Abstract
Abscisic acid (ABA) regulates many aspects of plant growth and development, including inhibition of root elongation and seed germination. We performed an ABA resistance screen to identify factors required for ABA response in root elongation inhibition. We identified two classes of Arabidopsis thaliana AR mutants that displayed ABA-resistant root elongation: those that displayed resistance to ABA in both root elongation and seed germination and those that displayed resistance to ABA in root elongation but not in seed germination. We used PCR-based genotyping to identify a mutation in ABA INSENSITIVE2 (ABI2), positional information to identify mutations in AUXIN RESISTANT1 (AUX1) and ETHYLENE INSENSITIVE2 (EIN2), and whole genome sequencing to identify mutations in AUX1, AUXIN RESISTANT4 (AXR4), and ETHYLENE INSENSITIVE ROOT1/PIN-FORMED2 (EIR1/PIN2). Identification of auxin and ethylene response mutants among our isolates suggested that auxin and ethylene responsiveness were required for ABA inhibition of root elongation. To further our understanding of auxin/ethylene/ABA crosstalk, we examined ABA responsiveness of double mutants of ethylene overproducer1 (eto1) or ein2 combined with auxin-resistant mutants and found that auxin and ethylene likely operate in a linear pathway to affect ABA-responsive inhibition of root elongation, whereas these two hormones likely act independently to affect ABA-responsive inhibition of seed germination.
Keywords: mutant screen, plant hormones, epistasis, whole genome sequencing
Plant growth and development are dynamic processes that allow for adaptation to the environment to which they are restricted. Plants adjust growth in response to internal and external stimuli through the activities of plant hormones. The phytohormones abscisic acid (ABA), auxin, and ethylene play key roles in regulating these growth responses (Vanstraelen and Benková 2012).
ABA controls many growth and stress responses, including seed dormancy onset and maintenance, germination inhibition, stomatal opening, and shoot and root growth (Cutler et al. 2010). Both genetic and chemical genomic screens in Arabidopsis thaliana have revealed many genes required for ABA biosynthesis and response (Cutler et al. 2010). Nuclear ABA perception occurs through the PYRABACTIN RESISTANCE/PYRABACTIN RESISTANCE1-LIKE/REGULATORY COMPONENT OF ABA RECEPTOR 1 (PYR/PYL/RCAR1) family (Ma et al. 2009; Park et al. 2009). PYR/PYL/RCAR proteins directly bind and regulate protein phosphatase type 2C (PP2C) proteins, such as ABA INSENSITIVE1 (ABI1) and ABI2, which have long been implicated in ABA signaling (Leung et al. 1997). Under high ABA concentrations, these PP2C proteins are inactivated by PYR/PYL/RCAR, allowing for the phosphorylation of SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2 (SnRK2) family members, which phosphorylate downstream ABA signaling components to mediate a response (Nakashima et al. 2009). In addition, auxin and ethylene signaling components have been identified as necessary for full ABA responsiveness.
Auxin is an essential plant hormone that regulates plant cell division and elongation (Perrot-Rechenmann 2010) to control nearly every aspect of plant growth and development, including root elongation, embryo patterning, vascularization, cell division and elongation, and many others (Mockaitis and Estelle 2008). Genetic studies in Arabidopsis have uncovered components of the auxin signaling pathway, and much is known about auxin biosynthesis (Korasick et al. 2013), transport (Petrášek and Friml 2009), and response (Sauer et al. 2013). TRANSPORT INHIBITOR RESPONSE1 (TIR1) and the related AUXIN SIGNALING F-BOX (AFB) family members, which are components of SCF E3 ubiquitin ligases (Parry and Estelle 2006), bind auxin (Dharmasiri et al. 2005; Kepinski and Leyser 2005), allowing for interaction with AUXIN/INDOLE-3-ACETIC ACID INDUCIBLE (Aux/IAA) transcriptional repressors. The interaction of SCFTIR1/AFB with Aux/IAA repressor proteins results in the ubiquitylation (Dos Santos Maraschin et al. 2009) and subsequent degradation of the multimeric (Korasick et al. 2014) Aux/IAA repressors, thus allowing expression of auxin-responsive genes.
Although the roles of ABA and auxin in plant growth are distinct, sensitivity to auxin correlates with sensitivity to ABA. For example, mutations in auxin response genes such as AUXIN RESISTANT1 (AUX1), AUXIN RESISTANT1 (AXR1), IBA RESPONSE5 (IBR5), and TIR1, which were identified for conferring auxin resistance or resistance to auxin transport inhibitors (Lincoln et al. 1990; Bennett et al. 1996; Ruegger et al. 1998; Monroe-Augustus et al. 2003), also confer ABA resistance (Tiryaki and Staswick 2002; Monroe-Augustus et al. 2003; Strader et al. 2008a). Conversely, the sax1 mutant displays hypersensitivity to both auxin and ABA (Ephritikhine et al. 1999). Interestingly, the ABA-regulated AP2 domain transcription factor ABI4 was recently shown to repress expression of the auxin transporter PIN1, and accordingly abi4 mutants display enhanced root auxin transport (Shkolnik-Inbar and Bar-Zvi 2011). These connections suggest that auxin responsiveness is required for plants to be able to fully respond to ABA.
Ethylene is a gaseous phytohormone that affects many aspects of plant growth and development, including fruit ripening, senescence, and leaf abscission (Kendrick and Chang 2008). The rate-limiting step in ethylene biosynthesis is controlled by 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) enzymes (Chae and Kieber 2005). A subset of these enzymes are targeted for degradation by the ETHYLENE OVERPRODUCER1 (ETO1) E3 ubiquitin ligase; mutation of ETO1 results in increased stability of these ACS enzymes and ethylene hyperproduction (Chae and Kieber 2005). Ethylene is perceived by a family of transmembrane histidine kinase receptors, including ETHYLENE RECEPTOR1 (ETR1), ETR2, ETHYLENE RESPONSE SENSOR1 (ERS1), ERS2, and ETHYLENE INSENSITIVE4 (EIN4) (Kendrick and Chang 2008). In the absence of ethylene, these receptors activate the Raf-like protein kinase CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) (Kieber et al. 1993), which represses the EIN2 ethylene response activator (Alonso et al. 1999). In the presence of ethylene, the ethylene receptors fail to activate CTR1, relieving repression of EIN2 and allowing for EIN2 activation of the EIN3 family of transcription factors (Kendrick and Chang 2008). Many auxin-resistant mutants display ethylene resistance in root elongation. Well-characterized examples include aux1 (Pickett et al. 1990), ibr5 (Strader et al. 2008b), and tir1 (Alonso et al. 2003), suggesting that auxin responsiveness is necessary for full ethylene responsiveness.
ABA and ethylene interactions are complex and not well-characterized. Many ethylene signaling mutants simultaneously display resistance to ABA in root elongation assays and hypersensitivity to ABA in seed germination assays (Gazzarrini and McCourt 2003). In addition, ABA and ethylene appear to interact to regulate plant stress responses. For example, ABA limits ethylene production in water-stressed plants (Sharp 2002; Sharp and Lenoble 2002), whereas ethylene antagonizes stomatal response to ABA under oxidative stress (Wilkinson and Davies 2009). Intriguingly, ABA inhibits transcription of the ethylene biosynthesis gene ACC OXIDASE (ACO) during seed germination in Lepidium sativum (Linkies et al. 2009) to limit ethylene production, suggesting a mechanism for ABA regulation of ethylene for seed germination regulation. However, the molecular details of ethylene inhibition of seed germination and ABA-responsive root elongation remain largely unknown.
In addition to ABA–auxin and ABA–ethylene interactions, ethylene and auxin display extensive interactions (Stepanova and Alonso 2009; Muday et al. 2012). Each hormone stimulates the biosynthesis of the other; ethylene stimulates IAA production (Růžička et al. 2007; Swarup et al. 2007) and auxin stimulates ethylene production (Tsuchisaka and Theologis 2004). Also, many ethylene signaling mutants are auxin-resistant, and many auxin signaling mutants are ethylene-resistant (Stepanova et al. 2007), suggesting that some facets of auxin signaling require ethylene response and some aspects of ethylene signaling require auxin response. Further supporting this connection, biosynthesis mutants with decreased IAA are mildly ethylene-resistant (Stepanova et al. 2005; Stepanova et al. 2008). From these auxin–ethylene interactions, a model has emerged (Stepanova and Alonso 2009) for ethylene regulation of root elongation in which ethylene promotes auxin biosynthesis in root meristems (Růžička et al. 2007; Swarup et al. 2007), followed by auxin transport from the meristem to root elongation zones in an ethylene-regulated manner (Růžička et al. 2007; Swarup et al. 2007) where auxin simultaneously inhibits cell elongation and promotes ethylene responsiveness (Stepanova et al. 2007).
To identify factors required for ABA-responsive root elongation, we performed a genetic screen in Arabidopsis thaliana for mutants that display ABA resistance in root elongation assays. We characterized 21 mutant isolates in detail and found that 12 of these isolates displayed resistance to ABA in both root elongation and seed germination assays, whereas nine isolates displayed ABA-resistant root elongation and ABA-sensitive seed germination. We used PCR analysis, recombination mapping, and whole genome sequencing to identify mutations in auxin, ethylene, and ABA signaling components defective in seven ABA Root Resistance (AR) mutants. Because we identified auxin and ethylene signaling components in our mutants, we examined AR mutant responses to auxin and ethylene in root elongation assays. We found that many of our isolates also displayed resistance to these hormones. We further characterized ABA responses in known auxin and ethylene mutants and found that all examined auxin and ethylene-resistant mutants displayed resistance to ABA in root elongation, whereas only the auxin-resistant mutants displayed ABA resistance in seed germination assays. Additionally, auxin and ethylene mutants were nonadditive in ABA-responsive root elongation assays, consistent with the possibility that auxin and ethylene act in a linear pathway to regulate ABA-responsive root elongation. Conversely, auxin and ethylene mutants were additive in ABA-responsive seed germination inhibition assays, suggesting that auxin and ethylene act independently to regulate ABA-responsive inhibition of seed germination. Our results illuminate auxin–ethylene interactions regulating ABA response.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana accessions Colombia (Col-0) and Landsberg erecta (Ler-0) were used as wild-type for experiments, as indicated in the figure legends. Surface-sterilized (Last and Fink 1988) seeds were plated on plant nutrient medium (PN) (Haughn and Somerville 1986), supplemented with 0.5% (w/v) sucrose (PNS), solidified with 0.6% (w/v) agar. The (±)-ABA, IAA, and 2,4-D stocks were dissolved in 100% ethanol and ACC stocks were dissolved in 50% ethanol. Ethanol-supplemented media were used as controls with all treatments normalized to the same ethanol content. Seedlings were grown at 22° under continuous illumination through yellow long-pass filters to decrease indolic compound breakdown (Stasinopoulos and Hangarter 1990) unless otherwise indicated.
Mutant isolation and nomenclature
Arabidopsis thaliana Col-0 seeds were mutagenized with ethyl-methansulfonate (EMS) (Normanly et al. 1997). M2 seeds were surface-sterilized and plated on Whatman 3M filter paper on top of PNS medium at ∼1500 seeds per 150-mm plate. After 4 d of growth under continuous illumination through yellow long-pass filters, germinated seedlings and filter paper were transferred to PNS supplemented with 10 µM ABA. After an additional 4 d of growth under continuous illumination through yellow long-pass filters, putative ABA root elongation mutants with long roots were selected, transferred to unsupplemented medium to recover, moved to soil, genotyped for the abi1-1 and abi2-1 mutations, and allowed to self-fertilize. M3 progeny were retested for resistance to ABA in root elongation assays.
Double mutant isolation
Generation of eto1-1 ein2-1, eto1-1 ibr5-1, eto1-1 tir1-1, eto1-1 axr1-3, and eto1-1 aux1-7 has previously been described (Strader et al. 2010). The ein2-1 (Alonso et al. 1999) mutant was crossed to ibr5-1 (Monroe-Augustus et al. 2003), tir1-1 (Ruegger et al. 1998), axr1-3 (Estelle and Somerville 1987), and aux1-7 (Maher and Martindale 1980). Double mutants were identified by PCR analysis of F2 progeny. Amplification of EIN2 with EIN2-1 (5′-TTCTCCATGCTAACAATCTTCTCCACAGG-3′) and the derived cleaved amplified polymorphic sequence primer (Neff et al. 2002) EIN2-BsmAI (5′-AGAGTTGGATGTAAAGTACTCTACGTCT-3′; altered residue underlined) results in a 186-bp product with one BsmAI site in ein2-1 and no sites in wild-type. Amplification of IBR5 with T1O3.4-1 (5′-CCTAATTTCCTCCGTCTGTGAAATCAAGGG-3′) and T1O3.4-6 (5′-CAAGGCAAAACCCTAACTAAACAAACCG-3′) results in a 463-bp product with one AciI site in wild-type and no sites in ibr5-1. Amplification of TIR1 with TIR1-3 (5′-TTGAAGAGATAAGGCTGAAGAGGATGG-3′) and TIR1-6 (5′-AAACCGGAACACGATTATATGGGATGATG-3′) results in a 488-bp product with one DpnII site in wild-type and two sites in tir1-1. Amplification of AXR1 with AXR1-15 (5′-TCTCATATGTACTTTTCCTCGTCCTCTTCAC-3′) and the derived cleaved amplified polymorphic sequence primer (Neff et al. 2002) AXR1-AccI (5′-AAACCAACTTAACGTTTGCATGTCG-3′; altered residue underlined) results in a 185-bp product with on AccI site in wild-type and no sites in axr1-3. Amplification of AUX1 with AUX1-3 (5′-CATGGGTCAACAAAGCTTTGGATTTTGTCC-3′) and AUX1-4 (5′-TTCGTGACTTTTACTCCCTTCACGTATACG-3′) results in a 464-bp product with two DpnII sites in wild-type and three sites in aux1-7.
Phenotypic assays
All phenotypic assays were performed at least three times, and the presented results are representative of these assays. For auxin-responsive and 1-aminocyclopropane-1carboxylic acid (ACC)–responsive root elongation assays, stratified seeds were grown for 8 d under continuous illumination through yellow long-pass filters on the indicated hormone concentrations and primary root lengths were measured. For ABA-responsive root elongation assays, imbibed seeds were incubated at 4′ for 2 d then plated on unsupplemented PNS medium and incubated at 22° under continuous illumination through yellow long-pass filters for 4 d to allow efficient germination. Seedlings then were transferred to medium supplemented with either ethanol or the indicated concentration of ABA and total primary root lengths were measured after an additional 4 d of growth under continuous illumination through yellow long-pass filters.
For ABA-responsive seed germination and seedling development assays, seeds from plants grown simultaneously under continuous light were afterripened at room temperature for 1 to 2 months before assessment of seed germination properties. Sterile, imbibed seeds were incubated at 4° for 2 d in the dark and then plated on PN supplemented with ethanol (Mock) or the indicated concentrations of ABA and grown under white light. Seeds were examined at the indicated time points and seeds with an emerged, elongating radicle were counted.
Identification of the abi2-11, aux1-53, aux1-99, and ein2-291 mutations
The abi2-11 mutation (in the Col-0 background) was identified in isolate AR11 by PCR-based genotyping M2 individuals identified in the AR mutant screen. PCR amplification with ABI2-1 (5′-ACGGTGAATCTAGGGTTACTTTAC-3′) and ABI2-2 (5′ -ACTCCGGTTTCTCCTTCACTATC-3′) results in a 586-bp product with one NcoI site in wild-type and no sites in abi2-1 or abi2-11.
The aux1-53 and aux1-99 mutations (in the Col-0 background) were identified in isolates AR53 and AR99, respectively, by recombination mapping. AR53 and AR99 were crossed to Landsberg erecta and resultant F2 progeny were selected for exhibiting a long root on ABA. DNA from ABA-resistant individuals was scored using PCR-based polymorphic markers (Konieczny and Ausubel 1993; Bell and Ecker 1994). The AUX1 gene within the AR53 and AR99 mapping intervals was PCR-amplified and sequenced from AR53 and AR99 genomic DNA.
The ein2-291 mutation (in the Col-0 background) was identified in isolate AR291 by recombination mapping. AR291 was crossed to Landsberg erecta and resultant F2 were selected for exhibiting a long root on ABA. DNA from ABA-resistant individuals was scored using PCR-based polymorphic markers (Konieczny and Ausubel 1993; Bell and Ecker 1994). The EIN2 gene within the AR291 mapping interval was PCR-amplified and sequenced from AR291 genomic DNA.
Identification of the aux1-116, axr4-241, and eir1-211 mutations by whole genome sequencing
The aux1-116, axr4-241, and eir1-211 mutations were identified in isolates AR116, AR241, and AR211, respectively, by whole genome sequencing. AR116 was crossed to wild type (Col-0) and resultant F2 progeny were selected for exhibiting a long root on ABA, moved to soil, and allowed to self-fertilize. Resultant F3 progeny were retested for ABA resistance in root elongation. Tissues from 500 seedlings from each of seven ABA-resistant F3 pools from the AR116 backcross were combined for genomic DNA extraction. Tissue from pooled M4 seedlings was used for AR211 and AR241 genomic DNA extractions.
For genomic DNA extraction, tissue was ground with mortar and pestle under liquid nitrogen to a fine powder and transferred to 7 mL extraction buffer (110 mM Tris, pH 8.0; 55 mM ethylenediaminetetraacetic acid, pH 8.0; 1.54 M NaCl, 1.1% cetyl trimethyl ammonium bromide) prewarmed to 65° in a 15-mL polypropylene conical tube. Tubes were capped and vortexed to incorporate ground tissue; 0.7 mL 20% sodium dodecyl sulfate was added to each sample and mixed by inversion. Samples were incubated in a 65° water bath for 2 hr with occasional inversion. Samples were then cooled to room temperature and 24:1 chloroform:isoamyl alcohol was added to the 14-mL mark on the conical tube. Samples were mixed by inversion for 15 min, and then phases were separated by centrifugation for 15 min at 3000 rpm. After centrifugation, the top (aqueous) phase was removed to a new 15-mL polypropylene conical tube and the chloroform:isoamyl extraction was repeated. The top (aqueous) phase from the second extraction was removed to a new 15-mL polypropylene conical tube and 0.6 volumes isopropanol were added. DNA was precipitated by gentle rocking and collected by centrifugation for 5 min at 3000 rpm. The supernatant was discarded and DNA was resuspended in 4 mL TE buffer (100 mM Tris, 10 mM ethylenediaminetetraacetic acid; pH 8.0). RNaseA was added to a final concentration of 10 µg/mL and incubated for 1 hr at 37°. After the RNaseA digest, 8 mL chloroform was added to each sample and mixed by inversion for 10 min, and then phases were separated by centrifugation for 15 min at 3000 rpm. After centrifugation, the top (aqueous) phase was removed to a new 15-mL polypropylene conical tube and 0.1 volume 3 M sodium acetate (pH 5.2) and 2 volumes 95% chilled ethanol added. DNA was precipitated by gentle rocking and collected by centrifugation for 20 min at 2000 rpm. The supernatant was discarded and 3 mL 70% ethanol was placed on top of the precipitated DNA. After 1 min, ethanol was removed and the tube was inverted to drain and dry the DNA pellet. DNA resuspended in 500 µL TE buffer.
Genomic DNA was sequenced using by the Washington University Genome Technology Assistance Center (GTAC; https://gtac.wustl.edu). Libraries were prepared using an Illumina Genomic DNA kit and sequenced on an Illumina HiSequation 2000 using multiplexing in a 100-bp paired end run.
Reads were aligned to the Arabidopsis thaliana Col-0 reference genome with Arabidopsis Information Resource 10 gene annotations (The Arabidopsis Genome Initiative 2000) using Novoalign (Novocraft, http://novocraft.com). SNPs were identified using SAMtools (Li et al. 2009) and annotated using snpEFF (Cingolani et al. 2012) to predict the effects of variants on genes. We then identified homozygous, canonical EMS-induced changes (G-to-A or C-to-T) from each population. Recombination mapping in the backcross using markers based on identified homozygous EMS-induced mutations was used to determine linkage of identified mutations to phenotype in AR241 and AR211 (Supporting Information, Table S1).
Results
Isolation of mutants with reduced ABA responsiveness in root elongation
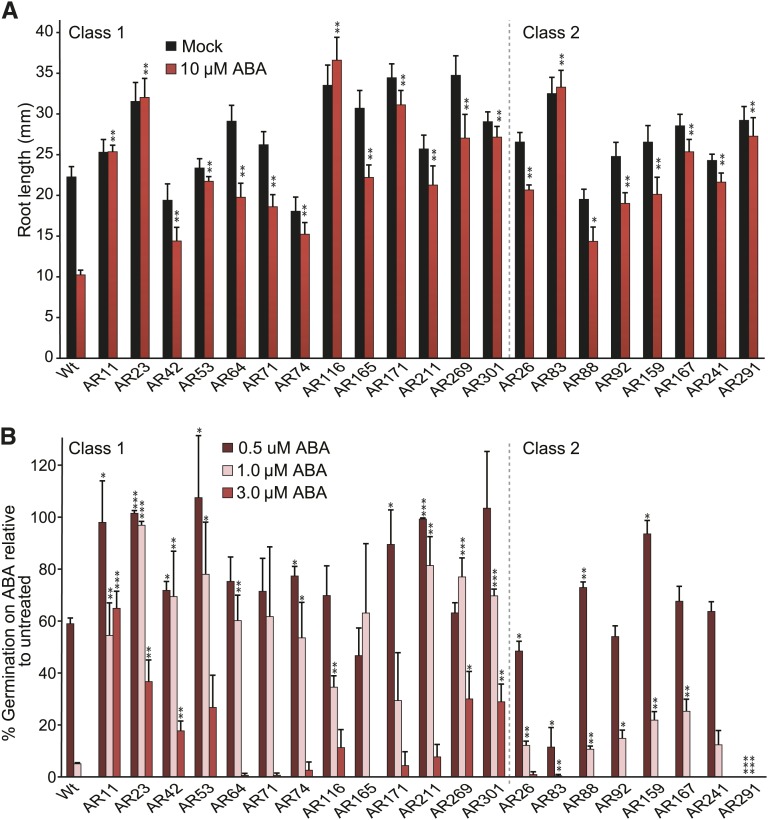
ABA inhibits both seed germination and primary root elongation in Arabidopsis (Cutler et al. 2010). To isolate mutants displaying ABA-resistant root growth, we generated 56 pools of EMS-mutagenized Col-0 seed and screened approximately 280,000 resultant M2 progeny for seedlings with reduced ABA-responsiveness in root elongation. We plated stratified M2 seeds on filter paper–lined unsupplemented media and grew them for 4 d to allow for seed germination. We then transferred the newly germinated seedlings atop filter papers to media supplemented with 10 µM ABA. After an additional 4 d of growth, we selected 316 putative mutants from 56 independent pools exhibiting a long root on ABA. Of these, 76 died and eight were infertile. Progeny of the 232 remaining putative mutants were rescreened for resistance to ABA in root elongation; 168 of these mutants displayed longer roots than wild-type in response to ABA. Twenty-one of the ABA-resistant lines were characterized in detail. Because each mutant came from a separate seed pool (Table 1), the mutants represent 21 independent mutagenic events. All 21 isolates demonstrated strong ABA resistance in root elongation assays (Figure 1A), whereas a subset of the isolates failed to demonstrate strong ABA resistance in seed germination (Figure 1B). To classify our mutants, we considered seed germination of more than 25% in the presence of 1 µM ABA after 72 hr of inhibition to be strong ABA resistance. We used these differences in ABA-responsive seed germination to divide our isolates into two mutant classes: class 1 mutants displayed strong resistance to the inhibitory effects of ABA in both root elongation and seed germination, whereas class 2 mutants displayed resistance to the inhibitory effects of ABA on root elongation and hypersensitivity, wild-type sensitivity, or mild resistance to ABA in seed germination inhibition (Table 1). Isolates AR11, AR23, AR42, AR53, AR64, AR71, AR74, AR116, AR165, AR171, AR211, AR269, and AR301 displayed strong ABA resistance in seed germination assays and were placed into class 1. AR26, AR88, AR92, AR159, AR167, and AR241 displayed mild resistance to ABA, and AR83 and AR291 displayed ABA hypersensitivity in seed germination assays and were placed into class 2. Interestingly, the separation of ABA responsiveness in root elongation and seed germination in class 2 mutants is consistent with the possibility that they carry defects in ABA response pathways specific to root length response that are independent from germination response.
Table 1. Characterization of mutants.
| Class | Isolate | M2 Pool | Response in Root Elongation Assays | ABA-Responsive Seed Germinationd | Causative Mutation | ||
|---|---|---|---|---|---|---|---|
| ABAa | ACCb | 2,4-Dc | |||||
| — | Wt | — | •••• | •••• | •••• | •••• | |
| 1 | AR11 | 3 | • | •••• | •••• | •• | abi2-11 |
| 1 | AR23 | 6 | • | • | • | • | |
| 1 | AR42 | 10 | •• | •• | ••• | •• | |
| 1 | AR53 | 12 | • | • | • | • | aux1-53 |
| 1 | AR64 | 15 | ••• | •••• | •••• | •• | |
| 1 | AR71 | 16 | •• | •••• | •••• | •• | |
| 1 | AR74 | 17 | •• | •••• | •••• | •• | |
| 1 | AR116 | 27 | • | • | • | ••• | aux1-116 |
| 1 | AR165 | 34 | •• | ••• | • | •• | |
| 1 | AR171 | 35 | • | •••• | •••• | ••• | |
| 1 | AR211 | 42 | •• | • | •••• | • | eir1-211 |
| 1 | AR269 | 48 | •• | • | • | • | iaa16-1 |
| 1 | AR301 | 53 | • | • | • | •• | |
| 2 | AR26 | 7 | •• | •••• | ••• | •••• | |
| 2 | AR83 | 19 | • | • | ••• | ••••• | |
| 2 | AR88 | 20 | •• | •••• | •••• | •••• | |
| 2 | AR92 | 21 | •• | ••• | •••• | •••• | |
| 2 | AR159 | 33 | •• | •••• | •••• | •••• | |
| 2 | AR167 | 35 | • | •••• | •••• | •••• | |
| 2 | AR241 | 44 | • | •••• | ••• | •••• | axr4-241 |
| 2 | AR291 | 50 | • | • | •••• | ••••• | ein2-291 |
| Known mutants | |||||||
| 1 | ibr5-1 | — | •• | ••• | ••• | • | ibr5-1 |
| 1 | tir1-1 | — | ••• | ••• | ••• | • | tir1-1 |
| 1 | axr1-3 | — | ••• | ••• | • | ••• | axr1-3 |
| 1 | aux1-7 | — | • | • | • | • | aux1-7 |
| 2 | ein2-1 | — | • | • | ••• | ••••• | ein2-1 |
| — | eto1-1 | — | ••• | • | •••• | ••• | eto1-1 |
| 2 | etr1-1 | — | • | • | ••• | Se | etr1-1 |
| 1 | abi1-1 | — | • | •••• | •••• | Rf | abi1-1 |
| 1 | abi2-1 | — | •• | •••• | •••• | Rf | abi2-1 |
| 1 | abi3-1 | — | •••• | •••• | •••• | Rf | abi3-1 |
Relative ABA responsiveness on 10 μM ABA vs. mock-treated. •••• = 0–60%; ••• = 60–70%; •• = 71–85%; and • = 86–100%.
Relative ACC responsiveness on 100 nM ACC vs. mock-treated. •••• = 0–70%; ••• = 70–80%; •• = 80–90%; and • = 90–100%.
Relative 2,4-D responsiveness on 100 nM 2,4-D vs. mock-treated. •••• = 0–50%; ••• = 50–65%; •• = 65–85%; and • = 85–100%.
Relative ABA inhibition of seed germination on 1 μM ABA vs. mock-treated. ••••• = 0–5%; •••• = 5–25%; ••• = 25–50%; •• = 50–75%; and • = 75–100%.
Beaudoin et al. (2000). S = Sensitive.
Koornneef et al. (1984). R = Resistant.
Figure 1.
AR mutant responses to ABA in root elongation and seed germination. (A) Mean primary root lengths (±SE; n ≥ 15) of Col-0 (Wt) and AR mutant seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with ethanol (mock) or 10 µM ABA. Asterisks indicate that the mutant root lengths were significantly longer than wild-type root lengths on 10 µM ABA (*P ≤ 0.01; **P ≤ 0.001) in two-tailed t tests assuming unequal variance. (B) Mean normalized percentage (±SE; n = 3) of Col-0 (Wt) and AR mutant seeds after 5 d grown under yellow-filtered light at 22° on medium supplemented with 0.3 µM ABA, 1.0 µM ABA, or 3 µM ABA. Asterisks indicate that the mutant germination percentages were significantly different from wild-type germination percentages (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001) in two-tailed t tests assuming unequal variance.
PCR-based genotyping to identify a mutation in ABI2
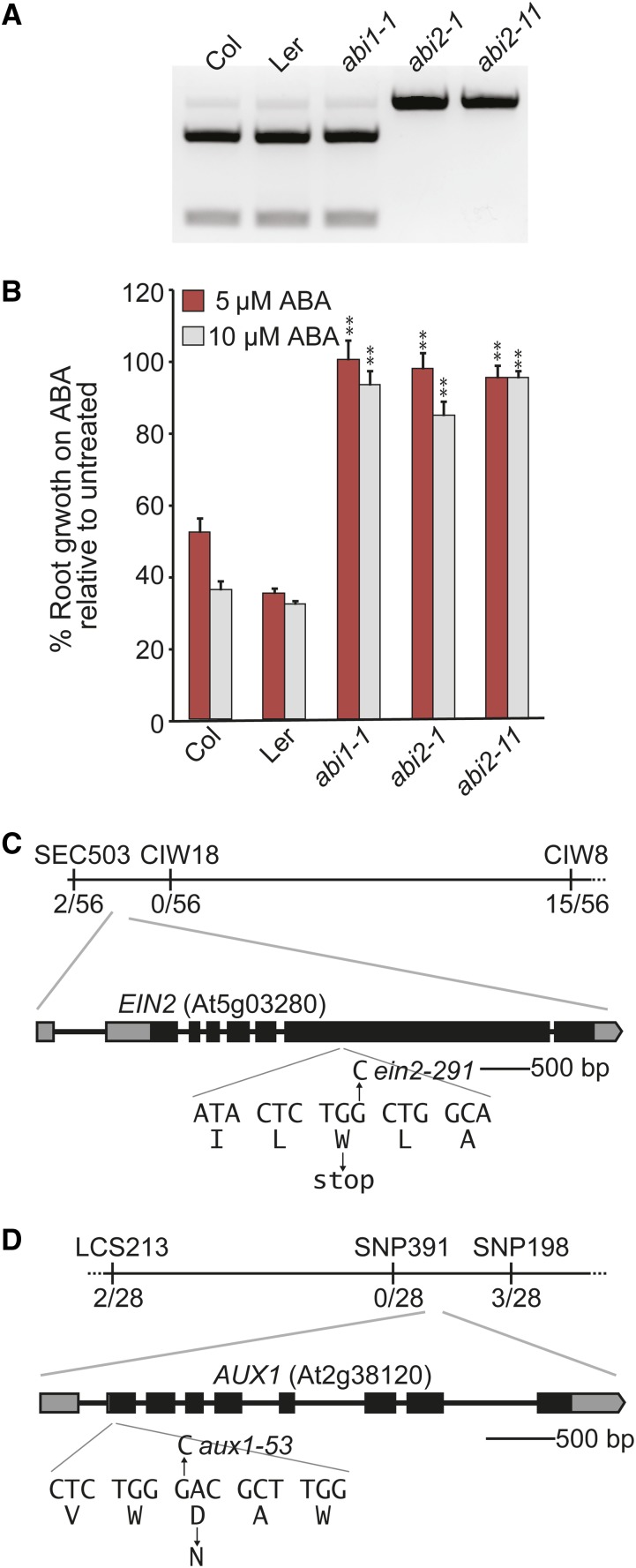
The gain-of-function abi1-1 and abi2-1 mutants display resistance to ABA in root elongation assays (Koornneef et al. 1984); therefore, we expected to identify alleles of abi1-1 and abi2-1 in our screen. We examined M2 isolates for the abi1-1 and abi2-1 lesions by PCR-based genotyping. Both abi1-1 and abi2-1 alleles result from similar C-to-T transitions that result in the loss of an NcoI site in ABI1 and ABI2 (Leung et al. 1997). Although we did not isolate an abi1-1 allele among our isolates, AR11, a class 1 mutant (Table 1), carried a C-to-T transition in ABA INSENSITIVE2/At5g57050 identical to the abi2-1 mutant in the Ler ecotype (Figure 2A). We therefore named the AR11 isolate abi2-11. abi2-11 displayed ABA resistance in root elongation assays similar to abi1-1 and abi2-1 (Figure 2B). Because both abi1-1 and abi2-1 are in the Ler ecotype, the abi2-11 allele in the Col-0 ecotype may be a useful tool for future genetic studies.
Figure 2.
Identification of AR mutant defects in ABA, ethylene, and auxin response factors. (A) Amplification of ABI2 from wild-type ecotypes Col-0 and Ler-0, abi1-1, abi2-1, and abi2-11, followed by an NcoI digest, demonstrates that abi2-1 and abi2-11 contain the same mutation that removes the NcoI restriction site from the sequence. (B) Mean normalized primary root lengths (±SE; n = 15) of Col-0, Ler-0, abi1-1, abi2-1, and abi2-11 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 5 or 10 µM ABA compared to seedlings grown on medium supplemented with ethanol (mock). Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths (**P ≤ 0.001) in two-tailed t tests assuming unequal variance. (C) Recombination mapping with PCR-based markers SEC503, CIW18, and CIW8 localized the ABA resistance mutation in AR291 between SEC503 and CIW8 with 2/56 north and 15/56 south recombinants. Examination of the EIN2 (At5g03280) gene in this region revealed a G-to-A mutation at position 1881 in AR291 DNA that results in Trp455-to-stop. (D) Recombination mapping with PCR-based markers LCS213, SNP391, and SNP198 localized the ABA resistance mutation in AR53 between LCS213 and SNP198 with 2/28 north and 3/28 south recombinants. Examination of the AUX1 (At2g38120) gene in this region revealed a G-to-A mutation at position 148 in AR53 DNA that results in an Asp50-to-Arg substitution.
Positional cloning to identify mutations in AUX1 and EIN2
We used ABA resistance in root elongation to map the recessive mutation in AR291 to a region on the upper arm of chromosome 5 between SEC503 and CIW8 (Figure 2C). This region contains ETHYLENE INSENSITIVE2/At5g03280 (EIN2). Mutations in EIN2 have previously been described to display ABA resistance in root elongation and hypersensitivity to ABA in seed germination assays (Beaudoin et al. 2000; Ghassemian et al. 2000), making a mutation in EIN2 a reasonable candidate for conferring the ABA resistance found in AR291, which is resistant to ABA in root elongation (Figure 1A) and hypersensitive to ABA in seed germination (Figure 1B) assays. We PCR-amplified and sequenced EIN2 from AR291 genomic DNA and identified a G-to-A base change at position 1881 (where the A of the ATG is at position 1) that causes a Trp455-to-stop mutation. We named the identified mutation in AR291 ein2-291.
We further used ABA resistance in root elongation to map the recessive mutation in AR53 to a region on the lower arm of chromosome 2 between the mapping markers LCS213 and SNP198 (Figure 2D). This region contains AUXIN RESISTANT1/At2g38120 (AUX1). Mutations in AUX1 have previously been described to display ABA resistance in root elongation (Strader et al. 2008a) and seed germination (Belin et al. 2009) assays, making a mutation in AUX1 a reasonable candidate for conferring the ABA resistance found in AR53. We PCR-amplified and sequenced AUX1 from AR53 genomic DNA and identified a G-to-A base change at position 148 (where the A of the ATG is at position 1) that causes a D50-to-N mutation. We named the identified mutation in AR53 aux1-53.
Whole genome sequencing to identify mutations in AUX1, AXR4, and EIR1/PIN2
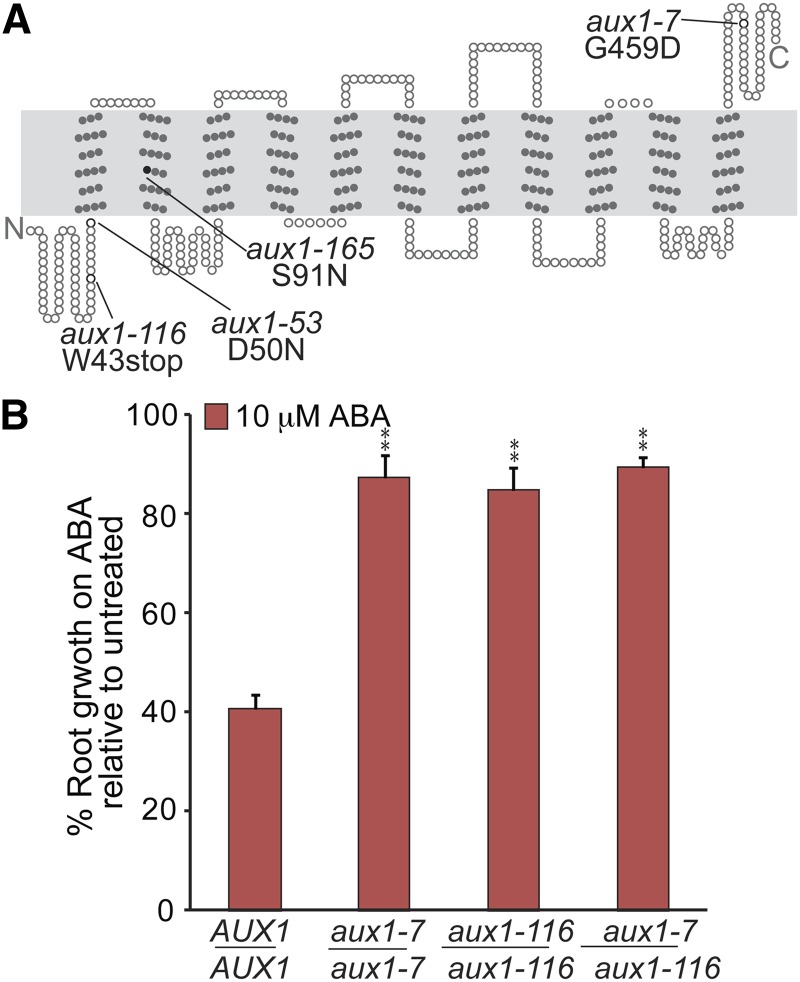
We isolated genomic DNA from seven pooled backcrossed lines (bulk segregants) of AR116 and used whole genome sequencing to quickly identify the potential causative EMS mutation. Because EMS typically causes G-to-A or C-to-T base pair changes, and because we assumed the causative mutation would be homozygous in the population, we mined the sequencing data for homozygous G-to-A or C-to-T base pair changes and found a total of 8 homozygous, EMS-related mutations not present in our wild-type sequence that clustered on the long arm of chromosome 2 (Table 2). This region included a promising candidate gene, AUXI/At2g38120, encoding an auxin influx carrier (Bennett et al. 1996; Marchant et al. 1999) that is necessary for ABA-responsive root elongation (Strader et al. 2008a). The EMS-generated mutation in AUX1 causes a G-to-A base pair change in the first exon of the coding sequence, resulting in a tryptophan-to-stop mutation at amino acid 43 of the protein (Figure 3A). We crossed AR116 to aux1-7 and found that the F1 progeny displayed resistance to ABA in root elongation assays similar to the two parents (Figure 3B), confirming that AR116 ABA resistance phenotypes are caused by the mutation identified in AUX1. We named this allele aux1-116.
Table 2. Homozygous EMS-related mutations in the AR116 exome.
| Chromosome | Locationa | Reference Sequence | Mutant Sequence | Gene | Amino Acid Change | Codon Change | Annotated Gene |
|---|---|---|---|---|---|---|---|
| 2 | 14625394 | G | A | At2g34680 | L/L | ctC/ctT | AIR9 |
| 2 | 15000961 | G | A | At2g35690 | G/E | gGa/gAa | ACX5 |
| 2 | 15674636 | G | A | At2g37360 | P/L | cCa/cTa | ABCG2 |
| 2 | 15674636 | G | A | At2g37362 | W/stop | tGg/tAg | Potential natural antisense gene, locus overlaps with AT2G37360 |
| 2 | 15973621 | G | A | At2g38120 | W/stop | tgG/tgA | AUX1 |
| 2 | 16725401 | G | A | At2g40050 | N/N | aaC/aaT | Cysteine/histidine-rich C1 domain family protein |
| 2 | 17850781 | G | A | At2g42890 | R/K | aGa/aAa | AML2 |
| 2 | 19007638 | G | A | At2g46290 | S/S | tcC/tcT | Transducin/WD40 repeat-like superfamily protein |
Base pair location on chromosome 2 in TAIR v10.
Figure 3.
Whole genome sequencing of AR116 uncovered a new allele of aux1. (A) Examination of AUX1 in AR116 revealed a G-to-A mutation in the first exon, which causes a Trp43-to-stop mutation. The AR53 causative mutation (aux1-53), identified by recombination mapping (Figure 2D), is also shown. (B) aux1-116 is allelic to aux1-7. Complementation test showing mean normalized primary root length (±SE; n ≥ 13) of Col-0 wild-type (AUX1/AUX1), aux1-7/aux1-7, aux1-116/aux1-116, and aux1-7/aux1-116 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 10 µM ABA. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on ABA (**P ≤ 0.001) in two-tailed t tests assuming unequal variance.
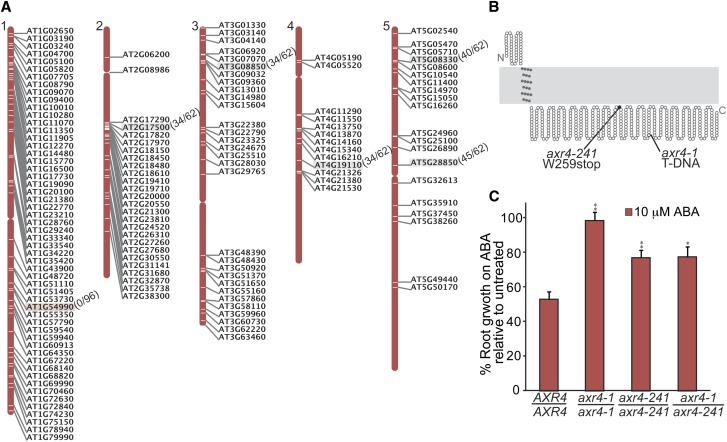
To identify the causative mutation in AR241, we isolated genomic DNA from pooled M4 seedlings and used whole genome sequencing to quickly identify the potential causative EMS mutation. Because this mutant had not been backcrossed to reduce the number of background mutations, our sequencing data revealed 144 homozygous EMS-related mutations not present in our wild-type sequence that were found throughout the genome (Figure 4A, Table S1). We then crossed AR241 to Col-0 and F2 progeny were selected for the ABA root elongation resistance phenotype. CAPS-based and dCAPS-based PCR genotyping primers were designed for identified mutations (Table S2) to determine linkage between ABA-resistant root elongation and the identified mutations. Genotyping for the AUXIN RESISTANT4/At1g54990 (AXR4) (Dharmasiri et al. 2006) mutation revealed that all of the individuals tested were homozygous at the AXR4 locus, suggesting this may be the locus responsible for the AR241 phenotype. The EMS-generated mutation in AXR4 causes a G-to-A base pair change in the first exon of the coding sequence, resulting in a tryptophan-to-stop mutation at amino acid 259 of the protein (Figure 4B). We crossed AR241 to axr4-1 (Hobbie and Estelle 1995) and found that the F1 progeny displayed resistance to ABA in root elongation assays similar to the two parents (Figure 4C), confirming that AR241 ABA resistance phenotypes are caused by the mutation identified in AXR4, and we named this new allele axr4-241.
Figure 4.
Whole genome sequencing of AR241 revealed a new allele of axr4. (A) Map positions of homozygous EMS-caused mutations identified in AR241. Genomic DNA from pooled M4 seedlings was sequenced and examined for G/C to A/T transitions typically associated with EMS mutagenesis in splice sites and coding sequences (see Table S1 for list of mutations identified). Approximate map positions of the identified mutations are shown to the right of each chromosome. PCR-based markers (Table S2) were designed for a subset of identified mutations and used to localize the AR241 causative mutation near At1g54990 with 0/96 recombination events in a population of AR241 backcrossed lines. Recombination events in examined backcrossed lines are listed to the right of gene names for each PCR-based marker. (B) Examination of AXR4/At1g54990 in AR241 revealed a G-to-A mutation in the first exon causing a Trp259-to-stop mutation. (C) axr4-241 is allelic to axr4-1. Complementation test showing mean normalized primary root lengths (±SE; n ≥ 10) of Col-0 wild-type (AXR4/AXR4), axr4-1/axr4-1, axr4-241/axr4-241, and axr4-1/axr4-241 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 10 µM ABA. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on ABA (*P ≤ 0.01; **P ≤ 0.001) in two-tailed t tests assuming unequal variance.
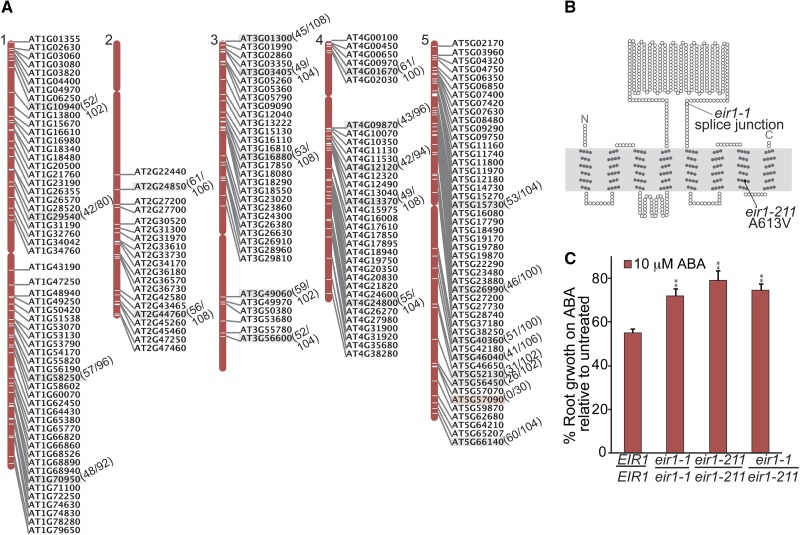
To identify the causative mutation in AR211, we isolated genomic DNA from pooled M4 seedlings and used whole genome sequencing to quickly identify the potential causative EMS mutation. Because this mutant had not been backcrossed to reduce the number of background mutations, our sequencing data revealed 195 homozygous EMS-related mutations not present in our wild-type sequence that were found throughout the genome (Figure 5, Table S3). We then crossed AR211 to Col-0 and selected F2 progeny for the ABA root elongation resistance phenotype. CAPS-based and dCAPS-based PCR genotyping primers (Table S2) were designed to screen these seedlings for the presence of the EMS mutations identified in the genome sequencing. Genotyping for the ETHYLENE INSENSITIVE ROOT1/PIN-FORMED 2/At5g57090 (EIR1/PIN2) (Luschnig et al. 1998) mutation revealed that all of the individuals tested were homozygous at the EIR1 locus, suggesting this may be the locus responsible for the AR211 phenotype. The EMS-generated mutation in EIR1 causes a C-to-T base pair change in the eighth exon of the coding sequence, resulting in an alanine-to-valine missense mutation at amino acid 613 of the protein (Figure 5B). We crossed AR211 to eir1-1 and found that the F1 progeny displayed resistance to ABA in root elongation assays similar to the two parents (Figure 5C), confirming that AR211 ABA resistance phenotypes are caused by the mutation identified in EIR1/PIN2, and we named this new allele eir1-211.
Figure 5.
Whole genome sequencing of AR211 reveals a new allele of eir1. (A) Map positions of homozygous EMS-caused mutations identified in AR211. Genomic DNA from pooled M4 seedlings was sequenced and examined for G/C to A/T transitions typically associated with EMS mutagenesis in splice sites and coding sequences (see Table S3 for list of mutations identified). Approximate map positions of the identified mutations are shown to the right of each chromosome. PCR-based markers (Table S2) were designed for a subset of identified mutations and used to localize the AR211 causative mutation near At5g57090 with 0/30 recombination events in a population of AR211 backcrossed lines. Recombination events in examined backcrossed lines are listed to the right of gene names for each PCR-based marker. (B) Examination of EIR1/PIN2 in AR211 revealed a C-to-T mutation in the eighth exon causing an Ala613-to-Val mutation. (C) eir1-211 is allelic to eir1-1. Complementation test showing mean normalized primary root lengths (±SE; n = 15) of Col-0 wild-type (EIR1/EIR1), eir1-1/eir1-1, eir1-211/eir1-211, and eir1-1/eir1-211 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with 10 µM ABA compared to mock-treated (ethanol) seedlings. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on ABA (**P ≤ 0.001) in two-tailed t tests assuming unequal variance.
AR mutants display varied responses to ACC and auxin
Because the causative mutations initially found in the AR isolates disrupted known components of ABA, ethylene, and auxin pathways, we examined AR mutant responsiveness to ethylene and auxin in root elongation assays to determine if the remaining isolates might also be defective in ethylene or auxin signaling. Primary root lengths of the selected AR mutants were measured in response to the ethylene precursor ACC (Figure 6A) and the synthetic auxin 2,4-D (Figure 6B).
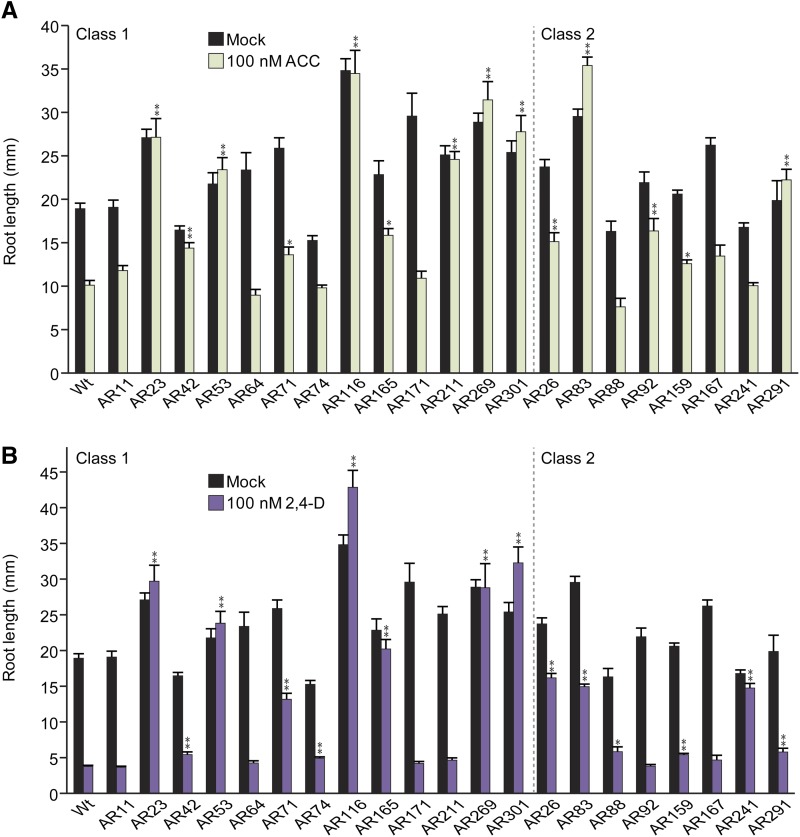
Figure 6.
AR mutant responses to ACC and 2,4-D. (A) Mean primary root lengths (±SE; n ≥ 12) of 8-day-old Col-0 (Wt) and AR mutant seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or 100 nM ACC. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on ACC (*P ≤ 0.01; **P ≤ 0.001) in two-tailed t tests assuming unequal variance. (B) Mean primary root lengths (±SE; n ≥ 12) of 8-day-old Col-0 (Wt) and AR mutant seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock) or 100 nM 2,4-D. Asterisks indicate that the mutant normalized root lengths were significantly longer than wild-type normalized root lengths on 2,4-D (*P ≤ 0.01; **P ≤ 0.001) in two-tailed t tests assuming unequal variance.
Class 1 AR mutants could be subdivided into categories based on hormone responses in root elongation. The class 1 isolates AR11 (defective in ABI2), AR64, and AR171 displayed wild-type sensitivity to ACC (Figure 6A) and 2,4-D (Figure 6B) in root elongation assays, consistent with roles for the defective genes in AR11 (abi2-11), AR64, and AR171 specifically in ABA response. The class 1 isolate AR53, defective in the auxin transporter AUX1, displayed resistance to ACC (Figure 6A) and 2,4-D (Figure 6B), consistent with previous results suggesting that AUX1 activity is necessary for response to these hormones in root elongation assays (Pickett et al. 1990). Likewise, the class 1 isolates AR23, AR42, AR116, AR165, AR269 (defective in IAA16) (Rinaldi et al. 2012), and AR301 displayed resistance to ACC (Figure 6A) and 2,4-D (Figure 6B), consistent with the possibility that they may also be defective in components of auxin response.
Class 2 AR mutants could be subdivided into categories based on hormone responses in root elongation assays. The class 2 isolate AR291 (defective in EIN2) displayed resistance to ACC (Figure 6A) (Romano et al. 1995; Fujita and Syono 1996; Alonso et al. 1999) and hypersensitivity to ABA in seed germination assays (Figure 1B). Similarly, the class 2 isolate AR83 displayed strong resistance to ACC (Figure 6A) and hypersensitivity to ABA in seed germination assays (Figure 1B), suggesting that this isolate may also be defective in a component of the ethylene response pathway. Intriguingly, isolates AR88 and AR167, both of which display strong ABA resistance in root elongation (Figure 1A), display wild-type sensitivity to ABA in seed germination assays (Figure 1B) and wild-type sensitivity to ACC (Figure 6A) and 2,4-D (Figure 6B) in root elongation assays, consistent with the possibility that these mutants are defective in components of the ABA response pathway specific to root responses.
Auxin and ethylene mutants display altered ABA-responsive root elongation
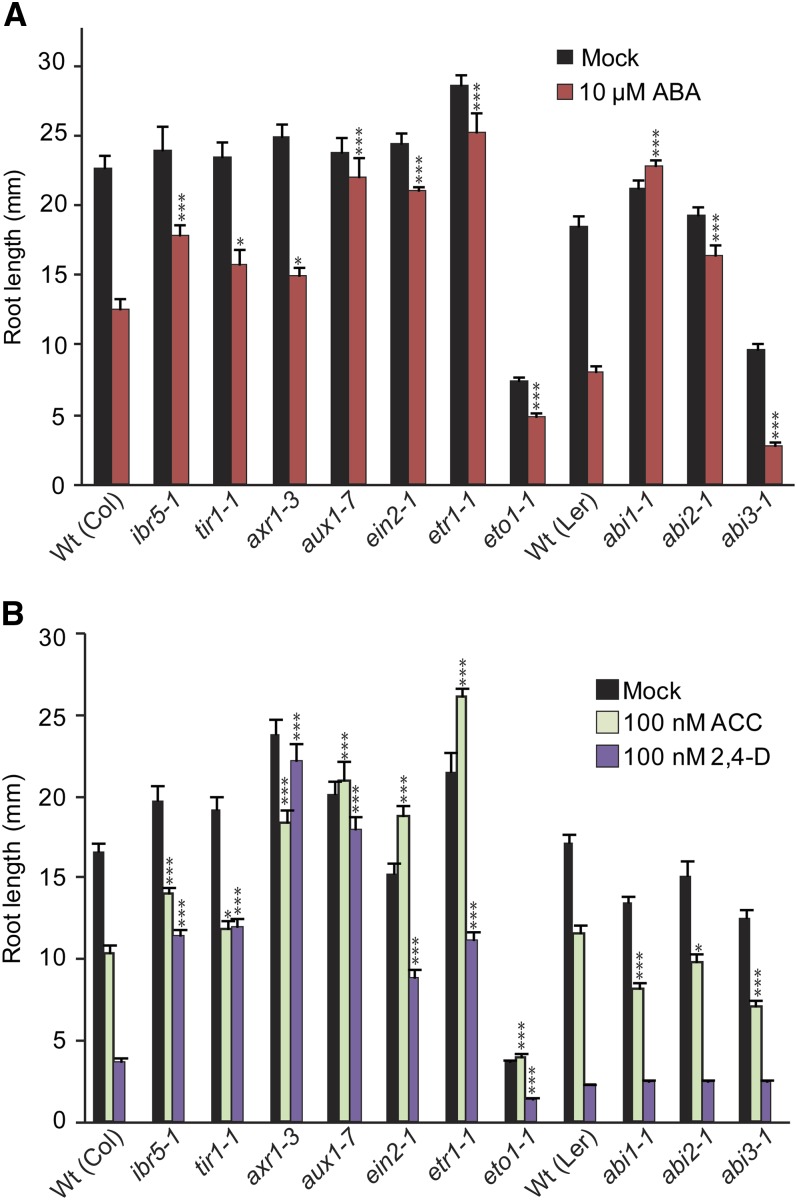
Because we identified AR mutants with defects in components of auxin, ethylene, and ABA response, we examined several known auxin, ethylene, and ABA mutants for comparison to our unknown isolates. We examined hormone responses of the protein phosphatase mutant ibr5-1 (Monroe-Augustus et al. 2003), the auxin co-receptor mutant tir1-1 (Ruegger et al. 1998), RUB activation mutant axr1-3 (Lincoln et al. 1990), the auxin transport mutant aux1-7 (Pickett et al. 1990), the ethylene response mutant ein2-1 (Alonso et al. 1999), the ethylene overproducing mutant eto1-1 (Guzmán and Ecker 1990), and the ethylene receptor mutant etr1-1 (Bleeker et al. 1988), all in the Col-0 ecotype, and compared their responses to the hormone responses of the protein phosphatase mutant abi1-1 (Finkelstein 1994), the protein phosphatase mutant abi2-1 (Finkelstein 1994), and the ABA transcription factor mutant abi3-1 (Giraudat et al. 1992), all in the Ler-0 ecotype. We found that, similar to previous reports, the auxin-resistant mutants ibr5-1 (Monroe-Augustus et al. 2003), tir1-1 (Strader et al. 2008a), axr1-3 (Monroe-Augustus et al. 2003), and aux1-7 (Strader et al. 2008a) exhibited ABA-resistant root growth (Figure 7A), consistent with a requirement for intact auxin responsiveness for ABA-responsive inhibition of root elongation. We also found that, similar to previous reports, the ethylene-resistant mutants ein2-1 (Beaudoin et al. 2000) and etr1-1 (Beaudoin et al. 2000) displayed ABA-resistant root growth, suggesting that intact ethylene responses are also required for ABA-responsive inhibition of root elongation.
Figure 7.
Responses of previously characterized auxin, ethylene, and ABA mutants to hormone treatment. (A) Mean primary root lengths (±SE; n = 16) of Col-0, ibr5-1, tir1-1, axr1-3, aux1-7, ein2-1, eto1-1, etr1-1, Ler-0, abi1-1, abi2-1, and abi3-1 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with ethanol (mock) or 10 µM ABA. Asterisks indicate that the mutant root lengths were significantly different than wild-type root lengths on ABA (*P ≤ 0.05; ***P ≤ 0.001) in two-tailed t tests assuming unequal variance. (B) Mean primary root lengths (±SE; n = 16) of 8-day-old Col-0, ibr5-1, tir1-1, axr1-3, aux1-7, ein2-1, eto1-1, etr1-1, Ler-0, abi1-1, abi2-1, and abi3-1 seedlings grown under yellow-filtered light at 22° on medium supplemented with ethanol (mock), 100 nM ACC, or 100 nM 2,4-D. Asterisks indicate that the mutant root lengths were significantly different than wild-type root lengths on ABA (*P ≤ 0.05; ** P ≤ 0.01; *** P ≤ 0.001) in two-tailed t tests assuming unequal variance.
To complete our characterization, we examined root elongation responses of ibr5-1, tir1-1, axr1-3, aux1-7, ein2-1, etr1-1, eto1-1, abi1-1, abi2-1, and abi3-1 to the ethylene precursor ACC and the synthetic auxin 2,4-D. As expected, the examined auxin mutants ibr5-1, tir1-1, axr1-3, and aux1-7 displayed resistance to 2,4-D (Figure 4B). In addition, the ethylene-resistant mutants ein2-1 and etr1-1 displayed resistance to 2,4-D (Figure 7B), consistent with previous reports that intact ethylene responses are necessary for full response to exogenous auxins in root elongation assays (Fujita and Syono 1996; Růžička et al. 2007; Stepanova et al. 2007). Also consistent with previous reports, we found that ibr5-1 (Strader et al. 2008b), tir1-1 (Alonso et al. 2003), axr1-3 (Timpte et al. 1995), and aux1-7 (Pickett et al. 1990) displayed resistance to the ethylene precursor ACC (Figure 7B). ein2-1 and etr1-1 displayed strong resistance to ACC and 2,4-D (Figure 7B), consistent with previous reports that mutation of EIN2 results in resistance to ACC (Alonso et al. 1999; Stepanova et al. 2007) and the auxin transport inhibitor 1-N-naphthylphthalamic acid (Fujita and Syono 1996), and that etr1-1 displays resistance to ACC (Chang et al. 1993; Beaudoin et al. 2000; Růžička et al. 2007). Conversely, the ABA signaling mutants abi1-1, abi2-1, and abi3-1 displayed wild-type responses in root elongation to the tested concentrations of 2,4-D (Figure 7B), suggesting that ABA responsiveness does not affect auxin responsiveness in root elongation assays. Intriguingly, abi1-1, abi2-1, and abi3-1 displayed mild hypersensitivity to the inhibitory effects of ACC on root elongation (Figure 7).
Auxin and ethylene mutants exhibit altered ABA response in seed germination
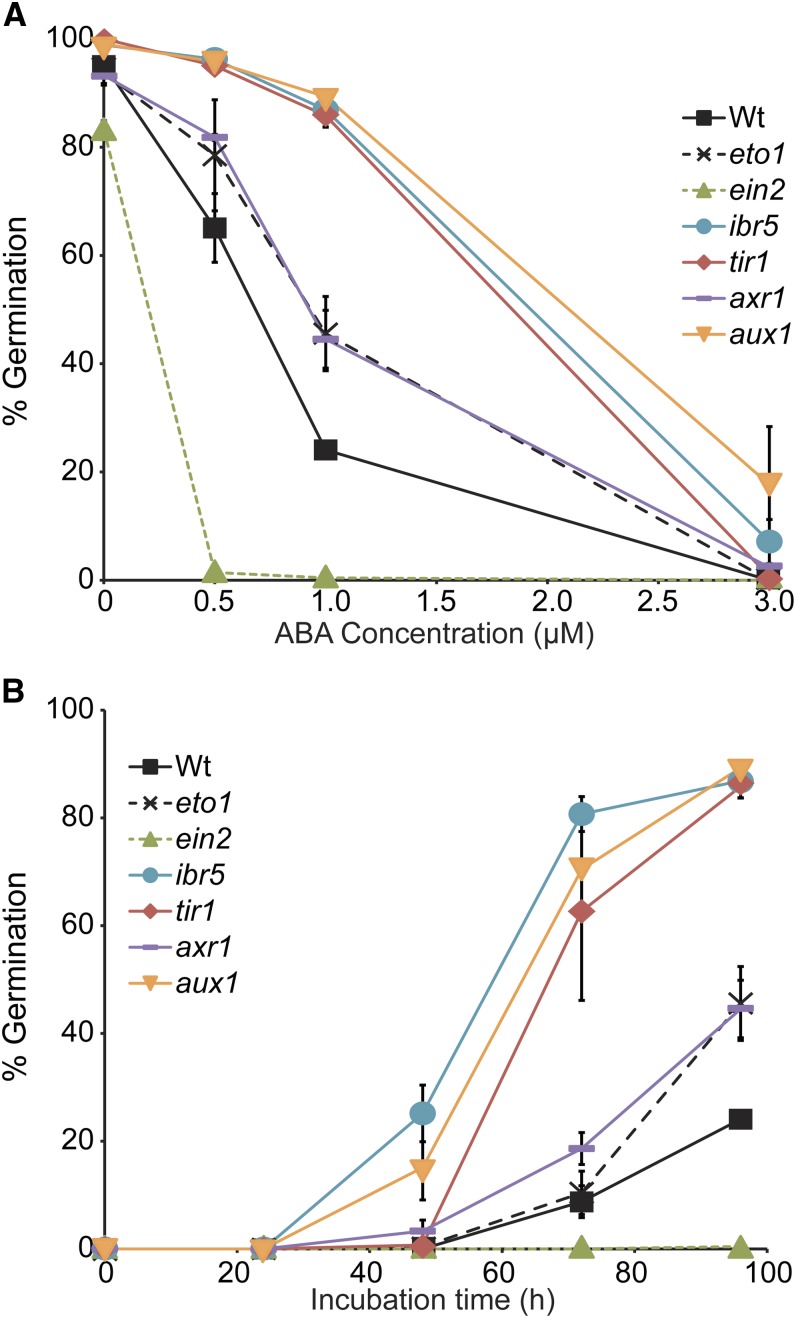
Because our AR mutants could be separated into two classes based on ABA-responsive seed germination, and because the examined auxin and ethylene mutants displayed ABA-resistant root elongation, we examined the auxin and ethylene mutants for altered ABA-responsive seed germination. We found that the auxin mutants ibr5-1, tir1-1, and aux1-7 demonstrated strong resistance to ABA inhibition of germination (Figure 8), consistent with previous reports (Monroe-Augustus et al. 2003; Belin et al. 2009). Conversely, the ethylene-resistant mutant ein2-1 demonstrated enhanced sensitivity to ABA in seed germination assays (Figure 8), consistent with previous reports that reduced ethylene sensitivity results in increased ABA sensitivity in seed germination assays (Beaudoin et al. 2000; Ghassemian et al. 2000). These results suggest that ABA-responsive seed germination can be an efficient means of separating ethylene response mutants and auxin response mutants phenotypically.
Figure 8.
Responses of previously characterized auxin and ethylene mutants to the inhibitory effects of ABA on seed germination. (A) Percentage (±SE; n = 3) of Col-0 (Wt), eto1-1, ein2-1, ibr5-1, tir1-1, axr1-3, and aux1-7 seed germination on medium supplemented with the indicated concentrations of ABA after 96 hr of growth under white light. eto1-1, ein2-1, ibr5-1, tir1-1, axr1-3, and aux1-7 seed germination percentages on 1 µM ABA were significantly different (P ≤ 0.02) from Wt seed germination percentage on 1 µM ABA. (B) Percentage (±SE; n = 3) of Col-0 (Wt), eto1-1, ein2-1, ibr5-1, tir1-1, axr1-3, and aux1-7 seed germination on medium supplemented with 1 µM ABA after the indicated time of growth under white light. eto1-1, ein2-1, ibr5-1, tir1-1, axr1-3, and aux1-7 seed germination percentages were significantly different (P ≤ 0.02) from Wt seed germination after 96 hr of incubation.
Effects of combining auxin and ethylene mutations on ABA responsiveness
To further our understanding of the crosstalk between auxin and ethylene response in altering ABA responsiveness, we examined eto1-1 and ein2-1 double mutant combinations with the auxin-resistant mutants ibr5-1, tir1-1, axr1-3, and aux1-7. For these studies, we used the previously described (Strader et al. 2010) double mutants of the ethylene overproducing mutant eto1-1 (Guzmán and Ecker 1990) with ibr5-1 (Monroe-Augustus et al. 2003), tir1-1 (Ruegger et al. 1998), axr1-3 (Estelle and Somerville 1987), and aux1-7 (Maher and Martindale 1980). To generate double mutants with the strong ethylene-resistant mutant ein2-1 (Alonso et al. 1999), we crossed ein2-1 to ibr5-1, tir1-1, axr1-3, and aux1-7 and identified double mutants using PCR-based polymorphic markers. In addition, we characterized the eto1-1 ein2-1 double mutant (Strader et al. 2010) to verify whether phenotypes observed in eto1-1 are caused by increased ethylene signaling rather than altered ACC levels in the eto1-1 mutant (Figure 9 and Figure 10), which may contribute to ethylene-independent signaling events (Tsang et al. 2011).
Figure 9.
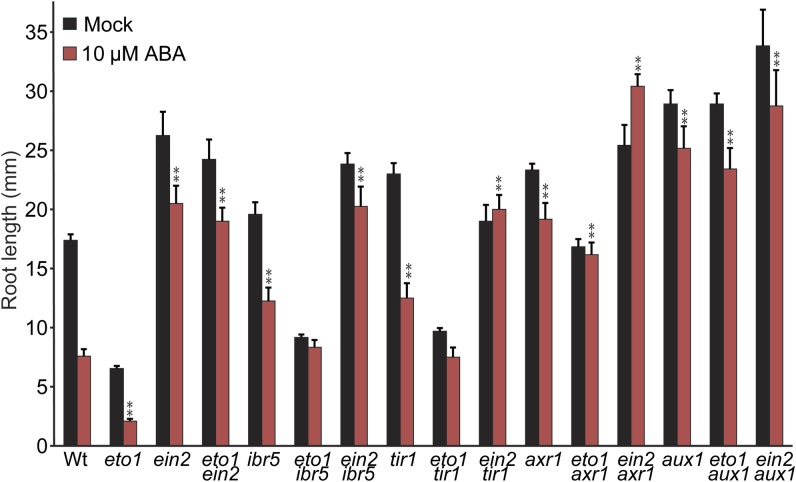
Auxin–ethylene double mutant analysis of ABA-responsive inhibition of primary root elongation. Mean primary root lengths (±SE; n ≥ 12) of Col-0, eto1, ein2, eto1 ein2, ibr5, eto1 ibr5, ein2 ibr5, tir1, eto1 tir1, ein2 tir1, axr1, eto1 axr1, aux1, eto1 aux1, and ein2 aux1 seedlings grown under yellow-filtered light at 22° for 4 d on unsupplemented medium, followed by 4 d on medium supplemented with ethanol (mock) or 10 µM ABA. eto1 ibr5, eto1 tir1, eto1 axr1, and eto1 aux1 root lengths were significantly (P ≤ 0.001) longer than the eto1 root length on ABA. ein2 ibr5, ein2 tir1, and ein2 aux1 root lengths were statistically indistinguishable from the ein2 root length on ABA, whereas ein2 axr1 root lengths were significantly longer than either ein2 or axr1 root lengths (P ≤ 0.001) on ABA.
Figure 10.
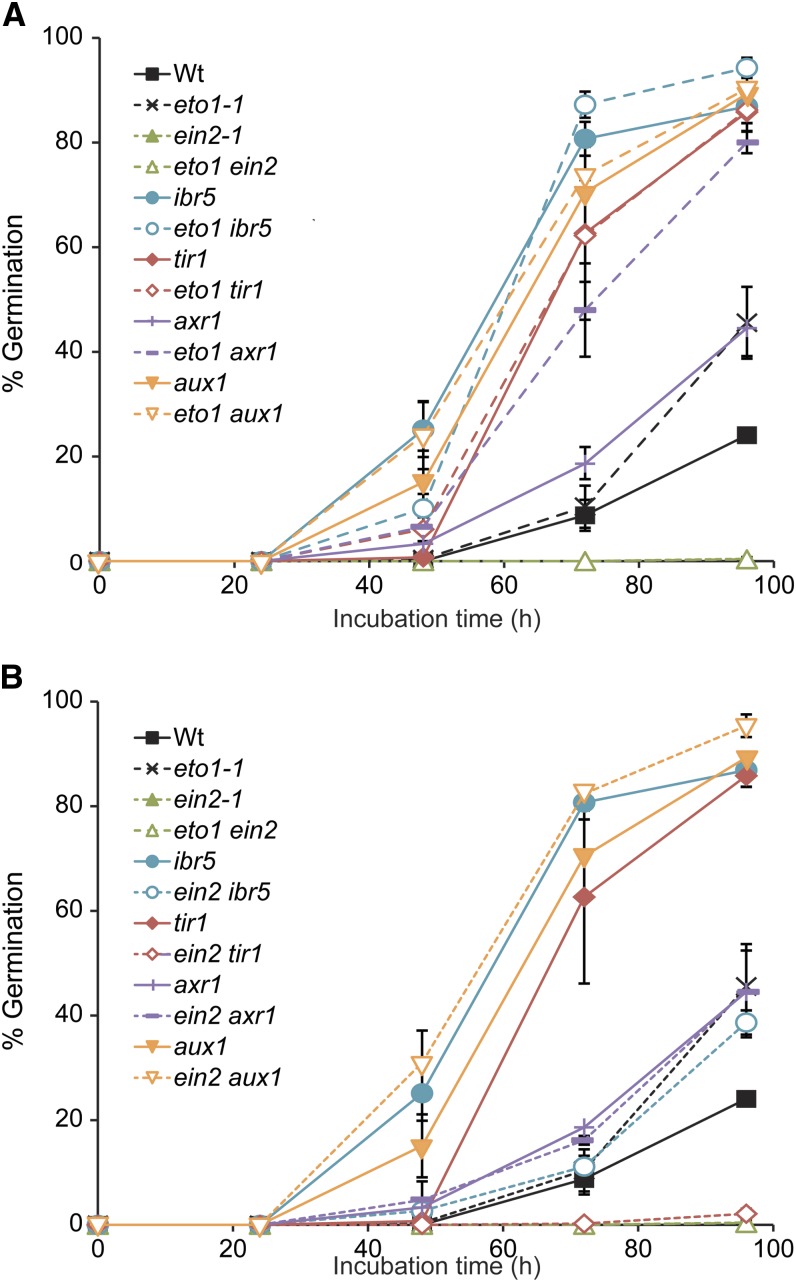
Auxin–ethylene double mutant analysis of ABA-responsive inhibition of seed germination. (A) Percentage seed germination (±SE; n = 3) at the indicated time points of Col-0 (Wt), eto1-1, ein2-1, eto1-1 ein2-1, ibr5, eto1 ibr5, tir1-1, eto1 tir1, axr1, eto1 axr1, aux1, and eto1 aux1 seeds grown on medium supplemented with 1 µM ABA. (B) Percentage seed germination (±SE; n = 3) at the indicated time points of Col-0 (Wt), eto1-1, ein2-1, eto1-1 ein2-1, ibr5, ein2 ibr5, tir1-1, ein2 tir1, axr1, ein2 axr1, aux1, and ein2 aux1 seeds grown on medium supplemented with 1 µM ABA under white light. ein2 ibr5, ein2 axr1, and ein2 aux1 germination percentages were significantly longer than the ein2 germination percentage (P ≤ 0.01) on ABA after 96 hr of incubation.
We examined ABA-responsive root elongation phenotypes of eto1-1 and ein2-1 combined with the auxin-resistant mutants ibr5-1, tir1-1, axr1-3, and aux1-7. eto1-1 did not suppress the ABA-resistant root elongation of ein2-1, ibr5-1, tir1-1, axr1-3, or aux1-7 (Figure 9), suggesting that ethylene overproduction cannot compensate for reduced auxin responsiveness in modulating ABA-responsive root elongation. In addition, the ein2 ibr5, ein2 tir1, and ein2 aux1 double mutants display ABA-resistant root elongation similar to the ein2 parent (Figure 9), suggesting that ethylene and auxin response may act linearly to modulate ABA-responsive root elongation. The ein2 axr1 double mutant displays greater ABA-resistant root elongation than either parent, suggesting EIN2 and AXR1 may contribute to ABA responsiveness in root elongation using at least partially distinct mechanisms, consistent with the pleiotropic nature of the axr1 mutant (Leyser et al. 1993), which potentially affects the activity of hundreds of SCF complexes. Results from examining ABA-responsive root elongation phenotypes of these ethylene–auxin double mutants are consistent with auxin and ethylene acting in a linear pathway to affect ABA-responsive inhibition of primary root elongation. Further, because ethylene overproduction by eto1 could not compensate for the reduced ABA sensitivity in auxin-resistant mutants (Figure 9), auxin is likely downstream of ethylene in this response pathway.
In addition to inhibiting root elongation, ABA inhibits seed germination. Interestingly, auxin and ethylene have been reported to have opposite effects on ABA-responsive inhibition of seed germination and seedling development; auxin-resistant mutants display resistance to ABA in seed germination (Monroe-Augustus et al. 2003; Belin et al. 2009; Rinaldi et al. 2012), whereas ethylene-resistant mutants display hypersensitivity to ABA in seed germination (Subramani 1992; Beaudoin et al. 2000; Ghassemian et al. 2000). We therefore examined ABA responsiveness in seed germination (Figure 10) assays in our mutants. The eto1-1 mutant, which overproduces ethylene, displayed mild resistance to the inhibitory effects of ABA on seed germination (Figure 8 and Figure 10A). In addition, eto1-1 mildly enhanced the ABA resistance in seed germination of the auxin-resistant mutants ibr5-1, axr1-3, and aux1-7 (Figure 10A). Conversely, the ein2-1 mutant, defective in ethylene response, displayed hypersensitivity to the inhibitory effects of ABA on seed germination (Figure 8 and Figure 10B) (Beaudoin et al. 2000; Ghassemian et al. 2000). ein2 partially to fully suppressed the ABA resistance in seed germination displayed by the weak auxin response mutants ibr5-1 and tir1-1 (Figure 10D). Intriguingly, ein2-1 failed to suppress ABA-resistant seed germination of the strong auxin transport mutant aux1-7 or the pleiotropic mutant axr1-3 (Figure 10D). The additive nature of combining altered ethylene production or response with altered auxin response on ABA-responsive seed germination suggests that auxin and ethylene independently affect ABA regulation of seed germination.
Discussion
Whole genome sequencing to identify causative mutations
In this study, we demonstrate the ease of using whole genome sequencing of backcrossed bulk segregants to quickly identify causative mutations. We found that sequencing backcrossed bulk segregants, rather than outcrossed bulk segregants, facilitates identification of a small interval of candidate mutations (Table 2 and Thole and Strader, unpublished data). We were curious how many mutations would be present without backcrossing and therefore sequenced nonbackcrossed pooled M4 seedlings for AR241 and AR211. For each of these mutants, homozygous EMS-caused mutations were littered throughout the genome, with 144 mutations identified in AR241 and 195 mutations identified in AR211. We then used recombination mapping to determine which of these mutations were linked to the ABA resistance phenotype. Although we were successful in identifying the causative mutations by sequencing nonbackcrossed mutant lines, we have decided to use sequencing of backcrossed bulk segregants in the future for mutant identification because the small number of homozygous mutations in these pools facilitates identification of the causative mutation.
Disruption of many genes can result in reduced ABA responsiveness in root elongation assays
Although ABA has long been known to inhibit root elongation, our understanding of mechanisms regulating ABA inhibition of root elongation remains incomplete. As may be expected, many components of the ABA signaling pathway are required for ABA inhibition of root elongation. For example, the pyr1 pyl1 pyl2 pyl4 quadruple mutant of ABA receptors displays strong ABA resistance in root elongation (Park et al. 2009) and the gain-of-function abi1-1 and abi2-1 mutations in protein phosphatases required for ABA perception display strong ABA resistant root elongation (Koornneef et al. 1984) and higher order loss-of-function mutations in this gene family results in ABA hypersensitivity in root elongation inhibition (Rubio et al. 2009). Downstream of ABA perception, the SnRK2.2 and SnRK2.3 protein kinases (Fujii et al. 2007), the ABA INSENSITIVE5 transcription factor (Lopez-Molina et al. 2001), and the calcium-dependent protein kinases CPK4 and CPK11 (Zhu et al. 2007) regulate ABA-responsive inhibition of root elongation. Additionally, G-proteins (Pandey et al. 2006) and the nuclear protein X1 (Kim et al. 2009) negatively regulate the inhibitory effects of ABA on root elongation and PROLINE-RICH EXTENSIN-LIKE RECEPTOR KINASE4 (Bai et al. 2009), and the NADPH oxidases AtrbohD and AtrbohF (Jiao et al. 2013) positively regulate ABA-responsive inhibition of root elongation.
To identify additional factors required for ABA-responsive inhibition of root elongation, we screened for mutations that conferred ABA-resistant root elongation (AR mutants). In this study, we characterized the ABA responsiveness of 21 AR lines and found that we could separate ABA-responsive root elongation inhibition from ABA-responsive seed germination inhibition in our mutants. In our screen, we identified alleles of ABI2 (Figure 2), EIN2 (Figure 2), AUX1 (Figure 2 and Figure 3), AXR4 (Figure 4), EIR1/PIN2 (Figure 5), and IAA16 (Rinaldi et al. 2012) that could confer ABA resistance in root elongation assays.
EIN2 is a positive regulator of ethylene responses. The N-terminus of ER-associated EIN2 (Bisson et al. 2009) contains 12 transmembrane helices and shows weak homology to mammalian NRAMP (natural resistance-associated macrophage protein) metal transporters, whereas its cytosolic C-terminus carries a nuclear localization signal. CTR1-dependent phosphorylation of EIN2 (Chen et al. 2011; Ju et al. 2012) determines whether the C-terminus of EIN2 is cleaved from its N-terminal transmembrane domain to allow translocation to the nucleus (Ju et al. 2012; Qiao et al. 2012; Wen et al. 2012) for activation of downstream ethylene-response transcription factors. EIN2 alleles have previously been identified in screens for enhanced response to ABA (era3) (Ghassemian et al. 2000) and for suppression of abi1-1 (Beaudoin et al. 2000). Interestingly, ein2 mutants display both hypersensitivity to ABA in seed germination assays and resistance to ABA in root elongation assays (Figure 7 and Figure 8) (Beaudoin et al. 2000; Ghassemian et al. 2000), suggesting that ethylene dampens ABA-responsive inhibition of seed germination and enhances ABA-responsive inhibition of root elongation. Thus far, molecular connections between ethylene effects on ABA responsiveness are not well-characterized.
We identified two alleles of auxin resistant1 (aux1) in our screen for mutants displaying resistance to the inhibitory effects of ABA on root elongation (Figure 2 and Figure 3). AUX1 is a major influx carrier of auxin; therefore, aux1 mutants are defective in many processes that require auxin transport, including ethylene responses (Stepanova and Alonso 2009; Muday et al. 2012) and ABA responses. aux1 displays resistance to the ABA-inducible ProDc3:GUS expression (Rock and Sun 2005), to the inhibitory effects of ABA on root elongation (Strader et al. 2008a), and to the inhibitory effects of ABA on postgerminative growth (Belin et al. 2009), suggesting that auxin transport is necessary for ABA-responsive inhibition of these processes.
We also identified an allele of auxin resistant4 (aux4) in our screen (Figure 4). The ER-localized AXR4 protein is necessary for targeting the AUX1 auxin influx carrier to the plasma membrane (Dharmasiri et al. 2006). axr4 displays resistance to ABA-inducible ProDc3:GUS expression (Rock and Sun 2005) and mild resistance to the inhibitory effects of ABA on root elongation (Figure 1A and Figure 4C) (Hobbie and Estelle 1994). Additionally, axr4 displays an interaction with ABA by principle component analysis of root system architecture (Ristova et al. 2013). Because AXR4 is required for proper AUX1 targeting to the plasma membrane, we would expect axr4 mutants to phenocopy aux1 mutants. Intriguingly, unlike the aux1 mutant or many other auxin resistant mutants, axr4 displays wild-type sensitivity to the ethylene precursor ACC in root elongation assays (Figure 6A) (Hobbie and Estelle 1994), suggesting that AXR4 is not required for AUX1 activity in response to ethylene.
EIR1/PIN2 is a polarly localized auxin efflux carrier (Feraru and Friml 2008). We identified an allele of ethylene insensitive root1/pin-formed2 (eir1/pin2) in our screen for mutants displaying resistance to the inhibitory effects of ABA on root elongation (Figure 4). eir1 displays strong resistance to the inhibitory effects of ABA on postgerminative growth (Belin et al. 2009) and moderate resistance to the inhibitory effects of ABA on root elongation (Figure 1A and Figure 5C) and seed germination (Figure 1B), consistent with the possibility that auxin efflux is necessary for ABA-responsive inhibition of these processes. In addition, eir1/pin2 was previously described for resistance to the growth-inhibitory effects of ethylene (Roman et al. 1995; Luschnig et al. 1998), suggesting that EIR1 is required for ethylene responses, in addition to being required for ABA responses. Indeed, auxin transport from the root meristem to the root elongation zone is required for ethylene responses (Stepanova and Alonso 2009; Muday et al. 2012) and also appears to be required for the inhibitory effects of ABA on root elongation.
Because we identified components of the ABA, ethylene, and auxin pathways, the remaining unidentified AR mutants could represent factors in the ABA, auxin, or ethylene signaling cascades or in an unanticipated response pathway. In particular, it will be interesting to further characterize class 2 AR mutants (Table 1), which appear to play stronger roles in responses to the inhibitory effects of ABA on root elongation than on seed germination.
Auxin–ethylene epistasis in ABA response
Because we identified factors required for both auxin and ethylene response in our screen for factors necessary for ABA-responsive root elongation, we became interested in understanding the auxin–ethylene interaction in modulating ABA responses. Previous auxin–ethylene interaction studies have been complicated by auxin-related ethylene inhibitor off-target effects; the ethylene biosynthesis inhibitor aminoethyoxyvinylglycine (AVG) also inhibits auxin biosynthesis (Soeno et al. 2010), whereas the ethylene receptor inhibitor AgNO3 promotes auxin efflux independently of its effects on ethylene responsiveness (Strader et al. 2009). To circumvent these issues, we examined double mutants between the ethylene overproducing mutant eto1 with auxin-resistant mutants (Strader et al. 2010) and double mutants between the strong ethylene-resistant mutant ein2 and auxin-resistant mutants. We used these double mutants to examine auxin–ethylene epistasis in modulating ABA-responsive root elongation and ABA-responsive seed germination.
Decreased auxin or ethylene sensitivity results in decreased ABA-responsive inhibition of root elongation. Because double mutants between ein2 and auxin-resistant mutants do not display additive phenotypes (Figure 9), auxin and ethylene likely act in a linear pathway to affect ABA-responsive inhibition of primary root elongation. In addition, ethylene overproduction by eto1 did not compensate for the reduced ABA sensitivity displayed by auxin-resistant mutants (Figure 9), suggesting that auxin acts downstream of ethylene in regulating ABA-responsive root elongation inhibition. This suggestion that auxin and ethylene act in a linear pathway to affect ABA-responsive root elongation is consistent with other reports that auxin acts downstream of ethylene to regulate root cell expansion (Strader et al. 2010) and that root ethylene responses require intact auxin responses (Růžička et al. 2007; Stepanova et al. 2007).
Decreased auxin responsiveness results in decreased ABA responsiveness in seed germination inhibition assays (Figure 8) (Monroe-Augustus et al. 2003; Belin et al. 2009; Rinaldi et al. 2012), whereas decreased ethylene responsiveness results in increased ABA responsiveness in seed germination inhibition assays (Figure 8) (Beaudoin et al. 2000; Ghassemian et al. 2000; Subbiah and Reddy 2010). Conversely, increasing ethylene production results in decreased ABA responsiveness in seed germination inhibition assays (Beaudoin et al. 2000; Ghassemian et al. 2000; Subbiah and Reddy 2010). The additive nature of combining altered ethylene production or response with altered auxin response or transport on ABA-responsive seed germination (Figure 10) suggests that auxin and ethylene independently affect ABA regulation of seed germination. Future experiments to uncover the molecular nature of these complex interactions should provide interesting insight into differential hormone crosstalk at various developmental time points and tissue types.
Supplementary Material
Acknowledgments
We are grateful to Mark Estelle for axr1-3 and aux1-7, the Arabidopsis Biological Resource Center at Ohio State University for ein2-1, eto1-1, and tir1-1, Sarah Burkhart for marker development, Michael Hilliard for technical assistance, and Tara Enders, Elizabeth Frick, David Korasick, and Marta Michniewicz Paciorek for critical comments on the manuscript. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine for help with genomic analysis. The Center is partially supported by the National Cancer Institute Cancer Center Support Grant (P30 CA91842) to the Siteman Cancer Center and by ITCTS/CTSA (UL1 TR000448) from the National Center for Research Resources, a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. This research was supported by the NIH (R00 GM089987).
Footnotes
Communicating editor: M. Estelle
Literature Cited
- Alonso J. M., Hirayama T., Roman G., Nourizadeh S., Ecker J. R., 1999. EIN2, a bifunctional transducer of ethylene and stress response in Arabidopsis. Science 284: 2148–2152. [DOI] [PubMed] [Google Scholar]
- Alonso J. M., Stepanova A. N., Solano R., Wisman E., Ferrari S., et al. , 2003. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L., Zhou Y., Song C. P., 2009. Arabidopsis proline-rich extensin-like receptor kinase 4 modulates the early event toward abscisic acid response in root tip growth. Plant Signal. Behav. 4: 1075–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N., Serizet C., Gosti F., Giraudat J., 2000. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin C., Megies C., Hauserova E., Lopez-Molina L., 2009. Abscisic acid represses growth of the Arabidopsis embryonic axis after germination by enhancing auxin signaling. Plant Cell 21: 2253–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C. J., Ecker J. R., 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144. [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Marchant A., Green H. G., May S. T., Ward S. P., et al. , 1996. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273: 948–950. [DOI] [PubMed] [Google Scholar]
- Bisson M. M., Bleckmann A., Allekotte S., Groth G., 2009. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem. J. 424: 1–6. [DOI] [PubMed] [Google Scholar]
- Bleeker A., Estelle M., Somerville C., Kende H., 1988. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241: 1086–1089. [DOI] [PubMed] [Google Scholar]
- Chae H. S., Kieber J. J., 2005. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 10: 291–296. [DOI] [PubMed] [Google Scholar]
- Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M., 1993. Arabidopsis ethylene-response gene ETR1: similarity of product to two-component regulators. Science 262: 539–544. [DOI] [PubMed] [Google Scholar]
- Chen R., Binder B. M., Garrett W. M., Tucker M. L., Chang C., et al. , 2011. Proteomic responses in Arabidopsis thaliana seedlings treated with ethylene. Mol. Biosyst. 7: 2637–2650. [DOI] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang Le L., Coon M., Nguyen T., et al. , 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R., 2010. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M., 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S., Swarup R., Mockaitis K., Dharmasiri N., Singh S. K., et al. , 2006. AXR4 is required for localization of the auxin influx facilitator AUX1. Science 312: 1218–1220. [DOI] [PubMed] [Google Scholar]
- Dos Santos Maraschin F., Memelink J., Offringa R., 2009. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. 59: 100–109. [DOI] [PubMed] [Google Scholar]
- Ephritikhine G., Fellner M., Vannini C., Lapous D., Barbier-Brygoo H., 1999. The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18: 303–314. [DOI] [PubMed] [Google Scholar]
- Estelle M. A., Somerville C., 1987. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206: 200–206. [Google Scholar]
- Feraru E., Friml J., 2008. PIN polar targeting. Plant Physiol. 147: 1553–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., 1994. Maternal effects govern variable dominance of two abscisic acid response mutations in Arabidopsis thaliana. Plant Physiol. 105: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K., 2007. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Syono K., 1996. Genetic analysis of the effects of polar auxin transport inhibitors on root growth in Arabidopsis thaliana. Plant Cell Physiol. 37: 1094–1101. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., McCourt P., 2003. Cross-talk in plant hormone signalling: What Arabidopsis mutants are telling us. Ann. Bot. (Lond.) 91: 605–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghassemian M., Nambara E., Cutler S., Kawaide H., Kamiya Y., et al. , 2000. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat J., Hauge B. M., Valon C., Smalle J., Parcy F., et al. , 1992. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4: 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P., Ecker J. R., 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughn G. W., Somerville C., 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434. [Google Scholar]
- Hobbie L., Estelle M., 1994. Genetic approaches to auxin action. Plant Cell Environ. 17: 525–540. [DOI] [PubMed] [Google Scholar]
- Hobbie L., Estelle M., 1995. The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7: 211–220. [DOI] [PubMed] [Google Scholar]
- Jiao Y., Sun L., Song Y., Wang L., Liu L., et al. , 2013. AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2+ signalling and auxin response of roots in Arabidopsis. J. Exp. Bot. 64: 4183–4192. [DOI] [PubMed] [Google Scholar]
- Ju C., Yoon G. M., Shemansky J. M., Lin D. Y., Ying Z. I., et al. , 2012. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA 109: 19486–19491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick M. D., Chang C., 2008. Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S., Leyser O., 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. [DOI] [PubMed] [Google Scholar]
- Kieber J., Rothenberg M., Roman G., Feldmann K. A., Ecker J., 1993. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72: 427–441. [DOI] [PubMed] [Google Scholar]
- Kim M. J., Shin R., Schachtman D. P., 2009. A nuclear factor regulates abscisic acid responses in Arabidopsis. Plant Physiol. 151: 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F. M., 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef M., Reuling G., Karssen C. M., 1984. The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61: 377–383. [Google Scholar]
- Korasick D. A., Enders T. A., Strader L. C., 2013. Auxin biosynthesis and storage forms. J. Exp. Bot. 64: 2541–2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick D. A., Westfall C. S., Lee S. G., Nanao M. H., Dumas R., et al. , 2014. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 111: 5427–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last R. L., Fink G. R., 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310. [DOI] [PubMed] [Google Scholar]
- Leung J., Merlot S., Giraudat J., 1997. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 9: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H. M. O., Lincoln C. A., Timpte C., Lammer D., Turner J., et al. , 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164. [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C., Britton J. H., Estelle M., 1990. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkies A., Muller K., Morris K., Tureckova V., Wenk M., et al. , 2009. Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21: 3803–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.-H., 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98: 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C., Gaxiola R. A., Grisafi P., Fink G. R., 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12: 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., et al. , 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324: 1064–1068. [DOI] [PubMed] [Google Scholar]
- Maher E. P., Martindale S. J. B., 1980. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem. Genet. 18: 1041–1053. [DOI] [PubMed] [Google Scholar]
- Marchant A., Kargul J., May S. T., Muller P., Delbarre A., et al. , 1999. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 18: 2066–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockaitis K., Estelle M., 2008. Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24: 55–80. [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M., Zolman B. K., Bartel B., 2003. IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15: 2979–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday G. K., Rahman A., Binder B. M., 2012. Auxin and ethylene: collaborators or competitors? Trends Plant Sci. 17: 181–195. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., et al. , 2009. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Neff M. M., Turk E., Kalishman M., 2002. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18: 613–615. [DOI] [PubMed] [Google Scholar]
- Normanly J., Grisafi P., Fink G. R., Bartel B., 1997. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9: 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Chen J. G., Jones A. M., Assmann S. M., 2006. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 141: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., et al. , 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Estelle M., 2006. Auxin receptors: a new role for F-box proteins. Curr. Opin. Cell Biol. 18: 152–156. [DOI] [PubMed] [Google Scholar]
- Perrot-Rechenmann C., 2010. Cellular responses to auxin: division vs. expansion. Cold Spring Harb. Perspect. Biol. 2: a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J., Friml J., 2009. Auxin transport routes in plant development. Development 136: 2675–2688. [DOI] [PubMed] [Google Scholar]
- Pickett F. B., Wilson A. K., Estelle M., 1990. The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94: 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H., Shen Z., Huang S. S., Schmitz R. J., Urich M. A., et al. , 2012. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi M. A., Liu J., Enders T. A., Bartel B., Strader L. C., 2012. A gain-of-function mutation in IAA16 confers reduced responses to auxin and abscisic acid and impedes plant growth and fertility. Plant Mol. Biol. 79: 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D., Rosas U., Krouk G., Ruffel S., Birnbaum K. D., et al. , 2013. RootScape: a landmark-based system for rapid screening of root architecture in Arabidopsis. Plant Physiol. 161: 1086–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock C. D., Sun X., 2005. Crosstalk between ABA and auxin signaling pathways in roots of Arabidopsis thaliana (L.) Heynh. Planta 222: 98–106. [DOI] [PubMed] [Google Scholar]
- Roman G., Lubarsky B., Kieber J. J., Rothenberg M., Ecker J. R., 1995. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics 139: 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano C. P., Robson P. R. H., Smith H., Estelle M., Klee M., 1995. Transgene-mediated auxin overproduction in Arabidopsis: Hypocotyl elongation phenotype and interactions with the hy6–1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 27: 1071–1083. [DOI] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M. B., Galle A., et al. , 2009. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150: 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Dewey E., Gray W. M., Hobbie L., Turner J., et al. , 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12: 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růžička K., Ljung K., Vanneste S., Podhorska R., Beeckman T., et al. , 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M., Robert S., Kleine-Vehn J., 2013. Auxin: simply complicated. J. Exp. Bot. 64: 2565–2577. [DOI] [PubMed] [Google Scholar]
- Sharp R. E., 2002. Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ. 25: 211–222. [DOI] [PubMed] [Google Scholar]
- Sharp R. E., Lenoble M. E., 2002. ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53: 33–37. [PubMed] [Google Scholar]
- Shkolnik-Inbar D., Bar-Zvi D., 2011. Expression of ABSCISIC ACID INSENSITIVE 4 (ABI4) in developing Arabidopsis seedlings. Plant Signal. Behav. 6: 694–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K., Goda H., Ishii T., Ogura T., Tachikawa T., et al. , 2010. Auxin biosynthesis inhibitors, identified by a genomics-based approach, provide insights into auxin biosynthesis. Plant Cell Physiol. 51: 524–536. [DOI] [PubMed] [Google Scholar]
- Stasinopoulos T. C., Hangarter R. P., 1990. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Alonso J. M., 2009. Ethylene signaling and response: where different regulatory modules meet. Curr. Opin. Plant Biol. 12: 548–555. [DOI] [PubMed] [Google Scholar]
- Stepanova A. N., Hoyt J. M., Hamilton A. A., Alonso J. M., 2005. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Yun J., Likhacheva A. V., Alonso J. M., 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A. N., Robertson-Hoyt J., Yun J., Benavente L. M., Xie D. Y., et al. , 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Strader L. C., Monroe-Augustus M., Bartel B., 2008a The IBR5 phosphatase promotes Arabidopsis auxin responses through a novel mechanism distinct from TIR1-mediated repressor degradation. BMC Plant Biol. 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Monroe-Augustus M., Rogers K. C., Lin G. L., Bartel B., 2008b Arabidopsis iba response5 (ibr5) suppressors separate responses to various hormones. Genetics 180: 2019–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Beisner E. R., Bartel B., 2009. Silver ions increase auxin efflux independently of effects on ethylene response. Plant Cell 21: 3585–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader L. C., Chen G. L., Bartel B., 2010. Ethylene directs auxin to control cell expansion. Plant J. 64: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah V., Reddy K. J., 2010. Interactions between ethylene, abscisic acid and cytokinin during germination and seedling establishment in Arabidopsis. J. Biosci. 35: 451–458. [DOI] [PubMed] [Google Scholar]
- Subramani S., 1992. Targeting of proteins into the peroxisomal matrix. J. Membr. Biol. 125: 99–106. [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G. T., et al. , 2007. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Timpte C., Lincoln C., Pickett F. B., Turner J., Estelle M., 1995. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 8: 561–569. [DOI] [PubMed] [Google Scholar]
- Tiryaki I., Staswick P. E., 2002. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 130: 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang D. L., Edmond C., Harrington J. L., Nuhse T. S., 2011. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 156: 596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Theologis A., 2004. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136: 2982–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanstraelen M., Benková E., 2012. Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28: 463–487. [DOI] [PubMed] [Google Scholar]
- Wen X., Zhang C., Ji Y., Zhao Q., He W., et al. , 2012. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22: 1613–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S., Davies W. J., 2009. Ozone suppresses soil drying- and abscisic acid (ABA)-induced stomatal closure via an ethylene-dependent mechanism. Plant Cell Environ. 32: 949–959. [DOI] [PubMed] [Google Scholar]
- Zhu S. Y., Yu X. C., Wang X. J., Zhao R., Li Y., et al. , 2007. Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.