Abstract
The essential neurotransmitter acetylcholine functions throughout the animal kingdom. In Caenorhabditis elegans, the acetylcholine biosynthetic enzyme [choline acetyltransferase (ChAT)] and vesicular transporter [vesicular acetylcholine transporter (VAChT)] are encoded by the cha-1 and unc-17 genes, respectively. These two genes compose a single complex locus in which the unc-17 gene is nested within the first intron of cha-1, and the two gene products arise from a common pre-messenger RNA (pre-mRNA) by alternative splicing. This genomic organization, known as the cholinergic gene locus (CGL), is conserved throughout the animal kingdom, suggesting that the structure is important for the regulation and function of these genes. However, very little is known about CGL regulation in any species. We now report the identification of an unusual type of splicing regulation in the CGL of C. elegans, mediated by two pairs of complementary sequence elements within the locus. We show that both pairs of elements are required for efficient splicing to the distal acceptor, and we also demonstrate that proper distal splicing depends more on sequence complementarity within each pair of elements than on the sequences themselves. We propose that these sequence elements are able to form stem-loop structures in the pre-mRNA; such structures would favor specific splicing alternatives and thus regulate CGL splicing. We have identified complementary elements at comparable locations in the genomes of representative species of other animal phyla; we suggest that this unusual regulatory mechanism may be a general feature of CGLs.
Keywords: cholinergic gene locus, VAChT, vesicular acetylcholine transporter, ChAT, choline acetyltransferase
ACETYLCHOLINE (ACh) is an essential and widespread neural and neuromuscular transmitter. ACh is synthesized by the enzyme choline acetyltransferase (ChAT) and packaged into synaptic vesicles by the vesicular ACh transporter (VAChT). In Caenorhabditis elegans, ChAT and VAChT are encoded by the cha-1 and unc-17 genes, respectively (Alfonso et al. 1993, 1994b). We have previously shown that the two genes compose a single complex locus (Figure 1A) in which unc-17 is nested within the first intron of cha-1, and two distinct gene products arise from a common premessenger RNA (pre-mRNA) through an unusual type of alternative splicing (Figure 1B) (Alfonso et al. 1994a). Thus, the sequential steps of ACh synthesis and vesicle loading are encoded by separate genes in a single complex transcription unit. Subsequent studies suggest that the nested structure of these genes, referred to as the cholinergic gene locus (CGL), is present in all metazoans having cholinergic neurons (Eiden 1998). Furthermore, the genes encoding the biosynthetic enzymes and vesicular transporters for other neurotransmitters are not organized in a comparable manner, suggesting that the specific organization of the CGL is important for the regulation of cholinergic gene expression.
Figure 1.
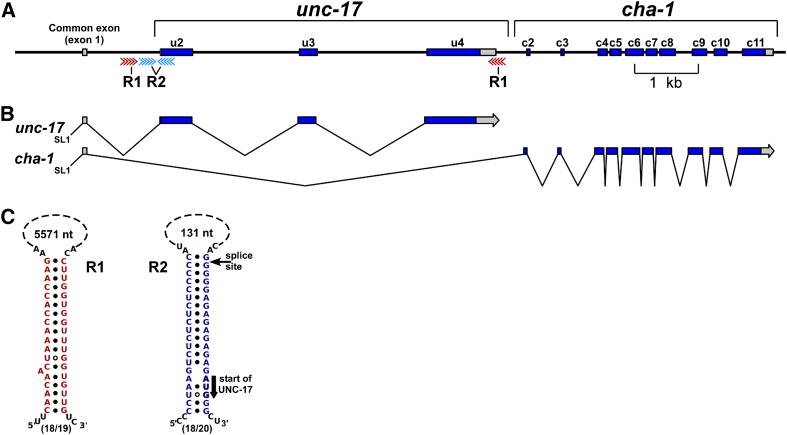
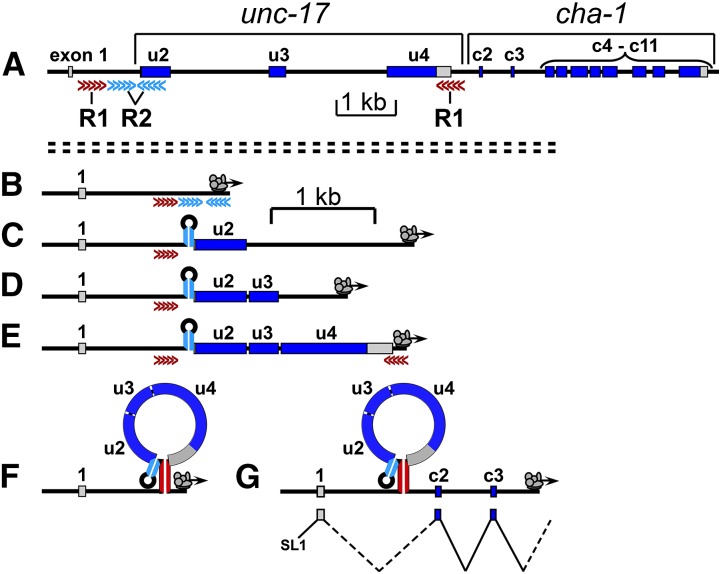
Splicing of the CGL in C. elegans. (A) Genomic organization. cha-1 encodes choline acetyltransferase (ChAT), the ACh biosynthetic enzyme; unc-17 encodes the vesicular acetylcholine transporter (VAChT), which loads ACh into synaptic vesicles. Gray regions are UTRs. The red and blue arrows correspond to R1 and R2 complementary sequence elements, respectively. u2, unc-17 exon 2; c3, cha-1 exon 3, etc. (B) Splicing pattern and transcript structures. SL1 is the leader sequence trans-spliced onto many C. elegans mRNAs. (C) Presumed R1 and R2 stem-loop structures in C. elegans. The solid black circles represent standard RNA (A-U and G-C) pairing; the circles with white centers represent G-U pairing. The thin arrow indicates the splice site at the 5′ end of exon u2; the thick arrow indicates the UNC-17 initiation (AUG) codon. The size of each putative loop is shown, and the fraction of paired nucleotides in each stem is given below each structure. R1 and R2 stem structures from seven other Caenorhabditis species are shown in Figure S1 and Figure S2.
Although many types and mechanisms of alternative splicing have been described (Matlin et al. 2005; Stamm et al. 2005), the splicing pattern of the unc-17/cha-1 locus in C. elegans is somewhat unusual: To produce a cha-1 transcript, the splice from the common exon (exon 1) to the first cha-1-specific exon (exon c2) requires skipping all three unc-17 exons, implying that there is a mechanism for coordinated exon skipping. A model of such a mechanism is discussed in greater detail below.
Except for the identification of two upstream transcription factor binding sites (Wenick and Hobert 2004; Kratsios et al. 2012), the regulation of the C. elegans locus has not been characterized. We therefore constructed a dual-reporter plasmid that mimics the splicing features of unc-17 and cha-1 and used derivatives of this plasmid to analyze the roles of specific CGL sequence elements in splicing regulation. Our results indicate that sequence elements capable of forming stem-loop structures in the pre-mRNA regulate CGL alternative splicing. We have identified comparable sequence elements in the CGLs of many other animal species, suggesting that the formation of RNA secondary structures is a conserved feature of CGL splicing.
Materials and Methods
Strains and strain maintenance
Standard laboratory methods for C. elegans are described by Brenner (1974). Worms were grown on NGM-L medium (Sun and Lambie 1997), modified by the addition of streptomycin and mycostatin to reduce contamination, and seeded with the streptomycin-resistant Escherichia coli strain OP50/1 (Johnson et al. 1988).
The cn355 mutant was originally isolated and identified as an unc-17 allele by Ryuji Hosono (Kanazawa University, Kanazawa, Japan); a molecular description of cn355 is presented below in Results, with additional details in Supporting Information, File S1. The pha-1(e2123) mutant was obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis). A complete list of strains is included in File S1.
Sequence analysis
Accession numbers for genomic sequences are presented in File S1. Some complementary DNAs (cDNAs) were provided by Yuji Kohara (National Institute of Genetics, Mishima, Japan). The genomic sequence encompassing the C. elegans CGL was downloaded from WormBase (www.WormBase.org). Sequence analysis utilized Vector NTI software (Life Technologies, Carlsbad, CA) or the Lasergene Suite (DNASTAR, Madison, WI). RNA structures were analyzed with Mfold (Zuker 2003) or Sfold (Ding and Lawrence 2003; Ding et al. 2005). The criteria for R1-like elements (in addition to complementarity) were that they flank the complete VAChT coding sequence but do not include any part of the ChAT coding sequence; the criterion for R2-like elements was that they flank or overlap the splice acceptor site of the first VAChT coding exon. The prediction of 3′-cleavage sites was based on the results of Mangone et al. (2010) that cleavage sites in C. elegans are ∼19 nucleotides downstream of the first base of the AAUAAA polyadenylation signal.
Reporter constructs and transgenic methods
Plasmids containing the GFP sequence were derived from the pPD95.67 plasmid (gift of Andrew Fire, Stanford University, Stanford, CA). A plasmid (pAA64) containing a modified mCherry gene (McNally et al. 2006; Green et al. 2008) was provided by Anjon Audhya and Karen Oegema (Ludwig Institute for Cancer Research, La Jolla, CA). Specific sequence changes were introduced into plasmids, using a modified version of Stratagene’s QuikChange procedure. DNA sequencing was performed at the Oklahoma Medical Research Foundation DNA Sequencing Core Facility, using oligonucleotide primers obtained from IDT (Coralville, IA). Additional details of plasmid construction are presented in File S1.
Fragments for microinjection were generated by linking the 3.2-kb unc-17/cha-1 promoter (amplified from a cloned genomic fragment) to PCR fragments amplified from dual-reporter constructs through the SOEing technique (Horton et al. 1989; Hobert 2002). Transgenic nematodes were generated by microinjection of DNA (Mello and Fire 1995). The final DNA concentration of each injection mix was adjusted to ∼50 ng/µl by addition of pBluescript plasmid DNA. The transformation marker was the pBX plasmid (Heinke and Ralf Schnabel, Max-Plank-Institute fur Biochemie, Martinsried, Germany), which rescues the temperature-sensitive lethality of pha-1(e2123) mutants (Granato et al. 1994).
Immunofluorescence staining
Nematodes were fixed and stained using a freeze-fracture procedure (Mullen et al. 2006). Antibodies used in this study include mouse monoclonal antibodies to CHA-1 (mAbs 1402 and 1414) and chicken polyclonal antibodies to UNC-17 (C96) (Duerr et al. 2008). Secondary antibodies [F(ab′)2 fragments] were obtained from Jackson ImmunoResearch (West Grove, PA).
Microscopy and imaging
Live worms were immobilized in polyacrylamide with 0.05% sodium azide. Confocal images were collected on a Leica TCS NT confocal microscope. Lower-resolution images were collected with a 40× Plan Fluotar 1.0 NA oil immersion objective, at 512 × 512 or 1024 × 1024 pixels, with 0.5-μm Z steps. Higher-resolution images were collected with a 63× Plan APO 1.4 NA oil immersion objective, at 512 × 512 pixels, with 4× zoom, and 0.2-μm Z steps. Images were cropped to size, assembled, and annotated using Adobe Photoshop CS2. Digital manipulations were limited to rotating and cropping (Photoshop Bicubic), as well as minor brightness adjustment for publication.
Quantification of mCherry and GFP fluorescence was performed on maximum-projection images of L1 larvae collected with identical imaging parameters. Animals carrying the wild-type dual reporter were imaged using conditions such that the total brightness of the mCherry and GFP fluorescence was approximately equal. Animals carrying mutant dual reporters were then imaged under the same conditions. Images were imported into ImageJ and mean pixel intensities were determined for the mCherry and GFP fluorescence images. For animals carrying the wild-type dual reporter, the ratio of mCherry to GFP fluorescence was normalized to a value of 1. The ratio of mCherry to GFP fluorescence for animals carrying mutant dual reporters was then normalized against the wild-type value so that deviation from the wild-type ratio could be quantified.
Results
Complementary sequence elements may promote alternative splicing
Careful inspection of the C. elegans unc-17/cha-1 genomic region revealed the presence of two pairs of complementary sequence elements. We designated these element pairs R1 and R2 (Figure 1). Comparable sequences are present in the genomes of seven other Caenorhabditis species (Figure S1 and Figure S2).
The two R1 elements flank the unc-17 coding exons (Figure 1). The 5′-R1 element is in the first unc-17 intron, and the 3′ element is ∼3.5 kb downstream in the unc-17 3′-UTR. The R1 elements vary from 19 to 62 nucleotides in length among the different Caenorhabditis species (Figure S1A). Pairing of the two R1 RNA sequences would produce a 19- to 62-bp stem, depending on the species, positioned at or near the site of unc-17 pre-mRNA cleavage and polyadenylation (see below). Apart from a conserved core motif (AAACCACCAAC), the R1 sequences from the eight Caenorhabditis species share little sequence similarity (Figure S1B).
Both R2 elements are located near the beginning of the unc-17 coding sequence (Figure 1). In C. elegans, the 5′-R2 element is ∼100 bp upstream of the first unc-17-specific exon (u2), and the 3′ element overlaps the splice acceptor of this exon. In C. elegans transcripts, the R2 elements are predicted to match each other at 18 of 20 nucleotides; in other Caenorhabditis species, the R2 elements range in size from 16 to 24 nucleotides (Figure S2A). These complementary elements could form a 16- to 24-bp stem, with an ∼100-nucleotide loop in which the stem is positioned at or next to the splice acceptor site at the beginning of the first unc-17 coding exon (Figure 1). The putative R2 elements are conserved among Caenorhabditis species and contain a 16-nucleotide (UCUGCGUCUCUCUCCC) core sequence (Figure S2B).
There are two RNA processing steps that would preclude the production of a cha-1 transcript. The first of these is the splice from the common exon 1 to the first unc-17-specific exon (u2), and the second step is cleavage and polyadenylation at the end of the unc-17 3′-UTR. These two sites of RNA processing correspond closely to the two sites identified by the putative R2 and R1 stem structures, respectively. Blocking or inhibiting RNA processing at either site would presumably reduce or prevent the production of unc-17 mRNA and favor the production of cha-1 mRNA. This suggests a model in which the pre-mRNA is able to form secondary structures, and these structures, perhaps together with specific RNA-binding proteins, regulate the splicing choice (see below).
A two-color (dual) splicing reporter for the CGL
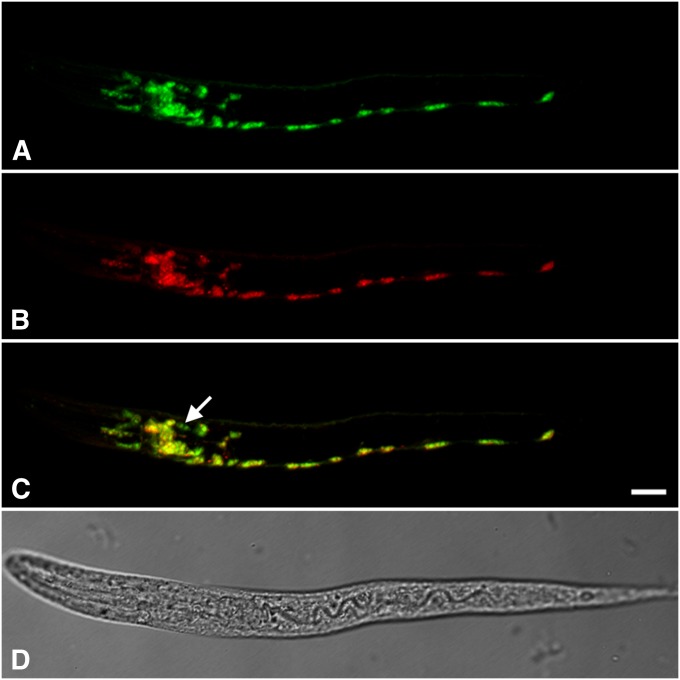
To test the roles of these complementary sequences, we constructed a dual-reporter plasmid (Figure 2; plasmid construction is described in File S1). This plasmid contains the unc-17/cha-1 genomic region, with the UNC-17 and CHA-1 coding sequences and introns replaced by GFP and mCherry, respectively. The GFP coding sequence also contains three introns, similar to the structure of the unc-17 gene. A 3.5-kb unc-17/cha-1 promoter was fused to the construct by PCR; the resulting 7.3-kb PCR product was used to generate transgenic nematodes. Animals carrying this construct express both fluorescent proteins in most or all of the known cholinergic neurons (Figure 3).
Figure 2.
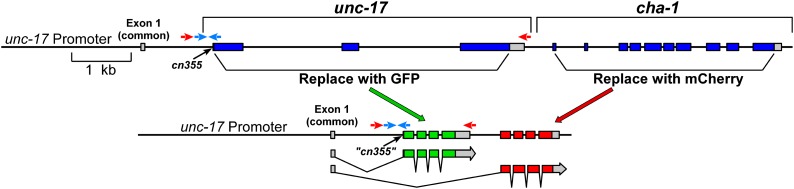
Structure of a “dual-reporter” transgene to mimic CGL structure and evaluate alternative cholinergic splicing. Construction details are provided in File S1.
Figure 3.
Expression of a “wild-type” dual-reporter transgene in C. elegans. Shown are images of an L1 animal; anterior is to the left, and ventral is down. Bar, 10 µm. (A) GFP fluorescence. (B) mCherry fluorescence. (C) Merged image of A and B, adjusted so that the ratio of red to green for the whole animal is ∼1.0. Even so, some cells appear greenish yellow and others reddish yellow, indicating that the red/green ratio is not uniform in all cholinergic neurons. The arrow points to a pair of cells in the head that appear to express only the GFP reporter. (D) Transmitted light image of the same animal.
Validation of the dual-reporter constructs
We used several approaches to verify that the presence of both fluorescent proteins was due to alternative splicing, rather than cryptic transcription initiation between the GFP and mCherry genes. We verified the structures of the GFP and mCherry transcripts by RACE, and both types of transcript included the common exon (exon 1) as depicted in Figure 2. We also tested a “promoterless” version of the dual reporter lacking the common exon and all upstream sequences; transgenic animals carrying this construct exhibited extremely limited fluorescence. These data support our belief that the reporter results reflect alternative splicing.
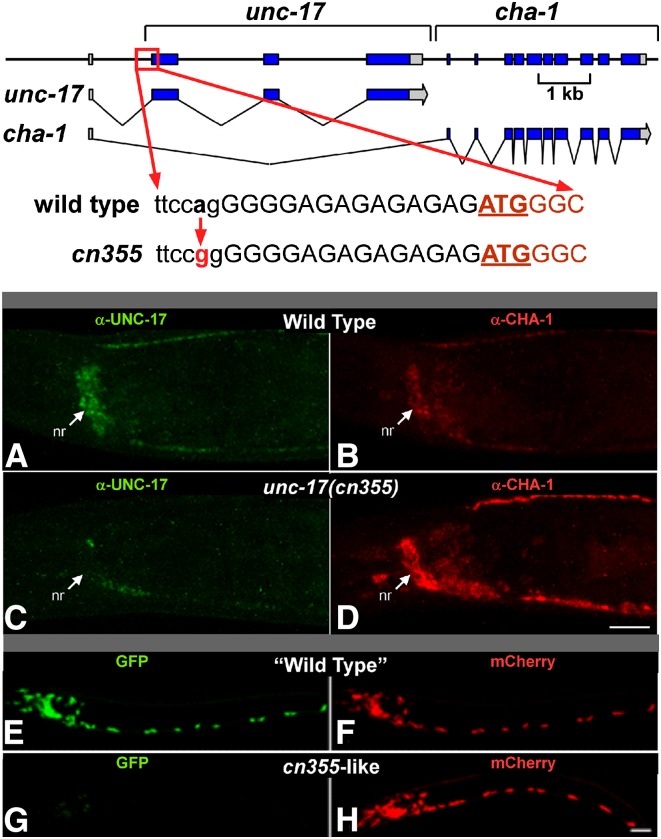
cn355 is a viable unc-17 mutation associated with an A to G transition that disrupts the unc-17 exon 2 splice acceptor site by changing TTCCAG to TTCCGG (Figure 4 and File S1). This is the critical alternative splice acceptor for generating unc-17 mRNA (Figure 1). We found that UNC-17 immunoreactivity in cn355 mutants is dramatically reduced, while ChAT immunoreactivity is greater than in wild-type animals (Figure 4, A–D). We have also found that ChAT enzymatic activity is elevated in cn355 mutants (J. Rand, unpublished results), indicating an increased level of functional ChAT. Thus, cn355 has the properties of a splicing mutant with reduced efficiency of the unc-17-specific splice and increased efficiency of the cha-1-specific splice.
Figure 4.
Validation of the unc-17–cha-1 dual reporter. Top: The unc-17(cn355) a > g mutation disrupts the splice acceptor site (in lowercase) required for production of the endogenous unc-17 transcript; this is predicted to reduce UNC-17 expression. The UNC-17 initiation codon is underlined. There are several weak upstream splice acceptor sequences; it is likely that collectively these account for the low level of residual unc-17 transcripts present in cn355 animals. Middle (A–D): Wild-type (A and B) and cn355 mutant (C and D) animals were double stained under identical conditions with anti-UNC-17 (A and C) and anti-CHA-1 (B and D) antibodies. Shown are head regions of young adults. nr, nerve ring; anterior is to the left, and ventral is down. Bar, 15 µm. Compared to wild type, cn355 homozygotes have reduced UNC-17 expression and increased CHA-1 expression. Bottom (E–H): When introduced into the dual reporter, the cn355 mutation had comparable effects on expression. L1 animals were imaged so that the ratio of red to green fluorescence (R/G) was ∼1.0 with the “wild-type” transgene (E and F). Green is GFP (E and G, corresponding to unc-17) and red is mCherry (F and H, corresponding to cha-1); anterior is to the left, and ventral is down. Bar, 10 µm.
As a test of our approach, we introduced the cn355 mutation into the dual reporter. We observed that the GFP fluorescence was greatly reduced, and mCherry fluorescence was significantly increased (Figure 4, E–H). When we introduced an analogous mutation that alters the alternative splice acceptor of exon c2 (critical for generating mCherry/cha-1), we observed the reciprocal results—increased GFP fluorescence and decreased mCherry fluorescence (data not shown). The requirement for a splice site upstream of the mCherry gene provides additional evidence against any significant transcriptional initiation between the GFP and mCherry genes. The alternative splicing of the dual reporter thus appears to be an accurate surrogate for unc-17/cha-1 splicing.
R1 element complementarity is important for splicing of the downstream transcript
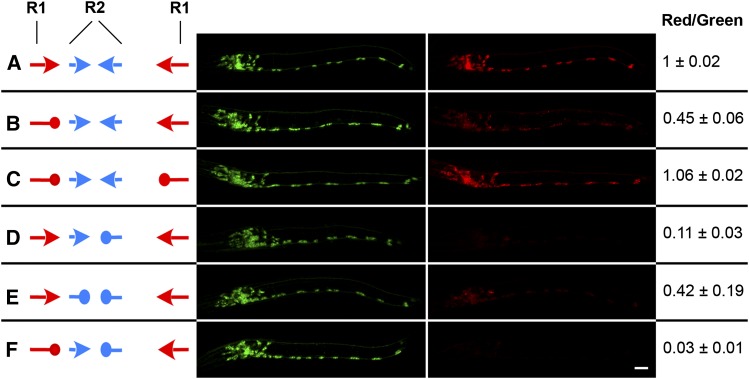
To test the role(s) of the complementary sequence elements in alternative splicing of the CGL, we made specific alterations to the R1 and/or R2 sequences in the dual reporter and assessed the ratio of mCherry to GFP fluorescence in the resulting transgenic animals. We found that replacing one of the R1 elements with a scrambled version of the sequence (Table S1A) led to a significant (∼55%) decrease in the mCherry/GFP ratio (Figure 5B). This suggests that the R1 elements are important for the production of the downstream (mCherry/cha-1) transcript. However, replacing both of the R1 elements with complementary versions of the scrambled sequence restored mCherry expression to essentially wild-type levels (Figure 5C). We conclude that R1 complementarity, rather than precise sequence, promotes production of the downstream transcript. This result suggests that R1 stem-loop formation inhibits 3′-end processing of potential GFP/unc-17 RNA transcripts, thereby promoting the formation of mCherry/cha-1 transcripts.
Figure 5.
R1 and R2 element pairs are required for alternate splicing of unc-17 and cha-1 transcripts. Left (A–F): Wild-type R1 and R2 elements are indicated as red and blue arrows, respectively, and scrambled elements as red and blue “tennis racquets.” Center: Confocal images of transgenic L1 animals, each carrying the dual-reporter construct diagrammed to its left. Green is GFP (corresponding to unc-17) and red is mCherry (corresponding to cha-1). Animals were imaged so that the wild-type ratio of red to green fluorescence (R/G) was ∼1.0. Anterior is to the left, and ventral is down. Bar, 10 μm. Right: Ratio of red to green fluorescence for the transgenic animals shown, presented as the mean ± SD of two to four individuals of each strain. Additional details are in Materials and Methods.
R2 complementarity and sequence are both important for alternative splicing
Comparable experiments on the R2 sequences led to somewhat different results. Replacing a single R2 element with a scrambled sequence resulted in a significant (∼89%) decrease in the relative amount of mCherry fluorescence (Figure 5D), demonstrating that the R2 elements also play a role in the alternative splicing. However, when we replaced both of the R2 elements with complementary versions of the scrambled sequence, mCherry fluorescence was only partially restored (Figure 5E). Therefore, both R2 sequence and complementarity appear to be important for mCherry/cha-1 transcript splicing. In addition, simultaneous disruption of an R1 and an R2 element has more severe consequences than disruption of either single element (Figure 5F); it therefore appears that the R1 and R2 element pairs work in concert to promote mCherry/cha-1 transcript production.
Generality of putative stem-loop structures
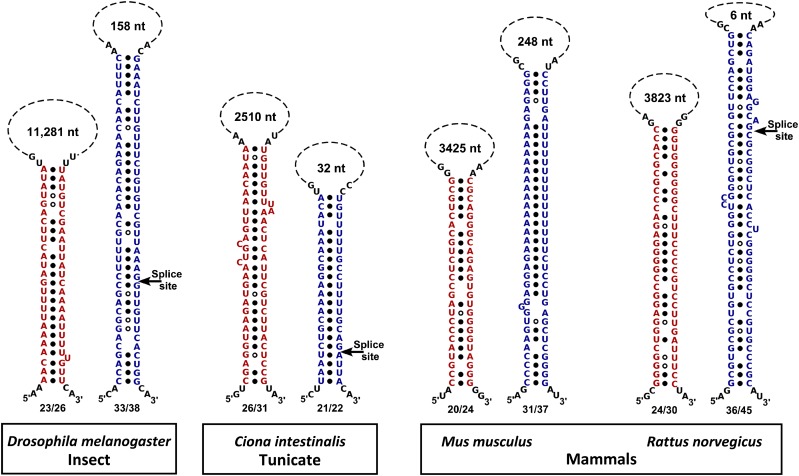
We have identified R1 and R2 elements associated with CGLs in all members of the genus Caenorhabditis that we investigated (Figure S1 and Figure S2). The size of the R2 elements is relatively constant (Figure S2), whereas the length of the R1 elements varies substantially among these species (Figure S1). We also identified similarly positioned R1- and R2-like complementary sequence pairs in the CGLs of other nematode species, although the specific AAACCACCAAC R1 and UCUGCGUCUCUCUCCC R2 core sequences were limited to the genus Caenorhabditis. In addition, we identified similarly positioned complementary elements in the CGLs of representative species of platyhelminthes, insects, hemichordates, tunicates, teleosts, and mammals (Figure 6 and Figure S3). Our observations suggest that the presence of sequence elements capable of forming such stem-loop structures may be a general feature of CGL architecture in most animal species.
Figure 6.
Putative stem-loop alignments of the R1-like (red) and R2-like (blue) sequences from the cholinergic loci of four representative species from three animal phyla. (Alignments for three additional species/phyla are shown in Figure S3.) The fractional pairing of the stem is given below each stem; the length of the putative loop is shown above each stem. Presumed splice acceptor sites at the 5′ end of the first VAChT coding exons are indicated with arrows.
Discussion
Roles of RNA stem-loop structures in splicing
We have demonstrated that the R1 and R2 elements play important roles in determining the relative levels of GFP/unc-17 and mCherry/cha-1. Disrupting either pair of elements significantly decreased the production of the distal (mCherry/cha-1) transcript. The R1 and R2 element pairs appear to act in concert to inhibit the production of GFP/unc-17 transcripts (Figure 5F), thus potentiating the mCherry/cha-1-specific splice.
Among the Caenorhabditis species, the sizes and sequences of the R2 elements are relatively similar, and the conserved 16-nucleotide R2 core sequence represents most or all of the R2 elements. In contrast, the R1 elements display considerable variation in size and sequence, and the conserved 11-nucleotide R1 core sequence represents only a small part of the total R1 elements (Figure S1 and Figure S2). These observations are consistent with our data that R1 function depends primarily on complementarity while R2 function depends on both complementarity and sequence and suggest that R1 and R2 function may involve somewhat different mechanisms for increasing utilization of the distal splice site.
RNA stem-loop structures have been implicated in the regulation of alternative splicing in other systems. A well-studied example is the alternative splicing of the mammalian fibroblast growth factor receptor 2 (FGFR2); tissue-specific alternative splicing is mediated by double-stranded RNA stems and RNA-binding proteins (Muh et al. 2002; Newman et al. 2006). However, the stem-loop structure does not appear to be directly involved in splice regulation, but rather serves to bring the splice donor and acceptor sites into proximity; artificial juxtaposition of these sites eliminated the requirement for an RNA stem structure (Baraniak et al. 2003).
Possible biological functions of these structural elements
We envision several possible ways in which R1- and R2-like stem-loop structures could play a role in alternative splicing and its regulation. These include (a) facilitating coordinated exon skipping, (b) enhancing the efficiency of long splices, and (c) providing a point of regulation to control the ratio of ChAT and VAChT proteins.
(a) Facilitation of coordinated exon skipping
A noteworthy aspect of CGL splicing is that to produce a cha-1 transcript, the splice from the common exon (exon 1) to the first cha-1-specific exon (exon c2) requires skipping all three unc-17 coding exons (Figure 1), implying that there is a mechanism for coordinated exon skipping. In most nematode species, the VAChT coding sequence consists of two or three exons; however, the need for a reliable mechanism for coordinated exon skipping is especially pronounced in nematodes such as Meloidogyne hapla and Globodera pallida, in which the VAChT genes consist of 10 and 13 coding exons, respectively (Figure S4). We identified potential R1- and R2-like sequence elements in both of these species (Figure S5).
A few other genes with multiple nested exons and coordinated exon skipping have been described in C. elegans: unc-60 encodes two alternate versions (UNC-60A and -B) of the actin-binding protein ADF (actin depolymerizing factor)/cofilin (McKim et al. 1994), avr-14 (gbr-2) encodes two similar subunits (AVR-14A and -B) of a glutamate-gated chloride channel (Laughton et al. 1997; Dent et al. 2000), and unc-49 encodes three similar subunits (UNC-49A, -B, and -C) of a GABA-gated chloride channel (Bamber et al. 1999). Transcripts from each of these loci share the first exon(s); the remaining unc-60A exons are nested in the long first intron of unc-60B, the remaining avr-14A exons are nested in a long avr-14B intron, the remaining unc-49A exons are nested in a long unc-49B intron, and most of unc-49B, in turn, is nested in a long unc-49C intron (McKim et al. 1994; Laughton et al. 1997; Bamber et al. 1999; Dent et al. 2000).
However, although these loci have similar genomic architecture, there are significant differences. The splicing events at the unc-60, avr-14, and unc-49 loci result in isoforms of a single protein, which share common coding sequences. In contrast, unc-17 and cha-1 transcripts share only a single noncoding exon (exon 1). The organization of the CGL is also conserved across phyla, whereas the UNC-60 and UNC-49 proteins are encoded by separate genes in higher phyla, and AVR-14 channels are present only in invertebrates. The unc-60A (nonmuscle) and unc-60B (muscle) transcripts are tissue specific with mutually exclusive expression (Ono et al. 1999, 2003); whereas avr-14A and avr-14B are coordinately expressed in a subset of neurons (Laughton et al. 1997; Dent et al. 2000), unc-49B and unc-49C are coordinately expressed in muscles (Bamber et al. 1999), and cha-1 and unc-17 are coordinately expressed in cholinergic neurons (Duerr et al. 2008). A splicing mechanism has been described for the unc-60 locus that does not involve stem-loop structures (Ohno et al. 2012; Kuroyanagi 2013), and we have been unable to identify R1- and R2-like complementary sequence elements at comparable locations in unc-60 or in the avr-14 and unc-49 loci (Table S1B). Therefore, complementary sequence elements do not appear to be a general feature of coordinated exon skipping in C. elegans.
(b) Improved efficiency of long splices
Another possible advantage of stem-loop structures is that they bring distant splice donor and acceptor sites into proximity; presumably, this would increase the efficiency of long splices. We note that the splice necessary to produce a cha-1 transcript is nominally 6.9 kb; this is much longer than most C. elegans splicing events (Kent and Zahler 2000). However, we have not identified complementary elements associated with other long splices in C. elegans (Table S1C), suggesting that stem-loop structures are not absolutely necessary for splicing over long distances.
It is also possible that stem-loop structures are useful where there is a significant disparity in the distance between the splice donor and the alternate splice acceptors. In such cases, there is likely to be preferential splicing to the proximal acceptor site; stem-loop structures could provide a compensatory mechanism to enhance splicing to the distal acceptor.
(c) Regulation of alternative splicing
Although it is clear that transgenic expression of the dual reporter leads to both GFP and mCherry fluorescence in most or all cholinergic neurons, we also observe that some cholinergic neurons preferentially express either GFP or mCherry (Figure 3C). This is consistent with previously published data that cholinergic neurons express CHA-1 and UNC-17 proteins in different ratios (Duerr et al. 2008); such results imply that the ratio of ChAT to VAChT is differentially regulated in specific neurons.
The immunostaining data in Figure 4, A–D, demonstrate that not only is UNC-17 abundance reduced in cn355 homozygotes, an expected consequence of the splice-site disruption, but also CHA-1 abundance is elevated. We observed comparable results when we introduced the cn355 mutation into the dual reporter: The GFP fluorescence was greatly reduced, and mCherry fluorescence was significantly increased (Figure 4, E–H). It appears that there is competition between the alternate splicing patterns, and decreasing the production of one of the mRNAs increases the production of the other. This mode of alternative splicing would make the splice choice a particularly sensitive site for regulation, and the R1 and R2 element pairs provide logical sites for such regulation.
A model of CGL splicing in C. elegans
The nature and the position of the R1 and R2 stem-loop structures suggest a model of how they might function. The model is also informed by results from another alternately spliced locus (see below). We propose the following sequence of steps for the production of cha-1 transcripts (Figure 7). During transcription, when the RNA polymerase complex reaches the downstream R2 (blue) element, the double-stranded R2 stem forms; this blocks the exon u2 acceptor site and impedes splicing from exon 1 to exon u2 (Figure 7, B and C). As exons u3 and u4 are transcribed, the introns preceding them are rapidly removed by standard splicing mechanisms (Figure 7, D and E); this removes potential interfering splice sites and also reduces the distance between the two R1 elements. When the RNA polymerase complex reaches the downstream R1 (red) element, the double-stranded R1 stem forms at or near the site(s) of unc-17 3′-end processing (Figure 7, E and F). The R1 stem structure reduces the probability or rate of 3′-end processing, possibly through an inhibitory interaction with the 3′-cleavage/polyadenylation complex. It also shortens the effective distance between the cha-1-specific splice donor and acceptor, increasing the efficiency of cha-1 splicing. As transcription then proceeds past the cha-1 exons, it becomes possible to splice from exon 1 to exon c2 (Figure 7G).
Figure 7.
Proposed sequence of events for production of cha-1 transcripts. (A) Genomic structure of the cholinergic (unc-17–cha-1) locus (for reference). (B and C) During transcription, when the RNA polymerase complex reaches the downstream R2 (blue) element, the double-stranded R2 stem forms; this blocks the exon u2 acceptor site and reduces the probability of splicing from exon 1 to exon u2. (D and E) As exons u3 and u4 are transcribed, the preceding introns are removed by standard splicing mechanisms. (E and F) When the RNA polymerase complex reaches the downstream R1 (red) element, the double-stranded R1 stem forms; this blocks the site of unc-17 3′-end processing and reduces the probability of cleavage and polyadenylation. (G) As transcription then proceeds into the cha-1 region, it becomes possible to splice from exon 1 to exon c2. Note that different scale bars are used for diagram A and diagrams B–G.
Cholinergic neurons need to express both unc-17 and cha-1, and each pre-mRNA can be processed to generate only one type of mRNA. We conclude that R2 inhibition of splicing and R1 inhibition of 3′-end processing are not absolute, but rather reduce the probability of splicing and 3′-end processing, respectively. Therefore, if each pair of complementary elements produced only partial inhibition, it would explain why disruption of both sets of elements simultaneously (Figure 5F) has more extreme consequences than disruption of either one by itself (Figure 5, B and D).
As noted previously, several other C. elegans genes (including unc-60, avr-14, and unc-49) have alternative transcripts and multiple nested exons; of these genes, detailed analysis of the splicing pattern has been reported for only the unc-60 locus. Production of the muscle-specific (distal) unc-60B transcript requires the activity of two muscle-specific RNA-binding proteins, SUP-12 and ASD-2. These proteins bind rapidly and cooperatively to target sequences near the splice acceptor site at the end of the first unc-60A intron, thus preventing the excision of that intron (Ohno et al. 2012; Kuroyanagi 2013). As transcription proceeds through the remaining unc-60A exons, they are rapidly spliced, and when the unc-60B exons are transcribed, exon 1 is able to undergo splicing to exon 2B.
Thus, the overall sequence of events for unc-60 is similar to what we propose for the CGL, although different mechanisms are employed to inhibit splicing to the proximal acceptor. Nevertheless, the requirements for “coordinated exon skipping” in general likely involve a mechanism to prevent or delay excision of the first intron, followed by the rapid excision of the subsequent introns. The rapid excision might merely require introns with canonical or near-canonical splice site sequences; consistent with this notion, we find that the splice sites flanking the unc-17 internal exons are good matches to the C. elegans consensus (Kent and Zahler 2000).
Trans-acting splicing factors
The question arises whether the proposed R1 and R2 stem-loop structures are sufficient by themselves to modulate splicing or if they function together with trans-acting factors. Presumably, if a stem-loop structure were properly positioned, it might be sufficient for steric inhibition of binding by RNA processing factors. In each of the Caenorhabditis species, the putative R2 stem-loop structure is positioned across or very close to the exon u2 splice acceptor site (Figure S2); these data are consistent with a model in which the R2 stem-loop structure is sufficient to inhibit unc-17-specific splicing. However, such a model suggests that blocking the splice acceptor site requires only proper positioning of the R2 stem and the specific R2 sequence is essentially irrelevant; thus, our data demonstrating the importance of both R2 complementarity and sequence for production of mCherry (and presumably CHA-1) (Figure 5, D and E) implicate sequence-specific factors in R2 function.
There is also evidence against a model of R1 stems overlapping and physically obstructing the site of unc-17 transcript 3′-end processing. Although a few of the polyadenylated C. elegans unc-17 cDNAs terminate within the downstream R1 sequence element, the majority terminate ∼35 nucleotides upstream of the R1 stem sequence (Mangone et al. 2010). This heterogeneity may be related to the lack of a canonical AAUAAA polyadenylation signal within the unc-17 3′-UTR. Four of the seven other Caenorhabditis species also lack a canonical AAUAAA sequence between the unc-17 termination codon and exon c2, and we therefore expect heterogeneity in the 3′-end processing sites of unc-17 transcripts from these species. Even in species containing an AAUAAA sequence, the predicted 3′-cleavage site in C. japonica is ∼41 nucleotides upstream of the R1 stem, and the predicted 3′-cleavage site in C. brenneri is ∼429 nucleotides downstream of the R1 stem (Mangone et al. 2010). Such variability makes it unlikely that R1 stem structures cover the 3′-cleavage site for all unc-17 transcripts in all Caenorhabditis species; rather, it suggests a model in which factors bound to the R1 stem structure block the cleavage and polyadenylation of the putative unc-17 transcript.
Approximately 300 genes in the C. elegans genome encode putative RNA-binding proteins (R. Barstead and J. Rand, unpublished observations); such proteins might be involved in recognizing and/or stabilizing RNA secondary structures (to promote production of cha-1 mRNA) or denaturing these structures (to promote production of unc-17 mRNA). To date, loss-of-function mutations in ∼12 C. elegans genes have been shown to affect alternative splicing of specific gene products (Barberan-Soler et al. 2011). We tested a subset of these mutants (unc-75, exc-7, mec-8, sup-12, sym-2, and fox-1), but did not observe any differences in the ratio of mCherry to GFP fluorescence, suggesting that these gene products do not participate in unc-17/cha-1 alternative splicing. However, the dual-reporter strain should be a useful tool for the isolation and analysis of mutations affecting unc-17/cha-1 splicing, and in a preliminary screen we have isolated a mutant affecting the mCherry/GFP ratio. This and related results will be the subject of a future publication.
Supplementary Material
Acknowledgments
We are grateful to Ryuji Hosono for the unc-17(cn355) strain, John McManus for sequencing assistance, and Jim Henthorn for assistance with confocal imaging. Some strains were obtained from the Caenorhabditis Genetics Center, which was supported by the National Center for Research Resources. These studies were supported by National Institutes of Health grants GM038679 and NS072923 (to J.B.R.).
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173765/-/DC1.
Communicating editor: O. Hobert
Literature Cited
- Alfonso A., Grundahl K., Duerr J. S., Han H.-P., Rand J. B., 1993. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261: 617–619. [DOI] [PubMed] [Google Scholar]
- Alfonso A., Grundahl K., McManus J. R., Asbury J. M., Rand J. B., 1994a Alternative splicing leads to two cholinergic proteins in Caenorhabditis elegans. J. Mol. Biol. 241: 627–630. [DOI] [PubMed] [Google Scholar]
- Alfonso A., Grundahl K., McManus J. R., Rand J. B., 1994b Cloning and characterization of the choline acetyltransferase structural gene (cha-1) from C. elegans. J. Neurosci. 14: 2290–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamber B. A., Beg A. A., Twyman R. E., Jorgensen E. M., 1999. The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J. Neurosci. 19: 5348–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak A. P., Lasda E. L., Wagner E. J., Garcia-Blanco M. A., 2003. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol. Cell. Biol. 23: 9327–9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan-Soler S., Medina P., Estella J., Williams J., Zahler A. M., 2011. Co-regulation of alternative splicing by diverse splicing factors in Caenorhabditis elegans. Nucleic Acids Res. 39: 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent J. A., Smith M. M., Vassilatis D. K., Avery L., 2000. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Lawrence C. E., 2003. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 31: 7280–7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Chan C. Y., Lawrence C. E., 2005. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr J. S., Han H.-P., Fields S. D., Rand J. B., 2008. Identification of major classes of cholinergic neurons in the nematode Caenorhabditis elegans. J. Comp. Neurol. 506: 398–408. [DOI] [PubMed] [Google Scholar]
- Eiden L. E., 1998. The cholinergic gene locus. J. Neurochem. 70: 2227–2240. [DOI] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R., 1994. Genesis of an organ: molecular analysis of the pha-1 gene. Development 120: 3005–3017. [DOI] [PubMed] [Google Scholar]
- Green, R. A., A. Audhya, A. Pozniakovsky, A. Dammermann, H. Pemble et al., 2008 Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo, pp. 179–218 in Methods in Cell Biology: Fluorescent Proteins, edited by F. S. Kevin. Academic Press, New York/London/San Diego. [DOI] [PubMed] [Google Scholar]
- Hobert O., 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730. [DOI] [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R., 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68. [DOI] [PubMed] [Google Scholar]
- Johnson C. D., Rand J. B., Herman R. K., Stern B. D., Russell R. L., 1988. The acetylcholinesterase genes of C. elegans: identification of a third gene (ace-3) and mosaic mapping of a synthetic lethal phenotype. Neuron 1: 165–173. [DOI] [PubMed] [Google Scholar]
- Kent W. J., Zahler A. M., 2000. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 10: 1115–1125. [DOI] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M., Hobert O., 2012. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H., 2013. Switch-like regulation of tissue-specific alternative pre-mRNA processing patterns revealed by customized fluorescence reporters. Worm 2: e23834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughton D. L., Lunt G. G., Wolstenholme A. J., 1997. Alternative splicing of a Caenorhabditis elegans gene produces two novel inhibitory amino acid receptor subunits with identical ligand binding domains but different ion channels. Gene 201: 119–125. [DOI] [PubMed] [Google Scholar]
- Mangone M., Manoharan A. P., Thierry-Mieg D., Thierry-Mieg J., Han T., et al. , 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F., Smith C. W. J., 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6: 386–398. [DOI] [PubMed] [Google Scholar]
- McKim K. S., Matheson C., Marra M. A., Wakarchuk M. F., Baillie D. L., 1994. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242: 346–357. [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., McNally F. J., 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C., Fire A., 1995. DNA transformation, pp. 451–482 in Caenorhabditis elegans: Modern Biological Analysis of an Organism, edited by Epstein H. F., Shakes D. C. Academic Press, San Diego. [Google Scholar]
- Muh S. J., Hovhannisyan R. H., Carstens R. P., 2002. A non-sequence-specific double-stranded RNA structural element regulates splicing of two mutually exclusive exons of fibroblast growth factor receptor 2 (FGFR2). J. Biol. Chem. 277: 50143–50154. [DOI] [PubMed] [Google Scholar]
- Mullen G. P., Mathews E. A., Saxena P., Fields S. D., McManus J. R., et al. , 2006. The Caenorhabditis elegans snf-11 gene encodes a sodium-dependent GABA transporter required for clearance of synaptic GABA. Mol. Biol. Cell 17: 3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman E. A., Muh S. J., Hovhannisyan R. H., Warzecha C. C., Jones R. B., et al. , 2006. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA 12: 1129–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno G., Ono K., Togo M., Watanabe Y., Ono S., et al. , 2012. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet. 8: e1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K., Parast M., Alberico C., Benian G. M., Ono S., 2003. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116: 2073–2085. [DOI] [PubMed] [Google Scholar]
- Ono S., Baillie D. L., Benian G. M., 1999. UNC-60B, an ADF cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm S., Ben Ari S., Rafalska I., Tang Y. S., Zhang Z. Y., et al. , 2005. Function of alternative splicing. Gene 344: 1–20. [DOI] [PubMed] [Google Scholar]
- Sun A. Y., Lambie E. J., 1997. gon-2, a gene required for gonadogenesis in Caenorhabditis elegans. Genetics 147: 1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick A. S., Hobert O., 2004. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell 6: 757–770. [DOI] [PubMed] [Google Scholar]
- Zuker M., 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.