Abstract
MicroRNAs (miRNAs) comprise a class of small, regulatory noncoding RNAs (ncRNAs) with pivotal roles in post-transcriptional gene regulation. Since their initial discovery in 1993, numerous miRNAs have been identified in mammalian genomes, many of which play important roles in diverse cellular processes in development and disease. These small ncRNAs regulate the expression of many protein-coding genes post-transcriptionally, thus adding a substantial complexity to the molecular networks underlying physiological development and disease. In part, this complexity arises from the distinct gene structures, the extensive genomic redundancy, and the complex regulation of the expression and biogenesis of miRNAs. These characteristics contribute to the functional robustness and versatility of miRNAs and provide important clues to the functional significance of these small ncRNAs. The unique structure and function of miRNAs will continue to inspire many to explore the vast noncoding genome and to elucidate the molecular basis for the functional complexity of mammalian genomes.
Introduction
In comparative genomic studies, the number of protein-coding genes within a given genome does not correlate well with the developmental and pathological complexity of the organism (1) . With the identification of numerous transcripts from the noncoding genome, which greatly exceeds protein-coding genes in number and diversity, recent studies have led to a reassessment of genomic information content (2, 3). For example, more than 80% of the human genome can be transcribed, yet only less than 2% of our genome contains protein-coding capacity (2). It is increasingly clear that the diversity of the non-coding genome correlates well with the functional complexity of a given organism (1). The vast noncoding transcriptome contains numerous noncoding RNAs (ncRNAs) that act as integral components of the molecular networks in development and disease (4). Although we are only starting to understand the realm of ncRNA biology, a frequent mode of action for ncRNAs is to form RNA-protein complexes to regulate gene expression at the level of gene transcription, RNA processing, RNA degradation and protein translation (5). It is most likely that the base-pairing between ncRNAs and other nucleic acids confers the specificity of such gene regulation (5).
One of the most studied classes of ncRNAs is microRNAs (miRNAs), a class of small regulatory ncRNAs with pivotal roles in post-transcriptional gene regulation (6–9). Nascent transcripts from miRNA genes contains one or multiple stem-loop structure(s), which are processed sequentially by the microprocessor complex (DGCR8/Drosha) and Dicer to yield mature miRNA duplexes (10). One strand of the mature miRNA duplex is incorporated into the effector complex, RISC (RNA Induced Silencing Complex), which recognizes multiple messenger RNA (mRNA) targets through imperfect base-pairing, and mediates post-transcriptional repression through combined mechanisms of mRNA degradation and translational repression (11).
Since the identification of the first miRNA using forward genetics assays in worms (12, 13), more than two thousand miRNAs have been identified in human cells to date, regulating nearly all essential cellular processes in development and disease. The biogenesis of miRNAs, the regulation of post-transcriptional gene regulation by miRNAs and the biological functions of specific miRNAs are the subject of multiple extensive reviews (9–11, 14–17), which we will not reiterate here.
Despite the relatively short history of the miRNA field, both miRNA antagonists and miRNA mimics have exhibited therapeutic potential (18–20)and recently entered clinical trials for treating human diseases. Nevertheless, efforts to characterize mammalian miRNA functions in vivo has not been straightforward, as many individual miRNA knockout mouse strains do not exhibit strong developmental phenotypes (21). Here, we will focus on the particular functionality of mammalian miRNAs conferred by their distinct gene structure, genomic organization and expression regulation, and will discuss the challenge we face to elucidate the functional significance of these small ncRNAs.
Functional importance of miRNAs in mammalian development and physiology
miRNAs exhibit unique gene structure and genome organization that distinguish them from most protein-coding genes in the mammalian genomes. One of the central questions in the miRNA field is to what extent these small ncRNAs play an essential or important role in development and disease. Targeted deletion of key miRNA biogenesis machineries in mice, including Dgcr8 and Dicer, invariably leads to profound developmental defects in many cell types and contexts (22–24). Mutations of miRNA biogenesis machineries have also been identified in pathological conditions, such as several cancer types. In addition, it is increasingly clear that numerous signaling pathways that establish cell fate and/or function — including the Hippo pathway, the transforming growth factor β (TGF-β) pathway, estrogen receptor (ER) signaling, the p38 mitogen-activated protein kinase (MAPK) pathway, and the p53-dependent DNA damage response — converge on controlling global or specific miRNA biogenesis (25–27). This regulation of miRNA synthesis in part involves a direct interaction between Microprocessor-associated helicase proteins, p68 and p72, and a component in each of the aforementioned signaling pathways (25–27). Thus, these findings clearly indicate the functional importance of miRNAs as a whole in diverse developmental and pathological processes.
The initial, systematic functional characterization of individual miRNAs was performed in C. elegans, in which most individual miRNA knockout worms are viable without obvious developmental defects (28). A subsequent study has focused on redundant miRNA families and generated mutant worm strains deficient for all members of each family (29). Yet 12 out of 15 miRNA family mutations fail to generate obvious developmental defects (29), leading some to question the extent of functional importance of redundant miRNA families. The story is somewhat different in mammals; mouse knockout studies have so far been performed on a limited number of miRNAs. Although individual miRNA-knockout mice often do not yield obvious developmental phenotype (Tables 1 and 2), emerging evidence has implicated the functional importance of redundant miRNA families in normal development and stress response. Of the eight mammalian miRNA families with essential developmental functions, six exhibit extensive genomic redundancy, and their functional importance can only be revealed when most or all of the redundancy is removed (Table 1). The redundant, polycistronic miRNAs are particularly important. Although only limited effort so far has been dedicated to study their functions in vivo by removing all or most paralogous loci, an essential developmental function has been revealed in all such studies (30–33).
Table 1. Mouse knockout phenotype for redundant miRNAs.
A summary of the phenotypes observed for mouse knockout studies to date examining miRNAs that exhibit redundancy. Dark pink, polycistronic miRNAs; light pink, moncistronic miRNAs
| miRNA family | Family members | Phenotypic characterization | Comments | References | |
|---|---|---|---|---|---|
| Polycistronic miRNAs | miR-17–92 |
miR-17–92 miR-106a-363 miR-106b-25 |
miR-17–92−/− mice exhibit postnatal lethality, lung hypoplasia, defective B-cell development, and ventricular septal defect in heart. miR- 106 a −363−/− mice and miR-106b-25−/− mice are viable, without any obvious phenotype. miR-17–92−/−; miR-106b-25−/− DKO mice and miR-17–92−/−; miR-106a-363−/−; miR-106b-25−/− TKO mice exhibit stronger phenotype than miR-17–92−/− mice, with embryonic lethality before E15.5, severe cardiac developmental defects and excessive apoptosis in multiple tissues. miR-92a−/− mice exhibit reduced body weight in embryonic and postnatal development, along with skeletal defect. |
Essential for development |
31,68,104 |
| miR-15/16 |
miR-15a/16-1 miR-15b/16-2 |
miR-15a/16-1 mice are viable, but they exhibit indolent, B cell-autonomous, clonal lymphoproliferative disorders, recapitulating the phenotypical spectrum of human CLL patients. |
64 | ||
| miR-34/449 |
miR-34a miR-34b/34c miR-449 |
miR-34a−/− mice are viable. They exhibit decreased bone mass, increased osteoclast differentiation and decreased osteoblast differentiation. miR-34a−/− mouse embryonic fibroblasts have increased efficiency in somatic reprogramming. miR-34b/34c−/− and miR- 449−/− mice are viable, without any obvious phenotype.miR-34a−/−; miR-34b/34c−/− DKO mice are viable. miR-34a−/−; miR-34b/34c−/− DKO cells have increased efficiency in somatic reprogramming. miR-34a−/−; miR-34b/34c−/−; miR-449−/− TKO mice exhibit postnatal lethality, infertility and respiratory dysfunction due to defective motile ciliogenesis. miR-34b/34c−/−; miR-449−/− DKO mice exhibit partial perinatal lethality, defective forebrain development, infertility and impaired motile ciliogenesis. |
Essential for development |
30, 77–79, 105, 106 |
|
| miR-1/133 |
miR-1a-1/133a-2 miR-1a-2/133a-1 miR-206/133b |
miR-1a-1−/− and miR-1a-2−/− mice phenotypically resemble each other, both exhibiting partial perinatal lethality, with subtle defects in cardiac morphogenesis. miR-1a-2−/−; miR-1a-1−/− DKO mice exhibit complete postnatal lethality due to strong cardiac defects, including ventricular septal defects and misalignment of the aorta over the ventricular septum. miR-133a-1−/− mice and miR-133a-2−/− mice are viable without obvious phenotype, miR-133a-1−/−; miR-133a-2−/− DKO mice exhibit lethal ventricular-septal defects in embryos and neonates with ~50% penetrance. Survived adult animals succumb to dilated cardiomyopathy and heart failure. miR-1a-1/133a-2−/− and miR-1a-2/133a-1−/− mice are viable without major developmental defects. miR-1a-1/133a-2−/−; miR-1a-2/133a-1−/− mice are embryonic lethal before E11.5, with aberrant heart development. miR-206−/− mice are viable, with normal neuromuscular synapses during development. However, deficiency of miR-206 in the ALS mouse model accelerates disease progression. |
Essential for development |
32, 66, 67, 107, 108 |
|
| miR-290–295 | No true paralogs. miR-17–92 and miR 302b-367 miRNA polycistrons contain many homologous miRNAs. |
miR-290–295−/− mice exhibit embryonic lethality from E11.5-E18.5 with a 70% penetrance. Survived adult have impaired female fertility due to a defective migration of primordial germ cells |
Essential for development |
33 | |
| miR-200 |
miR-200b/200a/429 miR-200c/141 |
mi R −141/200c−/− mice are viable, but exhibit impaired enamel formation. miR-200b−/−; miR-429−/− mice are viable but exhibit female infertility and impaired ovulation due to aberrant synthesis of luteinizing hormone. |
21, 62, 109 | ||
| miR-181 |
miR-181a-1/181b-1 miR-181a-2/181b-2 miR-181c/181d |
miR-181a-1/181b-1−/− mice are viable, yet display impaired early thymocyte development. These mice inhibit Notch1 oncogene- induced T-cell acute lymphoblastic leukemia (T-ALL). miR-181a-2/181b-2−/− and miR-181c/181d−/− mice are viable, with only subtle T- cell development phenotype. In miR-181a-1/181b-1−/−; miR-181a-2/181b-2−/− DKO mice, the inhibitory effects of miR-181a-1/181b-1 deletion on Notch-induced T-ALL are diminished. |
110 | ||
| miR-124 |
miR-124-1 miR-124-2 miR-124-3 |
miR-124a-1 mice are viable. Yet they display small brain size, axonal mis-sprouting of dentate gyrus granule cells and decreased cone photoreceptor cells. |
63 | ||
| Let-7 |
Let-7b, Let7-a2 Let-7b, Let-7c1 Let-c2, Let-7d Let-7e, Let-7f1 Let-7f2, Let-7g Let-7i, miR-98 |
Conditional KO for the bicistronic Let-7c2 and Let-7b miRNAs in intestinal epithelial exhibit very modest intestinal hypertrophy and t h icker small intestine. |
111 | ||
| miR-30 |
miR-30b/30d miR-30a miR-30c-1/30e miR-30c-2 |
miR-30b/30d−/− mice are viable. | 21 | ||
| miR-10 |
miR-10a miR-10b miR-99a/99b miR-100 |
miR-10a−/− mice are viable. | Essential for stress response |
112 | |
| miR-29 |
miR-29a/29b-1 miR-29b-2/29c |
miR-29a/29b-1−/− mice are viable but show reduced lifespan and Thymic involution. miR-29b-2/29c−/− mice are viable. | 113, 114 | ||
| monocistronic miRNAs |
miR-128 |
miR-128-1 miR-128-2 |
Mice deficient for miR-128-2, which yields 80% of total miR-128, are viable, but show increased motor activity and fatal epilepsy. Mice deficient for miR-128-1, which generates 20% of total miR-128 expression, are viable without overt developmental defects. |
Essential for development |
115 |
| miR-9 |
miR-9-1 miR-9-2 miR-9-3 |
miR-9-2 ; miR-9-3 DKO mice exhibit postnatal lethality at 1 week, with severe growth retardation, small cerebral hemispheres and olfactory bulbs. |
Essential for development |
116 | |
| miR-146 |
miR-146a miR-146b |
miR-146a−/− mice are viable but display spontaneous inflammation as they age, HSC exhaustion and hematopoietic neoplasms. | Essential for stress stress |
21,117–119 | |
| miR-208 |
miR-208a miR-208b miR-499 |
miR-208a−/− mice are viable, yet exhibit reduced cardiac hypertrophy in response to stress. miR-499−/− and miR-208b−/− mice are viable without any obvious phenotype, yet miR-208b−/−; miR-499−/− DKO mice exhibit defective specification of soleus muscle fiber. |
120 | ||
| miR-130 |
miR-130a miR-130b miR-130c |
miR-130a−/− mice are viable. | 21 | ||
| miR-7 |
miR-7a-1 miR-7a-2 miR-7b |
miR-7a-2−/− mice are viable. | 21 | ||
Table 2. Mouse knockout phenotype for non-redundant miRNAs.
A summary of the phenotypes observed for mouse knockout studies to date examining miRNAs that have no known redundancy. Dark pink, polycistronic miRNAs; light pink, moncistronic miRNAs.
| miRNA family | Phenotypic characterization | Comments | References | |
|---|---|---|---|---|
| Polycistronic miRNAs | miR-143/145 |
miR-143/145−/− mice are viable, but exhibit a variety of smooth muscle defects. Vascular smooth muscle cells are reduced in size and fail to acquire contractile phenotype due to dysregulated actin dynamics, leading to reduced blood pressure and impaired vascular remodeling after injury. Additionally, intestinal development is normal in these mice, yet epithelial regeneration after injury was defective due to dysfunction of smooth muscle and myofibroblasts. miR-143−/− mice have no obvious phenotype, while miR-145−/− mice phenocopy miR-143/145−/−, both exhibiting impaired injury- induced vascular remodeling. |
Essential for injury response |
42, 121–123 |
| miR-183/96/182 |
miR-183/96/182 mice are viable, but the postnatal functional differentiation and synaptic connectivity of photoreceptors are defective, leading to progressive retinal degeneration. miR-182−/− mice are viable without obvious phenotype. |
124, 125 | ||
| miR-132/212 |
miR-132/212 mice are viable. While the overt neuronal morphology is normal in these mice, they exhibit defects in synaptic transmission and plasticity in hippocampus and neocortex. |
126 | ||
| miR-144/451 |
miR-144/451 and miR-451 mice phenotypically resemble each other. Both are viable, yet exhibit erythroid hyperplasia, splenomegaly, and a mild anemia. |
127 | ||
| R-296/298 | miR-296/298−/− mice are viable. | 21 | ||
| miR-497–195 | miR-497–195−/− mice are viable. | 21 | ||
| miR-654–376b | miR-654–376b−/− mice are viable. | 21 | ||
| monocistronic miRNAs | miR-205 | miR-205−/− mice exhibit neonatal lethality with severe skin defects, due to impaired expansion of hair follical stem cells. | Essential for development |
21, 128 |
| miR-126 |
miR-126 mice exhibit embryonic and perinatal lethality at a 40% penetrance, with leaky vessels and frequent hemorrhage due to the loss of vascular integrity and defects in endothelial cell proliferation, migration, and angiogenesis. |
Essential for development |
129, 130 | |
| miR-214 |
miR-214 mice are viable, with normal cardiac structure and function at baseline. However, in response to ischemia/reperfusion injury, these mice exhibit loss of cardiac contractility, increased apoptosis, and excessive fibrosis. |
Essential for Injury response |
131 | |
| miR-150 | miR-150−/− mice are viable, yet exhibit B1 cell expansion and an enhanced humoral immune response. | 132 | ||
| miR-155 |
miR-155−/− mice are viable, yet they exhibit defective adaptive immunity. These mice have impaired T cell–dependent antibody responses and increased Th2 polarization and amplified Th2 cytokine production in T cells. They also display defective TNF and LT-α |
133, 134 | ||
| miR-21 |
miR-21 mice are viable and appear developmentally normal. These mice exhibit an improved survival rate in DSS-induced fatal colitis through protecting against inflammation and tissue injury. |
135 | ||
| miR-223 |
miR-223 mice are viable, yet they exhibit neutrophil hyperactivation, causing spontaneous inflammatory lung pathology, and exaggerated tissue destruction after endotoxin challenge. |
136 | ||
| miR-375 |
miR-375 mice are viable, but are hyperglycemic. They exhibit increased total pancreatic alpha-cell numbers, fasting and fed plasma glucagon levels, and increased gluconeogenesis and hepatic glucose output. |
137 | ||
| miR-675 |
miR-675 resides in the exon1 of a non-coding RNA H19, a maternally imprinted gene. Mice deficient for H19 exon1 are viable, yet show defective muscle regeneration that can be rescued by exogenous miR-675 expression. |
138 | ||
| miR-122 |
miR-122−/− mice are viable and developmentally normal. However, they develop steatohepatitis, liver fibrosis, and hepatocellular carcinoma when they are aged, due to impaired fat metabolism and cell proliferation in hepatocytes. In addition, miR-122 plays a non- canonical role in the life cycle of hepatitis C virus (HCV) by promoting the viral replication. |
Essential for stress response |
99, 101 | |
| miR-140 |
miR-140 mice are viable, yet exhibit a mild growth retardation due to impaired skeletal development. In addition, these mice also exhibit Osteoarthritis-like changes upon aging, due to defective cartilage homeostasis. |
139 | ||
| miR-33 | miR-33−/− mice are viable, yet exhibit higher serum HDL cholesterol level, and enhanced cholesterol efflux in macrophages. | 140 | ||
| miR-22 |
miR-22 mice are viable, yet they exhibit impaired cardiac intracellular calcium homeostasis and increased sensitivity to hemodynamic stress. |
141, 142 | ||
| miR-210 | miR-210−/− mice are viable. | 21 | ||
| miR-688 | miR-688−/− mice are viable. | 21 | ||
Polycistronic miRNAs harbor complex functional interactions
Unlike eukaryotic mRNAs that are mainly monocistronic, a substantial portion of miRNA genes tend to be organized in clusters to generate polycistronic precursors. 25% of human miRNAs and 31% of mouse miRNAs are predicted to be derived from polycistronic precursors (34), where a single transcript contains multiple stem-loop structures and yields multiple mature miRNAs. Polycistronic miRNA components are co-regulated transcriptionally; yet each encoded miRNA is individually processed by the biogenesis machinery, and has a distinct sequence, target specificity and functional readout. Among the polycistronic miRNAs, some contain a tandem of miRNA homologs working in concert, likely generated by local duplication of an existing miRNA locus during evolution. Others consist of miRNAs with different mature sequences and biological functions, potentially harboring a complex mode of interaction. Still others contain both homologous and heterologous miRNA components, conferring both a redundancy and a versatility that is rarely seen for single miRNA genes.
The small size of miRNAs, and the imperfect base-pairing for their target recognition, enable each miRNA to regulate many mRNA targets (9). miRNA polycistrons further expand the functional capacity of individual miRNAs, yielding a greater capacity for gene regulation through the functional coordination of multiple co-transcribed miRNAs. This internal interaction among miRNA polycistronic components could constitute a unique molecular basis for a pleiotropic function in development and disease. Indeed, for the reported miRNAs which, when deleted in mice, lead to essential developmental and physiological phenotypes, a significant portion are polycistronic miRNAs (Table 1).
Many polycistronic mammalian miRNAs are conserved among vertebrate genomes. When aligning the homologous miRNA polycistrons, their conserved structures are centered on regions that encode the precursor miRNA (pre-miRNA), whereas the regions between pre-miRNA sequences are often divergent. Interestingly, polycistronic miRNAs are often redundant within a particular genome, likely resulting from genomic duplication events during evolution (35). Among the paralogous miRNA polycistrons, not only is sequence conserved for most miRNA components, the organization of the polycistronic components is also largely preserved (36). Thus, both the individual miRNA components and their genomic organization are likely to impact on their functions.
Cooperative interactions within polycistronic miRNAs
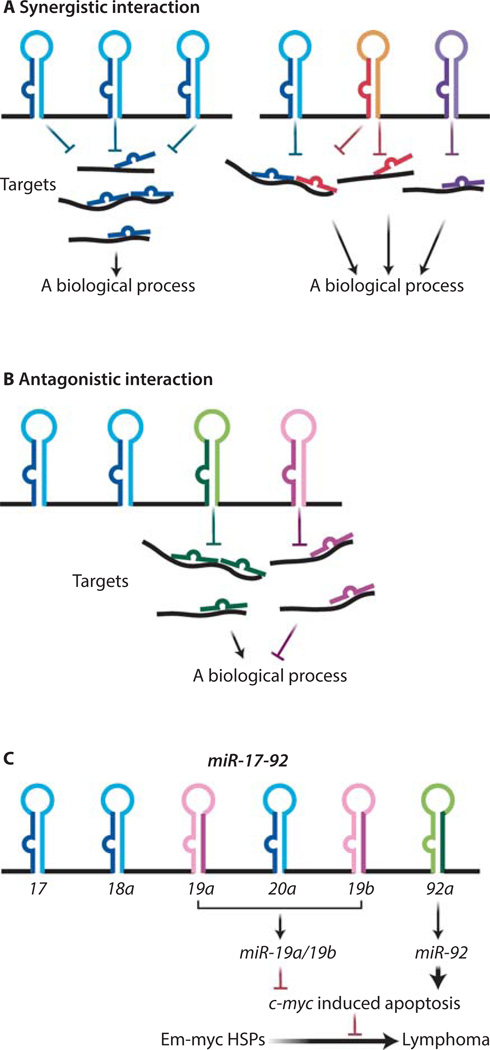
The polycistronic miRNAs bear striking structural resemblance to prokaryotic operons, in which multiple protein-coding genes under the same transcriptional control act cooperatively to regulate a specific biological process. This analogy has led many to speculate that multiple co-transcribed polycistronic miRNA components could also act synergistically to achieve functional robustness. Not surprisingly, polycistronic miRNAs that solely contain homologous components are likely to exhibit cooperative interactions to confer functional robustness, and such homologous miRNAs frequently share the same seed sequences and thus the same or similar target specificity and biological functions (Fig. 1A). For example, the miR-221/222 dicistron contains two homologous miRNAs, miR-221 and miR-222, each containing the same seed region. Overexpression of miR-221/222 has been observed in a large number of malignancies. Both miRNAs cooperatively promote proliferation and block TRAIL (TNF-related apoptosis-inducing ligand)-induced apoptosis by activating the PI3K (phosphatidylinositide 3-kinase)–AKT pathway, and they promote the cell cycle by repressing tumor suppressor genes such as those encoding p27, p57, PTEN (phosphatase and tensin homolog) and TIMP3 (Metalloproteinase inhibitor 3) (37, 38). Similarly, miR-221 and miR-222 act cooperatively to repress expression of the gene encoding the c-Kit receptor (CD117) in germ cells to maintain the undifferentiated state of mammalian spermatogonia (39), as well as in erythroleukemia cells to inhibit erythropoiesis (40). Polycistronic miRNAs containing heterologous components can also exhibit a similar cooperative interaction, because these co-transcribed miRNAs with distinct mature sequences can still target the same mRNA(s) at different sites, or target different components in the same pathway (Figure 1A).
Figure 1. Polycistronic miRNAs harbor complex functional interactions among its components.
(A) Cooperative functional interaction occurs within a polycistronic miRNA. Some polycistronic miRNAs contain a tandem of homologous components that frequently share the same seed sequences, thus they often have the same targets and biological function (right panel). Other miRNA clusters consist of heterologous miRNAs that act on different target sites to synergically regulate one biological process (left panel). (B) miRNA polycistrons can harbor a functional antagonism among the encoded components. (C) A schematic illustration of the miR-19:miR-92 antagonism in regulating the oncogenic cooperation between miR-17–92 and c-Myc. Whereas miR-19 miRNAs repressed c-Myc-induced apoptosis to promote oncogenesis, miR-92 exhibits an opposite effect to promote c-Myc induced apoptosis.
Despite the cooperative interactions among the polycistronic miRNA components, the extent of the functional contribution of each miRNA can be variable in a cell-type and context-dependent manner. For example, some miRNA polycistrons could harbor one functionally dominant miRNA component that acts cooperatively with the remaining component(s). The evolutionarily conserved miR-143/145 dicistron encodes miR-143 and miR-145, which exhibit no sequence homology. Although these two miRNAs likely act on different target sites, they share a surprising number of common targets for actin dynamics, cytoskeletal function, and injury-induced phenotypic switching of smooth muscle cells (41, 42). Deficiency of miR-145, but not of miR-143, perturbs the formation of actin stress fibers in mice and blocks vascular smooth muscle cell response to vessel injury (42). Mice deficient for miR-143/145 largely phenocopy miR-145−/− mice, yet with a slightly stronger manifestation on a subset of the defects (42). These findings support a cooperative mode of interactions between miR-143 and miR-145, with miR-145 as the dominant player. Similarly, the dicistronic miR-144/451 miRNA also harbors a component, miR-451, that is functionally dominant in erythroid differentiation. During differentiation through this lineage, miR-451 plays a major regulatory role, at least in part by down-regulating the chaperone protein 14-3-3ζ, which coordinates signal transduction downstream of hematopoietic growth factor receptor signaling (43).
Antagonistic interactions within polycistronic miRNAs
While cooperation is the predominant mode of interaction among polycistronic miRNA components, some miRNA polycistrons also harbor a functional antagonism. Such functional antagonism does not exist in isolation nor does it manifest as a stereotypical mechanism; instead, it is cell type and context dependent, often contributing to the functional complexity of miRNA polycistrons. The first characterized example involves two muscle specific miRNA clusters, miR-1-1/miR-133a-2 and miR-1–2/133a-1. In mouse C2C12 myoblast cells, miR-1 promotes differentiation by inhibiting myoblast proliferation through the repression of HDAC4 (histone deacetylase 4), whereas miR-133a counteracts this effect by promoting myoblast proliferation through the regulation of SRF (serum response factor) (44). Another well-characterized miRNA polycistron that harbors internal antagonism is miR-17–92, a potent oncomir cluster that encodes six mature miRNAs belonging to four families (miR-17/20, miR-18, miR-19a/19b and miR-92) (45). In a mouse model of Burkitt’s lymphoma, where miR-17–92 exhibits potent cooperation with the transcription factor c-Myc to promote B-cell lymphomagenesis, the miR-19 family components are both necessary and sufficient to mediate the oncogenic effect of miR-17–92 (45, 46), while the miR-92 component acts as an internal inhibitor to dampen this oncogenic activity (47) (Fig. 1C). This effect of miR-92 is, at least in part, mediated by enhancing c-Myc dosage to impose a strong coupling between excessive proliferation and p53-dependent apoptosis. The miR-19:miR-92 antagonistic interaction converges on the regulation of cell survival during B-cell transformation, with miR-19 promoting cell survival and miR-92 enhancing apoptosis upon aberrant c-Myc upregulation (47) (Fig. 1C). In this case, functional crosstalk between oncogene and tumor suppressor pathways is hardwired into the unique polycistronic miRNA structure, limiting detrimental oncogenic signaling in cells with inappropriate miR-17–92 induction.
A number of mechanisms could regulate the extent of the antagonism within a miRNA polycistron and the intricate balance it confers. For example, the miR-19: miR-92 antagonism is disrupted during c-Myc-induced oncogenesis due to a greater increase of miR-19 over miR-92 (47). Since polycistronic miRNA components are co-regulated transcriptionally, their differential expression regulation under a specific context is possibly mediated through differential miRNA biogenesis and/or miRNA turnover. Multiple discoveries have shed light on the identification of miRNA-specific post-transcriptional regulations that open a plethora of mechanisms by which individual miRNAs can be differentially expressed (Fig. 2). Emerging evidence has demonstrated that both the tertiary structure of miRNA precursors and trans-acting RNA binding proteins could impact the canonical miRNA biogenesis, resulting in cell type- and context-dependent, differential miRNA production from different polycistronic components encoded by the same precursor (48–51). Additionally, a number of RNA binding proteins are also identified to regulate miRNA stability via a sequence dependent manner (52, 53). Thus, although polycistronic miRNA components are co-regulated transcriptionally, differential expression clearly modulates their relative abundance, yielding unusual functional complexity and robustness through their functional interaction.
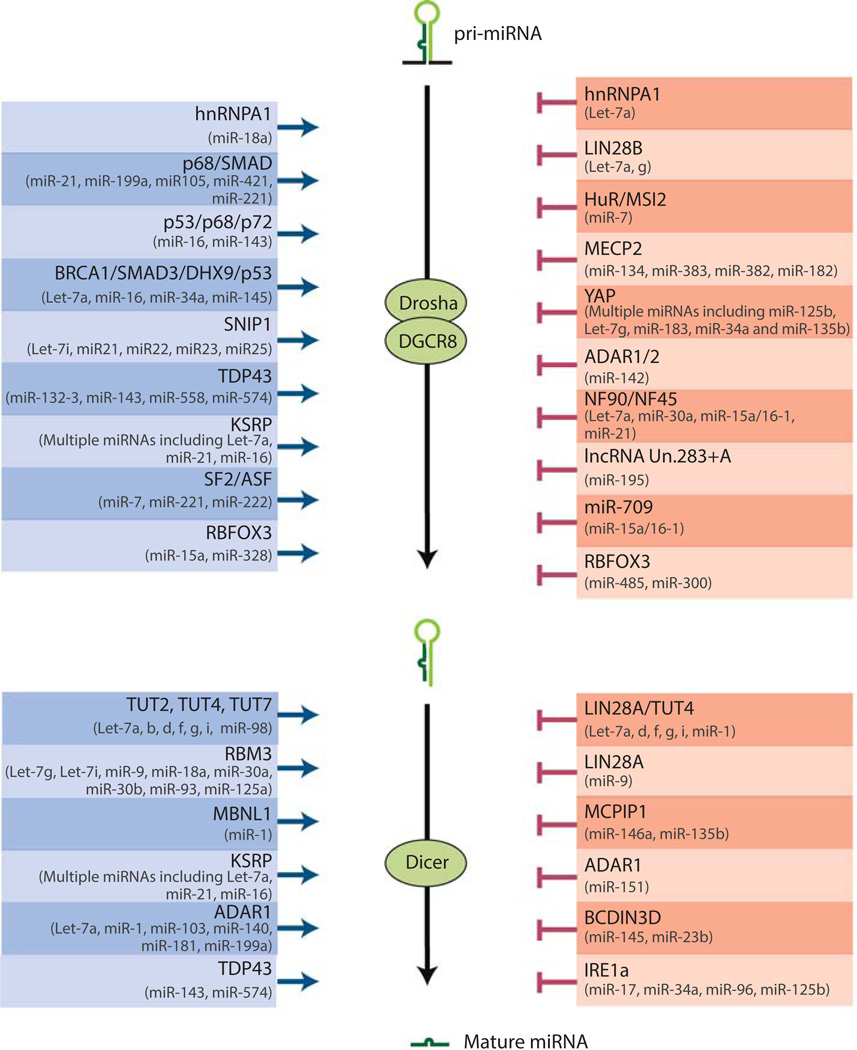
Figure 2. Diverse mechanisms regulate miRNA-specific biogenesis.
A schematic illustration summarizes the biogenesis regulators of specific miRNAs, with positive regulators on the left and negative regulators on the right (26, 50, 87, 92, 89, 144, 145, 98, 97, 91, 146, 147, 94, 96, 93, 148–152, 90, 153–158, 88, 95, 159, 160). miRNAs whose biogenesis is controlled by each regulator also are listed.
Cell type- and context-dependent versatility of polycistronic miRNA functions
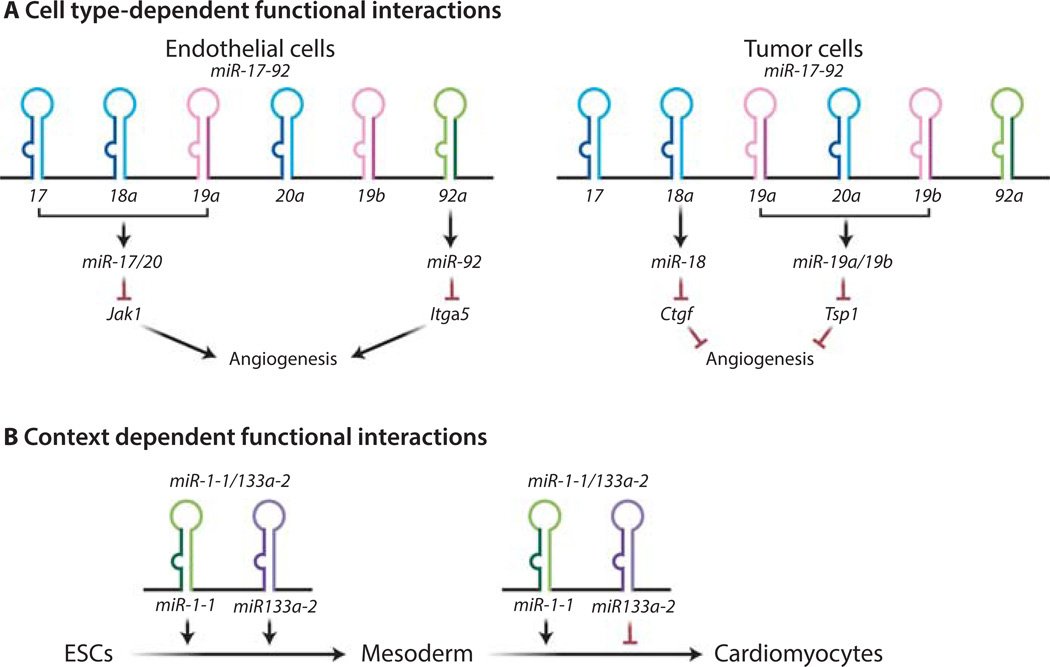
In a polycistronic miRNA, the functional readout of the individual miRNA components is cell type- and context-dependent. For example, miR-17–92 components exhibits opposing effects on the regulation of angiogenesis in two different cell types (54–56). In colon cancer cells that over-express MYC, miR-18 and miR-19 synergistically induce angiogenesis by repressing TSP1 and CTGF, respectively (54). In endothelial cells, however, miR-17/20 and miR-92 cooperatively mediate anti-angiogenic effects by downregulating Janus kinase (JAK) and the integrin subunit alpha5, respectively (55, 56). Thus, two non-overlapping subsets of miR-17–92 components generate opposing effects on a single biological process (angiogenesis) in two different cell types (Fig. 3A). Additionally, it is not uncommon that a specific biological context also determines which polycistronic component(s) plays a key function. The oncogenic potential of miR-17–92 is evident both in cooperation with c-Myc and in cooperation with RAS oncogene. Whereas miR-19 mediates the oncogenic collaboration with c-Myc through the repression of the tumor suppressor PTEN (46), the miR-17/20 miRNAs are responsible for cooperating with RAS by repressing the cyclin-dependent kinase inhibitor p21 (57). This is due to the distinct effects of miR-19 and miR-17/20 on repressing c-Myc-induced apoptosis and bypassing RAS-induced senescence, respectively (46, 57).
Figure 3. Cell type- and context-dependent versatility of polycistronic miRNA functions.
(A) The miR-17–92 functions on angiogenesis depend on cell types. miR-18a and miR-19 repress the abundance of several secreted molecules in tumor cells (54), thereby promoting angiogenesis through a cell non-autonomous mechanism. In contrast, in endothelial cells miR-17/20 and miR-92a suppress the abundance of JAK and the integrin subunit α5, respectively, thus inducing an anti-angiogenic effect through a cell autonomous mechanism (55, 56). (B) The mode of functional interaction between miR-1 and miR-133 depends on the biological contexts. Although miR-1 and miR-133 synergistically promote differentiation from embryonic stem cells to mesoderm, they exhibit an functional antagonism in regulating the muscle-specific differentiation from mesoderm (58).
Clearly, a distinct molecular pathway can act downstream of each polycistronic miRNA component, constituting the molecular basis of a pleiotropic effect. The exact mode of functional interaction within miRNA polycistrons reflects the crosstalk among these downstream signaling pathways, and could vary greatly depending on cell types and biological context. One of the best characterized examples is the cardiomyocyte-enriched miR-1/133a polycistron, which regulates ES cell differentiation into cardiomyocytes (58). It is known that the myogenic differentiation factors, including MyoD, serum response factor (SRF), and myocyte enhancer factor (MEF2), work in part through miR-1 and miR-133a to control skeletal and cardiac muscle cell specification. For example, MEF2 directly regulates miR-1/133a expression through a tissue-specific enhancer within an intron located between the miR-1 and miR-133a coding regions (59). The co-transcribed miR-1 and miR-133a components exhibit different modes of interaction in a stage-specific manner during ES cell differentiation to cardiomyocytes (Fig. 3B). In the first phase, when ES cells differentiate into mesoderm, miR-1 and miR-133a cooperate to suppress ectodermal and endodermal cell fates (58). In the second phase, when the embryonic mesoderm is differentiated into cardiomyocytes, miR-1 and miR-133a antagonize each other, with miR-1 promoting and miR-133a repressing this process (58). This internal antagonism is, at least in part, due to miR-1 regulation of the Notch ligand Delta and the transcription factor Hand2 (58, 60). Thus, the polycistronic gene structure of a miRNA can confer cell type- and context-dependent crosstalk among multiple downstream pathways, conferring unusual versatility and plasticity.
Functional Robustness conferred by miRNAs
To date, deletion of most individual miRNAs in mice failed to cause an obvious defect; however, important phenotypes often emerge when compound miRNA knockout mice are generated, thereby partially or completely removing the redundancy in the mouse genome (Tables 1 and 2). These findings suggest that mammals are generally resistant to moderate perturbation of individual miRNA expression. The unique gene structures, genomic organization and expression regulation of miRNAs all contribute to the molecular basis for their functional robustness, thus careful experimental strategies are required to elucidate their biological significance. Here, we will discuss how genomic redundancy, polycistronic gene structure and expression regulation all contribute to the unusual functional robustness of some miRNAs.
Genomic redundancy of miRNAs
The redundancy of miRNA families is more extensive in mammalian genomes than invertebrate genomes. This redundancy not only comes from polycistronic miRNAs that contain homologous components but also from multiple paralogous miRNA loci in a given genome (34). Among the limited miRNA families conserved among invertebrate and vertebrate genomes, expansion throughout higher order genomes is frequently observed. The let-7 family contains four members in worms, yet there are fourteen different Let-7 miRNAs in mice (61). Similarly, miR-34 family only has one member in worms, and there are six copies of miR-34 miRNAs in mammals (30). This increased miRNA redundancy during evolution likely reflects the functional importance of such miRNAs in mammalian development and physiology. In mouse knockout studies, a significant portion of miRNA deficiency phenotypes result from single or compound deletion of redundant miRNA family member(s) (Table 1). From a limited number of studies, these redundant miRNA loci appear not to be simply functional replicates of each other. Instead, they can undergo distinct expression regulation and engage complex feedback mechanisms to trigger functional compensation and confer robustness.
The complete removal of all members of a redundant miRNA family through the generation of knockout mice is a time consuming endeavor. Thus, most functional studies to date have focused on mice with the deletion of one miRNA or a subset of homologous miRNAs. Nevertheless, a wide spectrum of developmental and physiological phenotypes has emerged from such investigations (Tables 1 and 2). The miR-200 family contains five homologous miRNAs located in two polycistronic clusters, miR-200a/200b/429 and miR-200c/141. Deletion of miR-200b and miR-429 alone within miR-200a/200b/429 results in female infertility due to ovulatory failure (62). The miR-124 family encodes three identical, neuron specific miRNAs, including miR-124-1, miR-124-2 and miR-124-3. Mice deficient for miR-124-1 alone exhibit small brain size, aberrant axonal sprouting in dentate gyrus granule cells, and increased cell death in retinal cones (63). miR-15a/16-1, which is frequently deleted in B-cell chronic lymphocytic leukemia (CLL), is part of a redundant miRNA family with two distinct loci. miR-15a/16-1 deletion in mice causes indolent, B cell-autonomous, clonal lymphoproliferative disorders, recapitulating the phenotypical spectrum of human CLL patients (64). Protein kinase Cα (PKCα)-dependent signaling represses miR-15a, and this regulation, at least in some cells, is part of a feed-forward loop that promotes DNA synthesis, because miR-15a modulates cyclin E expression (65). Despite these compelling observations, the essential function of such redundant miRNA families is yet to be better revealed, as the typical experimental approaches underestimate the extent of the phenotype, due to functional compensation from the remaining miRNA homolog(s).
There are only a limited number of studies in which redundant miRNAs are studied systematically in knockout mice (30, 31, 66, 67). Not surprisingly, synergistic cooperation could occur among redundant miRNA paralogs, whereby only the complete removal of all redundant family members will yield an evident phenotype. For example, miR-133a-1 and miR-133a-2 encode identical, muscle-specific miRNAs, located within the miR-1–2/133a-1 and miR-1-1/133a-2 miRNA dicistrons, respectively. Deficiency of miR-133a-1 or miR-133a-2 alone fails to cause any specific phenotype, whereas miR-133a double knockout mice exhibit lethal ventricular-septal defects in embryos or neonates with 50% penetrance (67).
In addition to a synergistic interaction among redundant miRNA loci, some families contain multiple loci that have different functional contributions to a given biological process. Among the three individual loci encoding members of the miR-17–92 family (miR-17–92, miR-106a-363 and miR-106b-25), miR-17–92 clearly has the most prominent developmental role, as the single deletion of miR-17–92, but not that of the other paralogs, exhibits an obvious developmental phenotype, including postnatal lethality, lung hypoplasia, skeletal defects and aberrant apoptosis during pro- to pre-B transition (31, 68). Although miR-106b-25 deletion alone does not display any obvious defects, it clearly enhances the developmental phenotype of miR-17–92−/− mice, with the combined deletion producing embryonic lethality at embryonic day 15.5 (E15.5) with severe lung and heart defects (31). In contrast, deletion of miR-106a-363, either alone or in combination with its paralogs, did not yield any obvious phenotype, possibly due to its low or undetectable expression level (31).
An even more complex interaction is found in redundant miRNAs, where different subsets of paralogs preferentially regulate distinct biological processes. The best studied example is the let-7 family of miRNAs in worms. Let-7 was initially identified through forward genetic screen in worms as an important regulator for the larval-to-adult transition (69).Three additional let-7 family members exist in worms (miR-48, miR-84 and miR-241). Whereas let-7 predominantly promotes the larval-to-adult transition through the downregulation of hbl-1, lin-41 and daf-12, miR-48, miR-84 and miR-241 collectively repress hbl-1 and regulate the early larval developmental timing at the L2 to L3 transition (69–75). Sequence differences in the 3′ end of the let-7 family microRNAs could contribute to the functional specificity of different let-7 loci (75). More importantly, the functional specificity of each let-7 locus is due, at least in part, to the differential temporal and special expression regulation. let-7 itself is enriched in late larval stages to regulate larva-to-adult transition; in comparison, all other let-7 paralogs peak during early larval stages, thus predominantly promoting early larval developmental progression (69, 75). Because the mammalian let-7 family contains more than a dozen paralogs, it is likely to have even more complex interactions to regulate multiple distinct biological processes.
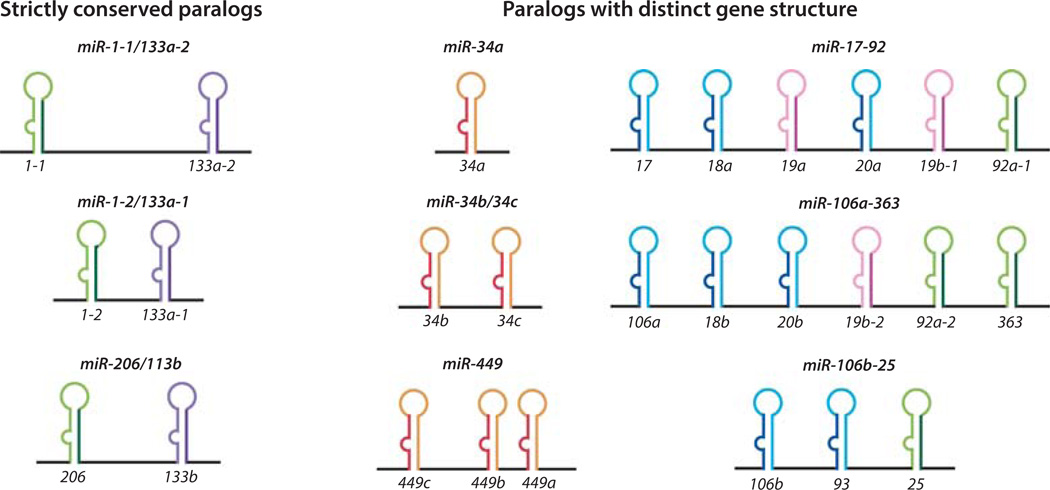
The monocistronic and polycistronic miRNAs often exhibit different patterns of redundancy. For monocistronic miRNAs, the manifestation of genomic redundancy is straightforward, as the related loci each encode a single miRNA that is similar in mature sequences. However, for polycistronic miRNAs, genomic redundancy can be more complicated (Fig. 4). In some cases, individual miRNA loci contain different numbers of the homologous miRNAs. A well-characterized example is the miR-34/449 miRNA family, whose three paralogs in mammals contain one, two and three miR-34/449 miRNAs respectively (30) (Fig. 4) . In other cases, redundant polycistronic miRNAs only contain a subset of components that are conserved. One of the best example is the miR-17–92 family, where only three out of six miR-17–92 components are conserved in its paralog miR-106b-25 (31, 76) (Fig. 4). It has been speculated that such paralogous miRNA polycistrons arise from genome duplication, and subsequently can be subjected to local deletion/duplication of specific component(s) (35). Thus, it is conceivable that the molecular composition of miRNA paralogs could also contribute to their functional specificity.
Figure 4. Distinct genomic organization of paralogous polycistronic miRNA loci.
miR-1-1/133a-2, miR-1-2/133a-1 and miR-206/133b are strictly conserved paralogs. miR-34a, miR-34b/34c and miR-449 are paralogs with distinct genes structure. They contain homologous miRNAs that are present in different copy numbers. miR-17–92, miR-106a-363 and miR-106b-25 are also paralogs that harbor distinct gene structures. They are composed of both homologous and heterologous miRNAs, and only a subset of components is conserved between the paralogs.
Paralogous miRNA loci provide functional redundancy not only by regulating overlapping targets, but also by providing compensatory expression. This is clearly illustrated by the miR-34/449 miRNA family, in which the loss-of-function of all three family members (miR-34a, miR-34b/34c and miR-449) results in strong motile ciliogenesis defects (30, 77) (see further discussion below), yet mice deficient for individual loci display no such phenotype (78, 79). In addition to the functional redundancy conferred by sequence homology, compensatory expression among miR-34/449 family members also reinforces this redundancy. A recent study reported that miR-449 was induced in miR-34b/34c deficient mice, whereas miR-34b and miR-34c induction was observed in miR-449 deficient mice (77). Although the precise signals that mediate this intricate compensatory expression regulation are not known, the identification of this feedback response highlights the extent of functional redundancy in miRNA genes.
Predominant cell type specific expression of particular miRNA families
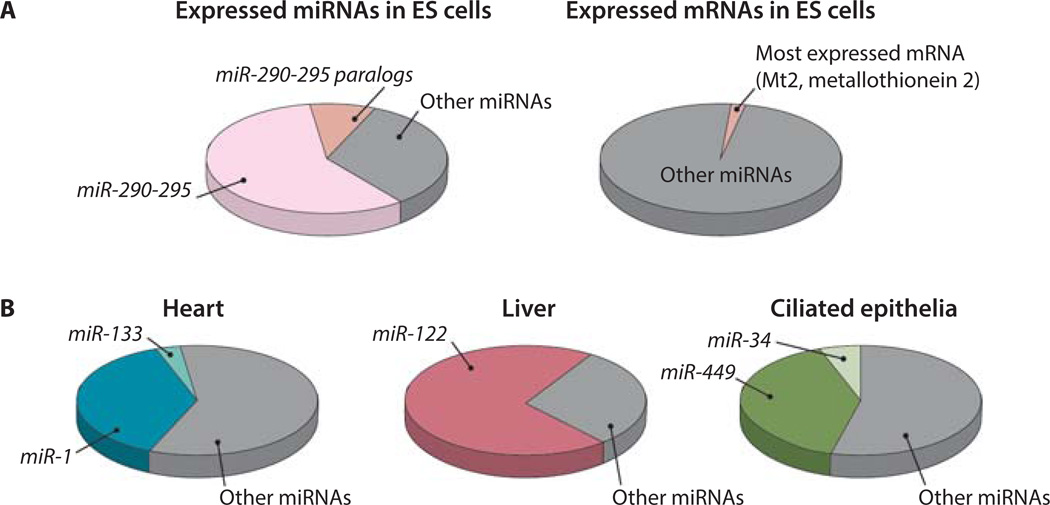
In addition to genomic redundancy, miRNA expression regulation also plays an important role in their functional robustness. Some miRNA families exhibit predominant, cell type-specific expression patterns. Predominantly expressed miRNAs can constitute a significant portion of total expressed miRNAs, ranging from 30 to 70% as reported in the literature. This is in sharp contrast to protein-coding genes, where the most abundant transcript often constitutes 1 to −2% of the total transcriptome in a given cell type (Fig. 5A).
Figure 5. The dominant expression of miRNAs in specific cell types.
(A) The distribution of the most abundant miRNA and mRNA in the transcriptome. miR-290–295 and its homologs constitute ~70% of total expressed miRNAs in ES cells (81). In contrast, the most abundant mRNA in ES cells constitutes 1.3% of the total transcriptome (Risso et al., personal communication). (B) The dominantly expressed miRNA families in heart tissue, hepatocytes and ciliated epithelia (GEO :GSM539871) (84, 85).
Previous studies have identified a number of such predominantly expressed, tissue specific miRNAs. miR-122 is among the first characterized tissue specific miRNAs, accounting for nearly 70% of all expressed miRNAs in hepatocytes (80). In mouse ES cells, components from the miR-290–295 polycistron make up ~70% of total miRNA populations, and their related homologs in the mouse genome constitute yet another 10% of miRNA expression (Fig. 5A) (81–83). In frog ciliated epithelia, miR-34/449 family miRNAs, encoded by three individual genomic loci, collectively account for half of all expressed miRNAs (84). in striated muscle in heart, the two homologous murine miR-1/133 dicistrons generate more than 40% of expressed miRNA (Fig. 5B) (85). Finally, the miR-124 miRNA and its variants constitute 25 to 48% of total expressed miRNAs in a variety of regions in the brain (80). Although predominantly expressed miRNAs exist in specific cell types, it is worth noting that the exact percentages reported may vary from different studies. These numbers are calculated on the basis of miRNA-sequencing experiments, in which the percentage of a miRNA among the total expressed miRNAs is determined not only by its abundance but also by its cloning efficiency (86). Nevertheless, it is clear that miRNAs from a single family could constitute the majority of expressed miRNAs in specific cell types.
It remains largely unknown how a single miRNA family predominates among all expressed mature miRNAs. In addition to transcriptional regulation, it is speculated that regulation of miRNA biogenesis of also plays an important role to yield differential production of different miRNAs post-transcriptionally (Figure 2). At least in some well-studied examples, RNA binding proteins can promote or inhibit the processing of specific miRNAs at different steps of the canonical miRNA biogenesis pathway (50, 87–90) (Figure 2). For example, the RNA binding protein TDP-43 (TAR DNA-binding protein 43) associates with terminal loop sequences in a small set of miRNAs and promote their processing by favoring their interaction with both Drosha in the nucleus and Dicer in the cytoplasm (89). Interestingly, some RNA binding proteins, such as the RNA binding proteins RBFOX3 (RNA binding protein fox-1 homolog 3) and hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1), exhibit opposite effects on the biogenesis of different miRNAs, acting as positive regulators for some but negative regulators for others (50, 91, 92). In addition to RNA binding proteins, miRNA biogenesis can also be regulated by ADAR deaminases through RNA editing (93), by classical DNA binding proteins, such as p53, SMAD and BRCA1, through direct interactions (94–96), or by non-coding RNAs (97, 98). Taken together, the plethora of mechanisms underlying miRNA biogenesis is likely to contribute to the predominant expression of specific miRNA families in certain cell types.
Predominant expression and genomic redundancy confer functional robustness
In many cases, this cell type-specific, predominant miRNA expression pattern is associated with genomic redundancy, and is likely to constitute an important molecular basis to confer functional robustness. Not surprisingly, the deletion of an entire family of such predominantly expressed miRNAs often gives rise to important developmental phenotypes (Table 1). One of the most characterized examples comes from studies of the miR-34/449 miRNA family that consists of six homologous miRNAs at three genomic loci, miR-34a, miR-34b/34c and miR-449. The genomic redundancy of miR-34/449 miRNAs and their predominant expression in multi-ciliated epithelia (nearly 50% of expressed miRNAs) suggests functional significance to ciliogenesis (Fig. 5B) (84). Consistently, deletion of all three miR-34/449 loci in mice lead to impaired motile ciliogenesis in multiple cell types, giving rise to partial postnatal lethality, strong respiratory dysfunction, and male/female infertility (30). Interestingly, not all miR-34/449 loci are equally enriched in ciliated epithelia, and their functional importance in ciliogenesis seems to be correlated with the extent of their expression, rather than their target specificity (30). Consistent with the more dominant expression of miR-34b/c and miR-449 polycistrons in multiciliated cells, one functional allele of miR-34b/34c or miR-449 is largely sufficient to support proper motile ciliation in otherwise miR-34/449-deficient cells (30). In contrast, having two intact miR-34a alleles in the absence of miR-34b/34c and miR-449 could not sustain normal ciliogenesis (30). The different functions among the three miR-34/449 genes could result from differential regulation of their expression in ciliated epithelia, particularly at the level of mature miRNAs.
Given the extensive redundancy exhibited by some miRNA families, it could be technically challenging to generate a knockout model to remove the entire family of miRNAs that collectively exhibit a predominant expression pattern. Nevertheless, deleting one or a subset of loci could render important insights in some cases. Deletion of miR-290–295 impairs the cell proliferation and differentiation potential of ES cells in vitro, and results in embryonic lethality with incomplete penetrance (33). It is curious why removing miR-290–295, which makes up nearly 70% ES cell expressed miRNAs, fails to generate a stronger phenotype that resembles Dicer- or Dgcr8-null ES cells. It turns out that miRNAs homologous to miR-290–295 components, including miR-17, miR-20, miR-302 miRNAs, constitute another 10% of ES cell expressed miRNAs to provide functional compensation (81). Similarly, the miR-124 family, including the most highly expressed miRNAs in neurons, consists of three genomic loci that yield three, identical mature miR-124 species. miR-124-1 knockout mice exhibit defective cell survival and axon outgrowth of dentate gyrus neurons and retinal cones, a relatively mild neuronal phenotype (63). It is conceivable that the complete removal of all miR-124 loci would generate a stronger neuronal defect. For cell type-specific, predominantly expressed miRNA families, functional redundancy can be particularly robust, as the redundant miRNA loci could each confer a nearly complete functional compensation due to their high level expression.
While most dominantly expressed miRNAs play an essential role in normal development, some regulate pathways important for physiologic homeostasis. For example, miR-122 makes up nearly 70% of miRNA expression in hepatocytes (Fig. 5B), yet mice with constitutive deletion or liver-specific deletion do not generate an overt developmental phenotype in liver (99). Nevertheless, miR-122 still regulate important physiological responses, particularly in response to various stresses. miR-122 is regulated in a circadian manner by orphan nuclear receptor REV-ERBα and appears to regulate the circadian expression of a large number of transcripts in murine hepatocytes (100). Additionally, as miR-122−/− mice age, a number of hepatic phenotypes also start to emerge, including steatohepatitis, liver fibrosis, and hepatocellular carcinoma, indicating that miR-122 constitutes an important checkpoint for fat metabolism and cell proliferation in hepatocytes (101, 102). This effect of miR-122 is mediated, at least in part, by its repression of the chemokine (C-C motif) ligand 2 (CCL2) and Klf6 (101, 102). Finally, miR-122 plays a non-canonical role in the life cycle of hepatitis C virus (HCV) by promoting viral replication (103).
Concluding remarks
The complexity of the miRNA transcriptome and the functional interplay both within its own members and with protein coding genes underscore the vast functionality of the noncoding gene landscape. Beyond the structure-function relationships intrinsic to miRNAs, their specialized genomic organization and expression regulation add additional layers of complexity to their biological functions. Additionally, transcriptional and epigenetic control, miRNA biogenesis and turnover, while incompletely understood, likely collaborate to establish highly dynamic miRNA output during tissue specification and maintenance. The important roles miRNAs play in mammalian development and disease will continue to drive investigations to explore the vast noncoding genome and understand the developmental and physiologic complexities resulting from the intricate interactions between noncoding RNAs and protein-coding genes.
Acknowledgments
We would like to thank members of the He Lab for their help and inspiring discussions, particularly Andrew Modzelewski for carefully proofreading the text.
Funding: L.H. acknowledges an R01 and an R21 grant from NCI (R01 CA139067, 1R21CA175560-01), a CIRM new faculty award (RN2-00923-1), a TRDRP research grant (21RT-0133), and a research scholar award from American Cancer Society (ACS, 123339-RSG-12-265-01-RMC). V.O. is supported by a Leukemia and Lymphoma Society special fellow award (LLS, 3423-13). A.C.M. acknowledges an R01 grant from NIH (R01HL098608) and an R01 grant from NHLBI (R01HL098608).
Footnotes
Note: At the time of preparing our review article, Vidigal et. al. also published a review article that summarizes the phenotype of miRNA knockout studies in mice (143).
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Mattick JS. Legislative Activity: RNA regulation: a new genetics? 2004;5:1662–1666. [Google Scholar]
- 2.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerstein MB, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L Rinn J, Chang HY. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.L He, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. (80−.) [DOI] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 14.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 17.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat. Rev. Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front. Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haussecker D, Kay MA. miR-122 continues to blaze the trail for microRNA therapeutics. Mol. Ther. 2010;18:240–242. doi: 10.1038/mt.2009.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park CY, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein E, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 23.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuellar TL, et al. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori M, et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 2014;156:893–906. doi: 10.1016/j.cell.2013.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong S, et al. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci. Signal. 2013;6:ra16. doi: 10.1126/scisignal.2003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr. Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song R, et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature. 2014;510:115–120. doi: 10.1038/nature13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9:e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medeiros LA, et al. Mir-290–295 deficiency in mice results in partially penetrant embryonic lethality and germ cell defects. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14163–14168. doi: 10.1073/pnas.1111241108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J. Mol. Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 36.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.le Sage C, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedde M, et al. Nat. Cell Biol. in press doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 39.Yang Q-E, Racicot KE, V Kaucher A, Oatley MJ, Oatley JM. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felli N, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cordes KR, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xin M, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patrick DM, et al. Defective erythroid differentiation in miR-451 mutant mice mediated by 14–3-3zeta. Genes Dev. 2010;24:1614–1619. doi: 10.1101/gad.1942810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J-F, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive V, et al. miR-19 is a key oncogenic component of mir-17–92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olive V, et al. A component of the mir-17–92 polycistronic oncomir promotes oncogene-dependent apoptosis. Elife. 2013;2:e00822. doi: 10.7554/eLife.00822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaulk SG, et al. Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis. RNA Biol. 2011;8:1105–1114. doi: 10.4161/rna.8.6.17410. [DOI] [PubMed] [Google Scholar]
- 49.Chakraborty S, Mehtab S, Patwardhan A, Krishnan Y. Pri-miR-17–92a transcript folds into a tertiary structure and autoregulates its processing. RNA. 2012;18:1014–1028. doi: 10.1261/rna.031039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 51.Heinrich E-M, et al. Regulation of miR-17–92a cluster processing by the microRNA binding protein SND1. FEBS Lett. 2013;587:2405–2411. doi: 10.1016/j.febslet.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Chen A-J, et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459–1472. doi: 10.1101/gad.189001.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dews M, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006;38:1060–1055. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonauer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. (80−.) [DOI] [PubMed] [Google Scholar]
- 56.Doebele C, et al. Members of the microRNA-17–92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–4950. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 57.Hong L, et al. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010;70:8547–8557. doi: 10.1158/0008-5472.CAN-10-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivey KN, et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu N, et al. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. U. S. A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 61.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Hasuwa H, Ueda J, Ikawa M, Okabe M. miR-200b and miR-429 function in mouse ovulation and are essential for female fertility. Science. 2013;341:71–73. doi: 10.1126/science.1237999. [DOI] [PubMed] [Google Scholar]
- 63.Sanuki R, et al. miR-124a is required for hippocampal axogenesis and retinal cone survival through Lhx2 suppression. Nat. Neurosci. 2011;14:1125–1134. doi: 10.1038/nn.2897. [DOI] [PubMed] [Google Scholar]
- 64.Klein U, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 65.Cohen EEW, et al. A feed-forward loop involving protein kinase Calpha and microRNAs regulates tumor cell cycle. Cancer Res. 2009;69:65–74. doi: 10.1158/0008-5472.CAN-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao Y, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 67.Liu N, et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Pontual L, et al. Germline deletion of the miR-17~92 cluster causes skeletal and growth defects in humans. Nat. Genet. 2011;43:1026–1030. doi: 10.1038/ng.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 70.Abrahante JE, et al. The Caenorhabditis elegans hunchback-like Gene lin-57/hbl-1 Controls Developmental Time and Is Regulated by MicroRNAs. Dev. Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 71.Lin S-Y, et al. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev. Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 72.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol. Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 73.Vella MC, Choi E-Y, Lin S-Y, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ. The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev. Cell. 2005;8:321–330. doi: 10.1016/j.devcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 75.Abbott AL, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olive V, Jiang I, He L. mir-17–92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42:1348–1354. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu J, et al. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Choi YJ, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011;13:1353–1360. doi: 10.1038/ncb2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Concepcion CP, et al. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lagos-Quintana M, et al. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 81.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs siRNAs, and other Microprocessor-independent Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ciaudo C, et al. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 2009;5:e1000620. doi: 10.1371/journal.pgen.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jouneau A, et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA. 2012;18:253–264. doi: 10.1261/rna.028878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marcet B, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 85.Rao PK, et al. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ. Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michlewski G, Guil S, Semple CA, Cáceres JF. Posttranscriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trabucchi M, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pilotte J, Dupont-Versteegden EE, Vanderklish PW. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS One. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim KK, Yang Y, Zhu J, Adelstein RS, Kawamoto S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat. Struct. Mol. Biol. 2014;21:901–910. doi: 10.1038/nsmb.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Michlewski G, Cáceres JF. Antagonistic role of hnRNP A1 and KSRP in the regulation of let-7a biogenesis. Nat. Struct. Mol. Biol. 2010;17:1011–1018. doi: 10.1038/nsmb.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Suzuki HI, et al. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
- 95.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J. Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang R, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liz J, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol. Cell. 2014;55:138–147. doi: 10.1016/j.molcel.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 99.Wen J, Friedman JR. miR-122 regulates hepatic lipid metabolism and tumor suppression. J. Clin. Invest. 2012;122:2773–2776. doi: 10.1172/JCI63966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gatfield D, et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–1326. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsu S-H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tsai W-C, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 104.Penzkofer D, et al. Phenotypic Characterization of miR-92a−/− Mice Reveals an Important Function of miR-92a in Skeletal Development. PLoS One. 2014;9:e101153. doi: 10.1371/journal.pone.0101153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krzeszinski JY, et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014 doi: 10.1038/nature13375. advance on (available at http://dx.doi.org/10.1038/nature13375) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Bao J, et al. MicroRNA-449 and microRNA-34b/c function redundantly in murine testes by targeting E2F transcription factor-retinoblastoma protein (E2F–pRb) pathway. J. Biol. Chem. 2012;287:21686–21698. doi: 10.1074/jbc.M111.328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heidersbach A, et al. microRNA-1 regulates sarcomere formation and suppresses smooth muscle gene expression in the mammalian heart. Elife. 2013;2 doi: 10.7554/eLife.01323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams AH, et al. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–1554. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao H, et al. The Pitx2:miR-200c/141:noggin pathway regulates Bmp signaling and ameloblast differentiation. Development. 2013;140:3348–3359. doi: 10.1242/dev.089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fragoso R, et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012;8:e1002855. doi: 10.1371/journal.pgen.1002855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Madison BB, et al. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27:2233–2245. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stadthagen G, et al. Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet. 2013;9:e1003913. doi: 10.1371/journal.pgen.1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Smith KM, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J. Immunol. 2012;189:1567–1576. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Papadopoulou AS, et al. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-α receptor. Nat. Immunol. 2012;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tan CL, et al. MicroRNA-128 governs neuronal excitability and motor behavior in mice. Science. 2013;342:1254–1258. doi: 10.1126/science.1244193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shibata M, Nakao H, Kiyonari H, Abe T, Aizawa S. MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 2011;31:3407–3422. doi: 10.1523/JNEUROSCI.5085-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao JL, et al. NF-kappaB dysregulation in microRNA-146a–deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. U. S. A. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao JL, Rao DS, O’Connell RM, Garcia-Flores Y, Baltimore D. MicroRNA-146a acts as a guardian of the quality and longevity of hematopoietic stem cells in mice. Elife. 2013;2:e00537. doi: 10.7554/eLife.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 121.Chivukula RR, et al. An essential mesenchymal function for miR-143/145 in intestinal epithelial regeneration. Cell. 2014;157:1104–1116. doi: 10.1016/j.cell.2014.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Elia L, et al. The knockout of miR-143 and −145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boettger T, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J. Clin. Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lumayag S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E507–E516. doi: 10.1073/pnas.1212655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jin Z-B, et al. Targeted deletion of miR-182, an abundant retinal microRNA. Mol. Vis. 2009;15:523–533. [PMC free article] [PubMed] [Google Scholar]
- 126.Remenyi J, et al. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One. 2013;8:e62509. doi: 10.1371/journal.pone.0062509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rasmussen KD, et al. The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 2010;207:1351–1358. doi: 10.1084/jem.20100458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang D, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 2013;15:1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuhnert F, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 131.Aurora AB, et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca2+ overload and cell death. J. Clin. Invest. 2012;122:1222–1232. doi: 10.1172/JCI59327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 133.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Thai T-H, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 135.Shi C, et al. MicroRNA-21 knockout improve the survival rate in DSS induced fatal colitis through protecting against inflammation and tissue injury. PLoS One. 2013;8:e66814. doi: 10.1371/journal.pone.0066814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 137.Poy MN, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev. 2014;28:491–501. doi: 10.1101/gad.234419.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miyaki S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24:1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gurha P, et al. Targeted deletion of microRNA-22 promotes stress-induced cardiac dilation and contractile dysfunction. Circulation. 2012;125:2751–2761. doi: 10.1161/CIRCULATIONAHA.111.044354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gurha P, et al. microRNA-22 promotes heart failure through coordinate suppression of PPAR/ERR-nuclear hormone receptor transcription. PLoS One. 2013;8:e75882. doi: 10.1371/journal.pone.0075882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2014 doi: 10.1016/j.tcb.2014.11.004. (available at http://www.ncbi.nlm.nih.gov/pubmed/25484347) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Choudhury NR, et al. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013;27:24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cheng T-L, et al. MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev. Cell. 2014;28:547–560. doi: 10.1016/j.devcel.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 146.Wu H, et al. A splicing-independent function of SF2/ASF in microRNA processing. Mol. Cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sakamoto S, et al. The NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol. Cell. Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xhemalce B, Robson SC, Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;151:278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Upton J-P, et al. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 152.Rau F, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat. Struct. Mol. Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 153.Heo I, et al. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 154.Nowak JS, Choudhury NR, de Lima Alves F, Rappsilber J, Michlewski G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat. Commun. 2014;5:3687. doi: 10.1038/ncomms4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Choudhury NR, et al. Trim25 Is an RNA-Specific Activator of Lin28a/TuT4-Mediated Uridylation. Cell Rep. 2014;9:1265–1272. doi: 10.1016/j.celrep.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 157.Ota H, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Piskounova E, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]