Abstract
Members of the flavobacterial genus Polaribacter thrive in response to North Sea spring phytoplankton blooms. We analyzed two respective Polaribacter species by whole genome sequencing, comparative genomics, substrate tests and proteomics. Both can degrade algal polysaccharides but occupy distinct niches. The liquid culture isolate Polaribacter sp. strain Hel1_33_49 has a 3.0-Mbp genome with an overall peptidase:CAZyme ratio of 1.37, four putative polysaccharide utilization loci (PULs) and features proteorhodopsin, whereas the agar plate isolate Polaribacter sp. strain Hel1_85 has a 3.9-Mbp genome with an even peptidase:CAZyme ratio, eight PULs, a mannitol dehydrogenase for decomposing algal mannitol-capped polysaccharides but no proteorhodopsin. Unlike other sequenced Polaribacter species, both isolates have larger sulfatase-rich PULs, supporting earlier assumptions that Polaribacter take part in the decomposition of sulfated polysaccharides. Both strains grow on algal laminarin and the sulfated polysaccharide chondroitin sulfate. For strain Hel1_33_49, we identified by proteomics (i) a laminarin-induced PUL, (ii) chondroitin sulfate-induced CAZymes and (iii) a chondroitin-induced operon that likely enables chondroitin sulfate recognition. These and other data suggest that strain Hel1_33_49 is a planktonic flavobacterium feeding on proteins and a small subset of algal polysaccharides, while the more versatile strain Hel1_85 can decompose a broader spectrum of polysaccharides and likely associates with algae.
Introduction
The genus Polaribacter belongs to the family Flavobacteriaceae of the phylum Bacteroidetes. To date, 12 Polaribacter species have been described with validly published names, P. atrinae (Hyun et al., 2014), P. butkevichii, P. dokdonensis, P. filamentus, P. franzmannii, P. glomeratus, P. irgensii, P. gangjinensis, P. porphyrae, P. reichenbachii, P. sejongensis and P. huanghezhanensis (Bernardet, 2010; Parte, 2014). The first described Polaribacter species were psychrophilic isolates from Arctic and Antarctic sea ice (Gosink et al., 1998), but ever since, Polaribacter have also been identified in temperate marine habitats, both in seawater (Nedashkovskaya et al., 2005; Yoon et al., 2006; Fukui et al., 2013) and sediments (Li et al., 2014). Based on in situ detection and 16S rRNA gene sequence analysis, it has been suggested that the Polaribacter genus consists of cold- and warm-water-adapted phylotypes (Gómez-Pereira et al., 2010; Díez-Vives et al., 2013). Polaribacter species have also been isolated from red (Fukui et al., 2013) and green macroalgae (Nedashkovskaya et al., 2013) and were found to thrive during diatom-dominated phytoplankton blooms in the North Sea (Teeling et al., 2012) and the Antarctic Southern Ocean (Williams et al., 2013). Association of Polaribacter spp. with phytoplankton was also observed in the open North Atlantic (Gómez-Pereira et al., 2012) and the coastal North Sea (Bennke et al., 2013) and has been inferred in a recent study on an Antarctic polynya (Kim et al., 2013). Because of their presence in cold and temperate waters, in oligotrophic and copiotrophic nutrient regimes within open ocean and coastal zones, members of the genus Polaribacter have recently been recognized as ‘widely distributed in marine habitats' (Nedashkovskaya et al., 2013).
Marine Bacteroidetes, in general, and marine Flavobacteriia, in particular, have been ascribed to act as degraders of biopolymers, such as proteins and polysaccharides (for example, Thomas et al., 2011). This view is supported by high Flavobacteriia abundances in nutrient-rich habitats, such as aggregates of particulate organic matter (Williams et al., 2013) or in the microbiota of marine algae and invertebrates (Di Camillo et al., 2012; Dong et al., 2012). Also, so far sequenced genomes of marine Flavobacteriia have revealed clear adaptations towards biopolymer degradation (for example, Bauer et al., 2006; González et al., 2008; Woyke et al., 2009; Mann et al., 2013), as they feature high proportions of peptidase and glycoside hydrolase (GH) genes. Fernández-Gómez et al. (2013) have shown that marine Bacteroidetes tend to have higher numbers of peptidase than GH genes, whereas the number in non-marine Bacteroidetes is about even. However, depending on lifestyle, there is considerable variation in the gene numbers for peptidases and carbohydrate-active enzymes (CAZymes; Lombard et al., 2014). Planktonic Flavobacteriia such as Polaribacter sp. MED152 and Dokdonia donghaensis MED134 are subjected to rare feast and extended famine conditions. Both strains feature relatively small genomes of around 3.0 Mbp, with almost twice as many peptidases than CAZymes, and can gain additional energy via the light-dependent proton-pumping membrane protein proteorhodopsin, as has been inferred for Polaribacter sp. MED152 (González et al., 2008) and verified for Dokdonia donghaensis MED134 (González et al., 2011). In contrast, macroalgae-colonizing Flavobacteriia such as Zobellia galactanivorans DsijT (Barbeyron et al., 2001) and Formosa agariphila KMM 3901T (Nedashkovskaya et al., 2006) live under more stable conditions with ample nutrient supply. Both feature much larger genomes of 5.5 and 4.2 Mbp, respectively, and feature more CAZymes than peptidases but are notably devoid of proteorhodopsin.
Pure culture studies on Polaribacter strains have demonstrated growth on biopolymers, such as aesculin, casein, starch and gelatin (Gosink et al., 1998; Nedashkovskaya et al., 2005; Yoon et al., 2006; Lee et al., 2011; Fukui et al., 2013; Kim et al., 2013; Nedashkovskaya et al., 2013). Growth on algal polysaccharides has not yet been investigated in detail, but Polaribacter co-occurrences with phytoplankton and metagenome data support this capability (Gómez-Pereira et al., 2012; Teeling et al., 2012).
So far, limited genomic data are available for members of the Polaribacter genus. A complete genome is present only for Polaribacter sp. MED152 (3.0 Mbp; González et al., 2008), an isolate from northwestern Mediterranean surface water. Furthermore, draft genomes are available for two isolates from Antarctic sea ice, P. irgensii 23-PT (∼2.8 Mbp; two scaffolds, seven contigs) and P. franzmannii 301T (∼6.9 Mbp; 56 scaffolds, 69 contigs).
In 2010, two novel Polaribacter species were isolated from surface seawater nearshore the North Sea island Helgoland during a diatom-dominated spring phytoplankton bloom (Hahnke and Harder, 2013; Hahnke et al., 2014). Strain Hel1_85 (formerly Hel_I_85) was isolated from agar plates and strain Hel1_33_49 in liquid culture. Strain Hel1_33_49 did not grow on agar plates, indicating distinct lifestyles of both strains.
In this study, we report almost complete genomes of these Polaribacter strains. Annotations were combined with growth experiments on various substrates, including a suite of different carbohydrates and polysaccharides, to elucidate the metabolic potential and associated niches of both strains. These experiments were complemented by comparative genomics with 25 additional Flavobacteriaceae genomes and by proteomics of Hel1_33_49 cultures grown on the non-substituted algal polysaccharide laminarin as well as the sulfated polysaccharide chondroitin sulfate.
Material and methods
Substrate tests
Various substrates (2.0 g l−1; Supplementary Table S1) were tested at 12 °C in marine mineral medium (10 ml) as described previously (Mann et al., 2013), including kappa- and iota-carrageenan, amorphous cellulose, glycogen, laminarin and xanthan. Volumes of 100 μl of either Polaribacter sp. Hel1_33_49 or Hel1_85 stock culture were added as inocula. Optical density measurements (660 nm) were taken for 2 weeks for monosaccharides and disaccharides and 8 weeks for polysaccharides and compared with inoculated control cultures without saccharide supplements and non-inoculated, saccharide-supplemented media.
DNA extraction, genome sequencing and annotation
Cells were grown on HaHa_100 medium (Hahnke et al., 2014) at 12 °C in the dark until the late exponential phase and subsequently pelleted by centrifugation (2500 g, 60 min). Wet cell pellets of 0.5 g were dissolved in 10 ml sodium phosphate buffer (pH 7.8) and incubated with 30 U α-amylase (10065, Sigma-Aldrich, Hamburg, Germany) at 37 °C for 60 min to remove glycoconjugates. Cells were subsequently centrifuged (30 000 g, 20 min), and DNA was extracted according to the protocol of Zhou et al. (1996).
Sequencing was performed at LGC Genomics (Berlin, Germany) using the 454 GS FLX Ti platform (454 Life Sciences, Branfort, CT, USA). The Hel1_33_49 genome was assembled from 733 179 reads from a standard shotgun library and 139 390 reads from a 3-kbp mate pair library (451.7 Mbp; 148 × coverage) using Newbler v2.6. The initial assembly comprised 38 contigs, 31 of which could be joined into 10 scaffolds. The remaining gaps were closed by PCR and Sanger sequencing yielding circular assemblies of 2 894 172 bp and 118 580 bp (total: 3 012 752 bp). The latter does not carry any known plasmid genes and likely constitutes a repeat-bound integral piece of the chromosome. The Hel1_85 draft genome was created the same way with 646 110 reads from a standard shotgun library and 135 989 reads of a 3-kbp mate pair library (394.9 Mbp; 100 × coverage). The initial assembly comprised 83 contigs, 63 of which could be joined into 46 scaffolds. Subsequent gap closure yielded four larger contigs of 1 642 720, 1 466 007, 752 210 and 53 131 bp (total: 3 914 068 bp). The remaining gaps consist of four almost identical 16S-23S-5S rRNA operons that could not be traversed in several targeted gap closure approaches.
Gene prediction and annotation (including the phylogeny-guided CAZyme annotations provided in Supplementary Table S2) were performed as described previously (Mann et al., 2013). Annotated genome sequences were submitted to NCBI's Genbank with the accession numbers JPDI00000000 (Hel1_33_49) and JPDS00000000 (Hel1_85).
Comparative analysis
We selected a total of 27 Flavobacteriaceae genomes from marine or aquatic habitats for comparative analysis (Supplementary Table S3). CAZymes were predicted using searches against the CAZy (Lombard et al., 2014), Pfam (Finn et al., 2014) and dbCAN (Yin et al., 2012) databases, and peptidases were predicted using similarity searches against the MEROPS database (Rawlings et al., 2014). The Pfam database was also used for the prediction of sulfatase (⩽E-13), bacteriorhodopsin (⩽E-5) and mannitol dehydrogenase (⩽E-5) frequencies.
Phylogenetic analysis
16S rRNA gene sequences of Polaribacter strains Hel1_33_49, Hel1_85 and all Flavobacteriaceae type strains of the ‘The All-Species Living Tree' project (LTPs_111_SSU; Yarza et al., 2010) were aligned in ARB (Ludwig et al., 2004) using the SINA aligner (Pruesse et al., 2012) and manually corrected according to secondary structure information. Bootstrapped maximum likelihood trees were constructed from sequences >1300 bp with and without a 40% conservation filter using RAxML (Stamatakis and Ott, 2008) with the GTR substitution model and 1000 repetitions. Average nucleotide identity and digital DNA-DNA hybridization were calculated using JSpecies (Richter and Rosselló-Móra, 2009) and the Genome-to-Genome Distance Calculator (Auch et al., 2010), respectively.
Proteomics
Polaribacter sp. Hel1_33_49 was grown at 12 °C in modified HaHa_100 medium with 0.1 g l−1 peptone, 0.1 g l−1 casamino acids, 200 μM NH4Cl and 16 μM KH2PO4. Laminarin (L9634, Sigma-Aldrich Chemie GmbH, Taufkirchen, Germany) or chondroitin 4-sulfate (27042, Sigma-Aldrich) were used as carbon source, and D-mannose served as control (concentrations: 2.0 g l−1; Supplementary Figure S1). The strain was adapted by three passages into the respective medium (1% inoculum), followed by cultivation in 100 ml medium. All growth experiments were carried out in triplicates. Cells were harvested by centrifugation (15 min; 13 000 g; 4 °C), and the resulting pellets were stored at −80 °C until use.
Cell pellets were washed in TE buffer (10 mM Tris-HCl pH 7.5, 10 mM EDTA pH 8.0) containing ‘cOmplete Protease Inhibitor' (Roche, Berlin, Germany). Soluble proteins were extracted by sonication (2 × 25 s), and the protein concentration was determined as described by Bradford (1976). Fifteen micrograms of protein were loaded onto 10% polyacrylamide sodium dodecyl sulfate mini gels, separated (60 min, 150 V) and stained with Roti-Blue quick (Carl Roth, Karlsruhe, Germany). Subsequently, entire gel lanes were excised, and proteins were digested as described by Heinz et al. (2012).
Peptides were subjected to a reversed-phase C18 column chromatography on a nano ACQUITY-UPLC (Waters Corporation, Milford, MA, USA) and separated as described by Otto et al. (2010). Mass spectrometry (MS) and MS/MS data were determined using an online-coupled LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). MS spectra were searched against a target-decoy protein sequence database, including sequences of Polaribacter sp. Hel1_33_49 and of common laboratory contaminants. Validation of MS/MS-based peptide and protein identifications was performed with Scaffold (http://www.proteomesoftware.com) using the SEQUEST filter settings described by Heinz et al. (2012). Normalized spectral abundance factors (NSAFs) were calculated (Zybailov et al., 2006) using Scaffold's ‘exclusive spectrum count' for each protein, which allows to compare relative protein abundances between individual samples.
Results and Discussion
Phylogeny
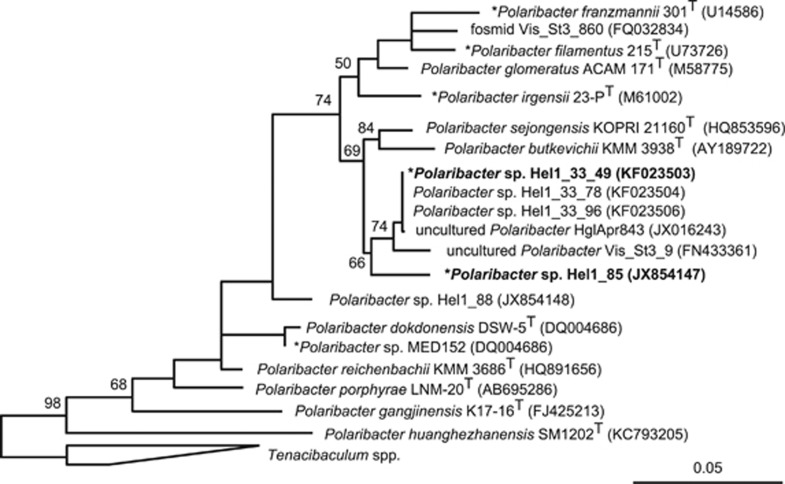
With three rRNA operons in Hel1_33_49 and four in Hel1_85, both genomes feature about one rRNA operon per megabase pair (Supplementary Table S4). The Hel1_33_49 16S rRNA gene sequence was nearly identical to previously obtained 16S rRNA gene sequences from Helgoland bacterioplankton (99.8% identity), whereas the Hel1_85 16S rRNA sequence was distinct and exhibited a weaker similarity to a sequence from the North Atlantic (97.8% identity) (Figure 1). Both strains have a 16S rRNA identity of 98.3% and an average nucleotide identity of 80% (Supplementary Table S5). They constitute distinct species that are closer related to each other than to the sequenced Polaribacter strains MED152 and 23-PT.
Figure 1.
Maximum likelihood tree based on Polaribacter spp. 16S rRNA gene sequences. Strains Hel1_33_49 and Hel1_85 are depicted in bold. Bootstrap values are based on 1000 sub-samplings. Polaribacter species with sequenced genomes are marked by an asterisk (*). Outgroup: Tenacibaculum spp. Bar: 0.05 substitutions per nucleotide position.
Degradation of biopolymers
GH and polysaccharide utilization loci (PUL) gene repertoires
CAZymes and, in particular, GHs are centrally important for polysaccharide decomposition. A comparison of the GH repertoires of strains Hel1_33_49 and Hel1_85 with 25 other sequenced marine Flavobacteriaceae comprising 133 GH families (Supplementary Table S3) revealed GH13, GH2, GH16, GH3, GH43 and GH29 as the families with the highest mean abundances (Supplementary Figure S2). No significant differences in GH abundances were detected between Polaribacter and other Flavobacteriaceae members (Supplementary Figure S3).
Polysaccharide degradation and uptake genes in Bacteroidetes are often co-located in operon-like PULs (Sonnenburg et al., 2010). PULs comprise a characteristic gene tandem coding for a SusD-like substrate-binding protein and a TonB-dependent receptor (TBDR) together with CAZyme genes and often contain genes for transport, transcriptional regulation, peptidases and sulfatases. Strains Hel1_33_49 and Hel1_85 contain 5 and 11 SusD/TBDR gene pairs, respectively, 4 and 7 of which are part of putative PULs (Supplementary Figures S4 and S5). A single PUL in Hel1_33_49 and three in Hel1_85 contain sulfatase genes (Supplementary Figures S4C and S5E, G and H). These PULs constitute the largest in both strains with up to 49 genes.
Both strains contain genes for β-galactosidases (Hel1_33_49: 1; Hel1_85: 3) and α-glucosidases (Hel1_33_49: 1; Hel1_85: 4). Such genes are typically induced by dissolved polymeric glucose. In addition, strain Hel1_85 can likely degrade xylan as it does grow on xylose and features four β-xylosidase genes, three D-xylose proton-symporters (xylE) and one hexuronate transporter. In contrast, strain Hel1_33_49 is notably devoid of these genes and does not grow on xylose. Furthermore, five putative PL7 alginate lyase genes were identified in strain Hel1_85, three in a single PUL (Supplementary Figure S5C). The polysaccharide alginate is found in brown alga cell walls, and alginate lyase excreting Polaribacter have been described before as abundant members of epiphytic bacterial communities on Laminaria sp. brown alga (Dong et al., 2012).
The storage polysaccharide of brown algae is laminarin, a β-glucan of β-1,3-linked glucose residues with occasional β-1,6-linked glucose side chains. These relatively short polysaccharides of ∼25 residues are terminated by 1-linked D-mannitol at the reducing end (for example, Read et al., 1996). Strain Hel1_85 contains a mannitol 2-dehydrogenase gene, which is absent from strain Hel1_33_49. Mannitol 2-dehydrogenase allows conversion of D-mannitol from mannitol-capped laminarin to D-fructose 6-phosphate that can subsequently be funneled into the Embden–Meyerhof–Parnas pathway. Presence of this gene in strain Hel1_85 hence supports possible association with brown algae. Diatoms in contrast feature chrysolaminarin as main storage polysaccharides (Beattie et al., 1961). Chrysolaminarin has a similar structure than laminarin, but reducing ends are not capped by mannitol residues.
Carbohydrate and polysaccharide utilization
Growth studies using a suite of different carbohydrate and polysaccharide substrates revealed that strains Hel1_33_49 and Hel1_85 both grew on D-mannose and D-maltose, but differed in the utilization of other tested monosaccharides, disaccharides and trisaccharides. As predicted, strain Hel1_85 grew on mannitol, and strain Hel1_33_49 did not. Neither strain could use D-trehalose, L-rhamnose and N-acetyl-D-glucosamine as is also true for their closest type strains (Supplementary Table S1). Both strains were able to grow on the polysaccharides cellulose, carboxymethyl cellulose, laminarin, agar, xanthan, kappa- and iota-carrageenan, glycogen and chondroitin sulfate. A surprising observation was that cells of both Polaribacter isolates increased in lengths from approximately 0.8 to 3–4 μm during growth on laminarin or chondroitin sulfate. Laminarin and chondroitin sulfate are readily metabolized by marine microbial communities (Arnosti, 2008; Arnosti et al., 2011) and were thus selected for further tests with strain Hel1_33_49.
Decomposition of laminarin
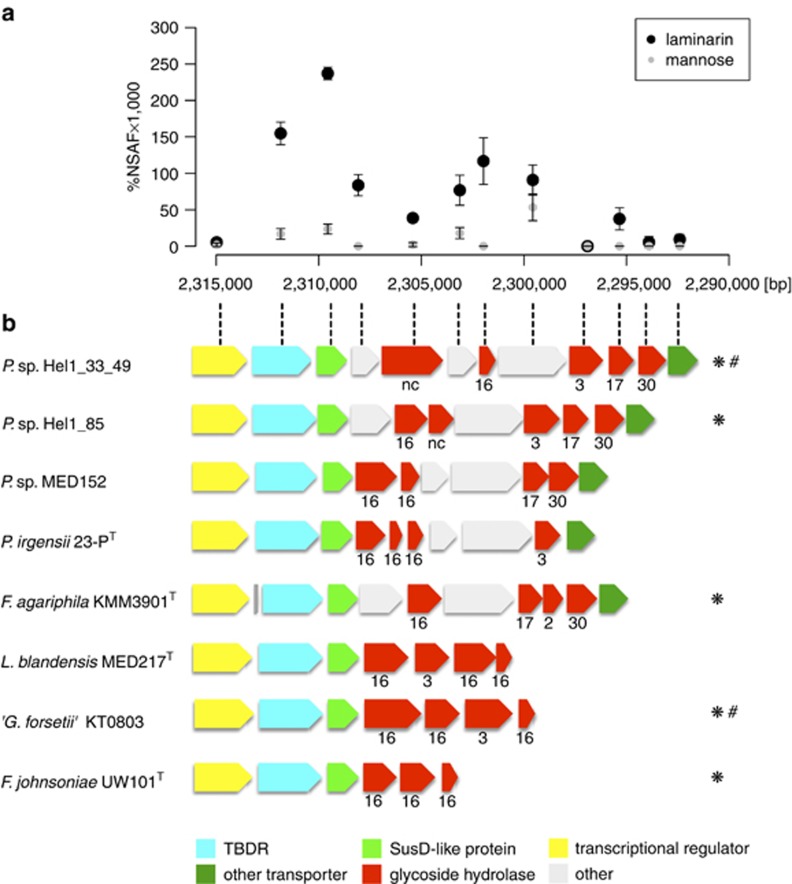
Both strains Hel1_33_49 and Hel1_85 grow on laminarin and contain a putative laminarin-specific PUL (Supplementary Figures S4D and S5B) with GH16 (Labourel et al., 2014), GH17 and one putative GH30 family β-1,6-glucanase but lack GH55 family β-1,3-glucanases (Ishida et al., 2009).
Proteomic analysis of strain Hel1_33_49 on laminarin supplemented medium revealed a significant upregulation of the respective PUL vs D-mannose-supplied controls (Figure 2a, Supplementary Table S6). Syntenic PULs were also found in other Flavobacteriaceae (Figure 2b). Induction of this PUL by laminarin was recently demonstrated in a detailed proteomics approach for ‘Gramella forsetii' KT0803 (Kabisch et al., 2014), which has been isolated earlier from the same North Sea sampling site as strain Hel1_33_49 (Eilers et al., 2001).
Figure 2.
Laminarin-induced locus of Polaribacter sp. Hel1_33_49: (a) Differential expression of the genes in laminarin- vs mannose-supplied cells. Ordinate: NSAFs; abscissa: position in the Hel1_33_49 genome (large contig; Supplementary Figure S4D). (b) Gene arrangement of the locus as well as similar loci in other Flavobacteriaceae, including Polaribacter sp. Hel1_85 (see Supplementary Figure S5B); numbers indicate GH families (nc=not classified); species that have been shown to use laminarin in cultivation experiments are marked by an asterisk (*); species where laminarin usage is supported by proteomics are marked by a hash (#).
Decomposition of chondroitin sulfate
Previous work indicates that members of Polaribacter are able to degrade sulfated algal polysaccharides. First, Gómez-Pereira et al. (2012) obtained a fosmid (S3_860) from the North Atlantic Ocean that harbored a 16S rRNA gene affiliating with Polaribacter together with a partial PUL with five sulfatase genes. Additionally, Polaribacter sulfatase expression was found to increase during a diatom-dominated phytoplankton bloom in the North Sea (Teeling et al., 2012). However, to date sequenced Polaribacter genomes contain only two to three sulfatase genes (Supplementary Table S4). In contrast, strains Hel1_33_49 and Hel1_85 harbor 12 and 33 sulfatases, respectively, with as many as 9 (Supplementary Figure S4C) and 8 (Supplementary Figure S5G) sulfatases in a single PUL. Strain Hel1_85 furthermore features a PUL-like gene cluster without a susD-like gene in which 14 sulfatases are co-located with GHs (Supplementary Figure S5D). Large numbers of diverse GH genes in these sulfatase-rich PULs indicate that they target more than one type or rather heterogeneous sulfated polysaccharides. This is supported by growth of both Helgoland strains on sulfated polysaccharides, such as kappa- and iota-carrageenan, agar and xanthan (Supplementary Table S1).
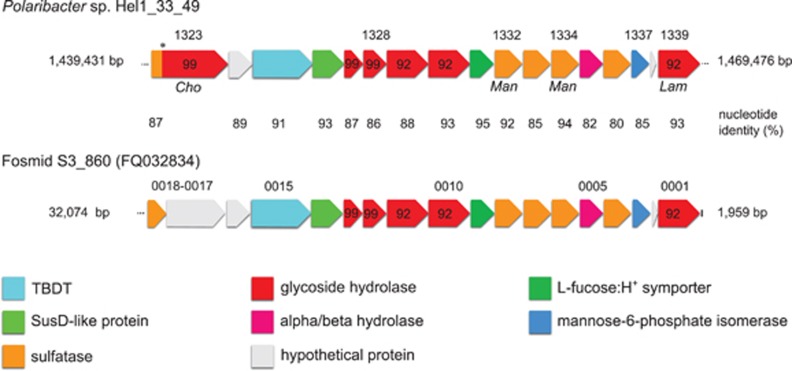
The sole sulfatase-containing PUL of the North Sea isolate Hel1_33_49 (Supplementary Figure 3C) exhibits a strong synteny with the partial PUL from the Atlantic fosmid S3_860 (Figure 3). In both PULs, three GH92 α-1,2-mannosidases are clustered with five sulfatases. A recent study on the human gut microbe Bacteroides thetaiotaomicron indicated that its arsenal of family GH92 exo-acting enzymes decomposes α-mannoside-containing N-glycans and glycoproteins from land plant cell walls (Zhu et al., 2010). It is possible that Polaribacter sp. Hel1_33_49 can utilize similarly structured but sulfated polysaccharides from marine algae. Furthermore, the presence of such highly similar PULs in distinct Polaribacter species from the North Sea and Atlantic Ocean (97.6% 16S rRNA sequence identity) indicates that this PUL is widespread and functionally important in marine systems.
Figure 3.
Synteny between a PUL in Polaribacter sp. strain Hel1_33_49 (Supplementary Figure S4C) and a partial PUL on fosmid S3_860 from the Atlantic Ocean identified by Gómez-Pereira et al. (2012). Cho, Man and Lam indicate proteins that were only detected in chondroitin sulfate-, mannose- and laminarin-supplied cells, respectively. Numbers above genes represent gene locus tags; numbers in genes represent CAZyme family affiliations. The asterisk (*) indicates a possible sequencing error mimicking a gene fusion.
Chondroitin sulfate is a heterogeneous polysaccharide composed of alternating N-acetylgalactosamine and glucuronic acid that carries sulfate moieties at various positions. It is not a common substrate in surface seawater, but it is readily metabolized by microbial communities and might mimic polysaccharides of similar composition, such as transparent exopolymers of diatoms that are known to contain high proportions of glucuronic acids and sulfated monosaccharides (for example, Zhang et al., 2008). It has been shown that Polaribacter abundance of up to 7% of the bacterioplankton (Gómez-Pereira et al., 2010) coincided with peaking potential for chondroitin sulfate decomposition in the North Atlantic Ocean (Arnosti et al., 2012). However, the so far sequenced Polaribacter genomes do not contain any known chondroitinase genes. Therefore, chondroitin sulfate decomposition in Polaribacter might not be mediated by a specific PUL but rather initiated by distinct sulfatases that desulfate chondroitin sulfate to a glycosaminoglycan composed of N-acetylgalactosamine and glucuronic acid, which is subsequently decomposed.
Proteomic analyses (Supplementary Table S6) of chondroitin sulfate-grown Hel1_33_49 cultures revealed differential expression of one sulfotransferase, three sulfatases and one gene with sulfatase and GH99 domains. The latter resides in the sulfatase-containing PUL with high synteny to fosmid S3_860 (Figure 3) and likely represents two distinct genes that are artificially fused due to a sequencing error. This is supported by an amino-acid identity of ca. 95% of the N-terminal sulfatase and of ca. 90% of the C-terminal GH (PHEL49_1323) with corresponding genes on the fosmid sequence. Expression of this sulfatase/GH99 locus was only detected with chondroitin sulfate (1.3%NSAF × 1000) and not with laminarin or mannose. A second sulfatase (PHEL49_1477) was detected only with chondroitin sulfate (7.0%NSAF × 1000) or laminarin (2.3%NSAF × 1000) but not with mannose. The remaining two sulfatases (PHEL49_1332/1334) were present only in mannose-supplied cultures, whereas the sulfotransferase (PHEL49_498) was higher expressed with chondroitin sulfate (12 × ) and mannose (7 × ) than with laminarin. Ten GHs (including families GH13, GH16, GH65, GH74 and GH99), eight GTs (including families GT2, GT4 and GT19) and two non-GH carbohydrate-binding proteins (CBM4 and CBM50) were more than twofold upregulated with chondroitin sulfate, whereas five GHs (including families GH13, GH16, GH31 and GH81) were more than twofold downregulated vs mannose-supplied controls. Furthermore, glutamate synthase and glutamine synthetase were downregulated with chondroitin sulfate, and the assimilatory nitrate/nitrite reductases were only expressed in laminarin-supplied cultures. This suggests that strain Hel1_33_49 can effectively scavenge nitrogen from the chondroitin sulfate glycosaminoglycan backbone, whereas it needs additional nitrogen when grown on nitrogen-free laminarin.
One of the two mannose-6-phosphate isomerase and a putative mannan endo-1,4-β-mannosidase (GH26) were downregulated with chondroitin sulfate vs mannose-supplied controls. Surprisingly, these enzymes were also expressed to similar extents in laminarin-grown cultures as in the controls (%NSAF × 1000: 31.2 vs 38.8 and 43.6 vs 41.7), which indicates co-regulation of the laminarin and mannose metabolism. Such co-regulations for naturally co-occurring saccharides have been described before, for example, for the β-1,3-glucan laminarin and α-1,4-glucans in ‘Gramella forsetii' KT0803 (Kabisch et al., 2014).
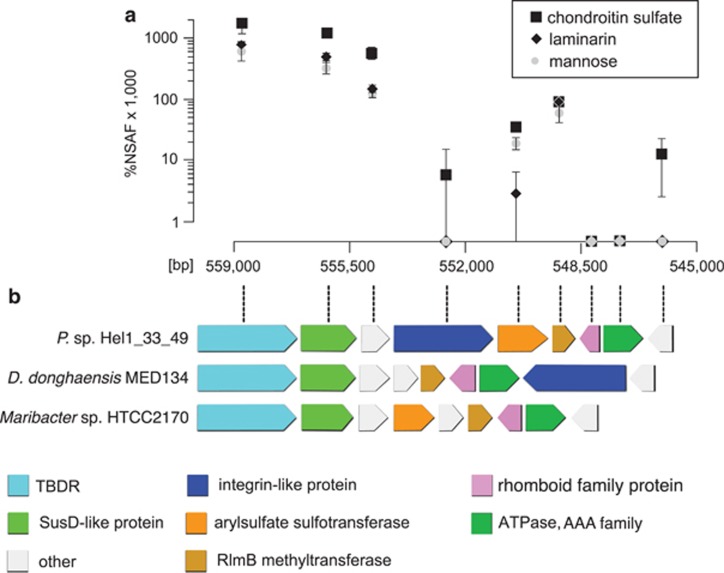
Hel1_33_49 cultures grown on chondroitin sulfate showed an unexpected upregulation of a PUL-like operon without known CAZymes (Figure 4) composed of genes for a TBDR (2.8 × ), a SusD-like protein (3.5 × ), an integrin-like protein (only detected with chondroitin sulfate), an arylsulfate sulfotransferase (1.8 × ), a methyltransferase (1.5 × ) and a conserved hypothetical protein (4.6 × ). The N-terminal half of the integrin-like protein resembles the human cartilage acidic protein CRTAC1 (Steck et al., 2007), which might specifically recognize chondroitin. Such integrin-like proteins are also present in strain Hel1_85 and Dokdonia donghaensis MED134 where they are also co-localized with TBDRs/SusD-like proteins. Conversely, the arylsulfate sulfotransferase of Hel1_33_49 has not been found in any other Polaribacter strain so far.
Figure 4.
Chondroitin sulfate-induced locus in Polaribacter sp. Hel1_33_49. (a) Differential expression with chondroitin sulfate and laminarin vs mannose. Ordinate: NSAFs; abscissa: position in the Hel1_33_49 genome (large contig). (b) Gene arrangement of the locus as well as similar loci in other Flavobacteriaceae.
Protein utilization
Peptidases of 90 different MEROPS families (Rawlings et al., 2014) were detected among the 27 investigated Flavobacteriaceae genomes (Supplementary Figure S6, Supplementary Table S3) dominated by cysteine (C), metallo (M) and serine (S) families (Supplementary Figure S7). Twenty-seven MEROPS families were common to all Flavobacteriaceae, with the highest average gene numbers in families S09 (7.0 Mbp−1), S33 and M38 (both 2.8 Mbp−1). Mesoflavibacter zeaxanthinifaciens S86 (Oh et al., 2011a) and Flavobacteriaceae bacterium S85 (Oh et al., 2011b) had the highest (68.0 Mbp−1) and lowest (28.7 Mbp−1) peptidase density, respectively. Both were isolated from seawater in Micronesia but seem to exhibit a pronounced niche separation.
Sequenced Polaribacter strains (excluding the fragmentary P. franzmannii 301T genome) have 161–167 peptidases. In comparison to the other investigated Flavobacteriaceae, Polaribacter have elevated proportions of serine (S) peptidases, and strain Hel1_33_49 also has an elevated proportion of metallo (M) peptidases. Members of these families mediate the degradation and uptake of extracellular proteins, suggesting that digestion of external proteins and subsequent uptake of oligopeptides and amino acids constitutes an important carbon and nitrogen source for both Helgoland strains. Kirchman (2000) estimated that ∼50% of bacterial production in the oceans is supported by amino acids as a key nitrogen source. This agrees with the finding that strain Hel1_33_49 did not grow in medium without yeast extract, casamino acids or peptone and due to a lack of detectable asparagine synthetase is likely auxotrophic for this amino acid.
There are notable differences in the peptidase profiles of Polaribacter sp. strain Hel1_33_49 and Hel1_85. For example, strain Hel1_33_49 contains more M38 and S09 peptidases than strain Hel1_85, which in turn has more S08 and S41 peptidases. However, MEROPS family assignments are of limited value in linking peptidase profiles to environmental processes. Distinct peptidases with varying specificities are known to act on polypeptides (for example, M01), oligopeptides (for example, S09, M20) and carboxy/aminopeptidases (for example, M14/M42). Strains Hel1_33_49 and Hel1_85 have six and eight exported family M01 aminopeptidases, respectively. Elevated aminopeptidase activities have been reported for marine bacteria that grow on high-molecular weight dissolved organic nitrogen as sole nitrogen source (Korth et al., 2012; Calleja et al., 2013). Polaribacter sp. Hel1_85 has 27 signal peptide-containing S09 prolyl oligopeptidases that cannot degrade peptides >30 amino acids and are likely used to decompose external oligopeptides prior to uptake.
Light-dependent proteins
Bacterial rhodopsins are light sensors or light-dependent ion pumps. The latter can either translocate protons or sodium ions across the cytoplasmic membrane, which contributes to the chemiosmotic potential that can be used, for example, for ATP production, or function as chloride importers as has been shown for the marine flavobacterium Nonlabens marinus S1–08T (Yoshizawa et al., 2014). Rhodopsin genes have been estimated to be present in about 50% of the North Sea bacterioplankton, most of which absorbs green light (Riedel et al., 2010). González et al. (2008) hypothesized that during oligotrophic conditions Polaribacter sp. MED152 uses surplus ATP from its proton-pumping rhodopsin for anaplerotic carbon dioxide fixation via pyruvate and phosphoenolpyruvate carboxylase. This is supported by recent stable isotope experiments that demonstrated active assimilation of dissolved inorganic carbon in Polaribacter in the northeast Pacific (DeLorenzo et al., 2012).
The rhodopsin light-absorbing cofactor retinal is produced from oxidative cleavage of β-carotene by 15,15'-monooxygenases. Both rhodopsin and 15,15'-monooxygenase genes are present in Polaribacter strains MED152, 23-PT and Hel1_33_49 but notably absent from strain Hel1_85. The Leu105 of the Hel1_33_49 rhodopsin sequence indicates a 490-nm light absorption maximum (Man et al., 2003). Its sequence resembles that of the Polaribacter isolate SA4–10 (99.7% amino-acid identity; Supplementary Figure S8), for which rhodopsin functionality has been demonstrated (Yoshizawa et al., 2012).
Strain Hel1_33_49 expressed its rhodopsin when grown on chondroitin sulfate even in the dark (Supplementary Table S6), similar to Dokdonia sp. PRO95, which expresses rhodopsin independent of light conditions and organic substrate supply (Riedel et al., 2010). In contrast, no rhodopsin expression was observed when strain Hel1_33_49 was grown on laminarin or mannose. This suggests that rhodopsin expression in Hel1_33_49 depends on the availability of specific substrates. The effect of light on rhodopsin expression has been demonstrated for marine model bacteria (Gómez-Consarnau et al., 2007; Riedel et al., 2010; Kimura et al., 2011; Steindler et al., 2011) and in field studies (Lami et al., 2009; Poretsky et al., 2009). Possible effects of nutrient availability on rhodopsin expression are still not well studied, but it was recently shown by Akram et al. (2013) that nutrient limitation and not light exposure regulates differential rhodopsin expression in Vibrio sp. AND4.
Photolyases and cryptochromes are further light-dependent proteins in Polaribacter strain Hel1_33_49. González et al. (2008) identified a set of three different DNA photolyase/cryptochrome genes in the Polaribacter sp. MED152 genome, which might function as blue light photoreceptors and mediate gene expression levels. We found that the respective locus has a strong synteny among the three rhodopsin-containing Polaribacter strains Hel1_33_49, MED152 and 23-PT, with amino-acid sequence identities >86% among all DNA photolyase/cryptochrome genes (Supplementary Figure S9).
Niche separation
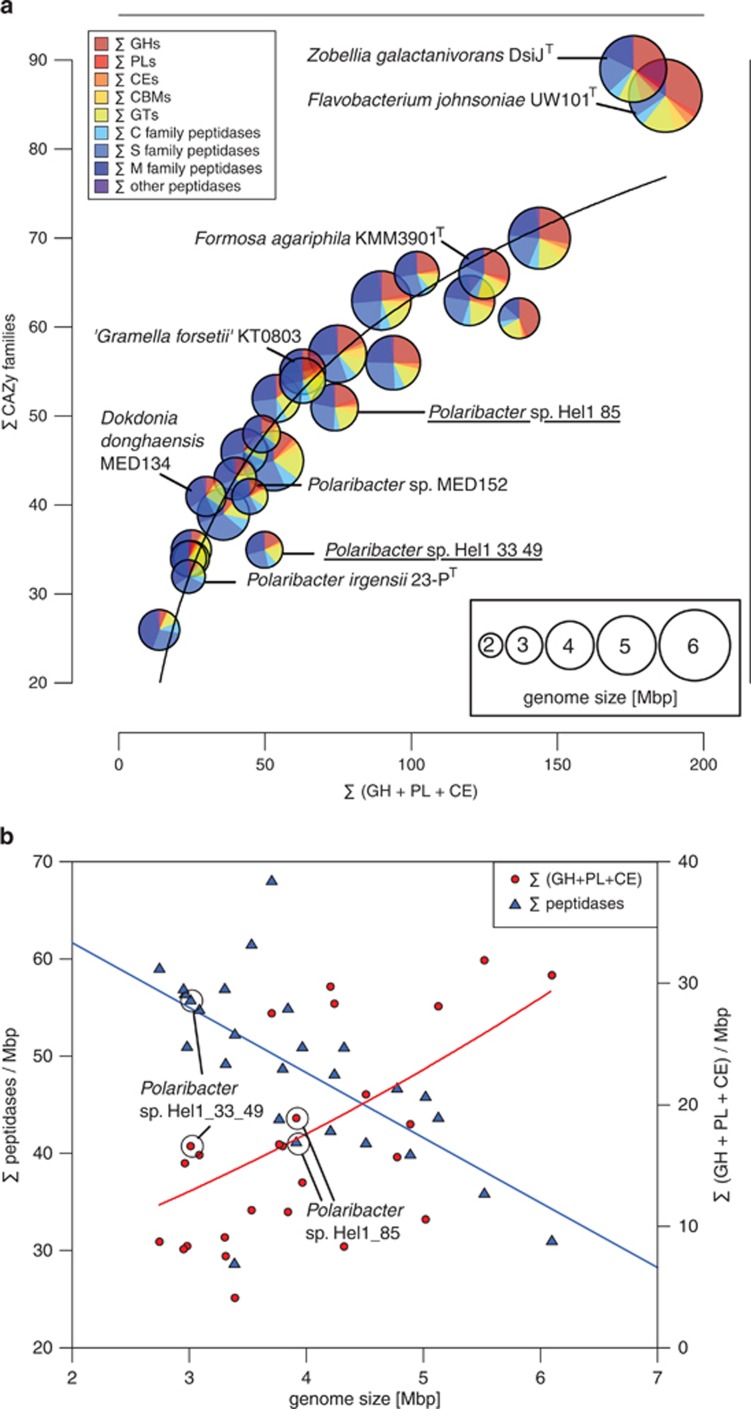
Comparisons of the CAZyme and peptidase profiles of the 27 analyzed Flavobacteriaceae revealed pronounced differences between both North Sea Polaribacter strains (Figure 5a). Strain Hel1_33_49 has a smaller genome with less CAZymes representing less families and, based on manual CAZyme annotations, a peptidase:CAZyme ratio of 1.37. In contrast, strain Hel1_85 has a larger genome with more CAZymes representing more families an even peptidase:CAZyme ratio. In general, small Flavobacteriaceae genomes tend to have a higher peptidase density than larger genomes, while the opposite trend is true for the number of CAZymes with degradation functions (Figure 5b). Although Hel1_33_49 and Hel1_85 feature similar CAZyme densities, they exhibit notably different peptidase densities.
Figure 5.
Comparison of 27 Flavobacteriaceae genomes based on automated annotations (Supplementary Table S3): (a) Interdependency of the number of degradative CAZymes and the number of CAZyme families (trend line: logarithmic regression). Genomes are represented by circles with radii proportional to genome sizes. Relative proportions of CAZymes vs peptidases are shown for each genome. (b) Densities of CAZymes with degradation functions and peptidases vs genome size (trend lines: second-order polynominal regressions).
Flavobacteriaceae species with high peptidase:CAZyme ratios include known planktonic species such as Polaribacter sp. MED152 and thus are likely adapted towards carbohydrate-depleted marine habitats and preferentially feed on proteins. By contrast, CAZyme-rich species with lower peptidase proportions include known algae-associated species such as Zobellia galactanivorans DsijT and Formosa agariphila KMM 3901T, reflecting that these species have an ample supply of algal polysaccharides. Further commonalities among these species are (i) the tendency for larger genomes, (ii) the absence of rhodopsins, and (iii) often presence of mannitol dehydrogenases (Supplementary Figure S10).
Although the characteristics of strain Hel1_33_49 match those of typical planktonic Flavobacteriaceae, Hel1_85 lies in between those of planktonic and algae-associated species (Figure 5) and might thrive under both conditions. Algae-association of Polaribacter sp. Hel1_85 is supported by the fact that it can use laminarin, D-mannitol and alginate as carbon sources, which are typical constituents of brown algae, its high number of sulfatases and its lack of rhodopsin genes.
In situ abundance
We used the genomes of the two North Sea Polaribacter strains for read-recruitment with 26 bacterioplankton metagenomes that were obtained in 2008–2012 from the North Sea Polaribacter's isolation site (Teeling et al., 2012) and the English Channel (Gilbert et al., 2010). The proportion of recruited reads at ⩾95% sequence identity by each genome was <1% for all metagenomes (Supplementary Table S7 and Supplementary Text). However, Polaribacter abundances in the North Sea are dynamic and peak during spring phytoplankton blooms (Teeling et al., 2012). It is thus likely that higher abundances of either of the two Polaribacter strains were missed by any of the metagenome sampling dates. Read abundances were consistently higher for Polaribacter sp. Hel1_33_49 than for Hel1_85, which supports genomic analyses indicating that the former is better equipped for a planktonic lifestyle than the latter.
Concluding remarks
Polaribacter sp. Hel1_33_49 and Hel1_85 were isolated from the same sample but exhibit detectable differences with respect to their genomes, physiologies and inferred lifestyles. Strain Hel1_33_49 constitutes what might be considered a typical pelagic representative with a smaller genome, rhodopsin, a limited number of PULs in conjunction with a higher peptidase:CAZyme ratio and no mannitol dehydrogenase. Strain Hel1_85, in contrast, has a larger genome with more PULs in conjunction with a lower peptidase:CAZyme ratio, features a mannitol dehydrogenase but no rhodopsin and seems better equipped to cope with low oxygen conditions as they might occur in biofilms (see Supplementary Text). This species possibly constitutes a Polaribacter representative that can associate with algae, which, however, has to be confirmed in a future in situ localization study. Strain Hel1_85 also has a higher density of histidine kinases and response regulators than strain Hel1_3_49 and thus seems to be the more adaptive and versatile of the two (Supplementary Table S4).
Both strains grow on laminarin and chondroitin sulfate, and for strain Hel1_33_49 we identified (i) a laminarin-dependent PUL, (ii) chondroitin sulfate-induced CAZymes and (iii) a novel operon that might enable recognition of chondroitin sulfate-like substrates. In comparison to the genomic data from Polaribacter spp., it is evident that the genus Polaribacter is not only broad in terms of phylogeny but also in terms of genome sizes, physiological traits and associated ecological niches.
Acknowledgments
We thank Nils Öster, Lennart Kappelmann and Karen Krüger for bioinformatics and Meghan Chafee for critical reading. The work conducted by the US Department of Energy Joint Genome Institute is supported under contract no. DE-AC02-05CH11231. This study was funded by the National Natural Science Foundation of China (31370508), the Chinese Academy of Sciences and the Max Planck Society.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akram N, Palovaara J, Forsberg J, Lindh MV, Milton DL, Luo H, et al. Regulation of proteorhodopsin gene expression by nutrient limitation in the marine bacterium Vibrio sp. AND4. Environ Microbiol. 2013;15:1400–1415. doi: 10.1111/1462-2920.12085. [DOI] [PubMed] [Google Scholar]
- Arnosti C. Functional differences between Arctic seawater and sedimentary microbial communities: contrasts in microbial hydrolysis of complex substrates. FEMS Microb Ecol. 2008;66:343–351. doi: 10.1111/j.1574-6941.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- Arnosti C, Steen AD, Ziervogel K, Ghobrial S, Jeffrey WH. Latitudinal gradients in degradation of marine dissolved organic carbon. PLoS One. 2011;6:e28900. doi: 10.1371/journal.pone.0028900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnosti C, Fuchs BM, Amann R, Passow U. Contrasting extracellular enzyme activities of particle-associated bacteria from distinct provinces of the North Atlantic Ocean. Front Microbiol. 2012;3:425. doi: 10.3389/fmicb.2012.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk H-P, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2:117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeyron T, L'Haridon S, Corre E, Kloareg B, Potin P. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int J Syst Evol Microbiol. 2001;51:985–997. doi: 10.1099/00207713-51-3-985. [DOI] [PubMed] [Google Scholar]
- Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, et al. Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii' reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8:2201–2213. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- Beattie A, Hirst EL, Percival E. Studies on the metabolism of the Chrysophyceae. Comparative structural investigations on leucosin (chrysolaminarin) separated from diatoms and laminarin from the brown algae. Biochem J. 1961;79:531–537. doi: 10.1042/bj0790531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennke CM, Neu TR, Fuchs BM, Amann R. Mapping glycoconjugate-mediated interactions of marine Bacteroidetes with diatoms. Syst Appl Microbiol. 2013;36:417–425. doi: 10.1016/j.syapm.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Bernardet J-F.2010Class II. Flavobacteriia class. novIn: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, et al (eds). Bergey's Manual of Systematic Bacteriology. The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes Springer: New York, NY, USA; 106–314. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Calleja ML, Batista F, Peacock M, Kudela R, McCarthy MD. Changes in compound specific δ15N amino acid signatures and d/l ratios in marine dissolved organic matter induced by heterotrophic bacterial reworking. Mar Chem. 2013;149:32–44. [Google Scholar]
- DeLorenzo S, Bräuer SL, Edgmont CA, Herfort L, Tebo BM, Zuber P. Ubiquitous dissolved inorganic carbon assimilation by marine bacteria in the Pacific Northwest coastal ocean as determined by stable isotope probing. PLoS One. 2012;7:e46695. doi: 10.1371/journal.pone.0046695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Camillo CG, Luna GM, Bo M, Giordano G, Corinaldesi C, Bavestrello G. Biodiversity of prokaryotic communities associated with the ectoderm of Ectopleura crocea (Cnidaria, Hydrozoa) PLoS One. 2012;7:e39926. doi: 10.1371/journal.pone.0039926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Vives C, Gasol JM, Acinas SG. Spatial and temporal variability among marine Bacteroidetes populations in the NW Mediterranean Sea. Syst Appl Microbiol. 2013;37:68–78. doi: 10.1016/j.syapm.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Dong S, Yang J, Zhang XY, Shi M, Song XY, Chen XL, et al. Cultivable alginate lyase-excreting bacteria associated with the Arctic brown alga Laminaria. Mar Drugs. 2012;10:2481–2491. doi: 10.3390/md10112481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R. Isolation of novel pelagic bacteria from the German bight and their seasonal contributions to surface picoplankton. Appl Environ Microbiol. 2001;67:5134–5142. doi: 10.1128/AEM.67.11.5134-5142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Gómez B, Richter M, Schüler M, Pinhassi J, Acinas SG, González JM, et al. Ecology of marine Bacteroidetes: a comparative genomics approach. ISME J. 2013;7:1026–1037. doi: 10.1038/ismej.2012.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:27. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, Abe M, Kobayashi M, Saito H, Oikawa H, Yano Y, et al. Polaribacter porphyrae sp. nov. isolated from the red alga Porphyra yezoensis and emended descriptions of the genus Polaribacter and two Polaribacter species. Int J Syst Evol Microbiol. 2013;63:1665–1672. doi: 10.1099/ijs.0.041434-0. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Thomas S, Cummings D, Temperton B, et al. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic' study of seasonal and diel temporal variation. PLoS One. 2010;5:e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R, et al. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature. 2007;445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- Gómez-Pereira PR, Fuchs BM, Alonso C, Oliver MJ, van Beusekom JE, Amann R. Distinct flavobacterial communities in contrasting water masses of the North Atlantic Ocean. ISME J. 2010;4:472–487. doi: 10.1038/ismej.2009.142. [DOI] [PubMed] [Google Scholar]
- Gómez-Pereira PR, Schüler M, Fuchs BM, Bennke C, Teeling H, Waldmann J, et al. Genomic content of uncultured Bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environ Microbiol. 2012;14:52–66. doi: 10.1111/j.1462-2920.2011.02555.x. [DOI] [PubMed] [Google Scholar]
- González JM, Fernández-Gómez B, Fernàndez-Guerra A, Gómez-Consarnau L, Sánchez O, Coll-Lladó M, et al. Genome analysis of the proteorhodopsin-containing marine bacterium Polaribacter sp. MED152 (Flavobacteria) Proc Natl Acad Sci USA. 2008;105:8724–8729. doi: 10.1073/pnas.0712027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Pinhassi J, Fernández-Gómez B, Coll-Lladó M, González-Velázquez M, Puigbò P, et al. Genomics of the proteorhodopsin-containing marine flavobacterium Dokdonia sp. Strain MED134. Appl Environ Microbiol. 2011;77:8676–8686. doi: 10.1128/AEM.06152-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink JJ, Woese CR, Staley JT. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus' as Polaribacter glomeratus comb. nov. Int J Syst Bacteriol. 1998;48:223–235. doi: 10.1099/00207713-48-1-223. [DOI] [PubMed] [Google Scholar]
- Hahnke RL, Harder J. Phylogenetic diversity of Flavobacteria isolated from the North Sea on solid media. Syst Appl Microbiol. 2013;36:497–504. doi: 10.1016/j.syapm.2013.06.006. [DOI] [PubMed] [Google Scholar]
- Hahnke RL, Bennke CM, Fuchs BM, Mann AJ, Rhiel E, Teeling H, et al. 2014Dilution cultivation of marine heterotrophic bacteria abundant after a spring phytoplankton bloom in the North Sea Environ Microbioldoi: 10.1111/1462-2920.12479 [DOI] [PubMed]
- Heinz E, Williams TA, Nakjang S, Noel CJ, Swan DC, Goldberg AV, et al. The genome of the obligate intracellular parasite Trachipleistophora hominis: new insights into microsporidian genome dynamics and reductive evolution. PLoS Pathog. 2012;8:e1002979. doi: 10.1371/journal.ppat.1002979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun DW, Shin NR, Kim MS, Kim PS, Jung MJ, Kim JY, et al. Polaribacter atrinae sp. nov., isolated from the intestine of a comb pen shell, Atrina pectinata. Int J Syst Evol Microbiol. 2014;64:1654–1661. doi: 10.1099/ijs.0.060889-0. [DOI] [PubMed] [Google Scholar]
- Ishida T, Fushinobu S, Kawai R, Kitaoka M, Igarashi K, Samejima M. Crystal structure of glycoside hydrolase family 55 β-1,3-glucanase from the basidiomycete Phanerochaete chrysosporium. J Biol Chem. 2009;284:10100–10109. doi: 10.1074/jbc.M808122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabisch A, Otto A, König S, Becher D, Albrecht D, Schuler M, et al. Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes ‘Gramella forsetii' KT0803. ISME J. 2014;8:1492–1502. doi: 10.1038/ismej.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-C, Oh HW, Kim H, Park D-S, Hong SG, Lee HK, et al. Polaribacter sejongensis sp. nov., isolated from Antarctic soil, and emended descriptions of the genus Polaribacter, Polaribacter butkevichii and Polaribacter irgensii. Int J Syst Evol Microbiol. 2013;63:4000–4005. doi: 10.1099/ijs.0.047100-0. [DOI] [PubMed] [Google Scholar]
- Kimura H, Young CR, Martinez A, DeLong EF. Light-induced transcriptional responses associated with proteorhodopsin-enhanced growth in a marine flavobacterium. ISME J. 2011;5:1641–1651. doi: 10.1038/ismej.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman DL.2000Uptake and regeneration of inorganic nutrients by marine heterotrophic bacteriaIn: Kirchman DL (ed). Microbial Ecology of the Oceans Wiley-Liss: Hoboken, NJ, USA; 261–288. [Google Scholar]
- Korth F, Deutsch B, Liskow I, Voss M. Uptake of dissolved organic nitrogen by size-fractionated plankton along a salinity gradient from the North Sea to the Baltic Sea. Biogeochemistry. 2012;111:347–360. [Google Scholar]
- Labourel A, Jam M, Jeudy A, Hehemann JH, Czjzek M, Michel G. The beta-glucanase ZgLamA from Zobellia galactanivorans evolved a bent active site adapted for efficient degradation of algal laminarin. J Biol Chem. 2014;289:2027–2042. doi: 10.1074/jbc.M113.538843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lami R, Cottrell MT, Campbell BJ, Kirchman DL. Light-dependent growth and proteorhodopsin expression by Flavobacteria and SAR11 in experiments with Delaware coastal waters. Environ Microbiol. 2009;11:3201–3209. doi: 10.1111/j.1462-2920.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- Lee YS, Lee DH, Kahng HY, Jung JS. Polaribacter gangjinensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2011;61:1425–1429. doi: 10.1099/ijs.0.024869-0. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang XY, Liu C, Lin CY, Xu Z, Chen XL, et al. Polaribacter huanghezhanensis sp. nov., isolated from Arctic fjord sediment, and emended description of the genus Polaribacter. Int J Syst Evol Microbiol. 2014;64:973–978. doi: 10.1099/ijs.0.056788-0. [DOI] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22:1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann AJ, Hahnke RL, Huang S, Werner J, Xing P, Barbeyron T, et al. The genome of the alga-associated marine flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl Environ Microbiol. 2013;79:6813–6822. doi: 10.1128/AEM.01937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedashkovskaya O, Kim S, Lysenko A, Kalinovskaya N, Mikhailov V, Kim I, et al. Polaribacter butkevichii sp. nov., a novel marine mesophilic bacterium of the family Flavobacteriaceae. Curr Microbiol. 2005;51:408–412. doi: 10.1007/s00284-005-0105-z. [DOI] [PubMed] [Google Scholar]
- Nedashkovskaya OI, Kim SB, Vancanneyt M, Snauwaert C, Lysenko AM, Rohde M, et al. Formosa agariphila sp. nov., a budding bacterium of the family Flavobacteriaceae isolated from marine environments, and emended description of the genus Formosa. Int J Syst Evol Microbiol. 2006;56:161–167. doi: 10.1099/ijs.0.63875-0. [DOI] [PubMed] [Google Scholar]
- Nedashkovskaya O, Kukhlevskiy A, Zhukova N. Polaribacter reichenbachii sp. nov.: a new marine bacterium associated with the green alga Ulva fenestrata. Curr Microbiol. 2013;66:16–21. doi: 10.1007/s00284-012-0200-x. [DOI] [PubMed] [Google Scholar]
- Oh C, Heo S-J, De Zoysa M, Affan A, Jung W-K, Park H-S, et al. Whole-genome sequence of the xylanase-producing Mesoflavibacter zeaxanthinifaciens strain S86. J Bacteriol. 2011;193:5557. doi: 10.1128/JB.05793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C, Kwon Y-K, Heo S-J, De Zoysa M, Affan A, Lee Y, et al. Complete genome sequence of strain S85, a novel member of the family Flavobacteriaceae. J Bacteriol. 2011;193:6107. doi: 10.1128/JB.05993-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto A, Bernhardt J, Meyer H, Schaffer M, Herbst FA, Siebourg J, et al. Systems-wide temporal proteomic profiling in glucose-starved Bacillus subtilis. Nat Commun. 2010;1:137. doi: 10.1038/ncomms1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parte AC. LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 2014;42:D613–D616. doi: 10.1093/nar/gkt1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read SM, Currie G, Bacic A. Analysis of the structural heterogeneity of laminarin by electrospray-ionisation-mass spectrometry. Carbohydr Res. 1996;281:187–201. doi: 10.1016/0008-6215(95)00350-9. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel T, Tomasch J, Buchholz I, Jacobs J, Kollenberg M, Gerdts G, et al. Constitutive expression of the proteorhodopsin gene by a Flavobacterium strain representative of the proteorhodopsin-producing microbial community in the North Sea. Appl Environ Microbiol. 2010;76:3187–3197. doi: 10.1128/AEM.02971-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, et al. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell. 2010;141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, Ott M. Efficient computation of the phylogenetic likelihood function on multi-gene alignments and multi-core architectures. Philos Trans R Soc Lond B Biol Sci. 2008;363:3977–3984. doi: 10.1098/rstb.2008.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck E, Bräun J, Pelttari K, Kadel S, Kalbacher H, Richter W. Chondrocyte secreted CRTAC1: a glycosylated extracellular matrix molecule of human articular cartilage. Matrix Biol. 2007;26:30–41. doi: 10.1016/j.matbio.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One. 2011;6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, et al. The role of planktonic Flavobacteria in processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol. 2013;15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- Woyke T, Xie G, Copeland A, González JM, Han C, Kiss H, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer KH, Glöckner FO, et al. Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol. 2010;33:291–299. doi: 10.1016/j.syapm.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Kang S-J, Oh T-K. Polaribacter dokdonensis sp. nov., isolated from seawater. Int J Syst Evol Microbiol. 2006;56:1251–1255. doi: 10.1099/ijs.0.63820-0. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Kawanabe A, Ito H, Kandori H, Kogure K. Diversity and functional analysis of proteorhodopsin in marine Flavobacteria. Environ Microbiol. 2012;14:1240–1248. doi: 10.1111/j.1462-2920.2012.02702.x. [DOI] [PubMed] [Google Scholar]
- Yoshizawa S, Kumagai Y, Kim H, Ogura Y, Hayashi T, Iwasaki W, et al. Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria. Proc Natl Acad Sci USA. 2014;111:6732–6737. doi: 10.1073/pnas.1403051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu C, Santschi PH. Chemical composition and 234Th (IV) binding of extracellular polymeric substances (EPS) produced by the marine diatom Amphora sp. Mar Chem. 2008;112:81–92. [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Suits MDL, Thompson AJ, Chavan S, Dinev Z, Dumon C, et al. Mechanistic insights into a Ca2+-dependent family of α-mannosidases in a human gut symbiont. Nat Chem Biol. 2010;6:125–132. doi: 10.1038/nchembio.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.