Abstract
Virulent Agrobacterium tumefaciens strains integrate their T-DNA into the plant genome where the encoded agrobacterial oncogenes are expressed and cause crown gall disease. Essential for crown gall development are IaaH (indole-3-acetamide hydrolase), IaaM (tryptophan monooxygenase) and Ipt (isopentenyl transferase), which encode enzymes for the biosynthesis of auxin (IaaH, IaaM) and cytokinin (Ipt). Although these oncogenes are well studied as the tumor-inducing principle, nothing is known about the regulation of oncogene expression in plant cells. Our studies show that the intergenic regions (IGRs) between the coding sequences (CDS) of the three oncogenes function as promoters in plant cells. These promoters possess a eukaryotic sequence organization and cis-regulatory elements for the binding of plant transcription factors. WRKY18, WRKY40, WRKY60 and ARF5 were identified as activators of the Ipt promoter whereas IaaH and IaaM is constitutively expressed and no transcription factor further activates their promoters. Consistent with these results, the wrky triple mutant plants in particular, develops smaller crown galls than wild-type and exhibits a reduced Ipt transcription, despite the presence of an intact ARF5 gene. WRKY40 and WRKY60 gene expression is induced by A. tumefaciens within a few hours whereas the ARF5 gene is transcribed later during crown gall development. The WRKY proteins interact with ARF5 in the plant nucleus, but only WRKY40 together with ARF5 synergistically boosts the activation of the Ipt promoter in an auxin-dependent manner. From our data, we propose that A. tumefaciens initially induces WRKY40 gene expression as a pathogen defense response of the host cell. The WRKY protein is recruited to induce Ipt expression, which initiates cytokinin-dependent host cell division. With increasing auxin levels triggered by ubiquitous expression of IaaH and IaaM, ARF5 is activated and interacts with WRKY40 to potentiate Ipt expression and balance cytokinin and auxin levels for further cell proliferation.
Author Summary
Crown gall development requires the expression of agrobacterial genes in the plant host. These genes are transferred by the T-DNA of the plant pathogen Agrobacterium tumefaciens and include the oncogenes IaaH, IaaM and Ipt, which, according to the tumor-inducing principle, are essential for crown gall development. The oncogenes are involved in auxin and cytokinin production. This results, when at appropriate hormone ratios, in enhanced cell proliferation. The T-DNA transformation process and the encoded oncogene enzymes have been intensively studied, but knowledge of oncogene expression in plant cells and the regulatory host factors is missing. We set out to fill this gap, providing evidence that expression of the Ipt gene is host-cell controlled, whereas the IaaH and IaaM genes are ubiquitously expressed at low levels in T-DNA transformed tissue. This is achieved by A. tumefaciens, which first hijacks transcription factors of the plant pathogen response pathway to activate Ipt oncogene expression and initiates cell proliferation. With increasing auxin levels during the infection process, a transcription factor of the auxin-signaling pathway is recruited, potentiating Ipt gene expression. Thus, for crown gall development, two host-signaling pathways are combined through the interaction of transcription factors that adjust the ratio of cytokinin to auxin.
Introduction
Agrobacterium tumefaciens is a pathogenic bacterium that infects several plant species. A region in the tumor inducing (Ti) plasmid, the transfer DNA (T-DNA), is integrated into the plant genome causing crown gall disease [1]. There are essentially two groups of genes encoded on the T-DNA of virulent A. tumefaciens strains [2]. The first is responsible for producing opines, so providing a carbon and nitrogen source for A. tumefaciens, with the second group expressing the oncogenes required for crown gall development. These oncogenes include IaaH, IaaM, Ipt, gene 6b and gene 5. It is assumed that although gene 6b and gene 5 are expendable, IaaH, IaaM and Ipt are crucial for crown gall development [3–5]. IaaH and IaaM code for enzymes that catalyze biosynthesis of auxin and Ipt mediates cytokinin biosynthesis [5,6]. IaaM encodes a tryptophan monooxygenase that converts tryptophan (Trp) into indole-3-acetamide (IAM), and IaaH an indole-3-acetamide hydrolase, converts IAM into indole-3-acetic acid (IAA) [7–9]. Ipt (isopentenyl transferase) catalyzes the rate-limiting step in cytokinin biosynthesis [2,5,10]. Cytokinins can also be synthesized in A. tumefaciens cells by the chromosomal encoded miaA enzyme [11,12] and the trans-zeatin synthesizing (tzs) enzyme encoded on the nopaline-type pTi-plasmid [13–15]. A. tumefaciens secretes auxin and cytokinin from the cells to initiate crown gall development [16] and pretreatment of plant tissues with auxin and cytokinin promotes A. tumefaciens-mediated transformation efficiency [14,17,18]. Very recently it was shown that cytokinins secreted by A. tumefaciens repress a Myb transcription factor in host plant cells, resulting in an enhanced transformation efficiency [18].
The increased production of auxin and cytokinin in T-DNA transformed plant cells expressing the IaaH, IaaM and Ipt oncogenes induces cell proliferation and differentiation [19,20]. Therefore, a T-DNA harboring plant cell needs to initiate transcription of the three oncogenes in order to express their function. In eukaryotic cells, the RNA polymerase II complex mediates transcription of mRNAs from protein-coding genes. This complex recognizes the TATA box and the transcription start site (TSS) [21] within upstream promoter regions that drive the expression of the downstream coding sequence (CDS). These two sequence features build the core promoter and this is sufficient to transcribe a gene [21]. TATA boxes were predicted to be present 5’ upstream of the CDS of the IaaH, IaaM and Ipt oncogenes [22–24]. In addition to initiation of transcription by the RNA polymerase II complex, expression of eukaryotic genes is usually regulated by transcription factors. These bind to regulatory sequence elements localized in the promoter regions of many eukaryotic genes and are oriented in a sense or anti-sense direction distant from the TSS [21]. For the Ipt gene of the octopine Ti plasmid pTiAch5, a 184 bp fragment upstream of the CDS is sufficient for transcription in plant cells [25]. In particular, the region between −185 and −139 bp from the translational start codon are essential [26]. Within that region, the 30 bp sequence cyt-1 binds an as yet unknown protein from tobacco nuclear protein extracts, designated CBF (cyt-1 binding factor) [27]. This suggests that expression of the agrobacterial oncogenes can be regulated by host transcription factors that await discovery.
A well-known response of plants to microbial pathogens is the microbe associated molecular pattern (MAMP)-induced innate immunity response, which includes expression of several WRKY transcription factors [28]. The expression profiles of 72 WRKY genes in Arabidopsis revealed that 49 genes are responsive to salicylic acid (SA) and pathogen treatment [29]. The WRKY transcription factor binding elements, the W-boxes (TGAC), are present in many defense related gene promoters [28]. In addition to the induction of pathogen defense responses, crown gall development requires cell proliferation and differentiation, such as vascularization [30]. These developmental programs are synergistically controlled by auxin and cytokinin signaling pathways that lead to changes in the regulation of gene expression. The expression of some auxin responsive factor (ARF) genes is induced by auxin, particularly in developing embryos and vascular tissues [31]. ARFs are known to induce the transcription of genes in an auxin-dependent manner by binding to auxin response elements (AuxREs) in auxin responsive promoters [31,32]. The regulation of ARF function involves auxin/indole acetic acid (Aux/IAA) proteins and TIR1 (transport inhibitor response 1) [33,34]. Aux/IAA proteins interact and repress the transcriptional activity of ARFs [35,36]. The F-box auxin receptor TIR1 is part of the SCFTIR ubiquitin ligase complex [37,38]. At increasing auxin concentrations, Aux/IAA proteins are recognized and ubiquitinylated by the SCFTIR complex and subsequently degraded by the 26S proteasome [39,40]. The de-repressed ARF proteins can activate target promoters.
This study focuses on the transcriptional regulation of the A. tumefaciens genes IaaH, IaaM and Ipt in the host plant. The intergenic regions between the CDSs of IaaH, IaaM and Ipt of the virulent T-DNA of A. tumefaciens strain C58 (pTiC58, AE007871) showed promoter activity in Arabidopsis cells. The IaaH and IaaM genes involved in auxin biosynthesis in T-DNA transformed cells, were ubiquitously expressed at low levels. In contrast, the Ipt promoter was activated by the transcription factor WRKY40 (AT1G80840), a transcription factor that responded rapidly to A. tumefaciens infection. WRKY40 together with ARF5 (AT1G19850), which is part of an auxin-dependent signaling pathway, boosted Ipt promoter activity in an auxin dependent manner. This enhanced activity correlated with cis-regulatory elements such as W-boxes and AuxREs in the Ipt promoter and the protein interaction of WRKY40 with ARF5. Our findings suggest that A. tumefaciens recruits the WRKY-dependent pathogen defense pathway to activate Ipt gene expression. This can be substantially increased when the auxin-dependent developmental process mediated by ARF5 is switched on.
Results
The intergenic regions between the oncogenes function as promoters in plant cells
To discover how the expression of the agrobacterial oncogenes IaaH, IaaM and Ipt is regulated in plant cells, we analyzed the structure of the T-DNA region of the nopaline-type Ti plasmid pTiC58. The CDS of the three oncogenes are sequentially arranged and interrupted by two non-coding intergenic regions (IGR1 and IGR2; Fig. 1A). The IaaM and Ipt genes are transcribed from the sense strand and the IaaH gene is encoded on the opposite strand. If IGR1 functions as a promoter for both the IaaH and IaaM oncogenes, it must be a bidirectional promoter: one direction being 5’ upstream of the TSS of the IaaH CDS (IGR1a) and the other, 5’ upstream of IaaM (IGR1b).
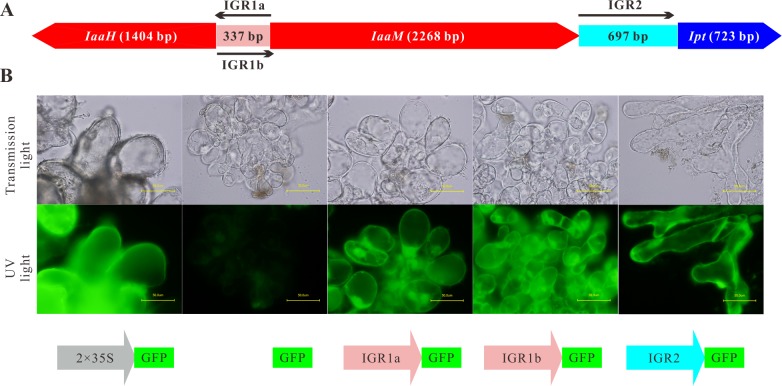
Figure 1. IGR1 and IGR2 function as promoters in Arabidopsis cells.
(A) Arrangement of the coding sequences of the IaaH, IaaM and Ipt oncogenes and the intergenic regions (IGRs) in the T-DNA region of the Ti plasmid of A. tumefaciens strain C58, pTiC58. (B) Arabidopsis crown gall callus cells expressing the green fluorescing protein (GFP) under the control of IGR1a (IGR1a::GFP), IGR1b (IGR1b::GFP) and IGR2 (IGR2::GFP). IGR1 was used in two orientations; one is upstream of IaaH CDS (IGR1a) and the other upstream of IaaM (IGR1b). The universal cauliflower mosaic virus promoter was used as a positive control (2× CaMV35S::GFP) and the GFP CDS without promoter, as the negative control (GFP). Images show crown gall callus cells in the transmission microscopy (top row) and the UV light mode (bottom row, excitation: 490 nm, emission: 510 nm). The UV-light intensity used for excitation is the same for both pictures. Bars, 50 μm.
To prove whether the IGRs function as promoters in plant cells, the complete IGR sequences were fused with the CDS of the green fluorescent protein (GFP) in a binary vector. The IGR1a and IGR1b sequences included the 5’ untranslated regions (5’ UTR) of both the IaaH and IaaM genes, whereas IRG2 contained the 3’ UTR of IaaM and 5’ UTR of the Ipt gene. We generated stable transformed Arabidopsis crown gall tumor cell lines by infecting Arabidopsis root segments with the virulent A. tumefaciens strain C58, which, in addition to their pTiC58, harbor a binary vector with the IGR::GFP constructs. Detection of GFP fluorescence in the IGR1a::GFP, IGR1b::GFP and IGR2::GFP crown gall cell lines demonstrated that the IGRs drive GFP expression, so function as promoters in plant cells (Fig. 1B). Furthermore, as the IGR1 sequence is a bidirectional promoter, it can drive transcription of both the IaaH and IaaM genes.
Since IGR1a, IGR1b and IGR2 all function as promoters in eukaryotic cells, their sequences should contain the core promoter elements, such as the initiator (Inr) sequence and TATA box. To localize these in the promoters, we determined the TSSs of the IaaH, IaaM and Ipt genes using the 5’ rapid amplification of cDNA ends (5’ RACE) assay, finding that the translational start codon of the IaaH, IaaM and Ipt CDSs are at positions +12 bp, +26 bp and +44 bp in respect to the TSS (Table 1). Upstream of the TSSs, the typical eukaryotic Inr box (YYANWYY, TSS is underlined, Y = C/T, W = A/T, N = A/G/C/T) was present in the three promoter sequences. This is in agreement with the plant specific “YR Rule” (YR, TSS is underlined, Y = C/T, R = A/G [41,42]). The TATA boxes, the binding sites for the general transcription factor complex, are found in the promoter regions −25 bp to −35 bp and another feature of many eukaryotic promoters, the CAAT boxes, are localized approximately −70 bp upstream of the TSSs within the oncogene promoter regions (Table 1).
Table 1. Cis-regulatory sequence elements within the promoter of the oncogenes IaaH, IaaM and Ipt encoded on the T-DNA of the Ti plasmid from A. tumefaciens strain C58, pTiC58.
| Promoter (sequence) | IaaH(−1 to −301) | IaaM(−1 to −301) | Ipt fwd(−1 to −654) | Iptrev (−1 to −654) |
|---|---|---|---|---|
| Positions of core promoter sequence elements | ||||
| Inr box (YYANWYY) | −2 CCAAACC +5 | −2 CTACACA +5 | −2 CTAATCC +5 | |
| Start coden (ATG) | +12 ATG | +26 ATG | +44 ATG | |
| TATA box(TATAAA) | −36 TATATT −31 1 | −32 TAAATA −27 2 | −29 TATAAC −24 34, 56 | |
| CAAT box(GGNCAATCN) | −66 CCAAT −62 1 | −75 CCATT −71 2 | −72 GGTAAAGCC −64 3 −49 AAGGAATCT −41 4 , 5 | |
To ascertain whether the regulatory promoter elements of pTiC58 are conserved, we performed a sequence alignment with the promoter and 5’ untranslated regions (5’ UTRs) of the three oncogenes from different Ti plasmids. We compared the upstream sequences of the three oncogene CDSs of the Ti plasmids from two nopaline-types (pTiC58, pTiSAKURA), three octopine-types (pTiA6NC, pTiAch5, pTi15955) and one agropine-type (pTiBo542). The alignment shows that the TSSs (arrows), TATA boxes and CAAT boxes of the promoters for IaaH (S1 Fig.), IaaM (S2 Fig.) and Ipt (S3 Fig.) are conserved between the pTi plasmids of the different A. tumefaciens strains. In contrast, two TATA boxes are present 5’ upstream of the CDS in the Ipt genes from the octopine Ti plasmids (S3 Fig.). In the Ipt promoter of pTiC58, two CAAT boxes were predicted (S3 Fig.), one of which (GGTAAAGCC, from −72 to −64 bp) is conserved and also found in other nopaline type and in the octopine type pTi plasmids, but not in the agropine type Ipt promoter where no CAAT box was predicted. The second CAAT box (AAGGAATCT, −49 to −41 bp) is specific for the Ipt promoters of the nopaline type Ti-plasmids (S3 Fig.). Cis-regulatory binding elements for transcription factors were also determined in the IaaH, IaaM and Ipt promoters on the Watson and Crick strand using PLACE (http://www.dna.affrc.go.jp/PLACE/index.html) [43–45]. Several binding elements for different transcription factor families including MYB, DOF, WRKY, bHLH, ARR1 and ARF, were localized within the Ipt promoter (Table 1). In the IaaH and IaaM promoters, the binding element for the ARR1 (AT3G16857) transcription factor was dominant and there were eight ARR1 elements altogether.
To identify potential transcription factors that may be involved in enhancing the expression of the oncogenes, we analyzed existing microarray data of Arabidopsis crown galls [20,46], based on the Arabidopsis transcription factors listed in the Plant Transcription Factor Database v3.0 [47] (http://planttfdb.cbi.pku.edu.cn/index.php?sp=Ath). A total of 151 transcription factor genes were found to be differentially transcribed in inflorescence stems inoculated with the virulent A. tumefaciens strain C58 compared to non-inoculated stems (S1 Table; fold change ≥ 2 or ≤ 0.5, p value < 0.01). As early as three hours post inoculation (hpi), three of these genes were up-regulated: WRKY53 (AT4G23810, 2.47 fold), WRKY40 (2.22 fold), and NAC102 (AT5G63790, 2.18 fold). WRKY53 was also up-regulated by the disarmed A. tumefaciens strain GV3101 (2.37 fold) 3 hpi. Six days post inoculation (dpi), the expression of six transcription factor genes was up- or down-regulated (S1 Table). In Arabidopsis crown gall material of A. tumefaciens strain C58, 141 transcription factor genes were transcriptionally changed compared to reference tissue 35 days post wounding (dpw). Amongst these, 74 genes were up-regulated, with 67 down-regulated (S1 Table) and all belong to various families including WRKYs, MYBs, DOFs, and NACs. The DNA binding elements and the microarray data both suggest that the MYB, DOF, WRKY, bHLH, ARR1 and ARF transcription factors are potential candidates for involvement in the regulation of Ipt expression, while ARR1 could regulate transcription of the IaaH or IaaM genes. The core promoter sequence elements could contribute to the basal expression of the three oncogenes in plant cells, whereas the binding sites for transcription factors might function in enhancing their transcription.
WRKY18, WRKY40, WRKY60 and ARF5 activate the Ipt oncogene promoter
To begin to study the regulation of onocgene expression, we first used quantitative real-time PCR (qRT-PCR). We assessed the relative transcript numbers of IaaH, IaaM and Ipt genes in 25-day-old Arabidopsis thaliana crown galls induced by the virulent A. tumefaciens strain C58, finding that the transcript levels of IaaH and IaaM were much lower compared to those of the Ipt gene in the crown galls (Fig. 2A). The high-throughput protoplast transactivation (PTA) system was then used [48] to identify transcription factors that could activate the three oncogene promoters in plant cells. To do so, the complete promoters of IaaH (IGR1a, 337 bp), IaaM (IGR1b, 337 bp) and Ipt (IGR2, 697 bp) of the pTiC58-encoded oncogenes (Fig. 1A) were fused with the CDS of the firefly luciferase (LUC) reporter gene. The plasmids containing the oncogene promoter-LUC constructs were transfected into Arabidopsis mesophyll protoplasts, either alone, or together with a second plasmid containing the CDS of a transcription factor fused to the constitutive cauliflower mosaic virus (CaMV35S) promoter. The relative luminescence, a measure for the oncogene promoter activity since it drives luciferase gene expression, was then determined. Mesophyll protoplasts transfected only with the oncogene promoter-LUC constructs showed the same pattern of promoter activity as that determined for the relative transcript numbers in crown galls (Fig. 2B). The Ipt promoter induced a higher relative luminescence than the IaaH and IaaM promoters.
Figure 2. Transcripts of oncogenes in crown galls and activity of oncogene promoters in protoplasts.
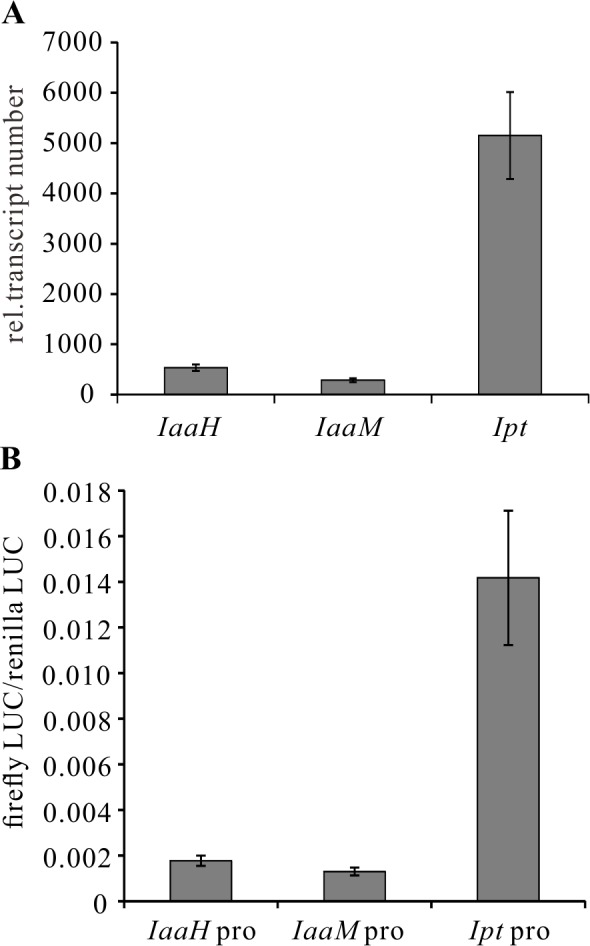
(A) Relative abundance of IaaH, IaaM and Ipt transcripts in crown gall tumors 25 days after inoculation of A. tumefaciens strain C58 into Arabidopsis inflorescence stems. Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. (B) Relative luciferase activity (firefly LUC/renilla LUC) driven by oncogene promoters (IaaH pro, IaaM pro and Ipt pro). Relative luciferase activity is calculated by firefly luminescence/renilla luminescence. Bars show mean values (±SD) of three independent experiments.
Next, a library containing the CDS of more than 400 transcription factors was screened. Among the included family members, WRKY, AP2/ERF, bHLH, bZIP, DOF, MYB and NAC, only WRKY18 (AT4G31800), WRKY40, WRKY60 (AT2G25000) and ARF5 were found to specifically activate the Ipt promoter in protoplasts (Fig. 3A). Protoplasts co-transfected with the WRKY or ARF effector and the Ipt-promoter-LUC reporter constructs exhibited a significantly higher promoter activity (reflected by luciferase activity) compared to the control samples that only harbored the reporter. Despite several attempts, no transcription factor was found to activate the IaaH and IaaM promoters. Comparison of the three WRKYs alone and in combination both showed that WRKY40 exerts the strongest impact on Ipt promoter-driven luciferase expression (S4 Fig.). Even all three WRKYs together did not increase the relative luminescence more than WRKY40 alone. This observation points towards a dominant role for WRKY40 in Ipt promoter regulation.
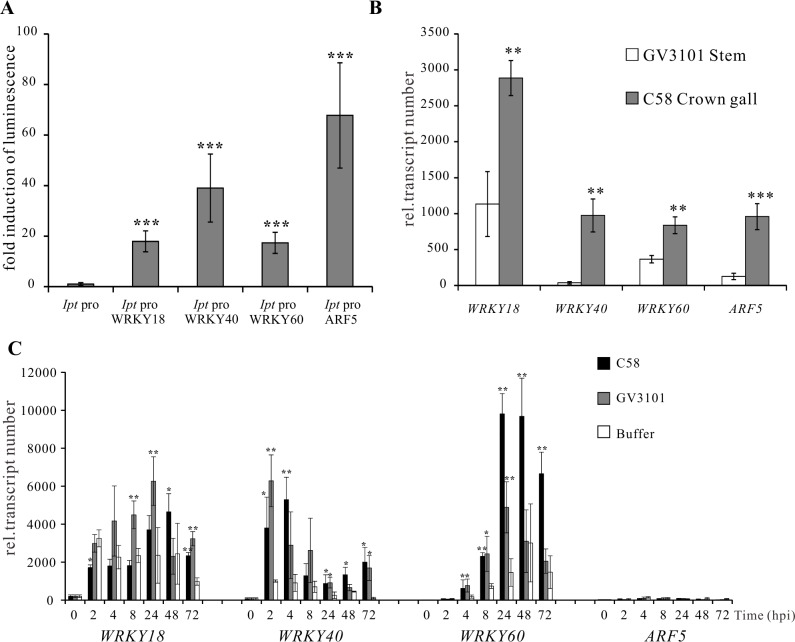
Figure 3. Activation of the Ipt promoter and gene expression of WRKY18, WRKY40, WRKY60 and ARF5.
(A) Fold induction of Ipt promoter-driven luminescence (Ipt pro) by WRKY18, WRKY40, WRKY60 and ARF5 in Arabidopsis mesophyll protoplasts transfected with two plasmid types. One harbors the Ipt promoter upstream of the firefly luciferase coding sequence (CDS) and the other, the universal cauliflower mosaic virus promoter (CaMV35S) upstream of a transcription factor CDS. The relative luminescence induced by the Ipt promoter in the absence of a transcription factor expression plasmid was set to 1. Bars show mean values (±SD) of three independent experiments. (B) Relative transcript numbers of WRKY18, WRKY40, WRKY60 and ARF5 genes in crown galls 25 days after inoculation with the virulent A. tumefaciens strain C58 (C58 Crown gall) and the disarmed strain (GV3101 Stems). (C) Time-dependent expression of the WRKY18, WRKY40, WRKY60 and ARF5 genes upon infiltration of five-week-old Arabidopsis leaves with suspension (OD600 1.0) of strain C58 and GV3101 as well as an Agromix buffer as control. Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. ** P<0.01 *** P<0.001 (Student’s t-test).
The transcript numbers of WRKY18, WRKY40, WRKY60 and ARF5 genes in crown gall tissues of A. tumefaciens strain C58 were determined using qRT-PCR. In agreement with the published microarray data [20,46], the transcript levels were clearly elevated in crown gall tumors compared to inflorescence stems inoculated with the disarmed A. tumefaciens strain GV3101 (Fig. 3B). It is already known that WRKY18, WRKY40 and WRKY60 are induced early after bacterial and fungal pathogen infection [49,50]. To analyze the impact of A. tumefaciens on gene induction, we analyzed the time-dependent expression of the three WRKY genes in Arabidopsis thaliana (Col-0) leaf tissues infiltrated with either the virulent A. tumefaciens strain C58, the disarmed strain GV3101 or buffer as a control. The qRT-PCR results demonstrated that the three WRKY genes responded to a certain degree to the infiltrated buffer solution at all analyzed time points (2 hpi to 72 hpi), indicating that they respond to wounding (Fig. 3C). The transcript levels of WRKY18 began to increase significantly at 8 hpi after infiltration by strain GV3101. The WRKY40 and WRKY60 genes were significantly induced by both A. tumefaciens strains as early as 2 and 4 hpi, respectively (Fig. 3C). In contrast, transcription of the ARF5 gene was still very low after 72 hpi, suggesting that this gene is not responsive to A. tumefaciens or wounding at the time points analyzed (Fig. 3C). The gene expression patterns imply that at the very beginning of A. tumefaciens infection (2 to 4 hpi), WRKY40 and WRKY60 genes are already expressed.
WRKY and ARF transcription factors bind respectively to specific DNA sequences, W-box (TGAC) and AuxRE (TGTCNC or TGTCTN). Sequence analysis of the two IGRs of pTiC58 revealed that seven W-boxes (one W-box is localized in the 5’ UTR of the Ipt gene) and five AuxREs are located in IGR2 (Table 1, 2), which are equally distributed along the promoter sequence (S5 Fig.). IGR1 drives expression of IaaH and IaaM and contains only one W-box and AuxRE sequence motif, and this is more closely localized upstream of the IaaM than that of the IaaH TATA box. Sequence comparisons of IGR1 and IGR2 regions illustrate that W-boxes and AuxREs are also conserved in the T-DNA regions of several A. tumefaciens strains (Table 2). Similar to the pTiC58, the majority of these elements are enriched in the Ipt promoters whereas only one or two of them are located in the IaaH and IaaM promoter sequences. From this in silico result, it can be concluded that the Ipt oncogenes, rather than IaaH and IaaM of the different A. tumefaciens strains are regulated by WRKY and ARF transcription factors in planta.
Table 2. Number of WRKY-boxes (W-boxes) and auxin response elements (AuxREs) within the intergenic regions (IGRs) of the tumor inducing (Ti) plasmids from different A. tumefaciens strains.
| IGR1
a
|
IGR2
b
|
||||
|---|---|---|---|---|---|
| pTi plasmid | W-box (TGAC) | AuxRE (TGTCNC or TGTCTN) | W-box (TGAC) | AuxRE (TGTCNC or TGTCTN) | |
| Nopaline | pTiC58 | 1 | 1 | 7 | 5 |
| pTiT37 | ? | ? | 5 | 5 | |
| pTiSAKURA | 2 | 1 | 6 | 5 | |
| Octopine | pTi15955 | 2 | 1 | 7 | 5 |
| pTiA6NC | 2 | 1 | 7 | 5 | |
| pTiAch5 | 2 | 1 | 7 | 5 | |
| Agropine | pTiBo542 | 2 | 1 | 6 | 3 |
Sequences are from the Genbank database (http://www.ncbi.nlm.nih.gov/genbank/). ”?”, IGR1 of pTiT37 is not present in the Genbank database.
a IGR1 is localized between the coding sequences of IaaH and IaaM
b IGR2 is localized between the coding sequences ofIaaM and Ipt.
WRKY18, WRKY40 and WRKY60 mutants display an impaired crown gall development
To unravel the role of WRKY18, WRKY40 and WRKY60 in A. tumefaciens-mediated crown gall development, we performed a crown gall growth assay with mutant plants of the three WRKY genes inoculated with the tumorigenic A. tumefaciens strain C58, determining the crown gall weights 25 days later. All mutant genotypes developed smaller crown galls than the wild-type Col-0 (Fig. 4A, B), with the double mutant wrky18/wrky40 and the triple mutant wrky18/40/60 developing the smallest crown galls. The triple mutant was most resistant to crown gall development; about 30% of the mutant plants did not development any crown gall material at all after 25 days. Unfortunately, the role of the ARF5-mediated auxin signaling pathway on crown gall development could not be analyzed due to the strong developmental phenotypes of arf5 mutant plants [51,52].
Figure 4. Arabidopsis wrky mutants develop smaller crown galls.
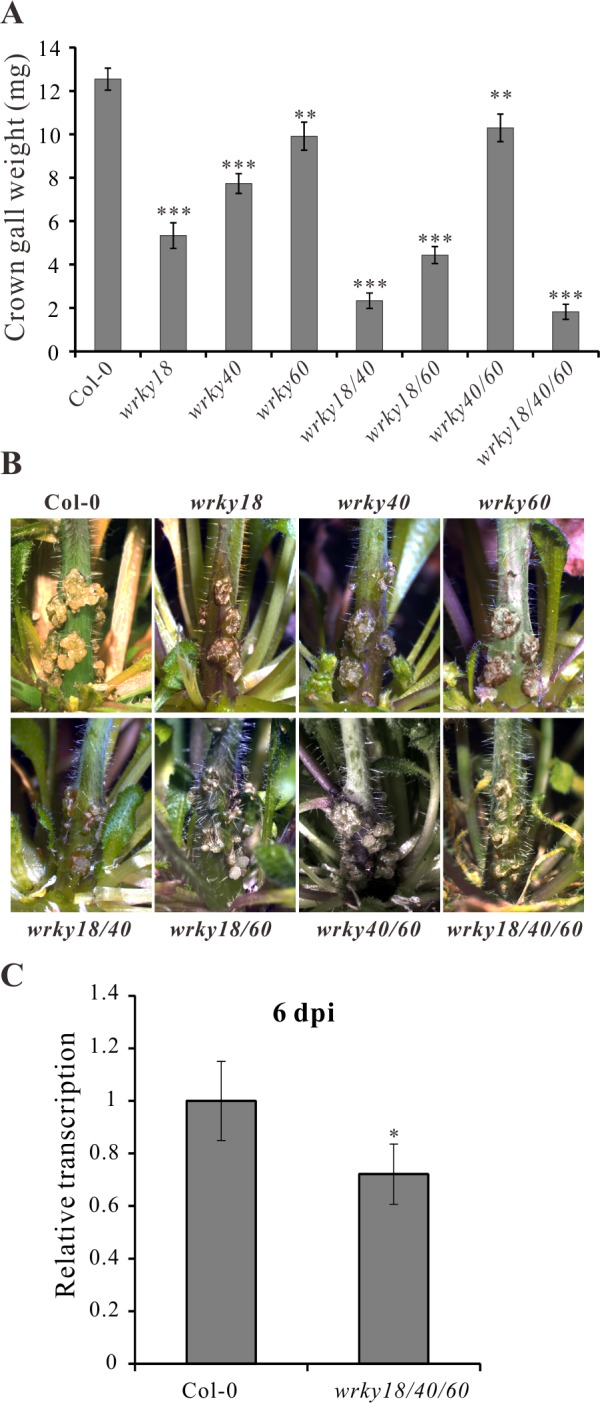
(A) Crown gall weights of wrky18, wrky40, wrky60 mutants and the wild type Col-0 25 days after inoculation of Arabidopsis inflorescence stems with the tumorigenic A. tumefaciens strain C58. Bars show mean values of crown gall weight (±SE) separated from the stems of at least 40 plants from each genotype. (B) Representative pictures of the stems of the different genotypes 25 days after inoculation of A. tumefaciens. (C) Relative transcript numbers of the Ipt oncogene in stems of the wild-type plant Col-0 and the wrky18/40/60 triple mutant 6 days post inoculation (6 dpi) of A. tumefaciens strain C58. Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. * P<0.05; ** P<0.01; *** P<0.001 (Student’s t-test).
If WRKY18, WRKY40 and WRKY60 activate the Ipt promoter, it would be expected that Ipt oncogene expression would be altered in the WRKY mutant plants. To investigate this, we used quantitative RT-PCR to measure the relative transcript numbers of the Ipt oncogene in Arabidopsis crown gall material of the wrky mutants inoculated with A. tumefaciens strain C58. Compared to crown galls from the wild-type (Col-0) plants, the Ipt transcript levels were similar in crown galls from the wrky18, wrky40 and wrky60 mutants (S6A Fig.). Due to this similarity, i.e., no obvious impact of WRKY on long term Ipt gene expression in crown galls, earlier time points of C58 Arabidopsis stem inoculations were analyzed. At 2 dpi, the Ipt transcript levels were far too low to reliably quantify differences (S6B Fig.). Only at 6 dpi did Ipt transcription reach a measureable level (S6B Fig.) and showed in the triple mutant (wrky18/40/60) a moderate reduction compared to the wild-type (Fig. 4C). The moderate reduction of Ipt transcription may be due to the function of ARF5, which is still expressed in the wrky triple mutant. This assumption is supported by the observation that in crown galls of the wrky single mutants gene expression of ARF5 was elevated and that of IAA12, an inhibitor of ARF5 function, was reduced (S6C Fig.).
WRKY40 and ARF5 proteins interact and synergistically potentiate Ipt promoter activity
The PTA data revealed that the Ipt promoter can be activated by WRKY18, WRKY40, WRKY60 and ARF5. To test whether these transcription factors cooperatively regulate the Ipt promoter, we co-expressed the WRKY40 protein with ARF5 in the presence of the Ipt promoter-LUC construct in Arabidopsis mesophyll protoplasts. The Ipt promoter-driven luciferase activity was clearly higher, particularly in the presence of ARF5 and WRKY40 compared to ARF5 or WRKY40 alone (Fig. 5A). In contrast, expression of ARF5 together with WRKY18 or WRKY60 did not further enhance the Ipt promoter activity. This also indicates that WRKY40 is more important than WRKY18 and WRKY60 for activating the Ipt promoter.
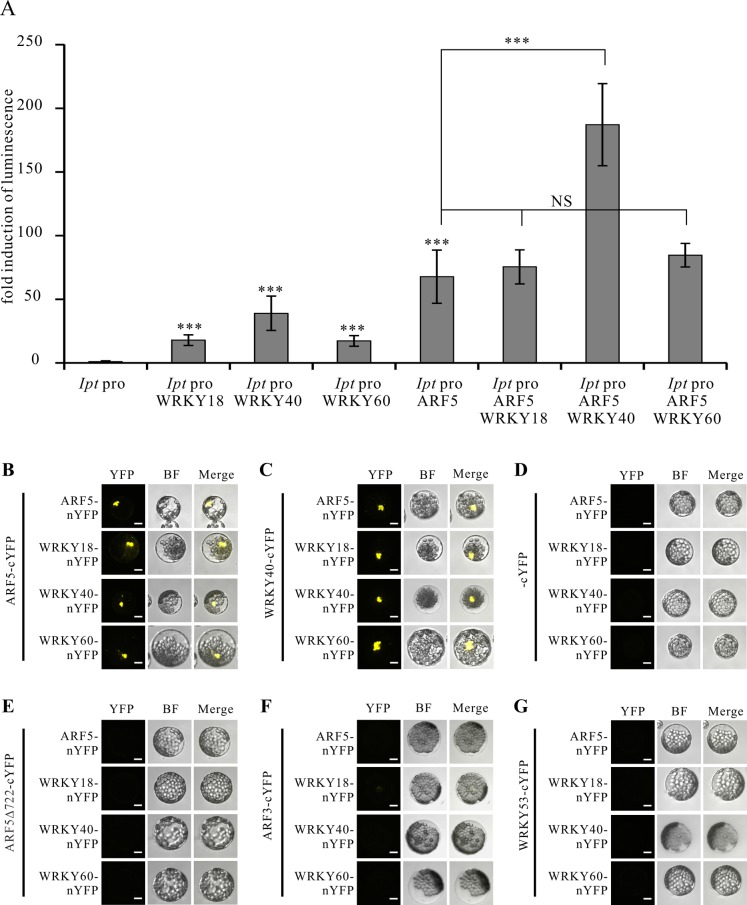
Figure 5. WRKY40 and ARF5 protein interaction potentiates Ipt promoter activity.
(A) Fold induction of Ipt promoter-driven luminescence in the presence of WRKY18, WRKY40, WRKY60 and ARF5 transcription factor expression plasmids in the protoplast transactivation system. The relative luminescence induced by the Ipt promoter in protoplasts without transfection of any of the transcription factor expression plasmids was set to 1. Bars show mean values (±SD) of three independent experiments *** P<0.01 (Student’s t-test). NS: not significant. (B) Bimolecular fluorescence (BiFC) assay with ARF5-cYFP and ARF5-nYFP, WRKY18-nYFP, WRKY40-nYFP, WRKY60-nYFP, (C) with WRKY40-cYFP, (D) with cYFP, (E) with a C-terminal deletion of ARF5 (ARF5Δ722-cYFP), (F) with ARF3-cYFP and (G) with WRKY53-cYFP in Arabidopsis mesophyll protoplasts. YFP, image in fluorescence mode of reconstituted yellow fluorescent proteins; BF, images in bright filed mode; merge, overlay of YFP with the corresponding BF image. Bars, 10 μm.
These results imply that the WRKY40 and ARF5 proteins interact to synergistically activate Ipt gene expression. This was tested using the Bimolecular Fluorescence Complementation (BiFC) assay to study protein interactions between the WRKYs and ARF5. The C-terminal half of the yellow fluorescent protein (cYFP) was fused to the C-terminus of the ARF5 and WRKY40 proteins to express ARF5- and WRKY40-cYFP fusion proteins, respectively. The N-terminal half of YFP (nYFP) was fused to the C-terminus of the three WRKY proteins as well as to ARF5 to generate WRKY18-, WRKY40-, WRKY60-nYFP and ARF5-nYFP. Observation of YFP-mediated fluorescence demonstrates that both WRKY40 and ARF5 interacted with themselves and with all the other expressed genes, when transiently co-expressed in Arabidopsis mesophyll protoplasts (Fig. 5B, C). The fluorescence signal was always restricted to the nucleus. The free cYFP construct was used as negative control, and showed no YFP fluorescence when co-expressed with the WRKY-nYFPs and ARF5-nYFP in protoplasts (Fig. 5D).
It has been reported that the domain III and IV at the C-terminus of the ARF5 protein is important for dimerization and protein-protein-interaction [53–55]. To prove whether these domains are required for the interaction with the WRKY proteins, we fused a C-terminal deletion of ARF5 (1–722 aa) to cYFP (ARF5Δ722-cYFP) and co-expressed them with either ARF5-nYFP or the three WRKY-nYFPs. Although stable [53], the truncated ARF5Δ722 protein was unable to interact with the intact ARF5 protein or with WRKY18, WRKY40 and WRKY60 (Fig. 5E). This indicates that the domains III and IV are not only required for self-interaction, but also for interaction with the three WRKYs. The specificity of the interactions between ARF5 and the three WRKYs was confirmed by co-expressing ARF3 (AT2G33860)-cYFP, which naturally lacks domain III and IV, and WRKY53-cYFP, expressed early after infection with A. tumefaciens strain C58 (3 hpi; S2 Table) [20,53]. Neither ARF3 nor WRKY53 interacted with ARF5, WRKY18, WRKY40, and WRKY60, thus verifying that the interactions between the WRKYs and ARF5 are specific (Fig. 5F, G).
The PTA assays indicate that WRKY40 has a stronger potential to activate the Ipt promoter than WRKY18 and WRKY60 (Fig. 3A and 5A). This implies that WRKY40 regulates the Ipt promoter directly. We therefore analyzed binding of WRKY40 to the Ipt promoter using the electrophoretic mobility shift assay (EMSA). The recombinant WRKY40 protein fused to six histidine amino acids at the N-terminus (6×His-WRKY40) was expressed and purified from E. coli and a 50 bp fragment (−184 bp to −135 bp) of the Ipt promoter, which contains three of the six W-boxes located in the promoter region, was radioactively labeled and served as a probe for EMSA (Fig. 6A). Only a weak band of the shifted Ipt promoter fragment (Fig. 6B, WRKY40-Ipt complex) was observed in the presence of 150 ng purified recombinant 6×His-WRKY40 protein, but a doubled amount of the His-tagged WRKY40 protein (300 ng) exhibited a much stronger band. Addition of unlabeled Ipt promoter fragments as competitor to the reaction mixture significantly reduced the binding of WRKY40 to the labeled Ipt promoter probe. Thus, the WRKY40 protein binds to the Ipt probe in vitro, suggesting that the Ipt promoter is a direct target of the WRKY40 transcription factor in plant cells.
Figure 6. WRKY40 binds to the Ipt promoter in vitro.
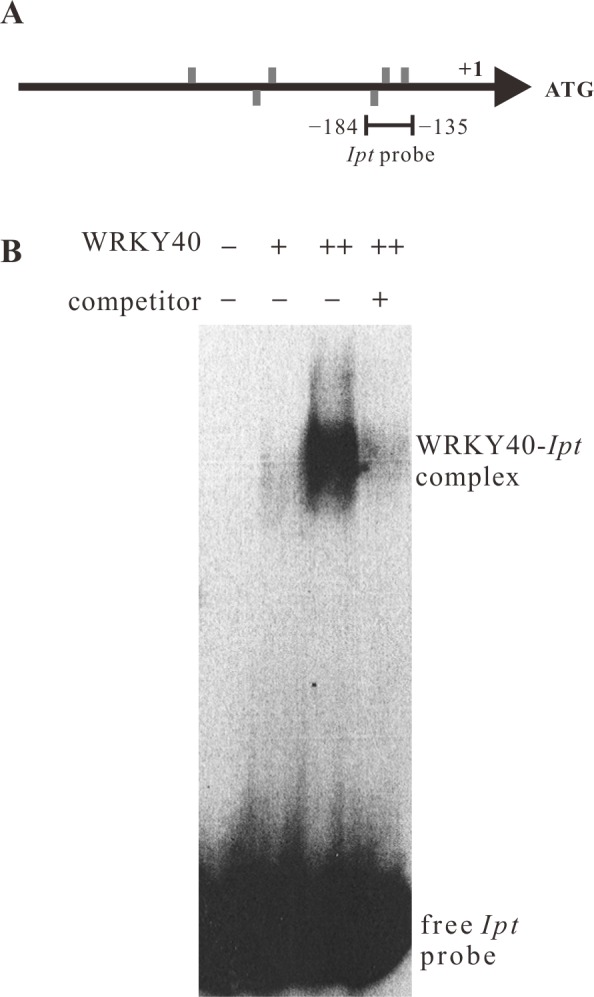
(A) Positions of W-boxes (TGAC, grey bars) in the sense (above the line) and anti-sense strand (below the line) of the Ipt promoter. The line below the Ipt promoter (−184 bp to −135 bp) indicates the fragment used as Ipt probe for electrophoretic mobility shift assay (EMSA). (B) EMSA with the labeled Ipt promoter probe in the absence (−) and in the presence of 150 ng (+) or 300 ng (++) of purified recombinant histidine-tagged WRKY40 protein. Competitor indicates without (−) and with (+) addition of unlabeled Ipt promoter probe. The WRKY40-Ipt complex indicates binding of WRKY40 protein to the labeled Ipt probe and the free Ipt probe no protein binding.
The ARF5-mediated auxin-signaling pathway induces Ipt, but not IaaH and IaaM gene expression
That ARF5 enhances the WRKY40-mediated activation of the Ipt promoter suggests that the auxin signaling pathway is involved in regulating Ipt expression. Previous studies have shown that the levels of unconjugated IAA in infected Arabidopsis stems are more than two-fold higher six days after inoculation with A. tumefaciens strain C58 compared to non-inoculated plant stems [20]. We found that crown galls accumulate four times more unconjugated IAA than control tissues and the total level of cytokinins in Arabidopsis crown gall tissues infected with A. tumefaciens strain C58 are 10 times higher than in crown gall-free stem tissues (8414 vs. 849 ng/g dry weight). The dominant cytokinin forms in Arabidopsis crown gall tissues were zeatin conjugates, including zeatin nucleotide (3657 vs. 308 ng/g dry weight) and zeatin riboside (2294 vs. 76 ng/g dry weight). The content of free zeatin was also higher in crown gall tissues than in mock-inoculated stems (544 vs. 34 ng/g dry weight).
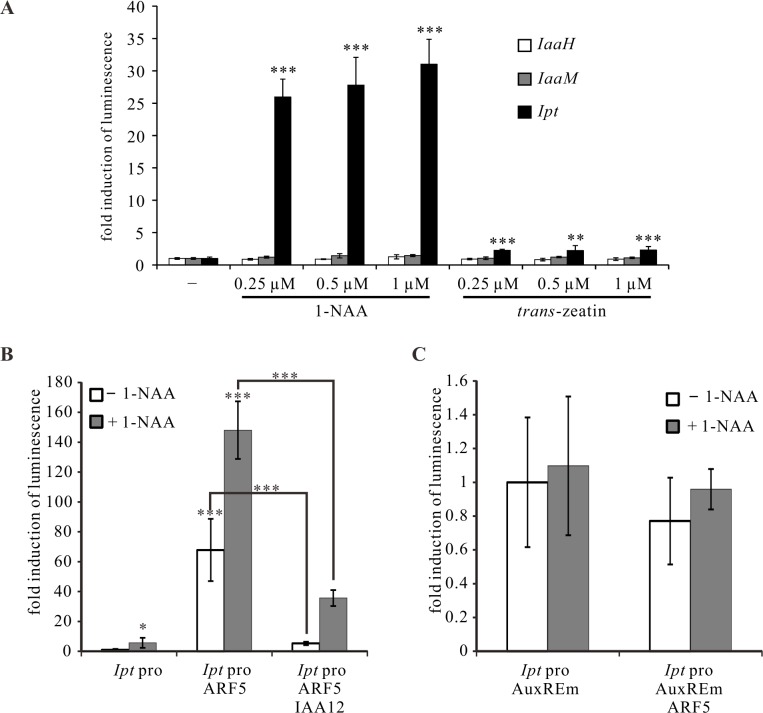
Based on these results, we used the PTA system to analyze the impact of auxin and cytokinin on IaaH, IaaM and Ipt promoter activity. The Ipt promoter was highly activated by the bioactive auxin type 1-naphthaleneacetic acid (1-NAA) and the cytokinin type trans-zeatin (Fig. 7A), with the latter much less effective. Increasing concentrations of auxin and cytokinin had no strong enhancing effect on the activity of the three oncogene promoters (Fig. 7A). The Ipt promoter sequence contains five auxin response elements (AuxREs, TGTCNC or TGTCTN) for binding of ARF transcription factors, whereas only one AuxRE is present in the bidirectional IaaH and IaaM promoter sequence (Table 1, S5 Fig.) and ARF transcription factors usually regulate their target genes in an auxin-dependent manner [33,34]. Thus, we analyzed the regulatory effect of ARF5 on the Ipt promoter in the presence of auxin in the PTA system. ARF5 activated the Ipt promoter, an activation that was even stronger when the mesophyll protoplasts were treated with auxin (1-NAA, Fig. 7B). Mutations in the AuxREs (sense TGTCNC or TGTCTN, anti-sense GNGACA or NAGACA) in the Ipt promoter abolished the auxin induction and the enhancing effect of ARF5 (Fig. 7C). It is known that auxin/indole-3-acetic acid (Aux/IAA) proteins can inhibit ARF mediated promoter activation and the repressor of ARF5 is IAA12 (also known as BODENLOS, BDL, AT1G04550) [54]. When we co-transfected Arabidopsis mesophyll protoplasts with the ARF5 and IAA12 plasmid constructs, a significant reduction in the Ipt promoter-driven luciferase activity was found compared to protoplasts transfected with only ARF5 (Fig. 7B). Nonetheless, the level of the Ipt promoter activity was not as low as it was in the absence of any transcription factor, indicating that not all ARF5 proteins are inhibited by IAA12.
Figure 7. ARF5 activates the Ipt promoter in an auxin-dependent manner.
(A) Fold induction of oncogene promoter-driven luminescence in Arabidopsis mesophyll protoplasts treated with auxin (1-NAA) or cytokinin (trans-zeatin) of different concentrations. Protoplasts were transfected with the IaaH, IaaM, and Ipt promoter-luciferase reporter constructs, and then incubated with different concentrations of 1-NAA or trans-zeatin overnight. (B) Fold induction of Ipt promoter-driven luminescence in Arabidopsis mesophyll protoplasts (Ipt pro) in the presence of the transcription factor expressing plasmids ARF5 (Ipt pro ARF5) or ARF5 plus IAA12 (Ipt pro ARF5 IAA12) with (+ 1-NAA) and without auxin (− 1-NAA) addition. (C) Mutations in the five auxin responsive elements (AuxREm, TGTCNC to TGGCNC and TGTCTN to TGGCTN) of the Ipt promoter abolish the ARF5- and auxin-dependent luminescence induction. The relative luminescence of intact or mutated Ipt promoters in the absence of any transcription factor expression plasmids and auxin treatment was set to 1. Bars show mean values (±SD) of three independent experiments. * P<0.05; ** P<0.01; *** P<0.001 (Student’s t-test).
In addition to the W-boxes and AuxREs, the IaaH, IaaM and Ipt promoters also contain ARR1 binding elements (GATT; Table 1), suggesting that the three oncogenes are regulated by type-B ARR transcription factors to mediate cytokinin signaling. The ARR1 gene is expressed at low levels in crown gall tissue of the virulent A. tumefaciens strain C58 and in stems infected with the disarmed strain GV3101 according to real time PCR measurements (S7A Fig.). ARR4 (AT1G10470), a type A transcription factor gene, was strongly expressed in crown gall tumors (S7A Fig.). The ability of both the ARR1 and ARR4 transcription factors to activate the IaaH, IaaM and Ipt promoters was tested in the PTA system. Neither ARR1 nor ARR4 significantly increased luciferase activity driven by the three oncogene promoters, even in the presence of trans-zeatin (S7B Fig.). Hence, the ARF5-mediated auxin signaling pathway, but not that of cytokinin, regulates Ipt expression, whereas the expression of IaaH and IaaM is not affected by either of the two signaling pathways.
Discussion
The plant pathogen, Agrobacterium tumefaciens takes advantage of the host transcriptional machinery to express its own T-DNA encoded oncogenes IaaH, IaaM and Ipt in plant cells. Expression of the oncogenes results in increased production of the phytohormones auxin and cytokinin, which induce uncontrolled cell proliferation and crown gall development. The T-DNA transformation process and the roles of the encoded oncogene enzymes is far better understood [10,56,57] than the regulation of oncogene expression in plant cells. We therefore examined this regulation, asking whether the expression of the IaaH, IaaM and Ipt oncogenes is regulated by host transcription factors and how oncogene expression is coordinated to obtain tumor-inducing auxin/cytokinin levels in a T-DNA transformed cell.
Agrobacterium tumefaciens utilizes a transcription factor of the pathogen defense pathway to induce Ipt oncogene expression
Expression of a gene in a eukaryotic cell requires general sequence features (e.g. TATA, CAAT) and potentially cis-regulatory elements for the binding of transcription factors. For the Ipt promoter of the octopine Ti plasmid pTiAch5, previous studies have shown that it binds CBF, a protein of unknown function from tobacco nuclear protein extracts [25–27]. This implies that at the least, expression of the Ipt oncogene is regulated by plant derived transcription factors. Nonetheless, using the PTA screening system we found that no transcription factor activated the IaaH and IaaM promoters of pTiC58. This may be because the transcription factor collection used for screening did not cover all the encoded Arabidopsis transcription factors; candidates for binding to the IaaH and IaaM promoters may have been missed. However, the very few cis-regulatory elements in the relatively short promoter sequence and the low level of transcription in crown galls, in addition to the low promoter activity in protoplasts, suggest that the IaaH and IaaM genes are not strongly activated by transcription factors, but instead are constitutively expressed at low basal levels. In contrast, the Ipt oncogene promoter of pTiC58 contains several W-boxes and is activated by the WRKY18, WRKY40 and WRKY60 proteins. The impaired crown gall growth on the wrky18, wrky40 and wrky60 mutant plants indicates that these WRKY transcription factors have a positive effect on crown gall development. The three WRKYs are paralogous transcription factors that cooperatively regulate biotic and abiotic stress responses in Arabidopsis [49,58–63] and the respective wrky mutants are known to be more resistant to biotrophic pathogens such as Pseudomonas syringae and powdery mildew Golovinomyces orontii [50]. Hence, the smaller crown galls on these wrky mutants may result from both fewer transformation events due to the stronger resistance response towards biotrophic pathogens and/or from reduced Ipt expression due to the loss of wrky function. Unfortunately, these two processes are not easy to separate in infection-based assays.
It is known that transcription of WRKY40 and WRKY60 is induced by fungal and bacterial pathogens [49,50]. Likewise, A. tumefaciens inoculation induced their transcription within two hours, indicating that they are expressed quite early in response to this pathogen. Thus, it is conceivable that the WRKYs are needed to trigger Ipt oncogene expression from the very beginning in a T-DNA transformed cell, so these pathogen responsive genes are already expressed when the T-DNA enters the host cell. Consequently, a reduction in Ipt promoter activity can be observed early on in the wrky triple mutant, vanishing at later infection stages. The relatively moderate difference in Ipt gene expression between the wrky triple mutant and wild-type most likely results from the increased expression of ARF5 and reduced expression of its inhibitor IAA12 in the mutant background. Thus, A. tumefaciens hijacks a host transcription factor, which is part of the plant pathogen defense machinery, to initiate expression of its own oncogene in the host cell.
Auxin, but not cytokinin signaling is important for inducing Ipt oncogene expression
A. tumefaciens and T-DNA transformed plant cells produce auxin and cytokinin [13,20]. Cytokinin affects cell division, essential for cell proliferation and initiation of crown gall development. Only the activity of the Ipt promoter, not that of the IaaH and IaaM genes, increased upon application of trans-zeatin, the dominant cytokinin in Arabidopsis crown galls. Eight binding elements for the ARR1 transcription factor are located in the bidirectional promoter of IaaH/IaaM and seven in the Ipt promoter. ARR1 is a type-B ARR transcription factor that activates transcription of cytokinin responsive genes [64,65]. Nonetheless, the activity of all three oncogene promoters was not influenced either by ARR1 or ARR4, even in the presence of trans-zeatin. This indicates that cytokinin signaling does not have a dominant role in oncogene expression. The auxin type 1-NAA was much more effective than trans-zeatin in activating the Ipt promoter, but again, not for the promoters of IaaH or IaaM. Elevated levels of free IAA are detectable in infected tissues six days after inoculation with A. tumefaciens strain C58 [20] and at the same infection stage, expression of the ARF5 gene begins to increase, as shown in the microarray data (1.49 fold, P value = 0.006) [20,46]. The Ipt promoter contains five AuxREs and is activated by 1-NAA and by the auxin response factor ARF5 upon release from inhibition by IAA12 in an auxin-dependent manner. Expression of the ARF5 gene is induced by auxin [66] and the elevated auxin levels in plant tissues infected and T-DNA transformed by A. tumefaciens most likely induce ARF5 gene expression and de-repress the ARF5 protein by proteolysis of IAA12. The release of ARF5 inhibition in the presence of auxin leads finally to activation of the Ipt promoter in the T-DNA transformed plant cell and may contribute to the moderate differences of Ipt transcript numbers in the wrky mutants and wild-type. Taken together, the results indicate that auxin is an important factor in regulating Ipt oncogene expression, which exerts its function through the auxin-sensitive transcription factor ARF5.
WRKY40 and ARF5 synergistically boost Ipt gene expression, thereby integrating host pathogen defense and auxin signaling
Our study shows that WRKY40 binds directly to the Ipt promoter in vitro and has the strongest effect on Ipt promoter activation in plant cells, an activation that increases even further in the presence of the ARF5 transcription factor. It has been shown that WRKY transcription factors specifically interact with different kinds of proteins [67] and WRKY18, WRKY40 and WRKY60 interact with each other and themselves [49], a result confirmed in this study. Moreover, WRKY18, WRKY40, and WRKY60 interact with ARF5. Most ARFs contain four important domains, except for ARF3, ARF13 and ARF17, which lack domain III and IV and ARF23, which has only domain I [31]. Domain III and IV are localized at the C-terminus of ARF proteins and are important for dimerization and interaction with Aux/IAA proteins [53]. According to our study, the domain III and IV of ARF5 seem to be required for the interaction with the three WRKY transcription factors. The interaction of ARF5 with WRKY40, but not that with WRKY18 and WRKY60, greatly enhances the activation of the Ipt promoter, so emphasizing the role of WRKY40 as the most important transcriptional activator of Ipt gene expression. Moreover, the WRKY40-ARF5 interaction links two signaling pathways for the regulation of Ipt gene expression: the ARF5-dependent auxin and WRKY-mediated pathogen defense pathway. Both pathways are activated in the host plant upon infection with A. tumefaciens and synergistically boost expression of the Ipt gene in T-DNA transformed cells.
Conclusion
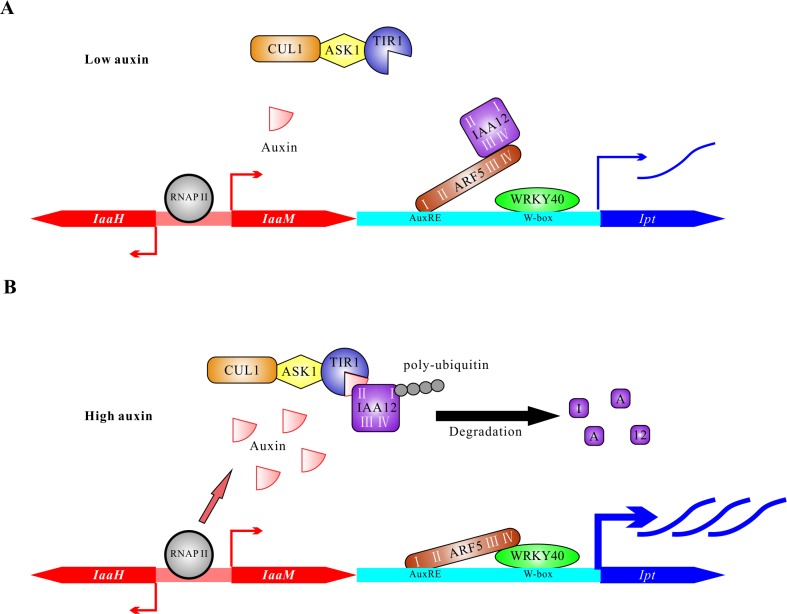
This study suggests a bifactorial regulation of oncogene expression in T-DNA transformed plant host cells (Fig. 8). Just after A. tumefaciens infection, auxin and cytokinin levels are as low as in an untransformed plant cell. The WRKY40 gene is soon expressed in response to infection, and the protein binds to W-boxes in the Ipt promoter to induce gene expression (Fig. 8A). Under low auxin conditions, ARF5 interacts with IAA12, so is inactivated. Over time, the auxin concentration increases in the T-DNA transformed cell, the result of the ubiquitous expression of IaaH and IaaM, driven by binding the RNA polymerase II complex to the promoter and additional auxin that can be secreted from the A. tumefaciens cells into the apoplast. Under high auxin concentrations, the ARF5 inhibitor IAA12 is poly-ubiquitinylated and degraded, thus releasing the transcription factor ARF5. The de-repressed ARF interacts via domain III and IV with WRKY40, resulting in strong expression of the Ipt oncogene. Taken together, this transcription factor interaction integrates two signaling pathways: the WRKY-based pathogen defense pathway for initial induction of Ipt gene expression and later, the auxin signaling pathway to boost Ipt expression. Moreover, the alterations in Ipt expression levels may be a mechanism to fine-tune the cytokinin to auxin ratios in a transformed plant cell. The appropriate auxin/cytokinin balance is an important mechanism to control whether a crown gall will proliferate or grow and differentiate.
Figure 8. Proposed model on the regulation of oncogene expression in host cells.
(A) The plant cells have low auxin levels at the early stage of transformation by A. tumefaciens. The bidirectional promoter between CDSs of IaaH and IaaM is recognized by RNA polymerase II complex (RNAP II) to induce basal levels of transcription. Under low auxin conditions, the transcriptional activity of ARF5 is largely blocked by IAA12 via the interaction of domain III and IV. The transcription factor WRKY40 binds to W-boxes (TGAC) located in the Ipt promoter and activates transcription of the Ipt gene. (B) T-DNA transformed plant cells contain elevated auxin levels six days after inoculation due to the constitutive activity of the IaaH and IaaM enzymes and secretion by A. tumefaciens cells. High concentration of auxin mediates the interaction between TIR1 and IAA12. Poly-ubiquitination of IAA12 by the complex of TIR1, SKP (ASK1) and cullin1 (CUL1) induces its degradation and releases ARF5 to bind to the AuxREs (TGTCNC or TGTCTN) in the Ipt promoter and form a complex with WRKY40 via domain III and IV. The ARF5-WRKY40 complex finally potentiates activation of the Ipt promoter and promotes expression of the Ipt oncogene.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the genetic background of the wrky18 (GABI-Kat 328G03), wrky40 (SLAT_N40001), and wrky60 (SALK_120706) mutants [58]. Plants were grown on soil and cultivated in growth chambers (Percival AR-66L2, Perry, USA) with 12 h light (ca. 120 μmol·m−2·s−1 fluorescent white light, TL70, Philips, Eindhoven, Netherlands) at 22°C and 12 h dark at 16°C. The crown gall callus cell culture was generated by inoculating A. thaliana root segments of ecotype Wassilewskija (WS-2) with the virulent Agrobacterium tumefaciens strain C58 and cultivated on MS agar plates [1× MS basal salts including vitamins and MES buffer (Murashige and Skoog medium, Duchefa Biochemie, Haarlem, Netherlands), 10 g/L sucrose, 100 mg/L myo-Inositol (Duchefa Biochemie, Haarlem, Netherlands), 7.5 g/L plant agar (Duchefa Biochemie, Haarlem, Netherlands), pH 5.7] without the addition of phytohormones, but with 100 mg/L ticarcillin disodium/clavulanate potassium (Duchefa Biochemie, Haarlem, Netherlands). The GFP expressing crown gall cell cultures were generated in the same manner except the A. tumefaciens strain C58 was used. This harbored, in addition to its pTiC58 plasmid, the binary vector pMDC206 [68] with the IaaH, IaaM and Ipt promoter-green fluorescent protein (GFP) constructs inserted in the T-DNA region. The antibiotic hygromycin (30 mg/L) was added to the agar medium for selection of transformed cells. All callus cultures were transferred to fresh media every three weeks. The crown gall cell suspension cell cultures were grown in the dark at 22°C with gentle shaking at 160 rpm, and transferred to fresh medium [1× MS basal salts including vitamins and MES buffer (Murashige and Skoog medium, Duchefa Biochemie, Haarlem, Netherlands), 20 g/L sucrose, 100 mg/L myo-Inositol (Duchefa Biochemie, Haarlem, Netherlands), pH 5.7] at a 1:2 dilution (v/v) twice a week.
Agrobacterium tumefaciens strains, cultivation and plant inoculation procedures
The virulent A. tumefaciens strain C58 nocc (nopaline catabolism, number 584; Max Planck Institute for Plant Breeding, Cologne, Germany) and the disarmed derivative of C58, strain GV3101 (pMP90) were used for plant inoculations. The strains were cultivated on YEB-agar plates (5 g/L yeast extract, 5 g/L tryptone, 5 g/L sucrose, 50mM MgSO4, and 15 g/L agar) at 28°C for 2 days. GV3101 was cultivated in the presence of rifampicin (10 mg/L) and gentamicin (25 mg/L). Before plant inoculation, the A. tumefaciens strains were transferred into King’s liquid medium (20 g/L protease peptone, 1.5 g/L K2HPO4, 10 mL/L glycerol, 600 μM MgSO4) and grown overnight at 28°C and 140 rpm. King’s medium was removed by pelleting the bacteria three times at 8000 rpm for 1 min and resuspension in Agromix buffer (0.01 M MgCl2, 0.01 M MES pH 5.6). For recovery, the resuspended cells were cultured at 28°C and 140 rpm for 2 to 3 hours. The optical density (OD600) was measured at 600 nm (NanoDrop 2000c UV-Vis Spectrophotometer, Thermo, Waltham, USA) and adjusted to OD600 1.0 for leaf infiltrations and OD600 0.5 for inflorescence stem inoculations. A. tumefaciens suspensions were infiltrated into the abaxial side of 5-week-old Arabidopsis (Col-0) leaves by tightly pressing the orifice of a 1 mL syringe onto the leaf surface. For induction of crown gall growth, young inflorescence stems (3 to 10 cm) of A. thaliana plants were inoculated by injecting A. tumefaciens suspensions four times with a 5 mL syringe and a needle attached to it. Crown galls were separated from the inflorescence stems 25 days after inoculation with a scalpel using a dissecting microscope (Leica MZ6) and their weight was immediately determined. Leaves infiltrated or stems inoculated with A. tumefaciens strain GV3101 served as reference.
Construction of recombinant plasmids
For construction of the promoter-GFP fusions (IGR1a::GFP, IGR1b::GFP and IGR2::GFP), the vector pMDC206 was used, which contains the coding sequence (CDS) of GFP including an intron [68]. The promoter sequences of the intergenic regions (Fig. 2A) between the IaaH and IaaM CDS (IGR1, 337 bp) and between the IaaM and Ipt CDS (IGR2, 697 bp) of the pTiC58 plasmid were inserted upstream of the GFP CDS using Gateway cloning technology [68]. IGR1 was cloned in both directions (IGR1a and IGR1b, Fig. 2B). The ubiquitous cauliflower mosaic virus (2× CaMV35S) promoter was used as a positive control. To construct the plasmids for the Bimolecular Fluorescence Complementation (BiFC) assay and the luciferase reporter constructs, the pSAT vector was altered to be used in the USER cloning strategy as described in [69,70]. For the BiFC assay, the ubiquitin 10 (UBQ10) promoter and CDS of the C-terminal half (Venus, 156–239) and N-terminal half (Venus, 1–173aa, I152L) of the yellow fluorescent protein (cYFP, nYFP) were inserted into the pSAT vector. The full CDSs, excluding the stop codon of WRKY18, WRKY40, WRKY53, WRKY60, ARF3 and ARF5, and of the C-terminal deletion of ARF5 (1–722 aa), were inserted before the C-terminus of the cYFP or nYFP to generate the fusion proteins WRKY-cYFP, WRKY-nYFP, ARF-cYFP, ARF-nYFP and ARF5Δ722-cYFP. To generate the IaaH, IaaM, Ipt promoter-firefly luciferase reporter constructs (IaaH promoter-LUC, IaaM promoter-LUC and Ipt promoter-LUC), DNA fragments of the luciferase reporter CDS and the CaMV-terminator were introduced into the pSAT vector first, then the sequences of IGR1a, IGR1b and IGR2 (Fig. 2B) were added upstream of the luciferase reporter CDS. To express a histidine-tagged WRKY40 protein in E. coli cells, full length CDS including the stop codon was cloned into the vector pET28b (Novagen Merck Millipore, Darmstadt, Germany) at the NdeI and XhoI restriction enzymes sites. This resulted in expression of a WRKY40 protein fused at its N-terminus with 6× histidine amino acids (6×His-WRKY40). For site-specific mutagenesis of the AuxREs in the Ipt promoter (Ipt promoter AuxREm), the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, USA) was used. All primer sequences used are listed in S2 Table.
5’ rapid amplification of cDNA ends (5’ RACE)
For analysis of the transcription start sites of the IaaH, IaaM and Ipt oncogenes of A. tumefaciens strain C58 in plant cells, the mRNA extracted from crown gall callus cells was used. The mRNA was extracted from approximately 50 mg crown gall callus material by using Dynabeads Oligo (dT)25 (Invitrogen, Carlsbad, USA) following the manufacturer’s protocol. First-strand cDNA was generated by using SMARTScribe Reverse Transcriptase, the SMARTer II A Oligonucleotide primer and the 5’ RACE CDS primer A (Clontech, Otsu, Japan). The fragments of the 5’ ends of the oncogene cDNAs were amplified using DreamTaq DNA Polymerase (Fermentas, Thermo, Waltham, USA) and the Universal Primer A Mix (UPM) and the gene specific primers (IaaH reverse, IaaM reverse and Ipt reverse, S2 Table). The resulting PCR products were cloned using the pGEM-T Easy Vector (Promega, Fitchburg, USA) and transformed into the E. coli strain MRF (Agilent Technologies, Santa Clara, USA). At least three independent clones were sequenced to determine the transcription start site of each gene.
Reverse transcription polymerase chain reaction (RT-PCR) and quantitative real-time PCR (qRT-PCR)
Total RNA from approximate 50 mg plant tissue was extracted by using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Before reverse transcription, about 500 to 1000 ng of total RNA extracted from Arabidopsis tissue was digested by DNase I (Fermentas, Thermo, Waltham, USA) for 30 min at 37°C. DNase digestion was terminated by the addition of 25 mM EDTA and subsequent incubation at 70°C for 10 min. First strand cDNA synthesis was performed using oligo(dT) 18 primers (Fermentas, Thermo, Waltham, USA) and the Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Thermo, Waltham, USA). Quantitative RT-PCR with the plant cDNA samples was performed as described in [20]. The primer sequences used are listed in S2 Table.
Protoplast transactivation (PTA) system and luminescence measurements
The Arabidopsis mesophyll protoplast isolation and transfection procedures were performed as described in [48,71]. For transfection, 30 μL protoplast suspension (approximately 1×104 cells), 1 μg plasmid DNA of oncogene promoter-LUC constructs (IaaH promoter-LUC, IaaM promoter-LUC and Ipt promoter-LUC) and 1 μg of the expression plasmids containing the CaMV35S::transcription factor constructs of the transcription factor library [48] were combined in each well of a microtiter plate (Nunc U96; MicroWell Polypropylene Plates, Thermo, Waltham, USA). As an internal standard, 1 μg plasmid expressing the Renilla luciferase driven by the CaMV35S promoter (CaMV35S::Renilla LUC) was co-transfected. The protoplast suspension mixture was incubated overnight in the dark and at room temperature. The following day, a dual luciferase measurement was performed using the Renilla-Juice BIG Kit and Beetle-Juice BIG Kit (PJK GmbH, Kleinblittersdorf, Germany). The protoplasts settled at the bottom of the wells by gravity, then the supernatant was removed from the protoplast suspensions and 20 μL Lysis-Juice 2 (Renilla-Juice BIG KIT) was added to each well and mixed by pipetting. After 15 min on ice, the microtiter plate was centrifuged (4000 rpm for 10 min). An aliquot of 10 μL of the supernatant was transferred into the wells of two new microtiter plates. As substrate for the two types of luciferase enzymes, 50 μL Renilla-Juice for renilla luciferase (CaMV35S::Renilla LUC) and 50 μL Beetle-Juice for firefly luciferase (IaaH promoter-LUC, IaaM promoter-LUC and Ipt promoter-LUC) were added via the liquid handling robotic device and the luminescence was measured by the Robion Solaris plate reader luminometer (STRATEC Biomedical Systems AG, Birkenfeld, Germany). The relative luminescence intensity was calculated from the values of Firefly-LUC versus Renilla-LUC. The relative luminescence intensity calculated from the oncogene promoter-LUC constructs (IaaH promoter-LUC, IaaM promoter-LUC and Ipt promoter-LUC) in the absence of any expression plasmids containing the CaMV35S::transcription factor constructs or phytohormone treatments was set to 1. The fold induction in luminescence represents the relative activity induced by certain transcription factors or treatments.
Protein expression and electrophoretic mobility shift assays (EMSA)
Protein synthesis was induced in the bacterial suspension of the transgenic E. coli SoluBL21 strain (Genlantis, San Diego, USA) expressing the 6×His-WRKY40 fusion protein by adding 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD600 0.6 overnight at 16°C. Purification of the histidine-tagged WRKY40 protein was performed according to the protocol from Novagen (Merck Millipore, Darmstadt, Germany). To generate the 50 bp of the Ipt promoter probe used in EMSA, two complementary oligonucleotides were synthesized by Sigma (Sigma Aldrich, St. Louis, USA). The two oligonucleotides were mixed at a 1:1 molar ratio in annealing buffer (10 mM Tris, pH8.0, 1 mM EDTA, 50 mM NaCl). The mixture was incubated at 95°C for 5 min and slowly cooled to room temperature and incubated overnight. The double-stranded oligonucleotides were purified from an 3% (w/v) agarose gel after electrophoresis, then radioactively labeled by using T4 polynucleotide kinase (Fermentas, Thermo, Waltham, USA) and [gamma-32P] adenosine 5’-triphosphate (ATP; Hartmann Analytic GmbH, Braunschweig, Germany). About 5 ng labeled probe and 150−300 ng 6×His-WRKY40 protein were mixed in DNA-protein binding buffer [10 mM Tris-HCl pH8.0, 0.5 mM ZnSO4, 0.25 mM DTT, 0.1 μg/μL poly [dI-dC], 5% (v/v) glycerol]. The binding reaction mixture was incubated on ice for 30 min and separated in a 6% (w/v) native polyacrylamide gel [45 mM Tris-borate, 1 mM EDTA, pH 8.6, 15% (v/v) Rotiphorese Gel 40 (29:1; Roth, Karlsruhe, Germany), 0.1% (w/v) ammonium persulfate (APS), 0.5% (v/v) TEMED] at 4°C for 3 h at 200 V in 0.5 × TBE buffer (45 mM Tris-borate and 1 mM EDTA; pH 8.6). The gel was fixed in 5% acetic acid for 10 min and dried for approximately 1 h (gel drying systems, Bio-Rad, Hercules, USA), exposed at −70°C to an x-ray film (Eastman Kodak, Rochester, USA) overnight and then developed.
Cytokinin analysis
Crown gall materials used for cytokinin analysis were obtained from Wassilewskija (WS-2) stems inoculated with A. tumefaciens strain C58. The analysis was performed as described in [19].
Bimolecular fluorescence complementation (BiFC) assay and microscopy
For the BiFC assay, 20 μg of each cYFP and nYFP protein fusion constructs (WRKY-cYFP, WRKY-nYFP, ARF-cYFP, ARF-nYFP and ARF5Δ722-cYFP) were transfected into mesophyll protoplasts using the PEG-calcium transfection method [71]. After incubation for 16–18 h in the dark at room temperature, protoplasts were inspected and images were taken using a confocal laser scanning microscope (Leica TCS SP5II, Leica Wetzlar, Germany).
Fluorescing plant cells and tissues were inspected and documented using an epifluorescence microscope (BZ 8000K, Biozero, Keyence, Osaka, Japan) and the software program (BZ observation application). For the inspection of intact plants, a dissecting microscope (Leica MZ6, Leica, Wetzlar, Germany) was used and pictures of crown galls were taken using a Leica DFC500 camera (Leica, Wetzlar, Germany).
Accession numbers of Arabidopsis genes
The Arabidopsis gene indexes (AGI) of genes mentioned in the text are AT2G33860 (ARF3), AT1G19850 (ARF5), AT3G16857 (ARR1), AT1G10470 (ARR4), AT1G04550 (IAA12), AT5G63790 (NAC102), AT4G31800 (WRKY18), AT1G80840 (WRKY40), AT4G23810 (WRKY53) and AT2G25000 (WRKY60). AGI codes are from The Arabidopsis Information Resource database (TAIR, http://www.arabidopsis.org).
Supporting Information
Nucleotide sequences of indole-3-acetamide hydrolase (IaaH) promoters including the 5’ untranslated regions of T-DNAs from the nopaline-type (pTiC58, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. The arrow indicates the position of the transcription start sites (TSS) +1. Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences.
(PDF)
Nucleotide sequences of tryptophan monooxygenase (IaaM) promoters including the 5’ untranslated regions of T-DNAs from the T-DNAs of the nopaline-type (pTiC58, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. An arrow indicates the position of the transcription start sites (TSS) +1. Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences.
(PDF)
Nucleotide sequences of isopentenyl transferase (Ipt) promoters including the 5’ untranslated regions of T-DNAs from the nopaline-type (pTiC58, pTiT37, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. An arrow indicates the position of the transcription start sites (TSS). Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences. The framed “TATAAA” sequence indicates the TATA box, only conserved in the octopine type Ti plasmids. The sequence from −150 to −91 shows the conserved region, which is named as Ipt element.
(PDF)
Fold induction of Ipt promoter-driven luminescence in the presence of WRKY18, WRKY40 and WRKY60 transcription factor expression plasmids in the protoplast transactivation system. The relative luminescence induced by the Ipt promoter in protoplasts without transfection of any of the transcription factor expression plasmids was set to 1. Bars show mean values (±SD) of three independent experiments.
(PDF)
Positions of TATA boxes (TATAAA, blue bars), auxin responsive elements (AuxREs, TGTCNC or TGTCTN, green bars), W-boxes (TGAC, red bars) and transcript start site (TSS arrow) in the sense (above the line) and anti-sense strand (below the line) of the intergenic region (IGR1) and IGR2.
(PDF)
(A) Relative transcript numbers of the Ipt oncogene in 25-day-old crown galls of wild-type plants and wrky18, wrky40 and wrky60 single mutants, (B) in stems of wild-type plants 2 days and 6 days post-inoculation (2 dpi and 6 dpi) of A. tumefaciens strain C58 and (C) of ARF5 and IAA12 in crown gall tumors of the wild-type Col-0 and wrky single mutants. Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. NS: not significant. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; NS: not significant (Student’s t-test).
(PDF)
(A) Relative transcript numbers of the ARR1 and ARR4 genes in crown galls 25 days after inoculation with the virulent A. tumefaciens strain C58 (C58 Crown gall) and in stems inoculated with the disarmed strain GV3101 (GV3101 Stems). Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. (B) Fold induction of IaaH, IaaM, Ipt promoter-driven luminescence in Arabidopsis mesophyll protoplasts transfected with ARR1 and ARR4 transcription factor expression plasmids and in the presence or absence of trans-zeatin. The relative luminescence in the absence of ARR1, ARR4 expression plasmids and trans-zeatin was set to 1. Bars show mean values (±SD) of three independent experiments.
(PDF)
Analyses are based on existing microarray data [20,46] using the Plant Transcription Factor Database v3.0 [47] (http://planttfdb.cbi.pku.edu.cn/index.php?sp=Ath) for annotation. Genes are listed according to the following criteria: Fold change (FCh) ≥ 2 or ≤ 0.5, p value < 0.01. hpi: hours post inoculation; dpi: days post inoculation.
(PDF)
(PDF)
Acknowledgments
The wrky mutants were kindly provided by Imre E. Somssich (Department of Plant Microbe Interaction, Max Planck Institute for Plant Breeding Research, Cologne, Germany). Special thanks go to Edgar Serfling (Institute of Pathology, University of Wuerzburg, Wuerzburg, Germany) for help with the EMSA experiment. We also like to thank Veselov Dmitry and Guzel R. Kudoyarova (Institute of Biochemistry and Genetics, Ufa Scientific Centre of Russian Academy of Sciences, Ufa, Russia) for cytokinin analysis. For critical reading the manuscript, we thank Tracey A. Cuin (Julius-von-Sachs-Institute, Molecular Plant Physiology and Biophysics, University of Wuerzburg, Germany). This publication was funded by the University of Wuerzburg in the funding programme Open Access Publishing.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was financially supported by the Deutsche Forschungsgemeinschaft, GRK1342 (TP A7), SFB 567 (TP B5) and the China Scholarship Council (CSC) [to YZ]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, et al. (1977) Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11: 263–271. [DOI] [PubMed] [Google Scholar]
- 2. Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, et al. (2000) The bases of crown gall tumorigenesis. J Bacteriol 182: 3885–3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garfinkel DJ, Simpson RB, Ream LW, White FF, Gordon MP, et al. (1981) Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27: 143–153. [DOI] [PubMed] [Google Scholar]
- 4. Joos H, Inze D, Caplan A, Sormann M, Van Montagu M, et al. (1983) Genetic analysis of T-DNA transcripts in nopaline crown galls. Cell 32: 1057–1067. [DOI] [PubMed] [Google Scholar]
- 5. Britton M, Escobar M, Dandekar A (2008) The Oncogenes of Agrobacterium Tumefaciens and Agrobacterium Rhizogenes. In: Tzfira T, Citovsky V, editors. Agrobacterium: From Biology to Biotechnology: Springer New York; pp. 523–563. [Google Scholar]
- 6. Garfinkel DJ, Nester EW (1980) Agrobacterium tumefaciens mutants affected in crown gall tumorigenesis and octopine catabolism. J Bacteriol 144: 732–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schroder G, Waffenschmidt S, Weiler EW, Schroder J (1984) The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur J Biochem 138: 387–391. [DOI] [PubMed] [Google Scholar]
- 8. Thomashow LS, Reeves S, Thomashow MF (1984) Crown gall oncogenesis: evidence that a T-DNA gene from the Agrobacterium Ti plasmid pTiA6 encodes an enzyme that catalyzes synthesis of indoleacetic acid. Proc Natl Acad Sci U S A 81: 5071–5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomashow MF, Hugly S, Buchholz WG, Thomashow LS (1986) Molecular basis for the auxin-independent phenotype of crown gall tumor tissues. Science 231: 616–618. [DOI] [PubMed] [Google Scholar]
- 10. Escobar MA, Dandekar AM (2003) Agrobacterium tumefaciens as an agent of disease. Trends Plant Sci 8: 380–386. [DOI] [PubMed] [Google Scholar]
- 11. Gray J, Wang J, Gelvin SB (1992) Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J Bacteriol 174: 1086–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gray J, Gelvin SB, Meilan R, Morris RO (1996) Transfer RNA Is the Source of Extracellular Isopentenyladenine in a Ti-Plasmidless Strain of Agrobacterium tumefaciens. Plant Physiol 110: 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akiyoshi DE, Regier DA, Gordon MP (1987) Cytokinin production by Agrobacterium and Pseudomonas spp. J Bacteriol 169: 4242–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hwang HH, Wang MH, Lee YL, Tsai YL, Li YH, et al. (2010) Agrobacterium-produced and exogenous cytokinin-modulated Agrobacterium-mediated plant transformation. Mol Plant Pathol 11: 677–690. 10.1111/j.1364-3703.2010.00637.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang HH, Yang FJ, Cheng TF, Chen YC, Lee YL, et al. (2013) The Tzs protein and exogenous cytokinin affect virulence gene expression and bacterial growth of Agrobacterium tumefaciens. Phytopathology 103: 888–899. 10.1094/PHYTO-01-13-0020-R [DOI] [PubMed] [Google Scholar]
- 16. Morris RO (1986) Genes Specifying Auxin and Cytokinin Biosynthesis in Phytopathogens. Annual Review of Plant Physiology 37: 509–538. [Google Scholar]
- 17. Chateau S, Sangwan RS, Sangwan-Norreel BS (2000) Competence of Arabidopsis thaliana genotypes and mutants for Agrobacterium tumefaciens-mediated gene transfer: role of phytohormones. J Exp Bot 51: 1961–1968. [DOI] [PubMed] [Google Scholar]
- 18. Sardesai N, Lee LY, Chen H, Yi H, Olbricht GR, et al. (2013) Cytokinins secreted by agrobacterium promote transformation by repressing a plant myb transcription factor. Sci Signal 6: ra100 10.1126/scisignal.2004518 [DOI] [PubMed] [Google Scholar]
- 19. Veselov D, Langhans M, Hartung W, Aloni R, Feussner I, et al. (2003) Development of Agrobacterium tumefaciens C58-induced plant tumors and impact on host shoots are controlled by a cascade of jasmonic acid, auxin, cytokinin, ethylene and abscisic acid. Planta 216: 512–522. [DOI] [PubMed] [Google Scholar]
- 20. Lee CW, Efetova M, Engelmann JC, Kramell R, Wasternack C, et al. (2009) Agrobacterium tumefaciens promotes tumor induction by modulating pathogen defense in Arabidopsis thaliana. Plant Cell 21: 2948–2962. 10.1105/tpc.108.064576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper GM (2000) The Cell: A Molecular Approach. 2nd edition.
- 22. Goldberg SB, Flick JS, Rogers SG (1984) Nucleotide sequence of the tmr locus of Agrobacterium tumefaciens pTi T37 T-DNA. Nucleic Acids Res 12: 4665–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Klee H, Montoya A, Horodyski F, Lichtenstein C, Garfinkel D, et al. (1984) Nucleotide sequence of the tms genes of the pTiA6NC octopine Ti plasmid: two gene products involved in plant tumorigenesis. Proc Natl Acad Sci U S A 81: 1728–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nester EW, Gordon MP, Amasino RM, Yanofsky MF (1984) Crown Gall—a Molecular And Physiological Analysis. Annual Review Of Plant Physiology And Plant Molecular Biology 35: 387–413. [Google Scholar]
- 25. de Pater BS, de Kam RJ, Hoge JH, Schilperoort RA (1987) Effects of mutations in the TATA box region of the Agrobacterium T-cyt gene on its transcription in plant tissues. Nucleic Acids Res 15: 8283–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neuteboom ST, Hulleman E, Schilperoort RA, Hoge JH (1993) In planta analysis of the Agrobacterium tumefaciens T-cyt gene promoter: identification of an upstream region essential for promoter activity in leaf, stem and root cells of transgenic tobacco. Plant Mol Biol 22: 923–929. [DOI] [PubMed] [Google Scholar]
- 27. Neuteboom ST, Stoffels A, Hulleman E, Memelink J, Schilperoort RA, et al. (1993) Interaction between the tobacco DNA-binding activity CBF and the cyt-1 promoter element of the Agrobacterium tumefaciens T-DNA gene T-CYT correlates with cyt-1 directed gene expression in multiple tobacco tissue types. Plant J 4: 525–534. [DOI] [PubMed] [Google Scholar]
- 28. Eulgem T, Somssich IE (2007) Networks of WRKY transcription factors in defense signaling. Curr Opin Plant Biol 10: 366–371. [DOI] [PubMed] [Google Scholar]
- 29. Dong J, Chen C, Chen Z (2003) Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol Biol 51: 21–37. [DOI] [PubMed] [Google Scholar]
- 30. Ullrich CI, Aloni R (2000) Vascularization is a general requirement for growth of plant and animal tumours. Journal Of Experimental Botany 51: 1951–1960. [DOI] [PubMed] [Google Scholar]
- 31. Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460. [DOI] [PubMed] [Google Scholar]
- 32. Ulmasov T, Hagen G, Guilfoyle TJ (1997) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868. [DOI] [PubMed] [Google Scholar]
- 33. Hayashi K (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975. [DOI] [PubMed] [Google Scholar]
- 34. Peer WA (2013) From perception to attenuation: auxin signalling and responses. Curr Opin Plant Biol 16: 561–568. 10.1016/j.pbi.2013.08.003 10.1016/j.pbi.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 35. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13: 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- 38. Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- 39. Calderon Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, et al. (2012) A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. 10.1038/nchembio.926 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276. [DOI] [PubMed] [Google Scholar]
- 41. Yamamoto YY, Ichida H, Abe T, Suzuki Y, Sugano S, et al. (2007) Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res 35: 6219–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, et al. (2007) Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics 8: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Prestridge DS (1991) SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci 7: 203–206. [DOI] [PubMed] [Google Scholar]
- 44. Higo K, Ugawa Y, Iwamoto M, Higo H (1998) PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res 26: 358–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Deeken R, Engelmann JC, Efetova M, Czirjak T, Muller T, et al. (2006) An integrated view of gene expression and solute profiles of Arabidopsis tumors: a genome-wide approach. Plant Cell 18: 3617–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jin J, Zhang H, Kong L, Gao G, Luo J (2014) PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42: D1182–1187. 10.1093/nar/gkt1016 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wehner N, Hartmann L, Ehlert A, Bottner S, Onate-Sanchez L, et al. (2011) High-throughput protoplast transactivation (PTA) system for the analysis of Arabidopsis transcription factor function. Plant J 68: 560–569. 10.1111/j.1365-313X.2011.04704.x 10.1111/j.1365-313X.2011.04704.x [DOI] [PubMed] [Google Scholar]
- 49. Xu X, Chen C, Fan B, Chen Z (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pandey SP, Roccaro M, Schon M, Logemann E, Somssich IE (2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 64: 912–923. 10.1111/j.1365-313X.2010.04387.x 10.1111/j.1365-313X.2010.04387.x [DOI] [PubMed] [Google Scholar]
- 51. Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17: 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, et al. (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319. [DOI] [PubMed] [Google Scholar]
- 54. Lau S, De Smet I, Kolb M, Meinhardt H, Jurgens G (2011) Auxin triggers a genetic switch. Nat Cell Biol 13: 611–615. 10.1038/ncb2212 10.1038/ncb2212 [DOI] [PubMed] [Google Scholar]
- 55. Krogan NT, Ckurshumova W, Marcos D, Caragea AE, Berleth T (2012) Deletion of MP/ARF5 domains III and IV reveals a requirement for Aux/IAA regulation in Arabidopsis leaf vascular patterning. New Phytol 194: 391–401. 10.1111/j.1469-8137.2012.04064.x 10.1111/j.1469-8137.2012.04064.x [DOI] [PubMed] [Google Scholar]
- 56. Citovsky V, Kozlovsky SV, Lacroix B, Zaltsman A, Dafny-Yelin M, et al. (2007) Biological systems of the host cell involved in Agrobacterium infection. Cell Microbiol 9: 9–20. [DOI] [PubMed] [Google Scholar]
- 57. Pitzschke A, Hirt H (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29: 1021–1032. 10.1038/emboj.2010.8 10.1038/emboj.2010.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, et al. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103. [DOI] [PubMed] [Google Scholar]
- 59. Chen H, Lai Z, Shi J, Xiao Y, Chen Z, et al. (2010) Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10: 281 10.1186/1471-2229-10-281 10.1186/1471-2229-10-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu ZQ, Yan L, Wu Z, Mei C, Lu K, et al. (2012) Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J Exp Bot 63: 6371–6392. 10.1093/jxb/ers293 10.1093/jxb/ers293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206. [DOI] [PubMed] [Google Scholar]
- 62. Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7: 491–498. [DOI] [PubMed] [Google Scholar]
- 63. Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12: 9–26. [DOI] [PubMed] [Google Scholar]
- 64. Sakai H, Honma T, Aoyama T, Sato S, Kato T, et al. (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521. [DOI] [PubMed] [Google Scholar]
- 65. Taniguchi M, Sasaki N, Tsuge T, Aoyama T, Oka A (2007) ARR1 directly activates cytokinin response genes that encode proteins with diverse regulatory functions. Plant Cell Physiol 48: 263–277. [DOI] [PubMed] [Google Scholar]
- 66. Wenzel CL, Schuetz M, Yu Q, Mattsson J (2007) Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J 49: 387–398. [DOI] [PubMed] [Google Scholar]
- 67. Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, et al. (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6: 287–300. [DOI] [PubMed] [Google Scholar]
- 68. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, Halkier BA (2006) Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res 34: e122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, et al. (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516. [DOI] [PubMed] [Google Scholar]
- 71. Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- 72. Heidekamp F, Dirkse WG, Hille J, van Ormondt H (1983) Nucleotide sequence of the Agrobacterium tumefaciens octopine Ti plasmid-encoded tmr gene. Nucleic Acids Res 11: 6211–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. de Pater BS, Klinkhamer MP, Amesz PA, de Kam RJ, Memelink J, et al. (1987) Plant expression signals of the Agrobacterium T-cyt gene. Nucleic Acids Res 15: 8267–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lichtenstein C, Klee H, Montoya A, Garfinkel D, Fuller S, et al. (1984) Nucleotide sequence and transcript mapping of the tmr gene of the pTiA6NC octopine Ti-plasmid: a bacterial gene involved in plant tumorigenesis. J Mol Appl Genet 2: 354–362. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequences of indole-3-acetamide hydrolase (IaaH) promoters including the 5’ untranslated regions of T-DNAs from the nopaline-type (pTiC58, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. The arrow indicates the position of the transcription start sites (TSS) +1. Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences.
(PDF)
Nucleotide sequences of tryptophan monooxygenase (IaaM) promoters including the 5’ untranslated regions of T-DNAs from the T-DNAs of the nopaline-type (pTiC58, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. An arrow indicates the position of the transcription start sites (TSS) +1. Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences.
(PDF)
Nucleotide sequences of isopentenyl transferase (Ipt) promoters including the 5’ untranslated regions of T-DNAs from the nopaline-type (pTiC58, pTiT37, pTiSAKURA), octopine-type (pTi15955, pTiA6NC, pTiAch5) and agropine-type (pTiBo542) Ti plasmids. An arrow indicates the position of the transcription start sites (TSS). Negative numbers indicate the nucleotide positions upstream and positive numbers downstream of the TSSs. TATA box and CAAT box sequences are written above the aligned sequences. The framed “TATAAA” sequence indicates the TATA box, only conserved in the octopine type Ti plasmids. The sequence from −150 to −91 shows the conserved region, which is named as Ipt element.
(PDF)
Fold induction of Ipt promoter-driven luminescence in the presence of WRKY18, WRKY40 and WRKY60 transcription factor expression plasmids in the protoplast transactivation system. The relative luminescence induced by the Ipt promoter in protoplasts without transfection of any of the transcription factor expression plasmids was set to 1. Bars show mean values (±SD) of three independent experiments.
(PDF)
Positions of TATA boxes (TATAAA, blue bars), auxin responsive elements (AuxREs, TGTCNC or TGTCTN, green bars), W-boxes (TGAC, red bars) and transcript start site (TSS arrow) in the sense (above the line) and anti-sense strand (below the line) of the intergenic region (IGR1) and IGR2.
(PDF)
(A) Relative transcript numbers of the Ipt oncogene in 25-day-old crown galls of wild-type plants and wrky18, wrky40 and wrky60 single mutants, (B) in stems of wild-type plants 2 days and 6 days post-inoculation (2 dpi and 6 dpi) of A. tumefaciens strain C58 and (C) of ARF5 and IAA12 in crown gall tumors of the wild-type Col-0 and wrky single mutants. Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. NS: not significant. * p-value < 0.05; ** p-value < 0.01; *** p-value < 0.001; NS: not significant (Student’s t-test).
(PDF)
(A) Relative transcript numbers of the ARR1 and ARR4 genes in crown galls 25 days after inoculation with the virulent A. tumefaciens strain C58 (C58 Crown gall) and in stems inoculated with the disarmed strain GV3101 (GV3101 Stems). Relative transcript numbers were quantified by qRT-PCR and normalized to 10,000 molecules of ACTIN2/8. Bars show mean values (±SD) of three independent samples. (B) Fold induction of IaaH, IaaM, Ipt promoter-driven luminescence in Arabidopsis mesophyll protoplasts transfected with ARR1 and ARR4 transcription factor expression plasmids and in the presence or absence of trans-zeatin. The relative luminescence in the absence of ARR1, ARR4 expression plasmids and trans-zeatin was set to 1. Bars show mean values (±SD) of three independent experiments.
(PDF)
Analyses are based on existing microarray data [20,46] using the Plant Transcription Factor Database v3.0 [47] (http://planttfdb.cbi.pku.edu.cn/index.php?sp=Ath) for annotation. Genes are listed according to the following criteria: Fold change (FCh) ≥ 2 or ≤ 0.5, p value < 0.01. hpi: hours post inoculation; dpi: days post inoculation.
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.