Abstract
Fusarium head blight is a prevalent disease of bread wheat (Triticum aestivum L.), which leads to considerable losses in yield and quality. Quantitative resistance to the causative fungus Fusarium graminearum is poorly understood. We integrated transcriptomics and metabolomics data to dissect the molecular response to the fungus and its main virulence factor, the toxin deoxynivalenol in near-isogenic lines segregating for two resistance quantitative trait loci, Fhb1 and Qfhs.ifa-5A. The data sets portrait rearrangements in the primary metabolism and the translational machinery to counter the fungus and the effects of the toxin and highlight distinct changes in the metabolism of glutamate in lines carrying Qfhs.ifa-5A. These observations are possibly due to the activity of two amino acid permeases located in the quantitative trait locus confidence interval, which may contribute to increased pathogen endurance. Mapping to the highly resolved region of Fhb1 reduced the list of candidates to few genes that are specifically expressed in presence of the quantitative trait loci and in response to the pathogen, which include a receptor-like protein kinase, a protein kinase, and an E3 ubiquitin-protein ligase. On a genome-scale level, the individual subgenomes of hexaploid wheat contribute differentially to defense. In particular, the D subgenome exhibited a pronounced response to the pathogen and contributed significantly to the overall defense response.
Keywords: Fusarium head blight, resistance QTL, Fhb1, Qfhs.ifa-5A, deoxynivalenol
Bread wheat (Triticum aestivum, 2n = 6x = 42, AABBDD) is one of the most important food crops worldwide, providing approximately 20% of the daily human calorie consumption (Brenchley et al. 2012). Increasing nutritional demands by a growing world population and environmental stresses present major challenges for wheat research and breeders. One of the most prevalent diseases on wheat and other small grain cereals is Fusarium head blight (FHB). The disease is mainly caused by the hemibiotrophic fungus Fusarium graminearum, which thrives under humid and temperate conditions, leading to large economic losses (McMullen et al. 2008; Pirgozliev et al. 2003). The most severe effect of FHB is the contamination of grains with mycotoxins such as deoxynivalenol (DON), which remain in the food chain and constitute a threat to the health of animals and humans (Pestka 2010). DON is a potent inhibitor of protein biosynthesis and, although its presence is not required to establish the infection site, it is essential for the pathogen to breach the barrier from the initially infected spikelet and its spread into the surrounding tissue (Jansen et al. 2005).
The wheat defense response includes a plethora of well-described mechanisms, including the biosynthesis of phenolics, polyamines, and other secondary metabolites, cell wall fortification, as well as countermeasures to reduce oxidative stress and to inactivate DON (reviewed in Kazan et al. 2012; Walter et al. 2010). Little is known on how the adaptations in the primary metabolism contribute to resistance against F. graminearum. Schwachtje and Baldwin (2008) discussed roles for the primary metabolism that surpass its function in nutrient acquisition for the costly defense response. These may act in defense signaling, contribute to defense by themselves, or work toward shifting recourses between infected/noninfected tissues to increase tolerance. In particular, the production of secondary metabolites is strongly linked to the expression of genes providing carbon, nitrogen, and sulfur equivalents (Aharoni and Galili 2011). Therefore, changes in the primary and secondary metabolisms need to be equally considered when observing plant/pathogen interactions.
Although bread wheat is considered susceptible to FHB, a diverse collection of resistant germplasm has been identified. More than 100 quantitative trait loci (QTL) have been described to contribute in varying extents to resistance against FHB (Buerstmayr et al. 2009). Yet, none of the underlying molecular mechanisms has been resolved to date. Two major and reproducible QTL derive from the Chinese spring wheat cultivar Sumai-3: Fhb1, located on the short arm of chromosome 3B, confers high resistance against spreading of the disease (type II) (Anderson et al. 2001; Buerstmayr et al. 2002), whereas Qfhs.ifa-5A, on 5AS, mainly confers resistance against initial infection (type I) (Buerstmayr et al. 2003).
A small number of studies investigated the differential transcriptional response to the pathogen in lines differing in the presence of Qfhs.ifa-5A (Kugler et al. 2013; Schweiger et al. 2013). In contrast, Fhb1 has been investigated widely and was introduced successfully into US elite breeding material (Jin et al. 2013). Fhb1-containing lines exhibited an improved ability to transform DON into the nontoxic DON-3-glucoside (Lemmens et al. 2005). Still, several transcriptomic and metabolomic studies that compared lines segregating for Fhb1 did not lead to the identification of a causal gene responsive for this mechanism (Gunnaiah et al. 2012; Jia et al. 2009; Kugler et al. 2013; Schweiger et al. 2013; Walter et al. 2008; Warth et al. 2015; Xiao et al. 2013; Zhuang et al. 2013)
A comparison of results between all these studies is challenging because they show little overlap due to the different investigated germplasms, sampling/inoculation procedures, and statistical methods used. Moreover, transcriptomic studies, including our own (Kugler et al. 2013; Schweiger et al. 2013), were long impeded by incomplete and frequently changing reference gene sets and incomplete gene annotations for bread wheat. All these factors have made it difficult to gain a complete picture of the transcriptomic response to the pathogen and to make a comparison between different studies. Recently, a comprehensive wheat survey sequence gene set has become available by the International Wheat Genome Sequencing Consortium (IWGSC) (Mayer et al. 2014). This reference provides a nearly complete mapping resource for transcriptomic studies. It comprises about 99,000 high-confidence genes allocated to the corresponding subgenomes and chromosome arms in version 2.2 of the annotation. To a large extent, genes were also linearly ordered (Mayer et al. 2009).
We have used the corresponding newly available gene models to revisit the data from Kugler et al. (2013), which describe the transcriptional response to F. graminearum in four near-isogenic lines (NILs) segregating for Fhb1 and Qfhs.ifa-5A. In this study we combined a gene coexpression network with differential gene expression analysis and metabolomics measurements, which were obtained from a time-course series. We identified QTL and treatment specific network components, which aided in the reconstruction of alterations in the bread wheat primary metabolism in response to the pathogen and identified pathway components showing distinct changes for lines harboring Qfhs.ifa-5A. Likely candidate genes for either QTL emerged from the analysis of QTL-specific modules in our network. With the bread wheat genome sequence at hand, it is now possible to also address the subgenome-specific contributions in the pathogen response on a genome-wide scale, which highlights a prominent role of the D subgenome.
Materials and Methods
Plant experiments
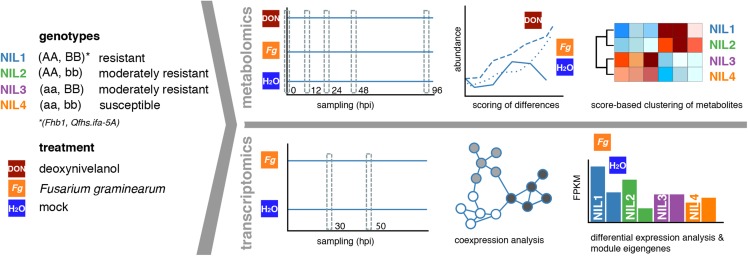
The procedures to capture the metabolomics data (Warth et al. 2015) and the transcriptomics data (Kugler et al. 2013) used similar plant material, growth conditions, and inoculation and sampling procedures with F. graminearum spore suspensions or DON (metabolomics dataset only), which also were described in the respective references. The metabolomics data set generated from F. graminearum−inoculated plants comprises novel data generated similarly as described in Warth et al. (2015) for the DON-treated samples. The employed BC5F2 NILs have the susceptible German spring wheat cultivar Remus as the recurrent parent. They harbor both (NIL1), either (NIL2: Fhb1, NIL3: Qfhs.ifa-5A), or neither of the two resistance QTL, which were introgressed from CM-82036, a Mexican spring wheat line. Metabolomics samples were taken at time points 0, 12, 24, 48, and 96 hours past infection/inoculation (hpi). Transcriptomics samples were taken at 30 and 50 hpi (Figure 1).
Figure 1.
Source material, experimental conditions, and analyses. Each near-isogenic line (blue: NIL1, green: NIL2, purple: NIL3, orange: NIL4) contains either the resistant or susceptible alleles of Fhb1 (AA, aa) and Qfhs.ifa-5A (BB, bb). Plants were either inoculated with F. graminearum spore suspensions, DON, or water as control. Samples were collected at different time points as indicated by gray, dashed boxes and subjected to RNA-sequencing (transcriptomics) or gas chromatography-mass spectrometry analysis (metabolomics). Transcriptomics data were characterized by the use of a coexpression network approach and differential expression analysis. Metabolomics data were characterized by the calculation of a Bayes factor score and clustering of these scores.
The metabolomics experiments have been conducted in a light- and temperature-controlled greenhouse in spring 2012 in full compliance with the Metabolomics Standards Initiative (Sumner et al. 2007). The transcriptomics experiment was conducted under comparable controlled light and climatic conditions in a growth chamber because of limited greenhouse space in Fall 2011. In the transcriptomics experiment 12 florets per head (the two basal florets of six central spikelets) were inoculated at anthesis with 10 μL of a F. graminearum conidia spore suspension (strain IFA65, concentration 50,000 conidia/mL) or mock by cautiously inserting a droplet onto the generative part of each floret. Similarly, for the metabolomics experiment 20 florets per head (from 10 central spikeles) were inoculated to yield sufficient tissue for analysis with either 10 µL of a F. graminearum conidia spore suspension (10,000 macroconidia/mL), DON (5 g/L in water), or mock. The treated heads were moistened and covered in plastic bags for 24 hr to provide humid conditions for the pathogen. Only palea and lemma of the inoculated florets were sampled, including the respective part of the rachis at the indicated time points and shock frozen in liquid nitrogen.
Metabolite analysis
After milling, extraction, and evaporation of the samples, an online two-step derivatization procedure was performed with the use of methoxyamine hydrochloride and N-methyl-N-trimethylsilyl trifluoroacetamide. Analytes were separated and detected on an Agilent 7890A gas chromatograph coupled to a 5975C inert XL MSD detector (Agilent, Waldbronn, Germany). Raw data were processed with the MetaboliteDetector software (Hiller et al. 2009). Gas chromatography with electron impact mass spectrometry spectra and retention indices of recognized features were compared with an in-house library, which was established with commercially available reference standards. Hence, most metabolites reported herein can be regarded as “level 1 – identified compounds” (Sumner et al. 2007). Chromatography and mass spectrometry features for the 59 identified metabolites as well as information on the identification level can be found in supporting information, Table S1. Data normalization was performed before statistical analysis with wheat aggregate QC samples and the internal standard nonadecanoic acid methylester. Missing value imputation and outlier testing was performed by tailored in-house R scripts (Warth et al. 2015). A multivariate evaluation of the DON-treated and mock samples (Warth et al. 2015) and the corresponding metadata is publically available via the MetaboLights metabolomics data repository (Salek et al. 2013) (accession number MTBLS112). The metadata for F. graminearum infected samples also is provided through the MetaboLights database (accession number MTBLS153). In Stegle et al. (2010) a Gaussian process (GP) based on two conditions test (GP2S) was introduced for detecting differential expression between two conditions. We applied GP2S tests on the metabolite time series data for a pair-wise comparison of treatment effects. The GP2S test compares two models: The first model assumes that the time series measurement in both conditions control and treatment are samples drawn from a shared distribution. The alternative model describes the time series in both conditions as sampled from two independent distributions. We marked scores larger than log(3)=1.099 as indications for substantial differences in treatments.
Gene expression analysis
Extracted RNA from sampled tissues was sequenced on an Illumina HiSeq2000 by an external sequencing provider (GATC, Konstanz, Germany). A detailed description of the generation of the RNA-seq data are given in Kugler et al. (2013). The corresponding data can be downloaded from Arrayexpress database under accession number E-MTAB-1729 (Kolesnikov et al. 2014).
A total of 1800 million single-end reads were mapped against the IWGSC bread wheat reference assembly. One replicate was removed after quality control (NIL2, M50, replicate 3). Reads were mapped against the reference by the use of TopHat (Trapnell et al. 2012) and Bowtie (Langmead and Salzberg 2012) with default parameters and keeping only unambiguously matched reads (Table S2). Mapped reads were transformed to FPKM values and normalized using Cufflinks (Trapnell et al. 2012). To test for differential expression [false-discovery rate (FDR)-adjusted P < 0.1, absolute log2 fold change >1], we applied the edgeR package in R (Robinson et al. 2010) on the raw counts as extracted with HTSeq (Anders et al. 2015) for the IWGSC high confidence genes. We tested mock-treated samples against F. graminearum-inoculated samples for all four NILs at both time points (Table S3) and for differences between NIL4 and the other NILs (Table S4). Deviations from the expected A, B, and D subgenome distribution (Mayer et al. 2014) were assessed with χ2 goodness-of-fit tests against 10,000 random multinomial distributions. A gene coexpression network comprising 18,948 genes was constructed using the weighted correlation network analysis (WGCNA) method (Langfelder and Horvath 2008), after a pseudocount transformation l2FPKM=(log2(FPKM+1)), a coefficient of variation filter (CV > 0.4), and keeping genes that surpassed a minimum expression level defined by the 5% percentile of all expressed genes The model was fit to a power law distribution (network type unsigned; power = 11), and the data clustered based on the Topological Overlap Matrix (Langfelder et al. 2008) using the cutreeDynamic method (minClusterSize = 50; deepSplit = 2; pamRespectsDendro = FALSE, merging close modules at 0.9; Figure S1). Intramodular hub genes were defined by the top 10% percentile of the intramodular connectivity. Eigengenes were calculated using the WGCNA package (Langfelder and Horvath 2008).
Enrichment of Gene Ontology (GO) terms was assessed with the Bioconductor package GOstats using conditional hypergeometric tests (Falcon and Gentleman 2007). Key enzymes were extracted by using their A. thaliana counterparts (Table S5) and sequence homology searches based on blastp (Altschul et al. 1990) with at least 60% sequence coverage. The 8605 gene triplets were based on a reciprocal best hit criterion in pairwise subgenome matches similar to the approaches in Pfeifer et al. (2014) and Mayer et al. (2014) and applying an identity threshold of 90%. For the gene triplet expression analysis gene triplets were concatenated into a triplet matrix as described in Mayer et al. (2014) and Pfeifer et al. (2014). The triplet coexpression network was inferred analogical to the overall network (CV threshold 0.7; lower expression 10% percentile; network type “signed hybrid”, beta = 8; minimal module size= 20; no merging of modules; Figure S2). The significance of differences in the subgenome-wise expression were quantified with a Kruskal-Wallis test and subsequent pair-wise Mann-Whitney U tests, as followed by an FDR adjustment of P values.
Data availability
Data availability is as follows: RNA-seq: EBI Arrayexpress (E-MTAB-1729); gas chromatography–mass spectrometry: MetaboLights (MTBLS112, MTBLS153).
Results
Integrated data analysis of the wheat response against F. graminearum and DON
The aim of this study was to explore QTL-associated differences in the response of four bread wheat NILs (NIL1−4) after inoculation with F. graminearum or DON by combining gene expression and metabolomics measurements (Figure 1). The investigated four BC5F2 NILs share the common susceptible genetic background of the recurrent parent Remus but are different for introgressed resistance QTL Fhb1 and Qfhs.ifa-5A from the donor line CM-82036, providing them with distinct resistance levels (Schweiger et al. 2013). To describe differences in transcript abundances, we used the recent IWGSC bread wheat genome sequence assembly (Mayer et al. 2014) as a mapping reference for RNA-seq measurements (Kugler et al. 2013). The RNA-seq data comprised samples from two time points, 30 and 50 hpi, with either F. graminearum spore suspensions or mock treatment. To complement expression profiles with functional output, we integrated gas chromatography-mass spectrometry−derived metabolite measurements from a dense time course of similar inoculation experiments on these NILs with the fungus and additionally with its major toxin DON (Warth et al. 2015).
To gain insights into system-wide expression patterns we first used the WGCNA approach (Langfelder and Horvath 2008) for grouping genes into sets of similar expression patterns. Genes within these groups show strongly correlated expression, which indicates common regulatory mechanisms or concerted actions. The distinct expression profiles of modules are represented by the module eigengene (Langfelder and Horvath 2007), which correspond to the first principal component of the module expression matrix and which can be regarded as a representative for the gene expression in a module. Putative functional characterizations of modules regarding F. graminearum-resistance and QTL activity were derived by integration of differential expression information, GO enrichments, and the corresponding module eigengene. Additionally, by quantifying intramodular connectivity, we also extracted hub genes for each module. On the basis of their expression profiles and their functional characteristics, we selected six network modules to be of special interest for subsequent investigations (Figure 2). Processed transcriptomic data and analysis results described in this study are available at the project’s RNASeqExpressionBrowser (Nussbaumer et al. 2014a) Web site (http://pgsb.helmholtz-muenchen.de/cgi-bin/db2/BOKUnils/index.cgi). The metabolomic data set for DON-treated NILs (Warth et al. 2015) and F. graminearum-inoculated NILs comprised 59 metabolites mainly derived from the primary metabolism sampled at 0, 12, 24, 48, and 96 hpi (Table S6). The differential abundance of metabolites among DON, F. graminearum, and mock treatment was quantified by the use of a Gaussian processes-based score, which was calculated for each metabolite reflecting differences between two conditions in metabolite abundance over time (Stegle et al. 2010). The scoring compares two alternative models in which the data from the two conditions is either explained by one single (shared) process or by two independent processes (one for each condition). The two alternatives (shared or independent model) can then be compared, with larger scores indicating more pronounced differences between the conditions (Table S7).
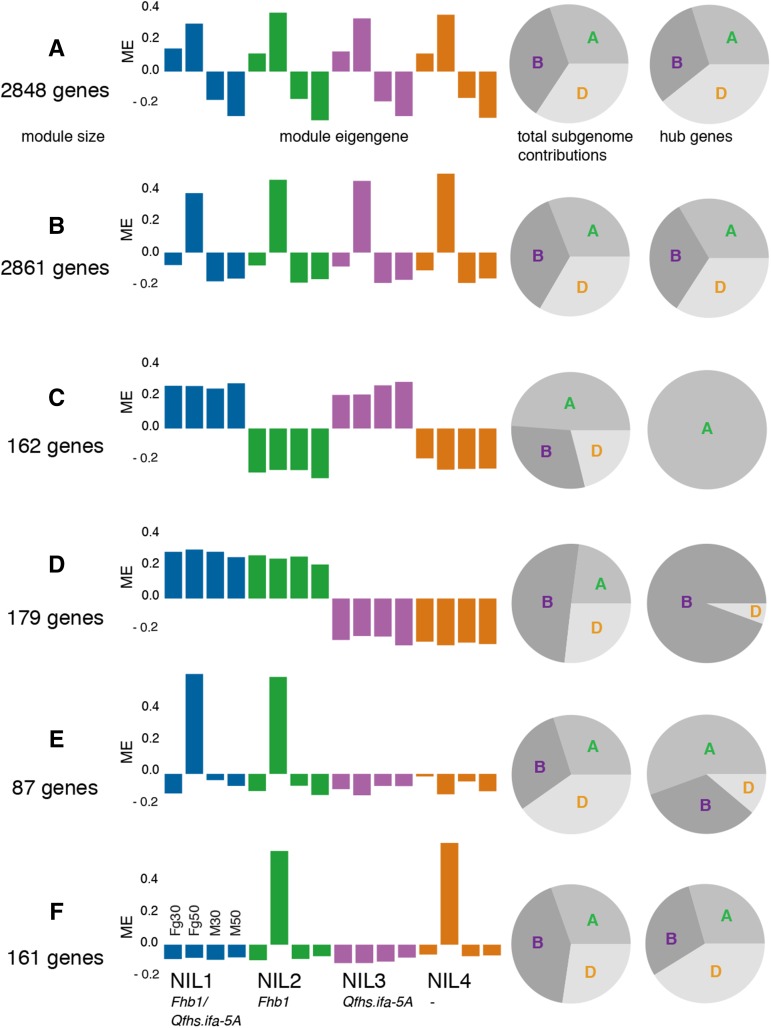
Figure 2.
Coexpression network modules. RNA-seq data were clustered into modules by inferring a weighted coexpression network. (A−F) Selected modules characterized by a general response to the fungus or are specific for QTL. The module eigengene panels (left) summarize the module-wise expression (Fg: treatment with F. graminearum, M: mock treatment, 30: 30 hpi, 50: 50 hpi; blue: NIL1, green: NIL2, purple: NIL3, orange: NIL4). Pie charts give the ratios of genes contributed by the individual subgenomes for the entire module (left) and for intramodular hub genes (right).
Increased turnover rates in a restructured primary metabolism fuel a broad defense response
We first investigated the coexpression network for gene functions in response to the pathogen by looking into functional enrichments and characteristic expression patterns. The two largest modules of the coexpression network, module A (2848 genes) and module B (2861 genes), grouped genes with strong general responses to the pathogen. Both were highly enriched for F. graminearum-responsive genes across all NILs regardless of individual resistance levels (one-sided Fisher’s test; FDR-adjusted P < 0.05; Figure 2, A and B and Figure S3). Genes in module A were differentially expressed in F. graminearum-inoculated samples at 30 hpi and 50 hpi (Figure 2A), whereas genes in module B were more specific for 50 hpi (Figure 2B). Although the subgenome distribution for all genes in these two modules showed no obvious bias, the distribution of the hub genes showed a slight overrepresentation of genes from the D subgenome.
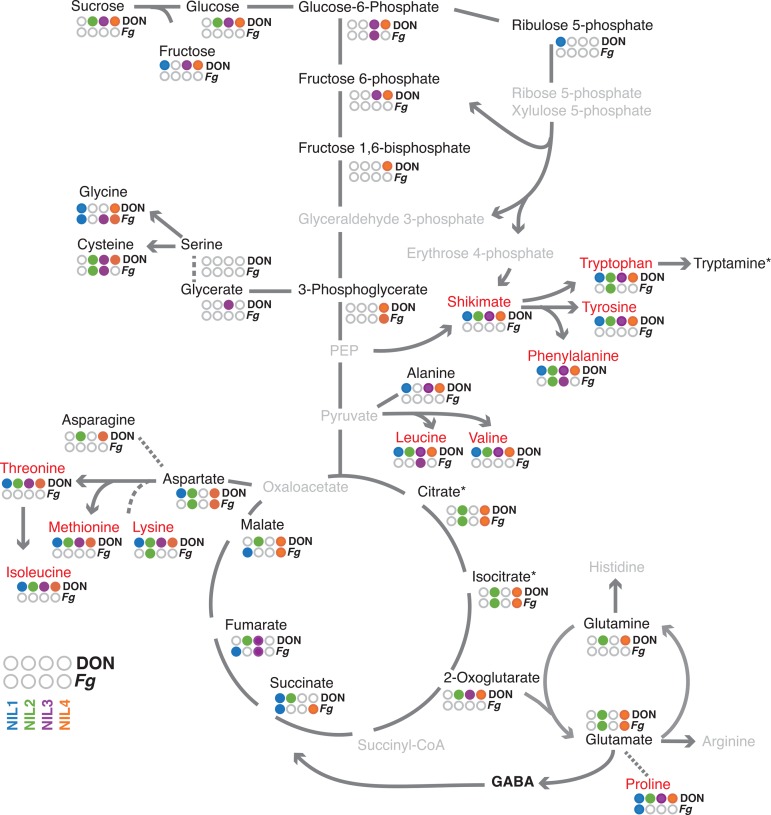
Overrepresented GO terms in module A reflected a broad response to the pathogen. These comprise functions in signaling, the defense against oxidative stress, the biosynthesis of tryptophan, and defense-associated secondary metabolites such as phenylpropanoids. Moreover this group included chitinases, proteinase inhibitors, and efflux pumps (Table S8). Several GO terms corresponded to the primary metabolism, which include a strongly up-regulated sucrose-phosphatase (Traes_1DS_9AE5A76AC in GO:0009312) and genes involved in the biosynthesis of thiamin (GO:0009229). Sucrose phosphatases mediate the last step in the biosynthesis of sucrose, the plants main transport form of carbohydrates. We also found elevated sucrose levels in our metabolomics experiment although only for DON-treated samples (Figure S4 and Figure S5). Abundant sucrose is broken down to fuel the tricarboxylic acid cycle (TCA) predominantly via glycolysis. The increased biosynthesis of thiamin relates to key enzymes in glycolysis and the TCA, pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase, which require thiamine diphosphate as cofactor.
To further expand on these observations we evaluated changes in transcript and metabolite abundances associated with F. graminearum/DON treatments in the respiratory chain and in the metabolism of glutamate. We extracted the expression profiles of all genes encoding for respective protein functions and generated eigengenes for each group of genes as the representative expression value. The expression of genes in glycolysis and the TCA cycle was strongly associated with F. graminearum−inoculated samples (Figure 3 and Table S9). In addition, the expression of genes encoding key enzymes in the pentose-phosphate pathway, glucose-6-phosphate dehydrogenase, and gluconate-6-phosphate dehydrogenase were strongly linked to pathogen treatment. The pentose-phosphate pathway provides an alternative route for the breakdown of hexoses into glycerinaldehyde-3-phosphate. It also generates nicotinamide adenine dinucleotide phosphate and erythrose-4-phosphate required in the shikimate pathway and ultimately for the production of phenylpropanoids. Despite increased abundances in transcript levels the corresponding metabolite levels in the glycolysis and pentose-phosphate pathway remain mostly unchanged (Figure 3, Figure S4, and Figure S5).
Figure 3.
Changes in the primary metabolism in response to F. graminearum and DON. Metabolites quantified by gas chromatography-mass spectrometry are set in black, and nonmeasured and nondetected metabolites are set in gray. Treatment-responsive metabolites (DON and F. graminearum (Fg); Table S2) in the individual lines are indicated by color (blue: NIL1, green: NIL2, purple: NIL3, orange: NIL4). Levels of metabolites set in red were strongly changed in response to DON or F. graminearum (average score greater than 10). Genes with significantly changed transcript abundances are indicated by arrows/lines set in gray, whereas dashed lines indicate no significantly changed transcript in any line. *Citrate and *isocitrate could not be distinguished due to chromatographic coelution. *Tryptamine levels were at or below the methods detection limit in most samples and thus could not be safely quantified.
Differences for Qfhs.ifa-5A in the TCA and the metabolism of glutamate
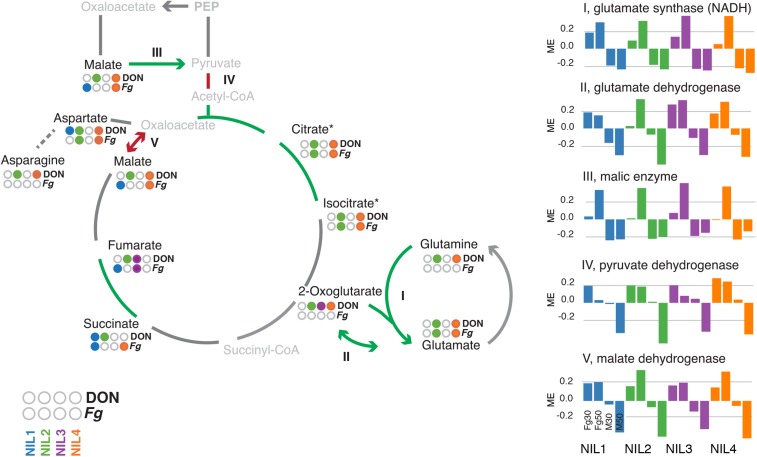
The metabolism of glutamate as a means to procure nitrogen for the biosynthesis of amines including amino acids depends on the availability of the TCA intermediate 2-oxoglutarate, which serves as a link to the TCA cycle. Genes of the TCA cycle and the metabolism of glutamate were in general responsive to the pathogen, yet distinct differences existed for lines harboring Qfhs.ifa-5A−related differences (Figure 4): Ammonia assimilation is mediated by glutamine synthetases (GS) and glutamate synthases (GOGAT), which generate glutamate from 2-oxoglutarate via glutamine. In plants the cytosolic (GS1) and the chloroplastic (GS2) isoenzymes for GS are regulated differentially with respect to tissue and external stimuli, assuring timely acquisition of ammonium from different sources (Miflin and Habash 2002). Three cytosolic GS1 genes showed increased transcript abundances in response to the pathogen and also exhibited by far the greatest expression rates as compared with 11 remaining wheat GS genes (Figure S6). For the GOGAT, we found a similar response for five NADH-dependent isozymes but not for cytosolic ferredoxin-dependent GOGAT (I in Figure 4). The expression of these genes was more strongly associated to the earlier infection time point 30 hpi in lines containing Qfhs.ifa-5A. A closer inspection of interrelated genes showed greater transcript abundances for many of them at 30 hpi for Qfhs.ifa-5A: This included most of the genes encoding TCA cycle steps as well as glutamate dehydrogenases and malic enzymes (II and III in Figure 4 and Table S9). Cytosolic malic enzymes provide additional pyruvate to the TCA cycle. The required malate originates from oxaloacetate, which most likely stems from the also strongly F. graminearum−responsive phosphoenolpyruvate carboxylase (Table S6 and Table S9). Yet, the largest QTL-effects were observed for pyruvate dehydrogenases and malate dehydrogenases (IV and V in Figure 4), which did not exhibit earlier expression for the QTL but showed strongly reduced expression levels at 50 hpi in lines harboring Qfhs.ifa-5A. Abundance pattern of metabolite intermediates for these pathways reflected the observed differences in expression levels: Glutamate and aspartate levels were changed after DON and F. graminearum treatment only for lines without the QTL (NIL2 and NIL4), whereas glutamine and asparagine levels in these lines were only changed in response to DON (Figure 3 and Figure 4). Within the TCA cycle malate levels (in response to DON) and citrate/isocitrate levels were similarly affected (Figure 4).
Figure 4.
QTL-associated differential transcript and metabolite abundances in the glutamate metabolism. Metabolites quantified by GC-MS are set in black, and nonmeasured and nondetected metabolites are set in gray. Treatment responsive (DON and F. graminearum (Fg); l Table S2) metabolites in the individual lines are indicated by color (blue: NIL1, green: NIL2, purple: NIL3, orange: NIL4). Green arrows/lines highlight genes with increased expression at 30 hpi for Qfhs.ifa-5A, red lines represents decreased expression at 50 hpi for Qfhs.ifa-5A. These expression differences are visualized for the isogene families by eigengene values (I-V). The individual NILs are distinguished by color. The four bars per line represent F. graminearum−inoculated samples at 30 and 50 hpi and mock-treated samples at 30 and 50 hpi from left to right.
Protein ubiquitination, elevated levels of tRNA ligases and amino acids characterize the specific response to the ribotoxic effect of DON
Module B was specific for genes expressed 50 hpi with the pathogen (Figure 2B). Independent studies demonstrated that at this time point the fungus has switched from the initial biotrophic growth to the formation of infection hyphae (Pritsch et al. 2000) and started to produce greater levels of DON (Audenaert et al. 2013). DON inhibits protein biosynthesis by interaction with the ribosomal 60S subunit and induces apoptosis via a mitogen-activated protein kinase-activated ribotoxic stress response (Pestka 2010). Accordingly, GO terms in module B were significantly enriched for terms relating to “oxidation reduction” (GO:0006979) and “translation” (GO:0006412) (l Table S8). The latter predominantly included genes encoding ubiquitin-60S ribosomal protein L40 fusion proteins. These fusion proteins act in ribosomal assembly and free ubiquitin acts in targeting nonfunctional proteins and unfinished peptide chains to the proteasome (Finley et al. 1989). Both mechanisms may be an active response to the effects of DON. Highly enriched genes within this module encoded for translation initiation factors (GO:0006413) and tRNA ligases (i.e., GO:0043039, GO:0034660), which mediate the transfer of amino acids to the expanding peptide chain in translation. All tRNA ligases showed strong associations to F. graminearum-inoculated samples with the exception of glycine tRNA ligases where none of the encoding genes were greater expressed in response to the pathogen (Table S6 and Table S9). We recorded high scores for most proteinogenic amino acids in the DON/mock comparison indicating higher abundances in the DON-inoculated samples, which has been a major finding in our previous analysis (Warth et al. 2015). Several proteinogenic amino acids were also found changed in F. graminearum-inoculated samples albeit less pronounced (Figure 3, Figure S4, and Figure S5). These elevated levels derive from increased biosynthesis as reflected by the increased abundance of transcripts corresponding to most key amino acid biosynthesis genes (Figure 3, Table S6, and Table S9) except for asparagine synthase (asparagine), pyrroline-5- carboxylate reductase (proline), 3-phosphoserine phosphatase (serine) and dihydrodipicolinate synthase (lysine).
QTL-specific modules and candidate genes emerging from the coexpression network
We identified several modules specific for either QTL: Modules C (162 genes, Qfhs.ifa-5A) and D (179 genes, Fhb1) showed significant expression differences between lines differing in the respective QTL (Figure 2, C and D). A chromoWIZ analysis showed that both modules were enriched for genes located on chromosomal regions harboring the respective QTL (Figure S7; Nussbaumer et al. 2014b). Hub genes were almost entirely located on the A and the B subgenomes harboring the respective QTL (Figure 2, C and D).
Hub gene expression profiles in module D characterized constitutively expressed genes for Fhb1-containing lines regardless of treatment or time point. Among the greatest expressed genes mapping to chromosome 3B are several protein kinases (Traes_3B_9985A569B, Traes_3B_ADCF93AE0, Traes_3B_E81A8FACB), a leucine-rich receptor protein (Traes_3B_CE31EE51C) and beta-fructofuranosidase (Traes_3B_61E72DF24), and a gene encoding an unknown protein (Traes_3B_908129DB2). Additionally, Traes_3DS_2BD8DC857, encoding an F-box protein, did not map to chromosome 3B of the FHB susceptible reference cultivar Chinese Spring (Figure S8).
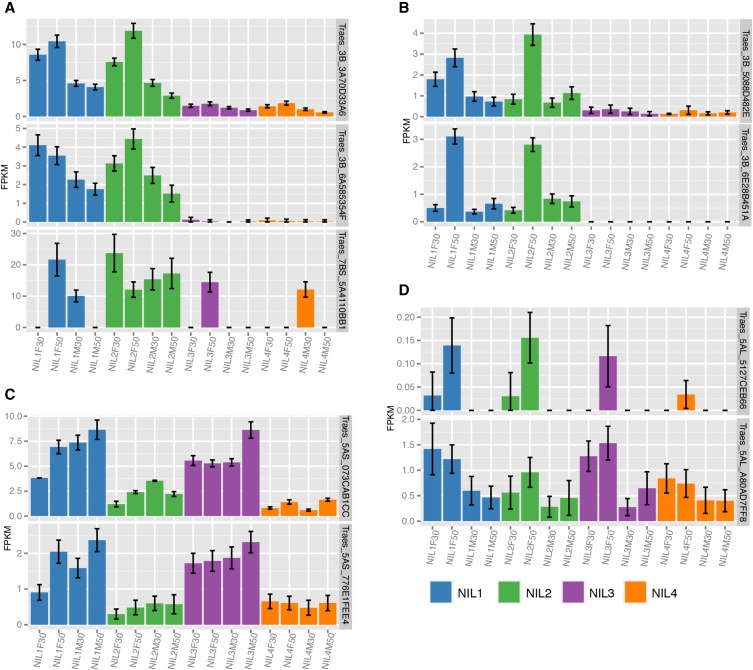
Several genes in module D also showed differential expression in response to F. graminearum: Traes_3B_3A70D33A6, a receptor-like protein kinase, and Traes_3B_6A585354F, a protein kinase, were significantly changed in response to the pathogen at 50 hpi and thus should also be considered likely candidates. Traes_7BS_5A4110BB1, a highly expressed and Fhb1-specific MATE-efflux pump, maps to chromosome 7B and could be a likely downstream target of Fhb1-activity (Figure 5A).
Figure 5.
Gene expression profiles of QTL candidates. (A) Fhb1-associated and F. graminearum−responsive genes in module D; (B) Fhb1-associated and F. graminearum-responsive genes mapped to chromosome 3B in module E; (C) Qfhs.ifa-5A−associated constitutively expressed amino acid permeases in module C and (D) Qfhs.ifa-5A−associated, F. graminearum−responsive genes. Means of FPKM values are given for each tested experimental condition (NIL1-4, F ... F. graminearum−inoculated, M ... mock-treated, 30/50 ... 30/50 hai).
A second Fhb1-related module E (87 genes) included genes greater expressed in Fhb1 lines at 50 hpi after inoculation with the pathogen (Figure 2E). Most genes within module E exhibited low transcript abundances, and we found no enrichment for 3B-mapped genes. Potentially, this module includes downstream targets of the QTL activity. Two genes emerge from this list as they were more abundant in response to the pathogen and mapped onto 3BS: Traes_3B_5088D482E a SINA-like 11 E3 ubiquitin-protein ligase, and Traes_3B_6E28B451A (unknown protein, Figure 5B). Of these, Traes_3B_5088D482E is the only gene in our analysis, differentially expressed also for 30 hpi in Fhb1 containing lines. GO enrichments for either module D or module E did not allow deducing biological functions of Fhb1-related downstream targets.
Module C includes genes with significant expression differences between lines differing in Qfhs.ifa-5A (NIL1 and NIL3 compared with NIL4). In contrast to genes in module D, none of the 162 genes within module C were also changed for F. graminearum treatment. The set of 5A-mapped genes therein shows a constitutive expression pattern for Qfhs.ifa-5A and contains 50 genes (Table S10). Among these we identified two amino acid permeases (AAP, isoforms 8, Traes_5AS_073CAB1CC and 6, Traes_5AS_776E1FEE4, Figure 5C). However, Traes_5AS_776E1FEE4 showed overall low expression levels.
Our network contained no module for genes with pathogen-specific expression changes for Qfhs.ifa-5A similar to module E for Fhb1, which matches previous observations suggesting a constitutive mode of action for the QTL (Kugler et al. 2013; Schweiger et al. 2013). Potentially, QTL-associated and pathogen-responsive genes also could be too small in numbers to form a module by themselves. Consequently, such genes could have been included in one of the pathogen-responsive modules. Two genes were significantly changed for Qfhs.ifa-5A and F. graminearum-treatment: Traes_5AL_5127CEB66 module B, flavin-containing monooxygenase) and Traes_5AL_A80AD7FF8 (module A, calmodulin-binding protein, Figure 5D). Alternatively, a susceptibility factor could be encoded in non-Qfhs.ifa-5A lines. Such genes and the downstream targets of such a factor might be included in module F (161 genes), specific for F. graminearum-inoculated NIL2 and NIL4 samples at 50 hpi (Figure 2F).
A pronounced role of the D subgenome in the response to F. graminearum
Two hybridization steps have resulted in three homoealleles for many of the functional genes in allohexaploid bread wheat (Marcussen et al. 2014). The consequent functional plasticity enabled reprogramming the individual subgenomes contributions to best meet environmental challenges including the response to pathogens (Feldman and Levy 2012). We hypothesized that redundant functions within homeoalleles are regulated differentially also in response to pathogen attack. Copies from single subgenomes may contribute in an additive manner, may have diverging expression patterns, have either been shut down, giving rise to expression dominance from one or two subgenomes, or alternatively been subjected to subfunctionalization. We observed an unbalanced genome-wide distribution of F. graminearum−responsive greater expressed genes. Significantly more genes than expected were differentially expressed in the D subgenome and fewer in the A subgenome (Figure S9).
After these observations, we hypothesized redundant functions within homeoalleles are regulated differentially. To inspect differential contributions to a putative similar functionality by subgenomes, we employed a set of 8605 homeologous gene triplets (25,815 genes). Triplet genes are genes with a mutual best match to genes in the other two subgenomes. They have been used previously for characterizing gene expression bias in different wheat organs and the developing endosperm (Mayer et al. 2014; Pfeifer et al. 2014). In our data 1384 (16%) triplets had at least one member residing in the coexpression network. Depending on the genotype, up to 15% of all triplets included at least one pathogen-responsive differentially expressed gene at 50 hpi. Only for 25% of these triplets all three copies were differentially expressed (Figure S10A), suggesting a tight regulation of the resource-intensive defense response. All subgenomes contributed equally to the number of differentially expressed genes in triplets with only one pathogen-responsive member at 30 hpi (Figure S10B). At 50 hpi, we found a significant deviation from the expected distribution for lines lacking Qfhs.ifa-5A (NIL 2 and NIL 4). Here the contribution of subgenome A dominated over contributions from subgenomes B and D.
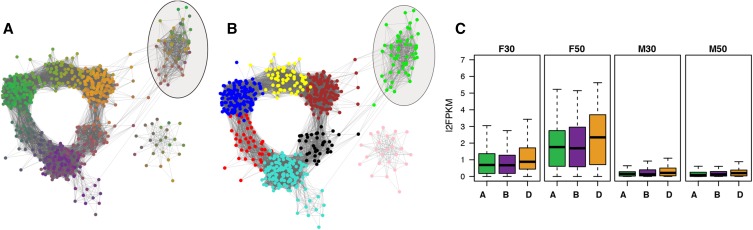
To investigate some of the dynamics in the observed gene expression bias, we inferred a gene coexpression network for triplet genes. Expression bias between the three subgenomes was then captured by a weighted average and nodes were colored according to this average (Figure 6A). The network contained eight modules with distinct expression patterns (Figure 6B and Figure S11). Most of the modules were defined by the expression in one dominant subgenome or by the pair-wise domination of the AB, BD, or respectively the AD subgenomes. One module of the triplet network included triplets with strong transcriptional responses to F. graminearum at 30 hpi and 50 hpi (highlighted in Figure 5, A and B). For genes within this module, responses were more pronounced for triplet members in the D subgenome, whereas the A and B subgenome contributed about equally in expression strength (Figure 6C). With respect to the D subgenome dominance, no differences between the four NILs were observed within this module (Figure S12). To investigate whether the more-pronounced reaction of the D genome is also reflected by the greater expression of pathogenesis-related genes, we observed expression differences for genes encoding NB-ARC domains (IPR002182). Although the number of gene family members are dominated by the A and the B subgenome, genes from the D subgenome are significantly greater expressed than homeoalleles from the A subgenome and in many cases also than those from the B subgenome (Figure S13 and Table S11). Similar observations were made for the NBS-LRR genes, although the differences were not as pronounced (Figure S14 and Table S11). Overall, our observations indicate a pronounced role of the D subgenome in response to F. graminearum.
Figure 6.
Coexpression analysis of homeologous triplet genes. A set of conserved triplet genes, with one copy per subgenome (A, B, and D) was used to investigate genome-specific expression behavior and dosage effects in a triplet-based coexpression network. (A) Coloring the network nodes by expression contributions from individual genomes highlighted regions where the combined triplet expression is dominated by a single or two genomes. (B) The triplet network was split into triplet network modules with specific expression patterns for genome expression bias (Figure S11). (C) The boxplots show subgenome-wise expression strength of triplet members in a F. graminearum-responsive triplet module (highlighted in A and B) under the given conditions for NIL1 (Fg: treatment with F. graminearum; M: mock treatment; 30: 30hpi; 50: 50 hpi). Within this module, expression in response to the fungus was dominated by the D subgenome, which was also observed for NIL2, NIL3, and NIL4 (Figure S12).
Discussion
Combined analysis of metabolomics and transcriptomics data
The recent release of the bread wheat reference genome sequence and annotation includes an almost-complete wheat gene set sorted into chromosome-arms (Mayer et al. 2014). Both features, the completeness of the resource, and the possibility to assign these genes to genomic regions surpasses by far any previous reference gene sets as a mapping reference for RNA-seq studies. This significantly improved mapping reference, combined with metabolomics data after inoculation with DON (Warth et al. 2015) and novel metabolomics data after inoculation with F. graminearum, was the main motivation for this study and for revisiting existing data. All of the included experiments used the same F. graminearum isolate and bread wheat near-isogenic material (differing in Fhb1 and Qfhs.ifa-5A) and are based on similar protocols for infection and tissue harvesting. Differences existed in the applied amount of spores between the transcriptomics experiment (500 conidia spores/floret) and metabolomics experiment (100 conidia spores/floret). Both concentrations suffice to successfully establish infection. In many biological reactions a stress trigger level must be reached to initiate a process. In complex processes it is often advantageous that all consecutive steps of a response occur automatically and are more or less “programmed.” Applying sufficient conidia to initiate the plant response probably will trigger the whole process. Therefore we do not expect significant differences in the recorded metabolomics and transcriptomics data sets (an example for phenylalanine and is given in Figure S15). Comparable transcriptomics and metabolomics studies have used a wide range of concentrations from 200 to 1000 conidia/floret to elicit a defense response to FHB (Jia et al. 2009; Gottwald et al. 2012; Foroud et al. 2012; Zhuang et al. 2013; Schweiger et al. 2013; Steiner et al. 2009; Gunnaiah et al. 2012; Diethelm et al. 2012). However, we cannot fully exclude the possibility that the observed metabolite changes, i.e., for amino acids may have been recorded earlier at greater inoculum concentrations. The Bayes score reporting differences in metabolite abundances between treatments considers all time points by comparing differences between models of individual treatments. Possibly, earlier changes resulting from greater concentrations would have influenced the time course models resulting in higher scores indicating even larger differences to the control mock models. However these changes would not alter the reported results here, as difference in score may also reflects the delay in/of metabolite changes but certainly highlights the presence of underlying metabolism.
In a previous transcriptomic study (Schweiger et al. 2013) we used very early and late time points for expression analysis (8, 24, and 72 hpi). Although the early time points provided only few differentially expressed genes, at 72 hpi a large and general response was detected for the susceptible lines. Apparently, resistance relevant reactions likely happen before 72 hpi. In the present study we chose two earlier time points, 30 and 50 hpi, to better capture the resistant reaction with two time points that encompass the onset of the production of larger amounts of DON (Pritsch et al. 2000, Audenaert et al. 2013). Fhb1 is closely linked to resistance against DON and given the investigated NILs segregating for Fhb1 these time points are appropriate to investigate related changes. The present metabolomics data describe five time points covering a time span of 96 hr and thus embraces both critical time points captured by RNAseq. Although the individual time points (0, 12, 24, 48, 96 hpi) do not overlap perfectly with 30 and 50 hpi measured in the transcriptomics experiment, they allow modeling a dynamic behavior which can be brought into context with the measured gene expression data. We approached this by avoiding direct comparisons between data sets at specific time points, but compare single time point observations for the RNAseq data with the time course-derived Bayesian score. As such the scores are time-point independent and provide a very reliable means to report changes in relation to treatment.
We analyzed the gathered data on different tiers: I. Given that our metabolomics analysis focused on metabolites derived from the well-annotated primary metabolism, we were able to integrate genotype-specific changes in metabolite abundances with differential transcript abundances of genes involved in the respiratory chain, glutamate metabolism, and amino acid biosynthesis. II. Homeoalleles of hexaploid bread wheat can be identified in the chromosome-sorted IWGSC gene set. We made use of these to investigate differences in the subgenome-wise contributions to defense response. III. Compared with our previous analysis (Kugler et al. 2013), the gene coexpression network here includes significantly more genes, as a result of the IWGSC bread wheat reference genome sequence, and allows allocating genes to their corresponding genomes. Therefore, we can provide a more complete and detailed picture on the genome-wide pathogen response and the corresponding dynamics.
In line with our previous observations in Kugler et al. (2013) two network modules, representing an early and a late response to the pathogen, were found in the coexpression network. Given the improved gene annotation, we could now further refine these data and provide a more comprehensive functional description of the corresponding genes and the respective pathways, while also taking subgenome contributions into account. For instance, using the current bread wheat annotation, we identified a pathogen-responsive network module (B) enriched for WRKY transcription factors (one-sided Fisher’s test based on the human readable description line; FDR-adjusted P < 0.001). In our previous study, such enrichment was also observed for such a generally pathogen responsive network module. Moreover, expanding in comparison with the previous approach, we were able to detect and characterize QTL-specific modules, which were investigated for F. graminearum-responsive genes mapping to chromosome-arms harboring the respective QTL.
Increases in respiration and amino acid biosynthesis as a response to F. graminearum and DON
The plant defense against pathogens requires the de novo synthesis of a plethora of secondary metabolites and defense-related proteins as well as the fortification of cell walls and antagonizing the effects of oxidative stress (Walter et al. 2010). As such, it is energy intensive and requires elevated rates in respiration (Bolton 2009). Mounting a successful defense response may rely on the efficiency of these mechanisms to supply the required substrates, but alterations in the primary metabolism have also been suggested to contribute to defense by themselves (Schwachtje and Baldwin 2008). For instance, glucose and hexokinase activity induce PR genes in Arabidopsis thaliana (Xiao et al. 2000), whereas silencing of the hexokinase 1 in Nicotiana benthamiana led to increased levels of H2O2 and programmed cell death−associated transcripts (Kim et al. 2006). Also, much evidence has been gathered for similar mechanisms involving the metabolism of several amino acids as well as lipids and the photorespiratory chain (Rojas et al. 2014). Our analysis aimed to reconstruct such activities in the bread wheat respiratory chain, in glutamate metabolism and in the biosynthesis of amino acids.
We found glycolysis, the pentosephosphate pathway, and the TCA cycle strongly induced in response to the pathogen, which might be fueled by the invertase-mediated cleavage of sucrose into glucose and fructose. The greater turnover rates in these pathways were visible in increased transcript abundances despite the—in many cases—unchanged pathway intermediate metabolites. In this respect, the response to F. graminearum is similar to the general model of the plant primary metabolism under pathogen attack (Bolton 2009). Upon carbon starvation, plants may procure additional carbon from abundant amino acids for respiration (Araújo et al. 2011), but this seems not to be the case in the wheat/F. graminearum interaction. Warth et al. (2015) have reported increased amino acid abundances in response to DON, hypothesizing that this may either be due to the increased biosynthesis of amino acids or that amino acids stem from degradation of unfinished peptide chains as a consequence of the ribotoxic effect of DON. In the gene expression data, we found strong indications for the increased biosynthesis of amino acids, likely reflecting the efforts of keeping up the protein biosynthesis to counteract the effect of DON.
Key biosynthesis genes for all amino acids except proline, lysine, serine, and asparagine were more abundant in response to the pathogen. Although we did not find evidence for the increased biosynthesis of serine or proline in the transcriptomic data, DON-inoculated samples showed strong differences in the abundance of serine and proline compared with mock treatment as indicated by the Bayes score (highlighted in red in Figure 4). Levels of both amino acids are strongly increased after treatment with DON (Warth et al. 2015). In particular, proline was reported to be more abundant under different stresses (Sharma and Dietz 2006). Proline accumulated in tissues surrounding hypersensitive lesions caused by Pseudomonas syringae on Arabidopsis thaliana, leading to the assumption that it may play a role in quenching free radicals (Fabro et al. 2004). It also holds a role in nitrogen transport in the phloem, as its levels are possibly linked to GS1 activity (Brugière et al. 1999). Other proteinogenic amino acids such as the phenolic amino acids and amino acids derived from aspartate were also highly changed in response to DON. Yet, aspartate itself, as well as the functionally linked pools of asparagine, glutamine and glutamate, were largely unchanged. Glutamate pools remain largely unchanged under stress conditions (Forde and Lea 2007), and we speculate that this could also be true for the closely linked aspartate. Glutamine and asparagine levels in contrast can be subject to change due to active influx of these compounds as nitrogen-sources into the sink tissue (Masclaux-Daubresse et al. 2006). Other amino acids with less pronounced changes such as serine or alanine may in fact be remobilized into the respiratory chain as suggested by (Araújo et al. 2011). Concerning the suggested two alternative models for increased amino acid abundances (Warth et al. 2015) our combination of transcriptomic and metabolomic data provided clear evidence that a higher tRNA-ligase activity is supported by increased amino acid biosynthesis - yet not all amino acids biosynthesis pathways are similarly affected.
Increased activity in the glutamate metabolism: a possible role for Qfhs.ifa-5A−mediated resistance?
Our previous study (Kugler et al. 2013) found the activity of glutamate regulated Ca2+ channels associated with Qfhs.ifa-5a. Now using the almost complete bread wheat genes, we were able to fully reconstruct changes in the glutamate metabolism during defense. The F. graminearum-responsive GS1 and NADPH-dependent GOGAT genes are not part in metabolizing newly synthesized ammonium from photosynthesis but facilitate the transport of nitrogen in the form of glutamine through the phloem into sink/infected tissue (Masclaux-Daubresse et al. 2010). Seifi et al. (2013) suggested two different roles in pathogen defense for the metabolism of glutamate, which may either act toward depleting infected tissue from nitrogen compounds to prevent these from being scavenged by the pathogen or it may assist the cell to endure the disease by hauling energy equivalents into the infected tissue. The latter is characterized by the increased activity of the GOGAT/GS cycle, the GABA shunt, and glutamate deyhdrogenase genes, whereas aspartate transaminases and asparagine synthases are less active. Our data suggest the bread wheat defense against F. graminearum aims to endure the disease by strengthening the TCA cycle and supplying carbon/nitrogen for the biosynthesis of secondary metabolites: GS1-generated glutamine enters the infected tissue as an additional carbon and nitrogen source. In sink tissue glutamine is metabolized to glutamate by NADH-dependent GOGAT and then further decomposed into 2-oxoglutarate by the also highly F. graminearum-responsive glutamate dehydrogenases (GDHs). Although GDH can perform the reverse reaction to additionally assimilate ammonium under given conditions, the more likely reaction is the deamination of glutamate into 2-oxoglutarate as an anaplerotic reaction to fuel the TCA cycle (Masclaux-Daubresse et al. 2006). Abundant ammonium from GDH activity may be reused by GS1 located in the infected tissue.
Several of these described genes showed differential expression patterns for lines differing in Qfhs.ifa-5A. Transcript abundances for NADPH-GOGAT and GDH genes were greater in F. graminearum−treated samples at 30 hpi for lines harboring Qfhs.ifa-5A. Potentially, these lines react earlier to the influx of glutamine and provide earlier an increased amount of 2-oxoglutatare to the TCA cycle (II, III, and V in Figure 4). Such differences also were observed for several TCA cycle genes (aconitases, citrate synthases, succinate dehydrogenases) as well as malic enzymes, which provide additional pyruvate for the TCA cycle. Similarly, associated metabolite levels are changed in response to DON and the fungus only for lines lacking this earlier reaction, which we observed for the Qfhs.ifa-5A-lines (Figure 4). TCA intermediate substances are subject to high turnover rates and concentrations tend to be stable (Sweetlove et al. 2010). The required increased flux in response to the pathogen seems to be more efficiently met by the earlier action of Qfhs.ifa-5A-lines. In contrast, the adaptation to F. graminearum in non-Qfhs.ifa-5A lines leads to the observed changes in pool levels. However, the large variances in the measurements of metabolites do not allow a further interpretation of the present data. Not all genes changed for Qfhs.ifa-5A act earlier in response to the pathogen: Pyruvate dehydrogenases and malate dehydrogenases were less strongly changed at 50 hpi in response to F. graminearum in Qfhs.ifa-5A containing lines (Figure 4). How these expression patterns fit into the proposed mechanism remains unclear. For a more detailed interpretation of these observations more comprehensive, preferably longitudinal expression profiles will be needed.
The observed changes for the TCA cycle and glutamate metabolism could contribute to a greater “endurance” in Qfhs.ifa-5A lines and thus be part the resistance mechanism encoded by the QTL. Among the genes constitutively changed for the QTL encoded on 5A in module C two amino acid permeases (AAP, isoforms 6, Traes_5AS_776E1FEE4 and 8, Traes_5AS_073CAB1CC) could contribute to the greater influx of amino acids from the phloem and provide substrates for the observed QTL-associated changes. In A. thaliana AtAAP6 regulates the phloem amino acid composition and AtAAP8 is involved in seed development (Tegeder and Rentsch 2010). AtAAP6 was reported to be expressed in sink tissue with a high affinity for neutral amino acids and other amino acids with acidic side chains (Hunt et al. 2010). Also AtAAP8 has a high affinity to aspartate and glutamate (Schmidt et al. 2007). Because of the potentially large pericentromeric introgression harboring the QTL many other genes that show constitutive expression differences cannot be ruled out as putative candidates. This list (Table S10) also includes candidates from our previous studies (Schweiger et al. 2013, Kugler et al. 2013) including a lipid transfer protein, which shows among the highest expression differences.
Narrowing down single-gene candidates is aggravated by the susceptible Chinese Spring mapping reference
We have made use of a combination of coexpression patterns, differential expression analysis, and chromosome location to narrow down the list of candidates for Fhb1. Most genes specifically expressed for Fhb1 exhibited a constitutive expression pattern (module D). Although many of these genes map to chromosome 3B a closer inspection showed that none of the modules hub genes mapped within close vicinity of the genomic region carrying the susceptible Fhb1 locus of cultivar Chinese Spring (contig ctg0954 [GenBank:FN564434] (Choulet et al. 2010). It is unclear how large the introgressed region carrying Fhb1 is. Possibly several of the reported genes are distant from the locus and QTL unrelated: Mapping against the complete 3B chromosome sequence (Choulet et al. 2014) placed Traes_3B_3A70D33A6, a receptor-like protein kinase, at position 10.095.372 bp and Traes_3B_6A585354F, a protein kinase, at position 9.817.008 bp, which is about 17 Mbp distal to the Fhb1 marker Umn10 (27.605.772 bp). Several other genes in module D mapped to ctg0954, but only three within the QTL confidence interval between flanking markers Sts32 and Sts189. All three are only weakly expressed and corresponding transcripts are more abundant in lines harboring the susceptible QTL allele (Traes_3B_07980E2CE, Traes_3B_0D8C9A632, Traes_3B_CDF3C9680). Similarly, candidates from module E, which groups Fhb1-specific and F. graminearum−responsive genes (Traes_3B_5088D482E, 19.748.084 bp, SINA-like 11 E3 ubiquitin-protein ligase and Traes_3B_6E28B451A, 4.483.234 bp, unknown) are too distant from the marker to be considered candidate genes based on the Chinese Spring reference.
The absence of closely mapped genes does not necessarily mean that the elusive Fhb1 gene remains unrecognized by the IWGSC wheat gene set, which is also based on the susceptible Chinese Spring cultivar. It may be that the resistance gene is only present in the resistant genotype and would not map onto the contig. Likely reads derived from genes not represented in the gene set will map to close homologs or in case of hexaploid wheat to the next homoelog on sister chromosomes should they exist. For example Traes_3DS_2BD8DC857 specifically expressed for Fhb1 containing lines maps to the homoelogous region of Fhb1 on chromosome 3D. Possibly, the susceptible reference genotype Chinese Spring lacks such a gene in the Fhb1 region and 3B-specific reads map to this putative homeolog. The possible absence of this gene in the susceptible cultivar Chinese Spring may derive from pseudogenization or small chromosomal rearrangements (Bennetzen and Ramakrishna 2002), which may occur even between different varieties of the same species (Feuillet and Salse 2009). Liu et al. have compared the synteny of the genetically mapped locus of Fhb1 to the rice and barley physical maps and did find evidence for rearrangements based on marker collinearity in this region (Liu et al. 2006). Possibly, the gene content and/or gene order of the genomic region containing the resistant Fhb1 locus does not follow the established Chinese Spring reference cultivar. This would also allow to hypothesize that candidate genes that mapped in this study distant to the Fhb1 marker Umn10 in Chinese Spring could in fact be located within the QTL confidence interval in Fhb1-containing genotypes.
On the other hand, this study has considered high-confidence gene models only. Low confidence genes in wheat comprise a large number, more than 100,000, of putatively fragmented gene models, pseudogenes, and repeat associated elements for which little evidence for functionally expressed gene products exist. In Schweiger et al. (2013) we have measured gene expression using the Affymetrix wheat GeneChip, yielding four differentially expressed transcripts mapping to the region of Fhb1. Two of these genes corresponded to low confidence genes (Ta.6066.2.S1_a_at: Ta3bMIPSv2Loc009233; Ta.22694.1.A1_at: Ta3bMIPSv2Loc008006) and only one, Ta.28185.1.S1_at, had homology to a high confidence gene (Traes_3B_05EEE7D3F1). This is also the only gene mapping to the QTL confidence interval between markers sts32 and sts189. All genes are expressed in a constitutive manner for the absence/presence of the introgressed QTL region, which matches the reported findings. Traes_3B_05EEE7D3F1 is higher expressed for the susceptible QTL allele. One of the reported differentially expressed probe sets (TaAffx.12498.2.S1_at) was not included in either data set. Possibly these low confidence genes are relevant for the QTL activity and should be considered once the genomic region of a Fhb1-containing cultivar is resolved and available.
Genetic approaches to map these genes in materials segregating for Fhb1 are required to narrow down this list of candidates. Zhuang et al. (2013) have mapped the expression traits of one out of 47 FHB investigated resistance candidate genes in an expression QTL study to the Fhb1 locus (10.1094/MPMI-10-12-0235-R). However, a BLAST survey of this putative resistance gene designated WFhb1_c1 showed that the IWGSC gene set does not include a homolog on chromosome 3B and also their mapped Fhb1 interval spanning more than 16 cM is large.
Imbalances in subgenome expression contribution
Polyploidization events present a form of “genomic shock,” which leads to increased transposable element activity and epigenetic silencing (Wendel 2000). Such effects may also be reflected in the expression patterns and the interplay between the A, B, and D subgenomes. An imbalance in the number of disease-resistance genes has been described for tetraploid and hexaploid wheat, with the highest number of genes stemming from the B subgenome (Feldman et al. 2012; Fahima et al. 2006; Peng et al. 2003). Based on the IWGSC annotation, most members of two defense-related gene families were encoded on the A and B subgenomes (Mayer et al. 2014). This distribution does not correspond to findings in our data, where in terms of gene expression contributions to the defense response from the D and the B subgenomes dominated over contributions from the A subgenome. Such expression observations might be affected by differences in total gene numbers on the subgenomes. To address this we eliminated this bias by considering only the 1:1:1 homoelogous triplet genes in the triplet gene coexpression network, which showed that genes from the D subgenome are more abundant in response to the pathogen than their A and B counterparts. From these observations and the hub gene-specific subgenome distribution in modules A and B we reason that subgenome D contributions may play a decisive role in overall resistance to F. graminearum. This hypothesis could relate to the overall high susceptibility to FHB of tetraploid durum wheat (Triticum turgidum ssp. durum, 2n = 4x = 28, AABB). Only single lines have been described which harbor intermediate levels resistance to FHB (Huhn et al. 2012; Prat et al. 2014). Because durum and bread wheat share the same ancestral A and B subgenomes, the added resistance in bread wheat may stem from D subgenome contributions. Although, most of the relevant resistance QTL in T. aestivum have been mapped to the A or B subgenome (Buerstmayr et al. 2009), resistances encoded on the D subgenome may well play decisive roles: The D subgenome is much less polymorphic due to its evolutionary only recent addition to wheat and thus resistance contributors may not be segregating in mapping populations. An indication for its relevance comes from Aegilops tauschii, the contributor of the D subgenome to wheat, which has been used widely in the generation of synthetic hexaploid wheats from crosses with tetraploid species such as Triticum turgidum. The addition of the D subgenome has improved resistances against a variety of biotic and abiotic stresses including resistance against FHB in comparison to the tetraploid parental line (Mujeeb-Kazi et al. 2008). Whether these resistance genes are in effect in T. aestivum remains to be shown. However, we also observed a slight bias toward the A genome for triplets, where only a single copy was responsive to the pathogen. Overall, it appears that the response to the pathogen is distributed between the different subgenomes, and more data is needed to generalize findings of subgenome bias in the context of bread wheat pathogen response.
Supplementary Material
Acknowledgments
We acknowledge the technical assistance of Denise Schöfbeck and Bernhard Kluger during greenhouse experiments and sample preparation and Nora Neumann for the support in metabolite data processing. We thank Gerhard Adam for helpful suggestions and discussions on the manuscript. This work was supported by the Austrian Science Fund (FWF) (SFB F3705, F3706, F3711) and by the Deutsche Forschungsgemeinschaft (DFG) (SFB 924). The authors declare no conflict of interest.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.021550/-/DC1
Communicating editor: E. Akhunov
Literature Cited
- Aharoni A., Galili G., 2011. Metabolic engineering of the plant primary–secondary metabolism interface. Curr. Opin. Biotechnol. 22: 239–244. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W., 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. A., Stack R., Liu S., Waldron B., Fjeld A., et al. , 2001. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor. Appl. Genet. 102: 1164–1168. [Google Scholar]
- Araújo W. L., Tohge T., Ishizaki K., Leaver C. J., Fernie A. R., 2011. Protein degradation—an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16: 489–498. [DOI] [PubMed] [Google Scholar]
- Audenaert K., Vanheule A., Höfte M., Haesaert G., 2013. Deoxynivalenol: a major player in the multifaceted response of Fusarium to its environment. Toxins (Basel) 6: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Ramakrishna W., 2002. Numerous small rearrangements of gene content, order and orientation differentiate grass genomes. Plant Mol. Biol. 48: 821–827. [DOI] [PubMed] [Google Scholar]
- Bolton M. D., 2009. Primary metabolism and plant defense-fuel for the fire. Mol. Plant Microbe Interact. 22: 487–497. [DOI] [PubMed] [Google Scholar]
- Brenchley R., Spannagl M., Pfeifer M., Barker G. L., D’Amore R., et al. , 2012. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugière N., Dubois F., Limami A. M., Lelandais M., Roux Y., et al. , 1999. Glutamine synthetase in the phloem plays a major role in controlling proline production. The Plant Cell Online 11: 1995–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstmayr H., Lemmens M., Hartl L., Doldi L., Steiner B., et al. , 2002. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (Type II resistance). Theor. Appl. Genet. 104: 84–91. [DOI] [PubMed] [Google Scholar]
- Buerstmayr H., Steiner B., Hartl L., Griesser M., Angerer N., et al. , 2003. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor. Appl. Genet. 107: 503–508. [DOI] [PubMed] [Google Scholar]
- Buerstmayr H., Ban T., Anderson J. A., 2009. QTL mapping and marker‐assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 128: 1–26. [Google Scholar]
- Choulet F., Wicker T., Rustenholz C., Paux E., Salse J., et al. , 2010. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. The Plant Cell Online 22: 1686–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choulet F., Alberti A., Theil S., Glover N., Barbe V., et al. , 2014. Structural and functional partitioning of bread wheat chromosome 3B. Science 345: 1249721. [DOI] [PubMed] [Google Scholar]
- Diethelm M., Rhiel M., Wagner C., Mikolajewski S., Groth J., et al. , 2012. Gene expression analysis of four WIR1-like genes in floret tissues of European winter wheat after challenge with G. zeae. Euphytica 186: 103–114. [Google Scholar]
- Fabro G., Kovács I., Pavet V., Szabados L., Alvarez M. E., 2004. Proline accumulation and AtP5CS2 gene activation are induced by plant-pathogen incompatible interactions in Arabidopsis. Mol. Plant Microbe Interact. 17: 343–350. [DOI] [PubMed] [Google Scholar]
- Fahima T., Cheng J., Peng J., Nevo E., Korol A., 2006. Asymmetry distribution of disease resistance genes and domestication synrome QTLs in tetraploid wheat genome in 8th International Congress of Plant Molecular Biology, Adelaide, Australia, August 20–25, 2006. [Google Scholar]
- Falcon S., Gentleman R., 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23: 257–258. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., 2012. Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Levy A. A., Fahima T., Korol A., 2012. Genomic asymmetry in allopolyploid plants: wheat as a model. J. Exp. Bot. 63: 5045–5059. [DOI] [PubMed] [Google Scholar]
- Feuillet C., Salse J., 2009. Comparative genomics in the triticae, p. 451 in Genetics and Genomics of the Triticeae, edited by Feuillet C., Muehlbauer G. J. Springer Science + Business Media, New York, NY. [Google Scholar]
- Finley D., Bartel B., Varshavsky A., 1989. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–401. [DOI] [PubMed] [Google Scholar]
- Forde B. G., Lea P. J., 2007. Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58: 2339–2358. [DOI] [PubMed] [Google Scholar]
- Foroud N., Ouellet T., Laroche A., Oosterveen B., Jordan M., et al. , 2012. Differential transcriptome analyses of three wheat genotypes reveal different host response pathways associated with Fusarium head blight and trichothecene resistance. Plant Pathol. 61: 296–314. [Google Scholar]
- Gunnaiah R., Kushalappa A. C., Duggavathi R., Fox S., Somers D. J., 2012. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One 7: e40695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller K., Hangebrauk J., Jäger C., Spura J., Schreiber K., et al. , 2009. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal. Chem. 81: 3429–3439. [DOI] [PubMed] [Google Scholar]
- Huhn M. R., Elias E. M., Ghavami F., Kianian S. F., Chao S., et al. , 2012. Tetraploid Tunisian wheat germplasm as a new source of Fusarium head blight resistance. Crop Sci. 52: 136–145. [Google Scholar]
- Hunt E., Gattolin S., Newbury H. J., Bale J. S., Tseng H.-M., et al. , 2010. A mutation in amino acid permease AAP6 reduces the amino acid content of the Arabidopsis sieve elements but leaves aphid herbivores unaffected. J. Exp. Bot. 61: 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C., Von Wettstein D., Schäfer W., Kogel K.-H., Felk A., et al. , 2005. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 102: 16892–16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Cho S., Muehlbauer G. J., 2009. Transcriptome analysis of a wheat near-isogenic line pair carrying Fusarium head blight-resistant and-susceptible alleles. Mol. Plant Microbe Interact. 22: 1366–1378. [DOI] [PubMed] [Google Scholar]
- Jin F., Zhang D., Bockus W., Baenziger P. S., Carver B., et al. , 2013. Fusarium head blight resistance in US winter wheat cultivars and elite breeding lines. Crop Sci. 53: 2006–2013. [Google Scholar]
- Gottwald S., Samans B., Lück S., Friedt W., 2012. Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of Fusarium head blight resistance in wheat? BMC Genomics 13: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K., Gardiner D. M., Manners J. M., 2012. On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance. Mol. Plant Pathol. 13: 399–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Lim J.-H., Ahn C. S., Park K., Kim G. T., et al. , 2006. Mitochondria-associated hexokinases play a role in the control of programmed cell death in Nicotiana benthamiana. Plant Cell 18: 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y. A., et al. , 2014. ArrayExpress update—simplifying data submissions. Nucleic Acids Res. 43(Database Issue): D1113–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler K. G., Siegwart G., Nussbaumer T., Ametz C., Spannagl M., et al. , 2013. Quantitative trait loci-dependent analysis of a gene co-expression network associated with Fusarium head blight resistance in bread wheat (Triticum aestivum L.). BMC Genomics 14: 728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S., 2007. Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S., 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Zhang B., Horvath S., 2008. Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R. Bioinformatics 24: 719–720. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens M., Scholz U., Berthiller F., Dall’Asta C., Koutnik A., et al. , 2005. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol. Plant Microbe Interact. 18: 1318–1324. [DOI] [PubMed] [Google Scholar]
- Liu S., Zhang X., Pumphrey M. O., Stack R. W., Gill B. S., et al. , 2006. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct. Integr. Genomics 6: 83–89. [DOI] [PubMed] [Google Scholar]
- Marcussen T., Sandve S. R., Heier L., Spannagl M., Pfeifer M., et al. , 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345: 1250092. [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Pageau K., Lelandais M., Grandjean O., et al. , 2006. Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 140: 444–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Daniel-Vedele F., Dechorgnat J., Chardon F., Gaufichon L., et al. , 2010. Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann. Bot. 105: 1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F., Taudien S., Martis M., Šimková H., Suchánková P., et al. , 2009. Gene content and virtual gene order of barley chromosome 1H. Plant Physiol. 151: 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer K. F., Rogers J., Doležel J., Pozniak C., Eversole K., et al. , 2014. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 1251788. [DOI] [PubMed] [Google Scholar]
- McMullen M., Halley S., Schatz B., Meyer S., Jordahl J., et al. , 2008. Integrated strategies for Fusarium head blight management in the United States. Cereal Res. Commun. 36: 563–568. [Google Scholar]
- Miflin B. J., Habash D. Z., 2002. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53: 979–987. [DOI] [PubMed] [Google Scholar]
- Mujeeb-Kazi A., Gul A., Farooq M., Rizwan S., Ahmad I., 2008. Rebirth of synthetic hexaploids with global implications for wheat improvement. Crop Pasture Sci. 59: 391–398. [Google Scholar]
- Nussbaumer T., Kugler K. G., Bader K. C., Sharma S., Seidel M., et al. , 2014a RNASeqExpressionBrowser—a web interface to browse and visualize high-throughput expression data. Bioinformatics 30: 2519–2520. [DOI] [PubMed] [Google Scholar]
- Nussbaumer T., Kugler K. G., Schweiger W., Bader K. C., Gundlach H., et al. , 2014b chromoWIZ: a web tool to query and visualize chromosome-anchored genes from cereal and model genomes. BMC Plant Biol. 14: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Ronin Y., Fahima T., Röder M. S., Li Y., et al. , 2003. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 100: 2489–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka J. J., 2010. Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch. Toxicol. 84: 663–679. [DOI] [PubMed] [Google Scholar]
- Pfeifer M., Kugler K. G., Sandve S. R., Zhan B., Rudi H., et al. , 2014. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345: 1250091. [DOI] [PubMed] [Google Scholar]
- Pirgozliev S. R., Edwards S. G., Hare M. C., Jenkinson P., 2003. Strategies for the control of Fusarium head blight in cereals. Eur. J. Plant Pathol. 109: 731–742. [Google Scholar]
- Prat N., Buerstmayr M., Steiner B., Robert O., Buerstmayr H., 2014. Current knowledge on resistance to Fusarium head blight in tetraploid wheat. Mol. Breed. 34: 1689–1699. [Google Scholar]
- Pritsch C., Muehlbauer G. J., Bushnell W. R., Somers D. A., Vance C. P., 2000. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol. Plant Microbe Interact. 13: 159–169. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas C. M., Senthil-Kumar M., Tzin V., Mysore K. S., 2014. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salek R. M., Haug K., Conesa P., Hastings J., Williams M., et al. , 2013. The MetaboLights repository: curation challenges in metabolomics. Database 2013: bat029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Stransky H., Koch W., 2007. The amino acid permease AAP8 is important for early seed development in Arabidopsis thaliana. Planta 226: 805–813. [DOI] [PubMed] [Google Scholar]
- Schwachtje J., Baldwin I. T., 2008. Why does herbivore attack reconfigure primary metabolism? Plant Physiol. 146: 845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger W., Steiner B., Ametz C., Siegwart G., Wiesenberger G., et al. , 2013. Transcriptomic characterization of two major Fusarium resistance quantitative trait loci (QTLs), Fhb1 and Qfhs. ifa‐5A, identifies novel candidate genes. Mol. Plant Pathol. 14: 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifi H. S., Van Bockhaven J., Angenon G., Höfte M., 2013. Glutamate metabolism in plant disease and defense: friend or foe? Mol. Plant Microbe Interact. 26: 475–485. [DOI] [PubMed] [Google Scholar]
- Sharma S. S., Dietz K.-J., 2006. The significance of amino acids and amino acid-derived molecules in plant responses and adaptation to heavy metal stress. J. Exp. Bot. 57: 711–726. [DOI] [PubMed] [Google Scholar]
- Stegle O., Denby K. J., Cooke E. J., Wild D. L., Ghahramani Z., et al. , 2010. A robust Bayesian two-sample test for detecting intervals of differential gene expression in microarray time series. J. Comput. Biol. 17: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner B., Kurz H., Lemmens M., Buerstmayr H., 2009. Differential gene expression of related wheat lines with contrasting levels of head blight resistance after Fusarium graminearum inoculation. Theor. Appl. Genet. 118: 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner L. W., Amberg A., Barrett D., Beale M. H., Beger R., et al. , 2007. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove L. J., Beard K. F., Nunes-Nesi A., Fernie A. R., Ratcliffe R. G., 2010. Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15: 462–470. [DOI] [PubMed] [Google Scholar]
- Tegeder M., Rentsch D., 2010. Uptake and partitioning of amino acids and peptides. Mol. Plant 3: 997–1011. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S., Nicholson P., Doohan F. M., 2010. Action and reaction of host and pathogen during Fusarium head blight disease. New Phytol. 185: 54–66. [DOI] [PubMed] [Google Scholar]
- Walter S., Brennan J. M., Arunachalam C., Ansari K. I., Hu X., et al. , 2008. Components of the gene network associated with genotype-dependent response of wheat to the Fusarium mycotoxin deoxynivalenol. Funct. Integr. Genomics 8: 421–427. [DOI] [PubMed] [Google Scholar]
- Warth B., Parich A., Bueschl C., Schoefbeck D., Neumann N. K. N., et al. , 2015. GC–MS based targeted metabolic profiling identifies changes in the wheat metabolome following deoxynivalenol treatment. Metabolomics 11: 722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F., 2000. Genome evolution in polyploids, pp. 225–249 in Plant Molecular Evolution, edited by Goyle J. J., Gaut B. S. Springer, The Netherlands. [PubMed] [Google Scholar]
- Xiao J., Jin X., Jia X., Wang H., Cao A., et al. , 2013. Transcriptome-based discovery of pathways and genes related to resistance against Fusarium head blight in wheat landrace Wangshuibai. BMC Genomics 14: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Sheen J., Jang J.-C., 2000. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol. Biol. 44: 451–461. [DOI] [PubMed] [Google Scholar]
- Zhuang Y., Gala A., Yen Y., 2013. Identification of functional genic components of major Fusarium head blight resistance quantitative trait loci in wheat cultivar Sumai 3. Mol. Plant Microbe Interact. 26: 442–450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data availability is as follows: RNA-seq: EBI Arrayexpress (E-MTAB-1729); gas chromatography–mass spectrometry: MetaboLights (MTBLS112, MTBLS153).