Abstract
The fibroblast growth factor (FGF) signal transduction pathway serves as one of the key regulators of early metazoan development, displaying conserved roles in the specification of endodermal, mesodermal, and neural fates during vertebrate development. FGF signals also regulate gastrulation, in part, by triggering epithelial to mesenchymal transitions in embryos of both vertebrates and invertebrates. Thus, FGF signals coordinate gastrulation movements across many different phyla. To help understand the breadth of FGF signaling deployment across the animal kingdom, we have examined the presence and expression of genes encoding FGF pathway components in the anthozoan cnidarian Nematostella vectensis. We isolated three FGF ligands (NvFGF8A, NvFGF8B, and NvFGF1A), two FGF receptors (NvFGFRa and NvFGFRb), and two orthologs of vertebrate FGF responsive genes, Sprouty (NvSprouty), an inhibitor of FGF signaling, and Churchill (NvChurchill), a Zn finger transcription factor. We found these FGF ligands, receptors, and response gene expressed asymmetrically along the oral/aboral axis during gastrulation and in a developing chemosensory structure of planula stages known as the apical tuft. These results suggest a conserved role for FGF signaling molecules in coordinating both gastrulation and neural induction that predates the Cambrian explosion and the origins of the Bilateria.
Keywords: Gastrulation, Neurogenesis, Evolution of development
Introduction
Fibroblast growth factors (FGFs) were originally isolated from vertebrate brain and pituitary fibroblasts for their roles in angiogenesis, mitogenesis, cellular differentiation, migration, and tissue-injury repair (Itoh and Ornitz 2004; Ornitz and Itoh 2001; Popovici et al. 2005). FGFs signal through fibroblast growth factor receptors (FGFRs), which are membrane-associated class IV receptor tyrosine kinases (RTKs). The interaction of FGF ligands with these receptors are stabilized by co-binding of heparin or heparin sulfate proteoglycans, which prevent thermal denaturation and proteolysis (Itoh and Ornitz 2004; Popovici et al. 2005). Ligand binding results in receptor dimerization and autophosphorylation of several intracellular tyrosines, which leads to activation of the GTPase Ras, resulting in a cascade of kinase signaling including Raf, mitogen-activated and extracellular signal-regulated kinase (MEK), and mitogen-activated protein kinase (MAPK; Nutt et al. 2001). FGF signals often work in concert with other important pathways, such as transforming growth factor-β (TGF-β), Wnt, Hedgehog, and Notch (Gerhart 1999). In both invertebrates and vertebrates, FGF signaling functions in body plan patterning, including mesoderm and neural induction and coordinate cell movements during gastrulation (Bertrand et al. 2003; De Robertis and Kuroda 2004; Isaacs et al. 1994; Popovici et al. 2005; Rossant et al. 1997; Sheng et al. 2003; Sivak et al. 2005; Stathopoulos et al. 2004).
FGF signaling complexity in the Metazoa
Genome duplications within the vertebrates (Huang and Stern 2005; Itoh and Ornitz 2004; Popovici et al. 2005; Powers et al. 2000) have made it difficult to decipher the molecular machinery underlying FGF signaling due to the diversity and sheer number of FGF ligands and receptors among the various vertebrate lineages. As many as 22 FGF ligands and 4 receptors have been identified in the human genome, with alternative splice forms of the receptors adding significantly to their complexity (Satou et al. 2002). The scope of FGF pathway members has been investigated in lower chordates that diversified before vertebrate genome duplications, and this has provided a clearer picture of FGF family evolution within the deuterostomes. In the urochordate ascidians Ciona intestinalis and Ciona savigny (whose genomes have been sequenced) there are six FGF ligands and one FGF receptor (Imai et al. 2002, 2003).
Within the protostomes, FGF ligand and receptor diversity has been explored in the ecdysozoan model systems of Caenorhabditis elegans and Drosophila melanogaster, where only a few FGFs and one or two receptors have been isolated. In C. elegans, two FGF ligands (egl-17 and LET-756) and a single FGFR (egl-15) have been described (Branda and Stern 2000). Likely due to a lack of sampling, no FGF ligands have been described for any lophotrochozoan (e.g., molluscs, annelids, platyhelminths).
Conserved roles of FGF signaling
FGFs and the control of branching morphogenesis
Mutant analysis in Drosophila has revealed that an FGF ligand (branchless) and its receptor (breathless) regulate tracheal development (Klambt et al. 1992; Sutherland et al. 1996). In mouse, FGF signaling through FGF-10 and FGFR2-IIIb direct branching morphogenesis of the lungs (Min et al. 1998; Shishido et al. 1997; Stathopoulos et al. 2004), and although mammalian lungs and insect trachea may not be directly homologous, these data suggest a conserved role for FGF signaling in controlling branching morphogenesis in organisms as diverse as flies and mammals.
FGFs in gastrulation
FGF signaling during fly, vertebrate, and ascidian gastrulation suggests a more conserved role for FGFs in protostome and deuterostome development. In Drosophila, a second receptor (heartless) and pair of FGF ligands (pyramus and thisbe) coordinate gastrulation movements and mesoderm induction (Huang and Stern 2005; Stathopoulos et al. 2004). Heartless is required for cell-autonomous spreading of mesodermal cells, as they migrate dorsolaterally under the neurogenic ectoderm, which expresses the FGF ligands pyramus and thisbe (Stathopoulos et al. 2004). Functional tests in ascidian urochordates have shown that FGF signals induce mesodermal mesenchymal cells (Bertrand et al. 2003). Roles for FGF signaling in vertebrate gastrulation are well known. FGF4 and FGF8 are required during mouse gastrulation for specification of endodermal and mesodermal derivatives and cell migration through the primitive streak (Amaya et al. 1993; Isaacs et al. 1994; Schulte-Merker and Smith 1995). These FGFs also specify mesodermal fates and regulate gastrulation movements in the frog, Xenopus (Isaacs et al. 1994), zebrafish (Griffin et al. 1995), and chick (Yang et al. 2002). Signaling through FGFR1 orchestrates epithelial–mesenchymal transformation (EMT) during mouse gastrulation by activating a genetic network, which includes snail and E-cadherin genes (Ciruna and Rossant 2001).
FGFs in neural induction
Neural induction is another function for FGF signaling in bilaterian development. In Drosophila, the FGFR heartless is required for neurogenesis, functioning in both glia migration and morphogenesis as a downstream mediator of Neuroglian cell-adhesion molecules (Forni et al. 2004; Garcia-Alonso et al. 2000). In C. elegans, FGF signaling affects axon outgrowth via egl-15 (Bulow et al. 2004). In deuterostomes, FGF signaling has been implicated in neural induction in ascidians and all vertebrate models tested thus far (Bertrand et al. 2003; Darras and Nishida 2001; De Robertis and Kuroda 2004; Imai et al. 2002). Although no data exists on FGF ligand expression in the Lophotrochozoa, FGFRs have, however, been implicated in a role in neurogenesis of the platyhelminth Dugesia japonica (Cebria et al. 2002; Mineta et al. 2003), suggesting that a conserved role for FGF signaling in neural induction at least extends to the Lophotrochozoa.
FGF signaling in non-bilaterians
Modern molecular phylogenetics suggest that cnidarians, and possibly placozoans, represent the closest extant animals related to the triploblastic, bilaterally symmetric bilaterians (protostomes and deuterostomes; Collins et al. 2005; Wallberg et al. 2004). Recent molecular evidence suggests that these diverse groups use many of the same signal transduction pathways and transcription factors during early development (Kusserow et al. 2005; Miller et al. 2005; Technau et al. 2005; Technau and Scholz 2003). Two recent studies have examined the FGF signaling pathway in Cnidaria. Sudhop et al. (2004) showed that an FGFR-like gene kringelchen is expressed during bud detachment during asexual reproduction of the hydrozoan Hydra vulgaris and a recent expressed sequence tag (EST) survey of the anthozoans Nematostella vectensis and Acropora millepora yielded several members of FGF signaling pathways (Technau et al. 2005). Although these findings suggests that the evolution of FGF signal transduction in the Metazoa predates the evolution of the Bilateria, their expression or role in development has not been characterized.
We examined the expression of FGF pathway components, including three ligands (NvFGF8A, NvFGF8B, and NvFGF1A), two receptors (NvFGFRa and NvFGFRb), and two targets of FGF signaling, NvChurchill (an ortholog of the zinc finger transcription factor Churchill) and NvSprouty (a feedback inhibitor of FGF signaling), isolated from the genome of the starlet sea anemone N. vectensis, an anthozoan cnidarian. We find these genes variously expressed in the invaginating endoderm during gastrulation, the pharynx, and the developing apical tuft (a chemo-sensory structure). Our results suggest a role for FGF signaling in gastrulation and neurogenesis that predates the protostome/deuterostome split.
Materials and methods
Isolation of genes from N. vectensis
Two assemblies of the N. vectensis genome (Sullivan et al. 2006; http://www.stellabase.org) and Joint Genome Institute (http://www.genome.jgi-psf.org/Nemve1/Nemve1.home.html) and available ESTs (NCBI) were searched using TBLASTN (BLAST, basic local alignment search tool) parameters to isolate potential members of the FGF signal transduction pathway. Gene-specific primers were then designed for 5′ and 3′ rapid amplification of cDNA ends (RACE) with annealing temperatures between 68 and 70°C. RACE was performed using the Smart Race cDNA amplification kit (BD Biosciences Clontech). RACE products were cloned in a plasmid vector (p-GEM T Easy, Promega) and sequenced at Macrogen (South Korea). Of the 13 predicted FGF ligands in the N. vectensis genome, seven were isolated by polymerase chain reaction. The remaining six FGF ligands are predicted and referenced by their protein identification number from the Joint Genome Institute N. vectensis assembly. Overlapping 5′- and 3′-RACE fragments were aligned and submitted to GenBank as composite transcripts. (EF068140-EF068151). Gene-specific primer sequences are available upon request.
Linkage analysis
An assembly of the N. vectensis genome (Joint Genome Institute) was searched using nucleotide sequences of all isolated FGF ligands and NvSprouty utilizing BLASTN search parameters. Where multiple genes were found to occupy the same scaffold, further BLAST searches were conducted to determine whether any of the FGF ligands were located near each other or the Sprouty homolog NvSprouty.
Phylogenetic analyses
Phylogenetic analysis of the FGF ligands, receptors, and NvSprouty were performed to determine orthology. N. vectensis genes were analyzed via BLASTX searches of the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/) to build an alignment. Amino acid alignments were then made using MacVector (ClustalW) and corrected by hand for obvious alignment errors. A Bayesian phylogenetic analysis was conducted using MrBayes 3.1 (Ronquist and Huelsenbeck 2003) using the “WAG+G” amino acid model option with four independent runs of 1 million generations each, sampled every 100 generations with four chains. A summary “consensus tree” was produced in MrBayes, from the last 9,500 trees of each run (38,000 total trees) representing 3.8 million stationary generations. Posterior probabilities were calculated from this “consensus”. Additionally, maximum likelihood (using PHYML [Guindon and Gascuel 2003]) with the WAG+G model of evolution (selected via ProtTest [Abascal et al. 2005]) using 1,000 bootstrap replicates and neighbor joining (using mean AA distances with 1,000 bootstrap replicates in PAUP* v4.0b10 [Swofford 1998]) analyses were conducted. Nexus alignment files can be found in the supplemental information (S7, S8, S9, S10).
In situ hybridization
In situ hybridizations using 1–3 kb digoxygenin-labeled antisense ribonucleotide probes were performed to follow transcript distribution as previously described (Martindale et al. 2004). Probe concentrations ranged from 1.0–2.0 ng/μl, and hybridizations were performed at 60°C for 24–48 h. Alkaline phosphatase reaction products were visualized with nitro blue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl phosphate (NBT-BCIP). Specimens were photographed on a Zeiss Axioplan and AxioImager with a Nikon Coolpix 990 digital camera. Detailed protocols are available upon request (mqmartin@hawaii.edu).
Results
The identification of FGF signaling pathway genes
Directed searches of an assembly of the N. vectensis genome ([Sullivan et al. 2006] and Joint Genome Institute) resulted in the identification of 13 potential FGF ligands, three potential FGFRs, and two known targets of the FGF signaling pathway, Sprouty and Churchill. Phylogenetic analyses of these genes were performed to determine orthology (Fig. 1, and S1, S2, S3, S4) and to gain a better understanding about the evolution of the FGF signaling pathway. A recent phylogenetic analysis of the FGF superfamily suggests grouping FGF ligands into eight classes (FGF-A to -H; Popovici et al. 2005). A Bayesian phylogenetic analysis of the FGF core domain (Fig. 1) from bilaterian and N. vectensis FGFs suggests that N. vectensis possesses four clear orthologs to the FGF-D class (NvFGF8A, NvFGF8B, 208072, and 204532). The nine remaining N. vectensis FGFs (NvFGF1A–1E, 212165, 211797) fail to group with any other bilaterian FGFs with significant support values and likely represent cnidarian-specific FGFs (Fig. 1, S1).
Fig. 1.

Molecular phylogeny FGF signaling pathway members. Previous studies have identified eight classes of FGFs within the Metazoa (Popovici et al. 2005). We identified 13 putative genes that possessed FGF core domains within the N. vectensis genome. Of the 13, a Bayesian analysis confirms the orthology for four FGF ligands (blue arrows) all within the FGF-D class (FGF8/17/18). The remaining nine ligands (black arrows) in the N. vectensis genome appear to cluster together and may either represent cnidarian-specific FGF groups or belong to one of the established classes, but the phyloge netic relationship has been obscured. N. vectensis sequences are shown in bold with arrows. Boxes demark those FGF ligands where expression patterns have been determined. Numbers above branches indicate posterior probabilities, whereas numbers below branches indicate bootstrap support from a maximum likelihood analysis. Clades have been condensed down for illustrative purposes. See supplementary information for the complete tree
Genomic searches for FGFRs identified three RTKs that were potential FGFRs. Previous phylogenetic analyses of the FGFRs suggested an ancestral state of one receptor in bilaterians, with one receptor in nematodes, two related receptors in Drosophila, one receptor in ascidians, and four in vertebrates (FGFR1–4; Itoh and Ornitz 2004). The presence of three potential receptors in the N. vectensis genome runs contrary to previously proposed evolutionary scenarios (Itoh and Ornitz 2004). However, a Bayesian phylogenetic analysis of the tyrosine kinase catalytic domain shows that two of the N. vectensis receptors, NvFGFRa and NvFGFRb, cluster together with 100% posterior probability, forming a sister group to a Hydra and a C. elegans FGFR (S2). The third potential FGFR isolated from N. vectensis (NvFGFRc) seems most similar to a vascular endothelial growth factor receptor from another hydrozoan, Podocoryne carnea, and may not be a true FGFR.
We also cloned orthologs of Churchill, an FGF target gene in vertebrates, and Sprouty, an FGF and epidermal growth factor (EGF, also RTK) target gene in Drosophila and vertebrates. Churchill encodes a zinc finger transcription factor (Sheng et al. 2003), and Sprouty encodes a ring finger ubiquitin ligase that acts as an intracellular inhibitor of FGF signaling (Kim and Bar-Sagi 2004; Nutt et al. 2001; Sivak et al. 2005). In a Bayesian phylogenetic analysis, NvSprouty shows a sister group relationship to both Drosophila and vertebrate Sprouty genes with 100% posterior probability (S3), and NvChurchill shows a sister group relationship to the urchin ortholog of Churchill with 100% posterior probability (S4).
Genomic linkage of FGF and Sprouty genes in the N. vectensis genome
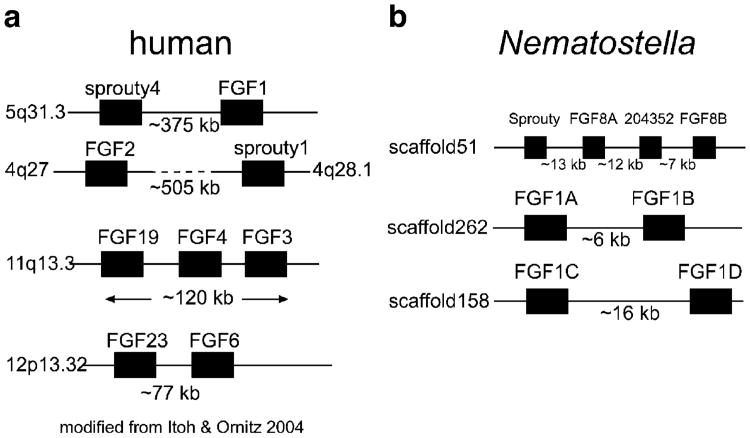
The regulatory relationship between FGF ligands and Sprouty genes may be an ancient one that extends to close physical linkage of their genes. In the human genome, FGF1/2 class genes are closely linked to Sprouty orthologs and also with each other (Fig. 2a; Itoh and Ornitz 2004; Popovici et al. 2005). The conservation of the gene order among FGF classes, as well as FGFRs, in the human genome has lead to speculation that an ancestral FGF cluster arose via large-scale genome duplication events. Searching the available N. vectensis genomic assembly (Joint Genome Institute) has revealed linkage between N. vectensis FGF pathway members. The NvSprouty gene is closely linked to the three of four NvFGF8-class orthologs separated by ∼13 kb from NvFGF8A along a contig of ∼1.2 Mb (Fig. 2b). The two other FGF-D class orthologs (204532 and NvFGF8B) are separated by ∼12 and ∼7 kb, respectively. The close linkage of these genes in a cnidarian suggests that the association between FGF ligands and a potential target is an ancient one, predating the Bilateria. Additionally, two other pairs of putative FGF ligands are linked in the N. vectensis genome (NvFGF1A and NvFGF1B are ∼6 kb apart, and NvFGF1C and NvFGF1D are ∼16 kb apart; Fig. 2b). This linkage may reflect recent duplications in N. vectensis.
Fig. 2.
Ancient genomic linkage for FGF and Sprouty genes. a Gene loci maps for human FGF and Sprouty show linkage in the human genome. FGF1/2 (FGF-A class) genes are linked to Sprouty orthologs in the human (a) and mouse genomes (data not shown; Itoh and Ornitz 2004). Members of FGF-B (FGF3), FGF-C (FGF4 and FGF6) and FGF-G (FGF19 and FGF23) classes also show linkage in mammalian genomes. The approximate distances separating linked genes in kilobases (kb) is shown below each linkage group. b In the FGF-D class, three of four genes (NvFGF8A, 204532, and NvFGF8B) are all linked in the Nematostella genome, with NvFGF8A also closely linked to the Sprouty ortholog, NvSprouty, suggesting that this association between FGF and Sprouty genes is an ancient one, predating the cnidarian–bilaterian split (∼600 MYA). There is also evidence of linkage between other FGF genes in Nematostella, with two pairs of FGFs closely associated on two different genomic scaffolds
NvFGF8A is expressed during gastrulation and during apical tuft formation
We determined the spatio-temporal localization of FGF signaling pathway molecules in N. vectensis embryogenesis by in situ hybridization with antisense RNA probes. NvFGF8A transcripts are not detected during cleavage or blastula stages of development (Fig. 3a,b) but first appear localized to the invaginating blastopore during gastrulation (Fig. 3c–e). This expression persists at the blastopore and expands into the developing pharynx during gastrulation, but the expression is absent in endodermal derivatives post-gastrulation (Fig. 3f). At the end of gastrulation, NvFGF8A is expressed in the pharynx and a few endoderm cells, as well as ectodermal cells at the base of the apical tuft, a chemosensory structure of the planula larva (Pang et al. 2004; Fig. 3g). This pattern persists throughout planula and early polyp stages of development (Fig. 3h–i). An additional domain of expression arises during early polyp development, where NvFGF8A transcripts appear in a group of ectodermal cells at the aboral end of the growing primary mesenteries and adjoining body wall endoderm (Fig. 3i,j). NvFGF8B expression was only detectable during planula and polyp stages in a few endodermal cells at the aboral end below the apical tuft (S5).
Fig. 3.

NvFGF8A is expressed during gastrulation and during apical tuft formation. In situ hybridization of an antisense RNA probe towards NvFGF8A shows that NvFGF8A is not expressed during cleavage (a) or blastula (b) stages. Transcripts first appear at the onset of gastrulation, with staining visible in the blastopore (c–e). Expression remains restricted orally, to the developing pharynx in pharyngeal ectoderm (f–h). During planula and polyp development, transcripts are also detectable at the base of the apical tuft, in the ectoderm (g–j). During tentacle bud formation, expression is also detected in a few cells at the tips of the growing mesenteries and in the accompanied body wall endoderm (h–j). All embryo views are lateral, with the asterisk denoting the blastopore and future oral side, except (a) and (b), which are cleavage and blastula stage embryos, and (e), which is an oral view
NvFGF1A and NvFGFRa are co-expressed at the aboral pole
The orphan FGF ligand NvFGF1A and a receptor NvFGFRa are expressed in similar patterns. Expression of both genes begins during gastrulation, localized at the aboral pole of the embryo (Fig. 4a,e). The initial expression domain of the receptor NvFGFRa (Fig. 4e), however, is broader than that of the ligand NvFGF1A (Fig. 4a), but by the end of gastrulation (Fig. 4b,f) and throughout planula (Fig. 4c,g) and polyp (Fig. 4d,h) stages, the ligand and receptor are expressed exclusively in similar domains in aboral ectoderm at the base of the apical tuft.
Fig. 4.

Co-expression of an FGF ligand, NvFGF1A, and a receptor, FGFRa at the aboral pole during development. NvFGF1A is first expressed during gastrulation where transcripts accumulate at the aboral pole in a broad domain (a). This domain of NvFGF1A expression becomes restricted to fewer cells as development proceeds (b–d). During planula and polyp development, expression can only be found in the ectodermal cells that give rise to the apical tuft (c–d). e–h NvFGFRa is expressed in a pattern nearly identical to that of NvFGF1A, except the initial expression domain of NvFGF1A is broader during gastrulation and early planula development (e, f). All embryo views are lateral, with the asterisk denoting the blastopore and future oral side
NvFGFRb is expressed orally and aborally during development
In contrast to NvFGFRa, the second N. vectensis FGFR gene, NvFGFRb, displays a more dynamic expression pattern, with transcripts absent at blastula and early gastrula stages (Fig. 5a) and first appearing in the pharynx at the planula stage (Fig. 5b). Shortly thereafter, during late planula and early polyp development, endodermal NvFGFRb expression expands into the primary mesenteries and is detectable at the base of the apical tuft at the aboral end (Fig. 5c,d). This aboral staining is transient, as transcripts disappear during later polyp development (Fig. 5e). As the polyp develops, pharyngeal NvFGFRb expression expands into the developing tentacular (Fig. 5d) and inter-tentacular endoderm (Fig. 5e), the latter forming a symmetrical expression pattern (Fig. 5f).
Fig. 5.
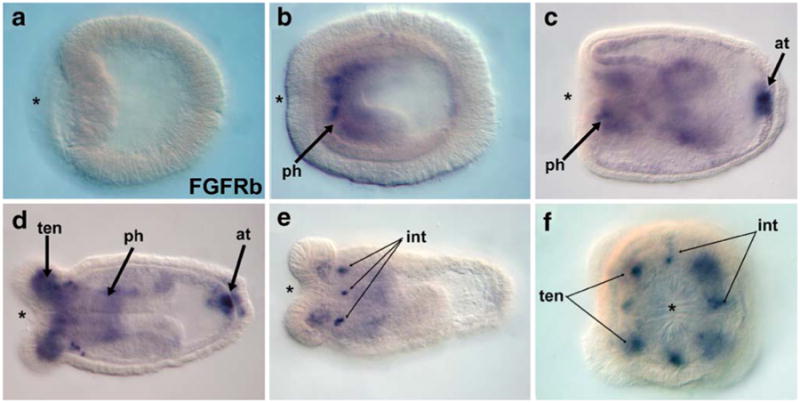
Expression of the receptor, FGFRb at the oral and aboral poles. NvFGFRb is not expressed during gastrulation (a), but transcripts are first visible in the developing pharynx of the planula (b). During later planula development, expression can also be visualized in a few cells in the endoderm below the apical tuft (arrow). Later, during polyp formation, NvFGFRb is expressed in the endoderm of the growing tentacles (d, f). Expression is also visible in discrete cells forming a symmetric pattern in the intra-tentacle endoderm (e, f). The asterisk denotes the blastopore and future oral pole (ph pharynx; at apical tuft; ten tentacle; int inter-tentacular endoderm). All embryo views are lateral except (f), which is an oral view
NvSprouty, a potential target and inhibitor, is co-expressed with FGF ligands and receptors
NvSprouty is expressed in domains that overlap NvFGF8A, NvFGF8B, and NvFGF1A. This expression is dynamic, with transcripts appearing during blastula stages in a subset of cells (Fig. 6a) that continue to express NvSprouty at the blastopore during gastrulation (Fig. 6b,c). NvSprouty expression initiates in the blastula before FGF ligand and receptor gene expression (Fig. 6a). This is somewhat unusual because Sprouty genes are typically induced in response to FGF or EGF ligands (Nutt et al. 2001; Sivak et al. 2005; perhaps EGF or cryptic levels of FGF are expressed in the blastula). As gastrulation proceeds, a second domain of expression is initiated at the aboral pole in adjoining ectoderm and endoderm (Fig. 6b). This bipolar pharyngeal-apical tuft pattern of expression continues throughout development in both germ layers, (Fig. 6d–f). During early polyp development (tentacle bud stages), oral expression expands to include the ectoderm and endoderm of the growing tentacle (data not shown). Tentacle expression persists throughout polyp development (Fig. 6e,f), although it is later confined to a few endodermal cells at the tips of the tentacles (Fig. 6f).
Fig. 6.

Expression of two potential targets of FGF signaling, NvSprouty and NvChurchill. The expression profile of NvSprouty largely follows that of NvFGF8A and NvFGF1A during embryogenesis, with a few notable exceptions. NvSprouty is first expressed in presumptive endoderm during blastula-stages (a). During gastrulation, NvSprouty is expressed broadly in the blastopore and invaginating endoderm as well as at the apical tuft (b, c). In planula development, the oral/aboral expression pattern remains, although it expands orally such that expression is seen in the ectoderm and endoderm of the apical tuft (d, e) in the ectoderm and endoderm of growing tentacles (e, f) and in a broad domain in the pharynx (d–f). Transcripts to the zinc finger transcription factor NvChurchill are not detectable during cleavage (data not shown) and gastrulation (g). Expression begins during planula stages (h) in body wall endoderm near the oral pole. During polyp formation, expression is restricted to a ring of cells in the pharyngeal endoderm (i–l), where it remains expressed throughout juvenile stages (k, l). The asterisk denotes the blastopore and future oral pole. (ph pharynx; at apical tuft; ten tentacle; t. ect tentacle ectoderm; t. end tentacle endoderm, b. end body-wall endoderm) All embryo views are lateral, except (c), (j), and (l), which are oral views
Expression of NvChurchill, another potential target of FGF signaling, is restricted to a ring in the pharyngeal endoderm
In N. vectensis, the Churchill ortholog, NvChurchill (S4), is not expressed until after gastrulation is complete (Fig. 6g). During planula stages, transcripts are localized to body wall endoderm along the oral half of the embryo (Fig. 6h). During tentacle bud formation in early polyps, transcripts are detectable in a ring of pharyngeal endoderm (Fig. 6i,j) that persists throughout polyp stages (Fig. 6k,l).
Discussion
Evolution of the FGF family
With the identification of FGF signal transduction components in N. vectensis and other cnidarians (Sudhop et al. 2004; Technau et al. 2005), all eumetazoans surveyed possess FGF pathways. Thus, the FGF signal transduction system is an ancient one (Itoh and Ornitz 2004; Popovici et al. 2005), predating the cnidarian/bilaterian divergence during the pre-Cambrian (∼600 MYA). Although no data have addressed the presence of FGF pathways in other nonbilaterians (e.g., ctenophores, placozoans, and sponges), genomic sequencing efforts within these phyla should soon shed light on this issue. The broad question of whether FGF signaling is metazoan specific remains unanswered, but an RTK has been isolated from a choanoflagellate, a taxa considered to be the outgroup to the Metazoa (King and Carroll 2001; King et al. 2003). Thus, an evolutionary antecedent of the FGF pathway may have been in place before the metazoan radiation (King and Carroll 2001).
FGF receptor evolution
Our identification of 13 potential FGF ligands, two receptors, and two predicted downstream target genes suggests that FGF signaling has diversified within the Cnidaria. Concerning FGF receptors, previous work predicted one FGFR in the protostome/deuterostome ancestor (Itoh and Ornitz 2004), and our data support this hypothesis, with the two N. vectensis FGFRs (NvFGFRa and NvFGFRb) likely having arisen through a cnidarian-specific independent duplication event (S2). N. vectensis possesses orthologs of the canonical downstream RTK signal transduction pathway (GRB1–SOS–ras–MEK–ERK; S6), demonstrating that signals from FGFRs to their nuclear target genes likely travel an ancient pathway that is conserved across Metazoa.
Ligand expansion in the Cnidaria
Phylogenetic analyses (Fig. 1, S1) suggest that many of the FGF ligands we identified may be either N. vectensis or cnidarian specific, or that their orthologs may have been lost in the Bilateria. We were only able to show clear orthology for four of 13 potential ligand families, all within the FGF-D (FGF8/17/18) class (Fig. 1). Due to low conservation of amino acid sequence identity within the core FGF domain and the relatively short length of the domain itself (∼120 amino acids), phylogenetic reconstruction of relationships within FGFs has been difficult (Itoh and Ornitz 2004; Popovici et al. 2005).
The remainder of FGFs in N. vectensis may represent a unique cnidarian-specific FGF class, as we were unable to establish orthology with bilaterian classes (Fig. 1). Four of these nine orphan FGFs show linkage in the N. vectensis genome (Fig. 2b), suggesting that they arose by tandem duplication events over cnidarian evolution. Previous work on FGF family evolution has predicted two or three FGFs present during early metazoan evolution (Itoh and Ornitz 2004), with as many as eight proto-FGFs present in the protostome/deuterostome ancestor (Popovici et al. 2005). Our data suggests that one FGF member was present in the cnidarian-bilaterian ancestor. The absence of cnidarian members of FGF-B, -C, -E, -F, -G, and -H classes suggests that these classes may be bilaterian specific, or were lost in the cnidarian lineage.
Sprouty can serve as a target and inhibitor for FGF signaling
Sprouty was first identified in Drosophila, where it has been shown to inhibit several different RTK signaling pathways, including EGFs and FGFs (Hacohen et al. 1998). Sprouty, originally thought to be secreted, has subsequently been found to act intracellularly, where it inhibits the Ras/MAPK pathway, although it has been shown to act at different levels of the signaling pathway in different contexts (Kim and Bar-Sagi 2004; Nutt et al. 2001). In frogs, FGF signaling controls mesoderm induction through the activation of brachyury (Xbra) and helps coordinate convergent extension movements along with noncanonical Wnt signaling. The two frog Sprouty orthologs have been shown to inhibit convergent extension by serving as antagonists of FGF-dependent calcium signaling rather than affecting the MAPK pathway that leads to mesoderm induction (Kim and Bar-Sagi 2004; Nutt et al. 2001). Xenopus Sprouty2 is expressed in an overlapping but broader domain than that of Xenopus FGF8, both during gastrulation and in anterior neural structures (Nutt et al. 2001). Expression of NvSprouty also seems to parallel that of FGF ligands, in that it is expressed both orally and aborally throughout development in broader but similar domains to that of the FGF8 orthologs (NvFGF8A, NvFGF8B) and the orphan FGF, NvFGF1A (Fig. 7). However, NvSprouty is temporally expressed before both NvFGF8 class genes in both presumptive endoderm and aborally marking both the future blastopore and apical tuft, respectively, suggesting that it may be under the activation of a different signaling pathway during early development. The later co-expression of ligands and NvSprouty suggests that the FGF-Sprouty feedback loop may be conserved between cnidarians and bilaterians, an observation supported by genomic linkage between vertebrate Sprouty and FGF1/2 genes and N. vectensis NvSprouty and NvFGF8 class genes (Fig. 2).
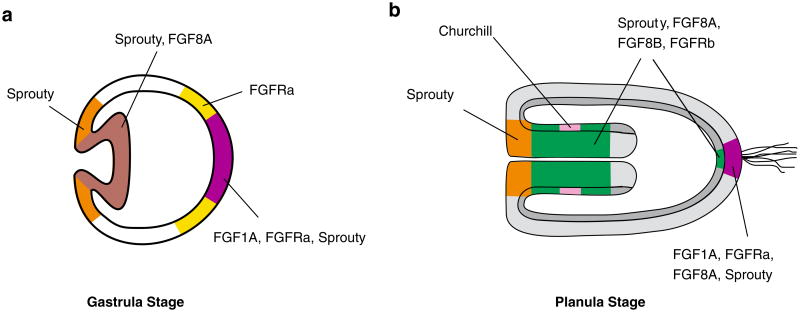
Fig. 7.
Summary of expression of FGF signaling molecules during gastrulation and planula development in N. vectensis. During gastrulation (a), FGF ligands are expressed at the blastopore and in invaginating endoderm (NvFGFA8) and at the aboral pole (NvFGF1A). An FGF receptor NvFGFRa is also expressed at the aboral pole in a slightly broader domain than that of the ligand NvFGF1A. NvSprouty, a potential downstream target and known inhibitor of the pathway, is expressed in oral and aboral domains coincident with ligand and receptor localization. During planula development (b), FGF ligands, receptors, and an inhibitor continue to show restricted expression orally in the pharynx (NvFGF8A, NvFGFRb, NvChurchill and NvSprouty) and at the base of the apical tuft in endoderm (NvFGFRb, NvFGF8A, NvFGF8B, and NvSprouty) and ectoderm (NvFGF1A, NvFGFRa, NvFGF8A, and NvSprouty). The expression profiles of FGF pathway genes suggest a role in both gastrulation and the induction of a known neural structure, the planula's apical tuft
FGF signaling in cnidarians
It has recently been shown in N. vectensis that TGF-β signaling may be occurring in a planar autocrine fashion, with ligands and downstream components (Smads) co-expressed in presumptive endoderm during and after gastrulation (Matus et al. 2006b). From expression data alone, it appears possible that FGF signaling in N. vectensis may be occurring in either a planar or trans-epithelial fashion. At the oral pole, during gastrulation and pharynx development, a ligand (NvFGF8A), a receptor (NvFGFRb), and a potential target and inhibitor (NvSprouty) are all co-expressed in the endoderm, suggesting that planar signaling may be occurring (Fig. 7a). Aborally, FGF ligands, receptors, and NvSprouty are localized to both endodermal and ectodermal cells at the base of the apical tuft. Although it seems likely that NvFGF1A and NvFGFRa, which share an ectodermal aboral expression domain throughout embryogenesis, form a ligand/receptor pair, another receptor (NvFGFRb) is expressed in the endoderm and could be receiving the FGF signal, whereas NvFGF8B is expressed exclusively within the endoderm at the base of the apical tuft, and NvFGF8A is expressed in both germ layers at the base of the apical tuft (Fig. 7b). Even in a model system work, it has been difficult to predict FGF receptor/ligand pairing, and further work will be needed to predict specific ligand/receptor pairings for FGF signaling in N. vectensis.
Although FGF signaling is well characterized in epithelial–mesenchymal signaling in a variety of different contexts in both protostomes (Huang and Stern 2005) and deuterostomes (Martin 1998; Min et al. 1998; Zhang et al. 2006), cnidarians only have two definitive germ layers, an outer ectoderm and an inner endoderm. It may be that the germ-layer organization of cnidarians and the lack of true mesenchymal derivatives in N. vectensis precludes epithelial-mesenchymal signaling in general. Although there are reports of EMT occurring in N. vectensis (Kraus and Technau 2006), more recent work indicates that EMT does not occur during N. vectensis gastrulation (Magie et al., personal communication). The expression of “mesodermal” genes in the endoderm of N. vectensis (Fritzenwanker et al. 2004; Martindale et al. 2004; Technau 2001) suggests that the endoderm may be a precursor to bilaterian endoderm and mesoderm. The evolution of signaling systems that segregated receptors and ligands to endoderm and mesoderm, respectively, likely predated the evolution of mesoderm. If this is the case, then it is not surprising that cell–cell signaling can occur in a planar fashion during N. vectensis development.
A role for FGFs in gastrulation in cnidarians
FGF signaling has been implicated in playing a role in coordinating gastrulation movements and the induction of mesoderm in both vertebrates (Isaacs et al. 1994; Rossant et al. 1997) and flies (Stathopoulos et al. 2004). The onset of expression of NvSprouty, a likely target of FGF signaling (Nutt et al. 2001; Sivak et al. 2005), in the blastula (Fig. 6a), and the blastoporal and pharyngeal expression of both NvSprouty and NvFGF8A suggest that FGF signaling may be important in coordinating gastrulation and endoderm development in N. vectensis, implicating an ancient conservation of FGF signaling that predates the cnidarian-bilaterian divergence.
A role for FGFs in neural induction in cnidarians
FGF pathway members are deployed in a bipolar fashion during N. vectensis development, with ligands, receptors, an inhibitor, and response gene expressed in the pharynx (NvFGF8A, NvFGFRb, NvSprouty, and NvChurchill, respectively) and at the apical tuft (NvFGF8A, NvFGF8B, NvFGF1A, NvFGFRa, NvFGFRb, and NvSprouty; Fig. 7). It has recently been shown that the pharynx of cnidarians asymmetrically expresses several of the genes found in the vertebrate “organizer”, including TGF-β antagonists NvNoggin1 and NvFollistatin and a transcription factor NvGsc (Matus et al. 2006a), and that some of these genes are also expressed at the aboral pole at the base of the apical tuft, a chemosensory structure (Pang et al. 2004). FGF signaling alone in ascidians (Bertrand et al. 2003; Miya and Nishida 2003) or together with TGF-β antagonism in vertebrates (De Robertis and Kuroda 2004; Delaune et al. 2005; Koshida et al. 2002) coordinate neural induction. It seems likely then that the expression of FGF signaling pathway genes in the pharynx and apical tuft may to be playing a role in neural induction in N. vectensis. Because Churchill genes have been shown to be downstream of neural induction pathways in FGF signaling in vertebrates (Sheng et al. 2003), the pharyngeal endodermal ring of NvChurchill expression, which may correspond to the location of a cnidarian circumoral nerve ring (Nielsen 2005), supports a role for FGF signaling in neural patterning in cnidarians. However, a better understanding of the organization of the cnidarian nervous system is needed.
FGF signaling coordinates gastrulation and neurogenesis
From work in bilaterian model systems, a given FGF ligand can have multiple conserved developmental roles, including the induction of both mesoderm and neural tissue and the coordination of gastrulation movements (Bertrand et al. 2003; Sheng et al. 2003). In ascidians, CiFGF9/16/20 is the ligand responsible for inducing both mesenchyme and notochord in the vegetal hemisphere, inducing neural fates in the animal hemisphere (Bertrand et al. 2003; Sheng et al. 2003). In chick development, the FGF-responsive zinc finger gene, Churchill, acts as a switch to regulate the transition between gastrulation and neurulation by activating Smad-interacting protein-1, which inhibits the FGF-dependent induction of mesoderm fate determination genes, such as brachyury and Tbx6L (Sheng et al. 2003). Differences in the downstream transcriptional machinery present within particular cells could allow the same FGF signal to carry out diverse functions, such as mesoderm or neural induction. The pharyngeal endodermal expression of NvChurchill (Figs. 6g–1 and 7) is located within the domain of NvFGF8A expression and may be involved with the induction of the circumpharyngeal nerve ring found in most anthozoans (Nielsen 2005).
Conclusions
Along with Wnt, TGF-β, Hedgehog, and Notch, the FGF signal transduction pathway is a key player in the early development of animals (Gerhart 1999). It is involved in the regulation of a myriad of developmental processes, especially in mediating gastrulation movements, as well as mesoderm and neural induction in organisms as diverse as flies and vertebrates. We have shown the first evidence of expression of FGF signaling pathway members during development in N. vectensis, an anthozoan cnidarian. FGF orthologs appear to be involved in both gastrulation, pharynx development, and the formation of the apical tuft, suggesting that the link between FGF signaling in gastrulation and neural induction is an ancient one, predating the Cambrian explosion and the cnidarian/bilaterian divergence. The development of functional techniques in N. vectensis will be vital to elucidate whether FGF signaling is directly involved in the coordination of these events, which seems likely as suggested by the spatio-temporal expression pattern of the FGF signal transduction pathway genes reported here.
Supplementary Material
Footnotes
Electronic supplementary material: Supplementary material is available in the online version of this article at http://dx.doi.org/10.1007/s00427-006-0122-3 and is accessible for authorized users.
Contributor Information
David Q. Matus, Kewalo Marine Lab, Pacific Bioscience Research Centre, University of Hawai'i, 41 Ahui Street, Honolulu, HI 96813, USA
Gerald H. Thomsen, Department of Biochemistry and Cell Biology, Center for Developmental Genetics, Stony Brook University, Stony Brook, NY 11794-5215, USA
Mark Q. Martindale, Email: mqmartin@hawaii.edu, Kewalo Marine Lab, Pacific Bioscience Research Centre, University of Hawai'i, 41 Ahui Street, Honolulu, HI 96813, USA.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Amaya E, Stein PA, Musci TJ, Kirschner MW. FGF signaling in the early specification of mesoderm in Xenopus. Development. 1993;118:477–487. doi: 10.1242/dev.118.2.477. [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115:615–627. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Branda CS, Stern MJ. Mechanisms controlling sex myoblast migration in Caenorhabditis elegans hermaphrodites. Dev Biol. 2000;226:137–151. doi: 10.1006/dbio.2000.9853. [DOI] [PubMed] [Google Scholar]
- Bulow HE, Boulin T, Hobert O. Differential functions of the C. elegans FGF receptor in axon outgrowth and maintenance of axon position. Neuron. 2004;42:367–374. doi: 10.1016/s0896-6273(04)00246-6. [DOI] [PubMed] [Google Scholar]
- Cebria F, Kobayashi C, Umesono Y, Nakazawa M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sanchez Alvarado A, Agata K. FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature. 2002;419:620–624. doi: 10.1038/nature01042. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Collins AG, Cartwright P, McFadden CS, Scheirwater B. Phylogenetic context and basal metazoan model systems. Integr Comp Biol. 2005;45:585–594. doi: 10.1093/icb/45.4.585. [DOI] [PubMed] [Google Scholar]
- Darras S, Nishida H. The BMP signaling pathway is required together with the FGF pathway for notochord induction in the ascidian embryo. Development. 2001;128:2629–2638. doi: 10.1242/dev.128.14.2629. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Forni JJ, Romani S, Doherty P, Tear G. Neuroglian and FasciclinII can promote neurite outgrowth via the FGF receptor Heartless. Mol Cell Neurosci. 2004;26:282–291. doi: 10.1016/j.mcn.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Fritzenwanker JH, Saina M, Technau U. Analysis of forkhead and snail expression reveals epithelial–mesenchymal transitions during embryonic and larval development of Nematostella vectensis. Dev Biol. 2004;275:389–402. doi: 10.1016/j.ydbio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Garcia-Alonso L, Romani S, Jimenez F. The EGF and FGF receptors mediate neuroglian function to control growth cone decisions during sensory axon guidance in Drosophila. Neuron. 2000;28:741–752. doi: 10.1016/s0896-6273(00)00150-1. [DOI] [PubMed] [Google Scholar]
- Gerhart J. 1998 Warkany lecture: signaling pathways in development. Teratology. 1999;60:226–239. doi: 10.1002/(SICI)1096-9926(199910)60:4<226::AID-TERA7>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Huang P, Stern MJ. FGF signaling in flies and worms: more and more relevant to vertebrate biology. Cytokine Growth Factor Rev. 2005;16:151–158. doi: 10.1016/j.cytogfr.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. Early embryonic expression of FGF4/6/9 gene and its role in the induction of mesenchyme and notochord in Ciona savignyi embryos. Development. 2002;129:1729–1738. doi: 10.1242/dev.129.7.1729. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. A twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development. 2003;130:4461–4472. doi: 10.1242/dev.00652. [DOI] [PubMed] [Google Scholar]
- Isaacs HV, Pownall ME, Slack JM. eFGF regulates Xbra expression during Xenopus gastrulation. EMBO J. 1994;13:4469–4481. doi: 10.1002/j.1460-2075.1994.tb06769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families. Trends Genet. 2004;20:563–569. doi: 10.1016/j.tig.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol. 2004;5:441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci USA. 2001;98:15032–15037. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- Klambt C, Glazer L, Shilo BZ. Breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Koshida S, Shinya M, Nikaido M, Ueno N, Schulte-Merker S, Kuroiwa A, Takeda H. Inhibition of BMP activity by the FGF signal promotes posterior neural development in zebrafish. Dev Biol. 2002;244:9–20. doi: 10.1006/dbio.2002.0581. [DOI] [PubMed] [Google Scholar]
- Kraus Y, Technau U. Gastrulation in the sea anemone Nematostella vectensis occurs by invagination and immigration: an ultrastructural study. Dev Genes Evol. 2006;216:119–132. doi: 10.1007/s00427-005-0038-3. [DOI] [PubMed] [Google Scholar]
- Kusserow A, Pang K, Sturm C, Hrouda M, Lentfer J, Schmidt HA, Technau U, von Haeseler A, Hobmayer B, Martindale MQ, Holstein TW. Unexpected complexity of the Wnt gene family in a sea anemone. Nature. 2005;433:156–160. doi: 10.1038/nature03158. [DOI] [PubMed] [Google Scholar]
- Martin GR. The roles of FGFs in the early development of vertebrate limbs. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- Martindale MQ, Pang K, Finnerty JR. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa) Development. 2004;131:2463–2474. doi: 10.1242/dev.01119. [DOI] [PubMed] [Google Scholar]
- Matus DQ, Pang K, Marlow H, Dunn CW, Thomsen GH, Martindale MQ. Molecular evidence for deep evolutionary roots of bilaterality in animal development. Proc Natl Acad Sci USA. 2006a;103:11195–11200. doi: 10.1073/pnas.0601257103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus DQ, Thomsen GH, Martindale MQ. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr Biol. 2006b;16:499–505. doi: 10.1016/j.cub.2006.01.052. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21:536–539. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineta K, Nakazawa M, Cebria F, Ikeo K, Agata K, Gojobori T. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc Natl Acad Sci USA. 2003;100:7666–7671. doi: 10.1073/pnas.1332513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya T, Nishida H. An Ets transcription factor, HrEts, is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev Biol. 2003;261:25–38. doi: 10.1016/s0012-1606(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Nielsen C. Larval and adult brains. Evol Dev. 2005;7:483–489. doi: 10.1111/j.1525-142X.2005.05051.x. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol. 2001;2(3) doi: 10.1186/gb-2001-2-3-reviews3005. REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K, Matus DQ, Martindale MQ. The ancestral role of COE genes may have been in chemoreception: evidence from the development of the sea anemone, Nematostella vectensis (phylum Cnidaria; class Anthozoa) Dev Genes Evol. 2004;214:134–138. doi: 10.1007/s00427-004-0383-7. [DOI] [PubMed] [Google Scholar]
- Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionary history of the FGF superfamily. Bioessays. 2005;27:849–857. doi: 10.1002/bies.20261. [DOI] [PubMed] [Google Scholar]
- Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7:165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rossant J, Ciruna B, Partanen J. FGF signaling in mouse gastrulation and anteroposterior patterning. Cold Spring Harbor Symp Quant Biol. 1997;62:127–133. [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. Fgf genes in the basal chordate Ciona intestinalis. Dev Genes Evol. 2002;212:432–438. doi: 10.1007/s00427-002-0266-8. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Smith JC. Mesoderm formation in response to Brachyury requires FGF signalling. Curr Biol. 1995;5:62–67. doi: 10.1016/s0960-9822(95)00017-0. [DOI] [PubMed] [Google Scholar]
- Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- Shishido E, Ono N, Kojima T, Saigo K. Requirements of DFR1/Heartless, a mesoderm-specific Drosophila FGF-receptor, for the formation of heart, visceral and somatic muscles, and ensheathing of longitudinal axon tracts in CNS. Development. 1997;124:2119–2128. doi: 10.1242/dev.124.11.2119. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Tam B, Ronshaugen M, Frasch M, Levine M. Pyramus and thisbe: FGF genes that pattern the mesoderm of Drosophila embryos. Genes Dev. 2004;18:687–699. doi: 10.1101/gad.1166404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhop S, Coulier F, Bieller A, Vogt A, Hotz T, Hassel M. Signalling by the FGFR-like tyrosine kinase, Kringelchen, is essential for bud detachment in Hydra vulgaris. Development. 2004;131:4001–4011. doi: 10.1242/dev.01267. [DOI] [PubMed] [Google Scholar]
- Sullivan JC, Ryan JF, Watson JA, Webb J, Mullikin JC, Rokhsar D, Finnerty JR. StellaBase: the Nematostella vectensis genomics database. Nucleic Acids Res. 2006;34:D495–D499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland, MA: 1998. [Google Scholar]
- Technau U. Brachyury, the blastopore and the evolution of the mesoderm. Bioessays. 2001;23:788–794. doi: 10.1002/bies.1114. [DOI] [PubMed] [Google Scholar]
- Technau U, Scholz CB. Origin and evolution of endoderm and mesoderm. Int J Dev Biol. 2003;47:531–539. [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, Gordon PM, Saina M, Grasso LC, Hayward DC, Sensen CW, Saint R, Holstein TW, Ball EE, Miller DJ. Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Wallberg A, Thollesson M, Farris JS, Jondelius U. The phylogenetic position of the comb jellies (Ctenophora) and the importance of taxonomic sampling. Cladistics. 2004;20:558–578. doi: 10.1111/j.1096-0031.2004.00041.x. [DOI] [PubMed] [Google Scholar]
- Yang X, Dormann D, Munsterberg AE, Weijer CJ. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, Ornitz DM. Reciprocal epithelial–mesenchymal FGF signaling is required for cecal development. Development. 2006;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.