ABSTRACT
The giant panda evolved from omnivorous bears. It lives on a bamboo-dominated diet at present, but it still retains a typical carnivorous digestive system and is genetically deficient in cellulose-digesting enzymes. To find out whether this endangered mammalian species, like other herbivores, has successfully developed a gut microbiota adapted to its fiber-rich diet, we conducted a 16S rRNA gene-based large-scale structural profiling of the giant panda fecal microbiota. Forty-five captive individuals were sampled in spring, summer, and late autumn within 1 year. Significant intraindividual variations in the diversity and structure of gut microbiota across seasons were observed in this population, which were even greater than the variations between individuals. Compared with published data sets involving 124 gut microbiota profiles from 54 mammalian species, these giant pandas, together with 9 captive and 7 wild individuals investigated previously, showed extremely low gut microbiota diversity and an overall structure that diverged from those of nonpanda herbivores but converged with those of carnivorous and omnivorous bears. The giant panda did not harbor putative cellulose-degrading phylotypes such as Ruminococcaceae and Bacteroides bacteria that are typically enriched in other herbivores, but instead, its microbiota was dominated by Escherichia/Shigella and Streptococcus bacteria. Members of the class Clostridia were common and abundant in the giant panda gut microbiota, but most of the members present were absent in other herbivores and were not phylogenetically related with known cellulolytic lineages. Therefore, the giant panda appears not to have evolved a gut microbiota compatible with its newly adopted diet, which may adversely influence the coevolutionary fitness of this herbivore.
IMPORTANCE
The giant panda, an endangered mammalian species endemic to western China, is well known for its unique bamboo diet. Unlike other herbivores that have successfully evolved anatomically specialized digestive systems to efficiently deconstruct fibrous plant matter, the giant panda still retains a gastrointestinal tract typical of carnivores. We characterized the fecal bacterial communities from a giant panda population to determine whether this animal relies on its symbiotic gut microbiota to cope with the complex carbohydrates that dominate its diet, as is common in other herbivores. We found that the giant panda gut microbiota is low in diversity and highly variable across seasons. It also shows an overall composition typical of bears and entirely differentiated from other herbivores, with low levels of putative cellulose-digesting bacteria. The gut microbiota of this herbivore, therefore, may not have well adapted to its highly fibrous diet, suggesting a potential link with its poor digestive efficiency.
INTRODUCTION
The giant panda (Ailuropoda melanoleuca) is one of the most intriguing herbivorous mammalian species in evolutionary history. Phylogenetically, it is considered a “bear” species within the family of Ursidae (1, 2), where other members are carnivores or omnivores. However, highly fibrous bamboo leaves and stems dominate its diet, with the addition of fresh, soft bamboo shoots in spring and summer. The giant panda spends up to 14 h daily consuming a remarkable quantity of bamboo (3), which can reach 12.5 kg each day (4). It was estimated that the ancient omnivorous giant panda started to eat bamboo at least 7 million years ago (MYA) and became an exclusively bamboo-eating mammalian species at about 2 to 2.4 MYA (5). This dietary switch was probably associated with several mutations in the giant panda genome, including the pseudogenization of the umami taste receptor gene T1R1 since about 4.2 MYA (6, 7), and defects of dopamine metabolism in its appetite-reward system (8). To adapt to its highly specialized food source, the giant panda has developed a suite of unique morphological characteristics, including powerful jaws and teeth (9) and an enlarged radial sesamoid (also known as the “pseudothumb”) (10–12). Most herbivores have evolved an elongated foregut or hindgut to lengthen gut retention times of otherwise indigestible plant polysaccharides, mostly cellulose and hemicellulose. The giant panda, however, retains a simple stomach, degenerate cecum, and short, straight colon with a rapid transit time, all of which are typical of a carnivore’s gastrointestinal tract (13). Despite the dietary switch, the giant panda did not evolve any enzymes for bamboo digestion, and it still retains all necessary enzyme homologs for a carnivorous digestive system (6). Therefore, the giant panda appears to have no alternative but to rely on symbiotic gut microbes to adapt to its highly fibrous diet (6).
Previous studies have suggested that the diversity, structure, and function of the mammalian gut microbiome are mainly shaped by adaptation to diet (14–16). Highly diverse cellulolytic obligate anaerobes such as lineages within Bacteroidales, Clostridiales, Fibrobacterales, and Spirochaetales colonize most herbivores’ gastrointestinal tracts, providing microbial fermentation to enhance nutrient absorption (17–20). In contrast, the gut microbiota of omnivores and carnivores, particularly within the mammalian order Carnivora, are dominated by the facultative anaerobes Enterobacteriaceae and Enterococcus (21, 22). For the giant panda, Enterobacteriaceae and Streptococcus have been identified as predominant members of its gut microbiota by traditional culture-dependent methods (23–25) and 16S rRNA gene clone library analysis (26). Metagenomic sequencing of three fecal samples from wild giant pandas identified cellulolytic genes, suggesting that the giant panda gut microbiome might be capable of digesting cellulose in its bamboo diet (27). However, because of the particularly low digestibility of bamboo dry matter (<20%), it was suggested that the giant panda may rely on utilization of bamboo cellular contents rather than the cell wall constituents (28).
To elucidate whether the giant panda with its carnivorous digestive system has evolved a gut microbiota adapted to its herbivorous diet, we performed a large-scale dynamic structural profiling of fecal samples from cub, juvenile, and adult giant panda populations across three seasons and compared this data set with those of other herbivores, omnivores, and phylogenetically related carnivores (14, 16, 17, 27). We found that the giant panda harbors a carnivore-like gut microbiota with poor diversity and extensive seasonal variations. This gut microbiota structure appears to deviate from the general rule that “adaptation of the microbiota to diet is similar across different mammalian lineages” (16).
RESULTS
Overall gut microbiota structure of the giant panda.
We collected a total of 121 giant panda fecal samples from 24 adults, 16 juveniles, and 5 unweaned cubs, in which 21 females and 12 males completed sampling in three seasons, the spring (between March and May, here referred to as T1), summer (August, here referred to as T2), and late autumn (between November and December, here referred to as T3), within 1 year. Bar-coded pyrosequencing of the V3 region of bacterial 16S rRNA genes generated a data set consisting of 97,156 reads. After removal of chloroplast sequences, PyNAST (Python Nearest Alignment Space Termination) alignment (29) was performed and 92,819 sequences were grouped into 781 operational taxonomic units (OTUs) at a threshold of 97% sequence identity. Although the OTU-level rarefaction curves did not reach stable values (see Fig. S1A to D in the supplemental material), the Shannon diversity index, which considers both the microbial richness and evenness, plateaued in all samples, indicating that although additional rare phylotypes would likely be detected by deeper sequencing, most of the microbial diversity present in these fecal samples had already been captured at the current sequencing depth (see Fig. S1E to H).
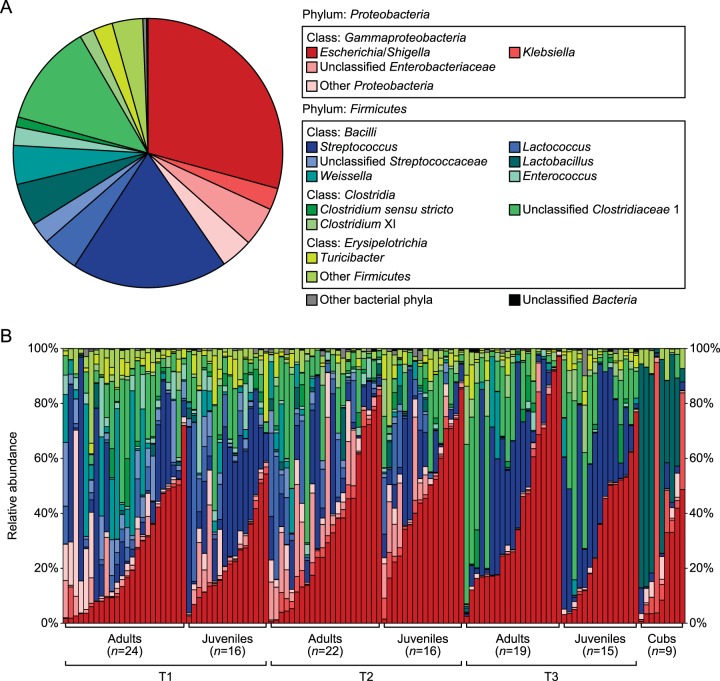
Across all the samples, 99.84% and 77.78% of the total sequences were assigned into 9 phyla and 109 genera, respectively. Firmicutes and Proteobacteria constituted the two dominant phyla in the giant panda gut microbiota (contributing 58.96% and 40.42% of the total sequences, respectively) (Fig. 1A). Other detected phyla in this study included Bacteroidetes (282 sequences), Actinobacteria (67 sequences), Fusobacteria (64 sequences), TM7 (4 sequences), Planctomycetes (2 sequences), Acidobacteria (1 sequence) and Cyanobacteria (1 sequence). At the genus level, there were 10 taxa that each occupied more than 1% of the total sequences. Escherichia/Shigella was the most predominant genus (29.26% of the total sequences) in the giant panda gut microbiota. Klebsiella (2.61% [Fig. 1A]) was another dominant genus in Proteobacteria. In the class Bacilli from Firmicutes, Streptococcus (18.76%) was more abundant than Lactobacillus (5.13%), Weissella (4.67%), Lactococcus (4.38%), and Enterococcus (2.23%) (Fig. 1A). In Clostridia, 85.20% of the reads belonged to Clostridiaceae 1 but were mostly unassignable at the genus level; only 8.50% of the reads within in this family were identified as Clostridium sensu stricto (accounting for 1.19% of the total sequences [Fig. 1A]). Besides the predominance of Clostridiaceae 1, Peptostreptococcaceae were also well represented in Clostridia, in which Clostridium XI reached an overall abundance of 1.72% in these samples (Fig. 1A). Turicibacter, a genus from another class of Firmicutes, Erysipelotrichia, composed 2.36% of the giant panda gut microbiota (Fig. 1A). Each of the 10 predominant genera showed wide abundance variation across the collected samples (Fig. 1B). For example, Escherichia/Shigella, the only genus that was present in all of the adult, juvenile, and cub samples, ranged in abundance from 0.60% to 97.11% across samples (Fig. 1B).
FIG 1 .
Genus-level gut microbiota composition of the giant panda population. (A) The 10 most dominant genera (>1% of the total sequences) in the 121 giant panda samples. (B) Relative contribution of these dominant genera in each sample. Samples are grouped according to the sampling time and the age range and are then arranged by the proportion of the most dominant genus, Escherichia/Shigella.
Dramatic intra- and interindividual structural variations in the giant panda gut microbiota.
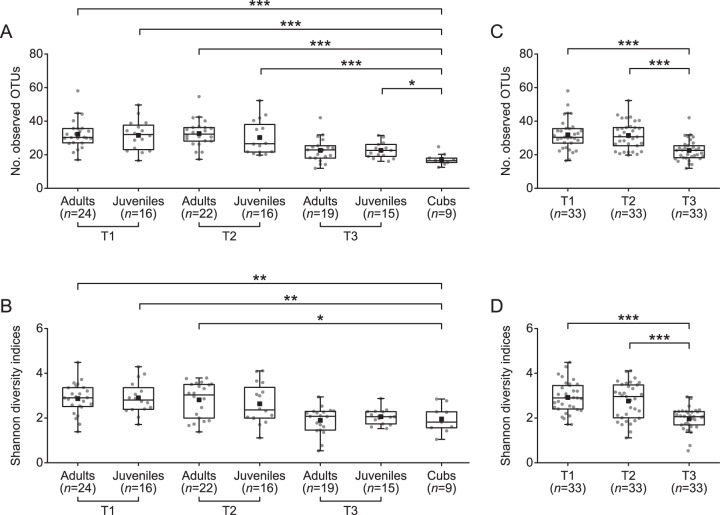
The alpha diversities of the adult and juvenile samples were similar to each other in every sampling season, as measured by their numbers of observed OTUs and Shannon diversity indices, and they dropped to almost as low as those of cub samples in T3 (Fig. 2A and B). Comparison between the paired T1, T2, and T3 samples from the 33 adults and juveniles sampled in all three seasons confirmed their significant changes in alpha diversity across seasons: neither their numbers of observed OTUs nor the Shannon diversity indices showed significant difference between T1 and T2 samples, but both measurements decreased drastically in the last sampling season (Fig. 2C and D). We also noticed that 7 of the 10 dominant genera fluctuated markedly across seasons, except Escherichia/Shigella, Clostridium sensu stricto, and Turicibacter (see Fig. S2A to J in the supplemental material). Klebsiella was the only genus that was significantly enriched in T2 but diminished in the other two seasons (see Fig. S2B), whereas Streptococcus and Clostridium XI were more abundant in T3 (see Fig. S2C and I). Apart from Streptococcus, the other four dominant genera from Bacilli (i.e., Lactococcus, Lactobacillus, Weissella, and Enterococcus) were less abundant in T3 than in T1 or T2 (see Fig. S2D to G).
FIG 2 .
Variations in alpha diversity of the giant panda gut microbiota. (A and B) Comparisons of the number of observed OTUs (A) and Shannon diversity indices (B) among adult, juvenile, and cub samples by Mann-Whitney test. The adult and juvenile samples collected in T1, T2, and T3 are compared with the cub samples, respectively. (C and D) Comparisons of the number of observed OTUs (C) and Shannon diversity indices (D) among the T1, T2, and T3 samples by paired sample Wilcoxon signed-rank test. Only the 33 individuals that were sampled in all three seasons are included. Both alpha diversity metrics were calculated upon the rarified OTU subsets, using 440 sequences per sample with 1,000 replications. In all panels, boxes represent the interquartile range (IQR) between the first and third quartiles. The lines and squares inside boxes represent the median and mean, respectively. Whiskers denote the lowest and highest values within 1.5× IQR from the first and third quartiles, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (with Bonferroni post hoc test).
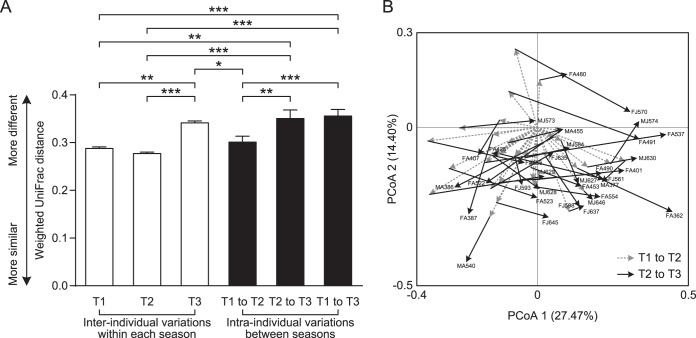
The remarkable changes in the diversity and composition of the giant panda gut microbiota across individuals and seasons led us to assess the extent of interindividual structural variations within each season and intraindividual variations across seasons. While the diversity was lower in T3 (Fig. 2C and D), the interindividual weighted UniFrac distances between different individuals increased significantly in this sampling season (Fig. 3A). This giant panda population also manifested dramatic intraindividual variations from T1/T2 to T3, which were significantly larger than that from T1 to T2. Notably, the intraindividual variations from T1/T2 to T3 were even higher than the interindividual variations within T1 and T2 (Fig. 3A). The principal coordinate analysis (PCoA)-based trajectory plot also revealed that the gut microbiota structure of each individual became more and more dissimilar over seasons (Fig. 3B). Moreover, such seasonal structural shifts seemed individual specific, resulting in a “radial” trajectory pattern (Fig. 3B). Such large interindividual and even larger intraindividual structural variations were also supported by the unweighted UniFrac distances, which concern only the occurrence of phylotypes rather than their abundance (see Fig. S3A and B in the supplemental material). These significant changes in the gut microbiota structure from T1 to T3 were also verified by permutational multivariate analysis of variance (PERMANOVA; P = 0.0001 for both the weighted and unweighted UniFrac distances) (30).
FIG 3 .
Inter- and intraindividual variations of the gut microbiota of the 33 giant pandas that were sampled in all three seasons. (A) Interindividual variations were determined by average weighted UniFrac distances between individuals in T1, T2, or T3, respectively, while intraindividual variations were determined by distances between paired T1 and T2, T2 and T3, and T1 and T3 samples, respectively. Mean values ± standard errors of the means are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test with 1,000 Monte Carlo permutations). (B) Trajectory of the gut microbiota structure of each giant panda individual across seasons. The principal coordinate analysis (PCoA) scores of T1 samples were set to zero, and the relative scores of T2 and T3 samples corresponding to their paired T1 samples were plotted. The animal identifiers are shown. A, adult; J, juvenile; F, female; M, male.
Concordant with their similar levels of alpha diversity (Fig. 2A and B), the adult and juvenile samples also tended to share similar overall gut microbiota structures in each season (PERMANOVA, P > 0.05 for both weighted and unweighted UniFrac distances, except the “unweighted” comparison in T3, which reported a P value of 0.0266). However, the cub samples harbored a gut microbiota structure significantly distinct from those of adults and juveniles (PERMANOVA, P = 0.0001 for both UniFrac distances; cubs were compared with adults and juveniles separately), in which Lactobacillus (contributing 52.31% of the total sequences in cub samples) outnumbered Escherichia/Shigella (Fig. 1B).
Comparison of gut microbiota structure of the captive and wild giant pandas with that of other mammals.
To compare the gut microbiota of the giant panda with that of many other mammalian species, we combined our pyrosequencing data set with the previously released 16S rRNA gene clone library data from 8 captive and 7 wild giant pandas (27) and three other mammal data sets encompassing 128 animals representing 57 species from 13 taxonomic orders, which also included one captive giant panda sample, and those from its close relatives such as bears and red pandas, typical carnivores such as lions and cheetahs, and distantly related herbivores such as ruminants, horses, rabbits, and kangaroos (14, 16, 17) (see Table S1 in the supplemental material). In total, 137 giant panda samples were included in the analysis. After removal of chloroplast sequences, 176,568 sequences were extracted from the four published data sets and were incorporated into the multistep OTU picking procedure with the 92,819 sequences generated in this study (see Fig. S4). At a 97% sequence identity threshold, 229,288 out of 269,387 sequences were assigned into 8,646 Greengenes reference OTUs and 3,075 additional OTUs (see Fig. S4 and Table S1). Of the 40,099 discarded sequences, 95.11% were contributed by the pyrosequencing data set from the 39 mammal samples (16). For the 137 giant panda samples, most sequences were retained after OTU picking (98.12% ± 1.06%). To avoid discarding giant panda samples, we excluded only the 19 samples that retained fewer than 140 sequences after the OTU picking; a total of 245 samples with 227,388 sequences were thus included in the following comparative analysis (see Table S1).
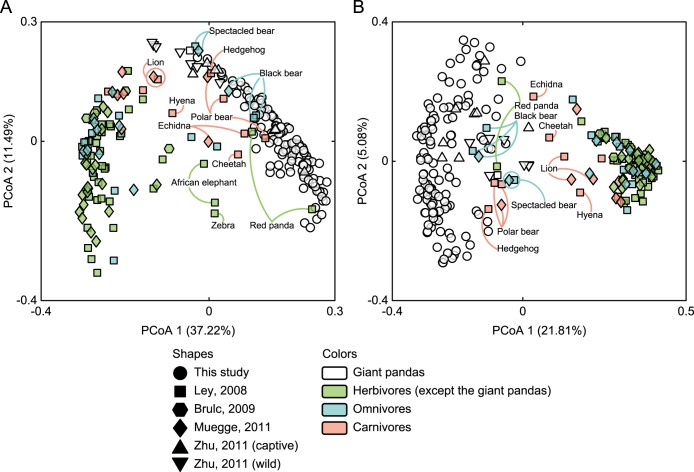
The obtained OTU sets were evenly resampled to generate the rarified OTU subset at the depth of 140 sequences per sample. The giant panda had significantly lower alpha diversity than did other herbivores, omnivores, and carnivores (see Fig. S5A and B in the supplemental material). Moreover, both the captive and wild giant pandas were completely differentiated from all the nonpanda herbivores (i.e., all herbivores included in this study except the giant pandas and red pandas) but closely clustered with the red pandas, bears, and some of the other carnivores along the first two UniFrac principal coordinates (PCs) (Fig. 4A and B), indicating that the giant pandas harbored a distinctive gut microbiota structure compared with that of nonpanda herbivores. The reliability of these findings was substantiated by the observation that multiple samples from the same mammalian species derived from different data sets tended to cluster together on the UniFrac hierarchical clustering trees (see Fig. S6A and B).
FIG 4 .
Comparison of gut microbiota structure of captive and wild giant pandas with that of other herbivores, omnivores, and carnivores. Principal coordinate analysis (PCoA) score plots based on the weighted (A) and unweighted (B) UniFrac distances, respectively. In both panels, the shapes of data points indicate the source of the samples, whereas the colors differentiate the giant panda samples from those of other herbivores, omnivores, and carnivores.
To address the potential bias resulting from the heterogeneity of sequencing depth across these data sets and the considerable number of discarded sequences, we constructed a new data set by pyrosequencing the V3 region of bacterial 16S rRNA genes in fecal samples from Asian black bears, Siberian tigers, chimpanzees, golden snub-nosed monkeys, hoolock gibbons, blue peafowls, and giant pandas (samples from each mammalian species were pooled prior to pyrosequencing, as described in Materials and Methods) and compared their microbiota structure with that of the giant panda population described in this study. Each sample yielded at least 2,300 sequences after removal of chloroplast sequences and PyNAST alignment (median = 3,665 sequences, ranging from 2,300 to 4,995 sequences). Combined with our first pyrosequencing data set of the giant panda gut microbiota, 1,446 de novo OTUs were delineated at 97% sequence identity. Using the rarified OTU subset (440 sequences per sample), UniFrac PCoA (see Fig. S7A and B in the supplemental material) revealed that the newly sequenced giant panda sample clustered together with other giant panda samples. The omnivorous bear and carnivorous tiger samples showed gut microbiota structure similar to that of these giant pandas, but samples from other omnivores and herbivores diverged markedly (see Fig. S7A and B).
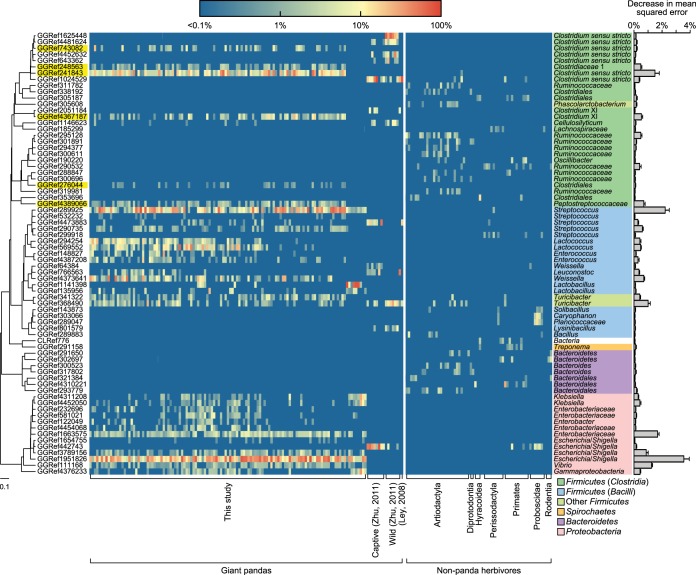
Random forests (31), a powerful supervised machine-learning technique that has been widely used for high-dimensional data mining, including gut microbiome analysis (32–34), was employed to characterize which phylotypes were involved in the deviation of the giant panda gut microbiota (n = 137) from the nonpanda herbivores (n = 64). The discriminating model constructed by random forests demonstrated its high predictability (baseline error = 31.84%, 10-fold cross-validation error = 2.96% ± 2.55%). Seventy-one OTUs with random forests variable importance scores of more than 0.0005 were considered predictive signatures that were highly distinguishable between the giant pandas and nonpanda herbivores (Fig. 5). Of these key phylotypes, 42 were enriched in the giant pandas but depleted in most nonpanda herbivores (and also in most carnivores and omnivores [data not shown]), including OTUs from Clostridium sensu stricto, Streptococcus, and Enterobacteriaceae. In contrast, 29 OTUs commonly occurring in nonpanda herbivores, particularly the animals within the order Artiodactyla, were consistently absent in the giant pandas. Most of these OTUs belonged to Ruminococcaceae and Bacteroidetes.
FIG 5 .
Heat map of the 71 OTU-level phylotypes identified as key variables for differentiation between gut microbiota structure of the giant pandas and that of nonpanda herbivores by random forests. The OTUs are arranged according to their phylogenetic positions (left) determined by the Greengenes reference tree. The six Clostridia OTUs enriched in the 121 giant panda gut microbiota samples collected in the current study are highlighted in yellow. The deepest level of confident taxonomic annotation of these OTU lineages was obtained by the Ribosomal Database Project classifier (see the legend at the bottom right corner for the color key). The OTU importance score was determined by estimating the increase in mean squared error when that OTU was removed from the set of predictors. The abundance of the evenly rarified OTU subset (using 140 sequences per sample) is log transformed in the heat map.
Identification of putatively cellulolytic Clostridia in the giant panda gut.
As a previous study indicated that the giant panda gut microbiota harbored Clostridium-related lineages that might be involved in cellulose digestion and utilization (27), we inferred the putatively cellulolytic Clostridia phylotypes in the 121 giant panda samples collected in this study on the phylogenetic tree (Materials and Methods). We focused on Clostridia OTUs that were present in at least 10 samples, including the 16 OTUs picked by the de novo method and 10 OTUs assigned to the Greengenes reference sequence collection (see Fig. S8 in the supplemental material, with prefix “Denovo” or “GGRef” on the tree, respectively). Of these OTUs, Denovo653 and GGRef241843 were the most prevalent and predominant in their respective data sets (see Fig. S8 for their abundance and prevalence). However, there was no evidence that their nearest neighbors contribute to cellulose digestion.
Of the 10 GGRef Clostridia OTUs, 6 were selected as key giant panda phylotypes by random forests (Fig. 5; see also Fig. S8 in the supplemental material), because they were almost entirely absent in all the other herbivores, omnivores, and carnivores. GGRef743082 (see Fig. S8 for its abundance), which was detected in 36 giant panda samples, showed a close relationship with Clostridium sartagoforme, a chitinolytic bacterium able to grow on hemicellulose (35). Clostridium sartagoforme was also the nearest neighbor for Denovo209, which occurred in 20 giant panda samples (see Fig. S8 for its abundance). Denovo58 and GGRef276044 (see Fig. S8 for their abundance and prevalence) could be related to Clostridium cellulosi, which was identified as a cellulolytic thermophile (36). Additionally, Denovo704 (see Fig. S8 for its abundance), which was identified from 18 samples, was closely associated with Cellulosilyticum ruminicola and Cellulosilyticum lentocellum, both of which show significant capability to degrade cellulosic materials (37). However, none of these giant-panda-specific, putatively cellulolytic lineages was shared by a majority of the investigated giant panda samples (see Fig. S8 for their prevalence). Instead, they were not detected from 83 and 63 samples in the de novo and reference-based OTU data sets, respectively.
DISCUSSION
The giant panda is regarded as a national treasure in China due to its delightful appearance, unusual dietary behavior, low fecundity rate, and endangered status. Massive efforts have been invested in conserving this iconic species. Accumulating evidence emphasizes the tight interactions between the gut microbiota and animal health (38–40), but until now, only a few fecal samples had been collected to explore the characteristics of the giant panda gut microbiota. In this study, Firmicutes and Proteobacteria overwhelmingly dominated the 121 fecal samples from the investigated giant panda population, with a shortage of other well-known mammalian microbiome phyla such as Bacteroidetes, which is normally abundant in the bovine rumen, the tammar wallaby foregut, and the human gut (17, 19, 41). This relatively simple composition was in line with previous studies on the giant panda gut microbiota carried out by traditional molecular ecological techniques (23–27). Our large-scale comparative analysis also confirmed that the giant panda shows extremely low gut microbial diversity compared with other mammalian species (14, 27), even its close carnivorous relatives, indicating that this animal harbors a simple gut microbiota. In theory, a high level of diversity provides “functional redundancy” that helps an ecosystem retain its resistance, resilience, and stability after environmental stresses (42, 43). A gut microbiota with high diversity is generally considered beneficial for the host health (44–46). For the giant panda, whether the low diversity of its gut microbiota has any impact on its highly fragile lifestyle remains to be addressed.
We have identified excessive variations within the giant panda gut microbiota across individuals and seasons, which may exemplify the association between microbial diversity and ecosystem stability. At the last time of sampling (T3), the diversity within the giant panda gut microbiota was significantly decreased, while the variations between individuals were increased. Surprisingly, this interindividual variation was smaller than the intraindividual structural variations between seasons. These pronounced structural shifts across seasons included significant abundance fluctuations in particular genera, such as Streptococcus and Lactobacillus. In contrast, Escherichia/Shigella, Clostridium sensu stricto, and Turicibacter did not exhibit season-associated variations. These results are congruent with another recent study on the association of the abundance variations of specific gut bacterial taxa in one pair of adult male and female giant pandas with their seasonal dietary change in preference for bamboo plant parts (47).
A previous study of three polar bears similarly showed that the overall gut microbiota structure fluctuated within 1 month (22). Healthy human adults, in contrast, show temporal stability of human gut microbiota, with variation in the gut microbiota being generally much greater between unrelated individuals than within a particular individual over time (48, 49). Indeed, the potential for intraindividual structural variation in humans appeared too slight to overcome the interindividual variations during short-term controlled feeding with specific high-fat/low-fiber or low-fat/high-fiber diets (50). For the giant panda, the food source fluctuates little, although the parts of the bamboo plant that were consumed do vary with the seasons. One recent nutritional geometry-based study revealed that the migratory behavior of the wild giant pandas, including their seasonal food choices, could be associated with the concentrations of nitrogen, phosphorus, and calcium in the different parts of two bamboo species (51). Notably, the extraction efficiencies of these nutrients were differentiated, such as the selective excretion of calcium in the giant panda’s feces (51). In our study, bamboo shoots were absent from the giant pandas’ diet at the last time of sampling (T3) when the largest variation in the gut microbiota diversity and structure occurred. The seasonal availability of particular bamboo parts is therefore probably an important factor in affecting the giant panda gut microbiota. Considering the evidence that each giant panda underwent individual-specific seasonal variation in its overall gut microbiota structure (Fig. 3B; see also Fig. S3B in the supplemental material) while in seemingly stable health, the connections among the seasonal dietary variation, the responses from the gut microbiota, and the giant panda health may represent an exceptional paradigm in host-gut microbiota interactions (52) and thus warrant further study.
Age is generally considered an important factor to shape the gut microbiota. In this study, the diversity of the gut microbiota of the cub giant panda was significantly lower than that of juveniles and adults, with a simple and distinct microbial composition that probably reflects the unweaned status of these cubs as well as the immaturity of their gut microbiota. The maturation of the human gut microbiota during the first 3 years of life concordantly shows increased phylogenetic diversity with age (34). Compared with the predominance of Bifidobacterium in the human infant gut microbiota throughout the first year of life (34), Lactobacillus dominated the fecal communities of the nine giant panda cub samples in this study. It remains to be determined whether the dominance of this genus serves a physiologically beneficial role in the gut microbial development of the infant giant panda.
Given the observed structural variation in the giant panda gut microbiota, a large number of giant panda samples, including both captive and wild individuals, are required to compare with those of other herbivores, omnivores, and carnivores to evaluate the adaptation of its gut microbiota structure to the bamboo diet. In this study, we demonstrate the “carnivore-like” features of the gut microbiota structure of the giant panda. This is consistent with a previous study based on the comparison of one captive giant panda fecal microbiota sample with those of other mammals, which also showed that the overall structure of the giant panda gut microbiota clustered with those of carnivorous and omnivorous bears and two red pandas (14). This carnivore-like structure could be further summarized as the enrichment of lineages from the family Enterobacteriaceae, the genus Streptococcus, and Clostridium sensu stricto, with a dearth of putative fiber-degrading lineages from Ruminococcaceae and Bacteroidetes that commonly occur in nonpanda herbivores. Enterobacteriaceae were also prominent in the fecal samples from grizzly bears, one close relative of the giant panda (21). The prevalence of these phylotypes may represent a common feature in the gut microbiota of the giant panda (53), bears (21), and other carnivores. These Gram-negative, facultative anaerobes are often considered endotoxin-producing opportunistic pathogens and have shown a putatively causative role in the development of obesity in humans (54, 55). Because the giant panda is highly susceptible to enteric diseases, the question of whether the proliferation of these potentially pathogenic bacteria in its gut also has negative impacts on its health merits further exploration.
The most important question in the study of the giant panda gut microbiota is whether its carnivore-like structure can still effectively utilize cellulose—the major component of its highly fibrous diet. In this study, the predominance of noncellulolytic bacteria such as Escherichia/Shigella and Streptococcus left little room for the putative polysaccharide-degrading specialists such as some Clostridium-related species (56–58). These cellulolytic candidates were detectable in only a limited number of the samples. In addition, the genus Clostridium sensu stricto was not significantly enriched at the last time of sampling (T3), when only highly fibrous bamboo leaves and stems were available, further suggesting that this genus is not sensitive to the host’s seasonal dietary changes. These results were consistent with the previously reported extremely low number of cellulase and hemicellulase genes in the fecal microbiomes of three wild giant panda samples (8 genes comprising merely 2% of the total number of glycoside hydrolases) (27), even less than in the reported human gut microbiome (which contains 404 cellulase and endohemicellulase genes comprising 7% of glycoside hydrolases) (59). Thus, this nearly negligible contribution of the gut microbiota to cellulose digestion, which corresponds with the fact that the giant panda is a fairly inefficient digester of bamboo (28), argues against the previously hypothesized importance of the gut microbiota for bamboo digestion (6). Further quantitative assessments will be helpful to provide a complete picture of the actual population size of the cellulose-digesting bacteria in the giant panda gut microbiota and to complement the current interpretation of the abundance data from metagenomic sequences (27).
In the present study, we delineate a carnivore-like, simple, and highly variable gut microbiota in the giant panda with an apparent deficiency in cellulose-digesting lineages. Although studies of its genetic diversity, population structure, and demographic history suggest that the giant panda is not a species at an evolutionary dead end (60), the peculiar characteristics of its gut microbiota may put it at high risk of extinction. Unlike other mammalian species that have evolved gut microbiota (and also digestive system anatomies) optimized for their specific diets, the aberrant coevolution of the giant panda, its dietary preferences, and its gut microbiota remains enigmatic. Building upon the current observations, further well-designed studies combining metagenomics, metatranscriptomics, metaproteomics, and metabonomics may provide valuable insights to biological scientists and wildlife conservationists about how to improve the giant panda’s digestive functions, nutritional status, and physiological condition, perhaps via targeted modulation of the structure and metabolism of the gut microbiota.
MATERIALS AND METHODS
Sampling.
A total of 45 healthy, captive-born giant pandas housed in the Chengdu Research Base of Giant Panda Breeding were sampled, including 5 cubs (aged <0.5 years, unweaned), 7 juvenile females and 9 juvenile males (aged 2 to 5 years), and also 17 adult females and 7 adult males (aged 6 to 22 years). In total, 112 fecal samples were collected from juvenile and adult individuals in T1, T2, and T3 within 1 year. One adult female (sample identifier [ID] FA494) was sampled twice (in March and May, respectively) in T1. The ingestion and health status of each giant panda were monitored daily by veterinarians, and they received no antibiotics or dietary supplements in the 2 months prior to sampling. At least 10 kg of bamboo (mainly Bashania fargesii) and bamboo shoots (available only in T1 and T2) constituted the staple diet for each juvenile and adult individual every day. Specifically, about 70% of the dietary components were stems in T1 and more than 90% were leaves in T2 and T3. A small amount of steamed bread (500 to 800 g, including 80% corn and 20% wheat) was also provided for each juvenile and adult individual every day. The total weight of the feces was about 10 to 15 kg per day per juvenile or adult individual. An additional 9 fecal samples were collected from the 5 cubs between August and December. Fresh milk from the female giant pandas was the only diet for the cubs. Fresh fecal samples were collected immediately after defecation and frozen at −80°C for subsequent use.
Fecal samples from 7 mammalian species maintained in Yunnan Wild Animal Park were also collected, including Asian black bear (Ursus thibetanus), Siberian tiger (Panthera tigris altaica), chimpanzee (Pan troglodytes), golden snub-nosed monkey (Rhinopithecus roxellana), hoolock gibbon (Hoolock leuconedys), blue peafowl (Pavo cristatus), and the giant panda. For each mammalian species, multiple fecal samples were collected from adult individuals and pooled, to represent the “typical” fecal microbiota for each species.
DNA extraction, PCR amplification, and pyrosequencing.
All the fecal samples were subjected to the same procedures for DNA extraction and PCR amplification by the same laboratory staff. To dissociate microbes from plant residue, 150 g of each fecal sample was pretreated at Chengdu Research Base of Giant Panda Breeding prior to DNA extraction according to the procedure described previously (26) with slight improvements: the fecal sample was first suspended in 500 ml of sterile 0.05 M phosphate-buffered saline (PBS; pH 7.4). This mixture was vortexed vigorously, and large plant particles were filtered out. The suspension was centrifuged at low speed (200 × g) for 5 min to sediment the residual coarse particles, and this process was repeated two more times, with the supernatants being collected and pooled. Microbial biomass was collected and washed three times by centrifugation at 9,000 × g for 4 min with 20 ml of sterile PBS. The cell pellets were finally resuspended in 10 ml of sterile PBS and were stored as 1-ml aliquots at −80°C for transportation and subsequent DNA extraction. Genomic DNA was extracted by using the InviMag stool DNA kit (Invitek, Germany) with agitation in a mini-bead beater (BioSpec Products, Bartlesville, OK) as follows: microbial biomass was collected from the pretreated cell suspension by centrifugation at 9,000 × g for 3 min, and the cell pellets were resuspended in 1.2 ml of lysis buffer P of the kit. The cell suspension was then transferred to a 2-ml screw-cap tube containing 0.3 g zirconia beads (0.1 mm; BioSpec Products, Inc., USA). After agitation by bead beating for 1 min at maximum speed, DNA was extracted by following the manufacturer’s instructions for bacterial DNA extraction using the KingFisher instrument (Invitek, Germany) and stored at −20°C for further analysis. The extracted DNA from each sample was used as the template to amplify the V3 region of 16S rRNA genes. PCR amplification, bar-coded pyrosequencing of the PCR amplicons, and quality control of raw data were performed according to the procedures described previously (61).
De novo OTU picking and statistical analysis.
All raw reads were sorted into different samples according to sample-unique 6-base bar codes. After removal of bar codes and primers, the Ribosomal Database Project (RDP) naive Bayesian rRNA classifier (v2.6 with 16S rRNA training set 9) was used for taxonomic assignment of all the sequences at an 80% confidence level (62), and sequences assigned to chloroplasts were excluded from subsequent analysis. All the retained sequences were processed using the QIIME (Quantitative Insights into Microbial Ecology) package (v1.8.0) (63). The sequences were aligned using the PyNAST aligner (29) with the Greengenes core set (64) and then classified into OTUs at a threshold of 97% sequence identity using UCLUST (65). The representative sequence for each OTU was selected using default parameters and was imported into the latest Greengenes ARB database (66) to construct a phylogenetic tree through the ARB parsimony insertion tool (67) (hypervariable regions were masked by the lanemaskPH filter).
Rarified OTU subsets (440 sequences were used per sample) were generated to calculate the alpha diversity in QIIME. The phylogenetic tree was subsequently used for UniFrac-based beta diversity analysis (68). Student’s t test with 1,000 Monte Carlo permutations was performed on the UniFrac distance matrix (46) to determine whether the UniFrac distances between individuals were significantly different among T1, T2, and T3 samples and whether the distances between paired T1 and T2, T2 and T3, or T1 and T3 samples from respective individuals were different. PERMANOVA was performed to test whether the gut microbiota structure was significantly different by using the method implemented in the R “vegan” package (69), and the P values were obtained with 9,999 permutations. The Mann-Whitney test and paired sample Wilcoxon signed-rank test were used for univariate statistical analysis. The Bonferroni correction was used to adjust the multiple comparisons. All the univariate statistical analysis was conducted using OriginPro 8.5.1 (OriginLab Corporation, Northampton, MA) and MatLab R2012b (The MathWorks, Natick, MA).
Multistep OTU picking for combined data sets and statistical analysis.
Previously published 16S rRNA gene data sets used in this study included clone library data generated from 85 mammalian fecal samples (17,760 near-full-length sequences) (14), 4 bovine rumen samples (3,617 near-full-length sequences) (17), and 15 giant panda fecal samples (85 OTUs selected from 5,522 near-full-length sequences as representative sequences at a 97% identity threshold) (27) and pyrosequencing data generated from 39 mammalian fecal samples (149,675 V1-V2 region sequences) (16). For the giant panda clone library data set (27), we readded the original sequence counts to the respective OTU representative sequences downloaded from the GenBank database (accession numbers from JF920308 to JF920392). After exclusion of chloroplast sequences by RDP classifier at an 80% confidence level, all the retained sequences were combined with our data set generated from the giant panda population.
As the combined data were composed of sequences from different regions of the 16S rRNA gene, a Greengenes reference-based OTU picking procedure (48, 70) rather than a de novo method was first applied by using the QIIME package. In this step, the latest Greengenes reference database (66) that had been prebinned at 97% sequence identity (released in August 2013) was used as the reference database. Sequences were assigned to OTUs based on their best match to a reference sequence by using UCLUST at a 97% identity threshold (65), and sequences that did not match any reference sequences were discarded. The discarded sequences from the three published clone library studies (14, 17, 27) that passed the PyNAST alignment were then grouped into OTUs at 97% sequence identity by performing the de novo OTU picking method. These de novo OTUs served as the second reference sequence collection to recover the remaining sequences discarded by the initial Greengenes reference OTU picking. After merging the OTU data generated through the multistep OTU picking procedure, the rarified OTU subsets were generated for alpha and beta diversity analysis. Random forest analysis (31) was applied to discriminating the giant panda gut microbiota from that of the nonpanda herbivores, using the “randomForest” package in R (71) with 1,000 trees and all default settings. The generalization error was estimated using 10-fold cross-validation. The expected “baseline” error obtained by a classifier that simply predicts the most common category label was also included.
Identification of putatively cellulolytic phylotypes within Clostridia in the giant panda gut.
Because the Greengenes reference database used in the multistep OTU picking procedure was a subset of the Greengenes ARB database, which was also used to construct the phylogenetic tree of the OTUs classified by a de novo method, it was possible to merge all OTUs classified by the two procedures and infer their phylogenetic information in ARB. For the OTUs present in our pyrosequencing data set, those that had been assigned to the clade of the class Clostridia by RDP classifier with a minimal occurrence in 10 samples were picked, and their nearest known strain neighbors were selected from the tree manually for phylogenetic analysis.
Sequence data accession numbers.
The pyrosequencing reads generated from the giant panda population and the seven mammalian species described in this study have been deposited in the sequence read archive (SRA) at the NCBI under the accession numbers SRP050128 and SRP050129, respectively.
SUPPLEMENTAL MATERIAL
OTU-level alpha diversity of the gut microbiota of the giant panda population. (A to D) Rarefaction curves of T1 (A), T2 (B), T3 (C), and cub (D) samples. (E to H) Shannon diversity index curves of T1 (E), T2 (F), T3 (G), and cub (H) samples. Download
Abundance variations in the 10 dominant genera (>1% of total sequences), Escherichia/Shigella (A), Klebsiella (B), Streptococcus (C), Lactococcus (D), Lactobacillus (E), Weissella (F), Enterococcus (G), Clostridium sensu stricto (H), Clostridium XI (I), and Turicibacter (J), across seasons, compared by paired sample Wilcoxon signed-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (with Bonferroni post hoc test). Only the 33 individuals that were sampled in all three seasons are included. See Fig. 2 for definition of box-and-whisker plot. Download
Inter- and intraindividual variations of the gut microbiota of the 33 giant pandas that were sampled in all three seasons. (A) Interindividual variations were determined by average unweighted UniFrac distances between individuals in T1, T2, or T3, respectively, while intraindividual variations were determined by distances between paired T1 and T2, T2 and T3, and T1 and T3 samples, respectively. Mean values ± standard errors of the means are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test with 1,000 Monte Carlo permutations). (B) Trajectory of the gut microbiota structure of each giant panda individual across seasons. The principal coordinate analysis (PCoA) scores of T1 samples were set to zero, and the relative scores of T2 and T3 samples corresponding to their paired T1 samples were plotted. The animal identifiers are shown. A, adult; J, juvenile; F, female; M, male. Download
Pipeline of the multistep OTU picking procedure. GGRef denotes the reference-based OTU picking against the Greengenes reference database, whereas CLRef denotes the reference-based OTU picking against the 3,075 OTUs derived from the GGRef-discarded sequences from the three clone library data sets. NFL, near full length. Download
The gut microbiota of the captive and wild giant pandas (n = 137) had lower numbers of observed OTUs (A) and Shannon diversity indices (B) than did those of other herbivores (n = 66), omnivores (n = 26), and carnivores (n = 16) by Mann-Whitney test (with Bonferroni post hoc test). See Fig. 2 for definition of box-and-whisker plot. NS, not significant (P > 0.05). Download
UPGMA (unweighted-pair group method using average linkages) hierarchical clustering analysis based on the weighted (A) and unweighted (B) UniFrac distance. The scale bar represents a distance of 0.1. Samples are color coded as described in Fig. 4. Red clades indicate the samples of the same nonpanda mammalian species from the published data sets that clustered together. Download
Comparison of gut microbiota structure of the giant panda population described in this study with that of the newly sampled herbivores, omnivores, and carnivores. PCoA score plots based on the weighted (A) and unweighted (B) UniFrac distances are shown. Download
Phylogeny of Clostridia-related lineages identified by the de novo and multistep OTU classification procedures. Only the OTUs present in at least 10 out of the 121 giant panda samples are included. The prefix “Denovo” represents the OTUs classified by the de novo method (highlighted by the blue text), whereas “GGRef” represents the OTUs classified by the reference-based approach, with numerals representing the Greengenes prokMSA identifier (highlighted by the red text). OTUs classified by the de novo method were inserted into preestablished Greengenes phylogenetic tree in ARB. The six OTUs selected as key phylotypes to discriminate the gut microbiota of giant pandas from that of nonpanda herbivores are highlighted in yellow. Accession numbers of reference sequences from the Greengenes database are noted in parentheses. The abundance range, abundance median (of only samples in which the respective OTU was detected), and the prevalence of each OTU among the 121 giant panda samples are listed in square brackets. Download
Basic information and the reference-based OTU picking statistics of the gut microbiota data sets from the 121 giant panda samples collected in this study and the 143 published mammal samples
ACKNOWLEDGMENTS
This study was supported by Project 31121064 of the National Natural Science Foundation of China (NSFC) and Key Project 31330005 and 30730005 of the NSFC, Chengdu Giant Panda Breeding Research Foundation (CPF08002), the Bureau of Science and Technology of Chengdu City (11GGYB341SF-027), the National Key Technology R&D Program (2012BAC01B06), and the Chengdu Urban and Rural Construction Commission.
We thank Hong Liu and other staff at the Chengdu Research Base of Giant Panda Breeding for assistance with sample collection. We thank the reviewers and appreciate their constructive comments on this study.
Footnotes
Citation Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, Zhang X, Huang H, Bridgewater LC, Jiang Y, Jiang C, Zhao L, Pang X, Zhang Z. 2015. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. mBio 6(3):e00022-15. doi:10.1128/mBio.00022-15.
REFERENCES
- 1.Arnason U, Gullberg A, Janke A, Kullberg M. 2007. Mitogenomic analyses of caniform relationships. Mol Phylogenet Evol 45:863–874. doi: 10.1016/j.ympev.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 2.Krause J, Unger T, Noçon A, Malaspinas AS, Kolokotronis SO, Stiller M, Soibelzon L, Spriggs H, Dear PH, Briggs AW, Bray SC, O’Brien SJ, Rabeder G, Matheus P, Cooper A, Slatkin M, Pääbo S, Hofreiter M. 2008. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol 8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Viswanathan VK. 2010. What is black and white and a puzzle all over? Gut Microbes 1:129–130. doi: 10.4161/gmic.1.3.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schaller GB, Hu J, Pan W, Zhu J. 1985. The giant pandas of Wolong, 1st ed. University of Chicago Press, Chicago, IL. [Google Scholar]
- 5.Jin C, Ciochon RL, Dong W, Hunt RM, Liu J, Jaeger M, Zhu Q. 2007. The first skull of the earliest giant panda. Proc Natl Acad Sci U S A 104:10932–10937. doi: 10.1073/pnas.0704198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, Zhang Z, Zhang Y, Wang W, Li J, Wei F, Li H, Jian M, Li J, Zhang Z, Nielsen R, Li DW, Gu WJ, Yang ZT, Xuan ZL, Ryder OA, Leung FCC, Zhou Y, Cao JJ, Sun X, Fu YG, Fang XD, Guo XS, Wang B, Hou R, Shen FJ, Mu B, Ni PX, Lin RM, Qian WB, Wang GD, Yu C, Nie WH, Wang JH, Wu ZG, Liang HQ, Min JM, Wu Q, Cheng SF, Ruan J, Wang MW, Shi ZB, Wen M, Liu BH, Ren XL, Zheng HS, Dong D, Cook K, Shan G, Zhang H, Kosiol C, Xie XY, Lu ZH, Zheng HC, Li YR, Steiner CC, Lam TTY, Lin SY, Zhang QH, Li GQ, Tian J, Gong TM, Liu HD, Zhang DJ, Fang L, Ye C, Zhang JB, Hu WB, Xu AL, Ren YY, Zhang GJ, Bruford MW, Li QB, Ma LJ, Guo YR, An N, Hu YJ, Zheng Y, Shi YY, Li ZQ, Liu Q, Chen YL, Zhao J, Qu N, Zhao SC, Tian F, Wang XL, Wang HY, Xu LZ, Liu X, Vinar T, Wang YJ, Lam TW, Yiu SM, Liu SP, Zhang HM, Li DS, Huang Y, Wang X, Yang GH, Jiang Z, Wang JY, Qin N, Li L, Li JX, Bolund L, Kristiansen K, Wong GKS, Olson M, Zhang XQ, Li SG, Yang HM, Wang J. 2010. The sequence and de novo assembly of the giant panda genome. Nature 463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao H, Yang JR, Xu H, Zhang J. 2010. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol 27:2669–2673. doi: 10.1093/molbev/msq153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin K, Xue C, Wu X, Qian J, Zhu Y, Yang Z, Yonezawa T, Crabbe MJ, Cao Y, Hasegawa M, Zhong Y, Zheng Y. 2011. Why does the giant panda eat bamboo? A comparative analysis of appetite-reward-related genes among mammals. PLoS One 6:e22602. doi: 10.1371/journal.pone.0022602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bininda-Emonds ORP. 2004. Phylogenetic position of the giant panda, p 11–35. In Lindburg D, Baragona K (ed), Giant panda: biology and conservation, 1st ed. University of California Press, Oakland, CA. [Google Scholar]
- 10.Endo H, Sasaki N, Yamagiwa D, Uetake Y, Kurohmaru M, Hayashi Y. 1996. Functional anatomy of the radial sesamoid bone in the giant panda (Ailuropoda melanoleuca). J Anat 189:587–592. [PMC free article] [PubMed] [Google Scholar]
- 11.Endo H, Yamagiwa D, Hayashi Y, Koie H, Yamaya Y, Kimura J. 1999. Role of the giant panda’s “pseudo-thumb.” Nature 397:309–310. doi: 10.1038/16830. [DOI] [PubMed] [Google Scholar]
- 12.Salesa MJ, Antón M, Peigné S, Morales J. 2006. Evidence of a false thumb in a fossil carnivore clarifies the evolution of pandas. Proc Natl Acad Sci U S A 103:379–382. doi: 10.1073/pnas.0504899102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beijing Zoo, Beijing University, Beijing Agricultural University, Beijing Second Medical College, Beijing Natural History Museum, Shaanxi Zoology Institute 1986. Morphology of the giant panda. Systematic anatomy and organ-histology, 1st ed, p 189–195. Science Press, Beijing, China. [Google Scholar]
- 14.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. 2011. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brulc JM, Antonopoulos DA, Miller ME, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci U S A 106:1948–1953. doi: 10.1073/pnas.0806191105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen F, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 19.Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, McHardy AC, Cheng JF, Hugenholtz P, McSweeney CS, Morrison M. 2010. Adaptation to herbivory by the tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci U S A 107:14793–14798. doi: 10.1073/pnas.1005297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toyoda A, Iio W, Mitsumori M, Minato H. 2009. Isolation and identification of cellulose-binding proteins from sheep rumen contents. Appl Environ Microbiol 75:1667–1673. doi: 10.1128/AEM.01838-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab C, Cristescu B, Northrup JM, Stenhouse GB, Gänzle M. 2011. Diet and environment shape fecal bacterial microbiota composition and enteric pathogen load of grizzly bears. PLoS One 6:e27905. doi: 10.1371/journal.pone.0027905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwab C, Gänzle M. 2011. Comparative analysis of fecal microbiota and intestinal microbial metabolic activity in captive polar bears. Can J Microbiol 57:177–185. doi: 10.1139/W10-113. [DOI] [PubMed] [Google Scholar]
- 23.Hirayama K, Kawamura S, Mitsuoka T, Tashiro K. 1989. The faecal flora of the giant panda (Ailuropoda melanoleuca). J Appl Bacteriol 67:411–415. doi: 10.1111/j.1365-2672.1989.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang ZH, He GX, Wang XL, Zhong SL, Zhang AJ, Li GH. 1995. The study on the giant panda’s intestinal flora. Acta Theriol Sin 15:170–175. [Google Scholar]
- 25.Tan Z, Pao N, Lai Y, Zhang HM, Li DS, Liu CJ. 2004. The study on the normal intestinal microflora of the giant panda returned to wild and the giant panda in captivity. J Sichuan Univ (Nat Sci Ed) 41:1276–1279. [Google Scholar]
- 26.Wei G, Lu H, Zhou Z, Xie H, Wang A, Nelson K, Zhao L. 2007. The microbial community in the feces of the giant panda (Ailuropoda melanoleuca) as determined by PCR-TGGE profiling and clone library analysis. Microb Ecol 54:194–202. doi: 10.1007/s00248-007-9225-2. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L, Wu Q, Dai J, Zhang S, Wei F. 2011. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc Natl Acad Sci U S A 108:17714–17719. doi: 10.1073/pnas.1017956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dierenfeld ES, Hintz HF, Robertson JB, Van Soest PJ, Oftedal OT. 1982. Utilization of bamboo by the giant panda. J Nutr 112:636–641. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. doi: 10.1046/j.1442-9993.2001.01070.x. [DOI] [Google Scholar]
- 31.Breiman L. 2001. Random forests. Mach Learn 45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 32.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 33.Lee JW, Lee JB, Park M, Song SH. 2005. An extensive comparison of recent classification tools applied to microarray data. Comput Stat Data Anal 48:869–885. doi: 10.1016/j.csda.2004.03.017. [DOI] [Google Scholar]
- 34.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simůnek J, Hodrová B, Bartonová H, Kopecný J. 2001. Chitinolytic bacteria of the mammal digestive tract. Folia Microbiol (Praha) 46:76–78. doi: 10.1007/BF02825892. [DOI] [PubMed] [Google Scholar]
- 36.He YL, Ding YF, Long YQ. 1991. Two cellulolytic Clostridium species: Clostridium cellulosi sp. nov. and Clostridium cellulofermentans sp. nov. Int J Syst Bacteriol 41:306–309. doi: 10.1099/00207713-41-2-306. [DOI] [PubMed] [Google Scholar]
- 37.Cai S, Dong X. 2010. Cellulosilyticum ruminicola gen. nov., sp. nov., isolated from the rumen of yak, and reclassification of Clostridium lentocellum as Cellulosilyticum lentocellum comb. nov. Int J Syst Evol Microbiol 60:845–849. doi: 10.1099/ijs.0.014712-0. [DOI] [PubMed] [Google Scholar]
- 38.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee WJ, Hase K. 2014. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol 10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 40.Zhao L. 2013. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol 11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 41.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Konopka A. 2009. What is microbial community ecology? ISME J 3:1223–1230. doi: 10.1038/ismej.2009.88. [DOI] [PubMed] [Google Scholar]
- 43.Naeem S, Li S. 1997. Biodiversity enhances ecosystem reliability. Nature 390:507–509. doi: 10.1038/37348. [DOI] [Google Scholar]
- 44.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, Almeida M, Quinquis B, Levenez F, Galleron N, Gougis S, Rizkalla S, Batto JM, Renault P, ANR MicroObes Consortium, Doré J, Zucker JD, Clément K, Ehrlich SD. 2013. Dietary intervention impact on gut microbial gene richness. Nature 500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 45.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, MetaHIT Consortium, Bork P, Wang J, Ehrlich SD, Pedersen O. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 46.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams CL, Willard S, Kouba A, Sparks D, Holmes W, Falcone J, Williams CH, Brown A. 2013. Dietary shifts affect the gastrointestinal microflora of the giant panda (Ailuropoda melanoleuca). J Anim Physiol Anim Nutr 97:577–585. doi: 10.1111/j.1439-0396.2012.01299.x. [DOI] [PubMed] [Google Scholar]
- 48.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R. 2011. Moving pictures of the human microbiome. Genome Biol 12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. 2011. Linking long-term dietary patterns with gut microbial enterotypes. Science 334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nie Y, Zhang Z, Raubenheimer D, Elser JJ, Wei W, Wei F. 2015. Obligate herbivory in an ancestrally carnivorous lineage: the giant panda and bamboo from the perspective of nutritional geometry. Funct Ecol 29:26–34. doi: 10.1111/1365-2435.12302. [DOI] [Google Scholar]
- 52.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X, Yan Q, Xia X, Zhang Y, Li D, Wang C, Chen S, Hou R. 2013. Serotypes, virulence factors, and antimicrobial susceptibilities of vaginal and fecal isolates of Escherichia coli from giant pandas. Appl Environ Microbiol 79:5146–5150. doi: 10.1128/AEM.01367-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 55.Fei N, Zhao L. 2013. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J 7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 57.Stackebrandt E, Hippe H. 2001. Taxonomy and systematics, p 19–48. In Bahl H, Dürre P (ed), Clostridia: biotechnology and medical applications, 1st ed. Wiley-VCH Verlag, Weinheim, Germany. [Google Scholar]
- 58.Susan L. 2005. Degradation of polymers, p 101–131. In Dürre P (ed), Handbook on clostridia, 1st ed. CRC Press, Boca Raton, FL. [Google Scholar]
- 59.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu HM, Yu C, Li ST, Jian M, Zhou Y, Li YR, Zhang XQ, Li SG, Qin N, Yang HM, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang B, Li M, Zhang Z, Goossens B, Zhu L, Zhang S, Hu J, Bruford MW, Wei F. 2007. Genetic viability and population history of the giant panda, putting an end to the “evolutionary dead end”? Mol Biol Evol 24:1801–1810. doi: 10.1093/molbev/msm099. [DOI] [PubMed] [Google Scholar]
- 61.Zhang CH, Li SF, Yang L, Huang P, Li WJ, Wang SY, Zhao GP, Zhang MH, Pang XY, Yan Z, Liu Y, Zhao LP. 2013. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. J Commun 4:2163. doi: 10.1038/ncomms3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, Phan R, Andersen GL. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. 2013. vegan: community ecology package. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 70.Hamady M, Lozupone C, Knight R. 2010. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liaw A, Wiener M. 2002. Classification and regression by randomForest. R News 2:18–22. http://www.r-project.org/doc/Rnews/Rnews_2002-3.pdf. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
OTU-level alpha diversity of the gut microbiota of the giant panda population. (A to D) Rarefaction curves of T1 (A), T2 (B), T3 (C), and cub (D) samples. (E to H) Shannon diversity index curves of T1 (E), T2 (F), T3 (G), and cub (H) samples. Download
Abundance variations in the 10 dominant genera (>1% of total sequences), Escherichia/Shigella (A), Klebsiella (B), Streptococcus (C), Lactococcus (D), Lactobacillus (E), Weissella (F), Enterococcus (G), Clostridium sensu stricto (H), Clostridium XI (I), and Turicibacter (J), across seasons, compared by paired sample Wilcoxon signed-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (with Bonferroni post hoc test). Only the 33 individuals that were sampled in all three seasons are included. See Fig. 2 for definition of box-and-whisker plot. Download
Inter- and intraindividual variations of the gut microbiota of the 33 giant pandas that were sampled in all three seasons. (A) Interindividual variations were determined by average unweighted UniFrac distances between individuals in T1, T2, or T3, respectively, while intraindividual variations were determined by distances between paired T1 and T2, T2 and T3, and T1 and T3 samples, respectively. Mean values ± standard errors of the means are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test with 1,000 Monte Carlo permutations). (B) Trajectory of the gut microbiota structure of each giant panda individual across seasons. The principal coordinate analysis (PCoA) scores of T1 samples were set to zero, and the relative scores of T2 and T3 samples corresponding to their paired T1 samples were plotted. The animal identifiers are shown. A, adult; J, juvenile; F, female; M, male. Download
Pipeline of the multistep OTU picking procedure. GGRef denotes the reference-based OTU picking against the Greengenes reference database, whereas CLRef denotes the reference-based OTU picking against the 3,075 OTUs derived from the GGRef-discarded sequences from the three clone library data sets. NFL, near full length. Download
The gut microbiota of the captive and wild giant pandas (n = 137) had lower numbers of observed OTUs (A) and Shannon diversity indices (B) than did those of other herbivores (n = 66), omnivores (n = 26), and carnivores (n = 16) by Mann-Whitney test (with Bonferroni post hoc test). See Fig. 2 for definition of box-and-whisker plot. NS, not significant (P > 0.05). Download
UPGMA (unweighted-pair group method using average linkages) hierarchical clustering analysis based on the weighted (A) and unweighted (B) UniFrac distance. The scale bar represents a distance of 0.1. Samples are color coded as described in Fig. 4. Red clades indicate the samples of the same nonpanda mammalian species from the published data sets that clustered together. Download
Comparison of gut microbiota structure of the giant panda population described in this study with that of the newly sampled herbivores, omnivores, and carnivores. PCoA score plots based on the weighted (A) and unweighted (B) UniFrac distances are shown. Download
Phylogeny of Clostridia-related lineages identified by the de novo and multistep OTU classification procedures. Only the OTUs present in at least 10 out of the 121 giant panda samples are included. The prefix “Denovo” represents the OTUs classified by the de novo method (highlighted by the blue text), whereas “GGRef” represents the OTUs classified by the reference-based approach, with numerals representing the Greengenes prokMSA identifier (highlighted by the red text). OTUs classified by the de novo method were inserted into preestablished Greengenes phylogenetic tree in ARB. The six OTUs selected as key phylotypes to discriminate the gut microbiota of giant pandas from that of nonpanda herbivores are highlighted in yellow. Accession numbers of reference sequences from the Greengenes database are noted in parentheses. The abundance range, abundance median (of only samples in which the respective OTU was detected), and the prevalence of each OTU among the 121 giant panda samples are listed in square brackets. Download
Basic information and the reference-based OTU picking statistics of the gut microbiota data sets from the 121 giant panda samples collected in this study and the 143 published mammal samples