Abstract
Aims: Obesity is characterized by a low-grade systemic inflammatory state and adipose tissue (AT) dysfunction, which predispose individuals to the development of insulin resistance (IR) and metabolic disease. However, a subset of obese individuals, referred to as metabolically healthy obese (MHO) individuals, are protected from obesity-associated metabolic abnormalities. Here, we aim at identifying molecular factors and pathways in adipocytes that are responsible for the progression from the insulin-sensitive to the insulin-resistant, metabolically unhealthy obese (MUHO) phenotype. Results: Proteomic analysis of paired samples of adipocytes from subcutaneous (SC) and omental (OM) human AT revealed that both types of cells are altered in the MUHO state. Specifically, the glutathione redox cycle and other antioxidant defense systems as well as the protein-folding machinery were dysregulated and endoplasmic reticulum stress was increased in adipocytes from IR subjects. Moreover, proteasome activity was also compromised in adipocytes of MUHO individuals, which was associated with enhanced accumulation of oxidized and ubiquitinated proteins in these cells. Proteasome activity was also impaired in adipocytes of diet-induced obese mice and in 3T3-L1 adipocytes exposed to palmitate. In line with these data, proteasome inhibition significantly impaired insulin signaling in 3T3-L1 adipocytes. Innovation: This study provides the first evidence of the occurrence of protein homeostasis deregulation in adipocytes in human obesity, which, together with oxidative damage, interferes with insulin signaling in these cells. Conclusion: Our results suggest that proteasomal dysfunction and impaired proteostasis in adipocytes, resulting from protein oxidation and/or misfolding, constitute major pathogenic mechanisms in the development of IR in obesity. Antioxid. Redox Signal. 23, 597–612.
Introduction
Understanding fat cell biology has gained intensive scientific attention during the past decades as the prevalence of obesity is increasing worldwide and constitutes a major public health issue (2). It is well established that adipocyte hypertrophy and expansion of fat mass results in adipose tissue (AT) fibrosis and dysfunction, causing the activation of inflammatory cascades in adipocytes [i.e., c-Jun NH2-terminal kinase and nuclear factor κB (NF-κB)] and the secretion of pro-inflammatory cytokines, which, in turn, trigger AT infiltration by macrophages and other immune cells (8, 36, 42, 64). Low-grade chronic inflammation and adipocyte dysfunction induce insulin resistance (IR), thus increasing the risk for the development of type 2 diabetes, cardiovascular disease, hepatic steatosis, hypertension, and cancer (21, 53). At the molecular level, obesity-related pathogenic factors causing adipocyte dysfunction include lipid and adipokine deregulation, mitochondrial malfunction, increased autophagy, and induction of cell stress processes [oxidative-, nitrosative-, and endoplasmic reticulum (ER)-stress] (8, 25).
Innovation.
Increasing evidence demonstrates that impairment of protein homeostasis and proteotoxicity are major contributors to the pathogenesis of multiple disorders, including neurodegenerative diseases and other age-related diseases. Nonetheless, whether proteasome function is dysregulated in adipocytes in obesity remained to be determined. Here, we propose a model in which proteasome dysfunction in adipocytes contributes to the accumulation of oxidatively damaged and unfolded proteins, leading to the activation of inflammatory pathways and impaired insulin signaling. We suggest that proteasome impairment along with oxidative damage and endoplasmic reticulum stress in adipocytes contribute to the onset and/or progression of insulin resistance in obesity.
A subset of obese individuals, known as “metabolically healthy obese” (MHO) subjects, seems to be resistant to obesity-associated metabolic complications (17). These individuals, which comprise between 20 and 30% of the adult obese population, are mainly characterized by (i) a lean-like adipokine pattern (i.e., normal adiponectin levels), (ii) reduced central body fat and AT fibrosis, (iii) diminished AT macrophage infiltration and inflammatory profile, and (iv) lower ectopic fat deposition (17, 39, 48), as compared with metabolically unhealthy obese (MUHO) individuals. Likewise, activation of pro-inflammatory signaling effectors challenging insulin sensitivity, such as NF-κB, in omental AT has been also proposed as a discriminative factor of MHO and MUHO individuals (6). However, the molecular components and processes underlying the favorable metabolic profile of MHO individuals are still poorly understood.
We, therefore, sought to (i) identify novel AT markers associated with the MUHO phenotype by comparing the proteomic profiles of mature adipocytes from human subcutaneous (SC) and omental (OM) fat of morbidly obese individuals with different degrees of insulin sensitivity, and (ii) to investigate the potential contribution of protein homeostasis deregulation to the pathogenesis of IR in obesity.
Results
Baseline clinical and metabolic characteristics of subjects
The clinical characteristics of normoglycemic (NG) (n=18) and insulin-resistant (IR) (n=13) morbidly obese subjects (body mass index [BMI]>30 kg/m2) are shown in Table 1. Both groups were matched by age, BMI, and percent body fat. Average blood pressure values were similar in the two groups, yet some subjects in the IR group exhibited hypertension (4 out of 13 subjects). Fasting plasma glucose and insulin levels were significantly higher in insulin-resistant individuals. These subjects also exhibited higher homeostasis model assessment of insulin resistance (HOMA-IR) values and waist circumference than NG individuals. Plasma cholesterol, FFA, and triglycerides were higher in IR individuals than in NG subjects, although differences did not reach statistical significance.
Table 1.
Anthropometric and Analytical Characteristics of Subjects
| NG (n=18) | IR (n=13) | |
|---|---|---|
| Sex (men/women) | 7/11 | 4/9 |
| Age (years) | 39±6.2 | 37±5.4 |
| BMI (kg/m2) | 51.0±10.7 | 55.0±13.4 |
| Waist circumference (cm) | 136.5±20.3 | 157.7±20.0a |
| Sistolic pressure (mm/Hg) | 127.4±6.6 | 129.0±17.5 |
| Diastolic pressure (mm/Hg) | 77.8±11.7 | 77.1±14.5 |
| Cholesterol (mg/dl) | 180.3±28.3 | 194.8±31.7 |
| LDL (mg/dl) | 120.3±25.6 | 127.2±30.2 |
| HDL (mg/dl) | 49.8±19.4 | 39.8±6.8 |
| Triglycerides (mg/dl) | 97.0±31.9 | 117.5±37.5 |
| FFA (mM) | 62.5±14.7 | 80.3±25.9 |
| Glucose (mg/dl) | 92.0±9.3 | 106.8±6.5b |
| Insulin (mU/L) | 10.2±3.8 | 29.3±8.1b |
| HbA1c (% [mmol/mol]) | 4.85 (25.0)±1.7 (17.7) | 6.34 (40)±0.51 (5.1)a |
| HOMA-IR | 2.30±0.87 | 7.61±2.08b |
Data represent the mean±SD.
p<0.01.
p<0.001.
BMI, body mass index; LDL, low-density lipoprotein; HDL, high-density lipoprotein; FFA, free fatty acid; HbA1c, glycated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance.
Proteomic profile of adipocytes from NG and IR individuals
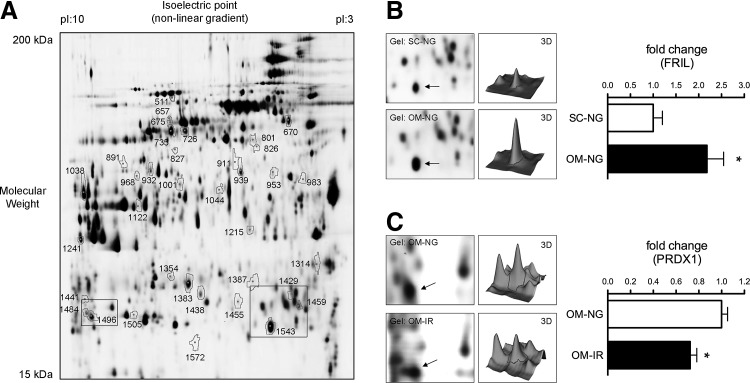
A multi-comparative proteomic analysis was employed to identify adipocyte proteins that are differentially expressed between SC and OM fat of NG versus IR individuals. From the 1687 protein spots identified in the master gel, 49 spots were differentially expressed between groups (p<0.05) (Fig. 1A–C). Among them, 40 spots were identified by MALDI-TOF protein identification (Table 2). Of the identified proteins, 11 proteins were found to be differentially expressed between SC and OM adipocytes in NG subjects (3 upregulated and 8 down-regulated), and 3 proteins were downregulated in SC adipocytes of IR individuals. Proteomic comparison of adipocytes from NG and IR subjects revealed significant changes in 20 proteins (12 upregulated and 8 downregulated) and 6 proteins (3 upregulated and 3 downregulated) in SC and OM adipocytes, respectively. Of note, 16 out of the 20 differentially expressed proteins in SC adipocytes of NG and IR subjects were also regulated in OM adipocytes, although differences did not reach statistical significance (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars).
FIG. 1.
Two-dimensional DIGE analysis of human adipocytes. (A) Representative image of a 2D-DIGE gel of protein extracts from subcutaneous (SC) and omental (OM) adipocytes from normoglycemic (NG) and insulin-resistant (IR) subjects. Spots correspond to the proteins in Table 2. (B, C) Magnifications of the rectangle–delimited areas in (A) and 3D-fluorescence intensity profiles of the proteins FRIL (B) and PRDX1 (C). Spots corresponding to FRIL and PRDX1 in the gels are indicated by arrows. Histograms show quantitative fold changes in protein expression. Mean±SEM of n=4 individuals/group; *p<0.05 versus NG.
Table 2.
Proteins Identified by MALDI-MS/MS Differentially Expressed in Mature Subcutaneous and Omental Adipocytes from Normoglycemic and Insulin-Resistant Morbidly Obese Individuals
| DIGE | Protein | Mascot | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Spota | T-testb | Av ratioc | Accession numberd | Symbol | Name | Mw (kDa)/pIe | Pepf | % coverg | Expecth |
| Proteomic comparison of SC vs. OM adipocytes from NG subjects | |||||||||
| 1429 | 0.027 | 1.60 | CAA00975 | APOA1 | Apolipoprotein A-I | 30.7/5.5 | 21 | 54 | 2.0e-023 |
| 1505 | 0.015 | 1.43 | NP_000468 | ALBU | Serum albumin | 71.3/5.9 | 15 | 21 | 1.0e-023 |
| 1572 | 0,025 | 1.97 | Q13418 | ILK | Integrin-linked protein kinase | 51.8/8.3 | 9 | 16 | 4.0e-009 |
| 1543 | 0.040 | −2.18 | NP_000137 | FRIL | Ferritin light chain | 20.0/5.5 | 21 | 65 | 2.0e-067 |
| 726 | 0.018 | −2.45 | NP_000680 | AL1A1 | Retinal dehydrogenase 1 | 55.4/6.3 | 21 | 38 | 2.0e-037 |
| 735 | 0.029 | −2.56 | NP_000680 | AL1A1 | Retinal dehydrogenase 1 | 55.4/6.3 | 25 | 54 | 1.6e-045 |
| 953 | 0.026 | −1.37 | NP_001814 | KCRB | Creatine kinase B-type | 42.9/5.3 | 13 | 34 | 6.4e-010 |
| 1354 | 0.043 | −1.48 | NP_002620 | PGAM1 | Phosphoglycerate mutase 1 | 28.9/6.6 | 9 | 45 | 0.00018 |
| 1484 | 0.029 | −1.42 | NP_000467 | KAD1 | Adenylate kinase isoenzyme 1 | 21.7/8.7 | 18 | 65 | 8.1e-028 |
| NP_000468 | ALBU | Serum albumin | 71.3/5.9 | 9 | 15 | 1.3e-009 | |||
| 1459 | 0.025 | −1.99 | P63261 | ACTG | Actin, cytoplasmic 2 | 42.1/5.3 | 11 | 34 | 2.0e-020 |
| 968 | 0.050 | −1.50 | NP_062800 | IVD | Isovaleryl-CoA dehydrogenase, mitochondrial | 46.8/8.4 | 16 | 34 | 3.1e-006 |
| Proteomic comparison of SC vs. OM adipocytes from IR subjects | |||||||||
| 670 | 0.046 | −1.72 | NP_955472 | CH60 | 60 kDa heat shock protein, mitochondrial | 61.1/5.7 | 23 | 53 | 2.5e-049 |
| 675 | 0.049 | −1.77 | NP_000680 | AL1A1 | Retinal dehydrogenase 1 | 55.4/6.3 | 19 | 36 | 8.1e-029 |
| 801 | 0.007 | −2.31 | P21399 | ACOC | Cytoplasmic aconitate hydratase | 98.8/6.2 | 20 | 18 | 1.0e-026 |
| Proteomic comparison of SC adipocytes of IR vs. NG subjects | |||||||||
| 826 | 0.038 | 1.32 | NP_000169 | GSHB | Glutathione synthetase | 52.5/5.6 | 13 | 31 | 1.3e-009 |
| 1438 | 0.038 | 2.66 | P28161 | GSTM2 | Glutathione S-transferase Mu 2 | 25.8/6.0 | 30 | 83 | 1.3e-048 |
| 1387 | 0.025 | 2.14 | NP_006397 | PRDX4 | Peroxiredoxin-4 | 30.7/5.8 | 16 | 58 | 2.5e-031 |
| 924 | 0.039 | 1.91 | Q99536 | VAT1 | Synaptic vesicle membrane protein VAT-1 | 42.1/5.8 | 13 | 40 | 9.0e-005 |
| 1484 | 0.003 | 2.81 | NP_000467 | KAD1 | Adenylate kinase isoenzyme 1 | 21.7/8.7 | 18 | 65 | 8.1e-028 |
| NP_000468 | ALBU | Serum albumin | 71.3/5.9 | 9 | 15 | 1.3e-009 | |||
| 511 | 0.020 | 5.71 | NP_000399 | GPDM | Glycerol-3-phosphate dehydrogenase, mitochondrial | 81.3/7.5 | 24 | 34 | 2.0e-014 |
| 827 | 0.010 | 2.34 | NP_000009 | ACADV | Very long-chain specific acyl-CoA dehydrogenase, mitochondrial | 70.7/8.9 | 17 | 29 | 1.0e-028 |
| 983 | 0.041 | 23.42 | NP_003371 | VIME | Vimentin | 53.6/5.0 | 31 | 56 | 2.0e-040 |
| 1314 | 0.044 | 3.25 | CAA39992 | CALB2 | Calretinin | 31.6/5.0 | 11 | 36 | 1.1e-005 |
| 1122 | 0.026 | 2.30 | AAA18024 | FIBB | Fibrinogen beta chain | 56.5/8.5 | 27 | 43 | 1.6e-020 |
| 911 | 0.034 | 4.35 | Q9P2R7 | SUCB1 | Succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial | 50.6/7.0 | 18 | 29 | 5.1e-016 |
| 1441 | 0.050 | 3.30 | Q9Y6M9 | NDUB9 | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 | 22.0/8.5 | 10 | 48 | 4.0e-011 |
| 1455 | 0.043 | −1.55 | P48668 | K2C6C | Keratin, type II cytoskeletal 6C | 60.2/8.0 | 18 | 31 | 6.4e-023 |
| NP_009193 | PARK7 | Protein DJ-1/PARK7 | 20.0/6.3 | 9 | 60 | 0.0011 | |||
| 1241 | 0.024 | −2.60 | P15559 | NQO1 | NAD(P)H dehydrogenase [quinone] 1 | 30.9/8.9 | 16 | 44 | 3.2e-026 |
| 670 | 0.004 | −2.46 | NP_955472 | CH60 | 60 kDa heat shock protein, mitochondrial | 61.1/5.7 | 23 | 53 | 2.5e-049 |
| 1001 | 0.043 | −1.88 | NP_006588 | HSP7C | Heat shock cognate 71 kDa protein | 71.0/5.3 | 17 | 29 | 4.0e-025 |
| NP_005336 | HSP71 | Heat shock 70 kDa protein 1A/1B | 70.2/5,4 | 14 | 23 | 3.2e-016 | |||
| 1038 | 0.036 | −2.33 | CAG46678 | ALDOA | Fructose-bisphosphate aldolase A | 39.8/8.3 | 23 | 67 | 1.0e-050 |
| 932 | 0.036 | −1.66 | P11310 | ACADM | Medium-chain specific acyl-CoA dehydrogenase, mitochondrial | 47.0/8.6 | 27 | 51 | 1.0e-046 |
| 939 | 0.016 | −5.20 | NP_001257 | EST1 | Liver carboxylesterase 1 | 62.7/6.1 | 20 | 41 | 5.1e-034 |
| 657 | 0.050 | −3.59 | AAB29178 | PGM1 | Phosphoglucomutase-1 | 61.6/6.3 | 20 | 37 | 2.0e-016 |
| Proteomic comparison of OM adipocytes of IR vs. NG subjects | |||||||||
| 1572 | 0,025 | 1.9 | Q13418 | ILK | Integrin-linked protein kinase | 51.8/8.3 | 9 | 16 | 4.0e-009 |
| 1044 | 0.019 | 1.99 | CAA36267 | CO6A3 | Collagen alpha-3 (VI) chain | 34.5/8.9 | 1 | 4 | 0.00068 |
| 711 | 0.046 | 3.39 | NP_005304 | PDIA3 | Protein disulfide-isomerase A3 | 57.1/5.9 | 15 | 33 | 6.4e-013 |
| 1215 | 0.017 | −1.58 | NP_689953 | 3HIDH | 3-hydroxyisobutyrate dehydrogenase, mitochondrial | 35.7/8.3 | 14 | 33 | 5.1e-009 |
| 1496 | 0.014 | −1.38 | CAG28580 | PRDX1 | Peroxiredoxin-1 | 22.3/8.2 | 19 | 68 | 4.0e-046 |
| CAG46588 | PRDX2 | Peroxiredoxin-2 | 22.0/6.7 | 3 | 16 | 2.2e-006 | |||
| 1383 | 0.050 | −1.66 | NP_004896 | PRDX6 | Peroxiredoxin-6 | 25.1/6.0 | 26 | 83 | 1.6e-052 |
Spot numbers correspond to those in Figure 1A.
p-value of the Student's t-test. Only those proteins with a p≤0.05 were considered.
Average volume ratio (a,b,c were calculated by the DeCyder analysis).
Accession numbers from the NCBInr database.
Theoretical molecular weight (kDa) and isoelectric point (pI).
Pep. corresponds to the number of peptides identified.
Coverage of all peptide sequences matched to the identified protein (%).
Mascot expected value sequence (f,g,h were provided by Mascot).
The PANTHER classification analysis revealed that the identified proteins belonged to four major functional groups: (i) catalytic activity: SUCB1, 3HIDH, GSHB, VAT1, ACADM, PRDX6, GSTM2, ACADV, NDUB9, PRDX4, PGM1, ALDOA, PRDX2, KAD1, EST1, PRDX1, ILK, DJ1, and PDIA3; (ii) antioxidant activity: PRDX1, PRDX2, PRDX4, and PRDX6; (iii) binding: FIBB, EST1, CALB2, and DJ1; and (iv) structural molecule activity: CO6A3, K2C6C, and VIME (Supplementary Fig. S2). Statistical differences in the expression of proteins with antioxidant (PRDX1, PRDX2, PRDX4, and PRDX6) or oxidoreductase (PRDX1, PRDX2, 3HIDH, VAT1, PRDX4, ACADV, ACADM, PRDX6, and NDUB9) activities were found between NG and IR individuals (p<0.05).
Enhanced oxidative stress and failure of antioxidant defenses in adipocytes from IR subjects.
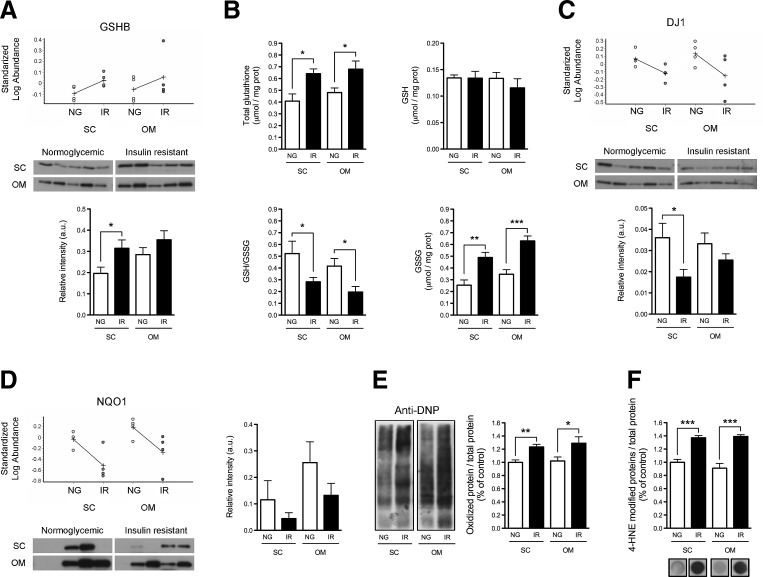
To confirm and extend the data on the oxidative status of adipocytes, we characterized the glutathione synthetase (GSHB)/glutathione system in SC and OM adipocytes from additional NG and IR subjects to those employed for the 2D-DIGE study. Consistent with the 2D-DIGE observations, GSHB expression was higher in SC adipocytes from IR individuals than from NG subjects (Fig. 2A). Accordingly, total glutathione levels were increased in both SC and OM adipocytes of IR subjects (Fig. 2B). Nevertheless, while the amount of reduced glutathione (GSH) was similar in NG and IR adipocytes, oxidized glutathione (GSSG) was significantly enhanced and the GSH/GSSG ratio was markedly reduced in adipocytes from IR subjects (Fig. 2B).
FIG. 2.
Analysis of antioxidant defenses and oxidative stress in SC and OM adipocytes from NG and IR subjects. (A) GSHB expression in isolated SC and OM adipocytes from NG and IR subjects. Standardized log. abundance of the spot was determined by DeCyder analysis of 2D-DIGE data (upper panels). Values indicate increased (+) or decreased (−) levels relative to the internal standards (n=4 individuals/group). Immunoblot images (middle panel) and quantification of GSHB expression (lower panel) are shown (n=5 individuals/group). (B) Total glutathione, reduced (GSH) and oxidized (GSSG) glutathione, and GSH/GSSG ratio in NG and IR adipocytes (n=5 individuals/group). (C, D) Protein expression of DJ1 (C) and NQO1 (NAD(P)H dehydrogenase [quinone] 1) (D) in SC and OM adipocytes from NG and IR subjects (n=4 individuals/group for 2D-DIGE and n=5 individuals/group for immunoblotting studies). (E, F) Quantification of carbonylated proteins (DNP-protein adducts; E) and 4-HNE-modified proteins (F: dot blots) in NG and IR adipocytes (n=5 individuals/group); protein levels in SC-NG adipocytes were set to 1 and used as reference for data normalization. *p<0.05, **p<0.01 , and ***p<0.001 versus NG.
We then analyzed the protein content of several antioxidant proteins identified by 2D-DIGE, including DJ1 (57) and the quinone reductase, NQO1 (18). Quantitative immunoblotting confirmed that the protein levels of DJ1 were decreased in SC adipocytes of IR subjects as compared with NG individuals (Fig. 2C). A similar trend was observed in OM adipocytes, although differences did not reach statistical significance (Fig. 2C). As observed by 2D-DIGE, protein levels of NQO1 were numerically lower in both SC and OM adipocytes of IR individuals compared with NG subjects, although differences did not reach statistical significance due to interindividual variability within groups (Fig. 2D).
In view of these results, we analyzed oxidative-induced damage on adipocyte proteins. As shown in Figure 2E and F, both the levels of carbonylated proteins and 4-Hydroxynonenal (4-HNE)-modified proteins were significantly increased in SC and OM adipocytes of IR individuals as compared with NG subjects. A positive correlation in carbonylated protein content was found between SC and OM adipocytes (Supplementary Fig. S3A).
IR is associated with impaired protein folding and decreased proteasome activity in adipocytes
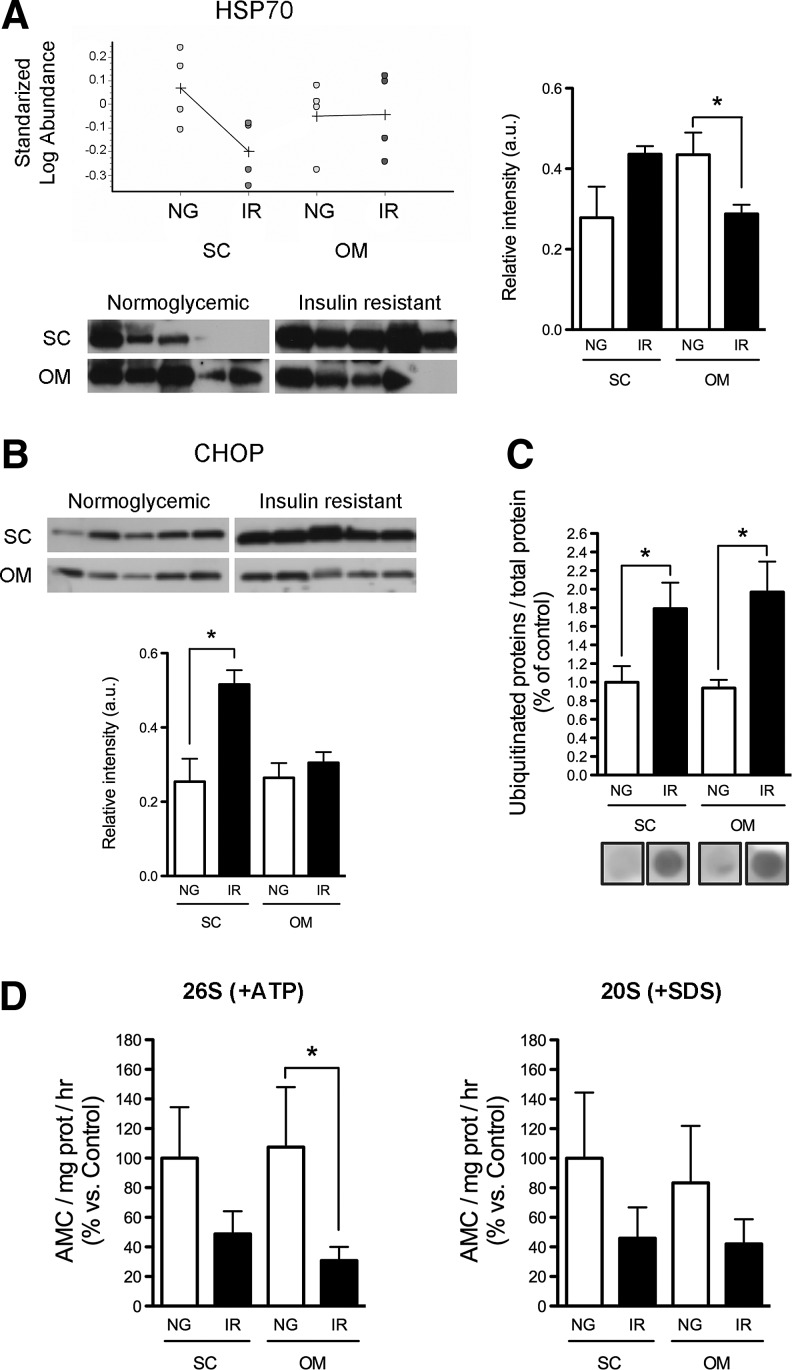
2D-DIGE analysis also revealed the differential expression of several molecular chaperones between groups (Table 2 and Supplementary Fig. S1). Quantitative immunoblotting showed that the protein levels of heat shock protein 70 (Hsp70/Hsc70) were decreased in OM adipocytes of IR subjects as compared with NG subjects (Fig. 3A). Contrary to that shown by 2D-DIGE, HSP70 levels in SC adipocytes of NG were not significantly different from those observed in IR subjects, likely due to interindividual variability (Fig. 3A).
FIG. 3.
Analysis of the protein folding, ER stress and degradation machineries in SC and OM adipocytes from NG and IR subjects. (A) HSP70 protein content in SC and OM adipocytes of NG and IR subjects (n=5 individuals/group). Standardized log. abundance of the spots in the 2D-DIGE gels (upper panels; n=4 individuals/group). (B) Protein content of the ER marker, CHOP, in adipocytes of NG and IR subjects (n=5 individuals/group). (C) Quantification of ubiquitinated proteins in NG and IR adipocytes (n=5 individuals/group); protein levels in SC-NG adipocytes in the dot blots were set to 1 and used as reference for data normalization. (D) Measurement of the chymotrypsin-like (ChT-L) peptidase activity of the 26S proteasome (+ATP) (left panel) and 20S proteasome (+SDS) (right panel) in SC and OM adipocytes of NG and IR subjects (n=5 individuals/group). Data are expressed as a percentage of values in SC-NG adipocytes (100%). *p<0.05 versus NG.
We also investigated ER stress in SC and OM adipocytes of the two groups of subjects. As shown in Figure 3B, protein levels of the ER stress marker, C/EBP homologous protein (CHOP), were significantly higher in SC adipocytes of IR individuals than in IR subjects. In all, these data were suggestive of dysregulated protein folding in IR adipocytes. In line with this, we observed higher levels of ubiquitinated proteins in adipocytes from IR subjects (Fig. 3C). Ubiquitinated protein content in SC and OM adipocytes was positively correlated (Supplementary Fig. S3B). Therefore, we next analyzed the capacity of IR adipocytes to preserve protein homeostasis and, in particular, we determined the activity of the proteasome. As shown in Figure 3D, IR adipocytes showed lower chymotrypsin-like (ChT-L) peptidase activities of the 26S and 20S proteasomes than NG adipocytes.
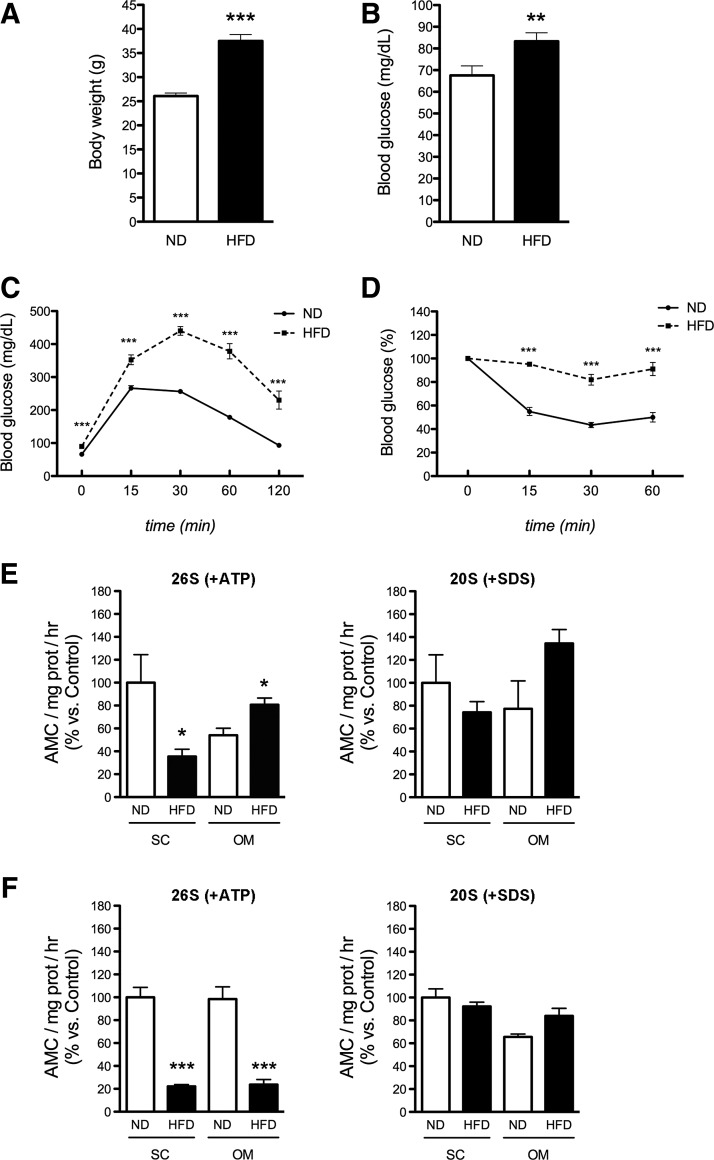
To validate the results obtained in human adipocytes, we examined the activity of the proteasome in adipocytes from mice fed a normal diet (ND) or a high-fat diet (HFD). As shown in Figure 4A, body weight was significantly higher in mice fed an HFD compared with ND-fed mice. Fasting plasma glucose levels were also increased in obese HFD-fed mice (Fig. 4B). Moreover, glucose (ipGTT; Fig. 4C) and insulin tolerance (ITT; Fig. 4D) tests demonstrated that mice fed an HFD exhibited lower glucose tolerance and insulin sensitivity than ND-fed animals. We next analyzed the activity of the proteasome in mature and in vitro differentiated adipocytes isolated from both SC and visceral AT of both groups. Our data showed that ChT-L peptidase activity of the 26S proteasome was significantly decreased in mature SC adipocytes of mice fed an HFD as compared with their ND counterparts (Fig. 4E). In contrast, the activity of the 26S proteasome was increased in mature visceral adipocytes of obese mice (Fig. 4E); 26S activity was markedly reduced in both SC and visceral in vitro differentiated adipocytes from HFD-fed mice (Fig. 4F).
FIG. 4.
Characterization of the proteasome system in mature adipocytes and in vitro differentiated adipocytes from ND and HFD mice. Body weight (A) and fasting glucose (B) of mice after 14 weeks of free access to a normal diet (ND) or a high-fat diet (HFD) (n=10). (C, D) Intraperitoneal glucose tolerance test (ipGTT; C) and insulin tolerance test (ITT; D) in ND and HFD mice (n=10). (E) Chymotrypsin-like (ChT-L) peptidase activity of the 26S proteasome (+ATP) (left panel) and 20S proteasome (+SDS) (right panel) in mature adipocytes isolated from subcutaneous and visceral fat of mice on ND or HFD (n=3; 3–4 animals/pool). (F) Chymotrypsin-like (ChT-L) peptidase activity of the 26S proteasome (+ATP) (left panel) and 20S proteasome (+SDS) (right panel) in in vitro differentiated adipocytes from SVFCs of subcutaneous and visceral fat of mice on ND and HFD (n=3; 3–4 animals/pool). *p<0.05, **p<0.01, and ***p<0.001 versus mice on ND.
Activation of c-Jun N-terminal kinase in adipocytes of IR subjects
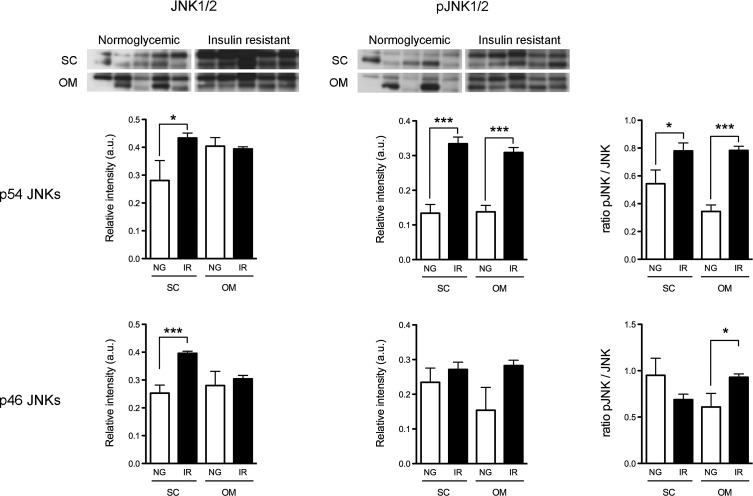
We examined the expression of p54/p46 JNK1/2 and its phosphorylated forms in SC and OM adipocytes of NG and IR subjects. Figure 5 shows that either p54 JNKs or p46 JNKs protein levels were higher in SC adipocytes of IR subjects than in NG subjects. Notably, pJNK/JNK ratios were increased in OM adipocytes of IR subjects. SC adipocytes of IR individuals also exhibited increased pJNK/JNK ratio for p46. Together, these results indicated that IR was associated with an increase in JNK activity (Fig. 5).
FIG. 5.
Total JNK, pJNK, and pJNK/JNK ratio [p54 (upper panels) and p46 (lower panels) JNK1/2] in adipocytes of NG and IR subjects. Mean±SEM of samples from five individuals/group. *p<0.05, and ***p<0.001 versus SC-NG group.
Proteasome inhibition impairs insulin signaling in 3T3-L1 cells
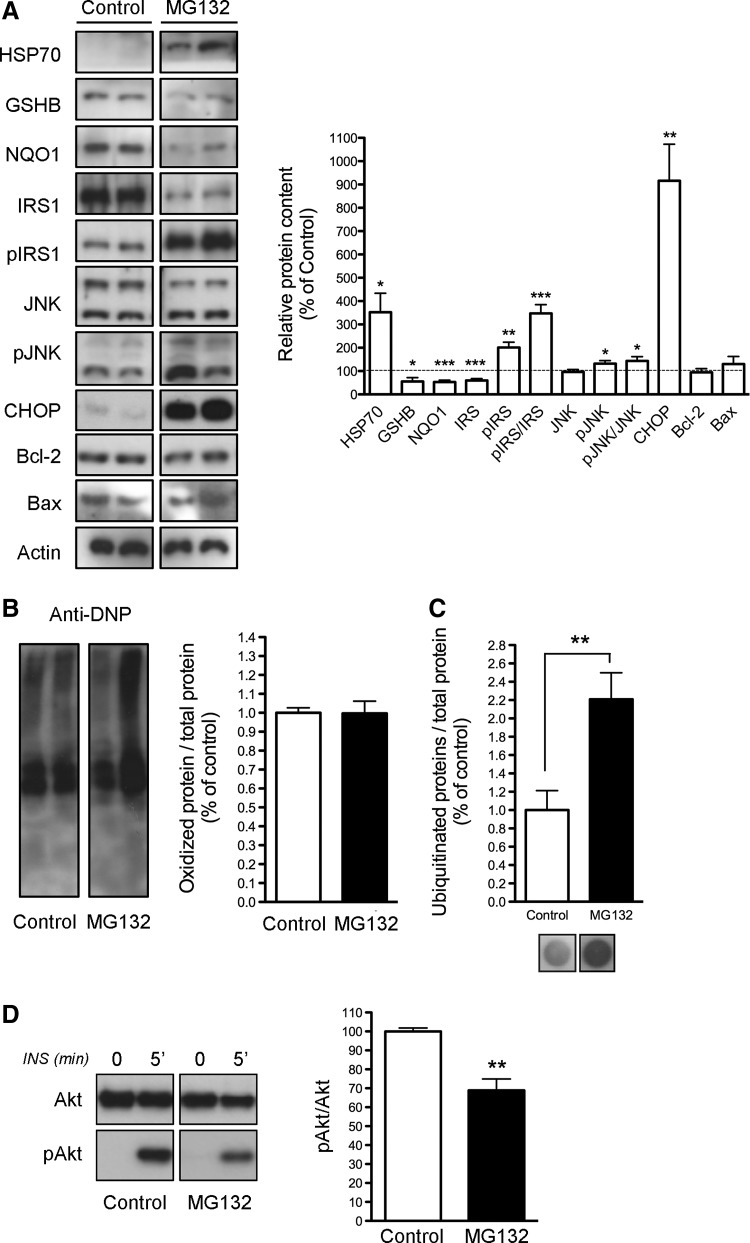
To determine whether proteasomal dysfunction could mediate IR in adipocytes, we examined the effects of proteasome inhibition by treating 3T3-L1 cells with MG132 for 24 h. It is noteworthy that chronic exposure to insulin has been shown to induce degradation of the insulin receptor substrate 1 (IRS-1) by the proteasome (56), and that proteasome inhibition under chronic insulin stimulation evokes tyrosine phosphorylation of IRS-1 and glucose uptake in adipocytes (51). Nevertheless, whether proteasomal dysfunction impairs insulin signaling has not been yet examined. As shown in Figure 6A, 3T3-L1 cells exhibited enhanced HSP70 levels but reduced protein content of GSHB and NQO1 after 24 h of MG132 treatment. Notably, CHOP protein levels were significantly higher in MG132-treated cell cultures than in control cultures (Fig. 6A). However, MG132 did not modify the protein content of the pro-apoptotic factor Bax or the anti-apoptotic factor Bcl-2 (Fig. 6A). Measurement of DNP-protein adducts showed no differences between MG132-treated and untreated 3T3-L1 cells (Fig. 6B), while the proteasome inhibitor enhanced levels of ubiquitinated proteins (Fig. 6C).
FIG. 6.
Effects of proteasome inhibition on 3T3-L1 adipocytes. (A) Representative immunoblots of HSP70, GSHB, NQO1, IRS-1, phosphor-Ser307IRS-1, JNK, phosphoJNK (pJNK), CHOP, Bcl-2, and Bax (left panel) and quantification of protein levels (right panel) in 3T3-L1 adipocytes cultured for 24 h in the absence (Control) or presence of the proteasome inhibitor, MG132 (n=5). Data were normalized for β-actin. (B, C) Quantification of oxidized proteins (B), and ubiquitinated proteins (C) in MG132-treated cell and control cells. Protein levels in control cells were set to 1 and used as reference for data normalization. (D) Effects of MG132-induced proteasome inhibition in the response to insulin in 3T3-L1 cells. Cells were incubated for 24 h in the absence or presence of MG132 and then challenged with 100 nM insulin for 5 min. Cell lysates were analyzed for Akt and phospho-Akt (pAkt) content. Mean±SEM of four independent experiments. *p<0.05, **p<0.01, and ***p<0.001 versus control.
MG132 treatment also reduced the expression of IRS-1 while increasing IRS-1 phosphorylation at Ser307 (Fig. 6A). Notably, the pJNK/JNK ratio was enhanced in cells exposed to MG132 (Fig. 6A). In all, these data supported the notion that inhibition of the proteasome could impair insulin signaling. Indeed, insulin-induced phosphorylation of Akt was significantly reduced in MG132-treated cells when compared with control cells (Fig. 6D).
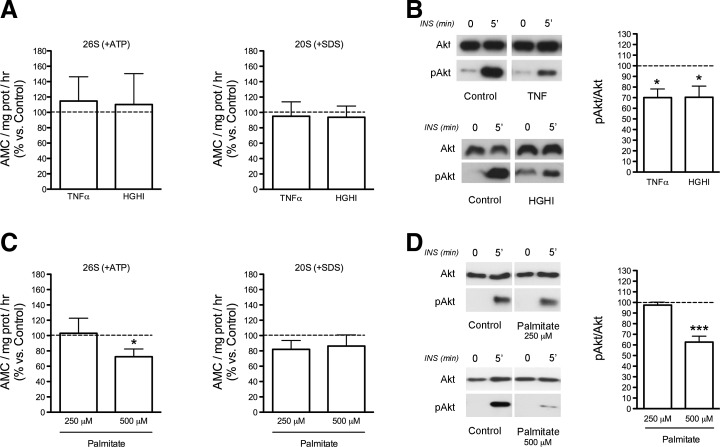
We then analyzed the activity of the proteasome in differentiated 3T3-L1 cells exposed to different treatments known to induce IR, including TNFα, a combination of high glucose and high insulin (HGHI), or palmitate (34). These experiments showed that exposure of 3T3-L1 adipocytes for 24 h to TNFα or HGHI had no effect on the ChT-L peptidase activity of the 26S and 20S proteasomes (Fig. 7A), despite both models exhibiting diminished insulin-induced phosphorylation of Akt (Fig. 7B). In contrast, 3T3-L1 cells treated with 500 μM palmitate, but not with 250 μM, exhibited significant reductions in both the activity of the 26S proteasome and insulin-induced Akt phosphorylation as compared with control cells (Fig. 7C, D).
FIG. 7.
Effects of exposure of 3T3-L1 adipocytes on TNFα (5 nM; 24 h), to a combination of high glucose and high insulin (4.5 gr/L 100 nM, respectively; 24 h) (HGHI), or to palmitate (250 and 500 μM; 18 h). (A and C) Chymotrypsin-like peptidase activity of the 26S proteasome (+ATP) (left panels) and 20S proteasome (+SDS) (right panels) in 3T3-L1 adipocytes in response to TNFα or HGHI (A), and palmitate (C). (B and D) Insulin response (pAkt/Akt ratios) of 3T3-L1 cells exposed to TNFα or HGHI (B), and palmitate (D). After the treatments, control and TNFα-, HGHI-, and palmitate-treated 3T3-L1 cells were stimulated with insulin (100 nM) for 5 min. Cell lysates were analyzed for Akt and pAkt content. Data are expressed as a percentage of values in control cultures (100%). Mean±SEM of five independent experiments. *p<0.05, and ***p<0.001 versus control.
Discussion
It has been proposed that metabolically healthy obesity, as compared with MUHO, is characterized by an improved adipogenic capacity and a healthy AT expansion, along with a low inflammatory profile (48). This study provides evidence demonstrating that defective protein folding and degradation, and the accumulation of oxidized and unfolded/misfolded proteins in adipocytes may contribute to the development of IR in obesity.
Here, we report the first comparative analysis of the proteome of paired SC and OM mature adipocytes of obese subjects with different degrees of insulin sensitivity. In agreement with the long established association between oxidative stress and IR in obesity (62), our 2D-DIGE data from isolated adipocytes indicated the existence of significant differences in cellular oxidoreductase and antioxidant activities between MHO and MUHO individuals. In particular, when compared with adipocytes from NG subjects, IR adipocytes exhibited diminished levels of several antioxidant enzymes, including NQO1 and DJ1 (18, 57), as well as a lower GSH/GSSG ratio, which is a hallmark of oxidative stress (43). Together, these results support the notion that, contrarily to obese NG individuals, adipocytes from obese IR subjects are unable to cope with increased levels of oxidative stress in obesity.
Of note, both SC and OM adipocytes from IR subjects exhibited proteomic features of increased oxidative stress and impaired antioxidant defenses. Indeed, our proteomic and biochemical data suggest that SC and OM adipocytes undergo similar molecular rearrangements in the transition from the NG to the IR state, supporting the notion that adipocyte cell damage in both SC and OM fat may contribute to metabolic disease. We cannot comment on the relative association of whole OM or SC fat depots with the IR obese phenotype, inasmuch as our analysis did not include the other major component of AT, the stromal–vascular fraction (68), and we have no information on abdominal fat distribution in our subjects. Notwithstanding these limitations, our observations are in accordance with previous genomic data on human AT, demonstrating that molecular adaptations in abdominal SC AT prove as discriminating as those in OM AT with respect to obesity and metabolic complications (32).
When cellular antioxidant and repair pathways fail to restore protein oxidative damage, cells activate additional defense mechanisms to prevent the accumulation of oxidized proteins, which are toxic for cells and threaten cell viability (20). The major cellular proteolytic system responsible for the removal of oxidized proteins is the proteasomal system, predominantly the ATP/ubiquitin-independent 20S proteasome (47). Recently, a biphasic oxidative stress-dependent regulation of the proteasome proteolytic machinery has been proposed, with mild oxidative stress activating preexisting 20S proteasomes, and prolonged oxidative damage causing decreased proteolysis (3). Here, we found that the activity of the 20S proteasome was not upregulated in IR adipocytes but, instead, tended to diminish as compared with that observed in NG adipocytes. In line with this, our data revealed elevated protein carbonylation and 4-HNE-modified protein levels in human adipocytes from both fat depots under IR conditions, which is consistent with previous studies in both AT and isolated adipocytes, and further supports the link between enhanced protein carbonylation and obesity, IR, and diabetes (15). Specifically, carbonylation and 4-HNE modification of different proteasome subunits have been shown to suppress their activity (3). In this scenario, the increased levels of carbonylated and 4-HNE-modified proteins found in IR adipocytes might contribute, at least in part, to prevent the activation of the 20S proteasome in IR adipocytes, with this, in turn, causing accumulation of oxidatively modified proteins in these cells.
In accordance to that observed for the 20S proteasome, the activity of the 26S proteasome, which is composed by the core 20S proteasome and two 19S ATPase regulatory complexes (33), was also reduced in IR adipocytes. In line with these findings, we observed that the 26S proteasome was also decreased in freshly isolated mature adipocytes, and in in vitro differentiated adipocytes of SC fat from IR mice induced by an HFD, as well as in palmitate-induced insulin-resistant 3T3-L1 adipocytes. The 26S proteasome is more vulnerable to oxidative damage than the 20S proteasome (50). In particular, it has been reported that sustained oxidative stress causes the dissociation of the 20S core from the regulatory subunits in the 26S proteasome, thus resulting in the loss of the 26S proteasome function (65). The 26S proteasome is responsible for removing misfolded proteins that have been previously tagged with ubiquitin, including both cytosolic proteins and proteins translocated from the ER during conditions of ER stress, as well as oxidized ubiquitinated proteins (55). The 26S proteasome also degrades structurally abnormal proteins in a ubiquitin-independent manner (69). Indeed, the ubiquitin/26S proteasome system (UPS) represents the major intracellular pathway for selective extralysosomal protein degradation and thus, it is essential for protein quality control in the cells (55).
Together with the proteasome system, molecular chaperones play a major role in cellular protein homeostasis by assisting refolding of misfolded proteins or unfolded/partially folded newly synthesized proteins, which, if incorrectly folded, are finally targeted for degradation (12). Interestingly, it has been reported that IR and hyperglycemic individuals have reduced expression of HSP72 in muscle, and that heat shock therapy, transgenic overexpression, or pharmacological activation of this chaperone protects against obesity-induced IR (13, 30). Along with the reduction in the activity of the 26S proteasome, we also show that OM adipocytes of IR subjects express diminished levels of HSP70, which binds and helps proteins fold to their functional state (29). Moreover, these changes were accompanied by the accumulation of ubiquitinated proteins in adipocytes of IR obese individuals. When viewed together, our data indicate that IR is associated with dysfunction of the proteolytic and protein-folding machinery in adipocytes, which leads to enhanced accumulation of abnormal proteins (oxidized and/or misfolded), thus linking proteotoxicity to impaired insulin response. Interestingly, it has been shown that proteasome inhibition induces the activation of JNK in several cell systems (19, 23, 37). In line with these observations, levels of phosphorylated JNK were upregulated in both OM and SC adipocytes of IR obese subjects as compared with those found in their counterparts from NG subjects, which is in agreement with the involvement of the kinase in IR (24).
In all, our data indicate that the transition from the insulin-sensitive to the insulin-resistant phenotype is linked to an imbalance in protein homeostasis in adipocytes. Noteworthy, it has been recently demonstrated that decreased proteasomal activity in mice aggravates HFD-induced obesity and hepatic steatosis (63), and that proteasome disruption further exacerbates IR in skeletal muscle cells from type 2 diabetic patients (4). Moreover, the expression of several components of the UPS is coordinately regulated in the liver of obese individuals, and proteasome dysregulation has been shown to mediate obesity-induced ER stress and IR in hepatocytes (41, 58).
Notably, human SC adipocytes exhibited both decreased 26S proteasome activity and increased levels of the ER stress marker, CHOP. In this scenario, our studies in 3T3-L1 cells demonstrate that proteasome dysfunction negatively regulates insulin response in adipocytes and this occurs through proteasome-mediated effects on key components of the insulin signaling pathway. Specifically, treatment of 3T3-L1 cells with the proteasome inhibitor, MG132, reduced IRS-1 levels, increased Ser307 IRS-1 phosphorylation, and inhibited insulin-induced phosphorylation of Akt, all of which are hallmark features of IR (60, 61, 67). Remarkably, JNK activation, which has been demonstrated to promote IRS-1 phosphorylation in Ser307 residues (1), was upregulated in MG132-treated 3T3-L1 cells, as it was also observed in human adipocytes from IR individuals. Likewise, proteasome inhibition induced a robust increase in CHOP protein levels in 3T3-L1 cells. It is thus likely that other cellular stress processes that are triggered in response to proteasome inhibition, including oxidative stress (16) and/or ER stress (41) (our results), directly or indirectly (i.e., via JNK activation) contribute toward negatively regulating insulin response in IR adipocytes.
It is now accepted that proteasome complexes are regulated by various mechanisms under different physiological and pathological conditions [reviewed in Kaake et al. and Schmidt and Finley (28, 54)]. In an attempt to dissect the factors and pathways responsible for the inhibition of the proteasome in IR adipocytes, we examined the activity of the proteasome in 3T3-L1 adipocytes after induction of IR by a variety of treatments. Specifically, the in vitro insulin-resistance models tested included exposure of 3T3-L1 cells to TNFα, a combination of HGHI, or palmitate (34), as a means to recapitulate the conditions observed in our human and/or animal models of MUHO, that is, inflammation, a combination of hyperglucemia and hyperinsulinemia, and systemic saturated fatty acid excess, respectively. Intriguingly, palmitate, but not TNFα or HGHI, inhibited the activity of the 26S proteasome in 3T3-L1 cells. It has been reported that palmitate exposure alters multiple cellular processes, including from insulin signaling to fatty acid metabolism, autophagy, or apoptosis in several cell types (9, 14, 26). Importantly, this saturated fatty acid has been recently shown to modify genes regulating ubiquitin and proteasome functions in human pancreatic islets (14).
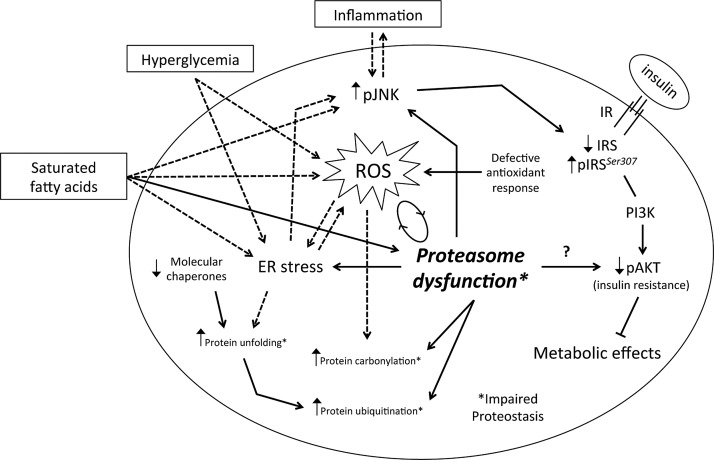
In adipocytes, palmitate has been shown to impair insulin signaling by inducing both oxidative and ER stress as well as inflammation (22, 31, 40). However, when we examined the effects of H2O2 and the ER stressor, thapsigargin, on 3T3-L1 cells, we observed that, at least at the conditions tested (1 and 50 μM, respectively, for 24 h), these compounds were unable to mimic the effects of palmitate on either 26S proteasome activity or pAkt/Akt ratio (data not shown). In all, these results, together with our data on TNFα and HGHI in 3T3-L1 cells, suggest that inhibition of proteasome activity in IR might be mediated by the combined activation of several cellular stress processes, which, in turn, are differentially activated by distinct environmental insults. In this vein, a growing body of evidence suggests that cellular stress pathways are closely linked and impact each other at various stages to functionally impair insulin response in metabolic disease (8, 10, 52, 71). As summarized in Figure 8, our data support the occurrence of a feed-forward cascade or a vicious cycle of cellular stress responses in adipocytes in obesity-associated IR, in which proteasome malfunction would impact other central pathways that contribute to IR.
FIG. 8.
Schematic representation of the intracellular pathways altered in adipocytes of IR individuals. Impairment of proteasome activity aggravates the effects caused by the inability of adipocytes to cope with oxidative stress and excess protein-folding load. The vicious cycle between proteasome dysfunction and oxidative stress in obesity worsens: (i) cellular protein homeostasis (i.e., ER stress and disruption of protein-folding responses), and (ii) the inflammatory state, which negatively impacts insulin responsiveness in adipocytes. The model is reproduced based on published molecular mechanisms (dashed arrows), and the main findings are shown in this article (solid arrows; see Discussion section).
Our study presents some limitations. First, it is limited by small sample size. However, the subjects were well matched for baseline anthropometric and clinical characteristics. Another limitation is that, as mentioned earlier, no information is available on the distribution of OM and SC intra-abdominal fat and the regional distribution of body fat, both of which are markers of metabolic health (66). This information could provide a more integrated view of the contribution of adipocyte dysfunction to the development of IR in obesity.
In conclusion, our data suggest that proteasome dysfunction exacerbates the effects caused by the inability of adipocytes to activate efficient adaptive antioxidant and protein-folding responses to increased oxidative stress and ER stress occurring in obesity. Based on this and published data, we propose a model in which the proteasome would play a major role in maintaining adequate insulin responsiveness in adipocytes (Fig. 8). Thus, proteasome dysfunction and disruption of cell proteostasis in adipocytes appear as important contributors to the onset and/or progression of IR in obesity.
Materials and Methods
Subjects
Subjects were recruited at the Lipids and Atherosclerosis Unit of the Reina Sofia University Hospital (Cordoba, Spain). Blood samples were obtained by venipuncture after an overnight fast. Serum measurements of cholesterol, triglycerides, high-density lipoprotein cholesterol, glucose, and insulin were carried out as previously described (70). Subjects were not on insulin therapy or medication that was likely to influence endogenous insulin levels. Cardiovascular disease, diabetes, arthritis, acute inflammatory disease, and infectious or renal diseases were considered exclusion criteria. Obese subjects (BMI >30 kg/m2) were subclassified into two groups [normoglycemia (NG): Glu <100 mg/dl and HbA1c <5.7%; and impaired fasting glucose (IFG): Glu 100–126 mg/dl and HbA1c: 5.7–6.4%] following the criteria of the Expert Committee on the Diagnosis and Classification of Diabetes (5). IFG individuals exhibited significantly higher HOMA-IR values than NG individuals, as well as other clinic and plasma parameters that are within the cutoff points for identifying IR in hyperinsulinemic-euglycemic clamp studies (5, 59), and they will be referred to as insulin-resistant (IR) subjects. All participants gave written informed consent, and the study was reviewed and approved by the ethics and research committee of the Reina Sofia University Hospital.
Isolation of mature adipocytes
Paired OM and SC abdominal AT samples were obtained from individuals undergoing bariatric surgery. Fresh AT samples were dispersed by collagenase digestion to isolate mature adipocytes as previously described (44). Floating adipocytes were frozen in liquid nitrogen and stored at −80°C until further processing.
Protein extraction
For proteomic studies, human adipocytes were homogenized in cold urea/thiourea buffer [7 M urea, 2 M thiourea, 4% CHAPS, 45 mM Tris, pH 8.4, and Complete protease inhibitors (Roche)]. The homogenate was incubated for 30 min on ice and centrifuged at 13,500 rpm for 30 min. The aqueous phase between the upper lipid phase and the lower cellular debris phase was collected. Proteins were precipitated using the 2D-CleanUp Kit (GE Healthcare) and quantified using the RC/DC Protein Assay (Bio-Rad Laboratories).
2D-DIGE and data analysis
Proteins were labeled according to the manufacturer's instructions (GE Healthcare). Briefly, 50 μg of protein from SC and OM adipocytes of NG and IR subjects were randomly labeled with 400 pmol of the N-hydroxysuccinimide esters of Cy3 or Cy5 fluorescent cyanine dyes. An internal standard consisting of a mixture of equal amounts of all the samples was labeled with Cy2 dye and combined with the Cy3- or Cy5-labeled samples. The labeling reaction was performed on ice for 30 min and then quenched by adding 1 μl of lysine (10 mM) on ice for 10 min. Paired Cy2-, Cy3-, and Cy5-labeled samples (150 μg) were diluted in Rehydratation buffer (GE Healthcare) to a final volume of 340 μl containing 0.8% (v/v) IPG buffer. Isoelectric focusing onto immobilized pH gradient strips (18 cm, pH 3–10 NL) and 2D-electrophoresis in 11% Tris-glycine gels were performed following the procedure previously described (45). Gels were scanned with a Typhoon 9400 scanner (GE Healthcare) at 100 μm resolution using appropriate excitation and emission wavelengths.
Relative quantification of differentially expressed proteins was performed using DeCyder software version 7.0 (GE Healthcare). After differential In-gel Analysis using the internal standard as a reference, spot abundance was measured in each image. Statistical analysis was carried out to determine protein expression changes. p-values lower than 0.05 as calculated from Student's t-test were considered significant.
MALDI-TOF-MS analysis
Spots were excised automatically in a ProPic station (Genomic Solutions), digested automatically, and analyzed in a 4800 MALDI-ToF/ToF Analyzer (Applied Biosystems/MDS SCIEX) to obtain the MALDI-MS and MALDI-MS/MS spectra as previously reported (45).
MS and MS/MS spectra data were combined through the GPS Explorer Software v3.6 using Mascot software v2.2 (Matrix Science) to search against a nonredundant human database (SwissProt release 56.0), with 30 ppm precursor tolerance, and 0.2 Da MS/MS fragment tolerance. Analysis was limited to peptides of six or more amino acids and maximum one missed cleavage site. Oxidation of methionine was searched as variable modification, and carbamidomethylation of cysteine was set as fixed modification. All spectra and database results were manually inspected in detail. Protein scores greater than 56 were accepted as significant (p<0.05), considering positive the identification when protein score CI% (Confidence Interval) was above 98. In the case of MS/MS spectra, total ion score CI% was above 95.
3T3-L1 cell culture and treatment
3T3-L1 adipocytes were differentiated as previously described (49). Differentiated 3T3-L1 adipocytes (day 9) were exposed for 24 h to Dulbecco's modified Eagle's medium (DMEM) alone or containing the proteasome inhibitor MG132 (50 μM). Thereafter, control and MG132-treated 3T3-L1 cells were stimulated with insulin (100 nM) for 5 min.
In another set of experiments, differentiated 3T3-L1 adipocytes were exposed for 24 h to the inflammatory cytokine TNFα (5 nM) or to a combination of high glucose (4.5 gr/L) and high insulin (100 nM) (HGHI). Finally, the effects of 18-h exposure of 3T3-L1 cells to palmitate (sodium salt; 250 and 500 μM) were also explored. Palmitate solutions containing 2% fatty acid-free bovine serum albumin were prepared in DMEM containing no serum. After the treatments, control and TNFα-, HGHI-, and palmitate-treated 3T3-L1 cells were stimulated with insulin (100 nM) for 5 min.
At the end of the experiments, 3T3-L1 cells were processed for immunoblotting as previously described (49).
Quantitative immunoblotting
Mature adipocytes isolated from additional NG and IR subjects distinct from those employed for 2D-DIGE analysis were disrupted in buffer containing 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton-X-100, 1 mM EDTA, and 1 μg/ml anti-protease cocktail. Thirty microgram of protein per sample were loaded into 4–20% precasted SDS-PAGE gels and transferred to nitrocellulose membranes using the Trans-Blot Turbo Transfer System (Bio-Rad). For dot-blot assays, 10 μL aqueous samples containing 10 μg of protein were laid in dots onto nitrocellulose membranes. Membranes were stained with Ponceau to ensure equal sample loading (Supplementary Fig. S4) and processed for immunoblotting with specific antibodies (Supplementary Table S1). Ponceau staining was employed as a loading control (27, 45).
Analysis of oxidative markers
Total glutathione and reduced (GSH) glutathione were quantified in human adipocytes, and the GSH/GSGG ratio was calculated as previously described (11).
Protein carbonyl levels were determined by 2,4-dinitrophenylhydrazine (DNPH) derivatization and immunoblotting using an anti-DNP antibody (Supplementary Table S1) (7). 4-HNE-modified proteins were analyzed by dot-blot using an anti-4-HNE antibody (Supplementary Table S1) (16).
Proteasome activity assay
Proteasome activity of adipocytes was measured by using synthetic fluorogenic substrates (N-Suc-LLVY-AMC, ZLLE-AMC, and Boc-LRR-AMC) as previously described (35). Fluorescence of the released AMC products was kinetically followed for 1 h in a FlexStation 3 microplate reader (Molecular Devices) (excitation, 355 nm; emission, 460 nm). Parallel reactions containing 25 μM MG132 were run as controls for nonspecific peptide hydrolysis, and fluorescence background was subtracted from activity values. Proteasome peptidase activity was calculated as AMC values per hour and mg of protein.
Analysis of proteasome activity in adipocytes of lean and obese mice
Four-week-old male C57BL/6J mice were housed (five per cage) under a 12 h light/12 h dark cycle, in a temperature-controlled room (22°C), with food and water ad libitum. After weaning, mice were fed a standard chow diet, and after 4 weeks, they were divided into two groups (10 animals per group) with a similar average body weight and assigned either to a standard chow global diet (ND, #2918, 6.2% fat, 18% protein; 3.1 kcal/g; Harlan Laboratories) or to an HFD (D12492, 60 kcal% fat, 20 kcal% carbohydrates and 20 kcal% protein; 5.24 kcal/g; Research Diets) during 14 weeks. Blood glucose levels were measured with an Accucheck glucometer (Roche) after an intraperitoneal injection of either 2 mg/g D-glucose (Sigma) or 0.75 U/kg insulin (Sigma-Aldrich) as described (46). Tests were performed after 13 weeks of feeding mice with ND or HFD. All experiments were conducted under approval of the Animal Care and Use Committee of the University of Santiago de Compostela and the University of Córdoba (Spain). All the experiments were performed in agreement with the Rules of Laboratory Animal Care and International Law on Animal Experimentation.
Animals were sacrificed, and SC and visceral AT samples were dispersed by collagenase digestion to isolate mature adipocytes and stromal vascular fraction cells (SVFCs) as described (44). Cells (mature adipocytes or SVFCs) from 3 or 4 animals were pooled together for further analysis. Mature adipocytes were frozen in liquid nitrogen and stored to −80°C until analysis. SVFCs were seeded in 35-mm plastic plates (30,000 cells/plate) and grown in adipocyte medium (DMEM/F-12 [1:1] (Invitrogen), 16 μM biotin, 18 μM panthotenate, 100 μM ascorbate, and antibiotic-antimycotic) supplemented with 10% newborn calf serum (NCS) at 37°C in a humidified atmosphere with 95% air: 5% CO2. After 4 days, the medium was changed to adipocyte medium supplemented with 3% NCS, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 0.1 μM dexamethasone, 1 μM BRL49653, and 10 μg/ml insulin. After a 3-day induction period, cells were cultured in differentiation medium lacking IBMX and BRL49653 for the remaining 7 days of adipocyte differentiation. Thereafter, proteasomal activities in the cell samples were measured as described for human adipocytes.
Statistical analysis
Correlations were analyzed using Pearson's correlation test. Statistical differences were determined using Student's t-test. Values were considered significant at p<0.05. Data are expressed as mean±SEM. Proteins identified by the proteomic study were analyzed using the PANTHER database (www.pantherdb.org/) for classification according to their function (38). The Bonferroni correction for multiple testing was used for the calculation of PANTHER p-values. Statistical analyses were performed using SSPS statistical software (version 13.0).
Supplementary Material
Abbreviations Used
- 4-HNE
4-hydroxynonenal
- AT
adipose tissue
- BMI
body mass index
- ChT-L
chymotrypsin-like
- DMEM
Dulbecco's modified Eagle's medium
- ER
endoplasmic reticulum
- FFA
free fatty acid
- GSH
reduced glutathione
- GSHB
glutathione synthetase
- GSSG
oxidized glutathione
- GT
total glutathione
- HbA1c
glycated hemoglobin
- HDL
high-density lipoprotein
- HFD
high-fat diet
- HGHI
high glucose and high insulin
- HOMA-IR
homeostasis model assessment of insulin resistance
- IR
insulin resistance
- IRS-1
insulin receptor substrate 1
- JNK
c-Jun NH2-terminal kinase
- LDL
low-density lipoprotein
- MHO
metabolically healthy obese
- MUHO
metabolically unhealthy obese
- ND
normal diet
- NF-κB
nuclear factor κB
- NG
normoglycemic
- NQO1
NADPH quinone oxidoreductase 1
- OM
omental
- proteostasis
protein homeostasis
- SC
subcutaneous
- SVFC
stromal vascular fraction cell
- UPS
ubiquitin/26S proteasome system
Acknowledgments
The authors thank Jana Alonso [Proteomic platform of the Health Research Institute of Santiago (IDIS), University of Santiago de Compostela, Spain] and the Proteomics Facilities of the IMIBIC/University of Córdoba-SCAI (ProteoRed, PRB2-ISCIII, supported by grant PT13/0001) for their help with mass spectrometry studies. This study is integrated into Chromosome 16 Spanish Human Proteome Project. The authors are grateful to Dr. José Manuel Villalba (University of Córdoba, Spain) for his generous gifts of antibodies. They thank Laura Molero and Encarnación Palomo (Dept. of Cell Biology, Physiology, and Immunology; IMIBIC/Reina Sofia University Hospital, University of Cordoba, CIBERobn, Spain) for their technical assistance with cell cultures. This work was supported by MINECO/FEDER (BFU2010-17116; BFU2013-44229-R), J. Andalucia/FEDER (CTS-6606; PI-0200/2013), and CIBERobn (Instituto de Salud Carlos III), Spain.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Aguirre V, Uchida T, Yenush L, Davis R, and White MF. The c-Jun NH(2)-terminal kinase promotes insulin resistance during association with insulin receptor substrate-1 and phosphorylation of Ser(307). J Biol Chem 275: 9047–9054, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Ahima RS. Digging deeper into obesity. J Clin Invest 121: 2076–2079, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aiken CT, Kaake RM, Wang X, and Huang L. Oxidative stress-mediated regulation of proteasome complexes. Mol Cell Proteomics 10: R110 006924, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Khalili L, de Castro Barbosa T, Ostling J, Massart J, Cuesta PG, Osler ME, Katayama M, Nystrom AC, Oscarsson J, and Zierath JR. Proteasome inhibition in skeletal muscle cells unmasks metabolic derangements in type 2 diabetes. Am J Physiol Cell Physiol 307: C774–C787, 2014 [DOI] [PubMed] [Google Scholar]
- 5.American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care 35 Suppl 1: S64–S71, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbarroja N, Lopez-Pedrera R, Mayas MD, Garcia-Fuentes E, Garrido-Sanchez L, Macias-Gonzalez M, El Bekay R, Vidal-Puig A, and Tinahones FJ. The obese healthy paradox: is inflammation the answer? Biochem J 430: 141–149, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Barcelo-Batllori S, Kalko SG, Esteban Y, Moreno S, Carmona MC, and Gomis R. Integration of DIGE and bioinformatics analyses reveals a role of the antiobesity agent tungstate in redox and energy homeostasis pathways in brown adipose tissue. Mol Cell Proteomics 7: 378–393, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Bluher M. Adipose tissue dysfunction contributes to obesity related metabolic diseases. Best Pract Res Clin Endocrinol Metab 27: 163–177, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Cao J, Dai DL, Yao L, Yu HH, Ning B, Zhang Q, Chen J, Cheng WH, Shen W, and Yang ZX. Saturated fatty acid induction of endoplasmic reticulum stress and apoptosis in human liver cells via the PERK/ATF4/CHOP signaling pathway. Mol Cell Biochem 364: 115–129, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Cao SS. and Kaufman RJ. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid Redox Signal 21: 396–413, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardona F, Tunez I, Tasset I, Montilla P, Collantes E, and Tinahones FJ. Fat overload aggravates oxidative stress in patients with the metabolic syndrome. Eur J Clin Invest 38: 510–515, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Chen B, Retzlaff M, Roos T, and Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 3: a004374, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung J, Nguyen AK, Henstridge DC, Holmes AG, Chan MH, Mesa JL, Lancaster GI, Southgate RJ, Bruce CR, Duffy SJ, Horvath I, Mestril R, Watt MJ, Hooper PL, Kingwell BA, Vigh L, Hevener A, and Febbraio MA. HSP72 protects against obesity-induced insulin resistance. Proc Natl Acad Sci U S A 105: 1739–1744, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cnop M, Abdulkarim B, Bottu G, Cunha DA, Igoillo-Esteve M, Masini M, Turatsinze JV, Griebel T, Villate O, Santin I, Bugliani M, Ladriere L, Marselli L, McCarthy MI, Marchetti P, Sammeth M, and Eizirik DL. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 63: 1978–1993, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Curtis JM, Hahn WS, Long EK, Burrill JS, Arriaga EA, and Bernlohr DA. Protein carbonylation and metabolic control systems. Trends Endocrinol Metab 23: 399–406, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dasuri K, Zhang L, Ebenezer P, Fernandez-Kim SO, Bruce-Keller AJ, Szweda LI, and Keller JN. Proteasome alterations during adipose differentiation and aging: links to impaired adipocyte differentiation and development of oxidative stress. Free Radic Biol Med 51: 1727–1735, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denis GV. and Obin MS. ‘Metabolically healthy obesity’: origins and implications. Mol Aspects Med 34: 59–70, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinkova-Kostova AT. and Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys 501: 116–123, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans JL, Maddux BA. and Goldfine ID. The molecular basis for oxidative stress-induced insulin resistance. Antioxid Redox Signal 7: 1040–1052, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Grune T. and Davies KJ. Breakdown of oxidized proteins as a part of secondary antioxidant defenses in mammalian cells. Biofactors 6: 165–172, 1997 [DOI] [PubMed] [Google Scholar]
- 21.Guh DP, Zhang W, Bansback N, Amarsi Z, Birmingham CL, and Anis AH. The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Public Health 9: 88, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo W, Wong S, Xie W, Lei T, and Luo Z. Palmitate modulates intracellular signaling, induces endoplasmic reticulum stress, and causes apoptosis in mouse 3T3-L1 and rat primary preadipocytes. Am J Physiol Endocrinol Metab 293: E576–E586, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Hideshima T, Mitsiades C, Akiyama M, Hayashi T, Chauhan D, Richardson P, Schlossman R, Podar K, Munshi NC, Mitsiades N, and Anderson KC. Molecular mechanisms mediating antimyeloma activity of proteasome inhibitor PS-341. Blood 101: 1530–1534, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, and Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature 420: 333–336, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140: 900–917, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishii M, Maeda A, Tani S, and Akagawa M. Palmitate induces insulin resistance in human HepG2 hepatocytes by enhancing ubiquitination and proteasomal degradation of key insulin signaling molecules. Arch Biochem Biophys 566: 26–35, 2015 [DOI] [PubMed] [Google Scholar]
- 27.Jimenez-Gomez Y, Mattison JA, Pearson KJ, Martin-Montalvo A, Palacios HH, Sossong AM, Ward TM, Younts CM, Lewis K, Allard JS, Longo DL, Belman JP, Malagon MM, Navas P, Sanghvi M, Moaddel R, Tilmont EM, Herbert RL, Morrell CH, Egan JM, Baur JA, Ferrucci L, Bogan JS, Bernier M, and de Cabo R. Resveratrol Improves Adipose Insulin Signaling and Reduces the Inflammatory Response in Adipose Tissue of Rhesus Monkeys on High-Fat, High-Sugar Diet. Cell Metab 18: 533–545, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaake RM, Kao A, Yu C, and Huang L. Characterizing the dynamics of proteasome complexes by proteomics approaches. Antioxid Redox Signal 21: 2444–2456, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampinga HH. and Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11: 579–592, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kavanagh K, Flynn DM, Jenkins KA, Zhang L, and Wagner JD. Restoring HSP70 deficiencies improves glucose tolerance in diabetic monkeys. Am J Physiol Endocrinol Metab 300: E894–E901, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy A, Martinez K, Chuang CC, LaPoint K, and McIntosh M. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J Nutr 139: 1–4, 2009 [DOI] [PubMed] [Google Scholar]
- 32.Klimcakova E, Roussel B, Marquez-Quinones A, Kovacova Z, Kovacikova M, Combes M, Siklova-Vitkova M, Hejnova J, Sramkova P, Bouloumie A, Viguerie N, Stich V, and Langin D. Worsening of obesity and metabolic status yields similar molecular adaptations in human subcutaneous and visceral adipose tissue: decreased metabolism and increased immune response. J Clin Endocrinol Metab 96: E73–E82, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, and Finley D. Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507, 2002 [DOI] [PubMed] [Google Scholar]
- 34.Lo KA, Labadorf A, Kennedy NJ, Han MS, Yap YS, Matthews B, Xin X, Sun L, Davis RJ, Lodish HF, and Fraenkel E. Analysis of in vitro insulin-resistance models and their physiological relevance to in vivo diet-induced adipose insulin resistance. Cell Rep 5: 259–270, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin-Clemente B, Alvarez-Castelao B, Mayo I, Sierra AB, Diaz V, Milan M, Farinas I, Gomez-Isla T, Ferrer I, and Castano JG. alpha-Synuclein expression levels do not significantly affect proteasome function and expression in mice and stably transfected PC12 cell lines. J Biol Chem 279: 52984–52990, 2004 [DOI] [PubMed] [Google Scholar]
- 36.Mathis D. Immunological Goings-on in Visceral Adipose Tissue. Cell Metabolism 17: 851–859, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meriin AB, Gabai VL, Yaglom J, Shifrin VI, and Sherman MY. Proteasome inhibitors activate stress kinases and induce Hsp72. Diverse effects on apoptosis. J Biol Chem 273: 6373–6379, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Mi H, Muruganujan A, Casagrande JT, and Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muller MJ, Lagerpusch M, Enderle J, Schautz B, Heller M, and Bosy-Westphal A. Beyond the body mass index: tracking body composition in the pathogenesis of obesity and the metabolic syndrome. Obes Rev 13 Suppl 2: 6–13, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Nguyen MT, Satoh H, Favelyukis S, Babendure JL, Imamura T, Sbodio JI, Zalevsky J, Dahiyat BI, Chi NW, and Olefsky JM. JNK and tumor necrosis factor-alpha mediate free fatty acid-induced insulin resistance in 3T3-L1 adipocytes. J Biol Chem 280: 35361–35371, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Otoda T, Takamura T, Misu H, Ota T, Murata S, Hayashi H, Takayama H, Kikuchi A, Kanamori T, Shima KR, Lan F, Takeda T, Kurita S, Ishikura K, Kita Y, Iwayama K, Kato K, Uno M, Takeshita Y, Yamamoto M, Tokuyama K, Iseki S, Tanaka K, and Kaneko S. Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver. Diabetes 62: 811–824, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouchi N, Parker JL, Lugus JJ, and Walsh K. Adipokines in inflammation and metabolic disease. Nat Rev Immunol 11: 85–97, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owen JB. and Butterfield DA. Measurement of oxidized/reduced glutathione ratio. Methods Mol Biol 648: 269–277, 2010 [DOI] [PubMed] [Google Scholar]
- 44.Peinado JR, Jimenez-Gomez Y, Pulido MR, Ortega-Bellido M, Diaz-Lopez C, Padillo FJ, Lopez-Miranda J, Vazquez-Martinez R, and Malagon MM. The stromal-vascular fraction of adipose tissue contributes to major differences between subcutaneous and visceral fat depots. Proteomics 10: 3356–3366, 2010 [DOI] [PubMed] [Google Scholar]
- 45.Peinado JR, Quiros PM, Pulido MR, Marino G, Martinez-Chantar ML, Vazquez-Martinez R, Freije JM, Lopez-Otin C, and Malagon MM. Proteomic profiling of adipose tissue from Zmpste24-/- mice, a model of lipodystrophy and premature aging, reveals major changes in mitochondrial function and vimentin processing. Mol Cell Proteomics 10: M111 008094, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Sieira S, Martinez G, Porteiro B, Lopez M, Vidal A, Nogueiras R, and Dieguez C. Female Nur77-deficient mice show increased susceptibility to diet-induced obesity. PLoS One 8: e53836, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pickering AM. and Davies KJ. Degradation of damaged proteins: the main function of the 20S proteasome. Prog Mol Biol Transl Sci 109: 227–248, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Primeau V, Coderre L, Karelis AD, Brochu M, Lavoie ME, Messier V, Sladek R, and Rabasa-Lhoret R. Characterizing the profile of obese patients who are metabolically healthy. Int J Obes (Lond) 35: 971–981, 2011 [DOI] [PubMed] [Google Scholar]
- 49.Pulido MR, Diaz-Ruiz A, Jimenez-Gomez Y, Garcia-Navarro S, Gracia-Navarro F, Tinahones F, Lopez-Miranda J, Fruhbeck G, Vazquez-Martinez R, and Malagon MM. Rab18 dynamics in adipocytes in relation to lipogenesis, lipolysis and obesity. PLoS One 6: e22931, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinheckel T, Sitte N, Ullrich O, Kuckelkorn U, Davies KJ, and Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem J 335 (Pt 3): 637–642, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rondinone CM. and Kramer D. Proteasome inhibitors regulate tyrosine phosphorylation of IRS-1 and insulin signaling in adipocytes. Biochem Biophys Res Commun 296: 1257–1263, 2002 [DOI] [PubMed] [Google Scholar]
- 52.Samuel VT. and Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell 148: 852–871, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schelbert KB. Comorbidities of obesity. Prim Care 36: 271–285, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Schmidt M. and Finley D. Regulation of proteasome activity in health and disease. Biochim Biophys Acta 1843: 13–25, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shang F. and Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic Biol Med 51: 5–16, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun XJ, Goldberg JL, Qiao LY, and Mitchell JJ. Insulin-induced insulin receptor substrate-1 degradation is mediated by the proteasome degradation pathway. Diabetes 48: 1359–1364, 1999 [DOI] [PubMed] [Google Scholar]
- 57.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, and Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep 5: 213–218, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takamura T, Misu H, Matsuzawa-Nagata N, Sakurai M, Ota T, Shimizu A, Kurita S, Takeshita Y, Ando H, Honda M, and Kaneko S. Obesity upregulates genes involved in oxidative phosphorylation in livers of diabetic patients. Obesity (Silver Spring) 16: 2601–2609, 2008 [DOI] [PubMed] [Google Scholar]
- 59.Tam CS, Xie W, Johnson WD, Cefalu WT, Redman LM, and Ravussin E. Defining insulin resistance from hyperinsulinemic-euglycemic clamps. Diabetes Care 35: 1605–1610, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanti JF, Gremeaux T, van Obberghen E, and Le Marchand-Brustel Y. Serine/threonine phosphorylation of insulin receptor substrate 1 modulates insulin receptor signaling. J Biol Chem 269: 6051–6057, 1994 [PubMed] [Google Scholar]
- 61.Tanti JF. and Jager J. Cellular mechanisms of insulin resistance: role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr Opin Pharmacol 9: 753–762, 2009 [DOI] [PubMed] [Google Scholar]
- 62.Tiganis T. Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol Sci 32: 82–89, 2011 [DOI] [PubMed] [Google Scholar]
- 63.Tomaru U, Takahashi S, Ishizu A, Miyatake Y, Gohda A, Suzuki S, Ono A, Ohara J, Baba T, Murata S, Tanaka K, and Kasahara M. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am J Pathol 180: 963–972, 2012 [DOI] [PubMed] [Google Scholar]
- 64.Trayhurn P. Hypoxia and adipose tissue function and dysfunction in obesity. Physiol Rev 93: 1–21, 2013 [DOI] [PubMed] [Google Scholar]
- 65.Wang X, Yen J, Kaiser P, and Huang L. Regulation of the 26S proteasome complex during oxidative stress. Sci Signal 3: ra88, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White UA. and Tchoukalova YD. Sex dimorphism and depot differences in adipose tissue function. Biochim Biophys Acta 1842: 377–392, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wing SS. The UPS in diabetes and obesity. BMC Biochem 9 Suppl 1: S6, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, and Chen H. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 112: 1821–1830, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yano M, Kanesaki Y, Koumoto Y, Inoue M, and Kido H. Chaperone activities of the 26S and 20S proteasome. Curr Protein Pept Sci 6: 197–203, 2005 [DOI] [PubMed] [Google Scholar]
- 70.Yubero-Serrano EM, Delgado-Casado N, Delgado-Lista J, Perez-Martinez P, Tasset-Cuevas I, Santos-Gonzalez M, Caballero J, Garcia-Rios A, Marin C, Gutierrez-Mariscal FM, Fuentes F, Villalba JM, Tunez I, Perez-Jimenez F, and Lopez-Miranda J. Postprandial antioxidant effect of the Mediterranean diet supplemented with coenzyme Q10 in elderly men and women. Age (Dordr) 33: 579–590, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang K. Integration of ER stress, oxidative stress and the inflammatory response in health and disease. Int J Clin Exp Med 3: 33–40, 2010 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.