An unusual multidomain glutaredoxin and its protein partner affect plant development and flowering time in relation to photoperiod in Arabidopsis.
Abstract
Glutaredoxins (GRXs) catalyze the reduction of protein disulfide bonds using glutathione as a reductant. Certain GRXs are able to transfer iron-sulfur clusters to other proteins. To investigate the function of Arabidopsis (Arabidopsis thaliana) GRXS17, we applied a strategy combining biochemical, genetic, and physiological approaches. GRXS17 was localized in the nucleus and cytosol, and its expression was elevated in the shoot meristems and reproductive tissues. Recombinant GRXS17 bound Fe2S2 clusters, a property likely contributing to its ability to complement the defects of a Baker’s yeast (Saccharomyces cerevisiae) strain lacking the mitochondrial GRX5. However, a grxs17 knockout Arabidopsis mutant exhibited only a minor decrease in the activities of iron-sulfur enzymes, suggesting that its primary function is as a disulfide oxidoreductase. The grxS17 plants were sensitive to high temperatures and long-day photoperiods, resulting in elongated leaves, compromised shoot apical meristem, and delayed bolting. Both environmental conditions applied simultaneously led to a growth arrest. Using affinity chromatography and split-Yellow Fluorescent Protein methods, a nuclear transcriptional regulator, the Nuclear Factor Y Subunit C11/Negative Cofactor 2α (NF-YC11/NC2α), was identified as a GRXS17 interacting partner. A mutant deficient in NF-YC11/NC2α exhibited similar phenotypes to grxs17 in response to photoperiod. Therefore, we propose that GRXS17 interacts with NF-YC11/NC2α to relay a redox signal generated by the photoperiod to maintain meristem function.
Glutaredoxins (GRXs) are small oxidoreductases structurally related to thioredoxins (TRXs) and present in most organisms (Rouhier et al., 2008; Meyer et al., 2009). Their capacity to reduce disulfide bonds is usually dependent on glutathione (GSH) and relies on a four-residue active-site motif comprising at least one redox-active Cys (Rouhier et al., 2006). In higher plants, GRXs are encoded by multigene families and subdivided into four classes (Couturier et al., 2009). GRXs from classes I and II are present in all photosynthetic organisms and possess, in most cases, the motifs CPXC and CGFS as active sites, respectively. GRXs from class III are restricted to terrestrial plants and display a CCXX motif. Class IV GRXs are present in both green algae and terrestrial plants and composed of three domains: one N-terminal GRX module carrying a CXXC/S motif followed by two domains of unknown function.
Through their biochemical function as disulfide reductases, GRXs are thought to alter the activity of metabolic enzymes and transcriptional factors (Michelet et al., 2005; Murmu et al., 2010; Couturier et al., 2014). They also participate in the regeneration of thiol-dependent antioxidant enzymes (Rouhier et al., 2001; Gama et al., 2007; Tarrago et al., 2009; Couturier et al., 2011). Also, other functions have been proposed for classes I and II GRXs owing to their capacity to bind iron-sulfur clusters (Rouhier et al., 2007, 2010; Bandyopadhyay et al., 2008). For instance, oxidized GSH promotes iron-sulfur cluster disassembly from human GRX2 and restores disulfide reductase activity; therefore, class I GRXs may constitute redox sensors (Lillig et al., 2005). GRXs belonging to class II, also named monothiol GRXs, seem intimately linked to iron metabolism. Those present in mitochondria participate in iron-sulfur cluster assembly, most likely as iron-sulfur transfer proteins from scaffold proteins to acceptor proteins (Rodríguez-Manzaneque et al., 2002; Mühlenhoff et al., 2003; Bandyopadhyay et al., 2008). In addition, nucleocytosolic monothiol GRXs participate in iron sensing and trafficking in Baker’s yeast (Saccharomyces cerevisiae) and animals (Ojeda et al., 2006; Pujol-Carrion et al., 2006; Kumánovics et al., 2008; Mühlenhoff et al., 2010; Haunhorst et al., 2013). However, it is not known if GRXs play a role in iron-sulfur cluster assembly or iron sensing in plants.
Recently, essential roles of plant GRXs have been unveiled in developmental processes and stress responses. Several Arabidopsis (Arabidopsis thaliana) GRXs from class III participate in the tolerance to photooxidative stress (Laporte et al., 2012) and defense against pathogens (Ndamukong et al., 2007; La Camera et al., 2011). Others are required for proper reproductive development through interaction with basic Leu zipper-type TGA transcription factors (Xing and Zachgo, 2008; Murmu et al., 2010). Concerning class I GRXs, an Arabidopsis mutant deficient in both GRXC1 and C2 has a lethal phenotype because of impaired embryo development (Riondet et al., 2012). Among the four class II GRXs (S14, S15, S16, and S17), the plastidial S14 isoform participates in arsenic tolerance in a hyperaccumulating fern (Pteris vittata; Sundaram et al., 2008) and is induced in response to high temperature in Arabidopsis (Sundaram and Rathinasabapathi, 2010). Tomato (Solanum lycopersicum) plants silenced for the expression of GRXS16 encoding another plastid-localized GRX display increased sensitivity to osmotic stress (Guo et al., 2010). GRXS14 and GRXS15 are presumed to participate in responses to oxidative stress (Cheng et al., 2006; Cheng, 2008). Concerning GRXS17, Arabidopsis knockout plants growing at 28°C exhibited impaired primary root growth, impaired flowering, and altered sensitivity to auxin (Cheng et al., 2011). Consistently, ectopic expression of Arabidopsis GRXS17 in tomato plants resulted in enhanced thermotolerance (Wu et al., 2012).
In this work, we examined the physiological role of Arabidopsis GRXS17, which belongs to class II and has three CGFS active sites, in relation to its biochemical functions. Recombinant GRXS17 incorporated Fe2S2 clusters and complemented the Baker’s yeast grx5 mutant. However, in plants, GRXS17 had a minor role in iron-sulfur cluster metabolism, because the activities of cytosolic iron-sulfur-dependent enzymes were not substantially altered in grxs17 mutant plants. In fact, grxS17 plants exhibited severe developmental defects as a consequence of a perturbed shoot meristem, specifically at elevated temperatures and in long-day conditions. We show that GRXS17 interacts with the Nuclear Factor Y Subunit C11/Negative Cofactor 2α (NF-YC11/NC2α), which is also involved in the control of plant development as a function of photoperiod duration. Our data indicate that GRXS17 plays an important role in meristem maintenance and suggest that this role is fulfilled through the relay of a redox-dependent signal to NF-YC11/NC2α.
RESULTS
Expression of GRXS17 in Arabidopsis Plant Organs and Subcellular Localization
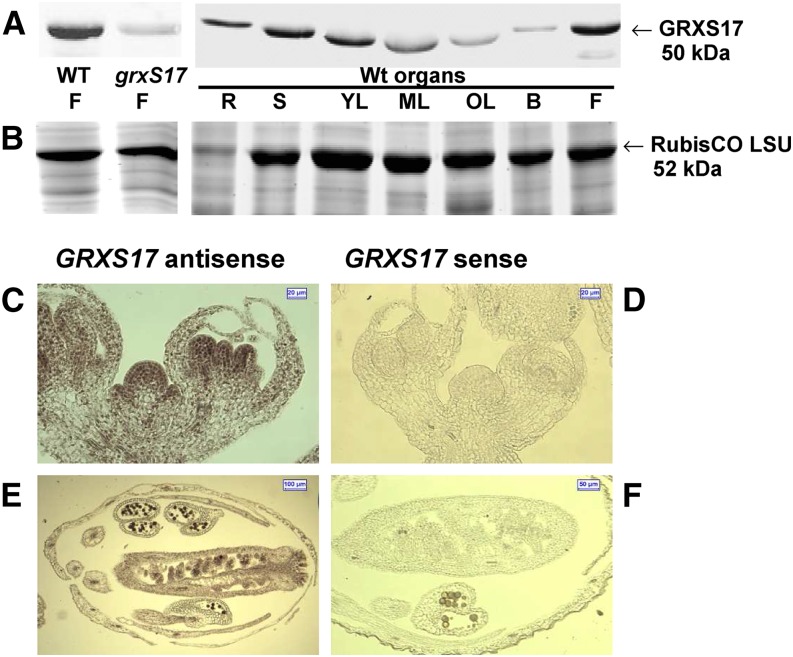
To investigate the function of GRXS17 in planta, we first analyzed its expression pattern. Previously published quantitative reverse transcription (RT)-PCR and promoter-GUS fusion data showed high GRXS17 expression in growing leaves and anthers of Arabidopsis (Cheng et al., 2011). To gain more detail, the GRXS17 protein abundance in various organs was determined using a serum raised against the whole recombinant protein. The serum specifically recognized a protein at approximately 50 kD as shown by western-blot analysis of flower protein extracts from the wild type, and the signal was absent in homozygous grxS17 plants (Fig. 1A; Supplemental Fig. S1). GRXS17 was substantially more abundant in stems, young leaves, and flowers. Note that the electrophoretic mobility of GRXS17 varies in leaf samples because of the proximity of Rubisco, which appears as a light-gray background band in grxS17 extracts (Fig. 1, A and B). In situ hybridization was performed on shoot apical meristem (SAM) and flowers of Arabidopsis wild-type plants. A strong digoxigenin staining was found in all meristematic cells, particularly stem cells (Fig. 1, C and D), pollen, and ovules (Fig. 1, E and F). These data reveal that GRXS17 is expressed in very different cell types localized in meristematic areas or reproductive and vascular organs.
Figure 1.
Expression of GRXS17 in Arabidopsis plants. A and B, Western analysis of GRXS17 abundance. A, Western-blot analysis in flowers from wild-type and grxS17 plants and organs of wild-type plants grown under standard conditions (20 µg per lane). The Rubisco Large Subunit (LSU) appears as a light-gray background band in the grxS17 lane. B, Floral bud; F, flower; ML, mature leaf; OL, old leaf; R, root from adult plant; S, stem from bolting plant; WT, wild type; YL, young leaf. B, Loading controls: Coomassie Blue-stained gels (50-kD region). C to F, RNA in situ hybridization. Longitudinal sections of wild-type meristem inflorescences (C and D) and flower buds (E and F). Hybridizations were performed with antisense (C and E) or sense (D and F) GRXS17 cDNA probes.
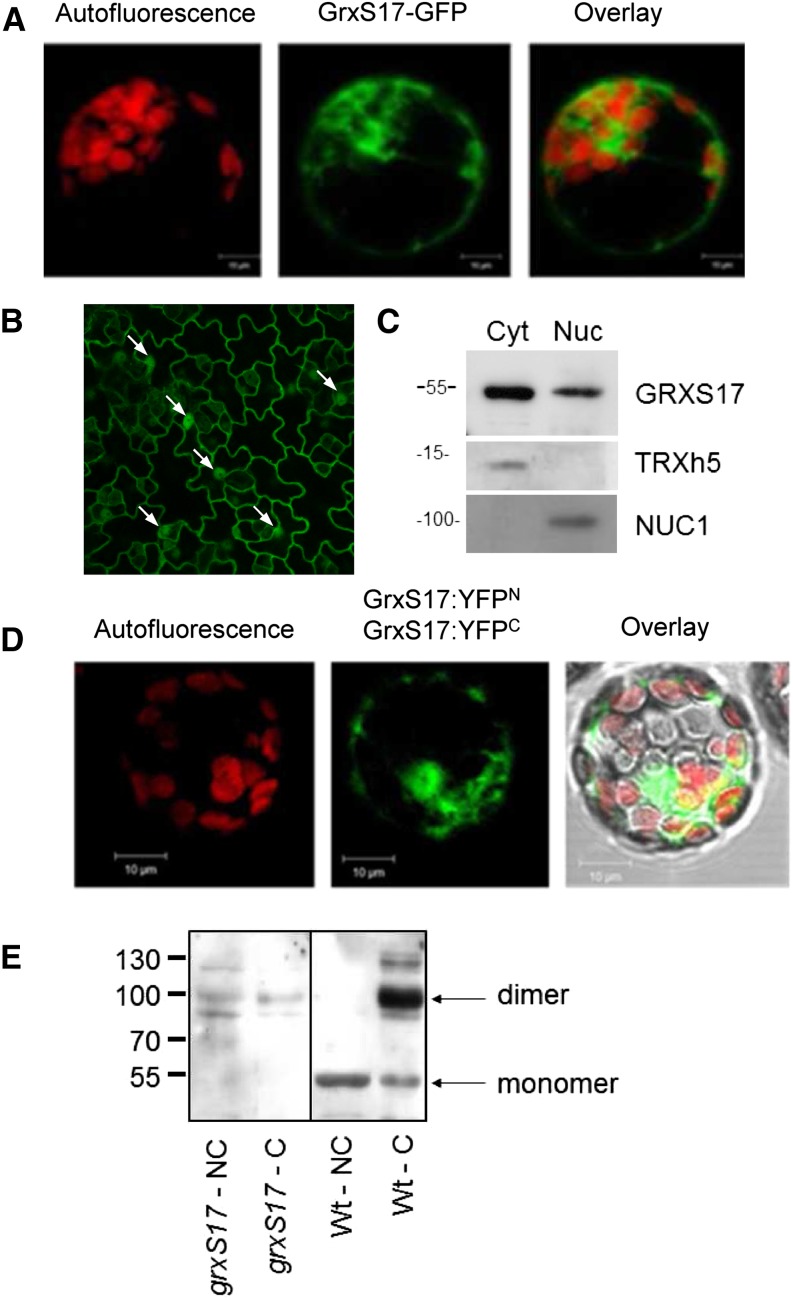
To determine the subcellular localization of GRXS17, transient expression in protoplasts and stable expression of P35S:GRXS17:GFP fusion were undertaken. The results indicated that the protein is targeted to both cytosol and nucleus (Fig. 2, A and B). The nuclear localization is surprising considering the absence of a recognizable nuclear localization signal in GRXS17 sequence and the size of the fusion protein, which should be too big to freely diffuse through nuclear pores. Therefore, we prepared nuclear and cytosolic fractions from Arabidopsis inflorescences. Their relative purity was verified using sera against cytosolic and nuclear markers TRXh5 (Marchal et al., 2014) and Nucleolin1 (NUC1; Pontvianne et al., 2010), respectively. GRXS17 was detected in both fractions in agreement with GRXS17-GFP localization (Fig. 2C). No signal for GRXS17 was detected in mitochondrial or chloroplastic extracts (Supplemental Fig. S2).
Figure 2.
Subcellular localization and dimerization of GRXS17. A, Transient expression of a GRXS17:GFP fusion in Arabidopsis mesophyll protoplasts. Autofluorescence of chlorophyll indicates chloroplasts. B, Stable expression of a GRXS17:GFP fusion in Arabidopsis leaves. White arrows indicate nuclei. C, Immunodetection of GRXS17, TRXh5, and nucleolin NUC1 in cytosolic (Cyt) and nuclear (Nuc) extracts of Arabidopsis flower buds. D, BiFC assay of GRXS17. Vectors encoding C- and N-terminal split-YFP fusions with GRXS17 were cotransformed into Arabidopsis mesophyll protoplasts. E, Immunodetection of GRXS17 in grxS17 and wild-type extracts cross-linked (C) or not (NC) with DMP. WT, Wild type.
Using bimolecular fluorescence complementation (BiFC; i.e. transient expression of two GRXS17 constructs fused to one-half Yellow Fluorescent Proteins (YFPs) in Arabidopsis protoplasts), we observed YFP signal confirming the nucleocytosolic localization and indicating that GRXS17 forms dimers in both compartments (Fig. 2D; Supplemental Fig. S3). The ability of GRXS17 to dimerize in vivo was further investigated in crude leaf extracts incubated with the cross-linker dimethyl pimelimidate/2 HCl (DMP). Upon nonreducing SDS-PAGE and western-blot analysis, a single band at 50 kD was apparent in untreated wild-type samples, and an additional 100-kD band, corresponding to a GRXS17 dimer, was specifically observed in the cross-linked extract (Fig. 2E). These data indicate that the GRX dimer is not formed by disulfide bridging.
Development of the grxS17 Mutant in Response to Photoperiod
To analyze the physiological function of AtGRXS17, we isolated homozygous grxS17 knockout plants from the Arabidopsis SALK_021301 line (Supplemental Fig. S1) and transformed this line with the GRXS17 complementary DNA (cDNA) under the control of the Cauliflower mosaic virus-35S promoter. Two independent grxS17 complemented lines, termed 3.3 and 17.8, were generated, and transgene expression was confirmed at the protein level (Supplemental Fig. S1). The phenotype characteristics of grxS17 mutant and complemented lines were investigated under various light and temperature conditions. When cultivated under standard conditions (22°C/18°C day-night regime, 8-h photoperiod, and 200 µmol photons m−2 s−1), all lines showed a similar development (Fig. 3A). Transfer of 2.5-week-old seedlings to 28°C and standard light for 2.5 weeks strongly impaired development of all genotypes, which displayed thin and elongated leaves. In addition, the grxS17 mutant failed to form new leaves, which was partially or entirely complemented in lines 3.3 and 17.8, respectively (Fig. 3B). These data indicate that GRXS17 is required for maintenance of the SAM at high temperature, consistent with the previously reported thermosensitivity of the grxS17 line (Cheng et al., 2011).
Figure 3.
Growth and development of plants modified in GRXS17 expression as a function of photoperiod and temperature. A, Five-week-old plants grown in standard conditions (8-h photoperiod and 200 µmol photons m−2 s−1) at 22°C. B, Plants grown for 2.5 weeks in standard conditions and transferred to 28°C (8-h photoperiod and 200 µmol photons m−2 s−1) for 2.5 weeks. The arrow indicates the absence of young leaves. C, Plants grown in long-day photoperiod conditions (16 h and 200 µmol photons m−2 s−1) at 22°C. D, Plants grown for 2 weeks in standard conditions and transferred to continuous light (200 µmol photons m−2 s−1 and 22°C) for 3 weeks. E, Plants grown for 2 weeks in standard conditions and transferred to 28°C and long-day photoperiod (16 h and 200 µmol photons m−2 s−1) for 2.5 weeks. F, Plants grown for 3 weeks in standard conditions and transferred to 15°C and long-day photoperiod (16 h and 200 µmol photons m−2 s−1) for 4 weeks. grxS17, Homozygous SALK_021301 plants; grxS17 3.3 and 17.8, two independent grxS17 lines expressing GRXS17; LD, long (16-h) day; SD, short (8-h) day; WT, wild type.
When plants were grown at 22°C and moderate light but under long-day photoperiod (16-h-day/8-h-night cycle), we observed that 4-week-old grxS17 plants displayed elongated and thickened lamina (Fig. 3C). The development of the main floral spike (raceme) was delayed, which entirely failed to form when plants were shifted to continuous light (Fig. 3D). To analyze whether the phenotype appearance is caused by day length or related to the total light inception, plants grown for 2 weeks under standard conditions were transferred to high-light (500 µmol photons m−2 s−1), 22°C, and short-day conditions. After 3 weeks, there was no change in grxS17 development in this light regime (Supplemental Fig. S4A), indicating that photoperiod duration is the primary determinant for the observed phenotype. When plants were grown under the same high-light intensity under long days, grxS17 exhibited strongly impaired development (Supplemental Figs. S4B and S5G). It is worth mentioning that the temperature measured at the plant level is elevated by 2°C (24°C) in high-light conditions, thus possibly explaining the more severe phenotype in long days at high light compared with moderate light. Under this light regime, there was no visual evidence of photooxidative damage in grxS17 leaves (Supplemental Fig. S4B). This was confirmed by autoluminescence imaging (Supplemental Fig. S6), which allows recording of the photon emission associated with lipid peroxidation (Havaux et al., 2006). When combining high temperature and long day, we observed that grxS17 growth stopped after a few days (Fig. 3E). Interestingly, the growth and the reproductive development of grxS17 plants cultivated for 4 weeks in long-day conditions at 15°C were not modified (Fig. 3F). Similarly, when young plants were transferred to long-day conditions at 15°C and high light (500 µmol photons m−2 s−1), no alteration was noticed in grxS17 (Supplemental Fig. S4C). Altogether, these data reveal that plants deficient in GRXS17 display sensitivity to a long-day regime in a temperature-dependent manner.
When grxS17 plants were grown under long-day and high-light conditions, a significant delay of bolting was observed. These plants only formed secondary floral spikes after some time (Supplemental Fig. S5, B, D, F, and G). In short-day conditions, floral development was even accelerated (Supplemental Fig. S5, A, C, and E), whereas grxS17 vegetative growth was not affected. Complemented lines exhibited contrasting phenotypes under short day: one line (3.3) flowering like the mutant and the other (17.8) flowering like the wild type. The difference could originate from the much higher GRXS17 amount in the latter (Supplemental Fig. S1). Collectively, these data point to the central role of GRXS17 in conveying environmental variations, such as temperature and day length, to coordinate the flowering response in plants.
The SAM Is Compromised in the grxS17 Mutant
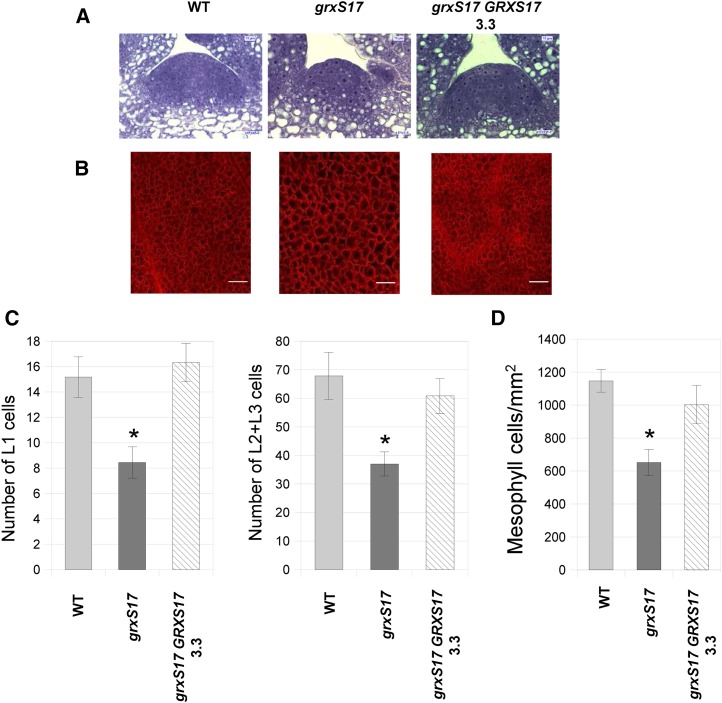
Because floral induction depends on the transition of the SAM from vegetative to reproductive fate (Levy and Dean, 1998), we performed histological analysis of SAM in grxS17 mutants. The SAM overall structure was not altered in the mutant grown in short-day conditions (Supplemental Fig. S7). On the contrary, when grown under long days, the meristem area was smaller in grxS17 compared with the wild type (Fig. 4A). The cell numbers in L1, L2, and L3 layers were 45% lower in grxS17 than in the wild type, revealing impairment in the division of stem cells (Fig. 4C). Moreover, the size of meristematic cells was noticeably increased in the mutant (Fig. 4A), suggesting that the lower cell division rates are associated with increased cell expansion. These changes in meristematic cell size and numbers are consistent with the high GRXS17 expression level observed in the meristem (Fig. 1C) and likely lead to the impaired development of grxS17 plants observed under conditions of long-day photoperiod and/or high temperature (Fig. 3; Supplemental Fig. S5). Histological analysis of mesophyll cells in plants grown under long-day and high-light conditions revealed a reduced cell density in mutant plants (652 ± 79 cells mm−2) compared with the wild type (1,148 ± 68 cells mm−2) and a much larger cell size (Fig. 4, B and D). These data indicate that GRXS17 is required for cell division under long-day conditions.
Figure 4.
Structure of SAM and leaves in grxS17 plants. A, Histological structure of the SAM stained by toluidine blue in 7-d-old wild-type, grxS17, and grxS17 GRXS17 (line 3.3) plants grown in long-day conditions (16 h) and high light (500 µmol photons m−2 s−1). B, Observation of mesophyll cells in leaves of 3-week-old plants grown under long-day/high-light conditions. C, Number of L1, L2, and L3 layer stem cells in the SAM cross sections shown in A. Ten sections per genotype were analyzed. D, Density of mesophyll cells in the sections shown in B (n = 12). WT, Wild type. *, Value significantly different from wild-type value with P < 0.05 (Student’s t test).
AtGRXS17 Architecture and Capacity to Bind Iron-Sulfur Clusters
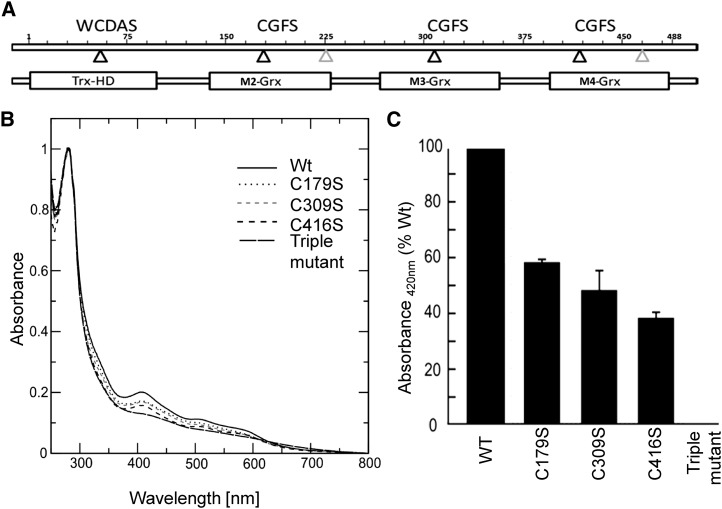
To investigate the biochemical function of AtGRXS17 and how this could affect meristem development, we first analyzed the capacity of the protein to bind iron-sulfur clusters. Arabidopsis GRXS17 possesses an N-terminal TRX-like domain with a WCDAS motif in place of the canonical WCGPC active site followed by three GRX domains containing CGFS motifs (Fig. 5A). This architecture is unique to land plants, because mammalian, fungal, and algal homologs consist of one TRX and maximally, two GRX domains (Couturier et al., 2009a, 2009b). The GRX modules of GRXS17 share 62% to 65% identity and are subsequently referred to as M2, M3, and M4. From secondary structure prediction and three-dimensional structure modeling, the four AtGRXS17 domains all adopt a classical TRX fold and are connected by long linker sequences (Supplemental Fig. S8). The capacity of recombinant AtGRXS17 to incorporate iron-sulfur clusters, like other Arabidopsis CGFS GRXs (Bandyopadhyay et al., 2008), was analyzed after anaerobic in vitro reconstitution mediated by the Cys desulfurase IscS in the presence of GSH. Indeed, upon purification of GRXS17, the oxygen-sensitive iron-sulfur clusters are lost; therefore, reconstitution of the clusters guarantees a sufficient amount of holo-GRXS17 for spectroscopy analysis. The UV-visible spectrum of the reconstituted GRXS17 showed absorbance peaks at 320 nm and around 420 nm, similar to other iron-sulfur cluster-coordinating GRXs and typical for Fe2S2 clusters (Fig. 5B). Estimation of the iron content in a freshly reconstituted wild-type protein indicated the presence of 2.48 ± 0.58 iron atoms per monomer. To investigate the contribution of each domain to cluster binding, the active-site cysteines were replaced by Ser either individually or in all three GRX domains. Whereas the triple-Cys mutant (C179/309/416S) did not incorporate any iron-sulfur cluster upon in vitro reconstitution, variants carrying one single substitution all incorporated between 40% and 60% of clusters as assessed by relative absorbance measurements at 420 nm (Fig. 5C). Because each of the active-site cysteines of the GRX subunits contributed to Fe2S2 incorporation, these data—together with the quantification of iron—indicate that AtGRXS17 dimers incorporate three Fe2S2 clusters in vitro, involving each GRX domain. Furthermore, the stoichiometry indicates that the GSH included in the reconstitution assay acts as an iron-sulfur cluster ligand as described for all other CGFS GRXs.
Figure 5.
Incorporation of iron-sulfur clusters into recombinant wild-type and mutated AtGRXS17. A, Domain structure of GRXS17. Positions of active-site cysteines are indicated by black triangles, and positions of other Cys are indicated by gray triangles. M2-GRX, M3-GRX, and M4-GRX are three monothiol-GRX domains. TRX-HD, TRX-like homology domain; WCDAS. B, Absorption spectra of GRXS17 and Cys mutants. UV-visible absorption spectra were recorded immediately after in vitro reconstitution in anaerobic conditions. The active-site cysteines of each GRX domain were individually or together substituted by Ser (M2:C179S, M3:C309S, M4:C416S, and C179/309/416S). C, Relative absorption at 420 nm of GRXS17 mutants. WT, Wild type.
Arabidopsis GRXS17 Rescues Most Baker’s Yeast grx5 Mutant Phenotypes
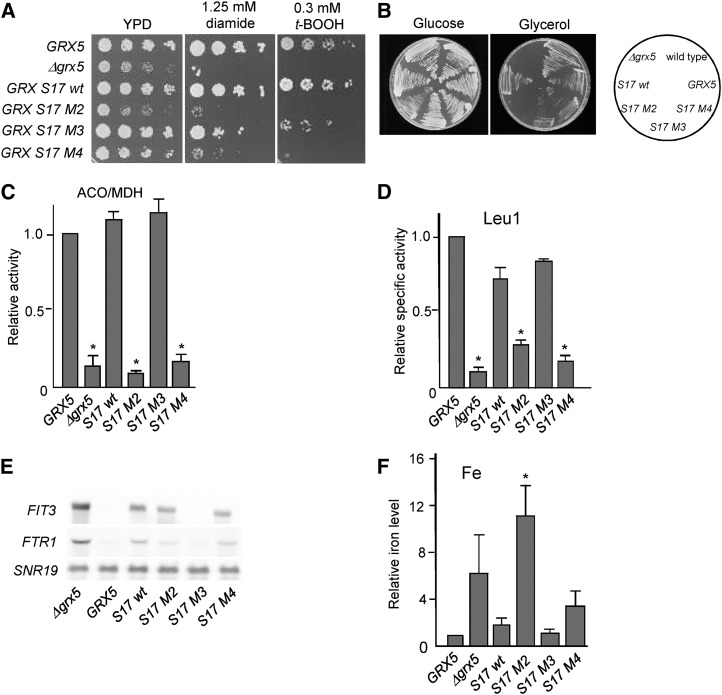
To further investigate a possible role of GRXS17 as an iron-sulfur cluster transfer protein, its capacity to rescue the defects of a Baker’s yeast grx mutant was examined. The entire protein or the three individual GRX domains were fused to the Grx5 mitochondrial targeting sequence and a C-terminal hemaglutinin (HA) tag. The constructs were expressed in Baker’s yeast, and the localization of the Arabidopsis proteins in the mitochondrial matrix was confirmed by western-blot analysis (Supplemental Fig. S9). Phenotype studies indicated that only the entire protein (the wild type) and the M3 module, to a lesser extent, rescued the sensitivity to two externally added oxidants, for which Baker’s yeast grx5 mutant cells are hypersensitive (Rodríguez-Manzaneque et al., 2002): tert-butyl hydroperoxide (causing general oxidative damage on cellular macromolecules) and diamide (specific oxidant of thiol groups; Fig. 6A). When grown under obligate respiratory conditions (glycerol as the carbon source), both the M3 and M4 modules in addition to the entire GRXS17 molecule totally or partially rescued the Baker’s yeast grx5-defective phenotype (Fig. 6B). The M3 module also fully rescued, like GRXS17, the ability to express active aconitase holoenzyme (Fig. 6C) and mostly restored isopropylmalate isomerase (Leu-1) activity (Fig. 6D). These two iron-sulfur-containing enzymes are located in mitochondria and cytosol, respectively, and both are dependent on the mitochondrial iron-sulfur cluster assembly pathway. We then checked the expression of iron uptake genes by determining the transcript levels of two reporter genes of the Activation of ferrous transport1 regulon, FTR1 (for Fe Transporter1) and FIT3 (for Facilitator of Iron Transport3). Of the three modules, only M3 restored repression of both genes in grx5 cells, whereas the entire GRXS17 molecule was unable to repress FTR1 and FIT3 expression (Fig. 6E), indicating that one component participating in iron status signaling is still deficient and that this could be caused by some steric incompatibility linked to the modular architecture of GRXS17. Accordingly, the M3 domain suppressed iron accumulation in grx5. Of note, expression of the entire GRXS17 protein also prevented iron accumulation (Fig. 6F). To summarize, the entire AtGRXS17 rescued most grx5 phenotypes, notably the activities of iron-sulfur-containing enzymes.
Figure 6.
Rescue of the Baker’s yeast grx5 mutant defects by AtGRXS17. A, Sensitivity to tert-butyl hydroperoxide (t-BOOH) or diamide. The indicated Baker’s yeast strains were spotted on YPD plates at 10× serial dilutions and grown for 3 d at 30°C on YPD plates. B, Growth on Glc (YPD plates) or glycerol (YPGly plates) after 3 d at 30°C. C, Relative ratio between aconitase and malate dehydrogenase (MDH) activities normalized with respect to the ratio in the wild-type strain in exponential cultures at 30°C in yeast extract-peptone-Gal medium. D, Relative specific Leu-1 activity. E, Northern-blot analysis of FIT3 and FTR1 mRNA levels from exponential cultures at 30°C. Loading control: SNR19 (for Small Nuclear RNA). F, Relative iron content from exponential cultures at 30°C in YPD medium. Mean ± sd (n = 3). ACO, Aconitase; WT, wild type. *, Statistically significant differences (Student’s t test, P < 0.05) compared with wild-type (GRX5) cultures.
GRXS17 Plays a Minor Role in Maintaining the Activity of Cytosolic Iron-Sulfur Enzymes
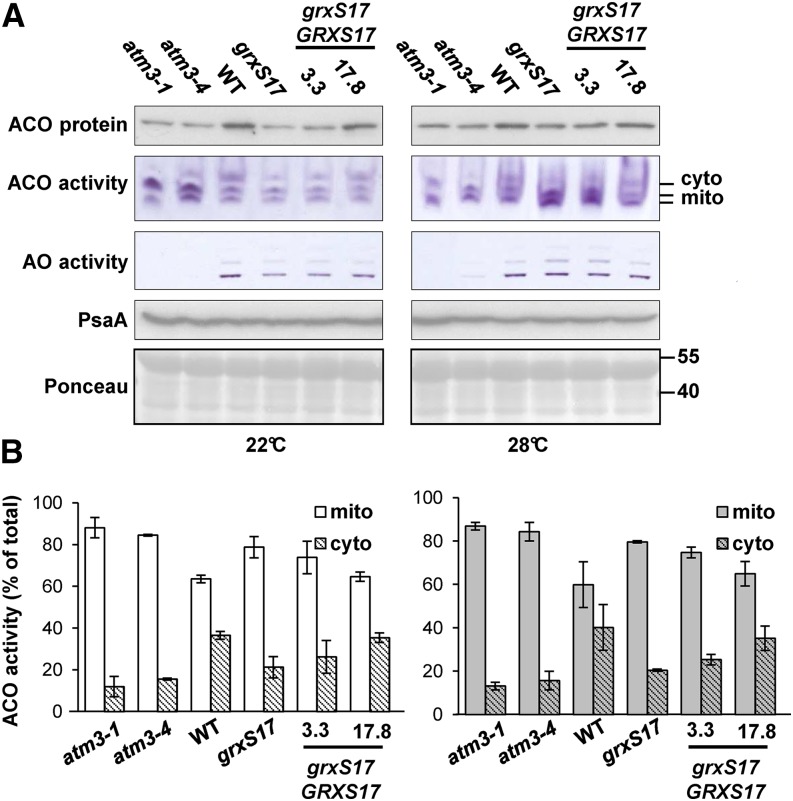
Next, we investigated whether GRXS17 fulfils a role in iron-sulfur cluster assembly in Arabidopsis. Therefore, activities and/or abundance of iron-sulfur enzymes were analyzed, including aconitases (one Fe4S4), aldehyde oxidases (two Fe2S2, FAD, and molybdenum cofactors), and PSI (three Fe4S4). All measurements were performed using 2-week-old seedlings grown under conditions where mild phenotypic changes are visible (22°C and 16 h of light) or conditions leading to severely impaired growth in grxS17 (plants shifted to 28°C and 16 h of light; Supplemental Fig. S10A). Two mutant alleles of ABC TRANSPORTER OF THE MITOCHONDRIA3/ABCB25 (ATM3) encoding a transporter that provides persulfide for cytosolic iron-sulfur cluster assembly (Schaedler et al., 2014) were used for comparison. The atm3-1 and atm3-4 mutants are strong and weak mutant alleles, respectively, that have significantly decreased activities of aldehyde oxidases and cytosolic aconitase (Bernard et al., 2009). Protein extracts from leaf samples were separated by native gel electrophoresis followed by in-gel activity staining. Part of the same protein extract was subjected to denaturing SDS-PAGE and western blotting to estimate protein levels of aconitase and PSI. As expected for a cytosolic protein, GRXS17 is not required for the maturation of mitochondrial (aconitases) and plastidial (PSI) iron-sulfur proteins (Fig. 7A). In grxS17 plants grown at 22°C, the amount of total aconitase protein was approximately 40% of the wild type, corresponding to a similar decrease in activity of the cytosolic isoform (Fig. 7). Aldehyde oxidase activity was decreased to approximately 50% in grxS17 at this temperature compared with less than 5% in atm3-4 (Fig. 7A, left; Supplemental Fig. S10B). In the complemented 3.3 line, the activities at 22°C of cytosolic aconitase and aldehyde oxidase were partially restored, and they were fully restored in the 17.8 line. In grxS17 plants grown at 28°C, the activities of the mitochondrial aconitase isoforms were increased, and total aconitase protein levels were close to wild-type levels (Fig. 7, right). Furthermore, there was no difference in aldehyde oxidase activity at this temperature (Supplemental Fig. S10B, right). Compared with the atm3 alleles, neither of which is a knockout, the grxS17 knockout mutant displayed a relatively moderate decrease in cytosolic iron-sulfur enzymes in environmental conditions that leads to strongly impaired development. Taken together, these data suggest that GRXS17, despite its capacity to bind iron-sulfur clusters in vitro and rescue the Baker’s yeast grx5 mutant, does not play a critical function in de novo synthesis of iron-sulfur clusters in planta.
Figure 7.
Activities and levels of iron-sulfur enzymes in plants modified in GRXS17 expression. The wild type, a grxS17 knockout mutant, and two complemented lines (3.3 and 17.8) were grown for 14 d under standard conditions (22°C) or transferred to 28°C after 7 d (Supplemental Fig. S10). Mutant alleles in the atm3-1 and atm3-4 were used for comparison. A, Activities of aconitase (ACO) and aldehyde oxidase (AO) isozymes visualized as a formazan precipitate in a native gel assay. Cytosolic (cyto) and mitochondrial (mito) isoforms of ACO are indicated. Equal protein amounts (160 μg for ACO and 60 μg for AO) were loaded per lane. Immunoblotting of ACO and PsaA (a subunit of PSI) in Arabidopsis seedlings (10 μg of protein per lane) and loading control (Ponceau). B, Relative activities of cyto and mito ACO isoforms from A quantified using ImageJ software. The values are given as percentages of the sum of the band intensities per lane (mean ± sd). The data are representative of three independent experiments. WT, Wild type.
Identification of a Nuclear Factor Interacting with GRXS17
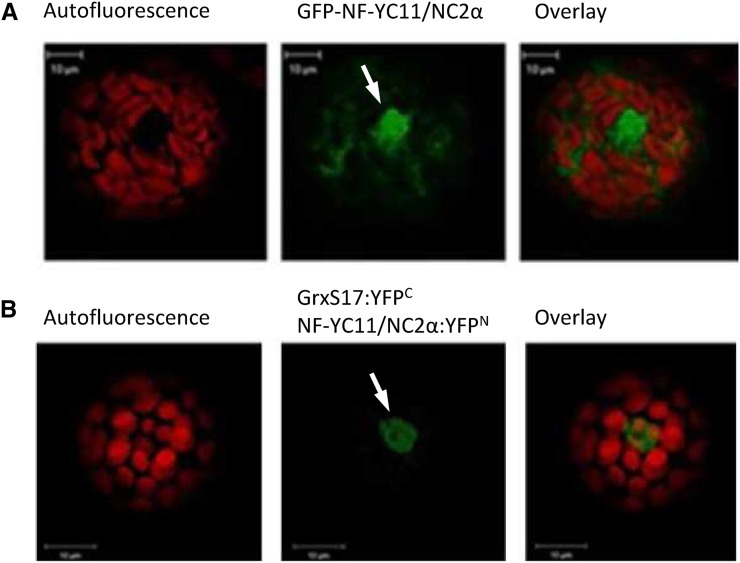
To explore other possibilities for how GRXS17 functions in meristem development, we performed affinity chromatography using a nickel matrix loaded with His-tagged GRXS17 to identify interacting proteins. After applying a crude leaf extract, the bound proteins were eluted with dithiothreitol (DTT) and identified by mass spectrometry. A number of proteins were repeatedly isolated in 15 independent affinity experiments (Table I). Interestingly, the predicted or experimentally determined localization of most proteins (e.g. cytosol or nucleus) is comparable with that of GRXS17. In accordance with a role of GRXS17 in redox signaling pathways and the known interaction of GRX3/protein kinase C-interacting cousin of thioredoxin with protein kinase C in animal cells, one transcription factor and one kinase were identified. The At3g12480 gene product NF-YC11/NC2α isolated in 7 of 15 experiments displayed high peptide sequence coverage. This partner was selected for deeper investigations, because proteins of the NF-Y family are known nuclear factors regulating developmental processes (Kumimoto et al., 2010). Transient expression of NF-YC11/NC2α-GFP in Arabidopsis protoplasts indicated that the protein is localized in both cytosol and nucleus (Fig. 8A), like GRXS17. The interaction between both proteins was further confirmed using BiFC exclusively in the nucleus (Fig. 8B; Supplemental Fig. S3).
Table I. Putative interaction partners of AtGRXS17.
The list includes the proteins identified at least four times in 15 independent affinity chromatography experiments with His-tagged AtGRXS17. CY, Cytosol; ER, endoplasmic reticulum; ES, extracellular space; MI, mitochondria; NU, nucleus; PL, plastid; PM, plasma membrane; PX, peroxisome.
| Accession No. | Protein Name | Function | Subcellular Localizationa | Sequence Length | Protein Identificationb | Sequence Coveragec |
|---|---|---|---|---|---|---|
| Amino acids | Times | % | ||||
| At4g04950 | GRXS17 | Cell redox homeostasis | CY, NU, PL | 488 | 15 | 90 |
| At1g50570 | Calcium-dependent lipid-binding family protein | Unknown | NU | 388 | 13 | 68 |
| At3g12480 | NF-YC11/NC2α | Transcription factor | CY, MI, NU, PL | 293 | 7 | 59 |
| At4g25860 | Oxysterol-binding protein-related protein 4A | Sterol transport | CY | 386 | 11 | 56 |
| At3g13460 | Calcineurin B-Like-Interacting Protein Kinase1 interacting protein Evolutionarily Conserved C-terminal Region2 | Unknown | CY, NU, PL | 667 | 6 | 52 |
| At2g39960 | Microsomal signal peptidase | Peptidase activity | CY, ER, MI, PM | 192 | 5 | 45 |
| At1g13440 | GapC2 | Glycolysis | CY, MI, NU, PL, PX | 338 | 9 | 41 |
| At5g46570 | Brassinosteroid-signaling kinase2 | Protein phosphorylation | CY, PM | 492 | 5 | 41 |
| At4g31180 | Asp-tRNA ligase-like protein | Aspartyl-tRNA synthetase activity | CY, PL | 270 | 4 | 31 |
| At5g11870 | Alkaline phytoceramidase | Hydrolase activity | ES, MI, PL, PM | 270 | 8 | 28 |
| At5g54050 | Cys/His-rich C1 domain-containing protein | Protein-disulfide reductase activity | ES, NU | 580 | 8 | 21 |
| At5g20830 | Suc synthase1 | UDP-glycosyltransferase activity | CY, MI | 808 | 4 | 19 |
| At1g24510 | T-complex protein1 subunit-ε | Chaperone activity | CY, MI, PM | 535 | 6 | 18 |
| At4g22030 | Probable F-box protein | Unknown | CY, MI, PL | 626 | 5 | 12 |
Subcellular localization predicted by the BAR Cell eFP Browser.
Proteins were identified because of the presence of their peptides in the indicated number of experiments.
Identified peptides with a MOWSE score higher than 15 were used to calculate the total sequence coverage.
Figure 8.
Localization of NF-YC11/NC2α and interaction with GRXS17. Isolated Arabidopsis mesophyll protoplasts were transiently transformed with an NF-YC11/NC2α:GFP fusion (A) or split-YFP constructs containing GRXS17 or NF-YC11/NC2α (B). White arrows indicate nuclei.
Development of nf-yc11/nc2α Plants as a Function of Photoperiod
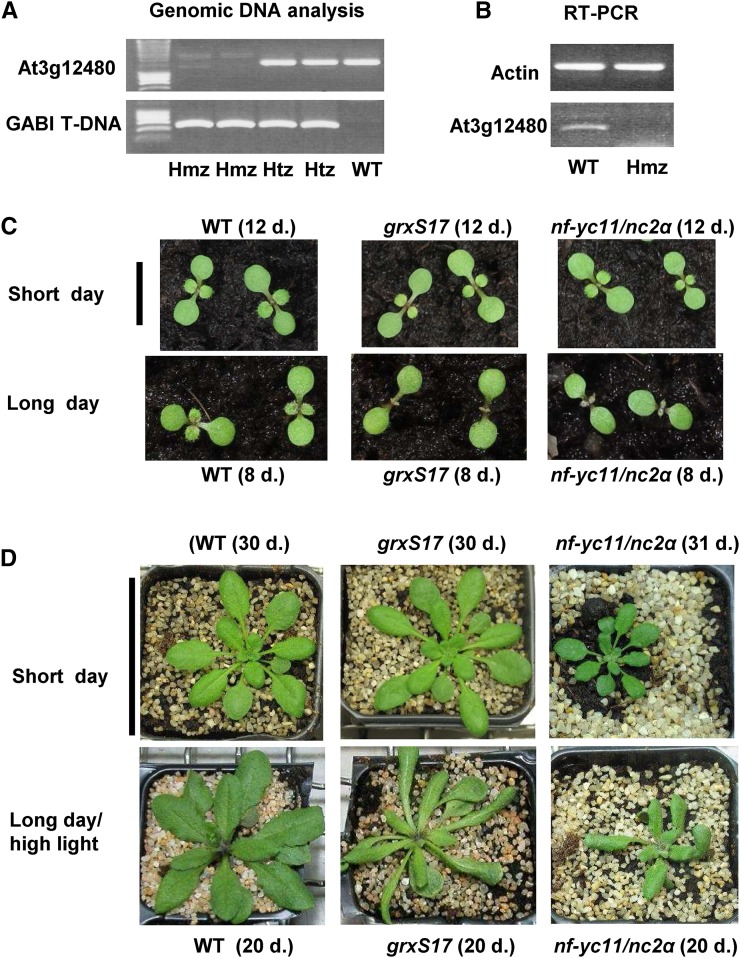
We isolated knockout plants for NF-YC11/NC2α expression from the German Plant Genomics Research Program-Kat 042E02 line (Kleinboelting et al., 2012), and only 4% as opposed to the expected 25% homozygous nf-yc11/nc2α plants were recovered in the T2 progeny (Fig. 9, A and B). When grown under short-day conditions and moderate light, nf-yc11/nc2α plants showed slow and altered development, with a compact rosette, small leaves, and a disorganized floral spike (Fig. 9D; Supplemental Fig. S11C). We transferred 33-d-old nf-yc11/nc2α plants grown in short- to long-day conditions and observed a strongly reduced development of the main spike and many secondary inflorescences compared with the wild type (Fig. 3C; Supplemental Fig. S11C). We then compared the development of wild-type, nf-yc11/nc2α, and grxS17 seedlings under short- and long-day conditions (Fig. 9C). We did not observe any difference in germination and cotyledon development for the three genotypes in both photoperiod conditions. Whereas in the short-day photoperiod, the first two leaves appeared at the same time in the three lines, the development of these two leaves was substantially delayed in nf-yc11/nc2α and grxS17 plants in long-day conditions (Fig. 9C). We analyzed the phenotype of nf-yc11/nc2α plants after sowing in long-day photoperiod and high-light conditions. We observed that growth and development of these plants were more perturbed than in control conditions (Fig. 9D). Particularly, nf-yc11/nc2α plants remained much smaller than wild-type and grxS17 plants. They exhibited elongated and distorted leaves, a trait characteristic shared by the grxS17 mutant. In these long-day conditions, no main floral spike was observed, and only secondary small spikes developed (Supplemental Fig. S11C). Taken together, these observations indicate that NF-YC11/NC2α participates in plant developmental processes in relation to the photoperiod duration and reveal that nf-yc11/nc2α and grxS17 plants share similar developmental characteristics in long-day conditions.
Figure 9.
Characterization of an nf-yc11/nc2α mutant and comparison of the growth of grxS17 and nf-yc11/nc2α plants as a function of photoperiod duration. A, PCR analysis of genomic DNA of At3g12480 and transfer DNA in wild-type, heterozygous, and homozygous plants of the German Plant Genomics Research Program-042E02 mutant line. B, RT-PCR analysis of wild-type and homozygous plants. C, Seedling development in short- and long-day conditions (8 and 16 h, respectively) at moderate light (200 µmol photons m−2 s−1) and 22°C. The seedlings were grown simultaneously in the same conditions, and the original photographs are shown in Supplemental Figure S10. D, Vegetative growth of plants in standard light conditions (short day, 8-h photoperiod and moderate light at 200 µmol photons m−2 s−1) and long-day and high-light conditions (16-h photoperiod and 500 µmol photons m−2 s−1) at 22°C. The plant age is indicated for each genotype and culture condition. Hmz, Homozygous; Htx, heterozygous; WT, wild type. Bar in C = 1 cm. Bar in D = 6 cm.
DISCUSSION
Physiological Function of GRXS17 in Relation to Its Biochemical Properties
In this work, we showed that GRXS17 is a central element for plant development in relation to environmental factors, such as photoperiod and temperature, and we investigated whether a major function in iron-sulfur protein biogenesis may underpin its physiological role. Previous studies indicated that multidomain GRX orthologs from Baker’s yeast and vertebrates bind iron-sulfur clusters and modify the activity of iron-responsive transcriptional regulators (Ojeda et al., 2006; Pujol-Carrion et al., 2006; Kumánovics et al., 2008; Mercier and Labbé, 2009; Jbel et al., 2011), affecting the intracellular iron distribution through the maturation of most iron-containing proteins (Mühlenhoff et al., 2010; Haunhorst et al., 2013). Accordingly, we showed that AtGRXS17 is able to bind Fe2S2 clusters by in vitro reconstitution experiments and complement the defects in iron-sulfur cluster maturation of the Baker’s yeast grx5 strain (Figs. 5 and 6). However, we found that GRXS17 in Arabidopsis does not play a major role in iron-sulfur protein biogenesis. Although grxS17 seedlings do have decreased activities of cytosolic aconitase and aldehyde oxidases in control conditions (Fig. 7), the effect is far less severe than that observed in the atm3 mutant lines (Bernard et al., 2009). Moreover, the aldehyde oxidase activity, which depends on two Fe2S2 clusters, is not decreased in grxS17 plants at 28°C, a temperature condition leading to a severe phenotype. Therefore, our data indicate that GRXS17 is not involved in de novo biosynthesis of cytosolic Fe2S2 clusters, which is in agreement with the viability of the knockout mutant. Indeed, because many cytosolic and nuclear iron-sulfur proteins are essential, mutants in iron-sulfur cluster assembly are generally embryo lethal (Balk and Schaedler, 2014). An alternative explanation is that the function of GRXS17 is redundant or compensated for by another component of the iron-sulfur cluster assembly pathway. However, because GRXS17 is the only class II GRX present in cytosol and nucleus, it is unlikely that such a function is fulfilled by another GRX. Taking into consideration the in vitro capacity of GRXS17 to bind iron-sulfur clusters and the variations observed in aconitase activity in grxS17 plants, we can speculate that it protects iron-sulfur proteins from oxidative stress and destruction of clusters. Such a hypothesis will need further investigation.
We assumed a function of GRXS17 in connection with its redox properties and hypothesized that it participates in signaling pathways related to the changes in the cellular redox status occurring in response to environmental variations. Of note, AtGRXS17 has been recently reported as prone to hydrogen peroxide-induced Cys sulfenation (Waszczak et al., 2014). Redox changes might affect the subcellular localization of AtGRXS17 or the set of interacting partners through posttranslational redox modifications. Concerning the first point, our experiments (GFP fusion and cellular fractionation) show that GRXS17 is localized in both nucleus and cytosol (Fig. 2), and a former study indicated that high temperature induces GRXS17 translocation from cytosol to nucleus (Wu et al., 2012). Thus, in response to environmental signals or a specific physiological state, the GRXS17 function might be associated with nucleocytosolic shuttling. With regard to 14 putative GRXS17 interaction partners identified by affinity chromatography, their localization is consistent with an interaction in vivo. Note that BOLA2, a possible transcriptional regulator interacting with GRXS17 in binary yeast two-hybrid and BiFC experiments (Couturier et al., 2014), was not isolated. This could originate from a low abundance or the type of plant material used in this work. Nevertheless, the data gained from affinity experiments clearly show the ability of GRXS17 to interact with different types of partners and thus, possibly modify their conformation or regulate their activity through posttranslational redox modification.
GRXS17: A Hub Integrating Hormonal and Redox Signals?
Consistent with the pleiotropic phenotype of grxS17 plants, western data revealed the presence of the GRXS17 protein in all organs, particularly those containing actively dividing or elongating cells. In addition, in situ hybridization showed a high transcript level in apical meristem, and histological analyses of plants grown in long-day conditions specifically revealed a larger cell size in both meristems and leaves of grxS17 plants. All of these data suggest a crucial role of GRXS17 in meristem activity and cell division. Several studies provided evidence for a tight relationship between intracellular redox status, disulfide reductases, and development (Considine and Foyer, 2014). For instance, the development of root meristems is influenced by changes in the overall redox status and auxin content/distribution (Vernoux et al., 2000; Jiang et al., 2003; Yu et al., 2013). Another example is the Arabidopsis NADPH-dependent thioredoxin reductase A (ntra), NADPH-dependent thioredoxin reductase B (ntrb), and cadmium-sensitive2 (cad2), ntra ntrb cad2 triple mutant, which is defective in TRX reduction and GSH synthesis and exhibits strongly impaired reproductive development in relation to altered auxin metabolism (Bashandy et al., 2010). The plastidial TRXm3 is essential for meristem maintenance in Arabidopsis through a role in symplastic permeability (Benitez-Alfonso et al., 2009), and the nucleoredoxin1 participates in the establishment of pollen fertility (Qin et al., 2009; Marchal et al., 2014). With regard to GRXs, the fate of germ cells in maize (Zea mays) anthers is determined by the redox status by a GRX, termed MALE STERILE CONVERTED ANTHER1, presumed to play a role in gene transcription (Kelliher and Walbot, 2012). The nuclear class III GRXs, ROXY1 and ROXY2, are required for proper development of floral organs in Arabidopsis, likely through interaction with TGA transcription factors (Xing and Zachgo, 2008; Hong et al., 2012). Altogether, these reports support the view that disulfide reductases finely control plant development.
Arabidopsis grxS17 plants display hypersensitivity to high temperature and altered auxin-mediated signaling pathways in roots (Cheng et al., 2011). This work unveils another function for GRXS17 in integrating photoperiod signals for proper development. Temperature is, however, an important determinant, because the phenotype is evident in long-day conditions at 22°C and 28°C but not at 15°C. In this study, we mainly investigated the phenotype of aerial parts of plants grown on soil and observed an altered shape of the leaves, which turned thick and elongated and displayed a reduced number of large cells. Looking for mutants with a similar phenotype (Bensmihen et al., 2008), we noticed that drl1 and elo mutants exhibit elongated leaves and a strongly reduced number of larger palisade cells compared with the wild type (Nelissen et al., 2003, 2005). DEFORMED ROOTS AND LEAVES1 (DRL1) regulates RNA polymerase II-mediated transcription through the elongator complex, which is composed of several ELONGATED (ELO) proteins and displays histone acetyltransferase activity preferentially in regions of auxin-related genes (Nelissen et al., 2010). Importantly, the floral development of drl1 plants is delayed, and the root development of both elo and drl1 mutants is substantially reduced (Nelissen et al., 2003). Interestingly, by investigating the genes coexpressed with GRXS17 using Genevestigator (Hruz et al., 2008), a high correlation was found with ELO2 (At5g13680). It is, thus, tempting to hypothesize that the grxS17 phenotype is linked to defects in ELO- and/or DRL1-dependent transcription mechanisms.
Possible Roles of GRXS17 in Interaction with the NF-YC11/NC2α Nuclear Factor
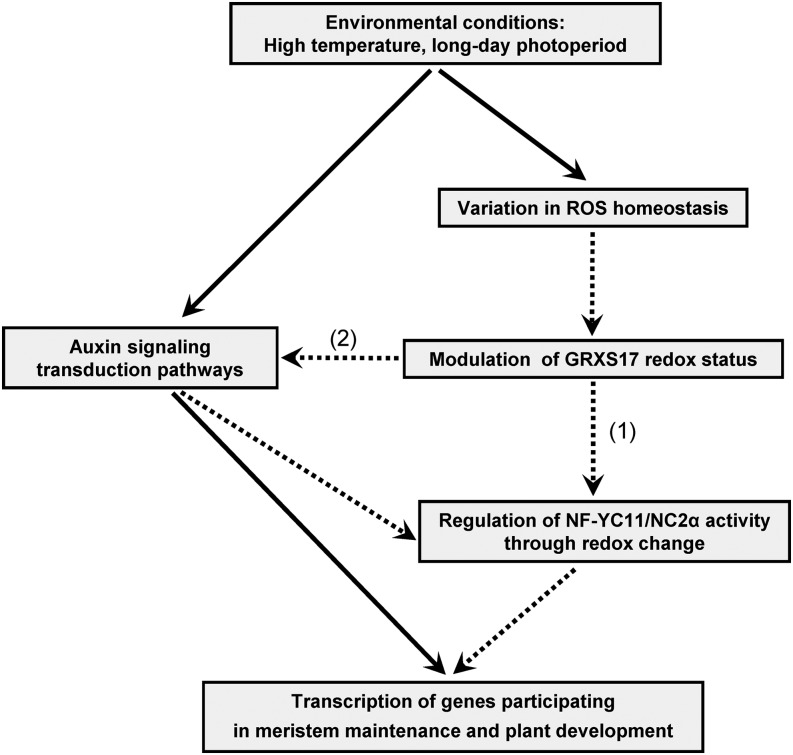
Among the GRXS17 partners identified using affinity chromatography, we focused on the nuclear factor NF-YC11/NC2α. Because the interaction between this factor and the GRX was confirmed by BiFC experiments in Arabidopsis protoplasts, a physiologically relevant interaction can be assumed. The Arabidopsis NF-Y family comprises three main types (A–C). Initially named CCAAT-box-binding factor or Heme Activator Protein (HAP), they usually form trimeric complexes of A, B, and C subunits binding to CCAAT-promoter sequences and transcriptionally regulating genes participating in plant development and stress responses (Dolfini et al., 2012; Laloum et al., 2013). In animal cells, they are central to cell-cycle progression (Benatti et al., 2011), and in fungi, the corresponding trimeric complex formed by HAP2, HAP3, and HAP5 includes a fourth HAP4 subunit. Interestingly, in Schizosaccharomyces pombe, the function and subcellular localization of HAP4, which participates in iron homeostasis, are under the control of the multidomain GRX4 (Mercier and Labbé, 2009). In relation to the grxS17 phenotype, it is worth mentioning that AtNF-YC11 specifically interacts with the Arabidopsis nuclear factor Y subunit B3 (AtNF-YB3), an isoform controlling flowering time (Kumimoto et al., 2008). To date, the only evidence for redox control was obtained for a mammalian NF-YB, which has an association to NF-YC that is dependent on the reduction of an intermolecular disulfide bond (Nakshatri et al., 1996). Interestingly, all plant NF-YC11 orthologs exhibit a unique N-terminal sequence clearly distinguishing them from other NF-YCs. On this basis, they have been reclassified as homologs to NC2α factors (Petroni et al., 2012). NC2α together with another factor called NC2β form a tight heterodimer able to associate with the TATA-binding complex and act as a transcription repressor as shown in Baker’s yeast and rice (Oryza sativa; Kim et al., 1997; Song et al., 2002). Two Cys residues are present in the NF-YC11/NC2α N-terminal extension (Supplemental Fig. S12). Of note, one is strictly conserved in plant orthologs and also, human NC2α protein, whereas the position of the second varies in Dicotyledons and Monocotyledons. Unfortunately, all attempts to produce recombinant NF-YC11/NC2α failed, which precluded investigations on a possible redox-mediated interaction with GRXS17. The data gained from the characterization of the Arabidopsis nf-yc11/nc2α mutant line (Fig. 9; Supplemental Fig. S11) revealed that the nuclear factor is a central element for proper plant development. In short-day conditions, nf-yc11/nc2α plants display developmental defects (slow growth and altered floral spikes). Most interestingly, comparing the phenotypes of grxS17 and nf-yc11/nc2α plants, we noticed that both mutants share common photoperiod-dependent characteristics, such as delayed appearance of the first two leaves, abnormal leaf shapes, and impaired flowering (Fig. 9; Supplemental Fig. S11). This phenotype resemblance, which is revealed in long- but not short-day conditions, gives further credence to a concerted action of GRXS17 and NF-YC11/NC2α in the control of plant development in relation to environmental conditions. We might, thus, speculate that GRXS17 modulates the NF-YC11/NC2α function by controlling its redox state, and we propose a working model illustrating such a role for GRXS17 (Fig. 10). Based on the resemblance of grxS17 and nf-yc11/nc2α mutants, it is conceivable that GRXS17-mediated redox changes modify the capacity of AtNF-YC11/NC2α to bind to an NC2β subunit (AtNF-YB11-13), ultimately resulting in the modification of the transcription level of genes involved in meristem maintenance and plant developmental programs, such as those related to auxin action (Fig. 10). Taken collectively, these data lead us to propose that Arabidopsis GRXS17 relays environmental signals, possibly through subtle changes in the cellular/nuclear redox state, and then, enters this information into the control of gene transcription to initiate essential developmental steps. The use of mutant lines expressing mutated GRXS17 and NF-YC11/NC2α forms will help to determine the precise mechanisms underlying the functions of these two key actors in plant development in relation to the presence and the redox status of their cysteines.
Figure 10.
Suggested role of GRXS17 during plant development in connection with environmental conditions and auxin-related mechanisms. Dotted lines indicate the proposed steps in signal transduction involving GRXS17. The model is based on the data presented in this work (1) and those reported in Cheng et al., 2011 (2). ROS, Reactive oxygen species.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia-0 plants were grown in standard conditions under an 8-h photoperiod and a photon flux density of 200 µmol photons m−2 s−1 at 22°C. Other conditions of light (500 µmol photons m−2 s−1), temperature (15°C or 28°C), and photoperiod (16 h or continuous light) were applied in controlled growth chambers either from sowing or on 2- to 3-week-old plants grown under standard conditions.
Transformation of Arabidopsis Plants
The full-length GRXS17 cDNA (At4g04950) was cloned into the pB2GW7 vector (GATEWAY; Invitrogen). After transformation using Agrobacterium tumefaciens C58 strain (Clough and Bent, 1998), homozygous lines (T2) were obtained from resistance segregation assays. Leaf genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen) to perform PCR using appropriate primers (Supplemental Table S1), Taq DNA Polymerase (Life Technologies), and the GeneAmp PCRSystem 2700 (Applied Biosystems). RT-PCR was performed using Sensiscript III (Life Technologies) after leaf RNA extraction (NucleoSpin; Macherey-Nagel).
Expression of the Recombinant Wild Type and Cys-Mutant GRXS17
AtGRXS17 cDNA was cloned into the pET-16b vector (Novagen; Merck Biosciences) for expression in Escherichia coli BL21(DE3)-pLysS. The protein was purified by nickel-chelate chromatography (GE Healthcare). Site-directed mutagenesis was performed using the QuikChange II protocol (Stratagene) and appropriate primers (Supplemental Table S3).
In Vitro Reconstitution Assay of Iron-Sulfur Clusters and Analytical Measurements
GRXS17 (50 µm) was reconstituted in vitro by incubation with Fe(NH4)2(SO4)2:Cys:GSH:pyridoxal phosphate:E. coli IscS in the molar ratio of 1 GRX:6:9:10:0.04:0.02 in 50 mm sodium phosphate buffer, pH 8.0, 300 mm NaCl, and 5 mm DTT under argon atmosphere for 2 h. After centrifugation (4 min at 4°C at 13,000 rpm), UV-visible spectra were recorded with a Shimadzu UV-2100 Spectrophotometer. Iron content was determined after reconstitution of 100 µm GRXS17 according to Fish (1988).
Cytology and in Situ Hybridization
Meristem cross sections were prepared using a microtome (Leica RM2255) from tissues fixed with formaldehyde/glutaraldehyde, embedded in hydroxyethyl methacrylate (Technovit 7100; Heraeus Kulzer), and counterstained with toluidine blue. Measurements of mesophyll cells were performed after propidium iodide staining (5 ng mL−1). Confocal microscopic observations were carried out using the Axio Observer Z1 Microscope with the LSM 700 Scanning Module, the ZEN 2010 software (Zeiss), and the propidium iodide (566–1,000 bp) filters. In situ hybridization was performed as in Bashandy et al. (2010). After fixation, dehydration, and embedding in paraffin wax, sample sections (7 mm thick) were attached to precoated glass slides (DAKO). Probes were synthesized using digoxigenin-UTP (Boehringer Mannheim). Immunodetection was performed using an antidigoxigenin antibody coupled to alkaline phosphatase.
Transient Transformation of Protoplasts with GFP and Split-YFP Constructs
Protoplasts were isolated from leaves of 6-week-old Arabidopsis plants for transient expression of fusion proteins (Wojtera-Kwiczor et al., 2012). cDNAs encoding GRXS17 and NF-YC11/NC2α were cloned in the sense direction into the pGFP2-vector for C-terminal GFP fusion. The GRXS17 and NF-YC11/NC2α cDNAs were inserted in frame with YFP N- and C-terminal parts in pSPYNE and pSPYCE vectors obtained from Jörg Kudla (University of Muenster). Combinations of empty vectors and fusion constructs were used as controls. GFP and chlorophyll autofluorescence were visualized with excitation at 488 nm and emission at 500 to 530 and 650 to 710 nm, respectively, using the cLSM 510 META (Carl Zeiss). YFP was visualized with excitation at 514 nm and emission at 535 to 590 nm.
Affinity Chromatography and Electrospray Ionization Mass Spectrometry
His-tagged GRXS17 was bound to a nickel-nitrilotriacetic acid column and used as affinity matrix. Leaves of 5-week-old plants grown under short-day conditions were homogenized in 20 mL of 50 mm Bicine, pH 7.8, 100 mm Suc, and 50 mm NaCl. After filtration through Miracloth and centrifugation (10 min at 6,000g at 4°C and 50 min at 4°C at 100,000g), the clarified supernatant (30 mg of protein) was applied to the matrix and incubated for 2 h at 4°C. Nonbound material was removed by washing the column four times with 10 mL of 20 mm Bicine, pH 7.8. Elution was achieved with 4 mL of the same buffer containing 150 mm DTT. After tryptic digestion (50 µg of proteins per analysis), the fragments were separated by reverse-phase HPLC and analyzed by electrospray ionization-mass spectrometry (Holtgrefe et al., 2008). Results were analyzed using the Brucker Daltronics software.
Biochemical Methods
Soluble proteins were prepared from plant material, separated by SDS-PAGE, and electrotransferred onto a nitrocellulose membrane (Pall Corporation; Rey et al., 2005). Protein cross-linking was achieved using DMP (Thermo Fisher Scientific; Riondet et al., 2012). Polyclonal antibodies were raised in rabbit against His-tagged AtGRXS17 (Genecust). Immunodetection of AtGRXS17 was carried out using primary antibodies diluted 1:1,000 and the goat anti-rabbit Alexa Fluor 680 IgG at 1:10,000 (Invitrogen). Bound antibodies were revealed at 680 nm using the Odyssey Infrared Imager (LiCor). For immunodetection of aconitase (Bernard et al., 2009) and PsaA (Agrisera), horseradish peroxidase-conjugated secondary antibodies and chemiluminescence were used. Autoluminescence imaging of lipid peroxidation was performed as in Collin et al. (2008).
Baker’s Yeast Plasmids, Strains, and Growth Conditions
The GRXS17 entire sequence and each GRX module, M2 (S17167–252), M3 (S17297–383), and M4 (S17404–488), were cloned in frame in the Baker’s yeast (Saccharomyces cerevisiae) plasmid pMM221. pMM221 contains the Baker’s yeast GRX5 mitochondrial targeting sequence plus a C-terminal 3HA/His-6 tag under the control of the doxycycline-regulatory tetO2 promoter (Supplemental Tables S1 and S2; Molina et al., 2004). pMM54 contains a Baker’s yeast GRX5-3HA construction under its endogenous promoter (Rodríguez-Manzaneque et al., 2002). Strains are described in Supplemental Table S3. Plasmids were linearized by ClaI previous to chromosomal integration. Samples were taken from cultures grown exponentially (Molina et al., 2004) for at least 10 generations at 30°C. Sensitivity to oxidants was determined on yeast extract-peptone-dextrose plates by spotting 1:5 serial dilutions of exponential cultures and recording growth after 2 d at 30°C. Subfractionation of mitochondria was performed as in Bandyopadhyay et al. (2008). Northern-blot analyses using Baker’s yeast RNA were performed with digoxigenin (Bellí et al., 1998). Gene probes were generated by PCR from genomic DNA using appropriate oligonucleotides (Supplemental Table S3).
Enzyme Activity Determinations
Aconitase and malate dehydrogenase were assayed in extracts from Baker’s yeast growing exponentially in yeast extract-peptone-Gal medium (Robinson et al., 1987). Isopropylmalate isomerase activity was determined in extracts prepared from cells growing exponentially in synthetic complete medium supplemented with the specific auxotrophy requirement (Pierik et al., 2009). In the case of Leu, only one-third of the standard concentration was added into the medium to allow growth. In-gel activity assays for aldehyde oxidase and aconitase were as previously described (Bernard et al., 2009).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Molecular characterization of Arabidopsis plants modified in GRXS17 expression and GRXS17 protein abundance.
Supplemental Figure S2. Subcellular localization of GRXS17.
Supplemental Figure S3. Controls of the bimolecular fluorescence complementation assay of GRXS17 and NF-YC11/NC2α.
Supplemental Figure S4. Growth of Arabidopsis plants modified in GRXS17 expression as a function of light and temperature.
Supplemental Figure S5. Floral development of Arabidopsis plants modified in GRXS17 expression as a function of photoperiod and light intensity.
Supplemental Figure S6. Autoluminescence in Arabidopsis plants modified in GRXS17 expression.
Supplemental Figure S7. Structure of the shoot apical meristem in the grxS17 mutant.
Supplemental Figure S8. Hypothetical structure of GRXS17.
Supplemental Figure S9. Analysis of the GRXS17 forms expressed in yeast.
Supplemental Figure S10. Growth phenotypes and quantification of aldehyde oxidase enzyme activity.
Supplemental Figure S11. Development of nf-yc11/nc2α plants as a function of light environment.
Supplemental Figure S12. Sequence alignment of plant, human, and yeast proteins containing an NC2α domain.
Supplemental Table S1. List of primers used in the study.
Supplemental Table S2. Plasmids used for experiments on Baker’s yeast.
Supplemental Table S3. Baker’s yeast strains.
Supplementary Material
Acknowledgments
We thank the Groupe de Recherche Appliquée en Phytotechnologie (Commissariat à l’Energie Atomique, Institut de Biologie Environnementale et Biotechnologie, and Service de Biologie Végétale et Microbiologie Environnementales) for technical assistance with controlled growth chambers; Patricia Henri (Commissariat á l'Energie Atomique, Institut de Biologie Environnementale et Biotechnologie, and Service de Biologie Végétale et Microbiologie Environnementales) for valuable technical assistance; Delphine Cerveau, Brigitte Ksas, and Dr. Michel Havaux (Commissariat á l'Energie Atomique, Institut de Biologie Environnementale et Biotechnologie, and Service de Biologie Végétale et Microbiologie Environnementales) for helpful assistance in phenotype characterization and imaging experiments; and Drs. Mohammed Bendahmane and Michiel Vandenbussche (Laboratoire de Reproduction et Développement des Plantes-Unité Mixte de Recherche 5667 Ecole Normale Supérieure-Centre National de la Recherche Scientifique-Institut National de la Recherche Agronomique-Université de Lyon) for fruitful discussion about NF-Y transcription factors.
Glossary
- BiFC
bimolecular fluorescence complementation
- cDNA
complementary DNA
- DMP
dimethyl pimelimidate/2 HCl
- DTT
dithiothreitol
- GSH
glutathione
- RT
reverse transcription
- SAM
shoot apical meristem
Footnotes
This work was supported by the Agence Nationale de la Recherche (grant nos. 2010 BLAN–1616 to C.R., N.B., S.T., J.G.-M., F.G., J.-P.R., N.R., and P.R.; BLAN 12–BSV6–0011 to C.R. and J.-P.R.; and JC07–204825 to N.R. and P.R.) and the German Research Foundation (grant nos. SPP 1710 BE3259/5–1 to C.B. and SCHE217/16–1 to R.S.).
References
- Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65: 125–153 [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, et al. (2008) Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J 27: 1122–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashandy T, Guilleminot J, Vernoux T, Caparros-Ruiz D, Ljung K, Meyer Y, Reichheld JP (2010) Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellí G, Garí E, Piedrafita L, Aldea M, Herrero E (1998) An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res 26: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatti P, Dolfini D, Viganò A, Ravo M, Weisz A, Imbriano C (2011) Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res 39: 5356–5368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D (2009) Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc Natl Acad Sci USA 106: 3615–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, Hanna AI, Langlade NB, Micol JL, Bangham A, Coen ES (2008) Mutational spaces for leaf shape and size. HFSP J 2: 110–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DG, Cheng Y, Zhao Y, Balk J (2009) An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol 151: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH. (2008) AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Lett 582: 848–854 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelson RS, Hirschi KD (2006) AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. J Biol Chem 281: 26280–26288 [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Liu X, Wu Q, Thompson SM, Lin J, Chang J, Whitham SA, Park S, Cohen JD, et al. (2011) Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J Biol Chem 286: 20398–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Collin VC, Eymery F, Genty B, Rey P, Havaux M (2008) Vitamin E is essential for the tolerance of Arabidopsis thaliana to metal-induced oxidative stress. Plant Cell Environ 31: 244–257 [DOI] [PubMed] [Google Scholar]
- Considine MJ, Foyer CH (2014) Redox regulation of plant development. Antioxid Redox Signal 21: 1305–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Jacquot JP, Rouhier N (2009) Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell Mol Life Sci 66: 2539–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Ströher E, Albetel AN, Roret T, Muthuramalingam M, Tarrago L, Seidel T, Tsan P, Jacquot JP, Johnson MK, et al. (2011) Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron-sulfur cluster binding into glutaredoxins. J Biol Chem 286: 27515–27527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier J, Wu HC, Dhalleine T, Pégeot H, Sudre D, Gualberto JM, Jacquot JP, Gaymard F, Vignols F, Rouhier N (2014) Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1. Mol Plant 7: 187–205 [DOI] [PubMed] [Google Scholar]
- Dolfini D, Gatta R, Mantovani R (2012) NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol 47: 29–49 [DOI] [PubMed] [Google Scholar]
- Fish WW. (1988) Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol 158: 357–364 [DOI] [PubMed] [Google Scholar]
- Gama F, Keech O, Eymery F, Finkemeier I, Gelhaye E, Gardestrom P, Dietz KJ, Rey P, Jacquot JP, Rouhier N (2007) The mitochondrial type II peroxiredoxin from poplar. Physiol Plant 129: 196–206 [Google Scholar]
- Guo Y, Huang C, Xie Y, Song F, Zhou X (2010) A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 232: 1499–1509 [DOI] [PubMed] [Google Scholar]
- Haunhorst P, Hanschmann EM, Bräutigam L, Stehling O, Hoffmann B, Mühlenhoff U, Lill R, Berndt C, Lillig CH (2013) Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol Biol Cell 24: 1895–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havaux M, Triantaphylidès C, Genty B (2006) Autoluminescence imaging: a non-invasive tool for mapping oxidative stress. Trends Plant Sci 11: 480–484 [DOI] [PubMed] [Google Scholar]
- Holtgrefe S, Gohlke J, Starmann J, Druce S, Klocke S, Altmann B, Wojtera J, Lindermayr C, Scheibe R (2008) Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol Plant 133: 211–228 [DOI] [PubMed] [Google Scholar]
- Hong L, Tang D, Zhu K, Wang K, Li M, Cheng Z (2012) Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell 24: 577–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jbel M, Mercier A, Labbé S (2011) Grx4 monothiol glutaredoxin is required for iron limitation-dependent inhibition of Fep1. Eukaryot Cell 10: 629–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Meng YL, Feldman LJ (2003) Quiescent center formation in maize roots is associated with an auxin-regulated oxidizing environment. Development 130: 1429–1438 [DOI] [PubMed] [Google Scholar]
- Kelliher T, Walbot V (2012) Hypoxia triggers meiotic fate acquisition in maize. Science 337: 345–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Na JG, Hampsey M, Reinberg D (1997) The Dr1/DRAP1 heterodimer is a global repressor of transcription in vivo. Proc Natl Acad Sci USA 94: 820–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumánovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, et al. (2008) Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J Biol Chem 283: 10276–10286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto RW, Adam L, Hymus GJ, Repetti PP, Reuber TL, Marion CM, Hempel FD, Ratcliffe OJ (2008) The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 228: 709–723 [DOI] [PubMed] [Google Scholar]
- Kumimoto RW, Zhang Y, Siefers N, Holt BF III (2010) NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J 63: 379–391 [DOI] [PubMed] [Google Scholar]
- La Camera S, L’haridon F, Astier J, Zander M, Abou-Mansour E, Page G, Thurow C, Wendehenne D, Gatz C, Métraux JP, et al. (2011) The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J 68: 507–519 [DOI] [PubMed] [Google Scholar]
- Laloum T, De Mita S, Gamas P, Baudin M, Niebel A (2013) CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci 18: 157–166 [DOI] [PubMed] [Google Scholar]
- Laporte D, Olate E, Salinas P, Salazar M, Jordana X, Holuigue L (2012) Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J Exp Bot 63: 503–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Dean C (1998) The transition to flowering. Plant Cell 10: 1973–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Vergnolle O, Lönn ME, Hudemann C, Bill E, Holmgren A (2005) Characterization of human glutaredoxin 2 as iron-sulfur protein: a possible role as redox sensor. Proc Natl Acad Sci USA 102: 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C, Delorme-Hinoux V, Bariat L, Siala W, Belin C, Saez-Vasquez J, Riondet C, Reichheld JP (2014) NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol Plant 7: 30–44 [DOI] [PubMed] [Google Scholar]
- Mercier A, Labbé S (2009) Both Php4 function and subcellular localization are regulated by iron via a multistep mechanism involving the glutaredoxin Grx4 and the exportin Crm1. J Biol Chem 284: 20249–20262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP (2009) Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu Rev Genet 43: 335–367 [DOI] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Marchand C, Collin V, Decottignies P, Tsan P, Lancelin JM, Trost P, Miginiac-Maslow M, Noctor G, et al. (2005) Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc Natl Acad Sci USA 102: 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina MM, Bellí G, de la Torre MA, Rodríguez-Manzaneque MT, Herrero E (2004) Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J Biol Chem 279: 51923–51930 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U, Gerber J, Richhardt N, Lill R (2003) Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J 22: 4815–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, et al. (2010) Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab 12: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li S, Xu M, Khan M, Malcolmson C, Fobert PR, Zachgo S, Hepworth SR (2010) Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol 154: 1492–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Bhat-Nakshatri P, Currie RA (1996) Subunit association and DNA binding activity of the heterotrimeric transcription factor NF-Y is regulated by cellular redox. J Biol Chem 271: 28784–28791 [DOI] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C (2007) SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J 50: 128–139 [DOI] [PubMed] [Google Scholar]
- Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, Dyer T, Lust S, Inzé D, Van Lijsebettens M (2003) DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 15: 639–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, De Groeve S, Fleury D, Neyt P, Bruno L, Bitonti MB, Vandenbussche F, Van der Straeten D, Yamaguchi T, Tsukaya H, et al. (2010) Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc Natl Acad Sci USA 107: 1678–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, Micol JL, Van Montagu M, Inzé D, Van Lijsebettens M (2005) The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA 102: 7754–7759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojeda L, Keller G, Mühlenhoff U, Rutherford JC, Lill R, Winge DR (2006) Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J Biol Chem 281: 17661–17669 [DOI] [PubMed] [Google Scholar]
- Petroni K, Kumimoto RW, Gnesutta N, Calvenzani V, Fornari M, Tonelli C, Holt BF III, Mantovani R (2012) The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24: 4777–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik AJ, Netz DJ, Lill R (2009) Analysis of iron-sulfur protein maturation in eukaryotes. Nat Protoc 4: 753–766 [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Abou-Ellail M, Douet J, Comella P, Matia I, Chandrasekhara C, Debures A, Blevins T, Cooke R, Medina FJ, et al. (2010) Nucleolin is required for DNA methylation state and the expression of rRNA gene variants in Arabidopsis thaliana. PLoS Genet 6: e1001225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA (2006) Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J Cell Sci 119: 4554–4564 [DOI] [PubMed] [Google Scholar]
- Qin Y, Leydon AR, Manziello A, Pandey R, Mount D, Denic S, Vasic B, Johnson MA, Palanivelu R (2009) Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, pointing to genes critical for growth in a pistil. PLoS Genet 5: e1000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Cuiné S, Eymery F, Garin J, Court M, Jacquot JP, Rouhier N, Broin M (2005) Analysis of the proteins targeted by CDSP32, a plastidic thioredoxin participating in oxidative stress responses. Plant J 41: 31–42 [DOI] [PubMed] [Google Scholar]
- Riondet C, Desouris JP, Montoya JG, Chartier Y, Meyer Y, Reichheld JP (2012) A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant Cell Environ 35: 360–373 [DOI] [PubMed] [Google Scholar]
- Robinson JB Jr, Inman L, Sumegi B, Srere PA (1987) Further characterization of the Krebs tricarboxylic acid cycle metabolon. J Biol Chem 262: 1786–1790 [PubMed] [Google Scholar]
- Rodríguez-Manzaneque MT, Tamarit J, Bellí G, Ros J, Herrero E (2002) Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol Biol Cell 13: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Jacquot JP (2006) Genome-wide analysis of plant glutaredoxin systems. J Exp Bot 57: 1685–1696 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Johnson MK, Jacquot JP (2010) Glutaredoxins: roles in iron homeostasis. Trends Biochem Sci 35: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Faÿ E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127: 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot JP (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu Rev Plant Biol 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Rouhier N, Unno H, Bandyopadhyay S, Masip L, Kim SK, Hirasawa M, Gualberto JM, Lattard V, Kusunoki M, Knaff DB, et al. (2007) Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci USA 104: 7379–7384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaedler TA, Thornton JD, Kruse I, Schwarzländer M, Meyer AJ, van Veen HW, Balk J (2014) A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J Biol Chem 289: 23264–23274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Solimeo H, Rupert RA, Yadav NS, Zhu Q (2002) Functional dissection of a Rice Dr1/DrAp1 transcriptional repression complex. Plant Cell 14: 181–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram S, Rathinasabapathi B (2010) Transgenic expression of fern Pteris vittata glutaredoxin PvGrx5 in Arabidopsis thaliana increases plant tolerance to high temperature stress and reduces oxidative damage to proteins. Planta 231: 361–369 [DOI] [PubMed] [Google Scholar]
- Sundaram S, Rathinasabapathi B, Ma LQ, Rosen BP (2008) An arsenate-activated glutaredoxin from the arsenic hyperaccumulator fern Pteris vittata L. regulates intracellular arsenite. J Biol Chem 283: 6095–6101 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Zaffagnini M, Marchand C, Le Maréchal P, Rouhier N, Lemaire SD, Rey P (2009) Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. J Biol Chem 284: 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Wilson RC, Seeley KA, Reichheld JP, Muroy S, Brown S, Maughan SC, Cobbett CS, Van Montagu M, Inzé D, et al. (2000) The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waszczak C, Akter S, Eeckhout D, Persiau G, Wahni K, Bodra N, Van Molle I, De Smet B, Vertommen D, Gevaert K, et al. (2014) Sulfenome mining in Arabidopsis thaliana. Proc Natl Acad Sci USA 111: 11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtera-Kwiczor J, Groß F, Leffers HM, Kang M, Schneider M, Scheibe R (2012) Transfer of a redox-signal through the cytosol by redox-dependent microcompartmentation of glycolytic enzymes at mitochondria and actin cytoskeleton. Front Plant Sci 3: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Lin J, Liu JZ, Wang X, Lim W, Oh M, Park J, Rajashekar CB, Whitham SA, Cheng NH, et al. (2012) Ectopic expression of Arabidopsis glutaredoxin AtGRXS17 enhances thermotolerance in tomato. Plant Biotechnol J 10: 945–955 [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S (2008) ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J 53: 790–801 [DOI] [PubMed] [Google Scholar]
- Yu X, Pasternak T, Eiblmeier M, Ditengou F, Kochersperger P, Sun J, Wang H, Rennenberg H, Teale W, Paponov I, et al. (2013) Plastid-localized glutathione reductase2-regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 25: 4451–4468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.