Abstract
Random fluctuations in gene expression lead to wide cell-to-cell differences in RNA and protein counts. Most efforts to understand stochastic gene expression focus on local (intrinisic) fluctuations, which have an exact theoretical representation. However, no framework exists to model global (extrinsic) mechanisms of stochasticity. We address this problem by dissecting the sources of stochasticity that influence the expression of a yeast heat shock gene, SSA1. Our observations suggest that extrinsic stochasticity does not influence every step of gene expression, but rather arises specifically from cell-to-cell differences in the propensity to transcribe RNA. This led us to propose a framework for stochastic gene expression where transcription rates vary globally in combination with local, gene-specific fluctuations in all steps of gene expression. The proposed model better explains total expression stochasticity than the prevailing ON-OFF model and offers transcription as the specific mechanism underlying correlated fluctuations in gene expression.
Graphical abstract
Introduction
The processes underlying gene expression produce remarkable cell-to-cell heterogeneity of RNA and protein counts between genetically identical cells (Chabot et al., 2007; Elowitz et al., 2002; Newman et al., 2006; Ozbudak et al., 2002; Raj et al., 2006; Raser and O’Shea, 2004; Stewart-Ornstein et al., 2012). This heterogeneity arises, in part, from random molecular collisions, which introduce local, ‘intrinsic’ fluctuations in transcription and translation that act independently on individual genes within the same cell. In contrast, global ‘extrinsic’ factors, such as changes in the number of transcription factors or ribosomes, act on many genes simultaneously and induce correlated fluctuations between genes in the same cell. To quantify and separate global (extrinsic) effects from local (intrinsic) mechanisms investigators quantify the covariance between two identical reporter genes in single cells (Elowitz et al., 2002; Raser and O’Shea, 2004). The covariance between identical genes captures extrinsic sources of variance, while intrinsic mechanisms decouple their expression. The relative positioning of the two reporter genes defines whether a particular mechanism is labeled intrinsic or extrinsic in a given experiment. The theoretical basis of intrinsic noise has been studied extensively (Elgart et al., 2011; Elowitz et al., 2002; Gillespie, 1976; Paulsson, 2005; Shahrezaei et al., 2008) yielding a consensus model–the ON-OFF model–which seems necessary to explain higher than expected variability in RNA and protein levels levels (Blake et al., 2003; Blake et al., 2006; Golding et al., 2005; Harper et al., 2011; Lionnet and Singer, 2012; Raj et al., 2006; Raj and van Oudenaarden, 2008; Raser and O’Shea, 2004; Suter et al., 2011).
No such model exists for capturing extrinsic stochasticity. For example, differences in cell volume (Becskei et al., 2005; Mogno et al., 2010; Newman et al., 2006; Stewart-Ornstein et al., 2012), cell cycle position (Zenklusen et al., 2008; Zopf et al., 2013), mitochondrial content (Guantes et al., 2015), and co-transcriptional regulation (Gandhi et al., 2011; Stewart-Ornstein et al., 2012) contribute to extrinsic stochasticity, but it remains unclear how to incorporate these nonspecific effects into the intrinsic-only ON-OFF model.
Given that both intrinsic and extrinsic sources of noise contribute substantially to stochastic gene expression (Elowitz et al., 2002; Stewart-Ornstein et al., 2012; Volfson et al., 2006), it is crucial to understand how the interaction between intrinsic and extrinsic factors generates total expression stochasticity. Here, the lack of a theoretical framework for handling extrinsic noise represents a serious limitation. Rather than modeling both sources of variance together, previous investigations separated total variance into its intrinsic and extrinsic components, and then analyzed only the intrinsic component (Carey et al., 2013; Dadiani et al., 2013; Newman et al., 2006; Raser and O’Shea, 2004; Shalem et al., 2013). New mechanistic models of stochastic gene expression will be necessary to analyze sources of extrinsic variance, such as changes in cell volume.
Changes in cell volume occur in predictable ways across the cell cycle and introduce a large portion of extrinsic variance in gene expression (Becskei et al., 2005; Mogno et al., 2010; Newman et al., 2006; Padovan-Merhar et al., 2015; Stewart-Ornstein et al., 2012; Zenklusen et al., 2008; Zopf et al., 2013). The physiological changes associated with particular stages of the cell cycle also generate extrinsic differences between genetically identical cells. How are the impacts of these changes mediated? One possibility is that the rate of every step in gene expression–transcription, translation, along with RNA and protein degradation–varies as the protein effectors involved change in abundance through the cell cycle. Alternatively, extrinsic contributions may operate mainly at one particular step in gene expression.
To distinguish between these possibilities we propose a theoretical model that incorporates both intrinsic and extrinsic sources of noise in a unified framework. We show that this hybrid model faithfully captures both intrinsic and extrinsic noise, and predicts the shape of the full stochastic expression distribution. We conclude that our hybrid model broadly captures the underlying mechanisms that generate noise in gene expression.
Results
Intrinsic and extrinsic stochasticity are mechanistically related
Intrinsic and extrinsic sources of stochasticity are often treated as orthogonal contributors to expression variability, since total variance is the sum of intrinsic and extrinsic variance (Elowitz et al., 2002; Newman et al., 2006).
| (1) |
However, intrinsic and extrinsic variances share a more nuanced relationship (Hilfinger and Paulsson, 2011; Lei, 2009; Shahrezaei et al., 2008). We represent that relationship using the definition of variance and the double expectation theorem E[E[X ∣ e]] = E[X], yielding Eq. 2. Here, e represents any extrinsic factor on which the random variable X depends.
| (2) |
Eq. 2 demonstrates a key insight: intrinsic variance explicitly depends on extrinsic factors e. It is therefore not clear whether intrinsic and extrinsic variance can be considered separately. This led us to explore how intrinsic and extrinsic sources of variability might be brought together in a single model.
We started with the simplest mechanistic model of gene expression (Fig. 1A). This model captures transcription and translation at rates km and kp, along with the degradation of RNA (dm) and protein (dp), which occurs in proportion to the amount of each molecule present. The stochastic formulation of this model is a chemical master equation (Eq. 3), with Nm and Np representing the number of mRNAs and proteins respectively.
| (3) |
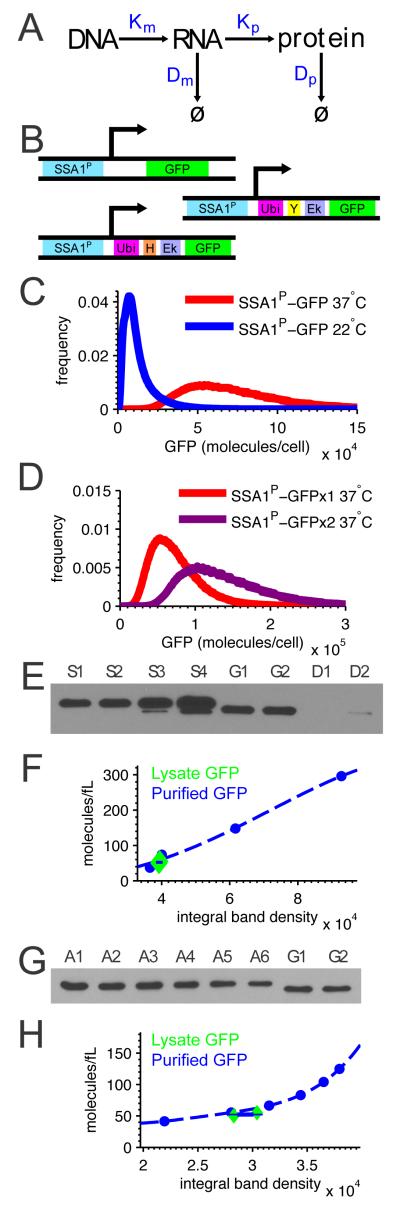
Figure 1. Experimental setup and validation.
A Hybrid model of intrinsic and extrinsic noise represents transcription (Km), translation (Km), and protein (Dp) and RNA (Dm) degradation with each step’s rate being a random variable (capital letter). B Gene constructs are, from top to bottom, SSA1P-GFP, SSA1P-Y-GFP, and SSA1P-H-GFP. C Basal GFP expression at 22° C and 37 ° C of BY4743 diploid yeast strains with single chromosomal integrants of SSA1P-GFP demonstrates temperature sensitive expression. D Expression of one-copy versus two-copy diploid SSA1P-GFP strains. E Western blot of autofluorescent controls with increasing concentrations of purified GFP spiked into GFP-negative lysate as standards (S1-S4), two replicates of GFP purified from lysate (G1, G2) from SSA1P-GFP strain replicates, and resuspended insoluble debris from the same samples (D1, D2). F Density integrals for the standards (blue) plotted versus the calculated number of molecules of spiked-in purified GFP. Blue dashed line is a third order polynomial best fit. G1 and G2 (green triangles) are plotted as Western blot band density versus FCS-measured particle count. G Western blot of finer dilutions of autofluorescent standard S3 (from first blot) as new standards (A1-A6), and the same two replicates of GFP from lysate (G1, G2). H Density integrals for the standards (blue) plotted versus the calculated number of molecules of spiked-in purified GFP. Blue dashed line is a third order polynomial best fit. G1 and G2 (green triangles) are plotted as Western blot band density versus FCS-measured particle count. See also Figures S1 and S4.
Assuming steady state yields solutions to the model’s first and second moments:
| (4) |
| (5) |
A key caveat is that this model only represents intrinsic sources of variability. This follows since Eq. 3 equally describes the behavior of all genes whether they lie in the same cell or different cells; therefore all genes act completely independently and by definition will not display correlated fluctuations. To introduce correlated (extrinsic) fluctuations within single cells, one approach would be to replace the rate constants of this model with time-varying concentrations, affinities and interactions that determine the average rate of each step. In such a model, two genes within the same cell would experience the same cell-specific number of polymerases, transcription factors, ribosomes, and decay factors, but these variables would change from cell to cell.
We represent this idea by assuming that extrinsic fluctuations (e.g. the changes in the number of polymerases in a cell over time) operate on a slower timescale than intrinsic fluctuations (the thermodynamic fluctuations in bimolecular interactions) (Rosenfeld et al., 2005). In this regime, each parameter in the intrinsic-only model is replaced by a random variable: km to Km, kp to Kp dm to Dm, and dp to Dp, where, for example, each individual cell experiences a specific transcription rate km drawn from Km (Fig. 1A). The variability in the rate constants themselves may be linked, necessitating a joint distribution across all parameter values, P(Km = km, Kp = kp, Dm = dm, Dp = dp).
Despite the simplifying assumption of fixed extrinsic rates for each cell, a model incorporating these factors yields a quagmire even for mean expression:
| (6) |
In this worst case scenario, experimentally extracting P(km,kp, dm, dp) would require instantaneous estimates of all four rate constants in single cells. The complexity of such a system has led the majority of studies to isolate intrinsic stochasticity by itself (Carey et al., 2013; Dadiani et al., 2013; Newman et al., 2006; Raser and O’Shea, 2004; Shalem et al., 2013). Alternatively, if extrinsic noise can be represented by only a single parameter that varies widely from cell to cell then the theory simplifies considerably. However, it is not clear if a model in which a single parameter controls the majority of extrinsic stochasticity will capture the cell-to-cell variability observed in real expression distributions. If one parameter is primarily responsible for extrinsic noise, it is still not clear which parameter is the best candidate. Thus, it may be that every step in gene expression introduces significant extrinsic noise, or that extrinsic variation in only one parameter captures most cell-to-cell variability. We used the SSA1 promoter in yeast as a model to understand where one particular gene falls on this spectrum.
SSA1, a model heat shock inducible promoter
We quantified stochastic expression of a GFP reporter gene driven by the SSA1 promoter in S. cerevisiae. SSA1 belongs to the HSP70 family of chaperone proteins, which respond to stress conditions including heat shock (Hahn and Young, 2011; Slater et al., 1987; Young and Craig, 1993). We fused the promoter to a fast maturing GFP variant (Cormack et al., 1996; Iizuka et al., 2011) and integrated this reporter gene into the genome at the HIS3 locus.
Protein half-life sets the timescale of stochastic fluctuations and contributes substantially to the balance of intrinsic and extrinsic noise (Wang et al., 2008). We therefore assembled two destabilized GFP mutants by exploiting the N-end pathway (Hackett et al., 2006; Varshavsky, 1996). N-end amino acid identity determines protein stability; we selected tyrosine (moderately destabilizing) and histidine (very destabilizing) as our N-end amino acids (Varshavsky, 1996) and integrated these constructs at the HIS3 locus (Fig. 1B).
We measured the cell-to-cell distribution of GFP produced by each construct using flow cytometry. The constructs behaved as expected, with mean expression the highest for SSA1P-GFP, then SSA1P-Y-GFP, and then SSA1P-H-GFP. All strains also expressed higher at 37 ° C than 22 ° C (Fig. 1C, Fig. S1B-F).
Flow cytometry enables us to record gene expression variability among single cells. However, the units of this measurement, arbitrary units of fluorescence, are problematic since stochastic models of gene expression operate in units of protein number (Np, Eq. 3). We therefore used Fluorescence Correlation Spectroscopy (FCS) and quantitative western blotting to calibrate fluorescence distributions in units of absolute protein. Our estimates of the absolute molecule count to fluorescence conversion factor were concordant between these two methods (Fig. 1E-H). Details of both calibration experiments are in the Experimental Procedures and in the Supplement.
Intrinsic and extrinsic variance both contribute to steady state SSA1P-GFP expression
With a protocol for calibrating our cytometer measurements in units of protein count, we quantified stochastic expression of our reporter constructs. Steady state measurements were obtained at 22 ° C and 37 ° C. The mean steady state expression level in the untagged diploid single-copy strain was 80958±5160 GFP molecules per cell at 37 ° C , which decreased by a factor of 10 at 22 ° C (Fig. 1C). As expected, the destabilized reporters SSA1P-Y-GFP and SSA1P-H-GFP showed lower steady state expression at 4921.5±257 and 4170.4±193 proteins per cell, respectively (Table S1, Figs. S1B-G). All errors are 99% confidence intervals based on biological replicates’ fluorescence measurements. The absolute counts scale with an error of ± 22 % (99% CI, see Experimental Procedures) which do not affect relative comparisons, as all fluorescence measurements are multiplied by a common conversion factor.
We measured extrinsic and intrinsic noise by comparing expression between diploid strains containing either one or two copies of the same reporter gene on homologous chromosomes (Chabot et al., 2007; Stewart-Ornstein et al., 2012) (Fig. 1D). Details can be found in the Supplement under the heading “Estimation of intrinsic and extrinsic noise by 1- and 2-copy reporter strains.” This approach avoids difficulties with reporter equivalence in dual color experiments (Chabot et al., 2007; Stewart-Ornstein et al., 2012). At 37 °C intrinsic noise comprises 7 % of SSA1P-GFP expression stochasticity (Table S1). At 22 ° C , the balance shifts to 15% intrinsic variance. The destabilized reporters SSA1P-Y-GFP and SSA1P-H-GFP generate variance with larger contributions from intrinsic variance–as high as 45% for SSA1P-H-GFP at 22 ° C. These results are consistent with previous observations that lower expression shifts the source of variance toward intrinsic noise (Bar-Even et al., 2006; Newman et al., 2006; Stewart-Ornstein et al., 2012). Nonetheless, even at the very lowest expression of a few thousand proteins per cell, more than 50% of expression variability originates extrinsically. This observation underlines the need for quantitative models that represent both sources of stochasticity.
RNA and protein degradation rates constant across cell cycle
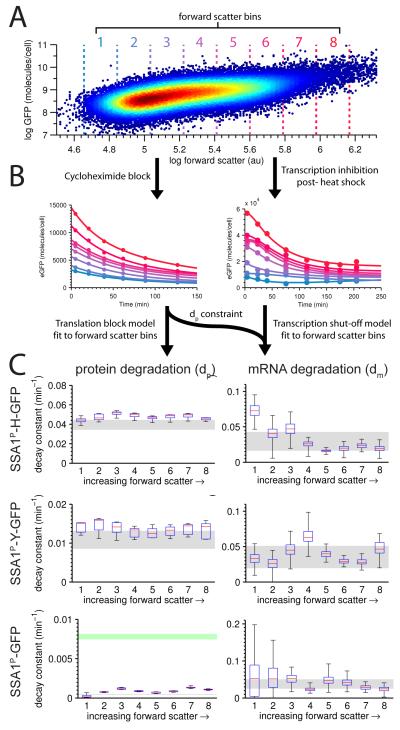
We first tested whether extrinsic factors might contribute to variability through fluctuations in the rates of mRNA and protein degradation. To do so we measured bulk degradation rate constants across different phases of the cell cycle, a cellular process that contributes substantially to extrinsic noise in gene expression (Becskei et al., 2005; Mogno et al., 2010; Newman et al., 2006; Stewart-Ornstein et al., 2012; Zenklusen et al., 2008; Zopf et al., 2013). The cell cycle is an attractive model of extrinsic noise because the expression of genes tends to rise as the cell grows (Fig. 2A), naturally titrating the levels of candidate extrinsic factors (Mogno et al., 2010; Newman et al., 2006; Stewart-Ornstein et al., 2012).
Figure 2. RNA and protein degradation rates conditioned on cell size.
A Single-cell SSA1P-Y-GFP expression measured by flow cytometry in one replicate (37 ° C) plotted against forward scatter. The same forward scatter bin parameters were used across all experiments. B Protein decay constants were obtained by tracking bin-specific (color-coded to match A, above) decay in the presence of the translation-blocking agent cycloheximide. RNA decay constants were obtained by tracking transcriptionally-mediated inhibition of SSA1 expression after heat shock, and constrained by the protein decay rate. C Estimated protein (left column) and RNA (right column) decay constants for each forward scatter bin. Red lines are median rate constant estimates, blue lines are 25th and 75th percentiles, and whiskers mark the full range. Gray regions indicate decay constants estimated on the bulk data rather than binned by forward scatter (99% CI). For SSA1P-GFP protein degradation (bottom left), the green region represents the 99% CI for expected degradation assuming dilution from growth alone. See also Figure S2.
To measure protein degradation of GFP we blocked translation using cycloheximide when the cells were at 37 ° C steady state and recorded the initial decline in signal over time by flow cytometry. As expected, fluorescence from SSA1P-H-GFP declines at a faster rate than SSA1P-Y-GFP, yielding bulk d p values of .039±.0044min −1 and .011±.00085min −1 respectively. Untagged GFP degraded at a rate well below the dilution rate, with the degradation rate by dilution alone corresponding to 9 0 minute division cycles (bulk d p = .0078±.00035min −1).
We adopted a drug-free approach to measure RNA degradation. Heat shocking cells by shifting their temperature from 22 ° C to 37 ° C elicits a large pulse of RNA expression (Slater et al., 1987). This pulse is followed by canonical, HSF1-mediated transcriptional deactivation 20 minutes after heat shock (Young and Craig, 1993). To take advantage of this natural, partial transcriptional shutoff, we fit an incomplete shutoff model of RNA and protein degradation to the decay curve of expression as it relaxes toward 37 ° C equilibrium. The model was parameterized using protein degradation rates obtained from the cycloheximide experiment. RNA degradation rates for each of our constructs were: .029±.0067min −1 (SSA1P-H-GFP), .035±.0024min −1 (SSA1P-Y-GFP), and .038±.013min −1 (SSA1P-GFP). The remarkable agreement between these three estimates for RNA degradation suggests that the slight differences in RNA sequence of the three constructs mediate very little change in the RNA degradation rate.
With bulk decay rates in hand we tested the extent to which dm and dp vary across cell size by partitioning cells into bins by forward scatter size (Fig. 2A) and applying the same degradation models to each bin (Fig. 2B). For all three constructs the bins show a remarkable lack of trend (Fig. 2C, left panels), indicating that dp is invariant across cell size. SSA1P-GFP degradation rates derived from cycloheximide block experiments are substantially less than the normal growth rate, demonstrating that N-end methionine GFP degrades primarily by dilution alone. On the whole we observe very little extrinsic variability in dp across cells that vary widely in volume.
We then fit RNA degradation curves across the same forward scatter bins. All plots show more variability than for protein degradation rates, yet trend less than a factor of two away from the bulk rate constant (Fig. 2C, right panels). RNA degradation across cell size is also invariant.
Our data show that both RNA and protein degradation rates vary minimally with cell size and cell cycle, and indicate that extrinsic variance from the cell cycle must be mediated through transcription or translation rates.
Hybrid model predicts expression distribution without fitting
Two lines of evidence elsewhere suggest transcription rate (km) is the primary source of extrinsic variance. First, if km were static from cell-to-cell, intrinsic variance would account for all of the variance in RNA count distributions. In contrast, the largest study of intrinsic and extrinsic variance to date demonstrates that extrinsic variance contributes 20-90% of total variance at the RNA level (Gandhi et al., 2011). Second, genes regulated by the same transcription factors MSN2/4 covary in their expression fluctuations (Stewart-Ornstein et al., 2012), a finding consistent with an extrinsically varying Km. These data led us to focus on a hybrid model where km is the only extrinsically varying parameter within forward scatter bins. We derived analytical forms of mean expression along with intrinsic and extrinsic variance for an extrinsically varying Km.
| (7) |
| (8) |
| (9) |
This trio of equations is directly solvable, without fitting, using the experimental measurements we made. Flow cytometry, calibrated by fluorescence correlation spectroscopy, yielded E[Np] in units of proteins/cell and permitted measurement of variance in units of proteins 2. The single and double copy strains enabled quantification of intrinsic and extrinsic variance as described above, and the degradation rates were obtained by cycloheximide block and transcriptionally mediated inhibition near 37 ° C steady state (above). Thus, this set of equations can be solved directly, yielding the translation rate kp and the first two moments of the extrinsically varying distribution of Km:
| (10) |
| (11) |
| (12) |
By solving these equations we obtained values for all of the parameters in the hybrid intrinsic-extrinsic model. Our goal was to determine the extent to which this hybrid model captures the whole distribution of observed expression stochasticity. To generate expression distributions from this model we first needed to make a principled choice about the Km distribution itself.
We hypothesized that the shape of Km is determined by cell-to-cell variability in the concentration of a protein, or proteins, and therefore, that Km would take the shape of typical protein expression distributions. To get an idea of which distributions empirically fit the shape of expression stochasticity, we fit all common statistical distributions to each expression distribution in a random promoter library fused to GFP (Data S2) (Mogno et al., 2010). In 80% of 288 expression distributions the top rank was the generalized extreme value distribution with zero scale parameter (Gumbel, or type-I extreme value distribution). The Gumbel distribution was ranked in the top 3 in 100% of fits. Previously, Gamma distributions were noted to well-approximate expression variability (Taniguchi et al., 2010) in E. coli, however, Gamma fits ranked in the top 3 in only 5 of 288 distributions.
Broad agreement of generic gene expression distributions with the Gumbel distribution led us to hypothesize that Km would also be Gumbel distributed. Gumbel distributions have two parameters, μ and σ, which are uniquely specified by the mean and variance of Km above (see Supplement under the heading “Km as a Generalized Extreme Value distribution”).
Using this framework we solved our model for expression data across different forward scatter bins. For each bin we measured mean expression and both intrinsic and extrinsic variance. We accounted for minimal changes in dm and dp across the cell cycle by measuring bin-specific values for both (2). With five observables for each bin, we solved the model for the two unknown parameters of Km–E[Km] and Var[Km]–and for the translation rate kp. The mean and variance of Km were then used to deduce the Gumbel parameters μk and σk for each bin. Parameter sets are reported in Data S3. These results are a complete model-based characterization of the full expression distribution taking into account the contributions from both intrinsic and extrinsic stochasticity, and yield testable predictions about SSA1 transcription and translation rates.
How well do these resulting parameter sets correspond to experimental observations? Although mean, intrinsic and extrinsic variance of our measured distributions are guaranteed to be correct, mean and variance do not uniquely determine the shape of gene expression distributions (Huh and Paulsson, 2011; Sherman and Cohen, 2014). In contrast, four statistical moments very nearly recapitulate gene expression distributions (Sherman and Cohen, 2014). The large number of cells measured enabled us to obtain reproducible estimates of the skewness and kurtosis for each bin. We then sought to test whether the model parameterized above, for each bin, is consistent with these higher moments.
We used the experimentally parameterized model to simulate expression distributions from each forward scatter bin (see Supplement). Our simulated mean, intrinsic variance and extrinsic variance were in agreement with our experimental quantities, as expected since these quantities were used to generate the parameters. We then computed total distribution skewness and kurtosis–quantities not used for any fitting–to determine whether these total moments agree with those from our experimental data. Like total variance, total skewness and kurtosis each comprise intrinsic and extrinsic terms, in addition to other covariant terms.
| (13) |
| (14) |
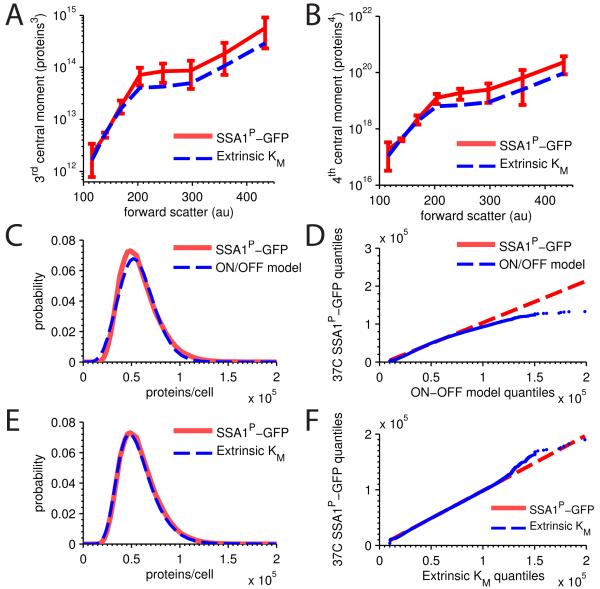
Inserting the estimated distributions for each simulated conditional moment into Eqs. 13 and 14 yielded predicted values of total skewness and kurtosis that we could compare to our experimental data. We found that the model almost exactly predicts higher order distribution moments (Fig. 3A and B), indicating our model likely reproduces, without fitting, the entire distribution comprised of both intrinsic and extrinsic noise (Sherman and Cohen, 2014).
Figure 3. Extrinsic Km model distributional predictions.
SSA1P-GFP single copy measured expression (red, 99% CI) versus extrinsic Km model predicted (blue) A central skewness and B central kurtosis. C measured SSA1P-GFP distribution versus simulated distribution for the best fit ON-OFF model for an example forward scatter bin, along with a D QQ-plot of the same data. E measured SSA1P-GFP distribution versus simulated distribution for extrinsic Km model for an example forward scatter bin, along with a F QQ-plot of the same data. RNA decay constants. See also Figure S3.
Although we were encouraged that our model matched the higher order moments without fitting, it was still possible that all models of gene expression would also yield close agreement with higher moments. We compared our results to the best available alternative model, the widely accepted ON-OFF model, which, although it captures only intrinsic noise, has been used to model total stochasticity previously (Golding et al., 2005; Neuert et al., 2013; Raser and O’Shea, 2004; So et al., 2011; Suter et al., 2011). However, using an established fitting procedure for the ON-OFF model (Sherman and Cohen, 2014) we found no solutions that returned the correct higher order moments of the expression distribution across the forward scatter bins. To confirm this non-result, we relaxed the fitting constraints by fitting on only mean and variance, which did yield some candidate solutions (Fig. S3A). When we Gillespie-simulate these candidate parameter sets, we find, as expected, that they fail to capture the full distribution shape. In particular, the ON-OFF model, though having one additional parameter than the extrinsic Km model we propose, fails to reproduce both peak position and tail behavior (Fig. 3C and D).
In contrast, Gillespie-simulation of our model demonstrates excellent agreement between the predicted distribution and the measured distribution, as expected given the agreement between the first four moments. This exercise led us to conclude that agreement between mean and variances does not guarantee that the rest of the distribution will fall into place. On the contrary, while no parameter set exists where the ON-OFF model can capture the SSA1P-GFP distribution, our model, with one less parameter, provides excellent agreement using only mean and the two variances.
Although experimental data here and elsewhere (Gandhi et al., 2011; Stewart-Ornstein et al., 2012) suggests a crucial role for extrinsically varying transcription rates, it is unclear whether translation rates also contribute. In the Supplement we show that a model of extrinsically varying translation rates Kp yields similar moment predictions (Fig. S3B and C), leaving open the possibility that translation may also vary extrinsically alone, or in combination with transcription rates.
Model predicts N-end degron also mediates differences in translation, but not transcription
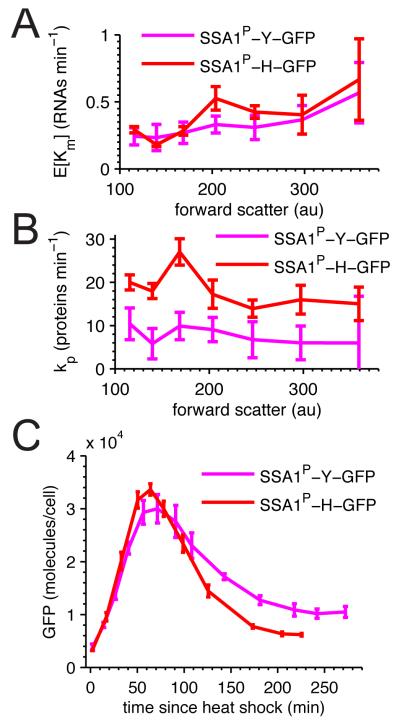
We next used our hybrid extrinsic Km model to analyze stochastic expression in the destabilized constructs, SSA1P-H-GFP and SSA1P-Y-GFP. Because the mechanisms of N-end mediated degradation are well understood we expected that the predicted parameters for these constructs would be the same, except for the protein degradation rate. We found close agreement between the three constructs’ bulk transcription rates (E[Km]): SSA1P-GFP, SSA1P-Y-GFP, and SSA1P-H-GFP were .35±.04min −1, .33±.05min −1, and .40±.05min −1 respectively. Unexpectedly, the predicted bulk translation rates substantially differed from one another. SSA1P-GFP translated most efficiently at 73.27±4.0min −1, while the destabilized constructs translated at slower rates of 7.69±2.34min −1 for SSA1P-Y-GFP and 18.16±1.38min −1 for SSA1P-H-GFP.
The difference in translation rate, but not transcription rate between the destabilized reporter constructs warranted further investigation across the cell cycle. Overall SSA1P-H-GFP and SSA1P-Y-GFP GFP transcribed similarly even across cells of different sizes (Fig. 4A), indicating that the two nucleotide differences in the N-end sequence have little effect on the transcription rate. In contrast SSA1P-H-GFP consistently translates faster than SSA1P-Y-GFP (Fig. 4B). These data suggest that SSA1P-H-GFP is both translated and degraded faster than SSA1P-Y-GFP.
Figure 4. SSA1P-H-GFP and SSA1P-Y-GFP differ in translation rate but not transcription rate.
SSA1P-H-GFP (red) and SSA1P-Y-GFP (magenta) predicted single-copy A average transcription rates and B translation rates across forward scatter. Error bars are SEM. C Heat shock induction comparison of SSA1P-H-GFP (red) and SSA1P-Y-GFP (magenta) single-copy strains. All errors are 99% CI. See also Figure S4.
If SSA1P-H-GFP is translated faster than SSA1P-Y-GFP, a prediction made from steady state observations, then we expect to observe differences in the dynamics of their expression during heat shock. Since SSA1P-H-GFP degrades 2-3 faster than SSA1P-Y-GFP, the null expectation (translation rates equal) is that during heat shock SSA1P-H-GFP expression should express 2-3 times less than SSA1P-Y-GFP at all points. In contrast, we observed that SSA1P-H-GFP exhibited an equal or slightly faster rise in expression during heat shock compared to SSA1P-Y-GFP, with peak SSA1P-H-GFP expression exceeding peak SSA1P-Y-GFP expression (Fig. 4C). Following the initial rise SSA1P-H-GFP signal degrades more rapidly, as expected by the more destabilized N-end tag. These temporal results support the steady-state prediction that apparent translation rates differ between SSA1P-H-GFP and SSA1P-Y-GFP.
Rise in mid-cycle transcription rate is driven extrinsically
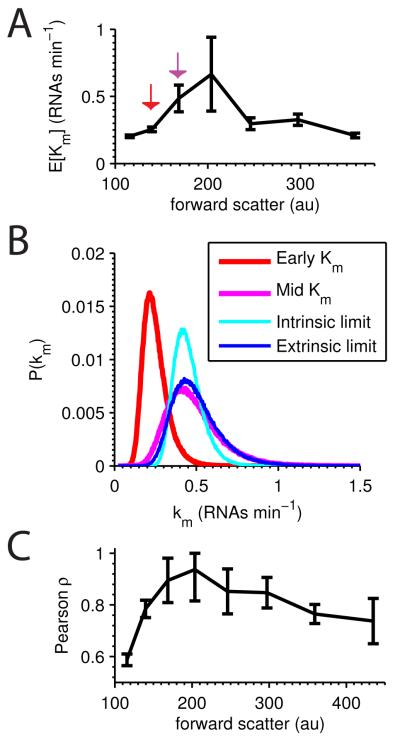
Although average transcription rates were consistent for all three constructs, SSA1P-GFP exhibits a different pattern across forward scatter bins. Consistent with previous observations (Zopf et al., 2013) SSA1P-GFP has a distinct mid-cycle peak in transcription rate (Fig. 5A). From bin 2 to 3 the average transcription rate more than doubles, consistent with the appearance of a second active gene during DNA replication. To what extent are the newly replicated genes coherently expressed (correlated)? If the two copies exhibit independent fluctuations, and Km is the model-fit distribution of transcription rates in bin 2, then the distribution in bin 3 should be (the intrinsic limit). In the extrinsic limit, expression of two identical genes would be perfectly correlated, and the distribution in bin 3 would be 2Km. Graphically we see in Fig. 5B that the extrinsic limit better approximates the experimentally measured distribution for bin 3, suggesting a transcriptionally-mediated extrinsic factor drives highly correlated fluctuations in the new gene copy.
Figure 5. SSA1P-GFP transcriptional rate distribution across the cell cycle.
A Average transcriptional rate (EKm) for SSA1P-GFP single-copy strain across forward scatter; error bars are SEM. B In red and magenta are the distributions of Km with means EKm corresponding to the red and magenta arrowed points in A. From the red to magenta distribution, the average transcription rate doubles. Given the red distribution, the expected intrinsic limit is plotted in cyan, and the extrinsic limit in blue, while the magenta distribution represents the experimentally derived behavior. C Pearson’s ρ across the forward scatter represents the correlation between identical SSA1P-GFP copies in diploid cells; error bars are SEM.
We cannot directly measure correlation between the old and the newly replicated gene copies, but the one- and two-copy experiments permit direct quantification of correlation between two identical genes in single cells across the cell cycle. Since extrinsic variance (Ev) is equal to the covariance, Pearson’s correlation ρ=Ev/σ2, where σ2 is expression variance. As seen in Fig. 5C, expression correlation rises to near unity midway through the cell cycle.
Extrinsic Km model explains super-Poissonian RNA distributions
Single-molecule RNA FISH experiments show that genes’ RNA count distributions exhibit both Poisson and super-Poisson statistics (Gandhi et al., 2011; Taniguchi et al., 2010; Zenklusen et al., 2008). The observation of super-Poisson distributions led investigators to hypothesize an ON and OFF state for genes, implying bursty gene expression. Crucially, this model represents an intrinsic-only process, predicting that RNA-level expression of identical genes in single cells would be perfectly independent.
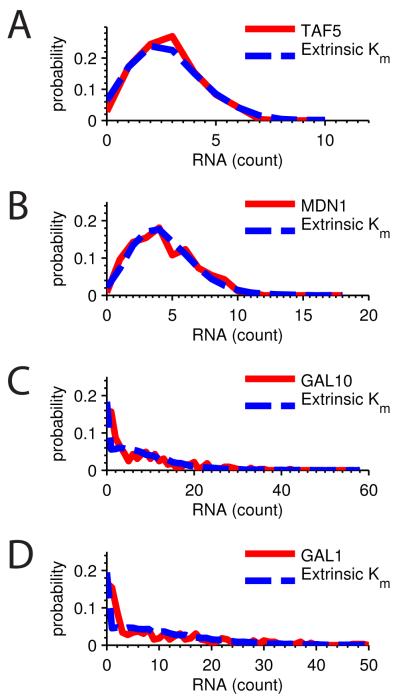
In contrast to this prediction a recent study showed correlation of expression at the RNA level among non-identical genes, indicating significant extrinsic stochasticity acting at the RNA level. Our model predicts that wider-than-expected RNA distributions derive from cell-to-cell variation in the transcription rate. Consistent with this prediction our model captures both Poisson and super-Poisson RNA distributions in this study (Fig. 6A-D). Super-Poisson expression patterns can therefore be explained as cell-to-cell variability in the rate of transcription without invoking discrete ON and OFF promoter states.
Figure 6. Extrinsic model consistent with RNA count distributions.
All figures are RNA count distributions from Gandhi et al. (Gandhi et al., 2011). Extrinsic Km model distributions (blue) were determined from the first and second moments and plotted against experimental data (red). Distributions are A TAF5 (μ=σ2), B MDN1 (μ=σ2), C GAL10 (μ=σ2), and D GAL1 (μ=σ2).
Discussion
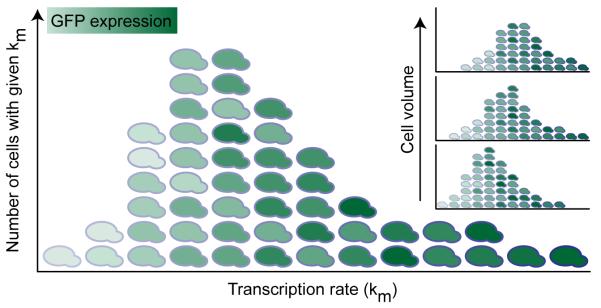
The prevailing ON-OFF model postulates that many genes have an active and a quiescent state, which leads to transcriptional bursting. Key support for the ON-OFF model originates from studies demonstrating transcriptional bursting by time-lapse microscopy (Chubb et al., 2006; Golding et al., 2005). We have proposed an alternative model in which cells vary in their propensity to transcribe. The resulting model, a hybrid intrinsic model with extrinsically varying transcription rate (depicted graphically in Fig. 7), recapitulates the full expression distribution shape without fitting, and does not necessitate incorporation of ON and OFF states. In contrast, the ON-OFF model fails to capture total expression stochasticity, primarily by underestimating the tail or skewness seen experimentally.
Figure 7. Graphical representation of the hybrid model.
Distribution shape represents the extrinsic Km distribution. Overall expression increases with increasing km (x-axis), but in addition, each column demonstrates expression heterogeneity representing the intrinsic stochasticity conditioned on km. (Inset) Each Km distribution is itself conditioned on cell volume via forward scatter, with increasing volume corresponding to Km distributions with rising means, and varying shapes.
Although transcriptional bursting likely occurs for some genes, a large fraction of stochastic gene expression arises from mechanisms acting coherently on genes distantly positioned within single cells. This conclusion is supported by studies in yeast demonstrating large extrinsic contributions to expression stochasticity (Raser and O’Shea, 2004; Stewart-Ornstein et al., 2012; Volfson et al., 2006). Intrinsic fluctuations may be more prominent in mammalian cells (Levesque and Raj, 2013; Raj et al., 2006), although recent advances in single-cell RNA-seq provide broad evidence of correlated fluctuations among genes in single mammalian cells (Klein et al., 2015; Marinov et al., 2014). Given that transcriptional bursting has been directly observed for some genes we suggest either that bursting contributes only a modest fraction of total gene stochasticity, or that bursting is highly coordinated between distantly positioned genes, driven, perhaps, by extreme extrinsic fluctuations in transcription factor concentrations. We have shown that for SSA1, cell-to-cell differences in the propensity to transcribe best explains the mechanism by which distant genes express in a correlated fashion. Further work will help elucidate whether such a mechanism is specific to heat shock genes, S. cerevisiae, or represents a general biological principle.
Future work will focus on two caveats to our approach. First, it was unclear how to select a specific distribution for the random variable Km. We confronted this issue by assuming that Km might vary as a single protein. Based on a variety of measured protein distributions we arrived at the Gumbel distribution, however, any highly skewed distribution might replace the Gumbel in our model. This is consistent with the fat-tailed distribution recently proposed by Rosenfeld et al. to represent ensemble transcription rates in vivo (Rosenfeld et al., 2015). The true distributional form of Km should reflect the fact that the transcription rate is a complex cis-regulatory function, one that is saturable and depends on the concentration of multiple trans-acting molecules. Second, once we eliminated extrinsically varying degradation rates, evidence from outside work pointed us toward an extrinsically varying transcription (Km) model. Nevertheless, an extrinsically varying Kp model is also consistent with our data (Supplement, Fig. S3B,C), leaving open the possibility that translation may also contribute to extrinsic stochasticity. We anticipate that hybrid models that incorporate both intrinsic and extrinsic sources of variability will be important tools for unraveling the mechanisms underlying stochastic gene expression in many systems.
Experimental Procedures
S. cerevisiae strains were generated in the BY4743 background. Flow cytometry was accomplished with a Cytomics FC500 MPL (Beckman Coulter, Brea, CA). A single cytometer profile was used for all experiments, and Flow-Check Fluorospheres (Beckman Coulter, 6605359) calibration before every experiment demonstrated <3% drift over the course of all measurements (3 months). Instrument noise was also approximated with Flow Check beads and found to be negligible in comparison to biological noise (Fig. S4).
Calibration of flow cytometry fluorescence in units of absolute protein per cell was obtained as follows. A general assumption that justifies the use of fluorescent reporters is that a fixed constant kct relates the fluorescence (Fi) of the ith cell to the actual number of fluorescent proteins Pi inside. Thus, kctFi = Pi, or using the mean, kct = ⟨Pi⟩/⟨Fi⟩. To determine kct we simultaneously performed flow cytometry on a concentrated culture of cells to measure ⟨Fi⟩, determined cell concentration using a Nexcelom Cellomter X2 cell counter, and lysed the cells. After removing the insoluble protein debris, which did not contain measurable amounts of GFP (Fig. 1E D1-D2), we measured the concentration of fluorescent molecules ⟨Pi⟩ in the lysate by fluorescence correlation spectroscopy (Crick et al., 2006; Magde et al., 1972). Combining these numbers and tracking volume changes we obtained estimates of GFP molecules per cell (see Supplement for details). For our instrument we find kct = 1826±403 proteins/a.u. based on four biological replicates evaluated on different days. Technical replicates split immediately before cellular lysis yielded little error, with one day resulting in kct=1416±67 proteins/a.u.
We validated our FCS calibration with Western blotting of the same samples (Fig. 1E-H) (Garcia and Phillips, 2011). To obtain absolute quantification by Western blot we spiked purified GFP at different concentrations into our autofluorescent control samples (Fig. 1E, S1-S4, G, A1-A6). Samples G1 and G2 represent technical replicates split at the point of cytometry, and thus capture technical error introduced from the lysis steps, sample handling, and quantification by Western or FCS. Close agreement between FCS measurements and corresponding Western blot quantifications (Fig. 1F, H) gave us confidence that FCS provides a reliable quantitation of GFP molecule count. Additional methods may be found in the Supplement.
Supplementary Material
Acknowledgments
The authors thank G. Rieckh and the Cohen lab for critical reviews of the manuscript, E. Elson and T. Pryse for sharing their FCS expertise and imaging platform, Saumil Gandhi for sharing the raw RNA-FISH data, and H. Garcia for helpful conversations about western blot quantification. HL was supported by NIH Grants NIHR01GM38542 and F30CA171595; MSS was supported by NIH GM07200, GM092910, and HG000045.
Footnotes
Author Contributions
Conceptualization, MSS and BAC; Formal Analysis, MSS; Investigation, MSS and KL; Validation, MSS and HL; Writing – Original Draft, MSS and BAC; Writing – Review & Editing, MSS, BAC, and KL; Funding Acquisition, BAC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bar-Even Arren, Paulsson Johan, Maheshri Narendra, Carmi Miri, O’Shea Erin, Pilpel Yitzhak, Barkai Naama. Noise in protein expression scales with natural protein abundance. Nature genetics. 2006;38(6):636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- Becskei Attila, Kaufmann Benjamin B, van Oudenaarden Alexander. Contributions of low molecule number and chromosomal positioning to stochastic gene expression. Nature genetics. 2005;37(9):937–44. doi: 10.1038/ng1616. [DOI] [PubMed] [Google Scholar]
- Blake William J, Balázsi Gábor, Kohanski Michael, Isaacs Farren J, Murphy Kevin F, Kuang Yina, Cantor Charles R, Walt David R, Collins James J. Phenotypic consequences of promoter-mediated transcriptional noise. Molecular cell. 2006;24(6):853–65. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Blake William J, KAErn Mads, Cantor Charles R, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422(6932):633–7. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Carey Lucas B, van Dijk David, Sloot Peter M a, Kaandorp Jaap a, Segal Eran. Promoter sequence determines the relationship between expression level and noise. PLoS biology. 2013;11(4):e1001528. doi: 10.1371/journal.pbio.1001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot Jeffrey R, Pedraza Juan M, Luitel Prashant, van Oudenaarden Alexander. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature. 2007;450(7173):1249–52. doi: 10.1038/nature06395. [DOI] [PubMed] [Google Scholar]
- Chubb Jonathan R, Trcek Tatjana, Shenoy Shailesh M, Singer Robert H. Transcriptional pulsing of a developmental gene. Current biology : CB. 2006;16(10):1018–25. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173(1 Spec No):33–8. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Crick Scott L, Jayaraman Murali, Frieden Carl, Wetzel Ronald, Pappu Rohit V. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(45):16764–9. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadiani Maya, van Dijk David, Segal Barak, Field Yair, Ben-Artzi Gil, Raveh-Sadka Tali, Levo Michal, Kaplow Irene, Weinberger Adina, Segal Eran. Two DNA-encoded strategies for increasing expression with opposing effects on promoter dynamics and transcriptional noise. Genome research. 2013;23(6):966–76. doi: 10.1101/gr.149096.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgart Vlad, Jia Tao, Fenley Andrew T, Kulkarni Rahul. Connecting protein and mRNA burst distributions for stochastic models of gene expression. Physical biology. 2011;8(4):046001. doi: 10.1088/1478-3975/8/4/046001. [DOI] [PubMed] [Google Scholar]
- Elowitz Michael B, Levine Arnold J, Siggia Eric D, Swain Peter S. Stochastic gene expression in a single cell. Science (New York, N.Y.) 2002;297(5584):1183–6. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Gandhi Saumil J, Zenklusen Daniel, Lionnet Timothée, Singer Robert H. Transcription of functionally related constitutive genes is not coordinated. Nature structural & molecular biology. 2011;18(1):27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Hernan G, Phillips Rob. Quantitative dissection of the simple repression input-output function. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12173–12178. doi: 10.1073/pnas.1015616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie Daniel T. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. Journal of Computational Physics. 1976;22(4):403–434. [Google Scholar]
- Golding Ido, Paulsson Johan, Zawilski Scott M, Cox Edward C. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123(6):1025–36. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Guantes Raul, Rastrojo Alberto, Neves Ricardo, Lima Ana, Aguado Begoña, Iborra Francisco J. Global variability in gene expression and alternative splicing is modulated by mitochondrial content. Genome Research. 2015;25(5):633–644. doi: 10.1101/gr.178426.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett Elizabeth a, Esch R Keith, Maleri Seth, Errede Beverly. A family of destabilized cyan fluorescent proteins as transcriptional reporters in S. cerevisiae. Yeast (Chichester, England) 2006;23(5):333–49. doi: 10.1002/yea.1358. [DOI] [PubMed] [Google Scholar]
- Hahn Steven, Young Elton T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189(3):705–36. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper Claire V, Finkenstädt Bärbel, Woodcock Dan J, Friedrichsen Sönke, Semprini Sabrina, Ashall Louise, Spiller David G, Mullins John J, Rand David a, Davis Julian R E, White Michael R H. Dynamic analysis of stochastic transcription cycles. PLoS biology. 2011;9(4):e1000607. doi: 10.1371/journal.pbio.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfinger Andreas, Paulsson Johan. Separating intrinsic from extrinsic fluctuations in dynamic biological systems. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(29):12167–72. doi: 10.1073/pnas.1018832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh Dann, Paulsson Johan. Non-genetic heterogeneity from stochastic partitioning at cell division. Nature genetics. 2011;43(2):95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka Ryo, Yamagishi-Shirasaki Mai, Funatsu Takashi. Kinetic study of de novo chromophore maturation of fluorescent proteins. Analytical biochemistry. 2011;414(2):173–8. doi: 10.1016/j.ab.2011.03.036. [DOI] [PubMed] [Google Scholar]
- Klein Allon M, Mazutis Linas, Akartuna Ilke, Tallapragada Naren, Veres Adrian, Li Victor, Peshkin Leonid, Weitz David A, Kirschner Marc W. Droplet Barcoding for Single-Cell Transcriptomics Applied to Embryonic Stem Cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Jinzhi. Stochasticity in single gene expression with both intrinsic noise and fluctuation in kinetic parameters. Journal of theoretical biology. 2009;256(4):485–92. doi: 10.1016/j.jtbi.2008.10.028. [DOI] [PubMed] [Google Scholar]
- Levesque Marshall J, Raj Arjun. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nature methods. 2013;10(3):246–8. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet Timothée, Singer Robert H. Transcription goes digital. EMBO reports. 2012;13(4):313–21. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde Douglas, Elson Elliot, Webb W. Thermodynamic Fluctuations in a Reacting System–Measurement by Fluorescence Correlation Spectroscopy. Physical Review Letters. 1972 Sep;29:705–708. [Google Scholar]
- Marinov Georgi K, Williams Brian A, McCue Ken, Schroth Gary P, Gertz Jason, Myers Richard M, Wold Barbara J. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome research. 2014;24(3):496–510. doi: 10.1101/gr.161034.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogno Ilaria, Vallania Francesco L M, Mitra Robi D, Cohen Barak A. TATA is a modular component of synthetic promoters. Genome research. 2010;20(10):1391–7. doi: 10.1101/gr.106732.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuert Gregor, Munsky Brian, Tan Rui Zhen, Teytelman Leonid, Khammash Mustafa, van Oudenaarden Alexander. Systematic identification of signal-activated stochastic gene regulation. Science (New York, N.Y.) 2013;339(6119):584–7. doi: 10.1126/science.1231456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman John R S, Ghaemmaghami Sina, Ihmels Jan, Breslow David K, Noble Matthew, DeRisi Joseph L, Weissman Jonathan S. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441(7095):840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Ozbudak Ertugrul M, Thattai Mukund, Kurtser Iren, Grossman Alan D, van Oudenaarden Alexander. Regulation of noise in the expression of a single gene. Nature genetics. 2002;31(1):69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- Padovan-Merhar Olivia, Nair Gautham P, Biaesch Andrew G, Mayer Andreas, Scarfone Steven, Foley Shawn W, Wu Angela R, Churchman L. Stirling, Singh Abhyudai, Raj Arjun. Single Mammalian Cells Compensate for Differences in Cellular Volume and DNA Copy Number through Independent Global Transcriptional Mechanisms. Molecular Cell. 2015;58(2):339–352. doi: 10.1016/j.molcel.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson Johan. Models of stochastic gene expression. Physics of Life Reviews. 2005;2(2):157–175. [Google Scholar]
- Raj Arjun, Peskin Charles S, Tranchina Daniel, Vargas Diana Y, Tyagi Sanjay. Stochastic mRNA synthesis in mammalian cells. PLoS biology. 2006;4(10):e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj Arjun, van Oudenaarden Alexander. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135(2):216–26. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser Jonathan M, O’Shea Erin K. Control of stochasticity in eukaryotic gene expression. Science (New York, N.Y.) 2004;304(5678):1811–4. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld Liat, Kepten Eldad, Yunger Sharon, Shav-Tal Yaron, Garini Yuval. Single-site transcription rates through fitting of ensemble-averaged data from fluorescence recovery after photobleaching: A fat-tailed distribution. Physical Review E. 2015;92(3):032715. doi: 10.1103/PhysRevE.92.032715. [DOI] [PubMed] [Google Scholar]
- Rosenfeld Nitzan, Young Jonathan W, Alon Uri, Swain Peter S, Elowitz Michael B. Gene regulation at the single-cell level. Science (New York, N.Y.) 2005;307(5717):1962–5. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Shahrezaei Vahid, Ollivier Julien F, Swain Peter S. Colored extrinsic fluctuations and stochastic gene expression. Molecular systems biology. 2008;4(196):196. doi: 10.1038/msb.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem Ophir, Carey Lucas, Zeevi Danny, Sharon Eilon, Keren Leeat, Weinberger Adina, Dahan Orna, Pilpel Yitzhak, Segal Eran. Measurements of the Impact of 3′ End Sequences on Gene Expression Reveal Wide Range and Sequence Dependent Effects. PLoS Computational Biology. 2013;9(3):e1002934. doi: 10.1371/journal.pcbi.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman Marc S, Cohen Barak a. A computational framework for analyzing stochasticity in gene expression. PLoS computational biology. 2014;10(5):e1003596. doi: 10.1371/journal.pcbi.1003596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater Michael R, Craig Elizabeth a, Slatert Michael R. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Molecular and cellular biology. 1987;7(5):1906–16. doi: 10.1128/mcb.7.5.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So Lok-Hang, Ghosh Anandamohan, Zong Chenghang, Sepúlveda Leonardo, Segev Ronen, Golding Ido. General properties of transcriptional time series in Escherichia coli. Nature genetics. 2011;43(6):554–60. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein Jacob, Weissman Jonathan S, El-Samad Hana. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Molecular cell. 2012;45(4):483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter David M, Molina Nacho, Gatfield David, Schneider Kim, Schibler Ueli, Naef Felix. Mammalian genes are transcribed with widely different bursting kinetics. Science (New York, N.Y.) 2011;332(6028):472–4. doi: 10.1126/science.1198817. [DOI] [PubMed] [Google Scholar]
- Taniguchi Yuichi, Choi Paul J, Li Gene-Wei, Chen Huiyi, Babu Mohan, Hearn Jeremy, Emili Andrew, Xie X Sunney. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science (New York, N.Y.) 2010;329(5991):533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky a. The N-end rule: functions, mysteries, uses. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(22):12142–9. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volfson D, Marciniak J, Blake WJ, Ostroff N, Tsimring LS, Hasty J. Origins of extrinsic variability in eukaryotic gene expression. Nature. 2006;439:861–4. doi: 10.1038/nature04281. [DOI] [PubMed] [Google Scholar]
- Wang Xiao, Errede Beverly, Elston Timothy C. Mathematical analysis and quantification of fluorescent proteins as transcriptional reporters. Biophysical journal. 2008;94(6):2017–26. doi: 10.1529/biophysj.107.122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young Michael R MR, Craig Elizabeth A EA. Saccharomyces cerevisiae HSP70 heat shock elements are functionally distinct. Molecular and cellular biology. 1993;13(9) doi: 10.1128/mcb.13.9.5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen Daniel, Larson Daniel R, Singer Robert H. Single-RNA counting reveals alternative modes of gene expression in yeast. Nature structural & molecular biology. 2008;15(12):1263–71. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zopf CJ, Quinn Katie, Zeidman Joshua, Maheshri Narendra. Cell-cycle dependence of transcription dominates noise in gene expression. PLoS computational biology. 2013;9(7):e1003161. doi: 10.1371/journal.pcbi.1003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.