Abstract
AMP-activated protein kinase (AMPK) is a central cellular energy sensor and regulator of energy homeostasis, and a promising drug target for the treatment of diabetes, obesity, and cancer. Here we present low-resolution crystal structures of the human α1β2γ1 holo-AMPK complex bound to its allosteric modulators AMP and the glycogen-mimic cyclodextrin, both in the phosphorylated (4.05 Å) and non-phosphorylated (4.60 Å) state. In addition, we have solved a 2.95 Å structure of the human kinase domain (KD) bound to the adjacent autoinhibitory domain (AID) and have performed extensive biochemical and mutational studies. Together, these studies illustrate an underlying mechanism of allosteric AMPK modulation by AMP and glycogen, whose binding changes the equilibria between alternate AID (AMP) and carbohydrate-binding module (glycogen) interactions.
Keywords: AMPK, adenine nucleotides, glycogen, diabetes
Introduction
AMPK is an evolutionarily conserved, heterotrimeric serine/threonine protein kinase. Its kinase activity is activated by energy stress (i.e., a drop in the ratio of ATP to AMP/ADP), allowing AMPK to upregulate ATP-generating pathways and inhibit energy-consuming programs by phosphorylating numerous cellular targets1,2,3. Due to its important roles in glucose uptake and metabolism, as well as growth and proliferation, AMPK is a promising target for the treatment of diabetes, obesity, and cancer4.
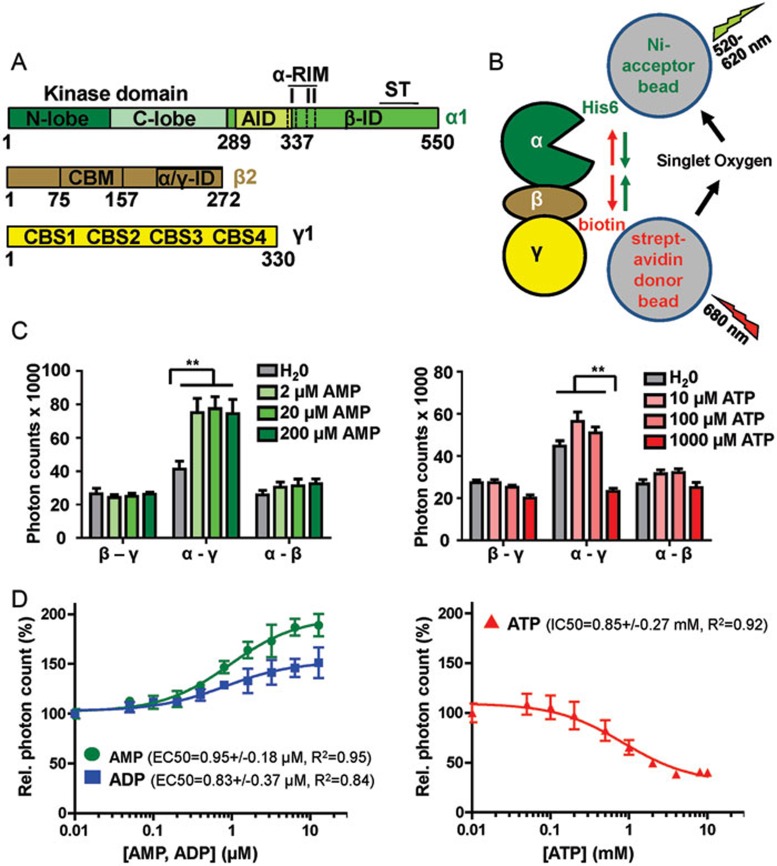
AMPK consists of an α-subunit with an N-terminal kinase domain (KD) and a regulatory C-terminus, a glycogen-binding β-subunit, and an AMP/ADP/ATP-binding γ-subunit (Figure 1A). The regulatory C-terminus of the α-subunit contains an autoinhibitory domain (AID)5,6 sandwiched between the KD and a sensor (αRIM) loop, a β-subunit interaction domain (β-ID)5, and a serine/threonine-rich loop (ST loop), which mediates phosphorylation and signaling by PI3K/Akt and glycogen synthase kinase 37,8 (Figure 1A). AMPK activity is greatly enhanced by phosphorylation of its kinase activation loop by upstream kinases, including LKB19,10,11 and CaMKKβ12,13,14. AMP binding to the γ-subunit increases net activation loop phosphorylation by stimulating physical interaction of LKB1 and AMPK with the scaffolding protein axin15,16, and by decreasing activation loop accessibility to phosphatases17, the latter of which is dependent on the presence of the carbohydrate-binding module (CBM) of the β-subunit18. In addition, AMP binding also directly activates AMPK kinase activity2,19,20. ADP appears to protect weakly against activation loop dephosphorylation21,22,23, has no effect on direct activation of kinase activity21,22,23, and has been proposed to affect phosphorylation by upstream kinases24, although the latter has recently been challenged22. In contrast, ATP is thought to counter the stimulating effect of AMP by competing with AMP for binding to the regulatory γ-subunit22. The γ-subunit contains four cystathionine β-synthetase (CBS)-type adenine nucleotide-binding motifs, of which CBS-1, CBS-3, and CBS-4 can competitively bind AMP, ADP, or ATP. In contrast, CBS-2 is incompetent of nucleotide binding and it is thus unoccupied21,25.
Figure 1.
AMP and ATP induce opposing conformational changes in AMPK. (A) Schematic presentation of the three subunits of the AMPK isoform α1β2γ1 (human AMPK has two alternative α-subunits, two β-subunits, and three γ-subunits that can form a total of 12 different isoforms). Amino acid positions are indicated below domain boundaries. AID, autoinhibitory domain; CBM, carbohydrate-binding module; CBS, cystathionine β-synthetase motif-type adenine nucleotide binding site; αRIM, regulatory subunit-interacting motif loop, which contains two motifs, αRIM-1 (I) and αRIM-2 (II); α/γ-ID, C-terminal α-subunit and β-subunit interaction domain; β-ID, C-terminal β-subunit interaction domain; ST, ST-loop. (B) Cartoon of AlphaScreen luminescence proximity assay for AMPK with a His6-tag at the N-terminus of the α-subunit and a biotin tag at the N-terminus of the γ-subunit. The tags bind to streptavidin donor and Ni-acceptor nanobeads, respectively, and bring donor- and acceptor-beads into close proximity. Donor beads contain a photosensitizer that upon activation at 680 nm converts ambient oxygen into singlet oxygen. With decreasing distance (green arrows), more short-lived (t1/2 = 4 μsec) singlet oxygen will diffuse to the acceptor beads to transfer energy from the singlet oxygen to thioxene derivatives in the acceptor beads, resulting in increased light emission at 520-620 nm. Since the energy transfer between beads is chemically mediated by singlet oxygen, signals are insensitive to bead orientation. (C) Adenine nucleotides modulate AlphaScreen signals of AMPK with selectively tagged subunits (β-γ: α1-biotin-β2-His6-γ1, α-γ: His6-α1-β2-biotin-γ1, α-β: His6-α1-biotin-β2-γ1; n = 3; error bars, SD). **P < 0.01 (Student's t-test). (D) Dose-response curves of the effect of adenine nucleotides on the AlphaScreen signal (n = 3; error bars, SD). Note that concentration of AMP and ADP are in μM and of ATP in mM.
Important insights of how AMP regulates AMPK activity came from the crystal structure of an almost full length mammalian AMPK complex bound to two molecules of AMP (at CBS-3 and CBS-4), which only lacked the CBM and the flexible ST-loop of the α-subunit (“AMPKΔCBM”)21. This structure identified a loop from the α-subunit, originally named α-hook and recently reassigned and renamed αRIM26,27, which directly binds AMP at CBS-3, suggesting that this loop functions as adenine nucleotide sensor21,26. In addition to the CBS-3/AMP binding to the αRIM-2 segment of the αRIM loop, the N-terminus of the loop folds back and is connected to the AID. In the AMP-bound structure, the C-terminus of the AID and the N-terminus of the αRIM loop formed an interaction surface, termed αRIM-1, that bound to the unoccupied CBS-2 site of the γ-subunit27. In contrast, in the structure of an inactive fission yeast KD-AID fragment, the AID bound to the hinge region of the KD to arrest the KD in an inactive, open conformation28. While fission yeast AMPK lacks the αRIM sensor loop and has a relatively weakly conserved AID, mutational data together with the combined structures suggest that AMP directly activates the kinase activity by inducing αRIM-2 to bind AMP-occupied CBS-3. That in turn is believed to stabilize the interaction of the AID-αRIM with the γ-subunit, thereby pulling the AID away from the inhibitory interaction with the KD26,27,28.
In addition to adenine nucleotides, glycogen and the non-physiological glycogen-mimic cyclodextrin may also regulate AMPK29. While Polekhina et al.30 did not detect an effect of glycogen on the activity of purified rat liver AMPK, McBride et al.31 reported that glycogen and the glycogen-mimic cyclodextrin mildly inhibit the catalytic activity of recombinant, E. coli-expressed AMPK, and more substantially inhibit the activity of immunoprecipitated native rat liver AMPK. The reported glycogen regulation occurred both in the presence and absence of AMP and was dependent on its binding to the carbohydrate-binding loop of the CBM31. Finally, AMPK can also be activated by pharmacological compounds that bind to the CBM outside of the carbohydrate-binding loop and that activate AMPK by a mechanism different from that of its known physiological regulators. Recent structures of AMPK bound to compounds 991 and A769662 revealed that they bind to both the CBM and KD to mediate CBM-KD interaction, and that they may stabilize the KD in an active conformation by inducing formation of an α-helix, termed C-interacting helix, that packs against the regulatory αC-helix of the KD18. Moreover, A769662 can synergize with AMP to fully activate AMPK in a CBM-dependent manner, even in the absence of phosphorylation by upstream kinases32.
To determine the molecular and structural basis of AMPK regulation by its physiological allosteric modulators, we solved the crystal structure of non-phosphorylated and active phosphorylated holo-AMPK bound to AMP and the glycogen-mimic cyclodextrin as well as the structure of the inactive human AMPK KD-AID complex. In combination with biochemical and mutational data, our structures recapture a mechanistic model of AMPK regulation by AMP and ATP, in which AMP binding to CBS-3 induces the interaction of AID-αRIM and γ-subunit and leads to AMPK activation by releasing AID inhibition of the kinase domain. ATP counters the AMP effect not only by competitively binding to the γ-subunit, but also by destabilizing the interaction between the AID-αRIM and the γ-subunit independently of adenine nucleotide binding to CBS-3. Furthermore, our structures reveal an allosteric mechanism by glycogen, whose binding to the CBM destabilizes the CBM-KD interaction. Together, our results provide a comprehensive framework for understanding AMPK modulation by adenine nucleotides and carbohydrates.
Results
AMP and ATP induce distinct conformational changes in AMPK
We adapted an AlphaScreen luminescence proximity assay to detect nucleotide-dependent conformational changes in AMPK subunit geometry. AlphaScreen measures a singlet oxygen-mediated luminescence energy transfer between biotinylated proteins bound to streptavidin-coated nanobeads and His6-tagged proteins bound to Ni-chelating acceptor beads. This assay is highly sensitive to changes in distance, but in contrast to FRET has extremely low background interference because excitation and emission are uncoupled (Figure 1B). We expressed and purified AMPK with His6- and biotin-tags introduced into subunits of the complex. As shown in Figure 1C, addition of 2 μM AMP to AMPK doubled the luminescence signal for AMPK with N-terminally His6-tagged α-subunit and N-terminally biotin-tagged γ-subunit (indicating a decreased tag distance, consistent with measurements by SAXS33), and addition of 1 mM ATP decreased the signal by a factor of two (indicating an increase in distance). In contrast, AMP and ATP only slightly affected signals for the β-γ and α-β subunit interactions. These results suggest that ATP not only competes for the activator AMP, but may also actively inhibit AMPK by inducing a conformation distinct from that induced by AMP. Second, while AMP and ATP bind the two readily exchangeable adenine nucleotide-binding sites (CBS1 and CBS3) of AMPK with overall similar affinities21, conformational changes are induced by almost three orders of magnitude lower AMP concentration (EC50 = 0.95 μM) than ATP concentration (IC50 = 0.85 mM; Figure 1D), which reflects physiological concentrations of ATP in the mM and of AMP in the μM range (see Discussion).
The AID binds to the γ-subunit and the CBM binds to the KD in the structure of phosphorylated, active AMP- and cyclodextrin-bound AMPK
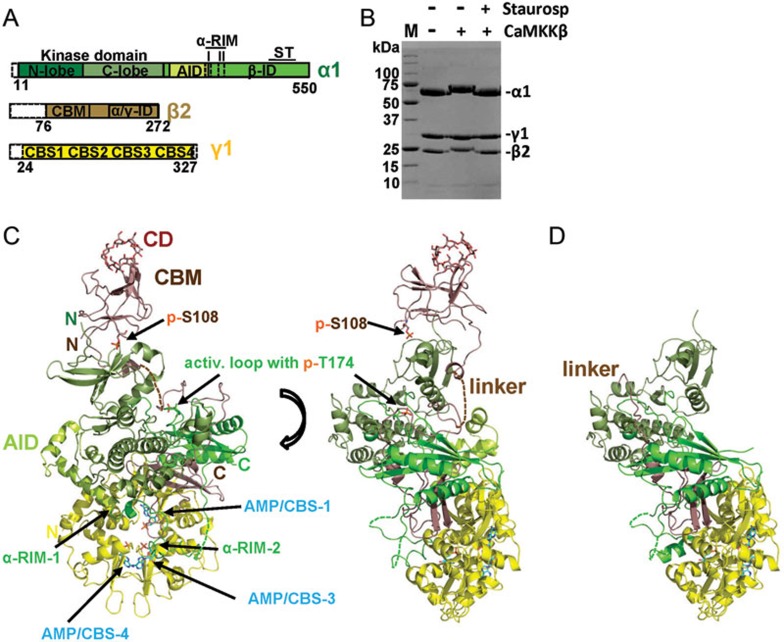
To gain insight into the mechanism of AMPK modulation by AMP and glycogen, we co-crystallized holo-AMPK that contains the CBM, which is required both for glycogen binding30 and for the AMP-dependent protection against dephosphorylation18, with AMP and the glycogen-mimic cyclodextrin. We could overcome the recalcitrance of holo-AMPK to crystallization in the absence of pharmacological activators by extensive construct screening and by altering three flexible solvent-exposed side chains (E471G/E474A/K476A) at the N-terminus of the ST-loop of the α-subunit (see Materials and Methods and Figure 2A). These residues lie at the crystal-packing interface and their substitutions reduced the entropic cost of crystal formation without affecting subunit interactions, catalytic activity, or the modulation of kinase activity by AMP or glycogen (Supplementary information, Figure S1). We solved the structure of phosphorylated, terminally trimmed human AMPK (Figure 2A and 2B) bound to AMP and the glycogen mimic30,31,34,35 cyclodextrin, at 4.05 Å (Figure 2C). While the resolution is low, it allows clear identification of ligands (Supplementary information, Figure S2), all secondary structure elements, and bulky amino acid side chains with excellent structural geometry and crystallographic statistics (Table 1 and Supplementary information, Figure S8).
Figure 2.
Structure overview of human AMPK bound to AMP, cyclodextrin and staurosporine. (A) Schematic presentation of the crystallization construct, using the same color code as in the following structure panels. Unstructured termini omitted in the crystallization construct are shown as white dashed boxes. (B) Coomassie-stained SDS-PAGE gel of purified AMPK prior and after phosphorylation by CaMKKβ, in the presence and absence of the kinase inhibitor staurosporine. M: protein standards, with molecular weights indicated in kDa. (C) Structure overview of phosphorylated AMPK in two different orientations. (D) Structure overview of non-phosphorylated AMPK. AID, autoinhibitory domain; CBM, carbohydrate-binding module; CBS, cystathionine β-synthetase motif-type adenine nucleotide binding site; CD, cyclodextrin; αRIM, regulatory subunit-interacting motif; α/γ-ID, C-terminal α-subunit and β-subunit interaction domain; β-ID, C-terminal β-subunit interaction domain; ST, ST-loop.
Table 1. Data collection and refinement statistics.
| p-AMPK | AMPK | KD-AID | |
|---|---|---|---|
| PDB code | 4RER | 4REW | 4RED |
| Data collection | |||
| Space group | P3221 | P3221 | I4122 |
| Cell dimensions | |||
| a, b, c (Å) | 132.6, 132.6, 195.4 | 126.9, 126.9, 188.8 | 119.5, 119.5, 215.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 30-4.05 (4.19-4.051) | 30-4.6 (4.68-4.6) | 30-2.95 |
| Rmerge (%) | 18.6 (63.91) | 9.9 (69.0) | 9.6 (76.0) |
| I/σI | 7.6 (2.01) | 20.0 (3.4) | 32.2 (3.9) |
| Completeness (%) | 99.3 (93.01) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 13.1 (5.01) | 8.2 (8.5) | 17.3 (16.9) |
| Refinement | |||
| Resolution (Å) | 30-4.05 | 30-4.6 | 30-2.95 |
| No. reflections | 16 566 | 10 260 | 14 927 |
| Rwork/Rfree (%) | 22.5/25.9 | 19.8/24.4 | 23.0/27.5% |
| No. atoms in asu | |||
| Protein | 7 607 | 6 502 | 5 291 |
| Ligand/ion | 196 | 102 | 0 |
| Water | 0 | 0 | 21 |
| B-factors | |||
| Protein | 143.6 | 230.3 | 64.4 |
| Ligand/ion | 129.7 | 160.0 | NA |
| Water | NA | NA | 58.6 |
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.005 | 0.004 | 0.009 |
| Bond angles (°) | 0.964 | 0.916 | 1.58 |
1Values in parentheses are for highest-resolution shell.
The structure revealed that AMP bound to all three functional adenine nucleotide-binding sites of the γ-subunit (Figure 2C and Supplementary information, Figure S2). As in the structures of AMP-bound AMPKΔCBM21,26 and AMP- and compound 991-bound AMPK18, the AID is dissociated from the KD and instead binds to CBS-2 of the γ-subunit. While the CBM packed against the γ-subunit in a truncated yeast AMPK complex lacking most of the α-subunit36, the cyclodextrin-bound CBM in our structure of human holo-AMPK was bound to the KD (Figure 2C and Supplementary information, Figure S3). This overall arrangement is similar to that of compound 991-bound AMPK18, indicating that the CBM can associate with the KD even in the absence of pharmacological CBM-KD stabilizers. However, there are also several important differences. First, the conformation of the CBM is significantly more extended and cyclodextrin binds to the carbohydrate-binding loop distal from the interface and the compound 991-binding site. Second, we do not see complete helix in the linker region corresponding to the C-interacting helix due to partial disorder in this region (Supplementary information, Figure S3A). Third, the B-factor of the CBM (249.0) is much higher than that of the remainder of AMPK (159.8; 991-bound AMPK: 80.5/75.6), indicating that the CBM is more dynamic in cyclodextrin-bound AMPK (similar to the dynamic AID, B-factor = 255.8).
An important component of the interaction is autophosphorylated S108 in the CBM, whose phosphate group is in a distance consistent with the formation of salt bridges with K31 and K33 from the KD (Supplementary information, Figure S3B; note that in the AMPK/991 structure, the lysine corresponding to K31 makes predominantly an ionic contact with the carboxyl group of compound 99118). Mutation of S108 reduces the ability of compounds 991 and A769662 to activate AMPK in the absence of AMP18,32 and significantly decreases AMPK activity in the absence of pharmacological KD-CBM stabilizers32,37, consistent with the importance of S108 phosphorylation for CBM-KD interaction and AMPK activity.
The CBM is dissociated from the KD in the structure of non-phosphorylated AMPK
We were also able to co-crystallize the same AMPK construct bound with AMP, cyclodextrin, and staurosporine in the non-phosphorylated state and solve its structure at a resolution of 4.60 Å (Figure 2D and Supplementary Information, Figure S9). The overall structure is very similar to that of phosphorylated AMPK and is stabilized in a largely active kinase conformation by high affinity binding of the non-hydrolysable ATP-competitive kinase inhibitor staurosporine. Consistent with the importance of CBM phosphorylation for CBM-KD binding, we failed to detect CBM density at the KD interface while the core has solid density, indicating that the CBM is dissociated from the KD in non-phosphorylated AMPK (Figure 2D). In contrast to the CBM, the C-terminal part of the CBM linker is resolved and points away from the KD interface seen in the structure of phosphorylated AMPK (compare Figure 2C and 2D). Small additional changes are a modest shift of the αC helix, indicating a more open conformation for non-phosphorylated AMPK, and small changes in the αRIM-2 position.
Carbohydrate binding modulates CBM interactions
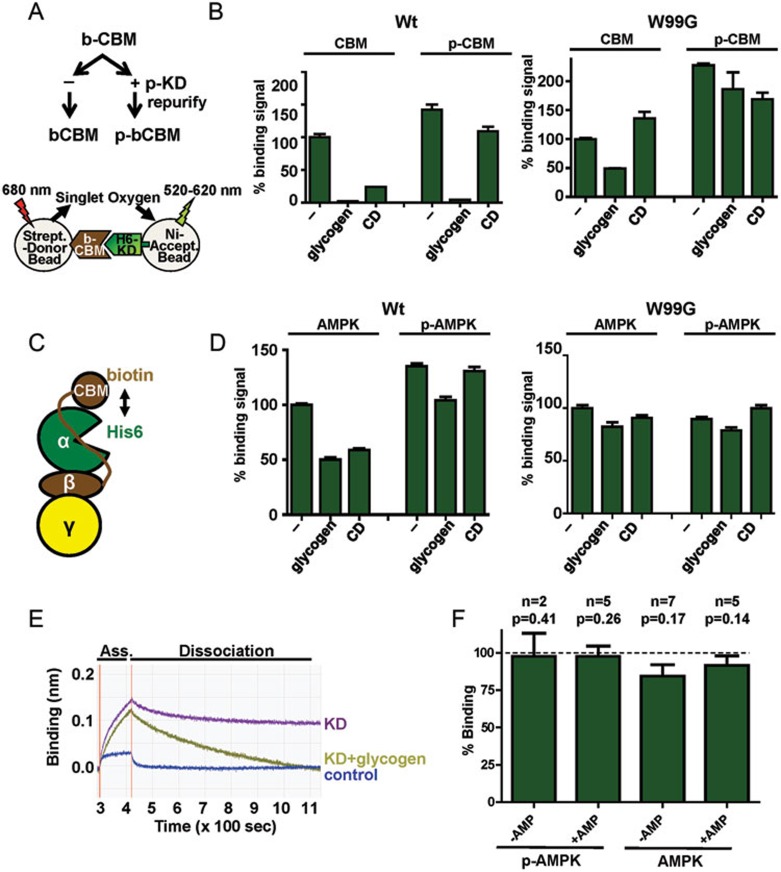
We biochemically analyzed the CBM-KD interaction by AlphaScreen assay using the isolated CBM and KD. For these experiments we split a purified biotin-CBM prep in two aliquots. One aliquot was phosphorylated by the KD in trans, the other was left untreated (Figure 3A and Supplementary information, Figure S4). Samples from both aliquots were then analyzed for binding His6-KD in the presence and absence of glycogen and cyclodextrin (Figure 3B). In the absence of carbohydrates, both phosphorylated and non-phosphorylated CBM bound to the KD, while the binding signal of phosphorylated CBM was stronger than that of non-phosphorylated CBM. Unexpectedly, addition of glycogen almost abolished this interaction, while cyclodextrin greatly reduced the interaction with non-phosphorylated CBM and mildly with phosphorylated CBM. This observation was confirmed by biolayer interferometry, using an Octed Red instrument (Figure 3E). The effect of glycogen and cyclodextrin on the interaction was dependent on their binding to the CBM as mutation of W99, a key residue for cyclodextrin/glycogen binding31,34, greatly blunted the signal reduction. Together, these results support that phosphorylated CBM binds the KD more potently than non-phosphorylated CBM and they demonstrate that binding of carbohydrates to the CBM strongly reduces CBM-KD binding, especially for the weaker interaction with non-phosphorylated CBM.
Figure 3.
Carbohydrates destabilize the CBM-KD interaction. (A) Experimental outline. b-, biotinylated, p-, phosphorylated. (B) AlphaScreen interaction between phosphorylated and non-phosphorylated wild-type (WT) and mutant (W99G) biotin-CBM and the isolated His6-KD in trans (n = 3; error bars, SD). (C) Cartoon presentation of the AlphaScreen experiment in the context of holo-AMPK. (D) Glycogen and cyclodextrin reduce the AlphaScreen signal for holo-AMPK containing biotinylated CBM and His6-tagged KD (n = 3; error bars, SD). (E) Interaction of the CBM with KD by biolayer interferometry. Biotinylated CBM was immobilized to the biosensor of an Octed Red instrument. Addition of the untagged KD in the presence or absence of glycogen resulted in an increase in mass due to CBM binding (association), followed by a decrease after washing away unbound protein due to dissociation of the interaction. (F) Effect of glycogen on the kinase activity of phosphorylated and non-phosphorylated AMPK in the presence and absence of 200 μM AMP. Activity was determined by radioactive kinase assays as shown in Supplementary information, Figure S1 and represented as % activity in the presence of glycogen relative to the absence of glycogen (error bars, SD; n, number of replicate experiments using independent AMPK preps; P, probability for observed inhibition occurring by chance (one-sided, paired t-test); glycogen, 1 mg/ml glycogen, CD, 2 mM cyclodextrin.
We also analyzed the effect of carbohydrates on the CBM-KD interaction in the context of phosphorylated and non-phosphorylated holo-AMPK (Figure 3C and 3D). In the case of holo-AMPK, dissociation of the CBD-KD interaction results in a reduction, rather than abolishment, of the AlphaScreen signal since CBM and KD remain components of the same complex. Similarly, both glycogen and CD caused a significant (P < 0.01) signal decrease in the context of non-phosphorylated AMPK and a smaller decrease in the context of phosphorylated AMPK, indicating that carbohydrate binding modulates the CBM-KD interaction also in the context of the AMPK complex (Figure 3D).
When we analyzed the effect of glycogen on AMPK catalytic activity using radioactive kinase assays, we failed to detect an effect in the context of phosphorylated AMPK and saw potential very subtle (10%-20%) inhibition in the context of non-phosphorylated AMPK (Figure 3F and Supplementary information, Figure S1B). While the trend toward subtle inhibition by glycogen is clear, the effect is not statistically significant (P > 0.1) with the small extent of inhibition and the limited number of independent AMPK preparations tested.
The human AID can form a compact domain that stabilizes the KD in an inactive conformation
Recent structural studies have revealed the conformational changes of the AID as a key determining factor in regulating AMPK activity26,27. Consistent with the reassigned structure of AMP-bound AMPKΔCBM26, the KD in our structure adopts a closed conformation characteristic for active protein kinases, in which the AID is not directly bound to the KD, but instead packs against the γ-subunit (Figure 2C). In contrast, in the structure of a yeast KD-AID fragment, the AID is bound to the hinge region of the KD, which adopted an inactive, “open” conformation28. Based on this structure and mutational analysis of human AMPK, Chen et al.28 hypothesized that binding of the AID stabilizes the open KD conformation and that direct activation of AMPK catalytic activity is mediated at least in part by dissociation of this inhibitory KD-AID interaction.
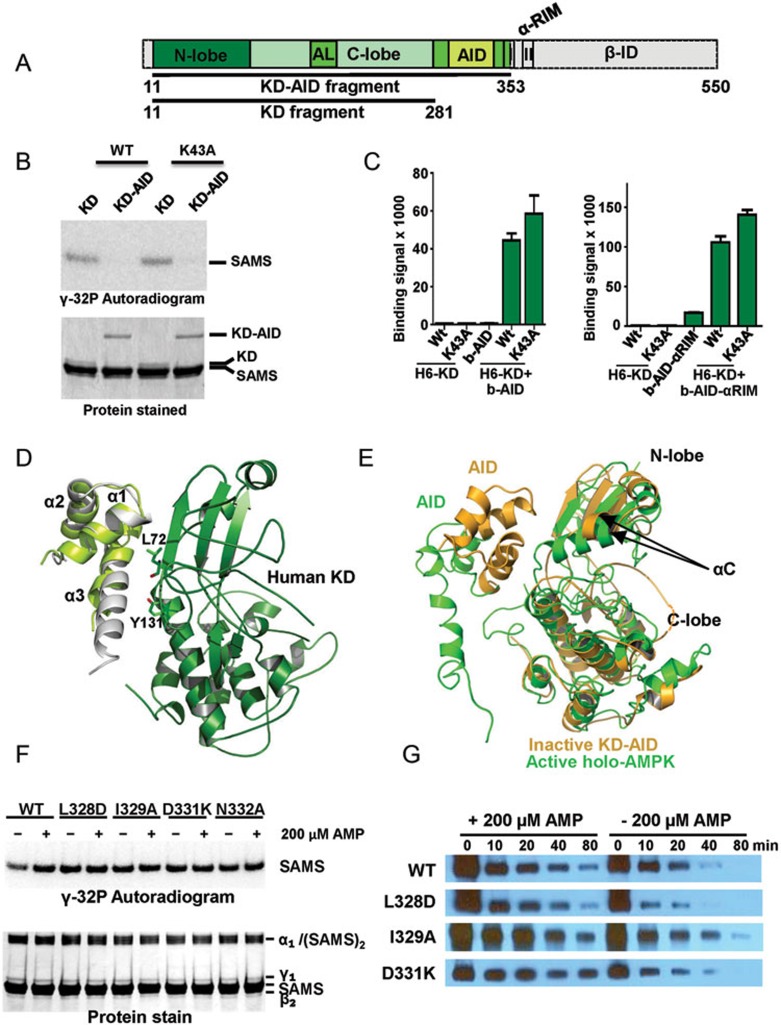
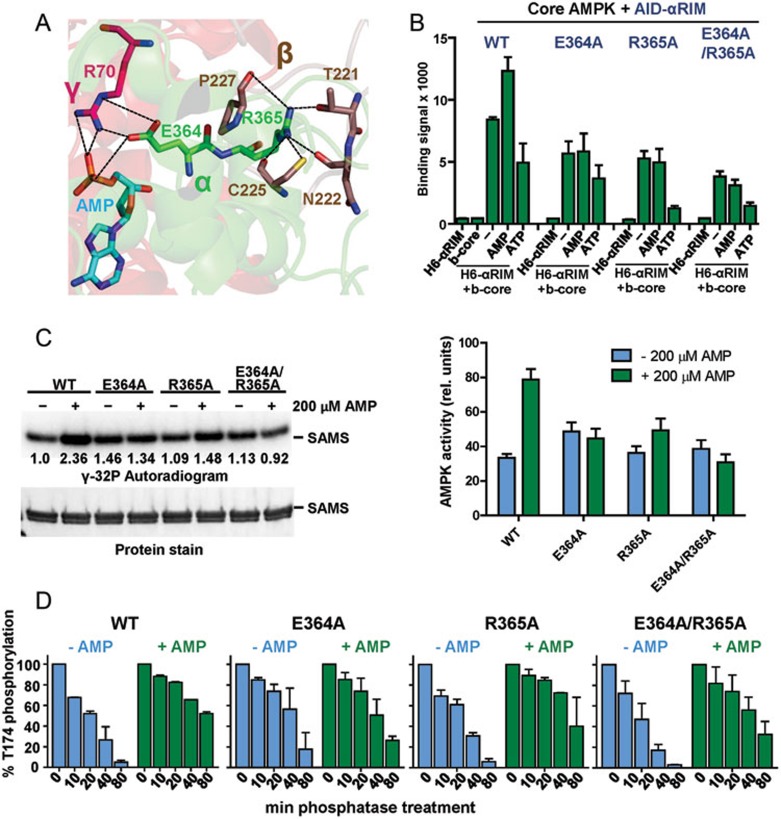
In Figure 4B we recapitulated the inhibitory effect of the AID on the catalytic activity of the AMPK KD in kinase assays using [γ-32P]-ATP and a His6GST fusion peptide from the AMPK substrate acetyl-CoA carboxylase (SAMS peptide38). Note that the presence of the AID reduced the kinase activity by at least one order of magnitude. In order to test whether the human AID can also directly bind to the KD, we employed an Alpha-Screen assay using a purified His6-tagged KD with either biotinylated AID or biotinylated AID-αRIM (Figure 4C). In both cases, the isolated domains interacted robustly with each other, suggesting that in the absence of constraints the human AID binds and inhibits the KD. The original KD-AID structure was solved from the evolutionarily distant yeast S. pombe, which has not yet been demonstrated to be regulated by AMP, because the human KD-AID fragment was recalcitrant to crystallization36. Consistently, we were unable to crystallize any of several different constructs encompassing the wild-type human KD and AID. Based on our experience with holo-AMPK, we introduced entropy-reducing mutations into flexible, solvent-exposed surface residues of AMPK α1 (11-353). One of them, KD-AID K43A, readily formed crystals that diffracted X-rays to a resolution of 2.95 Å (Table 1 and Supplementary Information, Figure S10). The K43A mutation did not affect kinase activity, interaction between KD and AID, or inhibition of kinase activity by the AID (Figure 4B and 4C).
Figure 4.
The human AID can bind the KD hinge to keep the KD in an inactive, open conformation. (A) Schematic presentation of the human α1-subunit. Numbers indicate amino acid positions. The KD-AID fragment used for crystallization (α1 11-353) is shown in color. AL, activation loop; I, αRIM-1; II, αRIM-2. (B) The AID inhibits AMPK KD catalytic activity. Wild-type (WT) and K43A recombinant KD (α1 11-281) and KD-AID (α1 11-353) were incubated with His6GST-SAMS peptide substrate and [γ-32P]-ATP. Terminated reactions were separated by SDS-PAGE and subjected to autoradiography. (C) AlphaScreen interaction between wild-type (WT) and K43A His6-tagged KD and biotinylated AID (α1[282-353]), left) or AID-αRIM (α1[282-392]), right). (D) Structure model of the human inhibitory cis KD-AID complex. The three α-helices of the human AID are shown in light green, overlaid on the structure of the yeast AID (PDB, 3H4J) in grey. Key AID-interacting residues of the KD are shown in stick presentation. (E) Superposition of the KD-AID inhibitory complex from D (pale orange) with the KD-AID conformation in the context of activated holo-AMPK (green; detail from Figure 2D). αC:αC helix, whose position tilted toward the kinase C-lobe, is critical for the correct positioning of ATP in the substrate-binding cleft. (F) Mutations that disrupt the KD-AID interaction make AMPK constitutively active. Wild-type (WT) and mutant KD-AID (α1 11-353) were incubated with His6GST-SAMS peptide (SAMS) substrate and [γ-32P]-ATP. Terminated reactions were separated by SDS-PAGE and subjected to autoradiography. See Supplementary information, Figure S7 for repeat experiments and SD. (G) Mutations that disrupt the KD-AID interaction do not affect AMP-dependent stabilization of activation loop phosphorylation. Phosphorylated WT and mutant AMPK were incubated with human PP2Cα in the absence or presence of 0.2 mM AMP for the indicated amount of time, separated by SDS-PAGE, and subjected to immunoblotting with an antibody specific for T174-phosphorylated AMPK. See Supplementary information, Figure S7 for repeat experiments and SD.
The structure of the human KD-AID fragment shows a dimeric arrangement, in which helix-α3 and the loop region of the AID from one monomer interact with the N-lobe and hinge region of the KD of the other monomer, such that the KD-AID interaction forms the dimer interface (Supplementary information, Figure S5A). The observed dimer formation is due to crystal packing, while the physiological interaction occurs in cis (within one monomer) (Supplementary information, Figure S5B), as demonstrated by size exclusion chromatography at moderate AMPK concentrations (Supplementary information, Figure S5C) and by mutational analysis (Supplementary information, Figure S6). The mutational analysis demonstrates that the interaction occurs between AID α3 and the KD hinge, as in yeast and as shown in the structure model in Figure 4D (see legend of Supplementary information, Figure S6), with mutations in the interface (KD: L72A, Y131A; AID: L328A, I329A, D331K, N332A) relieving the inhibitory effects of the AID.
AMP binding shifts the equilibrium between AID-KD and AID-γ-subunit interactions
Overlay of the structures of active, phosphorylated AMP-bound holo-AMPK with that of the inactive KD-AID fragment (Figure 4E) clearly shows two alternative AID conformations: the AID with helix α3 either bound to the KD in the inactive, open conformation or being released from the inhibitory KD interaction and bound to the γ-subunit. In agreement with previous results28, mutations that disrupt the interaction between the KD and the AID in the context of the KD-AID fragment abolished AMP regulation of the kinase activity in the context of full-length AMPK and render AMPK constitutively active (Figure 4F and Supplementary information, Figure S7A), providing strong support that the same inhibitory AID-KD interaction also occurs in the context of full-length AMPK. Together with the two AID and KD conformations observed (Figure 4E), these data support that activation by AMP induces a conformational change in AMPK that releases the inhibitory KD-AID interaction to allow the KD to adopt the active, closed conformation. Importantly, the same interface mutations that abolished AMP regulation of kinase activity did not significantly affect AMP-mediated protection against activation loop dephosphorylation (Figure 4G and Supplementary information, Figure S7B), indicating that activation loop accessibility to phosphatases is not regulated by AID interaction.
AMP and ATP regulate the interaction between AID-αRIM and the core complex
Consistent with previous structures of AMPK complexes, the αRIM-2 in active AMPK sticks into the AMP-occupied CBS-3 pocket and αRIM-2 E364 and R365 form a network of interactions with AMP, R70 of the γ-subunit, and several residues of a C-terminal loop of the β-subunit (Figure 5A)25,26,18,39. It has been proposed that ATP binding to CBS-3 would interfere with this interaction21, although it is not obvious from the structure (Figure 5A). Moreover, in the presence of high concentrations of ATP, ATP binding to CBS-4 and CBS-1 would sterically interfere with ATP occupancy at CBS-3 as shown in the structure of the ATP-occupied AMPK core complex40.
Figure 5.
Adenine nucleotide-mediated conformational changes in AMPK depend on inducible α-RIM-CBS3 and AID-KD interactions. (A) Interaction network between AMP, CBS-3 R70, αRIM-2 E364 and R365, and 4 residues from an induced loop of the β-subunit. AMP and key amino acids are shown in stick presentation with bonds indicated as dashed lines. (B) AMP increases and ATP decreases the interaction between AID-αRIM and core AMPK(α1[395-550]-β2[187-272]-γ1). AlphaScreen interaction between biotinylated core AMPK and wild-type (WT) and mutant His6GST-AID-αRIM in the presence and absence of adenine nucleotides. (C) Mutations that disrupt the α-RIM-CBS3 interaction abolish direct AMP regulation of AMPK catalytic activity. Radioactive kinase assay showing 32P incorporation into SAMS substrate (left, representative autoradiogram; right, quantification of 32P incorporation based on three independent experiments (error bars, SD). (D) Mutations that disrupt the α-RIM-CBS3 interaction reduce AMP-mediated protection against activation loop dephosphorylation. Bars represent relative AMPK T174 phosphorylation as determined by immunoblot, following the indicated time of incubation with PP2Cα (error bars, SD; n = 3).
To directly test the prediction that AMP- and ATP-binding modulate the AID-αRIM interaction with core AMPK (α1[395-550]-β2[187-272]-γ1), we analyzed the interaction between biotinylated core complex and His6-tagged AID-αRIM fragment (α1 282-392). As shown in Figure 5B, the core complex in the absence of added nucleotides is capable of interacting with the AID-αRIM fragment and this signal increased in the presence of AMP and decreased in the presence of ATP, providing direct evidence that the AID-αRIM can indeed function as “adenine nucleotide sensor”. Moreover, mutations of the key two αRIM-2 residues, E364 and to a lesser degree R365, made the interaction between the AID-αRIM and the core complex AMP-independent (Figure 5B), and, in agreement with Chen et al.40, abolished or strongly reduced the ability of AMP to increase allosteric kinase activity (Figure 5C) and to protect against activation loop dephosphorylation (Figure 5D). Therefore, AMP/CBS-3 sensing by αRIM-2 is required for regulation of both kinase activity and accessibility of the phosphorylated activation loop, whereas the interaction between AID and KD is only required for regulation of kinase activity.
In contrast, the same two mutations did not affect the ability of ATP to weaken the AID-αRIM interaction with the core complex, indicating that ATP does not predominantly signal through CBS-3/αRIM-2. Since the interaction of AID-αRIM with core AMPK is mediated by both αRIM-2 and αRIM-1, ATP may indirectly affect the CBS-2/αRIM-1 interaction as has been suggested27. Collectively, these results indicate that the interaction between AID-αRIM and core AMPK is stabilized by CBS-3/AMP-αRIM-2 binding and destabilized by ATP independent of nucleotide binding to CBS-3.
Discussion
In this paper we present the structures of AMP- and cyclodextrin-bound holo-AMPK in the phosphorylated and non-phosphorylated state and of an KD-AID complex in the autorepressed state. Together with biochemical and mutational data these structures provide important new insight into the complex modulation of AMPK by adenine nucleotides and glycogen. First, using a modified AlphaScreen assay we could detect distinct adenine nucleotide-induced conformational changes in the context of holo-AMPK. In contrast to kinase assays, which cannot independently assess the effect of ATP because ATP is also a substrate of the reaction, this assay allowed us to demonstrate that AMP and ATP induce distinct conformational changes in AMPK. In the presence of AMP, inter-subunit distances decreased, indicating a more compact state that correlates with less accessibility of the activation loop to phosphatases. Conversely, ATP induced a more extended and accessible state. Strikingly, the concentration of AMP to induce the AMPK conformational change is almost three orders of magnitude lower than that of ATP (Figure 1D). Competition experiments suggested that both CBS-1 and CBS-3 bind AMP and ATP with similar affinities21, while CBS-4 appears to bind AMP with much higher affinity than ATP21,40. This suggests that CBS-4 has a critical role in adenine nucleotide discrimination, in agreement with the essential roles of both CBS-3 and CBS-4 for allosteric AMPK regulation as suggested by mutational studies16,40.
Second, we could directly demonstrate that AMP and ATP modulate the strength of the interaction between the AID-αRIM module and the AMPK core complex. Importantly, this interaction also occurs independently of adenine nucleotides. AMP and ATP do not cause all-or-nothing effects, but induce moderate equilibrium changes that in turn affect the equilibrium between AID-KD and AID-core AMPK interactions. These relatively subtle equilibrium changes are functionally reflected by a relatively small activity difference induced by AMP in the context of holo-AMPK (Figures 4F and 5C) compared to an at least 10-fold difference between the activity of the isolated KD and KD-AID (Figure 4B, an all-or-nothing effect in the absence of the competing AID-core AMPK interaction).
Both the αRIM-CBS-3 and the AID-KD interface play critical roles in adenine nucleotide-mediated allosteric kinase activation: mutations of the αRIM2-CBS-3 interface make AMPK largely constitutively inactive (resembling the activity of wild-type AMPK in the absence of AMP, Figure 5C) while mutations of the KD-AID interface render AMPK largely constitutively active (resembling the activity of wild-type AMPK in the presence of AMP, Figure 4F). In contrast, only mutations of the αRIM2-CBS-3 interface reduced the effect of AMP on protection against activation loop dephosphorylation. While the activation loop in AMP-bound holo-AMPK has limited accessibility due to the packing of the CBM linker and the C-termini of the α- and β-subunits18,27 (see Figure 2C), full understanding of the underlying mechanism of AMP-mediated activation loop protection will require structures of holo-AMPK both in AMP- and ATP-bound form.
Fourth, the ability of ATP to induce a distinct conformational change relative to nucleotide-free AMPK (Figure 1C and 1D) suggested that ATP does not only function as AMP competitor, but instead also as active allosteric modulator. We corroborated this finding by demonstrating that mutation of key αRIM-2 residues abolished AMP-dependent, but not ATP-dependent, regulation of AMPK (Figure 5B), indicating that ATP does not predominantly signal through αRIM-2/CBS-3. Since ATP destabilizes the AID-αRIM1/2 interaction with core AMPK (Figure 5B), binding of the larger ATP likely indirectly destabilizes the interaction of αRIM-1 with the nucleotide-free CBS-2.
Finally, we present evidence that the CBM is a dynamic domain, whose interaction with the KD is modulated by carbohydrate binding and by phosphorylation. While the cyclodextrin-bound CBM is dissociated from the KD in our structure of non-phosphorylated AMPK, it is bound to the KD in phosphorylated AMPK. These structure snapshots are consistent with our binding studies using the isolated CBM and KD, in which cyclodextrin strongly reduced the interaction with non-phosphorylated CBM, but only weakly reduced the interaction with phosphorylated CBM. Two additional lines of evidence further support the dynamic interaction between CBM and KD. First, CBM-containing holo-AMPK in the absence of surface entropy-reducing mutations or internal deletions crystallized only in the presence of CBM-KD interaction-stabilizing compounds18. Second, CBM S108 is efficiently cis-autophosphorylated in holo-AMPK32, yet when bound to the KD, it lies outside of the substrate-binding groove of the KD39, implying that the non-phosphorylated CBM must be capable to adopt a different position in order to become phosphorylated39.
Compounds 991 and A769662 stabilize the CBM-KD interaction in β1 subunit-containing AMPK complexes and induce formation of an α-helix in the flexible linker C-terminal to the CBM that packs against and shifts the position of the αC-helix of the KD18. The position of the αC-helix is critical for ATP binding and is regulated in many protein kinases41, including by parallel intramolecular packing in a class of AMPK homologous plant stress kinases42. The strong activating effect of compounds 991 and A769662 and the inhibitory effects of mutating an αC-binding residue from the C-interacting helix27 therefore suggests that these compounds activate AMPK by stabilizing a selective αC position. Formation of the C-interacting helix likely requires a CBM-KD interaction as this region is not resolved when the CBM is dissociated from the KD (cyclodextrin-bound non-phosphorylated AMPK), and is incompletely formed when the interaction is destabilized (cyclodextrin-bound phosphorylated CBM). While stabilizing the CBM-KD interaction and inducing formation of the C-interacting helix is associated with AMPK activation, destabilizing the interaction by mutating S108 in β1-containing AMPK complexes reduces AMPK activity37.
By analogy, the strong destabilizing effect of glycogen on CBM-KD interaction could explain the inhibitory effect of glycogen on AMPK kinase activity that was reported by McBride et al.31. While we failed to detect a clear inhibitory effect of glycogen on the activity of AMPK (consistent with Polekhina et al.30), glycogen-mediated CBM-KD destabilization (Figure 3B, 3D and 3E) may require cellular cofactor(s) or other (e.g., β1-subunit containing) AMPK isoforms to cause inhibition of kinase activity.
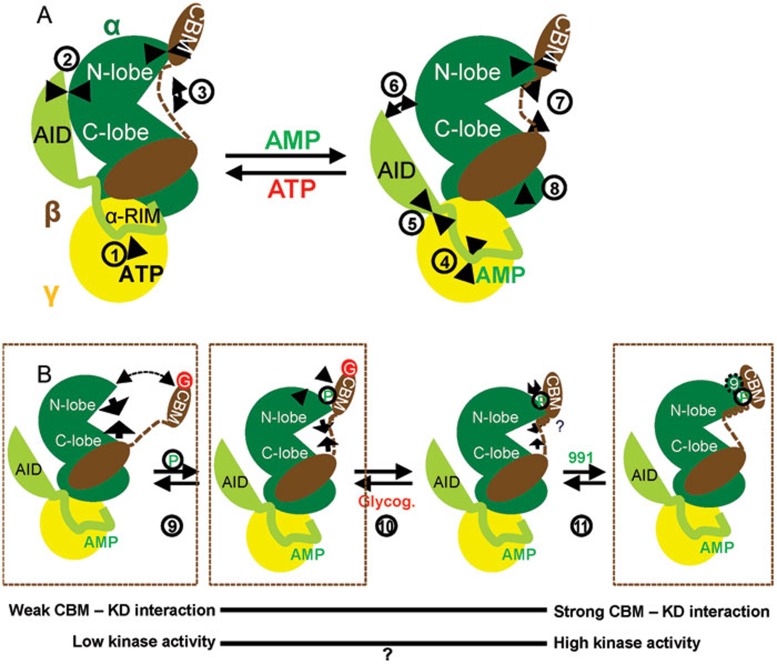
In summary, our data illustrate AMPK as a molecular machine with four interconnected moveable parts: the CBM, the αRIM, the AID, and the KD N- and C-lobes. We present a cartoon model of AMPK modulation by adenine nucleotides (Figure 6, panel a) and by glycogen (Figure 6, panel b). In the presence of ATP (Figure 6A, left), the interaction of the AID-αRIM with core AMPK is weakened (1), shifting the equilibrium toward AID release from CBS-2 to engage in the inhibitory interaction with the KD (2). The AID-KD interaction in turn locks the KD in an open, inactive conformation (3), resulting in low catalytic activity. In addition, release of the αRIM-2/CBS-3 interaction causes conformational changes that make AMPK less compact and the kinase activation loop more accessible to phosphatases. In the presence of AMP (right), the AID-αRIM interaction with core AMPK becomes stabilized (4), shifting the equilibrium toward AID binding to CBS-2 (5) and concomitant release from the KD (6). Release from autoinhibition allows the N- and C-lobes of the KD to move toward each other to adopt a closed, active conformation (7). AMP binding to CBS-3 also causes incompletely understood conformational changes (8) to make AMPK more compact and thereby the activation loop less accessible to phosphatases. While adenine nucleotides allosterically regulate the kinase activity by modulating the interaction of the AID with the hinge region of the KD, glycogen binding and CBM phosphorylation modulate the interaction of the CBM with the N-lobe of the KD (Figure 6B). Phosphorylation (9), release of carbohydrate binding (10), and the binding of pharmacological activators and their hypothesized physiological counterparts (11) increasingly shift the binding equilibrium toward a more stable CBM-KD interaction, formation and positioning of the C-interacting helix in the CBM, and concomitant repositioning of the αB and αC helices in the KD, as illustrated by biochemical analysis and by structure snapshots of three of these states. While the S108A mutation in the β1 subunit-containing CBM (CBM non-phosphorylatable without affecting KD phosphorylation) reduces kinase activity37 and CBM-KD interaction-stabilizing compounds 991 and A769662 increase kinase activity18,43, effects of glycogen on AMPK activity remain controversial (question mark in Figure 6B) and require additional investigation.
Figure 6.
Cartoon model of allosteric AMPK regulation. (A) Regulation by adenine nucleotides. (B) Regulation by glycogen and compound 991. The α-subunit of AMPK is shown in green, with the AID-αRIM module in light green, the β-subunit in brown, and the γ-subunit in yellow. Arrows indicate the directions of allosteric modulator-induced movements within the AMPK complex, as referred to in the text by the numbers in circles next to the arrows. P, CBM phosphorylation; G, glycogen; 9, compound 991. See text for details.
While this paper was in revision, Calabrese et al.39 published the structure of a phosphorylated, CBM-containing α1β1γ1-AMPK complex in the absence of carbohydrates and pharmacological activators (PDB code: 4QFG). In agreement with our model in Figure 6B, the CBM in this structure adopts a compact, KD-bound conformation with an interface that has features of both cyclodextrin-bound and pharmacological activator-bound AMPK (well-resolved C-interacting helix, as in 991-bound AMPK, but in a position that is similar to that of the partially disordered helix in the structure of cyclodextrin-bound phosphorylated AMPK, αB-helix position similar to 991-bound AMPK and αC-helix position similar to cyclodextrin-bound AMPK).
Materials and Methods
Protein preparation and crystallization of holo-AMPK and AMPK KD-AID
To identify the most suitable crystallization construct, we first cloned and expressed all 12 different isoforms of human AMPK in a pET28 (Novagen)-derived tri-cistronic bacterial expression plasmid, similar to previously published constructs44. The α-subunits in these constructs were N-terminally His6-tagged, while the β- and γ-subunits remained untagged. E.coli BL21 (DE3) cells transformed with these expression plasmids were grown in 2-4 L LB broth at 16 °C to an OD600 of ∼1.0 and induced with 50 μM IPTG for 18 h. Cells were harvested, resuspended in 150 ml extract buffer (50 mM sodium phosphate, pH 8.0, 15% sucrose, 500 mM NaCl, 25 mM imidazole, 3 mM β-mercaptoethanol), and lysed by passing through a French Press with pressure set at 900 Pa. Lysates were centrifuged at 18 000 rpm in a Sorvall SS34 rotor for 30 min, and supernatants were loaded on a 10 ml Nickel HP column. The columns were washed with 150 ml 95% extract buffer/5% buffer B (50 mM sodium phosphate, pH 8.0, 15% sucrose, 500 mM NaCl, 500 mM imidazole, 3 mM β-mercaptoethanol) and eluted with 100% buffer B. The eluted His6-tagged AMPK was further purified by chromatography using a HiLoad 26/60 Superdex 200 gel filtration column in 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, 10 mM DTT. Of the 12 different isoforms, the four constructs that contained the γ1-subunit expressed and purified well, with the two α1-containing constructs yielding the highest expression levels. Since the γ1-subunit in the α1β1γ1 complex purified slightly substochiometrically, we identified human α1β2γ1 as the most stable and best expressed isoform. As anticipated, none of the 12 purified complexes crystallized under any of the conditions tested, indicating that the complexes required further engineering.
Based on existing crystal structures, homology comparison between AMPK sequences from different species, and disorder and secondary structure predictions, we designed the trimmed construct AMPK α1(11-550)-β2(76-272)-γ1(24-327) shown in Figure 2A. Notably, this complex had increased proteolytic stability and decreased aggregation relative to full-length AMPK. Crystallization further required introduction of surface entropy reduction (SER) mutations, which replaced solvent-exposed clusters of charged, high-entropy residues (K, E, and Q) with the small, low-entropy residues A and G. We generated 16 different sets of mutant constructs that were each expressed, purified in the phosphorylated and non-phosphorylated state, and complexed with either (i) AMP, (ii) AMP and the kinase inhibitor staurosporine, or (iii) AMP, staurosporine, and cyclodextrin.
We quantitatively phosphorylated a part of the purified AMPK complex by overnight (16 h) incubation with CaMKKβ in 0.2 mM ATP, 0.2 mM AMP, 2 mM CaCl2, 10 mM DTT, and 1 μM calmodulin at room temperature. The phosphorylated AMPK protein was repurified by chromatography using a HiLoad 26/60 Superdex 200 gel filtration column in 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, and 10 mM DTT. Phosphorylated and non-phosphorylated AMPK monomer fractions were incubated with 3 mM staurosporine and 4 mg/ml β-cyclodextrin hydrate for 3 h at 4 °C, spin-concentrated to ∼7 mg/ml, and incubated with 5 mM AMP on ice for 1 h. Both AMPK preparations with mutations E471G/E474A/K476A in the α-subunit complexed with AMP, staurosporine, and cyclodextrin crystallized and diffracted.
Crystals of phosphorylated AMPK were grown at ∼18 °C in sitting drops containing 0.3 μl of purified protein at 6 mg/ml and 0.2 μl of well solution in 0.1 M HEPES, pH 7.8, 12% PEG 4000, 10% 2-propanol (v/v) and 0.19 mM 7-cyclohexyl-1-heptyl-β-D-maltoside. Rod-shaped crystals of about 60 μm in length appeared in 1-2 days. Crystals were cryo-protected in 20% 2-methyl-2,4-pentanediol before flash freezing in liquid nitrogen. Crystals of non-phosphorylated AMPK were grown at ∼18 °C in sitting drops containing 0.22 μl of purified protein at 10.5 mg/ml and 0.2 μl of well solution in 0.1 M N-acetamido-iminodiacetic acid, pH 6.8, 9% 2-methyl-2,4-pentanediol, and 11.5 mM C-HEGA -11. Crystals of about 100-150 μm in length appeared in 1-2 days. Crystals were cryo-protected in 30% 2-methyl-2,4-pentanediol before flash freezing in liquid nitrogen.
Mutant K43A human AMPK KD-AID [α1(11-353)] was expressed as a fusion protein with a His6 tag at the N-terminus in E. coli BL21 (DE3). Cells were grown to an OD600 of ∼1, followed by induction with 100 μM of IPTG at 16 °C. After overnight incubation, the cells were harvested and resuspended in buffer A (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 15% sucrose, 20 mM imidazole and 3 mM β-mercaptoethanol). Cells were lysed using the French Press with pressure set at 1 000 Pa. Lysates were centrifuged at 18 000 rpm for 40 min in a Sorvall S34 rotor. The supernatants were loaded onto a pre-equilibrated 10-ml Ni-NTA agarose column. The column was washed with 150 ml 94% buffer A/6% buffer B (50 mM sodium phosphate, pH 8.0, 500 mM NaCl, 15% sucrose, 500 mM imidazole, 3 mM β- mercaptoethanol) and eluted with 100% buffer B. The eluted protein was further purified by Superdex 200 gel filtration chromatography (Amersham Biosciences) in 10 mM Tris, pH 8.0, 5 mM magnesium chloride, 100 mM sodium chloride, 1 mM EDTA, and 1 mM DTT, and concentrated to 8-9 mg/ml for crystallization.
Human AMPK α1 (11-353) K43A crystals were grown at ∼20 °C in hanging drops containing 0.5 M HNa2PO4/0.5 M HK2PO4, and 0.1 M Tris hydrochloride, pH 8.5, and 0.1 M H(NH4)2PO4. Crystals were mounted and soaked in the mother liquor solution supplemented with 20% ethylene glycol prior to flash freezing with liquid nitrogen.
His6-tagged wild-type and mutant human AMPK used for biochemical analysis were expressed and purified following the same method used to purify protein for crystallization except that all buffers contained 10% glycerol (v/v). To generate biotinylated proteins, we expressed the β- and γ-subunits of full-length AMPK, the AID (α1[282-353]), AID-αRIM (α1[282-392]), CBM (β2[75-163]), and core AMPK complex (α1[395-550]-β2[187-272]-γ1) as avitag fusions from the first T7 polymerase-driven expression unit of a pETDuet (Novagen) derivative vector. The 14 amino acid avitag45 functions as defined biotinylation site that in combination with the biotin ligase BirA expressed from the second expression unit is quantitatively biotinylated in vivo in E. coli in the presence of 40 μM biotin. His6-KD was phosphorylated by overnight incubation with CaMKKβ in 0.2 mM ATP, 0.2 mM AMP, 2 mM CaCl2, 10 mM DTT and 1 μM calmodulin at room temperature. Phosphorylated His6-KD was repurified by chromatography using a HiLoad 26/60 Superdex 200 gel filtration column in 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, and 10 mM DTT. Wild-type and W99G biotin-CBM were phosphorylated by incubation with the phosphorylated His6-KD at a CBM:KD ratio of 50:1 for 4 h at room temperature. Phosphorylated CBMs were separated from His6-KD by Nickel HP column chromatography and further purified by size exclusion chromatography in 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 10% glycerol.
Purification of CamKKβ, His6GST-SAMS, and PP2Cα
CamKKβ was expressed as N-terminally MBP-tagged protein from a pMAL-derived bacterial expression plasmid. Expression and purification of CamKKβ were carried out as for AMPK, with the difference that maltose agarose instead of Ni-agarose chromatography was used as first purification step. The maltose agarose column was washed with buffer containing 20 mM Tris, pH 8.0, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, and 10% glycerol and eluted with the same buffer in the presence of 20 mM maltose. SAMS and human PP2Cα(aa1-340) were expressed from pET24 vectors as fusion proteins with a His6GST tag at the N-terminus in E. coli BL21 (DE3). Cells were grown and purified as described for AMPK KD-AID, with the addition of 10% glycerol in the gel filtration buffer. Proteins were concentrated to 5-10 mg/ml and stored at −80 °C.
Structure determination and refinement
All diffraction data sets were collected at 100 K using an X-ray beam with Rayomics 300 CCD detectors at the ID-E beamline of sector-21(LS-CAT) at the Advanced Photon Source at Argonne National Laboratory. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package46. The resolution cutoff is based on the criteria of l/sigma ratio = 2.0 in the highest resolution shell as guided by the HKL2000 package. Molecular replacements were performed by using the Collaborative Computational Project 4 (CCP4) program Phaser47 with the mammalian AMPK complex structure (PDB: 2Y94) and the human α2 kinase domain (PDB: 2H6D) as search models. Programs O48 and Coot49 were used to manually fit the protein models. Model refinement was performed with CNS50 and Phenix program package51. All figures were prepared using PyMOL (DeLano Scientific).
AlphaScreen assays
Protein-protein interactions and conformational changes were assessed by luminescence-proximity AlphaScreen technology as described previously52. Protein-protein interaction reactions contained 100 nM recombinant His6-tagged proteins bound to nickel-acceptor beads and 100 nM recombinant biotin-tagged proteins bound to streptavidin donor beads. In the reactions assessing conformational changes, 100 nM recombinant biotin-γ1(residues 24-327)-β2 (residues 76-272)-His6-α1(residues 11-550) was attached to nickel-chelated acceptor beads and streptavidin-coated donor beads. All reactions were performed in buffer containing 50 mM MOPS, pH 7.4, 50 mM NaF, 50 mM CHAPS, and 0.1 mg/ml bovine serum albumin. Where present, bovine liver glycogen (Sigma) had a final concentration of 0.5 mg/ml (∼3 mM glucose units) and cyclodextrin of 2 mM. With the exception of the dose-response curves in Figure 1, AMP concentrations were fixed at 0.2 mM and ATP concentrations at 1 mM.
Kinase activity assays
1 μM non-phosphorylated His6-AMPK, His6-α1 KD, or KD-AID or 10 nM phosphorylated AMPK were incubated with 14 μM His6GST-SAMS in 10 mM Tris, pH 8.0, 5 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, 250 μM unlabeled ATP and 2 μCi [γ-32P]-ATP for 10-20 min at room temperature in a total volume of 15 μl. Reactions were terminated by boiling in SDS sample buffer and subjected to Tricine SDS-PAGE. Gels were stained with Coomassie Blue and destained with 10% v/v methanol/10% v/v acetic acid, dried, and subjected to autoradiography using a FLA-5000 phosphor imager (Fuji) and quantitated using ImageGauge software.
Biolayer interferometry
Binding curves were measured by biolayer interferometry using an Octet Red instrument (ForteBio). For the biolayer interferometry assay, a layer of molecules attached to the tip of the biosensor creates an interference pattern at the detector. Any change in the number of molecules bound due to protein-protein interactions causes a measured shift in the interferometric profile. When this shift is measured over a period of time and its magnitude plotted as a function of time, a classic association/dissociation curve is obtained. However, due to rebinding after dissociation, Octed Red is known to underestimate dissociation rates53. A streptavidin sensor was used to bind biotinylated CBM in kinetic buffer (10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, 10 mM DTT). The protein-loaded biosensors were incubated sequentially with kinetic buffer in the baseline step and the purified His6-KD in solution in the association step and kinetic buffer in the dissociation step to detect their binding to biotin-CBM. Assays were performed in the presence or absence of 0.5 mg/ml bovine liver glycogen. The control is biotinylated CBM without KD.
Phosphatase assay and immunoblotting
7 μM phosphorylated AMPK protein were incubated with 1 μM H6GST-PP2Cα and 0.2 mM AMP in 10 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, and 10 mM DTT in a total volume of 20 μl for 0, 10, 20, 40, or 80 min at room temperature. Reactions were started by adding PP2Cα and stopped by addition of SDS sample buffer. Reactions corresponding to ∼80 ng protein were separated by 4%-20% gradient SDS-PAGE and subjected to immunoblotting with an AMPK-α1 p-T174 specific antibody (Abcam) following standard protocols. Densitometric analysis was performed with Quantity One software (BioRad).
Mutagenesis
Site-directed mutagenesis was carried out using the QuikChange method (Agilent). Mutations and all plasmid constructs were confirmed by sequencing.
Acknowledgments
This work was supported by Van Andel Research Institute (HEX and KM), by the National Institute of Health (grant R01 GM104212 to KM), by the National Basic Research Program of China (973 Program; 2011CB504004 and 2010CB945500 to D Wu), by the National Natural Science Foundation of China (to D Wang) and by an overseas PhD scholarship from the National University of Singapore to MHET. We thank Stephanie Grant for administrative support and staff members of the Life Science Collaborative Access Team of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of APS was supported by the Office of Science of the US Department of Energy, under contract no. DE-AC02-06CH11357.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
AMPK α1(E471G/E474A/K476A) is active in subunit interactions, kinase activity, and modulation of kinase activity by adenine nucleotides and glycogen.
Electron density omit maps of ligands in the 4.05 Å structure of phosphorylated AMPK: (a) AMP, (b) staurosporine, (c) cyclodextrin.
Close-up views of the CBM–KD interface and conformation.
CBM phosphorylation by AMPK KD in trans.
Crystal packing stabilizes a KD–AID trans dimer.
KD and AID form a monomeric cis-complex at moderate protein concentrations.
Mutations at the KD–AID interface abolish AMP-dependent allosteric kinase activation, but not protection against activation loop dephosphorylation.
Molprobity statistics (top) and polygon diagram (bottom) of the p-AMPK model.
Molprobity statistics (top) and polygon diagram (bottom) of the model of non-phosphorylated AMPK.
Molprobity statistics (top) and polygon diagram (bottom) of the KD-AID model.
References
- 1Hardie DG, Ross FA, Hawley SA. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol 2012; 13:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev 2009; 89:1025–1078. [DOI] [PubMed] [Google Scholar]
- 3Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 2011; 13:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Hardie DG, Ross FA, Hawley SA. AMP-activated protein kinase: a target for drugs both ancient and modern. Chem Biol 2012; 19:1222–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Crute BE, Seefeld K, Gamble J, Kemp BE, Witters LA. Functional domains of the alpha1 catalytic subunit of the AMP-activated protein kinase. J Biol Chem 1998; 273:35347–35354. [DOI] [PubMed] [Google Scholar]
- 6Pang T, Xiong B, Li JY, et al. Conserved alpha-helix acts as autoinhibitory sequence in AMP-activated protein kinase alpha subunits. J Biol Chem 2007; 282:495–506. [DOI] [PubMed] [Google Scholar]
- 7Hawley SA, Ross FA, Gowans GJ, Tibarewal P, Leslie NR, Hardie DG. Phosphorylation by Akt within the ST loop of AMPK-alpha1 down-regulates its activation in tumour cells. Biochem J 2014; 459:275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Suzuki T, Bridges D, Nakada D, et al. Inhibition of AMPK catabolic action by GSK3. Mol Cell 2013; 50:407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Hawley SA, Boudeau J, Reid JL, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol 2003; 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Shaw RJ, Kosmatka M, Bardeesy N, et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci USA 2004; 101:3329–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Woods A, Johnstone SR, Dickerson K, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol 2003; 13:2004–2008. [DOI] [PubMed] [Google Scholar]
- 12Hawley SA, Pan DA, Mustard KJ, et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab 2005; 2:9–19. [DOI] [PubMed] [Google Scholar]
- 13Woods A, Dickerson K, Heath R, et al. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab 2005; 2:21–33. [DOI] [PubMed] [Google Scholar]
- 14Hurley RL, Anderson KA, Franzone JM, Kemp BE, Means AR, Witters LA. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem 2005; 280:29060–29066. [DOI] [PubMed] [Google Scholar]
- 15Zhang YL, Guo H, Zhang CS, et al. AMP as a low-energy charge signal autonomously initiates assembly of AXIN-AMPK-LKB1 complex for AMPK activation. Cell Metab 2013; 18:546–555. [DOI] [PubMed] [Google Scholar]
- 16Oakhill JS, Chen ZP, Scott JW, et al. beta-Subunit myristoylation is the gatekeeper for initiating metabolic stress sensing by AMP-activated protein kinase (AMPK). Proc Natl Acad Sci USA 2010; 107:19237–19241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Davies SP, Helps NR, Cohen PT, Hardie DG. 5′-AMP inhibits dephosphorylation, as well as promoting phosphorylation, of the AMP-activated protein kinase. Studies using bacterially expressed human protein phosphatase-2C alpha and native bovine protein phosphatase-2AC. FEBS Lett 1995; 377:421–425. [DOI] [PubMed] [Google Scholar]
- 18Xiao B, Sanders MJ, Carmena D, et al. Structural basis of AMPK regulation by small molecule activators. Nat Commun 2013; 4:3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Yeh LA, Lee KH, Kim KH. Regulation of rat liver acetyl-CoA carboxylase. Regulation of phosphorylation and inactivation of acetyl-CoA carboxylase by the adenylate energy charge. J Biol Chem 1980; 255:2308–2314. [PubMed] [Google Scholar]
- 20Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 2007; 8:774–785. [DOI] [PubMed] [Google Scholar]
- 21Xiao B, Sanders MJ, Underwood E, et al. Structure of mammalian AMPK and its regulation by ADP. Nature 2011; 472:230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Gowans GJ, Hawley SA, Ross FA, Hardie DG. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab 2013; 18:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Carling D, Clarke PR, Zammit VA, Hardie DG. Purification and characterization of the AMP-activated protein kinase. Copurification of acetyl-CoA carboxylase kinase and 3-hydroxy-3-methylglutaryl-CoA reductase kinase activities. Eur J Biochem 1989; 186:129–136. [DOI] [PubMed] [Google Scholar]
- 24Oakhill JS, Steel R, Chen ZP, et al. AMPK is a direct adenylate charge-regulated protein kinase. Science 2011; 332:1433–1435. [DOI] [PubMed] [Google Scholar]
- 25Kemp BE, Oakhill JS, Scott JW. AMPK structure and regulation from three angles. Structure 2007; 15:1161–1163. [DOI] [PubMed] [Google Scholar]
- 26Chen L, Xin FJ, Wang J, et al. Conserved regulatory elements in AMPK. Nature 2013; 498:E8–10. [DOI] [PubMed] [Google Scholar]
- 27Xin FJ, Wang J, Zhao RQ, Wang ZX, Wu JW. Coordinated regulation of AMPK activity by multiple elements in the alpha-subunit. Cell Res 2013; 23:1237–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Chen L, Jiao ZH, Zheng LS, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature 2009; 459:1146–1149. [DOI] [PubMed] [Google Scholar]
- 29Carling D. Branching out on AMPK Regulation. Cell Metab 2009; 9:7–8. [DOI] [PubMed] [Google Scholar]
- 30Polekhina G, Gupta A, Michell BJ, et al. AMPK beta subunit targets metabolic stress sensing to glycogen. Curr Biol 2003; 13:867–871. [DOI] [PubMed] [Google Scholar]
- 31McBride A, Ghilagaber S, Nikolaev A, Hardie DG. The glycogen-binding domain on the AMPK beta subunit allows the kinase to act as a glycogen sensor. Cell Metab 2009; 9:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Scott JW, Ling N, Issa SM, et al. Small molecule drug A-769662 and AMP synergistically activate naive AMPK independent of upstream kinase signaling. Chem Biol 2014; 21:619–627. [DOI] [PubMed] [Google Scholar]
- 33Riek U, Scholz R, Konarev P, et al. Structural properties of AMP-activated protein kinase: dimerization, molecular shape, and changes upon ligand binding. J Biol Chem 2008; 283:18331–18343. [DOI] [PubMed] [Google Scholar]
- 34Polekhina G, Gupta A, van Denderen BJ, et al. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure 2005; 13:1453–1462. [DOI] [PubMed] [Google Scholar]
- 35Koay A, Woodcroft B, Petrie EJ, et al. AMPK beta subunits display isoform specific affinities for carbohydrates. FEBS Lett 2010; 584:3499–3503. [DOI] [PubMed] [Google Scholar]
- 36Amodeo GA, Rudolph MJ, Tong L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 2007; 449:492–495. [DOI] [PubMed] [Google Scholar]
- 37Warden SM, Richardson C, O'Donnell J, Jr., Stapleton D, Kemp BE, Witters LA. Post-translational modifications of the beta-1 subunit of AMP-activated protein kinase affect enzyme activity and cellular localization. Biochem J 2001; 354:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Woods A, Salt I, Scott J, Hardie DG, Carling D. The alpha1 and alpha2 isoforms of the AMP-activated protein kinase have similar activities in rat liver but exhibit differences in substrate specificity in vitro. FEBS Lett 1996; 397:347–351. [DOI] [PubMed] [Google Scholar]
- 39Calabrese MF, Rajamohan F, Harris MS, et al. Structural basis for AMPK Activation: natural and synthetic ligands regulate kinase activity from opposite poles by different molecular mechanisms. Structure 2014; 22:1161–1172. [DOI] [PubMed] [Google Scholar]
- 40Chen L, Wang J, Zhang YY, et al. AMP-activated protein kinase undergoes nucleotide-dependent conformational changes. Nat Struct Mol Biol 2012; 19:716–718. [DOI] [PubMed] [Google Scholar]
- 41Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA 2006; 103:17783–17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Ng LM, Soon FF, Zhou XE, et al. Structural basis for basal activity and autoactivation of abscisic acid (ABA) signaling SnRK2 kinases. Proc Natl Acad Sci USA 2011; 108:21259–21264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Cool B, Zinker B, Chiou W, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab 2006; 3:403–416. [DOI] [PubMed] [Google Scholar]
- 44Neumann D, Woods A, Carling D, Wallimann T, Schlattner U. Mammalian AMP-activated protein kinase: functional, heterotrimeric complexes by co-expression of subunits in Escherichia coli. Protein Expr Purif 2003; 30:230–237. [DOI] [PubMed] [Google Scholar]
- 45Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res 1998; 26:1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A 2003; 59:228–234. [DOI] [PubMed] [Google Scholar]
- 47McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr 1996; 52:829–832. [DOI] [PubMed] [Google Scholar]
- 49Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 50Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 51Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 2008; 64:61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Melcher K, Ng LM, Zhou XE, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009; 462:602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Abdiche Y, Malashock D, Pinkerton A, Pons J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal Biochem 2008; 377:209–217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
AMPK α1(E471G/E474A/K476A) is active in subunit interactions, kinase activity, and modulation of kinase activity by adenine nucleotides and glycogen.
Electron density omit maps of ligands in the 4.05 Å structure of phosphorylated AMPK: (a) AMP, (b) staurosporine, (c) cyclodextrin.
Close-up views of the CBM–KD interface and conformation.
CBM phosphorylation by AMPK KD in trans.
Crystal packing stabilizes a KD–AID trans dimer.
KD and AID form a monomeric cis-complex at moderate protein concentrations.
Mutations at the KD–AID interface abolish AMP-dependent allosteric kinase activation, but not protection against activation loop dephosphorylation.
Molprobity statistics (top) and polygon diagram (bottom) of the p-AMPK model.
Molprobity statistics (top) and polygon diagram (bottom) of the model of non-phosphorylated AMPK.
Molprobity statistics (top) and polygon diagram (bottom) of the KD-AID model.