Abstract
Background. Klebsiella pneumoniae causes serious infections and healthcare burdens in humans. We have previously reported that the deficiency of autophagy-related gene (Atg) 7 in macrophages (murine alveolar macrophage cell line [MH-S]) induced irregular host immunity against K. pneumoniae and worsened pathologic effects in the lung. In the current study, we investigated the molecular mechanism by which Atg7 influenced K. pneumoniae-induced inflammatory responses.
Methods. Expression levels of Atg7, ubiquitin (Ub), and tumor necrosis factor (TNF) α and phosphorylation of IκBα (p-IκBα) were determined with immunoblotting. Ubiquitylation of p-IκBα was determined with immunoprecipitation.
Results. We noted an interaction between Atg7 and p-IκBα, which was decreased in MH-S after K. pneumoniae infection, whereas the interaction between Ub and p-IκBα was increased. Knock-down of Atg7 with small interfering RNA increased p-IκBα ubiquitylation, promoted nuclear factor κB translocation into the nucleus, and increased the production of TNF-α. Moreover, knock-down of Ub with lentivirus-short hairpin RNA Ub particles decreased binding of p-IκBα to Ub and inhibited TNF-α expression in the primary alveolar macrophages and lung tissue of atg7-knockout mice on K. pneumoniae infection.
Conclusions. Loss of Atg7 switched binding of p-IκBα from Atg7 to Ub, resulting in increased ubiquitylation of p-IκBα and intensified inflammatory responses against K. pneumoniae. Our findings not only reveal a regulatory role of Atg7 in ubiquitylation of p-IκBα but also indicate potential therapeutic targets for K. pneumoniae control.
Keywords: autophagy, alveolar macrophage, ubiquitin, IκBα/NF-κB pathway, TLR4
Bacterial infection is a major cause of morbidity and mortality, imposing a financial burden over $40 billion in the United States [1]. The gram-negative bacterium Klebsiella pneumoniae is the third most common microorganism isolated from intensive care units in the United States [2]. Despite great interest and rapid progress in understanding its pathogenesis in the last decade, the molecular mechanism by which K. pneumoniae is eliminated from the lung by alveolar macrophages remains largely unknown.
Autophagy is critical for cell survival, development, and homeostasis; thus, autophagy defect may lead to various disorders, including cancer and inflammatory diseases [3, 4]. Among the 3 known types of autophagy, macroautophagy (hereafter autophagy) is the most studied and is responsible for the largest degradation events in autophagy [5]. Critical autophagy factors, encoded by >30 autophagy-related genes (Atgs), regulate autophagosome initiation, elongation, and maturation at different stages. Of the known 32 Atg proteins [6], Atg7 is responsible for elongation of autophagosomes. It has been involved in multiple physiologic and pathologic conditions [7, 8], such as viral and bacterial infection [9–11]. Recently, we have identified a role of Atg7 in down-regulating nuclear factor (NF) κB expression and proinflammatory cytokines during K. pneumoniae infection [12]. However, the underlying mechanism by which Atg7 modulates inflammatory responses has not been fully explored.
NF-κB is a family of heterodimeric transcription factors involved in a variety of physiologic and pathologic processes, especially inflammatory and immune response. In the resting state, NF-κB binds to inhibitory proteins of the κB family (IκB) and is sequestered in the cytoplasm. On stimulation, IκB is phosphorylated, and phosphorylated IκB is subsequently ubiquitylated and degraded by 26S proteasome, thus allowing NF-κB to translocate into the nucleus. This will initiate the expression of a number of downstream proinflammatory cytokine genes, such as tumor necrosis factor (TNF) α and interleukin 6 [13, 14].
As a major degradation system, the ubiquitin (Ub)–proteasome system (UPS) has also been implicated in inflammatory responses (eg, regulation of NF-κB pathways) [15]. In addition to UPS, another intracellular degradation system, the autophagy-lysosome system, plays a role in modulating inflammatory processes. Clinical data also suggest the existence of interactions between UPS and autophagy during pathogenesis of many diseases [16]. However, it remains unknown how autophagy and Ub proteasomes regulate K. pneumoniae–mediated inflammatory responses.
Having found that Atg7 altered NF-κB signaling pathway during K. pneumoniae infection in vivo [17], we demonstrated that atg7 deficiency in cells led to a switch in phosphorylated IκBα (p-IκBα) binding from Atg7 to Ub, thus increased ubiquitylation of p-IκBα. In mice, loss of Atg7 increased binding of p-IκBα to Ub and increased expression of TNF-α, whereas infection of atg7 knockout (KO) mice by lentivirus-short hairpin RNA (shRNA) Ub decreased binding of p-IκBα to Ub and inhibited inflammatory responses. In the current study, we further investigated the mechanisms by which Atg7 negatively regulates ubiquitylation of p-IκBα to limit NF-κB-initiated inflammatory responses against K. pneumoniae.
METHODS AND MATERIALS
Animal Handling
The atg7-KO mice (in a C57BL/6J background) were kindly provided by Dr Youwen He (Duke University) [18], the Atg7flox/flox mice were generated as reported elsewhere [7, 19], and the tlr4-KO mice were kindly provided by Dr Jyotika Sharma (University of North Dakota). Mice were maintained in the animal facility at the University of North Dakota, and the animal experiments were performed under National Institutes of Health guidelines and approved by the institutional animal care and use committee. Mice were given ketamine (45 mg/kg) and intranasally infected with 5 × 105 colony-forming units (CFUs) of K. pneumoniae per mouse (6 mice per group). After bronchoalveolar lavage, lungs were obtained for cell biology assays or fixed in 10% formalin for histologic analysis.
Cell Culture
The murine alveolar epithelial cell line (MLE-12) and murine alveolar macrophage cell line (MH-S) were obtained from the American Type Culture Collection and maintained as reported elsewhere [12]. HEK-Blue Toll-like receptor (TLR) 4 cells were kindly provided by Dr Matthew L. Nilles (University of North Dakota) and were originally bought from InvivoGen. These cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum, 50 U/mL penicillin, 50 mg/mL streptomycin, 100 mg/mL normocim, and 2 mmol/L L-glutamine. The U937 and THP1 cells were kindly provided by Drs Archana Dhasarathy and Matthew L. Nilles, respectively (University of North Dakota). These cells were maintained in Roswell Park Memorial Institute 1640 medium with 10% fetal bovine serum and penicillin-streptomycin. Immunostaining, Western blotting, and coimmunoprecipitation are described in detail in the online Supplementary Material.
Bacterial Strains and Infection
The K. pneumoniae strain (American Type Culture Collection 43816 serotype II strain) was provided by Dr V. Miller (University of North Carolina at Chapel Hill) [20]. Bacteria were grown overnight in Luria-Bertani broth at 37°C with shaking at 180 rpm. Optical density was measured at 600 nm (0.1 optical density, 1 × 108 bacterial cells/mL). Mammalian cells were infected by K. pneumoniae with a 10:1 (bacteria-cell) ratio [12].
Cell Transfection
Cells were transfected with Atg7 small interfering RNA (siRNA; Invitrogen), IκBα, TLR4, or Ub siRNA (Santa Cruz), using LipofectAmine 2000 reagent (Invitrogen) in serum-free Roswell Park Memorial Institute 1640 medium according to the manufacturer's instructions [21]. The transfection efficiency of these siRNAs is shown in Supplementary Figure 1A–D. The Ub-hemagglutinin plasmid was kindly provided by Dr Ron Hay (University of Dundee). Tandem green fluorescent protein (GFP)-red fluorescent protein (RFP)-microtubule-associated protein 1 light chain 3 beta (LC3) plasmids were transfected to MH-S cells for 24 hours, as described elsewhere [22]. The tandem RFP-GFP-LC3 plasmid was generated and kindly provided by Dr Tamotsu Yoshimori of Osaka University, Japan [23]. After K. pneumoniae infection, the cells were observed with confocal fluorescence microscopy.
In Vivo Transduction
Mice were anesthetized. The lentiviral particles of Ub shRNA (Santa Cruz, sc-36770-V; 10 μL per mouse) was delivered intranasally [24]. Thirty minutes before infection, all viral supernatants were mixed with LipofectAmine 2000 (5% final vol/vol; Invitrogen) to increase in transduction efficiency [25]. The infection efficiency is shown in Supplementary Figure 1E.
Electrophoretic Mobility Shift Assay
Nuclear extracts from the cells with different treatment were isolated with a nuclear extraction kit according to the manufacturer's instructions (Thermofisher). Oligonucleotide labeling and binding reactions were performed by using the reagent supplied in the NF-κB electrophoretic mobility shift assay (Gel Shift Assay System; Thermofisher). The membrane was visualized with a digital imaging system (Bio-Rad). The specificity of the bands has been confirmed by adding an excess amount of cold oligonucleotide to the reaction mixture.
Phagocytosis Assay
MH-S cells were plated in 24-well plates and grown overnight. The cells were treated with the antibiotic-free medium immediately followed by K. pneumoniae infection. After 1 hour of incubation at 37°C, the wells were washed and treated with 100 µg/mL polymyxin B for 1 hour to kill extracellular bacteria [26, 27]. After washing with phosphate-buffered saline 3 times, the cells were lysed in 1% Triton X-100. The CFUs were then counted to quantify phagocytosis [12].
Histopathologic Analysis
Lung tissues were fixed in 10% formalin using a routine histologic procedure. The fixed tissue samples were processed for standard hematoxylin-eosin staining and examined for morphologic differences after infection [28].
Statistical Analysis
All experiments were performed in triplicate and repeated ≥3 times. Data were presented as mean percentage changes compared with control (and standard deviations) from 3 independent experiments. Group means were compared using 1-way analysis of variance and posttest (Bonferroni selected multiple comparison or Tukey post-hoc test; Prism software 5.0), and differences were considered significant at P < .05 [29].
RESULTS
Disassociation of Atg7 From p-IκBα After K. pneumoniae Infection in MH-S Cells
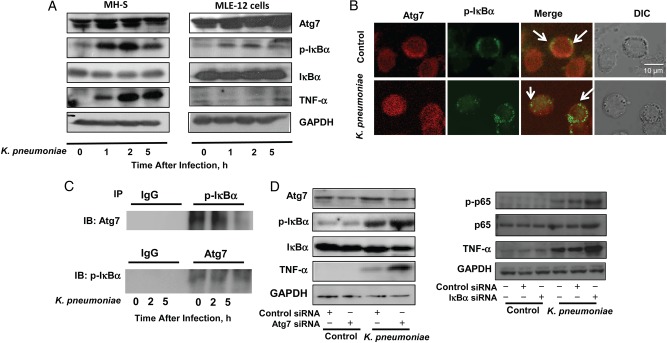
To define the regulatory role of Atg7 in the inflammatory responses during infection, we infected alveolar macrophage MH-S cells and murine lung epithelial MLE-12 cells with K. pneumoniae for different times (0, 1, 2, and 5 hours) and found that Atg7 expression and p-IκBα activity were increased in a time-dependent manner (Figure 1A). Meanwhile, the expression of IκBα was not significantly increased but was somewhat increases in MLE-12 cells at later times (2 and 5 hours), indicating that IκBα protein levels are not dramatically altered by infection. Next, we probed the molecular interaction between these 2 proteins using confocal laser scanning fluorescence microscopy and found that Atg7 was colocalized with p-IκBα at resting, but this codistribution became less after 2 hours of K. pneumoniae infection (see arrows, Figure 1B). We then identified the interaction between Atg7 and p-IκBα by immunoprecipitation assay (Figure 1C). Similar to fluorescence microscopic results, the binding between these 2 proteins was decreased after K. pneumoniae infection, also proportionally to duration of infection (typically 5 hours) (Figure 1C). These data together indicated that K. pneumoniae infection caused time-dependent disassociation of Atg7 from p-IκBα.
Figure 1.
Autophagy-related gene (Atg) 7 interacted with phosphorylated IκBα (p-IκBα) in cells from the murine alveolar macrophage cell line (MH-S). A, Expression levels of Atg7, p-IκBα, and tumor necrosis factor (TNF) α were increased with time after Klebsiella pneumoniae infection (0, 1, 2, or 5 hours) in MH-S and murine alveolar epithelial cell line (MLE-12) cells, as assessed by immunoblotting (IB). Glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was used as a loading control. B, Colocalization of Atg7 and p-IκBα was observed with fluorescence microscopy. MH-S cells were infected with K. pneumoniae at a multiplicity of infection (MOI) of 10:1 for 2 hours. C, Interaction between p-IκBα and Atg7, as detected with a coimmunoprecipitation assay. MH-S cells were infected with K. pneumoniae at an MOI of 10:1 for 0, 2, 5 hours. D, Left, p-IκBα and TNF-α levels were determined with immunoblotting. MH-S cells were transfected with Atg7 small interfering RNA (siRNA) or control (scrambled) siRNA. After 24 hours, cells were infected with K. pneumoniae at an MOI of 10:1 for 1 hour. Right, Nuclear factor κB and TNF-α expression was determined with IB. MH-S cells were transfected with IκBα siRNA or control siRNA. After 24 hours, cells were infected with K. pneumoniae at an MOI of 10:1 for 1 hour. Data were representative of 3 experiments. Abbreviations: +, with; −, without; DIC, differential interference contrast; IgG, immunoglobulin G; IP, immunoprecipitation.
Alteration of NF-κB Pathway by Silencing of Atg7 or IκBα in MH-S Cells
To define the role of Atg7 in NF-κB signaling, we found that down-regulating Atg7 by siRNA silencing led to an increase in p-IκBα levels after K. pneumoniae infection (Figure 1D, left). To elucidate the impact of IκBα activity on NF-κB signaling, we transfected IκBα siRNA into MH-S cells. After successful knock-down of IκBα with siRNA (Supplementary Figure 1B), the phosphorylation of NF-κB subunit p65 (ser536) was found to be increased with K. pneumoniae infection, suggesting that IκBα sits upstream of the NF-κB pathway. Furthermore, knock-down of IκBα increased protein levels and secretion of TNF-α, as assessed by immunoblotting (Figure 1D, right) and enzyme-linked immunosorbent assay (ELISA), respectively (Supplementary Figure 2A). These data indicated that IκBα was critical for the regulation of host inflammatory responses to K. pneumoniae infection via NF-κB signaling.
In addition, protein levels and secretion of TNF-α were higher in Atg7 siRNA–transfected MH-S cells than in scrambled siRNA-transfected controls, as detected by immunoblotting (Figure 1D, left) and ELISA, respectively (Supplementary Figure 2A). Thus, Atg7 silencing may be attributable to dysregulated proinflammatory responses through the Atg7/IκBα/NF-κB axis.
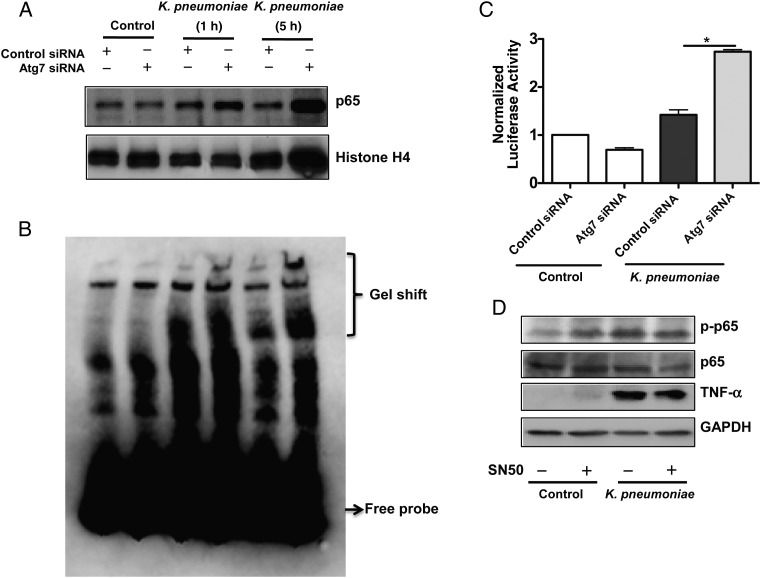
NF-κB is a master transcriptional factor for initiating inflammatory responses. We isolated the nuclear fraction of MH-S cells at 1 or 5 hours after K. pneumoniae infection and found that NF-κB expression was significantly increased in Atg7 siRNA–silenced cells compared with controls after K. pneumoniae infection (Figure 2A). We then carried out an electrophoretic mobility shift assay to study the potential NF-κB activation and found that ostensible shifts of NF-κB occurred in Atg7 siRNA cells after K. pneumoniae challenging (Figure 2B). We also noted that the NF-κB luciferase reporter activity was significantly increased in Atg7 siRNA–transfected MH-S cells compared with control siRNA–transfected cells (Figure 2C). We also used an NF-κB inhibitor (SN50, 1.8 μmol/L) to validate the activation and function of NF-κB and found that SN50 inhibited levels and secretion of and TNF-α after K. pneumoniae infection with immunoblotting (Figure 2D) and ELISA, respectively (Supplementary Figure 2B).
Figure 2.
Nuclear factor (NF) κB translocated into nuclei in autophagy-related gene (Atg) 7–silenced cells. A, NF-κB levels were increased in the Atg7-silencing cells. Cells from the murine alveolar macrophage cell line (MH-S) were transfected with Atg7 small interfering RNA (siRNA) or control (scrambled) siRNA. After 24 hours, the cells were infected with Klebsiella pneumoniae for 1 hour and 5 hours. Nuclear fractions were isolated from cells by a nuclear extraction kit (Thermofisher). B, Electrophoretic mobility shift assay was performed in MH-S cell nuclear extracts using the biotin-labeled probe (Thermofisher), which contains only a single copy of the 21–base pair element. C, Increased luciferase reporter activity of NF-κB in Atg7 siRNA–transfected MH-S cells. MH-S cells were transfected with Atg7 siRNA or control siRNA. After 24 hours, cells were transfected with luciferase reporter NF-κB plasmid. After 24 hours of transfection, cells were infected with K. pneumoniae at a multiplicity of infection (MOI) of 10:1 for 1 hour. Cell-permeable NF-κB inhibitor (SN50) (1.8 µmol/L) was used to pretreat cells for 0.5 hour before infection. *P < .001 (1-way analysis of variance and Bonferroni selected multiple comparison test). D, Tumor necrosis factor (TNF) α expression was decreased after inhibiting NF-κB with SN50 (1.8 µmol/L). MH-S cells were infected with K. pneumoniae at an MOI of 10:1 for 1 hour. Data were representative of 3 experiments. Abbreviations: +, with; −, without; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Ubiquitylation of p-IκBα With K. pneumoniae Infection
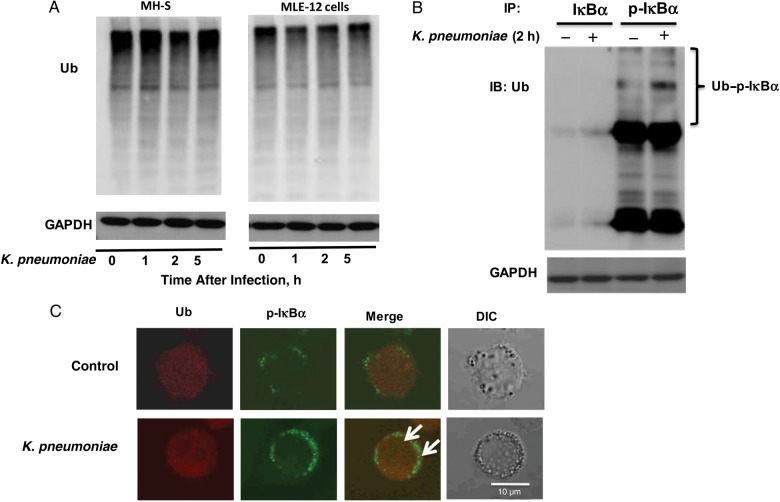
Within the eukaryotic cell, there are 2 main intracellular protein degradation pathways: the UPS and autophagy. In the current study, we determined whether UPS also contributed to the degradation of p-IκBα and observed that expression of Ub was not significantly influenced after K. pneumoniae infection at different time points in either immune active murine lung epithelial MLE-12 cells or MH-S cells (Figure 3A). However, the association between p-IκBα and Ub became significantly increased after K. pneumoniae infection, as detected using coimmunoprecipitation in MH-S cells (Figure 3B). In addition, colocalization of p-IκBα and Ub was revealed by fluorescence imaging (Figure 3C, arrows), suggesting a potential interaction between p-IκBα and Ub.
Figure 3.
The phosphorylated IκBα (p-IκBα) was ubiquitylated after Klebsiella pneumoniae infection. A, Expression of ubiquitin (Ub) protein was not significantly changed after K. pneumoniae infection at different time points in either the murine alveolar epithelial cell line (MLE-12) or the murine alveolar macrophage cell line (MH-S). Cells were infected with K. pneumoniae for 1, 2 or 5 hours. B, Interaction between p-IκBα and Ub was significantly increased after K. pneumoniae infection. MH-S cells were infected with K. pneumoniae for 2 hours and then were collected for coimmunoprecipitation assay. C, Colocalization of Ub and p-IκBα was observed by fluorescence microscopy (arrows). MH-S cells were infected with K. pneumoniae for 2 hours. Data were representative of 3 experiments. Abbreviations: +, with; −, without; DIC, differential interference contrast; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IP, immunoprecipitation.
Ubiquitylation of p-IκBα Increased by Atg7 Silencing
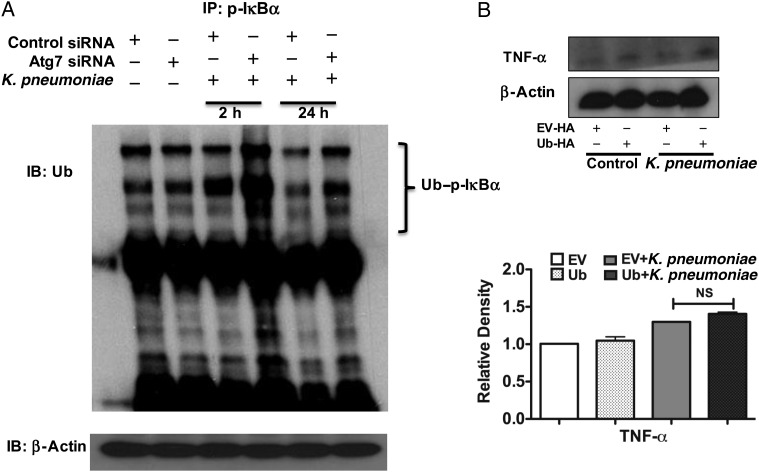
Because our results showed that p-IκBα could interact with either Atg7 or Ub, we sought to elucidate whether there is a competition between Atg7 and Ub during their interaction with p-IκBα. To approach this question, we knocked down Atg7 with specific siRNA and found an increased interaction between p-IκBα and Ub in the MH-S cells on K. pneumoniae infection (Figure 4A). To confirm the role of Ub in K. pneumoniae–infected cells, we transfected Ub plasmid (Ub-hemagglutinin) to overexpress Ub and found that overexpression of Ub led to increased expression (Figure 4B) and secretion (Supplementary Figure 2C) of TNF-α compared with vector-transfected controls; however, the change was not significant. Moreover, in human macrophage THP1 and U937 cells, we observed similar interaction of p-IκBα and Ub (Supplementary Figure 3). We also attained similar results with another gram-negative bacterium, Pseudomonas aeruginosa PAO1 strain, or lipopolysaccharide derived from K. pneumoniae-treated MH-S cells (Supplementary Figure 4), which showed that silencing Atg7 significantly increased the ubiquitylation of p-IκBα.
Figure 4.
Autophagy-related gene (Atg) 7 silencing increased the ubiquitylation of phosphorylated IκBα (p-IκBα). A, Knocking known Atg7 with specific small interfering RNA (siRNA) increased the interaction between p-IκBα and ubiquitin (Ub) compared with control siRNA-silenced cells after Klebsiella pneumoniae infection. Cells from the murine alveolar macrophage cell line (MH-S) were infected with K. pneumoniae for the indicated hours. B, Overexpression of Ub increased the expression of tumor necrosis factor (TNF) α. The Ub–hemagglutinin (HA) plasmid was kindly provided by Dr Ron Hay (University of Dundee). MH-S cells were transfected with Ub-hemagglutinin. After 24 hours, cells were infected with K. pneumoniae for 2 hours. Data are representative of 3 experiments. NS, not significant (1-way analysis of variance and Bonferroni selected multiple comparison test). Abbreviations: +, with; −, without; EV, empty vector; IB, immunoblotting; IP, immunoprecipitation.
To examine whether Atg7 plays a specific role in p-IκBα ubiquitylation, we knocked down other Atgs, for example, Atg5 and Beclin 1. Importantly, knock-down of Atg5 and Beclin 1 by siRNA did not significantly affect the ubiquitylation of p-IκBα (Supplementary Figure 5A), suggesting a specific role for Atg7 in regulating the ubiquitylation of p-IκBα. However, 3-methyladenine pretreatment increased ubiquitylation of p-IκBα after K. pneumoniae infection (Supplementary Figure 5B).
Deubiquitylation of p-IκBα
To further analyze the role of ubiquitylation at the molecular level, we employed Ub siRNA to examine the level of Ub in K. pneumoniae infection in MH-S cells. We demonstrated that knock down of Ub decreased the ubiquitylation of p-IκBα (Figure 5A) as well as the expression of TNF-α (Figure 5B). In addition, we used a deubiquitinase (usp30) to assess the role of Ub in immunity to K. pneumoniae infection in vitro models. We found that usp30 expression was significantly decreased after K. pneumoniae infection in MH-S macrophage cells but not in MLE-12 epithelial cells (Supplementary Figure 6A). We further transfected MH-S cells with usp30 siRNA and noticed that TNF-α expression (Supplementary Figure 6B) and secretion (Supplementary Figure 2A) was significantly decreased with K. pneumoniae infection. We also used a proteasome inhibitor (MG132) to evaluate the downstream effects of ubiquitylation. MG132 treatment alone could not alter the ubiquitylation status of p-IκBα (Supplementary Figure 7A). However, after knock-down of Atg7, pretreament with MG132 increased the ubiquitylation of p-IκBα (Supplementary Figure 7B).
Figure 5.
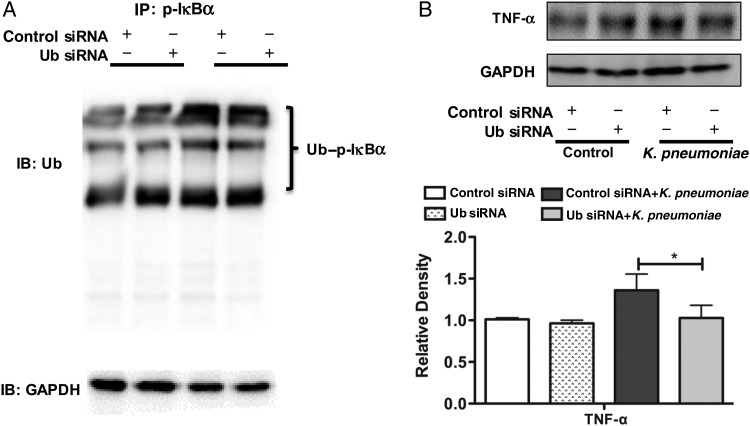
Silencing ubiquitin (Ub) with small interfering RNA (siRNA) inhibited inflammatory responses. A, Interaction between phosphorylated IκBα and Ub was significantly decreased after Klebsiella pneumoniae infection after knock-down of Ub. Cells were transfected with Ub siRNA. At 24 hours after transfection, cells from the murine alveolar macrophage cell line (MH-S) were infected with K. pneumoniae for 2 hours and then collected for the coimmunoprecipitation assay. B, Silencing Ub followed by K. pneumoniae infection decreased the expressions of tumor necrosis factor (TNF) α. MH-S cells were transfected with Ub siRNA. After 24 hours, the cells were infected with K. pneumoniae for 2 hours. Data were representative of 3 experiments. *P < .05 (1-way analysis of variance and Bonferroni selected multiple comparison test). Abbreviations: +, with; −, without; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IP, immunoprecipitation.
Effect of Ub Silencing on Ubiquitylation of p-IκBα and Inflammatory Responses In Vitro and In Vivo
To clearly characterize the role of ubiquitylation of p-IκBα in the inflammatory responses, we simultaneously knocked down both Ub and Atg7 with specific siRNAs in MH-S cells. We found that after K. pneumoniae infection, the increased ubiquitylation of p-IκBα in Atg7 siRNA–silencing cells was drastically blunted in the Ub and Atg7 dual knock-down group (Figure 6A). Similarly, expression of TNF-α was significantly decreased when both Ub and Atg7 were knocked down (Figure 6B). To detect whether Atg7 regulates p-IκBα in vivo, we infected wild-type mice or atg7-KO mice with lentivirus-shUb particles and evaluated lung injury histologically. At 24 hours after K. pneumoniae infection, we observed a decrease in inflammatory cell infiltration in lentivirus-shUb–infected atg7-KO mice compared with wild-type mice (Figure 7A). The secretion of TNF-α in the bronchoalveolar lavage fluid was significantly decreased in lentivirus-shUb–infected atg7-KO mice compared with vector control–infected atg7-KO mice (Figure 7B). We also isolated primary alveolar macrophages from different groups of mice and showed that ubiquitylation of p-IκBα and expression of TNF-α were significantly decreased in lentivirus-shUb–infected atg7-KO mice, whereas ubiquitylation of p-IκBα and expression of TNF-α were markedly increased in atg7-KO mice (Figure 7C). Similar results were also found in the lung tissue of such mice (Figure 7D).
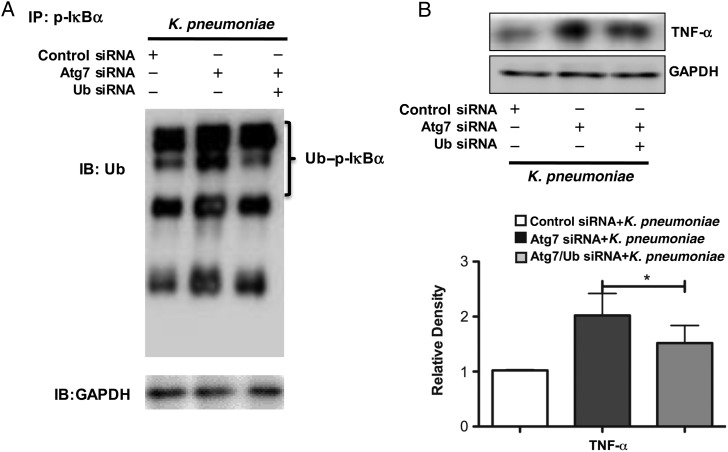
Figure 6.
Dual knock-down of ubiquitin (Ub) and autophagy-related gene (Atg) 7 decreased the ubiquitylation of phosphorylated IκBα (p-IκBα) and inflammatory responses in the murine alveolar macrophage cell line (MH-S). A, Interaction between p-IκBα and Ub is significantly decreased after Klebsiella pneumoniae infection after knock-down of both Ub and Atg7, compared with Atg7 small interfering RNA (siRNA)–silencing cells. Cells were transfected with Atg7 siRNA for 24 hours. At 24 hours after transfection, MH-S cells were transfected with Ub siRNA. After another 24 hours, the cells were infected by K. pneumoniae for 2 hours and then collected for the coimmunoprecipitation assay. B, Dual knock-down of Ub and Atg7 followed by K. pneumoniae infection decreased the expressions of tumor necrosis factor (TNF) α compared with the Atg7 siRNA group. After 24 hours, cells were infected with K. pneumoniae for 2 hours. Data were representative of 3 experiments. *P < .05 (1-way analysis of variance and Bonferroni selected multiple comparison test). Abbreviations: +, with; −, without; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; IP, immunoprecipitation.
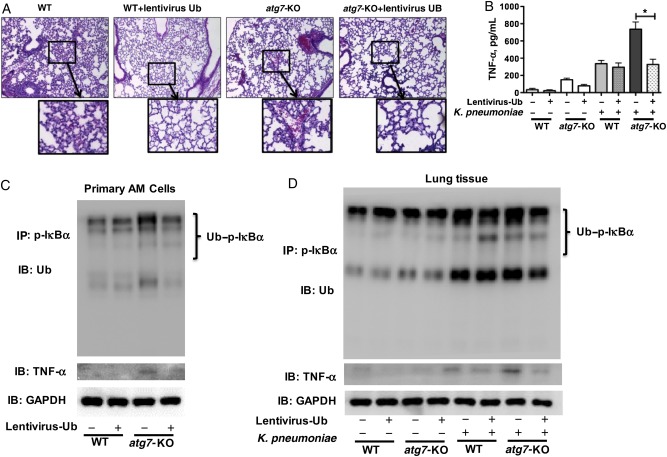
Figure 7.
Lentivirus-shUb–infected atg7-knockout (KO) mice exhibited decreased inflammatory responses after Klebsiella pneumoniae infection. A, Decreased lung injury and inflammation was detected histologically. Mice were infected with lentivirus-short hairpin RNA Ub. After 3 days, mice were infected 5 × 105 colony-forming units per mouse for 24 hours. Mice were dissected, their lungs were embedded in formalin, and sections were analyzed with hematoxylin-eosin staining. Data were representative of 6 mice per group. B, Tumor necrosis factor (TNF) α in the bronchoalveolar lavage fluid was significantly decreased in lentivirus-shUb–infected atg7-KO mice, as detected with enzyme-linked immunosorbent assay. *P < .001 (1-way analysis of variance and Bonferroni selected multiple comparison test). C, D, Ubiquitylation of phosphorylated IκBα and the expression of TNF-α were significantly decreased in the lentivirus-shUb particle–infected alveolar macrophage (AM) cells (C) and lung tissue (D) of atg7-KO mice compared with that of lentivirus control vector–infected atg7-KO mice after K. pneumoniae infection. After infection, alveolar macrophages or lung tissue were isolated and lysed in immunoprecipitation (IP) lysis buffer. Half of the lysate was used for the coimmunoprecipitation assay, and the rest was used to determine the protein expressions of TNF-α and GAPDH. Data were representative of 3 experiments. Abbreviations: +, with; −, without; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IB, immunoblotting; Ub, ubiquitin; WT, wild type.
Role of TLR4 in Controlling Autophagy After K. pneumoniae Infection
To identify the upstream signals of Atg7, we examined several innate immunity players and identified that TLR4 was involved in autophagy pathways during K. pneumoniae infection (data not shown). After knocking down TLR4 with specific siRNA, we found that Atg7 expression and LC3 conversion from LC3-I to LC3-II (LC3-phosphatidylethanolamine conjugate) were significantly decreased (Supplementary Figure 8A, left). These results were further confirmed by using TLR4 neutralizing antibodies (Supplementary Figure 8A, right). To evaluate the effect of TLR4 on autophagy, we cotransfected a tandem RFP-GFP-LC3 plasmid and TLR4 siRNA into MH-S cells and revealed that TLR4 silencing significantly decreased the formation of LC3-II puncta (Supplementary Figure 8B), indicating that blocking TLR4 could reduce autophagy after K. pneumoniae infection. The CFU counts demonstrated that down-regulated levels of TLR4 led to decreased bacterial phagocytosis and bactericidal activity (Supplementary Figure 8C).
To further confirm the impact of TLR4 on Atg7, we used the HEK-Blue TLR4 cell line, which stably overexpressed TLR4 (InvivoGen), and we found that Atg7 activation by K. pneumoniae infection was partially blocked by TLR4 silencing or neutralizing antibody (Supplementary Figure 8D). To convincingly validate these data and indicate physiologic relevance, we used tlr4-KO mice to evaluate the role of TLR4. We found that these mice also manifested decreased Atg7 expression 24 hours after K. pneumoniae infection (Supplementary Figure 8E), which indeed suggests a critical role of TLR4 in Atg7 signaling and its associated host defense against K. pneumoniae infection by providing enhanced bactericidal activity. To summarize the findings of this study, we illustrated the signaling pathway we demonstrated in atg7-KO mice and siRNA-silenced cells (Supplementary Figure 8F).
DISCUSSION
Atg7 has been demonstrated to affect host defense against various pathogens [30–32]. In this study, we explored the mechanism by which Atg7 regulates inflammatory responses during gram-negative bacterial infection. Importantly, we reveal a new mechanism whereby Atg7 regulates inflammation by inhibiting ubiquitylation degradation of p-IκBα in both cells and mice after K. pneumoniae infection. NF-κB activation may be triggered to augment inflammatory responses after disassociation of the normally suppressed p-IκBα from an Atg7–p-IκBα complex. This signaling pathway is critically initiated and controlled by TLR4, as determined using overexpressing stable cell lines and KO mice.
Ubiquitylation and autophagy are 2 major protein degradation systems in the eukaryotic cells involved in a variety of cellular processes. Both play essential roles in cellular protein homeostasis and control many cell functions, including cell growth, proliferation, apoptosis, and immune response [33]. They are usually considered independent of each other because of their differences in constitutes and degradation mechanisms. However, there is also evidence that immune response may be associated with both degradation systems to affect different disease processes in various models [34, 35]. Inhibition of proteasomal activities used to induce cell death has been shown elsewhere to activate autophagy, indicating a coordinated and complementary relationship between these 2 important protein degradation systems [16, 36]. On the other hand, in U87MG glioma cells, increasing autophagy may decrease the activity of the UPS [34].
In our K. pneumoniae infection model, we found that Atg7 deficiency disrupted the balance between the UPS and autophagy systems, skewed the reaction toward the former. We observed that p-IκBα ubiquitylation was increased in Atg7 silencing cells, suggesting that p-IκBα degradation was augmented. The degradation of p-IκBα allowed the release of NF-κB to initiate transcriptional activity of proinflammatory cytokines (eg, TNF-α). However, we cannot exclude the involvement of other cytokines or signaling pathways.
The degradation of IκBα kinase is the key regulatory mechanism in NF-κB activation and may be modulated either by an autophagy pathway [14] or by a ubiquitylation pathway [37]. The ability of autophagy to inhibit inflammatory responses by influencing IκBα/NF-κB signaling has also been documented in other disease models. For instance, the liver in beclin1-mutant mice exhibits increased apoptosis and NF-κB activation owing to the accumulation of p62 [38]. Defects in Atg7 contribute to the pathogenesis of obesity by activating the I kappa B kinase β pathway [39]. Ub is required for the phosphorylation and degradation of IκBα both in vitro and in vivo [40]. Here, we reveal a role of Ub in Atg7-modulated inflammation during bacterial infection, which explains the critical regulation of inflammatory responses by Atg7. At this time, however, we cannot exclude another possibility, that is, regulation of IKK-IκBα complex. IκBα could be phosphorylated through activation of IKK and recognized by beta-transducin repeats-containing proteins after p-IκBα ubiquitylation [41].
Other studies have shown that TLR4 signaling can induce autophagy in leukocytes and positively influence microbial clearance and NF-κB signaling [42, 43], and these findings led us to propose a role of TLR4 in autophagy after K. pneumoniae infection. To identify upstream immune molecules of Atg7, we found that TLR4, the pattern recognition receptor of gram-negative bacterial lipopolysaccharide, is critically involved in Atg7 signaling. We also found that knocking down TLR4 led to an increased superoxide production after K. pneumoniae infection (data not shown), which might be a link between TLR4 and the autophagy pathway. Our findings indicate that TLR4 may be a potential sensor of autophagy in K. pneumoniae infection.
In conclusion, we identified an important mechanism for Atg7-associated innate immunity against gram-negative bacterium K. Pneumoniae, showing that atg7 deficiency intensified proinflammatory responses via a ubiquitylation mechanism. Collectively, these observations unravel ubiquitylation of p-IκBα as a critical molecular process by which Atg7 negatively regulates inflammatory response to K. pneumoniae, indicating potentially novel targets for therapeutic interventions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank S. Rolling of the University of North Dakota imaging core for help with confocal imaging, Dr Ron Hay at University of Dundee for providing the Ub-hemagglutinin plasmid, and Tamotsu Yoshimori at Osaka University, Japan, for providing the tandem RFP-GFP-LC3 plasmid.
Financial support. This project was supported by Flight Attendant Medical Research Institute (grant 103007) and the National Institutes of Health (grants R01 AI109317-01A1, R15 AI101973-01, and R03 AI097532-01A1 to M. W.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Moore TA, Standiford TJ. The role of cytokines in bacterial pneumonia: an inflammatory balancing act. Proc Assoc Am Physicians 1998; 110:297–305. [PubMed] [Google Scholar]
- 2.Jones RN. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 2010; 51(suppl 1):S81–7. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463–77. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861–73. [DOI] [PubMed] [Google Scholar]
- 6.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010; 221:3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komatsu M, Waguri S, Ueno T et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuma A, Hatano M, Matsui M et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032–6. [DOI] [PubMed] [Google Scholar]
- 9.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology 2011; 53:406–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenz HD, Vierstra RD, Nurnberger T, Gust AA. ATG7 contributes to plant basal immunity towards fungal infection. Plant Signal Behav 2011; 6:1040–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue J, Nishiumi S, Fujishima Y et al. Autophagy in the intestinal epithelium regulates Citrobacter rodentium infection. Arch Biochem Biophys 2012; 521:95–101. [DOI] [PubMed] [Google Scholar]
- 12.Ye Y, Li X, Wang W et al. Atg7 deficiency impairs host defense against Klebsiella pneumoniae by impacting bacterial clearance, survival and inflammatory responses in mice. Am J Physiol Lung Cell Mol Physiol 2014; 307:L355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beutler B, Cerami A. The biology of cachectin/TNF—a primary mediator of the host response. Annu Rev Immunol 1989; 7:625–55. [DOI] [PubMed] [Google Scholar]
- 14.Sha WC. Regulation of immune responses by NF-κB/Rel transcription factor. J Exp Med 1998; 187:143–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J, Chen ZJ. Regulation of NF-κB by ubiquitination. Curr Opin Immunol 2013; 25:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojcik S. Crosstalk between autophagy and proteasome protein degradation systems: possible implications for cancer therapy. Folia Histochem Cytobiol 2013; 51:249–64. [DOI] [PubMed] [Google Scholar]
- 17.Consumption of raw or unpasteurized milk and milk products by pregnant women and children. Pediatrics 2014; 133:175–9. [DOI] [PubMed] [Google Scholar]
- 18.Jia W, Pua HH, Li QJ, He YW. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol 2011; 186:1564–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2001; 2:292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005; 58:1054–73. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Zhou X, Ye Y et al. Lyn regulates inflammatory responses in Klebsiella pneumoniae infection via the p38/NF-κB pathway. Eur J Immunol 2014; 44:763–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan K, Huang C, Fox J et al. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci 2012; 125:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452–60. [DOI] [PubMed] [Google Scholar]
- 24.Wilson AA, Kwok LW, Porter EL et al. Lentiviral delivery of RNAi for in vivo lineage-specific modulation of gene expression in mouse lung macrophages. Mol Ther 2013; 21:825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson AA, Murphy GJ, Hamakawa H et al. Amelioration of emphysema in mice through lentiviral transduction of long-lived pulmonary alveolar macrophages. J Clin Invest 2010; 120:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan S, Audet A, Huang H, Chen LJ, Wu M. Cholesterol-rich membrane rafts and Lyn are involved in phagocytosis during Pseudomonas aeruginosa infection. J Immunol 2008; 180:2396–408. [DOI] [PubMed] [Google Scholar]
- 27.Yuan K, Huang C, Fox J et al. Elevated inflammatory response in caveolin-1-deficient mice with Pseudomonas aeruginosa infection is mediated by STAT3 protein and nuclear factor κB (NF-κB). J Biol Chem 2011; 286:21814–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu M, Hussain S, He YH, Pasula R, Smith PA, Martin WJ II. Genetically engineered macrophages expressing IFN-γ restore alveolar immune function in scid mice. Proc Natl Acad Sci U S A 2001; 98:14589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu M, Huang H, Zhang W et al. Host DNA repair proteins in response to Pseudomonas aeruginosa in lung epithelial cells and in mice. Infect Immun 2011; 79:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia K, Thomas C, Akbar M et al. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A 2009; 106:14564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy 2009; 5:937–45. [DOI] [PubMed] [Google Scholar]
- 32.Amer AO, Byrne BG, Swanson MS. Macrophages rapidly transfer pathogens from lipid raft vacuoles to autophagosomes. Autophagy 2005; 1:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knecht E, Aguado C, Carcel J et al. Intracellular protein degradation in mammalian cells: recent developments. Cell Mol Life Sci 2009; 66:2427–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Errafiy R, Aguado C, Ghislat G et al. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS One 2013; 8:e83318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korolchuk VI, Menzies FM, Rubinsztein DC. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett 2010; 584:1393–8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Li W, Wang C et al. Inhibition of autophagy enhances apoptosis induced by proteasome inhibitor bortezomib in human glioblastoma U87 and U251 cells. Mol Cell Biochem 2014; 385:265–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent IκBα phosphorylation marks the NF-κB inhibitor for degradation via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A 1995; 92:10599–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathew R, Karp CM, Beaudoin B et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell 2009; 137:1062–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng Q, Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. J Biol Chem 2011; 286:32324–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell 1996; 84:853–62. [DOI] [PubMed] [Google Scholar]
- 41.Kanarek N, London N, Schueler-Furman O, Ben-Neriah Y. Ubiquitination and degradation of the inhibitors of NF-κB. Cold Spring Harb Perspect Biol 2010; 2:a000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neal MD, Sodhi CP, Dyer M et al. A critical role for TLR4 induction of autophagy in the regulation of enterocyte migration and the pathogenesis of necrotizing enterocolitis. J Immunol 2013; 190:3541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity 2007; 27:135–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.