Abstract
Acetylation of transcriptional regulators is normally dynamically regulated by nutrient status but is often persistently elevated in nutrient-excessive obesity conditions. We investigated the functional consequences of such aberrantly elevated acetylation of the nuclear receptor FXR as a model. Proteomic studies identified K217 as the FXR acetylation site in diet-induced obese mice. In vivo studies utilizing acetylation-mimic and acetylation-defective K217 mutants and gene expression profiling revealed that FXR acetylation increased proinflammatory gene expression, macrophage infiltration, and liver cytokine and triglyceride levels, impaired insulin signaling, and increased glucose intolerance. Mechanistically, acetylation of FXR blocked its interaction with the SUMO ligase PIASy and inhibited SUMO2 modification at K277, resulting in activation of inflammatory genes. SUMOylation of agonist-activated FXR increased its interaction with NF-κB but blocked that with RXRα, so that SUMO2-modified FXR was selectively recruited to and trans-repressed inflammatory genes without affecting FXR/RXRα target genes. A dysregulated acetyl/SUMO switch of FXR in obesity may serve as a general mechanism for diminished anti-inflammatory response of other transcriptional regulators and provide potential therapeutic and diagnostic targets for obesity-related metabolic disorders.
Keywords: acetylation, NF-κB, PIASy, steatosis, SUMO2
Introduction
Recent global acetylome studies have revealed that over 2,000 functionally diverse proteins, ranging from histones to non-histone proteins such as transcriptional regulators and metabolic enzymes, are direct targets of acetylation (Choudhary et al, 2009; Zhao et al, 2010). Acetylation level of a particular protein is determined by the opposite reactions of acetylation and deacetylation, and regulated by cellular levels of the acetyl donor, acetyl-CoA, and NAD+, the essential cofactor of Sirtuin deacetylases (Canto et al, 2010; Cai & Tu, 2011; Guarente, 2011). Increasing evidence indicates that acetylation levels of many transcriptional regulators, including FXR, SREBP-1c, ChREBP, FoxO1, NF-κB, and PGC-1α, are normally dynamically regulated in response to nutrient/energy status for metabolic adaptation, but are persistently elevated in nutrient-excessive obesity (Rodgers et al, 2005; Kemper et al, 2009; Bricambert et al, 2010; Ponugoti et al, 2010; Walker et al, 2010; Choi et al, 2013). However, the functional role of such aberrantly elevated acetylation of gene regulators in obesity remains unknown.
Chronic inflammation, as well as metabolic dysfunction, is a key feature in obesity (Hotamisligil, 2006; Olefsky & Glass, 2010; Chawla et al, 2011; Lumeng & Saltiel, 2011). Nuclear receptors (NRs) are key regulators that control transcriptional programs of metabolism and inflammation (Evans et al, 2004; Glass & Saijo, 2010; Calkin & Tontonoz, 2012), and their transcriptional signaling pathways are profoundly modulated by post-translational modifications (PTMs) such as protein acetylation and small ubiquitin-like modifier (SUMO) modification (Kemper, 2011; Treuter & Venteclef, 2011). In recent studies, the critical role of SUMO modification in the anti-inflammatory action of the NRs, PPARγ, LXR, LRH-1, and Nurr1 has been described (Pascual et al, 2005; Ghisletti et al, 2007; Saijo et al, 2009; Venteclef et al, 2010). Pascual et al showed that agonist-activated SUMOylated PPARγ is targeted to the NcoR corepressor complex at inflammatory genes and prevents the clearance of the corepressor complex (Pascual et al, 2005). Venteclef et al recently showed that SUMOylation of LRH-1 or LXR is important for their recruitment to hepatic acute-phase response genes and that a stoichiometric subunit of the NcoR complex, GPS2, acts as a novel molecular mediator between SUMOylated NRs and the NcoR complex (Venteclef et al, 2010). Despite recent advances in understanding the role of SUMO modification of NRs in anti-inflammatory actions, it is not clear whether SUMOylation of NRs is decreased in obesity, where inflammatory responses are chronically activated.
The bile acid NR, FXR, plays a key role in maintaining cholesterol and bile acid levels and is also important in regulating fatty acid and glucose metabolism (Fiorucci et al, 2009; Lefebvre et al, 2009; Modica et al, 2010; Calkin & Tontonoz, 2012). In addition to these metabolic functions, FXR has recently been shown to have anti-inflammatory functions. Agonist-activated FXR inhibited NF-κB target inflammatory genes in hepatocytes (Wang et al, 2008) and also acted as an important modulator of innate immunity in the intestine (Vavassori et al, 2009). Consistent with these findings, our genome-scale liver ChIP-seq analysis has identified inflammatory genes as potential direct targets repressed by agonist-activated FXR in lean mice, and intriguingly, FXR binding at these inflammatory genes was substantially decreased in obese mice (Lee et al, 2012). We previously showed that FXR acetylation is normally dynamically regulated by nutrient status during feeding/fasting cycles but is persistently elevated in diet-induced obese mice (Kemper et al, 2009). However, it is not known whether elevated acetylation of FXR underlies the decreased occupancy and diminished anti-inflammatory action of FXR in obesity.
Using FXR as a model, we investigated the functional consequences of elevated acetylation of transcriptional regulators in diet-induced obese mice. We show that acetylation of FXR at K217 in obese mice inhibits SUMO2 modification at K277, which alters transcriptional pathways promoting hepatic inflammation and metabolic dysfunction. From biochemical studies using the SUMO-defective K277 mutant, along with ChIP, gel mobility shift assay, and structural modeling analysis, we provide compelling evidence that agonist-activated SUMO2-modified FXR is selectively recruited to NF-κB target inflammatory genes in response to inflammatory signals and trans-represses these genes in an RXRα-independent manner, without entering the transcriptional pathway regulating classical FXR/RXRα target genes.
Results
Acetylation of hepatic FXR at K217 is highly elevated in obesity
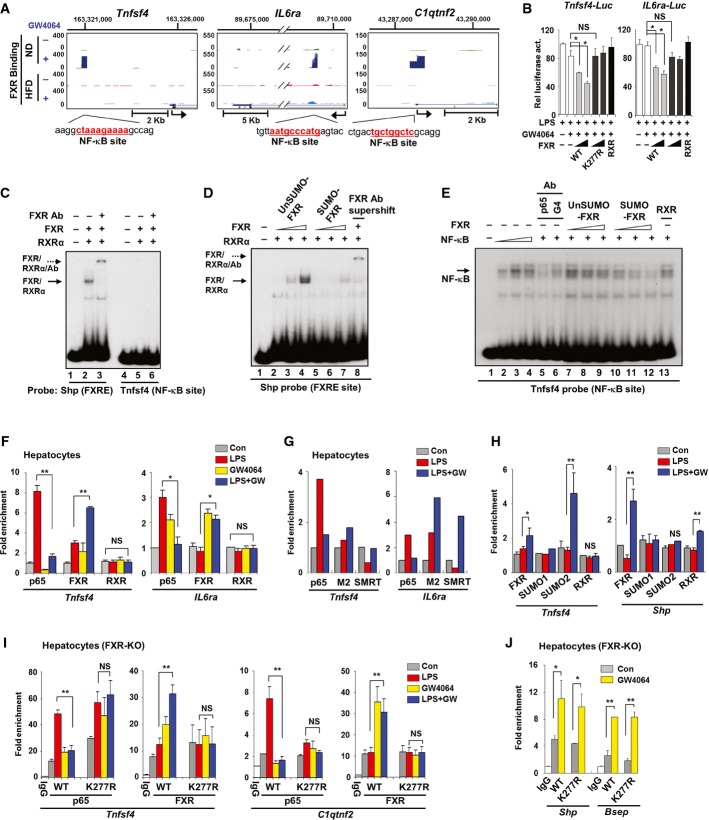
To explore the functional roles of elevated acetylation of FXR in obesity (Kemper et al, 2009), we first identified acetylation site(s) of hepatic FXR in diet-induced obese mice. Flag-FXR was expressed by adenoviral infection in lean mice fed a normal diet (ND) or obese mice fed a high-fat diet (HFD), and hepatic flag-FXR was isolated and subjected to tandem mass spectrometry (Fig1A). FXR acetylated at K217 was detected in diet-induced obese mice, but not in lean mice (Fig1B). To confirm these results, an antibody specific for FXR acetylated at K217 (Ac-K217) was developed (Supplementary Fig S1). Flag-tagged wild-type (WT) FXR or K217R-FXR was adenovirally expressed in livers of lean or diet-induced obese mice. Acetylation of flag-FXR was nearly undetectable in lean mice but was readily detected in obese mice by both the pan-acetyl-Lys and Ac-K217-specific antibodies (Fig1C). In addition, in mice expressing the K217R mutant, detection of acetylation of FXR by the pan-acetyl-Lys antibody was substantially reduced, suggesting that K217 is the major FXR acetylation site in obese mice (Fig1D), which is consistent with the MS/MS analysis (Fig1B). Acetylation of endogenous hepatic FXR at K217 was markedly increased by 12–16 weeks of feeding mice a HFD (Fig1E), providing further evidence for the correlation of acetylation of FXR with obesity. Further, in vivo IHC studies using the Ac-K217 antibody showed that Ac-K217 FXR levels are elevated in HFD mice regardless of nutrient states (Fig1F). These in vivo proteomic, biochemical, and IHC studies demonstrate that acetylation of FXR at K217 is elevated in dietary obese mice.
Figure 1. FXR is acetylated at K217 in diet-induced obese mouse liver.
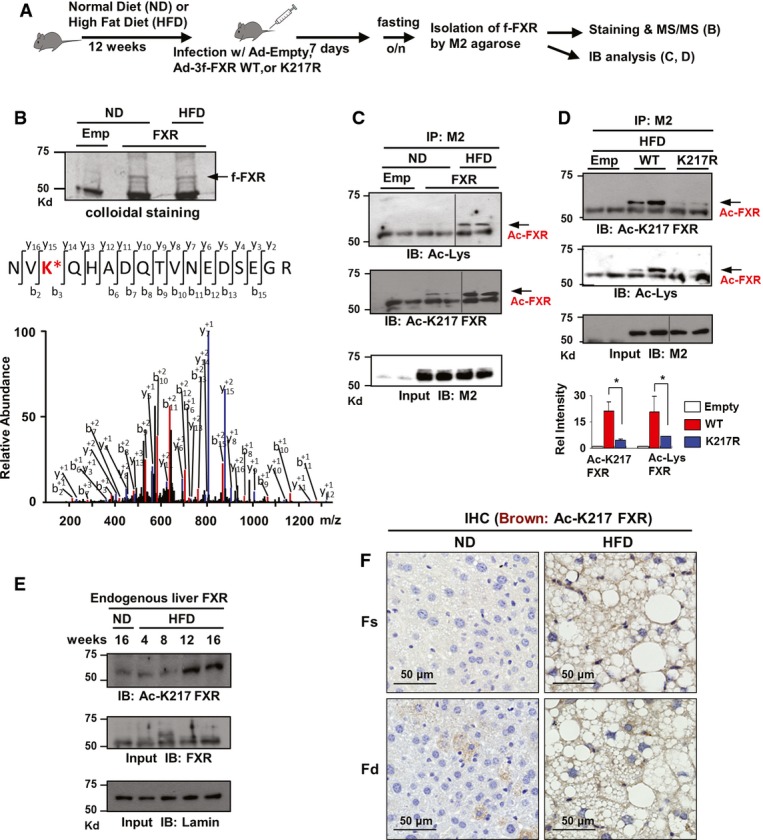
- Experimental outline.
- An FXR peptide modified by acetylation at Lys 217 (NV KQHADQTVNEDSEGR) was identified in an MS2 spectrum and manually confirmed by a subsequent MS3 scan. Emp: control empty Ad-Track vector.
- FXR acetylated at K217 of FXR in mice fed a ND or HFD was detected by IP/IB analysis using an Ac-K217-specific antibody. Duplicates are shown.
- Acetylated FXR or FXR acetylated at K217 in livers of dietary obese mice was detected by IP/IB. Relative levels of acetylated FXR or FXR acetylated at K217 are shown on the right. Values are presented as mean ± SEM (n = 3); two-tailed Student's t-test, *P < 0.05.
- Livers from three mice were pooled, liver extracts were prepared, and the effects of feeding a HFD on acetylation of FXR at K217 as a function of time were detected by IB.
- Histology of the liver stained for Ac-K217 by IHC to detect acetylated FXR in fasting/feeding of normal (ND) and HFD mice.
Source data are available online for this figure.
Elevated Ac-K217 of FXR in obesity promotes hepatic inflammation
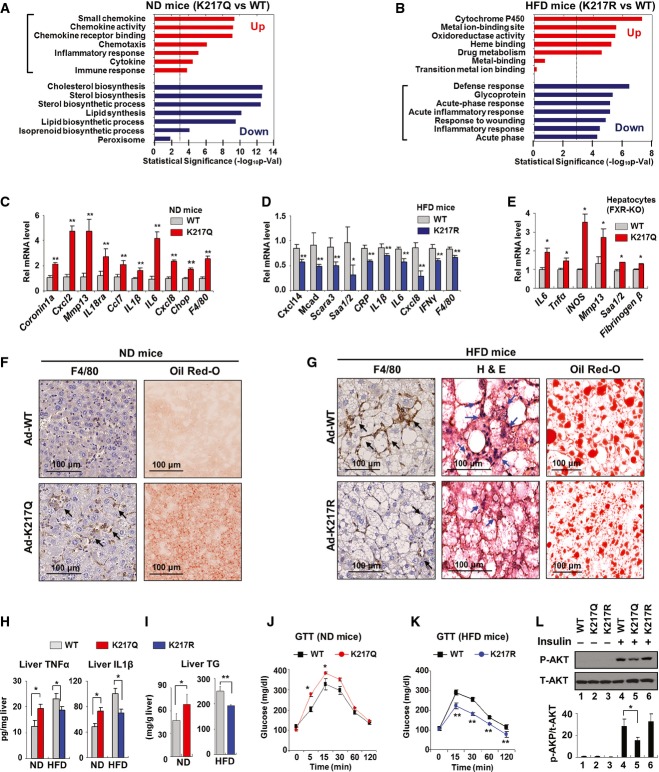
To explore the functional consequences of acetylation of FXR at K217 in vivo, an Ac-mimic K217Q mutant was expressed in lean mice that normally have low levels of acetylated FXR. Conversely, an Ac-defective K217R mutant was expressed in dietary obese mice that have abnormally elevated levels of acetylated FXR. Then, global gene expression in these mice was analyzed by microarray. A group of genes involved in proinflammatory responses were identified by gene ontology analysis as the most upregulated genes, and sterol/lipid metabolic genes were the most downregulated genes in lean mice expressing the K217Q mutant compared to mice expressing FXR-WT (Fig2A). The upregulated genes encoded chemokines, such as Mmp13, Cxcl2, Cxcl8, and Cxcl14, and cytokines, including IL1β and IL6 (Supplementary Table S1). Conversely, inflammatory genes were the most downregulated genes in obese mice expressing the K217R mutant compared to mice expressing FXR-WT, while genes involved in mitochondrial functions were the most upregulated genes (Fig2B, Supplementary Table S2). These results suggest that Ac-K217 of FXR might be involved in transcriptional activation of inflammatory responses in liver. The changes in mRNA levels of selected proinflammatory genes were confirmed by qRT–PCR (Fig2C and D). Hepatic expression of metabolic genes, including gluconeogenic genes, Pepck, and G-6-pase, and lipogenic genes, Srebp-1c, Fas, Dgat1, and Scd1, were also significantly altered by mice expressing the Ac-defective or Ac-mimic K217 mutant (Supplementary Fig S2). To eliminate the confounding effects of endogenous FXR in liver, the effects of expression of FXR-WT or the K217Q mutant on inflammatory genes were also determined in hepatocytes isolated from FXR-knockout (KO) mice with similar results (Fig2E).
Figure 2. Acetylation of FXR at K217 in obesity promotes hepatic inflammation.
- A, B FXR-WT or K217 mutants were expressed in livers of lean (ND) (A) or obese (HFD) (B) mice as indicated, and Illumina microarray was performed followed by gene ontology analysis.
- C, D The mRNA levels of selected inflammatory genes were confirmed by qRT–PCR for lean (ND) (C) and obese (HFD) (D) mice (n = 4).
- E Hepatocytes were isolated from FXR-KO mice and infected with Ad-FXR-WT or the Ad-K217Q mutant, and the mRNA levels of inflammatory genes were measured by qRT–PCR.
- F, G Histology of the liver of lean (ND) (F) and obese (HFD) (G) mice stained for F4/80 by IHC to detect macrophages and by H&E, or with Oil Red-O to detect liver fat. Infiltrated macrophages are indicated by arrows.
- H, I Cytokine and TG levels in liver extracts of lean (ND) (H) and obese (HFD) (I) mice (n = 4).
- J, K Glucose tolerance test (GTT): Serum glucose levels of lean (ND) (J) and obese (HFD) (K) mice were measured at the indicated times after injection i.p. of glucose (n = 4).
- L Ad-FXR-WT and K217 mutants were expressed in hepatocytes isolated from lean mice and treated with insulin for 30 min, and phospho- and total AKT levels were measured by IB (n = 3).
Data information: Values are presented as mean ± SEM; two-tailed Student's t-test, *P < 0.05, **P < 0.005.
Macrophage infiltration into liver is a sign of local hepatic inflammation in obesity (Hotamisligil, 2006; Olefsky & Glass, 2010; Chawla et al, 2011; Lumeng & Saltiel, 2011), which results in selective insulin resistance, such as sustained hepatic lipogenesis but impaired inhibition of gluconeogenesis. Notably, both mRNA levels (Fig2C and D) and protein levels detected by IHC (Fig2F and G) of the macrophage marker, F4/80, were increased in livers of lean mice expressing the K217Q Ac-mimic mutant and decreased in obese mice expressing the K217R Ac-defective mutant. Consistent with these results, levels of the cytokines, TNFα and IL1β, in liver extracts were significantly increased in lean mice expressing the K217Q mutant and reduced in obese mice expressing the K217R mutant (Fig2H). The amounts of liver triglycerides (TG) (Fig2I) and the size of lipid droplets (Fig2G) were reduced in obese mice expressing the K217R mutant. Excess lipid accumulation in non-adipose tissues is associated not only with local inflammation, but also with impaired insulin signaling and insulin resistance (Hotamisligil, 2006; Olefsky & Glass, 2010; Chawla et al, 2011; Lumeng & Saltiel, 2011). In dietary obese mice expressing the Ac-defective K217R mutant, glucose tolerance was improved (Fig2K). Conversely, in lean mice expressing the Ac-mimic K217Q mutant, glucose (Fig2J) and insulin (Supplementary Fig S3) tolerance was decreased. Further, expression of the Ac-mimic K217Q mutant significantly decreased p-AKT levels in response to insulin treatment in hepatocytes from lean mice (Fig2L), suggesting a role of FXR acetylation in impaired insulin signaling. We also examined expression of genes related to fat accumulation and inflammation as a function of time after expression of Ac-mimic K217Q in hepatocytes. In general, increased expression of inflammatory genes appeared to occur before increases in lipogenic genes (Supplementary Fig S4). Although the Q mutation may not completely mimic acetylated lysine, these findings from in vivo studies using the K217Q mutant, together with those using the Ac-defective K217R mutant, indicate that FXR acetylation at K217 in obese mice promotes hepatic inflammatory responses, which is associated with liver steatosis and impaired insulin signaling.
Agonist-activated FXR is SUMO2 modified at K277 in vivo
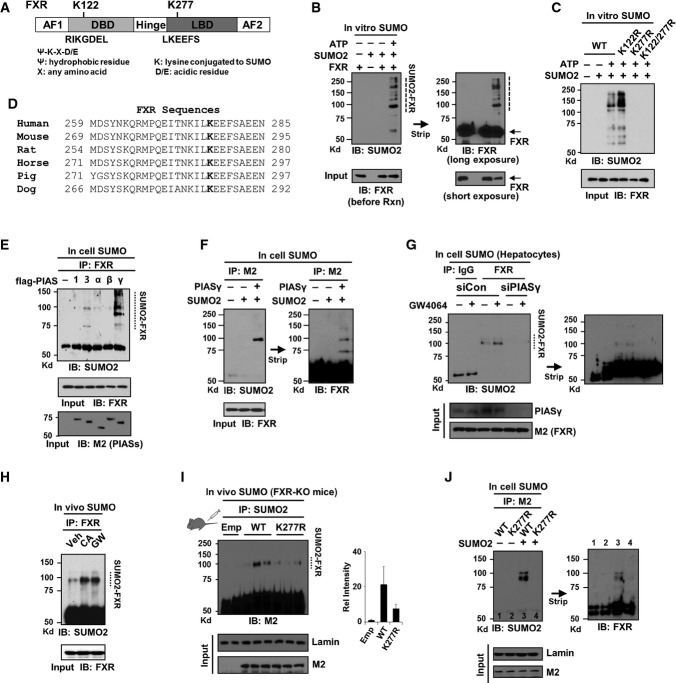
SUMOylation of the lipid-sensing metabolic NRs, PPARγ, LXR, and LRH-1 is critical for their anti-inflammatory function (Pascual et al, 2005; Ghisletti et al, 2007; Saijo et al, 2009; Venteclef et al, 2010). Since Ac-K217 of FXR promotes hepatic inflammatory responses (Fig2), we hypothesized that elevated acetylation of FXR in obesity inhibits SUMOylation, which would contribute to the diminished anti-inflammatory action of FXR. To test this idea, we first examined whether FXR is SUMOylated. Two consensus SUMO motifs are present at K122 and K277 (Fig3A). K277 was reported as a potential FXR SUMO site (Vavassori et al, 2009) but not directly tested by mutation of K277, nor was tested whether SUMOylation was by SUMO1 or SUMO2. We therefore performed in vitro SUMOylation assays using purified SUMO components and flag-FXR-WT and mutant proteins. SUMO2-modified FXR was detected (Fig3B), and mutation of K277, but not K122, abolished the SUMO2 modification (Fig3C), indicating that K277 is the major SUMO2 site in FXR. K277 and surrounding sequences are highly conserved in mammals (Fig3D). Notably, in in vitro studies, FXR was also SUMO1 modified, but mutation of either K122 or K277 only partially reduced SUMO1-FXR levels (Supplementary Fig S5A), suggesting that FXR is SUMO1 modified at multiple sites as recently reported (Balasubramaniyan et al, 2013). However, in in-cell SUMO studies, SUMOylation of agonist-activated FXR was selectively increased with SUMO2, but not with SUMO1, in response to inflammatory signals (Supplementary Fig S5B). In this study, we therefore examined the role of SUMO2 in the anti-inflammatory function of FXR.
Figure 3. SUMO2 modification of agonist-activated FXR at K277 represses NF-κB target inflammatory genes.
- A Two consensus SUMO sites in FXR.
- B, C In vitro SUMO assay: Flag-FXR-WT or FXR mutants were incubated with purified SUMO components as indicated, and SUMO2 modification of FXR was detected by IB.
- D Alignment of the FXR region containing K277 from various species.
- E, F In-cell SUMO assay: COS-1 cells were transfected with flag-FXR and with E3 SUMO ligase as indicated, and SUMOylated FXR was detected by IP/IB.
- G In-cell SUMO assay: Flag-FXR-WT and siRNA targeting PIASy were expressed in hepatocytes isolated from lean mice and treated with GW4064 for 30 min, and SUMOylated FXR was detected by IP/IB.
- H Mice were treated with GW4064 or fed chow containing 0.5% CA for 3 h, and SUMO2-modified endogenous hepatic FXR was detected by IP/IB. Veh: Mice fed a normal diet and treated with vehicle, DMSO.
- I Mice were infected with Ad-FXR-WT or the K277R mutant and treated with GW4064, and SUMO2-FXR levels were detected by IP/IB. Values are presented as mean ± SEM (n = 3 mice).
- J COS-1 cells were infected with adenoviral vectors and transfected with SUMO2 plasmid as indicated, and SUMO2-FXR levels were detected by IP/IB. The membrane was stripped and reprobed with FXR antibody.
Source data are available online for this figure.
SUMO2-FXR levels were substantially increased by overexpression of one of the PIAS family E3 SUMO ligases, PIASy (Palvimo, 2007) in cells (Fig3E and F), and conversely, downregulation of PIASy by siRNA nearly abolished SUMO2 modification of FXR (Fig3G), suggesting that PIASy may act as an E3 ligase for SUMO2 modification of FXR. Levels of SUMO2-modified endogenous FXR in liver were markedly increased in mice after activation of FXR by its natural agonist cholic acid (CA) or the synthetic agonist GW4064 (Fig3H). In primary mouse hepatocytes, SUMO2-FXR levels were increased by treatment with GW4064, CDCA, or UDCA, but not with LCA or an FXR antagonist guggulsterone (Supplementary Fig S6). To establish that hepatic FXR is SUMO2 modified at K277 in vivo, FXR-WT or the K277R mutant was adenovirally expressed in FXR-KO mice, and SUMO2-FXR levels in liver extracts were measured. Importantly, SUMO2-FXR levels were substantially decreased in mice expressing the K277R mutant (Fig3I, Supplementary Fig S7), and similar results were observed in in-cell SUMO studies (Fig3J). These biochemical studies in vitro, in cells, and in vivo, utilizing the SUMO-defective mutant, identify K277 as the major SUMO2 site in FXR.
Inverse correlation between acetyl- and SUMO2-FXR levels during the development of obesity
To test whether acetylation of FXR in obesity might inhibit SUMOylation, we first examined the effects of feeding a HFD as a function of time on acetylation and SUMOylation of FXR, as well as on inflammatory and lipogenic gene expression. Feeding a HFD for 4–16 weeks (Fig4A) resulted in gradual increases in mRNA levels of proinflammatory genes, IL1, IL6, iNOS, and Tnfα, a macrophage marker, F4/80, an ER stress gene, Chop, lipogenic genes, Srebp-1c, Fas, and Scd1, and an acetyltransferase, p300, while mRNA levels of Sirt1 deacetylase decreased (Fig4B). Feeding a HFD resulted in modest but significant decreases in mRNA levels of SUMO2 and PIASy, but substantial increases in SUMO1 mRNA levels (Fig4B). Acetylation of FXR was elevated with time in mice after 8–16 weeks of feeding a HFD (Fig4C), and acetylation of other NRs, LXR, ERα, and PXR, and a key lipogenic activator, SREBP-1c, was also increased (Supplementary Fig S8). SUMOylation of FXR decreased with time and was nearly undetectable after 12 weeks corresponding to the time that acetylation of FXR was substantially increased (Fig4C). Notably, levels of acetylated hepatic proteins in general were increased, whereas levels of SUMO2-modified proteins were decreased with time of feeding a HFD (Fig4D). In line with these findings, a recent study showed that SUMOylation of SREBP-1c by PIASy decreased its lipogenic activity and that knockdown of PIASy promoted hepatic lipogenesis in lean mice, while overexpression of PIASy attenuated steatosis in genetic obese mice (Lee et al, 2014). These results indicate that acetylation levels of the tested transcriptional regulators are increased in obesity and that there is a strong inverse correlation between the levels of acetyl- and SUMO2-FXR during the development of obesity.
Figure 4. Inverse correlation between acetyl- and SUMO2-FXR levels during the development of diet-induced obesity.
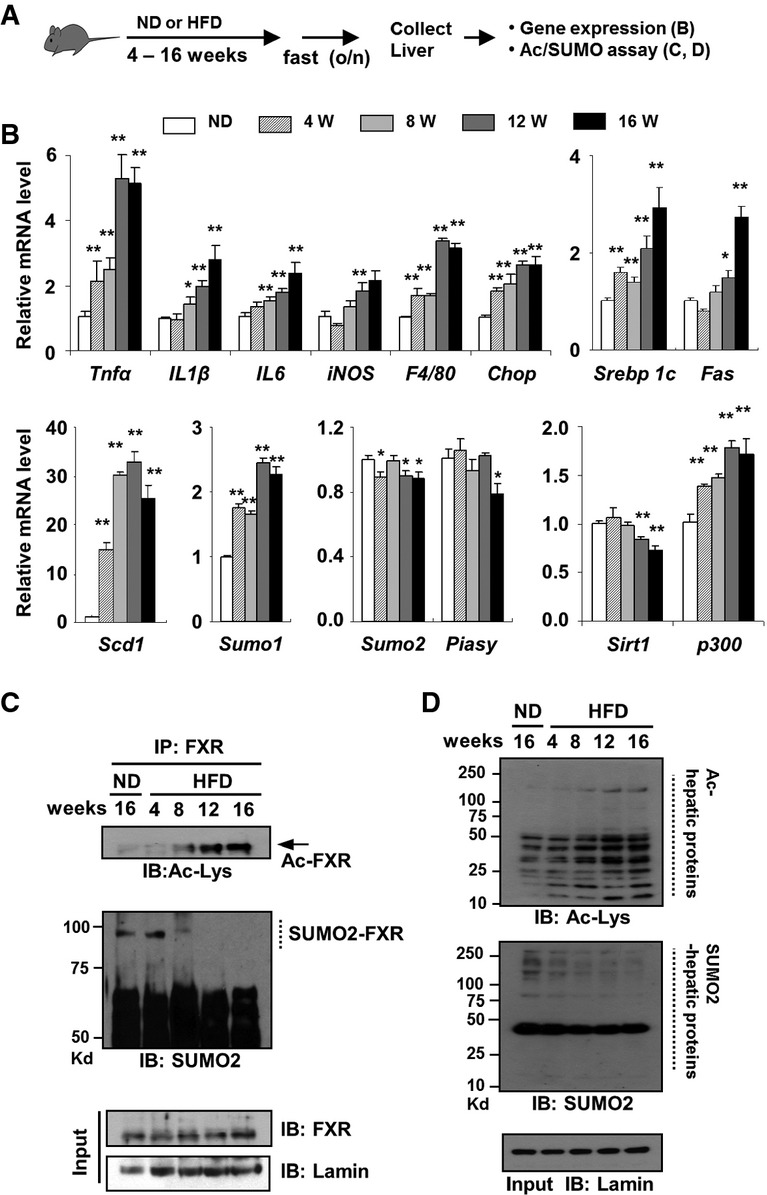
- A Experimental outline.
- B–D Effects of feeding a HFD as a function of time on mRNA levels of the indicated genes (B), levels of SUMO2-modified FXR (C), and levels of acetylated and SUMO2-modified proteins (D) in mouse liver (pooled from 3 mice). Values in (B) are presented as mean ± SEM; two-tailed Student's t-test, *P < 0.05, **P < 0.005, n = 3.
Acetylation of FXR at K217 inhibits its SUMO2 modification, partly by blocking the interaction with PIASy
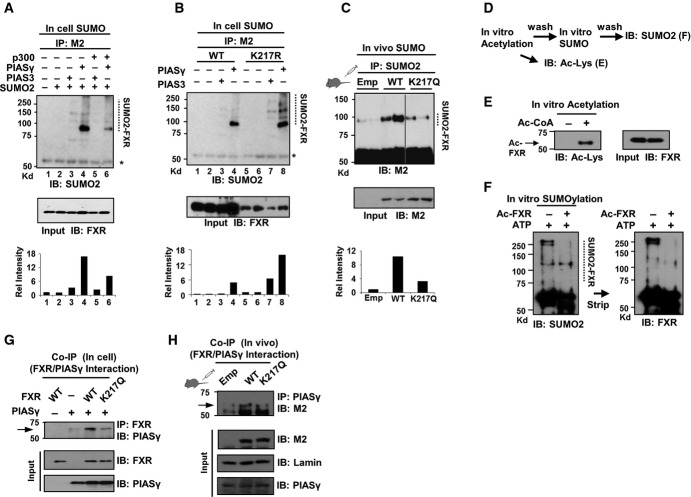
We next examined whether acetylation of FXR inhibits its SUMO2 modification. Overexpression of p300, which increases FXR acetylation (Kemper et al, 2009), decreased SUMO2-FXR levels in in-cell SUMO assays (Fig5A). SUMO2-FXR levels were increased in cells expressing the Ac-defective K217R (Fig5B), and conversely, SUMO2-FXR levels were decreased in mice expressing the Ac-mimic K217Q (Fig5C). Consistent with these results, in vitro SUMOylation assays using acetylated FXR demonstrated that acetylation of FXR decreased its SUMOylation (Fig5D–F). Intriguingly, the interaction of FXR with the SUMO E3 ligase, PIASy, was reduced by the Ac-mimic K217Q mutation in cells and in mice (Fig5G and H), suggesting that acetylation of FXR inhibits SUMOylation, at least in part, by inhibiting its interaction with PIASy. These results, taken together, suggest that elevated acetylation of FXR inhibits its SUMO2 modification in obesity by inhibiting the interaction with PIASy. We also observed that SUMO2 modification of FXR at K277 appears to inhibit its acetylation at K217 in in vitro assays (data not shown), but this observation, however, likely has little physiological relevance due to only a small minor fraction of SUMO target proteins that is SUMOylated at steady state and the dynamic nature of SUMO modifications (Hay, 2005).
Figure 5. Acetylation of FXR at K217 inhibits its SUMO2 modification, partly by blocking the interaction with PIASy.
- A, B In-cell SUMO assay: COS-1 cells were transfected with the plasmids as indicated, and levels of SUMO2-FXR were detected by IP/IB. At the bottom, levels of SUMO2-FXR relative to total FXR are shown.
- C Mice were infected with Ad-FXR-WT or the K217Q mutant and treated with GW4064, and SUMO2-FXR levels were detected by IP/IB (n = 3 mice).
- D Experimental outline for in vitro acetylation and SUMOylation.
- E, F In vitro assay: Purified flag-FXR-WT was incubated with purified p300 and acetyl-CoA as indicated, and acetylated FXR was detected by IB (E). Effects of acetylation of FXR on its SUMOylation (F).
- G, H Effects of the Ac-mimic K217Q mutation on the interaction of FXR with PIASy in COS-1 cells (G) or in liver extracts pooled from three mice (H) as detected by CoIP.
Source data are available online for this figure.
SUMO2-FXR selectively represses NF-κB target inflammatory genes
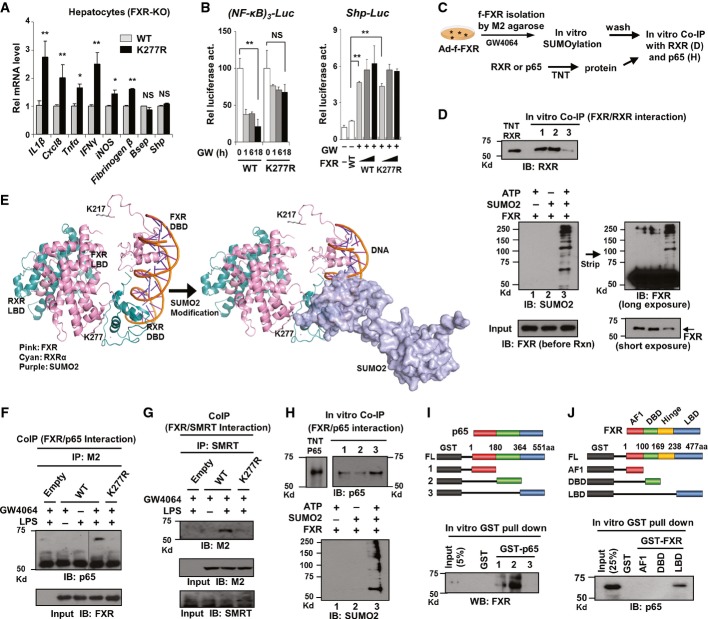
We next examined the effect of SUMO2 modification of FXR on expression of inflammatory genes. Activation of FXR signaling by GW4064 treatment decreased expression of inflammatory genes, IL6 and iNOS, in mouse liver in vivo, in hepatocytes, and in HepG2 cells (Supplementary Fig S9), as previously reported (Wang et al, 2008). In hepatocytes isolated from FXR-KO mice, the mRNA levels of the inflammatory genes, including IL1β and Tnfα, were significantly higher in cells expressing the K277R mutant compared to cells expressing FXR-WT (Fig6A). In contrast, expression of well-known FXR/RXRα target genes, Shp and Bsep, was unchanged (Fig6A), suggesting that SUMOylation of agonist-activated FXR may selectively inhibit expression of NF-κB target inflammatory genes. Supporting the idea that SUMO-FXR has a role in anti-inflammation, downregulation of the SUMO ligase PIASy by siRNA resulted in increased inflammatory gene expression but no effects on expression of the FXR target gene, Shp (Supplementary Fig S10A). Consistent with these results, in luciferase reporter assays, expression of FXR-WT, but not the SUMO-defective K277R mutant, decreased NF-κB transactivation (Fig6B, left). In contrast, expression of FXR-WT or the K277R mutant resulted in similar increases in Shp promoter-driven luciferase activity (Fig6B, right). These results suggest that SUMO2-FXR selectively represses inflammatory genes without inhibiting genes targeted by the FXR/RXR heterodimer.
Figure 6. Sumoylation of FXR blocks its interaction with RXRα and increases its interaction with NF-κB.
- A Effects of expression of either FXR-WT or FXR-K277R on the mRNA levels of the indicated genes in hepatocytes isolated from FXR-KO mice. Values are presented as mean ± SEM (n = 6); two-tailed Student's t-test, **P < 0.005, NS, not significant.
- B Reporter assays: HepG2 cells were transfected with plasmids as indicated and treated with GW4064, and luciferase activities were measured. Values are presented as mean ± SEM (n = 3); two-tailed Student's t-test, *P < 0.05, **P < 0.005, NS, not significant.
- C Experimental outline for in vitro CoIP studies.
- D Effects of SUMOylation of FXR on its interaction with RXRα.
- E FXR modeling analysis: The predicted structure of FXR and RXR bound to DNA is shown. The positions of K217 and K277 in FXR are indicated. K277 in the LBD of FXR is located in the interface with the RXR DBD (left). SUMO2 modification (dimers of SUMO2 are shown) of the LBD of FXR would block the interaction of FXR with RXRα (right). Modeling was done using the RXR/PPARγ/DNA (3DZU) model with PPARγ replaced by FXR (1OSV).
- F, G Effects of the mutation, K277R, on the interaction of FXR with the p65 subunit of NF-κB (F) or SMRT (G) detected by CoIP.
- H Effects of SUMOylation of FXR on its interaction with p65. FXR was SUMOylated in vitro, and the interaction of SUMO-FXR with p65 was detected by CoIP.
- I, J A schematic of the domains of p65 or FXR that were fused to GST is shown on the top. The interaction between FXR and p65 or their domains was detected by GST pull-down followed by IB.
Source data are available online for this figure.
SUMOylation of FXR increases its interaction with NF-κB but blocks the interaction with RXRα
An important mechanistic question is how SUMO2-FXR is selectively recruited to NF-κB inflammatory genes and inhibits these genes without affecting the classical FXR/RXRα target genes. Recent studies have shown that the SUMOylated NRs, PPARγ and LXR, repress inflammatory genes in an RXRα-independent manner (Pascual et al, 2005; Venteclef et al, 2010), but the direct effects of SUMOylation on interaction of the NRs with RXRα were not examined. To address this question for FXR, purified flag-FXR was SUMO2 modified in vitro, and then the SUMO-FXR was incubated with purified RXRα, and the interaction between FXR and RXRα was analyzed by IB experiments (Fig6C). Strikingly, the interaction of FXR with RXRα was substantially decreased (Fig6D, top, lane 3) by SUMO2 modification of FXR (Fig6D, bottom, lane 3).
To understand the structural basis for the inhibition of the interaction of FXR and RXRα by SUMOylation, models of the FXR/RXR interaction were examined. Since the structure of full-length FXR has not been reported, FXR was substituted for PPARγ (Chandra et al, 2008). The SUMO site, K277, is located in the helix 2 region of the LBD which is near the DBD of RXRα, whereas the acetylation site, K217, is in the hinge region (Fig6E, left). In the model, SUMO2 modification at K277 in FXR would sterically hinder the interaction between FXR and the DBD of RXRα (Fig6E, right). Similar conclusions were reached in structural modeling studies utilizing an LXR/RXRα/DNA complex (Lou et al, 2014) (Supplementary Fig S11). Since the hinge region of FXR is longer than that of these NRs, prediction of the position of K217 in FXR is uncertain and speculative. However, Ac-K217 may result in conformational changes that affect binding of SUMO machinery, including PIASy as shown in our study (Fig5G and H), which would result in inhibition of SUMO2 modification.
We further tested whether SUMOylation of FXR also alters its interaction with NF-κB or SMRT, a known corepressor recruited to NF-κB target inflammatory genes (Pascual et al, 2005; Ghisletti et al, 2009). In CoIP studies, treatment with LPS markedly increased the interaction of agonist-activated FXR with an NF-κB subunit, p65, or SMRT, but this interaction was largely abolished by the SUMO-defective K277R mutation (Fig6F and G). Importantly, the increased interaction of SUMO2-FXR with NF-κB was also detected in vitro (Fig6H, Supplementary Fig S10B), in sharp contrast to the decreased interaction with RXRα (Fig6D). Further, GST pull-down assays suggested that FXR and p65 directly interact through the LBD of FXR and the central domain of p65 (Fig6I and J) and that NF-κB also directly interacts with SUMO2 but not with SUMO1 protein in vitro (Supplementary Fig S12). These studies demonstrate that SUMOylation of FXR increases its interaction with p65 but blocks its interaction with RXRα. These findings provide a molecular basis for selective trans-repression of NF-κB target genes by SUMO2-FXR, without entering the transcriptional pathway regulating FXR/RXRα target genes such as Shp.
SUMO2 modification of FXR is important for its recruitment and decreased NF-κB occupancy at inflammatory genes
We recently analyzed genome-scale hepatic FXR binding sites in lean and dietary obese mice by ChIP-seq (Lee et al, 2012). FXR binding peaks were detected at inflammatory genes, including Tnfsf4, C1qtnf2, and IL6ra, in lean mice treated with GW4064, but intriguingly, these FXR peaks were not detected in obese mice (Fig7A, Supplementary Fig S13). FXR DNA binding IR1 motifs were not detected, but NF-κB binding motifs were present within the FXR binding peaks (Fig7A). A DNA fragment that contained the major FXR peak in Tnfsf4 or IL6ra was inserted into a luciferase reporter vector. Expression of FXR-WT inhibited these reporters, but the expression of the K277R mutant did not, which suggests that SUMO2 modification at K277 is important for FXR repression of these genes (Fig7B). Inhibition of these inflammatory genes was still observed when a FXR mutant lacking DNA binding was used, suggesting that FXR represses these genes without directly binding to DNA (Supplementary Fig S14). These results are consistent with our findings that SUMOylation of agonist-activated FXR is important for increased interaction with NF-κB and inhibition of NF-κB target inflammatory genes (Fig6H) and that SUMO2-FXR levels are reduced in obese mice (Fig4C).
Figure 7. SUMO2 modification of FXR at K277 is important for increased FXR recruitment and decreased NF-κB occupancy at inflammatory genes.
- A FXR binding near inflammatory genes in mice fed a ND or HFD treated i.p. with vehicle (−) or GW4064 (+) for 1 h. The sequence of an NF-κB binding site present within the FXR binding peak is shown.
- B Luciferase reporter assays using direct FXR target inflammatory gene promoters (n = 3–6).
- C–E Gel mobility shift assays. Purified flag-FXR was incubated with flag-RXRα and the radiolabeled oligonucleotide probe containing the FXRE or NF-κB binding site from the SHP or TNFsf4 promoter, respectively (C). Increasing amounts of flag-FXR or SUMOylated flag-FXR were incubated with purified RXRα (D) or whole-cell extracts containing NF-κB (E), and the complexes were detected by gel electrophoresis.
- F ChIP assay of hepatocytes from WT mice treated as indicated. Occupancy of endogenous p65, FXR, and RXR was detected by qPCR.
- G ChIP assay of hepatocytes from WT mice infected with Ad-flag-FXR-WT and treated as indicated.
- H ChIP assay of hepatocytes from WT mice treated as indicated.
- I, J ChIP assays of hepatocytes from FXR-KO mice infected with Ad-FXR-WT or Ad-K277R and treated as indicated.
Data information: In (F, H?J), three independent ChIP assays were performed per experiment. Values are presented as mean ± SEM; two-tailed Student's t-test, *P < 0.05, **P < 0.005, NS, not significant.
To determine the effect of SUMO2 modification of FXR on binding of NF-κB and FXR/RXRα to DNA, we performed gel mobility shift assays (Fig7C). Two oligonucleotides with sequences either from the Shp gene containing the FXR site or from the Tnfsf4 gene containing the NF-κB site were used as probes (Fig7A). Purified FXR, incubated with RXRα, effectively bound to the Shp, but not the Tnfsf4, probe (Fig7C). Incubation of increasing amounts of FXR resulted in a progressive increase in binding of FXR/RXRα to the Shp probe (Fig7D lanes 2–4), but binding was substantially decreased by SUMOylation of FXR (Fig7D, lanes 5–7). With the Tnfsf4 probe, addition of increasing amounts of nuclear extracts overexpressing the NF-κB proteins (p65 and p50) with the Tnfsf4 probe, resulted in the formation of an NF-κB/DNA complex, and this complex was inhibited by p65 antibody, but not by Gal4DBD antibody (Fig7E, lane 4–6). Interestingly, addition of SUMO2-FXR markedly decreased NF-κB binding (Fig7E, lanes 10–12), whereas unSUMO-FXR did not affect the binding (Fig7E, lanes 7–9). The quality of partially purified FXR and RXRα and also SUMO2-FXR and NF-κB used in gel mobility shift assay was confirmed (Supplementary Fig S15A–D). About 40% of FXR was SUMOylated under these conditions (Supplementary Fig S15E and F). These results suggest that purified SUMO2-FXR may inhibit NF-κB binding to DNA and that SUMO2-FXR may not be recruited to FXR/RXRα binding sites as shown above because of its impaired interaction with RXRα (Fig6D).
We next examined the effects of GW4064 and LPS treatment on the occupancy of FXR, RXRα, and p65 at Tnfsf4 and IL6ra genes in a normal chromatin context by ChIP assay. In mouse hepatocytes, treatment with LPS increased p65 occupancy, but cotreatment with GW4064 reversed the increase and also increased occupancy of endogenous FXR at these two genes while occupancy of RXR was not affected by the treatments (Fig7F). Similar results were observed in hepatocytes expressing flag-FXR-WT (Fig7G). Treatment with LPS did not affect occupancy of SUMO2, but cotreatment with GW4046 resulted in increased SUMO2 levels at Tnfsf4, IL6ra, and C1qtnf2, but not at Shp and Bsep, while SUMO1 occupancy was not changed at any of the genes (Fig7H, Supplementary Fig S16). These results suggest that SUMO2-FXR is specifically recruited to the inflammatory genes, but not to FXR/RXRα target genes. LPS treatment also decreased occupancy of the corepressor SMRT, but cotreatment with GW4064 restores SMRT occupancy (Fig7G), which correlated with repression of these inflammatory genes (Supplementary Fig S17).
To directly establish the role of the SUMO2 modification of FXR on the occupancy of FXR and p65 at inflammatory genes, we performed ChIP assays utilizing the SUMO2-defective K277R mutant. In hepatocytes isolated from FXR-KO mice, treatment with LPS increased p65 occupancy at the Tnfsf4 and C1qtnf2 genes and this increased occupancy was significantly reduced by treatment with GW4064 in cells expressing FXR-WT, but not in cells expressing the K277R mutant (Fig7I). Further, occupancy of FXR at these genes was increased after treatment with GW4064, but not in cells expressing the K277R mutant (Fig7I). In sharp contrast with decreased occupancy of the K277R mutant at inflammatory genes compared to FXR-WT (Fig7I), occupancies of both FXR-WT and the K277R mutant were similar at the known FXR/RXRα target genes, Shp and Bsep (Fig7J). These protein/DNA interaction studies by gel shift and ChIP assays, together with molecular, biochemical, and modeling studies above (Figs5 and 6), provide compelling evidence that SUMO2 modification of FXR is important for its increased interaction with NF-κB, while interaction with RXRα is blocked, which results in the recruitment of SUMO2-FXR selectively to NF-κB target genes and trans-repression of these genes without affecting expression of FXR/RXR target genes.
Discussion
In this study, we show that elevated acetylation of FXR at K217 in diet-induced obese mice inhibited SUMO2 modification at K277, which resulted in altered transcriptional pathways promoting hepatic inflammation and metabolic dysfunction (Fig8). Expression of an Ac-mimic mutant in lean mice resulted in elevated liver cytokine and fat levels, while conversely, expression of an Ac-defective mutant in dietary obese mice was associated with beneficial outcomes, including ameliorated hepatic inflammation, decreased liver cytokine and fat levels, and increased glucose tolerance. Elevated acetylation of FXR in obesity thus appears to play a causative role in hepatic inflammation and abnormal metabolism. Acetylation levels of FXR increased after 8–12 weeks of feeding a HFD, while the SUMO2 levels of FXR were substantially reduced. Intriguingly, acetylation levels of all other tested transcriptional regulators, LXR, PXR, ER, PXR, and SREBP-1c, and hepatic proteins in general also increased. Dysregulation of an acetyl/SUMO switch, such as identified for FXR, therefore may serve as a general mechanism for diminished SUMO-dependent anti-inflammatory functions of other transcriptional regulators in obesity.
Figure 8. Model depicting the role of a dysregulated acetyl/SUMO switch of FXR in obesity.
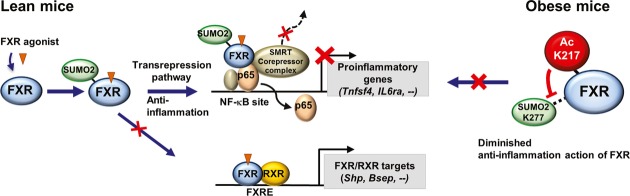
Under normal conditions, a fraction of agonist-activated FXR is modified by SUMO2 at K277 by the E3 SUMO ligase PIASy and regulates an anti-inflammatory transcriptional pathway in response to inflammatory signaling. SUMO2-FXR increases its interaction with NF-κB and SMRT and blocks its interaction with RXRα, which results in selective trans-repression of inflammatory genes without entering transcriptional pathway regulating FXR/RXRα target genes (left). In contrast, in diet-induced obese mice, FXR is highly and constitutively acetylated at K217, which inhibits SUMO2 modification at K277, at least in part, by blocking its interaction with PIASy and results in diminished SUMO2-dependent anti-inflammatory action (right).
An important question is the underlying cause of the hyperacetylation of FXR and other transcriptional regulators in obesity. Recent evidence suggests that a functional imbalance between HATs and HDACs may play a role. We have shown that acetylation of FXR is tightly regulated by p300 and SIRT1 under physiological conditions, which is important for dynamic transcriptional regulation, but that in obesity, SIRT1 levels and activity are low, resulting in persistently elevated acetylation of FXR (Kemper et al, 2009). Further, FXR acetylation was elevated in liver-specific SIRT1-KO mice (Purushotham et al, 2012), and conversely, acetylation of FXR as well as that of SREBP-1c, PGC-1α, and NF-κB was decreased by overexpression of SIRT1 (Kemper et al, 2009; Ponugoti et al, 2010; Choi et al, 2013). Intriguingly, expression of the corepressor complex subunits, GPS2 and SMRT, was reduced in adipocytes from obese individuals, and their expression was restored by weight loss after gastric bypass surgery (Toubal et al, 2013). In other studies, hepatic overexpression of p300 and consequently hyperacetylation of ChREBP resulted in liver steatosis, insulin resistance, and inflammation (Bricambert et al, 2010). In the present study, mRNA levels of p300 were progressively elevated, whereas those of SIRT1 were decreased during the development of diet-induced obesity. Importantly, cellular NAD+ levels are decreased and NAD+-dependent SIRT1 activity as well as its expression is substantially decreased in obesity (Yoshino et al, 2011; Choi et al, 2013). All these findings suggest that the balance of expression and activities of HATs and HDACs is disrupted in obesity possibly due to aberrant cellular signaling, which leads to persistently elevated acetylation of transcriptional regulators, which in turn causes altered transcriptional regulation promoting hepatic inflammation and metabolic dysfunction.
In the present study, we show that agonist-activated FXR is SUMOylated and selectively trans-represses NF-κB target inflammatory genes without entering the transcriptional pathway regulating classical FXR/RXRα target genes and that elevated acetylation of FXR in obesity inhibits its SUMO2 modification and attenuates its anti-inflammatory functions (Fig8). We also show that the differential interaction of SUMO2-FXR with NF-κB and RXRα contributes to the selective repression of inflammatory genes by SUMOylated FXR. Utilizing in vitro synthesized SUMO2-FXR, we showed that SUMO2 modification of FXR increased its interaction with NF-κB, while its interaction with RXRα was blocked. Supporting these findings, modeling studies indicated that SUMOylation would sterically block the interaction of FXR with RXRα. Further, in ChIP assays, activation of FXR-WT, but not the SUMO-defective K277 mutant, with GW4064 increased its recruitment to NF-κB target genes with a concomitant decrease in occupancy of NF-κB, indicating that SUMO2 modification was important for FXR recruitment. In vitro gel shift assays utilizing SUMOylated FXR further support this conclusion.
It has been shown that SUMOylated NRs inhibit inflammatory responses by preventing clearance of SMRT/NcoR corepressor complex at the inflammatory genes (Pascual et al, 2005; Venteclef et al, 2010). In this study, we also show that SUMOylated FXR exerts anti-inflammatory responses by inhibiting NF-κB binding to DNA as well as preventing clearance of SMRT. Interaction of SUMO-FXR with NF-kB appears to be important for recruitment of FXR to inflammatory genes, but once FXR is recruited, the occupancy of NF-kB is decreased, whereas that of SUMO2-FXR is increased. FXR is likely retained at the genes by interactions with the corepressor complex after NF-κB dissociates. It was also shown that trans-repression of inflammatory pathways by SUMO-NRs is linked to SUMO-dependent interactions with the SUMO-interacting motif (SIM)-containing corepressor complex components, coronin or GPS2 (Venteclef et al, 2010; Huang et al, 2011). We observed that GPS2 appears to be important for increased occupancy of FXR at the IL6Ra gene but not at the Tnfsf4 and C1qtnf2 genes, whereas the occupancy of p65 was not markedly affected by GPS2 (Supplementary Fig S18). In addition to promoting inflammatory responses, elevated acetylation of FXR should also affect metabolic pathways by altering expression of FXR/RXR target genes since DNA binding of FXR is inhibited by acetylation of FXR (Kemper et al, 2009), which is consistent with decreased FXR cistrome numbers in obese mice (Lee et al, 2012). It will be interesting to see how acetylation of FXR impacts global binding and metabolic dysfunction in obesity.
The role of PTMs in gene-specific regulation in physiology and disease and their high potential for treating obesity and diabetes were demonstrated in recent studies of PPARγ and FoxO1. Aberrant phosphorylation of PPARγ by cdk5 in obesity was shown to selectively regulate a subset of genes, and anti-diabetic agonists of PPARγ had therapeutic benefits with fewer side effects by blocking the obesity-induced phosphorylation (Choi et al, 2010). Analysis of the effects of FoxO1 acetylation mutations in knock-in mice showed that acetylation of FoxO1 is critically involved in organismal survival during development and in maintaining metabolic homeostasis and that acetylation of FoxO1 mediates these functions by selective regulation of gene expression (Banks et al, 2011). Although PTMs of proteins have been extensively studied, there is little evidence establishing the physiological/pathological relevance of PTMs and the mechanisms underlying PTM-mediated selective gene regulation. The present study, for the first time, demonstrates the relevance in vivo of a functional antagonism between acetylation and SUMOylation of FXR in obesity, at least in mouse liver, and further identified a mechanism for the selective trans-repression of inflammatory genes without affecting FXR/RXRα target genes. Since FXR is expressed in many non-hepatic tissues, including intestine, kidney, endothelial cells, and macrophages, the dysregulated acetyl/SUMO switch identified for hepatic FXR in obesity may also play a role in the pathogenesis of inflammatory diseases in other tissues, such as inflammatory bowel disease and atherosclerosis, as well as, liver steatohepatitis. Targeting the dysregulated acetyl/SUMO switch of FXR may provide novel therapeutic options and diagnostic markers for obesity-related inflammatory and metabolic disorders.
Materials and Methods
Materials and reagents
Antibodies for FXR (H-130 and C-20), RXRα (D-20), LXR (H-144), ERα (F-10), PXR (A-20), SREBP-1c (H-160), p65 (C-20), SUMO1 (D-11), and SIRT1 (H-300) were purchased from Santa Cruz Biotech; for acetyl-lysine (9441S), SUMO2 (4971S), and PIASy (4392S) from Cell Signaling; and for F4/80 (AP10243PU-M) from Acris Antibodies, Inc. M2 antibody, M2 agarose, trichostatin A (TSA), nicotinamide (Nam), LPS (from Escherichia coli 0111:B4) and N-ethylmaleimide (NEM) were purchased from Sigma Inc. GW4064 was purchased from Tocris Bioscience. TNFα was purchased from R&D Systems, Inc. Ac-K217-specific FXR antibody was developed by Covance.
Animal experiments
To develop dietary obese mice, 8-week-old C57BL/6J male mice were fed a HFD (42% fat, Harlan Teklad) for 4–16 weeks (3 mice per group). Mice were fasted overnight, GW4064 (30 mg/kg in corn oil) or vehicle was injected i.p., and 6 h later, livers were collected. For cholic acid (CA) feeding experiments, mice were fasted for at least 6 h and fed a normal diet (ND) or chow supplemented with 0.5% CA for 3 h from 5 PM to 8 PM before sacrifice. For adenoviral experiments, adenovirus (2.5–5.0 × 108 active viral particles in 100 μl saline) was injected via the tail vein, and one week later, livers were collected (4 mice per group). At the viral dose used, there were only modest changes in mRNA levels of the inflammatory genes, Cxcl8, IL1β, and Tnfα (Supplementary Fig S19). For the glucose tolerance test (GTT), fasted mice (4 mice per group) were injected i.p. with 2 g/kg glucose (Sigma Inc.) and blood glucose levels were measured using an Accu-chek Aviva glucometer (Roche Inc., Nutley, NJ, USA). All animal use and adenoviral protocols were approved by the Institutional Animal Care and Biosafety Committees.
Adenoviral vector constructions
The SUMO-defective flag-FXR mutant was constructed by site-directed mutagenesis (Stratagene, Inc.) and confirmed by sequencing. Flag-FXR in this manuscript refers to 3flag-human FXR. Construction, amplification, purification, and tittering of Ad-3flag-FXR have been previously described (Fang et al, 2008; Kemper et al, 2009).
Tandem mass spectrometry analysis
Flag-FXR was expressed in mice fed (3 mice per group) a ND or HFD by tail vein injection of Ad-3flag-human FXR. FXR mutants were produced by site-directed mutagenesis and incorporated into adenoviral vectors. One week after injection, mice were treated with GW4064 and sacrificed 3 h later. Flag-FXR protein was purified by binding to M2 agarose and subjected to MS/MS analysis as described previously (Kemper et al, 2009). MS/MS spectra were screened against the FXR sequence using SEQUEST (Thermo Finnigan, San Jose, CA, USA), and the identified acetylated peptides were confirmed by manual inspection of the MS2 and MS3 spectra.
Gene expression profiling by Illumina microarray
FXR-WT or the FXR-K217Q mutant was expressed in lean mice, and FXR-WT or the FXR-K217R mutant was expressed in obese mice by adenoviral infection (3 mice per group). One week after infection, mice were treated with GW4064 (30 mg/kg in corn oil) overnight before sacrifice, and hepatic expression was analyzed by Illumina microarray. The microarray raw data were processed using the bioconductor package bead array, and differentially expressed genes were identified using SAM (significance analysis of microarray) with false discovery rate set at 0.05. The differentially expressed genes were further characterized by gene ontology analysis using DAVID.
The microarray raw data have been submitted to the GEO database, Accession Number GSE62414, and are accessible at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62414.
Quantification of mRNA
RNA was isolated from liver (4 mice per group), primary mouse hepatocytes, or cultured cells, and the amount of mRNA for each gene was determined by qRT–PCR with an iCycler iQ (Bio-Rad) and normalized to that of 36B4 mRNA. The list of qRT–PCR primer sequences is shown in Supplementary Table S3.
Oil Red-O staining, immunohistochemistry (IHC), and liver cytokine detection
Frozen liver sections were stained with Oil Red-O, and paraffin-embedded liver sections were stained with hematoxylin and eosin (H&E) and then analyzed by confocal microscopy. For IHC studies, paraffin-embedded liver sections were incubated with Ac-K217 or F4/80 antibody for overnight at 4°C. Signaling was then amplified (HRP/DAB detection IHC kit, Abcam) and stained with hematoxylin, and stained slides were imaged with a NanoZoomer Scanner (Hamamatsu). Livers were homogenized in RIPA buffer, and the levels of the cytokines, IL-1β and TNFα, were measured by ELISA (eBioscience, Inc.).
In-cell and in vivo acetylation assays
COS-1 cells were treated with 200 nM of GW4064 and deacetylase inhibitors (500 nM trichostatin A and 10 mM of nicotinamide) for 2–5 h. For in vivo studies, livers were collected from mice fasted overnight. Flag-FXR or endogenous FXR was immunoprecipitated from freshly prepared cell or liver extracts at 4°C for 3 h in 50 mM Tris–HCl, pH 7.6, 150 mM NaCl, 5 mM EDTA, 1% NP-40, and 0.1% SDS. FXR acetylated at K217 was detected using the Ac-K217 FXR-specific antibody by immunoblotting (IB). Increased FXR levels were not observed in contrast to earlier studies, showing that stability of FXR was increased by acetylation in cells incubated with full media and also treated with insulin and glucose (Kemper et al, 2009; Ponugoti et al, 2010) probably because of different experimental conditions including fasting overnight in the present study.
In vitro, in-cell, and in vivo SUMO assay
For the in vitro SUMO assay, all SUMO components were highly purified as previously described (Wu & Chiang, 2009). Flag-FXR was expressed in COS-1 cells and isolated by binding to M2 agarose. Reactions that contained recombinant SUMO2 protein, E1 (SAE1/SAE2) activating enzyme, E2 (Ubc9) conjugation enzyme, and flag-FXR bound to M2 agarose in 20 μl of reaction buffer were incubated at 30°C for 1 h. The only difference between SUMO and unSUMO-FXR reactions is the presence or absence of ATP. The agarose beads were washed before being used for the gel mobility shift and CoIP analyses. SUMOylation was confirmed by IB using SUMO2 or SUMO1 antibody. For in-cell assays, COS-1 cells were transfected with plasmids for SUMO components. Cells were lysed in IP buffer containing 50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, protease inhibitors, and 20 mM NEM; the proteins were incubated with antibodies and then bound to protein G agarose beads; the beads were washed three times; and bound proteins were analyzed by IB. For in vivo assays, flag-FXR or endogenous FXR in mouse liver was immunoprecipitated at 4°C for 3 h in IP buffer containing 20 mM NEM, and SUMO-FXR was detected by IB.
CoIP and GST pull-down assays
For CoIP assays, cells were transfected with the relevant expression plasmids, including the PIAS SUMO ligases (Palvimo, 2007), or infected with adenoviral vectors. Cell extracts were prepared by brief sonication of cell pellets in CoIP buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.5% NP-40, 5% glycerol) supplemented with protease inhibitors, DTT, and phosphatase inhibitors. After centrifugation, the supernatant was incubated with 1–2 μg of antibodies for 2 h and 30 μl of a 25% protein G agarose slurry was added. One hour later, samples were washed with CoIP buffer three times and bound proteins were detected by IB. The in vitro GST pull-down assay was performed as described previously (Fang et al, 2007; Kemper et al, 2009; Ponugoti et al, 2010), and protein interaction was detected by IB.
Construction of TNFsf4-luc and IL6Ra-luc and reporter assays
A KpnI/NheI or NheI/XhoI genomic DNA fragment containing the major FXR binding peak in Tnfsf4 (−4,464 to −4,994) or IL6Ra (+34,593 to +35,236), respectively, was inserted into the pGL3-SV40 luc plasmid. HepG2 cells were cotransfected with 200 ng luciferase reporter plasmids along with 300 ng of CMV β-galactosidase plasmids and 10–50 ng of expression plasmids for flag-FXR-WT or the FXR mutants. Luciferase activities were normalized to β-galactosidase activities.
Isolation of hepatocytes and ChIP assays
FXR-WT or FXR mutants were adenovirally expressed in primary hepatocytes that were prepared from FXR-KO mice. In preliminary experiments, the amounts of adenovirus needed to express FXR at levels similar to those in WT hepatocytes were determined (Supplementary Fig S19). Adenoviral infection did not increase mRNA levels of tested inflammatory genes in hepatocytes (Supplementary Fig S19). Hepatocytes were treated with GW4064 for 3 h and then with LPS (Sigma, Inc.) for 30 min. ChIP assays in triplicate were performed as previously described (Kemper et al, 2009; Ponugoti et al, 2010; Seok et al, 2013). Primer sequences for ChIP qPCR are shown in Supplementary Table S3.
FXR modeling analysis
FXR modeling was based on either the RXRα/PPARγ/DNA (3DZU) structure (Chandra et al, 2008) or the RXRα/LXR/DNA (4NQA) structure (Lou et al, 2014) with the FXR structure (1OSV) substituted for the PPARγ or LXR structure. The NMR solution structure (2AWT) was used for SUMO2.
Statistical analyses
Values are presented as mean ± SEM. Differences between two groups were considered significant at P < 0.05 using the two-tailed Student's t-test.
Gel mobility shift assays
Gel mobility shift assays were performed as previously described (Kemper et al, 2009). Briefly, double-stranded oligonucleotides (32-mers) that contain the FXRE and NF-κB site from the Shp and Tnfsf4 genes, respectively, were labeled with [γ-32P]ATP and incubated with purified FXR and RXRα, and whole-cell extracts containing NF-κB, and in vitro SUMOylated FXR in the presence of 100 ng poly(dIdC) and 10 μg BSA. Binding reactions were analyzed by electrophoresis in 6% non-denaturing polyacrylamide gels in a low ionic Tris-acetate buffer with recirculation at 4°C.
Acknowledgments
We thank Ron Hay at University of Dundee for kindly providing SUMO expression plasmids and Peter Tontonoz at UCLA for (NF-κB site)3-tk-luc plasmid. This study was supported by grants from an AHA post-doctoral fellowship to DHK (14POST20420006), NSF grants (DMS-1440037 and DMS-1228288) to PM, NIH (CA103867), CPRIT (RP110471 and RP140367), and Welch Foundation (I-1805) to CMC, the Academy of Finland to JP, and NIH grants (DK62777 and DK95842) to JKK.
Author contributions
D-HK and JKK designed research; D-HK, SK, DR, and DT performed experiments; ZX performed proteomic analysis; S-YW, C-MC, JJP, and L-FC provided critical reagents and technical expertise; EZ and HEX performed structural modeling analysis; XS and PM performed computational analysis; D-HK, ZX, and BK analyzed data; and D-HK, BK, and JKK wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Information
Review Process File
Source Data for Figure 1 C part a
Source Data for Figure 1 C part b
Source Data for Figure 1 D
Source Data for Figure 3 I
Source Data for Figure 5 C
Source Data for Figure 6 F
Source Data for Figure 6 H
Source Data for Figure 6 I
References
- Balasubramaniyan N, Luo Y, Sun AQ, Suchy FJ. SUMOylation of the farnesoid X receptor (FXR) regulates the expression of FXR target genes. J Biol Chem. 2013;288:13850–13862. doi: 10.1074/jbc.M112.443937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks AS, Kim-Muller JY, Mastracci TL, Kofler NM, Qiang L, Haeusler RA, Jurczak MJ, Laznik D, Heinrich G, Samuel VT, Shulman GI, Papaioannou VE, Accili D. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 2011;14:587–597. doi: 10.1016/j.cmet.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricambert J, Miranda J, Benhamed F, Girard J, Postic C, Dentin R. Salt-inducible kinase 2 links transcriptional coactivator p300 phosphorylation to the prevention of ChREBP-dependent hepatic steatosis in mice. J Clin Invest. 2010;120:4316–4331. doi: 10.1172/JCI41624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Tu BP. On acetyl-CoA as a gauge of cellular metabolic state. Cold Spring Harb Symp Quant Biol. 2011;76:195–202. doi: 10.1101/sqb.2011.76.010769. [DOI] [PubMed] [Google Scholar]
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Jiang LQ, Deshmukh AS, Mataki C, Coste A, Lagouge M, Zierath JR, Auwerx J. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab. 2010;11:213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Bluher M, Griffin PR, Spiegelman BM. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SE, Fu T, Seok S, Kim DH, Yu E, Lee KW, Kang Y, Li X, Kemper B, Kemper JK. Elevated microRNA-34a in obesity reduces NAD levels and SIRT1 activity by directly targeting NAMPT. Aging Cell. 2013;12:1062–1072. doi: 10.1111/acel.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Evans RM, Barish GD, Wang YX. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–361. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The p300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008;283:35086–35095. doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Mencarelli A, Palladino G, Cipriani S. Bile-acid-activated receptors: targeting TGR5 and farnesoid-X-receptor in lipid and glucose disorders. Trends Pharmacol Sci. 2009;30:570–580. doi: 10.1016/j.tips.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Guarente L. The logic linking protein acetylation and metabolism. Cell Metab. 2011;14:151–153. doi: 10.1016/j.cmet.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, Rosenfeld MG, Glass CK. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–418. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper JK, Xiao Z, Ponugoti B, Miao J, Fang S, Kanamaluru D, Tsang S, Wu S, Chiang CM, Veenstra TD. FXR acetylation is normally dynamically regulated by p300 and SIRT1 but constitutively elevated in metabolic disease states. Cell Metab. 2009;10:392–404. doi: 10.1016/j.cmet.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper JK. Regulation of FXR transcriptional activity in health and disease: Emerging roles of FXR cofactors and post-translational modifications. Biochim Biophys Acta. 2011;1812:842–850. doi: 10.1016/j.bbadis.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Seok SM, Yu P, Kim K, Smith Z, Rivas-Astroza M, Zhong S, Kemper JK. Genomic analysis of hepatic Farnesoid X Receptor (FXR) binding sites reveals altered binding in obesity and direct gene repression by FXR. Hepatology. 2012;56:108–117. doi: 10.1002/hep.25609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GY, Jang H, Lee JH, Huh JY, Choi S, Chung J, Kim JB. PIASy-mediated sumoylation of SREBP1c regulates hepatic lipid metabolism upon fasting signaling. Mol Cell Biol. 2014;34:926–938. doi: 10.1128/MCB.01166-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev. 2009;89:147–191. doi: 10.1152/physrev.00010.2008. [DOI] [PubMed] [Google Scholar]
- Lou X, Toresson G, Benod C, Suh JH, Philips KJ, Webb P, Gustafsson JA. Structure of the retinoid X receptor alpha-liver X receptor beta (RXRalpha-LXRbeta) heterodimer on DNA. Nat Struct Mol Biol. 2014;21:277–281. doi: 10.1038/nsmb.2778. [DOI] [PubMed] [Google Scholar]
- Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest. 2011;121:2111–2117. doi: 10.1172/JCI57132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal. 2010;8:e005. doi: 10.1621/nrs.08005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–1408. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, Rose DW, Willson TM, Rosenfeld MG, Glass CK. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–763. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponugoti B, Kim DH, Xiao Z, Smith Z, Miao J, Zang M, Wu SY, Chiang CM, Veenstra TD, Kemper JK. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J Biol Chem. 2010;285:33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham A, Xu Q, Lu J, Foley JF, Yan X, Kim DH, Kemper JK, Li X. Hepatic deletion of SIRT1 decreases hepatocyte nuclear factor 1alpha/farnesoid X receptor signaling and induces formation of cholesterol gallstones in mice. Mol Cell Biol. 2012;32:1226–1236. doi: 10.1128/MCB.05988-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. A Nurr1/CoREST pathway in microglia and astrocytes protects dopaminergic neurons from inflammation-induced death. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seok S, Kanamaluru D, Xiao Z, Ryerson D, Choi SE, Suino-Powell K, Xu HE, Veenstra TD, Kemper JK. Bile acid signal-induced phosphorylation of small heterodimer partner by protein kinase Czeta is critical for epigenomic regulation of liver metabolic genes. J Biol Chem. 2013;288:23252–23263. doi: 10.1074/jbc.M113.452037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubal A, Clement K, Fan R, Ancel P, Pelloux V, Rouault C, Veyrie N, Hartemann A, Treuter E, Venteclef N. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. J Clin Invest. 2013;123:362–379. doi: 10.1172/JCI64052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuter E, Venteclef N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim Biophys Acta. 2011;1812:909–918. doi: 10.1016/j.bbadis.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J Immunol. 2009;183:6251–6261. doi: 10.4049/jimmunol.0803978. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, Steffensen KR, Treuter E. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, Israelian K, Westphal CH, Rodgers JT, Shioda T, Elson SL, Mulligan P, Najafi-Shoushtari H, Black JC, Thakur JK, Kadyk LC, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SY, Chiang CM. Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J. 2009;28:1246–1259. doi: 10.1038/emboj.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File
Source Data for Figure 1 C part a
Source Data for Figure 1 C part b
Source Data for Figure 1 D
Source Data for Figure 3 I
Source Data for Figure 5 C
Source Data for Figure 6 F
Source Data for Figure 6 H
Source Data for Figure 6 I