Abstract
Latent Epstein–Barr virus (EBV) infection has a substantial role in causing many human disorders. The persistence of these viral genomes in all malignant cells, yet with the expression of limited latent genes, is consistent with the notion that EBV latent genes are important for malignant cell growth. While the EBV-encoded nuclear antigen-1 (EBNA-1) and latent membrane protein-2A (LMP-2A) are critical, the EBNA-leader proteins, EBNA-2, EBNA-3A, EBNA-3C and LMP-1, are individually essential for in vitro transformation of primary B cells to lymphoblastoid cell lines. EBV-encoded RNAs and EBNA-3Bs are dispensable. In this review, the roles of EBV latent genes are summarized.
Introduction
A physican by name Burkitt was the first to describe a unique lymphoma. Epstein and Barr then discovered virus particles in cultured lymphoblasts from Burkitt's lymphoma (BL) in 1964.1 The Epstein–Barr virus (EBV) infection is ubiquitous in adult humans.2, 3, 4 Higher titer of EBV antibody was evident in BL, lymphoproliferative diseases (LPDs), Hodgkin's lymphoma (HL), endemic nasopharyngeal carcinoma (NPC) and infectious mononucleosis.5, 6, 7, 8, 9, 10, 11, 12, 13 EBV primarily infects the human oropharynx epithelial cells, and then replicates and spreads to B cells, resulting in latent infection in B cells, epithelial cells and natural killer/T cells after extensive host T-cell immune surveillance.14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33 Latent EBV infection substantially causes many human malignancies. In immunocompetent people, EBV likely causes ~20% of BL in the developed world, almost all African BL, 50% of HL, 10% gastric carcinomas (GCs), almost all endemic NPC, certain fractions of diffuse large B-cell lymphoma and T-cell lymphoma, multiple sclerosis and systemic lupus erythematosus (SLE).5, 6, 7, 8, 9, 10, 11, 12, 13, 34, 35 In the absence of normal T-cell immune responses, EBV-infected B-lymphocyte proliferations can cause LPD, similar to posttransplant LPD. The persistence of EBV genomes in all cells of these malignancies, even in people with otherwise normal immune responses, is consistent with the notion that EBV genomes are important for malignant cell growth.
EBV latent infection
Latent EBV genomes express five EBV-encoded nuclear antigens (EBNA) and two latent membrane proteins (LMPs), namely EBV-encoded small RNA (EBER) and non-transcribed BART (BamHI-A region rightward transcript) RNAs. Primary EBV infection establishes typically three distinct latent infection statuses from the initial infection as a non-integrated episome: latency types III, II and I depending on the viral gene expression pattern.27, 28, 29, 30, 31, 32, 33 Actively proliferating (posttransplantation) lymphoproliferative diseases and in vitro EBV infection-mediated establishment of the lymphoblastoid cell line (LCL) show type III latency, in which most latent genes are expressed (EBER1/2 RNA, EBNA-leader protein (EBNA-LP), EBNA-2, EBNA-3ABC, EBNA-1, LMP-2A/B, LMP-1 protein, BART RNA). HL and NPC display type II latency (EBER1/2 RNA, EBNA-1, LMP-2A/B, LMP-1 (type IIa) or EBNA-2 (type IIb), BART RNA) and BL shows type I latency (EBER1/2 RNA, EBNA-1, LMP-2A/B, BART RNA). Although EBNA-1 and LMP-2A play a critical role, EBNA-LP, EBNA-2, EBNA-3A, EBNA-3C and LMP-1 are individually essential for in vitro transformation of primary B cells to LCLs.36, 37, 38
The EBV's role in cell growth is most evident in latency III EBV-associated posttransplant LPD, as EBNA-2, EBNA-LP, EBNA-3A and EBNA-3C in latency III infection coordinately upregulate cMyc expression and cell proliferation, and EBV LMP-1 enhances cell survival.39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58 Furthermore, EBV's role is also evident in latency II-infected HL and NPC, where LMP-1 and LMP-2 expression likely contributes to cell survival by activation of nuclear factor-κB (NF-κB) and phosphatidyl inositol 3 kinase (PI3K) pathways.59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75 Moreover, EBERs are expressed in latency types III, II and I and are implicated in the survival of latency I BL cells.61, 76, 77, 78, 79 Thus, EBV gene expression is likely critical for the growth and survival of EBV-associated malignancies (see Table 1).
Table 1. Roles of EBV-encoded latent genes.
| Latent genes | Roles |
|---|---|
| EBNA-1 | Sequence-specific DNA-binding protein to EBV element; sequence-nonspecific chromosome association protein; transactivator of viral latent genes and host genes; responsible for episome replication, segregation and persistence of viral genome; involved in p53 degradation and oncogenesis |
| EBNA-LP | Transcriptional coactivator of EBNA-2-dependent viral and cellular gene transcription; primarily indirectly associates with host DNA sites located at or near the transcriptional start; associates with cellular transcriptional (co)factors and EBNA-2; dismisses repressor complex from promoter or enhancer sites; is essential for EBV-mediated B-cell transformation |
| EBNA-2 | Together with EBNA-LP cooperatively activates viral and cellular gene transcription for transformation; primarily indirectly associates with host DNA sites located at the enhancer or intergenic region; associates with cellular transcriptional (co)factors and EBNA-LP; is critical for EBV-mediated B-cell transformation |
| EBNA-3A | A coactivator of EBNA-2, EBNA-3A and EBNA-3C associations with RBPJ inhibit RBPJ recruitments to DNA; downregulate cMyc transcription and block EBNA-2 activation effects; and induce CDKN2 and chemokines. Induces G1 arrests, which is essential for EBV-mediated B-cell transformation |
| EBNA-3B | A coactivator of EBNA-2; dispensable for B-cell transformation; viral tumor suppressor; and upregulates CXCL10. EBNA-3B-knockout induces DLBCL-like tumors |
| EBNA-3C | Coactivates with EBNA-2 host CXCR4 and CXCL12 genes; induces CDKN2, chemokines and aurora kinase B; mediates RB degradation; attenuates H2AX expression and overcomes EBV-infection-mediated DNA damage response; promotes cell proliferation; induces G1 arrests; essential for EBV-mediated B-cell transformation |
| LMP-1 | Mimics the constitutively active form of CD40, a major EBV-encoded oncogene; activates NF-κB, JNK and p38 pathways; is critical for EBV-mediated B-cell transformation, a major EBV-encoded oncogene; activates NF-κB, JNK and p38 pathways; and induces EMT of NPC and acquisition of CSC-like properties |
| LMP-2A | Mimics constitutively active, antigen-independent BCR signaling through constitutive activation of the ERK/MAPK pathway224; blocks antigen-dependent BCR signaling; induces B-cell lymphoma in transgenic condition; is important but not essential for in vitro primary B-lymphocyte growth transformation; rescues the LMP-1-generated impairment in germinal center in the response to antigen in animals; confers resting B cells sensitive to NF-κB inhibition and apoptosis; suppresses differentiation and promotes epithelial cell spreading and motility in epithelial cells; and enriches cancer stem cell-like population |
| EBER | Most abundant EBV-encoded noncoding RNAs; augments colony formation and induces growth; confers cells resistance to PKR-dependent apoptosis; induces cytokines and modulates innate immune response; binds to La, PKR, L22, PRR and RIG-I; and EBER-mediated RIG-I activation likely contributes to EBV oncogenesis. EBER blockades of PKR-mediated phosphorylation of eIF2α results in blockage of eIF2α-mediated inhibition of protein synthesis and resistance to IFNα-induced apoptosis |
| miRNAs | Transcribed from BART and BHRF1; validated targets include Bim, BRUCE, CXCL11, DICER1, PUMA; has a role in sustaining latently infected cells. BHRF1 miRNA and BART miRNAs interfere with apoptosis. The miR-BART15-3p promoted apoptosis 331 |
Abbreviations: BART, BamHI-A region rightward transcript; BHRF1, BamHI fragment H rightward open reading frame 1; CSC, cancer stem cell; DLBCL, diffuse large B-cell lymphoma; EBER, EBV-encoded nuclear antigen; EBV, Epstein–Barr virus; eIF2α, eukaryotic initiation factor 2α EMT, epithelial–mesenchymal transition; ERK, extracellular signal-regulated kinase; IFN, interferon; JNK, c-Jun N-terminal kinase; LMP, latent membrane protein; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor-κB; NPC, nasopharyngeal carcinoma; LP, leader protein; PKR, RNA-dependent protein kinase; PRR, pattern-recognition receptors; RBPJ, recombination signal-binding immunoglobulin κJ region; RIG-1, retinoic acid-inducible gene I.
EBV-encoded nuclear antigen-1
EBNA-1 roles
EBNA-1 is expressed in all forms of latent EBV infection; it is essential for efficient EBV genome replication, persistence and transcription in dividing cells80, 81, 82, 83 and binds to and uses nucleolin and nucleophosmin (NPM) for EBNA-1-dependent transcriptional activation and genome persistence.84, 85 EBNA-1 is the only nuclear EBV antigen expressed in both latent and lytic modes of infection and contributes to the latent infection in multiple ways. EBNA-1 suppresses spontaneous lytic reactivation in latent infection status;86 however, it interacts with and disrupts promyelocytic leukemia (PML) nuclear bodies and also promotes lytic infection. EBNA-1 induces a family of microRNAs (let-7 microRNAs (miRNAs)), which in turn decreases the level of the cellular protein Dicer and inhibits the reactivation of latent EBV and may increase metastasis.87 EBNA-1 in NPC and GC induces the loss of PML nuclear bodies, and decreased p53 activation and apoptosis in response to DNA damage.86, 88
EBNA-1 binds to viral DNA elements and cellular promoters,89, 90 activates EBV viral Cp and Wp promoters, inhibits Qp promoters,91 upregulates STAT1 (signal transducers and activators of transcription 1), whose expression correlates with major histocompatibility complex class I and II increase, downregulates tumor growth factor-β signaling pathways, reduces SMAD2, a tumor growth factor-β signaling mediator protein tyrosine phosphatase receptor K,92, 93 upregulates CCL20 in HL,94 inhibits the canonical NF-κB pathway by inhibiting IKK (IκB kinase) phosphorylation in NPC95 and enhances activity of the AP-1 transcription factor (TF) in NPC cells by EBNA-1 binding to the promoters of c-Jun and ATF296 (see Table 1).
Domains of EBNA-1
EBNA-1 encodes 641 amino acids (a.a.) from a prototype EBV strain.97 EBNA-1 a.a. 2–30 have no known function and are dispensable for replication, DNA binding, transactivation and persistence.98 Both arginine-glycine (RG)1 (a.a. 33–89) and RG2 (a.a. 328–386)99, 100, 101, 102, 103, 104, 105, 106 are necessary, sufficient and essential for efficient association of EBNA-1 with host chromosomes and EBNA-1-dependent transcription of latent genes, and for EBV oriP (an Origin of Plasmid replication) genome persistence. An almost inseparable dimerization domain (DD) and oriP DNA-binding domain (a.a. 459–607) bind specifically to EBV oriP, an enhancer of the transcription and origin of viral genome replication, and thereby brings to chromosomes. The dimerization domain/DNA-binding domain has central functions in DNA binding, transcription, persistence and replication.
RG1 and RG2 are separated by an irregular hydrophobic glycine-alanine repeats domain.107, 108, 109 Deletion of the entire glycine-alanine repeats has no discernible effect on EBNA-1 abundance or functional interaction with oriP. The glycine-alanine domain minimizes translation,110 binds to proteasomes and inhibits EBNA-1 proteolysis.111, 112, 113 As a consequence of both decreased synthesis and very slow degradation, EBNA-1 peptides are poorly presented in the context of major histocompatibility complex class I. Cells expressing EBNA-1 are therefore partially protected from recognition by CD8 cytotoxic T lymphocytes.112, 113, 114, 115, 116, 117
The EBNA-1 dimerization and DNA-binding domain (a.a. 459–607), were crystallized, bound to cognate DNA sites and resolved at 2.2 Å.104, 106 EBNA-1's essential role in EBV episome replication, transcription and persistence requires EBNA-1 homodimerization and DNA binding.106 This domain mediates EBNA-1 interaction with oriP and supplementary sequence for replication (Rep*), and also EBV Qp, the promoter for EBNA-1 transcription in latencies I and II.82, 118, 119, 120, 121, 122, 123 EBNA-1 a.a. 379–386 is a nuclear localization sequence;99 K379 and R380 are essential components and S385 phosphorylation has an upregulatory effect on nuclear import, whereas S383 and S386 phosphorylation inhibits nuclear import.124 EBNA-1 a.a. 379–641 is also a dominant-negative inhibitor of EBNA-1 interaction with cognate DNA, resulting in decreased EBNA-1-dependent transcription and episome maintenance.125, 126, 127, 128, 129, 130 Dominant-negative EBNA-1 proteins and EBNA-1 antisense oligonucleotide or RNA interference inhibition of EBNA-1 result in EBV genome loss and abrogation of tumor cell growth and survival, indicating that EBNA-1 inhibition is a valid target for prevention or treatment of EBV-associated diseases.
EBNA-1 binds to viral element and host chromosomes to tether for replication and maintenance of genome
EBV episomes persist in dividing malignant and non-malignant cells through EBNA-1 interaction with multiple cognate sites in EBV oriP DNA.81, 82, 122, 131, 132 OriP comprises a family of repeats and a dyad symmetry. EBNA-1 interaction with oriP enables EBV DNA replication once per cell cycle.133, 134, 135, 136, 137 The family of repeats and dyad symmetry are required for efficient episome persistence and transcriptional activation in infected cells.80, 81, 82, 83, 91, 131, 138, 139, 140, 141 The family of repeats is an EBNA-1-dependent enhancer,91, 141, 142, 143, 144, 145 whereas dyad symmetry is the site of initiation of EBV episome DNA replication (see Table 1).
EBNA-1-interacting proteins
EBNA-1 RG1/2 interactions with hEBP2 (human EBNA-1-binding protein 2), P32/TAP (protein 32KD/HIV TAT-associating protein), Nap1, Karyopherin a2, PRMT5 and PRMT1 (protein methyl transferase-5 and -1), nucleolin and NPM146, 147, 148, 149, 150, 151, 152 are implicated in transcriptional activation (hEBP2, p32/TAP, Karyopherin, PRMT5, PRMT1, nucleolin and NPM) or episome maintenance (hEBP2, Nap1, nucleolin and NPM). EBNA-1 a.a. 395–450 binds to host USP7 (ubiquitin-specific protease 7)148 and forms a quaternary complex with USP7, GMPSC and EBV oriP DNA, 153 and this interaction alters histone modification at oriP, disrupts p53 and also the PML levels.154 EBNA-1 a.a. 387–394 interacts with the host CK2 kinase α, α′ and β, and this interaction leads to the disruption of PML bodies. EBNA-1 also associates with PML proteins. The EBNA-1–CK2 complex phosphorylates PML proteins and triggers the polyubiquitylation and degradation of PML.155 EBNA-1 also binds to NAP1, template-activating factor-Iβ/SET, CK2 and PRMT5.148 EBNA-1 interacts with NPM, heterogeneous ribonucleoproteins and La protein.156 EBNA-1 association with NPM contributes to the EBNA-1 transactivation function.84
EBNA-LP and EBNA-2
EBNA-2 and EBNA-LP are coexpressed soon after EBV infection in B cells,39 are essential for B-cell transformation to LCL and LCL outgrowth41, 42, 157 and cooperatively activate viral and cellular gene transcriptions for transformation.158, 159 Both LP and EBNA-2 associate with the transcriptional factor and the linking factors bound to upstream DNA elements of cMyc and also cMyc-regulated genes, forming a long-range DNA looping, which ultimately leads to cell cycle entry for proliferation.39, 160, 161
Recombination signal-binding immunoglobulin κJ region (RBPJ) protein associates with the NCoR repressor and is thus inherently a transcription repressor. Host DNA carries ~20 000 and ~10 000 sites, where LP or EBNA-2 and RBPJ bind (LP or EBNA-2 sites and RBPJ sites, respectively). A considerable fraction of LP sites were colocalized with EBNA-2 sites. LP and EBNA-2 sites are primarily located at or near the transcriptional start site, whereas EBNA-2 sites are more at the enhancer or intergenic region. LP sites were enriched for sites of B-cell TFs including YY1, SP1, PAX5, BATF, IRF4, ETS1, RAD21, PU.1, CTCF, RBPJ, ZNF143, SMC3, NF-κB, TBLR and EBF. The CTCF as a transcription insulator associates with YY1, RAD21 and SMC3 to mediate long-range chromatin interactions (DNA linking) and promoter derepression.162 In addition, LP sites were marked by RNAPII and histone acetylase P300, and also by activated histone tags such as H3K4me3, H3K27ac, H2Az and H3K9ac, indicative of LP sites being activated transcriptional sites. EBA2 induces cMyc transcription within 24 h after EBV infection of resting B cells (see Table 1).
EBV-encoded nuclear antigen-LP
By costimulation of EBNA-2-dependent transcription, LP coactivates EBNA-2 transcriptional activation,163 associates with EBNA-2, HA95 and Hsp70/72,164, 165 associates with and relocates 14-3-3 and histone deacetylase 4,166 displaces Sp100 and Hp1α from ND10 bodies and disrupts matrix-associated deacetylase bodies, dismisses repressor complex (NCoR/HA95) from promoter or enhancer sites and shuttles them from the nuclei to the cytoplasm.158, 160, 164, 166 This LP dismissal of NCoR and RBPJ repressors reduces the occupancy of repressors NCoR and RBPJ at EBNA-2 sites without altering EBNA-2 occupancy. However, LP and EBNA-2 do not affect each other's association with the enhancer or promoter.158 These multiple complexes load on or near promoter sites and increase activated marks on the histone, leading to transcriptional activation for EBV-dependent efficient cell transformation166 (see Table 1).
EBV-encoded nuclear antigen-2
The EBNA-2 does not directly bind to DNA but instead associates with viral (LP) and cellular factors (RBPJ transcriptional repressor and ZNF143)167 for transcriptional activation;168 it associates with NCoR-deficient RBPJ and increases RBPJ binding to DNA, recruiting cellular TFs to EBNA-2 sites in the enhancer or promoter clustered with RBPJ EBF, ETS1, ZNF143, PU.1, NF-κB and RUNX1 sites.158, 161
Similar to LP, EBNA-2 adds up the activation mark H3K4me1 on the histone, depletes the nucleosome, recruits transcriptional factors, coactivators and histone acetylases161, 167, 169, 170 and links the EBNA-2 site to target promoters by associating with RBPJ and other factors (see Table 1).
Latent membrane protein-1
LMP-1 roles
LMP-1 and LMP-2A mimic CD40 and B-cell receptor (BCR) signaling, respectively, on B cells. EBV infection rescues BCR-negative, proapoptotic germinal center B cells from apoptosis.171 LMP-1 is expressed in LCLs, HLs and undifferentiated NPCs but not GCs, and also during EBV replication; it is a major EBV-encoded oncogene and activates NF-κB, c-Jun N-terminal kinase (JNK) and p38 pathways;54, 57, 58, 172 it transforms primary rodent fibroblasts and is essential for EBV-mediated transformation; it induces an anchorage-independent growth with increased tumor formation after subcutaneous inoculation into nude mice, and also has effects on epithelial cell differentiation;60, 173, 174, 175, 176, 177, 178, 179, 180, 181, 182, 183, 184, 185, 186, 187, 188 it upregulates surface molecules ICAM1, LFA1, CD40, CD21 and CD23 and downregulates CD10 expression, membrane ruffling and adhesion.189, 190, 191, 192, 193, 194, 195, 196, 197, 198, 199, 200
LMP-1 is a major EBV-encoded oncogene and activates NF-κB, JNK and p38 pathways in vitro and in vivo; it increases the telomerase activity via cMyc induction201 and promotes migration of NPC cells;202 it induces epithelial–mesenchymal transition of NPC and acquisition of cancer stem cell-like properties202 and inhibits LKB1-AMPK1 tumor suppressor pathways in NPC through the phosphorylation of LKB1 at serine 428, with subsequent suppression of the phosphorylation of AMPK.203 LMP-1 induces a proapoptotic Bmi-1 (Bcl-2-interacting mediator of cell death) in HL cells, which is downregulated by EBNA-3A and EBNA-3C.204, 205 LMP-1 induces IL8 expression through the NF-κB binding site, which may contribute in part to angiogenesis in NPC.206 LMP-1 induces a proapoptotic Bmi-1 in HL cells, which is downregulated by EBNA-3A and EBNA-3C.204, 205 LMP-1 induces IL-8 expression through the NF-κB binding site, which may contribute in part to angiogenesis in NPC.206 In LMP-1-nonexpressing GC, BARF1 likely has a growth promoter activity via activation of NF-κB in GC207 (see Table 1).
LMP1 structure, domain and interactions
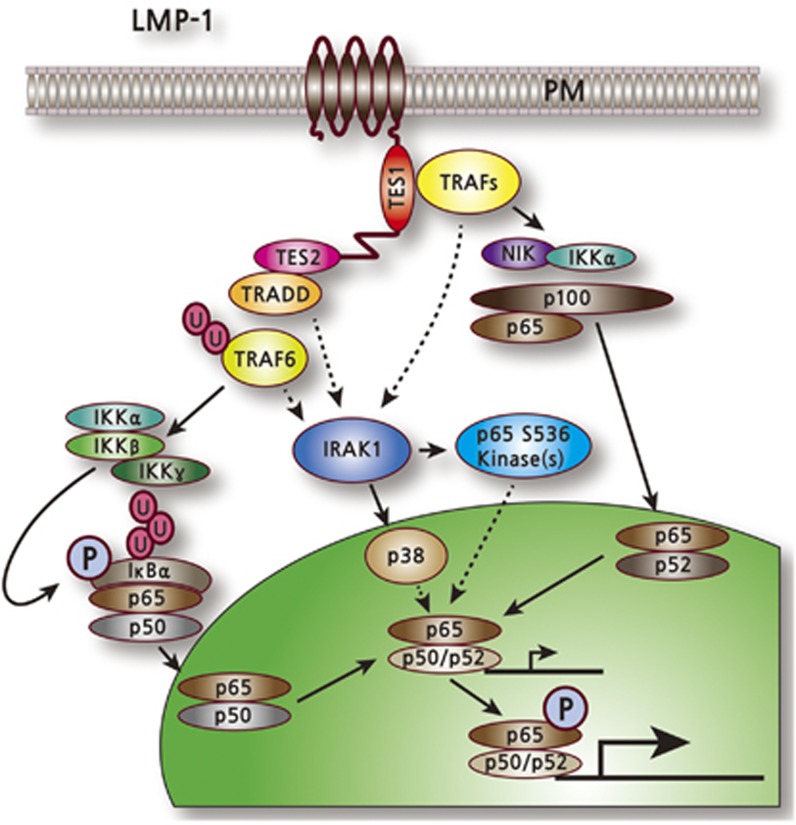
The key LMP-1 functional domains are: (i) six transmembrane domains (TM1–6), which mediate raft association, constitutive aggregation and constitutive signaling; and (ii) two transformation effector sites (TES1 and TES2). LMP-1 oligomerizes on the plasma membrane through TM1 interaction with TM3–6, forming a ligand-independent signaling complex. TM1–4 is important for wild-type LMP-1 C-terminus-mediated NF-κB activation, whereas TM3–4, TM5–6 or TM3–6 is dispensable.208 LMP-1 (also LMP-2) is palmitoylated at cysteine residues, but palmitoylation is not required for raft association or signaling (Figure 1).209
Figure 1.
Latent membrane protein-1 (LMP1)-mediated activation of natural factor-κB (NF-κB) signaling.
LMP-1 C-terminus domains have two transformation effecter sites (TES 1 and TES2),which mediate tumor necrosis factor receptor signaling. TES1 and TES2 are required for efficient NF-κB- and EBV-mediated B-cell transformation. The PQQAT motif in TES1 associates with TRAF1, 2, 3 and 5, to which CD40 binds and thus provides mechanisms for LMP-1 to act as a constitutively active CD40 decoy for TRAFs. The TES1 interaction with these TRAFs induces an NF-κB noncanonical pathway by phosphorylating NIK, IKKα and p100, which in turn process p100 to p52. TES1 is required for long-term outgrowth, whereas TES2 associates with TRADD and functionally links to TRAF6. TES2 is essential for the initial phase of transformation and activates the classical NF-κB pathway. The direct or indirect association of TRADD with TRAF6 activates TRAF6 E3 ligase, TAK1 and TAK1-like kinase. The TAK1 kinases activate IKKβ, which phosphorylates IκBα, leading to IκBα ubiquitylation and degradation, and release of p50/p65 complexes to the nucleus. Both TES1 and TES2, possibly through TRAF3 and TRAF6, respectively, also induce IRAK1-mediated activation of p38 and other kinase(s) that phosphorylate p65. IRAK1 is required for both p38 activation and p65/RelA phosphorylation. LMP-1 activation of NF-κB is largely IRAK1 and TRAF6 dependent, whereas tumor necrosis factor receptor activation of NF-κB is largely IRAK1 and TRAF6 independent (Figure 1).70, 172, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219
Latent membrane protein-2A
LMP-2A roles in B cells
LMP-2A mimics constitutively active, antigen-independent, BCR signaling. It can also block an antigen-dependent BCR signaling that can be experimentally initiated by surface immunoglobulin crosslinking to increase calcium mobilization and by lytic reactivation of EBV;62, 220, 221 it binds BCR-associated kinases and Nedd4 family ubiquitin-protein ligases, which downmodulate LMP-2A activity by ubiquitinating LMP-2A,222 prevents BCR recruitment to lipid rafts, thereby abrogating BCR function, does not require palmitoylation to localize to buoyant complexes or for function,223 requires a cholesterol for LMP-2A trafficking and stability, provides a pre-BCR-like signal to developing B cells through constitutive activation of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway,224 and requires LMP-2A immunoreceptor tyrosine-based activation motif and PY motifs for an Ag-dependent BCR block and subsequent activation of the PI3K/AKT; in addition, β-catenin accumulation prevents the switch from latent to lytic reactivation.225
LMP-2A promotes B-cell growth,62 induces B lymphoma, has a transformation ability in vitro and in vivo, which was blocked by an immunoreceptor tyrosine-based activation motif LMP-2A mutant, the Syk inhibitor or Syk-specific small interfering RNA,226 and is important but not essential for in vitro primary B-lymphocyte growth transformation, latent infection and lytic virus replication in vitro43, 227, 228 but is essential for growth transformation of germinal center B cells, which do not express the genuine BCR because of deleterious somatic hypermutations in their immunoglobulin genes;221 it increases the prosurvival and anti-inflammatory cytokine IL-10 via PI3K,229 upregulates genes associated with cell cycle induction and inhibition of apoptosis and downregulates genes associated with B-cell-specific factors and immunity similarly to those in HRS cells of HL,230 and it counteracts the antiproliferative effect of the S10A mutant to promote the S-phase entry.231
LMP-2A rescues the LMP-1-generated impairment in germinal center in response to antigen in LMP-1/2A animals,232 makes resting B cells sensitive to NF-κB inhibition and apoptosis,233 potentiates cMyc to promote cell cycle progression and hyperproliferation by downregulating cyclin-dependent kinase inhibitor p27 (kip1) in a proteasome-dependent manner,231 bypasses p53 inactivation in a cMyc-induced lymphomagenesis model,234, 235 is downregulated by c-CBL ubiquitin ligase (E3), a critical negative regulator in the BCR signal pathway,236 cooperates with Notch 1 to alter B-cell identity in vivo,237 activates its own promoter and Notch promoter in an EBNA-2-independent manner,224 enhances the development of autoimmune diseases in transgenic LMP-2A expression in B cells,238 induces hypersensitivity to TLR stimulation, leading to the activation of autoantigen-reactive B cells through the BCR/TLR pathway,34 bypasses anergy induction in response to low levels of soluble antigen, induces NF-κB nuclear translocation independent of BCR crosslinking239 and transactivates the human endogenous retrovirus HERV-K18 superantigen240 (see Table 1).
LMP-2A in epithelial cells
LMP-2A suppresses differentiation,74 promotes epithelial cell spreading and motility together with LMP-2B,241 leads to the transcriptional repression of the hTERT gene,242 contributes to anoikis resistance,243 enriches cancer stem cell-like population from fibroblasts,244 enhances epithelial–mesenchymal transition in NPC via induction of metastatic tumor antigen (MTA1) and mammalian target of rapamycin signaling,245 induces the detected form of P63a (deltaP63) that impairs epithelial cell differentiation,246 activates DNA methyltransferase 1 leading to promoter hypermethylation of the PTEN gene in GC,247 is frequently detected in NPC and induces UDP-glucose dehydrogenase expression via ERK and PI3K/AKT pathways but not JNK and p38 pathways,248 does not induce anchorage-independent cell growth in a human keratinocyte cell line but does in a human GC cell line via the constitutive Ras/PI3K/AKT pathway67 and limits the interferon together with LMP-2B by targeting interferon receptors for degradation249(see Table 1).
Structure, domain and interaction
EBV encodes two nearly identical LMP-2 with the same TMs (LMP-2A and LMP-2B).250 The LMP-2A isoform has 12 TMs and extra 119 a.a.at the amino-terminal cytoplasmic signaling domain, whereas the LMP-2b isoform is identical but lacks the cytoplasmic signaling domain.250 LMP-2A/B are constitutively expressed primarily in the plasma membrane, and also in cytoplasmic location, in all EBV-infected cells.250, 251 LMP-2 associates with and is a substrate for a B-lymphocyte tyrosine kinase Lyn and Syk protein tyrosine kinases252 through the first 167 of the LMP-2A 497 a.a, colocalizes with the cellular tyrosine-phosphorylated proteins on the plasma membrane and is also serine and threonine phosphorylated.62, 253 Although in B cells LMP-2 is tyrosine phosphorylated by the Src family kinase (Lyn, Syk), in epithelial cells it is mediated by the C-terminal Src kinase, which is triggered by epithelial cell adhesion to extracellular matrix proteins.254 The immunoreceptor tyrosine-based activation motif contributes to LMP-2A phosphorylation and participates in signal transduction events in epithelial cells. The BCR block by LMP-2A is bypassed by raising intracellular-free Ca2+ levels with an ionophore or by activating protein kinase C with phorbol 12-myristate 13-acetate. LMP-2A, but not LMP-2B, mediates this effect on calcium mobilization.225 LMP-2A is secreted through exosomes similarly to LMP-1.222 Cholesterol depletion from the plasma membrane increases LMP-2A abundance and LMP-2A exosome secretion and also blocks endocytosis, phosphorylation and ubiquitylation of LMP-2A, indicating that cholesterol-dependent LMP-2A trafficking determines the fate of LMP-2A.222
Latent membrane protein-2B
LMP-2B interferes with LMP-2A functions, increases lytic activation from its latent forms upon BCR crosslinking, lowers the threshold of BCR crosslinking required to induce lytic EBV infection, colocalizes with LMP-2A and restores LMP-2A-mediated Ca2+ mobilization upon BCR crosslinking. Collectively, LMP-2B negatively regulates LMP-2A, the function in preventing the switch from latent to lytic EBV replication.255, 256
EBNA-3 family
EBNA-3A, EBNA-3B and EBNA-3C gene families have the same promoter, similar gene structures, are similarly regulated and regulate host transcription. Each has a domain for binding to RBPJ, a cellular sequence-specific DNA-binding TF that mediates EBNA-2 or Notch binding to DNA.257 All EBNA-3 families are coactivators of EBNA-2. EBNA-3C functions as a coactivator and corepressor. The coactivation activities EBNA-3A and EBNA-3B are around half that of EBNA-3C.258 Although EBNA-3B is dispensable for B-cell transformation, both EBNA-3A and EBNA-3C are essential.49, 50, 259 Despite the similarity, EBNA-3C deletion can only be rescued by 3C but not by EBNA-3A or EBNA-3B expression in the restoration of LCL growth, and EBNA-3A deletion can only be rescued by EBNA-3A.49, 50, 260, 261
In contrast to EBNA-2, which tethers to DNA via the RBPJ bridge, EBNA-3A and EBNA-3C associations with RBPJ inhibits RBPJ recruitments to DNA, downregulates cMyc transcription and blocks EBNA-2 activation effects.46, 262, 263 EBNA-3C residues a.a. 130–159 bind to IRF4 or IRF8,264 and coactivate the EBV LMP-1 promoter with EBNA-2 through an SPI1 site in the absence of RBPJ258, 265 (see Table 1).
EBV-encoded nuclear antigen-3A
Both EBNA-3A and EBNA-3C repress the EBNA-2-activated transcription by direct interaction with RBPJ proteins, a cellular DNA-binding factor known to recruit EBNA-2 to EBNA-2-responsive genes. EBNA-3A represses contiguous clusters arrayed in the human genome by polycomb group-mediated epigenetic silencing.266 The CXCL10 and CXCL9 chemokines and their receptors (CXCR3/4) can control herpesvirus infections. EBNA-3A associates with intergenic enhancers located between CXCL10 and CXCL9 and displaces the transactivator EBNA-2, leading to a rapid transcriptional shutdown, which is also because of a delayed gain of polycomb group histone marks.266
A Bim is a cellular inducer of apoptosis. In the absence of Bim, EBNA-3A and EBNA-3C provide no survival advantage.205 The level of Bim is a critical regulator of B-cell survival and reduced expression is a major determinant of LPD in mice and humans. cMyc can induce apoptosis via Bim. EBNA-3A and EBNA-3C likely repress Bim expression without altering Bim protein or RNA stability, but through reduced histone acetylation and increased DNA methylation on the Bim promoter, which was preceded by polycomb protein-mediated repression.267
EBNA-3A binds to the cMyc-interacting DNA-binding zinc-finger protein-1. EBNA-3A interaction with cMyc-interacting DNA-binding zinc-finger protein-1 prevents cMyc-interacting DNA-binding zinc-finger protein-1 from binding to a coactivator, NPM, resulting in a decrease in CDKN2B transcription.268 EBNA-3A or EBNA-3C inactivation in LCLs induces G1 arrests resulting from EBNA-3A/C-mediated induction of CDKN2A p16INK4A expression.260, 261, 269, 270, 271, 272 Because EBNA-2 activates cMyc expression through RBPJ, and associates less stably with RBPJ compared with EBNA-3A, EBNA-3B or EBNA-3C, some EBNA-3 effects on transcription and LCL growth may be in limitation of EBNA-2 access to RBPJ (10–14, 18–21). EBNA-3A or EBNA-3C association with RBPJ, but not with the adenovirus E1a C-terminal binding protein, is essential for LCL growth.260, 261, 269, 270, 273 Similar to EBNA-3C, EBNA-3A interacts with many cellular partners, including PU.1, Spi-B, histone deacetylase 1, DP103, prothymosin-α, p300, Nm23-H1 and SUMO1, as well as SUMO3, cyclin A, SCF-Skp2 ubiquitin ligase, pRb, Chk2, Mdm2 and MRS18-2. Some of these interactions repress CDKN2A p16INK4A or p14ARF for enabling LCL growth.270. EBNA-3A and EBNA-3C cooperatively repress a transcription of the p16INK4A and p14ARF tumor suppressors, allowing cell cycle entry270 (see Table 1).
EBV-encoded nuclear antigen-3B
Among six latency-associated EBNAs, only EBNA-3B is completely dispensable for B-cell transformation in vitro and could be a tumor suppressor. In contrast to EBNA-3A and EBNA-3C, both of which repress transcriptions of tumor suppressors p14ARF, p16INK4Aand chemokine CXCL10, EBNA-3B upregulates CXCL10 and has a growth inhibitory role. EBNA-3B knockout induces diffuse large B-cell lymphoma-like tumors in humanized NOD/SCID/γc−/− mice reconstituted with the human immune system with the expansion of EBV-specific T cells. The B cells infected with EBNA-3B knockout EBV expand more rapidly and secrete less T-cell chemoattractant CXCL10, leading to inefficient recruitment of T cells in vitro and T-cell-mediated killing in vivo. Natural human B lymphoma cell lines from patients with truncated EBNA-3B EBV exhibited similar genotypic and phenotypic characteristics, including reduced CXCL10 secretion. Importantly, EBNA-3B-mutated B-cell lymphomas were frequently found. EBNA-3B is the EBV-encoded tumor suppressor whose inactivation drives lymphomagenesis and immune evasion274 (see Table 1).
EBV-encoded nuclear antigen-3C
EBNA-3C through N-terminal a.a. 50–400 is essential for LCL growth;50, 273 it coactivates the EBV LMP-1 promoter with EBNA-2 and host CXCR4 and CXCL12 gene expression but represses the EBV C promoter.265, 275, 276.
EBNA-3C associates with SUMO-1, P300, prothymolysin (ProTalpha), histone deacetylase 1/2, metastatic suppressor NM23-H1 through EBNA-3C glutamine- and proline-rich domain, corepressor mSinA and NCoR, SCF-Skp2, cyclin A/D1277 and cMyc, Gemin3 (also called DDX20 or DP103), p53, p53 regulatory proteins, the inhibitor of growth family proteins ING4/5, IRF4/8, aurora kinase B, H2AX and Pim-1;258, 264, 278, 279, 280, 281, 282 it regulates chromatin remodeling via recruitment of histone (de)acetyltransferases, facilitates cell cycle entry, stabilizes Geminin3 and cMyc, induces the Mdm2-mediated p53 degradation and represses p53-dependent transactivation on its downstream genes p21 and Bax, as well as p53- and E2F-mediated apoptosis in part through targeted regulation of interferon regulatory factors 4 and 8.
EBNA-3C also mediates the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase, upregulates aurora kinase B transcription, increases aurora kinase B protein stability by reducing ubiquitylation of aurora kinase B and attenuates H2AX expression, stabilizes Pim-1 and Pim-1-mediated proteasomal degradation of the cell cycle inhibitor p21/WAF1, promoting cell proliferation, upregulates TCL1A and ITGA4, downregulates JAG1 and NCALD and cooperates with EBNA-3A in repressing Bim, a proapoptotic Bcl-2 family protein.264, 267, 273, 283, 284, 285, 286, 287
EBNA-3C coactivation of EBNA-2 requires PU.1 site, but not RBPJ binding sites, in the LMP-1 promoter. The expression of chemokine CXCL12 and its receptor contributes to EBV-positive peripheral blood mononuclear cell growth in mice with severe combined immunodeficiency disease.288 EBNA-3A- and EBNA-3C-mediated B-cell transformation is primarily through transcriptional deregulation of host genes. EBNA-3C and EBNA-3A repress p14ARF and p16INK4A transcription, which help in LCL growth. Depletion of p14ARF and p16INK4A or knockout of p16INK4A supports LCL growth in the absence of EBNA-3C.270, 272 Repressive activities of EBNA-3A and EBNA-3C are associated with histone modifications: EBNA-3A induces repressive histone mark H3K27me3, which is installed by polycomb group proteins at the CXCL10 and CXCL9 chemokine genes,266 whereas EBNA-3C-mediated histone modifications are important for p14ARF and p16INK4A repression.289
Similar to EBNA-2 and LP, EBNA-3C regulates the viral and cellular gene transcription through interactions with cellular proteins including RBPJ264, 265, 290 at 13 000 promoter and enhancer sites (called 3C sites). The 13 000 3C sites are located on EBV LMP-1, BIM and ITGA4 promoters and were highly colocalized with AICE (IRF4/BATF complex), EICE (IRF4/SPI1) and RUNX3. EBNA-3C interactions with AICE and EICE sites drive LCL proliferation.291 EBNA-3C recruits Sin3A repressive complexes (Sin3A, histone deacetylases 1 and 2 and RBPJ) to the p14ARF promoter to mediate p14ARF, and p16INK4A repression in cooperation with EBNA-3A.272 EBNA-3C overcomes p16(INK4a) increase-driven proliferation block after EBV infection. In p16(INK4a)-null cells, functional EBNA-3C is dispensable for the outgrowth of LCLs.272 EBNA-3C functions as a gene regulator in combination with TFs, mostly AICEs, EICEs and RUNX3.290, 291, 292 EBV uses B-cell TFs to drive cell cycle entry for persistence or virus replication (see Table 1).
EBV-encoded RNA
EBV genomes abundantly express noncoding EBV-encoded RNAs (called EBER1 and EBRE2). EBERs are transcribed by host RNA polymerase III as small non coding nonpolyadenylated RNAs.293, 294, 295, 296 The role of EBERs in EBV-induced B-lymphocyte transformation has been contradictory. Earlier reports described nonessential roles of EBER for B-lymphocyte transformation.48, 297 However, a critical role was also demonstrated.298 EBER expression augments colony formation and induces growth in in vitro or in vivo tumorigenesis,79, 299, 300, 301 resistance to RNA-dependent protein kinase (PKR)-dependent apoptosis302 and cytokines including IL-10, IL-9, IGF1 and IL-6,303, 304, 305, 306 and modulates innate immune response.307, 308
EBERs binds to La,293 PKR, ribosomal protein L22 (also called as EAP),309 pattern-recognition receptors, retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated gene-5307 and AU-rich element binding factor 1.310 EBER-mediated RIG-I activation likely contributes to EBV oncogenesis.307
EBERs in complex with La release from cells308 and bind to the dephosphorylated PKR, which is double-stranded RNA-dependent and an interferon (IFN)-inducible serine/threonine kinase.311, 312 Antibody to La is implicated in SLE.293 Viral infection-induced IFNs activate PKR, which phosphorylates the α-subunit of the protein synthesis initiation factor eukaryotic initiation factor 2, leading to translational inhibition. EBER blockades of PKR-mediated phosphorylation of eukaryotic initiation factor 2α result in the blockage of eukaryotic initiation factor 2α-mediated inhibition of protein synthesis and resistance to IFNα-induced apoptosis.78, 313, 314 Most EBERs establish stable complexes with L22 in vivo, thereby modulating protein translation.315 L22 and PKR compete for EBER binding and L22 interferes with EBER inhibition of PKR and EBER-induced gene expression.316 Interaction of EBERs with RIG-I, AU-rich element binding factor 1 and pattern-recognition receptors could activate host innate immune responses.317 EBER double-stranded RNA structures also activate RIG-mediated NF-κB and IRF-3 signaling and subsequently type I IFN induction. EBV latent infection is maintained by counterbalancing to IFN-mediated viral clearance through PKR inhibition. EBER induction of anti-inflammatory and growth-promoting cytokine IL-10 promotes cell growth and this process is a RIG-I-mediated IRF3-dependent but largely NF-κB-independent process (see Table 1).
EBV-encoded miRNAs
EBV genomes express many miRNAs from two regions of EBV's genome: BART and BHRF1 (BamHI fragment H rightward open reading frame 1). The EBV genome transcribes at least 25 pre-miRNAs that encode 40 short single-stranded RNAs.318 These miRNAs were expressed in a variety of EBV-infected malignant cells with abundance of individual miRNA being largely cell type specific. The BART transcript encodes miRNA. Although BART miRNA expression occurs in almost all types of EBV-associated latency cells, BHRF1-encoded miRNAs are quite restricted.319, 320, 321, 322
Many of the EBV miRNA targets were validated. Cellular targets of EBV miRNAs include Bim (BCL2L11), which is targeted by BART-9, -11 and -12, BRUCE by BART15-3p, CASP3 by BART1-3p, CLEC2D by BART̀1-3p, CAPRIN2 by BART13-3p, CXCL11 by BHRF1-3, DICER1 by BART6-5p, DAZAP2, DICE1, IPO7, PDE7A and PELI1 by BART-3, LY75 and SP100 by BART1-5p, PDCD1LG2 by BHRF1-2-5p, BART1-5p and 15-3p, PUMA by BART-5, T-bet(TBX21) by BART-20-5p,TOMM22 by BART-16, NLRP3 by BART-15 and ZNF451 by BHRF1-1.322 CXCL-11, miR-BHRF1–3 target, is a chemokine that is induced by IFN-responsive reactive T cells and binds CXCR3, a common chemokine receptor for many chemokines expressed on T cells.323 The miR-BART2-5p targets a stress-induced natural killer cell ligand, MICB, allowing EBV-infected cells to escape recognition and subsequent elimination.324, 325
Most EBV miRNAs have the ability to sustain latently infected cells. BHRF1 miRNA facilitates progressive growth, in vitro transformation of infected cells and acute systemic EBV infection but not the overall oncogenic potential of EBV in vivo.326, 327, 328 In addition, BHRF1 and BART miRNAs prevent primary B cells or BLs, respectively, from apoptosis.327, 329. In contrast, miR-BART15-3p promoted apoptosis.330 Given that most of the EBV infections persist for a lifetime with asymptomatic penetration, viral miRNAs should also participate, at least in part, in the evasion from host immune surveillance (see Table 1).
Appendix
EBV-induced immediate hyperproliferation of host cell mimics and induces strong ATM/Chk2-mediated DNA damage response, resulting in acute attenuation of infected B-cell growth, which should be bypassed or suppressed for efficient and ultimate immortalization by an EBV antigen. Biochemical and genetic study demonstrated that EBNA3C may function in overcoming the growth arrest.331 Despite its high stability as a dimer in high salt condition, it has been recently shown that EBNA-1 DNA-binding and transactivation activity could be targeted by small molecules or peptides identified by high-throughput cell-based or in silico screens.332, 333, 334, 335, 336
Acknowledgments
This study was supported by a grant from the National R&D Program for Cancer Control, Ministry for Health and Welfare, Republic of Korea (1120010), and by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0012393).
The authors declare no conflict of interest.
References
- Epstein M, Achong B, Barr Y. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet. 1964;1:702–703. doi: 10.1016/s0140-6736(64)91524-7. [DOI] [PubMed] [Google Scholar]
- Henle G, Henle W. Immunofluorescence in cells derived from Burkitt's lymphoma. J Bacteriol. 1966;91:1248–1256. doi: 10.1128/jb.91.3.1248-1256.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle G, Henle W. Observations on childhood infections with the Epstein–Barr virus. J Infect Dis. 1970;121:303–310. doi: 10.1093/infdis/121.3.303. [DOI] [PubMed] [Google Scholar]
- Henle G, Henle W, Clifford P, Diehl V, Kafuko GW, Kirya BG, et al. Antibodies to Epstein–Barr virus in Burkitt's lymphoma and control groups. J Natl Cancer Inst. 1969;43:1147–1157. [PubMed] [Google Scholar]
- Henle W, Henle G. The relation of the Epstein–Barr virus to Burkitt's lymphoma. Zentralbl Bakteriol (Orig A) 1972;220:40–46. [PubMed] [Google Scholar]
- Henle W, Henle G. Epstein–Barr virus-related serology in Hodgkin's disease. Natl Cancer Inst Monogr. 1973;36:79–84. [PubMed] [Google Scholar]
- Henle W, Henle G. Evidence for an oncogenic potential of the Epstein–Barr virus. Cancer Res. 1973;33:1419–1423. [PubMed] [Google Scholar]
- Henle W, Henle G. The Epstein–Barr virus (EBV) in Burkitt's lymphoma and nasopharyngeal carcinoma. Ann Clin Lab Sci. 1974;4:109–114. [PubMed] [Google Scholar]
- Henle W, Henle G. Epstein–Barr virus and human malignancies. Cancer. 1974;34 (Suppl:1368–1374. doi: 10.1002/1097-0142(197410)34:8+<1368::aid-cncr2820340806>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Henle W, Henle G. The sero-epidemiology of Epstein–Barr virus. Adv Pathobiol. 1976;5:5–17. [PubMed] [Google Scholar]
- Henle W, Henle G. Evidence for an etiologic relation of the Epstein–Barr virus to human malignancies. Laryngoscope. 1977;87:467–473. doi: 10.1288/00005537-197704000-00001. [DOI] [PubMed] [Google Scholar]
- Geser A, de The G, Lenoir G, Day NE, Williams EH. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt's lymphoma. Int J Cancer. 1982;29:397–400. doi: 10.1002/ijc.2910290406. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Zhang LG, Wu YC, Huang YS, Huang NQ, Li JY, et al. Prospective studies on nasopharyngeal carcinoma in Epstein–Barr virus IgA/VCA antibody-positive persons in Wuzhou City, China. Int J Cancer. 1985;36:545–547. doi: 10.1002/ijc.2910360505. [DOI] [PubMed] [Google Scholar]
- Sixbey JW, Vesterinen EH, Nedrud JG, Raab-Traub N, Walton LA, Pagano JS. Replication of Epstein–Barr virus in human epithelial cells infected in vitro. Nature. 1983;306:480–483. doi: 10.1038/306480a0. [DOI] [PubMed] [Google Scholar]
- Sixbey JW, Nedrud JG, Raab-Traub N, Hanes RA, Pagano JS. Epstein–Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- Sixbey JW, Lemon SM, Pagano JS. A second site for Epstein–Barr virus shedding: the uterine cervix. Lancet. 1986;2:1122–1124. doi: 10.1016/s0140-6736(86)90531-3. [DOI] [PubMed] [Google Scholar]
- Sixbey JW, Shirley P, Sloas M, Raab-Traub N, Israele V. A transformation-incompetent, nuclear antigen 2-deleted Epstein–Barr virus associated with replicative infection. J Infect Dis. 1991;163:1008–1015. doi: 10.1093/infdis/163.5.1008. [DOI] [PubMed] [Google Scholar]
- Gan YJ, Chodosh J, Morgan A, Sixbey JW. Epithelial cell polarization is a determinant in the infectious outcome of immunoglobulin A-mediated entry by Epstein–Barr virus. J Virol. 1997;71:519–526. doi: 10.1128/jvi.71.1.519-526.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Middeldorp J, Thorley-Lawson DA. Epstein–Barr virus infection in ex vivo tonsil epithelial cell cultures of asymptomatic carriers. J Virol. 2004;78:12613–12624. doi: 10.1128/JVI.78.22.12613-12624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Clark D, Sixbey JW, Rickinson AB. Epstein–Barr virus receptors on human pharyngeal epithelia. Lancet. 1986;1:240–242. doi: 10.1016/s0140-6736(86)90776-2. [DOI] [PubMed] [Google Scholar]
- Young LS, Lau R, Rowe M, Niedobitek G, Packham G, Shanahan F, et al. Differentiation-associated expression of the Epstein–Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J Virol. 1991;65:2868–2874. doi: 10.1128/jvi.65.6.2868-2874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchley AT, De Souza YG, Williams DM, Greenspan D, Greenspan JS. Bromodeoxyuridine incorporation and Ki 67 expression in oral hairy leukoplakia. Oral Dis. 1998;4:9–15. doi: 10.1111/j.1601-0825.1998.tb00248.x. [DOI] [PubMed] [Google Scholar]
- Walling DM, Clark NM, Markovitz DM, Frank TS, Braun DK, Eisenberg E, et al. Epstein–Barr virus coinfection and recombination in non-human immunodeficiency virus-associated oral hairy leukoplakia. J Infect Dis. 1995;171:1122–1130. doi: 10.1093/infdis/171.5.1122. [DOI] [PubMed] [Google Scholar]
- Webster-Cyriaque J, Middeldorp J, Raab-Traub N. Hairy leukoplakia: an unusual combination of transforming and permissive Epstein–Barr virus infections. J Virol. 2000;74:7610–7618. doi: 10.1128/jvi.74.16.7610-7618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling DM, Ling PD, Gordadze AV, Montes-Walters M, Flaitz CM, Nichols CM. Expression of Epstein–Barr virus latent genes in oral epithelium: determinants of the pathogenesis of oral hairy leukoplakia. J Infect Dis. 2004;190:396–399. doi: 10.1086/422039. [DOI] [PubMed] [Google Scholar]
- Walling DM, Ray AJ, Nichols JE, Flaitz CM, Nichols CM. Epstein–Barr virus infection of Langerhans cell precursors as a mechanism of oral epithelial entry, persistence, and reactivation. J Virol. 2007;81:7249–7268. doi: 10.1128/JVI.02754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A, Lindahl T. Epstein–Barr virus genomes with properties of circular DNA molecules in carrier cells. Proc Natl Acad Sci USA. 1975;72:1477–1481. doi: 10.1073/pnas.72.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G, Agathanggelou A, Herbst H, Whitehead L, Wright DH, Young LS. Epstein–Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J Pathol. 1997;182:151–159. doi: 10.1002/(SICI)1096-9896(199706)182:2<151::AID-PATH824>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Niedobitek G, Kremmer E, Herbst H, Whitehead L, Dawson CW, Niedobitek E, et al. Immunohistochemical detection of the Epstein–Barr virus-encoded latent membrane protein 2A in Hodgkin's disease and infectious mononucleosis. Blood. 1997;90:1664–1672. [PubMed] [Google Scholar]
- Niedobitek G, Agathanggelou A, Steven N, Young LS. Epstein–Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol Pathol. 2000;53:37–42. doi: 10.1136/mp.53.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth J, Hansmann ML, Rajewsky K, Kuppers R. Epstein–Barr virus-infected B cells expanding in germinal centers of infectious mononucleosis patients do not participate in the germinal center reaction. Proc Natl Acad Sci USA. 2003;100:4730–4735. doi: 10.1073/pnas.2627966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza TA, Stollar BD, Sullivan JL, Luzuriaga K, Thorley-Lawson DA. Peripheral B cells latently infected with Epstein–Barr virus display molecular hallmarks of classical antigen-selected memory B cells. Proc Natl Acad Sci USA. 2005;102:18093–18098. doi: 10.1073/pnas.0509311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichalk LL, Thorley-Lawson DA. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein–Barr virus in vivo. J Virol. 2005;79:1296–1307. doi: 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Nicholas MW, Conway KL, Sen P, Diz R, Tisch RM, et al. EBV latent membrane protein 2A induces autoreactive B cell activation and TLR hypersensitivity. J Immunol. 2006;177:2793–2802. doi: 10.4049/jimmunol.177.5.2793. [DOI] [PubMed] [Google Scholar]
- Geser A, Lenoir GM, Anvret M, Bornkamm G, Klein G, Williams EH, et al. Epstein–Barr virus markers in a series of Burkitt's lymphomas from the West Nile District, Uganda. Eur J Cancer Clin Oncol. 1983;19:1393–1404. doi: 10.1016/0277-5379(93)90009-t. [DOI] [PubMed] [Google Scholar]
- Henle W, Diehl V, Kohn G, Zur Hausen H, Henle G. Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967;157:1064–1065. doi: 10.1126/science.157.3792.1064. [DOI] [PubMed] [Google Scholar]
- Pope J. Establishment of cell lines from peripheral leukocytes in infectious mononucleosis. Nature. 1967;216:810–811. doi: 10.1038/216810a0. [DOI] [PubMed] [Google Scholar]
- Pope JH, Horne MK, Scott W. Transformation of huiman foetal leukocytes in vitro by filtrates of a human leukaemic line containing herpes-like virus. Int J Cancer. 1968;3:857–866. doi: 10.1002/ijc.2910030619. [DOI] [PubMed] [Google Scholar]
- Alfieri C, Birkenbach M, Kieff E. Early events in Epstein–Barr virus infection of human B lymphocytes. Virology. 1991;181:595–608. doi: 10.1016/0042-6822(91)90893-g. [DOI] [PubMed] [Google Scholar]
- Cohen JI, Wang F, Kieff E. Epstein–Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, Wang F, Mannick J, Kieff E. Epstein–Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick JB, Cohen JI, Birkenbach M, Marchini A, Kieff E. The Epstein–Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Miller CL, Miao XQ, Tomkinson B, Kieff E. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein–Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J Virol. 1993;67:2006–2013. doi: 10.1128/jvi.67.4.2006-2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Miller CL, Tomkinson B, Miao XQ, Kieff E. Deletion of DNA encoding the first five transmembrane domains of Epstein–Barr virus latent membrane proteins 2A and 2B. J Virol. 1993;67:5068–5074. doi: 10.1128/jvi.67.8.5068-5074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini A, Tomkinson B, Cohen JI, Kieff E. BHRF1, the Epstein–Barr virus gene with homology to Bc12, is dispensable for B-lymphocyte transformation and virus replication. J Virol. 1991;65:5991–6000. doi: 10.1128/jvi.65.11.5991-6000.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, et al. Epstein–Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Tomkinson B, Kieff E. An Epstein–Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J Virol. 1994;68:1449–1458. doi: 10.1128/jvi.68.3.1449-1458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein–Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson B, Kieff E. Use of second-site homologous recombination to demonstrate that Epstein–Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J Virol. 1992;66:2893–2903. doi: 10.1128/jvi.66.5.2893-2903.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson B, Robertson E, Kieff E. Epstein–Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Kieff E. Epstein–Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Li H, Johannsen E, Davidson D, Longnecker R, et al. An Epstein–Barr virus that expresses only the first 231 LMP1 amino acids efficiently initiates primary B-lymphocyte growth transformation. J Virol. 1999;73:10525–10530. doi: 10.1128/jvi.73.12.10525-10530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye KM, Izumi KM, Mosialos G, Kieff E. The Epstein–Barr virus LMP1 cytoplasmic carboxy terminus is essential for B-lymphocyte transformation; fibroblast cocultivation complements a critical function within the terminal 155 residues. J Virol. 1995;69:675–683. doi: 10.1128/jvi.69.2.675-683.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Cahir McFarland ED, Riley EA, Rizzo D, Chen Y, Kieff E. The residues between the two transformation effector sites of Epstein–Barr virus latent membrane protein 1 are not critical for B-lymphocyte growth transformation. J Virol. 1999;73:9908–9916. doi: 10.1128/jvi.73.12.9908-9916.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Cahir McFarland ED, Ting AT, Riley EA, Seed B, Kieff ED. The Epstein–Barr virus oncoprotein latent membrane protein 1 engages the tumor necrosis factor receptor-associated proteins TRADD and receptor-interacting protein (RIP) but does not induce apoptosis or require RIP for NF-kappaB activation. Mol Cell Biol. 1999;19:5759–5767. doi: 10.1128/mcb.19.8.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Kaye KM, Kieff ED. Epstein–Barr virus recombinant molecular genetic analysis of the LMP1 amino-terminal cytoplasmic domain reveals a probable structural role, with no component essential for primary B-lymphocyte growth transformation. J Virol. 1994;68:4369–4376. doi: 10.1128/jvi.68.7.4369-4376.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Kaye KM, Kieff ED. The Epstein–Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc Natl Acad Sci USA. 1997;94:1447–1452. doi: 10.1073/pnas.94.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi KM, Kieff ED. The Epstein–Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF- kappaB. Proc Natl Acad Sci USA. 1997;94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, et al. Role of NF-kappa B in cell survival and transcription of latent membrane protein 1-expressing or Epstein–Barr virus latency III-infected cells. J Virol. 2004;78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahir-McFarland ED, Davidson DM, Schauer SL, Duong J, Kieff E. NF-kappa B inhibition causes spontaneous apoptosis in Epstein–Barr virus-transformed lymphoblastoid cells. Proc Natl Acad Sci USA. 2000;97:6055–6060. doi: 10.1073/pnas.100119497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto N, Maeda A, Kobayashi K, Hayashi K, Oka T, Takahashi K, et al. Epstein–Barr virus infection to Epstein–Barr virus-negative nasopharyngeal carcinoma cell line TW03 enhances its tumorigenicity. Lab Invest. 2000;80:303–312. doi: 10.1038/labinvest.3780035. [DOI] [PubMed] [Google Scholar]
- Miller CL, Burkhardt AL, Lee JH, Stealey B, Longnecker R, Bolen JB, et al. Integral membrane protein 2 of Epstein–Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. Epstein–Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- Caldwell RG, Brown RC, Longnecker R. Epstein–Barr virus LMP2A-induced B-cell survival in two unique classes of EmuLMP2A transgenic mice. J Virol. 2000;74:1101–1113. doi: 10.1128/jvi.74.3.1101-1113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart R, Ruf IK, Sample J, Longnecker R. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J Virol. 2000;74:10838–10845. doi: 10.1128/jvi.74.22.10838-10845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels N, Merchant M, Pappu R, Chan AC, Longnecker R, Wienands J. Epstein–Barr virus latent membrane protein 2A (LMP2A) employs the SLP-65 signaling module. J Exp Med. 2001;194:255–264. doi: 10.1084/jem.194.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Longnecker R. Epstein–Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt pathway. J Virol. 2007;81:9299–9306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathmanathan R, Prasad U, Sadler R, Flynn K, Raab-Traub N. Clonal proliferations of cells infected with Epstein–Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–698. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- Fries KL, Miller WE, Raab-Traub N. Epstein–Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J Virol. 1996;70:8653–8659. doi: 10.1128/jvi.70.12.8653-8659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WE, Mosialos G, Kieff E, Raab-Traub N. Epstein–Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-kappaB activation. J Virol. 1997;71:586–594. doi: 10.1128/jvi.71.1.586-594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F, Bendt KM, Raab-Traub N. Epstein–Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J Virol. 2000;74:10681–10689. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JA, Klingelhutz AJ, Raab-Traub N. Epstein–Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J Virol. 2003;77:12276–12284. doi: 10.1128/JVI.77.22.12276-12284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everly DN, Jr, Mainou BA, Raab-Traub N. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein–Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J Virol. 2004;78:13470–13478. doi: 10.1128/JVI.78.24.13470-13478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JA, Raab-Traub N. Roles of the ITAM and PY motifs of Epstein–Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of {beta}-catenin signaling. J Virol. 2005;79:2375–2382. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Raab-Traub N. Induction of EGFR expression by EBV LMP1 CTAR1 is mediated by NF-{kappa}B p50 homodimer/BCL-3 complexes. J Virol. 2007;81:12954–12961. doi: 10.1128/JVI.01601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano J, Takada K. Role of bcl-2 in Epstein–Barr virus-induced malignant conversion of Burkitt's lymphoma cell line Akata. J Virol. 2001;75:1561–1564. doi: 10.1128/JVI.75.3.1561-1564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Nanbo A, Takada K. Replacement of the Epstein–Barr virus plasmid with the EBER plasmid in Burkitt's lymphoma cells. J Virol. 2001;75:9977–9982. doi: 10.1128/JVI.75.20.9977-9982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Inoue K, Adachi-Takasawa K, Takada K. Epstein–Barr virus RNA confers resistance to interferon-alpha-induced apoptosis in Burkitt's lymphoma. EMBO J. 2002;21:954–965. doi: 10.1093/emboj/21.5.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf IK, Rhyne PW, Yang C, Cleveland JL, Sample JT. Epstein–Barr virus small RNAs potentiate tumorigenicity of Burkitt lymphoma cells independently of an effect on apoptosis. J Virol. 2000;74:10223–10228. doi: 10.1128/jvi.74.21.10223-10228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D, Yates J, Sugden B. A putative origin of replication of plasmids derived from Epstein–Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985;5:1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein–Barr virus. Mol Cell Biol. 1985;5:410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein–Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein–Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Malik-Soni N, Frappier L. Nucleophosmin contributes to the transcriptional activation function of the Epstein–Barr virus EBNA1 protein. J Virol. 2014;88:2323–2326. doi: 10.1128/JVI.02521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CD, Chen YL, Min YL, Zhao B, Cheng CP, Kang MS, et al. The nuclear chaperone nucleophosmin escorts an Epstein–Barr virus nuclear antigen to establish transcriptional cascades for latent infection in human B cells. PLoS Pathogen. 2012;8:e1003084. doi: 10.1371/journal.ppat.1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachandran N, Wang X, Frappier L. Functions of the Epstein–Barr virus EBNA1 protein in viral reactivation and lytic infection. J Virol. 2012;86:6146–6158. doi: 10.1128/JVI.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri S, Pan Q, Blencowe BJ, Claycomb JM, Frappier L. Epstein–Barr virus EBNA1 protein regulates viral latency through effects on let-7 microRNA and dicer. J Virol. 2014;88:11166–11177. doi: 10.1128/JVI.01785-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachandran N, Sarkari F, Frappier L. Epstein–Barr nuclear antigen 1 contributes to nasopharyngeal carcinoma through disruption of PML nuclear bodies. PLoS Pathogen. 2008;4:e1000170. doi: 10.1371/journal.ppat.1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaan A, Haviv I, Urban AE, Schulz VP, Hartman S, Zhang Z, et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc Natl Acad Sci USA. 2009;106:22421–22426. doi: 10.1073/pnas.0911676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresang LR, Vereide DT, Sugden B. Identifying sites bound by Epstein–Barr virus nuclear antigen 1 (EBNA1) in the human genome: defining a position-weighted matrix to predict sites bound by EBNA1 in viral genomes. J Virol. 2009;83:2930–2940. doi: 10.1128/JVI.01974-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B, Warren N. A promoter of Epstein–Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood VH, O'Neil JD, Wei W, Stewart SE, Dawson CW, Young LS. Epstein–Barr virus-encoded EBNA1 regulates cellular gene transcription and modulates the STAT1 and TGFbeta signaling pathways. Oncogene. 2007;26:4135–4147. doi: 10.1038/sj.onc.1210496. [DOI] [PubMed] [Google Scholar]
- Flavell JR, Baumforth KR, Wood VH, Davies GL, Wei W, Reynolds GM, et al. Down-regulation of the TGF-beta target gene, PTPRK, by the Epstein–Barr virus encoded EBNA1 contributes to the growth and survival of Hodgkin lymphoma cells. Blood. 2008;111:292–301. doi: 10.1182/blood-2006-11-059881. [DOI] [PubMed] [Google Scholar]
- Baumforth KR, Birgersdotter A, Reynolds GM, Wei W, Kapatai G, Flavell JR, et al. Expression of the Epstein–Barr virus-encoded Epstein–Barr virus nuclear antigen 1 in Hodgkin's lymphoma cells mediates up-regulation of CCL20 and the migration of regulatory T cells. Am J Pathol. 2008;173:195–204. doi: 10.2353/ajpath.2008.070845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine R, Dawson CW, Hu C, Shah KM, Owen TJ, Date KL, et al. Epstein–Barr virus-encoded EBNA1 inhibits the canonical NF-kappaB pathway in carcinoma cells by inhibiting IKK phosphorylation. Mol Cancer. 2010;9:1. doi: 10.1186/1476-4598-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil JD, Owen TJ, Wood VH, Date KL, Valentine R, Chukwuma MB, et al. Epstein–Barr virus-encoded EBNA1 modulates the AP-1 transcription factor pathway in nasopharyngeal carcinoma cells and enhances angiogenesis in vitro. J Gen Virol. 2008;89:2833–2842. doi: 10.1099/vir.0.2008/003392-0. [DOI] [PubMed] [Google Scholar]
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, et al. DNA sequence and expression of the B95-8 Epstein–Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Yates JL, Camiolo SM. Dissection of DNA replication and enhancer activation function of Epstein–Barr virus nuclear antigen 1. Cancer Cells. 1988;6:197–205. [Google Scholar]
- Ambinder RF, Mullen MA, Chang YN, Hayward GS, Hayward SD. Functional domains of Epstein–Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah WA, Ambinder RF, Hayward GS, Hayward SD. Binding of EBNA-1 to DNA creates a protease-resistant domain that encompasses the DNA recognition and dimerization functions. J Virol. 1992;66:3355–3362. doi: 10.1128/jvi.66.6.3355-3362.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith K, Bendell L, Frappier L. Identification of EBNA1 amino acid sequences required for the interaction of the functional elements of the Epstein–Barr virus latent origin of DNA replication. J Virol. 1993;67:3418–3426. doi: 10.1128/jvi.67.6.3418-3426.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L, Goldsmith K, Bendell L. Stabilization of the EBNA1 protein on the Epstein–Barr virus latent origin of DNA replication by a DNA looping mechanism. J Biol Chem. 1994;269:1057–1062. [PubMed] [Google Scholar]
- Bochkarev A, Barwell JA, Pfuetzner RA, Furey W, Jr, Edwards AM, Frappier L. Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein EBNA 1. Cell. 1995;83:39–46. doi: 10.1016/0092-8674(95)90232-5. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM. Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein, EBNA1, bound to DNA. Cell. 1996;84:791–800. doi: 10.1016/s0092-8674(00)81056-9. [DOI] [PubMed] [Google Scholar]
- Summers H, Barwell JA, Pfuetzner RA, Edwards AM, Frappier L. Cooperative assembly of EBNA1 on the Epstein–Barr virus latent origin of replication. J Virol. 1996;70:1228–1231. doi: 10.1128/jvi.70.2.1228-1231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The 2.2A structure of a permanganate-sensitive DNA site bound by the Epstein–Barr virus origin binding protein, EBNA1. J Mol Biol. 1998;284:1273–1278. doi: 10.1006/jmbi.1998.2247. [DOI] [PubMed] [Google Scholar]
- Heller M, Flemington E, Kieff E, Deininger P. Repeat arrays in cellular DNA related to the Epstein–Barr virus IR3 repeat. Mol Cell Biol. 1985;5:457–465. doi: 10.1128/mcb.5.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M, Henderson A, Ripley S, Van Santen V, Kieff E, et al. The IR3 repeat in Epstein–Barr virus DNA has homology to cell DNA, encodes part of a messenger RNA in EBV transformed cells but does not mediate integration of Epstein–Barr virus DNAIn: Prasad U (eds)Nasopharyngeal Carcinoma: Current Concepts University of Malaya: Kuala Lumpur; 1983177–202. [Google Scholar]
- Heller M, van Santen V, Kieff E. Simple repeat sequence in Epstein–Barr virus DNA is transcribed in latent and productive infections. J Virol. 1982;44:311–320. doi: 10.1128/jvi.44.1.311-320.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Manoury B, Fahraeus R. Self-inhibition of synthesis and antigen presentation by Epstein–Barr virus-encoded EBNA1. Science. 2003;301:1371–1374. doi: 10.1126/science.1088902. [DOI] [PubMed] [Google Scholar]
- Trivedi P, Masucci MG, Winberg G, Klein G. The Epstein–Barr-virus-encoded membrane protein LMP but not the nuclear antigen EBNA-1 induces rejection of transfected murine mammary carcinoma cells. Int J Cancer. 1991;48:794–800. doi: 10.1002/ijc.2910480527. [DOI] [PubMed] [Google Scholar]
- Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, et al. Inhibition of antigen processing by the internal repeat region of the Epstein–Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- Levitskaya J, Sharipo A, Leonchiks A, Ciechanover A, Masucci MG. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein–Barr virus nuclear antigen 1. Proc Natl Acad Sci USA. 1997;94:12616–12621. doi: 10.1073/pnas.94.23.12616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharipo A, Imreh M, Leonchiks A, Imreh S, Masucci MG. A minimal glycine–alanine repeat prevents the interaction of ubiquitinated I kappaB alpha with the proteasome: a new mechanism for selective inhibition of proteolysis. Nat Med. 1998;4:939–944. doi: 10.1038/nm0898-939. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Heessen S, Lindsten K, Jellne M, Masucci MG. Inhibition of proteasomal degradation by the gly-Ala repeat of Epstein–Barr virus is influenced by the length of the repeat and the strength of the degradation signal. Proc Natl Acad Sci USA. 2000;97:8381–8385. doi: 10.1073/pnas.140217397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Sharipo A, Masucci MG. Avoiding proteasomal processing: the case of EBNA1. Curr Top Microbiol Immunol. 2002;269:23–36. doi: 10.1007/978-3-642-59421-2_2. [DOI] [PubMed] [Google Scholar]
- Fogg MH, Kaur A, Cho YG, Wang F. The CD8+ T-cell response to an Epstein–Barr virus-related gammaherpesvirus infecting rhesus macaques provides evidence for immune evasion by the EBNA-1 homologue. J Virol. 2005;79:12681–12691. doi: 10.1128/JVI.79.20.12681-12691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw JM, Yates JL. Replication from oriP of Epstein–Barr virus requires exact spacing of two bound dimers of EBNA1 which bend DNA. J Virol. 2001;75:10603–10611. doi: 10.1128/JVI.75.22.10603-10611.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SC, Kang MS, Kieff E. Maintenance of Epstein–Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc Natl Acad Sci USA. 2001;98:1865–1870. doi: 10.1073/pnas.031584698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton T, Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein–Barr virus plasmid origin of DNA replication. J Virol. 1992;66:489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton T, Sugden B. Retention of plasmid DNA in mammalian cells is enhanced by binding of the Epstein–Barr virus replication protein EBNA1. J Virol. 1994;68:4067–4071. doi: 10.1128/jvi.68.6.4067-4071.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins DR, Milman G, Hayward SD, Hayward GS. Sequence-specific DNA binding of the Epstein–Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985;42:859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Yates JL, Camiolo SM, Bashaw JM. The minimal replicator of Epstein–Barr virus oriP. J Virol. 2000;74:4512–4522. doi: 10.1128/jvi.74.10.4512-4522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura R, Sekimoto T, Ito S, Harada S, Yamagata H, Masai H, et al. Nuclear import of Epstein–Barr virus nuclear antigen 1 mediated by NPI-1 (Importin alpha5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J Virol. 2006;80:1979–1991. doi: 10.1128/JVI.80.4.1979-1991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Hung SC, Kieff E. Epstein–Barr virus nuclear antigen 1 activates transcription from episomal but not integrated DNA and does not alter lymphocyte growth. Proc Natl Acad Sci USA. 2001;98:15233–15238. doi: 10.1073/pnas.211556598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q, Flemington EK. siRNAs against the Epstein–Barr virus latency replication factor, EBNA1, inhibit its function and growth of EBV-dependent tumor cells. Virology. 2006;346:385–393. doi: 10.1016/j.virol.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Nasimuzzaman M, Kuroda M, Dohno S, Yamamoto T, Iwatsuki K, Matsuzaki S, et al. Eradication of Epstein–Bbarr virus episome and associated inhibition of infected tumor cell growth by adenovirus vector-mediated transduction of dominant-negative EBNA1. Mol Ther. 2005;11:578–590. doi: 10.1016/j.ymthe.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Frappier L. Functional analyses of the EBNA1 origin DNA binding protein of Epstein–Barr virus. J Virol. 2000;74:4939–4948. doi: 10.1128/jvi.74.11.4939-4948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B. Dominant-negative inhibitors of EBNA-1 of Epstein–Barr virus. J Virol. 1997;71:1766–1775. doi: 10.1128/jvi.71.3.1766-1775.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kapoor P, Frappier L. Separation of the DNA replication, segregation, and transcriptional activation functions of Epstein–Barr nuclear antigen 1. J Virol. 2002;76:2480–2490. doi: 10.1128/jvi.76.5.2480-2490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton S, Levine AJ. Mapping genetic elements of Epstein–Barr virus that facilitate extrachromosomal persistence of Epstein–Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985;5:2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CH, Hayward SD, Rawlins DR. Interaction of the lymphocyte-derived Epstein–Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J Virol. 1989;63:101–110. doi: 10.1128/jvi.63.1.101-110.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampar B, Tanaka A, Nonoyama M, Derge JG. Replication of the resident repressed Epstein–Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc Natl Acad Sci USA. 1974;71:631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams A. Replication of latent Epstein–Barr virus genomes in Raji cells. J Virol. 1987;61:1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T, Adams A, Bjursell G, Bornkamm GW, Kaschka-Dierich C, Jehn U. Covalently closed circular duplex DNA of Epstein–Barr virus in a human lymphoid cell line. J Mol Biol. 1976;102:511–530. doi: 10.1016/0022-2836(76)90331-4. [DOI] [PubMed] [Google Scholar]
- Nonoyama M, Pagano JS. Replication of viral deoxyribonucleic acid and breakdown of cellular deoxyribonucleic acid in Epstein–Barr virus infection. J Virol. 1972;9:714–716. doi: 10.1128/jvi.9.4.714-716.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JL, Guan N. Epstein–Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991;65:483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B, Warren N. Plasmid origin of replication of Epstein–Barr virus, oriP, does not limit replication in cis. Mol Biol Med. 1988;5:85–94. [PubMed] [Google Scholar]
- Kirchmaier AL, Sugden B. Plasmid maintenance of derivatives of oriP of Epstein–Barr virus. J Virol. 1995;69:1280–1283. doi: 10.1128/jvi.69.2.1280-1283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson GS, Gibson TJ, Barrell BG. The BamHI F region of the B95-8 Epstein–Barr virus genome. Virology. 1985;147:99–109. doi: 10.1016/0042-6822(85)90230-2. [DOI] [PubMed] [Google Scholar]
- Reisman D, Sugden B. trans activation of an Epstein–Barr viral transcriptional enhancer by the Epstein–Barr viral nuclear antigen 1. Mol Cell Biol. 1986;6:3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokenski DA, Yates JL. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein–Barr virus. J Virol. 1989;63:2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahn TA, Sugden B. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein–Barr virus LMP gene. J Virol. 1995;69:2633–2636. doi: 10.1128/jvi.69.4.2633-2636.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglielli MT, Woisetschlaeger M, Speck SH. OriP is essential for EBNA gene promoter activity in Epstein–Barr virus- immortalized lymphoblastoid cell lines. J Virol. 1996;70:5758–5768. doi: 10.1128/jvi.70.9.5758-5768.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A, Tyree C, Sugden B. The plasmid replicon of EBV consists of multiple cis-acting elements that facilitate DNA synthesis by the cell and a viral maintenance element. EMBO J. 1998;17:6394–6403. doi: 10.1093/emboj/17.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shire K, Ceccarelli DF, Avolio-Hunter TM, Frappier L. EBP2, a human protein that interacts with sequences of the Epstein–Barr virus nuclear antigen 1 important for plasmid maintenance. J Virol. 1999;73:2587–2595. doi: 10.1128/jvi.73.4.2587-2595.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Kremmer E, Lautscham G, Mueller-Lantzsch N, Grasser FA. Epstein–Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin alpha2. J Biol Chem. 1997;272:3999–4005. doi: 10.1074/jbc.272.7.3999. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Zeghouf M, Wu H, Tellam J, Athanasopoulos V, Greenblatt J, et al. Protein profiling with Epstein–Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J Biol Chem. 2003;278:29987–29994. doi: 10.1074/jbc.M303977200. [DOI] [PubMed] [Google Scholar]
- Kim AL, Maher M, Hayman JB, Ozer J, Zerby D, Yates JL, et al. An imperfect correlation between DNA replication activity of Epstein–Barr virus nuclear antigen 1 (EBNA1) and binding to the nuclear import receptor, Rch1/importin alpha. Virology. 1997;239:340–351. doi: 10.1006/viro.1997.8874. [DOI] [PubMed] [Google Scholar]
- Van Scoy S, Watakabe I, Krainer AR, Hearing J. Human p32: a coactivator for Epstein–Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology. 2000;275:145–157. doi: 10.1006/viro.2000.0508. [DOI] [PubMed] [Google Scholar]
- Wang Y, Finan JE, Middeldorp JM, Hayward SD. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein–Barr virus. Virology. 1997;236:18–29. doi: 10.1006/viro.1997.8739. [DOI] [PubMed] [Google Scholar]
- Shire K, Kapoor P, Jiang K, Hing MN, Sivachandran N, Nguyen T, et al. Regulation of the EBNA1 Epstein–Barr virus protein by serine phosphorylation and arginine methylation. J Virol. 2006;80:5261–5272. doi: 10.1128/JVI.02682-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari F, Sanchez-Alcaraz T, Wang S, Holowaty MN, Sheng Y, Frappier L. EBNA1-mediated recruitment of a histone H2B deubiquitylating complex to the Epstein–Barr virus latent origin of DNA replication. PLoS Pathogen. 2009;5:e1000624. doi: 10.1371/journal.ppat.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari F, Wang X, Nguyen T, Frappier L. The herpesvirus associated ubiquitin specific protease, USP7, is a negative regulator of PML proteins and PML nuclear bodies. PLoS One. 2011;6:e16598. doi: 10.1371/journal.pone.0016598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachandran N, Cao JY, Frappier L. Epstein–Barr virus nuclear antigen 1 Hijacks the host kinase CK2 to disrupt PML nuclear bodies. J Virol. 2010;84:11113–11123. doi: 10.1128/JVI.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik-Soni N, Frappier L. Proteomic profiling of EBNA1-host protein interactions in latent and lytic Epstein–Barr virus infections. J Virol. 2012;86:6999–7002. doi: 10.1128/JVI.00194-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RJ, Wang D, Young LS, Wang F, Rowe M, Kieff E, et al. Epstein–Barr virus-specific cytotoxic T-cell recognition of transfectants expressing the virus-coded latent membrane protein LMP. J Virol. 1988;62:3747–3755. doi: 10.1128/jvi.62.10.3747-3755.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Zhao B, Calderwood MA, Sommermann T, Johannsen E, Kieff E. EBV nuclear antigen EBNALP dismisses transcription repressors NCoR and RBPJ from enhancers and EBNA2 increases NCoR-deficient RBPJ DNA binding. Proc Natl Acad Sci USA. 2011;108:7808–7813. doi: 10.1073/pnas.1104991108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Moses SC, Tan J, Kremmer E, Ling PD. The Epstein–Barr virus EBNA-LP protein preferentially coactivates EBNA2-mediated stimulation of latent membrane proteins expressed from the viral divergent promoter. J Virol. 2005;79:4492–4505. doi: 10.1128/JVI.79.7.4492-4505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Zhou H, Zhao B, Kharchenko PV, Lowry E, Wong L, et al. Epstein–Barr virus nuclear antigen leader protein localizes to promoters and enhancers with cell transcription factors and EBNA2. Proc Natl Acad Sci USA. 2013;110:18537–18542. doi: 10.1073/pnas.1317608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Zou J, Wang H, Johannsen E, Peng CW, Quackenbush J, et al. Epstein–Barr virus exploits intrinsic B-lymphocyte transcription programs to achieve immortal cell growth. Proc Natl Acad Sci USA. 2011;108:14902–14907. doi: 10.1073/pnas.1108892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- Harada S, Kieff E. Epstein–Barr virus nuclear protein LP stimulates EBNA-2 acidic domain- mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han I, Harada S, Weaver D, Xue Y, Lane W, Orstavik S, et al. EBNA-LP associates with cellular proteins including DNA-PK and HA95. J Virol. 2001;75:2475–2481. doi: 10.1128/JVI.75.5.2475-2481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick JB, Tong X, Hemnes A, Kieff E. The Epstein–Barr virus nuclear antigen leader protein associates with hsp72/hsc73. J Virol. 1995;69:8169–8172. doi: 10.1128/jvi.69.12.8169-8172.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portal D, Rosendorff A, Kieff E. Epstein–Barr nuclear antigen leader protein coactivates transcription through interaction with histone deacetylase 4. Proc Natl Acad Sci USA. 2006;103:19278–19283. doi: 10.1073/pnas.0609320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CW, Xue Y, Zhao B, Johannsen E, Kieff E, Harada S. Direct interactions between Epstein–Barr virus leader protein LP and the EBNA2 acidic domain underlie coordinate transcriptional regulation. Proc Natl Acad Sci USA. 2004;101:1033–1038. doi: 10.1073/pnas.0307808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein–Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein–Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Wang F, Thut CJ, Kieff E. The Epstein–Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancao C, Altmann M, Jungnickel B, Hammerschmidt W. Rescue of ‘crippled' germinal center B cells from apoptosis by Epstein–Barr virus. Blood. 2005;106:4339–4344. doi: 10.1182/blood-2005-06-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein–Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- Wang D, Liebowitz D, Kieff E. The truncated form of the Epstein–Barr virus latent-infection membrane protein expressed in virus replication does not transform rodent fibroblasts. J Virol. 1988;62:2337–2346. doi: 10.1128/jvi.62.7.2337-2346.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichwal VR, Sugden B. Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein–Barr virus. Oncogene. 1988;2:461–467. [PubMed] [Google Scholar]
- Moorthy RK, Thorley-Lawson DA. All three domains of the Epstein–Barr virus-encoded latent membrane protein LMP-1 are required for transformation of rat-1 fibroblasts. J Virol. 1993;67:1638–1646. doi: 10.1128/jvi.67.3.1638-1646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LF, Chen F, Zheng X, Ernberg I, Cao SL, Christensson B, et al. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene. 1993;8:1575–1583. [PubMed] [Google Scholar]
- Kondo S, Wakisaka N, Schell MJ, Horikawa T, Sheen TS, Sato H, et al. Epstein–Barr virus latent membrane protein 1 induces the matrix metalloproteinase-1 promoter via an Ets binding site formed by a single nucleotide polymorphism: enhanced susceptibility to nasopharyngeal carcinoma. Int J Cancer. 2005;115:368–376. doi: 10.1002/ijc.20849. [DOI] [PubMed] [Google Scholar]
- Wakisaka N, Pagano JS. Epstein–Barr virus induces invasion and metastasis factors. Anticancer Res. 2003;23:2133–2138. [PubMed] [Google Scholar]
- Lo AK, Liu Y, Wang XH, Huang DP, Yuen PW, Wong YC, et al. Alterations of biologic properties and gene expression in nasopharyngeal epithelial cells by the Epstein–Barr virus-encoded latent membrane protein 1. Lab Invest. 2003;83:697–709. doi: 10.1097/01.lab.0000067480.44925.10. [DOI] [PubMed] [Google Scholar]
- Kim KR, Yoshizaki T, Miyamori H, Hasegawa K, Horikawa T, Furukawa M, et al. Transformation of Madin–Darby canine kidney (MDCK) epithelial cells by Epstein–Barr virus latent membrane protein 1 (LMP1) induces expression of Ets1 and invasive growth. Oncogene. 2000;19:1764–1771. doi: 10.1038/sj.onc.1203502. [DOI] [PubMed] [Google Scholar]
- Nicholson LJ, Hopwood P, Johannessen I, Salisbury JR, Codd J, Thorley-Lawson D, et al. Epstein–Barr virus latent membrane protein does not inhibit differentiation and induces tumorigenicity of human epithelial cells. Oncogene. 1997;15:275–283. doi: 10.1038/sj.onc.1201187. [DOI] [PubMed] [Google Scholar]
- Sheu LF, Chen A, Meng CL, Ho KC, Lee WH, Leu FJ, et al. Enhanced malignant progression of nasopharyngeal carcinoma cells mediated by the expression of Epstein–Barr nuclear antigen 1 in vivo. J Pathol. 1996;180:243–248. doi: 10.1002/(SICI)1096-9896(199611)180:3<243::AID-PATH655>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Miller WE, Earp HS, Raab-Traub N. The Epstein–Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol. 1995;69:4390–4398. doi: 10.1128/jvi.69.7.4390-4398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedobitek G, Fahraeus R, Herbst H, Latza U, Ferszt A, Klein G, et al. The Epstein–Barr virus encoded membrane protein (LMP) induces phenotypic changes in epithelial cells. Virchows Arch B. 1992;62:55–59. doi: 10.1007/BF02899665. [DOI] [PubMed] [Google Scholar]
- Dawson CW, Rickinson AB, Young LS. Epstein–Barr virus latent membrane protein inhibits human epithelial cell differentiation. Nature. 1990;344:777–780. doi: 10.1038/344777a0. [DOI] [PubMed] [Google Scholar]
- Fahraeus R, Rymo L, Rhim JS, Klein G. Morphological transformation of human keratinocytes expressing the LMP gene of Epstein–Barr virus. Nature. 1990;345:447–449. doi: 10.1038/345447a0. [DOI] [PubMed] [Google Scholar]
- Carter KL, Cahir-McFarland E, Kieff E. Epstein–Barr virus-induced changes in B-lymphocyte gene expression. J Virol. 2002;76:10427–10436. doi: 10.1128/JVI.76.20.10427-10436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbach M, Josefsen K, Yalamanchili R, Lenoir G, Kieff E. Epstein–Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J Virol. 1993;67:2209–2220. doi: 10.1128/jvi.67.4.2209-2220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbach M, Liebowitz D, Wang F, Sample J, Kieff E. Epstein–Barr virus latent infection membrane protein increases vimentin expression in human B-cell lines. J Virol. 1989;63:4079–4084. doi: 10.1128/jvi.63.9.4079-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, et al. Induction of bcl-2 expression by Epstein–Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Hu HM, Opipari AW, Wang F, Dixit VM. The Epstein–Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J Biol Chem. 1992;267:24157–24160. [PubMed] [Google Scholar]
- Liebowitz D, Kieff E. Epstein–Barr virus latent membrane protein: induction of B-cell activation antigens and membrane patch formation does not require vimentin. J Virol. 1989;63:4051–4054. doi: 10.1128/jvi.63.9.4051-4054.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JM, Veis D, Korsmeyer SJ, Sugden B. Latent membrane protein of Epstein–Barr virus induces cellular phenotypes independently of expression of Bcl-2. J Virol. 1993;67:5269–5278. doi: 10.1128/jvi.67.9.5269-5278.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng M, Lundgren E. Transient expression of the Epstein–Barr virus LMP1 gene in human primary B cells induces cellular activation and DNA synthesis. Oncogene. 1992;7:1775–1782. [PubMed] [Google Scholar]
- Peng M, Lundgren E. Transient expression of the Epstein–Barr virus LMP1 gene in B-cell chronic lymphocytic leukemia cells, T cells, and hematopoietic cell lines: cell-type-independent-induction of CD23, CD21, and ICAM-1. Leukemia. 1993;7:104–112. [PubMed] [Google Scholar]
- Rowe M, Peng-Pilon M, Huen DS, Hardy R, Croom-Carter D, Lundgren E, et al. Upregulation of bcl-2 by the Epstein–Barr virus latent membrane protein LMP1: a B-cell-specific response that is delayed relative to NF-kappa B activation and to induction of cell surface markers. J Virol. 1994;68:5602–5612. doi: 10.1128/jvi.68.9.5602-5612.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liebowitz D, Wang F, Gregory C, Rickinson A, Larson R, et al. Epstein–Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J Virol. 1988;62:4173–4184. doi: 10.1128/jvi.62.11.4173-4184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, et al. Epstein–Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Brooks L, Busson P, Wang F, Charron D, Kieff E, et al. Epstein–Barr virus (EBV) latent membrane protein 1 increases HLA class II expression in an EBV-negative B cell line. Eur J Immunol. 1994;24:1467–1470. doi: 10.1002/eji.1830240635. [DOI] [PubMed] [Google Scholar]
- Yang J, Deng X, Deng L, Gu H, Fan W, Cao Y. Telomerase activation by Epstein–Barr virus latent membrane protein 1 is associated with c-Myc expression in human nasopharyngeal epithelial cells. J Exp Clin Cancer Res. 2004;23:495–506. [PubMed] [Google Scholar]
- Liu HP, Chen CC, Wu CC, Huang YC, Liu SC, Liang Y, et al. Epstein–Barr virus-encoded LMP1 interacts with FGD4 to activate Cdc42 and thereby promote migration of nasopharyngeal carcinoma cells. PLoS Pathogen. 2012;8:e1002690. doi: 10.1371/journal.ppat.1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AK, Lo KW, Ko CW, Young LS, Dawson CW. Inhibition of the LKB1-AMPK pathway by the Epstein–Barr virus-encoded LMP1 promotes proliferation and transformation of human nasopharyngeal epithelial cells. J Pathol. 2013;230:336–346. doi: 10.1002/path.4201. [DOI] [PubMed] [Google Scholar]
- Dutton A, Woodman CB, Chukwuma MB, Last JI, Wei W, Vockerodt M, et al. Bmi-1 is induced by the Epstein–Barr virus oncogene LMP1 and regulates the expression of viral target genes in Hodgkin lymphoma cells. Blood. 2007;109:2597–2603. doi: 10.1182/blood-2006-05-020545. [DOI] [PubMed] [Google Scholar]
- Anderton E, Yee J, Smith P, Crook T, White RE, Allday MJ. Two Epstein–Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: clues to the pathogenesis of Burkitt's lymphoma. Oncogene. 2008;27:421–433. doi: 10.1038/sj.onc.1210668. [DOI] [PubMed] [Google Scholar]
- Yoshizaki T, Horikawa T, Qing-Chun R, Wakisaka N, Takeshita H, Sheen TS, et al. Induction of interleukin-8 by Epstein–Barr virus latent membrane protein-1 and its correlation to angiogenesis in nasopharyngeal carcinoma. Clin Cancer Res. 2001;7:1946–1951. [PubMed] [Google Scholar]
- Chang MS, Kim DH, Roh JK, Middeldorp JM, Kim YS, Kim S, et al. Epstein–Barr virus-encoded BARF1 promotes proliferation of gastric carcinoma cells through regulation of NF-kappaB. J Virol. 2013;87:10515–10523. doi: 10.1128/JVI.00955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui T, Luftig M, Soni V, Kieff E. Latent infection membrane protein transmembrane FWLY is critical for intermolecular interaction, raft localization, and signaling. Proc Natl Acad Sci USA. 2004;101:278–283. doi: 10.1073/pnas.2237224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Izumi KM, Kieff E. Epstein–Barr virus latent-infection membrane proteins are palmitoylated and raft-associated: protein 1 binds to the cytoskeleton through TNF receptor cytoplasmic factors. Proc Natl Acad Sci USA. 2001;98:4675–4680. doi: 10.1073/pnas.081075298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Kaye KM, Devergne O, Harada JN, Izumi KM, Yalamanchili R, Kieff E, et al. Tumor necrosis factor receptor associated factor 2 is a mediator of NF-kappa B activation by latent infection membrane protein 1, the Epstein–Barr virus transforming protein. Proc Natl Acad Sci USA. 1996;93:11085–11090. doi: 10.1073/pnas.93.20.11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Mizushima S, Azuma S, Kobayashi N, Tojo T, Suzuki K, et al. Identification of TRAF6, a novel tumor necrosis factor receptor- associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J Biol Chem. 1996;271:28745–28748. doi: 10.1074/jbc.271.46.28745. [DOI] [PubMed] [Google Scholar]
- Ansieau S, Scheffrahn I, Mosialos G, Brand H, Duyster J, Kaye K, et al. Tumor necrosis factor receptor-associated factor (TRAF)-1, TRAF-2, and TRAF-3 interact in vivo with the CD30 cytoplasmic domain; TRAF-2 mediates CD30-induced nuclear factor kappa B activation. Proc Natl Acad Sci USA. 1996;93:14053–14058. doi: 10.1073/pnas.93.24.14053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Eliopoulos AG, Dawson CW, Mosialos G, Floettmann JE, Rowe M, Armitage RJ, et al. CD40-induced growth inhibition in epithelial cells is mimicked by Epstein–Barr virus-encoded LMP1: involvement of TRAF3 as a common mediator. Oncogene. 1996;13:2243–2254. [PubMed] [Google Scholar]
- Devergne O, Cahir McFarland ED, Mosialos G, Izumi KM, Ware CF, Kieff E. Role of the TRAF binding site and NF-kappaB activation in Epstein–Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–7908. doi: 10.1128/jvi.72.10.7900-7908.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devergne O, Hatzivassiliou E, Izumi KM, Kaye KM, Kleijnen MF, Kieff E, et al. Association of TRAF1, TRAF2, and TRAF3 with an Epstein–Barr virus LMP1 domain important for B-lymphocyte transformation: role in NF-kappaB activation. Mol Cell Biol. 1996;16:7098–7108. doi: 10.1128/mcb.16.12.7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylla BS, Hung SC, Davidson DM, Hatzivassiliou E, Malinin NL, Wallach D, et al. Epstein–Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-kappaB through a pathway that includes the NF-kappaB-inducing kinase and the IkappaB kinases IKKalpha and IKKbeta. Proc Natl Acad Sci USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J, et al. Epstein–Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA. 2003;100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanArsdale TL, VanArsdale SL, Force WR, Walter BN, Mosialos G, Kieff E, et al. Lymphotoxin-beta receptor signaling complex: role of tumor necrosis factor receptor-associated factor 3 recruitment in cell death and activation of nuclear factor kappaB. Proc Natl Acad Sci USA. 1997;94:2460–2465. doi: 10.1073/pnas.94.6.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Lee JH, Kieff E, Burkhardt AL, Bolen JB, Longnecker R. Epstein–Barr virus protein LMP2A regulates reactivation from latency by negatively regulating tyrosine kinases involved in sIg-mediated signal transduction. Infect Agents Dis. 1994;3:128–136. [PubMed] [Google Scholar]
- Mancao C, Hammerschmidt W. Epstein–Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood. 2007;110:3715–3721. doi: 10.1182/blood-2007-05-090142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Longnecker R. Cholesterol is critical for Epstein–Barr virus latent membrane protein 2A trafficking and protein stability. Virology. 2007;360:461–468. doi: 10.1016/j.virol.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman RB, Longnecker R. LMP2A does not require palmitoylation to localize to buoyant complexes or for function. J Virol. 2004;78:10878–10887. doi: 10.1128/JVI.78.20.10878-10887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LJ, Longnecker R. EBV LMP2A provides a surrogate pre-B cell receptor signal through constitutive activation of the ERK/MAPK pathway. J Gen Virol. 2008;89:1563–1568. doi: 10.1099/vir.0.2008/001461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CL, Lee JH, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein–Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci USA. 1994;91:772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Kawaguchi Y. Role of the immunoreceptor tyrosine-based activation motif of latent membrane protein 2A (LMP2A) in Epstein–Barr virus LMP2A-induced cell transformation. J Virol. 2014;88:5189–5194. doi: 10.1128/JVI.03714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochford R, Miller CL, Cannon MJ, Izumi KM, Kieff E, Longnecker R. In vivo growth of Epstein–Barr virus transformed B cells with mutations in latent membrane protein 2 (LMP2) Arch Virol. 1997;142:707–720. doi: 10.1007/s007050050113. [DOI] [PubMed] [Google Scholar]
- Speck P, Kline KA, Cheresh P, Longnecker R. Epstein–Barr virus lacking latent membrane protein 2 immortalizes B cells with efficiency indistinguishable from that of wild-type virus. J Gen Virol. 1999;80 (Part 8:2193–2203. doi: 10.1099/0022-1317-80-8-2193. [DOI] [PubMed] [Google Scholar]
- Incrocci R, McCormack M, Swanson-Mungerson M. Epstein–Barr virus LMP2A increases IL-10 production in mitogen-stimulated primary B-cells and B-cell lymphomas. J Gen Virol. 2013;94:1127–1133. doi: 10.1099/vir.0.049221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis T, Dyck P, Longnecker R. Epstein–Barr virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed–Sternberg cells of Hodgkin lymphoma. Blood. 2003;102:4166–4178. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- Fish K, Chen J, Longnecker R. Epstein–Barr virus latent membrane protein 2A enhances MYC-driven cell cycle progression in a mouse model of B lymphoma. Blood. 2014;123:530–540. doi: 10.1182/blood-2013-07-517649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrazo AC, Chauchard M, Raab-Traub N, Longnecker R. Epstein–Barr virus LMP2A reduces hyperactivation induced by LMP1 to restore normal B cell phenotype in transgenic mice. PLoS Pathogen. 2012;8:e1002662. doi: 10.1371/journal.ppat.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Mungerson M, Bultema R, Longnecker R. Epstein–Barr virus LMP2A imposes sensitivity to apoptosis. J Gen Virol. 2010;91:2197–2202. doi: 10.1099/vir.0.021444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging KT, Amick AC, Longnecker R. Epstein–Barr virus LMP2A bypasses p53 inactivation in a MYC model of lymphomagenesis. Proc Natl Acad Sci USA. 2009;106:17945–17950. doi: 10.1073/pnas.0907994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultema R, Longnecker R, Swanson-Mungerson M. Epstein–Barr virus LMP2A accelerates MYC-induced lymphomagenesis. Oncogene. 2009;28:1471–1476. doi: 10.1038/onc.2008.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Longnecker R. The c-Cbl proto-oncoprotein downregulates EBV LMP2A signaling. Virology. 2009;385:183–191. doi: 10.1016/j.virol.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LJ, Longnecker R. Epstein–Barr virus latent membrane protein 2A exploits Notch1 to alter B-cell identity in vivo. Blood. 2009;113:108–116. doi: 10.1182/blood-2008-06-160937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson-Mungerson M, Longnecker R. Epstein–Barr virus latent membrane protein 2A and autoimmunity. Trends Immunol. 2007;28:213–218. doi: 10.1016/j.it.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Swanson-Mungerson MA, Caldwell RG, Bultema R, Longnecker R. Epstein–Barr virus LMP2A alters in vivo and in vitro models of B-cell anergy, but not deletion, in response to autoantigen. J Virol. 2005;79:7355–7362. doi: 10.1128/JVI.79.12.7355-7362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutkowski N, Chen G, Calderon G, Huber BT. Epstein–Barr virus latent membrane protein LMP-2A is sufficient for transactivation of the human endogenous retrovirus HERV-K18 superantigen. J Virol. 2004;78:7852–7860. doi: 10.1128/JVI.78.14.7852-7860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Young LS, Dawson CW. The Epstein–Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J Virol. 2005;79:1789–1802. doi: 10.1128/JVI.79.3.1789-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Liu C, Lindvall C, Xu D, Ernberg I. Epstein–Barr virus latent membrane 2A (LMP2A) down-regulates telomerase reverse transcriptase (hTERT) in epithelial cell lines. Int J Cancer. 2005;113:284–289. doi: 10.1002/ijc.20594. [DOI] [PubMed] [Google Scholar]
- Iwakiri D, Minamitani T, Samanta M. Epstein–Barr virus latent membrane protein 2A contributes to anoikis resistance through ERK activation. J Virol. 2013;87:8227–8234. doi: 10.1128/JVI.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya T, Kikuchi Y, Kunita A, Ishikawa S, Matsusaka K, Hino R, et al. Enrichment of stem-like cell population comprises transformation ability of Epstein–Barr virus latent membrane protein 2A for non-transformed cells. Virus Res. 2013;174:108–115. doi: 10.1016/j.virusres.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Lin Z, Wan X, Jiang R, Deng L, Gao Y, Tang J, et al. EBV-encoded LMP2A promotes EMT in nasopharyngeal carcinoma via MTA1 and mTOR signaling induction. J Virol. 2014;88:11872–11885. doi: 10.1128/JVI.01867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotheringham JA, Mazzucca S, Raab-Traub N. Epstein–Barr virus latent membrane protein-2A-induced DeltaNp63alpha expression is associated with impaired epithelial-cell differentiation. Oncogene. 2010;29:4287–4296. doi: 10.1038/onc.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino R, Uozaki H, Murakami N, Ushiku T, Shinozaki A, Ishikawa S, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- Pan YR, Vatsyayan J, Chang YS, Chang HY. Epstein–Barr virus latent membrane protein 2A upregulates UDP-glucose dehydrogenase gene expression via ERK and PI3K/Akt pathway. Cell Microbiol. 2008;10:2447–2460. doi: 10.1111/j.1462-5822.2008.01221.x. [DOI] [PubMed] [Google Scholar]
- Shah KM, Stewart SE, Wei W, Woodman CB, O'Neil JD, Dawson CW, et al. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Kieff E. A second Epstein–Barr virus membrane protein (LMP2) is expressed in latent infection and colocalizes with LMP1. J Virol. 1990;64:2319–2326. doi: 10.1128/jvi.64.5.2319-2326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszewski-Flick MJ, Rowe DT. Minimal protein domain requirements for the intracellular localization and self-aggregation of Epstein–Barr virus latent membrane protein 2. Virus Genes. 2007;35:225–234. doi: 10.1007/s11262-007-0118-8. [DOI] [PubMed] [Google Scholar]
- Rovedo M, Longnecker R. Epstein–Barr virus latent membrane protein 2A preferentially signals through the Src family kinase Lyn. J Virol. 2008;82:8520–8528. doi: 10.1128/JVI.00843-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R, Druker B, Roberts TM, Kieff E. An Epstein–Barr virus protein associated with cell growth transformation interacts with a tyrosine kinase. J Virol. 1991;65:3681–3692. doi: 10.1128/jvi.65.7.3681-3692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F, Longnecker R, Raab-Traub N. Epithelial cell adhesion to extracellular matrix proteins induces tyrosine phosphorylation of the Epstein–Barr virus latent membrane protein 2: a role for C-terminal Src kinase. J Virol. 1999;73:4767–4775. doi: 10.1128/jvi.73.6.4767-4775.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner MP, Berger C, Zauner L, Sigrist JA, Weber M, Longnecker R, et al. Latent membrane protein 2B regulates susceptibility to induction of lytic Epstein–Barr virus infection. J Virol. 2008;82:1739–1747. doi: 10.1128/JVI.01723-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovedo M, Longnecker R. Epstein–Barr virus latent membrane protein 2B (LMP2B) modulates LMP2A activity. J Virol. 2007;81:84–94. doi: 10.1128/JVI.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson ES, Lin J, Kieff E. The amino-terminal domains of Epstein–Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(kappa) J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Johannsen E, Robertson E, Kieff E. Epstein–Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J Virol. 2002;76:232–242. doi: 10.1128/JVI.76.1.232-242.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Divisconte M, Jiang X, Quink C, Wang F. Epstein–Barr virus with the latent infection nuclear antigen 3B completely deleted is still competent for B-cell growth transformation in vitro. J Virol. 2005;79:4506–4509. doi: 10.1128/JVI.79.7.4506-4509.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Johannsen E, Illanes D, Cooper A, Kieff E. Epstein–Barr virus nuclear protein EBNA3A is critical for maintaining lymphoblastoid cell line growth. J Virol. 2003;77:10437–10447. doi: 10.1128/JVI.77.19.10437-10447.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Wu Y, Ishikawa S, Kanda T, Iwakiri D, Takada K. Epstein–Barr virus nuclear protein EBNA3C is required for cell cycle progression and growth maintenance of lymphoblastoid cells. Proc Natl Acad Sci USA. 2006;103:19500–19505. doi: 10.1073/pnas.0604919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Johannsen E, Maruo S, Cahir-McFarland E, Illanes D, Davidson D, et al. EBNA3A association with RBP-Jkappa down-regulates c-myc and Epstein–Barr virus-transformed lymphoblast growth. J Virol. 2003;77:999–1010. doi: 10.1128/JVI.77.2.999-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Marshall DR, Sample CE. A conserved domain of the Epstein–Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jkappa. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Lu J, Cai Q, Saha A, Jha HC, Dzeng RK, et al. The EBV latent antigen 3C inhibits apoptosis through targeted regulation of interferon regulatory factors 4 and 8. PLoS Pathogen. 2013;9:e1003314. doi: 10.1371/journal.ppat.1003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Sample CE. Epstein–Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein–Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J Virol. 2000;74:5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth-Hertle ML, Scholz BA, Erhard F, Glaser LV, Dolken L, Zimmer R, et al. Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9. PLoS Pathogen. 2013;9:e1003638. doi: 10.1371/journal.ppat.1003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos K, Smith P, Anderton E, Middeldorp JM, White RE, Allday MJ. Epstein–Barr virus latency in B cells leads to epigenetic repression and CpG methylation of the tumour suppressor gene Bim. PLoS Pathogen. 2009;5:e1000492. doi: 10.1371/journal.ppat.1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazot Q, Deschamps T, Tafforeau L, Siouda M, Leblanc P, Harth-Hertle ML, et al. Epstein–Barr virus nuclear antigen 3A protein regulates CDKN2B transcription via interaction with MIZ-1. Nucleic Acids Res. 2014;42:9700–9716. doi: 10.1093/nar/gku697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Wu Y, Ito T, Kanda T, Kieff ED, Takada K. Epstein–Barr virus nuclear protein EBNA3C residues critical for maintaining lymphoblastoid cell growth. Proc Natl Acad Sci USA. 2009;106:4419–4424. doi: 10.1073/pnas.0813134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. Epstein–Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci USA. 2011;108:1919–1924. doi: 10.1073/pnas.1019599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertle ML, Popp C, Petermann S, Maier S, Kremmer E, Lang R, et al. Differential gene expression patterns of EBV infected EBNA-3A positive and negative human B lymphocytes. PLoS Pathogen. 2009;5:e1000506. doi: 10.1371/journal.ppat.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalska L, White RE, Parker GA, Turro E, Sinclair AJ, Paschos K, et al. Induction of p16(INK4a) is the major barrier to proliferation when Epstein–Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines. PLoS Pathogen. 2013;9:e1003187. doi: 10.1371/journal.ppat.1003187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sakakibara S, Maruo S, Zhao B, Calderwood MA, Holthaus AM, et al. Epstein–Barr virus nuclear protein 3C domains necessary for lymphoblastoid cell growth: interaction with RBP-Jkappa regulates TCL1. J Virol. 2009;83:12368–12377. doi: 10.1128/JVI.01403-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RE, Ramer PC, Naresh KN, Meixlsperger S, Pinaud L, Rooney C, et al. EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest. 2012;122:1487–1502. doi: 10.1172/JCI58092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allday MJ, Farrell PJ. Epstein–Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68:3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Mar JC, Maruo S, Lee S, Gewurz BE, Johannsen E, et al. Epstein–Barr virus nuclear antigen 3C regulated genes in lymphoblastoid cell lines. Proc Natl Acad Sci USA. 2011;108:337–342. doi: 10.1073/pnas.1017419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Robertson ES. Functional modulation of the metastatic suppressor Nm23-H1 by oncogenic viruses. FEBS Lett. 2011;585:3174–3184. doi: 10.1016/j.febslet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Lan K, Subramanian C, Robertson ES. Epstein–Barr virus nuclear antigen 3C recruits histone deacetylase activity and associates with the corepressors mSin3A and NCoR in human B-cell lines. J Virol. 2003;77:4261–4272. doi: 10.1128/JVI.77.7.4261-4272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Halder S, Upadhyay SK, Lu J, Kumar P, Murakami M, et al. Epstein–Barr virus nuclear antigen 3C facilitates G1–S transition by stabilizing and enhancing the function of cyclin D1. PLoS Pathogen. 2011;7:e1001275. doi: 10.1371/journal.ppat.1001275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Bamidele A, Murakami M, Robertson ES. EBNA3C attenuates the function of p53 through interaction with inhibitor of growth family proteins 4 and 5. J Virol. 2011;85:2079–2088. doi: 10.1128/JVI.02279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha HC, A J MP, Saha A, Banerjee S, Lu J, Robertson ES. Epstein-Barr virus essential antigen EBNA3C attenuates H2AX expression. J Virol. 2014;88:3776–3788. doi: 10.1128/JVI.03568-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Lu J, Cai Q, Sun Z, Jha HC, Robertson ES. EBNA3C augments Pim-1 mediated phosphorylation and degradation of p21 to promote B-cell proliferation. PLoS Pathogen. 2014;10:e1004304. doi: 10.1371/journal.ppat.1004304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha HC, Lu J, Saha A, Cai Q, Banerjee S, Prasad MA, et al. EBNA3C-mediated regulation of aurora kinase B contributes to Epstein–Barr virus-induced B-cell proliferation through modulation of the activities of the retinoblastoma protein and apoptotic caspases. J Virol. 2013;87:12121–12138. doi: 10.1128/JVI.02379-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Murakami M, Kumar P, Bajaj B, Sims K, Robertson ES. Epstein–Barr virus nuclear antigen 3C augments Mdm2-mediated p53 ubiquitination and degradation by deubiquitinating Mdm2. J Virol. 2009;83:4652–4669. doi: 10.1128/JVI.02408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Saha A, Murakami M, Kumar P, Knight JS, Cai Q, et al. Epstein–Barr virus nuclear antigen 3C targets p53 and modulates its transcriptional and apoptotic activities. Virology. 2009;388:236–247. doi: 10.1016/j.virol.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Sharma N, Robertson ES. Epstein–Barr virus latent antigen 3C can mediate the degradation of the retinoblastoma protein through an SCF cellular ubiquitin ligase. Proc Natl Acad Sci USA. 2005;102:18562–18566. doi: 10.1073/pnas.0503886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Lu J, Morizur L, Upadhyay SK, Aj MP, Robertson ES. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathogen. 2012;8:e1002573. doi: 10.1371/journal.ppat.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovan E, Tosello V, Indraccolo S, Cabrelle A, Baesso I, Trentin L, et al. Chemokine receptor expression in EBV-associated lymphoproliferation in hu/SCID mice: implications for CXCL12/CXCR4 axis in lymphoma generation. Blood. 2005;105:931–939. doi: 10.1182/blood-2004-03-0799. [DOI] [PubMed] [Google Scholar]
- Skalska L, White RE, Franz M, Ruhmann M, Allday MJ. Epigenetic repression of p16(INK4A) by latent Epstein–Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP. PLoS Pathogen. 2010;6:e1000951. doi: 10.1371/journal.ppat.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan MJ, Wood CD, Ojeniyi O, Cooper TJ, Kanhere A, Arvey A, et al. Modulation of enhancer looping and differential gene targeting by Epstein–Barr virus transcription factors directs cellular reprogramming. PLoS Pathogen. 2013;9:e1003636. doi: 10.1371/journal.ppat.1003636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Willox B, Zhou H, Holthaus AM, Wang A, Shi TT, et al. Epstein–Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A. Proc Natl Acad Sci USA. 2014;111:421–426. doi: 10.1073/pnas.1321704111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos K, Parker GA, Watanatanasup E, White RE, Allday MJ. BIM promoter directly targeted by EBNA3C in polycomb-mediated repression by EBV. Nucleic Acids Res. 2012;40:7233–7246. doi: 10.1093/nar/gks391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Andrews NC, Miller G, Steitz JA. Two small RNAs encoded by Epstein–Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo L. Identification of transcribed regions of Epstein–Barr virus DNA in Burkitt lymphoma-derived cells. J Virol. 1979;32:8–18. doi: 10.1128/jvi.32.1.8-18.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D. Epstein–Barr virus-encoded RNAs: key molecules in viral pathogenesis. Cancers (Basel) 2014;6:1615–1630. doi: 10.3390/cancers6031615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa MD, Gottlieb E, Lerner MR, Steitz JA. Striking similarities are exhibited by two small Epstein–Barr virus-encoded ribonucleic acids and the adenovirus-associated ribonucleic acids VAI and VAII. Mol Cell Biol. 1981;1:785–796. doi: 10.1128/mcb.1.9.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorovic G, Bosshard R, Karstegl CE, White RE, Pattle S, Chiang AK, et al. Cellular gene expression that correlates with EBER expression in Epstein–Barr Virus-infected lymphoblastoid cell lines. J Virol. 2011;85:3535–3545. doi: 10.1128/JVI.02086-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Kanda T, Takada K. Critical role of Epstein–Barr Virus (EBV)-encoded RNA in efficient EBV-induced B-lymphocyte growth transformation. J Virol. 2005;79:4298–4307. doi: 10.1128/JVI.79.7.4298-4307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano J, Maruo S, Kurozumi K, Oda T, Takada K. Oncogenic role of Epstein–Barr virus-encoded RNAs in Burkitt's lymphoma cell line Akata. J Virol. 1999;73:9827–9831. doi: 10.1128/jvi.73.12.9827-9831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houmani JL, Davis CI, Ruf IK. Growth-promoting properties of Epstein–Barr virus EBER-1 RNA correlate with ribosomal protein L22 binding. J Virol. 2009;83:9844–9853. doi: 10.1128/JVI.01014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repellin CE, Tsimbouri PM, Philbey AW, Wilson JB. Lymphoid hyperplasia and lymphoma in transgenic mice expressing the small non-coding RNA, EBER1 of Epstein–Barr virus. PLoS One. 2010;5:e9092. doi: 10.1371/journal.pone.0009092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Takada K. The role of Epstein–Barr virus-encoded small RNAs (EBERs) in oncogenesis. Rev Med Virol. 2002;12:321–326. doi: 10.1002/rmv.363. [DOI] [PubMed] [Google Scholar]
- Kitagawa N, Goto M, Kurozumi K, Maruo S, Fukayama M, Naoe T, et al. Epstein–Barr virus-encoded poly(A)(−) RNA supports Burkitt's lymphoma growth through interleukin-10 induction. EMBO J. 2000;19:6742–6750. doi: 10.1093/emboj/19.24.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Eizuru Y, Tokunaga M, Takada K. Autocrine growth of Epstein–Barr virus-positive gastric carcinoma cells mediated by an Epstein–Barr virus-encoded small RNA. Cancer Res. 2003;63:7062–7067. [PubMed] [Google Scholar]
- Iwakiri D, Sheen TS, Chen JY, Huang DP, Takada K. Epstein–Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene. 2005;24:1767–1773. doi: 10.1038/sj.onc.1208357. [DOI] [PubMed] [Google Scholar]
- Yang L, Aozasa K, Oshimi K, Takada K. Epstein–Barr virus (EBV)-encoded RNA promotes growth of EBV-infected T cells through interleukin-9 induction. Cancer Res. 2004;64:5332–5337. doi: 10.1158/0008-5472.CAN-04-0733. [DOI] [PubMed] [Google Scholar]
- Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakiri D, Zhou L, Samanta M, Matsumoto M, Ebihara T, Seya T, et al. Epstein–Barr virus (EBV)-encoded small RNA is released from EBV-infected cells and activates signaling from Toll-like receptor 3. J Exp Med. 2009;206:2091–2099. doi: 10.1084/jem.20081761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toczyski DP, Steitz JA. The cellular RNA-binding protein EAP recognizes a conserved stem-loop in the Epstein–Barr virus small RNA EBER 1. Mol Cell Biol. 1993;13:703–710. doi: 10.1128/mcb.13.1.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Pimienta G, Steitz JA. AUF1/hnRNP D is a novel protein partner of the EBER1 noncoding RNA of Epstein–Barr virus. RNA. 2012;18:2073–2082. doi: 10.1261/rna.034900.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PA, Sharp NA, Clemens MJ. Translational control by the Epstein–Barr virus small RNA EBER-1. Reversal of the double-stranded RNA-induced inhibition of protein synthesis in reticulocyte lysates. Eur J Biochem. 1990;193:635–641. doi: 10.1111/j.1432-1033.1990.tb19381.x. [DOI] [PubMed] [Google Scholar]
- McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze MG, Wambach M, Wong ML, Garfinkel M, Meurs E, Chong K, et al. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp TV, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud CG, et al. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein–Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok V, Mitton-Fry RM, Grech A, Steitz JA. Multiple domains of EBER 1, an Epstein–Barr virus noncoding RNA, recruit human ribosomal protein L22. RNA. 2006;12:872–882. doi: 10.1261/rna.2339606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia A, Vyas J, Laing KG, Clemens MJ. Ribosomal protein L22 inhibits regulation of cellular activities by the Epstein–Barr virus small RNA EBER-1. Eur J Biochem. 2004;271:1895–1905. doi: 10.1111/j.1432-1033.2004.04099.x. [DOI] [PubMed] [Google Scholar]
- Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. Mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, et al. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathogen. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein–Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009;386:387–397. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, et al. microRNA profiling in Epstein–Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011;39:1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. Expression profile of microRNAs in Epstein–Barr virus-infected AGS gastric carcinoma cells. J Virol. 2014;88:1389–1393. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzembayeva M, Hayes M, Sugden B. Multiple functions are mediated by the miRNAs of Epstein–Barr virus. Curr Opin Virol. 2014;7C:61–65. doi: 10.1016/j.coviro.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- Feederle R, Haar J, Bernhardt K, Linnstaedt SD, Bannert H, Lips H, et al. The members of an Epstein–Barr virus microRNA cluster cooperate to transform B lymphocytes. J Virol. 2011;85:9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein–Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathogen. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A, Linnstaedt SD, Esoda C, Krisko JF, Martinez-Torres F, Delecluse HJ, et al. A cluster of virus-encoded microRNAs accelerates acute systemic Epstein–Barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J Virol. 2013;87:5437–5446. doi: 10.1128/JVI.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereide DT, Seto E, Chiu YF, Hayes M, Tagawa T, Grundhoff A, et al. Epstein–Barr virus maintains lymphomas via its miRNAs. Oncogene. 2014;33:1258–1264. doi: 10.1038/onc.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein–Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J Virol. 2013;87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein–Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Song KA, Kieff E, Kang MS. Small molecule and peptide-mediated inhibition of Epstein–Barr virus nuclear antigen 1 dimerization. Biochem Biophys Res Commun. 2012;424:251–256. doi: 10.1016/j.bbrc.2012.06.095. [DOI] [PubMed] [Google Scholar]
- Lee EK, Kim SY, Noh KW, Joo EH, Zhao B, Kieff E, et al. Small molecule inhibition of Epstein–Barr virus nuclear antigen-1 DNA binding activity interferes with replication and persistence of the viral genome. Antiviral Res. 2014;104:73–83. doi: 10.1016/j.antiviral.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MS, Lee EK, Soni V, Lewis TA, Koehler AN, Srinivasan V, et al. Roscovitine inhibits EBNA1 serine 393 phosphorylation, nuclear localization, transcription, and episome maintenance. J Virol. 2011;85:2859–2868. doi: 10.1128/JVI.01628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S, Messick T, Schultz DC, Reichman M, Lieberman PM. Development of a high-throughput screen for inhibitors of Epstein–Barr virus EBNA1. J Biomol Screen. 2010;15:1107–1115. doi: 10.1177/1087057110379154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Thompson S, Schultz DC, Zhu W, Jiang H, Luo C, et al. Discovery of selective inhibitors against EBNA1 via high throughput in silico virtual screening. PLoS One. 2010;5:e10126. doi: 10.1371/journal.pone.0010126. [DOI] [PMC free article] [PubMed] [Google Scholar]