Abstract
Terpenoids are the largest group of small-molecule natural products, with more than 60,000 compounds made from isopentenyl diphosphate (IPP) and its isomer dimethylallyl diphosphate (DMAPP). As the most diverse group of small-molecule natural products, terpenoids play an important role in the pharmaceutical, food, and cosmetic industries. For decades, Escherichia coli (E. coli) and Saccharomyces cerevisiae (S. cerevisiae) were extensively studied to biosynthesize terpenoids, because they are both fully amenable to genetic modifications and have vast molecular resources. On the other hand, our literature survey (20 years) revealed that terpenoids are naturally more widespread in Bacillales. In the mid-1990s, an inherent methylerythritol phosphate (MEP) pathway was discovered in Bacillus subtilis (B. subtilis). Since B. subtilis is a generally recognized as safe (GRAS) organism and has long been used for the industrial production of proteins, attempts to biosynthesize terpenoids in this bacterium have aroused much interest in the scientific community. This review discusses metabolic engineering of B. subtilis for terpenoid production, and encountered challenges will be discussed. We will summarize some major advances and outline future directions for exploiting the potential of B. subtilis as a desired “cell factory” to produce terpenoids.
Keywords: Biosynthesis, Metabolic engineering, Terpenoids, Bacillus subtilis
Introduction
Nature provides an infinite treasure of complex molecules (Wilson and Danishefsky 2006) which have served as leads and scaffolds for drug discovery in the past centuries (Newman and Cragg 2007; Newman and Cragg 2012; Newman et al. 2003). Numerous reports have detailed their diverse structures and biological functions. The largest and most diverse class of small-molecule natural products is the terpenoids, also known as isoprenoids or terpenes (Köksal et al. 2011). The Dictionary of Natural Products describes approximately 359 types of terpenoids, which comprise 64,571 compounds (as of May 2015). Since these terpenoids account for ca. 24.11 % (64,571 of 267,783) of all natural products (recorded in the dictionary, http://dnp.chemnetbase.com/) and are required for biological functions in all living creatures, they indisputably play a dominant role in both the scientific community and the commercial world (Breitmaier 2006).
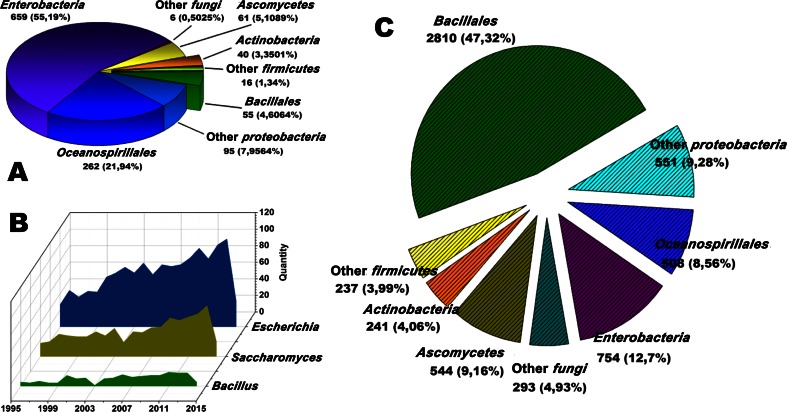
Along with a growing attraction for sustainable production, great interest has been expressed in biotechnological production of chemical products in general and terpenoids in particular. Since the 1990s, the interest in biosynthesizing terpenoids has skyrocketed, especially for desperately needed efficacious drugs such as artemisinin (Chang et al. 2007; Martin et al., 2003; Newman et al., 2006; Paddon et al. 2013; Ro et al. 2006; Tsuruta et al. 2009; Westfall et al. 2012) and taxol (Ajikumar et al. 2010; Jiang et al. 2012). In the past 20 years, most research has focused on using Escherichia coli, the host with the most advanced genetic tools, for biosynthesis of terpenoids (Fig. 1). Intensive experimentation in Escherichia coli (E. coli) has led to high yield production of some isoprenoids. However, uncertainty still looms around some aspects such as genetic engineering, characterization, reliability, quantitative strategy, and independence of biological modules (Kwok 2010). More options are needed to validate and optimize cell factories for terpenoid production. According to PubMed data, in comparison to other microorganisms, Bacillales (47.32 %) naturally possess more genes and proteins related to terpenoid biosynthesis pathways (Fig. 1), but surprisingly, little research effort has been devoted to the study of Bacillales as factories for natural products.
Fig. 1.
Percent of terpenoid biosynthesis related articles and terpenoid related gene reports, by source. a Percent of terpenoid biosynthesis related articles, by source. b Publication amount of terpenoid biosynthesis related articles, by year. c Percent of terpenoid related gene reports, by source
In the mid-1990s, it was discovered that Bacillus subtilis, a member of Bacillales that has a fast growth rate and is considered generally recognized as safe (GRAS) (FDA 1997; Schallmey et al. 2004; Widner et al. 2005), has inherent MEP pathway genes (Kuzma et al. 1995; Takahashi et al. 1998). The interest rose in B. subtilis as it has been used extensively for the industrial production of proteins (Westers et al. 2004; Sauer et al. 1998; Stockton and Wyss 1946). In addition, it was also reported that Bacillus is the highest isoprene producer among all tested microorganisms including E. coli, Pseudomonas aeruginosa, and Micrococcus luteus. The reported isoprene production rate (B. subtilis ATCC 6051) is 7 to 13 nmol per gram cells per hour (Kuzma et al. 1995). This high yield makes it a promising microbial host for terpenoid biosynthesis (Julsing et al. 2007; Wagner et al. 2000). Furthermore, B. subtilis has a wide substrate range and is able to survive under harsh conditions. Owing to its innate cellulases, it can even digest lignocellulosic materials and use the pentose sugars as its carbon source, hence decreasing the cost of biomass pretreatment (Maki et al. 2009; Ou et al. 2009). Here, we review major progress in metabolic engineering of B. subtilis for synthesizing terpenoids. The related pathway enzymes, genetic engineering reports, terpenoid detection methods, and their advantages and challenges will be summarized and discussed. We hope to provide a comprehensive review for exploiting the potential of B. subtilis as a cell factory for terpenoid production.
Inherent terpenoid biosynthetic pathways of B. subtilis
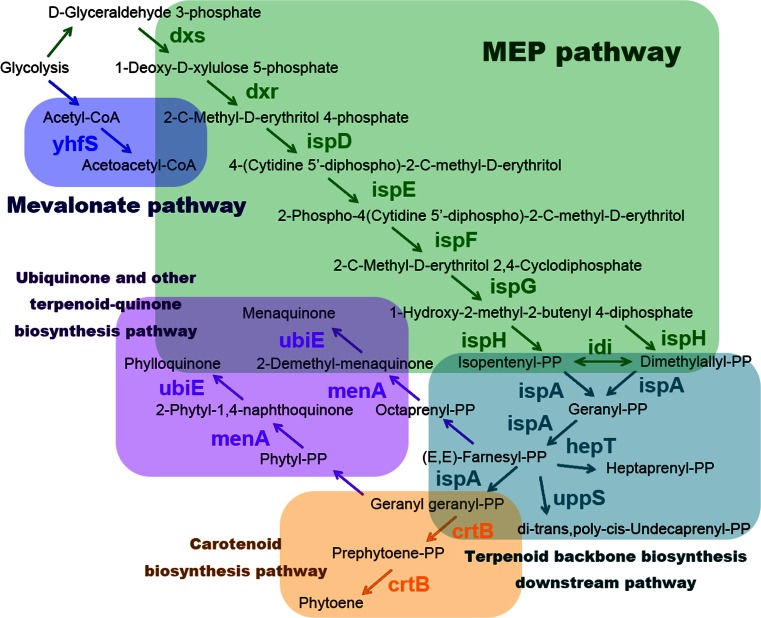
Terpenoids are synthesized based on isoprene (C5) units. In terpenoid biosynthetic pathways, IPP and DMAPP (C5 unit, diphosphate isoprene forms) are the basic terpenoid building blocks, generated by the Mevalonate and MEP pathways (the terpenoid backbone biosynthesis upstream pathways). The terpenoid backbone downstream pathway is responsible for biosynthesis of geranyl diphosphate (GPP), farsenyl diphosphate (FPP), and geranylgeranyl diphosphate (GGPP), which are the precursors of monoterpenoids (C10), sesquiterpenoids (C15), and diterpenoids (C20), respectively. B. subtilis has 15 inherent enzymes, belonging to five terpenoid biosynthesis pathways: two terpenoid backbone biosynthesis upstream pathways (the mevalonate pathway and MEP pathway), the terpenoid backbone biosynthesis downstream pathway, carotenoid biosynthesis pathway, and ubiquinone and other terpenoid-quinone biosynthesis pathway (Table 1, Fig. 2). For decades, isoprene yield has been considered the bottleneck for all terpenoid biosynthesis. Thus, to construct a cell platform which can produce and tolerate high amounts of isoprene and downstream intermediates is crucial. Since B. subtilis possesses all of the eight MEP pathway enzymes and can naturally produce high amounts of isoprene, it appears to be an ideal choice to utilize overexpression mutants of these enzymes to increase isoprene production.
Table 1.
B. subtilis inherent terpenoid biosynthesis enzymes
| Inherent pathways | EC number | Strains |
|---|---|---|
| Mevalonate pathway | 2.3.1.9 |
Bacillus subtilis subsp. Subtilis 168 Bacillus subtilis subsp. Subtilis RO-NN-1 Bacillus subtilis subsp. Subtilis BSP1 Bacillus subtilis subsp. Subtilis 6051-HGW Bacillus subtilis subsp. Subtilis BAB-1 Bacillus subtilis subsp. Subtilis AG1839 Bacillus subtilis subsp. Subtilis JH642 Bacillus subtilis subsp. Subtilis OH 131.1 Bacillus subtilis subsp. spizizenii W23 Bacillus subtilis subsp. spizizenii TU-B-10 Bacillus subtilis subsp. Natto BEST195 Bacillus subtilis BSn5 Bacillus subtilis QB928 Bacillus subtilis XF-1 Bacillus subtilis PY79 |
| MEP/DOXP pathway | 2.2.1.7, 1.1.1.267, 2.7.7.60, 2.7.1.148, 4.6.1.12, 1.17.7.1, 1.17.1.2, 5.3.3.2 | |
| Terpenoid backbone biosynthesis (downstream) | 2.5.1.1, 2.5.1.10, 2.5.1.29, 2.5.1.30, 2.5.1.31 | |
| Ubiquinone and other terpenoid-quinone biosynthesis | 2.5.1.74, 2.1.1.163, 2.5.1.- | |
| Carotenoid biosynthesis | 2.5.1.32 |
• Detailed information can be found at KEGG website, http://www.kegg.jp/
• Underlined enzymes (B. subtilis): functional parameters can be found at the BRENDA website, http://brenda-enzymes.info/index.php
• 2.3.1.9, acetyl-CoA acetyltransferase, yhfS
• 2.2.1.7, 1-deoxy-D-xylulose-5-phosphate synthase, dxs
• 1.1.1.267, 1-deoxy-D-xylulose 5-phosphate reductoisomerase, dxr
• 2.7.7.60, 2-D-methyl-D-erythritol 4-phosphate cytidylyltransferase, ispD
• 2.7.1.148, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, ispE
• 4.6.1.12, 2-D-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ispF
• 1.17.7.1, (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase, ispG
• 1.17.1.2, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, ispH
• 5.3.3.2, isopentenyl-diphosphate delta-isomerase, idi
• 2.5.1.1, 2.5.1.10, 2.5.1.29, geranylgeranyl diphosphate synthase, type II, ispA
• 2.5.1.30, heptaprenyl diphosphate synthase component 2, hepT
• 2.5.1.31, undecaprenyl diphosphate synthase, uppS
• 2.5.1.74, 2.5.1.-, 1,4-dihydroxy-2-naphthoate octaprenyltransferase, menA
• 2.1.1.163, demethylmenaquinone methyltransferase, ubiE
• 2.5.1.32, phytoene synthase, crtB
Fig. 2.
B. subtilis inherent terpenoid biosynthesis pathways
However, there are few reports on the B. subtilis MEP pathway. Most of the MEP pathway studies are based on E. coli. Withers and Keasling have described the MEP pathway of E. coli briefly (Withers and Keasling 2007). Kuzuyama and Seto (Kuzuyama and Seto 2012) clearly illustrated the enzymes and reactions involved in the MEP pathway. Carlsen summarized MEP pathway reactions and cofactors in a table (Carlsen et al. 2013). More details can be found in Zhao’s review (Zhao et al. 2013). As the kinetics of the MEP pathway enzymes are still unknown, it is unclear which step represents the largest barrier. Thus, the lack of knowledge about the kinetic parameters of the key enzymes is the main obstacle facing metabolic engineering of the MEP pathway in B. subtilis to produce terpenoids. Besides that, the low number of reports about using the B. subtilis MEP pathway to produce terpenoids highlights the need for more research in this area.
Here, we summarize information about the MEP pathway:
The initial enzyme in the MEP pathway is 1-deoxy-d-xylulose-5-phosphate synthase (dxs), which forms 1-deoxy-d-xylulose 5-phosphate (DXP) by the condensation of d-glyceraldehyde 3-phosphate (GAP) and pyruvate. This enzyme is not only specific for the MEP pathway but also plays a role in thiamine metabolism (Sprenger et al. 1997), which shares the flux with the MEP pathway. Gene knockout results (Julsing et al. 2007) suggest that overexpressing dxs may result in a significant improvement in terpenoid production without notable toxicity to the host cell (Zhao et al. 2011; Zhou et al. 2013b). Previous studies in other bacteria also supported the theory that dxs may be the first rate-limiting step of the MEP pathway, as overexpressing dxs can increase isoprenoid production (Estévez et al. 2001; Kim et al. 2006; Xue and Ahring 2011). Moreover, compared to the mevalonate pathway, the theoretical mass yield of terpenoids from glucose is 30 % from DXP, 5 % higher than the yield from MVA (Rude and Schirmer 2009; Whited et al. 2010), which emphasizes the importance of dxs in the MEP pathway.
The enzymes 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase (ispD), 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (ispE), and 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (ispF) are required to convert MEP to 2-C-methyl-d-erythritol 2,4-cyclodiphosphate (MECDP) (Herz et al. 2000; Kuzuyama et al. 2000a; Kuzuyama et al. 2000b; Lüttgen et al. 2000; Rohdich et al. 1999). In most organisms containing MEP pathway homologs, the genes encoding ispD and ispF are neighbors on the chromosome with the ispE at a distal location. They are also regarded as key enzymes in the MEP pathway (Ajikumar et al. 2010; Lu et al. 2014; Yuan et al. 2006; Zhou et al. 2013b). IspD and ispF are essential for cell survival due to their significant impact on cell wall biosynthesis and depletion (Campbell and Brown 2002). IspE has also been identified as crucial for survival of pathogenic bacteria and essential in Mycobacterium smegmatis (Eoh et al. 2009).
The most controversial enzymes in the MEP pathway are 1-deoxy-d-xylulose-5-phosphate reductoisomerase (dxr) and isopentenyl-diphosphate delta-isomerase (idi). Some researchers consider them as key enzymes in the MEP pathway (Berthelot et al. 2012; Soliman et al. 2011; Sun et al. 1998; Xue et al. 2015), while others find that they are not essential, at least in some cases (Fox and Poulter 2005; Lagarde et al. 2000; Sangari et al. 2010; Xue and Ahring 2011; Zhao et al. 2011). As far as we know now, there are two families of idi, B. subtilis possesses type 2 idi, which was considered as a nonessential enzyme in the bacillus MEP pathway (Julsing et al. 2007; Takagi et al. 2004).
Other important enzymes in the MEP pathway are (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (ispG) and 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (ispH), but their catalytic mechanisms are still unclear (Zhao et al. 2013). The enzyme ispH catalyzes the 2H+∕2e− reduction of hydroxy-2-methyl-2-butenyl-4-diphosphate (HMBDP) producing an approximately 5:1 mixture of IPP and DMAPP in return (Wang et al. 2010). This enzyme and ispG are deemed essential enzymes for cell survival (Liu et al. 2012; Rohmer 2008). It has been reported that ispG can effectively reduce the efflux of methylerythritol cyclodiphosphate (MECDP), resulting in a significant increase in downstream terpenoid production (Zhou et al. 2012). Additional information on the bio-organometallic chemistry of ispG and ispH can be found in Wang’s review (Wang and Oldfield 2014).
Genetic engineering of B. subtilis
Most of the knowledge about the MEP pathway was obtained from research in E. coli and other bacteria. Therefore, research into the progress of genetic engineering of MEP pathway enzymes in B. subtilis can provide more direct support for utilizing B. subtilis as a microbial host for terpenoid biosynthesis.
Wagner first described the phases of isoprene formation during growth and sporulation of B. subtilis (Wagner et al. 1999). They found that isoprene formation is linked to glucose catabolism, acetoin catabolism, and sporulation. One possible mechanism is that isoprene is a metabolic overflow metabolite released when flow of carbon to higher isoprenoids is restricted. This phenomenon can be illustrated as follows: (a) when cells are rapidly metabolizing the available carbon sources, isoprene is released; (b) when less carbon is available during transitions in carbon assimilation pathways, isoprene production declines; and (c) when cell growth ceases and spore formation is initiated, production of isoprene continues. In 2000, it was confirmed that isoprene is a product of the MEP pathway in B. subtilis (Wagner et al. 2000). It was also reported that isoprene release might be used as a barometer of central carbon flux changes during the growth of Bacillus strains (Shirk et al. 2002). Besides that, the activity of isoprene synthase (ISPS) was studied by using permeabilized cells. When grown in a bioreactor, B. subtilis cells released isoprene in parallel with the ISPS activity (Sivy et al. 2002).
In order to gain more insight into the MEP pathway of B. subtilis, conditional knockouts of the MEP pathway genes of B. subtilis were constructed, then the amount of emitted isoprene was analyzed. The results show that the emission of isoprene is severely decreased without the genes encoding dxs, ispD, ispF, or ispH, indicating their importance in the MEP pathway. In addition, idi has been proven not to be essential for the B. subtilis MEP pathway (Julsing et al. 2007). Xue and Ahring first tried to enhance isoprene production by modifying the MEP pathway in B. subtilis. They overexpressed the dxs and dxr genes. The strain that overexpressed dxs showed a 40 % increase in isoprene yield compared to the wild-type strain, whereas in the dxr overexpression strain, the isoprene level was unchanged. Furthermore, they studied the effect of external factors and suggested that 1 % ethanol inhibits isoprene production, but the stress factors heat (48 °C), salt (0.3 M), and H2O2 (0.005 %) can induce the production of isoprene. In addition, they found that these effects are independent of SigB, which is the general stress-responsive alternative sigma factor of B. subtilis (Xue and Ahring 2011). Hess et al. co-regulated the terpenoid pathway genes in B. subtilis. Transcriptomics results showed that the expression levels of dxs and ispD are positively correlated with isoprene production, while on the other hand, the expression levels of ispH, ispF, ispE, and dxr are inversely correlated with isoprene production. Moreover, their results supported Xue’s conclusions about the effect of external factors (Hess et al. 2013).
In 2009, Yoshida et al. first successfully transcribed and transfected crtM and crtN genes into B. subtilis to direct the carbon flux from the MEP pathway to C30 carotenoid biosynthesis and successfully produced 4,4′-diapolycopene and 4,4′-diaponeurosporene (Yoshida et al. 2009). Thereafter, Maeda reported a method to produce glycosylated C30 carotenoic acid by introducing Staphylococcus aureus (S. aureus) crtP and crtQ genes into B. subtilis, together with crtM and crtN (Maeda 2012). Later, Zhou overexpressed dxs and idi genes along with introducing ads (ads encodes the synthase which cyclizes farnesyl diphosphate into amorphadiene) in B. subtilis and got the highest yield of amorphadiene (∼20 mg/L) at shake-flask scale. They thought that the lack of genetic tools for fine-tuning the expression of multiple genes is the bottleneck in production of terpenoids in B. subtilis. So they modified B. subtilis genes by using a two-promoter system to independently control the expression levels of two gene cassettes (Zhou et al. 2013a). After that, Xue et al. systematically studied the B. subtilis MEP pathway enzymes (Xue et al. 2015). A series of synthetic operons expressing MEP pathway genes were analyzed by using the level of C30 carotenoid production as a measure of the effect of those modulations. All of the overexpressed gene constructs showed higher production of carotenoids compared to wild type. Dxs and dxr (8-fold and 9.2-fold increase in carotenoid production) have been validated as the most productive part of the MEP pathway genes in this study.
Other reports are related to C35 terpenoids and their enzymes, which were found in B. subtilis, like heterodimeric enzyme, heptaprenyl diphosphate synthase (HepS and HepT), and tetraprenyl-β-curcumene synthase (YtpB), which are responsible for forming long prenyl diphosphate chains (C35) (Sato et al. 2011). As Heider noted in his review, B. subtilis has not yet been a major focus to produce carotenoids (Heider et al. 2014). Furthermore, we cannot find other research about terpenoid biosynthesis in B. subtilis. Since B. subtilis possesses many advantages as mentioned above in the introduction section, biosynthesis of terpenoids via the B. subtilis MEP pathway could be both an opportunity and a challenge.
Detection and metabolomics methods for engineering terpenoid pathway
As is known, most metabolic engineering work is improved by using a combination of random and targeted approaches. Mariët and Renger (Wilson and Danishefsky 2006) pointed out that the selection of these targets has depended at best on expert knowledge but to a great extent also on “educated guesses” and “gut feeling.” Consequently, time and money are wasted on irrelevant targets or only a minor improvement result. Along with the development of systems biology, metabolomics, a technology that includes non-targeted, holistic metabolite analysis of the cellular and/or environmental changes combined with multivariate data analysis tools is being increasingly used to replace empirical approaches for targeted natural product biosynthesis (Newman et al. 2006; Paddon et al. 2013). Gregory’s group (Ajikumar et al. 2010) has used metabolomics analysis of their previous strains, leading them to identify a noticeable metabolite by-product that inversely correlated with taxadiene accretion. This hint helped them to achieve approximately 1 g per liter taxadiene from E. coli.
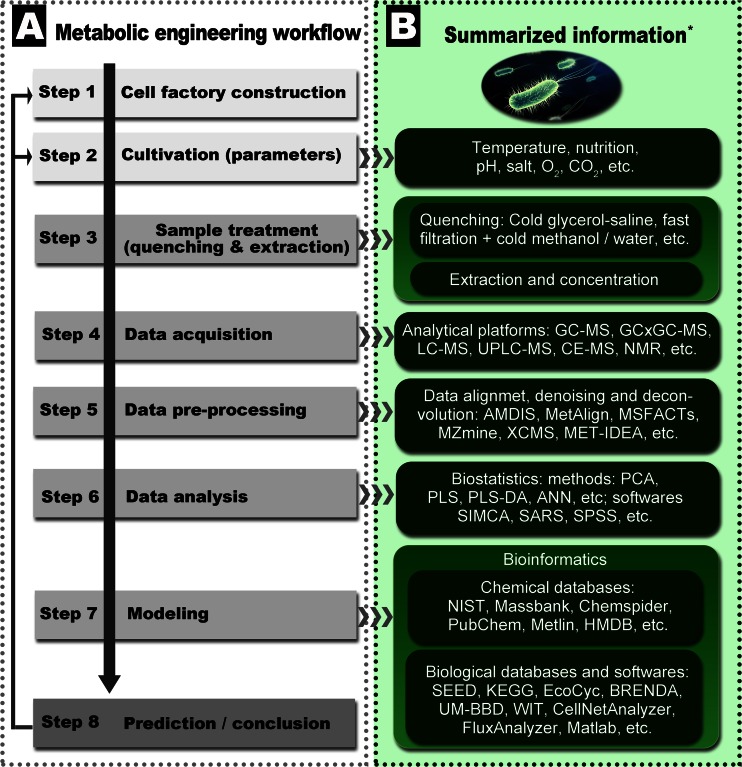
Because the research on the Bacillus MEP pathway is still at an early stage, it is urgent to develop guidelines for unbiased selection of the best rational design approach to engineering the terpenoid. The newest developments of metabolomics, meta-omics, computer, and mathematic sciences offer more options for not only unbiased selection and ranking methods but also high-throughput and more precise prediction models that enable a mechanistic description of microbial metabolic pathways (Breitmaier 2006; Martin et al., 2003). Scheme 1 summarizes the workflow, essential reports, and resources for the study of terpenoid microbial metabolomics.
Scheme 1.
Flowchart and resources for terpenoid microbial metabolomics study. a Microbial metabolic engineering workflow. b Related information of each step for microbial metabolic engineering. * Selected resources: 1. MS data of B. subtilis metabolites (Coulier et al. 2006; Koek et al. 2006; Soga et al. 2003). 2. The metabolomics standards initiative (Fiehn et al. 2007). 3. Microbial metabolomics study examples for terpenoid biosynthesis (Paddon and Keasling 2014; Zhou et al. 2012). 4. Databases, software packages, and protocols (Thiele and Palsson 2010) and http://omictools.com/. 5. Genome-scale data of reconstructed B. subtilis metabolic net (impact of single-gene deletions on growth in B. subtilis) (Oh et al. 2007). 6. Comparative microbial metabolomics study of E. coli, B. subtilis, and S. cerevisiae (van der Werf et al. 2007). 7. The complete genome sequence of B. subtilis (Kunst et al. 1997). 8. Constraint-based modeling methods (Bordbar et al. 2014). 9. Software applications for flux balance analysis (including a software comparative list) (Lakshmanan et al. 2012). 10. Sample treatment methods (Jia et al. 2004; Larsson and Törnkvist 1996; Maharjan and Ferenci 2003; van der Werf et al. 2007; Villas-Bôas and Bruheim 2007)
To observe and optimize the terpenoid biosynthesis pathways, detection methods are also crucial. The techniques that are currently employed in the study of microbially produced terpenoids are usually gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-MS (LC-MS). Other techniques such as nuclear magnetic resonance (NMR) (Hecht et al. 2001) and Raman spectroscopic analysis (de Oliveira et al. 2010) are also used in terpenoid analysis, although compared with MS-based coupling techniques, they are less sensitive and/or reliable. Most likely, the currently existing methods for the quantitative determination of terpenoids in bacteria are sufficient. There are numerous articles about quantifying and identifying terpenoids (esp. carotenoids, see Foppen’s tables (Foppen 1971)) in plants, microorganisms, and other organisms. Most of these methods can be applied in B. subtilis.
In 1995, Kuzma discovered that B. subtilis can produce isoprene efficiently (Kuzma et al. 1995). This Colorado research group focused on the isoprene biosynthesis mechanism in B. subtilis. They used GC, GC-MS, HPLC, 13C, and 2H labeling methods, non-radioactive methods, and online chemical-ionization mass spectrometry (CIMS) to measure isoprene and MEP pathway metabolites (Custer et al. 2003; Fisher et al. 2001; Kuzma et al. 1995; Shirk et al. 2002; Wagner et al. 2000; Wagner et al. 1999). Table 2 summarizes their methods, as well as more recent methods to detect and analyze B. subtilis terpenoid metabolites.
Table 2.
Detection and analysis reports of B. subtilis terpenoid pathway metabolites
| Method | Compound | Characteristic | Reference |
|---|---|---|---|
| GC-MS | Isoprene | Rt = 16.5 min, m/z 39, 53, 67 | Kuzma et al. 1995 |
| GC | Isoprene | Wagner et al. 1999 | |
| GC-MS (13C, 2H labeling) | Isoprene | Common substrate m/z 39, 53, 67; Substrate: U-[13C6] glucose m/z 42, 57, 72; 1-[13C] pyruvate m/z 40, 54, 68; 2-[13C] pyruvate m/z 40, 55, 69; 3-[13C] pyruvate m/z 40, 54, 69. |
Wagner et al. 2000 |
| GC | DMAPP | Fisher et al. 2001 | |
| GC | Isoprene | Rt = 2.6 min | Shirk et al. 2002 |
| HPLC | Acetoin | Rt = 5.5 min, 354 nm | |
| Kits | Glucose | ||
| Kits | Lactic, pyruvic acids | ||
| CIMS | (M + H)+(H2O)n: | Custer et al. 2003 | |
| Acetaldehyde | m/z 63 (n = 1) | ||
| Acetoin | m/z 89 (n = 0) | ||
| Acetone | m/z 77 (n = 1) | ||
| 2,3-Butanediol | m/z 91 (n = 0) | ||
| Butanol | m/z 111 (n = 2) | ||
| 2-Butanone | m/z 91 (n = 1) | ||
| Butyraldehyde | m/z 91 (n = 1) | ||
| Butyl acetate | m/z 135 (n = 1) | ||
| Diacetyl | m/z 123 (n = 2) | ||
| Dimethyl sulfide | m/z 63 (n = 0) | ||
| Ethanol | m/z 83 (n = 2) | ||
| Ethyl acetate | m/z 107 (n = 1) | ||
| Isoamyl alcohol | m/z 107 (n = 1) | ||
| Isoprene | m/z 69 (n = 0) | ||
| GC | Isoprene | Julsing et al. 2007 | |
| HPLC | 4,4′-Diapolycopene | Rt = 26.8 min, Absorption: 293, 443, 472, 501 nm | Yoshida et al. 2009 |
| 4,4′-Diaponeurosporene | Rt = 28.9 min, Absorption: 266, 415, 439, 469 nm | ||
| MALDI-TOF MS | 4,4′-Diapolycopene | m/z 399.9 | |
| 4,4′-Diaponeurosporene | m/z 401.9 | ||
| GC-MS | Isoprene | Rt = 1.9 min | Xue and Ahring 2011 |
| HPLC | Glycosyl 4,4′-diaponeurosporenoate | Rt = 10.0 min, Absorption: 282, 469 nm | Barredo 2012 |
| 4,4′-Diapolycopene | Absorption: 293, 443, 472, 501 nm | ||
| 4,4′-Diaponeurosporene | Rt = 14.4 min, Absorption: 265, 414, 441, 469 nm | ||
| UPLC-MS | DXP | Rt = 5.6 min, m/z 213.0170 | Tsuruta et al. 2009; Zhou et al. 2012; Zhou et al. 2013a |
| MEP | Rt = 5.2 min, m/z 215.0330 | ||
| CDP-ME | Rt = 6.2 min, m/z 520.0730 | ||
| CDP-MEP | Rt = 7.3 min, m/z 600.0390 | ||
| MEC | Rt = 6.6 min, m/z 276.9884 | ||
| HMBPP | Rt = 7.0 min, m/z 260.9920 | ||
| GC-MS | Trans-Caryophyllene | Rt = 3.4 min, m/z 189, 204 | |
| Amorpha-4,11-diene | Rt = 3.5 min, m/z 189, 204 | ||
| LC-Fourier transform MS | (Untargeted metabolomics study) | Cho et al. 2014 | |
As is the case for biosynthesis of different chemical compounds, genetic modification often leads to dead ends. The difficulties in metabolic engineering of bacteria for terpenoid production normally are not terpenoid detection but problems in the complex metabolic net (Baidoo and Keasling 2013). Although the latest reports (Zhou et al. 2012; Zhou et al. 2013a) describe a promising method that can simultaneously detect MEP pathway intermediates, the repeatability is not as good for CDP-MEP as for the other intermediates, especially when the amount of CDP-MEP in bacteria is very low (summarized MS information of MEP pathway metabolites can be found in Table 3). In addition, even if the reported methods are sufficient to analyze all the MEP pathway intermediates, it is still difficult to predict and identify the unknown mechanisms for improving terpenoid production and other relevant compounds due to the fact that all of the MEP pathway enzymes are also involved in other metabolic activities (http://www.kegg.jp/). Cho’s untargeted metabolomics study (Cho et al. 2014) may have pointed out a direction that can help solve some of these problems, whereas few untargeted metabolomics research for B. subtilis metabolic pathway study can be found online. As mentioned above, integrated metabolomics studies and constraint-based models might orient future study for biosynthesis of terpenoids (see Scheme 1). The current state of analysis methods, which can be integrated into metabolomics researches and be used in terpenoid biosynthesis studies, raises questions about the following issues: (1) detailed preparation work such as reproducible growth of B. subtilis, sampling, and quenching methods, which can be used in metabolomics studies to elucidate the mechanisms of the MEP pathway; (2) extraction methods that maintain the original structure of intermediates and subsequently allow the identification of those compounds and their accurate quantification; (3) extraction coupled quantification methods that can be used to quantify minor components from small-scale bacterial cultures to reduce the workload; and (4) data pre-processing, biostatistics, and bioinformatics methods for big data analysis, integration, and modeling that can reflect the cell bio-net, narrow the research scope, target the key products, genes, and enzymes, and finally lead us to further improvements.
Table 3.
MS information of B. subtilis inherent terpenoid pathway intermediates
| Compound | Formula | Mass | ESI-Q-TOF | ||
|---|---|---|---|---|---|
| Mode | CE (V) | m/z | |||
| G3P | C3H7O6P | 169.9980 | + | 40 | 80.9730, 62.9631, 98.9823, 45.0347 |
| DXP | C5H11O7P | 214.0242 | − | 0 | 213.0167, 96.9695, 138.9795,78.9592 |
| MEP | C5H13O7P | 216.0399 | |||
| CDP-ME | C14H25N3O14P2 | 521.0812 | |||
| CDP-ME2P | C14H26N3O17P3 | 601.0475 | |||
| MECDP | C5H12O9P2 | 277.9957 | + | 40 | 98.9830, 83.0480, 55.0538, 65.0394, 80.9733, 43.0536 |
| HMBDP | C5H12O8P2 | 262.0007 | |||
| IPP | C5H12O7P2 | 246.0058 | − | 0 | 244.9979, 78.9591 |
| DMAPP | C5H12O7P2 | 246.0058 | − | 0 | 244.9979, 78.9592 |
| GPP | C10H20O7P2 | 314.0684 | − | 0 | 313.0629, 78.9593 |
| FPP | C15H28O7P2 | 382.1310 | |||
| GGPP | C20H36O7P2 | 450.1936 | |||
| PPDP | C40H68O7P2 | 722.4440 | |||
| Phytoene | C40H64 | 544.5008 | |||
| HepPP | C35H60O7P2 | 654.3814 | |||
| UDPP | C55H92O7P2 | 926.6318 | |||
| PDP | C20H42O7P2 | 456.2406 | |||
| OPP | C40H68O7P2 | 722.4440 | |||
| 2-Phytyl-1,4-naphthoquinone | C30H44O2 | 436.3341 | |||
| 2-Demethylmenaquinone | C50H70O2 | 702.5376 | |||
| Phylloquinone | C31H46O2 | 450.3498 | + | 40 | 187.0749, 57.0703, 43.0550, 71.0856, 171.0799, 199.0758, 105.0326, 157.0650 |
| Menaquinone | C41H56O2 | 580.4280 | |||
• Data sources: http://www.hmdb.ca/, http://www.massbank.jp/index.html?lang=en, http://www.chemspider.com/, https://metlin.scripps.edu/index.php, http://pubchem.ncbi.nlm.nih.gov/
• G3P d-glyceraldehyde 3-phosphate
• DXP deoxy-d-xylulose 5-phosphate
• MEP 2-C-methyl-d-erythritol 4-phosphate
• CDP-ME 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol
• CDP-ME2P phospho-4(Cytidine 5′-diphospho)-2-C-methyl-d-erythritol
• MECDP 2-C-methyl-d-erythritol 2,4-Cyclodiphosphate
• HMBDP hydroxy-2-methyl-2-butenyl 4-diphosphate
• IPP isopentenyl-PP
• DMAPP dimethylallyl-PP
• GPP geranyl-PP
• FPP (E,E)-farnesyl-PP
• GGPP geranylgeranyl-PP
• PPDP prephytoene-PP.
• HepPP heptaprenyl-PP
• UDPP di-trans, poly-cis-undecaprenyl-PP
• PDP phytyl-PP
• OPP octaprenyl-PP
Summary
B. subtilis offers new opportunities and good prospects for terpenoid biosynthesis. This review provides a brief account of metabolic engineering of B. subtilis for terpenoid production, summarizing our understanding of B. subtilis, the MEP pathway, and related techniques. While the mevalonate pathway and terpenoid biosynthesis in E. coli have been studied for decades, research on the Bacillus MEP pathway is still at an early stage. That is why, at this point, there is no sufficient data on Bacillus yield to make a fair comparison with published yields of terpenoids in E. coli and other cell factories. However, theoretically, B. subtilis has the potential to be optimized as a high-yield-producing cell factory. The advantages of studying terpenoid biosynthesis in B. subtilis include (1) its fast growth rate and ability to survive under harsh conditions, (2) its GRAS status, (3) its wide substrate range and inherent MEP pathway genes, (4) the fact that it is a naturally high isoprene producer, (5) its clear genetic background, abundant genetic tools, and (6) its innate cellulases, which can digest lignocellulosic materials and use the breakdown products as its carbon source, which would decrease large-scale production costs. Still, B. subtilis share some of the features of other gram-positive bacteria like plasmid instability. Also, there are some B. subtilis-specific engineering challenges that need to be explored. The catalytic mechanisms of two MEP pathway enzymes (IspG, IspH) in B. subtilis are unclear yet. The importance of DXR and IDI in the MEP pathway is controversial. DXS has been generally regarded as the essential rate-limiting enzyme, but even the functional parameters of DXS in B. subtilis have not yet been reported. Many questions regarding the mechanism of the MEP pathway, the interactions of related enzymes and metabolites, and the kinetic parameters of MEP pathway enzymes in B. subtilis remain unanswered. Obviously, the organism is promising and the questions are fascinating. There is thus significant reason for detailed investigations of terpenoid biosynthesis via the B. subtilis MEP pathway, particularly in metabolic engineering where there is not yet sufficient knowledge about the precise mechanisms or the effects of co-regulation of the enzymes.
Acknowledgments
The authors gratefully acknowledge support from EU FP-7 grant 289540 (PROMYSE). Z.G. acknowledges financial support from the China Scholarship Council (201408330157). I.I.A. is a recipient of Erasmus Mundus Action 2, Strand 1, Fatima Al Fihri project ALFI1200161 scholarship and is on study leave from Faculty of Pharmacy, Alexandria University. They also thank Dr. Diane Black, Language Centre, University of Groningen for carefully reading the manuscript and correcting the English.
Compliance with ethical standards
ᅟ
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Zheng Guan and Dan Xue contributed equally to this work
References
- Ajikumar PK, Xiao W-H, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science (New York, NY) 2010;330(6000):70–74. doi: 10.1126/science.1191652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidoo EE, Keasling JD. Microbial metabolomics: welcome to the real world! Metabolomics. 2013;9(4):755–756. [Google Scholar]
- Barredo J-L. Microbial carotenoids from bacteria and microalgae. Methods Mol Biol. 2012;892:133–141. [Google Scholar]
- Berthelot K, Estevez Y, Deffieux A, Peruch F. Isopentenyl diphosphate isomerase: a checkpoint to isoprenoid biosynthesis. Biochimie. 2012;94(8):1621–1634. doi: 10.1016/j.biochi.2012.03.021. [DOI] [PubMed] [Google Scholar]
- Bordbar A, Monk JM, King ZA, Palsson BO. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15(2):107–120. doi: 10.1038/nrg3643. [DOI] [PubMed] [Google Scholar]
- Breitmaier E (2006) Terpenes: flavors, fragrances, pharmaca, pheromones. John Wiley & Sons
- Campbell TL, Brown ED (2002) Characterization of the depletion of 2-C-methyl-D-erythritol-2,4-cyclodiphosphate synthase in Escherichia coli and Bacillus subtilis. J Bacteriol 184(20):5609–5618 [DOI] [PMC free article] [PubMed]
- Carlsen S, Ajikumar PK, Formenti LR, Zhou K, Phon TH, Nielsen ML, Lantz AE, Kielland-Brandt MC, Stephanopoulos G (2013) Heterologous expression and characterization of bacterial 2-C-methyl-D-erythritol-4-phosphate pathway in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97(13):5753–5769. doi:10.1007/s00253-013-4877-y [DOI] [PubMed]
- Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD. Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol. 2007;3(5):274–277. doi: 10.1038/nchembio875. [DOI] [PubMed] [Google Scholar]
- Cho K, Evans BS, Wood BM, Kumar R, Erb TJ, Warlick BP, Gerlt JA, Sweedler JV (2014) Integration of untargeted metabolomics with transcriptomics reveals active metabolic pathways. Metabolomics 2014(August). doi:10.1007/s11306-014-0713-3 [DOI] [PMC free article] [PubMed]
- Coulier L, Bas R, Jespersen S, Verheij E, van der Werf MJ, Hankemeier T. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal Chem. 2006;78(18):6573–6582. doi: 10.1021/ac0607616. [DOI] [PubMed] [Google Scholar]
- Custer TG, Wagner WP, Kato S, Bierbaum VM, Fall R. Potential of on-line CIMS for bioprocess monitoring. Biotechnol Prog. 2003;19(4):1355–1364. doi: 10.1021/bp025730k. [DOI] [PubMed] [Google Scholar]
- de Oliveira VE, Castro HV, Edwards HG, de Oliveira LFC. Carotenes and carotenoids in natural biological samples: a Raman spectroscopic analysis. J Raman Spectrosc. 2010;41(6):642–650. [Google Scholar]
- Eoh H, Narayanasamy P, Brown AC, Parish T, Brennan PJ, Crick DC (2009) Expression and characterization of soluble 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase from bacterial pathogens. Chem Biol 16(12):1230–1239 [DOI] [PMC free article] [PubMed]
- Estévez JM, Cantero A, Reindl A, Reichler S, León P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem. 2001;276(25):22901–22909. doi: 10.1074/jbc.M100854200. [DOI] [PubMed] [Google Scholar]
- FDA FDA proposed simplified. GRAS notification system - 62 FR 18937. 1997;62(74):18938–18964. [Google Scholar]
- Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, Sumner LW, Goodacre R, Hardy NW, Taylor C. The metabolomics standards initiative (MSI) Metabolomics. 2007;3(3):175–178. [Google Scholar]
- Fisher AJ, Rosenstiel TN, Shirk MC, Fall R. Nonradioactive assay for cellular dimethylallyl diphosphate. Anal Biochem. 2001;292(2):272–279. doi: 10.1006/abio.2001.5079. [DOI] [PubMed] [Google Scholar]
- Foppen FH. Tables for the identification of carotenoid pigments. Chromatogr Rev. 1971;14(3):133–298. doi: 10.1016/0009-5907(71)80012-1. [DOI] [PubMed] [Google Scholar]
- Fox DT, Poulter CD (2005) Mechanistic studies with 2-C-methyl-D-erythritol 4-phosphate synthase from Escherichia coli. Biochemistry 44(23):8360–8368. doi:10.1021/bi047312p [DOI] [PubMed]
- Hecht S, Eisenreich W, Adam P, Amslinger S, Kis K, Bacher A, Arigoni D, Rohdich F. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci. 2001;98(26):14837–14842. doi: 10.1073/pnas.201399298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heider SAE, Peters-Wendisch P, Wendisch VF, Beekwilder J, Brautaset T. Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl Microbiol Biotechnol. 2014;98(10):4355–4368. doi: 10.1007/s00253-014-5693-8. [DOI] [PubMed] [Google Scholar]
- Herz S, Wungsintaweekul J, Schuhr CA, Hecht S, Lüttgen H, Sagner S, Fellermeier M, Eisenreich W, Zenk MH, Bacher A (2000) Biosynthesis of terpenoids: YgbB protein converts 4-diphosphocytidyl-2C-methyl-D-erythritol 2-phosphate to 2C-methyl-D-erythritol 2, 4-cyclodiphosphate. Proc Natl Acad Sci 97(6):2486–2490 [DOI] [PMC free article] [PubMed]
- Hess BM, Xue J, Markillie LM, Taylor RC, Wiley HS, Ahring BK, Linggi B. Coregulation of terpenoid pathway genes and prediction of isoprene production in using transcriptomics. PLoS One. 2013;8(6):e66104. doi: 10.1371/journal.pone.0066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Liu B-F, Terabe S, Nishioka T. Two-dimensional separation method for analysis of Bacillus subtilis metabolites via hyphenation of micro-liquid chromatography and capillary electrophoresis. Anal Chem. 2004;76(5):1419–1428. doi: 10.1021/ac035039b. [DOI] [PubMed] [Google Scholar]
- Jiang M, Stephanopoulos G, Pfeifer BA. Downstream reactions and engineering in the microbially reconstituted pathway for Taxol. Appl Microbiol Biotechnol. 2012;94(4):841–849. doi: 10.1007/s00253-012-4016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julsing MK, Rijpkema M, Woerdenbag HJ, Quax WJ, Kayser O. Functional analysis of genes involved in the biosynthesis of isoprene in Bacillus subtilis. Appl Microbiol Biotechnol. 2007;75(6):1377–1384. doi: 10.1007/s00253-007-0953-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-J, Kim M-D, Choi J-H, Kim S-Y, Ryu Y-W, Seo J-H. Amplification of 1-deoxy-D-xyluose 5-phosphate (DXP) synthase level increases coenzyme Q(10) production in recombinant Escherichia coli. Appl Microbiol Biotechnol. 2006;72(5):982–985. doi: 10.1007/s00253-006-0359-9. [DOI] [PubMed] [Google Scholar]
- Koek MM, Muilwijk B, van der Werf MJ, Hankemeier T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal Chem. 2006;78(4):1272–1281. doi: 10.1021/ac051683+. [DOI] [PubMed] [Google Scholar]
- Köksal M, Hu H, Coates RM, Peters RJ, Christianson DW. Structure and mechanism of the diterpene cyclase ent-copalyl diphosphate synthase. Nat Chem Biol. 2011;7(7):431–433. doi: 10.1038/nchembio.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini A, Alloni G, Azevedo V, Bertero M, Bessieres P, Bolotin A, Borchert S. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390(6657):249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kuzma J, Nemecek-Marshall M, Pollock WH, Fall R. Bacteria produce the volatile hydrocarbon isoprene. Curr Microbiol. 1995;30(2):97–103. doi: 10.1007/BF00294190. [DOI] [PubMed] [Google Scholar]
- Kuzuyama T, Seto H. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc Japan Acad Ser B-Phys Biol. 2012;88(3):41–52. doi: 10.2183/pjab.88.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzuyama T, Takagi M, Kaneda K, Dairi T, Seto H (2000a) Formation of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol from 2-C-methyl-D-erythritol 4-phosphate by 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett 41(5):703–706
- Kuzuyama T, Takagi M, Kaneda K, Watanabe H, Dairi T, Seto H (2000b) Studies on the nonmevalonate pathway: conversion of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol to its 2-phospho derivative by 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase. Tetrahedron Lett 41(16):2925–2928
- Kwok R. Five hard truths for synthetic biology. Nat News. 2010;463(7279):288–290. doi: 10.1038/463288a. [DOI] [PubMed] [Google Scholar]
- Lagarde D, Beuf L, Vermaas W. Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl Environ Microbiol. 2000;66(1):64–72. doi: 10.1128/aem.66.1.64-72.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan M, Koh G, Chung BK, Lee D-Y (2012) Software applications for flux balance analysis. Briefings in bioinformatics:bbs069 [DOI] [PubMed]
- Larsson G, Törnkvist M. Rapid sampling, cell inactivation and evaluation of low extracellular glucose concentrations during fed-batch cultivation. J Biotechnol. 1996;49(1):69–82. doi: 10.1016/0168-1656(96)01534-9. [DOI] [PubMed] [Google Scholar]
- Liu Y-L, Guerra F, Wang K, Wang W, Li J, Huang C, Zhu W, Houlihan K, Li Z, Zhang Y, Nair SK, Oldfield E. Structure, function and inhibition of the two- and three-domain 4Fe-4S IspG proteins. Proc Natl Acad Sci U S A. 2012;109(22):8558–8563. doi: 10.1073/pnas.1121107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Ye L, Xu H, Xie W, Gu J, Yu H (2014) Enhanced production of coenzyme Q(10) by self-regulating the engineered MEP pathway in Rhodobacter sphaeroides. Biotechnol Bioeng 111(4):761–769. doi:10.1002/bit.25130 [DOI] [PubMed]
- Lüttgen H, Rohdich F, Herz S, Wungsintaweekul J, Hecht S, Schuhr CA, Fellermeier M, Sagner S, Zenk MH, Bacher A (2000) Biosynthesis of terpenoids: YchB protein of Escherichia coli phosphorylates the 2-hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Proc Natl Acad Sci 97(3):1062–1067 [DOI] [PMC free article] [PubMed]
- Maeda I (2012) Genetic modification in Bacillus subtilis for production of C30 carotenoids. Methods in molecular biology (Clifton, NJ) 892:197–205 doi:10.1007/978–1–61779–879-5_11 [DOI] [PubMed]
- Maharjan RP, Ferenci T. Global metabolite analysis: the influence of extraction methodology on metabolome profiles of Escherichia coli. Anal Biochem. 2003;313(1):145–154. doi: 10.1016/s0003-2697(02)00536-5. [DOI] [PubMed] [Google Scholar]
- Maki M, Leung KT, Qin W. The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci. 2009;5(5):500. doi: 10.7150/ijbs.5.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin VJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70(3):461–477 [DOI] [PubMed]
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75(3):311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66(7):1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- Newman JD, Marshall J, Chang M, Nowroozi F, Paradise E, Pitera D, Newman KL, Keasling JD. High-level production of amorpha-4, 11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng. 2006;95(4):684–691. doi: 10.1002/bit.21017. [DOI] [PubMed] [Google Scholar]
- Oh Y-K, Palsson BO, Park SM, Schilling CH, Mahadevan R. Genome-scale reconstruction of metabolic network in Bacillus subtilis based on high-throughput phenotyping and gene essentiality data. J Biol Chem. 2007;282(39):28791–28799. doi: 10.1074/jbc.M703759200. [DOI] [PubMed] [Google Scholar]
- Ou MS, Mohammed N, Ingram L, Shanmugam K. Thermophilic Bacillus coagulans requires less cellulases for simultaneous saccharification and fermentation of cellulose to products than mesophilic microbial biocatalysts. Appl Biochem Biotechnol. 2009;155(1–3):76–82. doi: 10.1007/s12010-008-8509-4. [DOI] [PubMed] [Google Scholar]
- Paddon C, Westfall P, Pitera D, Benjamin K, Fisher K, McPhee D, Leavell M, Tai A, Main A, Eng D. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496(7446):528–532. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Paddon CJ, Keasling JD. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12(5):355–367. doi: 10.1038/nrmicro3240. [DOI] [PubMed] [Google Scholar]
- Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(7086):940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH (1999) Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci 96(21):11758–11763 [DOI] [PMC free article] [PubMed]
- Rohmer M. From molecular fossils of bacterial hopanoids to the formation of isoprene units: discovery and elucidation of the methylerythritol phosphate pathway. Lipids. 2008;43(12):1095–1107. doi: 10.1007/s11745-008-3261-7. [DOI] [PubMed] [Google Scholar]
- Rude MA, Schirmer A. New microbial fuels: a biotech perspective. Curr Opin Microbiol. 2009;12(3):274–281. doi: 10.1016/j.mib.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Sangari FJ, Perez-Gil J, Carretero-Paulet L, Garcia-Lobo JM, Rodriguez-Concepcion M. A new family of enzymes catalyzing the first committed step of the methylerythritol 4-phosphate (MEP) pathway for isoprenoid biosynthesis in bacteria. Proc Natl Acad Sci U S A. 2010;107(32):14081–14086. doi: 10.1073/pnas.1001962107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Yoshida S, Hoshino H, Tanno M, Nakajima M, Hoshino T (2011) Sesquarterpenes (C35 terpenes) biosynthesized via the cyclization of a linear C35 isoprenoid by a tetraprenyl-β-curcumene synthase and a tetraprenyl-β-curcumene cyclase: identification of a new terpene cyclase. J Am Chem Soc 133(25):9734–9737. doi:10.1021/ja203779h [DOI] [PubMed]
- Sauer U, Cameron DC, Bailey JE. Metabolic capacity of Bacillus subtilis for the production of purine nucleosides, riboflavin, and folic acid. Biotechnol Bioeng. 1998;59(2):227–238. [PubMed] [Google Scholar]
- Schallmey M, Singh A, Ward OP. Developments in the use of Bacillus species for industrial production. Can J Microbiol. 2004;50(1):1–17. doi: 10.1139/w03-076. [DOI] [PubMed] [Google Scholar]
- Shirk MC, Wagner WP, Fall R. Isoprene formation in Bacillus subtilis: a barometer of central carbon assimilation in a bioreactor? Biotechnol Prog. 2002;18(5):1109–1115. doi: 10.1021/bp0255412. [DOI] [PubMed] [Google Scholar]
- Sivy TL, Shirk MC, Fall R. Isoprene synthase activity parallels fluctuations of isoprene release during growth of Bacillus subtilis. Biochem Biophys Res Commun. 2002;294(1):71–75. doi: 10.1016/S0006-291X(02)00435-7. [DOI] [PubMed] [Google Scholar]
- Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J Proteome Res. 2003;2(5):488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- Soliman SS, Tsao R, Raizada MN. Chemical inhibitors suggest endophytic fungal paclitaxel is derived from both mevalonate and non-mevalonate-like pathways. J Nat Prod. 2011;74(12):2497–2504. doi: 10.1021/np200303v. [DOI] [PubMed] [Google Scholar]
- Sprenger GA, Schorken U, Wiegert T, Grolle S, de Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci U S A. 1997;94(24):12857–12862. doi: 10.1073/pnas.94.24.12857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton JR, Wyss O. Proteinase production by Bacillus subtilis. J Bacteriol. 1946;52(2):227. [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Cunningham FX, Gantt E. Differential expression of two isopentenyl pyrophosphate isomerases and enhanced carotenoid accumulation in a unicellular chlorophyte. Proc Natl Acad Sci U S A. 1998;95(19):11482–11488. doi: 10.1073/pnas.95.19.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Kaneda K, Shimizu T, Hayakawa Y, Seto H, Kuzuyama T. Bacillus subtilis ypgA gene is fni, a nonessential gene encoding type 2 isopentenyl diphosphate isomerase. Biosci Biotechnol Biochem. 2004;68(1):132–137. doi: 10.1271/bbb.68.132. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Kuzuyama T, Watanabe H, Seto H (1998) A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-elythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci U S A 95(17):9879–9884. doi:10.1073/pnas.95.17.9879 [DOI] [PMC free article] [PubMed]
- Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5(1):93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta H, Paddon CJ, Eng D, Lenihan JR, Horning T, Anthony LC, Regentin R, Keasling JD, Renninger NS, Newman JD. High-level production of amorpha-4, 11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One. 2009;4(2):e4489. doi: 10.1371/journal.pone.0004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf MJ, Overkamp KM, Muilwijk B, Coulier L, Hankemeier T. Microbial metabolomics: toward a platform with full metabolome coverage. Anal Biochem. 2007;370(1):17–25. doi: 10.1016/j.ab.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Villas-Bôas SG, Bruheim P. Cold glycerol–saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal Biochem. 2007;370(1):87–97. doi: 10.1016/j.ab.2007.06.028. [DOI] [PubMed] [Google Scholar]
- Wagner WP, Helmig D, Fall R. Isoprene biosynthesis in Bacillus subtilis via the methylerythritol phosphate pathway. J Nat Prod. 2000;63(1):37–40. doi: 10.1021/np990286p. [DOI] [PubMed] [Google Scholar]
- Wagner WP, Nemecek-Marshall M, Fall R. Three distinct phases of isoprene formation during growth and sporulation of Bacillus subtilis. J Bacteriol. 1999;181(15):4700–4703. doi: 10.1128/jb.181.15.4700-4703.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Oldfield E. Bioorganometallic chemistry with IspG and IspH: structure, function, and inhibition of the Fe4S4 proteins involved in isoprenoid biosynthesis. Angew Chem Int Ed. 2014;53(17):4294–4310. doi: 10.1002/anie.201306712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang K, Liu Y-L, No J-H, Li J, Nilges MJ, Oldfield E. Bioorganometallic mechanism of action, and inhibition, of IspH. Proc Natl Acad Sci U S A. 2010;107(10):4522–4527. doi: 10.1073/pnas.0911087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PJ, Pitera DJ, Lenihan JR, Eng D, Woolard FX, Regentin R, Horning T, Tsuruta H, Melis DJ, Owens A. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc Natl Acad Sci. 2012;109(3):E111–E118. doi: 10.1073/pnas.1110740109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westers L, Westers H, Quax WJ. Bacillus subtilis as cell factory for pharmaceutical proteins: a biotechnological approach to optimize the host organism. Bioinorg Chem Appl. 2004;1694(1):299–310. doi: 10.1016/j.bbamcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- Whited GM, Feher FJ, Benko DA, Cervin MA, Chotani GK, McAuliffe JC, LaDuca RJ, Ben-Shoshan EA, Sanford KJ. Technology update: development of a gas-phase bioprocess for isoprene-monomer production using metabolic pathway engineering. Ind Biotechnol. 2010;6(3):152–163. [Google Scholar]
- Widner B, Behr R, Von Dollen S, Tang M, Heu T, Sloma A, Sternberg D, DeAngelis PL, Weigel PH, Brown S. Hyaluronic acid production in Bacillus subtilis. Appl Environ Microbiol. 2005;71(7):3747–3752. doi: 10.1128/AEM.71.7.3747-3752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RM, Danishefsky SJ. Applications of total synthesis to problems in neurodegeneration: fascinating chemistry along the way. Acc Chem Res. 2006;39(8):539–549. doi: 10.1021/ar068018n. [DOI] [PubMed] [Google Scholar]
- Withers ST, Keasling JD. Biosynthesis and engineering of isoprenoid small molecules. Appl Microbiol Biotechnol. 2007;73(5):980–990. doi: 10.1007/s00253-006-0593-1. [DOI] [PubMed] [Google Scholar]
- Xue D, Abdallah II, de Haan IE, Sibbald MJ, Quax WJ. Enhanced C30 carotenoid production in Bacillus subtilis by systematic overexpression of MEP pathway genes. Appl Microbiol Biotechnol. 2015;99(14):5907–5915. doi: 10.1007/s00253-015-6531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J, Ahring BK (2011) Enhancing isoprene production by genetic modification of the 1-deoxy-D-xylulose-5-phosphate pathway in Bacillus subtilis. Appl Environ Microbiol 77(7):2399–2405. doi:10.1128/aem.02341-10 [DOI] [PMC free article] [PubMed]
- Yoshida K, Ueda S, Maeda I. Carotenoid production in Bacillus subtilis achieved by metabolic engineering. Biotechnol Lett. 2009;31(11):1789–1793. doi: 10.1007/s10529-009-0082-6. [DOI] [PubMed] [Google Scholar]
- Yuan LZ, Rouviere PE, Larossa RA, Suh W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab Eng. 2006;8(1):79–90. doi: 10.1016/j.ymben.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Zhao L, Chang WC, Xiao Y, Liu HW, Liu P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu Rev Biochem. 2013;82:497–530. doi: 10.1146/annurev-biochem-052010-100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Qin B, Li Y, Sun Y, Su S, Xian M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl Microbiol Biotechnol. 2011;90(6):1915–1922. doi: 10.1007/s00253-011-3199-1. [DOI] [PubMed] [Google Scholar]
- Zhou K, Zou R, Stephanopoulos G, Too H-P. Metabolite profiling identified methylerythritol cyclodiphosphate efflux as a limiting step in microbial isoprenoid production. PLoS One. 2012;7(11):e47513. doi: 10.1371/journal.pone.0047513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Zou R, Zhang C, Stephanopoulos G, Too HP. Optimization of amorphadiene synthesis in Bacillus subtilis via transcriptional, translational, and media modulation. Biotechnol Bioeng. 2013;110(9):2556–2561. doi: 10.1002/bit.24900. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Nambou K, Wei L, Cao J, Imanaka T, Hua Q. Lycopene production in recombinant strains of Escherichia coli is improved by knockout of the central carbon metabolism gene coding for glucose-6-phosphate dehydrogenase. Biotechnol Lett. 2013;35(12):2137–2145. doi: 10.1007/s10529-013-1317-0. [DOI] [PubMed] [Google Scholar]