Abstract
Many studies have investigated genotoxic effects of high Se diets but very few have addressed the genotoxicity of Se deprivation and its consequences in germ cells and none in somatic cells. To address these data gaps, C57BL/6 male mice were subjected to Se deprivation starting in the parental generation, i.e. before conception. Mice were given a diet of either low (0.01mg Se/kg diet) or normal (0.23mg Se/kg diet) Se content. Ogg1-deficient (Ogg1 −/−) mice were used as a sensitive model towards oxidative stress due to their reduced capacity to repair oxidised purines. Ogg1 −/− mice also mimic the repair characteristics of human post-meiotic male germ cells which have a reduced ability to repair such lesions. The genotoxicity of Se deficiency was addressed by measuring DNA lesions with the alkaline single cell gel electrophoresis (+ Fpg to detect oxidised DNA lesions) in somatic cells (nucleated blood cells and lung cells) and male germ cells (testicular cells). Total Se concentration in liver and GPx activity in plasma and testicular cells were measured. Gene mutation was evaluated by an erythrocyte-based Pig-a assay. We found that Se deprivation of F1 from their conception and until early adulthood led to the induction of DNA lesions in testicular and lung cells expressed as significantly increased levels of DNA lesions, irrespective of the mouse genotype. In blood cells, Se levels did not appear to affect DNA lesions or mutant cell frequencies. The results suggest that the testis was the most sensitive tissue. Thus, genotoxicity induced by the low Se diet in the spermatozoal genome has potential implications for the offspring.
Introduction
The geographical distribution of Selenium (Se) varies throughout the world: parts of Canada, USA, South America, China and Russia are described as selenious areas (1), whereas New Zealand and European countries including Scandinavia have poor Se levels (2). In general, Se concentrations in the soil are relatively high in coastal areas and decrease with distance from the sea (3). This leads to decreased Se concentrations in plants and animal products from Se poor areas that in turn influence the dietary Se intake of humans. Consequently, human Se blood levels vary around the world [(4), reviewed in (5)].
Laboratory rodents as well as epidemiological studies have been used to investigate the effects of Se supplementation at high levels [reviewed by (6)]. Among the few studies that considered low Se intake, the focus was on male fertility in rodents (7–9) and humans (10, 11). Hence, data on genotoxic effects of Se deficiency is very limited for germ cells (12) and no information could be found for somatic cells of Se deprived animals. Investigation of possible adverse effects such as pre-mutagenic DNA lesions and mutations (genotoxic effects) of dietary Se levels deviating from the recommended daily intake level is important. Possible consequences of genotoxic effects include increased risk of cancer as well as germ line mutations that could potentially affect the next generation. The latter mechanism has been suggested in a recent human study showing that the parental lifestyle is a potential source of germ line mutations transmitted to offspring (13).
Se is an essential trace element that is incorporated into selenoproteins and is crucial for these proteins’ catalytic activity. Selenoproteins have diverse functions in the body. They act as antioxidant agents (glutathione peroxidase, GPx), have redox activity (thioredoxin reductases), are involved in production of active thyroid hormones (iodothyronine deiodinases), act as Se transporters (selenoprotein P, SePP), and have anti-inflammatory properties (selenoprotein S) (5). GPx’s catalyse reactions reducing H2O2 (GPx1, GPx3, GPx4, GPx7, GPx8) or lipid hydroperoxides (GPx1, GPx3, GPx4) [reviewed in (14)], thus protecting DNA and other biomolecules from reactive oxygen species (ROS). The thioredoxin reductase system plays an important role in scavenging ROS (1). In this way selenoproteins may reduce the formation of DNA lesions. To estimate the recommended daily Se intake for humans, functional biomarkers such as the maximum GPx-activity in plasma (15) or the SePP concentration in plasma (16, 17) are used. The current recommendation (based on SePP) for the Nordic countries is a dietary intake of 50 µg/day for women and 60 µg/day for men (with a lower and upper intake level for both genders of 20 and 300 µg/day) (18). The SePP concentration in seminal fluid has also been suggested as a useful biomarker for the determination of sperm quality (19).
Given the important functions of selenoproteins, one may hypothesise that Se deficiency could aggravate negative effects induced by other stressors such as environmental contaminants and ionising radiation, e.g. via elevated ROS formation. As part of an extensive study of the combined effects of chronic low dose rate gamma irradiation and Se deficiency in mice (to be presented in a separate publication), an initial experiment was carried out to establish suitable feeding and breeding conditions as well as endpoints. The lack of studies on the genotoxicity of dietary Se deficiency, combined with the perceived biological relevance of these initial results, justified this report.
For this experiment, C57BL/6 mice were deprived of Se over two generations, i.e. Se depletion started in the parental generation, resulting in a Se deficiency of F1 from conception and lasting until early adulthood. This simulates a situation similar to chronic low Se intake in humans. Different compartments representing somatic cells (whole blood and lung cells) and germ cells (testicular cells) were investigated taking into account the different DNA repair capacities in somatic cells compared to male germ cells (20–23). We have previously shown that mouse and human male germ cells differ in their ability to remove oxidised purines such as 8-oxo-7,8-dihydroguanine (8-oxoG) (21). Therefore, we used two different genotypes of mice: the Ogg1-knockout mouse model (Ogg1 −/−) (24) with a reduced ability to repair 8-oxoG thus mimicking the repair characteristics of human testicular cells, and their functional wild type as control. We measured DNA lesions in whole blood and lung cells and testicular cells from mice fed on different Se diets. Pig-a mutant phenotype cells were measured in peripheral blood erythrocytes. Further parameters such as Gpx activities, total Se levels and spermatid head counts were determined.
Materials and methods
Reagents
Lympholyte®-Mammal cell separation reagent was purchased from CedarLane, Burlington, ON, Canada. Anti-PE MicroBeads, LS+ Positive Selection Columns and QuadroMACS™ Separator were purchased from Miltenyi Biotec GmbH, Bergisch Gladbach, Germany. CountBright™ Absolute Counting Beads were from Invitrogen, Life Technologies™, Carlsbad, CA, USA. Heat-inactivated fetal bovine serum (FBS) was from PAA Laboratories, Pasching, Austria. Anticoagulant Solution, Buffered Salt Solution, Nucleic Acid Dye Solution (SYTO®13), Anti-CD24-PE and Anti-CD61-PE were from the Prototype In Vivo Mouse MutaFlow® Kit, Litron Laboratories, Rochester, NY, USA. CK14 ceramic (zirconium oxide) beads and Glutathione Peroxidase Kit (cat. no. 703102) were purchased from Cayman Chemical, Ann Arbor, MI, USA. N-Nitroso-N-ethylurea (ENU, cat. no. 3385), DL-Dithiothreitol (DTT, cat. no. D0632) and Ethylenediaminetetraacetic acid disodium salt dihydrate (Na2EDTA, cat. no. ED2SS) were purchased from Sigma-Aldrich Norway AS, Oslo, Norway. Low melting point agarose (NuSieve®GTG®Agarose) and Gelbond® films were purchased from Lonza, Rockland, ME, USA. SYBR®Gold Nucleic Acid Gel Stain (10000 × concentrate in DMSO) was from Life Technologies™, Carlsbad, CA, USA. Dimethyl sulfoxide (DMSO, cat no. 1029521000) was from Merck, Darmstadt, Germany.
Animals
Wild-type C57BL/6 mice (Ogg1 +/+) with BigBlueTM background were originally purchased from Stratagene (Stratagene, La Jolla, CA, USA). Ogg1 gene knockout mice (deficient of the 8-oxoG-DNA-Glycosylase, Ogg1 −/−, mixed BL6/SV129 background) (24) were generously provided by University of Oslo, Norway. Mice were back-crossed in-house with Ogg1 +/+ BigBlueTM C57BL/6 mice for nine generations, to achieve isogenic strains with similar genetic background (C57BL/6). Homozygote (Ogg1 −/−) and heterozygote (Ogg1 +/–) genotypes were bred in-house and litter mates were used in this study. We have previously shown (unpublished data) that the repair capacity of oxidised purines is similar in the Ogg1 +/− and the Ogg1 +/+ genotype. Animals were kept at a 12h light/dark cycle, controlled temperature (20–24°C) and humidity (55±10%), following the European convention, Appendix A (25). A total of 28 male animals were used in this study and allocated to normal Se diet (6 mice of each genotype) or low Se diet (8 mice of each genotype) (see Diet). The mice were 8–11 weeks old at time of sacrifice. Two to six mice were kept per individually ventilated plastic cage (Innovive, San Diego, CA, USA) with Nestpack bedding (Datesand Ltd., Manchester, UK). Two additional mice were treated with ENU (IP injection, 22mg/kg bw on three consecutive days, total dose of 66mg/kg bw) and used as positive control in the Pig-a mutation assay. The experiments were approved by the National Experimental Animal Board of Norway.
Study design
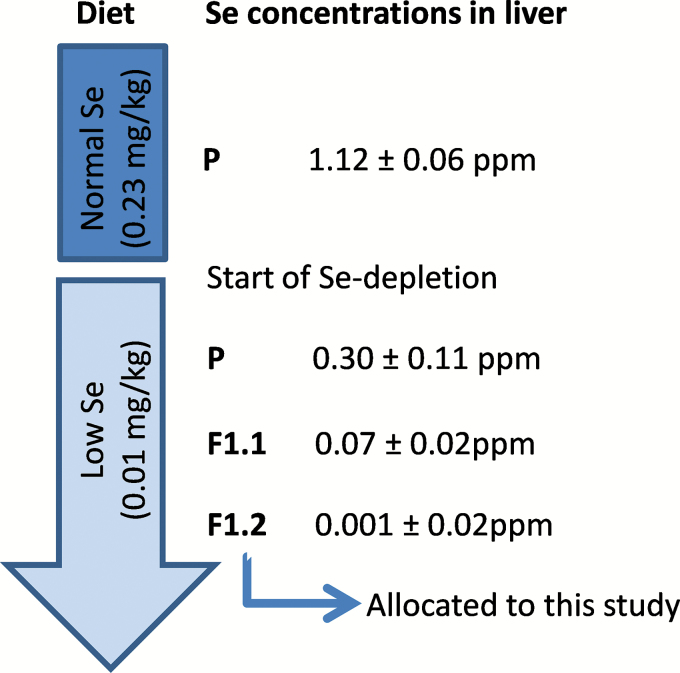
This was a pilot study to investigate genotoxic effects of a low Se diet and to establish endpoints, logistics and proper Se depletion in mice as a basis for a more comprehensive study. To study the effect of Se deficiency starting in utero and lasting until early adulthood, mice were kept on Se-depleted feed during two generations. For confirmation of the feed status, total Se levels in livers were measured during the depletion revealing that the second or later litter (second generation, F1.2) were Se-depleted and suitable for inclusion in the study (Figure 1). This means that parents (P, male and female) received low Se diet for at least 3 weeks before conception of the offspring used in this study (2. litter, F1.2). The complete study, to be reported elsewhere, was designed to investigate the impact of Se deficiency also on spermatogenesis, and therefore only male offspring (F1.2) was studied. It is known that Se is important for sperm motility and avoidance of oxidative stress (GPx).
Figure. 1.
Study design for Se depletion over two generations. Diets are indicated as normal Se diet (0.23mg/kg) and low Se diet (0.01mg/kg) in the vertical arrow. The generations of mice are indicated as follows: P – parental; offspring (F1) of the first litter (F1.1), or second litter (F1.2). The total Se level in liver (ppm, mean ± SD) for each generation is indicated. Parents were given low Se diet for at least 3 weeks before conception of the F1.2 which were used for genotoxicity and mutation studies.
Diet
Mice were randomly assigned to groups fed either with a normal Se diet (0.23mg Se/kg diet, cat. no. HT2019) or a low Se diet (0.01mg Se/kg diet, based on torula yeast, cat. no. TD.92163) whereupon Se concentrations are based on dry weight. Table I shows the main nutrient composition of the diets (both purchased from Harlan Teklad, Harlan Laboratories, Indianapolis, IN, USA) with ground wheat and corn as the only Se sources. In both wheat and corn Se is mainly present as selenomethionine (26–28). Forage and Se-free water (type I water acidised with 2mM HCl to prevent bacterial growth) were available ad libitum for all mice throughout the study.
Table I.
Composition of normal and low Se diet.
| Normal Se dieta | Low Se dietb | |
|---|---|---|
| Selenium (mg/kg) | 0.23 | 0.01 |
| Energy (kcal/g) | 3.3 | 3.9 |
| Calories from protein (%) | 23.0 | 16.2 |
| Calories from carbohydrate (%) | 55.0 | 69.5 |
| Calories from fat (%) | 22.0 | 14.3 |
aHT2019, Harlan Teklad, USA.
bTD.92163, Harlan Teklad, USA.
Tissue and blood sampling
Blood samples (100 µl) were taken from the saphenous vein using a 21-G needle and a heparinised capillary tube (Fisherbrand®, Pittsburgh, PA, USA) 2 days prior to kill. Sixty µl of free-flowing blood for Pig-a gene mutation assay was added to 100 µl anticoagulant (supplied in the Prototype In Vivo Mouse MutaFlow® Kit, Litron Laboratories), mixed well and kept at room temperature. For the single cell gel electrophoresis (SCGE) assay 30 µl of free-flowing blood was added to 100 µl anticoagulant, mixed well and kept on ice. Further processing was performed within 2h.
Mice were killed by cervical dislocation. Liver, lung, testes and epididymis (caput epididymis, cauda epididymis/vas deferens) were harvested and quickly frozen, either in tubes in liquid N2 (epididymis) or wrapped in aluminium foil between blocks of dry ice (liver, lung, testicle). A piece of the liver and one half of a testis without capsule were kept in ice-cold Merchant’s buffer (0.14M NaCl, 1.47mM KH2PO4, 2.7mM KCl, 8.1mM Na2HPO4, 10mM Na2EDTA) for SCGE until further processing with a mechanical technique for isolation of nuclei/cells from tissue (29) as follows: tissue was cut into small pieces in 1ml ice-cold Merchant’s buffer and then transferred to a small cylindrical tube with a stainless steel screen (0.4mm) at its end. The tissue was pressed through the screen with a modified plastic plunger. The resulting crude nuclei/cell suspension was filtered through cotton gauze (only for testis) and nylon mesh (100 µm). The majority of the nuclei/cells in the testicular cell preparation are male germ cells, judging from earlier studies in our laboratory showing that more than 50% (52–69% in this study) of a testicle cell suspension consists of haploid germ cells, i.e. round and elongated spermatids (unpublished data). All samples were kept on ice at all times and processing was performed within 20min.
Total Se level in liver
For the determination of the total Se level in liver, samples of about 100mg of tissues were freeze dried and digested as described (30). In short, concentrations of Se were determined using ICP-MS (Agilent 8800) with neutral gain, where the quadropoles (Q) are set to scan with a fixed mass difference between Q1 and Q2. In the measurement of Se-78 the Q1 was set to 78, then oxygen was used as a reaction gas in the collision cell and Q2 measured on mass to charge (m/z) ratio 94 (78+16). The results were consistent among the measured isotopes of Se (76, 78, 79 and 82), and 78 Se (94 m/z) was chosen for further calculations. Bovine Liver (NIST 1577 b) was used as certified reference material for Se. Indium was used as an internal standard correcting for loss during sample preparation, sample introduction and possible matrix effects on the ICP-MS.
GPx activity
Glutathione peroxidase (EC 1.11.1.9) activities were measured in samples of testicle, caput epididymis and plasma following the instructions provided with the Glutathione Peroxidase Kit (Cayman Chemical). The assay does not distinguish between the various GPxs (e.g. Se-dependent and -independent GPx) but rather measures the activity of all GPxs. Testis and caput epididymis were homogenised by adding the tissue to tubes filled with CK14 ceramic beads and buffer (50mM Tris–HCl, 5mM Na2EDTA, 1mM DTT) using Precelly®24 (Bertin Technologies, USA) at 5000rpm for 2×20 s. The samples were centrifuged for 15min (10 000 × g at 4°C) to remove debris. The GPx activity was indirectly measured as the decrease of NADPH at 340nm for 6min in plasma and supernatant (testis and caput epididymis).
Testicular spermatid head count
Following the protocol by Seung et al. (31) testes were homogenised (Ultra Turrax T8, Jahnke & Kunkel, Staufen, Germany) in 5ml DMSO/saline solution (10% DMSO, 0.9% NaCl) for 1min at 7500rpm. Testicular spermatid heads were stained with Trypan Blue and counted manually by means of a haemocytometer. One testis from each group (Ogg1 +/− and Ogg1 −/− on normal and low Se diet, respectively) was analysed.
SCGE
A high throughput version of the alkaline SCGE (32) was used, with minor modifications.
Blood samples (nucleated blood cells, i.e. leukocytes) were analysed without further purification. Cell density of blood and tissue suspension was checked under the microscope and the titre adjusted to a final concentration of approximately 106 cells/ml. The homogenous nuclei/cell suspensions or the whole blood/anticoagulant suspensions were diluted 1:10 in 0.75% low melting point agarose at 37°C; triplicates (3×4 µl) from each individual sample were then immediately applied to a cold GelBond® film as described (32). Lysis was performed overnight in lysis buffer (2.5M NaCl, 0.1M Na2EDTA, 0.01M Tris-base, 0.2M NaOH, 0.034M N-Lauroylsarcosine, 10% DMSO, 1% TritonX-100, pH 10). Films were transferred to an enzyme buffer (0.04M Hepes, 0.1M KCl, 0.5mM Na2EDTA, pH 7.6) for 60min at 4°C. For enzyme treatment, films were immersed in fresh pre-warmed enzyme buffer with 0.2mg/ml BSA and 0.02 µg/ml Formamidopyrimidine DNA glycosylase (crude Fpg extract, a bacterial homologue of the eukaryotic Ogg1 detecting oxidative DNA lesions) (33). The Fpg concentration was optimised based on titration experiments using a photoactivated compound (Ro12-9786) as recommended (34). After 60min treatment (+/− Fpg) at 37°C, films were transferred to electrophoresis solution (0.3M NaOH, 0.001M Na2EDTA, pH > 13) for 40min to unwind the DNA, followed by 20min alkaline electrophoresis at 8°C, 25V and 0.8V/cm in fresh electrophoresis solution, with circulation.
After electrophoresis, films were rinsed in water (1min) and in neutralisation solution (0.32M Tris–HCl, 0.08M Tris-base, pH 7.5) for 2×5min, and dried after fixation in ethanol (5min in <70% and 90min in 96% EtOH). For scoring of comets, DNA was stained by immersing the films in TE-buffer (1mM Na2EDTA, 10mM Tris–HCl, pH 8.0) plus SYBR®Gold Nucleic Acid Gel Stain (10000 × diluted). Fifty randomly chosen comets per replicate were scored using 20 × magnification in an Olympus BX51 microscope (light source: Olympus BH2-RFL-T3, Olympus optical Co., Ltd; camera: A312f-VIS, BASLER, Ahrensburg, Germany) and the Comet IV analysis software (Perceptive Instruments Ltd, Bury St. Edmunds, UK) to quantify the relative amount of fluorescing DNA in the comet tail vs that of the whole comet (% tail DNA) as a measure of the level of DNA damage in the individual cell. For the lung, fewer than 50 comets per sample could be scored due to incorrect dilutions. Data for Fpg-sensitive sites (Fpg-ss) were obtained by subtracting % tail DNA of samples in the control film [single strand breaks (ssb) and alkali labile sites (als)] from % tail DNA of samples in the Fpg-treated film.
Pig-a gene mutation assay
The Pig-a gene mutation assay was performed as described in the Prototype In Vivo Mouse MutaFlow® Kit instruction manual. Blood/anticoagulant mixture (60 + 100 µl) was carefully placed on top of 3ml Lympholyte®-Mammal and centrifuged (20min, 800 × g, room temperature) to isolate leuko- and platelet-depleted red blood cells (RBC) and reticulocytes (RET, immature red blood cells). After rinsing the pellet with PBS three times without disturbing the blood cells (RBCs and RETs), cells were first incubated with anti-CD24-PE [labelling wild type (wt) erythrocytes]/anti-CD61-PE (labelling platelets) and subsequently with anti-PE magnetic particles (binding to the wt-erythrocytes and platelets). Incubations lasted for 20min and were separated by washing/centrifugation steps. Afterwards a small fraction of each sample was taken and added to a Nucleic Acid Dye/Counting Bead Solution and incubated at 37°C for 30min. This ‘pre-column’ sample provides information about the cell-to-bead-ratio. The majority of the sample was transferred to a magnetic column (Miltenyi Biotec) to selectively remove CD24 positive cells and CD61 positive platelets. The eluate (enriched CD24-negative erythrocytes and reticulocytes, i.e. RBCCD24- and RETCD24-, respectively) was concentrated in a further centrifugation step, washed and subsequently incubated with 300 µl Nucleic Acid Dye/Counting Bead Solution at 37°C for 30min. This so-called ‘post-column’ sample gives information about the mutant-to-bead-ratio. All pre- and post-column samples were analysed using a flow cytometer (BD LSRII, FACSDiva Software, BD Bioscience, San Jose, CA, USA) and their ratios are used to calculate the phenotypic mutant cell frequency as described previously for rats (35). The total numbers of reticulocyte and erythrocyte equivalents are derived from the cell-to-bead ratios in the pre-column sample and the number of counting beads observed in the post-column sample. On average this was 128×106 RBCs and 7.3×106 RETs per sample.
It should be noted that the Prototype MutaFlow® protocol as used herein (version120301) has been modified since this study was completed. In the current protocol, post-column samples are no longer incubated at 37°C for 30min, a step that is now known to lyse mouse erythrocytes, especially RNA-negative erythrocytes. Thus, the RBCCD24- results presented herein should be interpreted with caution.
Statistical analysis
The raw data of the SCGE was processed with the help of the Comet Assay Spreadsheet Generator Version 1.3.1 (Perceptive Instruments Ltd). Subsequently the % TI (tail intensity, i.e. % tail DNA) of 50 comets were summarised as median (per gel) and 3 gels per animal (technical replicates) were summarised as mean as suggested by Bright et al. (36). The data for Pig-a mutation assay was processed and calculated as described by Dertinger et al. (35) using Microsoft Excel 2010.
Further statistical analysis was performed in JMP Pro 11 (Statistical Analysis System Institute Inc., Cary, NC, USA).
Pig-a mutation assay data were analysed with a generalised linear model assuming Poisson distribution; SCGE data were analysed with two-way ANOVA in order to identify impacts of diet, genotype or interactions of both.
Weight data (body weight and relative testis weight) were analysed with the non-parametric Wilcoxon-Rank-Sum test and are given as median with its 25th and 75th quartile.
Total Se levels measured in liver, GPx activity and testicular sperm head count, were analysed with a t-test; data is given as the mean (± SD).
Results
In order to confirm Se depletion in mice fed with the low Se diet (Table I) total Se levels (liver) were analysed in parents and their offspring (F1.1 and F1.2). Only offspring of the second or later litter showed complete Se depletion (i.e. no further reduction of total Se level, data not shown) and were considered adequate to be included in the study (Figure 1). The body weight of the Se-depleted mice was significantly lower compared to the weight of those on the normal Se diet: 21.5g (21.0; 22.0) versus 23.5g (22.3; 26.0), respectively; P = 0.0050. Some of the mice (offspring) on the low Se diet showed signs of ataxia.
GPx activities and total Se level
In general, the GPx activities were lower in plasma than in tissues (Table II). Biological material from each investigated organ was pooled for mice in the same group, i.e. six and eight mice per genotype on normal and low Se diet, respectively. The data showed a significant drop in GPx activity of mice on the low Se diet in testicle, caput epididymis and plasma (Table II), P < 0.0001. The decrease in GPx activity was less pronounced in testis (86%) compared to caput epididymis and plasma (98%). The GPx activity was similar in both mouse genotypes.
Table II.
Glutathione peroxidase (GPx) activity in testis, caput epididymis and plasma.
| nmol/min/ml | ||||
|---|---|---|---|---|
| Normal Se diet | Low Se diet | |||
| Ogg1 +/− | Ogg1 −/− | Ogg1 +/− | Ogg1 −/− | |
| Testis | 123.1 | 128.0 | 16.6 (13.5%)a | 18.4 (14.4%)a |
| Caput epididymis | 148.7 | 133.9 | 3.1 (2.1%)a | 3.1 (2.3%)a |
| Plasma | 85.5 | 96.9 | 1.7 (2.0%)a | 4.3 (4.4%)a |
Mouse tissues were pooled in each group before analysis, i.e. six and eight mice per genotype on normal and low Se diet, respectively.
aRelative activity compared to normal diet of same genotype
A significant decrease (Table III, P < 0.001) of the total Se level in liver of Se-depleted mice was measured. The mean levels were 0.030 (± 0.005) and 3.958 (± 0.340) mg Se/kg liver in mice on low and normal Se diet, respectively.
Table III.
Testicular spermatid head count, relative testis weight, bodyweight and total Se levels in liver of mice on normal and low Se diet.
| Diet | Genotype | n | Spermatid heads/g testisa | Relative testis weight (%) | Bodyweight (g) | Se level in liver (ppm) |
|---|---|---|---|---|---|---|
| Normal Se | Ogg1 +/− | 6 | 149.7×106 (100%)b | 1.17 | 25 | 4.016 |
| Ogg1 −/− | 6 | 114.8×106 (77%)b | 1.22 | 22.5 | 3.900 | |
| Low Se | Ogg1 +/− | 8 | 103.8×106 (69%)b | 1.10 | 22.0 | 0.029 |
| Ogg1 −/− | 8 | 92.7×106 (62%)b | 1.03 | 21.0 | 0.032 |
aOne representative testis per group.
bNumber of testicular spermatid heads per gram testis (%), relative to the number in Ogg1 +/− mice on normal Se diet.
Testicular spermatid head count and relative testis weight
In mice fed the low Se diet, the number of testicular spermatid heads was reduced by up to 62% (Table III). On both normal Se diet and low Se diet the Ogg1 deficient mice had fewer testicular spermatid heads than the functional wild type. The relative testis weights were significantly reduced compared to mice given the normal Se diet: 1.05% (0.98; 1.22) vs 1.21% (1.17; 1.28), respectively; P = 0.028 (Table III).
Single cell gel electrophoresis (SCGE)
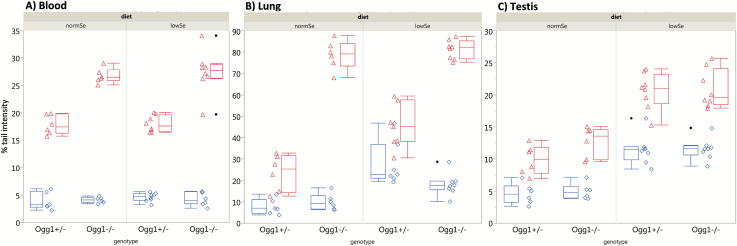
The low Se diet caused a significant increase (Table IV, P < 0.0001) in the level of DNA damage (ssb/als) in testicular cells (Figure 2), independent of the genotype. The same trend is observed in lung cells; however, the level of ssb/als appears to be also dependent on the interaction of diet and genotype. The DNA damage level in nucleated blood cells remained unchanged, regardless of diet or genotype, at a level below 6% TI in all groups (Figure 2). These levels were within the historical background level of nucleated blood cells of Ogg1 +/− and Ogg1 −/− observed in our laboratory.
Table IV.
P values of two-way ANOVA for alkaline SCGE data.
| Tissue | Treatment | Diet | Genotype | Diet × genotype |
|---|---|---|---|---|
| Blood | ssb/alsa | 0.3145 | 0.9748 | 0.5829 |
| Fpgb | 0.7682 | <0.0001 | 0.7009 | |
| Fpg-ssc | 0.8577 | <0.0001 | 0.4637 | |
| Lung | ssb/alsa | <0.0001 | 0.1518 | 0.0258 |
| Fpgb | 0.0006 | <0.0001 | 0.0043 | |
| Fpg-ssc | 0.4113 | <0.0001 | 0.1527 | |
| Testis | ssb/alsa | <0.0001 | 0.7570 | 0.8571 |
| Fpgb | <0.0001 | 0.1353 | 0.2909 | |
| Fpg-ssc | 0.0174 | 0.1790 | 0.3309 |
Bold values are significant (P < 0.05).
aSingle strand breaks and alkali labile sites.
bSingle strand breaks, alkali labile sites and oxidative DNA lesions detected by Fpg.
cFpg-sensitive sites, i.e. oxidative DNA lesions only.
Figure 2.
Results of the alkaline SCGE assay. The different genotypes (Ogg1 +/−; Ogg1 −/−) and diets (normSe and lowSe) are indicated. Each symbol (diamond or triangle) represents, for one animal, the mean % TI. Boxplot: median with quartile. Whiskers indicate the lowest/highest value still within 1.5 IQR (inter quartile range) of the 25th and 75th quartile. Solid circles are outliers. Diamonds and boxplots left to the diamonds show ssb/als; triangles and box plots right to the triangles show oxidative DNA lesions after Fpg-treatment for blood (A), lung (B) and testis (C).
Oxidative DNA lesions, i.e. mainly oxidised purines, are revealed in the SCGE by incubating nucleoids with Fpg before electrophoresis. Oxidative DNA lesions were evident in all three tissues (blood, lung and testicle) and groups (Figure 2). In testicular cells, the low Se diet was associated with a significant increase of oxidative DNA lesions compared to controls (normal Se diet) with P < 0.0001, independent of the genotype. The lack of Ogg1 caused a significant increase of DNA lesions in both blood and lung cells (P < 0.0001, Table IV).
Pig-a gene mutation assay
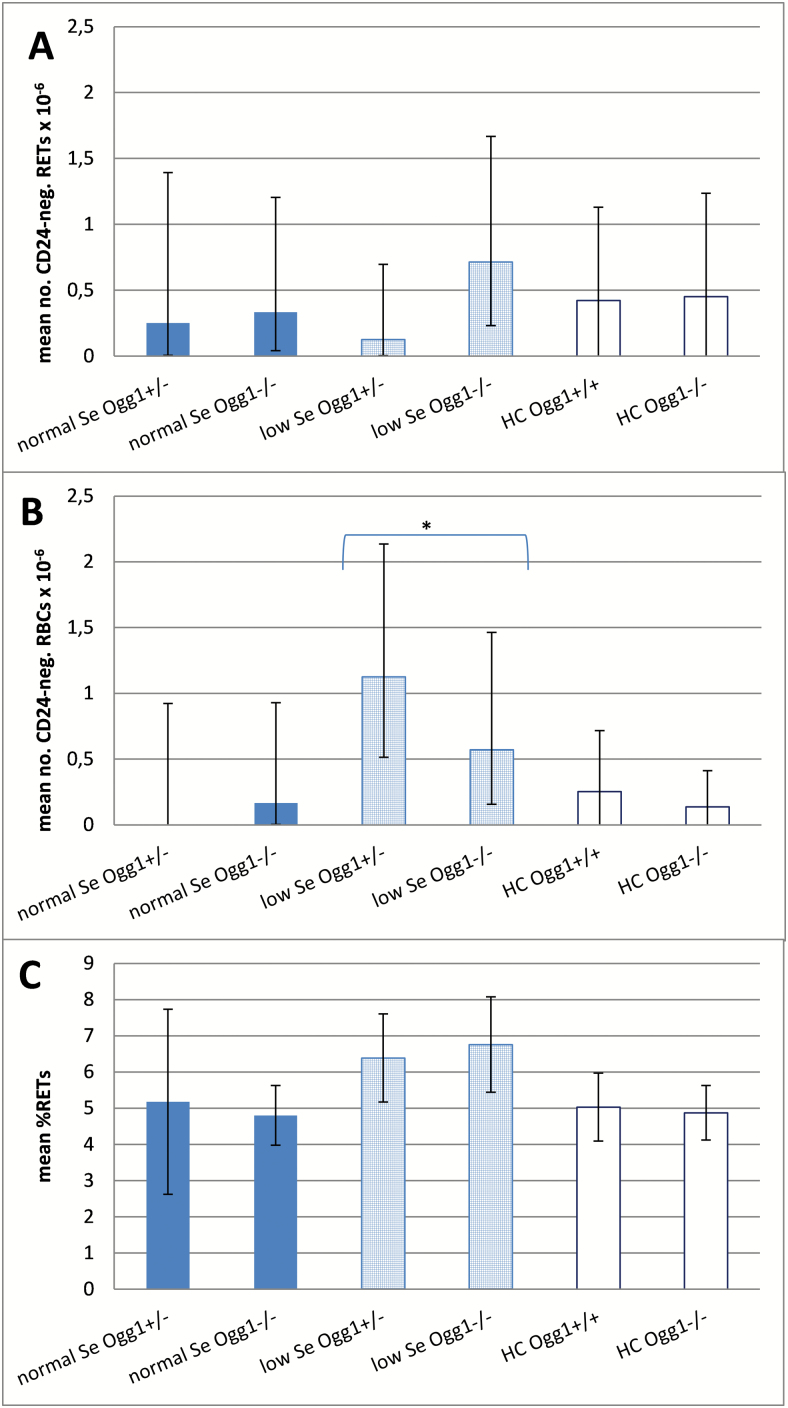
The frequency of RETCD24−, RBCCD24−, and RET were determined in peripheral blood samples. The generalised linear model was applied to identify impacts of diet, genotype and a combination of both (Figure 3). Only the factor diet had an impact: a significantly higher frequency of RBCCD24− was measured in mice on the low Se diet with P = 0.0081 (Figure 3B). The mean RETCD24− value of the Ogg1 −/− mice was also increased compared to mice on the normal Se diet (Figure 3A), but this increase did not attain statistical significance. Mice on low Se diet had a non-significant increase of % RET (Figure 3C). The genotype influenced neither the Pig-a gene mutation levels nor the mean % RET level, and no interaction between genotype and diet was observed. The measured levels of mutant phenotype cells did not differ significantly from the historical control data obtained in our laboratory (shaded bars in Figure 3). Conversely, mice treated with the positive control agent ENU showed markedly higher mean values for each parameter: 14.7% RET, 179.9 RETCD24− × 10–6 and 33.4 RBCCD24− × 10–6.
Figure. 3.
Mutant phenotype reticulocyte (RETCD24−) and mutant phenotype erythrocyte (RBCCD24−) frequencies per 106 cells are graphed in panels A and B, respectively. The percentage of reticulocytes (% RET) are shown in panel C. Values are given as group mean with lower and upper 95% confidence interval (Poisson distribution) for the different diets (normal Se: plain dark shaded bars; low Se: light shaded bars with squared pattern) and genotypes (Ogg1 +/−; Ogg1 −/−). White bars are historical controls (HC) in our laboratory (mean ± SD) of Ogg1 +/+ and Ogg1 −/− on a normal Se diet with n = 23 and 10, respectively. Asterisk indicates significant impact of the factor diet (P = 0.0081).
Discussion
Low Se blood levels are associated with different types of cancer (e.g. lung and prostate) (37–39) whereas there is a lack of knowledge concerning the potential of low Se levels to cause genotoxic effects. We explored consequences of a two generational low Se diet with respect to induced pre-mutagenic DNA lesions and mutations in somatic (blood, lung) and reproductive (testicle) tissue. In order to simulate a life-long Se deficiency, mice received a low Se diet during two generations, starting in the parental generation and sustained during fertilisation, gestation, postnatally and until early adulthood of the offspring (Figure 1).
This study was initially intended to establish the experimental design to obtain Se depletion in mice, endpoints and logistics, as a basis for a comprehensive study of potential interactions between a Se deficient status and chronic oxidative stress (chronic low dose-rate gamma irradiation). The observed genotoxic effects of the two-generational Se depletion are of probable biological relevance and are thus presented here despite some experimental shortcomings regarding the composition of diets (Table I).
Overall, the results indicate that Se deprivation is genotoxic since higher endogenous levels of DNA lesions were observed in both testicular cells and lung cells compared to mice on a normal Se diet. In blood cells no significant or biologically relevant Se-dependent induction of DNA lesions (SCGE) or mutagenic effects (Pig-a assay) were observed. While the mean mutation frequency in red blood cells (RBCCD24−) was significantly elevated in mice on the Se-deficient diet, this effect was not observed in the RET subpopulation. Furthermore, the mean frequency was within our historical control distribution. This suggests that the statistically significant effect may be of limited biological significance. Even so, as noted previously, it is now known that the assay as performed was not fully optimised for maximal recovery of RBCCD24− cells. Thus, while it would appear that genotoxicity resulting from Se deficiency is best detected in non-blood tissues, further experimentation using a more fully optimised Pig-a assay is necessary to form a firmer conclusion.
In lung cells, the genotoxic effect of Se deficiency—expressed as increased levels of ssb/als—was clearly evident (Figure 2B). This is interesting since there is an association between lung cancer and low levels of Se in serum (37). Se-dependent Fpg-ss in the lung were not found. The high spontaneous levels of Fpg-ss in lung cells of Ogg1-deficient mice were in the upper detectable range of the SCGE assay, which may have limited our ability to detect modest changes in Fpg-ss levels.
Testicular cells showed significantly increased levels of both ssb/als and Fpg-ss in mice on low Se diet, independently of the genotype (Figure 2C). This suggests that Se depletion starting in the parental generation before gestation and continued until early adulthood may lead to a pre-mutagenic effect in the F1.2 male germ cells. Disturbance of germ cell genomic integrity may potentially have important implications for the offspring.
One of the main functions of Se is to scavenge ROS. ROS attack DNA, cellular membranes and lipids, causing lipid peroxidation (LPO) that in turn gives rise to LPO-induced DNA lesions. The testis has an abundance of highly unsaturated fatty acids (40) and is hence prone to LPO. Also the lung has been shown to be susceptible to LPO (41). Products of LPO interact with DNA, forming several types of adducts such as etheno adducts (ethenoadenine and –cytosine) (42) and exocyclic mutagenic adducts (malondialdehyde-deoxyguanosine, M1dG) (43). The substrate specificity of Fpg (as used in the SCGE assay) reveals oxidised bases such as 8-oxoG or derivatives of etheno adducts) (44). M1dG, however, is not a substrate for Fpg. The repair of this adduct requires nucleotide excision repair which is not efficient in rodent testicular cells (22, 23) whereas oxidised bases are readily removed in rodent male germ cells via base excision repair (20). Others have reported increased oxidative stress measured as elevated levels of LPO in testis and sperm from mice given a low Se diet (7). Taking into account that the levels of major endogenous antioxidant enzymes (Gpx) are significantly reduced in the tissues of mice on low Se diet (Table II), the elevated DNA lesion levels observed in testis and lung cells may be explained in part by induction of LPO-derived DNA lesions. In particular, GPx4 (phospholipid hydroperoxide GPx) is primarily expressed in the testes as active peroxidase with an anti-oxidative function in spermatids (40), specifically reducing lipid peroxides (45). Later during spermiogenesis (i.e. the final stage of spermatogenesis during which the differentiation into spermatozoa takes place) GPx4 loses its peroxidase activity and is incorporated into the spermatozoa mid-piece as a structural protein that is important for the motility of spermatozoa (46, 47). It has been shown recently that GPx4 knockout male mice are infertile (48).
Since Ogg1 −/− mice lack at least one mechanism for removing 8-oxoG, they are likely to accumulate oxidative DNA lesions as shown for spontaneously induced endogenous DNA lesions (24). These mice could represent a more sensitive model for these lesions than the functional wild type (Ogg1 +/−). A similar accumulation of spontaneous oxidative DNA lesions was apparent in blood and lung cells with a significant increase of Fpg-ss in Ogg1 −/− compared to Ogg1 +/− (Table IV). In testis, the loss of Ogg1 seems to have only a moderate impact on the level of spontaneously induced oxidative DNA lesions in Ogg1 −/− mice (Figure 2); this may be related to the reduced intratesticular oxygen pressure (49). We expected that the low Se diet would cause oxidative stress expressed as higher levels of oxidative DNA lesions in the Ogg1 −/− model compared to Ogg1 +/−. In testicular cells, the Ogg1 −/− mice on low Se diet had elevated levels of DNA lesions compared to the same mice on a normal Se diet; however, this increase was similar to the diet-dependent increase in the Ogg1 +/− mice.
Taken together, the increased levels of DNA damage can be ascribed to the diet alone rather than the genotype of the mice. One explanation might be the existence of other antioxidants and ROS-scavenging molecules present in each tissue. Also alternative DNA enzymes or pathways might be triggered to ensure the genome integrity.
Previously it has been shown that a low Se diet has an adverse effect on the male reproductive system (8, 9). Also our results suggest that low Se diet affect the testicular function (Tables 2 and 3). The level of Se in seminal plasma is associated with the dietary Se supply (50). Since seminal SePP concentrations correlate positively with sperm density (19), a decreased spermatid head count can be expected in mice on low Se diet (Table III). Also, it seems the absence of Ogg1 plays a role in the production of spermatozoa: spermatid head counts for Ogg1 −/− mice were reduced compared to Ogg1 +/− mice on normal and low Se diet. The observed effects are reflected in male fertility: a low Se diet that started in utero and was retained until early adulthood caused sterility in male mice (unpublished data). Oxidative stress caused by a Se-deficient diet (51) is expected to contribute to reduced male fertility, and indeed DNA damage in spermatozoa is reported to correlate with the presence of 8-hydroxy-7,8-dihydro-2′-deoxyguanosine (8-OHdG) (52). The corresponding nucleobase 8-hydroxy-7,8-dihydroguanine (8-oxoG) is the most prevalent oxidative DNA lesion and recently shown to give rise to de novo germ line mutations in mice (53).
The connection between Se deficiency and various types of cancers has led to the suggestion that Se supplementation could be used as preventive treatment (54, 55). However, the beneficial effect in humans seem dependent on the initial Se plasma level [summarised in (1)] demanding more studies at suboptimal Se levels.
In conclusion, we have shown that Se deprivation during two generations leads to genotoxic effects in mice. These effects are tissue dependent: male germ cells seemed to be more susceptible to the induction of genotoxic effects than somatic cells, highlighting the importance of studying multiple tissue compartments and germ cells as well as somatic cells when studying Se deprivation. The genotoxic effects reported herein may have potential implications for male fertility and de novo mutations possibly transmitted to offspring and warrant further investigation.
Funding
This work was supported by the Research Council of Norway (RCN) through its Centres of Excellence funding scheme (223268/F50), the Bioernæringsprogrammet (190704), and EURATOM (RCN 217297, EU FP7 NoE DoReMi No. 249689). Costs associated with Prototype MutaFlow Kits were defrayed with a grant from the National Institute of Health/National Institute of Environmental Health Sciences (NIEHS; R44ES018017). The contents are solely the responsibility of the authors, and do not necessarily represent the official views of the NIEHS.
Conflict of interest statement
SDD is an employee of Litron Laboratories; Litron holds patents covering flow cytometric methods for scoring GPI anchor-deficient erythrocytes and sells kits based on this technology (In Vivo MicroFlow(R)).
Acknowledgements
The authors wish to thank Kari G. Løken, Victor Ong and Henrik Rasmussen for the animal care and Dag M. Eide for his help with statistical analysis. Ogg1 gene knockout mice were generously provided by Dr. A. Klungland, University of Oslo, Norway.
References
- 1. Fairweather-Tait S. J., Bao Y., Broadley M. R., Collings R., Ford D., Hesketh J. E., Hurst R. (2011). Selenium in human health and disease. Antioxid. Redox Signal., 14, 1337–1383. [DOI] [PubMed] [Google Scholar]
- 2. Gissel-Nielsen G. (1984). Selenium in soils and plants and its importance in livestock and human nutrition. Adv. Agron., 37, 397–459. [Google Scholar]
- 3. Låg J. (1998). Geomedical aspects of selenium: Norwegian investigations. J. Environ. Pathol. Toxicol. Oncol., 17, 229–232. [PubMed] [Google Scholar]
- 4. Meltzer H. M., Norheim G., Bibow K., Myhre K., Holm H. (1990). The form of selenium determines the response to supplementation in a selenium replete population. Eur. J. Clin. Nutr., 44, 435–446. [PubMed] [Google Scholar]
- 5. Rayman M. P. (2012). Selenium and human health. Lancet, 379, 1256–1268. [DOI] [PubMed] [Google Scholar]
- 6. Valdiglesias V., Pásaro E., Méndez J., Laffon B. (2010). In vitro evaluation of selenium genotoxic, cytotoxic, and protective effects: a review. Arch. Toxicol., 84, 337–351. [DOI] [PubMed] [Google Scholar]
- 7. Sánchez-Gutiérrez M., García-Montalvo E. A., Izquierdo-Vega J. A., Del Razo L. M. (2008). Effect of dietary selenium deficiency on the in vitro fertilizing ability of mice spermatozoa. Cell Biol. Toxicol., 24, 321–329. [DOI] [PubMed] [Google Scholar]
- 8. Behne D., Weiler H., Kyriakopoulos A. (1996). Effects of selenium deficiency on testicular morphology and function in rats. J. Reprod. Fertil., 106, 291–297. [DOI] [PubMed] [Google Scholar]
- 9. Watanabe T., Endo A. (1991). Effects of selenium deficiency on sperm morphology and spermatocyte chromosomes in mice. Mutat. Res., 262, 93–99. [DOI] [PubMed] [Google Scholar]
- 10. Bleau G., Lemarbre J., Faucher G., Roberts K. D., Chapdelaine A. (1984). Semen selenium and human fertility. Fertil. Steril., 42, 890–894. [PubMed] [Google Scholar]
- 11. Oldereid N. B., Thomassen Y., Purvis K. (1998). Selenium in human male reproductive organs. Hum. Reprod., 13, 2172–2176. [DOI] [PubMed] [Google Scholar]
- 12. Shalini S., Bansal M. P. (2008). Dietary selenium deficiency as well as excess supplementation induces multiple defects in mouse epididymal spermatozoa: understanding the role of selenium in male fertility. Int. J. Androl., 31, 438–449. [DOI] [PubMed] [Google Scholar]
- 13. Linschooten J. O., Verhofstad N., Gutzkow K., Olsen A. K., Yauk C., Oligschläger Y., Brunborg G., van Schooten F. J., Godschalk R. W. (2013). Paternal lifestyle as a potential source of germline mutations transmitted to offspring. FASEB J., 27, 2873–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brigelius-Flohé R., Maiorino M. (2013). Glutathione peroxidases. Biochim. Biophys. Acta, 1830, 3289–3303. [DOI] [PubMed] [Google Scholar]
- 15. Thomson C. D. (2004). Assessment of requirements for selenium and adequacy of selenium status: a review. Eur. J. Clin. Nutr., 58, 391–402. [DOI] [PubMed] [Google Scholar]
- 16. Xia Y., Hill K. E., Li P., Xu J., Zhou D., Motley A. K., Wang L., Byrne D. W., Burk R. F. (2010). Optimization of selenoprotein P and other plasma selenium biomarkers for the assessment of the selenium nutritional requirement: a placebo-controlled, double-blind study of selenomethionine supplementation in selenium-deficient Chinese subjects. Am. J. Clin. Nutr., 92, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burk R. F., Hill K. E. (2005). Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu. Rev. Nutr., 25, 215–235. [DOI] [PubMed] [Google Scholar]
- 18. NNR (2012). Nordic Nutrition Recommendations 2012, Part 1. Summary, principles and use. Nordic Nutrition Recommendations, Copenhagen.
- 19. Michaelis M., Gralla O., Behrends T., Scharpf M., Endermann T., Rijntjes E., Pietschmann N., Hollenbach B., Schomburg L. (2014). Selenoprotein P in seminal fluid is a novel biomarker of sperm quality. Biochem. Biophys. Res. Commun., 443, 905–910. [DOI] [PubMed] [Google Scholar]
- 20. Olsen A. K., Bjørtuft H., Wiger R., Holme J., Seeberg E., Bjørås M., Brunborg G. (2001). Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res., 29, 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olsen A. K., Duale N., Bjørås M., Larsen C. T., Wiger R., Holme J. A., Seeberg E. C., Brunborg G. (2003). Limited repair of 8-hydroxy-7,8-dihydroguanine residues in human testicular cells. Nucleic Acids Res., 31, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jansen J., Olsen A. K., Wiger R., Naegeli H., de Boer P., van Der Hoeven F., Holme J. A., Brunborg G., Mullenders L. (2001). Nucleotide excision repair in rat male germ cells: low level of repair in intact cells contrasts with high dual incision activity in vitro. Nucleic Acids Res., 29, 1791–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu G., Spivak G., Mitchell D. L., Mori T., McCarrey J. R., McMahan C. A., Walter R. B., Hanawalt P. C., Walter C. A. (2005). Nucleotide excision repair activity varies among murine spermatogenic cell types. Biol. Reprod., 73, 123–130. [DOI] [PubMed] [Google Scholar]
- 24. Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D. E. (1999). Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U. S. A., 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. European Convention (2006). European convention for the protection of vertebrate animals used for experimental and other scientific purposes, Appendix A. .
- 26. Govasmark E., Brandt-Kjelsen A., Szpunar J., Bierla K., Vegarud G., Salbu B. (2010). Bioaccessibility of Se from Se-enriched wheat and chicken meat. Pure Appl. Chem., 82, 461–471. [Google Scholar]
- 27. Wolf W. R., Goldschmidt R. J. (2007). Updated estimates of the selenomethionine content of NIST wheat reference materials by GC-IDMS. Anal. Bioanal. Chem., 387, 2449–2452. [DOI] [PubMed] [Google Scholar]
- 28. Beilstein M. A., Whanger P. D., Yang G. Q. (1991). Chemical forms of selenium in corn and rice grown in a high selenium area of China. Biomed. Environ. Sci., 4, 392–398. [PubMed] [Google Scholar]
- 29. Brunborg G., Holme J. A., Søderlund E. J., Omichinski J. G., Dybing E. (1988). An automated alkaline elution system: DNA damage induced by 1,2-dibromo-3-chloropropane in vivo and in vitro. Anal. Biochem., 174, 522–536. [DOI] [PubMed] [Google Scholar]
- 30. Brandt-Kjelsen A., Govasmark E., Haug A., Salbu B. (2014). Turnover of Se in adequately fed chickens using Se-75 as a tracer. J. Anim. Physiol. Anim. Nutr. (Berl)., 98, 547–558. [DOI] [PubMed] [Google Scholar]
- 31. Seung H., Wolfe G., Rocca M. (2003). Performing a testicular spermatid head count. Curr. Protoc. Toxicol., Chapter 16, Unit16.7. [DOI] [PubMed] [Google Scholar]
- 32. Gutzkow K. B., Langleite T. M., Meier S., Graupner A., Collins A. R., Brunborg G. (2013). High-throughput comet assay using 96 minigels. Mutagenesis, 28, 333–340. [DOI] [PubMed] [Google Scholar]
- 33. Boiteux S., O’Connor T. R., Lederer F., Gouyette A., Laval J. (1990). Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem., 265, 3916–3922. [PubMed] [Google Scholar]
- 34. Collins A. R., Oscoz A. A., Brunborg G., Gaivão I., Giovannelli L., Kruszewski M., Smith C. C., Stetina R. (2008). The comet assay: topical issues. Mutagenesis, 23, 143–151. [DOI] [PubMed] [Google Scholar]
- 35. Dertinger S. D., Bryce S. M., Phonethepswath S., Avlasevich S. L. (2011). When pigs fly: immunomagnetic separation facilitates rapid determination of Pig-a mutant frequency by flow cytometric analysis. Mutat. Res., 721, 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bright J., Aylott M., Bate S., Geys H., Jarvis P., Saul J., Vonk R. (2011). Recommendations on the statistical analysis of the Comet assay. Pharm. Stat., 10, 485–493. [DOI] [PubMed] [Google Scholar]
- 37. Knekt P., Marniemi J., Teppo L., Heliövaara M., Aromaa A. (1998). Is low selenium status a risk factor for lung cancer? Am. J. Epidemiol., 148, 975–982. [DOI] [PubMed] [Google Scholar]
- 38. Brinkman M., Reulen R. C., Kellen E., Buntinx F., Zeegers M. P. (2006). Are men with low selenium levels at increased risk of prostate cancer? Eur. J. Cancer, 42, 2463–2471. [DOI] [PubMed] [Google Scholar]
- 39. Jaworska K., Gupta S., Durda K., et al. (2013). A low selenium level is associated with lung and laryngeal cancers. PLoS One, 8, e59051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Guerriero G., Trocchia S., Abdel-Gawad F. K., Ciarcia G. (2014). Roles of reactive oxygen species in the spermatogenesis regulation. Front. Endocrinol. (Lausanne)., 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim K. S., Suh G. J., Kwon W. Y., Kwak Y. H., Lee K., Lee H. J., Jeong K. Y., Lee M. W. (2012). Antioxidant effects of selenium on lung injury in paraquat intoxicated rats. Clin. Toxicol. (Phila)., 50, 749–753. [DOI] [PubMed] [Google Scholar]
- 42. el Ghissassi F., Barbin A., Nair J., Bartsch H. (1995). Formation of 1,N6-ethenoadenine and 3,N4-ethenocytosine by lipid peroxidation products and nucleic acid bases. Chem. Res. Toxicol., 8, 278–283. [DOI] [PubMed] [Google Scholar]
- 43. Jeong Y. C., Swenberg J. A. (2005). Formation of M1G-dR from endogenous and exogenous ROS-inducing chemicals. Free Radic. Biol. Med., 39, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 44. Speina E., Ciesla J. M., Wojcik J., Bajek M., Kusmierek J. T., Tudek B. (2001). The pyrimidine ring-opened derivative of 1,N6-ethenoadenine is excised from DNA by the Escherichia coli Fpg and Nth proteins. J. Biol. Chem., 276, 21821–21827. [DOI] [PubMed] [Google Scholar]
- 45. Koulajian K., Ivovic A., Ye K., Desai T., Shah A., Fantus I. G., Ran Q., Giacca A. (2013). Overexpression of glutathione peroxidase 4 prevents β-cell dysfunction induced by prolonged elevation of lipids in vivo. Am. J. Physiol. Endocrinol. Metab., 305, E254–E262. [DOI] [PubMed] [Google Scholar]
- 46. Kehr S., Malinouski M., Finney L., et al. (2009). X-ray fluorescence microscopy reveals the role of selenium in spermatogenesis. J. Mol. Biol., 389, 808–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohé L. (1999). Dual function of the selenoprotein PHGPx during sperm maturation. Science, 285, 1393–1396. [DOI] [PubMed] [Google Scholar]
- 48. Imai H., Hakkaku N., Iwamoto R., et al. (2009). Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem., 284, 32522–32532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Free M. J., Schluntz G. A., Jaffe R. A. (1976). Respiratory gas tensions in tissues and fluids of the male rat reproductive tract. Biol. Reprod., 14, 481–488. [DOI] [PubMed] [Google Scholar]
- 50. Li P., Zhong Y., Jiang X., Wang C., Zuo Z., Sha A. (2012). Seminal plasma metals concentration with respect to semen quality. Biol. Trace Elem. Res., 148, 1–6. [DOI] [PubMed] [Google Scholar]
- 51. Zhang Q., Chen L., Guo K., et al. (2013). Effects of different selenium levels on gene expression of a subset of selenoproteins and antioxidative capacity in mice. Biol. Trace Elem. Res., 154, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aitken R. J., Smith T. B., Jobling M. S., Baker M. A., De Iuliis G. N. (2014). Oxidative stress and male reproductive health. Asian J. Androl., 16, 31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ohno M., Sakumi K., Fukumura R., et al. (2014). 8-oxoguanine causes spontaneous de novo germline mutations in mice. Sci. Rep., 4, 4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Nicastro H. L., Dunn B. K. (2013). Selenium and prostate cancer prevention: insights from the selenium and vitamin E cancer prevention trial (SELECT). Nutrients, 5, 1122–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Etminan M., FitzGerald J. M., Gleave M., Chambers K. (2005). Intake of selenium in the prevention of prostate cancer: a systematic review and meta-analysis. Cancer Causes Control, 16, 1125–1131. [DOI] [PubMed] [Google Scholar]