Abstract
Members of the GATA protein family play important roles in lineage specification and transdifferentiation. Previous reports show that some members of the GATA protein family can also induce pluripotency in somatic cells by substituting for Oct4, a key pluripotency-associated factor. However, the mechanism linking lineage-specifying cues and the activation of pluripotency remains elusive. Here, we report that all GATA family members can substitute for Oct4 to induce pluripotency. We found that all members of the GATA family could inhibit the overrepresented ectodermal-lineage genes, which is consistent with previous reports indicating that a balance of different lineage-specifying forces is important for the restoration of pluripotency. A conserved zinc-finger DNA-binding domain in the C-terminus is critical for the GATA family to induce pluripotency. Using RNA-seq and ChIP-seq, we determined that the pluripotency-related gene Sall4 is a direct target of GATA family members during reprogramming and serves as a bridge linking the lineage-specifying GATA family to the pluripotency circuit. Thus, the GATA family is the first protein family of which all members can function as inducers of the reprogramming process and can substitute for Oct4. Our results suggest that the role of GATA family in reprogramming has been underestimated and that the GATA family may serve as an important mediator of cell fate conversion.
Keywords: GATA transcription factors, lineage specifier, iPSCs
Introduction
For years, pluripotency-associated factors and their rivals, lineage specifiers, have been generally considered to determine the identities of pluripotent and differentiated cells, respectively. In addition to Yamanaka factors (OSKM), several other pluripotency-associated factors have been identified as mediators of cellular reprogramming1,2,3,4. Recently, a few lineage specifiers that were previously considered rivals to pluripotency were reported to substitute for particular Yamanaka factors5,6. This finding suggests a “seesaw” model wherein pluripotency-associated proteins, such as Yamanaka factors, can function as lineage specifiers and differentially direct cell fate. Pluripotency is maintained as a consequence of the balance of different lineage-specifying forces5,7.
Among these lineage specifiers, GATA3, GATA4, and GATA6 have the strongest ability to substitute for Oct4 in reprogramming5. GATA3, GATA4, and GATA6 can inhibit the overrepresented ectodermal lineage markers to facilitate successful reprogramming, highlighting the fine-tuned balance of the different lineage-specifying forces required for pluripotency maintenance. However, the mechanism that links the lineage-specifying cues to the activation of pluripotency remains a “black box”.
GATA3, GATA4, and GATA6 belong to the GATA family of transcription factors, which are important for development and differentiation of multiple mesendodermal lineages. Members of this family, which are all related by a degree of amino acid sequence identity within their zinc-finger DNA-binding domains, are characterized by their ability to bind the DNA sequence “GATA”8. Given the role of GATA1/2/5 in reprogramming, it is intriguing to investigate whether other three GATA family members function as inducers for pluripotency reprogramming.
In this study, we found that all six members of the GATA transcription factor family could substitute for Oct4 and reprogram mouse somatic cells to pluripotency. Additionally, all six members could inhibit ectodermal lineage markers such as Dlx3 and Lhx5. This is consistent with a previous study in which Oct4 and its substitutes inhibited ectodermal lineage markers in the process of pluripotency induction5. A single-site mutation in the conserved DNA-binding region of the GATA family proteins hampered the reprogramming process. In addition, using the secondary MEF induction system, we found that the GATA family could activate transcription factors, such as Sall4, which are important regulators in the pluripotency network. This study provides evidence that lineage specifiers can directly activate particular pluripotency-associated factors. Additionally, our results suggest that the GATA transcription factor family is the first protein family of which all members act as inducers of reprogramming. Together, this study indicates the importance of GATA family in reprogramming which has been underestimated and increases our understanding of the interaction of lineage specifiers with pluripotency-associated factors.
Results
GATA family can enhance reprogramming in place of Oct4
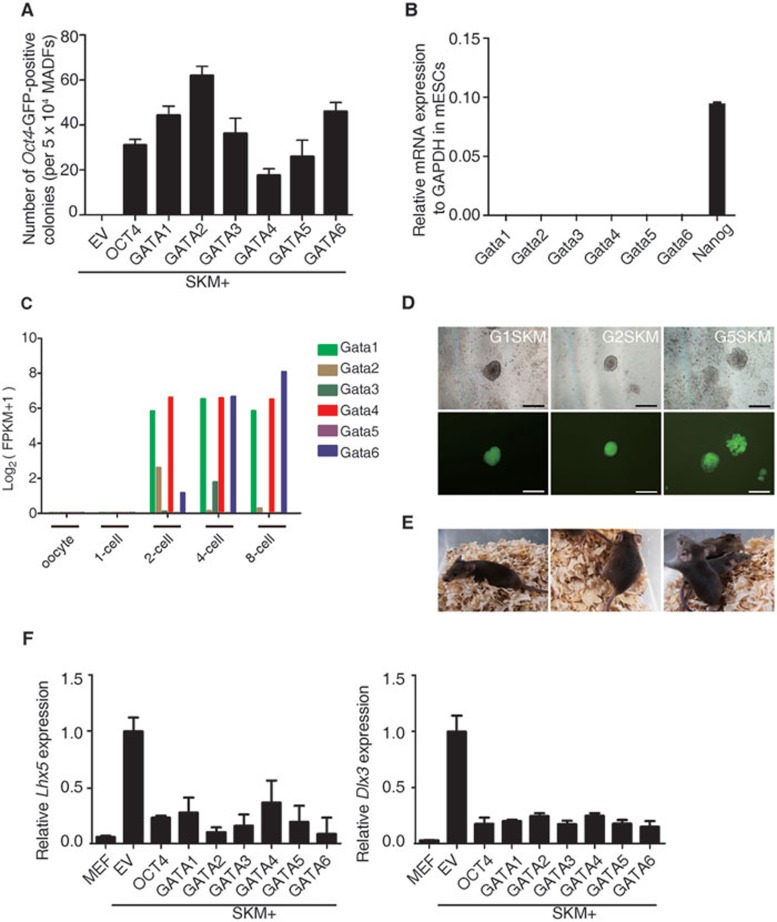
There are six members of the GATA transcription factor family: GATA1, GATA2, GATA3, GATA4, GATA5, and GATA6. During development, each GATA factor shows a specific and regulated expression pattern. GATA1/2/3 are prominently expressed in the hematopoietic system. GATA4/5/6 are not expressed in hematopoietic cells, although they play crucial roles in the formation and differentiation of mesendodermal lineages such as lung, heart, and hepatocytes9,10,11. GATA4 is used for the transdifferentiation of somatic cells into cardiomyocytes and hepatocytes12,13. In addition to their known roles in lineage specification and transdifferentiation, it is important to investigate whether GATA family members can function as inducers for reprogramming of pluripotency. In addition to GATA3, GATA4, and GATA6, which were identified in our previous report5, we tested GATA1, GATA2, and GATA5. We used mouse somatic cells containing a green fluorescent protein (GFP) reporter driven by an Oct4 promoter and enhancer. The human GATA family of transcription factors was inserted into Dox-inducible lentiviral vectors. We found that together with SOX2, KLF4, and c-MYC (SKM), all of the GATA transcription factors could facilitate reprogramming of mouse adult dermal fibroblasts (MADFs) and mouse embryonic fibroblasts (MEFs) to iPSCs in the absence of Oct4 (Figures 1A and 2F). The reprogramming efficiencies of the different GATA transcription factors varied. Among the six GATA transcription factors, GATA4 had a relatively lower reprogramming efficiency during primary infection, whereas the other five had efficiencies comparable to or higher than Oct4. Even proteins closely related to Oct4 could not substitute for Oct4 in somatic cell pluripotency reprogramming14. Thus, the GATA transcription factor family is the first protein family of which all members have been identified to induce pluripotency in mouse somatic cells.
Figure 1.
GATA family members can substitute for OCT4 in pluripotency reprogramming. (A) The reprogramming assay that determines the ability of GATA family members to enhance reprogramming in the absence of OCT4. The Oct4-GFP-positive colonies were counted at 9 days post induction. Induction with empty vector (EV) plus SKM was used as a negative control. Error bars indicate SD (n = 3). (B) Quantitative RT-PCR analysis of the expression of GATA family members, and Nanog in mouse ESCs (R1). R1 cells were cultured in 2i medium. (C) Re-analysis of the expression of GATA family members in early mouse embryo development from published data15. (D) The generation of iPS colonies with G1SKM (left), G2SKM (middle), and G5SKM (right) from Oct4-GFP MADFs. Phase (upper panel) and GFP images (lower panel) of primary iPS colonies. Scale bar, 500 μm. (E) Germline transmission mice (agouti) from G1SKM (left), G2SKM (middle), and G5SKM (right) are depicted. (F) Quantitative RT-PCR analysis of the expression of endogenous Lhx5 (left) and Dlx3 (right) relative to the expression by SKM induction.
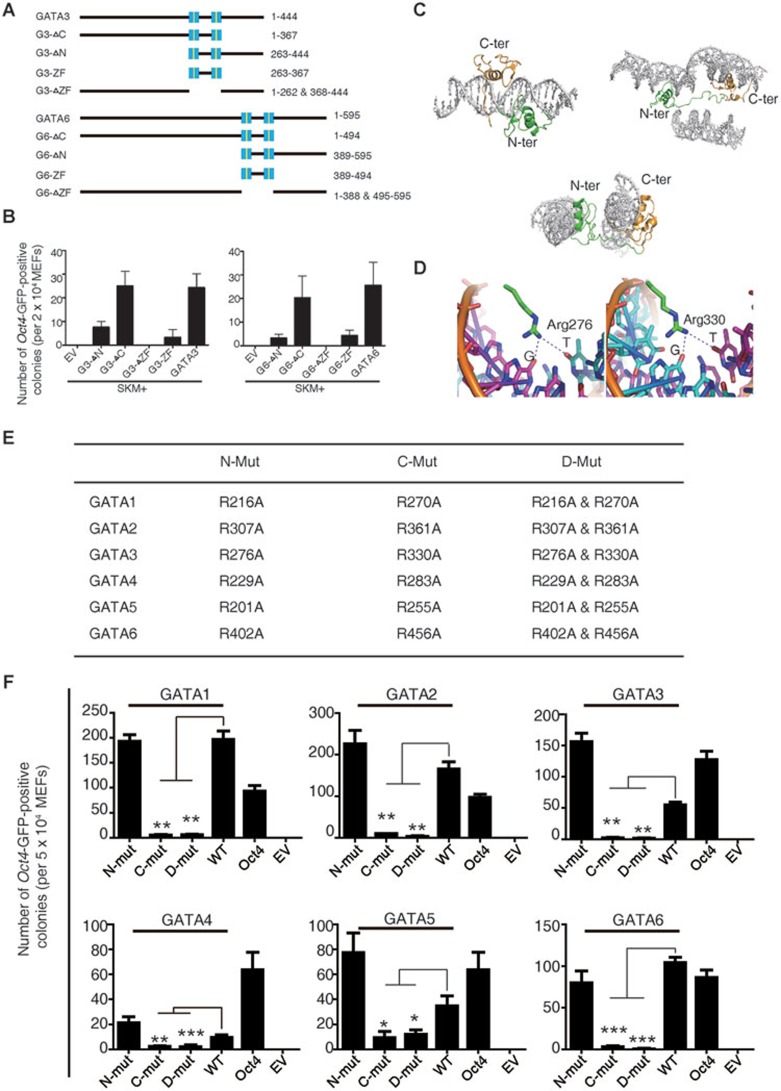
Figure 2.
The GATA DNA-binding domain is critical for GATA-mediated pluripotency reprogramming. (A) Schematic diagrams illustrating various GATA3 and GATA6 deletion mutants. Zinc fingers are highlighted in blue squares. (B) The reprogramming assay that determines the abilities of GATA3 and GATA6 deletion mutants to induce pluripotency. The Oct4-GFP colonies were counted at 9 days post induction. Induction with EV plus SKM was used as a negative control. Error bars indicate SD (n = 3). (C) Overall structure of GATA3/DNA complexes16. Three complexes are listed. The N-terminal zinc finger and the linker are colored in green and the C-terminal zinc finger and C-tail are colored in gold. (D) DNA recognition by the GATA3 DNA-binding domain16. Hydrogen-bonding interactions between Arg276 (N-terminal zinc finger), Arg330, and 3 bp (GAT) of the binding site at their major groove core. (E) Mutation strategy of GATA family members. Alanine was used to substitute for Arginine. (F) Reprogramming assay that determines the abilities of the GATA family member zinc-finger mutants to induce pluripotency, including N-terminal zinc-finger mutants (N-mut), C-terminal zinc-finger mutants (C-mut), and double mutants (D-mut). WT represents wild-type GATA family members. The Oct4-GFP colonies were counted at 9 days post induction. Induction with EV plus SKM was used as a negative control. Error bars indicate SD (n = 3).
Next, we tested the expression of GATA transcription factors in mouse embryonic stem cells (mESCs). Unlike pluripotency-associated factors such as Nanog, GATA transcription factors are not highly enriched in mESCs (Figure 1B). We further analyzed the expression of the GATA transcription factors that were identified during early embryonic development in a previous report15 and found that they were also expressed at early embryonic stages (Figure 1C), indicating that GATA transcription members play important roles in early development.
GATA transcription factor-reprogrammed iPSCs are fully pluripotent
In our previous report, GATA3-, GATA4-, and GATA6-reprogrammed iPSCs were shown to be pluripotent5. We characterized the iPSCs reprogrammed by other GATA transcription factors for pluripotency. The GATA1-, GATA2-, and GATA5-reprogrammed iPSCs began to express Oct4-GFP at 5-6 days post induction and expressed the pluripotency markers NANOG and REX1 (Figure 1D and Supplementary information, Figure S1A). No cross contamination was detected (Supplementary information, Figure S1B). Importantly, we successfully obtained germline-transmitted mice from GATA1-, GATA2-, and GATA5-reprogrammed iPSCs (Figure 1E). Together with the previous report, these results demonstrate that iPSCs generated using GATA transcription factors are pluripotent.
GATA family members inhibit ectodermal lineage markers in reprogramming
Our previous work and Montserrat's work showed that the balance between different lineage-specifying forces during reprogramming could direct final cell fate5,6. We examined whether other GATA transcription factors, i.e., GATA1, GATA2, and GATA5, could inhibit ectodermal lineage markers such as Lhx5 and Dlx3 in the same manner as GATA3, GATA4, and GATA6. Consistent with our previous report, we found that all GATA family members could inhibit ectodermal lineage markers during reprogramming, ensuring that no single lineage-specifying force dominated the others, thereby preserving pluripotency induction (Figure 1F and Supplementary information, Figure S2).
GATA DNA-binding domain is critical for GATA-mediated reprogramming
We next asked why all six GATA transcription factors were capable of enhancing reprogramming. To investigate this question, we examined the structurally conserved domain of the six proteins. We hypothesized that the GATA DNA-binding domain, which is conserved across all six GATA transcription factors, might be related to the shared function of these family members9. We found two conserved zinc-finger domains in the GATA family (Figure 2A and Supplementary information, Figure S3), and using deletion fragments of GATA3 and GATA6 as examples, we found that the fragments containing the two zinc fingers were able to induce reprogramming (Figure 2B). We narrowed our focus to the two zinc-finger regions and found that deletion of these regions abolished the ability of both proteins to induce reprogramming of cellular pluripotency. The overexpression of a fragment of the two zinc-finger regions together with SOX2, KLF4 and c-MYC, induced pluripotency, although at a low efficiency (Figure 2B). Taken together, these results indicate that the two zinc-finger domains are critical for GATA-mediated reprogramming.
Based on the previously reported structure of GATA316 (Figure 2C and 2D), we tested whether the DNA-binding site in each zinc finger was critical for GATA transcription factor-mediated pluripotency reprogramming. Mutation of the conserved putative DNA-binding site16 within the N-terminal zinc finger, which recognizes guanine, had little effect on reprogramming. In contrast, mutation of the conserved putative DNA-binding site within the C-terminal zinc finger hindered GATA-mediated reprogramming. More importantly, all members of the GATA protein family share this same characteristic (Figure 2E and 2F, Supplementary information, Figure S4A and S4B). Furthermore, we found that mutants of GATA family members can barely inhibit the overrepresented ectodermal genes (Supplementary information, Figure S5). These results suggest that the DNA-binding site in the C-terminal zinc finger of GATA transcription factors is critical for successful reprogramming.
GATA family can activate the pluripotency-associated gene Sall4 in pluripotency reprogramming
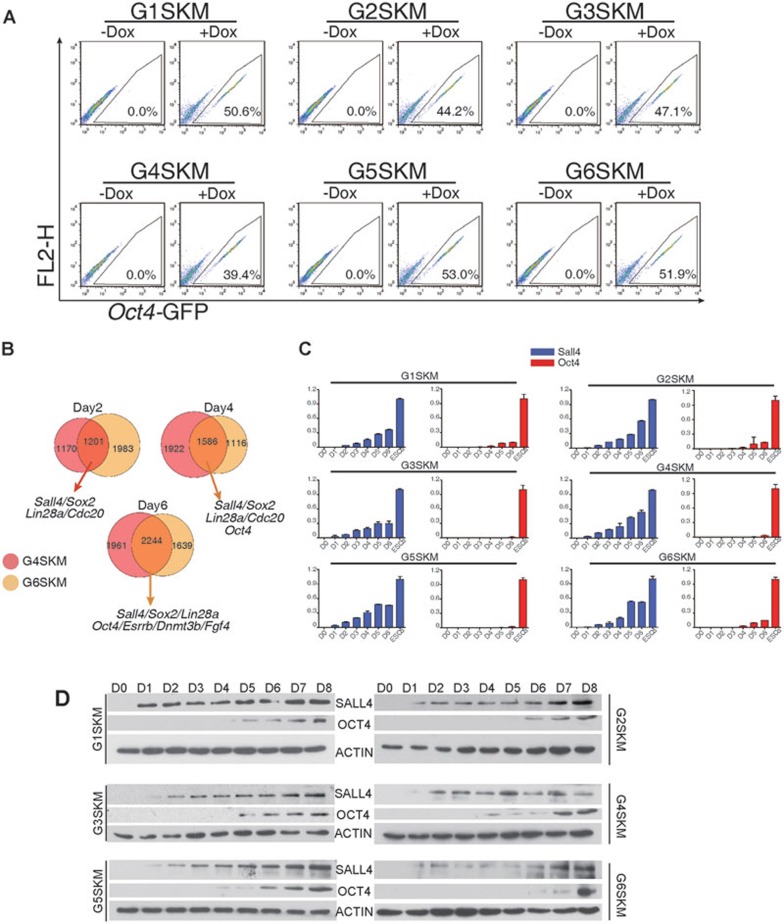
We established a genetically homogeneous secondary reprogramming system using GATA transcription factor-reprogrammed iPSCs. We infected fibroblasts with dox-inducible lentiviruses, reprogrammed fibroblasts by dox addition, selected iPSCs and then produced chimeric mice. Fibroblasts were obtained from these chimeric mice17. Different GATA transcription factors induced pluripotency with varying levels of efficiency. Oct4-GFP-positive cells emerged 4-5 days after the addition of dox. Approximately 20%-50% of the Oct4-GFP-positive cells were obtained using FACS analysis 9 days after the addition of dox, and representative results were shown in Figure 3A. Therefore, this technique serves as a useful tool to analyze the molecular events of GATA-mediated reprogramming.
Figure 3.
The GATA family members can activate Sall4 for pluripotency reprogramming. (A) The flow cytometric analysis of GFP in GATA-secondary MEFs. Representative results from three independent experiments are shown. (B) Venn diagram illustrating the overlap between the differentially expressed genes compared with day 0 in GATA4- and GATA6-mediated secondary MEF reprogramming. (C) Quantitative RT-PCR analysis of the relative expression of endogenous Sall4 and Oct4 during GATA-secondary MEF reprogramming. Error bars indicate SD (n = 3). (D) Western blot analysis of SALL4 and OCT4 in GATA-secondary MEF reprogramming. Actin was used as a loading control.
To find the potential targets of GATA family members in reprogramming, we used GATA4- and GATA6-mediated reprogramming as examples. We performed RNA-seq to analyze mRNA dynamics on days 2, 4, and 6 of GATA-mediated reprogramming (Supplementary information, Tables S1–S4). We found the activation of several pluripotency-associated genes by day 2 in shared targets of GATA4 and GATA6, including Sall4, Sox2 and Lin28a, but not Oct4 (Figure 3B, Supplementary information, Figure S6 and Table S6). Sall4 is an important regulator of pluripotency and differentiation and is a key factor in amphibian limb regeneration18,19,20,21. Sall4 also directly interacts with Gata4 and Gata6 in early embryonic development19. Furthermore, Sall4 was reported to be a transcriptional activator of Oct4 and to be able to partially replace Oct4 in mouse somatic reprogramming20,22, which was confirmed (Supplementary information, Figure S7A and S7B). Of the pluripotency-related factors that were activated 2 days after induction using GATA transcription factors together with SKM, we focused on Sall4 (Figure 3B). To further validate the results obtained from the RNA-seq data, we examined the expression of Sall4 in all GATA-mediated reprogramming. We found that Sall4 was activated shortly after induction with exogenous GATA family members, while Oct4 expression was negligible until the emergence of iPSCs at 4 or 5 days after induction (Figure 3C and 3D). These results suggest that GATA transcription factors may act to replace Oct4 through the activation of endogenous Sall4.
Sall4 is a bridge linking lineage-specifying GATA family members to the pluripotency circuit
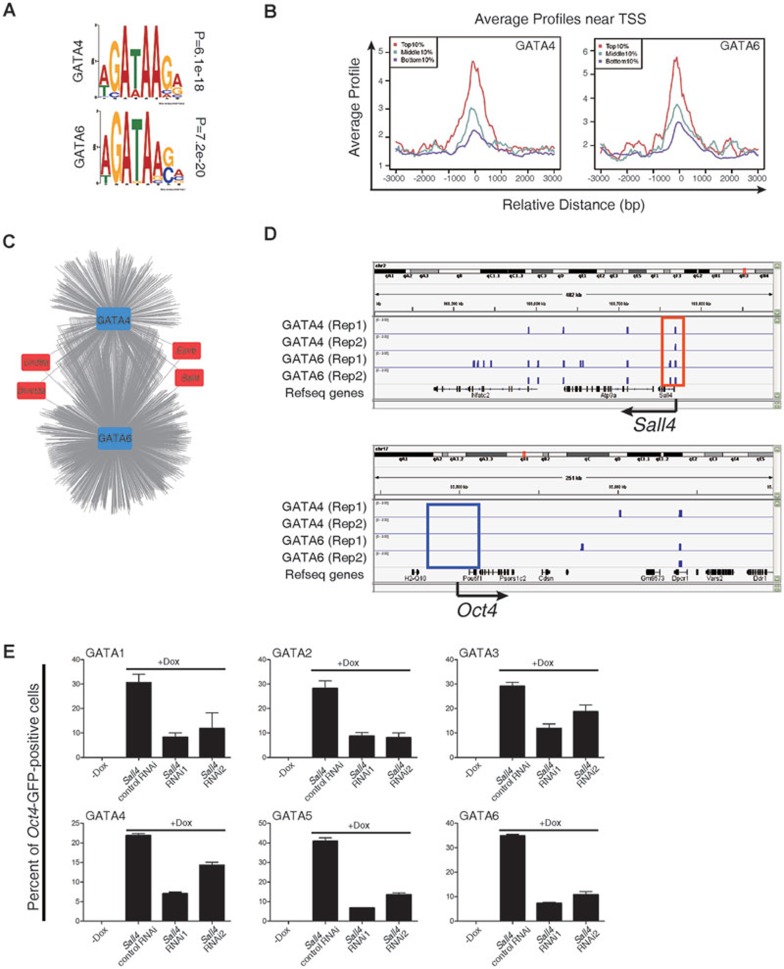
To further investigate the direct targets of GATA family members in reprogramming, we performed ChIP-seq using GATA4- and GATA6-secondary MEFs (Supplementary information, Table S5). We analyzed the direct targets of GATA4 and GATA6 at day 6 post induction; we found that they contained the “GATA” binding motif (Figure 4A) and that the highly expressed genes were correlated with the GATA-binding signals around the TSS of genes (Figure 4B). To comprehensively identify the functional targets of GATA4 and GATA6, we collated a list of genes that could directly bind GATA4 and GATA6 by ChIP-seq and examined their expression by RNA-seq during reprogramming. We found putative direct targets, including some core pluripotency-associated genes (Figure 4C). In addition, by comparing the results obtained from the RNA-seq and ChIP-seq, we found that both GATA4 and GATA6 bound directly to Sall4 promoters, but not to Oct4, Sox2, or Nanog promoters, indicating that Sall4 is a direct target of the GATA family in pluripotency reprogramming (Figure 4D, Supplementary information, Figure S8 and Table S7). These results also indicate that the GATA family can function to replace Oct4 by avoiding direct activation of endogenous Oct4.
Figure 4.
Sall4 as a bridge linking the lineage-specifying GATA family to the pluripotency circuit. (A) GATA4 and GATA6 motifs were predicted using the Multiple Em for Motif Elicitation software. ChIP-seq data were generated using GATA4- and GATA6-secondary MEFs. (B) The average ChIP enrichment signals around the TSS of the genes. The red, cyan and purple colors indicate the average ChIP enrichment signals of the top 10%, middle 10%, and bottom 10% of expressed genes from day-6 RNA-seq data. (C) A model for the regulatory interactions of differentially expressed genes reconstructed from binding profiles and expression data. Particular pluripotency-associated factors and epigenetic regulators are highlighted. (D) ChIP-seq binding profiles at the Sall4 and Oct4 loci using secondary MEFs. (E) The flow cytometric analysis of GFP in GATA-secondary MEFs before and after knockdown of the endogenous Sall4 expression. Scrambled shRNA was used as a control. Error bars indicate SD (n = 3).
To further confirm that GATA transcription factors activated endogenous Sall4 to enhance reprogramming in the absence of Oct4, we performed knockdown experiments to investigate whether Sall4 is required for GATA-mediated activation of endogenous Oct4 and subsequent reprogramming. We found that knockdown of Sall4 in all GATA family member-induced reprogramming hindered the reprogramming process (Figure 4E and Supplementary information, Figure S9). Taken together, these results suggest that GATA transcription factors can enhance reprogramming by directly activating endogenous Sall4 and that Sall4 serves as a bridge linking lineage-specifying GATA family members to the pluripotency circuit.
Discussion
It is known that only a few members of the Oct4, Sox2, and Klf4 protein families can be used for reprogramming of cellular pluripotency14. After the first discovery that lineage specifiers could substitute for key pluripotency factors5, we further confirmed that not only some but all members of the GATA family had the ability to substitute for Oct4, the most important pluripotency factor23,24. Thus, we have described the first protein family that can substitute for Oct4 and function as inducers of the reprogramming process. We now show that the GATA family of transcription factors had a previously underestimated role in the restoration of pluripotency, in addition to their important roles in lineage specification and transdifferentiation. Together, these results indicate that GATA family members may be important mediators of the cell fate transition in lineage specification, transdifferentiation and reprogramming to pluripotency.
Sall4 has been described as a “star” factor of pluripotency and plays an important role in differentiation and pluripotency18,19,22,25. In addition, Sall4 is important in the maintenance of the primitive endodermal lineage by interacting with primitive endoderm lineage markers such as Gata4, Gata6, and Sox1719. Sall4 is also a key factor in amphibian limb regeneration21. Furthermore, Sall4 also regulates cell fate decisions in hepatic stem/progenitor cells and hematopoietic lineages26,27. We previously proposed a “seesaw” model to suggest that the pluripotent state is a fine-tuned balance between competing differentiation forces. However, the mechanisms that link lineage-specifying cues and the activation of the pluripotency circuit remain unclear5,28,29. We found that the introduction of exogenous GATA family members could directly and rapidly activate Sall4 rather than Oct4. We suggest that Sall4 serves as a bridge linking the lineage-specifying circuit to the pluripotency circuit. In addition to the mutual inhibition of lineage-specifying forces by lineage specifiers and pluripotency factors, we found evidence that activation of key pluripotency factors by lineage specifiers could be a complementary mechanism for pluripotency reprogramming (Figure 5). Despite the key roles of Sall4 in reprogramming and development, we believe that there are other factors that may be involved in activation of the pluripotency circuit by lineage specifiers. In a previous report, the pluripotency-associated factors Sall4, Lin28a, Esrrb, together with Nanog or Dppa2, could induce pluripotency in mouse somatic cells. A late hierarchic phase was proposed for the induction of pluripotency, where Sox2 was the upstream factor in the gene expression hierarchy20. In our study, Sall4, Sox2, and lin28a were found to be activated by GATA-induced reprograming at 2 days post induction, which can explain how the hierarchic pluripotency circuit could be restored after the forced expression of lineage specifiers in somatic cells. Concurrently, the precarious balance between these factors to successfully obtain stable pluripotency may also be important. Once one is dominant or overrepresented, it is plausible to end up with another lineage state instead of a pluripotent state. It is likely that reprogramming factors play multiple roles in the process and that there are still other undiscovered relationships and functions of the GATA family, for example, whether GATA family members function as pioneer factors to alter the landscape of chromatin accessibility and whether the GATA family can function together with epigenetic regulators (Figure 5)30. These questions warrant further study to uncover the mysteries of cellular reprogramming.
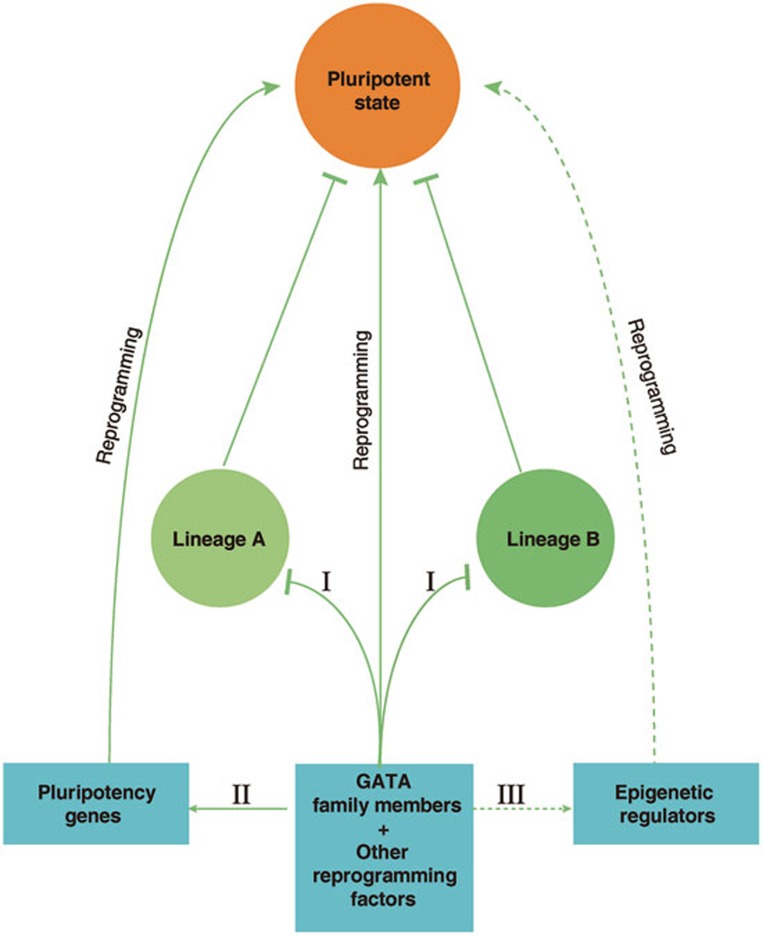
Figure 5.
Diagram illustrating the roles of GATA family members in pluripotency reprogramming. (I) The balance of lineage-specifying forces5; (II) direct activation of pluripotency-associated genes; (III) potential interaction with epigenetic regulators.
Materials and Methods
Mice
The transgenic mouse strain C57BL/6J-Tg(GOFGFP)11Imeg/Rbrc (OG) was purchased from the RIKEN Bioresource Center. Offspring carrying Oct4 promoter-driven GFP were obtained by crossbreeding OG with mice from an ICR background. iPSC-derived mice were generated as previously described5. All animal experiments were conducted in accordance with the Animal Protection Guidelines of Peking University, China.
Cell culture
MEFs and 293T cells were cultured in DMEM/High Glucose (Hyclone) supplemented with 10% fetal bovine serum (FBS; Hyclone). iPSCs and mESCs were grown on feeders of Mitomycin C-treated MEFs in mESC culture medium (80% KnockOut DMEM (Gibco), 10% KnockOut serum replacement (Gibco), 10% FBS (embryonic stem cell-screened; Hyclone), 100 μg/ml streptomycin, 100 U/ml penicillin, 1 mM L-glutamine, 55 μM β-mercaptoethanol, nonessential amino acids, plus 1 μM PD0325901, 3 μM CHIR99021 and LIF (Millipore)). iPSCs and ESCs were passaged using Trypsin-EDTA (Invitrogen), and the culture medium was changed daily.
iPSC generation
The dox-inducible lentiviral system was used as previously described5. The cDNAs of human GATA family of transcription factors were obtained from Origene Co., Ltd and inserted into the dox-inducible lentiviral system.
Briefly, 293T cells cultured in 100-mm dishes were co-transfected with 5 μg each of pMDLg/pRRE, RSV-Rev, and VSV-G vectors and 15 μg of the corresponding lentiviral vector using the Ca3(PO4)2 method. The medium was changed 12 h after transfection and incubated for an additional 36 h before virus collection. The virus-containing supernatant was filtered through 0.45-μm filters.
MEFs were seeded at a density of 5 × 104 cells per well in 6-well plates. On the day after seeding, the cells were infected with virus-containing supernatant at an appropriate MOI and supplemented with 10 ng/μl Polybrene (Sigma). The virus- and Polybrene-containing medium was changed to fibroblast medium 12 h after infection, and the cells were incubated for an additional 12 h. The expression of exogenous genes was induced by replacement of the culture medium on the infected cells with induction medium (80% KnockOut DMEM (Gibco), 10% KnockOut serum replacement (Gibco), 10% FBS (embryonic stem cell-screened; Hyclone), 100 μg/ml streptomycin, 100 U/ml penicillin, 1 mM L-glutamine, 55 μM β-mercaptoethanol, nonessential amino acids, and 1 μg/ml dox). The induction medium was changed every 3 days.
Characterization of iPSCs
The chimera experiment was performed as previously described5. For immunofluorescence, cultured cells were washed using PBS and immediately fixed in 4% PFA for 15 min. Fixed cells were blocked for 1 h at room temperature in PBS containing 2.5% donkey serum and 0.2% Triton X-100. Samples were then incubated with primary antibodies at room temperature for 2 h, followed by secondary antibodies at room temperature for 1 h. Total RNA of cultured cells was extracted using the RNeasy Plus Mini Kit (QIAGEN) and converted to cDNA using the EasyScript Reverse Transcriptase (TransGen Biotech). Genomic DNA from cultured cells was isolated using the DNeasy Blood and Tissue Kit (QIAGEN), and PCR was performed to detect the corresponding genome-inserted exogenous genes.
Western blotting
Cells were collected and washed with PBS, lysed in RIPA buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, plus Protease Inhibitor Cocktails (Thermo Scientific)) at 4 °C for 45 min. The cell lysates were boiled in protein loading buffer and centrifuged at 14 000× g. The protein supernatants were separated on a 10% SDS-PAGE gel by electrophoresis using the recommended time. The separated proteins were then immediately transferred to a PVDF membrane (Millipore), and the membrane was blocked with 5% skim milk in TBST at room temperature for 1 h. Antibodies were dissolved in TBST containing 3% BSA and 0.2% Triton X-100. The membrane was incubated with primary antibodies overnight at 4 °C, washed in TBST, incubated with secondary antibodies at room temperature for 1 h, and washed with TBST. The proteins on membrane were detected with the Luminata Classico Western HRP substrate (Millipore). The antibodies used for western blotting included rabbit anti-Sall4 (1:1 000; ab157172, Abcam), rabbit anti-Oct4 (1:1 000; ab19857, Abcam), rabbit anti-β-actin (1:3 000; 4970, Cell Signaling), anti-rabbit IgG-HRP (1:3 000; 7074, Cell Signaling), and anti-mouse IgG-HRP (1:3 000; 7076, Cell Signaling).
Flow cytometry analysis
Cultured cells were collected using trypsin-EDTA treatment and resuspended in PBS containing 3% FBS. Endogenous Oct4-GFP was used for sorting on a FACSCalibur instrument (BD Bioscience).
RNA-seq
Total RNA was extracted from each cell line using TRIzol reagent according to the manufacturer's instructions. After mRNA was enriched using oligo(dT) magnetic beads, ∼1 μg of mRNA was fragmented. Isolated RNA fragments of ∼200-250 bp were separated by electrophoresis and prepared for cDNA synthesis through end repair, 3′ end adenylation, and adapter ligation. The cDNA fragments ranging from 250-300 bp were excised by electrophoresis for sequencing on a HiSeq2000 (Illumina).
The generated sequencing reads were aligned to a reference sequence (GRCm38/mm10, downloaded from Ensembl database, ftp.ensembl.org) using TopHat alignment software tools31. Only uniquely aligned reads were used for transcript assembly with Cufflinks software32. Read counts for each gene were calculated, and the expression values of each gene were normalized using FPKM (fragments per kilobase of exon model per million mapped reads). The results of differential gene expression were visualized and analyzed using the Bioconductor function “CummeRbund” in the R programming language33. Hierarchical clustering was performed in R using the “heatmap” package34. In addition, the “VennDiagram” package in R language was used to display the Venn diagram.
ChIP-seq
Approximately 150 million cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The crosslinking was then quenched by adding 125 mM glycine buffer and incubating the samples for 5 min at room temperature. After washing with ice-cold PBS, the cell pellet was resuspended in 250 μl SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8) and incubated for 15 min on ice. Samples were sonicated to obtain DNA fragments between 100-200 bp, and debris was removed by centrifugation at 13 000 rpm for 10 min at 4 °C. The resulting supernatant was transferred to a new tube and diluted 10-fold with ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8, 167 mM NaCl). Protein A-agarose beads (100 μl) were added and incubated for 1 h at 4 °C with rotation to pre-clear the samples. After centrifugation for 5 min at 3 000 rpm, the supernatant was collected into a new tube. Then, 1 μg of antibody (anti-GATA4 (AF2606, R&D Systems) or anti-GATA6 (AF1700, R&D Systems) in 2% BSA) was added for overnight incubation at 4 °C on a rotating wheel. The immunoprecipitated pellet was obtained by adding 500 μl of Protein A-agarose beads and incubating for 1 h at 4 °C. The pellet was then washed with low-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris, pH 8, 0.5 M NaCl), LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% NaDOC, 1 mM EDTA, 10 mM Tris, pH 8), and TE buffer (1 mM EDTA, 10 mM Tris, pH 8). Immunoprecipitates were eluted with elution buffer (0.2% SDS, 0.1 M NaHCO3), and cross-links were reversed overnight at 65 °C in 0.2 M NaCl. DNA was RNase-treated and purified for sequencing.
After the ChIP-seq library was constructed, a HiSeq2000 sequencer (Illumina) was used to generate 101-base sequences. Sequencing reads were also aligned to the reference sequence (GRCm38/mm10) using MACS software35. The results generated by MACS were loaded into IGV for visualization36. Multiple Em for Motif Elicitation (MEME) was used to search the GATA4 and GATA6 motifs37. PeakAnnotator was used to annotate the information of each peak generated by MACS38, and the average ChIP enrichment signals around TSS were displayed using the Cis-regulatory Element Annotation System (CEAS).
Site-directed mutagenesis of GATA genes
Partially overlapping primers were designed using a previously reported method39. Wild-type GATA plasmids were used as templates, and PCR was performed using PrimeSTAR HS DNA Polymerase (TaKaRa), followed by DpnI restriction enzyme treatment to remove the methylated DNA templates. Bacteria were transformed, and single colonies were picked after 12 h. The mutations were identified by DNA sequencing.
Knockdown
Lentiviral vectors containing a puromycin resistance gene were used to knock down Sall4 expression according to the manufacturer's protocol. Prior to infection with the reprogramming genes, the cells were selected for 6 days with 2 μg/ml puromycin to eliminate uninfected cells.
Accession number
RNA-seq and ChIP-seq data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE57849.
Acknowledgments
We thank Yang Zhao and Jun Xu for helpful discussions. This work was supported by the National Basic Research Program of China (973 Program; 2012CB966401), the National Natural Science Foundation of China (91319305), the National Science and Technology Major Project (2013ZX10001003), the Ministry of Science and Technology of China (2013DFG30680) and the Ministry of Education of China (111 Project) to HD, and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA01040407), the National Natural Science Foundation of China (91019024 and 31100558) and the Hundred Talents Program of the Chinese Academy of Sciences to YS.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Characterization of GATA-reprogrammed iPSCs.
Quantitative RT-PCR analysis of the expression of endogenous ECT related genes relative to the expression of SKM.
A schematic diagram to illustrate zinc-finger domains in GATA family members.
Strategies for mutation of GATA family members.
Quantitative RT-PCR analysis of the expression of endogenous Dlx3 and Lhx5 genes relative to the expression in SKM, G1D, G2D, G3D, G4D, G5D, and G6D represent corresponding GATA-Double mutations.
Venn diagram illustrating the overlap genes during secondary MEFs reprogramming.
Sall4 substitutes for Oct4 in MEF reprogramming to pluripotency.
ChIP-seq binding profiles for Sox2 and Nanog loci using secondary MEFs.
Western blot analysis of SALL4 knockdown in different GATA family member-mediated secondary MEF reprogramming.
Number of reads (total reads, mapped reads, unique reads) in RNA-seq (GATA4)
Distribution of all expressed genes in different expression levels (GATA4)
Number of reads (total reads, mapped reads, unique reads) in RNA-seq (GATA6)
Distribution of all expressed genes in different expression levels (GATA6)
Number of reads (total reads, mapped reads, unique reads) in ChIP-seq
References
- 1Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature 2013; 502:462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663–676. [DOI] [PubMed] [Google Scholar]
- 3Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861–872. [DOI] [PubMed] [Google Scholar]
- 4Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007; 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 5Shu J, Wu C, Wu Y, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013; 153:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Montserrat N, Nivet E, Sancho-Martinez I, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell 2013; 13:341–350. [DOI] [PubMed] [Google Scholar]
- 7Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell 2011; 8:363–369. [DOI] [PubMed] [Google Scholar]
- 8Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 2002; 12:416–422. [DOI] [PubMed] [Google Scholar]
- 9Chlon TM, Crispino JD. Combinatorial regulation of tissue specificationregulation of tissue specification by GATA and FOG factors. Development 2012; 139:3905–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Weiss MJ, Orkin SH. GATA transcription factors — key regulators of hematopoiesis. Exp Hematol 1995; 23:99–107. [PubMed] [Google Scholar]
- 11Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 2000; 275:38949–38952. [DOI] [PubMed] [Google Scholar]
- 12Ieda M, Fu JD, Delgado-Olguin P, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010; 142:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011; 475:390–393. [DOI] [PubMed] [Google Scholar]
- 14Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol 2008; 26:101–106. [DOI] [PubMed] [Google Scholar]
- 15Xue Z, Huang K, Cai C, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013; 500:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Chen Y, Bates DL, Dey R, et al. DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep 2012; 2:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Wernig M, Lengner CJ, Hanna J, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol 2008; 26:916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 2009; 14:683–694. [DOI] [PubMed] [Google Scholar]
- 19Lim CY, Tam WL, Zhang J, et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 2008; 3:543–554. [DOI] [PubMed] [Google Scholar]
- 20Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012; 150:1209–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Neff AW, King MW, Mescher AL. Dedifferentiation and the role of sall4 in reprogramming and patterning during amphibian limb regeneration. Dev Dyn 2011; 240:979–989. [DOI] [PubMed] [Google Scholar]
- 22Zhang J, Tam WL, Tong GQ, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol 2006; 8:1114–1123. [DOI] [PubMed] [Google Scholar]
- 23van den Berg DL, Snoek T, Mullin NP, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 2010; 6:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Pardo M, Lang B, Yu L, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 2010; 6:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Elling U, Klasen C, Eisenberger T, Anlag K, Treier M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci USA 2006; 103:16319–16324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Oikawa T, Kamiya A, Kakinuma S, et al. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009; 136:1000–1011. [DOI] [PubMed] [Google Scholar]
- 27Paik EJ, Mahony S, White RM, et al. A Cdx4-Sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis. Stem Cell Reports 2013; 1:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Shu J, Deng H. Lineage specifiers: new players in the induction of pluripotency. Genomics Proteomics Bioinformatics 2013; 11:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Ben-David U, Nissenbaum J, Benvenisty N. New balance in pluripotency: reprogramming with lineage specifiers. Cell 2013; 153:939–940. [DOI] [PubMed] [Google Scholar]
- 30Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 2011; 25:2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009; 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004; 5:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009; 37:W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 2010; 11:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characterization of GATA-reprogrammed iPSCs.
Quantitative RT-PCR analysis of the expression of endogenous ECT related genes relative to the expression of SKM.
A schematic diagram to illustrate zinc-finger domains in GATA family members.
Strategies for mutation of GATA family members.
Quantitative RT-PCR analysis of the expression of endogenous Dlx3 and Lhx5 genes relative to the expression in SKM, G1D, G2D, G3D, G4D, G5D, and G6D represent corresponding GATA-Double mutations.
Venn diagram illustrating the overlap genes during secondary MEFs reprogramming.
Sall4 substitutes for Oct4 in MEF reprogramming to pluripotency.
ChIP-seq binding profiles for Sox2 and Nanog loci using secondary MEFs.
Western blot analysis of SALL4 knockdown in different GATA family member-mediated secondary MEF reprogramming.
Number of reads (total reads, mapped reads, unique reads) in RNA-seq (GATA4)
Distribution of all expressed genes in different expression levels (GATA4)
Number of reads (total reads, mapped reads, unique reads) in RNA-seq (GATA6)
Distribution of all expressed genes in different expression levels (GATA6)
Number of reads (total reads, mapped reads, unique reads) in ChIP-seq