Background: KSHV LANA binds virus DNA to mediate episome maintenance.
Results: Mutating the novel LANA DNA binding domain (DBD) positive electrostatic patch engenders replication and episome persistence deficiencies.
Conclusion: The LANA positive patch, possibly acting through a cell partner, is important for episome maintenance.
Significance: Strategies that interfere with LANA positive patch function may allow future disruption of KSHV latency.
Keywords: cancer; DNA viruses; herpesvirus; site-directed mutagenesis; structure-function; tumor virus; viral protein; Kaposi sarcoma-associated herpesvirus (KSHV),; episome; latency-associated nuclear antigen (LANA)
Abstract
Kaposi sarcoma-associated herpesvirus (KSHV) has a causative role in several human malignancies. KSHV latency-associated nuclear antigen (LANA) mediates persistence of viral episomes in latently infected cells. LANA mediates KSHV DNA replication and segregates episomes to progeny nuclei. The structure of the LANA DNA binding domain was recently solved, revealing a positive electrostatic patch opposite the DNA binding surface, which is the site of BET protein binding. Here we investigate the functional role of the positive patch in LANA-mediated episome persistence. As expected, LANA mutants with alanine or glutamate substitutions in the central, peripheral, or lateral portions of the positive patch maintained the ability to bind DNA by EMSA. However, all of the substitution mutants were deficient for LANA DNA replication and episome maintenance. Mutation of the peripheral region generated the largest deficiencies. Despite these deficiencies, all positive patch mutants concentrated to dots along mitotic chromosomes in cells containing episomes, similar to LANA. The central and peripheral mutants, but not the lateral mutants, were reduced for BET protein interaction as assessed by co-immunoprecipitation. However, defects in BET protein binding were independent of episome maintenance function. Overall, the reductions in episome maintenance closely correlated with DNA replication deficiencies, suggesting that the replication defects account for the reduced episome persistence. Therefore, the electrostatic patch exerts a key role in LANA-mediated DNA replication and episome persistence and may act through a host cell partner(s) other than a BET protein or by inducing specific structures or complexes.
Introduction
Kaposi sarcoma (KS)2-associated herpesvirus (KSHV or human herpesvirus 8), a gamma-2 herpesvirus, has a causative role in KS, primary effusion lymphoma, and multicentric Castleman disease, an aggressive lymphoproliferative disorder (1–6). KSHV establishes lifelong latency in its human host and persists in latently infected cells in the form of multiple copy, circularized, extrachromosomal episomes (7, 8). Episome persistence in proliferating cells requires replication of DNA before each cell division and segregation of episomes to daughter nuclei. Persistence of latent infection requires the expression of the KSHV latency-associated nuclear antigen (LANA) (9, 10).
LANA, an 1162-amino acid protein, is encoded by ORF73 of the viral genome and is one of a small subset of viral genes expressed during latent infection (11–14). LANA is a multifunctional protein; it regulates a variety of cellular activities including gene transcription, DNA replication, and signaling pathways involved in cell proliferation (15–27).
LANA mediates episome persistence by promoting KSHV DNA replication before each cell division and ensuring segregation of episomes to progeny nuclei. C-terminal LANA self-associates to bind specific sequence in KSHV terminal repeat (TR) DNA to mediate its replication (10, 17, 28–30, 32–36). LANA tethers KSHV DNA to mitotic chromosomes during cell division by simultaneously binding KSHV episomes and metaphase chromosomes; this tethering mechanism ensures the segregation of KSHV genomes to progeny nuclei (9, 37–39). Although the C-terminal LANA DNA binding domain (DBD) binds TR DNA, N- and C-terminal LANA each bind mitotic chromosomes. N-terminal LANA associates with chromosomes through amino acids 1–23 binding to histones H2A/H2B (40). C-terminal LANA can also bind to chromosomes, with concentration at pericentromeric and peritelomeric regions. However, N-terminal LANA is the dominant chromosome association region (41–44).
We and others recently solved the x-ray crystal structure of the murine gamma herpesvirus 68 (MHV68) or KSHV LANA C-terminal DNA binding domain (45–48). The LANA DBD shares common structural features with the Epstein-Barr virus EBNA1 and papillomavirus E2 episome maintenance proteins; however, LANA contains a unique positive electrostatic charged patch on its dorsal surface, opposite the DNA binding interface, which is absent from the EBNA1 and E2 proteins. The positively charged LANA dorsal face has an important role in MHV68 establishment of latent infection in mice, as substitution mutations that altered the peripheral, but not central dorsal positive patch charge resulted in substantial deficiency in expansion of latent infection (45, 47). The dorsal positive patch is also notable for being a site of interaction with BET (bromodomain and extra terminal domain) family proteins (21, 45–47, 49–51), including BRD2, BRD3, and BRD4, which interact with acetylated histones.
To investigate if the dorsal positive patch has a role in LANA-mediated episome persistence, we generated a panel of substitution mutants that targeted either the central, peripheral, or lateral regions of the positive patch. We found that mutations of each of the positive patch regions resulted in deficiencies in LANA-mediated DNA replication and episome persistence and that mutating the LANA peripheral positive patch engendered the most severe deficiencies. There was a close correlation of deficiencies in episome persistence and DNA replication, consistent with replication defects accounting for the episome maintenance deficiencies. Some mutations in the LANA dorsal patch reduced BET protein binding; however, the reduction in binding did not correlate with the deficiencies in episome persistence.
Experimental Procedures
Cell Lines
Uninfected BJAB B lymphoma cells were cultured in RPMI medium containing 10% bovine growth serum (Hyclone) and 15 μg/ml gentamicin. KSHV-infected BCBL-1 cells were cultured in RPMI medium containing 20% BGS and 15 μg/ml gentamicin. 293T cell were maintained in DMEM supplied with 10% BGS and 15 μg/ml gentamicin.
Plasmids
pT7 LANA encodes LANA with an N-terminal T7 epitope tag (52). LANA containing substitution mutations in the dorsal positively charged patch of the DNA binding domain were each cloned into pSG5 (Stratagene) with an engineered N-terminal T7 epitope tag to generate LANA K1044A, LANA K1044E, LANA K1109A/K1113A/K1114A, LANA K1109E/K1113E/K1114E, LANA K1138A/K1140A/K1141A, or LANA K1138E/K1140E/K1141E. Mutations were first introduced into pSG5 T7 LANA Δ33–888 (52) using overlapping, reverse orientation primers containing the mutated sequence (Table 1) to amplify the entire LANA Δ33–888 plasmid. The PCR amplification product was then digested 1 h with DpnI to digest template DNA before transformation into Escherichia coli bacteria. LANA Δ33–888 containing each mutation was digested with restriction enzymes AscI and NruI to remove the central Δ33–888 LANA sequence. An AscI/NruI fragment from full-length LANA was then inserted into each digested plasmid to generate full-length LANA containing each of the C-terminal positive patch mutations. p8TR contains eight copies of the KSHV TR element and was described previously (9, 44). Brd2 and Brd3 were amplified from cDNA generated from 293T cells using the primers listed in Table 1. Brd4 was amplified from plasmid pCDNA Brd4 (49) using the primers listed in Table 1. The Brd PCR products were each digested with the enzymes indicated in Table 1 and then cloned in-frame into EGFP-C1 (Clontech). All PCR generated constructs were sequence-confirmed.
TABLE 1.
Oligonucleotides
F, forward; R, reverse.
| Name | Sequence |
|---|---|
| K1044A Fa | AGACGCTTTTTGGGAGCGGATGGAAGACGAGAT |
| K1044A R | ATCTCGTCTTCCATCCGCTCCCAAAAAGCGTCT |
| K1044E F | AGACGCTTTTTGGGAGAGGATGGAAGACGAGAT |
| K1044E R | ATCTCGTCTTCCATCCTCTCCCAAAAAGCGTCT |
| K1109A/K1113A/K1114A F | TATATGTGTATTGTCAGAACGCAGACACAAGTGCGGCAGTACAAATGGCCCGCCTAGC |
| K1109E/K1113R/K1114A R | GCTAGGCGGGCCATTTGTACTGCCGCACTTGTGTCTGCGTTCTGACAATACACATATA |
| K1109E/K1113E/K1114E F | TATATGTGTATTGTCAGAACGAAGACACAAGTGAGGAAGTACAAATGGCCCGCCTAGC |
| K1109R/K1113E/K1114E R | GCTAGGCGGGCCATTTGTACTTCCTCACTTGTGTCTTCGTTCTGACAATACACATATA |
| K1138A/K1140A/1141A F | ACCTACAATCTTCCATAGTTGCGTTTGCAGCGCCCCTGCCATTAACCCAGCC |
| K1138A/K1140A/1141A R | GGCTGGGTTAATGGCAGGGGCGCTGCAAACGCAACTATGGAAGATTGTAGGT |
| K1138E/K1140E/1141E F | ACCTACAATCTTCCATAGTTGAGTTTGAAGAGCCCCTGCCATTAACCCAGCC |
| K1138E/K1140E/1141E R | GGCTGGGTTAATGGCAGGGGCTCTTCAAACTCAACTATGGAAGATTGTAGGT |
| LBS1Fb | GGATTCCCGCCCGGGCATGGGGCCG |
| LBS1R | GGTACGGCCCCATGCCCGGGCGGGA |
| LBS1 6TT7 Fc | GGATTCCCGTTCGGGCATGGGGCCG |
| LBS1 6TT7 R | GGTACGGCCCCATGCCCGAACGGGA |
| BRD2 EcoRI Fd | CATCATGAATTCGATGCTGCAAAACGTGACTC |
| BRD2 BamHI R | CATCATGGATCCGCCTGAGTCTGAATCACTGG |
| BRD3 Bgl II F | CATCATAGATCTATGTCCACCGCCACGACAGT |
| BRD3 EcoRI R | CATCATGAATTCTTCTGAGTCACTGCTGTCAG |
| BRD4 HindIII F | CATCATAAGCTTCGATGTCTGCGGAGAGCGGCCC |
| BRD4 BamHI R | CATCATGGATCCGAAAAGATTTTCTTCAAATATTGAC |
a Nucleotides encoding substitution mutations are indicated by bold and italics.
b LANA high affinity binding site (LBS1) is indicated in bold and underlined.
c LBS1 (indicated in bold and underlined) is mutated with substitution mutations at positions 6 and 7 (indicated by italics) (54).
d Restriction enzyme sites are underlined.
Generation of LANA Expressing Cell Lines
To generate BJAB cells stably expressing LANA or LANA-containing mutations in the dorsal positive patch, 30 μg of pSG5 LANA or 30 μg each of the pSG5 LANA positive patch mutants was co-transfected with 5 μg of a plasmid encoding the hygromycin resistance gene. 10 × 106 BJAB cells were transfected in 400 μl of RPMI medium supplemented with 10% BGS using a Bio-Rad electroporator at 200 V and 960 microfarads. Forty-eight hours after transfection cells were seeded at a density of 200 cells per well in 96-well microtiter plates and placed under selection with hygromycin. Hygromycin-resistant cells that grew out were screened for LANA expression. LANA-expressing stable cells were subcloned to generate clonal cell lines.
Episome Maintenance Experiments
10 × 106 BJAB cell lines stably expressing WT or mutated LANA were each transfected with 30 μg of p8TR, which encodes for G418 resistance, using a Bio-Rad electroporator as above. Transfected cells were maintained in RPMI medium containing 10% BGS for 72 h before seeding at 1000, 100, 10, or 1 cell/well in 96-well microtiter plates in medium containing 600 μg/ml G418 (Gibco).
G418-resistant cell lines were expanded and assessed for the presence of episomes by Gardella gel (53). Cells were loaded in agarose gel wells containing sodium dodecyl sulfate and DNase-free protease (Sigma). Cell lysis occurs as electrophoresis begins in the Tris borate-EDTA buffer. The gel was run overnight at 110 V, and DNA was transferred to a nylon membrane. Episomal DNA was detected by 32P-labeled TR probe.
Electrophoretic Mobility Shift Assay
LANA or LANA mutants were in vitro-translated using TNT coupled reticulocyte system (Promega). High affinity LANA binding site 1 (LBS1) (10, 28) oligonucleotides (Table 1) were annealed and 32P-radiolabeled with Klenow polymerase (New England BioLabs). Similar amounts of in vitro-translated LANA, as assessed by Western blot with anti-T7.Tag antibody (Novagen), were incubated with LBS1 probe for 30 min at 4 °C in EMSA buffer (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 1 mm EDTA, 20 μg/ml poly(dI-dC), 0.1 mm DTT, and 10% glycerol). For the competition controls, a 50-fold excess of unlabeled LBS1 oligonucleotide or 50-fold excess unlabeled LBS1 oligonucleotide containing the substitution mutations 6C→T/7C→T, which abolish LANA binding (54), was added to the incubation. For the electrophoretic mobility supershift assay, 1 μg of T7.Tag antibody (Novagen) was included with the incubation. The reaction was mixed with loading buffer (95% formamide, 10 mm EDTA, 0.1% xylene cyanol, and 0.1% bromphenol blue), and bound complexes were resolved in a 3.5% non-denaturing TBE-polyacrylamide gel after electrophoresis for 1 h at 300 V. In addition, loading buffer lacking xylene cyanol and bromphenol blue was also used with WT LANA reactions to assess if these dyes affect LANA DNA binding. The gel was dried on Whatman paper, and signal was captured on Kodak Biomax MS film (Eastman Kodak Co.).
Fluorescence Microscopy
Cells were metaphase-arrested by overnight incubation with 1 μg/ml colcemid (Calbiochem). Metaphase-arrested cells were swollen for 5 min in hypotonic buffer (1% sodium citrate, 10 mm CaCl2 10 mm MgCl2) and spread onto slides by cytospin (Thermoshandon). The cells were fixed in 4% paraformaldehyde and incubated with primary anti-LANA monoclonal antibody (ABI Inc., LN53) and secondary anti-rat Alexa Fluor 488 (Invitrogen, A11006). Cells were counterstained with propidium iodide (Molecular probes) at 1 μg/ml. Images were captured using a Zeiss Axioskop, PCM2000, with C-imaging software (Compix, Inc.).
Immunoprecipitation and Immunoblot Analysis
For immunoprecipitation experiments, 5 μg of LANA or LANA mutant DNA was co-transfected with 5 μg plasmid DNA encoding GFP, GFP-BRD2, GFP-BRD3, or GFP-BRD4 into 10-cm dishes containing 293T cells at 70% confluence using polyethyleneimine (PEI) solution (20 μg of PEI/10 μg of DNA) and incubated at 37 °C for 48 h. Cells were then incubated in lysis buffer (20 mm Tris pH 7.9, 137 mm NaCl, 1% Triton X-100, 10 mm CaCl2, 2 mm EDTA, 10% glycerol, 2000 units/ml micrococcal nuclease) and subjected to centrifugation. Dynabeads Protein G (Invitrogen) were incubated with rabbit anti-GFP polyclonal antibody in lysis buffer for 30 min at 37 °C and washed 3 times with lysis buffer. The anti-GFP antibody-bound Dynabeads were then incubated with cell lysate supernatant overnight at 4 °C. Input (5%) and immunoprecipitates were analyzed by Western blot with mouse monoclonal anti-GFP JL8 (Clontech) or human anti-LANA serum.
DNA Replication Assay
DNA replication assays were performed by real time PCR as previously described (55, 56). Briefly, 10 × 106 BJAB cells expressing LANA or LANA mutants were each transfected with 5 μg of p8TR-gB using Amaxa Nucleofactor program O-17 and solution V (Amaxa), and cells were then cultured in RPMI medium containing 10% BGS. 5 × 106 cells were harvested for low molecular weight Hirt DNA at 24 or 72 h post transfection. DNA harvested at 72 h was incubated with DpnI and ExoIII to digest non-replicated plasmid. Quantitation of total p8TR-gB at 24 h or replicated p8TR-gB at 72 h was performed by real time PCR. Total DNA levels were used to normalize for transfection efficiencies, and -fold replication over BJAB control was calculated.
Results
LANA Dorsal Positive Electrostatic Patch Mutants Bind TR DNA
LANA mediates KSHV episome persistence by acting on TR DNA. The C-terminal LANA DNA binding domain is essential for this process. C-terminal LANA directly binds a specific sequence in TR DNA, allowing LANA to mediate DNA replication and to tether KSHV episomes to mitotic chromosomes. Because the dorsal positive electrostatic patch is a distinctive LANA feature that is absent from the structurally homologous EBV EBNA1 gamma-1 herpesvirus DNA binding domain, we asked whether these residues are important for LANA's episome maintenance function.
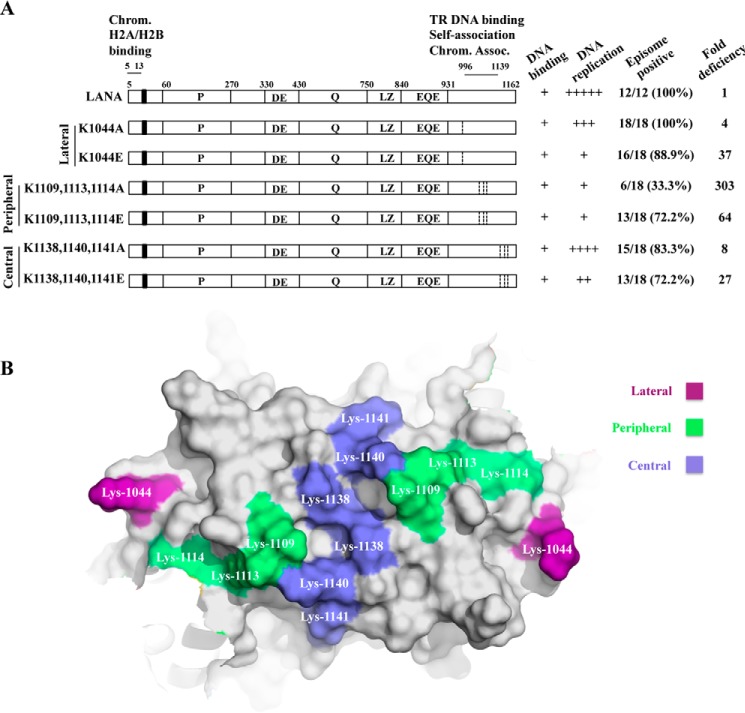
The LANA dorsal positive patch can be spatially divided into three regions: lateral, peripheral, and central (Fig. 1B). To investigate these regions, we independently mutated each by substituting the positively charged lysine residues either with alanine or to a negative charge with glutamate in full-length LANA (Fig. 1A). LANA K1044A and LANA K1044E contain substitutions in the lateral region, LANA K1109A/K1113A/K1114A and LANA K1109E/K1113E/K1114E contain substitutions in the peripheral region, and LANA K1138A/K1140A/K1141A and LANA K1138E/K1140E/K1141E are mutated in the central region.
FIGURE 1.
LANA dorsal positive patch mutants. A, a schematic diagram is shown of LANA and LANA mutants. The proline-rich region (P), aspartate and glutamate (DE) region, glutamine (Q) region, putative leucine zipper (LZ), and glutamate and glutamine region (EQE) are indicated. The N-terminal nuclear localization signal is indicated by a wide vertical bar. Residues 5–13 bind the surface of histones H2A/H2B. LANA 996–1139 self-associates to bind TR DNA and can also independently associate with mitotic chromosomes. Capabilities for DNA binding, DNA replication, episome maintenance, and -fold deficiency for episome maintenance are summarized at the right. Ratios indicate the number of G418-resistant cell lines that contain episomes over the total number of G418-resistant cell lines tested by the Gardella gel, and percentages are shown in parenthesis. The numbers used for the ratios were derived from two independent experiments. -Fold deficiency was determined based on comparisons of the calculated numbers of cells necessary to seed per microtiter well to obtain episome containing cell outgrowth in 63.2% of wells. B, surface representation of the positively charged dorsal patch of a dimer of the KSHV LANA DBD (PDB ID code 2YPY) (47) viewed in PyMOL (v.0.99; Schrödinger). The lateral, peripheral, and central regions of the electrostatic patch are colored purple, green, and blue, respectively, and individual residues are indicated.
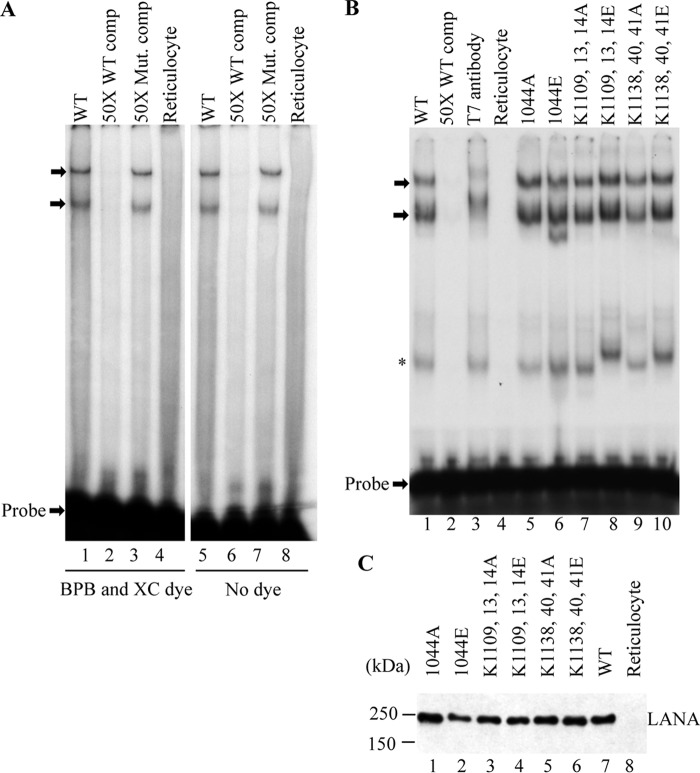
We investigated the ability of each LANA mutant to bind KSHV TR DNA. Because the positive patch is on the opposite face from the DNA binding surface in C-terminal LANA, we expected DNA binding to remain intact. However, because it remained possible that substitution of these residues could result in conformational changes that might affect DNA binding, we used EMSA to assess the ability of each mutant to bind its recognition sequence in TR DNA. As expected (10), incubation of in vitro translated LANA with TR probe resulted in two prominent shifted complexes (Fig. 2, A and B, arrows). These complexes are specific and were effectively competed with a 50-fold excess of unlabeled oligonucleotide (Fig. 2, A and B, lanes 2) but not with a 50-fold excess of unlabeled oligonucleotide containing substitution mutations at position 6 and 7 (6C→T/7C→T) that abolished LANA binding (54) (Fig. 2A, lanes 3 and 7). The bromphenol blue and xylene cyanol loading dyes used to visualize gel migration did not affect DNA binding as omitting these dyes did not alter LANA complexes (compare Fig. 2, A, lanes 1–4, with lanes 5–8). Incubation with anti-T7 antibody, which detects the N-terminal LANA T7 epitope tag, resulted in a supershift of the two complexes (Fig. 2B, lane 3), demonstrating that these complexes contain LANA. A third, minor complex (Fig. 2B, asterisk) migrated more quickly in some incubations and was specific for LANA as it was not present after incubation of the probe with reticulocyte lysate alone (Fig. 2B, lane 4) and was competed by excess cold competitor (Fig. 2B, lane 2). This complex likely contains a C-terminal LANA truncation as incubation with T7 antibody did not result in a supershifted complex (Fig. 2B, lane 3), indicating the absence of N-terminal LANA. Incubation of in vitro translated LANA mutants (Fig. 2B) with TR probe resulted in complexes that migrated similar to the LANA complexes (Fig. 2B, lanes 5–10). Therefore, the mutations in the dorsal patch region did not disrupt LANA DNA binding.
FIGURE 2.
Mutation of the dorsal patch does not abolish DNA binding as assessed by EMSA. A, radiolabeled LBS1 probe was incubated with rabbit reticulocyte lysate (lanes 4 and 8) or in vitro translated LANA (lanes 1–3 and 5–7). Samples were run either with (lanes 1–4) or without (lanes 5–8) bromphenol blue (BPB) and xylene cyanol (XC) tracking dye. A 50-fold excess of unlabeled LBS1 competitor was included in lanes 2 and 6. A 50-fold excess of unlabeled LBS1 competitor containing substitution mutations at positions 6 and 7 (6C→T/7C→T), which abolishes LANA binding, was included in lanes 3 and 7. After incubation, complexes were resolved in a nondenaturing polyacrylamide gel. B, LANA (lanes 1–3) or LANA mutant (lanes 5–10) complexes were resolved after incubation with LBS1 probe. Anti-T7 epitope antibody, which binds to the N-terminal LANA T7 tag, was included in lane 3. LBS1 probe was incubated with reticulocyte lysate in lane 4. Arrows indicate full-length LANA complexes in panels A and B. The asterisk indicates C-terminal LANA truncation complexes. Brightness and contrast were uniformly adjusted with Adobe Photoshop. C, immunoblot of in vitro translated LANA or LANA mutants with anti-T7 antibody which detects the N-terminal LANA T7 epitope tag.
LANA Dorsal Patch Mutants Are Deficient for TR DNA Replication
We assessed the role of the LANA dorsal positive patch in DNA replication. During latent infection, the KSHV genome must replicate with each cell division in order to persist in progeny cells. LANA mediates DNA replication after binding to KSHV TR sequence through its C-terminal domain (28, 32, 34–36, 57).
To assess DNA replication, BJAB cells or BJAB cells stably expressing LANA or each of the LANA mutants were transfected with p8TR-gB, a plasmid containing eight terminal repeat elements. p8TR-gB, purified from Dam methylase-positive bacteria, contains Dam-methylated DNA. The Dam methylation results in susceptibility to DpnI digestion. After replication in mammalian cells, which lack Dam methylase, DNA is resistant to DpnI digestion. Therefore, detection of DpnI resistant TR DNA allows for assessment of LANA DNA replication.
Fig. 3 shows the normalized -fold replication for LANA and each LANA mutant over the BJAB control. As expected, LANA efficiently mediated p8TR-gB replication at a 7.2-fold greater than BJAB control. Notably, all the positive patch mutants were reduced for TR DNA replication. The central region mutants, LANA K1138A/K1140A/K1141A and LANA K1138E/K1140E/K1141E, were only modestly reduced and had a 5.5- and 2.6-fold replication over control. The peripheral patch mutants, LANA K1109A/K1113A/K1114A and LANA K1109E/K1113E/K1114E, were most severely reduced and had 2.0- and 1.4-fold replication over control. The lateral patch mutants, LANA K1044A and LANA K1044E, had an intermediate phenotype, replicating DNA at levels 3.6- and 1.9-fold over control. The lower replication of the mutants was not due to reduced protein expression as all were expressed at levels at least equivalent to WT LANA (Fig. 3B). Therefore, mutation of the LANA positive patch resulted in deficient DNA replication that was most severe after mutation of the peripheral patch and least deficient after mutation of the central region.
FIGURE 3.

Dorsal positive patch mutations reduce LANA DNA replication. A, BJAB cells stably expressing LANA or LANA mutants were transfected with p8TR-gB, which contains eight copies of the TR element. Hirt DNA was isolated after 24 h to assess transfection efficiencies and again at 72 h to assess replicated DNA. The amounts of p8TR-gB DNA isolated were quantitated by real time PCR. The -fold replication for LANA or LANA mutants is shown relative to BJAB, which lacks LANA. Averages from three independent experiments are shown. Error bars indicate S.D. B, Western blot for LANA expression or tubulin (below) is shown for BJAB cells (lane 9), BJAB cells expressing LANA (lane 7), or BJAB cells expressing LANA mutants (lanes 1–6). BCBL1 is a KSHV-infected primary effusion lymphoma cell line. The multiple LANA bands are due to initiation of translation at downstream noncanonical initiation sites within LANA and also from an isoform resulting from a noncanonical polyadenylation site that results in C-terminal-truncated LANA (31, 68).
LANA Positive Patch Mutants Are Reduced for Episome Persistence
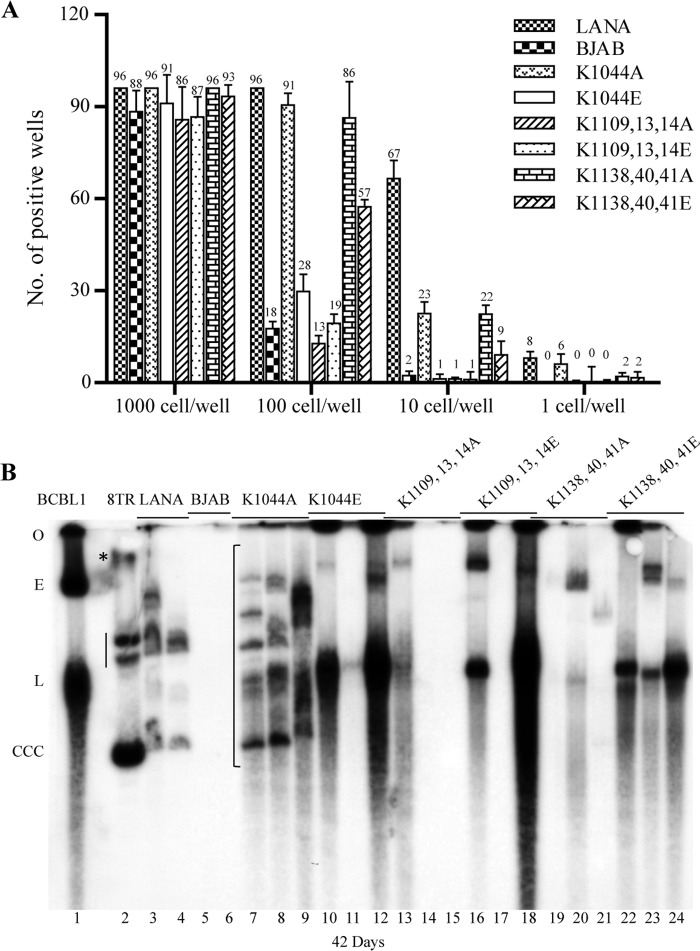
We investigated the role of the LANA DBD dorsal positive patch in episome persistence. BJAB cells or BJAB cells stably expressing each LANA mutant were transfected with p8TR, which contains 8 copies of the KSHV TR element, and 3 days later seeded into 96-well microtiter plates at 1000, 100, 10, or 1 cell per well and placed under G418 selection, for which the plasmid vector encodes resistance. From three experiments, BJAB cells had an average of 88, 18, 2, and 0 wells with G418-resistant outgrowth after seeding at 1000, 100, 10, or 1 cell/well (Fig. 4A). In contrast, LANA expressing cells had G418-resistant outgrowth in 96, 96, 86, and 6 wells after seeding at 1000, 100, 10, or 1 cell/well. The higher outgrowth of the LANA expressing cells is due to the much higher efficiency of episome persistence compared with integration, which is required for p8TR persistence in BJAB cells lacking LANA.
FIGURE 4.
Disruption of the positive electrostatic patch on the LANA dorsal face leads to episome persistence deficiency. A, limiting dilution G418-resistant outgrowth was performed after transfection of p8TR DNA. BJAB cells or BJAB cells stably expressing LANA or LANA mutants were transfected with p8TR plasmid and 72 h later seeded into 96-well microtiter plates at different cell concentrations. Well outgrowth was recorded after 21 days of G418 selection. Results are the averages of three independent experiments. Error bars indicate S.D. B, Gardella gel analysis for episomal DNA was performed after 42 days of G418 selection. G418-resistant cell lines were expanded from microtiter wells, and ∼1 × 106 cells were loaded in each lane of the Gardella gel. Gardella gel containing KSHV-infected BCBL1 primary effusion lymphoma cells (lane 1), p8TR plasmid DNA (lane 2), G418-resistant, p8TR-transfected, BJAB cells stably expressing LANA (lanes 3 and 4), BJAB cells (lanes 5 and 6), or BJAB cells expressing LANA mutants (lanes 7–24). G418-resistant, LANA-expressing cell lines expanded from microtiter plates seeded at 100 cells/well were used for all Gardella assays. The gel origin (O), p8TR covalently closed, circular (CCC) DNA, p8TR nicked plasmid DNA (asterisk), and BCBL1 episomal (E) and linear (L) forms (linear, a result of lytic replication) are indicated at the left. The vertical bar adjacent to lane 2 indicates two forms of the p8TR plasmid DNA that co-migrate with certain episomal forms in the BJAB/LANA and BJAB/LANA K1044A lanes. The bracket indicates positions of episomal bands for LANA mutants.
Each of the LANA mutants had deficient G418-resistant outgrowth. The central patch mutants were moderately reduced and LANA K1138A/K1140A/K1141A and LANA K1138E/K1140E/K1141E had outgrowth in 96, 86, 22, and 2 wells or 93, 57, 9, and 2 wells, respectively, after seeding at 1000, 100, 10, or 1 cell/well. The lateral patch mutants, LANA K1044A and LANA K1044E, were also moderately reduced and had outgrowth in 96, 91, 23, and 6 wells or 91, 28, 1, and 0 wells, respectively, after seeding at 1000, 100, 10, or 1 cell/well. The peripheral patch mutants were most severely affected, and LANA K1109A/K1113A/K1114A and LANA K1109E/K1113E/K1114E had outgrowth in 86, 13, 1, and 0 wells, or 87, 19, 1, and 0 wells, respectively, after seeding at 1000, 100, 10, or 1 cell/well. Therefore, G418-resistant outgrowth was moderately reduced for the central and lateral patch mutants, and more severely reduced for the peripheral patch mutants, consistent with moderate episome maintenance deficiencies for the central and lateral patch mutants, and a more severe deficiency for the peripheral patch mutants.
We directly assessed for the presence of episomal DNA in the G418-resistant cell lines. BJAB cells expressing LANA or LANA mutants were investigated by the Gardella gel (53). In a Gardella gel, live cells are lysed in the loading wells at the start of the gel run. Episomal DNA as large as several hundred kilobases migrates into the gel, whereas chromosomal DNA remains at the gel origin. Episomal DNA is then detected by Southern blot. BCBL1 (Fig. 4B, lane 1), a KSHV-infected primary effusion lymphoma cell line had a slowly migrating band, which represents episomal DNA (Fig. 4B, indicated by E), and a faster migrating band (Fig. 4B, indicated by L), which represents linear, replicating DNA. As expected, BJAB cells (Fig. 4B, lanes 5–6), which lack LANA expression, did not contain episomal DNA; in two experiments 0/12 (0%) G418-resistant BJAB cell lines had episomes. In contrast, LANA-expressing cells (Fig. 4B, lanes 3 and 4) had episomes in all lanes; in two experiments, 12 of 12 (100%) G418-resistant LANA cell lines had episomal DNA. The lateral patch mutant LANA K1044A (Fig. 4B, lanes 7–9) also had episomal DNA in all lanes; in two experiments 18/18 (100%) G418-resistant LANA K1044A cell lines had episomes. Similar to LANA K1044A, LANA K1044E had episomal DNA in all lanes (Fig. 4B, lanes 10–12); in two experiments 16 of 18 (89%) G418-resistant LANA K1044E cell lines had episomes. The central patch mutant LANA K1138A/K1140A/K1141A had episomal DNA in two of three lanes (Fig. 4B, lanes 19–21), and in two experiments 15 of 18 (83%) of LANA K1138A/K1140A/K1141A cell lines had episomes. LANA K1138E/K1140E/K1141E had episomes in all three lanes (Fig. 4B, lanes 22–24); in two experiments 13 of 18 (72%) of LANA K1138E/K1140E/K1141E cell lines had episomes. The peripheral patch mutant LANA K1109A/K1113A/K1114A had episomal DNA in one of three lanes (Fig. 4B, lanes 13–15), and in two experiments 6 of 18 (33%) mutant LANA K1109A/K1113A/K1114A cell lines had episomes. Last, LANA K1109E/K1113E/K1114E had episomal DNA in two of three lanes (Fig. 4B, lanes 16–18), and in two experiments 13 of 18 (72%) cell lines had episomes.
Notably, the migration pattern of episomal DNA for all mutants except LANA K1044A differed from that of LANA. Both LANA and LANA K1044A had episomal DNA bands that co-migrated with the circular, covalently closed form of the plasmid p8TR DNA as well as bands that co-migrated with slower migrating forms of the plasmid p8TR DNA (Fig. 4B, lane 2, vertical line) that migrated between nicked (Fig. 4B, lane 2, asterisk) and ccc p8TR plasmid DNA. In contrast, the other LANA mutants lacked these bands. In addition, bands with much more intense signal compared with the LANA and LANA K1044A lanes were present for the other mutants (except LANA K1138A/K1140A/K1141A, which lacked the more intense bands.) Smears of even faster migrating DNA were also present in some lanes, such as for LANA K1044E and LANA K1109E/K1113E/K1114E, and may be due to partially replicated or degraded DNA. Very slowly migrating bands, which migrated similar to the ∼200kb BCBL1 episome, were also present for the mutants. The multiple different migrating episomal forms are due to recombination events, including expansion of the TR elements and multimers of the p8TR plasmid, which vary in each independently derived cell line. The proportion of slowly migrating DNA compared with episomes that co-migrate with input p8TR plasmid increases with time under G418 selection and is due to selection for such recombinants (10, 52).
To allow comparison of episome maintenance efficiencies of the different LANA mutants, we calculated the number of cells necessary to seed per well to obtain episome containing cell outgrowth in 63.2% of wells. Using the Poisson distribution, if there is one episome-containing cell per well of a microtiter plate at the time of cell seeding, it is expected that 36.8% of wells will have no episome-containing cells. Therefore, nonlinear regression analyses were used with the G418-resistant limiting dilution cell outgrowth data (Fig. 4A) to predict how many cells are required to seed each well of a microtiter plate to obtain episome-containing cell outgrowth in 63.2% of wells. If the percent of G418-resistant cell lines containing episomes was <100% (Fig. 1), these percentages, determined from Gardella gel analyses after seeding at 100 cells/well were used to determine the number of episome containing wells after seeding at 100 cells/well; using these outgrowth numbers, the Poisson distribution was used to estimate episome-containing well outgrowth numbers at lower and higher cell seeding concentrations before nonlinear regression analyses. The values for numbers of cells seeded per well needed for outgrowth of episome-containing cells in 63.2% of wells were 8, 33, 291, 2393, 503, 62, and 216 for LANA, LANA K1044A, LANA K1044E, LANA K1109A/K1113A/K1114A, LANA K1109E/K1113E/K1114E, LANA K1138A/K1140A/K1141A, and LANA K1138E/K1140E/K1141E, respectively (Table 2). Comparison of the values to that of WT LANA indicated deficiencies of 4-, 37-, 303-, 64-, 8-, and 27-fold for LANA K1044A, LANA K1044E, LANA K1109A/K1113A/K1114A, LANA K1109E/K1113E/K1114E, LANA K1138A/K1140A/K1141A, and LANA K1138E/K1140E/K1141E, respectively (Table 2). For BJAB cells, which require integration of transfected DNA for G418 resistance, 375 cells/well were necessary to obtain outgrowth in 63.2% of wells, which is 42-fold less efficient than LANA's ability to effect G418-resistant outgrowth through episome persistence. Notably, the efficiency of episome maintenance for the peripheral patch mutants, LANA K1109A/K1113A/K1114A and LANA K1109E/K1113E/K1114E, each were lower than the efficiency of integration as observed in BJAB cells. Therefore, although these mutants were still capable of episome maintenance, the rate of successful episome maintenance events was dramatically reduced, indicating a high level of deficiency compared with LANA.
TABLE 2.
Efficiency of episome maintenance
| LANA | 63.2% Value | Relative deficiency |
|---|---|---|
| WT LANA | 8 | 1 |
| K1044A | 33 | 4 |
| K1044E | 291 | 37 |
| K1109/1113/1114A | 2393 | 303 |
| K1109/1113/1114E | 503 | 64 |
| K1138/1140/1141A | 62 | 8 |
| K1138/1140/1141E | 216 | 27 |
LANA Positive Patch Mutants Concentrate to Dots along Mitotic Chromosomes in the Presence of Episomes
We investigated whether the LANA dorsal positively charged patch exerts a role in LANA's ability to concentrate to dots along mitotic chromosomes in cells containing episomes. LANA distributes broadly over mitotic chromosomes but concentrates to dots at sites of episomal DNA (9). As expected, in cells with episomes, LANA (green) concentrated to dots along mitotic chromosomes (red) (overlay of green and red generates yellow dots) (Fig. 5A). Similarly, in G418-resistant cells containing episomes, each LANA mutant also concentrated to dots along mitotic chromosomes (Fig. 5, panels C–H). Therefore, all LANA positive patch mutants concentrated to dots in cells with episomal DNA.
FIGURE 5.
LANA dorsal positive patch mutants associate with mitotic chromosomes. Control BJAB cells or BJAB cells expressing LANA or LANA mutants were metaphase-arrested with colcemid. LANA (green) was detected by anti-T7 antibody, and chromosomes (red) were counterstained with propidium iodide. Overlay of green and red generates yellow. Magnification, ×630.
Central and Peripheral, but Not Lateral, Positive Patch Mutants Reduce LANA Association with BRD2, BRD3, and BRD4
Because the LANA dorsal positive patch exerts a role in interaction with BET family proteins (21, 45–47, 49–51), we assessed the association of BRD2, BRD3, and BRD4 with each LANA positive patch mutant. As expected, GFP-BRD2, GFP-BRD3, or GFP-BRD4 each efficiently co-immune-precipitated LANA after co-transfection into 293T cells (Fig. 6, A and B). The lateral positive patch mutants, LANA K1044A and LANA K1044E, each maintained the ability to co-precipitate with BRD2 (Fig. 6B), BRD3 (Fig. 6C), or BRD4 (Fig. 6D) with efficiencies similar to LANA. In contrast, the central or peripheral positive patch mutants, LANA K1109A/K1113A/K1114A, LANA K1109E/K1113E/K1114E, LANA K1138A/K1140A/K1141A, and LANA K1138E/K1140E/K1141E each were reduced for the ability to co-precipitate with BRD2 (Fig. 6B), BRD3 (Fig. 6C), or BRD4 (Fig. 6D) when compared with that of LANA. Therefore, the central and peripheral positive patch, but not the lateral patch, mediates BET family protein binding.
FIGURE 6.
Mutation of the peripheral or central, but not lateral, dorsal positive patch reduces LANA association with BET proteins. A, LANA was co-expressed with GFP or GFP fused with BRD2, BRD3, or BRD4 by transient transfection of 293T cells. After immunoprecipitation (IP) with anti-GFP antibody, a Western blot was performed for GFP (bottom panel) or for co-precipitating LANA (center panel). The blot showing LANA input is shown at the top. Ig heavy chain is indicated in the center panel. Asterisks denote GFP or the full-length GFP-BRD fusions. LANA or LANA mutants were also co-expressed with GFP-BRD2 (panel B), GFP-BRD3 (panel C), or GFP-BRD4 (panel D). Immunoprecipitations with anti-GFP followed by Western blot for LANA or GFP were then performed for each GFP-BRD fusion. Blots showing input LANA for each co-transfection are also shown. The multiple LANA bands are due to initiation of translation at downstream, noncanonical initiation sites and also from a noncanononical polyadenylation site that results in C-terminal truncation of LANA (31, 68).
Discussion
This work demonstrates that the dorsal electrostatic positive patch exerts roles in KSHV LANA-mediated DNA replication and episome persistence. Although mutations in the lateral, peripheral, or central regions of the positive patch each exerted effects on replication and episome persistence, mutation of the peripheral region resulted in the most severe deficiencies. Overall, the reductions in episome persistence efficiencies closely corresponded to the relative DNA replication deficiencies of the mutants, consistent with these replication defects accounting for the reduced episome maintenance. The reduced DNA replication was not due to the inability of LANA to efficiently bind DNA as all mutants similarly bound the TR DNA binding site as assessed by EMSA (Fig. 2).
The finding that mutations in the peripheral region of the dorsal positive patch resulted in larger deficiencies in episome maintenance and DNA replication compared with mutations in the central patch is supported by other work. In work published after these experiments were initiated, Domsic et al. (46) used a 7-day plasmid maintenance assay that examined Southern blot intensity of 8TR plasmids to assess the same triple alanine or glutamate substitutions of the central or peripheral region as used here. Our approach used G418-resistant limiting dilution cell outgrowth and Gardella gel analyses to quantify episome maintenance efficiency relative to LANA. Both studies similarly found that the mutations reduced maintenance and that the peripheral patch mutants were more deficient than the central region mutants (46). Domsic et al. (46) found that charge reversal with glutamate resulted in greater deficiency compared with alanine substitution for both the central and peripheral patch. Results here differed in that although glutamate substitution resulted in greater episome maintenance deficiency for the central patch, alanine substitution resulted in greater deficiency for the peripheral patch (Fig. 1). Here, we also assessed the lateral patch and found a moderate episome maintenance deficiency for these mutants. Domsic et al. (46) showed peripheral patch mutants were generally more reduced for DNA replication compared with the central patch mutants, consistent with results here. In contrast to the work here or by Domsic et al. (46), which both used triple amino acid substitutions to mutate either the central or peripheral patch, Hellert et al. (47) examined single amino acid substitutions in the central or peripheral patch, and also constructed a mutant with substitutions in both. Substitution of single peripheral (K1109A) or central (K1138A) patch residues did not reduce DNA replication, although substitution of a different central (K1141A) patch residue did reduce replication (47). Although K1109A and K1138A did not reduce replication in isolation, a mutant with both these substitutions was deficient.
These findings suggest that DNA replication and episome maintenance deficiencies may underlie the observed reductions in viral latency after infection of mice with MHV68 containing LANA peripheral positive patch mutations. Alanine or glutamate substitution mutations of the corresponding residues in the MHV68 LANA positive patch resulted in substantial reduction in expansion of latent infection when the peripheral patch was mutated but not when the central patch was mutated (45, 47). Because MHV68 LANA mediates MHV68 episome persistence using mechanisms similar to KSHV LANA (58), it is likely that these MHV68 LANA mutants are also deficient for DNA replication and episome persistence.
Notably, after episome establishment, even those positive patch mutants with substantial episome maintenance deficiencies were capable of maintaining episomes for well over a month and likely can do so indefinitely. It is likely that selection for recombination events in the TR DNA results in compensatory changes that allow long term persistence for these mutants, as discussed previously (52, 56, 59). Episomal DNA for these mutants often migrated much more slowly than input covalently closed, circular TR plasmid DNA, sometimes co-migrating with BCBL1 KSHV viral genomes, which are ∼200 kb (Fig. 4) (33, 60). These changes are due to recombination events resulting in duplication of TR elements or head to tail multimerization of input TR plasmids (10). In addition, such mutants often had much larger amounts of episomal TR DNA compared with that for LANA (Fig. 4). It is likely that amplification of the TR elements or certain critical TR components allows enhanced persistence in the presence of LANA deficiency.
These results indicate that the positive patch exerts an important role in LANA-mediated DNA replication. LANA lacks enzymatic function and recruits host cell machinery to replicate DNA. It is possible that the positive patch interacts with a key replication factor. Origin recognition complex (ORC) proteins, responsible for pre-replication complex formation, are recruited to TR DNA in a LANA-dependent manner, and ORC1 to ORC6 each interact with C-terminal LANA (36, 61, 62). Replication factor C, the DNA polymerase clamp (proliferating cell nuclear antigen (PCNA)) loader allows processive DNA replication and is recruited to TR DNA by direct interaction with LANA to mediate replication. However, LANA residues 262–320, rather than the C-terminal domain, are critical for replication factor C interaction (63). Topoisomerase IIβ, which facilitates DNA replication by inducing double stranded breaks, interacts with LANA N-terminal amino acids 1–32 and not the C-terminal domain (64). Structure-specific recognition protein 1 (SSRP1), which was shown to exert a role in LANA replication, and replication proteins A1 and A2 also associate with LANA, but their binding regions within LANA have not been identified (65, 66). In addition, HBO1, a histone acetyltransferase important for replication licensing, and minichromosome maintenance complex (MCM), a DNA helicase important for replication, are both recruited to the TR DNA region in a LANA-dependent manner, although they have not been shown to bind LANA (62). Ubiquitin-specific protease USP7 interacts with LANA residues 971–986 upstream of the positive patch, and deletion of the USP7 interacting sequence enhanced LANA replication (67). It is possible that one of these or another replication factor may interact with the positive patch to mediate LANA DNA replication.
The dorsal positive patch was recently shown to mediate interaction of C-terminal LANA with DNA, independent of DNA sequence (48). Therefore, it is possible that the LANA DBD can bind its specific TR recognition sequence along its ventral surface and simultaneously bind additional DNA regardless of sequence along its dorsal surface. It is possible that such DNA binding through the dorsal positive patch, perhaps inducing specific three-dimensional structures, may exert an important role in DNA replication.
Although mutation of the peripheral and central positive patch reduced BET protein binding, the diminished binding did not correlate with the deficiencies in LANA-mediated DNA replication or episome persistence. For instance, despite similar reductions in BET protein interaction, the peripheral patch mutants were more compromised for episome persistence than were the central patch mutants. In other work reduced BRD4 binding to LANA was similarly observed with triple substitution central patch mutants. However, in contrast to results here, there was little to no loss of BRD4 binding when the same triple substitution peripheral patch mutants were tested (46). In addition, work found that the peripheral patch mutation K1109A and the central patch mutations K1138A or K1140A each reduced BRD2 and BRD4 binding and that combined K1109A/K1138A mutations completely abolished BRD2 and BRD4 binding. The central patch mutation K1141A also reduced BRD2 and BRD4 binding, although to a lesser extent (47). The peripheral patch mutation K1113A reduced BRD2, but not BRD4 binding (47). The persistence of a modest level of LANA binding to BET proteins in most mutants despite positive patch mutations likely relates to the two distinct contact points between BET proteins and C-terminal LANA. The positive patch interacts with a conserved serine-rich region of BET proteins, whereas residues 1125–1129 interact with the upstream BET protein ET domain (47). Consequently, it is expected that although the positive patch mutants will have reduced interaction with the BET serine-rich region, the ET domain can continue to interact with LANA 1125–1129.
Our results demonstrate that all positive patch mutants were capable of concentrating to LANA dots along mitotic chromosomes (Fig. 5). Dot formation is due to LANA concentration at sites of KSHV episomes (9). In other work, Hellert et al. (47) found that the LANA dorsal positive patch exerted a role in higher order LANA oligomerization as K1109A (peripheral patch), K1113A (peripheral patch), K1138A (central patch), or K1109A/K1138A (peripheral and central patch) each reduced the ability of the LANA C-terminal domain to precipitate LANA from cell extracts. In addition, the K1109A/K1138A mutant was impaired for the ability to form LANA dots after transient transfection of LANA and DNA containing four TR elements into HeLa cells, although the K1109A or K1138A mutants maintained the ability to form dots. This finding differs from our results as we did not observe loss of LANA dots in any positive patch mutants that stably maintained episomes. It is possible that these differences could be due to the different experimental approaches used or to the differences in LANA mutations. For instance, K1109A/K1138A contains mutations in both the peripheral and central positive patch region, but the mutants used here independently mutated the peripheral or central positive patch, and no mutant contained substitutions in both the peripheral and central regions.
This work shows that the LANA C-terminal domain dorsal positive electrostatic patch exerts roles in LANA-mediated DNA replication and episome persistence. Future work should elucidate whether the dorsal positive patch exerts its effects through interactions with a particular host cell partner(s) or through another mechanism, such as interacting with DNA in a sequence independent manner.
Author Contributions
S. L. and K. M. K. designed and coordinated the study and wrote the paper. At the initiation of this work, R. P., B. C., J. P. S., M. A. C., and C. E. M. provided unpublished structural information and designed the positive patch mutations. S. L., M. T., and F. J. performed experiments and analyzed the results. All authors contributed to conceiving the paper, analyzed the results, and approved the final version of the manuscript.
This work was supported in part by National Institutes of Health Grants CA082036 (NCI) and DE025208 (NIDCR; both to K. M. K.), Harvard Medical School-Portugal Program in Translational research and Information (to J. P. S., M. A. C., C. E. M., and K. M. K.), and Brigham and Women's Hospital Research Institute (to K. M. K.). The authors declare that they have no conflicts of interest with the contents of this article.
- KS
- Kaposi sarcoma
- KSHV
- Kaposi sarcoma-associated herpesvirus
- LANA
- latency-associated nuclear antigen
- DBD
- DNA binding domain
- TR
- terminal repeat
- MHV68
- murine gamma herpesvirus 68
- BET
- bromodomain and extra terminal domain
- LBS1
- LANA binding site 1.
References
- 1.Cesarman E., Chang Y., Moore P. S., Said J. W., and Knowles D. M. (1995) Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332, 1186–1191 [DOI] [PubMed] [Google Scholar]
- 2.Dupin N., Fisher C., Kellam P., Ariad S., Tulliez M., Franck N., van Marck E., Salmon D., Gorin I., Escande J. P., Weiss R. A., Alitalo K., and Boshoff C. (1999) Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. U.S.A. 96, 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupin N., Diss T. L., Kellam P., Tulliez M., Du M. Q., Sicard D., Weiss R. A., Isaacson P. G., and Boshoff C. (2000) HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 95, 1406–1412 [PubMed] [Google Scholar]
- 4.Moore P. S., and Chang Y. (1995) Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection [see comments]. N. Engl. J. Med. 332, 1181–1185 [DOI] [PubMed] [Google Scholar]
- 5.Oksenhendler E., Duarte M., Soulier J., Cacoub P., Welker Y., Cadranel J., Cazals-Hatem D., Autran B., Clauvel J. P., and Raphael M. (1996) Multicentric Castleman's disease in HIV infection: a clinical and pathological study of 20 patients. AIDS 10, 61–67 [PubMed] [Google Scholar]
- 6.Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d'Agay M.-F., Clauvel J.-P., Raphael M., Degos L., and Sigaux F. (1995) Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86, 1276–1280 [PubMed] [Google Scholar]
- 7.Cesarman E., Moore P. S., Rao P. H., Inghirami G., Knowles D. M., and Chang Y. (1995) In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell Lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86, 2708–2714 [PubMed] [Google Scholar]
- 8.Decker L. L., Shankar P., Khan G., Freeman R. B., Dezube B. J., Lieberman J., and Thorley-Lawson D. A. (1996) The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184, 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballestas M. E., Chatis P. A., and Kaye K. M. (1999) Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284, 641–644 [DOI] [PubMed] [Google Scholar]
- 10.Ballestas M. E., and Kaye K. M. (2001) Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75, 3250–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szekely L., Kiss C., Mattsson K., Kashuba E., Pokrovskaja K., Juhasz A., Holmvall P., and Klein G. (1999) Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin- associated nuclear bodies. J. Gen. Virol. 80, 2889–2900 [DOI] [PubMed] [Google Scholar]
- 12.Kedes D. H., Lagunoff M., Renne R., and Ganem D. (1997) Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Invest. 100, 2606–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellam P., Boshoff C., Whitby D., Matthews S., Weiss R. A., and Talbot S. J. (1997) Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1, 19–29 [PubMed] [Google Scholar]
- 14.Rainbow L., Platt G. M., Simpson G. R., Sarid R., Gao S.-J., Stoiber H., Herrington C. S., Moore P. S., and Schulz T. F. (1997) The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71, 5915–5921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An F. Q., Compitello N., Horwitz E., Sramkoski M., Knudsen E. S., and Renne R. (2005) The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus modulates cellular gene expression and protects lymphoid cells from p16 INK4A-induced cell cycle arrest. J. Biol. Chem. 280, 3862–3874 [DOI] [PubMed] [Google Scholar]
- 16.Di Bartolo D. L., Cannon M., Liu Y. F., Renne R., Chadburn A., Boshoff C., and Cesarman E. (2008) KSHV LANA inhibits TGF-β signaling through epigenetic silencing of the TGF-β type II receptor. Blood 111, 4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber A. C., Shu M. A., Hu J., and Renne R. (2001) DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75, 7882–7892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knight J. S., Cotter M. A. 2nd, and Robertson E. S. (2001) The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276, 22971–22978 [DOI] [PubMed] [Google Scholar]
- 19.Lu F., Day L., Gao S. J., and Lieberman P. M. (2006) Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 80, 5273–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami Y., Yamagoe S., Noguchi K., Takebe Y., Takahashi N., Uehara Y., and Fukazawa H. (2006) Ets-1-dependent expression of vascular endothelial growth factor receptors is activated by latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus through interaction with Daxx. J. Biol. Chem. 281, 28113–28121 [DOI] [PubMed] [Google Scholar]
- 21.Ottinger M., Christalla T., Nathan K., Brinkmann M. M., Viejo-Borbolla A., and Schulz T. F. (2006) Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80, 10772–10786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shamay M., Krithivas A., Zhang J., and Hayward S. D. (2006) Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U.S.A. 103, 14554–14559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang J., Gordon G. M., Müller M. G., Dahiya M., and Foreman K. E. (2003) Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen induces expression of the helix-loop-helix protein Id-1 in human endothelial cells. J. Virol. 77, 5975–5984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma S. C., Borah S., and Robertson E. S. (2004) Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 78, 10348–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye F. C., Zhou F. C., Yoo S. M., Xie J. P., Browning P. J., and Gao S. J. (2004) Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78, 11121–11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim K. Y., Huerta S. B., Izumiya C., Wang D. H., Martinez A., Shevchenko B., Kung H. J., Campbell M., and Izumiya Y. (2013) Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A. J. Virol. 87, 6782–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell M., Chang P. C., Huerta S., Izumiya C., Davis R., Tepper C. G., Kim K. Y., Shevchenko B., Wang D. H., Jung J. U., Luciw P. A., Kung H. J., and Izumiya Y. (2012) Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Biol. Chem. 287, 5806–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garber A. C., Hu J., and Renne R. (2002) Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277, 27401–27411 [DOI] [PubMed] [Google Scholar]
- 29.Cotter M. A. 2nd, Subramanian C., and Robertson E. S. (2001) The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxyl terminus. Virology 291, 241–259 [DOI] [PubMed] [Google Scholar]
- 30.Fejér G., Medveczky M. M., Horvath E., Lane B., Chang Y., and Medveczky P. G. (2003) The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts preferentially with the terminal repeats of the genome in vivo and this complex is sufficient for episomal DNA replication. J. Gen. Virol. 84, 1451–1462 [DOI] [PubMed] [Google Scholar]
- 31.Canham M., and Talbot S. J. (2004) A naturally occurring C-terminal truncated isoform of the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus does not associate with viral episomal DNA. J. Gen. Virol. 85, 1363–1369 [DOI] [PubMed] [Google Scholar]
- 32.Hu J., Garber A. C., and Renne R. (2002) The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76, 11677–11687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagunoff M., and Ganem D. (1997) The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). Virology 236, 147–154 [DOI] [PubMed] [Google Scholar]
- 34.Grundhoff A., and Ganem D. (2003) The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77, 2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Komatsu T., Ballestas M. E., Barbera A. J., Kelley-Clarke B., and Kaye K. M. (2004) KSHV LANA1 binds DNA as an oligomer and residues N-terminal to the oligomerization domain are essential for DNA binding, replication, and episome persistence. Virology 319, 225–236 [DOI] [PubMed] [Google Scholar]
- 36.Lim C., Sohn H., Lee D., Gwack Y., and Choe J. (2002) Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76, 10320–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krithivas A., Fujimuro M., Weidner M., Young D. B., and Hayward S. D. (2002) Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76, 11596–11604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piolot T., Tramier M., Coppey M., Nicolas J. C., and Marechal V. (2001) Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75, 3948–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cotter M. A. 2nd, and Robertson E. S. (1999) The latency-associated nuclear antigen tethers the Kaposi's sarcoma- associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264, 254–264 [DOI] [PubMed] [Google Scholar]
- 40.Barbera A. J., Chodaparambil J. V., Kelley-Clarke B., Joukov V., Walter J. C., Luger K., and Kaye K. M. (2006) The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311, 856–861 [DOI] [PubMed] [Google Scholar]
- 41.Kelley-Clarke B., De Leon-Vazquez E., Slain K., Barbera A. J., and Kaye K. M. (2009) Role of Kaposi's sarcoma-associated herpesvirus C-terminal LANA chromosome binding in episome persistence. J. Virol. 83, 4326–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley-Clarke B., Ballestas M. E., Komatsu T., and Kaye K. M. (2007) Kaposi's sarcoma herpesvirus C-terminal LANA concentrates at pericentromeric and peri-telomeric regions of a subset of mitotic chromosomes. Virology 357, 149–157 [DOI] [PubMed] [Google Scholar]
- 43.Kelley-Clarke B., Ballestas M. E., Srinivasan V., Barbera A. J., Komatsu T., Harris T. A., Kazanjian M., and Kaye K. M. (2007) Determination of Kaposi's sarcoma-associated herpesvirus C-terminal latency-associated nuclear antigen residues mediating chromosome association and DNA binding. J. Virol. 81, 4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbera A. J., Ballestas M. E., and Kaye K. M. (2004) The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 N terminus is essential for chromosome association, DNA replication, and episome persistence. J. Virol. 78, 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Correia B., Cerqueira S. A., Beauchemin C., Pires de Miranda M., Li S., Ponnusamy R., Rodrigues L., Schneider T. R., Carrondo M. A., Kaye K. M., Simas J. P., and McVey C. E. (2013) Crystal structure of the gamma-2 herpesvirus LANA DNA binding domain identifies charged surface residues which impact viral latency. PLoS Pathog. 9, e1003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domsic J. F., Chen H. S., Lu F., Marmorstein R., and Lieberman P. M. (2013) Molecular basis for oligomeric-DNA binding and episome maintenance by KSHV LANA. PLoS Pathog. 9, e1003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hellert J., Weidner-Glunde M., Krausze J., Richter U., Adler H., Fedorov R., Pietrek M., Rückert J., Ritter C., Schulz T. F., and Lührs T. (2013) A structural basis for BRD2/4-mediated host chromatin interaction and oligomer assembly of Kaposi sarcoma-associated herpesvirus and murine γ-herpesvirus LANA proteins. PLoS Pathog. 9, e1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellert J., Weidner-Glunde M., Krausze J., Lünsdorf H., Ritter C., Schulz T. F., and Lührs T. (2015) The 3D structure of Kaposi sarcoma herpesvirus LANA C-terminal domain bound to DNA. Proc. Natl. Acad. Sci. U.S.A. 112, 6694–6699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.You J., Srinivasan V., Denis G. V., Harrington W. J. Jr., Ballestas M. E., Kaye K. M., and Howley P. M. (2006) Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80, 8909–8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viejo-Borbolla A., Ottinger M., Brüning E., Bürger A., König R., Kati E., Sheldon J. A., and Schulz T. F. (2005) Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79, 13618–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ottinger M., Pliquet D., Christalla T., Frank R., Stewart J. P., and Schulz T. F. (2009) The interaction of the Gammaherpesvirus 68 orf73 protein with cellular BET proteins affects the activation of cell cycle promoters. J. Virol. 83, 4423–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De León Vázquez E., and Kaye K. M. (2011) The internal Kaposi's sarcoma-associated herpesvirus LANA regions exert a critical role on episome persistence. J. Virol. 85, 7622–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardella T., Medveczky P., Sairenji T., and Mulder C. (1984) Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50, 248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srinivasan V., Komatsu T., Ballestas M. E., and Kaye K. M. (2004) Definition of sequence requirements for latency-associated nuclear antigen 1 binding to Kaposi's sarcoma-associated herpesvirus DNA. J. Virol. 78, 14033–14038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De León Vázquez E., and Kaye K. M. (2011) Rapid and quantitative assessment of KSHV LANA-mediated DNA replication. Arch Virol 156, 1323–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vázquez Ede L., Carey V. J., and Kaye K. M. (2013) Identification of KSHV LANA regions important for episome segregation, replication and persistence. J. Virol. 87, 12270–12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim C., Choi C., and Choe J. (2004) Mitotic chromosome-binding activity of latency-associated nuclear antigen 1 is required for DNA replication from terminal repeat sequence of Kaposi's sarcoma-associated herpesvirus. J. Virol. 78, 7248–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Habison A. C., Beauchemin C., Simas J. P., Usherwood E. J., and Kaye K. M. (2012) Murine Gammaherpesvirus 68 LANA acts on terminal repeat DNA to mediate episome persistence. J. Virol. 86, 11863–11876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De León Vázquez E., Juillard F., Rosner B., and Kaye K. M. (2014) A short sequence immediately upstream of the internal repeat elements is critical for KSHV LANA-mediated DNA replication and impacts episome persistence. Virology 448, 344–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russo J. J., Bohenzky R. A., Chien M.-C., Chen J., Yan M., Maddalena D., Parry J. P., Peruzzi D., Edelman I. S., Chang Y., and Moore P. S. (1996) Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U.S.A. 93, 14862–14867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verma S. C., Choudhuri T., Kaul R., and Robertson E. S. (2006) Latency-associated nuclear antigen (LANA) of Kaposi's sarcoma-associated herpesvirus interacts with origin recognition complexes at the LANA binding sequence within the terminal repeats. J. Virol. 80, 2243–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stedman W., Deng Z., Lu F., and Lieberman P. M. (2004) ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J. Virol. 78, 12566–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun Q., Tsurimoto T., Juillard F., Li L., Li S., De León Vázquez E., Chen S., and Kaye K. (2014) Kaposi's sarcoma-associated herpesvirus LANA recruits the DNA polymerase clamp loader to mediate efficient replication and virus persistence. Proc. Natl. Acad. Sci. U.S.A. 111, 11816–11821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purushothaman P., McDowell M. E., McGuinness J., Salas R., Rumjahn S. M., and Verma S. C. (2012) Kaposi's sarcoma-associated herpesvirus-encoded LANA recruits topoisomerase IIβ for latent DNA replication of the terminal repeats. J. Virol. 86, 9983–9994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu J., Liu E., and Renne R. (2009) Involvement of SSRP1 in latent replication of Kaposi's sarcoma-associated herpesvirus. J. Virol. 83, 11051–11063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shamay M., Liu J., Li R., Liao G., Shen L., Greenway M., Hu S., Zhu J., Xie Z., Ambinder R. F., Qian J., Zhu H., and Hayward S. D. (2012) A protein array screen for KSHV LANA interactors links LANA to TIP60, PP2A activity, and telomere shortening. J. Virol. 86, 5179–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jäger W., Santag S., Weidner-Glunde M., Gellermann E., Kati S., Pietrek M., Viejo-Borbolla A., and Schulz T. F. (2012) The ubiquitin-specific protease USP7 modulates the replication of Kaposi's sarcoma-associated herpesvirus latent episomal DNA. J. Virol. 86, 6745–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Toptan T., Fonseca L., Kwun H. J., Chang Y., and Moore P. S. (2013) Complex alternative cytoplasmic protein isoforms of the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 generated through noncanonical translation initiation. J. Virol. 87, 2744–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]