Abstract
Oncogenic K-Ras mutation occurs frequently in several types of cancers including pancreatic and lung cancers. Tumors with K-Ras mutation are resistant to chemotherapeutic drugs as well as molecular targeting agents. Although numerous approaches are ongoing to find effective ways to treat these tumors, there are still no effective therapies for K-Ras mutant cancer patients. Here we report that K-Ras mutant cancers are more dependent on K-Ras in anchorage independent culture conditions than in monolayer culture conditions. In seeking to determine mechanisms that contribute to the K-Ras dependency in anchorage independent culture conditions, we discovered the involvement of Met in K-Ras-dependent, anchorage independent cell growth. The Met signaling pathway is enhanced and plays an indispensable role in anchorage independent growth even in cells in which Met is not amplified. Indeed, Met expression is elevated under anchorage-independent growth conditions and is regulated by K-Ras in a MAPK/ERK kinase (MEK)-dependent manner. Remarkably, in spite of a global down-regulation of mRNA translation during anchorage independent growth, we find that Met mRNA translation is specifically enhanced under these conditions. Importantly, ectopic expression of an active Met mutant rescues K-Ras ablation-derived growth suppression, indicating that K-Ras mediated Met expression drives “K-Ras addiction” in anchorage independent conditions. Our results indicate that enhanced Met expression and signaling is essential for anchorage independent growth of K-Ras mutant cancer cells and suggests that pharmacological inhibitors of Met could be effective for K-Ras mutant tumor patients.
Keywords: K-Ras mutations, Met expression, anchorage independent, K-Ras addiction, MAPK/ERK kinase
Introduction
K-Ras mutations are frequently found in pancreatic, lung and colorectal tumors [1-3]. These mutations are predominantly somatic missense mutations at position 12, 13, or 61 that impair the GTPase activity of K-Ras and result in constitutive activation of downstream signaling pathways including the mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K) pathways. Even after three decades of research, many questions remain as to how mutant K-Ras contributes to tumor initiation and progression. There are currently no effective treatments for cancers with mutant K-Ras. Patients with K-Ras mutant tumors are associated with resistance to chemotherapy, radiation and epidermal growth factor receptor (EGFR) targeted therapies [4-8], and tend to have poor overall survival [9-12]. Numerous research efforts to develop therapeutic agents to directly inhibit oncogenic Ras have thus far been unsuccessful [13], and the focus has therefore been on targeting druggable molecules in the pathways downstream of Ras. In this regard, combination therapies with mitogen-activated protein kinase kinase (MEK) and PI3K pathway inhibitors have been shown to be effective in mouse tumor models harboring mutant K-Ras [14-16], and some combinations of these inhibitors are already in clinical trials.
The dependency on K-Ras for growth in monolayer culture conditions varies widely between different K-Ras mutant cell lines as evaluated by the growth suppression rate after K-Ras depletion with RNA interference (RNAi). While K-Ras dependency has been shown to correlate with elevated expression of K-Ras and E-cadherin, the contribution of two major K-Ras downstream signaling molecules, Akt and MAPK, to K-Ras dependency is unclear [17, 18]. Compared with in vitro culture conditions, however, K-Ras mutant cells are known to be more broadly dependent on K-Ras in vivo [19-21]. Cells change the strength of many signaling pathways in response to different culture conditions, suggesting that the importance of specific signaling pathways for survival or proliferation would change in response to distinct environmental changes [22-24]. Recent data has shown that pancreatic cancer cells cultured in anchorage independent conditions express higher levels of stem cell markers and show higher tumorigenicity in vivo than cells in adherent conditions [25], suggesting that anchorage independent culture conditions are more reflective of in vivo tumor growth. Thus, the use of an in vitro anchorage independent culture model may identify more relevant in vivo signaling pathways downstream of K-Ras.
Hepatocyte growth factor (HGF) and its receptor Met regulate various signaling pathways that contribute to physiological processes such as embryonic development, organ regeneration and wound healing [26]. Deregulation of this signaling pathway frequently occurs in many different types of cancers via Met mutation or overexpression in the tumor, or HGF overexpression in the surrounding stroma, resulting in the promotion of tumor growth, invasion and metastasis [27, 28]. Moreover, increased HGF/Met signaling is known to cause resistance to many small molecule inhibitors, such as the BRAF inhibitor vemurafenib (PLX4032) and several receptor tyrosine kinase (RTK) inhibitors, including the EGFR inhibitors gefitinib and erlotinib, the Her2/EGFR inhibitor lapatinib, and the anaplastic lymphoma kinase inhibitor TAE684 [29]. Currently, several small molecule compounds and antibodies targeting HGF/Met are under clinical development, including the Met kinase inhibitor cabozantinib, which was recently approved by the FDA for the treatment of medullary thyroid cancer.
In this report, we compared K-Ras mutant tumor cells for their dependency on K-Ras during growth in monolayer culture conditions and in anchorage independent culture conditions and found that cells were more dependent on K-Ras in anchorage independent conditions. Analysis comparing the activation state and dependencies of various signaling pathways between these culture conditions revealed that Met plays a critical role in proliferation and drives, at least in part, the enhanced K-Ras dependency observed specifically in anchorage independent culture conditions. Further analysis revealed that K-Ras/MEK signaling regulates Met mRNA expression, while anchorage independent culture conditions promotes increased translation of Met mRNA. Thus, our results uncover novel modes of regulation underlying Met expression, which is critical for anchorage-independent growth of K-Ras mutant tumor cells. These findings suggest that pharmacological inhibitors of Met could have significant therapeutic potential for the treatment of K-Ras mutant cancers.
Materials and Methods
Reagents and cell culture
PHA-665752, XL-184, MK2206, GSK-1120212 and BKM120 were from Selleckchem. 4EGI-1 was from Calbiochem. Human and mouse HGF, human basic FGF and human EGF were from Peprotech and Sigma-Aldrich. Antibodies were obtained from: Met, pMetY1003, Y1234/Y1235, Y1349), pAKT(S473), pERK(Y202/Y204), ERK, pMEK, MEK, EGFR, Cyclin D1, eIF4E and eIF4G antibodies from Cell Signaling Technology; actin and K-RAS antibodies from Sigma; AKT antibody from Millipore. K-Raslox (Hras−/−Nras−/−KRaslox/loxRERTert/ert) mouse embryonic fibroblasts (MEFs) were kindly provided by the laboratory of Mariano Barbacid (CNIO, Madrid, Spain). All other cell lines were obtained from the American Type Culture Collection, Life Technologies and Japan Health Science Foundation. No cell line authentication was performed. Human pancreatic cancer cell lines (Capan-1, CFPAC1, Panc8.13, BxPC3), 293FT and mouse MEFs were maintained in DMEM (Life Technologies) supplemented with 10% FBS (Atlanta Biologicals) and 500 U/mL Penicillin and 500 μg/mL Streptomycin (Life Technologies). Human pancreatic cancer cell line Suit-2, human colorectal cancer cell line HCT116 and human lung cancer cell line A549 were maintained in RPMI (Life Technologies) supplemented with 10% FBS and 500 U/mL Penicillin and 500 μg/mL Streptomycin. To generate “Rasless” MEFs, 4-Hydroxytamoxifen (4-OHT) (Sigma) was used at a final concentration of 600 nM for 12 days.
siRNA transfection
Cells were transfected with the appropriate siRNAs (Thermo Scientific) at a final concentration of 1-10 nM using Lipofectamine RNAiMAX reagent (Life Technologies) according to the manufacturer’s instructions.
Vector construction and lentivirus production
Full-length wild-type human Met in pDONR223 (Addgene) and all Ras cDNAs were cloned into pLenti CMV/TO Puro or pLenti CMV Blast (Addgene) by LR reaction (Life Technologies). Met or Ras active mutants were generated by PCR amplification. Lentivirus was produced by UCSF ViraCore or by co-transfection of the corresponding lentiviral vector with a packaging system into 293FT cells using the standard protocol for Lipofectamine 2000 (Invitrogen). Cells were infected with lentiviruses in the presence of polybrene (Millipore) at a final concentration of 8 μg/mL.
Data analysis of Met expression and K-Ras mutation status
Met mRNA expression levels in 807 cell lines with or without K-Ras mutations were analyzed using the cell line encyclopedia. Comparison of normalized Met mRNA expression levels in K-RAS mutant versus wild-type samples in the pancreatic TCGA project. Data obtained from http://www.cbioportal.org
Growth assay
Cells were seeded at 1.25-2.5 × 103 cells/well (monolayer) or 2.5-5 × 103 cells/well (anchorage independent) in 96 well plates (monolayer, Becton Dickinson) or 96 well Ultra Low Attachment plates (anchorage independent, Corning). After incubation for indicated time periods, Cell Titer Glo (Promega) was added in each well and the mixture was transferred to 96 well white plates (Corning). Luminescence was analyzed by GLOMAX (Promega).
Western blot analysis
Cells were lysed in 1% Triton lysis buffer {20 mM Tris pH 7.5, 135 mM NaCl, 1% Triton X-100, 10% Glycerol, 1 mM EDTA} supplemented with protease inhibitor cocktail (Sigma) and phosphatase inhibitor cocktails (Sigma) and cleared by centrifugation (13,000 rpm, 10 minutes). Protein concentration was measured using a Bio-Rad modified Bradford Protein Assay (Bio-Rad). Lysates were boiled in 1× SDS sample buffer. Equivalent protein quantities were run on SDS-PAGE (NuPAGE; Invitrogen), transferred to nitrocellulose (iBlot; Invitrogen), and probed with the indicated primary antibodies. Subsequently, primary antibodies were detected with secondary antibodies conjugated to horseradish peroxidase (GE health care). Blots were visualized with an enhanced chemiluminescence system, according to the manufacturer’s instructions (GE health care).
Real-time PCR
RNA was extracted with RNeasy mini kit using DNase I (Qiagen), and cDNA was synthesized with High-capacity cDNA Reverse Transcription Kits (Applied Biosystems). SYBR Green PCR Master Mix with M×3000p qPCR system (Agilent Technologies) was used for analysis of cDNA by real-time PCR. The expression of each gene was normalized to the expression of β-actin by the standard curve method. The primer sets were as follows: Met AGCGTCAACAGAGGGACCT and GCAGTGAACCTCCGACTGTATG; β-actin ACTGGGACGACATGGAGAAAAT and TGAAGGTCTCAAACATGATCTGG; eIF4E GGTTGCTAACCCAGAACAC and CACTTCGTCTCTGCTGTTTG; eIF4G1 CCCGAAAAGAACCACGCAAG and TTCCCCTCGATCCTTATCAGC; eIF4G2 AGGGCAAAACGCTCAGAAATG and TCCTGAAGATTGCATCATGTCG; eIF4G3 CCTAGAGCTACCATCCCGAAC and GGGCCACTATGACGGTACTG.
Polysome fractionation and analysis of polysomal associated mRNA
Monolayer and anchorage-independent cell cultures were harvested 72hrs after plating and incubated with 100 μg/ml cyclohexamide (Sigma) in PBS for 10 minutes on ice. Cells were pelleted and lysed in 10mM Tris-HCl pH8, 140 mM NaCl, 1.5mM MgCl2, 0.25% NP-40, 0.1% Triton-X 100, 50mM DTT, 150μg/ml cyclohexamide, 640U/ml RNasin for 30 minutes. Lysates were cleared by centrifugation for 5 minutes at 9,300×g and loaded onto a 10-50% sucrose gradient. Loaded sucrose gradients were spun at 37,000 rpm for 2.5 hours at 4°C in a Beckman L8-70M ultracentrifuge. Sucrose gradients were then fractionated on an ISCO gradient fractionation system to assess polysome profiles and collect polysomal mRNA. RNA was isolated from gradient fractions using TRIzol Reagent (Invitrogen) and Pure Link RNA mini kits (Invitrogen) according to the manufacturer’s protocol. Purified RNA was reverse-transcribed to cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). cDNA samples were diluted 1:4 and 1ul of template was used in a SYBR green qPCR assay (Biorad) run on a MyiQ2 Real-Time PCR Detection System (Biorad) to determine Met mRNA expression relative to β-actin. The primer sets used were as follows: Met AGCGTCAACAGAGGGACCT and GCAGTGAACCTCCGACTGTATG; β-actin GCAAAGACCTGTACGCCAAC and AGTACTTGCGCTCAGGAGGA.
Statistical analysis
All analyses for statistically significant differences were performed with the 2-tailed unpaired Student’s t test. p-values less than 0.05 were considered significant. All error bars shown in the figures in this article are S.D.
Results
K-Ras mutant cell lines are more dependent on K-Ras in anchorage independent than in monolayer culture conditions
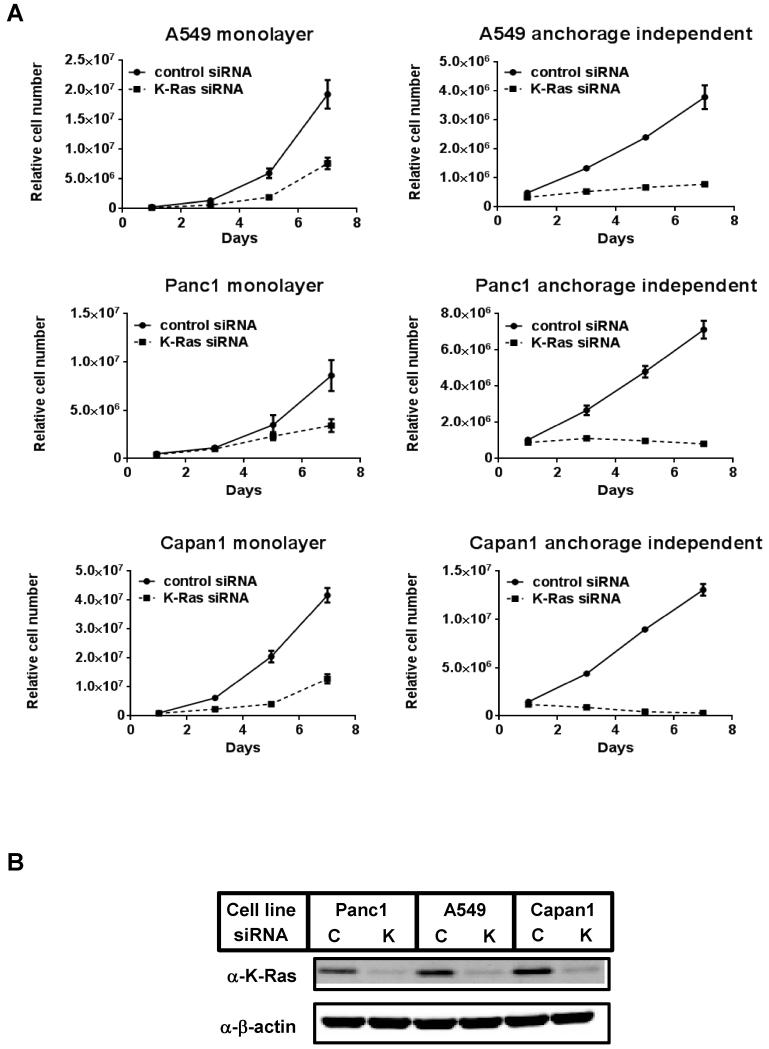
To understand the alterations of K-Ras dependencies in response to environmental changes in K-Ras mutant cancer cells, we first introduced a small interfering RNA (siRNA) targeting K-Ras to cells and cultured them in monolayer and anchorage independent culture conditions. Consistent with previous reports [17, 18], in monolayer conditions, the growth of A549 and Panc1 cells was relatively resistant to K-Ras depletion, whereas Capan1 cell growth was decreased markedly upon K-Ras knockdown, reflecting the different degrees of K-Ras requirements among these cell lines in this culture condition. Interestingly, K-Ras depletion resulted in striking growth suppression in all three cell lines in anchorage independent culture conditions (Fig. 1A and 1B). To exclude the possibility of off-target effects of the siRNA, three additional siRNAs targeting different sequences of K-Ras were also used for proliferation assays and gave similar results (Supplementary Fig. 1). These results suggest that in K-Ras mutant cancer cells, where K-Ras may be dispensable for cell proliferation in monolayer conditions, K-Ras plays an indispensable role in cell proliferation in anchorage independent conditions.
Figure 1. Effect of K-Ras knock-down on proliferation of K-Ras mutant cell lines in monolayer and anchorage independent culture conditions.
(A) Cell proliferation after siRNA-mediated K-Ras ablation in K-Ras mutated A549, Panc1 and Capan1 cells. One day after transfection of control or K-Ras siRNA, cells were seeded on normal cell culture plates (monolayer) or ultra-low attachment plates (anchorage independent). Relative viable cell numbers were estimated by quantitating ATP present in each well after the indicated time points. Data were shown as the mean ± S.D. of quadruplicate samples.
(B) The same cell as in (A) were collected after 96 hours of siRNA transfection. Cell lysates were immunoblotted with the indicated antibodies, Repeated experiments gave similar results. C: control siRNA, K: K-Ras siRNA.
Met signaling is up-regulated in anchorage independent culture conditions
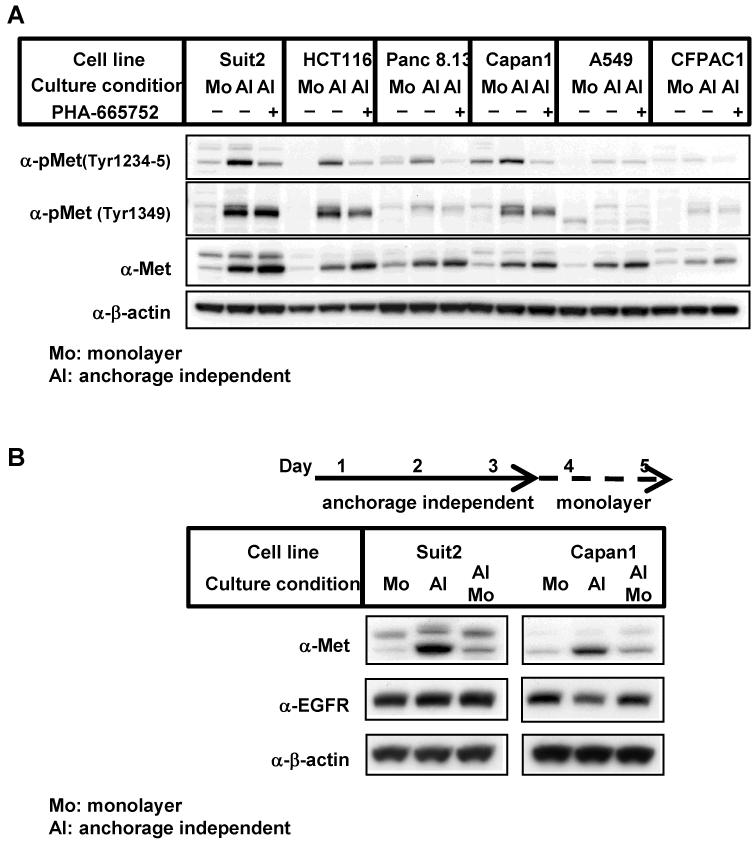
To identify differences in the signaling profiles between monolayer and anchorage independent culture conditions, the expression and phosphorylation of several different signaling proteins were analyzed. Strikingly, compared to monolayer conditions, Met protein expression and phosphorylation was found to be up-regulated in anchorage independent conditions in several pancreatic, colorectal and lung cancer cell lines harboring mutant K-Ras (Fig. 2A). In addition, Met phosphorylation could be inhibited by addition of the Met specific inhibitor, PHA-665752, in line with the activation of Met signaling in anchorage independent culture conditions. Importantly, all of these cell lines were found to be more dependent on K-Ras for growth in anchorage independent culture conditions than monolayer conditions (Supplementary Fig. 2).
Figure 2. Increased Met protein and phosphorylation in K-Ras mutant tumor cells cultured in anchorage independent culture conditions.
(A) Cells were cultured in monolayer conditions (Mo) or in anchorage independent conditions (AI) for 72 h. In some experiments, cells were treated with 1 μM PHA-665752 for the last 24 h of the incubation. Cell lysates were immunoblotted with the indicated antibodies. Repeated experiments gave similar results.
(B) Cells were cultured as in (A). In some experiments, cells cultured for 72 h in anchorage independent culture conditions (AI) were harvested and then seeded on normal cell culture plates (monolayer: Mo). After an additional 48 h of incubation, cells were harvested and immunoblotted with the indicated antibodies.
To exclude the possibility that the increase in Met in anchorage independent conditions is caused by the selection of cells with higher Met expression, we cultured cells in anchorage independent conditions for 3 days, and then switched them to monolayer conditions and incubated for an additional 2 days. Met protein levels significantly increased after 3 days of anchorage independent culture, and decreased after an additional 2 days in monolayer cultures (Figure 2B). Notably, EGFR expression was not altered by the change in culture conditions for either cell line. These results suggest that the culture condition itself alters Met protein expression levels.
Increased contribution of HGF/Met signaling to cell proliferation in anchorage independent culture conditions
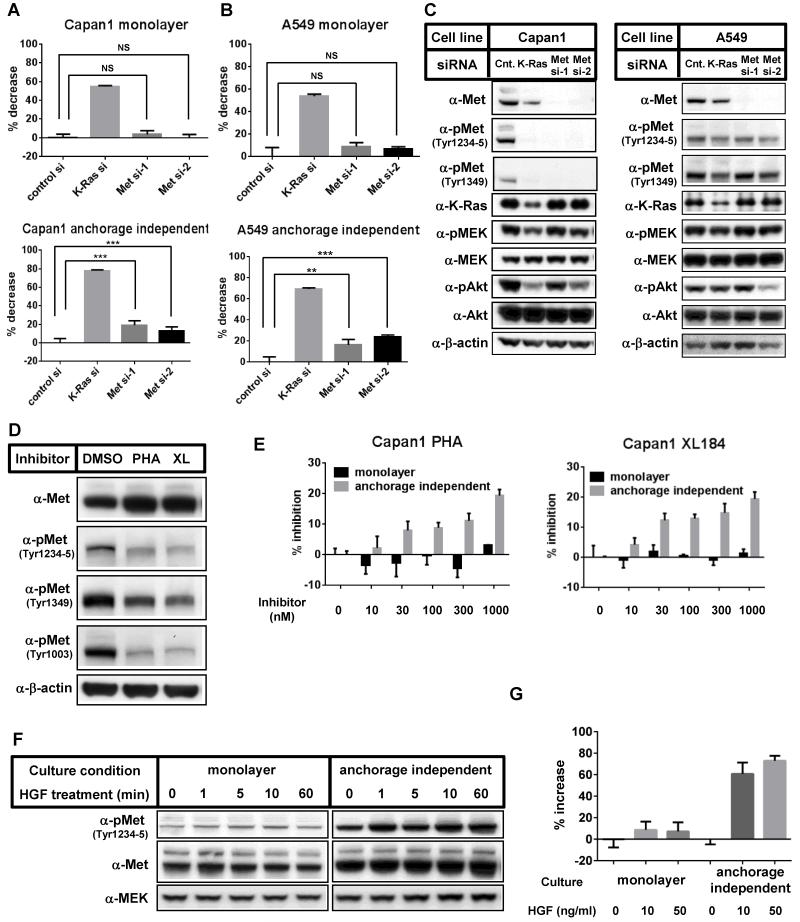
Given the increase in protein and phosphorylation levels of Met in the anchorage independent conditions, we hypothesized that Met signaling may play a more significant role for cell proliferation in anchorage independent culture conditions than in monolayer culture conditions (Fig. 1A). To address this, we introduced two siRNAs targeting different Met sequences (Met si-1 and Met si-2) and compared their effects on cell proliferation in monolayer and anchorage independent conditions (Fig. 3A and 3B). Met knockdown efficiently suppressed Met expression in Capan1 and A549 cells (Fig. 3C). Growth of both Capan1 and A549 cells was more sensitive to Met ablation under anchorage independent culture conditions than monolayer culture conditions, though the growth inhibitory effect was weaker than that seen with K-Ras ablation (Fig. 3A and 3B). In monolayer conditions, A549 cells contain lower levels of Met protein than Met amplified cells, and are resistant to Met inhibitors [30]. However, we found that anchorage independent culture of A549 cells induces Met expression and renders them sensitive to Met depletion. Similar results were observed with CFPAC1 and ASPC1 cells (data not shown).
Figure 3. Involvement of Met-driven signaling pathways in the growth of cells in anchorage independent culture conditions.
(A-B) Capan1 (A) or A549 (B) cells were transfected with control siRNA, K-Ras siRNA, or Met siRNAs. 24 h after transfection, cells were harvested and seeded onto normal cell culture plates (monolayer) or ultra-low attachment plates (anchorage independent), and relative cell growth was analyzed 3 days (Capan1) or 4 days (A549) after cell seeding. Data were normalized with each control siRNA transfected sample and shown as the mean ± S.D. of quadruplicate samples. NS; not significant, *; p < 0.05, **; p < 0.01, ***; p < 0.0001 (C) The effects of each siRNA were confirmed by Western blot analysis 3 days after siRNA transfection. (D) Capan1 cells were incubated with medium containing vehicle (DMSO), 500 nM PHA-665752 (PHA) or 500 nM XL184 (XL) in monolayer culture conditions for 2 days. Cell lysates were immunoblotted with the indicated antibodies. Repeated experiments gave similar results. (E) Capan1 cells were seeded on normal cell culture plates (monolayer) or ultra-low attachment plates (anchorage independent) and incubated with the indicated concentrations of PHA-665752 (left panel) or XL184 (right panel) for 4 days. Relative viable cell numbers were determined by quantitating ATP present in each well. Data were shown as the mean ± S.D. of triplicate samples. Repeated experiments gave similar results. (F) Capan1 cells were cultured either in monolayer or anchorage independent culture conditions for 3 days, then the cells were treated with 10 ng/mL of HGF. At the indicated time points, cells were collected and immunoblotted with the indicated antibodies.
(G) Capan1 cells were plated on normal cell culture plates (monolayer) or ultra-low attachment plates (anchorage independent) and cultured for 72 h in the medium containing 0, 10 or 50 ng/mL of HGF. Relative viable cell numbers were determined by quantitating ATP present in each well. Data were shown as the mean ± S.D. of quadruplicate samples.
We next sought to determine whether pharmacological inhibition of Met would, similar to Met knockdown, affect cell proliferation more significantly under anchorage independent conditions than monolayer conditions. Capan1 cells were incubated with different concentrations of the Met specific inhibitor PHA-665752 or XL-184 (cabozantinib), a dual VEGFR2 and Met inhibitor that has recently been approved by the FDA. Both compounds inhibited Met phosphorylation (Fig. 3D) and, consistent with a previous report [30], increased Met protein expression levels in cells upon prolonged exposure. Strikingly, both compounds inhibited cell growth in a dose dependent manner only when cells were cultured in anchorage independent conditions (Fig. 3E).
In tumors, Met signaling can be enhanced by increased Met expression, activating Met mutations or HGF overexpression. The main source of HGF is not thought to be the cancer cells themselves but instead the surrounding stroma [27]. In line with this, HGF mRNA or HGF protein expression was below detection levels in Capan1 and Suit2 cells (data not shown). Therefore, in order to further explore the significance of Met signaling pathways, we next stimulated cells with recombinant human HGF in monolayer and anchorage independent culture conditions. Stimulation of the cells with human HGF resulted in the phosphorylation of Met in both conditions (Fig. 3F). However, the degree of HGF-induced Met phosphorylation was significantly higher in anchorage independent culture conditions than in monolayer culture conditions (Fig. 3F). Consistent with these results, HGF promoted proliferation of cells cultured in anchorage independent culture conditions significantly more than in monolayer conditions (Fig. 3G and Supplementary Fig. 3A) and Met kinase inhibitor treatment suppressed this effect (Supplementary Fig. 3A), indicating that the HGF/Met signaling pathway supports anchorage independent growth of K-Ras mutant cancer cells. In contrast, EGF stimulation slightly enhanced cell growth to similar degrees in monolayer and anchorage independent culture conditions (Supplementary Fig. 3B and 3C), consistent with the observation that EGFR levels were identical between the two conditions (Fig. 2B). Together, these observations strongly suggest that HGF/Met signaling contributes to K-Ras-dependent cell proliferation specifically in anchorage independent culture conditions.
K-Ras/MEK signaling regulates Met expression
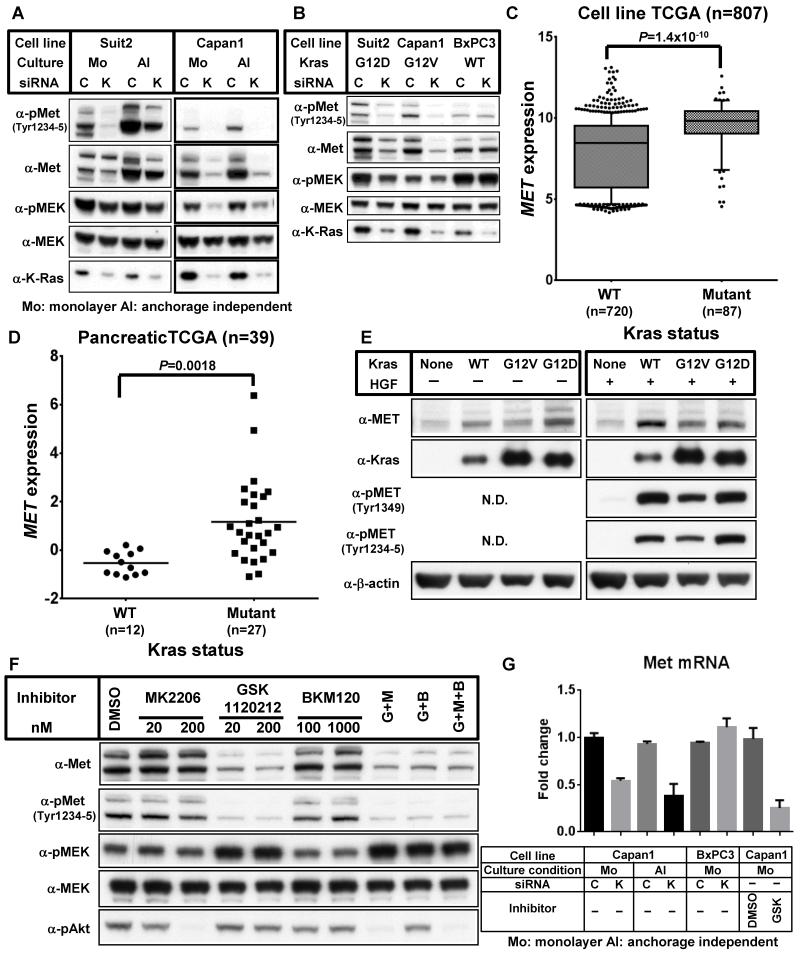
Our results demonstrate that both K-Ras and Met play an essential role in anchorage independent cell growth of K-Ras mutant cancer cells. Although K-Ras is a known effector of Met, other mechanisms linking K-Ras and Met signaling may play a role in anchorage independent cell growth. To examine the relationship between K-Ras and Met we knocked down either K-Ras or Met and examined their effects on the expression of each other. As shown in Fig. 4A, K-Ras ablation resulted in the down-regulation of Met protein and phosphorylation levels both in monolayer and anchorage independent culture conditions. In contrast, knockdown of Met did not affect K-Ras protein expression (Fig. 3C). These results suggest that K-Ras regulates Met expression.
Figure 4. K-Ras regulates Met expression via MEK.
(A) Suit2 and Capan1 cells were transfected with either control siRNA (C) or K-Ras siRNA (K). 24 h after transfection, cells were harvested and seeded on normal cell culture plates (Monolayer: Mo) or ultra-low attachment plates (anchorage independent: AI). 72 h after seeding, cells were harvested and immunoblotted with the indicated antibodies. (B) 24 h after transfection of control siRNA (C) or K-Ras siRNA (K), Suit2, Capan1 and BxPC3 cells were cultured in monolayer culture conditions for additional 72 h. Repeated experiments gave similar results. (C) Each cell line was grouped as K-Ras wild-type (WT) or K-Ras mutant (Mutant) and their relative Met mRNA levels plotted. Data is shown as 10-90% percentile. 807 cell lines were analyzed. (D) Comparison of normalized Met mRNA expression levels in K-RAS mutant versus wild-type samples in the pancreatic TCGA project. (E) Rasless MEFs that had been transduced with nothing (None) or wild-type K-Ras (WT), G12V K-Ras (G12V) or G12D K-Ras (G12D) were seeded onto normal culture plates. 24 h after plating, cells were stimulated with (+) or without (−) 20 ng/mL mouse HGF for 10 min. Cell lysates were immunoblotted with the indicated antibodies. (F) Capan1 cells were seeded on normal culture plates. After 24 of incubation, cells were treated with the indicated concentrations of the inhibitors (G: GSK 1120212 (200 nM), M: MK2206 (200 nM), B: BKM120 (1000 nM)) and 48 h later cells were harvested and immunoblotted with the indicated antibodies. (G) Capan1 and BxPC3 cells were transfected with either control siRNA (C) or K-Ras siRNA (K). 24 h after transfection, cells were harvested and seeded on normal cell culture plates (monolayer: Mo) or ultra-low attachment plates (anchorage independent: AI) and incubated for an additional 72 h. In some experiments, Capan1 cells were harvested and seeded onto normal cell culture plates. 24 h after seeding, cells were treated with medium containing vehicle (DMSO) or 100 nM GSK 1120212 (GSK) for 72 h. Relative Met mRNA expression levels were evaluated by qPCR. Data were shown as the mean ± S.D. of triplicate samples.
To assess the effect of K-Ras activity on Met expression, we focused our analysis on pancreatic cancer cell lines harboring mutant K-Ras (Suit2: G12D and Capan1: G12V) or wild-type K-Ras (BxPC3). K-Ras knockdown reduced the phosphorylation levels of MEK in K-Ras mutant cell lines, but not in cells with wild-type K-Ras (Fig. 4B). Similarly, K-Ras ablation decreased the expression levels of Met and phospho-Met only in K-Ras mutant cells. Importantly, an expanded analysis of 807 cancer cell lines confirmed the correlation between Met expression levels and K-Ras activation status, showing that, in general, cell lines with mutant K-Ras express higher levels of Met mRNA than cells with wild-type K-Ras (Fig. 4C). Moreover, analysis of human patient samples demonstrated that pancreatic cancers with mutant K-Ras also express higher levels of Met mRNA than those with wild-type K-Ras (Fig. 4D).
To further investigate the role of K-Ras on Met expression, we used “Rasless” MEFs completely devoid of all three Ras isoforms (H-, N-, and K-Ras) and re-introduced exogenous wild-type or mutant (G12D or G12V) forms of K-Ras. Ectopic expression of either wild-type or mutant K-Ras in the Rasless cells elevated Met protein levels (Fig. 4E). Interestingly, MEFs ectopically expressing K-Ras, but not Rasless MEFs, enhanced Met phosphorylation levels drastically after HGF stimulation (Fig. 4E). Together, these findings strongly suggest that K-Ras-driven signaling is essential for Met expression and function.
To identify intermediaries in the K-Ras/Met axis, cells were incubated with small molecule kinase inhibitors targeting Akt (MK2206), MEK1/2 (GSK 1120212) and/or PI3K (BKM120). Only inhibition of MEK1/2 significantly decreased Met protein levels, indicating that K-Ras-driven MEK signaling is responsible for Met expression (Fig. 4F). In addition, qPCR analysis revealed that K-Ras knockdown or MEK inhibitor treatment decreased Met mRNA expression (Fig. 4G). Moreover, cells were more sensitive to MEK1/2 inhibition in anchorage independent culture conditions than in monolayer conditions, consistent with an enhanced requirement for Met during anchorage-independent growth (Supplementary Fig. 4). Collectively, these results demonstrate that the K-Ras/MEK signaling pathway positively regulates Met mRNA levels.
Ectopic expression of active Met rescues K-Ras ablation-induced growth suppression
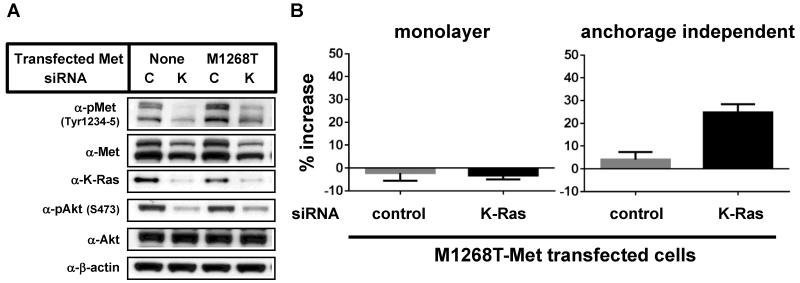
Our observations that cells with mutant K-Ras required both K-Ras and Met for proliferation in anchorage-independent conditions and that K-Ras/MEK signaling positively regulated Met expression prompted us to examine the biological significance of crosstalk between these two pathways. To directly evaluate the role of Met in K-Ras mutant cancer cells, a lentiviral system was used to express a constitutively active M1268T Met mutant. The M1268T mutation in Met was originally identified in papillary renal carcinomas and is reported to cause enhanced Met phosphorylation and activation [28, 31]. Met(M1268T) expression increased basal phosphorylation levels of Met and partially restored the phosphorylation levels of Met and Akt after K-Ras knock-down in Capan1 cells (Fig. 5A). Importantly, expression of Met(M1268T) promoted the growth of K-Ras depleted cells only in anchorage independent culture conditions (Fig. 5B). Similar results were obtained using A549 cells (Supplementary Fig. 5). This data suggests that K-Ras mediated Met expression contributes to K-Ras mediated cell proliferation in anchorage independent culture conditions.
Figure 5. Transfection of active Met overcomes K-Ras knock down-mediated growth suppression.
(A) Untransfected Capan1 cells or Capan1 cells transfected with Met(M1268T) were transfected with either control siRNA (C) or K-Ras siRNA (K). 24 h after transfection, cells were harvested and plated on normal cell culture plates. After incubation for an additional 24 h, cells were harvested and immunoblotted with the indicated antibodies. For immunoblot analysis (A), cells were cultured in normal plates and collected 1 day after cell seeding. (B) Untransfected Capan1 cells or Capan1 cells that had been transfected with Met(M1268T) were treated as described in (A). After 72 h of incubation, viable cell were counted. Data were normalized with each parental sample data and shown as the mean ± S.D. of quadruplicate samples.
Met translation is specifically enhanced in anchorage independent culture conditions
Although K-Ras and Met contributed to cell proliferation more in anchorage independent culture conditions than in monolayer culture conditions, knock-down of K-Ras decreased Met protein levels both in anchorage independent and monolayer culture conditions (Fig. 4A), indicating that K-Ras/MEK-mediated regulation of Met mRNA expression does not change between these two conditions. To understand the mechanism by which Met protein levels increase specifically in anchorage independent conditions, we first tested whether the half-life of Met protein is altered between the two culture conditions. Capan1 and Suit2 cells were cultured in anchorage independent or monolayer conditions for 3 days, and then de novo protein synthesis was inhibited with cycloheximide (CHX). Immunoblot analysis at different time points after CHX treatment showed similar profiles for Met half-life in anchorage independent and monolayer culture conditions, indicating that anchorage independent culture does not alter the turnover of Met protein (Supplementary Fig.6A). Furthermore, qPCR analysis showed no difference in Met mRNA levels between cells cultured in anchorage independent or monolayer conditions (Fig. 4G).
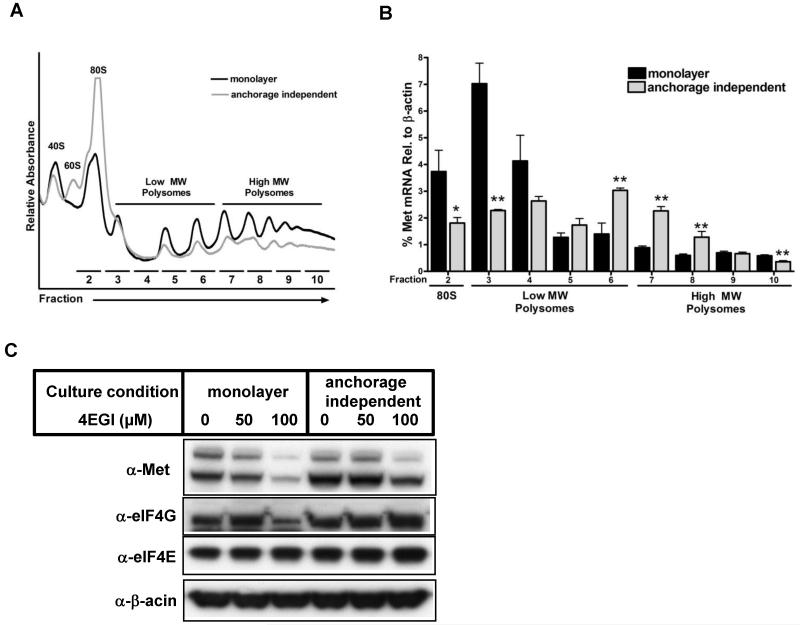
Previous reports indicated that Met expression could be positively regulated by cap-dependent translation through increased expression of the cap-binding protein eIF4E [32]. Therefore, in order to test the possibility that cap-dependent Met translation is enhanced during anchorage-independent culture, we assessed the effect of anchorage-independent conditions on both global mRNA translation as well as Met specific mRNA translation. Interestingly, analysis of polysome profiles uncovered a profound decrease of global translation under anchorage-independent growth conditions, where there was a relative loss of actively translating polysomes and accumulation of monosomes (Fig. 6A). Remarkably, however, we found that Met mRNA translation was instead enhanced under anchorage-independent growth conditions, as revealed by a shift in Met mRNA association towards more actively translating polysomal fractions (Fig. 6B and Supplementary Fig. 6B). As enhanced Met translation has previously been attributed to increased eIF4E expression, we next evaluated mRNA expression levels of the eIF4F components, eIF4E, eIF4G1, eIF4G3 and eIF4G2. Despite enhanced Met translation in anchorage-independent growth conditions, we found that eIF4E mRNA levels were unaffected by culture conditions, although there was an increase in eIF4G3 expression (Supplementary Fig. 6C). Consistent with previous work [32], we found that inhibition of eIF4E-dependent translation with the small molecule 4EGI-1 decreased Met protein levels. However, the decrease was more significant in cells cultured in monolayer conditions, indicating that Met expression is less dependent on eIF4E when cells are cultured in anchorage independent conditions (Fig. 6C). These results suggest that Met translation is specifically increased in cells cultured in anchorage independent conditions, perhaps in part through cap-independent mechanisms, to sustain higher Met protein levels necessary for anchorage-independent growth.
Figure 6. Met translation is enhanced in anchorage independent culture conditions.
(A) Representative polysome profiles showing relative levels of 40S ribosomal subunit (40S), 60S ribosomal subunit (60S), monosomes (80S), low molecular weight polysomes, and high molecular weight polysomes in monolayer and anchorage independent culture conditions for Capan1 cells. (B) qPCR analysis of Met mRNA levels in polysomal fractions for Capan1 cells under monolayer and anchorage independent culture conditions. (C) Capan1 cells were seeded on normal cell culture plates (monolayer) or ultra-low attachment plates (anchorage independent) in medium containing the indicated concentration of 4EGI-1 for 2 days. Cell lysates were immunoblotted with the indicated antibodies.
Together, our data support a model in which mutant K-Ras contributes to Met expression, which is increased in anchorage independent culture conditions via increased translation, causing increased Met-driven proliferation and a stronger dependence on K-Ras.
Discussion
The ability of cells to grow in anchorage independent and sphere forming culture conditions is known to reflect some characteristics of tumor cells, including self-renewal ability [25, 33]. Thus, in many cases, anchorage independent cell culture may be a better model for cancer studies. Here we have demonstrated that cancer cells with active K-Ras mutations are more dependent on K-Ras driven signaling in anchorage independent culture than in monolayer culture (Fig. 1A). In investigating the mechanisms for this distinction in K-Ras dependency, we discovered that Met plays a pivotal role in anchorage independent conditions, potentially acting upstream of the Akt pathway (Fig. 5A). Met translation and activation were increased when cells were cultured in anchorage independent conditions (Fig. 2), leading to a stronger dependence of cells for Met, as seen by attenuated cell proliferation upon Met knock-down or Met inhibitor treatment (Fig. 3). Furthermore, we found Met expression levels to be dependent on K-Ras/MEK signaling (Fig. 4).
Although we could not detect HGF expression in our cancer cells lines (data not shown), we observed an increase in phospho-Met levels in anchorage independent culture conditions (Fig. 2). RTKs such as Met are known to transduce signals in the absence of their ligands by phosphorylating each other when overexpressed, which is likely the cause for the increase in phospho-Met levels in this study. The impact of Met knock-down or Met inhibition on cell proliferation was smaller than the impact of K-Ras knock-down (Fig. 3A and 3B), indicating that K-Ras driven Met expression contributes only partially to K-Ras dependency under in vitro culture conditions in the absence of HGF. In vivo, however, HGF is secreted from the surrounding tumor stroma and from organs, including the liver. The finding that HGF enhanced cell growth only in anchorage independent conditions (Fig. 3) suggests that cancer cells in vivo are more likely to be dependent on Met signaling downstream of HGF (secreted from non-tumor cells), which would go completely undetected in standard monolayer culture conditions. Consistent with this hypothesis, a recent report demonstrated that pancreatic cancer cells with high Met expression exhibit cancer stem cell-like properties [34]. Moreover, the pancreatic tumor cells with high Met expression showed higher tumorigenicity compared to cells with low Met expression or Met negative cells, and XL-184 treatment significantly inhibited in vivo tumor growth. [34]. Although the xenograft model from this study could not evaluate the effect of HGF produced outside of tumor cells, it has been reported that mouse HGF cannot activate human Met [35], thus our observation that Met signaling is enhanced in anchorage independent conditions may explain tumor growth suppression by XL-184 in vivo.
Recently, it has been reported that EGFR signaling contributes to Met expression in lung cancer cell lines [36]. This work demonstrates that EGFR-mediated signaling pathways can regulate Met at multiple levels, including mRNA expression and protein stability. Considering that K-Ras is a downstream effector of EGFR, it is probable that Met is regulated at both the level of mRNA and protein stability in K-Ras driven cancer cells. Although we did not examine the effect of K-Ras ablation or MEK inhibitor treatment on Met protein stability, it is likely that EGFR activating mutation, EGF stimulation, and Ras activation all promote Met protein expression through similar mechanisms. In line with this this, the transfection of N-Ras into Rasless MEFs also induced high Met expression (data not shown).
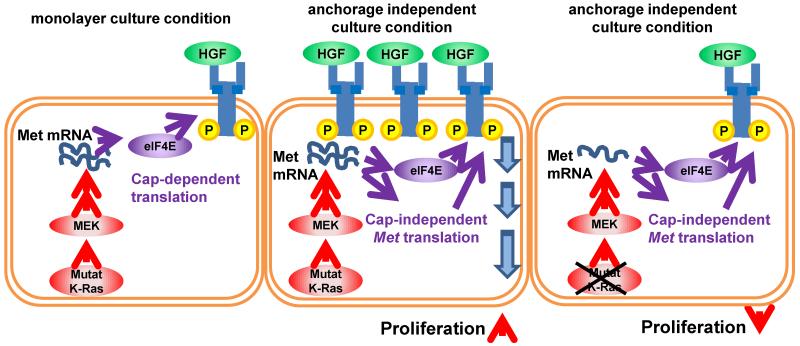
Although both Met mRNA expression and protein stability are regulated similarly in monolayer and anchorage independent culture conditions, cells cultured in anchorage independent conditions contain higher Met protein (Fig. 2, Fig. 3 and Supplementary Fig. 6). Our data indicate that this can be at least partially attributed to differences in Met mRNA translation, which we found to be increased in anchorage-independent conditions. Interestingly, Met translation was enhanced under anchorage-independent culture conditions despite a marked decreases in global mRNA translation. These results, along with the finding that Met expression is less sensitive to eIF4E inhibition under anchorage-independent conditions, suggests that Met mRNA may be translated, at least in part, through cap-independent mechanisms. It is possible that under anchorage-independent growth conditions there is a mechanism for selective ribosome recruitment to Met mRNA, perhaps through an internal ribosome entry site (IRES). Although the mechanism remains unclear, it is tempting to speculate that increased expression of eIF4G3 could be involved, as eIF4G family members have been shown to promote IRES-dependent translation of certain mRNA transcripts [37-41]. Importantly, K-Ras regulates Met expression through MEK signaling in both monolayer and anchorage independent culture conditions, however, in anchorage independent conditions, Met expression is elevated by enhanced translation, leading to increased Met-driven cell proliferation. Thus, in anchorage independent culture conditions, the depletion of K-Ras leads to down regulation of Met mRNA and subsequent growth suppression (Fig. 7).
Figure 7. K-Ras-mediated regulation of Met expression and cell proliferation.
K-Ras/MEK signaling pathway positively regulates Met mRNA levels. Met mRNA is translated by the eIF4F complex (eIF4E and eIF4G) via cap-dependent translation (left). In anchorage-independent culture conditions, enhanced Mettranslation increases Met expression. Increased Met expression results in increased cell proliferation (middle). In anchorage independent culture conditions under K-Ras ablation, Met protein expression is decreased due to Met mRNA down-regulation even with enhanced Met translation. Down-regulation of Met signaling results in the suppression of cell proliferation.
The discovery of drugs that are effective against K-Ras mutant cancers is critical. Current initiatives to develop therapeutic agents to directly inhibit Ras are ongoing, and initiatives to inhibit pathways downstream of K-Ras, such as PI3K and MEK inhibitor combinations, are promising. Clinical trials for Met inhibitors are ongoing in a wide range of cancers, although previous clinical trials for Met inhibitors only targeted patients with noted Met amplifications. Our results suggest that Met inhibitors may be especially effective against K-Ras mutant cancers.
Supplementary Material
Acknowledgments
Grant Support:
This work was supported by Daiichi-Sankyo and partially supported by NIH R01 CA184624 (to Davide Ruggero).
Footnotes
Disclosure of Potential Conflicts of Interest:
S. Fujita-Sato is an employee of Daiichi-Sankyo
Authors’ Contributions:
S. Fujita-Sato, M. Truitt, D. Ruggero and F. McCormick designed research; S. Fujita-Sato, J. Galeas, M. Truitt, C. Pitt, A. Urisman, S. Bandyopadhyay, D. Ruggero and F. McCormick carried out research and analyzed data; S. Fujita-Sato, M. Truitt, D. Ruggero and F. McCormick edited the manuscript.
References
- 1.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11:761–74. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–31. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 4.Winder T, Lenz HJ. Molecular predictive and prognostic markers in colon cancer. Cancer Treat Rev. 2010;36:550–6. doi: 10.1016/j.ctrv.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Garassino MC, Borgonovo K, Rossi A, Mancuso A, Martelli O, Tinazzi A, et al. Biological and clinical features in predicting efficacy of epidermal growth factor receptor tyrosine kinase inhibitors: a systematic review and meta-analysis. Anticancer Res. 2009;29:2691–701. [PubMed] [Google Scholar]
- 6.Bernhard EJ, Stanbridge EJ, Gupta S, Gupta AK, Soto D, Bakanauskas VJ, et al. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000;60:6597–600. [PubMed] [Google Scholar]
- 7.Sklar MD. Increased resistance to cis-diamminedichloroplatinum(II) in NIH 3T3 cells transformed by ras oncogenes. Cancer Res. 1988;48:793–7. [PubMed] [Google Scholar]
- 8.Winton T, Livingston R, Johnson D, Rigas J, Johnston M, Butts C, et al. Vinorelbine plus cisplatin vs. observation in resected non-small-cell lung cancer. N Engl J Med. 2005;352:2589–97. doi: 10.1056/NEJMoa043623. [DOI] [PubMed] [Google Scholar]
- 9.Meng D, Yuan M, Li X, Chen L, Yang J, Zhao X, et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer. 2013;81:1–10. doi: 10.1016/j.lungcan.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 10.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–8. [PubMed] [Google Scholar]
- 11.Ihle NT, Byers LA, Kim ES, Saintigny P, Lee JJ, Blumenschein GR, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst. 2012;104:228–39. doi: 10.1093/jnci/djr523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mascaux C, Iannino N, Martin B, Paesmans M, Berghmans T, Dusart M, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92:131–9. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bollag G, Zhang C. Drug discovery: Pocket of opportunity. Nature. 2013;503:475–6. doi: 10.1038/nature12835. [DOI] [PubMed] [Google Scholar]
- 14.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14:1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28:1075–83. doi: 10.1200/JCO.2009.25.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebi H, Corcoran RB, Singh A, Chen Z, Song Y, Lifshits E, et al. Receptor tyrosine kinases exert dominant control over PI3K signaling in human KRAS mutant colorectal cancers. J Clin Invest. 2011;121:4311–21. doi: 10.1172/JCI57909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardessy N, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi XH, Liang ZY, Ren XY, Liu TH. Combined silencing of K-ras and Akt2 oncogenes achieves synergistic effects in inhibiting pancreatic cancer cell growth in vitro and in vivo. Cancer Gene Ther. 2009;16:227–36. doi: 10.1038/cgt.2008.82. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann I, Weiss A, Elain G, Schwaederle M, Sterker D, Romanet V, et al. K-RAS mutant pancreatic tumors show higher sensitivity to MEK than to PI3K inhibition in vivo. PLoS One. 2012;7:e44146. doi: 10.1371/journal.pone.0044146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Jiang G, Yang F, Wang J. Knockdown of mutant K-ras expression by adenovirus-mediated siRNA inhibits the in vitro and in vivo growth of lung cancer cells. Cancer Biol Ther. 2006;5:1481–6. doi: 10.4161/cbt.5.11.3297. [DOI] [PubMed] [Google Scholar]
- 22.Pickl M, Ries CH. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2009;28:461–8. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- 23.Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res Treat. 2010;122:35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaviraghi M, Tunici P, Valensin S, Rossi M, Giordano C, Magnoni L, et al. Pancreatic cancer spheres are more than just aggregates of stem marker-positive cells. Biosci Rep. 2011;31:45–55. doi: 10.1042/BSR20100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9:314–26. doi: 10.1038/nrclinonc.2012.71. [DOI] [PubMed] [Google Scholar]
- 27.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt L, Duh FM, Chen F, Kishida T, Glenn G, Choyke P, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 29.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–9. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama R, Aoyama A, Yamori T, Qi J, Oh-hara T, Song Y, et al. Cytotoxic activity of tivantinib (ARQ 197) is not due solely to c-MET inhibition. Cancer Res. 2013;73:3087–96. doi: 10.1158/0008-5472.CAN-12-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffers M, Schmidt L, Nakaigawa N, Webb CP, Weirich G, Kishida T, et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci USA. 1997;94:11445–50. doi: 10.1073/pnas.94.21.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Fan S, Koo J, Yue P, Chen ZG, Owonikoko TK, et al. Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer. Cancer Biol Ther. 2012;13:272–80. doi: 10.4161/cbt.18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–8. [PubMed] [Google Scholar]
- 34.Li C, Wu JJ, Hynes M, Dosch J, Sarkar B, Welling TH, et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology. 2011;141:2218–2227. doi: 10.1053/j.gastro.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Rong S, Bodescot M, Blair D, Dunn J, Nakamura T, Mizuno K, et al. Tumorigenicity of the met proto-oncogene and the gene for hepatocyte growth factor. Mol Cell Biol. 1992;12:5152–8. doi: 10.1128/mcb.12.11.5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breindel JL, Haskins JW, Cowell EP, Zhao M, Nguyen DX, Stern DF. EGF receptor activates MET through MAPK to enhance non-small cell lung carcinoma invasion and brain metastasis. Cancer Res. 2013;73:5053–65. doi: 10.1158/0008-5472.CAN-12-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hayashi S, Nishimura K, Fukuchi-Shimogori T, Kashiwagi K, Igarashi K. Increase in cap- and IRES-dependent protein synthesis by overproduction of translation initiation factor eIF4G. Biochem Biophys Res Commun. 2000;277:117–23. doi: 10.1006/bbrc.2000.3637. [DOI] [PubMed] [Google Scholar]
- 38.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, et al. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11:903–8. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 39.de la Parra C, Otero-Franqui E, Martinez-Montemayor M, Dharmawardhane S. The soy isoflavone equol may increase cancer malignancy via up-regulation of eukaryotic protein synthesis initiation factor eIF4G. J Biol Chem. 2012;287:41640–50. doi: 10.1074/jbc.M112.393470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 41.De Gregorio E, Preiss T, Hentze MW. Translation driven by an eIF4G core domain in vivo. EMBO J. 1999;18:4865–74. doi: 10.1093/emboj/18.17.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.