Abstract
Molecular sub-classification is rapidly informing the clinical management of medulloblastoma. However, the disease remains associated with poor outcomes and therapy-associated late-effects, and the majority of patients are not characterized by a validated prognostic biomarker. Here, we investigated the potential of epigenetic DNA methylation for disease sub-classification, particularly in formalin-fixed biopsies, and to identify biomarkers for improved therapeutic individualization. Tumor DNA methylation profiles were assessed, alongside molecular and clinical disease features, in 230 patients primarily from the SIOP-UKCCSG PNET3 clinical trial. We demonstrate by cross-validation in frozen training and formalin-fixed test sets that medulloblastoma comprises four robust DNA methylation subgroups (termed WNT, SHH, G3 and G4), highly related to their transcriptomic counterparts, and which display distinct molecular, clinical and pathological disease characteristics. WNT patients displayed an expected favorable prognosis, while outcomes for SHH, G3 and G4 were equivalent in our cohort. MXI1 and IL8 methylation were identified as novel independent high-risk biomarkers in cross-validated survival models of non-WNT patients, and were validated using non-array methods. Incorporation of MXI1 and IL8 into current survival models significantly improved the assignment of disease-risk; 46% of patients could be classified as ‘favorable-risk’ (>90% survival) compared to 13% using current models, while the high-risk group was reduced to 16% from 30%. DNA methylation profiling enables the robust sub-classification of four disease sub-groups in frozen and routinely-collected/archival formalin-fixed biopsy material, and the incorporation of DNA methylation biomarkers can significantly improve disease-risk stratification. These findings have important implications for future risk-adapted clinical disease management.
Keywords: Subgroups, medulloblastoma, methylation, prognosis, biomarkers
Introduction
Medulloblastoma is the most common malignant pediatric brain tumor accounting for ~10% of childhood cancer deaths[37]. Current therapies are associated with variable survival outcomes and life-long cognitive and neuro-endocrine late-effects in the majority of survivors[37]. Four consensus molecular disease variants characterized by their gene expression profiles are recognized (WNT, SHH (Sonic Hedgehog), Group 3 and Group 4), which display distinct clinical, pathological and molecular biologic features[48]. Robust biomarkers of favorable-risk (β-catenin; WNT variant biomarker; ~10% of patients) and high-risk (MYC amplification; ~5%) have been identified and validated in large/trials-based series[14-16,36,43]. These will be used to assign reduced treatments to WNT patients in first international clinical trials of biologically-stratified therapy[37]. Despite these advances, the majority of medulloblastomas are not characterized by a validated prognostic biomarker and there remains a major unmet need for improved biological characterization to inform further therapeutic refinements, particularly in non-WNT patients.
Studies of DNA methylation profiles in adult brain tumors have demonstrated great promise in sub-classifying disease[34], and predicting clinical outcome; MGMT status predicts temozolomide sensitivity in glioblastoma[18]. However, studies of methylation events in medulloblastoma have been restricted to specific genes and/or modestly-sized cohorts[1,10,20,25,27]. The wider role of DNA methylation patterns in medulloblastoma, and their clinical impact, remain unknown.
We report a first examination of medulloblastoma DNA methylation patterns, using an extensive primary tumor cohort (n=230) drawn primarily from the SIOP/UKCCSG PNET3 clinical trial[49]. We aimed to assess the ability of DNA methylation patterns to sub-classify medulloblastoma, especially the validity of such sub-grouping in formalin-fixed/paraffin-embedded (FFPE) material. DNA methylation events are far more stable in comparison to RNA and methylation-based sub-grouping may prove more amenable to clinical application in standard or archival biopsy material than RNA-based methods. Finally, we assessed the ability of individual methylation events to improve outcome prediction. We establish methylation events as clinically useful biomarkers and demonstrate how their incorporation into current risk-stratification schemes significantly improves the accuracy of survival prediction, allowing the re-classification of disease-risk in the majority of patients and providing a potential basis for future therapeutic individualization aimed at improved outcomes and reduced late-effects.
Materials and methods
DNA methylation profiling of medulloblastoma cohorts
DNA methylation events were profiled using the Illumina GoldenGate Cancer Panel I microarray[2], according to manufacturer’s instructions. This array measures the DNA methylation status at 1505 loci in 807 genes, as a β-score ranging from zero (fully unmethylated) to one (fully methylated). All samples were subjected to array quality control (QC)[4,12], alongside assessments of intra- and inter-replicate reproducibility, and quantitative accuracy, in extensive validation experiments versus independent bisulfite sequencing estimations of methylation status (Supplementary Methods, Supplementary Table S1 and Supplementary Figure S1).
230 primary medulloblastomas passed QC measures following array analysis. Patients were split into training (n=100; DNA (n=100) and RNA (n=88) extracted from fresh-frozen tumor material) and test cohorts (n=130; DNA from FFPE tissue, derived exclusively from patients enrolled on the International Society of Pediatric Oncology (SIOP)/United Kingdom Children’s Cancer Study Group (UKCCSG) PNET3 clinical trial[49]). Pathology was reviewed according to WHO criteria[28], and metastatic (M) stage determined using Chang’s criteria[6]. Prognostic models were developed using the combined cohort (n=191) aged 3.0-16.0 years at diagnosis, representing 136 PNET3, and 55 training cohort patients. Cohort demographics are summarized in Table 1.
Table 1. Clinical, pathological and molecular features of medulloblastoma cohorts.
M−, M0-1; M+, M2-4; CLA, classic; DN, desmoplastic / nodular; LCA, large-cell anaplastic; RTX, radiotherapy; CTX, chemotherapy. The single tumor with extensive nodular (MBEN) pathology was included in the DN group. Percentages shown are based on patients with available data. NA, data not available; NC, non-classifiable.
| Demographic | Cohort |
|||
|---|---|---|---|---|
| Training (n=100) | Test (n=130) | Combined survival (n=191) | ||
|
| ||||
| Tissue type | Frozen | 94 (94% ) | 0 (0%) | 55 (29%) |
| FFPE | 6 (6%) | 130 (100%) | 136 (71%) | |
|
| ||||
| Gender | Male (M) | 62 (62%) | 78 (60%) | 113 (59%) |
| Female (F) | 38 (38%) | 52 (40%) | 78 (41%) | |
| M:F ratio | 1.6:1 | 1.5:1 | 1.4:1 | |
|
| ||||
| Age at diagnosis (years) | Median (range) | 7.9 (0.1 – 43.0) | 8.4 (3.1 – 15.6) | 8.5 (3.1 – 15.8) |
|
| ||||
| < 3 years | 15 (15%) | 0 (0%) | 0 (0%) | |
| ≥ 3 years | 85 (85%) | 130 (100%) | 191 (100%) | |
|
| ||||
| Pathology variant | CLA | 72 (72%) | 110 (85%) | 157 (82%) |
| DN | 18 (18%) | 9 (7%) | 16 (8%) | |
| LCA | 10 (10%) | 11 (8%) | 18 (9%) | |
|
| ||||
| Metastatic stage | M− | 75 (81%) | 105 (81%) | 154 (81%) |
| M+ | 18 (19%) | 25 (19%) | 37 (19%) | |
| NA | 7 | 0 | 0 | |
|
| ||||
| Treatment | RTX-alone | 8 (11%) | 63 (48%) | 68 (36%) |
| RTX + CTX | 65 (89%) | 67 (52%) | 121 (64%) | |
| NA | 27 | 0 | 2 | |
|
| ||||
| Follow-up (surviving patients (years)) | Median (range) | 5.0 (0.1 – 15.5) | 10.1 (0.1 – 14.9) | 8.9 (0.1 – 14.9) |
|
| ||||
| CTNNB1 | No mutation | 87 (90%) | 118 (93%) | 168 (90%) |
| Mutation | 10 (10%) | 9 (7%) | 18 (10%) | |
| NA | 3 | 3 | 5 | |
|
| ||||
| Chromosome 6 | No loss | 89 (89%) | 121 (94%) | 172 (91%) |
| Loss | 11 (11%) | 8 (6%) | 18 (9%) | |
| NA | 0 | 1 | 1 | |
|
| ||||
| Chromosome 17p | No loss | 25 (74%) | 102 (79%) | 121 (78%) |
| Loss | 9 (26%) | 27 (21%) | 35 (22%) | |
| NA | 66 | 1 | 35 | |
|
| ||||
| MYC | No amplification | 82 (98%) | 128 (98%) | 187 (98%) |
| Amplification | 2 (2%) | 2 (2%) | 4 (2%) | |
| NA | 23 | 0 | 0 | |
|
| ||||
| MYCN | No amplification | 80 (95%) | 125 (96%) | 184 (96%) |
| Amplification | 4 (5%) | 5 (4%) | 7 (4%) | |
| NA | 23 | 0 | 0 | |
|
| ||||
| Expression subgroup | GeXP mRNA signature: | |||
| WNT | 6 (7%) | 0 (0%) | 5 (10%) | |
| SHH | 19 (22%) | 0 (0%) | 7 (14%) | |
| WNT/SHH-independent | 63 (72%) | 0 (0%) | 39 (76%) | |
| NA | 12 | 130 | 140 | |
|
| ||||
| Nanostring mRNA signature: | ||||
| WNT | 5 (8%) | 0 (0%) | 4 (13%) | |
| SHH | 19 (32%) | 0 (0%) | 8 (25%) | |
| Group 3 | 12 (20%) | 0 (0%) | 6 (19%) | |
| Group4 | 24 (40%) | 0 (0%) | 14 (44%) | |
| NA | 40 | 130 | 159 | |
|
| ||||
| Immunohistochemistry: | ||||
| WNT | 5 (100%) | 16 (14%) | 21 (18%) | |
| SHH | 0 (0%) | 23 (20%) | 23 (19%) | |
| WNT/SHH-independent | 0 (0%) | 76 (66%) | 74 (63%) | |
| NA | 95 | 15 | 73 | |
|
| ||||
| Methylation subgroup | WNT | 10 (11%) | 18 (15%) | 28 (16%) |
| SHH | 21 (23%) | 29 (24%) | 36 (20%) | |
| G3 | 19 (20%) | 25 (20%) | 36 (20%) | |
| G4 | 43 (46%) | 51 (41%) | 79 (44%) | |
| NC | 7 | 7 | 12 | |
Assessment of medulloblastoma molecular features
Established molecular correlates of medulloblastoma, encompassing markers of the WNT subgroup (CTNNB1 mutation[15], chromosome 6 loss[24]), MYC and MYCN amplification[43], and chromosome 17 status[24], were assessed as previously described. Medulloblastoma expression subgroup was assessed by immunohistochemistry[13,15], and using GeXP[44] and Nanostring[33] mRNA expression signature assays.
Bioinformatic and statistical analyses
A non-negative matrix factorization (NMF)[3] consensus clustering approach was used to identify robust and reproducible DNA methylation subgroups, and to validate these across training and test cohorts[47] (Supplementary Methods, Supplementary Figure S2). Pearson’s correlations between methylation probe status and training cohort metagene values were calculated to identify subgroup-specific methylation changes.
The potential of methylation markers as prognostic biomarkers was assessed in non-WNT patients within the combined survival cohort (n=163). Probes were first selected for variability and bimodality (Supplementary Methods). The prognostic potential of the 250 most bimodal probes was assessed using multivariate Cox proportional hazards models (Supplementary Methods).
First, we estimated the number of probes which could optimally improve the prognostic ability of predictive Cox proportional hazards models, when added to a base model containing the mandatory markers M+ disease, large-cell/anaplastic (LCA) histology and MYC/MYCN gene amplification. Briefly, 1-5 probes significant (p<0.05) in univariate analysis were added to the base model, using stepwise forward likelihood ratio testing. Using leave-one-out cross-validation (LOOCV), 5-year survival was estimated for each excluded patient, and area-under-the-curve (AUC) of time-dependent receiver operator characteristic (ROC) curves, and -log10 p values of the included covariates were calculated. ROC curve p values and 95% AUC confidence intervals (CIs) were calculated using bootstrapped-case cross-validation with bias correction[30] (Supplementary Methods).
The prognostic utilities of the selected methylation probe covariates were tested in univariate and multivariate Cox proportional hazards models. The methylation status of the selected prognostic methylation probes, as well as their relationship to neighboring CpG dinucleotides were validated using bisulfite sequencing with specific PCR primer sets (Supplementary Table S1), as previously described[26]. A clinically-applicable risk-stratification scheme was developed following application of cut-offs to binarize the selected prognostic methylation probes. The predictive accuracy of the identified models was assessed using LOOCV[46], and by calculating AUC[17] (Supplementary Methods).
All bioinformatic and statistical analyses were performed using R (v2.13)[39]. Survival analyses were performed using progression-free survival (PFS) times, defined as time elapsed from diagnosis to first event, which was tumor recurrence or progression.
Results
Medulloblastoma comprises four major DNA methylation subgroups
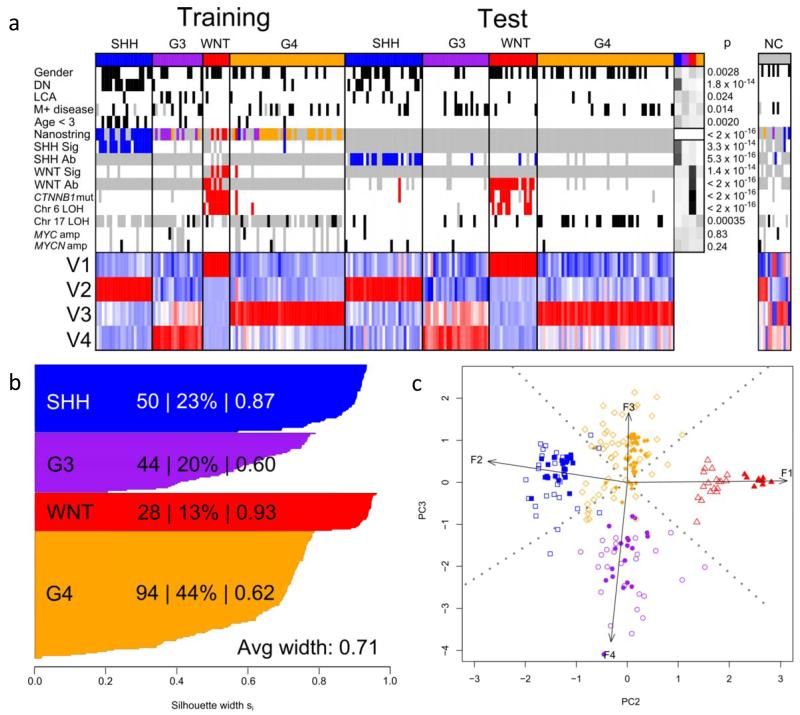
We first examined whether our tumor cohort (n=230; Table 1) could be sub-classified based on its DNA methylation patterns. The cohort was split into training and test sets containing frozen tumor (n=100) and FFPE-derived (n=130; 9.4-17.1 years storage time) samples respectively. This distinction was chosen deliberately to allow us to assess the efficacy of methylation sub-grouping on archival diagnostic FFPE material, which cannot be sub-grouped reliably using transcriptomic methods[33]. Metagenes were derived from the training set using NMF and clustered using K-means. Different possible numbers of metagenes and clusters (both 2-6) were tested and cross-validated by iterative resampling (i=100), and the number of metagenes (four) and clusters (four) which gave the most consistent and robust classification was selected (Figure 1 and Supplementary Figure S2).
Figure 1. DNA methylomics classifies medulloblastoma into four major subgroups.
a. NMF-based consensus clustering of the training cohort (n=100) identifies 4 subgroups that are validated in the test cohort (n=130). Methylation subgroup membership (SHH, blue; G3, purple; WNT, red; G4, orange) and clinico-pathological and molecular subgroup-correlates are shown. Female gender, DN histology, LCA histology, M+ disease and age <3 years are marked in black. Nanostring assay shows transcriptomic subgroup assignment (WNT, red; SHH, blue; Group 3, purple; Group 4, orange). SHH GeXP mRNA signature positivity (SHH Sig) and SHH GAB1 IHC positivity (SHH Ab) are labeled blue. WNT GeXP mRNA signature positivity (WNT Sig), β-catenin IHC nuclear positivity (WNT Ab), CTNNB1 mutation and chromosome 6 loss (Chr6 loss) are labeled red. Chromosome 17 loss (Chr17 loss), MYC amplification and MYCN amplification are labeled black. Missing data are labeled grey. ‘Residuals’ panel displays chi-squared test residuals that indicate any over- (black) or under- (white) representation of each correlate across subgroups, and p values are shown for these relationships. Non-classifiable (NC; n = 14) tumors are also shown. Magnitudes of the 4 defining metagenes (V1 to V4) are shown; highly expressed metagenes are red, lowly expressed are blue. b. Silhouette plots demonstrate correct subgroup assignment (score >0) for 216/216 classified samples from the training and test cohorts. For each subgroup, the number of members, the percentage of cluster members and average silhouette (si) width are shown. c. Bi-plot of combined training and test datasets demonstrates reproducibility of DNA methylomic clusters across datasets. Arrows show projections of 4 metagenes along second and third principal components, labeled with their metagene number. For all clusters, training set samples are shown as filled shapes, with test samples as empty shapes (WNT, red triangles; SHH, blue squares; G3, purple circles; G4, orange diamonds).
Next, metagene values derived from the training set were projected onto the test set of FFPE-derived samples, and a subgroup support vector machine (SVM) classifier trained on the frozen tumor cohort applied to assign sub-group membership within this test dataset. 126 of 130 FFPE samples were assigned with high confidence, assessed using modified Brier scores[47] and metagene patterns were similar in both cohorts. Overall, 216/230 (94%) tumors could be assigned to a subgroup, demonstrating a positive silhouette score (a measure of strength of clustering), and reproducible subgroup assignment (assigned to the same sub-group in >80% iterative replicates). Using PCA to represent metagene values, each sub-group can be shown as distinct (Figure 1). A subset of samples (n=10) could not be assigned reproducibly (i.e. in >80% of iterative replicates) to any subgroup, or had negative silhouette scores indicating poorly clustered tumors (n=4); these were designated non-classifiable (NC). NC tumors showed no significant molecular or clinico-pathological differences to the classified cohort (Figure 1).
Medulloblastoma DNA methylation and gene expression subgroups are closely related
To investigate the relationship between the identified methylation-dependent subgroups and previously reported gene expression subgroups[7,22,32,48,51], we tested for significant association between DNA methylation subgroups and established expression subgroup markers in our combined training/test cohorts (Figure 1)[7,22,32,48,51]. One methylation subgroup was characterized by markers of WNT-pathway activation. 17/19 CTNNB1 mutations occurred in this subgroup alongside significant enrichment for chromosome 6 loss and nuclear accumulation of β-catenin (χ2 tests, all p<2×10−16)[8,13,14]. A second subgroup showed significant enrichment for tumors with high protein expression of SHH markers (GAB1/YAP1)[13] (χ2 test, p=5.3×10−16) and both groups were respectively significantly associated with WNT and SHH subgroup (χ2 tests, both p<2×10−14) mRNA expression signatures determined independently by both GeXP and Nanostring assays[33,44]. Finally, the remaining two methylation subgroups correlated significantly with Group 3 (8/10 tumors tested) or Group 4 (21/27 tumors tested) transcriptomic subgroups determined by Nanostring assay[33] (χ2 test, p<2×10−16).
On this basis, the DNA methylation subgroups were designated WNT-associated (WNT; n=28 (13%)), SHH-associated (SHH; n=50 (23%)), Group 3-associated (G3; n=44 (20%)), and Group 4-associated (G4; n=94 (44%)).
Methylation subgroups have distinct clinical, pathological and genomic features, but survival is equivalent in non-WNT subgroups
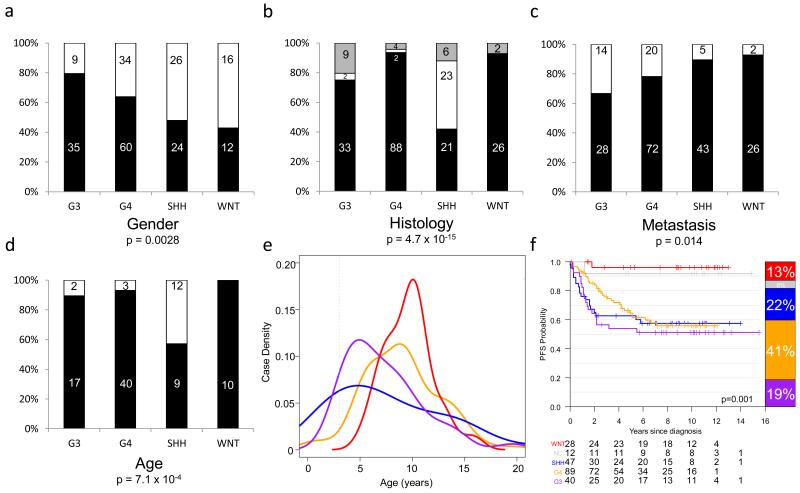
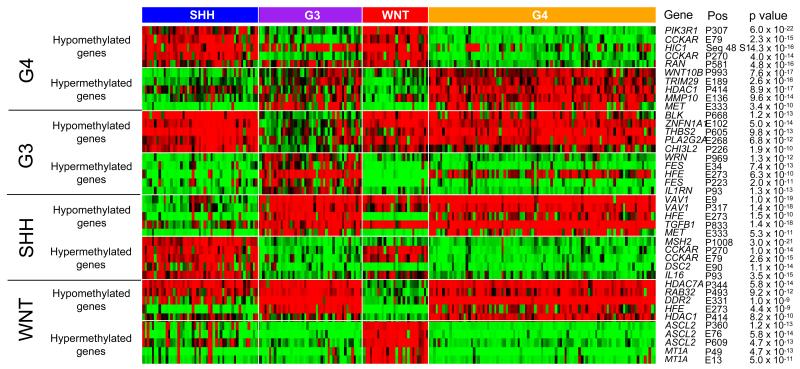
Clinico-pathological features of the methylation subgroups in the combined test/training cohorts were wholly consistent with those observed for their medulloblastoma gene expression subgroup counterparts[7,22,32,48,51] (Figure 2). For example, the methylation-derived SHH subgroup showed a significant enrichment for desmoplastic/nodular (DN) pathology and infant (aged <3 at diagnosis) disease (χ2 tests, p=4.7×10−15 and p=7.1×10−4, respectively). Both G3 and G4 tumors showed a significant male excess (80% and 64% male, respectively, (χ2 test, p=0.0028), and G3 tumors were enriched for LCA histology (9/44 tumors (20%), χ2 test, p=0.02) and metastasis (14/42 tumors (33%), χ2 test, p=0.014). Consistent with previous findings, G4 was enriched for loss of chromosome 17p relative to G3 (Fisher’s Exact Test, p=0.032), while MYC and MYCN amplifications were observed in all non-WNT subgroups. Importantly, a series of gene-specific hyper- and hypo-methylation events associated with each subgroup were identified (Figure 3 and Supplementary Table S2).
Figure 2. Clinical, pathological and outcome correlates of medulloblastoma methylation subgroups.
a-d. Number and percentage incidence are shown for each disease feature across methylation subgroups. a. Gender – male, black; female – white. b. Pathology – classic, black; LCA, grey; DN, white. c. Metastasis – M− black; M+, white. d. Age (training cohort only - test cohort did not contain infant tumors) – black, ≥3 years at diagnosis; white, <3 years. P values are from chi-squared tests. e. Age at diagnosis distribution for each subgroup. f. Kaplan-Meier plots for subgroup PFS, including NC tumors. P value, assessed by Log-Rank test, is shown; barplots to the right show subgroup incidence and at-risk tables are shown below each curve. WNT, red; SHH, blue; G3, purple; G4, orange; NC, grey. Combined data from training and test cohorts are shown except where indicated, with analyses based on patients with available data.
Figure 3. Gene-specific DNA methylation differences between medulloblastoma subgroups.
The top 5 most significant hyper- and hypo-methylation changes associated with each subgroup (methylated, red; unmethylated, green; partially-methylated, black). For each probe, gene name, position (P, promoter region; E, exonic region; number is distance from transcriptional start site) and BH corrected p values, assessed by Mann-Whitney U tests, are shown.
Finally, WNT subgroup tumors showed the favorable outcomes previously reported for WNT patients (identified by β-catenin IHC) within PNET3[13-15] (Figure 2; Log-rank test, χ2=18.2, p=0.001). However, despite the increased incidence of high-risk disease features in G3, all non-WNT subgroups showed equivalent PFS rates (Figure 2) within this trials-based cohort. Of note, NC patients were associated with favorable outcomes.
MXI1 and IL8 methylation predict poor prognosis in non-WNT medulloblastomas
In view of the equivalent survival rates observed for all non-WNT methylomic subgroups, we next investigated the prognostic potential of individual DNA methylation events across the non-WNT subgroups. A cohort of 191 patients aged 3.0-16.0 at diagnosis based on the PNET3 clinical trial, including 163 non-WNT patients, was selected for analysis (Table 1). Established prognostic features, previously identified in PNET3 and other trials (MYC family amplification, LCA histology, metastatic disease, WNT subgroup[14-16,23,42,43,50]), were first validated and found to be significant, and PNET3 and non-PNET3 patients behaved equivalently within this cohort (Supplementary Figure S3). Females were additionally associated with improved survival, although their relationship to prognosis in previous studies has been inconsistent[9,14,43]. A Cox model with MYC or MYCN amplification, LCA histology and metastatic disease as covariates was thus considered to be the base survival model, representing the current clinical paradigm. We then systematically assessed the ability of methylation markers to augment the base model and improve risk prediction in non-WNT patients.
The 250 most bimodal methylation probes were assessed for their ability to significantly improve accuracy of survival prediction (Supplementary Figure S4). To determine the optimal number of methylation probes to include in a putative risk model, methylation probes with univariate significance (p<0.05) were added, using forward likelihood ratio testing, to the base Cox model. Using LOOCV, the predicted survival of the excluded sample was recorded and the accuracy and significance of the cross-validated risk models, after augmenting the base model with between 1 and 5 methylation probes, were evaluated by calculating survival accuracy at 5 years using a cross-validated AUC (from ROC analysis) and p value.
Adding methylation probes to the base model always improved accuracy and significance of survival prediction, regardless of number of added probes. Adding a combination of two methylation markers was chosen as optimal, and conferred a large increase in AUC relative to the base model, whilst maintaining low covariate p values and minimizing additional model complexity (Supplementary Figure S4).
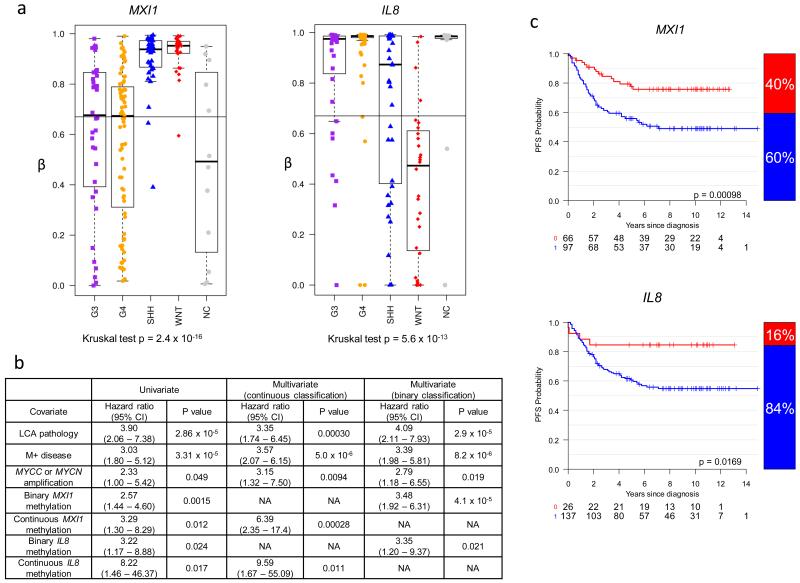
Having chosen the optimal number of genes to be added to the model, the genes were ranked according to their combinatorial ability to increase accuracy of survival prediction. This was evaluated as before, however, in order to correct for cohort and marker selection bias and calculate confidence intervals, the prediction accuracy (AUC) was now evaluated by bias-corrected LOOCV (see Supplementary methods). We found that MXI1 (probe P1269; 163/163 folds) and IL8 (P83; 161/163 folds) were selected in >99% of cross-validation iterations and demonstrated a significantly improved survival risk prediction (AUC = 0.733, 95%CI 0.551–0.844, p=0.023) (Supplementary Figure S4).
We next divided the MXI1 and IL8 methylation values into thirds (top third, beta≥0.67, hyper-methylated; middle/lower thirds, beta<0.67, partially/hypo-methylated); this was compatible with the array-based distribution of methylation scores (Figure 4) and the distribution of survival-risk. MXI1 and IL8 methylation were both significantly associated with a worse prognosis in binary-classified log-rank tests, and as both continuous and binary variables in univariate and multivariate Cox models, in non-WNT patients (Figure 4). Moreover, binary-classified, cross-validated survival models demonstrated similar predictive power to models constructed using continuous methylation variables (AUC=0.776, 95%CI 0.608–0.883, p=0.005) (Figure 4).
Figure 4. MXI1 and IL8 methylation are poor prognosis biomarkers in non-WNT medulloblastomas.
a. Distributions of methylation across subgroups for MXI1 and IL8 probes. Black line at β=0.67 shows binary cut-off used for assessment of prognostic relationships. b. Prognostic relationships of MXI1 and IL8 as binary and continuous variables in univariate and multivariate Cox proportional hazard models of non-WNT patients (n=163). CI, confidence interval. c. Binary classification of MXI1 and IL8 is prognostic in log-rank tests of non-WNT patients (n=163). Kaplan-Meier plots show survival curves. Bar plots to right of curve show binary distribution. Methylated tumors, blue; unmethylated tumors, red. At-risk tables are shown below each curve.
Subgroups show differential patterns of IL8 and MXI1 methylation
Partial or hypomethylation of MXI1 was primarily observed in G3 and G4 medulloblastomas, whilst hypomethylation of IL8 was most commonly observed in the WNT and SHH subgroups (Figure 4). Significant relationships between array and bisulfite sequencing estimations of methylation status were observed for both MXI1 (linear, Spearman’s ρ=0.79, p=8.3×10−9) and IL8 (exponential, Spearman’s ρ=0.83, p=2.5×10−10), and their methylation at the array-assessed CpG residues reflected wider methylation patterns at adjacent CpG residues (Supplementary Figure S5).
Incorporation of DNA methylation markers significantly improves medulloblastoma risk-stratification models
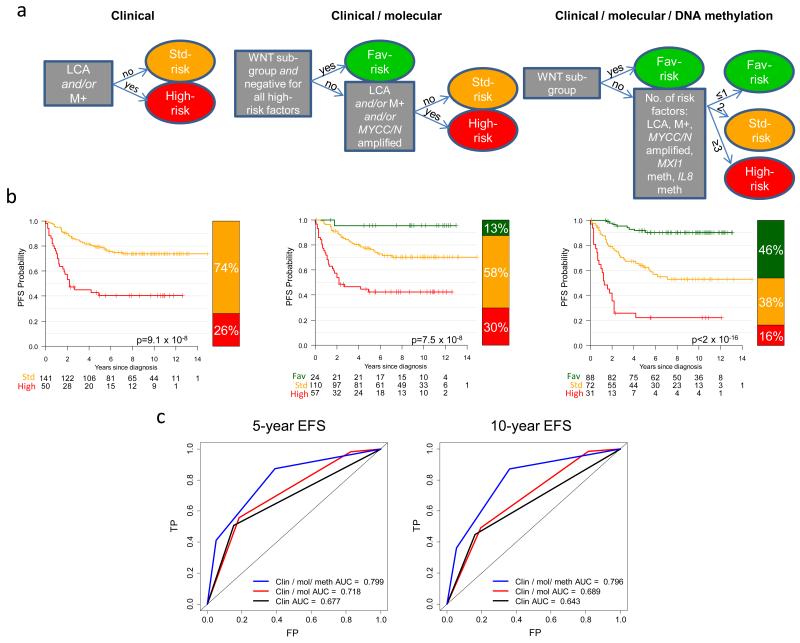
Finally, we derived a novel risk-classification scheme which provides precedent for how DNA methylation biomarkers could form the basis of improved disease prognostication. Each risk factor (M+ disease, LCA histology, MYC family amplification, and binary methylation of IL8 and MXI1), conferred approximately equivalent hazards (ratios 2.79-4.09) in Cox models of non-WNT medulloblastomas (Figure 4 and Supplementary Figure S6). Based on their even risk-distribution, a risk-stratification model was tested whereby non-WNT patients with ≤1 risk-factor were classified as favorable-risk, 2 risk-factors as standard-risk and ≥3 risk-factors as poor-risk. All WNT tumors were classified as favorable-risk. This model achieved a significantly superior prediction of outcome (AUC=0.799 at 5-years, p=4.7×10−11) compared to both current clinical stratification (AUC=0.677, p=0.0001) and clinical/molecular schema (AUC=0.718, p=1.9×10−6) which will form the basis of the forthcoming PNET5 clinical trial[14,37] (Figure 5). The addition of MXI1 and IL8 DNA methylation biomarkers enabled classification of 46% of patients as favorable-risk (5-year PFS 92%, 95%CI 86-98%), compared to 13% in the PNET5 model (5-year PFS 95%, 95% CI 87-100%). Likewise, the proportion of high-risk patients is reduced to 16% (5-year PFS 22%, 95%CI 11-43%) compared to 30% using the PNET5 model (5-year PFS 43%, 95% CI 31-58%).
Figure 5. Risk-stratification using DNA methylation markers in 191 medulloblastoma patients aged 3.0 to 16.0 at diagnosis.
a. Current clinical (first panel) and clinical / molecular (second panel) models were assessed alongside a novel model incorporating DNA methylation biomarkers (third panel), and used to derive Kaplan-Meier plots (b). LCA, large-cell/anaplastic pathology; M+, metastatic disease; MXI1 and IL8 meth, methylated (β≥0.67). Low-risk, green; standard-risk, amber; high-risk, red. P values from log-rank tests are shown. c. Time-dependent ROC curves for all three models at 5 and 10 years. The area under the curve (AUC) is shown for each model. FP, false positive rate; TP, true positive rate. Fav, favorable.
Discussion
This first comprehensive investigation of gene-specific DNA methylation profiles in medulloblastoma clearly demonstrates the potential of epigenetics to support advances in clinical management of this heterogeneous disease.
We have shown using cross-validated class-discovery approaches that medulloblastoma comprises four major DNA methylation subgroups. Independent assessments of subgroup status using a range of alternative assays (expression signatures, IHC, mutational and genomic markers) demonstrate a close relationship to previously described gene expression subgroups[7,22,32,48,51]. Importantly, DNA methylomics allowed the robust discrimination of subgroup status in 95% (129/136) of FFPE biopsies, independent of sample storage time (9.4-17.1 years; median 13.0) and outperformed transcriptomic methods (68% FFPE biopsies classifiable[33]). This offers distinct advantages for the sub-classification of patients in routine practice or from archival cohorts. As a proof-of-concept, we identified minimal signatures capable of identifying disease subgroups (65-probe signature) and assigning WNT subgroup membership (5 probes)(Supplementary Methods, Supplementary Figures 7,8 and 9) which demonstrate how diagnostic methylation signatures could be applied in clinical assay development. Finally, a subset of samples (~6%; frozen and FFPE biopsies) could not be classified confidently, indicating recognition of molecular ‘non-classified’ patients may be important for accurate definition of clinical and molecular associations. Further investigations will be required to determine whether these represent true biological effects or technical (e.g. sample/assay) limitations.
Subgroup-specific DNA methylation events were more common than DNA sequence mutations reported in recent whole-genome mutational studies[19,35,38,40]. Our data identify candidate epigenetic events with potential roles in tumorigenesis, particularly in the poorly characterized Groups 3 and 4, and any role(s) in the disease, or as a basis for future therapies, now need to be considered. While the genome-wide approach to assay DNA methylation used in this study does not lend itself to direct comparison with previous candidate-gene driven studies of methylation in medulloblastoma, the previously reported relationship between desmoplastic/nodular pathology and COL1A2 methylation[1] was also observed in our dataset (p=1.4 × 10−5, χ2 test).
Our study is distinguished by its use of a centrally reviewed, trials-based cohort to make assessments of molecular events, subgroup status and prognostic relationships using high-throughput data. In this group of non-infant patients (3-16 years at diagnosis), only WNT patients displayed significantly different outcomes. Despite reports from recent retrospective studies in group-wide medulloblastoma cohorts[7,32], Group 3 patients did not have a worse outcome in our cohort, consistent with a recent retrospective meta-analysis of 7 independent studies, which showed survival equivalence for all non-WNT subgroups in non-infant patients aged 4-16 at diagnosis[21]. Group 3 tumors were associated with a poor prognosis in infant patients <4 years of age at diagnosis in this meta-analysis[21], suggesting any overall survival differences for Group 3 tumors may be influenced by patient age or treatment received; these observations now require further examination in large, clinically-controlled studies.
We demonstrate here that addition of DNA methylation biomarkers significantly improves survival prediction for non-WNT medulloblastomas arising in patients aged 3-16 at diagnosis. Incorporation of MXI1 and IL8 methylation status alongside currently used molecular, pathological and clinical variables[37] markedly improved stratification of disease-risk. Almost half of patients (46%) could be assigned to a favorable-risk group (>90% 5yr PFS) and could potentially be considered for therapy de-escalation aimed at reduced late-effects, whilst the most intensive therapies could potentially now be limited to a much smaller group of high-risk patients (16%; 22% 5yr PFS). The biomarker discovery and validation strategies employed here used case resampling and cross-validation to prevent overfitting of risk models, to give an unbiased estimate of prognostic performance according to best practice as outlined by Simon and Subramanian[46]. Our methodologies conformed to the REMARK criteria for novel biomarker development[29], including the validation of array-based findings using alternative methods (bisulfite sequencing). The powerful performance of MXI1 and IL8 as novel prognostic biomarkers selected from initial high-dimensional datasets suggests future discoveries using emerging higher-resolution DNA methylation analysis technologies will pay further dividends.
MXI1 is a negative regulator of the MYC family of proteins[53]. We have previously reported its allelic loss and mutation[45], and MXI1 methylation may represent an additional mechanism for the disruption of MYC pathways in medulloblastoma[5]. MXI1 methylation was not associated with membership of Group 3, large-cell / anaplastic disease or MYC amplification in our study (data not shown). Likewise, IL8 has potential involvement in chemokine signaling and angiogenic processes in tumor development[31]. Thus, although outside the scope of the present study, the functional contributions of MXI1 and IL8 to medulloblastoma tumorigenesis and clinical behavior now require urgent investigation.
In summary, we have demonstrated that DNA methylation profiling enables the robust sub-classification of four disease sub-groups in frozen and routinely-collected or archival FFPE biopsy material. Moreover, we show that incorporation of DNA methylation biomarkers can significantly improve upon current disease-risk stratification schemes. These findings have important implications for future risk-adapted clinical disease management in medulloblastoma.
Supplementary Material
Supplementary Table S1. Primer sequences for bisulfite sequencing of differentially methylated and prognostic loci. Sequences are listed 5′ - 3′.
Supplementary Table S2: The top 10 most correlative and anti-correlative probes that define each metagene represent novel biomarkers for the methylation subgroups of medulloblastoma. Anti-correlative probes are shown with a white background. Correlative probes are shown with a grey background. Probe name, gene name, Pearson correlation, β-scores of group and non-group members, as well as the difference in β-values between group and non-group members are given. P values, calculated using Mann-Whitney tests comparing one subgroup versus others, with a Bonferroni correction for multiple hypothesis testing, are shown.
Supplementary Figure S1. DNA methylation array data quality, reproducibility and quantitative accuracy. a. Intra- and inter-array replicates demonstrate high reproducibility. b. Bland-Altman plot showing direct comparison between bisulfite sequencing estimation of methylation and Golden Gate array-estimated methylation status of 18 samples at 7 loci (ASCL2, CCKAR, COL1A2, HFE, MSH2, NOS2A, SPDEF). The x-axis shows the average score from the two estimations of β-value by bisulfite sequencing and array analysis, and the y-axis shows the difference between them. Horizontal dotted lines are plotted at the mean difference and at 2 standard deviations of the difference. c. Density plot shows distribution of deviation between bisulfite sequencing and array estimates of methylation. A blue line indicates the modal value for deviation between estimates. Estimates more than two standard deviations from the mean deviation are shown in red.
Supplementary Figure S2. Identification of DNA methylation-dependent medulloblastoma subgroups using NMF and consensus clustering. a. Flow-chart shows consensus-clustering procedure to identify optimal combinations of metagenes and clusters in the training dataset. b. The average percentage assignment of samples to the same cluster over 100 iterations is shown as a 3D surface plot for each tested combination of metagenes and clusters. Data peaks represent optimal combinations of metagenes and clusters. c. The data shown in the surface plot is tabulated. The chosen optimal number of 4 metagenes / 4 clusters is highlighted red. d. A SVM classifier of the training cohort H matrix perfectly recapitulates the group assignments by k-means clustering.
Supplementary Figure S3. Survival relationships for molecular and clinico-pathological variables within the survival cohort (n=191). Each panel shows a Kaplan-Meier plot, a bar plot showing group membership and an at-risk table. P values are from log-rank tests. PNET3 and age-matched - PNET3, PNET3 trial patients; Age match, age-matched non-trials patients. MYC / MYCN amp - 0, no amplification; 1, amplification of MYC or MYCN. LCA - 1, LCA pathology; 0, non-LCA. M stage - M−, 0; M+, 1; Gender - 0, male; 1, female. WNT - 0, non-WNT subgroup; 1 - WNT subgroup.
Supplementary Figure S4. Identification of bi-modal methylation markers for assessment in survival models. a. The bimodality index[52] was applied to identify candidate biomarkers for assessment of their prognostic ability to integrate into existing disease survival models. The first column shows the three most bimodal probes, the second column the three least bimodal probes, for illustration. b. Selection of optimal additional numbers of prognostic methylation probes to a base model consisting of MYC family amplification, metastatic disease and LCA histology. Left hand y axis shows cross-validated AUC for adding 0-5 methylation probes (red). Right-hand y axis shows -log 10 p value for the selected covariates (blue). c. Cross-validated ROC curves for adding two methylation probes to the base survival model. d. Cross-validated ROC curves for adding two binary-classified methylation probes to the base survival model shows equivalent AUC to model using methylation probes as continuous variables.
Supplementary Figure S5. Assessment of MXI1 and IL8 methylation status by bisulfite sequencing. a,b. Scatterplots showing estimates of DNA methylation at the MXI1 (P1269; a) and IL8 (P83; b) probe loci, derived by independent bisulfite sequencing and methylation array analysis. c,d. Methylation status of CpG dinucleotides within the MXI1 (c) and IL8 (d) CpG islands determined by bisulfite sequencing analysis. Positions of individual CpG residues and the array-probes are shown; estimated methylation status is represented by lollipop plots (white, <20% methylation; one-quarter black, 20-40%; half black, 40-60%, three-quarter black, 60-80%; black, >80%). For MXI1, the patterns of methylation observed at the array probe site are similar to those across the CpG region assessed. IL8 shows consistency of methylation status across the first two CpG island sites including the array-probe site.
Supplementary Figure S6. Development of a cumulative model for medulloblastoma risk-stratification. a. Nomogram of risk-factors identified in a Cox proportional hazards model derived from non-WNT patients (n=163; Figure 4B), demonstrates similar magnitudes of hazards. Risk boundaries are shown (Low, low-risk; Std, standard risk; Poor, poor risk), defined by the total number of points conferred by risk-factor positivity and delineated by blue lines. In the illustrated stratification scheme, the absence of any risk-factor, or positivity for a single risk-factor, would confer membership of the low-risk group. Positivity for any combination of two risk-factors would confer membership of the standard-risk group and tumors with positivity for any combination of three or more risk-factors would be classified as high-risk. b. The number of risk factors in the non-WNT survival cohort (n=163) is associated with survival. Kaplan-Meier curves are shown for each occurrence of risk factor frequency (0, green; 1, dark green; 2, orange; 3, red; 4, dark red). Bar plot showing group membership and at-risk table is shown below Kaplan-Meier plot. For information, the survival for the WNT cohort (n=28) is shown in blue. c. Co-occurrence of risk factors in the non-WNT survival cohort. 5-way Venn diagram shows risk-factor occurrence for 163 tumors. 5/163 (3%) tumors were negative for all risk factors.
Supplementary Figure S7. Development and validation of a minimal DNA methylation signature for assessment of WNT subgroup status. a. Recursive feature elimination SVM[11] identifies smallest cross-validation error to distinguish WNT and non-WNT methylomic subgroups using a 5-probe classifier. b. SVM classifier predicts class membership with high (probability >0.8) confidence in all but one previously-classified samples. NC samples remain difficult to assign. c. Stacked bar plot showing the vote for each class from the 5-probe subgroup classifier. Prior class assignment (Figure 2) is shown at the top of the bar plot; tumors previously classified as WNT are shown in red, non-WNT white. For each sample, the probabilities for membership of each class using the 5-probe signature are colored in the same way. A blue line separates training and test cohorts. d. Confusion matrix demonstrates one false-negative mis-classification error of WNT-subgroup assignment by applying classifier.
Supplementary Figure S8. Development and validation of a minimal DNA methylation signature for assessment of medulloblastoma methylation subgroup status. a. Recursive feature elimination SVM[11] identifies smallest cross-validation error to distinguish between 4 methylomic subgroups using a 65 probe classifier. b. SVM classifier predicts class membership with high (probability >0.8) confidence in all previously classified samples. NC samples remain difficult to assign. c. Stacked bar plot showing the vote for each class from the 65-probe subgroup classifier. Prior class assignment (Figure 2) is shown at the top of the bar plot; tumors previously classified as SHH are shown in blue, G3 purple, WNT red and G4 orange. For each sample, the probabilities for membership of each class using the 65-probe signature are colored in the same way. A white line separates training and test cohorts. d. Confusion matrix demonstrates perfect recapitulation of subgroup membership by applying classifier.
Supplementary Figure S9. Minimal DNA methylation signatures for medulloblastoma subgroup classification and assessment of risk-stratification markers (MXI1/IL8). a. Minimal 5-probe methylomic classifier shows different methylation patterns in WNT and non-WNT samples. MXI1 and IL8 probe status are shown as continuous and binarized variables. Binarized variables are shown in black (methylated; beta > 0.67) and white (unmethylated; beta ≤ 0.67). b. PCA of selected probes demonstrates separation of WNT from non-WNT tumors. Covariance spheroids have been plotted at 95% confidence intervals for the assigned groups. c. Minimal sixty-five probe methylomic classifier showing signature methylation patterns across assigned subgroups. d. PCA of selected probes demonstrates separation between the previously assigned classes. Covariance spheroids for the assigned groups are again plotted at 95% confidence intervals. Heatmaps show methylation status (methylated, red; unmethylated, green; partially-methylated, black.
Acknowledgements
This work was supported by grants from The Brain Tumour Charity, Cancer Research UK, The Katie Trust and North of England Children’s Cancer Research. Medulloblastomas investigated in this study include samples provided by the UK Children’s Cancer and Leukaemia Group (CCLG) as part of CCLG-approved biological study BS-2007-04. This study was conducted with ethics committee approval from Newcastle / North Tyneside REC (study reference 07/Q0905/71).
References
- 1.Anderton JA, Lindsey JC, Lusher ME, Gilbertson RJ, Bailey S, Ellison DW, Clifford SC. Global analysis of the medulloblastoma epigenome identifies disease-subgroup-specific inactivation of COL1A2. Neuro Oncol. 2008;10:981–994. doi: 10.1215/15228517-2008-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet J-P, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci U S A. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns JM, Dunning MJ, Ritchie ME, Russell R, Lynch AG. BASH: a tool for managing BeadArray spatial artefacts. Bioinformatics. 2008;24:2921–2922. doi: 10.1093/bioinformatics/btn557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cascon A, Robledo M. MAX and MYC: a heritable breakup. Cancer Res. 2012;72:3119–3124. doi: 10.1158/0008-5472.CAN-11-3891. [DOI] [PubMed] [Google Scholar]
- 6.Chang CH, Housepian EM, Herbert C., Jr. An operative staging system and a megavoltage radiotherapeutic technic for cerebellar medulloblastomas. Radiology. 1969;93:1351–1359. doi: 10.1148/93.6.1351. [DOI] [PubMed] [Google Scholar]
- 7.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clifford SC, Lusher ME, Lindsey JC, Langdon JA, Gilbertson RJ, Straughton D, Ellison DW. Wnt/Wingless pathway activation and chromosome 6 loss characterize a distinct molecular sub-group of medulloblastomas associated with a favorable prognosis. Cell Cycle. 2006;5:2666–2670. doi: 10.4161/cc.5.22.3446. [DOI] [PubMed] [Google Scholar]
- 9.Curran EK, Sainani KL, Le GM, Propp JM, Fisher PG. Gender affects survival for medulloblastoma only in older children and adults: A study from the surveillance epidemiology and end results registry. Pediatr Blood Cancer. 2008;52:60–64. doi: 10.1002/pbc.21832. [DOI] [PubMed] [Google Scholar]
- 10.Diede SJ, Guenthoer J, Geng LN, Mahoney SE, Marotta M, Olson JM, Tanaka H, Tapscott SJ. DNA methylation of developmental genes in pediatric medulloblastomas identified by denaturation analysis of methylation differences. Proc Natl Acad Sci U S A. 2010;107:234–239. doi: 10.1073/pnas.0907606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan KB, Rajapakse JC, Wang H, Azuaje F. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans Nanobioscience. 2005;4:228–234. doi: 10.1109/tnb.2005.853657. [DOI] [PubMed] [Google Scholar]
- 12.Dunning MJ, Smith ML, Ritchie ME, Tavare S. R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 13.Ellison DW, Dalton J, Kocak M, Nicholson SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, Taylor RE, Bailey S, Clifford SC, Gilbertson RJ. Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121:381–396. doi: 10.1007/s00401-011-0800-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellison DW, Kocak M, Dalton J, Megahed H, Lusher ME, Ryan SL, Zhao W, Nicholson SL, Taylor RE, Bailey S, Clifford SC. Definition of disease-risk stratification groups in childhood medulloblastoma using combined clinical, pathologic, and molecular variables. J Clin Oncol. 2011;29:1400–1407. doi: 10.1200/JCO.2010.30.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, Pearson AD, Clifford SC. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. doi: 10.1200/JCO.2005.01.5479. [DOI] [PubMed] [Google Scholar]
- 16.Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- 17.Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–344. doi: 10.1111/j.0006-341x.2000.00337.x. [DOI] [PubMed] [Google Scholar]
- 18.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 19.Jones DT, Jager N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29:3017–3024. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- 21.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS One. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamont JM, McManamy CS, Pearson AD, Clifford SC, Ellison DW. Combined histopathological and molecular cytogenetic stratification of medulloblastoma patients. Clin Cancer Res. 2004;10:5482–5493. doi: 10.1158/1078-0432.CCR-03-0721. [DOI] [PubMed] [Google Scholar]
- 24.Langdon JA, Lamont JM, Scott DK, Dyer S, Prebble E, Bown N, Grundy RG, Ellison DW, Clifford SC. Combined genome-wide allelotyping and copy number analysis identify frequent genetic losses without copy number reduction in medulloblastoma. Genes Chromosomes Cancer. 2006;45:47–60. doi: 10.1002/gcc.20262. [DOI] [PubMed] [Google Scholar]
- 25.Lindsey JC, Anderton JA, Lusher ME, Clifford SC. Epigenetic events in medulloblastoma development. Neurosurg Focus. 2005;19:E10. doi: 10.3171/foc.2005.19.5.11. [DOI] [PubMed] [Google Scholar]
- 26.Lindsey JC, Lusher ME, Anderton JA, Bailey S, Gilbertson RJ, Pearson AD, Ellison DW, Clifford SC. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 27.Lindsey JC, Lusher ME, Anderton JA, Gilbertson RJ, Ellison DW, Clifford SC. Epigenetic deregulation of multiple S100 gene family members by differential hypomethylation and hypermethylation events in medulloblastoma. Br J Cancer. 2007;97:267–274. doi: 10.1038/sj.bjc.6603852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies. J Clin Oncol. 2005;23:9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- 30.Missiaglia E, Williamson D, Chisholm J, Wirapati P, Pierron G, Petel F, Concordet JP, Thway K, Oberlin O, Pritchard-Jones K, Delattre O, Delorenzi M, Shipley J. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J Clin Oncol. 2012;30:1670–1677. doi: 10.1200/JCO.2011.38.5591. [DOI] [PubMed] [Google Scholar]
- 31.Ning Y, Manegold PC, Hong YK, Zhang W, Pohl A, Lurje G, Winder T, Yang D, LaBonte MJ, Wilson PM, Ladner RD, Lenz HJ. Interleukin-8 is associated with proliferation, migration, angiogenesis and chemosensitivity in vitro and in vivo in colon cancer cell line models. Int J Cancer. 2011;128:2038–2049. doi: 10.1002/ijc.25562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins CE, French P, Rutka JT, Pfister S, Taylor MD. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Northcott PA, Shih DJ, Remke M, Cho YJ, Kool M, Hawkins C, et al. Rapid, reliable, and reproducible molecular sub-grouping of clinical medulloblastoma samples. Acta Neuropathol. 2012;123:615–626. doi: 10.1007/s00401-011-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfister SM, Remke M, Benner A, Mendrzyk F, Toedt G, Felsberg J, et al. Outcome Prediction in Pediatric Medulloblastoma Based on DNA Copy-Number Aberrations of Chromosomes 6q and 17q and the MYC and MYCN Loci. J Clin Oncol. 2009;27:1627–1636. doi: 10.1200/JCO.2008.17.9432. [DOI] [PubMed] [Google Scholar]
- 37.Pizer BL, Clifford SC. The potential impact of tumour biology on improved clinical practice for medulloblastoma: progress towards biologically driven clinical trials. Br J Neurosurg. 2009;23:364–375. doi: 10.1080/02688690903121807. [DOI] [PubMed] [Google Scholar]
- 38.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; City: 2011. [Google Scholar]
- 40.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53–65. [Google Scholar]
- 42.Rutkowski S, von Bueren A, von Hoff K, Hartmann W, Shalaby T, Deinlein F, et al. Prognostic Relevance of Clinical and Biological Risk Factors in Childhood Medulloblastoma: Results of Patients Treated in the Prospective Multicenter Trial HIT’91. Clin Cancer Res. 2007;13:2651–2657. doi: 10.1158/1078-0432.CCR-06-1779. [DOI] [PubMed] [Google Scholar]
- 43.Ryan SL, Schwalbe EC, Cole M, Lu Y, Lusher ME, Megahed H, et al. MYC family amplification and clinical risk-factors interact to predict an extremely poor prognosis in childhood medulloblastoma. Acta Neuropathol. 2012;123:501–513. doi: 10.1007/s00401-011-0923-y. [DOI] [PubMed] [Google Scholar]
- 44.Schwalbe EC, Lindsey JC, Straughton D, Hogg TL, Cole M, Megahed H, Ryan SL, Lusher ME, Taylor MD, Gilbertson RJ, Ellison DW, Bailey S, Clifford SC. Rapid diagnosis of medulloblastoma molecular subgroups. Clin Cancer Res. 2011;17:1883–1894. doi: 10.1158/1078-0432.CCR-10-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott DK, Straughton D, Cole M, Bailey S, Ellison DW, Clifford SC. Identification and analysis of tumor suppressor loci at chromosome 10q23.3-10q25.3 in medulloblastoma. Cell Cycle. 2006;5:2381–2389. doi: 10.4161/cc.5.20.3360. [DOI] [PubMed] [Google Scholar]
- 46.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform. 2011;12:203–214. doi: 10.1093/bib/bbr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamayo P, Scanfeld D, Ebert BL, Gillette MA, Roberts CWM, Mesirov JP. Metagene projection for cross-platform, cross-species characterization of global transcriptional states. Proc Natl Acad Sci U S A. 2007;104:5959–5964. doi: 10.1073/pnas.0701068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor MD, Northcott PA, Korshunov A, Remke M, Cho YJ, Clifford SC, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123:465–472. doi: 10.1007/s00401-011-0922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor RE, Bailey CC, Robinson K, Weston CL, Ellison D, Ironside J, Lucraft H, Gilbertson R, Tait DM, Walker DA, Pizer BL, Imeson J, Lashford LS. Results of a Randomized Study of Preradiation Chemotherapy Versus Radiotherapy Alone for Nonmetastatic Medulloblastoma: The International Society of Paediatric Oncology/United Kingdom Children’s Cancer Study Group PNET-3 Study. J Clin Oncol. 2003;21:1581–1591. doi: 10.1200/JCO.2003.05.116. [DOI] [PubMed] [Google Scholar]
- 50.Taylor RE, Bailey CC, Robinson KJ, Weston CL, Walker DA, Ellison D, Ironside J, Pizer BL, Lashford LS. Outcome for patients with metastatic (M2-3) medulloblastoma treated with SIOP/UKCCSG PNET-3 chemotherapy. Eur J Cancer. 2005;41:727–734. doi: 10.1016/j.ejca.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 51.Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, Gajjar A, Gilbertson RJ. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Wen S, Symmans WF, Pusztai L, Coombes KR. The bimodality index: a criterion for discovering and ranking bimodal signatures from cancer gene expression profiling data. Cancer Inform. 2009;7:199–216. doi: 10.4137/cin.s2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Primer sequences for bisulfite sequencing of differentially methylated and prognostic loci. Sequences are listed 5′ - 3′.
Supplementary Table S2: The top 10 most correlative and anti-correlative probes that define each metagene represent novel biomarkers for the methylation subgroups of medulloblastoma. Anti-correlative probes are shown with a white background. Correlative probes are shown with a grey background. Probe name, gene name, Pearson correlation, β-scores of group and non-group members, as well as the difference in β-values between group and non-group members are given. P values, calculated using Mann-Whitney tests comparing one subgroup versus others, with a Bonferroni correction for multiple hypothesis testing, are shown.
Supplementary Figure S1. DNA methylation array data quality, reproducibility and quantitative accuracy. a. Intra- and inter-array replicates demonstrate high reproducibility. b. Bland-Altman plot showing direct comparison between bisulfite sequencing estimation of methylation and Golden Gate array-estimated methylation status of 18 samples at 7 loci (ASCL2, CCKAR, COL1A2, HFE, MSH2, NOS2A, SPDEF). The x-axis shows the average score from the two estimations of β-value by bisulfite sequencing and array analysis, and the y-axis shows the difference between them. Horizontal dotted lines are plotted at the mean difference and at 2 standard deviations of the difference. c. Density plot shows distribution of deviation between bisulfite sequencing and array estimates of methylation. A blue line indicates the modal value for deviation between estimates. Estimates more than two standard deviations from the mean deviation are shown in red.
Supplementary Figure S2. Identification of DNA methylation-dependent medulloblastoma subgroups using NMF and consensus clustering. a. Flow-chart shows consensus-clustering procedure to identify optimal combinations of metagenes and clusters in the training dataset. b. The average percentage assignment of samples to the same cluster over 100 iterations is shown as a 3D surface plot for each tested combination of metagenes and clusters. Data peaks represent optimal combinations of metagenes and clusters. c. The data shown in the surface plot is tabulated. The chosen optimal number of 4 metagenes / 4 clusters is highlighted red. d. A SVM classifier of the training cohort H matrix perfectly recapitulates the group assignments by k-means clustering.
Supplementary Figure S3. Survival relationships for molecular and clinico-pathological variables within the survival cohort (n=191). Each panel shows a Kaplan-Meier plot, a bar plot showing group membership and an at-risk table. P values are from log-rank tests. PNET3 and age-matched - PNET3, PNET3 trial patients; Age match, age-matched non-trials patients. MYC / MYCN amp - 0, no amplification; 1, amplification of MYC or MYCN. LCA - 1, LCA pathology; 0, non-LCA. M stage - M−, 0; M+, 1; Gender - 0, male; 1, female. WNT - 0, non-WNT subgroup; 1 - WNT subgroup.
Supplementary Figure S4. Identification of bi-modal methylation markers for assessment in survival models. a. The bimodality index[52] was applied to identify candidate biomarkers for assessment of their prognostic ability to integrate into existing disease survival models. The first column shows the three most bimodal probes, the second column the three least bimodal probes, for illustration. b. Selection of optimal additional numbers of prognostic methylation probes to a base model consisting of MYC family amplification, metastatic disease and LCA histology. Left hand y axis shows cross-validated AUC for adding 0-5 methylation probes (red). Right-hand y axis shows -log 10 p value for the selected covariates (blue). c. Cross-validated ROC curves for adding two methylation probes to the base survival model. d. Cross-validated ROC curves for adding two binary-classified methylation probes to the base survival model shows equivalent AUC to model using methylation probes as continuous variables.
Supplementary Figure S5. Assessment of MXI1 and IL8 methylation status by bisulfite sequencing. a,b. Scatterplots showing estimates of DNA methylation at the MXI1 (P1269; a) and IL8 (P83; b) probe loci, derived by independent bisulfite sequencing and methylation array analysis. c,d. Methylation status of CpG dinucleotides within the MXI1 (c) and IL8 (d) CpG islands determined by bisulfite sequencing analysis. Positions of individual CpG residues and the array-probes are shown; estimated methylation status is represented by lollipop plots (white, <20% methylation; one-quarter black, 20-40%; half black, 40-60%, three-quarter black, 60-80%; black, >80%). For MXI1, the patterns of methylation observed at the array probe site are similar to those across the CpG region assessed. IL8 shows consistency of methylation status across the first two CpG island sites including the array-probe site.
Supplementary Figure S6. Development of a cumulative model for medulloblastoma risk-stratification. a. Nomogram of risk-factors identified in a Cox proportional hazards model derived from non-WNT patients (n=163; Figure 4B), demonstrates similar magnitudes of hazards. Risk boundaries are shown (Low, low-risk; Std, standard risk; Poor, poor risk), defined by the total number of points conferred by risk-factor positivity and delineated by blue lines. In the illustrated stratification scheme, the absence of any risk-factor, or positivity for a single risk-factor, would confer membership of the low-risk group. Positivity for any combination of two risk-factors would confer membership of the standard-risk group and tumors with positivity for any combination of three or more risk-factors would be classified as high-risk. b. The number of risk factors in the non-WNT survival cohort (n=163) is associated with survival. Kaplan-Meier curves are shown for each occurrence of risk factor frequency (0, green; 1, dark green; 2, orange; 3, red; 4, dark red). Bar plot showing group membership and at-risk table is shown below Kaplan-Meier plot. For information, the survival for the WNT cohort (n=28) is shown in blue. c. Co-occurrence of risk factors in the non-WNT survival cohort. 5-way Venn diagram shows risk-factor occurrence for 163 tumors. 5/163 (3%) tumors were negative for all risk factors.
Supplementary Figure S7. Development and validation of a minimal DNA methylation signature for assessment of WNT subgroup status. a. Recursive feature elimination SVM[11] identifies smallest cross-validation error to distinguish WNT and non-WNT methylomic subgroups using a 5-probe classifier. b. SVM classifier predicts class membership with high (probability >0.8) confidence in all but one previously-classified samples. NC samples remain difficult to assign. c. Stacked bar plot showing the vote for each class from the 5-probe subgroup classifier. Prior class assignment (Figure 2) is shown at the top of the bar plot; tumors previously classified as WNT are shown in red, non-WNT white. For each sample, the probabilities for membership of each class using the 5-probe signature are colored in the same way. A blue line separates training and test cohorts. d. Confusion matrix demonstrates one false-negative mis-classification error of WNT-subgroup assignment by applying classifier.
Supplementary Figure S8. Development and validation of a minimal DNA methylation signature for assessment of medulloblastoma methylation subgroup status. a. Recursive feature elimination SVM[11] identifies smallest cross-validation error to distinguish between 4 methylomic subgroups using a 65 probe classifier. b. SVM classifier predicts class membership with high (probability >0.8) confidence in all previously classified samples. NC samples remain difficult to assign. c. Stacked bar plot showing the vote for each class from the 65-probe subgroup classifier. Prior class assignment (Figure 2) is shown at the top of the bar plot; tumors previously classified as SHH are shown in blue, G3 purple, WNT red and G4 orange. For each sample, the probabilities for membership of each class using the 65-probe signature are colored in the same way. A white line separates training and test cohorts. d. Confusion matrix demonstrates perfect recapitulation of subgroup membership by applying classifier.
Supplementary Figure S9. Minimal DNA methylation signatures for medulloblastoma subgroup classification and assessment of risk-stratification markers (MXI1/IL8). a. Minimal 5-probe methylomic classifier shows different methylation patterns in WNT and non-WNT samples. MXI1 and IL8 probe status are shown as continuous and binarized variables. Binarized variables are shown in black (methylated; beta > 0.67) and white (unmethylated; beta ≤ 0.67). b. PCA of selected probes demonstrates separation of WNT from non-WNT tumors. Covariance spheroids have been plotted at 95% confidence intervals for the assigned groups. c. Minimal sixty-five probe methylomic classifier showing signature methylation patterns across assigned subgroups. d. PCA of selected probes demonstrates separation between the previously assigned classes. Covariance spheroids for the assigned groups are again plotted at 95% confidence intervals. Heatmaps show methylation status (methylated, red; unmethylated, green; partially-methylated, black.