Abstract
Diverse environmental stimuli and a complex network of regulatory factors are known to modulate expression of Vibrio cholerae's principal virulence factors. However, there is relatively little known about how metabolic factors impinge upon the pathogen's well-characterized cascade of transcription factors that induce expression of cholera toxin and the toxin-coregulated pilus (TCP). Here, we used a transposon insertion site (TIS) sequencing-based strategy to identify new factors required for expression of tcpA, which encodes the major subunit of TCP, the organism's chief intestinal colonization factor. Besides identifying most of the genes known to modulate tcpA expression, the screen yielded ptsI and ptsH, which encode the enzyme I (EI) and Hpr components of the V. cholerae phosphoenolpyruvate phosphotransferase system (PTS). In addition to reduced expression of TcpA, strains lacking EI, Hpr, or the associated EIIAGlc protein produced less cholera toxin (CT) and had a diminished capacity to colonize the infant mouse intestine. The PTS modulates virulence gene expression by regulating expression of tcpPH and aphAB, which themselves control expression of toxT, the central activator of virulence gene expression. One mechanism by which PTS promotes virulence gene expression appears to be by modulating the amounts of intracellular cyclic AMP (cAMP). Our findings reveal that the V. cholerae PTS is an additional modulator of the ToxT regulon and demonstrate the potency of loss-of-function TIS sequencing screens for defining regulatory networks.
INTRODUCTION
Cholera remains a threat to public health in many parts of the world (1). This severe and sometimes lethal dehydrating diarrheal disease is caused by Vibrio cholerae, a curved Gram-negative rod. The El Tor biotype of V. cholerae is the cause of the ongoing seventh pandemic of cholera. Humans develop cholera after ingestion of water or food contaminated with the pathogen. Following ingestion, V. cholerae colonizes the small intestine where the bacteria produce cholera toxin (CT), an AB5 type toxin that induces a marked secretory response by enterocytes that accounts for the diarrhea characteristic of cholera. The toxin-coregulated pilus (TCP), a bundle-forming pilus whose production is coregulated with cholera toxin (2), is the chief V. cholerae colonization factor. The pilus enables V. cholerae microcolony formation in the intestine and may also promote adhesion to enterocytes (3).
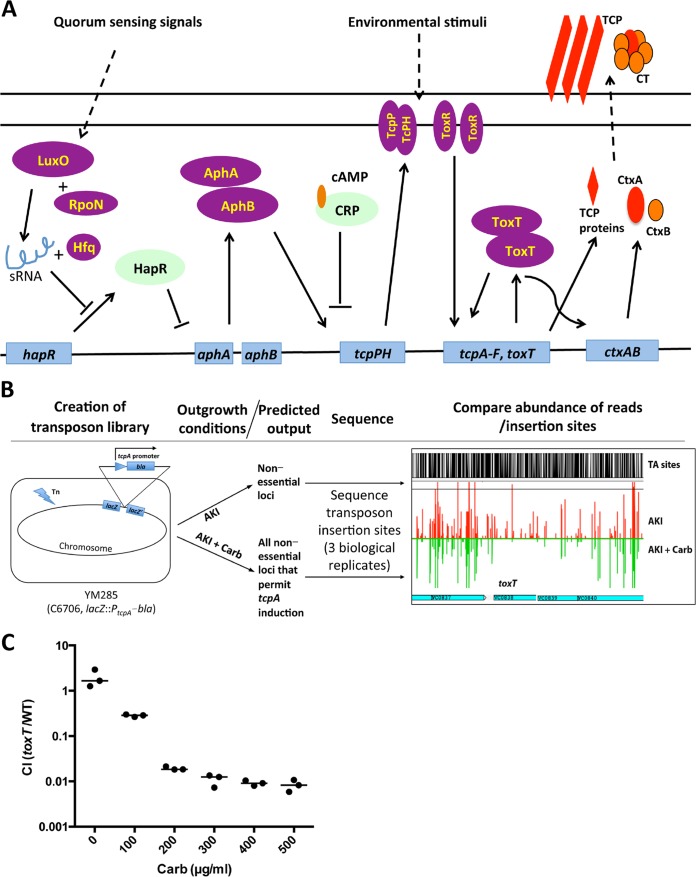
Extensive in vitro and in vivo studies have revealed that sophisticated regulatory processes control V. cholerae virulence gene expression (schematized in Fig. 1A) (reviewed in reference 4). Environmental signals trigger a virulence regulatory cascade, mediated by the membrane-imbedded proteins ToxR and TcpP/TcpH, that ultimately leads ToxT, an AraC-type transcription factor (5, 6), to directly activate the transcription of the genes encoding cholera toxin (ctxAB), TCP biogenesis, and several accessory colonization factors. ToxR can also directly activate ctxAB expression. Transcription of tcpPH requires AphA and AphB, which enable linkage of the ToxT regulon to quorum sensing (7, 8). At high cell densities, quorum sensing pathways repress aphA expression, thereby turning off virulence gene expression (9). The ToxT regulon is not active in El Tor V. cholerae grown in standard culture medium, such as LB broth; however, it can be induced through use of rich medium and microaerobic growth conditions, collectively termed AKI conditions (10, 11). The host intestinal environment also prompts marked induction of the ToxT regulon (12, 13). Environmental cues that are present in the intestine, including pH, bicarbonate (14), reduced oxygen tension (15), bile (16), and unsaturated fatty acids (17), as well as temperature (18), have been shown to modulate virulence gene expression by diverse mechanisms.
FIG 1.
A TIS sequencing-based strategy for identification of regulators of tcpA expression. (A) Simplified schematic of regulation of V. cholerae virulence gene expression. (B) A transposon library was created in YM285, a strain harboring a lacZ::PtcpA-bla fusion, which yields resistance to carbenicillin (Carb) when the tcpA promoter is active. The library was grown under AKI conditions, which induce the tcpA promoter, in either the absence or presence of Carb. The sites and abundance of transposon insertions under the two conditions were compared. In principle, mutants that are present only in the culture lacking Carb correspond to genes/loci that are critical for TcpA expression. An example of an Artemis screenshot of the abundance of reads in toxT in AKI (red) versus AKI-Carb (green) medium is shown at the right. The height of the red and green bars correlates with the number of reads. TA sites (shown in black) are potential transposon insertion sites. (C) Relative growth (expressed as a competitive index [CI]) of a toxT mutant (toxT) strain versus wt V. cholerae grown in AKI medium with different amounts of carbenicillin. sRNA, small RNA.
Metabolic signals also modulate V. cholerae virulence, but there is relatively little knowledge of the molecular bases for this regulation. The cyclic AMP (cAMP) receptor protein, CRP, along with cAMP, likewise a central modulator of cellular metabolism that controls utilization of carbon and energy sources (19), inhibits virulence gene expression by repressing tcpPH expression (20). The cAMP-CRP complex binds to sites in the tcpPH promoter that overlap AphA and AphB binding sites, competing with the capacity of these two activators to bind (21). Also, it was recently reported that disruption of tricarboxylic acid (TCA) cycle function elevates toxT transcription, perhaps by modulating acetyl-coenzyme A (CoA) (or its derivatives) levels (22).
Here, we carried out a transposon insertion site (TIS) sequencing-based screen to identify new regulators of TcpA expression. In addition to most known regulators, our screen yielded components of the V. cholerae phosphoenolpyruvate (PEP) phosphotransferase system (PTS). PTSs are multicomponent protein phosphotransfer cascades that enable the concomitant phosphorylation and import of various sugars. Typically, PTSs are composed of one inner membrane-spanning protein and four soluble proteins. Two of the cytoplasmic components, enzyme I (EI) and Hpr, usually contribute to the import of most PTS sugars, whereas the membrane-spanning EIIC proteins and their associated EIIA and EIIB components are generally specific for one or few substrates (23). As the name suggests, in the PTS phosphorylation cascade, phosphoenolpyruvate (PEP) serves as the initial phosphoryl donor for enzyme I (EI), which in turn phosphorylates HPr, which then passes the phosphoryl group to one of several sugar-specific EIIA proteins. Finally, after EIIA passes the phosphoryl group to its cognate EIIB, the latter protein transfers the phosphoryl group to a sugar bound to the cognate EIIC, facilitating its import. PTS proteins have regulatory functions in addition to mediating the import of sugars, and their regulatory activities are determined by their phosphorylation states (23, 24). For example, the phosphorylated form of Escherichia coli EIIAGlc, the glucose-specific EIIA protein, promotes the activity of adenylate cyclase (which generates cAMP), whereas its nonphosphorylated form inhibits non-PTS sugar transporters (23, 24). Additionally, PTSs have been implicated in control of diverse processes, including chemotaxis, biofilm formation, and virulence in some pathogens (23).
Our studies reveal that the V. cholerae EI, Hpr, and EIIAGlc proteins are required for robust TcpA and cholera toxin production in vitro. Moreover, strains lacking these PTS proteins have a diminished capacity to colonize the infant mouse intestine. The PTS promotes expression of tcpPH and aphAB and, through them, toxT to induce tcpA and ctx expression. PTS appears to promote virulence gene expression in part by modulating accumulation of intracellular cAMP.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All V. cholerae strains described in this study are derivatives of the El Tor Peruvian clinical isolate C6706 and are listed in Table 1. Bacteria were cultivated in Luria-Bertani (LB) broth unless otherwise noted. To induce production of TcpA and CT, V. cholerae cultures were grown in freshly prepared AKI medium (1.5% Bacto peptone, 0.4% yeast extract [Difco], 0.5% NaCl, 0.3% NaHCO3) for 4 h of static growth followed by 2 h of growth with vigorous rotation, as previously described (10). When appropriate, the medium was supplemented with streptomycin (Sm; 200 μg/ml), carbenicillin (AKI-Carb; 50 μg/ml), kanamycin (Km; 50 μg/ml), or l-arabinose (0.04%, wt/vol).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Vibrio cholerae strains | ||
| C6706 | Wild-type strain, El Tor biotype, O1 serogroup, lacZ+, Smr | Lab collection |
| YM081 | C6706 ΔtcpA lacZ+ | Lab collection |
| MKW1144 | C6706 ΔctxAB; Peru-NT | 56 |
| YM285 | C6706 lacZ::PtcpA-bla | This study |
| YM171 | C6706 lacZ::PtcpA-gfp | This study |
| YM287 | C6706 Tn-toxT lacZ::PtcpA-bla | This study |
| MKW1014 | O395 ΔtoxT | Lab collection |
| QW086 | C6706 ΔptsI | This study |
| QW088 | C6706 ΔptsH | This study |
| QW180 | C6706 ΔVC0964 | This study |
| QW208 | C6706 ΔptsI lacZ::PtcpA-gfp | This study |
| QW210 | C6706 ΔptsH lacZ::PtcpA-gfp | This study |
| QW126 | C6706 lacZ::PctxAB-gfp | This study |
| QW213 | C6706 ΔptsI lacZ::PctxAB-gfp | This study |
| QW215 | C6706 ΔptsH lacZ::PctxAB-gfp | This study |
| QW170 | C6706 ptsI+; ΔptsI with ptsI reversed into the same locus | This study |
| QW171 | C6706 ptsH+; ΔptsH with ptsH reversed into the same locus | This study |
| QW199 | C6706 ΔcyaA | This study |
| QW200 | C6706 Δcrp | This study |
| QW174 | C6706 ΔcpdA | This study |
| QW236 | C6706 ΔcyaA ΔptsI | This study |
| QW241 | C6706 Δcrp ΔptsI | This study |
| QW178 | C6706 ΔcpdA ΔptsI | This study |
| QW239 | C6706 ΔcyaA ΔptsH | This study |
| QW278 | C6706 Δcrp ΔptsH | This study |
| QW176 | C6706 ΔcpdA ΔptsH | This study |
| QW182 | C6706 ΔptsP (VC0672) | This study |
| QW184 | C6706 Δfpr (VCA0518) | This study |
| QW264 | C6706 ΔVC1820 | This study |
| QW313 | C6706 ΔVCA0245 | This study |
| QW189 | C6706 ΔVC1822 | This study |
| QW187 | C6706 ΔVC2013 | This study |
| QW191 | C6706 ΔVC0995 | This study |
| QW193 | C6706 ΔVCA1045 | This study |
| QW311 | C6706 ΔVC0910 | This study |
| QW330 | PtsI(H189A) | This study |
| QW332 | VC0964(H91D) | This study |
| QW334 | PtsH(H15A) | This study |
| Escherichia coli strains | ||
| DH5α λpir | λpir lysogen Δ(ara-leu) araD Δ(lacX74) phoA20 thi-1 rpoB argE(Am) recA1 | Lab collection |
| SM10 λpir | thi thr leu tonA lacy supE recA::RP4-2-Tc::Mu λpir Kanr | Lab collection |
| Plasmids | ||
| pCVD442 | Suicide plasmid, pir dependent, R6K, SacBR, Carbr | 26 |
| pJZ111 | Suicide plasmid, pir dependent, R6K, SacBR, Carbr | Lab collection |
| pYB742 | pSC189 derivative plasmid carrying mariner transposon, pir dependent, R6K, Cmr | Lab collection |
| pBAD-ptsI | pBAD-TOPO carrying a fragment of VC0965 (ptsI) | 45 |
| pBAD-ptsH | pBAD-TOPO carrying a fragment of VC0966 (ptsH) | 44 |
| pBAD-ptsI (H189A) | pBAD-TOPO carrying VC0965 with a mutation H189A | 45 |
| pBAD-ptsH (H15A) | pBAD-TOPO carrying VC0966 with a mutation H15A | 44 |
| pToxT | pBAD24 derivative plasmid expression toxT | Lab collection |
| pYM294 | pJZ111 carrying PtcpA fused to bla | This study |
| pYM137 | pJZ111 carrying PtcpA fused to gfp | This study |
| pYM93 | pJZ111 carrying PctxAB fused to gfp | This study |
| pCVD-ΔptsI | pCVD442 carrying an unmarked, in-frame deletion in VC0965 | This study |
| pCVD-ΔptsH | pCVD442 carrying an unmarked, in-frame deletion in VC0966 | This study |
| pCVD-ΔVC0964 | pCVD442 carrying an unmarked, in-frame deletion in VC0964 | This study |
| pCVD-ptsI | pCVD442 carrying an intact gene fragment of VC0965 | This study |
| pCVD-ptsH | pCVD442 carrying an intact gene fragment of VC0966 | This study |
| pCVD-ΔcyaA | pCVD442 carrying an unmarked, in-frame deletion in cyaA | This study |
| pCVD-Δcrp | pCVD442 carrying an unmarked, in-frame deletion in crp | This study |
| pCVD-ΔcpdA | pCVD442 carrying an unmarked, in-frame deletion in cpdA | This study |
| pCVD-ΔVC0672 | pCVD442 carrying an unmarked, in-frame deletion in VC0672 | This study |
| pCVD-ΔVCA0518 | pCVD442 carrying an unmarked, in-frame deletion in VCA0518 | This study |
| pCVD-ΔVC1820 | pCVD442 carrying an unmarked, in-frame deletion in VCA1820 | This study |
| pCVD-ΔVCA0245 | pCVD442 carrying an unmarked, in-frame deletion in VCA0245 | This study |
| pCVD-ΔVC1822 | pCVD442 carrying an unmarked, in-frame deletion in VC1822 | This study |
| pCVD-ΔVC2013 | pCVD442 carrying an unmarked, in-frame deletion in VC2013 | This study |
| pCVD-ΔVC0995 | pCVD442 carrying an unmarked, in-frame deletion in VC0995 | This study |
| pCVD-ΔVCA1045 | pCVD442 carrying an unmarked, in-frame deletion in VCA1045 | This study |
| pCVD-ΔVC0910 | pCVD442 carrying an unmarked, in-frame deletion in VC0910 | This study |
| pCVD-PtsI(H189A) | pCVD442 carrying intact VC0965 with a mutation H189A | This study |
| pCVD-Ptsh(H15A) | pCVD442 carrying intact VC0966 with a mutation H15A | This study |
| pCVD-VC0964(H91D) | pCVD442 carrying intact VC0964 with a mutation H91D | This study |
Strain construction.
All cloning was carried out using isothermal assembly (25). Allele replacements were created with the sacB-containing suicide vector pCVD442 (26) as described previously (27). The promoter fusion constructs PtcpA-bla, PtcpA-gfp, and PctxAB-gfp were cloned into the suicide vector pJZ111 (a kind gift from Jun Zhu) in place of the Plac-green fluorescent protein (GFP) sequence by isothermal assembly. The primers used for generating these constructs as well as the names of the resulting vectors are shown in Table S1 in the supplemental material.
Transposon-insertion sequencing.
TIS sequencing was performed as described previously (28). In brief, a highly saturated transposon library of ∼126,000 unique insertion mutants (insertions at >65% of TA sites) was generated in strain YM285 (C6706 lacZ::PtcpA-bla) by conjugation using pYB742, a modified version of the transposon delivery vector pSC189 carrying the resistance gene to chloramphenicol instead of ampicillin (Table 1). The transposon library was scraped and resuspended in LB broth, and ∼108 CFU were inoculated into 5 ml of AKI medium (input cells) or AKI medium containing 500 μg/ml Carb (output cells) in triplicates. TcpA expression was induced by incubating the cultures at 37°C without shaking for 4 h, followed by shaking at 37°C for 2 h. After this outgrowth, 1 ml each of input and output cultures was plated onto 245-mm LB plates containing 200 μg/ml Sm and 50 μg/ml Km, and lawns of bacteria were grown overnight at 30°C.
The input and output libraries were scraped from the plates and resuspended in 10 ml of LB medium, and 2 ml of cells was used for genomic DNA extraction. Ten micrograms of genomic DNA was sheared to ∼300-bp fragments using acoustic disruption on an M220 focused ultrasonicator (Covaris, Woburn, MA). The DNA ends were repaired, the transposon junctions were amplified, adaptor sequences were attached, and the library was sequenced on a MiSeq instrument (Illumina, San Diego, CA) as described previously (28, 29). Insertion sites were identified and analyzed as described previously (30, 31). Results were visualized using Artemis (32). Three separate input and output libraries were sequenced and analyzed, respectively.
The reads for the input and output replicates were combined, and then each pooled data set was normalized to the same number of total reads (∼3.5 million). The reads for each gene in the input data set were compared to the reads at the same locus in the output data set using a Mann-Whitney U rank sum statistical test. Using the statistical results from known genes that are required for TcpA induction, candidates of interest were defined as those with a P value of <0.005.
Immunoblotting for TcpA and CT.
Overnight V. cholerae cultures were diluted 1:100 and inoculated into freshly prepared AKI medium and grown as described above to induce production of TcpA and CT. TcpA and CT levels were assayed for whole-cell lysates and concentrated culture supernatants, respectively. For preparation of cell lysates, 1 ml of culture was centrifuged and resuspended in 1 volume of phosphate-buffered saline (PBS; pH 7.4) to normalize the culture densities based on the optical density at 600 nm (OD600). The cell-free culture supernatants from 5 ml of culture were concentrated 20 times with Amicon Ultra filters (Millipore, Billerica, MA) at 4°C and normalized to the OD600 of the cultures. One hundred microliters of the adjusted cell lysates or supernatants was mixed with 100 μl of Novex sample buffer (Life Technologies, Grand Island, NY) supplemented with dithiothreitol (DTT) and then boiled for 10 min. The resulting suspensions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 12% Bis-Tris gel (NuPAGE; Life Technologies). The proteins were transferred to a nitrocellulose membrane using an iBlot system (Life Technologies), and after being blocked with 5% skimmed milk solution, blots were incubated first with 1:2,000 dilutions of rabbit polyclonal anti-TcpA or anti-CT (C3062; Sigma, St. Louis, MO) antiserum and then with a 1:10,000 dilution of anti-rabbit peroxidase-conjugated IgG secondary antibody (Sigma). For loading controls, the same blots from the cell lysates were stripped and then incubated with a 1:20,000 dilution of mouse monoclonal anti-RNA polymerase alpha (RNAP alpha) (4RA2 antibody; Santa Cruz Biotechnology, Dallas, TX) and then with a 1:10,000 dilution of anti-mouse peroxidase-conjugated IgG secondary antibody (Sigma). Reacting bands were visualized using a SuperSignal chemiluminescent kit (Thermo Fisher Scientific, Inc., Waltham, MA).
Microscopy and flow cytometry analysis.
AKI-induced cells were immobilized on pads containing 1% agarose, 10% LB medium, and PBS and were visualized using a Zeiss Axio Imager 2 microscope equipped with a Plan Neofluor 100×/1.3 oil Ph3 objective and a Hamamatsu Orca ER 1394 camera. Images were processed with ImageJ (version 1.49j10). For flow cytometry, bacterial cultures were fixed with 1% paraformaldehyde on ice for at least 10 min. Then, the fixed cells were diluted into PBS and assayed for fluorescence using a FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). The wild-type ([wt] GFP-negative) strain was used as the negative control to determine the level of autofluorescence.
Intestinal tissue from mice inoculated with lacZ::PtcpA-gfp-labeled wild-type (YM285) or PTS mutant strains was analyzed via confocal microscopy (n = 3 per assay). Tissue samples from the medial and distal intestines were fixed in PBS with 4% paraformaldehyde for 2 h at room temperature (RT), placed in PBS with 20% sucrose for 2 h at 4°C, mounted in 22-oxacalcitriol (OCT), and then frozen at −80°C before being sectioned on a cryostat. For staining, frozen sections were washed in PBS for 5 to 15 min at RT, blocked in blocking buffer (1% bovine serum albumin [BSA], 5% normal goat serum in PBS) for 1 h at RT, stained with a primary anti-O1 mouse antiserum (1:100) and anti-GFP-Alexa 488 (1:100) in PBS for 3 h at RT, washed three times in PBS, stained with anti-mouse IgG-Dylight 594 for 40 min at RT, washed three times in PBS, counterstained with 4′,6′-diamidino-2-phenylindole (DAPI; 1 μg/ml) and phalloidin-Alexa Fluor 647 (1:100) (Life Technologies) for 20 min at RT, and washed twice in PBS. Slides were mounted in Prolong Gold (Life Technologies) and imaged with a Nikon confocal microscope using a 20× or 60× objective. Multiple images were collected per section and analyzed with ImageJ.
Quantitative reverse transcriptase PCR (qRT-PCR).
Total RNA was isolated from AKI-induced V. cholerae cultures using TRIzol regent (Life Technologies) and treated with DNase I for 1 h with a Turbo DNA-free kit (Life Technologies). Reverse transcription was performed using a Superscript III first-strand synthesis system (Life Technologies) according to the manufacturer's protocol. Five micrograms of total RNA was used for the reverse-transcription reaction mixture with random hexamers. The synthesized cDNA was subjected to real-time PCR amplification using a Fast SYBR green master mix kit (Life Technologies) with specific primer pairs (see Table S1 in the supplemental material) in a total volume of 20 μl. The amplification and data analysis with ΔΔCT (where CT is threshold cycle) method were performed using a StepOnePlus system (Life Technologies). gyrB mRNA was measured and used for normalization (33).
cAMP assay.
A quantitative assay of intracellular cyclic AMP (cAMP) concentration was performed using a modified version of a previously described protocol (34). Briefly, overnight cultures of wild-type and mutant V. cholerae strains grown under AKI conditions were used for these assays. After determination of the samples' OD600s, 6-ml aliquots of cells were collected by centrifugation (12,000 × g for 3 min) and washed twice with ice-cold PBS (pH 7.0). Half of each sample was resuspended in 0.1 N HCl (500 μl), vortexed five times during a 15-min incubation on ice, and centrifuged (12,000 × g for 5 min). The supernatant was used for cAMP determination using an immunoassay (Cyclic AMP EIA kit; Cayman Chemical Co., Ann Arbor, MI) according to the manufacturer's protocol. The remaining sample was resuspended in PBS (500 μl), subjected to three freeze-thaw cycles, and centrifuged (12,000 × g for 3 min), and the supernatant was used for total protein determination (bicinchoninic acid [BCA] protein assay; Thermo Fisher). Assay results were used to calculate the intracellular cAMP concentration based on the estimated cellular volume for the bacteria per milligram of protein (4.92 μl/mg of total cellular protein) (34). The assays were performed in duplicate on three biologic replicates.
Infection experiments.
In competition experiments, infant mice were inoculated with ∼1:1 mixtures of mutant and wt (a lacZ-negative derivative of C6706) strains as previously described (35). To generate the inocula, ∼20-h LB cultures were diluted 1:1000 and then mixed 1:1. Five-day-old suckling mice (Charles River, Wilmington, MA) were orally inoculated with 50 μl of the mixtures. Mice were sacrificed after 24 h, and the small intestines were homogenized using a tissue grinder. Dilutions of the homogenate were then plated on agar plates containing streptomycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to distinguish lacZ-negative wild-type and lacZ-positive (lacZ+) mutant and wild-type strains. For single-infection assays, ∼2 × 105 cells were inoculated into each mouse, and after 24 h, the small intestine samples were prepared and processed as described above.
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal protocols were reviewed and approved by the Harvard Medical Area Standing Committee on Animals (protocol 04316).
RESULTS
A TIS sequencing-based approach to identify novel regulators of TcpA expression.
We set out to harness the power of TIS sequencing to search for novel regulators of TcpA expression. To this end, we constructed an El Tor V. cholerae strain harboring a chromosome-encoded selectable reporter of tcpA expression. The reporter, a fusion of the tcpA promoter (513 bp upstream of the start codon) to a carbenicillin (Carb) resistance marker (PtcpA-bla) was introduced into the lacZ locus of V. cholerae strain C6706, yielding YM285 (Fig. 1B). Subsequently, a high-density Himar transposon (Tn) insertion library was created in YM285, and the library was grown under AKI conditions, which induce expression of TcpA, in either the absence or presence of Carb. In principle, mutants that are present only in the culture lacking Carb correspond to genes/loci that are critical for TcpA expression. Such mutants can be identified through comparison of the Tn insertion sites in the Carb-selected and unselected cultures using high-throughput sequencing (Fig. 1B).
Initially, to validate our strategy and to estimate the optimal concentration of Carb for the screen, we carried out competitive growth experiments between YM285 and a toxT transposon mutant carrying the PtcpA-bla construct (YM287) in AKI medium supplemented with Carb (AKI-Carb medium). YM285 (lacZ mutant) and YM287 (carrying lacZ in its transposon) were discriminated using LB plates supplemented with X-Gal. Since ToxT is a critical activator of tcpA expression under these conditions (5, 6), we reasoned that poor survival of the toxT mutant (measured in competitive growth experiments as a competitive index [CI]) would provide a gauge of the maximal effect we would observe when we screened the transposon library. We recovered equal amounts of the toxT mutant and the wt strain in the absence of Carb; however, there was ∼100-fold growth deficiency of the toxT mutant relative to growth of YM285 at Carb concentrations higher than 200 μg/ml (Fig. 1C). Consequently, the subsequent genome-wide screen was performed in the presence of 500 μg/ml Carb.
We compared the number of reads mapping to each V. cholerae gene in the cultures grown in AKI versus AKI-Carb using Mann Whitney U test-based statistical analysis (29, 31). In total, there were 34 genes with statistically fewer reads (P value < 0.005) in the AKI-Carb cultures (see Table S2 in the supplemental material). Notably, nearly all of the genes encoding factors known to act upstream of and to promote tcpA expression were found to be underrepresented in the library grown in the presence of Carb (Table 2), validating the approach. For example, there is a marked paucity of insertions in toxT in the AKI-Carb cultures compared to the number in AKI cultures (Fig. 1B), as well as in tcpPH, aphAB, and toxR, which encode factors that promote expression of ToxT (Table 2) (4–7, 36, 37). Many genes on the list, including ptsI, cpdA, pta, pstA-1, pstC-1, tolQ, mukF, and mukB, have been reported to be required for robust V. cholerae intestinal colonization; e.g., they have been identified in previous signature-tagged mutagenesis (STM) screens (38, 39), but the mechanisms by which they attenuate colonization have not been reported. Since these genes answered our screen, they likely induce tcpA expression and thus may promote V. cholerae intestinal colonization by facilitating expression of its major colonization factor. In addition, there were fewer insertions in several genes, including pbp1a, lpoA, and csiV, whose inactivation diminishes V. cholerae peptidoglycan synthesis and thereby diminishes growth in the presence of a cell wall-acting antibiotic like Carb but are likely unrelated to TCP production (27, 40). Finally, the list included 12 additional loci that have not been linked to TcpA expression or V. cholerae colonization previously (see Table S2 in the supplemental material). We selected three of these genes—ptsH, aspK, and cafA—along with ptsI for further study (Table 2, bold genes).
TABLE 2.
Subset of loci identified in TIS sequencing-based screen for factors that promote TcpA expression
| Locusa | Gene name(s) | P valueb | TIGR annotation |
|---|---|---|---|
| VC0838 | tcpN, toxT | 0.00001 | TCP pilus virulence regulatory protein |
| VC1049 | aphB | 1.0 × 10−10 | Transcriptional regulator, lysR family |
| VC2647 | aphA | 1.0 × 10−10 | PadR family transcriptional regulator |
| VC0826 | tcpP | 1.0 × 10−10 | Toxin coregulated pilus biosynthesis protein P |
| VC0827 | tcpH | 1.0 × 10−10 | Toxin coregulated pilus biosynthesis protein H |
| VC0984 | toxR | 0.00006 | Regulatory protein ToxT |
| VC1021 | luxO | 1.0 × 10−10 | LuxO repressor protein |
| VC0347 | hfq | 0.00073 | Host factor I |
| VC2529 | rpoN | 1.0 × 10−10 | RNA polymerase sigma-54 factor |
| VC0034 | tcpG, dsbA | 0.00025 | Thiol:disulfide interchange protein |
| VC0965 | ptsI | 1.0 × 10−10 | Phosphoenolpyruvate protein phosphotransferase, EI |
| VC0966 | ptsH | 0.00009 | Phosphocarrier protein HPr |
| VC0547c | aspK | 0.00103 | Aspartokinase, alpha and beta subunits |
| VC0419c | cafA | 1.0 × 10−10 | RNase G/cytoplasmic axial filament protein |
Genes in bold represent loci that have not been previously associated with tcpA expression and that were subjected to additional analyses in this study; genes not in bold have been previously linked to tcpA expression.
Determined by a Mann-Whitney U test; limit of detection, P < 1.0 × 10−10.
Genes that proved to be false positives.
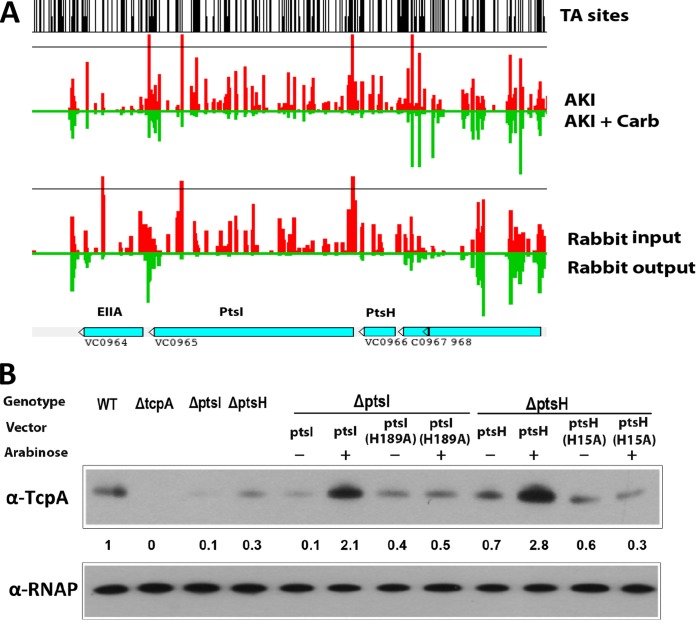
In attempting to validate hits from our screen, we found that strains with transposon insertions (41) and/or deletions of aspK and cafA produced wt levels of TcpA in AKI medium, suggesting that these are false positives (see Fig. S1 in the supplemental material). In contrast, the ptsH mutant exhibited reduced TcpA expression under AKI conditions (see below). ptsH is found upstream of ptsI, another gene that answered our screen; the two encode critical components (Hpr and EI, respectively) of the V. cholerae PTS system (42, 43). Both genes, in addition to being significantly under represented in AKI-Carb medium versus AKI medium (Fig. 2A), were recently found to be essential for growth in the infant rabbit intestine (29) (Fig. 2A). Furthermore, using an STM screen, Merrell et al. (38) found that a ptsI mutant had reduced growth in the mouse intestine compared to growth in vitro although the basis for the strain's attenuated colonization was not investigated. In addition, a ptsI mutant was found to have a markedly reduced capacity to grow in the adult gnotobiotic mouse intestine (44), in which the importance of known virulence regulators (e.g., ToxT) and colonization factors (e.g., TCP) has not been reported. Our identification of both ptsI and ptsH in our screen as factors that promote TcpA expression, along with the previous observations linking ptsI to intestinal colonization, prompted us to focus on these genes below.
FIG 2.
PtsH and PtsI promote TcpA expression in AKI medium. (A) Artemis screenshot of abundance of reads in ptsH and ptsI detected in AKI (red) versus AKI-Carb medium (green) and in input (red) and output (green) from infant rabbits (data from Pritchard et al. [29]). (B) TcpA expression detected by immunoblotting in the indicated strains. The numbers correspond to densitometry measurements normalized to the value of the wt strain. Levels of RNAP alpha serve as loading controls.
ptsI and ptsH are required for robust TcpA expression.
Initially, strains with in-frame deletions of ptsI and ptsH were generated to further validate and explore the role of these two PTS genes in expression of TcpA under AKI conditions. Immunoblotting with anti-TcpA serum showed that, in AKI medium, TcpA expression in the ptsI and ptsH mutants was reduced to ∼10% and 30% of the amounts observed in the wt strain, respectively (Fig. 2B). These defects in TcpA expression could be complemented using arabinose-inducible PtsI and PtsH expression vectors, establishing that the absence of PtsI and PtsH accounts for the reduced production of TcpA in the respective mutants. Both PtsI and PtsH undergo phosphorylation at conserved histidine residues, and their phosphorylation is critical for their function in the PTS phosphorelay and often for their interactions with proteins that are not part of the PTS cascade (23, 43). In contrast to the wt proteins, arabinose-inducible PtsI with the amino acid change H189A [PtsI(H189A)] and PtsH(H15A), which are incapable of phosphorylation (44, 45), were unable to complement the defect in TcpA expression in the ptsI and ptsH mutants in AKI media (Fig. 2B), suggesting that the phosphorylation of PtsI and PtsH is critical for their role in promoting TcpA production.
The ptsI and ptsH mutants formed slightly smaller colonies than wt cells on LB agar plates, so we investigated if their growth is deficient in AKI medium. In AKI medium, the growth of the two PTS mutants was similar to that of the wt strain (see Fig. S2 in the supplemental material). Thus, failure to recover the ptsI and ptsH mutants in the initial screen does not simply reflect a reduced growth rate by these mutants during the selection.
EI, Hpr, and EIIAGlc mutants exhibit defective intestinal colonization.
We used suckling mice to investigate the capacity of the ptsI and ptsH deletion mutants to survive and proliferate in the murine small intestine. When one-to-one mixtures of each mutant and the wt were used to inoculate 5-day-old CD1 mice in in vivo competition experiments, 10 to 40 times more wt CFU were recovered from intestinal homogenates 24 h later, indicating that the two mutants have a reduced capacity to survive and/or proliferate in the intestine compared with that of the wt (Fig. 3B). Mutants that harbored chromosomal versions of the nonphosphorylatable alleles of ptsI [ptsI(H189A)] or ptsH [ptsH(H15A)] exhibited colonization defects similar to those of the deletion mutants (Fig. 3B), suggesting that the capacity of these signaling proteins to be phosphorylated is required for their contribution to intestinal colonization. However, the colonization defects of these PTS mutants was not nearly as profound as the defect of a ΔtcpA mutant (Fig. 3B), suggesting that their in vivo production of TCP (an essential colonization factor) is not sufficiently reduced to abrogate colonization to the extent seen with the tcpA mutant.
FIG 3.
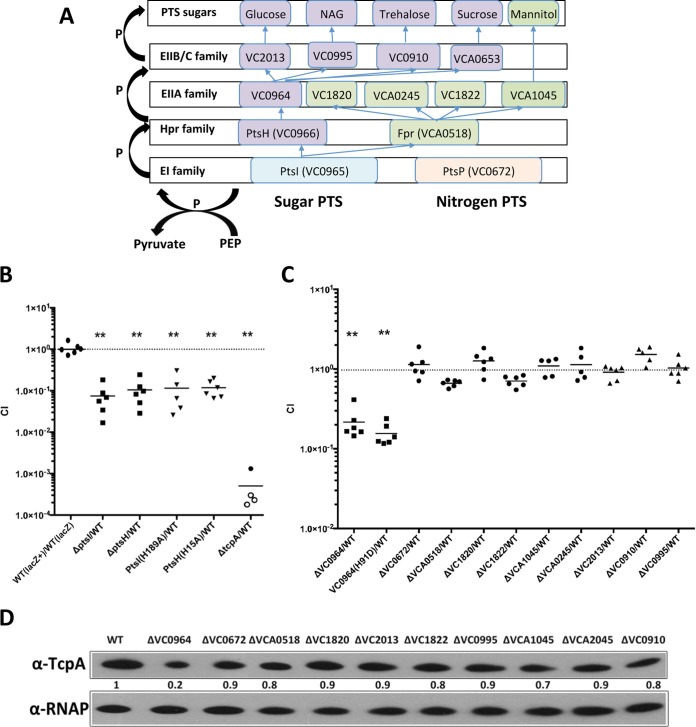
Intestinal colonization of PTS mutants. (A) Schematic of a subset of V. cholerae PTS components (42). (B and C) Suckling mouse intestinal colonization of various PTS deletion mutants assessed using competition assays with a lacZ-negative derivative of the wild-type strain [wt (lacZ)]. Competitive indices (CIs) represent the output ratio (number of mutant CFU/number of wt CFU) divided by the input ratio. Each point represents a single animal, and the horizontal lines represent geometric means. Open symbols mark the limits of detection for animals from which no mutants were recovered. **, P < 0.01, based on ANOVA followed by Bonferroni's multiple-comparison posttest comparing the data with the corresponding wt (lacZ+)/wt (lacZ) samples. (D) TcpA expression detected by immunoblotting in the indicated strains. Levels of RNAP alpha serve as loading controls.
Like many proteobacteria, V. cholerae encodes PTS components in addition to ptsI, which encodes the EI protein, and ptsH, which encodes the Hpr protein (Fig. 3A) (42). These include ptsP (VC0672), a putative nitrogen-related EI protein (42), and two additional HPr homologues, fpr (VCA0518), a putative fructose-specific Hpr-like protein (42), and a putative nitrogen Hpr protein (VC2533). Finally, V. cholerae encodes at least 20 EII family proteins. The specific sugars transported by the EII proteins have not been completely defined, but Houot and colleagues have shown that EIIAGlc (VC0964), which is encoded in the same cluster as ptsH and ptsI, is required for sucrose, trehalose, and N-acetylglucosamine (NAG) transport and contributes to glucose transport and that VCA1045 mediates mannitol transport (44). Although our screen did not yield other components of the V. cholerae PTS system, we generated deletion mutants of additional PTS genes to assess whether transport mediated by particular EIIA family proteins (VC0964, VC1820, VCA0245, VC1822, and VCA1045) or EIIB/EIIC proteins (VC2013, VC0995, and VC0910) (42) (Fig. 3A) is linked to the requirement for ptsI and pstH for intestinal colonization. In addition, we constructed strains with deletions in the nitrogen EI (VC0672) and the fructose Hpr component (VCA0518). With the exception of the VC0964 deletion mutant and a nonphosphorylatable VC0964 H91D mutant, none of these mutants exhibited attenuated intestinal colonization in competition assays versus the wt strain (Fig. 3C). Since the EIIAGlc homologue VC0964 has been implicated in transport of NAG, sucrose, and trehalose, as well as glucose, intestinal transport of one or more of these sugars may be important for V. cholerae intestinal colonization. In contrast, transport of mannitol and fructose, as well as the activity of the nitrogen PTS, appears to be dispensable for colonization (Fig. 3C). In concordance with the colonization capacities of the PTS mutant panel, only the ΔVC0964 mutant showed significantly reduced TcpA expression on immunoblots of cells grown under AKI conditions (Fig. 3D). Thus, sugar transport mediated by EIIAGlc, which works in conjunction with EI/Hpr, may promote intestinal colonization by inducing Tcp expression; in addition, it is possible that the sugars imported by these PTS proteins promote robust V. cholerae growth in the intestine.
The absence of PTS diminishes the fraction of cells that express tcpA.
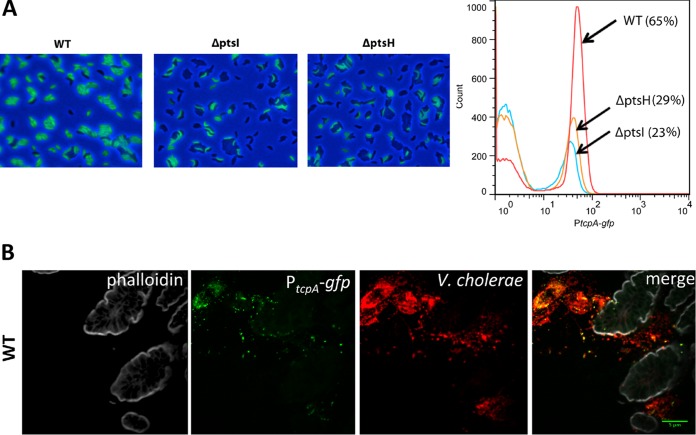
Previous studies have revealed that even under TcpA-inducing conditions, such as growth in AKI media, TCP expression is heterogeneous and not detectable in all cells (46). We used fluorescence microscopy and flow cytometry to decipher whether the decreased TcpA evident in the immunoblots of lysates of the PTS mutants (Fig. 2B), which represents a population-wide average, arises from reduced TcpA per cell or from a decrease in the fraction of cells which produce TcpA. For these experiments, a PtcpA-gfp transcriptional fusion was introduced into the lacZ locus of the wt and ΔptsI and ΔptsH mutant strains. After growth under AKI conditions, a lower fraction of cells expressing GFP was detected in the mutant strains, and agglutination (perhaps mediated by TCP) appeared to be reduced in the mutants as well (Fig. 4A). Flow cytometry confirmed that the fractions of GFP-positive (GFP+) cells in the ΔptsI (23%) and ΔptsH (29%) mutants were considerably lower than the fraction in the wt (65%) (Fig. 4A) and also showed that there was a slight reduction in the intensity of GFP fluorescence in the mutants compared with the wt level. Thus, the absence of PtsI and PtsH markedly reduces the fraction of cells that induce TcpA expression in response to AKI conditions and has only a marginal effect on the magnitude of tcpA induction in cells that do respond.
FIG 4.
A smaller fraction of cells express tcpA in PTS mutants. (A) GFP expression from a chromosomal PtcpA-gfp reporter in the wt and ptsI and ptsH deletion mutants after growth under AKI conditions was monitored with epifluorescence microscopy (left) or flow cytometry (right). The figure shows representative results from three replicates. (B) Heterogeneous expression of tcpA in wt V. cholerae is also seen during infection of suckling mice. V. cholerae cells in the small intestine (red) were detected by immunofluorescence, and tcpA expression (green) was detected from the chromosomal PtcpA-gfp reporter. Tissue sections were counterstained with phalloidin (gray) to visualize actin.
Heterogenous expression of TcpA has also been detected in infection studies by Nielsen and colleagues, who used ligated rabbit ileal loops to study tcpA expression in vivo (46). We wondered whether the reduced fractions of ptsI and ptsH mutant cells expressing TcpA under AKI conditions might also be present in vivo and provide an explanation of why these mutants exhibited reduced intestinal colonization. To investigate this possibility, we first carried out single-infection studies in infant mice using the wt strain harboring the chromosomal PtcpA-gfp reporter to test whether the heterogeneity of TcpA expression in vivo was detectable with fluorescence microscopy. Sections from the medial and distal parts of the small intestine, the principal sites of V. cholerae intestinal colonization (35, 47), revealed that there is heterogeneous expression of TcpA in vivo (Fig. 4B; see also Fig. S3 in the supplemental material). It is difficult to quantify the fraction of V. cholerae cells, detected by immunofluorescence with an anti-O1 antiserum (red in Fig. 4B), that expressed TcpA (green in Fig. 4B), but in many sections, less than half of the V. cholerae cells were TcpA+ (Fig. 4B; see also Fig. S3). Since our previous findings revealed that there is uniform intraintestinal expression of gfp driven from a chromosomal Plac-gfp construct (47), heterogenous expression is not a general phenomenon of this experimental setup. Thus, heterogenous TcpA expression is apparent in infant mice as well as in ligated ileal loops, suggesting that the previously observed heterogeneity in the latter system is not attributable to the closed nature of this model. We carried out similar single-infection experiments with the ptsI and ptsH mutants but, as discussed below (see Fig. 6D), the severe colonization defect of these strains precluded an analysis of the heterogeneity of TcpA expression in vivo.
FIG 6.
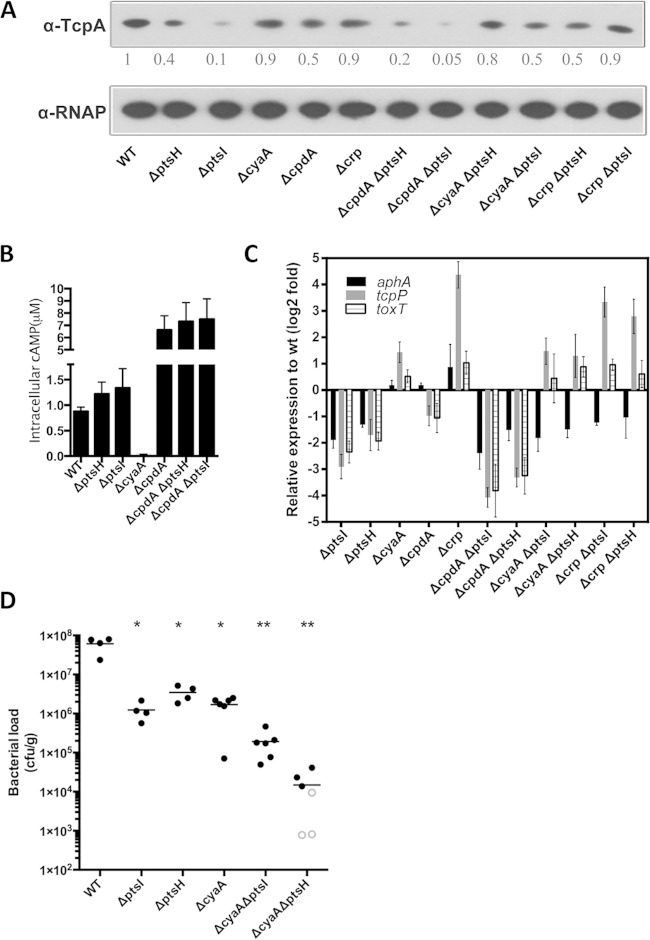
In the absence of PTS, cAMP-CRP represses TcpA expression under AKI conditions. (A) TcpA expression detected by immunoblotting in the indicated strains. The numbers correspond to densitometry measurements normalized to the value of the wt strain. Levels of RNAP alpha serve as loading controls. (B) Cyclic AMP concentrations in the wt and ptsI- and ptsH-related mutants cultured under AKI conditions. Data represent three independent assays, and each sample was assayed in duplicate. Values are reported as the means ± standard deviations. All mutants have cAMP levels significantly different from the wt level (P < 0.01, unpaired t test compared to the wt). (C) qRT-PCR measurements of the transcription of aphA, tcpP, and toxT after AKI induction in ptsI- and ptsH-related mutants relative to wt levels. gyrB expression was used as an internal control. Triplicate experiments were conducted, and the relative expression levels compared to the wt strain are shown as means ± standard deviations. (D) Intestinal colonization (bacterial load in intestinal homogenates) of the indicated strains after single inoculation of suckling mice. Open symbols mark the limits of detection for animals from which no mutants were recovered. *, P < 0.05; **, P < 0.01 (Mann-Whitney test in a comparison with wt levels).
PTS acts upstream of ToxT to promote expression of cholera toxin and TcpA.
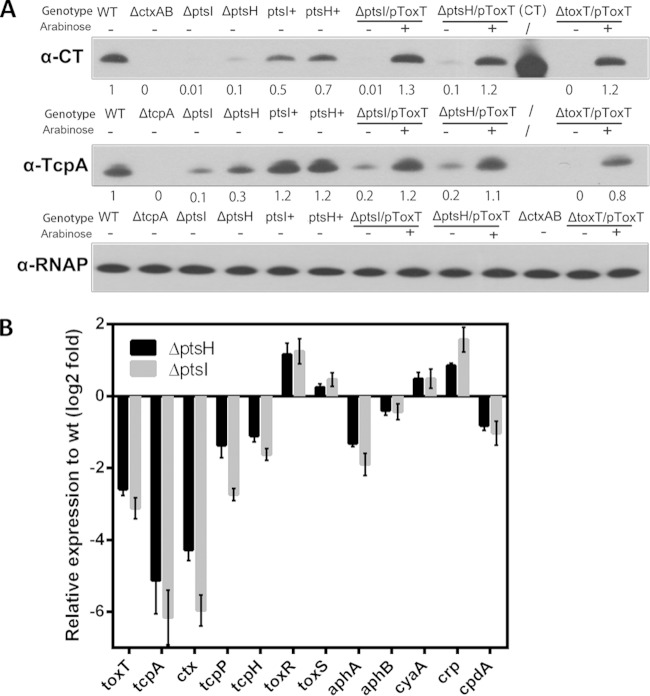
To address how deletions in either ptsI or ptsH interrupt activation of TcpA, we first tested whether cholera toxin (CT) production is reduced in the ΔptsI and ΔptsH mutant strains. Defective CT production would suggest that ptsI and ptsH do not act at the tcpA promoter but at some upstream point in the ToxR virulence cascade (Fig. 1A). Immunoblotting of concentrated supernatants from the wt and PTS mutant strains grown under AKI conditions revealed a marked reduction in CT in the PTS mutants compared with the wt level (Fig. 5A). To confirm that the defect in CT production in the ΔptsI and ΔptsH mutant strains was attributable to the absence of ptsI and ptsH, we generated revertants where the ptsI and ptsH genes were put back into their respective native loci on the chromosome. The revertants (ptsI+ and ptsH+ in Fig. 5A) produced amounts of CT (and TcpA) comparable to those of the wt strains.
FIG 5.
The V. cholerae PTS acts upstream of toxT to modulate TcpA and CT expression. (A) CT subunit A and TcpA expression detected by immunoblotting in the indicated strains. Numbers correspond to densitometry measurements normalized to the value of the wt strain. Purified CT is used as a control. Levels of RNAP alpha serve as loading controls. The ptsI+ and ptsH+ strains are revertants derived from the ptsI and ptsH deletion mutants. (B) qRT-PCR measurements of the transcription of the indicated genes after AKI induction in ptsI and ptsH deletion mutants relative to the wt. gyrB expression was used as an internal control. Triplicate experiments were conducted, and the relative expression levels compared to the level of the wt strain are shown as means ± standard deviations.
A chromosomal PctxA-gfp transcriptional fusion (566 bp upstream of the ctxA start codon fused with gfp) was engineered and introduced into wild-type and PTS mutant strains to quantify the fraction of cells expressing ctxA after induction under AKI conditions. Consistent with the Western analysis results, ctxA reporter activity was reduced relative to that of the wt strain in the absence of either ptsI or ptsH. In the flow cytometry assay, ∼68% of wt cells were found to express the PctxA-gfp fusion, whereas only 21% and 26% of the ΔptsI and ΔptsH mutant cells, respectively, expressed the fusion (see Fig. S4 in the supplemental material). The magnitude of the reduction in the fraction of cells expressing the PctxA-gfp fusion in the absence of the PTS genes is very similar to that observed above with the PtcpA-gfp fusion (Fig. 4A), suggesting that PTS regulates a factor that modulates expression of both virulence genes in similar ways.
Diminished production of both CT and TcpA in the PTS mutants could result from reduced amounts/activity of ToxT, the transcription activator that directly regulates production of both factors (5, 6). An arabinose-inducible toxT expression vector was introduced into the PTS mutants to explore this possibility. Ectopic expression of ToxT in the ΔptsI and ΔptsH mutant strains restored production of CT and TcpA to wild-type levels (Fig. 5A). These observations are consistent with the idea that defective induction of ToxT expression/activity explains the reduced production of CT and TcpA in the PTS mutants under AKI conditions.
To begin to investigate how ptsI and ptsH promote ToxT-dependent activation of CT and TcpA expression, we compared the transcript levels of several components of the V. cholerae virulence cascade in the wt and PTS mutant strains. Our qRT-PCR assays showed that ToxT transcripts were >2-fold less abundant in the PTS mutants than in the wt under AKI conditions, consistent with our supposition above that PTS promotes expression of toxT (Fig. 5B), and that expression of ctxA and tcpA was reduced to a greater extent (∼6-fold). In addition, we detected reduced levels of transcripts of tcpP, tcpH, and aphA (Fig. 5B), suggesting that ptsI and ptsH promote expression of toxT by augmenting expression of these ToxT activators. In contrast, the defect in toxT expression in the absence of the PTS genes cannot be attributed to defective toxR or toxS expression since there was a slight increase in the levels of their transcripts in the PTS mutants.
Since cAMP-CRP is known to repress tcpPH expression, we also used qRT-PCR to measure the expression of crp, cyaA (which encodes adenylate cyclase), and cpdA (which encodes cAMP phosphodiesterase, a cAMP hydrolase) in the PTS mutants. Expression of cyaA and crp was elevated, and cpdA was reduced in the mutants (Fig. 5B), suggesting that the reduced expression of tcpPH in the PTS mutants could result from higher levels of cAMP-CRP in these cells.
PTS regulation of TcpA expression is partially dependent on cAMP-CRP.
To further explore the possibility that elevated cAMP-CRP in the PTS mutants might account for their diminished TcpA expression, we generated strains with ptsI or ptsH mutations along with deletions in cyaA, crp, or cpdA and performed epistasis analyses. TcpA expression in the double mutants was compared to that observed in wt cells and in the single mutants after growth under AKI conditions. Providing additional validation for our TIS sequencing screen, in which cpdA mutants were underrepresented after selection (see Table S2 in the supplemental material), the cpdA mutant had a modest reduction in TcpA expression in immunoblots and flow cytometry with the PtcpA-gfp reporter (Fig. 6A; see also Fig. S5), whereas the single cyaA and crp mutants exhibited TcpA expression similar to that of the wt strain. Thus, in a PTS-positive background, wt levels of cAMP-CRP appear to have minimal influence on TcpA expression under AKI conditions. However, deletion of cyaA or crp in the ptsI or ptsH single mutant increased TcpA expression although not to wild type levels (Fig. 6A; see also Fig. S5 in the supplemental material). These findings suggest that in the absence of EI or Hpr, cAMP-CRP represses TcpA expression. Consistent with this idea, in the EI or Hpr mutants, intracellular cAMP concentrations were significantly higher than the concentration in wt cells (Fig. 6B). Thus, it appears that the absence of PtsH or PtsI leads to increases in cAMP-CRP that are sufficiently large to repress expression of tcpA. Deletion of cpdA in the ptsH and ptsI mutants leads to further increases in cAMP levels (Fig. 6B), as well as a further reduction in TcpA (Fig. 6A).
To further define the regulatory interplay between cAMP-CRP, PTS, and TcpA expression, we used qRT-PCR to assess the transcript abundance of the key regulators AphA and TcpP in a variety of our mutants. As previously noted, both aphA and tcpP expression were significantly downregulated in ΔptsI and ΔptsH mutants compared to the wt levels (Fig. 5B and 6C). Deletion of cpdA in these backgrounds caused a further reduction in tcpP transcripts, consistent with the elevated level of the repressive cAMP-CRP. In contrast, deletion of either cyaA or crp in the ptsH and ptsI mutants resulted in increased tcpP transcripts, while levels of aphA transcripts remained low. Coupled with our previous analyses of TCP production in these backgrounds, our analyses suggest that disruption of the PTS in V. cholerae reduces production of TCP by reducing expression of several of its activators (TcpP, AphA, and ToxT) but that increased expression of TcpP and ToxT (e.g., by removing transcriptional repression by cAMP-CRP) is largely sufficient to restore wt levels of TCP production to the pts mutants. However, our results also suggest that parameters in addition to tcpP and toxT expression are important for controlling TcpA production in the double mutants lacking ptsI or ptsH and cyaA or crp since the transcript levels of tcpP and toxT, which are higher in these mutants than wt levels, do not lead to TcpA production that is higher than the wt level. It is not clear from these analyses why aphA transcript abundance is altered in the PTS mutants. The fact that disruption of crp or cyaA in the ptsI and ptsH mutants restored their production of TcpA to nearly wt levels allowed us to explore whether the colonization deficiency of the ptsI and ptsH mutants in infant mice was solely due to reduced production of TCP or whether it might also reflect an independent need for specific sugar transport pathways in vivo. A single-infection protocol, rather than a competitive assay, was used for these experiments to avoid any possibility of complementation in trans by a competing strain. Disruption of cyaA was used as the means of elevating TCP production as disruption of crp had a far more marked effect on bacterial growth (see Fig. S6 in the supplemental material). The ΔcyaA strain, like the ΔptsI and ΔptsH mutants, exhibited marked colonization deficits (∼40 to 50 times fewer CFU recovered from the intestine) compared with colonization of the wt strain (Fig. 6D). However, despite the ability of the ΔcyaA ΔptsI and ΔcyaA ΔptsH double mutants to produce TCP at nearly wt levels (at least under AKI conditions), these mutants had a more severe colonization deficit in vivo than the ΔcyaA strain (Fig. 6D), arguing against the idea that the colonization defect of the PTS mutants is solely attributable to defective TcpA expression. However, it is difficult to draw a robust conclusion from these experiments since the pathways for TCP induction in vivo may not precisely parallel those under AKI growth conditions.
DISCUSSION
Since the discovery of ToxR 30 years ago (48, 49), many studies have elucidated the large set of regulatory factors that V. cholerae employs to coordinate expression of its virulence factors during infection. This extensive body of work has revealed that many environmental and cellular factors modulate the cascade of transcription factors that ultimately lead to the expression and activity of ToxT, the key transcription activator that induces expression of genes for biosynthesis of TCP and cholera toxin. Here, we utilized a high-throughput TIS sequencing-based strategy to identify new regulators of TcpA expression. We identified many of the known regulators of TcpA (Table 1; see also Table S2 in the supplemental material), lending validity to our approach. ptsI and ptsH, linked genes encoding the EI and Hpr components of the V. cholerae PTS, also answered the screen, and additional experiments established that these proteins, along with EIIAGlc, are indeed required for robust TcpA expression in AKI medium and for intestinal colonization of infant mice. The V. cholerae PTS promotes expression of tcpPH, aphA, and aphB, all upstream activators of toxT, as well as of toxT itself (Fig. 5B). The mechanisms by which PTS promotes expression of these upstream activators require additional investigation; however, elevated levels of cAMP-CRP in the PTS mutants (Fig. 6B) appear to contribute by augmenting repression of tcpPH expression. Thus, in aggregate our work reveals that the V. cholerae PTS is an additional modulator of the ToxT regulon and suggests a new link between V. cholerae cell metabolism and the control of virulence.
PTSs have been implicated in control of virulence in several additional Gram-negative and Gram-positive pathogens (23, 50, 51). Notably, at least in some cases, similar to our findings in V. cholerae, PTSs have been found to regulate the expression/activity of critical virulence regulators, e.g., Mga in Streptococcus pyogenes (52) and PrfA in Listeria monocytogenes (53), through various mechanisms. Deciphering the precise mechanism(s) by which the V. cholerae PTS modulates toxT activation is complex because the PTS impinges on many aspects of the control of cell metabolism (23, 24). In addition to importing sugars, PTS proteins interact with and modulate the activity of a variety of transcription factors and enzymes, including adenylate cyclase and CRP (23, 43). Providing further complexity, the binding partners of PTS proteins differ depending on their phosphorylation states, which reflect the amounts of PTS sugars in the environment as well as the ratio of the intracellular pool of PEP to pyruvate (24). We found that mutants harboring nonphosphorylatable forms of EI and Hpr have defects in TcpA expression and intestinal colonization that are similar to those of the ptsI and ptsH null mutants (Fig. 2B and 3B), suggesting that the phosphorylation events are also essential for the PTS modulation of TCP expression.
In part, the reduced expression of tcpA in the ptsI and ptsH mutants likely reflects the increased levels of cAMP in these strains (Fig. 6B). In conjunction with CRP, cAMP represses expression of tcpP, a key transcription activator required for induction of toxT, the master regulator of virulence gene expression in V. cholerae. Levels of cAMP rise in response to nutrient depletion, and thus their increase in the ptsI and ptsH mutants may reflect their inability to utilize a subset of nutrients. cAMP-CRP has also been proposed to regulate the quorum sensing regulator HapR, which governs expression of aphA, another activator of toxT (54). However, in contrast to tcpP, expression of aphA did not rise in conjunction with the reduction of cAMP in strains lacking ptsI or ptsH and cyaA or crp, suggesting that an alternate process may underlie the PTS mutants' reduced expression of aphA.
Several factors likely contribute to the colonization defect of the EI, Hpr, and EIIAGlc mutants. The reduction in expression of TcpA and presumably other ToxT-regulated colonization factors that is observed under AKI conditions likely occurs in vivo as well and could in principal be the sole explanation for the attenuated colonization of the PTS mutants. However, our analysis of the PTS mutants also lacking cyaA or crp, which produce TcpA at close to wt levels, suggests that decreased TCP production is not the only factor underlying reduced growth of the V. cholerae PTS mutants in vivo. Mutants lacking EI, Hpr, or EIIAGlc are unable to import NAG, sucrose, and trehalose and have a reduced capacity to import glucose (44) (Fig. 3A), so the impaired in vivo growth of these mutants may also reflect reliance by V. cholerae on one or more of these sugars as a significant carbon source during infection. Our results indicate that V. cholerae growth in the infant mouse intestine does not depend on utilization of mannitol or fructose since strains harboring deletions in the genes enabling their import (VCA1045 and VCA0518) were not attenuated (Fig. 3A and C). Similarly, our observations indicate that the activity of the nitrogen PTS is dispensable for intestinal colonization (Fig. 3A and C).
Using a ligated ileal loop model, Nielsen and colleagues discovered that there is heterogenous (bifurcated) expression of tcpA and ctx, particularly late in infection in this model (46). Our observation that only a fraction of V. cholerae cells express tcpA during infection of infant mice demonstrates that bifurcation of expression of V. cholerae virulence factors is not explained by the closed nature of ileal loops but, instead, is likely a feature common to V. cholerae growth in the intestine (Fig. 4B; see also Fig. S3 in the supplemental material). Nielsen et al. hypothesized that the bimodal expression of V. cholerae virulence genes is mediated by a bistable switch that governs toxT expression (46). In their experiments, they added bicarbonate to LB-grown exponential-phase cultures to induce tcpA expression and observed that a subset of cells shut off TCP production in a cAMP- and CRP-dependent manner as cultures entered stationary phase (46). In contrast to the findings of Nielsen et al., we found that under AKI conditions, using a different strain, deletion of cyaA or crp did not have a major influence on tcpA expression (Fig. 6A). Notably, we found that the ptsI and ptsH mutants diminished the fraction of cells that expressed tcpA and ctxA by nearly one-third but did not abolish the bifurcated expression pattern of these virulence genes. Intriguingly, combination of a cyaA or crp deletion with the PTS mutation elevated the fraction of cells expressing tcpA compared to that in the PTS mutants (Fig. 6A; see also Fig. S5 in the supplemental material), raising the possibility that under AKI conditions the levels of the cAMP-CRP complex modify the set point of bistability. Since the levels of this transcription factor can influence the expression of many genes that directly (e.g., tcpPH) or indirectly (e.g., rpoS) control toxT expression, in the PTS mutant background cAMP-CRP levels may adjust the site of the “fulcrum” of the bistable switch governing ToxT expression.
Finally, our simple loss-of-function TIS sequencing-based approach to define the set of genes that constitute the pathway leading to tcpA expression should be easily adaptable for determining additional virulence-regulatory networks in other pathogens as well as other regulatory pathways. McDonough and colleagues used a similar approach to identify the regulators of xds in V. cholerae (55). Thus, identifying regulatory factors that are required for expression of a gene of interest is as simple as engineering a strain containing a selectable marker (e.g., an antibiotic resistance gene, as we used here) or potentially a screenable marker (e.g., GFP) fused to a promoter of interest. While transposon insertion mutagenesis approaches have been used for many years, the application of high-throughput sequencing to transposon insertion mutagenesis enables robust loss-of-function screens that rely on cell growth as their output. Given adequate density of the transposon libraries, such screens can potentially provide complete definition of regulatory pathways in a single experiment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paula Watnick from Boston Children's Hospital for generously providing plasmids for the complementation experiments and Yoshiharu Yamaichi for construction of the modified pSC189-Cm plasmid.
Research in the Waldor laboratory is supported by Howard Hughes Medical Institute (HHMI) and National Institutes of Health grants (AI R37-042347 to M.K.W.; 5F32 GM108355-02 to M.C.C.). Q.W. is supported by a fellowship from the China Scholarship Council (201308310080) and Shanghai Rising-Star Program (13QA1401000).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00411-15.
REFERENCES
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim MS, Ng D, Zong Z, Arvai AS, Taylor RK, Tainer JA, Craig L. 2010. Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol Microbiol 77:755–770. doi: 10.1111/j.1365-2958.2010.07244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matson JS, Withey JH, DiRita VJ. 2007. Regulatory networks controlling Vibrio cholerae virulence gene expression. Infect Immun 75:5542–5549. doi: 10.1128/IAI.01094-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiRita VJ, Parsot C, Jander G, Mekalanos JJ. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higgins DE, Nazarend E, DiRita VJ. 1992. The virulence gene activator ToxT from Vibio cholerae is a member of AraC family of transcriptional activators. J Bacteriol 174:6974–6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 8.Kovacikova G, Lin W, Skorupski K. 2010. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol 192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin W, Kovacikova G, Skorupski K. 2007. The quorum sensing regulator HapR downregulates the expression of the virulence gene transcription factor AphA in Vibrio cholerae by antagonizing Lrp- and VpsR-mediated activation. Mol Microbiol 64:953–967. doi: 10.1111/j.1365-2958.2007.05693.x. [DOI] [PubMed] [Google Scholar]
- 10.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. 1986. Culture conditions for stimulating cholera toxin production by Vibrio cholerae O1 El Tor. Microbiol Immunol 30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 11.Kanjilal S, Citorik R, LaRocque RC, Ramoni MF, Calderwood SB. 2010. A systems biology approach to modeling Vibrio cholerae gene expression under virulence-inducing conditions. J Bacteriol 192:4300–4310. doi: 10.1128/JB.00182-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SH, Hava DL, Waldor MK, Camilli A. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625–634. doi: 10.1016/S0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 13.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abuaita BH, Withey JH. 2009. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect Immun 77:4111–4120. doi: 10.1128/IAI.00409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. 2011. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci U S A 108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowden MJ, Skorupski K, Pellegrini M, Chiorazzo MG, Taylor RK, Kull FJ. 2010. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc Natl Acad Sci U S A 107:2860–2865. doi: 10.1073/pnas.0915021107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber GG, Kortmann J, Narberhaus F, Klose KE. 2014. RNA thermometer controls temperature-dependent virulence factor expression in Vibrio cholerae. Proc Natl Acad Sci U S A 111:14241–14246. doi: 10.1073/pnas.1411570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popovych N, Tzeng S-R, Tonelli M, Ebright RH, Kalodimos CG. 2009. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci U S A 106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skorupski K, Taylor RK. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci U S A 94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovacikova G, Skorupski K. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol 41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 22.Minato Y, Fassio SR, Wolfe AJ, Hase CC. 2013. Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology 159:792–802. doi: 10.1099/mic.0.064865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deutscher J, Aké FMD, Derkaoui M, Zébré AC, Cao TN, Bouraoui H, Kentache T, Mokhtari A, Milohanic E, Joyet P. 2014. The bacterial phosphoenolpyruvate: carbohydrate phosphotransferase system: regulation by protein phosphorylation and phosphorylation-dependent protein-protein interactions. Microbiol Mol Biol Rev 78:231–256. doi: 10.1128/MMBR.00001-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev 70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 26.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59:4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dörr T, Lam H, Alvarez L, Cava F, Davis BM, Waldor MK. 2014. A novel peptidoglycan binding protein crucial for Pbp1A-mediated cell wall biogenesis in Vibrio cholerae. PLoS Genet 10:e1004433. doi: 10.1371/journal.pgen.1004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, Waldor MK. 2013. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res 41:9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog 8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang YJ, Reddy MC, Ioerger TR, Rothchild AC, Dartois V, Schuster BM, Trauner A, Wallis D, Galaviz S, Huttenhower C, Sacchettini JC, Behar SM, Rubin EJ. 2013. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Pang B, Jiang X, Kan B. 2014. Selection of reference genes for gene expression analysis in Vibrio cholerae. Chin J Zoonoses 30:433–438. [Google Scholar]
- 34.Fuchs EL, Brutinel ED, Klem ER, Fehr AR, Yahr TL, Wolfgang MC. 2010. In vitro and in vivo characterization of the Pseudomonas aeruginosa cyclic AMP (cAMP) phosphodiesterase CpdA, required for cAMP homeostasis and virulence factor regulation. J Bacteriol 192:2779–2790. doi: 10.1128/JB.00168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelichio MJ, Spector J, Waldor MK, Camilli A. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun 67:3733–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DiRita VJ, Neely M, Taylor RK, Bruss PM. 1996. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc Natl Acad Sci U S A 93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merrell DS, Hava DL, Camilli A. 2002. Identification of novel factors involved in colonization and acid tolerance of Vibrio cholerae. Mol Microbiol 43:1471–1491. doi: 10.1046/j.1365-2958.2002.02857.x. [DOI] [PubMed] [Google Scholar]
- 39.Chiang SL, Mekalanos JJ. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol 27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 40.Dörr T, Moll A, Chao MC, Cava F, Lam H, Davis BM, Waldor MK. 2014. Differential requirement for Pbp1a and Pbp1b in in vivo and in vitro fitness of Vibrio cholerae. Infect Immun 82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cameron DE, Urbach JM, Mekalanos JJ. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J Bacteriol 192:3055–3067. doi: 10.1128/JB.00213-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pickering BS, Smith DR, Watnick PI. 2012. Glucose-specific enzyme IIA has unique binding partners in the Vibrio cholerae biofilm. mBio 3(6):e00228-12. doi: 10.1128/mBio.00228-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect Immun 78:1482–1494. doi: 10.1128/IAI.01356-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Houot L, Watnick PI. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J Bacteriol 190:311–320. doi: 10.1128/JB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog 6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Millet YA, Alvarez D, Ringgaard S, Andrian von UH, Davis BM, Waldor MK. 2014. Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog 10:e1004405. doi: 10.1371/journal.ppat.1004405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller VL, Taylor RK, Mekalanos JJ. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 49.Miller VL, Mekalanos JJ. 1985. Genetic analysis of the cholera toxin-positive regulatory gene toxR. J Bacteriol 163:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mazé A, Glatter T, Bumann D. 2014. The central metabolism regulator EIIAGlc switches Salmonella from growth arrest to acute virulence through activation of virulence factor secretion. Cell Rep 7:1426–1433. doi: 10.1016/j.celrep.2014.04.022. [DOI] [PubMed] [Google Scholar]
- 51.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 52.Hondorp ER, Hou SC, Hause LL, Gera K, Lee CE, McIver KS. 2013. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A Streptococcus. Mol Microbiol 88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mertins S, Joseph B, Goetz M, Ecke R, Seidel G, Sprehe M, Hillen W, Goebel W, Muller-Altrock S. 2007. Interference of components of the phosphoenolpyruvate phosphotransferase system with the central virulence gene regulator PrfA of Listeria monocytogenes. J Bacteriol 189:473–490. doi: 10.1128/JB.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 55.McDonough E, Lazinski DW, Camilli A. 2014. Identification of in vivo regulators of the Vibrio cholerae xds gene using a high-throughput genetic selection. Mol Microbiol 92:302–315. doi: 10.1111/mmi.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rui H, Ritchie JM, Bronson RT, Mekalanos JJ, Zhang Y, Waldor MK. 2010. Reactogenicity of live-attenuated Vibrio cholerae vaccines is dependent on flagellins. Proc Natl Acad Sci U S A 107:4359–4364. doi: 10.1073/pnas.0915164107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.