Highlight
Seed storage proteins protect seed against oxidative stress during seed ageing.
Key words: Arabidopsis, carbonylation, proteomics, reactive oxygen species, seed longevity, seed storage proteins.
Abstract
Proteomics approaches have been a useful tool for determining the biological roles and functions of individual proteins and identifying the molecular mechanisms that govern seed germination, vigour and viability in response to ageing. In this work the dry seed proteome of four Arabidopsis thaliana genotypes, that carry introgression fragments at the position of seed longevity quantitative trait loci and as a result display different levels of seed longevity, was investigated. Seeds at two physiological states, after-ripened seeds that had the full germination ability and aged (stored) seeds of which the germination ability was severely reduced, were compared. Aged dry seed proteomes were markedly different from the after-ripened and reflected the seed longevity level of the four genotypes, despite the fact that dry seeds are metabolically quiescent. Results confirmed the role of antioxidant systems, notably vitamin E, and indicated that protection and maintenance of the translation machinery and energy pathways are essential for seed longevity. Moreover, a new role for seed storage proteins (SSPs) was identified in dry seeds during ageing. Cruciferins (CRUs) are the most abundant SSPs in Arabidopsis and seeds of a triple mutant for three CRU isoforms (crua crub cruc) were more sensitive to artificial ageing and their seed proteins were highly oxidized compared with wild-type seeds. These results confirm that oxidation is involved in seed deterioration and that SSPs buffer the seed from oxidative stress, thus protecting important proteins required for seed germination and seedling formation.
Introduction
Ecologically, seeds represent a critical stage in the survival of higher plants. Seeds are also important for biodiversity conservation, especially for plants producing orthodox seeds, as they provide desiccation tolerance and permit propagation after long-term dry storage. The germination ability of seeds changes over the seed life span in dry storage conditions. In dormant seeds, the first phase after harvest is reflected by a period in which seeds gradually lose dormancy; during this so called after-ripening (AR) period seeds gain full germination ability. Thereafter, dry seeds slowly deteriorate and lose vigour during storage, which ultimately results in germination failure. It is of both ecological and agronomical relevance to understand the mechanisms governing seed vigour loss during ageing.
Seed longevity is affected by storage conditions, including temperature and humidity (related to seed moisture content). It has been shown that both low temperature and low seed moisture content prolong seed life span during storage (Walters, 1998; Walters et al., 2005). Besides long-term storage (natural ageing), accelerated ageing [controlled deterioration test (CDT), artificial ageing], in which seeds are stored at high temperature and relative humidity, can be used to study seed longevity (Tesnier et al., 2002; Rajjou et al., 2008). It is debated whether accelerated ageing mimics natural ageing (Schwember and Bradford, 2010; Groot et al., 2012). However, for Arabidopsis seeds, accelerated ageing largely results in the same regulating factors as those identified for natural ageing (Bentsink et al., 2000; Clerkx et al., 2004b ). Seed longevity is strongly determined by genetic components. Quantitative trait loci (QTLs) for seed longevity after both natural and artificial ageing were identified in A. thaliana (Bentsink et al., 2000; Clerkx et al., 2004b ; Nguyen et al., 2012), barley (Hordeum vulgare) (Nagel et al., 2009), lettuce (Lactuca sativa) (Schwember and Bradford, 2010), oilseed rape (Nagel et al., 2011), rice (Oryza sativa) (Miura et al., 2002; Sasaki et al., 2005; Zeng et al., 2006; Xue et al., 2008; Hang et al., 2014) and wheat (Triticum aestivum) (Landjeva et al., 2009). In addition, several Arabidopsis mutants and over-expression lines show altered seed longevity phenotypes. Mutations in seed maturation and dormancy genes, such as LEAFY COTYLEDON1 and ABSCISIC ACID INTENSITIVE3 (ABI3), lead to dramatic reduction in seed viability (Ooms et al., 1993; Clerkx et al., 2004a ; Sugliani et al., 2009). Debeaujon et al. (2000) showed that testa-defective mutants, including the transparent testa (tt) and the aberrant testa shape, have reduced seed longevity. The non-dormant dog1 (delay of germination1) mutant (Bentsink et al., 2006) and DOG1-Cvi (Cape Verde Islands) transformed into Ler (Landsberg erecta) also showed reduced seed longevity phenotypes (Nguyen et al., 2012). Vitamin E, an antioxidant preventing non-enzymatic lipid oxidation, has been proven to promote seed longevity since mutants in vitamin E synthesis genes (vte1 and vte2) showed decreased seed longevity (Sattler et al., 2004). DNA LIGASE4 and 6 are necessary to maintain genome integrity, as revealed by the high sensitivity of the lig6 mutant and the lig6 lig4 double mutant to seed ageing (Waterworth et al., 2010). Tobacco (Nicotiana tabacum) seeds over-expressing HaHSFA9 (a heat shock transcription factor isolated from sunflower Helianthus annuus) accumulate elevated levels of heat shock proteins and are more tolerant to CDT (Prieto-Dapena et al., 2006). It has been shown that HaHSFA9 enhances longevity of seeds through functional interaction with a DROUGHT-RESPONSIVE ELEMENT-BINDING FACTOR2 (Almoguera et al., 2009). Bueso et al. (2014) showed that the Arabidopsis isl1-1D mutant, over-expressing ATHB25 (Homeobox25/Zinc finger protein domain), exhibited enhanced seed longevity due to the increased expression of (GIBBERELLIC ACID 3-OXIDASE2), a GA synthesis gene, and thus elevated GA1 and GA4 content. The authors suggested a connection between GA and seed longevity through the reinforcement of the seed coat. Over-expression of PROTEIN-L-ISOASPARTATE METHYLTRANSFERASE1 (PIMT1) enhances seed longevity and germination vigour in Arabidopsis (Oge et al., 2008). In addition, Arabidopsis seeds overexpressing PIMT1 or PIMT2 from chickpea (Cicer arietinum) were remarkably less sensitive to CDT (Verma et al., 2013). Besides PIMT1, which repairs age-induced damage to aspartyl and asparaginyl residues, METHIONINE SULFOXIDE REDUCTASES (MSRs) also repair damaged proteins at methionine residues. Recently, Chaletain et al. (2013) reported that MSR abundance and enzyme activity are strongly linked to seed longevity in Medicago truncatula and Arabidopsis.
Proteomics approaches have been a useful tool for determining the biological roles and functions of individual proteins and identifying the molecular mechanisms that govern seed germination, vigour and viability in response to ageing (Job et al., 2005; Rajjou et al, 2007; 2008). Post-translational modification (PTM) of proteins in dry seeds plays a central role in dormancy release, metabolism resumption, and ageing processes (Arc et al., 2011). These researchers also demonstrated that the accumulation of oxidized (carbonylated) proteins in dry seed is associated with ageing and might induce loss-of-function of proteins and enzymes. Therefore, detoxification of reactive oxygen species (ROS) that result in oxidative stress and maintenance of redox homeostasis are crucial for seed vigour (Rajjou et al., 2008). Due to their abundance in seeds, seed storage proteins (SSPs) are a primary target for oxidation (Davies, 2005). Arabidopsis contains two major SSPs, cruciferins and napins. Cruciferins in Arabidopsis are 12S globulins encoded by four paralogous genes AT5G44120 (CRUA), AT1G03880 (CRUB), AT4G28520 (CRUC) and AT1G03890 (CUPIN). Napins are referred as 2S albumins and belong to a family with five members. Their abundance in Arabidopsis seeds and wide range of PTMs can result in marked changes of SSPs during ageing (Job et al., 2005; Wan et al., 2007). This, together with the fact that SSPs are suggested to contributed to seed germination vigour and support early seedling growth when mobilized upon germination (Muntz et al. 2001), might imply a role for SSPs in seed longevity.
Overall seed longevity is a complex trait and a better understanding of the underlying molecular mechanisms is required. In a previous study, we identified 12 GAAS (GERMINATION ABILITY AFTER STORAGE) loci controlling seed longevity after natural ageing in Arabidopsis (Nguyen et al., 2012). To investigate seed longevity mechanisms influenced by the GAAS loci, here we performed proteome analyses on dry seeds at two physiological states, after-ripened (AR) and 4-year-old (aged) seeds, from four lines with different seed longevities. T-DNA insertion lines for a subset of protein candidates were investigated for their role in seed longevity after accelerated ageing. These analyses revealed that loss of crucifernins and napins reduced seed longevity and identified a role for cruciferins in buffering oxidation during ageing.
Materials and methods
Plant material
The four A. thaliana genotypes, namely Ler, NILGAAS1(-Cape Verde Islands (Cvi)), NILGAAS2(-Antwerp (An-1)) and NILGAAS5 (-Shakdara (Sha)), were originally developed as NILDOG2, NILDOG22 and NILDOG1, respectively (Bentsink et al., 2010; Nguyen et al., 2012). Those genotypes were grown in a randomized complete block design with replicates in soil as described in Bentsink et al. (2010). Seeds of four plants per replicate were bulked. Proteome analyses were conducted for the four genotypes at two physiological stages, fully AR and 4-year-old (aged) seeds. Fully AR seeds are competent to germinate 100% while aged seeds have germination ability reduced; in this study germination phenotype of those seeds were assessed (Fig. 1).
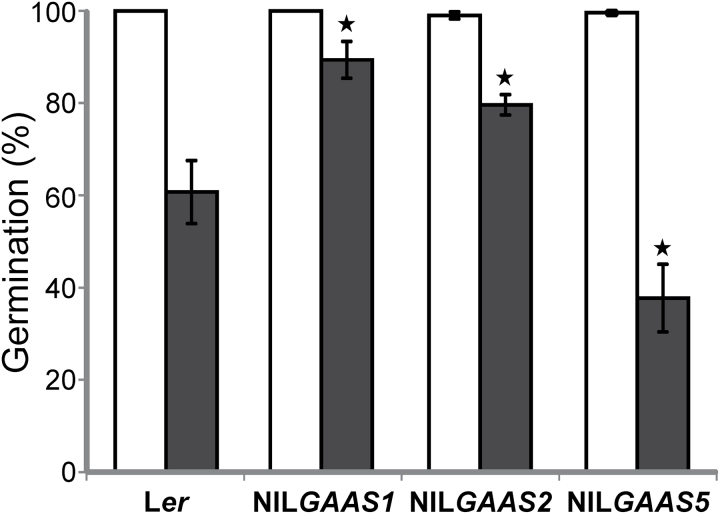
Fig. 1.
Seed germination after seed dry storage. The germination (%) of Ler and the near-isogenic lines NILGAAS1, NILGAAS2 and NILGAAS5 were analysed for after-ripened seeds (open bars) and naturally-aged seeds after four years of storage (filled bars). Averages of four biological replicates with standard errors are presented. The asterisks indicate significant differences between aged NILs and Ler (P<0.05).
Fully AR seeds were stored in 1.5ml microcentrifuge tubes at −80ºC. Aged seeds came from the same harvest, but were stored in cellophane bags (HERA, papierverarbeitung, Germany) under ambient conditions (∼50% relative humidity and 21°C) for four years. Four biological replicates were used in the proteomic analyses.
The single, double and triple T-DNA insertion lines of cruciferin SSPs and the RNAi line of napin SSP family gene were obtained from Withana-Gamage et al. (2013). T-DNA insertion lines (Supplementary Table S1) for candidate genes were screened for homozygous insertions and grown with wild-type Columbia (Col) under greenhouse conditions using rock wool (Grodan, the Netherlands) supplemented with a Hyponex solution 1g l-1 (www.hyponex.co.jp), in a randomized complete block design with four replicates per genotype.
Germination and dormancy assays
Germination assays were performed according to Joosen et al. (2010) over a period of 7 d. Briefly, samples of 50−200 seeds each were sown on two layers of blue germination paper (5.6ʹʹ×8ʹʹ Blue Blotter Paper; Anchor Paper Company, http://www.seedpaper.com) equilibrated with 43ml of demineralized water in a plastic tray (15×21cm) (Joosen et al., 2010). Trays were piled and wrapped in a transparent plastic bag and incubated at 22°C under continuous light (30W m-2). Pictures of the germination trays were taken twice daily over 7 d. Automatic scoring of germination and statistical analysis were conducted using the Germinator package (Joosen et al., 2010).
Dormancy was calculated as days of seed dry storage required to reach 50% germination as described in He et al. (2014).
Artificial ageing
Artificial ageing was used to evaluate seed longevity of the T-DNA insertion lines. Approximately 200 seeds were placed into an opened 1.5ml eppendorf tube and stored above a saturated NaCl solution in a closed tank that has a ventilator to ensure equal humidity inside the tank monitored by Lascar data logger (80−85% relative humidity and temperature of 40°C) for 0−10 d. After treatment, germination assays were performed as described above.
Total soluble protein extracts
Thirty milligrams of dry seeds of each sample (four biological replicates) were ground with a mortar and pestle in liquid nitrogen for ∼1min. Extraction buffer and protease inhibitor (Rajjou et al. 2008), were added to the seed powder, followed by further grinding for 2min. The solution was placed in 1.5ml microcentrifuge tube and incubated with DNase I 53 units/ml, RNase A 4.9 Kunitz units/ml, and DTT 14mM at 4°C for 1h on rotator (Labinco) at 10rpm. Soluble material extract was collected as supernatant after centrifugation at 14 000rpm at 4°C for 10min.
2D gel electrophoresis
Protein separation was performed with 20 µl of protein extract, equivalent to ∼150 µg of protein. 2D gel electrophoresis was conducted as by Rajjou et al. (2008, 2011), adapted for gel strips forming an immobilized nonlinear pH gradient from 3 to 11 (Immobilized DryStrip pH 3–11 NL, 24cm; GE Healthcare).
Comparison of proteome profiles
2D gels were stained with silver nitrate according to Rajjou et al. (2008). Stained gels were placed within two layers of cellophane membrane stretched on cassette frames for drying. Images of dry gels were obtained with an Epson Perfection V700 scanner. Quantitative image analysis was conducted using Progenesis Samespot software (v3.2, NonLinear Dynamics) to quantify proteins spots and to detect changes in protein accumulation.
The PCA was performed based on the abundance of the differentially-accumulated protein spots.
Pair-wise statistics were used to detect protein spots that significantly changed in abundance. Two categories were defined; physiological state and genotype and four physiological state comparisons were made between the protein profiles of the two physiological states (aged versus AR) within each genotype. The AR seed proteome profiles of the NILs were also compared to that the Ler genetic background. Protein spots were considered to have been significantly different in abundance if they were higher than or equal to 1.5-fold up or down accumulated and when the P-value was equal or smaller than 0.05 according to one-way ANOVA test (Progenesis Samespot software) between the means of the four replicates.
Protein identification
Due to the high reproducibility of 2D protein patterns, many proteins could be identified based on their position on gel and comparison to reference maps (http://www.seed-proteome.com; Galland et al., 2012). Other proteins spots were excised from the gels, digested with trypsin and identified by LC-MS/MS as described by Arc et al. (2012). Peptide sequences were submitted to the XTandem Pipeline (http://pappso.inra.fr/bioinfo/xtandempipeline/) databases to retrieve the full protein sequence and the gene annotation.
T-DNA insertion genotype analyses
Genomic DNA was isolated from leaves using a modification of the method of Cheung et al. (1993). Briefly, 0.5cm diameter leaf sample was ground in 1ml of extraction buffer containing 2M NaCl, 200mM Tris-HCl (pH 8), 70mM EDTA and 20mM Na2S2O5. The grinding was conducted with a stainless steel ball at 30 Hz for 1min in a 96-well plate shaker (Mo Bio Laboratory). Samples were then incubated at 65°C for 1h. Supernatants were collected after centrifugation at 13 000rpm for 10min in 1.5ml eppendorf tubes. DNA was precipitated by adding isopropanol and 10M NH4Ac with ratio of 1:0.5:1 to the supernatant. This mixture was incubated at room temperature for 15min and then centrifuged for 20min at 13 000rpm. The DNA pellet was retrieved and rinsed with 1ml of 70% ethanol followed by centrifugation for 5min at 13000rpm to recover the pellet. After drying, the DNA pellet was dissolved in 50 µl distilled water.
Homozygous T-DNA insertion lines were screened with gene-specific primers and T-DNA border-specific primers (Supplementary Table S1). T-DNA plants that amplified only the insertion product were considered to be homozygous mutants.
Polymerase chain reactions (PCR) were performed in a 12.5 µl volume containing ∼30ng DNA, 25 µM of each dNTP, 25ng of forward and reverse primers, 0.05U of DNA polymerase (Firepol, Solis BioDyne), and 312.5 µM of MgCl2. The reaction protocol was as follows: denaturation at 95°C for 5min followed by 30 s at 95°C, 30 s annealing at 52 to 57°C (dependent upon the primer pair) and a 45 s to 2min extension (dependent upon the length of the product) at 72°C; this cycle was repeated for 35 times and ended with a final incubation for 10min at 72°C.
The amplified products were separated by agarose gel electrophoresis at concentrations from 1.5 % and higher (w/v) depending on size of differences.
Detection of protein carbonylation
Carbonylated protein profiles were determined by 1D PAGE of total protein extract followed by derivatization with 2,4-dinitrophenylhydrazine and immunological detection of the DNP adducts with monoclonal anti-DNP antibody (OxyBlot Oxidized Protein Detection Kit; Chemicon) as described previously (Job et al., 2005).
Results and discussion
Effect of natural ageing on seed germination ability
The germination ability of AR and 4-year-old (aged) seeds from four genotypes was investigated. The genotypes used are the Arabidopsis background line Ler and three near isogenic lines (NILs) that contain introgression fragments of Cvi, An-1 and Sha accessions, at the position of the earlier identified seed longevity QTL, NILGAAS1-Cvi, NILGAAS2-An-1, and NILGAAS5-Sha, respectively. Upon storage, all genotypes showed a significant reduction in germination percentage, but with a different level of sensitivity to ageing (Fig. 1). NILGAAS1 and NILGAAS2 had better seed longevity (Gmax=89.3% and 79.6%, respectively) compared with Ler (Gmax=60.7%), whereas NILGAAS5 was less storable (Gmax=37.7%). These results confirmed the seed longevity phenotypes described by Nguyen et al. (2012).
Seed dry storage affects the proteome in a genotype-specific manner
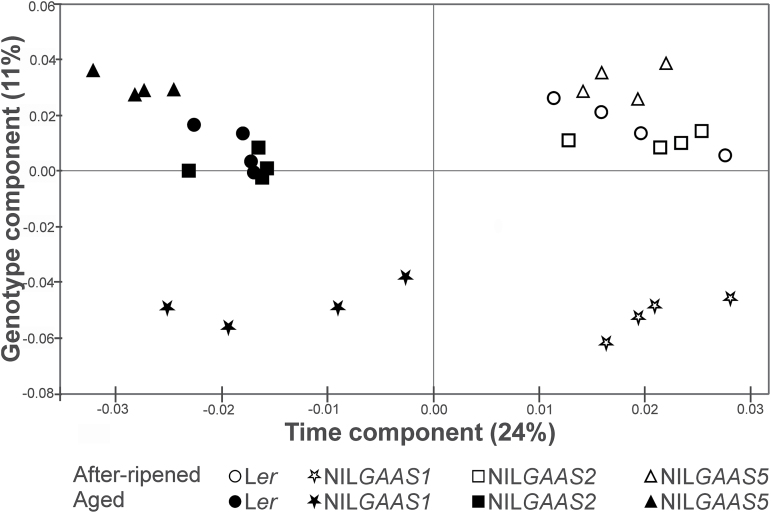
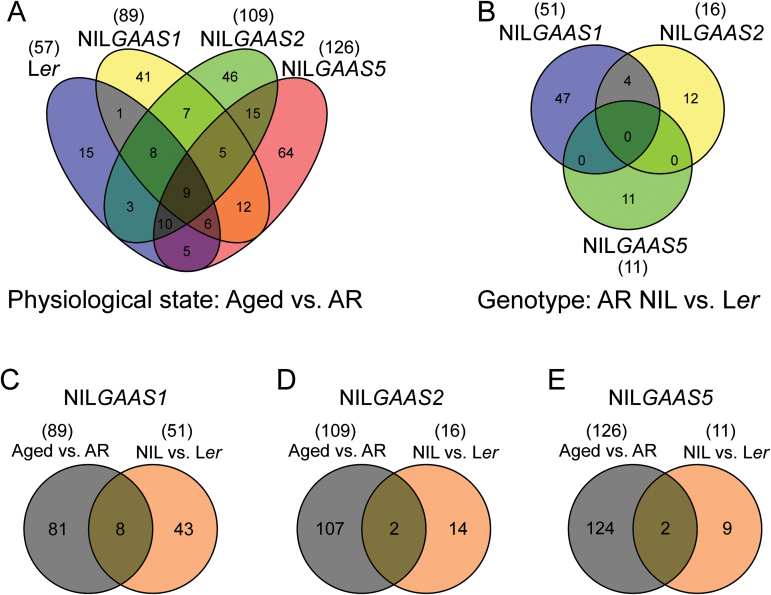
Proteomic profiling for AR and aged seeds was performed to identify mechanisms and modifications associated with the loss of germination ability during seed dry storage. Total soluble protein extracts were separated using 2D PAGE (Fig. 2A) and seven pair-wise comparisons were made (Supplementary Fig. S1). Pair-wise comparisons between the two physiological states (aged versus AR) for each genotype revealed protein spots that were affected by ageing (57 for Ler, 89 for NILGAAS1, 109 for NILGAAS2, and 126 for NILGAAS5). The three NILs contain introgression fragments at different genome positions and exhibit different levels of seed longevity. The differences in seed longevity could result from proteome variation already present in the AR seeds, which can be revealed by pair-wise comparisons between the NILs and Ler. Comparison with the AR Ler seed proteome allowed the identification of 51, 16 and 11 genotype-specific protein spots for NILGAAS1, NILGAAS2 and NILGAAS5, respectively. Some of these protein spots (8, 2 and 2, respectively) overlapped with the aged versus AR comparison (Table 1A). A total of 309 differentially accumulated protein spots were detected in the seven pair-wise comparisons (Supplementary Fig. S1). Principal component analysis (PCA) on the 309 protein spots separated the samples into two groups which represented the two physiological states (AR and aged seeds; Fig. 3). The time component (storage) accounted for 24% of the variation and the genotype component explained 11%. NILGAAS1 was the most distinct genotype, and separation of its aged seeds in the PCA might reflect its better longevity on the time component. NILGAAS5 is the least storable genotype (Fig. 3). The aged versus AR physiological state comparisons for the four genotypes showed a total of 247 protein spots whose abundance changed significantly (P≤0.05) upon ageing (Fig. 4A). A large number of the 247 spots were genotype specific (15 for Ler, 41 for NILGAAS1, 46 for NILGAAS2, and 64 for NILGAAS5) (Fig. 4A). The three genotype comparisons led to the identification of 74 altered protein spots (Fig. 4B), of which most were unique to the genotypes (47 for NILGAAS1, 12 for NILGAAS2, and 11 for NILGAAS5) (Fig. 4B). The four spots in common between NILGAAS1 and NILGAAS2 (Fig. 4B; Supplementary Table S2) when compared with Ler at the AR state might play a role in seed longevity, because both genotypes are more storable than Ler.
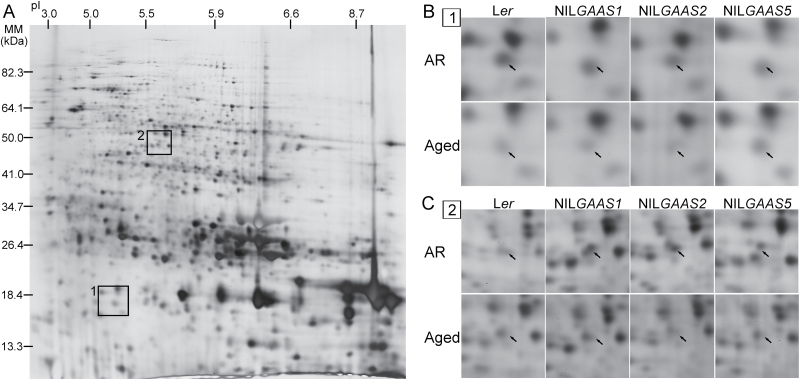
Fig. 2.
2D gel separation of seed proteins and the effect of ageing on seed protein abundance. (A) 2D gel of total soluble proteins from dry seeds stained with silver nitrate. The areas indicated on the gel (1 and 2) are enlarged in panels B and C. (B) Area 1 selected on the gel depicting the abundance of protein spot ID1345 that contains RPS12C and TPX1 proteins for the four genotypes (Ler and the near-isogenic lines NILGAAS1, NILGAAS2 and NILGAAS5) at two physiological states [after-ripened (AR) and aged]. (C) Area 2 showing the change in abundance of protein spot ID0667, corresponding to VTE1 protein, for the four genotypes at two physiological states. Arrows indicate the position of the proteins.
Table 1.
Genotype specific protein spots that were identified in (A) both physiological state and genotype comparisons and (B) in either physiological state or genotype comparison for near-isogenic lines NILGAAS1, NILGAAS2 and NILGAAS5 (Fig. 4C−E)
The table displays protein spots based on the seven comparisons. Spot ID, the gene corresponding to the protein underlying the spot, molecular weight (MW in kD) and the theoretical (Th) and experimental (Exp) isoelectric point (pI), are presented respectively. Furthermore the relative abundance (fold change) of the spots in both types of comparisons (Fig. 1; physiological state and genotype) is indicated. Positive fold changes indicate higher abundances, and negative lower abundances. Spots in bold exhibit seed longevity up or seed longevity down protein profile. Fold changes in bold indicate statistically significant changes. NG1, NILGAAS1; NG2, NILGAAS2; NG5, NILGAAS5. Spots that were identified based on comparison to the reference protein map (http://www.seed-proteome.com) are labelled with R. n.i., not identified.
| A | Spot ID | Gene | Protein | MW (kDa) | pI | Relative abundance (fold change) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Th | Exp | Th | Exp | Physiological state: Aged vs. AR | Genotype AR: NIL vs. Ler | |||||||||
| Ler | NG1 | NG2 | NG5 | NG1 | NG2 | NG5 | ||||||||
| NILGAAS1 | ||||||||||||||
| ID1279 | n.i. | n.i. | n.i. | 22.82 | n.i. | 8.68 | -1.0 | 2.5 | -1.1 | 1.5 | -2.6 | 1.3 | -1.3 | |
| eID0255 R | AT1G03890 | Cupin family protein | 49.67 | 29.33 | 5.45 | 5.63 | -1.0 | 1.9 | 1.0 | 1.0 | -2.3 | -1.0 | -1.0 | |
| ID0426 | n.i. | n.i. | n.i. | 65.51 | n.i. | 5.41 | 1.2 | 1.8 | 1.2 | 1.4 | -1.7 | -1.4 | -1.2 | |
| ID1104 | AT1G03880 | Cruciferin B | 50.56 | 27.38 | 7.0 | 5.61 | 1.1 | 1.5 | 1.0 | 1.1 | 1.9 | 1.1 | 1.0 | |
| AT5F35590 | 20S proteasome alpha subunit | 27.29 | 5.66 | |||||||||||
| AT1G03890 | Cupin family protein | 49.67 | 5.45 | |||||||||||
| eID0138 R | AT4G28520 | Cruciferin C | 58.24 | 25.40 | 6.99 | 5.81 | 1.9 | 1.5 | 1.5 | 1.6 | 2.3 | 1.3 | 1.0 | |
| ID0994 | AT1G54870 | Oxidoreductase family protein | 36.76 | 32.75 | 8.76 | 5.65 | 2.3 | 1.7 | 2.0 | 2.4 | 1.5 | 1.2 | 1.3 | |
| AT4G28520 | Cruciferin C | 58.24 | 6.99 | |||||||||||
| ID0715 | AT1G03880 | Cruciferin B | 50.56 | 47.74 | 7.00 | 6.02 | -1.2 | -1.6 | -1.3 | -1.6 | 1.6 | -1.0 | 1.1 | |
| AT1G74960 | Fatty acid biosynthesis 1 | 57.60 | 7.93 | |||||||||||
| ID0537 | AT2G14170 | Aldehyde dehydrogenase 6B2 | 53.40 | 58.77 | 5.79 | 5.68 | -2.0 | -3.1 | -2.3 | -2.0 | 1.5 | 1.3 | 1.1 | |
| AT5G08670 | ATP synthase beta chain 1 | 59.63 | 6.53 | |||||||||||
| AT5G08680 | ATP synthase beta chain | 59.86 | 6.45 | |||||||||||
| AT5G08690 | ATP synthase beta chain 2 | 59.71 | 6.60 | |||||||||||
| NILGAAS2 | ||||||||||||||
| ID0458 | AT3G20050 | T-complex protein 1 alpha subunit | 59.23 | 63.81 | 6.22 | 5.87 | -1.2 | 1.1 | 1.5 | 1.1 | -1.4 | -2.0 | -1.2 | |
| ID0955 | n.i. | n.i. | n.i. | 38.44 | n.i. | 7.29 | 1.4 | -1.7 | -2.1 | -1.8 | 1.4 | 1.7 | 1.5 | |
| NILGAAS5 | ||||||||||||||
| eID0228 | AT5G19510 | Elongation factor EF1B | 24.20 | 43.24 | 4.17 | 3.88 | -1.6 | 1.3 | 1.2 | 1.7 | -1.3 | -1.1 | -1.6 | |
| ID1146 | AT2G31670 | Unknown protein | 28.86 | 25.36 | 6.96 | 5.10 | 1.1 | 1.3 | 1.4 | 1.8 | -1.0 | -1.3 | -1.5 | |
| B | Spot ID | Gene | Protein | MW (kD) | pI | Relative abundance (fold change) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Th | Exp | Th | Exp | Physiological state: Aged vs. AR | Genotype AR: NIL vs. Ler | |||||||||
| Ler | NG1 | NG2 | NG5 | NG1 | NG2 | NG5 | ||||||||
| NILGAAS1 | ||||||||||||||
| ID0765 | AT3G52880 | Monodehydroascorbate reductase 1 | 50.16 | 43.31 | 8.38 | 6.08 | -1.3 | -1.3 | -1.4 | 1.3 | 2.6 | -1.0 | -1.6 | |
| ID0712 | AT5G28840 | GDP-D-mannose 3ʹ,5ʹ-epimerase | 42.76 | 45.14 | 6.15 | 5.78 | 1.2 | 1.2 | -1.0 | -1.0 | 1.9 | -1.1 | 1.0 | |
| ID0734 | AT5G28840 | GDP-D-mannose 3ʹ,5ʹ-epimerase | 42.76 | 44.99 | 6.15 | 5.74 | 1.2 | 1.7 | 1.2 | 1.5 | 1.1 | -1.1 | -1.1 | |
| ID0951 | AT1G18080 | Receptor for activated C kinase 1A | 35.75 | 38.56 | 7.81 | 7.13 | -1.1 | -1.6 | -1.3 | -1.8 | 1.6 | 1.4 | 1.4 | |
| NILGAAS2 | ||||||||||||||
| ID0206 | AT5G52300 | Responsive to dehydration 29B | 65.97 | 94.21 | 4.81 | 5.05 | -1.5 | -1.2 | -1.7 | -1.5 | -1.3 | -1.2 | 1.1 | |
| ID0196 R | AT5G52300 | Responsive to dehydration 29B | 65.97 | 94.21 | 4.81 | 5.00 | -1.4 | -1.9 | 1.0 | 1.1 | 1.2 | -1.6 | -1.5 | |
| ID0762 | AT2G15430 | RNA polymerase II | 35.46 | 42.82 | 4.39 | 4.46 | -1.3 | -1.1 | -1.6 | -1.1 | -1.4 | -1.1 | -1.3 | |
| NILGAAS5 | ||||||||||||||
| ID1505 | AT4G27160 | Napin AT2S3 | 18.76 | 9.85 | 7.87 | 8.69 | 1.5 | 1.1 | -1.2 | 1.7 | -1.0 | 1.2 | 1.2 | |
| ID0632 | AT5G38470 | Radiation sensitive 23D | 40.07 | 50.00 | 4.29 | 4.60 | 1.1 | -1.1 | -1.0 | -1.1 | 1.0 | -1.1 | -2.0 | |
| ID0258 | AT3G15670 | Late embryogenesis abundant protein | 24.19 | 26.57 | 9.43 | 8.95 | -1.2 | -1.0 | 1.1 | 1.1 | 1.0 | -1.1 | -1.9 | |
| ID1144 | AT3G15670 | Late embryogenesis abundant protein | 24.19 | 29.73 | 9.43 | 7.79 | -1.1 | -1.2 | -1.1 | -1.5 | 1.0 | 1.0 | 1.2 | |
| ID0976 | AT3G17520 | Late embryogenesis abundant protein | 32.56 | 36.11 | 5.02 | 5.54 | 1.2 | 1.2 | 1.4 | 1.6 | 1.0 | -1.1 | -1.2 | |
| ID0448 | AT2G19900 | NADP-dependent malic enzyme 1 | 64.28 | n.i. | 6.73 | n.i. | -1.3 | -1.7 | -1.7 | -1.7 | 1.1 | -1.1 | -1.0 | |
Fig. 3.
Principal component analysis (PCA) for proteome profiles of Ler and the near-isogenic lines NILGAAS1, NILGAAS2 and NILGAAS5. PCA was performed on the differentially accumulated protein spots in the seven comparisons (n=309).
Fig. 4.
Classification of differentially expressed protein spots. (A) Intersection of proteins that were differentially expressed between the two physiological states, aged versus after-ripened (AR), within Landsberg erecta (Ler) and the near-isogenic lines NILGAAS1, NILGAAS2 or NILGAAS5 (n=247). (B) Intersection of proteins that were differentially expressed between genotypes, each NIL versus Ler (n=74). (C, D, E) Intersection of proteins that were differential expressed in the genotypes NILGAAS1, NILGAAS2 and NILGAAS5 respectively in the earlier comparisons (as mentioned in A and B).
Genotype-specific protein spots include those differentially expressed in only one genotype derived from physiological comparisons (aged versus AR seeds; Fig 4A), and those identified in genotype comparisons (AR seeds of NILs versus Ler; Fig 4B). Overlapping spots of both comparison types are presented in Fig. 4C−E and Table 1A (8 for NILGAAS1, 2 for NILGAAS2 and 2 for NILGAAS5). Generally, these results indicate that seed dry storage affects the proteome in a genotype-specific manner. Furthermore, since the NILs used in this study possess different introgressed genomic regions at seed longevity QTLs (Nguyen et al., 2012), several distinct pathways might be involved in seed ageing which could be manifested as unique elements in their respective seed proteomes. These differences can either be causes or consequences of ageing and one cannot exclude the differences in the AR seed proteomes are caused by the nature of the introgression in the NILs and are therefore unrelated to seed longevity.
Diverse genetic pathway involved in seed longevity of the different genotypes
To identify the factors underlying the protein differences both reference mapping and mass spectrometry (MS) analysis was performed. Approximately 70% of the differentially accumulated protein spots could be identified. Seed proteins are targets of various modifications (reviewed by Arc et al., 2011), including carbonylation and S-nitrosylation, especially in dry storage. As a consequence of these modifications, proteins can be represented by several isoforms with different molecular weight (MW) and/or isoelectric point (pI). Thus multiple spots of the same protein were identified.
Investigation of the identified proteins in the four genotypes, Ler, NILGAAS1, NILGAAS2 and NILGAAS5 revealed that possible diverse genetic pathways are involved in seed longevity which is reflected by proteins for which the isoforms are significantly altered in abundance in a genotype-specific manner.
Mechanism related to seed ageing in NILGAAS1
NILGAAS1 is the most storable line of the four tested genotypes (Fig. 1). NILGAAS1 has 41 unique protein spots that accumulate differently in aged compared to AR seeds (Fig. 4A), and 47 when comparing AR seeds of NILGAAS1 to that of Ler (Fig. 4B). Eight spots were at the intersect of both comparison types for NILGAAS1 (Fig. 4C; Table 1A), of which two, ID0715 and ID1104, contained CRUB. NILGAAS1 carries the truncated CRUB Cvi allele (Hou et al., 2005) resulting in a CRUB α-subunit with a lower MW compared to that from the Ler allele, which might account for the identification of CRUB in NILGAAS1.
Dry seeds are well equipped to confront oxidative stress during storage due to their low water content and reduced metabolic activity. Auto-oxidation leading to accumulation of ROS occurs in dry seed where proteins are the major targets of oxidative damage because of their abundance and high affinity with radicals (Davies, 2005). To control ROS-induced damage, seeds have detoxification mechanisms to scavenge or inactivate ROS. We found two proteins that are involved in the ascorbate antioxidant metabolism pathway in NILGAAS1. Protein spot ID0765 (Table 1B) corresponds to MONODEHYDROASCOBATE REDUCTASE1 (MDAR1), a well known antioxidant enzyme that removes hydrogen peroxide at the ascorbate-glutathione cycle (Bailly, 2004). T-DNA insertion mutant analysis for seed longevity showed that mdar1.1 and mdar1.2 did not differ in seed longevity compared to Col (Supplementary Fig. S2). The lack of a phenotype might be due to redundancy since the MDAR gene family contains five members. Over-expression of AtMDAR1 in tobacco conferred enhanced tolerance to ozone, salt and osmotic stresses (Eltayeb et al., 2007) suggesting that the ascorbate-glutathione cycle may play a role in seed ageing. GDP-D-MANNOSE 3′,5′-EPIMERASE (GME) is a key enzyme involved in ascorbate (vitamin C) synthesis in plants (Wolucka et al., 2001; Wolucka and Van Montagu, 2003). NILGAAS1 AR seeds had higher levels of GME (ID0712), which likely explains the higher resistance to ageing of NILGAAS1 compared to Ler seeds (Table 1B). The GME isoform ID0734 was found more abundant in aged than AR seeds, which could indicate GME modification during ageing. Homozygous GME T-DNA insertion lines could not be isolated indicating that GME is an essential protein (Supplementary Table S1).
RECEPTOR FOR ACTIVATED C KINASE1 (RACK1) isoform ID0951 was more abundant in AR seeds of NILGAAS1 than that of Ler (Table 1B), which might also contribute to the better seed longevity of this NIL. RACK1 is a multi-function protein that plays a regulatory role in diverse signal transduction pathways and its transcripts are present during seed germination (Chen et al., 2006; Guo et al., 2009). In Arabidopsis, the translation initiation factor eIF6-2 interacts with RACK1, a negative regulator of ABA response and positive regulator of GA signalling (Guo et al., 2011; Fennell et al., 2012). It was demonstrated that ABA inhibited RACK1 and eIF6 gene expression (Guo et al., 2011). Thus seed germination of a rack1A T-DNA insertion line was examined, however, did not significantly differ from that of wild-type Col (Supplementary Fig. S2).
Mechanism involved in seed ageing expressed in NILGAAS2
NILGAAS2, the second most storable genotype, had 46 unique protein spots that differentially accumulated when aged and AR seeds were compared (Fig. 4A) and 12 when AR seeds of NILGAAS2 were compared to that of Ler (Fig. 4B). Two spots were common in both comparisons (Fig. 4D; Table 1A). Although no specific pathway was identified, there are interesting proteins that might be involved in seed longevity in this NIL.
Spot ID0458 (Table 1A) corresponds to the T-complex Protein 1 α Subunit (TCP1). Several members of the TCP1 protein family were described to be up-accumulated during Arabidopsis seed dormancy release, suggesting that they could play a central role in seed germination (Arc et al., 2012). RESPONSIVE TO DEHYDRATION29B (RD29B) was identified in two protein spots, ID0206 and ID0196 (Table 1B). The increased level of RD29B could be a marker for increased seed longevity.
It was noted that de novo transcription is not required for germination since seeds are able to germinate until radical protrusion in the presence of α-amanitin (a transcription inhibitor targeting RNA POLYMERASE II); however subsequent seedling growth was prevented (Rajjou et al., 2004). Newly synthesized transcripts might also be necessary for germination of aged seed. The low abundance of RNA POLYMERASE II (AT2G15430, ID0762), 1.6-fold less abundant after ageing of NILGAAS2 seeds (Table 1B), may contribute to reduced seed germination after storage. Homozygous T-DNA insertion lines for RNA POLYMERASE II could not be isolated (Supplementary Table S1).
Seed longevity mechanisms in NILGAAS5
NILGAAS5 is the most sensitive genotype to ageing in this study (Fig. 1). The reduced seed longevity in NILGAAS5 is caused by the DOG1-Cvi allele, as was revealed by complementation cloning (Nguyen et al., 2012). DOG1 protein accumulates during seed maturation and remains stable throughout seed storage, however it is modified during after-ripening (Nakabayashi et al., 2012). We did not identify DOG1, likely because it is rather stable and not modified anymore at later stages during dry storage. NILGAAS5 had 64 unique protein spots that differentially accumulated when aged and AR seeds were compared (Fig. 4A), and 11 when AR seeds of NILGAAS5 were compared to that of Ler (Fig. 4B). Two spots, eID0228 and ID1146, encoding elongation factor EF1B and an unknown protein (Fig. 4E; Table 1A) were common to both comparisons.
One of the proteins differentially accumulating in NILGAAS5 is RADIATION SENSITIVE23D (RAD23D), its protein spot (ID0632) is down regulated compared to Ler (Table 1B). RAD23D was suggested to participate in DNA damage repair since the two carrot (Daucus carota) RAD23 isoforms rescue the UV-sensitive phenotype of the rad23 deletion mutant in yeast (Saccharomyces cerevisiae) (Sturm and Lienhard, 1998). During storage seeds are subjected to DNA damage and genome instability, which are considered to be a main cause of reduced germination after ageing. The maintenance of a functional DNA repair complex is essential for long-term survival (reviewed by Rajjou et al., 2008). However, since RAD23D is located in the introgression region of NILGAAS5, we cannot exclude the possibility that it affects seed longevity independently from DOG1.
Another group of differentially accumulating proteins are the identified LATE EMBRYOGENESIS ABUNDANT FAMILY-4 PROTEINS; AT3G17520 in spot ID0976 and AT3G15670 in spot ID0258 and ID1144 (Table 1B). The association of LEA AT3G15670 and seed germination after ageing was examined by T-DNA insertion mutant analysis (Supplementary Table S1); however, the lea mutant exhibited similar seed longevity compared to Col (Supplementary Fig. S2). NADP-DEPENDENT MALIC ENZYME1 (NADP-ME1) in spot ID0448 (Table 1B) was lower abundant in aged compared with AR seeds of NILGAAS5, which makes it a possible marker for seed longevity.
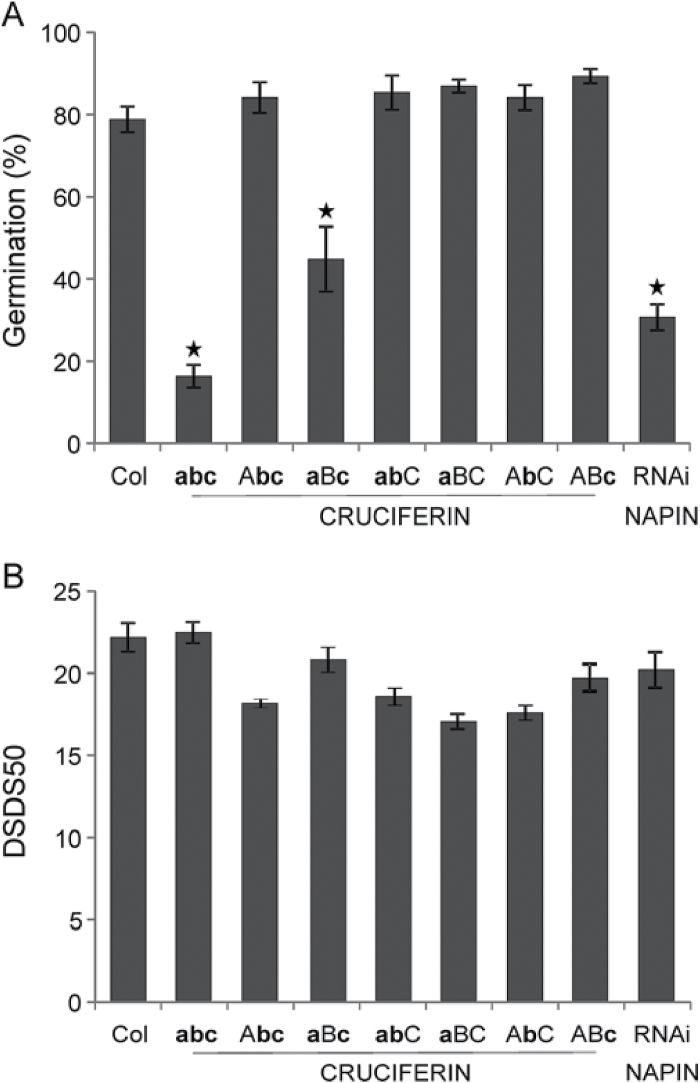
AT2S3, one of the five 2S albumin or napin isoforms (Krebbers et al., 1988; Van der Klei et al., 1993), was more abundant in aged NILGAAS5 (the most sensitive genotype to ageing) (Table 1B). The AT2S3 protein spot ID1505 might be a degradation product due to its altered MW and pI (Table 1B). To examine if napins could affect seed longevity, a napin RNAi line was analysed for seed longevity (Withana-Gamage et al., 2013) (Supplementary Table S3). The line, which is depleted of napins and has a reduced protein content mainly in the endosperm, was more sensitive to ageing than wild-type Col (Fig. 5).
Fig. 5.
The effect of seed storage proteins (SSPs) on seed longevity and seed dormancy.(A) Seed longevity presented as germination (%) of different SSP knock-out lines was measured after 10 d of artificial ageing. The lines include the wild-type Col, as well as single (crua, aBC; crub, AbC and cruc, ABc), double (crub cruc, Abc; crua cruc, aBc and crua crub, abC) and triple (crua crub cruc, abc) knock-out lines of cruciferins, and an RNAi napin line that is depleted of napins. (B) Seed dormancy presented as days of seed dry storage required to reach 50% germination (DSDS50) of Col and different SSP knock-out lines.
Pathways/proteins that are generally affected during storage
Energy metabolism and seed longevity
Carbohydrate metabolism is important for germinating seeds since it provides energy and intermediate metabolites for seed germination and seedling establishment. The metabolic pathways related to energy production are glycolysis, the oxidative pentose phosphate pathway (OPP), fermentation, the tricarboxylic acid cycle (TCA), the glyoxylate cycle, and the electron transport chain (Supplementary Fig. S3). The abundance of several protein isoforms that encode for enzymes in these pathways were altered in the seed proteome upon storage (Table 2). Similar changes in the seed proteome upon ageing has also been found previously (Rajjou et al., 2008). In order to investigate the importance of these enzymes for seed longevity we have investigated T-DNA insertion lines of these genes for their seed longevity behaviour. The list of T-DNA insertion mutants that was tested is provided in Supplementary Table S1. For none of the mutants investigated seed longevity phenotypes have been revealed. It is possible that these enzymes are not important for seed longevity however we expect that the lack of phenotypes are caused by gene redundancy. For example, for the glycolysis enzyme UDP GLUCOSE URIDYLYLTRANSFERASE (UGP) we could only obtain one homozygous mutant (ugp1) and this mutant did not show a seed longevity phenotype (Supplementary Fig. S2), however there are two UGPs in Arabidopsis and these likely act redundantly.
Table 2.
Differentially expressed protein spots upon ageing that are related to translation, energy metabolism, reactivation of cell activity, redox homeostasis, and ABA signalling
The table displays protein spots based on the seven comparisons. Spot ID, the gene corresponding to the protein underlying the spot, molecular weight (MW in kD) and the theoretical (Th) and experimental (Exp) isoelectric point (pI), are presented respectively. Furthermore the relative abundance (fold change) of the spots in both types of comparisons (Fig. 1; physiological state and genotype) is indicated. Positive fold changes indicate higher abundances, and negative lower abundances. Fold changes in bold indicate the statistically significant changes. NG1, NILGAAS1; NG2, NILGAAS2; NG5, NILGAAS5. Spots that were identified based on comparison to the reference protein map (http://www.seed-proteome.com) are labelled with R. n.i., not available.
| Spot ID | Gene | Protein | MW (kD) | pI | Relative abundance (fold change) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Th | Exp | Th | Exp | Physiological state: Aged vs. AR | Genotype AR: NIL vs. Ler | ||||||||
| Ler | NG1 | NG2 | NG5 | NG1 | NG2 | NG5 | |||||||
| Translation | |||||||||||||
| ID1345 | AT2G32060 | Ribosomal protein 40S subunit | 15.33 | 16.25 | 5.70 | 5.07 | -1.7 | -2.5 | -1.5 | 1.1 | -1.1 | -1.3 | -1.5 |
| ID0667 R | AT1G57720 | Elongation factor EF1Bγ | 46.40 | 49.95 | 5.40 | 5.54 | -1.4 | -1.1 | -1.5 | -1.5 | 1.2 | 1.4 | 1.2 |
| eID0228 R | AT5G19510 | Elongation factor EF1B | 24.20 | 43.24 | 4.17 | 3.88 | -1.6 | 1.3 | 1.2 | 1.7 | -1.3 | -1.1 | -1.6 |
| ID0885 | AT1G30230 | Elongation factor EF1β | 28.77 | 38.67 | 4.36 | 3.98 | -1.3 | -1.0 | -1.5 | -1.7 | -1.2 | 1.1 | 1.2 |
| ID1226 | AT1G07930 | Elongation factor EF1α | 49.50 | 25.11 | 9.64 | 7.81 | -1.2 | -1.6 | 1.0 | -1.1 | 1.4 | 1.1 | -2.1 |
| AT5G60390 | Elongation factor EF1α | 49.50 | 9.64 | ||||||||||
| Energy metabolism | |||||||||||||
| ID0621 | AT5G17310 | UGP-1 glucose uridylytransferase 2 | 51.92 | 55.53 | 5.80 | 5.80 | 1.3 | 1.5 | 1.5 | 1.3 | -1.1 | -1.0 | -1.1 |
| ID0603 | AT3G03250 | UGP-1 glucose uridylytransferase 1 | 51.74 | 56.94 | 5.98 | 5.71 | 1.1 | -1.1 | -1.4 | -1.5 | 1.3 | 1.4 | 1.3 |
| AT5G17310 | UGP-1 glucose uridylytransferase 2 | 51.92 | 5.80 | ||||||||||
| ID0832 | AT1G13440 | Glyceraldehyde-3-P dehydrogenase C2 | 36.91 | 43.15 | 7.22 | 6.05 | 1.9 | 1.7 | 1.7 | 1.3 | -1.0 | 1.2 | 1.4 |
| AT3G04120 | Glyceraldehyde-3-P dehydrogenase C1 | 36.91 | 7.15 | ||||||||||
| ID0997 | AT1G13440 | Glyceraldehyde-3-P dehydrogenase C2 | 36.91 | 33.67 | 7.22 | 5.72 | 1.7 | 1.7 | 1.9 | 2.7 | 1.2 | 1.1 | -1.1 |
| AT3G04120 | Glyceraldehyde-3-P dehydrogenase C1 | 36.91 | 7.15 | ||||||||||
| eID0214 | AT2G45290 | Transketolase | 79.92 | 81.80 | 6.55 | 5.77 | -2.0 | -1.8 | -2.3 | -1.8 | -1.0 | -1.1 | -1.1 |
| eID0096 | AT3G60750 | Transketolase | 79.97 | 78.47 | 6.32 | 5.45 | -1.6 | -1.9 | -1.6 | -1.9 | 1.2 | 1.1 | 1.2 |
| eID0275 | AT5G54960 | Pyruvate decarboxylase 2 | 65.82 | 64.35 | 5.84 | 5.54 | -1.7 | -1.5 | -1.7 | -2.0 | 1.2 | 1.4 | 1.2 |
| ID1180 | AT1G04410 | Malate dehydrogenase 1 | 35.57 | 25.20 | 6.51 | 5.64 | 1.6 | -1.0 | 1.5 | 2.0 | 1.3 | -1.2 | -1.2 |
| ID0448 | AT2G19900 | NADP-dependent malic enzyme 1 | 64.28 | n.i. | 6.73 | n.i. | -1.3 | -1.7 | -1.7 | -1.7 | 1.1 | -1.1 | -1.0 |
| ID0526 | AT5G08670 | ATP synthase β chain 1 | 59.63 | 59.34 | 6.53 | 5.35 | 1.2 | 1.3 | 1.2 | 1.6 | 1.0 | 1.0 | -1.1 |
| AT5G08680 | ATP synthase β chain | 59.86 | 6.45 | ||||||||||
| AT5G08690 | ATP synthase β chain 2 | 59.71 | 6.60 | ||||||||||
| ID0535 | AT5G08680 | ATP synthase β chain | 59.86 | 58.73 | 6.45 | 5.49 | -1.3 | -1.5 | 1.2 | 1.3 | 1.0 | -1.3 | -1.4 |
| ID0538 | AT5G08670 | ATP synthase β chain 1 | 59.63 | 58.22 | 6.53 | 5.51 | -1.3 | -1.3 | -1.5 | -1.4 | 1.1 | 1.3 | 1.1 |
| ID0556 | AT5G08670 | ATP synthase β chain 1 | 59.63 | 57.44 | 6.53 | 5.49 | 1.0 | -1.6 | 1.1 | 1.0 | 1.9 | 1.4 | 1.4 |
| Reactivation of cell | |||||||||||||
| ID0693 | AT2G36880 | Methionine adenosyltransferase 3 | 42.50 | 46.03 | 6.09 | 5.73 | -1.1 | 1.6 | -1.3 | -1.4 | 1.3 | 1.1 | 1.2 |
| ID0694 R | AT4G01850 | Methionine adenosyltransferase 2 | 43.25 | 47.58 | 5.94 | 5.65 | -1.1 | -1.0 | -1.6 | -1.1 | -1.0 | 1.3 | 1.3 |
| Redox homeostasis | |||||||||||||
| ID1345 | AT1G65980 | Thioredoxin-dependent peroxidase 1 | 17.43 | 16.25 | 5.00 | 5.06 | -1.7 | -2.5 | -1.5 | 1.1 | -1.1 | -1.3 | -1.5 |
| ID0667 | AT4G32770 | Vitamin E deficient 1 | 54.72 | 49.95 | 6.26 | 5.54 | -1.4 | -1.1 | -1.5 | -1.5 | 1.2 | 1.4 | 1.2 |
| ABA signalling | |||||||||||||
| eID0200 | AT1G43890 | Responsive to abscisic acid 18 | 23.53 | 18.53 | 5.43 | 5.79 | 1.2 | 1.4 | 1.7 | 1.7 | -1.1 | -1.4 | -1.3 |
The effect of ageing on translation capacity and protein metabolism
Protein translation is essential for seed germination, since the presence of the translation inhibitor cycloheximide prevented radicle protrusion (Rajjou et al., 2004). Furthermore, aged seeds were strongly affected in their translation capacity (Rajjou et al., 2008). This research also demonstrated that many proteins involved in protein metabolism were highly carbonylated in deteriorated seeds. Consistent with previous studies, we observed that the levels of elongation factor EF1B family proteins were lower in aged compared to AR seeds (Table 2). Moreover, spot ID1345, corresponding to the ribosomal protein 40S subunit RPS12C, was reduced in abundance after storage of Ler, NILGAAS1 and NILGAAS2 seeds (Fig. 2B; Table 2).
Reactivation of cellular activity
Changes in the abundance of METHIONINE ADENOSYLTRANSFERASE (MAT) were observed in both NILGAAS1 (MAT3, ID0693; Table 2) and NILGAAS2 (MAT2, ID0694; Table 2) after ageing. MAT participates in S-adenosylmethionine (Ado-Met) biosynthesis and is important for reactivating cellular activity in germinating seeds (Ravanel et al., 1998; Gallardo et al., 2002). This indicates genotype-specific differences in the dependency on pathways related to Ado-Met metabolism in seed longevity.
Redox homeostasis and antioxidants in seed longevity
Seed storage and germination are coupled to extensive changes in the redox state of seed proteins and even in the dry state seed proteins are subjected to various types of PTMs which include redox modifications (Arc et al., 2011; Rajjou et al., 2012). Thioredoxin has a vital role in redox processes, in which it transforms essential proteins from the oxidized to the reduced form and in the process retrieves the molecular function of those proteins. THIOREDOXIN-DEPENDENT PEROXIDASE1 (TPX1) (ID1345) abundance declined in aged compared to AR seeds of Ler, NILGAAS1, NILGAAS2 (Fig. 2B; Table 2). TPX1 is a thioredoxin-dependent peroxidases type II B (Prx IIB), one of the six isoforms in this family, which has a wide range of redox buffering activities (Rouhier and Jacquot, 2005). Thus, TPX1 might be very important for retaining the redox balance during ageing and germination. To investigate the role of TPX1, seed longevity for the tpx1 mutant was analysed; however, it was similar to the wild-type Col accession (Supplementary Fig. S2). The lack of a visible phenotype for tpx1 could be due to the nature of the T-DNA insertion (3ʹ-UTR region of the gene) (Supplementary Table S1) or due to redundancy of enzymatic antioxidant systems in seeds, so that missing one might not have obvious effects.
Vitamin E is another antioxidant important for seed longevity since the vitamin E deficient1 (vte1) mutant seeds were more sensitive to artificial ageing than those of the wild type (Sattler et al., 2004). In this study, VTE1 (ID0667) abundance was lower in aged seeds of NILGAAS2 and NILGAAS5 (Fig. 2C; Table 2) than AR seeds, which is in agreement with previous studies suggesting a role of VTE1 in seed longevity.
The role of seed storage proteins in ageing
Many of the identified protein spots were SSP 12S globulin fragments, a predominant type of SSP referred to as cruciferin (Pang et al., 1988). A set of single, double and triple knock-out lines for CRUA, CRUB and CRUC (Withana-Gamage et al., 2013) (Supplementary Table S3) was analysed to study the role of cruciferins in seed longevity under artificial ageing. Cruciferin single knock-out mutants lacking one of the cruciferin isoforms did not differ in seed longevity compared to wild-type Col (Fig. 5). The crua cruc double mutant lacking both CRUA and CRUC exhibited reduced seed longevity, whereas seed longevity for the crua crub and crub cruc double mutants was unaffected. CRUB is poorly transcribed and CRUB is the least abundant cruciferin isoform (Withana-Gamage et al., 2013), thus the crua cruc double mutant has very low levels of cruciferin. This explains why double mutants that have eliminated CRUB, but retained the more abundant CRUA or CRUC isoforms, did not show reduced seed longevity. The effect of cruciferin content on seed longevity was even more apparent in the crua crub cruc triple mutant which was very sensitive to artificial ageing (Fig. 5A). The role of cruciferins on seed longevity cannot be explained by reduced protein levels since the crua cruc double mutant has wild type protein levels in both the endosperm and the embryo (Withana-Gamage et al., 2013). This phenomenon is referred to as seed proteome rebalancing and involves a general increase in the production of other seed proteins to compensate for the loss of a major SSP (Herman, 2014). Defects in seed development that lead to reduced seed longevity often result in reduced seed dormancy levels as well, examples are the abi3-5, dog1-1 and tt mutants (Ooms et al., 1993; Debeaujon et al., 2000; Clerkx et al., 2004a ; Sugliani et al., 2009). However, despite the reduction in SSPs and reduced seed longevity, seed dormancy as measured by days of seed dry storage required to reach 50% germination was unaffected in the cru mutants (Fig. 5B). Thus, the loss of germination in the mutant is not caused by the lack of protein reserves. It is possible that the localization and distribution of the proteins is important since the crua crub cruc triple mutant contains very small protein storage vesicles with very little protein within (Withana-Gamage et al., 2013). The influences of SSPs can also be due to their modification because SSPs are subject to a wide range of PTMs (Job et al., 2005; Wan et al., 2007), suggesting that the effects of PTMs on SSPs play a role in seed longevity.
SSPs function as oxidation buffers in seed longevity
Although several T-DNA mutants were tested for seed longevity, SSP mutants showed the most severe phenotype, especially the cruciferin triple mutant crua crub cruc and the napin RNAi line. Therefore, we further investigated the seed longevity mechanism provided by SSPs using these two mutants. SSPs were reported to be massively oxidized, especially in the form of carbonylation, during seed germination in Arabidopsis (Job et al., 2005) and in pea (Pisum sativum) (Barba-Espín et al., 2011). Different roles for SSPs in seed germination have been proposed due to its affinity to carbonylation: (i) Role in reserve mobilization. Carbonylated SSPs are easily destabilized from larger complexes, since they are more susceptible to proteolysis to remobilize resources for seed germination and seedling establishment. (ii) It was also suggested that the abundance of SSPs makes them an efficient scavenging system for ROS that are actively generated during seed germination (reviewed by El-Maarouf-Bouteau et al., 2013). During long-term storage, SSPs are often carbonylated, which is an irreversible form of oxidation leading to deterioration, in dry aged seeds of Arabidopsis (Rajjou et al., 2007; 2008) and beech (Fagus sylvatica) (Kalemba and Pukacka, 2014). The high abundance of SSPs might protect other proteins that are important for germination from oxidation, suggesting a role for SSPs in ROS-buffering during seed dry storage. We examined this hypothesis by investigation of the carbonylation pattern of AR and aged SSP mutant seeds in comparison with that of wild-type seeds.
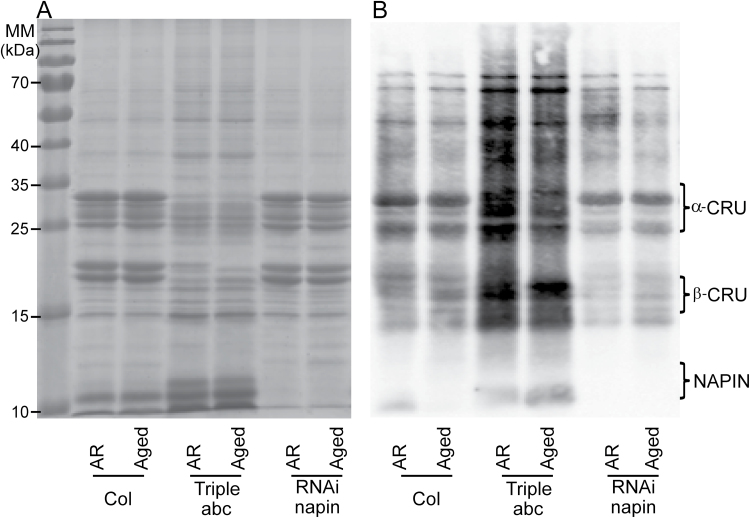
1D-PAGE analysis of total protein extracts confirmed the reduction of cruciferin and napin proteins in dry seeds of the crua crub cruc and RNAi-napin mutant, respectively (Fig. 6A). Carbonylation of seed proteins was significant in both AR and aged seeds, with cruciferin being a major target in the wild-type and RNAi-napin lines (Fig. 6B). The cruciferin mutant exhibited a different protein carbonylation pattern in which carbonylation levels increased for the remaining proteins compared to the wild-type profile. In addition, there is a slight reduction in protein oxidation profiles comparing aged to AR seeds of all three genotypes, but also this effect was the strongest in the crua crub cruc mutant. Our result provides the first proof that SSPs, mainly cruciferins, are buffers for oxidative stress especially in dry seeds during storage. The carbonylated proteins in SSP mutants are interesting, since they will reveal new insights on the elements important for seed longevity.
Fig. 6.
Protein carbonylation of seed proteins in after-ripened (AR) artificially aged seeds (Aged). (A) 1D gel electrophoresis stained with Coomassie Brilliant Blue of total seed protein extracts from Col, the triple cruciferin mutant abc (crua crub cruc) and the napin mutant (RNAi-napin). (B) Carbonylated proteins as detected by immunodetection of protein-bound DNP after derivatization with hydrazine.
Conclusions
The naturally and artificially aged material used in this analysis allowed the molecular processes involved in seed ageing to be examined. In addition, the use of genetic material with different levels of seed longevity increased the likelihood that novel seed longevity factors would be identified. Despite being metabolically quiescent, the dry seed proteome was greatly altered upon ageing. Proteins that appear to be involved in seed longevity that were common to all genotypes included SSPs, proteins related to translation and energy metabolism (glycolytic and TCA pathways), and vitamin E synthesis (VTE1). These results indicated the importance of protection and maintenance of functional energetic and metabolic pathways, as well as antioxidant systems, for seed longevity. Interestingly, the different genotypes also expressed specific seed longevity pathways.
Our work presents the first evidence of ROS buffering by SSPs in dry seeds. This was revealed by analysing cruciferin double and triple mutants. We could not prove the participation of the other identified proteins in seed longevity, probably due to gene redundancy. Over-expression analyses of the identified genes or RNAi lines targeting whole gene families may be a better way to examine the effect of these proteins.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Schematic presentation of the experiment.
Supplementary Fig. S2. Seed longevity of T-DNA insertion mutants after artificial ageing.
Supplementary Fig. S3. Schematic representation of energy metabolism-related pathways affected during seed storage.
Supplementary Table S1. The T-DNA insertion lines of the selected candidate genes.
Supplementary Table S2. Protein spots that show an overlapping abundance pattern in after-ripened seeds of NILGAAS1 and NILGAAS2 in comparison with that of Ler.
Supplementary Table S3. The nomenclature of seed storage proteins.
Acknowledgements
We thank Magdalena Gamm (Department of Molecular Plant Physiology, Utrecht University) for providing seeds of the rack1A T-DNA insertion line. We are also grateful to Fred van Eeuwijk (Department of Biometrics, Wageningen University) for discussions on statistical analysis of the data. This work was supported by the Dutch Technology Foundation (STW), which is the applied science division of the Netherlands Organization for Scientific Research and the Technology Program of the Ministry of Economic Affairs (to TPN and LB).
Glossary
Abbreviations:
- AR
after-ripening
- CDT
controlled deterioration test
- CRUs
cruciferins
- GA
gibberellic acid
- MW
molecular weight
- pI
isoelectric point
- PTM
post-translational modification
- QTL
quantitative trait loci
- ROS
reactive oxygen species
- SSP
seed storage protein.
References
- Almoguera C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano J. 2009. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biology doi: 10.1186/1471-2229-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arc E, Chiban K, Grappin P, Jullien M, Godin B, Cueff G, Valot B, Balliau T, Job D, Rajjou L. 2012. Cold stratification and exogenous nitrates entail similar functional proteome adjustments during Arabidopsis seed dormancy release. Journal of Proteome Research 11, 5418–5432. [DOI] [PubMed] [Google Scholar]
- Arc E, Galland M, Cueff G, Godin B, Lounifi I, Job D, Rajjou L. 2011. Reboot the system thanks to protein post-translational modifications and proteome diversity: how quiescent seeds restart their metabolism to prepare seedling establishment. Proteomics 11, 1606–1618. [DOI] [PubMed] [Google Scholar]
- Bailly C. 2004. Active oxygen species and antioxidants in seed biology. Seed Science Research 14, 93–107. [Google Scholar]
- Barba-Espín G, Diaz-Vivancos P, Job D, Belghazi M, Job C, Hernández JA. 2011. Understanding the role of H2O2 during pea seed germination: a combined proteomic and hormone profiling approach. Plant, Cell & Environment 34, 1907–1919. [DOI] [PubMed] [Google Scholar]
- Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M. (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis . Plant Physiol 124, 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Hanson J, Hanhart CJ, et al. 2010. Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proceedings of the National Academy of Sciences, USA 107, 4264–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L, Jowett J, Hanhart CJ, Koornneef M, 2006. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis . Proceedings of the National Academy of Sciences, USA 103, 17042–17047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Muñoz-Bertomeu J, Campos F, Brunaud V, Martínez L, Sayas E, Ballester P, Yenush L, Serrano R. 2014. Arabidopsis thaliana HOMEOBOX25 uncovers a role for Gibberellins in seed longevity. Plant Physiology 164, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain E, Le Gall S, Hundertmark M, Leprince O, Satour P, Deligny-Penninck S, Rogniaux H, Buitink J. 2012. Temporal profiling of the heat stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant, Cell & Environment 35, 1440–1455. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. 2006. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis . Journal of Experimental Botany 57, 2697–2708. [DOI] [PubMed] [Google Scholar]
- Cheung WY, Hubert N, Landry BS. 1993. A simple and rapid DNA microextraction method for plant, animal, and insect suitable for RAPD and other PCR analyses. PCR Methods and Applications 3, 69–70. [DOI] [PubMed] [Google Scholar]
- Clerkx EJM, Blankestijn-De Vries H, Ruys GJ, Groot SPC, Koornneef M. 2004. a . Genetic differences in seed longevity of various Arabidopsis mutants. Physiologia Plantarum 121, 448–461. [Google Scholar]
- Clerkx EJM, El-Lithy ME, Vierling E, Ruys GJ, Blankestijin-De Vries H, Groot SPC, Vreugdenhil D, Koornneef M. 2004. b . Analysis of natural allelic variation of Arabidopsis seed germination and seed longevity traits between the accessions Landsberg erecta and Shakdara, using a new recombinant inbred line population. Plant Physiology 135, 432−443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MJ. 2005. The oxidative environment and protein damage. Biochimica et Biophysica Acta 1703, 93–109. [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. 2000. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis . Plant Physiology 122, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarouf-Bouteau H, Meimoun P, Job C, Job D, Bailly C. 2013. Role of protein and mRNA oxidation in seed dormancy and germination. Frontiers in Plant Science 4, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K. 2007. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225, 1255–1264. [DOI] [PubMed] [Google Scholar]
- Fennell H, Olawin A, Mizanur RM, Izumori K, Chen JG, Ullah H. 2012. Arabidopsis scaffold protein RACK1A modulates rare sugar D-allose regulated gibberellin signaling. Plant Signaling & Behavior 7, 1407–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galland M, Job D, Rajjou L. 2012. The seed proteome web portal. Frontiers in Plant Science 3, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. 2002. Importance of methionine biosynthesis for Arabidopsis seed germination and seedling growth. Physiologia Plantarum 116, 238–247. [DOI] [PubMed] [Google Scholar]
- Groot SPC, Surki AA, de Vos RCH, Kodde J. 2012. Seed storage at elevated partial pressure of oxygen, a fast method for analysing seed ageing under dry conditions. Annals of Botany 110, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG. 2011. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiology 155, 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JJ, Wang JB, Xi L, Huang WD, Liang JS, Chen JG. 2009. RACK1 is a negative regulator of ABA responses in Arabidopsis . Journal of Experimental Botany 60, 3819–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang TN, Lin Q, Liu L, Liu X, Liu S, Wang W, Li L, He N, Liu Z, Jiang L, Wan J. 2014. Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions. Euphytica doi:10.1007/s10681-014-1304-0. [Google Scholar]
- He H, de Souza Vidigal D, Snoek LB, Schnabel S, Nijveen H, Hilhorst H, Bentsink L. 2014. Interaction between parental environment and genotype affects plant and seed performance in Arabidopsis . Journal of Experimental Botany doi:10.1093/jxb/eru378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman EM. 2014. Soybean seed proteome rebalancing. Frontiers in Plant Science 5, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou A, Liu K, Catawatcharakul N, Tang X, Nguyen V, Keller WA, Tsang EW, Cui Y. 2005. Two naturally occurring deletion mutants of 12S seed storage proteins in Arabidopsis thaliana . Planta 222, 512–520. [DOI] [PubMed] [Google Scholar]
- Job C, Rajjou L, Lovigny Y, Belghazi M, Job D. 2005. Patterns of protein oxidation in Arabidopsis seeds and during germination. Plant Physiology 138, 790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosen RVL, Kodde J, Willems LAJ, Ligterink W, van der Plas LHW, Hilhorst HWM. 2010. GERMINATOR: a software package for high-throughput scoring and curve fitting of Arabidopsis seed germination. Plant Journal 62, 148–159. [DOI] [PubMed] [Google Scholar]
- Kalemba EM, Pukacka S. 2014. Carbonylated proteins accumulated as vitality decreases during long-term storage of beech (Fags sylvatica L.) seeds. Trees 28, 503–515. [Google Scholar]
- Krebbers E, Herdies L, De Clercq A, Seurinck J, Leemans J, Van Damme J, Segura M, Gheysen G, Van Montagu M, Vandekerckhove J. 1988. Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiology 87, 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landjeva S, Lohwasser U, Börner A. 2009. Genetic mapping within the wheat D genome reveals QTL for germination, seed vigour and longevity, and early seedling growth. Euphytica 171, 129–143. [Google Scholar]
- Miura K, Lin SY, Yano M, Nagamine T. 2002. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theoretical and Applied Genetics 104, 981–986. [DOI] [PubMed] [Google Scholar]
- Muntz K, Belozersky MA, Dunaevsky YE, Schlereth A, Tiedemann J. 2001. Stored proteinases and the initiation of storage protein mobilization in seeds during germination and seedling growth. Journal of Experimental Botany 52, 1741–1752. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Bartsch M, Xiang Y, Miatton E, Pellengahr S, Yano R, Seo M, Soppe WJJ. 2012. The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel M, Vogel H, Landjeva S, Buck-Sorlin G, Lohwasser U, Scholz U, Börner A. 2009. Seed conservation in ex situ genebanks—genetic studies on longevity in barley. Euphytica 170, 5–14. [Google Scholar]
- Nagel M, Rosenhauer M, Willner E, Snowdon RJ, Friedt W, Börner A. 2011. Seed longevity in oilseed rape (Brassica napus L.)—genetic variation and QTL mapping. Plant Genetic Resources 9, 260–263. [Google Scholar]
- Nguyen TP, Keizer P, van Eeuwijk F, Smeekens S, Bentsink L. 2012. Natural variation for seed longevity and seed dormancy are negatively correlated in Arabidopsis . Plant Physiology 160, 2083–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oge L, Bourdais G, Bove J, Collet B, Godin B, Granier F, Boutin JP, Job D, Jullien M, Grappin P. 2008. Protein repair L-isoaspartyl methyltransferase 1 is involved in both seed longevity and germination vigor in Arabidopsis . Plant Cell 20, 3022–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms JJJ, Léon-Kloosterziel KM, Bartels D, Koornneef M, Karssen CM. 1993. Acquisition of desiccation tolerance and longevity in seeds of Arabidopsis thaliana: a comparative study using abscisic acid-insensitive abi3 mutants. Plant Physiology 102, 1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang PP, Pruitt RE, Meyerowitz EM. 1988. Molecular-cloning, genomic organization, expression and evolution of 12S-seed storage protein genes of Arabidopsis thaliana . Plant Molecular Biology 11, 805–820. [DOI] [PubMed] [Google Scholar]
- Prieto-Dapena P, Castano R, Almoguera C, Jordano J. 2006. Improved resistance to controlled deterioration in transgenic seeds. Plant Physiology 142, 1102–1112.16998084 [Google Scholar]
- Rajjou L, Belghazi M, Catusse J, et al. 2011. Proteomics and posttranslational proteomics of seed dormancy and germination. Methods in Molecular Biology 773, 215–236. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. 2012. Seed germination and vigor. Annual Review of Plant Biology 63, 507–533. [DOI] [PubMed] [Google Scholar]
- Rajjou L, Gallardo K, Debeaujon I, Vandekerckhove J, Job C, Job D. 2004. The effect of alpha-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiology 134, 1598–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Lovigny Y, Groot SPC, Belghaz M, Job C, Job D. 2008. Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiology 148, 620–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Miche L, Huguet R, Job C, Job D. 2007. The use of proteome and transcriptome profiling in the understanding of seed germination and identification of intrinsic markers determining seed quality, germination efficiency and early seedling vigour. In: Navie SC, Adkins SW, Ashmore S, eds. Seeds: Biology, Development and Ecology . Oxfordshire, CAB International, 149–158. [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R. 1998. The specific features of methionine biosynthesis and metabolism in plants. Proceedings of the National Academy of Sciences, USA 95, 7805–7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Jacquot JP. 2005. The plant multigenic family of thiol peroxidases. Free Radical Biology & Medicine 38, 1413–1421. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Fukuta Y, Sato T. 2005. Mapping of quantitative trait loci controlling seed longevity of rice (Oryza sativa L.) after various periods of seed storage. Plant Breeding 124, 361–366. [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D. 2004. Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16, 1419–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwember AR, Bradford KJ. 2010. Quantitative trait loci associated with longevity of lettuce seeds under conventional and controlled deterioration storage conditions. Journal of Experimental Botany 61, 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Lienhard S. 1998. Two isoforms of plant RAD23 complement a UV-sensitive rad23 mutant in yeast. Plant Journal 13, 815–821. [DOI] [PubMed] [Google Scholar]
- Sugliani M, Rajjou L, Clerkx EJM, Koornneef M, Soppe WJJ. 2009. Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3). New Phytologist 184, 898–908. [DOI] [PubMed] [Google Scholar]
- Tesnier K, Strookman-Donkers HM, Van Pijlen JG, Van der Geest AHM, Bino RJ, Groot SPC. 2002. A controlled deterioration test for Arabidopsis thaliana reveals genetic variation in seed quality. Seed Science Technology 30, 149–165. [Google Scholar]
- Van der Klei H, Vandamme J, Casteels P, Krebbers E. 1993. A 5th 2S albumin isoform is present in Arabidopsis thaliana . Plant Physiology 101, 1415–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Kaur H, Petla BP, Rao V, Saxena SC, Majee M. 2013. PROTEIN L-ISOASPARTYL METHYLTRANSFERASE2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiology 161, 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters C. 1998. Understanding the mechanisms and kinetics of seed aging. Seed Science Research 8, 223–244. [Google Scholar]
- Walters C, Hill LM, Wheeler LJ. 2005. Dying while dry: kinetics and mechanisms of deterioration in desiccated organisms. Integrative and Comparative Biology 45, 751–758. [DOI] [PubMed] [Google Scholar]
- Wan LL, Ross ARS, Yang JY, Hegedus DD, Kermode AR. 2007. Phosphorylation of the 12S globulin cruciferin in wild-type and abi1-1 mutant Arabidopsis thaliana (thale cress) seeds. Biochemistry Journal 404, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Masnavi G, Bhardwaj RM, Jiang Q, Bray CM, West CE. 2010. A plant DNA ligase is an important determinant of seed longevity. Plant Journal 63, 848–860. [DOI] [PubMed] [Google Scholar]
- Withana-Gamage TS, Hegedus DD, Qiu X, Yu PQ, May T, Lydiate D, Wanasundara JPD. 2013. Characterization of Arabidopsis thaliana lines with altered seed storage protein profiles using synchrotron-powered FT-IR spectromicroscopy. Journal of Agricultural and Food Chemistry 61, 901–912. [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Persiau G, Van Doorsselaere J, Davey MW, Demol H, Vandekerckhove J, Van Montagu M, Zabeau M, Boerjan W. 2001. Partial purification and identification of GDP-mannose 3′,5′-epimerase of Arabidopsis thaliana, a key enzyme of the plant vitamin C pathway. Proceedings of the National Academy of Sciences, USA 98, 14843–14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M. 2003. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. Journal of Biological Chemistry 278, 47483–47490. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhang SQ, Yao QH, Peng RH, Xiong AS, Li X, Zhu WM, Zhu YY, Zha DS. 2008. Identification of quantitative trait loci for seed storability in rice (Oryza sativa L.) Euphytica 164, 739–744. [Google Scholar]
- Zeng DL, Guo LB, Xu YB, Yasukumi K, Zhu LH, Qian Q. 2006. QTL analysis of seed storability in rice. Plant Breeding 125, 57–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.