Abstract
The extracellular matrix (ECM) is a highly dynamic structure that not only provides a physical framework for cells within connective tissues, but also imparts instructive signals for development, tissue homeostasis, and basic cell functions through its composition and ability to exert mechanical forces. The ECM of tissues is composed of, in addition to proteoglycans and hyaluronic acid, a number of proteins, most of which are generated after alternative splicing of their pre-mRNA. However, the precise function of these protein isoforms is still obscure in most cases. Fibronectin (FN), one of the main components of the ECM, is also one of the best-known examples of a family of proteins generated by alternative splicing, having at least 20 different isoforms in humans. Over the last number of years, considerable progress on elucidating the functions of the alternatively spliced FN isoforms has been achieved with the essential development of key engineered mouse strains. Here we summarise the phenotypes of the mouse strains having targeted mutations in the FN gene, which may lead to novel insights linking function of alternatively spliced isoforms of fibronectin to human pathologies.
Keywords: pulmonary fibrosis, arterial thrombosis, alternative splicing, fibronectin isoforms, atherosclerosis, mouse models, transgenic mice
Introduction
The complexity of higher eukaryotes is achieved with a finite number of genes, which generate protein diversity by a number of mechanisms. Among them, the most prominent one is alternative splicing of the pre-mRNA. It is estimated that over 60% of genes undergo alternative splicing [1]. In most cases, it occurs in a tissue-specific and developmentally regulated manner and is also regulated by different stimuli. However, in the majority of cases the in vivo functions of protein isoforms generated by alternative splicing remain not well defined.
Fibronectin (FN) is an excellent model in which to study gene function regulated by alternative splicing. FN is a multifunctional glycoprotein found in plasma and in the extracellular matrix (ECM) of tissues. It is expressed by multiple cell types and plays a key role in cell adhesive and migratory behavior. Indeed, fundamental processes such as embryogenesis, hemostasis, wound healing and maintenance of tissue integrity are all processes that depend on interactions between cells and the ECM [2]. The critical importance of FN in vivo was conclusively demonstrated by the lethal effect of the murine FN null mutation: mice lacking FN died from severe defects in embryonic development [3] (Table 1). In humans, clinical significance of FN is evidenced by recent data demonstrating that certain types of glomerulopathy result from mutations in the FN gene [4].
Table 1.
| Animal Model: | Main Phenotype: | Ref. |
|---|---|---|
| FN null (FN promoter & 1st exon deletion) |
|
[3] [131] [132] [93] |
| FN null EDB (EDB-cDNA fusion) |
|
[121] |
| pFN KO (conditional “floxed” FN gene) |
|
[82] [92] [133] [134] [135] |
| EDB null (deletion of EDB exon) |
|
[79] [88] [93] |
| EDA null/EDA knock in (deletion of floxed EDA exon/optimization of EDA splicing sites) |
|
[78] [11] [136] [112] [111] [120] [97] [86] |
| EDA null (deletion of EDA exon) |
|
[80] [88] [93] |
| EDA&EDB KO Deletion of EDA and EDB exon from the same allele) |
|
[81] |
| RGD (mutation of RGD to RGE) |
|
[62] |
Description of the main phenotypes observed in the different mouse models having targeted mutations in the FN gene.
Although FN was one of the first genes reported to undergo alternative splicing [5–7] and is one of the best studied models of alternative splicing [8–31], the in vivo relevance of alternatively spliced isoforms were poorly understood prior to development of mouse models having targeted mutations of the FN gene. Conclusive evidence for the role of FN in some of the processes mentioned previously was obtained taking advantage of the gene targeting methodology, which remains the best approach to understand the in vivo function of specific gene products. Here we will discuss the recent advances in our understanding of the function of FN isoforms, largely derived from the use of mouse models having targeted mutations of the FN gene.
Fibronectin protein structure
FN is present only among vertebrates and its appearance in evolution correlates with the emergence of organisms with endothelial cell-lined vasculature [32, 33]. The functional FN molecule consists of two similar or identical subunits of 220–250 kDa that are held together by two disulfide-bonds near their carboxyl-termini forming a dimer (Figure 1A). Each monomer is comprised of a combination of three different types of homologous repeating domains, termed Type I, II, and III [2]. The fifteen Type III modules constitute the largest part of the FN polypeptide and are clustered in the central part of the protein (Figure 1A). Individual FN Type III repeats have a high degree of structural homology, despite displaying only 20–40% identity in amino acid sequence. They are composed of ~ 90 amino acids organized in seven anti-parallel β strands with exposed loops between these strands and without disulfide bonds [34] (Figure 1B and C). Domains may undergo structural modifications exposing cryptic sites induced by mechanical forces producing “stretching” of the FN molecule, by proteolysis [35, 36], or by incorporation of one (or both) of the alternatively spliced Type III domains (Extra Domains A and B, denominated EDA, EIIIA or EDI and EDB, EIIIB or EDII, respectively) [37].
Figure 1. Fibronectin primary structure.
The scheme shows a representation of a fibronectin dimer and its interactions (Panel A). The different types of homologies (twelve Type I, two Type II and fifteen Type III) are represented. Numbering of Type III homologies excludes EDA and EDB domains. Type I, II and III domains are constituted of 40, 60 and 90 aa, respectively. Constitutive (RGD) and alternatively spliced (LDV), synergy (PHSRN) and EDA (EDGIHEL) cell-binding sites are indicated, together with their integrin receptor partners. EDA and EDB splicing is similar in all species (either total inclusion or exclusion), while that of the IIICS region is species-specific (five variants in humans, three in rodents, and two in chickens). Type III homologies are organized in seven anti-parallel β strands (Spatial and planar representations are shown in Panels B and C, respectively).
Broadly, two variations of FN exist: plasma FN (pFN), a dimeric and soluble form secreted by hepatocytes directly into the circulation, lacking the alternatively spliced EDA and EDB sequences, and cellular FN (cFN), found in the ECM of tissues as a multimeric form assembled into fibrils which contain variable proportions of the EDA and EDB domains. FN fibrils are prominent in loose connective tissue, granulation tissue, embryonic basement membranes and on the surface of numerous cell types in culture [2, 38].
FN gene structure and alternative splicing
FN is produced after transcription of a single gene, composed of 47 exons that spans over 90 Kbp in the genome. Type III domains of FN are encoded by two exons with the exceptions of the EDA and EDB, and the 9th Type III domain, each of which are encoded by a single exon. Multiple FN mRNAs, and consequently multiple protein isoforms, can arise due to alternative splicing within a single pre-mRNA (Figure 1A). The EDA and EDB exons can be included or excluded from FN mRNA [5, 7, 17, 27], while the Type III Connecting Segment (IIICS) element (also termed the Variable, or V, region) undergoes a more complicated splicing pattern: it can be completely included (V120) or excluded (V0), or partially included, according to the species (Figure 1). Adding to the complexity, in zebrafish (a more distant species in evolution), two FN genes are present (termed FN1a and FN1b) [39, 40]. FN1a undergoes no alternative splicing (and thus includes EDA, EDB, and the V region constitutively), while the EDB and V regions are alternatively spliced in FN1b [39]. Interestingly, the complexity of V-region alternative splicing and the total number of FN isoforms seems to correlate with the evolutionary scale. In fact, only one V-region variant is found in the FN1a gene of zebrafish and two in the FN1b gene (for a total of five FN isoforms, from two independent genes) [39], whereas two V variants are present in frogs [41] and chickens (producing up to six FN isoforms) [25], three in rats and mice (with twelve FN isoforms) [7], and five in humans (generating up to twenty FN isoforms) [42].
Inclusion of the alternatively spliced regions is elevated during embryonic development [2] and decreases substantially after birth and with aging [2, 11, 26, 43]. However, the “embryonic” splicing pattern is temporally re-established in adult life in certain circumstances such as tissue repair, tissue fibrosis and angiogenesis. An excellent example is skin wound healing, where the inclusion of the EDA and EDB domains is increased in the cells at the base of the wound [16]. Similarly, both exons are up-regulated in regenerating liver, but with different kinetics [44–46]. The re-appearance of EDA cFN has been also observed in lung fibrosis, prior to the appearance of collagen. These tissue repair situations are also characterized by the differentiation and activation of myofibroblasts. These processes require TGFβ which promotes EDA exon incorporation into the mature pre-mRNA [47, 48] through an as-yet unknown mechanism.
Alternative splicing of EDA and EDB is modulated by members of the family of splicing regulators denoted SR proteins (for Serine- and Arginine-rich proteins) through distinctive mechanisms (Figure 2). In the case of EDA, regulatory sequences are located within the exon itself; in the case of EDB, distinct regulatory sequences are located within the exon and within the downstream intron.
Figure 2. Mechanisms of EDA and EDB alternative splicing.
Panel A: EDA exon splicing is positively regulated by the splicing factor SF2/ASF, which recognizes the exonic splicing enhancer (ESE) displayed in a loop region of stem-loop structure of the FN pre-mRNA. Skipping of the EDA exon is favored by hnRNP A1. Panel B: The splicing factor SRp40 promotes EDB exon inclusion by binding to a purine-rich sequence (ESE) within the EDB exon itself, or by recognizing the intronic splicing enhancers (ISE) located in the adjacent downstream intron. The presence of weak splice sites favors skipping of the EDA and EDB exons.
The EDA exon is one of the first reported examples where regulatory sequences are located within the exon itself [49]. It contains a purine rich region recognized by SR proteins, notably SF2/ASF, enhancing exon recognition and subsequent inclusion into the mature mRNA (Figure 2A) [9, 10, 12, 22]. As additional steps of regulation and complexity, the activity of SR proteins is modulated in response to extracellular stimuli, by SR protein kinases [50, 51], and by PKB/Akt [52]. The efficiency of exon recognition is affected by the promoter type and velocity of the RNA polymerase II that transcribes the gene [12, 13, 53, 54].
The FN pre-mRNA is characterized by a particular secondary structure in the region of the EDA exon, which assures proper display of the exon splicing enhancer (ESE) region in a loop region of a stem-loop structure [23]. This particular secondary structure of the mRNA is stabilized by a nearby sequence (ESS, exonic splicing silencer). On the contrary, the recognition of the EDB exon depends on the presence of TGCATG repeats in the downstream intron (ISE, intronic splicing enhancers) [18, 19]. The recognition of these elements by SRp40 enhances EDB inclusion into the mRNA (Figure 2B) [55]. However, in regenerating liver, a second mechanism of EDB regulation has been identified which is functionally similar to EDA: the increased inclusion of EDB is mediated by SRp40 recognition of a specific purine rich sequence present within the EDB exon [46].
Less attention has been paid to the mechanisms regulating alternative splicing of the IIICS region. One regulator of alternative splicing, termed mammalian homolog of suppressor-of-white-apricot (SWAP), influences IIICS splicing in a way that favors complete skipping of the IIICS region, generating the IIICS-0 form; in contrast, the SF2/ASF splicing factor stimulates inclusion of the complete IIICS region [56]. However, the sequences involved in the regulation of the IIICS region remain to be determined.
Interactions of FN with cellular receptors
FN interacts with cellular receptors known as integrins through specific sites of the protein. Integrins are cell-surface heterodimeric transmembrane receptors consisting of an α and a β subunit that link the ECM with the intracellular actin cytoskeleton and regulate specific signal transduction cascades. Although a large number of integrins can bind FN [57], the classic fibronectin receptor is the α5β1 integrin. This integrin recognizes the well known RGD cell-binding site located in a mobile and exposed loop region [34] in the 10th Type III repeat, along with a synergy sequence (PHSRN) located in the adjacent 9th Type III module [58–60] (See Figure 1A). The RGD sequence is the prototypic integrin recognition sequence, found in other ECM proteins such as tenascin, fibrinogen, thrombospondin, vitronectin and von Willebrand factor (vWF) [61]. The functional importance of the RGD sequence has been confirmed recently through experiments demonstrating that knock-in mice with an RGD to RGE mutation in the FN gene die at embryonic day 10 (E10) (Table 1) resembling the phenotype of α5 integrin null animals [62]. However, FN from these animals can be assembled into fibrils in vivo and in vitro, suggesting that RGD is dispensable for this function.
In addition to its cell binding properties, FN also binds other glycoproteins, including other ECM molecules, as well as components of the complement and coagulation systems [2, 38]. Notably, FN binds quite strongly to fibrin and fibrinogen shortly after tissue injury, and along with activated platelets makes up the bulk of the provisional hemostatic clot. The FN-fibrin meshwork undergoes covalent cross-linking to stabilize the clot and to allow for reparative cell migration. Moreover, within the ECM, FN can cross-link collagens, heparan sulfate proteoglycans, and itself to provide a stable matrix platform on and in which cells reside.
Other cell-binding sites are present in alternatively spliced regions of FN, conferring the capacity to regulate cell-binding ability and affinity by alternative splicing. In vitro experiments have determined that the EDGIHEL sequence present in the EDA domain (Figure 3) is recognized by the α4β1 and α9β1 integrins [63–66]. A second cell-binding region is present in the IIICS segment, which contains the LDV and REDV sequences, recognized by the leukocyte α4β1 and α4β7 integrins [67]. In contrast, the cellular receptor(s) for the EDB domain remain largely unknown.
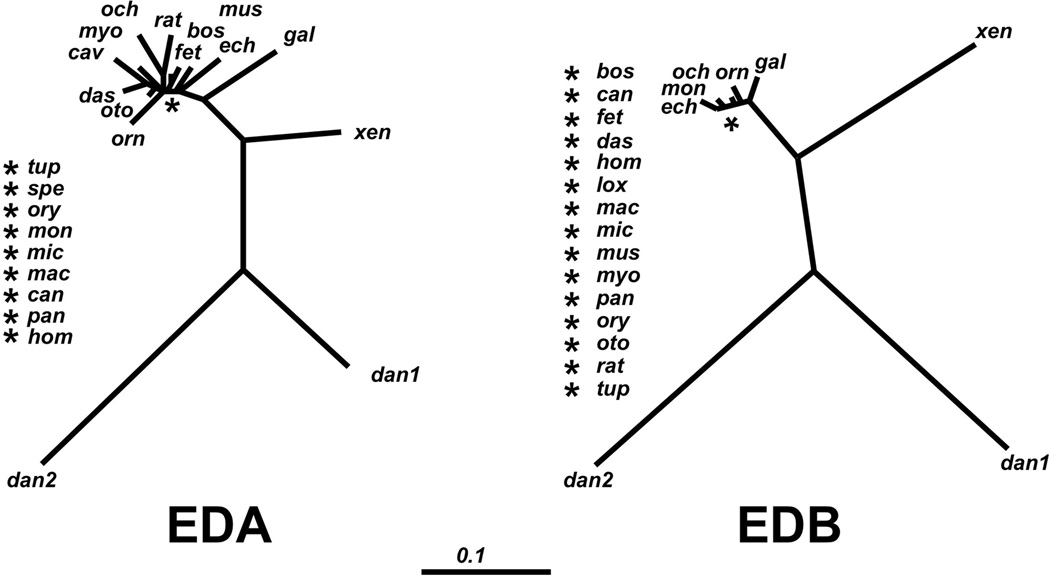
Figure 3. Unrooted phylogenetic diagram showing the relationships of the EDA and EDB domains among vertebrates.
The tree was constructed by comparing the amino acid sequence of the EDA and EDB domains with the ClustalW2 software (http://www.ch.embnet.org/software/ClustalW.html) by the neighbour-joining method [137]. The dnd file generated was plotted using the TreeViewPPC package. The abbreviations are described in Legend to Figures 3 and 4.
Both the EDA and the EDB exons show a very high degree of homology among vertebrates. Indeed, they have 53.3% and 71.4% of amino acid sequence identity, and 87.8% and 94.5% similarity for EDA and EDB, respectively, among 22 vertebrate species ranging from humans to frogs (see Supplementary Figures 1 and Supplementary 2), in contrast to the lower conservation observed with the only other single-exon Type III domain, the 9th Type III repeat (41.1% identity and 75.5% similarity). Amino acid sequence identity decreases to 30% and 38.5% (and 63.3% and 79.1% similarity) for the EDA and EDB, respectively, if we consider the two FN genes of zebrafish. The evolutionary distance of the EDA and EDB among vertebrates is schematically shown in Figure 3.
The high level of conservation of the EDA and EDB sequences during evolution and of their tightly regulated patterns of splicing, in contrast with their low homology within a species (only 28% homology between human EDA and EDB amino acid sequences), strongly suggests a conserved but distinct role of these domains.
Functions of the EDA domain
Numerous functions have been ascribed to the EDA domain, including cell adhesion [68, 69], wound healing [16, 70], matrix assembly [71], dimer formation [72], protein secretion [73], cytokine-dependent matrix metalloproteinase expression [74], cell differentiation [45, 75], tissue injury and inflammation [76, 77], cell cycle progression and mitogenic signal transduction [69]. However, most of these studies were done using in vitro cell culture systems. Thus, the development of genetically modified animal models was a requisite to specifically address these issues in vivo. The importance of correct EDA splicing is strongly supported by the observation that mice either lacking or constitutively expressing the EDA exon displayed a substantially reduced lifespan [78] (Table 1).
Elevated levels of EDA cFN are found in plasma and affected tissues of patients with certain disorders such as psoriasis, rheumatoid arthritis, diabetes and cancer, although the functional role of EDA cFN in these disease states is still obscure. Embryonic development is not possible without FN [3], but mice bearing single deletions of EDB or EDA are born normally [78–80]. In contrast, simultaneous deletion of both the EDA and EDB exons in the FN gene leads to embryonic lethality (albeit with incomplete penetrance) at E10.5 displaying severe cardiovascular defects (Table 1) (see below and [81]).
EDA cFN domain has an essential role in lung fibrosis
Fibronectin is a moderately abundant protein in blood (approximately 300–400 µg/ml and 580 µg/ml for humans and mice, respectively). Following tissue injury, pFN is extravasated from injured vessels and forms, together with platelets and fibrin, the hemostatic plug. This FN-rich clot serves as a provisional matrix to support cell migration during the wound closure process. However, mice lacking pFN undergo normal healing of cutaneous wounds [82] suggesting that cFN produced locally a the site of injury is sufficient to guide a normal healing process. Indeed, the importance of the EDA domain in cutaneous healing in vivo was shown in mice lacking the EDA domain (EDA−/−), which demonstrated abnormal healing, ulceration and inflammation at the sites of wounding [78].
An important functional role for the EDA domain in the in vitro differentiation of fibroblasts into myofibroblasts has been suggested [45, 75]. Myofibroblast differentiation and ultimate removal, when dysregulated in pathological situations, may lead to tissue fibrosis. One such threatening form of fibrosis in the lung is idiopathic pulmonary fibrosis (IPF). IPF is largely untreatable and patients ultimately die from unrelenting ECM deposition in the lung resulting in progressive respiratory failure [83, 84]. In IPF, EDA cFN is deposited before collagens in regions of active fibrosis [85], which correlates with increased expression of markers of fibroblast activation (α-smooth muscle actin [α-SMA]).
The link between EDA cFN and myofibroblast differentiation in vivo has been recently demonstrated using the well-described intratracheal bleomycin model of lung fibrosis. In EDA−/− mice receiving intratracheal bleomycin, lung fibrosis was completely prevented, suggesting that EDA cFN is necessary for the development of pulmonary fibrosis [86]. Failure to develop lung fibrosis in EDA−/− mice correlated with diminished activation of latent TGF-β and decreased lung fibroblast responsiveness to active TGF-β in vitro (Figure 4 and Table 1). Notably, EDA−/− lung fibroblasts could undergo TGF-β-induced myofibroblast differentiation, but only when plated on a matrix containing EDA cFN (Figure 4) [86]. These findings provide important new clues to the mechanisms of tissue fibrosis. The observation that EDA cFN is important for latent TGF-β activation and myofibroblast differentiation supports the previously recognized crucial role of TGF-β and the ECM in lung fibrosis. Further, it implicates EDA cFN in the pathogenesis of lung fibrosis. Of note, when the same EDA engineered mice were used in an allergen-induced chronic asthma model, we again observed that EDA cFN is critical for myofibroblast differentiation and subsequent airway fibrosis [87].
Figure 4. Role of EDA cFN in lung fibrosis.
Fibroblast differentiation into myofibroblasts occurs only on the presence of EDA cFN that could be either produced and secreted by the fibroblast (EDAwt/wt fibroblast, Panel A) or already present in the ECM, since EDA−/− fibroblasts differentiate into myofibroblasts only when plated on a EDA cFN (Panels A and B). EDA cFN secreted by the cells or already present in the ECM activates latent TGF β (Panels A and B) and fibroblast differentiation proceeds, leading to lung fibrosis. In the absence of EDA cFN (produced either by the EDA−/− cells or by the ECM prepared from EDA−/− fibroblasts, Panel C), the absence of latent TGF β activation limits differentiation into myofibroblasts preventing lung fibrosis.
The role of the EDA or EDB domains in α-SMA expression appears to be less important for non-fibroblast cells in other experimental models such as vasculogenesis (Table 1). In fact, analysis of α−SMA in pericytes around blood vessels showed no differences between EDA-null, EDB-null and control mice, suggesting that neither EDA nor EDB are required for α−SMA expression in this model [88]. In addition, neovascularization in retinas, pancreatic tumors and transplanted melanomas were not affected by the absence of the EDA or EDB splice variants [88]. However, mice bearing a targeted deletion of both EDA and EDB exons display a reduced number of α−SMA positive cells immediately surrounding the dorsal aorta, perhaps caused by a delay in the recruitment or differentiation of these cells [81] (see below). Based on the above studies, it is clear that the EDA domain is important for lung myofibroblast differentiation during experimental and possibly human lung fibrosis and that the absence of EDA cFN protects against lung fibrosis.
The role of FN and FN isoforms in thrombosis
Plasma FN participates in blood coagulation and thrombosis as a ligand of platelet surface receptors, and cross-links to fibrin. Platelets store a mixture of cFN (synthesized by megakaryocyte precursors) and pFN (endocytosed from plasma) in their α−granules [82, 89], which is released and deposited on platelet surfaces only after activation by thrombin. pFN plays a role in thrombosis and hemostasis, an observation supported by numerous reports [see references in [90]]. However, conclusive in vivo evidence of its role derives from three important observations: First, mice lacking both fibrinogen and vWF, key molecules involved in platelet aggregation and thrombus formation, still formed thrombi following ferric-chloride arterial injury, thereby suggesting that FN could be a ligand supporting platelet aggregation [91]. Second, conditional pFN knockout mice formed thrombi that continuously shed platelets (or groups of platelets) in the same model of arterial injury, reducing their growth and delaying the occlusion time of the vessels [92] (Table 1). Finally, FN-null heterozygous mice (having reduced levels of pFN) also displayed a striking reduction in thrombus initiation and growth following ferric-chloride arterial injury, a defect rescued after adding pFN back into the bloodstream [93] (Table 1). Together, these results indicate that pFN increases the stability of adherent platelet aggregates that form in response to experimental vascular injury. Since there is a considerable range in pFN concentration in humans and increases in its levels have been associated with coronary artery disease [94, 95], these reports established an experimental link between pFN concentration and arterial disease in larger populations [96].
As might be expected, since pFN does not include the EDA or EDB domains, no differences in vascular thrombosis were observed in the EDA−/− and EDB−/− mouse strains [93]. However, somewhat surprisingly, the constitutive inclusion of EDA into the FN molecule in EDA+/+ mice conferred pro-thrombotic activity, adding an extra level of complexity to fibronectin-platelet interactions [97]. These animals, despite having only 20% of normal pFN levels, developed occlusive thrombi more quickly in ferric-chloride damaged arteries and were more susceptible to pulmonary emboli after infusion of collagen and epinephrine into the bloodstream (Table 1). In vitro, platelets from EDA+/+ mice covered collagen-coated surfaces to a greater degree than wild-type platelets [97].
It is unclear why circulating EDA cFN may be pro-thrombotic, although different lines of evidence point towards a major conformational change in the FN molecule when the alternatively spliced domains are included which could result in enhanced FN functions. Normally, pFN circulates in blood in a closed, non-active form with the RGD cell-binding loop unavailable for cell binding (Figure 5A). However, structural studies suggest that insertion of an extra domain may induce a conformational change that affects the exposure of the RGD site [98] or other epitopes [37], thereby enhancing the potential interaction between circulating FN with surface integrins on blood-borne effector cells [99–102] (Figure 5B). Since EDA cFN is more potent in promoting cell adhesion and spreading than FN lacking EDA [100], it is possible that circulating EDA cFN is able to serve as a nidus for platelet and fibrin(ogen) aggregation, resulting in enhanced thrombosis and vascular occlusion.
Figure 5. Schemes of the possible conformational changes of FN by the inclusion of the extra domains EDA and EDB.
In the “closed” or “compact” conformation of pFN the RGD loop and nearby synergy sites are not available for interaction by integrins (Panel A). The inclusion of the EDA domain in EDA cFN (Panel B) triggers a conformational change enhancing the exposition of the RGD loop and the synergy site, and the binding of integrin receptors (indicated). The model postulates that inclusion of the EDB domain may also enhance the exposition of the RGD loop and the synergy site (Panel C). Asterisks in pFN indicate the expected position of the EDB (left asterisks) and EDA (right asterisks) domains if inserted (Panel A).
The inclusion of the EDA domain may trigger a global conformational change in the FN molecule resulting in enhanced platelet integrin-FN interaction via αIIbβ3 or other RGD-binding integrins. This hypothesis is indirectly supported by the observation that specific EDA-binding integrins (the α9β1 and α4β1 integrins) are not present on platelets. The main platelet integrin receptor involved in thrombus formation is αIIbβ3, although αVβ3 and α5β1 may also bind the RGD sequence. Global conformational changes (which may also occur due to forces applied during the platelet activating process) result in enhanced RGD binding by integrins, and may indeed be responsible for pro-thrombotic properties of EDA cFN [35, 36, 103]. If true, then one would expect a similar finding in mice constitutively expressing EDB cFN (Figure 7C). This hypothesis has yet to be tested. However, the concept that EDA and EDB deficiency results in impaired RGD binding and FN function may account for the embryonic lethality of EDA−/−/EDB−/− double-mutant mice [81].
These observations confer pro-thrombotic activity to EDA cFN and suggest that increased plasma EDA cFN levels might be a risk factor for thrombosis. Indeed, elevated plasma EDA cFN levels are present in patients with diabetes [104], vascular injury [105], and rheumatoid vasculitis [106], all conditions which predispose to vessel thrombosis. Moreover, one recent study suggests that patients with acute stroke receiving tissue plasminogen activator (t-PA) for thrombolysis may be at increased risk for brain hemorrhage in the setting of elevated plasma cFN levels [107], indicating that circulating cFN may also predict worse outcomes for patients with vascular diseases.
Secretion of pFN from hepatocytes
pFN is synthesized by hepatocytes [108]. While the EDA and EDB exons are spliced out from the pre-mRNA in normal liver, a fraction of mRNA contains the IIICS region. Interestingly, soluble FN is secreted as a heterodimer in which one subunit must contain the IIICS segment and the other may not, as IIICS-0/IIICS+ and IIICS+/IIICS+ combinations [109]. Although IIICS-0 subunits are synthesized, IIICS-0 homodimers seem to be degraded along the secretory pathway [109]. Of particular note, IIICS-0 homodimers, much like circulating EDA cFN dimers, possess enhanced pro-thrombotic properties [110]. In vivo and in vitro, EDA+/EDA+ dimers produced by EDA+/+ hepatocytes are not efficiently secreted into either plasma or culture medium and appear to be selectively degraded, perhaps due to an unfolded or misfolded conformation [111]. Consequently, plasma FN concentrations in EDA+/+ mice are only 20–25% of that found in control animals [78, 111].
It is tempting to suggest that negative selection for the pro-thrombotic forms of pFN has occurred through evolution. As such, hepatocytes may have developed at least two mechanisms to avoid the secretion of EDA+ or IIICS-0/IIICS-0 FN isoforms into the bloodstream. The first one is the total exclusion of the EDA exon from the FN mRNA by the splicing machinery [6], and the second one is the apparent intracellular degradation of these pro-thrombotic isoforms [109, 111]. The pro-thrombotic properties of EDA+ and IIICS-0 FNs may help explain the splicing patterns of pFN and cFN. It will be interesting to determine whether the EDB+ FN isoform also possesses pro-thrombotic properties.
Mice lacking regulated splicing of the EDA exon have defects in behaviour and motor-coordination
Mice lacking regulated splicing of the EDA exon show no evident morphological alterations in the brain [78, 112]. Surprisingly, though, each mutant strain displayed different CNS-related defects, suggesting a different role for each FN isoform (Table 1). EDA+/+ mice displayed a reduction in horizontal exploratory activity, but not vertical exploratory activity in the open field test. Conversely, in the same test, EDA−/− mice showed a decrease in vertical exploratory activity but not in the horizontal one [112]. Moreover, only the EDA−/− mice showed significant impairment in the accelerating rotarod test, a test that requires enhanced motor coordination skills and gives an idea of impaired cerebellar function. Therefore, regulated splicing of the EDA exon appears to be essential for normal brain function and behaviour in mice, and perhaps in other species. Although the reasons are unclear, constitutive presence or absence of EDA cFN may affect fine-tuning during the migration of neurons, axons and dendrites in the central nervous system. This is consistent with the previously-reported role for FN in regeneration and migration of axons in the central nervous system [113, 114]. On the contrary, no behavioral alterations were observed in EDB−/− mice [79], suggesting perhaps a less critical role for this isoform in brain development and function.
Although further anatomical, behavioral and electrophysiological studies are necessary to better understand the neurological basis of the motor impairments described above, these behavioral alterations may represent a disadvantage for mice living under conditions where natural selection works. While purely speculative, this might account in part for the strikingly high degree of conservation of the EDA and EDB exons among species (see Supplementary Figures 1 and 2) and the conserved pattern of expression of these splice variants [2, 20, 99].
EDA cFN in atherosclerosis
Abundant evidence links FN and its isoforms to the development of atherosclerosis. Atherogenesis involves a series of events where strong interaction with ECM is required, such as recruitment of blood monocytes to the arterial intima, maturation to tissue macrophages, lipid accumulation leading to foam cell formation, and smooth muscle cell migration from the arterial wall [115]. Notably, lipid accumulation does not occur and foam cells do not form without stable monocyte interactions with tissues, suggesting the importance of specific ECM-dependent signaling [116]. Considerable amounts of FN are present in the normal arterial wall, and it is strictly devoid of the EDA and EDB domains. However, in atherosclerotic lesions and in experimentally induced thickening of the aorta, there is a marked increase in total and EDA cFN adjacent to smooth muscle cells [117, 118]. One possible role for EDA cFN in this context may be to activate toll-like receptor (TLR) 4 [77], thereby triggering nuclear translocation of nuclear factor (NF)-κB, a molecule central to inflammation and atherogenesis [119], However, the precise role of EDA cFN in the development and progression of atherosclerosis was unclear prior to elegant studies in EDA−/− mice crossed with the atherosclerosis-prone ApoE−/− mice [80] (Table 1).
In these studies, ApoE−/−/EDA−/− mice developed smaller atherosclerotic lesions in the aortic tree than ApoE−/− control animals. In an in vitro foam cell assay, macrophages derived from ApoE−/−/EDA−/− mice accumulated less modified cholesterol than control macrophages, suggesting a role for EDA cFN in plasma lipid metabolism and foam cell formation. However, a recent report questions whether it is the regulation of FN splicing, and not the splice variant produced, as being more important in atherosclerosis [120]. In this series of experiments, aged EDA−/− or EDA+/+ mice on a pure C57Bl/6 genetic background were fed an atherogenic diet (Table 1). As one might expect, EDA−/− mice developed smaller and fewer atherosclerotic lesions associated with decreased macrophage cholesterol uptake. Surprisingly, however, EDA+/+ mice also demonstrated a significant reduction both in lesion size and frequency, as well as macrophage cholesterol uptake [120]. The similar findings between EDA−/− and EDA+/+ mice support the possibility that a common mechanism related to the genetically introduced abrogation of EDA alternative splicing might affect some of the processes involved in plaque formation. Although the molecular mechanisms for these observations have not yet been defined, they highlight the critical role of alternative splicing regulation in disease pathogenesis.
Functional role of the EDB domain
Although more than 20 years have passed since the identification of the EDB exon [17], the biological function(s) of FN isoforms containing this domain remain unknown. One attempt to elucidate the role of the EDB exon in the early years of gene targeting produced, unfortunately, complete null mutations of the FN gene (due to the presence of the neomycin cassette in the flanking intron) showing a phenotype similar to that previously observed after the targeted mutation of the FN gene [121]. Subsequently, a second strain of EDB−/− mouse was generated [79]; these mice developed normally and were fertile. Interestingly, in this strain no significant phenotype was observed in vivo, even after analyzing a number of different models where the EDB was previously thought to participate, such as behaviour, angiogenesis, thrombosis, organogenesis, tumorigenesis, and healing of bone fractures [79, 88, 93]. However, a mild in vitro effect in matrix assembly and proliferation was observed in EDB−/− embryonic fibroblasts (MEF) [79] (Table 1). These cells grew slower and produced fibrils that were shorter and thinner than those deposited by control MEF. The putative role for EDB in FN matrix assembly agrees with a previous report showing that EDB cFN is incorporated more efficiently into the ECM [71]. In an analogous manner, EDA+/+ MEF grow at a faster rate than those prepared from EDA−/− or WT embryos (A. F. Muro, unpublished observations), supporting previous in vitro data that favor a role for the EDA domain in cell proliferation [69]. However, no in vivo correlate has been identified in either EDB−/−, EDA−/−, or EDA+/+ animals [78, 79] suggesting that other influences on cell proliferation may compensate for a minor modulatory role of EDA and EDB cFN.
Despite the absence of an obvious phenotype in the EDB−/− mice, the extremely high degree of amino acid sequence conservation among species (~95% among 22 different vertebrates, see Figure 4) highly suggests an important function for this segment that may become evident under other pathophysiologic conditions or in response to external stress. Indeed, a similar situation occurs in tenascin-C knockout mice. In these animals, lack of tenascin-C results in no abnormalities in development, life span, or fertility [122, 123]. However, well-defined phenotypes were observed using specific models, such as corneal wounding, behavioural tests [122–125], and more acute examination of brain neurochemistry and neuromuscular junctions [126–128], to name but a few. Thus, further investigation into the role of EDB cFN in pathophysiologic or stressed models is necessary to better identify potential roles for this splice isoform.
EDA and EDB combined (double) knockout mice
EDA cFN and EDB cFN are highly expressed in the presence of tissue and vascular remodeling. They are found around embryonic vessels but are absent in normal arteries and veins of adult humans or mice [16, 129]. The domains reappear during pathological tissue regeneration and angiogenesis, such as in the processes of liver regeneration, wound healing and tumor development.
With the aim of determining in vivo EDA and EDB functions, a mouse strain with the simultaneous deletion of both domains was generated recently [81]. Mice harboring the EDA−/−/EDB−/− double mutation were embryonic lethal with about 80% penetrance, in a mixed c129 and C57Bl/6 genetic background (Table 1). No defect in production or cell-surface association of FN was observed, suggesting that lack of EDA and EDB together accounted for the defect in viability. Most of the affected embryos had a variety of cardiovascular defects, reminiscent of, but milder than, the phenotype observed in FN-null embryos [3]. Detailed analysis of EDA and EDB cFN expression showed their presence around the dorsal aorta in wild-type littermates at E9.5 and E10.5, associated with α−SMA expression, suggesting that EDA and EDB cFN may play a role in the development of aortic vessels during embryogenesis. EDA−/−/EDB−/− double mutants had a reduction in the number of α−SMA positive cells at E9.5 with an abnormal rounded morphology, but relatively equal numbers of α-SMA-expressing cells by E10.5, suggesting perhaps a defect in recruitment or in differentiation [81].
Instead of forming distinct vessel tubes, small vessels in EDA−/−/EDB−/− double mutant mice were not formed properly. The formation of endothelial cell sheets instead of tubes may be due to defective endothelial apical-basolateral polarity in the absence of EDA and EDB [81]. Polarized secretion of both FN and EDA cFN isoforms have been reported in endothelial cells and in airway epithelial cells, respectively [73, 130]. Therefore, vascular defects of the double-mutant mice could be related to a defect in endothelial cell polarity resulting in aberrant endothelial tube formation.
Since the absence of either the EDA or EDB exons did not produce any obvious defects in angiogenesis or in the levels of α−SMA positive cells around blood vessels in embryos or in regions of neo-angiogenesis [78–80, 88, 120], it is reasonable to hypothesize that the roles of the EDA and EDB domains may be different, although they may have some redundant function. Furthermore, the presence of only one domain is sufficient to attain normal blood vessel development, which is absent after the concerted deletion of both domains.
While the phenotypic ramifications of knocking out both the EDA and EDB domains of FN are not fully understood, we are able to appreciate some of the implications of this model. First, it is clear that FN, and more specifically the EDA and EDB domains, are critical for heart and blood vessel development. Indeed, the presence of at least one of the extra domains is required for normal cardiac and vascular development. Further, it appears that EDA and/or EDB cFN is necessary for the recruitment or differentiation of vessel-associated α−SMA cells. It is also possible that genetic background of the animal may influence the role of FN in development; FN-null mice created on pure C57Bl/6 mice had less severe defects in cardiac development, whereas the same mutation on a 129/Sv background resulted in severe defects in myocardial and endocardial morphogenesis [131, 132]. Further detailed in vivo and in vitro analysis of this strain will provide insights in the molecular mechanisms underlying the roles of the EDA and EDB domains in the process described above.
Concluding remarks
The ECM is clearly a dynamic structure that provides instructive signals to cells during homeostasis as well as during disease states. FN, a major component of the ECM, has been extensively studied in an attempt to determine the functions of the greater than 20 possible isoforms in humans. Through rigorous investigation, it is becoming clear that the alternatively spliced EDA and EDB domains possess distinct properties that allow for variation in function. Transgenic mice bearing targeted mutations in the FN gene display, in most cases, clear phenotypes; however, these phenotypes may be drastically different under conditions of stress or disease. The strikingly high conservation of both exons during vertebrate evolution and the conserved patterns of splicing in different organisms strongly support the idea of evolutionarily conserved functions. Moreover, it suggests that regulated splicing of the FN gene confers adaptive advantages in mice, and perhaps in humans.
More precise definition of the functions of the alternatively spliced FN isoforms will await further detailed in vivo and in vitro studies of the existing models and the generation of new engineered mouse strains having isoform-specific mutations in the FN gene. Those analyses will help us to better elucidate the molecular interactions and signaling pathways activated by alternatively-spliced isoforms of FN, the role of alternative splicing mechanisms in modulating cell and organism phenotype, and possibly the identity of currently unknown mechanisms associating FN functions and polymorphisms to pathological states in humans.
Supplementary Material
Acknowledgments
to Fabiola Porro and Emanuele Buratti for critical discussion. This work is supported, in part, by National Institutes of Health Grant HL083085, a T. Franklin Williams Geriatrics Research Development Award from the Association of Specialty Professors and the CHEST Foundation and from the Martin E. Galvin Fund and Quest for Breath Foundation (all to E.S.W.).
References
- 1.Kornblihtt AR. Promoter usage and alternative splicing. Curr Opin Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. Fibronectins. 1st edn. New York: Springer-Verlag; 1990. [Google Scholar]
- 3.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 4.Castelletti F, Donadelli R, Banterla F, Hildebrandt F, Zipfel PF, Bresin E, et al. Mutations in FN1 cause glomerulopathy with fibronectin deposits. Proc Natl Acad Sci U S A. 2008;105:2538–2543. doi: 10.1073/pnas.0707730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kornblihtt AR, Vibe-Pedersen K, Baralle FE. Human fibronectin: molecular cloning evidence for two mRNA species differing by an internal segment coding for a structural domain. Embo J. 1984;3:221–226. doi: 10.1002/j.1460-2075.1984.tb01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kornblihtt AR, Vibe-Pedersen K, Baralle FE. Human fibronectin: cell specific alternative mRNA splicing generates polypeptide chains differing in the number of internal repeats. Nucleic Acids Res. 1984;12:5853–5868. doi: 10.1093/nar/12.14.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzbauer JE, Tamkun JW, Lemischka IR, Hynes RO. Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell. 1983;35:421–431. doi: 10.1016/0092-8674(83)90175-7. [DOI] [PubMed] [Google Scholar]
- 8.Barone MV, Henchcliffe C, Baralle FE, Paolella G. Cell type specific trans-acting factors are involved in alternative splicing of human fibronectin pre-mRNA. Embo J. 1989;8:1079–1085. doi: 10.1002/j.1460-2075.1989.tb03476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buratti E, Muro AF, Giombi M, Gherbassi D, Iaconcig A, Baralle FE. RNA folding affects the recruitment of SR proteins by mouse and human polypurinic enhancer elements in the fibronectin EDA exon. Mol Cell Biol. 2004;24:1387–1400. doi: 10.1128/MCB.24.3.1387-1400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo CA, et al. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chauhan AK, Iaconcig A, Baralle FE, Muro AF. Alternative splicing of fibronectin: a mouse model demonstrates the identity of in vitro and in vivo systems and the processing autonomy of regulated exons in adult mice. Gene. 2004;324:55–63. doi: 10.1016/j.gene.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 12.Cramer P, Caceres JF, Cazalla D, Kadener S, Muro AF, Baralle FE, et al. Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 13.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.ffrench-Constant C, Hynes RO. Patterns of fibronectin gene expression and splicing during cell migration in chicken embryos. Development. 1988;104:369–382. doi: 10.1242/dev.104.3.369. [DOI] [PubMed] [Google Scholar]
- 15.ffrench-Constant C, Hynes RO. Alternative splicing of fibronectin is temporally and spatially regulated in the chicken embryo. Development. 1989;106:375–388. doi: 10.1242/dev.106.2.375. [DOI] [PubMed] [Google Scholar]
- 16.ffrench-Constant C, Van de Water L, Dvorak HF, Hynes RO. Reappearance of an embryonic pattern of fibronectin splicing during wound healing in the adult rat. J Cell Biol. 1989;109:903–914. doi: 10.1083/jcb.109.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutman A, Kornblihtt AR. Identification of a third region of cell-specific alternative splicing in human fibronectin mRNA. Proc Natl Acad Sci U S A. 1987;84:7179–7182. doi: 10.1073/pnas.84.20.7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huh GS, Hynes RO. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh GS, Hynes RO. Regulation of alternative pre-mRNA splicing by a novel repeated hexanucleotide element. Genes Dev. 1994;8:1561–1574. doi: 10.1101/gad.8.13.1561. [DOI] [PubMed] [Google Scholar]
- 20.Kornblihtt AR, Pesce CG, Alonso CR, Cramer P, Srebrow A, Werbajh S, et al. The fibronectin gene as a model for splicing and transcription studies. Faseb J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- 21.Kornblihtt AR, Umezawa K, Vibe-Pedersen K, Baralle FE. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. Embo J. 1985;4:1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavigueur A, La Branche H, Kornblihtt AR, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 23.Muro AF, Caputi M, Pariyarath R, Pagani F, Buratti E, Baralle FE. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol Cell Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muro AF, Iaconcig A, Baralle FE. Regulation of the fibronectin EDA exon alternative splicing. Cooperative role of the exonic enhancer element and the 5' splicing site. FEBS Lett. 1998;437:137–141. doi: 10.1016/s0014-5793(98)01201-0. [DOI] [PubMed] [Google Scholar]
- 25.Norton PA, Hynes RO. Alternative splicing of chicken fibronectin in embryos and in normal and transformed cells. Mol Cell Biol. 1987;7:4297–4307. doi: 10.1128/mcb.7.12.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagani F, Zagato L, Vergani C, Casari G, Sidoli A, Baralle FE. Tissue-specific splicing pattern of fibronectin messenger RNA precursor during development and aging in rat. J Cell Biol. 1991;113:1223–1229. doi: 10.1083/jcb.113.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarzbauer JE, Patel RS, Fonda D, Hynes RO. Multiple sites of alternative splicing of the rat fibronectin gene transcript. Embo J. 1987;6:2573–2580. doi: 10.1002/j.1460-2075.1987.tb02547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srebrow A, Blaustein M, Kornblihtt AR. Regulation of fibronectin alternative splicing by a basement membrane-like extracellular matrix. FEBS Lett. 2002;514:285–289. doi: 10.1016/s0014-5793(02)02382-7. [DOI] [PubMed] [Google Scholar]
- 29.Tamkun JW, Schwarzbauer JE, Hynes RO. A single rat fibronectin gene generates three different mRNAs by alternative splicing of a complex exon. Proc Natl Acad Sci U S A. 1984;81:5140–5144. doi: 10.1073/pnas.81.16.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vibe-Pedersen K, Kornblihtt AR, Baralle FE. Expression of a human alpha-globin/fibronectin gene hybrid generates two mRNAs by alternative splicing. Embo J. 1984;3:2511–2516. doi: 10.1002/j.1460-2075.1984.tb02165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vibe-Pedersen K, Magnusson S, Baralle FE. Donor and acceptor splice signals within an exon of the human fibronectin gene: a new type of differential splicing. FEBS Lett. 1986;207:287–291. doi: 10.1016/0014-5793(86)81506-x. [DOI] [PubMed] [Google Scholar]
- 32.Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- 33.Whittaker CA, Bergeron KF, Whittle J, Brandhorst BP, Burke RD, Hynes RO. The echinoderm adhesome. Dev Biol. 2006;300:252–266. doi: 10.1016/j.ydbio.2006.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Main AL, Harvey TS, Baron M, Boyd J, Campbell ID. The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- 35.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Forced unfolding of the fibronectin type III module reveals a tensile molecular recognition switch. Proc Natl Acad Sci U S A. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carnemolla B, Leprini A, Allemanni G, Saginati M, Zardi L. The inclusion of the type III repeat ED-B in the fibronectin molecule generates conformational modifications that unmask a cryptic sequence. J Biol Chem. 1992;267:24689–24692. [PubMed] [Google Scholar]
- 38.Mosher DF. Fibronectin. 1st edn. New York: Academic Press; 1989. [Google Scholar]
- 39.Sun L, Zou Z, Collodi P, Xu F, Xu X, Zhao Q. Identification and characterization of a second fibronectin gene in zebrafish. Matrix Biol. 2005;24:69–77. doi: 10.1016/j.matbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Q, Liu X, Collodi P. Identification and characterization of a novel fibronectin in zebrafish. Exp Cell Res. 2001;268:211–219. doi: 10.1006/excr.2001.5291. [DOI] [PubMed] [Google Scholar]
- 41.DeSimone DW, Norton PA, Hynes RO. Identification and characterization of alternatively spliced fibronectin mRNAs expressed in early Xenopus embryos. Dev Biol. 1992;149:357–369. doi: 10.1016/0012-1606(92)90291-n. [DOI] [PubMed] [Google Scholar]
- 42.Umezawa K, Kornblihtt AR, Baralle FE. Isolation and characterization of cDNA clones for human liver fibronectin. FEBS Lett. 1985;186:31–34. doi: 10.1016/0014-5793(85)81333-8. [DOI] [PubMed] [Google Scholar]
- 43.Magnuson VL, Young M, Schattenberg DG, Mancini MA, Chen DL, Steffensen B, et al. The alternative splicing of fibronectin pre-mRNA is altered during aging and in response to growth factors. J Biol Chem. 1991;266:14654–14662. [PubMed] [Google Scholar]
- 44.Caputi M, Melo CA, Baralle FE. Regulation of fibronectin expression in rat regenerating liver. Nucleic Acids Res. 1995;23:238–243. doi: 10.1093/nar/23.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jarnagin WR, Rockey DC, Koteliansky VE, Wang SS, Bissell DM. Expression of variant fibronectins in wound healing: cellular source and biological activity of the EIIIA segment in rat hepatic fibrogenesis. J Cell Biol. 1994;127:2037–2048. doi: 10.1083/jcb.127.6.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du K, Peng Y, Greenbaum LE, Haber BA, Taub R. HRS/SRp40-mediated inclusion of the fibronectin EIIIB exon, a possible cause of increased EIIIB expression in proliferating liver. Mol Cell Biol. 1997;17:4096–4104. doi: 10.1128/mcb.17.7.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balza E, Borsi L, Allemanni G, Zardi L. Transforming growth factor beta regulates the levels of different fibronectin isoforms in normal human cultured fibroblasts. FEBS Lett. 1988;228:42–44. doi: 10.1016/0014-5793(88)80580-5. [DOI] [PubMed] [Google Scholar]
- 48.Borsi L, Castellani P, Risso AM, Leprini A, Zardi L. Transforming growth factor-beta regulates the splicing pattern of fibronectin messenger RNA precursor. FEBS Lett. 1990;261:175–178. doi: 10.1016/0014-5793(90)80664-5. [DOI] [PubMed] [Google Scholar]
- 49.Mardon HJ, Sebastio G, Baralle FE. A role for exon sequences in alternative splicing of the human fibronectin gene. Nucleic Acids Res. 1987;15:7725–7733. doi: 10.1093/nar/15.19.7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y, Manley JL. A complex signaling pathway regulates SRp38 phosphorylation and pre-mRNA splicing in response to heat shock. Mol Cell. 2007;28:79–90. doi: 10.1016/j.molcel.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 51.Prasad J, Manley JL. Regulation and substrate specificity of the SR protein kinase Clk/Sty. Mol Cell Biol. 2003;23:4139–4149. doi: 10.1128/MCB.23.12.4139-4149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blaustein M, Pelisch F, Tanos T, Munoz MJ, Wengier D, Quadrana L, et al. Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT. Nat Struct Mol Biol. 2005;12:1037–1044. doi: 10.1038/nsmb1020. [DOI] [PubMed] [Google Scholar]
- 53.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 54.de la Mata M, Kornblihtt AR. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol. 2006;13:973–980. doi: 10.1038/nsmb1155. [DOI] [PubMed] [Google Scholar]
- 55.Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarkissian M, Winne A, Lafyatis R. The mammalian homolog of suppressor-of-white-apricot regulates alternative mRNA splicing of CD45 exon 4 and fibronectin IIICS. J Biol Chem. 1996;271:31106–31114. doi: 10.1074/jbc.271.49.31106. [DOI] [PubMed] [Google Scholar]
- 57.Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002;115:3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 58.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 59.Pytela R, Pierschbacher MD, Ruoslahti E. Identification and isolation of a 140 kd cell surface glycoprotein with properties expected of a fibronectin receptor. Cell. 1985;40:191–198. doi: 10.1016/0092-8674(85)90322-8. [DOI] [PubMed] [Google Scholar]
- 60.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 61.Plow EF, Haas TA, Zhang L, Loftus J, Smith JW. Ligand binding to integrins. J Biol Chem. 2000;275:21785–21788. doi: 10.1074/jbc.R000003200. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi S, Leiss M, Moser M, Ohashi T, Kitao T, Heckmann D, et al. The RGD motif in fibronectin is essential for development but dispensable for fibril assembly. J Cell Biol. 2007;178:167–178. doi: 10.1083/jcb.200703021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liao YF, Gotwals PJ, Koteliansky VE, Sheppard D, Van De Water L. The EIIIA segment of fibronectin is a ligand for integrins alpha 9beta 1 and alpha 4beta 1 providing a novel mechanism for regulating cell adhesion by alternative splicing. J Biol Chem. 2002;277:14467–14474. doi: 10.1074/jbc.M201100200. [DOI] [PubMed] [Google Scholar]
- 64.Liao YF, Wieder KG, Classen JM, Van De Water L. Identification of two amino acids within the EIIIA (ED-A) segment of fibronectin constituting the epitope for two function-blocking monoclonal antibodies. J Biol Chem. 1999;274:17876–17884. doi: 10.1074/jbc.274.25.17876. [DOI] [PubMed] [Google Scholar]
- 65.Shinde AV, Bystroff C, Wang C, Vogelezang MG, Vincent PA, Hynes RO, et al. Identification of the Peptide Sequences within the EIIIA (EDA) Segment of Fibronectin That Mediate Integrin {alpha}9{beta}1-dependent Cellular Activities. J Biol Chem. 2008;283:2858–2870. doi: 10.1074/jbc.M708306200. [DOI] [PubMed] [Google Scholar]
- 66.Yamada KM. Fibronectins: structure, functions and receptors. Curr Opin Cell Biol. 1989;1:956–963. doi: 10.1016/0955-0674(89)90065-3. [DOI] [PubMed] [Google Scholar]
- 67.Hynes RO. Integrins: a family of cell surface receptors. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 68.Xia P, Culp LA. Adhesion activity in fibronectin's alternatively spliced domain EDa (EIIIA): complementarity to plasma fibronectin functions. Exp Cell Res. 1995;217:517–527. doi: 10.1006/excr.1995.1117. [DOI] [PubMed] [Google Scholar]
- 69.Manabe R, Oh-e N, Sekiguchi K. Alternatively spliced EDA segment regulates fibronectin-dependent cell cycle progression and mitogenic signal transduction. J Biol Chem. 1999;274:5919–5924. doi: 10.1074/jbc.274.9.5919. [DOI] [PubMed] [Google Scholar]
- 70.Clark RA, Winn HJ, Dvorak HF, Colvin RB. Fibronectin beneath reepithelializing epidermis in vivo: sources and significance. J Invest Dermatol. 1983;80(Suppl):26s–30s. [PubMed] [Google Scholar]
- 71.Guan JL, Trevithick JE, Hynes RO. Retroviral expression of alternatively spliced forms of rat fibronectin. J Cell Biol. 1990;110:833–847. doi: 10.1083/jcb.110.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peters JH, Sporn LA, Ginsberg MH, Wagner DD. Human endothelial cells synthesize, process, and secrete fibronectin molecules bearing an alternatively spliced type III homology (ED1) Blood. 1990;75:1801–1808. [PubMed] [Google Scholar]
- 73.Wang A, Cohen DS, Palmer E, Sheppard D. Polarized regulation of fibronectin secretion and alternative splicing by transforming growth factor. J Biol Chem. 1991;266:15598–15601. [PubMed] [Google Scholar]
- 74.Saito S, Yamaji N, Yasunaga K, Saito T, Matsumoto S, Katoh M, et al. The fibronectin extra domain A activates matrix metalloproteinase gene expression by an interleukin-1-dependent mechanism. J Biol Chem. 1999;274:30756–30763. doi: 10.1074/jbc.274.43.30756. [DOI] [PubMed] [Google Scholar]
- 75.Serini G, Bochaton-Piallat ML, Ropraz P, Geinoz A, Borsi L, Zardi L, et al. The fibronectin domain ED-A is crucial for myofibroblastic phenotype induction by transforming growth factor-beta1. J Cell Biol. 1998;142:873–881. doi: 10.1083/jcb.142.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Satoi S, Hiramatsu Y, Kitade H, Kwon AH, Matsui K, Miyashita K, et al. Different responses to surgical stress between extra domain A+ and plasma fibronectins. Clin Exp Pharmacol Physiol. 1999;26:225–229. doi: 10.1046/j.1440-1681.1999.03019.x. [DOI] [PubMed] [Google Scholar]
- 77.Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, Rose J, et al. The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem. 2001;276:10229–10233. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 78.Muro AF, Chauhan AK, Gajovic S, Iaconcig A, Porro F, Stanta G, et al. Regulated splicing of the fibronectin EDA exon is essential for proper skin wound healing and normal lifespan. J Cell Biol. 2003;162:149–160. doi: 10.1083/jcb.200212079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fukuda T, Yoshida N, Kataoka Y, Manabe R, Mizuno-Horikawa Y, Sato M, et al. Mice Lacking the EDB Segment of Fibronectin Develop Normally but Exhibit Reduced Cell Growth and Fibronectin Matrix Assembly in Vitro. Cancer Res. 2002;62:5603–5610. [PubMed] [Google Scholar]
- 80.Tan MH, Sun Z, Opitz SL, Schmidt TE, Peters JH, George EL. Deletion of the alternatively spliced fibronectin EIIIA domain in mice reduces atherosclerosis. Blood. 2004;104:11–18. doi: 10.1182/blood-2003-09-3363. [DOI] [PubMed] [Google Scholar]
- 81.Astrof S, Crowley D, Hynes RO. Multiple cardiovascular defects caused by the absence of alternatively spliced segments of fibronectin. Dev Biol. 2007;311:11–24. doi: 10.1016/j.ydbio.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakai T, Johnson KJ, Murozono M, Sakai K, Magnuson MA, Wieloch T, et al. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nat Med. 2001;7:324–330. doi: 10.1038/85471. [DOI] [PubMed] [Google Scholar]
- 83.Thannickal VJ, Toews GB, White ES, Lynch JP, 3rd, Martinez FJ. Mechanisms of pulmonary fibrosis. Annu Rev Med. 2004;55:395–417. doi: 10.1146/annurev.med.55.091902.103810. [DOI] [PubMed] [Google Scholar]
- 84.White ES, Lazar MH, Thannickal VJ. Pathogenetic mechanisms in usual interstitial pneumonia/idiopathic pulmonary fibrosis. J Pathol. 2003;201:343–354. doi: 10.1002/path.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuhn C, McDonald JA. The roles of the myofibroblast in idiopathic pulmonary fibrosis. Ultrastructural and immunohistochemical features of sites of active extracellular matrix synthesis. Am J Pathol. 1991;138:1257–1265. [PMC free article] [PubMed] [Google Scholar]
- 86.Muro AF, Moretti FA, Moore BB, Yan M, Atrasz RG, Wilke CA, et al. An essential role for fibronectin extra type III domain A in pulmonary fibrosis. Am J Respir Crit Care Med. 2008;177:638–645. doi: 10.1164/rccm.200708-1291OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kohan M, Muro AF, Bader R, Breuer R, Berkman N. EDA-Fibronectin Modulates Myofibroblast Differentiation and Promotes Airway Fibrosis in a Murine Model of Chronic Allergen-Induced Airway Remodeling. Am J Respir Crit Care Med. 2008;177:A496. [Google Scholar]
- 88.Astrof S, Crowley D, George EL, Fukuda T, Sekiguchi K, Hanahan D, et al. Direct test of potential roles of EIIIA and EIIIB alternatively spliced segments of fibronectin in physiological and tumor angiogenesis. Mol Cell Biol. 2004;24:8662–8670. doi: 10.1128/MCB.24.19.8662-8670.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paul JI, Schwarzbauer JE, Tamkun JW, Hynes RO. Cell-type-specific fibronectin subunits generated by alternative splicing. J Biol Chem. 1986;261:12258–12265. [PubMed] [Google Scholar]
- 90.Cho J, Mosher DF. Role of fibronectin assembly in platelet thrombus formation. J Thromb Haemost. 2006;4:1461–1469. doi: 10.1111/j.1538-7836.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- 91.Ni H, Denis CV, Subbarao S, Degen JL, Sato TN, Hynes RO, et al. Persistence of platelet thrombus formation in arterioles of mice lacking both von Willebrand factor and fibrinogen. J Clin Invest. 2000;106:385–392. doi: 10.1172/JCI9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ni H, Yuen PS, Papalia JM, Trevithick JE, Sakai T, Fassler R, et al. Plasma fibronectin promotes thrombus growth and stability in injured arterioles. Proc Natl Acad Sci U S A. 2003;100:2415–2419. doi: 10.1073/pnas.2628067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matuskova J, Chauhan AK, Cambien B, Astrof S, Dole VS, Piffath CL, et al. Decreased plasma fibronectin leads to delayed thrombus growth in injured arterioles. Arterioscler Thromb Vasc Biol. 2006;26:1391–1396. doi: 10.1161/01.ATV.0000216282.58291.c6. [DOI] [PubMed] [Google Scholar]
- 94.Orem C, Celik S, Orem A, Calapoglu M, Erdol C. Increased plasma fibronectin levels in patients with acute myocardial infarction complicated with left ventricular thrombus. Thromb Res. 2002;105:37–41. doi: 10.1016/s0049-3848(01)00414-5. [DOI] [PubMed] [Google Scholar]
- 95.Song KS, Kim HK, Shim W, Jee SH. Plasma fibronectin levels in ischemic heart disease. Atherosclerosis. 2001;154:449–453. doi: 10.1016/s0021-9150(00)00490-1. [DOI] [PubMed] [Google Scholar]
- 96.Mosher DF. Plasma fibronectin concentration: a risk factor for arterial thrombosis? Arterioscler Thromb Vasc Biol. 2006;26:1193–1195. doi: 10.1161/01.ATV.0000223342.15969.7a. [DOI] [PubMed] [Google Scholar]
- 97.Chauhan AK, Kisucka J, Cozzi MR, Walsh MT, Moretti FA, Battiston M, et al. Prothrombotic effects of fibronectin isoforms containing the EDA domain. Arterioscler Thromb Vasc Biol. 2008;28:296–301. doi: 10.1161/ATVBAHA.107.149146. [DOI] [PubMed] [Google Scholar]
- 98.Leahy DJ, Aukhil I, Erickson HP. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 99.ffrench-Constant C. Alternative splicing of fibronectin--many different proteins but few different functions. Exp Cell Res. 1995;221:261–271. doi: 10.1006/excr.1995.1374. [DOI] [PubMed] [Google Scholar]
- 100.Manabe R, Ohe N, Maeda T, Fukuda T, Sekiguchi K. Modulation of cell-adhesive activity of fibronectin by the alternatively spliced EDA segment. J Cell Biol. 1997;139:295–307. doi: 10.1083/jcb.139.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson KJ, Sage H, Briscoe G, Erickson HP. The compact conformation of fibronectin is determined by intramolecular ionic interactions. J Biol Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 102.Niimi T, Osawa M, Yamaji N, Yasunaga K, Sakashita H, Mase T, et al. NMR structure of human fibronectin EDA. J Biomol NMR. 2001;21:281–284. doi: 10.1023/a:1012947209393. [DOI] [PubMed] [Google Scholar]
- 103.Krammer A, Craig D, Thomas WE, Schulten K, Vogel V. A structural model for force regulated integrin binding to fibronectin's RGD-synergy site. Matrix Biol. 2002;21:139–147. doi: 10.1016/s0945-053x(01)00197-4. [DOI] [PubMed] [Google Scholar]
- 104.Kanters SD, Banga JD, Algra A, Frijns RC, Beutler JJ, Fijnheer R. Plasma levels of cellular fibronectin in diabetes. Diabetes Care. 2001;24:323–327. doi: 10.2337/diacare.24.2.323. [DOI] [PubMed] [Google Scholar]
- 105.Peters JH, Maunder RJ, Woolf AD, Cochrane CG, Ginsberg MH. Elevated plasma levels of ED1+ ("cellular") fibronectin in patients with vascular injury. J Lab Clin Med. 1989;113:586–597. [PubMed] [Google Scholar]
- 106.Voskuyl AE, Emeis JJ, Hazes JM, van Hogezand RA, Biemond I, Breedveld FC. Levels of circulating cellular fibronectin are increased in patients with rheumatoid vasculitis. Clin Exp Rheumatol. 1998;16:429–434. [PubMed] [Google Scholar]
- 107.Castellanos M, Sobrino T, Millan M, Garcia M, Arenillas J, Nombela F, et al. Serum cellular fibronectin and matrix metalloproteinase-9 as screening biomarkers for the prediction of parenchymal hematoma after thrombolytic therapy in acute ischemic stroke: a multicenter confirmatory study. Stroke. 2007;38:1855–1859. doi: 10.1161/STROKEAHA.106.481556. [DOI] [PubMed] [Google Scholar]
- 108.Tamkun JW, Hynes RO. Plasma fibronectin is synthesized and secreted by hepatocytes. J Biol Chem. 1983;258:4641–4647. [PubMed] [Google Scholar]
- 109.Schwarzbauer JE, Spencer CS, Wilson CL. Selective secretion of alternatively spliced fibronectin variants. J Cell Biol. 1989;109:3445–3453. doi: 10.1083/jcb.109.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wilson CL, Schwarzbauer JE. The alternatively spliced V region contributes to the differential incorporation of plasma and cellular fibronectins into fibrin clots. J Cell Biol. 1992;119:923–933. doi: 10.1083/jcb.119.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moretti FA, Chauhan AK, Iaconcig A, Porro F, Baralle FE, Muro AF. A major fraction of fibronectin present in the extracellular matrix of tissues is plasma-derived. J Biol Chem. 2007;282:28057–28062. doi: 10.1074/jbc.M611315200. [DOI] [PubMed] [Google Scholar]
- 112.Chauhan AK, Moretti FA, Iaconcig A, Baralle FE, Muro AF. Impaired motor coordination in mice lacking the EDA exon of the fibronectin gene. Behav Brain Res. 2005;161:31–38. doi: 10.1016/j.bbr.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 113.Tom VJ, Doller CM, Malouf AT, Silver J. Astrocyte-associated fibronectin is critical for axonal regeneration in adult white matter. J Neurosci. 2004;24:9282–9290. doi: 10.1523/JNEUROSCI.2120-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang Z, Suzuki R, Daniels SB, Brunquell CB, Sala CJ, Nishiyama A. NG2 glial cells provide a favorable substrate for growing axons. J Neurosci. 2006;26:3829–3839. doi: 10.1523/JNEUROSCI.4247-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 116.Wesley RB, 2nd, Meng X, Godin D, Galis ZS. Extracellular matrix modulates macrophage functions characteristic to atheroma: collagen type I enhances acquisition of resident macrophage traits by human peripheral blood monocytes in vitro. Arterioscler Thromb Vasc Biol. 1998;18:432–440. doi: 10.1161/01.atv.18.3.432. [DOI] [PubMed] [Google Scholar]
- 117.Glukhova MA, Frid MG, Shekhonin BV, Vasilevskaya TD, Grunwald J, Saginati M, et al. Expression of extra domain A fibronectin sequence in vascular smooth muscle cells is phenotype dependent. J Cell Biol. 1989;109:357–366. doi: 10.1083/jcb.109.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Magnusson MK, Mosher DF. Fibronectin: structure, assembly, and cardiovascular implications. Arterioscler Thromb Vasc Biol. 1998;18:1363–1370. doi: 10.1161/01.atv.18.9.1363. [DOI] [PubMed] [Google Scholar]
- 119.Xu XH, Shah PK, Faure E, Equils O, Thomas L, Fishbein MC, et al. Toll-like receptor-4 is expressed by macrophages in murine and human lipid-rich atherosclerotic plaques and upregulated by oxidized LDL. Circulation. 2001;104:3103–3108. doi: 10.1161/hc5001.100631. [DOI] [PubMed] [Google Scholar]
- 120.Babaev VR, Porro F, Linton MF, Fazio S, Baralle FE, Muro AF. Absence of regulated splicing of fibronectin EDA exon reduces atherosclerosis in mice. Atherosclerosis. 2008 doi: 10.1016/j.atherosclerosis.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Georges-Labouesse EN, George EL, Rayburn H, Hynes RO. Mesodermal development in mouse embryos mutant for fibronectin. Dev Dyn. 1996;207:145–156. doi: 10.1002/(SICI)1097-0177(199610)207:2<145::AID-AJA3>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 122.Saga Y, Yagi T, Ikawa Y, Sakakura T, Aizawa S. Mice develop normally without tenascin. Genes Dev. 1992;6:1821–1831. doi: 10.1101/gad.6.10.1821. [DOI] [PubMed] [Google Scholar]
- 123.Forsberg E, Hirsch E, Frohlich L, Meyer M, Ekblom P, Aszodi A, et al. Skin wounds and severed nerves heal normally in mice lacking tenascin-C. Proc Natl Acad Sci U S A. 1996;93:6594–6599. doi: 10.1073/pnas.93.13.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fukamauchi F, Mataga N, Wang YJ, Sato S, Youshiki A, Kusakabe M. Abnormal behavior and neurotransmissions of tenascin gene knockout mouse. Biochem Biophys Res Commun. 1996;221:151–156. doi: 10.1006/bbrc.1996.0561. [DOI] [PubMed] [Google Scholar]
- 125.Mackie EJ, Tucker RP. The tenascin-C knockout revisited. J Cell Sci. 1999;112(Pt 22):3847–3853. doi: 10.1242/jcs.112.22.3847. [DOI] [PubMed] [Google Scholar]
- 126.Fukamauchi F, Aihara O, Kusakabe M. The effects of central administration of neuropeptide Y on behavior of tenascin-gene knockout mice. Neuropeptides. 1998;32:461–464. doi: 10.1016/s0143-4179(98)90072-5. [DOI] [PubMed] [Google Scholar]
- 127.Fukamauchi F, Mataga N, Wang YJ, Sato S, Yoshiki A, Kusakabe M. Tyrosine hydroxylase activity and its mRNA level in dopaminergic neurons of tenascin gene knockout mouse. Biochem Biophys Res Commun. 1997;231:356–359. doi: 10.1006/bbrc.1997.6101. [DOI] [PubMed] [Google Scholar]
- 128.Cifuentes-Diaz C, Velasco E, Meunier FA, Goudou D, Belkadi L, Faille L, et al. The peripheral nerve and the neuromuscular junction are affected in the tenascin-C-deficient mouse. Cell Mol Biol (Noisy-le-grand) 1998;44:357–379. [PubMed] [Google Scholar]
- 129.Peters JH, Hynes RO. Fibronectin isoform distribution in the mouse. I. The alternatively spliced EIIIB, EIIIA, and V segments show widespread codistribution in the developing mouse embryo. Cell Adhes Commun. 1996;4:103–125. doi: 10.3109/15419069609010766. [DOI] [PubMed] [Google Scholar]
- 130.Kowalczyk AP, Tulloh RH, McKeown-Longo PJ. Polarized fibronectin secretion and localized matrix assembly sites correlate with subendothelial matrix formation. Blood. 1990;75:2335–2342. [PubMed] [Google Scholar]
- 131.George EL, Baldwin HS, Hynes RO. Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood. 1997;90:3073–3081. [PubMed] [Google Scholar]
- 132.Astrof S, Kirby A, Lindblad-Toh K, Daly M, Hynes RO. Heart development in fibronectin-null mice is governed by a genetic modifier on chromosome four. Mech Dev. 2007;124:551–558. doi: 10.1016/j.mod.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 133.Yi M, Sakai T, Fassler R, Ruoslahti E. Antiangiogenic proteins require plasma fibronectin or vitronectin for in vivo activity. Proc Natl Acad Sci U S A. 2003;100:11435–11438. doi: 10.1073/pnas.1635112100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Aszodi A, Hunziker EB, Brakebusch C, Fassler R. Beta1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;17:2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nyberg P, Sakai T, Cho KH, Caparon MG, Fassler R, Bjorck L. Interactions with fibronectin attenuate the virulence of Streptococcus pyogenes. Embo J. 2004;23:2166–2174. doi: 10.1038/sj.emboj.7600214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fededa JP, Petrillo E, Gelfand MS, Neverov AD, Kadener S, Nogues G, et al. A polar mechanism coordinates different regions of alternative splicing within a single gene. Mol Cell. 2005;19:393–404. doi: 10.1016/j.molcel.2005.06.035. [DOI] [PubMed] [Google Scholar]
- 137.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.