Abstract
Avian pathogenic Escherichia coli (APEC) is a major pathogen that causes avian colibacillosis and is associated with severe economic losses in the chicken-farming industry. Here, bacteriophage KBNP1315, infecting APEC strain KBP1315, was genomically and functionally characterized. The evolutionary relationships of KBNP1315 were analyzed at the genomic level using gene (protein)-sharing networks, the Markov clustering (MCL) algorithm, and comparative genomics. Our network analysis showed that KBNP1315 was connected to 30 members of the Autographivirinae subfamily, which comprises the SP6-, T7-, P60-, phiKMV-, GAP227- and KP34-related groups. Network decomposition suggested that KBNP1315 belongs to the SP6-like phages, but our comparison of putative encoded proteins revealed that key proteins of KBNP1315, including the tail spike protein and endolysin, had relative low levels of amino acid sequence similarity with other members of the SP6-like phages. Thus KBNP1315 may only be distantly related to the SP6-like phages, and (based on the difference in endolysin) its lysis mechanism may differ from theirs. To characterize the lytic functions of the holin and endolysin proteins from KBNP1315, we expressed these proteins individually or simultaneously in E. coli BL21 (DE3) competent cell. Interestingly, the expressed endolysin was secreted into the periplasm and caused a high degree of host cell lysis that was dose-dependently delayed/blocked by NaN3-mediated inhibition of the SecA pathway. The expressed holin triggered only a moderate inhibition of cell growth, whereas coexpression of holin and endolysin enhanced the lytic effect of endolysin. Together, these results revealed that KBNP1315 appears to use a pin-holin/signal-arrest-release (SAR) endolysin pathway to trigger host cell lysis.
Introduction
Avian colibacillosis, which is caused by strains of avian pathogenic Escherichia coli (APEC), is a severe bird disease that affects the poultry industry and is responsible for enormous economic losses worldwide [1, 2]. Escherichia coli is typically a commensal microorganism in the intestinal tract of chickens, but APEC strains are extraintestinal pathogenic E. coli (ExPEC) whose infection is characterized by various symptoms, such as airsacculitis, polyserositis, enteritis, cellulitis, salpingitis, omphilitis, coligranuloma, egg peritonitis and septicemia [1–3]. Other ExPEC strains have been isolated from human urinary tract infections (UTIs) and neonatal meningitis [2, 4, 5]. In addition, the antibiotic treatment capable of controlling this pathogen has been imprudently used, leading to the appearance of multi-antibiotic resistant APEC stains [6–9]. Therefore, we urgently need an alternative agent or strategy for treating antibiotic-resistant E. coli.
Bacteriophages (or phages), predators of bacteria, are the most abundant organisms on the planet and contribute to the reservoir of genetic diversity in the microbial environment [10, 11]. Recently, phage genomics have been advancing as a result of the remarkable development in phage genome sequencing, which contributes to the progress of genetic, biotechnological and clinical tools [12, 13]. However, because the process of phage evolution is being affected by frequent horizontal gene transfer (HGT), phage genomes show pervasive mosaicism and it is difficult to characterize them by classical taxonomic study based on hierarchical classification [10, 11, 13]. To overcome the limitation of the classic approach, Lima-Mendez et al. [11] proposed a reticulated network model representing the phage-phage relationship in consideration of shared gene (protein) contents, including the evolutionary reconstructions of individual gene phylogeny; this model gives a clear understanding of the true evolutionary history of phage genomes [10, 14].
Bacteriophage SP6 is a well-characterized member of the Autographivirinae subfamily (which infects Salmonella enterica Typhimurium LT2), which is closely related to the Escherichia coli phages, K1-5, K5 and K1E [15]. Collectively, these phages were traditionally considered very similar to the T7-like phages. However, Scholl et al. proposed the novel notion that these phages are separate from the T7 group, and instead form a novel genus called the “SP6-like phages” [16]. Several proteins of these phages have been functionally characterized, including those related to phage replication (RNA polymerase) and host specificity determination (tail spike) [17, 18]. Although the phages of this group have been studied in detail, only a single functional study has examined the host lysis proteins of a putative member of the SP6 group: the Erwinia amylovora phage, Era103 [19]. To better understand the host lysis mechanisms of this group, further functional studies of lytic proteins are needed [19].
At the final stage of their life cycle, the double-stranded DNA bacteriophages generally use two proteins, holin and endolysin, to lyse the host bacterium by degrading the inner membrane and peptidoglycan layer of the bacterial membrane, respectively [20, 21, 22]. This canonical mechanism was traditionally considered to be a universal strategy through which phages escape from their host bacteria [23]. However, other lytic mechanisms have since been reported, including that involving pin-holin and SAR (signal-anchor-release) endolysin, which are seen in phages P1, 21, phiKMV, and JG068. The endolysins of these phages possess N-terminal SAR domains that are exported to the periplasm via the host sec system by virtue of an N-terminal signal sequence. There, the positively charged anchors of the SAR domain tether the exported endolysin to the cytoplasmic membrane in an inactive form. Pin-holin (which differs from canonical holin) then forms a small hole that induces membrane depolarization and releases the tethered endolysin into the periplasm in an active form [24–27]. Similarly, the endolysin of Oenococcus oeni bacteriophage fOg44 has a typical signal sequence with the peptidase recognition site, and it is activated by the proteolytic cleavage of the N-terminal signal peptide [28].
Here, we report a genomic analysis of the novel APEC phage, KBNP1315, and the functional characterization of its lysis-cassette. To understand the evolution of this phage, we used gene (protein)-sharing network and comparative genomic analyses to assess its relationships with its relatives. In addition, we characterized the lytic proteins, holin and endolysin, of KBNP1315 to provide novel insights into the host lysis strategy of the SP6-like phages.
Materials and Methods
Bacterial strain and characterization
An E. coli strain was isolated from a chicken farm located in Yesan, South Korea. Phages were isolated from sewage samples obtained from the same chicken farm from which the host bacterium was isolated. The isolated bacteria was identified by using MALDI-TOF MS (Bruker Daltonik GmbH, Leipzig, Germany) and named “KBP1315.” To determine its pathotype, the APEC-specific genes, fimC, tsh, ironN, fyuA, irp2, and stx2, were detected by PCR. In addition, a disk diffusion method was used to evaluate the antimicrobial susceptibility of E. coli KBP1315 on LB agar plates [28].
Bacteriophage isolation
Phages were isolated from sewage samples obtained from the same chicken farm from which the host bacterium was isolated. Briefly, each sewage sample was centrifuged at 5,000×g for 10 min, and then filtered through a 0.22 membrane (Merk Millipore, MA, USA). The phage solution was cultured with E. coli KBP1315 and a plaque assay was performed [29]. After incubation for 24 h at 30°C, a single plaque was picked from plaque assay, eluted in LB broth and combined with a host culture again. Following several rounds of plaque assay and single plaque selection, bacteriophage KBNP1315 was isolated. To purify phage KBNP1315, the phage particles were propagated up to 200 ml and centrifuged at 3,000×g for 30 min at 4°C. The supernatant was carefully collected, treated with 10% polyethylene glycol (PEG) for 1 hr, and centrifuged at 16,000×g for 15 min at 4°C. The precipitate was resuspended in TM buffer (10 mM Tris-HCL, 100 mM NaCl, 10 mM MgCl2, and 10 mM CaCl2, pH 7.5) and purified by CsCl density gradient equilibrium centrifugation, as previously described [30].
Transmission electron microscopy (TEM)
Morphological analysis of purified phage KBNP1315 was performed using TEM, as previously described by Jang et al. [10]. Purified phage solution (8 μl) was dropped onto 400-mesh Formvar carbon-coated copper grids (Ted Pella Inc., CA, USA). After 2 min, the grids were stained with 8 μl of 2% aqueous uranyl acetate (Merck, Darmstadt, Germany) and examined with a Philips TECNAI F12 FEI transmission electron microscope (FEI, OR, USA) at an accelerating voltage of 120 kV. KBNP1315 was morphologically classified according to the classification scheme of the International Committee on Taxonomy of Viruses (ICTV; http://www.ictvdb.org/).
Genomic sequencing and bioinformatic analysis
KBNP1315 DNA was prepared for sequencing as previously described [31], using proteinase K digestion followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation. The genomic sequence of KBNP1315 was obtained using an FLX Titanium genome sequencer (Roche, Mannheim, Germany) according to the manufacturer's standard procedures. All reads were assembled with Newbler Assembler, version 2.3 (454 Life Sciences, Branford, CT). Sequencing and assembly were performed by ChunLab Inc. (Seoul, South Korea), and open reading frames (ORFs) were predicted and annotated using Rapid Annotation Subsystem Technology, version 2.0 (RAST; http://rast.nmpdr.org). The potential gene products of the ORFs were compared with the proteins in the nonredundant GenBank databases using BLASTp or PSI-BLAST (http://www.ncbi.nlm.nih.gov/blast/) [32], with an E values of < 10-4. The functional assignment of each ORF was predicted using HHpred, version 2.0 (http://toolkit.tuebingen.mpg.de/hhpred) [33]. To search for tRNA genes, we used tRNAscan-SE, version 1.2.1 (http://lowelab.ucsc.edu/tRNAscan-SE/) [34]. The PHIRE program, version 1.00 (http://www.agr.kuleuven.ac.be/logt/PHIRE.htm) [35], was used to identify phage-specific promoters under the following default parameters: string length, 20; degeneracy, 4; dominanNum, 4; and window size, 20. The ARNold program (http://rna.igmors.u-psud.fr/toolbox/arnold/) [36] was used to predict rho-independent transcription terminators.
Protein family clustering
To cluster the protein sequences predicted from the KBNP1315 genome (n = 62) into orthologous families, we performed pairwise similarity comparisons using BLAST in the A CLAssification of Mobile genetic Elements database, version 0.4 (http://aclame.ulb.ac.be), using an E-value cutoff of 0.0001 [37]. Thirty-four potential gene products of KBNP1315 were found to be associated with protein families in the “viruses” database (as of Jan, 2015), while the remaining proteins were assigned to uncharacterized protein families. The unassigned protein sequences were compared with the protein sequences in the HMMER database (E < 10-4) [38]. Because the ACLAME database lacked information on recently-identified phages [10], we further directly examined protein families from the following putative relatives of KBNP1315: SP6 (GenBank accession number AY370673), K1E (AM084415), K1-5 (NC_008152), ACG-c91 (NC_019403), Era103 (NC_09014), phiEa1H (FQ482084), phiEa100 (NC_019926), PP1 (NC_019542), Vc1 (KJ502657), T7 (GU071091), T3 (NC_003298), YeO3-12 (NC_001271), phiA1122 (NC_004777), gh-1 (NC_004665), Berlin (NC_008694), K1F (NC_007456), phiKMV (NC_005045), VP4 (NC_007149), P60 (NC_003390), P-SSP7 (NC_006882), Syn5 (NC_009531), LKA1 (NC_009936), LKD16 (NC_009935), KP34 (NC_013649), F19 (NC_023567), VP93 (NC_012662), GAP227 (NC_020078), phiAS7 (NC_019528), and phi80-18 (NC_019911). A subset of predicted protein sequences (n = 564) retrieved from ACG-c91, phiEa1H, phiEa100, PP1, Vc1, KP34, F19, VP93, GAP227, phiAS7, and phi80-18 were subjected to clustering by comparison with the ACLAME and HMMER databases, as described for KBNP1315.
Network construction, analysis, and decomposition into clusters
The results of pairwise comparisons were used to generate a matrix in which the rows represented mobile genetic elements (MGEs) and the columns represented the protein families. We then determined the similarities of KBNP1315, ACG-c91, PP1, phiEa1H, phiEa100, VC1, KP34, F19, VP93, GAP227, phiAS7, and phi80-18 to their respective related MGEs, as previously described [11]. We determined the phage-phage similarity (the Sig score) as the minus logarithmic score by multiplying the hypergeometric similarity P-value by the total number of pairwise comparisons [11].
MGEs with Sig values > 1 were used to construct a new network of bacteriophage genome-encoded protein-sharing relationships among KBNP1315 and its potential relatives. The network was visualized with the Cytoscape software, version 3.0.2 (http://cytoscape.org/) [39], which uses an edge-weighted spring-embedded model in which MGEs that share more protein families are placed closer together in the network. We then estimated the topological properties of the network with the Network Analyzer 2.7 Cytoscape plug-in [11].
The Markov clustering (MCL) algorithm was used to assign phages into clusters. The main parameter of MCL algorithm is called “inflation”, which influences the number and size of the clusters. We tested all inflation values from 1.2 to 5 by steps of 0.2 and analyzed each clustering result to maximize the cluster homogeneity. We then calculated the membership as the proportion of edges (or the “weight”) that linked each node to each cluster, using the Network Analysis Tools (NeAt; http://rsat.ulb.ac.be/neat/) [40].
Functional characterization of the lysis cassette of KBNP1315
The primers and plasmids used throughout this study are listed in S1 Table. To construct plasmids encoding the putative holin and endolysin genes of KBNP1315, the phage DNA was PCR amplified with specific primers hol F and -R, and endolysin F and -R. The obtained PCR products were cloned into the pET-DEST42 expression vector (Invitrogen, CA, USA) to generate pDEST-hol and pDEST-lys. To clone holin and endolysin together, pDEST-hol, including the T7 promoter and terminator, was amplified with primers insert F and -R. The pDEST-lys was linearized by PCR using primers linearizing F and -R, which contained 16 bp of overlapping homologous sequences found in the amplified fragment of pDEST-hol. The PCR products were purified and fused to form a circular plasmid containing both ORFs, using an EZ-fusion cloning kit (Enzynomics, Dajeon, Korea). All constructs were verified by DNA sequencing analysis. The generated plasmids were transformed to E. coli BL21 (DE3), and expression was induced with 0.3 or 1 mM IPTG. To block the SecA secretion system, 0.2, 1, or 5 mM sodium azide (NaN3) was added at the time of induction. For our growth pattern analysis, the optical density was measured by spectrometry at 600 nm every 30 minutes or 1 hr.
Nucleotide sequence accession numbers
The sequences of bacteriophage KBNP1315 have been deposited in GenBank under accession number KJ749827.
Results
Determination of E. coli KBP1315 as an APEC strain
To determine whether E. coli strain KBP1315 represented an APEC strain, we performed PCR amplification of six well-characterized virulence genes [41–43]: hlyF (hemolysin), papC (p-fimbria), iucD, fyuA, irp2, and iroN (iron uptake-related gene). All six APEC-virulence genes were identified in this bacterium (S1 Fig).
Phage morphology
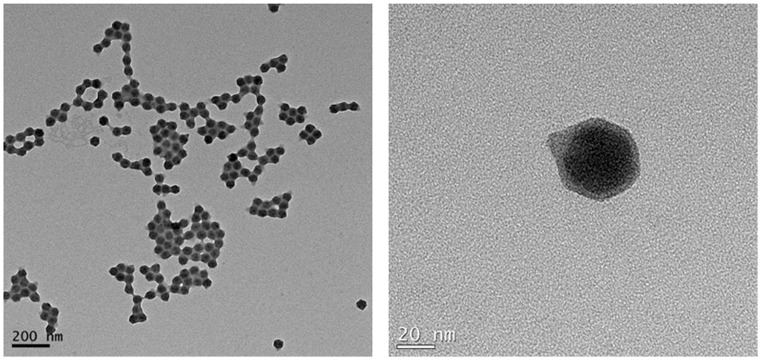
TEM-based showed that phage KBNP1315 has an icosahedral capsid and a short tail (Fig 1). The phage head was 47.71 ± 1.51 nm in diameter, while its tail was 11.9 ± 1.64 nm in length. Based on these morphological characteristics, we propose that KBNP1315 belongs to the C1 morphotype of the Podoviridae family [44].
Fig 1. Morphology of bacteriophage KBNP1315.
Transmission electron micrograph (TEM) of purified phage particles negatively stained with 2% uranyl acetate.
Genome sequencing and annotation
The genome of KBNP1315 is a linear dsDNA of 45,403 bp, including 208 bp of direct terminal repeats (GC content, 45.89%). Sixty-two open reading frames (ORFs) were predicted using an automatic analysis tool available through the RAST server. Annotation of the predicted proteins by BLASTp, PSI-BLAST, and HHpred identified only 27 of the ORFs. They were categorized into the following functional groups: a) transcription/translation (ORF 6, ORF 9, and ORF 21); b) DNA replication/modification (ORF 13, ORF 22, ORF 26, ORF 27, ORF 28, ORF 29, ORF 31); c) phage morphogenesis (ORF 37, ORF 38, ORF 39, ORF 40, ORF 41, ORF 44, ORF 45, ORF 47, ORF 48, ORF 49, ORF 56); d) host lysis (ORF 51); and e) nucleotide metabolism (ORF 11, ORF 20, ORF 30, ORF 35, and ORF 50). Seven phage-specific promoters were predicted, all of which were predicted to function in the rightward direction. We identified three rho-independent transcription terminators. No tRNA motif was predicted (S2 Table).
Pairwise comparison of KBNP1315
Pairwise comparison of the predicted ORFs of KBNP1315 and its relatives (SP6, Vc1, K1E, K1-5, ACG-c91, Era103, phiEa1H, Ea100, PP1, T7, T3, YeO3-12, gh-1, K1F, Berlin, phiA1122, VP4, P60, Syn5, P-SSP7, phiAS7, GAP227, phi80-18, phiKMV, LKA1, LKD16, KP34, F19 and VP93) against 11,475 protein families from 457 virus (phage) genomes allowed us to classify 1,366 protein sequences into 531 protein families, including 178 uncharacterized protein families (S3 Table) [45]. The resulting matrix (in which columns represented protein families and rows represented phage genomes) revealed that KBNP1315 shared the most protein families with phages belonging to the Autographivirinae subfamily, especially the SP6- and T7-like phages [46]. Among these phage groups, we identified seven protein families common to the genomes of KBNP1315 and its relatives (more than 29 phages), namely families vir:14 (putative DNA polymerase function), f:v:81 (terminase large subunit), f:v:99 (portal protein), f:v:100 (exonuclease), f:v:120 (RNA polymerase) and f:v:133 and 134 (tail fibers). Our findings suggest that these sets of genetic components appear to have been conserved during the evolution of the Autographivirinae phages. Notably, RNA polymerase was previously reported as signature for the Autographivirinae phages [47].
Network construction, analysis and decomposition
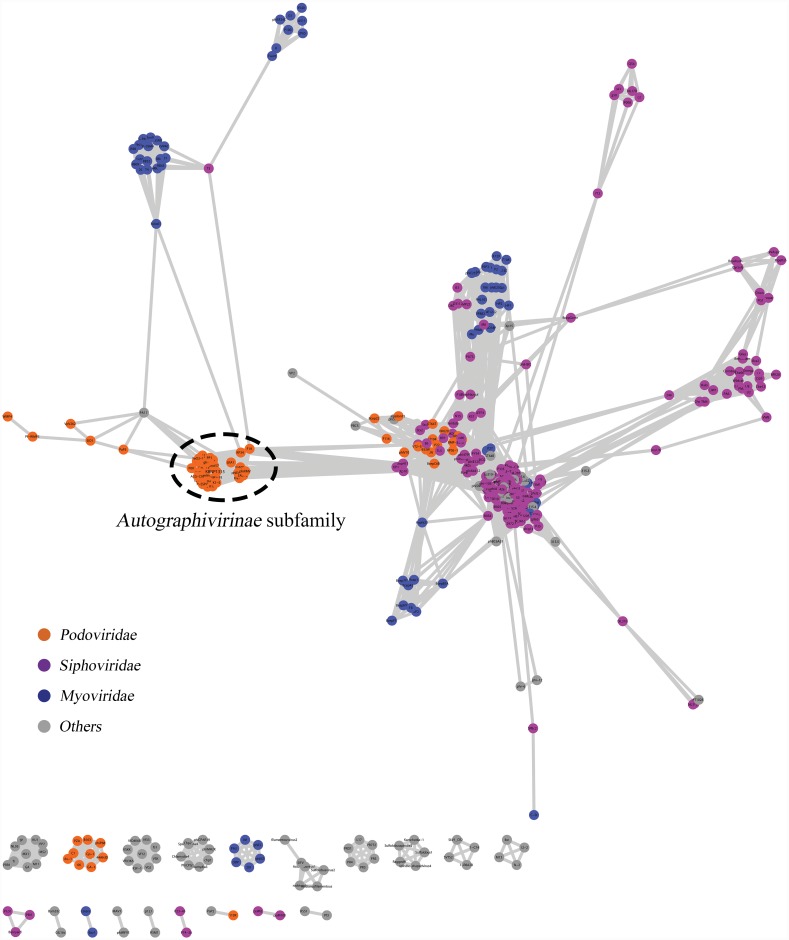
Based on the number of protein families shared between the considered phages, we established a weighted network in which each node represented the phage and each edge represented the phage-phage similarity (called the Sig score) [10, 11]. To analyze the relationships among the nodes, we computed network topological values, including the clustering coefficient (CC, which represents the interconnectivity scale of a given node with the neighbors) and the betweenness centrality (BC, which indicates the extent which a given node lies on the shortest path between all the other nodes) [48]. The resulting network consisted of 431 nodes belonging to Podoviridae, Myoviridae, Siphoviridae, and uncharacterized/other phages; 6,823 edges; and 20 networks of different sizes (Fig 2). Notably, KBNP1315 was located in the peripheral region of the largest network that comprised a dense cluster of phages belonging to the Autographivirinae subfamily (SP6-, T7-, phiKMV-, and P60-) and putative members of this subfamily (GAP227, phiAS7, phi80-18, KP34, F19, and VP93) [46, 47, 49]. In terms of network topologies, the phages, KBNP1315 and its relatives formed a highly interconnected cluster with an average CC value of 0.94 (absolute cohesiveness = 1) (data not shown). Among the highly interconnected members of Autographivirinae, some nodes (e.g., those representing phiKMV, LKD16, LKA1, phiEa1H, Ea100, Era103, KP34, and F19) were slightly closer to the center of network, perhaps reflecting that these phages had multiple connections with other phages.
Fig 2. Protein-sharing network of KBNP1315.
A network for KBNP1315 was produced using Cytoscape 3.1.2. Nodes represent phages, while edges indicate significant relationships between nodes. Each node is colored in accordance with its ICTV classification.
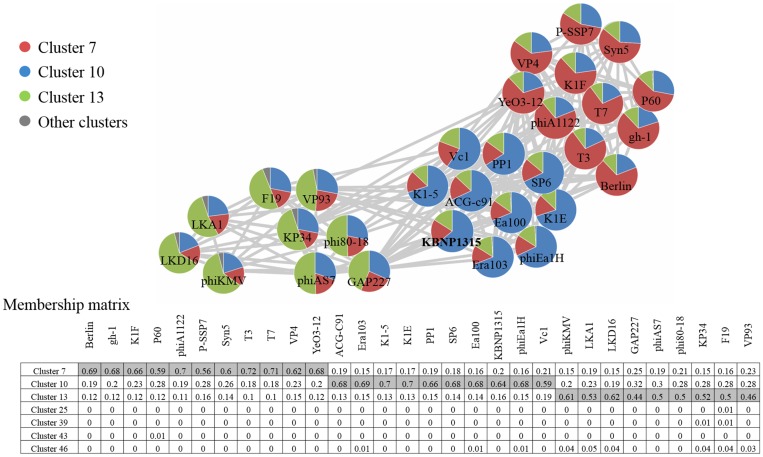
The MCL algorithm was used to clarify subgroups of the Autographivirinae phages as this has been a useful approach for extracting groups of densely connected phage genomes (i.e., clusters) from such networks [40, 50]. As a result, MCL clustering with an inflation value of 2.6 (data not shown) produced a total of 64 clusters (S4 Table). As a result, the 30 phages of the Autographivirinae group including KBNP1315 were classified into clusters 7, 10, and 13, comprising SP6-like phages, T7-like phages plus three unassigned phages (P60, Syn5, and P-SSP7) and phiKMV-like phages plus KP34 like phages, respectively. We further analyzed the membership of each node within these three clusters to represent how strongly each node is related to each cluster [11, 40]. The obtained membership matrix is represented by an independent network and the included phages all showed the highest membership to their respective clusters, supporting that these phages were appropriately clustered by MCL algorithm (Fig 3). Interestingly, KBNP1315 exhibited a much higher membership to cluster 10 (0.64) compared to cluster 7 (0.2) or 13 (0.16), indicating that it has a strong relationship with the SP6-like phages. On the other hand, the 11 phage members of cluster 7 showed overall high memberships to cluster 7 (0.68 on average), with the exceptions of P60, Syn5, and P-SSP7 (less than or equal to 0.6), suggesting that the latter three phages have a more distant relationship with the T7-like phages. Also, nine phages including phiKMV-, KP34- and GAP227-like phages classified into cluster 13 revealed high degrees of mosaicism and chimerism, reflecting that they shared complicated relationships with other members of the network (Fig 2). In particular, KP34 and F19 were assigned to five and six clusters, respectively; this is consistent with their high BC value and provides additional evidence that they may function as bridges between unrelated phages.
Fig 3. Autographivirinae subfamily phages are represented as an independent network.
Each node is illustrated as a pie chart, with the wedges representing the proportion of edges connected with nodes belonging to each cluster. The left panel shows the clusters in different colors. The lower panel represents the membership matrix. The rows indicate phages related to Autographivirinae subfamily and the columns represent the clusters that share wedges with the nodes more than 0.01. Shared boxes indicate the clusters into which the phages were assigned by the MCL algorithm.
Comparative genome analysis of KBNP1315
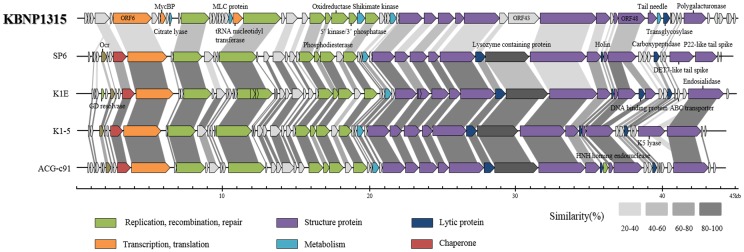
To better understand the evolutionary relationship between KBNP1315 and the SP6-like phages, the genome of KBNP1315 was compared with its closest relative genomes (SP6, K1E, K1-5, and ACG-c91). As shown in Fig 4, the genome of KBNP1315 was found to have a gene order similar to those of the other SP6-like phages, but its ORFs had relatively low sequence identities with those of the other SP6-like phages: 23 to 58% with SP6 (average, 41%) and 25 to 64% with K1E (40%) (Fig 4 and S5 Table). Interestingly, our comparison of SP6 phage that infects a different host (Salmonella enterica Typhimurium LT2) versus several coliphages (K1E, K1-5, and ACG-c91) revealed higher average similarities (74, 72, and 71%, respectively) than those with KBNP1315 (S5 Table). This suggests that KBNP1315 may have shared a common ancestor with the SP6-like phages, but diverged from them some time ago [13, 16].
Fig 4. Comparative genome analysis among KBNP1315 and select relatives (KBNP1315, SP6, K1E, K1-5, and ACG-c91).
Each ORF is represented by a color assigned according to the protein function. The degree of amino acid similarity between paired ORFs was analyzed by BLASTp search, and the percentage of similarity is represented in color from pale gray to dark gray.
In addition, comparative analysis revealed a unique feature with respect to the host specificity of KBNP1315. Deduced ORF 56, which is encoded at the far right end of the genome, is a putative tail spike protein that is likely to play an important role in recognizing the host outer membrane. Our HHpred analysis revealed that ORF 56 had strong similarities to the endo-polygalacturonase from Erwinia carotovora ssp. carotovora and this enzyme is totally different from those of its relatives (SP6; DET7-liketail spike and P22-like tail spike/ K1E, K1-5, ACG-c91; endosialidase, K5 lyase (only K1-5)).
We further identified the separated position of host cell lysis genes of KBNP1315, including holin, endolysin, Rz and Rz1 (Fig 4). ORFs 46, 51, 54, and 55 of KBNP1315 encode putative versions of holin, endolysin, Rz, and Rz1, respectively. Despite the conserved position of these lysis genes, the putative holin, endolysin, Rz and Rz1 sequences of KBNP1315 share low amino acid similarities with those of the other SP6-like phages, with the putative endolysin-encoding sequence failing to show any similarity. We therefore speculated that KBNP1315 may use a different strategy to lyse the host bacterial cell wall at the end of the infection cycle, compared to the other SP6-like phages. Consistent with this, most phages with known lytic mechanisms have their lysis genes encoded in canonical serial order [24], whereas those of KBNP1315 are encoded separately. To better understand the mechanisms through which KBNP1315 lyses the host cell, we focused on the functions of holin and endolysin, which cleave the inner cell membrane and the peptidoglycan layer, respectively, of the bacterial cell wall.
Functional analysis of the lysis proteins of KBNP1315
Our analysis with the SignalP 4.1 server revealed that ORF 51 possesses a putative signal sequence and a cleavage site embedded in the C-region, but has no putative positive anchor region at its N-terminus (S2 Fig). Thus, we propose that ORF 51 encodes a putative SAR endolysin that should be secreted in a membrane-tethered form via the host sec system and be released via signal peptidase cleavage [23].
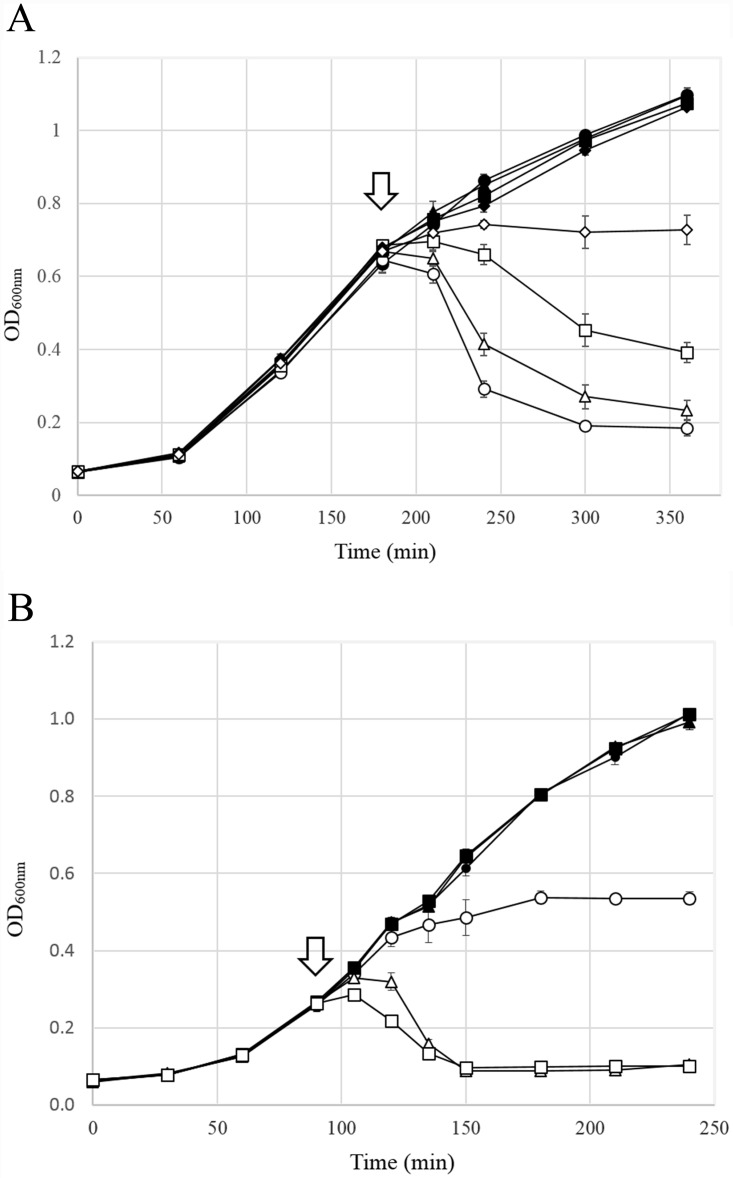
To evaluate the involvement of the host sec system in the exportation of the putative SAR endolysin of KNBP1315, we induced the expression of this endolysin in E.coli BL21 (DE3) competent cells using 1 mM IPTG, added sodium azide (NaN3, at 0.1, 0.5, and 1 mM; to inhibit the ATPase activity of SecA) to induced or non-induced cultures, and observed cell growth by monitoring the absorbance at 600 nm. In the NaN3-free induction group, the addition of 1 mM IPTG resulted in cell lysis: the optical density decreased slightly from 0.645 to 0.607 over the first 30 minutes after induction, and then decreased rapidly from 0.607 to 0.292 over the next 30 minutes due to rapid cell lysis (Fig 5A). In contrast, the NaN3-treated induction groups showed dose-dependent delays in lysis (Fig 5A). The 0.1 mM NaN3-treated group showed a lysis pattern similar to that of the NaN3-untreated group, although there was a slight post-induction decline in OD. Compared with the untreated control group, in which rapid lysis was observed 30 minutes post-induction, the 0.5 mM NaN3-treated group showed a delayed lysis pattern, with lysis first observed at 30 minutes post-induction and drastically increased (as reflected by a sharp decrease in OD) over the succeeding 30 minutes. The group treated with 1 mM NaN3 showed a complete blockade of cell lysis for at least 3 hours post-induction, and did not have any noticeable effect on non-induced cells. Based on these results, we conclude that KBNP1315 ORF51 acts as a typical SAR endolysin [24] by causing host cell lysis via the host sec pathway.
Fig 5. Growth curves of E.coli BL21 cells expressing two putative lysis proteins of KBNP1315.
(A) ORF 51 (putative endolysin) was expressed in E.coli BL21 in the presence of different concentrations of NaN3. IPTG (1 mM) was added at 90 min (arrow) in the induction group (open symbols). The non-induction group is represented with closed symbols. NaN3 was simultaneously applied at concentrations of 0 mM (circle), 0.1 mM (triangle), 0.5 mM (square), and 1 mM (diamond). (B) Growth curves of induction (open symbols) and non-induction (closed symbols) groups expressing ORF46/pDEST42 (circle), ORF51/pDEST42 (triangle), and ORF46/ORF51/pDEST42 (square) are shown. Induction was performed by the addition of 0.3 mM IPTG (arrow).
Next, we analyzed the function of the putative holin protein in KBNP1315. The deduced amino acid sequence of ORF 46 was predicted to possess a single transmembrane domain (residues 35 to 57) and a putative N-out, C-in topology, suggesting that the gene product of ORF 46 has a structure similar to that of the class III holins (S3 Fig) [51]. To characterize the holin-like protein of KBNP1315, we cloned ORF 46 and/or ORF 51 into the pDEST42 vector and transformed the resulting plasmids to E. coli BL21. To examine the lytic function of the putative holin protein, we gradually expressed ORF 46 with and without ORF 51 using a low concentration of IPTG (0.3 mM). The expression of ORF 46 alone resulted in a moderate decrease of optical density at two hours post-induction (OD = 0.563) (Fig 5B). In contrast, the expression of the putative endolysin (ORF 51) alone yielded a dramatic decrease in optical density at 90 min post-induction (OD = 0.089). The coexpression of both ORF 46 and ORF 51 was associated with an immediate response (a slight decrease in growth) following the addition of IPTG. There was no difference in the time taken to achieve complete lysis between the endolysin- and holin/endolysin-expressing groups; however, the optical density of the coexpression group was more strongly decreased at 60 minutes post-induction (ODORF46,51 = 0.219, ODORF51 = 0.320). Although the difference between the induced and non-induced conditions in terms of optical density after 60 min was greater for the group treated with the higher concentration of IPTG (1 mM) compared to the 0.3 mM induction condition, the two induction conditions yielded similar patterns (data not shown). These observations suggest that holin may not be an essential protein for host lysis in KBNP1315, but rather it appears to work synergistically with endolysin to enhance host lysis.
Discussion
In this study, we newly purified a coliphage and designated it KBNP1315, as it was shown to infect avian pathogenic E. coli strain KBP1315. We then performed genomic and functional characterizations of this new phage. Pairwise comparison and a mathematically modeled gene (protein)-sharing network revealed that KBNP1315 has several informative evolutionary connections with phages belonging to the Autographivirinae subfamily. Interestingly, the examined phage genomes showed strong conservation of essential genes involved in DNA replication, transcription, packaging and morphogenesis, including those encoding putative RNA polymerase, DNA polymerase, portal protein, terminase large subunit, exonuclease and tail fiber proteins (S3 Table). This allowed the phages to densely connect with one another and form a highly interconnected cluster in the generated protein-sharing network. These observations suggest that the conserved homologs have undergone flow among the studied phage genomes beyond the genus level. Thus, our findings suggest that these phages have a common ancestor and have taken advantage of their shared genetic pool to exchange functional genetic elements during their evolution [52].
Further analysis of the assignment of various nodes to different clusters on the basis of MCL clustering allowed us to classify the members of the Autographivirinae subfamily into three subgroups. Especially, KBNP1315 showed its highest membership to cluster 11 which included members of the SP6-like phages. Since the highest value of ICCC helps in identifying clusters of densely related phages consisting of unique genetic module-shared descendants, this result reveals that KBNP1315 is one of the member of SP6-like phages and most likely shares vertically inherited modules with the SP6-like phages [11]. However, although MCL clustering showed a close relationship among KBNP1315 and SP6-like phages, comparative genomic analysis provided three differences in this phage’s genome from those of other SP6-like phages studied herein. First, although KBNP1315 shares a conserved gene order with other members in this group, the amino acid identities are low, possibly indicating that KBNP1315 is the most widely divergent member of the SP6-like phages [16]. Second, KBNP1315 encodes a unique tail spike protein for attachment to the host membrane. As tail spikes give phages their infectious specificity, the observation that the tail spike of KBNP1315 (endo-polygalacturonase) differs from that of other coli phages (e.g., endosialiase, K5 lyase) within the SP6-like phages supports the notion that the former has developed a disparate strategy for host recognition [18, 53, 54]. Third, the holin, endolysin and Rz/Rz1-like proteins of KBNP1315 also differ strongly from those of the other SP6-like phages, indicating that the former uses a different host lysis mechanism [21, 54, 55]. Collectively, these observations may suggest that KBNP1315 shares a common ancestor with the SP6-like phages but diverged a long time ago, and that KBNP1315 uses atypical strategies for infection and host lysis, and appears to have taken advantage of the HGT process to adjust to various environmental factors.
Our observation that ORF 51 could trigger host cell lysis independent of holin function confirms that KBNP1315 uses a SAR endolysin in its lysis system. Although the catalytic triad (E-8aa-D/C-5aa-T) found in the endolysin sequences from P1, 21, phiKMV, Era103 and T4 is not conserved in ORF 51 (S2 Fig), the enzymatic activity of the latter is comparable with those of SAR endolysins containing these catalytic residues [23, 24, 25]. Most SAR endolysins (roughly 85%) contain the lysozyme-type catalytic domain; however, the lack of conserved catalytic residues in ORF 51 may indicate that it has a different type of catalytic domain. Indeed, our HHpred analysis indicated that, unlike the catalytic domains of P1, 21, phiKMV, Era103 and T4 endolysin, that of ORF51 is related to the soluble lytic transglycosylase (SLT) domain found in Burkholderia, Campylobacter, Escherichia, and Pseudomonas phage endolysins [56]. We found that expression of ORF51 triggered host lysis without the initial delay observed from several other SAR endolysins; this may reflect the unique structural features of its SAR domain that appears to lack the positively charged anchor and the signal peptide cleavage site [23, 24, 27, 57]. We therefore conclude that the SAR endolysin of KBNP1315 is easily released from the cytoplasmic membrane by the signal peptidase, and possesses quite a different PG-degrading enzyme from other SAR endolysins. On the other hand, another putative lytic protein, encoded by ORF46, moderately inhibited cell growth and had a synergistic effect on SAR endolysin-mediated lysis of host cells. In addition, the secondary structure of ORF 46 (an N-out, C-in topology with a single TMD-spanning α-helix domain spanning residues 33–57) is similar to that of gp5 of mycobacteriophage Ms6, which forms lesions that are actually too small to translocate the canonical endolysins from phage λ and Ms6 [51]. Although the exact role of ORF 46 remains elusive, we suggest that its protein product functions as a pin-holin. Together, our findings strongly suggest that KBNP1315 uses a pin-holin/SAR endolysin pathway for host lysis. We also provide the first experimental evidence that holin and endolysin can have synergistic lytic activity in SP6-like phages.
In summary, we herein used gene (protein)-sharing network and comparative analyses to perform an evolutionary study of novel phage KBNP1315. Our results strongly suggest that this phage is a distantly related member of SP6-like phages with unique genetic features, tail spike and lytic proteins revealing its atypical strategies for infection and host lysis. Moreover, our functional study of two lytic proteins of KBNP1315 demonstrates their important roles for host lysis and we believe that this work could contribute to the development of a novel antimicrobial approach for controlling avian colibacillosis.
Supporting Information
(PPTX)
(PPTX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
This work was made possible by a Korea Research Foundation grant funded by the Ministry of Science, ICT and Future Planning of South Korea (NRF- 2013R1A1A1059608).
Data Availability
GenBank under accession number KJ749827.
Funding Statement
This work was made possible by a Korea Research Foundation grant funded by the Ministry of Science, ICT and Future Planning of South Korea (NRF- 2013R1A1A1059608). KBNP Technology Institute, KBNP, Inc., provided support in the form of salaries for authors KHK and THK, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1. Barnes H, Vaillancourt J, Gross W. Colibacillosis. Diseases of poultry. 2003;12:691–732. [Google Scholar]
- 2. Kabir S. Avian colibacillosis and salmonellosis: a closer look at epidemiology, pathogenesis, diagnosis, control and public health concerns. International journal of environmental research and public health. 2010;7(1):89–114. 10.3390/ijerph7010089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oliveira A, Sereno R, Azeredo J. In vivo efficiency evaluation of a phage cocktail in controlling severe colibacillosis in confined conditions and experimental poultry houses. Veterinary microbiology. 2010;146(3):303–8. [DOI] [PubMed] [Google Scholar]
- 4. Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, et al. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Applied and environmental microbiology. 2008;74(22):7043–50. 10.1128/AEM.01395-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Fakhr MK, Nolan LK. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151(6):2097–110. [DOI] [PubMed] [Google Scholar]
- 6. Kim TE, Jeong YW, Cho SH, Kim SJ, Kwon HJ. Chronological study of antibiotic resistances and their relevant genes in Korean avian pathogenic Escherichia coli isolates. Journal of clinical microbiology. 2007;45(10):3309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahman M, Samad M, Rahman M, Kabir S. In vitro antibiotic sensitivity and therapeutic efficacy of experimental salmonellosis, colibacillosis and pasteurellosis in broiler chickens. Bangladesh Journal of Veterinary Medicine. 2004;2(2):99–102. [Google Scholar]
- 8. Cloud S, Rosenberger J, Fries P, Wilson R, Odor E. In vitro and in vivo characterization of avian Escherichia coli. I. Serotypes, metabolic activity, and antibiotic sensitivity. Avian diseases. 1985:1084–93. [PubMed] [Google Scholar]
- 9. Bass L, Liebert CA, Lee MD, Summers AO, White DG, Thayer SG, et al. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli . Antimicrobial Agents and Chemotherapy. 1999;43(12):2925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jang HB, Fagutao FF, Nho SW, Park SB, Cha IS, Yu JE, et al. Phylogenomic Network and Comparative Genomics Reveal a Diverged Member of the ϕKZ-Related Group, Marine Vibrio Phage ϕJM-2012. Journal of virology. 2013;87(23):12866–78. 10.1128/JVI.02656-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Molecular biology and evolution. 2008;25(4):762–77. 10.1093/molbev/msn023 [DOI] [PubMed] [Google Scholar]
- 12. Brüssow H, Hendrix RW. Phage genomics: small is beautiful. Cell. 2002;108(1):13–16. [DOI] [PubMed] [Google Scholar]
- 13. Hatfull GF. Bacteriophage genomics. Current opinion in microbiology. 2008;11(5):447–53. 10.1016/j.mib.2008.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dagan T. Phylogenomic networks. Trends in microbiology. 2011;19(10):483–491. 10.1016/j.tim.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 15. Dobbins AT, George M, Basham DA, Ford ME, Houtz JM, Pedulla ML, et al. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. Journal of bacteriology. 2004;186(7):1933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scholl D, Kieleczawa J, Kemp P, Rush J, Richardson C, Merril C, et al. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. Journal of molecular biology. 2004;335(5):1151–71. [DOI] [PubMed] [Google Scholar]
- 17. Butler ET, Chamberlin M. Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. Journal of Biological Chemistry. 1982;257(10):5772–8. [PubMed] [Google Scholar]
- 18. Stummeyer K, Schwarzer D, Claus H, Vogel U, Gerardy-Schahn R, Muhlenhoff M. Evolution of bacteriophages infecting encapsulated bacteria: Lessons from Escherichia coli K1-specific phages. Molecular microbiology. 2006;60(5):1123–35. [DOI] [PubMed] [Google Scholar]
- 19. Kuty GF, Xu M, Struck DK, Summer EJ, Young R. Regulation of a phage endolysin by disulfide caging. Journal of bacteriology. 2010;192(21):5682–7. 10.1128/JB.00674-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang I-N, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annual Reviews in Microbiology. 2000;54(1):799–825 [DOI] [PubMed] [Google Scholar]
- 21. Young R. Phage lysis: do we have the hole story yet? Current opinion in microbiology. 2013;16(6):790–7. 10.1016/j.mib.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Young R, Wang N, Roof WD. Phages will out: strategies of host cell lysis. Trends in microbiology. 2000;8(3):120–8. [DOI] [PubMed] [Google Scholar]
- 23. Xu M, Struck DK, Deaton J, Wang N, Young R. A signal-arrest-release sequence mediates export and control of the phage P1 endolysin. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(17):6415–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Briers Y, Peeters LM, Volckaert G, Lavigne R. The lysis cassette of bacteriophage фKMV encodes a signal-arrest-release endolysin and a pinholin. Bacteriophage. 2011;1(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lynch KH, Abdu AH, Schobert M, Dennis JJ. Genomic characterization of JG068, a novel virulent podovirus active against Burkholderia cenocepacia . BMC genomics. 2013;14(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park T, Struck DK, Dankenbring CA, Young R. The pinholin of lambdoid phage 21: control of lysis by membrane depolarization. Journal of bacteriology. 2007;189(24):9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sao-Jose C, Parreira R, Vieira G, Santos MA. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin Behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal Cells. Journal of bacteriology. 2000;182(20):5823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murray PR, Baron EJ, Pfaller M, Tenover F, Yolken RH, Morgan DR. Manual of Clinical Microbiology (6th edn). Trends in microbiology. 1995;3(11):449–449. [Google Scholar]
- 29. Clokie MR, Kropinski AM. Bacteriophages: Springer; 2009. [Google Scholar]
- 30.Adams MH. Bacteriophages. Bacteriophages. 1959.
- 31. Green MR, Sambrook J. Molecular cloning: a laboratory manual: Cold Spring Harbor Laboratory Press; New York; 2012. [Google Scholar]
- 32. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33(suppl 2):W244–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic acids research. 2005;33(suppl 2):W686–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lavigne R, Sun W, Volckaert G. PHIRE, a deterministic approach to reveal regulatory elements in bacteriophage genomes. Bioinformatics. 2004;20(5):629–35. [DOI] [PubMed] [Google Scholar]
- 36. Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA biology. 2011;8(1):11–3. [DOI] [PubMed] [Google Scholar]
- 37. Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic acids research. 2004;32(suppl 1):D45–D9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic acids research. 2011:gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brohée S, Faust K, Lima-Mendez G, Sand O, Vanderstocken G, Deville Y, van Helden J. NeAT: a toolbox for the analysis of biological networks, clusters, classes and pathways. Nucleic acids research. 2008;36(suppl 2):W444–W51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janben T, Schwarz C, Preikschat P, Voss M, Philipp H-C, Wieler LH. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. International Journal of Medical Microbiology. 2001;291(5):371–8. [DOI] [PubMed] [Google Scholar]
- 42. Schubert S, Rakin A, Karch H, Carniel E, Heesemann J. Prevalence of the "High-Pathogenicity Island" of Yersinia species among Escherichia coli strains that are pathogenic to humans. Infection and immunity. 1998;66(2):480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Johnson TJ, Siek KE, Johnson SJ, Nolan LK. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. Journal of bacteriology. 2006;188(2):745–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ackermann HW. Frequency of morphological phage descriptions in the year 2000. Archives of virology. 2001;146(5):843–57. [DOI] [PubMed] [Google Scholar]
- 45. Lima-Mendez G, Toussaint A, Leplae R. A modular view of the bacteriophage genomic space: identification of host and lifestyle marker modules. Research in microbiology. 2011;162(8):737–46. 10.1016/j.resmic.2011.06.006 [DOI] [PubMed] [Google Scholar]
- 46. Francki RIB, Fauquet C, Knudson D, Brown F. Classification and Nomenclature of Viruses: Fifth Report of the International Committee on Taxonomy of Viruses Virology Division of the International Union of Microbiological Societies: Springer Science & Business Media; 2012. [Google Scholar]
- 47. Lavigne R, Seto D, Mahadevan P, Ackermann HW, Kropinski AM. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Research in microbiology. 2008;159(5):406–14. 10.1016/j.resmic.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 48. Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24(2):282–4. [DOI] [PubMed] [Google Scholar]
- 49. Grose JH, Casjens SR. Understanding the enormous diversity of bacteriophages: the tailed phages that infect the bacterial family Enterobacteriaceae . Virology. 2014;468:421–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Dongen SM. Graph clustering by flow simulation. PhD Thesis. 2001.
- 51. Catalão MJ, Gil F, Moniz-Pereira J, Pimentel M. Functional analysis of the holin-like proteins of mycobacteriophage Ms6. Journal of bacteriology. 2011;193(11):2793–803. 10.1128/JB.01519-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world'sa phage. Proceedings of the National Academy of Sciences. 1999;96(5):2192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Scholl D, Adhya S, Merril CR. Bacteriophage SP6 is closely related to phages K1-5, K5, and K1E but encodes a tail protein very similar to that of the distantly related P22. Journal of bacteriology. 2002;184(10):2833–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Leiman PG, Battisti AJ, Bowman VD, Stummeyer K, Mühlenhoff M, Gerardy-Schahn R, et al. The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. Journal of molecular biology. 2007;371(3):836–49. [DOI] [PubMed] [Google Scholar]
- 55. Shin H, Lee JH, Yoon H, Kang DH, Ryu S. Genomic investigation of lysogen formation and host lysis systems of the Salmonella temperate bacteriophage SPN9CC. Applied and environmental microbiology. 2014;80(1):374–84. 10.1128/AEM.02279-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oliveira H, Melo LD, Santos SB, Nóbrega FL, Ferreira EC, Cerca N, et al. Molecular aspects and comparative genomics of bacteriophage endolysins. Journal of virology. 2013;87(8):4558–70. 10.1128/JVI.03277-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun Q, Kuty GF, Arockiasamy A, Xu M, Young R, Sacchettini JC. Regulation of a muralytic enzyme by dynamic membrane topology. Nature structural & molecular biology. 2009;16(11):1192–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX)
(PPTX)
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
GenBank under accession number KJ749827.