270.Xu Y, Potapova O, Leschziner AE, Grindley ND, Joyce CM. Contacts between the 5′ nuclease of DNA polymerase I and its DNA substrate. J Biol Chem. 2001;276:30167–30177. doi: 10.1074/jbc.M100985200. [DOI] [PubMed] [Google Scholar]
- 271.Yajko DM, Valentine MC, Weiss B. Mutants of Escherichia coli with altered deoxyribonucleases. II. Isolation and characterization of mutants for exonuclease I. J Mol Biol. 1974;85:323–343. doi: 10.1016/0022-2836(74)90367-2. [DOI] [PubMed] [Google Scholar]
- 272.Yajko DM, Weiss B. Mutations simultaneously affecting endonuclease II and exonuclease III in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:688–692. doi: 10.1073/pnas.72.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yamagata A, Kakuta Y, Masui R, Fukuyama K. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc Natl Acad Sci USA. 2002;99:5908–5912. doi: 10.1073/pnas.092547099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Young Park A, Elvin CM, Hamdan SM, Wood RJ, Liyou NE, Hamwood TE, Jennings PA, Dixon NE. Hydrolysis of the 5′-p-nitrophenyl ester of TMP by oligoribonucleases (ORN) from Escherichia coli, Mycobacterium smegmatis, and human. Protein Expr Purif. 2008;57:180–187. doi: 10.1016/j.pep.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 275.Yu D, Deutscher MP. Oligoribonuclease is distinct from the other known exoribonucleases of Escherichia coli. J Bacteriol. 1995;177:4137–4139. doi: 10.1128/jb.177.14.4137-4139.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yu M, Souaya J, Julin DA. Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J Mol Biol. 1998;283:797–808. doi: 10.1006/jmbi.1998.2127. [DOI] [PubMed] [Google Scholar]
- 277.Yu M, Souaya J, Julin DA. The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc Natl Acad Sci USA. 1998;95:981–986. doi: 10.1073/pnas.95.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Zahradka K, Buljubasić M, Petranović M, Zahradka D. Roles of ExoI and SbcCD nucleases in “reckless” DNA degradation in recA mutants of Escherichia coli. J Bacteriol. 2009;191:1677–1687. doi: 10.1128/JB.01877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Zaman MM, Boles TC. Chi-dependent formation of linear plasmid DNA in exonuclease-deficient recBCD+ strains of Escherichia coli. J Bacteriol. 1994;176:5093–5100. doi: 10.1128/jb.176.16.5093-5100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhang J, Xing X, Herr AB, Bell CE. Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks. Structure. 2009;17:690–702. doi: 10.1016/j.str.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhang XJ, Julin DA. Isolation and characterization of the C-terminal nuclease domain from the RecB protein of Escherichia coli. Nucleic Acids Res. 1999;27:4200–4207. doi: 10.1093/nar/27.21.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 284.Zhuo S, Clemens JC, Stone RL, Dixon JE. Mutational analysis of a Ser/Thr phosphatase. Identification of residues important in phosphoesterase substrate binding and catalysis. J Biol Chem. 1994;269:26234–26238. [PubMed] [Google Scholar]
- 285.Zissler J, Signer E, Sachaefer F. The role of recombination in the growth of bacteriophage lambda. In: Hershey AD, editor. Bacteriophage Lambda. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1971. pp. 455–475. [Google Scholar]
- 286.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Zuo Y, Deutscher MP. Mechanism of action of RNase T. I. Identification of residues required for catalysis, substrate binding, and dimerization. J Biol Chem. 2002;277:50155–50159. doi: 10.1074/jbc.M207706200. [DOI] [PubMed] [Google Scholar]
- 288.Zuo Y, Deutscher MP. Mechanism of action of RNase T. II. A structural and functional model of the enzyme. J Biol Chem. 2002;277:50160–50164. doi: 10.1074/jbc.M207707200. [DOI] [PubMed] [Google Scholar]
- 289.Zuo Y, Deutscher MP. The DNase activity of RNase T and its application to DNA cloning. Nucleic Acids Res. 1999;27:4077–4082. doi: 10.1093/nar/27.20.4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zuo Y, Deutscher MP. The physiological role of RNase T can be explained by its unusual substrate specificity. J Biol Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- 291.Zuo Y, Wang Y, Malhotra A. Crystal structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing. Structure. 2005;13:973–984. doi: 10.1016/j.str.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 292.Zuo Y, Zheng H, Wang Y, Chruszcz M, Cymborowski M, Skarina T, Savchenko A, Malhotra A, Minor W. Crystal structure of RNase T, an exoribonuclease involved in tRNA maturation and end turnover. Structure. 2007;15:417–428. doi: 10.1016/j.str.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]